Abstract
Microglia play multiple roles in such processes as brain development, homeostasis, and pathology. Due to their diverse mechanisms of functions, the complex sub-classifications, and the large differences between different species, especially compared with humans, very different or even opposite conclusions can be drawn from studies with different research models. The choice of appropriate research models and the associated tools are thus key ingredients of studies on microglia. Mice are the most commonly used animal models. In this review, we summarize in vitro and in vivo models of mouse and human-derived microglial research models, including microglial cell lines, primary microglia, induced microglia-like cells, transgenic mice, human-mouse chimeric models, and microglial replacement models. We also summarize recent developments in novel single-cell and in vivo imaging technologies. We hope our review can serve as an efficient reference for the future study of microglia.
Keywords: Microglial cell lines, Primary microglia, Induced microglia-like cells, Transgenic mice, Human-mouse chimeric models, Microglial replacement, Single-cell technology, In vivo imaging
Introduction
Microglia are tissue-resident macrophages in the brain parenchyma, accounting for more than 5% of all glial cells in the human brain [1]. The proportion of microglia in the normal adult mouse brain varies from 5% to 12% depending on the region [2]. Unlike other neurons and glial cells in the brain that develop from the ectoderm, microglia are derived from the yolk sac [3]. Taking mice as an example, their microglia originate from the erythro-myeloid progenitors of the yolk sac around embryonic day 7.25 (E7.25), earlier than the appearance of other glial cells, and have the potential for primitive erythropoiesis [4]. They migrate into the central nervous system (CNS) when neural progenitor cells begin to divide and form neurons, just before definitive hematopoiesis begins [5, 6]. Mononuclear cells derived from blood do not enter the brain through the blood-brain barrier (BBB) under normal physiological conditions. Under neuroinflammatory and neurodegenerative pathological conditions, peripheral myeloid cells can invade the brain through the BBB [7].
Microglial activation plays a major role in the brain’s immune response and is an important indicator of immune activation and neuroinflammation [8]. In addition to participating in immune regulation, microglia contribute largely to brain development during embryonic and perinatal periods. They are pioneers in the developing brain and regulate the establishment of neural circuits. Microglia can regulate the growth and development of dopaminergic axons and the migration of interneurons (especially parvalbumin-positive (PV+) interneurons) [9]. The absence or dysfunction of microglia can lead to an imbalance of the neural distribution at birth and postnatal inhibition/excitation balance disorder. Microglia regulate neurogenesis by actively promoting neuronal cell death and eliminating neural precursor cells (NPCs) in the developing CNS [10–12]. Microglia also support the proliferation of NPCs and promote neuronal survival by releasing trophic factors [8, 13]. Besides, microglia play a key role in synaptic density and homeostasis regulation by engulfing synapses, which can mediate synapse elimination [14]. Furthermore, microglia are vital in the pathological processes of neuropsychiatric disorders such as depression, schizophrenia, neurodevelopment disorders such as autism spectrum disorders, neurodegenerative diseases such as Alzheimer's disease (AD), and also stroke [15–19]. In short, microglia are involved in the development of the brain and the processes of various CNS diseases. The study on the role and mechanism of microglia has received a lot of attention recently.
However, there are still many unresolved questions and obstacles in the field of microglial research. There are many controversies and uncertainties about the conclusions of existing research in this field, such as the development and classification of microglia, the mechanisms of microglial action in different physiological processes, the determinants of CNS-associated macrophages (CAMs) differentiating from microglia in the development, function, and dynamics. The differences between microglia of different species also bring uncertainty to the research results. Although microglia of different species do share some basic similarities, research using animal models sometimes do not encompass the complexity of the human condition [20]. So, choosing the appropriate research program and model is the key to studying microglia. Our review comprehensively introduces the current models and tools used in microglial research, from traditional 2D culture to 3D culture and then to in vivo models, from single-cell technology to in vivo imaging technology. The applicability and advantages and disadvantages of these tools are discussed. The combination of these emerging technologies provides the possibility to explore the spatio-temporal characteristics of microglia and their heterogeneity in different physiological and pathological conditions at the single-cell level. Our discussion may inspire readers to develop new research approaches to microglia and answer those key questions.
In Vitro Microglial Models
Microglial Cell Lines
The characterization of human cells has been limited due to the restricted availability of primary sources of human microglia. So human immortalized microglial cell lines are important tools for studying the characteristics of microglia. The human microglia clone 3 cell line (HMC3 or HMC-3), also under the name of CHME-3, CHME-5, or C13-NJ among different laboratories, was established through SV40-dependent immortalization of a human fetal brain-derived primary microglial cell culture [21]. HMC3 cells retain most of the original antigenic properties and express many specific microglial markers. They strongly express the microglia/macrophage marker IBA1 (ionized calcium binding adapter molecule 1) at rest, and MHC II (major histocompatibility complex class II), CD68, CD11b, and other markers after being activated. Other commonly used human-derived microglial cell lines include HMO6 and HM1900. These cells also retain a similar antigenic profile and similar functional properties. They are capable of responding to a pattern of chemokines and inflammatory stimuli, regulating the expression of typical activation markers of microglia. Mouse immortalized microglial cell lines have also been established. The SIM-A9 cell line is a spontaneously immortalized cell line derived from mouse primary microglial cell cultures [22]. It expresses microglial markers such as CD68 and IBA1 and exhibits phagocytic activity under inflammatory stimulation. Other mouse immortalized microglial cell lines include BV-2, N9, and C8-B4. Using immortalized microglial cell lines for research can shorten the experimental period and cost of microglial cell cultures, and the cell uniformity is high. However, microglial cell lines are in vitro models. And after conversion to immortalization, their characteristics also change somewhat compared with in vivo microglia.
Primary Microglial Cells
Primary mouse microglia are widely used in microglia-associated studies. The current acquisition of primary mouse microglia is based on the cultivation of mixed glial cells [23]. After isolating the target brain region and digesting it into a single-cell suspension, cells are cultured in the glial culture medium and gradually adhere to the wall and fuse. The mixed glial cells are cultured for about 14 days, and the upper microglia are harvested by gentle trypsin digestion or shaking on a shaker. This method has strict requirements on the time window. Usually, newborn mice at postnatal 1–4 days are taken. But microglia are not fully mature at this time; they behave differently from adult microglia. A second method has been optimized with the addition of immuno-magnetic cell sorting steps [24], which select microglia by specific antibodies to recognize microglia-specific surface antigens such as CD11b. This second method can also be used for the screening and subsequent cultivation of adult mouse microglia. Another method for isolating and culturing primary microglia of adult mice is purifying the digested mixed brain cells by density gradient centrifugation and adding GM-CSF [granulocyte-macrophage colony-stimulating factor, or colony stimulating factor 2 (CSF2)] to the culture medium [25]. The purity of primary microglia obtained by these methods is generally 95%–99%. Caution should be taken that there are other subsets of macrophages residing in the brain: CNS border-associated macrophages, which are also called CAMs, in the meninges, choroid plexus, and perivascular spaces [26]. These cells share several myeloid lineage-related properties in the CNS, and may be mixed into the microglia during the experimental manipulation and affect the research results. The serum is also an important factor affecting the culture of microglia because serum cannot penetrate the BBB and serum has drastic effects on cell morphology and function, leaving microglia in an activated state of inflammation. So, serum-free medium is recommended in microglial cell culture to mimic the in vivo state. Primary microglial cell culture has been used for decades to study their function. However, cultured primary microglia is only a poor model to investigate ramified microglia. Various electrophysiological and genetic studies have provided evidence that primary microglia lose many of their functions compared to cells in vivo and are more closely related to activated peripheral macrophages [27].
Human brain tissue is usually derived from elderly and diseased autopsy brains when such material is available. Immunohistochemical staining for certain microglial markers (like Iba1 and Ki-M1P) on autopsy brain tissue, can detect changes in the number and activity of microglia and help to characterize the pathogenic role of microglia in these diseases [28]. Alternative sources to autopsy include surgically excised and fetal brain tissues [29–31], but these tissues are more difficult to obtain due to ethical issues. Isolating microglia from human brain tissue through a rapid autopsy program can be used in primary culture and subsequent research [32]. After enzymatic digestion, using density gradient centrifugation to separate the cell layer containing microglia, and then pure microglia can be obtained by their characteristic strong and rapid attachment to plastic culture surfaces while the other cell types require coating matrices to attach. These isolated microglia can be phenotypically heterogeneous, so experiments using cells from different preparation procedures and brain regions may produce discordant results. When there are enough cases, using the same cell acquisition procedure from the same brain regions, one can still study the impact of age, disease, and treatment on the morphology and function of microglia.
Induced Microglia-like Cells
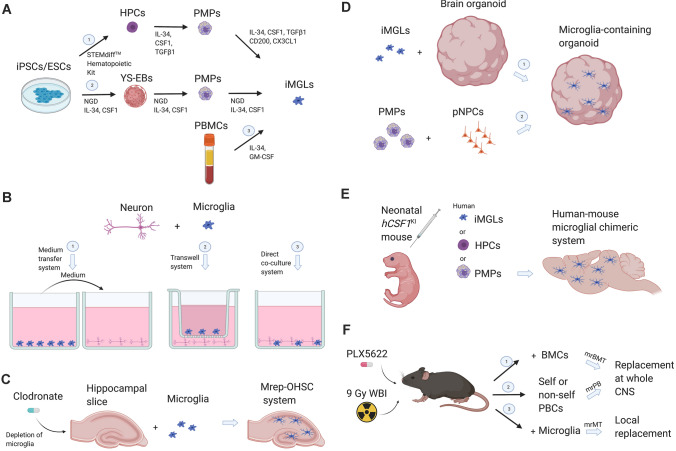
Since the technology of human induced-pluripotent stem cells (iPSCs) came out in 2007 [33, 34], they have been widely used in disease modeling, drug screening, and disease treatment. iPSCs possess multiple differentiation potentials and their expansion can be scaled up easily in vitro. But it was not until the last few years that human induced microglia-like cell (iMGL), also named pluripotent stem cell-derived microglia-like cell (pMGL) technology was realized (Fig. 1A).
Fig. 1.
Toolkits for studying microglial cells. A Strategies for developing human induced microglia-like cells: iPSCs/ESCs are differentiated into HPCs (top) or YS-EBs (middle) first to recapitulate the true ontogeny of microglia. Or, iMGLs are directly generated from PBMCs (bottom). B 2D co-culture systems of microglia and neurons have three different levels of cell-to-cell communication: Left, a conditioned medium transfer system in which no cell-to-cell contact or communication is possible; Center, a transwell system that allows cell-contact-independent communication through diffusible soluble factors; and Right, a co-culture system that permits direct contact of microglia and neurons. C Refilling the ramified microglia isolated from adult mouse brains into OHSC with microglia depleted, creates the Mrep-OHSC system. This system provides a situation that is very close to in vivo. Replenished microglia can integrate into the tissue and exhibit characteristics like their counterparts in the mouse brain. D Microglia-containing brain organoid system. Mature iMGLs are integrated into brain organoids that have been cultured for a long time. Or, pNPCs and PMPs are co-cultured to undergo differentiation and maturation together. In this 3D system, induced microglia-like cells extend varying degrees of ramified processes that resemble microglia in vivo. E The human-mouse chimeric microglial system provides a tool to study the role of human microglia in vivo. F Three efficient strategies for microglial replacement: microglial replacement by bone marrow transplantation (mrBMT), microglial replacement by peripheral blood (mrPB), and microglial replacement by microglial transplantation (mrMT). Abbreviations: NGD, serum-free neuroglial differentiation medium; YS-EBs, yolk sac embryoid bodies; PMPs, primitive macrophage progenitors or microglial precursors; pNPCs, primitive neural progenitor cells; iMGLs, human induced microglia-like cells; OHSCs, organotypic hippocampal slice cultures; Mrep-OHSCs, microglia-replenished OHSCs; iPSCs, induced pluripotent stem cells; ESCs, embryonic stem cells; PBMCs, peripheral blood mononuclear cells; BMCs, bone marrow cells; PBCs, peripheral blood cells; PLX5622, a CSF1R inhibitor; WBI, whole-body irradiation. The figure was created with BioRender.com.
One technique is based on the belief that only when iPSCs are differentiated into hematopoietic progenitor cells (iHPCs) first and then differentiated into microglia, they can recapitulate the true ontogeny of microglia [35]. This two-step protocol effectively generates iMGLs from iPSCs in more than 5 weeks. First of all, iPSCs are grown in defined conditions with several hematopoietic growth factors under certain oxygen concentrations (5% for 4 days and 20% for 6 days). This yields primitive CD43+/CD235a+/CD41+ iHPCs by 10 days with a > 90% purity. These iHPCs represent the early primitive hematopoietic cells derived from the yolk sac that give rise to microglia. Secondly, CD43+ iHPCs are cultured in serum-free differentiation medium containing CSF-1, IL-34, and TGFβ1. On day 14, induced cells present an early commitment to the fate of microglia and are similar to the developing microglial progenitor cells found in vivo. On day 38, iMGLs seem to resemble human microglia rather than monocytes or macrophages by cytospin/Giemsa staining. As iMGLs mature, they also become more branched. Transcriptome analysis shows that the iMGLs induced by this method are highly similar to primary human microglia. In contrast to their respective iPSCs, genomic integrity is also maintained during differentiation. This approach outlines the ontogenesis of microglia and can be easily scaled up for high-content screening. Another two-stage protocol also first differentiates iPSCs into iHPCs, then into microglia-like cells by co-culture with astrocytes [36]. The cells induced in this way also have phenotypic, transcriptional, and in vitro functional characteristics of brain-derived microglia. Compared with the previous method, this method does not require specific changes in oxygen concentration during the induction process of iHPCs, which is more feasible for most laboratories.
The second scheme is to induce iPSCs to differentiate into embryoid bodies (EBs) first, and then continue to culture and isolate iMGLs [37]. Uniform clumps of iPSCs are transferred in ultra-low attachment plates and cultured in a fully defined serum-free neuroglial differentiation (NGD) medium. The components and concentrations of this medium match human cerebrospinal fluid and extra IL-34 and CSF1 are added. Formed EBs have two types of structure. One is composed of compact phase-bright neuralized spheroids, and the other is composed of large, expanding cystic bodies. The second type of EB is positive for VE-cadherin, c-kit CD41, and CD235a, which have been identified as markers of early yolk sac myelogenesis in mice. Therefore, these EBs are referred to as yolk sac EBs (YS-EBs). Large domains of the YS-EBs are positive for myeloid transcription factor PU.1, which is necessary for microglial differentiation and maintenance. Every 5 days, the YS-EBs are triturated gently and cells of interest are selected for further culture. Attached cells are monitored for morphological characteristics of microglial precursors. iMGLs generated by this method are believed to faithfully recapitulate the expected ontogeny and characteristics of their in vivo counterparts, and they highly resemble primary human microglia. But the yield of this scheme is low.
There is also an alternative approach which directly induces iMGLs from peripheral blood mononuclear cells (PBMCs) simply by supplementing IL-34 and GM-CSF into PBMC culture medium [18, 38]. Cells can be harvested or used for functional assays in 10 days. This approach does not aim at representing the ontogeny of microglia. Cluster analysis shows that the iMGLs generated by this method are the closest to the gene expression of human fetal primary microglia. This approach is simple and easy to operate, and does not need to repeat the complicated development process.
There are other reports on how to induce microglia-like cells [39–43]. For a further overview of methods for generating induced microglia-like cells, see Table 1. But actually, there is no gold standard for successful generation of induced microglia yet and no iMGLs can recapitulate all aspects of microglial characteristics. iMGL models based on patient-derived iPSCs can better reflect the patient’s genetic background, but they can still not reflect the epigenetic or environmental factors’ contribution to the pathophysiology of diseases. This is a rapidly developing field, and as single-cell sequencing provides more detailed information, researchers can determine the most accurate models that resemble a certain state of interest. But human iPSC-derived microglia under 2D culture condition are immature, and can best represent embryonic microglia. That is the rational microglia-containing brain organoid, and human-mouse microglial chimeric models are needed.
Table 1.
Strategies for establishing human induced microglia-like cells
| Name (reference) | Process of induction | Characteristics (including strength and weakness) | Additional information |
|---|---|---|---|
|
pMGLs (Muffat et al. 2016 [37]) |
1. Form YS-EBs from iPSCs/ESCs 2. Microglial differentiation and maintenance |
∙ Resemble primary human fetal microglia ∙ Resemble yolk sac ontogeny ∙ Can embed in neuronal co-culture environment ∙ The organotypic 3D culture system cannot resemble in vivo environment |
Cited in tMGs established by Claes et al. 2019 [43] (the last row in this table) |
|
iMGLs (Abud et al. 2017 [35]) |
1. Differentiate iPSCs to iHPCs 2. Differentiate iHPCs to iMGLs |
∙ Resemble primary human microglia ∙ Resemble hematopoietic ontogeny ∙ Need hypoxic conditions (5%) ∙ Can integrate within 3D brain organoids ∙ Can engraft into MITRG mice and act as microglia in the development |
Associated kits are commercially available from STEMCELL Technologies Inc.; Improved into iPS-microglia 2.0 by McQuade et al. 2018 [42] (row 6 in this table, from the same lab); Cited in tMGs established by Claes et al. 2019 [43] (the last row in this table) |
|
iPS-MG (Pandya et al. 2017 [36]) |
1. Differentiate iPSCs to iPS-HPCs 2. Co-culture with astrocytes and differentiate to iPS-MG |
∙ Resemble primary human fetal microglia ∙ Resemble hematopoiesis ontogeny ∙ Need co-culture with astrocytes to induce final differentiation ∙ 2D culture system cannot resemble in vivo environment |
|
|
iPSC-MG (Douvaras et al. 2017 [39]) |
1. Differentiate ESCs/iPSCs to myeloid progenitors 2. Continue to differentiate into ramified microglia |
∙ Resemble primary human microglia ∙ Resemble primitive hematopoiesis ontogeny ∙ monoculture system cannot resemble in vivo environment |
Cited in human-mouse microglial chimeras established by Svoboda et al. 2019 [86] |
|
co-pMG (Haenseler et al. 2017 [40]) |
1. Form defined-size EBs from iPSCs 2. pMacpre differentiate from EBs 3. Differentiate iPSCs to pNeurons 4. Co-culture pMacpre with pNeurons |
∙ Resemble primary human fetal microglia ∙ Resemble yolk sac ontogeny ∙ Need co-culture with neurons ∙ 2D culture system cannot resemble in vivo environment |
Cited in human-mouse microglial chimeras established by Xu et al. 2020 [85]; Cited in microglia-containing brain organoids established by Xu et al. 2020 [50] |
|
iPS-microglia 2.0 (McQuade et al. 2018 [42]) |
1. Simplified differentiation of iPSCs/ESCs to HPCs 2. Updated differentiation of HPCs to iPS-microglia 2.0 |
∙ Equivalent to previously developed iPSC-microglia (Abud et al. 2017 [35]) ∙ Do not need hypoxia or cell sorting ∙ Can engraft into MITRG mice and enable in vivo study of human microglia |
Cited in human-mouse microglial chimeras established by Hasselmann et al. 2019 [82] (from the same lab) |
|
oMG (Ormel et al. 2018 [49]) |
1. iPSC generation 2. Microglia-containing organoid differentiation |
∙ Resemble primary human adult microglia ∙ Innately developing within cerebral organoids can mimic the CNS microenvironment ∙ Heterogeneity is high and uncontrollable |
|
|
hiMG (Sellgren et al. 2017 [38]) |
1. Preparation of PBMCs from whole blood 2. Generation of hiMG from PBMCs |
∙ Resemble primary human fetal microglia ∙ Do not represent the ontogeny of microglia ∙ The method is quick and convenient ∙ 2D culture system cannot resemble in vivo environment |
|
|
tMGs (Claes et al. 2019 [43]) |
1. Differentiate hPSCs to monocytes 2. Transdifferentiate monocytes to tMGs |
∙ Resemble primary human microglia ∙ Do not represent the ontogeny of microglia |
Cited in human-mouse microglial chimeras established by Mancuso et al. 2019 [84] (from the same lab) |
Abbreviations: ESCs, embryonic stem cells; pMGLs, pluripotent stem cell-derived microglia-like cells; iHPCs, iPSC-derived human hematopoietic progenitor cells; iMGLs, human microglia-like cells; iPS-HPCs, induced pluripotent stem cell-derived hematopoietic progenitor-like cells; iPS-MG, induced pluripotent stem cell-derived microglia-like cells; iPSC-MG, induced pluripotent stem cell-derived microglia; hiMG, human induced microglia-like cells; co-pMG, co-culture PSC microglia; pMacpre, PSC-derived macrophage precursors; pNeurons, PSC-derived cortical neurons; oMG, organoid-grown microglia; EBs, embryoid bodies; YS-EBs, yolk sac embryoid bodies; PBMCs, peripheral blood mononuclear cells; tMGs, transdifferentiated microglia-like cells.
Co-culture of Microglial Cells and Other Cells/Tissues
The co-culture model of microglia and neurons can be used in studying the interaction between microglia and neurons. Microglia and neurons can be co-cultured with three experimental systems involving different levels of cell-to-cell communication [44, 45]: (1) a conditioned medium transfer system in which no cell-to-cell contact or communication is possible, (2) a transwell system that allows cell-contact-independent communication only through diffusible soluble factors, and (3) a co-culture system that permits direct contact of microglia and neurons (Fig. 1B). These strategies can be used in investigating the roles of soluble and/or cell-associated chemokines in neuron-microglia interactions.
The ex vivo brain slice can retain important cell-to-cell interactions and is an important model for studying the activation and migration of parenchymal microglia [46]. However, the preparation of ex vivo brain slices causes trauma, especially damage of neuronal axons, resulting in microglia in brain slices having many of the same pathological damages that occur with serum exposure. Organotypic hippocampal slice culture (OHSC) is a well-accepted model in which to study different neurobiological aspects very close to the in vivo situation [47]. The Masuch team refilled the ramified microglia isolated from adult mouse brains into OHSC with microglia depleted, creating microglia-replenished OHSC (Mrep-OHSC) (Fig. 1C). Replenished microglia can integrate into the tissue, and the degree of ramification is no different from their counterparts in the mouse brain. Studies suggest that these replenished microglia maintain their original functions and properties as in vivo. This model is a unique tool for constructing chimeric brain slices allowing study of the function of different phenotypes of in vivo-like microglia in a tissue culture environment.
As noted in the last section, iPSC technologies are constantly evolving. And then, a new class of in vitro system—the 3D culture of brain organoids—has been improved [48]. This brain organoid can contain a variety of randomly distributed structures similar to different regions of the brain, or it can be directionally differentiated into the structure of a specific brain region. They can reproduce the process of the early development of the human brain. These models have been quickly applied to the study of human brain development and related diseases, and have achieved many results. Generally, iPSCs in vitro are stimulated to develop EBs. EBs are then subjected to neural induction in a minimal medium that only supports the development of neuroectoderm. The neuroectodermal tissues then gradually develop into mature brain organoids by enormous self-organizing capacity. This process determines that the brain organoid models do not contain microglia derived from the yolk sac. Therefore, researchers attempt to fuse brain organoids and microglia from various sources to build models for studying the role of microglia in human brain development, neurodegenerative diseases, and neuropsychiatric diseases (Fig. 1D). Abud et al. transplanted iMGLs into 3D human brain organoids to resemble microglia in the brain environment [35]. The limitation of their model is that integration of mature microglia and the organoid at a stage when neurons have already developed, which may not reflect the early embryonic development when microglial progenitors and neural progenitor cells (NPCs) interact with each other and undergo differentiation and maturation together. There is also a report that microglia can innately develop within a brain organoid model and display certain microglial characteristics [49]. However, due to the absence of SMAD (small mothers against decapentaplegic signaling) inhibition, the heterogeneity of cell types in these organoids is high and uncontrollable. Recently, Xu et al. generated developmentally appropriate and brain region-specific microglia-containing brain organoids by co-culturing hPSC-derived primitive macrophage progenitors (PMPs) and primitive neural progenitor cells (pNPCs) [50]. This model better mimics early neurodevelopmental processes observed in in vivo brain development and the ratio of microglia can be adjusted by controlling the starting number of pNPCs and PMPs. At present, the relevant technology threshold still is relatively high. And due to the problem of high heterogeneity of brain organoids, such technology needs to be further improved.
In Vivo Microglial Models
Labeling of Microglia
Visualization is an important way to study microglia in vivo. Iba1 is a specific marker for microglia. Iba1-EGFP transgenic mice use an Iba1 promoter fragment to control the expression of fluorescent reporter genes, which can effectively mark in vivo microglia [51]. The expression of EGFP can be detected from embryonic day 10.5 (E10.5) onwards, and strong EGFP signals appear in the yolk sac and the CNS ~E11.5. In recent years, Cx3cr1+/GFP transgenic mice have also been widely used to investigate the functions of microglia and other myeloid cells in the CNS [52]. CX3CR1 is the receptor of fractalkine and has been shown to be of great importance for the development of the CNS. The Cx3cr1+/GFP mouse line knocks the EGFP gene into the gene locus of Cx3cr1 and can strongly label microglia. The colony-stimulating factor 1 receptor (CSF1R), also known as macrophage colony-stimulating factor receptor (MCSFR) and cluster of differentiation 115 (CD115), is a cell-surface protein encoded by the Csf1r gene and plays a vital role in the development and maintenance of macrophages. The “MacGreen” (Csf1r-EGFP) mouse line, which uses the Csf1r gene promoter element to control EGFP expression [53], as well as the “MacBlue” (Csf1r-Gal4VP16/UAS-ECFP) mouse line [54], both can be used to label microglia in vivo. However, whether it is IBA1, CX3CR1, or CSF1R, they are not expressed exclusively in microglia. That means fluorescent reporter genes are not only expressed in microglia but also in other cells of the monophagocytic system [55]. TMEM119 is a newly identified microglia-specific marker in both mice and humans [56]. It has better specificity to target microglia. The Tmem119-EGFP mouse line was generated and was shown to completely and faithfully label parenchymal microglia rather than other brain macrophages [57], and Tmem119 mRNA is expressed throughout microglial development. The Tmem119-EGFP line can be used for early developmental studies of microglia.
Depletion of Microglia
The depletion of microglia is an effective model in which to study the regulatory function and activity of microglia in neurodevelopment. A model for knocking out microglia in vivo is CSF1R-deficient (Csf1r−/−) mice [58]. microglia of this mouse line are practically completely knocked out during development, and these mice develop a disturbed brain architecture during the post-natal period. But these mice die a few weeks after birth. Surprisingly, it has been reported that CSF1R is also expressed in some neurons and has a protective effect on excitotoxic injury [59]. Therefore, Csf1r−/− mice may not only be devoid of microglia, but some neurons are also directly affected. The CD11b-HSVTK transgenic mouse is another model for ablating microglia [60]. CD11b is also known as ITGAM, αM integrin, or complement receptor 3 (CR3), and is specifically expressed in myeloid cells such as macrophages. This model uses the CD11b promoter to control the expression of herpes simplex virus thymidine kinase (HSVTK) so that HSVTK is exclusively expressed by microglia in the brain. Thymidine kinase can convert ganciclovir (GCV) into a cytotoxic kinase that can cause cell suicide. This means that ganciclovir exposure can induce the suicide of HSVTK-expressing microglia in CD11b-HSVTK transgenic mice. However, CD11b is also expressed in the circulatory system. Systemic ganciclovir administration can cause hematopoietic toxicity and lead to fatal aplastic anemia, which should be prevented by transplanting bone marrow cells from wild-type mice. However, this bone marrow chimeric mouse needs to receive whole-body radiation before transplantation, which causes damage to the BBB [61], so this model may not reflect the normal physiological state. One alternative method of ganciclovir administration is by intraventricular injection to achieve partial removal of microglia in the cortex [62]. However, it should be noted that, as an antiviral drug, ganciclovir itself has an effect on the immune response. It has been shown to significantly inhibit the proliferation and activity of microglia in certain diseases, which may have an impact on microglial research [63]. Another method of selectively depleting microglia involves the use of DTR transgenic mice in which the human diphtheria toxin receptor (DTR) is expressed under the control of the CD11b promoter [64]. Localized injection of diphtheria toxin into the mice selectively ablates the microglia expressing human DTR. The number of microglia decreases significantly 12–24 h after diphtheria toxin treatment, but then gradually increases and returns to control levels at 36 h. Besides, transgenic mice targeting CD11c and PU.1 may be potential models for in vivo microglial study [65, 66]. However, due to their limitations, these models have not been widely used in the study of microglia.
There are also methods of using drugs to knock out microglia. Clodronate is such a drug which mediates apoptosis, but itself cannot penetrate cell membranes. It can be encapsulated in liposomes and then be engulfed by phagocytes. Once engulfed, the liposomes degrade and clodronate is released. This method was first used to selectively delete macrophages [67] but has now been used to deplete microglia. Clodronate liposomes are injected directly into the lateral ventricle of late embryonic rats, or clodronate liposomes are added to the culture medium of brain slices in vitro [10, 68]. The knock-out rate of microglia can reach 90% and 95%, respectively. CSF1R inhibitors are novel choices which can effectively deplete microglia in the CNS. PLX3397 is a small molecule CSF1R inhibitor. By oral gavage for consecutive 21 days, it can cross the mouse BBB and deplete ~90% of CD11b+CD45int microglia without significantly altering the number of monocytes or macrophages [69]. PLX5622 is another CSF1R inhibitor. It can achieve acute and near-complete microglial depletion within 3 days [70]. These pharmacological approaches to specifically deplete microglia have been used more commonly. It is worth noting that, since CSF1R is also expressed by myeloid cells including monocytes and macrophages, as CSF1R inhibitors, PLX5622 and PLX3397, have off-target effects, not only targeted on microglia.
Altering Gene Expression of Microglia
Alteration of gene expression in microglia in vivo is usually achieved through the Cre-loxP recombinase system. Using CD11b-Cre transgenic mice to hybridize with mice whose target gene is marked by loxP sites, can specifically knock out the target gene in cells expressing CD11b [71, 72]. Also, CD11c-Cre, Csf1r-iCre, Csf1r-Mer-iCre-Mer, Cx3cr1Cre, and other transgenic mice may become potential research models for altering microglial gene expression [73–75]. This technique of altering microglial gene expression has the same problem as microglial labeling and knockout, that is, it is not specific to microglia. Besides, approaches of genetic targeting, including the approaches noted above, contain hidden risks. The integration process of inserting or deleting sequences can cause unpredictable consequences like mutations in coding regions and altering the expression of other genes, affecting non-coding genomic elements [76]. A 2016 paper identified the Sall1CreER line as another specific genetic mouse line for microglia-specific manipulation [77]. Sall1 was reported to be critical in maintaining the microglial core signature. In this mouse line, the Cre recombinase is under the control of the Sall1 promoter and is expressed under tamoxifen induction. It should be noted that although Sall1 is expressed largely by microglia but not peripheral myeloid cells or other adult CNS-resident cells, its expression can be detected in liver, kidney, and neuronal and glial progenitors in the CNS during embryogenesis. The Tmem119-CreERT2 line was also established to control gene expression in both adult and early postnatal microglia [57]. This line has better specificity but the Cre activity of this mouse line is still not 100% specific in microglia. There is low Cre activity in the choroid plexus and blood monocytes. Researchers have newly identified hexosaminidase subunit beta (Hexb) as a core gene stably expressed by microglia both during homeostasis and disease [78]. They applied CRISPR/Cas9 genome editing to develop new transgenic mouse models for visualizing and altering the gene expression of microglia under the control of the Hexb promoter. These models can discriminate microglia from CAMs at the genetic level and stably monitor microglial behavior in vivo.
Recombinant viruses are important tools for manipulating gene expression in in vivo models and can achieve cell type-specific gene expression regulation in a short time. In neuroscience, viral targeting strategies have been successful in neurons, astrocytes, and oligodendrocytes. Although microglia are refractile to viruses, there have also been numerous attempts to achieve the viral transduction of microglia. Lentiviruses and adeno-associated viruses (AAVs) are preferentially selected because they have low immunogenicity. One study demonstrated that a capsid-modified rAAV6 expresses the transgene under the control of microglia-specific promoters (F4/80 or CD68) [79]. This rAAV6 capsid variant has triple mutations (Y731F/Y705F/T492V) which prevent proteasomal degradation when AAV escapes the endosomal compartment and has high tropism for monocytes. Researchers report a high transduction efficiency in primary microglia, but low specificity when injected into the mouse brain. One approach for enhancing viral selectivity toward a specific type of cell is by posttranscriptional regulation. Via inserting complementary miRNA target sites into the transgene cassette, transgene messenger RNA degrades specifically in cells expressing the miRNA. It has been reported that murine microglia lack microRNA-9 (miR-9) activity, whereas most other cells with a neuroectodermal origin in the brain express miR-9. Injection of miR-9-regulated lentivirus vectors into the striatum of adult rats induces expression of the GFP reporter gene mainly in ramified microglia [80], 75% of GFP-expressing cells co-label with the microglial marker IBA1. In general, it is still difficult to achieve high efficiency and high specificity for microglial transduction in vivo and available approaches are very limited. As a type of immune cell, the resistance to virus infection of microglia may be due to their functions of detecting, engulfing, and destroying pathogens similar to macrophages [81]. Better viral vectors need to be designed so that can be used efficiently in microglial transduction.
Human-Mouse Chimeric Model
For a long time, there has been no experimental platform for systematic research and analysis of human microglia in vivo. The human-mouse microglial chimeric model emerged and has become an alternative solution (Fig. 1E). In 2019, Mathew Blurton-Jones’s lab improved a chimeric method to study human microglia based on their previous study [82]. They used MITRG humanized immunodeficient mice to establish this chimeric model. This mouse line was constructed by knocking the genes of humanized M-CSFh, IL-3/GM-CSFh, and TPOh into BRG (Balb/c Rag2−/−Il2rg−/−) immunodeficient mice through homologous recombination technology [83]. Since the survival of microglia is CSF1R signal-dependent, xenotransplantation of iPSC-derived iHPCs into the early postnatal brain of this MITRG mouse can cause their environment-dependent differentiation into microglia. They showed that the expression of hCSF2 (hGM-CSF) and hTPO are not necessary for xenotransplanted microglia (xMG); hCSF1 is both necessary and sufficient to enable long-term survival of xMG in the mouse brain. These xMGs have the transcriptome characteristics of human microglia in vivo and are responsive to both acute and chronic injuries. But a small population of the transplanted iHPCs was found to differentiate into other CNS macrophages. At a similar time, Bart De Strooper’s lab also reported a chimeric model established by using other types of human cells and mouse lines [84]. hESCs were differentiated into microglia first before being transplanted. And the recipient Rag2−/−Il2rγ−/−hCSF1KI mice (hCSF1KI) were pretreated with the CSF1R inhibitor BLZ945 to remove nearly half of the host microglia. Although most transplanted cells of this chimeric model mimic primary human microglia at the transcriptome level, some cells also showed a CAM expression profile. The hPSC-derived microglial chimeric mouse brain model developed by Peng Jiang’s lab is also based on the human CSF1 knock-in mouse line [85]. The transplanted cells are hPSC-derived primitive macrophage progenitors (PMPs). They tracked these engrafted cells for a longer time of 6 months. The vast majority of hPSC-derived PMPs differentiated into microglia and a relatively small population developed into CAMs or remained as progenitors. hPSC-derived microglia showed gradual maturation in a spatiotemporal manner. Rudolf Jaenisch’s lab used NSG mice (NOD scid gamma mice), NSG-T mice (NSG mice carrying the humanized IL3, SCF, and GM-CSF genes; NSG-triples), and NSG-Q mice (NSG-T mice also carrying the humanized CSF1 gene; NSG-quads) as recipients of their chimeric model [86]. And they also confirmed that human CSF1 is crucial for the survival and integration of transplanted human cells. The donor cells they used were hiPSC-derived iMPs (induced microglial precursors) or iMGs (induced microglia-like cells) which carry a GFP reporter for tracking. They found that iMPs more efficiently integrate into the mouse brain than iMGs. They explored the maturation process of transplanted iMPs. But they did not mention whether transplanted iMPs differentiated into other CNS macrophages. In summary, human-mouse chimeric models are new and potential tools for detecting the in vivo function of patient-derived or specific genetically modified microglia.
Replacement of Microglia
As shown above, the genetic manipulation of microglia has great limitations. Some scholars believe that exogenous microglial replacement may be an effective solution to this problem and a potential clinical treatment strategy. Varvel et al. established an irradiation-independent microglial replacement system via intracerebroventricular GCV treatment for 2 weeks in CD11b-HSVTK transgenic mice and assessed the peripheral origin of engrafted cells [87]. But in their further study, this model failed in AD therapy [88]. This raised the problem that whether myeloid-derived microglial replacement is not an effective therapeutic approach or whether those replaced microglia are exclusively from the bloodstream. Parabiosis is a surgical union of two organisms allowing sharing of the blood circulation [89]. Ajami et al. established a mouse blood-chimeric model by joining GFP-expressing transgenic mice and C57BL/6 wild-type mice in parabiosis [90]. They showed that the replacement of microglia by circulating precursors cannot be induced under physiological conditions. Huang et al. further validated that BBB disruption is not a sufficient prerequisite for blood cells differentiating into microglia and repopulated microglia are derived from residual microglia by using the symbiotic mouse model [91]. Another method of microglial replacement is through bone marrow transplantation (BMT). Derecki et al. transplanted wild-type bone marrow into a lethally irradiated mouse model of Rett syndrome that resulted in engraftment of bone-marrow-derived microglia-like myeloid cells into brain parenchyma and helped to keep the disease from developing [92]. But a validation study does not support BMT-derived microglial replacement as therapy for Rett syndrome [93]. Although attempts to replace microglia are less successful in the treatment of diseases, researchers are still trying. Recently, Xu et al. developed three improved strategies and named them microglial replacement by bone marrow transplantation (mrBMT), microglial replacement by peripheral blood (mrPB), and microglial replacement by microglial transplantation (mrMT), which can effectively increase the replacement rate of microglia [94] (Fig. 1F). The prerequisite for the successful replacement of microglia is to make a microglia-free niche. The strategies in this study were to utilize the CSF1R inhibitor PLX5622 before giving 9 Gy whole-body irradiation (WBI) treatment. Because CSF1R is essential for microglial survival, two-week administration of PLX5622 can fully deplete CNS-resident microglia. mrBMT and mrPB can replace microglia-like cells at the CNS-wide scale, and the replacement rate can reach 92% and 80%, respectively. However, transcriptome analysis showed that the microglia-like cells derived from these two methods exhibited characteristics closer to those of macrophages. mrMT can replace endogenous microglia with exogenous microglia in specific brain regions of interest, with a local replacement rate of >50%. Transcriptome analysis shows that mrMT cells retain microglia-like characteristics. The researchers tested replacement cells of these 3 strategies by intraperitoneal injection of lipopolysaccharide and confirmed that these cells retain the environmental surveillance function as CNS-resident immune cells. But no other functions of microglia were tested. And the researchers did not test the therapeutic potentials on diseases. The clinical application of microglial replacement may still have a long way to go.
Tools for Microglial Studies
Single-Cell Techniques
Microglia participate in a wide range of physiological and pathological processes in the brain. From development to aging, from homeostasis to disease, microglia are highly heterogeneous. It is thought that different subpopulations of microglia may play a role in different events, and the expression profile of microglia also changes in different responses. However, in the past, the classification of microglia was relatively simple, mainly based on their morphology, density, surface marker expression, and electrophysiological characteristics. Single-cell RNA sequencing (scRNA-seq) is a technology that has developed rapidly in recent years. It can analyze the complex heterogeneity of cell populations by accurately identifying single cells and labeling them. This method prevents the biologically relevant signals of a single cell from being obscured by the average measurement data of a large number of cells. Single-cell transcriptome sequencing of brain tissues from different regions, different developmental stages, and different physiological and pathological characteristics can obtain spatio-temporal specific and disease-related expression profiles of microglia, helping to describe the development, migration, and response processes of microglia, and may discover new subtypes of microglia [95]. The discovery of new biomarkers, such as the Hexb gene noted above, also benefited from the use of massively parallel single-cell sequencing technology [78]. Single-cell sequencing can be used not only for transcriptome analysis but also for studying epigenetic modifications and protein-protein interactions. The problem with this technique is that a single cell suspension is needed first. For tissue cells, the cell state and acute expression may change due to environmental changes and cell damage caused by the process of cell dissociation. And fresh human brain tissue is difficult to obtain. Single-nucleus RNA sequencing (snRNA-seq) is an alternative to scRNA-seq as it allows transcriptomic profiling of frozen human brain tissue. Studies have used snRNA-seq to assess changes in expression in multiple cell lineages from frozen post-mortem brain tissue of multiple sclerosis (MS) and AD and found transcriptomic changes in the microglia in these diseases [96, 97]. Gerrits et al. compared cellular versus nuclear transcriptomes from fresh and frozen human brain samples and demonstrated that microglial nuclear RNAs obtained from CNS tissue are a reliable proxy for microglial gene expression [98]. But Thrupp et al. put forward that snRNA-seq is not suitable for the detection of microglial activation genes in human frozen biopsy tissues [99]. They demonstrated that although there are only a small set of genes (1.1% of the gene population) is depleted in nuclei relative to cells in human microglia by using snRNA and scRNA sequencing, this population is enriched for microglial activation genes. Further improvements in snRNA-seq library preparation may possibly acquire better sensitivity and resolution of the nuclear transcriptome.
Another emerging single-cell technology is cytometry by time-of-flight mass spectrometry (CyTOF) [100]. This is a revolutionary technology merging conventional flow cytometry and mass spectrometry with more than 50 different surface markers, which can deeply profile the immune phenotype of small samples at the single-cell level. Böttcher et al. used this method to simultaneously measure multiple samples from different donors and brain regions, and compare them with cells from other compartments at the same time to study the heterogeneity of microglia [101]. This single-cell technique can help establish a more comprehensive molecular view of microglia, making it possible to identify novel markers, pathways, and regulators that are critical to their development, health, and disease. Moreover, by a combination of scRNA-seq with CyTOF, Sankowski et al. identified diverse functional states of human microglia and demonstrated microglial spatial diversity during homeostasis and disease [102].
While scRNA-seq and CyTOF give a comprehensive molecular characterization of a population of cells, they do not provide the in situ spatial information of the brain during homeostasis and disease progression. Both changes in molecular profiling and morphological transformations serve as a read-out of microglial functional changes. Salamanca et al. developed an automated pipeline named Microglia and Immune Cells Morphologies Analyser and Classifier (MIC-MAC) [103]. It specializes in accurately reconstructing and classifying 3D microglial morphologies at the single-cell level in more complex human postmortem samples. In the future, by combining this unbiased high-throughput imaging technology with other single-cell technologies, the spatial information of heterogenous microglia in health and disease can be more enriched.
In Vivo Imaging Techniques
Live imaging can help to elucidate the precise functions and responses of cells in vivo. Transcranial two-photon imaging of GFP-labeled microglia reveals their rapid dynamics and response to traumatic injuries in the mouse brain [104]. Wu's lab has further revealed that the mechanism of microglia-neuron communication depends on neuronal NMDA receptors and microglial P2Y12 receptors through two-photon time-lapse imaging techniques [105]. They also documented a daily rearrangement of the microglial landscape using chronic in vivo two-photon imaging and show that the microglial landscape can be modulated by various pathological states [106]. A recent study identified an interaction site between microglial processes and neuronal cell bodies by in vivo two-photon imaging [107]. Combined with STORM super-resolution microscopy, high-resolution electron tomography, and other techniques, they further discovered that microglia-neuron junctions have a specialized nanoarchitecture optimized for purinergic signaling. Three-photon imaging is a revolutionary non-invasive method for investigating deep brain structures in live and behaving animals [108]. It can be a potential tool for in vivo imaging of subcortical microglia within an intact mouse brain.
Calcium imaging is an method based on calcium shifts operated by different intracellular and extracellular mechanisms [109]. It has been widely used in studies in various brain cells. However, the knowledge about in vivo calcium signaling in microglia has been lacking since microglia largely resist attempts of in vivo labeling that are routinely used for in vivo calcium imaging of other cell types [110]. Tvrdik et al. used a Cre-dependent conditional mouse reporter of calcium, which facilitates the deployment of genetically encoded calcium indicators, to cross with the Iba1(Aif1)-IRES-Cre mouse line [111]. Microglial calcium signals have been recorded through high-speed intravital two-photon laser scanning microscopy. This method effectively reflects changes in the intracellular free calcium concentration in large microglial cell populations.
Studies on human in vivo microglia have mainly been carried out by in vivo imaging. Activated microglia express a series of pro-inflammatory cytokines and certain receptors on their surface, including the 18 kDa translocator protein (TSPO), which has been identified as the peripheral-type benzodiazepine receptor (PBR). In mammalian brains, the expression of TSPO turns out to be very low. However, under conditions of local inflammatory responses caused by brain injuries, neoplasms, or infections, the expression of TSPO appears to be upregulated. This makes TSPO a potentially ideal and sensitive biomarker of brain injury. Therefore, translocator protein positron emission tomography (TSPO-PET) imaging has been developed and widely used to track microglial activation. Positive signals can be found in the early stages in patients with AD and other psychoses [112]. However, translocator protein is not expressed in microglia exclusively but also in reactive astrocytes and other proliferating cells. This has led to the development of numerous second-generation TSPO ligands such as the radioligand [11C]PBR28 [113]. The total gray matter [11C]PBR28 binding ratio is used as a marker of microglial activity. [11C]PBR28 combined with PET imaging technology shows improved affinity and nonspecific binding properties.
Outlook
There are many classic and emerging research tools and technologies in the field of microglial research. In addition to rodents, animal models used to study microglia include non-human primates, other mammals, zebrafish, chickens, and Drosophila [114–117]. Human microglia have some basic similarities to their animal counterparts. For instance, they have some similar protein expression, such as IBA-1, transcription factor PU.1, adaptor protein DAP12, and M-CSF receptor. But microglia of these animal models, including rodent models, have some properties of their own, and human microglia differ from these animals in many ways. For example, in vitro cultures of primary mouse microglia do not adhere to the wall but on the upper layer of astrocytes, while human primary microglia directly adhere to the surface of the culture dish [25, 32]. The proliferation of rodent microglia cultured in vitro is also inconsistent compared to humans, and evaluating the proliferation of human microglia in vivo is more complicated [20]. Due to biochemical differences, microglia in rodents and humans have different responses to drugs. In certain cases, the effects of drugs may even be in opposite directions. Taking valproic acid (VPA), a neuroactive drug clinically used to treat bipolar disorder and epilepsy, as an example, rodent studies have found that VPA selectively kills microglia through a caspase-3-dependent mechanism, while primary microglia in humans fail to activate caspase 3 or induce apoptosis. VPA can also increase the phagocytosis of rodent microglia, but it inhibits the phagocytic capacity of human primary microglia [118, 119]. A cross-species single-cell analysis revealed that human microglia exhibit significant heterogeneity compared to all other mammals, including primates [117]. The origin of microglia in different species is not unique as well. In mice, microglia are derived from primitive macrophages emanating from the embryonic yolk sac during development and sustain the microglial population locally by self-renewal [8, 120]. Zebrafish embryonic microglia initiate from the rostral blood island, which is similar to the yolk sac, while adult microglia in zebrafish arise from the ventral wall of the dorsal aorta [121]. Human hematopoiesis also starts in the yolk sac [122]. And human microglia originate similar to their rodent counterparts [123]. Therefore, it is very important and challenging to choose appropriate research tools and to improve the existing animal models. Only then the research results in alternative models can be translated into humans and even applied to clinical practice.
The patient-derived iMGL is an emerging tool with great potential to study the pathogenesis and therapeutic targets of microglia-related CNS diseases. Microglial markers and morphological identification, phagocytosis assay, migration assay, and cytokine and chemokine profile analysis are traditional experiments for microglial identification. But these methods are not precise enough at the transcriptome level and mainly target the inflammatory response function of microglia. Transcriptomic analysis, especially single-cell techniques, can better identify and analyze the microglia in different states. The iMGL models have been used to study mental illnesses such as schizophrenia and AD. It has been found that the abnormal engulfment and increased inflammatory responses of brain microglia may be the cause of various mental illnesses [18, 35]. But patient-derived iMGLs under traditional 2D culture are immature and can only represent embryonic microglia. The microglia-containing brain organoid and human-mouse microglial chimeric models can partially solve this problem by providing an environment that complex cell-to-cell or cell-matrix interactions exist. The 3D and in vivo models are becoming superior tools to study microglia in human CNS diseases [35, 50, 82, 84–86].
To study the origin and development of microglia, the role and mechanism of microglia in normal physiology and disease, the ultimate target is to carry out microglia-related drug development and clinical treatment. Eliminating the genetically defective microglia and transplanting genetically modified microglia is a possible direction for the treatment of microglia-related CNS diseases. Efficient microglial replacement has been achieved in mouse models though there is still a long way to go before it can be applied to humans. Metachromatic leukodystrophy (MLD) is a neurodegenerative lysosomal storage disease (LSD). A phase I/II clinical trial for MLD patients has shown that after transplantation of the genetically modified hematopoietic stem cells into the patients’ CNS, these HSCs differentiate into microglia and then delay disease progress [124]. Microglial replacement has also been exploited to deliver therapeutics to X-linked adrenoleukodystrophy (X-ALD) and mucopolysaccharidosis (MPS) [125]. Transdifferentiation is another possible option. The approaches of converting glial cells like astrocytes into neurons of different subtypes in vivo have been achieved and have become a potential treatment option for some neurodegenerative diseases [126, 127]. When nerve injury occurs, innate microglia can rapidly multiply and recruit to the injury site. Based on these properties of microglia, if transdifferentiation of microglia into functional neurons in vivo could be realized, it would be a potential direction for the treatment of neurological damage and neurodegenerative diseases in the future.
Auxiliary tools for microglial research include mitochondrial energy metabolism analysis, genomic, proteomic, expression profile analysis, chemogenetic and optogenetic tools, electrophysiological recording, and behavioral analysis. Electrophysiological behaviors of microglia have been described both in cultured primary microglia and in situ microglia in acutely isolated brain slices [128, 129]. The ramified in situ microglia are distinct from their cultured counterparts. Even in the same acute cerebral slices from an AD mouse model, microglia show plaque-associated electrophysiological heterogeneity [130]. Chemogenetic and optogenetic tools are highly prevalent in neuronal studies. But these techniques have not been widely applied to microglia, perhaps due to microglial resistance to viral vectors. Recently, Yi et al. used transgenic mice to enable selective expression of inhibitory Designer Receptors Exclusively Activated by Designer Drugs (Gi DREADD) in microglia [131]. This chemogenetic approach in microglia can inhibit neuroinflammation and neuropathic pain in mice. In the future, with the development of effective targeting strategies for microglia, opto/chemogenetic tools could be broadly used in microglial research. Combination of RNAseq profiling, in situ hybridization, or mass cytometry together with high-throughput imaging technologies can help to enrich the spatial information of heterogenous microglia at the single-cell level. Studies of microglia can also benefit from various interdisciplinary approaches, like physics, mathematics, chemistry, information science, and artificial intelligence, to help develop new technologies, models, and tools. We expect significant progress and breakthroughs will be made in the field of microglial research in the near future.
Acknowledgments
This review was supported by grants from the National Key Research and Development Program of China (2017YFC0909200), the National Natural Science Foundation of China (81671336), Shanghai Key Laboratory of Psychotic Disorders (YG2016ZD06), and the Shanghai Mental Health Center (2019-YJ06).
Conflict of interest
All authors claim that there is no conflict of interest.
References
- 1.Pelvig DP, Pakkenberg H, Stark AK, Pakkenberg B. Neocortical glial cell numbers in human brains. Neurobiol Aging. 2008;29:1754–1762. doi: 10.1016/j.neurobiolaging.2007.04.013. [DOI] [PubMed] [Google Scholar]
- 2.Lawson LJ, Perry VH, Dri P, Gordon S. Heterogeneity in the distribution and morphology of microglia in the normal adult mouse brain. Neuroscience. 1990;39:151–170. doi: 10.1016/0306-4522(90)90229-W. [DOI] [PubMed] [Google Scholar]
- 3.Tay TL, Savage JC, Hui CW, Bisht K, Tremblay MÈ. Microglia across the lifespan: From origin to function in brain development, plasticity and cognition. J Physiol. 2017;595:1929–1945. doi: 10.1113/JP272134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ginhoux F, Greter M, Leboeuf M, Nandi S, See P, Gokhan S, et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 2010;330:841–845. doi: 10.1126/science.1194637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hirbec H, Déglon N, Foo LC, Goshen I, Grutzendler J, Hangen E, et al. Emerging technologies to study glial cells. Glia. 2020;68:1692–1728. doi: 10.1002/glia.23780. [DOI] [PubMed] [Google Scholar]
- 6.Prinz M, Jung S, Priller J. Microglia biology: One century of evolving concepts. Cell. 2019;179:292–311. doi: 10.1016/j.cell.2019.08.053. [DOI] [PubMed] [Google Scholar]
- 7.Cronk JC, Filiano AJ, Louveau A, Marin I, Marsh R, Ji E, et al. Peripherally derived macrophages can engraft the brain independent of irradiation and maintain an identity distinct from microglia. J Exp Med. 2018;215:1627–1647. doi: 10.1084/jem.20180247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nayak D, Roth TL, McGavern DB. Microglia development and function. Annu Rev Immunol. 2014;32:367–402. doi: 10.1146/annurev-immunol-032713-120240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Squarzoni P, Oller G, Hoeffel G, Pont-Lezica L, Rostaing P, Low D, et al. Microglia modulate wiring of the embryonic forebrain. Cell Rep. 2014;8:1271–1279. doi: 10.1016/j.celrep.2014.07.042. [DOI] [PubMed] [Google Scholar]
- 10.Cunningham CL, Martínez-Cerdeño V, Noctor SC. Microglia regulate the number of neural precursor cells in the developing cerebral cortex. J Neurosci. 2013;33:4216–4233. doi: 10.1523/JNEUROSCI.3441-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mallat M, Marín-Teva JL, Chéret C. Phagocytosis in the developing CNS: More than clearing the corpses. Curr Opin Neurobiol. 2005;15:101–107. doi: 10.1016/j.conb.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 12.Marín-Teva JL, Cuadros MA, Martín-Oliva D, Navascués J. Microglia and neuronal cell death. Neuron Glia Biol. 2011;7:25–40. doi: 10.1017/S1740925X12000014. [DOI] [PubMed] [Google Scholar]
- 13.Chounchay S, Noctor SC, Chutabhakdikul N. Microglia enhances proliferation of neural progenitor cells in an in vitro model of hypoxic-ischemic injury. EXCLI J . 2020;19:950–961. doi: 10.17179/excli2020-2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schafer DP, Lehrman EK, Kautzman AG, Koyama R, Mardinly AR, Yamasaki R, et al. Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron. 2012;74:691–705. doi: 10.1016/j.neuron.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hong S, Beja-Glasser VF, Nfonoyim BM, Frouin A, Li S, Ramakrishnan S, et al. Complement and microglia mediate early synapse loss in Alzheimer mouse models. Science. 2016;352:712–716. doi: 10.1126/science.aad8373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim HJ, Cho MH, Shim WH, Kim JK, Jeon EY, Kim DH, et al. Deficient autophagy in microglia impairs synaptic pruning and causes social behavioral defects. Mol Psychiatry. 2017;22:1576–1584. doi: 10.1038/mp.2016.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brites D, Fernandes A. Neuroinflammation and depression: Microglia activation, extracellular microvesicles and microRNA dysregulation. Front Cell Neurosci. 2015;9:476. doi: 10.3389/fncel.2015.00476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sellgren CM, Gracias J, Watmuff B, Biag JD, Thanos JM, Whittredge PB, et al. Increased synapse elimination by microglia in schizophrenia patient-derived models of synaptic pruning. Nat Neurosci. 2019;22:374–385. doi: 10.1038/s41593-018-0334-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qin C, Zhou LQ, Ma XT, Hu ZW, Yang S, Chen M, et al. Dual functions of microglia in ischemic stroke. Neurosci Bull. 2019;35:921–933. doi: 10.1007/s12264-019-00388-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith AM, Dragunow M. The human side of microglia. Trends Neurosci. 2014;37:125–135. doi: 10.1016/j.tins.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 21.Dello Russo C, Cappoli N, Coletta I, Mezzogori D, Paciello F, Pozzoli G, et al. The human microglial HMC3 cell line: Where do we stand? A systematic literature review. J Neuroinflammation. 2018;15:259. doi: 10.1186/s12974-018-1288-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagamoto-Combs K, Kulas J, Combs CK. A novel cell line from spontaneously immortalized murine microglia. J Neurosci Methods. 2014;233:187–198. doi: 10.1016/j.jneumeth.2014.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Floden AM, Combs CK. Microglia repetitively isolated from in vitro mixed glial cultures retain their initial phenotype. J Neurosci Methods. 2007;164:218–224. doi: 10.1016/j.jneumeth.2007.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gordon R, Hogan CE, Neal ML, Anantharam V, Kanthasamy AG, Kanthasamy A. A simple magnetic separation method for high-yield isolation of pure primary microglia. J Neurosci Methods. 2011;194:287–296. doi: 10.1016/j.jneumeth.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moussaud S, Draheim HJ. A new method to isolate microglia from adult mice and culture them for an extended period of time. J Neurosci Methods. 2010;187:243–253. doi: 10.1016/j.jneumeth.2010.01.017. [DOI] [PubMed] [Google Scholar]
- 26.Utz SG, See P, Mildenberger W, Thion MS, Silvin A, Lutz M, et al. Early fate defines microglia and non-parenchymal brain macrophage development. Cell 2020, 181: 557–573.e18. [DOI] [PubMed]
- 27.Biber K, Owens T, Boddeke E. What is microglia neurotoxicity (Not)? Glia. 2014;62:841–854. doi: 10.1002/glia.22654. [DOI] [PubMed] [Google Scholar]
- 28.Bergner CG, van der Meer F, Winkler A, Wrzos C, Türkmen M, Valizada E, et al. Microglia damage precedes major myelin breakdown in X-linked adrenoleukodystrophy and metachromatic leukodystrophy. Glia. 2019;67:1196–1209. doi: 10.1002/glia.23598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagai A, Mishima S, Ishida Y, Ishikura H, Harada T, Kobayashi S, et al. Immortalized human microglial cell line: Phenotypic expression. J Neurosci Res. 2005;81:342–348. doi: 10.1002/jnr.20478. [DOI] [PubMed] [Google Scholar]
- 30.Williams K, Bar-Or A, Ulvestad E, Olivier A, Antel JP, Yong VW. Biology of adult human microglia in culture: Comparisons with peripheral blood monocytes and astrocytes. J Neuropathol Exp Neurol. 1992;51:538–549. doi: 10.1097/00005072-199209000-00009. [DOI] [PubMed] [Google Scholar]
- 31.McLarnon JG, Helm J, Goghari V, Franciosi S, Choi HB, Nagai A, et al. Anion channels modulate store-operated calcium influx in human microglia. Cell Calcium. 2000;28:261–268. doi: 10.1054/ceca.2000.0150. [DOI] [PubMed] [Google Scholar]
- 32.Lue LF, Beach TG, Walker DG. Alzheimer's disease research using human microglia. Cells. 2019;8:838. doi: 10.3390/cells8080838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 34.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 35.Abud EM, Ramirez RN, Martinez ES, Healy LM, Nguyen CHH, Newman SA, et al. iPSC-derived human microglia-like cells to study neurological diseases. Neuron 2017, 94: 278–293.e9. [DOI] [PMC free article] [PubMed]
- 36.Pandya H, Shen MJ, Ichikawa DM, Sedlock AB, Choi Y, Johnson KR, et al. Differentiation of human and murine induced pluripotent stem cells to microglia-like cells. Nat Neurosci. 2017;20:753–759. doi: 10.1038/nn.4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muffat J, Li Y, Yuan B, Mitalipova M, Omer A, Corcoran S, et al. Efficient derivation of microglia-like cells from human pluripotent stem cells. Nat Med. 2016;22:1358–1367. doi: 10.1038/nm.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sellgren CM, Sheridan SD, Gracias J, Xuan D, Fu T, Perlis RH. Patient-specific models of microglia-mediated engulfment of synapses and neural progenitors. Mol Psychiatry. 2017;22:170–177. doi: 10.1038/mp.2016.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Douvaras P, Sun B, Wang M, Kruglikov I, Lallos G, Zimmer M, et al. Directed differentiation of human pluripotent stem cells to microglia. Stem Cell Reports. 2017;8:1516–1524. doi: 10.1016/j.stemcr.2017.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haenseler W, Sansom SN, Buchrieser J, Newey SE, Moore CS, Nicholls FJ, et al. A highly efficient human pluripotent stem cell microglia model displays a neuronal-co-culture-specific expression profile and inflammatory response. Stem Cell Reports. 2017;8:1727–1742. doi: 10.1016/j.stemcr.2017.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beutner C, Roy K, Linnartz B, Napoli I, Neumann H. Generation of microglial cells from mouse embryonic stem cells. Nat Protoc. 2010;5:1481–1494. doi: 10.1038/nprot.2010.90. [DOI] [PubMed] [Google Scholar]
- 42.McQuade A, Coburn M, Tu CH, Hasselmann J, Davtyan H, Blurton-Jones M. Development and validation of a simplified method to generate human microglia from pluripotent stem cells. Mol Neurodegener. 2018;13:67. doi: 10.1186/s13024-018-0297-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Claes C, Van Den Daele J, Boon R, Schouteden S, Colombo A, Monasor LS, et al. Human stem cell-derived monocytes and microglia-like cells reveal impaired amyloid plaque clearance upon heterozygous or homozygous loss of TREM2. Alzheimers Dement. 2019;15:453–464. doi: 10.1016/j.jalz.2018.09.006. [DOI] [PubMed] [Google Scholar]
- 44.Zujovic V, Taupin V. Use of cocultured cell systems to elucidate chemokine-dependent neuronal/microglial interactions: Control of microglial activation. Methods. 2003;29:345–350. doi: 10.1016/S1046-2023(02)00358-4. [DOI] [PubMed] [Google Scholar]
- 45.Roqué PJ, Costa LG. Co-culture of neurons and microglia. Curr Protoc Toxicol 2017, 74: 11.24.1–11.24.17. [DOI] [PMC free article] [PubMed]
- 46.Petersen MA, Dailey ME. Diverse microglial motility behaviors during clearance of dead cells in hippocampal slices. Glia. 2004;46:195–206. doi: 10.1002/glia.10362. [DOI] [PubMed] [Google Scholar]
- 47.Masuch A, van der Pijl R, Füner L, Wolf Y, Eggen B, Boddeke E, et al. Microglia replenished OHSC: A culture system to study in vivo like adult microglia. Glia. 2016;64:1285–1297. doi: 10.1002/glia.23002. [DOI] [PubMed] [Google Scholar]
- 48.Lancaster MA, Knoblich JA. Generation of cerebral organoids from human pluripotent stem cells. Nat Protoc. 2014;9:2329–2340. doi: 10.1038/nprot.2014.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ormel PR, Vieira de Sá R, van Bodegraven EJ, Karst H, Harschnitz O, Sneeboer MAM, et al. Microglia innately develop within cerebral organoids. Nat Commun 2018, 9: 4167. [DOI] [PMC free article] [PubMed]
- 50.Xu R, Boreland AJ, Li X, Erickson C, Jin M, Atkins C, et al. Developing human pluripotent stem cell-based cerebral organoids with a controllable microglia ratio for modeling brain development and pathology. bioRxiv 2020. 10.1101/2020.10.09.331710. [DOI] [PMC free article] [PubMed]
- 51.Hirasawa T, Ohsawa K, Imai Y, Ondo Y, Akazawa C, Uchino S, et al. Visualization of microglia in living tissues using Iba1-EGFP transgenic mice. J Neurosci Res. 2005;81:357–362. doi: 10.1002/jnr.20480. [DOI] [PubMed] [Google Scholar]
- 52.Jung S, Aliberti J, Graemmel P, Sunshine MJ, Kreutzberg GW, Sher A, et al. Analysis of fractalkine receptor CX3CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol Cell Biol. 2000;20:4106–4114. doi: 10.1128/MCB.20.11.4106-4114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sasmono RT, Oceandy D, Pollard JW, Tong W, Pavli P, Wainwright BJ, et al. A macrophage colony-stimulating factor receptor-green fluorescent protein transgene is expressed throughout the mononuclear phagocyte system of the mouse. Blood. 2003;101:1155–1163. doi: 10.1182/blood-2002-02-0569. [DOI] [PubMed] [Google Scholar]
- 54.Ovchinnikov DA, van Zuylen WJ, DeBats CE, Alexander KA, Kellie S, Hume DA. Expression of Gal4-dependent transgenes in cells of the mononuclear phagocyte system labeled with enhanced cyan fluorescent protein using Csf1r-Gal4VP16/UAS-ECFP double-transgenic mice. J Leukoc Biol. 2008;83:430–433. doi: 10.1189/jlb.0807585. [DOI] [PubMed] [Google Scholar]
- 55.Wieghofer P, Prinz M. Genetic manipulation of microglia during brain development and disease. Biochim Biophys Acta. 2016;1862:299–309. doi: 10.1016/j.bbadis.2015.09.019. [DOI] [PubMed] [Google Scholar]
- 56.Bennett ML, Bennett FC, Liddelow SA, Ajami B, Zamanian JL, Fernhoff NB, et al. New tools for studying microglia in the mouse and human CNS. Proc Natl Acad Sci U S A. 2016;113:E1738–E1746. doi: 10.1073/pnas.1525528113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kaiser T, Feng G. Tmem119-EGFP and Tmem119-CreERT2 transgenic mice for labeling and manipulating microglia. eNeuro 2019, 6. 10.1523/eneuro.0448-18.2019. [DOI] [PMC free article] [PubMed]
- 58.Erblich B, Zhu L, Etgen AM, Dobrenis K, Pollard JW. Absence of colony stimulation factor-1 receptor results in loss of microglia, disrupted brain development and olfactory deficits. PLoS One. 2011;6:e26317. doi: 10.1371/journal.pone.0026317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Luo J, Elwood F, Britschgi M, Villeda S, Zhang H, Ding Z, et al. Colony-stimulating factor 1 receptor (CSF1R) signaling in injured neurons facilitates protection and survival. J Exp Med. 2013;210:157–172. doi: 10.1084/jem.20120412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Heppner FL, Greter M, Marino D, Falsig J, Raivich G, Hövelmeyer N, et al. Experimental autoimmune encephalomyelitis repressed by microglial paralysis. Nat Med. 2005;11:146–152. doi: 10.1038/nm1177. [DOI] [PubMed] [Google Scholar]
- 61.Mildner A, Schmidt H, Nitsche M, Merkler D, Hanisch UK, Mack M, et al. Microglia in the adult brain arise from Ly-6ChiCCR2+ monocytes only under defined host conditions. Nat Neurosci. 2007;10:1544–1553. doi: 10.1038/nn2015. [DOI] [PubMed] [Google Scholar]
- 62.Simard AR, Soulet D, Gowing G, Julien JP, Rivest S. Bone marrow-derived microglia play a critical role in restricting senile plaque formation in Alzheimer's disease. Neuron. 2006;49:489–502. doi: 10.1016/j.neuron.2006.01.022. [DOI] [PubMed] [Google Scholar]
- 63.Ding Z, Mathur V, Ho PP, James ML, Lucin KM, Hoehne A, et al. Antiviral drug ganciclovir is a potent inhibitor of microglial proliferation and neuroinflammation. J Exp Med. 2014;211:189–198. doi: 10.1084/jem.20120696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ueno M, Fujita Y, Tanaka T, Nakamura Y, Kikuta J, Ishii M, et al. Layer V cortical neurons require microglial support for survival during postnatal development. Nat Neurosci. 2013;16:543–551. doi: 10.1038/nn.3358. [DOI] [PubMed] [Google Scholar]
- 65.Goldmann T, Wieghofer P, Müller PF, Wolf Y, Varol D, Yona S, et al. A new type of microglia gene targeting shows TAK1 to be pivotal in CNS autoimmune inflammation. Nat Neurosci. 2013;16:1618–1626. doi: 10.1038/nn.3531. [DOI] [PubMed] [Google Scholar]
- 66.McKercher SR, Torbett BE, Anderson KL, Henkel GW, Vestal DJ, Baribault H, et al. Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. EMBO J 1996, 15: 5647–5658. [PMC free article] [PubMed]
- 67.van Rooijen N, van Nieuwmegen R. Elimination of phagocytic cells in the spleen after intravenous injection of liposome-encapsulated dichloromethylene diphosphonate. An enzyme-histochemical study. Cell Tissue Res. 1984;238:355–358. doi: 10.1007/BF00217308. [DOI] [PubMed] [Google Scholar]
- 68.Faustino JV, Wang X, Johnson CE, Klibanov A, Derugin N, Wendland MF, et al. Microglial cells contribute to endogenous brain defenses after acute neonatal focal stroke. J Neurosci. 2011;31:12992–13001. doi: 10.1523/JNEUROSCI.2102-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li MS, Li ZG, Ren HL, Jin WN, Wood K, Liu Q, et al. Colony stimulating factor 1 receptor inhibition eliminates microglia and attenuates brain injury after intracerebral hemorrhage. J Cereb Blood Flow Metab. 2017;37:2383–2395. doi: 10.1177/0271678X16666551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Acharya MM, Green KN, Allen BD, Najafi AR, Syage A, Minasyan H, et al. Elimination of microglia improves cognitive function following cranial irradiation. Sci Rep. 2016;6:31545. doi: 10.1038/srep31545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ferron M, Vacher J. Targeted expression of Cre recombinase in macrophages and osteoclasts in transgenic mice. Genesis. 2005;41:138–145. doi: 10.1002/gene.20108. [DOI] [PubMed] [Google Scholar]
- 72.Boillée S, Yamanaka K, Lobsiger CS, Copeland NG, Jenkins NA, Kassiotis G, et al. Onset and progression in inherited ALS determined by motor neurons and microglia. Science. 2006;312:1389–1392. doi: 10.1126/science.1123511. [DOI] [PubMed] [Google Scholar]
- 73.Caton ML, Smith-Raska MR, Reizis B. Notch-RBP-J signaling controls the homeostasis of CD8- dendritic cells in the spleen. J Exp Med. 2007;204:1653–1664. doi: 10.1084/jem.20062648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Deng L, Zhou JF, Sellers RS, Li JF, Nguyen AV, Wang Y, et al. A novel mouse model of inflammatory bowel disease links mammalian target of rapamycin-dependent hyperproliferation of colonic epithelium to inflammation-associated tumorigenesis. Am J Pathol. 2010;176:952–967. doi: 10.2353/ajpath.2010.090622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Qian BZ, Li JF, Zhang H, Kitamura T, Zhang JH, Campion LR, et al. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature. 2011;475:222–225. doi: 10.1038/nature10138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wieghofer P, Knobeloch KP, Prinz M. Genetic targeting of microglia. Glia. 2015;63:1–22. doi: 10.1002/glia.22727. [DOI] [PubMed] [Google Scholar]
- 77.Buttgereit A, Lelios I, Yu X, Vrohlings M, Krakoski NR, Gautier EL, et al. Sall1 is a transcriptional regulator defining microglia identity and function. Nat Immunol. 2016;17:1397–1406. doi: 10.1038/ni.3585. [DOI] [PubMed] [Google Scholar]
- 78.Masuda T, Amann L, Sankowski R, Staszewski O, Lenz M, D Errico P, et al. Novel Hexb-based tools for studying microglia in the CNS. Nat Immunol 2020, 21: 802–815. [DOI] [PubMed]
- 79.Rosario AM, Cruz PE, Ceballos-Diaz C, Strickland MR, Siemienski Z, Pardo M, et al. Microglia-specific targeting by novel capsid-modified AAV6 vectors. Mol Ther Methods Clin Dev. 2016;3:16026. doi: 10.1038/mtm.2016.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Åkerblom M, Sachdeva R, Quintino L, Wettergren EE, Chapman KZ, Manfre G, et al. Visualization and genetic modification of resident brain microglia using lentiviral vectors regulated by microRNA-9. Nat Commun. 2013;4:1770. doi: 10.1038/ncomms2801. [DOI] [PubMed] [Google Scholar]
- 81.Maes ME, Colombo G, Schulz R, Siegert S. Targeting microglia with lentivirus and AAV: Recent advances and remaining challenges. Neurosci Lett. 2019;707:134310. doi: 10.1016/j.neulet.2019.134310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hasselmann J, Coburn MA, England W, Figueroa Velez DX, Kiani Shabestari S, Tu CH, et al. Development of a chimeric model to study and manipulate human microglia in vivo. Neuron 2019, 103: 1016–1033.e10. [DOI] [PMC free article] [PubMed]
- 83.Rongvaux A, Willinger T, Martinek J, Strowig T, Gearty SV, Teichmann LL, et al. Development and function of human innate immune cells in a humanized mouse model. Nat Biotechnol. 2014;32:364–372. doi: 10.1038/nbt.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mancuso R, Van Den Daele J, Fattorelli N, Wolfs L, Balusu S, Burton O, et al. Stem-cell-derived human microglia transplanted in mouse brain to study human disease. Nat Neurosci. 2019;22:2111–2116. doi: 10.1038/s41593-019-0525-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xu RJ, Li XX, Boreland AJ, Posyton A, Kwan K, Hart RP, et al. Human iPSC-derived mature microglia retain their identity and functionally integrate in the chimeric mouse brain. Nat Commun. 2020;11:1577. doi: 10.1038/s41467-020-15411-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Svoboda DS, Barrasa MI, Shu J, Rietjens R, Zhang S, Mitalipova M, et al. Human iPSC-derived microglia assume a primary microglia-like state after transplantation into the neonatal mouse brain. Proc Natl Acad Sci U S A. 2019;116:25293–25303. doi: 10.1073/pnas.1913541116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Varvel NH, Grathwohl SA, Baumann F, Liebig C, Bosch A, Brawek B, et al. Microglial repopulation model reveals a robust homeostatic process for replacing CNS myeloid cells. Proc Natl Acad Sci U S A. 2012;109:18150–18155. doi: 10.1073/pnas.1210150109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Varvel NH, Grathwohl SA, Degenhardt K, Resch C, Bosch A, Jucker M, et al. Replacement of brain-resident myeloid cells does not alter cerebral amyloid-β deposition in mouse models of Alzheimer's disease. J Exp Med. 2015;212:1803–1809. doi: 10.1084/jem.20150478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kamran P, Sereti KI, Zhao P, Ali SR, Weissman IL, Ardehali R. Parabiosis in mice: A detailed protocol. J Vis Exp. 2013 doi: 10.3791/50556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ajami B, Bennett JL, Krieger C, Tetzlaff W, Rossi FM. Local self-renewal can sustain CNS microglia maintenance and function throughout adult life. Nat Neurosci. 2007;10:1538–1543. doi: 10.1038/nn2014. [DOI] [PubMed] [Google Scholar]
- 91.Huang Y, Xu Z, Xiong S, Sun F, Qin G, Hu G, et al. Repopulated microglia are solely derived from the proliferation of residual microglia after acute depletion. Nat Neurosci. 2018;21:530–540. doi: 10.1038/s41593-018-0090-8. [DOI] [PubMed] [Google Scholar]
- 92.Derecki NC, Cronk JC, Lu Z, Xu E, Abbott SB, Guyenet PG, et al. Wild-type microglia arrest pathology in a mouse model of Rett syndrome. Nature. 2012;484:105–109. doi: 10.1038/nature10907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang J, Wegener JE, Huang TW, Sripathy S, De Jesus-Cortes H, Xu P, et al. Wild-type microglia do not reverse pathology in mouse models of Rett syndrome. Nature. 2015;521:E1–E4. doi: 10.1038/nature14444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Xu Z, Rao Y, Huang Y, Zhou T, Feng R, Xiong S, et al. Efficient strategies for microglia replacement in the central nervous system. Cell Rep. 2020;32:108041. doi: 10.1016/j.celrep.2020.108041. [DOI] [PubMed] [Google Scholar]
- 95.Li Q, Cheng Z, Zhou L, Darmanis S, Neff NF, Okamoto J, et al. Developmental heterogeneity of microglia and brain myeloid cells revealed by deep single-cell RNA sequencing. Neuron 2019, 101: 207–223.e10. [DOI] [PMC free article] [PubMed]
- 96.Schirmer L, Velmeshev D, Holmqvist S, Kaufmann M, Werneburg S, Jung D, et al. Neuronal vulnerability and multilineage diversity in multiple sclerosis. Nature. 2019;573:75–82. doi: 10.1038/s41586-019-1404-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mathys H, Davila-Velderrain J, Peng Z, Gao F, Mohammadi S, Young JZ, et al. Single-cell transcriptomic analysis of Alzheimer's disease. Nature. 2019;570:332–337. doi: 10.1038/s41586-019-1195-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gerrits E, Heng Y, Boddeke EWGM, Eggen BJL. Transcriptional profiling of microglia; current state of the art and future perspectives. Glia. 2020;68:740–755. doi: 10.1002/glia.23767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Thrupp N, Sala Frigerio C, Wolfs L, Skene NG, Fattorelli N, Poovathingal S, et al. Single-nucleus RNA-seq is not suitable for detection of microglial activation genes in humans. Cell Rep. 2020;32:108189. doi: 10.1016/j.celrep.2020.108189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Masuda T, Sankowski R, Staszewski O, Prinz M. Microglia heterogeneity in the single-cell era. Cell Rep. 2020;30:1271–1281. doi: 10.1016/j.celrep.2020.01.010. [DOI] [PubMed] [Google Scholar]
- 101.Böttcher C, Schlickeiser S, Sneeboer MAM, Kunkel D, Knop A, Paza E, et al. Human microglia regional heterogeneity and phenotypes determined by multiplexed single-cell mass cytometry. Nat Neurosci. 2019;22:78–90. doi: 10.1038/s41593-018-0290-2. [DOI] [PubMed] [Google Scholar]
- 102.Sankowski R, Böttcher C, Masuda T, Geirsdottir L, Sagar, Sindram E, et al. Mapping microglia states in the human brain through the integration of high-dimensional techniques. Nat Neurosci 2019, 22: 2098–2110. [DOI] [PubMed]
- 103.Salamanca L, Mechawar N, Murai KK, Balling R, Bouvier DS, Skupin A. MIC-MAC: An automated pipeline for high-throughput characterization and classification of three-dimensional microglia morphologies in mouse and human postmortem brain samples. Glia. 2019;67:1496–1509. doi: 10.1002/glia.23623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Davalos D, Grutzendler J, Yang G, Kim JV, Zuo Y, Jung S, et al. ATP mediates rapid microglial response to local brain injury in vivo. Nat Neurosci. 2005;8:752–758. doi: 10.1038/nn1472. [DOI] [PubMed] [Google Scholar]
- 105.Eyo UB, Peng J, Swiatkowski P, Mukherjee A, Bispo A, et al. Neuronal hyperactivity recruits microglial processes via neuronal NMDA receptors and microglial P2Y12 receptors after status epilepticus. J Neurosci. 2014;34:10528–10540. doi: 10.1523/JNEUROSCI.0416-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Eyo UB, Mo M, Yi MH, Murugan M, Liu J, Yarlagadda R, et al. P2Y12R-dependent translocation mechanisms gate the changing microglial landscape. Cell Rep. 2018;23:959–966. doi: 10.1016/j.celrep.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cserép C, Pósfai B, Lénárt N, Fekete R, László ZI, Lele Z, et al. Microglia monitor and protect neuronal function through specialized somatic purinergic junctions. Science. 2020;367:528–537. doi: 10.1126/science.aax6752. [DOI] [PubMed] [Google Scholar]
- 108.Horton NG, Wang K, Kobat D, Clark CG, Wise FW, Schaffer CB, et al. In vivo three-photon microscopy of subcortical structures within an intact mouse brain. Nat Photonics. 2013;7:205–209. doi: 10.1038/nphoton.2012.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.de Melo Reis RA, Freitas HR, de Mello FG. Cell calcium imaging as a reliable method to study neuron-glial circuits. Front Neurosci. 2020;14:569361. doi: 10.3389/fnins.2020.569361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Brawek B, Garaschuk O. Microglial calcium signaling in the adult, aged and diseased brain. Cell Calcium. 2013;53:159–169. doi: 10.1016/j.ceca.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 111.Tvrdik P, Kearns KN, Sharifi KA, Sluzewski MF, Acton ST, Kalani MYS. Calcium imaging of microglial network activity in stroke. Methods Mol Biol. 2019;2034:267–279. doi: 10.1007/978-1-4939-9658-2_19. [DOI] [PubMed] [Google Scholar]
- 112.Edison P, Donat CK, Sastre M. In vivo imaging of glial activation in Alzheimer's disease. Front Neurol. 2018;9:625. doi: 10.3389/fneur.2018.00625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bloomfield PS, Selvaraj S, Veronese M, Rizzo G, Bertoldo A, Owen DR, et al. Microglial activity in people at ultra high risk of psychosis and in schizophrenia: An [11C]PBR28 PET brain imaging study. Am J Psychiatry. 2016;173:44–52. doi: 10.1176/appi.ajp.2015.14101358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kuil LE, Oosterhof N, Ferrero G, Mikulasova T, Hason M, Dekker J, et al. Zebrafish macrophage developmental arrest underlies depletion of microglia and reveals Csf1r-independent metaphocytes. Elife 2020, 9: e53403. [DOI] [PMC free article] [PubMed]
- 115.Orczykowski ME, Calderazzo SM, Shobin E, Pessina MA, Oblak AL, Finklestein SP, et al. Cell based therapy reduces secondary damage and increases extent of microglial activation following cortical injury. Brain Res. 2019;1717:147–159. doi: 10.1016/j.brainres.2019.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Stanhope BA, Jaggard JB, Gratton M, Brown EB, Keene AC. Sleep regulates glial plasticity and expression of the engulfment receptor draper following neural injury. Curr Biol 2020, 30: 1092–1101.e3. [DOI] [PubMed]
- 117.Geirsdottir L, David E, Keren-Shaul H, Weiner A, Bohlen SC, Neuber J, et al. Cross-species single-cell analysis reveals divergence of the primate microglia program. Cell 2019, 179: 1609–1622.e16. [DOI] [PubMed]
- 118.Smith AM, Gibbons HM, Dragunow M. Valproic acid enhances microglial phagocytosis of amyloid-beta(1–42) Neuroscience. 2010;169:505–515. doi: 10.1016/j.neuroscience.2010.04.041. [DOI] [PubMed] [Google Scholar]
- 119.Gibbons HM, Smith AM, Teoh HH, Bergin PM, Mee EW, Faull RL, et al. Valproic acid induces microglial dysfunction, not apoptosis, in human glial cultures. Neurobiol Dis. 2011;41:96–103. doi: 10.1016/j.nbd.2010.08.024. [DOI] [PubMed] [Google Scholar]
- 120.Prinz M, Priller J. Microglia and brain macrophages in the molecular age: From origin to neuropsychiatric disease. Nat Rev Neurosci. 2014;15:300–312. doi: 10.1038/nrn3722. [DOI] [PubMed] [Google Scholar]
- 121.Xu J, Zhu L, He S, Wu Y, Jin W, Yu T, et al. Temporal-spatial resolution fate mapping reveals distinct origins for embryonic and adult microglia in zebrafish. Dev Cell. 2015;34:632–641. doi: 10.1016/j.devcel.2015.08.018. [DOI] [PubMed] [Google Scholar]
- 122.Tavian M, Péault B. Embryonic development of the human hematopoietic system. Int J Dev Biol. 2005;49:243–250. doi: 10.1387/ijdb.041957mt. [DOI] [PubMed] [Google Scholar]
- 123.Ginhoux F, Lim S, Hoeffel G, Low D, Huber T. Origin and differentiation of microglia. Front Cell Neurosci. 2013;7:45. doi: 10.3389/fncel.2013.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Biffi A, Montini E, Lorioli L, Cesani M, Fumagalli F, Plati T, et al. Lentiviral hematopoietic stem cell gene therapy benefits metachromatic leukodystrophy. Science. 2013;341:1233158. doi: 10.1126/science.1233158. [DOI] [PubMed] [Google Scholar]
- 125.Cartier N, Lewis CA, Zhang R, Rossi FM. The role of microglia in human disease: Therapeutic tool or target? Acta Neuropathol. 2014;128:363–380. doi: 10.1007/s00401-014-1330-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Janowska J, Gargas J, Ziemka-Nalecz M, Zalewska T, Buzanska L, Sypecka J. Directed glial differentiation and transdifferentiation for neural tissue regeneration. Exp Neurol. 2019;319:112813. doi: 10.1016/j.expneurol.2018.08.010. [DOI] [PubMed] [Google Scholar]
- 127.Qian H, Kang X, Hu J, Zhang D, Liang Z, Meng F, et al. Reversing a model of Parkinson's disease with in situ converted nigral neurons. Nature. 2020;582:550–556. doi: 10.1038/s41586-020-2388-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kettenmann H, Banati R, Walz W. Electrophysiological behavior of microglia. Glia. 1993;7:93–101. doi: 10.1002/glia.440070115. [DOI] [PubMed] [Google Scholar]
- 129.Boucsein C, Kettenmann H, Nolte C. Electrophysiological properties of microglial cells in normal and pathologic rat brain slices. Eur J Neurosci. 2000;12:2049–2058. doi: 10.1046/j.1460-9568.2000.00100.x. [DOI] [PubMed] [Google Scholar]
- 130.Plescher M, Seifert G, Hansen JN, Bedner P, Steinhäuser C, Halle A. Plaque-dependent morphological and electrophysiological heterogeneity of microglia in an Alzheimer's disease mouse model. Glia. 2018;66:1464–1480. doi: 10.1002/glia.23318. [DOI] [PubMed] [Google Scholar]
- 131.Yi MH, Liu YU, Liu K, Chen T, Bosco DB, Zheng J, et al. Chemogenetic manipulation of microglia inhibits neuroinflammation and neuropathic pain in mice. Brain Behav Immun. 2021;92:78–89. doi: 10.1016/j.bbi.2020.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]