Abstract
In the current study, we report the genome sequence of two different clinical isolates from India, Trichophyton indotineae UCMS-IGIB-CI12 and Trichophyton indotineae UCMS-IGIB-CI14. The resulting genome assembly achieved a 143-fold coverage in 824 contigs for T. indotineae UCMS-IGIB-CI12 and a 136-fold coverage in 904 contigs for T. indotineae UCMS-IGIB-CI14. Both the clinical isolates contain a c.1342G>A mutation corresponding to Ala448Thr amino acid substitution in erg1 and exhibit an intermittent drug response to terbinafine. Comparative genomics analysis with available genomes of Trichophyton interdigitale/Trichophyton mentagrophytes species complex revealed a similar genome architecture and identified large number of genes associated with virulence and pathogenicity, namely, lipases, proteases, LysM domain-containing factors, carbon metabolism enzymes and cytochrome P450 enzymes, in all the genomes. An analysis of single amino acid polymorphisms (SAPs) in the protein sequences of subtilisin and lipase enzyme families identified a higher frequency of SAPs in functionally important proteins, Sub3 and Sub6 and their possible use in multilocus phylogenetic analysis of T. interdigitale/T. mentagrophytes species complex. The whole genome sequences of T. indotineae clinical isolates provided in this report will, hence, serve as a key reference point for investigation of clinical strains and emerging drug resistance among dermatophytes originating from different parts of the world.
Keywords: Trichophyton spp.; Dermatophytes, whole-genome sequencing; Phylogenetic analysis
Dermatophytes are a group of keratinophilic filamentous fungi belonging to the genera Trichophyton, Epidermophyton and Microsporum that initiate infection with the attachment of arthroconidia to the stratum corneum and consume keratin, a main component of outer layer of skin, as its primary nutrient source (White et al. 2008; Baldo et al. 2012). Further spread of the developing hyphae through the keratin layer is aided by a repertoire of secretory keratinases, proteases and lipases (Martinez et al. 2012; Latka et al. 2015), that act as important virulence factors (Monod 2008; Peres et al. 2010). During recent years, there are many reports of drug resistance in several Trichophyton spp. among dermatophytes emerging from India and rest of the world. While majority of the recalcitrant infections indicate resistance to terbinafine, prevalence of resistance across azole family of drugs has also been reported, with most reports for resistance to either drug from Trichophyton mentagrophytes/Trichophyton interdigitale isolates (Yamada et al. 2017; Rudramurthy et al. 2018; Singh et al. 2018; Saunte et al. 2019; Ebert et al. 2020). Failure of allylamine and azole drugs to make interference in ergosterol synthesis pathway, the primary target of antifungal agents, sounds the alarm for revisiting the traditional molecular workflow and a need for change in traditional thinking in treatment of dermatophytosis. Availability of few whole genome sequences (wgs), across genomic resources of EMBL/GenBank/DDBJ has limited a genome-wide detailed analysis approach so far. We provide here the whole genome sequence of two clinical isolates from India belonging to T. mentagrophytes/T. interdigitale genotype VIII (Taghipour et al. 2019; Nenoff et al. 2019) and recently reannotated as Trichophyton indotineae (Kano et al. 2020; Tang et al. 2021). The wgs of T. indotineae clinical isolates from India and their comparison with available genomes of T. interdigitale/T. mentagrophytes species complex will aid in our understanding of pathogenesis of these dermatophytes, in better management of the disease and further evaluation of available therapeutic options.
The clinical isolates were collected from skin scrapings around the lesions and examined under the microscope using 10% KOH. A portion of the sample was cultured on Sabouraud’s dextrose agar with chloramphenicol (0.05 g/L), gentamicin (20 mg/L) and cycloheximide (0.5 g/L) at 25 °C for 3–4 weeks. After growth, the etiological agent was confirmed by the characteristic morphology of the colony and by studying the microscopic appearance of the fungus on Lacto Phenol Cotton Blue mount. Genotypic identification of the isolates was carried out by DNA extraction and PCR amplification of the region spanning nuclear ribosomal internal transcribed spacer (ITS) regions 1 to 2 of 18S rRNA using panfungal primers; ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) (White et al. 1990) followed by Sanger sequencing and comparison with available ITS sequences of dermatophytes in GenBank using BLAST. A 100% sequence similarity of the amplified 18S rRNA region was obtained with the ITS sequence of T. mentagrophytes genotype VIII (GenBank accession number: MH517560.1, Nenoff et al. 2019). T. mentagrophytes genotype VIII has recently been reclassified as a new species, i.e. T. indotineae, with characteristic three single nucleotide polymorphisms (SNPs) at position 94 (C), 125 (T) and 462 (T) in the ITS region. The presence of all three characteristic SNPs instead of 94A, 125C and 462C compared to the reference T. interdigitale 428.63 strain (Kano et al. 2020) (Fig. 1) confirmed the two isolates as T. indotineae and were designated as T. indotineae UCMS-IGIB-CI12 (abbreviated here as TiCI12) and T. indotineae UCMS-IGIB-CI14 (abbreviated here as TiCI14), respectively. The sequence of the internal transcribed spacer regions 1 to 2 of 18S rRNA of the two isolates TiCI12 and TiCI14 are deposited in GenBank with Accession numbers MW600527.1 and MW600653.1.
Fig. 1.
Comparison of ITS sequences of Trichophyton spp. Sequence alignment of internal transcribed spacer region (ITS1–ITS2) of 18S rRNA gene of T. indotineae UCMS-IGIB-CI12 (GenBank accession number: MW600527.1, this study) and T. indotineae UCMS-IGIB-CI14 (GenBank accession number: MW600653.1, this study), T. interdigitale NCCPF 800062 (T. mentagrophytes type VIII reference strain, Nenoff et al. (2019), GenBank accession number: MH517560.1) and T. interdigitale CBS 428.63 (T. interdigitale Type II reference strain, Nenoff et al. (2019), GenBank accession number: KT155896.1) showing the SNPs for T. indotineae (in red). The numbering convention for ITS sequence according to Kano et al. (2020) has been used for clarity and for maintaining reproducibility with their analysis
Several dermatophytes from the T. interdigitale/T. mentagrophytes species complex from India, including the recently reannotated T. indotineae, exhibit high level of terbinafine resistance associated with mutations in its molecular target, squalene epoxidase (SQLE/erg1) (Khurana et al. 2018; Rudramurthy et al. 2018; Singh et al. 2018, 2019, 2021; Shaw et al. 2020; Ebert et al. 2020; Gaurav et al. 2021). The presence of any mutations in erg1 of TiCI12 or TiCI14 was probed by sequencing of PCR amplified product of erg1 of TiCI12 and TiCI14 using forward primer, FP (5′ ATGGTTGTAGAGGCTCCTCCCTGC 3′) and reverse primer, RP (5′ CTAGCTTTGAAGTTCGGCAAATA 3′). A c.1342G>A mutation corresponding to Ala448Thr amino acid substitution was identified in erg1 of both the clinical isolates. Any change in minimum inhibitory concentration (MIC) due to these substitutions was assessed by an antifungal susceptibility test (AFST) to terbinafine as well as fluconazole by broth microdilution method as per Clinical and Lab Standards Institute (CLSI) M38-A2 guidelines, as defined earlier (Gaurav et al. 2021). T. mentagrophytes ATCC 18748 was included as a quality control strain for AFST. The in vitro antifungal susceptibility profile of TiCI12 and TiCI14 to these agents is summarized in Table 1. While MIC90 to fluconazole (16 μg/mL) was same in the control and clinical isolates, and in the range reported earlier for T. interdigitale/T. mentagrophytes (Rudramurthy et al. 2018; Singh et al. 2019), TiCI12 and TiCI14 showed variable tolerance to terbinafine, highlighting the need for detailed analysis of genomic features likely to be associated with virulence, pathogenicity and variability of these Indian strains.
Table 1.
MIC profile of indicated dermatophytes against different antifungal agents
| Strains | MIC90 (μg/mL) | |
|---|---|---|
| Terbinafine | Fluconazole | |
| T. mentagrophytes ATCC 18748 | 0.03 | 16 |
| TiCI12 | 0.06 | 16 |
| TiCI14 | > 8 | 16 |
For whole genome sequencing, the purified gDNA was end-repaired, adapter-ligated and enriched by following manufacturer’s protocol to make the library for sequencing using Nextra XT DNA Library Prep Kit. Library quantification was carried out using Agilent Bioanalyzer with high sensitivity DNA kit. The enriched libraries were pooled according to the unique adapter barcodes and sequencing was carried out on Illumina platform by 2 × 100 bp paired-end sequencing. Quality assessment of raw reads obtained in paired-end reads was done using FastQC-v.0.11.8 (Release Date: 04-October-2018) (Andrews 2010) followed by trimming to remove any duplicates and low quality reads using Trimmomatic v0.39 tools (Bolger et al. 2014). Further de novo assembly of filtered reads was done using SPAdes assembler v-3.13 (Bankevich et al. 2012) followed by Pilon correction (Walker et al. 2014) in order to make improvement within the draft assemblies. Assembly statistics and completeness was estimated using Assemblathon 3 and BUSCO v4.0.3 (Seppey et al. 2019) with lineage dataset i.e. fungi_odb10 (creation date: 2019-12-13). Finally, assembled fasta files were carried forward for structural and functional annotation using web-interface of AUGUSTUS (Stanke and Morgenstern 2005) and PANNZER2 (Protein ANNotation with Z-scoRE) (Törönen et al. 2018), respectively. The general features of TiCI12 and TiCI14 are summarized in Table 2 following the Minimal Information about any (X) Sequence (MIxS) standard checklist.
Table 2.
Standard checklist of the minimal information about any (X) sequence (MIxS) for Trichophyton indotineae UCMS-IGIB-CI12 and Trichophyton indotineae UCMS-IGIB-CI14
| Trichophyton indotineae UCMS-IGIB-CI12 | Trichophyton indotineae UCMS-IGIB-CI14 | |
|---|---|---|
| Classification |
Kingdom: Fungi Division: Ascomycota Class: Eurotiomycetes Order: Onygenales Family: Arthrodermataceae Genus: Trichophyton Species: T. indotineae Strain: UCMS-IGIB CI12 |
Kingdom: Fungi Division: Ascomycota Class: Eurotiomycetes Order: Onygenales Family: Arthrodermataceae Genus: Trichophyton Species: T. indotineae Strain: UCMS-IGIB CI14 |
| Environment | ||
|
Collection date Geographical location (latitude and longitude) Geographical location (country) Environment (biome) Environment (feature) Environmental (material) |
2014-09-25 28.7041 N 77.1025 E Delhi, India Homo sapiens Skin Lesion |
2014-09-30 28.7041 N 77.1025 E Delhi, India Homo sapiens Skin Lesion |
| MIxS data investigation | ||
|
Submitted to insdc Investigation type Project name BioProject BioSample Depth Ploidy Isolation and growth condition Sequencing method Assembly Finishing strategy |
JAATJQ000000000.1 (GenBank) Fungi Whole genome sequence of Trichophyton indotineae CI12 from India PRJNA604098 SAMN13951834 143.7× Diploid SDA with chloramphenicol, gentamicin and cycloheximide Illumina HiSeq SPAdes v. 3.13 assembler Draft, over 143.7-fold genome coverage, 824 contigs |
JAAQVJ000000000.1 (GenBank) Fungi Whole genome sequence of Trichophyton indotineae CI14 from India PRJNA604102 SAMN13951939 136.3× Diploid SDA with chloramphenicol, gentamicin and cycloheximide Illumina HiSeq SPAdes v. 3.13 assembler Draft, over 136.3-fold genome coverage, 904 contigs |
The total estimated size of the draft assembled genomes of TiCI12 and TiCI14 is 22.06 Mb and 22.04 Mb in 824 and 904 contigs with > 130-fold coverage for each genome and an N50 value of 57.4 kb and 54.0 kb for TiCI12 and TiCI14, respectively (Table 2). Despite the recent classification of members of T. interdigitale/T. mentagrophytes species complex into different genotype groups representing their geographical distribution (Nenoff et al. 2019; Taghipour et al. 2019) and the subsequent recent reannotation of T. mentagrophytes genotype VIII as T. indotineae (Kano et al. 2020; Tang et al. 2021), species boundaries between the two is difficult phylogenetically as T. mentagrophytes and T. interdigitale species are conspecific (Pchelin et al. 2019). Further, T. interdigitale and T. indotineae have been considered as anthropophilic clonal offshoots of T. mentagrophytes and the two names have been suggested to be retained primarily to maintain the epidemiological source of infection only; T. interdigitale for true anthropophilic and T. mentagrophytes for zoophilic infections (Symoens et al. 2011; de Hoog et al. 2017; Pchelin et al. 2019; Taghipour et al. 2019). Hence, considering the overall relatedness of members of T. interdigitale/T. mentagrophytes species complex, we decided to compare genomic features of all the available T. interdigitale/T. mentagrophytes strains in NCBI. In order to avoid heterogeneity among genomic datasets that may arise due to different sequencing platforms or different gene prediction pipelines, the complete genome assembly of all available T. interdigitale/T. mentagrophytes genomes were downloaded from NCBI and a structural and functional annotation was carried out using the same pipeline as described for TiCI12 and TiCI14, before comparative genomics analysis.
The achieved level of genome assembly for TiCI12 or TICI14 is in excellent agreement with the representative genome of RefSeq strain, T. interdigitale MR816 (Table 3). A total of 7581 and 7575 protein coding DNA sequences (cds) were predicted and annotated in the genomes of TiCI12 and TiCI14, respectively. A similar number, varying between 7579 and 7823 cds, were predicted for the other genomes except D15P152 (8488 cds) (Table 3). The somewhat higher predicted cds in D15P152, could possibly be due to the low coverage (5X), lower N50 (~ 5 kb) and large number of scaffolds (7988) for this genome (Table 3).
Table 3.
Overall genomic features of T. indotineae UCMS-IGIB-CI12 and T. indotineae UCMS-IGIB-CI14 and other available genomes in NCBI, of T. interdigitale/T. mentagrophytes species complex
| Strain name | Speciesa | GenBank accession number (wgs) | Total Sequence length (Mbp) | Depth of coverage (×) | N50 (bp) | Total number of scaffolds | Predicted cds (as per this study) | Reference |
|---|---|---|---|---|---|---|---|---|
| MR816 (RefSeq) | T. interdigitalea | AOKY00000000 | 22.47 | 56 | 58,103 | 956 | 7621 | Persinoti et al. (2018) |
| H6 | T. mentagrophytesa | AOKS00000000 | 21.94 | 91 | 18,239 | 2831 | 7823 | Persinoti et al. (2018) |
| M8436 | T. mentagrophytesa | FUFL00000000 | 22.59 | 35 | 27,151 | 3285 | 7674 | Gallo et al. (2017) |
| TiCI12 | T. indotineae | JAATJQ000000000 | 22.06 | 143.7 | 57,434 | 824 | 7581 | This study |
| TiCI14 | T. indotineae | JAAQVJ010000000 | 22.04 | 136.3 | 54,025 | 904 | 7575 | This study |
| D15P127 | T. mentagrophytesa | QQSR00000000 | 23.71 | 15 | 72,043 | 1842 | 7585 | Pchelin et al. (2019) |
| D15P135 | T. mentagrophytesa | QQSQ00000000 | 22.49 | 12 | 49,567 | 1462 | 7633 | Pchelin et al. (2019) |
| D15P152 | T. mentagrophytesa | QQSP00000000 | 23.12 | 5 | 5,051 | 7988 | 8488 | Pchelin et al. (2019) |
| D15P156 | T. mentagrophytesa | QQSO00000000 | 23.23 | 12 | 67,448 | 1442 | 7579 | Pchelin et al. (2019) |
| TIMM2789b | T. mentagrophytesa | BFBS00000000 | 24.07 | 160 | 81,080 | 16,543 | 7619 | Unpublishedc |
aSpecies classification is according to Pchelin et al. (2019)
bThis entry is flagged by NCBI as ‘Anomalous assembly’ due to contaminated reads (https://ncbi.nlm.nih.gov/assembly/GCA_003118255.1/). Hence was not considered further in this study
cAlshahni et al. (2018), Unpublished work
Presence of virulence genes encoded in the genomes of dermatophytes enables them to establish and maintain infection and survive on the outer cornified layers of skin of the host. A comparative analysis of virulence factors in the genomes using online prediction tools can reveal genomic features related to high pathogenicity of strains belonging to different genotypes and different geographical locations. PHI-base v.410 (The Pathogen-Host Interaction database, http://www.phi-base.org/) (Urban et al. 2017) is commonly used to predict the presence of broad spectrum of genes associated with virulence or pathogenicity. PHI-base v.410 did not identify any major difference in predicted pathogenicity, virulence or other effector genes in any of the dermatophytes and revealed a similar number of genes, varying between twenty to twenty seven, in all the genomes, with an identical number of twenty one pathogenicity and/or virulence factors in both TiCI12 or TiCI14 (Table 4).
Table 4.
Comparative distribution of predicted virulence factors in the genomes of T. interdigitale/T. mentagrophytes species complex
| Strain name | Virulence/pathogenicity factors (PHI-base) | Lipasesa | Peptidase | Subtilisin | LysM | Cytochrome-P450 | Carbohydrate active enzymes (dbCAN) |
|---|---|---|---|---|---|---|---|
| MR816 | 23 | 23 (5) | 167 | 16 | 6 | 30 | 140 |
| H6 | 22 | 23 (5) | 175 | 16 | 6 | 30 | 138 |
| M8436 | 22 | 22 (5) | 174 | 17 | 6 | 29 | 137 |
| TiCI12 | 21 | 24 (6) | 174 | 16 | 8 | 29 | 163 |
| TiCI14 | 21 | 24 (6) | 175 | 18 | 8 | 29 | 163 |
| D15P127 | 22 | 22 (5) | 170 | 17 | 6 | 29 | 139 |
| D15P135 | 21 | 23 (5) | 172 | 16 | 4 | 28 | 137 |
| D15P152 | 27 | 21 (4) | 191 | 18 | 6 | 35 | 117 |
| D15P156 | 20 | 22 (5) | 168 | 16 | 5 | 29 | 139 |
aThe predicted secretory subset of lipases are indicated in parenthesis
Secreted lipases and proteases are other important enzymes that not only serve as key virulence factors but also aid the dermatophytic fungi in surviving on the terminally differentiated keratinized layers of skin by deriving nutrients from it (Burmester et al. 2011; Achterman and White 2012; Martinez et al. 2012) and were next predicted in the genomes. A nearly identical number of lipases were predicted in all the genomes including TiCI12 and TiCI14 with help of lipase prediction server, LED v4.0.0 (https://led.biocatnet.de/sequence-browser) (Table 4). Dermatophytes also encode a large number of proteases in their genomes, with a key role of secretory subtilases in adherence and in initial stages of infection in the host cells (Kaufman et al. 2007; Latka et al. 2015). Among the 174 or 175 total peptidases predicted in the genomes of TiCI12 or TICI14 by the MEROPS server (https://www.ebi.ac.uk/merops/download_list.shtml) (Rawlings et al. 2018), a similar subset of 16 or 18 secreted subtilases were identified, which compares well with the 16–18 secreted subtilases among 167–191 predicted peptidases in all the strains (Table 4).
Among other key virulence factors, LysM domain-containing proteins help in binding of the fungi to N-linked oligosaccharides on the human skin glycoproteins (Kar et al. 2019) and have been reported to be enriched in pathogenic fungi. Genome annotation and function prediction identified eight LysM domain-containing proteins each in TiCI12 and TiCI14, compared to seven in the RefSeq T. mentagrophytes MR816 strain. Among other genes associated with virulence, proteins associated with carbohydrate metabolism were predicted as a set of ‘carbohydrate-active enzymes’ using dbCAN server (http://bcb.unl.edu/dbCAN/index.php) (Lombard et al. 2014). However, no clear distinguishing factor could be identified across different dermatophytes as the predicted set of genes was similar among all dermatophyte genomes (for instance, 163 proteins for TiCI12 or TiCI14 and 165 for RefSeq T. interdigitale MR816).
Cytochrome P450 family is another family of proteins that are abundant in fungi due to their roles not only in ergosterol biosynthesis but also in production of secretory secondary metabolites and detoxification of drugs and xenobiotics (Shin et al. 2018). Several cytochrome P450 enzymes (29 each in TiCI12 and TiCI14 and 30 in T. interdigitale MR816) were predicted by the Cytochrome P450 domain-containing proteins server (CYPED v6.0, CYtochrome P450 Engineering Database; https://cyped.biocatnet.de/sequence-browser) (Fischer et al. 2007), highlighting the key role of this family of enzymes in dermatophytic fungi.
Although a large number of potential factors associated with pathogenicity and/or virulence were identified in TiCI12 and TiCI14, there was no obvious difference in the predicted proteins in any group when compared to the purported zoophilic T. mentagrophytes or the anthropophilic T. interdigitale genomes (Table 4). Single nucleotide polymorphisms (SNPs) is a key method to identify any population/geographical/lineage/drug response or pathogenicity variability among different available genomes. The low number of nine wgs available from the genomic resources of EMBL/GenBank/DDBJ, however, limits a statistically significant analysis for SNPs across the genomes of T. interdigitale/T. mentagrophytes species complex. Among different types of SNPs in the genomes, the non-synonymous SNPs, designated as single amino-acid polymorphisms (or SAPs), (Kumar et al. 2009) would result in amino-acid changes and have a direct affect on function of the protein, providing important clues into mechanistic aspects of the disease. Owing to the key role of subtilisins and lipases in nutrient acquisition and survival on host cells, we analyzed SAPs in these two enzyme families in all the genomes. A multiple sequence alignment using CLUSTAL omega (Sievers et al. 2011), for the predicted subtilisins and lipases in all genomes helped identify SAPs in each protein when compared to T. interdigitale MR816 (Fig. 2). Predicted subtilisins with GenBank accession numbers KDB23473.1, KDB24326.1, KDB23788.1 and KDB27845.1 (Fig. 2A) and predicted lipases with GenBank accession numbers KDB26859.1 and KDB25940.1 (Fig. 2B) show largest number of SAPs in dermatophytes, as compared to RefSeq T. interdigitale MR816. Interestingly, TiCI12, TiCI14 and D15P135 (belonging to T. mentagrophytes genotype VIII, (Pchelin et al. 2019)) cluster together for both subtilase and lipase families on the basis of single amino acid change analysis (Fig. 2).
Fig. 2.
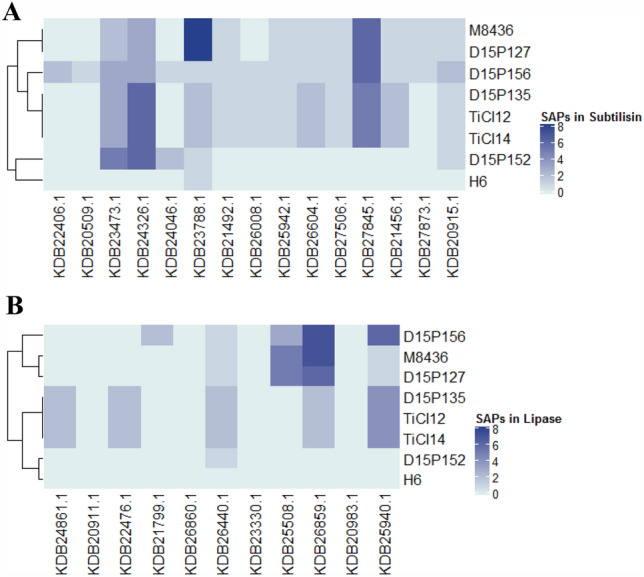
Heatmap for SAP analysis of (A) subtilisin and (B) lipase families. Corresponding homologous protein sequences of subtilisins and lipases were identified by local BLAST with T. interdigitale MR816 sequences as query in all the indicated genomes. Only full length sequences were used for mapping amino acid polymorphisms in a CLUSTAL omega multiple sequence alignment based SAP analysis. Gene accession numbers of T. interdigitale MR816 proteins used as a reference are indicated for A subtilisins and B lipases. T. interdigitale/T. mentagrophytes species included in the analysis are as mentioned in Table 3
The clustering in subtilase and lipase families of these genomes prompted us to investigate the possible use of the respective subtilase or lipase members for multilocus phylogenetic classification. Functional annotation of KDB23788.1 and KDB24326.1, showing large number of SAPs, identified them as Sub3 and Sub6 subtilisins, respectively. Both proteins play key roles in pathogenesis of dermatophytes. While Sub3 is one of the major subtilases secreted by dermatophytes at alkaline pH, Sub6 is the major protease secreted during infection (Méhul et al. 2016; Gräser et al. 2018).
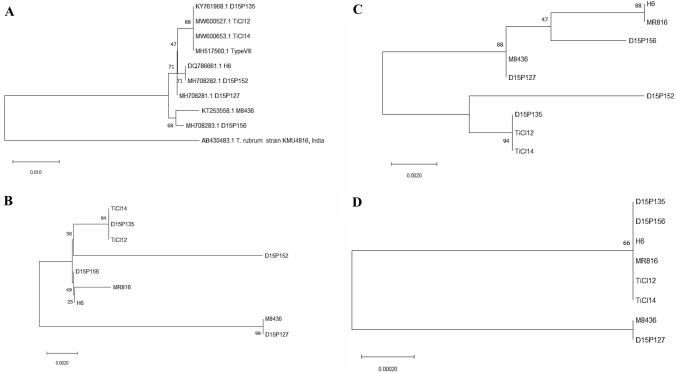
A maximum likelihood phylogenetic tree after 1000 bootstrap cycles was constructed for Sub3 (KDB23788.1), Sub6 (KDB24326.1) and transcription elongation factor 1-α (TEF1-α) and compared with an ITS-based phylogenetic tree. TiCI12, TiCI14 and D15P135 all cluster together in Sub3- and Sub6-based phylogenetic trees, similar to that of the ITS-based tree (Fig. 3). However, phylogenetic analysis with TEF1-α offered low diversity that could not distinguish among different members of T. interdigitale/T. mentagrophytes species complex, possibly due to only one identified SAP each in T. interdigitale M8436 and T. mentagrophytes D15P127 genomes when compared to T. interdigitale MR816 strain, suggesting low frequency of SAPs in housekeeping genes. Multilocus phylogenetic analysis earlier had also shown that although topologies of ITS and TEF1-α based trees were congruent, TEF1-α offers relative less diversity as compared to the ITS locus (Nenoff et al. 2019; Tang et al. 2021). It is hence provocative to propose that functionally important proteins (namely Sub3 and Sub6) offer higher diversity among amino acid sequences and may be considered in future multilocus studies for phylogenetic analysis and delineation of T. interdigitale/T. mentagrophytes species complex.
Fig. 3.
Maximum likelihood phylogenetic tree different for T. interdigitale/T. mentagrophytes. A maximum likelihood phylogenetic tree was constructed in MEGA (Kumar et al. 2018) after 1000 bootstrap cycles for A ITS locus, and protein sequences of B Sub3, C Sub6 and D TEF1-α. Only full length sequences were used for in the construction of maximum likelihood phylogenetic trees. As only a partial sequence of TEF1-α of D15P152 was identified (possibly due to the low coverage of this genome), it is not included in the TEF1-α-based phylogenetic tree in (D). T. interdigitale/T. mentagrophytes species included in the analysis are as listed in Table 3. The T. mentagrophytes genotype VIII reference strain (GenBank Accession number: MH517560.1), as reported by Nenoff et al. (2019) is included and indicated as Type VIII in the ITS tree in (A)
In conclusion, the overall architecture of the genomes of T. indotineae UCMS-IGIB-CI12 and T. indotineae UCMS-IGIB-CI14 from India were found to be similar to that of the RefSeq T. interdigitale MR816 strain with no major difference in the predicted gene families involved in virulence and/or infection. However, key members of proteases and lipases of TiCI12 and TiCI14 exhibited a higher frequency of SAPs. Phylogenetic analysis with Sub3 and Sub6 subtilases reveals a clustering of strains that was similar to that in ITS-based trees, and further analysis will help in their consideration in multilocus phylogenetic analysis in future. TiCI12 and TiCI14 harbor an Ala448Thr mutation in erg1 but exhibit a variable response to terbinafine. The whole genome sequences of clinical isolates provided in this report can serve as a key reference point for a thorough and detailed investigation of clinical strains originating from different parts of the world and in devising disease management policies towards emerging resistance cases in India.
Accession numbers
The sequence of the internal transcribed spacer regions 1–2 of 18S rRNA of Trichophyton indotineae UCMS-IGIB-CI12 and Trichophyton indotineae UCMS-IGIB-CI14 were deposited in GenBank with Accession numbers MW600527 and MW600653, respectively. The whole genome project of T. indotineae UCMS-IGIB-CI12 and T. indotineae UCMS-IGIB-CI14 has been deposited into GenBank and are available under the accession numbers JAATJQ000000000.1 and JAAQVJ000000000.1 for TiCI12 and TiCI14, respectively.
Acknowledgements
PK acknowledges Senior Research Fellowship from CSIR. The authors thank Vinod Scaria, S. Ramachandran and Debasis Dash for useful discussions. We thank CSIR-Institute of Genomics and Integrative Biology for providing the infrastructure and central instrument lab facility.
Abbreviations
- wgs
Whole genome sequence
- cds
Coding DNA sequences
- ITS
Internal transcribed spacer
- TiCI12
T. indotineae UCMS-IGIB-CI12
- TiCI14
T. indotineae UCMS-IGIB-CI14
- MIC
Minimum inhibitory concentration
- AFST
Antifungal susceptibility test
- SNP
Single nucleotide polymorphism
- SAP
Single amino-acid polymorphism
Author contributions
All the authors contributed to this work. PK, SD, SNB and BT conceptualized the work; PK, RT and RP performed the experiments; PK, SD, RT, RP and BT analyzed the data; SD and BT supervised the work; PK and BT wrote the paper. All authors reviewed and approved the manuscript.
Funding
This work was supported by funding from Department of Biotechnology (DBT), Government of India. (Grant number: BT/PR20790/MED/29/1130/2016).
Data availability
The sequence of the internal transcribed spacer regions 1–2 of 18S rRNA of the isolates TiCI12 and TiCI14 were deposited in NCBI with Accession numbers MW600527 and MW600653, respectively. The complete annotated genome assemblies of TiCI12 and TICI14 have also been deposited at GenBank and are available under the accession numbers JAATJQ000000000.1 and JAAQVJ000000000.1, respectively.
Code availability
Not applicable.
Declarations
Conflict of interest
The authors declare that they have no potential conflict of interest.
Ethical approval
This study was approved by the Institutional Human Ethics Committee of UCMS-GTB (IECHR/2016/28/2 dated 27/12/2016) and CSIR-IGIB (CSIR-IGIB/IHEC/2017-18 dated 30/05/2017).
References
- Achterman RR, White TC. Dermatophyte virulence factors: identifying and analyzing genes that may contribute to chronic or acute skin infections. Int J Microbiol. 2012;2012:358305. doi: 10.1155/2012/358305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S (2010) FastQC: a quality control tool for high throughput sequence data. Version FastQC-v.0.11.8. http://www.bioinformatics.babraham.ac.uk/projects/fastqc. Accessed 4 Oct 2018
- Baldo A, Monod M, Mathy A, et al. Mechanisms of skin adherence and invasion by dermatophytes. Mycoses. 2012;55:218–223. doi: 10.1111/j.1439-0507.2011.02081.x. [DOI] [PubMed] [Google Scholar]
- Bankevich A, Nurk S, Antipov D, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burmester A, Shelest E, Glöckner G, et al. Comparative and functional genomics provide insights into the pathogenicity of dermatophytic fungi. Genome Biol. 2011;12:R7. doi: 10.1186/gb-2011-12-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hoog GS, Dukik K, Monod M, et al. Toward a novel multilocus phylogenetic taxonomy for the dermatophytes. Mycopathologia. 2017;182:5–31. doi: 10.1007/s11046-016-0073-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert A, Monod M, Salamin K, et al. Alarming India-wide phenomenon of antifungal resistance in dermatophytes: a multicentre study. Mycoses. 2020;63:717–728. doi: 10.1111/myc.13091. [DOI] [PubMed] [Google Scholar]
- Fischer M, Knoll M, Sirim D, et al. The Cytochrome P450 Engineering Database: a navigation and prediction tool for the cytochrome P450 protein family. Bioinformatics. 2007;23:2015–2017. doi: 10.1093/bioinformatics/btm268. [DOI] [PubMed] [Google Scholar]
- Gallo JG, Woods M, Graham RM, Jennison AV. A severe transmissible Majocchi’s granuloma in an immunocompetent returned traveler. Med Mycol Case Rep. 2017;18:5–7. doi: 10.1016/j.mmcr.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaurav V, Bhattacharya SN, Sharma N, et al. Terbinafine resistance in dermatophytes: time to revisit alternate antifungal therapy. J Mycol Med. 2021;31:101087. doi: 10.1016/j.mycmed.2020.101087. [DOI] [PubMed] [Google Scholar]
- Gräser Y, Monod M, Bouchara J-P, et al. New insights in dermatophyte research. Med Mycol. 2018;56:2–9. doi: 10.1093/mmy/myx141. [DOI] [PubMed] [Google Scholar]
- Kano R, Kimura U, Kakurai M, et al. Trichophyton indotineae sp. nov.: a new highly terbinafine-resistant anthropophilic dermatophyte species. Mycopathologia. 2020;185:947–958. doi: 10.1007/s11046-020-00455-8. [DOI] [PubMed] [Google Scholar]
- Kar B, Patel P, Free SJ. Trichophyton rubrum LysM proteins bind to fungal cell wall chitin and to the N-linked oligosaccharides present on human skin glycoproteins. PLoS One. 2019;14:e0215034. doi: 10.1371/journal.pone.0215034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman G, Horwitz BA, Duek L, et al. Infection stages of the dermatophyte pathogen Trichophyton: microscopic characterization and proteolytic enzymes. Med Mycol. 2007;45:149–155. doi: 10.1080/13693780601113618. [DOI] [PubMed] [Google Scholar]
- Khurana A, Masih A, Chowdhary A, et al. Correlation of in vitro susceptibility based on MICs and squalene epoxidase mutations with clinical response to terbinafine in patients with tinea corporis/cruris. Antimicrob Agents Chemother. 2018;62:e01038–e1118. doi: 10.1128/AAC.01038-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, et al. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latka C, Dey SS, Mahajan S, et al. Genome sequence of a clinical isolate of dermatophyte, Trichophyton rubrum from India. FEMS Microbiol Lett. 2015;362:fnv039. doi: 10.1093/femsle/fnv039. [DOI] [PubMed] [Google Scholar]
- Lombard V, Golaconda Ramulu H, Drula E, et al. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42:D490–495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez DA, Oliver BG, Gräser Y, et al. Comparative genome analysis of Trichophyton rubrum and related dermatophytes reveals candidate genes involved in infection. mBbio. 2012;3:e00259–e1212. doi: 10.1128/mBio.00259-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méhul B, Gu Z, Jomard A, et al. Sub6 (Tri r 2), an onychomycosis marker revealed by proteomics analysis of Trichophyton rubrum secreted proteins in patient nail samples. J Investig Dermatol. 2016;136:331–333. doi: 10.1038/JID.2015.367. [DOI] [PubMed] [Google Scholar]
- Monod M. Secreted proteases from dermatophytes. Mycopathologia. 2008;166:285–294. doi: 10.1007/s11046-008-9105-4. [DOI] [PubMed] [Google Scholar]
- Nenoff P, Verma SB, Vasani R, et al. The current Indian epidemic of superficial dermatophytosis due to Trichophyton mentagrophytes-a molecular study. Mycoses. 2019;62:336–356. doi: 10.1111/myc.12878. [DOI] [PubMed] [Google Scholar]
- Pchelin IM, Azarov DV, Churina MA, et al. Species boundaries in the Trichophytonmentagrophytes / T. interdigitale species complex. Med Mycol. 2019;57:781–789. doi: 10.1093/mmy/myy115. [DOI] [PubMed] [Google Scholar]
- Peres NT, Sanches PR, Falcão JP, et al. Transcriptional profiling reveals the expression of novel genes in response to various stimuli in the human dermatophyte Trichophyton rubrum. BMC Microbiol. 2010;10:39. doi: 10.1186/1471-2180-10-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persinoti GF, Martinez DA, Li W, et al. Whole-genome analysis illustrates global clonal population structure of the ubiquitous dermatophyte pathogen Trichophyton rubrum. Genetics. 2018;208:1657–1669. doi: 10.1534/genetics.117.300573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings ND, Barrett AJ, Thomas PD, et al. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 2018;46:D624–D632. doi: 10.1093/nar/gkx1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudramurthy SM, Shankarnarayan SA, Dogra S, et al. Mutation in the squalene epoxidase gene of Trichophyton interdigitale and Trichophyton rubrum associated with allylamine resistance. Antimicrob Agents Chemother. 2018;62:e02522–e2617. doi: 10.1128/AAC.02522-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunte DML, Hare RK, Jørgensen KM, et al. Emerging terbinafine resistance in Trichophyton: clinical characteristics, squalene epoxidase gene mutations, and a reliable EUCAST method for detection. Antimicrob Agents Chemother. 2019;63:e01126–e1219. doi: 10.1128/AAC.01126-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seppey M, Manni M, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness. Methods Mol Biol. 2019;1962:227–245. doi: 10.1007/978-1-4939-9173-0_14. [DOI] [PubMed] [Google Scholar]
- Shaw D, Singh S, Dogra S, et al. MIC and upper limit of wild-type distribution for 13 antifungal agents against a Trichophyton mentagrophytes-Trichophyton interdigitale complex of Indian origin. Antimicrob Agents Chemother. 2020;64:e01964–e2019. doi: 10.1128/AAC.01964-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Kim J-E, Lee Y-W, Son H. Fungal cytochrome P450s and the P450 complement (CYPome) of Fusarium graminearum. Toxins. 2018;10:112. doi: 10.3390/toxins10030112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Masih A, Khurana A, et al. High terbinafine resistance in Trichophyton interdigitale isolates in Delhi, India harbouring mutations in the squalene epoxidase gene. Mycoses. 2018;61:477–484. doi: 10.1111/myc.12772. [DOI] [PubMed] [Google Scholar]
- Singh A, Masih A, Monroy-Nieto J, et al. A unique multidrug-resistant clonal Trichophyton population distinct from Trichophyton mentagrophytes/Trichophyton interdigitale complex causing an ongoing alarming dermatophytosis outbreak in India: genomic insights and resistance profile. Fungal Genet Biol. 2019;133:103266. doi: 10.1016/j.fgb.2019.103266. [DOI] [PubMed] [Google Scholar]
- Singh A, Singh P, Dingemans G, et al. Evaluation of DermaGenius® resistance real-time polymerase chain reaction for rapid detection of terbinafine-resistant Trichophyton species. Mycoses. 2021;64:721–726. doi: 10.1111/myc.13271. [DOI] [PubMed] [Google Scholar]
- Stanke M, Morgenstern B. AUGUSTUS: a web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. 2005;33:W465–467. doi: 10.1093/nar/gki458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symoens F, Jousson O, Planard C, et al. Molecular analysis and mating behaviour of the Trichophyton mentagrophytes species complex. Int J Med Microbiol. 2011;301:260–266. doi: 10.1016/j.ijmm.2010.06.001. [DOI] [PubMed] [Google Scholar]
- Taghipour S, Pchelin IM, Mahmoudabadi AZ, et al. Trichophyton mentagrophytes and T interdigitale genotypes are associated with particular geographic areas and clinical manifestations. Mycoses. 2019;62:1084–1091. doi: 10.1111/myc.12993. [DOI] [PubMed] [Google Scholar]
- Tang C, Kong X, Ahmed SA, et al. Taxonomy of the Trichophyton mentagrophytes/T. interdigitale species complex harboring the highly virulent, multiresistant genotype T. indotineae. Mycopathologia. 2021 doi: 10.1007/s11046-021-00544-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Törönen P, Medlar A, Holm L. PANNZER2: a rapid functional annotation web server. Nucleic Acids Res. 2018;46:W84–W88. doi: 10.1093/nar/gky350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban M, Cuzick A, Rutherford K, et al. PHI-base: a new interface and further additions for the multi-species pathogen-host interactions database. Nucleic Acids Res. 2017;45:D604–D610. doi: 10.1093/nar/gkw1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BJ, Abeel T, Shea T, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 2014;9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. PCR protocols. Amsterdam: Elsevier; 1990. Amplification and direct sequencing of fungal ribosomal rna genes for phylogenetics; pp. 315–322. [Google Scholar]
- White TC, Oliver BG, Gräser Y, Henn MR. Generating and testing molecular hypotheses in the dermatophytes. Eukaryot Cell. 2008;7:1238–1245. doi: 10.1128/EC.00100-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Maeda M, Alshahni MM, et al. Terbinafine resistance of Trichophyton clinical isolates caused by specific point mutations in the squalene epoxidase gene. Antimicrob Agents Chemother. 2017;61:e00115–e117. doi: 10.1128/AAC.00115-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The sequence of the internal transcribed spacer regions 1–2 of 18S rRNA of the isolates TiCI12 and TiCI14 were deposited in NCBI with Accession numbers MW600527 and MW600653, respectively. The complete annotated genome assemblies of TiCI12 and TICI14 have also been deposited at GenBank and are available under the accession numbers JAATJQ000000000.1 and JAAQVJ000000000.1, respectively.
Not applicable.