Abstract
Current therapies for treating heterogeneous cancers such as head and neck squamous cell carcinoma (HNSCC) are non-selective and are administered independent of response biomarkers. Therapy resistance subsequently emerges, resulting in increased cellular proliferation that is associated with loss of differentiation. Whether a cancer cell differentiation potential can dictate therapy responsiveness is still currently unknown. A multi-omic approach integrating whole-genome and whole-transcriptome sequencing with drug sensitivity was employed in a HNSCC mouse model, primary patients’ data, and human cell lines to assess the potential of functional differentiation in predicting therapy response. Interestingly, a subset of HNSCC with effective GRHL3-dependent differentiation was the most sensitive to inhibitors of PI3K/mTOR, c-Myc, and STAT3 signaling. Furthermore, we identified the GRHL3-differentiation target gene Filaggrin (FLG) as a response biomarker and more importantly, stratified HNSCC subsets as treatment resistant based on their FLG mutational profile. The loss of FLG in sensitive HNSCC resulted in a dramatic resistance to targeted therapies while the GRHL3-FLG signature predicted a favorable patient prognosis. This study provides evidence for a functional GRHL3-FLG tumor-specific differentiation axis that regulates targeted therapy response in HNSCC and establishes a rationale for clinical investigation of differentiation-paired targeted therapy in heterogeneous cancers.
Keywords: differentiation, targeted therapy, biomarkers, sensitivity, HNSCC, GRHL3, Filaggrin, STAT3, mTOR, c-MYC
Graphical abstract

This study is the first to identify a functional differentiation axis that dictates targeted therapy response in head and neck cancer (HNC). The axis also serves as a favorable prognostic biomarker for HNC patients. Differentiation-paired targeted therapy shapes as a promising novel avenue that may improve outcomes in heterogeneous HNC.
Introduction
Head and neck squamous cell carcinoma (HNSCC) is the sixth most prevalent cancer worldwide, with 600,000 new cases reported annually.1 HNSCC is recognized as a highly heterogeneous cancer due to patients being exposed to a variety of risk factors that include smoking tobacco, frequent alcohol use, chewing betel quid, radiation exposure, genetic predisposition, and infection with high-risk human papilloma viruses (HPVs).2,3 Despite this heterogeneity, current conventional HNSCC treatment regimens such as radiation and chemotherapy are non-selective and are administered independent of etiology or molecular drivers. While immunotherapy has recently shown effectiveness in subgroups of HNSCC with high Programmed Death-Ligand 1 (PD-L1) expression, additional biomarkers beyond PD-L1 are needed to inform the choice of therapy.4 The clinical introduction of the epidermal growth factor receptor (EGFR)-inhibitor Cetuximab for treating HNSCC ushered in a new era of targeted therapy. Unfortunately, only a small subset of patients responded to Cetuximab and showed increased overall survival,5 while STAT3 activation played a significant role in the resistance to this therapy.6 Similar data showed a STAT3-driven resistance mechanism for PI3K inhibitors7 that is independent of the presence or absence of PIK3CA activating mutations8 and reinforces the urgent need for additional stratification methods for the treatment of HNSCCs.
Large-scale genomic and transcriptomic sequencing of HNSCC tumors has shown very high (∼90%) inactivating mutations in tumor suppressor genes.9 While TP53 is the most frequently mutated gene (TP53Mut) in HNSCC (up to 83%), mutations of squamous differentiation genes (i.e., TP63, RIPK4, IRF6) are dominant and co-exist in the same cancer.3,9,10 These mutations are likely to drive more proliferative basal-like HNSCC phenotypes and correlate with poor patient survival.3,10 Surprisingly, the incidence of oncogene-activating mutations is low (∼20%), suggesting that dysregulation of differentiation acts as a primary driver in TP53Mut HNSCC. A tremendous challenge remains in translating genomic information into functional evidence for the separation of driver from passenger mutations, the identification of key impaired differentiation pathways, and the testing of biomarker-driven therapy responsiveness.9
Balanced proliferation and differentiation of squamous epithelial cells maintains tissue homeostasis and prevents transformation.11 Activation of differentiation programs results in terminal differentiation that effectively suppresses the proliferation of squamous cells.12,13 Interestingly, the majority of the head and neck differentiation factors such as p63, RIPK4, IRF6,3,10 and the tumor suppressor TP53 converge to regulate expression of the terminal differentiation factor Grainyhead-like 3 (GRHL3).14,15 GRHL3 is a highly conserved epidermal-specific developmental transcription factor that functions as a major tumor suppressor in mouse and human HNSCC.16,17 Germline deletion of Grhl3 in mouse embryos results in a markedly hyperproliferative oral and skin epithelium, dysregulated epidermal differentiation, and defective wound healing and skin barrier defects, with newborn pups dying of dehydration.11 Interestingly, epithelial-specific conditional deletion of Grhl3 (Grhl3cKO) in mice using a keratin (K)-14 driven Cre recombinase induced spontaneous head and neck tumor development.17 Furthermore, ∼90% of human tumors and HNSCC cell lines show reduced levels of GRHL3 downstream of the oncogenic microRNA-21 (miR-21),17 resulting in hyperactivation of phosphatidylinositol 3-kinase (PI3K)/mammalian target of rapamycin (mTOR) and c-Myc signaling and conferring a poor prognosis.18,19 Since inhibitors of PI3K/mTOR and c-Myc signaling are not universally effective in TP53Mut HNSCC, response biomarkers are required to stratify patients likely to derive benefit and exclude those unlikely to respond.1 These observations prompted the analysis of a functional GRHL3-dependent differentiation pathway to identify a clinically relevant molecular vulnerability in heterogeneous HNSCC.
Results
A GRHL3-linked differentiation signature balances oral epithelial proliferation and differentiation
To explore key regulators of oral epithelial homeostasis, we performed transcriptome sequencing of normal, dysplastic, and differentiated cultured oral keratinocytes and assigned differentially expressed genes (DEGs) to four clusters (Figure 1A). Cluster 1 (C1) genes were high in differentiated cells and low in dysplastic cells, with the inverse DEG signature observed for C4. Gene Ontology (GO) functional enrichment analysis illustrated that C1 (high in differentiated cells) was enriched for keratinocyte differentiation and keratinization, while C4 (high in dysplastic cells) was enriched for G1/S transition, RNA pol II transcription, and DNA replication. These transcriptomic transitions (Figure 1B) further support suppressed differentiation and concomitant activation of proliferation. Intriguingly, within DEGs significantly enriched in the top 5 GO biological processes during differentiation (Figure 1C), GRHL3 and GRHL1 were the only transcription factors significantly upregulated in the differentiated stage (Figure 1D, right). Meanwhile, GRHL3 was significantly downregulated in dysplastic cells but no change was observed for GRHL1 expression (Figure 1D, left). Analysis of gene-gene interactions between GRHL3 and DEGs in C1 and C4 revealed a direct association with differentiation-related genes while loss of GRHL3 was linked to proliferation-related factors, respectively (Figure S1A). Knockdown of GRHL3 using short hairpin RNA (shRNA) in differentiated normal human epidermal keratinocytes (Figure S1B) and heatmaps of gene expression (Figure S1C) confirm the in-silico analysis and indicate a shift to proliferation-related DEGs (Figure S1D). These data imply that GRHL3-dependent differentiation plays an important role in the regulation of oral epithelial homeostasis and its loss promotes oral hyperproliferation and transformation.
Figure 1.
A GRHL3-linked differentiation signature balances oral epithelial proliferation and differentiation
(A) Heatmap showing unsupervised hierarchical clustering of gene-expression clusters (C1 to C4) specifically expressed in dysplastic, normal, and differentiated oral epithelial cells, identified from the analysis of differentially expressed genes (DEGs) between the three stages. Top significantly enriched functional terms in each gene cluster are listed on the right. GO, Gene Ontology. (B) Principal-component analysis (PCA) plot showing gene expression transitions between dysplastic, normal, and differentiated oral epithelial cells. (C) Top 5 significantly enriched GO terms for the DEGs between normal and differentiated oral epithelial cells. (D) Volcano plots of DEGs between normal and differentiated cells (right) and between dysplastic and normal cells (left). Values with an absolute logFC > 2 and p value < 0.05 are highlighted in pink. DEGs related to epithelial differentiation are highlighted in purple and GRHL3 in red.
Loss of GRHL3 potentiates STAT3 activation in mouse and human HNSCC
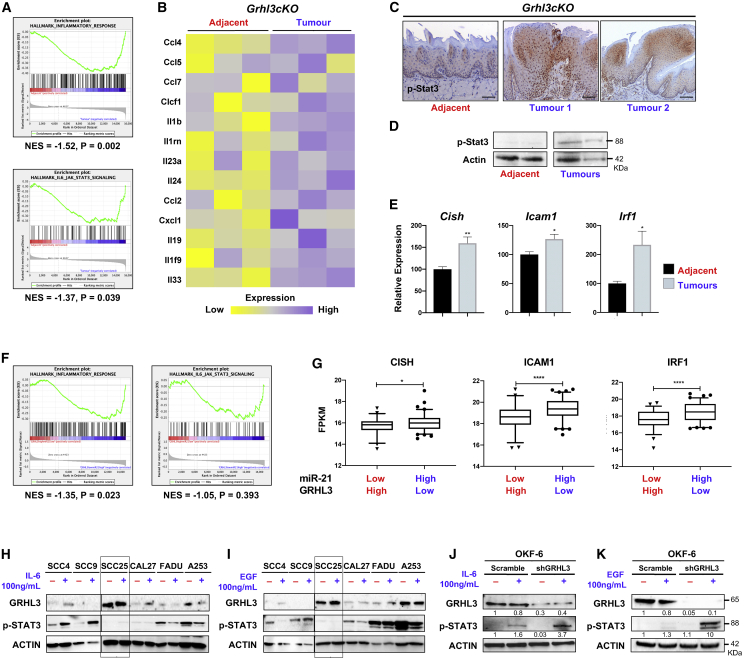
The hyperactivation of STAT3 signaling is a key resistance mechanism against therapies in HNSCC.7 Active STAT3 is known to upregulate miR-21 in HNSCC,20 and miR-21 directly targets and reduces the tumor suppressor GRHL3.16 Expression of miR-21 was assessed in oral epithelial tissue of gp130Y757F knockin mice, which express a mutant gp130 signal transducer resulting in constitutive Stat3 signaling.21 We found ∼4-fold upregulation of miR-21 in 6-month-old gp130Y757F mice compared to wild-type (WT) control animals (Figure S2A), demonstrating a conserved correlation between STAT3 activation and elevated miR-21 in humans and mice. However, Grhl3 levels were not reduced (Figure S2B) and the gp130Y757F mice did not develop spontaneous oral tumors. This indicates that an inverse correlation between Grhl3 levels and STAT3 activation is HNSCC-specific. We then examined normal whole-tongue tissue and spontaneous tumors from Grhl3-deficient mice17 using whole transcriptome sequencing. Interestingly, gene set enrichment analysis (GSEA) identified a significant enrichment for genes involved in the inflammatory response (Figure 2A) and cytokines and chemokines associated with STAT3 signaling in Grhl3cKO tumors (Figure 2B). Analysis by immunohistochemistry (IHC) revealed strong nuclear staining for p-Stat3 in Grhl3cKO tumors, whereas the normal adjacent cells were negative (Figure 2C). Western blot (WB) analysis demonstrated upregulation of active Stat3 (pY705) in tumor samples (Figure 2D) along with increased mRNA expression of the STAT3 target genes, Cish, Icam1, and Irf122 in the Grhl3cKO tumors (Figure 2E). The correlation between miR-21/GRHL3 expression and STAT3 pathway activation was further assessed in human samples using The Cancer Genome Atlas (TCGA) Research Network-HNSCC dataset (n = 279). Compared to normal adjacent oral tissues, the matched primary tumor samples displayed a significant ∼2-fold upregulation of miR-21 and ∼4-fold reduction in GRHL3 gene expression (Figure S2C). Pearson analysis revealed a significant (R = –0.26, p < 0.0001) negative correlation in human HNSCC (Figure S2D). Furthermore, a similar enrichment for genes involved in the inflammatory response and STAT3 signaling was identified in patients with high miR-21 and low GRHL3 expression (Figure 2F) along with significant overexpression of the STAT3 target genes CISH, ICAM1, and IRF1 compared to patients with low miR-21 and high GRHL3 expression (Figure 2G). Expression of these genes was not significantly altered between the group of patients with low miR-21 and low GRHL3 and those with high miR-21 and high GRHL3 (Figure S2E). These data indicate that loss of GRHL3 downstream of miR-21 correlates with the activation of STAT3 signaling in mouse and human HNSCCs.
Figure 2.
Loss of GRHL3 potentiates STAT3 activation in mouse and human HNSCC
(A) A significant enrichment for inflammatory genes is observed in Grhl3cKO tumors compared to normal adjacent tissues. The significance was empirically determined by 1,000 gene-set permutations (normal adjacent tissues, red; tumors, blue). (B) Expression of cytokines/chemokines is shown as a heatmap. (C) p-Stat3 IHC demonstrates hyperactive Stat3 in tumors but not in adjacent tissues from Grhl3cKO mice. Scale bars correspond to 50 μm. (D) WB for p-Stat3 on spontaneous tongue tumor tissues compared to adjacent control tissues from 6-month-old Grhl3cKO mice. (E) Upregulation of the STAT3 target genes Cish, Icam1, and Irf1 was quantitated by qPCR for tumors from Grhl3cKO mice normalized to Gapdh and relative to adjacent (normal) tissue. The results are presented as mean ± SEM. The statistical values were considered significant at p < 0.05. ∗p ≤ 0.05, and ∗∗p ≤ 0.01. (F) GSEA of the TCGA-HNSCC RNA-seq data. Patients were divided into two groups: high miR-21, low GRHL3 (n = 79, in blue), and low miR-21, high GRHL3 (n = 62, in red). The significance was empirically determined by 1,000 gene-set permutations. (G) Analyses of STAT3 target gene expression in the same patient groups. The comparison between the two groups was performed using an unpaired Student’s t test (∗p < 0.05; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001). Data are shown as mean ± SEM. Patients HNSCC with high miR-21, low GRHL3 expression show STAT3 pathway activation compared to patients with low miR-21, high GRHL3 expression. (H and I) WB analyses of GRHL3 expression and STAT3 activation in cell lines treated for 2 h with (H) 100 ng/mL IL-6 or (I) 100 ng/mL EGF. While STAT3 activity is undetectable in SCC25 (boxed), these cells do not respond to IL-6 or EGF treatment. (J and K) OKF-6 cells with shRNA-mediated GRHL3 knockdown respond to (J) IL-6 stimulation, or (K) EGF stimulation, and show a significant increase in STAT3 activation compared to cells transduced with a scrambled control. β-actin was used as a loading control.
To evaluate the effects of GRHL3 loss on STAT3 activation, we characterized the molecular profiles of human HNSCC cell lines cultured under normal conditions. Expression of GRHL3 was measured by qPCR for a series of cell lines (SCC4, CAL27, SCC25, SCC9, A253, and FaDu). The gene expression data mirrors the degree of heterogeneity observed in primary human HNSCC23 with the highest GRHL3 expression identified for SCC25 (Figure S2F, top panel). We next examined the commonly activated signaling pathways by WB (Figure S2F, lower panel). Consistent with previous findings, PI3K/mTOR signaling was hyperactive (phospho-S6 positive) and c-Myc was overexpressed in all HNSCC lines.17 Furthermore, SCC25 showed increased expression and activation of the translation initiation factor eIF4E, a downstream effector of PI3K/mTOR and c-Myc signaling.24 ERK and STAT3 were also examined under basal culture conditions.25 Compared to normal OKF6 cells, p-ERK was consistently low in all SCC lines examined. While p-STAT3 levels were variable, they were negative in SCC25 and SCC9 (Figure S2F) and these lines showed differential expression of pro-inflammatory genes related to STAT3 signaling (Figure S2G). We next stimulated the HNSCC lines with interleukin-6 (IL-6) and EGF to activate the STAT3 pathway, as this can induce adaptive resistance to targeted therapy.7 Interestingly, SCC25 was the only line non-responsive to 100 ng/mL of IL-6 (Figure 2H) or EGF (Figure 2I) (i.e., complete lack of p-STAT3), suggesting a potential inhibitory mechanism unique to SCC25, which has high GRHL3 expression and lacks STAT3 activation. To validate this, OKF-6 cells transduced with either a scrambled control or GRHL3 shRNA and treated with IL-6 (Figure 2J) or EGF (Figure 2K) showed a significant increase in p-STAT3 in shGRHL3 compared to control cells. Furthermore, we show that the SCC1 cancer cell line has comparable GRHL3 expression to the normal oral cell line OKF-6 (Figure S3A). Interestingly, shRNA-mediated knockdown of GRHL3 in SCC1 (Figure S3B) induced significant upregulation of p-STAT3 that was further increased following treatment with IL-6 but not EGF (Figures S3C and S3D). Collectively, our data support a role for GRHL3 in the negative regulation of IL-6/STAT3 signaling and indicate that loss of GRHL3 promotes hyperactive STAT3 in both normal and HNSCC cell lines.
Mutational analysis and drug sensitivity of HNSCC cell lines
We investigated the role of GRHL3 in acquisition of drug sensitivity by integrating whole-genome mutational analysis, whole-transcriptome and drug sensitivity data for HNSCC patient samples, and cell lines retrieved from the TCGA Research Network, Cancer Cell Line Encyclopedia (CCLE; Broad Institute), COSMIC, and Genomics of Drug Sensitivity in Cancer (GDSC) databases.26 The inhibitors of PI3K/AKT/mTOR and c-Myc downstream signaling were prioritized because these pathways are hyperactive in all the cell lines (Figure S2F). While a generally broad range of the half maximum inhibitory concentration (IC50) values was observed (Figure 3A), SCC25 showed the highest sensitivity to drugs in all of the classes. Data retrieved from the CCLE database showed that SCC25 harbors the lowest number of somatic mutations, while A253 and FaDu have the highest burden (Figure 3B). Interestingly, SCC9 is the only cell line with WT TP53 (Figure 3B, lower panel; Table S1) demonstrating sensitivity to most inhibitors while the other resistant TP53 mutant lines mirror patients with TP53 mutant variants who have poor treatment responses and reduced overall survival.27
Figure 3.
Mutational analysis and drug sensitivity of HNSCC cell lines
(A) Heatmap of drug sensitivity and resistance to inhibitors of STAT3, PI3K/AKT/mTOR, c-Myc, and their downstream kinases in the HNSCC cell lines. The drug log-IC50 was adapted from the Genomics of Drug Sensitivity in Cancer (GDSC) database. (B) The top panel shows the type and rate of mutations from the Cancer Cell Line Encyclopedia (CCLE) Cell Line mutation and COSMIC databases. The bottom panel depicts the most common somatic mutations in TCGA-HNSCC patients that were also present in the HNSCC cell lines. (C) qPCR analysis of the highly mutated genes (TP53, CDKN2A, and LRP1B) in the HNSCC cell lines treated with the selected inhibitors for 24 h. mRNA expression levels were normalized to untreated cells. The results are presented as mean ± SEM. The statistical values were considered significant at p < 0.05. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, and ∗∗∗∗p ≤ 0.0001.
To uncover predictive therapeutic vulnerabilities, we assessed 13 genes (ASTN1, BRD1, CARD10, CEP78, EPHB3, FANCM, LRRK2, MUC20, RYR3, SAP130, SRRM2, TMX4, and TTN) showing common hotspot mutations in the HNSCC lines (Figure S4A) for their basal expression. Gene-expression profiling did not show a pattern that correlates with their mutational status in the mutant cell lines (Figure S4B). Furthermore, treatment with the small molecule inhibitors did not consistently induce (or repress) their expression in treated cells (Figure S5).
Interestingly, within the TP53 mutant HNSCC cell lines, SCC25 was the most sensitive to all tested inhibitors (Figure 3A). This drug-sensitive cell line is the only one with a WT filaggrin (FLG) gene (Table S1), and FLG is within the top commonly mutated genes (∼16%) in TCGA Research Network-HNSCC patient samples (Figure 3B). To assess whether FLG is associated with SCC25’s sensitivity to small molecule inhibitors, we quantitated FLG expression after treatment with drugs targeting the PI3K/AKT/mTOR (Dactolisib and SEL201), c-Myc (JQ1 and CX-5461), and STAT3 (Niclosamide) signaling pathways. Strikingly, FLG expression was robustly induced in response to these inhibitors only in SCC25 compared to the other FLG mutant HNSCC cells (Figure 3C), and more importantly, FLG levels correlated with the drug sensitivity of SCC25 to each of the PI3K/AKT/mTOR and c-Myc inhibitors (Figure 3A). These findings suggest that WT FLG is a predictive biomarker of responsiveness to PI3K/AKT/mTOR and c-Myc inhibitors.
A functional GRHL3-FLG differentiation pathway promotes sensitivity to targeted therapy
FLG is an important terminal differentiation gene in squamous tissues and its deregulation results in defective epithelial barrier formation and atopic dermatitis.28 Under normal (non-stimulated) cell culture conditions, FLG expression was the highest in SCC25 (Figure S6A). To assess whether FLG-dependent differentiation plays a role in HNSCC, we induced cellular differentiation by treating the cell lines with 2 mM CaCl2 (24 h). Both GRHL3 and FLG were significantly upregulated following Ca2+-induced differentiation in SCC25 only, reflecting a functional differentiation response in these cells (Figures 4A and 4B). This was further confirmed by growing SCC25 to >100% confluency (14 days), which resulted in differentiation-dependent GRHL3-FLG induction (Figure S6B). However, the expression of other terminal differentiation markers, such as GRHL1 and TGM1, did not increase in response to calcium treatment, indicating that functional differentiation in SCC25 might be entirely restricted to (or only dependent on) GRHL3 and FLG expression (Figure S6C).
Figure 4.
A functional GRHL3-FLG pathway promotes sensitivity to targeted therapy
(A and B) qPCR expression of (A) GRHL3, and (B) FLG in HNSCC cell lines treated with 2 mM CaCl2 (24 h) and normalized to GAPDH, relative to control untreated cells show significant induction only in SCC25. (C) Illustration of the CRISPR-dCas9 GRHL3-activation SAM system with the potential GRHL3 binding site in FLG promoter. (D) Significant induction of GRHL3 and FLG mRNA, but not of TGM1 (not significant, n.s.) in 293T cells transduced with the CRISPR-dCas9 GRHL3-activation SAM system. mRNA expression levels were normalized to GAPDH relative to control empty sgRNA vector. (E) FLG expression following Dox (1 μM, 72 h) treatment of SCC25 cells transduced with an inducible lentivirus CRISPR-Cas9 system to mediate FLG knockdown. Two single guide RNAs (sgRNAs) were employed to target the FLG gene at different sites. Compared to control (Dox negative) cells, downregulation of FLG (~50%) was achieved with both sgFLG.1 and sgFLG.2 following Dox induction. (F) SCC25 cells grown for 96 h in the presence of Dox (1 μM) did not show any proliferative difference to untreated cells. (G) Downregulation of FLG renders the treatment-sensitive SCC25 resistant to all small molecule inhibitors. (H) WB analyses of p-STAT3 in SCC25 treated with the inhibitors at IC50 in the absence of FLG for 3 days. p-STAT3 was increased in cells treated with c-Myc and PI3K/mTOR inhibitors and absent in those treated with Niclosamide. β-actin was used as the loading control. The results are presented as mean ± SEM. The statistical values were considered significant at p < 0.05. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, and ∗∗∗∗p ≤ 0.0001.
We next sought to identify whether GRHL3 transcriptionally activates the FLG gene. The GRHL3 DNA consensus-binding site (5′-AACCGGTT-3′) has been conserved across 700 million years of evolution.29 Using the Jaspar database, we identified a potential GRHL3-binding site in the FLG promoter (5′-TACAGGTT-3′) located 173 bp upstream of the TSS, which is conserved in human and mouse (Figure 4C). To test whether GRHL3 activates FLG in vitro, we induced GRHL3 expression from its endogenous promoter using a two-component CRISPR-activation SAM system.30 A nuclease dead Cas9 (dCas9) fused to a VP64 transcriptional activator domain promoted GRHL3 overexpression in human HEK293T cells, leading to specific induction of its downstream FLG target, but not of its TGM1 target31 (Figure 4D).
Direct binding of GRHL3 to the FLG promoter was confirmed in vitro by electrophoretic mobility shift assay (EMSA; Figure S6D). Furthermore, RNA sequencing (RNA-seq) analysis of Grhl3-deficient mouse tongues showed reduced FLG levels (∼4 fold) only in tumors when compared to adjacent tissues and WT mice (Figure S6E). These data indicate that FLG is a critical downstream target of GRHL3 in HNSCC differentiation.
In parallel, we measured the enzymatic activity of the aldehyde dehydrogenase (ALDH) stem cell marker in the HNSCC cells using the AldeRed detection assay (Figures S7A and S7B). SCC25 and FaDu, which have the highest ALDH activity among the HNSCC lines, were grown on ultra-low attachment plates for 7 days. Compared to FaDu, SCC25 formed relatively small spheroids (Figure S7C), suggesting that functional differentiation may limit the 3D-growth of SCC25. We then examined whether loss of functional GRHL3-FLG-dependent differentiation affects SCC25 drug sensitivity. Dox-induced CRISPR-Cas932 with 2 separate single guide RNAs against FLG (sgFLG), confirmed by qPCR (Figure 4E), did not affect the proliferation of SCC25 (Figure 4F). Interestingly, the loss of FLG in SCC25 resulted in a dramatic increase in resistance to the PI3K/AKT/mTOR and c-Myc inhibitors compared to sensitive control cells (Figure 4G). WB analysis showed increased p-STAT3 in sgFLG cells treated with these inhibitors (Figure 4H). We next validated these findings in SCC1 cells that are FLG WT (CCLE and COSMIC databases). This cell line was able to differentiate in response to calcium treatment and showed significant upregulation of both GRHL3 and FLG (Figure S3E). When FLG was knocked down in SCC1 (Figure S3F), similar to SCC25, there was no effect on cell proliferation (Figure S3G), but importantly, a significant reduction in the sensitivity to PI3K/AKT/mTOR and c-Myc inhibitors was observed (Figure S3H). Moreover, increased resistance to these inhibitors correlated with increased p-STAT3 (Figure S3I) compared to control untreated cells. These results support the data in SCC25 and indicate that FLG is a biomarker for treatment response and its loss induces resistance to targeted therapy that correlates with STAT3 activation.
A GRHL3-FLG signature predicts favorable prognosis for HNSCC patients
Expression levels of GRHL3 and FLG were significantly higher in both moderate and well differentiated HNSCC compared to poorly differentiated patient HNSCC (Affymetrix array; Figure 5A), who have worse survival outcomes.33,34 Interestingly, expression levels of the terminal differentiation markers GRHL1 and TGM1 were not consistently correlated with moderate/well differentiated HNSCC compared to the poorly differentiated ones (Figure S6F), suggesting that increased expression of GRHL3 and FLG is favorably associated with tumor differentiation. To determine whether the functional GRHL3-FLG axis is clinically relevant, RNA-seq expression data of human HNSCC was extracted from the TCGA Research Network-HNSCC dataset and revealed that while a significant positive correlation exists for GRHL3 and FLG, their expression is negatively correlated to CISH, ICAM1, and IRF1 (Figure 5B). Supervised clustering showed that clusters with high or low GRHL3-FLG levels are inversely correlated with STAT3 target gene expression (CISH, ICAM1, and IRF1) in patient HNSCC (Figure 5C). Furthermore, the survival analyses of patients (n = 271) with primary HNSCC displayed no significant difference between GRHL3low (1/3 of patients with the lowest GRHL3 expression) and GRHL3high (compared to adjacent tissue) or between FLGWT and FLGMut; but importantly, a survival advantage of the GRHL3high-FLGWT cohort was evident in comparison to the GRHL3low-FLGMut patients (Figure 5D).
Figure 5.
A GRHL3-FLG signature predicts favorable prognosis for HNSCC patients
(A) The expression of GRHL3 and FLG is significantly higher in moderate (n = 38) and well-differentiated (n = 19) compared to poorly differentiated (n = 21) HNSCC samples (Affymetrix HG133 arrays) and normalized to GAPDH. The results are presented as mean ± SEM. The statistical values were considered significant at p < 0.05. The results are presented as mean ± SEM. The statistical values were considered significant at p < 0.05. ∗p ≤ 0.05, and ∗∗p ≤ 0.01. (B)Significant correlation between GRHL3 and FLG and STAT3-target (CISH, ICAM1, and IRF1) gene expression from the TCGA-HNSCC patient database (n = 279; Pearson’s correlation). (C) Supervised clustering of high GRHL3 and FLG levels shows a clear subtype (boxed) with an inverse correlation to STAT3-target gene (CISH, ICAM1, and IRF1) expression. Rows are centered and unit variance scaling is applied to rows. (D) Kaplan-Meier survival analyses of HNSCC patients stratified based on their GRHL3 expression level and FLG mutation status. The difference in survival rates between patients with GRHL3high (n = 24) and GRHL3low (n = 247) or between patients with FLGWT (n = 227) and FLGMut (n = 44) was n.s. The survival of the cohort with GRHL3high and FLGWT (n = 15) was significantly superior to that of patients with GRHL3low and FLGMut (n = 35; p = 0.04771, log-rank test).
Collectively, these data establish a functional GRHL3-FLG differentiation axis as a favorable prognostic predictor and a response biomarker in HNSCC and indicate that loss of GRHL3-FLG promotes an acquired STAT3-dependent resistance to targeted therapy.
Discussion
Many studies have focused on genomic biomarker-driven targeted therapies, which have not proven totally effective for predicting responsiveness. Key examples include EGFR-T790M mutations and resistance to EGFR inhibitors in EGFR mutant lung cancer,35 ESR1 mutations in estrogen-receptor-positive breast cancer treated with endocrine therapy36 and reversions of pathogenic mutations in BRCA1- and BRCA2-deficient cancers treated with poly-ADP ribose polymerase (PARP) inhibitors.37 Differentiation-paired targeted therapy has not been investigated, and whether it promotes therapy response remains to date largely unknown.38 Mutations in TP53 synergizes with the loss of differentiation pathways to facilitate tumor progression.39 In agreement with this, upregulation of TP53 was shown to induce differentiation and suppress cutaneous SCC in mice,40 and inhibitors of PI3K/mTOR signaling provided anti-tumor activity through the induction of WT TP53 in human xenograft and murine models of HNSCC.41 This is recapitulated in the TP53WT SCC9 cells that showed high sensitivity to multiple inhibitors compared to TP53Mut cells (harboring FLGMut; Figure 3A). Differentiation mechanisms are therefore promising response biomarkers for the stratification of therapies in both loss-of-function (LOF) and gain-of-function (GOF) TP53Mut HNSCC.
Substantial efforts have been made to stratify HNSCC patients into subtypes based on their HPV status, EGFR expression and activation, and CD8+ lymphocyte tumor infiltration.42,43 However, the survival rate of these patients is still significantly low. While HPV-positive patients generally show better responses to therapies and favorable prognosis,1 an inflammatory response may contribute to the resistance of HPV-negative HNSCC to therapies.44 These HNSCC are commonly induced by heavy smoking and alcohol consumption and have been associated with chronic inflammation, cytokine-activated receptors, and STAT3 pro-inflammatory signaling.45 Recent studies have shown that cytokines are also secreted by epithelial cells.7,13 Importantly, SCC25 cells are non-responsive to IL-6 (or EGF) and display minimal STAT3 phosphorylation. We propose that the developmental factor GRHL3, which is highly expressed in SCC25 cells, directly regulates STAT3-related phosphatase gene transcription. A GRHL3 consensus binding motif was identified in the type-1 protein phosphatase (PP1) regulatory subunit (PPP1R3F) genes’ proximal first intron, and this is conserved across the placental mammals. We showed direct binding of GRHL3 to the PPP1R3F putative regulatory region by a chromatin immunoprecipitation (ChIP) assay (Figures S8A and S8B). Moreover, reduced GRHL3 expression, both in vivo and in vitro, correlated with reduced PPP1R3F mRNA expression (Figures S8C and S8D). Therefore, a GRHL3-PPP1R3F-STAT3 regulatory mechanism may operate (Figure S9) since PP1 inhibition induces phosphorylation of STAT3.46
Our multi-omic approach integrating whole-genome, whole-transcriptome, and drug sensitivity data strongly implicates the downregulation of GRHL3 and FLG with STAT3 pathway activation. While STAT3 is active in most HNSCC samples, STAT3 GOF mutations have not been observed nor have activating mutations in upstream regulators.3,10 Conversely, FLG is mutated in ∼16% of HNSCC. Our findings demonstrate that loss of GRHL3-FLG-mediated differentiation in HNSCC potentiates a pre-existing STAT3 pro-inflammatory pathway, resulting in resistance to small molecule inhibitors. This is shown in cells initially sensitive to targeted therapies (SCC25) that became resistant along with an increase in p-STAT3 (Figures 4G and 4H). On the other hand, expression of FLGWT was significantly induced in sensitive cells treated with the inhibitors. Collectively, these data indicate that the terminal differentiation GRHL3-FLG axis predicts response to PI3K/AKT/mTOR and c-Myc inhibition, and its induction by targeted therapy dramatically reduces cell growth. Since Cetuximab-resistant SCC25 cells also express low GRHL3-FLG,47 this mechanism of action may be valid for the sensitization of HNSCC cells to a broader range of inhibitors.
Recently, immunotherapy has resulted in improved HNSCC patient survival by exploiting inhibitory checkpoint pathways that suppress anti-tumor T cell responses.48 However, only a minority of patients derive benefit for non-selective immunotherapies.49 Because Stat3 signaling was shown to regulate PD-1/PD-L1 expression50 and the anti-tumor immune response,51 future investigations will determine whether combining immunotherapy with differentiation-paired targeted therapy may provide greater therapeutic benefit.
In summary, this study identifies a functional GRHL3-FLG differentiation axis in a subset of HNSCC. GRHL3-FLGWT upregulation in response to targeted therapy efficiently reverses cancer cell proliferation, while disruption of this axis confers resistance to PI3K/AKT/mTOR and c-Myc inhibitors (Figure S9). These findings have exciting therapeutic implications with the potential to evaluate this axis as a predictive prognostic and response biomarker in the clinic for the therapeutic stratification of heterogeneous HNSCC, notably those with TP53 mutations for which there are currently modest clinical response rates to most available treatment options.
Materials and methods
RNA-seq analysis
3′ RNA-seq was conducted by the Molecular Genomics Core (PMCC, Melbourne). The total RNA quantity was measured using Qubit RNA HS (Thermo Fisher Scientific). 500 ng total RNA was used for library preparation according to standard protocols (QuantSeq 3′ mRNA-seq FWD, Lexogen). Indexed libraries were pooled and sequenced on a NextSeq500 (Illumina). 5–15 million single-end 75 bp reads were generated per sample. Sequenced reads were trimmed and aligned to hg38 genome via Cutadapt and HISAT2. Gene counts were obtained from featureCounts. Differential expression was performed using Limma. Genes were considered as DEGs if the absolute log-fold change was >2 and p value <0.05. All analysis packages were operated within the Galaxy suite environment. The functional enrichment analysis of annotated terms from GO was performed with the online tool DAVID using the human genome as the background. The gene-gene interaction analysis was completed using the GeneMania app in Cytoscape 3.
Drug-sensitivity assays
HNSCC cells grown at log phase were harvested, counted, and 8,000 cells seeded in 96-well plates. Cells were allowed to attach and grow overnight and were then treated for 3 days with drugs at IC50 values. To calculate the IC50 of each drug, we calculated the percentage of cell growth normalized to a vehicle control using the following equation: 100 × (T–T0) ⁄ (C–T0), where C is the optical density (OD) of the control. For SCC25, IC50 values were as follow for Niclosamide, 0.16 μM; JQ1, 0.8 μM; Dactolisib, 0.1 μM; SEL201, 4 μM; and CX-5461, 1.2 μM. Cell viability was measured using Alarma Blue according to the manufacturer’s instructions. To calculate the cell-killing rate, we normalized absorbance readings to controls (without cells or without treatment).
For gene-expression analysis, HNSCC cells were seeded in 12-well plates at a density of 100,000 cells/well and treated with drugs at IC25 or IC50 for 24 h followed by RNA extraction. For western blotting, SCC25 with/without reduced FLG were seeded in 6-well plates at a density of 200,000 cells/well and treated with drugs at IC50 for 72 h followed by protein extraction.
Drug sensitivity and proliferation of the HNSCC cells were measured using the Sulforhodamine B (SRB) assay. Cells were seeded into 96-well plates (3,000–8,000 cells/well) and 24 h later serial dilutions of the drugs dissolved in DMSO were added independently. One plate per cell line was used to estimate the OD at the start (T0) in the absence of any treatment (DMSO vehicle only). After 3 days, absorbance readings of treated plates were measured. The plates were fixed and then stained with SRB, and the OD values of cells exposed to each drug concentration were measured at a wavelength of 564 nm (T).
Animal studies
Grhl3cKO52 and gp130(Y757F)21 mice were maintained on a C57BL/6J background. All animal studies followed the National Health and Medical Research Council (NHMRC) Australian Code of Practice for the Care and Use of Animals for Scientific Purposes and were approved by the Animal Experimentation Ethics Committee at the PMCC (E587). Mice were euthanized by CO2 asphyxiation, the tongues dissected, and tumors and adjacent tissues removed for pathological and gene-expression analysis. gp130Y757F knockin mice21 were a kind gift from Dr. Michael Buchert (Olivia Newton-John Cancer Research Institute, Heidelberg, VIC, Australia).
Bioinformatic datasets and analyses
Publicly available datasets were retrieved from TCGA Research Network using the TCGA-biolinks R package. Gene expression (RNA-seq and miR-seq), mutation status, and patient survival information for 279 HNSCC samples were analyzed. Gene mutation profiling of the cell lines were retrieved from the CCLE (release date 02-Jan-2019). Publicly available HNSCC microarray data from ArrayExpress: E-TABM-302 based on Affymetrix HG133 arrays were retrieved from ArrayExpress. Publicly available RNA-seq data (GEO: GSE37049) of normal human keratinocytes transduced with shRNA GRHL3 were retrieved from Gene Expression Omnibus.
Statistical analyses
Statistical analysis was performed using Prism 8 (GraphPad). Statistical significance was assessed using the unpaired Student’s t test or ANOVA and multiple comparison test or log-rank (Mantel-Cox) test for survival analysis. The results are presented as mean ± SEM. The statistical values were considered significant at p < 0.05. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, and ∗∗∗∗p ≤ 0.0001.
Acknowledgments
The authors would like to acknowledge the assistance of Jason Li from the Peter MacCallum Cancer Centre Bioinformatics Core Facility. The RNA polymerase I inhibitor CX-546153 was kindly provided by Professor Rick Pearson (PMCC, Melbourne). The STAT3 inhibitor Niclosamide54 and the c-Myc inhibitor JQ1 were kindly provided by Dr. Rodney Luwor (Royal Melbourne Hospital, Melbourne). The MAP kinase-interacting serine/threonine kinases 1 and 2 (MNK1/2) inhibitor SEL20155 was provided by SELVITA through an MTA with the assistance of Dr. Luke Furic. This research was supported by a grant from the Australian National Health and Medical Research Council (NHMRC; APP1106697) and a Victorian Cancer Agency Mid-Career Fellowship (MCRF16017) to C.D.
Author contributions
Y.B. and C.D. designed the experiments. Y.B., Z.Z., J.B., and B.J.v.D. performed the in vitro and in vivo experiments. Y.B. and Z.Z. performed the experiments in silico. Y.B., B.J.v.D., and C.D. analyzed the data. Y.B., B.J.v.D., and C.D. wrote the manuscript. C.D. guaranteed the funding for this research.
Declaration of interest
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ymthe.2021.03.016.
Supplemental information
References
- 1.Tan F.H., Bai Y., Saintigny P., Darido C. mTOR Signalling in Head and Neck Cancer: Heads Up. Cells. 2019;8:333. doi: 10.3390/cells8040333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chi A.C., Day T.A., Neville B.W. Oral cavity and oropharyngeal squamous cell carcinoma--an update. CA Cancer J. Clin. 2015;65:401–421. doi: 10.3322/caac.21293. [DOI] [PubMed] [Google Scholar]
- 3.Stransky N., Egloff A.M., Tward A.D., Kostic A.D., Cibulskis K., Sivachenko A., Kryukov G.V., Lawrence M.S., Sougnez C., McKenna A. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burtness B., Harrington K.J., Greil R., Soulières D., Tahara M., de Castro G., Jr., Psyrri A., Basté N., Neupane P., Bratland Å., KEYNOTE-048 Investigators Pembrolizumab alone or with chemotherapy versus cetuximab with chemotherapy for recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-048): a randomised, open-label, phase 3 study. Lancet. 2019;394:1915–1928. doi: 10.1016/S0140-6736(19)32591-7. [DOI] [PubMed] [Google Scholar]
- 5.Xu M.J., Johnson D.E., Grandis J.R. EGFR-targeted therapies in the post-genomic era. Cancer Metastasis Rev. 2017;36:463–473. doi: 10.1007/s10555-017-9687-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sen M., Joyce S., Panahandeh M., Li C., Thomas S.M., Maxwell J., Wang L., Gooding W.E., Johnson D.E., Grandis J.R. Targeting Stat3 abrogates EGFR inhibitor resistance in cancer. Clin. Cancer Res. 2012;18:4986–4996. doi: 10.1158/1078-0432.CCR-12-0792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yun M.R., Choi H.M., Kang H.N., Lee Y., Joo H.S., Kim D.H., Kim H.R., Hong M.H., Yoon S.O., Cho B.C. ERK-dependent IL-6 autocrine signaling mediates adaptive resistance to pan-PI3K inhibitor BKM120 in head and neck squamous cell carcinoma. Oncogene. 2018;37:377–388. doi: 10.1038/onc.2017.339. [DOI] [PubMed] [Google Scholar]
- 8.Cai Y., Dodhia S., Su G.H. Dysregulations in the PI3K pathway and targeted therapies for head and neck squamous cell carcinoma. Oncotarget. 2017;8:22203–22217. doi: 10.18632/oncotarget.14729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cancer Genome Atlas Network Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Agrawal N., Frederick M.J., Pickering C.R., Bettegowda C., Chang K., Li R.J., Fakhry C., Xie T.X., Zhang J., Wang J. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333:1154–1157. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Darido C., Georgy S.R., Jane S.M. The role of barrier genes in epidermal malignancy. Oncogene. 2016;35:5705–5712. doi: 10.1038/onc.2016.84. [DOI] [PubMed] [Google Scholar]
- 12.Cangkrama M., Ting S.B., Darido C. Stem cells behind the barrier. Int. J. Mol. Sci. 2013;14:13670–13686. doi: 10.3390/ijms140713670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldie S.J., Cottle D.L., Tan F.H., Roslan S., Srivastava S., Brady R., Partridge D.D., Auden A., Smyth I.M., Jane S.M. Loss of GRHL3 leads to TARC/CCL17-mediated keratinocyte proliferation in the epidermis. Cell Death Dis. 2018;9:1072. doi: 10.1038/s41419-018-0901-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de la Garza G., Schleiffarth J.R., Dunnwald M., Mankad A., Weirather J.L., Bonde G., Butcher S., Mansour T.A., Kousa Y.A., Fukazawa C.F. Interferon regulatory factor 6 promotes differentiation of the periderm by activating expression of Grainyhead-like 3. J. Invest. Dermatol. 2013;133:68–77. doi: 10.1038/jid.2012.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brynczka C., Labhart P., Merrick B.A. NGF-mediated transcriptional targets of p53 in PC12 neuronal differentiation. BMC Genomics. 2007;8:139. doi: 10.1186/1471-2164-8-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Darido C., Georgy S.R., Wilanowski T., Dworkin S., Auden A., Zhao Q., Rank G., Srivastava S., Finlay M.J., Papenfuss A.T. Targeting of the tumor suppressor GRHL3 by a miR-21-dependent proto-oncogenic network results in PTEN loss and tumorigenesis. Cancer Cell. 2011;20:635–648. doi: 10.1016/j.ccr.2011.10.014. [DOI] [PubMed] [Google Scholar]
- 17.Georgy S.R., Cangkrama M., Srivastava S., Partridge D., Auden A., Dworkin S., McLean C.A., Jane S.M., Darido C. Identification of a Novel Proto-oncogenic Network in Head and Neck Squamous Cell Carcinoma. J. Natl. Cancer Inst. 2015;107:djv152. doi: 10.1093/jnci/djv152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Darido C., Georgy S.R., Cullinane C., Partridge D.D., Walker R., Srivastava S., Roslan S., Carpinelli M.R., Dworkin S., Pearson R.B., Jane S.M. Stage-dependent therapeutic efficacy in PI3K/mTOR-driven squamous cell carcinoma of the skin. Cell Death Differ. 2018;25:1146–1159. doi: 10.1038/s41418-017-0032-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li J., Huang H., Sun L., Yang M., Pan C., Chen W., Wu D., Lin Z., Zeng C., Yao Y. MiR-21 indicates poor prognosis in tongue squamous cell carcinomas as an apoptosis inhibitor. Clin. Cancer Res. 2009;15:3998–4008. doi: 10.1158/1078-0432.CCR-08-3053. [DOI] [PubMed] [Google Scholar]
- 20.Zhou X., Ren Y., Liu A., Jin R., Jiang Q., Huang Y., Kong L., Wang X., Zhang L. WP1066 sensitizes oral squamous cell carcinoma cells to cisplatin by targeting STAT3/miR-21 axis. Sci. Rep. 2014;4:7461. doi: 10.1038/srep07461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tebbutt N.C., Giraud A.S., Inglese M., Jenkins B., Waring P., Clay F.J., Malki S., Alderman B.M., Grail D., Hollande F. Reciprocal regulation of gastrointestinal homeostasis by SHP2 and STAT-mediated trefoil gene activation in gp130 mutant mice. Nat. Med. 2002;8:1089–1097. doi: 10.1038/nm763. [DOI] [PubMed] [Google Scholar]
- 22.Gaykalova D.A., Manola J.B., Ozawa H., Zizkova V., Morton K., Bishop J.A., Sharma R., Zhang C., Michailidi C., Considine M. NF-κB and stat3 transcription factor signatures differentiate HPV-positive and HPV-negative head and neck squamous cell carcinoma. Int. J. Cancer. 2015;137:1879–1889. doi: 10.1002/ijc.29558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iorio F., Knijnenburg T.A., Vis D.J., Bignell G.R., Menden M.P., Schubert M., Aben N., Gonçalves E., Barthorpe S., Lightfoot H. A Landscape of Pharmacogenomic Interactions in Cancer. Cell. 2016;166:740–754. doi: 10.1016/j.cell.2016.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Z., Feng X., Molinolo A.A., Martin D., Vitale-Cross L., Nohata N., Ando M., Wahba A., Amornphimoltham P., Wu X. 4E-BP1 Is a Tumor Suppressor Protein Reactivated by mTOR Inhibition in Head and Neck Cancer. Cancer Res. 2019;79:1438–1450. doi: 10.1158/0008-5472.CAN-18-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lakshmanachetty S., Balaiya V., High W.A., Koster M.I. Loss of TP63 Promotes the Metastasis of Head and Neck Squamous Cell Carcinoma by Activating MAPK and STAT3 Signaling. Mol. Cancer Res. 2019;17:1279–1293. doi: 10.1158/1541-7786.MCR-18-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barretina J., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehár J., Kryukov G.V., Sonkin D. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niehr F., Eder T., Pilz T., Konschak R., Treue D., Klauschen F., Bockmayr M., Türkmen S., Jöhrens K., Budach V., Tinhofer I. Multilayered Omics-Based Analysis of a Head and Neck Cancer Model of Cisplatin Resistance Reveals Intratumoral Heterogeneity and Treatment-Induced Clonal Selection. Clin. Cancer Res. 2018;24:158–168. doi: 10.1158/1078-0432.CCR-17-2410. [DOI] [PubMed] [Google Scholar]
- 28.Kypriotou M., Huber M., Hohl D. The human epidermal differentiation complex: cornified envelope precursors, S100 proteins and the ‘fused genes’ family. Exp. Dermatol. 2012;21:643–649. doi: 10.1111/j.1600-0625.2012.01472.x. [DOI] [PubMed] [Google Scholar]
- 29.Miles L.B., Dworkin S., Darido C. Alternative splicing and start sites: Lessons from the Grainyhead-like family. Dev. Biol. 2017;429:12–19. doi: 10.1016/j.ydbio.2017.06.018. [DOI] [PubMed] [Google Scholar]
- 30.Maeder M.L., Linder S.J., Cascio V.M., Fu Y., Ho Q.H., Joung J.K. CRISPR RNA-guided activation of endogenous human genes. Nat. Methods. 2013;10:977–979. doi: 10.1038/nmeth.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cangkrama M., Darido C., Georgy S.R., Partridge D., Auden A., Srivastava S., Wilanowski T., Jane S.M. Two Ancient Gene Families Are Critical for Maintenance of the Mammalian Skin Barrier in Postnatal Life. J. Invest. Dermatol. 2016;136:1438–1448. doi: 10.1016/j.jid.2016.02.806. [DOI] [PubMed] [Google Scholar]
- 32.Aubrey B.J., Kelly G.L., Kueh A.J., Brennan M.S., O’Connor L., Milla L., Wilcox S., Tai L., Strasser A., Herold M.J. An inducible lentiviral guide RNA platform enables the identification of tumor-essential genes and tumor-promoting mutations in vivo. Cell Rep. 2015;10:1422–1432. doi: 10.1016/j.celrep.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 33.Lindenblatt Rde.C., Martinez G.L., Silva L.E., Faria P.S., Camisasca D.R., Lourenço Sde.Q. Oral squamous cell carcinoma grading systems--analysis of the best survival predictor. J. Oral Pathol. Med. 2012;41:34–39. doi: 10.1111/j.1600-0714.2011.01068.x. [DOI] [PubMed] [Google Scholar]
- 34.Padma R., Kalaivani A., Sundaresan S., Sathish P. The relationship between histological differentiation and disease recurrence of primary oral squamous cell carcinoma. J. Oral Maxillofac. Pathol. 2017;21:461. doi: 10.4103/jomfp.JOMFP_241_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hata A.N., Niederst M.J., Archibald H.L., Gomez-Caraballo M., Siddiqui F.M., Mulvey H.E., Maruvka Y.E., Ji F., Bhang H.E., Krishnamurthy Radhakrishna V. Tumor cells can follow distinct evolutionary paths to become resistant to epidermal growth factor receptor inhibition. Nat. Med. 2016;22:262–269. doi: 10.1038/nm.4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Magnani L., Frige G., Gadaleta R.M., Corleone G., Fabris S., Kempe M.H., Verschure P.J., Barozzi I., Vircillo V., Hong S.P. Acquired CYP19A1 amplification is an early specific mechanism of aromatase inhibitor resistance in ERα metastatic breast cancer. Nat. Genet. 2017;49:444–450. doi: 10.1038/ng.3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lheureux S., Bruce J.P., Burnier J.V., Karakasis K., Shaw P.A., Clarke B.A., Yang S.Y., Quevedo R., Li T., Dowar M. Somatic BRCA1/2 Recovery as a Resistance Mechanism After Exceptional Response to Poly (ADP-ribose) Polymerase Inhibition. J. Clin. Oncol. 2017;35:1240–1249. doi: 10.1200/JCO.2016.71.3677. [DOI] [PubMed] [Google Scholar]
- 38.Goldie S.J., Chincarini G., Darido C. Targeted Therapy Against the Cell of Origin in Cutaneous Squamous Cell Carcinoma. Int. J. Mol. Sci. 2019;20:20. doi: 10.3390/ijms20092201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Thé H. Differentiation therapy revisited. Nat. Rev. Cancer. 2018;18:117–127. doi: 10.1038/nrc.2017.103. [DOI] [PubMed] [Google Scholar]
- 40.Guinea-Viniegra J., Zenz R., Scheuch H., Jiménez M., Bakiri L., Petzelbauer P., Wagner E.F. Differentiation-induced skin cancer suppression by FOS, p53, and TACE/ADAM17. J. Clin. Invest. 2012;122:2898–2910. doi: 10.1172/JCI63103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Herzog A., Bian Y., Vander Broek R., Hall B., Coupar J., Cheng H., Sowers A.L., Cook J.D., Mitchell J.B., Chen Z. PI3K/mTOR inhibitor PF-04691502 antitumor activity is enhanced with induction of wild-type TP53 in human xenograft and murine knockout models of head and neck cancer. Clin. Cancer Res. 2013;19:3808–3819. doi: 10.1158/1078-0432.CCR-12-2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chung C.H., Parker J.S., Karaca G., Wu J., Funkhouser W.K., Moore D., Butterfoss D., Xiang D., Zanation A., Yin X. Molecular classification of head and neck squamous cell carcinomas using patterns of gene expression. Cancer Cell. 2004;5:489–500. doi: 10.1016/s1535-6108(04)00112-6. [DOI] [PubMed] [Google Scholar]
- 43.Keck M.K., Zuo Z., Khattri A., Stricker T.P., Brown C.D., Imanguli M., Rieke D., Endhardt K., Fang P., Brägelmann J. Integrative analysis of head and neck cancer identifies two biologically distinct HPV and three non-HPV subtypes. Clin. Cancer Res. 2015;21:870–881. doi: 10.1158/1078-0432.CCR-14-2481. [DOI] [PubMed] [Google Scholar]
- 44.Lepikhova T., Karhemo P.R., Louhimo R., Yadav B., Murumägi A., Kulesskiy E., Kivento M., Sihto H., Grénman R., Syrjänen S.M. Drug-Sensitivity Screening and Genomic Characterization of 45 HPV-Negative Head and Neck Carcinoma Cell Lines for Novel Biomarkers of Drug Efficacy. Mol. Cancer Ther. 2018;17:2060–2071. doi: 10.1158/1535-7163.MCT-17-0733. [DOI] [PubMed] [Google Scholar]
- 45.Chen Z., Malhotra P.S., Thomas G.R., Ondrey F.G., Duffey D.C., Smith C.W., Enamorado I., Yeh N.T., Kroog G.S., Rudy S. Expression of proinflammatory and proangiogenic cytokines in patients with head and neck cancer. Clin. Cancer Res. 1999;5:1369–1379. [PubMed] [Google Scholar]
- 46.Zgheib C., Zouein F.A., Chidiac R., Kurdi M., Booz G.W. Calyculin A reveals serine/threonine phosphatase protein phosphatase 1 as a regulatory nodal point in canonical signal transducer and activator of transcription 3 signaling of human microvascular endothelial cells. J. Interferon Cytokine Res. 2012;32:87–94. doi: 10.1089/jir.2011.0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stein-O’Brien G., Kagohara L.T., Li S., Thakar M., Ranaweera R., Ozawa H., Cheng H., Considine M., Schmitz S., Favorov A.V. Integrated time course omics analysis distinguishes immediate therapeutic response from acquired resistance. Genome Med. 2018;10:37. doi: 10.1186/s13073-018-0545-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ferris R.L., Blumenschein G., Jr., Fayette J., Guigay J., Colevas A.D., Licitra L., Harrington K., Kasper S., Vokes E.E., Even C. Nivolumab for Recurrent Squamous-Cell Carcinoma of the Head and Neck. N. Engl. J. Med. 2016;375:1856–1867. doi: 10.1056/NEJMoa1602252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mroz E.A., Tward A.D., Hammon R.J., Ren Y., Rocco J.W. Intra-tumor genetic heterogeneity and mortality in head and neck cancer: analysis of data from the Cancer Genome Atlas. PLoS Med. 2015;12:e1001786. doi: 10.1371/journal.pmed.1001786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bu L.L., Yu G.T., Wu L., Mao L., Deng W.W., Liu J.F., Kulkarni A.B., Zhang W.F., Zhang L., Sun Z.J. STAT3 Induces Immunosuppression by Upregulating PD-1/PD-L1 in HNSCC. J. Dent. Res. 2017;96:1027–1034. doi: 10.1177/0022034517712435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Johnson D.E., O’Keefe R.A., Grandis J.R. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat. Rev. Clin. Oncol. 2018;15:234–248. doi: 10.1038/nrclinonc.2018.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Youssef M., Cuddihy A., Darido C. Long-Lived Epidermal Cancer-Initiating Cells. Int. J. Mol. Sci. 2017;18:1369. doi: 10.3390/ijms18071369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Poortinga G., Quinn L.M., Hannan R.D. Targeting RNA polymerase I to treat MYC-driven cancer. Oncogene. 2015;34:403–412. doi: 10.1038/onc.2014.13. [DOI] [PubMed] [Google Scholar]
- 54.Li Y., Li P.K., Roberts M.J., Arend R.C., Samant R.S., Buchsbaum D.J. Multi-targeted therapy of cancer by niclosamide: A new application for an old drug. Cancer Lett. 2014;349:8–14. doi: 10.1016/j.canlet.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhan Y., Guo J., Yang W., Goncalves C., Rzymski T., Dreas A., Żyłkiewicz E., Mikulski M., Brzózka K., Golas A. MNK1/2 inhibition limits oncogenicity and metastasis of KIT-mutant melanoma. J. Clin. Invest. 2017;127:4179–4192. doi: 10.1172/JCI91258. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.