Abstract
Mutations in USH2A are among the most common causes of syndromic and non-syndromic retinitis pigmentosa (RP). The two most recurrent mutations in USH2A, c.2299delG and c.2276G > T, both reside in exon 13. Skipping exon 13 from the USH2A transcript presents a potential treatment modality in which the resulting transcript is predicted to encode a slightly shortened usherin protein. Morpholino-induced skipping of ush2a exon 13 in zebrafish ush2armc1 mutants resulted in the production of usherinΔexon 13 protein and a completely restored retinal function. Antisense oligonucleotides were investigated for their potential to selectively induce human USH2A exon 13 skipping. Lead candidate QR-421a induced a concentration-dependent exon 13 skipping in induced pluripotent stem cell (iPSC)-derived photoreceptor precursors from an Usher syndrome patient homozygous for the c.2299delG mutation. Mouse surrogate mQR-421a reached the retinal outer nuclear layer after a single intravitreal injection and induced a detectable level of exon skipping until at least 6 months post-injection. In conclusion, QR-421a-induced exon skipping proves to be a highly promising treatment option for RP caused by mutations in USH2A exon 13.
Keywords: exon skipping, retinitis pigmentosa, USH2A, antisense oligonucleotide, zebrafish
Graphical abstract

Mutations in USH2A exon13 are a frequent cause of (non-)syndromic recessively inherited retinitis pigmentosa. The authors showed that skipping of this exon using antisense oligonucleotide (AON) restores usherin protein production and visual function in zebrafish models. Lead AON QR-421a-induced exon13 skipping can therefore be regarded as a promising future treatment option.
Introduction
Retinitis pigmentosa (RP) is a genetically and clinically heterogeneous disorder characterized by a progressive loss of visual function caused by the degeneration of the light-sensitive photoreceptor cells in the retina.1 Although being designated as an orphan disease with an overall prevalence of 1:4,000 individuals, RP is the most common type of inherited retinal dystrophy (IRD), affecting ∼125,000 patients within the European Union and almost two million individuals worldwide. As such, it imposes a significant burden on health care systems and society in general.
To date, mutations in over 100 genes are known to cause non-syndromic or syndromic RP (https://sph.uth.edu/Retnet/). It is estimated that autosomal recessively inherited RP (arRP) accounts for up to 60% of all RP cases.2 Mutations in USH2A collectively account for 7%–23% of arRP cases and can either result in non-syndromic arRP or in Usher syndrome (combination of RP and hearing impairment).2,3 The mutations in this gene are mostly private and evenly distributed throughout the gene. Three mutations, including c.2299delG, p.(Glu767fs); c.2276G > T, p.(Cys759Phe); and c.7595-2144A > G, p.(Lys2532Thrfs), are derived from a common ancestor and are therefore seen more frequently.4, 5, 6, 7 The c.2299delG and c.2276G > T mutations represent, respectively, 27.8% and 7.1% of all pathogenic USH2A alleles, and both reside in exon 13 of the USH2A gene.8
Although attempts have been made, clear genotype-phenotype correlations for USH2A mutations have been proven difficult to establish. Generally, nonsense mutations, frameshift mutations, or canonical splice site mutations in USH2A, either biallelic or combined with one missense allele, are associated with Usher syndrome type II, whereas the combination of two missense changes typically results in non-syndromic RP.9 The auditory phenotype of patients with Usher syndrome can be partially compensated by providing patients with hearing aids or cochlear implants.10 However, currently, no treatment options exist for the progressive loss of vision associated with mutations in USH2A.
The poor understanding of the physiological role(s) of the usherin protein in photoreceptor cells and the pathophysiological mechanism underlying USH2A-associated RP hampers the development of treatment that interferes with the disease mechanisms. The recent approval of Luxturna (voretigene neparvovec), a gene augmentation therapy for the treatment of patients with RPE65-associated retinal dystrophy,11,12 has led to a paradigm shift in the therapeutic approach to monogenic retinal diseases and provides hope for many visually impaired individuals worldwide. However, the development of an USH2A gene augmentation therapy is severely hampered by the size of the usherin-encoding sequence (15,606 nucleotides), which exceeds by a significant margin the cargo capacity of the currently used viral vehicles for gene delivery. An RNA therapy approach, for example, using intravitreal (IVT) delivery of synthetic antisense oligonucleotides (AON) to correct aberrant pre-mRNA splicing, could overcome this limitation for a subset of mutations. In vitro experiments demonstrated that AONs can be used to correct the aberrant pre-mRNA splicing caused by the c.7595-2144A > G mutation in USH2A, which leads to the inclusion of a pseudoexon in the mature USH2A transcript.7 The promise for clinical application of AON-based therapies for IRDs is currently under investigation (data not shown). Sepofarsen (QR-110), a candidate AON designed to treat patients with CEP290-associated Leber congenital amaurosis (LCA10), has demonstrated a significant improvement in visual acuity and other secondary endpoints in LCA10 patients following a single IVT delivery of sepofarsen.13
In this study, we explored AON-induced exon skipping as a potential treatment modality for patients with RP caused by mutations in exon 13 of the USH2A gene. As this exon consists of a multiplier of three nucleotides, skipping the exon does not disturb the open reading frame and could result in the production of a shortened protein with predicted residual function. With the use of use of our previously characterized ush2armc1 zebrafish model,14 we demonstrate that usherin lacking the amino acids encoded by exon 13 has sufficient residual function to prevent loss of retinal function. With the use of cellular models, we identified and validated AON QR-421a as candidate molecule for an exon-skipping therapy described above. Currently, QR-421a is being evaluated in the phase 1/2 clinical trial (Clinicaltrials.gov: NTC03780257).
Results
Formation of epidermal growth factor (EGF)-like fusion domain after targeted USH2A exon 13 skipping
Wild-type usherin is predicted to contain ten EGF-lam domains (http://smart.embl-heidelberg.de/). EGF-lam domains typically contain eight cysteine residues that interact in a pairwise fashion, through covalent disulfide bond, necessary for protein folding and stability. These EGF-lam domains in usherin harbor multiple protein-truncating mutations. Also, 22 out of the 80 cysteine residues in these EGF-lam domains have been found to be mutated in patients with USH2A-associated RP (USH2A LOVD mutation database, https://www.lovd.nl/USH2A), five of which reside within the protein region encoded by exon 13. Unpaired cysteine residues contain a reactive-free thiol group that can induce unwanted multimerization or crosslinking with other proteins.15,16 The in-frame skipping of exon 13 is predicted to result in the fusion of parts of EGF-lam domains 4 and 8 into a functionally related EGF-like domain (Figure 1A). EGF-like domains contain six cysteine residues that together create three disulfide bonds by cysteines 1 + 3, 2 + 4, and 5 + 8 in the fused EGF-like domain.17 There are 16 amino acids between the fifth and sixth cysteine residue in the EGF-like 4-8 fusion domain, which is different from the canonical spacing between cysteine residue 5 and 6 within EGF-like domains, namely 8 amino acids.18 However, 3D homology modeling predicted normal disulfide-bridge formation within the EGF-like 4-8 fusion domain (Figure 1B). In conclusion, molecular modeling warrants exploring the effect of exon 13 skipping at the level of visual function.
Figure 1.
In silico modeling of usherin after exon 13 skipping
(A) Schematic representation of the domain architecture of wild-type (WT) usherin and usherinΔexon 13. Individual EGF-lam domains are numbered. Skipping of exon 13 results in the exclusion of EGF-lam domains 5, 6, and 7, as well as the partial exclusion of EGF-lam domains 4 and 8. The remaining amino acids of EGF-lam domains 4 and 8 are predicted to form an EGF-like domain with six cysteine residues. The fusion site of this domain is located between the fifth cysteine residue of EGF-lam domain 4 and the eighth cysteine derived from EGF-lam 8. (B) 3D homology modeling predicts the formation of a stable EGF-like domain with normal disulfide bridge formations. The predicted structure of usherin EGF-lam domains 4 (left) and 8 (right) are shown. The amino acids that are encoded by USH2A exon 13 are depicted in gray and predicted to be absent after translation of USH2A Δexon 13 transcripts. Cysteine residues that are present in the EGF-like fusion domain are numbered and indicated in orange. The cysteine residues numbered 1 to 5 are derived from EGF-lam domain 4, whereas residue 8 is derived from EGF-lam domain 8.
AON-induced skipping of ush2a exon 13 in a mutant zebrafish model restores usherin protein expression and visual function
To validate exon 13 skipping as a potential therapeutic strategy, we employed our previously characterized ush2a zebrafish mutant (ush2armc1) that contains a frameshift-inducing mutation in exon 13.14 We previously reported that the electroretinogram (ERG) response is significantly reduced in homozygous ush2armc1 larvae and that the usherin protein is absent from the retina, indicating that ush2armc1 is a true null allele. The length of USH2A exon 13 is well conserved between human (642 nucleotides) and zebrafish (648 nucleotides), and the (spacing between) cysteine residues that are essential for EGF-lam domain formation are identical (Figure 2A). Following the previously published guidelines for AON design,19,20 six antisense phosphorodiamidate morpholino oligomers (PMOs) were designed to target the zebrafish ush2a exon 13 splice acceptor site, splice donor site, or exonic splice enhancer (ESE) motifs (Figure S1A; Table 1). The exon-skipping potential of the PMOs was first investigated by injecting the individual PMOs into the yolk of 1- to 2-cell-stage ush2armc1 embryos (Figure S1B). Combined delivery of a low dose of two of the most potent PMOs, targeting different regions of ush2a exon 13, resulted in a more efficient skipping of ush2a exon 13 than individually injected PMOs (Figures S2A and S2B). The combination of PMO1 and PMO2 appeared most potent after reverse transcriptase (RT)-PCR analysis, without leading to aberrations in overall body morphology, and was subsequently used to determine whether exon 13 skipping had an effect on the phenotypic outcome of the ush2armc1 mutant (Figures 2B and S2C).
Figure 2.
Morpholino antisense oligonucleotides (AONs) mediate ush2a exon 13 skipping, usherinΔexon 13 protein expression, and restoration of electroretinogram (ERG) in a mutant zebrafish model
(A) Amino acid alignment of the sequences encoded by human and zebrafish USH2A exon 13. The (partial) EGF-lam domains are indicated. The cysteine residues required for 3D topology of the EGF-lam domains (green) are completely conserved between zebrafish and human. (B) Phosphorodiamidate morpholino oligonucleotide (PMO)-induced skipping of ush2a exon 13 in zebrafish larvae. ush2armc1 mutant embryos were injected with a combination of PMO1 and PMO2 (1 ng of each). Investigation of ush2a pre-mRNA splicing at 3 days post-fertilization (dpf) revealed the skipping of ush2a exon 13 upon injection of PMOs targeting ush2a exon 13. Uninjected ush2armc1 mutant zebrafish larvae and WT larvae were used as controls. (C) Subcellular localization of usherin in horizontal cryosections of larval (5 dpf) zebrafish retinae. Usherin was visualized with anti-usherin antibodies directed against the intracellular C-terminal tail of zebrafish usherin (green signal). Nuclei were stained with DAPI (blue signal), and the connecting cilium is labeled using anti-centrin antibodies (red). In WT larvae, usherin is present at the photoreceptor periciliary membrane, adjacent to the connecting cilium. In homozygous ush2armc1 larvae, no specific usherin signal could be detected. PMO-induced ush2a exon 13 skipping in ush2armc1 mutant larvae resulted in partial restoration of usherinΔexon 13 expression with the correct subcellular localization in the retina. OS, outer segment; ONL, outer nuclear layer; OPL, outer plexiform layer; IPL, inner plexiform layer; wt: WT; ush2armc1, zebrafish with exon 13 mutation. (D) Scatterplot of the relative fluorescence intensity of anti-usherin staining in the periciliary region of all photoreceptors in the middle section of the larval zebrafish eye. The signal intensity is decreased in the ush2armc1 retina compared to WTs. Relative fluorescent signal intensity of anti-usherin staining is significantly increased in PMO-injected ush2armc1 mutants as compared to uninjected mutants (∗∗∗∗p < 0.0001, Kruskal-Wallis test followed by Dunn’s nonparametric post-test). (E) Average ERG b-wave traces from uninjected, control PMO-injected, exon 13 PMO-injected ush2armc1 larvae and WT controls at 5−6 dpf. PMO-induced skipping of ush2a exon 13 completely restored b-wave amplitudes in ush2armc1 larvae as compared to uninjected or control PMO-injected mutants. (F) Maximum b-wave amplitudes recorded in uninjected or control PMO-injected ush2armc1 larvae are significantly reduced as compared to ERG traces from age- and strain-matched WT controls. Maximum b-wave amplitudes recorded in PMO-injected ush2armc1 mutants are significantly improved as compared to ERG traces from uninjected or control PMO-injected ush2armc1 mutants and do not significantly differ from WTs (p > 0.99). Data are shown as mean ± SD, ∗p < 0.05, ∗∗p < 0.01, Kruskal-Wallis test followed by Dunn’s nonparametric post-test. (G) Quantification of ush2a Δexon 13 transcripts in uninjected and PMO-injected zebrafish larvae at 3 dpf.
Table 1.
Antisense oligonucleotide sequences used in the study
| Name | System | Sequence | Remark |
|---|---|---|---|
| PMO1 | zebrafish | 5′-GTTACAACGGTCACAGGTTAGACCTAAA-3′ | splice acceptor and SC35 motif 1 |
| PMO2 | zebrafish | 5′-CATGGGTCACAGCCACAGGAAATGC-3′ | SC35 motifs 3 and 4 |
| PMO3 | zebrafish | 5′-GGTAGGCAGATACACTGACCACTTA-3′ | SC35 motif 18 |
| PMO4 | zebrafish | 5′-AACGGTCACAGGTTAGACCTAAAAATAA-3′ | splice acceptor and SC35 motif 1 |
| PMO5 | zebrafish | 5′-GGATTACAGAACTGGTGCAGAGAAC-3′ | SC35 motifs 5 and 6 |
| PMO6 | zebrafish | 5′-AAGCACTAACCTGGTTTACAGGTTCCAC-3′ | splice donor and SC35 motifs 19 and 20 |
| Control PMO | zebrafish | 5′-CCTCTTACCTCAGTTACAATTTATAC-3′ | − |
| QR-421a | human | 5′-AGCUUCGGAGAAAUUUAAAUC-3′ | fully phosphorothioated backbone and 2′ O-methoxyethyl RNA bases |
| Control | human/mouse | 5′-AUAGUAACGGAUUGAGG-3′ | fully phosphorothioated backbone and 2′ O-methoxyethyl RNA bases |
| mQR-421a | mouse | 5′-AACUCUGGAGGAAUUUAAAUC-3′ | fully phosphorothioated backbone and 2′ O-methoxyethyl RNA bases |
We first determined whether skipping of zebrafish ush2a exon 13 in homozygous ush2armc1 mutant larvae resulted in the synthesis of a shortened usherin protein (usherinΔexon 13). Antibodies directed against the intracellular region of zebrafish usherin were used to stain unfixed cryosections of wild-type larvae, uninjected ush2armc1 larvae, and ush2armc1 larvae in which ush2a exon 13 skipping was induced by PMO injection (Figure 2C, green signal). The photoreceptor connecting cilium was labeled by antibodies against centrin (Figure 2C, red signal). In wild-type larvae, usherin localizes in the periciliary region, as expected. In the retina of uninjected ush2armc1 larvae, no usherin could be detected. In PMO-injected ush2armc1 larvae, a partial restoration of usherin expression was detected with an expected subcellular localization. The intensity of the anti-usherin fluorescence signals was quantified using an automated Fiji script. Skipping of ush2a exon 13 resulted in a small but statistically significant increase of the average fluorescence intensity in the periciliary region of photoreceptors as compared to uninjected larvae from the same clutch (34.98 ± 0.14 [Δexon 13; n = 10] versus 30.04 ± 0.16 [uninjected; n = 10]; p < 0.0001 [Kruskal-Wallis and Dunn’s nonparametric test]) (Figure 2D). This corroborated that exon 13 skipping resulted in the synthesis of usherinΔexon 13.
ERGs were subsequently recorded from ush2armc1 larvae that were injected with a combination of ush2a exon 13-targeting PMOs (n = 25) or with a standard control PMO (n = 14). Uninjected age- and strain-matched wild-types (n = 10) and ush2armc1 (n = 11) larvae were used as controls. Uninjected and control PMO-injected ush2armc1 mutant larvae demonstrated significantly reduced b-wave amplitudes as compared to age- and strain-matched wild-type larvae (p < 0.05 [uninjected], and p < 0.001 [control PMO-injected]; Kruskal-Wallis and Dunn’s nonparametric test) (Figures 2E and 2F). PMO-induced skipping of ush2a exon 13 from ush2armc1 larvae resulted in significantly increased b-wave amplitudes as compared to uninjected or control PMO-injected ush2armc1 larvae, which is indicative for a restoration of visual function. The ERG b-wave amplitudes recorded in ush2armc1 larvae after injection with exon 13-targeting PMOs were not significantly different from those recorded in age- and strain-matched wild-type larvae (p > 0.999) (Figures 2E and 2F). Quantitative RT-PCR (qRT-PCR) analysis of exon 13 skipping in larvae injected with low or high doses of PMOs revealed that increasing the PMO dose did not result in a significant gain in ush2a Δexon 13 transcripts but rather decreased the number of full-length ush2a transcripts (Figures 2G and S2D). At all tested doses of PMO, the levels of ush2a Δexon 13 transcripts ranged between 18% and 26% of the amount of total ush2a transcripts observed in wild-type zebrafish. Together, these data show that AON-induced skipping results in the formation and correct localization of an usherinΔexon 13 protein with sufficient residual function to rescue visual dysfunction in ush2armc1 zebrafish larvae.
Identification of lead oligonucleotide QR-421a
Based on the ability of usherinΔexon 13 to restore visual function in zebrafish, we aimed to develop AONs with the ability to induce skipping of exon 13 from human USH2A transcripts. Fourteen AONs were designed based on the bio-informatic analysis of the sequence of USH2A exon 13 and flanking intronic regions. Both the intron-exon boundaries and the ESE motifs within exon 13, identified using the SpliceAid webserver,21 were used as targets for AONs. With the use of in silico analysis, parameters for (lack of) secondary structure formation, thermodynamic properties, and sequence selectivity were taken into account to minimize potential off-target effects. The designed AONs were transfected in the retinoblastoma-derived WERI-Rb1 cell line22 at a concentration of 200 nM and screened for their potential to induce USH2A exon 13 skipping (Figure S3). Because of these analyses, the best-performing 21-mer RNA AON sequence was selected. For further preclinical development, the molecule was synthesized as an antisense RNA molecule with 2′-O-(2-methoxyethyl) ribose sugar modification and a fully phosphorothioated backbone. This candidate was named QR-421a thereafter.
QR-421a was screened for pro-inflammatory potential in silico, revealing the absence of known inflammatory motifs, and in vitro, using a human peripheral blood mononuclear cell (PBMC) activation assay. Gymnotic delivery of QR-421a at concentrations between 0.1 and 10 μM had no effect on PBMC viability and no statistically significant increase in cytokine release (Figure S4).
The target specificity of QR-421a was investigated using an in silico analysis. QR-421a showed no full complementarity to any mRNA, pre-mRNA, or DNA targets other than the anticipated region in USH2A. Partial complementarity to other genomic regions was only found with ≥2 mismatches. Only two off-target sequences were identified with 2 mismatches, residing in one intergenic and one intronic region, and are therefore not expected to influence gene expression or pre-mRNA splicing. Other hits with >2 mismatches are not considered biologically meaningful for a 21-mer splice modulation oligonucleotide, as a single mismatch in an AON was previously shown to already markedly decrease splice modulation efficiency.23 Hence, the risk for potential off-target splicing effects, due to the hybridization of QR-421a to targets other than the intended target, is considered negligible.
WERI-Rb1 cells were treated with QR-421a gymnotically or with the aid of a transfection reagent to provide pharmacodynamic proof of concept for the USH2A exon 13 skipping potential using transcript-specific qRT-digital droplet PCR (ddPCR) analysis. Upon QR-421a transfection, a dose-dependent skipping of USH2A exon 13 was induced, which was already evident at a concentration of 25 nM. An exon 13 skipping efficiency of ∼60% was reached upon transfection at the highest concentration tested (200 nM) (Figure 3A). After gymnotic delivery of QR-421a at concentrations ranging from 10 to 50 μM, exon 13 skipping efficiencies ranging from 10% to 17% were observed (Figure 3B). In both experiments, treatment of WERI-Rb1 cells with a control oligonucleotide did not induce skipping of exon 13, confirming that the observed exon-skipping potential is specific for QR-421a (Figures 3A and 3B). Amplification of USH2A exons 11 to 15 in QR-421a-treated WERI-Rb1 cells (200 nM) revealed mainly transcripts lacking exon 13, which was confirmed by Sanger sequencing but also two minor alternative products that were also identified in untreated WERI-Rb1 cells (Figures 3C and 3D). One of these fragments lacked both exons 12 and 13; the other fragment contained only exons 11 and 15. Altogether, these data show that QR-421a has the ability to enter proliferating WERI-Rb1 cells after transfection or even unaided, thereby inducing a concentration-dependent skipping of USH2A exon 13.
Figure 3.
QR-421a shows a concentration-dependent increase of USH2A exon 13 skipping in WERI-Rb1 cells
(A and B) WERI-Rb1 retinoblastoma cells were treated with different concentrations of QR-421a, using either (A) transfection or (B) gymnotic uptake. Untreated and control oligo-treated cells were included as negative controls. Exon-skipping level was determined by quantification of USH2A transcripts with and without exon 13 by RT-ddPCR. Treatment with QR-421a resulted in a significant concentration-dependent increase of USH2A Δexon 13 transcripts. Data are shown as mean ± SD. Two biological replicates per treatment condition. Asterisks indicate significant differences with scrambled control oligo-treated cells (∗∗∗p < 0.001; ∗∗∗∗p < 0.0001; one-way ANOVA followed by Dunnett’s multiple comparison test). (C) Representative image of exon 11-15 RT-PCR amplicons obtained with RNA isolated from untransfected and QR-421a-transfected WERI-Rb1 cells. QR-421a is able to induce skipping of USH2A exon 13 and does not increase the formation of other alternatively spliced USH2A transcripts. Of note, USH2A Δexon 12-13 and Δexon 12-14 transcripts are already present in untreated WERI-Rb1 cells and yield out-of-frame mRNA transcripts. (D) Sanger sequencing traces of the USH2A Δexon 13 amplicon shown in (C) confirm that the sequence of exon 13 is lacking from the transcript.
QR-421a treatment induces a concentration-dependent increase of USH2A exon 13 skipping in induced pluripotent stem cell (iPSC)-derived photoreceptor progenitor cells (PPCs)
PPCs, differentiated from iPSCs obtained from an USH2A patient with a homozygous c.2299delG mutation in exon 13, were used to assess the exon-skipping potential of QR-421a in a differentiated cell model with the appropriate genetic context. PPCs have been previously shown to be a valuable and clinically relevant tool for the evaluation of novel human-specific therapeutic strategies.24,25
Initially, patient-derived fibroblasts were reprogrammed into iPSCs and subsequently differentiated into PPCs. In order to validate that the cells had differentiated into PPCs, we assessed the expression levels of photoreceptor marker genes (CRX, NRL, OPN1SW, OPN1LW, and RHO) by qRT-PCR analysis after 90 days of differentiation. As expected, the expression levels of photoreceptor marker genes were all significantly increased as compared to iPSCs, whereas the expression of the iPSC-specific marker gene NANOG was simultaneously decreased (Figure 4A).
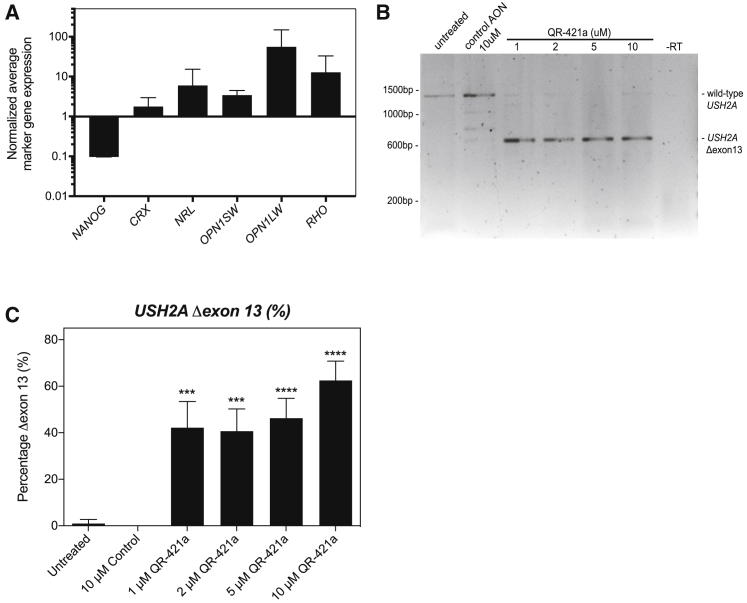
Figure 4.
QR-421a treatment induces a concentration-dependent increase of USH2A exon 13 skipping in iPSC-derived photoreceptor progenitor cells (PPCs) from a patient (USH2Ac.2299delG/c.2299delG)
(A) Gene-expression analysis indicates successful differentiation toward PPCs. The decrease in NANOG expression is indicative for loss of pluripotency, whereas the increased expression of photoreceptor markers CRX, NRL, OPN1SW, OPN1LW, and RHO is indicative of the successful differentiation toward photoreceptor cells. (B) RT-PCR analysis of USH2A exons 11 to 15 in untreated PPCs of a patient homozygous for the c.2299delG mutation only revealed an amplicon containing exons 11 to 15. Continuous treatment of PPCs with QR-421a (28 days) specifically induced the skipping of USH2A exon 13 from the transcript. There was no evidence of alternative splice site activation in exon 13 or skipping of multiple exons. (C) Quantitative analysis of USH2A exon 13 skipping by RT-ddPCR upon continuous treatment with QR-421a. Treatment was started after 3 months of differentiation and lasted for 28 days. One-half of the culture medium was refreshed every other day with medium containing QR-421a. Skipping of exon 13 was already observed at the lowest concentration and further increased with increasing concentrations. Asterisks indicate significant differences with scrambled control oligo-treated cells (∗p < 0.05; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001; mean ± SD of 3 samples per condition, one-way ANOVA followed by Dunnett’s multiple comparison test).
Patient-derived PPCs were treated with a stable concentration of QR-421a for 28 days using gymnotic delivery. Every 2 days, one-half of the culture medium was replaced with fresh medium containing a new dose of QR-421a. Untreated PPCs and PPCs treated with a control oligonucleotide (with the same chemistry and length but a random sequence) were used as negative controls. RT-PCR analysis of USH2A exons 11 to 15 revealed that, in contrast to previous analysis in patient-derived fibroblasts,26 no alternatively spliced USH2A transcripts could be detected in untreated PPCs homozygous for the c.2299delG mutation (Figure 4B). Results furthermore showed that QR-421a induced significant levels of exon 13 skipping at all concentrations tested (1−10 μM), whereas exons 12 and 14 were retained within the USH2A Δexon 13 transcript (Figures 4B and 4C). At a 1-μM concentration, exon 13 skipping was observed in 42% ± 11% (p = 0.001, Sidak’s multiple comparison test) of USH2A transcripts. This increased to 63% ± 8% (p < 0.0001, Sidak’s multiple comparison test) of transcripts lacking exon 13 when QR-421a was supplied at a 10-μM concentration (Figure 4C). No exon 13 skipping was detected in untreated or control oligonucleotide-treated PPCs, indicating that skipping of this exon was specifically induced by QR-421a.
Retinal uptake, efficacy, and duration of action of mQR-421a in wild-type mouse retina
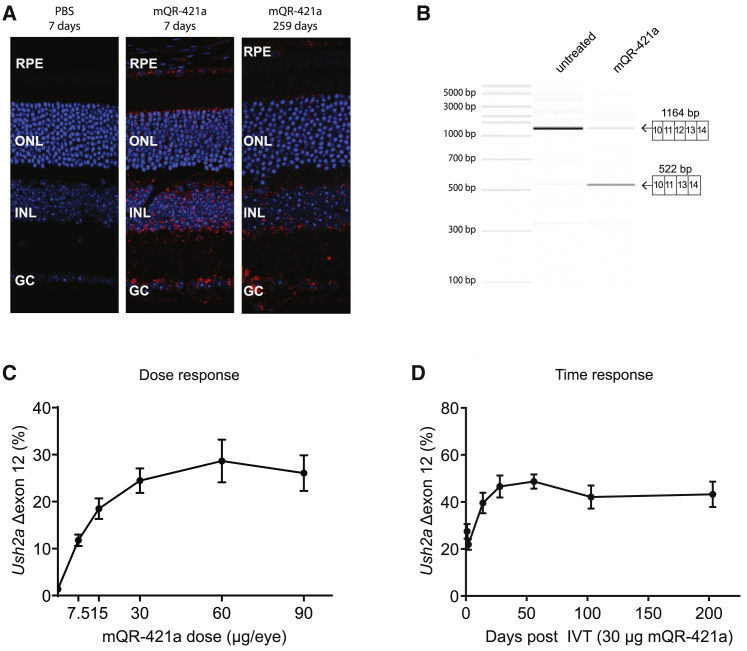
In the absence of a humanized exon 13 mutant mouse model, wild-type mice were explored as a model system to study the molecular in vivo efficacy of QR-421a. The USH2A gene is well conserved across species and is very similar in humans and mice. However, the mouse sequence has a few base differences at the QR-421a binding site; therefore, a mouse surrogate mQR-421a was used in the mouse studies. mQR-421a has the same chemistry, length, binding site, and sequence as QR-421a, the only difference being four bases that are changed to match the mouse sequence. mQR-421a was expected to mediate exon 12 (equivalent of human USH2A exon 13) skipping in a mouse Ush2a transcript (Ensembl: ENSMUST00000060479.13). To assess the in vivo uptake, wild-type C57BL/6J mice received a single bilateral IVT injection of 50 μg per eye (n ≥ 4 eyes/time point) of mouse surrogate oligo mQR-421a or PBS and were sacrificed 7 days or 259 days post-injection. mQR-421a was visualized in murine eye sections using a complementary mQR-421a Cy5-labeled probe on a confocal microscope (Figure 5A). The AON was observed in all of the layers of the retina, and the strongest signal was observed in the ganglion cell layer followed by the inner and outer nuclear layer in the order of relative proximity of these cell layers to the site of injection. There is a clear and abundant distribution of mQR-421a to the photoreceptor cell bodies, the pharmacological target site, with punctate perinuclear localization. No signal was observed in eyes injected with PBS. To investigate mQR-421a dose response in vivo, mice received bilateral IVT injections of 7.5, 15, 60, or 90 μg mQR-421a (12 eyes/dose) in a single dosing occasion and were maintained for 7 days. The control group received 30 μg control oligonucleotide. First, mQR-421a-mediated exon 12 skipping was visualized by amplifying the Ush2a transcripts using primers binding to exons 10 and 14 and analyzing the resulting PCR product on a bioanalyzer (Figure 5B). In untreated animals, only one prominent band corresponding to Ush2a transcripts with exon 12 was detected, whereas in mQR-421a-treated samples, two bands were detected, with the predominant band corresponding to transcripts without exon 12, confirming the mQR-421a-mediated exon 12 skipping in vivo. Interestingly, the untreated sample showed faint but visible bands corresponding to exon 12 skipping and exon 11 + 12 skipping, indicating the natural skipping of these exons, albeit at very low levels. Next, levels of Ush2a transcripts with and without exon 12 were quantified using isoform-specific RT-ddPCR assays, and the percentage of exon skipping was calculated. Results showed that mQR-421a induced Ush2a exon 12 skipping at all of the tested doses compared to the control AON (Figure 5C). Exon-skipping levels were dose dependent and ranged from 12% at the 7.5-μg dose to 29% at the 60-μg dose. In comparison, only <1.5% exon skipping was detected in the control AON-treated group. Doses higher than 60 μg did not result in an increase in exon skip; rather, a slight reduction was noticed. To study the duration of action in vivo, mice received bilateral IVT injections of 30 μg mQR-421a per eye in a single dosing occasion and were maintained for 1, 2, 14, 28, 56, 103, or 203 days. Levels of Ush2a transcripts with and without exon 12 were quantified using isoform-specific RT-ddPCR assays, and the percentage of exon skipping was calculated. Results showed that mQR-421a induced significant levels of Ush2a exon 12 skipping at all time points tested in this study (Figure 5D). Skipping levels increased with time, and highest skipping was detected at 56 days, and the skipping levels decreased slightly thereafter. An average of 25% and 20% exon skipping was detected, respectively, at 1 and 2 days post-dose. Exon skipping increased over time to approximately 40%, 50%, and 53% at 17, 28, and 56 days post-dose, respectively. At days 103 and 203 post-dose, skipping percentage decreased to 43% and 38%, respectively.
Figure 5.
Retinal uptake, efficacy, and duration of action of mQR-421a upon intravitreal (IVT) injection in WT mice
(A) Representative sections of untreated and mQR-421a-treated mouse retina. WT C57BL/6J mice received a single bilateral IVT injection of 50 μg of mQR-421a and were sacrificed to visualize the presence of mQR-421a at 7 and 259 days post-injection. mQR-421a was visualized using a Cy5-labeled FISH assay (red signal); cell nuclei were stained with DAPI (blue signal). GC, ganglion cell layer; INL, inner nuclear layer; RPE, retinal pigment epithelium. (B) Bioanalyzer (PCR) analysis of Ush2a exon 12 skipping in untreated and mQR-421a-treated mice. Ush2a transcripts were amplified using primers binding to exons 10 and 14. The top 1,164-base pair band corresponds to Ush2a transcripts with exon 12, and the bottom 522-base pair band corresponds to transcripts without exon 12. (C) Dose-dependent Ush2a exon 12 skipping by mQR-421a in WT C57BL/6J mice. Mice received a single bilateral IVT of 7.5, 15, 60, or 90 μg per eye of mQR-421a or 30 μg control AON. Ush2a transcripts with or without exon 12 were quantified by ddPCR and normalized by total Ush2a levels measured at exon 8-9, and percentage of exon 12 skipping was calculated. Mean ± SEM, n = 6 animals per dose per group. (D) Long duration of action of mQR-421a in mice retina. Mice received a single bilateral IVT of 30 μg mQR-421a per eye and followed for 1, 2, 14, 28, 56, 103, or 203 days. Ush2a transcripts with or without exon 12 were quantified by ddPCR and normalized by total Ush2a levels measured at exon 8-9, and percentage of exon 12 skipping was calculated. Mean ± SEM, n = 12 (1, 2, 14, and 28 day time points) or n = 8 (56, 103, and 203 day time point) eyes.
Discussion
Mutations in exon 13 of the USH2A gene, including the recurrent mutations c.2299delG and c.2276G > T, are estimated to underlie syndromic (Usher syndrome) and non-syndromic RP in approximately 16,000 individuals in the Western world. In this study, we used PMOs targeting zebrafish ush2a exon 13 to evaluate exon skipping as a therapeutic strategy for the future treatment of USH2A-associated RP. We show that skipping of ush2a exon 13 resulted in a partially restored expression of usherin protein in photoreceptors of ush2armc1 larvae. Furthermore, exon 13 skipping restored ERG b-wave amplitudes, which indicates improved retinal function. We established QR-421a, an AON drug candidate that induces skipping of human USH2A exon 13 in cellular models. Finally, the corresponding mouse AON showed a long in vivo duration of action in mouse retina following a single IVT treatment. Our study therefore provides proof of concept for exon skipping as a highly promising treatment option for USH2A-associated retinal degeneration as a consequence of mutations in exon 13.
In the inner ear, usherin is essential for the maturation of hair bundles that are located at the apex of hair cells,27 whereas in the retina, the large extracellular tails of usherin and ADGRV1 have been proposed to interact and together, bridge the gap between the opposing membranes of the photoreceptor connecting cilium and the periciliary region.28,29 In contrast to the situation in the inner-ear hair cells, usherin seems redundant for the initial development of photoreceptors28 and rather fulfills a post-developmental role. As such, usherin seems to be particularly important for photoreceptor maintenance.30 Therefore, therapeutic strategies that rescue the expression of functional usherin protein in the retina can potentially prevent or slow down the progression of photoreceptor degeneration and, as such, preserve visual function in patients.
The in-frame skipping of exons harboring pathogenic mutations has already been shown to have a particularly high therapeutic potential for large genes encoding (structural) proteins that contain a series of repetitive protein domains.31 Duchenne muscular dystrophy (DMD) is caused by mutations in the DMD gene. DMD encodes dystrophin, a structural linker protein consisting of a stretch of 24 spectrin-like domains flanked by protein-protein interaction domains that are used to connect the F-actin cytoskeleton to β-dystroglycan. Exon skipping was previously shown to restore the reading frame in patients suffering from DMD due to mutations in exon 51 and has the ability to restore the production of a functional dystrophin protein.32,33 Like dystrophin, usherin is also a large structural protein and contains repetitive EGF-lam and FN3 domains. The in-frame exon 13 of USH2A, containing the recurring mutations c.2299delG and c.2276G > T, encodes multiple EGF-lam domains that are proposed to form a stiff rod-like element.34,35
Functionality of the usherinΔexon 13 protein in the retina was assessed after the skipping of exon 13 in our recently published ush2armc1 zebrafish mutant, which has a homozygous protein-truncating mutation in exon 13.14 Microinjection of a combination of two PMO-based AONs in the yolk of single-cell-staged zebrafish embryos induced a transient skipping of exon 13 from the ush2a transcript. As a result, a small but significant increase in usherin protein at the photoreceptor periciliary membrane of PMO-treated ush2armc1 larvae was observed. This indicates that translation of ush2a Δexon 13 transcripts results in a shortened usherin protein that is able to properly localize in zebrafish photoreceptor cells. Furthermore, PMO-induced skipping of the mutated exon 13 resulted in completely restored ERG b-wave amplitudes, indicative of a restored visual function. Although PMOs are remarkably stable in zebrafish embryos, the intranuclear PMO concentration declines with the increasing number of nuclei during development.36 As such, the effect of PMO-induced gene knockdown at the transcriptional level or splice modulation are generally most prominent within the first 3 days of zebrafish development.37 The observed levels of exon 13 skipping (∼20% at 3 days post-fertilization [dpf]) already indicate that relatively few ush2a Δexon 13 transcripts are required to rescue the retinal defects in ush2armc1 larvae. As it is only useful to record ERGs in zebrafish larvae that have a functional retina (≥5 dpf),38 the complete restoration of ERG defects observed in PMO-treated ush2armc1 larvae, at 5−6 dpf, either suggests that even lower levels of ush2a Δexon 13 transcripts are sufficient for retinal function or that the encoded usherinΔexon 13 is relatively stable, at least until 5−6 dpf. Rods do not significantly contribute to the zebrafish ERG until 15 dpf, and therefore, all responses recorded in these larvae are expected to be cone derived.39,40 Patients with USH2A-associated RP often present with night blindness as the initial symptom of retinal dysfunction, indicating a primary dysfunction of the rods.41 However, it was recently reported that both rod and cone responses were markedly reduced in the ERGs of adolescent USH2a patients.42 Therefore, a restored ERG response in zebrafish ush2armc1 larvae upon exon 13 skipping is promising for a beneficial effect in patients. Furthermore, the functionality of the usherinΔexon 13 protein is also supported by a recent book chapter by Pendse et al.43, demonstrating that auditory function is not affected in Ush2aΔexon 13/Δexon13 nor in Ush2aΔexon 13/mut mice.
Following the therapeutic proof of concept obtained for ush2a exon 13 skipping in zebrafish photoreceptors, we present evidence for the pharmacodynamic potential of QR-421a, the lead-candidate AON for the future treatment of patients with RP due to mutations in exon 13 of the USH2A gene. QR-421a treatment resulted in a concentration-dependent USH2A exon 13 skipping in a retinoblastoma cell line. In general, retinal tissue displays a high degree of transcriptional activity and alternative splicing.44 Retinal organoids and PPC differentiated from patient-derived iPSCs provide an excellent platform to test therapeutic interventions for IRDs in vitro, as recently demonstrated by us24 and others.25 Treatment of PPCs, derived from a patient homozygous for the USH2A c.2299delG mutation with QR-421a, resulted in a dose-dependent skipping of exon 13, with no induction of unwanted, alternative exon-skipping events observed.
It is important that oligonucleotides intended for the treatment of retinal dystrophies are capable of accessing retinal cells in order to reach the intended target site. In this study, in vivo efficacy of a mouse surrogate, mQR-421a, was characterized in wild-type mice following IVT administration of the AON. Upon IVT dose, mQR-421a was distributed into all ocular tissues and most importantly, into photoreceptor cells, where it mediated dose-dependent Ush2a exon 13 skipping. mQR-421a also displayed a long duration of action with abundant skipping levels even 203 days after a single IVT injection. The ocular kinetics of mQR-421a are similar to those described for other large hydrophilic molecules, with the vitreous acting as a central compartment from which the oligonucleotide is rapidly taken up by the surrounding ocular tissue after IVT injection and from which it is slowly cleared via anterior and/or posterior routes.45 Sepofarsen, formerly known as QR-110, an AON designed for the CEP290-associated splice correction therapy to treat LCA, is similar to QR-421a in chemical composition and resulted in exon-skipping efficiencies in human iPSC-derived photoreceptor cells and uptake into rabbit and mouse photoreceptors.24 As an IVT injection of sepofarsen was recently shown to significantly improve visual acuity and a host of other retinal functional parameters in LCA10 patients,13 our data suggest that QR-421a has the appropriate physicochemical and pharmacological properties for future clinical applications.
A maximum percentage of ∼20% of ush2a Δexon 13 transcripts was observed in PMO-treated ush2armc1 zebrafish larvae relative to the total amount of ush2a transcripts in untreated strain- and age-matched wild-type larvae. As this relatively low percentage of exon 13 skipping still resulted in a complete restoration of ERG traces, it is tempting to speculate on the minimal amount of USH2A Δexon 13 transcripts needed for a detectable and durable therapeutic effect. Individuals that carry a heterozygous loss-of-function mutation in USH2A are asymptomatic, indicating that about 50% of wild-type usherin would be enough for a sustained retinal function. The work of Pendse et al.43 shows that the amount of protein produced from a single Ush2a Δexon 13 allele is indeed sufficient for normal usherin localization in photoreceptors, normal hair cell development, and normal auditory function in mice. The surprisingly low amount of usherinΔexon 13 protein detected in the retina of PMO-injected ush2armc1 larvae, nevertheless leading to complete restoration of retinal function, suggests that an even lower amount of usherinΔexon 13 protein can be sufficient for retinal function. Interestingly, studies in a humanized mouse model for USH1c showed that ∼20% of correctly spliced Ush1c transcripts, observed after the delivery of splice-correcting AONs at postnatal day 5, is sufficient to rescue auditory function up to 3 months post-injection.46 Based on this and what is known from other AONs acting through an exon-skipping mechanism,46, 47, 48 exon-skipping levels in the range of 10%–20% could potentially be enough to result in sufficient protein restoration to reach efficacious levels.
The age of onset and slow rate of progression of USH2A-associated RP leave ample opportunity for therapeutic intervention to halt the slow disease progression. All evidence indicates that ush2a transcripts lacking exon 13 encode a usherinΔexon 13 protein with sufficient residual function to rescue visual function. The splice-modulating functionality of QR-421a was demonstrated in vitro and in vivo for the treatment of RP, resulting from mutations in exon 13 of the USH2A gene. QR-421a is currently being investigated in a phase I/II clinical trial (ClinicalTrials.gov: NCT03780257), and to our knowledge, this is the first time that proof of concept for a molecular treatment that went into clinical trials was solely obtained in zebrafish, stressing the value of this model organism in translational science.
Materials and methods
Animals
ush2armc1 (c.2337_2342delinsAC; p.Cys780GlnfsTer32; Dona et al.14) and strain-matched wild-type Tüpfel long fin zebrafish were bred and raised under standard conditions.49 Both adult and larval zebrafish were kept at a light-dark regime of 14 h light:10 h darkness. All experiments were carried out in accordance with European guidelines on animal experiments (2010/63/EU). Zebrafish eggs were obtained from natural spawning and reared at 28.5°C in E3 embryo medium (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, and 0.33 mM MgSO4), supplemented with 0.1% methylene blue. Wild-type male and female C57BL/6J mice between 9 and 13 weeks (at start of treatment) were randomly assigned to different groups, with balanced age and gender distribution. Sample sizes were estimated based on in vitro data. Mice were group housed under pathogen-free conditions with reversed light-dark cycle and cage enrichment in standard open polysulfone type II cages at constant temperature and humidity according to recommendations of the Federation of European Laboratory Animal Science Associations. Water and standard chow were provided ad libitum. Mouse experiments were approved by the local Ethics Committee and conformed to the European Community regulations (EEC number 86/609).
In silico modeling of the effect of USH2A exon 13 skipping on the EGF-lam protein domain structure
Models of the separate EGF domains were built using the modeling script in the YASARA50 and WHAT IF51 algorithms with standard parameters. The separate domain sequences were used, resulting in a model for each domain based on PDB: 4AQS (domain 4, 45% sequence identity), 3TBD (domain 5, 46% sequence identity), 4AQS (domain 6, 44% sequence identity), 5LF2 (domain 7, 51% sequence identity), and 1KLO (domain 8, 41% sequence identity) (https://www.rcsb.org/). To create the fusion model of domains 4 and 8, we used the model for domain 4 as template and swapped the C-terminal residues for their corresponding residues in domain 8. A subsequent energy minimization was performed to remove big errors.
Zebrafish PMO design and microinjection
PMOs were designed by first assessing the target sequence for SRSF2 (SC35) ESE sites (threshold of 3.0) using the online ESE finder 3.0 tool.52 Zebrafish ush2a exon 13-targeting PMOs were synthesized by Gene Tools (USA). PMOs were dissolved in ultrapure water at a stock concentration of 50 μg/μL and stored at −20°C. One nanoliter containing 0.5−4 pg per PMO and 0.25% (v/v) phenol red was injected into the yolk of 1- to 2-cell-stage embryos with a Pneumatic PicoPump pv280 (World Precision Instruments). After injection, embryos were raised at 28.5°C in E3 embryo medium until analysis. PMO sequences are provided in Table 1.
Zebrafish ush2a transcript analysis
Total RNA was isolated from pools of 10−15 larvae per condition. Larvae were snap frozen on liquid nitrogen and subsequently homogenized in QIAzol reagent (QIAGEN; #79306) using a 25G needle. QIAzol extraction of total RNA was performed as per the manufacturer’s instruction. Total RNA was DNase treated using the NucleoSpin RNA Extraction Kit (Macherey-Nagel; #740955.50). 1 μg of total RNA was reverse transcribed using the SuperScript VILO Reverse Transcriptase Kit (Thermo Fisher Scientific; #11755050). PMO-induced alternative splicing of ush2a transcripts was analyzed by PCR amplification using primers in zebrafish ush2a exons 11 and 14 using Q5 HF DNA polymerase (New England Biolabs; #M0491L). Primer sequences are provided in Table 2. Amplified transcripts were visualized by agarose gel electrophoresis (1% agarose in 0.5 × Tris-borate-EDTA [TBE]) and subsequently validated using Sanger sequencing. Exon 13-skipping levels were determined using a qRT-PCR approach, including a standard curve of custom synthetic oligonucleotide templates (gBlocks; Integrated DNA Technologies), using primer pairs that specifically amplify exon 13-containing ush2a transcripts or Δexon 13 ush2a transcripts. Targets were amplificated using GoTaq DNA polymerase (Promega; #M3001) and analyzed on a QuantStudio 3 Real-Time PCR System (Applied Biosystems).
Table 2.
Primer/probe list
| Target | Species | Sequence |
|---|---|---|
| ush2a exon 11-14 | zebrafish | 5′-AGCGCTGTCGGAGTCTCTTC-3′ |
| 5′-CTGTGACCGGTCAGTGATGG-3′ | ||
| ush2a exon 12 + 13 | zebrafish | 5′-TGTATCTGCCTACCCACACG-3′ |
| 5′-CACACACACACTGCCCTGA-3′ | ||
| Δexon 13 ush2a | zebrafish | 5′-AGTGCAATCAGTGCCAACAC-3′ |
| 5′-CGGACAGGAAAAAACCGATTAC-3′ | ||
| NANOG | human | 5′-CCTGTGATTTGTGGGCCTG-3′ |
| 5′-CAGTCTCCGTGTGAGGCAT-3′ | ||
| CRX | human | 5′-GCCCCACTATTCTGTCAACG-3′ |
| 5′-CTTCAGAGCCACCTCCTCAC-3′ | ||
| NRL | human | 5′-GGCTCCACACCTTACAGCTC-3′ |
| 5′-AGCCAGTACAGCTCCTCCAG-3′ | ||
| OPN1SW | human | 5′-ACCATTGGTATTGGCGTCTC-3′ |
| 5′-GGAGAGAGGCACAATGAAGC-3′ | ||
| OPN1LW | human | 5′-GTGGTCACTGCATCCGTCTT-3′ |
| 5′-ACGGTCTCTGCTAGGTCAGC-3′ | ||
| RHO | human | 5′-TCATCATGGTCATCGCTTTC-3′ |
| 5′-CATGAAGATGGGACCGAAGT-3′ | ||
| GUSB | human | 5′-TGTTTCGGTTGGTTGCCTCC-3′ |
| 5′-GGTCCAGGTTTGTCCTCTGC-3′ | ||
| USH2A exon 11-15 | human | 5′-AGTTGGTGCAGATCCTTCGG-3′ |
| 5′-CTTGCACTGGGAACACAAGC-3′ | ||
| USH2A Δexon 13 | human | 5′-TGGGACAGTGGATGGAGATA-3′ |
| 5′-TGGCATTGCCTGGAGAAATA-3′ | ||
| 5′/6-FAM/ATTCAGGCCAGTGCAAGTGCAAAG-BHQ-1-3′ | ||
| Ush2a exon 10-14 | mouse | 5′-TTACCGACCTGTTGGTGCTG-3′ |
| 5′-CCATTCGAGGCTCCTGCTAC-3′ | ||
| Ush2a Δexon 12 | mouse | 5′-GACCGTGGATGGAGACATTAC-3′ |
| 5′-GCATTGCTGGGAGAACTGTA-3′ | ||
| 5′-/56-FAM/CAGTGCTCG/ZEN/TGCAAAGCGAATGTT/3IABkFQ/-3′ |
Zebrafish immunohistochemistry and quantification of fluorescent signal intensity
Per group, 10 zebrafish larvae were imbedded in Tissue-Tek O.C.T. (Optimal Cutting Temperature) compound (Sakura; #4583) without prior fixation. Unfixed cryosections were permeabilized using 0.01% Tween 20 (Merck; #8.22184) in PBS, rinsed, and then pre-incubated with blocking solution (10% normal goat serum; Brunschwig; #G-S-1000) and 2% bovine serum albumin (Sigma-Aldrich; #A7906) in PBS. Primary antibodies (rabbit anti-C-terminal zebrafish usherin [1:1000; Novus Biologicals; #27640002] and mouse anti-centrin [1:500; Millipore; #04-1624]) were diluted in blocking solution and incubated overnight at 4°C. After rinsing the sections three times with PBS, they were incubated for 1 h at room temperature with blocking solution containing the secondary antibodies (Alexa Fluor 568 goat anti-rabbit [1:800; Molecular Probes; #A11011] and Alexa Fluor 488 goat anti-mouse [1:800; Molecular Probes; #A11029]) and the nuclei staining 4′,6-diamidino-2-phenylindole (DAPI; diluted 1:8,000; Molecular Probes; #D1306). After a dip in ultrapure water, the sections were coverslipped with ProLong Gold Antifade Reagent (Life Technologies; #P39930; lot. #1737358). The sections were examined using a Zeiss Axio Imager Z2 microscope with ZEN 2012 software and photographed with a Zeiss Axiocam 506 mono camera. Images of the middle section of each eye, taken at identical exposure settings, were used for quantification of the fluorescent signal intensity of anti-usherin immunoreactivity using the Fiji version (v.)1.47 software.53 First, the area of the connecting cilia was selected and manually isolated from the picture based on the centrin immunofluorescence signal. Subsequently, a mask was made based on the centrin staining using the “find maxima” option (noise = 50) and dilated five times. To find the exact location of usherin immunofluorescence, the centrin mask and layer containing the usherin immunofluorescent signal were combined. Find maxima (noise = 10) was used to identify the usherin immunofluorescence within the centrin mask. The resulting mask was dilated three times, and touching objects were separated using the watershed option. Subsequently, the maximum gray value of the identified regions was measured on the original image of usherin immunofluorescence (“analyse particles” option; size = 0–50, pixel circularity = 0.00–1.00).
ERG recordings in zebrafish larvae
Larvae, of 5−6 dpf, were placed on a filter paper in the middle of a plastic recording chamber. The chamber contained 1% agarose, in which the reference electrode was inserted. The isolated eye was positioned to face the light source. Under visual control via a standard microscope equipped with red illumination (Stemi 2000C; Zeiss, Oberkochen, Germany), a glass microelectrode with an opening of approximately 20 μm at the tip was placed against the center of the cornea. This electrode was filled with E3 medium (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl, and 0.33 mM MgSO4), the same in which the embryos were raised and held. A custom-made stimulator was invoked to provide light pulses of 100 ms duration, with a light intensity of 7,000 lux using a ZEISS XBO 75W light source and a fast shutter (Uni-Blitz Model D122; Vincent Associates, Rochester, NY, USA), driven by a delay unit interfaced to the main ERG recording setup. Electronic signals were amplified 1,000 times by a pre-amplifier (P55 A.C. pre-amplifier; Astro-Med, Grass Technology) with a band pass between 0.1 and 100 Hz, digitized by DAQ Board NI PCI-6035E (National Instruments) via NI BNC-2090 accessories and displayed via a self-developed NI LabVIEW program.
AON delivery in a retinoblastoma cell line
The WERI-Rb1 (ATCC HTB-169) retinoblastoma cell line was obtained from ATCC. WERI-Rb1 cells were cultured in RPMI-1640 medium (Gibco; #21875034) supplemented with 10% fetal bovine serum (Bio-West; #S1810-500). Cells were maintained by addition of fresh medium or replacement of medium every 3 to 4 days. Cells were transfected with QR-421a (Table 1) using Lipofectamine 2000 transfection reagent (Invitrogen; #11668019). A ratio of 2:1 (volume:weight) between Lipofectamine 2000 and the AON was used. Both Lipofectamine 2000 and AON were prepared in Opti-MEM. Lipofectamine 2000 mixture was added to the AON mixture and incubated for 20 min at room temperature before adding the transfection complexes to the cells. Cells were incubated for 24 h at 37°C. For gymnotic delivery, QR-421a was directly added to the medium without any transfection reagent. Cells were incubated with QR-421a for 48 h at 37°C before harvesting for analysis. Two samples were treated per condition. AON, which is not complementary to the USH2A sequence but with similar chemistry and length as that of QR-421a, was used as a control.
PBMC immune assay
Buffy coats, the fraction of an anti-coagulated blood sample that contains most of the white blood cells and platelets following centrifugation of the blood (500 mL blood in 70 mL citrate phosphate dextrose coagulant), from 5 healthy human (consensual) blood donors, were obtained from Sanquin Blood Supply in Leiden (the Netherlands). PBMCs were isolated from each buffy coat within 24 h after blood collection. PBMCs were stimulated for 24 h with QR-421a at a concentration of 0.1 μM, 1 μM, and 10 μM; positive control R-848 (1 μM); or PBS (vehicle control) at 37°C under a 5% CO2 atmosphere. For every donor, all conditions were tested in triplicate in 96-well round-bottom microtiter plates. The total number of viable PBMCs per well was 105. R-848 (Resiquimod; InvivoGen; tlrl-R848), a potent Toll-like receptor (TLR)7/8 agonist, was selected as a positive control for its strong and robust immune-activating properties, inducing the production of pro-inflammatory cytokines. Also, R-848 acts upon the TLRs that are most likely to be involved in recognition of single-strand RNA, arguably making it the most relevant positive control for this purpose. After incubation, cell-culture supernatant was isolated following centrifugation (300 relative centrifugal force [RCF], 5 min, room temperature) for cytokine analysis. Cytokine levels in PBMC culture supernatants were measured using the MILLIPLEX MAP Human Cytokine/Chemokine Magnetic Bead Panel-Custom 12 Plex-Immunology Multiplex Assay (Millipore; #HCYTOMAG-60K). Analytes included were the following: interferon (IFN)-α2, IFN-γ, interleukin (IL)-1β, IL-10, IL-12 p70, IL-6, IL-8, IP-10, MCP-1, MIP-1α, MIP-1β, and tumor necrosis factor (TNF)-α. Assay plates were read on the Luminex MAGPIX platform (Luminex, San Francisco, CA, USA). Analysis of the Luminex data was performed in Bio-Plex Manager 6.1 software (Bio-Rad). Standard curves were fitted using 5 parameter logistic regression. Cytokine concentrations that were below the limit of detection (LOD), rendered “out of range <” by the analysis software, were imputed with a concentration value of ½ ⋅ LOD for calculation purposes and statistical analysis. The LOD values, which were empirically determined by the manufacturer of the Luminex kit, were derived from the technical data sheet. Statistical analysis of the cytokine data was performed using GraphPad Prism 7 software. Geometric means of replicate cytokine concentration values were first log transformed.
Differentiation and AON treatment of iPSC-derived PPCs
Following informed consent, a skin biopsy was obtained from a homozygous USH2A c.2299delG patient, and a primary fibroblast cell line was generated as previously described.54 Fibroblasts were reprogrammed using 4 lentiviruses expressing Oct3/4, Sox2, Klf4, and c-Myc. iPSC lines were generated on feeder cells (mouse embryonic fibroblasts) and subsequently maintained in Essential 8 medium (Life Technologies; #A1517001). After reaching confluence, iPSC clumps were digested with Accutase (Sigma-Aldrich; #A6964) and plated in a 12-well plate to form a monolayer. Upon reaching confluence, Essential-Flex E8 medium (Thermo Fisher Scientific; #A2858501) was changed into a differentiation medium consisting of DMEM/F12 (Gibco; #11320-033), supplemented with non-essential amino acids (NEAAs; Gibco; #11140-050), B27 supplements (Thermo Fisher Scientific; #1287010), N2 supplements (Thermo Fisher Scientific; #17502048), 100 ng/μL insulin-like growth factor-1 (IGF-1; Sigma-Aldrich; #I3769), 10 ng/μL recombinant fibroblast growth factor basic (βFGF; Sigma-Aldrich; #F0291), 10 μg/μL heparin (Sigma-Aldrich; #H3149-10KU), and 200 μg/mL human recombinant COCO (Bio-Techne; #3047-CC). The medium was changed every day for 90 days, after which, the cells were treated with different concentrations of AONs for 1 month. At the end of the 4th month, the cells were collected and characterized. PPCs were treated continuously with 1, 2, 5, or 10 μM QR-421a or 10 μM control oligo, after 90 days of differentiation, for 28 days. Every 2 days, 50% of culture medium was refreshed with fresh culture medium containing AON. qRT-PCR was used to evaluate the differentiation status at the end of the experiment. Total RNA was isolated from PPCs as described for the human USH2A transcript analysis. 1 μg of RNA was reverse transcribed using Superscript VILO (Life Technologies; #11756050). Expression of NANOG, CRX, NRL, OPN1SW, OPN1LW, and RHO was investigated using 10× diluted cDNA using GoTaq DNA polymerase (Promega; #M3001) and a CFX96 Touch Real-Time PCR Detection System (Bio-Rad). Data are normalized for the expression of the housekeeping gene GUSB. Primer sequences are provided in Table 2.
Human USH2A transcript analysis
Total RNA was isolated from the cells using the RNeasy Plus Mini Kit (QIAGEN; #74136), according to the manufacturer’s protocol, and cDNA was synthesized. To visualize AON-induced alternative splicing in WERI-Rb1 cells and human iPSC-derived PPCs, a PCR was performed using primers on USH2A exons 11 and 15. PCR fragments were gel extracted, purified using the NucleoSpin Gel and PCR Clean-up Kit (Macherey-Nagel; #740609.50), and subjected to Sanger sequencing. For the quantification of USH2A transcripts, RT-ddPCR was performed using the One-Step RT-ddPCR Advanced Kit for probes (Bio-Rad; #1864022). The final 20-μL reaction mix contained the following: 5 μL Supermix, 2 μL RT, 1 μL 300 mM DTT, 1× TaqMan gene-expression assays, or 450 nM USH2A forward and reverse primer each and 250 nM USH2A probe. Total USH2A (Hs01071797_m1; Applied Biosystems) and USH2A exon 13 (Hs01071797_m1; Applied Biosystems) levels were quantified using 50 or 100 ng RNA, respectively, in a multiplex manner using commercial TaqMan gene-expression assays. USH2A Δexon 13 levels were quantified in 50 or 100 ng RNA using an in-house-designed assay (Table 2). PCR reactions were dispersed into droplets using the QX200 droplet generator (Bio-Rad) according to the manufacturer’s instructions and transferred to a 96-well PCR plate. End-point PCR was performed in a T100 Thermocycler (Bio-Rad). The fluorescence of each droplet was quantified in the QX200 droplet reader (Bio-Rad). Each sample was analyzed in duplicate. Absolute quantification was performed in QuantaSoft software (Bio-Rad). Thresholds were manually set to distinguish between positive and negative droplets.
During the data analysis, average copy numbers of duplicate measurements per nanogram RNA input were calculated and normalized for total USH2A by dividing the target gene copy numbers by the total USH2A copy numbers. Percentage of USH2A Δexon 13 transcripts was expressed relative to the amount of exon 13-containing transcript of untreated samples. In case untreated samples were not available, control-treated samples were used. This method was used as multiple USH2A isoforms are present in the cells, and the splicing modulation may not only lead to exon 13 skipping (e.g., combined exon 12 and 13 skipping, partial exon 13 skipping).
Mouse IVT injection, necropsy, and collection of retinae
Animals were anesthetized using an intraperitoneal injection (10 μL/g body weight) of ketamine (10 mg/mL) and xylazine (0.5 mg/mL). Both eyes were then dilated with a topical mydriatic eye-drop mixture (1 drop each of tropicamide [1%] and phenylephrine hydrochloride [2.5%]) to visualize the intraocular injection needle during the IVT injection. The head of the mouse was secured in a custom-built nose bar and rotated horizontally to present the eye to the surgeon. Eyelids were retracted, and the eyes were held steady with forceps while IVT injection was performed under direct visual control through a Leica M80 stereomicroscope. Injections were made at an angle of approximately 45° below the lens (to prevent damage to the lens and retinal tissue) using a 2.5-μL Hamilton syringe (VWR; HAMI7632-01) with a Hamilton 32G, length 15 mm, point style 4/tap needle (VWR; HAMI7803-04) attached. The injection equipment was rinsed with ethanol and PBS between animals. For each injection, once the needle bevel was observed in the vitreous of the eye (through the dilated pupil), 1 μL of dosing substance was injected, immediately followed by an anterior counter-punch in the cornea before extracting the syringe. This counterpunch was performed in order to reduce intraocular pressure and prevent reflux of the injected substance. The injection needle was retracted, and vascular integrity was examined by looking for signs of intraocular bleeding. Both eyes were covered with eye ointment (Duodrops; Medpets) to protect the cornea during recovery. Mice were sacrificed at the designated times using CO2 asphyxiation, and the scleral tissue posterior to the limbus was grasped with 0.22 forceps to stabilize the globe (eye ball), using an 8.5-cm spring scissor (AgnTho’s; #05-230-085). A cross incision was made in the cornea, and a curved dressing forceps (AgnTho’s; #08-513-005) was inserted behind the lens toward the posterior aspect of the globe, partially closed and used to pull forward and remove the lens through the corneal cross incision that was made (to prevent damage of the eye wall). With the use of curved forceps, the cornea was peeled backward until the retina became visible as a yellow vascularized tissue. With the use of the forceps, the retina was carefully removed, and any strips of pigmented tissue were peeled away with forceps. The retina tissue was placed in a 2.0-mL tube, snap frozen in liquid nitrogen (for RNA analysis), or Hartmann’s fixative was added (for histology). AONs used for mouse studies are described in Table 1.
Mouse Ush2a transcript analysis
Mouse retinae were homogenized in the MagNA Lyser (Roche) in 350 μL RLT Plus buffer (QIAGEN;#74136) containing 1% beta mercaptoethanol. For homogenization, 1/5th of the beads (Roche;0335894100) per sample was used along with the following program: 2 times 5,000 rpm for 30 s each, 1 min cooling on ice in between. After homogenization, the samples were centrifuged at full speed for 3 min. Supernatant was used for RNA isolation with the RNeasy Plus Micro Kit (QIAGEN; #74034) according to the manufacturer’s protocol. RNA was eluted in 14 μL RNase-free water, and the concentrations were measured on the Nanodrop 2000. Samples were stored at −80°C until further use. For endpoint PCR analysis, RNA was converted into cDNA and amplified using the SuperScript IV One-Step RT-PCR System (Invitrogen; #12594100). A 50-μL reaction contained the following: 5 or 25 ng RNA template, 25 μL 2× Platinum SuperFi RT-PCR Master Mix, 0.5 μL SuperScript IV RT Mix, and 500 nM of each forward and reverse primer (Table 2). The reaction was run in a Thermocycler programmed for 10 min at 50°C for RT and 2 min at 98°C for RT inactivation and as an initial denaturation step, followed by 40 cycles of 10 s at 98°C, 10 s at 62°C, and 45 s at 72°C, and final extension at 72°C for 5 min. PCR products were analyzed on the Bioanalyzer (Agilent) using the Agilent DNA 7500 kit. For the quantification of Ush2a transcripts, One-Step RT-ddPCR analysis was used. Levels of Ush2a total (measured at exon 8-9) and Ush2a with exon 12 were quantified using 5 ng RNA input using commercial TaqMan gene-expression assays (Mm01316803_m1 and Mm00498761_m1; Applied Biosystems). Ush2a Δexon 12 levels were quantified using 25 ng RNA using a custom-designed assay (Table 2). RT-ddPCR was performed using the One-Step RT-ddPCR Advanced Kit for Probes (Bio-Rad; #1864022). The final 20-μL reaction mix contained the following: 5 μL Supermix, 2 μL RT, 1 μL 300 mM DTT, 1× TaqMan gene-expression assays, or 900 nM forward and reverse primers each and 250 nM probe. PCR reactions were dispersed into droplets using the QX200 droplet generator (Bio-Rad) according to the manufacturer’s instructions and transferred to a 96-well PCR plate (SOP-EQP-028). End-point PCR was performed in a T100 Thermocycler (Bio-Rad). The fluorescence of each droplet was quantified in the QX200 droplet reader (Bio-Rad). Each sample was analyzed in duplicate. Absolute quantification was performed in QuantaSoft software (Bio-Rad). Thresholds were manually set to distinguish between positive and negative droplets. Copy numbers were first normalized to correct for a different amount of RNA input. Percentage of exon 12 skipping was expressed relative to the total number of Ush2a (measured at exon 8-9) copies.
Visualization of mQR-421a localization in the mouse retina
A fluorescent in situ hybridization (FISH) assay was used for the detection of mQR-421a in murine ocular tissues to investigate localization and distribution. Eyes were collected and fixed in Hartmann’s fixative overnight at room temperature. The next day, lenses were removed, and the remaining eyeball was immediately processed to paraffin. Sections were cut and mounted on glass slides. Slides were hybridized with a fully complementary (to mQR-421a) Cy5-labeled, partly locked nucleic acid (LNA)-modified-probe (Cy5 5′-GATTTAAATTCCTCCAGAGTT-3′). DAPI containing medium was used to coverslip the slides and stain the nuclei. Microscopic images were taken using a confocal laser-scanning microscope (LSM, LSM800; Zeiss) using ZEN Blue software (Zeiss).
Acknowledgments
We are grateful to the patients for donating tissue for this study. We would like to acknowledge the Radboud University Zebrafish Facility and in particular, Tom Spanings and Antoon van der Horst for their excellent zebrafish husbandry. We would like to acknowledge Monica Tartjiono and Frits van der Ham for their assistance with histological analysis of the murine retina. We would also like to acknowledge Thomas Hoogenboezem for his assistance with PBMC assays. This study was financially supported by ProQR Therapeutics, the Foundation Fighting Blindness USA (grant PPA-0517-0717-RAD to E.v.W.), the Dutch Organisation for Scientific Research (Veni grant 016.136.091 to E.v.W.), the Gelderse Blinden Stichting, Stichting Ushersyndroom, and Stichting Klavertje2.
Author contributions
Conceptualization, K.D., H.C.v.D., E.d.V., J.J.T., G.P., P.A., and E.v.W.; writing, K.D., H.C.v.D., R.S., E.d.V., and E.v.W.; investigation and analyses: zebrafish experiments, R.S., M.D., J.Z., S.B., T.P., and E.d.V.; 3D modeling, H.V.; cell culture, PPC, and mouse experiments, S.A., R.P., H.L.C., I.A.S., L.B., I.S., and J.M.; supervision, K.D., S.C.F.N., H.K., W.B., J.J.T., L.V., C.d.B., G.P., P.A., and E.v.W.; funding acquisition, E.v.W.; review, editing, and approval of manuscript, all authors.
Declaration of interests
International patent applications have been filed by Stichting Katholieke Universiteit Nijmegen (WO2016/005514) and ProQR Therapeutics (WO2018/055134) describing methods and means regarding oligonucleotide therapy for USH2A-associated retinitis pigmentosa. Stichting Katholieke Universiteit Nijmegen has licensed the exclusive rights of the patent to ProQR Therapeutics. As the inventor, E.v.W. is entitled to a share of any future royalties paid to Stichting Katholieke Universiteit Nijmegen, should the therapy eventually be brought to the market. K.D., H.C.v.D., H.L.C., I.A.S., L.V., C.d.B., L.B., I.S., J.J.T., J.M., G.P., and P.A. were employed by ProQR Therapeutics during this project.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ymthe.2021.04.024.
Supplemental information
References
- 1.Hartong D.T., Berson E.L., Dryja T.P. Retinitis pigmentosa. Lancet. 2006;368:1795–1809. doi: 10.1016/S0140-6736(06)69740-7. [DOI] [PubMed] [Google Scholar]
- 2.McGee T.L., Seyedahmadi B.J., Sweeney M.O., Dryja T.P., Berson E.L. Novel mutations in the long isoform of the USH2A gene in patients with Usher syndrome type II or non-syndromic retinitis pigmentosa. J. Med. Genet. 2010;47:499–506. doi: 10.1136/jmg.2009.075143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rivolta C., Sweklo E.A., Berson E.L., Dryja T.P. Missense mutation in the USH2A gene: association with recessive retinitis pigmentosa without hearing loss. Am. J. Hum. Genet. 2000;66:1975–1978. doi: 10.1086/302926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pennings R.J.E., Te Brinke H., Weston M.D., Claassen A., Orten D.J., Weekamp H., Van Aarem A., Huygen P.L.M., Deutman A.F., Hoefsloot L.H. USH2A mutation analysis in 70 Dutch families with Usher syndrome type II. Hum. Mutat. 2004;24:185. doi: 10.1002/humu.9259. [DOI] [PubMed] [Google Scholar]
- 5.Aller E., Larrieu L., Jaijo T., Baux D., Espinós C., González-Candelas F., Nájera C., Palau F., Claustres M., Roux A.-F., Millán J.M. The USH2A c.2299delG mutation: dating its common origin in a Southern European population. Eur. J. Hum. Genet. 2010;18:788–793. doi: 10.1038/ejhg.2010.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vaché C., Besnard T., le Berre P., García-García G., Baux D., Larrieu L., Abadie C., Blanchet C., Bolz H.J., Millán J. Usher syndrome type 2 caused by activation of an USH2A pseudoexon: implications for diagnosis and therapy. Hum. Mutat. 2012;33:104–108. doi: 10.1002/humu.21634. [DOI] [PubMed] [Google Scholar]
- 7.Slijkerman R.W.N., Vaché C., Dona M., García-García G., Claustres M., Hetterschijt L., Peters T.A., Hartel B.P., Pennings R.J.E., Millán J.M. Antisense Oligonucleotide-based Splice Correction for USH2A-associated Retinal Degeneration Caused by a Frequent Deep-intronic Mutation. Mol. Ther. Nucleic Acids. 2016;5:e381. doi: 10.1038/mtna.2016.89. [DOI] [PubMed] [Google Scholar]
- 8.Baux D., Blanchet C., Hamel C., Meunier I., Larrieu L., Faugère V., Vaché C., Castorina P., Puech B., Bonneau D. Enrichment of LOVD-USHbases with 152 USH2A genotypes defines an extensive mutational spectrum and highlights missense hotspots. Hum. Mutat. 2014;35:1179–1186. doi: 10.1002/humu.22608. [DOI] [PubMed] [Google Scholar]
- 9.Hartel B.P., Löfgren M., Huygen P.L.M., Guchelaar I., Lo-A-Njoe Kort N., Sadeghi A.M., van Wijk E., Tranebjærg L., Kremer H., Kimberling W.J. A combination of two truncating mutations in USH2A causes more severe and progressive hearing impairment in Usher syndrome type IIa. Hear. Res. 2016;339:60–68. doi: 10.1016/j.heares.2016.06.008. [DOI] [PubMed] [Google Scholar]
- 10.Hartel B.P., van Nierop J.W.I., Huinck W.J., Rotteveel L.J.C., Mylanus E.A.M., Snik A.F., Kunst H.P.M., Pennings R.J.E. Cochlear Implantation in Patients With Usher Syndrome Type IIa Increases Performance and Quality of Life. Otol. Neurotol. 2017;38:e120–e127. doi: 10.1097/MAO.0000000000001441. [DOI] [PubMed] [Google Scholar]
- 11.Bennett J., Wellman J., Marshall K.A., McCague S., Ashtari M., DiStefano-Pappas J., Elci O.U., Chung D.C., Sun J., Wright J.F. Safety and durability of effect of contralateral-eye administration of AAV2 gene therapy in patients with childhood-onset blindness caused by RPE65 mutations: a follow-on phase 1 trial. Lancet. 2016;388:661–672. doi: 10.1016/S0140-6736(16)30371-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Russell S., Bennett J., Wellman J.A., Chung D.C., Yu Z.-F., Tillman A., Wittes J., Pappas J., Elci O., McCague S. Efficacy and safety of voretigene neparvovec (AAV2-hRPE65v2) in patients with RPE65-mediated inherited retinal dystrophy: a randomised, controlled, open-label, phase 3 trial. Lancet. 2017;390:849–860. doi: 10.1016/S0140-6736(17)31868-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cideciyan A.V., Jacobson S.G., Drack A.V., Ho A.C., Charng J., Garafalo A.V., Roman A.J., Sumaroka A., Han I.C., Hochstedler M.D. Effect of an intravitreal antisense oligonucleotide on vision in Leber congenital amaurosis due to a photoreceptor cilium defect. Nat. Med. 2019;25:225–228. doi: 10.1038/s41591-018-0295-0. [DOI] [PubMed] [Google Scholar]
- 14.Dona M., Slijkerman R., Lerner K., Broekman S., Wegner J., Howat T., Peters T., Hetterschijt L., Boon N., de Vrieze E. Usherin defects lead to early-onset retinal dysfunction in zebrafish. Exp. Eye Res. 2018;173:148–159. doi: 10.1016/j.exer.2018.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duering M., Karpinska A., Rosner S., Hopfner F., Zechmeister M., Peters N., Kremmer E., Haffner C., Giese A., Dichgans M., Opherk C. Co-aggregate formation of CADASIL-mutant NOTCH3: a single-particle analysis. Hum. Mol. Genet. 2011;20:3256–3265. doi: 10.1093/hmg/ddr237. [DOI] [PubMed] [Google Scholar]
- 16.Rutten J.W., Dauwerse H.G., Peters D.J.M., Goldfarb A., Venselaar H., Haffner C., van Ommen G.-J.B., Aartsma-Rus A.M., Lesnik Oberstein S.A. Therapeutic NOTCH3 cysteine correction in CADASIL using exon skipping: in vitro proof of concept. Brain. 2016;139:1123–1135. doi: 10.1093/brain/aww011. [DOI] [PubMed] [Google Scholar]
- 17.Wouters M.A., Rigoutsos I., Chu C.K., Feng L.L., Sparrow D.B., Dunwoodie S.L. Evolution of distinct EGF domains with specific functions. Protein Sci. 2005;14:1091–1103. doi: 10.1110/ps.041207005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Appella E., Weber I.T., Blasi F. Structure and function of epidermal growth factor-like regions in proteins. FEBS Lett. 1988;231:1–4. doi: 10.1016/0014-5793(88)80690-2. [DOI] [PubMed] [Google Scholar]
- 19.Aartsma-Rus A. Overview on AON design. Methods Mol. Biol. 2012;867:117–129. doi: 10.1007/978-1-61779-767-5_8. [DOI] [PubMed] [Google Scholar]
- 20.Slijkerman R., Kremer H., van Wijk E. Antisense Oligonucleotide Design and Evaluation of Splice-Modulating Properties Using Cell-Based Assays. Methods Mol. Biol. 2018;1828:519–530. doi: 10.1007/978-1-4939-8651-4_34. [DOI] [PubMed] [Google Scholar]
- 21.Piva F., Giulietti M., Nocchi L., Principato G. SpliceAid: a database of experimental RNA target motifs bound by splicing proteins in humans. Bioinformatics. 2009;25:1211–1213. doi: 10.1093/bioinformatics/btp124. [DOI] [PubMed] [Google Scholar]
- 22.McFall R.C., Sery T.W., Makadon M. Characterization of a new continuous cell line derived from a human retinoblastoma. Cancer Res. 1977;37:1003–1010. [PubMed] [Google Scholar]
- 23.Garanto A., Duijkers L., Tomkiewicz T.Z., Collin R.W.J. Antisense Oligonucleotide Screening to Optimize the Rescue of the Splicing Defect Caused by the Recurrent Deep-Intronic ABCA4 Variant c.4539+2001G>A in Stargardt Disease. Genes (Basel) 2019;10:452. doi: 10.3390/genes10060452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dulla K., Aguilà M., Lane A., Jovanovic K., Parfitt D.A., Schulkens I., Chan H.L., Schmidt I., Beumer W., Vorthoren L. Splice-Modulating Oligonucleotide QR-110 Restores CEP290 mRNA and Function in Human c.2991+1655A>G LCA10 Models. Mol. Ther. Nucleic Acids. 2018;12:730–740. doi: 10.1016/j.omtn.2018.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McDougald D.S., Kmiec E., Mills J.A. Personalized models reveal mechanistic and therapeutic insights into CEP290-associated Leber congenital amaurosis. Stem Cell Investig. 2016;3:65. doi: 10.21037/sci.2016.10.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lenassi E., Saihan Z., Bitner-Glindzicz M., Webster A.R. The effect of the common c.2299delG mutation in USH2A on RNA splicing. Exp. Eye Res. 2014;122:9–12. doi: 10.1016/j.exer.2014.02.018. [DOI] [PubMed] [Google Scholar]
- 27.Michalski N., Michel V., Bahloul A., Lefèvre G., Barral J., Yagi H., Chardenoux S., Weil D., Martin P., Hardelin J.-P. Molecular characterization of the ankle-link complex in cochlear hair cells and its role in the hair bundle functioning. J. Neurosci. 2007;27:6478–6488. doi: 10.1523/JNEUROSCI.0342-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maerker T., van Wijk E., Overlack N., Kersten F.F.J., McGee J., Goldmann T., Sehn E., Roepman R., Walsh E.J., Kremer H., Wolfrum U. A novel Usher protein network at the periciliary reloading point between molecular transport machineries in vertebrate photoreceptor cells. Hum. Mol. Genet. 2008;17:71–86. doi: 10.1093/hmg/ddm285. [DOI] [PubMed] [Google Scholar]
- 29.Sorusch N., Bauß K., Plutniok J., Samanta A., Knapp B., Nagel-Wolfrum K., Wolfrum U. Characterization of the ternary Usher syndrome SANS/ush2a/whirlin protein complex. Hum. Mol. Genet. 2017;26:1157–1172. doi: 10.1093/hmg/ddx027. [DOI] [PubMed] [Google Scholar]
- 30.Liu X., Bulgakov O.V., Darrow K.N., Pawlyk B., Adamian M., Liberman M.C., Li T. Usherin is required for maintenance of retinal photoreceptors and normal development of cochlear hair cells. Proc. Natl. Acad. Sci. USA. 2007;104:4413–4418. doi: 10.1073/pnas.0610950104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Niks E.H., Aartsma-Rus A. Exon skipping: a first in class strategy for Duchenne muscular dystrophy. Expert Opin. Biol. Ther. 2017;17:225–236. doi: 10.1080/14712598.2017.1271872. [DOI] [PubMed] [Google Scholar]
- 32.Mendell J.R., Rodino-Klapac L.R., Sahenk Z., Roush K., Bird L., Lowes L.P., Alfano L., Gomez A.M., Lewis S., Kota J., Eteplirsen Study Group Eteplirsen for the treatment of Duchenne muscular dystrophy. Ann. Neurol. 2013;74:637–647. doi: 10.1002/ana.23982. [DOI] [PubMed] [Google Scholar]
- 33.Mendell J.R., Goemans N., Lowes L.P., Alfano L.N., Berry K., Shao J., Kaye E.M., Mercuri E., Eteplirsen Study Group and Telethon Foundation DMD Italian Network Longitudinal effect of eteplirsen versus historical control on ambulation in Duchenne muscular dystrophy. Ann. Neurol. 2016;79:257–271. doi: 10.1002/ana.24555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beck K., Hunter I., Engel J. Structure and function of laminin: anatomy of a multidomain glycoprotein. FASEB J. 1990;4:148–160. doi: 10.1096/fasebj.4.2.2404817. [DOI] [PubMed] [Google Scholar]
- 35.Yurchenco P.D., Cheng Y.S. Self-assembly and calcium-binding sites in laminin. A three-arm interaction model. J. Biol. Chem. 1993;268:17286–17299. [PubMed] [Google Scholar]
- 36.Eisen J.S., Smith J.C. Controlling morpholino experiments: don’t stop making antisense. Development. 2008;135:1735–1743. doi: 10.1242/dev.001115. [DOI] [PubMed] [Google Scholar]
- 37.Bill B.R., Petzold A.M., Clark K.J., Schimmenti L.A., Ekker S.C. A primer for morpholino use in zebrafish. Zebrafish. 2009;6:69–77. doi: 10.1089/zeb.2008.0555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Biehlmaier O., Neuhauss S.C.F., Kohler K. Synaptic plasticity and functionality at the cone terminal of the developing zebrafish retina. J. Neurobiol. 2003;56:222–236. doi: 10.1002/neu.10243. [DOI] [PubMed] [Google Scholar]
- 39.Branchek T. The development of photoreceptors in the zebrafish, brachydanio rerio. II. Function. J. Comp. Neurol. 1984;224:116–122. doi: 10.1002/cne.902240110. [DOI] [PubMed] [Google Scholar]
- 40.Bilotta J., Saszik S., Sutherland S.E. Rod contributions to the electroretinogram of the dark-adapted developing zebrafish. Dev. Dyn. 2001;222:564–570. doi: 10.1002/dvdy.1188. [DOI] [PubMed] [Google Scholar]
- 41.Pierrache L.H.M., Hartel B.P., van Wijk E., Meester-Smoor M.A., Cremers F.P.M., de Baere E., de Zaeytijd J., van Schooneveld M.J., Cremers C.W.R.J., Dagnelie G. Visual Prognosis in USH2A-Associated Retinitis Pigmentosa Is Worse for Patients with Usher Syndrome Type IIa Than for Those with Nonsyndromic Retinitis Pigmentosa. Ophthalmology. 2016;123:1151–1160. doi: 10.1016/j.ophtha.2016.01.021. [DOI] [PubMed] [Google Scholar]
- 42.Sengillo J.D., Cabral T., Schuerch K., Duong J., Lee W., Boudreault K., Xu Y., Justus S., Sparrow J.R., Mahajan V.B., Tsang S.H. Electroretinography Reveals Difference in Cone Function between Syndromic and Nonsyndromic USH2A Patients. Sci. Rep. 2017;7:11170. doi: 10.1038/s41598-017-11679-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pendse N.D., Lamas V., Pawlyk B.S., Maeder M.L., Chen Z.-Y., Pierce E.A., Liu Q. In Vivo Assessment of Potential Therapeutic Approaches for USH2A-Associated Diseases. Adv. Exp. Med. Biol. 2019;1185:91–96. doi: 10.1007/978-3-030-27378-1_15. [DOI] [PubMed] [Google Scholar]
- 44.Farkas M.H., Grant G.R., White J.A., Sousa M.E., Consugar M.B., Pierce E.A. Transcriptome analyses of the human retina identify unprecedented transcript diversity and 3.5 Mb of novel transcribed sequence via significant alternative splicing and novel genes. BMC Genomics. 2013;14:486. doi: 10.1186/1471-2164-14-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Del Amo E.M., Rimpelä A.-K., Heikkinen E., Kari O.K., Ramsay E., Lajunen T., Schmitt M., Pelkonen L., Bhattacharya M., Richardson D. Pharmacokinetic aspects of retinal drug delivery. Prog. Retin. Eye Res. 2017;57:134–185. doi: 10.1016/j.preteyeres.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 46.Lentz J.J., Jodelka F.M., Hinrich A.J., McCaffrey K.E., Farris H.E., Spalitta M.J., Bazan N.G., Duelli D.M., Rigo F., Hastings M.L. Rescue of hearing and vestibular function by antisense oligonucleotides in a mouse model of human deafness. Nat. Med. 2013;19:345–350. doi: 10.1038/nm.3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rigo F., Seth P.P., Bennett C.F. Antisense oligonucleotide-based therapies for diseases caused by pre-mRNA processing defects. Adv. Exp. Med. Biol. 2014;825:303–352. doi: 10.1007/978-1-4939-1221-6_9. [DOI] [PubMed] [Google Scholar]
- 48.Charleston J.S., Schnell F.J., Dworzak J., Donoghue C., Lewis S., Chen L., Young G.D., Milici A.J., Voss J., DeAlwis U. Eteplirsen treatment for Duchenne muscular dystrophy: Exon skipping and dystrophin production. Neurology. 2018;90:e2146–e2154. doi: 10.1212/WNL.0000000000005680. [DOI] [PubMed] [Google Scholar]
- 49.Westerfield M. Fourth Edition. University of Oregon Press; 2000. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio)http://zfin.org/zf_info/zfbook/zfbk.html [Google Scholar]
- 50.Krieger E., Koraimann G., Vriend G. Increasing the precision of comparative models with YASARA NOVA--a self-parameterizing force field. Proteins. 2002;47:393–402. doi: 10.1002/prot.10104. [DOI] [PubMed] [Google Scholar]
- 51.Vriend G. WHAT IF: a molecular modeling and drug design program. J. Mol. Graph. 1990;8:52–56. doi: 10.1016/0263-7855(90)80070-v. [DOI] [PubMed] [Google Scholar]
- 52.Smith P.J., Zhang C., Wang J., Chew S.L., Zhang M.Q., Krainer A.R. An increased specificity score matrix for the prediction of SF2/ASF-specific exonic splicing enhancers. Hum. Mol. Genet. 2006;15:2490–2508. doi: 10.1093/hmg/ddl171. [DOI] [PubMed] [Google Scholar]
- 53.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sangermano R., Bax N.M., Bauwens M., van den Born L.I., De Baere E., Garanto A., Collin R.W.J., Goercharn-Ramlal A.S.A., den Engelsman-van Dijk A.H., Rohrschneider K. Photoreceptor Progenitor mRNA Analysis Reveals Exon Skipping Resulting from the ABCA4 c.5461-10T→C Mutation in Stargardt Disease. Ophthalmology. 2016;123:1375–1385. doi: 10.1016/j.ophtha.2016.01.053. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.