Abstract
The development of an aptamer-based therapeutic has rapidly progressed following the first two reports in the 1990s, underscoring the advantages of aptamer drugs associated with their unique binding properties. In 2004, the US Food and Drug Administration (FDA) approved the first therapeutic aptamer for the treatment of neovascular age-related macular degeneration, Macugen developed by NeXstar. Since then, eleven aptamers have successfully entered clinical trials for various therapeutic indications. Despite some of the pre-clinical and clinical successes of aptamers as therapeutics, no aptamer has been approved by the FDA for the treatment of cancer. This review highlights the most recent and cutting-edge approaches in the development of aptamers for the treatment of cancer types most refractory to conventional therapies. Herein, we will review (1) the development of aptamers to enhance anti-cancer immunity and as delivery tools for inducing the expression of immunogenic neoantigens; (2) the development of the most promising therapeutic aptamers designed to target the hard-to-treat cancers such as brain tumors; and (3) the development of “carrier” aptamers able to target and penetrate tumors and metastasis, delivering RNA therapeutics to the cytosol and nucleus.
Keywords: aptamer, cancer, RNA therapeutics, immunity, delivery, brain tumors, cancer stem cells
Graphical abstract
The review highlights the latest advances in the development of aptamer-based therapeutics and underscores their advantages for treatment of cancer. Owing to their unique binding properties, protein-binding aptamers are emerging as safe and effective macromolecules to manage even the hard-to-treat cancers by overcoming the limitations of several conventional therapeutics.
Enhancing anti-cancer immunity
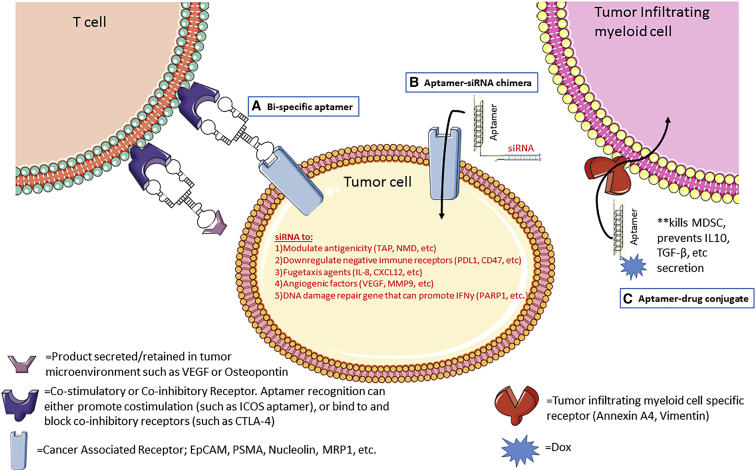
Despite the proven antitumor efficacy of immune-checkpoint blockade by the combined use of anti-cytotoxic T lymphocyte-associated antigen-4 (CTLA-4) and anti-programmed death-1 (PD-1) antibodies for the treatment of advanced melanoma, mesothelioma, and non-small cell lung cancer, only a small fraction of these tumors responds to treatment or progress after transient disease control. To improve immunotherapeutic effects, agonist and antagonist aptamers have been described. They have the unique advantage to be easily conjugated as bispecific and multispecific molecules targeting the tumor site and able to locally stimulate the immune response.1,2 One such proof-of-concept study demonstrated the ability to target immunomodulatory aptamers specific to targets secreted into the tumor microenvironment. Products like vascular endothelial growth factor (VEGF) and osteopontin are highly upregulated and retained in the tumor microenvironment, making them an attractive target to enhance anti-tumor immunity by allowing preferential enrichment of immunomodulatory drugs, like costimulatory 4-1BB aptamers.3 Recently, Soldevilla et al.4 demonstrated the efficacy of the combined treatment of an anti-CTLA-4 antibody with a tumor-targeted anti-inducible T cell costimulator (ICOS) aptamer in vitro and in vivo. With the multidrug resistance protein 1 (MRP1) being specifically in cancer stem cells (CSCs), the authors took advantage of a previously described MRP1-targeting aptamer to generate a MRP1-ICOS bi-specific aptamer (Figure 1A).5 The resulting aptamer holds two binding sites: one specific for the MRP1-expressing cancer cells and another one for ICOS, which can stimulate T lymphocytes in the proximity of the tumor. These findings underscore the potentiality of using aptamers in combined therapies involving anti-CTLA-4 blockade and an ICOS agonist.
Figure 1.
Schematic depicting approaches utilizing aptamers for immune-oncology applications
(A) A bi-specific aptamer can have dual specificity in which one moiety binds to and activates receptors on tumor-infiltrating T cells (e.g., ICOS or 4-1BB, depicted here as a dimer), whereas the second moiety acts as a tumor-targeting agent (e.g., MRP1) with the intention of limiting co-stimulation to tumor-infiltrating T cells. (B) A tumor cell marker-specific aptamer linked to an siRNA can downregulate the target of interest in a cell-specific manner. The described siRNA can modulate immunogenicity and enhance an anti-tumor immune response. (C) An aptamer specific to tumor-associated myeloid cells is conjugated to a drug (e.g., Dox), whereby the aptamer delivers the drug in a cell-specific manner, reducing toxicity and killing only cells of interest.
In addition, strong evidence suggesting that tumor neoantigens are recognized by the immune system and that their low expression would be a possible cause for the low responsiveness of tumors to immune-checkpoint blockade therapies is emerging.6 Having previously reported the induction of neoantigens by inhibiting the nonsense-mediated messenger RNA (mRNA) decay (NMD) pathway in tumor cells,7 Garrido et al.8 used an elegant and broadly applicable approach to selectively induce neoantigens in tumor cells as a promising approach to potentiate the responsiveness of patients to immunotherapies. As a targeting moiety for a small interfering (si)RNA specific for the transporter associated with antigen processing (TAP) expression, the authors took advantage of the previously characterized AS1411 aptamer.9 By folding into G-quadruplex, AS1411, originally discovered serendipitously by developing cancer-selective therapies, was shown to bind nucleolin. Nucleolin is an RNA-binding protein that is mainly localized in the nucleolus in normal cells, whereas it is found abundantly on the cell surface of most tumor cells where it can shuttle into the cytoplasm.9, 10, 11 Indeed, TAP acts as a heterodimer with an essential role in the major histocompatibility complex (MHC) class I peptide presentation. Its genetic ablation inhibits the MHC class I canonical processing. However, TAP inhibition leads to the upregulation of alternative pathways leading to the presentation of neoantigens, named T cell epitopes associated with impaired peptide processing (TEIPP) that can elicit effective CD8+ T cell responses.12, 13, 14, 15 The authors show that the AS1411-siTAP chimera (AsiC), named Nucl-TAP, exhibited high selectivity for tumor cells, leading to the targeted downregulation of TAP and increased antigenicity of tumor cells (Figure 1B). TAP downregulation promoted an antitumor immune response in mice, inhibiting tumor growth in the absence of measurable toxicity. Based on the antitumor response elicited in cancer cells upon TAP downregulation, Garrido and coworkers16 further investigated a novel anti-tumor strategy of vaccination against TEIPP induced by TAP downregulation. To this end, the authors took advantage of a CpG oligonucleotide as the targeting moiety for a TAP-specific siRNA. Short CpG oligodeoxyribonucleotides (ODNs) have been previously shown to preferentially target and activate in vivo the Toll-like receptor 9 (TLR9)-expressing dendritic cells (DCs). Upon binding to TLR9, the CpG-TAP siRNA is taken up by the receptor-expressing antigen-presenting cells, leading to TAP downregulation and the induction of TEIPP neo-antigens. The authors concluded that vaccination against TEIPP antigens is effective at enhancing the antitumor response elicited against tumor cells treated with the Nucl-TAP aptamer conjugate to induce TEIPP.16
Given the limited success of current immunotherapies against solid tumors, combinatorial and synergistic therapies might be required to fight the most difficult-to-treat tumors. A recent study by Zhang et al.17 set out to combine different aptamer-siRNA chimeras targeting different tumor cell regulatory pathways to enhance immunotherapy. In this study, the authors used the previously described murine and human targeting epithelial cell adhesion molecule (EpCAM) aptamer, a target frequently upregulated on triple-negative breast cancer (TNBC). They conjugated an siRNA against the following: (1) Upf2, a target regulating the aforementioned NMD pathway to induce neoantigens, (2) CD47, a cell-surface marker frequently upregulated on tumor cells that acts as a “don’t eat me signal” to the immune system, (3) myeloid cell leukemia 1 (MCL1), a TNBC dependency gene, or (4) poly(ADP-ribose) polymerase 1 (PARP1), a DNA damage repair gene that, when downregulated, can lead to genomic instability and initiate an innate interferon (IFN) response (Figure 1B). In this study, a cocktail of these four aptamer-siRNA chimeras inhibited tumor growth in the orthotopic murine 4T1 breast adenocarcinoma mouse model and in the genetically engineered ErbB2ΔEx16 transgenic mice. Furthermore, the authors provided mechanistic evidence that the cocktail therapy led to a significant increase in infiltrating CD8+ T cells and a more functional immune response.17
Developing aptamers for hard-to-treat cancers
There are a number of factors that make cancers hard to treat. These include the location of the tumor, such as those cancers arising in or spread to the brain, and tumor resistance mechanisms, such as overexpression of drug efflux pumps. Targeted drug therapies can, by the way that they typically enter cells, abrogate the issues around drug efflux pumps. Targeted therapies bind to cell-surface receptors and are then internalized via receptor-mediated endocytosis.18 If these targeted therapies carry drugs, then these drugs are internalized alongside the targeting ligand.19 By far, the most well-characterized aptamer-drug conjugate is the anthracycline, doxorubicin-HCl (Dox).
Dox has been used in the treatment of cancers for nearly 50 years and is still on the World Health Organization’s (WHO’s) list of essential medicines (WHO 2019).20 It has a known mechanism of action and can be effective in some patients, although it has the typical side effects of a non-targeted therapeutic, as well as dose-limiting cardiotoxicity. Dox works by intercalating into double-stranded (ds)DNA, typically in the nucleus, following successful cellular uptake. This mechanism of action can be very simply applied to the double-stranded stem region of an aptamer as long as the correct base pairs are present (G-C).21,22 This presents a very simplified and quick-to-assemble targeted therapeutic. The complex is very stable at physiological pH, making this an ideal aptamer drug conjugate, but the complex dissociates at acidic pH, similar to the pH observed in lysosomes.21,23 This latter property is, by far, the most important characteristic of this conjugate because if the drug is not released from the aptamer once inside the cell, then the drug remains inert and will have no cytotoxic effect. Dox is not the only drug that intercalates into dsDNA. Care must be taken to ensure that the drug chosen is able to detach from the aptamer to intercalate into the cellular DNA.
Intercalation is not the only method for utilizing aptamers for drug delivery but is possibly the simplest. As chemistries have developed over time, more and more “linkers” have been developed to allow for drugs to be attached to the aptamer.23 These linkers often require a simple chemical reaction to be coupled to other molecules, and with companies such as Baseclick licensing their technology, it is becoming simpler for researchers to attach cytotoxic payloads to aptamers. Indeed, with Dox intercalation having an effect on the tertiary structure of the aptamer and potentially affecting its specificity and sensitivity, the functionalization of an aptamer through click chemistry would likely have minor effects on these parameters.24,25
The choice of the biomarker is also an important consideration for targeting hard-to-treat cancers. Although it is often said that there are no perfect cell-surface receptors, because there are no unique biomarkers to cancer cells, some cell-surface receptors are overexpressed on cancer cells but have a low expression profile on normal cells. It is possible that tuning the binding affinity of the aptamer to its target could prevent binding to normal cells. This was quite nicely exemplified by Münz et al.26 who performed a side-by-side analysis of EpCAM antibodies. EpCAM was one of the first cancer cell biomarkers described and consequently, was the target of the first monoclonal antibody characterized for therapeutic applications. However, clinical trials demonstrated that patients had a poor tolerability due to pancreatitis. Further investigations into the antibody confirmed that it had a picomolar binding affinity and was able to bind to cells with a fairly low expression of EpCAM such as normal pancreatic cells. Next-generation antibodies with nanomolar binding affinity did not have the same poor tolerability. Due to the unique nature of aptamers, modifying binding regions so that they will only bind to the cancer cells with a higher expression rather than to all cells that express a particular biomarker is a fairly simple process.27 If the sensitivity can be adjusted while maintaining the specificity through changing some nucleotides in the loop or stem region of the aptamer, then fewer side effects will be associated with the treatment, and bioavailability at the cancer cells might be increased. With these considerations in mind, how may aptamers progress to enhance survivability with treatment-retardant cancers?
Leukemias
Although there has been a great deal of success in treating leukemias with small-molecule drugs and monoclonal antibodies, prognosis still remains poor for a number of patients. It is often thought that leukemias, being described as liquid tumors, are easier to treat than solid tumors due to a less complex environment. However, if that were the case, then the long-term survival rates would be much improved. The issues surrounding drug resistance in leukemic cells relate to the interplay among the cancerous cells, the bone marrow niche, and the extracellular matrix. This interaction leads to changes in cell signaling, which favors drug resistance and provides what has been termed a “safe haven” for the stem cells.28 An additional reason for failure with classical cytotoxic drugs is that they typically target rapidly dividing cells, and it is thought that once leukemic cells take up residence in the bone marrow niche, they become fairly quiescent. This suggests that targeting biomarkers on the cell surface for drug delivery would be highly effective. Although some leukemias have very well defined cell-surface markers, such as CD19 in B cell acute lymphoblastic leukemia (ALL), not all leukemias have such well-characterized markers,29 suggesting that a multipronged or synergistic targeting approach will be required in the future.17
Probably the most well-characterized aptamers to leukemia cells are those developed by the Tan group30 using cell-SELEX (systematic evolution of ligands by exponential enrichment). The sgc8 aptamer was originally selected against the T cell ALL cell line, CCRF-CEM, and shown to have a binding affinity of 0.8 nM. Further testing confirmed that sgc8 could also bind to other T cell ALL cell lines, suggesting a common biomarker to these cell lines that was not present on acute myeloid leukemia (AML) cell lines. Further studies suggested that sgc8 binds to the protein tyrosine kinase 7 (PTK7) protein31,32 and that it could deliver daunorubicin into a T cell ALL line and demonstrate specific cytotoxicity.33 Finally, this aptamer was truncated from 88 nucleotides to 41 nucleotides with no loss of binding affinity.34 Dox was then intercalated into this shortened aptamer, sgc8c, which reduced the binding affinity from 0.78 nM to 2 nM. However, this did not affect cytotoxicity, with a similar effect observed between free Dox and aptamer-delivered Dox to the CCRF-CEM cell line.25 Recently, through click chemistry, this aptamer has also been functionalized with radionuclides for imaging, although this could just as easily be used for therapeutic or theranostic applications.35 Although this study tested the aptamer in an in vivo B cell lymphoma model, it did show biodistribution in the bone, with sustained accumulation over 3 days, suggesting potential for future therapeutic delivery to tackle the leukemic cells in the bone marrow niche.
There are several more aptamers that have demonstrated promise for targeting leukemic cells that have been reviewed elsewhere.36 However, one worth highlighting in this review is the CD117 aptamer.37 CD117 is a biomarker that appears to correlate with patient prognosis, whereby a higher level of expression is associated with a poorer prognosis. This is similar to PTK7, which is involved in resistance mechanisms to chemotherapy.38 This would suggest that the sgc8 aptamer and this CD117 aptamer have the potential to treat “hard-to-treat” leukemias. Another reason to highlight this aptamer is that with the use of click chemistry to attach methotrexate (MTX) to the aptamer, Zhao et al.37 demonstrated a favorable result in patient samples. They demonstrated that, when incubated with primary bone marrow patient samples, the aptamer-MTX was able to kill the leukemic cells, whereas it had no effect on the healthy bone marrow cells.
Both Dox and MTX have been used in targeted delivery as a means to reduce the toxicity of free drug dosing and not necessarily for overcoming drug resistance outside of drug efflux pumps. Although a low cellular accumulation of long-chain polyglutamates has been associated with poor therapeutic outcome, enzyme mutations and cytogenetic aberrations also contribute to MTX resistance.39 Although MTX has been used for decades and has known toxicities, simply having a higher accumulation in specific cell populations does not fix the inherent issue of drug resistance associated with enzyme mutations.
One of the issues with hard-to-treat cancers is that the patient may appear in remission for an extended period but is actually relapsing. However, the current methods of detection are not sensitive enough to detect minimal residual disease (MRD). One very recent publication has detailed the generation of an aptamer specific to a patient diagnosed with multiple myeloma. Their aptamer isolation process is automatable and only requires a small amount of patient sera. Importantly, this aptamer was 2000-fold more sensitive than immunofixation electrophoresis and was able to detect MRD at 6 months, whereas the conventional methods failed to diagnose MRD for 25 months. Although only a proof-of-principle study, it suggests that aptamers could play a much larger role in personalized medicine in the future.40
Carcinomas
Additional factors should be considered for the treatment of solid tumors. The cancer lesion is a complex environment, consisting of not only tumor cells but also tumor-associated cells such as macrophages, lymphocytes, fibroblasts, blood vessels, and the lymphatic system.41 This creates a very dynamic environment that affects the bioavailability of drugs. The use of the term “active targeting” or “targeted therapy” is often a misnomer, as it is only from sheer luck that a drug evades the numerous hurdles to finally have an effect on the cancer cells in a solid tumor. Although the vasculature becomes leaky around solid tumors, there is sufficient back pressure from the lymphatic system that will prevent large particles from entering the tumor space. This does not mean that large molecules cannot target tumor cells, but only a small amount is able to get through. This is where aptamers demonstrate a clear benefit, as they are much smaller in size to their proteinaceous counterparts.42 Given that antibody-drug conjugates have shown some successes, it is only natural that, in the field of aptamer therapeutics, this would be the simplest mechanism of targeted drug delivery.
In addition to the physical barriers that prevent adequate treatment efficacy in solid tumors, the knowledge gains of the last 20 years have contributed to our understanding of the dynamics of cancer. Although the hallmarks of cancer have being collated very nicely in review articles by Hanahan and Weinberg,43,44 more information has been forthcoming regarding the genotyping of cancer cells. Targeting single molecular events has shown some positive effects but is unlikely to be the paradigm shift required to show positive benefits in all patients.45 This is due to redundant pathways being activated when one is targeted and to the intra- and inter-tumoral heterogeneity. Indeed, a study in 2018 found that less than 10% of cancer patients in the United States were eligible for genome-targeted therapy and that only about 50% of these patients demonstrated any benefit.46 For checkpoint inhibitors, these numbers were slightly better in that approximately 43% of patients were eligible. However, less than one-third benefited from this treatment.47 To put this into perspective, approximately 80% of US cancer patients are eligible for cytotoxic chemotherapy with an average response rate of 48%.48 For the patients that did respond, the benefit was durable, but, as we have highlighted, different approaches are required to offer precision medicine to all patients.17
Malignant melanoma is one of the more aggressive and treatment-resistant carcinomas, and the number of cases continues to rise.49 Dacarbazine was the first cytotoxic chemotherapy to be used for melanoma but only provided a 20% objective response rate.50 Newer genome-targeted therapies have also demonstrated similar response rates, and the development of resistance within 6 months of treatment has limited their use in metastatic melanoma, although, for the patients who do respond, this can be sustained.51 Combination therapy (nivolumab-plus-ipilimumab) has also been demonstrated to increase progression-free survival to 11.5 months.52 Noteworthy, dacarbazine has immunostimulatory effects, which may explain some similarities in treatment response between the initial chemotherapeutic treatment and recent immunotherapy trials.53 This suggests, as pointed out by the authors, that combining the old with the new may offer additional benefits. However, a phase II clinical trial of dacarbazine and ipilimumab was discontinued after all patients experienced serious adverse effects and liver toxicity.54 A separate study evaluated survival benefits from this combination and found the 5-year survival rate to be only 18%.55
One aptamer that has already been mentioned is the MRP1 aptamer.5 This aptamer was initially selected to the MRP1 before being attached to a previously selected CD28 aptamer that had shown agonist abilities.56 This was then tested in an animal model of drug-resistant melanoma alongside the GVAX (granulocyte-macrophage colony-stimulating factor [GM-CSF]-secreting vaccine) and P60 peptide to block forkhead box P3 (FOXP3). This combination resulted in higher T cell tumor infiltration, slower growth rate, and a longer survival: 50% still alive at 50 days versus control treatment reaching the endpoint at 28 days. This demonstrates the potential of aptamers for future trials, especially given that the targeting of stimulating aptamers to the tumor has resulted in reduced toxicity.3 This study also highlights the changing paradigm of developing targeting agents to biomarkers rather than specific tumors, a theme that has consistently been applied to aptamers in cancer theranostics.57 A similar aptamer, the F3B aptamer, developed to matrix metalloproteinase 9 (MMP9), overexpressed in a number of malignant tumors, has been radiolabeled and demonstrated a good uptake in the metastatic melanoma mouse model (2% injected dose [ID]/g versus 0.7% ID/g for the control aptamer).58 Another aptamer that can be used in multiple different cancers and potentially in patients with diabetes is the RAGE aptamer (targeting the receptor of advanced glycation end products).59 It suppressed liver metastasis and reduced tumor growth.60 A final example is the CD63 aptamer that has shown specificity to vemurafenib-resistant melanoma cells. The authors suggested that this aptamer could be used in conjugation with therapeutics to detect these resistant cells and make early decisions about treatment options.61 However, it could also be used in aptamer-drug conjugates in resistant cells. These aptamers have all been characterized in malignant melanoma models. Given the ubiquitous nature of these biomarkers, there is no reason to believe they could not be used in other hard-to-treat carcinomas, especially given the heterogeneity of biomarker expression. Similar to the very patient-specific study cited previously for myeloma, these aptamers and a combination of these aptamers could be tested against patient samples to offer a truly personal therapeutic regimen.
Brain tumors
Given the issues already apparent with treating solid tumors, brain tumors and brain metastases face an additional barrier, the blood brain barrier (BBB). This is incredibly important in maintaining the day-to-day health of our brains and preventing toxic substances from having a neurotoxic effect. However, it also prevents the vast majority of drugs passing into the brain from circulation. It is thought that the integrity of the BBB breaks down during the development of brain metastases, thereby allowing more cancer cells to spread. As an upside to this, it also means that, in the initial stages of treatment, drugs can more readily pass into the brain. As the metastatic lesions are reduced, however, the BBB starts to repair, and the bioavailability of drugs in the brain decreases. This typically leaves behind the more drug-resistant tumor cells, which are then mostly untreatable even if drugs do traverse the BBB. A similar situation is likely in primary brain cancers. It does have to be acknowledged that the non-specific diffusion of drugs into the brain will most likely have detrimental effects on the patient.41,62,63
Given that the number of patients presenting with brain metastasis continues to increase, the issues of how to circumvent the BBB have become imperative to solve. A number of strategies have been proposed, including, but not limited to, direct brain implantation, ultrasound, microbubbles, and transcytosis. As most of these would lead to toxic substances also traveling into the brain, they will not be considered here. Transcytosis is an interesting concept that highjacks the receptors on the BBB that deliver essential nutrients into the brain. There are a number of well-characterized receptors present on the BBB that have potential for transcytosing drugs into the brain. The transferrin receptor (TfR) is probably the most well characterized for its ability to improve the delivery of therapeutics or loaded nanocarriers across the BBB.64,65 The group headed by DeRosa at Carleton University exploited the safe targeting and high specificity advantages of aptamers to promote delivery from the peripheral injection site into the brain of two different dopamine aptamers: either a DNA homolog of a previously characterized RNA aptamer,66 named DBA, or a truncated monomer aptamer, DA20m.67 To this end, the authors directly conjugated a short DNA aptamer ligand for TfR, named TRA, to the surface of PEGylated liposome cargoes pre-loaded with dopamine aptamers (DAL-TRAM). Upon systemic administration, the efficacy of systemically administered DAL-TRAM was assessed by functional assays in cocaine-treated animals to attenuate hyperlocomotion and mitigating induced behavioral changes. They showed that chronic administration did not produce any apparent motor deficits or neuronal degeneration.
TfR is also overexpressed on cancer cells; thus, it is not without its own challenges to use for transcytosis into the brain for the treatment of tumors. In the scenario where a patient presents with classical metastatic spread to the bones, other organs, and the brain, the bioavailability in the brain will be much reduced. However, there is the potential for all metastases to be treated. In the scenario where the patient has undetected brain metastases, the reduction of these might be a side effect of the treatment, but the treatment would be seen as having limited efficacy in the other metastases. In the scenario where the patient has brain cancer, the same targeting modality would target the tumor cells after transcytosing into the brain. In all three scenarios, the treatment would be working as hypothesized. However, the binding affinity would need to be very finely tuned to ensure that it is capable of performing both roles.
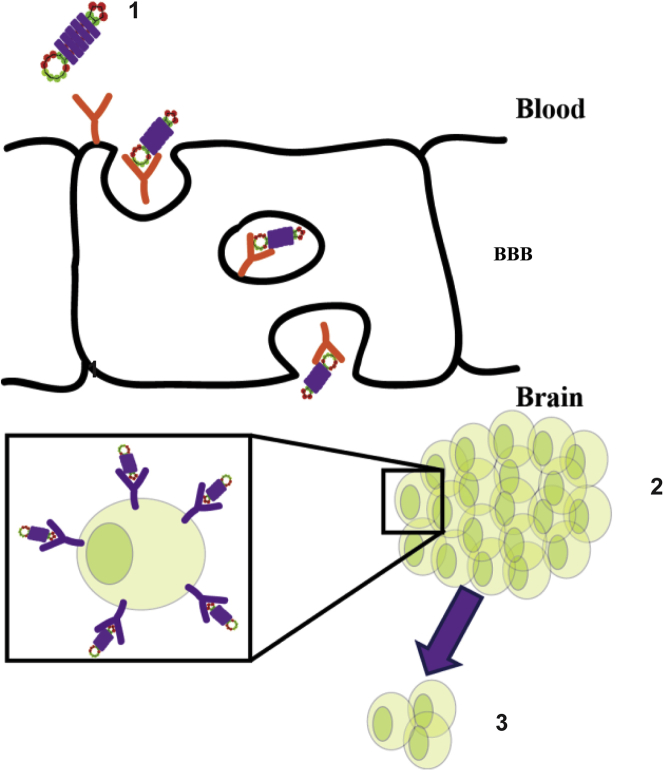
The use of a single aptamer targeting the TfR for the targeted treatment of cancer is therefore feasible but not necessarily the best approach. Recently, Macdonald et al.68 demonstrated that a TfR aptamer could be joined to an aptamer targeting EpCAM, a transmembrane receptor expressed in several epithelial cancer cells.69 Specifically, in this study, this bifunctional aptamer was used to target TNBC cells, and the functionality of the aptamer was confirmed in vitro using a cell line-based model of the BBB. This aptamer was also confirmed to retain functionality when Dox was intercalated into the stem region. An extension of this study also demonstrated that the aptamer-Dox conjugate could cross the BBB, in vivo, and specifically target cancer cells that had naturally metastasized into the brain (Figure 2). The authors postulated that this represents a platform technology whereby the EpCAM aptamer moiety could be switched for an aptamer targeting a different cell-surface receptor or target.70
Figure 2.
Schematic depicting a bifunctional aptamer transcytosing across the blood brain barrier (BBB)
The bifunctional aptamer-drug conjugate (1) binds to the transferrin receptor on the BBB and transcytoses across the BBB to carry its cargo to EpCAM+ cancer cells (2) leading to a reduction in tumor size (3).
Other receptors, such as the glucose transporter Glut1, the leptin hormone receptor LepR, CD98, and CD147, have been assessed for the ability to cross the BBB.63 However, to date there have been no known aptamers selected to these targets. Glut1, CD98, and CD147 would present the same caveats as the TfR, given that it is also expressed on cancer cells. LepR is also associated with breast cancer, with higher levels observed in obese patients, suggesting that care would be required if using this receptor for transcytosing the BBB and with the same caveats as previously mentioned.71 Like many other cancer cell targets, these receptors are expressed on normal cells as well, so the use of these receptors is highly contingent on the levels of expression among normal cells, endothelial cells on the BBB, and cancer cells. In terms of bioavailability in the brain, a study published in 2016 by Zuchero et al.72 compared antibody uptake into the brain targeting several of these receptors. They observed that Glut1 antibody levels were similar to TfR antibodies. However, CD147 antibody levels were approximately 2- to 4-fold higher, and CD98 antibody levels were about 10-fold higher than TfR antibodies. Interestingly, when they conjugated the anti-CD98 antibody to an anti-Beta-Secretase 1 (BACE1) antibody, the binding affinity of the anti-CD98 moiety decreased and led to increased accumulation in the brain. These results are consistent with those previously observed with an antibody targeting the TfR, suggesting that aptamers targeting receptors on the BBB for transcytosis should have a binding affinity >100 nM to their target.73
Although aptamers do not necessarily cross the BBB easily, a number of aptamers have been shown to have the ability to cross the BBB, either in vitro or in vivo, as recently reviewed by Bukari et al.63 One of the most recent derivations of the SELEX process, which is also one pertinent to the issue of crossing the BBB, is the selection of aptamers capable of crossing the BBB in vivo.74 In this study, the aptamer library was injected into the tail vein of a mouse and allowed to circulate for 1–3 h prior to the collection of aptamers that had penetrated into the brain. The brain distribution of several aptamers, including the A15 aptamer, was confirmed in vivo using in situ hybridization. The ability of the A15 aptamer to be internalized by endothelial cells was confirmed in vitro. However, no further study using this A15 aptamer has been published, although the initial publication is highly cited. This could be due to the modified RNA sequence being used, which, although ensuring a good half-life, would be supplanted by other simpler or cheaper aptamers.
Given that the number of patients presenting with brain metastases far exceeds the number of patients presenting with high-grade gliomas, it is not surprising that the number of aptamers specifically targeting these tumors is low.75 However, the known targets for these aptamers, including AXL, CD133, epidermal growth factor receptor (EGFR)vIII, ephrin receptor tyrosine kinase (RTK; Eph)A2, EphB3/2, nucleolin, platelet-derived growth factor receptor (PDGFR)-α, PDGFR-β, tenascin-C, and the TfR,76 represent a more diverse range than any other solid tumors. These aptamers have been eloquently reviewed.62,75,76 Results have varied with not all studies extending to in vivo results. When considering in vivo results, care must be taken in their interpretation due to how the cancer cells have been delivered to the brain. Direct implantation of cancer cells involves breaching the BBB and allowing most substances to cross over a leaky BBB. The BBB is also known to become leaky following establishment of tumors in the brain, so careful selection of controls is required to confirm that any drug or targeting ligand has breached the BBB as part of its mechanism of action and not because the BBB is damaged. It is important to confirm the aptamer’s ability to cross the BBB using well-established in vitro methods or in healthy animals and with a full range of controls, if possible, to ensure that future animal trials have a high rate of success.
Targeting CSCs
CSCs are present in several tumor types as a small cell population with an enhanced capacity to initiate tumor formation and resistance to conventional therapies. They present several stem-like characteristics such as the ability to undergo symmetric and asymmetric cell division and are believed to be a major cause of tumor relapse. Because of their ability to drive secondary therapeutic reagents into the tumor mass, aptamers are highly promising for CSCs targeting. Aptamers have been raised either for CSC-specific targets such as CD133, also known as prominin-1,77 or for tumor cell-surface markers highly expressed in various cancer types on both CSCs and non-stem cells such as the TfR, CD44, and EpCAM.78 With the use of an aptamer against CD133, Qiao and others79 have recently shown that doxorubicin-loaded aptamers (CD133-apt-Dox) can selectively deliver Dox into hepatic CSCs, inhibiting proliferation and diethyl nitrosamine-induced tumor growth in an immunocompetent mice model. Several aptamers have been selected to EpCAM,80, 81, 82 with one demonstrating similar results to the CD133 A15 aptamer used by Xiang et al.21 in an in vivo model of colon cancer. CD44 is another cell-surface receptor associated with CSCs for which aptamers have been generated.83 Interestingly, Zheng et al.84 combined the EpCAM aptamer with the CD44 aptamer and tested it using an in vivo model of ovarian cancer. They found that, whereas the EpCAM aptamer by itself had no effect, possibly due to its small size (19 nt), the CD44 aptamer and the bispecific CD44-EpCAM aptamer decreased tumor growth. Noteworthy, these three cell-surface receptors are overexpressed, either singly or as multiple markers in most solid tumors.78
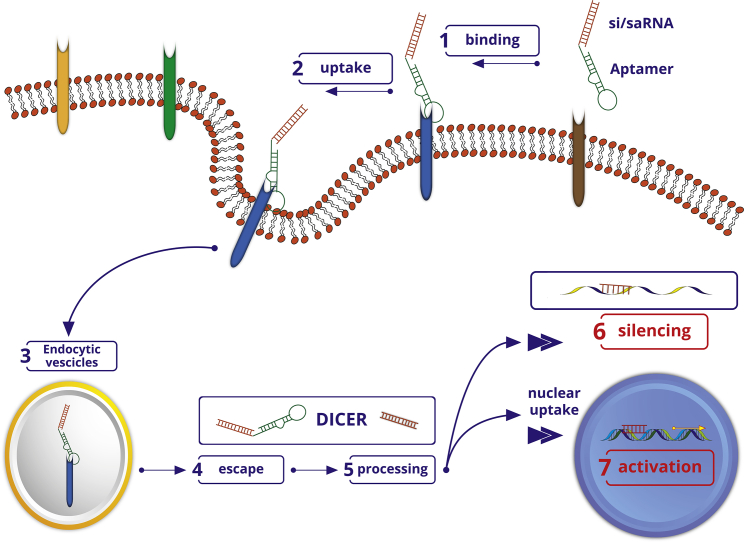
Several reports highlight an important linkage between the presence of glioblastoma (GBM) stem cells (GSCs) and resistance to conventional therapies.85,86 The dysregulated activity or expression of a few transcription factors and micro-RNAs, including the signal transducer and activator of transcription-3 (STAT3), is critical to initiate and sustain the GSC population.87,88 By using two different aptamer-siRNA chimeras able to selectively drive an siRNA against STAT3 to GBM cells, two independent reports have addressed the selective knockdown of STAT3 in brain tumor cells. To enhance the selectivity for brain cells, the group led by Rossi89 at the City of Hope National Medical Center generated an aptamer against the PDGFR-α, an RTK involved in neural crest development. The aptamer, named PDR3, was conjugated to an siRNA specific to STAT3 using a stick-based approach in which the aptamer and the siRNA are elongated with complementary GC-rich linkers, thereby maintaining the functional integrity of both the aptamer and the siRNA component (Figure 3).90 Upon efficient uptake in U251 GBM cells, the PD3-STAT3 AsiC inhibited target cell growth.89 Esposito et al.91 took advantage of the aptamer for a different isoform of the PDGFR, PDGFR-β, named Gint4.T, which was previously characterized and shown to be able to cross an in vitro model of BBB. Gint4.T was conjugated using a stick-based approach to a STAT3-specific siRNA. The treatment of patient-derived GSC lines with the Gint4.T-STAT3 AsiC in combination with a chimera containing the anti-Axl aptamer GL21.T conjugated to an antagonist for microRNA (miR)-10b (GL21.T-10b) efficiently inhibited tumor sphere formation of GSCs, cell propagation, and invasion.92,93 Recently, using differential cell-SELEX, Affinito et al.94,95 described a 2′ F Py-modified RNA aptamer that selectively binds patient-derived GSCs but not the same cells grown as adherent cells. The aptamer, named A40s, targets EphA2 on the cell surface of GSCs, inhibiting tumor growth stemness and migration. Upon binding to EphA2, A40s rapidly internalizes into target cells in a receptor-mediated manner, serving as a targeting carrier for the functional delivery of short non-coding RNAs such as miR-34c and anti-miR10b to the stem cell population.94,95 Given the intrinsic heterogeneity of GBM tumors, the development of aptamers for the combined targeting of multiple differentially expressed receptors in various tumor areas, including the GSC niche, appears to be a promising RNA-based therapeutic approach in gliomas.
Figure 3.
Schematic depicting aptamer-mediated selective targeting
The si/miRNA- and saRNA (orange)-aptamer chimeras bind through the aptamer moiety (green) to the target cell-surface receptor (blue) (1). Upon endocytosis, si/saRNA-aptamer chimeras are internalized into the cell (2). Uptake of si/saRNA-aptamer conjugates into intracellular vesicles, endosomes, or lysosomes (3). Molecules escape the endosomal compartment (4), si/saRNA-aptamer chimeras are recognized by the RNA interference (RNAi) machinery, and the si/saRNA part of the conjugate is cut off the aptamer by DICER (5). The si/saRNA is either loaded into the RNA-induced silencing complex for specific degradation of cytosolic messenger RNAs or (6) undergoes nuclear uptake for targeted gene activation (7).
Targeting nuclear functions
Over the last decades, the development of RNA drugs has greatly increased, uncovering the possibility to post-transcriptionally downregulate the expression levels of disease-associated proteins. For example, disease-associated proteins can be inhibited by exploiting the RNA interference (RNAi) cellular mechanism or by antisense oligonucleotide (ASO)-mediated inhibition of translation, thereby reducing disease severity or eliminating the disease. Furthermore, a number of therapeutic oligonucleotides have emerged that modulate nuclear functions such as splicing or mRNA transcription, including splice-switching oligonucleotides (SSOs), short guide RNAs, and short or small activating RNAs (saRNAs). In neoplastic diseases, RNA drugs with the potential to activate the expression of target genes have been developed to restore onco-suppressor gene levels, thus complementing the downregulation of tumor-promoting protein levels. However, to target nuclear functions of this class of therapeutic short RNAs, the development of delivery carriers is required.96,97
SSOs
Among aptamers that have been shown to promote nuclear uptake of drug conjugates (see below), the 26-mer G-rich DNA aptamer, AS1411, has been the first widely used for nuclear targeting into cancer cells because of its property to shuttle from cell surface into the nucleus of most tumor cells. A first evidence for its use as a targeting moiety for nuclear uptake was provided by Sullenger et al.98 who used the AS1411 aptamer to specifically deliver SSOs into nucleolin-expressing target cells. SSOs are single-stranded RNA oligonucleotides that are made complementary to a splice site or a splice enhancer within a target pre-mRNA. By binding to these sequences, SSOs interfere with the normal splicing process at the target sites giving rise to alternative splice transcripts and are therefore emerging as tools to generate phenotypic changes in cells.99 The authors took advantage of AS1411 to give the first proof of principle for aptamer-based SSO delivery to the nucleus of PC3/Luc 705 cells by using an SSO directed against the pre-mRNA of the luciferase reporter gene conjugated to the AS1411 aptamer. The AS1411 was shown to target tumor cells expressing nucleolin on the cell surface and to be efficiently translocated to the nucleus, its final site of action.100 In a recent study, Rao et al.101 used the AS1411 to drive Dox into the nuclei of drug-resistant MCF7 (MCF7/Adr) breast cancer-derived cells. Indeed, in breast cancer, the high levels of adenosine triphosphate-binding cassette transporter proteins are responsible for the frequent resistance to Dox treatment by increasing efflux across the cell surface. To enhance Dox accumulation in the nucleus, the authors designed an Apt-Dox conjugate able to internalize into MCF7/Adr and translocate to the nucleus. To this end, the AS1411 aptamer was conjugated to Dox at approximately 1.4 Dox molecules per aptamer. The Apt-Dox molecules were then loaded into liposomes to protect them from degradation by circulating nucleases and to prevent their rapid clearance from the bloodstream. Upon intracellular uptake of the liposome, the Apt-Dox conjugate is released and translocated into the nucleus driven by the aptamer AS1411. Although the results were obtained in vitro, the study provides a first evidence that a two-stage nano-vector can enhance nuclear uptake mediated by nucleolin.
saRNAs
The recent evidence that saRNAs are able to selectively enhance the expression of silenced tumor-suppressor genes in tumor cells has triggered a great hope in cancer medicine. Instead of genome engineering that permanently changes the target DNA gene sequences, RNA-mediated promoter activation has recently emerged as a novel strategy for gene activation in mammalian cells. saRNAs are duplex, ∼21-mer oligonucleotides that are imported into the nucleus in complex with Argonaute family members, like AGO2, and targeted to promoter regions within the chromosomal DNA, activating gene expression.102,103 To date, multiple genes have been successfully targeted by saRNAs, including the tumor-suppressor genes, dihydropyrimidinase-like 3 (DPYSL3)104 and CCAAT/enhancer-binding protein alpha (C/EBPα) genes.105
C/EBPα is a leucine zipper containing a transcriptional factor that regulates the expression of several genes, including p21, which is epigenetically silenced in several cancers.106 Habib et al.105 have recently addressed the reactivation of C/EBPα in cancer cells. The authors have developed bioinformatic tools to design specific saRNA sequences for activating transcription from targeted promoters (Figure 3). This approach allowed the identification of an saRNA, named C/EPBα-saRNA, targeting and able to activate the C/EBPα promoter. By using an experimental hepatocellular carcinoma (HCC) rat model, the authors demonstrated the efficacy of the systemic injection of C/EPBα-saRNA-dendrimer conjugates in reducing tumor burden and improving liver functions.97 More recently, to assess the antitumor effects of C/EBPα-saRNA in non-hepatic tumors, Yoon et al.107,108 developed an aptamer-based C/EPBα-saRNA-targeted delivery to pancreatic ductal adenocarcinoma (PDAC) experimental tumors. By using differential cell-SELEX, they selected two 2′-FY-containing RNA aptamers as targeting moieties for PDAC, named P19 and P1, which were conjugated to C/EBPα-saRNA, P19-C/EBPα-saRNA, and P1-C/EBPα-saRNA.
Both conjugates were delivered systemically into human pancreatic PANC-1 cancer cell subcutaneous xenografts. Tumor growth was compared to that of gemcitabine-treated xenografts over a period of 4 weeks. P19-C/EBPα-saRNA presented no toxicity to the host and was 30% more efficient to elicit the antitumor response when compared to the current standard of care, gemcitabine. The same group also developed a traceable in vivo model of hepatic xenografts with luciferase-expressing PANC-1 cells (PANC-Luc). As a targeting moiety, Yoon et al.107 developed an RNA aptamer, named TR14, for the human (h)TfR, a receptor that is overexpressed in several tumors, including PDAC. Once conjugated to the C/EBPα-saRNA, the TR14- C/EBPα-saRNA was injected into tumor-bearing mice via the tail vein. The aptamer-saRNA chimera was effectively targeted to the PANC-1 tumors, enhancing C/EBPA expression and reducing tumor burden, indicating that the saRNA cargo, once delivered to the nuclear compartment, maintains its functional integrity.
The DPYSL3 gene codes for a cytosolic phosphoprotein, also named collapsin response mediator protein-4 (CRMP4), which has been reported to act as a suppressor of invasion in several human cancers, including in metastatic prostate cancer. The DPYSL3 gene is transcribed by distinct promoter usage producing two transcripts that result in two distinct proteins (CRMP4a and CRMP4b). The most expressed is the shorter variant, DPYSL3v2/CRMP4a, which is consistently downregulated in prostate cancer tissues.104 With the aim of developing a therapeutic agent to effectively reduce prostate cancer cell migration and invasion, Li et al.104 took advantage of the saRNA approach to enhance promoter activation of the DPYSL3v2 variant, increasing the intracellular levels of the CRMP4a protein. Out of four activating candidates studied, the saV2-9-saRNA was shown to be the most effective in reducing in vitro cancer cell migration. The saV2-9-saRNA was then conjugated to the RNA aptamer A10-3.2, a widely used aptamer, which, by binding to the prostate-specific membrane antigen (PSMA), promotes PSMA-dependent cellular uptake in prostate cancer cells.109 The A10-3.2-saRNA conjugate was then analyzed in an orthotopic nude mouse model of prostate cancer for its antitumor efficacy. Results indicate the aptamer-saRNA conjugate was selectively delivered to prostate cancer cells; DPYSL3v2/CRMP4a expression was enhanced, and growth of distal lung metastasis was reduced in vivo.104
Although restricted to C/EBPα and DPYSL3, these studies provide a blueprint of the potential of aptamers to act as effective delivery moieties for saRNAs and epigenetic RNA drugs able to modulate local gene expression.
Aptamers as targeting ligands
Direct conjugation to therapeutic RNAs, siRNA/microRNA (miRNA)
The efficacy and safety of treatments with several anticancer drugs such as cytotoxic small drugs, tyrosine kinase inhibitors, or miRNA replacement therapies are hampered by off-target effects in surrounding healthy cells and tissues. The selective delivery of anticancer drugs to target cells that combine the reduction of severe, undesired effects in healthy tissues with a greater therapeutic efficacy has thus become a challenging need to increase the therapeutic efficacy of treatments and the quality of life of cancer patients. Since the first reports a decade ago, aptamers are now proving to be very attractive targeting carriers for various therapeutic molecules, providing high tissue penetration and intracellular uptake in tumors and in cells within the tumor microenvironment.7,110,111 Indeed, upon binding to the extracellular domain of the proper target receptor, aptamers undergo receptor-mediated uptake, driving internalization of secondary therapeutic reagents, including short therapeutic oligonucleotides and small molecules. Several reports have addressed the understanding of the internalization and transport processes that aptamers undergo upon binding to the cell surface that largely depends on the endocytic pathway of the receptor involved.18 In a recent study, Tan et al.112 used the single-particle tracking (SPT) technique to monitor in real time the specific endocytic pathways and intracellular transport of sgc8, a DNA aptamer that targets the PTK7, a receptor PTK shown to regulate the Wnt signaling pathway.113 By conjugating the sgc8 aptamer to the 5-fluorouracil drug (sgc8-5FU), the authors showed that, upon binding to PTK7, the aptamer, either alone or in the context of the conjugate, internalizes mainly via caveolin-mediated endocytosis although partially via clathrin-mediated endocytosis.112
In contrast to neoplastic cells that can frequently develop therapy-induced acquired resistance, cells from the tumor microenvironment are genetically stable, representing a valuable alternative for therapeutic targeting. Therefore, De La Fuente et al.114 addressed the selective delivery of the chemotherapeutic drug, Dox, to tumor-associated stromal cells as a strategy to suppress tumor growth (Figure 1C). To this end, the authors developed aptamers that selectively bind to tumor-infiltrating myeloid cells (TIMCs) that represent a great portion of the tumor microenvironment.114 TIMCs are key therapeutic targets involved in cancer cell growth and metastasis. They are responsible for the suppression of the immune response. A great advantage to developing drugs that target TIMC-associated antigens, with respect to heterogeneous neoplastic cells, comes from the fact that these cells stably express a unique phenotype that is similar in the tumor and in the metastasis. Therefore, the authors exploited the advantages of aptamers as targeting moieties because of their easy penetration into tumors, poor or no-immunogenicity, high stability, half-life in biological matrices, and affordable production costs. By using the myeloid-derived suppressor cell (MDSC)-derived mouse cell line, MSC-2,115 as the complex target for cell-SELEX, the authors identified four different RNA aptamers, named aptamers 3, 6, 11, and 14, as well as annexin A4 and vimentin as putative ligands for aptamers 3 and 11, respectively.114 The four aptamers selectively bind to tumor-infiltrating, but not to peripheral myeloid cells, from a variety of mouse or primary tumor specimens from human head and neck squamous cell carcinoma (HNSCC). To investigate the in vivo therapeutic efficacy, the authors then generated Dox-conjugated aptamers showing that the four aptamers are effective targeting moieties for the delivery of drugs to the tumor site. Dox conjugates were then administered intravenously (i.v.) using the 4T1-derived mammary metastatic carcinoma model. The authors demonstrated that the TIMC-targeting aptamers can deliver Dox to both the primary orthotopic tumor and metastatic sites, increasing the therapeutic index. Furthermore, because aptamers were shown to bind to different TIMC-specific targets, Dox conjugates were administered in combination, increasing the overall specificity. The authors showed the potential of this approach by demonstrating enhanced drug delivery and reduced toxicity of chemotherapeutic agents as compared with the systemic delivery.114
Aptamers as targeting moieties for loaded nanoparticles
Liposomes are spherical, self-closed vesicles formed by concentric lipid bilayers that surround an aqueous core phase. Aptamer-driven liposomes have been shown to be an effective approach for the selective delivery of therapeutic nucleic acids and drugs, improving the functional uptake of payloads to the target-affected cells.116,117 In a recent study, Fattal et al.83 described a liposome-based siRNA delivery system with a core composed of the siRNA:protamine complex and a shell designed for the active targeting of CD44-expressing cells. To this end, the authors took advantage of a previously reported aptamer, Apt1, raised against the CD44 receptor, a surface biomarker found overexpressed in various tumor types.118 Apt1 was post-inserted into neutral liposomes loaded with siRNA. The efficacy of the nanocarrier was then evaluated for the silencing of the reporter gene luciferase (luc2) in vitro and in vivo using an orthotopic MDA-MB-231 TNBC model.83
Dendrimers are synthetic symmetrical hyperbranched polymer nanoparticles that have extensively been studied as carriers for the effective delivery of small RNAs such as the cationic dendrimer in vitro and in vivo.119 The polyamide-amine family of dendrimers (PAMAM) is one of the most studied for its potential use in biomedicine. Making use of persistent luminescence nanoparticles (PLNPs), Zhang et al.120 reported the design of a PLNPs-PAMAM-aptamer/Dox nanoplatform for cancer theranostic applications. To this end, the authors used the AS1411 aptamer that binds to nucleolin on the cell surface of cancer cells as the targeting moiety. Owing to the presence of the PLNP, the biodistribution of the PLNPs-PAMAM-AS1411/Dox nanocomposite has then been monitored by in vivo luminescence imaging. The delivery of the anthracycline Dox selectively to the tumor site was demonstrated. The authors used HeLa tumor-bearing athymic nude mice as an experimental model to evaluate the effect of PLNPs-PAMAM-AS1411/Dox’s treatment on tumor growth inhibition. Results showed an enhanced therapeutic effect of the targeted nanocomposite over the free Dox or the untargeted nanocomposite.
DNA-based dendrimers have advantages over other polymers. They are easily functionalized due to GC-rich “stick” terminal linkers on their surface for annealing to complementary therapeutic nucleic acids.110 To improve target specificity, Jia et al.121 developed stable aptamer-conjugated, highly branched DNA dendrimers. The authors used the sgc8 aptamer as the targeting moiety. The sgc8 aptamer recognizes the catalytically defective PTK7, a receptor PTK that is upregulated in various cancers.30 The targeting aptamer was assembled by annealing to the dendrimer nanocomposite through the presence of the terminal sticky ends. The DNA dendrimers were then nick sealed to increase the stability. CCRF-CEM tumor-bearing mice were used to assess the therapeutic efficacy of sgc8-DNA dendrimers-Dox complexes. The nanocomposite showed in vivo-specific targeting of CCRF-CEM cells and uptake of doxorubicin to nuclei of target tumor cells, significantly inhibiting tumor growth.121
Aptamers in clinical trials
Aptamers as a therapeutic modality have had limited success in clinic. To date, only one aptamer has been US Food and Drug Administration (FDA) approved for the treatment of age-related macular degeneration (AMD) with no aptamer(s) approved for the treatment of cancer. Several properties of aptamers have limited their development into effective systemic therapies. Due to their inherent small size, aptamers are cleared quickly by the renal system. To circumvent this, aptamers have been PEGylated to increase their half-life. However, clinically, anti-peg antibodies can interfere with and inhibit the function of aptamer binding and activity and can lead to an anaphylactic reaction, which is mostly likely what contributed to the failure of Regado Bioscience’s clinical trial.122 Further work and enhanced approaches are needed to increase the half-life and pharmacokinetics (PK)/pharmacodynamics (PD) profile of aptamers. Additionally, although a lot of progress has been made to stabilize, prevent degradation, and reduce TLR recognition, there has been some evidence of hepatic toxicity, specifically when phosphorothioate-modified nucleotides are used.123
Among the aptamers that recently entered clinical trials for anticancer therapeutic indications (https://clinicaltrials.gov/), a promising molecule is represented by a variant of aptamers, named Spiegelmers. These aptamers are produced by NOXXON Pharma from non-natural L-nucleotides adopting a modified SELEX drug-discovery proprietary platform.124 The L-configuration confers Spiegelmers’ resistance to degradation by nucleases present in circulating plasma and non-appreciable immunogenic response, both considered critical features for nucleic acid therapeutics.125 NOXXON Pharma is developing Spiegelmers-neutralizing chemokines in the tumor microenvironment. A recent example of therapeutic Spiegelmer is the NOX-A12 (olaptesed pegol) that binds to the C-X-C motif chemokine ligand 12 (CXCL12), a chemokine ligand that is secreted by bone marrow stromal cells. Hoellenriegel et al.126 showed that NOX-A12 inhibits CXCL12-dependent activation of the chemokine receptor, CXCR4, interfering with intercellular signaling between tumor cells and their microenvironment. NOX-A12 is in a phase 1/2 combination trial in metastatic pancreatic and colorectal cancer together with the monoclonal antibody anti-PD-1, pembrolizumab (KEYTRUDA). In 2019, a second combination clinical trial of NOX-A12 with radiotherapy for brain cancer (GBM/glioma) treatment was initiated.
A search of the Clinical Trials database, using the search term “aptamer,” showed 47 clinical trials involving aptamers. A further restriction to cancer clinical trials limits this number to seven, as shown in Table 1. Delving further into these trials, two are observational, whereas one, listed as an intervention trial (ClinicalTrials.gov: NCT01830244), was, in fact, using the aptamer for follow-up review of biomarkers. Of the five remaining, one was withdrawn (ClinicalTrials.gov: NCT02780011), and one was terminated. The former was withdrawn due to lack of funding; it should be noted that this trial was using the aptamers in flow cytometry and not directly in the patient. Of the remaining clinical trials listed, two are listed as active but not yet recruiting (ClinicalTrials.gov: NCT04459468 and NCT02237183). The final trial (ClinicalTrials.gov: NCT03385148) is listed as recruiting; initial results of this trial indicate that the radiolabeled sgc8 aptamer with 68Gallium for diagnostic medical imaging can be used to distinguish between benign and malignant colorectal cancer.127 This study also demonstrated a favorable distribution profile and was safe and tolerated by patients. Given the number of aptamers that is being developed for both diagnostic and therapeutic applications, this suggests that there will be more clinical trials registered for these applications.128
Table 1.
Cancer aptamers in clinical trials
| NCT number/phase | Title | Conditions | Interventions | Outcome measures | Locations |
|---|---|---|---|---|---|
| ClinicalTrials.gov: NCT03385148 early phase I | the clinical application of 68Ga-labeled ssDNA aptamer sgc8 in healthy volunteers and colorectal patients | colorectal cancer | drug: 68Ga-sgc8 | diagnostic efficacy | China |
| ClinicalTrials.gov: NCT02780011 phase I | alisertib (MLN8237) and brentuximab vedotin for relapsed/refractory CD30-positive lymphomas and solid malignancies | CD30-positive lymphoma; CD30-positive solid tumor | drug: brentuximab vedotin; drug: alisertib | MTD; DLTs; RP2D; antitumor activity; area under the plasma concentration versus time curve | USA |
| ClinicalTrials.gov: NCT02237183 phase I | iloprost in preventing lung cancer in former smokers | lung carcinoma | drug: iloprost; other: placebo administration; other: quality-of-life assessment; other: questionnaire administration; descriptive statistics for the effect on serum proteins as quantitated by aptamer-based analysis | incidence of clinical toxicity; treatment compliance; response of airway histology; serum protein profiling; endobronchial-brushing gene expression; gene expression of dysplastic lesions; improvement in chronic obstructive pulmonary disease (COPD); whether the in vitro response of cultured airway epithelial progenitor cells to iloprost is a predictor of in vivo response in study subjects | USA |
| ClinicalTrials.gov: NCT01034410 phase II | a study of AS1411 combined with cytarabine in the treatment of patients with primary refractory or relapsed acute myeloid leukemia | acute myeloid leukemia | drug: AS1411; drug: cytarabine | comparison of 40 and 80 mg/kg/day of AS1411 in combination with cytarabine therapy or cytarabine alone for response rate; duration of emission; disease-free survival; overall survival; hematological recovery; safety; PD and PK | Australia, China, New Zealand, Taiwan, USA |
| ClinicalTrials.gov: NCT01830244 phase II | IST neoadjuvant Abraxane in newly diagnosed breast cancer | breast cancer | drug: nab-paclitaxel; post-biopsy aptamer assessment | pathological complete response in the breast; pathologic response rate in breast and axillary lymph nodes; rate of pathologic complete response and near-complete response in the breast combined; breast conservation rate; progression-free survival; safety and tolerability | Australia |
| ClinicalTrials.gov: NCT04459468 | identify proteomic biomarkers for outcome prediction of Lipiodol TACE treatment | hepatocellular carcinoma | drug: Lipiodol | predictive accuracy of proteomic biomarker(s) for overall survival in hepatocellular carcinoma patients; predictive accuracy of proteomic biomarker(s) for progression-free survival in hepatocellular carcinoma patients | USA |
| ClinicalTrials.gov: NCT02957370 | molecular biosensors for detection of bladder cancer | urinary bladder neoplasms | urinary “fingerprint” for urinary bladder neoplasms |
DLT, dose-limiting toxicity; IST, immunosuppressive therapy; MTD, maximum-tolerated dose; PK, pharmacokinetics; PD, pharmacodynamics; RP2D, recommended phase 2 dose; ssDNA, single-stranded DNA; TACE, transarterial chemoembolization.
Conclusions
RNA therapeutics is a rapidly growing field of biotherapeutics. These emerging modalities, including aptamers, siRNAs, and mRNA-based drugs, are based upon a powerful and versatile platform, which has nearly unlimited capacity to address unmet clinical needs. RNA therapeutics are destined to change the standard of care for many diseases. The number of RNA drugs under development and in clinical trials is growing rapidly. This rapid growth has been made possible by solving the problems surrounding stability, delivery, and immunogenicity of oligonucleotides. Although there is room for further improvement and innovation in each of these areas, the solutions have advanced to the point that RNA drugs are now feasible. Several dominating players in the RNA biopharma sector have emerged, and new, small biotech startups as well as academic groups with transformative ideas are also inching their way into the clinic.
More recently, the development of RNA drugs has largely focused on two modalities: (1) RNAi/anti-sense RNA where synthetic short oligonucleotides recognize and hybridize to complementary sequences in endogenous RNA transcripts and modulate their processing,129,130 and (2) mRNA where modified mRNAs encoding therapeutic proteins elicit their transient expression in the cytoplasm (for example, to replace defective or missing proteins or introduce antigens for vaccination).131,132 The realization of RNA as a drug required that several major hurdles be overcome. These hurdles include the following: (1) the rapid degradation of exogenous RNA by extracellular and intracellular RNases present in the serum and within tissues, (2) delivery of negatively charged RNA across the cellular membrane, which is hydrophobic in nature, and (3) strong immunogenicity of exogenous RNA, resulting in pronounced cell toxicity (i.e., cytokine storm) and impaired translation into therapeutic proteins (i.e., immune clearance).
These hurdles have been substantially overcome with recent advancements in RNA chemistry, biology, bioinformatics, manufacturing, and nanotechnology, thereby facilitating the recent rapid development of RNA therapeutics. Advantages of RNA-based drugs that are rapidly driving development include the following: (1) their ability to act on targets that are otherwise “undruggable” with small molecules or a protein-based drug; (2) their rapid and cost-effective development and manufacturing, in contrast to that of small molecules, recombinant proteins, and viral gene therapy approaches; and (3) the ability to rapidly alter the sequence of the RNA drug for personalized treatments or to adapt to an evolving pathogen (i.e., recent COVID-19 mRNA vaccines from Moderna and Pfizer/BioNTech).
The first approved and marketed RNA drug, patisiran (Onpattro; Alnylam Pharmaceuticals; FDA approval in August 2018) is an siRNA-based drug.133 Patisiran is used to treat adult patients with polyneuropathy caused by hereditary transthyretin (TTR)-mediated amyloidosis. This is a dsRNA that acts through the RNAi pathway and induces degradation of mRNA encoding TTR.133 Recently, another siRNA drug, givosiran (Givlaari; Alnylam Pharmaceuticals) received FDA approval in November 2019 for the treatment of acute hepatic porphyria.134 Givosiran targets aminolevulinate synthase 1 mRNA in the liver and reduces the levels of disease-causing neurotoxic intermediates aminolevulinic acid and porphobilinogen.134,135
The new kid on the block with respect to RNA therapeutics is synthetic or in vitro-translated mRNA. mRNA is uniquely engineered to mimic naturally occurring mRNAs.136,137 mRNA drugs consist of a single-stranded open reading frame flanked by untranslated regions (UTRs) and contain a 5′ cap for translation and a 3′ poly(A) tail for stability.137, 138, 139 The exogenously delivered mRNA is translated into protein in the cytoplasm and degrades within the cytoplasm typically within hours, thereby posing no risk of integration into the genome. There are several therapeutic modalities that utilize mRNA: (1) replacement therapy where the mRNA is administered to the patient to compensate for a defective/missing protein or to supply therapeutic proteins; (2) vaccination where the mRNA-encoding-specific antigen(s) are administered to elicit protective immunity; and (3) cell therapy where the mRNA is transfected into the cells ex vivo to alter cell phenotype or function, and then these cells are delivered into the patient.
In general, the development and manufacturing of RNA therapeutics (aptamers, siRNAs, and mRNAs) are relatively simple and a more cost-effective process in comparison to that of recombinant protein or small molecule drugs. In addition, RNA sequences can be easily modified allowing for personalization of RNA therapy.
Although aptamers were the first RNA drugs to be described, only one FDA-approved aptamer-based drug, pegaptanib (Macugen; Bausch + Lomb Pharmaceutical Retina Portfolio; FDA approval in December 2004) is on the market after more than 15 years.140 Although many other aptamer-based drugs are currently in clinical trials or in various stages of development (see section above),141 their entry into the clinical market has been slow when compared to the development pace of siRNAs and mRNA-based drugs. Despite these delays, aptamer aficionados vouch for the potential of aptamers to replace monoclonal antibodies in therapeutic and diagnostic applications because they can be produced via chemical synthesis, are more cost effective in manufacturing, are simpler to modify, and elicit little immunogenicity.142,143 However, despite the fact that aptamers have many advantages over antibodies, their clinical translation is challenging due to suboptimal pharmacokinetic properties (highly sensitive to nucleases and readily excreted by the kidneys) and complexity of selection techniques (a time-consuming process with low success rates).144 Many of these hurdles can be easily overcome with those same advancements in RNA chemistry, biology, bioinformatics, manufacturing, and nanotechnology that have enabled the entry of siRNAs and mRNAs into the clinic.145 Furthermore, in silico and artificial intelligence (AI)-based technologies are rapidly being applied to aptamer selection/design to enable the rapid and facile identification of aptamers against various therapeutic and diagnostic targets.
Acknowledgments
Author contributions
All authors contributed equally.
Declaration of interests
B.S. is co-founder of Sebastian Bio, which has intellectual property (IP) on the AS1411-siTAP chimera. P.H.G. is employed by Wave Life Sciences and receives salary and shares from Wave Life Sciences.
References
- 1.Pastor F., Berraondo P., Etxeberria I., Frederick J., Sahin U., Gilboa E., Melero I. An RNA toolbox for cancer immunotherapy. Nat. Rev. Drug Discov. 2018;17:751–767. doi: 10.1038/nrd.2018.132. [DOI] [PubMed] [Google Scholar]
- 2.Shu D., Zhang L., Bai X., Yu J., Guo P. Stoichiometry of multi-specific immune checkpoint RNA Abs for T cell activation and tumor inhibition using ultra-stable RNA nanoparticles. Mol. Ther. Nucleic Acids. 2021;24:426–435. doi: 10.1016/j.omtn.2021.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schrand B., Berezhnoy A., Brenneman R., Williams A., Levay A., Kong L.Y., Rao G., Zhou S., Heimberger A.B., Gilboa E. Targeting 4-1BB costimulation to the tumor stroma with bispecific aptamer conjugates enhances the therapeutic index of tumor immunotherapy. Cancer Immunol. Res. 2014;2:867–877. doi: 10.1158/2326-6066.CIR-14-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Soldevilla M.M., Villanueva H., Meraviglia-Crivelli D., Menon A.P., Ruiz M., Cebollero J., Villalba M., Moreno B., Lozano T., Llopiz D. ICOS Costimulation at the Tumor Site in Combination with CTLA-4 Blockade Therapy Elicits Strong Tumor Immunity. Mol. Ther. 2019;27:1878–1891. doi: 10.1016/j.ymthe.2019.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Soldevilla M.M., Villanueva H., Casares N., Lasarte J.J., Bendandi M., Inoges S., López-Díaz de Cerio A., Pastor F. MRP1-CD28 bi-specific oligonucleotide aptamers: target costimulation to drug-resistant melanoma cancer stem cells. Oncotarget. 2016;7:23182–23196. doi: 10.18632/oncotarget.8095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yarchoan M., Johnson B.A., 3rd, Lutz E.R., Laheru D.A., Jaffee E.M. Targeting neoantigens to augment antitumour immunity. Nat. Rev. Cancer. 2017;17:569. doi: 10.1038/nrc.2017.74. [DOI] [PubMed] [Google Scholar]
- 7.Pastor F., Kolonias D., Giangrande P.H., Gilboa E. Induction of tumour immunity by targeted inhibition of nonsense-mediated mRNA decay. Nature. 2010;465:227–230. doi: 10.1038/nature08999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garrido G., Schrand B., Rabasa A., Levay A., D’Eramo F., Berezhnoy A., Modi S., Gefen T., Marijt K., Doorduijn E. Tumor-targeted silencing of the peptide transporter TAP induces potent antitumor immunity. Nat. Commun. 2019;10:3773. doi: 10.1038/s41467-019-11728-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bates P.J., Reyes-Reyes E.M., Malik M.T., Murphy E.M., O’Toole M.G., Trent J.O. G-quadruplex oligonucleotide AS1411 as a cancer-targeting agent: Uses and mechanisms. Biochim. Biophys. Acta, Gen. Subj. 2017;1861(5 Pt B):1414–1428. doi: 10.1016/j.bbagen.2016.12.015. [DOI] [PubMed] [Google Scholar]
- 10.Yazdian-Robati R., Bayat P., Oroojalian F., Zargari M., Ramezani M., Taghdisi S.M., Abnous K. Therapeutic applications of AS1411 aptamer, an update review. Int. J. Biol. Macromol. 2020;155:1420–1431. doi: 10.1016/j.ijbiomac.2019.11.118. [DOI] [PubMed] [Google Scholar]
- 11.Santos T., Miranda A., Campello M.P.C., Paulo A., Salgado G., Cabrita E.J., Cruz C. Recognition of nucleolin through interaction with RNA G-quadruplex. Biochem. Pharmacol. 2021;189:114208. doi: 10.1016/j.bcp.2020.114208. [DOI] [PubMed] [Google Scholar]
- 12.Vigneron N., Ferrari V., Van den Eynde B.J., Cresswell P., Leonhardt R.M. Cytosolic Processing Governs TAP-Independent Presentation of a Critical Melanoma Antigen. J. Immunol. 2018;201:1875–1888. doi: 10.4049/jimmunol.1701479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marijt K.A., Blijleven L., Verdegaal E.M.E., Kester M.G., Kowalewski D.J., Rammensee H.G., Stevanović S., Heemskerk M.H.M., van der Burg S.H., van Hall T. Identification of non-mutated neoantigens presented by TAP-deficient tumors. J. Exp. Med. 2018;215:2325–2337. doi: 10.1084/jem.20180577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doorduijn E.M., Sluijter M., Marijt K.A., Querido B.J., van der Burg S.H., van Hall T. T cells specific for a TAP-independent self-peptide remain naïve in tumor-bearing mice and are fully exploitable for therapy. OncoImmunology. 2017;7:e1382793. doi: 10.1080/2162402X.2017.1382793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doorduijn E.M., Sluijter M., Querido B.J., Oliveira C.C., Achour A., Ossendorp F., van der Burg S.H., van Hall T. TAP-independent self-peptides enhance T cell recognition of immune-escaped tumors. J. Clin. Invest. 2016;126:784–794. doi: 10.1172/JCI83671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garrido G., Schrand B., Levay A., Rabasa A., Ferrantella A., Da Silva D.M., D’Eramo F., Marijt K.A., Zhang Z., Kwon D. Vaccination against Nonmutated Neoantigens Induced in Recurrent and Future Tumors. Cancer Immunol. Res. 2020;8:856–868. doi: 10.1158/2326-6066.CIR-20-0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y., Xie X., Yeganeh P.N., Lee D.-J., Valle-Garcia D., Meza-Sosa K.F., Junqueira C., Su J., Luo H.R., Hide W., Lieberman J. Immunotherapy for breast cancer using EpCAM aptamer tumor-targeted gene knockdown. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2022830118. e2022830118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoon S., Rossi J.J. Aptamers: Uptake mechanisms and intracellular applications. Adv. Drug Deliv. Rev. 2018;134:22–35. doi: 10.1016/j.addr.2018.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu G., Niu G., Chen X. Aptamer-Drug Conjugates. Bioconjug. Chem. 2015;26:2186–2197. doi: 10.1021/acs.bioconjchem.5b00291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tacar O., Sriamornsak P., Dass C.R. Doxorubicin: an update on anticancer molecular action, toxicity and novel drug delivery systems. J. Pharm. Pharmacol. 2013;65:157–170. doi: 10.1111/j.2042-7158.2012.01567.x. [DOI] [PubMed] [Google Scholar]
- 21.Xiang D., Shigdar S., Bean A.G., Bruce M., Yang W., Mathesh M., Wang T., Yin W., Tran P.H.-L., Al Shamaileh H. Transforming doxorubicin into a cancer stem cell killer via EpCAM aptamer-mediated delivery. Theranostics. 2017;7:4071–4086. doi: 10.7150/thno.20168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bagalkot V., Farokhzad O.C., Langer R., Jon S. An aptamer-doxorubicin physical conjugate as a novel targeted drug-delivery platform. Angew. Chem. Int. Ed. Engl. 2006;45:8149–8152. doi: 10.1002/anie.200602251. [DOI] [PubMed] [Google Scholar]
- 23.Moses J.E., Moorhouse A.D. The growing applications of click chemistry. Chem. Soc. Rev. 2007;36:1249–1262. doi: 10.1039/b613014n. [DOI] [PubMed] [Google Scholar]
- 24.Macdonald J., Denoyer D., Henri J., Jamieson A., Burvenich I.J.G., Pouliot N., Shigdar S. Bifunctional Aptamer-Doxorubicin Conjugate Crosses the Blood-Brain Barrier and Selectively Delivers Its Payload to Epithelial Cell Adhesion Molecule-Positive Tumor Cells. Nucleic Acid Ther. 2020;30:117–128. doi: 10.1089/nat.2019.0807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang Y.F., Shangguan D., Liu H., Phillips J.A., Zhang X., Chen Y., Tan W. Molecular assembly of an aptamer-drug conjugate for targeted drug delivery to tumor cells. ChemBioChem. 2009;10:862–868. doi: 10.1002/cbic.200800805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Münz M., Murr A., Kvesic M., Rau D., Mangold S., Pflanz S., Lumsden J., Volkland J., Fagerberg J., Riethmüller G. Side-by-side analysis of five clinically tested anti-EpCAM monoclonal antibodies. Cancer Cell Int. 2010;10:44. doi: 10.1186/1475-2867-10-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shigdar S., Macdonald J., O’Connor M., Wang T., Xiang D., Al Shamaileh H., Qiao L., Wei M., Zhou S.-F., Zhu Y. Aptamers as theranostic agents: modifications, serum stability and functionalisation. Sensors (Basel) 2013;13:13624–13637. doi: 10.3390/s131013624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cartledge Wolf D.M., Langhans S.A. Moving Myeloid Leukemia Drug Discovery Into the Third Dimension. Front Pediatr. 2019;7:314. doi: 10.3389/fped.2019.00314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haubner S., Perna F., Köhnke T., Schmidt C., Berman S., Augsberger C., Schnorfeil F.M., Krupka C., Lichtenegger F.S., Liu X. Coexpression profile of leukemic stem cell markers for combinatorial targeted therapy in AML. Leukemia. 2019;33:64–74. doi: 10.1038/s41375-018-0180-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shangguan D., Li Y., Tang Z., Cao Z.C., Chen H.W., Mallikaratchy P., Sefah K., Yang C.J., Tan W. Aptamers evolved from live cells as effective molecular probes for cancer study. Proc. Natl. Acad. Sci. USA. 2006;103:11838–11843. doi: 10.1073/pnas.0602615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shangguan D., Cao Z., Meng L., Mallikaratchy P., Sefah K., Wang H., Li Y., Tan W. Cell-specific aptamer probes for membrane protein elucidation in cancer cells. J. Proteome Res. 2008;7:2133–2139. doi: 10.1021/pr700894d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiao Z., Shangguan D., Cao Z., Fang X., Tan W. Cell-specific internalization study of an aptamer from whole cell selection. Chemistry. 2008;14:1769–1775. doi: 10.1002/chem.200701330. [DOI] [PubMed] [Google Scholar]
- 33.Taghdisi S.M., Abnous K., Mosaffa F., Behravan J. Targeted delivery of daunorubicin to T-cell acute lymphoblastic leukemia by aptamer. J. Drug Target. 2010;18:277–281. doi: 10.3109/10611860903434050. [DOI] [PubMed] [Google Scholar]
- 34.Shangguan D., Tang Z., Mallikaratchy P., Xiao Z., Tan W. Optimization and modifications of aptamers selected from live cancer cell lines. ChemBioChem. 2007;8:603–606. doi: 10.1002/cbic.200600532. [DOI] [PubMed] [Google Scholar]
- 35.Sicco E., Baez J., Ibarra M., Fernández M., Cabral P., Moreno M., Cerecetto H., Calzada V. Sgc8-c Aptamer as a Potential Theranostic Agent for Hemato-Oncological Malignancies. Cancer Biother. Radiopharm. 2020;35:262–270. doi: 10.1089/cbr.2019.3402. [DOI] [PubMed] [Google Scholar]
- 36.Yang S., Li H., Xu L., Deng Z., Han W., Liu Y., Jiang W., Zu Y. Oligonucleotide Aptamer-Mediated Precision Therapy of Hematological Malignancies. Mol. Ther. Nucleic Acids. 2018;13:164–175. doi: 10.1016/j.omtn.2018.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao N., Pei S.-N., Qi J., Zeng Z., Iyer S.P., Lin P., Tung C.-H., Zu Y. Oligonucleotide aptamer-drug conjugates for targeted therapy of acute myeloid leukemia. Biomaterials. 2015;67:42–51. doi: 10.1016/j.biomaterials.2015.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goto H., Grantham R.W., Mizuuchi M., Yamaguchi T., Teshima T., Reed J.C., Matsuzawa S.-i. PTK7 Is Involved in the Chemotherapy Resistance of T-Cell Acute Lymphoblastic Leukemia through Interaction with FADD-Death Domain. Blood. 2015;126:3701. [Google Scholar]
- 39.Wojtuszkiewicz A., Peters G.J., van Woerden N.L., Dubbelman B., Escherich G., Schmiegelow K., Sonneveld E., Pieters R., van de Ven P.M., Jansen G. Methotrexate resistance in relation to treatment outcome in childhood acute lymphoblastic leukemia. J. Hematol. Oncol. 2015;8:61. doi: 10.1186/s13045-015-0158-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tapia-Alveal C., Olsen T.R., Worgall T.S. Personalized immunoglobulin aptamers for detection of multiple myeloma minimal residual disease in serum. Commun. Biol. 2020;3:781. doi: 10.1038/s42003-020-01515-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Macdonald J., Henri J., Roy K., Hays E., Bauer M., Veedu R.N., Pouliot N., Shigdar S. EpCAM Immunotherapy versus Specific Targeted Delivery of Drugs. Cancers (Basel) 2018;10:19. doi: 10.3390/cancers10010019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiang D., Zheng C., Zhou S.-F., Qiao S., Tran P.H.-L., Pu C., Li Y., Kong L., Kouzani A.Z., Lin J. Superior Performance of Aptamer in Tumor Penetration over Antibody: Implication of Aptamer-Based Theranostics in Solid Tumors. Theranostics. 2015;5:1083–1097. doi: 10.7150/thno.11711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hanahan D., Weinberg R.A. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 44.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 45.Zugazagoitia J., Guedes C., Ponce S., Ferrer I., Molina-Pinelo S., Paz-Ares L. Current Challenges in Cancer Treatment. Clin. Ther. 2016;38:1551–1566. doi: 10.1016/j.clinthera.2016.03.026. [DOI] [PubMed] [Google Scholar]
- 46.Marquart J., Chen E.Y., Prasad V. Estimation of the Percentage of US Patients With Cancer Who Benefit From Genome-Driven Oncology. JAMA Oncol. 2018;4:1093–1098. doi: 10.1001/jamaoncol.2018.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haslam A., Prasad V. Estimation of the Percentage of US Patients With Cancer Who Are Eligible for and Respond to Checkpoint Inhibitor Immunotherapy Drugs. JAMA Netw. Open. 2019;2:e192535. doi: 10.1001/jamanetworkopen.2019.2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maldonado E.B., Parsons S., Chen E.Y., Haslam A., Prasad V. Estimation of US patients with cancer who may respond to cytotoxic chemotherapy. Future Sci. OA. 2020;6:FSO600. doi: 10.2144/fsoa-2020-0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paluncic J., Kovacevic Z., Jansson P.J., Kalinowski D., Merlot A.M., Huang M.L.H., Lok H.C., Sahni S., Lane D.J.R., Richardson D.R. Roads to melanoma: Key pathways and emerging players in melanoma progression and oncogenic signaling. Biochim. Biophys. Acta. 2016;1863:770–784. doi: 10.1016/j.bbamcr.2016.01.025. [DOI] [PubMed] [Google Scholar]
- 50.Serrone L., Zeuli M., Sega F.M., Cognetti F. Dacarbazine-based chemotherapy for metastatic melanoma: thirty-year experience overview. Journal of experimental & clinical cancer research. J. Exp. Clin. Canc. Res. 2000;19:21–34. [PubMed] [Google Scholar]
- 51.Wróbel S., Przybyło M., Stępień E. The Clinical Trial Landscape for Melanoma Therapies. J. Clin. Med. 2019;8:368. doi: 10.3390/jcm8030368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Larkin J., Chiarion-Sileni V., Gonzalez R., Grob J.-J., Rutkowski P., Lao C.D., Cowey C.L., Schadendorf D., Wagstaff J., Dummer R. Five-Year Survival with Combined Nivolumab and Ipilimumab in Advanced Melanoma. N. Engl. J. Med. 2019;381:1535–1546. doi: 10.1056/NEJMoa1910836. [DOI] [PubMed] [Google Scholar]
- 53.Ugurel S., Paschen A., Becker J.C. Dacarbazine in melanoma: from a chemotherapeutic drug to an immunomodulating agent. J. Invest. Dermatol. 2013;133:289–292. doi: 10.1038/jid.2012.341. [DOI] [PubMed] [Google Scholar]
- 54.Yamazaki N., Uhara H., Fukushima S., Uchi H., Shibagaki N., Kiyohara Y., Tsutsumida A., Namikawa K., Okuyama R., Otsuka Y., Tokudome T. Phase II study of the immune-checkpoint inhibitor ipilimumab plus dacarbazine in Japanese patients with previously untreated, unresectable or metastatic melanoma. Cancer Chemother. Pharmacol. 2015;76:969–975. doi: 10.1007/s00280-015-2870-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maio M., Grob J.J., Aamdal S., Bondarenko I., Robert C., Thomas L., Garbe C., Chiarion-Sileni V., Testori A., Chen T.T. Five-year survival rates for treatment-naive patients with advanced melanoma who received ipilimumab plus dacarbazine in a phase III trial. J. Clin. Oncol. 2015;33:1191–1196. doi: 10.1200/JCO.2014.56.6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pastor F., Soldevilla M.M., Villanueva H., Kolonias D., Inoges S., de Cerio A.L., Kandzia R., Klimyuk V., Gleba Y., Gilboa E., Bendandi M. CD28 aptamers as powerful immune response modulators. Mol. Ther. Nucleic Acids. 2013;2:e98. doi: 10.1038/mtna.2013.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lemery S., Keegan P., Pazdur R. First FDA Approval Agnostic of Cancer Site - When a Biomarker Defines the Indication. N. Engl. J. Med. 2017;377:1409–1412. doi: 10.1056/NEJMp1709968. [DOI] [PubMed] [Google Scholar]
- 58.Kryza D., Debordeaux F., Azéma L., Hassan A., Paurelle O., Schulz J., Savona-Baron C., Charignon E., Bonazza P., Taleb J. Ex Vivo and In Vivo Imaging and Biodistribution of Aptamers Targeting the Human Matrix MetalloProtease-9 in Melanomas. PLoS ONE. 2016;11:e0149387. doi: 10.1371/journal.pone.0149387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ojima A., Matsui T., Maeda S., Takeuchi M., Inoue H., Higashimoto Y., Yamagishi S. DNA aptamer raised against advanced glycation end products inhibits melanoma growth in nude mice. Lab. Invest. 2014;94:422–429. doi: 10.1038/labinvest.2014.5. [DOI] [PubMed] [Google Scholar]
- 60.Nakamura N., Matsui T., Ishibashi Y., Sotokawauchi A., Fukami K., Higashimoto Y., Yamagishi S.-I. RAGE-aptamer Attenuates the Growth and Liver Metastasis of Malignant Melanoma in Nude Mice. Mol. Med. 2017;23:295–306. doi: 10.2119/molmed.2017.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li H., Liu J., Xiao X., Sun S., Zhang H., Zhang Y., Zhou W., Zhang B., Roy M., Liu H. A Novel Aptamer LL4A Specifically Targets Vemurafenib-Resistant Melanoma through Binding to the CD63 Protein. Mol. Ther. Nucleic Acids. 2019;18:727–738. doi: 10.1016/j.omtn.2019.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hays E.M., Duan W., Shigdar S. Aptamers and Glioblastoma: Their Potential Use for Imaging and Therapeutic Applications. Int. J. Mol. Sci. 2017;18:2576. doi: 10.3390/ijms18122576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bukari B., Samarasinghe R.M., Noibanchong J., Shigdar S.L. Non-Invasive Delivery of Therapeutics into the Brain: The Potential of Aptamers for Targeted Delivery. Biomedicines. 2020;8:120. doi: 10.3390/biomedicines8050120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shi N., Pardridge W.M. Noninvasive gene targeting to the brain. Proc. Natl. Acad. Sci. USA. 2000;97:7567–7572. doi: 10.1073/pnas.130187497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shi N., Boado R.J., Pardridge W.M. Receptor-mediated gene targeting to tissues in vivo following intravenous administration of pegylated immunoliposomes. Pharm. Res. 2001;18:1091–1095. doi: 10.1023/a:1010910523202. [DOI] [PubMed] [Google Scholar]
- 66.Mannironi C., Di Nardo A., Fruscoloni P., Tocchini-Valentini G.P. In vitro selection of dopamine RNA ligands. Biochemistry. 1997;36:9726–9734. doi: 10.1021/bi9700633. [DOI] [PubMed] [Google Scholar]
- 67.McConnell E.M., Ventura K., Dwyer Z., Hunt V., Koudrina A., Holahan M.R., DeRosa M.C. In Vivo Use of a Multi-DNA Aptamer-Based Payload/Targeting System To Study Dopamine Dysregulation in the Central Nervous System. ACS Chem. Neurosci. 2019;10:371–383. doi: 10.1021/acschemneuro.8b00292. [DOI] [PubMed] [Google Scholar]
- 68.Macdonald J., Henri J., Goodman L., Xiang D., Duan W., Shigdar S. Development of a Bifunctional Aptamer Targeting the Transferrin Receptor and Epithelial Cell Adhesion Molecule (EpCAM) for the Treatment of Brain Cancer Metastases. ACS Chem. Neurosci. 2017;8:777–784. doi: 10.1021/acschemneuro.6b00369. [DOI] [PubMed] [Google Scholar]
- 69.Imrich S., Hachmeister M., Gires O. EpCAM and its potential role in tumor-initiating cells. Cell Adhes. Migr. 2012;6:30–38. doi: 10.4161/cam.18953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li X., Yang Y., Zhao H., Zhu T., Yang Z., Xu H., Fu Y., Lin F., Pan X., Li L. Enhanced in Vivo Blood-Brain Barrier Penetration by Circular Tau-Transferrin Receptor Bifunctional Aptamer for Tauopathy Therapy. J. Am. Chem. Soc. 2020;142:3862–3872. doi: 10.1021/jacs.9b11490. [DOI] [PubMed] [Google Scholar]
- 71.Hosney M., Sabet S., El-Shinawi M., Gaafar K.M., Mohamed M.M. Leptin is overexpressed in the tumor microenvironment of obese patients with estrogen receptor positive breast cancer. Exp. Ther. Med. 2017;13:2235–2246. doi: 10.3892/etm.2017.4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zuchero Y.J., Chen X., Bien-Ly N., Bumbaca D., Tong R.K., Gao X., Zhang S., Hoyte K., Luk W., Huntley M.A. Discovery of Novel Blood-Brain Barrier Targets to Enhance Brain Uptake of Therapeutic Antibodies. Neuron. 2016;89:70–82. doi: 10.1016/j.neuron.2015.11.024. [DOI] [PubMed] [Google Scholar]
- 73.Yu Y.J., Zhang Y., Kenrick M., Hoyte K., Luk W., Lu Y., Atwal J., Elliott J.M., Prabhu S., Watts R.J., Dennis M.S. Boosting brain uptake of a therapeutic antibody by reducing its affinity for a transcytosis target. Sci. Transl. Med. 2011;3:84ra44. doi: 10.1126/scitranslmed.3002230. [DOI] [PubMed] [Google Scholar]
- 74.Cheng C., Chen Y.H., Lennox K.A., Behlke M.A., Davidson B.L. In vivo SELEX for Identification of Brain-penetrating Aptamers. Mol. Ther. Nucleic Acids. 2013;2:e67. doi: 10.1038/mtna.2012.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nuzzo S., Brancato V., Affinito A., Salvatore M., Cavaliere C., Condorelli G. The Role of RNA and DNA Aptamers in Glioblastoma Diagnosis and Therapy: A Systematic Review of the Literature. Cancers (Basel) 2020;12:2173. doi: 10.3390/cancers12082173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Amero P., Khatua S., Rodriguez-Aguayo C., Lopez-Berestein G. Aptamers: Novel Therapeutics and Potential Role in Neuro-Oncology. Cancers (Basel) 2020;12:2889. doi: 10.3390/cancers12102889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shigdar S., Qiao L., Zhou S.F., Xiang D., Wang T., Li Y., Lim L.Y., Kong L., Li L., Duan W. RNA aptamers targeting cancer stem cell marker CD133. Cancer Lett. 2013;330:84–95. doi: 10.1016/j.canlet.2012.11.032. [DOI] [PubMed] [Google Scholar]
- 78.Shigdar S., Lin J., Li Y., Yang C.J., Wei M., Zhus Y., Liu H., Duan W. Cancer stem cell targeting: the next generation of cancer therapy and molecular imaging. Ther. Deliv. 2012;3:227–244. doi: 10.4155/tde.11.148. [DOI] [PubMed] [Google Scholar]
- 79.Zhou G., Da Won Bae S., Nguyen R., Huo X., Han S., Zhang Z., Hebbard L., Duan W., Eslam M., Liddle C. An aptamer-based drug delivery agent (CD133-apt-Dox) selectively and effectively kills liver cancer stem-like cells. Cancer Lett. 2021;501:124–132. doi: 10.1016/j.canlet.2020.12.022. [DOI] [PubMed] [Google Scholar]
- 80.Shigdar S., Lin J., Yu Y., Pastuovic M., Wei M., Duan W. RNA aptamer against a cancer stem cell marker epithelial cell adhesion molecule. Cancer Sci. 2011;102:991–998. doi: 10.1111/j.1349-7006.2011.01897.x. [DOI] [PubMed] [Google Scholar]
- 81.Song Y., Zhu Z., An Y., Zhang W., Zhang H., Liu D., Yu C., Duan W., Yang C.J. Selection of DNA aptamers against epithelial cell adhesion molecule for cancer cell imaging and circulating tumor cell capture. Anal. Chem. 2013;85:4141–4149. doi: 10.1021/ac400366b. [DOI] [PubMed] [Google Scholar]
- 82.Alshaer W., Ababneh N., Hatmal M., Izmirli H., Choukeife M., Shraim A., Sharar N., Abu-Shiekah A., Odeh F., Al Bawab A. Selection and targeting of EpCAM protein by ssDNA aptamer. PLoS ONE. 2017;12:e0189558. doi: 10.1371/journal.pone.0189558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Alshaer W., Hillaireau H., Vergnaud J., Mura S., Deloménie C., Sauvage F., Ismail S., Fattal E. Aptamer-guided siRNA-loaded nanomedicines for systemic gene silencing in CD-44 expressing murine triple-negative breast cancer model. J. Control. Release. 2018;271:98–106. doi: 10.1016/j.jconrel.2017.12.022. [DOI] [PubMed] [Google Scholar]
- 84.Zheng J., Zhao S., Yu X., Huang S., Liu H.Y. Simultaneous targeting of CD44 and EpCAM with a bispecific aptamer effectively inhibits intraperitoneal ovarian cancer growth. Theranostics. 2017;7:1373–1388. doi: 10.7150/thno.17826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bao S., Wu Q., McLendon R.E., Hao Y., Shi Q., Hjelmeland A.B., Dewhirst M.W., Bigner D.D., Rich J.N. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 86.Lathia J.D., Gallagher J., Myers J.T., Li M., Vasanji A., McLendon R.E., Hjelmeland A.B., Huang A.Y., Rich J.N. Direct in vivo evidence for tumor propagation by glioblastoma cancer stem cells. PLoS ONE. 2011;6:e24807. doi: 10.1371/journal.pone.0024807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sherry M.M., Reeves A., Wu J.K., Cochran B.H. STAT3 is required for proliferation and maintenance of multipotency in glioblastoma stem cells. Stem Cells. 2009;27:2383–2392. doi: 10.1002/stem.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Carro M.S., Lim W.K., Alvarez M.J., Bollo R.J., Zhao X., Snyder E.Y., Sulman E.P., Anne S.L., Doetsch F., Colman H. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463:318–325. doi: 10.1038/nature08712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yoon S., Wu X., Armstrong B., Habib N., Rossi J.J. An RNA Aptamer Targeting the Receptor Tyrosine Kinase PDGFRα Induces Anti-tumor Effects through STAT3 and p53 in Glioblastoma. Mol. Ther. Nucleic Acids. 2019;14:131–141. doi: 10.1016/j.omtn.2018.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Catuogno S., Esposito C.L., Giangrande P.H. Stick-based methods for aptamer-mediated siRNA targeted delivery. In: Ditzel H.J., Tuttolomondo M., Kauppinen S., editors. Design and Delivery of siRNA Therapeutics. Springer US; New York, NY: 2021. pp. 31–42. [DOI] [PubMed] [Google Scholar]
- 91.Esposito C.L., Nuzzo S., Kumar S.A., Rienzo A., Lawrence C.L., Pallini R., Shaw L., Alder J.E., Ricci-Vitiani L., Catuogno S., de Franciscis V. A combined microRNA-based targeted therapeutic approach to eradicate glioblastoma stem-like cells. J. Control. Release. 2016;238:43–57. doi: 10.1016/j.jconrel.2016.07.032. [DOI] [PubMed] [Google Scholar]
- 92.Esposito C.L., Nuzzo S., Catuogno S., Romano S., de Nigris F., de Franciscis V. STAT3 Gene Silencing by Aptamer-siRNA Chimera as Selective Therapeutic for Glioblastoma. Mol. Ther. Nucleic Acids. 2018;10:398–411. doi: 10.1016/j.omtn.2017.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Esposito C.L., Nuzzo S., Ibba M.L., Ricci-Vitiani L., Pallini R., Condorelli G., Catuogno S., de Franciscis V. Combined Targeting of Glioblastoma Stem-Like Cells by Neutralizing RNA-Biol.-Drugs for STAT3. Cancers. 2020;12:1434. doi: 10.3390/cancers12061434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Affinito A., Quintavalle C., Esposito C.L., Roscigno G., Vilardo C., Nuzzo S., Ricci-Vitiani L., De Luca G., Pallini R., Kichkailo A.S. The Discovery of RNA Aptamers that Selectively Bind Glioblastoma Stem Cells. Mol. Ther. Nucleic Acids. 2019;18:99–109. doi: 10.1016/j.omtn.2019.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Affinito A., Quintavalle C., Esposito C.L., Roscigno G., Giordano C., Nuzzo S., Ricci-Vitiani L., Scognamiglio I., Minic Z., Pallini R. Targeting Ephrin Receptor Tyrosine Kinase A2 with a Selective Aptamer for Glioblastoma Stem Cells. Mol. Ther. Nucleic Acids. 2020;20:176–185. doi: 10.1016/j.omtn.2020.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.de Franciscis V. Challenging cancer targets for aptamer delivery. Biochimie. 2018;145:45–52. doi: 10.1016/j.biochi.2017.09.014. [DOI] [PubMed] [Google Scholar]
- 97.Kwok A., Raulf N., Habib N. Developing small activating RNA as a therapeutic: current challenges and promises. Ther. Deliv. 2019;10:151–164. doi: 10.4155/tde-2018-0061. [DOI] [PubMed] [Google Scholar]
- 98.Kotula J.W., Pratico E.D., Ming X., Nakagawa O., Juliano R.L., Sullenger B.A. Aptamer-mediated delivery of splice-switching oligonucleotides to the nuclei of cancer cells. Nucleic Acid Ther. 2012;22:187–195. doi: 10.1089/nat.2012.0347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ming X., Alam M.R., Fisher M., Yan Y., Chen X., Juliano R.L. Intracellular delivery of an antisense oligonucleotide via endocytosis of a G protein-coupled receptor. Nucleic Acids Res. 2010;38:6567–6576. doi: 10.1093/nar/gkq534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mol A.A., Groher F., Schreiber B., Rühmkorff C., Suess B. Robust gene expression control in human cells with a novel universal TetR aptamer splicing module. Nucleic Acids Res. 2019;47:e132. doi: 10.1093/nar/gkz753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Li X., Wu X., Yang H., Li L., Ye Z., Rao Y. A nuclear targeted Dox-aptamer loaded liposome delivery platform for the circumvention of drug resistance in breast cancer. Biomed. Pharmacother. 2019;117:109072. doi: 10.1016/j.biopha.2019.109072. [DOI] [PubMed] [Google Scholar]
- 102.Janowski B.A., Younger S.T., Hardy D.B., Ram R., Huffman K.E., Corey D.R. Activating gene expression in mammalian cells with promoter-targeted duplex RNAs. Nat. Chem. Biol. 2007;3:166–173. doi: 10.1038/nchembio860. [DOI] [PubMed] [Google Scholar]
- 103.Corey D.R. RNA-Mediated Gene Activation: Identifying a Candidate RNA for Preclinical Development. Adv. Exp. Med. Biol. 2017;983:161–171. doi: 10.1007/978-981-10-4310-9_11. [DOI] [PubMed] [Google Scholar]
- 104.Li C., Jiang W., Hu Q., Li L.C., Dong L., Chen R., Zhang Y., Tang Y., Thrasher J.B., Liu C.B., Li B. Enhancing DPYSL3 gene expression via a promoter-targeted small activating RNA approach suppresses cancer cell motility and metastasis. Oncotarget. 2016;7:22893–22910. doi: 10.18632/oncotarget.8290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Reebye V., Sætrom P., Mintz P.J., Huang K.W., Swiderski P., Peng L., Liu C., Liu X., Lindkaer-Jensen S., Zacharoulis D. Novel RNA oligonucleotide improves liver function and inhibits liver carcinogenesis in vivo. Hepatology. 2014;59:216–227. doi: 10.1002/hep.26669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang H., Iakova P., Wilde M., Welm A., Goode T., Roesler W.J., Timchenko N.A. C/EBPalpha arrests cell proliferation through direct inhibition of Cdk2 and Cdk4. Mol. Cell. 2001;8:817–828. doi: 10.1016/s1097-2765(01)00366-5. [DOI] [PubMed] [Google Scholar]
- 107.Yoon S., Huang K.W., Andrikakou P., Vasconcelos D., Swiderski P., Reebye V., Sodergren M., Habib N., Rossi J.J. Targeted Delivery of C/EBPα-saRNA by RNA Aptamers Shows Anti-tumor Effects in a Mouse Model of Advanced PDAC. Mol. Ther. Nucleic Acids. 2019;18:142–154. doi: 10.1016/j.omtn.2019.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yoon S., Huang K.-W., Reebye V., Mintz P., Tien Y.-W., Lai H.-S., Sætrom P., Reccia I., Swiderski P., Armstrong B. Targeted Delivery of C/EBPα -saRNA by Pancreatic Ductal Adenocarcinoma-specific RNA Aptamers Inhibits Tumor Growth In Vivo. Mol. Ther. 2016;24:1106–1116. doi: 10.1038/mt.2016.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Dassie J.P., Liu X.Y., Thomas G.S., Whitaker R.M., Thiel K.W., Stockdale K.R., Meyerholz D.K., McCaffrey A.P., McNamara J.O., 2nd, Giangrande P.H. Systemic administration of optimized aptamer-siRNA chimeras promotes regression of PSMA-expressing tumors. Nat. Biotechnol. 2009;27:839–849. doi: 10.1038/nbt.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhou J., Rossi J. Aptamers as targeted therapeutics: current potential and challenges. Nat. Rev. Drug Discov. 2017;16:181–202. doi: 10.1038/nrd.2016.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Alshaer W., Hillaireau H., Fattal E. Aptamer-guided nanomedicines for anticancer drug delivery. Adv. Drug Deliv. Rev. 2018;134:122–137. doi: 10.1016/j.addr.2018.09.011. [DOI] [PubMed] [Google Scholar]
- 112.Lv C., Yang C., Ding D., Sun Y., Wang R., Han D., Tan W. Endocytic Pathways and Intracellular Transport of Aptamer-Drug Conjugates in Live Cells Monitored by Single-Particle Tracking. Anal. Chem. 2019;91:13818–13823. doi: 10.1021/acs.analchem.9b03281. [DOI] [PubMed] [Google Scholar]
- 113.Berger H., Wodarz A., Borchers A. PTK7 Faces the Wnt in Development and Disease. Front. Cell Dev. Biol. 2017;5:31. doi: 10.3389/fcell.2017.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.De La Fuente A., Zilio S., Caroli J., Van Simaeys D., Mazza E.M.C., Ince T.A., Bronte V., Bicciato S., Weed D.T., Serafini P. Aptamers against mouse and human tumor-infiltrating myeloid cells as reagents for targeted chemotherapy. Sci. Transl. Med. 2020;12:eaav9760. doi: 10.1126/scitranslmed.aav9760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Apolloni E., Bronte V., Mazzoni A., Serafini P., Cabrelle A., Segal D.M., Young H.A., Zanovello P. Immortalized myeloid suppressor cells trigger apoptosis in antigen-activated T lymphocytes. J. Immunol. 2000;165:6723–6730. doi: 10.4049/jimmunol.165.12.6723. [DOI] [PubMed] [Google Scholar]
- 116.Jiang F., Liu B., Lu J., Li F., Li D., Liang C., Dang L., Liu J., He B., Badshah S.A. Progress and Challenges in Developing Aptamer-Functionalized Targeted Drug Delivery Systems. Int. J. Mol. Sci. 2015;16:23784–23822. doi: 10.3390/ijms161023784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Fattal E., Hillaireau H., Ismail S.I. Aptamers in Therapeutics and Drug Delivery. Adv. Drug Deliv. Rev. 2018;134:1–2. doi: 10.1016/j.addr.2018.11.001. [DOI] [PubMed] [Google Scholar]
- 118.Ababneh N., Alshaer W., Allozi O., Mahafzah A., El-Khateeb M., Hillaireau H., Noiray M., Fattal E., Ismail S. In vitro selection of modified RNA aptamers against CD44 cancer stem cell marker. Nucleic Acid Ther. 2013;23:401–407. doi: 10.1089/nat.2013.0423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Biswas S., Torchilin V.P. Dendrimers for siRNA Delivery. Pharmaceuticals (Basel) 2013;6:161–183. doi: 10.3390/ph6020161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhang H.J., Zhao X., Chen L.J., Yang C.X., Yan X.P. Dendrimer grafted persistent luminescent nanoplatform for aptamer guided tumor imaging and acid-responsive drug delivery. Talanta. 2020;219:121209. doi: 10.1016/j.talanta.2020.121209. [DOI] [PubMed] [Google Scholar]
- 121.Le J., Xu J., Zheng J., Li B., Zheng T., Lu Y., Shen W., Kudryavtseva A.V., Katanaev V.L., Shao J., Jia L. One nanometer self-assembled aptamer-DNA dendrimers carry 350 doxorubicin: Super-stability and intra-nuclear DNA comet tail. Chem. Eng. J. 2020;388:124170. [Google Scholar]
- 122.Povsic T.J., Lawrence M.G., Lincoff A.M., Mehran R., Rusconi C.P., Zelenkofske S.L., Huang Z., Sailstad J., Armstrong P.W., Steg P.G., REGULATE-PCI Investigators Pre-existing anti-PEG antibodies are associated with severe immediate allergic reactions to pegnivacogin, a PEGylated aptamer. J. Allergy Clin. Immunol. 2016;138:1712–1715. doi: 10.1016/j.jaci.2016.04.058. [DOI] [PubMed] [Google Scholar]
- 123.Bruno J.G. Potential Inherent Stimulation of the Innate Immune System by Nucleic Acid Aptamers and Possible Corrective Approaches. Pharmaceuticals (Basel) 2018;11:62. doi: 10.3390/ph11030062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Eulberg D., Klussmann S. Spiegelmers: biostable aptamers. ChemBioChem. 2003;4:979–983. doi: 10.1002/cbic.200300663. [DOI] [PubMed] [Google Scholar]
- 125.Vater A., Klussmann S. Turning mirror-image oligonucleotides into drugs: the evolution of Spiegelmer(®) therapeutics. Drug Discov. Today. 2015;20:147–155. doi: 10.1016/j.drudis.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 126.Hoellenriegel J., Zboralski D., Maasch C., Rosin N.Y., Wierda W.G., Keating M.J., Kruschinski A., Burger J.A. The Spiegelmer NOX-A12, a novel CXCL12 inhibitor, interferes with chronic lymphocytic leukemia cell motility and causes chemosensitization. Blood. 2014;123:1032–1039. doi: 10.1182/blood-2013-03-493924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zhang M., Wang Z., Wang S., Yang W., Wang J., Chen X. The first application of 68Ga labeled ssDNAAptamer Sgc8 in Colorectal Patients. J. Nucl. Med. 2018;59(Suppl 1):53. [Google Scholar]
- 128.Khalid U., Vi C., Henri J., Macdonald J., Eu P., Mandarano G., Shigdar S. Radiolabelled Aptamers for Theranostic Treatment of Cancer. Pharmaceuticals (Basel) 2018;12:2. doi: 10.3390/ph12010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Stephenson M.L., Zamecnik P.C. Inhibition of Rous sarcoma viral RNA translation by a specific oligodeoxyribonucleotide. Proc. Natl. Acad. Sci. USA. 1978;75:285–288. doi: 10.1073/pnas.75.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Shen X., Corey D.R. Chemistry, mechanism and clinical status of antisense oligonucleotides and duplex RNAs. Nucleic Acids Res. 2018;46:1584–1600. doi: 10.1093/nar/gkx1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wolff J.A., Malone R.W., Williams P., Chong W., Acsadi G., Jani A., Felgner P.L. Direct gene transfer into mouse muscle in vivo. Science. 1990;247:1465–1468. doi: 10.1126/science.1690918. [DOI] [PubMed] [Google Scholar]
- 132.Ulmer J.B., Geall A.J. Recent innovations in mRNA vaccines. Curr. Opin. Immunol. 2016;41:18–22. doi: 10.1016/j.coi.2016.05.008. [DOI] [PubMed] [Google Scholar]
- 133.Kristen A.V., Ajroud-Driss S., Conceição I., Gorevic P., Kyriakides T., Obici L. Patisiran, an RNAi therapeutic for the treatment of hereditary transthyretin-mediated amyloidosis. Neurodegener. Dis. Manag. 2019;9:5–23. doi: 10.2217/nmt-2018-0033. [DOI] [PubMed] [Google Scholar]
- 134.Alnylam Pharmaceuticals . 2019. Alnylam Announces Approval of GIVLAARI™ (givosiran) by the U.S. Food and Drug Administration (FDA)https://www.businesswire.com/news/home/20191120005849/en/Alnylam-Announces-Approval-of-GIVLAARI%E2%84%A2-givosiran-by-the-U.S.-Food-and-Drug-Administration-FDA [Google Scholar]
- 135.Alnylam Pharmaceuticals . 2019. Introducing GIVLAARI™ (givosiran). Now Approved to treat acute hepatic porphyria (AHP) in adults.https://www.givlaari.com/sites/default/files/2020-02/givlaari-patient-brochure.pdf [Google Scholar]
- 136.Jirikowski G.F., Sanna P.P., Maciejewski-Lenoir D., Bloom F.E. Reversal of diabetes insipidus in Brattleboro rats: intrahypothalamic injection of vasopressin mRNA. Science. 1992;255:996–998. doi: 10.1126/science.1546298. [DOI] [PubMed] [Google Scholar]
- 137.Conry R.M., LoBuglio A.F., Wright M., Sumerel L., Pike M.J., Johanning F., Benjamin R., Lu D., Curiel D.T. Characterization of a messenger RNA polynucleotide vaccine vector. Cancer Res. 1995;55:1397–1400. [PubMed] [Google Scholar]
- 138.Whisenand J.M., Azizian K.T., Henderson J.M., Shore S., Shin D., Lebedev A., McCaffrey A.P., Hogrefe R.L. TriLink BioTechnologies; San Diego, CA: 2017. Considerations for the Design and cGMP Manufacturing of mRNA Therapeutics. [Google Scholar]
- 139.Shin H., Park S.-J., Yim Y., Kim J., Choi C., Won C., Min D.-H. Recent Advances in RNA Therapeutics and RNA Delivery Systems Based on Nanoparticles. Adv. Ther. 2018;1:1800065. [Google Scholar]
- 140.Ng E.W.M., Shima D.T., Calias P., Cunningham E.T., Jr., Guyer D.R., Adamis A.P. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat. Rev. Drug Discov. 2006;5:123–132. doi: 10.1038/nrd1955. [DOI] [PubMed] [Google Scholar]
- 141.Kaur H., Bruno J.G., Kumar A., Sharma T.K. Aptamers in the Therapeutics and Diagnostics Pipelines. Theranostics. 2018;8:4016–4032. doi: 10.7150/thno.25958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Damase T.R., Miura T.A., Parent C.E., Allen P.B. Application of the Open qPCR Instrument for the in Vitro Selection of DNA Aptamers against Epidermal Growth Factor Receptor and Drosophila C Virus. ACS Comb. Sci. 2018;20:45–54. doi: 10.1021/acscombsci.7b00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Bauer M., Strom M., Hammond D.S., Shigdar S. Anything You Can Do, I Can Do Better: Can Aptamers Replace Antibodies in Clinical Diagnostic Applications? Molecules. 2019;24:4377. doi: 10.3390/molecules24234377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Nimjee S.M., White R.R., Becker R.C., Sullenger B.A. Aptamers as Therapeutics. Annu. Rev. Pharmacol. Toxicol. 2017;57:61–79. doi: 10.1146/annurev-pharmtox-010716-104558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Amero P., Lokesh G.L.R., Chaudhari R.R., Cardenas-Zuniga R., Schubert T., Attia Y.M., Montalvo-Gonzalez E., Elsayed A.M., Ivan C., Wang Z. Conversion of RNA Aptamer into Modified DNA Aptamers Provides for Prolonged Stability and Enhanced Antitumor Activity. J. Am. Chem. Soc. 2021;143:7655–7670. doi: 10.1021/jacs.9b10460. [DOI] [PubMed] [Google Scholar]