Abstract
Inflammation is regulated by the host and is a protective response activated by the evolutionarily conserved immune system in response to harmful stimuli, such as dead cells or pathogens. cGAS-STING pathway is a vital natural sensor of host immunity that can defend various tissues and organs against pathogenic infection, metabolic syndrome, cellular stress and cancer metastasis. The potential impact of cGAS-STING pathway in hepatic ischemia reperfusion (I/R) injury, alcoholic/non-alcoholic steatohepatitis (ASH), hepatic B virus infection, and other liver diseases has recently attracted widespread attention. In this review, the relationship between cGAS-STING pathway and the pathophysiological mechanisms and progression of liver diseases is summarized. Additionally, we discuss various pharmacological agonists and antagonists of cGAS-STING signaling as novel therapeutics for the treatment of liver diseases. A detailed understanding of mechanisms and biology of this pathway will lay a foundation for the development and clinical application of therapies for related liver diseases.
Keywords: DNA-sensing, cyclic GMP-AMP synthase, stimulator of interferon genes, liver diseases, cyclic guanosine monophosphate-adenosine monophosphate
Introduction
As a first line of defense, the innate immune system identifies extracellular or intracellular pathogenic-associated molecular patterns (PAMPs) and damage-associated molecular patterns (DAMPs) through pattern recognition receptors (PRRs) (Tang et al., 2012; Gong et al., 2020; Zindel and Kubes, 2020). Transmembrane receptors and cytoplasmic receptors, including NOD-like receptors (NLRs), toll-like receptors (TLRs), C-type lectin receptors (CLRs), and RIG-I-like receptors (RLRs), are the main susceptor for pathogens or danger signals (Kim et al., 2016; Zhu et al., 2018; McKernan, 2020). These receptors’ signal domains are located in the cytoplasm and initiate a signal cascade reaction to produce a series of gene products related to immune and inflammatory responses after binding with ligands. When a pathogenic organism enters and replicates within the cell, cytoplasmic sensors that activate different signaling pathways are activated. For example, NLRs can activate inflammasomes and trigger downstream signaling pathways by identifying various microbial molecules, toxins and damaged cells, and induce inflammatory responses and interleukin 1β (IL-1β) and IL-18 production (Davis et al., 2011; Man and Kanneganti, 2015). Cytoplasmic RLRs trigger signal cascades by recognizing virus and host-derived RNAs in the cytoplasm, leading to the secretion of type I interferon and inflammatory cytokines (Kawai and Akira, 2008; Kato et al., 2011; Kato and Fujita, 2015). Cyclic guanosine monophosphate (GMP)-adenosine monophosphate (AMP) synthase (cGAS) is a cellular DNA receptor that primarily recognizes viruses, bacteria and endogenous double-stranded DNA (dsDNA) irrespective of the sequence to activate innate immune responses and induce interferon expression and pro-inflammatory cytokines (Hopfner and Hornung, 2020; Wan et al., 2020).
Hepatitis viruses, especially hepatitis B virus (HBV), are DNA viruses and the leading cause of chronic viral hepatitis. In addition, the function of the liver is highly dependent on mitochondria for the production of ATP for biosynthesis and detoxification effects (Rui, 2014). There is growing evidence that most liver diseases are characterized by severe mitochondrial dysfunction (Paradies et al., 2014; Mansouri et al., 2018), which is manifested not only by the destruction and depletion of mitochondrial DNA (mtDNA), but also by the release of mtDNA (Malik and Czajka, 2013; Bose and Beal, 2016; Sharma et al., 2019). Moreover, the occurrence of liver diseases may induce necrosis, apoptosis, pyroptosis and other death processes in liver parenchymal and non-parenchymal cells (NPCs), resulting in DNA damage (Najimi et al., 2009; Chen et al., 2018; Xu et al., 2021). cGAS has now also been found to interact with various endogenous self-DNAs, including nuclear and mitochondrial cytoplasmic DNA, cytoplasmic micronucleus DNA, and nuclear chromatin. The inhibitory or activating effect of the cGAS-stimulator of interferon genes (STING) pathway plays a vital role in the occurrence and development of non-alcoholic fatty liver disease (NAFLD), alcoholic liver disease (ALD), viral hepatitis, liver fibrosis, liver cancer and other liver diseases. In this review, we discuss cGAS-STING signaling in liver diseases, and the profound implications of this pathway for novel therapeutic approaches against liver diseases.
The cGas-Sting Signaling Pathway
DNA Sensing cGAS
In 2013, James Chen used liquid chromatography-mass spectrometry to discover cyclic guanosine monophosphate-adenosine monophosphate (cGAMP), a substance that can activate cells to produce type I IFN (Wu et al., 2013). cGAMP functions as an endogenous second messenger and binds to STING to induce type I IFN in response to cytosolic DNA (Wu et al., 2013; Kato et al., 2017). Then, through protein purification and mass spectrometry, a nucleotidyl transferase family cGAMP synthase (cGAS) was identified (Sun et al., 2013). It was found that DNA in the cytoplasm can activate cGAS, and cGAMP catalyzed by cGAS can activate downstream transcription factor interferon regulatory factor 3 (IRF3) and induce the expression of STING dependent IFNβ (Cai et al., 2014; Chen Q. et al., 2016). Functional studies of cGAS mutants have shown that the conserved active sites G212, S213, E225, and D227 are necessary for cGAS activity, and mutations C396A and C397A decrease cGAS activity in humans (Civril et al., 2013). In addition to exogenous DNA, endogenous mtDNA can also be recognized by cGAS after entering the cytoplasm and triggering the immune response (Aarreberg et al., 2019; Maekawa et al., 2019; Huang et al., 2020). mtDNA is a circular double strand of about 17 Kbp, which can encode various enzymes and related ribosomal RNA and transport RNA required for oxidative phosphorylation, and has characteristics similar to prokaryotic nucleic acids (Banoth and Cassel, 2018). Under normal circumstances, mtDNA exists in the mitochondrial matrix, but it can be released into the cytosol or into circulation during cellular stress or mitochondrial injury, and then bind with DNA receptor cGAS to activate the cGAS-STING signaling pathway and enhance type I IFN responses (West et al., 2015).
cGAS-STING Activation
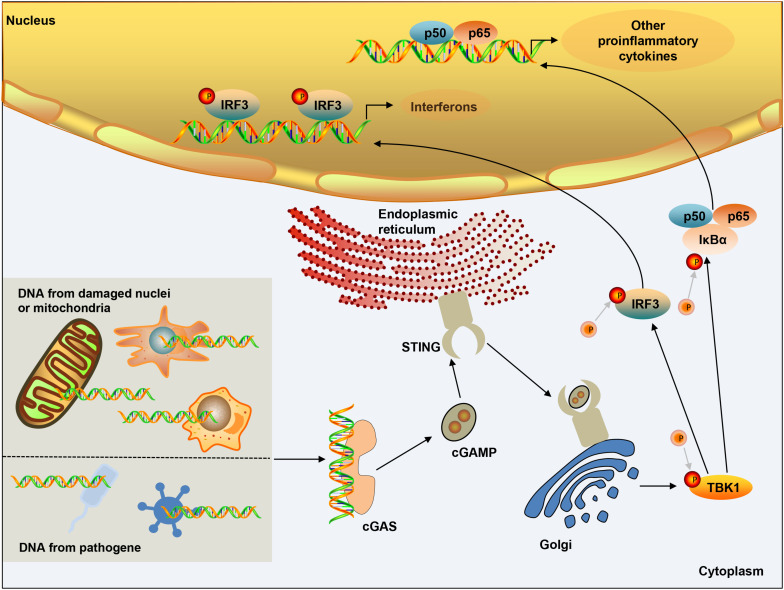
DNA in the cytoplasm is a powerful activator of the type I interferon response. In the normal physiological state of the body, DNA exists in the nucleus and mitochondria, and nucleases in the cytoplasm and endolysosomal compartments can quickly degrade DNA. However, when the organism is infected, the increase in the amount of intracellular DNA can be detected, which is closely related to the activation of the cGAS pathway. cGAS is a member of the nucleotide transferase (NTase) enzyme family, consisting of one NTase domain and two major DNA-binding domains (Sun et al., 2013). In the normal physiological state, DNA exists in the nucleus and mitochondria, and nucleases in the cytoplasm and endolysosomal compartments can quickly degrade DNA. Cytosolic DNA is required for cGAS activation, therefore, in the absence of DNA, cGAS is maintained an autoinhibited state (Civril et al., 2013; Li X. et al., 2013; Zhang X. et al., 2014). Once bound to DNA, cGAS can form a 2:2 complex that induces conformational changes at the active site and further catalyzes the synthesis of cGAMP from ATP and GTP (Ablasser et al., 2013a; Gao et al., 2013; Li X. D. et al., 2013). As a second messenger, cGAMP binds to the adaptor protein STING on the endoplasmic reticulum (ER) membrane, and induces a conformational change that may lead to the activation of STING (Shang et al., 2019). STING is transferred from the ER to the intermediate compartment of the ER-Golgi and Golgi apparatus, and then activates TANK-binding kinase 1 (TBK1) (Zhao et al., 2019). TBK1 then undergoes phosphorylation of itself and STING, followed by the IRF3 transcription factor. IRF3 dimerizes and enters the nucleus, triggering the production of type I interferon (Zhang et al., 2019). STING can also activate IKK kinase, then phosphorylate the IkB family of inhibitors of the transcription factor NF-kB (Ishikawa and Barber, 2008), and finally induce inflammatory cytokines such as tumor necrosis factor (TNF) and IL-6 (Figure 1). The cGAS-STING pathway activation has been shown to activate canonical and – in cancer cells – non-canonical nuclear factor kB (NF-kB), mitogen-activated protein (MAP) kinases, and signal transducer and activator of transcription (STAT) transcription factors (McWhirter et al., 2009; Bakhoum et al., 2018). In addition, after the activation of cGAS-STING signaling pathway can induce apoptosis (Zierhut et al., 2019; Li et al., 2021; Reinert et al., 2021), pyroptosis (Gaidt et al., 2017; Swanson et al., 2017), necroptosis (Schock et al., 2017), autophagy (Gui et al., 2019; Liu D. et al., 2019), and cell senescence (Yu et al., 2015; Glück and Guey, 2017; Yang et al., 2017).
FIGURE 1.
cGAS-STING signaling activation. DNA from microbial pathogens, damaged nuclei, or mitochondria binds and activates cGAS, and catalyzes the synthesis of cGAMP. cGAMP functions as a second messenger and binds to the ER-membrane adaptor STING and induces a conformational change that presumably results in the activation of STING. STING traffics from the ER to an ER-Golgi intermediate compartment and the Golgi apparatus, and then activates TBK1. TBK1 phosphorylates the transcription factor IRF3. STING also activates NF-kB, which enters the nucleus, where it functions together with IRF3 to induce the expression of type I IFNs and many other genes that mediate immune and inflammatory responses.
Regulation of the cGAS-STING Pathway of Cytosolic DNA Sensing
Regulation of cGAS Activation
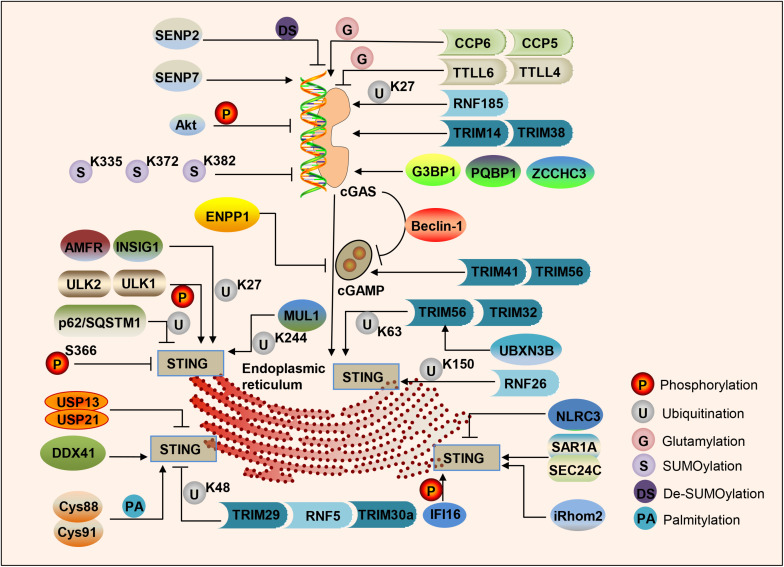
cGAS binds to the sugar-phosphate backbone of dsDNA independently of the DNA sequence (Li X. et al., 2013; Zhang X. et al., 2014). Oxidized DNA is more resistant to cellular nucleases, leading to stronger induction of interferon (Gehrke et al., 2013). Subsequent research has found that single-stranded DNA that can form an internal double-stranded structure can induce the activation of cGAS (Herzner et al., 2015). Various studies have shown that the post-translational modification of cGAS can adjust its enzymatic activity. The ER ubiquitin ligase RNF185 specifically catalyzes the polyubiquitination of cGAS’s K27 linkage and promotes cGAS activity (Wang et al., 2017). Ser305 in human cGAS (Ser291 in mouse cGAS) can be phosphorylated by the kinase Akt, and this phosphorylation can inhibit the activity of cGAS (Seo et al., 2015). Type I IFN induces TRIM14 to accelerate cGAS stabilization by recruiting USP14 to cleave the cGAS ubiquitin chain at lysine 414 of cGAS (Chen M. et al., 2016). cGAS can be polyglutamylated by the enzyme TTLL6 and monoglutamylated by TTLL4, and those modifications inhibit the activity of cGAS (Xia et al., 2016). The carboxypeptidases CCP6 and CCP5 can reverse this modification and activate cGAS activity (Xia et al., 2016). Small ubiquitin-like modifiers (SUMO) are coupled to lysine residues 335, 372, and 382 of cGAS to inhibit cGAS-DNA binding, oligomerization, and nucleotidyl transferase activity. The ubiquitin ligase TRIM38 can target cGAS for SUMOylation, which prevents its polyubiquitination and degradation and increases its stability (Hu et al., 2016). In contrast, cGAS is De-SUMOylated through SENP2 and then degraded through proteasome pathways (Hu et al., 2016). Similarly, SENP7 reverses this inhibition by catalyzing the De-SUMOlation of cGAS and enhances cGAS activation (Cui et al., 2017; Figure 2).
FIGURE 2.
Regulation of the cGAS-STING pathway of cytosolic DNA sensing. cGAS, cGAMP and STING are dynamically regulated by various modifications in response to DNA attacks, such as phosphorylation, ubiquitination, SUMOylation, glutamylation, palmitoylation, and the protein–protein binding in cells.
In addition to post-translational modification processes such as (De)ubiquitination, (De)glutamination, and (De)SUMOylation, protein–protein binding can also regulate the activity of cGAS. G3BP1 binds to the N-terminus of cGAS, increases DNA-binding affinity and primes cGAS for effective activation (Liu Z. S. et al., 2019). PQBP1 binds to cGAS via the PQBP1 WW domain and enhances cGAMP production (Yoh et al., 2015). ZCCHC3 directly binds to dsDNA to enhance the binding of cGAS and dsDNA (Lian et al., 2018). In addition, cGAS directly interacts with Beclin-1 autophagy protein, leading to inhibition of the synthesis of cGAMP, enhancing autophagy-mediated DNA degradation of cytoplasmic pathogens, and preventing excessive activation of cGAS and continuous immune stimulation (Liang et al., 2014; Figure 2).
Regulation of cGAMP Synthesis and Production
As a second messenger, the amount of cGAMP is regulated by its synthesis and decay rate. The extracellular enzyme ecto-nucleotide pyrophosphatase/phosphodiesterase (ENPP1) can degrade 2′3′-cGAMP with high specificity (Li et al., 2014). cGAMP can transfer between cells through gap junctions, allowing virus-infected cells to warn uninfected nearby cells to activate the interferon pathway to fight infection (Ablasser et al., 2013b), which is especially crucial for generating immune defenses in virus-infected cells. The E3 ubiquitin ligases TRIM41 and TRIM56 actively regulate cGAS-mediated cGAMP synthesis by promoting the monoubiquitination of cGAS (Liu et al., 2018; Seo et al., 2018; Figure 2).
Regulation of STING by Post-translational Modifications
TANK-binding kinase 1 phosphorylates several serine and threonine residues of STING, including Ser366. This phosphorylation of STING plays a crucial role in the subsequent phosphorylation of IRF3 by TBK1 (Liu S. et al., 2015). Phosphorylation of S366 can inhibit STING function, and UNC-51-like kinases ULK1 and 2 are responsible for phosphorylating STING on S366 (Konno et al., 2013). STING degradation can be mediated by p62/SQSTM1, which is phosphorylated by TBK1, leading to ubiquitination of STING to autophagosomes (Prabakaran et al., 2018). An E3 ubiquitin ligase RNF26 can promote K11-linked polyubiquitination of STING at lysine150 (Qin et al., 2014). Ubiquitin protein E3 ligases TRIM56 and TRIM32 can promote Lys63 polyubiquitination of STING, thereby enhancing its downstream pathway activation (Tsuchida et al., 2010; Zhang et al., 2012). The ER localization E3 ligase complex composed of AMFR and INSIG1 promotes K27-linked STING polyubiquitination, TBK1 recruitment, and interferon induction (Wang et al., 2014). UBXN3B interacts with STING and its E3 ligase TRIM56, and promotes STING’s ubiquitination, dimerization, trafficking, and subsequent recruitment and phosphorylation TBK1 (Yang et al., 2018). MUL1 can catalyze the K63-linked polyubiquitination of K224 on STING and facilitate STING trafficking (Ni and Konno, 2017). In contrast, E3 ligases TRIM29, RNF5, and TRIM30a can promote the K48 polyubiquitination of STING, which is degraded by proteasomes, finally leading to inhibition of the DNA-sensing pathway (Zhong et al., 2009; Wang et al., 2015; Xing et al., 2017). NLRC3 is related to both STING and TBK1, and prevents STING-TBK1 interaction and downstream type I interferon production (Zhang L. et al., 2014). CYLD partially accumulates with STING, and selectively interacts with its K48-linked polyubiquitin chains, specifically removing them from STING (Zhang et al., 2018). USP13 and USP21 negatively regulate the polyubiquitin chain from STING and prevent the recruitment of TBK1 to the signal complex, thereby negatively regulating the cellular antiviral response (Chen et al., 2017; Sun et al., 2017). Moreover, STING is palmitoylated at Cys88 and Cys91 of the Golgi apparatus, and the activation of STING can be blocked by palmitoylation inhibitor 2-bromopalmitate (2-BP) or by mutation of two cysteine residues (Cys88/91) to inhibit palmitoylation of STING (Mukai et al., 2016; Figure 2).
Furthermore, protein–protein binding also regulates STING trafficking and activation. SAR1A and SEC24C facilitate STING trafficking and are essential for STING signaling (Gui et al., 2019). A member of the DEXDc family of helicases, DDX41, binds to DNA and STING and localizes in the cytoplasm together with STING (Zhang et al., 2011; Parvatiyar et al., 2012). Interferon-γ-induced protein 16 (IFI16) interacts with STING to promote STING phosphorylation and translocation (Almine et al., 2017). In addition, IFI16 promotes the recruitment and activation of TBK1 in the STING complex (Jønsson et al., 2017). Inactive rhomboid protein 2 (iRhom2) recruits the translocon-related protein TRAPβ to the STING complex to promote the trafficking of STING. iRhom2 also recruits the deubiquitinating enzyme EIF3S5 to maintain the stability of STING by removing its K48-linked polyubiquitin chain (Luo et al., 2016). An NLR protein NLRX1 blocks the interaction of DNA-sensing adaptor STING with TBK1 (Guo et al., 2016). The Ca2+ sensor stromal interaction molecule 1 (STIM1) regulates the type I IFN response by retaining the adaptor protein STING at the ER (Srikanth et al., 2019; Figure 2).
In addition, the cGAS-STING signaling pathway is also regulated by various intracellular mechanisms. Rongvaux et al. found that activation of pro-apoptotic Bcl-2 family members Bak and Bax, which are located on the mitochondrial membrane, can trigger the release of mtDNA from mitochondria into the cytoplasm and activate the cGAS-STING signaling pathway (Rongvaux et al., 2014). However, the apoptosis-related caspase cascade proteins caspase-9, Apaf-1, or caspase-3/7 can inhibit Bak/Bax-mediated cGAS pathway activation and mtDNA-induced innate immune responses (White et al., 2014). Recently, Banerjee et al. found that Gasdermin D can inhibit the type I IFN response mediated by cGAS in macrophages. In addition, the negative regulation of Gasdermin D on IFN-β is independent of pyroptosis and inflammasome products IL-1β and IL-18 (Banerjee et al., 2018).
cGas-Sting Pathway in Liver Diseases
Hepatic I/R Injury
Liver ischemia reperfusion (I/R) is a complex process that can cause severe liver injury and even multiple organ dysfunction. During ischemia, the complex interaction between ER stress, oxidative stress, and inflammation leads to a large amount of hepatocyte loss during reperfusion, as well as a large number of cell death responses resulting in necrosis and apoptosis. The hepatotoxicity of I/R is caused by complex interactions between hepatocytes, tissue-resident immune cells, and immune cells recruited to the injury site. In this case, ER stress response, lipid signaling, and DAMPs released by dead hepatocytes play a major role in liver I/R (Brenner et al., 2013).
Enhanced protein levels of p-STING and p-TBK1 are observed in aged Kupffer cells (KCs) post-I/R and mtDNA stimulation, and aging facilitates STING overactivation, resulting in increased proinflammatory cytokines/chemokines production of macrophages, which ultimately aggravates liver I/R injury (Zhong et al., 2020). Targeting STING may be a key strategy for the treatment of liver I/R. STING siRNA abrogates the detrimental role of aging in aggravating liver I/R injury and intrahepatic inflammation. A specific inhibitor of STING, C-176, inhibits immoderate secretion of proinflammatory cytokines/chemokines in mtDNA-stimulated aged mice bone marrow-derived macrophages (Zhong et al., 2020). MicroRNA (miR)-24-3p downregulates of STING signaling and reduces expression of p-IRF3, serum inflammatory factors, and aminotransferase levels in liver I/R mice (Shen et al., 2020). In addition, distant organs (lung and liver) suffer severely by intestinal I/R injury, including increased levels of IL-6, IFN-β, and histopathological scores (Wu et al., 2021). However, STING–/– ameliorates intestinal I/R-induced distant organ damage. This harmful effect relies on excessive lipid peroxidation, which has been confirmed by the levels of 4-hydroxynonenal and the malondialdehyde (Wu et al., 2021).
However, one study has also shown that I/R is not associated with activation of the STING pathway. In a non-lethal model of segmental (70%) hepatic warm I/R, cGAS knockout (cGAS–/–) mice have significantly increased serum ALT levels and areas of necrosis in liver sections compared with wild-type (WT) mice (Lei and Deng, 2018). Surprisingly, STING deficiency (STINGgt/gt) does not phenocopy cGAS–/– mice, suggesting a protective effect of cGAS in liver I/R independent of STING (Lei and Deng, 2018). Moreover, cGAS–/– mice have increased cell death and decreased autophagy induction as well as increased apoptosis under hypoxic conditions, which may be due to increased mitochondrial dysfunction and subsequent increased oxidative stress damage (Lei and Deng, 2018). Therefore, whether the cGAS-STING signaling pathway plays a role in liver I/R still needs to be further explored and studied.
Alcoholic Liver Disease
Alcoholic liver disease is the most common type of chronic liver disease worldwide including fatty liver, alcoholic hepatitis (AH), and cirrhosis with complications. ALD can gradually develop from alcoholic fatty liver (AFL) to alcoholic steatohepatitis (ASH), and is characterized by liver inflammation. Chronic ASH can eventually lead to fibrosis and cirrhosis, and in some cases even hepatocellular carcinoma (HCC). At present, fundamental treatments for ALD include abstinence, nutritional support, corticosteroids, and anti-TNF antibodies (Gao and Bataller, 2011; Louvet and Mathurin, 2015).
A few studies have shown that targeting the cGAS-STING pathway is a therapeutic strategy for ALD and is expected to play a vital role in its treatment. The expression of IRF3 and its related genes (cGAS, TBK1, IKKε, and STING) is positively correlated with the severity of ALD. One study has shown that pathogen DNA, produced by excessive drinking of alcohol, contributes to IRF3 activation in hepatic cells (Wang et al., 2011). Alcohol-fed mice have increased expression of cGAS-STING in the liver, while mice deficient in cGAS and IRF3 genes show a protective effect against ALD (Luther et al., 2020). The destruction of the predominant hepatic gap junction Connexin 32 (Cx32) in ALD impairs the expression of IRF3-stimulated genes, leading to reduced liver damage (Luther et al., 2020). Ethanol induces ER stress and triggers the association of IRF3 with the ER adaptor STING, as well as subsequent p-IRF3. Activated IRF3 is related to the proapoptotic molecule Bax and promotes hepatocyte apoptosis. Deficiency of STING inhibits the phosphorylation of IRF3 under ethanol or ER stress, and deletion of IRF3 prevents hepatocyte apoptosis (Petrasek et al., 2013). In summary, cGAS-STING is considered to be a key factor in the onset of ALD and a potential therapeutic target.
Non-alcoholic Fatty Liver Disease and Non-alcoholic Steatohepatitis
Non-alcoholic fatty liver disease is one of the most important causes of liver disease worldwide, and may become the main cause of end-stage liver disease in the next few decades (Byrne and Targher, 2015; Younossi et al., 2018). NAFLD can occur in both adults and children, and it has a variable course but can lead to cirrhosis and liver cancer (Friedman et al., 2018). In addition, studies have shown that the clinical burden of NAFLD is not limited to liver-related morbidity and mortality, and growing evidence shows that NAFLD is a multi-system disease that affects extrahepatic organs and regulatory pathways (Byrne and Targher, 2015; Rinella and Sanyal, 2016). Non-alcoholic steatohepatitis (NASH) is considered to be a progressive form of NAFLD, characterized by liver steatosis, inflammation, hepatocellular damage, and varying degrees of fibrosis (Schuster et al., 2018). These causes of liver inflammation may come from outside the liver (such as adipose tissue or intestines) (Gäbele et al., 2011; Bijnen et al., 2018), or from inside organs (due to lipotoxicity, innate immune responses, cell death pathways, mitochondrial dysfunction, and ER stress) (Gao et al., 2018; Peng et al., 2018; Yu et al., 2019; Li et al., 2020), both of which contribute to the development of NAFLD/NASH. Therefore, innovative potential NAFLD/NASH therapies include four main pathways (Sumida and Yoneda, 2018): targeting liver fat accumulation; targeting oxidative stress, inflammation, and apoptosis; targeting intestinal microbiomes and metabolic endotoxemia; and targeting hepatic fibrosis. The contribution of cGAS-STING activation in NAFLD/NASH has therefore received substantial interest.
In clinical NAFLD/NASH patients, higher levels of STING in macrophages have been reported. The protein level of STING in non-parenchymal liver cells, including macrophages/KCs and endothelial cells, in liver tissues of NAFLD patients is higher than that in non-NAFLD patients (Luo et al., 2018). The number of STING + cells in the livers of NASH patients is increased with the severity of inflammation and the fibrosis stage (Wang et al., 2020). Compared with controls, the numbers of STING+/CCR2+ and STING+/S100A9+ macrophages in the livers of NASH patients are increased significantly, and they are positively correlated with the liver inflammation grade and fibrosis stage (Wang et al., 2020).
In animal models, cGAS-STING is observed to be involved in the formation of NAFLD/NASH. The expressions of STING and its downstream factor IRF3 are increased significantly in the livers of high-fat diet (HFD) mice (Li and Su, 2020). In HFD-fed mouse livers and free fatty acid (FFA)-induced LO2 cells, STING and IRF3 expression are up-regulated (Qiao et al., 2018). After HFD treatment, STINGgt mice and mice with destroyed myeloid cells (myeloid cell-specific STING disruption) show less liver steatosis, inflammation, and/or fibrosis, and after transplantation of bone marrow cells from mice in the control group to STINGgt mice, the severity of steatosis and inflammation after HFD can be restored (Luo et al., 2018). These results indicate that STING is specifically present in myeloid cells and can aggravate HFD-induced NAFLD. Moreover, the accumulation of gut microbial DNAs encapsulated with intestinal extracellular vesicles can activate cGAS-STING-mediated inflammatory signaling, which is pathogenic in the development of tissue inflammation and metabolic disorders (Luo et al., 2021).
Therefore, targeting cGAS-STING may be an effective way to treat NAFLD/NASH. In a NAFLD/NASH mouse model induced by methionine-choline deficient (MCD) and HFD, STING deficiency reduces liver steatosis, fibrosis and inflammation (Luo et al., 2018; Yu et al., 2019). Knock-out/down of STING or IRF3 can significantly reduce FFA-induced liver inflammation and apoptosis (Qiao et al., 2018). Meanwhile, HFD-fed mouse hepatocyte mtDNA induces the expression of TNF-α and IL-6 in cultured KCs, which can be attenuated by STING deficiency (Yu et al., 2019). Remdesivir (RDV) functions as an inhibitor of STING, inhibiting STING and IRF3 expression significantly in HFD-induced NAFLD (Li and Su, 2020). Chronic exposure to DMXAA, a mouse STING agonist, can cause liver steatosis and inflammation in WT mice, but not in STING-deficient mice (Yu et al., 2019). Through various mouse models and NAFLD/NASH patients, we have further confirmed that STING plays a harmful role in NAFLD/NASH. In summary, pharmacological inhibition of STING activation results in potent therapeutic effects in NAFLD/NASH, encouraging the development of novel therapeutics against this target.
Hepatitis B Virus
Hepatitis B virus infection affects approximately 350 million people and is a major public health problem worldwide. HBV is the main cause of liver cirrhosis and HCC (Megahed and Zhou, 2020). The current non-therapeutic treatment is mainly nucleos(t)ide analogs (NAs), which can profoundly but not completely inhibit the DNA synthesis of viral reverse transcriptase (Pierra Rouviere et al., 2020). Increasing evidence indicates that HBV interacts with hepatocyte innate immune signaling pathways and inhibits innate immunity. Therefore, an in-depth understanding of the interaction between HBV virus and innate immunity is important for the development of new therapeutic strategies for the treatment of HBV infection.
Naked circle HBV DNA is sensed in a cGAS-dependent manner in liver cancer cell lines, immunocompetent myeloid cells and human primary hepatocytes (Verrier et al., 2018; Lauterbach-Rivière et al., 2020). When the cGAS-STING pathway is activated by dsDNA or cGAMP, whether in cell culture or in a mouse model, HBV replication is significantly inhibited (He et al., 2016). However, hepatocytes did not produce type 1 IFN upon foreign DNA stimulation or HBV infection, and mice lacking STING or cGAS show the same ability to control infection in an adenovirus-HBV model (Thomsen et al., 2016). Meanwhile, HBV infection suppresses the expression and function of cGAS in cell culture models and humanized mice, indicating that HBV exploits multiple strategies to circumvent the perception of cGAS and its effector pathways and antiviral activity (Verrier et al., 2018). Knockout of cGAS in human peripheral blood mononuclear cells leads to an increase in intracellular HBV DNA levels (He et al., 2016). Targeting of the cGAS-STING pathway may therefore be an effective strategy for treating HBV. Treatment of immortalized mouse liver cells that support HBV replication with cGAMP or small molecule pharmacological STING agonists significantly reduces viral DNA in a STING- and Janus kinase 1-dependent manner (Guo et al., 2017). DMXAA has been found to induce a strong cytokine response dominated by type I IFNs in macrophages. By reducing the number of cytoplasmic virus nucleocapsids, DMXAA effectively inhibits HBV replication in mouse hepatocytes (Guo et al., 2015). In addition, in an HBV hydrodynamic mouse model, intraperitoneal injection of DMXAA significantly induces the expression of IFN-stimulated genes and reduces HBV DNA replication intermediates in the mouse liver (Guo et al., 2015).
In terms of mechanistic research, the viral polymerase (Pol) of HBV can inhibit the activation of STING-stimulated IRF3 and the induction of IFN-β (Liu Y. et al., 2015). In addition, the reverse transcriptase and RNase H domains of Pol have been identified as the cause of this inhibition (Liu Y. et al., 2015). Moreover, Pol has been shown to be physically related to STING and significantly reduces the K63-linked polyubiquitination of STING through its reverse transcriptase domain, which provides a new mechanism for HBV to fight against the innate DNA sensing pathway (Liu Y. et al., 2015). The methylation frequency of the STING promoter in chronic hepatitis B patients is significantly higher, and the level of STING mRNA is significantly lower than that in healthy controls (Wu et al., 2018). Hypermethylation of the STING promoter and the resulting transcriptional inhibition of STING weaken the effect of STING in inhibiting HBV replication and reduce the effectiveness of antiviral therapy (Wu et al., 2018).
It is necessary to develop small-molecule human cGAS or STING agonists as immunotherapeutic drugs for the treatment of chronic hepatitis B. Daunorubicin (DNR), a topoisomerase II poison, triggers an endogenous cGAS-dependent innate immune response and subsequently inhibits the production of HBV in NKNT-3/NTCP cells (Imai et al., 2018). GV1001 inhibits HBV replication and hepatitis B surface antigen (HBsAg) secretion in a dose-dependent manner (Choi et al., 2020). GV1001, a telomerase-derived 16-mer peptide, promotes the release of oxidized DNA into the cytoplasm mediated by mtDNA stress and produces IFN-I-dependent anti-HBV effects through the STING-IRF3 axis (Choi et al., 2020). The anti-HBV effect of GV1001 is through translocation of extracellular heat shock proteins into the cytosol, leading to mtDNA stress mediated by the escape of phagosomes (Choi et al., 2020). Importantly, the specific introduction of STING expression in hepatocytes reconstructs the DNA sensing pathway, thereby improving the control of HBV in vivo (Thomsen et al., 2016). Therefore, activating the cGAS-STING pathway can induce the antiviral cytokine response to HBV.
Liver Fibrosis
Liver fibrosis is the excessive accumulation of extracellular matrix proteins including collagen, and is common in most types of chronic liver disease (Bataller and Brenner, 2005). Advanced liver fibrosis can lead to cirrhosis, liver failure, and portal hypertension, and often requires liver transplantation (Zhou et al., 2014; Schuppan et al., 2018; Parola and Pinzani, 2019). Emerging anti-fibrosis therapies aim to inhibit the accumulation of fibroblasts and/or prevent the deposition of extracellular matrix proteins. Under stimulation by the main fibrogenic cytokine, transforming growth factor-β1 (TGF-β1), the expression of IRF3 in HSCs increases significantly (Ni et al., 2016). In addition, the LX-2 cell line transfected with IRF3-siRNA reduces the type I collagen (Col1a1) and α-smooth muscle actin (α-SMA) expression (Ni et al., 2016). Acute administration of CCl4 in WT mice results in early ER stress, IRF3 activation, and type I IFN induction, followed by hepatocyte apoptosis and liver damage, with liver fibrosis occurring after repeated administration of CCl4 (Iracheta-Vellve et al., 2016). In addition, Yong et al. (2021) found that activation of the cGAS/STING pathway caused by mitochondrial localization of TAR DNA-binding protein 43 (TDP-43) may be a pathogenic mechanism of CCl4-induced liver fibrosis. STING might therefore be a therapeutic target for liver fibrosis. IRF3 or STING deficiency can prevent acute or chronic CCl4 hepatocyte death and fibrosis; however, mice deficient in type I IFN receptor or TLR4 signal adaptor TRAM or TRIF are not protected from hepatocyte death and/or fibrosis, indicating that the pro-apoptotic effect of IRF3 is independent of TLR signaling in fibrosis (Iracheta-Vellve et al., 2016). IRF3 is associated with STING in the presence of ER stress and combines hepatocyte apoptosis with liver fibrosis, indicating that innate immune signals regulate the outcomes of liver fibrosis by regulating hepatocyte death in the liver (Iracheta-Vellve et al., 2016).
Hepatocellular Carcinoma and Liver Metastasis
Liver cancer is a global challenge that threatens human health, and HCC accounts for approximately 90% of liver cancer cases and is its most common form. HBV and HCV infection are the main risk factors for the development of HCC. NASH associated with metabolic syndrome or diabetes is also a common risk factor in the West (Llovet et al., 2021). In HCC, immuno-oncology treatment methods have attracted much attention, including the use of checkpoint inhibitors, tyrosine kinase inhibitors or anti-VEGF therapy, and even a combination of two immunotherapies. However, we still need to optimize combination strategies to maximize the potential of these methods (Finn and Zhu, 2020).
In recent years, a large number of studies have reported a connection between the cGAS-STING signaling pathway and anti-tumor immunity, cancer progression, the tumor microenvironment, and pharmacological strategies for cancer treatment (Jiang et al., 2020; Kwon and Bakhoum, 2020; Wang, 2020). Prognostic biomarkers in HCC have been identified: the overexpressions of XRCC5, IRF3, TRIM21, STAT6, DDX41, TBK1, XRCC6, TREX1, PRKDC, and STING are significantly related to the clinical stage and pathological grades of HCC (Qi et al., 2020). Higher expressions of IFI16, STAT6, NLRC3, and STING mRNA are associated with a favorable overall survival rate (Qi et al., 2020). The cGAS-STING pathway is significantly related to the infiltration of B cells, CD4 + T cells, CD8 + T cells, macrophages, neutrophils, and dendritic cells in HCC (Qi et al., 2020). Compared with non-tumor tissues, the expression of STING protein in tumor tissues is significantly reduced, and the intensity of STING staining in HCC patients is negatively correlated with tumor size, tumor invasion tumor node metastasis (TNM) stage, and overall survival (Bu et al., 2016). The combined expression of STING and TNM staging has a better prognostic effect for overall survival in HCC patients (Bu et al., 2016). In the late stages of HCC induced by diethylnitrosamine, STINGgt/gt mice show larger tumors (Thomsen et al., 2020). Treatment of mice after HCC development with 3′3′-cAIMP, a cyclic dinucleotide STING agonist, effectively reduces the tumor size in cGAS–/– mice, which cannot generate 2′3′-cGAMP (Thomsen et al., 2020). In addition, Lactobacillus rhamnosus GG induces cGAS/STING-dependent IFN-β production and improves the response to anti-programmed cell death 1 (PD-1) immunotherapy (Si et al., 2021). Therefore, regulation by STING pathway agonists may reverse the development of HCC and is expected to be an effective treatment strategy.
Liver metastasis remains a major obstacle to the successful treatment of malignant diseases, especially of gastrointestinal cancers and other malignant tumors, such as breast cancer and melanoma. The main cause of mortality from colorectal cancer is the development of liver metastases. Nanoencapsulation improves the half-life of encapsulated cGAMP by 40-fold, allowing for sufficient accumulation of cGAMP in tumors and activation of the STING pathway in the tumor microenvironment. In a B16-F10 melanoma model, nanoparticle delivery also changes the biodistribution profile, resulting in increased cGAMP accumulation and STING activation in the liver and spleen (Wehbe et al., 2020), which may be an important strategy for the treatment of liver metastases. E7766, a macrocyclic bridging STING agonist with a novel topology, shows extensive pan-genotypic activity in all major human STING variants. In a mouse liver metastasis model, E7766 has shown strong anti-tumor activity and a durable immune memory response (Kim et al., 2021).
In conclusion, STING agonists has an independent prognostic value and may be a potential target for HCC and liver metastasis immunotherapy. Clarifying the molecular mechanisms of STING agonists against HCC or liver metastases, searching for and screening out predictive markers that are suitable for STING agonist treatment, and selecting appropriate therapeutic doses to improve the effect of STING agonists on tumors and reduce their adverse effects are crucial for the clinical development of STING agonists. In addition, combined application with STING agonists can not only reduce the dosage and side effects of radiotherapy, chemotherapy, tumor vaccines, immune checkpoint inhibitors, chimeric antigen receptor T-cell (CAR-T) immunotherapy or other types of treatments, but also can synergistically enhance anti-tumor effects through different mechanisms.
Other Liver Diseases
Other liver diseases have also been shown to be associated with the cGAS-STING pathway. In a two-thirds hepatectomy mouse model, the combined deficiency of MAVS and STING resulted in severely impaired hepatocyte proliferation and delayed the recovery of liver mass, indicating that the innate immune cytoplasmic nucleic acid sensor promotes liver regeneration (Schulze et al., 2018). Radiation causes dsDNA accumulation, and in NPCs, dsDNA rapidly activates the cGAS-STING pathway, leading to the production and release of IFN-I, accompanied by hepatocyte damage (Du et al., 2020). cGAS- or STING-deficient mice have alleviated radiation-induced liver steatosis and inflammation (Du et al., 2020). Moreover, clinically irradiated human liver tissue peri-HCC liver tissues show higher expression of STING and IFNβ than unirradiated tissue (Du et al., 2020). Washingtonia robusta leaves has been found to restore elevated liver index, ALT, albumin, cholesterol, and reactive oxygen species levels, and ameliorates the increase in STING expression in γ-radiation-induced oxidative hepatotoxicity (Selim et al., 2020). When acetaminophen is excessive, hepatocytes release a large amount of DNA, but these cells do not primarily sense DNA; however, liver NPCs quickly perceive this environmental interference and activate multiple DNA sensing pathways (Araujo and Antunes, 2018). Liver NPCs synthesize and release IFN-1, accompanied by hepatocyte necrosis. Ablation of IFN-1 in interferon α/β receptor (IFNAR–/–) mice delays APAP-mediated hepatic necrosis and inhibits the IFN-1 sensing pathway (Araujo and Antunes, 2018). In addition, although not a DNA virus, HCV virus NS4B protein inhibits STING and STING-induced IFN-β activation (Ding et al., 2013; Nitta et al., 2013; Yi et al., 2016). Interestingly, the expression of NS4B blocks the protein interaction between STING and Cardif, which is required for the activation of IFN-β(Nitta et al., 2013).
Conclusion
As outlined in this review, the cGAS-STING pathway is a vital component of most acute and chronic liver diseases and contributes to liver I/R injury, ALD, NAFLD, NASH, HBV, fibrosis, and even HCC. Understanding the structural features and functions of cGAS-STING will provide new insights for the treatment of liver diseases based on this pathway through novel methods and agents. However, important questions remain in our understanding of how the liver regulates the cGAS-STING pathway to drive disease. Therefore, additional in-depth research on the cGAS-STING pathway will contribute to clinical treatments using cGAS-STING-related activators and inhibitors. In addition, when developing agonists or inhibitors that regulate the cGAS-STING signaling pathway, its potential adverse drug reactions are worthy of attention. For example, when using STING agonists for tumor immunotherapy, the treatment window and toxic side effects must be considered to avoid excessive activation of the STING pathway, which may cause severe inflammation, such as abnormally elevated cytokine levels (Gangadhar and Vonderheide, 2014; Cui et al., 2019). When using STING inhibitors to treat autoimmune diseases, excessive inhibition of the STING pathway may lead to a weakened immune system and significantly increase the risk of pathogen infection and tumor occurrence. The development of drugs that inhibit or promote the cGAS-STING signaling pathway has broad prospects for treating HCC, ALD, NASH, HBV, and other liver diseases, giving it a major role in basic immunology, tumor biology, and clinical treatment.
Author Contributions
ZW, NC, and ZL: investigation, validation, visualization, and writing – original draft. XZ and GX: investigation, formal analysis, and data curation. JT, XX, and ZB: conceptualization, methodology, validation, resources, supervision, project administration, and funding acquisition. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
Funding. This work was supported by the National Natural Science Foundation of China (81874368 and 81630100), the National Science and Technology Major Project “Key New Drug Creation and Manufacturing Program” (2017ZX09301022 and 2018ZX09101002-001-002).
References
- Aarreberg L. D., Esser-Nobis K., Driscoll C., Shuvarikov A., Roby J. A., Gale M., Jr. (2019). Interleukin-1β Induces mtDNA Release to Activate Innate Immune Signaling via cGAS-STING. Mol. Cell 74 801.e–815.e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ablasser A., Goldeck M., Cavlar T., Deimling T., Witte G., Röhl I., et al. (2013a). cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature 498 380–384. 10.1038/nature12306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ablasser A., Schmid-Burgk J. L., Hemmerling I., Horvath G. L., Schmidt T., Latz E., et al. (2013b). Cell intrinsic immunity spreads to bystander cells via the intercellular transfer of cGAMP. Nature 503 530–534. 10.1038/nature12640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almine J. F., O’Hare C. A., Dunphy G., Haga I. R., Naik R. J., Atrih A., et al. (2017). IFI16 and cGAS cooperate in the activation of STING during DNA sensing in human keratinocytes. Nat. Comm. 8:14392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araujo A. M., Antunes M. M. (2018). Liver Immune Cells Release Type 1 Interferon Due to DNA Sensing and Amplify Liver Injury from Acetaminophen Overdose. Cells 7:88. 10.3390/cells7080088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhoum S. F., Ngo B., Laughney A. M., Cavallo J. A., Murphy C. J., Ly P., et al. (2018). Chromosomal instability drives metastasis through a cytosolic DNA response. Nature 553 467–472. 10.1038/nature25432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee I., Behl B., Mendonca M., Shrivastava G., Russo A. J., Menoret A., et al. (2018). Gasdermin D Restrains Type I Interferon Response to Cytosolic DNA by Disrupting Ionic Homeostasis. Immunity 49 413.e–426.e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banoth B., Cassel S. L. (2018). Mitochondria in innate immune signaling. Transl. Res. 202 52–68. 10.1016/j.trsl.2018.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bataller R., Brenner D. A. (2005). Liver fibrosis. J. Clin. Invest. 115 209–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bijnen M., Josefs T., Cuijpers I., Maalsen C. J., van de Gaar J., Vroomen M., et al. (2018). Adipose tissue macrophages induce hepatic neutrophil recruitment and macrophage accumulation in mice. Gut 67 1317–1327. 10.1136/gutjnl-2016-313654 [DOI] [PubMed] [Google Scholar]
- Bose A., Beal M. F. (2016). Mitochondrial dysfunction in Parkinson’s disease. J. Neurochem. 139(Suppl. 1), 216–231. [DOI] [PubMed] [Google Scholar]
- Brenner C., Galluzzi L., Kepp O., Kroemer G. (2013). Decoding cell death signals in liver inflammation. J. Hepatol. 59 583–594. 10.1016/j.jhep.2013.03.033 [DOI] [PubMed] [Google Scholar]
- Bu Y., Liu F., Jia Q. A., Yu S. N. (2016). Decreased Expression of TMEM173 Predicts Poor Prognosis in Patients with Hepatocellular Carcinoma. PLoS One 11:e0165681. 10.1371/journal.pone.0165681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne C. D., Targher G. (2015). NAFLD: a multisystem disease. J. Hepatol. 62 S47–S64. [DOI] [PubMed] [Google Scholar]
- Cai X., Chiu Y. H., Chen Z. J. (2014). The cGAS-cGAMP-STING pathway of cytosolic DNA sensing and signaling. Mol. Cell 54 289–296. 10.1016/j.molcel.2014.03.040 [DOI] [PubMed] [Google Scholar]
- Chen M., Meng Q., Qin Y., Liang P., Tan P., He L., et al. (2016). TRIM14 Inhibits cGAS Degradation Mediated by Selective Autophagy Receptor p62 to Promote Innate Immune Responses. Mol. Cell 64 105–119. 10.1016/j.molcel.2016.08.025 [DOI] [PubMed] [Google Scholar]
- Chen Q., Sun L., Chen Z. J. (2016). Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nat. Immunol. 17 1142–1149. 10.1038/ni.3558 [DOI] [PubMed] [Google Scholar]
- Chen S., Ren Z., Yu D., Ning B., Guo L. (2018). DNA damage-induced apoptosis and mitogen-activated protein kinase pathway contribute to the toxicity of dronedarone in hepatic cells. Environ. Mol. Mutagen 59 278–289. 10.1002/em.22173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Wang L., Jin J., Luan Y., Chen C., Li Y., et al. (2017). p38 inhibition provides anti-DNA virus immunity by regulation of USP21 phosphorylation and STING activation. J. Exp. Med. 214 991–1010. 10.1084/jem.20161387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y. M., Kim H., Lee S. A., Lee S. Y., Kim B. J. A. (2020). Telomerase-Derived Peptide Exerts an Anti-Hepatitis B Virus Effect via Mitochondrial DNA Stress-Dependent Type I Interferon Production. Front. Immunol. 11:652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civril F., Deimling T., de Oliveira Mann C. C., Ablasser A., Moldt M., Witte G., et al. (2013). Structural mechanism of cytosolic DNA sensing by cGAS. Nature 498 332–337. 10.1038/nature12305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X., Zhang R., Cen S., Zhou J. (2019). STING modulators: Predictive significance in drug discovery. Eur. J. Med. Chem. 182:111591. 10.1016/j.ejmech.2019.111591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y., Yu H., Zheng X., Peng R., Wang Q., Zhou Y., et al. (2017). SENP7 Potentiates cGAS Activation by Relieving SUMO-Mediated Inhibition of Cytosolic DNA Sensing. PLoS Pathog. 13:e1006156. 10.1371/journal.ppat.1006156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis B. K., Wen H., Ting J. P. (2011). The inflammasome NLRs in immunity, inflammation, and associated diseases. Annu. Rev. Immunol. 29 707–735. 10.1146/annurev-immunol-031210-101405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q., Cao X., Lu J., Huang B., Liu Y. J., Kato N., et al. (2013). Hepatitis C virus NS4B blocks the interaction of STING and TBK1 to evade host innate immunity. J. Hepatol. 59 52–58. 10.1016/j.jhep.2013.03.019 [DOI] [PubMed] [Google Scholar]
- Du S., Chen G., Yuan B., Hu Y., Yang P., Chen Y., et al. (2020). DNA sensing and associated type 1 interferon signaling contributes to progression of radiation-induced liver injury. Cell Mol. Immunol. 18 1718–1728. 10.1038/s41423-020-0395-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R. S., Zhu A. X. (2020). Evolution of Systemic Therapy for Hepatocellular Carcinoma. Hepatology 73 150–157. 10.1002/hep.31306 [DOI] [PubMed] [Google Scholar]
- Friedman S. L., Neuschwander-Tetri B. A., Rinella M., Sanyal A. J. (2018). Mechanisms of NAFLD development and therapeutic strategies. Nat. Med. 24 908–922. 10.1038/s41591-018-0104-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gäbele E., Dostert K., Hofmann C., Wiest R., Schölmerich J., Hellerbrand C., et al. (2011). DSS induced colitis increases portal LPS levels and enhances hepatic inflammation and fibrogenesis in experimental NASH. J. Hepatol. 55 1391–1399. 10.1016/j.jhep.2011.02.035 [DOI] [PubMed] [Google Scholar]
- Gaidt M. M., Ebert T. S., Chauhan D., Ramshorn K., Pinci F., Zuber S., et al. (2017). The DNA Inflammasome in Human Myeloid Cells Is Initiated by a STING-Cell Death Program Upstream of NLRP3. Cell 171 1110.e–1124.e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangadhar T. C., Vonderheide R. H. (2014). Mitigating the toxic effects of anticancer immunotherapy. Nat. Rev. Clin. Oncol. 11 91–99. 10.1038/nrclinonc.2013.245 [DOI] [PubMed] [Google Scholar]
- Gao B., Bataller R. (2011). Alcoholic liver disease: pathogenesis and new therapeutic targets. Gastroenterology 141 1572–1585. 10.1053/j.gastro.2011.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P., Ascano M., Wu Y., Barchet W., Gaffney B. L., Zillinger T., et al. (2013). Cyclic [G(2′,5′)pA(3′,5′)p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell 153 1094–1107. 10.1016/j.cell.2013.04.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Guo S., Zhang S., Liu A., Shi L., Zhang Y. (2018). Matrine attenuates endoplasmic reticulum stress and mitochondrion dysfunction in nonalcoholic fatty liver disease by regulating SERCA pathway. J. Transl. Med. 16:319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrke N., Mertens C., Zillinger T., Wenzel J., Bald T., Zahn S., et al. (2013). Oxidative damage of DNA confers resistance to cytosolic nuclease TREX1 degradation and potentiates STING-dependent immune sensing. Immunity 39 482–495. 10.1016/j.immuni.2013.08.004 [DOI] [PubMed] [Google Scholar]
- Glück S., Guey B. (2017). Innate immune sensing of cytosolic chromatin fragments through cGAS promotes senescence. Nat. Cell Biol. 19 1061–1070. 10.1038/ncb3586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong T., Liu L., Jiang W., Zhou R. (2020). DAMP-sensing receptors in sterile inflammation and inflammatory diseases. Nat. Rev. Immunol. 20 95–112. 10.1038/s41577-019-0215-7 [DOI] [PubMed] [Google Scholar]
- Gui X., Yang H., Li T., Tan X., Shi P., Li M., et al. (2019). Autophagy induction via STING trafficking is a primordial function of the cGAS pathway. Nature 567 262–266. 10.1038/s41586-019-1006-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F., Han Y., Zhao X., Wang J., Liu F., Xu C., et al. (2015). STING agonists induce an innate antiviral immune response against hepatitis B virus. Antimicrob. Agents Chemother. 59 1273–1281. 10.1128/aac.04321-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F., Tang L., Shu S., Sehgal M., Sheraz M., Liu B., et al. (2017). Activation of Stimulator of Interferon Genes in Hepatocytes Suppresses the Replication of Hepatitis B Virus. Antimicrob. Agents Chemother. 61 e771–e717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., König R., Deng M., Riess M., Mo J., Zhang L., et al. (2016). NLRX1 Sequesters STING to Negatively Regulate the Interferon Response, Thereby Facilitating the Replication of HIV-1 and DNA Viruses. Cell Host Microbe 19 515–528. 10.1016/j.chom.2016.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J., Hao R., Liu D., Liu X., Wu S., Guo S., et al. (2016). Inhibition of hepatitis B virus replication by activation of the cGAS-STING pathway. J. Gen. Virol. 97 3368–3378. 10.1099/jgv.0.000647 [DOI] [PubMed] [Google Scholar]
- Herzner A. M., Hagmann C. A., Goldeck M., Wolter S., Kübler K., Wittmann S., et al. (2015). Sequence-specific activation of the DNA sensor cGAS by Y-form DNA structures as found in primary HIV-1 cDNA. Nat. Immunol. 16 1025–1033. 10.1038/ni.3267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfner K. P., Hornung V. (2020). Molecular mechanisms and cellular functions of cGAS-STING signalling. Nat. Rev. Mol. Cell Biol. 21 501–521. 10.1038/s41580-020-0244-x [DOI] [PubMed] [Google Scholar]
- Hu M. M., Yang Q., Xie X. Q., Liao C. Y., Lin H., Liu T. T., et al. (2016). Sumoylation Promotes the Stability of the DNA Sensor cGAS and the Adaptor STING to Regulate the Kinetics of Response to DNA Virus. Immunity 45 555–569. 10.1016/j.immuni.2016.08.014 [DOI] [PubMed] [Google Scholar]
- Huang L. S., Hong Z., Wu W., Xiong S., Zhong M., Gao X., et al. (2020). mtDNA Activates cGAS Signaling and Suppresses the YAP-Mediated Endothelial Cell Proliferation Program to Promote Inflammatory Injury. Immunity 2020 475.e–486.e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai H., Dansako H., Ueda Y., Satoh S., Kato N. (2018). Daunorubicin, a topoisomerase II poison, suppresses viral production of hepatitis B virus by inducing cGAS-dependent innate immune response. Biochem. Biophys. Res. Commun. 504 672–678. 10.1016/j.bbrc.2018.08.195 [DOI] [PubMed] [Google Scholar]
- Iracheta-Vellve A., Petrasek J., Gyongyosi B., Satishchandran A., Lowe P., Kodys K., et al. (2016). Endoplasmic Reticulum Stress-induced Hepatocellular Death Pathways Mediate Liver Injury and Fibrosis via Stimulator of Interferon Genes. J. Biol. Chem. 291 26794–26805. 10.1074/jbc.m116.736991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H., Barber G. N. (2008). STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455 674–678. 10.1038/nature07317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M., Chen P., Wang L., Li W., Chen B., Liu Y., et al. (2020). cGAS-STING, an important pathway in cancer immunotherapy. J. Hematol. Oncol. 13:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jønsson K. L., Laustsen A., Krapp C., Skipper K. A., Thavachelvam K., Hotter D., et al. (2017). IFI16 is required for DNA sensing in human macrophages by promoting production and function of cGAMP. Nat. Comm. 8:14391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H., Fujita T. (2015). RIG-I-like receptors and autoimmune diseases. Curr Opin Immunol 37 40–45. 10.1016/j.coi.2015.10.002 [DOI] [PubMed] [Google Scholar]
- Kato H., Takahasi K., Fujita T. (2011). RIG-I-like receptors: cytoplasmic sensors for non-self RNA. Immunol. Rev. 243 91–98. 10.1111/j.1600-065x.2011.01052.x [DOI] [PubMed] [Google Scholar]
- Kato K., Omura H., Ishitani R., Nureki O. (2017). Cyclic GMP-AMP as an Endogenous Second Messenger in Innate Immune Signaling by Cytosolic DNA. Annu. Rev. Biochem. 86 541–566. 10.1146/annurev-biochem-061516-044813 [DOI] [PubMed] [Google Scholar]
- Kawai T., Akira S. (2008). Toll-like receptor and RIG-I-like receptor signaling. Ann. N. Y. Acad. Sci. 1143 1–20. 10.1196/annals.1443.020 [DOI] [PubMed] [Google Scholar]
- Kim D. S., Endo A., Fang F. G., Huang K. C., Bao X., Choi H. W., et al. (2021). E7766, a Macrocycle-Bridged Stimulator of Interferon Genes (STING) Agonist with Potent Pan-Genotypic Activity. ChemMedChem 16 1740–1743. [DOI] [PubMed] [Google Scholar]
- Kim Y. K., Shin J. S., Nahm M. H. (2016). NOD-Like Receptors in Infection, Immunity, and Diseases. Yonsei Med. J. 57 5–14. 10.3349/ymj.2016.57.1.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno H., Konno K., Barber G. N. (2013). Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sustained innate immune signaling. Cell 155 688–698. 10.1016/j.cell.2013.09.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon J., Bakhoum S. F. (2020). The Cytosolic DNA-Sensing cGAS-STING Pathway in Cancer. Cancer Discov. 10 26–39. 10.1158/2159-8290.cd-19-0761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauterbach-Rivière L., Bergez M., Mönch S., Qu B., Riess M., Vondran F. W. R., et al. (2020). Hepatitis B Virus DNA is a Substrate for the cGAS/STING Pathway but is not Sensed in Infected Hepatocytes. Viruses 12:592. 10.3390/v12060592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Z., Deng M. (2018). cGAS-mediated autophagy protects the liver from ischemia-reperfusion injury independently of STING. Am. J. Physiol. Gastrointest. Liver Physiol. 314 G655–G667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Liu W., Wang F., Hayashi T., Mizuno K., Hattori S., et al. (2021). DNA damage-triggered activation of cGAS-STING pathway induces apoptosis in human keratinocyte HaCaT cells. Mol. Immunol. 131 180–190. 10.1016/j.molimm.2020.12.037 [DOI] [PubMed] [Google Scholar]
- Li L., Yin Q., Kuss P., Maliga Z., Millán J. L., Wu H., et al. (2014). Hydrolysis of 2′3′-cGAMP by ENPP1 and design of nonhydrolyzable analogs. Nat. Chem. Biol. 10 1043–1048. 10.1038/nchembio.1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. D., Wu J., Gao D., Wang H., Sun L., Chen Z. J. (2013). Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science 341 1390–1394. 10.1126/science.1244040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Shu C., Yi G., Chaton C. T., Shelton C. L., Diao J., et al. (2013). Cyclic GMP-AMP synthase is activated by double-stranded DNA-induced oligomerization. Immunity 39 1019–1031. 10.1016/j.immuni.2013.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Wang T. X., Huang X., Li Y., Sun T., Zang S., et al. (2020). Targeting ferroptosis alleviates methionine-choline deficient (MCD)-diet induced NASH by suppressing liver lipotoxicity. Liver Intern. 40 1378–1394. 10.1111/liv.14428 [DOI] [PubMed] [Google Scholar]
- Li Y. N., Su Y. (2020). Remdesivir attenuates high fat diet (HFD)-induced NAFLD by regulating hepatocyte dyslipidemia and inflammation via the suppression of STING. Biochem. Biophys. Res. Commun. 526 381–388. 10.1016/j.bbrc.2020.03.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian H., Wei J., Zang R., Ye W., Yang Q., Zhang X. N., et al. (2018). ZCCHC3 is a co-sensor of cGAS for dsDNA recognition in innate immune response. Nat. Comm. 9:3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Q., Seo G. J., Choi Y. J., Kwak M. J., Ge J., Rodgers M. A., et al. (2014). Crosstalk between the cGAS DNA sensor and Beclin-1 autophagy protein shapes innate antimicrobial immune responses. Cell host & microbe 15 228–238. 10.1016/j.chom.2014.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Wu H., Wang C., Li Y., Tian H., Siraj S., et al. (2019). STING directly activates autophagy to tune the innate immune response. Cell Death Differ 26 1735–1749. 10.1038/s41418-018-0251-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Cai X., Wu J., Cong Q., Chen X., Li T., et al. (2015). Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 347:aaa2630. 10.1126/science.aaa2630 [DOI] [PubMed] [Google Scholar]
- Liu Y., Li J., Chen J., Li Y., Wang W., Du X., et al. (2015). Hepatitis B virus polymerase disrupts K63-linked ubiquitination of STING to block innate cytosolic DNA-sensing pathways. J. Virol. 89 2287–2300. 10.1128/jvi.02760-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z. S., Cai H., Xue W., Wang M., Xia T., Li W. J., et al. (2019). G3BP1 promotes DNA binding and activation of cGAS. Nat. Immunol. 20 18–28. 10.1038/s41590-018-0262-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z. S., Zhang Z. Y., Cai H., Zhao M., Mao J., Dai J., et al. (2018). RINCK-mediated monoubiquitination of cGAS promotes antiviral innate immune responses. Cell Biosci. 8:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llovet J. M., Kelley R. K., Villanueva A. (2021). Hepatocellular carcinoma. Nat. Rev. Dis. Primers 7:6. [DOI] [PubMed] [Google Scholar]
- Louvet A., Mathurin P. (2015). Alcoholic liver disease: mechanisms of injury and targeted treatment. Nat Rev. Gastroenterol. Hepatol. 12 231–242. 10.1038/nrgastro.2015.35 [DOI] [PubMed] [Google Scholar]
- Luo W. W., Li S., Li C., Lian H., Yang Q., Zhong B., et al. (2016). iRhom2 is essential for innate immunity to DNA viruses by mediating trafficking and stability of the adaptor STING. Nat. Immunol. 17 1057–1066. 10.1038/ni.3510 [DOI] [PubMed] [Google Scholar]
- Luo X., Li H., Ma L., Zhou J., Guo X., Woo S. L., et al. (2018). Expression of STING Is Increased in Liver Tissues From Patients With NAFLD and Promotes Macrophage-Mediated Hepatic Inflammation and Fibrosis in Mice. Gastroenterology 155 1971.e–1984.e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z., Ji Y., Gao H., Gomes D., Reis F. C., Bandyopadhyay G., et al. (2021). CRIg(+) Macrophages Prevent Gut Microbial DNA-Containing Extracellular Vesicle-Induced Tissue Inflammation and Insulin Resistance. Gastroenterology 160 863–874. 10.1053/j.gastro.2020.10.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luther J., Khan S., Gala M. K., Kedrin D., Sridharan G., Goodman R. P., et al. (2020). Hepatic gap junctions amplify alcohol liver injury by propagating cGAS-mediated IRF3 activation. Proc. Natl. Acad. Sci. U S A 117 11667–11673. 10.1073/pnas.1911870117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa H., Inoue T., Ouchi H., Jao T. M., Inoue R., Nishi H., et al. (2019). Mitochondrial Damage Causes Inflammation via cGAS-STING Signaling in Acute Kidney Injury. Cell Rep. 29 1261.e–1273.e. [DOI] [PubMed] [Google Scholar]
- Malik A. N., Czajka A. (2013). Is mitochondrial DNA content a potential biomarker of mitochondrial dysfunction? Mitochondrion 13 481–492. 10.1016/j.mito.2012.10.011 [DOI] [PubMed] [Google Scholar]
- Man S. M., Kanneganti T. D. (2015). Regulation of inflammasome activation. Immunol. Rev. 265 6–21. 10.1111/imr.12296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansouri A., Gattolliat C. H., Asselah T. (2018). Mitochondrial Dysfunction and Signaling in Chronic Liver Diseases. Gastroenterology 155 629–647. 10.1053/j.gastro.2018.06.083 [DOI] [PubMed] [Google Scholar]
- McKernan D. P. (2020). Pattern recognition receptors as potential drug targets in inflammatory disorders. Adv. Protein Chem. Struct. Biol. 119 65–109. 10.1016/bs.apcsb.2019.09.001 [DOI] [PubMed] [Google Scholar]
- McWhirter S. M., Barbalat R., Monroe K. M., Fontana M. F., Hyodo M., Joncker N. T., et al. (2009). A host type I interferon response is induced by cytosolic sensing of the bacterial second messenger cyclic-di-GMP. J. Exp. Med. 206 1899–1911. 10.1084/jem.20082874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megahed F. A. K., Zhou X. (2020). The Interactions between HBV and the Innate Immunity of Hepatocytes. Viruses 12:285. 10.3390/v12030285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai K., Konno H., Akiba T., Uemura T., Waguri S., Kobayashi T., et al. (2016). Activation of STING requires palmitoylation at the Golgi. Nat. Comm. 7:11932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najimi M., Smets F., Sokal E. (2009). Hepatocyte apoptosis. Methods Mol Biol 481 59–74. 10.1007/978-1-59745-201-4_6 [DOI] [PubMed] [Google Scholar]
- Ni G., Konno H. (2017). Ubiquitination of STING at lysine 224 controls IRF3 activation. Sci. Immunol 2:eaah7119. 10.1126/sciimmunol.aah7119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M. M., Xu T., Wang Y. R., He Y. H., Zhou Q., Huang C., et al. (2016). Inhibition of IRF3 expression reduces TGF-β1-induced proliferation of hepatic stellate cells. J. Physiol. Biochem. 72 9–23. 10.1007/s13105-015-0452-6 [DOI] [PubMed] [Google Scholar]
- Nitta S., Sakamoto N., Nakagawa M., Kakinuma S., Mishima K., Kusano-Kitazume A., et al. (2013). Hepatitis C virus NS4B protein targets STING and abrogates RIG-I-mediated type I interferon-dependent innate immunity. Hepatology 57 46–58. 10.1002/hep.26017 [DOI] [PubMed] [Google Scholar]
- Paradies G., Paradies V., Ruggiero F. M., Petrosillo G. (2014). Oxidative stress, cardiolipin and mitochondrial dysfunction in nonalcoholic fatty liver disease. World J. Gastroenterol. 20 14205–14218. 10.3748/wjg.v20.i39.14205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parola M., Pinzani M. (2019). Liver fibrosis: Pathophysiology, pathogenetic targets and clinical issues. Mol. Aspects Med. 65 37–55. 10.1016/j.mam.2018.09.002 [DOI] [PubMed] [Google Scholar]
- Parvatiyar K., Zhang Z., Teles R. M., Ouyang S., Jiang Y., Iyer S. S., et al. (2012). The helicase DDX41 recognizes the bacterial secondary messengers cyclic di-GMP and cyclic di-AMP to activate a type I interferon immune response. Nat. Immunol. 13 1155–1161. 10.1038/ni.2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng K. Y., Watt M. J., Rensen S., Greve J. W., Huynh K., Jayawardana K. S., et al. (2018). Mitochondrial dysfunction-related lipid changes occur in nonalcoholic fatty liver disease progression. J. Lipid Res. 59 1977–1986. 10.1194/jlr.m085613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrasek J., Iracheta-Vellve A., Csak T., Satishchandran A., Kodys K., Kurt-Jones E. A., et al. (2013). STING-IRF3 pathway links endoplasmic reticulum stress with hepatocyte apoptosis in early alcoholic liver disease. Proc. Natl. Acad. Sci. U S A 110 16544–16549. 10.1073/pnas.1308331110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierra Rouviere C., Dousson C. B., Tavis J. E. (2020). HBV replication inhibitors. Antiviral. Res. 179:104815. 10.1016/j.antiviral.2020.104815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabakaran T., Bodda C., Krapp C., Zhang B. C., Christensen M. H., Sun C., et al. (2018). Attenuation of cGAS-STING signaling is mediated by a p62/SQSTM1-dependent autophagy pathway activated by TBK1. EMBO J. 37:e97858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Z., Yan F., Chen D., Xing W., Li Q., Zeng W., et al. (2020). Identification of prognostic biomarkers and correlations with immune infiltrates among cGAS-STING in hepatocellular carcinoma. Biosci. Rep. 2020:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao J. T., Cui C., Qing L., Wang L. S., He T. Y., Yan F., et al. (2018). Activation of the STING-IRF3 pathway promotes hepatocyte inflammation, apoptosis and induces metabolic disorders in nonalcoholic fatty liver disease. Metabolism 81 13–24. 10.1016/j.metabol.2017.09.010 [DOI] [PubMed] [Google Scholar]
- Qin Y., Zhou M. T., Hu M. M., Hu Y. H., Zhang J., Guo L., et al. (2014). RNF26 temporally regulates virus-triggered type I interferon induction by two distinct mechanisms. PLoS Pathog. 10:e1004358. 10.1371/journal.ppat.1004358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinert L. S., Rashidi A. S., Tran D. N., Katzilieris-Petras G., Hvidt A. K., Gohr M., et al. (2021). Brain immune cells undergo cGAS/STING-dependent apoptosis during herpes simplex virus type 1 infection to limit type I IFN production. J. Clin. Invest. 2021:131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinella M. E., Sanyal A. J. (2016). Management of NAFLD: a stage-based approach. Nat. Rev. Gastroenterol. Hepatol. 13 196–205. 10.1038/nrgastro.2016.3 [DOI] [PubMed] [Google Scholar]
- Rongvaux A., Jackson R., Harman C. C., Li T., West A. P., de Zoete M. R., et al. (2014). Apoptotic caspases prevent the induction of type I interferons by mitochondrial DNA. Cell 159 1563–1577. 10.1016/j.cell.2014.11.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rui L. (2014). Energy metabolism in the liver. Comprehen. Physiol. 4 177–197. 10.1002/cphy.c130024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schock S. N., Chandra N. V., Sun Y., Irie T., Kitagawa Y. (2017). Induction of necroptotic cell death by viral activation of the RIG-I or STING pathway. Cell Death Differ 24 615–625. 10.1038/cdd.2016.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze S., Stöß C., Lu M., Wang B., Laschinger M., Steiger K. (2018). Cytosolic nucleic acid sensors of the innate immune system promote liver regeneration after partial hepatectomy. Sci. Rep. 8:12271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuppan D., Ashfaq-Khan M., Yang A. T., Kim Y. O. (2018). Liver fibrosis: Direct antifibrotic agents and targeted therapies. Matrix Biol. 6 435–451. 10.1016/j.matbio.2018.04.006 [DOI] [PubMed] [Google Scholar]
- Schuster S., Cabrera D., Arrese M., Feldstein A. E. (2018). Triggering and resolution of inflammation in NASH. Nat. Rev. Gastroenterol. Hepatol. 15 349–364. 10.1038/s41575-018-0009-6 [DOI] [PubMed] [Google Scholar]
- Selim N. M., El-Hawary S. S., El Zalabani S. M., Shamma R. N. (2020). Impact of Washingtonia robusta Leaves on Gamma Irradiation-Induced Hepatotoxicity in Rats and Correlation with STING Pathway and Phenolic Composition. Pharmaceuticals 13:320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo G. J., Kim C., Shin W. J. (2018). TRIM56-mediated monoubiquitination of cGAS for cytosolic DNA sensing. Nat. Comm. 9:613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo G. J., Yang A., Tan B., Kim S., Liang Q., Choi Y., et al. (2015). Akt Kinase-Mediated Checkpoint of cGAS DNA Sensing Pathway. Cell Rep. 13 440–449. 10.1016/j.celrep.2015.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang G., Zhang C., Chen Z. J., Bai X. C., Zhang X. (2019). Cryo-EM structures of STING reveal its mechanism of activation by cyclic GMP-AMP. Nature 567 389–393. 10.1038/s41586-019-0998-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma N., Pasala M. S., Prakash A. (2019). Mitochondrial DNA: Epigenetics and environment. Environ. Mol. Mutagen 60 668–682. 10.1002/em.22319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen A., Zheng D., Luo Y., Mou T., Chen Q., Huang Z., et al. (2020). MicroRNA-24-3p alleviates hepatic ischemia and reperfusion injury in mice through the repression of STING signaling. Biochem. Biophys. Res. Commun. 522 47–52. 10.1016/j.bbrc.2019.10.182 [DOI] [PubMed] [Google Scholar]
- Si W., Liang H., Bugno J., Xu Q., Ding X., Yang K., et al. (2021). Lactobacillus rhamnosus GG induces cGAS/STING- dependent type I interferon and improves response to immune checkpoint blockade. Gut 2021:323426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikanth S., Woo J. S., Wu B., El-Sherbiny Y. M. (2019). The Ca(2+) sensor STIM1 regulates the type I interferon response by retaining the signaling adaptor STING at the endoplasmic reticulum. Nat. Immunol. 20 152–162. 10.1038/s41590-018-0287-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumida Y., Yoneda M. (2018). Current and future pharmacological therapies for NAFLD/NASH. J. Gastroenterol. 53 362–376. 10.1007/s00535-017-1415-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H., Zhang Q., Jing Y. Y., Zhang M., Wang H. Y., Cai Z., et al. (2017). USP13 negatively regulates antiviral responses by deubiquitinating STING. Nat. Comm. 8:15534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Wu J., Du F., Chen X., Chen Z. J. (2013). Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339 786–791. 10.1126/science.1232458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson K. V., Junkins R. D., Kurkjian C. J., Holley-Guthrie E., Pendse A. A. (2017). A noncanonical function of cGAMP in inflammasome priming and activation. J. Exp. Med. 214 3611–3626. 10.1084/jem.20171749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D., Kang R., Coyne C. B., Zeh H. J., Lotze M. T. (2012). PAMPs and DAMPs: signal 0s that spur autophagy and immunity. Immunol. Rev. 249 158–175. 10.1111/j.1600-065x.2012.01146.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen M. K., Nandakumar R., Stadler D., Malo A., Valls R. M., Wang F., et al. (2016). Lack of immunological DNA sensing in hepatocytes facilitates hepatitis B virus infection. Hepatology 64 746–759. 10.1002/hep.28685 [DOI] [PubMed] [Google Scholar]
- Thomsen M. K., Skouboe M. K., Boularan C., Vernejoul F., Lioux T., Leknes S. L., et al. (2020). The cGAS-STING pathway is a therapeutic target in a preclinical model of hepatocellular carcinoma. Oncogene 39 1652–1664. 10.1038/s41388-019-1108-8 [DOI] [PubMed] [Google Scholar]
- Tsuchida T., Zou J., Saitoh T., Kumar H., Abe T., Matsuura Y., et al. (2010). The ubiquitin ligase TRIM56 regulates innate immune responses to intracellular double-stranded DNA. Immunity 33 765–776. 10.1016/j.immuni.2010.10.013 [DOI] [PubMed] [Google Scholar]
- Verrier E. R., Yim S. A., Heydmann L., El Saghire H., Bach C., Turon-Lagot V., et al. (2018). Hepatitis B Virus Evasion From Cyclic Guanosine Monophosphate-Adenosine Monophosphate Synthase Sensing in Human Hepatocytes. Hepatology 68 1695–1709. 10.1002/hep.30054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan D., Jiang W., Hao J. (2020). Research Advances in How the cGAS-STING Pathway Controls the Cellular Inflammatory Response. Nat. Rev. Mol. Cell Biol. 11:615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Huang L., Hong Z. (2017). The E3 ubiquitin ligase RNF185 facilitates the cGAS-mediated innate immune response. PLoS Pathog. 13:e1006264. 10.1371/journal.ppat.1006264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Liu X., Cui Y., Tang Y., Chen W., Li S., et al. (2014). The E3 ubiquitin ligase AMFR and INSIG1 bridge the activation of TBK1 kinase by modifying the adaptor STING. Immunity 41 919–933. 10.1016/j.immuni.2014.11.011 [DOI] [PubMed] [Google Scholar]
- Wang X., Rao H., Zhao J., Wee A., Li X., Fei R., et al. (2020). STING expression in monocyte-derived macrophages is associated with the progression of liver inflammation and fibrosis in patients with nonalcoholic fatty liver disease. Lab. Invest. 100 542–552. 10.1038/s41374-019-0342-6 [DOI] [PubMed] [Google Scholar]
- Wang Y. (2020). cGAS-STING pathway in cancer biotherapy. J. Hematol. Oncol. 19:136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Guo Y., Wang F., Sun S. (2011). Pathogen DNA also contributes to interferon regulatory factor 3 activation in hepatic cells: Implications for alcoholic liver diseases. Hepatology 53 1783–1784. 10.1002/hep.24089 [DOI] [PubMed] [Google Scholar]
- Wang Y., Lian Q., Yang B., Yan S., Zhou H., He L., et al. (2015). TRIM30α Is a Negative-Feedback Regulator of the Intracellular DNA and DNA Virus-Triggered Response by Targeting STING. PLoS Pathog. 11:e1005012. 10.1371/journal.ppat.1005012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehbe M., Wang-Bishop L., Becker K. W., Shae D., Baljon J. J., He X., et al. (2020). Nanoparticle delivery improves the pharmacokinetic properties of cyclic dinucleotide STING agonists to open a therapeutic window for intravenous administration. J. Control Release 330 1118–1129. 10.1016/j.jconrel.2020.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West A. P., Khoury-Hanold W., Staron M., Tal M. C., Pineda C. M., Lang S. M., et al. (2015). Mitochondrial DNA stress primes the antiviral innate immune response. Nature 520 553–557. 10.1038/nature14156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White M. J., McArthur K., Metcalf D., Lane R. M., Cambier J. C., Herold M. J., et al. (2014). Apoptotic caspases suppress mtDNA-induced STING-mediated type I IFN production. Cell 159 1549–1562. 10.1016/j.cell.2014.11.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. S., Zhao Q., Zhang J., Wang J. W., Qian Y., Fan Y. C., et al. (2018). Methylation status of the stimulator of interferon genes promoter in patients with chronic hepatitis B. Medicine 97:e13904. 10.1097/md.0000000000013904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Liu Q., Zhang X., Wu X., Zhao Y., Ren J. (2021). STING-dependent induction of lipid peroxidation mediates intestinal ischemia-reperfusion injury. Free Radic Biol. Med. 163 135–140. 10.1016/j.freeradbiomed.2020.12.010 [DOI] [PubMed] [Google Scholar]
- Wu J., Sun L., Chen X., Du F., Shi H., Chen C., et al. (2013). Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339 826–830. 10.1126/science.1229963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia P., Ye B., Wang S., Zhu X., Du Y., Xiong Z., et al. (2016). Glutamylation of the DNA sensor cGAS regulates its binding and synthase activity in antiviral immunity. Nat. Immunol. 17 369–378. 10.1038/ni.3356 [DOI] [PubMed] [Google Scholar]
- Xing J., Zhang A., Zhang H., Wang J., Li X. C., Zeng M. S., et al. (2017). TRIM29 promotes DNA virus infections by inhibiting innate immune response. Nat. Comm. 8:945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Zhou J., Che J., Wang H., Yang W., Zhou W., et al. (2021). Mitochondrial DNA enables AIM2 inflammasome activation and hepatocyte pyroptosis in non-alcoholic fatty liver disease. Am. J. Physiol. Gastrointest. Liver Physiol. 320 G1034–G1044. [DOI] [PubMed] [Google Scholar]
- Yang H., Wang H., Ren J., Chen Q., Chen Z. J. (2017). cGAS is essential for cellular senescence. Proc. Natl. Acad. Sci. U S A 114 E4612–E4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Wang L., Ketkar H., Ma J., Yang G., Cui S., et al. (2018). UBXN3B positively regulates STING-mediated antiviral immune responses. Nat. Comm. 9:2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi G., Wen Y., Shu C., Han Q., Konan K. V., Li P., et al. (2016). Hepatitis C Virus NS4B Can Suppress STING Accumulation To Evade Innate Immune Responses. J. Virol. 90 254–265. 10.1128/jvi.01720-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoh S. M., Schneider M., Seifried J., Soonthornvacharin S., Akleh R. E., Olivieri K. C., et al. (2015). PQBP1 Is a Proximal Sensor of the cGAS-Dependent Innate Response to HIV-1. Cell 161 1293–1305. 10.1016/j.cell.2015.04.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong H., Wang S., Song F. (2021). Activation of cGAS/STING pathway upon TDP-43-mediated mitochondrial injury may be involved in the pathogenesis of liver fibrosis. Liver Internat. 2021:14895. [DOI] [PubMed] [Google Scholar]
- Younossi Z., Anstee Q. M., Marietti M., Hardy T., Henry L., Eslam M., et al. (2018). Global burden of NAFLD and NASH: trends, predictions, risk factors and prevention. Nat. Rev. Gastroenterol. Hepatol. 15 11–20. 10.1038/nrgastro.2017.109 [DOI] [PubMed] [Google Scholar]
- Yu Q., Katlinskaya Y. V., Carbone C. J., Zhao B., Katlinski K. V., Zheng H., et al. (2015). DNA-damage-induced type I interferon promotes senescence and inhibits stem cell function. Cell reports 11 785–797. 10.1016/j.celrep.2015.03.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Liu Y., An W., Song J., Zhang Y., Zhao X. (2019). STING-mediated inflammation in Kupffer cells contributes to progression of nonalcoholic steatohepatitis. J. Clin. Invest. 129 546–555. 10.1172/jci121842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Shang G., Gui X., Zhang X., Bai X. C., Chen Z. J. (2019). Structural basis of STING binding with and phosphorylation by TBK1. Nature 567 394–398. 10.1038/s41586-019-1000-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Hu M. M., Wang Y. Y., Shu H. B. (2012). TRIM32 protein modulates type I interferon induction and cellular antiviral response by targeting MITA/STING protein for K63-linked ubiquitination. J. Biol. Chem. 287 28646–28655. 10.1074/jbc.m112.362608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Mo J., Swanson K. V., Wen H., Petrucelli A., Gregory S. M., et al. (2014). NLRC3, a member of the NLR family of proteins, is a negative regulator of innate immune signaling induced by the DNA sensor STING. Immunity 40 329–341. 10.1016/j.immuni.2014.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Wei N., Cui Y., Hong Z., Liu X., Wang Q., et al. (2018). The deubiquitinase CYLD is a specific checkpoint of the STING antiviral signaling pathway. PLoS Pathog. 14:e1007435. 10.1371/journal.ppat.1007435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Wu J., Du F., Xu H., Sun L., Chen Z., et al. (2014). The cytosolic DNA sensor cGAS forms an oligomeric complex with DNA and undergoes switch-like conformational changes in the activation loop. Cell Rep. 6 421–430. 10.1016/j.celrep.2014.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Yuan B., Bao M., Lu N., Kim T., Liu Y. J. (2011). The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat. Immunol. 12 959–965. 10.1038/ni.2091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Du F., Xu P., Shu C., Sankaran B., Bell S. L., et al. (2019). A conserved PLPLRT/SD motif of STING mediates the recruitment and activation of TBK1. Nature 569 718–722. 10.1038/s41586-019-1228-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong B., Zhang L., Lei C., Li Y., Mao A. P., Yang Y., et al. (2009). The ubiquitin ligase RNF5 regulates antiviral responses by mediating degradation of the adaptor protein MITA. Immunity 30 397–407. 10.1016/j.immuni.2009.01.008 [DOI] [PubMed] [Google Scholar]
- Zhong W., Rao Z., Rao J., Han G., Wang P., Jiang T., et al. (2020). Aging aggravated liver ischemia and reperfusion injury by promoting STING-mediated NLRP3 activation in macrophages. Aging Cell 19:e13186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W. C., Zhang Q. B., Qiao L. (2014). Pathogenesis of liver cirrhosis. World J. Gastroenterol. 20 7312–7324. 10.3748/wjg.v20.i23.7312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G., Xu Y., Cen X., Nandakumar K. S., Liu S., Cheng K. (2018). Targeting pattern-recognition receptors to discover new small molecule immune modulators. Eur. J. Med. Chem. 144 82–92. 10.1016/j.ejmech.2017.12.026 [DOI] [PubMed] [Google Scholar]
- Zierhut C., Yamaguchi N., Paredes M., Luo J. D., Carroll T., Funabiki H. (2019). The Cytoplasmic DNA Sensor cGAS Promotes Mitotic Cell Death. Cell 2019 302.e–315.e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zindel J., Kubes P. (2020). DAMPs, PAMPs, and LAMPs in Immunity and Sterile Inflammation. Annu. Rev. Pathol. 15 493–518. 10.1146/annurev-pathmechdis-012419-032847 [DOI] [PubMed] [Google Scholar]