Summary
• Plants transition through juvenile and adult phases of vegetative development in a process known as vegetative phase change (VPC). In poplars (genus Populus) the differences between these stages are subtle, making it difficult to determine when this transition occurs. Previous studies of VPC in poplars have relied on plants propagated in vitro, leaving the natural progression of this process unknown.
• We examined developmental morphology of seed-grown and in vitro derived Populus tremula × alba (clone 717–1B4), and compared the phenotype of these to transgenics with manipulated miR156 expression, the master regulator of VPC.
• In seed-grown plants, most traits changed from node-to-node during the first 3 months of development but remained constant after node 25. Many traits remained unchanged in clones over-expressing miR156, or were enhanced when miR156 was lowered, demonstrating their natural progression is regulated by the miR156/SPL pathway. The characteristic leaf fluttering of Populus is one of these miR156-regulated traits.
• Vegetative development in plants grown from culture mirrored that of seed-grown plants, allowing direct comparison between plants often used in research and those found in nature. These results provide a foundation for further research on the role of VPC in the ecology and evolution of this economically important genus.
Keywords: development, juvenile-to-adult transition, miR156, Populus (poplar), vegetative phase change
Introduction
During ontogeny, plants transition through distinct phases, which is reflected in morphological variation in the organs produced across development. These developmental transitions include the visually obvious transition between vegetative and reproductive growth as well as the often more subtle change between juvenile and adult vegetative stages, known as vegetative phase change (Ashby, 1948; Brink, 1962; Doorenbos, 1965; Poethig, 2013). In species with abrupt changes in vegetative morphology – termed heteroblastic – the distinction between the juvenile and adult phases is unambiguous (Zotz et al., 2011). For example, many species of Acacia transition from producing bipinnate compound leaves to producing a simple undivided leaf termed a phyllode (Walters & Bartholomew, 1984). Other examples include Eucalyptus globulus, which transitions from producing ovate, wax-covered leaves to producing lanceolate glaucous leaves (James & Bell, 2001) and Ipomoea caerulea, which transitions from producing monolobed to trilobed leaves (Ashby, 1948). Vegetative phase change is more difficult to identify in species that experience less drastic changes in leaf and shoot morphology. As a result, we know little about the duration of juvenile and adult phases in different taxa, or the relationship between this developmental transition and other aspects of plant life history, such as plant responses to biotic and abiotic conditions. However, even in these so-called homoblastic species (Goebel, 1900), organs produced early in shoot development differ from those produced later in development in a variety of morphological, anatomical and physiological traits (Feng et al., 2016). Identifying these traits and determining which are components of vegetative phase change as opposed to those regulated by other factors, such as plant size or chronological age, is an important problem in plant development.
In Arabidopsis and most other plants, the transition from the juvenile to the adult phase of vegetative development is regulated by two closely related microRNAs, miR156 and miR157 (Poethig, 2013; He et al., 2018). These miRNAs directly repress members of the Squamosa Promoter Binding Protein-Like (SPL) family of transcription factors (Rhoades et al., 2002; Schwab et al., 2005) which promote adult traits (Xu et al., 2016). During the juvenile phase, miR156/157 levels are high, but as plants age their expression declines, allowing for the increased expression of their SPL targets. This coordinated change in miR156 and SPL gene expression leads to the onset of adult traits, with different traits typically appearing at slightly different times in shoot development, depending on their sensitivity to the level of SPL proteins (He et al., 2018).
Vegetative phase change is likely to be of particular importance in long-lived perennial species such as trees, because the ability of these species to survive to reproductive competence requires that they cope with changing conditions over a long period of time. Poplars are of considerable economic and ecological importance and have become popular model systems for tree biology due to their ability to be propagated in culture and susceptibility to Agrobacterium-mediated transformation (Taylor, 2002). Leaf and shoot phenotypes of plants derived from culture and over-expressing miR156 have been described in clones of Populus × canendensis (Wang et al., 2011) and P. tremula × alba (Rubinelli et al., 2013). However, the significance of this information for vegetative phase change in Populus is still unknown because these studies lacked the earliest developmental stages and fine-scale timeline needed to capture vegetative phase change, and relied on gain-of-function transgenics. Here, we compare the vegetative phenotype of seed-grown P. tremula × alba with plants originating from explants in tissue culture across multiple time points during vegetative development, and identify phase-specific traits using both gain- and loss-of-function miR156 transgenics. We found that shoot development in plants propagated in vitro closely resembles the development of seed-grown plants, and that vegetative phase change occurs very early in both types of shoots. Most of the traits we examined changed significantly across shoot development and were sensitive to the level of miR156. One of these was leaf fluttering, a trait that has a significant effect on photosynthetic efficiency (Roden & Pearcy, 1993b). Our results provide basic information about the nature of vegetative phase change in P. tremula × alba and demonstrate that shoots regenerated in vitro can be used as models of primary shoot development in this species.
Materials and Methods
Plant transformation
Assembly of Ubi10:AthMIR156a and Ubi10:MIM156 constructs have been described previously (Feng et al., 2016). Additionally, a second MIM156 construct was designed with Golden Gate Cloning (Engler et al., 2014) and used for a later transformation after losing many of the original lines to a chamber malfunction. Further details of construct assembly can be found in Supporting Information Table S1.
Poplar line 717–1B4 was transformed using Agrobacterium tumefaciens (strain AGL1) following the protocol described by Filichkin et al. (2006). Briefly, leaf and stem explants were selected on callus induction, shoot induction, shoot elongation and rooting media using BASTA. A total of 30 Ubi10: AthMIR156a events and 25 Ubi10:MIM156 events were isolated. PCR was used to first screen for transgene insertion using two primer sets, each utilizing a primer to the Arabidopsis UBIQUITIN 10 promoter and either the MIR156a or MIM156 sequence (Table S2). Of these, we screened 28 Ubi:AthMIR156a lines and 19 Ubi10:MIM156 lines for levels of miR156 and miR157 (Fig. S1A,B). Based on levels of miR156, two lines for each construct type were selected (or based on availability in the case of event GG.MIM156–22). For the Ubi10:AthMIR156a and Ubi10:MIM156 constructs made with the pCambia 3300 backbone, the Ubi10:AthMIR156a construct had a higher rate of producing transgenic shoots (proportion of explants forming shoots was 52% vs 33.9%). We did not track efficiency for the other MIM156 construct.
Plant material and growth conditions
Populus tremula × alba line 717–1B4, two independent miR156 over-expressing (miR156OX) lines, 40 and 78, and two independent MIM156 lines, 22 and 84, were obtained by in vitro propagation and hardening on propagation media as described by Meilan & Ma (2006). Plants were then transplanted to Fafard-2 growing mix (Sungro Horticulture, Agawam, MA, USA) in 0.3-l pots in the glasshouse at the University of Pennsylvania (39.9493°N, 75.1995°W, 22.38 m above sea level) and kept in plastic bags for increased humidity for 2 wk. Plants were transferred to 4.2-litre pots with Fafard-52 growing mix 3 wk later and fertilized with Osmocote 14–14–14 (The Scotts Co., Marysville, OH, USA), supplemented by weekly applications of Peters 20–10–20 (ICL Fertilizers, Dublin, OH, USA). Plants were grown at 22–27°C on a 16 h photoperiod, with illumination provided by a combination of natural light and 400 W metal halide lamps (P.L. Light Systems, Ontario, Canada) with daily irradiances of 300–1500 lmol m−2 s−1 measured at the top of the canopy across the day. Planting of individuals was staggered across three months with the position of plants in the glasshouse randomized and individuals rotated frequently.
Populus tremula × alba seeds were purchased from Sheffield’s Seed Co. (Locke, NY, USA), and were germinated on a layer of vermiculite on top of Fafard-2 growing mix in 0.64-l pots in the glasshouse under conditions described above. Seedlings were transplanted 1 month after germination into Fafard-52 growing mix in 1.76-l pots, fertilized with Osmocote 14–14–14. They were transplanted to 4.2-l pots 3 months later. Planting of individuals was staggered across 5 months and individuals were rotated within the glasshouse frequently.
mRNA and smRNA abundance
For initial screening of miR156 and miR157 abundance, and follow-up validation of selected lines, shoot apices with leaf primordia less than 3 cm were used. For characterization of temporal patterns of miR156/157 and SPL abundance in seed-grown and culture derived plants, leaf primordia smaller than 1 cm were sampled and pooled with at least three leaves per replicate. For all plants, node or leaf positions are counted from the base of the plant where the first leaf produced is number 1. Primordia were collected across the plant’s lifespan to obtain samples for each of the measured leaf positions when they were at the same stage of development, frozen with liquid N2 and stored at −70°C until all samples were ready for RNA extraction. Total RNA was isolated using the Spectrum™ Plant Total RNA Kit (Sigma-Aldrich, St Louis, MO, USA), according to the manufacturer’s instructions. Small noncoding RNA (smRNA) levels were measured by real-time quantitative PCR (RT-qPCR), using primers specific for mature sequences of miR156, miR157 and miR159 in combination with the stem-loop RT primers described by Varkonyi-Gasic et al., (2007) (Table S2). miR159 was used as an endogenous control (Leichty & Poethig, 2019) and validated across time points in seed-grown and culture derived plants (Fig. S2). cDNA was synthesized using Invitrogen SuperScript III following the methods of Varkonyi-Gasic et al. (2007). Platinum Taq (Invitrogen, Waltham, MA, USA) was used with the Roche universal hydrolysis probe #21, and a three-step amplification protocol. For measuring mRNA abundance, total RNA was DNase-digested (Qiagen, Germantown, MD, USA) followed by cDNA synthesis using Invitrogen SuperScript III using each manufacturer’s protocol and a polyT primer. PtaACT2 or PtaCDC2 was used as the endogenous control (Pettengill et al., 2012). Resulting cDNA was quantified by qPCR using SYBR-Green Master Mix (Bimake, Houston, TX, USA) and primers specific to the endogenous control and SPL genes (PtSPL24 and PtSPL23 which each contain an miR156 target site and PtSPL4 and PtSPL7 which do not) (Table S2) with a three-step amplification protocol. Relative measures of abundance were calculated using the 2−ΔΔCt method (Livak & Schmittgen, 2001).
Stomatal impressions, epidermal traits, petiole morphology and abaxial trichome density
Stomatal density and epidermal cell size were measured using clear nail polish impressions of adaxial and abaxial surfaces of fully expanded leaves. Trichomes were removed from the abaxial side of leaves with tape when necessary, and three sections per leaf were imaged and photographed using an Olympus BX51 light microscope. Hand sections of leaf petioles within 1 cm of the leaf attachment site were imaged as described above. Trichome density on the abaxial side of the leaf was measured using an Olympus MVX10 dissecting scope. Measurements were made along the midvein, where the density was low enough to permit accurate counts. All measurements and counts were made using FIJI software (Schindelin et al., 2012).
Leaf vein densities
Sections from new fully expanded leaves were cleared using NaOH and bleach, and then stained with Safranin following the protocol described by Scoffoni & Sack (2013). Vein lengths of all visible vein orders were measured from microscope images using FIJI software (Schindelin et al., 2012), and vein densities were calculated following Scoffoni & Sack (2013).
Leaf shape and size
Fully expanded leaves were attached to paper using double-sided tape, labeled by node number increasing from the base of the plant and scanned. The scanned images were measured using the Morphological Analysis of Size and Shape (MASS) software (Chuanromanee et al., 2019). The SHAPE program (Iwata & Ukai, 2002) and FIJI (Schindelin et al., 2012) were used to determine the number of serrations and the perimeter of the leaf. The perimeter of the leaf was measured from the tops of the serrations so as not to include the added length from the serration indentations. Serrations were analyzed relative to the leaf perimeter so that the data were not influenced by differences in overall leaf size.
Leaf initiation rate, internode distance and branching
The leaf initiation rate was determined by counting the node of the uppermost visible leaf at least twice a week between 10 and 160 d of growth in seed-grown plants and between 32 and 55 d after transplant to soil in 717–1B4 lines. The number of branches at least 1 cm in length between nodes 1 and 25 was determined after 4 months of growth in soil.
Fluttering
To measure leaf fluttering, plants were placed 100 cm away from a fan, with the leaf to be measured facing perpendicular to the wind. Wind speeds at the sample leaf position were 1.5 m s−1 as measured with an anemometer. Leaf movement was filmed at 240 fps for 30 s (representative example in Video S1). Leaf movement was analyzed by following the movement of a mark placed at the tip of the leaf using the TRACKER video analysis and modeling tool v.5.0.7. The height and width of the petiole were measured using a digital caliper.
Statistical analysis
All statistical analyses were performed in JMP® PRO v.14.0.0 (SAS Institute Inc., Cary, NC, USA). Traits in seed-grown poplar across leaf positions were measured from a minimum of 42 individuals and fit with a linear or second-degree polynomial as appropriate with P-values and R2 values reported. Traits that were examined at multiple leaf positions in 717 wild-type, MIM156 lines 84 and 22, and miR156OX lines 40 and 78 were measured from a minimum of six individuals and compared by analysis of covariance (ANCOVA) and a Student’s t-test (α = 0.05) where leaf position and genotype were the main effects. Whole-plant measures of branching and internode length in these plants was compared by one-way ANOVA where genotype was the main effect and a Student’s t-test (α = 0.05), and leaf emergence was compared by ANCOVA with days after transplant and genotype as the main effects.
Results
Isolation of MIR156a over-expressing and MIM156 plants
For the lines with confirmed transgene insertions, the relative abundance of miR156 averaged 0.63 and 1.37 for the MIM156 and MIR156a lines, respectively (Fig. S1A,B). MIR156A lines 78 and 40 were selected as strong and moderate miR156 over-expressors, respectively (Fig. S1A). MIM156 line 84 was selected as a strong miR156 mimic, while MIM156 line 22 was selected to represent a transgenic line in which miR156 levels were not significantly different from the wild-type (Fig. S1A,B, t-test: P = 0.51). The levels of miR156-targeted SPL genes were consistent with the levels of miR156/157 in different transgenic lines. MIM156 line 84 had, respectively, 3- and 9-fold more PtSPL24 and PtSPL23 transcripts relative to the wild-type (Fig. S1C). As expected based on its levels of miR156, the transcripts of both PtSPL24 and PtSPL23 in MIM156 line 22 did not differ from the wild-type (Fig. S1C). Both MIR156A lines 40 and 78 showed significantly lower levels of these same genes when compared with the wild-type (Fig. S1C). To confirm the specificity of these transgenes, we also measured levels of PtSPL4 and PtSPL7 – genes with no predicted miR156 target site. In both cases, there was no clear pattern of transcript abundance across transgenic lines (Fig. S1C).
miR156/157 and PtSPL24 abundance in seed-grown plants and 717–1B4 lines
Patterns of miR156/157 abundance in the leaf primordia of seed-grown plants matched that previously reported for other species, such as Arabidopsis thaliana and Vachellia sp. (He et al., 2018; Leichty & Poethig, 2019). miR156/157 abundance dropped 7-fold between nodes 1 and 10, followed by a slower decline in successive nodes (Fig. 1a). We first determined that the control small RNA, miR159, was present at the same level in leaves 15 and 25 of seed-grown plants and the clone 717–1B4 (Fig. S2), and then used this transcript to normalize the level of miR156/157 in wild-type and transgenic genotypes of 717–1B4. The abundance of miR156/157 in the primordia of leaves 15 and 25 in wild-type 717–1B4 was similar to the amount of miR156/157 in these same leaves in seed-grown poplar (Fig. 1a, b), suggesting that miR156/157 are expressed at comparable levels at the same node in plants propagated by these methods. Leaves 15 and 25 of the miR156OX lines had, respectively 4.9 and 5.6 times the amount of miR156 as wild-type leaves, a level comparable to that of leaves 3 and 5 of seed-grown plants (Fig. 1a,b). Although the amount of miR156/157 in leaves 15 and 25 of the MIM156 lines was similar to that in 717–1B4 and seed-grown poplar (Fig. 1a,b), line 84 had about one-third the amount of miR156 at an earlier stage of development, before it was transferred to soil (Fig. S1A). Target mimics do not necessarily reduce the level of their target miRNAs (Franco-Zorrilla et al., 2007), so the significance of these MIM156 data is difficult to interpret. Regardless, the similarity between the amount of miR156/157 in MIM156 and wild-type lines shows that the transformation protocol and vector components have no independent effect on miR156/157 abundance.
Fig. 1.
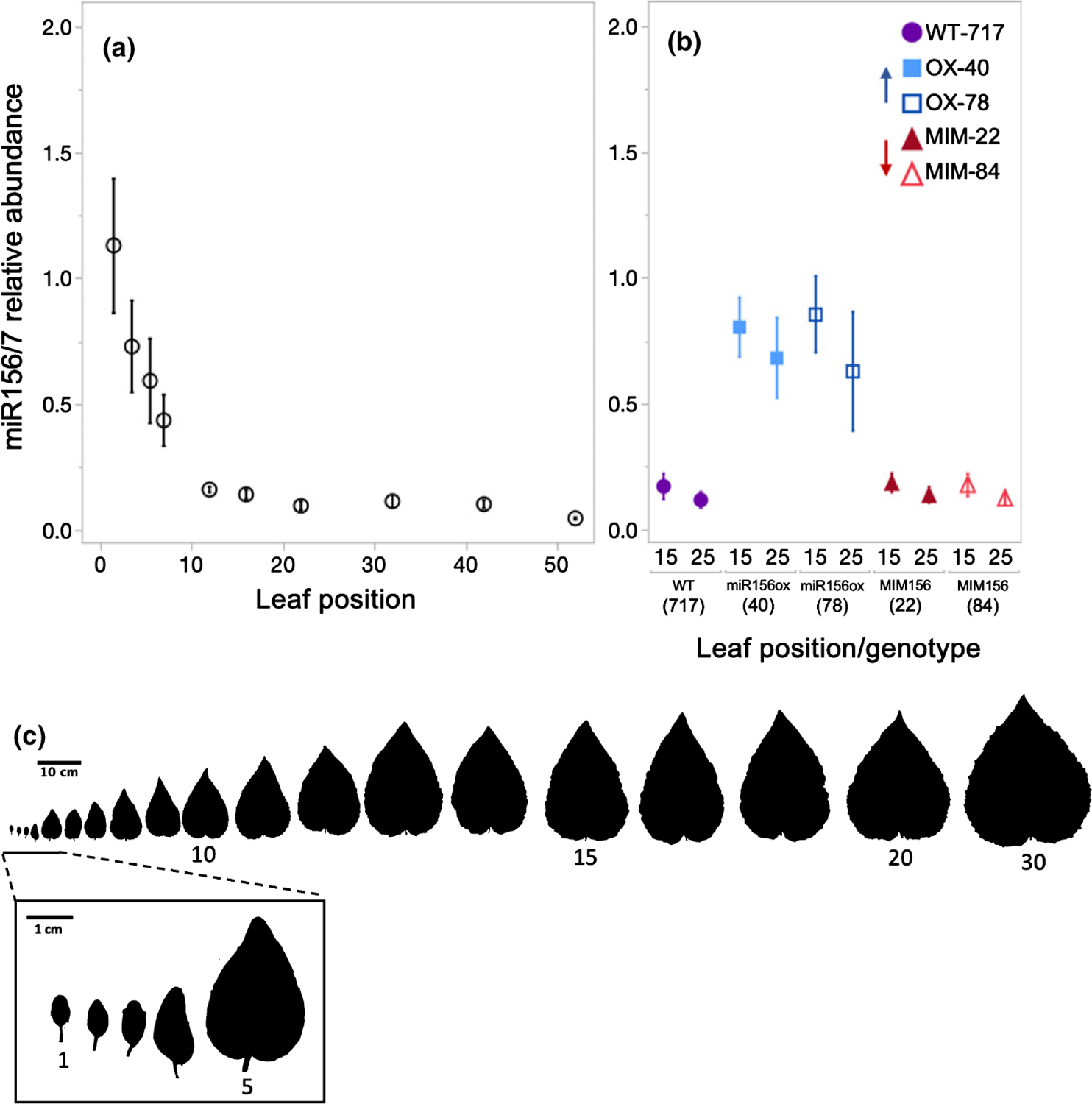
miR156/157 relative abundance in Populus tremula × alba leaf primordia grown from seed (a) or in transgenic and wild-type plants of the 717–1B4 background grown from tissue culture (b). miR156/157 abundance relative to the average of four replicates from seed-grown leaves 1 and 2. Data presented as means ± SE with seed-grown samples depicted by black open circles, 717 wild-type by purple circles, miR156a overexpression lines depicted by blue squares and MIM156 lines depicted by red triangles. Representative leaf shape for leaves 1–17, 20 and 30 from plants grown from seed (c).
To determine if the changes in miR156/157 abundance observed in seed- and culture-derived plants were large enough to have downstream effects on the expression of their SPL targets, we measured PtSPL24 abundance across nodes 1 and 34 in seed-grown plants and at nodes 15 and 25 of 717–1B4 lines. PtSPL24 abundance increased more than 11-fold between nodes 1 and 34 in seed-grown plants (Fig. 2a). In wild-type 717–1B4, PtSPL24 abundance increased 2-fold between nodes 15 and 25 (Fig. 2b). Across a similar span in seed-grown plants, PtSPL24 abundance increased by more than 1.5-fold between nodes 12 and 22, again indicating that the expression patterns of genes in the miR156/157-SPL pathway are comparable between similar nodes of plants propagated via seeds and cuttings. Leaves 15 and 25 in the miR156OX lines had 4- and 15-fold less PtSPL24 respectively, compared to these same nodes in the wild-type line. This is consistent with the levels of miR156/157 in these lines, and indicates that these levels of miR156/157 are functionally significant. The abundance of PtSPL24 in MIM156 line 22 was similar to that in the wild-type, consistent with the observation that the abundance of miR156 in this line is not significantly different from the wild-type. Interestingly, the abundance of PtSPL24 was more than 2-fold greater in leaf 15 of MIM156 line 84 compared to the wild-type, although the level of miR156/157 in this transgenic leaf is similar to the wild-type. This discrepancy between miR156 and PtSPL24 expression differences could be due to the nature of the target mimic method as discussed previously. There was no difference in the abundance of PtSPL24 at node 25 in MIM156 line 84 and wild-type plants. Together with the abundance of miR156 and PtSPL24 measured in young plants still in culture (Fig. S1), these data suggest that the MIM156 transgene has the greatest effect during the juvenile phase, when miR156 levels are naturally high.
Fig. 2.
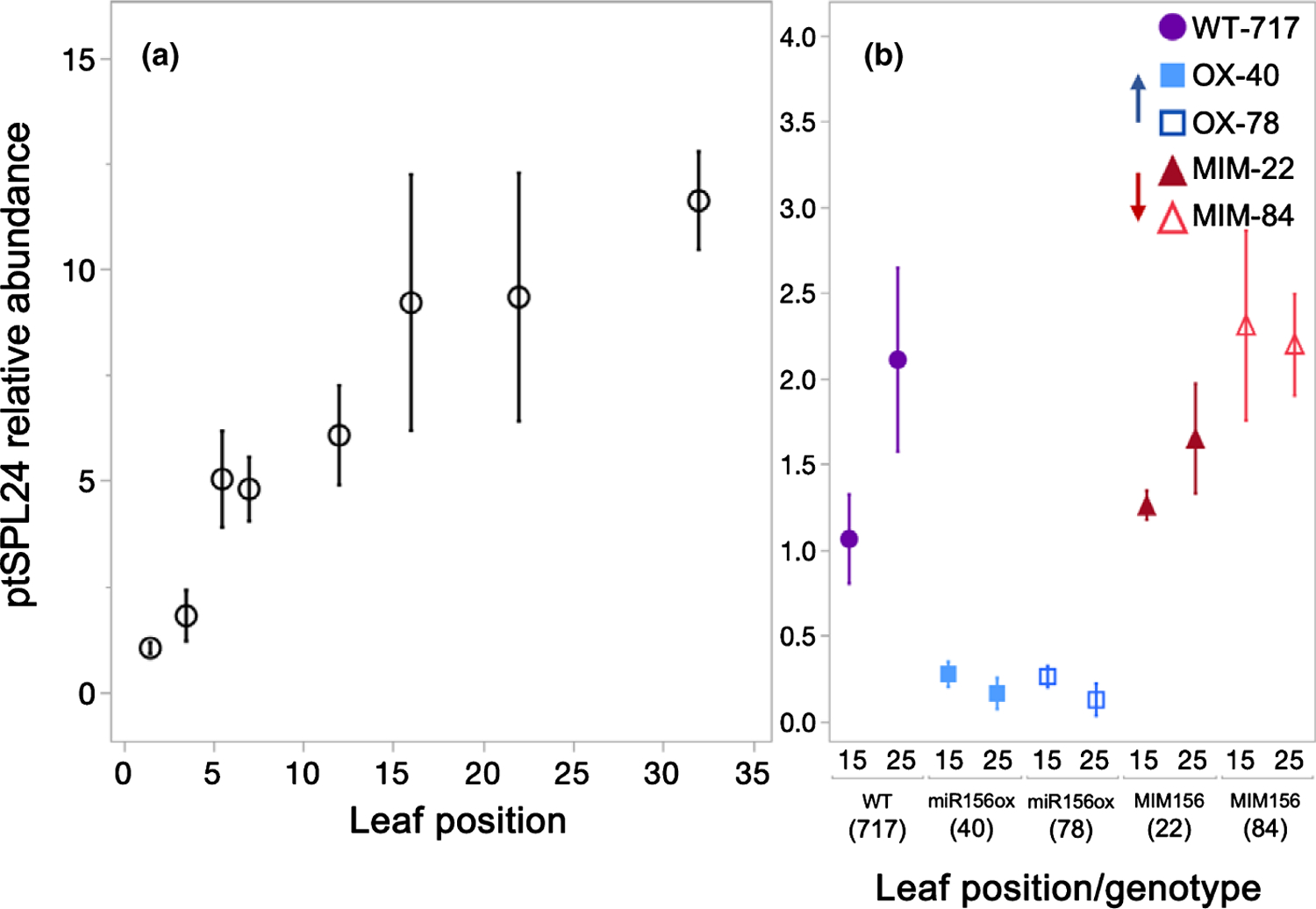
PtSPL24 relative abundance in Populus tremula × alba leaf primordia grown from seed (a) or in transgenic and wild-type plants of the 717–1B4 background grown from tissue culture (b). PtSPL24 abundance in seed-grown plants relative to seed-grown leaves 1 and 2 (a) or relative to the average wild-type 717 leaf 15 in plants from tissue culture (b). Data presented as means ± SE (n = 3) with seed-grown data depicted as black open circles, 717 wild-type by purple circles, miR156a overexpression lines depicted by blue squares and MIM156 lines depicted by red triangles.
Changes in miR156 abundance alter plant growth rate and form
Plants grown from seed produced leaves at a relatively slow rate for the first few weeks after planting, and transitioned to a faster, linear growth rate after producing 15–20 leaves (Fig. 3a). Leaf initiation in the wild-type and transgenic clones of 717–1B4 was delayed for 35–40 d after transplanting to soil, and then occurred at increasing rate over the next 2–3 wk of measurement. During this latter period, miR156OX lines had higher rates of leaf initiation than either the wild-type or MIM156 lines (Fig. 3b). miR156OX lines also had shorter internodes than either the wild-type or MIM156 lines (Fig. 3c) and produced significantly more axillary branches after 3 months than these genotypes (Fig. 3d; photographs of plants at this age in Fig. S3). The MIM156 lines were not significantly different from 717–1B4 for any of these traits, although these lines did have slightly longer internodes than 717–1B4.
Fig. 3.
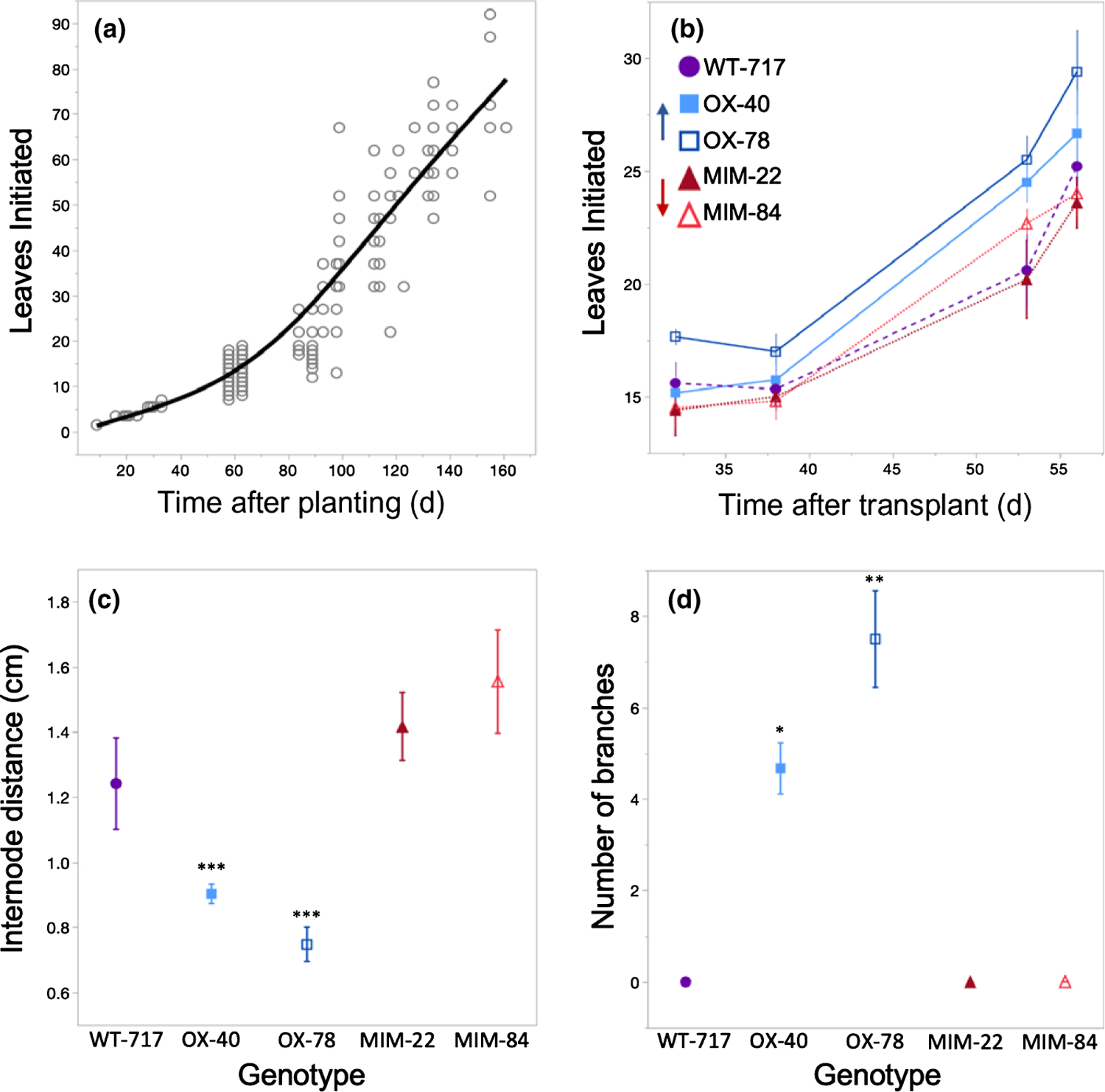
Number of leaves initiated by Populus tremula × alba grown from seed (a) and the 717–1B4 wild-type and four transgenic lines from tissue culture (n = 3–6) (b). Internode distance between nodes 10 and 20 (n = 6) (c), and number of branches of > 1 cm in length between nodes 1 and 25 after 4 months of growth (n = 6) (d) in the 717–1B4 and four transgenic lines. Data from seed-grown plants presented as individual biological replicates using grey open circles and a mean best-fit line in black or as means ± SE with 717 wild-type data depicted by purple circles, miR156a overexpression lines depicted by blue squares and MIM156 lines depicted by red triangles. Data in (a) show a second-degree polynomial fit with P < 0.0001 and R2 = 0.92 and an ANCOVA for (b) found P < 0.0001 for genotype and days after transplant but no significance for an interaction between effects. One-way ANOVA for (c) and (d) found P < 0.0001 with the results of a Student’s t-test between wild-type and mutants represented by: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Juvenile and adult leaves differ in leaf shape and size
The leaves of seed-grown plants became gradually rounder (decreased length : width (L : W) ratio) (Fig. 4a) and larger (Fig. 4c) from node 1 to node 25 and did not change significantly in shape or size after this point (Figs 1c, 4c). 717–1B4 displayed a similar trend, although the leaves of this clone reached their climax shape and size a few nodes earlier than seed-grown plants (Figs 4b,d, 5). MIM156 lines did not differ significantly in size from 717–1B4, but the leaves of line 84 were rounder than 717–1B4 beginning at node 15. By contrast, the leaves of miR156OX lines were significantly more elongated and smaller than wild-type leaves starting from node 15 (Student’s t-test P < 0.05). These results suggest that changes in leaf shape and size during shoot development in poplar are partly regulated by a decrease in the level of miR156.
Fig. 4.
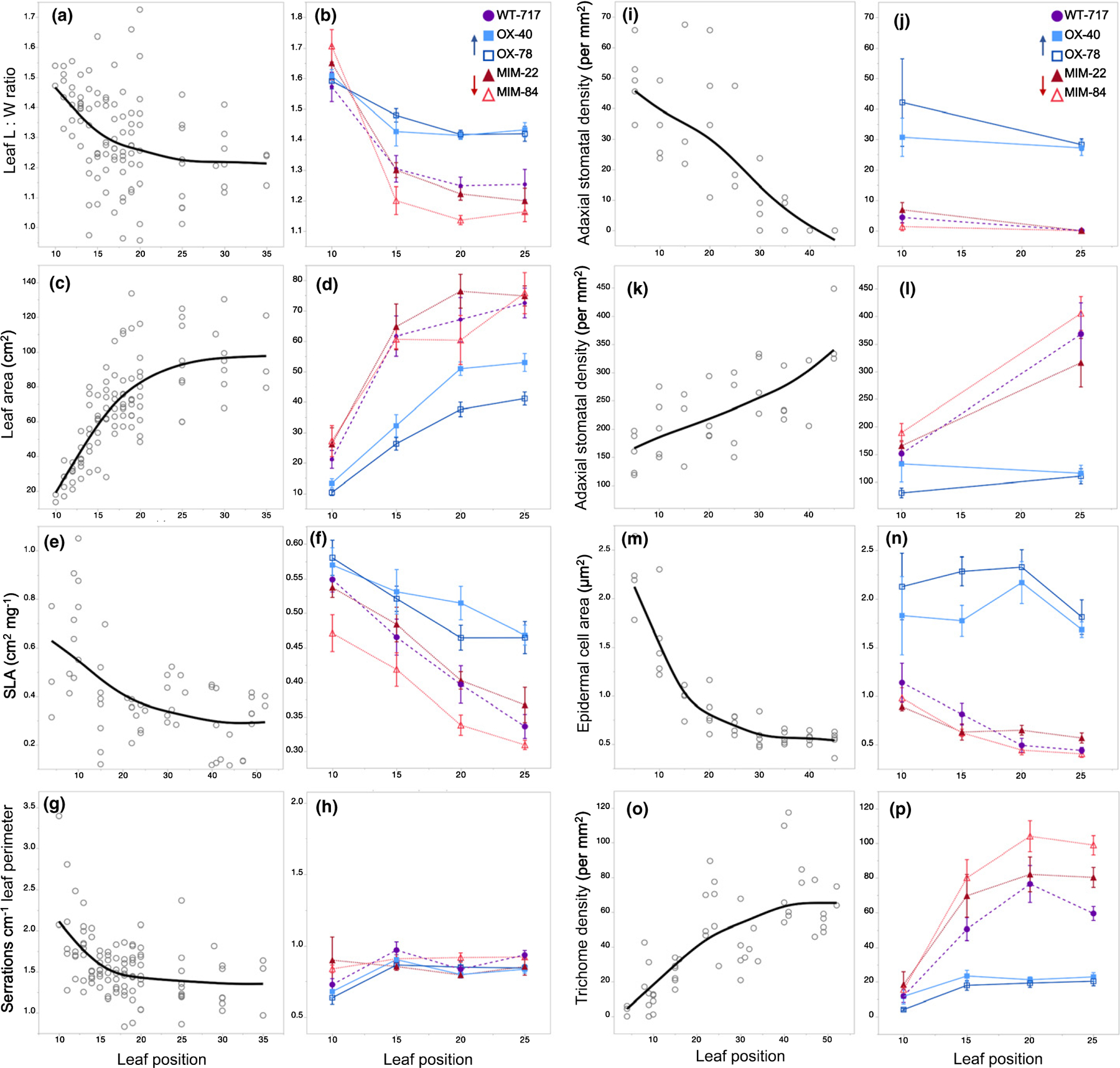
Leaf length : width (L : W) ratio, leaf area, specific leaf area (SLA), serrations per centimeter of leaf perimeter, adaxial stomatal density, abaxial stomatal density, adaxial epidermal cell area and abaxial trichome density of seed-grown P. tremula × alba (a, c, e, g, i, k, m, o) and the 717–1B4 and transgenic lines (b, d, f, h, j, l, n, p). Data from seed-grown plants presented as individual biological replicates using grey open circles and a mean best-fit line in black or as means ± SE with 717 wild-type data depicted by purple circles, miR156a overexpression lines depicted by blue squares and MIM156 lines depicted by red triangles (n = 5–11). Data in (a), (c), (e), (g), (m) and (o) show a second-degree polynomial relationship with P < 0.0001 and R2 values of 0.17, 0.6, 0.32, 0.25, 0.85 and 0.56, respectively. Data in (i) and (k) show a linear relationship with P < 0.0001 and R2 values of 0.58 and 0.46 respectively. ANCOVAs with leaf position and genotype as the effects for data in (b), (d), (f), (l), (n) and (p) found P < 0.001 for both leaf position and genotype. In (j), P < 0.001 for genotype but not significant (ns) for leaf position. In (h), P < 0.01 for leaf position but ns for genotype. For the interaction between genotype and leaf position, P < 0.05 for data in (b), (f), (l), and (p) and ns for (d), (h), (j) and (n).
Fig. 5.

Representative plants from the wild-type 717–1B4 Populus tremula × alba, miR156a overexpressor line 40 and MIM156 line 84 2 months following transplant from tissue culture. Leaves at positions 15 and 20 are noted. Representative leaf shape for fully expanded leaves from positions 10, 15, 20 and 25 in the 717–1B4 and transgenic poplar plants from tissue culture.
To obtain a comprehensive picture of the effect of miR156 on leaf morphology in P. tremula × alba, we also measured the specific leaf area (SLA), serrations, stomatal density, epidermal cell area, abaxial trichome density and petiole morphology of successive leaves. We examined every fifth leaf of seed-grown plants from germination to nodes 45–50, and every fifth leaf of 717–1B4 and transgenic lines from nodes 10 to 25. We observed greater variation for these traits in seed-grown plants compared to the culture-derived clones, probably because of the increased genetic variation in these open-pollinated plants.
SLA is the area of the leaf blade per unit mass and is determined by the thickness of the leaf blade and cell density. SLA impacts leaf physiology through changes in intraleaf light dynamics, CO2 diffusion and the distribution of leaf nitrogen (Parkhurst, 1994; Epron et al., 1995; Terashima & Hikosaka, 1995; Reich et al., 1998; Terashima et al., 2006; Evans et al., 2009), and therefore provides insights into how changes in leaf development alter function. SLA decreased in a curvilinear fashion from leaf 5 to leaf 40 in seed-grown plants and decreased linearly from leaf 10 to leaf 25 in 717–1B4 and the four transgenic lines (Fig. 4e,f). However, SLA decreased nearly linearly between leaf 10 and leaf 25 in seed-grown plants, so these data do not reveal if there is a significant difference in the pattern in seed-grown and clonally propagated plants. On the other hand, we found that – at nodes 15, 20 and 25 – lines over-expressing miR156 had a significantly higher SLA, and one MIM156 line (84) had a significantly lower SLA, than 717–1B4 at node 10 (Student’s t-test, P < 0.05 and P < 0.0001 for miR156OX lines at nodes 15 and 20, and node 25 respectively and P = 0.019 for line 84 node 10). These results suggest that SLA is a phase-specific trait and is regulated by miR156.
Leaf serrations are present on leaves of all measured positions for both seed- and culture-derived plants. To account for significant differences in leaf size, serration numbers are normalized to leaf perimeter. We found a slight decrease in the number of serrations per centimeter of leaf perimeter in seed-grown plants, decreasing from just over two at node 10 to around 1.5 at node 35 (Fig. 4g). 717–1B4 lines showed no phase-specific differences in the number of serrations (P = 0.1469) as leaves of all genotypes and all positions had between 0.64 and 0.97 serrations per centimeter of leaf perimeter (Fig. 4h). It is unclear why culture-derived plants have lower serration density than seed-grown plants, but these small differences in serrations are not likely to have any biological significance.
Stomatal density on both the adaxial and the abaxial side of the leaf changed across leaf positions, but in opposite directions (Fig. 4i,k). In seed-grown plants, adaxial stomatal density decreased linearly from leaf 5 to leaf 40, whereas abaxial stomatal density increased linearly over this interval; at every node, the density of stomata on the adaxial surface of the leaf was significantly less than that on the abaxial surface. Both 717–1B4 and the MIM156 lines had a very low density of adaxial stomata on leaf 10, and this number was reduced to nearly zero on leaf 25. By contrast, the density of abaxial stomata in these genotypes increased significantly between leaf 10 and leaf 25. In the miR156OX lines, the density of stomata on both the adaxial and the abaxial surfaces of leaves 10 and 25 was essentially identical to that of the basal-most leaves of seed-grown plants. Thus, miR156 has a role in regulating stomatal density on both sides of the leaf. However, other factors are also likely to contribute to the developmental changes in stomatal density (Berger & Altmann, 2000; Sugano et al., 2010), because this parameter changes linearly during shoot development, rather than in the curvilinear pattern characteristic of the change in miR156 levels (Fig. 4j,l).
We measured epidermal cell area by taking impressions of the adaxial epidermis, because the high density of trichomes made it impractical to measure this trait on the abaxial surface. In seed-grown plants, cell area decreased c. 4-fold over the first 30 leaves and remained relatively constant after this point (Fig. 4m). In 717–1B4, cell area decreased about 2-fold from leaf 10 to leaf 20, and decreased much less dramatically, if at all, between leaf 20 and leaf 25. The MIM156 lines were not significantly different from 717–1B4. By contrast, cell area in the miR156OX did not change significantly between leaf 10 and leaf 25 (Fig. 4n). These results suggest that epidermal cell area is a phase-specific trait.
The leaves of Populus alba appear white because of a dense layer of trichomes on the abaxial surface of the leaf blade, and this trait is also characteristic of P. tremula × alba (Fig. S4). All the leaves of P. tremula × alba produced abaxial trichomes, but trichome density increased dramatically during shoot growth (Fig. 4o,p). In seed-grown plants, abaxial trichome density increased more than 10-fold between leaf 5 and leaf 40 and increased by about 6-fold between leaf 10 and leaf 25 in 717–1B4. By contrast, abaxial trichome density remained at the same relatively low level from node 10 to node 25 in miR156OX, and it increased significantly more than 717–1B4 in both MIM156 lines (Student’s t-test; P < 0.01 and P < 0.0001 for lines 22 and 84 respectively). This result suggests that abaxial trichome production is a phase-specific trait in P. tremula × alba and is hyper-sensitive to the level of miR156.
Leaf vein density increases as plants transition from juvenile to adult
While working with the 717–1B4 lines, we noted differences in vasculature of leaves at the same position between wild-type and transgenic lines. Because vein density and patterning impact leaf structure and function (i.e. assimilation and herbivore resistance) (Brodribb et al., 2007; Blonder et al., 2018), we thought this trait was worth investigating. Leaf vein density increased 1.8-fold with increasing leaf position in 717–1B4 and in the MIM156 lines, although MIM156 line 84 had a significantly higher vein density than 717–1B4 in leaf 10 (Student’s t-test, P < 0.01). Similar results were obtained for the density of vein endings. In the miR156OX lines, vein density and the density of vein endings did not differ much between leaf 10 and leaf 25, and were identical to, or slightly lower than, the values observed for leaf 10 in 717–1B4 (Fig. 6).
Fig. 6.
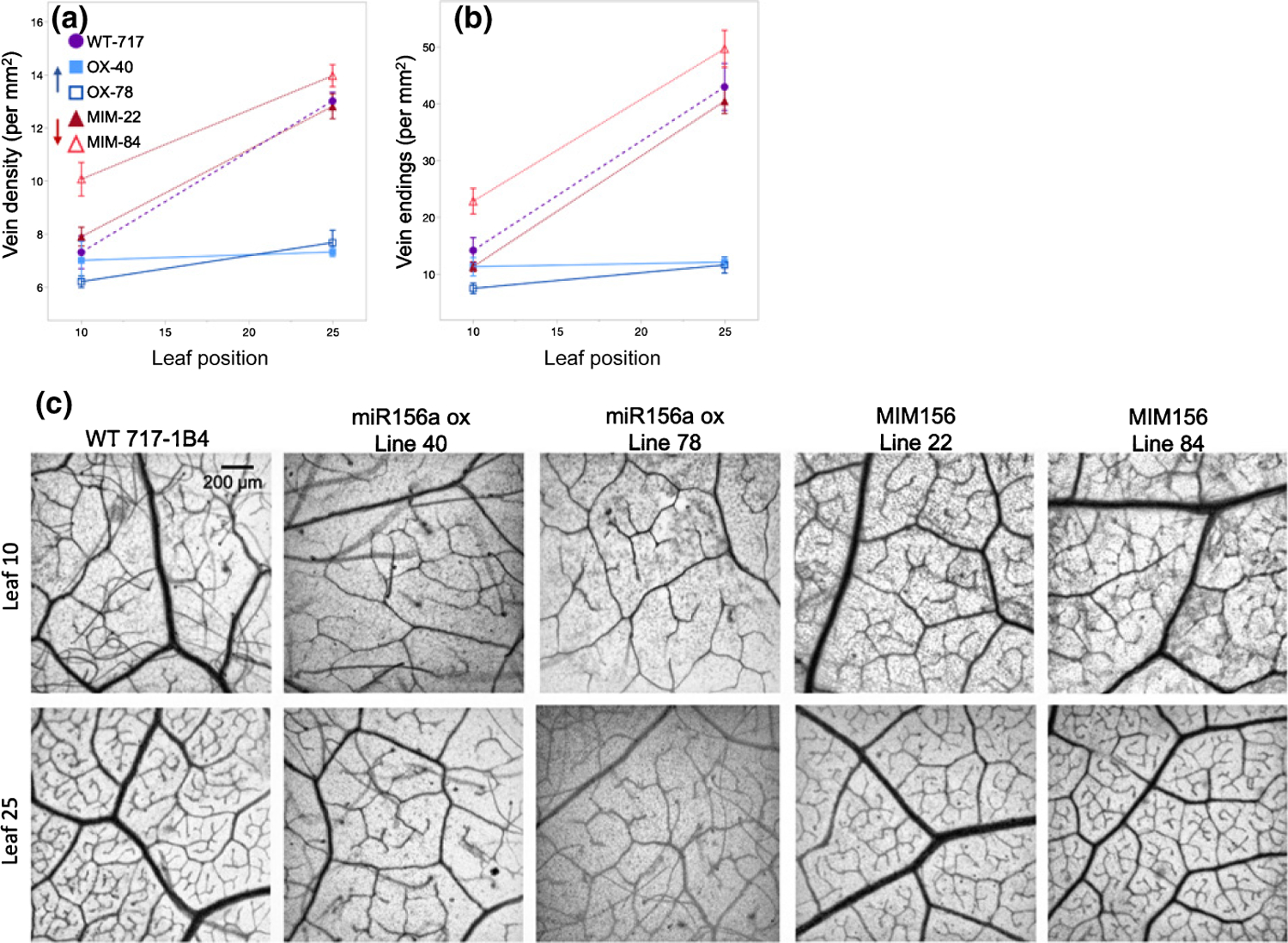
Total leaf vein density (a) and the number of vein endings (b) in the 717–1B4 and transgenic Populus tremula × alba. Data from seed-grown plants presented as individual biological replicates using grey open circles and a mean best-fit line in black or as means ± SE with 717 wild-type data depicted by purple circles, miR156a overexpression lines depicted by blue squares and MIM156 lines depicted by red triangles (n = 5–6). ANCOVA with leaf position and genotype as the effects found P < 0.0001 for both effects and the interaction for data in (a) and (b). Representative samples of leaves cleared and stained to visualized vein density and architecture in leaves 10 and 25 of 717–1B4 and transgenic poplar lines (c).
Petiole morphology changes during development leading to altered leaf fluttering behavior
Many species of poplar, including P. tremula × alba, exhibit leaf fluttering, a trait that has been attributed to the vertical elongation of the petiole (Niklas, 1991). In a previous study of Populus × canadensis, we found that the petioles of in vitro propagated plants became increasing elongated as the shoot developed, and that over-expression of miR156 suppressed this process (Wang et al., 2011). To determine if this phenomenon occurs naturally, we examined the L : W ratio of a cross-section of the petiole in seed-grown P. tremula × alba (Fig. 7a,d). We found that the petiole of leaves at node 10 was slightly broader than tall (L : W < 1), whereas the petioles of leaves at higher nodes became vertically elongated (L : W > 1), reaching their climax shape around node 40. In wild-type 717–1B4, and in the MIM156 and miR156OX lines, the petiole also became progressively elongated (Fig. 7b,c). However, the miR156OX lines had significantly fewer elongated petioles than the 717–1B4 and the MIM156 lines, and reached a maximum L : W ratio at an earlier node. We also examined the effect of these transgenes on petiole anatomy. In 717–1B4, the petioles of leaves at nodes 10 and 15 had an adaxial indentation, a large central vascular bundle and two smaller vascular bundles located in a horizontal plane above the central bundle. The petioles of leaves 20 and 25 lacked an adaxial indentation and had three vertically oriented vascular bundles that decreased in size from the abaxial to the adaxial side of the petiole. In the MIM156 lines, the vascular anatomy of leaf 15 was intermediate between that of leaf 10 and leaf 20 in 717–1B4, indicating that the transition to the adult shape is accelerated in these lines, despite the lack of a significant change in the L : W ratio of the petiole (Fig. 7c). Consistent with their L : W ratio, lines over-expressing miR156 retained a juvenile petiole morphology up to at least node 25. Petiole anatomy is therefore an obvious qualitative indicator of the phase identity of a leaf in P. tremula × alba.
Fig. 7.
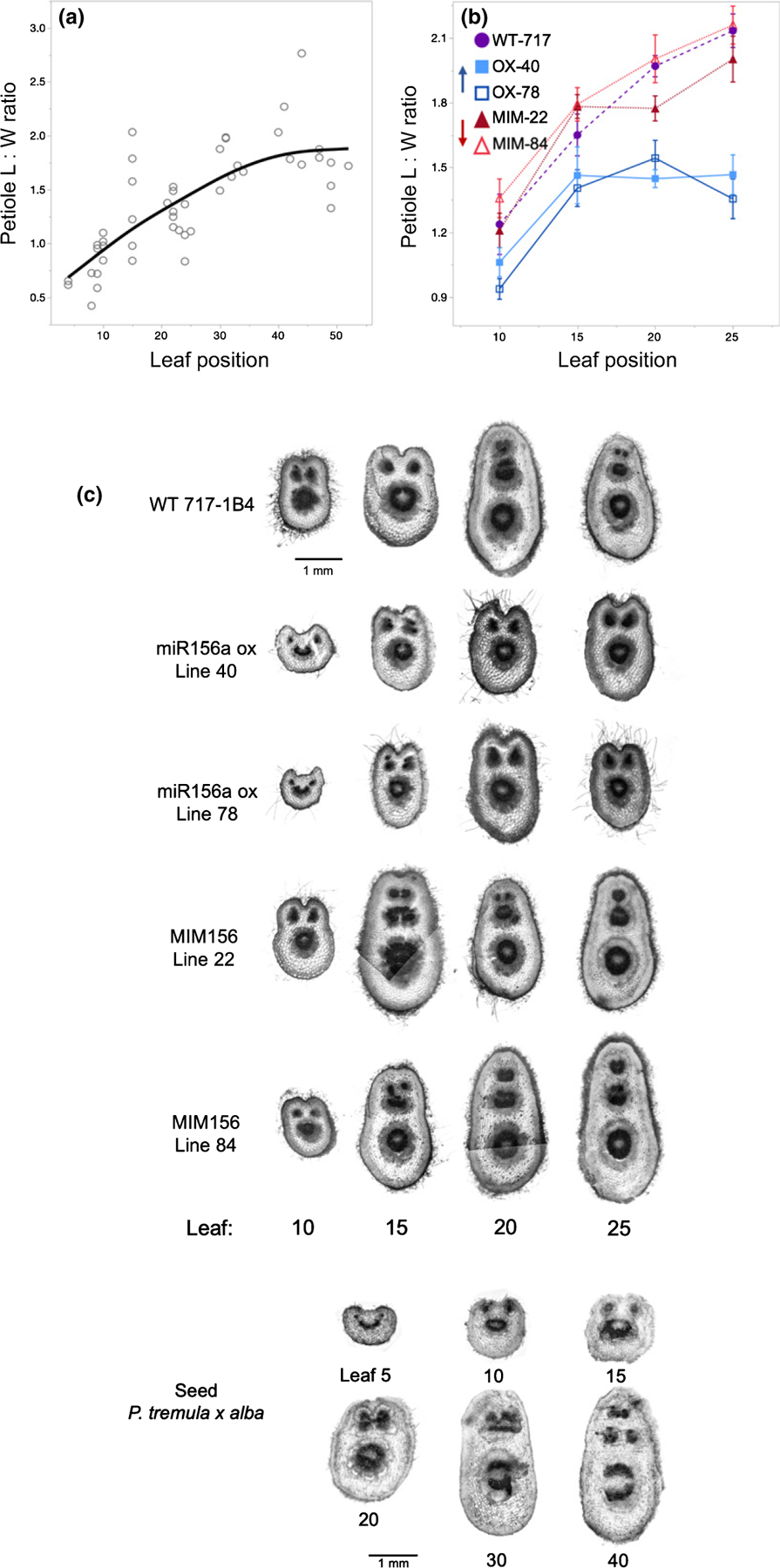
Petiole length : width (L : W) ratio from cross-sections made within 1 cm of leaf attachment in seed-grown (a) and 717–1B4 and transgenic Populus tremula × alba (b). Data from seed-grown plants presented as individual biological replicates using grey open circles and a mean best-fit line in black or as means ± SE with 717 wild-type data depicted by purple circles, miR156a overexpression lines depicted by blue squares and MIM156 lines depicted by red triangles (n = 7–12). Data from (a) show a second-degree polynomial relationship with P < 0.0001 and R2 = 0.62. For (b), ANCOVA with leaf position and genotype as the effects found P < 0.0001 for leaf position and genotype and P = 0.01 for the interaction. Representative samples of petiole cross-sections from leaves 10, 15, 20 and 25 of the 717–1B4 and four transgenic lines (c). Representative samples of petiole cross-sections from leaf positions between 5 and 40 in plants grown from seed (d).
To determine if this change in petiole morphology has an effect on the behavior of the leaf, we looked at two measures of leaf fluttering – the total distance moved by the leaf, and the maximum horizontal span of the leaf tip – when leaves were exposed to a constant wind velocity (Figs 8a, S5). There was a statistically significant (P < 0.0001) positive linear relationship between petiole L : W ratio and these fluttering measures in 717–1B4, the MIM156 and miR156OX lines. Leaves at basal node 10 in the 717–1B4 and in the MIM156 lines fluttered less than leaves at more apical node 25, whereas juvenile leaves at node 25 of the miR156OX lines fluttered less than leaves at node 25 of the 717–1B4 and in the MIM156 lines (Fig. 8b,c). Thus, the petiole morphology of juvenile and adult leaves of P. tremula × alba has a significant effect on their response to wind.
Fig. 8.
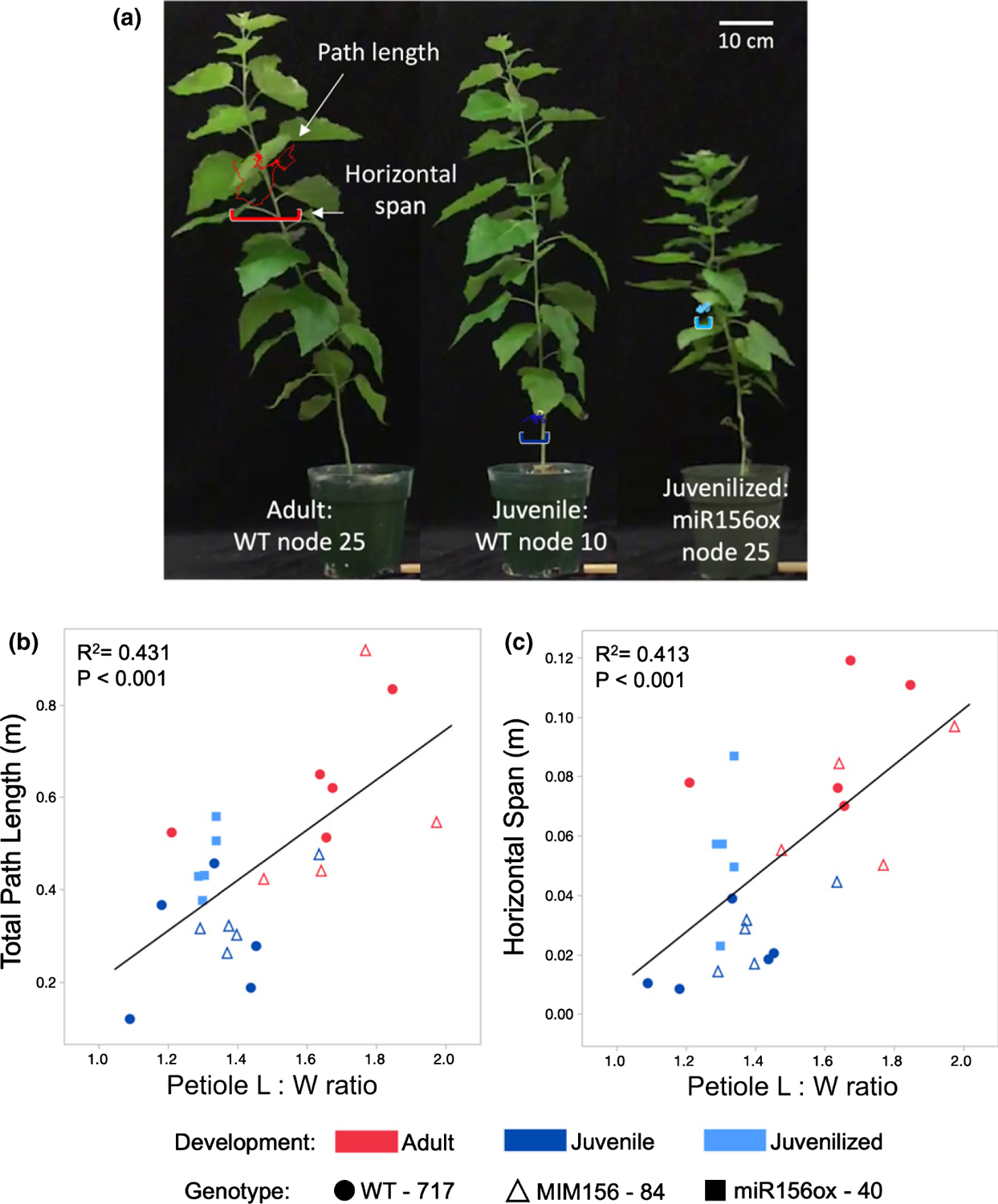
Diagram showing the path length and horizontal span measurements of leaf fluttering of an adult (WT 717–1B4 node 25), juvenile (WT 717–1B4 node 10) and juvenilized (miR156 overexpressor line 40 node 25) leaf of Populus tremula × alba. Quantification of leaf fluttering through total path length over a 1 s period (a) and maximum horizontal span of the tip of the leaf (b) and their relationship with petiole length : width ratio (L : W). 717–1B4 data are depicted by circles, miR156a overexpressor line 40 by squares and MIM156 line 84, by triangles. Leaf developmental stage is noted by color with adult leaves in red, juvenile in dark blue and juvenilized in light blue. Linear fit for (b) path length = −0.34288 + 0.544 (petiole L : W ratio) and (c) horizontal span = −0.0852 + 0.0939(petiole L : W ratio).
Discussion
Many plants lack visually abrupt changes in form during shoot development, making the onset of vegetative phase change difficult to observe. As a result, the existence of this phenomenon and its significance for plant ecology and evolution are often under-appreciated. In this study, we characterized the growth rate, developmental morphology and temporal pattern of miR156 and SPL expression in P. tremula × alba, an ecologically and economically important tree. We found that many leaf traits change during shoot development in plants grown from seeds, as well as in shoots arising de novo in culture, and that most of these changes are correlated with and sensitive to changes in the level of miR156. These data confirm and extend previous studies of the effect of miR156 on Populus development (Wang et al., 2011; Rubinelli et al., 2013), and provide a foundation for future research on the function of vegetative phase change in the life history of this woody species.
In nature, poplars propagate primarily from adventitious shoots, whereas clones used for research or commercial purposes are propagated from axillary buds or by inducing shoot regeneration in vitro (Taylor, 2002). The extent to which these plants recapitulate the development of the primary shoot is still unknown. We found by at least the 10th node, the development of a regenerated shoot is remarkably similar to that of seed-grown plants. This result suggests that the phase-identity of poplar cells is either reset during the dedifferentiation of the primary tissue sample, or during shoot regeneration, and is consistent with observations in Zea mays (Irish & Karlen, 1998; Orkwiszewski & Poethig, 2000). This observation also raises the long-standing question of whether differentiated traits can be maintained in tissue culture.
Although vegetative phase change sometimes involves traits unique to a particular species (Wang et al., 2011; Leichty & Poethig, 2019), analyses of plants with altered levels of miR156 have revealed a number of phase-specific traits that are common to phylogenetically diverse species. In Z. mays, A. thaliana, Nicotiana tabacum and now P. tremula × alba, these traits include patterns of trichome production, leaf shape and size, epidermal and stomatal cell traits, vein density, leaf initiation rate, and branching (Bongard-Pierce et al., 1996; Telfer et al., 1997; Feng et al., 2016; He et al., 2018). At least some of these traits are also regulated by miR156 in Populus × canadensis (Wang et al., 2011), rice (Xie et al., 2006), alfalfa (Aung et al., 2015), switchgrass (Fu et al., 2012), Lotus japonicus (Wang, 2014) and tomato (Zhang et al., 2011). This observation indicates that vegetative phase change is an ancient and fundamental aspect of shoot development, and also suggests that the differential expression of various traits during shoot development contributes to plant fitness and is therefore under natural selection (Leichty & Poethig, 2019).
Determining how phase-specific traits contribute to plant fitness is challenging. However, a number of studies have shown that plants over-expressing miR156 are more or less sensitive to various environmental stresses (Stief et al., 2014; Cui et al., 2014; Arshad et al., 2017; Kang et al., 2020), Despite this, there is still little evidence that juvenile and adult phases of wild-type plants display these same differences in sensitivity or how they might contribute to plant fitness. There are relatively few examples of phase-specific vegetative traits that are either known or expected to be selectively advantageous (Williams et al., 1998; Holeski et al., 2009; Leichty & Poethig, 2019). Our observation that adult leaves flutter more than juvenile leaves is another such example. In poplar, leaf fluttering is beneficial under high light and high heat environments, but is disadvantageous under low light conditions as it reduces photosynthesis (Roden & Pearcy, 1993a,b). This suggests that reduced leaf fluttering is advantageous early in development, when plants are more likely to be shaded, whereas increased fluttering is advantageous once plants are no longer in the understory.
Surprisingly, we found that progression of vegetative phase change in poplar, a woody perennial, is not very different from this process in Arabidopsis, an annual herb. In P. tremula × alba, the onset of adult traits begins within 3 months of growth, despite the much longer lifespan of this species. As in Arabidopsis (He et al., 2018), miR156 expression in poplar declines rapidly over the first few leaves, but it is not until later, when the decline in miR156 is more subtle, that adult morphological traits first appear. This result further supports the findings in Arabidopsis that miR156/157 expression in early leaves far exceeds the amount necessary to repress most SPL targets (He et al., 2018). Furthermore, these findings suggest that it is inappropriate to assume the timing of vegetative phase change is proportional to the lifespan of a species. As we continue to characterize this developmental transition across species with diverse life histories, we are likely to find large variations in the proportion of time spent in either the juvenile or the adult vegetative phase.
During shoot development, different morphological and physiological traits change in a coordinated fashion, but not necessarily at the same node or at the same rate (Hackett & Murray, 1997; Leichty & Poethig, 2019). Furthermore, under certain conditions, traits can become dissociated, resulting in leaves or shoots that have unusual combinations of juvenile and adult traits (Robbins, 1960; Doorenbos, 1965). In our experiments, this was apparent from the phenotype of transgenic lines expressing a miR156/miR157 target site mimic (MIM156). The stronger of these transgenes had a significant effect on leaf L : W ratio, SLA, epidermal cell area, petiole morphology and leaf vein density, but did not affect leaf initiation rate, stomatal density or petiole L : W ratio. These observations are consistent with studies in Arabidopsis, which show that different miR156-regulated SPL genes have different transcription patterns and sensitivity to miR156 abundance (Xu et al., 2016; He et al., 2018). Along with the evidence that SPL genes also have different loss-of-function phenotypes (Xu et al., 2016), these results suggest that variability in the expression of different phase-specific traits may arise from the differential response of functionally distinct SPL genes to changes in the level of miR156.
miR156 and its SPL targets control many economically important traits, including flowering time, shoot architecture, biomass and lignin production, pest and disease resistance, and abiotic stress responses (Wang & Wang, 2015). In the biofuel crop Panicum virgatum, for example, over-expressing miR156 leads to increases in biomass, digestibility and lignin production, a delay in flowering time, and altered susceptibility to the pathogen Puccinia emaculata (Chuck et al., 2011; Fu et al., 2012; Baxter et al., 2018). Similar effects of miR156 over-expression have been observed in the forage legumes Trifolium pretense (Zheng et al., 2016) and Medicago sativa (Aung et al., 2015). Poplars are fast-growing trees useful for a wide variety of industrial purposes, as well for the reforestation of degraded land (Taylor, 2002). The ability to engineer their chemistry and vegetative development through the manipulation of miR156 offers new opportunities for the improvement of this important group of plants.
Supplementary Material
Supporting Information
Additional Supporting Information may be found online in the Supporting Information section at the end of the article.
Table S1 Transformation constructs for MIM156 and miR156a transgenic P. tremula × alba lines.
Table S2 RT-qPCR primer sequences.
Fig. S1 The relative abundance of miR156 and SPL transcripts in leaf primordia of young transgenic and 717–1B4 wild-type P. tremula × alba grown in culture.
Fig. S2 Cq values from qPCR of miR159 at across-leaf positions.
Fig. S3 Representative plants from the wild-type 717–1B4, miR156a overexpressor line 40 and MIM156 line 84, 3 months following transplant from tissue culture.
Fig. S4 Leaf scans showing the adaxial (left) and abaxial (right) sides of adult, juvenile and juvenilized leaves of P. tremula × alba.
Fig. S5 Diagram showing representative fluttering paths for adult (node 25) and juvenile (node 10) leaves of MIM156 line 84.
Video S1 Representative samples of leaf fluttering in an miR156 overexpressor (left) and 717–1B4 wild-type line (right) of P. tremula × abla.
Acknowledgements
We thank Samara Gray and Joshua Darfler for their assistance in caring for the plants used in this study. This research was funded by the NSF Graduate Research Fellowship (DGE-1845298), University of Pennsylvania SAS Dissertation Research Fellowship and the Peachey Research Fund to EHL, NIH T32GM008216 to ARL and NIH GM51893 to RSP.
References
- Arshad M, Feyissa BA, Amyot L, Aung B, Hannoufa A. 2017. MicroRNA156 improves drought stress tolerance in alfalfa (Medicago sativa) by silencing SPL13. Plant Science 258: 122–136. [DOI] [PubMed] [Google Scholar]
- Ashby E 1948. Studies in the morphogenesis of leaves. II. The area, cell size and cell number of leaves of Ipomoea in relation to their position on the shoot. New Phytologist 47: 177–195. [Google Scholar]
- Aung B, Gruber MY, Amyot L, Omari K, Bertrand A, Hannoufa A. 2015. Ectopic expression of LjmiR156 delays flowering, enhances shoot branching, and improves forage quality in alfalfa. Plant Biotechnology Reports 9: 379–393. [Google Scholar]
- Baxter HL, Mazarei M, Dumitrache A, Natzke JM, Rodriguez M, Gou J, Fu C, Sykes RW, Turner GB, Davis MF et al. 2018. Transgenic miR156 switchgrass in the field: growth, recalcitrance and rust susceptibility. Plant Biotechnology Journal 16: 39–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger D, Altmann T. 2000. A subtilisin-like serine protease involved in the regulation of stomatal density and distribution in Arabidopsis thaliana. Genes and Development 14: 1119–1131. [PMC free article] [PubMed] [Google Scholar]
- Blonder B, Salinas N, Patrick Bentley L, Shenkin A, Porroa POC, Tejeira YV, Espinoza TEB, Goldsmith GR, Enrico L, Martin R et al. 2018. Structural and defensive roles of angiosperm leaf venation network reticulation across an Andes–Amazon elevation gradient. Journal of Ecology 106: 1683–1699. [Google Scholar]
- Bongard-Pierce DK, Evans MMS, Poethig RS. 1996. Heteroblastic features of leaf anatomy in maize and their genetic regulation. International Journal of Plant Sciences 157: 331–340. [Google Scholar]
- Brink RA. 1962. Phase change in higher plants and somatic cell heredity. The Quarterly Review of Biology 37: 1–22. [Google Scholar]
- Brodribb TJ, Feild TS, Jordan GJ. 2007. Leaf maximum photosynthetic rate and venation are linked by hydraulics. Plant Physiology 144: 1890–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuanromanee TS, Cohen JI, Ryan GL. 2019. Morphological analysis of size and shape (MASS): an integrative software program for morphometric analyses of leaves. Applications in Plant Sciences 7: e11288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck GS, Tobias C, Sun L, Kraemer F, Li C, Dibble D, Arora R, Bragg JN, Vogel JP, Singh S et al. 2011. Overexpression of the maize Corngrass1 microRNA prevents flowering, improves digestibility, and increases starch content of switchgrass. Proceedings of the National Academy of Sciences, USA 109: 995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L-G, Shan J-X, Shi M, Gao J-P, Lin H-X. 2014. The miR156-SPL9-DFR pathway coordinates the relationship between development and abiotic stress tolerance in plants. The Plant Journal 80: 1108–1117. [DOI] [PubMed] [Google Scholar]
- Doorenbos J 1965. Juvenile and adult phases in woody plants. Encyclopedia of Plant Physiology 15: 1222–1235. [Google Scholar]
- Engler C, Youles M, Gruetzner R, Ehnert T-M, Werner S, Jones JDG, Patron NJ, Marillonnet S. 2014. A Golden Gate modular cloning toolbox for plants. ACS Synthetic Biology 3: 839–843. [DOI] [PubMed] [Google Scholar]
- Epron D, Godard D, Cornic G, Genty B. 1995. Limitation of net CO2 assimilation rate by internal resistances to CO2 transfer in the leaves of two tree species (Fagus sylvatica L. and Castanea sativa Mill.). Plant, Cell & Environment 18: 43–51. [Google Scholar]
- Evans JR, Kaldenhoff R, Genty B, Terashima I. 2009. Resistances along the CO2 diffusion pathway inside leaves. Journal of Experimental Botany 60: 2235–2248. [DOI] [PubMed] [Google Scholar]
- Feng S, Xu Y, Guo C, Zheng J, Zhou B, Zhang Y, Ding Y, Zhang L, Zhu Z, Wang H et al. 2016. Modulation of miR156 to identify traits associated with vegetative phase change in tobacco (Nicotiana tabacum). Journal of Experimental Botany 67: 1493–1504. [DOI] [PubMed] [Google Scholar]
- Filichkin SA, Meilan R, Busov VB, Ma C, Brunner AM, Strauss SH. 2006. Alcohol-inducible gene expression in transgenic Populus. Plant Cell Reports 25: 660–667. [DOI] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, García JA, Paz-Ares J et al. 2007. Target mimicry provides a new mechanism for regulation of microRNA activity. Nature Genetics 39: 1033–1037. [DOI] [PubMed] [Google Scholar]
- Fu C, Sunkar R, Zhou C, Shen H, Zhang J-YY, Matts J, Wolf J, Mann DGJ, Stewart CN, Tang Y et al. 2012. Overexpression of miR156 in switchgrass (Panicum virgatum L.) results in various morphological alterations and leads to improved biomass production. Plant Biotechnology Journal 10: 443–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebel K 1900. Organography of Plants Part I. General Organography. (English Translation by Balfour IB). Oxford, UK: Clarendon Press. [Google Scholar]
- Hackett WP, Murray JR. 1997. Approaches to understanding maturation or phase change. In: Geneve RL, Preece JE, Merkle SA, eds. Biotechnology of ornamental plants. New York, NY, USA: CAB International, 73–86. [Google Scholar]
- He J, Xu M, Willmann MR, McCormick K, Hu T, Yang L, Starker CG, Voytas DF, Meyers BC, Poethig RS. 2018. Threshold-dependent repression of SPL gene expression by miR156/miR157 controls vegetative phase change in Arabidopsis thaliana. PLoS Genetics 14: e1007337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holeski LM, Kearsley MJC, Whitham TG. 2009. Separating ontogenetic and environmental determination of resistance to herbivory in cottonwood. Ecology 90: 2969–2973. [DOI] [PubMed] [Google Scholar]
- Irish EE, Karlen S. 1998. Restoration of juvenility in maize shoots by meristem culture. International Journal of Plant Sciences 159: 695–701. [Google Scholar]
- Iwata H, Ukai Y. 2002. SHAPE: a computer program package for quantitative evaluation of biological shapes based on elliptic Fourier descriptors. The Journal of Heredity 93: 384–385. [DOI] [PubMed] [Google Scholar]
- James SA, Bell DT. 2001. Leaf morphological and anatomical characteristics of heteroblastic Eucalyptus globulus ssp. globulus (Myrtaceae). Australian Journal of Botany 49: 259–269. [Google Scholar]
- Kang T, Yu CY, Liu Y, Song WM, Bao Y, Guo XT, Li B, Zhang HX. 2020. Subtly manipulated expression of ZmmiR156 in tobacco improves drought and salt tolerance without changing the architecture of transgenic plants. Frontiers in Plant Science 10: 1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leichty AR, Poethig RS. 2019. Development and evolution of age-dependent defenses in ant-acacias. Proceedings of the National Academy of Sciences, USA 116: 15596–15601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Meilan R, Ma C. 2006. Poplar (Populus spp.). In: Wang K, ed. Methods in Molecular Biology: Agrobacterium Protocols. Totowa, NJ, USA: Humana Press Inc., 143–151. [DOI] [PubMed] [Google Scholar]
- Niklas KJ. 1991. The elastic moduli and mechanics of Populus tremuloides (Salicaceae) petioles in bending. American Journal of Botany 78: 989–996. [Google Scholar]
- Orkwiszewski JAJ, Poethig RS. 2000. Phase identity of the maize leaf is determined after leaf initiation. Proceedings of the National Academy of Sciences, USA 97: 10631–10636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhurst DF. 1994. Diffusion of CO2 and other gases inside leaves. New Phytologist 126: 449–479. [DOI] [PubMed] [Google Scholar]
- Pettengill EA, Parmentier-Line C, Coleman GD. 2012. Evaluation of qPCR reference genes in two genotypes of Populus for use in photoperiod and low-temperature studies. BMC Research Notes 5: 366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poethig RS. 2013. Vegetative phase change and shoot maturation in plants. Current Topics in Developmental Biology 105: 125–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich PB, Ellsworth DS, Walters MB. 1998. Leaf structure (specific leaf area) modulates photosynthesis–nitrogen relations: evidence from within and across species and functional groups. Functional Ecology 12: 948–958. [Google Scholar]
- Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP. 2002. Prediction of plant microRNA targets. Cell 110: 513–520. [DOI] [PubMed] [Google Scholar]
- Robbins WJ. 1960. Further observations on juvenile and adult Hedera. American Journal of Botany 47: 485–491. [Google Scholar]
- Roden JS, Pearcy RW. 1993a. Effect of leaf flutter on the light environment of poplars. Oecologia 93: 201–207. [DOI] [PubMed] [Google Scholar]
- Roden JS, Pearcy RW. 1993b. The effect of flutter on the temperature of poplar leaves and its implications for carbon gain. Plant, Cell & Environment 16: 571–577. [Google Scholar]
- Rubinelli PM, Chuck G, Li X, Meilan R. 2013. Constitutive expression of the Corngrass1 microRNA in poplar affects plant architecture and stem lignin content and composition. Biomass and Bioenergy 54: 312–321. [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B et al. 2012. Fiji: An open-source platform for biological-image analysis. Nature Methods 9: 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D. 2005. Specific effects of microRNAs on the plant transcriptome. Developmental Cell 8: 517–527. [DOI] [PubMed] [Google Scholar]
- Scoffoni C, Sack L. 2013. Quantifying leaf vein traits. Prometheus Wiki. [WWW document] URL http://prometheuswiki.org/tiki-index.php?page=Quantifying+leaf+vein+traits [accessed1 May 2018].
- Stief A, Altmann S, Hoffmann K, Pant BD, Scheible W-R, Bäurle I. 2014. Arabidopsis miR156 regulates tolerance to recurring environmental stress through SPL transcription factors. Plant Cell 26: 1792–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugano SS, Shimada T, Imai Y, Okawa K, Tamai A, Mori M, Hara-Nishimura I. 2010. Stomagen positively regulates stomatal density in Arabidopsis. Nature 463: 241–244. [DOI] [PubMed] [Google Scholar]
- Taylor G 2002. Populus: Arabidopsis for forestry. Do we need a model tree? Annals of Botany 90: 681–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telfer A, Bollman KM, Poethig RS. 1997. Phase change and the regulation of trichome distribution in Arabidopsis thaliana. Development 124: 645–654. [DOI] [PubMed] [Google Scholar]
- Terashima I, Hanba YT, Tazoe Y, Vyas P, Yano S. 2006. Irradiance and phenotype: comparative eco-development of sun and shade leaves in relation to photosynthetic CO2 diffusion. Journal of Experimental Botany 57: 343–354. [DOI] [PubMed] [Google Scholar]
- Terashima I, Hikosaka K. 1995. Comparative ecophysiology of leaf and canopy photosynthesis. Plant, Cell & Environment 18: 1111–1128. [Google Scholar]
- Varkonyi-Gasic E, Wu R, Wood M, Walton EF, Hellens RP. 2007. Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 3: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters GA, Bartholomew DP. 1984. Acacia koa leaves and phyllodes: gas exchange, morphological, anatomical, and biochemical characteristics. Botanical Gazette 145: 351–357. [Google Scholar]
- Wang H, Wang H. 2015. The miR156/SPL module, a regulatory hub and versatile toolbox, gears up crops for enhanced agronomic traits. Molecular Plant 8: 677–688. [DOI] [PubMed] [Google Scholar]
- Wang J-W. 2014. Regulation of flowering time by the miR156-mediated age pathway. Journal of Experimental Botany 65: 4723–4730. [DOI] [PubMed] [Google Scholar]
- Wang JW, Park MY, Wang LJ, Koo Y, Chen XY, Weigel D, Poethig RS. 2011. MiRNA control of vegetative phase change in trees. PLoS Genetics 7: e1002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams WP, Davis FM, Buckley PM, Hedin PA, Baker GT, Luthe DS. 1998. Factors associated with resistance to fall Armyworm (Lepidoptera: Noctuidae) and southwestern Corn Borer (Lepidoptera: Crambidae) in corn at different vegetative stages. Journal of Economic Entomology 91: 1471–1480. [Google Scholar]
- Xie K, Wu C, Xiong L. 2006. Genomic organization, differential expression, and interaction of SQUAMOSA Promoter-Binding-Like Transcription Factors and microRNA156 in rice. Plant Physiology 142: 280–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Hu T, Zhao J, Park M-Y, Earley KW, Wu G, Yang L, Poethig RS. 2016. Developmental functions of miR156-regulated SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) genes in Arabidopsis thaliana. PLoS Genetics 12: e1006263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zou Z, Zhang J, Zhang Y, Han Q, Hu T, Xu X, Liu H, Li H, Ye Z. 2011. Over-expression of sly-miR156a in tomato results in multiple vegetative and reproductive trait alterations and partial phenocopy of the sft mutant. FEBS Letters 585: 435–439. [DOI] [PubMed] [Google Scholar]
- Zheng Q-L, Liu J, Goff BM, Dinkins RD, Zhu H. 2016. Genetic manipulation of miR156 for improvement of biomass production and forage quality in red clover. Crop Science 56: 1–7. [Google Scholar]
- Zotz G, Wilhelm K, Becker A. 2011. Heteroblasty – a review. Botanical Review 77: 109–151. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Additional Supporting Information may be found online in the Supporting Information section at the end of the article.
Table S1 Transformation constructs for MIM156 and miR156a transgenic P. tremula × alba lines.
Table S2 RT-qPCR primer sequences.
Fig. S1 The relative abundance of miR156 and SPL transcripts in leaf primordia of young transgenic and 717–1B4 wild-type P. tremula × alba grown in culture.
Fig. S2 Cq values from qPCR of miR159 at across-leaf positions.
Fig. S3 Representative plants from the wild-type 717–1B4, miR156a overexpressor line 40 and MIM156 line 84, 3 months following transplant from tissue culture.
Fig. S4 Leaf scans showing the adaxial (left) and abaxial (right) sides of adult, juvenile and juvenilized leaves of P. tremula × alba.
Fig. S5 Diagram showing representative fluttering paths for adult (node 25) and juvenile (node 10) leaves of MIM156 line 84.
Video S1 Representative samples of leaf fluttering in an miR156 overexpressor (left) and 717–1B4 wild-type line (right) of P. tremula × abla.


