Abstract
As the most abundant and reversible chemical modification in eukaryotic mRNA, the epitranscriptomic mark N6-methyladenine (m6A) regulates plant development and stress response. We have previously characterized that ALKBH10B is an Arabidopsis mRNA m6A demethylase and regulates floral transition. However, it is unclear whether ALKBH10B plays a role in abiotic stress response. Here, we found that the expression of ALKBH10B is increased in response to abscisic acid (ABA), osmotic, and salt stress. The alkbh10b mutants showed hypersensitive to ABA, osmotic, and salt stress during seed germination. Transcriptome analysis revealed that the expression of several ABA response genes is upregulated in alkbh10b-1 than that of wild type, indicating ALKBH10B negatively affects the ABA signaling. Furthermore, m6A sequencing showed that ABA signaling genes, including PYR1, PYL7, PYL9, ABI1, and SnRK2.2 are m6A hypermethylated in alkbh10b-1 after ABA treatment. Taken together, our work demonstrated that ALKBH10B negatively modulates ABA response during seed germination in Arabidopsis.
Keywords: Arabidopsis, N6-methyladenine, ALKBH10B, seed germination, ABA, osmotic stress
Introduction
N6-methyladenine (m6A) is the most abundant chemical modification in eukaryotic mRNA, and it is dynamically reversible (Jia et al., 2011; Zheng et al., 2013). Different protein complexes (“writer,” “eraser,” and “reader”) can “write,” “erase,” and “recognize” the m6A modification in mRNA. In mammals, the “writer” complex of RNA m6A mainly consists of three core subunits: METTL3 (methyltransferase like 3), METTL14, and WTAP (Wilms’ tumor 1-associating protein) (Liu et al., 2014; Ping et al., 2014). In addition, the m6A modification in RNA can be erased by its “eraser,” such as FTO (fat mass and obesity-associated protein) and ALKBH5 (alkylated DNA repair protein AlkB homolog 5) (Jia et al., 2011; Zheng et al., 2013). As an epitranscriptomic mark, m6A modification through its “reader,” such as the YTH family proteins, regulates RNA processing and metabolism, including alternative splicing, mRNA nuclear export, RNA stability maintenance, translation regulation, and pri-miRNA processing (Fustin et al., 2013; Wang et al., 2014, 2015; Alarcon et al., 2015; Meyer et al., 2015; Xiao et al., 2016).
RNA m6A modification is a conservative chemical modification. The mechanisms of “writing,” “erasing,” and “recognizing” of m6A in plants are similar to mammals. In Arabidopsis, the core subunits of RNA m6A methyltransferase complex includes MTA (homolog of human METTL3), MTB (homolog of human METTL14), and FIP37 (homolog of human WTAP). The core subunits of RNA m6A methyltransferase are essential proteins for embryonic development (Zhong et al., 2008; Shen et al., 2016; Ruzicka et al., 2017). In rice, the m6A methyltransferase are also essential for rice growth and development; mutation of OsFIP causes microspore degradation and affects its fertility (Zhang et al., 2019). There are 13 YTH domain proteins in Arabidopsis (Zhou et al., 2018). ECT2/ECT3/ECT4 and CPSF30-L have been confirmed as Arabidopsis m6A reader proteins. ECT2 promotes stability of m6A-modified mRNA and regulates trichome branching (Wei et al., 2018). ECT2/ECT3/ECT4 redundantly regulate the development of Arabidopsis epidermis (Arribas-Hernandez et al., 2018; Scutenaire et al., 2018; Wei et al., 2018). CPSF30-L recognizes m6A-modified FUE signal to mediate alternative polyadenylation and regulate flowering time, ABA response, and nitrogen metabolism (Hou et al., 2021; Song et al., 2021). The demethylases fine tune RNA m6A modification level, thus affect the dynamic state and reversibility of m6A. ALKBH9B and ALKBH10B are Arabidopsis RNA m6A demethylase and play important roles in regulating floral transition and viral infection of alfalfa mosaic virus (AMV). ALKBH10B mediates m6A demethylation in SPL3, SPL9, and FT transcripts and regulates their mRNA expression levels through m6A-mediated mRNA degradation pathway, thereby affects flowering time (Duan et al., 2017). ALKBH9B is driven to demethylate m6A in viral RNA of AMV through the specific interaction with the coat protein of AMV and positively regulates AMV accumulation and systemic invasion (Martinez-Perez et al., 2017). SIALKBH2 is identified as tomato m6A demethylase and regulates the ripening of tomato fruit. SIALKBH2 regulates the m6A level in the mRNA of DNA 5mC demethylase SIDML2 and affects its expression through m6A-mediated mRNA decay (Zhou et al., 2019).
Seed germination is the beginning of a new life cycle in higher plants, which is regulated by internal and environmental signals. The environmental signals perceived by seeds can be incorporated into endogenous hormonal signals. Abscisic acid (ABA) and gibberellic acid (GA) are regarded as the main hormones regulating seed germination (Seo et al., 2006; Shu et al., 2016). ABA positively regulates seed dormancy and inhibits seed germination, while GA acts antagonistically to release dormancy and to initiate seed germination (Shu et al., 2013). In this study, we found ABA, osmotic, and salt stress increase the expression of m6A demethylase ALKBH10B, which depends on ABI1. Further studies revealed that ALKBH10B plays a role in seed germination by negatively affecting ABA response genes. Moreover, we also found several ABA signaling genes with hypermethylated m6A peaks in the alkbh10b mutant, including PYR1, ABI1, SnRK2.2, PYL7, and PYL9. In all, this study revealed a negative regulatory role of ALKBH10B in ABA response during seed germination.
Materials and Methods
Plant Materials and Growth Conditions
Arabidopsis thaliana materials used in this study: alkbh10b-1 (Salk_004215), alkbh10b-2 (Salk_107289), and ALKBH10Bp:ALKBH10B/alkbh10b-1 were derived from previous study, all in the Columbia-0 ecotype (Col-0) background (Duan et al., 2017); abi1-1 (CS22) were in the Landsberg erecta (Ler) background.
All plants were grown at 22°C under long-day conditions (16 h light/8 h dark, white fluorescent tubes, 150–200 μmol m–2 s–1), and mature seeds of each genotype were collected at the same day, seeds were dried and stored at room temperature. For the seed germination assay, seeds were surfaced sterilized with 20% bleach for 10 min, then rinsed five times with sterile water, and plated on half strength Murashige and Skoog (MS) medium containing 0.8% sucrose, supplemented with various concentrations of ABA (0, 0.5, and 1 μM), 300 mM mannitol, or 150 mM NaCl, respectively. Plates were kept at 4°C in the dark for 3 days, and transferred to a culture room at 22°C under long-day conditions. Radicle emergence was scored after the indicated time intervals.
Quantitative Reverse Transcription-PCR Analysis
Seven-day-old seedlings of Col-0 were used to evaluate ALKBH10B expression under external signals treatment. Seven-day-old seedlings were transferred to solid 1/2 MS medium, medium with 300 mM mannitol, 150 mM NaCl, or liquid 1/2 MS medium, medium with 50 μM ABA or 50 μM MeJA, respectively. Seedlings were collected after treatment for different times. Total RNA was isolated with Trizol reagent, and 1 μg total RNA was used for reverse transcription by PrimeScript RT reagent kit with gDNA Eraser (Takara). qRT-PCR was performed using SYBR Green Master Mix (YEASEN) on a ViiA 7 Dx (Applied Biosystems). All qRT-PCRs were performed in triplicate using total RNA samples extracted from three independent biological replicates. The 2–ΔΔCT method was used to calculate the gene expression levels. At2G28390 was used as an internal control (Czechowski et al., 2005). All primers used are listed in Supplementary Table 2.
m6A Sequencing
Seven-day-old seedlings of alkbh10b-1 and Col-0 were treated with half strength liquid MS medium supplemented with 50 μM ABA for 3 h, and the total RNA were extracted. Poly (A)+ RNA was isolated from total RNA using oligo(dT)25 Dynabeads (Thermo Fisher Scientific). 5 μg poly(A)+ RNA was fragmented into 100–150 nt by Magnesium RNA fragmentation module (NEB). m6A immunoprecipitation and library preparation were performed using the EpiMark N6-Methyladenosine enrichment kit (NEB). Input and RNA eluted from m6A-IP were used to prepare libraries with NEBNext Ultra II RNA Library Prep Kit. Sequencing was performed on an Illumina HiSeq X Ten machine in pair-end mode with 150 bp per read (Genewiz).
Analysis of m6A-Seq Data
Sequence data were analyzed according to the procedure described by Song et al. (2021). Briefly, adapters and low-quality reads were trimmed by Cutadapt (v1.18) (Martin, 2011), clean reads were mapped to TAIR10 by HISAT2 (v2.1.0) (Pertea et al., 2016). PCR duplication were filtered by Picard Toolkit. The m6A peaks were identified using the MACS2 (Zhang et al., 2008) peak-calling algorithm based on enrichment criteria (IP/Input) ≥2 and FDR <0.05. Hypermethylated m6A sites were identified by MeTDiff (Cui et al., 2018) based on enrichment criteria fold change ≥2 and FDR <0.05. To evaluated the m6A distribution in transcripts, we divided TAIR10 transcripts into five-non-overlapping regions: 5′UTRs, Start_Codon (100-nucleotide window centered on the start codon), coding sequences (CDSs), Stop_Codon (100-nucleotide window centered on the stop codon) and 3′UTRs (Duan et al., 2017). We used a python script to assert subtypes of those RNA molecules contain hypermethylated m6A peak (scripts can be found in Github1). Candidate gene list was submitted to DAVID2 to perform GO ontology (GO) and KEGG pathway enrichment analyses.
Analysis of RNA-seq Data
The input data of m6A sequencing was used for transcriptome data analysis. Sequencing reads were trimmed and mapped to the reference genome (TAIR10) by Cutadapt (v1.18) and HISAT2 (v2.1.0), respectively. The differential expression genes (DEG) were identified by R package (DEseq2) (Love et al., 2014) with a cutoff criterion of fold change ≥1.5 and FDR <0.05. Gene ontology (GO) enrichment analyses were performed by using DAVID.
Results
ALKBH10B Expression is Induced by ABA, JA, Osmotic, and Salt Stress
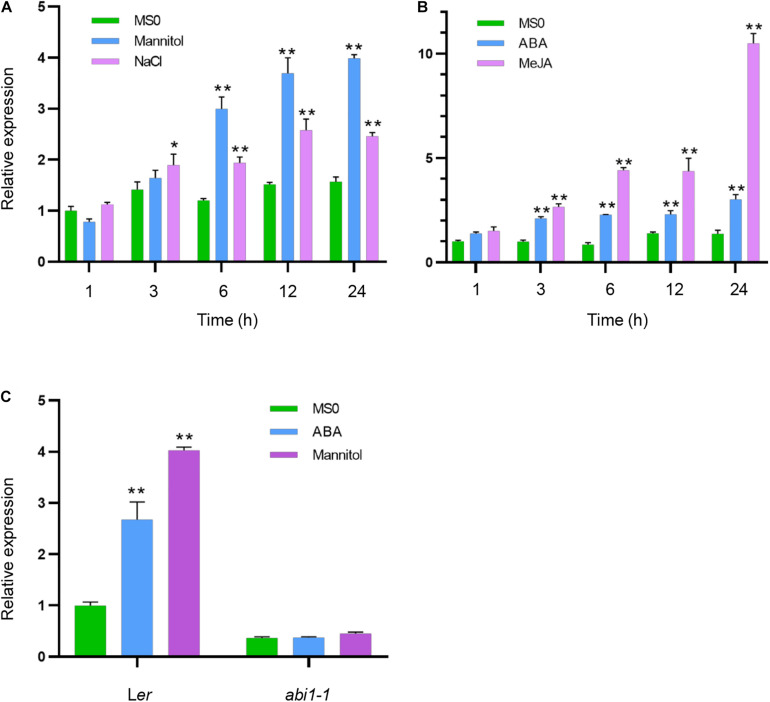
In our previous study, we have identified ALKBH10B as an mRNA m6A demethylase, which regulates floral transition in Arabidopsis (Duan et al., 2017). In addition to developmental function of ALKBH10B, we asked whether ALKBH10B is involved in abiotic stress response. We firstly examined whether the expression level of ALKBH10B is affected by osmotic and salt stress. Seven-day-old seedlings growing on 1/2 MS medium were transferred to solid medium containing 300 mM mannitol or 150 mM NaCl. Seedlings were collected after time-course treatment (1, 3, 6, 12, or 24 h). After treatment with mannitol for 6 h, or NaCl for 3 h, the expression of ALKBH10B was significantly increased than that of the control (Figure 1A). This is consistent with the data in AtGenExpress global stress expression database (Kilian et al., 2007). Both ABA and jasmonic acid (JA) are important phytohormones which regulate environment response (Ju et al., 2019). Next, we tested whether ALKBH10B expression is activated by ABA and methyl jasmonate (MeJA) treatment. The results showed that after treatment with ABA or MeJA for 3 h, the expression of ALKBH10B was significantly increased (Figure 1B).
FIGURE 1.
The expression of ALKBH10B is induced by virous external stimuli. (A,B) ALKBH10B expression in Col-0 plants in response to osmotic stress, salt stress, ABA, and MeJA. Seven-day-old seedlings were transferred to solid 1/2 MS medium, medium with 300 mM mannitol or 150 mM NaCl, or liquid 1/2 MS medium, medium with 50 μM ABA or 50 μM MeJA, respectively. After treated with the indicated time, seedlings were harvested followed by RNA extraction and cDNA synthesis for qRT-PCR. Asterisks indicate statistically significant differences as compared to the mock of every time-point (*P < 0.05; **P < 0.01; Student’s t-test). (C) ALKBH10B expression in abi1-1 mutant in response to ABA and mannitol. Seven-day-old seedlings were transferred to solid 1/2 MS medium or medium containing 50 μM ABA or 300 mM mannitol for 24 h, respectively. Then seedlings were harvested followed by qRT-PCR. The transcript level of ALKBH10B were normalized to the At2G28390 expression. Error bars represent the standard deviation of three replicates. Asterisks indicate statistically significant differences (*P < 0.05; **P < 0.01; Student’s t-test).
In ABA signaling pathway, ABA is perceived by the intracellular pyrabactin resistance 1 (PYR1) and PYR1-like (PYL)/regulatory component of ABA receptor (RCAR) proteins. From the transcriptome data of previous studies (Zhao et al., 2018), in pyl duodecuple mutant, pyr1pyl1/2/3/4/5/7/8/9/10/11/12, the induction of ALKBH10B expression by ABA or mannitol was much less in the mutant (Supplementary Figure 1). ABA insensitive 1 (ABI1), encodes group A protein phosphatase 2Cs (PP2C), which regarded as ABA co-receptor (Leung et al., 1994), is crucial for ABA signaling. The activation of ALKBH10B expression by ABA or mannitol was blocked in the abi1-1 mutant (Figure 1C). These results indicated that the expression of ALKBH10B is regulated by the PYR/PYL/RCAR-ABI1 mediated ABA signaling pathway.
ALKBH10B Modulates Seed Germination and Post-germination Development Responding to ABA, Salt, and Osmotic Stress
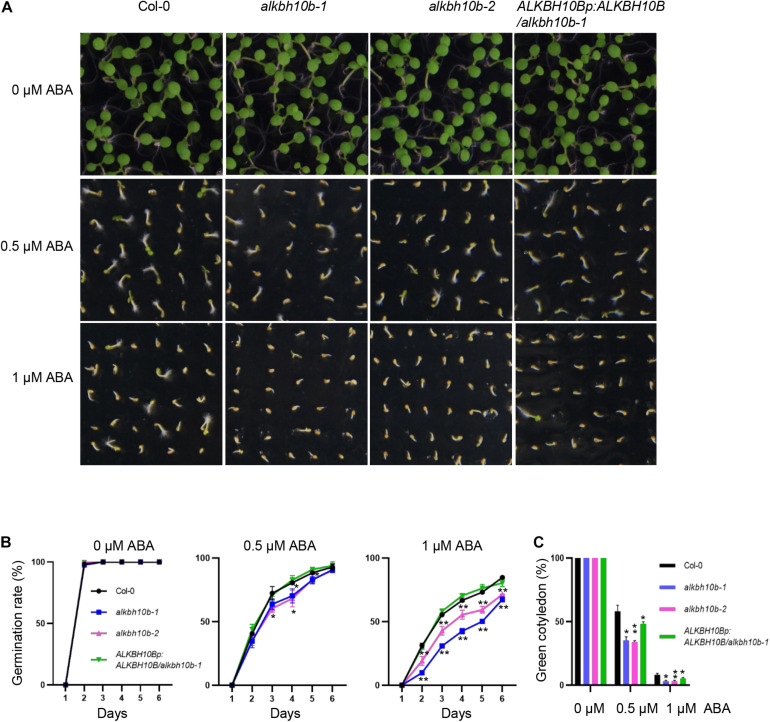
The expression of ALKBH10B is activated by ABA, suggesting that ALKBH10B plays potential roles in ABA response in Arabidopsis. To understand the roles of ALKBH10B in ABA response, two T-DNA insertion mutants: SALK_004215 (alkbh10b-1) and SALK_107289 (alkbh10b-2), as well as one complementation material (ALKBH10Bp:ALKBH10B/alkbh10b-1) arrived from our previous study were used in the following research. Seed germination is sensitive to ABA concentration; Therefore, we performed the seed germination assay of each genotype under ABA treatment. We germinated the seeds of alkbh10b-1, alkbh10b-2, ALKBH10Bp:ALKBH10B/alkbh10b-1, and wild type (Col-0) on 1/2 MS medium with or without ABA treatment. In the absence of ABA, the germination rate of different genotypes was similar (Figures 2A,B). In the presence of different concentrations of ABA, alkbh10b-1 and alkbh10b-2 seeds were more sensitive to ABA inhibition, both mutants showed lower germination rate compared to the wild type (Figures 2A,B). Complementation of ALKBH10B in alkbh10b-1 (ALKBH10Bp:ALKBH10B/alkbh10b-1) recovered the lower germination rate of alkbh10b-1 under ABA treatment (Figures 2A,B). In line with the germination rate data, alkbh10b-1 and alkbh10b-2 mutants also showed lower cotyledon greening rates after 10 days of germination (Figure 2C). These results indicated that ALKBH10B acts as a negative regulator of ABA signaling during seed germination and post-germination growth.
FIGURE 2.
ALKBH10B modulates ABA sensitivity during seed germination. The germination rates of indicated genotypes were recorded from 1–6 days after stratification in the presence of various ABA contents (0, 0.5, or 1.0 μM). Cotyledon-greening percentages of the 10th day were recorded. Three independent experiments were conducted, with at least 100 seeds per genotype in each replicate. Error bars represent the standard deviation of three replicates. Asterisks indicate statistically significant differences as compared to the mock of every time-point (*P < 0.05; **P < 0.01; Student’s t-test). (A) Photographs of seedings grown on different media at day 6 after stratification. (B) Seed germination rates of indicated genotypes grown on different medium. (C) Green cotyledon ratio at day 10 after stratification.
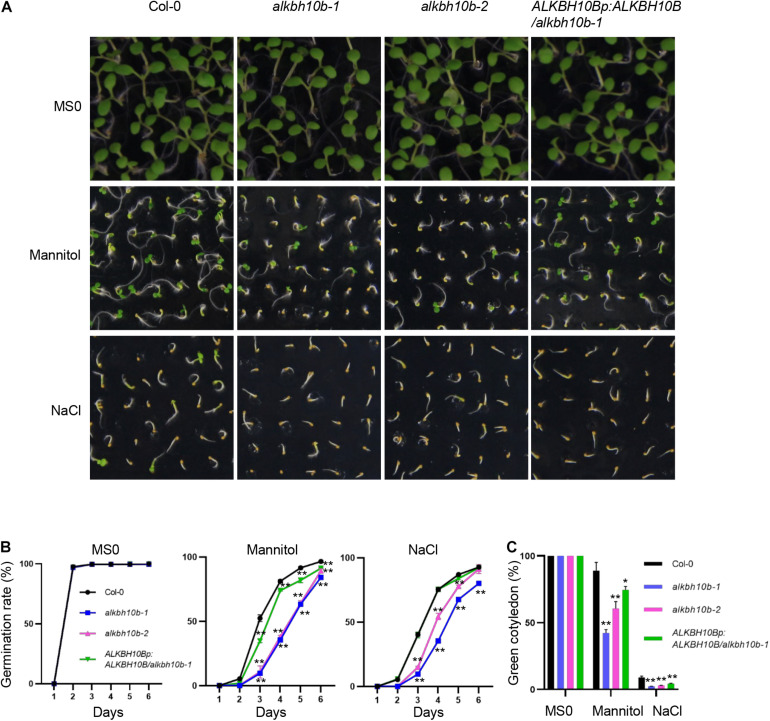
We also tested whether ALKBH10B affects the seed germination in the presence of osmotic and salt stress. The seeds were sown on 1/2 MS medium supplemented with 300 mM mannitol or 150 mM NaCl. Compared with the wild type seeds, germination and cotyledon greening ratios of alkbh10b-1 and alkbh10b-2 seeds were more inhibited under mannitol or NaCl treatment (Figures 3A–C), indicating that ALKBH10B plays roles in regulating seed germination and post-germination growth in response to osmotic and salt stress.
FIGURE 3.
ALKBH10B modulates osmotic and salt stress response during seed germination. The germination rates of indicated genotypes were recorded from 1–6 days after stratification in the presence of 300 mM mannitol or 150 mM NaCl. Cotyledon-greening percentages of the 8th day were recorded. Three independent experiments were conducted, with at least 100 seeds per genotype in each replicate. Error bars represent the standard deviation of three replicates. Asterisks indicate statistically significant differences as compared to the mock of every time-point (*P < 0.05; **P < 0.01; Student’s t-test). (A) Photographs of seedings grown on different media at day 6 after stratification. (B) Seed germination rates of indicated genotypes grown on different medium. (C) Green cotyledon ratio at day 8 after stratification.
Transcriptome Analysis of alkbh10b-1 After ABA Treatment
Since both alkbh10b mutants are more sensitive to ABA than Col-0, we speculated that ALKBH10B may affects the expression of some ABA response genes. Therefore, we performed RNA sequencing experiments to profile the transcriptome of alkbh10b-1 and the wild type after ABA treatment. Total RNA was isolated from 7-day-old seedlings treated with ABA for 3 h and used for RNA-seq library construction. We identified 312 upregulated genes and 272 downregulated genes (fold change ≥1.5 and FDR ≤0.05) (Figure 4A and Supplementary Dataset 2). Hierarchical clustering depicted differential expression profiles in all samples (Figure 4B). Then we performed GO analysis of these DEG using DAVID. The results showed that the upregulated genes were significantly enriched in cellular response to ABA stimulus, ABA activated signaling pathway, seed maturation and water deprivation (Figure 4C), which suggest that ALKBH10B may participates in ABA signaling pathway by affecting genes involved in ABA signaling pathway and plays a role in seed germination, which corresponds to the phenotype of alkbh10b mutation lines. Furthermore, downregulated genes were significantly enriched in the response of plant hormone signaling, including salicylic acid and JA (Figure 4C), which are import in various stress response and signal transduction. These results suggesting ALKBH10B plays a role in abiotic stress response and system defense.
FIGURE 4.
Differential expression genes identified in alkbh10b-1 compared with Col-0. (A) Volcano plots showing the differentially expressed genes identified in alkbh10b-1. (B) Heatmap plots showing the normalized RPKM of differentially expressed genes in alkbh10b-1. (C) Biological process enrichment analysis of the upregulated and downregulated genes in alkbh10b-1.
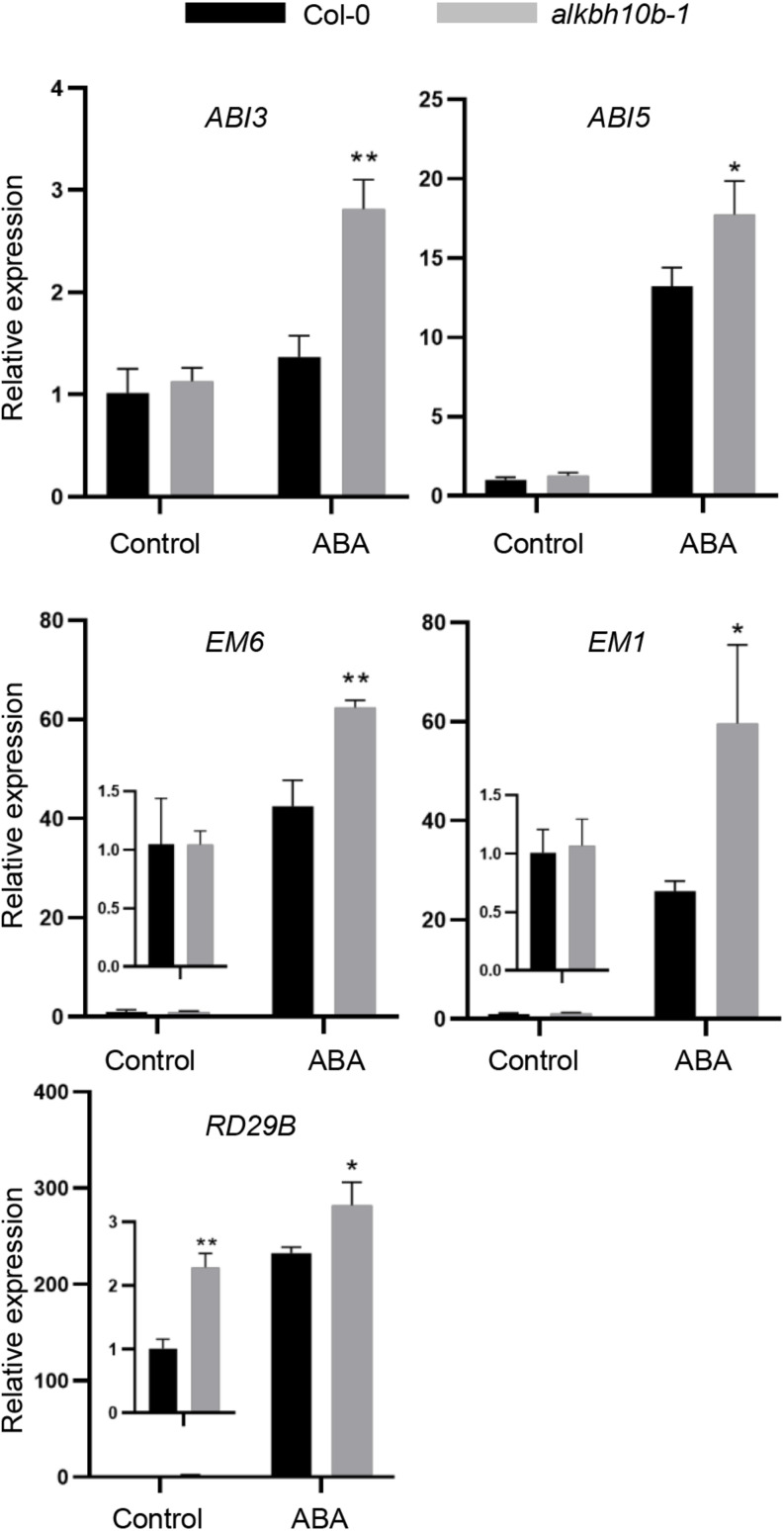
ALKBH10B Negatively Affects the Expression of ABA Response Genes
RNA-seq data revealed that the upregulated genes were significantly enriched in ABA response and seed maturation, which may explain why the alkbh10b mutants were more sensitive under ABA treatment. ABI3 and ABI5 are two important transcription factors in regulating seed maturation, germination, and early development of seedlings (Bensmihen et al., 2002; Lopez-Molina et al., 2002). We measured the transcript levels of ABI3, ABI5, and some of their downstream genes, including RD29B, EM6, and EM1 in Col-0 and alkbh10-1 using qRT-PCR under mock and ABA treatment. The results showed that the expression of ABI3, ABI5, EM6, EM1, and RD29B were upregulated in alkbh10b-1 after ABA treatment (Figure 5). Among them, the higher expression of EM6 and RD29B in alkbh10b-1 was consistent with the RNA-seq data; the higher expression of ABI3, ABI5, and EM1 were not identified in the RNA-Seq data because of their low expression or their insignificant difference between alkbh10b-1 and wild type. To better understand the role of ALKBH10B in seed germination, we detected the expression patterns of these ABA signaling-related genes in seeds germinated for 2.5 days on medium containing 0 or 0.5 μM ABA. The results showed that the expression of ABI3, ABI5, EM6, EM1, and RD29B in alkbh10b-1 was higher than that of the Col-0 on medium with or without ABA (Supplementary Figure 2). Above all, our results showed that ALKBH10B negatively affects the expression of ABA response genes.
FIGURE 5.
ALKBH10B regulates the expression of ABA response genes. The expression patterns of ABA signaling-related ABI3, ABI5, and RD29B, and embryogenesis-related EM1 and EM6 were determined in seven-day-old seedlings treated with 50 μM ABA for 3 h. The transcript level of each gene was normalized to that of At2G28390, and the relative level of each gene in mock-treated Col-0 was set to 1. Values are mean ± SD of three replications. Asterisks indicate statistically significant differences (*P < 0.05; **P < 0.01; Student’s t-test).
Transcriptome-Wide Analysis of mRNA m6A Modification in alkbh10b-1 After ABA Treatment
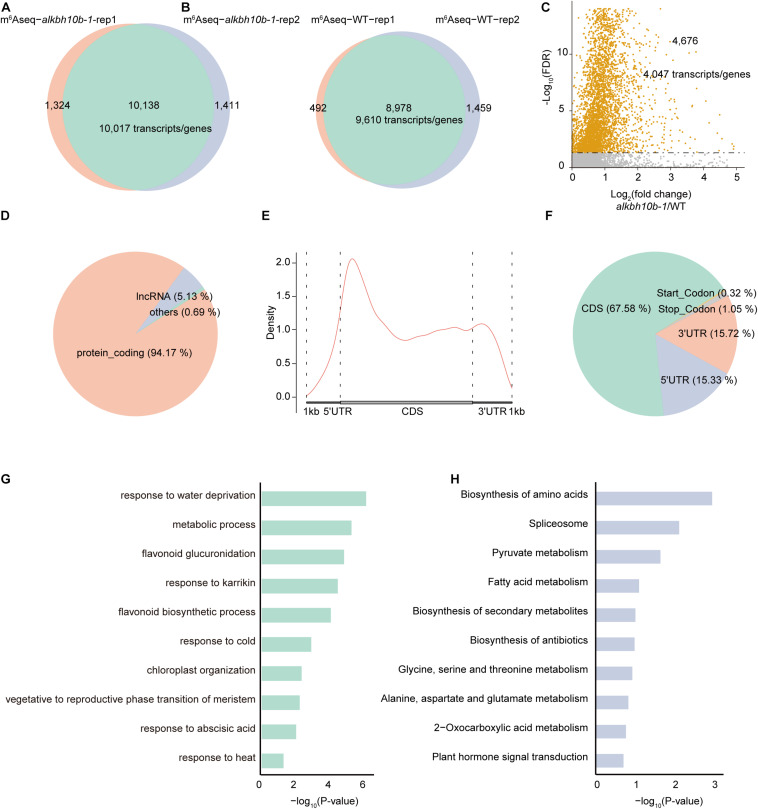
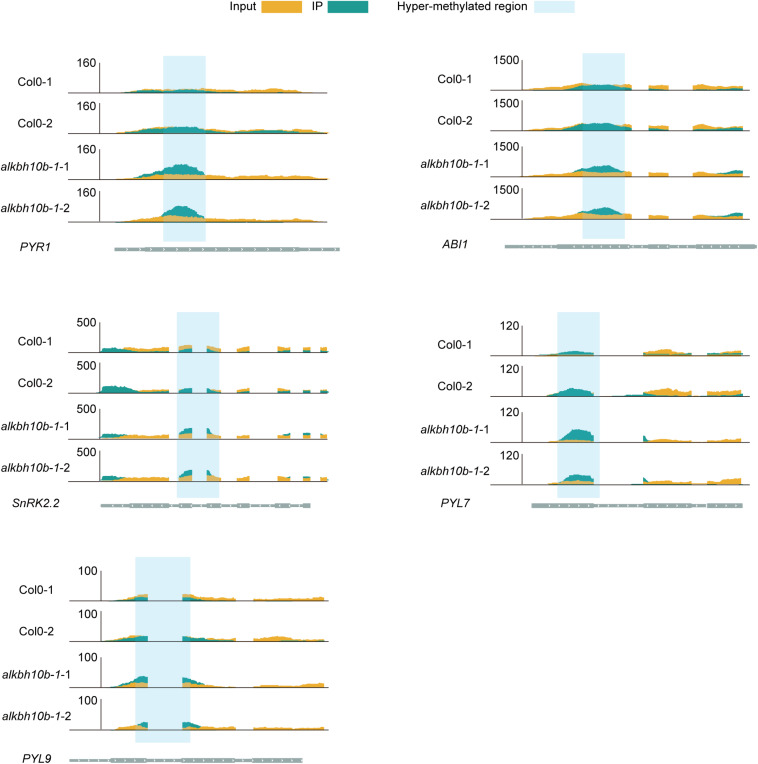
To investigate whether m6A methylation pattern is different in alkbh10b mutant compared to wild type under 50 μM ABA treatment, we performed m6A-seq in alkbh10b-1 and Col-0, respectively. After removing PCR duplication, approximately 10.0–21.6 million reads were mapped to TAIR10 genome with a more than 90% mapping rate (Supplementary Dataset 1). We used MACS2, a well-known peak caller, to identify peaks with a criteria of fold enrichment (IP/input) ≥2 and FDR ≤0.05. The m6A peaks identified in both replicates were used for further analysis. We identified 10,138 m6A peaks corresponding to 10,017 transcripts/genes in alkbh10b-1 mutant (Figure 6A), whereas 8,978 m6A peaks corresponding to 9,610 transcripts/genes in Col-0 (Figure 6B), indicating that loss of function of ALKBH10B caused an increase of m6A peak numbers. We subsequently identified hypermethylated m6A peaks in alkbh10b-1 mutant under ABA treatment using the MeTDiff R package software (FDR ≤ 0.05). Compared with WT, a total of 4,676 peaks were identified in alkbh10b-1, corresponding to 4,047 transcripts/genes (Figure 6C and Supplementary Dataset 3), which were mainly mRNA (94.17%), only ∼5.13% were lncRNA and a small proportion were other RNAs (Figure 6D). Then we examined the distribution profiles of these hypermethylated m6A peaks within the transcripts, and found that the density of these peaks tends to be enriched in CDS, near the start codon and 5′UTR (Figure 6E), which is consistent with the distribution of hypermethylated peaks in normal condition (Duan et al., 2017). To further locate these m6A peaks, we assigned these m6A peaks into five-non-overlapping regions and found that these hypermethylated peaks were dominantly enriched in CDS (67.58%), then 3′UTR (15.72%) and 5′UTR (15.33%) (Figure 6F). To explore the function of the hypermethylated m6A genes in alkbh10b-1 mutant under ABA treatment, we performed GO enrichment analysis using DAVID. The results revealed that genes with hypermethylated m6A peaks are enriched in various abiotic biological processes including response to water deprivation, cold, ABA, and heat (Figure 6G). KEGG pathway analysis showed that genes with hypermethylated m6A peaks are mainly enriched in biosynthesis of amino acids, pyruvate metabolism and plant hormone signal transduction (Figure 6H). In addition, several ABA signaling genes, including PYR1, PYL7, PYL9, ABI1, and SnRK2.2 were m6A hypermethylated in alkbh10b-1 under ABA treatment compared with WT (Figure 7). We also analyzed the m6A-seq data (Duan et al., 2017) between alkbh10b-1 and Col-0 under normal conditions. None of these genes were m6A hypermethylated in alkbh10b-1 under normal conditions (Supplementary Figure 3), suggesting m6A hypermethylation of these genes may specific to ABA treatment. These results indicated that ALKBH10B may regulates ABA response by affecting the m6A level of these ABA signaling genes.
FIGURE 6.
Characterization of m6A modification in Col-0 and alkbh10b-1 under ABA treatment. (A) Overlap of two biological replicates of m6A enriched peaks in alkbh10b-1. (B) Overlap of two biological replicates of m6A enriched peaks in Col-0. (C) Volcano plot showing the hypermethylated m6A peaks identified in alkbh10b-1 compared with Col-0. (D) Pie chart depicting RNA types of hypermethylated m6A peaks in alkbh10b-1. (E) Metagene profile showing the density of hypermethylated m6A peaks across the transcript. (F) Pie chart depicting the fraction of the hypermethylated m6A peaks in the five non-overlapping transcript segments [5′UTR, start codon, coding sequence (CDS), stop codon and 3′UTR]. (G) Biological process enrichment analysis of genes with hypermethylated m6A peaks. (H) Pathway analysis of genes with hypermethylated m6A peaks.
FIGURE 7.
Integrative genomics viewer (IGV) tracks showing the hypermethylated m6A peaks of several ABA signaling genes identified in alkbh10b-1.
Discussion
More and more studies have reported the importance of mRNA m6A modification in plant development, including embryo development (Zhong et al., 2008; Shen et al., 2016; Ruzicka et al., 2017), microspore development (Zhang et al., 2019), shoot apical meristem proliferation (Shen et al., 2016), trichome morphology (Arribas-Hernandez et al., 2018; Scutenaire et al., 2018; Wei et al., 2018), floral transition (Duan et al., 2017), and fruit ripping (Zhou et al., 2019). mRNA m6A modification also plays roles in biotic and abiotic stress response, for example, virus infection (Martinez-Perez et al., 2017), and salt stress response (Hu et al., 2021). Plant seeds need to identify and respond to different environmental factors to decide whether to germinate or not. Even after seed germination, the seedlings will feel the changes of the surrounding environment and inhibit growth in a process called post-germination developmental arrest. Abscisic acid (ABA) is an important hormone to balance early growth or inhibition. Here, we found that ALKBH10B, an mRNA m6A demethylase, modulates seed germination by regulating ABA response genes.
We demonstrated that ALKBH10B plays a role in seed germination through an ABA dependent manner based on the following evidence. First, the expression of ALKBH10B is induced by ABA, NaCl, and mannitol stress, however, in abi1-1, the inducibility of ALKBH10B is disappeared (Figure 1 and Supplementary Figure 1), suggesting that ALKBH10B is regulated by ABA signaling and work downstream of ABI1. Second, the seeds of alkbh10b mutants were more sensitive than that of the wild type in response to ABA, osmotic, and salt stress, and showed lower germination rate and cotyledons greening at the same day (Figures 2, 3). Third, RNA-seq data reveals that, ABA response genes were upregulated in alkbh10b-1 after ABA treatment. We found that transcription factors, ABI3 and ABI5, which are two well-known positive regulators of ABA signaling during seed germination, were upregulated in alkbh10b-1 after ABA treatment (Figures 4, 5). Fourth, some ABA response genes were m6A hypermethylated in alkbh10-1, including PYR1, PYL4, PYL7, PYL9, and ABI1, suggesting the role of ALKBH10B-mediated m6A demethylation in ABA signaling gene regulation. Taken together, our results indicate that ALKBH10B modulated seed germination by negatively affecting ABA response genes.
The mechanism of m6A-mediated gene regulated is complicated. So far, researchers have found virous mechanism in mammals, including RNA stability maintenance, alternative splicing, mRNA nuclear export, translation regulation, and pri-miRNA processing (Fustin et al., 2013; Wang et al., 2014, 2015; Alarcon et al., 2015; Meyer et al., 2015; Xiao et al., 2016). Recently, some reports found that RNA m6A modification can modulate chromatin accessibility in mammalian cells, mainly through its “writer” or “reader” proteins to recruit histone modification proteins to change the histone modification state (Li et al., 2020; Liu et al., 2020, 2021; Xu et al., 2021), increasing the complexity of m6A regulation mechanism. Although, we found ALKBH10B negatively regulates the expression of ABA signaling genes under ABA treatment, and found some ABA response genes were m6A hypermethylated, the specific mechanism of how ALKBH10B modulates the expression of ABA response genes needs further study.
Conclusion
Here, we found that the expression of ALKBH10B is induced in response to ABA, osmotic, and salt stress, which depends on ABI1. The alkbh10b mutants showed more sensitive to ABA, osmotic, and salt stress during seed germination than the wild type. The expression of several ABA response genes was upregulated in alkbh10b-1 than that of wild type, indicating ALKBH10B negatively affects the ABA signaling. Furthermore, mRNA m6A sequencing found that mRNA of ABA signaling genes, including PYR1, ABI1, PYL7, PYL9, and SnRK2.2 were m6A hypermethylated in alkbh10b-1 after ABA treatment. However, further studies are needed to explain the specific mechanism of how ALKBH10B modulating the expression of ABA response genes.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://bigd.big.ac.cn/gsa/s/uPCjOJ90, PRJCA005164.
Author Contributions
JT performed the experiments with the help of HD. JY analyzed the sequencing data. GJ and JT designed the experiments, interpreted the results, and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Qi Xie from the Institute of Genetics and Developmental Biology, Chinese Academy of Sciences for his generous share of abi1-1 mutant seeds.
Funding. This work was supported by the National Natural Science Foundation of China (Nos. 21822702, 21820102008, 92053109, and 21432002).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.712713/full#supplementary-material
References
- Alarcon C. R., Goodarzi H., Lee H., Liu X., Tavazoie S., Tavazoie S. F. (2015). HNRNPA2B1 is a mediator of m6A-dependent nuclear RNA processing events. Cell 162 1299–1308. 10.1016/j.cell.2015.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arribas-Hernandez L., Bressendorff S., Hansen M. H., Poulsen C., Erdmann S., Brodersen P. (2018). An m6A-YTH module controls developmental timing and morphogenesis in Arabidopsis. Plant Cell 30 952–967. 10.1105/tpc.17.00833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensmihen S., Rippa S., Lambert G., Jublot D., Pautot V., Granier F., et al. (2002). The homologous ABI5 and EEL transcription factors function antagonistically to fine-tune gene expression during late embryogenesis. Plant Cell 14 1391–1403. 10.1105/tpc.000869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X., Zhang L., Meng J., Rao M. K., Chen Y., Huang Y. (2018). MeTDiff: a novel differential RNA methylation analysis for MeRIP-Seq data. IEEE/ACM Trans. Comput. Biol. Bioinform. 15 526–534. 10.1109/tcbb.2015.2403355 [DOI] [PubMed] [Google Scholar]
- Czechowski T., Stitt M., Altmann T., Udvardi M. K., Scheible W. R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139 5–17. 10.1104/pp.105.063743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan H. C., Wei L. H., Zhang C., Wang Y., Chen L., Lu Z., et al. (2017). ALKBH10B is an RNA N6-methyladenosine demethylase affecting Arabidopsis floral transition. Plant Cell 29 2995–3011. 10.1105/tpc.16.00912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fustin J. M., Doi M., Yamaguchi Y., Hida H., Nishimura S., Yoshida M., et al. (2013). RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell 155 793–806. 10.1016/j.cell.2013.10.026 [DOI] [PubMed] [Google Scholar]
- Hou Y., Sun J., Wu B., Gao Y., Nie H., Nie Z., et al. (2021). CPSF30-L-mediated recognition of mRNA m6A modification controls alternative polyadenylation of nitrate signaling-related gene transcripts in Arabidopsis. Mol. Plant. 14 688–699. 10.1016/j.molp.2021.01.013 [DOI] [PubMed] [Google Scholar]
- Hu J., Cai J., Park S. J., Lee K., Li Y., Chen Y., et al. (2021). N6-methyladenosine mRNA methylation is important for salt stress tolerance in Arabidopsis. Plant J. 10.1111/tpj.15270 [Epub Online ahead of print]. [DOI] [PubMed] [Google Scholar]
- Jia G., Fu Y., Zhao X., Dai Q., Zheng G., Yang Y., et al. (2011). N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7 885–887. 10.1038/nchembio.687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju L., Jing Y., Shi P., Liu J., Chen J., Yan J., et al. (2019). JAZ proteins modulate seed germination through interaction with ABI5 in bread wheat and Arabidopsis. New Phytol. 223 246–260. 10.1111/nph.15757 [DOI] [PubMed] [Google Scholar]
- Kilian J., Whitehead D., Horak J., Wanke D., Weinl S., Batistic O., et al. (2007). The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 50 347–363. 10.1111/j.1365-313X.2007.03052.x [DOI] [PubMed] [Google Scholar]
- Leung J., Bouvier-Durand M., Morris P. C., Guerrier D., Chefdor F., Giraudat J. (1994). Arabidopsis ABA response gene ABI1: features of a calcium-modulated protein phosphatase. Science 264 1448–1452. 10.1126/science.7910981 [DOI] [PubMed] [Google Scholar]
- Li Y., Xia L., Tan K., Ye X., Zuo Z., Li M., et al. (2020). N6-methyladenosine co-transcriptionally directs the demethylation of histone H3K9me2. Nat. Genet. 52 870–877. 10.1038/s41588-020-0677-3 [DOI] [PubMed] [Google Scholar]
- Liu J., Dou X., Chen C., Chen C., Liu C., Xu M. M., et al. (2020). N6-methyladenosine of chromosome-associated regulatory RNA regulates chromatin state and transcription. Science 367 580–586. 10.1126/science.aay6018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Gao M., He J., Wu K., Lin S., Jin L., et al. (2021). The RNA m(6)A reader YTHDC1 silences retrotransposons and guards ES cell identity. Nature 591 322–326. 10.1038/s41586-021-03313-9 [DOI] [PubMed] [Google Scholar]
- Liu J., Yue Y., Han D., Wang X., Fu Y., Zhang L., et al. (2014). A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 10 93–95. 10.1038/nchembio.1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L., Mongrand S., McLachlin D. T., Chait B. T., Chua N. H. (2002). ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J. 32 317–328. 10.1046/j.1365-313x.2002.01430.x [DOI] [PubMed] [Google Scholar]
- Love M. I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. (2011). CUTADAPT removes adapter sequences from high-throughput sequencing reads. EMBO J. 17 10–12. 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- Martinez-Perez M., Aparicio F., Lopez-Gresa M. P., Belles J. M., Sanchez-Navarro J. A., Pallas V. (2017). Arabidopsis m6A demethylase activity modulates viral infection of a plant virus and the m6A abundance in its genomic RNAs. Proc. Natl. Acad. Sci.U. S. A. 114 10755–10760. 10.1073/pnas.1703139114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K. D., Patil D. P., Zhou J., Zinoviev A., Skabkin M. A., Elemento O., et al. (2015). 5’ UTR m6A promotes cap-independent translation. Cell 163 999–1010. 10.1016/j.cell.2015.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M., Kim D., Pertea G. M., Leek J. T., Salzberg S. L. (2016). Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11 1650–1667. 10.1038/nprot.2016.095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ping X. L., Sun B. F., Wang L., Xiao W., Yang X., Wang W. J., et al. (2014). Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 24 177–189. 10.1038/cr.2014.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzicka K., Zhang M., Campilho A., Bodi Z., Kashif M., Saleh M., et al. (2017). Identification of factors required for m6A mRNA methylation in Arabidopsis reveals a role for the conserved E3 ubiquitin ligase HAKAI. New Phytol. 215 157–172. 10.1111/nph.14586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scutenaire J., Deragon J. M., Jean V., Benhamed M., Raynaud C., Favory J. J., et al. (2018). The YTH domain protein ECT2 is an m6A reader required for normal trichome branching in Arabidopsis. Plant Cell 30 986–1005. 10.1105/tpc.17.00854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo M., Hanada A., Kuwahara A., Endo A., Okamoto M., Yamauchi Y., et al. (2006). Regulation of hormone metabolism in Arabidopsis seeds: phytochrome regulation of abscisic acid metabolism and abscisic acid regulation of gibberellin metabolism. Plant J. 48 354–366. 10.1111/j.1365-313X.2006.02881.x [DOI] [PubMed] [Google Scholar]
- Shen L., Liang Z., Gu X., Chen Y., Teo Z. W., Hou X., et al. (2016). N6-methyladenosine RNA modification megulates shoot stem cell fate in Arabidopsis. Dev. Cell 38 186–200. 10.1016/j.devcel.2016.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu K., Liu X. D., Xie Q., He Z. H. (2016). Two faces of one seed: hormonal regulation of dormancy and germination. Mol. Plant 9 34–45. [DOI] [PubMed] [Google Scholar]
- Shu K., Zhang H., Wang S., Chen M., Wu Y., Tang S., et al. (2013). ABI4 regulates primary seed dormancy by regulating the biogenesis of abscisic acid and gibberellins in arabidopsis. PLoS Genet. 9:e1003577. 10.1371/journal.pgen.1003577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song P., Yang J., Wang C., Lu Q., Shi L., Tayier S., et al. (2021). Arabidopsis N(6)-methyladenosine reader CPSF30-L recognizes FUE signals to control polyadenylation site choice in liquid-like nuclear bodies. Mol. Plant 14 571–587. 10.1016/j.molp.2021.01.014 [DOI] [PubMed] [Google Scholar]
- Wang X., Lu Z., Gomez A., Hon G. C., Yue Y., Han D., et al. (2014). N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505 117–120. 10.1038/nature12730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Zhao B. S., Roundtree I. A., Lu Z., Han D., Ma H., et al. (2015). N6-methyladenosine modulates messenger RNA translation efficiency. Cell 161 1388–1399. 10.1016/j.cell.2015.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L. H., Song P., Wang Y., Lu Z., Tang Q., Yu Q., et al. (2018). The m6A reader ECT2 controls trichome morphology by affecting mRNA stability in Arabidopsis. Plant Cell 30 968–985. 10.1105/tpc.17.00934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W., Adhikari S., Dahal U., Chen Y. S., Hao Y. J., Sun B. F., et al. (2016). Nuclear m6A reader YTHDC1 regulates mRNA splicing. Mol. Cell 61 507–519. 10.1016/j.molcel.2016.01.012 [DOI] [PubMed] [Google Scholar]
- Xu W., Li J., He C., Wen J., Ma H., Rong B., et al. (2021). METTL3 regulates heterochromatin in mouse embryonic stem cells. Nature 591 317–321. 10.1038/s41586-021-03210-1 [DOI] [PubMed] [Google Scholar]
- Zhang F., Zhang Y. C., Liao J. Y., Yu Y., Zhou Y. F., Feng Y. Z., et al. (2019). The subunit of RNA N6-methyladenosine methyltransferase OsFIP regulates early degeneration of microspores in rice. PLoS Genet. 15:e1008120. 10.1371/journal.pgen.1008120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., Bernstein B. E., et al. (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9:R137. 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Zhang Z., Gao J., Wang P., Hu T., Wang Z., et al. (2018). Arabidopsis duodecuple mutant of PYL ABA receptors reveals PYL repression of ABA-independent SnRK2 activity. Cell Rep. 23 3340–3351.e5. 10.1016/j.celrep.2018.05.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G., Dahl J. A., Niu Y., Fedorcsak P., Huang C. M., Li C. J., et al. (2013). ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 49 18–29. 10.1016/j.molcel.2012.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S., Li H., Bodi Z., Button J., Vespa L., Herzog M., et al. (2008). MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell 20 1278–1288. 10.1105/tpc.108.058883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Tian S., Qin G. (2019). RNA methylomes reveal the m6A-mediated regulation of DNA demethylase gene SlDML2 in tomato fruit ripening. Genome Biol. 20:156. 10.1186/s13059-019-1771-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Hu L., Jiang L., Liu S. (2018). Genome-wide identification and expression analysis of YTH domain-containing RNA-binding protein family in cucumber (Cucumis sativus). Genes Genom. 40 579–589. 10.1007/s13258-018-0659-3 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://bigd.big.ac.cn/gsa/s/uPCjOJ90, PRJCA005164.