Highlights
-
•
Protein arginine methylation is essential in multiple biological processes.
-
•
The family of PRMTs is a novel regulator of liver diseases.
-
•
Deregulation of PRMTs is correlated with HCC prognosis and clinical features.
-
•
PRMTs play a vital role in HCC malignancy, immune responses and metabolism.
-
•
PRMTs may represent druggable targets as novel strategies for HCC therapy.
Keywords: Protein arginine methyltransferases, Hepatocellular carcinoma, Therapeutic, Inhibitors
Abbreviations: HCC, hepatocellular carcinoma; PRMT, protein arginine methyltransferase; ADMA, asymmetric dimethylarginine; SDMA, symmetric dimethylarginine; MMA, monomethyl arginine; JMJD6, jumonji domain-containing protein 6; HBV, hepatitis B virus; HBx, hepatitis B virus X protein; CARM1, coactivator-associated arginine methyltransferase 1; LXRα, liver X receptor α; NAFLD, nonalcoholic fatty liver disease; TNM, tumor-node-metastasis; HNF4α, hepatocyte nuclear factor 4-alpha; EMT, epithelial-mesenchymal transition; MTDH, metadherin; 3’-UTR, 3’-untranslated region; ccRCC, clear cell renal cell carcinoma
Abstract
Hepatocellular carcinoma (HCC) is one of the most frequently diagnosed cancers with a high mortality rate worldwide. The complexity of HCC initiation and progression poses a great challenge to the diagnosis and treatment. An increasing number of studies have focused on the emerging roles of protein arginine methylation in cancers, including tumor growth, invasion, metastasis, metabolism, immune responses, chemotherapy sensitivity, etc. The family of protein arginine methyltransferases (PRMTs) is the most important proteins that mediate arginine methylation. The deregulation of PRMTs’ expression and functions in cancers have been gradually unveiled, and many PRMTs inhibitors are in preclinical and clinical investigations now. This review focuses predominantly on the aberrant expression of PRMTs, underlying mechanisms, as well as their potential applications in HCC, and provide novel insights into HCC therapy.
Introduction
Hepatocellular carcinoma (HCC) is one of the most prevalent cancers worldwide with a high mortality [1,2]. Though great progress in HCC diagnosis and therapy has been made during the past decades, the survival of HCC patients still remains poor, especially those diagnosed at advanced stage [3]. As a heterogeneous disease, the complexity of HCC initiation and progression poses a great challenge to the diagnosis and treatment. A more comprehensive and thorough understanding of regulatory mechanisms in HCC is of great importance for further investigations.
Protein arginine methylation is a vital post-translational modification that involves multiple biological processes, such as transcription, mRNA splicing and translation, DNA damage, cell fate determination, and signal transduction. The family of protein arginine methyltransferases (PRMTs) is the most important ‘writer’ to introduce methylation modifications on the arginine residues [4,5]. To date, nine members of the PRMTs family have been identified in humans based on their structures and functions. Increasing evidence has shown the aberrant expression and functions of PRMTs are associated with diverse types of cancers, including hematopoietic diseases, breast cancer, ovarian cancer, colorectal carcinoma, pancreatic cancer, and glioblastoma [6], [7], [8], [9], [10], [11], [12]. Over the last decade, the potential role of PRMTs in HCC has been gradually revealed, which may involve different mechanisms.
This review will focus on the roles and underlying mechanisms of PRMTs in HCC. Further, it will underscore the clinical significance of PRMTs in HCC, as well as emerging potential of such druggable targets as novel strategies for HCC therapy.
Overview of PRMTs
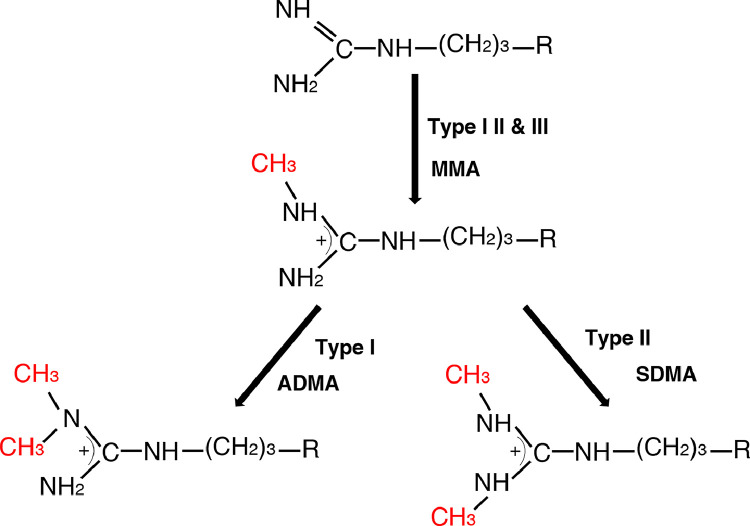
There are three different types of methylation modification generated during the process of arginine methylation: (I) asymmetric dimethylarginine (ADMA), (II) symmetric dimethylarginine (SDMA), and (III) monomethyl arginine (MMA) (Fig. 1). The formation of ADMA, SDMA and MMA in mammalian cells is found to be carried out mainly by PRMTs family, although several other proteins showing their functions as arginine methyltransferases, such as the putative arginine methyltransferases NDUFAF7 and Mettl23 [13,14]. In addition, recent studies have demonstrated the potential effect of another enzyme, Jumonji domain-containing protein 6 (JMJD6), which was initially identified as a phosphatidylserine receptor, on catalyzing histone arginine demethylation [15,16], and this reaction is dependent on the presence of cofactors, Fe (II) and 2‐oxoglutarate (2-OG). Several non‐histone proteins have also been found to be targets of JMJD6 for demethylation on their arginine residues, such as tumor necrosis factor receptor‐associated factor 6 (TRAF6), heat‐shock protein 70 (HSP70), and estrogen receptor α (ERα). However, the role of JMJD6 in demethylating arginine residues remains controversial. Lack of evidence for protein demethylation mediated by JMJD6 directly make us cannot rule out the possibility that JMJD6 may regulate the demethylation of these proteins indirectly. Additionally, many studies did not show the arginine demethylation activity of JMJD6. For instance, in the presence of oxygen, Fe (II) and 2-OG, JMJD6 was incubated with arginine-rich (RS) structural domain, then analyzed by mass spectrometry (MS). The results showed that JMJD6 was unable to produce demethylated arginine histone fragment peptides [17]. Another study on the crystal structure of JMJD6 was also skeptical of its arginine demethylation activity [18]. More studies are expected to further identify the effect of JMJD6 on arginine demethylation.
Fig. 1.
Types of methylation on arginine residues mediated by PRMTs.
MMA is generated by Type I, II and III PRMTs, followed by ADMA produced by Type I PRMTs and SDMA generated by Type II PRMTs.
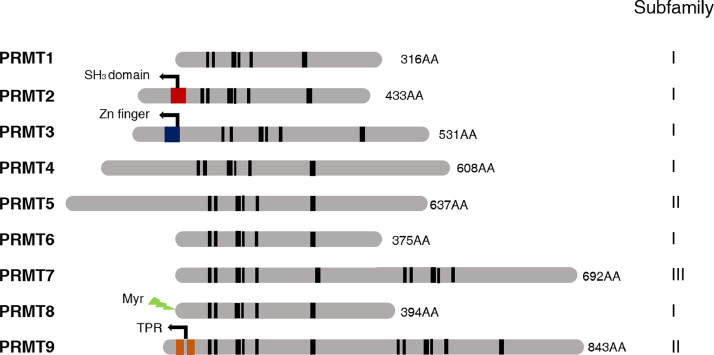
All nine PRMTs members possess signature motif I, post-I, motif II, post-II (double E loop), motif III, and the conserved THW loop, while several members harbor distinct domains (Fig. 2) [19,20]. PRMTs in human have been divided into three subfamilies according to the type of methylation they catalyze. All type I, II and III PRMTs are capable of generating MMA on one of the terminal guanidino nitrogen atoms. The subsequent formation of ADMA is catalyzed by type I enzymes (PRMT1, PRMT2, PRMT3, PRMT4, PRMT6 and PRMT8), and the generation of SDMA is catalyzed by type II enzymes (PRMT5, PRMT9) [4,19]. PRMT7 is the only type III PRMT specific for only catalyzing monomethyl arginine formation. It has no ability to catalyze SDMA formation directly. The effect of PRMT7 expression on cellular SDMA levels only occurs through the allosteric activation of PRMT5 via PRMT7 other monomethylated sites on the same substrate polypeptide [21].
Fig. 2.
The mammalian PRMTs.
All PRTMs possess conserved signature motifs I (VLD/EVGXGXG), post-I (V/IXG/AXD/E), motif II (F/I/VDI/L/K), post-II (double E loop), motif III (LR/KXXG), and THW loop (in black bars). Other motifs include SH3 domain, Zn Finger domain, myristoylation motif (Myr), tetratricopeptide repeats (TPR).
PRMTs can dynamically regulate chromatin structures, and function as coregulators that co-activate or co-repress gene transcription and expression. As stated, histone H4R3me2a, H3R2me2s, H3R17me2a, H3R26me2a are usually considered as activation marks, while H3R2me2a, H3R8me2a, H3R8me2s, H4R3me2s are repressive marks. Non-histone proteins can also be modified by PRMTs, which may associated with the changes in protein activities, stabilities, expression [22,19]. For example, ribosomal protein S2 (rpS2) a substrate for PRMT3 in mammals, and PRMT3 is able to inhibit rpS2 ubiquitination to modify the rpS2 protein level in cells [23,24]. PRMTs have an impact on a diverse range of cellular processes, such as cell proliferation and differentiation, transcription, DNA damage and repair, signal transduction, mRNA processing [4,5,19].
The role of PRMTs in benign liver diseases
The main risk factors for HCC are chronic infection with hepatitis B virus (HBV) or hepatitis C virus (HCV), heavy alcohol intake, and excess body weight. PRMTs have been reported to be involved in benign liver diseases, such as viral hepatitis, alcoholic liver disease (ALD), and non-alcoholic fatty liver disease (NAFLD). In the context of virus infection, PRMT1, the first protein arginine methyltransferase identified in the end of 1990s that accounting for approximately 85% of all cellular PRMT activity, overexpression of which inhibits HBV transcription via histone 4 (H4) methylation in HepG2 cells and the binding of HBx to PRMT1 might abolish the inhibitory effect of PRMT1 on HBV transcription [25], [26], [27]. PRMT5 has been reported to suppress HBV replication partially through mediating symmetrically demethylation of H4 at position R3 (H4R3me2s) on the cccDNA minichromosome [28]. PRMT6 seems to be a potential susceptibility gene for HBV-related HCC in a pilot two-phase genome-wide association study (GWAS) [29]. Besides, PRMTs also play a role in hepatic metabolism. PRMT1-mediated arginine methylation of FoxO1 contributes to the increase in hepatic glucose production in mouse models of genetic PRMT1 haploinsufficiency and transient depletion of hepatic PRMT1 in diabetic db/db mice. In the mouse models of alcohol induced liver injury, PRMT1 protect hepatocytes from oxidative stress response [30,31]. PRMT3 is involved in hepatic lipogenesis by interacting with LXRα, and palmitic acid treatment induces PRMT3 translocation to the nucleus accompanied by increased transcriptional activity of LXRα in mouse models of NAFLD [32], [33], [34]. PRMT6 promotes fasting-induced activation of the gluconeogenic program dependent on CRTC2, and couples glucose availability with mitochondria biogenesis by mediating SIRT7 methylation [35,36]. Furthermore, PRMT5 knockdown boosts the expression of PPARα and PGC-1α in mouse models fed with high-fat diet, with a concomitant increase in hepatic mitochondrial biogenesis, indicating its effect on redox reactions [37]. These studies indicate that distinct PRMTs perform distinct roles in distinct benign liver diseases that may progress to HCC.
Expression and clinical significance of PRMTs in HCC
As shown in Table 1, the deregulation of PRMTs expression is often observed in HCC. The expression of PRMT1, PRMT2, and PRMT5 in HCC tissues is significantly higher than that in matched nontumor tissues both at transcription and protein levels [38], [39], [40], [41], [42], [43], [44]. Additionally, compared with in matched noncancerous tissues, the expression of PRMT9 is higher in HCC tissues at protein level, whereas there is no difference at transcription level [45]. The expression of PRMT4 at protein level and the expression of PRMT6 at both levels has been found to be downregulated in human HCC tissues [46], [47], [48]. Intriguingly, in a model of DEN-induced HCC, the expression of PRMT4 is elevated in the early stage of hepatocarcinogenesis, which suggests that PRMT4 may play different roles in different stages of HCC initiation and progression [49]. However, this phenomenon is only observed in animal models, and more explorations should be conducted in human.
Table 1.
The expression and clinical relevance of PRMTs in HCC.
| PRMTs | Expression in HCC tissues | Prognostic values (high vs low) | Correlation with clinical characteristics | References |
|---|---|---|---|---|
| PRMT1 | Increased | Poor | Microvascular invasion, tumor differentiation, tumor size, portal vein tumor thrombus (PVTT), small lesions | [39,40,55] |
| PRMT2 | Increased | Poor | Tumor size, histological grade, tumor-node-metastasis (TNM) stage | [41] |
| PRMT3 | N.A | N.A | N.A | N.A |
| PRMT4 | Decreased | N.A | N.A | [46] |
| PRMT5 | Increased | Poor | Tumor size, UICC pathological stage, α-fetoprotein (AFP), tumor differentiation, microscopic hepatic invasion | [42], [43], [44] |
| PRMT6 | Decreased | N.A | Age, vascular invasion, intraoperative rupture | [47] |
| PRMT7 | N.A | N.A | N.A | N.A |
| PRMT8 | N.A | N.A | N.A | N.A |
| PRMT9 | Increased | Poor | Hepatitis B virus antigen (HBsAg), vascular invasion, tumor differentiation, TNM stage | [45] |
Importantly, the aberrant expression of some PRMTs have been demonstrated to be associated with the prognosis and clinicopathological features of patients with HCC. PRMT1 has been considered as a prognostic biomarker for HCC, as its overexpression is strongly correlated with poor prognosis in cohorts of HCC patients from different regions and adverse clinicopathological features, such as microvascular invasion presence, worse tumor differentiation, lager tumor volumes and more portal vein tumor thrombus (PVTT) [39,40]. Besides, it is also an independent risk factor for both disease-free survival and overall survival of HCC patients [40]. High PRMT2 expression is related to larger tumor size, poor histological grade, advanced tumor-node-metastasis (TNM) stage, as well as shorter survival time [41]. Similarly, the elevated expression of PRMT5 also has a positive correlation with poor prognosis and aggressive clinicopathological characters, including worse tumor differentiation, more frequent microvascular invasion, larger tumor size and higher serum α-fetoprotein levels [43,44,50]. Zhang et al. have shown that PRMT9 expression level has a relationship with HBsAg, vascular invasion, tumor differentiation, and TNM stage, and patients with positive PRMT9 expression have shorter survival time than those with negative PRMT9 expression [45]. These findings suggest that PRMTs might act as potential prognostic biomarkers for HCC patients.
The role of PRMTs in HCC malignant phenotypes
Sustaining proliferation, activation of invasion and metastasis have been considered as hallmarks of cancer cells [51]. Several previous studies have revealed the roles that PRMTs play in HCC. PRMT1 inhibition results in reduced HNF4α expression and hepatocyte proliferation, thus promoting alcohol-induced HCC progression, which suggests that PRMT1 seems to perform a protective role in the context of alcohol-induced HCC [52,53,31]. Nevertheless, PRMT1 functions as an oncogene in other contexts of HCC. PRMT1 promotes HCC proliferation, invasion and metastasis via activating STAT3 signaling, and STAT3 inhibitor can reverse the effects mediated by PRMT1 [40]. Epithelial-mesenchymal transition (EMT) has been considered as a vital program involved in cancer cell metastasis [54]. PRMT1 is able to drive EMT program in HCC cells through TGF-β1/Smad pathway, as its overexpression increases the expression of mesenchymal markers (Vimentin, Snail and N-cadherin) and decreases the expression of epithelial marker E-cadherin [55]. PRMT2 has been reported to facilitate HCC growth and inhibit cell apoptosis by regulating BCL2 expression. Mechanically, PRMT2 overexpression results in H3R8 asymmetric methylation (H3R8me2a) enrichment at the BCL2 promoter, which increases its accessibility to STAT3 and promotes BCL2 gene expression [41]. PRMT5 promotes HCC proliferation through affecting cell cycle partially by decreasing BTG2 expression. The regulation of BTG2 expression by PRMT5 is related to ERK phosphorylation, as treatment with a selective ERK1/2 inhibitor remarkably reverses the effect of PRMT5 on BTG2 expression [42]. Besides, when glucose induction in HCC cells, PRMT5 competitively interacts with CDK4 at R24 to drive G1/S cell cycle progression and tumor growth [56]. Subcellular localization of PRMT5 is also pivotal for HCC growth and metastasis. In the process of metadherin (MTDH)-mediated HCC metastasis, PRMT5 translocates from the nucleus to the cytoplasm, accompanied by nucleus localization of β-catenin, thus activating the WNT-β-catenin signaling pathway [57]. PRMT6 functions as a suppressor in HCC, and it methylates CRAF at R100 that decreases its RAS binding potential, then its downstream MEK/ERK signaling is inhibited [47]. Moreover, PRMT9 has been shown to enhance HCC invasion and metastasis by inducing EMT through PI3K/Akt/GSK-3b/Snail signaling [45].
The role of PRMTs in HCC immune responses
HCC is a type of inflammation-associated cancer, in which local and systemic immune responses play an important role [58,59]. Increasing evidence has shown the significance of PRMTs in immune response [60,61]. For example, PRMT1 is essential for proliferation, activation, and differentiation of human and mouse peripheral B cells. PRMT5 negatively modulates cGAS-mediated antiviral immune response by blocking the DNA binding capability of cGAS in mice [62,63]. We have mentioned above that PRMT1 might be a protector in alcohol-associated HCC, but in the perspective of immunology, PRMT1-dependent macrophage IL-6 secretion has been identified to be an important mechanism of alcohol-induced HCC progression. PRMT1 knockout results in the decrease in both IL-10 and IL-6 cytokines expression and tumor growth in mice of DEN-induced HCC [64]. The polymorphism rs975484 in the PRMT1 gene promoter regulates the expression of some immune checkpoint genes in HCC, such as PD-L1 and PD-L2, which indicates the polymorphism of the gene may be linked to immunotherapy of HCC [65]. Furthermore, PRMT5 inhibition induces lymphocytes infiltration and the expression of several MHC II molecule family components, including H2-Ab1, H2-Aa, and Cd74, which might enhance the anti-tumor immune responses [66].
The role of PRMTs in HCC metabolism
Metabolism reprogramming has been recognized as an emerging hallmark of cancer [51,67]. The liver plays a critical part in coordinating a variety of metabolic activities. Metabolic abnormalities are involved in both HCC initiation and malignant progression [68,69]. Recently, more and more studies have recognized the functions of PRMTs in HCC metabolism. PRMT4 has been reported to be correlated with glucose abnormality in HCC, it methylates GAPDH at R234 to inhibit its catalytic activity and Warburg effect, thus delaying HCC growth [46]. It has been demonstrated that PRMT5 is strongly correlated with metabolism regulation in cancers [70], [71], [72]. In HCC, PRMT5 enhances lipid biosynthesis and HCC growth by induce methylation of SREBP1α both in vivo and in vitro. PRMT5 promotes SREBP1α transcriptional activity through mediating its symmetrically dimethylation at R321. Moreover, PRMT5-induced methylation of SREBP1α induced by PRMT5 prevented its phosphorylation at S430 by GSK3β, followed by disassociation from Fbw7 (FBXW7) and evasion from degradation via ubiquitin-proteasome pathway [73]. PRMT6 is another member of PRMTs family that has been shown to be associated with metabolism deregulation of HCC. The interaction between PRMT6 and CRAF regulates aerobic glycolysis by altering ERK-mediated nuclear relocalization of PKM2. PRMT6 methylates BAG5 at R15 and R24 to promote the degradation of its interacting partner HSC70, an identified autophagy player. Under oxygen or nutrient-derived conditions, PRMT6 is downregulated to induce autophagy in HCC and PRMT6 expression negatively correlates with enhanced autophagic flux in cells. [74,75].
Regulatory mechanisms underlying PRMTs deregulation in HCC
Posttranslational modifications are of great significance in regulation of PRMTs, such as phosphorylation and ubiquitylation [4]. For instance, the methyltransferase activity of PRMT4 is suppressed by phosphorylation at Serine 228 (S228), and phosphorylation at S217 promotes PRMT4 cytoplasmic localization to impair its enzymatic activity [76,77]. PRMT1 can be polyubiquitylated for proteasome degradation by FBXL17 at Lysine 117(K117) [78]. Noteworthy, the activity and function of PRMTs can be regulated by automethylation, a special type of posttranslational modification mediated by themselves. Automethylation of PRMT4 at R551 has an impact on transcription and pre-mRNA splicing modulated by itself [79]. PRMT6 automethylation at R35 enhances its stability, while R58 automethylation of PRMT8 attenuates its activity by influencing R73 automethylation [80,81]. In the context of HCV infection, upregulation of protein phosphatase 2A (PP2A) has been reported to modulates NS3 helicase activity via inhibiting PRMT1 enzymatic activity [82]. Notably, the stability of PRMT4 is regulated by the SKP2-containing SCF (SKP1-cullin1-F-box protein) E3 ubiquitin ligase in the nucleus, but not in the cytoplasm. Under glucose deprivation induced autophagy, the activation of AMPKα2, reduction of SKP2, and subsequent upregulation of CARM1, are observed in the nucleus of HCC cells, suggesting a potential role of nuclear AMPK-SKP2-CARM1 signaling in the induction of autophagy in HCC cells [83,84]. Moreover, the activity of PRMT5 is regulated by phosphorylation at Threonine 80 (T80) by RhoA-associated protein kinase (ROK) and myosin phosphatase (MP). PRMT5-specific SDMA modification on arginine residues of histone 2A (H2A) and H4 is considered as a repressing gene expression mark, which can be increased by silencing of myosin phosphatase target subunit-1 (MYPT1), and it leads to a global change in the expression of genes associated with cell growth, proliferation and death [85].
Besides, the role of specific lncRNAs (long non-coding RNA) and miRNAs (microRNAs) in the regulation of PRMTs’ expression has also been investigated. MiR-503 suppresses the expression of PRMT1 by target 3’-untranslated region (3’-UTR) of PRMT1 mRNA, thus remarkably inhibiting invasion and migration in HCC cells [86]. MiR24-2 targeting 3’UTR of PRMT7 mRNA to inhibit the translational ability and the methylation of H4R3 mediated by PRMT7, which promotes liver cancer stem cells malignant progression [87]. Amplification of LINC01138 promotes HCC proliferation, invasion and metastasis through physically interacting with PRMT5 and protecting it from degradation by ubiquitin-proteasome pathway in HCC [88].
It has been recently demonstrated that PRMT5 can be regulated by MYC, a vital oncogene driving the genesis of many human cancers, in HCC. The level of urinary SDMA is increased in a MYC-dependent manner in DEN-induced HCC mouse model and patients with HCC, and PRMT5 is identified as a direct MYC target gene [66,89].
Applications of PRMT inhibitors in HCC
Unlike the irreversibility of genetic mutations, reversibility of epigenetic modifications shed light to cancer therapy [90,91]. Since the first PRMT inhibitor generated in 2004, AMI-1, which inhibits all type I PRMTs, the development and application of PRMTs inhibitors has undergone great progress in the past less than two decades [92]. Recently, many selective and potent PRMT inhibitors targeting PRMT3 (SGC707), PRMT4 (TP-064, EZM2302), PRMT5 (GSK591, LLY-283), PRMT6 (EPZ020411), and PRMT7 (SGC3027) with membrane permeability have been developed. Their efficacy in cellular and animal models have highlighted the therapeutic and pharmacological potential [93,94]. For instance, in breast cancers, PRMT4 inhibitor (TP-064) treatment inhibits a subset of multiple myeloma cell lines proliferation by affecting cell cycle [95]. A recent study has shown that the pharmacological inhibition of PRMT1 by a novel potent inhibitor DCPT1061 suppresses clear cell renal cell carcinoma (ccRCC) cell proliferation and sensitizes ccRCC to sunitinib treatment. Combination of DCPT1061 and sunitinib exhibited a striking anti-proliferative effect in ccRCC cell models and xenograft animal models [96].
Importantly, a growing number of PRMT inhibitors are in clinical trials, further indicating the possibility of clinical applications of these PRMT inhibitors, especially PRMT5 inhibitors for their efficacy in hematological diseases. GSK3368715, a PRMT5 inhibitor, is in Phase I clinical trial for relapsed or refractory diffuse large B cell lymphoma and selected solid tumors with deletion of the methylthioadenosine phosphorylase (MTAP) gene (NCT03666988) [97]. Besides, GSK3326595 and JNJ-64619178, another two small molecule inhibitors targeting PRMT5, have also be in clinical trials for hematological diseases and some solid tumors now (NCT02783300, NCT03614728, CT03573310) [98].
Here, we focus our attention on applications of PRMT inhibitors in HCC. Despite the great advances in PRMT inhibitors development, their applications in the field of HCC remains limited. MS023, a type I PRMT inhibitor, has been found to impair tumorigenesis in HCC mouse models [41]. Moreover, Zheng et al. have found that targeting PRMT5 activity by DW14800 treatment, a novel potent PRMT5 inhibitor that binds to the substrate binding site of PRMT5 with high activity against PRMT5, is able to inhibit HCC malignancy. DW14800 treatment decreases the occupation of H4R3me2s on the promoter of HNF4α to promote HNF4α expression, thus resulting in the reduced abilities of proliferation and invasion in HCC [99]. In another recent study, Luo et al. have noticed that PRMT5 inhibitor treatment, GSK3326595, increase the infiltration of CD45.1+leukocytes, natural killer (NK) cells, CD4+T cells, CD8+T cells, dendritic cells (DC) and monocytes in HCC tumors of mouse models. Combination of GSK3326595 with anti-PD-1 immune checkpoint (ICT) therapy significantly improved therapeutic effects in HCC, which provides a novel strategy to attenuate the resistance of ‘immune‐cold’ tumor to ICT [66].
Conclusions and future prospects
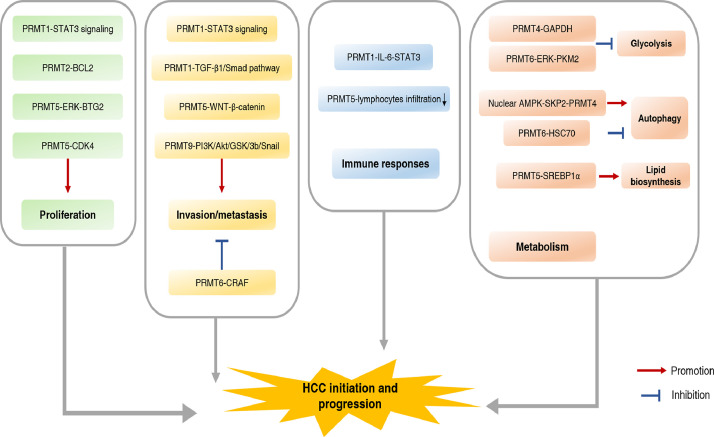
Primary liver cancer ranks as the sixth most frequently diagnosed cancer and the third leading cause of cancer death worldwide in 2020, and HCC accounts for 75%-85% of these cases [2]. Posttranslational modification of HCC is a rapidly growing and emerging area of research, contributing to our comprehensive understanding of the pathophysiological and molecular mechanisms underlying HCC tumorigenesis and progression. Many studies have reported the distinct roles of PRMTs in benign liver diseases that may progress to HCC, which indicates targeting PRMTs might play a role in HCC prevention. More importantly, above studies have also demonstrated that the deregulation of PRMTs expression or functions are involved in HCC, suggesting their potential as prognostic markers for HCC. PRMTs regulates but not limited to HCC growth, invasion, metastasis, immune response, and metabolism both dependent or independent on arginine methylation (Table 2) (Fig. 3). Therefore, it is worth considering several PRMTs as potential targets for HCC. Furthermore, the expression and activity of PRMTs can be regulated at transcriptional, posttranscriptional, and posttranslational levels. As mentioned in the review, studies on PRMTs in HCC are limited. There has been lack of investigations on the role of PRMTs in different context of HCC. In addition, previous studies have reported the alternative splicing, subcellular localization and the scope of isoforms within some PRMTs, whether they have an effect on HCC remains to be explored [94].
Table 2.
The role of PRMTs in HCC.
| PRMTs | Subcellular localization | Substrates or relevant signalings | Effects in HCC | References |
|---|---|---|---|---|
| PRMT1 | Cytoplasm, nucleus | HNF4α | A protector in alcohol-induced HCC | [52,53,100] |
| STAT3 signaling | Promotes proliferation, invasion, metastasis | [40] | ||
| TGF-β1/Smad pathway | Drives EMT program | [55] | ||
| IL-6-STAT3 | Immune environment formation in alcohol-induced HCC | [64] | ||
| PRMT2 | Cytoplasm, nucleus | H3R8me2a at the BCL2 promoter | Promotes proliferation, and inhibits apoptosis | [41] |
| PRMT3 | Cytoplasm | N.A | N.A | N.A |
| PRMT4 | Cytoplasm, nucleus | GAPDH at R234 | Inhibits Warburg effect and tumor growth | [46] |
| Nuclear AMPK-SKP2-CARM1 | Induces autophagy in HCC cells | [83] | ||
| PRMT5 | Cytoplasm, nucleus | ERK-BTG2 | Promotes proliferation | [42] |
| CDK4 at R24 | Promotes proliferation | [56] | ||
| WNT-β-catenin | Promotes metastasis | [43] | ||
| SREBP1α at R321 | Promotes lipid biosynthesis and tumor growth | [73] | ||
| PRMT6 | Nucleus | CRAF at R100 | Inhibits tumor-initiation, metastasis, and therapy resistance | [47] |
| ERK-PKM2 | Inhibits aerobic glycolysis | [74] | ||
| HSC70 | Inhibits autophagy | [75] | ||
| PRMT7 | Cytoplasm, nucleus | N.A | N.A | N.A |
| PRMT8 | Membrane | N.A | N.A | N.A |
| PRMT9 | Cytoplasm | PI3K/Akt/GSK-3b/Snail | Promotes invasion and metastasis | [45] |
Fig. 3.
The role of PRMTs in HCC.
The feasibility of the class of enzymes associated with arginine modification for drug development, coupled with the rapid development of emerging biological technologies, is revealing novel opportunities for HCC therapy. To date, the PRMTs inhibitors are predominantly utilized in hematological diseases, and adverse effects and efficacy of most PRMTs inhibitors to be further confirmed. The next decades will focus on the development and discovery of more specific and selective PRMT inhibitors useful for treating specific cancers. The utility of these compounds, together with the analysis of PRMTs’ functions will help to resolve the different and overlapping functions of these epigenetic enzymes in HCC. Furthermore, it is also important to develop more PRMT inhibitors with safety and efficacy for clinical applications, as well as conduct intensive clinical trials on existing drugs.
Declaration of Competing Interest
The authors declare that they have no conflict of interest.
Acknowledgments
Author contributions
Yu Lei, Ping Han, and Dean Tian contribute to conception and writing.
Funding
The work was supported by grants from the National Natural Science Foundation of China No. 81772610, No. 81974071.
Acknowledgements
None.
Data availability statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
Contributor Information
Ping Han, Email: hanzhouping@163.com.
Dean Tian, Email: datian@tjh.tjmu.edu.cn.
References
- 1.Villanueva A. Hepatocellular Carcinoma. N. Engl. J. Med. 2019;380:1450–1462. doi: 10.1056/NEJMra1713263. [DOI] [PubMed] [Google Scholar]
- 2.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021 doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 3.Forner A., Reig M., Bruix J. Hepatocellular carcinoma. Lancet. 2018;391:1301–1314. doi: 10.1016/S0140-6736(18)30010-2. [DOI] [PubMed] [Google Scholar]
- 4.Jarrold J., Davies C.C. PRMTs and arginine methylation: cancer's best-kept secret? Trends Mol. Med. 2019;25:993–1009. doi: 10.1016/j.molmed.2019.05.007. [DOI] [PubMed] [Google Scholar]
- 5.Bedford M.T., Clarke S.G. Protein arginine methylation in mammals: who, what, and why. Mol. Cell. 2009;33:1–13. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He X., Zhu Y., Lin Y.C., Li M., Du J., Dong H., Sun J., Zhu L., Wang H., Ding Z., Zhang L., Zhang L., Zhao D., Wang Z., Wu H., Zhang H., Jiang W., Xu Y., Jin J., Shen Y., Perry J., Zhao X., Zhang B., Liu S., Xue S.L., Shen B., Chen C.W., Chen J., Khaled S., Kuo Y.H., Marcucci G., Luo Y., Li L. PRMT1-mediated FLT3 arginine methylation promotes maintenance of FLT3-ITD(+) acute myeloid leukemia. Blood. 2019;134:548–560. doi: 10.1182/blood.2019001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Z., Wang D., Lu J., Huang B., Wang Y., Dong M., Fan D., Li H., Gao Y., Hou P., Li M., Liu H., Pan Z.Q., Zheng J., Bai J. Methylation of EZH2 by PRMT1 regulates its stability and promotes breast cancer metastasis. Cell Death Differ. 2020;27:3226–3242. doi: 10.1038/s41418-020-00615-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karakashev S., Zhu H., Wu S., Yokoyama Y., Bitler B.G., Park P.H., Lee J.H., Kossenkov A.V., Gaonkar K.S., Yan H., Drapkin R., Conejo-Garcia J.R., Speicher D.W., Ordog T., Zhang R. CARM1-expressing ovarian cancer depends on the histone methyltransferase EZH2 activity. Nat. Commun. 2018;9:631. doi: 10.1038/s41467-018-03031-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yin X.K., Wang Y.L., Wang F., Feng W.X., Bai S.M., Zhao W.W., Feng L.L., Wei M.B., Qin C.L., Wang F., Chen Z.L., Yi H.J., Huang Y., Xie P.Y., Kim T., Wang Y.N., Hou J.W., Li C.W., Liu Q., Fan X.J., Hung M.C., Wan X.B. PRMT1 enhances oncogenic arginine methylation of NONO in colorectal cancer. Oncogene. 2021;40:1375–1389. doi: 10.1038/s41388-020-01617-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsu M.C., Tsai Y.L., Lin C.H., Pan M.R., Shan Y.S., Cheng T.Y., Cheng S.H., Chen L.T., Hung W.C. Protein arginine methyltransferase 3-induced metabolic reprogramming is a vulnerable target of pancreatic cancer. J. Hematol. Oncol. 2019;12:79. doi: 10.1186/s13045-019-0769-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dong F., Li Q., Yang C., Huo D., Wang X., Ai C., Kong Y., Sun X., Wang W., Zhou Y., Liu X., Li W., Gao W., Liu W., Kang C., Wu X. PRMT2 links histone H3R8 asymmetric dimethylation to oncogenic activation and tumorigenesis of glioblastoma. Nat. Commun. 2018;9:4552. doi: 10.1038/s41467-018-06968-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stopa N., Krebs J.E., Shechter D. The PRMT5 arginine methyltransferase: many roles in development, cancer and beyond. Cellular Mol. Life Sci. 2015;72:2041–2059. doi: 10.1007/s00018-015-1847-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rhein V.F., Carroll J., Ding S., Fearnley I.M., Walker J.E. NDUFAF7 methylates arginine 85 in the NDUFS2 subunit of human complex I. J. Biol. Chem. 2013;288:33016–33026. doi: 10.1074/jbc.M113.518803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hatanaka Y., Tsusaka T., Shimizu N., Morita K., Suzuki T., Machida S., Satoh M., Honda A., Hirose M., Kamimura S., Ogonuki N., Nakamura T., Inoue K., Hosoi Y., Dohmae N., Nakano T., Kurumizaka H., Matsumoto K., Shinkai Y., Ogura A. Histone H3 methylated at arginine 17 is essential for reprogramming the paternal genome in zygotes. Cell Rep. 2017;20:2756–2765. doi: 10.1016/j.celrep.2017.08.088. [DOI] [PubMed] [Google Scholar]
- 15.Yang J., Chen S., Yang Y., Ma X., Shao B., Yang S., Wei Y., Wei X. Jumonji domain-containing protein 6 protein and its role in cancer. Cell Prolif. 2020;53:e12747. doi: 10.1111/cpr.12747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang B., Chen Y., Zhao Y., Bruick R.K. JMJD6 is a histone arginine demethylase. Science. 2007;318:444–447. doi: 10.1126/science.1145801. [DOI] [PubMed] [Google Scholar]
- 17.Webby C.J., Wolf A., Gromak N., Dreger M., Kramer H., Kessler B., Nielsen M.L., Schmitz C., Butler D.S., Yates J.R., 3rd, Delahunty C.M., Hahn P., Lengeling A., Mann M., Proudfoot N.J., Schofield C.J., Bottger A. Jmjd6 catalyses lysyl-hydroxylation of U2AF65, a protein associated with RNA splicing. Science. 2009;325:90–93. doi: 10.1126/science.1175865. [DOI] [PubMed] [Google Scholar]
- 18.Mantri M., Krojer T., Bagg E.A., Webby C.J., Butler D.S., Kochan G., Kavanagh K.L., Oppermann U., McDonough M.A., Schofield C.J. Crystal structure of the 2-oxoglutarate- and Fe(II)-dependent lysyl hydroxylase JMJD6. J. Mol. Biol. 2010;401:211–222. [PubMed] [Google Scholar]
- 19.Yang Y., Bedford M.T. Protein arginine methyltransferases and cancer. Nat. Rev. Cancer. 2013;13:37–50. doi: 10.1038/nrc3409. [DOI] [PubMed] [Google Scholar]
- 20.Feng Y., Hadjikyriacou A., Clarke S.G. Substrate specificity of human protein arginine methyltransferase 7 (PRMT7): the importance of acidic residues in the double E loop. J. Biol. Chem. 2014;289:32604–32616. doi: 10.1074/jbc.M114.609271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jain K., Clarke S.G. PRMT7 as a unique member of the protein arginine methyltransferase family: A review. Arch. Biochem. Biophys. 2019;665:36–45. doi: 10.1016/j.abb.2019.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blanc R.S., Richard S. Arginine methylation: the coming of age. Mol. Cell. 2017;65:8–24. doi: 10.1016/j.molcel.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 23.Swiercz R., Person M.D., Bedford M.T. Ribosomal protein S2 is a substrate for mammalian PRMT3 (protein arginine methyltransferase 3. Biochem. J. 2005;386:85–91. doi: 10.1042/BJ20041466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi S., Jung C.R., Kim J.Y., Im D.S. PRMT3 inhibits ubiquitination of ribosomal protein S2 and together forms an active enzyme complex. Biochim. Biophys. Acta. 2008;1780:1062–1069. doi: 10.1016/j.bbagen.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 25.Lin W.J., Gary J.D., Yang M.C., Clarke S., Herschman H.R. The mammalian immediate-early TIS21 protein and the leukemia-associated BTG1 protein interact with a protein-arginine N-methyltransferase. J. Biol. Chem. 1996;271:15034–15044. doi: 10.1074/jbc.271.25.15034. [DOI] [PubMed] [Google Scholar]
- 26.Tang J., Frankel A., Cook R.J., Kim S., Paik W.K., Williams K.R., Clarke S., Herschman H.R. PRMT1 is the predominant type I protein arginine methyltransferase in mammalian cells. J. Biol. Chem. 2000;275:7723–7730. doi: 10.1074/jbc.275.11.7723. [DOI] [PubMed] [Google Scholar]
- 27.Benhenda S., Ducroux A., Riviere L., Sobhian B., Ward M.D., Dion S., Hantz O., Protzer U., Michel M.L., Benkirane M., Semmes O.J., Buendia M.A., Neuveut C. Methyltransferase PRMT1 is a binding partner of HBx and a negative regulator of hepatitis B virus transcription. J. Virol. 2013;87:4360–4371. doi: 10.1128/JVI.02574-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang W., Chen J., Wu M., Zhang X., Zhang M., Yue L., Li Y., Liu J., Li B., Shen F., Wang Y., Bai L., Protzer U., Levrero M., Yuan Z. PRMT5 restricts hepatitis B virus replication through epigenetic repression of covalently closed circular DNA transcription and interference with pregenomic RNA encapsidation. Hepatology. 2017;66:398–415. doi: 10.1002/hep.29133. [DOI] [PubMed] [Google Scholar]
- 29.Qu L.S., Jin F., Guo Y.M., Liu T.T., Xue R.Y., Huang X.W., Xu M., Chen T.Y., Ni Z.P., Shen X.Z. Nine susceptibility loci for hepatitis B virus-related hepatocellular carcinoma identified by a pilot two-stage genome-wide association study. Oncol. Lett. 2016;11:624–632. doi: 10.3892/ol.2015.3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choi D., Oh K.J., Han H.S., Yoon Y.S., Jung C.Y., Kim S.T., Koo S.H. Protein arginine methyltransferase 1 regulates hepatic glucose production in a FoxO1-dependent manner. Hepatology. 2012;56:1546–1556. doi: 10.1002/hep.25809. [DOI] [PubMed] [Google Scholar]
- 31.Zhao J., Adams A., Weinman S.A., Tikhanovich I. Hepatocyte PRMT1 protects from alcohol induced liver injury by modulating oxidative stress responses. Sci. Rep. 2019;9:9111. doi: 10.1038/s41598-019-45585-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim D.I., Park M.J., Lim S.K., Park J.I., Yoon K.C., Han H.J., Gustafsson J.A., Lim J.H., Park S.H. PRMT3 regulates hepatic lipogenesis through direct interaction with LXRalpha. Diabetes. 2015;64:60–71. doi: 10.2337/db13-1394. [DOI] [PubMed] [Google Scholar]
- 33.Hoekstra M., Nahon J.E., de Jong L.M., Kroner M.J., de Leeuw L.R., Van Eck M. Inhibition of PRMT3 activity reduces hepatic steatosis without altering atherosclerosis susceptibility in apoE knockout mice. Biochimica et biophysica acta. Mol. Basis Dis. 2019;1865:1402–1409. doi: 10.1016/j.bbadis.2019.02.012. [DOI] [PubMed] [Google Scholar]
- 34.Nahon J.E., Groeneveldt C., Geerling J.J., van Eck M., Hoekstra M. Inhibition of protein arginine methyltransferase 3 activity selectively impairs liver X receptor-driven transcription of hepatic lipogenic genes in vivo. Br. J. Pharmacol. 2018;175:3175–3183. doi: 10.1111/bph.14361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Han H.S., Jung C.Y., Yoon Y.S., Choi S., Choi D., Kang G., Park K.G., Kim S.T., Koo S.H. Arginine methylation of CRTC2 is critical in the transcriptional control of hepatic glucose metabolism. Sci. Signal. 2014;7:ra19. doi: 10.1126/scisignal.2004479. [DOI] [PubMed] [Google Scholar]
- 36.Yan W.W., Liang Y.L., Zhang Q.X., Wang D., Lei M.Z., Qu J., He X.H., Lei Q.Y., Wang Y.P. Arginine methylation of SIRT7 couples glucose sensing with mitochondria biogenesis. EMBO Rep. 2018;19 doi: 10.15252/embr.201846377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang L., Liu J., Zhang X.O., Sibley K., Najjar S.M., Lee M.M., Wu Q. Inhibition of protein arginine methyltransferase 5 enhances hepatic mitochondrial biogenesis. J. Biol. Chem. 2018;293:10884–10894. doi: 10.1074/jbc.RA118.002377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gou Q., He S., Zhou Z. Protein arginine N-methyltransferase 1 promotes the proliferation and metastasis of hepatocellular carcinoma cells. Tumour Biol. 2017;39 doi: 10.1177/1010428317691419. [DOI] [PubMed] [Google Scholar]
- 39.Ryu J.W., Kim S.K., Son M.Y., Jeon S.J., Oh J.H., Lim J.H., Cho S., Jung C.R., Hamamoto R., Kim D.S., Cho H.S. Novel prognostic marker PRMT1 regulates cell growth via downregulation of CDKN1A in HCC. Oncotarget. 2017;8:115444–115455. doi: 10.18632/oncotarget.23296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang X.P., Jiang Y.B., Zhong C.Q., Ma N., Zhang E.B., Zhang F., Li J.J., Deng Y.Z., Wang K., Xie D., Cheng S.Q. PRMT1 promoted HCC growth and metastasis in vitro and in vivo via activating the STAT3 signalling pathway. Cellular Physiol. Biochem. 2018;47:1643–1654. doi: 10.1159/000490983. [DOI] [PubMed] [Google Scholar]
- 41.Hu G., Yan C., Xie P., Cao Y., Shao J., Ge J. PRMT2 accelerates tumorigenesis of hepatocellular carcinoma by activating Bcl2 via histone H3R8 methylation. Exp. Cell Res. 2020;394 doi: 10.1016/j.yexcr.2020.112152. [DOI] [PubMed] [Google Scholar]
- 42.Jiang H., Zhu Y., Zhou Z., Xu J., Jin S., Xu K., Zhang H., Sun Q., Wang J., Xu J. PRMT5 promotes cell proliferation by inhibiting BTG2 expression via the ERK signaling pathway in hepatocellular carcinoma. Cancer Med. 2018;7:869–882. doi: 10.1002/cam4.1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang B., Dong S., Li Z., Lu L., Zhang S., Chen X., Cen X., Wu Y. Targeting protein arginine methyltransferase 5 inhibits human hepatocellular carcinoma growth via the downregulation of beta-catenin. J. Transl. Med. 2015;13:349. doi: 10.1186/s12967-015-0721-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jeon J.Y., Lee J.S., Park E.R., Shen Y.N., Kim M.Y., Shin H.J., Joo H.Y., Cho E.H., Moon S.M., Shin U.S., Park S.H., Han C.J., Choi D.W., Gu M.B., Kim S.B., Lee K.H. Protein arginine methyltransferase 5 is implicated in the aggressiveness of human hepatocellular carcinoma and controls the invasive activity of cancer cells. Oncol. Rep. 2018;40:536–544. doi: 10.3892/or.2018.6402. [DOI] [PubMed] [Google Scholar]
- 45.Jiang H., Zhou Z., Jin S., Xu K., Zhang H., Xu J., Sun Q., Wang J., Xu J. PRMT9 promotes hepatocellular carcinoma invasion and metastasis via activating PI3K/Akt/GSK-3beta/Snail signaling. Cancer Sci. 2018;109:1414–1427. doi: 10.1111/cas.13598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhong X.Y., Yuan X.M., Xu Y.Y., Yin M., Yan W.W., Zou S.W., Wei L.M., Lu H.J., Wang Y.P., Lei Q.Y. CARM1 methylates GAPDH to regulate glucose metabolism and is suppressed in liver cancer. Cell Rep. 2018;24:3207–3223. doi: 10.1016/j.celrep.2018.08.066. [DOI] [PubMed] [Google Scholar]
- 47.Chan L.H., Zhou L., Ng K.Y., Wong T.L., Lee T.K., Sharma R., Loong J.H., Ching Y.P., Yuan Y.F., Xie D., Lo C.M., Man K., Artegiani B., Clevers H., Yan H.H., Leung S.Y., Richard S., Guan X.Y., Huen M.S.Y., Ma S. PRMT6 regulates RAS/RAF binding and MEK/ERK-mediated cancer stemness activities in hepatocellular carcinoma through CRAF methylation. Cell Rep. 2018;25:690–701. doi: 10.1016/j.celrep.2018.09.053. e698. [DOI] [PubMed] [Google Scholar]
- 48.Qin Q.F., Li X.J., Li Y.S., Zhang W.K., Tian G.H., Shang H.C., Tang H.B. AMPK-ERK/CARM1 signaling pathways affect autophagy of hepatic cells in samples of liver cancer patients. Front. Oncol. 2019;9:1247. doi: 10.3389/fonc.2019.01247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Osada S., Suzuki S., Yoshimi C., Matsumoto M., Shirai T., Takahashi S., Imagawa M. Elevated expression of coactivator-associated arginine methyltransferase 1 is associated with early hepatocarcinogenesis. Oncol. Rep. 2013;30:1669–1674. doi: 10.3892/or.2013.2651. [DOI] [PubMed] [Google Scholar]
- 50.Shimizu D., Kanda M., Sugimoto H., Shibata M., Tanaka H., Takami H., Iwata N., Hayashi M., Tanaka C., Kobayashi D., Yamada S., Nakayama G., Koike M., Fujiwara M., Fujii T., Kodera Y. The protein arginine methyltransferase 5 promotes malignant phenotype of hepatocellular carcinoma cells and is associated with adverse patient outcomes after curative hepatectomy. Int. J. Oncol. 2017;50:381–386. doi: 10.3892/ijo.2017.3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 52.Zhao J., Adams A., Roberts B., O'Neil M., Vittal A., Schmitt T., Kumer S., Cox J., Li Z., Weinman S.A., Tikhanovich I. Protein arginine methyl transferase 1- and Jumonji C domain-containing protein 6-dependent arginine methylation regulate hepatocyte nuclear factor 4 alpha expression and hepatocyte proliferation in mice. Hepatology. 2018;67:1109–1126. doi: 10.1002/hep.29587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao J., O'Neil M., Schonfeld M., Komatz A., Weinman S.A., Tikhanovich I. Hepatocellular protein arginine methyltransferase 1 suppresses alcohol-induced hepatocellular carcinoma formation by inhibition of inducible nitric oxide synthase. Hepatology Commun. 2020;4:790–808. doi: 10.1002/hep4.1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Diepenbruck M., Christofori G. Epithelial-mesenchymal transition (EMT) and metastasis: yes, no, maybe? Curr. Opin. Cell Biol. 2016;43:7–13. doi: 10.1016/j.ceb.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 55.Wei H., Liu Y., Min J., Zhang Y., Wang J., Zhou M., Xiong E., Yu G., Zhou H., He J., Zeng J., Gong A., Xu M. Protein arginine methyltransferase 1 promotes epithelial-mesenchymal transition via TGF-beta1/Smad pathway in hepatic carcinoma cells. Neoplasma. 2019;66:918–929. doi: 10.4149/neo_2018_181226N999. [DOI] [PubMed] [Google Scholar]
- 56.Yang H., Zhao X., Zhao L., Liu L., Li J., Jia W., Liu J., Huang G. PRMT5 competitively binds to CDK4 to promote G1-S transition upon glucose induction in hepatocellular carcinoma. Oncotarget. 2016;7:72131–72147. doi: 10.18632/oncotarget.12351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhu K., Peng Y., Hu J., Zhan H., Yang L., Gao Q., Jia H., Luo R., Dai Z., Tang Z., Fan J., Zhou J. Metadherin-PRMT5 complex enhances the metastasis of hepatocellular carcinoma through the WNT-beta-catenin signaling pathway. Carcinogenesis. 2020;41:130–138. doi: 10.1093/carcin/bgz065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Prieto J., Melero I., Sangro B. Immunological landscape and immunotherapy of hepatocellular carcinoma. Nature Rev. Gastroenterol. Hepatol. 2015;12:681–700. doi: 10.1038/nrgastro.2015.173. [DOI] [PubMed] [Google Scholar]
- 59.Greten T.F., Duffy A.G., Korangy F. Hepatocellular carcinoma from an immunologic perspective. Clin. Cancer Res. 2013;19:6678–6685. doi: 10.1158/1078-0432.CCR-13-1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Parry R.V., Ward S.G. Protein arginine methylation: a new handle on T lymphocytes? Trends Immunol. 2010;31:164–169. doi: 10.1016/j.it.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 61.Mowen K.A., David M. Unconventional post-translational modifications in immunological signaling. Nat. Immunol. 2014;15:512–520. doi: 10.1038/ni.2873. [DOI] [PubMed] [Google Scholar]
- 62.Infantino S., Light A., O'Donnell K., Bryant V., Avery D.T., Elliott M., Tangye S.G., Belz G., Mackay F., Richard S., Tarlinton D. Arginine methylation catalyzed by PRMT1 is required for B cell activation and differentiation. Nat. Commun. 2017;8:891. doi: 10.1038/s41467-017-01009-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ma D., Yang M., Wang Q., Sun C., Shi H., Jing W., Bi Y., Shen X., Ma X., Qin Z., Lin Y., Zhu L., Zhao Y., Cheng Y., Han L. Arginine methyltransferase PRMT5 negatively regulates cGAS-mediated antiviral immune response. Sci. Adv. 2021;7 doi: 10.1126/sciadv.abc1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhao J., O'Neil M., Vittal A., Weinman S.A., Tikhanovich I. PRMT1-dependent macrophage IL-6 production is required for alcohol-induced HCC progression. Gene Expr. 2019;19:137–150. doi: 10.3727/105221618X15372014086197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schonfeld M., Zhao J., Komatz A., Weinman S.A., Tikhanovich I. The polymorphism rs975484 in the protein arginine methyltransferase 1 gene modulates expression of immune checkpoint genes in hepatocellular carcinoma. J. Biol. Chem. 2020;295:7126–7137. doi: 10.1074/jbc.RA120.013401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Luo Y., Gao Y., Liu W., Yang Y., Jiang J., Wang Y., Tang W., Yang S., Sun L., Cai J., Guo X., Takahashi S., Krausz K.W., Qu A., Chen L., Xie C., Gonzalez F.J. MYC-protein arginine methyltransferase 5 axis defines the tumorigenesis and immune response in hepatocellular carcinoma. Hepatology. 2021 doi: 10.1002/hep.31864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Martinez-Outschoorn U.E., Peiris-Pages M., Pestell R.G., Sotgia F., Lisanti M.P. Cancer metabolism: a therapeutic perspective, nature reviews. Clin. Oncol. 2017;14:11–31. doi: 10.1038/nrclinonc.2016.60. [DOI] [PubMed] [Google Scholar]
- 68.Gebhardt R. Metabolic zonation of the liver: regulation and implications for liver function. Pharmacol. Ther. 1992;53:275–354. doi: 10.1016/0163-7258(92)90055-5. [DOI] [PubMed] [Google Scholar]
- 69.De Matteis S., Ragusa A., Marisi G., De Domenico S., Casadei Gardini A., Bonafe M., Giudetti A.M. Aberrant metabolism in hepatocellular carcinoma provides diagnostic and therapeutic opportunities. Oxidative Med. Cellular Longevity. 2018;2018 doi: 10.1155/2018/7512159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mavrakis K.J., McDonald E.R., 3rd, Schlabach M.R., Billy E., Hoffman G.R., deWeck A., Ruddy D.A., Venkatesan K., Yu J., McAllister G., Stump M., deBeaumont R., Ho S., Yue Y., Liu Y., Yan-Neale Y., Yang G., Lin F., Yin H., Gao H., Kipp D.R., Zhao S., McNamara J.T., Sprague E.R., Zheng B., Lin Y., Cho Y.S., Gu J., Crawford K., Ciccone D., Vitari A.C., Lai A., Capka V., Hurov K., Porter J.A., Tallarico J., Mickanin C., Lees E., Pagliarini R., Keen N., Schmelzle T., Hofmann F., Stegmeier F., Sellers W.R. Disordered methionine metabolism in MTAP/CDKN2A-deleted cancers leads to dependence on PRMT5. Science. 2016;351:1208–1213. doi: 10.1126/science.aad5944. [DOI] [PubMed] [Google Scholar]
- 71.Qin Y., Hu Q., Xu J., Ji S., Dai W., Liu W., Xu W., Sun Q., Zhang Z., Ni Q., Zhang B., Yu X., Xu X. PRMT5 enhances tumorigenicity and glycolysis in pancreatic cancer via the FBW7/cMyc axis. Cell Commun. Signaling: CCS. 2019;17:30. doi: 10.1186/s12964-019-0344-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cao L., Wu G., Zhu J., Tan Z., Shi D., Wu X., Tang M., Li Z., Hu Y., Zhang S., Yu R., Mo S., Wu J., Song E., Li M., Song L., Li J. Genotoxic stress-triggered beta-catenin/JDP2/PRMT5 complex facilitates reestablishing glutathione homeostasis. Nat. Commun. 2019;10:3761. doi: 10.1038/s41467-019-11696-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu L., Zhao X., Zhao L., Li J., Yang H., Zhu Z., Liu J., Huang G. Arginine Methylation of SREBP1a via PRMT5 promotes De novo lipogenesis and tumor growth. Cancer Res. 2016;76:1260–1272. doi: 10.1158/0008-5472.CAN-15-1766. [DOI] [PubMed] [Google Scholar]
- 74.Wong T.L., Ng K.Y., Tan K.V., Chan L.H., Zhou L., Che N., Hoo R.L.C., Lee T.K., Richard S., Lo C.M., Man K., Khong P.L., Ma S. CRAF Methylation by PRMT6 regulates aerobic glycolysis-driven hepatocarcinogenesis via ERK-dependent PKM2 nuclear relocalization and activation. Hepatology. 2020;71:1279–1296. doi: 10.1002/hep.30923. [DOI] [PubMed] [Google Scholar]
- 75.Che N., Ng K.Y., Wong T.L., Tong M., Kau P.W., Chan L.H., Lee T.K., Huen M.S., Yun J.P., Ma S. PRMT6 deficiency induces autophagy in hostile microenvironments of hepatocellular carcinoma tumors by regulating BAG5-associated HSC70 stability. Cancer Lett. 2021;501:247–262. doi: 10.1016/j.canlet.2020.11.002. [DOI] [PubMed] [Google Scholar]
- 76.Higashimoto K., Kuhn P., Desai D., Cheng X., Xu W. Phosphorylation-mediated inactivation of coactivator-associated arginine methyltransferase 1. PNAS. 2007;104:12318–12323. doi: 10.1073/pnas.0610792104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Feng Q., He B., Jung S.Y., Song Y., Qin J., Tsai S.Y., Tsai M.J., O'Malley B.W. Biochemical control of CARM1 enzymatic activity by phosphorylation. J. Biol. Chem. 2009;284:36167–36174. doi: 10.1074/jbc.M109.065524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lai Y., Li J., Li X., Zou C. Lipopolysaccharide modulates p300 and Sirt1 to promote PRMT1 stability via an SCF(Fbxl17)-recognized acetyldegron. J. Cell Sci. 2017;130:3578–3587. doi: 10.1242/jcs.206904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kuhn P., Chumanov R., Wang Y., Ge Y., Burgess R.R., Xu W. Automethylation of CARM1 allows coupling of transcription and mRNA splicing. Nucleic Acids Res. 2011;39:2717–2726. doi: 10.1093/nar/gkq1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Singhroy D.N., Mesplede T., Sabbah A., Quashie P.K., Falgueyret J.P., Wainberg M.A. Automethylation of protein arginine methyltransferase 6 (PRMT6) regulates its stability and its anti-HIV-1 activity. Retrovirology. 2013;10:73. doi: 10.1186/1742-4690-10-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dillon M.B., Rust H.L., Thompson P.R., Mowen K.A. Automethylation of protein arginine methyltransferase 8 (PRMT8) regulates activity by impeding S-adenosylmethionine sensitivity. J. Biol. Chem. 2013;288:27872–27880. doi: 10.1074/jbc.M113.491092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Duong F.H., Christen V., Berke J.M., Penna S.H., Moradpour D., Heim M.H. Upregulation of protein phosphatase 2Ac by hepatitis C virus modulates NS3 helicase activity through inhibition of protein arginine methyltransferase 1. J. Virol. 2005;79:15342–15350. doi: 10.1128/JVI.79.24.15342-15350.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wei X., Li X., Yan W., Zhang X., Sun Y., Zhang F. SKP2 promotes hepatocellular carcinoma progression through nuclear AMPK-SKP2-CARM1 signaling transcriptionally regulating nutrient-deprived autophagy induction. Cellular Physiol. Biochem. 2018;47:2484–2497. doi: 10.1159/000491622. [DOI] [PubMed] [Google Scholar]
- 84.Shin H.J., Kim H., Oh S., Lee J.G., Kee M., Ko H.J., Kweon M.N., Won K.J., Baek S.H. AMPK-SKP2-CARM1 signalling cascade in transcriptional regulation of autophagy. Nature. 2016;534:553–557. doi: 10.1038/nature18014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sipos A., Ivan J., Becsi B., Darula Z., Tamas I., Horvath D., Medzihradszky K.F., Erdodi F., Lontay B. Myosin phosphatase and RhoA-activated kinase modulate arginine methylation by the regulation of protein arginine methyltransferase 5 in hepatocellular carcinoma cells. Sci. Rep. 2017;7:40590. doi: 10.1038/srep40590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li B., Liu L., Li X., Wu L. miR-503 suppresses metastasis of hepatocellular carcinoma cell by targeting PRMT1. Biochem. Biophys. Res. Commun. 2015;464:982–987. doi: 10.1016/j.bbrc.2015.06.169. [DOI] [PubMed] [Google Scholar]
- 87.Wang L., Li X., Zhang W., Yang Y., Meng Q., Wang C., Xin X., Jiang X., Song S., Lu Y., Pu H., Gui X., Li T., Xu J., Li J., Jia S., Lu D. miR24-2 Promotes Malignant Progression of Human Liver Cancer Stem Cells by Enhancing Tyrosine Kinase Src Epigenetically. Mol. Ther. 2020;28:572–586. doi: 10.1016/j.ymthe.2019.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li Z., Zhang J., Liu X., Li S., Wang Q., Di C., Hu Z., Yu T., Ding J., Li J., Yao M., Fan J., Huang S., Gao Q., Zhao Y., He X. The LINC01138 drives malignancies via activating arginine methyltransferase 5 in hepatocellular carcinoma. Nat. Commun. 2018;9:1572. doi: 10.1038/s41467-018-04006-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dang C.V. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jones P.A., Issa J.P., Baylin S. Targeting the cancer epigenome for therapy. Nature reviews. Genetics. 2016;17:630–641. doi: 10.1038/nrg.2016.93. [DOI] [PubMed] [Google Scholar]
- 91.Dawson M.A., Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 92.Cheng D., Yadav N., King R.W., Swanson M.S., Weinstein E.J., Bedford M.T. Small molecule regulators of protein arginine methyltransferases. J. Biol. Chem. 2004;279:23892–23899. doi: 10.1074/jbc.M401853200. [DOI] [PubMed] [Google Scholar]
- 93.Guccione E., Richard S. The regulation, functions and clinical relevance of arginine methylation. Nature Rev. Mol. Cell Biol. 2019;20:642–657. doi: 10.1038/s41580-019-0155-x. [DOI] [PubMed] [Google Scholar]
- 94.Wang S.M., Dowhan D.H., Muscat G.E.O. Epigenetic arginine methylation in breast cancer: emerging therapeutic strategies. J. Mol. Endocrinol. 2019;62:R223–R237. doi: 10.1530/JME-18-0224. [DOI] [PubMed] [Google Scholar]
- 95.Nakayama K., Szewczyk M.M., Dela Sena C., Wu H., Dong A., Zeng H., Li F., de Freitas R.F., Eram M.S., Schapira M., Baba Y., Kunitomo M., Cary D.R., Tawada M., Ohashi A., Imaeda Y., Saikatendu K.S., Grimshaw C.E., Vedadi M., Arrowsmith C.H., Barsyte-Lovejoy D., Kiba A., Tomita D., Brown P.J. TP-064, a potent and selective small molecule inhibitor of PRMT4 for multiple myeloma. Oncotarget. 2018;9:18480–18493. doi: 10.18632/oncotarget.24883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang J., Wang C., Xu P., Li X., Lu Y., Jin D., Yin X., Jiang H., Huang J., Xiong H., Ye F., Jin J., Chen Y., Xie Y., Chen Z., Ding H., Zhang H., Liu R., Jiang H., Chen K., Yao Z., Luo C., Huang Y., Zhang Y., Zhang J. PRMT1 is a novel molecular therapeutic target for clear cell renal cell carcinoma. Theranostics. 2021;11:5387–5403. doi: 10.7150/thno.42345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fedoriw A., Rajapurkar S.R., O'Brien S., Gerhart S.V., Mitchell L.H., Adams N.D., Rioux N., Lingaraj T., Ribich S.A., Pappalardi M.B., Shah N., Laraio J., Liu Y., Butticello M., Carpenter C.L., Creasy C., Korenchuk S., McCabe M.T., McHugh C.F., Nagarajan R., Wagner C., Zappacosta F., Annan R., Concha N.O., Thomas R.A., Hart T.K., Smith J.J., Copeland R.A., Moyer M.P., Campbell J., Stickland K., Mills J., Jacques-O'Hagan S., Allain C., Johnston D., Raimondi A., Scott M.Porter, Waters N., Swinger K., Boriack-Sjodin A., Riera T., Shapiro G., Chesworth R., Prinjha R.K., Kruger R.G., Barbash O., Mohammad H.P. Anti-tumor Activity of the Type I PRMT Inhibitor, GSK3368715, Synergizes with PRMT5 Inhibition through MTAP Loss. Cancer Cell. 2019;36:100–114. doi: 10.1016/j.ccell.2019.05.014. e125. [DOI] [PubMed] [Google Scholar]
- 98.Tao H., Yan X., Zhu K., Zhang H. Discovery of Novel PRMT5 Inhibitors by Virtual Screening and Biological Evaluations. Chem. Pharm. Bull. 2019;67:382–388. doi: 10.1248/cpb.c18-00980. [DOI] [PubMed] [Google Scholar]
- 99.Zheng B.N., Ding C.H., Chen S.J., Zhu K., Shao J., Feng J., Xu W.P., Cai L.Y., Zhu C.P., Duan W., Ding J., Zhang X., Luo C., Xie W.F. Targeting PRMT5 Activity Inhibits the Malignancy of Hepatocellular Carcinoma by Promoting the Transcription of HNF4alpha. Theranostics. 2019;9:2606–2617. doi: 10.7150/thno.32344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jeong J.M., Ju C. The protective function of PRMT1 in alcohol-induced hepatocellular carcinoma. Hepatology Commun. 2020;4:787–789. doi: 10.1002/hep4.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.