Abstract
Background
Esophageal squamous cell carcinoma (ESCC) remains one of the deadly cancer types. Comprehensively dissecting the molecular characterization and the heterogeneity of ESCC paves the way for developing more promising therapeutics.
Methods
Expression profiles of multiple ESCC datasets were integrated. ATAC-seq and RNA-seq were combined to reveal the chromatin accessibility features. A prognosis-related subtype classifier (PrSC) was constructed, and its association with the tumor microenvironment (TME) and immunotherapy was assessed. The key gene signature was validated in clinical samples. Based on the TME heterogeneity of ESCC patients, potential subtype-specific therapeutic agents were screened.
Findings
The common differentially expressed genes (cDEGs) in ESCC were identified. Up-regulated genes (HEATR1, TIMELESS, DTL, GINS1, RUVBL1, and ECT2) were found highly important in ESCC cell survival. The expression alterations of PRIM2, HPGD, NELL2, and TFAP2B were associated with chromatin accessibility changes. PrSC was a robust scoring tool that was not only associated with the prognosis of ESCC patients, but also could reflect the TME heterogeneity. TNS1high fibroblasts were associated with immune exclusion. TG-101348 and Vinorelbine were identified as potential subtype-specific therapeutic agents. Besides, the application of PrSC into two immunotherapy cohorts indicated its potential value in assessing treatment response to immunotherapy.
Interpretation
Our study depicted the multi-dimensional characterization of ESCC, established a robust scoring tool for the prognosis assessment, highlighted the role of TNS1high fibroblasts in TME, and identified potential drugs for clinical use.
Funding
A full list of funding bodies that contributed to this study can be found in the Acknowledgements section.
Keywords: Esophageal squamous cell carcinoma, RNA-seq, ATAC-seq, Molecular classification, Tumor stroma, Drug repurposing
Research in context.
Evidence before this study
Esophageal squamous cell carcinoma (ESCC) is one of the deadly cancer types worldwide. The molecular changes in ESCC as well as the heterogeneity among different ESCC patients define the prognosis and treatment response. We searched PubMed for research articles containing the terms "esophageal squamous cell carcinoma AND integrative analysis" without language or date restrictions. Several studies analyzing the molecular changes of ESCC were found. However, the samples enrolled in these studies were relatively limited, while integrating multiple ESCC datasets would further help identify the key changes in ESCC. Besides, several gene signatures for the prognosis assessment of ESCC have been reported, while the association between previous gene signatures and tumor microenvironment was less emphasized, and meanwhile, clinical validation of the gene signatures is lacking. We also searched PubMed for research articles containing the terms "esophageal squamous cell carcinoma AND drug screening", and found no studies that screened potential drugs for ESCC based on tumor microenvironment heterogeneity. Moreover, no web application that could help researchers mine the publicly available data of ESCC has been established.
Added value of this study
Through the integration of the expression profiling of ESCC patients, we identified the shared molecular changes in ESCC. By combing the ATAC-seq data with the RNA-seq data of ESCC patients, we revealed the potential transcriptional regulation characterization in ESCC. Besides, we established a survival-related subtype classifier called PrSC, which was able to capture the stromal heterogeneity in ESCC and predict the clinical response to immunotherapy. We identified a group of TNS1high fibroblasts in the stroma, which was associated with immune exclusion phenotype and poor prognosis of ESCC patients. In addition, based on the molecular heterogeneity of ESCC patients, we identified some subtype-specific therapeutic agents. We also built a web application (ESCCEXPRESS, http://www.bioinfo-zs.com/esccexpress/)
Implications of all the available evidence
The core molecular changes in ESCC were depicted. Stromal heterogeneity was significantly associated with the prognosis of ESCC patients. TNS1high fibroblasts were associated with immune exclusion, disease recurrence, and lymph node metastasis. TG-101348 and Vinorelbine were potential subtype-specific therapeutic agents.
Alt-text: Unlabelled box
1. Introduction
Esophageal cancer is one of the most commonly diagnosed cancer types and the 6th leading cause of cancer-related death worldwide [1]. Histologically, esophageal cancer can be categorized into two major subtypes: esophageal adenocarcinoma (EAC) and esophageal squamous cell carcinoma (ESCC), showing different epidemiology and geographic distribution [2,3]. EAC is more prevalent in Western countries, whereas ESCC is more common in East Asian countries, such as China, Indian, and Iran [4]. In China, more than 90% of esophageal cancer cases were ESCC [5,6]. ESCC is a highly aggressive form of squamous cell carcinoma with a 5-year survival rate of less than 20% [1]. Currently, surgical resection, radiotherapy, and chemotherapy are still the primary treatment strategies against ESCC, but the prognosis of patients remains poor due to the high recurrence rate and early metastasis.
In recent decades, although there have been great advances in cancer treatment strategies, such as the development and application of immunotherapy [7,8], limited progress has been made in developing therapeutics against ESCC. Although results from recent clinical trials of advanced ESCC patients after failure with first-line chemotherapy showed that there was a significant improvement in the overall survival (OS) in using Pembrolizumab and Nivolumab compared with chemotherapy, there were still a number of ESCC patients showing little or no reaction to immunotherapy [9,10]. Therefore, A comprehensive understanding of the common core molecular features of ESCC is indispensable for developing more promising therapies, and on the basis of that, identification of subclass patients who may benefit more from different therapeutic agents based on intrinsic heterogeneity could further facilitate the development of personalized treatment strategies.
High-throughput detection platform has already become one of the important tools to explore the biological alterations during carcinogenesis and tumor progression [11]. With the continuous accumulation of data, researchers can now explore the key characteristics of diseases in a larger cohort based on appropriate data integration strategy and obtain more convincing results supported by multiple pieces of evidence [12,13]. Such strategies have been widely applied to study the molecular features of various cancer types such as lung adenocarcinoma, hepatocellular carcinoma, and gastric cancer [14], [15], [16]. However, compared with other common cancer types, such approaches were less applied to study the molecular features of ESCC. In the present study, we adopted an integrative strategy to systematically dissect the multi-dimensional features of ESCC. Through the integration of the expression profiling of ESCC patients from multiple cohorts, we identified the shared molecular features in ESCC. By combing the ATAC-seq data with the RNA-seq data of ESCC patients, we also revealed the potential transcriptional regulation characterization in ESCC. On the other hand, we established a survival-related subtype classifier called PrSC, which was able to capture the stromal heterogeneity in ESCC and predict the clinical response to immunotherapy. Meanwhile, using multiplex fluorescent immunohistochemistry (mfIHC), we identified a group of TNS1high fibroblasts in the stroma, which was associated with immune exclusion phenotype and poor prognosis of ESCC patients. In addition, based on the molecular heterogeneity of ESCC patients, we identified some subtype-specific therapeutic agents. Finally, we also built a web application (ESCCEXPRESS, http://www.bioinfo-zs.com/esccexpress/), which provides several key functions for users to browse and mine the data we used in the current study.
2. Methods
2.1. Ethics
Patients donating surgical tissue provided informed consent. All diagnoses were confirmed by histological review by qualified pathologists. This study was approved by the ethics committee on human research of Zhongshan Hospital, Fudan University (B2020-332R; B2020-412R), and conducted in accordance with the principles of the Declaration of Helsinki.
2.2. ESCC patient samples
Three pairs of ESCC tumor tissues and patient-matched adjacent non-cancerous tissues (> 3 cm apart from tumor edge) were collected at the Department of Thoracic Surgery, Zhongshan Hospital, Fudan University, China, in 2020. Collected specimens were divided into two parts, one for ATAC-seq and another for RNA-seq. The samples were stored at -80 °C until use. In addition, tissue microarrays from 241 ESCC patients who underwent radical esophagectomy at Zhongshan Hospital between 2008 and 2009 were used to verify the expression and spatial distribution of TNS1. Clinical-pathological data and follow-up information of these patients were collected as previously described, including age, gender, tumor size, tumor depth, lymph node metastasis, TNM stage, histological grade, disease free survival (DFS), and overall survival (OS) [17]. The DFS was defined as the interval from surgery to new tumor event, whether it was a local recurrence or distant metastasis, and OS was defined as the duration of survival after surgery.
2.3. ATAC-seq
Nuclei were purified from frozen ESCC samples based on the previously described protocol [18]. The quality of the harvested nuclei was assessed using trypan blue staining. Next, 50,000 nuclei were prepared for transposition reactions. The Nextera DNA Library Preparation Kit (Illumina, Cat#: FC-121-1030) was used to perform the transposition reactions according to manufacturer's instruction. Nuclei were pelleted and resuspended with transposase at 37 °C for 30 min. After transposition, DNA fragments were purified with the MinElute PCR Purification Kit (Qiagen, Cat#: 28004). Then, samples were amplified using NEBNext High-Fidelity PCR Master Mix (New England Biolabs, Cat#: M0541S). The amplified libraries were purified with the MinElute PCR Purification Kit (Qiagen, Cat#: 28004) and sequenced on Illumina Novaseq 6000 using PE150.
After obtaining the raw fastq data, the adapter sequences and low-quality reads were removed using Trimmomatic [19]. Then, the quality of sequencing data was assessed with FastQC (https://www.Bioinformaticsbabraham.ac.uk/projects/fastqc/). The clean data were aligned to human genome (hg38) with Burrows-Wheeler Alignment tool (BWA) [20]. Multiply mapped reads were removed using SAMtools [21]. The bam file generated by the uniquely mapped reads were used for peak calling using MACS2 with q-value < 0.05 [22]. DeepTools was used for results visualization [23]. Differential peak analysis was conducted using DESeq2 R package [24]. Genomic features of peaks were annotated using ChIPseeker R package [25]. Tracks were visualized using the IGV (https://igv.org).
2.4. RNA-seq
Total RNA was extracted using Trizol (Thermo Fisher Scientific, Cat#: 15596026). RNA degradation and contamination were assessed on 1% agarose gels. NanoPhotometer spectrophotometer (IMPLEN) was used to check RNA purity. RNA concentration was measured using Qubit RNA Assay Kit (Life Technologies, Cat#: Q32855). RNA integrity was evaluated using the Agilent Bioanalyzer 2100 system (Agilent). Sequencing libraries were generated using NEBNext UltraTM RNA Library Prep Kit for Illumina (NEB, Cat#: E7775) following manufacturer's protocol. The library fragments were then purified using AMPure XP system (Beckman Coulter). Library quality was assessed on the Agilent 2100 Bioanalyzer. The prepared libraries were sequenced using Illumina Novaseq6000 platform and 150 bp paired-end reads were generated.
Raw data were also processed using Trimmomatic [19]. Then, the quality of data was assessed using FastQC. Reads were aligned to hg38 using the STAR [26]. Read counts were generated using HTSeq [27]. Differential gene expression analysis was conducted using DESeq2 R package [24].
2.5. Chromatin accessibility and gene expression correlation analysis
In order to assess the global relationship between ATAC-seq signals and transcription levels of the corresponding genes, genes were first categorized into high and low abundance groups based on their mRNA levels using the median value. Then, we extracted TSSs information of genes in high and low abundance groups respectively and compared the ATAC-seq signals 10 kb up- and down-stream surrounding these TSSs using deepTools [23].
2.6. Luciferase reporter assay
PRIM2 promoter (chr6: 57312805-57314804) was subcloned into the Kpn I and XhoI sites of pGL3-Basica vector (Promega, Cat#: E1751) to generate the PRIM2-P luciferase reporter. The peak26955 region (chr6: 57274167-57274877) was subcloned into the PRIM2-P luciferase reporter via SalI and BamHI sites to generate the luciferase reporter PRIM2-P-E. Transfection was conducted using FuGENE Transfection Reagent (Promega, Cat#: E2311). Luciferase activity was measured using the Steady-Glo Luciferase Assay system (Promega, Cat#: E2550) according to the manufacturer's instructions.
2.7. Multiplex fluorescent immunohistochemistry
Multiplex fluorescent immunohistochemistry (mfIHC) staining of TNS1 (Protein Tech, Cat#: 20054-1-AP), CD8 (Servicebio, Cat#: GB13068-2), and α-SMA (Servicebio, Cat#: GB111364) was performed. Slides were first deparaffinized and rehydrated, followed by antigen retrieval using sodium citrate retrieval solution (Servicebio, pH 6.0, Cat#: G1206). Next, endogenous peroxidase and nonspecific binding sites were blocked using 3% H2O2 (SCRC) and 3% BSA (Servicebio, Cat#: G5001) respectively. After that, first primary antibodies (CD8) and corresponding secondary antibodies marked with HRP were applied, followed by the adding of CY3-TSA solution (Servicebio, Cat#: G1223). Then, slides were microwave treated to remove the primary antibodies and secondary antibodies combined with tissue. The same process was conducted for the next two antibodies α-SMA (FITC, Cat#: G1222) and TNS1 (CY5, Cat#: G1224). After sequential reactions, slides were stained with DAPI (Servicebio, Cat#: G1012) and scanned using Pannoramic MIDI (3DHISTECH). CY5 was originally red, in order to distinguish it from CY3, we set it to pink. After mfIHC, we assessed the staining quality of each section and excluded those with relatively poor qualities. Finally, 222 patients (222 patients with OS information and 201 patients with DFS information) were included for subsequent statistical analysis.
The proportion of α-SMA+TNS1+ fibroblasts in the stroma was semi-quantified as follows: 0 (0% α-SMA+TNS1+ cells present in the stroma), 1 (1–10% of α-SMA+ cells in the stroma were α-SMA+TNS1+ positive), 2 (11–50% of α-SMA+ cells in the stroma were α-SMA+TNS1+ positive) or 3 (>50% of α-SMA+ cells in the stroma were α-SMA+TNS1+ positive). The staining intensity of TNS1 in α-SMA+TNS1+ cells was scored as follows: 0 (negative), 1 (weak), 2 (intermediate), or 3 (strong). The total score was calculated as the sum of the above two factors. Patients were classified into negative (0), weak (1-2), moderate (3-4) and strong (5-6) staining groups, respectively, and moderate and strong groups were defined as the TNS1high group, while the negative and weak groups were defined as the TNS1low group.
CD8+ T cell infiltration status was determined as follows: immune desert (the proportion of CD8+ T cells was less than 1% of all nucleated cells in a 5x magnification in a section), immune inflamed (not immune desert, at least 10% of CD8+ T cells infiltrated into the tumor mass), immune exclusion (not immune desert, less than 10% of CD8+ T cells penetrated into the parenchyma).
2.8. Public data collection and preprocessing
Gene Expression Omnibus (GEO) data repository was thoroughly queried for all eligible ESCC expression profiles, and a total of 7 datasets (GSE17351, GSE20347, GSE23400, GSE38129, GSE45670, GSE53625, and GSE77861) from different independent studies of ESCC were included. All datasets, except for GSE53625, were based on the Affymetrix platform, we therefore uniformly processed the raw CEL data of these datasets using the RMA method for background correction and normalization [28]. Besides, these 6 datasets were integrated into a new GEO ESCC meta cohort (Supplementary Figure 3) after batch effect removal using sva R package [29]. As for GSE53625, which was based on Agilent platform, we re-annotated the probe sets by mapping all sequences provided in GPL18109 annotation file to human genome (hg38) using SeqMap [30]. Probes that were uniquely mapped were kept, meanwhile, probes that were mapped to non-protein-coding transcripts were removed. All probes were converted to gene symbols, and for genes that have multiple probe-set signals, we averaged the values to obtain a single expression value. The clinical information of above datasets was downloaded using GEOquery R package [31], and among them, only GSE53625 contained detailed survival information.
For TCGA data, somatic mutation information was downloaded using TCGAbiolinks R package [32]. Fisher's exact test was applied to identify different mutated genes between S1 and S2 subtype patients. For copy number variation (CNV) analysis, GISTIC 2.0 was used to identify significantly amplified or deleted genomes [33]. DNA methylation data were collected from our previously developed web application, SMART App (http://bioinfo-zs.com/smartapp/), and the methylation burden of each sample was defined as the median value of all CpGs associated with this sample [34]. ATAC-seq data of TCGA patients were downloaded from GDC data portal (https://gdc.cancer.gov/about-data/publications/ATACseq-AWG) [35]. As for TCGA-ESCC gene expression data, Toil-recomputed transcripts per million data from Xena public data hubs were used [36]. The EAC data were removed based on corresponding pathological information and we ended up obtaining 92 tumor samples and 2 normal samples. Considering that there were only 2 normal samples in TCGA cohort, differential expression analysis comparing tumor vs. normal samples was not performed in TCGA data. Survival information of these patients was collected based on the previously published study [37]. ChIP-seq profiles of H3K27ac of ESCC cell lines were downloaded from GSE76859 [38].
2.9. Functional annotation of cDEGs
GO term enrichment analysis was conducted using clusterProfiler R package [39]. The impacts of cDEGs on various pathways were evaluated using SPIA algorithm [40]. In brief, the built-in bicor function of WGCNA R package was first used to estimate the transcriptomics-based biweight midcorrelations between each cDEG and all other genes [41]. Then, genes were ranked based on their absolute values, and the top 500 genes were fitted into SPIA algorithm to identify the pathways significantly impacted. This process was performed on both GSE53625 and GEO ESCC meta cohorts respectively, and pathways supported by both datasets were considered significant.
2.10. Construction of PrSC and transcriptome similarity analysis
The flowchart illustrating the construction of PrSC is shown in Supplementary Table 7. Specifically, Cox regression analysis was first performed to screen survival-related genes in GSE53625 and TCGA ESCC cohorts, respectively. Then, overlapped genes were fitted into the Least Absolute Shrinkage and Selection Operator (LASSO) Cox analysis (10-fold cross-validation) to further reduce dimensionality and select the most representative marker genes (glmnet R package). We chose the λ value which corresponded to the smallest partial likelihood deviance [42]. Based on the λ value, a total of 14 genes were kept, and each gene was automatically assigned with a coefficient, then the PrSC score was generated based on the formula we previously introduced [43]:
where Num represents the number of genes, is the expression level of genei, and is the LASSO coefficient of genei. The maximally selected log-rank statistics was used to determine the optimal cut-off value for classifying high- and low-score groups [44]. Minprop parameter was set to 30% to avoid assigning too few patients into a given group. Subsequently, subclass mapping (SubMap) analysis was performed to evaluate whether subtypes identified in different cohorts exhibited similar transcriptional features [45].
2.11. Gene set variation analysis and tumor microenvironment components estimation
To quantify the biological function differences between S1 and S2 subtype patients, gene set variation analysis (GSVA) was performed using GSVA R package [46]. The pathway information was downloaded from the MSigDB database (https://www.gsea-msigdb.org/). Then, differential analysis was conducted to determine the significantly enriched pathways in each subtype. Tumor microenvironment components were quantified using MCPcounter [47]. Besides, stromal and immune scores were inferred using ESTIMATE method [48].
2.12. Single-cell RNA sequencing data analysis
Single-cell RNA sequencing data of mouse esophageal cancer model were downloaded from GSA database (CRA002118) [49]. Seurat (v4.0.0) standardized workflow was performed [50]. Tns1high and Tns1low fibroblast groups were determined based on the median expression value of Tns1. The FindMarkers function in Seurat was used to identify differentially expressed genes (adjusted p value < 0.05). Top 10 genes were used as marker genes and the cell abundance was calculated based on the mean value of the marker genes [51]. Biological processes were inferred using GSVA method [46]. Cell-cell communications were inferred using CellChat [52]. Cells were annotated based on canonical markers [49].
2.13. Drug response data collection and processing
Drug response data of human cancer cell lines were collected from three independent datasets, including Cancer Therapeutics Response Portal (CTRPv.2.0, https://portals.broadinstitute.org/ctrp), Genomics of Drug Sensitivity in Cancer (GDSC1&2, https://www.cancerrxgene.org/), and PRISM (19Q4, https://depmap.org/portal/prism/) [53], [54], [55]. The area under the dose-response curve (AUC) values were used to assess the drug sensitivity, with lower values indicating higher sensitivities. Drugs with NA values in more than 20% of cell lines were discarded, then, we used k-nearest neighbors (KNN) method to impute the remaining missing values. Finally, 632 cancer cell lines and 408 drugs were kept in CTRP, 741 cancer cell lines and 282 drugs were kept in GDSC, and 440 cancer cell lines and 1291 drugs were kept in PRISM. The corresponding expression data of cancer cell lines from CTRP and PRISM datasets were obtained from Cancer Cell Line Encyclopedia (CCLE) [56], and the expression data of cancer cell lines from GDSC were collected from GDSC1000 resource (https://www.cancerrxgene.org/gdsc1000/).
2.14. Drug sensitivity estimation in ESCC patients
The ridge regression model wrapped in pRRophetic R package was utilized to estimate the drug sensitivity of ESCC patients [57]. In brief, this model was trained on expression data and drug response data of cancer cell lines, therefore allowing the prediction of drug sensitivity using the patients’ gene expression data. This method is shown better than other models and can reflect the actual clinical drug response in patients [58]. This model required three input files, the drug response data of cancer cell lines, the expression profiles of cancer cell lines, as well as the expression profiles of ESCC patients, which were preprocessed by filtering out genes with low variability (MAD < 0.5). After obtaining the predicted drug response data of ESCC patients, differential analysis was conducted between two subtypes.
2.15. ESCC cell line and cell viability assay
ESCC cell lines KYSE150 (RRID: CVCL_1348) and TE1 (RRID: CVCL_1759) were purchased from Shanghai Fuheng Biological Technology Co. Ltd. (Shanghai, China). KYSE410 (RRID: CVCL_1352) was purchased from Wuhan Procell Life Science and Technology Co., Ltd. (Wuhan, China). TE11 (RRID: CVCL_1761) and KYSE180 (RRID: CVCL_1349) were purchased from Shanghai Binsui Bio-Technology Co. Ltd. (Shanghai, China). KYSE70 (RRID: CVCL_1356) was provided by Shanghai Cancer Institute, State Key Laboratory of Oncogenes and Related Genes (Shanghai, China). STR authentication of cell lines was performed by vendors. Cells were maintained in Dulbecco's modified Eagle's medium (DMEM, Gibco, Cat#: 11995500BT) containing 10% fetal bovine serum (FBS, BIOIND, Cat#: 04-001-1ACS) at 37 °C in 5% CO2. The classification of these cell lines was conducted using nearest template prediction (NTP) method in Gene Pattern web application (https://cloud.genepattern.org/) [59]. TG-101348 (10 mM, Cat#: S2736) was purchased from Selleck Chemicals. Vinorelbine (10 mM, Cat#: HY-12053A) and ZM-447439 (10 mM, Cat#: HY-10128) were purchased from MedChemExpress. For cell viability assay, 6000 cells were seeded in 96-well plates and drugs were added after 24 h. Cells were treated with drugs for 48 h. Afterwards, cell viability was determined using CellTiter-Glo Luminescent Cell Viability Assay (Promega, Cat#: G7572). Luminescence was detected using Wallac Victor2 1420 MultiLabel Counter (PerkinElmer). Experiments were repeated at least three times.
2.16. Statistics
Differential gene expression analysis of microarray data was performed using limma R package [60]. Log-rank test was used to describe the survival difference. Cox regression analysis was performed using survival R package. The Wilcoxon rank sum test was applied to determine the statistical difference of the two groups. For comparisons of more than two groups, the Kruskal-Wallis test was applied. The correlation analysis was conducted using the Spearman method. For cell viability assay, the statistical significance of differences was determined by two-way ANOVA. All statistical tests were two-sided, and statistical significance was considered when p < 0.05. The website was built based on R Shiny framework. R (version 3.6.1) was used for all statistical analyses.
2.17. Role of the funding source
The funding bodies were not involved in the study design, implementation, the analysis, or the interpretation of data.
3. Results
3.1. Identification of common differentially expressed genes across different ESCC cohorts
The study design is shown in Fig. 1. Data integration is gradually becoming a preferred strategy to investigate the shared key features of diseases [51]. Although heterogeneity exists among different ESCC patients, revealing the common core characteristics in different ESCC patients is also vital for us to gain more insights into the molecular features of ESCC. Seven ESCC datasets representing different independent ESCC studies from the GEO data repository were first examined (Supplementary Table 1). Differentially expressed genes in each of the seven datasets were determined respectively (Supplementary Figure 1A). A total of 252 genes showed consistent expression alterations after the intersection, among which 178 genes were up-regulated and 74 genes were down-regulated. Next, the tumor and adjacent non-cancerous tissues of three ESCC patients who underwent surgical resection at Zhongshan hospital (Zs) were subjected to RNA-seq (Supplementary Figure 1A and Supplementary Table 2). Then, we integrated the differentially expressed genes identified in both the GEO data repository and patients at Zhongshan hospital to look for shared genes, and this identified a total of 112 common differentially expressed genes (cDEGs), with 69 genes up-regulated and 43 genes down-regulated (Supplementary Figure 1B and Supplementary Table 3). Chromosomal distribution of cDEGs demonstrated that chromosomes 1 and 3 containing the greatest number of dysregulated genes in ESCC. Interestingly, two genes (ELF4 and KIF4A) on the X chromosome were up-regulated, but not a single Y chromosome gene was captured (Supplementary Figure 2).
Fig. 1.
Study overview. Study strategies (Upper panel). 8 ESCC datasets were included to identify common differentially expressed genes (cDEGs). Functional enrichment analysis of cDEGs suggested changes in chromatin accessibility. ATAC-seq combined with RNA-seq revealed the chromatin accessibility features in ESCC and identified genes whose expression alterations were related to changes in chromatin accessibility. On the other hand, the investigation of the heterogeneity of ESCC revealed that the stromal difference is one of the important factors for the prognosis of ESCC patients. A group of TNS1high fibroblasts was associated with immune exclusion phenotype in ESCC. In addition, based on the molecular heterogeneity of ESCC patients, we identified some subtype-specific therapeutic agents. ESCCEXPRESS web application interface (Lower panel). A web application ESCCEXPRESS (http://www.bioinfo-zs.com/esccexpress/) was established to facilitate data mining. Users can check the expression alteration of the gene of interest across different ESCC datasets, explore whether the gene of interest is associated with the clinical outcomes of ESCC patients in different datasets, check the correlation between any two given genes, browse the baseline expression of the gene of interest in different ESCC cell lines, and check the importance of the gene in the survival of ESCC cell lines.
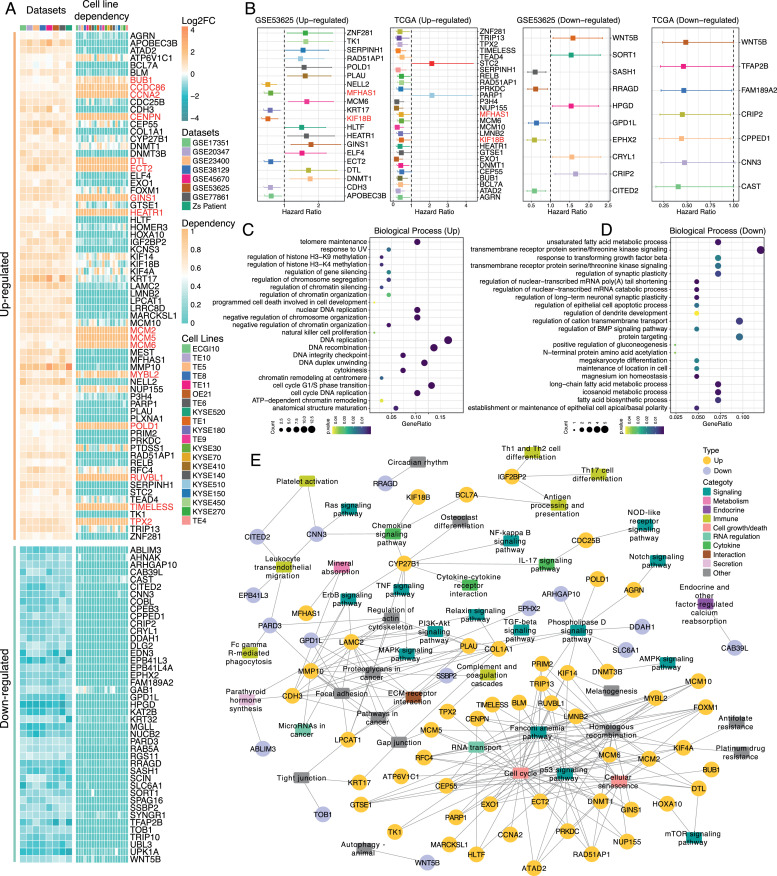
In order to further figure out whether these cDEGs were essential for the survival of ESCC cells, we next examined the dependency profiles of cDEGs across ESCC cell lines based on the genome-wide CRISPR-Cas9 loss-of-function data available from DepMap database (https://depmap.org/) [61]. Among cDEGs, genes that were highly important in ESCC cell survival were all up-regulated genes, including HEATR1, TIMELESS, DTL, GINS1, RUVBL1, and ECT2, while not a single down-regulated was essential in cell survival across ESCC cell lines (Fig. 2A). Next, we investigated the prognostic relevance of cDEGs in GSE53625 and TCGA cohorts (Fig. 2B) and found that MFHAS1 and KIF18B showed a consistent prognostic relevance in GSE53625 and TCGA cohorts, with higher expression associated with a better prognosis of ESCC patients.
Fig. 2.
Identification of cDEGs in ESCC and functional annotation.
A. Expression alterations of cDEGs across 8 ESCC cohorts and corresponding cell line dependency data across 20 ESCC cell lines. The heatmap on the left represented the Log2 fold-change of cDEGs. The heatmap on the right represented the cell line dependency value. A dependency value > 0.5 is considered significantly dependent. Genes that were highly associated with the cell survival were highlighted in red.
B. Survival analysis of cDEGs in GSE53625 and TCGA ESCC cohorts (Log-rank test, p < 0.05).
C. GO biological process enrichment analysis for genes that were up-regulated in cDEGs (p < 0.05).
D. GO biological process enrichment analysis for genes that were down-regulated in cDEGs (p < 0.05).
E. Pathways significantly associated with cDEGs (p < 0.05). Pathways were classified into different major categories. Yellow circles represent up-regulated genes, purple circles represent down-regulated genes.
To explore the potential biological functions of cDEGs in ESCC, GO term enrichment analysis was first performed. The up-regulated genes were mainly associated with DNA replication, histone modification, and chromatin regulation related biological processes, such as DNA recombination, DNA replication, histone H3−K9 methylation, and regulation of chromatin organization, whereas the down-regulated genes were predominantly enriched in metabolic related processes (Fig. 2C & D and Supplementary Figure 3A & B). Considering that GO term enrichment analysis is mainly based on known functions of input genes, and if the input genes are not well studied, the potential impacts of these genes on biological processes and pathways may not be fully reflected. Therefore, we here adopted SPIA algorithm [40], which took the expression features of input genes into consideration, to explore the impacts of cDEGs on various pathways in ESCC. Many cancer-related pathways were identified as highly correlated with cDEGs, including those that have functions in the immune system, signaling, cell growth/death, metabolism, endocrine, and secretion, cell interaction and RNA regulation (Fig. 2E and Supplementary Table 4). Consistent with GO enrichment analysis, most up-regulated genes were highly correlated cell cycle, homologous recombination and p53 signaling pathway. Besides, we found IGF2BP2, BCL7A, CYP27B1, CITED2, and CNN3 were involved in immune-related pathways such as Th17 cell differentiation, antigen processing and presentation, chemokine signaling pathway, and leukocyte transendothelial migration.
3.2. Chromatin accessibility features in ESCC
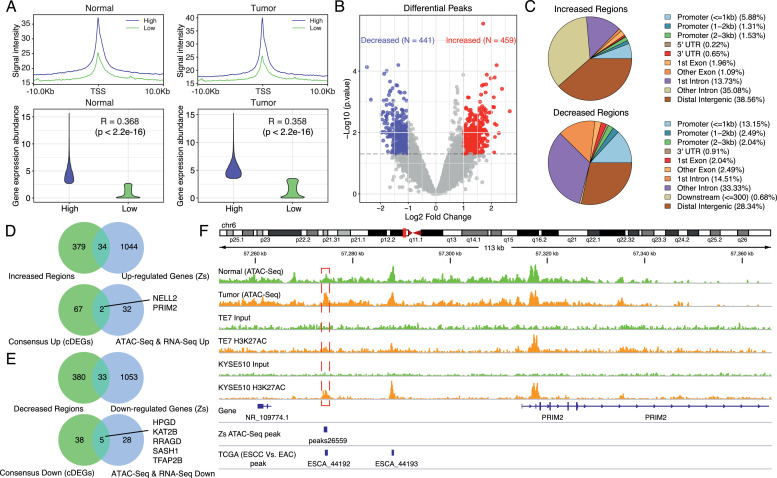
Chromatin remodeling play important roles in regulating chromatin accessibility and gene expression [35]. The aforementioned enrichment analysis revealed that histone modification, chromatin regulation, and transcription factor complex related processes were one of the enriched features in ESCC, suggesting alterations in epigenetic regulation and chromatin accessibility could be one of the contributing molecular mechanisms in ESCC. In order to explore the chromatin accessibility features in ESCC and identify whether there were certain genes in cDEGs whose expression alterations may result from chromatin accessibility changes, we further performed ATAC-seq on aforementioned ESCC samples collected at Zhongshan hospital (Supplementary Figure 5). For peaks identified in each sample, we first annotated their genomic locations and found they were mainly located at the promoter regions, followed by distal intergenic regions. Next, we focused on whether the degree of chromatin accessibility could affect the expression abundance of genes. We first used the median expression value to categorize the genes identified through matched RNA-seq data into high and low groups. Then, we compared the ATAC-seq signal intensities between high and low groups. Results revealed that in both tumor (correlation coefficient: 0.358) and normal (correlation coefficient: 0.368) samples, the level of ATAC-seq signals positively correlated with the expression abundance of the annotated genes (Fig. 3A).
Fig. 3.
Chromatin accessibility features in ESCC.
A. ATAC-seq signals at TSSs positively correlated with gene expression abundance. The left Fig.s showed the correlation between ATAC-seq signals at TSSs and gene expression in normal tissues, and the right Fig.s showed the correlation between ATAC-seq signals at TSSs and gene expression in tumor tissues. The significance level between the ATAC-seq signal and gene expression was shown (Spearman correlation, p < 0.05).
B. Identification of altered chromatin-accessible regions in ESCC. Up-regulated peaks were shown by red and down-regulated peaks were shown by blue (|Log2 fold-change| > 1, p < 0.05).
C. Annotation of altered chromatin-accessible regions. Upper: increased regions. Lower: decreased regions.
D. Identification of up-regulated genes that were associated with open chromatin regions. Upper: peak annotated genes were intersected with up-regulated genes from matched RNA-seq data. Lower: above genes were further overlapped with up-regulated genes in cDEGs.
E. Identification of down-regulated genes that were associated with closed chromatin regions. Upper: peak annotated genes were intersected with down-regulated genes from matched RNA-seq data. Lower: above genes were further overlapped with down-regulated genes in cDEGs.
F. Changes in chromatin accessibility upstream of PRIM2 and ChIP-Seq profiles (GSE76859) of TE7 and KYSE510 cells. Upper, track in green showed normalized and merged ATAC-seq signals in normal tissues and track in orange showed normalized and merged ATAC-seq signals in tumor tissues. Lower, ChIP-seq profiles of H3K27ac of ESCC cell lines (TE7 and KYSE510). Red boxes indicated changes supported by both Zs patients, ESCC cell lines, and TCGA ESCC patients.
Next, differential peak analysis was performed by comparing the ATAC-seq signals between tumor and normal samples to identify aberrant chromatin-accessible regions in ESCC. 459 increased chromatin-accessible regions and 441 decreased chromatin-accessible regions were identified (Fig. 3B and Supplementary Table 5). Subsequent annotation of the differentially accessible regions showed that these peaks were predominantly located at distal intergenic, intron, and promoter regions for both increased and decreased regions (Fig. 3C). To further explore which genes are significantly related to chromatin accessibility changes, we combined differential peak-annotated genes with differentially expressed genes from matched RNA-seq data to look for intersections. The results showed that 34 up-regulated genes were associated with increased ATAC-seq signals, whereas 33 down-regulated genes had decreased ATAC-seq signals (Fig. 3D & E). In order to obtain highly confident results, we further overlapped the above genes with the cDEGs, and this identified a total of 7 genes, among which NELL2 and PRIM2 (up-regulated genes) gained more open chromatin architecture, whereas regions around low expression genes, HPGD, KAT2B, RRAGD, SASH1, and TFAP2B, showed decreased chromatin signals. Based on that, we further compared the ATAC-seq signals of the 7 genes with the ATAC-seq data of esophageal cancer patients from TCGA to verify our findings. Because there were no normal samples in TCGA cohort, we here used EAC samples as the control. Finally, 4 genes (PRIM2, HPGD, NELL2, and TFAP2B) were identified (Fig. 3F & Supplementary Figure 6), whose peaks were identical in both TCGA and Zhongshan ESCC patients, suggesting chromatin accessibility changes of these regions were prevalent in ESCC and these changes were potential regulatory mechanisms of the expression of these genes.
We next investigated whether the peak26559 regulated the expression of PRIM2. We first examined the publicly available ChIP-seq profiles of H3K27ac of ESCC cell lines (TE7 and KYSE510). The results showed that there were also peaks at the peak26559 region (Fig. 3F). We next performed the luciferase reporter assay. Stronger transcription-enhancing activity was observed in TE1 and HEK293 cells transfected with PRIM2-P-E plasmid (containing the sequences of the PRIM2 promoter and peak26955) compared to PRIM2-P plasmid (containing the sequences of the PRIM2 promoter) (Supplementary Figure 7).
The above analyses revealed the shared key molecular changes and chromatin accessibility features in ESCC compared with normal tissues, and identified several genes whose expression alterations may result from changes in chromatin accessibility.
3.3. Stromal heterogeneity linked with distinct clinical outcomes of ESCC patients
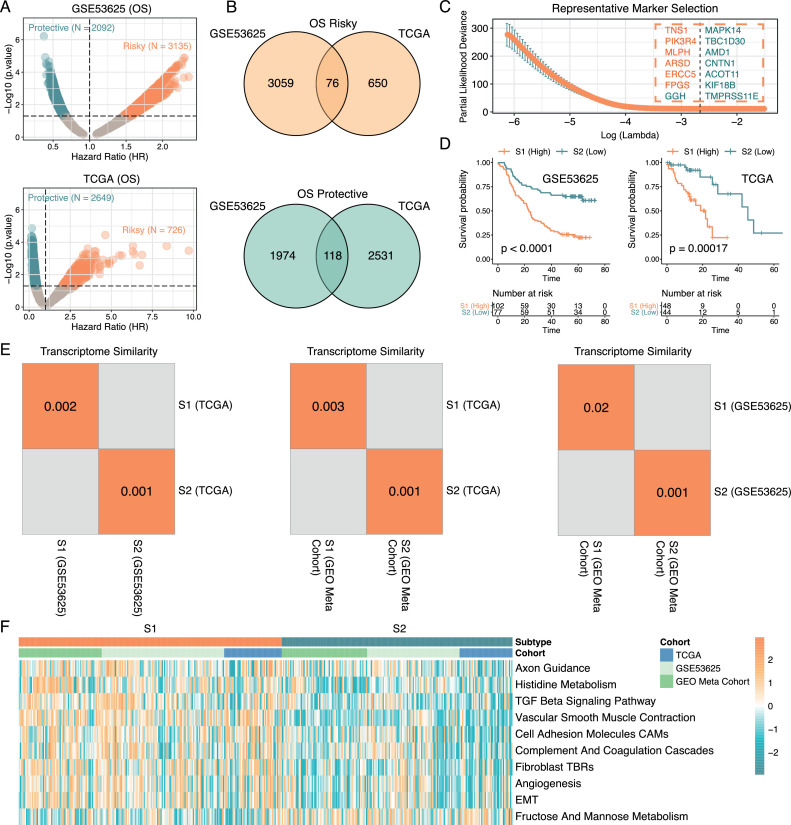
Previous studies have shown that molecular heterogeneities are tightly associated with therapeutic outcomes and prognosis of patients in various cancer types [14,62], we wondered whether there were distinct molecular phenotypes in ESCC that contributed to patients' survival differences. To answer this question, we first screened all survival-related genes in GSE53625 and TCGA ESCC cohorts separately (Fig. 4A & Supplementary Table 6), followed by the convergence of the survival-related genes identified in both cohorts. Eventually, 76 genes were classified as the risk factors and 116 genes were identified as protective factors in ESCC (Fig. 4B). To obtain the most representative survival-related marker genes, we further performed LASSO Cox regression analysis in the GSE53625 cohort to get the best combination of genes. As a result, a total of 14 genes were identified, including 6 risky and 8 protective genes (Fig. 4C & Supplementary Table 7). The KEGG pathway enrichment analysis of these 14 genes were shown in Supplementary Table 8. Based on the LASSO coefficients assigned to each of the 14 genes, we established a scoring system (termed here as Prognosis-related Subtype Classifier, PrSC). Based on optimal cut-off value generated using maximally selected log-rank statistics, ESCC patients were classified into high- and low-score groups. We here defined the high-score group as ESCC subtype 1 (S1) and the low-score group as ESCC subtype 2 (S2). Comparison of the survival outcomes between S1 (N = 102) and S2 (N = 77) patients in GSE53625 cohort revealed a significant difference. Subsequently, we applied PrSC into the TCGA cohort, and consistently, S1 (N = 48) patients displayed a poorer prognosis than S2 (N = 44) patients, which confirmed the stability of this scoring tool (Fig. 4D).
Fig. 4.
Identification of survival-related molecular subtypes in ESCC.
A. Screening of survival-related genes in GSE53625 (upper) and TCGA ESCC (lower) cohorts using Cox regression analysis. Risky genes were defined as genes with a hazard ratio (HR) > 1 whereas genes with HR < 1 were considered as protective genes (p < 0.05).
B. Overlapping of risky (upper) and protective (lower) genes identified in GSE53625 and TCGA ESCC cohorts.
C. Identification of representative marker genes using LASSO Cox regression analysis in GSE53625 cohort. Risky genes were colored in orange and protective genes were green.
D. S1/S2 subtype patients showed distinct OS differences in both GSE53625 (left) and TCGA ESCC (right) cohorts (Log-rank test, p < 0.05).
E. Submap analysis showing a significant correlation of classification among three ESCC cohorts. A p-value < 0.05 indicating a significant similarity.
F. The heatmap showing the biological pathway differences between S1 and S2 subtypes (Wilcoxon rank sum test, p <0.05).
Next, we assessed whether ESCC subtypes determined by PrSC had similar transcriptomic characterizations across different ESCC cohorts. Apart from GSE53625 and TCGA ESCC cohorts, we also enrolled a GEO ESCC meta cohort to cross-validate the transcriptomic similarities (Supplementary Figure 4). Using SubMap analysis [45], we found that S1 and S2 patients were highly correlated with corresponding subtypes in the above three cohorts, suggesting that PrSC was a robust tool that was able to capture the survival-related phenotypes in ESCC across multiple ESCC cohorts (Fig. 4E). Besides, to further depict the biological behaviour differences between S1 and S2 subtypes, functional annotations were performed using GSVA algorithm in GSE53625, TCGA ESCC, and GEO ESCC meta cohorts, respectively, and we here only considered biological behaviour differences supported by the above three datasets were core biological differences. The results showed that S1 group patients displayed significantly higher levels of stromal related activities such as TGF beta signaling pathway, vascular smooth muscle contraction, angiogenesis, as well as fibroblast TGF beta response signature (TBRs), whereas fructose and mannose metabolism was enriched in S2 group patients (Fig. 4F). To verify our findings, we also utilized the ESTIMATE algorithm to evaluate the stromal score [48]. Consistently, S1 patients showed a significantly higher level of stromal score than S2 patients and the tumor purity was also lower in S1 patients (Supplementary Figure 8). Moreover, another method, MCP-counter, which was able to quantify both immune cell and stromal cell populations in the tumor microenvironment was also used [47], and the results also substantiated a higher level of infiltration of fibroblasts in S1 patients (Supplementary Figure 9). It was noteworthy that S1 patients also tended to display a higher level of CD8+ T Cell. In most cases, the infiltration of immune cells such as CD8+ T cell in the tumor microenvironment (TME) was linked to a better prognosis [63], but immune-excluded phenotype also could show the presence of immune cells, while these immune cells could not penetrate into the parenchyma and thus this type of TME were considered T-cell suppressive [64]. Our results suggested that the stromal activation in the S1 subtype could inhibit the antitumor effect of immune cells.
We then compared the genomic features of patients in S1 and S2 subtypes based on available TCGA data. The somatic mutation landscapes in S1 and S2 subtypes were shown in Supplementary Figure 10A. The percentage of patients in S1 with mutations of genes including DIDO1, CREBBP, and ACACB were significantly higher than those in S2, whereas S2 patients had more KMT2D, FAT2, and LAMA5 mutations. Next, we compared the copy number variations (CNVs) in different groups (Supplementary Figure 10B). The distribution of the GISTIC score across all chromosomes indicated that S1 patients had more copy number gains at chromosomes 2 (2q31.2), 5 (5p15.33 and 5p12), 6 (6p22.3, 6p21.1, and 6p12.1), and 19 (19q13.12 and 19q13.43). While S2 patients showed more copy-number gains at chromosomes 4 (4q12 and 4q13.3), 7 (7q21.12 and 7q22.3), and 8 (8q24.21) (Supplementary Figure 10C & D). The investigation into the tumor mutation burden (TMB) between S1 and S2 subtype patients showed no significant difference (Supplementary Figure 10E), but interestingly, S1 subtype patients displayed a significantly higher level of DNA methylation burden than S2 subtype patients (Supplementary Figure 10F). In addition, the investigation into the chromatin accessibility features between S1 and S2 subtypes revealed that the accessibility of chromatin around the promoter regions was higher in the former (Supplementary Figure 10G). Meanwhile, we identified several extracellular matrix and fibroblast related genes, whose promoter regions were more accessible, and they were highly expressed in the S1 subtype (Supplementary Figure 10H).
These results supported by different ESCC cohorts and different algorithms indicated that the remodeling of stromal components was a crucial factor in determining the prognosis of ESCC patients.
3.4. TNS1high fibroblasts in the stroma linked with immune exclusion
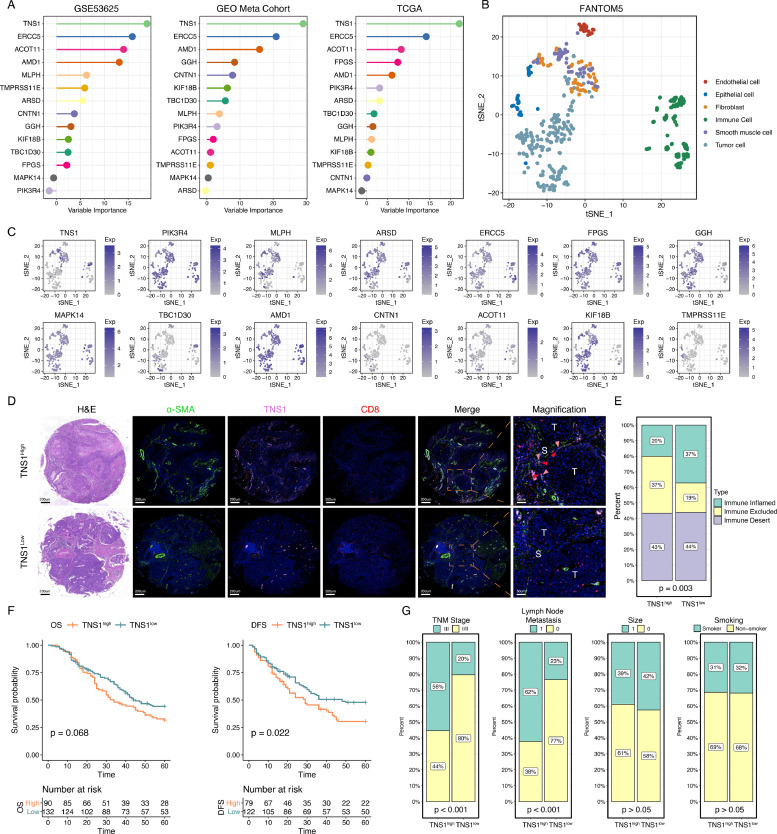
Next, we assessed each gene's contribution in PrSC in determining the S1/S2 subtypes using random forest algorithm (Fig. 5A). This analysis revealed that TNS1 was the most important gene that contributed to this classification, and therefore, we here chose TNS1 for further experimental validation. Before we conducted experimental validation, we first assessed the expression features of TNS1, i.e., whether it is predominantly expressed in tumor, immune, or stromal cells. We referred to the expression profiles of 119 immune cells, 197 tumor cells, 24 endothelial cells, 32 epithelial cells, 61 fibroblasts, and 49 smooth muscle cells from FANTOM5 [65] (Fig. 5B). The results demonstrated that TNS1 was predominantly expressed in fibroblasts and smooth muscle cells (Fig. 5C). Considering that S1 subtype patients showed a higher level of fibroblast infiltration (Fig. 4F & Supplementary Figure 9), we speculated that TNS1 was associated with the fibroblasts in the TME. To verify our speculation and validate the expression features of TNS1 in ESCC tumor microenvironment, we further collected the single-cell RNA sequencing data of mouse esophageal cancer model, including two pathological stages (carcinoma in situ, CIS, and invasive carcinoma, ICA) [49]. The CD45− non-immune cells were classified into four clusters, including endothelial cells, epithelial cells, fibroblasts, and myocytes (Supplementary Figure 11A). We first assessed the expression level of Tns1 in these cells. Consistently, Tns1 was predominantly expressed in fibroblasts (Supplementary Figure 11B). Next, we classified fibroblasts into two clusters (Tns1high and Tns1low groups) based on the median expression value of Tns1 (Supplementary Figure 11C & D). We compared the composition of Tns1high/low fibroblasts in CIS and ICA stages and found that the percentage of Tns1high fibroblasts tended to increase during pathological transition (Supplementary Figure 11E). Then, the marker genes for Tns1high fibroblasts were determined (Supplementary Figure 11F), based on which we quantified the cell abundance of Tns1high fibroblasts in GEO samples [51]. Subsequent survival analysis showed that ESCC patients with a higher proportion of Tns1high fibroblasts displayed a poorer OS (Supplementary Figure 11G). In addition, we compared the biological differences between Tns1high and Tns1low fibroblasts. Consistent with our findings from bulk gene expression profiles, Tns1high fibroblasts group showed stronger activities of angiogenesis, extracellular matrix (ECM) interaction, EMT, and axon guidance. While the effect of CD8+ T cell was weaker compared to Tns1low fibroblasts group (Supplementary Figure 11H).
Fig. 5.
The association between TNS1high fibroblasts and immune exclusion
A. Identification of TNS1 as the most important gene in contributing to S1/S2 classification using random forest method. The variable importance was based on Mean Decrease Accuracy.
B. t-SNE analysis of the expression profiles of immune cells, tumor cells, endothelial cells, epithelial cells, fibroblasts, and smooth muscle cells from the FANTOM5 project.
C. Expression levels of 14 genes illustrated as t-SNE plots.
D. Representative immunofluorescence images showing that TNS1high fibroblasts group patients (N = 90) showed a decreased infiltration level of CD8+ T cells than TNS1low fibroblasts group patients (N = 132) and the interaction between TNS1high fibroblasts and CD8+ T cells in the stroma. The red arrow represents CD8+ T cell, the pink arrow represents TNS1high fibroblasts. T, tumor; S, stroma.
E. TNS1high fibroblasts group showed a higher proportion of immune exclusion phenotype (Pearson's chi-square test, p < 0.05).
F. The association between TNS1high fibroblasts and patients’ prognosis (Log-rank test, p < 0.05).
G. The associations between TNS1high fibroblasts and clinical parameters (Pearson's chi-square test, p < 0.05).
Next, we performed multiplex fluorescent immunohistochemistry (mfIHC) on tissue microarrays (TMAs) of ESCC patients from Zs hospital. Included markers were α-SMA, a common cancer-associated fibroblasts marker [66], TNS1, and CD8. The results from mfIHC revealed that patients with a higher proportion of TNS1high fibroblasts in the stroma displayed a decreased infiltration level of CD8+ T cell in the tumor parenchyma and showed an immune exclusion phenomenon (Fig. 5D & E). These TNS1high fibroblasts resided near CD8+ T cells in the stroma, which suggested possible crosstalk between these two types of cells. Besides, patients with higher proportions of TNS1high fibroblasts in the stroma tended to show poorer OS, though not reaching statistical significance. But patients of TNS1high fibroblasts group did show a decreased disease-free survival (DFS) (Fig. 5F). In addition, TNS1high fibroblasts were associated with the advanced clinical stage and lymph node metastasis (Fig. 5G).
We further investigated how Tns1high fibroblasts interacted with CD8+ T cell in the TME based on aforementioned single-cell RNA sequencing data of mouse esophageal cancer model. We classified CD8+ T cells into 6 clusters (Supplementary Figure 12A), including four clusters of naive T cells (Tn), one cluster of cytotoxic T cell (Ct), and one cluster of exhausted T cell (Ex) according to canonical markers (Supplementary Figure 12B) [49,67,68]. Then, we used CellChat to infer the cell-cell communication network among CD8+ T cells and Tns1high fibroblasts [52]. This analysis revealed significant ligand-receptor and signaling interactions among these 7 cell groups (Supplementary Figure 12C). Notably, the number of interactions between cytotoxic T cells and Tns1high fibroblasts was quite prominent (Supplementary Figure 12D). Specifically, these two types of cells mainly interacted with each other via Cxcl12-Cxcr4, Fn1 − (Itga4+Itgb7), Fn1 − (Itga4+Itgb1), Fn1 − Cd44, Icam1 − Itgal, and Icam1 − (Itgal+Itgb2) pairs (Supplementary Figure 12E).
3.5. Subtype-specific therapeutic agents screening and validation
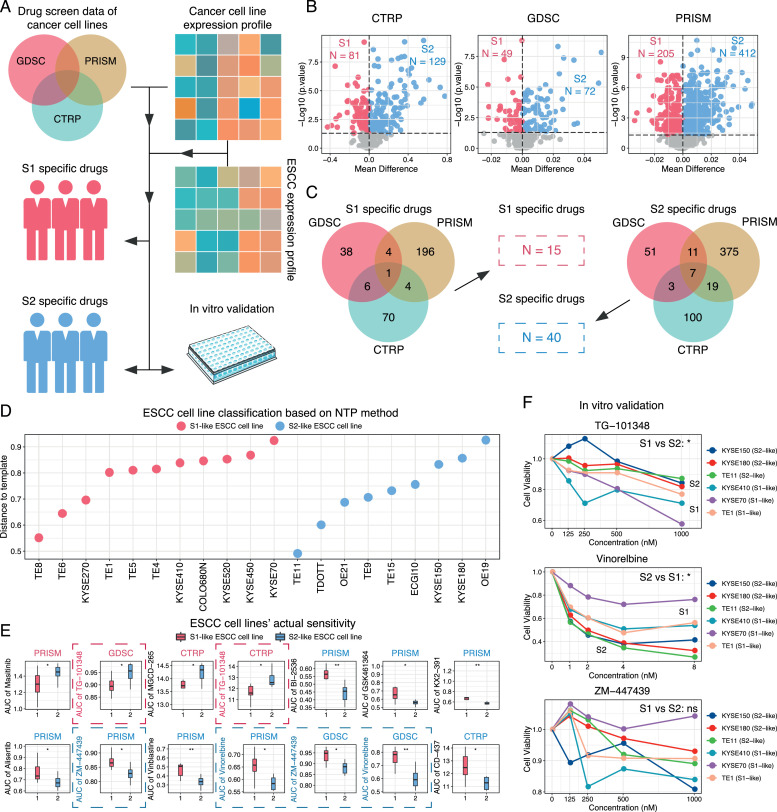
Considering that S1 and S2 subtype patients displayed distinct molecular features, exploring subtype-specific therapeutic agents for these individuals would be of great significance in determining personalized treatment strategies. Based on the S1 and S2 subtypes we identified in ESCC, we here adopted an integrative strategy to screen possible drugs that could be more suitable for the molecular features of these subtype patients (Fig. 6A). Specifically, gene expression data and drug response data of hundreds of cancer cell lines from three independent datasets (CTRP, GDSC, and PRISM) were collected [53], [54], [55]. Before we estimated potential drugs, we first assessed the expression features of these cancer cell lines. The results showed that blood cancer and skin cancer cell lines displayed distinct expression profiles from most solid cancer cell lines, so we excluded these cancer cell lines prior to subsequent analysis (Supplementary Figure 13). Next, we used the ridge regression model to infer the ESCC patients’ sensitivity to different drugs based on the gene expression data and drug response data in each dataset, separately. After obtaining the estimated sensitivity data of ESCC patients, differential analysis using estimated drug sensitivity data from CTRP, GDSC, and PRISM datasets between two subtypes was performed. As a result, 81 drugs in CTRP, 49 drugs in GDSC, and 205 drugs in PRISM were identified as potential S1-specific drugs, whereas 129 drugs in CTRP, 72 drugs in GDSC, and 412 drugs in PRISM were identified as potential S2-specific drugs (Fig. 6B). We then integrated these drugs and considered the ones that were identified in at least two datasets significant. As a result, 15 drugs were considered as S1-specific agents and 40 drugs were more suitable for S2 patients (Fig. 6C & Supplementary Table 9). It was noteworthy that among S1-specific agents, Nintedanib, which has anti-angiogenic and anti-fibrotic activities [69], was the most significant S1-specific drug that was supported by three datasets. Interestingly, S1 patients were indeed characterized by increased activities of angiogenesis and fibroblast infiltration, therefore, this consistency confirmed the feasibility of our drug screening strategy.
Fig. 6.
Identification of subtype-specific drugs.
A. A flowchart illustrating the process of subtype-specific drug identification. Human cancer cell line drug screen data from three independent datasets (CTRP, GDSC, and PRISM) were included, along with corresponding expression profiles. Based on cancer cell line data, the patients' sensitivities to various drugs were estimated and further validated via experiments.
B. S1/S2 specific drugs identified in each dataset (Wilcoxon rank sum test, p < 0.05).
C. Venn diagram showing S1 and S2 specific drugs.
D. Classification of ESCC cell lines into S1- and S2-like cells using NTP method (p < 0.05).
E. Evaluation of results using actual drug response data of ESCC cancer cell lines (Wilcoxon rank sum test, *: p < 0.05; **: p < 0.01).
F. In vitro validation of drug response between two subtypes using cell viability assay across 6 ESCC cell lines (Two-way ANOVA, *: p < 0.05; ns: not significant). Experiments were repeated at least three times.
The above analysis has identified a number of candidate drugs that targeted the molecular differences of ESCC subtype patients, we next wanted to know whether the tumor cells were sensitive to the estimated drugs. We first classified ESCC cell lines into S1- and S2-like cells based on their transcriptome similarities with ESCC subtype patients using NTP method [59] (Fig. 6D). We then compared the actual drug response data of the aforementioned S1/S2 specific agents between S1- and S2-like cells and found that S1-like cell lines were more sensitive to TG-101348, while S2-like cell lines were more sensitive to Vinorelbine and ZM-447439 (Fig. 6E). On the basis of that, we conducted in vitro experimental validation using 6 ESCC cell lines, TE1, KYSE70, KYSE410, TE11, KYSE150, and KYSE180. The results of the cell viability assay presented a relatively good agreement with our prediction except for ZM-447439 (Fig. 6F), suggesting that TG-101348 and Vinorelbine could be potentially promising agents for treating different subtypes of ESCC patients.
3.6. Exploring the role of PrSC in the pan-cancer cohort and its association with immunotherapy response
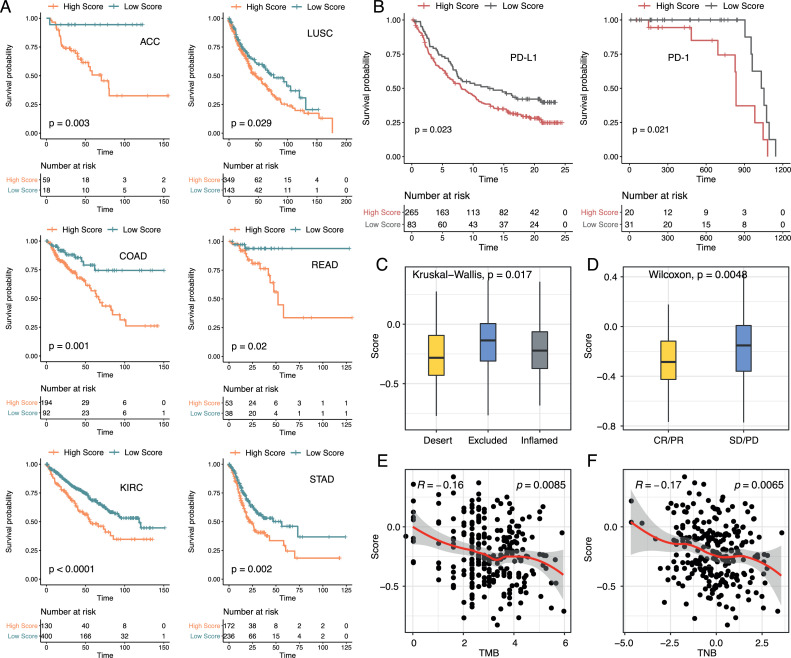
We next applied PrSC into the pan-cancer cohort to explore whether it could be extended to different types of cancer in predicting prognosis and reflecting the TME features. Of the 33 cancer types, PrSC was significantly related to the prognosis of patients in 6 cancer types, including ACC, COAD, KIRC, LUSC, READ, and STAD (Fig. 7A). Besides, high score patients in COAD, LUSC, READ, and STAD all exhibited higher activities of angiogenesis, EMT, and fibroblast TBRs, and a lower level of fructose and mannose metabolism (Supplementary Figure 14). It was worth mentioning that COAD, READ, and STAD all belonged to digestive tract malignancies, and LUSC was also squamous cell cancer, suggesting that the TME features captured by PrSC were not limited in ESCC, but also common in digestive tract malignancies and squamous cell carcinoma.
Fig. 7.
Application of PrSC into the pan-cancer cohort and two immunotherapy cohorts.
A. PrSC was significantly associated with the prognosis of patients in 6 cancer types (Log-rank test, p < 0.05).
B. Survival curves showing that the high PrSC score groups had a poorer prognosis than the low score groups in both PD-L1 (left) and PD-1 (right) treatment cohorts (Log-rank test, p < 0.05).
C. Distribution of the PrSC score in different immune phenotypes in the PD-L1 treatment cohort (Kruskal-Wallis test, p < 0.05).
D. Distribution of the PrSC score in different clinical response groups in the PD-L1 treatment cohort (Wilcoxon rank sum test, p < 0.05). SD, stable disease; PD, progressive disease; CR, complete response; PR, partial response.
E. The PrSC score was negatively correlated with TMB in the PD-L1 treatment cohort (Spearman correlation, p < 0.05)
F. The PrSC score was negatively correlated with TNB in the PD-L1 treatment cohort (Spearman correlation, p < 0.05).
On the other hand, previous studies have shown that stromal components can also affect immunotherapy outcomes [70,71]. Therefore, we speculated that S1/S2 subtypes may have different responses to immunotherapy. Based on two immunotherapy cohorts (IMvigor210 PD-L1 treatment cohort and Riaz. et al. melanoma PD-1 treatment cohort) [72,73], we here evaluated the ability of PrSC in predicting PD-1/PD-L1 treatment response. Interestingly, patients with low scores had a prominent survival advantage than high score patients in both cohorts (Fig. 7B). Besides, we found that immune excluded phenotype had the highest score than immune desert and inflamed phenotypes, and the score of non-responders was also higher (Fig. 7C & D). Moreover, PrSC was negatively correlated with TMB and neoantigen burden (Fig. 7E & F).
4. Discussion
Integrating multiple independent datasets to study the characteristics of diseases has gradually become a common and popular approach [16,74,75]. Through this method, researchers can obtain more reliable and meaningful results in a larger population and identify things that have been previously neglected.
The first part of this study was to examine the key molecular changes in ESCC compared with normal tissues. Seven independent ESCC expression datasets, along with the expression data of Zs hospital ESCC patients, were integrated to identify the aberrantly expressed genes in ESCC. By comparing the differentially expressed genes in tumor tissues, we identified a total of 112 cDEGs. These genes were abnormally expressed in the eight independent ESCC cohorts, which, to a great extent, indicated that the expression alterations of these genes were the shared features in ESCC and served as vital mechanisms during the pathogenesis of ESCC. Among these genes, in addition to some important genes that were previously identified to be involved in tumorigenesis, such as MCM5, MCM6, MCM10, and EXO1 [76], [77], [78], [79], [80], genes that were not previously reported to be involved in ESCC have also been identified, including PRIM2, KRT32, as well as CCDC86. Besides, it was also worth noting that IGF2BP2, a m6A reader [81], was also highly expressed in ESCC. Meanwhile, we found it was closely related to several immune-related pathways, such as antigen processing and presentation and T helper cell differentiation. Interestingly, our previous findings in lung adenocarcinoma revealed that IGF2BP2 was also associated with immune-related pathways [14], and this consistency may suggest that m6A modification could also be one of the crucial mechanisms involved in tumor immune regulation in ESCC.
The alteration of chromatin accessibility is one of the crucial mechanisms in regulating gene expression [82]. Here, we found some biological processes that can affect the chromatin accessibility were significantly enriched in ESCC, indicating that there may be changes in chromatin accessibility in ESCC. ATAC-seq is one of the popular methodologies for investigating the chromatin accessibility profiling in recent years, it has been applied to study the chromatin landscapes of many cancer types [35,83,84], but barely applied in ESCC. In the present study, we performed ATAC-seq to examine the chromatin accessibility features in ESCC. Via integrating the peak-annotated igenes supported by both Zs ESCC patients and TCGA ESCC patients with cDEGs, 4 genes, PRIM2, HPGD, NELL2, and TFAP2B, were finally identified. PRIM2 is a DNA primer enzyme which is involved in DNA replication regulation. Previous studies have shown that PRIM2 was upregulated in cervical cancer and lung cancer and can promote the proliferation of cancer cells and induce dihydroartemisinin resistance [85,86]. HPGD has been considered as a tumor suppressor in various malignancies [87,88]. Several previous studies have revealed microRNAs such as miR-21, miR-620, and miR-146b-3p can directly target HPGD and affect its expression [89], [90], [91]. Kawamata et al. reported that HPGD was down-regulated in human metastasizing esophageal cancer cell line [92]. Here, we observed that the promoter region of HPGD was less accessible in tumor tissues, which suggested that this change could be a vital regulation mechanism that contributed to the down-regulation of HPGD in ESCC. As for NELL2 and TFAP2B, the peaks associated with these two genes were at distal intron and downstream regions, respectively, and the specific regulating mechanisms may warrant further investigation.
We established a subtype classifier, PrSC, which was linked to the prognosis of ESCC patients and can reflect the TME heterogeneity in different ESCC cohorts. Among the genes in PrSC, TNS1 was the most important one that contributed to the classification, therefore, we further focused on TNS1. TNS1 belongs to the tensin family and is a key component of cellular adhesions [93]. Previous studies have shown that TNS1 was involved in tumorigenesis in several types of cancer [94,95]. A recent study from Liu et al. revealed that TNS1 was associated with tumor stroma in colorectal cancer and linked to poorer prognosis [96]. Here, the cell expression profiles from FANTOM5 [65] suggested that TNS1 was related to fibroblasts, subsequently, the results from single-cell RNA sequencing data analysis further confirmed the expression of TNS1 in fibroblasts and its clinical implications. Furthermore, the results from mfIHC on TMAs of ESCC patients revealed that TNS1high group was related to immune exclusion phenotype, and was significantly related to the patient's prognosis, clinical stage, and lymph node metastasis. These findings indicated that this type of fibroblasts in the tumor stroma was a crucial factor in determining the progression of ESCC. Targeting this type of fibroblasts may provide new ideas and prospects for ESCC treatment.
Identification of subgroup patients who may benefit more from certain drugs is essential for maximizing the effectiveness of therapies and improving the prognosis of patients. Considering that S1 and S2 subtype patients displayed significantly different molecular features, we speculated that this discrepancy could result in different responses to therapeutic agents. Through integrating hundreds of cancer cell lines' drug screen information, we identified some drugs that could target the molecular features of S1/S2 subtype patients. Interestingly, the drugs estimated for S1 patients were mainly associated with anti-angiogenic and anti-fibrotic activities, such as AZD4547, Foretinib, and Nintedanib [69,97,98]. In contrast, drugs that were more suitable for S2 patients were predominantly common chemotherapy and targeted therapy drugs, including Paclitaxel, Vinorelbine, and Gefitinib. On the basis of that, further screening for more promising drugs that could target the tumor cells in these two molecular subtypes identified 2 agents, TG-101348 and Vinorelbine. TG-101348 (Fedratinib) is a semi-selective inhibitor of JAK2 developed for treating myeloproliferative diseases such as myelofibrosis [99]. Phase 2 clinical trial of TG-101348 on patients with ruxolitinib-resistant or ruxolitinib-intolerant myelofibrosis showed that patients could obtain significant clinical benefit with TG-101348 [100]. Besides, a recent study from Liu et al. found that TG-101348 could exhibit KRAS-dependent anticancer activity in pancreatic ductal adenocarcinoma cell [101]. Vinorelbine is a common chemotherapy medication for the treatment of non-small cell lung cancer and other types of cancer [102,103]. A recent phase 3 clinical trial found that neoadjuvant chemoradiotherapy (Vinorelbine plus Cisplatin) was associated with significantly decreased recurrences in advanced ESCC compared with surgery alone, providing evidence of the application value of Vinorelbine in ESCC [104]. The identification of TG-101348 and Vinorelbine in the present study could provide more clues for the precise treatment of ESCC, but the efficacy still warrants rigorous clinical observation.
Finally, the application of PrSC into the pan-cancer cohort revealed its correlation with prognosis and stromal components in pan-digestive tract cancer and LUSC patients, which suggested possible stromal similarities of these cancer types. In addition, PrSC was able to reflect the prognosis of patients receiving immune checkpoint inhibitor therapy and correlated with immune excluded phenotype, which again confirmed its robustness. In addition, for such patients with stromal remodeling and immune exclusion, the combined application of anti-fibrotic/anti-angiogenic drugs and immunotherapy may bring more benefits, but the specific efficacy needs further research.
In conclusion, our study depicted the multi-dimensional characterization of ESCC, highlighted the indispensable role of stroma cells in shaping the complexity and heterogeneity of TME, and identified potential subtype-specific therapeutic agents for ESCC patients.
Declaration of Competing Interest
The authors declare that they have no competing interests.
Acknowledgments
Contributors
Conception and design: YL, DG, SZ and CLL; Methodology: YL, SZ and CLL; Data analysis: YL, FKX, FHC, YWC, DG, SZ and CLL; Website construction: YL; Manuscript writing: YL, YWC, FKX, DG, SZ and CLL. All authors read and approved the final manuscript.
Acknowledgements
This study was sponsored by Natural Science Foundation of Shanghai (21ZR1412600), Fuqing Scholar Student Scientific Research Program of Shanghai Medical College, Fudan University (FQXZ202105B), Shanghai Science and Technology Infrastructure Program (19441908800), and Shanghai Pujiang Program (2020PJD012). We would like to thank Shanghai Jiayin Biotechnology Ltd. (Shanghai, China) and Wuhan Servicebio technology Ltd. (Wuhan, China) for the technical support.
Data Sharing
Processed data generated in this study can be downloaded from the Supplementary files or ESCCEXPRESS (http://www.bioinfo-zs.com/esccexpress/). Raw sequencing data generated in this study are available at GEO data repository under the accession number GSE167488. Other public data used in this study can be downloaded from TCGA data repository (https://gdc.cancer.gov) and GEO data repository (https://www.ncbi.nlm.nih.gov/geo). The R code for result reproduction is available upon reasonable request.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2021.103510.
Contributor Information
Shu Zhang, Email: zhang.shu@zs-hospital.sh.cn.
Chunlai Lu, Email: lu.chunlai@zs-hospital.sh.cn.
Appendix. Supplementary materials
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 2.Arnold M, Soerjomataram I, Ferlay J, Forman D. Global incidence of oesophageal cancer by histological subtype in 2012. Gut. 2015;64(3):381–387. doi: 10.1136/gutjnl-2014-308124. [DOI] [PubMed] [Google Scholar]
- 3.Lagergren J, Smyth E, Cunningham D, Lagergren P. Oesophageal cancer. Lancet. 2017;390(10110):2383–2396. doi: 10.1016/S0140-6736(17)31462-9. [DOI] [PubMed] [Google Scholar]
- 4.Testa U, Castelli G, Pelosi E. Esophageal cancer: genomic and molecular characterization, stem cell compartment and clonal evolution. Medicines. 2017;4(3):67. doi: 10.3390/medicines4030067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu M, Zhang C, Cai H, Liu F, Liu Y, Li J. The willingness to change risky health behaviors among Chinese rural residents: what we learned from a population-based esophageal cancer cohort study. PLoS One. 2016;11(8) doi: 10.1371/journal.pone.0161999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun X, Zhao D, Liu Y, Liu Y, Yuan Z, Wang J. The long-term spatial-temporal trends and burden of esophageal cancer in one high-risk area: a population-registered study in Feicheng, China. PLoS One. 2017;12(3) doi: 10.1371/journal.pone.0173211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Riella LV, Paterson AM, Sharpe AH, Chandraker A. Role of the PD-1 pathway in the immune response. Am J Transplant. 2012;12(10):2575–2587. doi: 10.1111/j.1600-6143.2012.04224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boussiotis VA. Molecular and biochemical aspects of the PD-1 checkpoint pathway. N Engl J Med. 2016;375(18):1767–1778. doi: 10.1056/NEJMra1514296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kato K, Cho BC, Takahashi M, Okada M, Lin CY, Chin K. Nivolumab versus chemotherapy in patients with advanced oesophageal squamous cell carcinoma refractory or intolerant to previous chemotherapy (ATTRACTION-3): a multicentre, randomised, open-label, phase 3 trial. Lancet Oncol. 2019;20(11):1506–1517. doi: 10.1016/S1470-2045(19)30626-6. [DOI] [PubMed] [Google Scholar]
- 10.Kuo HY, Guo JC, Hsu CH. Anti-PD-1 immunotherapy in advanced esophageal squamous cell carcinoma: a long-awaited breakthrough finally arrives. J Formos Med Assoc. 2020;119(2):565–568. doi: 10.1016/j.jfma.2019.10.010. [DOI] [PubMed] [Google Scholar]
- 11.Stark R, Grzelak M, Hadfield J. RNA sequencing: the teenage years. Nat Rev Genet. 2019;20(11):631–656. doi: 10.1038/s41576-019-0150-2. [DOI] [PubMed] [Google Scholar]
- 12.Sun J, Zhang Z, Bao S, Yan C, Hou P, Wu N. Identification of tumor immune infiltration-associated lncRNAs for improving prognosis and immunotherapy response of patients with non-small cell lung cancer. J Immunother Cancer. 2020;8(1):e000110. doi: 10.1136/jitc-2019-000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y, Gu J, Xu F, Zhu Q, Ge D, Lu C. Transcriptomic and functional network features of lung squamous cell carcinoma through integrative analysis of GEO and TCGA data. Sci Rep. 2018;8(1):15834. doi: 10.1038/s41598-018-34160-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Y, Gu J, Xu F, Zhu Q, Chen Y, Ge D. Molecular characterization, biological function, tumor microenvironment association and clinical significance of m6A regulators in lung adenocarcinoma. Brief Bioinform. 2021;22(4):bbaa225. doi: 10.1093/bib/bbaa225. [DOI] [PubMed] [Google Scholar]
- 15.Gao Q, Zhu H, Dong L, Shi W, Chen R, Song Z. Integrated proteogenomic characterization of HBV-related hepatocellular carcinoma. Cell. 2019;179(5):1240. doi: 10.1016/j.cell.2019.10.038. [DOI] [PubMed] [Google Scholar]
- 16.Zeng D, Li M, Zhou R, Zhang J, Sun H, Shi M. Tumor microenvironment characterization in gastric cancer identifies prognostic and immunotherapeutically relevant gene signatures. Cancer Immunol Res. 2019;7(5):737–750. doi: 10.1158/2326-6066.CIR-18-0436. [DOI] [PubMed] [Google Scholar]
- 17.Xu F, Gu J, Wang L, Liu R, Yuan Y, Wang H. Up-regulation of EIF3e is associated with the progression of esophageal squamous cell carcinoma and poor prognosis in patients. J Cancer. 2018;9(7):1135–1144. doi: 10.7150/jca.22546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Corces MR, Trevino AE, Hamilton EG, Greenside PG, Sinnott-Armstrong NA, Vesuna S. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat Methods. 2017;14(10):959–962. doi: 10.1038/nmeth.4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N. The sequence alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramirez F, Dundar F, Diehl S, Gruning BA, Manke T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014;42(Web Server issue):W187–W191. doi: 10.1093/nar/gku365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu G, Wang LG, He QY. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 2015;31(14):2382–2383. doi: 10.1093/bioinformatics/btv145. [DOI] [PubMed] [Google Scholar]
- 26.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy–analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20(3):307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 29.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28(6):882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang H, Wong WH. SeqMap: mapping massive amount of oligonucleotides to the genome. Bioinformatics. 2008;24(20):2395–2396. doi: 10.1093/bioinformatics/btn429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davis S, Meltzer PS. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics. 2007;23(14):1846–1847. doi: 10.1093/bioinformatics/btm254. [DOI] [PubMed] [Google Scholar]
- 32.Colaprico A, Silva TC, Olsen C, Garofano L, Cava C, Garolini D. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016;44(8):e71. doi: 10.1093/nar/gkv1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12(4):R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Y, Ge D, Lu C. The SMART App: an interactive web application for comprehensive DNA methylation analysis and visualization. Epigenetics Chromatin. 2019;12(1):71. doi: 10.1186/s13072-019-0316-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Corces MR, Granja JM, Shams S, Louie BH, Seoane JA, Zhou W. The chromatin accessibility landscape of primary human cancers. Science. 2018;362(6413):eaav1898. doi: 10.1126/science.aav1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vivian J, Rao AA, Nothaft FA, Ketchum C, Armstrong J, Novak A. Toil enables reproducible, open source, big biomedical data analyses. Nat Biotechnol. 2017;35(4):314–316. doi: 10.1038/nbt.3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD. An Integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell. 2018;173(2):400–416. doi: 10.1016/j.cell.2018.02.052. e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang YY, Lin DC, Mayakonda A, Hazawa M, Ding LW, Chien WW. Targeting super-enhancer-associated oncogenes in oesophageal squamous cell carcinoma. Gut. 2017;66(8):1358–1368. doi: 10.1136/gutjnl-2016-311818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tarca AL, Draghici S, Khatri P, Hassan SS, Mittal P, Kim JS. A novel signaling pathway impact analysis. Bioinformatics. 2009;25(1):75–82. doi: 10.1093/bioinformatics/btn577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tibshirani R. Regression shrinkage and selection via the Lasso. J Royal Stat Soc: Ser B (Methodol) 1996;58(1):267–288. [Google Scholar]
- 43.Li Y, Ge D, Gu J, Xu F, Zhu Q, Lu C. A large cohort study identifying a novel prognosis prediction model for lung adenocarcinoma through machine learning strategies. BMC Cancer. 2019;19(1):886. doi: 10.1186/s12885-019-6101-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou R, Zeng D, Zhang J, Sun H, Wu J, Li N. A robust panel based on tumour microenvironment genes for prognostic prediction and tailoring therapies in stage I-III colon cancer. EBioMedicine. 2019;42:420–430. doi: 10.1016/j.ebiom.2019.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nat Genet. 2006;38(5):500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 46.Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Becht E, Giraldo NA, Lacroix L, Buttard B, Elarouci N, Petitprez F. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016;17(1):218. doi: 10.1186/s13059-016-1070-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoshihara K, Shahmoradgoli M, Martinez E, Vegesna R, Kim H, Torres-Garcia W. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yao J, Cui Q, Fan W, Ma Y, Chen Y, Liu T. Single-cell transcriptomic analysis in a mouse model deciphers cell transition states in the multistep development of esophageal cancer. Nat Commun. 2020;11(1):3715. doi: 10.1038/s41467-020-17492-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hao Y, Hao S, Andersen-Nissen E, Mauck WM, 3rd, Zheng S, Butler A. Integrated analysis of multimodal single-cell data. Cell. 2021;184(13):3573–3587.e29. doi: 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen Z, Zhou L, Liu L, Hou Y, Xiong M, Yang Y. Single-cell RNA sequencing highlights the role of inflammatory cancer-associated fibroblasts in bladder urothelial carcinoma. Nat Commun. 2020;11(1):5077. doi: 10.1038/s41467-020-18916-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jin S, Guerrero-Juarez CF, Zhang L, Chang I, Ramos R, Kuan CH. Inference and analysis of cell-cell communication using CellChat. Nat Commun. 2021;12(1):1088. doi: 10.1038/s41467-021-21246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rees MG, Seashore-Ludlow B, Cheah JH, Adams DJ, Price EV, Gill S. Correlating chemical sensitivity and basal gene expression reveals mechanism of action. Nat Chem Biol. 2016;12(2):109–116. doi: 10.1038/nchembio.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Iorio F, Knijnenburg TA, Vis DJ, Bignell GR, Menden MP, Schubert M. A landscape of pharmacogenomic interactions in cancer. Cell. 2016;166(3):740–754. doi: 10.1016/j.cell.2016.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Corsello SM, Nagari RT, Spangler RD, Rossen J, Kocak M, Bryan JG. Discovering the anti-cancer potential of non-oncology drugs by systematic viability profiling. Nat Cancer. 2020;1(2):235–248. doi: 10.1038/s43018-019-0018-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ghandi M, Huang FW, Jane-Valbuena J, Kryukov GV, Lo CC, McDonald ER., 3rd Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature. 2019;569(7757):503–508. doi: 10.1038/s41586-019-1186-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Geeleher P, Cox N, Huang RS. pRRophetic: an R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS One. 2014;9(9) doi: 10.1371/journal.pone.0107468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Geeleher P, Cox NJ, Huang RS. Clinical drug response can be predicted using baseline gene expression levels and in vitro drug sensitivity in cell lines. Genome Biol. 2014;15(3):R47. doi: 10.1186/gb-2014-15-3-r47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hoshida Y. Nearest template prediction: a single-sample-based flexible class prediction with confidence assessment. PLoS One. 2010;5(11):e15543. doi: 10.1371/journal.pone.0015543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dempster JM, Rossen J, Kazachkova M, Pan J, Kugener G, Root DE, et al. Extracting Biological Insights from the Project Achilles Genome-Scale CRISPR Screens in Cancer Cell Lines. bioRxiv. 2019:720243.doi: 10.1101/720243 [DOI]
- 62.Tirosh I, Izar B, Prakadan SM, Wadsworth MH, 2nd, Treacy D, Trombetta JJ. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352(6282):189–196. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tekpli X, Lien T, Rossevold AH, Nebdal D, Borgen E, Ohnstad HO. An independent poor-prognosis subtype of breast cancer defined by a distinct tumor immune microenvironment. Nat Commun. 2019;10(1):5499. doi: 10.1038/s41467-019-13329-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen DS, Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature. 2017;541(7637):321–330. doi: 10.1038/nature21349. [DOI] [PubMed] [Google Scholar]
- 65.Lizio M, Harshbarger J, Abugessaisa I, Noguchi S, Kondo A, Severin J. Update of the FANTOM web resource: high resolution transcriptome of diverse cell types in mammals. Nucleic Acids Res. 2017;45(D1):D737–DD43. doi: 10.1093/nar/gkw995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ohlund D, Handly-Santana A, Biffi G, Elyada E, Almeida AS, Ponz-Sarvise M. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J Exp Med. 2017;214(3):579–596. doi: 10.1084/jem.20162024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim N, Kim HK, Lee K, Hong Y, Cho JH, Choi JW. Single-cell RNA sequencing demonstrates the molecular and cellular reprogramming of metastatic lung adenocarcinoma. Nat Commun. 2020;11(1):2285. doi: 10.1038/s41467-020-16164-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lu T, Yang X, Shi Y, Zhao M, Bi G, Liang J. Single-cell transcriptome atlas of lung adenocarcinoma featured with ground glass nodules. Cell Discov. 2020;6:69. doi: 10.1038/s41421-020-00200-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yamasaki Y, Kuwana M. Nintedanib for the treatment of systemic sclerosis-associated interstitial lung disease. Expert Rev Clin Immunol. 2020;16(6):547–560. doi: 10.1080/1744666X.2020.1777857. [DOI] [PubMed] [Google Scholar]
- 70.Lakins MA, Ghorani E, Munir H, Martins CP, Shields JD. Cancer-associated fibroblasts induce antigen-specific deletion of CD8 (+) T Cells to protect tumour cells. Nat Commun. 2018;9(1):948. doi: 10.1038/s41467-018-03347-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yu Y, Schuck K, Friess H, Kong B. Targeting aggressive fibroblasts to enhance the treatment of pancreatic cancer. Expert Opin Ther Targets. 2021;25(1):5–13. doi: 10.1080/14728222.2021.1857727. [DOI] [PubMed] [Google Scholar]
- 72.Mariathasan S, Turley SJ, Nickles D, Castiglioni A, Yuen K, Wang Y. TGFbeta attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature. 2018;554(7693):544–548. doi: 10.1038/nature25501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Riaz N, Havel JJ, Makarov V, Desrichard A, Urba WJ, Sims JS. Tumor and microenvironment evolution during Immunotherapy with Nivolumab. Cell. 2017;171(4):934–949. doi: 10.1016/j.cell.2017.09.028. e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li G, He X, Zhang L, Ran Q, Wang J, Xiong A. Assessing ACE2 expression patterns in lung tissues in the pathogenesis of COVID-19. J Autoimmun. 2020;112:102463. doi: 10.1016/j.jaut.2020.102463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li Y, Xiao J, Bai J, Tian Y, Qu Y, Chen X. Molecular characterization and clinical relevance of m(6)A regulators across 33 cancer types. Mol Cancer. 2019;18(1):137. doi: 10.1186/s12943-019-1066-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Alison MR, Hunt T, Forbes SJ. Minichromosome maintenance (MCM) proteins may be pre-cancer markers. Gut. 2002;50(3):290–291. doi: 10.1136/gut.50.3.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Burger M. MCM2 and MCM5 as prognostic markers in colon cancer: a worthwhile approach. Dig Dis Sci. 2009;54(2):197–198. doi: 10.1007/s10620-008-0416-6. [DOI] [PubMed] [Google Scholar]
- 78.Zheng Y, Shi Y, Yu S, Han Y, Kang K, Xu H. GTSE1, CDC20, PCNA, and MCM6 synergistically affect regulations in cell cycle and indicate poor prognosis in liver cancer. Anal Cell Pathol. 2019;2019 doi: 10.1155/2019/1038069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wan W, Shen Y, Li Q. MCM10 acts as a potential prognostic biomarker and promotes cell proliferation in hepatocellular carcinoma: integrated bioinformatics analysis and experimental validation. Cancer Manag Res. 2020;12:9609–9619. doi: 10.2147/CMAR.S267493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sertic S, Quadri R, Lazzaro F, Muzi-Falconi M. EXO1: A tightly regulated nuclease. DNA Repair. 2020;93 doi: 10.1016/j.dnarep.2020.102929. [DOI] [PubMed] [Google Scholar]
- 81.Niu Y, Zhao X, Wu YS, Li MM, Wang XJ, Yang YG. N6-methyl-adenosine (m6A) in RNA: an old modification with a novel epigenetic function. Genomics Proteom Bioinform. 2013;11(1):8–17. doi: 10.1016/j.gpb.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128(4):707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 83.Voss TC, Hager GL. Dynamic regulation of transcriptional states by chromatin and transcription factors. Nat Rev Genet. 2014;15(2):69–81. doi: 10.1038/nrg3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr Protoc Mol Biol. 2015;109:21. doi: 10.1002/0471142727.mb2129s109. 9 1- 9 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liu D, Zhang XX, Xi BX, Wan DY, Li L, Zhou J. Sine oculis homeobox homolog 1 promotes DNA replication and cell proliferation in cervical cancer. Int J Oncol. 2014;45(3):1232–1240. doi: 10.3892/ijo.2014.2510. [DOI] [PubMed] [Google Scholar]
- 86.Yuan B, Liao F, Shi ZZ, Ren Y, Deng XL, Yang TT. Dihydroartemisinin inhibits the proliferation, colony formation and induces ferroptosis of lung cancer cells by inhibiting PRIM2/SLC7A11 Axis. Onco Targets Ther. 2020;13:10829–10840. doi: 10.2147/OTT.S248492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yan M, Rerko RM, Platzer P, Dawson D, Willis J, Tong M. 15-Hydroxyprostaglandin dehydrogenase, a COX-2 oncogene antagonist, is a TGF-beta-induced suppressor of human gastrointestinal cancers. Proc Natl Acad Sci U S A. 2004;101(50):17468–17473. doi: 10.1073/pnas.0406142101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gee JR, Montoya RG, Khaled HM, Sabichi AL, Grossman HB. Cytokeratin 20, AN43, PGDH, and COX-2 expression in transitional and squamous cell carcinoma of the bladder. Urol Oncol. 2003;21(4):266–270. doi: 10.1016/s1078-1439(02)00271-5. [DOI] [PubMed] [Google Scholar]
- 89.Yao S, Xu J, Zhao K, Song P, Yan Q, Fan W. Down-regulation of HPGD by miR-146b-3p promotes cervical cancer cell proliferation, migration and anchorage-independent growth through activation of STAT3 and AKT pathways. Cell Death Dis. 2018;9(11):1055. doi: 10.1038/s41419-018-1059-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Huang X, Taeb S, Jahangiri S, Korpela E, Cadonic I, Yu N. miR-620 promotes tumor radioresistance by targeting 15-hydroxyprostaglandin dehydrogenase (HPGD) Oncotarget. 2015;6(26):22439–22451. doi: 10.18632/oncotarget.4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.He Q, Chen Z, Dong Q, Zhang L, Chen D, Patel A. MicroRNA-21 regulates prostaglandin E2 signaling pathway by targeting 15-hydroxyprostaglandin dehydrogenase in tongue squamous cell carcinoma. BMC Cancer. 2016;16(1):685. doi: 10.1186/s12885-016-2716-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kawamata H, Furihata T, Omotehara F, Sakai T, Horiuchi H, Shinagawa Y. Identification of genes differentially expressed in a newly isolated human metastasizing esophageal cancer cell line, T.Tn-AT1, by cDNA microarray. Cancer Sci. 2003;94(8):699–706. doi: 10.1111/j.1349-7006.2003.tb01505.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bernau K, Torr EE, Evans MD, Aoki JK, Ngam CR, Sandbo N. Tensin 1 is essential for myofibroblast differentiation and extracellular matrix formation. Am J Respir Cell Mol Biol. 2017;56(4):465–476. doi: 10.1165/rcmb.2016-0104OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhou H, Zhang Y, Wu L, Xie W, Li L, Yuan Y. Elevated transgelin/TNS1 expression is a potential biomarker in human colorectal cancer. Oncotarget. 2018;9(1):1107–1113. doi: 10.18632/oncotarget.23275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Duan J, Wang L, Shang L, Yang S, Wu H, Huang Y. miR-152/TNS1 axis promotes non-small cell lung cancer progression through Akt/mTOR/RhoA pathway. BioSci Rep. 2021;41(1) doi: 10.1042/BSR20201539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liu JW, Yu F, Tan YF, Huo JP, Liu Z, Wang XJ. Profiling of tumor microenvironment components identifies five stroma-related genes with prognostic implications in colorectal cancer. Cancer Biother Radiopharm. 2020 doi: 10.1089/cbr.2020.4118. https://www.liebertpub.com/doi/10.1089/cbr.2020.4118 [DOI] [PubMed] [Google Scholar]
- 97.Phanhthilath N, Hakim S, Su C, Liu A, Subramonian D, Lesperance J. Mechanisms of efficacy of the FGFR1-3 inhibitor AZD4547 in pediatric solid tumor models. Invest New Drugs. 2020;38(6):1677–1686. doi: 10.1007/s10637-020-00933-2. [DOI] [PubMed] [Google Scholar]
- 98.Huynh H, Ong R, Soo KC. Foretinib demonstrates anti-tumor activity and improves overall survival in preclinical models of hepatocellular carcinoma. Angiogenesis. 2012;15(1):59–70. doi: 10.1007/s10456-011-9243-z. [DOI] [PubMed] [Google Scholar]
- 99.Blair HA. Fedratinib: first approval. Drugs. 2019;79(15):1719–1725. doi: 10.1007/s40265-019-01205-x. [DOI] [PubMed] [Google Scholar]
- 100.Harrison CN, Schaap N, Vannucchi AM, Kiladjian JJ, Tiu RV, Zachee P. Janus kinase-2 inhibitor fedratinib in patients with myelofibrosis previously treated with ruxolitinib (JAKARTA-2): a single-arm, open-label, non-randomised, phase 2, multicentre study. Lancet Haematol. 2017;4(7):e317–ee24. doi: 10.1016/S2352-3026(17)30088-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Liu LW, Hsieh YY, Yang PM. Bioinformatics data mining repurposes the JAK2 (Janus Kinase 2) inhibitor fedratinib for treating pancreatic ductal adenocarcinoma by reversing the KRAS (Kirsten Rat Sarcoma 2 viral oncogene homolog)-driven gene signature. J Pers Med. 2020;10(3):130. doi: 10.3390/jpm10030130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhong WZ, Wang Q, Mao WM, Xu ST, Wu L, Wei YC. Gefitinib versus vinorelbine plus cisplatin as adjuvant treatment for Stage II-IIIA (N1-N2) EGFR-Mutant NSCLC: final overall survival analysis of CTONG1104 Phase III trial. J Clin Oncol. 2021;39(7):713–722. doi: 10.1200/JCO.20.01820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lorusso V, Latorre A, Giotta F. Chemotherapy options beyond the first line in HER-negative metastatic breast cancer. J Oncol. 2020;2020 doi: 10.1155/2020/9645294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Liu S, Wen J, Yang H, Li Q, Chen Y, Zhu C. Recurrence patterns after neoadjuvant chemoradiotherapy compared with surgery alone in oesophageal squamous cell carcinoma: results from the multicenter phase III trial NEOCRTEC5010. Eur J Cancer. 2020;138:113–121. doi: 10.1016/j.ejca.2020.08.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.