Abstract
Scavenger receptor class A (SR-A) proteins are type II transmembrane glycoproteins that form homotrimers on the cell surface. This family has five known members (SCARA1 to 5, or SR-A1 to A5) that recognize a variety of ligands and are involved in multiple biological pathways. Previous reports have shown that some SR-A family members can bind modified low-density lipoproteins (LDLs); however, the mechanisms of the interactions between the SR-A members and these lipoproteins are not fully understood. Here, we systematically characterize the recognition of SR-A receptors with lipoproteins and report that SCARA1 (SR-A1, CD204), MARCO (SCARA2), and SCARA5 recognize acetylated or oxidized LDL and very-low-density lipoprotein in a Ca2+-dependent manner through their C-terminal scavenger receptor cysteine-rich (SRCR) domains. These interactions occur specifically between the SRCR domains and the modified apolipoprotein B component of the lipoproteins, suggesting that they might share a similar mechanism for lipoprotein recognition. Meanwhile, SCARA4, a SR-A member with a carbohydrate recognition domain instead of the SRCR domain at the C terminus, shows low affinity for modified LDL and very-low-density lipoprotein but binds in a Ca2+-independent manner. SCARA3, which does not have a globular domain at the C terminus, was found to have no detectable binding with these lipoproteins. Taken together, these results provide mechanistic insights into the interactions between SR-A family members and lipoproteins that may help us understand the roles of SR-A receptors in lipid transport and related diseases such as atherosclerosis.
Keywords: scavenger receptor class A, lipoproteins, SRCR domain, SCARA1, MARCO, SCARA3, SCARA4, SCARA5, apolipoprotein B (apoB)
Abbreviations: Ac-apoB, acetylated apoB; AcHDL, acetylated HDL; AcLDL, acetylated LDL; AcVLDL, acetylated VLDL; apoB, apolipoprotein B; BSA, bovine serum albumin; CC, coiled-coil; CL, collagen-like; FACS, fluorescence-activated cell sorting; FBS, fetal bovine serum; HDLs, high-density lipoproteins; LDLs, low-density lipoproteins; Ox-apoB, oxidized apoB; OxHDL, oxidized HDL; OxLDL, oxidized LDL; OxPC, oxidized phosphatidylcholine; OxVLDL, oxidized VLDL; SR, scavenger receptor; SR-A, scavenger receptor class A; SRCR, scavenger receptor cysteine-rich; VLDLs, very-low-density lipoproteins
The scavenger receptor (SR) was first identified in late 1970s because of its binding with the modified low-density lipoproteins (LDLs) (1). Over the past decades, a large number of SRs have been found with wide tissue distributions and play important roles in maintaining homeostasis and host defense by recognizing both self- and non–self-ligands (2, 3, 4), including proteins, carbohydrates (5, 6), and lipids (7, 8), and they are also associated with many diseases (9, 10, 11) such as autoimmune diseases (12), cardiovascular diseases (11, 13, 14), and cancer (9, 10, 12, 15).
SRs have been classified into more than ten subfamilies or classes based on the sequence and structural similarities (10, 16). Among them, scavenger receptor class A (SR-A) has five known members, including SCARA1 (CD204, SR-A, SR-A1, MSR, etc.), MARCO (SR-A2, SCARA2), SCARA3 (SR-A3, CSR), SCARA4 (SR-A4, COLEC12, SRCL, etc.), and SCARA5 (SR-A5, TESR) (17). SCARA1 and MARCO are highly expressed on macrophages and dendritic cells (18). SCARA4 is expressed on endothelial cells (19), granulocytes, and neutrophils (20). SCARA5 has relatively wide tissue distribution and mainly restricted to epithelial cells (21). The distribution of SCARA3 is still unclear, but its expression might be associated with UV irradiation and oxidative stress (22).
SR-A members are type II transmembrane proteins containing an N-terminal cytoplasmic region, a transmembrane helix, and a large C-terminal extracellular portion (Fig. 1A). The ectodomains of SR-A members form homotrimers on membrane surface and may have a coiled-coil (CC) region, a collagen-like (CL) region, and a C-terminal globular domain (17, 23, 24) (Fig. 1A). But each member has different structural features. The ectodomains of SCARA1 and SCARA5 contain a CC region, a CL region, and a scavenger receptor cysteine-rich (SRCR) domain (17, 24, 25). The ectodomain of MARCO is similar to that of SCARA1 and SCARA5 but lacks the CC region (26). SCARA4 also contains a CC region and a CL region but has a carbohydrate-recognition domain (CRD) at the C terminus instead of the SRCR domain (27, 28). The extracellular portion of SCARA3 only has a CC region and a CL region without the C-terminal globular domain (17, 23) (Fig. 1A).
Figure 1.
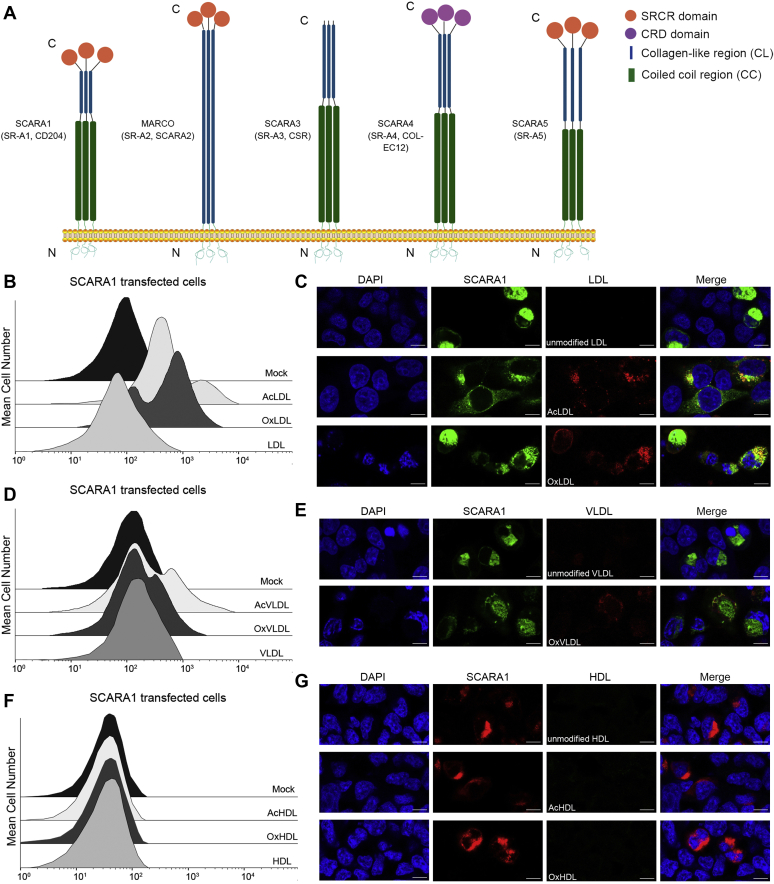
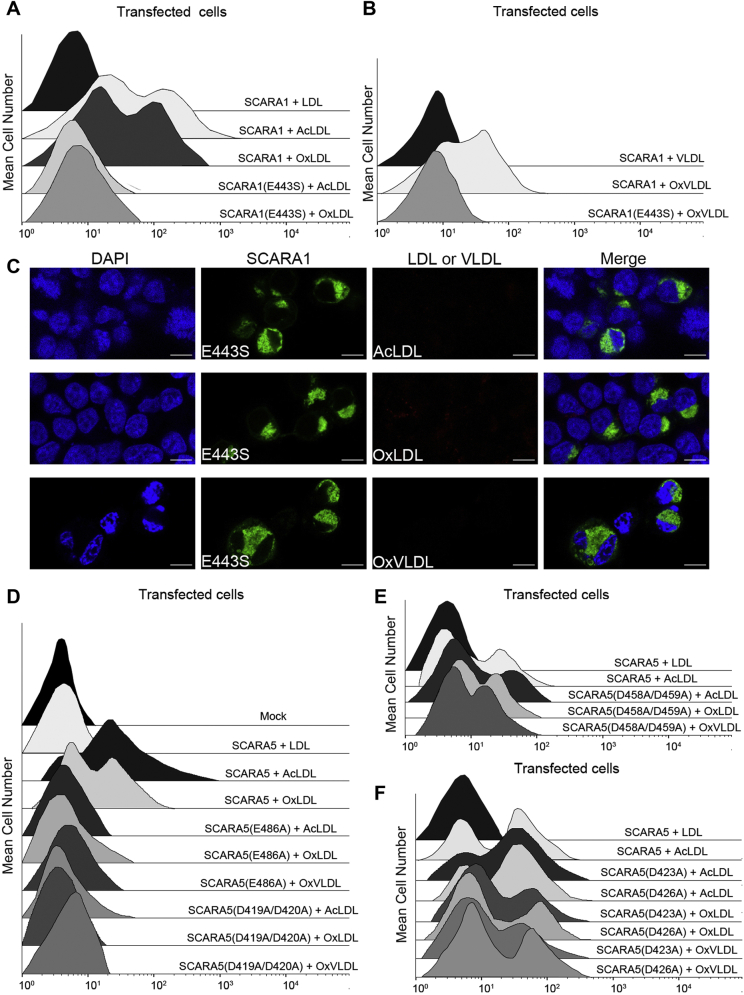
SCARA1 recognizes the modified LDL and VLDL.A, schematic models of the SR-A family members. B, FACS data of the hSCARA1-transfected HEK293 cells incubated with AcLDL, OxLDL, or LDL. Mock represents nontransfected cells. C, confocal fluorescent images of the hSCARA1 (with GFP tag)-transfected HEK293 cells incubated with LDL, AcLDL, or OxLDL (the scale bar represents 25 μm). D, FACS data of the hSCARA1-transfected HEK293 cells incubated with AcVLDL, OxVLDL, or VLDL. E, confocal fluorescent images of the hSCARA1 (with GFP-tag)-transfected HEK293 cells incubated with VLDL or OxVLDL (the scale bar represents 25 μm). F, FACS data of the hSCARA1-transfected HEK293 cells incubated with AcHDL, OxHDL, or HDL. G, confocal fluorescent images of the hSCARA1 (with mCherry tag)-transfected cells incubated with HDL, AcHDL, or OxHDL (the scale bar represents 25 μm). AcHDL, acetylated HDL; AcVLDL, acetylated VLDL; HDL, high-density lipoprotein; LDL, low-density lipoprotein; OxHDL, oxidized HDL; OxVLDL, oxidized VLDL; VLDL, very-low-density lipoprotein.
Despite the structural similarities, the functions of SR-A members appear to be diverse (2, 26, 29, 30, 31, 32, 33), and they are associated with a variety of diseases (9, 34, 35). Previous studies have shown that the ligand binding of the SR-A members usually occurs at the C-terminal globular domain. For example, the SRCR domains of SCARA1, MARCO, and SCARA5 have been reported to recognize different ligands such as spectrin (32), modified LDL (26), and ferritin (31); SCARA4 is able to bind the leukocyte cell surface glycan Lewis(x) via its C-terminal CRD (27). But the mechanisms of the receptor–ligand interactions of SR-A members are not fully understood.
Lipids in the human body such as cholesterol, phospholipids, and triglycerides are mainly transported by lipoproteins (36, 37, 38), which are roughly classified into five types: chylomicrons, very-low-density lipoproteins (VLDLs), intermediate-density lipoproteins, LDLs, and high-density lipoproteins (HDLs) according to their density and composition (37, 39, 40). Lipoproteins have a core of triglycerides surrounded by a layer of phospholipids, cholesterol, and apolipoproteins (41, 42). The main phospholipid components are phosphatidylcholine and sphingomyelin (42). Apolipoprotein B (apoB) is the primary structural component of LDLs, which is also found in VLDLs and intermediate-density lipoproteins, and required for the secretion of the lipoprotein particles (43, 44). HDLs have multiple apolipoproteins including apoA, apoC, and apoE (45). Lipoproteins are mainly regulated by the receptors such as the low-density lipoprotein receptor, very-low-density lipoprotein receptor, and high-density lipoprotein receptor, respectively (46, 47, 48, 49), and are used clinically to assess the risk of cardiovascular diseases (50, 51).
Lipoproteins may undergo modifications such as acetylation and oxidation and generate acetylated LDL (AcLDL) and oxidized LDL (OxLDL), respectively (52, 53). AcLDL can be generated in vivo by cholesterol acetyltransferase (54) and also be made in vitro (55). OxLDL can be prepared in vitro in the presence of micromolar concentrations of copper or iron (56, 57), which may also produce oxidized phosphatidylcholine (OxPC) during the process (58). The mechanisms regarding the generation of OxLDL in vivo are not entirely clear; one possibility is that LDL could be oxidized by ferritin in lysosomes where iron is released at acidic pH and induces LDL oxidation (59, 60). Excessive modified LDL could lead to the accumulation of cholesterol on the arterial wall, which may cause cardiovascular diseases such as atherosclerosis and other related diseases (61, 62, 63).
Over the past decades, several SRs from different classes have been found as receptors for the modified LDLs (64, 65, 66, 67, 68), but the mechanisms are not well understood. Here, we characterize the recognition of lipoproteins by the SR-A family members using biochemical and biophysical assays and identify the mechanisms of the interactions, which may help understand the roles of SR-A members in lipid transport and the related diseases.
Results
SCARA1 recognizes the modified LDL and VLDL through the C-terminal SRCR domain
SCARA1 has been reported to be able to internalize AcLDL and OxLDL (64, 69, 70), but the mechanism was unclear. Here, we transfected HEK293 cells with the full-length human SCARA1 (Fig. 1A), and the transfected cells were fed with the modified (acetylated or oxidized) or the unmodified LDL. The fluorescence-activated cell sorting (FACS) results showed that the SCARA1-transfected cells only bound to AcLDL and OxLDL, rather than the unmodified LDL (Fig. 1B). In parallel, we also tested the binding with VLDL and the results showed that the SCARA1-transfected cells only bound to acetylated VLDL (AcVLDL) and oxidized VLDL (OxVLDL) and had no binding to the unmodified VLDL (Fig. 1D), similar to the results of LDL. Furthermore, confocal microscopy was applied to monitor the internalization of LDL and VLDL, and the resulting images showed that only AcLDL and OxLDL could be internalized by the SCARA1 (fused with GFP tag) transfected cells (Fig. 1C). And it was also true for the modified VLDL particles (Fig. 1E), confirming that SCARA1 was a receptor for the modified LDL and VLDL. In parallel, the modified and the unmodified HDL particles were also applied for FACS and confocal microscopy, but no binding was detected for the SCARA1 (fused with mCherry tag)-transfected cells (Fig. 1, F and G), suggesting that SCARA1 was not a receptor for HDL, consistent with the results published before (17).
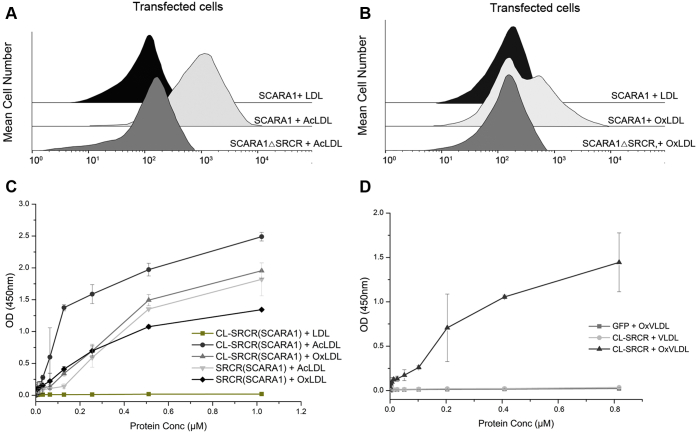
To identify the binding region of SCARA1 with the modified LDL, we transfected HEK293 cells with the full-length human SCARA1 and a deletion mutant without the C-terminal SRCR domain (SCARA1ΔSRCR), respectively. The transfected cells were incubated with LDL samples similarly as described above. The FACS results showed that unlike the cells transfected with the full-length SCARA1 that bound to both AcLDL and OxLDL, the SCARA1ΔSRCR-transfected cells had no binding to the modified LDL under similar conditions (Fig. 2, A and B), suggesting that the SRCR domain played a key role in recognizing the modified LDL. In addition, we expressed the CL-SRCR fragment and the SRCR domain of SCARA1 for ELISA. The data showed that both AcLDL and OxLDL were able to bind the purified fragments (Fig. 2C), and similar binding results were obtained for the modified VLDL (Fig. 2D), confirming that SCARA1 recognized the modified LDL and VLDL through its C-terminal SRCR domain. Previous data have shown that the ectodomain of SCARA1 could form homophilic trimers (18, 32, 71). Indeed, the ELISA results showed that the CL-SRCR fragment of SCARA1, which forms homotrimers (32), had stronger binding affinities to the modified LDL than the monomeric SRCR domain (Fig. 2C).
Figure 2.
SCARA1 recognizes the modified LDL and VLDL through its C-terminal SRCR domain.A, FACS data of the full-length hSCARA1 or the hSCARA1ΔSRCR-transfected HEK293 cells incubated with LDL or AcLDL. B, FACS data of the full-length hSCARA1 or the hSCARA1ΔSRCR-transfected HEK293 cells incubated with LDL or OxLDL. C, ELISA data of the interactions between the CL-SRCR fragment (homotrimer) and the SRCR domain (monomer) of hSCARA1 with the unmodified LDL or the modified LDL in the presence of Ca2+ (2 mM). D, ELISA data of the interactions between the CL-SRCR fragment of hSCARA1 with the unmodified VLDL and the modified VLDL in the presence of Ca2+ (2 mM). GFP was applied as a negative control. ELISA data are representative of three repeated experiments and presented as the mean ± SD. AcLDL, acetylated LDL; CL, collagen-like; LDL, low-density lipoprotein; OxLDL, oxidized LDL; SRCR, scavenger receptor cysteine-rich; VLDL, very-low-density lipoprotein.
MARCO and SCARA5 also interact with the modified LDL and VLDL via the SRCR domains
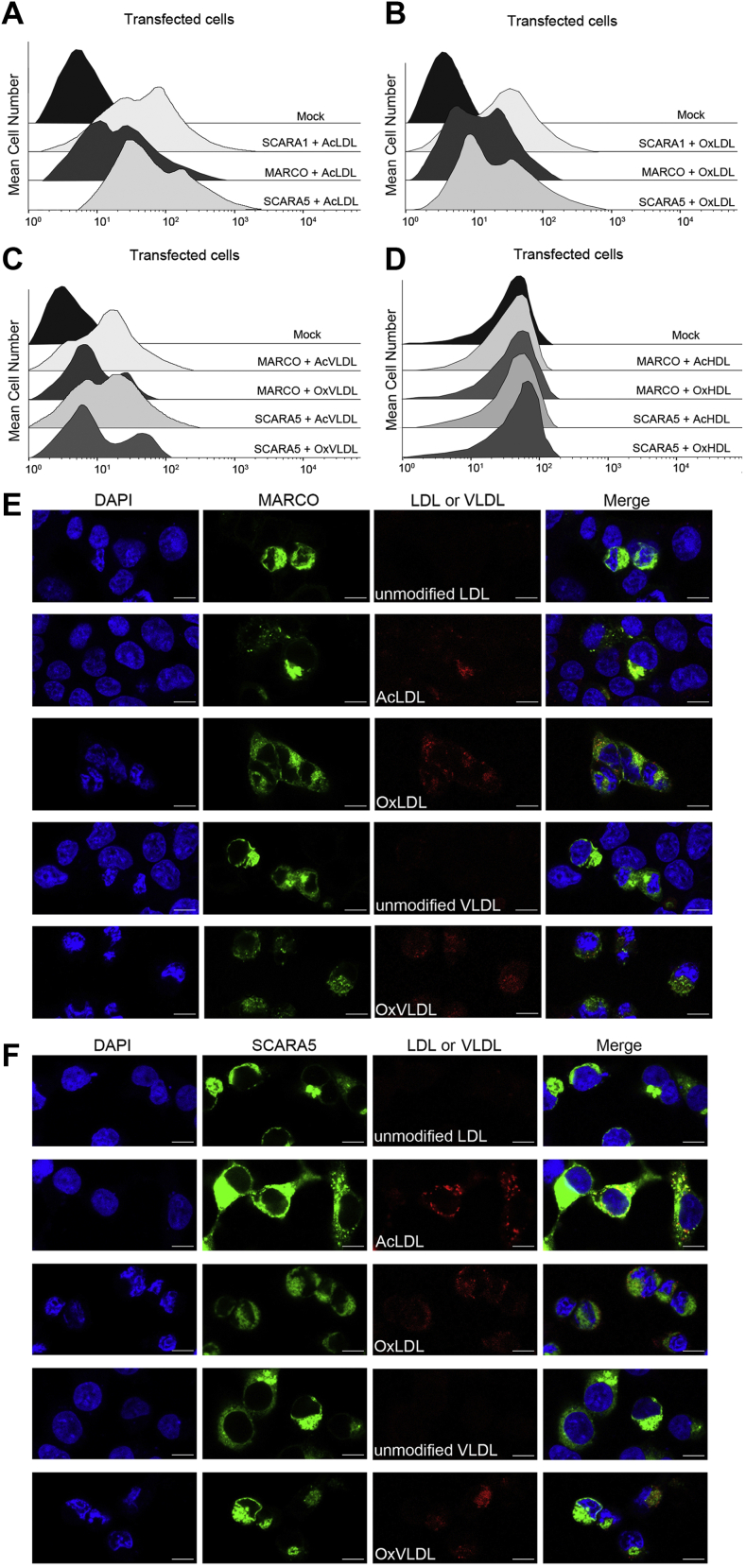
SCARA1, MARCO, and SCARA5 are the three SR-A members with the C-terminal SRCR domains (Fig. 1A). It has been shown that MARCO could bind AcLDL through the SRCR domain (26), but mouse SCARA5 was reported having no binding activity to the modified LDL (21). Here, we transfected HEK293 cells with human MARCO and SCARA5, respectively, and the FACS data showed that both MARCO and SCARA5 had binding signals with the modified LDL and VLDL, rather than the unmodified LDL and VLDL (Fig. 3, A–C). Moreover, both the modified and the unmodified LDL and VLDL were fed to the transfected cells for visualizing the internalization of the lipoproteins by confocal microscopy. The fluorescent images showed that both MARCO- and SCARA5-transfected cells could internalize the modified LDL and VLDL (Fig. 3, E and F), consistent with the FACS results. In parallel, we also tested the interactions of the modified HDL with MARCO and SCARA5, but no binding was detected for both of the receptors (Fig. 3D), which was similar to the results of SCARA1 (Fig. 1F). Because the structures of the SRCR domains of human and mouse SCARA5 are highly similar (31), the interactions between mSCARA5 and lipoproteins might need to be further explored.
Figure 3.
MARCO and SCARA5 interact with the modified LDL and VLDL via the SRCR domains.A, FACS data for the binding of AcLDL with the hSCARA1-, hMARCO-, or hSCARA5-transfected cells. Mock represents nontransfected cells. B, FACS data for the binding of OxLDL with the hSCARA1-, hMARCO-, or hSCARA5-transfected cells. C, FACS data for the binding of AcVLDL or OxVLDL with the hSCARA1-, hMARCO-, or the hSCARA5-transfected cells. D, FACS data of the hMARCO- or hSCARA5-transfected cells incubated with AcHDL or OxHDL. E, confocal fluorescent images of the hMARCO (with GFP tag) transfected cells incubated with the modified or unmodified LDL or VLDL (the scale bar represents 25 μm). F, confocal fluorescent images of the hSCARA5 (with GFP tag)-transfected cells incubated with the modified or unmodified LDL or VLDL (the scale bar represents 25 μm). AcHDL, acetylated HDL; AcLDL, acetylated LDL; AcVLDL, acetylated VLDL; LDL, low-density lipoprotein; OxHDL, oxidized HDL; OxLDL, oxidized LDL; OxVLDL, oxidized VLDL; SRCR, scavenger receptor cysteine-rich; VLDL, very-low-density lipoprotein.
Then, we expressed the SRCR domains of MARCO and SCARA5 fused with GFP in insect cells, and the purified proteins were applied for ELISA. The results showed that both SRCR domains exhibited binding activities to AcLDL (Fig. 4A), and the binding affinity of the SRCR domain of SCARA5 with AcLDL was similar to that of SCARA1, but stronger than that of MARCO (Fig. 4A). Similar binding characteristics were also observed for OxLDL (Fig. 4B) and OxVLDL (Fig. 4C). These results suggested that the SRCR domains of SCARA1, MARCO, and SCARA5 could all recognize the modified LDL and VLDL, and MARCO had relatively weaker affinities among the three molecules.
Figure 4.
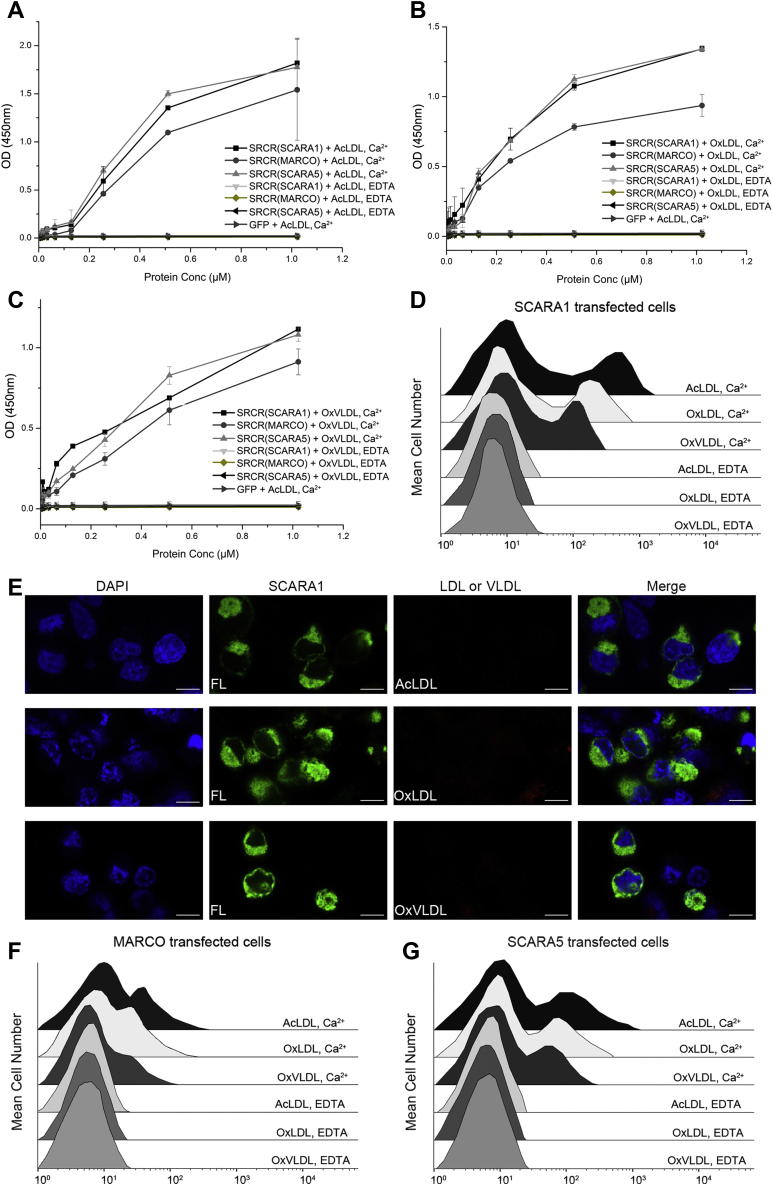
The interactions between the SRCR domains of SCARA1, MARCO, and SCARA5 with the modified LDL and VLDL are Ca2+dependent.A, ELISA data for the interactions of the SRCR domains of hSCARA1, hMARCO, or hSCARA5 with AcLDL in the presence of Ca2+ (2 mM) or EDTA (1 mM). B, ELISA data for the interactions of the SRCR domains of hSCARA1, hMARCO, or hSCARA5 with OxLDL in the presence of Ca2+ (2 mM) or EDTA (1 mM). C, ELISA data for the interactions of the SRCR domains of hSCARA1, hMARCO, or hSCARA5 with OxVLDL in the presence of Ca2+ (2 mM) or EDTA (1 mM). D, FACS data of the hSCARA1-transfected cells incubated with the modified LDL or VLDL in the presence of Ca2+ or EDTA. E, confocal fluorescent images of the hSCARA1 (with GFP-tag)-transfected cells incubated with the modified LDL or VLDL in the presence of EDTA (the scale bar represents 25 μm). F, FACS data of the hMARCO-transfected cells incubated with the modified LDL or VLDL in the presence of Ca2+ or EDTA. G, FACS data of the hSCARA5-transfected cells incubated with the modified LDL or VLDL in the presence of Ca2+ or EDTA. ELISA data are representative of three repeated experiments and presented as the mean ± SD. AcLDL, acetylated LDL; LDL, low-density lipoprotein; OxLDL, oxidized LDL; OxVLDL, oxidized VLDL; SRCR, scavenger receptor cysteine-rich; VLDL, very-low-density lipoprotein.
Interactions of SCARA1, MARCO, and SCARA5 with the modified LDL and VLDL are Ca2+ dependent
Previous reports have shown that the Ca2+-binding sites were important for the ligand binding of the SRCR domains (31, 32). To examine the roles of Ca2+ in the recognition of lipoproteins, FACS was applied, and the results showed that both the modified LDL and VLDL could bind SCARA1, MARCO, and SCARA5 in the presence of Ca2+, and the binding was eliminated by the addition of EDTA (Fig. 4, D, F and G). ELISA results also confirmed that the SRCR domains of SCARA1, MARCO, and SCARA5 could interact with the modified LDL and VLDL in the presence of Ca2+ and the interactions were abolished by EDTA (Fig. 4, A–C), indicating the importance of cation for the recognition. Furthermore, confocal microscopy showed that the cells transfected with SCARA1, MARCO, or SCARA5 were not able to internalize the modified LDL and VLDL by the addition of EDTA (Fig. 4E and Fig. S2), confirming the necessity of Ca2+ in the recognition of the modified LDL and VLDL.
Since the structure of the SRCR domain of human SCARA1 was unavailable, we expressed the domain (residues 348–451) in insect cells and purified by Ni-NTA affinity chromatography and size-exclusion chromatography (Fig. S1A). The purified fragment was crystalized and the structure was determined by molecular replacement using the SRCR domain of mouse SCARA1 (PDB entry: 6J02) (32) as a search model and refined to 2.0 Å resolution (Table S1). The SRCR domain of human SCARA1 had a typical SRCR fold (Fig. S1B), similar to other SRCR domains published before (25, 26, 32, 72). The sequence identity between the SRCR domains of human and mouse SCARA1 was 84%. Superposition of the structures of the two domains showed high similarities with RMSD of 0.43 Å for the backbones and 0.26 Å for the Cα atoms (Fig. S1B). The SRCR domain of hSCARA1 contained only one cation-binding site, which was modeled as Ca2+ as it was included in the sample buffer and coordinated with residues D376, D377, and E443 (Fig. S1C and Fig. 5).
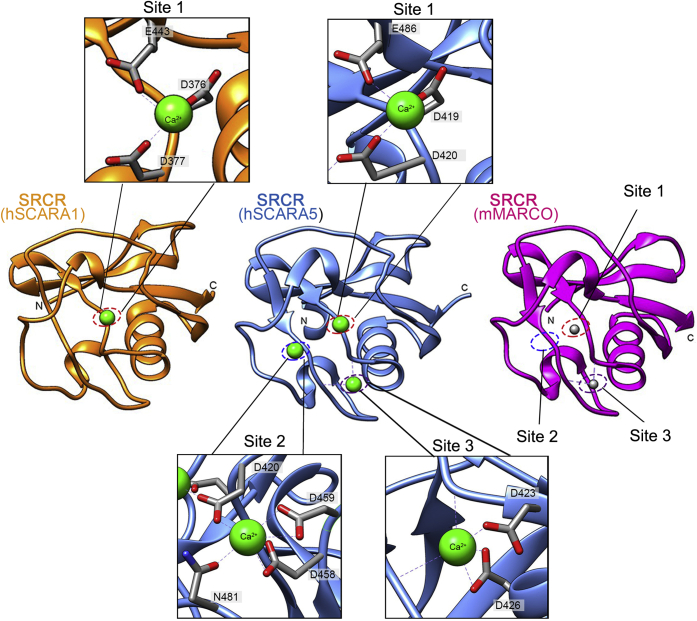
Figure 5.
Comparison of the crystal structures of the SRCR domains of SCARA1, SCARA5, and MARCO. Crystal structures of the SRCR domains of hSCARA1 (orange), hSCARA5 (blue, PDB ID:7C00), and mMARCO (magenta, PDB ID: 2OY3) are shown as ribbon diagrams. The cation-binding sites (dashed ovals) of hSCARA1 and hSCARA5 are zoomed in at insets (Ca2+ are shown as green spheres; Mg2+ are shown as gray spheres), respectively. SRCR, scavenger receptor cysteine-rich.
The SRCR domains of SCARA1, MARCO, and SCARA5 showed similar structural features, but differed in some loop regions (Fig. 5). Among them, the SRCR domains of SCARA1 and SCARA5 showed higher structural similarities with each other than with the SRCR domain of MARCO, which was consistent with the relatively higher sequence identity between the two molecules (31). The crystal structure of the SRCR domain of human SCARA5 showed three Ca2+-binding sites referred as site 1 (D419, D420, and E486), site 2 (D458, D459, N481, and D420) and site 3 (D423 and D426) (Fig. 5) (31). The only Ca2+-binding site of SCARA1 corresponded to the site 1 of SCARA5. MARCO had similar residues for the three binding sites with SCARA5, and two cations (Mg2+) were found at the binding site 1 and 3 in the crystal structure of mouse MARCO (Fig. 5) (26).
To further explore the roles of the Ca2+-binding sites on lipoprotein recognition, several single or double mutants of the Ca2+-coordinating residues were constructed, and the expression of the mutants on the cell surface was verified by FACS using antibodies against the WT receptors (Fig. S3, C and D). For SCARA1, a residue at the Ca2+-binding site was mutated (E443S), and the binding data showed that the mutant had no interactions with the modified LDL and VLDL in the presence of Ca2+ (Fig. 6, A and B). Similarly, the confocal images showed that the mutant was not able to internalize the modified LDL or VLDL particles (Fig. 6C), suggesting that the Ca2+-binding site was important for the recognition of the modified lipoproteins.
Figure 6.
Mutagenesis studies of the interactions between the SRCR domains of the SR-A members and the modified LDL and VLDL.A, FACS data of the WT or the mutant E443S of hSCARA1-transfected cells incubated with the unmodified or modified LDL in the presence of Ca2+. B, FACS data of the WT or the mutant E443S of hSCARA1-transfected cells incubated with the unmodified or modified VLDL in the presence of Ca2+. C, confocal fluorescent images of the mutant E443S of hSCARA1 (with GFP tag)-transfected cells incubated with the modified LDL or VLDL in the presence of Ca2+ (the scale bar represents 25 μm). D, FACS data of the mutant E486A or D419A/D420A of hSCARA5-transfected cells incubated with AcLDL, OxLDL, or OxVLDL in the presence of Ca2+. Mock represents nontransfected cells. E, FACS data of the mutant D458A/D459A of hSCARA5-transfected cells incubated with AcLDL, OxLDL, or OxVLDL in the presence of Ca2+. F, FACS data of the mutant D423A or D426A of hSCARA5-transfected cells incubated with AcLDL, OxLDL, or OxVLDL in the presence of Ca2+. AcLDL, acetylated LDL; LDL, low-density lipoprotein; OxLDL, oxidized LDL; OxVLDL, oxidized VLDL; SRCR, scavenger receptor cysteine-rich; VLDL, very-low-density lipoprotein.
The crystal structure of the SRCR domain of SCARA5 showed three Ca2+-binding sites (31); therefore, we constructed several mutants including E486A and D419A/D420A for site 1, D458A/D459A for site 2, D423A and D426A for site 3 (Fig. 5). The FACS results showed that the mutants for site 1, E486A and D419A/D420A, could reduce the interactions with the modified lipoproteins significantly (Fig. 6D), suggesting that this site was crucial for the interactions with the modified lipoproteins. Meanwhile, the mutant D458A/D459A for site 2 (Fig. 6E) and the mutants D423A and D426A for site 3 (Fig. 6F) had no obvious effect to the binding, suggesting that the Ca2+-binding sites 2 and 3 might not be involved in the recognition of the modified lipoproteins.
The SRCR domains of SCARA1, MARCO, and SCARA5 bind to the modified LDL and VLDL through apoB
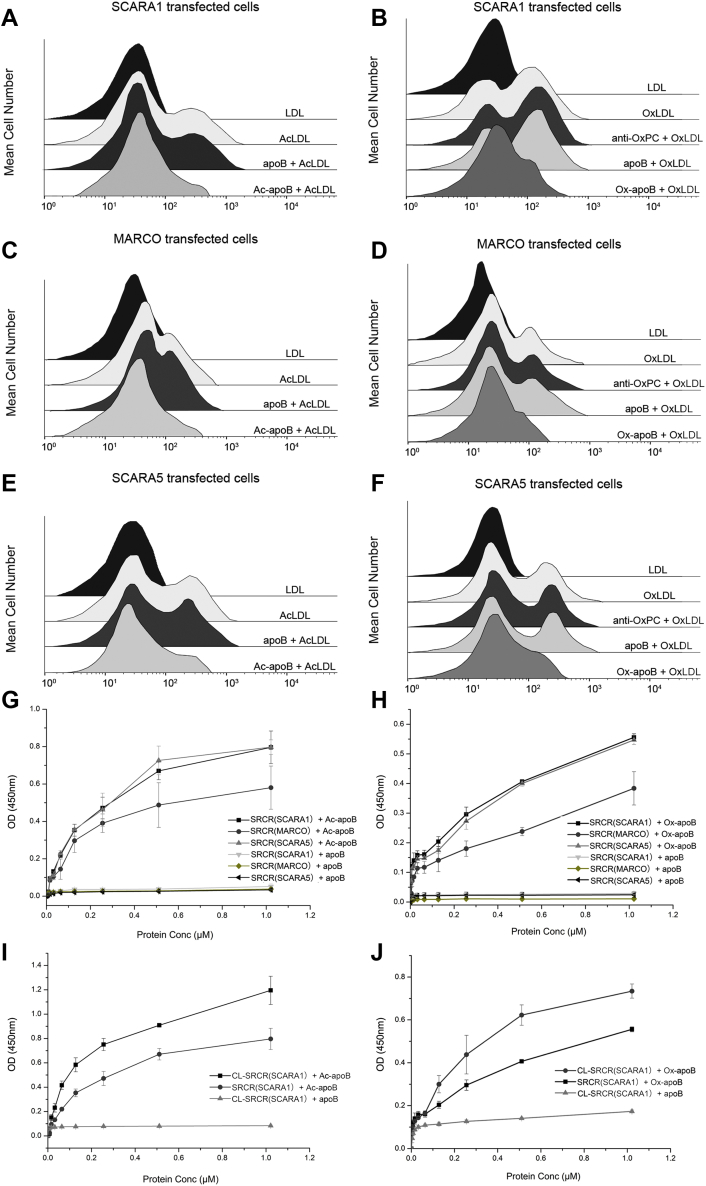
ApoB is the major apolipoprotein found in LDLs and VLDLs, but not in HDLs (44). Previous research showed that the recognition and transportation of LDLs by the low-density lipoprotein receptor might be achieved through apolipoproteins (73, 74). Here, we generated acetylated apoB (Ac-apoB) and oxidized apoB (Ox-apoB) in vitro using the similar procedures for preparing AcLDL (55) and OxLDL, respectively, and the AcLDL prepared in vitro showed similar binding activity with SCARA1 as the AcLDL isolated from human plasma (Fig. S3A). The FACS data showed that both Ac-apoB and Ox-apoB could inhibit the binding of SCARA1 with the modified LDL efficiently, but the unmodified apoB had no inhibition to the binding (Fig. 7, A and B). Similar inhibitory effects of Ac-apoB and Ox-apoB were also observed for the binding of the modified LDL with MARCO (Fig. 7, C and D) and SCARA5 (Fig. 7, E and F). ELISA results also confirmed that the SRCR domains of SCARA1, MARCO, and SCARA5 had binding activities with Ac-apoB and Ox-apoB, rather than the unmodified apoB (Fig. 7, G and H). Notably, the interactions of the SRCR domains of SCARA1 and SCARA5 with the modified apoB were stronger than that of MARCO (Fig. 7, G and H), consistent with the binding results for the modified LDL shown above (Fig. 4, A–C). In addition, the trimeric CL-SRCR fragment of SCARA1 also showed stronger binding affinities with Ac-apoB (Fig. 7I) and Ox-apoB (Fig. 7J) than the monomeric SRCR domain, similar to the binding of the modified LDL (Fig. 2C), indicating that the trimeric form could increase the binding affinity efficiently.
Figure 7.
SCARA1, MARCO, and SCARA5 interact with the modified lipoproteins via apoB.A, FACS data for the inhibition of Ac-apoB on the interaction of hSCARA1 with AcLDL. ApoB was applied as a control. B, FACS data for the inhibition of the Ox-apoB or anti-OxPC antibody on the interaction of hSCARA1 with OxLDL. C, FACS data for the inhibition of Ac-apoB on the interaction of hMARCO with AcLDL. D, FACS data for the inhibition of the Ox-apoB or anti-OxPC antibody on the interaction of hMARCO with OxLDL. E, FACS data for the inhibition of Ac-apoB on the interaction of hSCARA5 with AcLDL. F, FACS data for the inhibition of the Ox-apoB or anti-OxPC antibody on the interaction of hSCARA5 with OxLDL. G, ELISA data for the interactions of the SRCR domains of hSCARA1, hMARCO, or hSCARA5 with Ac-apoB or apoB. H, ELISA data for the interactions of the SRCR domains of hSCARA1, hMARCO, or hSCARA5 with Ox-apoB or apoB. I, ELISA data for the interactions of the trimeric CL-SRCR fragment or the monomeric SRCR domain of hSCARA1 with Ac-apoB. J, ELISA data for the interactions of the trimeric CL-SRCR fragment or the monomeric SRCR domain of hSCARA1 with Ox-apoB. ELISA data are representative of three repeated experiments and presented as the mean ± SD. AcLDL, acetylated LDL; CL, collagen-like; OxLDL, oxidized LDL; OxPC, oxidized phosphatidylcholine; SRCR, scavenger receptor cysteine-rich.
Because OxPC might also be generated during lipoprotein oxidation (58), we examined the role of lipids of lipoproteins in the interactions with SCARA1, MARCO, and SCARA5. The dot-blot assay showed that the anti-OxPC antibody could bind to the OxLDL prepared in vitro rather than the unmodified LDL (Fig. S3B), suggesting that OxPC was produced during the oxidation of LDL. However, the FACS data showed that the anti-OxPC antibody had no obvious inhibition to the binding of OxLDL with SCARA1, MARCO, and SCARA5 (Fig. 7, B, D and F), confirming that apoB was the major binding target of SCARA1, MARCO, and SCARA5 on the modified LDL, and lipids might not have a direct role on the recognition of the modified lipoproteins.
The interactions of lipoproteins with SCARA3 and SCARA4
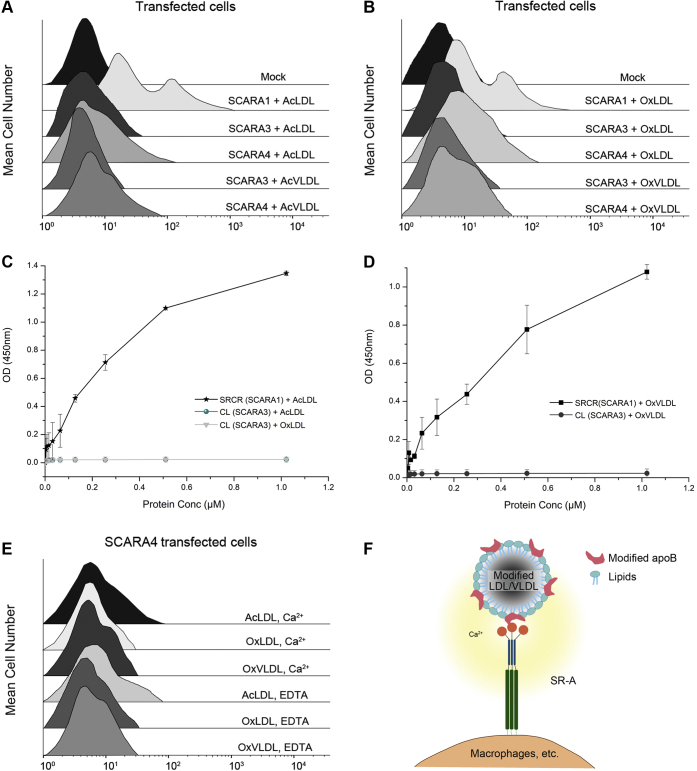
SCARA3 and SCARA4, the two SR-A members without the C-terminal SRCR domain (Fig. 1A), were also tested for lipoprotein binding. SCARA3 is unique among the SR-A members as it does not have a globular domain at the C-terminal end (Fig. 1A). The HEK293 cells transfected with SCARA3 or SCARA4 were fed with the modified LDL or VLDL following the similar procedures described above. The FACS data showed that SCARA3 had no binding with the modified lipoproteins (Fig. 8, A and B), and the ELISA results also confirmed that the CL fragment of SCARA3 had no binding affinity to AcLDL, OxLDL, or OxVLDL (Fig. 8, C and D). By contrast, the SCARA4-transfected cells showed weak binding with the modified LDL and VLDL (Fig. 8, A and B), but the binding was not affected by the addition of EDTA (Fig. 8E), suggesting the interactions were cation independent. This could be consistent with the previous reports showing that the binding of SCARA4 to the modified lipoproteins might occur through the CL region of the ectodomain rather than the C-terminal CRD (19, 75), thereby having a different mechanism with the Ca2+-dependent binding of SCARA1, MARCO, and SCARA5.
Figure 8.
Interactions of SCARA3 and SCARA4 with lipoproteins and a model for lipoprotein recognition by the SR-A members.A, FACS data of the hSCARA3- or hSCARA4-transfected cells incubated with AcLDL or AcVLDL. Mock represents nontransfected cells. B, FACS data of the hSCARA3- or hSCARA4-transfected cells incubated with OxLDL or OxVLDL. C, ELISA data for the interactions of the CL region of hSCARA3 with AcLDL or OxLDL in the presence of Ca2+ (2 mM). The SRCR domain of SCARA1 was applied as a control. D, ELISA data for the interactions of the CL region of hSCARA3 with OxVLDL in the presence of Ca2+ (2 mM). E, FACS data of the hSCARA4-transfected cells incubated with the modified LDL or VLDL in the presence of Ca2+ or EDTA. F, a cartoon representation of the recognition of the modified LDL/VLDL by the SR-A members SCARA1, MARCO, and SCARA5. ELISA data are representative of three repeated experiments and presented as the mean ± SD. AcLDL, acetylated LDL; AcVLDL, acetylated VLDL; apoB, apolipoprotein B; CL, collagen-like; LDL, low-density lipoprotein; OxLDL, oxidized LDL; OxVLDL, oxidized VLDL; SRCR, scavenger receptor cysteine-rich; VLDL, very-low-density lipoproteins.
Discussion
The SR-A members share similar structural features, but their ligands appear to be rather diverse (29, 30, 33). For SCARA1, the reported ligands include the modified LDL (1, 69), spectrin (32), ferritin (31), and factor F(x) (14). The interactions of SCARA1 with the modified LDL/VLDL, spectrin, and ferritin are all through its C-terminal SRCR domain, which is commonly found in SRs and usually contains divalent cation-binding sites for Ca2+ or Mg2+ that are associated with ligand recognition (76). Indeed, the Ca2+-binding site on the SRCR domain of SCARA1 is crucial for the binding with the modified LDL/VLDL and this site is also important for recognizing spectrin (32) and ferritin (31), suggesting that it might be a conserved mechanism for scavenging multiple targets. For SCARA5, it has the highest sequence identity with SCARA1 in the SR-A family and also retains the Ca2+-binding site, it is not surprising that human SCARA5 also interacts with the modified LDL/VLDL as SCARA1, and a similar binding mechanism is adopted for ferritin recognition (31). Therefore, SCARA1 and SCARA5 could be the two receptors sharing similar scavenging functions but having different tissue distributions. For MARCO, it also recognizes the modified LDL/VLDL in a Ca2+-dependent manner with relatively lower affinities compared with SCARA1 and SCARA5, which is similar to the situation when it interacts with ferritin (31), and the lower affinities for the ligands might be due to the different surface profile of MARCO (31). Because Ca2+ is crucial for the ligand recognition of these receptors, it is likely that there are clusters of residues such as Asp/Glu on the surface of ligands, which can coordinate with Ca2+ and mediate the interactions, but the details of the interactions need to be elucidated in the future. A distinct feature of SR-A family members is that they form homotrimers on the cell surface (16, 71), and trimerization can increase binding affinities with the ligands (32), this appears to be true for the recognition of the modified LDL/VLDL.
LDL particles have a core of triglyceride and cholesteryl ester molecules and a surface layer containing phospholipids and apolipoproteins (42, 43). It has been shown that oxidation of LDLs may lead to the modification of both apoB, the major apolipoprotein on the surface of the LDL/VLDL, and phospholipids (58). The data in this study suggest that the modified apoB is the binding target for SCARA1, MARCO, and SCARA5, and OxPC does not affect the interactions with the three SR-A members (Fig. 8F).
Atherosclerosis is known to be associated with the accumulation of plaques, which mainly composed of lipids and inflammatory debris from various cells including macrophages, endothelial cells, monocytes, smooth muscle cells, and lymphocytes on the arterial wall (77, 78, 79, 80, 81). Previous data have shown that SR-A members might be involved in the clearance of dead cells (9, 23, 32). Among them, SCARA1 can recognize dead cells by the interaction with spectrin, which is exposed during cell death (32). It is noteworthy that SCARA1 utilizes the same Ca2+-binding site on the SRCR domain for recognizing both spectrin and the modified LDL/VLDL; therefore, SCARA1 may have dual roles in the internalization of dead cells or cell debris and the modified lipoproteins. Interestingly, recent studies show that SCARA1, SCARA5, and MARCO can also recognize ferritin through the Ca2+-binding sites on the SRCR domains, and SCARA1 and SCARA5 have relatively high affinities with ferritins (31). Previous evidence has suggested that LDL could be oxidized by ferritin in lysosomes at acidic pH (59, 60). Therefore, it may not be a coincidence that the SR-A members can recognize these targets with the similar binding sites, especially for SCARA1, which is highly expressed on macrophages and known to be associated with atherosclerosis (8, 82, 83). In fact, previous reports have shown that lipoproteins, iron, and dead cells are linked in a number of diseases (84, 85, 86), implying that these receptors may be involved in the physiological or pathological processes. However, the exact roles of SCARA1 and other SRs in the diseases appear to be complex (87, 88) and have not been fully understood. The mechanistic information regarding the interactions between the SR-A members and lipoproteins identified in this study may help elucidate their roles in lipid transport and the related diseases in the future.
Experimental procedures
Protein expression and purification
The cDNAs of human SCARA1 to SCARA5 (GenBank: NM_138715.2, NM_006770, NM_016240.2, NM_130386 and NM_286133) were purchased from Sino Biological Inc or synthesized by Sangon Biotech. The CL-SRCR region of SCARA1 contains a CL region (residue 263–347 for human SCARA1) and an SRCR domain (residue 348–451 for human SCARA1). Constructs encoding the GFP-tagged SRCR domain and CL-SRCR region of SCARA1, SRCR domain of MARCO, CL region of SCARA3, and SRCR domain of SCARA5 were all subcloned into the pFastBac vectors (Invitrogen) with a melittin signal sequence and an N-terminal 8x His-tag for expression in insect cells. Sf9 cells were used for generating recombinant baculoviruses and High Five cells were used for protein production (Invitrogen). The baculovirus-infected cells were cultured for 3 days in a 27 °C humidified incubator. The supernatants of the infected High Five cells were buffer-exchanged with 50 mM Tris and 150 mM NaCl at pH 8.0 and then applied to Ni-NTA chromatography following the manufacturer's instructions (Ni-NTA Superflow, GE Healthcare). The imidazole eluates were further purified by gel-filtration chromatography with a Superdex 200 Increase 10/300 GL column (GE Healthcare).
The full-length SCARA1 to SCARA5 were subcloned into the pTT5 expression vectors fused with an mCherry tag or GFP tag at the N termini, respectively. The constructs were transiently transfected into HEK293F cells cultured with FreeStyle 293 Expression Medium (Gibco) in a humidified CO2 incubator at 37 °C for 24 h before flow cytometry and confocal microscopic analysis.
Crystallization and structural determination
The SRCR domain of hSCARA1 purified from the insect cell supernatant was buffer exchanged to a sample buffer (10 mM Tris, 150 mM NaCl, 2 mM CaCl2, pH 7.4) and concentrated to 10 mg/ml. Crystal screening was performed by the hanging-drop vapor diffusion method, and crystals were obtained at 16 °C in a solution containing 0.1 M Hepes (pH 7.5) and 20% PEG 8000. Diffraction data were collected at BL18U beamline at Shanghai Synchrotron Radiation Facility and processed using the HKL-3000 package (89). The structure was solved by molecular replacement using the structure of the SRCR domain of mouse SCARA1 (PDB entry: 6J02) as a search model with program PHASER (90). Coot (91) and PHENIX (92) were used for structural refinement. Structural superposition and the RMSD calculation between the SRCR domains of human and mouse SCARA1 were done using PyMOL 2.4.1 (93). The crystallographic statistics are listed in Table S1. Figures were made using UCSF Chimera (94).
Preparation of the modified lipoproteins
Lipoproteins (purity, 97%–98%), including Dil-LDL (20614ES76), Dil-AcLDL (20606ES76), Dil-OxLDL (20609ES76), LDL (20613ES05), AcLDL (20604ES05), VLDL (20617ES05), and HDL (20610ES05) were purchased from Yeasen. Purified human apoB (purity >95%) (MD-26-0010P) was purchased from RayBiotech. The lipoproteins mentioned above (LDL, AcLDL, VLDL, HDL) were isolated from human plasma.
AcLDL, AcVLDL, acetylated HDL (AcHDL), and Ac-apoB were also prepared in vitro following the protocol published before (55). Briefly, 0.3 ml of 0.15 M NaCl containing 5 mg of LDL/VLDL/HDL/apoB was added to 0.3 ml of a saturated solution of sodium acetate with continuous stirring in an ice-water bath. Then, acetic anhydride was added in multiple small aliquots over a period of 1 h with continuous stirring. After the addition of acetic anhydride equal to 1.5 times of protein used, the mixture was stirred for an additional 30 min. The reaction solution was then dialyzed against the buffer containing 0.15 M NaCl and 0.3 mM EDTA, pH 7.4, at 4 °C for 24 h.
OxLDL, OxVLDL, oxidized HDL (OxHDL), and Ox-apoB were prepared in vitro. Briefly, LDL/VLDL/HDL/apoB was diluted in a buffer containing 150 mM NaCl and 25 mM Tris, pH 7.4. Then, 1 μl of 1 M CuSO4 was diluted and divided into multiple small portions and added to the LDL/VLDL/HDL/apoB solutions gradually. The reaction solutions were then dialyzed against the buffer containing 150 mM NaCl and 25 mM Tris, pH 7.4, at 4 °C for 12 h.
Dot-blot assay
For OxPC detection assay, LDL, OxLDL, and phosphatidylcholine (840053C, Avanti Polar Lipids) were spotted onto nitrocellulose membranes (Whatman) according to manufacturer’s instructions. The membranes were air-dried at room temperature (RT) for 2 h and blocked with the blocking buffer (25 mM Hepes, 150 mM NaCl, 5% (w/v) bovine serum albumin (BSA), 0.1% Tween 20, pH 7.4) for at least 1 h. Then, the membranes were incubated with the mouse OxPC antibody (mouse E06 monoclonal antibody) (330001S, Avanti Polar Lipids, 330001S) for 1 h and then incubated with the goat anti-mouse IgG secondary antibody HRP conjugates (SAB, L3032-2) for 1 h and detected with the DAB reagent. Between every two steps, the membranes were washed six times (5 min each) with the washing buffer (25 mM Hepes, 150 mM NaCl, and 0.1% Tween 20, pH 7.4).
Flow cytometry
For the binding assays of hSCARA1 or hSCARA1ΔSRCR with lipoproteins, HEK293 cells were transiently transfected with the full-length hSCARA1 or hSCARA1ΔSRCR fused with a GFP tag. After 24 h, 5-μg Dil-tagged (wavelength: 565 nm) lipoprotein (Dil-LDL, Dil-AcLDL, Dil-OxLDL, Dil-VLDL, Dil-AcVLDL, Dil-OxVLDL) was added to the culture media containing 2 mM Ca2+. After 2 to 4 h, cells were washed three times with the washing buffer (25 mM Hepes, 150 mM NaCl, 0.1% Tween 20, 2 mM CaCl2, pH 7.4) and then washed twice with the cleaning buffer (25 mM Hepes, 150 mM NaCl, and 2 mM CaCl2, pH 7.4) for flow cytometry.
For the binding assays of hMARCO/hSCARA3 to 5 with lipoproteins, HEK293 cells were transiently transfected with the full-length hMARCO/hSCARA3 to 5 fused with an mCherry tag. After 24 h, 5 μg of lipoprotein (LDL, AcLDL, OxLDL, VLDL, AcVLDL, and OxVLDL) was added to the culture media containing 2 mM Ca2+. After 2 to 4 h, cells were washed and blocked similarly as described above. Then, cells were stained with the anti-apoB antibody (ab7616, Abcam) at RT for 1 h. After washing three times with the washing buffer (25 mM Hepes, 150 mM NaCl, 1% (w/v) BSA, 0.1% Tween 20, and 2 mM CaCl2, pH 7.4), the Donkey Anti-Goat IgG H&L (FITC) (ab6881, Abcam) was added and incubated for 1 h. After washing twice with the washing buffer (25 mM Hepes, 150 mM NaCl, 0.1% Tween 20, and 2 mM CaCl2, pH 7.4) and once with the cleaning buffer (25 mM Hepes, 150 mM NaCl, 2 mM CaCl2, pH 7.4), the cells were used for flow cytometry.
For the binding assays with HDL, HEK293 cells were transiently transfected with the full-length hSCARA1/hMARCO/hSCARA5 fused with an mCherry tag. After 24 h, 5-μg HDL/AcHDL/OxHDL was added to the culture media containing 2 mM Ca2+. After 2 to 4 h, cells were washed and blocked similarly as described above and then stained with the anti-apoA (for HDL) antibody (ab92487, Abcam) at RT for 1 h. After washing three times with the washing buffer (25 mM Hepes, 150 mM NaCl, 1% (w/v) BSA, 0.1% Tween 20, and 2 mM CaCl2, pH 7.4), the Donkey Anti-Goat IgG H&L (FITC) (ab6881, Abcam) was added and incubated for 1 h. After washing with the similar buffers described above, the cells were used for flow cytometry.
For the Ca2+-dependent binding assays, HEK293 cells were transiently transfected with the full-length hSCARA1/hMARCO/hSCARA5 fused with a GFP tag. After 24 h, 5-μg untagged lipoprotein (LDL, AcLDL, OxLDL, VLDL, and OxVLDL) was added to the culture media containing 2 mM Ca2+ or 1 mM EDTA. After 2 to 4 h, the cells were washed twice with the corresponding cleaning buffer (25 mM Hepes, 150 mM NaCl, 2 mM CaCl2, or 1 mM EDTA). Then, cells were blocked in the blocking buffer (25 mM Hepes, 150 mM NaCl, 5% (w/v) BSA, 0.1% Tween 20, 2 mM CaCl2, or 1 mM EDTA, pH 7.4) for 1 h and stained with the anti-apoB antibody (ab7616, Abcam) at RT for 1 h. After washing three times with the corresponding washing buffer (25 mM Hepes, 150 mM NaCl, 1% (w/v) BSA, 0.1% Tween 20, 2 mM CaCl2, or 1 mM EDTA, pH 7.4), the Donkey Anti-Goat IgG H&L (Alexa Fluor 594) (Abcam, ab150132) was added and incubated for 1 h. After washing twice with the corresponding washing buffer mentioned above and once with the cleaning buffer (25 mM Hepes, 150 mM NaCl, 2 mM CaCl2, or 1 mM EDTA, pH 7.4), the cells were used for flow cytometry.
For monitoring the expression of the receptors and their mutants, HEK293 cells were transiently transfected with the constructs including the WT hSCARA1/5, the mutants of hSCARA1(E443S and hSCARA1ΔSRCR), and the mutants of hSCARA5 (D419A/D420A, E486A, D458A/D459A, D423A, and D426A). After 24 h, the cells were stained with anti-SCARA1 antibody (ab217843, Abcam) and goat anti-rabbit IgG H&L Alexa Fluor 594 (ab150080, Abcam) for SCARA1 and its mutants or anti-SCARA5 antibody (ab106439, Abcam) and goat anti-rabbit IgG H&L Alexa Fluor 488 (ab150077, Abcam) for SCARA5 and its mutants following the similar procedures described above for flow cytometry.
For the binding assays of the mutants of hSCARA1/5 with lipoproteins, HEK293 cells were transiently transfected with the full-length hSCARA1/5 or the mutants of hSCARA1 (E443S and hSCARA1ΔSRCR) or the mutants of hSCARA5 (D419A/D420A, E486A, D458A/D459A, D423A, and D426A) fused with an mCherry tag. After 24 h, 5-μg untagged lipoprotein (LDL, AcLDL, OxLDL, VLDL, and OxVLDL) was added to the culture media containing 2 mM Ca2+. After 2 to 4 h, the cells were stained with the anti-apoB antibody (ab7616, Abcam) and Donkey Anti-Goat IgG H&L (Alexa Fluor 594) (ab150132, Abcam) following the similar procedures described above for flow cytometry.
For the apoB competitive binding assays, transfected HEK293 cells were incubated with the apoB or modified apoB (10 μg/ml) overnight and then 5 μg Dil-tagged lipoprotein (Dil-LDL, Dil-AcLDL, Dil-OxLDL) was added to the cells. After 3 h of incubation, the cells were washed three times with the washing buffer (25 mM Hepes, 150 mM NaCl, 0.5% Tween, and 2 mM CaCl2, pH 7.4) and then washed twice with the buffer (25 mM Hepes, 150 mM NaCl, and 2 mM CaCl2, pH 7.4) for flow cytometry.
For OxPC antibody inhibition assays, HEK293 cells were transfected with the GFP-tagged SCARA1, MARCO, and SCARA5, respectively. Then, 5 μg OxLDL prepared in vitro was incubated with 5 μg OxPC antibody (mouse E06 monoclonal antibody) (330001S, Avanti Polar Lipids) (95, 96) and added to the cells. After 3 h of incubation, the cells were blocked in the blocking buffer (25 mM Hepes, 150 mM NaCl, 5% (w/v) BSA, 0.1% Tween 20, and 2 mM CaCl2, pH 7.4) for 1 h and then stained with the anti-apoB (for LDL) antibody (ab7616, Abcam) at RT for 1 h. After washing three times with the washing buffer (25 mM Hepes, 150 mM NaCl, 1% (w/v) BSA, 0.1% Tween 20, 2 mM CaCl2, pH 7.4), the Donkey Anti-Goat IgG H&L Alexa Fluor 594 (ab150132, Abcam) was added and incubated for 1 h. After washing similarly as described above, the cells were used for flow cytometry.
FACS data were acquired using a Becton Dickinson FACSCalibur flow cytometer with CELLQuest software. Data analysis was performed using FlowJo software (Tree Star, Inc).
ELISA experiments
Lipoprotein (LDL, AcLDL, OxLDL) was coated onto 96-well plates with 1-μg protein per well at 4 °C overnight. The plates were then blocked with the blocking buffer (25 mM Hepes, 150 mM NaCl, 0.1% Triton X-100, and 5% (w/v) BSA, pH 7.4) at 37 °C for 3 h. The purified GFP-tagged mutant (the CL-SRCR fragment of SCARA1, the SRCR domain of SCARA1/MARCO/SCARA5, and the CL region of SCARA3) was serially diluted and added to each well in the binding buffer (25 mM Hepes, 150 mM NaCl, 2 mM CaCl2 or 1 mM EDTA, 0.1% Triton X-100, and 2 mg/ml BSA, pH 7.4). After 2 h of incubation at 37 °C, the plates were washed five times with the washing buffer (25 mM Hepes, 150 mM NaCl, 10 mM CaCl2 or 1 mM EDTA, 0.1% Triton X-100, pH 7.4) and then incubated with the mouse anti-GFP antibody (ab184601, Abcam) for 1 h. After washing with the washing buffer (25 mM Hepes, 150 mM NaCl, 10 mM CaCl2 or 1 mM EDTA, and 0.1% Triton X-100, pH 7.4), the plates were incubated with the goat anti-mouse IgG secondary antibody HRP conjugates (L3032-2, SAB) for 1 h. After washing three times with the washing buffer again, 100 μl of chromogenic substrate (1 μg/ml tetramethylbenzidine, 0.006% H2O2 in 0.05 M phosphate citrate buffer, pH 5.0) was added to each well and incubated for 30 min at 37 °C. Then, 50-μl H2SO4 (2.0 M) was added to each well to stop the reactions. The plates were read at 450 nm on a Synergy Neo machine (BioTek Instruments).
For the interactions of the SR-A mutants (the CL-SRCR fragment and the SRCR domain of SCARA1, the SRCR domains of MARCO and SCARA5) with apolipoproteins (apoB, Ac-apoB, Ox-apoB), about 0.5 μg of the purified apolipoprotein was coated onto each well of 96-well plates, and the proteins were serially diluted and added to each well. Mouse anti-GFP antibody (ab184601, Abcam) and goat anti-mouse IgG secondary antibody HRP conjugates (L3032-2, SAB) were used for binding detection following the similar procedures described above.
Confocal microscopy
HEK293 cells were transfected with the full-length SCARA1/MARCO/SCARA5 or the mutant of SCARA1(E443S) fused with GFP using 6-well plates. After 24 h of transfection, 5 μg of untagged lipoprotein (LDL, AcLDL, OxLDL, VLDL, and OxVLDL) was added to the plates with 2 mM Ca2+ or 1 mM EDTA. After 2 to 4 h of incubation, the cells were fixed by 4% paraformaldehyde in TBS (50 mM Tris and 150 mM NaCl, pH 7.4). After washing with the buffer (25 mM Hepes, 150 mM NaCl, 10 mM CaCl2, or 1 mM EDTA at pH 7.4), the cells were permeabilized with 0.25% Triton X-100 in the buffer (25 mM Hepes, 150 mM NaCl, 10 mM CaCl2, or 1 mM EDTA at pH 7.4). Then, cells were blocked in the blocking buffer (25 mM Hepes, 150 mM NaCl, 3% (w/v) BSA, 5% fetal bovine serum (FBS), 0.1% Triton X-100, and 2 mM CaCl2, pH 7.4) or (25 mM Hepes, 150 mM NaCl, 3% (w/v) BSA, 5% FBS, 0.1% Triton X-100, and 1 mM EDTA, pH 7.4) for 1 h and stained with the anti-apoB antibody (ab7616, Abcam) at RT for 1 h. After washing three times with the washing buffer (25 mM Hepes, 150 mM NaCl, 1% (w/v) BSA, 0.1% Triton X-100, 2 mM CaCl2, pH 7.4) or (25 mM Hepes, 150 mM NaCl, 1% (w/v) BSA, 0.1% Triton X-100, and 1 mM EDTA, pH 7.4), Donkey Anti-Goat IgG H&L (Alexa Fluor 594) (ab150132, Abcam) was added and incubated for 1 h. After washing three times with 0.1% Triton X-100 in the buffer (25 mM Hepes, 150 mM NaCl, 10 mM CaCl2, or 1 mM EDTA at pH 7.4), the cells were washed with the cleaning buffer (25 mM Hepes, 150 mM NaCl, 2 mM CaCl2, or 1 mM EDTA at pH 7.4) and incubated with 5 μM DAPI for 30 min. Then, the plates were washed again for confocal microscopy with a Leica SP8 microscope.
For the HDL-binding assay, HEK293 cells were transfected with the full-length hSCARA1 fused with an mCherry tag using 6-well plates. After 24 h of transfection, 5 μg of untagged lipoprotein (HDL, AcHDL, and OxHDL) was added to the plates with 2 mM Ca2+. After 2 to 4 h of incubation, the cells were fixed by 4% paraformaldehyde in TBS (50 mM Tris and 150 mM NaCl, pH 7.4). After washing with the buffer (25 mM Hepes, 150 mM NaCl, and 10 mM CaCl2 at pH 7.4), the cells were permeabilized with 0.25% Triton X-100 in the corresponding buffers mentioned above. Then, cells were blocked in the blocking buffer (25 mM Hepes, 150 mM NaCl, 3% (w/v) BSA, 5% FBS, 0.1% Triton X-100, and 2 mM CaCl2, pH 7.4) for 1 h and stained with the anti-apoA antibody (ab92487, Abcam) at RT for 1 h. After washing three times with the washing buffer (25 mM Hepes, 150 mM NaCl, 1% (w/v) BSA, 0.1% Triton X-100, and 2 mM CaCl2, pH 7.4), the Donkey Anti-Goat IgG H&L (FITC) (ab6881, Abcam) was added and incubated for 1 h. After washing three times with 0.1% Triton X-100 in the buffers (25 mM Hepes, 150 mM NaCl, and 10 mM CaCl2 at pH 7.4), the cells were washed with the cleaning buffer (25 mM Hepes, 150 mM NaCl, and 2 mM CaCl2 at pH 7.4) and incubated with 5 μM DAPI for 30 min. Then, the plates were washed again for confocal microscopy with a Leica SP8 microscope.
Data availability
The structure of the SRCR domain of human SCARA1 has been deposited in the PDB (www.rcsb.org) with PDB entry: 7DPX.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank the National Center for Protein Science Shanghai (The Integrated Laser Microscopy system and the Protein Expression and Purification system) for instrumental support and technical assistance. We also thank the beamline BL18U1 of National Facility for Protein Science Shanghai (NFPS) at Shanghai Synchrotron Radiation Facility for their assistance in X-ray diffraction data collection.
Author contributions
C. C. data curation; C. C. formal analysis; C. C. validation; C. C., E. Z., B. Y., Z. Z., and Y. W. investigation; C. C. and Y. H. methodology; C. C. and Y. H. writing–original draft; C. C. and Y. H. writing–review and editing; Y. L. and Y. H. resources; Y. L. and Y. H. supervision; Y. L. and Y. H. funding acquisition; Y. H. conceptualization; Y. H. project administration.
Funding and additional information
This work is supported by the National Natural Science Foundation of China (No. 91957102) to Y. H.
Edited by Dennis Voelker
Supporting information
References
- 1.Brown M.S., Goldstein J.L., Krieger M., Ho Y.K., Anderson R.G. Reversible accumulation of cholesteryl esters in macrophages incubated with acetylated lipoproteins. J. Cell Biol. 1979;82:597–613. doi: 10.1083/jcb.82.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peiser L., Gough P.J., Kodama T., Gordon S. Macrophage class A scavenger receptor-mediated phagocytosis of Escherichia coli: Role of cell heterogeneity, microbial strain, and culture conditions in vitro. Infect. Immun. 2000;68:1953–1963. doi: 10.1128/iai.68.4.1953-1963.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pombinho R., Sousa S., Cabanes D. Scavenger receptors: Promiscuous players during microbial pathogenesis. Crit. Rev. Microbiol. 2018;44:685–700. doi: 10.1080/1040841X.2018.1493716. [DOI] [PubMed] [Google Scholar]
- 4.Wang D., Sun B., Feng M., Feng H., Gong W., Liu Q., Ge S. Role of scavenger receptors in dendritic cell function. Hum. Immunol. 2015;76:442–446. doi: 10.1016/j.humimm.2015.03.012. [DOI] [PubMed] [Google Scholar]
- 5.Cavallari J.F., Anhe F.F., Foley K.P., Denou E., Chan R.W., Bowdish D.M.E., Schertzer J.D. Targeting macrophage scavenger receptor 1 promotes insulin resistance in obese male mice. Physiol. Rep. 2018;6 doi: 10.14814/phy2.13930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taylor M.E., Drickamer K. Paradigms for glycan-binding receptors in cell adhesion. Curr. Opin. Cell Biol. 2007;19:572–577. doi: 10.1016/j.ceb.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 7.Shen W.J., Asthana S., Kraemer F.B., Azhar S. Scavenger receptor B type 1: Expression, molecular regulation, and cholesterol transport function. J. Lipid Res. 2018;59:1114–1131. doi: 10.1194/jlr.R083121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu X.H., Fu Y.C., Zhang D.W., Yin K., Tang C.K. Foam cells in atherosclerosis. Clin. Chim. Acta. 2013;424:245–252. doi: 10.1016/j.cca.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 9.Canton J., Neculai D., Grinstein S. Scavenger receptors in homeostasis and immunity. Nat. Rev. Immunol. 2013;13:621–634. doi: 10.1038/nri3515. [DOI] [PubMed] [Google Scholar]
- 10.PrabhuDas M.R., Baldwin C.L., Bollyky P.L., Bowdish D.M.E., Drickamer K., Febbraio M., Herz J., Kobzik L., Krieger M., Loike J., McVicker B., Means T.K., Moestrup S.K., Post S.R., Sawamura T. A consensus definitive classification of scavenger receptors and their roles in health and disease. J. Immunol. 2017;198:3775–3789. doi: 10.4049/jimmunol.1700373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stephen S.L., Freestone K., Dunn S., Twigg M.W., Homer-Vanniasinkam S., Walker J.H., Wheatcroft S.B., Ponnambalam S. Scavenger receptors and their potential as therapeutic targets in the treatment of cardiovascular disease. Int. J. Hypertens. 2010;2010:646929. doi: 10.4061/2010/646929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peiser L., Mukhopadhyay S., Gordon S. Scavenger receptors in innate immunity. Curr. Opin. Immunol. 2002;14:123–128. doi: 10.1016/s0952-7915(01)00307-7. [DOI] [PubMed] [Google Scholar]
- 13.Sahebi R., Hassanian S.M., Ghayour-Mobarhan M., Farrokhi E., Rezayi M., Samadi S., Bahramian S., Ferns G.A., Avan A. Scavenger receptor class B type I as a potential risk stratification biomarker and therapeutic target in cardiovascular disease. J. Cell. Physiol. 2019;234:16925–16932. doi: 10.1002/jcp.28393. [DOI] [PubMed] [Google Scholar]
- 14.Muczynski V., Bazaa A., Loubiere C., Harel A., Cherel G., Denis C.V., Lenting P.J., Christophe O.D. Macrophage receptor SR-AI is crucial to maintain normal plasma levels of coagulation factor X. Blood. 2016;127:778–786. doi: 10.1182/blood-2015-05-647032. [DOI] [PubMed] [Google Scholar]
- 15.Yu X.F., Guo C.Q., Fisher P.B., Subjeck J.R., Wang X.Y. Scavenger receptors: Emerging roles in cancer biology and immunology. Adv. Cancer Res. 2015;128:309–364. doi: 10.1016/bs.acr.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.PrabhuDas M., Bowdish D., Drickamer K., Febbraio M., Herz J., Kobzik L., Krieger M., Loike J., Means T.K., Moestrup S.K., Post S., Sawamura T., Silverstein S., Wang X.Y., El Khoury J. Standardizing scavenger receptor nomenclature. J. Immunol. 2014;192:1997–2006. doi: 10.4049/jimmunol.1490003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bowdish D.M., Gordon S. Conserved domains of the class A scavenger receptors: Evolution and function. Immunol. Rev. 2009;227:19–31. doi: 10.1111/j.1600-065X.2008.00728.x. [DOI] [PubMed] [Google Scholar]
- 18.Kelley J.L., Ozment T.R., Li C.F., Schweitzer J.B., Williams D.L. Scavenger receptor-A (CD204): A two-edged sword in health and disease. Crit. Rev. Immunol. 2014;34:241–261. doi: 10.1615/critrevimmunol.2014010267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ohtani K., Suzuki Y., Eda S., Kawai T., Kase T., Keshi H., Sakai Y., Fukuoh A., Sakamoto T., Itabe H., Suzutani T., Gasawara M., Yoshida I., Wakamiya N. The membrane-type collectin CL-P1 is a scavenger receptor on vascular endothelial cells. J. Biol. Chem. 2001;276:44222–44228. doi: 10.1074/jbc.M103942200. [DOI] [PubMed] [Google Scholar]
- 20.Graham S.A., Antonopoulos A., Hitchen P.G., Haslam S.M., Dell A., Drickamer K., Taylor M.E. Identification of neutrophil granule glycoproteins as Lewis(x)-containing ligands cleared by the scavenger receptor C-type lectin. J. Biol. Chem. 2011;286:24336–24349. doi: 10.1074/jbc.M111.244772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang Y., Oliver P., Davies K.E., Platt N. Identification and characterization of murine SCARA5, a novel class A scavenger receptor that is expressed by populations of epithelial cells. J. Biol. Chem. 2006;281:11834–11845. doi: 10.1074/jbc.M507599200. [DOI] [PubMed] [Google Scholar]
- 22.Han H.J., Tokino T., Nakamura Y. CSR, a scavenger receptor-like protein with a protective role against cellular damage causedby UV irradiation and oxidative stress. Hum. Mol. Genet. 1998;7:1039–1046. doi: 10.1093/hmg/7.6.1039. [DOI] [PubMed] [Google Scholar]
- 23.Whelan F.J., Meehan C.J., Golding G.B., McConkey B.J., Bowdish D.M.E. The evolution of the class A scavenger receptors. BMC Evol. Biol. 2012;12:227. doi: 10.1186/1471-2148-12-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kodama T., Freeman M., Rohrer L., Zabrecky J., Matsudaira P., Krieger M. Type I macrophage scavenger receptor contains alpha-helical and collagen-like coiled coils. Nature. 1990;343:531–535. doi: 10.1038/343531a0. [DOI] [PubMed] [Google Scholar]
- 25.Hohenester E., Sasaki T., Timpl R. Crystal structure of a scavenger receptor cysteine-rich domain sheds light on an ancient superfamily. Nat. Struct. Biol. 1999;6:228–232. doi: 10.1038/6669. [DOI] [PubMed] [Google Scholar]
- 26.Ojala J.R.M., Pikkarainen T., Tuuttila A., Sandalova T., Tryggvason K. Crystal structure of the cysteine-rich domain of scavenger receptor MARCO reveals the presence of a basic and an acidic cluster that both contribute to ligand recognition. J. Biol. Chem. 2007;282:16654–16666. doi: 10.1074/jbc.M701750200. [DOI] [PubMed] [Google Scholar]
- 27.Feinberg H., Taylor M.E., Weis W.I. Scavenger receptor C-type lectin binds to the leukocyte cell surface glycan Lewis(x) by a novel mechanism. J. Biol. Chem. 2007;282:17250–17258. doi: 10.1074/jbc.M701624200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakamura K., Funakoshi H., Miyamoto K., Tokunaga F., Nakamura T. Molecular cloning and functional characterization of a human scavenger receptor with C-type lectin (SRCL), a novel member of a scavenger receptor family. Biochem. Biophys. Res. Commun. 2001;280:1028–1035. doi: 10.1006/bbrc.2000.4210. [DOI] [PubMed] [Google Scholar]
- 29.de Winther M.P., van Dijk K.W., Havekes L.M., Hofker M.H. Macrophage scavenger receptor class A: A multifunctional receptor in atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2000;20:290–297. doi: 10.1161/01.atv.20.2.290. [DOI] [PubMed] [Google Scholar]
- 30.Platt N., Haworth R., Darley L., Gordon S. The many roles of the class A macrophage scavenger receptor. Int. Rev. Cytol. 2002;212:1–40. doi: 10.1016/s0074-7696(01)12002-4. [DOI] [PubMed] [Google Scholar]
- 31.Yu B., Cheng C., Wu Y., Guo L., Kong D., Zhang Z., Wang Y., Zheng E., Liu Y., He Y. Interactions of ferritin with scavenger receptor class A members. J. Biol. Chem. 2020;295:15727–15741. doi: 10.1074/jbc.RA120.014690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng C., Hu Z., Cao L., Peng C., He Y. The scavenger receptor SCARA1 (CD204) recognizes dead cells through spectrin. J. Biol. Chem. 2019;294:18881–18897. doi: 10.1074/jbc.RA119.010110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Platt N., Gordon S. Is the class A macrophage scavenger receptor (SR-A) multifunctional? - the mouse's tale. J. Clin. Invest. 2001;108:649–654. doi: 10.1172/JCI13903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xie L., Li Q., Dong R., Zhao K., Feng Y., Bao Z., Zhou M. Critical regulation of inflammation via class A scavenger receptor. Int. J. Chron. Obstruct. Pulmon. Dis. 2018;13:1145–1155. doi: 10.2147/COPD.S153326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y., Wei Y., Jiang B., Chen L., Bai H., Zhu X., Li X., Zhang H., Yang Q., Ma J., Xu Y., Ben J., Christiani D.C., Chen Q. Scavenger receptor A1 prevents metastasis of non-small cell lung cancer via suppression of macrophage serum amyloid A1. Cancer Res. 2017;77:1586–1598. doi: 10.1158/0008-5472.CAN-16-1569. [DOI] [PubMed] [Google Scholar]
- 36.Camejo G. The interaction of lipids and lipoproteins with the intercellular matrix of arterial tissue: Its possible role in atherogenesis. Adv. Lipid Res. 1982;19:1–53. doi: 10.1016/b978-0-12-024919-0.50007-2. [DOI] [PubMed] [Google Scholar]
- 37.Ginsberg H.N. Lipoprotein physiology. Endocrinol. Metab. Clin. North Am. 1998;27:503–519. doi: 10.1016/s0889-8529(05)70023-2. [DOI] [PubMed] [Google Scholar]
- 38.Illingworth D.R. Lipoprotein metabolism. Am. J. Kidney Dis. 1993;22:90–97. doi: 10.1016/s0272-6386(12)70173-7. [DOI] [PubMed] [Google Scholar]
- 39.Lutomski C.A., Gordon S.M., Remaley A.T., Jarrold M.F. Resolution of lipoprotein subclasses by charge detection mass spectrometry. Anal. Chem. 2018;90:6353–6356. doi: 10.1021/acs.analchem.8b01127. [DOI] [PubMed] [Google Scholar]
- 40.Okazaki M., Usui S., Ishigami M., Sakai N., Nakamura T., Matsuzawa Y., Yamashita S. Identification of unique lipoprotein subclasses for visceral obesity by component analysis of cholesterol profile in high-performance liquid chromatography. Arterioscler. Thromb. Vasc. Biol. 2005;25:578–584. doi: 10.1161/01.ATV.0000155017.60171.88. [DOI] [PubMed] [Google Scholar]
- 41.Mahley R.W., Innerarity T.L., Rall S.C., Jr., Weisgraber K.H. Plasma lipoproteins: Apolipoprotein structure and function. J. Lipid Res. 1984;25:1277–1294. [PubMed] [Google Scholar]
- 42.Hevonoja T., Pentikainen M.O., Hyvonen M.T., Kovanen P.T., Ala-Korpela M. Structure of low density lipoprotein (LDL) particles: Basis for understanding molecular changes in modified LDL. Biochim. Biophys. Acta. 2000;1488:189–210. doi: 10.1016/s1388-1981(00)00123-2. [DOI] [PubMed] [Google Scholar]
- 43.Elovson J., Chatterton J.E., Bell G.T., Schumaker V.N., Reuben M.A., Puppione D.L., Reeve J.R., Jr., Young N.L. Plasma very low density lipoproteins contain a single molecule of apolipoprotein B. J. Lipid Res. 1988;29:1461–1473. [PubMed] [Google Scholar]
- 44.Olofsson S.O., Asp L., Boren J. The assembly and secretion of apolipoprotein B-containing lipoproteins. Curr. Opin. Lipidol. 1999;10:341–346. doi: 10.1097/00041433-199908000-00008. [DOI] [PubMed] [Google Scholar]
- 45.Plump A.S., Scott C.J., Breslow J.L. Human apolipoprotein A-I gene expression increases high density lipoprotein and suppresses atherosclerosis in the apolipoprotein E-deficient mouse. Proc. Natl. Acad. Sci. U. S. A. 1994;91:9607–9611. doi: 10.1073/pnas.91.20.9607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Soutar A.K., Knight B.L. Structure and regulation of the LDL-receptor and its gene. Br. Med. Bull. 1990;46:891–916. doi: 10.1093/oxfordjournals.bmb.a072445. [DOI] [PubMed] [Google Scholar]
- 47.Strickland D.K., Gonias S.L., Argraves W.S. Diverse roles for the LDL receptor family. Trends Endocrinol. Metab. 2002;13:66–74. doi: 10.1016/s1043-2760(01)00526-4. [DOI] [PubMed] [Google Scholar]
- 48.Howell B.W., Herz J. The LDL receptor gene family: Signaling functions during development. Curr. Opin. Neurobiol. 2001;11:74–81. doi: 10.1016/s0959-4388(00)00176-8. [DOI] [PubMed] [Google Scholar]
- 49.Slotte J.P., Oram J.F., Bierman E.L. Binding of high density lipoproteins to cell receptors promotes translocation of cholesterol from intracellular membranes to the cell surface. J. Biol. Chem. 1987;262:12904–12907. [PubMed] [Google Scholar]
- 50.Kostner G.M. Lipoprotein receptors and atherosclerosis. Biochem. Soc. Trans. 1989;17:639–641. doi: 10.1042/bst0170639. [DOI] [PubMed] [Google Scholar]
- 51.Kruth H.S. Fluid-phase pinocytosis of LDL by macrophages: A novel target to reduce macrophage cholesterol accumulation in atherosclerotic lesions. Curr. Pharm. Des. 2013;19:5865–5872. doi: 10.2174/1381612811319330005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Landers S.C., Lewis J.C. acLDL binding and endocytosis by macrophages and macrophage foam cells in situ. Exp. Mol. Pathol. 1993;59:38–50. doi: 10.1006/exmp.1993.1025. [DOI] [PubMed] [Google Scholar]
- 53.Parthasarathy S., Steinberg D., Witztum J.L. The role of oxidized low-density lipoproteins in the pathogenesis of atherosclerosis. Annu. Rev. Med. 1992;43:219–225. doi: 10.1146/annurev.me.43.020192.001251. [DOI] [PubMed] [Google Scholar]
- 54.Aviram M. Modified forms of low density lipoprotein and atherosclerosis. Atherosclerosis. 1993;98:1–9. doi: 10.1016/0021-9150(93)90217-i. [DOI] [PubMed] [Google Scholar]
- 55.Basu S.K., Goldstein J.L., Anderson G.W., Brown M.S. Degradation of cationized low density lipoprotein and regulation of cholesterol metabolism in homozygous familial hypercholesterolemia fibroblasts. Proc. Natl. Acad. Sci. U. S. A. 1976;73:3178–3182. doi: 10.1073/pnas.73.9.3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Leake D.S., Rankin S.M. The oxidative modification of low-density lipoproteins by macrophages. Biochem. J. 1990;270:741–748. doi: 10.1042/bj2700741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Steinbrecher U.P., Parthasarathy S., Leake D.S., Witztum J.L., Steinberg D. Modification of low density lipoprotein by endothelial cells involves lipid peroxidation and degradation of low density lipoprotein phospholipids. Proc. Natl. Acad. Sci. U. S. A. 1984;81:3883–3887. doi: 10.1073/pnas.81.12.3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Itabe H., Obama T., Kato R. The dynamics of oxidized LDL during atherogenesis. J. Lipids. 2011;2011:418313. doi: 10.1155/2011/418313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ojo O.O., Leake D.S. Low density lipoprotein oxidation by ferritin at lysosomal pH. Chem. Phys. Lipids. 2018;217:51–57. doi: 10.1016/j.chemphyslip.2018.09.016. [DOI] [PubMed] [Google Scholar]
- 60.Khalili A., Ghorbanihaghjo A., Rashtchizadeh N., Gaffari S. Association between serum ferritin and circulating oxidized low-density lipoprotein levels in patients with coronary artery disease. J. Cardiovasc. Thorac. Res. 2012;4:1–4. doi: 10.5681/jcvtr.2012.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schaftenaar F., Frodermann V., Kuiper J., Lutgens E. Atherosclerosis: The interplay between lipids and immune cells. Curr. Opin. Lipidol. 2016;27:209–215. doi: 10.1097/MOL.0000000000000302. [DOI] [PubMed] [Google Scholar]
- 62.Torzewski M. Enzymatically modified LDL, atherosclerosis and beyond: Paving the way to acceptance. Front. Biosci. 2018;23:1257–1271. doi: 10.2741/4642. [DOI] [PubMed] [Google Scholar]
- 63.Trpkovic A., Resanovic I., Stanimirovic J., Radak D., Mousa S.A., Cenic-Milosevic D., Jevremovic D., Isenovic E.R. Oxidized low-density lipoprotein as a biomarker of cardiovascular diseases. Crit. Rev. Clin. Lab. Sci. 2015;52:70–85. doi: 10.3109/10408363.2014.992063. [DOI] [PubMed] [Google Scholar]
- 64.Brown M.S., Goldstein J.L. Lipoprotein metabolism in the macrophage: Implications for cholesterol deposition in atherosclerosis. Annu. Rev. Biochem. 1983;52:223–261. doi: 10.1146/annurev.bi.52.070183.001255. [DOI] [PubMed] [Google Scholar]
- 65.Tamura Y., Osuga J., Adachi H., Tozawa R., Takanezawa Y., Ohashi K., Yahagi N., Sekiya M., Okazaki H., Tomita S., Iizuka Y., Koizumi H., Inaba T., Yagyu H., Kamada N. Scavenger receptor expressed by endothelial cells I (SREC-I) mediates the uptake of acetylated low density lipoproteins by macrophages stimulated with lipopolysaccharide. J. Biol. Chem. 2004;279:30938–30944. doi: 10.1074/jbc.M313088200. [DOI] [PubMed] [Google Scholar]
- 66.Van Berkel T.J., Van Velzen A., Kruijt J.K., Suzuki H., Kodama T. Uptake and catabolism of modified LDL in scavenger-receptor class A type I/II knock-out mice. Biochem. J. 1998;331:29–35. doi: 10.1042/bj3310029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kattoor A.J., Kanuri S.H., Mehta J.L. Role of Ox-LDL and LOX-1 in atherogenesis. Curr. Med. Chem. 2019;26:1693–1700. doi: 10.2174/0929867325666180508100950. [DOI] [PubMed] [Google Scholar]
- 68.Park Y.M. CD36, a scavenger receptor implicated in atherosclerosis. Exp. Mol. Med. 2014;46 doi: 10.1038/emm.2014.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Goldstein J.L., Ho Y.K., Basu S.K., Brown M.S. Binding site on macrophages that mediates uptake and degradation of acetylated low density lipoprotein, producing massive cholesterol deposition. Proc. Natl. Acad. Sci. U. S. A. 1979;76:333–337. doi: 10.1073/pnas.76.1.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kiyanagi T., Iwabuchi K., Shimada K., Hirose K., Miyazaki T., Sumiyoshi K., Iwahara C., Nakayama H., Masuda H., Mokuno H., Sato S., Daida H. Involvement of cholesterol-enriched microdomains in class A scavenger receptor-mediated responses in human macrophages. Atherosclerosis. 2011;215:60–69. doi: 10.1016/j.atherosclerosis.2010.10.019. [DOI] [PubMed] [Google Scholar]
- 71.Matsumoto A., Naito M., Itakura H., Ikemoto S., Asaoka H., Hayakawa I., Kanamori H., Aburatani H., Takaku F., Suzuki H., Kobari Y., Miyai T., Takahashi K., Cohen E.H., Wydro R. Human macrophage scavenger receptors: Primary structure, expression, and localization in atherosclerotic lesions. Proc. Natl. Acad. Sci. U. S. A. 1990;87:9133–9137. doi: 10.1073/pnas.87.23.9133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chappell P.E., Garner L.I., Yan J., Metcalfe C., Hatherley D., Johnson S., Robinson C.V., Lea S.M., Brown M.H. Structures of CD6 and its ligand CD166 give insight into their interaction. Structure. 2015;23:1426–1436. doi: 10.1016/j.str.2015.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yamamoto T., Davis C.G., Brown M.S., Schneider W.J., Casey M.L., Goldstein J.L., Russell D.W. The human LDL receptor: A cysteine-rich protein with multiple alu sequences in its mRNA. Cell. 1984;39:27–38. doi: 10.1016/0092-8674(84)90188-0. [DOI] [PubMed] [Google Scholar]
- 74.Phillips M.C. Apolipoprotein E isoforms and lipoprotein metabolism. IUBMB Life. 2014;66:616–623. doi: 10.1002/iub.1314. [DOI] [PubMed] [Google Scholar]
- 75.Mori K., Ohtani K., Jang S., Kim Y., Hwang I., Roy N., Matsuda Y., Suzuki Y., Wakamiya N. Scavenger receptor CL-P1 mainly utilizes a collagen-like domain to uptake microbes and modified LDL. Biochim. Biophys. Acta. 2014;1840:3345–3356. doi: 10.1016/j.bbagen.2014.08.015. [DOI] [PubMed] [Google Scholar]
- 76.Sarrias M.R., Gronlund J., Padilla O., Madsen J., Holmskov U., Lozano F. The scavenger receptor cysteine-rich (SRCR) domain: An ancient and highly conserved protein module of the innate immune system. Crit. Rev. Immunol. 2004;24:1–37. doi: 10.1615/critrevimmunol.v24.i1.10. [DOI] [PubMed] [Google Scholar]
- 77.Hansson G.K. Immune mechanisms in atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2001;21:1876–1890. doi: 10.1161/hq1201.100220. [DOI] [PubMed] [Google Scholar]
- 78.Paone S., Baxter A.A., Hulett M.D., Poon I.K.H. Endothelial cell apoptosis and the role of endothelial cell-derived extracellular vesicles in the progression of atherosclerosis. Cell. Mol. Life Sci. 2019;76:1093–1106. doi: 10.1007/s00018-018-2983-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bennett M.R., Sinha S., Owens G.K. Vascular smooth muscle cells in atherosclerosis. Circ. Res. 2016;118:692–702. doi: 10.1161/CIRCRESAHA.115.306361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Maguire E.M., Pearce S.W.A., Xiao Q. Foam cell formation: A new target for fighting atherosclerosis and cardiovascular disease. Vasc. Pharmacol. 2019;112:54–71. doi: 10.1016/j.vph.2018.08.002. [DOI] [PubMed] [Google Scholar]
- 81.Wigren M., Nilsson J., Kolbus D. Lymphocytes in atherosclerosis. Clin. Chim. Acta. 2012;413:1562–1568. doi: 10.1016/j.cca.2012.04.031. [DOI] [PubMed] [Google Scholar]
- 82.Durst R., Neumark Y., Meiner V., Friedlander Y., Sharon N., Polak A., Beeri R., Danenberg H., Erez G., Spitzen S., Ben-Avi L., Leitersdorf E., Lotan C. Increased risk for atherosclerosis of various macrophage scavenger receptor 1 alleles. Genet. Test. Mol. Biomarkers. 2009;13:583–587. doi: 10.1089/gtmb.2009.0048. [DOI] [PubMed] [Google Scholar]
- 83.Linton M.F., Fazio S. Class A scavenger receptors, macrophages, and atherosclerosis. Curr. Opin. Lipidol. 2001;12:489–495. doi: 10.1097/00041433-200110000-00003. [DOI] [PubMed] [Google Scholar]
- 84.Balla J., Vercellotti G.M., Jeney V., Yachie A., Varga Z., Jacob H.S., Eaton J.W., Balla G. Heme, heme oxygenase, and ferritin: How the vascular endothelium survives (and dies) in an iron-rich environment. Antioxid. Redox Signal. 2007;9:2119–2137. doi: 10.1089/ars.2007.1787. [DOI] [PubMed] [Google Scholar]
- 85.Ikeda Y., Suehiro T., Yamanaka S., Kumon Y., Takata H., Inada S., Ogami N., Osaki F., Inoue M., Arii K., Hashimoto K. Association between serum ferritin and circulating oxidized low-density lipoprotein levels in patients with type 2 diabetes. Endocr. J. 2006;53:665–670. doi: 10.1507/endocrj.k06-010. [DOI] [PubMed] [Google Scholar]
- 86.Michel J.B., Martin-Ventura J.L. Red blood cells and hemoglobin in human atherosclerosis and related arterial diseases. Int. J. Mol. Sci. 2020;21:6756. doi: 10.3390/ijms21186756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dai X.Y., Cai Y., Mao D.D., Qi Y.F., Tang C., Xu Q., Zhu Y., Xu M.J., Wang X. Increased stability of phosphatase and tensin homolog by intermedin leading to scavenger receptor A inhibition of macrophages reduces atherosclerosis in apolipoprotein E-deficient mice. J. Mol. Cell. Cardiol. 2012;53:509–520. doi: 10.1016/j.yjmcc.2012.07.006. [DOI] [PubMed] [Google Scholar]
- 88.Manning-Tobin J.J., Moore K.J., Seimon T.A., Bell S.A., Sharuk M., Alvarez-Leite J.I., de Winther M.P., Tabas I., Freeman M.W. Loss of SR-A and CD36 activity reduces atherosclerotic lesion complexity without abrogating foam cell formation in hyperlipidemic mice. Arterioscler. Thromb. Vasc. Biol. 2009;29:19–26. doi: 10.1161/ATVBAHA.108.176644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Minor W., Cymborowski M., Otwinowski Z., Chruszcz M. HKL-3000: The integration of data reduction and structure solution--from diffraction images to an initial model in minutes. Acta Crystallogr. D Biol. Crystallogr. 2006;62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 90.McCoy A.J. Acknowledging errors: Advanced molecular replacement with phaser. Methods Mol. Biol. 2017;1607:421–453. doi: 10.1007/978-1-4939-7000-1_18. [DOI] [PubMed] [Google Scholar]
- 91.Emsley P., Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 92.Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., McCoy A.J., Moriarty N.W., Oeffner R., Read R.J., Richardson D.C. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bramucci E., Paiardini A., Bossa F., Pascarella S. PyMod: Sequence similarity searches, multiple sequence-structure alignments, and homology modeling within PyMOL. BMC Bioinformatics. 2012;13 Suppl 4 doi: 10.1186/1471-2105-13-S4-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 95.Edelstein C., Pfaffinger D., Hinman J., Miller E., Lipkind G., Tsimikas S., Bergmark C., Getz G.S., Witztum J.L., Scanu A.M. Lysine-phosphatidylcholine adducts in kringle V impart unique immunological and potential pro-inflammatory properties to human apolipoprotein(a) J. Biol. Chem. 2003;278:52841–52847. doi: 10.1074/jbc.M310425200. [DOI] [PubMed] [Google Scholar]
- 96.Qin J., Goswami R., Balabanov R., Dawson G. Oxidized phosphatidylcholine is a marker for neuroinflammation in multiple sclerosis brain. J. Neurosci. Res. 2007;85:977–984. doi: 10.1002/jnr.21206. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The structure of the SRCR domain of human SCARA1 has been deposited in the PDB (www.rcsb.org) with PDB entry: 7DPX.