Abstract
The disuse of skeletal muscle, such as that which occurs during immobilization, can lead to the rapid loss of muscle mass, and a decrease in the rate of protein synthesis plays a major role in this process. Indeed, current dogma contends that the decrease in protein synthesis is mediated by changes in the activity of protein kinases (e.g. mTOR); however, the validity of this model has not been established. Therefore, to address this, we first subjected mice to 6, 24 or 72 h of unilateral immobilization and then used the SUnSET technique to measure changes in the relative rate of protein synthesis. The result of our initial experiments revealed that immobilization leads to a rapid (within 6 h) and progressive decrease in the rate of protein synthesis and that this effect is mediated by a decrease in translational efficiency. We then performed a deep mass spectrometry-based analysis to determine whether this effect could be explained by changes in the expression and/or phosphorylation state of proteins that regulate translation. From this analysis, we were able to quantify 4320 proteins and 15,020 unique phosphorylation sites, and surprisingly, the outcomes revealed that the rapid immobilization-induced decrease in protein synthesis could not be explained by changes in either the abundance, or phosphorylation state, of proteins. The results of our work not only challenge the current dogma in the field, but also provide an expansive resource of information for future studies that are aimed at defining how disuse leads to loss of muscle mass.
Keywords: atrophy, immobilization, protein synthesis, proteomics, skeletal muscle
Introduction
Comprising approximately 40% of total body mass, skeletal muscle is critical for the quality of life, and its loss is associated with an increased risk of morbidity, mortality and healthcare expenditures (Janssen et al. 2004; Zhou et al. 2010; Srikanthan & Karlamangla, 2011). Importantly, various types of mechanical unloading, such as immobilization, can lead to the loss of skeletal muscle mass. For instance, immobilization is often required to recover from an injury, and the rapid loss of muscle mass that occurs during immobilization can result in an ≈0.5% loss of muscle cross-sectional area per day in humans (de Boer et al. 2007). Thus, understanding how immobilization leads to the loss of muscle mass and developing strategies that prevent this effect is both a clinically and a fiscally significant goal.
Two factors that play a critical role in the regulation of skeletal muscle mass are the rate of protein synthesis and the rate of protein degradation, and the net balance between these processes dictates whether substantive alterations in muscle mass will occur (Goodman et al. 2011c). Indeed, it has been widely accepted that immobilization leads to a prominent decrease in the rate of protein synthesis, but whether immobilization induces a substantive increase in protein degradation remains a subject of debate (Symons et al. 2009; Bodine, 2013; Phillips & McGlory, 2014). Accordingly, we set out to develop a better understanding of how immobilization induces a decrease in protein synthesis.
Protein synthesis can be regulated through two major processes called translational capacity and translation efficiency (Nader et al. 2002; Figueiredo, 2019). Translational capacity refers to the concentration of the translation machinery (e.g. the ribosomes), whereas translational efficiency refers to the rate at which the ribosomes translate mRNA. In skeletal muscle, changes in translation efficiency can occur on a very acute time scale (within minutes), whereas changes in translational capacity often take days to even weeks to reach significance. For instance, several studies have shown that total RNA concentration (of which ≈80% is composed of rRNA) does not significantly change during the first 3 days of immobilization, yet a decrease in protein synthesis can be observed as early as 6 h after the onset of immobilization in rodents (Goldspink, 1977; Booth & Seider, 1979; You et al. 2015). Thus, the initial immobilization-induced decrease in protein synthesis appears to be mediated by a decrease in translational efficiency.
Changes in translational efficiency are primarily exerted through changes in the rate of translation initiation and/or elongation (Acevedo et al. 2018; Riba et al. 2019). Translation initiation is the process through which the ribosome becomes bound to the start codon of an mRNA, whereas elongation refers to events via which new amino acids get added to the growing polypeptide chain. Importantly, both of these processes can be regulated through changes in the expression of translation initiation and elongation factors. For instance, the mRNAs which encode the eukaryotic elongation factor 1 alpha (eEF1α) and eukaryotic elongation factor 2 (eEF2) both possess a 5′-terminal oligopyrimidine tract (5′-TOP) that exerts control over their expression and a decrease in their expression can lead to an impaired rate of translation elongation (Hamilton et al. 2006; Thoreen et al. 2012). The activity of the translation initiation and elongation factors can also be regulated by post-translational modifications, with changes in phosphorylation being the most prevalent (Hershey et al. 2012; Hizli et al. 2013). Indeed, the vast majority of proteins that are involved in translation initiation and elongation are phosphoproteins, and a number of specific phosphorylation sites have been shown to directly regulate the rate of initiation and elongation. For example, phosphorylation of the ribosomal protein S6 (RPS6) at serine 235/236, eukaryotic initiation factor 4E binding protein 1 (4E-BP1) at threonine 36/45, and eukaryotic initiation factor 4B (eIF4B) at serine 422 have all been shown to promote translation initiation (Pause et al. 1994; Gingras et al. 2001a; Holz et al. 2005; Roux et al. 2007; Dennis et al. 2012). On the other hand, phosphorylation of the eukaryotic elongation factor 2 (eEF2) at threonine 57 has been shown to inhibit translation elongation (Carlberg et al. 1990).
As alluded to above, translational efficiency can be controlled through changes in both the expression and the phosphorylation state of the proteins that are involved in translation initiation and elongation (Hamilton et al. 2006; Hershey et al. 2012). However, whether these types of alterations are responsible for the changes in translational efficiency that occur in response to immobilization has not been rigorously addressed. Thus, to fill this gap in knowledge, we set out to identify the proteomic and phosphoproteomic alterations that occur after the onset of immobilization. Specifically, we utilized our previously described mouse model of unilateral immobilization and collected muscles at 6, 24 and 72 h after the onset of immobilization (You et al. 2015). We first confirmed that our immobilization procedure led to a rapid (within 6 h) and progressive decrease in the relative rate of protein synthesis. Next, we performed a deep mass spectrometry-based analysis in which we were able to quantify 4320 proteins and 15,020 unique phosphorylation sites. Much to our surprise, the results from our analyses indicate that the rapid decrease in protein synthesis could not be explained by changes in the abundance or phosphorylation state of any proteins.
Materials and methods
Ethical approval
Male C57 (Jackson Laboratories, Bar Harbor, MA, USA) mice at 8–10 weeks of age were given food and water ad libitum and kept in a room that was maintained at 25°C with a 12-h light–dark cycle. Where indicated, mice were anaesthetized with 1–5% isoflurane mixed in oxygen and killed by cervical dislocation under anaesthesia. All animal experiments were approved by the Institutional Animal Care and Use Committee of the University of Wisconsin-Madison (#V005375). The study complies with the ethics policies of The Journal of Physiology.
Immobilization
Unilateral hindlimb immobilization was performed as previously described (You et al. 2015). In brief, mice were anaesthetized with isoflurane, and then a splint was used to fix the ankle in a plantar-flexed position and the knee in an extended position for 6, 24 or 72 h. Immobilization was always performed on the right hindlimb while the left hindlimb was untouched and used as a time-matched sham control. At the end of the immobilization period, the mice were anaesthetized and the plantar flexor muscles (i.e. gastrocnemius, soleus and plantaris) of each leg were collected as a single complex (i.e. the GSP complex). It should be noted that we have previously shown that our immobilization procedure leads to a significant decrease in the mass of gastrocnemius, soleus and plantaris muscles after 7 days (You et al. 2015). Moreover, previous studies in both mice and rats have shown that similar models on unilateral immobilization do not lead to a change in the mass of plantar flexor muscles in the untouched control limb (Lang et al. 2012; Kelleher et al. 2013).
Maximal-intensity contractions
As previously described, maximal-intensity contractions were elicited in mice that had not been subjected to any other kind of perturbation (Potts et al. 2017). At 1 h after the last set of contractions, the GSP complex was collected and immediately frozen in liquid nitrogen and then stored at −80°C.
Protein synthesis
The SUnSET technique was used to measure the relative rate of protein synthesis in a subset of mice. Importantly, the SUnSET technique uses puromycin to label nascent peptides and the measured relative rates of protein synthesis are based on the assumption that the experimental perturbations do not alter the equilibration kinetics of the free puromycin in the tissues being assessed (Goodman et al. 2011b). To perform these measurements, puromycin (MilliporeSigma, Burlington, MA, USA) was dissolved in diH2O to generate a 75 mM stock solution, and then 0.04 μmol g−1 body weight of puromycin in 200 μl of PBS was administered via an i.p. injection. At 30 min after injection, the GSP complex of each leg was collected and immediately frozen in liquid nitrogen and then stored at −80°C. The frozen muscles were homogenized in ice-cold buffer A [40 mM Tris (pH 7.5), 1 mM EDTA, 5 mM EGTA, 0.5% Triton X-100, 25 mM β-glycerolphosphate, 25 mM NaF, 1 mM Na3VO4, 10 μg ml−1 leupeptin and 1 mM phenylmethylsulfonyl fluoride] with a Polytron (PT 1200 E, Kinematica, Luceme, Switzerland) and then the homogenates were centrifuged at 6000 g for 1 min to remove bubbles and confirm complete homogenization. The sample was then thoroughly vortexed to resuspend insoluble material and an aliquot of the whole homogenate was subjected to western blot analysis to detect the amount of puromycin-labelled peptides. It should be noted that the muscles from the puromycin-injected mice were used exclusively for measuring relative rates of protein synthesis (i.e. they were not used for mass spectrometry or any of the orthogonal validations of the mass spectrometry-based results).
Western blot analysis
The protein concentration of individual samples was determined by a DC protein assay kit (Bio-Rad, Hercules, CA, USA). Equal amounts of protein from each sample were then dissolved in Laemmli buffer and then subjected to SDS-PAGE, transferred to polyvinylidene fluoride (PVDF), blocked with 5% milk TBST, and incubated with primary and secondary antibodies as previously described (You et al. 2019). The resulting blots were developed with a UVP Autochemi system (UVP, Upland, CA, USA) along with either an ECL-prime (Amersham, Piscataway, NJ, USA) or regular enhanced chemiluminescene (ECL) reagent (Pierce, Rockford, IL, USA). After the appropriate images were captured, Coomassie Blue staining was performed on the PVDF membrane to verify the equal protein loading and the images were quantified with ImageJ software (U.S. NIH, Bethesda, MD, USA).
Antibodies used in western blot analysis
Antibodies targeting 4E-BP1(1:2000, #9644), phospho 4E-BP1(37/46) (1:1000, #2855), eEF2 (1:3000, #2332), phospho eEF2(57) (1:1000, #2331), eIF2α (1:1000, #5324), phospho eIF2α(51) (1:1000, #3398), eIF4B (1:1000, #3592), phospho eIF4B(422) (1:1000, #3591), RPS6 (1:1000, #2217) and phospho RPS6(235/236) (1:1000, #2211) were purchased from Cell Signaling Technology (Danvers, MA, USA). Anti-puromycin (1:5000, 12D10) was purchased from MilliporeSigma. Anti-complement C3 (1:3000, 855463) was purchased from MP Biomedicals (Santa Ana, CA, USA). Peroxidase-labelled anti-mouse IgG2a (1:50 000, 115-035-206) was from Jackson ImmunoResearch Laboratories, Inc (West Grove, PA, USA). Peroxidase-labelled anti-rabbit (1:5000, PI-1000), peroxidase-labelled anti-mouse (1:5000, PI-2000) and peroxidase-labelled anti-goat (1:5000, PI-9500) were obtained from Vector Labs Inc. (Burlingame, CA, USA).
Analysis of total and rRNA
The GSP complex was pulverized in liquid nitrogen with a pre-chilled mortar and pestle. The powdered muscle was then separated into multiple aliquots and stored at −80°C. For RNA-based analyses, an aliquot of the powder was weighed, homogenized with a Polytron in ice-cold TRIzol (Ambion, Life Technologies, Grand Island, NY, USA) and then total RNA was isolated according to the manufacturer’s instructions. The resulting RNA pellet was resuspended in 4 μl of nuclease-free DEPC treated water (Genemate, Kaysville, UT, USA) per milligram of muscle. The purity (A260/A280) and concentration (A260) of the RNA in this solution was determined with a NanoDrop 2000 spectrometer (Thermo Scientific, Wilmington, DE, USA). The total amount of RNA per milligram of muscle was calculated by multiplying the RNA concentration by the volume of RNA solution and then dividing this value by the weight of the original muscle powder. In addition, 10 μl of the RNA solution was run on a 1% agarose gel and used to quantify the amount of 28S and 18S rRNA as well as to assess the RNA integrity as previously described (Goodman et al. 2011a). RNA from an equal weight of muscle was used to synthesize cDNA by using SuperScript III First-Strand Synthesis System (Invitrogen, Waltham, MA, USA) with random hexamer. Real-time PCR was performed by running Fast SYBR Green Master Mix on a StepOnePlus Real-Time PCR System (Applied Biosystems, Waltham, MA, USA) as detailed previously (You et al. 2018). The primer sequences for MuRF1 and Atrogin/MAFbx have been previously described (You et al. 2018). Additional primers used in this study included: 5′-TGTCTTGCCCCGCGTGTAAG-3′ (forward) and 5′-CGCTTACAAGAAACAGCGCG-3′ (reverse) for the precursor rRNA 5′ETS region, 5′-GTGGAGCGAGGTGTCTGGAG-3′ (forward) and 5′-AACGCGACAGCTAGGTACCC-3′ (reverse) for the precursor rRNA ITS1 region, and 5′-CCAGCCTCGTCCCGTAGAC-3′ (forward) and 5′-ATGGCAACAATCTCCACTTTGC-3′ (reverse) for glyceraldehyde-3-phosphate dehydrogenase as an internal control.
Sample preparation for mass spectrometry (MS)
Frozen GSP complexes were pulverized as described above and then an aliquot of the powder was homogenized with a Polytron in ice-cold buffer B [40 mM Tris (pH 7.5), 1 mM EDTA, 5 mM EGTA, 0.5% Triton X-100, with one PhosSTOP tablet (Roche, Mannheim, Germany) and one Complete Mini EDTA-Free Protease Inhibitor Cocktail Tablet (Roche) per 10 ml], or buffer C [8 m urea, 50 mM Tris (pH 8.0), with one PhosSTOP tablet and one Complete Mini EDTA-Free Protease Inhibitor Cocktail Tablet per 10 ml]. The resulting homogenates were centrifuged at 6000 g for 1 min to remove bubbles and confirm complete homogenization. For the muscles homogenized in buffer B, the samples were incubated at 4°C for 30 min and then further separated into supernatant and pellet fractions as previously described (Potts et al. 2017; Steinert et al. 2021). For all samples, a small aliquot was saved for western blot analysis with the remainder of the sample being devoted to subsequent MS analyses as described below.
Protein precipitation, enzymatic digestion and peptide desalting for MS
Proteins were precipitated by bringing the original sample solution to a 90% concentration of methanol by volume and then centrifuging the sample at 12 000 g for 5 min. The supernatant was discarded and the protein precipitate was resuspended in 8 m urea, 50 mM Tris (pH 8.0), 10 mM Tris(2 carboxyethyl)phosphine (TCEP), and 40 mM chloroacetamide and continuously shaken for 30 min to completely reduce and alkylate the proteins. The sample was diluted with 50 mM Tris (pH 8.0) to a concentration of 1.5 m urea and digested with trypsin (enzyme/protein = 1:50) at 37°C for 15 h. The reaction was quenched by adding 10% trifluoroacetic acid (TFA) to bring the pH of the sample to less than 2. Strata-X desalting columns (Phenomenex, Torrance, CA, USA) were prepared by flowing 1 ml of 100% acetonitrile (ACN) over the column, followed by 1 ml of 0.1% TFA. Each sample was spun down and the acidic supernatant was loaded on and gravity filtered through the Strata-X columns. The column-bound peptides were washed with 1 ml 0.1% TFA, then eluted into a fresh tube with 500 μl of 40% ACN and 0.1% TFA, and finally eluted with 300 μl of 80% ACN and 0.1% TFA. Vacuum centrifugation was used to dry the elution. A Pierce Quantitative Colorimetric Peptide Assay (Thermo Fisher Scientific) was executed to determine peptide concentrations before 10-plex TMT labelling.
TMT labelling
A total of 1 mg of peptides for each sample was incubated with 10-plex tandem mass tags (TMT) reagents according to the instructions of TMT 10-plex Isobaric label reagent set kit (Thermo Fisher Scientific) for the quantification of relative abundances of tryptic peptides. Samples were kept shaking for 3 h at room temperature and then quenched by using 5% hydroxylamine and kept shaking for 15 min at room temperature. An aliquot of all 10 samples was equally mixed across all channels and analysed using an Orbitrap Elite mass spectrometer (Thermo Fisher Scientific) to confirm complete TMT peptide labelling and compare peptide ratios in this ‘test mix’. The preliminary mixing ratios were the base for creating a final sample mix, where the 10 muscle samples were mixed at a 1:1 ratio. The final mixed sample comprising TMT-labelled peptides from all 10 samples was desalted using a Strata-X desalting column as described above. The mixed sample was then enriched with Immobilized Metal Affinity Chromatography (IMAC) Ti-IMAC magnetic beads (ReSyn Biosciences, Edenvale, South Africa). The enriched phosphopeptide sample and flow-through non-phosphopeptide sample were each further fractionated with reversed-phase HPLC to yield 12 total phosphopeptide fractions and 12 total non-phosphopeptide fractions. Each fraction was then dried using a vacuum centrifuge and resuspended in MS-grade water containing 0.2% formic acid for following MS analysis.
Nano-LC-MS/MS
Each fraction of the samples was analysed by an Orbitrap Elite mass spectrometer (Thermo Fisher Scientific) during a 90 min nano-LC separation with a Dionex UltiMate 3000 RSLCnano system (Thermo Fisher Scientific). Samples were analysed with an MS1 AGC target of 1 × 106 and maximum injection times of 50 ms. MS1 scans were analysed at 60,000 resolving power with a scan range from 300 to 1500 m/z. Precursor ions with charge states of +2 to +8 were selected for fragmentation and MS2 analysis. MS2 scans were collected with a quadrupole isolation window of 1.8 Th, and HCD fragmentation at 35% NCE. Product ions were analysed in the Orbitrap at 60,000 resolving power with an AGC target of 2 × 105 ions and 118 ms maximum injection times. Monoisotopic precursor selection and dynamic exclusion (60 s) were enabled.
MS data analysis
The RAW data files were searched using the COMPASS software suite (Wenger et al. 2011). Thermo RAW files were searched against a Mus musculus target-decoy database (UniProt, downloaded 14 August 2015). Phosphopeptide and peptide datasets were searched using a 50 ppm precursor mass tolerance and 0.02 Da fragment tolerance for b and y ions produced by HCD fragmentation. All fractions were searched with static carbadimomethyl of cysteine residues, static TMT 10-plex modifications of peptide N-termini and lysines, and dynamic methionine oxidation. Phosphopeptide fractions were searched with additional dynamic phosphorylation modifications of serine, threonine and tyrosine residues. The resulting peptide identifications were filtered to 1% false discovery rate (FDR). Peptides were then mapped back to their parent proteins and filtered to a 1% FDR at the protein level. 10-Plex TMT reporter ion signals were used for phosphopeptide and protein quantification.
Bioinformatics
The TMT reporter ion intensities for the phosphopeptides and proteins in each sample were normalized to the total reporter signal within each channel and then the values for each sample were expressed relative to the mean value observed in the 6 h control group. The normalized data were log2 transformed and then statistical comparisons between groups were performed with the moderated t test documented in the LIMMA package in RStudio (Smyth, 2004; Hoffman et al. 2015) and FDR corrected with the Benjamini–Hochberg method (Benjamini & Hochberg, 1995). The resulting data was then uploaded into Perseus V.1.6.0.7 (Tyanova & Cox, 2018) and annotated with gene ontology (GO) terms using the default database. Additional annotation for the term ‘regulation of translation’ was performed with information from the Mouse Genome Database (http://www.informatics.jax.org) retrieved on 28 August 2020. Cluster analyses were performed with the Mfuzz package in RStudio and the ‘mestimate’ function was used to determine the minimum number of clusters (Futschik & Carlisle, 2005; Kumar & Futschik, 2007). Enrichment analyses were performed in Perseus with the 1D annotation enrichment based on Mfuzz cluster membership scores, and the list of enriched GO terms was filtered through REVIGO using a similarity of 0.7. Network-based analyses were performed with Photon using the default settings along with mTOR as the anchor (Rudolph et al. 2016).
Statistical analysis
Statistical significance was determined by using the Student’s t test (two-tailed, paired), one-way ANOVA with Student–Newman–Keuls post hoc analysis, two-way repeated-measures ANOVA with planned comparisons, moderated t test, or pooled t test for mixed paired and unpaired samples (Guo & Yuan, 2017). Differences between groups were considered significant at P ≤ 0.05. All analyses except for pooled t tests were performed Biological replicates (n) are fromindependentGSPmuscle complexes and the type of statistical analysis that was performed for each experiment is indicated in the figure legends. Detailed statistical summaries can be found in the supplementary document, and all values are shown as mean ± SD.
Results
Immobilization induces a rapid and progressive decrease in protein synthesis
In this study, immobilization was always performed on the right hindlimb while the left hindlimb was untouched. The plantar flexor muscles from both hindlimbs were collected at 6, 24 or 72 h after the onset of immobilization, or 6 h after a sham immobilization procedure (i.e. the mice were anaesthetized but no immobilization was performed). As shown in Fig. 1, the outcomes revealed that the relative rate of protein synthesis in the contralateral (untouched) muscles from mice that had been subjected to 6 h of immobilization was significantly reduced when compared with muscles from the sham immobilization mice. This was a very important observation because it indicated that the events associated with having the immobilization device on one limb can lead to systemic alterations (e.g. the release of stress hormones) that impact the rate of protein synthesis (and probably other events) in the untouched limb. Therefore, to account for this effect, all of the remaining analyses in our study utilized time-matched contralateral muscles as the control condition. Of note, when using this approach, it was apparent that the relative rate of protein synthesis rapidly decreased (within 6 h) after the onset of immobilization, and that the magnitude of this effect became even larger after 24 and 72 h of immobilization (Fig. 1C).
Figure 1. Immobilization induces a rapid and progressive decrease in the relative rate of protein synthesis.
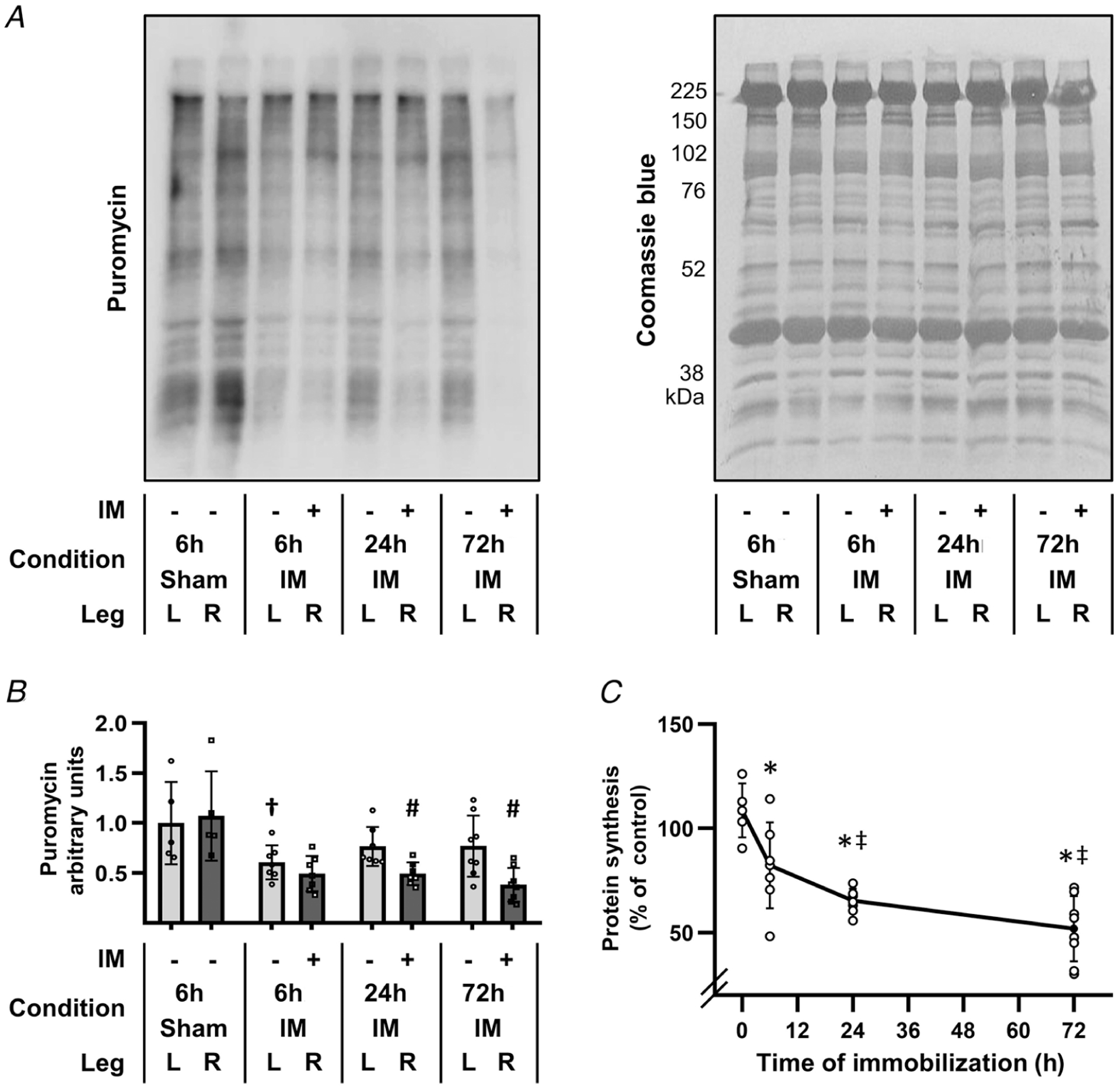
The right hindlimbs of mice were subjected to immobilization (IM) for 6, 24 or 72 h, or subjected to a 6 h sham control condition. All mice were injected with puromycin at 30 min prior to muscle collection for the measurement of protein synthesis. The plantar flexor muscles from both the left (L) and the right (R) hindlimbs were collected and subjected to western blot analysis for puromycin-labelled peptides (i.e. the relative rate of protein synthesis). A, representative western blot of puromycin-labelled peptides, and the subsequent Coomassie blue stain of total protein. B, graph showing the relative amount of puromycin-labelled peptides that were detected in the various conditions. C, graph illustrating the effect of immobilization on protein synthesis, which was determined by calculating the ratio between the amount of puromycin-labelled peptides in the R (immobilized or sham) muscle by the amount in the contralateral L (control) muscle. All R/L ratios were expressed as a percentage of the mean R/L ratio obtained in the 6 h sham control group (i.e. 0 h immobilization). Values in B and C are presented as the mean ± SD from n = 5–8 per group. Significantly different from, †time- and limb-matched sham control, #time-matched contralateral control, *0 h immobilization, and ‡6 h immobilization. Significance was determined by (B) repeated measures two-way ANOVA followed by planned comparison (interaction P < 0.001), or (C) one-way ANOVA followed by Student–Newman–Keuls post hoc analysis, P ≤ 0.05.
Immobilization does not induce a decrease in translational capacity
To assess whether the immobilization-induced decrease in protein synthesis was mediated by changes in translational capacity, we measured various markers of translational capacity including total RNA, 18S and 28S rRNA, as well as the ribosomal S6 protein (Kirby et al. 2015; Nakada et al. 2016). As shown in Fig. 2A–C, none of these markers were significantly altered by immobilization. We also measured pre-rRNA abundance by performing qPCR on the precursor rRNA 5′ external transcribed spacer (5′ ETS) region and normalized the results to GAPDH. As illustrated in Fig. 2D, immobilization did not alter the absolute levels of GAPDH and therefore verified its use as an appropriate internal control. It was also concluded that immobilization did not significantly alter the levels of the 5′ ETS (Fig. 2E). The validity of the qPCR results was further supported by measurements of MuRF1 and Atrogin which, consistent with other studies, revealed that immobilization led to a substantial increase in their expression (Fig. 2F and G) (Bodine et al. 2001; Okamoto et al. 2011). Thus, when taken together, the results of our analyses indicate that the rapid decrease in protein synthesis that follows the onset of immobilization is not mediated by a decrease in translational capacity.
Figure 2. Immobilization does not lead to a rapid decrease in translational capacity.
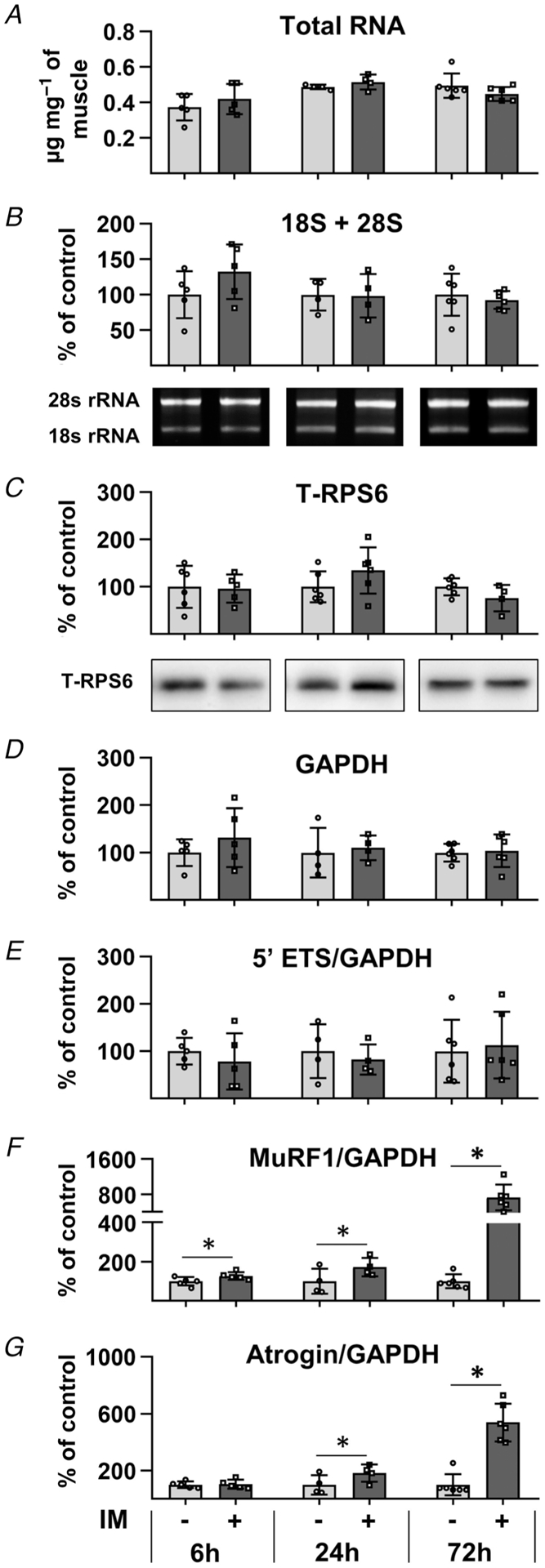
The right hindlimbs of mice were subjected to immobilization (IM) for 6, 24 or 72 h. The plantar flexor muscles from both the left (control) and right hindlimbs were collected and subjected to RNA or western blot analysis. A, total RNA to muscle weight ratio (μg mg−1). B, 18S + 28S rRNA to muscle weight ratio. C, total ribosomal protein S6 (RPS6) per μg of protein. D–G, cDNA of different samples was synthesized from RNA derived from 1.75 mg of muscle and subjected to qPCR analyses for 47S pre-rRNA 5′ ETS, 47S pre-rRNA ITS1, MuRF1, Atrogin and GAPDH. The level of expression of all transcripts, except for GAPDH, was normalized to GAPDH. The values in B–H are all expressed as a percentage of the mean value obtained in the time-matched control group. Bars are presented as the mean ± SD from n = 4–6 per group. *Significantly different from the time-matched contralateral control group. Significance was determined by paired t tests (A, B, D–G) or pooled t tests (C), P ≤ 0.05.
Identification of the proteomic and phosphoproteomic alterations that occur after the onset of immobilization
Immobilized and time-matched contralateral control muscles were subjected to deep MS-based analyses using the workflow described in Fig. 3. With this workflow we were able to perform quantitative analyses on 4320 different proteins (Supplementary Table 1), and 15,020 unique phosphopeptides (Supplementary Table 2). As reported in Supplementary Tables 1 and 2, all of the quantitative data from the MS analyses were initially normalized against the mean values obtained in the 6 h contralateral control samples so that differences between the treatments and time points could be assessed. Time-matched normalizations were then used to identify the proteins and phosphopeptides that were significantly affected by immobilization at each of the different time points (Fig. 4). Interestingly, the results of these comparisons revealed that the proteome was substantially altered after 72 h of immobilization (Fig. 4A). Surprisingly, and in sharp contrast to our expectations, the phosphoproteome was relatively unaffected by immobilization (Fig. 4B). Thus, to ensure that our phosphoproteomic analyses were able to successfully detect changes in protein phosphorylation, we included an internal positive control sample in a subset of the MS analyses. The internal positive control sample was derived from the plantar flexor muscles of a mouse that had been subjected to a bout of maximal-intensity contractions. Importantly, we have previously shown that maximal-intensity contractions can substantially alter the phosphoproteome, and a very similar effect was observed with the internal positive control sample (Fig. 5 and Supplementary Table 2) (Potts et al. 2017). Indeed, the results of GO term enrichment analyses on the positive control sample were remarkably similar to the results that were obtained when the same GO term enrichment analyses were performed on previously published phosphoproteomic data obtained from the tibialis anterior muscles of mice that had been subjected to a bout of maximal-intensity contractions (Fig. 5B) (Potts et al. 2017). Moreover, we used western blot analyses to perform orthogonal validations of the MS results, and the outcomes confirmed that the expression and phosphorylation state of several proteins which have been implicated in the regulation of translational efficiency were not significantly altered by immobilization (Fig. 4C and D) (Carlberg et al. 1990; Pause et al. 1994; Gingras et al. 2001a; Holz et al. 2005; Roux et al. 2007; Dennis et al. 2012; Hizli et al. 2013). Combined, these results establish that the lack of immobilization-induced alterations in the phosphoproteome were not simply the result of a technical flaw with the analyses.
Figure 3. Experimental workflow for mapping the proteomic and phosphoproteomic alterations that occur in response to immobilization.
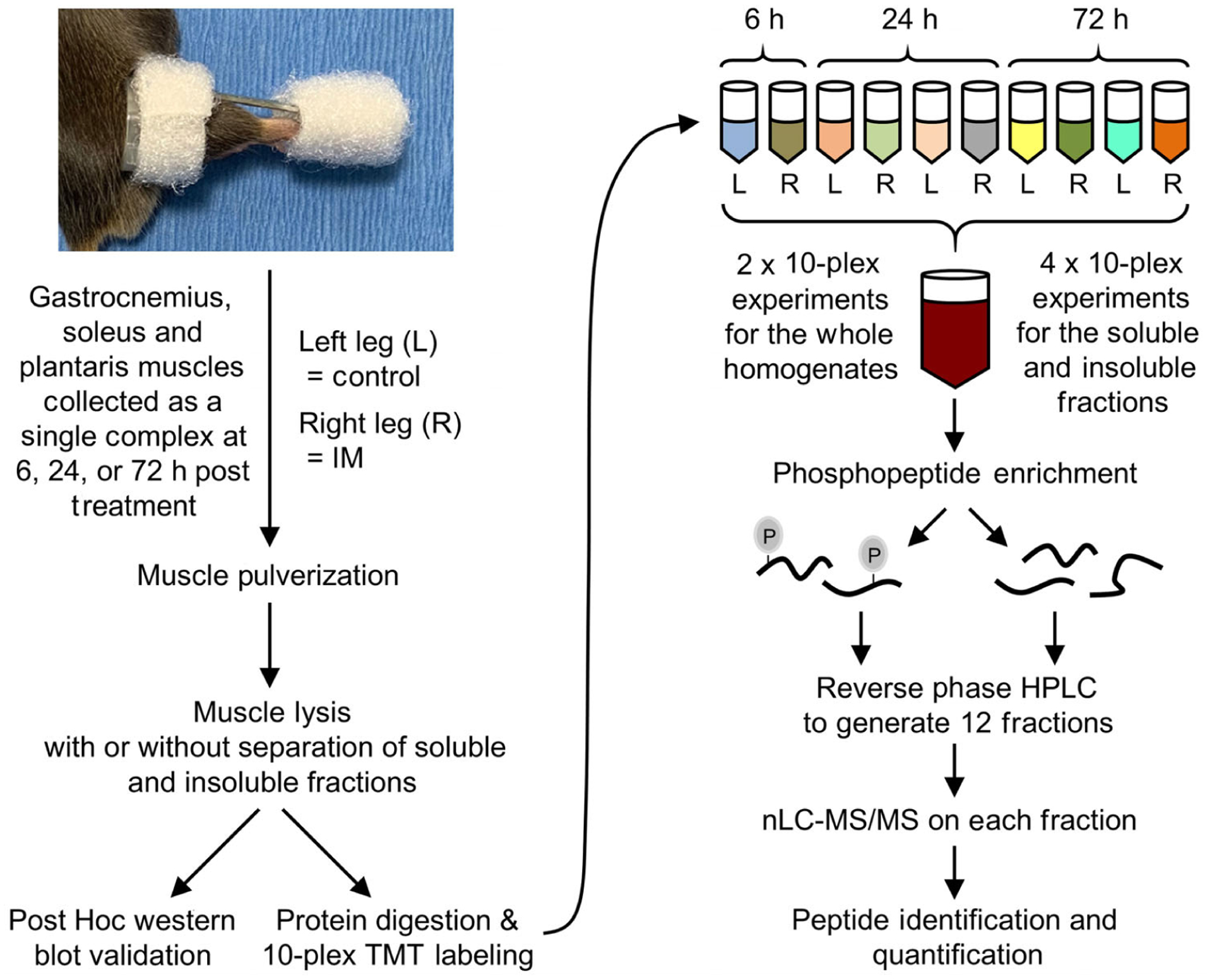
The right hindlimbs of mice were subjected to immobilization (IM) for 6, 24 or 72 h. The plantar flexor muscles (i.e. the gastrocnemius, soleus and plantaris) from both the left control (L) and right (R) hindlimbs were collected as a single complex and subjected to mass spectrometry (MS). Specifically, the muscles were pulverized and lysed, and then proteins from the whole homogenate, or soluble and insoluble fractions, were tryptically digested (see Methods for details). The resulting peptides from each sample were labelled with different tandem mass tags (TMT) and mixed to yield 10-plex pooled samples. Immobilized metal affinity chromatography was then used to enrich phosphopeptides and a total of 12 fractions for phosphopeptides and 12 fractions for the unbound (non-phospho) peptides was generated by reversed-phase HPLC. All fractions were analysed by nano-liquid chromatography-tandem MS spectrometry (nLC-MS/MS) and the relative quantity of the phosphopeptides and non-phosphorylated peptides in each sample was determined by the TMT reporter ions in the MS spectra.
Figure 4. The proteomic and phosphoproteomic alterations that occur after the onset of immobilization.
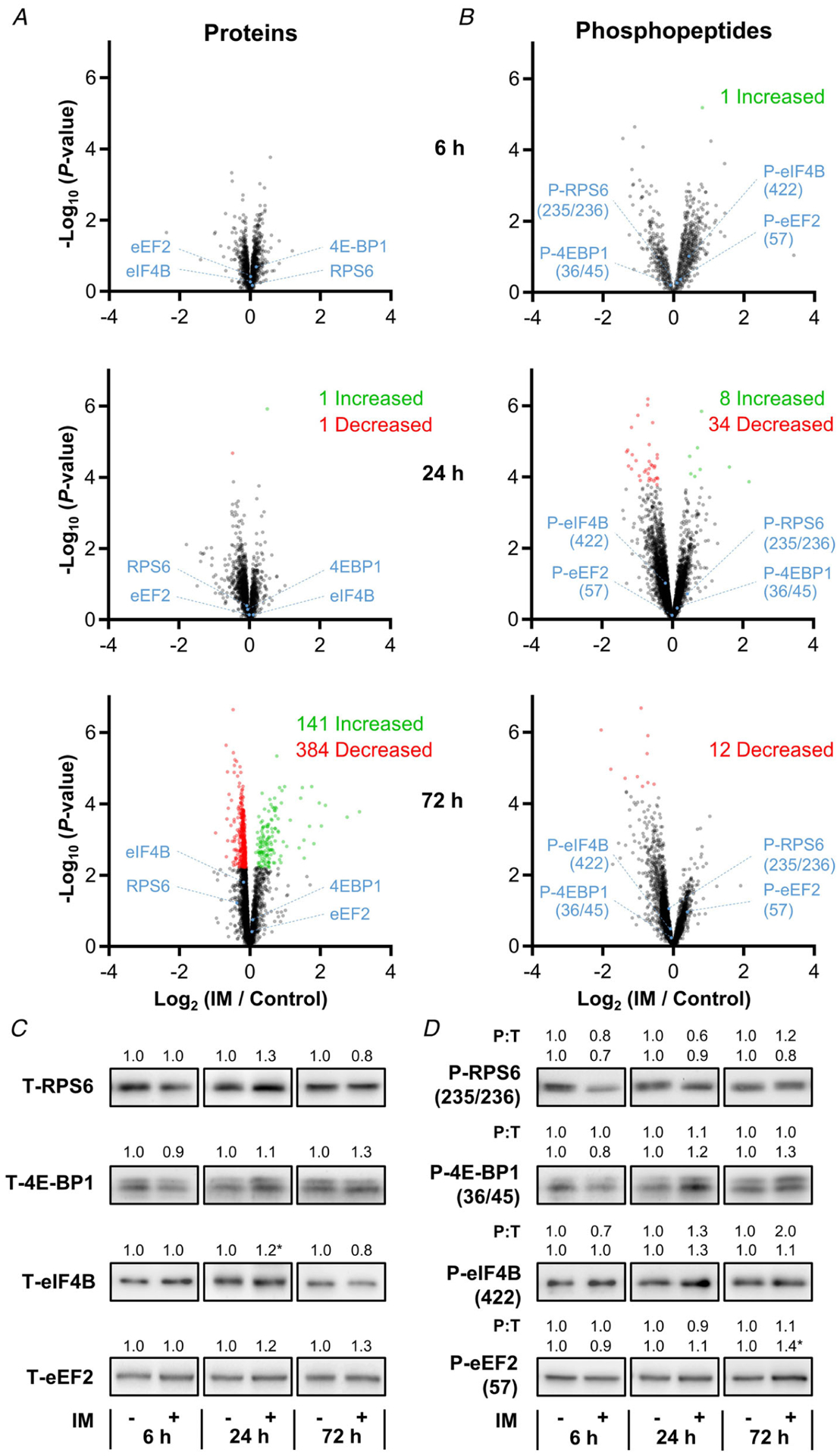
Mice were subjected to the workflow described in Figure 3. A and B, volcano plots of the proteomic and phosphoproteomic analyses in which the fold-change (immobilized/time-matched control) for each of the proteins (A) or phosphopeptides (B) was log2 transformed and then plotted against its corresponding −log10P-value that was derived from a moderated t test. Only data points with at least two valid values per group are shown in the plots. Statistically significant alterations (green and red data points) were identified by an FDR corrected P-value of q ≤ 0.05. C and D, representative western blots of total (T) RPS6, eIF2α, 4E-BP1, eIF4B and eEF2 (C), as well as phospho-(P) RPS6(235/236), eIF2α(51), 4E-BP1(36/45), eIF4B(422) and eEF2(57) (D). All values, including the phosphorylation state (i.e. P:T ratio), are group means and are expressed relative to the mean value obtained in the time-matched control samples, n = 3–7 per group. *Significantly different from the time-matched control as determined by a pooled t test, P ≤ 0.05.
Figure 5. Internal validation of the ability of the phosphoproteomic analyses to detect changes in protein phosphorylation.
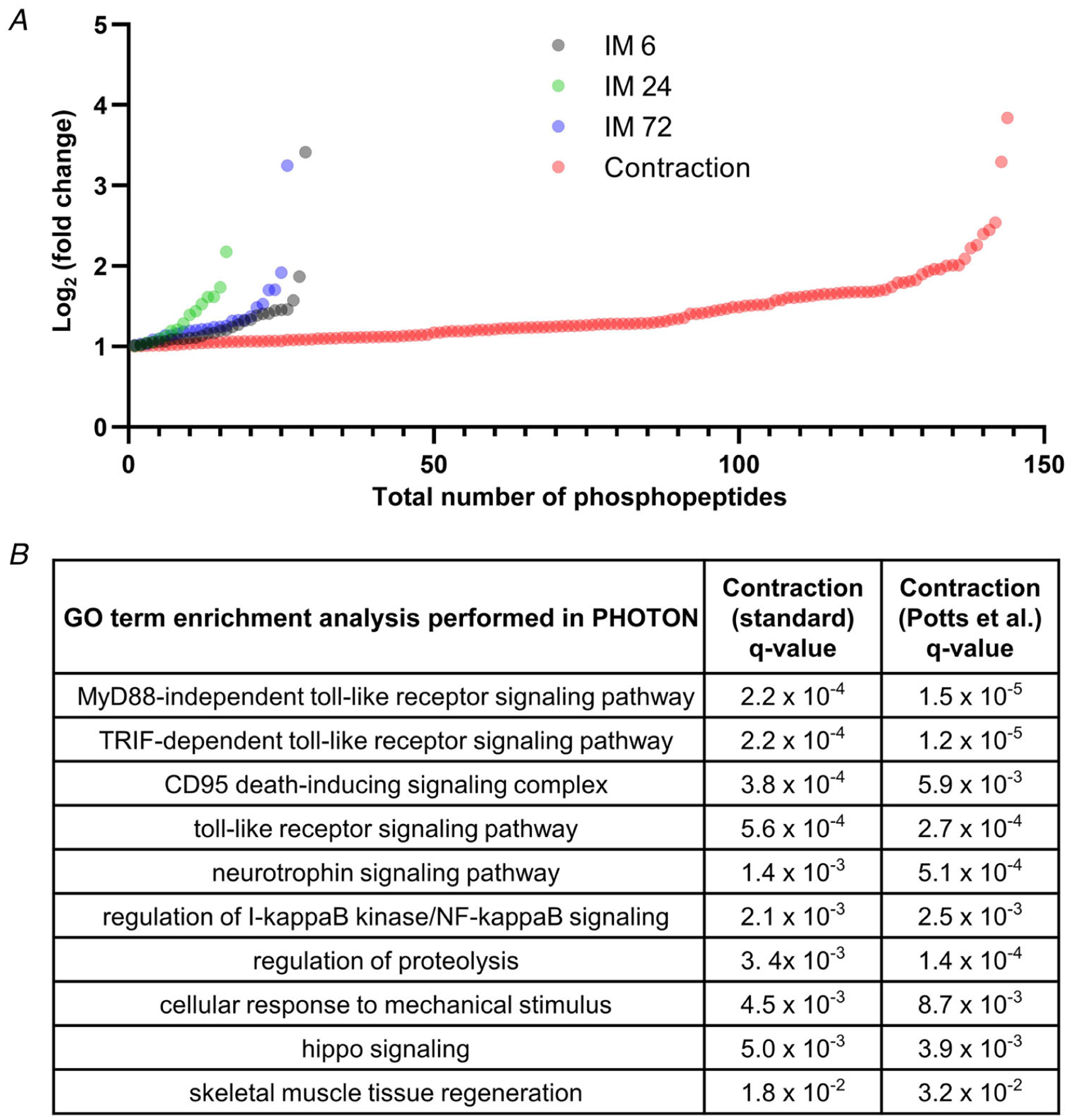
The plantar flexor muscles from a mouse that had been subjected to a bout of maximal-intensity contractions were included as an internal positive control in a subset of the MS analyses. The quantity of the phosphopeptides in this sample was expressed relative to the mean value of 6 h control samples that were present in the same MS analyses (see Supplementary Table 2). A, plot displaying all of the phosphopeptides from the MS analyses that revealed a greater than 2-fold increase. B, PHOTON was used to perform GO term enrichment analysis on the phosphoproteomic results that were obtained from the positive control sample, as well as on previously published phosphoproteomic data that were obtained from tibialis anterior muscles that had been subjected to a bout of maximal-intensity contractions (Potts et al. 2017).
Next, we set out to more precisely assess whether the results of our MS analyses had identified any immobilization-induced changes in the expression and/or phosphorylation state of the proteins that are involved in the regulation of protein synthesis. To accomplish this, we filtered the MS results so that only proteins and phosphopeptides from proteins that are annotated with the term ‘regulation of translation’ were retained. In total, the filtered results included 143 distinct proteins and 432 unique phosphopeptides. Consistent with the lack of immobilization-induced changes at the whole phosphoproteome level, none of the phosphopeptides in the filtered dataset were significantly altered by immobilization at any of the time points examined (Fig. 6). Moreover, no proteins revealed a significant alteration in expression after 6 or 24 h of immobilization. Thus, the rapid decrease in protein synthesis that follows the onset of immobilization does not appear to be mediated by changes in the expression and/or phosphorylation state of proteins that are involved in the regulation of protein synthesis.
Figure 6. The impact of immobilization on the expression and phosphorylation state of proteins that are involved in translation.
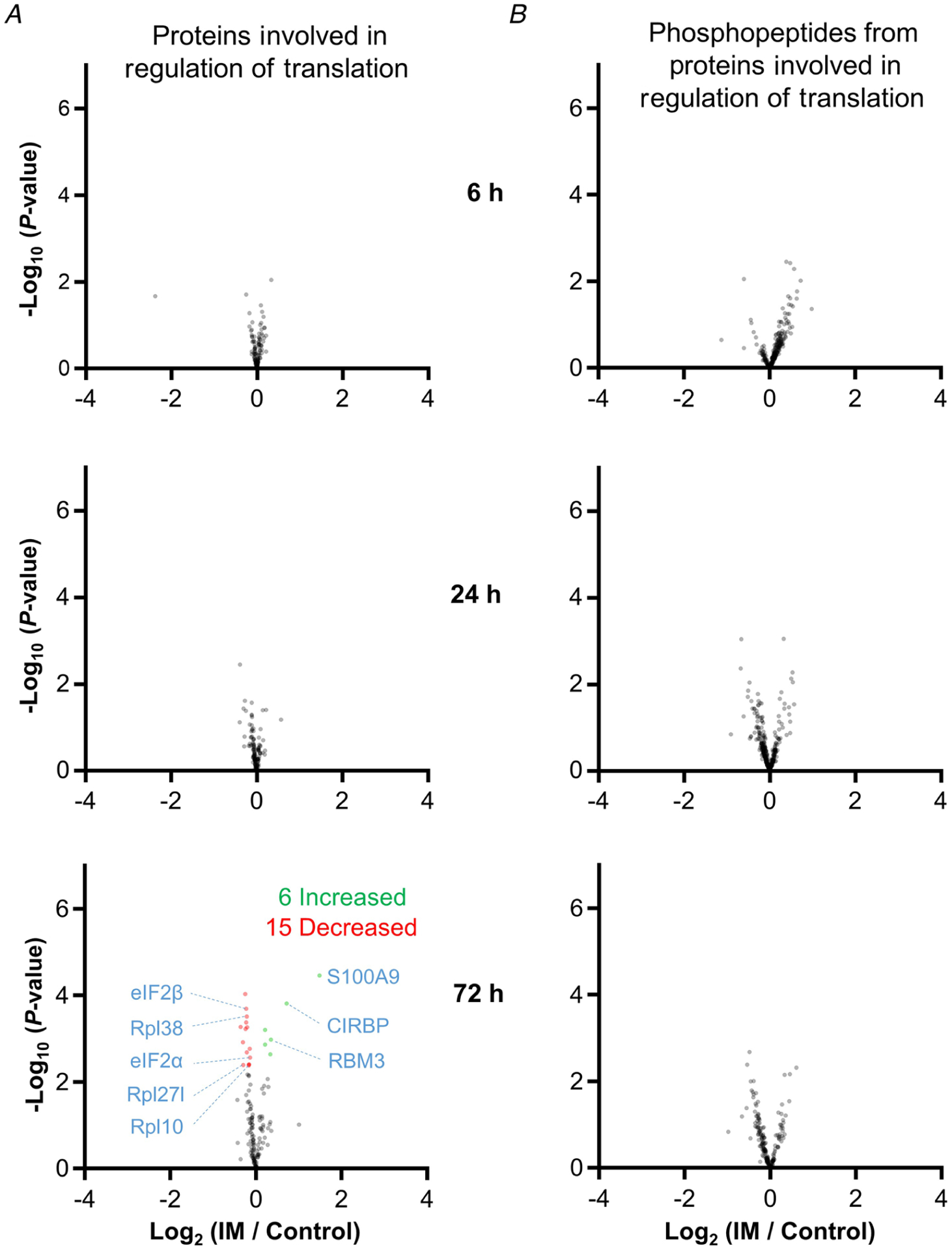
The volcano plots from Figure 4 were filtered so that only proteins (A), or phosphopeptides that were derived from proteins (B), that have been annotated with the term ‘translation’ were retained. Only data points with at least two valid values are shown in the plots. Statistically significant alterations (green and red data points) were identified by an FDR-corrected P-value of q ≤ 0.05.
It also bears mentioning that, although no proteins annotated with the term ‘regulation of translation’ were significantly altered after 6 or 24 h of immobilization, we did identify 21 proteins that showed a significant alteration in expression after 72 h of immobilization (six increased and 15 decreased) (Fig. 6). The six proteins that showed an increase were CIRBP, CNBP, HNRNPL, RBM3, Ptbp2 and S100A9. Interestingly, all of these proteins are known to promote translation (Mitchell et al. 2003; Smart et al. 2007; Huichalaf et al. 2009; Jia et al. 2014; Benhalevy et al. 2017; Lu et al. 2018; Venkata Subbaiah et al. 2019). The proteins that revealed a decrease in expression included eIF2α, eIF2β, Rpl10, Rpl27l and RPL38. Although these proteins are known to regulate translation, the decrease in their expression was very small (range 4–23%). Thus, it is unlikely that the change in expression of these proteins was a major driver of the decrease in protein that was observed after 72 h of immobilization.
Temporal analyses identify clusters of the proteins that may contribute to immobilization-induced loss of muscle mass
Significant changes in the expression and phosphorylation state of proteins that are known to regulate protein translation could not explain the rapid decrease in protein synthesis that occurred after the onset of immobilization; however, we did observe robust alterations in the expression of proteins at the level of the whole proteome. In fact, there were 525 different proteins (>12% of the proteome) that showed a significant alteration in expression after 72 h of immobilization, and we wondered if these changes could provide insight into the mechanisms that drive the immobilization-induced loss of muscle mass. Thus, to pursue this, we first used Mfuzz to identify the temporal nature through which the changes in protein expression occurred. Specifically, we collapsed all of the contralateral control values into a single group and then performed soft clustering on the Z-score normalized time-series data (Futschik & Carlisle, 2005; Kumar & Futschik, 2007). With this approach, we were able to identify two large clusters (Fig. 7A–C) and two small clusters (Fig. 8A–C) of proteins that showed similar temporal patterns in their expression level. We then used 1D annotation enrichment analyses to identify the top 10 most highly enriched GO terms within each cluster (Figs 7D and E and 8D and E).
Figure 7. Temporal analysis of the changes in protein expression that occur after the onset of immobilization.
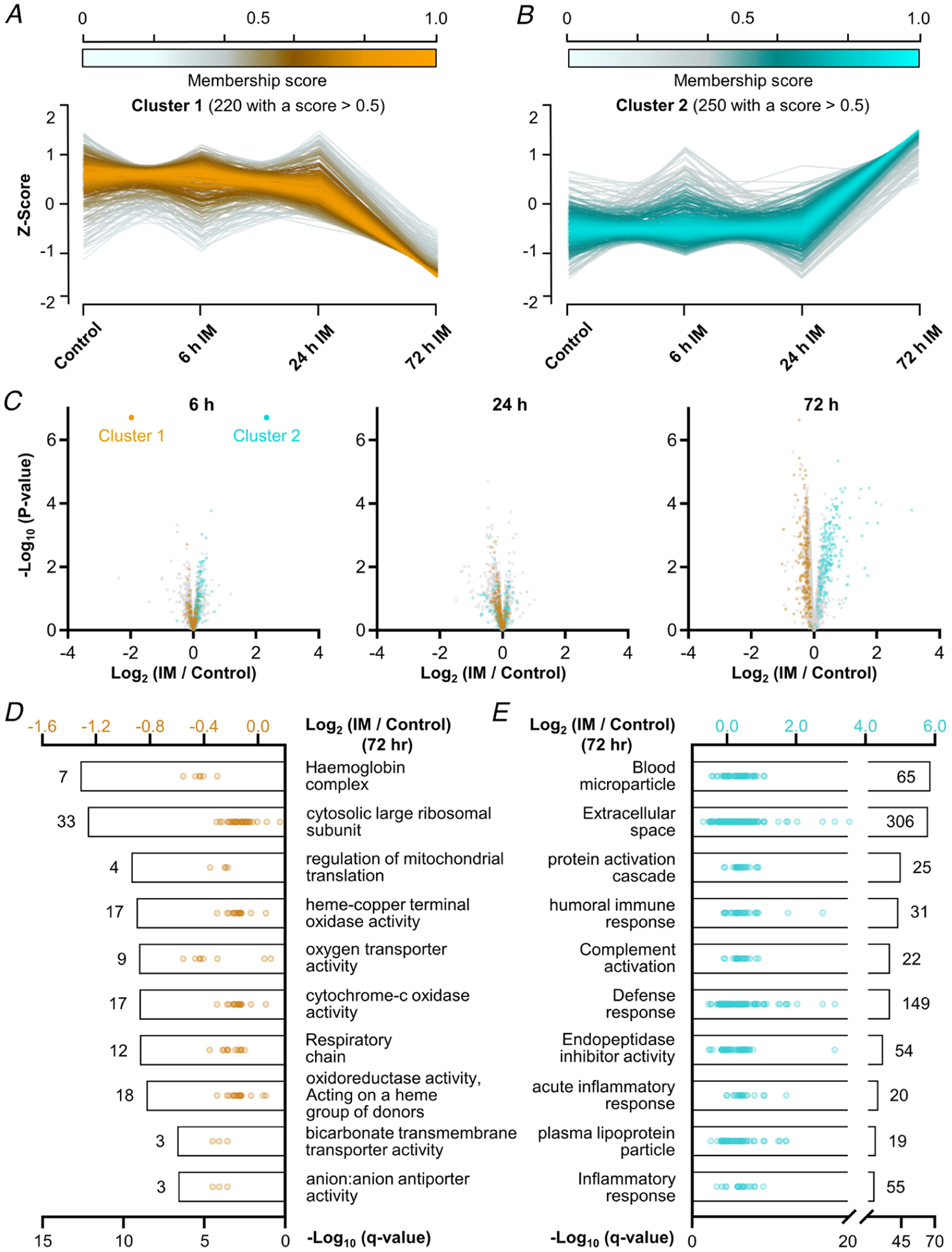
Soft clustering on Z-score normalized data proteomic data with Mfuzz led to the identification of four clusters (Figs 7 and 8). A and B, graphs showing the temporal pattern for the two largest clusters along with the associated membership score for each of the proteins in the cluster. C, volcano plots highlighting the position of cluster 1 (orange) and cluster 2 (cyan) proteins that possessed a membership score of > 0.5. Only data points with at least two valid values per group are shown in the plots. D and E, top 10 enriched GO terms in each cluster with the bars indicating the associated −log10 FDR-corrected P-value (i.e. q-value, bottom x-axis). The values indicate the number of proteins within each GO term, and the dots represent the log2 fold change of the individual proteins (top x-axis).
Figure 8. Temporal analysis of the changes in protein expression that occur after the onset of immobilization.
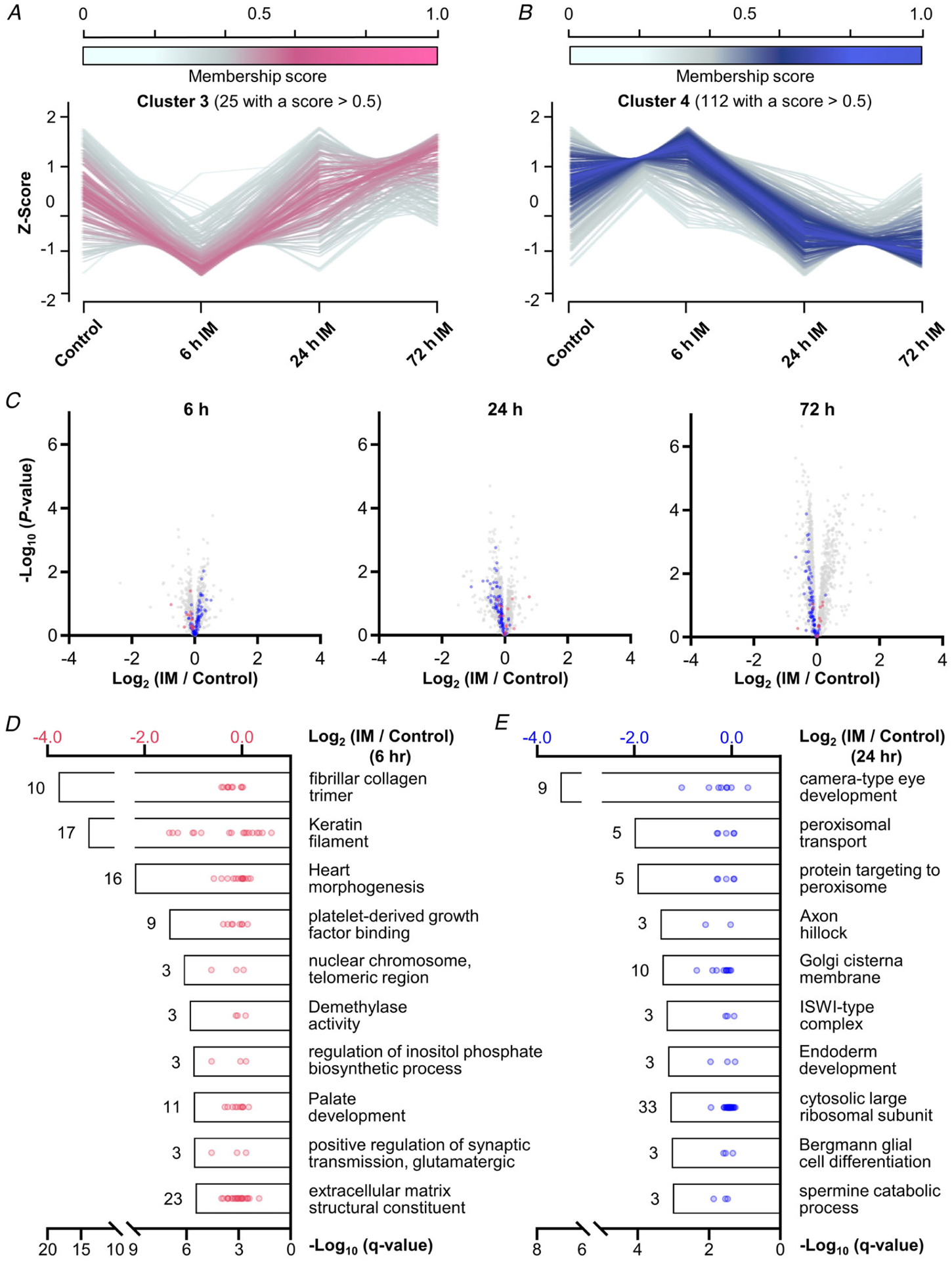
Soft clustering on Z-score normalized data proteomic data with Mfuzz led to the identification of four clusters. A and B, graphs showing the temporal pattern for clusters 3 and 4 along with the associated membership score for each of the proteins in the cluster. C, volcano plots highlighting the position of cluster 3 (purple) and cluster 4 (blue) proteins that possessed a membership score of > 0.5. Only data points with at least two valid values per group are shown in the plots. D and E, top 10 enriched GO term in each cluster with the bars indicating the associated −log10 FDR-corrected P-value (i.e. q-value, bottom x-axis). The values indicate the number of proteins within each GO term, and the dots represent the log2 fold change of the individual proteins (top x-axis).
The outcomes of the temporal analyses lead to several interesting observations. For instance, the proteins in cluster 2 revealed an abrupt increase in expression after 72 h of immobilization, and proteins annotated with the GO term ‘complement activation’ were dramatically over-represented in this cluster (FDR-corrected P < 1 × 10−36, Fig. 9A and B). This was interesting because members of the complement system circulate in the blood as inactive precursors and, once activated, they facilitate the localized production of pro-inflammatory molecules, as well as the formation of a membrane attack complex (MAC) that weakens the integrity of the cell membrane (Dunkelberger & Song, 2010; Noris & Remuzzi, 2013). Moreover, complement activation has been associated with the chronic inflammation that occurs in some models of muscle atrophy, and the level of transcripts that encode members of complement activation are elevated in several models of cachexia (Bonetto et al. 2011; Nosacka et al. 2020). Therefore, to further validate the proteomic observations, we first explored whether the increased prevalence of complement activation proteins might simply be due to an increased amount of blood in the muscle (impaired blood circulation, blood clot formation, etc.; Sachdeva et al. 2018). As illustrated in Fig. 9A and C, erythroid-specific proteins did not follow the cluster 2 temporal pattern but were instead found to be members of cluster 1 (Alvarez-Dominguez et al. 2017). This was an important observation because it provided support for the notion that the increased prevalence of complement proteins was not simply due to an increase in the presence of blood. Next, we performed western blot analysis on complement C3 as a means to orthogonally validate the proteomic data. Specifically, C3 is the central component of the complement system and, consistent with the proteomic data, the results of the western blots revealed a doubling in the amount of the C3α and β subunits after 72 h of immobilization (Fig. 9D, P ≤ 0.09) (Sahu & Lambris, 2001). Thus, when taken together, the results of our analyses indicate that immobilization leads to complement activation. Whether complement activation contributes to the loss of muscle mass is currently unknown and is certainly a topic that will be worthy of further investigation.
Figure 9. Immobilization leads to an increase in proteins that are involved in complement activation.
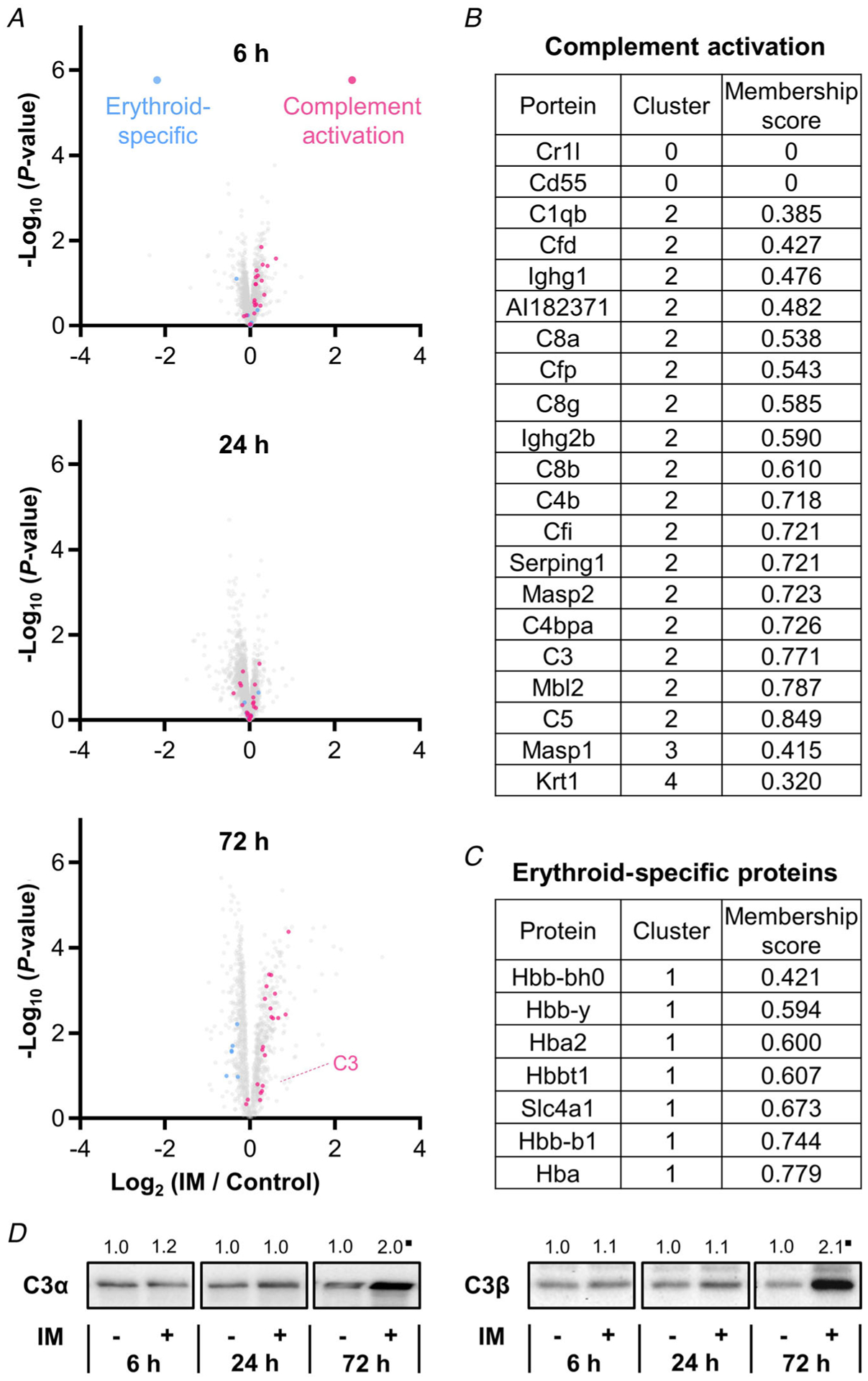
A, volcano plots highlighting the position of proteins that are annotated with the GO term ‘complement activation’ (pink) and ‘heme binding’ (blue). Only data points with at least two valid values per group are shown in the plots. B and C, list of all proteins annotated with the GO term ‘complement activation’ (B), and erythroid-specific proteins (C). D, representative western blots of complement component 3 (C3) α and β subunits. All values are group means expressed relative to the mean value obtained in the time-matched control samples; n = 5–7 per group. ∎Trend for significant difference from the time-matched control as determined by a pooled t test, P ≤ 0.09.
Discussion
In this study, we found that immobilization led to a rapid (within 6 h) and progressive decrease in the relative rate of protein synthesis (Fig. 1). Consistent with other studies in rodents, we also found that the rapid decrease in protein synthesis was not mediated by substantive changes in translational capacity (Fig. 2) (Goldspink, 1977; Booth & Seider, 1979; You et al. 2015). This is noteworthy because it indicates that the decrease in protein synthesis was largely mediated by a decrease in translational efficiency. Surprisingly, however, the results of our proteomic and phosphoproteomic analyses revealed that this effect could not be explained by changes in the abundance or phosphorylation state of proteins that have been implicated in the regulation of translation (Figs 4 and 6).
The lack of immobilization-induced changes in protein phosphorylation was highly unexpected and contradicts recent reviews which have argued that the immobilization-induced decrease in protein synthesis is mediated by a decrease in signalling by a protein kinase called the mechanistic target of rapamycin (mTOR) (Gao et al. 2018; Ji & Yeo, 2019). Indeed, it is well known that mTOR can regulate the phosphorylation state of several proteins that have been implicated in the regulation of translation (Gingras et al. 2001b; Hay & Sonenberg, 2004; Showkat et al. 2014; Saxton & Sabatini, 2017). However, the putative role that mTOR plays in the immobilization-induced decrease in protein synthesis is questionable. For instance, although some rodent-based studies have shown that immobilization can lead to the inhibition of mTOR signalling (You et al. 2010; Kelleher et al. 2013), there are also multiple studies which have shown that immobilization leads to the activation of mTOR signalling (You et al. 2015; Goodman et al. 2017; Docquier et al. 2019), or that it has no effect on mTOR signalling (Childs et al. 2003; Krawiec et al. 2005). Yet, unlike the highly variable responses that are observed with mTOR signalling, numerous studies have consistently shown that immobilization leads to a rapid decrease in the rate of protein synthesis in both rodents and humans (Goldspink, 1977; Booth & Seider, 1979; Kelleher et al. 2013; You et al. 2015; Kilroe et al. 2020). Thus, in our opinion, the notion that a decrease mTOR signalling serves as the primary driver of the immobilization-induced decrease in protein synthesis is not well founded.
In line with the aforementioned conclusion, the results of our study revealed that changes in mTOR signalling are not required for an immobilization-induced decrease in protein synthesis. Specifically, as shown in Fig. 4, our MS analyses detected several phosphorylation sites that are regulated by mTOR (e.g. serine 235/236 on RPS6, threonine 36/45 on 4E-BP1, serine 422 on eIF4B and threonine 57 on eEF2), and none of these sites were significantly altered at any time point following the onset of immobilization (Ryazanov & Davydova, 1989; Ferrari et al. 1991; Gingras et al. 1999; Wang et al. 2001; Raught et al. 2004; Hizli et al. 2013). Moreover, we used western blot analyses to perform an orthogonal validation of these MS results, and the outcomes confirmed that the aforementioned sites were not significantly altered by immobilization. In fact, the results of our study more broadly indicate that the rapid decrease in protein synthesis that occurs in response to immobilization is not mediated by changes in the abundance or phosphorylation state of any proteins.
Although it is well known that changes in phosphorylation can regulate the activity of proteins that control the rate of protein synthesis, there are a variety of other kinds of post-translational modifications that could also exert this effect, including sumoylation, glycosylation, ubiquitination, S-nitrosylation, methylation, acetylation, oxidation, lipidation and succinylation (Hess et al. 2005; Larsen et al. 2006; Wilkinson & Henley, 2010; Celi & Gabai, 2015; Jiang et al. 2018; Huang et al. 2019; Kumar et al. 2020). Indeed, it has been shown that the sumoylation of eIF4E can activate translation initiation, and O-GlyNAcylation of eIF4A1 can inhibit translation initiation via disruption of the eIF4F complex (Xu et al. 2010; Li et al. 2019). In addition to post-translational modifications, it appears that epigenetic/post-transcriptional modifications can also regulate translation. For instance, recent studies have shown that the acetylation of cytidines in mRNA can enhance translation and that the methylation of adenosines can inhibit translation (Slobodin et al. 2017; Arango et al. 2018). These examples are important to consider because they illustrate that non-traditional mechanisms could easily function as the primary driver of the immobilization-induced decrease in protein synthesis, and the results of our study indicate that role of such mechanisms warrants further investigation.
In addition to refining our understanding of the mechanisms that regulate the immobilization-induced decrease in protein synthesis, we also identified several proteome-based alterations that could potentially contribute to immobilization-induced loss of muscle mass. For instance, as described in the Results section, we identified a cluster of proteins (cluster 2) that showed an increase in expression after 72 h of immobilization, and this cluster of proteins was highly enriched with proteins that are involved in the activation of the complement system (Figs 7 and 9). We also identified a cluster of proteins (cluster 1) that showed a decrease in expression after 72 h of immobilization. Interestingly, this cluster of proteins was highly enriched with GO terms such as ‘cytochrome-c oxidase activity’, and ‘respiratory chain’ (Fig. 7). These GO terms are all associated with mitochondria and, as illustrated in Fig. 10, proteins that are annotated with the GO term ‘mitochondrial part’ revealed a small but extremely consistent decrease in expression after 72 h of immobilization (q = 0.002). This is noteworthy because previous studies have established that prolonged immobilization leads to a decrease in the quantity and function of mitochondria and that these changes probably contribute to immobilization-induced loss of muscle mass in both mice and humans (Abadi et al. 2009; Kang & Ji, 2013; Cannavino et al. 2014; Kang et al. 2016). Hence, we suspect that our proteomic dataset contains important clues about the mechanisms that contribute to the loss of factors such as mitochondria, and ultimately the loss of muscle mass.
Figure 10. Immobilization leads to a small but consistent decrease in mitochondrial proteins.
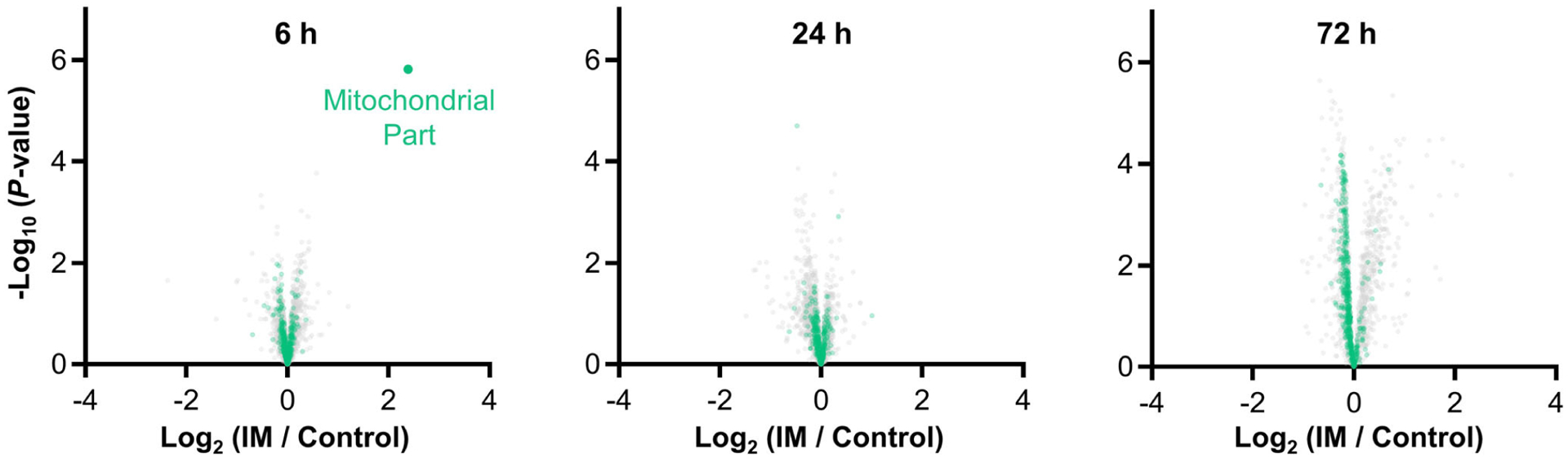
I Volcano plots from Fig. 4 were modified to highlight the position of proteins that are annotated with the GO term ‘mitochondrial part’. Only data points with at least two valid values per group are shown in the plots.
In summary, we have shown that immobilization leads to a rapid decrease in protein synthesis, and we used a deep MS-based analysis of the proteome and phosphoproteome to gain insight into the mechanisms that drive this effect. The validity of our analyses were supported by a carefully selected internal positive control, as well as various orthogonal analyses, and enabled us to firmly conclude that the rapid immobilization-induced decrease in protein synthesis is not mediated by changes in either the abundance, or phosphorylation state, of proteins. The outcomes of our work have not only challenged the current dogma in the field, but will also serve as an expansive resource of information for future studies that are aimed at defining how mechanical unloading leads to the loss of muscle mass.
Supplementary Material
Proteomic analysis summary document
Statistical Summary Document
Phosphoproteomic analysis summary document
Key points.
A decrease in protein synthesis plays a major role in the loss of muscle mass that occurs in response to immobilization.
In mice, immobilization leads to a rapid (within 6 h) and progressive decrease in the rate of protein synthesis and this effect is mediated by a decrease in translational efficiency.
Deep proteomic and phosphoproteomic analyses of mouse skeletal muscles revealed that the rapid immobilization-induced decrease in protein synthesis cannot be explained by changes in the abundance or phosphorylation state of proteins that have been implicated in the regulation of translation.
Funding
The research reported in this publication was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health under Award Numbers AR057347 and AR074932 to T.A.H. The research was also supported by the National Institutes of Health under Award Number GM108538 to J.J.C. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Biography

Kuan-Hung Lin completed his MSc at the National Taiwan University under the supervision of Dr Hsin-Fang Yang-Yen in the Department of Molecular Medicine. He is currently pursuing a PhD under the supervision of Dr Troy A. Hornberger (University of Wisconsin-Madison). His PhD research is primarily focused on studying the molecular mechanisms that regulate myogenesis.
Footnotes
Competing interests
J.J.C. is a consultant for Thermo Fisher Scientific; all other authors declare no conflicts of interest.
Data availability statement
All data needed to evaluate the conclusions or reperform analyses in the paper are present in the paper or the Supplementary Materials.
References
- Abadi A, Glover EI, Isfort RJ, Raha S, Safdar A, Yasuda N, Kaczor JJ, Melov S, Hubbard A, Qu X, Phillips SM & Tarnopolsky M (2009). Limb immobilization induces a coordinate down-regulation of mitochondrial and other metabolic pathways in men and women. PLoS One 4, e6518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acevedo JM, Hoermann B, Schlimbach T & Teleman AA (2018). Changes in global translation elongation or initiation rates shape the proteome via the Kozak sequence. Sci Rep 8, 4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Dominguez JR, Knoll M, Gromatzky AA & Lodish HF (2017). The super-enhancer-derived alncRNA-EC7/Bloodlinc potentiates red blood cell development in trans. Cell Rep 19, 2503–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arango D, Sturgill D, Alhusaini N, Dillman AA, Sweet TJ, Hanson G, Hosogane M, Sinclair WR, Nanan KK, Mandler MD, Fox SD, Zengeya TT, Andresson T, Meier JL, Coller J & Oberdoerffer S (2018). Acetylation of cytidine in mRNA promotes translation efficiency. Cell 175, 1872–1886.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhalevy D, Gupta SK, Danan CH, Ghosal S, Sun HW, Kazemier HG, Paeschke K, Hafner M & Juranek SA (2017). The human CCHC-type zinc finger nucleic acid-binding protein binds G-rich elements in target mRNA coding sequences and promotes translation. Cell Rep 18, 2979–2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y & Hochberg Y (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Methodol 57, 289–300. [Google Scholar]
- Bodine SC (2013). Disuse-induced muscle wasting. Int J Biochem Cell Biol 45, 2200–2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodine SC, Latres E, Baumhueter S, Lai VK, Nunez L, Clarke BA, Poueymirou WT, Panaro FJ, Na E, Dharmarajan K, Pan ZQ, Valenzuela DM, DeChiara TM, Stitt TN, Yancopoulos GD & Glass DJ (2001). Identification of ubiquitin ligases required for skeletal muscle atrophy. Science 294, 1704–1708. [DOI] [PubMed] [Google Scholar]
- Bonetto A, Aydogdu T, Kunzevitzky N, Guttridge DC, Khuri S, Koniaris LG & Zimmers TA (2011). STAT3 activation in skeletal muscle links muscle wasting and the acute phase response in cancer cachexia. PLoS One 6, e22538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth FW & Seider MJ (1979). Early change in skeletal muscle protein synthesis after limb immobilization of rats. J Appl Physiol Respir Environ Exerc Physiol 47, 974–977. [DOI] [PubMed] [Google Scholar]
- Cannavino J, Brocca L, Sandri M, Bottinelli R & Pellegrino MA (2014). PGC1-α over-expression prevents metabolic alterations and soleus muscle atrophy in hindlimb unloaded mice. J Physiol 592, 4575–4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlberg U, Nilsson A & Nygard O (1990). Functional properties of phosphorylated elongation factor 2. Eur J Biochem 191, 639–645. [DOI] [PubMed] [Google Scholar]
- Celi P & Gabai G (2015). Oxidant/antioxidant balance in animal nutrition and health: the role of protein oxidation. Front Vet Sci 2, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs TE, Spangenburg EE, Vyas DR & Booth FW (2003). Temporal alterations in protein signaling cascades during recovery from muscle atrophy. Am J Physiol Cell Physiol 285, C391–C398. [DOI] [PubMed] [Google Scholar]
- de Boer MD, Selby A, Atherton P, Smith K, Seynnes OR, Maganaris CN, Maffulli N, Movin T, Narici MV & Rennie MJ (2007). The temporal responses of protein synthesis, gene expression and cell signalling in human quadriceps muscle and patellar tendon to disuse. J Physiol 585, 241–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis MD, Jefferson LS & Kimball SR (2012). Role of p70S6K1-mediated phosphorylation of eIF4B and PDCD4 proteins in the regulation of protein synthesis. J Biol Chem 287, 42890–42899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Docquier A, Pavlin L, Raibon A, Bertrand-Gaday C, Sar C, Leibovitch S, Candau R & Bernardi H (2019). eIF3f depletion impedes mouse embryonic development, reduces adult skeletal muscle mass and amplifies muscle loss during disuse. J Physiol 597, 3107–3131. [DOI] [PubMed] [Google Scholar]
- Dunkelberger JR & Song WC (2010). Complement and its role in innate and adaptive immune responses. Cell Res 20, 34–50. [DOI] [PubMed] [Google Scholar]
- Ferrari S, Bandi HR, Hofsteenge J, Bussian BM & Thomas G (1991). Mitogen-activated 70K S6 kinase. Identification of in vitro 40 S ribosomal S6 phosphorylation sites. J Biol Chem 266, 22770–22775. [PubMed] [Google Scholar]
- Figueiredo VC (2019). Revisiting the roles of protein synthesis during skeletal muscle hypertrophy induced by exercise. Am J Physiol Regul Integr Comp Physiol 317, R709–R718. [DOI] [PubMed] [Google Scholar]
- Futschik ME & Carlisle B (2005). Noise-robust soft clustering of gene expression time-course data. J Bioinform Comput Biol 03, 965–988. [DOI] [PubMed] [Google Scholar]
- Gao Y, Arfat Y, Wang H & Goswami N (2018). Muscle atrophy induced by mechanical unloading: mechanisms and potential countermeasures. Front Physiol 9, 235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras AC, Gygi SP, Raught B, Polakiewicz RD, Abraham RT, Hoekstra MF, Aebersold R & Sonenberg N (1999). Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes Dev 13, 1422–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras AC, Raught B, Gygi SP, Niedzwiecka A, Miron M, Burley SK, Polakiewicz RD, Wyslouch-Cieszynska A, Aebersold R & Sonenberg N (2001a). Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes Dev 15, 2852–2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras AC, Raught B & Sonenberg N (2001b). Regulation of translation initiation by FRAP/mTOR. Genes Dev 15, 807–826. [DOI] [PubMed] [Google Scholar]
- Goldspink DF (1977). The influence of immobilization and stretch on protein turnover of rat skeletal muscle. J Physiol 264, 267–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman CA, Coenen AM, Frey JW, You JS, Barker RG, Frankish BP, Murphy RM & Hornberger TA (2017). Insights into the role and regulation of TCTP in skeletal muscle. Oncotarget 8, 18754–18772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman CA, Frey JW, Mabrey DM, Jacobs BL, Lincoln HC, You JS & Hornberger TA (2011a). The role of skeletal muscle mTOR in the regulation of mechanical load-induced growth. J Physiol 589, 5485–5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman CA, Mabrey DM, Frey JW, Miu MH, Schmidt EK, Pierre P & Hornberger TA (2011b). Novel insights into the regulation of skeletal muscle protein synthesis as revealed by a new nonradioactive in vivo technique. FASEB J 25, 1028–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman CA, Mayhew DL & Hornberger TA (2011c). Recent progress toward understanding the molecular mechanisms that regulate skeletal muscle mass. Cell Signal 23, 1896–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B & Yuan Y (2017). A comparative review of methods for comparing means using partially paired data. Stat Methods Med Res 26, 1323–1340. [DOI] [PubMed] [Google Scholar]
- Hamilton TL, Stoneley M, Spriggs KA & Bushell M (2006). TOPs and their regulation. Biochem Soc Trans 34, 12–16. [DOI] [PubMed] [Google Scholar]
- Hay N & Sonenberg N (2004). Upstream and downstream of mTOR. Genes Dev 18, 1926–1945. [DOI] [PubMed] [Google Scholar]
- Hershey JW (2010). Regulation of protein synthesis and the role of eIF3 in cancer. Braz J Med Biol Res 43, 920–930. [DOI] [PubMed] [Google Scholar]
- Hershey JW, Sonenberg N & Mathews MB (2012). Principles of translational control: an overview. Cold Spring Harb Perspect Biol 4, a011528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess DT, Matsumoto A, Kim SO, Marshall HE & Stamler JS (2005). Protein S-nitrosylation: purview and parameters. Nat Rev Mol Cell Biol 6, 150–166. [DOI] [PubMed] [Google Scholar]
- Hizli AA, Chi Y, Swanger J, Carter JH, Liao Y, Welcker M, Ryazanov AG & Clurman BE (2013). Phosphorylation of eukaryotic elongation factor 2 (eEF2) by cyclin A-cyclin-dependent kinase 2 regulates its inhibition by eEF2 kinase. Mol Cell Biol 33, 596–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman NJ, Parker BL, Chaudhuri R, Fisher-Wellman KH, Kleinert M, Humphrey SJ, Yang P, Holliday M, Trefely S, Fazakerley DJ, Stockli J, Burchfield JG, Jensen TE, Jothi R, Kiens B, Wojtaszewski JF, Richter EA & James DE (2015). Global phosphoproteomic analysis of human skeletal muscle reveals a network of exercise-regulated kinases and AMPK substrates. Cell Metab 22, 922–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holz MK, Ballif BA, Gygi SP & Blenis J (2005). mTOR and S6K1 mediate assembly of the translation preinitiation complex through dynamic protein interchange and ordered phosphorylation events. Cell 123, 569–580. [DOI] [PubMed] [Google Scholar]
- Huang KY, Hsu JB & Lee TY (2019). Characterization and identification of lysine succinylation sites based on deep learning method. Sci Rep 9, 16175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huichalaf C, Schoser B, Schneider-Gold C, Jin B, Sarkar P & Timchenko L (2009). Reduction of the rate of protein translation in patients with myotonic dystrophy 2. J Neurosci 29, 9042–9049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen I, Shepard DS, Katzmarzyk PT & Roubenoff R (2004). The healthcare costs of sarcopenia in the United States. J Am Geriatr Soc 52, 80–85. [DOI] [PubMed] [Google Scholar]
- Ji LL & Yeo D (2019). Cellular mechanism of immobilization-induced muscle atrophy: a mini review. Sport Med Health Sci 1, 19–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia J, Arif A, Terenzi F, Willard B, Plow EF, Hazen SL & Fox PL (2014). Target-selective protein S-nitrosylation by sequence motif recognition. Cell 159, 623–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Zhang X, Chen X, Aramsangtienchai P, Tong Z & Lin H (2018). Protein lipidation: occurrence, mechanisms, biological functions, and enabling technologies. Chem Rev 118, 919–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang C & Ji LL (2013). Muscle immobilization and remobilization downregulates PGC-1α signaling and the mitochondrial biogenesis pathway. J Appl Physiol 115, 1618–1625. [DOI] [PubMed] [Google Scholar]
- Kang C, Yeo D & Ji LL (2016). Muscle immobilization activates mitophagy and disrupts mitochondrial dynamics in mice. Acta Physiol 218, 188–197. [DOI] [PubMed] [Google Scholar]
- Kelleher AR, Kimball SR, Dennis MD, Schilder RJ & Jefferson LS (2013). The mTORC1 signaling repressors REDD1/2 are rapidly induced and activation of p70S6K1 by leucine is defective in skeletal muscle of an immobilized rat hindlimb. Am J Physiol Endocrinol Metab 304, E229–E236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilroe SP, Fulford J, Jackman S, Holwerda A, Gijsen A, van Loon L & Wall BT (2021). Dietary protein intake does not modulate daily myofibrillar protein synthesis rates or loss of muscle mass and function during short-term immobilization in young men: a randomized controlled trial. Am Am J Clin Nutr 113, 548–561. [DOI] [PubMed] [Google Scholar]
- Kirby TJ, Lee JD, England JH, Chaillou T, Esser KA & McCarthy JJ (2015). Blunted hypertrophic response in aged skeletal muscle is associated with decreased ribosome biogenesis. J Appl Physiol 119, 321–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawiec BJ, Frost RA, Vary TC, Jefferson LS & Lang CH (2005). Hindlimb casting decreases muscle mass in part by proteasome-dependent proteolysis but independent of protein synthesis. Am J Physiol Endocrinol Metab 289, E969–E980. [DOI] [PubMed] [Google Scholar]
- Kumar A, Narayanan V & Sekhar A (2020). Characterizing post-translational modifications and their effects on protein conformation using NMR spectroscopy. Biochemistry 59, 57–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar L & Futschik ME (2007). Mfuzz: a software package for soft clustering of microarray data. Bioinformation 2, 5–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang SM, Kazi AA, Hong-Brown L & Lang CH (2012). Delayed recovery of skeletal muscle mass following hindlimb immobilization in mTOR heterozygous mice. PLoS One 7, e38910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen MR, Trelle MB, Thingholm TE & Jensen ON (2006). Analysis of posttranslational modifications of proteins by tandem mass spectrometry. BioTechniques 40, 790–798. [DOI] [PubMed] [Google Scholar]
- Li X, Zhu Q, Shi X, Cheng Y, Li X, Xu H, Duan X, Hsieh-Wilson LC, Chu J, Pelletier J, Ni M, Zheng Z, Li S & Yi W (2019). O-GlcNAcylation of core components of the translation initiation machinery regulates protein synthesis. Proc Natl Acad Sci U S A 116, 7857–7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M, Ge Q, Wang G, Luo Y, Wang X, Jiang W, Liu X, Wu CL, Xiao Y & Wang X (2018). CIRBP is a novel oncogene in human bladder cancer inducing expression of HIF-1α. Cell Death Dis 9, 1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell SA, Spriggs KA, Coldwell MJ, Jackson RJ & Willis AE (2003). The Apaf-1 internal ribosome entry segment attains the correct structural conformation for function via interactions with PTB and unr. Mol Cell 11, 757–771. [DOI] [PubMed] [Google Scholar]
- Nader GA, Hornberger TA & Esser KA (2002). Translational control: implications for skeletal muscle hypertrophy. Clin Orthop Relat Res 403, S178–S187. [DOI] [PubMed] [Google Scholar]
- Nakada S, Ogasawara R, Kawada S, Maekawa T & Ishii N (2016). Correlation between ribosome biogenesis and the magnitude of hypertrophy in overloaded skeletal muscle. PLoS One 11, e0147284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noris M & Remuzzi G (2013). Overview of complement activation and regulation. Semin Nephrol 33, 479–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosacka RL, Delitto AE, Delitto D, Patel R, Judge SM, Trevino JG & Judge AR (2020). Distinct cachexia profiles in response to human pancreatic tumours in mouse limb and respiratory muscle. J Cachexia Sarcopenia Muscle 11, 820–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto T, Torii S & Machida S (2011). Differential gene expression of muscle-specific ubiquitin ligase MAFbx/Atrogin-1 and MuRF1 in response to immobilization-induced atrophy of slow-twitch and fast-twitch muscles. J Physiol Sci 61, 537–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pause A, Belsham GJ, Gingras AC, Donze O, Lin TA, Lawrence JC Jr & Sonenberg N (1994). Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature 371, 762–767. [DOI] [PubMed] [Google Scholar]
- Phillips SM & McGlory C (2014). CrossTalk proposal: The dominant mechanism causing disuse muscle atrophy is decreased protein synthesis. J Physiol 592, 5341–5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts GK, McNally RM, Blanco R, You JS, Hebert AS, Westphall MS, Coon JJ & Hornberger TA (2017). A map of the phosphoproteomic alterations that occur after a bout of maximal-intensity contractions. J Physiol 595, 5209–5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raught B, Peiretti F, Gingras AC, Livingstone M, Shahbazian D, Mayeur GL, Polakiewicz RD, Sonenberg N & Hershey JW (2004). Phosphorylation of eucaryotic translation initiation factor 4B Ser422 is modulated by S6 kinases. EMBO J 23, 1761–1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riba A, Di Nanni N, Mittal N, Arhné E, Schmidt A & Zavolan M (2019). Protein synthesis rates and ribosome occupancies reveal determinants of translation elongation rates. Proc Natl Acad Sci U S A 116, 15023–15032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux PP, Shahbazian D, Vu H, Holz MK, Cohen MS, Taunton J, Sonenberg N & Blenis J (2007). RAS/ERK signaling promotes site-specific ribosomal protein S6 phosphorylation via RSK and stimulates cap-dependent translation. J Biol Chem 282, 14056–14064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph JD, de Graauw M, van de Water B, Geiger T & Sharan R (2016). Elucidation of signaling pathways from large-scale phosphoproteomic data using protein interaction networks. Cell Syst 3, 585–593 e583. [DOI] [PubMed] [Google Scholar]
- Ryazanov AG & Davydova EK (1989). Mechanism of elongation factor 2 (EF-2) inactivation upon phosphorylation. Phosphorylated EF-2 is unable to catalyze translocation. FEBS Lett 251, 187–190. [DOI] [PubMed] [Google Scholar]
- Sachdeva A, Dalton M & Lees T (2018). Graduated compression stockings for prevention of deep vein thrombosis. Cochrane Database Syst Rev 11, CD001484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahu A & Lambris JD (2001). Structure and biology of complement protein C3, a connecting link between innate and acquired immunity. Immunol Rev 180, 35–48. [DOI] [PubMed] [Google Scholar]
- Saxton RA & Sabatini DM (2017). mTOR signaling in growth, metabolism, and disease. Cell 168, 960–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Showkat M, Beigh MA & Andrabi KI (2014). mTOR signaling in protein translation regulation: implications in cancer genesis and therapeutic interventions. Mol Biol Int 2014, 686984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slobodin B, Han R, Calderone V, Vrielink J, Loayza-Puch F, Elkon R & Agami R (2017). Transcription impacts the efficiency of mRNA translation via co-transcriptional N6-adenosine methylation. Cell 169, 326–337.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart F, Aschrafi A, Atkins A, Owens GC, Pilotte J, Cunningham BA & Vanderklish PW (2007). Two isoforms of the cold-inducible mRNA-binding protein RBM3 localize to dendrites and promote translation. J Neurochem 101, 1367–1379. [DOI] [PubMed] [Google Scholar]
- Smyth GK (2004). Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3, 1. [DOI] [PubMed] [Google Scholar]
- Srikanthan P & Karlamangla AS (2011). Relative muscle mass is inversely associated with insulin resistance and prediabetes. Findings from the third National Health and Nutrition Examination Survey. J Clin Endocrinol Metab 96, 2898–2903. [DOI] [PubMed] [Google Scholar]
- Steinert ND, Potts GK, Wilson GM, Klamen AM, Lin KH, Hermanson JB, McNally RM, Coon JJ & Hornberger TA (2021). Mapping of the contraction-induced phosphoproteome identifies TRIM28 as a significant regulator of skeletal muscle size and function. Cell Rep 34, 108796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symons TB, Sheffield-Moore M, Chinkes DL, Ferrando AA & Paddon-Jones D (2009). Artificial gravity maintains skeletal muscle protein synthesis during 21 days of simulated micro-gravity. J Appl Physiol 107, 34–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS & Sabatini DM (2012). A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 485, 109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyanova S & Cox J (2018). Perseus: A Bioinformatics platform for integrative analysis of proteomics data in cancer research. Methods Mol Biol 1711, 133–148. [DOI] [PubMed] [Google Scholar]
- Venkata Subbaiah KC, Wu J, Potdar A & Yao P (2019). hnRNP L-mediated RNA switches function as a hypoxia-induced translational regulon. Biochem Biophys Res Commun 516, 753–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Li W, Williams M, Terada N, Alessi DR & Proud CG (2001). Regulation of elongation factor 2 kinase by p90(RSK1) and p70 S6 kinase. EMBO J 20, 4370–4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenger CD, Phanstiel DH, Lee MV, Bailey DJ & Coon JJ (2011). COMPASS: a suite of pre- and post-search proteomics software tools for OMSSA. Proteomics 11, 1064–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson KA & Henley JM (2010). Mechanisms, regulation and consequences of protein SUMOylation. Biochem J 428, 133–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Vatsyayan J, Gao C, Bakkenist CJ & Hu J (2010). HDAC2 promotes eIF4E sumoylation and activates mRNA translation gene specifically. J Biol Chem 285, 18139–18143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You JS, Anderson GB, Dooley MS & Hornberger TA (2015). The role of mTOR signaling in the regulation of protein synthesis and muscle mass during immobilization in mice. Dis Model Mech 8, 1059–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You JS, Dooley MS, Kim CR, Kim EJ, Xu W, Goodman CA & Hornberger TA (2018). A DGKζ-FoxO-ubiquitin proteolytic axis controls fiber size during skeletal muscle remodeling. Sci Signal 11, eaao6847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You JS, McNally RM, Jacobs BL, Privett RE, Gundermann DM, Lin KH, Steinert ND, Goodman CA & Hornberger TA (2019). The role of raptor in the mechanical load-induced regulation of mTOR signaling, protein synthesis, and skeletal muscle hypertrophy. FASEB J 33, 4021–4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You JS, Park MN, Song W & Lee YS (2010). Dietary fish oil alleviates soleus atrophy during immobilization in association with Akt signaling to p70s6k and E3 ubiquitin ligases in rats. Appl Physiol Nutr Metab 35, 310–318. [DOI] [PubMed] [Google Scholar]
- Zhou X, Wang JL, Lu J, Song Y, Kwak KS, Jiao Q, Rosenfeld R, Chen Q, Boone T, Simonet WS, Lacey DL, Goldberg AL & Han HQ (2010). Reversal of cancer cachexia and muscle wasting by ActRIIB antagonism leads to prolonged survival. Cell 142, 531–543. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Proteomic analysis summary document
Statistical Summary Document
Phosphoproteomic analysis summary document
Data Availability Statement
All data needed to evaluate the conclusions or reperform analyses in the paper are present in the paper or the Supplementary Materials.


