Abstract
Purpose
Large-scale genome-wide association studies (GWAS) have reported important single nucleotide polymorphisms (SNPs) with significant associations with age-related macular degeneration (AMD). However, their role in disease development remains elusive. This study aimed to assess SNP–metabolite associations (i.e., metabolite quantitative trait loci [met-QTL]) and to provide insights into the biological mechanisms of AMD risk SNPs.
Design
Cross-sectional multicenter study (Boston, Massachusetts, and Coimbra, Portugal).
Participants
Patients with AMD (n = 388) and control participants (n = 98) without any vitreoretinal disease (> 50 years).
Methods
Age-related macular degeneration grading was performed using color fundus photographs according to the Age-Related Eye Disease Study classification scheme. Fasting blood samples were collected and evaluated with mass spectrometry for metabolomic profiling and Illumina OmniExpress for SNPs profiling. Analyses of met-QTL of endogenous metabolites were conducted using linear regression models adjusted for age, gender, smoking, 10 metabolite principal components (PCs), and 10 SNP PCs. Additionally, we analyzed the cumulative effect of AMD risk SNPs on plasma metabolites by generating genetic risk scores and assessing their associations with metabolites using linear regression models, accounting for the same covariates. Modeling was performed first for each cohort, and then combined by meta-analysis. Multiple comparisons were accounted for using the false discovery rate (FDR).
Main Outcome Measures
Plasma metabolite levels associated with AMD risk SNPs.
Results
After quality control, data for 544 plasma metabolites were included. Meta-analysis of data from all individuals (AMD patients and control participants) identified 28 significant met-QTL (β = 0.016–0.083; FDR q-value < 1.14 × 10–2), which corresponded to 5 metabolites and 2 genes: ASPM and LIPC. Polymorphisms in the LIPC gene were associated with phosphatidylethanolamine metabolites, which are glycerophospholipids, and polymorphisms in the ASPM gene with branched-chain amino acids. Similar results were observed when considering only patients with AMD. Genetic risk score–metabolite associations further supported a global impact of AMD risk SNPs on the plasma metabolome.
Conclusions
This study demonstrated that genomic–metabolomic associations can provide insights into the biological relevance of AMD risk SNPs. In particular, our results support that the LIPC gene and the glycerophospholipid metabolic pathway may play an important role in AMD, thus offering new potential therapeutic targets for this disease.
Keywords: AMD, Genetics, Metabolomic-genomic associations, Metabolomics
Abbreviations and Acronyms: AMD, age-related macular degeneration; BCAA, branched-chain amino acid; CFH, complement factor H; FDR, false discovery rate; GRS, genetic risk score; GWAS, genome-wide association study; met-QTL, metabolite quantitative trait loci; MS, mass spectrometry; PC, principal component; PE, phosphatidylethanolamine; SNP, single nucleotide polymorphism
Age-related macular degeneration (AMD) is a leading cause of blindness worldwide in people older than 50 years.1 Age-related macular degeneration susceptibility is determined by a combination of genetic and environmental risk factors. Support for a genetic contribution in AMD was proposed originally by classical epidemiologic and twin studies and became well established with the advent of genome-wide association studies (GWASs), which have identified 34 loci with more than 7000 single nucleotide polymorphisms (SNPs) linked to AMD risk, 52 of them with independent associations.2 Additional loci and candidate genes also have been proposed.3 Despite identifying strong associations between certain SNPs and AMD,2 GWAS studies do not provide information on the underlying disease mechanisms.4 To date, the biological consequences of most of the identified SNPs remain unclear. This knowledge gap has important consequences for clinical care. Currently, only advanced neovascular AMD can be treated.5 Limited treatment options exist to reduce AMD progression,6 and no existing treatments exist for all forms of dry AMD.
Metabolites are downstream of the genetic transcription process and may be associated with causal genes.7 Thus, they have been proposed as functional intermediates to investigate biological mechanisms in several conditions.8,9 Indeed, the study of SNP–metabolite associations can overcome some of the limitations of GWAS studies.10,11 Our group has reported12, 13, 14 that patients with AMD have a distinct plasma metabolomic profile as compared with control participants and that this profile changes with disease severity. The next step in this work is to analyze genomic–metabolomic associations, which may help to further our understanding of the functional role of AMD risk genes, as well as to provide insights into the underlying mechanisms of AMD. To our knowledge, only 1 study has been performed to date examining genomic–metabolomic associations in AMD.15 This well-designed study was performed by the Eye-Risk Consortium and had a large sample size, but used nuclear magnetic resonance spectroscopy instead of mass spectrometry (MS).16 Mass spectrometry measures a much broader range of metabolites and is becoming widely used in both clinical research and large-scale epidemiologic studies.16 Importantly, the 2 studies, that of the Eye-Risk Consortium and the current one, validate and complement each other.
In this study, we performed genomic–metabolomic analyses with the goal of increasing our understanding of the pathogenic mechanisms of AMD risk SNPs. To do so, we analyzed the association between established AMD risk SNPs and plasma metabolites measured by MS in our 2 cohorts: one from Boston, Massachusetts, and the other from Coimbra, Portugal.
Methods
This was a cross-sectional observational study that took place at 2 sites: the Department of Ophthalmology at Massachusetts Eye and Ear and Harvard Medical School, Boston, Massachusetts, and the Faculty of Medicine of the University of Coimbra, Coimbra, Portugal, in collaboration with the Association for Innovation and Biomedical Research on Light and Image and the Centro Hospitalar e Universitário de Coimbra, Coimbra, Portugal. The clinical protocol was conducted in accordance with the Health Insurance Portability and Accountability Act requirements and the tenets of the Declaration of Helsinki and was approved by the institutional review boards of Massachusetts Eye and Ear, Massachusetts General Brigham, the Faculty of Medicine of the University of Coimbra, the Association for Innovation and Biomedical Research on Light and Image, and the Portuguese National Data Protection Committee. All participants enrolled in the study provided written informed consent.
Eligibility Criteria
From January 2015 through July 2016, we recruited patients diagnosed with AMD as well as control participants with no evidence of AMD who were 50 years of age or older at both study sites (Boston, Massachusetts, and Coimbra, Portugal). At Massachusetts Eye and Ear, participants were recruited from the Retina Service and Comprehensive Ophthalmology and Optometry Services at their regular appointments. The Portuguese (the Faculty of Medicine of the University of Coimbra and the Association for Innovation and Biomedical Research on Light and Image) study population was derived from a population-based cohort study.17 All participants who had taken part in this project and had an established diagnosis of any stage of AMD were invited to participate in the current study. Those without signs of AMD in a prior evaluation17 also were invited and were included as control participants if they remained without the disease in the present evaluation (see criteria below).
For both cohorts, the exclusion criteria included: diagnosis of any other vitreoretinal disease, active uveitis or ocular infection, significant media opacities that precluded the observation of the ocular fundus, refractive error of 6 diopters or more of spherical equivalent, history of retinal surgery, history of any ocular surgery or intraocular procedure (such as laser therapy or intraocular injections) within the 90 days before enrolment, and diagnosis of diabetes mellitus.
Study Protocol
As described previously,13,14 all participants included in the study underwent a complete bilateral ophthalmologic examination, including a dilated fundus examination, and were imaged with 7-field nonstereoscopic color fundus photography using either a Topcon TRC-50DX (Topcon Corporation) or a Zeiss FF-450Plus (Carl Zeiss Meditec) camera. Additionally, a complete medical history was obtained, according to a standardized questionnaire, which included self-reported data on smoking habits (all those who ever smoked were included as smokers).
For all participants, 2 blood samples were collected: (1) 1 into a sodium–heparin tube, which was centrifuged within 30 minutes (1500 rpm for 10 minutes at 20° C) to obtain plasma for metabolomic analysis, and (2) the other one into an ethylenediaminetetraacetic acid tube that was used for DNA extraction. Overnight fasting was required, and samples were always collected in the morning. In Coimbra, Portugal, because the study visit was planned in advance, patients were informed about the fasting requirement a priori and sample collection always took place on the same day of the remaining study procedures. For the Boston cohort, a separate appointment for fasting blood collection frequently had to be scheduled because patients were recruited during their regular ophthalmic appointments. This collection appointment always occurred within a maximum of 1 month after study inclusion.
Age-Related Macular Degeneration Diagnosis and Staging
For AMD diagnosis and staging, 2 of 3 independent experienced graders analyzed field 2 color fundus photographs, according to the Age-Related Eye Disease Study classification system.18 In case of disagreement, a senior author (R.S. or D.H.) established the final categorization. Before grading, images were standardized using software developed by our group.19 Images obtained with Topcon cameras were evaluated with IMAGEnet 2000 software version 2.56 (Topcon Medical Systems), and those obtained with a Zeiss camera were observed using VISUPAC version 4.5.1 (Carl Zeiss Meditec).
We adopted the most recent Age-Related Eye Disease Study 2 definitions,18 namely (1) that the standard disc diameter equals 1800 μm (rather than 1500 μm), which affects the size of the Early Treatment Diabetic Retinopathy Study grid and of the standard drusen circles; and (2) that geographic atrophy is present if the lesion has a diameter of 433 μm or more (Age-Related Eye Disease Study circle I-2) and at least 2 of the following features are present: absence of retinal pigment epithelium pigment, circular shape, or sharp margins (meaning that involvement of the fovea is not a requirement). Therefore, the following groups were established and used for further assessments18,20: control, defined as the presence of drusen maximum size less than circle C0 and total area less than C1; early AMD, defined as drusen maximum size of C0 or more but less than C1 or presence of AMD characteristic pigment abnormalities in the inner or central subfields; intermediate AMD, defined as the presence drusen maximum size C1 or more or of drusen maximum size of C0 or more if the total area occupied is more than I2 for soft indistinct drusen and more than 02 for soft distinct drusen; and late AMD, defined as the presence of geographic atrophy according to the criteria described above (geographic atrophy or dry late AMD) or evidence of neovascular AMD (choroidal neovascularization or wet AMD).
Metabolomic Profiling and Data Processing
Fasting blood samples were collected into a sodium–heparin tube, which was centrifuged within 30 minutes (1500 rpm for 10 minutes at 20° C). Plasma aliquots of 1.5 ml (Boston) and 5 ml (Portugal) were transferred into sterile cryovials and stored at –80° C. When all participants had been recruited, plasma samples from Portugal were shipped to Massachusetts Eye and Ear in dry ice (through TNT Express, Inc.). Then, all samples (i.e., from both study locations) were shipped to Metabolon, Inc., also in dry ice (through TNT Express, Inc.). In both cases, frozen samples arrived frozen in less than 48 hours and were stored immediately at –80° C until processing. Nontargeted MS analysis was performed by Metabolon, using ultrahigh-performance liquid chromatography–tandem MS, according to previously published protocols.13 Samples from a pilot study (n = 120) were analyzed initially.13 Then, all the remaining samples underwent MS analysis simultaneously, with random samples from the initial batch included for data normalization.
Metabolite data were run through our standard quality control and data processing pipeline.21 Namely, metabolite levels were log-transformed to obtain approximately normal distributions and then pareto scaled to reduce the influence of metabolites with very high levels while keeping the data structure partially intact. These steps are important to allow for a standardized comparison of metabolite levels.22 We observed that 3 participants (2 from Portugal and 1 from Boston) had missing or undetectable levels for more than 30% of the metabolites and therefore were excluded. For all the remaining participants, the highest percentage of missing values was less than 7%. To ensure that only the most informative metabolites were included in the analyses, those metabolites with interquartile range levels of 0 were excluded.21 Missing values were imputed with half the minimum detected level for that metabolite.23 Additionally, 61 of the resulting metabolites (n = 605) were determined to be exogenous to humans (e.g., medications, food additives, and buffering agents) and were excluded from subsequent analyses because we were interested in investigating only endogenous metabolites that could be driving systemic biological characteristics. Thus, the final analyses included 544 endogenous metabolites (Supplemental Table 1). Among these, most showed 0 or a very low proportion of imputed data, with only 7% (n = 40) demonstrating missing values higher than 10% (maximum, 25%).
Genomic Profiling and Data Processing
For genetic profiling, venous blood samples were collected into ethylenediaminetetraacetic acid tubes. In Boston, the Massachusetts Eye and Ear Ocular Genomics Institute performed DNA extraction using the QIASymphony with QIASymphony DSP DNA Mini Kits (Qiagen). In Coimbra, Portugal, this was carried out by the Laboratorio de Citogenetica e Genomica of the Faculty of Medicine, University of Coimbra, using a GeneCatcher gDNA Blood Kit (Invitrogen). After extraction, the DNA was stored in the DNA accessioning room refrigerator under proper temperature conditions.
When all participants had been recruited, samples from Portugal were shipped to Massachusetts Eye and Ear (through TNT Express, Inc.). Then, all samples (i.e., from both study locations) were shipped to the Broad Genomics Institute. The Broad Genomics Institute completed whole genome genotyping on these samples using the OmniExpress array (HumanOmniExpress-24v1-1_A), which provides data on 713 014 SNPs. These data were shared with us through an encrypted website.
As part of our quality control procedures, we removed poorly genotyped variants with minor allele frequency of less than 1%, deviations from Hardy-Weinberg equilibrium with P values < 0.00001, and percentage of missing values of more than 5%. After this, 596 749 SNPs remained. Genotype imputation was performed using the 1,000 Genomes Project phase 3 as a reference panel. We excluded SNPs with minor allele frequency of less than 5% and SNPs with bad imputation quality (R2 < 0.8). This yielded a final total of 6 261 164 SNPs. Variant identifiers were established based on National Center for Biotechnology Information dbSNP version 137 and chromosomal position of the variants on the National Center for Biotechnology Information Reference Sequence Human Genome Build 19.
Statistical Analysis
Descriptive statistics were used to summarize the clinical and demographic characteristics of the included study population, including mean and standard deviation for continuous variables and percentages for dichotomous or categorical variables.
Our goal was to assess associations between known AMD risk SNPs and plasma metabolites using the data previously published by the International AMD Genomics Consortium,2 where more than 7000 genome-wide significant variants were identified and made available online. We determined that of these variants, 4795 genotyped and imputed SNPs (passing the genome-wide significance level across 34 independent genomic regions) were available in our population (Supplemental Table 2). These therefore were considered for statistical analyses (Fig 1). We then assessed metabolic quantitative trait loci (met-QTL) by testing for an association between the 544 metabolites and 4795 SNPs, where a met-QTL is defined as a DNA locus that correlates with variation of a metabolite.24 We first analyzed each cohort separately (i.e., Boston and Portugal). For each metabolite–gene pair, linear models adjusting for age, gender, smoking status (0 = nonsmoker; 1 = current or former smoker), and 10 SNP principal components (PCs); 10 metabolite PCs were computed. Of note, the 10 SNP PCs (representing potential population structure) and 10 metabolite PCs (representing nongenetic batch effect) corresponded to the top 10 PCs obtained from the linear combination of the original genetic (n = 6 000 000) and metabolomic (n = 544) data of 486 individuals using PC analysis. The top 10 SNP PCs were expected to adjust for population structure sufficiently in the Boston and Portugal samples, respectively. No significant associations were seen between PCs and metabolite measurements, supporting that our findings are unlikely to be driven by population stratification and admixture bias. The top 10 metabolite PCs (Supplemental Table 3) were determined to optimize the number of significant met-QTLs based on whole genome SNPs and all 544 metabolites. β-Coefficients were reported as estimates of effect size for each pair of the effect allele on the mean level of the metabolite.
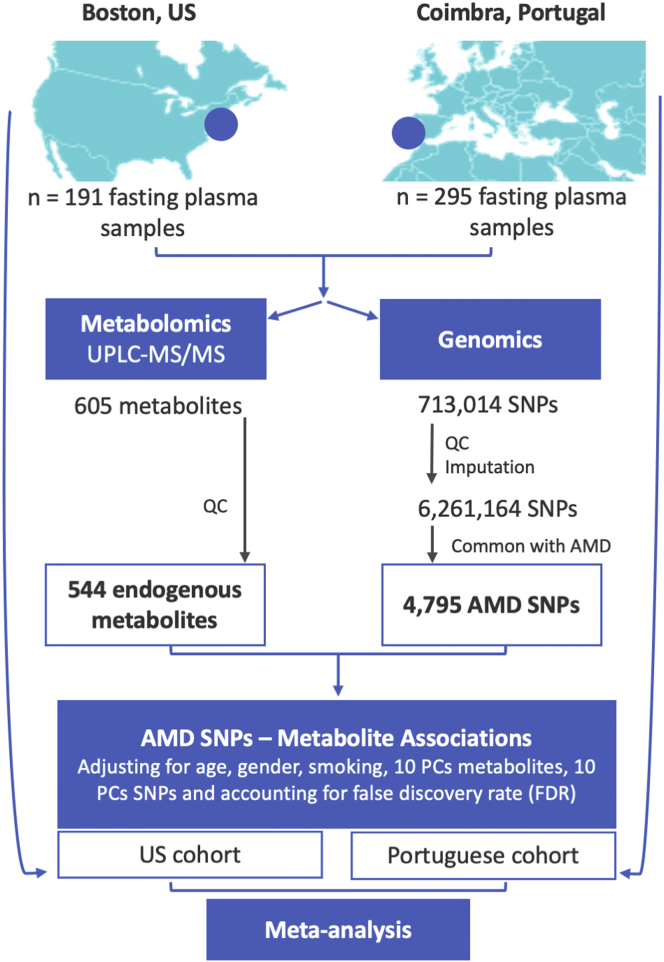
Figure 1.
Schematic representation of the study design. PC = principal component; QC = quality control; SNP = single nucleotide polymorphism; UPLC-MS/MS = ultra-performance liquid chromatography tandem mass spectrometry; US = United States.
After the analysis of each of the 2 cohorts separately, we combined their met-QTL results by performing a fixed-effect meta-analysis. For each met-QTL (SNP–metabolite pair), we computed the pooled inverse variance weighted β-coefficient.25 Then, the meta-analysis P values for each met-QTL were computed based on Z score. These analyses were performed considering all individuals included in this study (i.e., both AMD patients and control participants). Additionally, we performed sensitivity analysis to adjust for AMD stage and body mass index. To account for other potential unknown confounding factors, we also performed a separate analysis considering only patients with AMD and control participants. Figure 1 presents a schematic representation of our study design and data analyses.
All met-QTL analyses were performed using R package MatrixEQTL (R Foundation for Statistical Computing).26 We controlled for multiple comparisons using false discovery rate (FDR), which was calculated based on the Benjamini and Hochberg procedure27 accounting for all 4795 × 544 pairs. We chose FDR methodology because it has been considered appropriate for these type of analyses because the interdependence between metabolites results in nonindependence.21
Finally, with the goal of interpreting the cumulative effect of AMD risk SNPs on metabolomic profiles, we generated genetic risk scores (GRS). By definition, GRSs are aggregating results of GWAS significant genetic variants affecting a certain phenotype, which are generated to explain a larger fraction of the phenotype’s variance than any individual variant alone. These have been used in AMD to distinguish patients with AMD from control participants28 and to predict disease progression29 or drusen growth,30 but not in association with metabolomic profiles, which represent an intermediate phenotype. However, associations between GRS and metabolites have been performed in other fields.31 To calculate GRS, we used the traditional approach29 of summing the weighted allele dosage of the 4795 GWAS significant AMD SNPs available in our dataset after quality control, with the weights equal to the published effect size from the largest AMD GWAS study.2 Then, the sum of the dosage-weighted effect sizes was divided by the sum of effect sizes to produce the final standardized risk score. Associations between GRS and metabolites were determined using linear regression models with age, gender, smoking, and the first 10 PCs (both genetic and metabolomic) as covariates. These models initially were fitted on the 2 cohorts (Boston and Portugal) separately and then combined via meta-analysis, using the same methods described above. For further interpretation of the biological relevance of the identified metabolites linked with GRS, we performed a pathway analysis using Metaboanalyst version 4.0.32 This is based on (1) the number of metabolites that fall within Kyoto Encyclopedia of Genes and Genomes-defined metabolic pathways relative to the total number of metabolites that are within these pathways that we measured and (2) the positional importance of the metabolites within these pathways. This generates an enrichment P value and pathway impact score.
Data Availability
All data, including mass spectrometry and genetics data, as well as the code used for data analysis, are available on request.
Results
We included data from 388 patients with AMD and 98 control participants for a total of 486 individuals (193 from Boston and 293 from Portugal). Table 1 presents the clinical and demographic characteristics of the included study population.
Table 1.
Demographics and Clinical Characteristics of the 2 Study Cohorts
| Characteristic | AMD Patients | Control Participants | Total |
|---|---|---|---|
| Boston, Massachusetts | |||
| No. (%) | 148 (77) | 45 (23) | 193 (100) |
| Age (yrs), mean ± SD | 72.59 ± 7.88 | 67.11 ± 7.68 | 71.32 ± 8.15 |
| Female gender, no. (%) | 96 (65) | 29 (64) | 125 (65) |
| Smoking, no. (%) | |||
| Former smoker | 78 (53) | 19 (43) | 97 (51) |
| Nonsmoker | 65 (45) | 23 (52) | 88 (46) |
| Smoker | 3 (2) | 2 (5) | 5 (3) |
| Race, no. (%) | |||
| Asian | 2 (1) | 0 (0) | 2 (1.1) |
| Hispanic | 5 (4) | 1 (2.5) | 6 (3.3) |
| Black | 0 (0) | 1 (2.5) | 1 (0.6) |
| White | 136 (95) | 37 (95) | 173 (95) |
| AMD stage, no. (%) | NA | ||
| Early | 35 (24) | 35 (18) | |
| Intermediate | 63 (42) | 63 (33) | |
| Late | 50 (34) | 50 (26) | |
| Coimbra, Portugal | |||
| No. (%) | 240 | 53 | 293 (100) |
| Age (yrs), mean ± SD | 76.46 ± 7.96 | 68.62 ± 4.97 | 75.04 ± 8.08 |
| Female gender, no. (%) | 155 (65) | 35 (66) | 190 (65) |
| Smoking, no. (%) | |||
| Former smoker | 36 (15) | 10 (19) | 46 (15.7) |
| Nonsmoker | 203 (84.5) | 43 (81) | 246 (84) |
| Smoker | 1 (0.5) | 0 (0) | 1 (0.3) |
| Race, no. (%) | |||
| Asian | 0 (0) | 0 (0) | 0 (0) |
| Hispanic | 0 (0) | 0 (0) | 0 (0) |
| Black | 3 (1) | 0 (0) | 3 (1) |
| White | 237 (99) | 53 (100) | 290 (99) |
| AMD stage, no. (%) | NA | ||
| Early | 57 (24) | 57 (19) | |
| Intermediate | 130 (54) | 130 (44) | |
| Late | 53 (22) | 53 (18) |
AMD = age-related macular degeneration; NA = not applicable; SD = standard deviation.
Metabolite Quantitative Trait Loci of All Included Individuals
We initially analyzed met-QTL separately for the Boston and Portugal study cohorts (Supplemental Tables 4 and 5). With meta-analysis, 28 met-QTL reached an FDR q-value of < 0.05 (Table 2). As shown, these met-QTL corresponded to 2 genes (ASPM and LIPC). Single nucleotide polymorphisms in the LIPC gene were associated with 4 glycerophospholipids, more specifically, glycerophosphatidylethanolamines. The ASPM gene SNPs were associated with an amino acid belonging to leucine, isoleucine, and valine metabolites. Importantly, for all statistically significant met-QTL, the genetic effect on these metabolites showed a consistent direction across both cohorts, with the same signal for the β-coefficient (Table 2). Similar results were found also when accounting for body mass index and AMD stage (Supplemental Fig 1).
Table 2.
Genomic–Metabolomic Associations for All Included Participants Based on Significant False Discovery Rate (q < 0.05)
| Gene | Reference Singe Nucleotide Polymorphism Cluster Identification | Chromosome Position (Major/Minor) | Minor Allele Frequency† | Super Pathway | Subpathway | Metabolite | Portugal |
Boston |
Meta-analysis |
Age-Related Macular Degeneration‡ |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| β-Coefficient | P value | β-Coefficient | P Value | β-Coefficient | P Value | q Value | Odds Ratio | P Value | Effect Allele | Minor Allele Frequency | |||||||
| ASPM | rs201247969 | 1:196958216:G:GTT | 0.06 | Amino acid | Leucine; isoleucine and valine metabolism | 2-hydroxy-3-methylvalerate | 0.0847 | 6.18 × 10–5 | 0.0709 | 8.80 × 10–4 | 0.0778 | 2.15 × 10–7 | 2.24 × 10–2 | 1.46 | 2.48 × 10–30 | GTT | 0.067 |
| ASPM | rs199801555 | 1:197116704:GA:G | 0.07 | Amino acid | Leucine; isoleucine and valine metabolism | 2-hydroxy-3-methylvalerate | 0.0880 | 2.06 × 10–5 | 0.0779 | 1.96 × 10–4 | 0.0830 | 1.64 × 10–8 | 7.13 × 10–3 | 1.41 | 2.44 × 10–26 | G | 0.071 |
| ASPM | rs61819094 | 1:197119778:C:G | 0.07 | Amino acid | Leucine; isoleucine and valine metabolism | 2-hydroxy-3-methylvalerate | 0.0880 | 2.06 × 10–5 | 0.0779 | 1.96 × 10–4 | 0.0830 | 1.64 × 10–8 | 7.13 × 10–3 | 1.41 | 3.08 × 10–26 | G | 0.072 |
| LIPC | rs1601935 | 15:58671765:T:G | 0.39 | Lipid | PE | 1-palmitoyl-2-arachidonoyl-GPE (16:0/20:4)∗ | 0.0213 | 1.78 × 10–5 | 0.0223 | 7.31 × 10–4 | 0.0217 | 4.78 × 10–8 | 9.58 × 10–3 | 1.11 | 5.30 × 10–9 | T | 0.336 |
| LIPC | rs62001736 | 15:58674051:G:A (p) | 0.31 | Lipid | PE | 1-palmitoyl-2-arachidonoyl-GPE (16:0/20:4)∗ | 0.0203 | 4.16 × 10–5 | 0.0235 | 6.10 × 10–4 | 0.0214 | 9.87 × 10–8 | 1.51 × 10–2 | 0.90 | 1.70 × 10–9 | A | 0.287 |
| LIPC | rs2043082 | 15:58674308:G:A (p) | 0.36 | Lipid | PE | 1-palmitoyl-2-arachidonoyl-GPE (16:0/20:4)∗ | 0.0213 | 9.60 × 10–6 | 0.0174 | 8.19 × 10–3 | 0.0200 | 2.83 × 10–7 | 2.74 × 10–2 | 0.90 | 5.22 × 10–10 | A | 0.338 |
| LIPC | rs4775041 | 15:58674695:G:C (p) | 0.31 | Lipid | PE | 1-palmitoyl-2-arachidonoyl-GPE (16:0/20:4)∗ | 0.0212 | 1.98 × 10–5 | 0.0235 | 4.26 × 10–4 | 0.0220 | 3.27 × 10–8 | 8.52 × 10–3 | 0.90 | 2.46 × 10–9 | C | 0.287 |
| LIPC | rs80123226 | 15:58676032:A:T (p) | 0.31 | Lipid | PE | 1-palmitoyl-2-arachidonoyl-GPE (16:0/20:4)∗ | 0.0188 | 1.26 × 10–4 | 0.0221 | 7.87 × 10–4 | 0.0200 | 3.79 × 10–7 | 3.53 × 10–2 | 0.89 | 6.82 × 10–10 | T | 0.286 |
| LIPC | rs11858759 | 15:58676821:G:A (p) | 0.31 | Lipid | PE | 1-palmitoyl-2-arachidonoyl-GPE (16:0/20:4)∗ | 0.0200 | 4.87 × 10–5 | 0.0219 | 8.67 × 10–4 | 0.0207 | 1.55 × 10–7 | 1.83 × 10–2 | 0.89 | 5.90 × 10–10 | A | 0.287 |
| LIPC | rs10468017 | 15:58678512:C:T (p) | 0.3 | Lipid | PE | 1-palmitoyl-2-arachidonoyl-GPE (16:0/20:4)∗ | 0.0230 | 6.30 × 10–6 | 0.0242 | 2.86 × 10–4 | 0.0234 | 7.00 × 10–9 | 4.56 × 10–3 | 0.88 | 2.81 × 10–11 | T | 0.278 |
| LIPC | rs261290 | 15:58678720:C:T | 0.35 | Lipid | PE | 1-palmitoyl-2-arachidonoyl-GPE (16:0/20:4)∗ | 0.0225 | 3.67 × 10–6 | 0.0180 | 5.10 × 10–3 | 0.0209 | 7.40 × 10–8 | 1.29 × 10–2 | 1.13 | 1.24 × 10–11 | C | 0.327 |
| LIPC | rs7350789 | 15:58679668:G:A (p) | 0.35 | Lipid | PE | 1-palmitoyl-2-arachidonoyl-GPE (16:0/20:4)∗ | 0.0232 | 2.04 × 10–6 | 0.0186 | 2.54 × 10–3 | 0.0214 | 2.19 × 10–8 | 7.14 × 10–3 | 0.89 | 2.07 × 10–11 | A | 0.336 |
| LIPC | rs71884092 | 15:58679807:CAGA:C (p) | 0.35 | Lipid | PE | 1-palmitoyl-2-arachidonoyl-GPE (16:0/20:4)∗ | 0.0232 | 2.04 × 10–6 | 0.0186 | 2.54 × 10–3 | 0.0214 | 2.19 × 10–8 | 7.14 × 10–3 | 0.89 | 1.77 × 10–11 | C | 0.333 |
| LIPC | rs261291 | 15:58680178:T:C (p) | 0.36 | Lipid | PE | 1-palmitoyl-2-linoleoyl-GPE (16:0/18:2) | 0.0344 | 2.99 × 10–8 | 0.0106 | 1.87 × 10–1 | 0.0255 | 2.08 × 10–7 | 2.24 × 10–2 | 0.89 | 2.81 × 10–11 | C | 0.340 |
| LIPC | rs261291 | 15:58680178:T:C (p) | 0.36 | Lipid | PE | 1-palmitoyl-2-arachidonoyl-GPE (16:0/20:4)∗ | 0.0257 | 1.27 × 10–7 | 0.0215 | 5.06 × 10–4 | 0.0241 | 2.92 × 10–10 | 2.54 × 10–4 | 0.89 | 2.81 × 10–11 | C | 0.340 |
| LIPC | rs7177289 | 15:58680184:C:T (p) | 0.35 | Lipid | PE | 1-palmitoyl-2-arachidonoyl-GPE (16:0/20:4)∗ | 0.0229 | 3.12 × 10–6 | 0.0192 | 2.01 × 10–3 | 0.0215 | 2.49 × 10–8 | 7.22 × 10–3 | 0.89 | 2.65 × 10–11 | T | 0.336 |
| LIPC | rs2414577 | 15:58680638:T:C (p) | 0.4 | Lipid | PE | 1-palmitoyl-2-arachidonoyl-GPE (16:0/20:4)∗ | 0.0236 | 1.45 × 10–6 | 0.0175 | 6.17 × 10–3 | 0.0213 | 4.05 × 10–8 | 8.81 × 10–3 | 0.874301 | 4.31 × 10–15 | C | 0.366 |
| LIPC | rs2414578 | 15:58680639:T:C (p) | 0.4 | Lipid | PE | 1-palmitoyl-2-arachidonoyl-GPE (16:0/20:4)∗ | 0.0236 | 1.45 × 10–6 | 0.0175 | 6.17 × 10–3 | 0.0213 | 4.05 × 10–8 | 8.81 × 10–3 | 0.87 | 4.41 × 10–15 | C | 0.366 |
| LIPC | rs35853021 | 15:58680643:G:T (p) | 0.37 | Lipid | PE | 1-palmitoyl-2-arachidonoyl-GPE (16:0/20:4)∗ | 0.0241 | 1.89 × 10–6 | 0.0156 | 1.28 × 10–2 | 0.0208 | 1.33 × 10–7 | 1.66 × 10–2 | 0.89 | 1.43 × 10–11 | T | 0.339 |
| LIPC | rs2043085 | 15:58680954:C:T | 0.4 | Lipid | PE | 1-palmitoyl-2-arachidonoyl-GPE (16:0/20:4)∗ | 0.0263 | 2.96 × 10–8 | 0.0238 | 1.60 × 10–4 | 0.0254 | 2.09 × 10–11 | 2.73 × 10–5 | 1.14 | 6.68 × 10–14 | C | 0.370 |
| LIPC | rs2043085 | 15:58680954:C:T | 0.4 | Lipid | PE | 1-stearoyl-2-arachidonoyl-GPE (18:0/20:4) | 0.0126 | 1.07 × 10–3 | 0.0219 | 2.11 × 10–5 | 0.0159 | 2.39 × 10–7 | 2.40 × 10–2 | 1.14 | 6.68 × 10–14 | C | 0.370 |
| LIPC | rs1532085 | 15:58683366:G:A | 0.39 | Lipid | PE | 1-palmitoyl-2-arachidonoyl-GPE (16:0/20:4)∗ | 0.0279 | 1.87 × 10–9 | 0.0236 | 1.96 × 10–4 | 0.0264 | 1.81 × 10–12 | 4.72 × 10–6 | 1.13 | 3.28 × 10–12 | G | 0.367 |
| LIPC | rs1532085 | 15:58683366:G:A | 0.39 | Lipid | PE | 1-stearoyl-2-arachidonoyl-GPE (18:0/20:4) | 0.0134 | 4.18 × 10–4 | 0.0217 | 2.85 × 10–5 | 0.0162 | 1.05 × 10–7 | 1.52 × 10–2 | 1.13 | 3.28 × 10–12 | G | 0.367 |
| LIPC | rs35980001 | 15:58722590:G:GC (p) | 0.21 | Lipid | PE | 1-oleoyl-2-linoleoyl-GPE (18:1/18:2)∗ | 0.0374 | 3.74 × 10–3 | 0.0722 | 9.17 × 10–7 | 0.0525 | 6.12 × 10–8 | 1.14 × 10–2 | 0.88 | 2.42 × 10–10 | GC | 0.197 |
| LIPC | rs1077835 | 15:58723426:A:G (p) | 0.23 | Lipid | PE | 1-oleoyl-2-linoleoyl-GPE (18:1/18:2)∗ | 0.0367 | 3.47 × 10–3 | 0.0653 | 5.42 × 10–6 | 0.0491 | 2.07 × 10–7 | 2.24 × 10–2 | 0.88 | 7.64 × 10–11 | G | 0.205 |
| LIPC | rs1077834 | 15:58723479:T:C (p) | 0.22 | Lipid | PE | 1-oleoyl-2-linoleoyl-GPE (18:1/18:2)∗ | 0.0383 | 2.18 × 10–3 | 0.0645 | 6.18 × 10–6 | 0.0497 | 1.26 × 10–7 | 1.66 × 10–2 | 0.88 | 7.73 × 10–11 | C | 0.205 |
| LIPC | rs1800588 | 15:58723675:C:T (p) | 0.22 | Lipid | PE | 1-oleoyl-2-linoleoyl-GPE (18:1/18:2)∗ | 0.0334 | 8.65 × 10–3 | 0.0709 | 4.63 × 10–7 | 0.0503 | 9.82 × 10–8 | 1.51 × 10–2 | 0.87 | 3.23 × 10–11 | T | 0.204 |
| LIPC | rs2070895 | 15:58723939:G:A (p) | 0.22 | Lipid | PE | 1-oleoyl-2-linoleoyl-GPE (18:1/18:2)∗ | 0.0354 | 4.37 × 10–3 | 0.0675 | 2.01 × 10–6 | 0.0493 | 1.33 × 10–7 | 1.66 × 10–2 | 0.87 | 2.41 × 10–11 | A | 0.207 |
GPE = glycerophosphatidylethanolamine; PE = phosphatidylethanolamine.
Indicates a compound that has not been confirmed based on a standard, but we are confident in its identity.
(p) indicates that the minor allele is protective.
Refer to the results from the genome-wide association study by Fritsche et al (2015).4
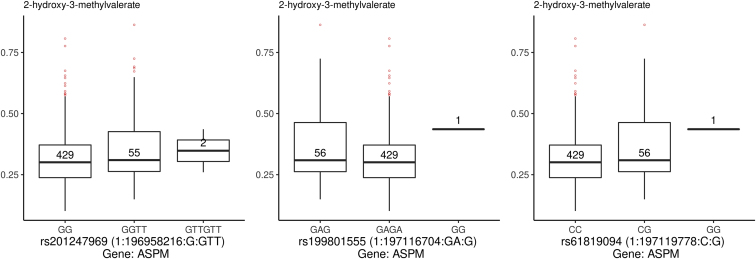
Boxplots were used to depict the metabolite levels for corresponding genotypes for the identified met-QTL (Figs 2 and 3). These demonstrated that for the SNPs in the ASPM gene, the presence of 2 minor alleles (risk allele for AMD2) was associated with higher levels of 2-hydroxy-methylvalerate (Fig 2 and always positive β-coefficient [0.078–0.083]; see Table 2). As shown, for 2 of the ASPM risk SNPs (rs19980155 and rs61819094), a single patient harbored the homozygote genotype GG. Therefore, we performed a sensitivity analysis removing this patient, which retained all previous statistically significant associations seen (q < 0.012), with a slightly different β-coefficient (for both rs19980155 and rs61819094 and 2-hydroxy-3-methylvalerate, β = 0.091, P = 4.79 × 10–9, and q = 0.025; for rs201247969 and 2-hydroxy-3-methylvalerate, β = 0.085, P = 7.51 × 10–8, and q = 0.012).
Figure 2.
Boxplots showing the metabolite quantitative trait loci for ASPM identified on meta-analysis based on a significant q-value (accounting for false discovery rate). In the x-axis, the polymorphisms for ASPM are shown with their major and minor alleles. In the y-axis, the normalized metabolite quantity is represented. The number in the center of each box plot corresponds to the number of individuals with a certain genotype for the polymorphism being represented.
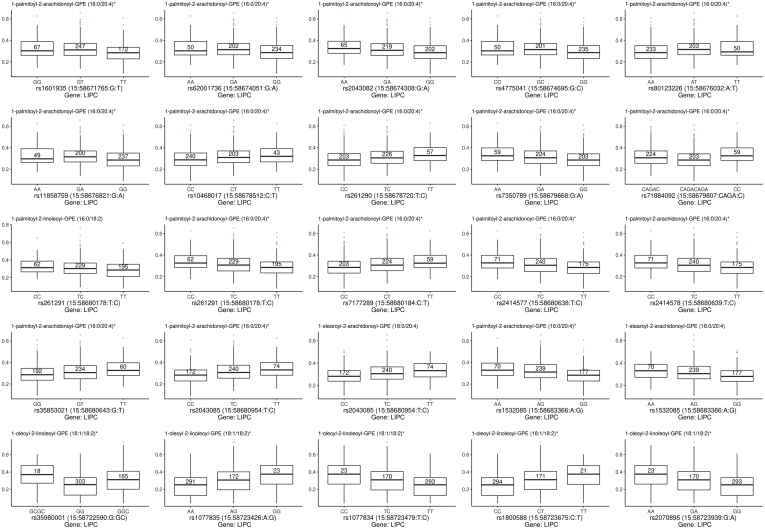
Figure 3.
Box plots showing the metabolite quantitative trait loci (met-QTL) for LIPC identified on meta-analysis based on a significant q-value (accounting for false discovery rate). In the x-axis, the polymorphisms for LIPC are shown with their major and minor alleles. In the y-axis, the normalized metabolite quantity is represented. The number in the center of each box plot corresponds to the number of individuals with a certain genotype for the polymorphism being represented.
For the LIPC gene, the presence of 2 minor alleles also was associated with higher levels of metabolites, all phosphatidylethanolamines (PEs). For some of these SNPs, the minor allele is the protective allele for AMD2; these are marked with a (p) in Table 2 and can be interpreted as follows: the minor allele was protective for AMD, but it was linked to a positive met-QTL (positive β-coefficient), and thus a higher level of a certain metabolite.
Metabolite Quantitative Trait Loci Analysis of Age-Related Macular Degeneration Patients and Control Participants
To account for potential unknown confounding factors, we performed an additional analysis including only AMD patients and only control participants. These analyses also initially were performed separately for each cohort (i.e., Boston and Portugal; Supplemental Tables 6, 7, 8, and 9) and then combined by meta-analysis. Meta-analysis of data from patients with AMD identified 6 met-QTL with FDR q < 0.05. These met-QTL corresponded to the same 2 genes (LIPC and ASPM) identified in the analysis including all individuals and 3 metabolites also in common with the analysis performed including all individuals (2 PEs and 1 essential amino acid). Table 3 details the list of met-QTL for AMD patients with FDR q < 0.05. As shown, estimates of the effect sizes for the minor alleles also were consistent across the 2 cohorts and were associated positively with the metabolite levels. For the control group, none of the met-QTL reached statistical significance with meta-analysis based on FDR.
Table 3.
Genomic–Metabolomic Associations for Patients with Age-Related Macular Degeneration Based on Significant False Discovery Rate (q < 0.05)
| Gene | Reference Singe Nucleotide Polymorphism Cluster Identification | Chromosome Position (Major/Minor) | Minor Allele Frequency† | Super Pathway | Subpathway | Metabolite | Portugal |
Boston |
Meta-analysis |
Age-Related Macular Degeneration‡ |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| β-Coefficient | P Value | β-Coefficient | P Value | β-Coefficient | P Value | q Value | Odds Ratio | P Value | Effect Allele | Minor Allele Frequency | |||||||
| ASPM | rs199801555 | 1:197116704:GA:G | 0.07 | Amino acid | Leucine; isoleucine and valine metabolism | 2-hydroxy-3-methylvalerate | 0.1182 | 4.19x10–6 | 0.0856 | 4.01x10–4 | 0.1009 | 9.96 × 10–9 | 5.20 × 10–3 | 1.418514 | 2.44 × 10–26 | G | 0.071 |
| ASPM | rs61819094 | 1:197119778:C:G | 0.07 | Amino acid | Leucine; isoleucine and valine metabolism | 2-hydroxy-3-methylvalerate | 0.1182 | 4.19 × 10–6 | 0.0856 | 4.01 × 10–4 | 0.1009 | 9.96 × 10–9 | 5.20 × 10–3 | 1.411552 | 3.08 × 10–26 | G | 0.072 |
| LIPC | rs261291 | 15:58680178:T:C (p) | 0.37 | Lipid | PE | 1-palmitoyl-2-arachidonoyl-GPE (16:0/20:4)∗ | 0.0257 | 7.88 × 10–7 | 0.0223 | 2.70 × 10–3 | 0.0246 | 8.13 × 10–9 | 5.20 × 10–3 | 0.890725 | 2.81 × 10–11 | C | 0.340 |
| LIPC | rs261291 | 15:58680178:T:C (p) | 0.37 | Lipid | PE | 1-palmitoyl-2-linoleoyl-GPE (16:0/18:2) | 0.0387 | 8.02 × 10–9 | 0.0125 | 2.08 × 10–1 | 0.0305 | 4.21 × 10–8 | 1.83 × 10–2 | 0.890725 | 2.81 × 10–11 | C | 0.340 |
| LIPC | rs2043085 | 15:58680954:C:T | 0.4 | Lipid | PE | 1-palmitoyl-2-arachidonoyl-GPE (16:0/20:4)∗ | 0.0274 | 9.03 × 10–8 | 0.0243 | 7.91 × 10–4 | 0.0264 | 2.94 × 10–10 | 3.84 × 10–4 | 1.13628 | 6.68 × 10–14 | C | 0.370 |
| LIPC | rs1532085 | 15:58683366:G:A | 0.39 | Lipid | PE | 1-palmitoyl-2-arachidonoyl-GPE (16:0/20:4)∗ | 0.0289 | 9.47 × 10–9 | 0.0237 | 1.25 × 10–3 | 0.0272 | 5.40 × 10–11 | 1.41 × 10–4 | 1.126336 | 3.28 × 10–12 | G | 0.367 |
GPE = glycerophosphatidylethanolamine; PE = phosphatidylethanolamine.
Indicates a compound that has not been confirmed based on a standard, but we are confident in its identity.
(p) indicates that the minor allele is protective.
Refer to the results from the genome-wide association study by Fritsche et al (2015).4
Association of Genetic Risk Scores and Metabolites
Meta-analysis of the associations between GRS and plasma metabolites identified 42 significant associations based on a nominal P value (P < 0.05). Table 4 shows that these associations comprised lipid metabolites (n = 27 [64%]), amino acids (n = 6 [14%]), nucleotides (n = 1 [2%]), and peptides (n = 1 [2%]), as well as cofactors and vitamins (n = 3 [7%]) and 1 carbohydrate (n = 1 [2%]). Interestingly, 3 of these metabolites—adenosine, the eicosanoid 12-HETE, and the amino acid S-adenosylhomocysteine—were common to those reported by our group in a previous study14 as differing significantly between AMD patients and control participants in a meta-analysis of plasma metabolomic profiles from the Boston and Portugal cohorts. Of note, reported associations consistently were more robust in the Boston population than in the Portugal cohort, as demonstrated by the P values and β-coefficients.
Table 4.
Metabolites Associated with Genetic Risk Scores
| Super Pathway | Subpathway | Metabolite | Human Metabolome Database Identifier | Portugal |
Boston |
Meta-analysis |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| Genetic Risk Score β-Coefficient | P Value | Genetic Risk Score β-Coefficient | P Value | Genetic Risk Score β-Coefficient | P Value | q Value | ||||
| Amino acid | Glutamate metabolism | Glutamine | HMDB00641 | –6.60E–06 | 0.297 | –1.19E–05 | 0.068 | –9.18E–06 | 0.042 | 0.587 |
| Amino acid | Leucine; isoleucine and valine metabolism | Isovalerate (i5:0) | HMDB00718 | 3.61E–05 | 0.043 | 6.33E–05 | 0.012 | 4.53E–05 | 0.002 | 0.406 |
| Amino acid | Methionine; cysteine; SAM and taurine metabolism | S-adenosylhomocysteine (SAH) | HMDB00939 | –3.27E–05 | 0.066 | –2.63E–05 | 0.182 | –2.98E–05 | 0.023 | 0.535 |
| Amino acid | Phenylalanine metabolism | Phenyl lactate (PLA) | HMDB00779 | 2.76E–05 | 0.224 | 3.57E–05 | 0.085 | 3.20E–05 | 0.036 | 0.587 |
| Amino acid | Tryptophan metabolism | C-glycosyl tryptophan | –1.82E–05 | 0.057 | –4.14E–05 | 0.022 | –2.34E–05 | 0.005 | 0.406 | |
| Amino acid | Urea cycle; arginine and proline metabolism | Argininate∗ | HMDB03148 | –5.24E–05 | 0.014 | –2.35E–06 | 0.938 | –3.59E–05 | 0.038 | 0.587 |
| Carbohydrate | Glycolysis; gluconeogenesis; and pyruvate metabolism | Glycerate | HMDB00139 | 1.53E–05 | 0.202 | 5.46E–05 | 0.002 | 2.78E–05 | 0.005 | 0.406 |
| Cofactors and vitamins | Ascorbate and aldarate metabolism | Oxalate (ethanedioate) | HMDB02329 | 1.58E–05 | 0.417 | 7.34E–05 | 0.002 | 3.91E–05 | 0.009 | 0.426 |
| Cofactors and vitamins | Ascorbate and aldarate metabolism | Threonate | HMDB00943 | 1.70E–05 | 0.469 | 9.21E–05 | 0.001 | 5.03E–05 | 0.004 | 0.406 |
| Cofactors and vitamins | Tocopherol metabolism | α-Tocopherol | HMDB01893 | 1.58E–05 | 0.085 | 9.40E–05 | 0.000 | 2.89E–05 | 0.001 | 0.298 |
| Energy | TCA cycle | Malate | HMDB00156 | 3.49E–05 | 0.050 | 4.08E–05 | 0.021 | 3.78E–05 | 0.002 | 0.406 |
| Energy | TCA cycle | Succinyl carnitine (C4-DC) | HMDB61717 | –5.11E–05 | 0.024 | –3.53E–05 | 0.058 | –4.16E–05 | 0.004 | 0.406 |
| Energy | TCA cycle | 2-Methylcitrate/homocitrate | 3.10E–05 | 0.074 | 2.06E–05 | 0.273 | 2.62E–05 | 0.039 | 0.587 | |
| Lipid | Androgenic steroids | 5α-Androstan-3β;17β-diol disulfate | HMDB00493 | 3.27E–05 | 0.534 | 0.000114847 | 0.026 | 7.50E–05 | 0.040 | 0.587 |
| Lipid | Diacylglycerol | Palmitoyl-arachidonyl-glycerol (16:0/20:4) [2]∗ | HMDB07112 | 5.41E–05 | 0.089 | 4.23E–05 | 0.239 | 4.89E–05 | 0.039 | 0.587 |
| Lipid | Diacylglycerol | Oleoyl-arachidonyl-glycerol (18:1/20:4) [1]∗ | HMDB07228 | 7.03E–05 | 0.001 | 3.18E–06 | 0.890 | 3.94E–05 | 0.011 | 0.426 |
| Lipid | Diacylglycerol | Stearoyl-arachidonyl-glycerol (18:0/20:4) [2]∗ | 7.45E–05 | 0.017 | 3.30E–05 | 0.192 | 4.94E–05 | 0.011 | 0.426 | |
| Lipid | Eicosanoid | 12-HETE | HMDB06111 | 6.81E–05 | 0.041 | 5.45E–05 | 0.141 | 6.20E–05 | 0.012 | 0.426 |
| Lipid | Fatty acid metabolism (acyl choline) | Arachidonyl choline | 4.26E–05 | 0.053 | 3.67E–05 | 0.123 | 3.98E–05 | 0.013 | 0.426 | |
| Lipid | Fatty acid metabolism (acyl carnitine) | Lauryl carnitine (C12) | HMDB02250 | –1.30E–05 | 0.585 | –7.54E–05 | 0.003 | –4.27E–05 | 0.013 | 0.426 |
| Lipid | Fatty acid metabolism (acyl carnitine) | Myristoleoylcarnitine (C14:1)∗ | –2.12E–05 | 0.322 | –6.93E–05 | 0.004 | –4.26E–05 | 0.007 | 0.426 | |
| Lipid | Fatty acid; dicarboxylate | Sebacate (C10-DC) | HMDB00792 | 7.51E–05 | 0.015 | 6.84E–06 | 0.859 | 4.88E–05 | 0.041 | 0.587 |
| Lipid | Fatty acid; dicarboxylate | 2-Hydroxyglutarate | HMDB00606 | 1.34E–05 | 0.557 | 6.67E–05 | 0.017 | 3.50E–05 | 0.047 | 0.612 |
| Lipid | Fatty acid; monohydroxy | 3-Hydroxylaurate | HMDB00387 | –2.34E–05 | 0.250 | –4.71E–05 | 0.039 | –3.39E–05 | 0.025 | 0.535 |
| Lipid | Long-chain fatty acid | Palmitate (16:0) | HMDB00220 | –1.64E–05 | 0.023 | –6.96E–06 | 0.400 | –1.24E–05 | 0.023 | 0.535 |
| Lipid | Long-chain fatty acid | Stearate (18:0) | HMDB00827 | –2.33E–05 | 0.010 | –1.16E–05 | 0.208 | –1.76E–05 | 0.006 | 0.406 |
| Lipid | Long-chain fatty acid | Oleate/vaccenate (18:1) | –2.41E–05 | 0.018 | –1.27E–05 | 0.250 | –1.88E–05 | 0.012 | 0.426 | |
| Lipid | Lysophospholipid | 1-Linoleoyl-GPI (18:2)∗ | –3.43E–05 | 0.133 | –3.35E–05 | 0.145 | –3.39E–05 | 0.036 | 0.587 | |
| Lipid | Medium-chain fatty acid | Caproate (6:0) | HMDB00535 | 5.30E–05 | 0.023 | 2.62E–05 | 0.540 | 4.70E–05 | 0.021 | 0.535 |
| Lipid | Monoacylglycerol | 1-Docosahexaenoylglycerol (22:6) | HMDB11587 | –5.70E–05 | 0.075 | –3.86E–05 | 0.152 | –4.62E–05 | 0.024 | 0.535 |
| Lipid | Phosphatidylcholine (PC) | 1;2-Dilinoleoyl-GPC (18:2/18:2) | HMDB08138 | –1.38E–05 | 0.368 | –3.14E–05 | 0.046 | –2.24E–05 | 0.040 | 0.587 |
| Lipid | Phosphatidylethanolamine (PE) | 1-Stearoyl-2-linoleoyl-GPE (18:0/18:2)∗ | HMDB08994 | –4.38E–05 | 0.011 | –6.24E–06 | 0.760 | –2.83E–05 | 0.031 | 0.587 |
| Lipid | Phosphatidylinositol (PI) | 1-Stearoyl-2-linoleoyl-GPI (18:0/18:2) | HMDB09809 | –2.71E–05 | 0.157 | –3.73E–05 | 0.045 | –3.24E–05 | 0.015 | 0.449 |
| Lipid | Pregnenolone steroids | Pregnanediol sulfate (C21H34O5S)∗ | –2.49E–05 | 0.331 | –5.24E–05 | 0.049 | –3.82E–05 | 0.038 | 0.587 | |
| Lipid | Secondary bile acid metabolism | Iso-ursodeoxycholate | HMDB00686 | 0.000115825 | 0.085 | 9.80E–05 | 0.319 | 0.000110161 | 0.047 | 0.612 |
| Lipid | Sphingomyelins | Stearoyl sphingomyelin (d18:1/18:0) | HMDB01348 | 1.21E–05 | 0.208 | 2.11E–05 | 0.082 | 1.56E–05 | 0.038 | 0.587 |
| Lipid | Sphingomyelins | Behenoyl sphingomyelin (d18:1/22:0)∗ | HMDB12103 | –2.21E–05 | 0.022 | –1.47E–05 | 0.170 | –1.88E–05 | 0.008 | 0.426 |
| Lipid | Sphingomyelins | Lignoceroyl sphingomyelin (d18:1/24:0) | –2.24E–05 | 0.126 | –2.56E–05 | 0.122 | –2.38E–05 | 0.029 | 0.587 | |
| Lipid | Sphingomyelins | Sphingomyelin (d18:2/23:1)∗ | 2.12E–05 | 0.073 | 2.07E–05 | 0.205 | 2.10E–05 | 0.028 | 0.578 | |
| Lipid | Sphingomyelins | Sphingomyelin (d17:2/16:0; d18:2/15:0)∗ | 3.21E–05 | 0.157 | 3.16E–05 | 0.075 | 3.18E–05 | 0.022 | 0.535 | |
| Nucleotide | Purine metabolism; adenine containing | Adenosine | HMDB00050 | 6.01E–05 | 0.346 | 0.000167068 | 0.016 | 0.000109649 | 0.019 | 0.535 |
| Peptide | γ-Glutamyl amino acid | γ-Glutamylvaline | HMDB11172 | –1.53E–05 | 0.168 | –2.51E–05 | 0.127 | –1.84E–05 | 0.045 | 0.612 |
GPC = glycerophosphatidylcholine; GPE = glycerophosphatidylethanolamine; GPI = glycerophosphatidylinositol; GRS = genetic risk score; HMDB = human metabolome database identifier; PT = Portugal; SAM = S-adenosylmethionine; TCA = tricarboxylic acid cycle.
Indicates a compound that has not been confirmed based on a standard, but we are confident in its identity.
Discussion
We present an assessment of the association between AMD risk SNPs and plasma metabolites in 2 cohorts of AMD patients and control participants that were combined by meta-analysis. Metabolomic profiling was performed using a state-of-the-art MS metabolomics platform with broad global coverage. Accounting for confounding factors and FDR, results from our meta-analysis showed 28 significant SNP–metabolite associations (met-QTL with a q < 0.05). The vast majority of these associations (n = 25) were with SNPs in the LIPC gene, and these were all observed with PE metabolites, which are glycerophospholipids. Similar results were observed when assessing only AMD patients.
The International AMD Genetics Consortium identified 34 loci and more than 7000 SNPs with associations with AMD risk, 52 of them representing independent variants.2 Yet, how these SNPs cause disease remains largely unknown. Because metabolites are downstream of genomic and transcriptomic process, and thus are closely related to disease phenotype,21 met-QTL can be a powerful approach to understanding the biological relevance of SNPs to a certain disease. In our work, met-QTL analyses demonstrated that AMD risk SNPs have strong associations with plasma metabolites. This observation also was reinforced by the observed GRS–metabolite associations. These results support the premise that integrating genetics with metabolomic information can provide insights into the mechanisms whereby AMD risk SNPs may cause disease.
Our study showed that the greatest number of highly significant met-QTL were seen with polymorphisms in LIPC. This is in agreement with published data suggesting a major role of LIPC in the pathogenesis of AMD,33,34 including a study conducted by the International AMD Genetics Consortium that used solely genetic data (GWAS statistics) and concluded that LIPC was one of the major genes driving statistical signals in multiple pathway databases.34 LIPC encodes a triglyceride lipase that breaks down triglycerides to diacyl- and monoacylglycerols and fatty acids, playing a role in the regulation of plasma lipids and in the glycerolipid and glycerophospholipid metabolism.35 In our work, all the LIPC polymorphisms showed highly significant associations with levels of PEs, which are glycerophospholipids and belong to the same metabolomic pathway influenced by LIPC. Importantly, these associations and their effect sizes were consistent across our 2 cohorts, and all LIPC polymorphisms showed a positive association with PE metabolite levels. Our previously published work assessing plasma metabolomic profiles in AMD13,14 identified glycerophospholipid metabolites as some of the major determinants distinguishing patients with AMD and control participants. Glycerophospholipids, including PE, are major components of biological cell membranes, providing structural stability, forming ion channels and receptors, and participating in the generation of second messengers in signal transduction.36,37 Among the glycerophospholipids in the vertebrate retina, PEs seem to be dominant and are involved in the transport of visual pigments.38 Indeed, a recent metabolomics study in human donor eyes described significant differences in PE metabolites (and other glycerophospholipids) in eyes with AMD compared with healthy eyes.39 A complete understanding of the role of glycerophospholipids (and lipids in general) in AMD pathogenesis remains lacking,40 but as shown, met-QTL may provide insights into these relationships.
A recent study by the Eye-Risk Consortium15 on associations between AMD genetic variants and metabolite levels also identified associations between LIPC and plasma lipid metabolites, some of them with the same polymorphisms observed in our study (rs2043085 and rs2070895). These SNP associations were seen with different lipids, which may be explained by the different methodologies and metabolomics platforms used in the 2 studies. The MS platform used in our study has high standards for metabolite identification based on 3 parameters: (1) retention time and index, (2) mass-to-charge ratio, and (3) chromatographic data including MS/MS spectral data with forward and reverse scores between the experimental data and authentic standards. The Eye-Risk Consortium used nuclear magnetic resonance spectroscopy for metabolomic profiling, which often is used as an initial approach, but has a lower sensitivity than MS.16 This lower sensitivity may explain why the Eye-Risk Consortium study detected only 146 metabolites in their large combined cohort,15 whereas MS identified 544 plasma metabolites in our study. It is also important to note that our study was designed prospectively, our samples from both sites were collected and processed according to the same predefined protocol, and all metabolomics profiling was carried out with the same Metabolon, Inc., MS platform. This is particularly relevant in metabolomics studies to ensure consistency of the metabolite measurements. Additionally, the Eye-Risk Consortium15 assessed only associations between plasma metabolites and the significant independent variants (n = 52 SNPs) that have been linked with AMD,2 whereas in our met-QTL analyses, we included more than 4000 AMD risk SNPs. We believe that our approach is advantageous because, although the 52 SNPs are the most significant (P < 5 × 10–8) independent variants reported by the International AMD Genetics Consortium, they may not represent the variables with the strongest associations with plasma metabolites.
Our meta-analysis also identified statistically significant met-QTL with polymorphisms in the ASPM gene. Despite its well-established relationship with AMD,2 to our knowledge, no previous associations between polymorphisms in this gene and metabolites have been documented. ASPM is located on chromosome 1 (1q31.3), has 28 exons, and encodes the abnormal spindle-like microcephaly-associated protein, which has been recognized as a centrosomal protein that modulates early neural development. More recently, the abnormal spindle-like microcephaly-associated protein also has been found to regulate cell division and proliferation in adult cells and is linked with a risk of multiple tumors.41 As described in our study, polymorphisms in the ASPM gene were linked with a branched-chain amino acid (BCAA) metabolite. Branched-chain amino acids are essential amino acids that modulate protein synthesis and degradation, as well as energy metabolism.42 Interestingly, despite coming only from the diet, the metabolism of BCAAs has an important heritability.24 Although no relationships have been described to date in the literature, one might hypothesize that ASPM may be involved in a pathway where BCAAs may serve as either energy or protein sources or may play a role as modulators. Further studies are needed to confirm this hypothesis.
Interestingly, despite the known strong associations between complement factor H (CFH) polymorphisms and AMD risk2 and the presence of multiple of these polymorphisms in our 2 cohorts, we did not find any significant met-QTL between CFH polymorphisms and plasma metabolites. This is in agreement with the results of the Eye-Risk Consortium.15
Our study has several limitations, including its relatively small sample size. This precluded us from analyzing specific met-QTL for the different forms of late AMD (i.e., choroidal neovascularization and geographic atrophy) and also limited our analysis to including only AMD patients and control participants. In this analysis, we did not observe any significant met-QTL based on our FDR statistical significance criteria for control participants, which is likely related to the small sample size of this group. The same was observed for GRS–metabolite association, for which we report nominal P values. Of note, even the statistically significant (based on FDR) met-QTL identified in this work do not imply causality. Additional work in larger and independent cohorts will be crucial to identifying potential causal variants and genes with refined analysis, such as Mendelian randomization and colocalization analysis. Multi-SNP models also can be useful in the future to evaluate the association between genetically determined gene expression and the risk of AMD.11 Another limitation of our study is the lack of data on the conventional plasma lipid measurements. Previous studies have shown that LIPC is linked to high-density lipoprotein and very low-density lipoprotein cholesterol levels,43 and phosphatidylethanolamines are known components of these lipoproteins (very low-density lipoprotein, high-density lipoprotein, and low-density lipoprotein).44 Thus, it would be interesting to analyze how conventional lipoprotein levels relate to our findings. For the met-QTL related to ASPM, further work needs to be performed to clarify the association with BCAA metabolites on a mechanistic level. Additionally, because BCAAs are essential amino acids derived from the diet, future work should consider analyzing the relationship between these and other met-QTL and dietary patterns, including also exogenous metabolites. The biological interpretation of our results also is limited by the currently available platforms for pathway analysis. Although MS is the metabolomics technique with the most available software for these assessments, the available platforms still lack information on many known metabolites or gene–metabolite associations. Finally, our study also is limited by its cross-sectional design, because it provides a snapshot of the metabolome of the patients studied. Another limitation is that both of our cohorts were nearly entirely White. This is related in part to the epidemiologic features of AMD45 and in part to the population served by both enrolling sites, 2 tertiary care hospitals.
In summary, our study supports that integrating genomics and metabolomics data can provide insights that may not be possible with genomics–phenotype data alone. In this work, we describe strong associations between AMD risk polymorphisms and the plasma metabolome, which contributes to the current understanding of the biological significance of AMD risk SNPs and the pathways by which they may contribute to the pathogenesis of AMD. In particular, our results showed that LIPC polymorphisms presented the highest number of associations with plasma metabolites, supporting previous work demonstrating LIPC as a key genetic driver in AMD. Importantly, we observed that SNPs in LIPC were linked to PEs, which are glycerophospholipids. This suggests that glycerophospholipid metabolism may have an important role in AMD, as previously proposed by our group.13,14 In conclusion, our work demonstrated that LIPC and the glycerophospholipid metabolites pathway likely are important in AMD pathogenesis and may represent a potential novel treatment target for this condition. We believe that this work moves us closer to the development of precision medicine for the diagnosis and treatment of AMD, and ultimately to reducing the burden of blindness resulting from AMD.
Acknowledgments
The authors thank Elizabeth Rossin, MD, PhD, for her support in the interpretation of the genetics data presented herein; and Matthew Gardiner, MD, Matt Goodman OD, Yan Jiang, OD, Stacey Brauner, MD, Peggy Chang, MD, and Christian Song, MD, for contributing to the recruitment of control participants for this study.
Manuscript no. 2021-36.
Footnotes
Supplemental material available atwww.ophthalmologyscience.org.
Disclosure(s): All authors have completed and submitted the ICMJE disclosures form.
The author(s) have made the following disclosure(s): I.L.: Patent - WO 2018/208970 A1
I.K.K.: Financial support – Novartis, Kodiak Sciences, Castle Biosciences, Biophytis; Nonfinancial support – Allergan
J.B.M.: Consultant – Alcon, Carl Zeiss Meditec, Inc., Heidelberg, Genentech, Allergan, Sunovion
D.G.V.: Grants/grants pending – NIH and foundations
R.S.: Consultant and Board member – Alimera, Allergan, Bayer, Novartis, Thea, Roche, NovoNordisk
J.W.M.: Consultant – Genentech/Roche, Sunovion, KalVista Pharmaceuticals, Ltd., ONL Therapeutics, LLC; Advisory board – Bausch + Lomb; Financial support – Lowy Medical Research Institute, Ltd., Heidelberg Engineering; Royalties – Massachusetts Eye and Ear/Valeant Pharmaceuticals; Equity owner – ONL Therapeutics, LLC; Patent – US 7,811,832 (royalties paid by ONL Therapeutics to Massachusetts Eye and Ear), US 5,798,349, US 6,225,303, US 6,610,679, CA 2,185,644, CA 2,536,069 (royalties paid by Valeant Pharmaceuticals to Massachusetts Eye and Ear), and Metabolomics for Biomarkers of AMD (pending)
D.H.: Patent – WO 2018/208970 A1
Supported by the Miller Retina Research Fund, Massachusetts Eye and Ear, Boston, Massachusetts; the Champalimaud Vision Award; the National Institutes of Health, Bethesda, Maryland (grant no.; R01EY030088-01A1); the National Eye Institute (D.G.V.); Research to Prevent Blindness, Inc., New York, New York (unrestricted departmental grant, D.G.V.); the Portuguese Foundation for Science and Technology/Harvard Medical School Portugal Program (grant no.: HMSP-ICJ/006/2013); the Commonwealth Unrestricted Grant for Eye Research; the Loefflers Family Foundation (D.V.G.); the Yeatts Family Foundation (D.V.G.); and the Alcon Research Institute (D.V.G.). None of these organizations had any role in the design or conduct of this research.
HUMAN SUBJECTS: Human subjects were included in this study. The human ethics committees at the Massachusetts Eye and Ear, Massachusetts General Brigham, the Faculty of Medicine of the University of Coimbra, the Association for Innovation and Biomedical Research on Light and Image, and the Portuguese National Data Protection Committee approved the study. All research complied with the Health Insurance Portability and Accountability Act (HIPAA) of 1996 and adhered to the tenets of the Declaration of Helsinki. All participants provided informed consent.
No animal subjects were included in this study.
Author Contributions:
Conception and design: Lains, Carreira, Silva, J.W.Miller, Lasky-Su, Liang, Husain
Analysis and interpretation: Lains, Zhu, Han, Chung, Yuan, Kelly, Gil, Katz, Nigalye, Kim, J.B.Miller, Carreira, Silva, Vavvas, J.W.Miller, Lasky-Su, Liang, Husain
Data collection: Lains, Zhu, Han, Chung, Yuan, Kelly, Gil, Katz, Nigalye, Kim, J.B.Miller, Silva, Vavvas, J.W.Miller, Lasky-Su, Liang, Husain
Obtained funding: Lains, Husain, Miller
Overall responsibility: Lains, Zhu, Han, Chung, Yuan, Kelly, Gil, Katz, Nigalye, Kim, J.B.Miller, Carreira, Silva, Vavvas, J.W.Miller, Lasky-Su, Liang, Husain
Contributor Information
Liming Liang, Email: Lliang@hsph.harvard.edu.
Deeba Husain, Email: Deeba_Husain@meei.harvard.edu.
Supplementary Data
Supplemental Table 3
References
- 1.Wong W.L., Su X., Li X., et al. Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: a systematic review and meta-analysis. Lancet Glob Heal. 2014;2(2):e106–e116. doi: 10.1016/S2214-109X(13)70145-1. [DOI] [PubMed] [Google Scholar]
- 2.Fritsche L.G., Igl W., Bailey J.N.C., et al. A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat Genet. 2016;48(2):134–143. doi: 10.1038/ng.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ratnapriya R., Acar İ.E., Geerlings M.J., et al. Family-based exome sequencing identifies rare coding variants in age-related macular degeneration. Hum Mol Genet. 2020;29(12):2022–2034. doi: 10.1093/hmg/ddaa057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Strunz T., Lauwen S., Kiel C., et al. A transcriptome-wide association study based on 27 tissues identifies 106 genes potentially relevant for disease pathology in age-related macular degeneration. Sci Rep. 2020;10(1):1584. doi: 10.1038/s41598-020-58510-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller J.W. Developing therapies for age-related macular degeneration: the art and science of problem-solving. The 2018 Charles L. Schepens, MD, Lecture. Ophthalmol Retin. 2019;3(10):900–909. doi: 10.1016/j.oret.2019.07.015. [DOI] [PubMed] [Google Scholar]
- 6.Miller J.W. Age-related macular degeneration revisited—piecing the puzzle. The LXIX Edward Jackson Memorial Lecture. Am J Ophthalmol. 2013;155(1):1–35. doi: 10.1016/j.ajo.2012.10.018. e13. [DOI] [PubMed] [Google Scholar]
- 7.Kraus W.E., Muoio D.M., Stevens R., et al. Metabolomic quantitative trait loci (mQTL) mapping implicates the ubiquitin proteasome system in cardiovascular disease pathogenesis. PLOS Genet. 2015;11(11) doi: 10.1371/journal.pgen.1005553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yazdani A., Yazdani A., Elsea S.H., et al. Genome analysis and pleiotropy assessment using causal networks with loss of function mutation and metabolomics. BMC Genomics. 2019;20(1):395. doi: 10.1186/s12864-019-5772-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park T.J., Lee H.S., Kim Y.J., Kim B.J. Identification of novel non-synonymous variants associated with type 2 diabetes-related metabolites in Korean population. Biosci Rep. 2019;39(10) doi: 10.1042/BSR20190078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suhre K., Gieger C. Genetic variation in metabolic phenotypes: study designs and applications. Nat Rev Genet. 2012;13(11):759–769. doi: 10.1038/nrg3314. [DOI] [PubMed] [Google Scholar]
- 11.Ndungu A., Payne A., Torres J.M., et al. A multi-tissue transcriptome analysis of human metabolites guides interpretability of associations based on multi-SNP models for gene expression. Am J Hum Genet. 2020;106(2):188–201. doi: 10.1016/j.ajhg.2020.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laíns I., Duarte D., Barros A.S., et al. Human plasma metabolomics in age-related macular degeneration (AMD) using nuclear magnetic resonance spectroscopy. PLoS One. 2017;12(5) doi: 10.1371/journal.pone.0177749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laíns I., Kelly R.S., Miller J.B., et al. Human plasma metabolomics study across all stages of age-related macular degeneration identifies potential lipid biomarkers. Ophthalmology. 2018;125(2):245–254. doi: 10.1016/j.ophtha.2017.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laíns I., Chung W., Kelly R.S., et al. Human plasma metabolomics in age-related macular degeneration: meta-analysis of two cohorts. Metabolites. 2019;9(7):127. doi: 10.3390/metabo9070127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Acar İ.E., Lores-Motta L., Colijn J.M., et al. Integrating metabolomics, genomics and disease pathways in age-related macular degeneration: the EYE-RISK Consortium. Ophthalmology. 2020;127(12):1693–1709. doi: 10.1016/j.ophtha.2020.06.020. [DOI] [PubMed] [Google Scholar]
- 16.Emwas A.-H.M. The strengths and weaknesses of NMR spectroscopy and mass spectrometry with particular focus on metabolomics research. Methods Mol Biol. 2015;1277:161–193. doi: 10.1007/978-1-4939-2377-9_13. [DOI] [PubMed] [Google Scholar]
- 17.Cachulo M. da L., Lobo C., Figueira J., et al. Prevalence of age-related macular degeneration in Portugal: the Coimbra Eye Study—report 1. Ophthalmologica. 2015;233(3–4):119–127. doi: 10.1159/000371584. [DOI] [PubMed] [Google Scholar]
- 18.Danis R.P., Domalpally A., Chew E.Y., et al. Methods and reproducibility of grading optimized digital color fundus photographs in the Age-Related Eye Disease Study 2 (AREDS2 report number 2) Invest Ophthalmol Vis Sci. 2013;54(7):4548–4554. doi: 10.1167/iovs.13-11804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsikata E., Laíns I., Gil J., et al. Automated brightness and contrast adjustment of color fundus photographs for the grading of age-related macular degeneration. Transl Vis Sci Technol. 2017;6(2):3. doi: 10.1167/tvst.6.2.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.The Age-Related Eye Disease Study system for classifying age-related macular degeneration from stereoscopic color fundus photographs: the Age-Related Eye Disease Study report number 6. Am J Ophthalmol. 2001;132(5):668–681. doi: 10.1016/s0002-9394(01)01218-1. [DOI] [PubMed] [Google Scholar]
- 21.Laíns I., Gantner M., Murinello S., et al. Metabolomics in the study of retinal health and disease. Prog Retin Eye Res. 2019;69:57–79. doi: 10.1016/j.preteyeres.2018.11.002. [DOI] [PubMed] [Google Scholar]
- 22.van den Berg R.A., Hoefsloot H.C.J., Westerhuis J.A., et al. Centering, scaling, and transformations: improving the biological information content of metabolomics data. BMC Genomics. 2006;7:142. doi: 10.1186/1471-2164-7-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zeleznik O.A., Eliassen A.H., Kraft P., et al. A prospective analysis of circulating plasma metabolites associated with ovarian cancer risk. Cancer Res. 2020;80(6):1357–1367. doi: 10.1158/0008-5472.CAN-19-2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hagenbeek F.A., Pool R., van Dongen J., et al. Heritability estimates for 361 blood metabolites across 40 genome-wide association studies. Nat Commun. 2020;11(1):39. doi: 10.1038/s41467-019-13770-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liptak T. On the combination of independent tests. Magy Tud Akad Mat Kut Int Kozl. 1958;3:171–197. [Google Scholar]
- 26.Shabalin A.A. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics. 2012;28(10):1353–1358. doi: 10.1093/bioinformatics/bts163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reiner A., Yekutieli D., Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics. 2003;19(3):368–375. doi: 10.1093/bioinformatics/btf877. [DOI] [PubMed] [Google Scholar]
- 28.Fritsche L.G., Chen W., Schu M., et al. Seven new loci associated with age-related macular degeneration. Nat Genet. 2013;45(4):433–439. doi: 10.1038/ng.2578. 439e1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heesterbeek T.J., de Jong E.K., Acar I.E., et al. Genetic risk score has added value over initial clinical grading stage in predicting disease progression in age-related macular degeneration. Sci Rep. 2019;9(1):6611. doi: 10.1038/s41598-019-43144-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Merle B.M.J., Rosner B., Seddon J.M. Genetic susceptibility, diet quality, and two-step progression in drusen size. Invest Ophthalmol Vis Sci. 2020;61(5):17. doi: 10.1167/iovs.61.5.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Battram T., Hoskins L., Hughes D.A., et al. Coronary artery disease, genetic risk and the metabolome in young individuals. Wellcome Open Res. 2019;3:114. doi: 10.12688/wellcomeopenres.14788.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chong J., Soufan O., Li C., et al. MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018;46(W1):W486–W494. doi: 10.1093/nar/gky310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burgess S., Davey Smith G. Mendelian randomization implicates high-density lipoprotein cholesterol–associated mechanisms in etiology of age-related macular degeneration. Ophthalmology. 2017;124(8):1165–1174. doi: 10.1016/j.ophtha.2017.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Waksmunski A.R., Grunin M., Kinzy T.G., et al. Pathway analysis integrating genome-wide and functional data identifies PLCG2 as a candidate gene for age-related macular degeneration. Invest Ophthalmol Vis Sci. 2019;60(12):4041–4051. doi: 10.1167/iovs.19-27827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gieger C., Geistlinger L., Altmaier E., et al. Genetics meets metabolomics: a genome-wide association study of metabolite profiles in human serum. PLoS Genet. 2008;4(11) doi: 10.1371/journal.pgen.1000282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Farooqui A.A., Horrocks L.A., Farooqui T. Glycerophospholipids in brain: their metabolism, incorporation into membranes, functions, and involvement in neurological disorders. Chem Phys Lipids. 2000;106(1):1–29. doi: 10.1016/s0009-3084(00)00128-6. [DOI] [PubMed] [Google Scholar]
- 37.Hopiavuori B.R., Agbaga M.-P., Brush R.S., et al. Regional changes in CNS and retinal glycerophospholipid profiles with age: a molecular blueprint. J Lipid Res. 2017;58(4):668–680. doi: 10.1194/jlr.M070714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fliesler S.J. Lipids and lipid metabolism in the eye. J Lipid Res. 2010;51(1):1–3. doi: 10.1194/jlr.E003533-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chistyakov D.V., Baksheeva V.E., Tiulina V.V., et al. Mechanisms and treatment of light-induced retinal degeneration-associated inflammation: Insights from biochemical profiling of the aqueous humor. Int J Mol Sci. 2020;21(3):704. doi: 10.3390/ijms21030704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Y., Wang M., Zhang X., et al. The association between the lipids levels in blood and risk of age-related macular degeneration. Nutrients. 2016;8(10):663. doi: 10.3390/nu8100663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang J., Lu M., Cui Q., et al. Overexpression of ASPM, CDC20, and TTK confer a poorer prognosis in breast cancer identified by gene co-expression network analysis. Front Oncol. 2019;9:310. doi: 10.3389/fonc.2019.00310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shimomura Y., Kitaura Y. Physiological and pathological roles of branched-chain amino acids in the regulation of protein and energy metabolism and neurological functions. Pharmacol Res. 2018;133:215–217. doi: 10.1016/j.phrs.2018.05.014. [DOI] [PubMed] [Google Scholar]
- 43.Tukiainen T., Kettunen J., Kangas A.J., et al. Detailed metabolic and genetic characterization reveals new associations for 30 known lipid loci. Hum Mol Genet. 2012;21(6):1444–1455. doi: 10.1093/hmg/ddr581. [DOI] [PubMed] [Google Scholar]
- 44.Kontush A., Lhomme M., Chapman M.J. Thematic review series. High density lipoprotein structure, function, and metabolism: unraveling the complexities of the HDL lipidome. J Lipid Res. 2013;54(11):2950–2963. doi: 10.1194/jlr.R036095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sobrin L., Seddon J.M. Nature and nurture—genes and environment—predict onset and progression of macular degeneration. Prog Retin Eye Res. 2014;40:1–15. doi: 10.1016/j.preteyeres.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 3
Data Availability Statement
All data, including mass spectrometry and genetics data, as well as the code used for data analysis, are available on request.