Abstract
The maintenance of telomere length supports repetitive cell division and therefore plays a central role in cancer development and progression. Telomeres are extended by either the enzyme telomerase or the alternative lengthening of telomeres (ALT) pathway. Here, we found that the telomere-associated protein SLX4IP dictates telomere proteome composition by recruiting and activating the E3 SUMO ligase PIAS1 to the SLX4 complex. PIAS1 SUMOylated the telomere-binding protein RAP1, which disrupted its interaction with the telomere-binding protein TRF2 and facilitated its nucleocytoplasmic shuttling. In the cytosol, RAP1 bound to IκB kinase (IKK), resulting in activation of the transcription factor NF-κB and its induction of Jagged-1 expression, which promoted Notch signaling and the institution of ALT. This axis could be targeted therapeutically in ALT-driven cancers and in tumor cells that develop resistance to anti-telomerase therapies. Our results illuminate the mechanisms underlying SLX4IP-dependent telomere plasticity and demonstrate the role of telomere proteins in directly coordinating intracellular signaling and telomere maintenance dynamics.
INTRODUCTION
During cell division, the genome loses DNA content as a consequence of asymmetric nascent strand synthesis (1). In addition, the exposed ends of linear chromosomes can be interpreted by the cell as double-strand breaks, thereby initiating aberrant DNA damage responses (DDRs) and chromosome end-to-end fusions culminating in widespread genomic instability (2). These end-replication and end-protection problems require that chromosome ends contain a DNA sequence that buffers against loss of genetic information while simultaneously shielding these regions from the DNA damage machinery. These parallel events are mediated by telomeres, which in humans are composed of a repetitive (TTAGGG)n sequence coated by a hexameric protein complex termed shelterin (3). Double-stranded telomere DNA is tightly bound by the telomere repeat-binding factors (TRF1 and TRF2) (4). This complex terminates in a 3′ single-stranded overhang that is bound by protection of telomeres 1 (POT1), which serves to sequester free DNA ends and prevent DDR activation (5). TRF1, TRF2, and POT1 further coordinate telomere protection by recruiting the remaining shelterin components: TRF1-interacting nuclear factor 2 (TIN2), TPP1 (also known as adrenocortical dysplasia), and repressor-activator protein 1 (RAP1; also known as TERF2IP) (6–8). TPP1 and TIN2 appear to play important roles in telomere length regulation (9, 10). RAP1 remains largely uncharacterized but is known to inhibit homologous recombination (HR) at telomeres, which, if left unchecked, serves as a source for both anomalous telomere extension and telomere DNA damage (11, 12).
In addition to shelterin, telomeres contain numerous proteins that are involved in regulating their length. Two primary telomere maintenance mechanisms (TMMs) exist: (i) telomerase, which is a ribonucleoprotein holoenzyme composed of a core reverse transcriptase (TERT) and RNA template for DNA synthesis (TR) (13); and (ii) Alternative Lengthening of Telomeres (ALT), which engages HR for telomere extension by a specialized replisome in a manner that resembles break-induced DNA synthesis (14, 15). ALT relies upon the coordinated actions of the HR proteins RAD51 and RAD52 (15, 16), DNA polymerase δ (14), the BLM helicase (17, 18), and the structure-specific endonuclease scaffold SLX4 (17–21). ALT and telomerase are normally suppressed in somatic cells; however, they can become operant under both physiologic and pathologic conditions. For instance, cancer cells must activate TMMs in order to achieve replicative immortality (22), leading to the acquisition of aggressive phenotypes, including metastasis and therapeutic resistance (23, 24). Thus, elucidating the molecular dynamics of telomere homeostasis is essential to understanding the fundamental hallmarks of cancer and their role in the development of clinically intractable disease.
Besides their roles in regulating telomere dynamics, telomere proteins possess a variety of extratelomeric functions, including genome organization (25–27), DNA damage repair (28), and signal transduction (29–31). Notably, RAP1 coordinates gene expression by binding to target chromatin regions (32). Moreover, loss of RAP1 produces metabolic derangements due to alterations in the expression of factors that regulate metabolism and mitochondrial function (33). In addition, RAP1 potentiates NF-κB signaling by binding and activating the IκB kinase (IKK) (34) and mediates additional cellular responses by modulating mitogen-activated protein kinase activity (35). By expanding their functional repertoire beyond protecting telomeres and maintaining genome stability, RAP1 and other telomere proteins influence signaling events that play critical roles in a host of processes, including tumor development and progression.
Previously, we characterized SLX4-interacting protein (SLX4IP) as a key mediator of breast cancer metastasis and therapeutic sensitivity by controlling the dynamic interplay between telomerase and ALT (36). However, the mechanisms whereby SLX4IP accomplishes these activities remain incompletely understood. In the present study, we show that SLX4IP helped to establish the composition of the telomere proteome, doing so by modulating the activity of the SLX4-associated E3 SUMO ligase Protein Inhibitor of Activated STAT1 (PIAS1). In particular, PIAS1 SUMOylated RAP1, which promoted its interaction with IKK and subsequent activation of NF-κB and Notch signaling. Ultimately, these signaling events helped to establish TMM identity and determine the sensitivity of cancer cells with distinct TMMs to therapies targeting NF-κB and Notch.
RESULTS
SLX4IP regulates the composition of the telomere proteome
SLX4IP oversees a dynamic interplay between ALT and telomerase (18, 36, 37). In order to better understand how SLX4IP dictates these events, we performed proteomics of isolated chromatin segments (PICh) (38) to quantitatively characterize the telomere proteomes of SLX4IP-proficient and -deficient U2OS osteosarcoma cells (36), which are a prototypical model of ALT (Fig. 1A and Data File S1). In doing so, we successfully enriched for telomere-associated proteins (Fig. 1B), with several of these factors exhibiting differential abundance as a function of SLX4IP expression, including TIN2 and RAP1 (Fig. 1C). Additionally, proteins involved in HR, including NBS1 and MRE11 were selectively localized to telomeres in ALT cells harboring wild-type SLX4IP expression levels. In contrast, proteins involved in DNA damage and double-strand break (DSB) repair, including Ku70, Ku80, poly(ADP-ribose) polymerase (PARP1), and replication protein A subunit 1 (RPA1), were enriched at the telomeres of SLX4IP-depleted cells. Protein-protein interaction analysis further revealed a preferential increase in the formation of RAP1:TRF2 complexes and assembly of Ku-containing DSB repair complexes upon SLX4IP depletion (Fig. 1D). These events may reflect a shift in telomeric RAP1 abundance (Fig. 1C) and its ability to recruit and inhibit the nonhomologous end-joining machinery at telomeres (39, 40). Lastly, we investigated the molecular functions that may be affected by the observed changes in telomere protein abundance. Using Gene Ontology (GO) enrichment, we found several signatures indicative of the core telomere protection complex. Importantly, this cohort contained proteins involved in unexpected processes, such as nucleotide binding and post-translational modification (PTM) by ubiquitin or ubiquitin-like proteins (Fig. 1E). Taken together, these findings provide insight into potential mechanisms whereby SLX4IP determines telomere proteome composition and TMM identity, namely via the recruitment and stabilization of RAP1.
Fig. 1. SLX4IP Regulates the Composition of the Telomere Proteome.
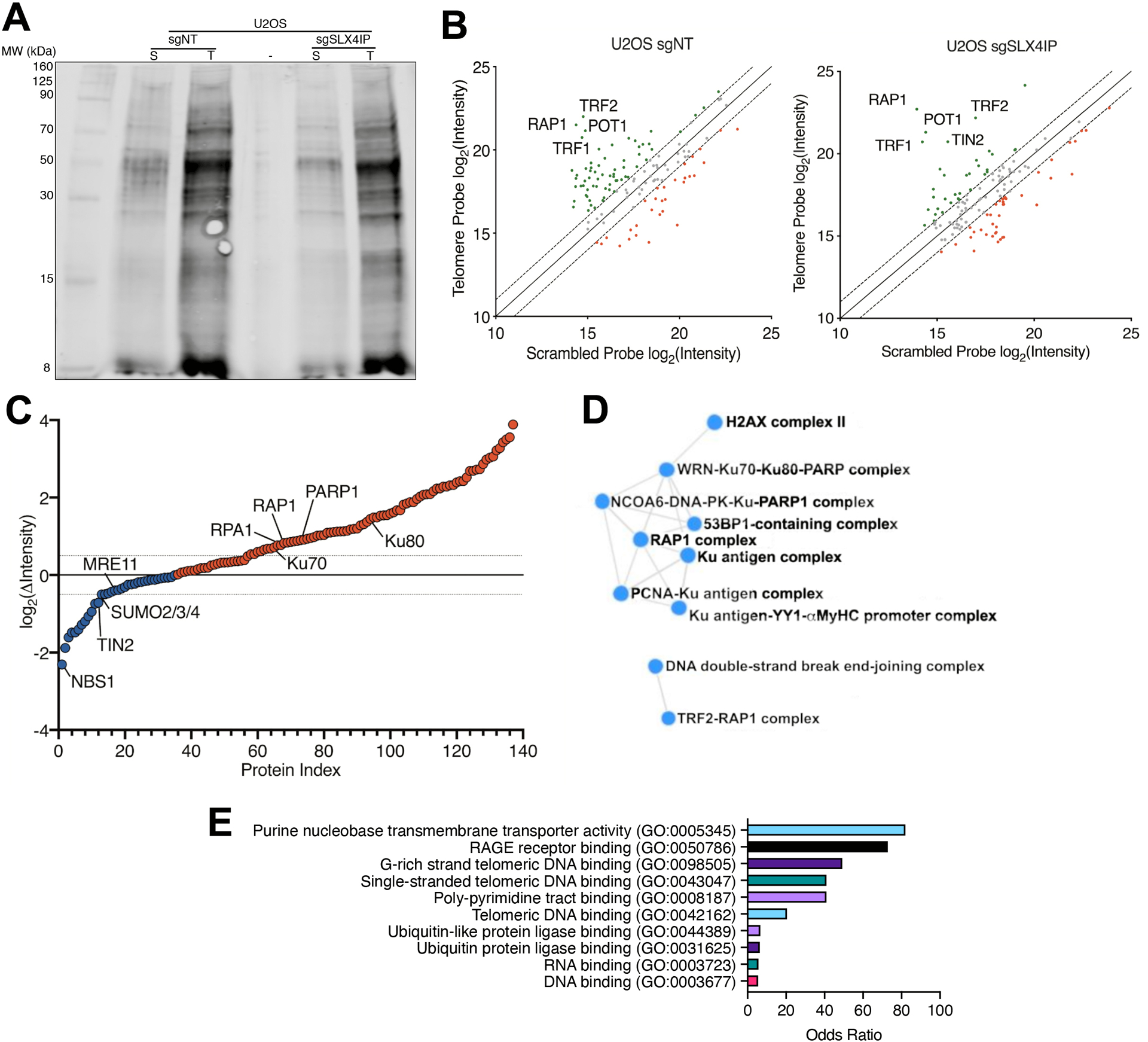
(A) Polyacrylamide gel showing proteins isolated from U2OS cells expressing a nontargeting CRISPR guide RNA (sgNT) or a guide RNA targeting SLX4IP (sgSLX4IP), using either a scrambled (S) or telomere-specific (T) LNA probe. n=1 technical replicate (exploratory). (B) Comparison of scrambled- versus telomere-bound protein abundance [represented as log2(peak intensity) from mass spectra] in parental (left) and SLX4IP-depleted (right) U2OS cells. Core shelterin proteins are labeled and are enriched in the telomere sample (green). Red points represent proteins that are enriched in the control sample. Dashed lines represent a relative protein abundance of ± 2. n=1 technical replicate per probe per cell line. (C) Waterfall plot depicting increased (red) and decreased (blue) protein abundance on SLX4IP-depleted U2OS telomeres. Highlighted proteins consist of core shelterin components; proteins associated with telomere maintenance and DNA repair; and SUMO isoforms. Dashed lines represent a relative protein abundance of ± 1.5. n=1 technical replicate. (D) Comprehensive Resource of Mammalian Protein Complexes (CORUM) network plot demonstrating RAP1-TRF2 stabilization stemming from SLX4IP depletion. In addition, SLX4IP-deficient telomeres possess a higher abundance of telomere recombination and repair factors. (E) Graph of Gene Ontology (GO) molecular function terms that are enriched among the cohort of proteins exhibiting differential abundance between parental and SLX4IP-depleted U2OS telomeres.
SLX4IP stimulates RAP1 SUMOylation by PIAS1
Although dynamic alterations to the telomere proteome likely underlie the function of SLX4IP in controlling telomere homeostasis (described above), the mechanisms whereby SLX4IP dictates telomere proteome composition remained unclear. We began to address this question by examining the known functions of the SLX4 complex (41). Notably, the SLX4 complex has functional ties with PTMs, including (i) phosphorylation through its interaction with the Polo-like kinase PLK1 (42); and (ii) ubiquitination and SUMOylation through its ubiquitin- and SUMO-binding properties, leading to the SUMOylation of XPF and of SLX4 itself (43–45). Intriguingly, our PICh analyses revealed differential abundance of multiple SUMO isoforms (SUMO2, −3, and −4), as well as a characteristic signature associated with ubiquitin-like protein ligation (Fig. 1, C and E). Moreover, SLX4IP possesses putative SUMO-interacting motifs (SIMs) (18), which led us to investigate the hypothesis that SLX4IP directs the SUMOylation of specific proteins that establish TMM identity.
Accordingly, SLX4IP influenced the SUMOylation of multiple target proteins, including core shelterin components (Fig. 2A and fig. S1A) that are known to undergo SUMOylation (46). In addition, we found that several other proteins identified by our PICh analyses were differentially SUMOylated in the presence of SLX4IP, including Ku70, PARP, and RPA1 (fig. S1B). Interestingly, SUMOylation of XPF, a member of the SLX4 complex known to undergo SLX4-dependent SUMOylation (43), was diminished by the loss of SLX4IP (fig. S1A). We focused our investigations on RAP1 because (i) its SUMOylation was significantly reduced upon SLX4IP depletion (Fig. 2A and fig. S1, C and D); and (ii) it plays a putative role in ALT by inhibiting HR at telomeres (11) and aiding in assembly of ALT-associated promyelocytic leukemia (PML) bodies (APBs) (47). Accordingly, RAP1 SUMOylation was lost in D2.OR cells rendered deficient in SLX4IP expression, an event that was restored upon SLX4IP rescue (Fig. 2B). Critically, SUMOylation of RAP1 was dependent upon the SUMO ligase activity associated with SLX4, as RAP1 SUMOylation was reduced in the presence of an SLX4 mutant that fails to promote XPF SUMOylation (SLX4-SIMmut; Fig. 2C) (43). These results implicate SLX4IP as an SLX4-dependent effector of SUMOylation whose targets encompass a diverse set of telomere-binding proteins, including RAP1.
Fig. 2. SLX4IP Stimulates RAP1 SUMOylation by PIAS1.
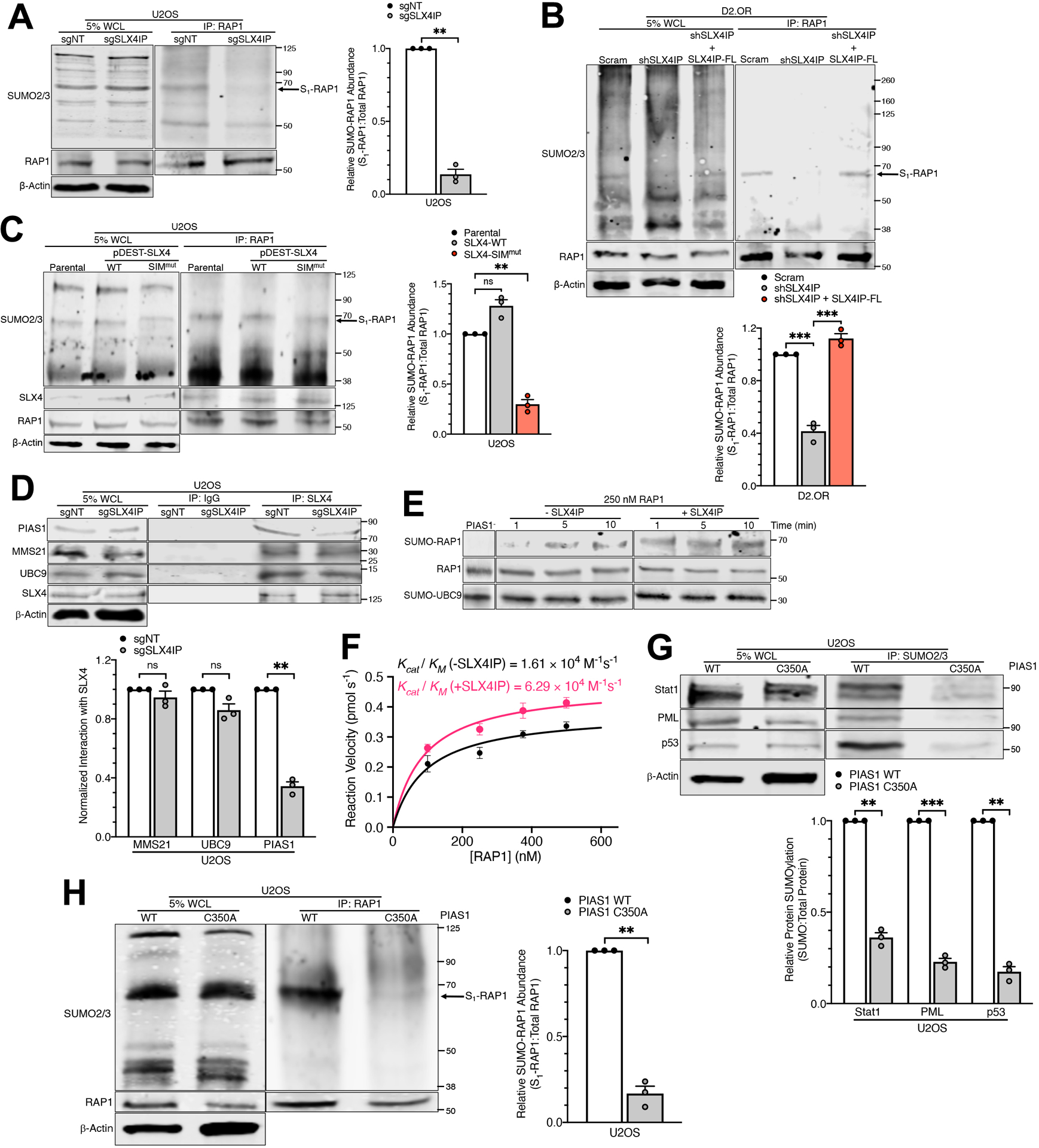
(A) Representative Western blot and accompanying quantitation of RAP1 SUMOylation (S1-RAP1) as a result of SLX4IP knockout in U2OS cells. WCL: whole-cell lysate. n=3 independent experiments. (B) Representative Western blot and accompanying quantitation of RAP1 SUMOylation following SLX4IP knockdown and reconstitution in D2.OR cells. n=3 independent experiments. (C) Representative Western blot and accompanying quantitation of RAP1 SUMOylation in U2OS cells expressing an SLX4 mutant containing mutated SUMO-interacting motifs (SIMmut). n=3 independent experiments. (D) Representative Western blot and accompanying quantitation of SLX4 complex formation with the E3 SUMO ligase PIAS1, E2 SUMO carrier UBC9, and E3 SUMO ligase MMS21 following SLX4IP depletion in U2OS cells. n=3 independent experiments. (E) Western blot demonstrating SLX4IP-dependent PIAS1 activation and RAP1 SUMOylation in a purified system. Nonlinear contrast adjustment applied to reduce background and aid visualization of SUMO-RAP1 bands. (F) Michaelis-Menten plot depicting SUMOylation rate as a function of RAP1 concentration. Reaction velocities were calculated using linear regression analysis of RAP1 SUMOylation over time at various starting concentrations of RAP1. Catalytic efficiency (Kcat/KM) for each condition was extrapolated from the best fit curves. The addition of SLX4IP stimulates PIAS1-directed SUMOylation of RAP1. n=3 independent experiments. (G) Representative Western blot and accompanying quantitation of PIAS1-dependent SUMOylation after introduction of the C350A mutation. Stat1, PML, and p53 are known targets of PIAS1. n=3 independent experiments. (H) Representative Western blot and accompanying quantitation of RAP1 SUMOylation following ablation of PIAS1 activity (C350A). n=3 independent experiments. **P<0.01, ***P<0.001, and ns: not significant, by Welch’s t test (A, D, G, and H) or one-way ANOVA (B and C).
Structural predictions suggest that SLX4IP is composed of a stable N-terminal domain (fig. S2A) and a highly disordered C-terminal domain (fig. S2B), neither of which is conserved among previously characterized SUMO ligases (fig. S2, C to E). Therefore, we determined whether SLX4IP regulates the recruitment or activity of the SUMOylation machinery. In doing so, we observed a SLX4IP-dependent reduction in SLX4 binding to the E3 SUMO ligase PIAS1 (Fig. 2D), which was previously found to regulate TRF2 stability (48). In contrast, SLX4IP depletion did not alter the ability of SLX4 to interact with (i) the E3 SUMO ligase MMS21, which regulates ALT via shelterin SUMOylation (46); or (ii) the E2 SUMO carrier protein UBC9, which has been shown to complex with SLX4 and facilitate its associated SUMO ligase activity (Fig. 2D) (43). In addition to recruitment, SLX4IP enhanced PIAS1-mediated SUMOylation of RAP1 (Fig. 2, E and F, and fig. S3). Conversely, expression of a catalytically inactive PIAS1 mutant (C350A; (49)) abrogated RAP1 SUMOylation (Fig. 2, G and H). Collectively, our findings establish SLX4 and SLX4IP as essential regulators of PIAS1 activity and assert PIAS1 as the catalytic component operant in SUMOylating RAP1.
RAP1 SUMOylation underlies SLX4IP-dependent telomere plasticity
As an initial step in determining the functional significance of RAP1 SUMOylation by PIAS1, we employed multiple SUMO site prediction tools (50, 51) to systematically identify potential sites of SUMOylation within the human RAP1 protein. These analyses independently revealed that lysine-240 (Lys240) is positioned within the strong consensus SUMOylation motif [ψ]-K-[x]-[D/E], where ψ is a hydrophobic, branched-chain amino acid and x is any amino acid (fig. S4A). Notably, Lys240 was previously identified as a potential site of SUMOylation in RAP1 through large-scale SUMO proteomic analyses (52–54). As such, we generated a putative SUMO-deficient RAP1 mutant (K240R) and assessed the impact of this mutation on SUMOylation and other lysine-directed PTMs. Notably, K240R-RAP1 exhibited a marked decrease in SUMOylation, with minimal alteration in acetylation and ubiquitination (Fig. 3A). Mutation of Lys240 also produced a reduction in RAP1 methylation, which could potentially impact RAP1 DNA binding or its interactions with other proteins (55). Nevertheless, our results demonstrate that RAP1 is SUMOylated at Lys240.
Fig. 3. RAP1 SUMOylation Underlies SLX4IP-dependent Telomere Plasticity.
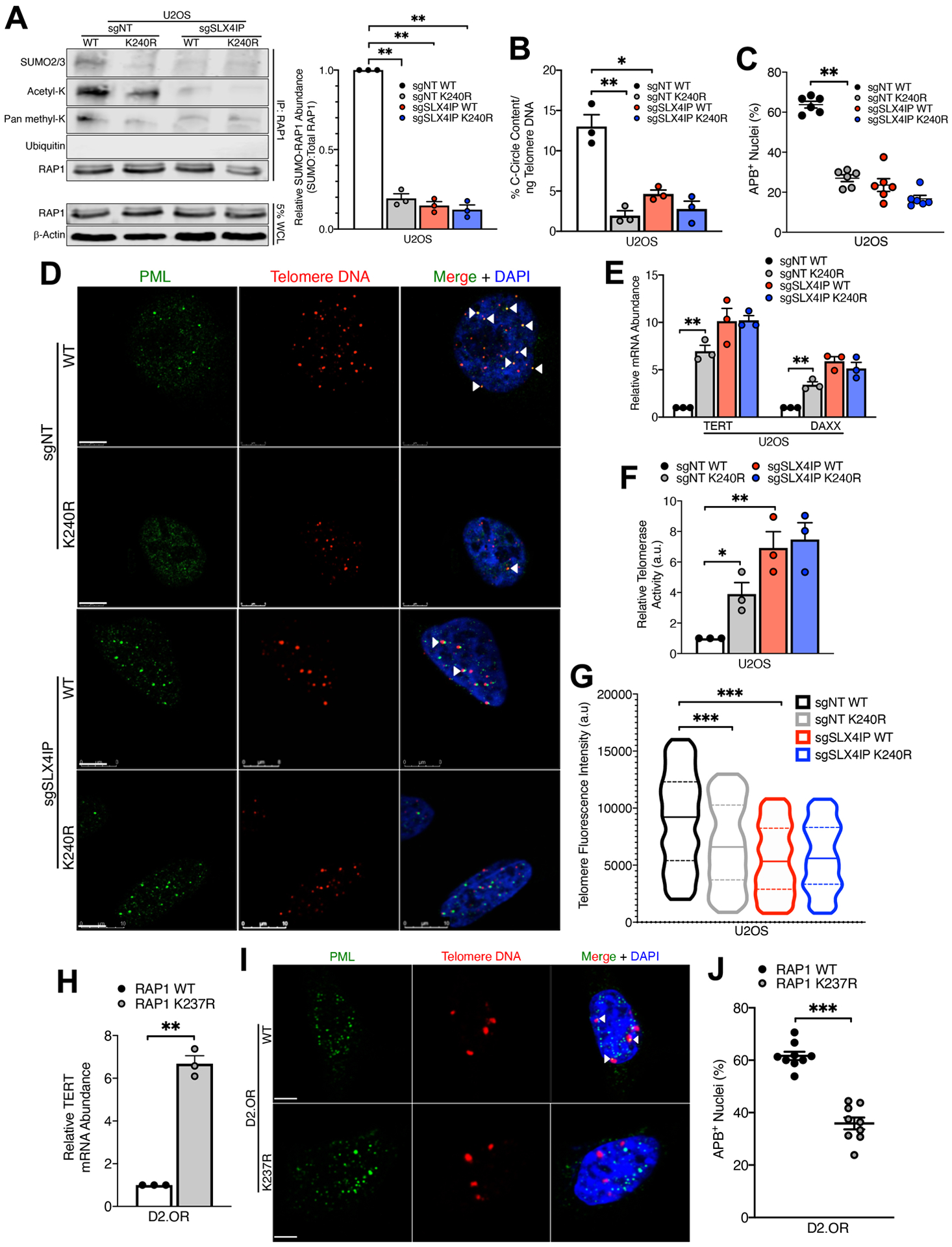
(A) Representative Western blot and accompanying SUMO quantitation of RAP1 SUMOylation upon introduction of the K240R mutation in parental and SLX4IP-depleted U2OS cell. n=3 independent experiments. (B) Quantitation of C-circle abundance in indicated U2OS derivatives by qRT-PCR. n=3 biological replicates. (C) Quantitation of APB abundance in indicated U2OS derivatives. n=6 visual fields (>90 nuclei per cell line). (D) Representative IF/FISH images showing APB formation in indicated U2OS derivatives. Scale bar: 5 μm. (E) qRT-PCR analysis of TERT and DAXX expression in indicated U2OS cell lines. Expression of each gene was normalized to parental U2OS cells expressing wild-type RAP1. n=3 biological replicates. (F) Telomerase activity in indicated U2OS derivatives, as determined by TRAP. n=3 independent experiments. (G) Violin plots showing quantitation of relative telomere length in indicated U2OS derivatives using Q-FISH. Solid lines indicate mean, and dashed lines indicate interquartile range. n>500 telomere foci per cell line. (H) qRT-PCR analysis of TERT expression in parental and RAP1 K237R-expressing D2.OR cells. n=3 biological replicates. (I) Representative IF/FISH images showing APB formation in parental and RAP1 K237R-expressing D2.OR derivatives. Scale bar: 5 μm. (J) Quantitation of APB abundance in parental and RAP1 K237R-expressing D2.OR cells. n=9 visual fields (>165 nuclei per cell line). *P<0.05, **P<0.01, and ***P<0.001 by Welch’s t test (C, E, H and J), one-way ANOVA (A, B, and F), or Kolmogorov-Smirnoff test (G).
Because SLX4IP functions as a molecular toggle between TMMs (36), we endeavored to determine whether this activity is regulated by PIAS1-mediated SUMOylation of RAP1. In doing so, we assayed markers of both ALT and telomerase, including (i) APBs; (ii) extrachromosomal telomere repeat-containing circular DNAs (C-circles); (iii) TERT expression and telomerase activity; and (iv) expression of the chromatin remodelers ATRX and DAXX, whose inactivation is associated with ALT (36). Significantly, U2OS cells expressing SUMO-deficient K240R-RAP1 harbored fewer C-circles (Fig. 3B) and APBs (Fig. 3, C and D) than their wild-type RAP1-expressing counterparts. Moreover, cells expressing K240R-RAP1 underwent an ALT-to-telomerase transcriptional reprogramming, as evidenced by increased TERT and DAXX expression to levels similar to those observed in telomerase-positive, SLX4IP-depleted cells (Fig. 3E). This stimulation of TERT expression produced a coincident increase in telomerase activity (Fig. 3F and fig. S5) and shorter telomeres (Fig. 3G), further indicating a transition from ALT to telomerase as the predominant TMM in cells expressing K240R-RAP1 (56, 57). Finally, the Lys240 SUMO motif in human RAP1 is highly conserved in mammals, including Lys237 in Mus musculus (fig. S4B). Importantly, engineering D2.OR cells to express K237R-RAP1 produced a sharp increase in TERT expression (Fig. 3H) and a dramatic decrease in APB abundance (Fig. 3, I and J), which serve as markers of TMM identity in these cells (36). Collectively, our findings connect the function of SLX4IP in modulating RAP1 SUMOylation to its role in dictating telomere plasticity.
RAP1 SUMOylation promotes activation of IKK and inhibition of telomerase
Given that SUMOylation plays a critical role in regulating the subcellular localization of proteins (58), we sought to elucidate the fate of SUMOylated RAP1 and examine its potential in mediating SLX4IP-dependent TMM plasticity. In the cytosol, RAP1 complexes with the β subunit of IKK (IKKβ), which activates IKK and NF-κB signaling (34). Accordingly, RAP1 underwent nucleocytoplasmic shuttling in parental U2OS cells; however, this cytoplasmic fraction was lost when SLX4IP expression or RAP1 SUMOylation were diminished (Fig. 4, A and B). Along the same lines, RAP1 interacted robustly with IKKβ in U2OS (Fig. 4C) and Saos-2 cells (fig. S6A), while SLX4IP depletion diminished this interaction and bolstered RAP1 binding to TRF2 and SLX4 (Fig. 4C). Moreover, IKK enzymatic activity was substantially decreased following SLX4IP ablation in U2OS cells (Fig. 4D). Notably, D2.OR cells exhibited a transcriptomic profile characteristic of elevated IKK activity, a state that was lost subsequent to SLX4IP knockdown (Fig. 4E and fig. S6B). Thus, the formation of highly active RAP1:IKK complexes is contingent upon the presence of SLX4IP.
Fig. 4. RAP1 SUMOylation Promotes Activation of IKK and Inhibition of Telomerase.
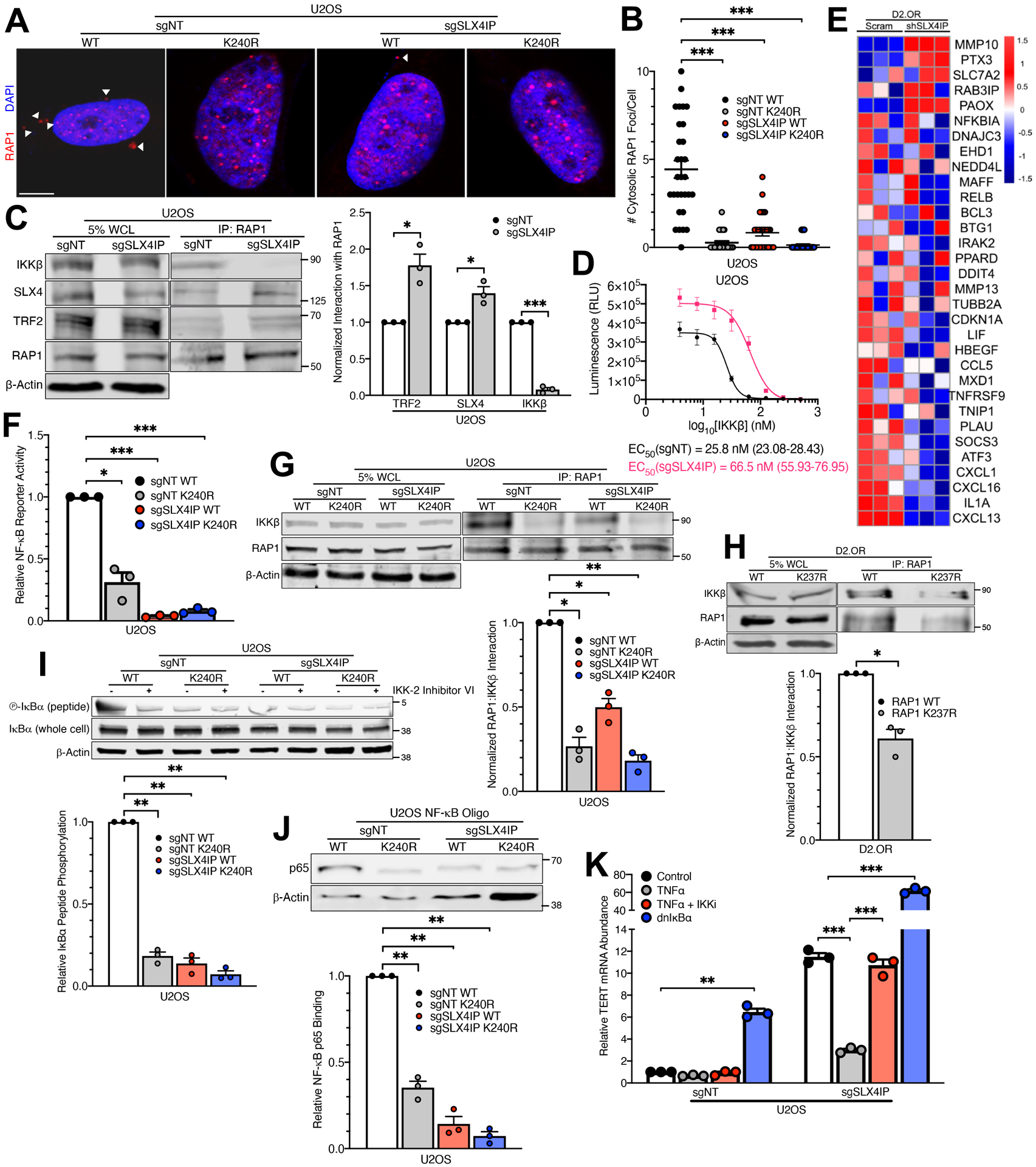
(A and B) Representative IF images (A) and quantitation (B) of RAP1 localization in indicated U2OS derivatives. Scale bar: 5 μm. n=30 cells per line. (C) Representative Western blot and accompanying quantitation showing RAP1 interaction with IKKβ, SLX4, and TRF2 in parental and SLX4IP-depleted U2OS cells. n=3 independent experiments. Images are taken from the same blot shown in Fig. 2A. (D) ATP consumption as a function of IKKβ concentration in parental and SLX4IP-depleted U2OS cells. IKKβ EC50 values indicate the [IKKβ] at which there has been a 50 percent reduction in luminescence intensity, which serves as a proxy for kinase activity. 95% confidence intervals are shown in parentheses. n=3 biological replicates. (E) Heatmap showing differential expression of genes in the Zhang Response to IKK Inhibitor and TNF gene set, which includes genes that are downregulated following administration of an IKK inhibitor, in parental and SLX4IP-depleted D2.OR cells. n=3 biological replicates. (F) NF-κB reporter activity in indicated U2OS derivatives. This assay quantifies luciferase activity downstream of the NF-κB-responsive MHC I promoter. n=3 biological replicates. (G and H) Representative Western blots and accompanying quantitation showing reliance of RAP1-IKKβ interaction on RAP1 SUMOylation in (G) U2OS and (H) D2.OR cells. n=3 independent experiments for each. (I) Representative Western blot and accompanying quantitation showing phosphorylation of an artificial IKK substrate (IκBα peptide) in indicated U2OS derivatives. Incubation with an IKK inhibitor (IKK-2 Inhibitor VI) serves as a negative control. n=3 independent experiments. (J) Representative Western blot and accompanying quantitation showing interaction between the NF-κB p65 subunit and a synthetic oligonucleotide containing a p65 consensus binding sequence (NF-κB oligo) in indicated U2OS derivatives. n=3 independent experiments. (K) qRT-PCR analysis of TERT expression in indicated U2OS cell lines that were pharmacologically or genetically manipulated to modulate NF-κB activity. Cells were either treated with TNFα with or without an IKK inhibitor (IKKi) or driven to ectopically express a dominant-negative IκBα isoform (dnIκBα) that inhibits NF-κB activity. n=3 biological replicates. *P<0.05, **P<0.01, and ***P<0.001 by Welch’s t test (C and H) or one-way ANOVA (B, F, G, I, J, and K).
Prompted by these findings, we next interrogated the functional connection between RAP1 SUMOylation, IKK signaling, and TMM identity. Remarkably, expression of SUMO-deficient K240R-RAP1 produced a marked decrease in NF-κB signaling (Fig. 4F). In addition, blocking SUMOylation diminished RAP1:IKKβ binding in both U2OS (Fig. 4G) and D2.OR cells (Fig. 4H). Disruption of RAP1 binding to IKKβ yielded a concomitant reduction in IKK activity (Fig. 4I and fig. S6C) and binding of the NF-κB subunit, p65, to its target DNA (Fig. 4J). Importantly, NF-κB signaling played a direct role in establishing TMM identity, as evidenced by a (i) sharp decrease in TERT expression following NF-κB stimulation with tumor necrosis factor α (TNFα), and (ii) a reciprocal increase following genetic or pharmacologic inhibition of NF-κB signaling (Fig. 4K). Collectively, our results indicate that PIAS1-mediated SUMOylation of RAP1 promotes NF-κB activation and telomerase repression, events subject to regulation by SLX4IP.
RAP1:IKK complexes drive ALT via an NF-κB:Notch signaling axis
In addition to targeting NF-κB, IKK phosphorylates several other intracellular signaling effectors, including β-catenin (59) and phosphoinositide 3-kinase (PI3K) (60). Furthermore, NF-κB exerts regulatory control over numerous signaling pathways, including transforming growth factor β (TGF-β) (61), Hedgehog (62), and Notch (63). Along these lines, we observed elevated expression of the Notch ligand Jagged-1 in parental U2OS (Fig. 5A) and Saos-2 cells (fig. S7A) compared to their SLX4IP-depleted counterparts, a phenomenon that was dependent upon both IKK and NF-κB (Fig. 5A) and resulted in increased Notch signaling (Fig. 5B). Moreover, parental D2.OR cells harbored a Notch-responsive gene signature, notably including Jagged-1, that was abrogated as a result of SLX4IP depletion (Fig. 5C and fig. S7B). Because parental U2OS and D2.OR cells possess greater RAP1 SUMOylation than their SLX4IP-deficient counterparts (above, Fig. 2), we set out to uncover whether this IKK:NF-κB:Notch signaling axis relied upon RAP1 SUMOylation. Interestingly, U2OS cells expressing SUMO-deficient K240R-RAP1 showed significantly decreased Jagged-1 expression (Fig. 5D) and, consequently, Notch signaling (Fig. 5E). Importantly, these findings mirror those that were observed when SLX4IP expression or IKK:NF-κB activity were disrupted, indicating that SLX4IP-directed SUMOylation of RAP1 promotes IKK-mediated activation of Notch signaling.
Fig. 5. RAP1-IKK Drives ALT via an NF-κB:Notch Signaling Axis.
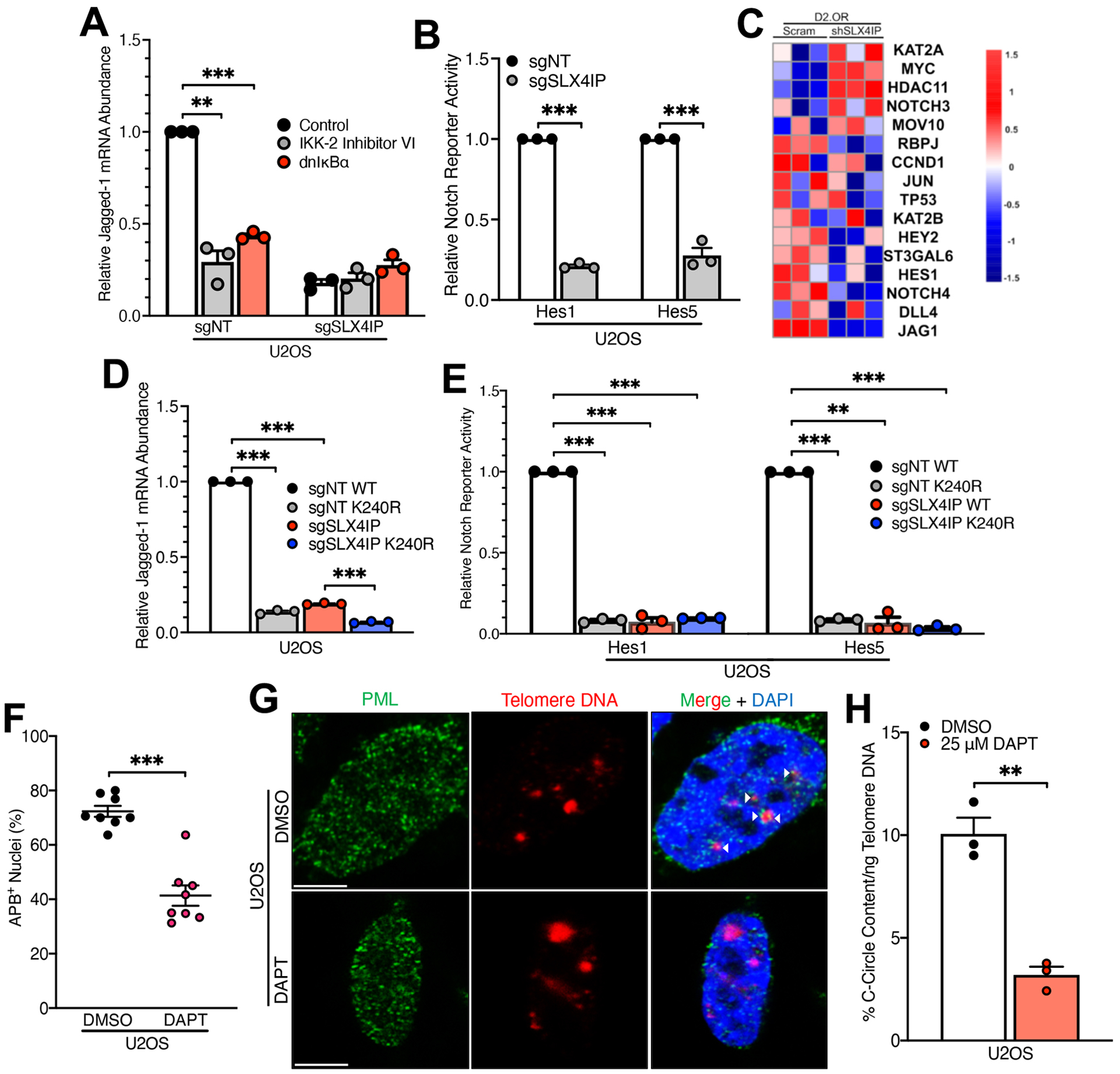
(A) qRT-PCR analysis of Jagged-1 expression in indicated U2OS derivatives treated with an IKK inhibitor or driven to ectopically express a dominant-negative IκBα isoform. n=3 biological replicates. (B) Notch reporter activity in indicated U2OS derivatives. This assay quantifies luciferase activity downstream of the HES1 and HES5 promoters, as indicated, which are Notch-responsive genes. n=3 biological replicates. (C) Heatmap showing differential expression of genes in the Reactome Signaling by Notch gene set in parental and SLX4IP-depleted D2.OR cells. (D and E) qRT-PCR analysis of Jagged-1 expression (D) or Notch reporter activity (E) in indicated U2OS derivatives. n=3 biological replicates. (F) Quantitation of APB abundance in parental U2OS cells treated with diluent (DMSO) or the γ-secretase inhibitor DAPT (25 μM). n=8 visual fields (>105 nuclei per cell line). (G) Representative IF/FISH images showing APB formation in U2OS cells subjected to the indicated treatments. Scale bar: 5 μm. (H) Quantitation of C-circle abundance by qRT-PCR in U2OS cells subjected to the indicated treatments. n=3 biological replicates. **P<0.01 and ***P<0.001 by Welch’s t test (B, F, and H) or one-way ANOVA (A, D, and E).
Because of the intimate connection between RAP1 SUMOylation and telomere homeostasis (above, Fig. 3), we next probed whether TMM identity is established via Notch signaling. At present, little is known about the identity of key signaling pathways that govern ALT. Intriguingly, we found that administration of the γ-secretase inhibitor, DAPT, to inactivate Notch signaling not only suppressed the assembly of APBs (Fig. 5, F and G and fig. S7, C and D), but also blocked the formation of C-circles (Fig. 5H). Mechanistically, Jagged-1-mediated activation of Notch resulted in epigenetic silencing of ATRX and DAXX in parental D2.OR cells (fig. S7, E and F), an event dependent upon the methyltransferase, EZH2 (fig. S7G). Taken together, these findings illuminate an NF-κB:Notch signaling cascade that coordinates multiple TMMs downstream of RAP1 SUMOylation and extratelomeric localization.
NF-κB and Notch can be therapeutically targeted in ALT-driven cancer cells
SLX4IP expression and TMM identity are associated with poor survival and metastatic progression in breast cancer patients. Additionally, these molecular markers can be used to guide the selection of therapies targeting individual TMMs (36). As such, we sought to assess the extent to which NF-κB and Notch signaling are related to the prognostic and therapeutic value of SLX4IP in cancer. Remarkably, SLX4IP showed a strong association with a Notch-driven transcriptional program (Fig. 6A) across a diverse array of human tumors. Likewise, both SLX4IP and PIAS1 expression are correlated with cancer cell sensitivity to IKK inhibition (Fig. 6B). Taken together, these results suggest that telomere plasticity mediated by SLX4IP and PIAS1 are intimately linked to NF-κB and Notch in human cancers.
Fig. 6. NF-κB and Notch Can be Therapeutically Targeted in ALT-driven Cancer Cells.
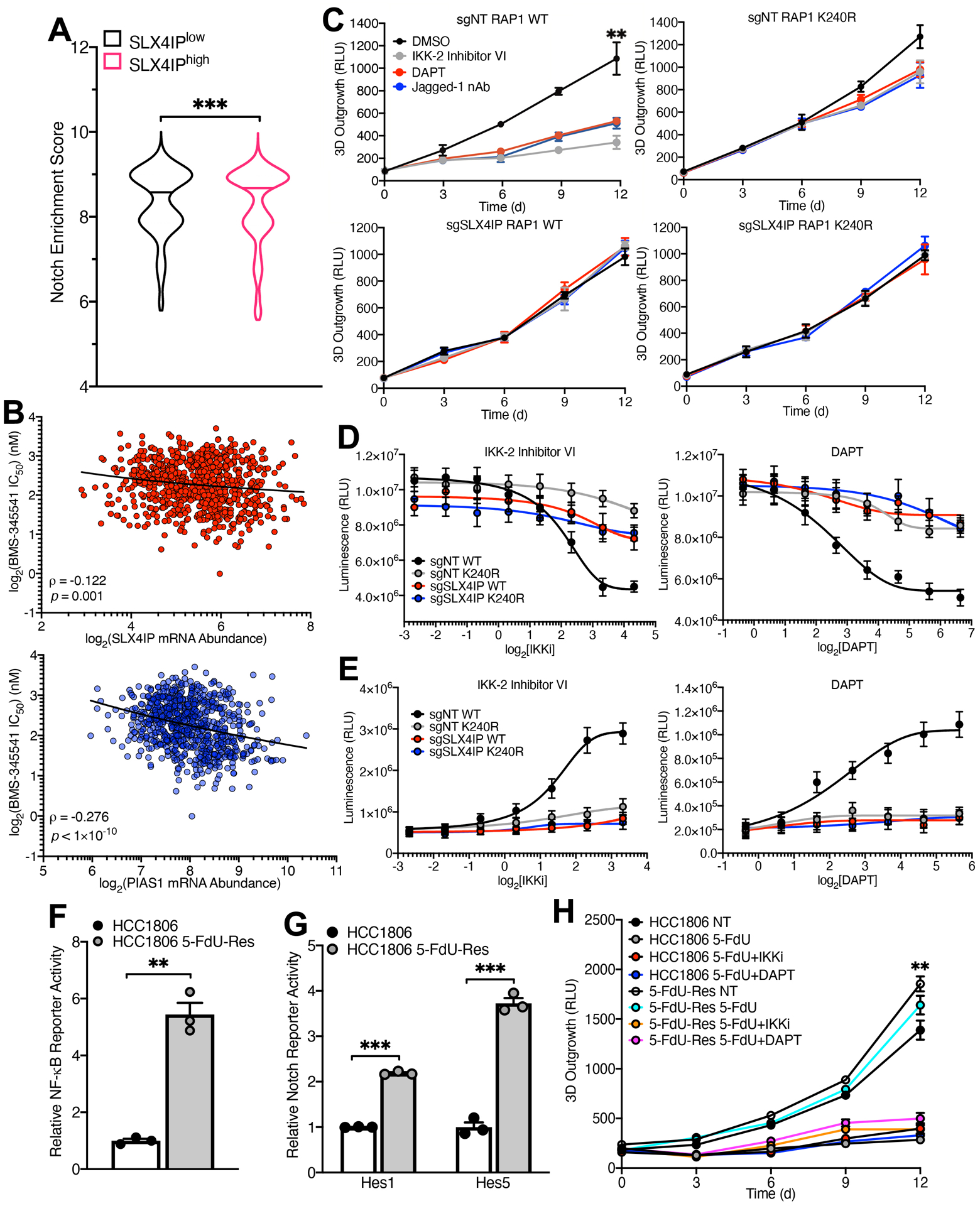
(A) Violin plot depicting pan-cancer enrichment analysis of a Notch-associated gene signature in TCGA specimens expressing high (SLX4IPhigh) and low (SLX4IPlow) levels of SLX4IP. Horizontal lines represent the median enrichment score for each group. n=4259 (SLX4IPlow), n=4207 (SLX4IPhigh). (B) Correlation (represented by Spearman’s ρ) of IKK inhibitor (BMS-345541) sensitivity with SLX4IP (top) and PIAS1 (bottom) expression across cancer cell lines (n=727). A lower IC50 value indicates increased sensitivity to treatment with BMS-345541. (C) Three-dimensional outgrowth of parental (top row) and SLX4IP-depleted (bottom row) U2OS cells expressing wild-type (left column) or SUMO-deficient (right column) RAP1. Cells were treated with diluent (DMSO), IKK-2 Inhibitor VI (1 μM), DAPT (25 μM), or Jagged-1 neutralizing antibody (nAb; 20 ng/mL) as indicated. n=3 biological replicates. (D) Viability of indicated U2OS derivatives treated with IKK-2 Inhibitor VI (left) or DAPT (right). n=3 independent experiments. (E) Caspase-3/7 activation in indicated U2OS derivatives treated as indicated. n=3 independent experiments. (F and G) NF-κB (F) and Notch (G) reporter activity in parental and 5-FdU-resistant (5-FdU-Res) HCC1806 cells. n=3 biological replicates for each. (H) Three-dimensional outgrowth of parental and 5-FdU-resistant HCC1806 cells treated with 5-FdU (500 nM) in combination with IKK-2 Inhibitor VI (1 μM) or DAPT (20 μM). n=3 biological replicates. **P<0.01 and ***P<0.001 by Mann-Whitney U test (A), Welch’s t test (F and G), or one-way ANOVA (C and H).
At least 15% of cancers activate ALT for telomere maintenance (64). However, therapies targeting ALT are not currently available to patients. Indeed, the efficacy of agents that have shown preclinical benefit against ALT-driven cancers remains controversial (65). Thus, there exists a substantial need for the development of new therapeutic approaches to target telomere homeostasis. With this in mind, we monitored the impact of drugs that inhibit IKK and Notch for their ability to impede the three-dimensional (3D) outgrowth and proliferation of U2OS cells, and to induce their apoptosis. Further, we analyzed the role of RAP1 SUMOylation in dictating sensitivity to these regimens. Notably, the 3D-outgrowth of parental U2OS cells was significantly impaired by administration of IKK-2 Inhibitor VI to inactivate IKK or by administration of either DAPT or neutralizing Jagged-1 antibodies to inactivate Notch (Fig. 6C). Moreover, inhibition of either IKK or Notch decreased the proliferation of U2OS cells (Fig. 6D) and increased their apoptosis (Fig. 6E). In stark contrast, expression of K240R-RAP1 or depletion of SLX4IP rendered U2OS cells resistant to IKK and Notch inhibitors (Fig. 6, C to E). Collectively, these results reinforce the mechanistic relationship between SLX4IP and RAP1 SUMOylation that underlie the activation of an IKK:NF-κB:Notch signaling axis; they also reveal therapeutic modalities that may be capable of targeting ALT-driven cancers, although additional work is needed to assess the general efficacy of these approaches in disrupting cancer cell growth as a function of TMM identity.
Finally, we previously determined that breast cancer cells upregulate SLX4IP expression and adopt ALT for telomere maintenance as a means to circumvent the anti-telomerase therapy 5-fluoro-2′-deoxyuridine (5-FdU) (36). Consequently, we endeavored to explore whether IKK or Notch emerged as viable therapeutic targets in the setting of 5-FdU resistance. Importantly, 5-FdU-resistant HCC1806 cells displayed hyperactivation of both the Notch (Fig. 6F) and NF-κB pathways (Fig. 6G). As a result, these cells acquired susceptibility to IKK and Notch inhibitors compared to their 5-FdU-sensitive counterparts (Fig. 6H). Interestingly, inhibiting IKK and Notch in parental HCC1806 cells did not exhibit synergy with 5-FdU, suggesting that these approaches may be of particular utility in recurrent, treatment-refractory disease.
DISCUSSION
in this study, we uncovered a previously unknown function of SLX4IP as a key regulator of the SUMO proteome. In doing so, we simultaneously revealed a mechanism whereby telomere proteins directly coordinate signaling events that institute specific telomere homeostatic programs, findings consistent with our previous work (36, 37). Nevertheless, this study focuses primarily on ALT-driven cancer cell lines, and the exact nature of the relationship between this pathway and specific TMMs in other cell types remains an active area of investigation (18). SUMOylation mediates a diverse array of processes, including nucleocytoplasmic shuttling (66–68), transcriptional activation and repression (69), and protein-protein interaction (70). Of note, SUMOylation plays an important part in telomere maintenance. For instance, SUMOylation of PML is necessary for the nucleation of PML nuclear bodies (71, 72), including APBs (73). Similarly, TRF1 and TRF2 SUMOylation are required for telomere extension in ALT cells (46). Moreover, the activity of specific SUMO ligases mobilizes the cellular response to replication stress (74), which is present in ALT in the context of break-induced telomere synthesis (75). It is noteworthy that previous investigations examining the connections between SUMOylation and telomere maintenance have focused on effects in situ at telomeres. Our results highlight the multifunctionality of telomere proteins as direct participants in signal transduction, thereby serving as determinants of telomere homeostasis in a telomere-independent manner and reveal the importance of SUMOylation in orchestrating these functions in malignant cells (Fig. 7).
Fig. 7. Proposed Model of IKK:NF-κB:Notch Signaling Control by SLX4IP and RAP1.
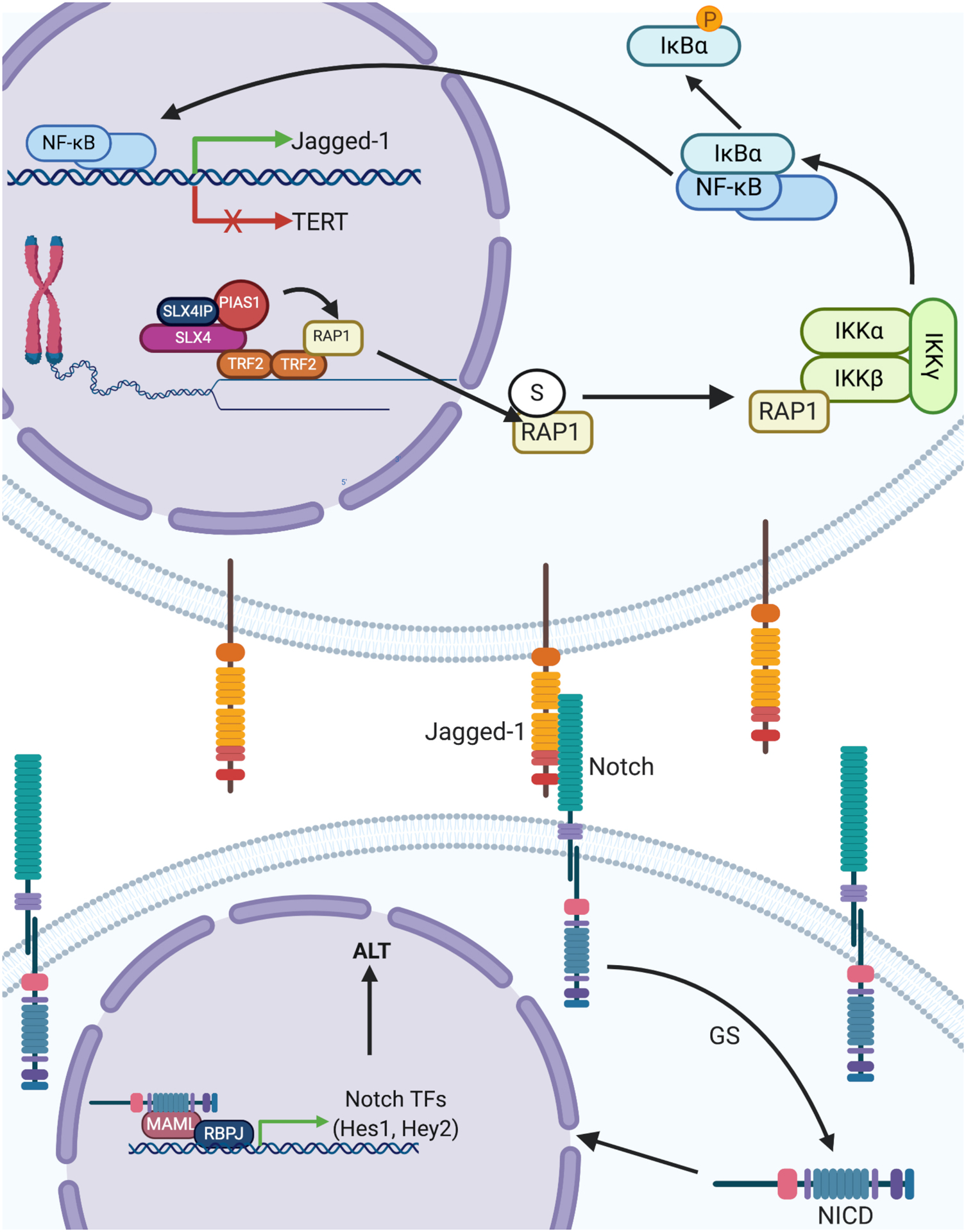
SLX4IP facilitates recruitment of PIAS1 to the SLX4 scaffold and promotes SUMOylation (S) of RAP1. SUMOylated RAP1 is exported from the nucleus to the cytoplasm, where it interacts with IKKβ (as part of the heterotrimeric IKK complex), stimulating NF-κB nuclear translocation via IκBα phosphorylation and degradation. Once in the nucleus, NF-κB increases Jagged-1 transcription while simultaneously suppressing TERT. Jagged-1 binds to Notch receptors on neighboring cells, which liberates the Notch intracellular domain (NICD) through activation of the γ-secretase (GS) pathway. NICD is a transcription factor that recognizes its targets by binding with additional transcriptional co-activators (RBPJ and MAML). These targets comprise a host of transcription factors (for example, Hes1 and Hey2) that directly coordinate the cellular Notch response. This response includes the modulation of ALT-associated gene expression as well as other features of ALT, such as APB formation. In the absence of SLX4IP, RAP1 fails to be SUMOylated and bind to IKKβ, which diminishes both NF-κB and Notch signaling, thereby removing repression of TERT and dampening Notch-driven activation of ALT. In this way, SLX4IP promotes ALT, and we hypothesize that it controls telomere plasticity through its role in directing PIAS1-mediated SUMOylation of RAP1.
In addition to SUMO, numerous PTMs contribute to the regulation of telomere stability. While telomeric chromatin is thought to be largely heterochromatic, chromatin architecture at telomeres may vary across cell types or developmental states (76). The degree of compaction of telomeric chromatin is heavily controlled by the balance of histone acetylation and methylation, which facilitate chromatin opening and closing, respectively. In turn, the compaction state of telomeric chromatin significantly impacts telomere length (77) and the replication and transcription of telomere and subtelomere DNA (78, 79). Apart from histones, several core telomere proteins, including telomerase and members of the shelterin complex, can be post-translationally modified to alter telomere maintenance in response to environmental cues. For example, TRF1 and TRF2 are phosphorylated by a host of kinases that link telomere homeostasis to DDR activation (80, 81), cell cycle progression (82), and mitotic fidelity (83). TRF1 is also subject to modification with poly(ADP-ribose) (PAR) by the PAR polymerase tankyrase-1, which inhibits TRF1 binding to telomere DNA and promotes telomere elongation (84). Moreover, the activity of telomerase and components of the ALT machinery are post-translationally regulated. Specifically, ubiquitination dictates both telomerase stability and activity (85), as well as telomere recombination and extension in ALT cells (86). Our results add to this body of knowledge (36) by uncovering the importance of RAP1 SUMOylation in telomere length regulation. These molecular insights not only increase our fundamental understanding of telomere dynamics, but they also shed light on how these dynamics unfold epigenetically to control processes that rely upon telomere lengthening, such as stem cell maintenance (87, 88) and embryonic development (89, 90).
Our findings assert RAP1 SUMOylation as an activator of NF-κB signaling, which in turn stimulates Notch through increasing expression of Jagged-1. Consequently, NF-κB and Notch provoke TMM plasticity by inhibiting telomerase, while simultaneously driving the emergence of ALT (Fig. 7). These results stand in contrast to previous reports demonstrating transcriptional activation of TERT by NF-κB (91–93). Clearly further studies designed to assess the contributions of other transcriptional co-regulators or parallel signaling events orchestrated by pro-inflammatory stimuli will be of value in deciphering this apparent conflict. More broadly, SUMOylation plays a critical role in various intracellular signaling pathways, including TGF-β (94, 95), Wnt/β-catenin (96, 97), Notch (98), insulin-like growth factor (99), and mitogenic signaling through AKT and ERK1/2 (100, 101). Interestingly, these pathways contribute extensively to processes in which telomere maintenance is essential, including development and tumorigenesis (102, 103). Thus, SUMOylation may serve dual functions in these contexts by governing telomere homeostasis and modulating cell signaling. In addressing this supposition, future studies need to (i) map the SUMO proteome as a function of SLX4IP abundance; (ii) identify the SUMO ligases regulated by SLX4IP and classify the signaling pathways impacted by their activity; and (iii) determine the regulatory roles of these pathways in telomere maintenance and tumor progression.
Both NF-κB and Notch signaling are intimately connected to cancer development and progression (104, 105). Previously, we demonstrated that SLX4IP expression and TMM identity can serve as potential prognostic and therapeutic markers for TMM-targeted treatment approaches in breast cancer (36). Here, we established the central importance NF-κB and Notch in dictating TMM identity and as potential therapeutic targets in cancer cells that harbor specific TMM or drug resistance profiles. Telomere-associated mutations and aberrant gene expression signatures are hallmarks of cancers that exhibit poor survival and therapeutic resistance, including high-grade gliomas (106) and aggressive breast cancers (36, 107). In the same vein, NF-κB signaling is associated with disease progression, cancer stem cell maintenance, and chemoresistance (108, 109). Incorporating NF-κB and Notch profiling into clinical decision-making paradigms and combinatorial treatment regimens may therefore provide much-needed benefit to patients suffering from the most aggressive and treatment-refractory cancers. Such advances would extend the fundamental molecular mechanisms illuminated by this study to provide important pathophysiologic and clinical insights.
MATERIALS AND METHODS
Cell lines and tissue culture
D2.OR cells were obtained from Fred Miller (Wayne State University, Detroit, MI) and cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS). U2OS and HCC1806 cells were obtained from ATCC and cultured in McCoy’s 5A media (U2OS) or RPMI-1640 medium (HCC1806) with 10% FBS. Saos-2 cells were obtained from ATCC and cultured in McCoy’s 5A medium supplemented with 20% FBS. Cell lines were authenticated using Short Tandem Repeat (STR) analysis (ATCC). All media were additionally supplemented with 1% penicillin-streptomycin prior to use. All cells were grown in a 37°C incubator with 5% CO2. Cell lines were engineered to stably express firefly luciferase by transfection with pNifty-CMV-luciferase, followed by zeocin selection (500 μg/mL). Depletion of SLX4IP, RAP1, and PIAS1 were achieved by lentiviral transduction of expression cassettes harboring both SpCas9 and a chimeric single guide RNA scaffold (pLentiCRISPRv2; Addgene), followed by selection with puromycin (5 μg/mL). sgRNA design was carried out using CHOPCHOP (110) in order to generate double-strand breaks (DSBs) in multiple exons and minimize off-target binding of sgRNAs. To confirm presence of repaired DSBs, genomic DNA was extracted from cells using the Quick gDNA Miniprep Kit (Zymo Research) per the manufacturer’s instructions, and the regions targeted by specific sgRNAs were PCR-amplified and subjected to Sanger sequencing. The SLX4 SIM mutant construct was generated as described (43) and was stably expressed in U2OS cells by lentiviral transduction, followed by selection with hygromycin (300 μg/mL).
Proteomics of isolated chromatin segments (PICh)
Telomere proteome characterization was accomplished using the previously described proteomics of isolated chromatin segments (PICh) workflow (38). U2OS cells (1 × 109 cells/sample) were crosslinked in 3.7% formaldehyde, equilibrated in sucrose buffer (0.3 M sucrose, 10 mM HEPES-NaOH pH 7.9, 1% Triton X-100, 3 mM CaCl2; 2 mM magnesium acetate), and dounced with a tight pestle. Cells were then equilibrated in glycerol buffer (25% glycerol, 10 mM HEPES-NaOH pH 7.9, 0.1 mM EDTA, 0.1 mM EGTA, 5 mM magnesium acetate) and lysed in PBS containing 0.5%Triton X-100 and RNaseA (1 mg/mL; QIAGEN) for 60 min at room temperature, followed by continuous agitation overnight at 4°C. The next day, cells were further lysed in high-salt lysis buffer (10 mM HEPES-NaOH pH 7.9, 100 mM NaCl, 2 mM EDTA, 1 mM EGTA, 0.2% SDS, 0.1% N-lauroylsarcosine sodium). Samples were sonicated (40 watts for 15 seconds, on ice for 45 seconds, 28 cycles) to mechanically lyse cells and shear genomic DNA. Chromatin was collected by centrifugation and applied to Sephacryl S-400 High Resolution columns (GE Healthcare). Flow-through was pre-cleared for 2 h at room temperature with MyOne Streptavidin C1 Dynabeads (ThermoFisher) that had been pre-equilibrated in low-salt lysis buffer (10 mM HEPES-NaOH pH 7.9, 30 mM NaCl, 2 mM EDTA, 1 mM EGTA, 0.2% SDS, 0.1% N-lauroylsarcosine sodium).
The following locked nucleic acid (LNA) probes were used for telomere chromatin capture (Fidelity Systems): Telomere: Desthiobiotin-C108-5′-TtAgGgTtAgGgTtAgGgTtAgGgt-3′; Scramble: Desthiobiotin-C108-5′-GaTgTgTgGaTgTggAtGtGgAtgTgg-3′, where capital letters are LNA residues and lowercase letters are DNA residues. A scrambled control was included for each cell line. LNA probes were used at a final concentration of 1 μM, and hybridization was conducted according to the following thermal profile: 25°C for 3 min, 70°C for 6 min, 38°C for 60 min, 60°C for 2 min, 38°C for 60 min, 60°C for 2 min, 38°C for 120 min. LNA-DNA hybrids were separated by centrifugation and diluted with Milli-Q water and MyOne Streptavidin C1 beads. Beads were incubated overnight at room temperature and subsequently washed six times with high-salt lysis buffer before being resuspended in elution buffer (12.5 mM biotin, 75% high-salt lysis buffer). Samples were incubated at room temperature for 1 h with shaking, followed by further incubation at 65°C for 10 min. The eluate was precipitated using trichloroacetic acid (18% v/v), and the pellet was cleared of organic solvent using acetone and resuspended into 50 μL crosslinking reversal solution (250 mM Tris pH 8.8, 2% SDS, 0.5 M β-mercaptoethanol) at 99°C for 25 min. 10% of the final protein yield was separated using a 12% Bis-Tris acrylamide gel (Bio-Rad) and visualized using Cyanine5 NHS ester (Lumiprobe) according to manufacturer’s instructions.
Mass spectrometry and proteomic identification
Protein isolates were subjected to 1D-SDS-PAGE approximately ~1 cm into the gel, which was excised and in-gel digested by Lys-C (111). Following digestion, LC-MS/MS was performed using the ThermoScientific Orbitrap Fusion Lumos mass spectrometry system. HPLC was carried out using a Dionex 15 cm × 75 μm id Acclaim Pepmap C18, 2 μm, 100 Å reversed-phase capillary chromatography column. Peptides eluted from the column in an acetonitrile/0.1% formic acid gradient (flow rate = 0.3 μL/min) were introduced into the microelectrospray ion source of the mass spectrometer, which was operated at 2.5 kV. Samples were analyzed using a data-dependent method with CID fragmentation. Proteins were identified by comparing all of the experimental peptide MS/MS spectra against the UniProt human database using the Andromeda search engine integrated into the MaxQuant version 1.6.3.3 (112, 113). Carbamidomethylation of cysteine was set as a fixed modification, whereas variable modifications included oxidation of methionine to methionine sulfoxide and acetylation of N-terminal amino groups. For peptide/protein identification, strict Lys-C specificity was applied, the minimum peptide length was set to 7, the maximum missed cleavage was set to 2, and the cutoff false discovery rate was set to 0.01. Match between runs (match time window: 0.7 min; alignment time window: 20 min) and label-free quantitation (LFQ) options were enabled. The LFQ minimum ratio count was set to 2. The remaining parameters were kept as default. Protein quantitation was accomplished using Perseus (114). LFQ values were log2-transformed, and missing values were imputed using the “Replace missing value from normal distribution” function on the entire matrix using default parameters.
SUMO site prediction
Human (accession: NP_061848.2) and mouse (accession: AAH17641.1) RAP1 amino acid sequences were curated from the National Center for Biotechnology Information (NCBI; https://www.ncbi.nlm.nih.gov/). Putative SUMOylation sites were identified using the prediction software GPS-SUMO (http://sumosp.biocuckoo.org/online.php) (51), the Joined Advanced SUMOylation Site and SIM Analyzer (JASSA) (http://www.jassa.fr/) (50), and SUMOplot (http://www.abcepta.com/sumoplot). Each program was run using its default parameters. Individual lysine residues in the human protein that emerged from at least two of these analyses included Lys9, Lys114, Lys212, and Lys240, which were adopted as candidates for site-directed mutagenesis. Conservation of SUMO motifs across species was assessed using the Pattern Hit-Initiated Basic Local Alignment Search Tool (PHI-BLAST).
RAP1 and PIAS1 mutagenesis
pLPC-hRAP1 (12) and pLPC-mRAP1 (115) were obtained from Addgene, while pLV-Hygro-hPIAS1 was synthesized using VectorBuilder. Utilizing the Agilent QuikChange Primer Design Tool, primers were designed to introduce the desired amino acid substitution as well as a synonymous point mutation in the protospacer adjacent motif (PAM) sequence to prevent CRISPR/Cas9 targeting. Site-directed mutagenesis was carried out using Q5 High-Fidelity DNA Polymerase (New England Biolabs) according to the manufacturer’s instructions. PCR reactions were subsequently incubated with DpnI (1 U/reaction; New England Biolabs) at 37°C for 2 h. Transformed PCR products were purified using the QIAprep Spin Miniprep Kit (QIAGEN) and subjected to Sanger sequencing to confirm mutagenesis. Oligonucleotide sequences for CRISPR sgRNAs and site-directed mutagenesis primers are provided in tables S1 and S2.
Protein structural prediction
Ab initio SLX4IP structural prediction was accomplished using QUARK (https://zhanglab.ccmb.med.umich.edu/QUARK/) (116). SLX4IP N- (residues 1–173) and C-terminal (residues 209–408) domains were defined by limited proteolysis, and corresponding amino acid sequences were analyzed using default parameters. Full-length structure models were assembled from fragments ranging in size from 1–20 amino acids using replica-exchange Monte Carlo (REMC) simulations guided by a composite knowledge-based force field. Primary sequences were first subjected to multiple sequence alignment using Position-Specific Iterated BLAST (PSI-BLAST) to predict secondary structures using neural network training of sequence-structure relationships from sequence alignment. Fragments from experimental structures with template modeling scores (TM-scores) >0.5 were excluded from comparison (117). Candidate fragment assemblies from REMC simulations underwent refinement to minimize root-mean-square deviation of atomic positions (RMSD). Three-dimensional conformations were modeled via minimization of the force field energy function, which includes terms corresponding to steric pairwise potential of backbone and side chain atoms, hydrogen bonding, solvent accessibility, backbone torsion potential, and secondary structure packing. Predicted SLX4IP domains were aligned to known crystal structures of the SUMO ligases PIAS2 (PDB: 4FO9) and RanBP2 (PDB: 4GA0) using UCSF Chimera.
Quantitative real-time PCR (qRT-PCR)
Cells were nonenzymatically isolated from 3D-culture (50,000 cells/well; 500 μL Cultrex/well in 12-well plate) using the Cultrex 3D-Culture Cell Harvesting Kit (Trevigen), and total RNA was extracted using the RNeasy Mini Kit (QIAGEN) according to the manufacturer’s protocol. One μg of total RNA was subsequently reverse-transcribed using the iScript cDNA Synthesis Kit (Bio-Rad) and subjected to quantitative real-time PCR using 1× iQ SYBR Green Supermix (Bio-Rad), as previously described (118) and using the primers listed in table S2. Changes in gene expression were determined using the ΔΔCt method.
Immunoblotting and immunoprecipitation (IP)
Cells were isolated from 3D-culture and homogenized on ice in RIPA lysis buffer (50 mM Tris-HCl, 150 mM NaCl, 6 mM sodium deoxycholate, 1.0% NP-40, 0.1% sodium dodecyl sulfate (SDS), pH 7.4) supplemented with protease inhibitor cocktail (Sigma-Aldrich) and phosphatase inhibitors (10 mM sodium orthovanadate, 40 mM β-glycerophosphate, 20 mM NaF). Lysates were cleared by centrifugation and subjected to immunoblot analysis as described (119). For co-immunoprecipitation, cells were solubilized with Buffer H (500 mM NaCl, 20 mM imidazole, 50 mM Na2HPO4/NaH2PO4 pH 7.6, 15% glycerol, 0.01% NP-40) supplemented with 1% Triton X-100 plus protease inhibitors, followed by immunoprecipitation of 700 μg protein as described (120) using IP-validated antibodies against RAP1 (Cell Signaling Technology), SLX4 (Bethyl Laboratories), or SUMO2/3 (Cell Signaling Technology) conjugated to Protein A Sepharose beads (GE Healthcare). For SUMO blots, lysis buffer (150 mM Tris-HCl pH 6.7, 5% SDS, 30% glycerol) was supplemented with the SUMO peptidase (SENP) inhibitor SUMO-2 aldehyde (Enzo Life Sciences), and gel electrophoresis was carried out under non-reducing conditions.
Chromatin immunoprecipitation (ChIP)
Protein-DNA crosslinking was achieved by incubating cells (2.5 × 106 cells in 10-cm plates) in 1% paraformaldehyde for 15 minutes at room temperature; crosslinking reactions were quenched with 100 mM glycine for 5 minutes. Cells were resuspended in RIPA lysis buffer, and genomic DNA shearing was performed by sonication (5 watts for 30 seconds, on ice for 30 seconds, 28 cycles) to yield DNA fragments ranging from 200–1000 bp in length. Antibodies against EZH2 and H3K27me3 were conjugated to Protein A/G Sepharose beads via overnight incubation at 4°C with continuous rotation. 25 μg of DNA and 0.2 μg antibody/μg DNA were used for each immunoprecipitation. Beads were washed three times with wash buffer (20 mM Tris-HCl pH 8.0, 150 mM NaCl, 2 mM EDTA, 1% Triton X-100, 0.1% SDS) prior to elution (100 mM NaHCO3, 1% SDS). Elution was performed at 65°C for 1 hour with periodic agitation. Crosslinks were reversed via overnight incubation at 65°C with RNase A (20 μg/mL; QIAGEN) and Proteinase K (2 mg/mL; Roche). Measurement of DNA binding was accomplished by qRT-PCR, and locus-specific occupancy (as a percentage of input DNA) was calculated in line with our previous studies (36, 119).
Notch and NF-κB reporter assays
Endogenous signaling activity was quantified using Notch- and NF-κB-responsive reporter constructs (121, 122). Briefly, cells were plated (50,000 cells/well in 12-well plate) and subsequently transfected with pcDNA3.1(+) containing Renilla luciferase, plus a plasmid expressing firefly luciferase (Fluc) downstream of the HES1 or HES5 (Notch) or MHC class I (NF-κB) promoters. 72 hours after transfection, cells were lysed using 1X Passive Lysis Buffer (Promega) at room temperature for 15 minutes, and lysates were transferred to a CELLSTAR white-walled 96-well plate (Greiner Bio-One). Fluc and Renilla activity were determined using the Dual-Glo Luciferase Assay System (Promega) according to the manufacturer’s protocol, and luminescence was detected with the GloMax-Multi detection system. For each condition, Fluc (reporter) signal intensity was normalized to Renilla (background) signal to control for differences in transfection efficiency.
NF-κB biotinylated oligonucleotide capture assay
NF-κB binding activity was measured by incubating 200 μg of protein extracted from U2OS cells expressing wild-type and SUMO-deficient RAP1 with 1 μM biotinylated double-stranded DNA oligonucleotide containing a repeat NF-κB consensus sequence. NF-κB binding was stimulated by pre-treatment with TNFα. Samples were incubated overnight at 4°C with continuous rotation, and the resulting NF-κB-oligonucleotide complexes were captured using streptavidin-agarose beads (ThermoFisher Scientific) and analyzed by immunoblotting against the NF-κB p65 subunit (Santa Cruz Biotechnology) according to our previous studies (123).
Immunofluorescence/fluorescence in situ hybridization (IF/FISH)
Cells were plated (30,000 cells/well in 12-well plate) directly on coverslips for 24 hours. Staining and hybridization were carried out as described (124). Briefly, cells were fixed with 4% paraformaldehyde for 15 minutes and permeabilized with 0.5% Triton X-100 for 10 minutes. Following blocking [1X maleic acid buffer (100 mM maleic acid, 150 mM NaCl, pH 7.5), 1X Blocking Reagent (Roche)], coverslips were incubated with anti-PML antibody (Santa Cruz Biotechnology) in 1% bovine serum albumin (BSA)/0.25% Triton X-100 overnight in a cold humidified chamber. Fluorescence detection was accomplished using a secondary antibody conjugated to Alexa Fluor 488 (Invitrogen). Cells were then re-fixed with 4% PFA and dehydrated in 70%, 90%, and 100% ethanol (2 minutes each) prior to hybridization with a telomere leading strand PNA probe [5′-(CCCTAA)3-3′] conjugated to cyanine-5 (Cy5-TelC; PNA Bio) overnight at room temperature. Cells were washed twice with PNA wash buffer A (70% formamide, 10 mM Tris-HCl pH 7.5, 1% BSA), counterstained with DAPI (1 μg/mL in PBS), and mounted with Fluoromount-G (Invitrogen) and sealed with fingernail polish. Images were captured using a Leica TCS SP8 STED confocal microscope (Light Microscopy Imaging Core, CWRU) and analyzed using Leica Application Suite X (LAS X).
Quantitative fluorescence in situ hybridization (Q-FISH)
Q-FISH was carried out according to established protocols (125). Briefly, U2OS cells passaged to 60 population doublings were treated with KaryoMAX Colcemid (100 ng/mL; Gibco) for 90 min prior to trypsinization. Nuclear lysis was accomplished via hypotonic swelling in 75 mM KCl at 37°C. DNA was fixed using methanol:acetic acid (3:1) overnight at 4°C. Fixed chromosomes were dropped onto ColorFrost Plus Microscope Slides (Fisher) and allowed to cure at room temperature overnight. The next day, the slides were re-fixed in 3.6% formaldehyde prior to incubation with RNase A (250 μg/mL; QIAGEN) and pepsin (1 mg/mL; Sigma-Aldrich) at 37°C. Slides were then dehydrated through a graded ethanol series, air-dried, and hybridized to a PNA probe mix (5′-Cy5-(CCCTAA)3-3′ for telomeres and 5′-FAM-ATTCGTTGGAAACGGGA-3′ for centromeres; PNA Bio). Denaturation was carried out at 70°C for 10 min, followed by overnight hybridization at room temperature in the dark. Subsequently, slides were washed twice with PNA wash buffer A (70% formamide, 10 mM Tris-HCl pH 7.5) and three times with PNA wash buffer B (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.8% Tween 20), counterstained with DAPI (50 ng/mL), and mounted using ProLong Gold Antifade Mountant (ThermoFisher). Images were captured using a Leica TCS SP8 STED confocal microscope. Single-telomere fluorescence intensity was detected using LAS X and normalized to contiguous centromere intensity.
C-circle amplification assay
Amplification and quantitation of telomeric extrachromosomal circles (C-circles) was performed as previously described (36, 126). Briefly, cells were isolated from 3D-culture, and total cellular DNA was extracted by incubating cells in Quick C-Circle Preparation (QCP) lysis buffer (10 mM Tris-HCl, 50 mM KCl, 2 mM MgCl2, 0.5% NP-40, 0.5% Tween-20, pH 8.5) containing QIAGEN Protease (0.05 U/mL) for 1 hour with continuous agitation. Where indicated, cells were first treated with DAPT (25 μM) for 96 h (~4 C-circle half-lives). DNA quantitation was performed with the QuantiFluor ONE dsDNA System (Promega), using Lambda DNA for standard curve generation. Circular DNAs were amplified via overnight rolling circle amplification using Φ29 DNA polymerase (New England Biolabs) according to the manufacturer’s protocol. Φ29-deficient reactions were performed for each sample. C-circle quantitation was accomplished using the standard curve method, using Saos-2 DNA to generate the standard curve. qRT-PCR was carried out according to the following thermal profile: 95°C for 15 min, 35 cycles of 95°C for 15 s, 54°C for 2 min, 72°C for 1 min. The ribosomal protein 36B4 was employed as a single-copy gene for normalization of linear chromosomal content.
Telomeric repeat amplification protocol (TRAP)
Real-time Q-TRAP assays were carried out as described previously (124). Briefly, cells were resuspended (at a density of 1,000 cells/μL) in ice-cold NP-40 lysis buffer (10 mM Tris-HCl pH 8.0, 1 mM MgCl2, 1 mM EDTA, 1% NP-40, 0.25 mM sodium deoxycholate, 10% glycerol, 150 mM NaCl, 5 mM β-mercaptoethanol, 0.1 mM AEBSF [4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride]). Lysates were incubated on ice for 30 min and centrifuged at 16,000g for 20 min at 4°C. Total protein was quantified using the Pierce BCA Protein Assay Kit (ThermoFisher). Each TRAP reaction contained 1 μg of protein lysate in 1× iQ SYBR Green Supermix supplemented with 1 mM EGTA, 100 ng of ACX primer (5′-GCGCGGCTTACCCTTACCCTTACCCTAACC-3′), and 100 ng of TS primer (5′-AATCCGTCGAGCAGAGTT-3′), which serves as a synthetic telomerase substrate. Reactions were incubated at 30°C for 30 min to allow for telomerase-mediated extension. The reaction mixture was then heated to 95°C for 10 min, and telomere DNA was amplified according to the following thermal profile: 95°C for 15 s, 60°C for 60 s, 40 cycles. Telomerase activity was quantified by generating a standard curve of log10[Protein] versus Ct value for serial dilutions of protein isolated from HEK293T cells and using the resulting best-fit equation to back-calculate relative telomerase activity (RTA) as described (127).
Protein expression and purification
Purified PIAS1 was purchased from Enzo Life Sciences. For SLX4IP, a bacterial expression construct was generated using Gibson assembly and included a glutathione-S-transferase (GST) tag fused at the N-terminus, followed by a TEV cleavage site. SLX4IP protein was overexpressed in BL21(DE3) E. coli and purified as described previously (128). Briefly, protein expression was induced by the addition of 0.1 mM IPTG at OD600 = 0.6, and cells were grown for an additional 18 hours at 18°C. Cells were lysed using French press and collected at 18,000g for 1 h. The soluble fraction was incubated with glutathione affinity beads pre-equilibrated with 50 mM Tris-HCl pH 8.0, 250 mM NaCl, 3 mM β-mercaptoethanol at 4°C for 1 h, followed by extensive washing. The beads were then incubated with TEV and dialyzed overnight at 4°C in 50 mM Tris-HCl pH 8.0, 50 mM NaCl, 1 mM DTT. The beads were pooled into a filter column, and the elution fraction was applied to a HiTrap HQ 5-mL column (GE Healthcare) pre-equilibrated with 50 mM Tris-HCl pH 8.0, 1 mM DTT.
In vitro SUMO ligase assay
SUMO ligation was carried out using the Abcam SUMOylation Assay Kit. The SUMO E1 (SAE1/SAE2) and E2 (UBC9) proteins and SUMO2/3 were mixed in a 1:1 molar ratio and incubated at 37°C for 1 h to facilitate E2~SUMO conjugation. RAP1 was isolated by immunoprecipitation from U2OS cells, eluted in ion exchange buffer (50 mM Tris-HCl pH 7.5, 250 mM NaCl), and titrated in to obtain kinetic parameters for RAP1 SUMOylation. Single-turnover ligation reactions (129) were catalyzed by the addition of recombinant PIAS1 (Enzo Life Sciences) and were carried out at 37°C for the indicated times. Proteins were separated by polyacrylamide gel electrophoresis and subjected to anti-SUMO2/3 immunoblotting. The addition of the small molecule SUMOylation Inhibitor III (2-(2,3,4-Trihydroxyphenyl)-4H-chromen-4-one; Calbiochem) served as a negative control. Reaction rates at various RAP1 concentrations were estimated by linear regression, and Kcat and KM values were calculated by plotting reaction rate against RAP1 concentration and performing hyperbolic fitting to the function (assuming Michaelis-Menten kinetics).
In vitro kinase assays
To measure ATP consumption, IKKβ complexes were captured from U2OS derivatives by immunoprecipitation, washed twice with PBS, and resuspended in 250 μL kinase assay buffer (40 mM Tris-HCl pH 7.5, 20 mM MgCl2 0.1 mg/mL BSA). Protein concentrations were determined spectroscopically using a NanoDrop. Purified IKKβ complexes were divided and incubated with either IKK-2 Inhibitor VI or diluent for 30 min at room temperature. Kinase reactions were set up using two-fold serial dilutions of IKKβ suspensions beginning at a concentration of 500 nM. Reactions were initiated by addition of synthetic IκB peptide substrate (200 nM; Enzo Life Sciences) and ATP (5 μM; Invitrogen) and were allowed to proceed for 1 h at room temperature with continuous rotation. Reactions were then mixed with an equal volume of Kinase Glo-Max reagent (Promega) and incubated at room temperature for 10 min. Luminescence was quantified over a 1-s integration window using the GloMax-Multi detection system. IKK activity was determined by subtracting the observed luminescence in inhibitor-treated samples from their diluent-containing counterparts. EC50 values were calculated using GraphPad Prism by plotting log10[IKKβ] versus normalized luminescence and performing nonlinear regression. For Western blotting (130), the amount of protein added to each reaction (vs) was calculated such that vs ⋅[IKK]s = vf ⋅ EC50(parental), where [IKK]s is the protein concentration in each sample and vf is the final reaction volume. Reaction mixtures were incubated with synthetic IκB substrate for 1 h at 37°C. Reactions were terminated by addition of 4× sample buffer and subsequently prepared for immunoblotting against anti-phospho-S32-IκB (Cell Signaling Technology).
Three-dimensional (3D) organotypic culture and outgrowth assay
Longitudinal 3D-outgrowth quantification was carried out by seeding 2,000 cells/well on top of solidified cushions of reconstituted basement extract (Cultrex; 50 μL/well in 96-well white-walled plates, Trevigen). Cells were cultured in appropriate media supplemented with 5% Cultrex, as well as the nucleoside analog 5-fluoro-2′-deoxyuridine (5-FdU; Sigma-Aldrich), IKK inhibitor 2-[(aminocarbonyl)amino]-5-phenyl-3-thiophenecarboxamide (IKK-2 Inhibitor VI; Calbiochem), γ-secretase inhibitor N-[N-(3,5-Difluorophenacetyl)-L-alanyl]-S-phenylglycine t-butyl ester (DAPT; Sigma-Aldrich), or Jagged-1 neutralizing antibody (MAB12771; R&D Systems) where indicated. Bioluminescent readings were obtained every 3 days by addition of D-luciferin potassium salt (Gold Biotechnology), followed by quantitation using the GloMax-Multi detection system (Promega). Longitudinal cell growth was normalized to an initial reading taken 24 hours post-plating, and media was replaced following each luminescence reading.
Proliferation assay
U2OS cells were plated (20,000 cells/well in white-walled 96-well plates) and treated with IKK-2 Inhibitor VI or DAPT for 72 h according to the following treatment schemes: IKK-2 Inhibitor VI: two-fold serial dilutions from 20 μM to 156 nM; DAPT: two-fold serial dilutions from 100 μM to 780 nM. An untreated control was also performed with each cell line. Cells were allowed to recover for 24 h in drug-free media prior to quantitation of proliferation using the CellTiter-Glo Cell Viability Assay (Promega) according to the manufacturer’s instructions.
Apoptosis assay
U2OS cells were plated (20,000 cells/well in white-walled 96-well plates) and treated with IKK-2 Inhibitor VI or DAPT for 72 h as described above. Cells were allowed to recover for 24 h in drug-free media prior to quantitation of caspase-3/7 activity using the Caspase-Glo 3/7 Assay (Promega) according to the manufacturer’s instructions. Data are reported for each cell line as fold change in luminescence intensity relative to untreated cells.
Gene set enrichment analysis (GSEA)
GSEA was performed using microarray data deposited in the Gene Expression Omnibus (GEO) under the accession number GSE125702. Analysis was carried out by querying significantly differentially expressed genes against the Molecular Signatures Database (MSigDB) collection C2 using GSEA software obtained from the Broad Institute. p-values were calculated as described (131). Individual microarray reads for each gene were extracted using the Affymetrix Transcriptome Analysis Console, and heatmaps were generated in R using the pheatmap package.
Gene expression analysis
mRNA expression values were curated from The Cancer Genome Atlas (TCGA) PanCancer Atlas using cBioPortal (https://www.cbioportal.org/) (132). The Notch-responsive gene signature was defined by those members of the Reactome Signaling by Notch gene set that were enriched in parental D2.OR cells. Expression values for these genes were compiled and subjected to single-sample GSEA (ssGSEA) using GenePattern (https://www.genepattern.org/) (133). ssGSEA was carried out using default parameters to obtain a Notch enrichment score for each sample, which was then plotted against matched SLX4IP expression. Samples were stratified by SLX4IP expression, using the median RPKM value from RNA sequencing across all samples as a cutoff. Outliers were removed using the ROUT method with FDR Q=1%.
In silico drug sensitivity analysis
Microarray and IC50 values were obtained from the Genomics of Drug Sensitivity in Cancer (GDSC) database (https://www.cancerrxgene.org/) (134). Extraction and correlation of gene expression and drug sensitivity were carried out using RStudio. Source code to perform this analysis is freely available through GitHub at https://github.com/jessicascarborough/gdsc-gene-drug-corr.
Statistical analysis
Unless otherwise noted, data are presented as mean ± SEM. Where mean was used as a measure of central tendency, statistical significance was determined using a Welch’s t test for two-group comparisons or one-way analysis of variance (ANOVA) for multiple-group comparisons. Telomere length distributions obtained by quantitative FISH were compared using a two-sample Kolmogorov-Smirnov test. For ATP consumption, empirical distributions were compared using Akaike’s information criterion to distributions generated by random sampling of experimental data. Michaelis-Menten parameters were compared using a nested model F test. Gene ontology p-values were calculated using Fisher’s exact test. For correlation analysis, significance was determined using Student’s t test by transforming Spearman’s ρ to a t-distributed test statistic as follows: . ssGSEA enrichment scores were compared using a Mann-Whitney U test. In all cases, two-sided p<0.05 was considered statistically significant, with post hoc multiple comparison corrections carried out using the Bonferroni-Dunn method (multiple t tests) or Dunnett’s test (ANOVA pairwise comparisons).
Supplementary Material
Acknowledgments:
We thank all members of the Schiemann and Taylor laboratories for their critical input throughout this project, including preparation of the manuscript. We additionally thank Belinda Willard and Ling Li for the acquisition of mass spectrometry data. We are grateful to Jean-Hugues Guervilly and Pierre-Henri L. Gaillard (Centre de Recherche en Cancérologie de Marseille, Aix Marseille Université) for their provision of SIM-mutant SLX4 expression constructs and their in-depth appraisal of the manuscript and to Fred Miller (Wayne State University) for supplying D2.OR cells. Lastly, we acknowledge Brendan Barton (Department of Pathology, CWRU) for supplying pheatmap source code and Ryan Gimple (Department of Medicine, University of California San Diego) for guidance on ssGSEA analysis.
Funding:
Research support was provided in part by the National Institutes of Health to W.P.S. (CA236273), D.J.T. (GM133841 and CA240993), and N.J.R. (T32 GM007250 and F30 CA213892). Additional support was graciously provided by the Case Comprehensive Cancer Center’s Research Innovation Fund, which is supported by the Case Council and Friends of the Case Comprehensive Cancer Center (to W.P.S.). The mass spectrometer used was purchased with an NIH Shared Instrumentation Grant (S10 RR031537). Microscopy experiments were performed in conjunction with the Case Light Microscopy Imaging Core, which is supported by an NIH Shared Instrumentation Grant (S10 OD016164).
Footnotes
SUPPLEMENTARY MATERIALS
Figures S1 to S7
Tables S1 and S2
Data File S1
Competing interests: D.J.T. reports personal and service fees provided by Rappta Therapeutics (with whom he has ownership interest in, provides consultation for, and has license agreements) but which are outside the submitted work. All other authors declare that they have no competing interests.
Data and materials availability:
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (135) under the accession number PXD024141. Source code to perform gene expression–drug sensitivity analysis is freely available through GitHub at https://github.com/jessicascarborough/gdsc-gene-drug-corr. All other data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials, and all cell lines and DNA constructs generated in this study are freely available upon request.
REFERENCES and Notes
- 1.Gerbi SA, Beginning at the end: DNA replication within the telomere. J Cell Biol 210, 177–179 (2015); published online EpubJul 20 ( 10.1083/jcb.201506078). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Lange T, How telomeres solve the end-protection problem. Science 326, 948–952 (2009); published online EpubNov 13 ( 10.1126/science.1170633). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Lange T, Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev 19, 2100–2110 (2005); published online EpubSep 15 ( 10.1101/gad.1346005). [DOI] [PubMed] [Google Scholar]
- 4.Stansel RM, de Lange T, Griffith JD, T-loop assembly in vitro involves binding of TRF2 near the 3’ telomeric overhang. EMBO J 20, 5532–5540 (2001); published online EpubOct 1 ( 10.1093/emboj/20.19.5532). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hockemeyer D, Sfeir AJ, Shay JW, Wright WE, de Lange T, POT1 protects telomeres from a transient DNA damage response and determines how human chromosomes end. EMBO J 24, 2667–2678 (2005); published online EpubJul 20 ( 10.1038/sj.emboj.7600733). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ye JZ, Donigian JR, van Overbeek M, Loayza D, Luo Y, Krutchinsky AN, Chait BT, de Lange T, TIN2 binds TRF1 and TRF2 simultaneously and stabilizes the TRF2 complex on telomeres. J Biol Chem 279, 47264–47271 (2004); published online EpubNov 5 ( 10.1074/jbc.M409047200). [DOI] [PubMed] [Google Scholar]
- 7.Li B, Oestreich S, de Lange T, Identification of human Rap1: implications for telomere evolution. Cell 101, 471–483 (2000); published online EpubMay 26 ( 10.1016/s0092-8674(00)80858-2). [DOI] [PubMed] [Google Scholar]
- 8.Rice C, Shastrula PK, Kossenkov AV, Hills R, Baird DM, Showe LC, Doukov T, Janicki S, Skordalakes E, Structural and functional analysis of the human POT1-TPP1 telomeric complex. Nat Commun 8, 14928 (2017); published online EpubApr 10 ( 10.1038/ncomms14928). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pike AM, Strong MA, Ouyang JPT, Greider CW, TIN2 Functions with TPP1/POT1 To Stimulate Telomerase Processivity. Mol Cell Biol 39, (2019); published online EpubNov 1 ( 10.1128/MCB.00593-18). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu M, Kiselar J, Whited TL, Hernandez-Sanchez W, Taylor DJ, POT1-TPP1 differentially regulates telomerase via POT1 His266 and as a function of single-stranded telomere DNA length. Proc Natl Acad Sci U S A 116, 23527–23533 (2019); published online EpubNov 19 ( 10.1073/pnas.1905381116). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rai R, Chen Y, Lei M, Chang S, TRF2-RAP1 is required to protect telomeres from engaging in homologous recombination-mediated deletions and fusions. Nat Commun 7, 10881 (2016); published online EpubMar 4 ( 10.1038/ncomms10881). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li B, de Lange T, Rap1 affects the length and heterogeneity of human telomeres. Mol Biol Cell 14, 5060–5068 (2003); published online EpubDec ( 10.1091/mbc.e03-06-0403). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmidt JC, Cech TR, Human telomerase: biogenesis, trafficking, recruitment, and activation. Genes Dev 29, 1095–1105 (2015); published online EpubJun 1 ( 10.1101/gad.263863.115). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dilley RL, Verma P, Cho NW, Winters HD, Wondisford AR, Greenberg RA, Break-induced telomere synthesis underlies alternative telomere maintenance. Nature 539, 54–58 (2016); published online EpubNov 3 ( 10.1038/nature20099). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang JM, Yadav T, Ouyang J, Lan L, Zou L, Alternative Lengthening of Telomeres through Two Distinct Break-Induced Replication Pathways. Cell Rep 26, 955–968 e953 (2019); published online EpubJan 22 ( 10.1016/j.celrep.2018.12.102). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sobinoff AP, Pickett HA, Alternative Lengthening of Telomeres: DNA Repair Pathways Converge. Trends Genet 33, 921–932 (2017); published online EpubDec ( 10.1016/j.tig.2017.09.003). [DOI] [PubMed] [Google Scholar]
- 17.Sobinoff AP, Allen JA, Neumann AA, Yang SF, Walsh ME, Henson JD, Reddel RR, Pickett HA, BLM and SLX4 play opposing roles in recombination-dependent replication at human telomeres. EMBO J 36, 2907–2919 (2017); published online EpubOct 2 ( 10.15252/embj.201796889). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Panier S, Maric M, Hewitt G, Mason-Osann E, Gali H, Dai A, Labadorf A, Guervilly JH, Ruis P, Segura-Bayona S, Belan O, Marzec P, Gaillard PHL, Flynn RL, Boulton SJ, SLX4IP Antagonizes Promiscuous BLM Activity during ALT Maintenance. Mol Cell, (2019); published online EpubAug 7 ( 10.1016/j.molcel.2019.07.010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Verma P, Dilley RL, Zhang T, Gyparaki MT, Li Y, Greenberg RA, RAD52 and SLX4 act nonepistatically to ensure telomere stability during alternative telomere lengthening. Genes Dev 33, 221–235 (2019); published online EpubFeb 1 ( 10.1101/gad.319723.118). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wan B, Yin J, Horvath K, Sarkar J, Chen Y, Wu J, Wan K, Lu J, Gu P, Yu EY, Lue NF, Chang S, Liu Y, Lei M, SLX4 assembles a telomere maintenance toolkit by bridging multiple endonucleases with telomeres. Cell Rep 4, 861–869 (2013); published online EpubSep 12 ( 10.1016/j.celrep.2013.08.017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilson JS, Tejera AM, Castor D, Toth R, Blasco MA, Rouse J, Localization-dependent and -independent roles of SLX4 in regulating telomeres. Cell Rep 4, 853–860 (2013); published online EpubSep 12 ( 10.1016/j.celrep.2013.07.033). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanahan D, Weinberg RA, Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011); published online EpubMar 4 ( 10.1016/j.cell.2011.02.013). [DOI] [PubMed] [Google Scholar]
- 23.Robinson NJ, Schiemann WP, Means to the ends: The role of telomeres and telomere processing machinery in metastasis. Biochim Biophys Acta 1866, 320–329 (2016); published online EpubDec ( 10.1016/j.bbcan.2016.10.005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinson NJ, Taylor DJ, Schiemann WP, Stem cells, immortality, and the evolution of metastatic properties in breast cancer: telomere maintenance mechanisms and metastatic evolution. J Cancer Metastasis Treat 5, (2019) 10.20517/2394-4722.2019.15). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takai KK, Hooper S, Blackwood S, Gandhi R, de Lange T, In vivo stoichiometry of shelterin components. J Biol Chem 285, 1457–1467 (2010); published online EpubJan 8 ( 10.1074/jbc.M109.038026). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaminker P, Plachot C, Kim SH, Chung P, Crippen D, Petersen OW, Bissell MJ, Campisi J, Lelievre SA, Higher-order nuclear organization in growth arrest of human mammary epithelial cells: a novel role for telomere-associated protein TIN2. J Cell Sci 118, 1321–1330 (2005); published online EpubMar 15 ( 10.1242/jcs.01709). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mendez-Bermudez A, Lototska L, Bauwens S, Giraud-Panis MJ, Croce O, Jamet K, Irizar A, Mowinckel M, Koundrioukoff S, Nottet N, Almouzni G, Teulade-Fichou MP, Schertzer M, Perderiset M, Londono-Vallejo A, Debatisse M, Gilson E, Ye J, Genome-wide Control of Heterochromatin Replication by the Telomere Capping Protein TRF2. Mol Cell 70, 449–461 e445 (2018); published online EpubMay 3 ( 10.1016/j.molcel.2018.03.036). [DOI] [PubMed] [Google Scholar]
- 28.Bradshaw PS, Stavropoulos DJ, Meyn MS, Human telomeric protein TRF2 associates with genomic double-strand breaks as an early response to DNA damage. Nat Genet 37, 193–197 (2005); published online EpubFeb ( 10.1038/ng1506). [DOI] [PubMed] [Google Scholar]
- 29.Ghosh A, Saginc G, Leow SC, Khattar E, Shin EM, Yan TD, Wong M, Zhang Z, Li G, Sung WK, Zhou J, Chng WJ, Li S, Liu E, Tergaonkar V, Telomerase directly regulates NF-kappaB-dependent transcription. Nat Cell Biol 14, 1270–1281 (2012); published online EpubDec ( 10.1038/ncb2621). [DOI] [PubMed] [Google Scholar]
- 30.Park JI, Venteicher AS, Hong JY, Choi J, Jun S, Shkreli M, Chang W, Meng Z, Cheung P, Ji H, McLaughlin M, Veenstra TD, Nusse R, McCrea PD, Artandi SE, Telomerase modulates Wnt signalling by association with target gene chromatin. Nature 460, 66–72 (2009); published online EpubJul 2 ( 10.1038/nature08137). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu N, Ding D, Hao W, Yang F, Wu X, Wang M, Xu X, Ju Z, Liu JP, Song Z, Shay JW, Guo Y, Cong YS, hTERT promotes tumor angiogenesis by activating VEGF via interactions with the Sp1 transcription factor. Nucleic Acids Res 44, 8693–8703 (2016); published online EpubOct 14 ( 10.1093/nar/gkw549). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martinez P, Thanasoula M, Carlos AR, Gomez-Lopez G, Tejera AM, Schoeftner S, Dominguez O, Pisano DG, Tarsounas M, Blasco MA, Mammalian Rap1 controls telomere function and gene expression through binding to telomeric and extratelomeric sites. Nat Cell Biol 12, 768–780 (2010); published online EpubAug ( 10.1038/ncb2081). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martinez P, Gomez-Lopez G, Garcia F, Mercken E, Mitchell S, Flores JM, de Cabo R, Blasco MA, RAP1 protects from obesity through its extratelomeric role regulating gene expression. Cell Rep 3, 2059–2074 (2013); published online EpubJun 27 ( 10.1016/j.celrep.2013.05.030). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Teo H, Ghosh S, Luesch H, Ghosh A, Wong ET, Malik N, Orth A, de Jesus P, Perry AS, Oliver JD, Tran NL, Speiser LJ, Wong M, Saez E, Schultz P, Chanda SK, Verma IM, Tergaonkar V, Telomere-independent Rap1 is an IKK adaptor and regulates NF-kappaB-dependent gene expression. Nat Cell Biol 12, 758–767 (2010); published online EpubAug ( 10.1038/ncb2080). [DOI] [PubMed] [Google Scholar]
- 35.Li CX, Lo CM, Lian Q, Ng KT, Liu XB, Ma YY, Qi X, Yeung OW, Tergaonkar V, Yang XX, Liu H, Liu J, Shao Y, Man K, Repressor and activator protein accelerates hepatic ischemia reperfusion injury by promoting neutrophil inflammatory response. Oncotarget 7, 27711–27723 (2016); published online EpubMay 10 ( 10.18632/oncotarget.8509). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robinson NJ, Morrison-Smith CD, Gooding AJ, Schiemann BJ, Jackson MW, Taylor DJ, Schiemann WP, SLX4IP and telomere dynamics dictate breast cancer metastasis and therapeutic responsiveness. Life Sci Alliance 3, (2020); published online EpubApr ( 10.26508/lsa.201900427). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mangosh TL, Awadallah WN, Grabowska MM, Taylor DJ, SLX4IP promotes telomere maintenance in androgen receptor-independent castration-resistant prostate cancer through ALT-like telomeric PML localization. Mol Cancer Res, (2020); published online EpubNov 13 ( 10.1158/1541-7786.MCR-20-0314). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dejardin J, Kingston RE, Purification of proteins associated with specific genomic Loci. Cell 136, 175–186 (2009); published online EpubJan 9 ( 10.1016/j.cell.2008.11.045). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sarthy J, Bae NS, Scrafford J, Baumann P, Human RAP1 inhibits non-homologous end joining at telomeres. EMBO J 28, 3390–3399 (2009); published online EpubNov 4 ( 10.1038/emboj.2009.275). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O’Connor MS, Safari A, Liu D, Qin J, Songyang Z, The human Rap1 protein complex and modulation of telomere length. J Biol Chem 279, 28585–28591 (2004); published online EpubJul 2 ( 10.1074/jbc.M312913200). [DOI] [PubMed] [Google Scholar]
- 41.Guervilly JH, Gaillard PH, SLX4: multitasking to maintain genome stability. Crit Rev Biochem Mol Biol 53, 475–514 (2018); published online EpubOct ( 10.1080/10409238.2018.1488803). [DOI] [PubMed] [Google Scholar]
- 42.Svendsen JM, Smogorzewska A, Sowa ME, O’Connell BC, Gygi SP, Elledge SJ, Harper JW, Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell 138, 63–77 (2009); published online EpubJul 10 ( 10.1016/j.cell.2009.06.030). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guervilly JH, Takedachi A, Naim V, Scaglione S, Chawhan C, Lovera Y, Despras E, Kuraoka I, Kannouche P, Rosselli F, Gaillard PHL, The SLX4 complex is a SUMO E3 ligase that impacts on replication stress outcome and genome stability. Mol Cell 57, 123–137 (2015); published online EpubJan 8 ( 10.1016/j.molcel.2014.11.014). [DOI] [PubMed] [Google Scholar]
- 44.Ouyang J, Garner E, Hallet A, Nguyen HD, Rickman KA, Gill G, Smogorzewska A, Zou L, Noncovalent interactions with SUMO and ubiquitin orchestrate distinct functions of the SLX4 complex in genome maintenance. Mol Cell 57, 108–122 (2015); published online EpubJan 8 ( 10.1016/j.molcel.2014.11.015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gonzalez-Prieto R, Cuijpers SA, Luijsterburg MS, van Attikum H, Vertegaal AC, SUMOylation and PARylation cooperate to recruit and stabilize SLX4 at DNA damage sites. EMBO Rep 16, 512–519 (2015); published online EpubApr ( 10.15252/embr.201440017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Potts PR, Yu H, The SMC5/6 complex maintains telomere length in ALT cancer cells through SUMOylation of telomere-binding proteins. Nat Struct Mol Biol 14, 581–590 (2007); published online EpubJul ( 10.1038/nsmb1259). [DOI] [PubMed] [Google Scholar]
- 47.Jiang WQ, Zhong ZH, Henson JD, Reddel RR, Identification of candidate alternative lengthening of telomeres genes by methionine restriction and RNA interference. Oncogene 26, 4635–4647 (2007); published online EpubJul 12 ( 10.1038/sj.onc.1210260). [DOI] [PubMed] [Google Scholar]
- 48.Her J, Jeong YY, Chung IK, PIAS1-mediated sumoylation promotes STUbL-dependent proteasomal degradation of the human telomeric protein TRF2. FEBS Lett 589, 3277–3286 (2015); published online EpubOct 24 ( 10.1016/j.febslet.2015.09.030). [DOI] [PubMed] [Google Scholar]
- 49.Kahyo T, Nishida T, Yasuda H, Involvement of PIAS1 in the sumoylation of tumor suppressor p53. Mol Cell 8, 713–718 (2001); published online EpubSep ( 10.1016/s1097-2765(01)00349-5). [DOI] [PubMed] [Google Scholar]
- 50.Beauclair G, Bridier-Nahmias A, Zagury JF, Saib A, Zamborlini A, JASSA: a comprehensive tool for prediction of SUMOylation sites and SIMs. Bioinformatics 31, 3483–3491 (2015); published online EpubNov 1 ( 10.1093/bioinformatics/btv403). [DOI] [PubMed] [Google Scholar]
- 51.Zhao Q, Xie Y, Zheng Y, Jiang S, Liu W, Mu W, Liu Z, Zhao Y, Xue Y, Ren J, GPS-SUMO: a tool for the prediction of sumoylation sites and SUMO-interaction motifs. Nucleic Acids Res 42, W325–330 (2014); published online EpubJul ( 10.1093/nar/gku383). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hendriks IA, D’Souza RC, Yang B, Verlaan-de Vries M, Mann M, Vertegaal AC, Uncovering global SUMOylation signaling networks in a site-specific manner. Nat Struct Mol Biol 21, 927–936 (2014); published online EpubOct ( 10.1038/nsmb.2890). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xiao Z, Chang JG, Hendriks IA, Sigurethsson JO, Olsen JV, Vertegaal AC, System-wide Analysis of SUMOylation Dynamics in Response to Replication Stress Reveals Novel Small Ubiquitin-like Modified Target Proteins and Acceptor Lysines Relevant for Genome Stability. Mol Cell Proteomics 14, 1419–1434 (2015); published online EpubMay ( 10.1074/mcp.O114.044792). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hendriks IA, Lyon D, Young C, Jensen LJ, Vertegaal AC, Nielsen ML, Site-specific mapping of the human SUMO proteome reveals co-modification with phosphorylation. Nat Struct Mol Biol 24, 325–336 (2017); published online EpubMar ( 10.1038/nsmb.3366). [DOI] [PubMed] [Google Scholar]
- 55.Lanouette S, Mongeon V, Figeys D, Couture JF, The functional diversity of protein lysine methylation. Mol Syst Biol 10, 724 (2014); published online EpubApr 8 ( 10.1002/msb.134974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boardman LA, Johnson RA, Viker KB, Hafner KA, Jenkins RB, Riegert-Johnson DL, Smyrk TC, Litzelman K, Seo S, Gangnon RE, Engelman CD, Rider DN, Vanderboom RJ, Thibodeau SN, Petersen GM, Skinner HG, Correlation of chromosomal instability, telomere length and telomere maintenance in microsatellite stable rectal cancer: a molecular subclass of rectal cancer. PLoS One 8, e80015 (2013) 10.1371/journal.pone.0080015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee M, Teber ET, Holmes O, Nones K, Patch AM, Dagg RA, Lau LMS, Lee JH, Napier CE, Arthur JW, Grimmond SM, Hayward NK, Johansson PA, Mann GJ, Scolyer RA, Wilmott JS, Reddel RR, Pearson JV, Waddell N, Pickett HA, Telomere sequence content can be used to determine ALT activity in tumours. Nucleic Acids Res 46, 4903–4918 (2018); published online EpubJun 1 ( 10.1093/nar/gky297). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ptak C, Wozniak RW, SUMO and Nucleocytoplasmic Transport. Adv Exp Med Biol 963, 111–126 (2017) 10.1007/978-3-319-50044-7_7). [DOI] [PubMed] [Google Scholar]
- 59.Sui Y, Liu Z, Park SH, Thatcher SE, Zhu B, Fernandez JP, Molina H, Kern PA, Zhou C, IKKbeta is a beta-catenin kinase that regulates mesenchymal stem cell differentiation. JCI Insight 3, (2018); published online EpubJan 25 ( 10.1172/jci.insight.96660). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Comb WC, Hutti JE, Cogswell P, Cantley LC, Baldwin AS, p85alpha SH2 domain phosphorylation by IKK promotes feedback inhibition of PI3K and Akt in response to cellular starvation. Mol Cell 45, 719–730 (2012); published online EpubMar 30 ( 10.1016/j.molcel.2012.01.010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bitzer M, von Gersdorff G, Liang D, Dominguez-Rosales A, Beg AA, Rojkind M, Bottinger EP, A mechanism of suppression of TGF-beta/SMAD signaling by NF-kappa B/RelA. Genes Dev 14, 187–197 (2000) [PMC free article] [PubMed] [Google Scholar]
- 62.Nakashima H, Nakamura M, Yamaguchi H, Yamanaka N, Akiyoshi T, Koga K, Yamaguchi K, Tsuneyoshi M, Tanaka M, Katano M, Nuclear factor-kappaB contributes to hedgehog signaling pathway activation through sonic hedgehog induction in pancreatic cancer. Cancer Res 66, 7041–7049 (2006); published online EpubJul 15 ( 10.1158/0008-5472.CAN-05-4588). [DOI] [PubMed] [Google Scholar]
- 63.Osipo C, Golde TE, Osborne BA, Miele LA, Off the beaten pathway: the complex cross talk between Notch and NF-kappaB. Lab Invest 88, 11–17 (2008); published online EpubJan ( 10.1038/labinvest.3700700). [DOI] [PubMed] [Google Scholar]
- 64.Henson JD, Reddel RR, Assaying and investigating Alternative Lengthening of Telomeres activity in human cells and cancers. FEBS Lett 584, 3800–3811 (2010); published online EpubSep 10 ( 10.1016/j.febslet.2010.06.009). [DOI] [PubMed] [Google Scholar]
- 65.Deeg KI, Chung I, Bauer C, Rippe K, Cancer Cells with Alternative Lengthening of Telomeres Do Not Display a General Hypersensitivity to ATR Inhibition. Front Oncol 6, 186 (2016) 10.3389/fonc.2016.00186). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Santiago A, Li D, Zhao LY, Godsey A, Liao D, p53 SUMOylation promotes its nuclear export by facilitating its release from the nuclear export receptor CRM1. Mol Biol Cell 24, 2739–2752 (2013); published online EpubSep ( 10.1091/mbc.E12-10-0771). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Imoto S, Ohbayashi N, Ikeda O, Kamitani S, Muromoto R, Sekine Y, Matsuda T, Sumoylation of Smad3 stimulates its nuclear export during PIASy-mediated suppression of TGF-beta signaling. Biochem Biophys Res Commun 370, 359–365 (2008); published online EpubMay 30 ( 10.1016/j.bbrc.2008.03.116). [DOI] [PubMed] [Google Scholar]
- 68.Terui Y, Saad N, Jia S, McKeon F, Yuan J, Dual role of sumoylation in the nuclear localization and transcriptional activation of NFAT1. J Biol Chem 279, 28257–28265 (2004); published online EpubJul 2 ( 10.1074/jbc.M403153200). [DOI] [PubMed] [Google Scholar]
- 69.Rosonina E, Akhter A, Dou Y, Babu J, Sri Theivakadadcham VS, Regulation of transcription factors by sumoylation. Transcription 8, 220–231 (2017); published online EpubAug 8 ( 10.1080/21541264.2017.1311829). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Geiss-Friedlander R, Melchior F, Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol 8, 947–956 (2007); published online EpubDec ( 10.1038/nrm2293). [DOI] [PubMed] [Google Scholar]
- 71.Shen TH, Lin HK, Scaglioni PP, Yung TM, Pandolfi PP, The mechanisms of PML-nuclear body formation. Mol Cell 24, 331–339 (2006); published online EpubNov 3 ( 10.1016/j.molcel.2006.09.013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McManus FP, Bourdeau V, Acevedo M, Lopes-Paciencia S, Mignacca L, Lamoliatte F, Rojas Pino JW, Ferbeyre G, Thibault P, Quantitative SUMO proteomics reveals the modulation of several PML nuclear body associated proteins and an anti-senescence function of UBC9. Sci Rep 8, 7754 (2018); published online EpubMay 17 ( 10.1038/s41598-018-25150-z). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brouwer AK, Schimmel J, Wiegant JC, Vertegaal AC, Tanke HJ, Dirks RW, Telomeric DNA mediates de novo PML body formation. Mol Biol Cell 20, 4804–4815 (2009); published online EpubNov ( 10.1091/mbc.E09-04-0309). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Galanty Y, Belotserkovskaya R, Coates J, Polo S, Miller KM, Jackson SP, Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature 462, 935–939 (2009); published online EpubDec 17 ( 10.1038/nature08657). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pan X, Drosopoulos WC, Sethi L, Madireddy A, Schildkraut CL, Zhang D, FANCM, BRCA1, and BLM cooperatively resolve the replication stress at the ALT telomeres. Proc Natl Acad Sci U S A 114, E5940–E5949 (2017); published online EpubJul 18 ( 10.1073/pnas.1708065114). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jezek M, Green EM, Histone Modifications and the Maintenance of Telomere Integrity. Cells 8, (2019); published online EpubFeb 25 ( 10.3390/cells8020199). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jones B, Su H, Bhat A, Lei H, Bajko J, Hevi S, Baltus GA, Kadam S, Zhai H, Valdez R, Gonzalo S, Zhang Y, Li E, Chen T, The histone H3K79 methyltransferase Dot1L is essential for mammalian development and heterochromatin structure. PLoS Genet 4, e1000190 (2008); published online EpubSep 12 ( 10.1371/journal.pgen.1000190). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Montero JJ, Lopez-Silanes I, Megias D, F. F. M, Castells-Garcia A, Blasco MA, TERRA recruitment of polycomb to telomeres is essential for histone trymethylation marks at telomeric heterochromatin. Nat Commun 9, 1548 (2018); published online EpubApr 18 ( 10.1038/s41467-018-03916-3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Caslini C, Connelly JA, Serna A, Broccoli D, Hess JL, MLL associates with telomeres and regulates telomeric repeat-containing RNA transcription. Mol Cell Biol 29, 4519–4526 (2009); published online EpubAug ( 10.1128/MCB.00195-09). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kishi S, Zhou XZ, Ziv Y, Khoo C, Hill DE, Shiloh Y, Lu KP, Telomeric protein Pin2/TRF1 as an important ATM target in response to double strand DNA breaks. J Biol Chem 276, 29282–29291 (2001); published online EpubAug 3 ( 10.1074/jbc.M011534200). [DOI] [PubMed] [Google Scholar]
- 81.Buscemi G, Zannini L, Fontanella E, Lecis D, Lisanti S, Delia D, The shelterin protein TRF2 inhibits Chk2 activity at telomeres in the absence of DNA damage. Curr Biol 19, 874–879 (2009); published online EpubMay 26 ( 10.1016/j.cub.2009.03.064). [DOI] [PubMed] [Google Scholar]
- 82.Lee TH, Tun-Kyi A, Shi R, Lim J, Soohoo C, Finn G, Balastik M, Pastorino L, Wulf G, Zhou XZ, Lu KP, Essential role of Pin1 in the regulation of TRF1 stability and telomere maintenance. Nat Cell Biol 11, 97–105 (2009); published online EpubJan ( 10.1038/ncb1818). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ohishi T, Hirota T, Tsuruo T, Seimiya H, TRF1 mediates mitotic abnormalities induced by Aurora-A overexpression. Cancer Res 70, 2041–2052 (2010); published online EpubMar 1 ( 10.1158/0008-5472.CAN-09-2008). [DOI] [PubMed] [Google Scholar]
- 84.Smith S, de Lange T, Tankyrase promotes telomere elongation in human cells. Curr Biol 10, 1299–1302 (2000); published online EpubOct 19 ( 10.1016/s0960-9822(00)00752-1). [DOI] [PubMed] [Google Scholar]
- 85.Kim JH, Park SM, Kang MR, Oh SY, Lee TH, Muller MT, Chung IK, Ubiquitin ligase MKRN1 modulates telomere length homeostasis through a proteolysis of hTERT. Genes Dev 19, 776–781 (2005); published online EpubApr 1 ( 10.1101/gad.1289405). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fan Q, Zhang F, Barrett B, Ren K, Andreassen PR, A role for monoubiquitinated FANCD2 at telomeres in ALT cells. Nucleic Acids Res 37, 1740–1754 (2009); published online EpubApr ( 10.1093/nar/gkn995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Benetti R, Gonzalo S, Jaco I, Schotta G, Klatt P, Jenuwein T, Blasco MA, Suv4-20h deficiency results in telomere elongation and derepression of telomere recombination. J Cell Biol 178, 925–936 (2007); published online EpubSep 10 ( 10.1083/jcb.200703081). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Marion RM, Strati K, Li H, Tejera A, Schoeftner S, Ortega S, Serrano M, Blasco MA, Telomeres acquire embryonic stem cell characteristics in induced pluripotent stem cells. Cell Stem Cell 4, 141–154 (2009); published online EpubFeb 6 ( 10.1016/j.stem.2008.12.010). [DOI] [PubMed] [Google Scholar]
- 89.Varela E, Schneider RP, Ortega S, Blasco MA, Different telomere-length dynamics at the inner cell mass versus established embryonic stem (ES) cells. Proc Natl Acad Sci U S A 108, 15207–15212 (2011); published online EpubSep 13 ( 10.1073/pnas.1105414108). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu L, Bailey SM, Okuka M, Munoz P, Li C, Zhou L, Wu C, Czerwiec E, Sandler L, Seyfang A, Blasco MA, Keefe DL, Telomere lengthening early in development. Nat Cell Biol 9, 1436–1441 (2007); published online EpubDec ( 10.1038/ncb1664). [DOI] [PubMed] [Google Scholar]
- 91.Yin L, Hubbard AK, Giardina C, NF-kappa B regulates transcription of the mouse telomerase catalytic subunit. J Biol Chem 275, 36671–36675 (2000); published online EpubNov 24 ( 10.1074/jbc.M007378200). [DOI] [PubMed] [Google Scholar]
- 92.Wu XQ, Yang Y, Li WX, Cheng YH, Li XF, Huang C, Meng XM, Wu BM, Liu XH, Zhang L, Lv XW, Li J, Telomerase reverse transcriptase acts in a feedback loop with NF-kappaB pathway to regulate macrophage polarization in alcoholic liver disease. Sci Rep 6, 18685 (2016); published online EpubJan 4 ( 10.1038/srep18685). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zuo QP, Liu SK, Li ZJ, Li B, Zhou YL, Guo R, Huang LH, NF-kappaB p65 modulates the telomerase reverse transcriptase in the HepG(2) hepatoma cell line. Eur J Pharmacol 672, 113–120 (2011); published online EpubDec 15 ( 10.1016/j.ejphar.2011.09.187). [DOI] [PubMed] [Google Scholar]
- 94.Lee PS, Chang C, Liu D, Derynck R, Sumoylation of Smad4, the common Smad mediator of transforming growth factor-beta family signaling. J Biol Chem 278, 27853–27863 (2003); published online EpubJul 25 ( 10.1074/jbc.M301755200). [DOI] [PubMed] [Google Scholar]
- 95.Liang M, Melchior F, Feng XH, Lin X, Regulation of Smad4 sumoylation and transforming growth factor-beta signaling by protein inhibitor of activated STAT1. J Biol Chem 279, 22857–22865 (2004); published online EpubMay 28 ( 10.1074/jbc.M401554200). [DOI] [PubMed] [Google Scholar]
- 96.Choi HK, Choi KC, Yoo JY, Song M, Ko SJ, Kim CH, Ahn JH, Chun KH, Yook JI, Yoon HG, Reversible SUMOylation of TBL1-TBLR1 regulates beta-catenin-mediated Wnt signaling. Mol Cell 43, 203–216 (2011); published online EpubJul 22 ( 10.1016/j.molcel.2011.05.027). [DOI] [PubMed] [Google Scholar]
- 97.Huang HJ, Zhou LL, Fu WJ, Zhang CY, Jiang H, Du J, Hou J, beta-catenin SUMOylation is involved in the dysregulated proliferation of myeloma cells. Am J Cancer Res 5, 309–320 (2015). [PMC free article] [PubMed] [Google Scholar]
- 98.Licciardello MP, Mullner MK, Durnberger G, Kerzendorfer C, Boidol B, Trefzer C, Sdelci S, Berg T, Penz T, Schuster M, Bock C, Kralovics R, Superti-Furga G, Colinge J, Nijman SM, Kubicek S, NOTCH1 activation in breast cancer confers sensitivity to inhibition of SUMOylation. Oncogene 34, 3780–3790 (2015); published online EpubJul ( 10.1038/onc.2014.319). [DOI] [PubMed] [Google Scholar]
- 99.Sehat B, Tofigh A, Lin Y, Trocme E, Liljedahl U, Lagergren J, Larsson O, SUMOylation mediates the nuclear translocation and signaling of the IGF-1 receptor. Sci Signal 3, ra10 (2010); published online EpubFeb 9 ( 10.1126/scisignal.2000628). [DOI] [PubMed] [Google Scholar]
- 100.de la Cruz-Herrera CF, Campagna M, Lang V, del Carmen Gonzalez-Santamaria J, Marcos-Villar L, Rodriguez MS, Vidal A, Collado M, Rivas C, SUMOylation regulates AKT1 activity. Oncogene 34, 1442–1450 (2015); published online EpubMar 12 ( 10.1038/onc.2014.48). [DOI] [PubMed] [Google Scholar]
- 101.Deng R, Zhao X, Qu Y, Chen C, Zhu C, Zhang H, Yuan H, Jin H, Liu X, Wang Y, Chen Q, Huang J, Yu J, Shp2 SUMOylation promotes ERK activation and hepatocellular carcinoma development. Oncotarget 6, 9355–9369 (2015); published online EpubApr 20 ( 10.18632/oncotarget.3323). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sever R, Brugge JS, Signal transduction in cancer. Cold Spring Harb Perspect Med 5, (2015); published online EpubApr 1 ( 10.1101/cshperspect.a006098). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Basson MA, Signaling in cell differentiation and morphogenesis. Cold Spring Harb Perspect Biol 4, (2012); published online EpubJun 1 ( 10.1101/cshperspect.a008151). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Aster JC, Pear WS, Blacklow SC, The Varied Roles of Notch in Cancer. Annu Rev Pathol 12, 245–275 (2017); published online EpubJan 24 ( 10.1146/annurev-pathol-052016-100127). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Xia L, Tan S, Zhou Y, Lin J, Wang H, Oyang L, Tian Y, Liu L, Su M, Wang H, Cao D, Liao Q, Role of the NFkappaB-signaling pathway in cancer. Onco Targets Ther 11, 2063–2073 (2018) 10.2147/OTT.S161109). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Liu J, Zhang X, Yan X, Sun M, Fan Y, Huang Y, Significance of TERT and ATRX mutations in glioma. Oncol Lett 17, 95–102 (2019); published online EpubJan ( 10.3892/ol.2018.9634). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Fallahpour S, Navaneelan T, De P, Borgo A, Breast cancer survival by molecular subtype: a population-based analysis of cancer registry data. CMAJ Open 5, E734–E739 (2017); published online EpubSep 25 ( 10.9778/cmajo.20170030). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kim SH, Ezhilarasan R, Phillips E, Gallego-Perez D, Sparks A, Taylor D, Ladner K, Furuta T, Sabit H, Chhipa R, Cho JH, Mohyeldin A, Beck S, Kurozumi K, Kuroiwa T, Iwata R, Asai A, Kim J, Sulman EP, Cheng SY, Lee LJ, Nakada M, Guttridge D, DasGupta B, Goidts V, Bhat KP, Nakano I, Serine/Threonine Kinase MLK4 Determines Mesenchymal Identity in Glioma Stem Cells in an NF-kappaB-dependent Manner. Cancer Cell 29, 201–213 (2016); published online EpubFeb 8 ( 10.1016/j.ccell.2016.01.005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Godwin P, Baird AM, Heavey S, Barr MP, O’Byrne KJ, Gately K, Targeting nuclear factor-kappa B to overcome resistance to chemotherapy. Front Oncol 3, 120 (2013) 10.3389/fonc.2013.00120). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Labun K, Montague TG, Krause M, Torres Cleuren YN, Tjeldnes H, Valen E, CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res 47, W171–W174 (2019); published online EpubJul 2 ( 10.1093/nar/gkz365). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M, In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc 1, 2856–2860 (2006) 10.1038/nprot.2006.468). [DOI] [PubMed] [Google Scholar]
- 112.Cox J, Mann M, MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26, 1367–1372 (2008); published online EpubDec ( 10.1038/nbt.1511). [DOI] [PubMed] [Google Scholar]
- 113.Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M, Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res 10, 1794–1805 (2011); published online EpubApr 1 ( 10.1021/pr101065j). [DOI] [PubMed] [Google Scholar]
- 114.Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, Mann M, Cox J, The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods 13, 731–740 (2016); published online EpubSep ( 10.1038/nmeth.3901). [DOI] [PubMed] [Google Scholar]
- 115.Sfeir A, Kabir S, van Overbeek M, Celli GB, de Lange T, Loss of Rap1 induces telomere recombination in the absence of NHEJ or a DNA damage signal. Science 327, 1657–1661 (2010); published online EpubMar 26 ( 10.1126/science.1185100). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Xu D, Zhang Y, Ab initio protein structure assembly using continuous structure fragments and optimized knowledge-based force field. Proteins 80, 1715–1735 (2012); published online EpubJul ( 10.1002/prot.24065). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhang Y, Skolnick J, Scoring function for automated assessment of protein structure template quality. Proteins 57, 702–710 (2004); published online EpubDec 1 ( 10.1002/prot.20264). [DOI] [PubMed] [Google Scholar]
- 118.La Belle Flynn A, Calhoun BC, Sharma A, Chang JC, Almasan A, Schiemann WP, Autophagy inhibition elicits emergence from metastatic dormancy by inducing and stabilizing Pfkfb3 expression. Nat Commun 10, 3668 (2019); published online EpubAug 14 ( 10.1038/s41467-019-11640-9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gooding AJ, Zhang B, Jahanbani FK, Gilmore HL, Chang JC, Valadkhan S, Schiemann WP, The lncRNA BORG Drives Breast Cancer Metastasis and Disease Recurrence. Sci Rep 7, 12698 (2017); published online EpubOct 5 ( 10.1038/s41598-017-12716-6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Galliher AJ, Schiemann WP, Beta3 integrin and Src facilitate transforming growth factor-beta mediated induction of epithelial-mesenchymal transition in mammary epithelial cells. Breast Cancer Res 8, R42 (2006) 10.1186/bcr1524). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Nishimura M, Isaka F, Ishibashi M, Tomita K, Tsuda H, Nakanishi S, Kageyama R, Structure, chromosomal locus, and promoter of mouse Hes2 gene, a homologue of Drosophila hairy and Enhancer of split. Genomics 49, 69–75 (1998); published online EpubApr 1 ( 10.1006/geno.1998.5213). [DOI] [PubMed] [Google Scholar]
- 122.Neil JR, Schiemann WP, Altered TAB1:I kappaB kinase interaction promotes transforming growth factor beta-mediated nuclear factor-kappaB activation during breast cancer progression. Cancer Res 68, 1462–1470 (2008); published online EpubMar 1 ( 10.1158/0008-5472.CAN-07-3094). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Neil JR, Tian M, Schiemann WP, X-linked inhibitor of apoptosis protein and its E3 ligase activity promote transforming growth factor-{beta}-mediated nuclear factor-{kappa}B activation during breast cancer progression. J Biol Chem 284, 21209–21217 (2009); published online EpubAug 7 ( 10.1074/jbc.M109.018374). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zeng X, Hernandez-Sanchez W, Xu M, Whited TL, Baus D, Zhang J, Berdis AJ, Taylor DJ, Administration of a Nucleoside Analog Promotes Cancer Cell Death in a Telomerase-Dependent Manner. Cell Rep 23, 3031–3041 (2018); published online EpubJun 5 ( 10.1016/j.celrep.2018.05.020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Cesare AJ, Heaphy CM, O’Sullivan RJ, Visualization of Telomere Integrity and Function In Vitro and In Vivo Using Immunofluorescence Techniques. Curr Protoc Cytom 73, 12 40 11–12 40 31 (2015); published online EpubJul 1 ( 10.1002/0471142956.cy1240s73). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lau LM, Dagg RA, Henson JD, Au AY, Royds JA, Reddel RR, Detection of alternative lengthening of telomeres by telomere quantitative PCR. Nucleic Acids Res 41, e34 (2013); published online EpubJan ( 10.1093/nar/gks781). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wege H, Chui MS, Le HT, Tran JM, Zern MA, SYBR Green real-time telomeric repeat amplification protocol for the rapid quantification of telomerase activity. Nucleic Acids Res 31, E3–3 (2003); published online EpubJan 15 ( 10.1093/nar/gng003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Cho US, Xu W, Crystal structure of a protein phosphatase 2A heterotrimeric holoenzyme. Nature 445, 53–57 (2007); published online EpubJan 4 ( 10.1038/nature05351). [DOI] [PubMed] [Google Scholar]
- 129.Yunus AA, Lima CD, Purification of SUMO conjugating enzymes and kinetic analysis of substrate conjugation. Methods Mol Biol 497, 167–186 (2009) 10.1007/978-1-59745-566-4_11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Morrison CD, Chang JC, Keri RA, Schiemann WP, Mutant p53 dictates the oncogenic activity of c-Abl in triple-negative breast cancers. Cell Death Dis 8, e2899 (2017); published online EpubJun 29 ( 10.1038/cddis.2017.294). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP, Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550 (2005); published online EpubOct 25 ( 10.1073/pnas.0506580102). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N, The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2, 401–404 (2012); published online EpubMay ( 10.1158/2159-8290.CD-12-0095). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP, GenePattern 2.0. Nat Genet 38, 500–501 (2006); published online EpubMay ( 10.1038/ng0506-500). [DOI] [PubMed] [Google Scholar]
- 134.Yang W, Soares J, Greninger P, Edelman EJ, Lightfoot H, Forbes S, Bindal N, Beare D, Smith JA, Thompson IR, Ramaswamy S, Futreal PA, Haber DA, Stratton MR, Benes C, McDermott U, Garnett MJ, Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res 41, D955–961 (2013); published online EpubJan ( 10.1093/nar/gks1111). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, Perez E, Uszkoreit J, Pfeuffer J, Sachsenberg T, Yilmaz S, Tiwary S, Cox J, Audain E, Walzer M, Jarnuczak AF, Ternent T, Brazma A, Vizcaino JA, The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res 47, D442–D450 (2019); published online EpubJan 8 ( 10.1093/nar/gky1106). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (135) under the accession number PXD024141. Source code to perform gene expression–drug sensitivity analysis is freely available through GitHub at https://github.com/jessicascarborough/gdsc-gene-drug-corr. All other data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials, and all cell lines and DNA constructs generated in this study are freely available upon request.


