Abstract
Tumor‐associated autoantibodies (TAAb) could be serological tumor markers. This study aims to discover novel TAAb signatures for breast cancer (BC) detection. The protein microarray was used to identify candidate TAAb, which were further validated in 1197 sera from BC, benign breast diseases (BD), and healthy controls (HC) by enzyme‐linked immunosorbent assay. In addition, 319 preoperative and postoperative sera were evaluated. A panel was determined using four different classifiers. Twelve TAAb were identified with frequencies of 15.8%‐59.2%; their levels were significantly decreased in postoperative sera compared to those in preoperative sera (P < .05). A panel with six TAAb was developed and evaluated. The area under the curve (AUC) was 0.879 (74.3% sensitivity, 91.9% specificity) and 0.865 (69.7% sensitivity, 91.7% specificity) for distinguishing BC from HC in the training set and test set, respectively. The panel had an AUC of .884 (71.2% sensitivity, 90.5% specificity) for discriminating BC from BD. For identifying BC from all controls (HC+BD), the AUC was .916 (78.9% sensitivity, 90.2% specificity). The AUC of the panel was .920 and .934 for distinguishing stage I‐II and age < 50 BC from HC, respectively. These identified TAAb have the potential to provide a non–invasive approach to detect BC.
Keywords: breast cancer, cancer driver genes, protein microarray, tumor‐associated antigens, tumor‐associated autoantibodies
This is the first study using a customized protein microarray consisting of proteins encoded by cancer driver genes to identify novel TAAb as serological biomarkers for BC. In screening 1197 sera samples, we identified 12 TAAb (ALK, BRCA2, CDKN2A, CEBPA, CEP55, FUBP1, GATA3, HIST1H3B, HRAS, PTCH1, p62, and RalA) that are potential biomarkers of BC. The serum levels of 12 TAAb in BC patients had decreased significantly 1 month after the operation. A panel with 6 TAAb (BRCA2, CEBPA, CEP55, FUBP1, HRAS, and RalA) was established. The panel shows superior performance in distinguishing breast cancer from healthy controls or benign breast diseases.

Abbreviations
- AUCs
area under the curves
- BC
breast cancer
- ELISA
enzyme‐linked immunosorbent assay
- GBDT
Gradient Boosting Decision Tree
- HC
healthy controls
- LASSO
the least absolute shrinkage and selection operator
- LR
logistic regression
- RF
random forest
- ROC
receiver operating characteristic
- SNR
signal to noise ratio
- SVM
support vector machine
- TAAbs
tumor‐associated autoantibodies
- TAAs
tumor‐associated antigens
1. INTRODUCTION
Breast cancer (BC) is among the most frequently diagnosed cancers in the world.1 It is also the leading cause of cancer death in women under the age of 45.2 Due to its heterogeneous attributes and complex biological processes, there are no typical symptoms and signs for most of BC patients in the early stages of disease.3 The current approaches for BC diagnosis, such as mammography and biopsy, cause discomfort and can be harmful to women, especially young women.3, 4, 5, 6 Thus, it is important to explore other approaches that could be applied for the early detection of BC. Detection of blood‐based biomarkers is a promising method that is attracting substantial interest from researchers.
Cancer antigen 15‐3 (CA15‐3) has been suggested as a potential serum marker for BC.7 However, it lacks the required sensitivity and specificity for early detection. Many studies have demonstrated that autoantibodies against tumor‐associated antigens (TAA), so called TAAb, could be used for BC detection.8, 9 As potential tumor biomarkers, TAAb have several advantages: (a) they appear in the blood of cancer patients much earlier than clinical diagnosis;10 and (b) the immune system has an amplification effect and memory immune response.11 Nevertheless, the detection of a single autoantibody does not appear to meet the requirements for clinical application. Therefore, identification of a panel of TAAb could improve the diagnostic value by achieving the higher sensitivity and specificity required for the early immunodiagnosis of BC.12, 13, 14, 15
To identify novel TAAb signatures, protein microarray technology has been of great interest to researchers.16, 17 Due to the advantages of rapidity, high sensitivity, and high‐throughput, protein microarray has been widely used as a screening technology for tumor serological markers.18 This technique is playing an increasingly important role in the study of protein‐protein interaction and the identification of biomarkers in various cancers.19
The development of cancer is related to many factors. Vogelstein et al. show that a typical tumor may contain two to eight “driver gene” mutations; the genes can be divided into 12 signal pathways, which regulate three core cell processes: cell fate, cell survival, and genome maintenance.20 Therefore, the development of BC may relate to mutations of multiple cancer driver genes and aberrant expression of proteins. Herein, we hypothesize that TAAbs against these antigens encoded by cancer driver genes are potential biomarkers for BC. Customized protein microarrays consisting of 154 proteins encoded by cancer driver genes were developed, aiming to screen the candidate TAAb for BC detection. Next, the two steps of ELISA validation were applied to identify potential TAAb for BC diagnosis. Finally, an optimal diagnostic model with a panel of TAAb was established for BC diagnosis.
2. MATERIALS AND METHODS
2.1. Sera from patients and controls
Sera from BC patients were collected from a hospital in Henan Province, China from June 2017 to April 2018. All specimens of BC were obtained prior to any treatment. Among them, 319 sera, including 120 preoperative sera and 199 postoperative sera, were obtained from 120 BC patients. Postoperative sera were collected within 1 month after surgery (average 16 days). Sera from age‐matched female healthy controls (HC) as well as women with benign breast diseases (BD) were obtained from Henan Key Laboratory of Tumor Epidemiology, China from December 2017 to April 2018. Detailed information for all samples is shown in Table 1. All sera were aliquoted and stored at −80°C until use. Informed consent forms were signed by all participants and the present study was approved by the Institutional Review Board of Zhengzhou University.
TABLE 1.
Characteristics of study subjects
| Characteristics | Identification of TAAb | TAAb detection | TAAb validation | ||||
|---|---|---|---|---|---|---|---|
| BC (%) | HC (%) | BC (%) | HC (%) | BC (%) | HC (%) | BD (%) | |
| Number | 27 | 27 | 120 | 120 | 279 | 279 | 200 |
| Female | 27 (100.0) | 27 (100.0) | 120 (100.0) | 120 (100.0) | 279 (100.0) | 279 (100.0) | 200 (100.0) |
| Age, years | |||||||
| Mean±SD | 39.7 ± 5.1 | 40.2 ± 10.5 | 49.1 ± 10.3 | 50.6 ± 11.4 | 47.2 ± 9.5 | 46.0 ± 11.2 | 37.7 ± 10.8 |
| Range | 21‐58 | 21‐58 | 29‐78 | 31‐81 | 25‐81 | 25‐76 | 21‐70 |
| <50 | 23 (85.2) | 21 (77.8) | 61 (50.8) | 56 (46.7) | 174 (62.4) | 182 (65.2) | 173 (86.5) |
| ≥50 | 4 (14.8) | 6 (22.2) | 59 (49.2) | 64 (53.3) | 105 (37.6) | 97 (34.8) | 27 (13.5) |
| ER+ | 18 (66.7) | NA | 62 (51.7) | NA | 149 (53.4) | NA | NA |
| PR+ | 16 (59.3) | NA | 58 (48.3) | NA | 143 (51.3) | NA | NA |
| HER‐2+ | 20 (70.4) | NA | 11 (9.2) | NA | 23 (8.2) | NA | NA |
| TNM stage | NA | NA | NA | NA | |||
| I | 4 (14.8) | 38 (31.7) | 92(33.3) | ||||
| II | 16 (59.3) | 31 (25.8) | 71 (25.4) | ||||
| III | 6 (22.2) | 26 (21.7) | 37 (13.3) | ||||
| IV | 1 (3.7) | 10 (8.3) | 11 (3.9) | ||||
| Unknown | 0 (0.0) | 15 (12.5) | 68 (24.4) | ||||
| Lymph node metastasis | NA | NA | NA | NA | |||
| Positive | 8 (29.4) | 8 (6.7) | 14 (5.0) | ||||
| Negative | 19 (70.4) | 112 (93.3) | 260 (93.2) | ||||
| Unknown | 2 (1.1) | 0 (0.0) | 5 (1.8) | ||||
| Histological type | NA | NA | NA | NA | |||
| Invasive | 27 (100.0) | 106 (88.3) | 250 (89.6) | ||||
| Non–invasive | 0 (0.0) | 8 (6.7) | 13 (4.7) | ||||
| Unknown | 0 (0.0) | 6 (5.0) | 16 (5.7) | ||||
BC, breast cancer; BD, breast benign disease; HC, healthy control; NA, not available.
2.2. Customized protein microarray
The layout of protein microarrays is shown in Table S1. Each focused HuProt array (BC‐Biotechnology) contains recombinant proteins that are encoded by 138 cancer‐driving genes.20 We also provided 11 proteins from our laboratory that have been demonstrated as promising biomarkers.21, 22 All 154 proteins (one protein may have more than one fragment) were purchased from CDI Laboratories. Focused protein microarrays were removed from the −80°C freezer and equalized at 4°C and room temperature, followed by incubation using 3% BSA for 3 hours at room temperature. A serum diluted with 1:50 was then added into each well of arrays (200 μL/well, 4°C overnight). After washing with PBS containing .05% Tween‐20 (PBST), microarrays were incubated in 1:1000 Alexa Fluor 532 conjugated goat anti–human IgG (Jackson ImmunoResearch) at room temperature for 1 hour in the dark. After washing with PBST, microarrays were rinsed with ddH2O and dried. Microarrays were scanned with LuxScan 10K‐A (CapitalBio) and analyzed using GenePix Pro 6.0 software (Molecular Devices).
For the scanned signal, the medians of foreground (F Median) and background (B Median) intensity of each protein were measured. The signal to noise ratio (SNR) (F median/B median) value was defined to control the background values between different samples. The test samples were repeated 30 times to evaluate the operational stability of various arrays at different times (Figure S1A). Candidate proteins were screened out by comparing the SNR value between BC and healthy controls using four criteria: (a) non–parametric or t test, P‐values ≤ .05; (b) receiver operating characteristic (ROC) analysis, area under the curve (AUC) > .600 and P‐values ≤ .05; (c) univariate and multivariate logistic regression (backward stepwise), P‐values ≤ .05; and (d) the differences between positive reaction rates of BC patients and positive reaction rates of healthy individuals are more than 10%, while the cut‐off value was defined as the mean plus 3SD.
2.3. ELISA assay
We adopt a relative quantitative ELISA methodology in this study. All purified recombinant proteins for ELISA testing were purchased from Cloud‐Clone. The 16 recombinant proteins (ALK, BRCA2, CDKN2A, CEBPA, CEP55, CSF1R, FGFR3, FUBP1, GATA3, GNAS, HIST1H3B, HRAS, IMP2/p62, PTCH1, RalA, and SRSF2) were diluted in carbonate buffer (pH = 9.6) to optimal concentrations of .250, .250, .500, 1.000, .250, .500, .500, .125, 1.000, .125, 1.000, .500, 1.000, .500, .250, and .125 μg/mL, respectively. The human IgG (Solarbio) was also diluted in carbonate buffer to final concentrations of 300.0, 150.0, 75.0, 37.5, 18.8, 9.4, 4.7, and .0 ng/mL to provide a standard curve. All the diluted proteins and human IgG were coated onto 96‐well flat‐bottom plates overnight at 4°C; then plates were blocked with 2% BSA in PBST overnight at 4°C. After washing with PBST, a serum dilution of 1:100 was added to each well except the wells for blank and human IgG. Then the plates were placed in 37°C water baths for 1 hour followed by washing with PBST. Plates were incubated with HRP‐conjugated goat anti–human IgG at 1:5000 diluted in 1% BSA, in 37°C water baths for 1 hour. A solution of 3,3ʹ,5,5ʹ‐tetramethyl benzidine (TMB)‐H2O2‐urea was used as a detecting agent and 50 μL 2 M sulfuric acid was added to each well as stopping solution. Optical density (OD) was read at 450 and 620 nm using a Multilabel Plate Reader (PerkinElmer).
The final OD values were defined as “OD450−OD620− blank.” The standard curve obtained by human IgG was used to calculate the relative concentration of each autoantibody and to normalize the data from different plates. Because we diluted serum at 1:100, after we calculated the relative concentration using the standard curve, it was multiplied by 100 as the final relative concentration.
2.4. Development of an optimal panel for breast cancer detection
The least absolute shrinkage and selection operator (LASSO) to remove less important TAAb and remaining TAAbs were used as input features for classification algorithms. A total of four classifiers, including a random forest (RF), a support vector machine (SVM), a logistic regression (LR), and a gradient boosting decision tree (GBDT), were constructed, and their performance was compared using 10‐fold cross‐validation within the training set. The classifier with the best performance was chosen. Then, the model based on less TAAb as a panel was built on the whole training dataset and tested on an independent test set. We also applied the panel to discern BC from BD. To further explore whether the final TAAb in the panel were specific to BC, we evaluated the final TAAb in esophageal cancer (EC), gastric cancer (GC), hepatocellular cell carcinoma (HCC), lung cancer (LC), and ovarian cancer (OC) based on the microarray data from our laboratory, which was obtained simultaneously with the data for this study.
2.5. Statistical analysis
The IBM SPSS Statistics 21, GraphPad Prism 5.0, and R version 3.6.1. were applied in this study. The Mann‐Whitney U test and Kruskal‐Wallis test were conducted to compare two or multiple groups (α value was corrected by Bonferroni correction). The diagnostic value of TAAb was evaluated using the ROC curve. The cut‐off value was defined at the point of the maximum Youden index, where the specificity was greater than 90%. The performance of the model based on the TAAb panel was indicated by the AUC. Moreover, the Gene Expression Profiling Interactive Analysis (GEPIA, http://gepia.cancer‐pku.cn/ , accessed 16 August 2020) was used to analyze the expression genes involved in this study. STRING (https://string‐db.org/, accessed July August 2020) was used to explore the functional protein association.
3. RESULTS
3.1. Study design
Four phases (Figure 1) were involved in this study: (a) a customized protein microarray with sera from 54 samples was used to preliminarily identify candidate TAAb by detecting bound IgG; (b) ELISA assay was conducted to detect candidate TAAb in 240 sera and potential TAAb were validated using 758 sera; (c) 319 sera from 120 BC patients were used to evaluate the change in TAAb expression among patients before and after surgery; and (d) all BC and healthy control samples were used to explore an optimal diagnostic model with a TAAb panel by performing the various classification algorithms mentioned in the “Materials and Methods.” Another 200 BD samples were used to evaluate the capability of differential diagnosis of the model. The characteristics of all participants enrolled in this study are shown in Table 1.
FIGURE 1.
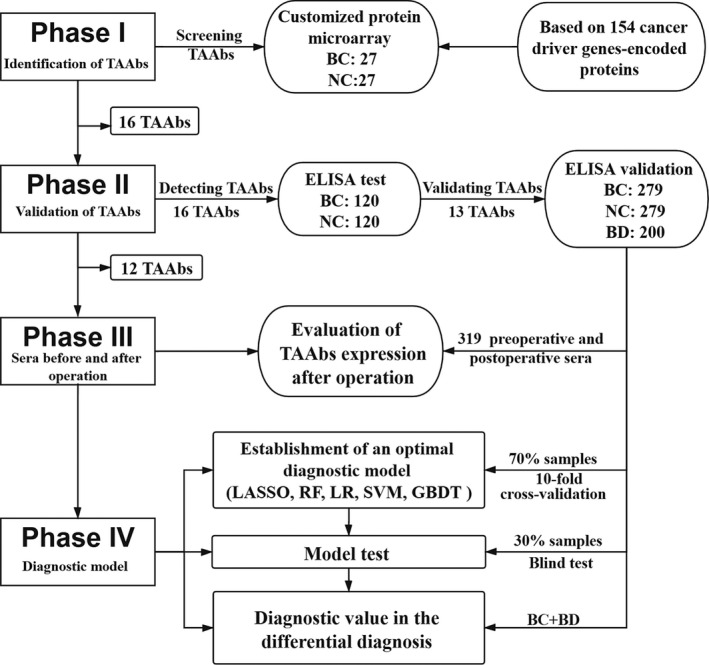
Study design. BC, breast cancer; BD, benign breast disease; GBDT, gradient boosting decision tree; HC, healthy controls; LASSO, least absolute shrinkage and selection operator; LR, logistic regression; RF, random forest; SVM, support vector machines; TAAbs, tumor‐associated autoantibodies
3.2. Discovery of tumor‐associated autoantibodies by customized protein microarray
Sixteen candidate TAAb (Table S2) were selected using the screening strategies described above. The expression signal and the AUC of the 16 TAAb (ALK, BRCA2, CDKN2A, CEBPA, CEP55, CSF1R, FGFR3, FUBP1, GATA3, GNAS, HIST1H3B, HRAS, PTCH1, p62, RalA, and SRSF2) in BC and control groups are shown in Figure 2. The AUC ranged from .613 to .734, and sensitivities ranged from 18.5% to 48.2% when the specificity was 92.6%. Moreover, the expression levels of the genes corresponding to 16 TAAb were queried in GEPIA and are displayed in Figure S1B. As shown in Figure S1C, we also analyzed the protein‐protein interaction (PPI) across 16 TAAs: 15 TAAs interacted with each others to some extent.
FIGURE 2.
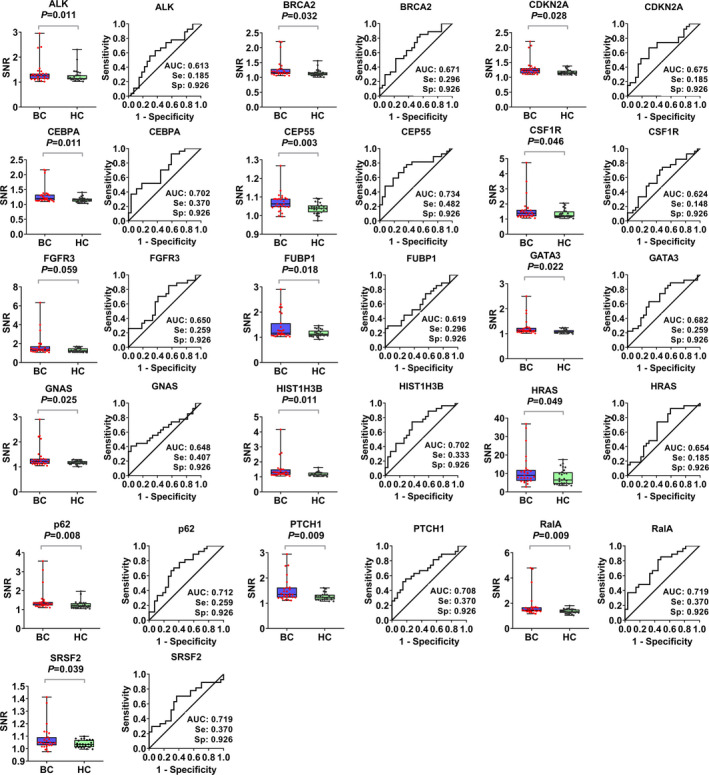
The signal to noise ratio (SNR) values and receiver operating characteristic (ROC) curves of 16 candidate tumor‐associated autoantibodies (TAAbs). For the scanned signal of protein microarrays, the median of foreground (F Median) and background (B Median) intensity of each protein was measured. The SNR (F median/B median) value was defined to control the background values between different samples. BC, breast cancer; HC, healthy controls. Se, sesitivity; Sp, specificity.
3.3. Validation of tumor‐associated autoantibodies by ELISA
In the preliminary detection of candidate TAAb among 240 sera by ELISA, the levels of 13 TAAb were significantly different between BC and healthy groups (Figure 3). These 13 differentially expressed TAAb were further validated in a large dataset (758 sera). The results (Figure 3) suggested that, regardless of whether healthy individuals or patients with benign breast disease served as the control group, the serum levels of 11 TAAb (ALK, BRCA2, CDKN2A, CEBPA, CEP55, FUBP1, GATA3, HRAS, PTCH1, p62, RalA) in the BC group were significantly higher than those in HC and BD groups (P < .01), and there was no difference between BD and healthy control groups (P > .01). The level of autoantibody against HIST1H3B was higher in the BC group than that in the HC group (P < .01), while no significant difference was found between BC and BD groups.
FIGURE 3.
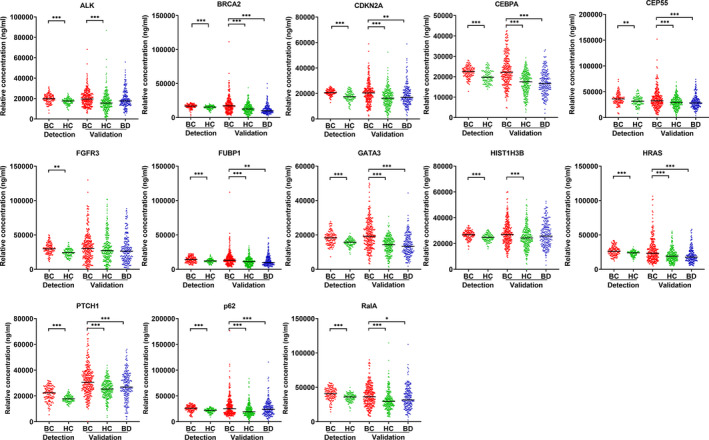
The relative concentration of individual tumor‐associated autoantibodies (TAAbs) among breast cancer, benign breast disease, and normal individuals in two datasets (TAAb detection set and TAAb validation set). BC, breast cancer; BD, benign breast disease; HC, healthy controls; TAAb, tumor‐associated autoantibodies. * P < .05, ** P < .01, *** P < .001 (Kruskal‐Wallis H test and Mann‐Whitney U test, Bonferroni correction)
The diagnostic value of validated TAAb was also evaluated. The range of AUC for distinguishing BC from healthy controls was .597‐.736 (Figure S2) and .600‐.724 (Figure S3) in two datasets, respectively. The AUC of 12 TAAb in discriminating BC from BD ranged from .530 to .758 (Figure S3). The diagnostic performance of the 12 TAAb among all BC and controls (HC and BD) is shown in Table 2 and Table 3. The positive rates of 12 TAAb in the BC group ranged from 20.8% to 49.1% at the corresponding cut‐off values; these values were higher than those in the healthy control group, which ranged from 9.5% to 10.0% (P < .05). In the BD group, the range of positive rates was 6.5%–27.0% (Table 3).
TABLE 2.
The diagnostic value of 12 TAAbs for identifying BC from healthy control (HC)
| TAAb | Cut‐off (ng/mL) | Positive(%) | χ2 | P | Se (%) | Sp (%) | YI | Accuracy (%) | FPR (%) | FNR (%) | PPV (%) | NPV (%) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BC | HC | ||||||||||||
| (n = 399) | (n = 399) | ||||||||||||
| ALK | 26 911 | 94 (23.6) | 40 (10.0) | 26.153 | <.001 | 23.6 | 90.0 | 0.1 | 16.8 | 10.0 | 76.4 | 70.2 | 54.1 |
| BRCA2 | 22 292 | 118 (29.6) | 38 (9.5) | 50.994 | <.001 | 29.6 | 90.5 | 0.2 | 19.5 | 9.5 | 70.4 | 75.7 | 56.2 |
| CDKN2A | 24 963 | 100 (25.1) | 40 (10.0) | 31.185 | <.001 | 25.1 | 90.0 | 0.2 | 17.5 | 10.0 | 74.9 | 71.5 | 54.6 |
| CEBPA | 23 449 | 147 (36.8) | 42 (10.5) | 76.437 | <.001 | 36.8 | 89.5 | 0.3 | 23.7 | 10.5 | 63.2 | 77.8 | 58.6 |
| CEP55 | 43 553 | 91 (22.8) | 40 (10.0) | 23.755 | <.001 | 22.8 | 90.0 | 0.1 | 16.4 | 10.0 | 77.2 | 69.5 | 53.8 |
| FUBP1 | 19 421 | 102 (25.6) | 42 (10.5) | 30.505 | <.001 | 25.6 | 89.5 | 0.2 | 18.0 | 10.5 | 74.4 | 70.9 | 54.6 |
| GATA3 | 19 858 | 196 (49.1) | 41 (10.3) | 144.197 | <.001 | 49.1 | 89.7 | 0.4 | 29.7 | 10.3 | 50.9 | 82.7 | 63.8 |
| HIST1H3B | 35 877 | 83 (20.8) | 41 (10.3) | 16.843 | <.001 | 20.8 | 89.7 | 0.1 | 15.5 | 10.3 | 79.2 | 66.9 | 53.1 |
| HRAS | 32 356 | 127 (31.8) | 40 (10.0) | 57.319 | <.001 | 31.8 | 90.0 | 0.2 | 20.9 | 10.0 | 68.2 | 76.1 | 56.9 |
| IMP2/p62 | 35 344 | 133 (33.3) | 40 (10.0) | 63.833 | <.001 | 33.3 | 90.0 | 0.2 | 21.7 | 10.0 | 66.7 | 76.9 | 57.4 |
| PTCH1 | 34 216 | 177 (44.4) | 39 (9.8) | 120.888 | <.001 | 44.4 | 90.2 | 0.3 | 27.1 | 9.8 | 55.6 | 81.9 | 61.9 |
| RalA | 49 331 | 117 (29.3) | 40 (10.0) | 47.014 | <.001 | 29.3 | 90.0 | 0.2 | 19.7 | 10.0 | 70.7 | 74.6 | 56.0 |
The cut‐off value was considered as the maximum Youden index at the point of more than 90% specificity.
FNR, false‐negative rate; FPR, false‐positive rate; NPV, negative predictive value; PPV, positive predictive value; Se, sensitivity; Sp, specificity; YI, Youden index.
TABLE 3.
The diagnostic value of 12 TAAbs for identifying BC from breast benign disease (BD)
| TAAb | Cut‐off (ng/mL) | Positive(%) | χ2 | P | Se (%) | Sp (%) | YI | Accuracy (%) | FPR (%) | FNR (%) | PPV (%) | NPV (%) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BC | BD | ||||||||||||
| (n = 399) | (n = 200) | ||||||||||||
| ALK | 26 911 | 94 (23.6) | 38 (19.0) | 1.612 | .204 | 23.6 | 82.0 | 0.1 | 22.0 | 18.0 | 76.4 | 71.2 | 51.8 |
| BRCA2 | 22 292 | 118 (29.6) | 13 (6.5) | 41.51 | <.001 | 29.6 | 93.5 | 0.2 | 21.9 | 6.5 | 70.4 | 82.0 | 57.0 |
| CDKN2A | 24 963 | 100 (25.1) | 34 (17.0) | 4.987 | .026 | 25.1 | 83.0 | 0.1 | 22.4 | 17.0 | 74.9 | 59.6 | 52.6 |
| CEBPA | 23 449 | 147 (36.8) | 22 (11.0) | 43.927 | <.001 | 36.8 | 89.0 | 0.3 | 28.2 | 11.0 | 63.2 | 77.0 | 58.5 |
| CEP55 | 43 553 | 91 (22.8) | 26 (13.0) | 8.152 | .004 | 22.8 | 87.0 | 0.1 | 19.5 | 13.0 | 77.2 | 63.7 | 53.0 |
| FUBP1 | 19 421 | 102 (25.6) | 32 (16.0) | 7.017 | .008 | 25.6 | 84.0 | 0.1 | 22.4 | 16.0 | 74.4 | 61.5 | 53.0 |
| GATA3 | 19 858 | 196 (49.1) | 36 (18.0) | 54.379 | <.001 | 49.1 | 82.0 | 0.3 | 38.7 | 18.0 | 50.9 | 73.2 | 61.7 |
| HIST1H3B | 35 877 | 83 (20.8) | 36 (18.0) | 0.657 | .418 | 20.8 | 82.0 | 0.0 | 19.9 | 18.0 | 79.2 | 53.6 | 50.9 |
| HRAS | 32 356 | 127 (31.8) | 26 (13.0) | 24.836 | <.001 | 31.8 | 87.0 | 0.2 | 25.5 | 13.0 | 68.2 | 71.0 | 56.1 |
| IMP2/p62 | 35 344 | 133 (33.3) | 42 (21.0) | 9.799 | .002 | 33.3 | 79.0 | 0.1 | 29.2 | 21.0 | 66.7 | 61.3 | 54.2 |
| PTCH1 | 34 216 | 177 (44.4) | 54 (27.0) | 16.948 | <.001 | 44.4 | 73.0 | 0.2 | 38.7 | 27.0 | 55.6 | 62.2 | 56.8 |
| RalA | 49 331 | 117 (29.3) | 32 (16.0) | 12.655 | <.001 | 29.3 | 84.0 | 0.1 | 24.9 | 16.0 | 70.7 | 64.7 | 54.3 |
The cut‐off value was considered as the maximum Youden index at the point of more than 90% specificity
FNR, false‐negative rate; FPR, false‐positive rate; NPV, negative predictive value; PPV, positive predictive value; Se, sensitivity; Sp, specificity; YI, Youden index.
3.4. Tumor‐associated autoantibodies decreased in sera of breast cancer patients after the operation
We obtained a total of 319 preoperative and postoperative sera from 120 BC patients in this study. As shown in Figure 4A, the preoperative levels of 12 TAAb were higher than those within 1 month after surgery (P < .05). Figure 4B depicts the changing’ trend of 12 TAAb in the sera of randomly selected patients.
FIGURE 4.
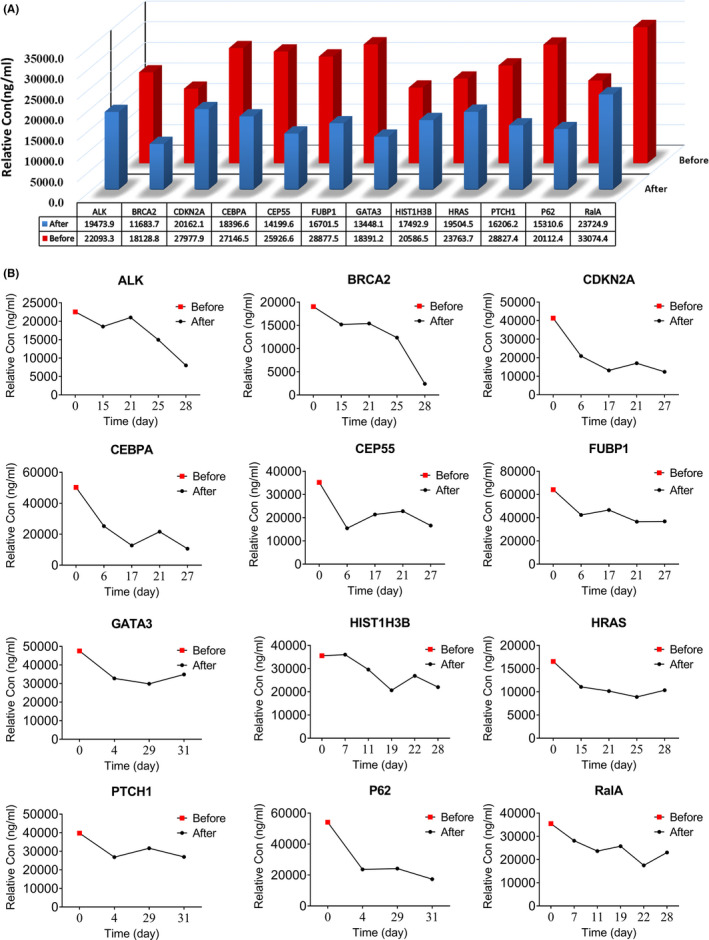
The preoperative and postoperative level of tumor‐associated autoantibodies (TAAbs) in the sera of breast cancer patients. A, Relative concentration levels (median) of individual TAAb between breast cancer (BC) patients before and after the operation (P <.05). B, Relative concentration of individual TAAb from randomly selected patients before and after the operation. TAAbs, tumor‐associated autoantibodies
We divided samples into subgroups based on age and stage. As shown in Figure S4, the level of TAAb to PTCH1 showed no significant difference (P > .05) between patients before and after surgery in the late stage (III‐IV), while in the early stage (I‐II), it was higher in patients before surgery than that in patients after surgery (P < .05). Moreover, the TAAb to CEBPA, GATA3, and PTCH1 showed higher expression levels in the preoperative patients aged ≥ 50 than those of aged < 50 (P < .01).
3.5. Establishment and validation of a panel with six tumor‐associated autoantibodies for breast cancer detection
A panel of six TAAb (CEBPA, CEP55, GATA3, HRAS, PTCH1, and RalA) was selected by LASSO. After 10‐fold cross‐validation within the training set, RF was chosen as the final model among four classifiers, as it showed the highest AUC of .879, with sensitivity of 74.3% and specificity of 91.9% (Figure 5A). The relative importance of the panel with six TAAb is shown in Figure 5E.
FIGURE 5.
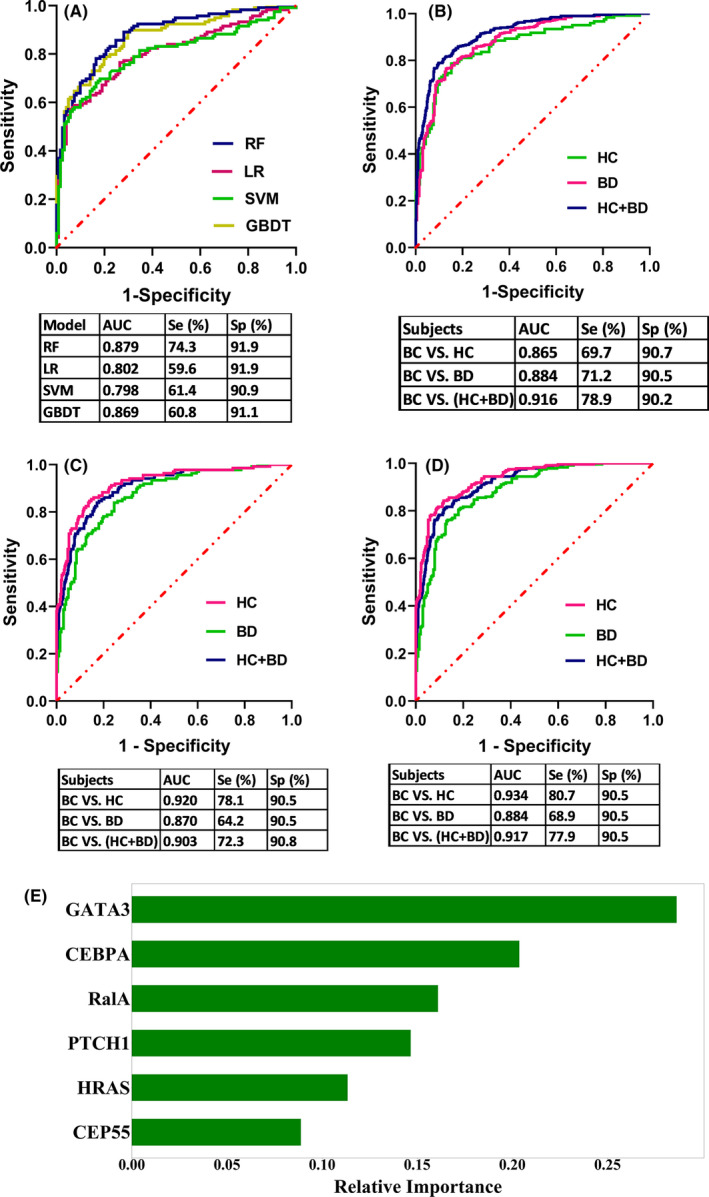
Performance of the panel with six tumor‐associated autoantibodies (TAAbs) to detect breast cancer. A, Receiver operating characteristic (ROC) curves from four models (RF, LR, SVM, GBDT) in a training set after 10‐fold cross‐validation. B, The performance of the RF model in distinguishing individuals with BC from HC (blind test set) or BD or all controls (HC+BD). C, Performance of the model for distinguishing individuals with early‐stage (I+II) BC from HC or BD or all controls (HC+BD). D, Performance of the model for distinguishing individuals with age < 50 BC from HC or BD or all controls (HC+BD). BC, breast cancer; BD, benign breast disease; GBDT, gradient boost decision tree; HC, healthy controls; LR, logistic regression; RF, random forest; SVM, support vector machines; TAAbs, tumor‐associated autoantibodies
Next, all samples in the training set were used to construct a model and the performance was evaluated in a blind test set. There was an AUC of .865 (69.7% sensitivity and 90.7% specificity) for BC detection. The performance of the model for differential diagnosis (BC vs BD) revealed an AUC of .884, a sensitivity of 71.2%, and a specificity of 90.5%. The PPV was 77.6%. When combining all controls (HC+BD) together, the model had an AUC of .916, and the sensitivity, specificity, and PPV were 78.9%, 90.2%, and 85.7%, respectively (Figure 5B).
The expressions of six TAAb in the panel were also verified with the data generated by protein microarray across other common cancers, such as esophageal cancer, gastric cancer, hepatocellular cell carcinoma, lung cancer, and ovarian cancer. As exhibited in Figure 6, autoantibodies to CEBPA, CEP55, GATA3, HRAS, and RalA showed no difference across these five cancers. Only elevated autoantibody to PTCH1 was detected in hepatocellular cell carcinoma and ovarian cancer. The results suggested that all TAAb in the panel except anti–PTCH1 are specific for BC.
FIGURE 6.
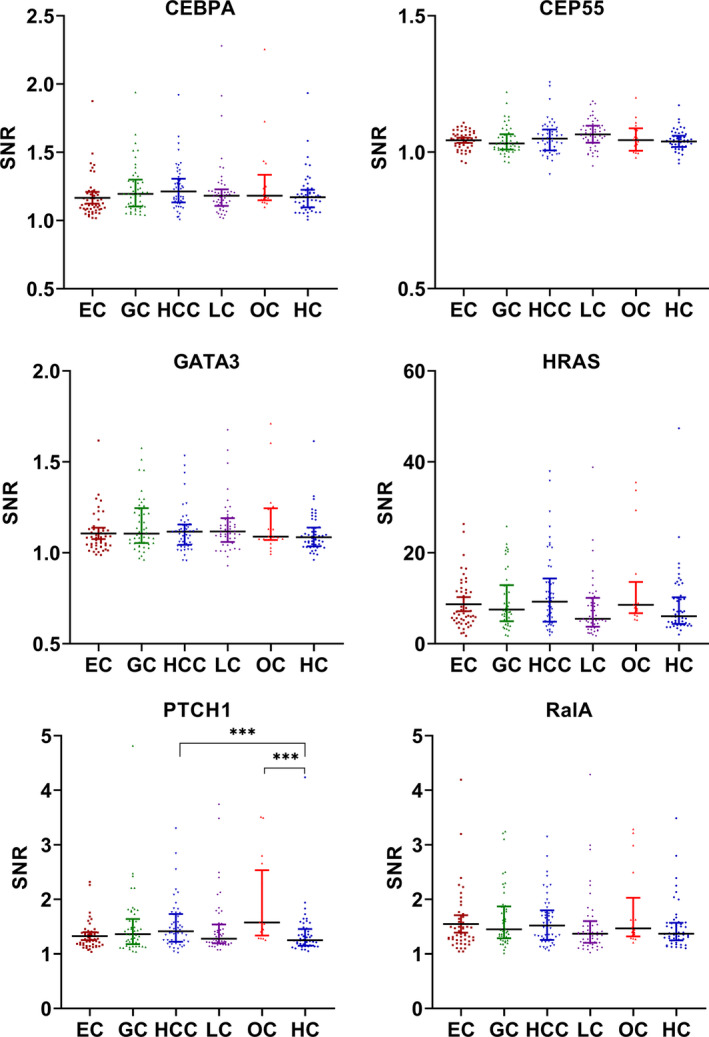
Scatter plot of a panel of six tumor‐associated autoantibodies (TAAbs) in common cancers (median with interquartile). EC, esophageal cancer; GC, gastric cancer; HC, healthy controls; HCC, hepatocellular cell carcinoma; LC, lung cancer; OC, ovarian cancer; SNR, signal to noise ratio; TAAb, tumor‐associated autoantibodies. *** P < .001
3.6. Performance of the panel with six tumor‐associated autoantibodies for breast cancer subgroups
In the early stage (I–II) of BC, as shown in Figure 5C, the model displayed AUC of .920 (BC vs HC with 78.1% sensitivity and 90.5% specificity), .870 (BC vs BD with 64.2% sensitivity and 90.5% specificity), and .903 (BC vs [HC+BD] with a sensitivity of 72.3% and specificity of 90.8%). In addition, for the diagnosis of BC patients under the age of 50, the model exhibited AUC of .934 (BC vs HC, 80.7% sensitivity and 90.5% specificity), .884 (BC vs BD, 68.9% sensitivity and 90.5% specificity), and .917 (BC vs [HC+BD], 77.9% sensitivity and 90.5% specificity) (Figure 5D). Table S3 showed that there was no significant difference between various clinical stages, ages, prognostic index, lymph node status, histological type, and family history of cancer (χ 2 test, P > .05). The predicted results of the model among BC subgroups are provided in Figure S5.
4. DISCUSSION
Protein microarray technology has shown broad application prospects in the biomedical field. It is able to simultaneously and rapidly discover a variety of biomarkers.23, 24 It is also a highly effective tool to profile novel autoantibodies in cancer with superior specificity.25 Using the customized protein microarrays, 16 candidate TAAb were screened out, including ALK, BRCA2, CDKN2A, CEBPA, CEP55, CSF1R, FGFR3, FUBP1, GATA3, GNAS, HIST1H3B, HRAS, PTCH1, p62 (IMP2), RalA, and SRSF2. These candidate TAAb can distinguish BC from healthy controls with a sensitivity range from 18.5% to 48.2% at 92.6% specificity. Except for HIST1H3B, the remaining TAA corresponding to 15 TAAb interact with each other, which means the novel biomarkers may be involved in cell cycle regulation and signal transduction.26 Furthermore, from the gene expression level, they could also distinguish BC from normal controls. The result indicates that this is a feasible method to screen TAAb by using the protein microarrays encoded by cancer driver genes. Moreover, it reveals that the proteins encoded by cancer driver genes have immunogenicity as well, which could trigger immune responses.
Of the identified novel biomarkers, several are associated with BC. As an important transcription factor, CEBPA regulates the proliferation and differentiation of various tissues and participates in the regulation of the expression or function of some cell cycle regulators, including BC cells.27 Gery et al found that CEBPA is related to the occurrence and development of BC.28 It was reported that PTCH1 is associated with early recurrence of BC and may become a powerful predictor of BC.29, 30 Studies have suggested that GATA3 is a member of the transcription regulatory factor family, which could be a useful immunohistochemical marker for BC.31, 32 CDKN2A/p16 has a meaningful association with the risk of BC and it is expected to be a candidate gene for BC.33 BRCA2 gene is a confirmed BC susceptibility gene,34 which could lead to the development of BC.35 HRAS is related to BC and could be a valuable screening biomarker for women.36 As the human far upstream element (FUSE) binding protein 1, FUBP1 overexpression was observed in many kinds of cancers.37, 38 A study found that CEP55 is a novel TAA for BC.39 ALK also plays an important role in cancer biology.40 All biomarkers selected in this study are closely implicated in the development of BC.
To the best of our knowledge, of the 16 TAAb, 3 (CDKN2A, p62, and RalA) have been reported as biomarkers for BC as well as other types of cancers.41, 42, 43 However, others have not been reported in the BC serological biomarkers study. Thus, we further investigated the presence and diagnostic value of them by indirect ELISA test. The ELISA tests were conducted in two steps: one for the preliminary detection of TAAb and the other for the validation of TAAb. We selected 12 potential TAAb (ALK, BRCA2, CDKN2A, CEBPA, CEP55, FUBP1, GATA3, HIST1H3B, HRAS, PTCH1, p62, and RalA). The serum levels of the 12 TAAb in the BC group were higher than those in the healthy control group and the benign breast disease group (P < .01). The 12 TAAb are potential serological biomarkers for BC and could be used in the diagnosis or differential diagnosis of BC. Moreover, serum samples are easy to obtain, and the detection method of antibody is comparatively simple with highly sensitive and non–invasive characteristics, which makes the detection of TAAb more practical. Based on our results, TAAb to CEBPA, PTCH1, GATA3, CDKN2A, and BRCA2 have relatively high diagnostic value for BC detection. The AUC range from .707 to .769.
Studies have shown that some circulating biomarkers emerge before clinical diagnosis of patients,10, 44, 45 while diminishing after surgery.46, 47, 48, 49, 50 Similarly, our findings indicated the reduction of 12 TAAb in patients’ sera after surgery. A previous study found that TAAb can indicate the malignant transformation of disease prior to clinical study and are promising biomarkers for early detection.44 Herein, the dynamic change of serum TAAb may reflect the changes in the tumor status of patients.51 Our results suggest that the 12 TAAb may be useful as predictors of tumor activity. However, the biological mechanism for the changes in serum TAAb in cancer patients before and after surgery remains unclear. In this study, the postoperative sera of BC patients were only collected within 1 month of the operation, with no follow‐up samples. Thus, we cannot verify the changes in the novel TAAb in the sera of BC patients in a lengthy time span after treatment.
Multiple tumor markers could improve the diagnostic performance and increase the efficiency of the test.52 A variety of machine learning (ML) techniques have been used in cancer research to develop predictive models and improve the accuracy of decision‐making.53 Therefore, in phase IV of this study, we applied different ML algorithms to choose a model with excellent diagnostic performance. To make the model more efficient and interpretable, we first used a feature selection technique LASSO to reduce the number of TAAb to be included. The inverse regularization parameter C was set as .1 to keep a reasonable set of TAAb. RF and GBDT showed comparable performance and outperformed the other two classifiers. Comprehensive studies have been conducted to compare ML and statistical methods with respect to their applications in cancer genomics; GBDT and RF are always among the most powerful.54, 55 As the model will finally be used on clinical data, where noise may arise, we chose RF because it is more tolerant to noises.
The RF model with the combination of six TAAb exhibited good diagnostic value both in training and blind test sets. In addition, the six‐TAAb panel could distinguish BC from breast benign diseases (AUC = .884, 71.2% sensitivity and 90.2% specificity). For early detection, the six‐TAAb panel showed an increasingly high AUC (AUC = .903 for stage I‐II, AUC = .917 for age < 50). The eminent performance of the RF model for different and unbalanced subgroups demonstrated its capability of handling different situations in the clinic. Consequently, the model based on six TAAb is well‐established, which is of great significance for its clinical application for early BC detection. Furthermore, five of the six TAAb are specific to BC among common cancers, which makes them ideal for BC detection. Nevertheless, there are some limitations of this study. Our study is a retrospective case‐control study and the prospective collection of specimens from a cohort is unavailable.56 Therefore, it is necessary to confirm autoantibody signatures in a large‐scale prospective study. In addition, there were no pre–diagnostic serial sera, so the expression status of these TAAb before the development of BC was unknown. Following up patients with precancerous breast lesions through the collection of their successive sera will be the focus of our future study.
In summary, this study found some novel TAAb that could be potential biomarkers for the detection of BC. These biomarkers can not only identify BC from healthy controls but also differentiate BC from benign breast disease. Furthermore, we developed a diagnostic model, the six‐TAAb combination, that performed well and is a promising potential method for BC detection. Further work is needed to determine the value of these markers in clinical application.
DISCLOSURE
The authors have no conflicts of interest to declare.
Supporting information
Figure S1
Figure S2
Figure S3
Figure S4
Figure S5
Table S1
Table S2
Table S3
Qiu C, Wang B, Wang P, et al. Identification of novel autoantibody signatures and evaluation of a panel of autoantibodies in breast cancer. Cancer Sci. 2021;112:3388–3400. 10.1111/cas.15021
Cuipeng Qiu, Bofei Wang, and Peng Wang contributed equally to this work.
Funding information
This study was funded by the Major Project of Science and Technology in Henan Province (No. 161100311400), the Zhengzhou Major Project for Collaborative Innovation (18XTZX12007), and the National Science and Technology Major Project of China (2018ZX10302205).
Contributor Information
Hua Ye, Email: yehua@zzu.edu.cn.
Jianying Zhang, Email: jianyingzhang@hotmail.com.
REFERENCES
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394‐424. [DOI] [PubMed] [Google Scholar]
- 2.DeSantis CE, Ma J, Gaudet MM, et al. Breast cancer statistics, 2019. CA Cancer J Clin. 2019;69:438‐451. [DOI] [PubMed] [Google Scholar]
- 3.Anastasiadi Z, Lianos GD, Ignatiadou E, Harissis HV, Mitsis M. Breast cancer in young women: an overview. Updates Surg. 2017;69:313‐317. [DOI] [PubMed] [Google Scholar]
- 4.Tagliafico AS, Piana M, Schenone D, Lai R, Massone AM, Houssami N. Overview of radiomics in breast cancer diagnosis and prognostication. Breast. 2020;49:74‐80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Srivastava S, Koay EJ. Cancer overdiagnosis: a biological challenge and clinical dilemma. Nat Rev Cancer. 2019;19:349‐358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saadatmand S, Geuzinge HA, Rutgers EJT, et al. MRI versus mammography for breast cancer screening in women with familial risk (FaMRIsc): a multicentre, randomised, controlled trial. Lancet Oncol. 2019;20:1136‐1147. [DOI] [PubMed] [Google Scholar]
- 7.Hayes DF, Zurawski VR Jr, Kufe DW. Comparison of circulating CA15‐3 and carcinoembryonic antigen levels in patients with breast cancer. J Clin Oncol. 1986;4:1542‐1550. [DOI] [PubMed] [Google Scholar]
- 8.Yadav S, Kashaninejad N, Masud MK, Yamauchi Y, Nguyen NT, Shiddiky MJA. Autoantibodies as diagnostic and prognostic cancer biomarker: Detection techniques and approaches. Biosens Bioelectron. 2019;139:111315. [DOI] [PubMed] [Google Scholar]
- 9.Rauf F, Anderson KS. Autoantibodies in early detection of breast cancer. Cancer Epidemiol Biomarkers Prev. 2020;29:2475‐2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang J, Chan EK. Autoantibodies to IGF‐II mRNA binding protein p62 and overexpression of p62 in human hepatocellular carcinoma. Autoimmun Rev. 2002;1:146‐153. [DOI] [PubMed] [Google Scholar]
- 11.Qiu J, Keyser B, Lin ZT, Wu T. Autoantibodies as potential biomarkers in breast cancer. Biosensors. 2018;8:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pagaza‐Straffon C, Marchat LA, Herrera L, et al. Evaluation of a panel of tumor‐associated antigens in breast cancer. Cancer Biomark. 2020;27:207‐211. [DOI] [PubMed] [Google Scholar]
- 13.Bassaro L, Russell SJ, Pastwa E, Somiari SA, Somiari RI. Screening for multiple autoantibodies in plasma of patients with breast cancer. Cancer Genomics Proteomics. 2017;14:427‐435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu W, De La Torre IG, Gutiérrez‐Rivera MC, et al. Detection of autoantibodies to multiple tumor‐associated antigens (TAAs) in the immunodiagnosis of breast cancer. Biosensors (Basel). 2015;36:1307‐1312. [DOI] [PubMed] [Google Scholar]
- 15.Qiu C, Wang P, Wang B, Shi J, Wang X, Li T. Establishment and validation of an immunodiagnostic model for prediction of breast cancer. Oncoimmunology. 2020;9:1682382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Emili AQ, Cagney G. Large‐scale functional analysis using peptide or protein arrays. Nat Biotechnol. 2000;18:393‐397. [DOI] [PubMed] [Google Scholar]
- 17.MacBeath G. Protein microarrays and proteomics. Nat Genet. 2002;32(Suppl):526‐532. [DOI] [PubMed] [Google Scholar]
- 18.Kijanka G, Murphy D. Protein arrays as tools for serum autoantibody marker discovery in cancer. J Proteomics. 2009;72:936‐944. [DOI] [PubMed] [Google Scholar]
- 19.Zhu H, Snyder M. Protein arrays and microarrays. Curr Opin Chem Biol. 2001;5:40‐45. [DOI] [PubMed] [Google Scholar]
- 20.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546‐1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dai L, Ren P, Liu M, Imai H, Tan EM, Zhang JY. Using immunomic approach to enhance tumor‐associated autoantibody detection in diagnosis of hepatocellular carcinoma. Clin Immunol. 2014;152:127‐139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qin J, Wang S, Shi J, et al. Using recursive partitioning approach to select tumor‐associated antigens in immunodiagnosis of gastric adenocarcinoma. Cancer Sci. 2019;110:1829‐1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee KB, Park SJ, Mirkin CA, Smith JC, Mrksich M. Protein nanoarrays generated by dip‐pen nanolithography. Science. 2002;295:1702‐1705. [DOI] [PubMed] [Google Scholar]
- 24.Gunawardana CG, Diamandis EP. High throughput proteomic strategies for identifying tumour‐associated antigens. Cancer Lett. 2007;249:110‐119. [DOI] [PubMed] [Google Scholar]
- 25.Zhang S, Liu Y, Chen J, et al. Autoantibody signature in hepatocellular carcinoma using seromics. Journal of Hematology & Oncology. 2020;13(1):85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fernández‐Madrid F, Maroun MC. Autoantibodies in breast cancer. Adv Clin Chem. 2014;64:221‐240. [DOI] [PubMed] [Google Scholar]
- 27.Liu LM, Sun WZ, Fan XZ, Xu YL, Cheng MB, Zhang Y. Methylation of C/EBPα by PRMT1 Inhibits Its tumor‐suppressive function in breast cancer. Cancer Res. 2019;79:2865‐2877. [DOI] [PubMed] [Google Scholar]
- 28.Gery S, Tanosaki S, Bose S, Bose N, Vadgama J, Koeffler HP. Down‐regulation and growth inhibitory role of C/EBPalpha in breast cancer. Clin Cancer Res. 2005;11:3184‐3190. [DOI] [PubMed] [Google Scholar]
- 29.Wang CY, Chang YC, Kuo YL, et al. Mutation of the PTCH1 gene predicts recurrence of breast cancer. Sci Rep. 2019;9:16359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Siddappa CM, Watson MA, Pillai SG, Trinkaus K, Fleming T, Aft R. Detection of disseminated tumor cells in the bone marrow of breast cancer patients using multiplex gene expression measurements identifies new therapeutic targets in patients at high risk for the development of metastatic disease. Breast Cancer Res Treat. 2013;137:45‐56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takaku M, Grimm SA, Wade PA. GATA3 in breast cancer: tumor suppressor or oncogene? Gene Expr. 2015;16:163‐168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krings G, Nystrom M, Mehdi I, Vohra P, Chen YY. Diagnostic utility and sensitivities of GATA3 antibodies in triple‐negative breast cancer. Hum Pathol. 2014;45:2225‐2232. [DOI] [PubMed] [Google Scholar]
- 33.Dadjoo P, Parvin SS, Yazdinezhad Y, et al. Association of cyclin‐dependent kinase inhibitor 2A/B with increased risk of developing breast cancer. J Cell Physiol. 2020;235:5141‐5145. [DOI] [PubMed] [Google Scholar]
- 34.Wooster R, Bignell G, Lancaster J, et al. Identification of the breast cancer susceptibility gene BRCA2. Nature. 1995;378:789‐792. [DOI] [PubMed] [Google Scholar]
- 35.Lima ZS, Ghadamzadeh M, Arashloo FT, Amjad G, Ebadi MR, Younesi L. Recent advances of therapeutic targets based on the molecular signature in breast cancer: genetic mutations and implications for current treatment paradigms. J Hematol Oncol. 2019;12(1):38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garrett PA, Hulka BS, Kim YL, Farber RA. HRAS protooncogene polymorphism and breast cancer. Cancer Epidemiol Biomarkers Prev. 1993;2:131‐138. [PubMed] [Google Scholar]
- 37.Debaize L, Troadec MB. The master regulator FUBP1: its emerging role in normal cell function and malignant development. Cell Mol Life Sci. 2019;76:259‐281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bettegowda C, Agrawal N, Jiao Y, et al. Mutations in CIC and FUBP1 contribute to human oligodendroglioma. Science. 2011;333:1453‐1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Inoda S, Hirohashi Y, Torigoe T, et al. Cep55/c10orf3, a tumor antigen derived from a centrosome residing protein in breast carcinoma. J Immunother. 2009;32:474‐485. [DOI] [PubMed] [Google Scholar]
- 40.Hallberg B, Palmer RH. The role of the ALK receptor in cancer biology. Ann Oncol. 2016;27(Suppl 3):iii4–iii15. [DOI] [PubMed] [Google Scholar]
- 41.Chen C, Huang Y, Zhang C, Liu T, Zheng HE, Wan S, et al. Circulating antibodies to p16 protein‐derived peptides in breast cancer. Mol Clin Oncol. 2015;3:591‐594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang JY, Casiano CA, Peng XX, Koziol JA, Chan EK, Tan EM. Enhancement of antibody detection in cancer using panel of recombinant tumor‐associated antigens. Cancer Epidemiol Biomarkers Prev. 2003;12:136‐143. [PubMed] [Google Scholar]
- 43.Liu W, Li Y, Wang B, Dai L, Qian W, Zhang JY. Autoimmune Response to IGF2 mRNA‐Binding Protein 2 (IMP2/p62) in Breast Cancer. Scand J Immunol. 2015;81:502‐507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tan EM, Zhang J. Autoantibodies to tumor‐associated antigens: reporters from the immune system. Immunol Rev. 2008;222:328‐340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bast RC, Lu Z, Han CY, Lu KH, Anderson KS. Biomarkers and strategies for early detection of ovarian cancer. Cancer Epidemiol Biomarkers Prev. 2020;29:2504‐2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun N, Sun S, Gao Y, et al. Utility of isocitrate dehydrogenase 1 as a serum protein biomarker for the early detection of non–small‐cell lung cancer: a multicenter in vitro diagnostic clinical trial. Cancer Sci. 2020;111:1739‐1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shen Q, Fan J, Yang XR, et al. Serum DKK1 as a protein biomarker for the diagnosis of hepatocellular carcinoma: a large‐scale, multicentre study. Lancet Oncol. 2012;13:817‐826. [DOI] [PubMed] [Google Scholar]
- 48.Peng YH, Xu YW, Guo H, et al. Combined detection of serum Dickkopf‐1 and its autoantibodies to diagnose esophageal squamous cell carcinoma. Cancer Med. 2016;5:1388‐1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu H, Deng Z, Wang H, et al. MGMT autoantibodies as a potential prediction of recurrence and treatment response biomarker for glioma patients. Cancer Med. 2019;8:4359‐4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li Y, Li CQ, Guo SJ, et al. Longitudinal serum autoantibody repertoire profiling identifies surgery‐associated biomarkers in lung adenocarcinoma. EBioMedicine. 2020;53:102674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ladd JJ, Chao T, Johnson MM, et al. Autoantibody signatures involving glycolysis and splicesome proteins precede a diagnosis of breast cancer among postmenopausal women. Cancer Res. 2013;73:1502‐1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang S, Qin J, Ye H, et al. Using a panel of multiple tumor‐associated antigens to enhance autoantibody detection for immunodiagnosis of gastric cancer. Oncoimmunology. 2018;7:e1452582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kourou K, Exarchos TP, Exarchos KP, Karamouzis MV, Fotiadis DI. Machine learning applications in cancer prognosis and prediction. Comput Struct Biotechnol J. 2015;13:8‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu B, Abbott T, Fishman D, et al. Comparison of statistical methods for classification of ovarian cancer using mass spectrometry data. Bioinformatics. 2003;19:1636‐1643. [DOI] [PubMed] [Google Scholar]
- 55.Lynch CM, Abdollahi B, Fuqua JD, et al. Prediction of lung cancer patient survival via supervised machine learning classification techniques. Int J Med Inform. 2017;108:1‐8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pepe MS, Feng Z, Janes H, Bossuyt PM, Potter JD. Pivotal evaluation of the accuracy of a biomarker used for classification or prediction: standards for study design. J Natl Cancer Inst. 2008;100:1432‐1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1
Figure S2
Figure S3
Figure S4
Figure S5
Table S1
Table S2
Table S3


