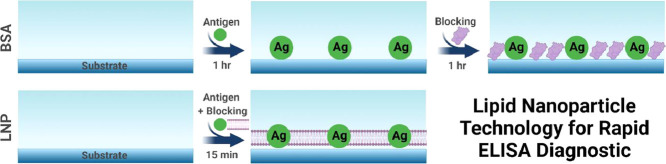
Abstract
The enzyme-linked immunosorbent assay (ELISA) is a widely used method for protein detection and relies on the specific capture of target proteins while minimizing the nonspecific binding of other interfering proteins and biomolecules. To prevent nonspecific binding events, blocking agents such as bovine serum albumin (BSA) protein, mixtures of proteins in media such as milk or serum, and/or surfactants are typically added to ELISA plates after probe attachment and before analyte capture. Herein, we developed a streamlined ELISA strategy in which readily prepared lipid nanoparticles are utilized as the blocking agent and are added together with the probe molecule to the ELISA plate, resulting in fewer processing steps, quicker protocol time, and superior detection performance compared to conventional BSA blocking. These measurement capabilities were established for coronavirus disease-2019 (COVID-19) antibody detection in saline and human serum conditions and are broadly applicable for developing rapid ELISA diagnostics.
Keywords: Enzyme-linked immunosorbent assay, ELISA, Blocking, Diagnostic, Lipid nanoparticle
Graphical abstract
1. Introduction
The enzyme-linked immunosorbent assay (ELISA) enables the sensitive detection of protein analytes through an enzymatically catalyzed chemiluminescent signal and is widely used in molecular biology and diagnostic applications [1]. There are a variety of ELISA configurations involving surface-adsorbed antigen or antibody probes in order to capture target antibodies or antigens, respectively, and a universal principle of ELISA measurements is to sensitively detect the target analyte while minimizing the nonspecific adsorption of other biomolecules, such as additional types of proteins in a complex medium [2], [3], [4]. To achieve this goal, a wide variety of blocking agents such as bovine serum albumin (BSA) protein, mixtures of proteins in milk or serum, and/or surfactants are utilized to passivate probe-functionalized surfaces, i.e., adsorb nonspecifically onto residual gaps in between probe molecules on the well surface prior to the addition of the analyte-containing sample [5], [6], [7]. While protein-based blocking agents have become a standard tool in ELISA protocols, emerging evidence from the nano- and microfluidics community supports that lipid bilayer coating technologies enable superior passivation performance to BSA protein coatings [8] and other polymeric options [9]. Combined with recent material advances in lipid nanoparticle fabrication for lipid bilayer coatings [10] in diagnostic application settings [11,12] and in post-fabrication repair [13], there is outstanding potential to explore lipid bilayer coatings for enhanced ELISA measurement capabilities.
Herein, we report the development of a streamlined ELISA measurement strategy in which lipid nanoparticles composed of zwitterionic phospholipids [14] are used as the blocking agent and can be added together with the probe molecule to the ELISA plate. This strategy relies on lipid bilayer nanostructures as templates to form effective lipid bilayer coatings and we demonstrate its applicability to coronavirus disease-2019 (COVID-19) antibody detection in saline and human serum. Key advantages of this new approach include fewer processing steps, quicker protocol time, and superior detection performance compared to conventional BSA blocking.
2. Materials and methods
2.1. Reagents
The ELISA Starter Kit (catalog no. K0331002) was obtained from Koma Biotech Inc. (Seoul, Republic of Korea) and stored at 4 °C until use. The kit included the 96-well, plastic ELISA plates, 50 mM carbonate coating buffer (pH 9.6), 1% BSA blocking solution in phosphate-buffered saline (PBS, pH 7.4), Tween-20 stock solution, 3,3′,5,5′-tetramethylbenzidine (TMB) solution, and 2 M H2SO4 stop solution. 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) phospholipids in chloroform were purchased from Avanti Polar Lipids (Alabaster, AL), and lauric acid (LA), normal human serum (NHS), and other reagents were obtained from Sigma-Aldrich (St. Louis, MO). Lipid nanoparticles were prepared in PBS (pH 7.4) and PBS with 0.05% Tween-20 (PBS-T) was used for washing steps. Recombinant COVID-19 nucleocapsid (N) protein antigen (catalog no. LIC-NP-04) and monoclonal primary antibody (clone 12D12), which binds to the COVID-19 N protein (referred to as anti-N antibody), in 10 mM PBS were manufactured and provided by LUCA AICell, Inc. (Anyang, Republic of Korea). A goat anti-human IgG secondary antibody with horseradish peroxidase (HRP) conjugate (catalog no. A18811) was obtained from Thermo Fisher Scientific (Waltham, MA).
2.2. Lipid nanoparticle preparation
Lipid nanoparticles (LNPs) were prepared by freeze-thaw-vortex processing, as previously described [15]. Dried films of DOPC lipid were hydrated in PBS only or in PBS containing LA in order to prepare DOPC and DOPC/LA lipid nanoparticles, respectively. The stock DOPC and LA concentrations were 1 and 0.5 mM, respectively, and hence the DOPC/LA molar ratio was 2. For both DOPC and DOPC/LA LNPs, the hydrated samples were subjected to 5 cycles of the following steps: 1 min exposure to liquid nitrogen; 5 min thaw in a water bath at 60 °C; and 30 s vortexing. The processed LNPs were then diluted in PBS to a final concentration of 0.063 mM DOPC lipid, which was used for experiments. This preparation protocol results in LNPs with a relatively broad size distribution around ~50–500 nm and the DOPC and DOPC/LA LNPs mainly consist of lamellar-phase vesicular and bicellar nanostructures, respectively [16,17]. In the DOPC/LA LNP case, the bicellar nanostructures are in equilibrium with LA monomers and/or micelles in the bulk solution as well [15].
2.3. ELISA measurements
Depending on the protocol, a 100 µL volume of 5 µg/mL N antigen in coating buffer (pH 9.6) was added to the ELISA plate wells and incubated at 37 °C for 1 h, or a 100 µL volume of 5 µg/mL N antigen together with 1% BSA or DOPC or DOPC/LA lipid nanoparticles (LNPs; fixed at 0.063 mM DOPC lipid concentration in the LNP cases) in PBS (pH 7.4) was added to the ELISA plate wells and incubated at 37 °C for 15 min. Then, each well was washed 5 times with 200 µL of PBS-T. In applicable cases, an additional blocking step was performed with 1% BSA or DOPC or DOPC/LA lipid nanoparticles (fixed at 0.063 mM DOPC lipid concentration) in PBS (pH 7.4) and incubated at 37 °C for 15 min or 1 h, followed by washing 5 times with 200 µL of PBS-T. A 100 µL volume of primary antibody in PBS-T or NHS was then incubated with the antigen-coated plates at 37 °C for 1 h, followed by washing 5 times with 200 µL of PBS-T. Next, 100 µL of the secondary antibody (1:20,000 dilution) in PBS-T was incubated with the antigen-coated plates at 37 °C for 1 h, followed by washing 5 times with 200 µL of PBS-T. Afterwards, 100 µL of TMB solution was added and incubated at room temperature for ~5 min, followed by adding 100 µL of stop solution. The absorbance was measured at 450 nm wavelength using a SpectraMax iD5 microplate reader (Molecular Devices, San Jose, CA). All reported data were normalized by subtracting the background signal corresponding to the same protocol run with no primary antibody (replaced by an incubation step in PBS or 1% NHS alone). The latter experiments served as the negative control and accounted for the potential nonspecific adsorption of secondary antibody onto the ELISA well plate (after the surface had been incubated in saline or serum as applicable).
2.4. Statistical analyses
Measurement data were compared by one-way analysis of variance (ANOVA) with Tukey's multiple comparisons test (versus BSA, indicated by *) and reported as multiplicity-adjusted P values. P < 0.05, P < 0.01, and P < 0.001 indicate the levels of statistical significance.
3. Results and discussion
3.1. Experimental strategy
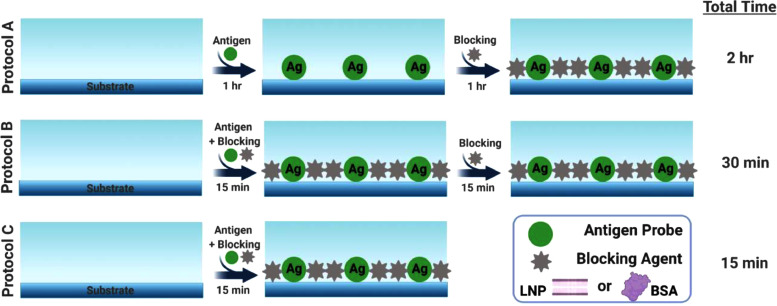
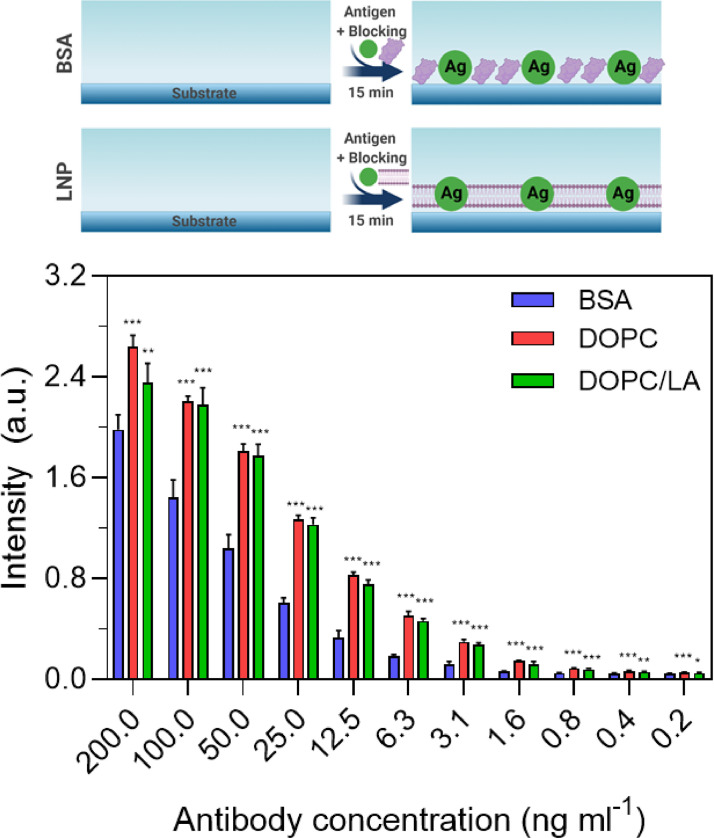
Fig. 1 presents the design strategy to develop a rapid ELISA measurement format and outlines three experimental protocols. Protocol A involves a classical design whereby the antigen coating step takes place first at 37 °C for 1 h, followed by adding the blocking agent at 37 °C for 1 h. Protocol B involves two distinct modifications, whereby the antigen and blocking agent are mixed together and added simultaneously at 37 °C while the incubation time is reduced from 1 hr to 15 min. An additional incubation step with blocking agent is then performed at 37 °C for 15 min. Protocol C is an abbreviated form of Protocol B and solely consists of adding the antigen and blocking agent together at 37 °C for 15 min.
Fig. 1.
Experimental protocols for developing rapid ELISA protocols based on replacing BSA proteins with lipid nanoparticles (LNPs) in order to reduce the number of processing steps and shorten the overall time for platform fabrication. All three protocols were tested with BSA proteins and DOPC or DOPC/LA LNPs as the blocking agent.
For the experiments, we utilized recombinant COVID-19 N protein as the model antigen in order to detect anti-N antibody, which is a widely used and sensitive biomarker for diagnostic purposes [18,19]. An HRP-conjugated secondary antibody was then added to generate the measurement readout. Three different types of blocking agents were tested [20]: (1) 1% BSA protein; (2) DOPC LNPs with 0.063 mM DOPC lipid concentration; and (3) DOPC/LA LNPs with 0.063 mM DOPC lipid concentration. For each protocol, we comparatively discuss the measurement results for detecting 0.2–200 ng/mL anti-N antibody in PBS (pH 7.4) in terms of the different blocking agents as described below.
3.2. Comparative evaluation of blocking agents
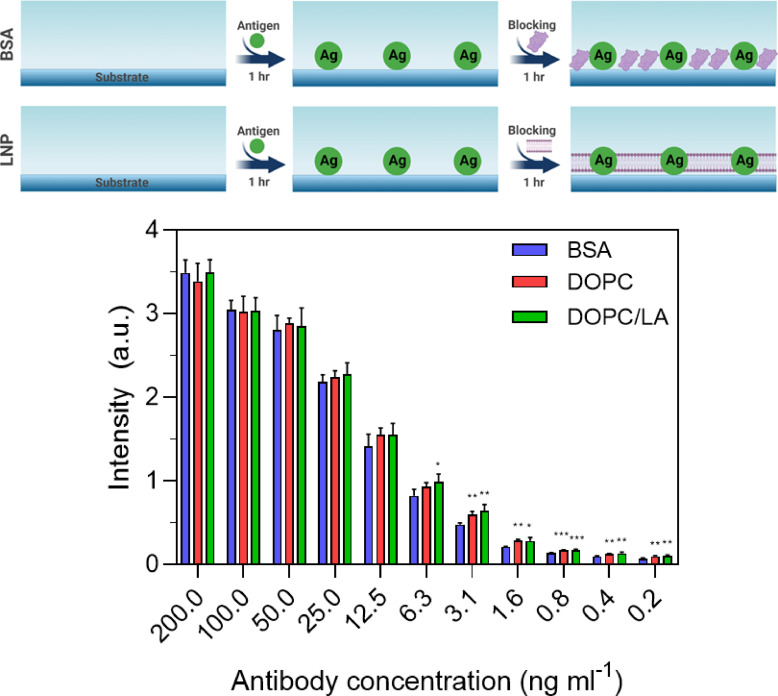
For Protocol A, the ELISA measurement response varied according to the anti-N antibody concentration across the tested concentration range (Fig. 2 ). At 12.5 ng/mL and higher antibody concentrations, all three types of blocking agents showed comparable performance, as indicated by similar absorbance intensity values. On the other hand, at lower antibody concentrations, higher absorbance intensity values were recorded when the DOPC or DOPC/LA LNPs were used as compared to the results obtained with BSA blocking. Hence, the LNP blocking agents could be potentially useful for high-sensitivity antibody detection at low concentrations on account of the relatively larger measurement responses. In addition, linear regression analysis indicated that the goodness of fit (r2) values were 0.70, 0.66, and 0.68 in the case of coatings formed from BSA, DOPC LNP, and DOPC/LA LNP blocking agents, respectively.
Fig. 2.
ELISA measurement responses for Protocol A. The absorbance intensity values are reported for anti-N antibody detection in PBS as a function of antibody concentration and for three different blocking strategies: BSA protein and DOPC or DOPC/LA lipid nanoparticles. Data are reported as mean ± standard deviation from n = 4 measurements.
For Protocol B, the ELISA measurement response again varied according to the anti-N antibody concentration across the tested concentration range (Fig. 3 ). Notably, in this protocol, the LNP blocking agents consistently yielded larger measurement responses than with BSA blocking. In some cases, there was a 2-fold greater or more increase in the measurement response due to replacing BSA with LNPs. This finding supports that the LNPs more readily form passivation coatings on the plate well surface while also facilitating high antigen uptake. By contrast, both BSA and the antigen are protein-based molecules and likely competitively bind to the surface through adsorption-related unfolding processes [21,22], which can diminish the binding signal. In addition, linear regression analysis indicated that the goodness of fit (r2) values were 0.96, 0.88, and 0.84 in the case of coatings formed from BSA, DOPC LNP, and DOPC/LA LNP blocking agents, respectively.
Fig. 3.
ELISA measurement responses for Protocol B. The absorbance intensity values are reported for anti-N antibody detection in PBS as a function of antibody concentration and for three different blocking strategies: BSA protein and DOPC or DOPC/LA lipid nanoparticles. Data are reported as mean ± standard deviation from n = 4 measurements.
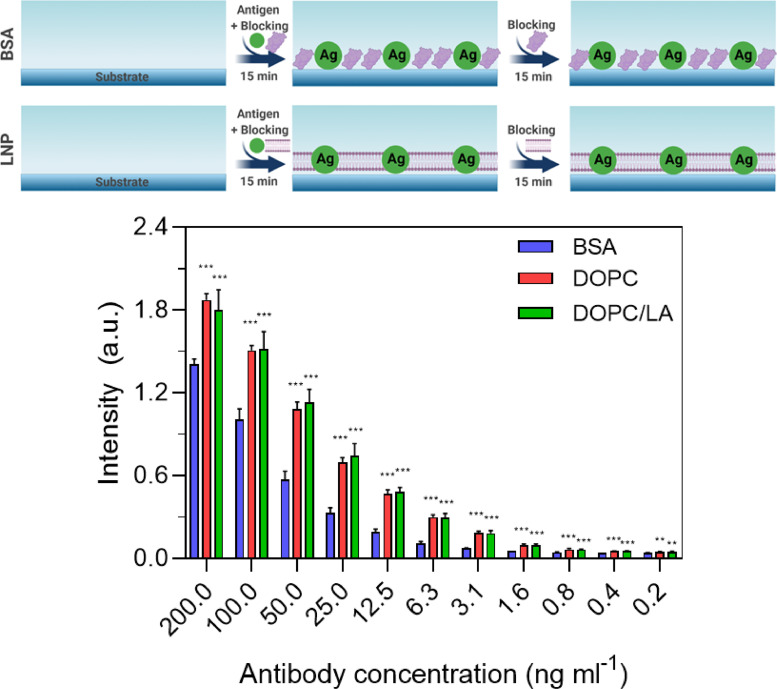
Similar results were also obtained with Protocol C and the LNP blocking agents outperformed BSA blocking across the tested antibody concentration range (Fig. 4 ). It should also be noted that the magnitudes of the absorbance intensity values were relatively larger than those observed in Protocol B, supporting that co-mixing the antigen and blocking agent for the addition step was sufficient to achieve high binding signals. In addition, linear regression analysis indicated that the goodness of fit (r2) values were 0.91, 0.80, and 0.75 in the case of coatings formed from BSA, DOPC LNP, and DOPC/LA LNP blocking agents, respectively.
Fig. 4.
ELISA measurement responses for Protocol C. The absorbance intensity values are reported for anti-N antibody detection in PBS as a function of antibody concentration and for three different blocking strategies: BSA protein and DOPC or DOPC/LA lipid nanoparticles. Data are reported as mean ± standard deviation from n = 4 measurements.
Together, these findings support that LNPs are effective replacements for BSA in conventional ELISA protocols and facilitate the development of streamlined protocols with a reduced number of steps and decreased processing time. Furthermore, the LNP blocking agents enabled higher sensitivity detection of low antibody concentrations.
3.3. Antibody detection in human serum
Using the three ELISA protocols and various blocking options, we proceeded to evaluate anti-N antibody detection in human serum. A normal human serum (NHS) specimen was spiked with 20 µg/mL anti-N antibody, which is within the range of clinically feasible conditions [23] for an individual infected with COVID-19 and was then diluted 1:100 fold and two-fold dilutions thereof for ELISA measurement (Fig. 5 ).
Fig. 5.
ELISA measurement responses for COVID-19 antibody detection in human serum. The data corresponds to (A) Protocol A, (B) Protocol B, and (C) Protocol C. The absorbance intensity values are reported for anti-N antibody detection in NHS as a function of antibody concentration and for three different blocking strategies: BSA protein and DOPC or DOPC/LA lipid nanoparticles. Data are reported as mean ± standard deviation from n=4 measurements.
For Protocol A, similar absorbance intensity values were recorded for 200 ng/mL antibody while antibody detection at lower concentrations was less discernable compared to the background signal. On the other hand, for Protocol B, the LNP blocking agents facilitated antibody-concentration-dependent signals down to 50 ng/mL antibody concentration, whereas appreciably smaller signals were observed in this case for BSA blocking and no signal was observed for 50 ng/mL antibody. Among the different protocols, the best performance was observed with Protocol C and the LNP blocking strategies yield relatively large measurement signals along with discernible antibody-concentration-dependent signals down to 25 ng/mL antibody concentration. Linear regression analysis indicated that the goodness of fit (r2) values were 0.81 and 0.76 in the case of coatings formed from DOPC LNP and DOPC/LA LNP blocking agents, respectively. In marked contrast, for Protocol C, BSA blocking was ineffective and there were nearly negligible measurement signals even at 200 ng/mL antibody concentration. Together, these results indicate that the streamlined ELISA protocol in combination with LNP blocking agents enabled superior anti-N antibody quantification in human serum compared to conventional ELISA with BSA blocking.
Of note, for the serum experiments, an additional series of negative control experiments were run, in which case there was no antigen on the surface and then anti-N and secondary antibodies were added sequentially following the same protocol in all other respects. The resulting absorbance intensity values (raw data) were around ~0.3 and ~0.2 for the coatings formed from BSA and LNP blocking agents, respectively. This finding is consistent with the ELISA testing results and supports that the lipid bilayer coatings outperform the protein coatings to prevent nonspecific adsorption of the antibodies in serum conditions.
4. Conclusions and outlook
In this study, we have demonstrated that LNP technology can enable the development of effective lipid bilayer blocking agents for ELISA applications. While protein-based blocking agents are the current standard in the field, our findings show that lipid bilayer coatings can be advantageous in terms of reducing the number of processing steps and time while also yielding superior detection capabilities in saline and human serum conditions. These findings are particularly important because there has been debate in the literature about whether blocking agents are necessary, especially when PBS-T is used for washing steps [24,25], and our findings support that blocking agents are in fact advantageous and advanced materials design can lead to improved ELISA detection performance. The ongoing COVID-19 pandemic has further highlighted the potential of LNP technologies for vaccine delivery applications [26] and there are numerous additional possibilities for LNPs in terms of coating applications. Such promise is aided by recent progress to develop LNPs using simple and scalable techniques such as freeze-thaw-vortex cycling, which we and others regularly use to prepare bicellar and vesicular nanostructures with the DOPC/LA and DOPC lipid compositions, respectively. The findings in this study establish proof-of-concept for a new ELISA format in terms of co-mixing the probe molecule and blocking agent during the addition process and with respect to utilizing lipid nanoparticles as blocking agents. Further optimization of these protocols can be explored for various ELISA types and solution conditions along with finetuning parameters such as the probe-to-lipid ratio, and can potentially open the door to a new class of molecular diagnostics.
Declaration of Competing Interest
N.-J.C. and J.A.J. are inventors on patents and patent applications related to supported lipid bilayer coatings, including U.S. patent no. 10,427,124, U.S. patent no. 10,787,470, and PCT patent no. US2019/047518. N.J.C. is a founder of, K.Y.Y. and S.H.L. are employees of, and J.A.J. is a scientific advisor to LUCA Health and LUCA AICell Inc., which are developing lipid-related diagnostic and therapeutic technologies. The other authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
This work was supported by the National Research Foundation of Singapore through a Proof-of-Concept grant (NRF2015NRF-POC0001-19), and by the National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (2020R1C1C1004385). In addition, this work was supported by the Brain Pool Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science and ICT (2019H1D3A1A01070318) and by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2021R1I1A1A01056302). This work was also supported by the SKKU Research Fellowship Program of Sungkyunkwan University. Schematic illustrations were created with BioRender.com under an academic lab subscription.
Data availability
The raw data required to reproduce these findings are available from the corresponding authors on reasonable request.
References
- 1.Crowther J.R. first ed. Humana Press; New York: 2001. The Elisa Guidebook. [Google Scholar]
- 2.Hosseini S., Vázquez-Villegas P., Rito-Palomares M., Martinez-Chapa S.O. Enzyme-Linked Immunosorbent Assay (ELISA) Springer; Singapore: 2018. Advantages, disadvantages and modifications of conventional ELISA; pp. 67–115. [Google Scholar]
- 3.Lichtenberg J.Y., Ling Y., Kim S. Non-Specific Adsorption Reduction Methods in Biosensing. Sensors. 2019;11:2488. doi: 10.3390/s19112488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin P.-H., Li B.-R. Antifouling strategies in advanced electrochemical sensors and biosensors. Analyst. 2020;4:1110–1120. doi: 10.1039/c9an02017a. [DOI] [PubMed] [Google Scholar]
- 5.Steinitz M. Quantitation of the blocking effect of tween 20 and bovine serum albumin in ELISA microwells. Anal. Biochem. 2000;2:232–238. doi: 10.1006/abio.2000.4602. [DOI] [PubMed] [Google Scholar]
- 6.Sweryda-Krawiec B., Devaraj H., Jacob G., Hickman J.J. A new interpretation of serum albumin surface passivation. Langmuir. 2004;6:2054–2056. doi: 10.1021/la034870g. [DOI] [PubMed] [Google Scholar]
- 7.Xiao Y., Isaacs S.N. Enzyme-linked immunosorbent assay (ELISA) and blocking with bovine serum albumin (BSA)—not all BSAs are alike. J. Immunol. Methods. 2012;1-2:148–151. doi: 10.1016/j.jim.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Persson F., Fritzsche J., Mir K.U., Modesti M., Westerlund F., Tegenfeldt J.O. Lipid-based passivation in nanofluidics. Nano Lett. 2012;5:2260–2265. doi: 10.1021/nl204535h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belling J.N., Heidenreich L.K., Park J.H., Kawakami L.M., Takahashi J., Frost I.M., Gong Y., Young T.D., Jackman J.A., Jonas S.J. Lipid-bicelle-coated microfluidics for intracellular delivery with reduced fouling. ACS Appl. Mater. Interfaces. 2020;41:45744–45752. doi: 10.1021/acsami.0c11485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jackman J.A., Cho N.-J. Supported lipid bilayer formation: beyond vesicle fusion. Langmuir. 2020;6:1387–1400. doi: 10.1021/acs.langmuir.9b03706. [DOI] [PubMed] [Google Scholar]
- 11.Wu J.-C., Tseng P.-Y., Tsai W.-S., Liao M.-Y., Lu S.-H., Frank C.W., Chen J.-S., Wu H.-C., Chang Y.-C. Antibody conjugated supported lipid bilayer for capturing and purification of viable tumor cells in blood for subsequent cell culture. Biomaterials. 2013;21:5191–5199. doi: 10.1016/j.biomaterials.2013.03.096. [DOI] [PubMed] [Google Scholar]
- 12.Kim J., Kim S., Ahn J., Lee J., Nam J.M. A lipid-nanopillar-array-based immunosorbent assay. Adv. Mater. 2020;26 doi: 10.1002/adma.202001360. [DOI] [PubMed] [Google Scholar]
- 13.Chung M., Kim B., Won J.I. Post-assembly filling of supported lipid bilayers by soluble lipid derivatives. Bull. Korean Chem. Soc. 2016;8:1354–1359. [Google Scholar]
- 14.Schlenoff J.B. Zwitteration: coating surfaces with zwitterionic functionality to reduce nonspecific adsorption. Langmuir. 2014;32:9625–9636. doi: 10.1021/la500057j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sut T.N., Park S., Yoon B.K., Jackman J.A., Cho N.-J. Supported lipid bilayer formation from phospholipid-fatty acid bicellar mixtures. Langmuir. 2020;18:5021–5029. doi: 10.1021/acs.langmuir.0c00675. [DOI] [PubMed] [Google Scholar]
- 16.Kolahdouzan K., Jackman J.A., Yoon B.K., Kim M.C., Johal M.S., Cho N.-J. Optimizing the formation of supported lipid bilayers from bicellar mixtures. Langmuir. 2017;33:5052–5064. doi: 10.1021/acs.langmuir.7b00210. [DOI] [PubMed] [Google Scholar]
- 17.Sut T.N., Jackman J.A., Yoon B.K., Park S., Kolahdouzan K., Ma G.J., Zhdanov V.P., Cho N.-J. Influence of NaCl concentration on bicelle-mediated SLB formation. Langmuir. 2019;35:10658–10666. doi: 10.1021/acs.langmuir.9b01644. [DOI] [PubMed] [Google Scholar]
- 18.Burbelo P.D., Riedo F.X., Morishima C., Rawlings S., Smith D., Das S., Strich J.R., Chertow D.S., Davey Jr R.T., Cohen J.I. Sensitivity in detection of antibodies to nucleocapsid and spike proteins of severe acute respiratory syndrome coronavirus 2 in patients with coronavirus disease. J. Infect. Dis. 2019;2(2020):206–213. doi: 10.1093/infdis/jiaa273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ng D.L., Goldgof G.M., Shy B.R., Levine A.G., Balcerek J., Bapat S.P., Prostko J., Rodgers M., Coller K., Pearce S. SARS-CoV-2 seroprevalence and neutralizing activity in donor and patient blood. Nat. Commun. 2020;1:1–7. doi: 10.1038/s41467-020-18468-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma G.J., Ferhan A.R., Jackman J.A., Cho N.-J. Conformational flexibility of fatty acid-free bovine serum albumin proteins enables superior antifouling coatings. Commun. Mater. 2020;1:45. [Google Scholar]
- 21.Rabe M., Verdes D., Seeger S. Understanding protein adsorption phenomena at solid surfaces. Adv. Colloid Interface Sci. 2011;1-2:87–106. doi: 10.1016/j.cis.2010.12.007. [DOI] [PubMed] [Google Scholar]
- 22.Kastantin M., Langdon B.B., Schwartz D.K. A bottom-up approach to understanding protein layer formation at solid–liquid interfaces. Adv. Colloid Interface Sci. 2014:240–252. doi: 10.1016/j.cis.2013.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keshavarz B., Wiencek J.R., Workman L.J., Straesser M.D., Muehling L.M., Canderan G., Drago F., Bonham C.A., Sturek J.M., Ramani C., McNamara C.A., Woodfolk J.A., Kadl A., Platts-Mills T.A.E., Wilson J.M. Quantitative measurement of IgG to severe acute respiratory syndrome coronavirus-2 proteins using immunocap. Int. Arch. Allergy Immunol. 2021;5:417–424. doi: 10.1159/000514203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mohammad K., Esen A. A blocking agent and a blocking step are not needed in ELISA, immunostaining dot-blots and western blots. J. Immunol. Methods. 1989;1:141–145. doi: 10.1016/0022-1759(89)90129-4. [DOI] [PubMed] [Google Scholar]
- 25.Ahirwar R., Bariar S., Balakrishnan A., Nahar P. BSA blocking in enzyme-linked immunosorbent assays is a non-mandatory step: a perspective study on mechanism of BSA blocking in common ELISA protocols. RSC Adv. 2015;121:100077–100083. [Google Scholar]
- 26.Igyártó B.Z., Jacobsen S., Ndeupen S. Future considerations for the mRNA-lipid nanoparticle vaccine platform. Curr. Opin. Virol. 2021:65–72. doi: 10.1016/j.coviro.2021.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw data required to reproduce these findings are available from the corresponding authors on reasonable request.