Key Words: astrocytes, glial cells, label-free proteomic analysis, mechanism, neurons, primary culture, reprogramming, spinal cord injury
Abstract
Our previous study has confirmed that astrocytes overexpressing neurogenic differentiation factor 1 (NEUROD1) in the spinal cord can be reprogrammed into neurons under in vivo conditions. However, whether they can also be reprogrammed into neurons under in vitro conditions remains unclear, and the mechanisms of programmed conversion from astrocytes to neurons have not yet been clarified. In the present study, we prepared reactive astrocytes from newborn rat spinal cord astrocytes using the scratch method and infected them with lentivirus carrying NEUROD1. The results showed that NEUROD1 overexpression reprogrammed the cultured reactive astrocytes into neurons in vitro with an efficiency of 13.4%. Using proteomic and bioinformatic analyses, 1952 proteins were identified, of which 92 were differentially expressed. Among these proteins, 11 were identified as candidate proteins in the process of reprogramming based on their biological functions and fold-changes in the bioinformatic analysis. Furthermore, western blot assay revealed that casein kinase II subunit alpha (CSNK2A2) and pinin (PNN) expression in NEUROD1-overexpressing reactive astrocytes was significantly increased, suggesting that NEUROD1 can directly reprogram spinal cord-derived reactive astrocytes into neurons in vitro, and that the NEUROD1-CSNK2A2-PNN pathway is involved in this process. This study was approved by the Animal Ethics Committee of Fujian Medical University, China (approval No. 2016-05) on April 18, 2016.
Chinese Library Classification No. R446; R741; Q811.4
Introduction
Spinal cord injury (SCI) is a common but devastating trauma of the central nervous system, and it has a severe socioeconomic burden worldwide. Glial cells, including astrocytes, microglia, and oligodendrocytes, proliferate in response to SCI and form a reactive glial scar (García et al., 2019). This glial scar impairs neuronal circuits, albeit also restricting the spread of inflammatory reactions (Cizkova et al., 2014; Liu et al., 2020; Oh et al., 2020). Neuronal regeneration after SCI is recognized as a notable challenge for spinal cord repair. Efforts have been made to use cell replacement therapy to generate neurons; for example, exogenous cells derived from embryonic stem cells or induced pluripotent stem cells have been trialed (Takahashi and Yamanaka, 2006; Takahashi et al., 2007; Sahni and Kessler, 2010). However, despite great potential, these cell transplantation approaches have been hampered by immunorejection, tumorigenesis, and differentiation uncertainty (Lee et al., 2013; Lukovic et al., 2014). Direct cell reprogramming, which is the process of converting cells from one cell type to another, may be applied as a regenerative therapy for SCI, allowing cell replacement in vivo. Lineage-specific transcription factors play an important role in cell reprogramming. Several studies have indicated that astrocytes in the mouse brain can be reprogrammed into neurons both in vitro and in vivo (Guo et al., 2014; Liu et al., 2015b; Brulet et al., 2017). Moreover, our pilot study has demonstrated that astrocytes in the rat spinal cord can be reprogrammed into neurons in vivo (Chen et al., 2017) by the overexpression of a single neurogenic transcription factor, neurogenic differentiation factor 1 (NEUROD1), which belongs to the basic helix-loop-helix family. Zhang et al. (2015) have reprogrammed human midbrain astrocytes into neurons both in vitro and in vivo by applying a cocktail of small molecules, although they failed to do so using human spinal cord astrocytes. More recently, Matsuda et al. (2019) demonstrated that NEUROD1 upregulation can convert microglia into neurons in mice with 25–35% efficiency; these authors also explored epigenetic remodeling during neuronal reprogramming. However, the underlying mechanisms of programmed conversion from astrocytes to neurons remain largely unknown. We hypothesized that several proteins might play a leading role in this reprogramming. A proteomic approach can be used to unravel the novel processes underlying reprogramming and regeneration.
The aims of the present study were therefore to reprogram rat spinal cord astrocytes into neurons in vitro via NEUROD1 overexpression, and to reveal the key proteins involved in NEUROD1-induced reprogramming from astrocytes to neurons using quantitative proteomic and bioinformatic approaches.
Materials and Methods
Animals
All efforts were made to minimize animal suffering. For the in vitro studies, we used 20 sex-unspecified wild-type Sprague-Dawley rats at postnatal days 1–3 (obtained from the Animal Experimental Center of Fujian Medical University, China, license No. SYXK (Min) 2012-0001). All experiments were carried out according to the Animal Experimental Guidelines of Fujian Medical University, which comply with the National Institutes of Health Guide for the Care and Use of Laboratory Animals, and were approved by the Animal Ethics Committee of Fujian Medical University, China (approval No. 2016-05) on April 18, 2016.
Primary astrocyte isolation and culture
Primary cultures of astrocytes were obtained from the spinal cords of newborn rats (postnatal days 1–3) using a previously published protocol (Yang et al., 2006). The animals were anesthetized by inhalation of 5% ketamine. After being sterilized, the rat spinal cords were carefully dissected using aseptic techniques. The animals were then sacrificed by cervical dislocation under anesthesia. The meninges and vessel tissues were stripped from the spinal cords in phosphate-buffered saline (PBS; Beyotime, Wuhan, China; Cat# C0221A). The spinal cords were then triturated and digested in trypsin (0.25%; Beyotime; Cat# C0201) at 37°C for 10 minutes. The supernatant was filtered after centrifugation. Next, the spinal cord tissue was again digested in trypsin under the aforementioned conditions, and then underwent centrifugation again. Finally, the cell pellet was suspended in Dulbecco’s modified Eagle medium/F12 (Hyclone, Waltham, MA, USA; Cat# SH30023.01) with 10% fetal bovine serum (Hyclone; Cat# SV30087.03), and cultured at 37°C, 5% CO2 , until the cells reach the required numbers. We renewed the medium every 2–3 days.
Preparation of reactive astrocytes by scratch insult
In SCI, astrocytes undergo a reactive response to injury and form glial scars, thus preventing neuronal growth (Guo et al., 2014). Reactive astrocytes can be obtained using a scratch insult technique (Yang et al., 2009). After the insult, the astrocytes convert to reactive astrocytes, which have larger soma, longer processes, and more connections between one another. When cultured astrocytes reached 85–90% confluence, an aseptic 10 μL pipette was used to scratch the confluent monolayers longitudinally and latitudinally at right angles to one another, with a 0.5 cm interval. Uniform speed and strength were maintained while scratching. After removing debris, the cells were cultured for a further 7 days. We used fresh maintenance medium (Dulbecco’s modified Eagle medium/F12 with 10% fetal bovine serum) to replace half of the medium every 3 days.
Identification of reactive astrocytes using immunofluorescence staining
For the detection of nestin-positive cells, cultured cells were fixed using 4% paraformaldehyde for 30 minutes (BBI Life Sciences, Shanghai, China; Cat# E672002-0500). After three rinses with PBS, the cells were permeabilized with 0.3% Triton X-100 (Beyotime; Cat# P0096) in PBS for 20 minutes, followed by three PBS washes. We then used blocking solution (5% bovine serum albumin; Odyssey, Xuzhou, China; Cat# 927-40000) to block the cells for 30 minutes at 20°C. Next, cells were incubated at 4°C overnight with monoclonal anti-nestin antibody (rabbit anti-mouse; 1:300; Thermo Fisher Scientific, Waltham, MA, USA; Cat# 14-5843-82), which is a marker of reactive astrocytes (Yang et al., 2006). After three 5-minute washes with PBS, the cells were incubated with secondary antibody (goat anti-rabbit IgG H&L (Alexa Fluor Plus 488); 1:200; Thermo Fisher Scientific; Cat# A32723) at 37°C for 1 hour, followed by 4′,6-diamidino-2-phenylindole (DAPI; 1:100; Beyotime; Cat# C1002) to stain the nuclei. Stained cells were visualized using a fluorescence microscope (Zeiss Axiovert 200M; Zeiss, Heidenheim, Germany).
Neuronal induction
We used lentivirus vectors to induce reprogramming from reactive astrocytes to neurons. The lentivirus was purchased from Hanbio Biotechnology Co., Ltd., Shanghai, China. Cultured reactive astrocytes were divided into three groups: the non-virus (NV) group, the green fluorescent protein (GFP) group, and the NEUROD1 group. The NV group was not infected with lentivirus. The GFP group was infected with lentivirus carrying GFP but not carrying any specific gene, and was used as the negative control. The NEUROD1 group was infected with lentivirus carrying both GFP and NEUROD1 genes. After 24 hours in lentivirus-containing medium, the culture medium was replaced by a medium suitable for neuronal growth, which consisted of Neurobasal medium (Gibco, Waltham, MA, USA, Cat# 21103049), B27 (Gibco; Cat# 17504044), NT3L-glutamine (BBI Life Sciences; Cat# E607004-0100), and penicillin/streptomycin/fungizone (Beyotime; Cat# C0222). The cells were cultured for 14 days. During the culture period, the medium was changed every 2–3 days.
Verifying neuronal induction
An experienced pathologist visualized the stained cells under a fluorescence microscope at 48 hours after infection. Green fluorescence was visualized if the infection was successful. Furthermore, total RNA was isolated following the manufacturer’s instructions (BBI Life Sciences; Cat# B518621). We used a spectrophotometer (Thermo Fisher Scientific, Bremen, Germany) to assess the RNA quality of samples. Next, we performed reverse transcription-polymerase chain reaction to verify the expression of the NEUROD1 gene (primer sequence: 5′-CCT TGC TAC TCC AAG ACC CAG A-3′). Glyceraldehyde-3-phosphate dehydrogenase was used as an endogenous control, to normalize samples (primer sequence: 5′-GTC TCC TCT GAC TTC AAC AGC G-3′).
Immunocytochemistry
At 7 days after infection, cells were fixed with 4% paraformaldehyde for further immunostaining studies. After three rinses with PBS, the cells were permeabilized with 0.3% Triton X-100 in PBS for 20 minutes, and then blocked with blocking solution (5% bovine serum albumin) for 30 minutes at room temperature. Next, cells were incubated at 4°C overnight with anti-doublecortin (DCX) antibody (rabbit anti-mouse; 1:300; Sigma, Darmstadt, Germany; Cat# SAB2501666), a marker of immature neurons (Guo et al., 2014). DAPI (Beyotime, Cat# C1002, 1:100, 5 mg/mL) was used to stain nuclei. The percentage of NEUROD1-transduced astrocytes (number of DCX-positive cells/total number of cells × 100) was calculated under a fluorescence microscope. An experienced pathologist randomly chose five fields and counted the number of positive cells per field. Cell counting was managed using Image-Pro Plus 6.0 (Media Cybernetics Co., Ltd., Silver Spring, MD, USA).
Proteomic analysis
At 7 days after infection, we extracted two protein samples from the GFP and NEUROD1 groups using lysis buffer (4% sodium dodecyl sulfate, 0.1 M dithiothreitol, 0.1 M Tris; pH 7.6). The protein was denatured at 95°C for 5 minutes, and then centrifuged at 12,000 × g for 30 minutes. Next, the supernatants were collected and protein concentration was determined by tryptophan fluorescence assay, as previous described (Wiśniewski, 2013). We used a filter-assisted sample preparation procedure to prepare the peptides (Wiśniewski et al., 2010; Chen et al., 2015). Experiments were performed on an Orbitrap Fusion mass spectrometer coupled with an EASY-nLC (Thermo Fisher Scientific). We loaded the total peptides onto a fused silica column with 3-μm ReproSil-Pur C18 beads (120 Å; Dr. Maisch GmbH, Ammerbuch, Germany), and they were separated with a 90-minute gradient at a flow rate of 300 nL/min. The parameters for the mass spectrum (MS)1 survey scan in the Orbitrap were 350–1500 m/z at a resolution of 60,000, with an automatic gain control (AGC) target of 400,000 and a maximum injection time of 50 ms. Precursor ions were filtered using monoisotopic precursor selection, charge state, and dynamic exclusion. The most intense precursors were subjected to higher-energy collisional dissociation fragmentation with a duty cycle of 3 seconds. The MS2 parameters were set as follows: 30% normalized collision energy, 15,000 resolution, 50,000 AGC target, 100 ms maximum injection time, and 1.6 m/z isolation width.
Bioinformatic analysis of the proteomic data
We analyzed the MS raw data using MaxQuant 1.6.0.1 software http://maxquant.org/). Carbamidomethyl cysteine was searched as a fixed modification, while N-acetylation of protein and oxidized methionine were set as variable modifications. We searched the MS and MS/MS peptide data against the UniProt database (release date: August 2019) to identify proteins. Label-free quantification (LFQ) was performed using MaxQuant as described previously (Cox et al., 2009). Protein abundance calculations were based on the normalized spectral protein intensity (LFQ intensity). We used the two-tailed Student’s t-test to calculate the P-value of log2 LFQ intensity for each protein between the GFP and NEUROD1 groups using the Perseus program in MaxQuant. Protein–protein interactions (PPI) and related biological processes were predicted using the STRING database. To search for more potential interactions, 0.4 was chosen as the medium confidence score. We reconstructed the PPI network using Cytoscape 3.6.1 software (https://cytoscape.org/).
Western blot to validate key proteins involved in reprogramming
After being infected by lentivirus for 14 days, reactive astrocytes in the GFP and NEUROD1 groups were lysed using radioimmunoprecipitation assay buffer (containing both protease and phosphatase inhibitors) for 30 minutes at 4°C, and then centrifuged at 12,000 × g for 15 minutes. The supernatant was collected for gel analysis. Next, we separated the lysates using sodium dodecyl sulfate-polyacrylamide gel electrophoresis on a 7.5% gel, and the lysates were then transferred onto a polyvinylidene difluoride membrane. Blocking was performed for 1 hour at room temperature with 10% fetal bovine serum in PBS. For the primary antibody incubation, antibodies including anti-casein kinase II subunit alpha (CSNK2A2; 1:1000; Invitrogen, Waltham, MA, USA; Cat# MA5-17062) and anti-pinin (PNN; 1:1000; Invitrogen; Cat# PA5-44563) were added to PBS with 0.05% Triton X-100, and the membrane was incubated overnight at 4°C. Next, the membrane was incubated with goat anti-rabbit IgG-horseradish peroxidase (1:2000; Abcam; Cat# ab6721) for 1 hour at 20°C. Signals were then detected on an Odyssey imaging system (LI-COR Biosciences, Lincoln, NE, USA).
Statistical analysis
All data were collected and processed using SPSS 23.0 (IBM, Armonk, NY, USA). The P-values for pairwise comparisons were calculated using the unpaired Student’s t-test and the Wilcoxon rank-sum test. For multiple comparisons, one-way analysis of variance was used, and further pairwise comparisons were carried out using the least significant difference test. Pearson’s chi-squared test or Fisher’s exact probability method was used for rate comparisons. A P-value less than 0.05 was considered to be statistically significant. We performed a post hoc power analysis for our hypothesis tests.
Results
Morphology and identification of primary astrocytes
The astrocyte-like cells multiplied rapidly and formed a confluent monolayer. Flat oligodendrocyte-like cells and microglia were rare. At 7 days after the scratch insult, almost all cells were immunopositive for nestin (Figure 1A).
Figure 1.
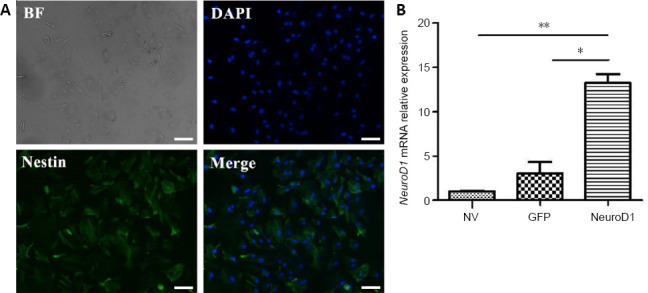
Identification of reactive astrocytes.
(A) Nestin (green, stained by Alexa Fluor Plus 488) immunopositivity detected by immunofluorescence staining. Nestin is a marker of reactive astrocytes. Scale bars: 5 µm. (B) NEUROD1 mRNA expression (relative to the NV group) detected by reverse transcription-polymerase chain reaction. Data are expressed as the mean ± SD. The experiment was repeated three times. *P < 0.05, **P < 0.01 (unpaired Student’s t-test). BF: Bright field; DAPI: 4′,6-diamidino-2-phenylindole; GFP: green fluorescent protein; NEUROD1: neurogenic differentiation factor 1; NV: non-virus group.
Neuron induction verified by NEUROD1 gene expression
After 72 hours of incubation with the NEUROD1 lentivirus, we carried out reverse transcription-polymerase chain reaction to verify the expression of NEUROD1. As shown in Figure 1B, NEUROD1 mRNA expression was significantly higher in the NEUROD1 group than in the GFP or NV groups (P < 0.05).
Reprogramming efficiency of the NEUROD1 lentivirus in primary astrocytes
At 7 days after infection by the NEUROD1 lentivirus, the percentage of reprogrammed neurons was investigated using immunocytochemistry. The percentage of NEUROD1-transduced astrocytes in the NEUROD1 group was markedly higher than those in the GFP and NV groups (P = 0.036 and P = 0.0006, respectively; Additional Table 1 and Figure 2). This finding indicates that spinal cord reactive astrocytes can be reprogrammed into neurons in vitro using NEUROD1, with an average efficiency of 13.4%.
Additional Table 1.
The percentage of DCX-positive cells in each group
| Groups | DCX-positive cells (%) | ||||
|---|---|---|---|---|---|
| NV | 2.6 | 0.3 | 5.1 | 1.7 | 0.3 |
| GFP | 1.0 | 0.9 | 2.7 | 9.6 | 1.8 |
| Neurod1 | 16.9 | 8.9 | 11.6 | 18.8 | 10.7 |
|
| |||||
| P-value | NV vs. NeuroD1 | 0.0006** | |||
| GFP vs. NeuroD1 | 0.0036** | ||||
| NV vs. GFP | 0.5367 | ||||
**P < 0.01, Power = 0.88. DCX: Doublecortin; GFP: green fluorescent protein; NeuroD1: neurogenic differentiation factor 1; NV: non-virus group.
Figure 2.
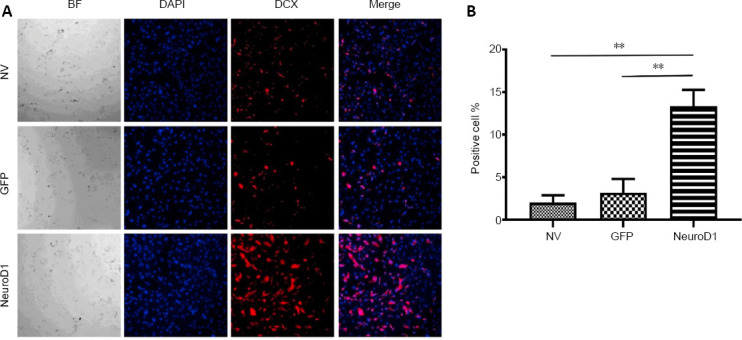
NEUROD1-induced reactive astrocytes at 7 days after infection (immunofluorescence staining).
(A) NEUROD1 overexpression led to significantly increased DCX immunopositivity (red). (B) Percentage of DCX positive cells. Scale bar: 10 µm. Data are expressed as the mean ± SD. The experiment was repeated three times. **P < 0.01 (unpaired Student’s t-test). BF: Bright field; DAPI: 4′,6-diamidino-2-phenylindole; DCX: doublecortin; GFP: green fluorescent protein; NEUROD1: neurogenic differentiation factor 1; NV: non-virus group.
Differentially expressed proteins in primary astrocytes with NEUROD1 lentivirus
Total protein from the cells was analyzed using nanoLC−MS/MS analysis. As shown in Table 1, 2814 proteins were identified across all samples, of which 1952 non-redundant proteins were identified. We evaluated the quality of the proteomic dataset and instrumental reproducibility. A box plot revealed that the medians of log2 LFQ intensity were at almost exactly the same level across all four samples, suggesting no biases toward any samples (Figure 3A). All of the correlation coefficients between each of two samples were higher than 0.95, indicating that the relative LFQ profiling was highly reproducible between any two samples (Figure 3B). After searching the dataset using cut-off values of P < 0.05 and fold change ≥ 1.5 (or ≤ 0.67), 92 proteins were identified as having significantly different expression between the two cohorts (Figure 3C). Unsupervised hierarchical clustering and correlation distance metrics were used to generate a heatmap of the 92 proteins (Figure 3D).
Table 1.
Identification of label-free quantitative proteomic analyses of astrocytes in the GFP and NEUROD1 groups
| Samples | MS | MS/MS | MS/MS identified | MS/MS identified (%) | Peptide sequences identified | Protein group |
|---|---|---|---|---|---|---|
| NEUROD1 group | ||||||
| Sample1 | 4341 | 51112 | 17167 | 41.61 | 14044 | 1715 |
| Sample2 | 4244 | 50626 | 17868 | 43.23 | 14640 | 1853 |
| GFP group | ||||||
| Sample1 | 4173 | 50215 | 16995 | 41.68 | 13941 | 1823 |
| Sample2 | 4206 | 51640 | 17197 | 41.35 | 14085 | 2068 |
GFP: Green fluorescent protein; MS: mass spectrum; NEUROD1: neurogenic differentiation factor 1.
Figure 3.
Bioinformatic analysis of differentially expressed proteins in primary astrocytes with NEUROD1 lentivirus.
(A) Box plots of average log2 protein intensity for each sample. (B) Correlation analysis between each two samples. Between each two samples, P < 0.05, r/R2 > 0.8. (C) Volcano plot of the differentially expressed proteins. (D) Heatmap of the significantly altered proteins. Red is increased and blue is decreased relative to the control group. A darker color represents a more significant fold-change. GFP: Green fluorescent protein; NEUROD1: neurogenic differentiation factor 1.
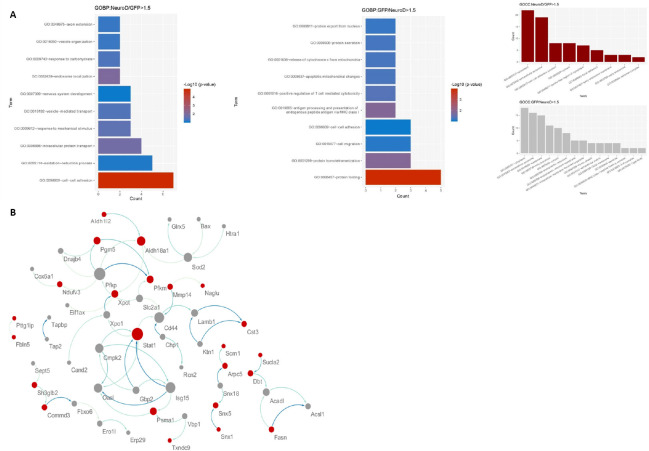
To investigate the biological significance of the differentially expressed proteins in neurons reprogrammed from reactive astrocytes by NEUROD1 overexpression, the cellular components and biological processes of the 92 proteins were explored by Gene Oncology Cellular Component (GOCC) and Gene Oncology Biological Process (GOBP) annotation using bioinformatic approaches (Figure 4A). To investigate the functional relationships among the 92 differentially expressed proteins between the NEUROD1 and GFP groups, a PPI network was developed based on STRING action scores (Figure 4B). Taking statistical significance in the form of P-values and fold change, GO annotation, and PPI network together, 11 proteins were selected as candidates (Table 2).
Figure 4.
Gene Oncology Cellular Component (GOCC), Gene Oncology Biological Process (GOBP) annotation, and protein-protein interaction analysis in primary astrocytes with NEUROD1 lentivirus.
To investigate the functional relationships among the 92 differentially expressed proteins between the NEUROD1 and GFP groups, a protein-protein interactions network was developed based on STRING action scores (A), GOBP, and GOCC. (B) Protein-protein interaction analysis. GFP: Green fluorescent protein; NEUROD1: neurogenic differentiation factor 1.
Table 2.
Protein candidates and corresponding genes
| Gene names | Protein names | Fold change | P | Related function |
|---|---|---|---|---|
| Csnk2a2 (B4F7A9) | Casein kinase II subunit alpha | 2.83 | 0.02 | GOBP: cerebral cortex development |
| Dclk1 (A0A0G2KB92) | Serine/threonine-protein kinase DCLK1 | 1.73 | 0.04 | GO:0007399~nervous system development/GO:0048675~axon extension |
| Stat1 (F1M9D6) | Signal transducer and activator of transcription 1-alpha/beta | 2.78 | 0.05 | GO:0098609~cell-cell adhesion |
| PNN (D3ZAY8) | Pinin, desmosome-associated protein | 1.8 | 0 | GO:0005737~cytoplasm |
| Xpo1 (Q80U96) | Exportin-1 | 0.64 | 0.02 | GO:0015030~Cajal body |
| Lima1 (F1LR10) | LIM domain and actin-binding protein 1 | 1.9 | 0.05 | GO:0098609~cell-cell adhesion |
| Myo5b (A0A0G2JVH1) | Unconventional myosin-Vb | 4.93 | 0.03 | GO:0032439~endosome localization |
| Lamtor1 (Q6P791) | Ragulator complex protein LAMTOR1 | 4.77 | 0.01 | GO:0070062~extracellular exosome |
| MapS1 (A0A0H2UHQ3) | Microtubule-associated protein 1S | 2.69 | 0.04 | GO:0007399~nervous system development |
| Sod2 (P07895) | Superoxide dismutase [Mn], mitochondrial | 0.64 | 0.01 | GO:0043209~myelin sheath |
| Cox6a1 (P10818) | Cytochrome c oxidase subunit 6A1, mitochondrial | 0.54 | 0.02 | GO:0043209~myelin sheath |
Validation of key proteins in the reprogramming process of primary astrocytes
The key proteins involving the reprogramming process were validated by western blot assay. CSNK2A2 is a kinase and PNN is the phosphorylation substrate of CSNK2A2 (Varjosalo et al., 2013; Boldt et al., 2016). Therefore, we chose to validate this pair of proteins from the 11 candidates listed in Table 2. Western blot analysis confirmed that both CSNK2A2 and PNN had significantly higher expression levels in the NEUROD1 group compared with the GFP group (P = 0.03 and P = 0.01, respectively; Figure 5). These results suggest that CSNK2A2 is probably involved in the NEUROD1-induced reprogramming from reactive astrocytes to neurons, likely by phosphorylating the substrate PNN.
Figure 5.

Western blot assay to validate the key proteins involved in the reprogramming process and to compare protein expression between the GFP and NEUROD1 groups.
Data are expressed as the mean ± SD. The experiment was repeated three times. *P < 0.05 (unpaired Student’s t-test). CSNK2A2: Casein kinase II subunit alpha; GAPDH: glyceraldehyde-3-phosphate dehydrogenase; GFP: green fluorescent protein; NEUROD1: neurogenic differentiation factor 1; PNN: pinin.
Discussion
In the present study, we reprogrammed cultured spinal cord-derived reactive astrocytes into neurons using NEUROD1 overexpression, with an average efficiency of 13.4%. We then used label-free proteomics to identify differentially expressed proteins between the cells overexpressing NEUROD1 and the control group. In total, 1952 proteins were identified, of which 92 had significantly altered expression. Of these, 11 proteins were identified as candidates to be involved in the reprogramming process, based on their biological functions and fold-changes in the bioinformatic analysis.
The conversion from one cell type to another can be achieved by the expression of lineage-specific transcription factors, and this technique has potential applications in regenerative therapy for SCI. A neurogenic basic helix-loop-helix transcription factor, NEUROD1, has been demonstrated to enable reprogramming from glial cells to neurons under a range of circumstances. Both Guo et al. (2014) and Brulet et al. (2017) have reported that reactive glial cells in the mouse cortex can be directly reprogrammed into neurons in vivo by overexpressing NEUROD1. Furthermore, Chen et al. (2020) successfully applied a NEUROD1 adeno-associated virus-based gene therapy for astrocyte-to-neuron conversion, for functional brain repair in an in vivo mouse model. In addition, our pilot study demonstrated that astrocytes in the rat spinal cord can be reprogrammed into neurons in vivo by overexpressing NEUROD1 (Chen et al., 2017). In in vitro studies, cultured astrocytes have also been reprogrammed into neurons using specific transcription factors. For example, Berninger et al. (2007) revealed that neurogenin-2 and Mash1 are able to reprogram astrocytes from the postnatal cerebral cortex. Moreover, Heinrich et al. (2010) reported that forced expression of neurogenin-2 directs cortical astroglia to become functional neurons; this was not only restricted to the postnatal stages, but was also achieved in terminally differentiated astroglia. Recently, more in vivo studies have reported that sex determining region Y-box 2 (SOX2) can reprogram astrocytes into neurons or neural progenitors in the spinal cord and brain (Su et al., 2014; Niu et al., 2015; Wang et al., 2016). However, whether astrocytes generated in the spinal cord can be reprogrammed into neurons via NEUROD1 expression remains largely unknown. In the present study, we illustrated that NEUROD1 upregulation can directly reprogram cultured spinal cord-derived reactive astrocytes into neurons.
Although NEUROD1 has been demonstrated to enable astrocytes to be reprogrammed into neurons both in vivo and in vitro, the mechanisms involved have not yet been clarified. Proteomics is one of the most promising methods for discovering the underlying mechanisms of SCI and identifying potential therapies (Liu et al., 2015a; Devaux et al., 2016; Didangelos et al., 2016; Tica et al., 2018). In our study, several proteins were identified that might play a significant role during neuronal reprogramming. CSNK2A2 is a catalytic subunit of a constitutively active serine/threonine–protein kinase complex that phosphorylates a large number of substrates, including PNN, in the process of astrocyte reprogramming. It has also been reported that CSNK2A2 is involved in the mechanisms underlying the neuroprotective effects of N-benzylhexadecanamide (Zhou et al., 2018). PNN is known to interact with CSNK2A2 (Varjosalo et al., 2013), and is reportedly expressed in neurons but not astrocytes (Hsu et al., 2011). Moreover, PNN has been demonstrated to play a key role in the reprogramming of human pluripotent stem cells (Kim et al., 2017). Therefore, CSNK2A2 is probably involved in the NEUROD1-induced reprogramming of reactive astrocytes to neurons, likely by phosphorylating the substrate PNN.
The current study is not without limitations. First, the sample size was relatively small. However, a post hoc power analysis showed acceptable power for each hypothesis test, supporting the reliability of our results. Second, it remains unknown whether CSNK2A2 is actually involved in the NEUROD1-induced reprogramming by phosphorylating PNN, and this should be addressed in future work. Third, although 11 proteins were identified as candidates in the bioinformatic analysis, only two were investigated further. In the future, western blots should be performed using the other nine candidate proteins, and functional studies should also be carried out. Furthermore, in future studies we will investigate the proteomic changes between the GFP group vs. only the DCX-positive cells in the NEUROD1 group because the majority of cells in this group did not appear to be reprogrammed.
In summary, this study demonstrated that NEUROD1 can directly reprogram cultured spinal cord-derived reactive astrocytes into neurons. Furthermore, we used a quantitative proteomic approach to investigate the mechanisms of this reprogramming from astrocytes to neurons, and identified several proteins that may play a significant role. However, it remains unclear whether the reprogrammed neurons are fully functional. In addition, more confirmatory studies are required to verify whether CSNK2A2 is involved in NEUROD1-induced reprogramming by phosphorylating PNN, and the phosphorylation site needs to be confirmed.
Additional file:
Additional Table 1: The percentage of DCX-positive cells in each group.
Footnotes
Conflicts of interest:The authors declare no competing interests.
Financial support:The study was supported by the Natural Science Foundation of Fujian Province, China, No. 2015J05153; Research Talents Training Project of Fujian Provincial Health Department, China, No. 2018-ZQN-29; and Joint Funds for the Innovation of Science and Technology of Fujian Province, China, No. 2018Y9002 (all to WHC). The funders had no roles in the study design, conduction of experiment, data collection and analysis, decision to publish, or preparation of the manuscript.
Institutional review board statement:The study was approved by the Animal Ethics Committee of Fujian Medical University (approval No. 2016-05) on April 18, 2016.
Copyright license agreement:The Copyright License Agreement has been signed by all authors before publication.
Data sharing statement:The mass spectrometry proteomics data were deposited into the ProteomeXchange Consortium64 via the PRIDE partner repository with identifier PXD016097.
Plagiarism check:Checked twice by iThenticate.
Peer review:Externally peer reviewed.
Open peer reviewer:Sanusi Mohammad Bello, King Faisal University, Saudi Arabia.
Funding:The study was supported by the Natural Science Foundation of Fujian Province, China, No. 2015J05153; Research Talents Training Project of Fujian Provincial Health Department, China, No. 2018-ZQN-29; and Joint Funds for the Innovation of Science and Technology of Fujian Province, China, No. 2018Y9002 (all to WHC).
P-Reviewer: Bello SM; C-Editor: Zhao M; S-Editors: Yu J, Li CH; L-Editors: Gardner B, Yu J, Song LP; T-Editor: Jia Y
References
- 1.Berninger B, Costa MR, Koch U, Schroeder T, Sutor B, Grothe B, Götz M. Functional properties of neurons derived from in vitro reprogrammed postnatal astroglia. J Neurosci. 2007;27:8654–8664. doi: 10.1523/JNEUROSCI.1615-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boldt K, van Reeuwijk J, Lu Q, Koutroumpas K, Nguyen TM, Texier Y, van Beersum SE, Horn N, Willer JR, Mans DA, Dougherty G, Lamers IJ, Coene KL, Arts HH, Betts MJ, Beyer T, Bolat E, Gloeckner CJ, Haidari K, Hetterschijt L, et al. An organelle-specific protein landscape identifies novel diseases and molecular mechanisms. Nat Commun. 2016;7:11491. doi: 10.1038/ncomms11491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brulet R, Matsuda T, Zhang L, Miranda C, Giacca M, Kaspar BK, Nakashima K, Hsieh J. NEUROD1 instructs neuronal conversion in non-reactive astrocytes. Stem Cell Reports. 2017;8:1506–1515. doi: 10.1016/j.stemcr.2017.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Q, Zhang A, Yu F, Gao J, Liu Y, Yu C, Zhou H, Xu C. Label-free proteomics uncovers energy metabolism and focal adhesion regulations responsive for endometrium receptivity. J Proteome Res. 2015;14:1831–1842. doi: 10.1021/acs.jproteome.5b00038. [DOI] [PubMed] [Google Scholar]
- 5.Chen W, Zhang B, Xu S, Lin R, Wang W. Lentivirus carrying the NeuroD1 gene promotes the conversion from glial cells into neurons in a spinal cord injury model. Brain Res Bull. 2017;135:143–148. doi: 10.1016/j.brainresbull.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 6.Chen YC, Ma NX, Pei ZF, Wu Z, Do-Monte FH, Keefe S, Yellin E, Chen MS, Yin JC, Lee G, Minier-Toribio A, Hu Y, Bai YT, Lee K, Quirk GJ, Chen G. A NeuroD1 AAV-based gene therapy for functional brain repair after ischemic injury through in vivo astrocyte-to-neuron conversion. Mol Ther. 2020;28:217–234. doi: 10.1016/j.ymthe.2019.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cizkova D, Le Marrec-Croq F, Franck J, Slovinska L, Grulova I, Devaux S, Lefebvre C, Fournier I, Salzet M. Alterations of protein composition along the rostro-caudal axis after spinal cord injury: proteomic, in vitro and in vivo analyses. Front Cell Neurosci. 2014;8:105. doi: 10.3389/fncel.2014.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cox J, Matic I, Hilger M, Nagaraj N, Selbach M, Olsen JV, Mann M. A practical guide to the MaxQuant computational platform for SILAC-based quantitative proteomics. Nat Protoc. 2009;4:698–705. doi: 10.1038/nprot.2009.36. [DOI] [PubMed] [Google Scholar]
- 9.Devaux S, Cizkova D, Quanico J, Franck J, Nataf S, Pays L, Hauberg-Lotte L, Maass P, Kobarg JH, Kobeissy F, Mériaux C, Wisztorski M, Slovinska L, Blasko J, Cigankova V, Fournier I, Salzet M. Proteomic analysis of the spatio-temporal based molecular kinetics of acute spinal cord injury identifies a time- and segment-specific window for effective tissue repair. Mol Cell Proteomics. 2016;15:2641–2670. doi: 10.1074/mcp.M115.057794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Didangelos A, Puglia M, Iberl M, Sanchez-Bellot C, Roschitzki B, Bradbury EJ. High-throughput proteomics reveal alarmins as amplifiers of tissue pathology and inflammation after spinal cord injury. Sci Rep. 2016;6:21607. doi: 10.1038/srep21607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.García E, Rodríguez-Barrera R, Buzoianu-Anguiano V, Flores-Romero A, Malagón-Axotla E, Guerrero-Godinez M, De la Cruz-Castillo E, Castillo-Carvajal L, Rivas-Gonzalez M, Santiago-Tovar P, Morales I, Borlongan C, Ibarra A. Use of a combination strategy to improve neuroprotection and neuroregeneration in a rat model of acute spinal cord injury. Neural Regen Res. 2019;14:1060–1068. doi: 10.4103/1673-5374.250627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo Z, Zhang L, Wu Z, Chen Y, Wang F, Chen G. In vivo direct reprogramming of reactive glial cells into functional neurons after brain injury and in an Alzheimer’s disease model. Cell Stem Cell. 2014;14:188–202. doi: 10.1016/j.stem.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heinrich C, Blum R, Gascón S, Masserdotti G, Tripathi P, Sánchez R, Tiedt S, Schroeder T, Götz M, Berninger B. Directing astroglia from the cerebral cortex into subtype specific functional neurons. PLoS Biol. 2010;8:e1000373. doi: 10.1371/journal.pbio.1000373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsu SY, Chen YJ, Ouyang P. Pnn and SR family proteins are differentially expressed in mouse central nervous system. Histochem Cell Biol. 2011;135:361–373. doi: 10.1007/s00418-011-0795-1. [DOI] [PubMed] [Google Scholar]
- 15.Kim YD, Lee J, Kim HS, Lee MO, Son MY, Yoo CH, Choi JK, Lee SC, Cho YS. The unique spliceosome signature of human pluripotent stem cells is mediated by SNRPA1, SNRPD1, and PNN. Stem Cell Res. 2017;22:43–53. doi: 10.1016/j.scr.2017.05.010. [DOI] [PubMed] [Google Scholar]
- 16.Lee AS, Tang C, Rao MS, Weissman IL, Wu JC. Tumorigenicity as a clinical hurdle for pluripotent stem cell therapies. Nat Med. 2013;19:998–1004. doi: 10.1038/nm.3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu W, Shang FF, Xu Y, Belegu V, Xia L, Zhao W, Liu R, Wang W, Liu J, Li CY, Wang TH. eIF5A1/RhoGDIα pathway: a novel therapeutic target for treatment of spinal cord injury identified by a proteomics approach. Sci Rep. 2015a;5:16911. doi: 10.1038/srep16911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Y, Miao Q, Yuan J, Han S, Zhang P, Li S, Rao Z, Zhao W, Ye Q, Geng J, Zhang X, Cheng L. Ascl1 converts dorsal midbrain astrocytes into functional neurons in vivo. J Neurosci. 2015b;35:9336–9355. doi: 10.1523/JNEUROSCI.3975-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu ZG, Li Y, Jiao JH, Long H, Xin ZY, Yang XY. MicroRNA regulatory pattern in spinal cord ischemia-reperfusion injury. Neural Regen Res. 2020;15(11):2123–2130. doi: 10.4103/1673-5374.280323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lukovic D, Stojkovic M, Moreno-Manzano V, Bhattacharya SS, Erceg S. Perspectives and future directions of human pluripotent stem cell-based therapies: lessons from Geron’s clinical trial for spinal cord injury. Stem Cells Dev. 2014;23:1–4. doi: 10.1089/scd.2013.0266. [DOI] [PubMed] [Google Scholar]
- 21.Matsuda T, Irie T, Katsurabayashi S, Hayashi Y, Nagai T, Hamazaki N, Adefuin AMD, Miura F, Ito T, Kimura H, Shirahige K, Takeda T, Iwasaki K, Imamura T, Nakashima K. Pioneer factor NeuroD1 rearranges transcriptional and epigenetic profiles to execute microglia-neuron conversion. Neuron. 2019;101:472–485.e7. doi: 10.1016/j.neuron.2018.12.010. [DOI] [PubMed] [Google Scholar]
- 22.Niu W, Zang T, Smith DK, Vue TY, Zou Y, Bachoo R, Johnson JE, Zhang CL. SOX2 reprograms resident astrocytes into neural progenitors in the adult brain. Stem Cell Reports. 2015;4:780–794. doi: 10.1016/j.stemcr.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oh JY, Hwang TY, Jang JH, Park JY, Ryu Y, Lee H, Park HJ. Muscovite nanoparticles mitigate neuropathic pain by modulating the inflammatory response and neuroglial activation in the spinal cord. Neural Regen Res. 2020;15:2162–2168. doi: 10.4103/1673-5374.282260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sahni V, Kessler JA. Stem cell therapies for spinal cord injury. Nat Rev Neurol. 2010;6:363–372. doi: 10.1038/nrneurol.2010.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Su Z, Niu W, Liu ML, Zou Y, Zhang CL. In vivo conversion of astrocytes to neurons in the injured adult spinal cord. Nat Commun. 2014;5:3338. doi: 10.1038/ncomms4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 27.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 28.Tica J, Bradbury EJ, Didangelos A. Combined transcriptomics, proteomics and bioinformatics identify drug targets in spinal cord injury. Int J Mol Sci. 2018;19:1461. doi: 10.3390/ijms19051461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Varjosalo M, Sacco R, Stukalov A, van Drogen A, Planyavsky M, Hauri S, Aebersold R, Bennett KL, Colinge J, Gstaiger M, Superti-Furga G. Interlaboratory reproducibility of large-scale human protein-complex analysis by standardized AP-MS. Nat Methods. 2013;10:307–314. doi: 10.1038/nmeth.2400. [DOI] [PubMed] [Google Scholar]
- 30.Wang LL, Su Z, Tai W, Zou Y, Xu XM, Zhang CL. The p53 pathway controls SOX2-mediated reprogramming in the adult mouse spinal cord. Cell Rep. 2016;17:891–903. doi: 10.1016/j.celrep.2016.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wiśniewski JR. Proteomic sample preparation from formalin fixed and paraffin embedded tissue. J Vis Exp. 2013:50589. doi: 10.3791/50589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wiśniewski JR, Nagaraj N, Zougman A, Gnad F, Mann M. Brain phosphoproteome obtained by a FASP-based method reveals plasma membrane protein topology. J Proteome Res. 2010;9:3280–3289. doi: 10.1021/pr1002214. [DOI] [PubMed] [Google Scholar]
- 33.Yang H, Cheng XP, Li JW, Yao Q, Ju G. De-differentiation response of cultured astrocytes to injury induced by scratch or conditioned culture medium of scratch-insulted astrocytes. Cell Mol Neurobiol. 2009;29:455–473. doi: 10.1007/s10571-008-9337-3. [DOI] [PubMed] [Google Scholar]
- 34.Yang H, Liang Z, Li J, Cheng X, Luo N, Ju G. Optimized and efficient preparation of astrocyte cultures from rat spinal cord. Cytotechnology. 2006;52:87–97. doi: 10.1007/s10616-006-9033-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang L, Yin JC, Yeh H, Ma NX, Lee G, Chen XA, Wang Y, Lin L, Chen L, Jin P, Wu GY, Chen G. Small molecules efficiently reprogram human astroglial cells into functional neurons. Cell Stem Cell. 2015;17:735–747. doi: 10.1016/j.stem.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou Y, Wang H, Guo F, Si N, Brantner A, Yang J, Han L, Wei X, Zhao H, Bian B. Integrated proteomics and lipidomics investigation of the mechanism underlying the neuroprotective effect of N-benzylhexadecanamide. Molecules. 2018;23:2929. doi: 10.3390/molecules23112929. [DOI] [PMC free article] [PubMed] [Google Scholar]