ABSTRACT
The latest outbreak of Zika virus (ZIKV) in the Americas was associated with significant neurologic complications, including microcephaly of newborns. We evaluated mechanisms that regulate ZIKV entry into human fetal astrocytes (HFAs). Astrocytes are key players in maintaining brain homeostasis. We show that the central mediator of canonical Wnt signaling, β-catenin, regulates Axl, a receptor for ZIKV infection of HFAs, at the transcriptional level. In turn, ZIKV inhibited β-catenin, potentially as a mechanism to overcome its restriction of ZIKV internalization through regulation of Axl. This was evident with three ZIKV strains tested but not with a laboratory-adapted strain which has a large deletion in its envelope gene. Finally, we show that β-catenin-mediated Axl-dependent internalization of ZIKV may be of increased importance for brain cells, as it regulated ZIKV infection of astrocytes and human brain microvascular cells but not kidney epithelial (Vero) cells. Collectively, our studies reveal a role for β-catenin in ZIKV infection and highlight a dynamic interplay between ZIKV and β-catenin to modulate ZIKV entry into susceptible target cells.
IMPORTANCE ZIKV is an emerging pathogen with sporadic outbreaks throughout the world. The most recent outbreak in North America was associated with small brains (microcephaly) in newborns. We studied the mechanism(s) that may regulate ZIKV entry into astrocytes. Astrocytes are a critical resident brain cell population with diverse functions that maintain brain homeostasis, including neurogenesis and neuronal survival. We show that three ZIKV strains (and not a heavily laboratory-adapted strain with a large deletion in its envelope gene) require Axl for internalization. Most importantly, we show that β-catenin, the central mediator of canonical Wnt signaling, negatively regulates Axl at the transcriptional level to prevent ZIKV internalization into human fetal astrocytes. To overcome this restriction, ZIKV downregulates β-catenin to facilitate Axl expression. This highlights a dynamic host-virus interaction whereby ZIKV inhibits β-catenin to promote its internalization into human fetal astrocytes through the induction of Axl.
KEYWORDS: astrocytes, β-catenin, Zika, ZIKV, infection, internalization, Axl, Tyro3, TCF4, brain, flavivirus, Wnt signaling
INTRODUCTION
Zika virus (ZIKV) is a mosquito-transmitted flavivirus originally discovered in Uganda in 1947 (1) which expanded into the Americas with outbreaks in 2014 and 2015. The viral genome consists of an 11-kb single-stranded positive-sense RNA encoding a single open reading frame between 5′ and 3′ untranslated regions (UTRs). Upon cell entry, the viral RNA is translated, and the polyprotein is cleaved by cellular and viral proteases. Cleavage of ZIKV polyprotein generates three structural proteins, capsid (C), envelope (E), and premembrane (prM), which form the virion. The remaining proteins are nonstructural, consisting of NS1, NS2A, NS2B, NS3, NS4A NS4B, and NS5, many of which act as transcriptional machinery within the host endoplasmic reticulum (ER). NS1, NS4A/B, and NS5 contribute to both pathogenesis and host cell immune evasion (2, 3).
Approximately 20% of ZIKV infections are symptomatic, with patients presenting with myalgia, arthralgia, rash, fever, conjunctivitis, and, in severe cases, Guillain-Barré syndrome (4). In newborns, the recent outbreak in the Americas was associated with congenital ZIKV syndrome, which represents significant neurologic abnormalities, including microcephaly (small brain), seizures, swallowing problems, hearing and sight abnormalities, and disrupted neurovasculature (5). Although the mechanisms that underlie these pathologies are yet to be established, ZIKV is neurotrophic and has a broad tropism for brain cells, including neural progenitor cells, microglia, and astrocytes (6–8). Astrocytes are the first cells to be infected in the brain, as demonstrated by ZIKV infection of immunocompetent mice; astrocytes subsequently disseminate the virus throughout the central nervous system (CNS) (9). Further, in vitro studies demonstrated productive ZIKV infection of astrocytes, which undergo prolonged viral shedding and may ultimately undergo apoptosis (10, 11). Astrocytes are a robust cell population in the brain. They play critical roles in fetal brain development, maintaining blood-brain barrier integrity, scavenging for excess neurotoxin neurotransmitters, regulating the ion concentration in the brain, storing glycogen to be converted to glucose, releasing neurotrophic factors, and even playing a role in immune regulation in the brain (12, 13). As such, their dysregulation and/or apoptosis in response to ZIKV infection can be a critical component of ZIKV-mediated neuropathogenesis; however, the mechanism(s) driving these effects is not entirely clear.
Wnt/β-catenin is an important pathway for the survival, development, and function of many organ systems, including the CNS (14). Dysregulation of this pathway is linked to neurodegenerative and psychiatric disorders, including Alzheimer’s disease, neuroHIV, and schizophrenia (15–17). Astrocytes robustly express the Wnt/β-catenin pathway, and their functions are intimately tied to this pathway (18). β-Catenin is a transcriptional coactivator and the central mediator in the canonical Wnt/β-catenin pathway. The pathway is initiated by the binding of Wnts (small, secreted glycoproteins) to seven transmembrane frizzled receptors and a coreceptor (low-density lipoprotein receptor-related protein 5/6 [LRP5/6]). This ligand/receptor binding leads to the destabilization of a proteasomal destruction complex, resulting in β-catenin accumulation. Stabilized β-catenin translocates to the nucleus and binds to T cell factor/lymphoid enhancer family (TCF/LEF) transcription factors to regulate target gene expression (19). β-Catenin is also a major structural component of the adherens junction protein complex linking N-cadherin to α-catenin and actin. The Wnt/β-catenin pathway has also been implicated in viral life cycles. It is a positive regulator of influenza virus replication (20, 21), and infection with West Nile virus leads to downregulation of the adherens and tight-junction proteins ZO-1, claudin-1, occludin, and β-catenin in brain endothelial cells, leading to higher levels of viral infiltration (22). Further, β-catenin forms a multiprotein complex with TCF4 and a nuclear matrix protein, SMAR1, on the HIV-1 promoter to pull the HIV DNA away from the nuclear matrix, preventing RNA polymerase II docking and suppressing HIV-1 transcription (19, 23, 24).
The host receptor(s) mediating ZIKV entry is not completely clear. Evidence suggests that the tyrosine kinases Tyro3, Axl, and Mer can mediate the binding and internalization of ZIKV into human astrocytes (7, 25–27). Further, ZIKV can utilize Axl to inhibit interferon-stimulated genes (ISGs) through activation of SOCS1, promoting astrocyte infection (27). ZIKV also uses Axl to infect human endothelial cells (28). However, Axl is not a bona fide universal ZIKV receptor, as it is not required for ZIKV infection of the neuronal progenitor (29) or for infection of mouse models (30, 31). We evaluated here the effect of various strains of ZIKV on the infection of human fetal astrocytes (HFAs). We used two Asian-lineage ZIKV strains (PRVABC59 and FLR [herein ZIKVPRV and ZIKVFLR, respectively) isolated from patients in Puerto Rico and Colombia, respectively, and two African-lineage strains (MR766 and IBH30656 [herein ZIKVMR and ZIKVIBH, respectively]). ZIKVMR was isolated from a sentinel rhesus monkey in Uganda and ZIKVIBH from human blood from a Nigerian patient. Both African-lineage strains were extensively passaged intracranially in animals. Our studies reveal that β-catenin regulates Axl expression at the transcriptional level, which is required for ZIKV internalization into human fetal astrocytes, except in an extensively lab-adapted strain. Further β-catenin-mediated regulation of Axl is important for infection of endothelial cells and not kidney epithelial cells, suggesting that the β-catenin/Axl axis is required for the infection of some brain cells. These findings demonstrate that β-catenin inhibits ZIKV internalization into human fetal astrocytes through regulation of Axl and that ZIKV in turn overcomes this restriction through inhibiting β-catenin.
RESULTS
ZIKV infects astrocytes and triggers cytopathic effects.
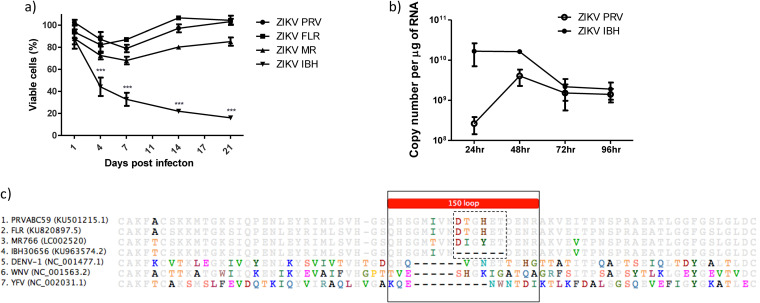
To determine the impact of diverse ZIKV strains on human fetal astrocytes (HFAs), HFAs were infected with one of four ZIKV strains (PRVABC59 [ZIKVPRV], FLR [ZIKVFLR], MR766 [ZIKVMR], or IBH30656 [ZIKVIBH]) at a multiplicity of infection (MOI) of 0.5, and cell viability was assessed at 1 to 21 days postinfection. Infection with the Asian-lineage strains representative of the American outbreak of ZIKVPRV and ZIKVFLR, as well as the African strain ZIKVMR, caused modest (∼15 to 30%) cell death compared to that of mock-infected cells at 7 days postinfection (dpi), followed by a slight recovery in viability for the remaining time points up to 21 dpi (Fig. 1a). Infection with ZIKVIBH caused severe cell death in that >70% of cells were not viable by 7 dpi, and by day 21, most of the cells were dead, with complete clearance of the plate of adherent cells (Fig. 1a). Given that ZIKVPRV, ZIKVFLR, and ZKIVMR behaved relatively the same with regard to the kinetics of modest cell death and that ZIKVIBH showed dramatic effects on cell death, we evaluated ZIKVIBH versus ZIKVPRV more closely. ZIKVIBH-infected cells had ∼10-fold more intracellular viral RNA than ZIKVPRV-infected cells at 24 h postinfection despite being inoculated at the same MOI (Fig. 1b), suggesting that ZIKVIBH is more proficient in infecting HFAs than ZIKVPR. At 48 h, however, intracellular viral RNA levels were similar between the two strains, which may be due to an accelerated rate of death by ZIKVIBH. Sequence comparison of partial ectodomain regions of envelope (E) proteins showed deletion of 6 amino acid residues in the 150 loop region of ZIKVIBH compared with the sequences of other Zika strains (Fig. 1c, dotted-line box). Similar deletions of 5 to 9 residues are found within the 150 loop region of other flaviviruses, such as the Dengue, yellow fever, and West Nile viruses (DENV, YFV, and WNV, respectively) (Fig. 1c, solid-line box).
FIG 1.
ZIKV infection of human fetal astrocytes (HFAs) and cytopathology. (a) HFAs were propagated in 48-well tissue culture plates to ∼90% confluence and infected with ZIKVPRV, ZIKVFLR, ZIKVMR, or ZIKVIBH at an MOI of 0.5. Twenty-four hours later, the medium was replaced and cells were cultured for 0 to 20 days. The viability of cells was measured by the MTS assay. (b) HFAs were infected with ZIKVPRV and ZIKVIBH at an MOI of 0.5 in a 24-well format, cells were harvested for RNA at the indicated times postinfection, and viral copy numbers were determined by qPCR. (c) A sequence alignment of partial E protein ectodomains from all four ZIKV strains, DENV-1, WNV, and YFV is depicted, with residues in color highlighting differences in amino acids to the 50% consensus. The solid box indicates the 150 loop region, and the dotted box indicates the 6-amino-acid region absent in ZIKVIBH. Asterisks denote a P of ≤0.001. Data represent means ± SEM and were analyzed by comparing each strain over time using the one-way ANOVA Bonferroni multiple-comparison test.
ZIKV inhibits β-catenin in HFAs.
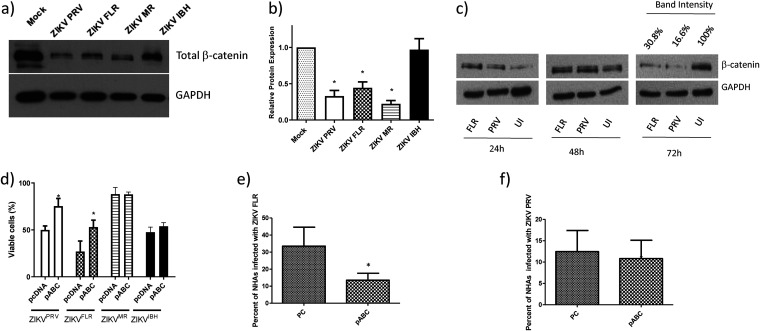
To investigate the role of β-catenin in ZIKV infection, we measured the protein expression of total β-catenin by Western blotting (WB) from HFAs infected with one of the four ZIKV strains at 3 dpi. Total β-catenin expression was significantly reduced (by 54 to 78%) in cultures infected with ZIKVPRV, ZIKVFLR, and ZIKVMR but not in cultures infected with ZIKVIBH (Fig. 2a and b). This downregulation of β-catenin occurred at later stages of infection (day 3 postinfection) (Fig. 2c). Since β-catenin is a pro-survival factor and positively regulates Bcl2 and BcL-xL (antiapoptotic proteins) (32), we assessed whether overexpression of active β-catenin can protect the cells from ZIKV-mediated cell death. Due to the limited cell death exhibited by three of the four strains, a higher virus MOI was used to expand the dynamic range of the survival assay. Overexpression of a constitutively active β-catenin in HFAs 24 h prior to ZIKV infection resulted in a significant increase in cell viability (at least 50% higher than that of mock-infected cells) in both ZIKVPRV- and ZIKVFLR-infected cells but not in ZIKVMR- or ZIKVIBH-infected cells (Fig. 2d). Further, infection of HFAs with ZIKV in the presence of pABC (β-catenin overexpression plasmid) significantly decreased (>50%) the number of ZIKVFLR-infected cells (Fig. 2e), and the amount of ZIKVPRV-infected cells trended toward a decrease of ∼20% (Fig. 2f). Notably, despite a higher MOI, ZIKVMR infection still resulted in minimal cell death, which may partially explain β-catenin’s inability to increase survival.
FIG 2.
ZIKV infection of HFAs inhibits β-catenin expression, and the induction of β-catenin rescues HFAs from ZIKV-mediated cell death. (a) HFAs were infected with the ZIKVPRV, ZIKVFLR, ZIKVMR, or ZIKVIBH strain for 4 days, cells were lysed for protein extraction and quantitation, and Western blotting was performed for total β-catenin. GAPDH was used as a loading control. (b) Band intensities from the WB were calculated using ImageJ, were normalized to GAPDH, and are represented as fold changes from results for uninfected cells. (c) HFAs were infected with ZIKVPRV or ZIKVFLR at an MOI of 0.5 for 1 to 4 days, cells were lysed for protein extraction and quantified by the BCA assay, and equal amounts of total cell proteins were processed for the detection of β-catenin and GAPDH by WB for 24 h to 96 h. (d) A constitutively active plasmid of β-catenin (pABC) or a control plasmid (pcDNA) were transfected into HFAs at 24 h posttransfection, the cells were infected with ZIKVPRV or ZIKVFLR at an MOI of 1.0, and cell viability was measured with MTS at day 4 postinfection. (e) pcDNA (control plasmid [PC]) or pABC was transfected into astrocytes; at 24 h posttransfection, the cells were infected with ZIKVFLR (e) or ZIKVPRV (f) at an MOI of 1.0, and 24 h later, the percentage of cells positive for ZIKV was calculated by intracellular flow cytometry using an antibody against ZIKA Env. Data were analyzed by FlowJo. Data are means ± SEM and were analyzed using a two-tailed one-sample t test. *, P ≤ 0.05.
Induction of β-catenin inhibits internalization of ZIKV in HFAs.
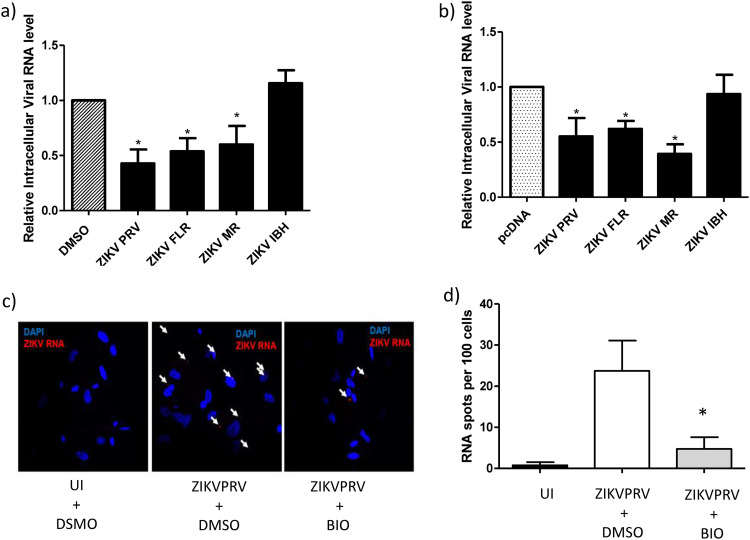
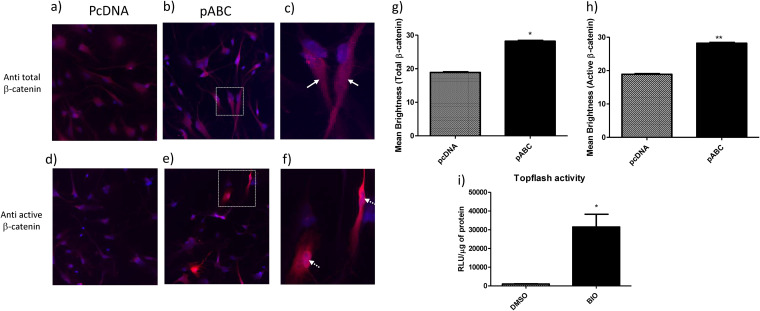
To determine the mechanism by which β-catenin impacts ZIKV infection, we evaluated at which stage in the ZIKV life cycle (early, middle, or late) β-catenin may exert its effect. β-Catenin was induced in HFAs either by transfection of a constitutively active β-catenin plasmid or by treatment of cells with 6-bromoindirubin-3′-oxime (BIO) at a 1 μM concentration. BIO activates β-catenin through inhibition of glycogen synthase kinase 3 α/β (GSK3α/β), which is part of the β-catenin destruction complex. Inhibition of GSK3α/β allows for the accumulation of stabilized β-catenin in the cytoplasm, followed by its translocation to the nucleus (33, 34). At a 1 μM concentration, BIO induces β-catenin robustly in human astrocytes without affecting cell viability (34). Twenty-four hours postinduction of β-catenin, cells were infected with various ZIKV strains at an MOI of 0.5 for 5 hours, washed, and trypsinized to remove bound virus, and the intracellular level of ZIKV RNA was measured by real-time PCR. Induction of β-catenin reduced the internalization of ZIKVPRV and ZIKVFLR by >50% (P < 0.05) and of ZIKVMR by ∼50% (P < 0.05) compared to that of a vehicle-treated infected control (Fig. 3a). Induction of β-catenin had no effect on the internalization of ZIKVIBH (Fig. 3a). Small-molecule inhibition of GSK3β can have off-target effects, including inhibition of GSK3β itself. Hence, to rule out this possibility, we repeated the experiments by inducing β-catenin via plasmid-based overexpression of active β-catenin. We observed similar results, with a >50% reduction in ZIKVPRV, ZIKVFLR, and ZIKVMR (P < 0.05) internalization in the presence of induced β-catenin, while ZIKVIBH internalization had no effect under similar conditions (Fig. 3b). Transfection of pABC into HFAs significantly increased cytoplasmic β-catenin and demonstrated a higher localization of the transcriptionally active form of β-catenin in the nucleus (Fig. 4a to h). Treatment with BIO at 1 μM along with transfection with a Wnt/β-catenin reporter plasmid (TOPFlash) induced luciferase reporter activity by approximately 31-fold (Fig. 4g). Further, we used RNAscope, a highly sensitive in situ hybridization technique, as an independent approach to quantify the internalization of ZIKV in HFAs with and without inhibition of β-catenin. Treatment with BIO followed by infection with ZIKVPRV and counting of RNA spots indicated significantly less ZIKV RNA in cells treated with BIO (2.75-fold reduction in ZIKV RNA spots per cell) (Fig. 3c and d).
FIG 3.
The induction of β-catenin inhibits ZIKV internalization in HFAs. (a) HFAs were incubated with 1 μM BIO or a vehicle (DMSO) for 24 h and infected with ZIKVPRV, ZIKVFLR, ZIKVMR, or ZIKVIBH at an MOI of 0.5 for 5 h, and then RNA was extracted and qPCR performed to measure intracellular ZIKA viral RNA. (b) HFAs were incubated with 200 ng of the control plasmid (pcDNA) or the constitutively active β-catenin construct (pABC) for 24 h and infected with the indicated viruses, and ZIKV intracellular RNA levels were measured by qPCR. (c) HFAs were seeded on coverslips in a 24-well plate, incubated with 1 μM BIO or the vehicle (DMSO) for 24 h, and infected with the indicated viruses at an MOI of 1.0 for 5 h, and then RNAscope was performed to visualize ZIKV RNA within cells. Arrows point to ZIKV RNA fluorescent signals (spots) in cells. UI, uninfected. (d) ZIKV RNA spots and nuclei (DAPI) were counted and analyzed using ImageJ, quantified by imaging 5 random fields per well, and analyzed using one-way ANOVA and Bonferroni’s multiple-comparison test. Data are means ± SEM and were analyzed using a two-tailed one-sample t test unless otherwise indicated. *, P ≤ 0.05.
FIG 4.
Active β-catenin is detected both in the nucleus and in the cytoplasm after induction using a constitutively active construct of β-catenin. (a) Astrocytes were plated on laminin-coated coverslips in a 24-well format, transfected with pcDNA (control plasmid) or pABC (constitutively active β-catenin) for 24 h, and stained with fluorescently labeled primary antibodies for total β-catenin (a to c) or active β-catenin (d to f) along with DAPI for the nucleus. Images were captured using a Keyence BZ-X810 microscope and analyzed using BZ-X800 Analyzer software. (c and f) Magnified images of the boxes in panels b and e, respectively. Arrows in panel c show the total β-catenin signal in the cytoplasm away from the nucleus (DAPI), and arrows in panel f indicate the active β-catenin signal in the nucleus. Signal intensities of at least 40 random spots were measured, and the mean brightness was calculated using BZ-X800 Analyzer software and plotted for total β-catenin (g) and active β-catenin (h). (i) Astrocytes were treated with DMSO or BIO for 24 h and transfected with TOPFlash (a Wnt pathway reporter plasmid) for 24 h. Cells were lysed, and clarified lysate was subjected to luciferase and BCA assays. Relative luciferase activity (RLU) was normalized to the total protein content and is plotted as RLU per microgram of protein. Data were analyzed using a two-tailed one-sample t test and are represented as means ± SEM. *, P ≤ 0.05; **, P < 0.01.
Induction of β-catenin negatively regulates the expression of Axl and Tyro3.
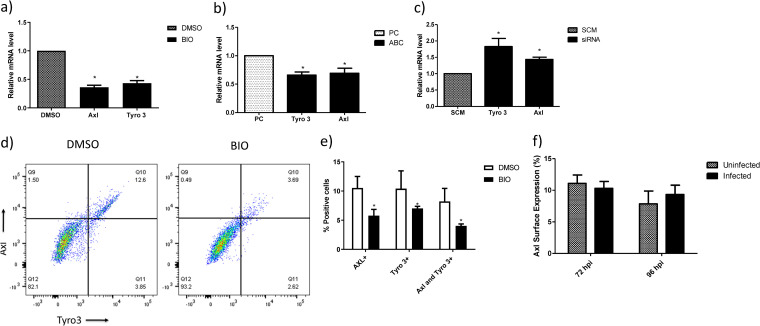
Given that ZIKV was reported to use the tyrosine receptor kinases Axl and Tyro3 to enter target cells (7, 25–27, 35, 36) and we showed inhibition of ZIKV internalization by induction of β-catenin, we assessed the relationship between β-catenin, Axl, and Tyro3. Induction of β-catenin, without infection, either by BIO (Fig. 5a) or overexpression of active β-catenin (Fig. 5b) significantly reduced the mRNA expression of Axl and Tyro3 in HFAs by 60 to 70%. Conversely, small interfering RNA (siRNA)-mediated knockdown of β-catenin increased the expression of Tyro3 and Axl mRNAs by ∼1.7- and ∼1.5-fold, respectively (Fig. 5c). The efficiency of siRNA knockdown (KD) of β-catenin in HFAs is routinely above 90%, as we reported previously (18, 34). Further, β-catenin induction led to a reduction in the surface expression of Axl and Tyro3 proteins, as demonstrated by flow cytometry. Specifically, doubly positive (Axl+ Tyro3+) cells and singly positive (Tyro3+ or Axl+) cells were reduced by approximately 2-fold (Fig. 4d and e). Downregulation of β-catenin by ZIKVFLR infection in HFAs after 72 h postinfection exhibited no change in the surface expression of Axl, as evaluated by flow cytometry (Fig. 5f).
FIG 5.
Induction of β-catenin downregulates Axl and Tyro3 transcription. (a) HFAs were incubated with 1 μM BIO or DMSO for 24 h, and the expression of Axl, Tyro3, and Mer was determined by qPCR. Data were normalized to GAPDH and are expressed as fold changes from results with the vehicle. (b) Constitutively active β-catenin (pABC) or a control plasmid (pcDNA) was transfected into HFAs and incubated for 24 h, and then the mRNA expression of Axl and Tyro 3 was determined by qPCR. Data are normalized to GAPDH and expressed as fold changes from results with the vehicle. (c) siRNA against β-catenin and scrambled siRNA (SCM) were transfected into HFAs and incubated for 72 h, and then the mRNA expression of Axl, Tyro3, and Mer was determined by qPCR. Data were normalized to GAPDH and are expressed as fold changes from results for the control siRNA. (d) Representative dot blot of the surface expression of Axl and Tyro3 on HFAs postincubation with 1 μM BIO or DMSO for 24 h. Data were acquired using an LSR II flow cytometer. (e) Flow data were analyzed using FlowJo and are represented as percentages of cells positive for Axl or Tyro3. (f) HFAs were infected with the ZIKVPRV and ZIKVFLR strains for 3 to 4 days, cells were lifted by sterile PBS-EDTA (5 mM) treatment (followed by gentle scraping or by Accutase treatment), stained with anti-Axl antibody or control IgG, and subjected to flow cytometry to measure the surface expression of Axl. Data were analyzed using FlowJo and are represented as percentages of cells positive for Axl. Data are means ± SEM and were analyzed using a two-tailed one-sample t test unless otherwise indicated. *, P ≤ 0.05.
Internalization of ZIKV in HFAs is dependent on Axl and not Tyro3.
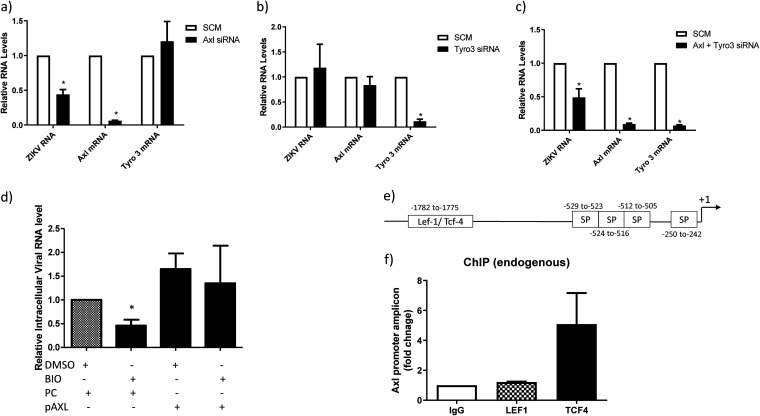
ZIKV internalization is not fully understood, with the viral receptor target being contingent on the cell type. Axl was described as an important receptor for ZIKV entry in astrocytes, with entry mediated by the binding of the Axl ligand growth arrest-specific 6 (Gas6), and when expressed, it transiently allows otherwise-nonpermissive cells to become permissive (7, 26, 27, 36). To determine if the downregulation of Axl and/or Tyro3 reduces ZIKV internalization, we used siRNAs to KD Axl and Tyro3. The targeted siRNAs were highly effective in the KD of their respective targets (Fig. 6a); Axl siRNA did not impact Tyro3 (Fig. 6a), and Tyro3 siRNA did not impact Axl (Fig. 6b), demonstrating specificity for their respective target. We next assessed whether KD of Axl can prevent the internalization of ZIKVPRV, as it is a strain which demonstrated downregulation of β-catenin. KD of Axl resulted in a >50% reduction in ZIKVPRV internalization (Fig. 6a), while KD of Tyro3 had no effect (Fig. 6b). A double KD of Axl and Tyro3 exhibited high efficiency in Axl and Tyro3 mRNA depletion and, as expected, resulted in an ∼50% reduction (P < 0.05) in ZIKV internalization, showing a specific correlation with the KD of Axl and the reduction in ZIKV internalization (Fig. 6c). Additionally, overexpression of Axl via a mammalian expression plasmid increased the entry of ZIKVPRV and alleviated BIO-mediated inhibition of ZIKV entry (Fig. 6d). Taken together, these results demonstrate that Axl and not Tyro3 is crucial for the internalization of ZIKVPRV in HFAs.
FIG 6.
Knockdown of Axl inhibits internalization of ZIKV in HFAs. HFAs were transfected with scrambled or siRNA against Axl (a), Tyro 3 (b), or both Axl and Tyro 3 (c) at 100 nM each for 72 h and infected with the indicated virus at an MOI of 0.5 for 5 h, and then RNA was measured by qPCR. Data are normalized to GAPDH and are expressed as fold changes from results for the scrambled siRNA. (d) HFAs were transfected with the control plasmid (pcDNA) (PC) or a mammalian expression plasmid containing the full-length cDNA for human Axl (pAxl). Twenty-four hours posttransfection, the cells were treated with BIO or DMSO and 24 h later infected with ZIKVFLR. Five hours postinfection, the cells were processed for the measurement of intracellular RNA levels for ZIKV and GAPDH by qPCR. (e) The promoter region (∼2 kb) for Axl was identified by a PubMed search. Binding sites for Sp1 (SP), Sp3, and TCFs/LEF were identified by using the PROMO tool. Arrows indicate the primers designed for ChIP. (f) HFAs were processed for ChIP using anti-TCF4, anti-LEF1, and control (IgG) antibodies to detect the biding of endogenous TCF4 or LEF1. Data are expressed as fold changes from results for IgG. Data are means ± SEM and were analyzed using a two-tailed one-sample t test. *, P ≤ 0.05.
β-Catenin interacts with TCF4 to repress Axl transcription.
Canonical β-catenin signaling involves β-catenin interaction with members of the TCF/LEF family of transcription factors (TCF1, TCF3, TCF4, and LEF1) to regulate gene expression. We identified and analyzed a 2-kb promoter region upstream of the Axl translation site and found a single consensus binding site for TCF/LEF transcription factors between positions −1782 and −1775 by PROMO analysis. Further binding sites for Sp transcription factors were also identified (37) (Fig. 6e). Chromatin immunoprecipitation (ChIP) followed by real-time PCR analysis of the promoter region indicated strong binding of TCF4 and not LEF1 specifically at the TCF/LEF binding site (Fig. 6f). Together, these results indicate that β-catenin negatively regulates Axl transcription by interacting specifically with TCF4 to repress Axl gene expression.
β-Catenin regulation of Axl transcription is cell type dependent.
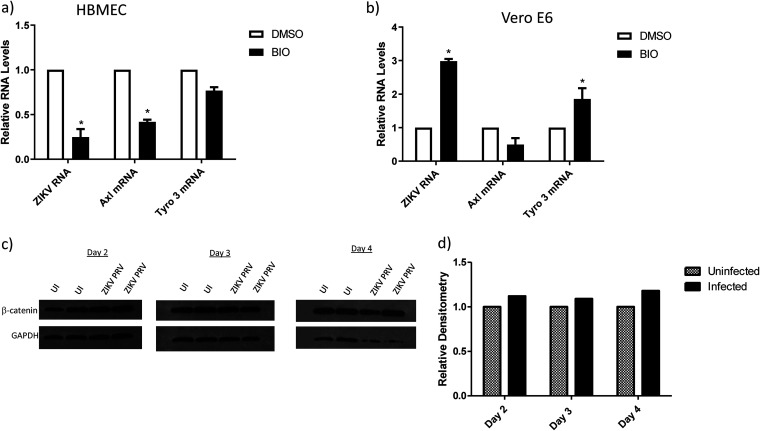
Recent reports have characterized additional cell receptors for ZIKV entry, such as integrin αvβ5, TIM, Mer, and DC-SIGN (35, 38, 39), depending on the cell type. Because β-catenin inhibited ZIKV internalization via downmodulation of Axl in HFAs, we examined whether activation of β-catenin in other ZIKV-susceptible cells would downregulate Axl and subsequently inhibit ZIKV internalization. Human brain microvascular endothelial cells (HBMECs), an endothelial cell line, are highly permissive to ZIKV infection (40, 41). Induction of β-catenin reduced the expression of Axl and ZIKV infection of HBMECs without altering Tyro3 expression (Fig. 7a). Interestingly, Vero cells that are regularly used to propagate ZIKV showed increased sensitivity toward ZIKV entry and Tyro3 expression with respect to β-catenin induction and no change in Axl expression (Fig. 7b). Further, ZIKV infection had no effect on β-catenin expression in these cells (Fig. 7c and d).
FIG 7.
The internalization of ZIKV in different cell types is not dependent on β-catenin and Axl. Human brain microvascular cells (HBMECs) (a) or Vero cells (b) were incubated with BIO (1 μM) or the vehicle (DMSO) for 24 h and infected with ZIKVPRV at an MOI of 0.5 for 5 h, and then ZIKV RNA was measured by qPCR. Data are normalized to GAPDH and expressed as fold changes from results for the vehicle. (c) Vero cells were infected with ZIKVPRV at an MOI of 0.5 for 1 to 4 days. Cells were lysed for protein extraction and quantified by the BCA assay, and equal amounts of total cell proteins were processed for the detection of β-catenin and GAPDH by WB on days 2 to 4. Band intensities from the WB in panel d were calculated using ImageJ, are normalized to GAPDH results, and are presented as fold changes from results for uninfected cells. Data are means ± SEM and were analyzed using a two-tailed one-sample t test. *, P ≤ 0.05.
DISCUSSION
ZIKV neuropathogenesis relies heavily on the infection of astrocytes after initial neuroinvasion (9, 42). Although ZIKV infection of astrocytes leads to significant cytopathology, it also supports prolonged viral shedding (10), which is correlated with worse patient outcomes (43). Viral RNA shedding has also been observed over weeks in urine and over months in semen (44, 45), which is likely due to productive infection of renal proximal tubular epithelial cells and human Sertoli cells (46, 47). In the brain, the continued shedding of ZIKV in astrocytes can drive neuropathogenesis either through direct cytopathic effects or indirect viral effects through induction of inflammatory responses in the brain. We show here that the contemporary and clinically relevant strains ZIKVPRV and ZIKVFLR and the African strain ZIKVMR infect astrocytes without major cell death for at least 21 days, confirming the persistent infection of astrocytes with ZIKV.
While the ZIKVPRV, ZIKVFLR, and ZIKVMR strains exhibited modest killing of HFAs, ZIKVIBH resulted in >80% loss in cell viability within 7 dpi. Many factors may underlie this observation. We observed that ZIKVIBH enters nearly 200 times more HFAs than ZIKVPRV, and this entry is independent of β-catenin and Axl. This robust entry may be directly linked to an enhanced rate of killing of astrocytes. ZIKVIBH was passaged in mouse brains 21 times, and this may have promoted viral entry efficiency and adaptation of heightened virulence (48). Other laboratory-adapted viruses, such as HIV-1 strains, have shown decreased dependence on cellular receptors for entry (49), which may explain how ZIKVIBH internalization is independent of Axl.
The envelope (E) protein of flaviviruses mediates virus entry and hence is an important target for neutralizing antibodies. The molecular structure of the ectopic part of ZIKV E protein with three main domains, β-barrel-shaped domain I, finger-like domain II, and immunoglobulin-like domain III, has been published (50, 51). An alignment of E proteins of the 4 strains used in this study shows a deletion of 6 amino acid residues in the ZIKVIBH strain (Fig. 1c). Atomic structure revealed that these 6 residues are part of an important region known as the 150 loop or glycan loop in domain I (50, 51). The 150 loop is a highly flexible loop formed by 18 residues, Q151 to R168, that include the glycosylation site N154 and may serve as a host attachment site for the virus. In addition to the 150 loop, several positively charged residues around this loop were identified to be important for ZIKV attachment. While most of the positively charged residues around this 150 loop are conserved in all 4 ZIKV strains, the deletion of 6 residues in the ZIKVIBH strain might result in a change in the net charge content around this loop, resulting in a structural modification favorable to attachment to the host. Alternatively, a sequence alignment of E proteins of the ZIKVIBH strain and other flaviviruses, such as DENV, WNV, and YFV, indicated similar deletions in the 150 loop (50), suggesting a possible structure conformation similarity between ZIKVIBH and other flaviviruses near the 150 loop region of the E protein. While only TAM (Tyro3, Axl, and Mer) phosphatidylserine transmembrane receptors were shown to be utilized by ZIKV, both the TIM (T cell, immunoglobulin, and mucin) and TAM receptors have been shown to be used by DENV for cell entry (50, 52). Collectively, one can speculate that, as with DENV, ZIKVIBH may have evolved to utilize both TIM and TAM receptors to enter host cells more robustly. Nevertheless, sequence comparison of all available (n = 200) ZIKV strains revealed that this 6-residue gap in the 150 loop is unique to ZIKVIBH and another strain passaged extensively in mouse brains, which may indicate that ZIKVIBH entry and the pathogenic phenotype observed here may not be representative of clinically relevant ZIKV strains (48) (Fig. 1c). Furthermore, other differences in the viral genome may account for these biologic effects, as some ZIKV mutations have been attributed to increased neuropathogenesis. Specifically, a point mutation in the prM protein is associated with microcephaly, and NS1 mutation is associated with ZIKV innate immunity evasion (53, 54), although ZIKVIBH does not carry these mutations. Due to the limitations in the availability of ZIKV strains, we cannot exclude the possibility that β-catenin may not uniformly impact the internalization of diverse ZIKV strains. Because of this, it will be important to determine whether β-catenin- and Axl-mediated astrocyte entry dependencies differ between clinical ZIKV strains or are instead lost during serial laboratory passage. Likewise, understanding whether strain differences are related to a clinical outcome or a result of laboratory adaptation can inform the potential therapeutic use of β-catenin to restrict ZIKV infection of human fetal astrocytes.
ZIKV internalization is a dynamic process utilizing the host machinery to enter target cells (55). Some of those factors described as binding factors for astrocytes are the receptor tyrosine kinases Axl and Tyro3 and their ligand Gas6 (7, 26, 27). In examining the expression of these putative receptors, we found that both Axl and Tyro3 were inhibited by overexpression of β-catenin. Our gene KD studies revealed that while Axl is crucial, Tyro3 is dispensable for ZIKVPRV entry into HFAs. Current understanding of the expression of Axl is that it is dependent on the transcription factors specificity protein 1 (Sp1) and Sp3 (37). We show here that in human fetal astrocytes, β-catenin regulates the expression of Axl through partnering with TCF4 (TCF7L2) at its consensus binding site, located at nucleotides −1782 to −1775 on the Axl promoter. Generally, β-catenin functions as a transcriptional coactivator in conjunction with TCF/LEF transcription factors in the Wnt/β-catenin pathway. However, in this case, along with other recently observed cases, such as the transcription of interleukin 6 (IL-6), IL-8, and HIV (23, 24, 34), β-catenin appears to act as a cosuppressor of gene transcription. Activation of β-catenin in other cell types also reduces the transcription of Axl and viral entry, such as in human endothelial cells (HBMECs) but not in kidney epithelial (Vero) cells. Ablation of Axl also did not inhibit the infection of neural progenitor cells in a cerebral brain organoid model (29). These findings indicate that ZIKV entry in cells is regulated by complex host gene regulation of specific receptors and binding factors, which may vary between tissues, cells, and/or developmental stages.
ZIKV infection is associated with hyperexcitability of neurons, and blocking the N-methyl-d-aspartate (NMDA) receptor protects neurons from ZIKV-mediated cell death (56, 57). While these adverse effects may be directly related to ZIKV infection of neurons, it may also be indirectly related to dysregulation of astrocytes. Astrocytes are key players in the maintenance of neuronal health. One of their primary functions is the conversion of glutamate through its uptake by excitatory amino acid transporter (EAAT2) on astrocytes and its conversion to glutamine through the enzymatic activity of glutamine synthetase (GS). Both EAAT2 and GS are regulated at the transcriptional level by β-catenin (18). Further, inhibition of β-catenin in astrocytes disrupts their functional integrity, especially as it relates to the glutamate/glutamine cycle and induction of inflammatory cytokines (18, 34, 58). Because of this, ZIKV-mediated downregulation of β-catenin can mediate neuroinflammation, as inhibition of β-catenin induces neuroinflammatory cytokines (IL-6) and disruption of glutamate uptake. Induction of β-catenin may be a pathway to protect against ZIKV-mediated cell death while also ameliorating and/or reducing ZIKV infection and neuropathogenesis.
Our studies demonstrate that β-catenin is a host restriction factor for ZIKV in human fetal astrocytes by inhibiting its internalization through downregulating Axl. Further, β-catenin can protect human fetal astrocytes from ZIKV-mediated cell death. This balance is often observed in virus-host interactions. For example, relevant to this pathway, β-catenin restricts HIV transcription, and HIV in turn inhibits β-catenin to overcome this restriction (24, 59). The balance in this virus-host dynamic interaction ultimately will guide the outcome of infection. Lastly, our study adds to a growing list of viruses that are regulated by β-catenin (e.g., HIV, influenza virus, West Nile virus), albeit at different stages of their life cycles (19–24). This underscores the wide reach of the Wnt/β-catenin pathway in impacting virus-host interactions (60).
MATERIALS AND METHODS
Cell culture and reagents.
Human fetal astrocytes (HFAs) (∼18-week gestation; Lonza Inc., Walkersville, MD) were cultured in the astrocyte growth medium (AGM) BulletKit (Lonza, Basel, Switzerland) supplemented with 0.3% heat-inactivated fetal bovine serum, 1 μl/ml ascorbic acid, 1 μl/ml recombinant human epidermal growth factor (rhEGF), 1 μl/ml AG-1000 (30 μg/ml gentamicin and 15 μg/ml amphotericin), 2.5 μl/ml insulin, and 10 μl/ml l-glutamine. Cells at an early passage (P), P2 to P6, were used for all experiments. Vero 76 (CRL-1587) and human brain microvascular endothelial cells (HBMECs) (ScienCell, Carlsbad, CA; no. 1000) were cultured in complete Dulbecco’s modified Eagle’s medium (cDMEM with 10% fetal bovine serum [FBS] and 1% penicillin/streptomycin). 6-Bromoindirubin-3′-oxime (BIO; no. 025M4611V), was purchased from Sigma (St. Louis, MO) and used at 1 μm.
Virus production and infection.
Strains PRVABC59 (NCBI accession no. KU501215.1), designated ZIKVPRV, FLR (accession no. KU820897.5), designated ZIKVFLR, IBH30656 (accession no. KU963574.2), designated ZIKVIBH, and MR766 (accession no. LC002520), designated ZIKVMR, were obtained from the ATCC (Manassas, VA). Viruses were propagated in Vero cells by infecting them at a multiplicity of infection (MOI) of 0.01, and supernatants were harvested 72 to 96 h postinfection, when significant cell lysis was observed. The supernatant was centrifuged at 5,000 rpm for 5 min, filtered with a 0.45-μm filter, aliquoted, and frozen at −80°C. Viral titers in Vero cells (96-well plate) were determined with a 50% tissue culture infective dose (TCID50). The number of TCID50s per milliliter was calculated by the Reed-Muench method, and viral titers were expressed as MOIs. For in vitro infection, HFAs were plated at 2 × 105 in 24-well plates, and at 96 h postseeding (∼90% confluence), cells were infected with various strains of ZIKV at the indicated MOIs. After 24 h, additional medium was added to the wells to a total volume of 0.5 ml/well. Infected cells were maintained in culture as indicated per experiment.
MTS and viral entry assays.
Cell viability was measured using the CellTiter 96 AQueous nonradioactive cell proliferation assay (Promega, Madison, WI). It is a colorimetric cell viability assay based on functional mitochondria within viable cells that are able to reduce a tetrazolium compound [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt; MTS] and an electron-coupling reagent, phenazine methosulfate (PMS). MTS is converted into a formazan product which absorbs at 490 nm, which was then detected using a Synergy HT microplate reader using Gen5 software (BioTek, Winooski, VT). Virus within the medium was inactivated by mixing the solution with 70% ethanol at a 2:1 ratio, and the assay was performed per the manufacturer’s instructions. To measure viral internalization, cells were treated as indicated for each experiment, inoculated with ZIKV at an MOI of 0.5 at 37°C for 5 h, washed with phosphate-buffered saline (PBS), and trypsinized to remove surface-bound virus. The cells were then suspended in PBS and collected by centrifugation at 1,500 rpm for 5 min, the supernatant was removed, and then the pellet was suspended in cell lysis buffer for RNA extraction using an RNeasy minikit (Qiagen, Hilden, Germany) and analyzed via reverse transcription-quantitative PCR (qPCR).
Western blotting and antibodies.
HFAs were lysed for 5 min at 37°C with radioimmunoprecipitation assay (RIPA) buffer containing protease inhibitors (Sigma). The lysate was centrifuged at 5,000 rpm for 5 min, and the protein concentration was determined with a Pierce (Waltham, MA) bicinchoninic acid (BCA) protein assay kit. Ten micrograms of cell lysates was loaded on a 10% SDS-PAGE gel for separation and then transferred to a 0.45-μm nitrocellulose membrane and blocked with superblock (Thermo Fisher Waltham, MA) containing Tween 20 (T20); then the primary antibody was added, and the mixture was incubated at 4°C overnight. The membrane was washed with Tris-buffered saline with 0.1% T20 (TBST) and incubated with an appropriate secondary antibody conjugated to horseradish peroxidase in superblock T20 for 1 h at room temperature (RT). The membrane was washed again with TBST and incubated for 3 min with the SuperSignal West Femto maximum-sensitivity substrate (Thermo Scientific). For β-catenin, the primary antibody (rabbit anti-human, no. C2206; Sigma) at a 1:20,000 dilution and secondary mouse anti-rabbit horseradish peroxidase (HRP) (Cell Signaling; no. 7074) at a 1:50,000 dilution were used. For GAPDH (glyceraldehyde-3-phosphate dehydrogenase), the primary antibody (rabbit, no. G9545; Sigma) at a 1:50,000 dilution and secondary mouse anti-rabbit HRP (Cell Signaling; no. 7074) at a 1:100,000 dilution were used. Band intensities were quantified using Image J version 2.1 (NIH, Bethesda, MD) and normalized to that of GAPDH, and data are represented as fold changes from the intensity of the control.
Plasmids and siRNA transfections.
To overexpress β-catenin, HFAs were transiently transfected with 200 ng (per well in a 24-well format) of a constitutively active β-catenin S33Y mutant construct (pABC, no. 19286; Addgene, Cambridge, MA) or a control plasmid (pcDNA3+, no. 10792; Addgene) for 24 h. Similarly, to overexpress Axl, HFAs were transfected with 200 ng of pAxl (pAxl, no. 105933; Addgene). To measure Wnt/β-catenin pathway activity, a β-catenin reporter plasmid, TOPFlash (M50 Super 8×, no. 12456; Addgene) was used. All plasmid transfections were performed using Lipofectamine 3000 (Invitrogen, Carlsbad, CA). All siRNAs used in these studies were purchased as ON-TARGET plus SMARTpool from Horizon Discovery (Cambridge, UK) and used at 100 nM. To reduce β-catenin levels, siRNA targeting β-catenin (L-003482-00) or a scrambled control siRNA (D-001810-10) was transfected into HFAs using Lipofectamine RNAiMAX (Invitrogen) as previously described (23). Knockdown (KD) of Tyro3 and Axl was achieved with respective siRNAs (L-003183-00 and L-003104-00).
Real-time PCR.
Total RNA was isolated from cells with an RNeasy minikit (Qiagen, Cambridge, MA). The A260 was measured using a NanoDrop system. An A260/280 of ≥1.8 was used as a qualitative measure, and 500 ng of total RNA was used for the indicated experiments. DNA contamination was removed by treatment with DNase 1 (Sigma) for 15 min at RT, followed by DNase denaturation for 15 min at 70°C. cDNA was generated using a qScript cDNA synthesis kit (Quantabio, Beverly, MA). Real-time PCR was performed with SSO fast SYBR Green 1 supermix (Bio-Rad, Hercules, CA) in a model 7900 real-time PCR system (Applied Biosystems, Forest City, CA) using SDS software v2.0.1. The PCR conditions were 50°C for 2 min and 95°C for 20s, followed by 40 cycles of 95°C for 15 s and 60°C for 60s. The amount of RNA was normalized to the amount of GAPDH using the ΔCT method, where CT is the threshold cycle, and data are represented as fold changes from the control. The primers used for amplification of ZIKV RNA were common to all ZIKV strains used in this study. They are ZV F (5′-CGC TGC CCA ACA CAA GGT-3′) and ZV R (5′-GCT CCC TTT GCC AAA AAG TCC ACA-3′), Axl primers F (5′-AAC CTT CAA CTC CTG CCT TC-3′) and R (5′-CCA TAA CGG GTC TCC TTC TTT C-3′), Tyro3 primers F (5′-GAG TGT ATG GAG GAC GTG TAT G-3′) and R (5′-GTT CCA TTC GCA GAC AAG TAA AG-3′), β-catenin primers F (5′-TCTTGCCCTTTGTCCCGCAAATCA-3′) and R (5′-TCCACAAATTGCTGTGTCCCAC-3′), and GAPDH primers F (5′-GGT GTG AAC CAT GAG AAG TAT GA-3′) and R (5′-GAG TCC CAC GAT ACC AAA G-3′). All primers were procured from Integrated DNA Technologies (Coralville, IA). Copy number was determined by generating a standard curve using a gBlocks DNA fragment conserved among all ZIKV strains (synthesized by Integrated DNA Technologies), which can be amplified with the above-mentioned common ZIKV primers.
ChIP.
ChIP was performed using the Imprint chromatin immunoprecipitation kit (Sigma) with antibodies for TCF4 and LEF1 from the TCF/LEF family antibody sampler kit (Cell Signaling, Danvers, MA) and a rabbit IgG control (Cell Signaling, Danvers, MA). Per immunoprecipitation, ∼1 × 106 to 2 × 106 cells and 4 μg of antibody were used. Precipitated DNA was washed and purified as per the manufacturer’s instructions. Samples were analyzed by qPCR as indicated above. Primers used to amplify Axl promoter regions were 5′-CACCTGTAATCCCAGCAACT-3′ (forward) and 5′-GAGACCCAGTCTTGCTTTGT-3′ (reverse). Data were normalized to IgG and are represented as fold changes from IgG.
RNAscope.
Approximately 100,000 HFAs were seeded onto coverslips. Twenty-four hours prior to infection, cells were treated with BIO at 1 μm or a vehicle (dimethyl sulfoxide [DMSO]) and incubated with ZIKV at an MOI of 1.0. Cells were then fixed with 4% paraformaldehyde (PFA) at RT for 30 min, washed with PBS, and then dehydrated using 1-min incubations with 50, 70, and 100% ethanol. Coverslips were immobilized on glass slides, rehydrated using ascending grades of ethanol, and permeabilized using PBS with 0.1% Tween 20 for 10 min at RT. Cells were treated with hydrogen peroxide for 10 min at RT and then incubated with a 1:5 dilution of RNAscope protease III (ACD Bio) at 40°C for 5 min. ZIKV RNA probe no. 467771 was hybridized for 2 h at 40°C, and then the signal was developed using the RNAscope 2.5 HD detection RED kit (ACD Bio, Newark, CA) according to the manufacturer’s instructions. Coverslips were mounted with ProLong Gold antifade reagent (Invitrogen, Carlsbad, CA) and imaged on a Keyence fluorescence microscope (Osaka, Japan). RNA spots and nuclei were counted using ImageJ version 2.1 (NIH, Bethesda, MA).
Immunofluorescence staining.
Approximately 100,000 HFAs were seeded onto coverslips in a 24-well format. Twenty-four hours later, cells were transfected with pcDNA or pABC for 24 h, washed twice gently with PBS, fixed with 4% PFA at RT for 15 min, washed thrice with PBS, and permeabilized with 250 μl/well PBS plus 0.2% Triton X-100 for 15 min at RT; then cells were washed thrice with PBS and blocked with PBS plus FBS (6%) plus (0.5%) Tween 20 for 1 h at RT. We then removed the blocking buffer and incubated the mixture with primary antibody in blocking buffer for 2 h at RT or overnight at 4°C. We washed the wells with wash buffer (PBS plus FBS [6%] plus [0.5%] Tween 20) for 5 min thrice and incubated the mixture with fluorescently labelled secondary antibodies in blocking buffer for 1 h at RT. Antibody was removed, and cells were washed with wash buffer for 5 min thrice. Coverslips were mounted with ProLong Gold antifade reagent, left in the dark for 5 min to dry, and imaged on a Keyence BZ-X810 microscope. Images from 40 random spots were quantified using BZ-X800 Analyzer software. For total β-catenin staining, rabbit anti-human primary antibody at a 1:1,500 dilution was used, and for active β-catenin staining (rabbit, no. 8814S; Cell Signaling), anti-human primary antibody at a 1:300 dilution was used. Appropriate fluorescently labeled secondary antibodies were used at a 1:500 dilution.
Flow cytometry.
Adherent HFAs were detached from the plate surface by EDTA treatment, followed by gentle scraping using a cell scraper. Cells were gently pipetted up and down in a 1-ml pipette for a few times to make single-cell suspensions, washed with PBS, stained with mouse anti human Axl-Alexa 488 (clone 108724R; R&D Systems, Minneapolis, MN) and/or mouse anti human Tyro3-Alexa 647 (clone 6201; R&D Systems) or with their appropriate isotype control antibodies for 30 min at RT, and washed twice in PBS. Data were collected on an LSR II flow cytometer with BD FACSDiva software (BD Biosciences, San Jose, CA) and analyzed using FlowJo Software version 10 (TreeStar, Ashland, OR).
Statistical analysis.
Statistical analyses were performed using Prism software (GraphPad Prism, San Diego, CA). The variables were compared using either one-way analysis of variance (ANOVA) and Bonferroni's multiple-comparison test or a two-tailed one-sample t test. All experiments were performed independently at least three times, and data are represented as means or fold changes with standard errors of means (SEM), with a P of ≤0.05 considered statistically significant.
Data availability.
Data are available at https://doi.org/10.17632/bbnxxpc9cr.1 and at the preview URL https://data.mendeley.com/datasets/bbnxxpc9cr/draft?a=b4d06d8d-8af7-45fa-b7c4-69f167b704a8.
ACKNOWLEDGMENTS
This work was supported by the Thomas J. Coogan, Sr., MD, endowment (L.A.-H.).
We thank Kiran Kondabagil (Department of Biosciences and Bioengineering, Indian Institute of Technology-Bombay, Mumbai, India) for assistance with sequence alignment and structure prediction.
O.A.J., S.D.N., and L.A.-H. designed the experiments, analyzed the data, and wrote the manuscript. O.A.J. and S.D.N. performed the experiments. S.D.N. and L.A.H. supervised the research. H.J.B. performed RNA scope experiments, analyzed data, and assisted in revising the manuscript. Y.A.A. assisted with flow experiments and analyses. M.S.S. supported IF staining. K.F.R. assisted with ChIP experiments. All authors edited the manuscript.
We declare that we have no competing interests.
Contributor Information
Lena Al-Harthi, Email: Lena_Al-Harthi@rush.edu.
Susana López, Instituto de Biotecnologia/UNAM.
REFERENCES
- 1.Dick GW, Kitchen SF, Haddow AJ. 1952. Zika virus. I. Isolations and serological specificity. Trans R Soc Trop Med Hyg 46:509–520. 10.1016/0035-9203(52)90042-4. [DOI] [PubMed] [Google Scholar]
- 2.Grant A, Ponia SS, Tripathi S, Balasubramaniam V, Miorin L, Sourisseau M, Schwarz MC, Sanchez-Seco MP, Evans MJ, Best SM, Garcia-Sastre A. 2016. Zika virus targets human STAT2 to inhibit type I interferon signaling. Cell Host Microbe 19:882–890. 10.1016/j.chom.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liang Q, Luo Z, Zeng J, Chen W, Foo SS, Lee SA, Ge J, Wang S, Goldman SA, Zlokovic BV, Zhao Z, Jung JU. 2016. Zika virus NS4A and NS4B proteins deregulate Akt-mTOR signaling in human fetal neural stem cells to inhibit neurogenesis and induce autophagy. Cell Stem Cell 19:663–671. 10.1016/j.stem.2016.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yun SI, Lee YM. 2017. Zika virus: an emerging flavivirus. J Microbiol 55:204–219. 10.1007/s12275-017-7063-6. [DOI] [PubMed] [Google Scholar]
- 5.Klase ZA, Khakhina S, Schneider ADB, Callahan MV, Glasspool-Malone J, Malone R. 2016. Zika fetal neuropathogenesis: etiology of a viral syndrome. PLoS Negl Trop Dis 10:e0004877. 10.1371/journal.pntd.0004877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lum FM, Low DK, Fan Y, Tan JJ, Lee B, Chan JK, Renia L, Ginhoux F, Ng LF. 2017. Zika virus infects human fetal brain microglia and induces inflammation. Clin Infect Dis 64:914–920. 10.1093/cid/ciw878. [DOI] [PubMed] [Google Scholar]
- 7.Retallack H, Di Lullo E, Arias C, Knopp KA, Laurie MT, Sandoval-Espinosa C, Mancia Leon WR, Krencik R, Ullian EM, Spatazza J, Pollen AA, Mandel-Brehm C, Nowakowski TJ, Kriegstein AR, DeRisi JL. 2016. Zika virus cell tropism in the developing human brain and inhibition by azithromycin. Proc Natl Acad Sci U S A 113:14408–14413. 10.1073/pnas.1618029113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang H, Hammack C, Ogden SC, Wen Z, Qian X, Li Y, Yao B, Shin J, Zhang F, Lee EM, Christian KM, Didier RA, Jin P, Song H, Ming GL. 2016. Zika virus infects human cortical neural progenitors and attenuates their growth. Cell Stem Cell 18:587–590. 10.1016/j.stem.2016.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van den Pol AN, Mao G, Yang Y, Ornaghi S, Davis JN. 2017. Zika virus targeting in the developing brain. J Neurosci 37:2161–2175. 10.1523/JNEUROSCI.3124-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Limonta D, Jovel J, Kumar A, Airo AM, Hou S, Saito L, Branton W, Ka-Shu Wong G, Mason A, Power C, Hobman TC. 2018. Human fetal astrocytes infected with Zika virus exhibit delayed apoptosis and resistance to interferon: implications for persistence. Viruses 10:646. 10.3390/v10110646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang Y, Li Y, Zhang H, Zhao R, Jing R, Xu Y, He M, Peer J, Kim YC, Luo J, Tong Z, Zheng J. 2018. Zika virus propagation and release in human fetal astrocytes can be suppressed by neutral sphingomyelinase-2 inhibitor GW4869. Cell Discov 4:19. 10.1038/s41421-018-0017-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schitine C, Nogaroli L, Costa MR, Hedin-Pereira C. 2015. Astrocyte heterogeneity in the brain: from development to disease. Front Cell Neurosci 9:76. 10.3389/fncel.2015.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gallo V, Deneen B. 2014. Glial development: the crossroads of regeneration and repair in the CNS. Neuron 83:283–308. 10.1016/j.neuron.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brafman D, Willert K. 2017. Wnt/beta-catenin signaling during early vertebrate neural development. Dev Neurobiol 77:1239–1259. 10.1002/dneu.22517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tapia-Rojas C, Inestrosa NC. 2018. Wnt signaling loss accelerates the appearance of neuropathological hallmarks of Alzheimer's disease in J20-APP transgenic and wild-type mice. J Neurochem 144:443–465. 10.1111/jnc.14278. [DOI] [PubMed] [Google Scholar]
- 16.Hoseth EZ, Krull F, Dieset I, Morch RH, Hope S, Gardsjord ES, Steen NE, Melle I, Brattbakk HR, Steen VM, Aukrust P, Djurovic S, Andreassen OA. 2018. Exploring the Wnt signaling pathway in schizophrenia and bipolar disorder. Transl Psychiatry 8:55. 10.1038/s41398-018-0102-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richards MH, Narasipura SD, Kim S, Seaton MS, Lutgen V, Al-Harthi L. 2015. Dynamic interaction between astrocytes and infiltrating PBMCs in context of neuroAIDS. Glia 63:441–451. 10.1002/glia.22763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lutgen V, Narasipura SD, Sharma A, Min S, Al-Harthi L. 2016. beta-Catenin signaling positively regulates glutamate uptake and metabolism in astrocytes. J Neuroinflammation 13:242. 10.1186/s12974-016-0691-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Al-Harthi L. 2012. Wnt/β-catenin and its diverse physiological cell signaling pathways in neurodegenerative and neuropsychiatric disorders. J Neuroimmune Pharmacol 7:725–730. 10.1007/s11481-012-9412-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.More S, Yang X, Zhu Z, Bamunuarachchi G, Guo Y, Huang C, Bailey K, Metcalf JP, Liu L. 2018. Regulation of influenza virus replication by Wnt/beta-catenin signaling. PLoS One 13:e0191010. 10.1371/journal.pone.0191010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hillesheim A, Nordhoff C, Boergeling Y, Ludwig S, Wixler V. 2014. Beta-catenin promotes the type I IFN synthesis and the IFN-dependent signaling response but is suppressed by influenza A virus-induced RIG-I/NF-kappaB signaling. Cell Commun Signal 12:29. 10.1186/1478-811X-12-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roe K, Kumar M, Lum S, Orillo B, Nerurkar VR, Verma S. 2012. West Nile virus-induced disruption of the blood-brain barrier in mice is characterized by the degradation of the junctional complex proteins and increase in multiple matrix metalloproteinases. J Gen Virol 93:1193–1203. 10.1099/vir.0.040899-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Narasipura SD, Henderson LJ, Fu SW, Chen L, Kashanchi F, Al-Harthi L. 2012. Role of beta-catenin and TCF/LEF family members in transcriptional activity of HIV in astrocytes. J Virol 86:1911–1921. 10.1128/JVI.06266-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henderson LJ, Narasipura SD, Adarichev V, Kashanchi F, Al-Harthi L. 2012. Identification of novel T cell factor 4 (TCF-4) binding sites on the HIV long terminal repeat which associate with TCF-4, beta-catenin, and SMAR1 to repress HIV transcription. J Virol 86:9495–9503. 10.1128/JVI.00486-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richard AS, Shim BS, Kwon YC, Zhang R, Otsuka Y, Schmitt K, Berri F, Diamond MS, Choe H. 2017. AXL-dependent infection of human fetal endothelial cells distinguishes Zika virus from other pathogenic flaviviruses. Proc Natl Acad Sci U S A 114:2024–2029. 10.1073/pnas.1620558114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meertens L, Labeau A, Dejarnac O, Cipriani S, Sinigaglia L, Bonnet-Madin L, Le Charpentier T, Hafirassou ML, Zamborlini A, Cao-Lormeau VM, Coulpier M, Misse D, Jouvenet N, Tabibiazar R, Gressens P, Schwartz O, Amara A. 2017. Axl mediates ZIKA virus entry in human glial cells and modulates innate immune responses. Cell Rep 18:324–333. 10.1016/j.celrep.2016.12.045. [DOI] [PubMed] [Google Scholar]
- 27.Chen J, Yang YF, Yang Y, Zou P, Chen J, He Y, Shui SL, Cui YR, Bai R, Liang YJ, Hu Y, Jiang B, Lu L, Zhang X, Liu J, Xu J. 2018. AXL promotes Zika virus infection in astrocytes by antagonizing type I interferon signalling. Nat Microbiol 3:302–309. 10.1038/s41564-017-0092-4. [DOI] [PubMed] [Google Scholar]
- 28.Liu S, DeLalio LJ, Isakson BE, Wang TT. 2016. AXL-mediated productive infection of human endothelial cells by Zika virus. Circ Res 119:1183–1189. 10.1161/CIRCRESAHA.116.309866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wells MF, Salick MR, Wiskow O, Ho DJ, Worringer KA, Ihry RJ, Kommineni S, Bilican B, Klim JR, Hill EJ, Kane LT, Ye C, Kaykas A, Eggan K. 2016. Genetic ablation of AXL does not protect human neural progenitor cells and cerebral organoids from Zika virus infection. Cell Stem Cell 19:703–708. 10.1016/j.stem.2016.11.011. [DOI] [PubMed] [Google Scholar]
- 30.Wang ZY, Wang Z, Zhen ZD, Feng KH, Guo J, Gao N, Fan DY, Han DS, Wang PG, An J. 2017. Axl is not an indispensable factor for Zika virus infection in mice. J Gen Virol 98:2061–2068. 10.1099/jgv.0.000886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hastings AK, Yockey LJ, Jagger BW, Hwang J, Uraki R, Gaitsch HF, Parnell LA, Cao B, Mysorekar IU, Rothlin CV, Fikrig E, Diamond MS, Iwasaki A. 2017. TAM receptors are not required for Zika virus infection in mice. Cell Rep 19:558–568. 10.1016/j.celrep.2017.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xie H, Huang Z, Sadim MS, Sun Z. 2005. Stabilized beta-catenin extends thymocyte survival by up-regulating Bcl-xL. J Immunol 175:7981–7988. 10.4049/jimmunol.175.12.7981. [DOI] [PubMed] [Google Scholar]
- 33.Meijer L, Skaltsounis AL, Magiatis P, Polychronopoulos P, Knockaert M, Leost M, Ryan XP, Vonica CA, Brivanlou A, Dajani R, Crovace C, Tarricone C, Musacchio A, Roe SM, Pearl L, Greengard P. 2003. GSK-3-selective inhibitors derived from Tyrian purple indirubins. Chem Biol 10:1255–1266. 10.1016/j.chembiol.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 34.Robinson K, Narasipura SD, Wallace J, Ritz EM, Al-Harthi L. 2020. β-Catenin and TCFs/LEF signaling discordantly regulate IL-6 expression in astrocytes. Cell Commun Signal 18:93. 10.1186/s12964-020-00565-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hamel R, Dejarnac O, Wichit S, Ekchariyawat P, Neyret A, Luplertlop N, Perera-Lecoin M, Surasombatpattana P, Talignani L, Thomas F, Cao-Lormeau V-M, Choumet V, Briant L, Desprès P, Amara A, Yssel H, Missé D. 2015. Biology of Zika virus infection in human skin cells. J Virol 89:8880–8896. 10.1128/JVI.00354-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nowakowski TJ, Pollen AA, Di Lullo E, Sandoval-Espinosa C, Bershteyn M, Kriegstein AR. 2016. Expression analysis highlights AXL as a Candidate Zika virus entry receptor in neural stem cells. Cell Stem Cell 18:591–596. 10.1016/j.stem.2016.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mudduluru G, Allgayer H. 2008. The human receptor tyrosine kinase Axl gene—promoter characterization and regulation of constitutive expression by Sp1, Sp3 and CpG methylation. Biosci Rep 28:161–176. 10.1042/BSR20080046. [DOI] [PubMed] [Google Scholar]
- 38.Tabata T, Petitt M, Puerta-Guardo H, Michlmayr D, Wang C, Fang-Hoover J, Harris E, Pereira L. 2016. Zika virus targets different primary human placental cells, suggesting two routes for vertical transmission. Cell Host Microbe 20:155–166. 10.1016/j.chom.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu Z, Mesci P, Bernatchez JA, Gimple RC, Wang X, Schafer ST, Wettersten HI, Beck S, Clark AE, Wu Q, Prager BC, Kim LJY, Dhanwani R, Sharma S, Garancher A, Weis SM, Mack SC, Negraes PD, Trujillo CA, Penalva LO, Feng J, Lan Z, Zhang R, Wessel AW, Dhawan S, Diamond MS, Chen CC, Wechsler-Reya RJ, Gage FH, Hu H, Siqueira-Neto JL, Muotri AR, Cheresh DA, Rich JN. 2020. Zika virus targets glioblastoma stem cells through a SOX2-integrin alphavbeta5 axis. Cell Stem Cell 26:187–204.e10. 10.1016/j.stem.2019.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Papa MP, Meuren LM, Coelho SVA, Lucas CGO, Mustafa YM, Lemos Matassoli F, Silveira PP, Frost PS, Pezzuto P, Ribeiro MR, Tanuri A, Nogueira ML, Campanati L, Bozza MT, Paula Neto HA, Pimentel-Coelho PM, Figueiredo CP, de Aguiar RS, de Arruda LB. 2017. Zika virus infects, activates, and crosses brain microvascular endothelial cells, without barrier disruption. Front Microbiol 8:2557. 10.3389/fmicb.2017.02557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mladinich MC, Schwedes J, Mackow ER. 2017. Zika virus persistently infects and is basolaterally released from primary human brain microvascular endothelial cells. mBio 8:e00952-17. 10.1128/mBio.00952-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ledur PF, Karmirian K, Pedrosa C, Souza LRQ, Assis-de-Lemos G, Martins TM, Ferreira J, de Azevedo Reis GF, Silva ES, Silva D, Salerno JA, Ornelas IM, Devalle S, Madeiro da Costa RF, Goto-Silva L, Higa LM, Melo A, Tanuri A, Chimelli L, Murata MM, Garcez PP, Filippi-Chiela EC, Galina A, Borges HL, Rehen SK. 2020. Zika virus infection leads to mitochondrial failure, oxidative stress and DNA damage in human iPSC-derived astrocytes. Sci Rep 10:1218. 10.1038/s41598-020-57914-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oliveira DB, Almeida FJ, Durigon EL, Mendes EA, Braconi CT, Marchetti I, Andreata-Santos R, Cunha MP, Alves RP, Pereira LR, Melo SR, Neto DF, Mesquita FS, Araujo DB, Favoretto SR, Safadi MA, Ferreira LC, Zanotto PM, Botosso VF, Berezin EN. 2016. Prolonged shedding of Zika virus associated with congenital infection. N Engl J Med 375:1202–1204. 10.1056/NEJMc1607583. [DOI] [PubMed] [Google Scholar]
- 44.Oliveira DBL, Durigon GS, Mendes EA. 2018. Persistence and intra-host genetic evolution of Zika virus infection in symptomatic adults: a special view in the male reproductive system. Viruses 10:615. 10.3390/v10110615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Campos RdM, Cirne-Santos C, Meira GLS, Santos LLR, de Meneses MD, Friedrich J, Jansen S, Ribeiro MS, da Cruz IC, Schmidt-Chanasit J, Ferreira DF. 2016. Prolonged detection of Zika virus RNA in urine samples during the ongoing Zika virus epidemic in Brazil. J Clin Virol 77:69–70. 10.1016/j.jcv.2016.02.009. [DOI] [PubMed] [Google Scholar]
- 46.Strange DP, Jiyarom B, Pourhabibi Zarandi N, Xie X. 2019. Axl promotes Zika virus entry and modulates the antiviral state of human Sertoli cells. mBio 10:e01372-19. 10.1128/mBio.01372-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen J, Yang YF, Chen J, Zhou X, Dong Z, Chen T, Yang Y, Zou P, Jiang B, Hu Y, Lu L, Zhang X, Liu J, Xu J, Zhu T. 2017. Zika virus infects renal proximal tubular epithelial cells with prolonged persistency and cytopathic effects. Emerg Microbes Infect 6:e77. 10.1038/emi.2017.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haddow AD, Schuh AJ, Yasuda CY, Kasper MR, Heang V, Huy R, Guzman H, Tesh RB, Weaver SC. 2012. Genetic characterization of Zika virus strains: geographic expansion of the Asian lineage. PLoS Negl Trop Dis 6:e1477. 10.1371/journal.pntd.0001477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kabat D, Kozak SL, Wehrly K, Chesebro B. 1994. Differences in CD4 dependence for infectivity of laboratory-adapted and primary patient isolates of human immunodeficiency virus type 1. J Virol 68:2570–2577. 10.1128/JVI.68.4.2570-2577.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dai L, Song J, Lu X, Deng YQ, Musyoki AM, Cheng H, Zhang Y, Yuan Y, Song H, Haywood J, Xiao H, Yan J, Shi Y, Qin CF, Qi J, Gao GF. 2016. Structures of the Zika virus envelope protein and its complex with a flavivirus broadly protective antibody. Cell Host Microbe 19:696–704. 10.1016/j.chom.2016.04.013. [DOI] [PubMed] [Google Scholar]
- 51.Kostyuchenko VA, Lim EX, Zhang S, Fibriansah G, Ng TS, Ooi JS, Shi J, Lok SM. 2016. Structure of the thermally stable Zika virus. Nature 533:425–428. 10.1038/nature17994. [DOI] [PubMed] [Google Scholar]
- 52.Meertens L, Carnec X, Lecoin MP, Ramdasi R, Guivel-Benhassine F, Lew E, Lemke G, Schwartz O, Amara A. 2012. The TIM and TAM families of phosphatidylserine receptors mediate dengue virus entry. Cell Host Microbe 12:544–557. 10.1016/j.chom.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yuan L, Huang XY, Liu ZY, Zhang F, Zhu XL, Yu JY, Ji X, Xu YP, Li G, Li C, Wang HJ, Deng YQ, Wu M, Cheng ML, Ye Q, Xie DY, Li XF, Wang X, Shi W, Hu B, Shi PY, Xu Z, Qin CF. 2017. A single mutation in the prM protein of Zika virus contributes to fetal microcephaly. Science 358:933–936. 10.1126/science.aam7120. [DOI] [PubMed] [Google Scholar]
- 54.Xia H, Luo H, Shan C, Muruato AE, Nunes BTD, Medeiros DBA, Zou J, Xie X, Giraldo MI, Vasconcelos PFC, Weaver SC, Wang T, Rajsbaum R, Shi PY. 2018. An evolutionary NS1 mutation enhances Zika virus evasion of host interferon induction. Nat Commun 9:414. 10.1038/s41467-017-02816-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Laureti M, Narayanan D, Rodriguez-Andres J, Fazakerley JK, Kedzierski L. 2018. Flavivirus receptors: diversity, identity, and cell entry. Front Immunol 9:2180. 10.3389/fimmu.2018.02180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gaburro J, Bhatti A, Sundaramoorthy V, Dearnley M, Green D, Nahavandi S, Paradkar PN, Duchemin JB. 2018. Zika virus-induced hyper excitation precedes death of mouse primary neuron. Virol J 15:79. 10.1186/s12985-018-0989-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Costa VV, Del Sarto JL, Rocha RF, Silva FR, Doria JG, Olmo IG, Marques RE, Queiroz-Junior CM, Foureaux G, Araujo JMS, Cramer A, Real A, Ribeiro LS, Sardi SI, Ferreira AJ, Machado FS, de Oliveira AC, Teixeira AL, Nakaya HI, Souza DG, Ribeiro FM, Teixeira MM. 2017. N-Methyl-d-aspartate (NMDA) receptor blockade prevents neuronal death induced by Zika virus infection. mBio 8:e00350-17. 10.1128/mBio.00350-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yu C, Narasipura SD, Richards MH, Hu XT, Yamamoto B, Al-Harthi L. 2017. HIV and drug abuse mediate astrocyte senescence in a beta-catenin-dependent manner leading to neuronal toxicity. Aging Cell 16:956–965. 10.1111/acel.12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Henderson LJ, Sharma A, Monaco MC, Major EO, Al-Harthi L. 2012. Human immunodeficiency virus type 1 (HIV-1) transactivator of transcription through its intact core and cysteine-rich domains inhibits Wnt/beta-catenin signaling in astrocytes: relevance to HIV neuropathogenesis. J Neurosci 32:16306–16313. 10.1523/JNEUROSCI.3145-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Al-Harthi L. 2012. Interplay between Wnt/beta-catenin signaling and HIV: virologic and biologic consequences in the CNS. J Neuroimmune Pharmacol 7:731–739. 10.1007/s11481-012-9411-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data are available at https://doi.org/10.17632/bbnxxpc9cr.1 and at the preview URL https://data.mendeley.com/datasets/bbnxxpc9cr/draft?a=b4d06d8d-8af7-45fa-b7c4-69f167b704a8.