ABSTRACT
The hemagglutinin (HA) surface protein is the primary immune target for most influenza vaccines. The neuraminidase (NA) surface protein is often a secondary target for vaccine designs. In this study, computationally optimized broadly reactive antigen (COBRA) methodology was used to generate the N1-I NA vaccine antigen that was designed to cross-react with avian, swine, and human influenza viruses of the N1 NA subtype. The elicited antibodies bound to NA proteins derived from A/California/07/2009 (H1N1)pdm09, A/Brisbane/59/2007 (H1N1), A/Swine/North Carolina/154074/2015 (H1N1), and A/Viet Nam/1203/2004 (H5N1) influenza viruses, with NA-neutralizing activity against a broad panel of HXN1 influenza strains. Mice vaccinated with the N1-I COBRA NA vaccine were protected from mortality and viral lung titers were lower when challenged with four different viral challenges (A/California/07/2009, A/Brisbane/59/2007, A/Swine/North Carolina/154074/2015, and A/Viet Nam/1203/2004). Vaccinated mice had little to no weight loss against both homologous, but also cross-NA, genetic clade challenges. Lung viral titers were lower than the mock-vaccinated mice and, at times, equivalent to the homologous control. Thus, the N1-I COBRA NA antigen has the potential to be a complementary component in a multiantigen universal influenza virus vaccine formulation that also contains HA antigens.
IMPORTANCE The development and distribution of a universal influenza vaccine would alleviate global economic and public health stress from annual influenza virus outbreaks. The influenza virus NA vaccine antigen allows for protection from multiple HA subtypes and virus host origins, but it has not been the focus of vaccine development. The N1-I NA antigen described here protected mice from direct challenge of four distinct influenza viruses and inhibited the enzymatic activity of an N1 influenza virus panel. The use of the NA antigen in combination with the HA antigen widens the breadth of protection against various virus strains. Therefore, this research opens the door to the development of a longer-lasting vaccine with increased protective breadth.
KEYWORDS: COBRA, neuraminidase inhibition, mice, influenza, N1, vaccine
INTRODUCTION
Influenza remains in the forefront of communicable diseases due to reoccurring global seasonal epidemics with pandemic potential. This negative-sense, single-stranded RNA virus contains an eight-segmented genome with the virion surface studded with viral hemagglutinin (HA) and neuraminidase (NA) glycoproteins. The sialic acid binding activity of the HA controls receptor binding specificity and thus host-cell fusion. Comparatively, the sialidase enzymatic activity of the NA contributes to cleavage of mucins, motility, release of progeny virions, and prevention of self-aggregation (1). During infection, virions can be neutralized by antibodies targeting one of these two proteins (1, 2). Viral isolates are classified by the HA and NA subtypes that are independently characterized based upon serological cross-reactivity (e.g., H1N1, H5N1, H3N2).
The N1 NA subtype can be matched with different HA subtypes and three distinct genetic NA clades are defined by phylogenetic analysis based on the NA nucleic acid sequences: the N1.1, N1.2, and N1.3 genetic clades which correspond to avian-like, human-like, and classical swine-like, respectively (3, 4). The N1.1 clade is the most diverse, and the N1.2 and N1.3 clades follow more closely a temporal evolution pattern. Prior to the pandemic in 2009, the N1 that dominated the human infections belonged to the human-like N1.2 clade. Further, the Eurasian swine viruses commonly contain the avian-like N1.1 clade NA protein. Thus, the 2009 pandemic NA originated from the Eurasian swine lineage with protein sequences more similar to the NA protein from highly pathogenic avian H5N1 viruses (N1.1) than to either the seasonal human (N1.2) or classical swine NA (N1.3). The classical swine NA proteins continue to circulate throughout the North American swine populations. Furthermore, through reassortment, some isolated swine-origin influenza viruses contain human-seasonal NA, which were introduced to the swine population through human interactions. Each of the three clades, i.e., N1.1 (highly pathogenic avian H5N1 influenza, 2009 pandemic H1N1 influenza), N1.2 (seasonal H1N1 influenza; e.g., A/Brisbane/59/2007), and N1.3 (H1N1 variant influenza), has been isolated from virus-infected humans. NA proteins from the three clades have been isolated from humans, indicating a potential for human adaptability and designating NA as a promising vaccine target (5, 6).
Indeed, vaccination remains the main method for prevention of influenza virus-induced disease. One of the most commonly used seasonal influenza virus vaccines for humans is a split-inactivated virus vaccine without removal of components of the virion. Although all of the components are present, the immunological response to the NA protein is limited. The structural integrity of the protein, the relative ratio of HA to NA, and immunodominance between antigens contribute to a minimal NA-specific immune response after vaccination (7, 8), even though influenza virus infection elicits NA antibodies (9). Split-virion vaccination increases antibody titers against the HA protein, but not against the NA protein (10). This differential antigenicity may be due to the vaccine preparation, but is difficult to determine due to the lack of methods for quantifying and standardizing NA protein content in vaccine doses. Despite the NA not being a standardized vaccine antigen, the clinical use of NA-inhibiting antivirals has led the reduction of influenza disease severity, duration, mortality, and hospitalization (11–14), and the anti-NA antibody titers are virus neutralizing (15–17) and meaningful serological correlates of protection in humans (18–20).
Approaches toward the development of a universal or broadly reactive influenza virus vaccine have focused on eliciting antibodies targeting the HA surface protein with less interest focused on the NA (21). However, with a broadly protective N1-based vaccine, both human seasonal and pandemic H1 epidemics can be managed, along with decreasing the potential for zoonotic pandemics from avian H5N1 and another swine-origin pandemic H1N1. The use of the NA as an immunogen crosses more HA subtypes and species-of-origin than the use of one HA subtype immunogen. Specific monoclonal antibodies (MAbs) bind conserved epitopes on human seasonal H1N1, human 2009 pandemic H1N1, and pandemic H5N1 NA proteins (22). Within the human host, the HA has higher nucleic acid substitution rates than NA (23); therefore, HA-specific protective antibodies are not as effective over many seasons compared to antibodies against NA. The use of an NA-based vaccine may potentially provide a longer-lasting vaccine antigen than current vaccination methods that rely primarily on immune responses against HA. The standardization of NA antigen in the influenza virus vaccine may contribute to increased vaccine efficacy. Clinical symptoms, peak viral titers, and viral shedding are inversely correlated with NA-inhibiting antibody titers (19).
In this report, the development and characterization of a computationally optimized broadly (COBRA) reactive NA antigen is described. Previously, the COBRA methodology was used to design broadly reactive HA-based vaccine candidates for H1, H2, H3, and H5 influenza A virus subtypes. These COBRA HA candidates are effective in mice, ferrets, chickens, and nonhuman primates (24–29). The design of a COBRA antigen requires variability in the target antigen amino acid sequence to produce a unique immunogen protein sequence after consecutive consensus layering. The diverse genetic clades of NA, antigenic drift, and influenza virus antigen sequencing surveillance contributes to the variability of the available wild-type NA sequences. N1 COBRA NA protein immunogens were designed using wild-type NA sequences from human, avian, and swine influenza isolates. The N1 COBRA and wild-type NA proteins are immunogenic as tetrameric soluble protein vaccines. They elicit broadly reactive antibodies across a panel of N1 viruses.
RESULTS
Design of N1 COBRA NA sequences.
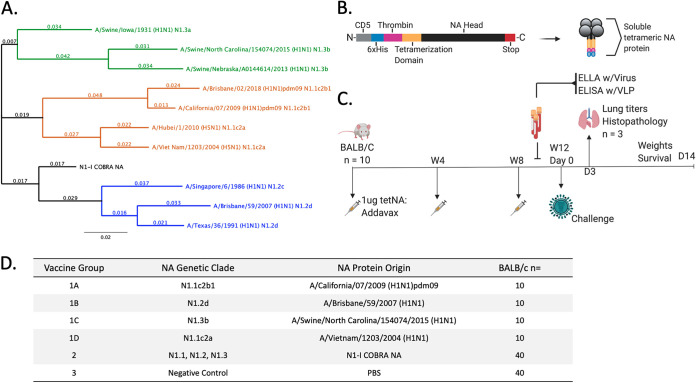
The computationally optimized broadly reactive NA antigen was designed from sequences obtained from the GISAID database (30). The N1-I COBRA NA antigen was designed using human, avian, and swine origin influenza virus NA sequences from 1990 to 2015 depending on host origin. The N1-I COBRA NA was phylogenetically located close to the branch point of the human-like NA clade N1.2 (Fig. 1A). When aligned to representative wild-type sequences, the N1-I COBRA NA had between 31 to 44 amino acids differences. The Sw/IA/1931 NA protein was the most similar to N1-I COBRA NA and the Bris/18 and Sw/NE/13 NA proteins were the least similar, with 44 amino acids different from the N1-I COBRA NA.
FIG 1.
Experimental groups and design used for influenza challenge. (A) Phylogenetic tree of truncated NA amino acid sequences (74 to 470). The N1-I COBRA NA (black) branched closer to the root of the tree than all other wild-type NA proteins. Representative viruses for each N1 clade (N1.1, orange; N1.2, blue; N1.3, green) were included. Scale bar = substitutions per site; total sites = 396 amino acids. (B) Cartoon schematic of soluble tetrameric NA protein used for vaccinations. From the N-terminal to C-terminal ends of the protein sequence, the domains were CD5 signal sequence (CD5; cleaved during protein processing), hexahistidine domain (6×His), thrombin cleavage domain (Thrombin), Staphylothermus marinus tetrabrachion domain (Tetramerization Domain), the NA head region of the select antigen (NA Head), and a double stop codon (Stop). (C) Vaccination and challenge regimen followed for each challenge. Female BALB/c mice (n = 10) were vaccinated with tetrameric soluble NA (tetNA) in a prime-boost-boost at 4-week intervals. Sera were collected after vaccinations and prior to challenge for serological assays (enzyme-linked lectin assay [ELLA]; enzyme-linked immunosorbent assay [ELISA] with virus-like particles [VLP]). Each of the four vaccine groups were challenged with one of the four challenge viruses. Over the course of infection, weights were monitored for up to 14 days, and lungs (n = 3) were harvested on day 3. (D) The vaccine groups for each challenge. Vaccine group 1 for each challenge varied and was the homologous control for the matched virus.
Soluble recombinant tetrameric NA protein vaccines are immunogenic.
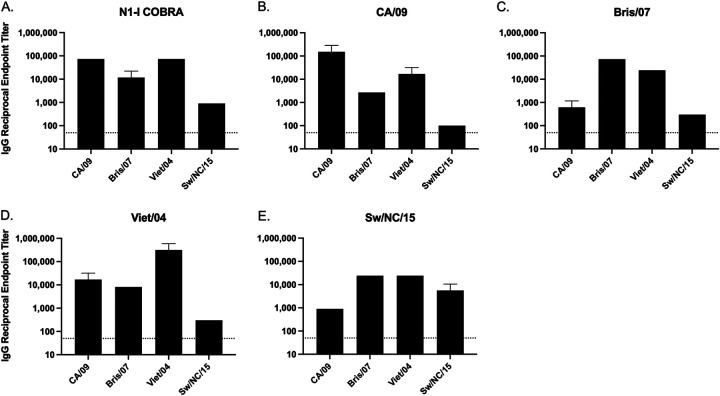
Sera was collected at week 12 (Fig. 1C) from mice (BALB/c) vaccinated at weeks 0, 4, and 8 with one of five tetrameric NA (tetNA) proteins (Fig. 1D). The adjuvanted tetNA soluble protein vaccines elicited an NA-specific antibody response and were confirmed to be immunogenic (Fig. 2). All NA vaccinations elicited IgG antibodies that bound to all four wild-type NA antigens presented in a tetramer formation on the surface of a virus-like particle. The N1-I COBRA NA group had endpoint titers of 1:1,000 or greater to each of the challenge virus NA proteins (Fig. 2A). The greatest titers were to CA/09 and Viet/04 NA antigens, and the lowest endpoint titer was to the Sw/NC/15 antigen. However, the binding of N1-I sera to Sw/NC/15 was comparable to how the wild-type Sw/NC/15 NA vaccination elicited endpoint titers of 1:890 and 1:5,620, respectively (Fig. 2E). Mice vaccinated with the Viet/04 NA had the highest anti-NA IgG titers when binding to the homologous protein of 1:316,000 (Fig. 2D). CA/09 and Viet/04 NA antigens elicited the highest binding toward the N1.1 clade proteins (Fig. 2B and D), whereas Bris/07 had higher titers to itself and Viet/04 (Fig. 2C). Sw/NC/15 NA antigen elicited antibodies bound at similar titers to Bris/07, Viet/04, and Sw/NC/15 NA protein. All vaccine antigens elicited antibodies that were able to bind to the Viet/04 NA protein. The mock-vaccinated group did not elicit detectable antibodies to the tested proteins (data not shown).
FIG 2.
Total IgG antibody binding of vaccinated mouse sera to tetrameric NA expressed on the surface of a virus-like particle. The homologous sera had the highest reciprocal endpoint titer for each challenge virus. The N1-I COBRA NA elicited binding to all four challenge NAs, and the mock vaccinations produced no measurable binding (not shown). Reciprocal serum endpoint titers were determined with raw serum starting from an initial dilution of 1:100 after a prime-boost-boost vaccination regimen with soluble tetNA. The endpoint titer was the last serum titer that produced an absorbance greater than 5 standard deviations above the background absorbance. Serum that did not produce an absorbance greater than the cutoff was defined as below the limit of detection (LOD), depicted as 50 reciprocal endpoint titer. The serum was analyzed in triplicate and the geometric mean reciprocal endpoint titer was calculated. Error bars represent the geometric standard deviation.
Soluble recombinant tetrameric NA protein vaccines elicit NA-inhibiting antibodies.
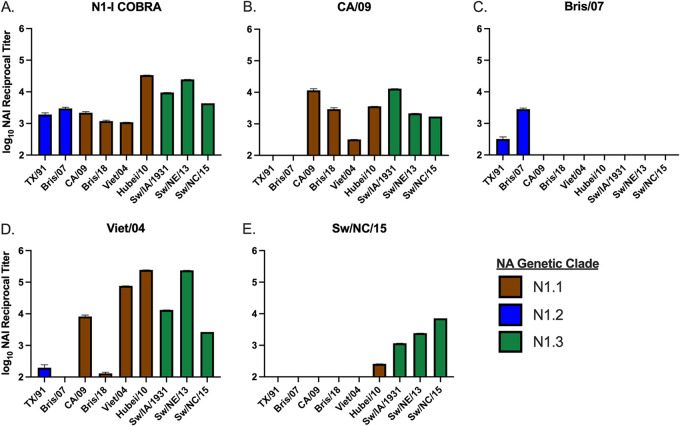
Collected antiserum was examined for the ability to inhibit the NA enzymatic activity through an enzyme-linked lectin assay (ELLA) (Fig. 3, Fig. S1 in the supplemental material). The ELLA measures the ability of NA to cleave sialic acids from the terminal ends of fetuin. Mice vaccinated with the COBRA N1-I NA vaccine had antibodies that inhibited all tested HXN1 viruses in the N1.1, N1.2, and N1.3 genetic clades (Fig. 3A). Mice vaccinated with CA/09 NA protein had broad NA inhibition with cross-reactivity in the N1.1 and swine-like N1.3 clades (Fig. 3B). Conversely, sera collected from mice vaccinated with the Bris/07 NA had the narrowest response, only inhibiting the human-like clade N1.2 H1N1 viruses (TX/91 and Bris/07) and no viruses from either clade N1.1 or N1.3 (Fig. 3C). Mice vaccinated with Viet/04 NA proteins had a NA inhibition activity pattern similar to CA/09-vaccinated mice (Fig. 3D). However, the CA/09 NA-vaccinated sera were able to inhibit Bris/18, and the Viet/04 sera had higher NA inhibition titers to Viet/04, Hubei/10, and Sw/NE/13 viruses. The Sw/NC/15 NA-elicited sera had a narrow breadth and inhibited the swine origin viruses and Hubei/10 (Fig. 3E). In conclusion, the COBRA N1-I NA protein elicited a broad inhibitory response mediated by antibody binding.
FIG 3.
Reciprocal NA inhibition (NAI) titers across a panel of N1 influenza viruses. The N1-I COBRA vaccinated sera (A) inhibited all representative viruses in the panel (B). The wild-type sera (B to E) elicited various levels of inhibition breadth from very broad (D) to narrow (C and E), but was not as broad as the N1-I COBRA. The enzyme-linked lectin assays (ELLAs) were performed with RDE-treated and heat-inactivated sera initially diluted to 1:100 followed by 2-fold dilutions. The limit of detection was defined as 2.0 log10 NAI reciprocal titer. The mean log10 NAI reciprocal titer that inhibited 50% of the NA activity with 95% confidence intervals was estimated with nonlinear regression of the ELLA dilution curves (Fig. S1 in the supplemental material).
Vaccinated mice challenged with human pandemic H1N1 influenza virus.
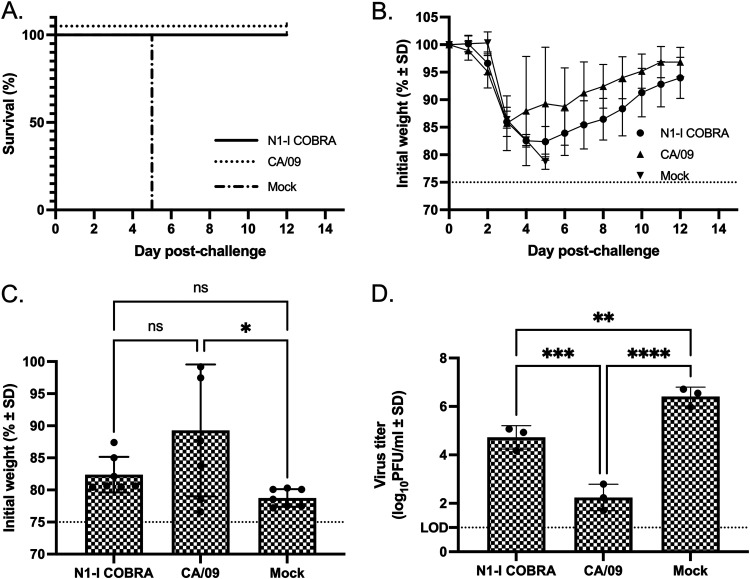
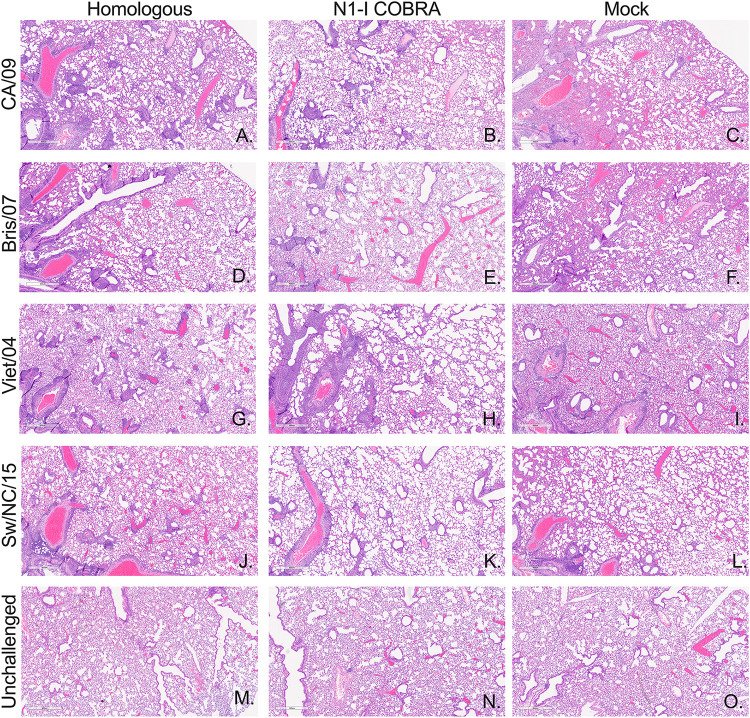
To determine if the antibody binding to an NA molecule correlates with inhibition of NA enzymatic activity and protection against infection, mice were challenged intranasally with CA/09 virus (Fig. 4). During the course of infection, all of the mock-vaccinated mice reached humane endpoints and were sacrificed by days 5 to 6 postinfection (Fig. 4A). The COBRA N1-I NA- and CA/09 NA-vaccinated mice survived challenge, but lost 14 to 17% of weight between days 3 to 5 postvaccination, but recovered almost all body weight by day 12 (Fig. 4B). At day 6, the average weight of the CA/09-vaccinated mice was not significantly different from the COBRA N1-I NA-vaccinated mice (Fig. 4C). The CA/09 NA-vaccinated mice had the lowest mean viral titer of 2.24 log10 PFU/ml on day 3 (Fig. 4D). The titer was significantly lower than the mean viral titer of 4.72 log10 PFU/ml from mice vaccinated with COBRA N1-I NA. Although greater than the CA/09 homologous control, the N1-I COBRA NA also had a significantly lower titer than the mock negative control with mean viral titer of 6.41 log10 PFU/ml (Fig. 4D). Formalin-fixed lung tissue stained with hemolysin and eosin (H&E) showed more inflammatory infiltration for the mock-vaccinated group in the CA/09 challenge than either the CA/09- (homologous) or the N1-I COBRA-vaccinated groups (Fig. 5A to C). Visually, the CA/09 and N1-I COBRA NA were similar in the amount of inflammatory reaction.
FIG 4.
A/California/07/2009 (H1N1)pdm09 challenge results after vaccination with NA antigens. (A and B) Survival (A) and weight loss (B) curves of mice postinfection are shown. (C) The day 5 peak weight loss of the CA/09-vaccinated mice was significantly different than the mock vaccinated. The variation of the CA/09 NA-vaccinated group was greater than the N1-I-vaccinated group. (D) The viral lung titers determined through plaque assay from lung tissue on day 3 postinfection. All error bars depict standard deviations, and the statistical analysis was conducted using a one-way ANOVA with Tukey’s multiple comparison. Not significant (ns); P value < 0.05 (*); P value < 0.01 (**); P value < 0.001 (***); P value < 0.0001 (****).
FIG 5.
Vaccine-specific inhibition of influenza inflammatory lung infiltration. Day 3 postinfection lungs were perfused and fixed with 10% buffered formalin. Representative images from hematoxylin and eosin (H&E)-stained sections are depicted. The vaccine groups along the horizontal axis include the homologous control vaccine groups in the first column with the appropriate vaccine per challenge virus listed in the vertical axis: CA/09 challenge (A to C), Bris/07 challenge (D to F), Viet/04 challenge (G to I), and Sw/NC/15 challenge (J to L). The unchallenged controls were age-matched unvaccinated, unchallenged mouse lungs (M to O). Each image represents separate individual mice. The magnification for all images was 4×, and the scale bar represents 0.6 mm.
Vaccinated mice challenged with human seasonal H1N1 influenza virus.
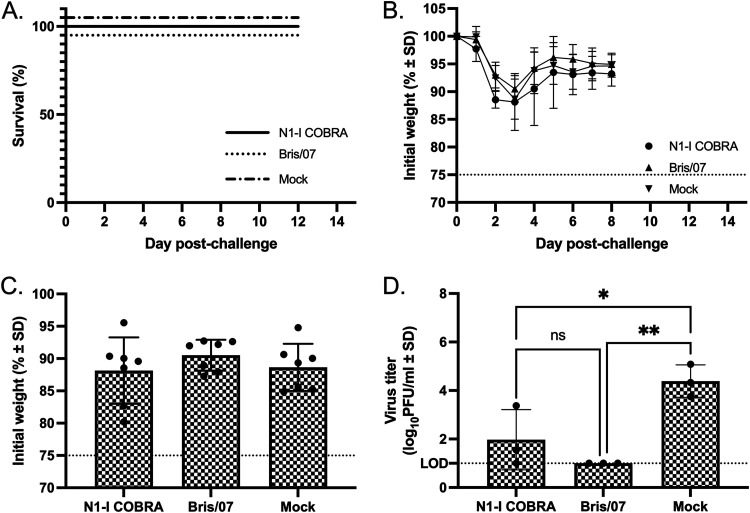
In addition to enzymatic inhibition of the human pandemic virus CA/09 NA, the COBRA N1-I NA also inhibited the human seasonal virus Bris/07 NA enzymatic activity. All Bris/07-challenged mouse groups did not reach humane endpoints, and all the mice survived challenge (Fig. 6A), with a peak weight loss at day 3 postinfection followed by quick recovery (Fig. 6B and C). However, there were significant differences in the lung viral titers on day 3 between the vaccinated groups (Fig. 6D). Mice vaccinated with the N1-I COBRA NA had a mean viral lung titer of 1.97 log10 PFU/ml that was not significantly different from the Bris/07 NA-vaccinated control group of 1.00 log10 PFU/ml (Fig. 6D). Furthermore, the N1-I COBRA NA-vaccinated mice had significantly lower mean viral lung titers than the mock-vaccinated mice with a mean titer of 4.39 log10 PFU/ml. In addition to the difference in viral lung titer, the mock-vaccinated lung H&E visualization showed greater inflammatory infiltration compared to the Bris/07- and N1-I COBRA NA-vaccinated mice (Fig. 5D to F). The Bris/07 and N1-I COBRA NA stained lung tissues were similar to each other and to the unchallenged mouse lung (Fig. 5M to O).
FIG 6.
A/Brisbane/59/2007 (H1N1)×PR8 challenge results after vaccination with NA antigens. (A and B) Survival (A) and weight loss (B) curves of mice postinfection. (C) The day 3 peak weight loss was not significantly different between any of the groups. (D) The viral lung titers were determined through plaque assay from lung tissue on day 3 postinfection. All error bars depict standard deviations, and the statistical analysis was conducted using a one-way ANOVA with Tukey’s multiple comparison. Not significant (ns); P value < 0.05 (*); P value < 0.01 (**).
Vaccinated mice challenged with avian H5N1 reassortant influenza virus.
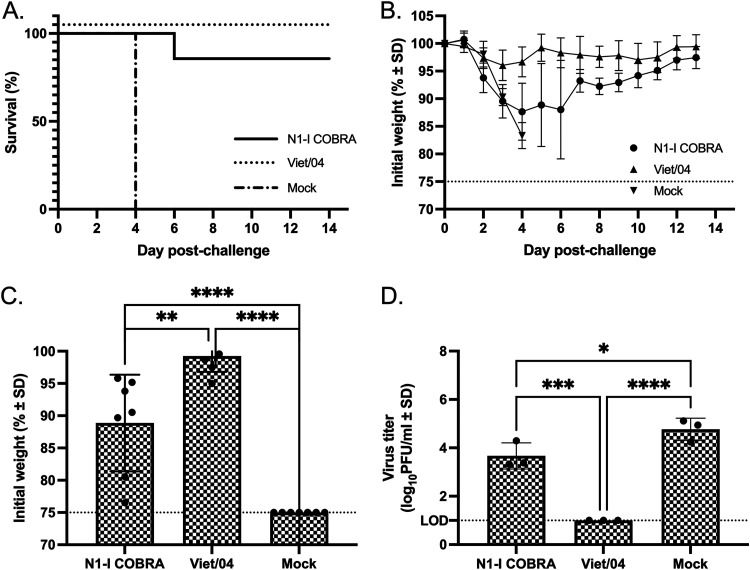
The reassortant H5N1 avian virus, Viet/04 challenge virus, contains the HA and NA gene segments originating from the highly pathogenic Viet/04 wild-type virus, but the multi-basic cleavage site of the HA is mutated to infer a low pathogenic phenotype. Mice vaccinated with Viet/04 NA and challenged with the Viet/04 virus containing the homologous NA protein all survived viral challenge (Fig. 7A) with little weight loss (Fig. 7B). In contrast, mock-vaccinated mice all reached humane endpoint by day 4 (Fig. 7A). Ninety percent of mice vaccinated with N1-I COBRA NA survived challenge (Fig. 7A), but these mice did lose between 10 and 15% of their body weight between days 4 to 6 postchallenge and then returned to full body weight by day 14 (Fig. 7B). At the peak of weight loss on day 5 postinfection, mice vaccinated with the N1-I COBRA NA had significantly more weight loss than mice vaccinated with the homologous Viet/04 NA, but had statistically less weight loss than mock-vaccinated mice (Fig. 7C). At day 3, mice vaccinated with the N1-I COBRA NA vaccine had 3.67 log10 PFU/ml mean lung viral titers that were significantly lower than viral titers in mock-vaccinated mice (Fig. 7D). The mock vaccinated mean lung titer was 4.77 log10 PFU/ml. The mock-vaccinated group challenged with Viet/04 had the greatest amount of infiltration of all groups of mice across all challenge viruses (Fig. 5). As such, the Viet/04- and N1-I COBRA NA-vaccinated groups had comparatively less infiltration (Fig. 5G to I).
FIG 7.
A/Viet Nam/1203/2004 (H5N1)×PR8 challenge results after vaccination with NA antigens. (A and B) Survival (A) and weight loss (B) curves of mice postinfection. (C) The day 5 peak weight loss was significantly different between all vaccination groups. The weights that reached humane endpoint before day 5 were analyzed at the limit of detection (75% body weight). (D) The viral lung titers determined through plaque assay from lung tissue on day 3 postinfection. All error bars depict standard deviations, and the statistical analysis was conducted using a one-way ANOVA with Tukey’s multiple comparison. P value < 0.05 (*); P value < 0.01 (**); P value < 0.001 (***); P value < 0.0001 (****).
Vaccinated mice challenged with swine-isolate H1N1 influenza virus.
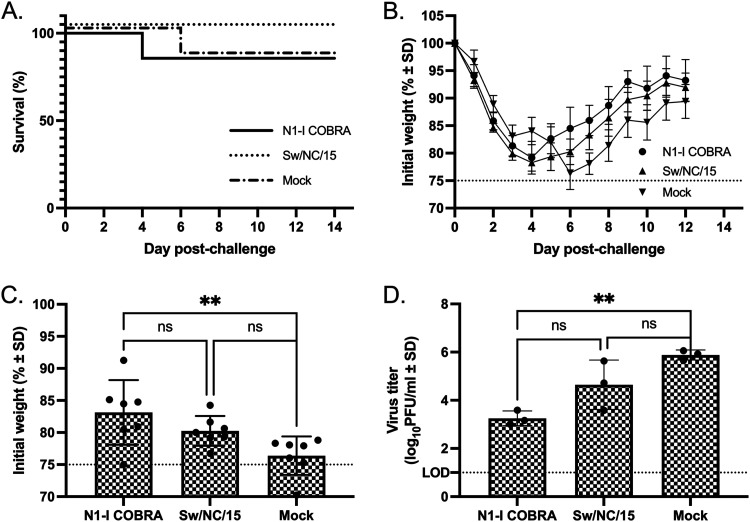
While all mice in the N1-I COBRA NA-, Sw/NC/15 NA-, and mock-vaccinated groups had significant weight loss following infection with Sw/NC/15 (Fig. 8B), few mice reached humane endpoint and were sacrificed (Fig. 8A). At day 6, the weight of the N1-I COBRA NA-vaccinated mice was not significantly different from the Sw/NC/15 NA-vaccinated mice, but was significantly higher than the mock-vaccinated mice (Fig. 8C). The weight at day 6 of the Sw/NC/15 NA-vaccinated mice was not statistically greater than the mock-vaccinated animals. Furthermore, mice vaccinated with the N1-I COBRA NA had lower lung titers on day 3 postinfection (mean viral titer of 3.25 log10 PFU/ml) compared to the mock-vaccinated mice (5.88 log10 PFU/ml) (Fig. 8D). There was no significant difference between the mean viral lung titer in N1-I COBRA NA-vaccinated mice and mice vaccinated with the Sw/NC/15 NA (4.64 log10 PFU/ml). The mice vaccinated with N1-I COBRA NA had less inflammatory infiltration than either the Sw/NC/15- or mock-vaccinated groups (Fig. 5J to L), and was more similar to the unchallenged controls (Fig. 5M to O).
FIG 8.
A/Swine/North Carolina/154074/2015 (H1N1) challenge results after vaccination with NA antigens. (A and B) Survival (A) and weight loss (B) curves of mice postinfection. (C) The day 6 peak weight loss was significantly different between the N1-I COBRA and mock groups. The weights that reached humane endpoint before day 6 were analyzed at the limit of detection (75% body weight). (D) Viral lung titers were determined through plaque assay from lung tissue on day 3 postinfection. All error bars depict standard deviations, and the statistical analysis was conducted using a one-way ANOVA with Tukey’s multiple comparison. Not significant (ns); P value < 0.01 (**).
DISCUSSION
Following influenza virus infection, the HA surface glycoprotein binds to sialic acids on the surface of ciliated epithelial cells lining the respiratory tract (31, 32). Following entry, genome replication, and viral protein production, nascent virions assemble and bud from the infected cell surfaces (33). The influenza virus NA is expressed on the surface of viral particles and infected cells and the protein assists in the budding and release of nascent virions from the cell surface (34–36). The NA sialidase enzymatic activity cleaves the host sialic acid receptors, allowing newly formed virions to efficiently bud from the cells surface, and also increases viral penetration through mucus (37, 38).
Both influenza virus HA and NA proteins are targets for the immune system to protect the host against infection and disease. Antibodies directed against HA bind to the protein and block receptor binding, viral fusion with and release from host cells, and NA activity (39–44). Several mechanism(s) may inhibit NA responses following antibody binding. In addition to steric hindrance from the anti-HA antibodies, anti-NA antibodies can bind multiple epitopes on the NA protein on the cell surface during budding and sterically hinder NA activity (18, 39). Bris/07 NA-specific elicited antibodies bound to the NA protein of Viet/04 (Fig. 2C), but they did not inhibit the NA enzymatic activity of the Viet/04 (H5N1) virus (Fig. 3C). Antibody binding to NA can inhibit the enzymatic activity of the molecule in cleaving sialic acid residues, thereby reducing virus release from infected cells. In this study, NA protein vaccinations were immunogenic and antigenic. The elicited anti-NA antibodies successfully bound NA proteins, as observed through enzyme-linked immunosorbent assay (ELISA) (Fig. 2), and blocked the NA activity of several N1 viral NA genetic clades, as observed through the ELLA assay (Fig. 3). NA antibodies can also target infected cells for destruction via the bound antibody Fc receptor to attract macrophages and other cells for antibody-dependent cellular cytotoxicity (ADCC) (45, 46), but those functions were not measured here.
Overall, influenza NA proteins are attractive targets in the development of a universal influenza virus vaccine (7, 47, 48). As outlined by the U.S. National Institutes of Allergy and Infectious Diseases, a next-generation influenza virus vaccine should be 75% effective against symptomatic influenza and virus-induced disease and elicit protective immunity that lasts for a minimum of 1 year (49). A broadly reactive NA antigen was generated from N1 NA amino acid sequences using COBRA algorithms (24) and termed N1-I. The N1-I COBRA NA elicited protective antibodies against four different HXN1 influenza viruses. These elicited antibodies inhibited sialidase enzymatic activity and inhibited virus replication of N1.1, N1.2, and N1.3 HXN1 influenza viruses (Fig. 3). In addition, these vaccines mitigated clinical signs of disease in vaccinated mice following challenge with four diverse influenza virus strains (Fig. 4 and 8). The ELLA assay quantifies the serological titers necessary to inhibit the influenza virus NA from cleaving 2,3- or 2,6-alpha sialic acids presented on fetuin, the host receptors utilized during infection (50). In humans and animal models, polyclonal NA inhibition titers have been correlated positively with increased influenza virus protection, similar to hemagglutination inhibition titers (20, 51, 52). Anti-NA antibodies are not able to inhibit the initial entry of the virus into the host cells, but they limit the viral spread during infection and contribute to immunity (15, 16, 19, 53).
Conserved epitopes on NA proteins that are responsible for eliciting broadly reactive antibodies have been identified using monoclonal antibodies (MAbs) (22, 54, 55). For example, there was a single conserved epitope on the H1N1 and H5N1 NA proteins that elicited cross-reactive antibodies against both H1N1 seasonal, H1N1pdm, and H5N1 NA proteins (22). Prophylactic treatment of mice with MAbs that bound these conserved epitopes resulted in less weight loss and mortality in virally challenged mice compared to nontreated mice. The N1-I COBRA NA immunogen retains this conserved epitope (amino acid residues 273, 338, and 339) and mice vaccinated with N1-I COBRA NA elicited antibodies that reduced the overall viral titer (Fig. 5D) as a result of potentially eliciting antibodies specific to this conserved NA epitope. However, this does not rule out that neutralization and protection against viral challenge are a result of binding to multiple NA epitopes on each NA protein. Total antibody binding may not always be associated with sialidase activity, since Viet/04 NA and Sw/NC/15 NA elicited sera bound to Bris/07, but did not inhibit Bris/07 NA enzymatic activity (Fig. 3E and F). The similar antibody binding and NA inhibition profiles of CA/09 and Viet/04 are a combined contribution of conserved epitope regions and originating from the same N1.1c2 clade (56).
There may be a direct correlation between antibody binding and severity of disease. Mice vaccinated with the Viet/04 NA and then challenged with the Viet/04 virus had little weight loss and undetectable lung virus titers (Fig. 6B to D). These mice had high anti-NA antibody titers (Fig. 2D). Mice vaccinated with NA derived from CA/09 or Sw/NC/15 had more weight loss and higher viral titers when challenged with their respective homologous viruses (Fig. 4 and 5), but had lower titer of antibodies that bound to each NA protein. Soluble tetramerized NA proteins were used to provide appropriate NA protein conformation to elicit protective antibodies, as previously described (57). However, differences in protein integrity and chemistry could have played a role in the variation in the magnitude of antibodies elicited (58, 59). It is unlikely that glycans shielded differential epitopes between NA proteins, since all NA proteins were predicted to express the same N-linked glycans (NetNGlyc 1.0) (60). Other factors, such as protein stability, folding, or hydrophobicity, may have contributed to the protein’s ability to elicit the immune responses. To further elucidate whether the lack of protection was due to protein integrity, different vaccine platforms can be used to deliver the NA antigen. Attenuated live reassortant viruses or virus-like particles that express NA antigens may be used in future studies to determine the contribution of the soluble protein format to these titer differences.
The addition of a standardized NA antigen component to a multiantigen influenza vaccine provides advantages and disadvantages. The inclusion of the NA protein may slow the overall antigenic drift of the vaccine, since the HA and NA antigens evolve independently of each other (61), and the NA protection is not dependent on the HA subtype of the infecting virus. However, with the addition, immunodominance between proteins may occur, and studies to determine the optimal ratios of protein will be necessary to overcome this, similar to previous vaccine optimization (62). Lastly, in a live-attenuated virus vaccine platform, the computationally derived NA protein will need to maintain stability and functionality to allow viral infection to be immunogenic. Before inclusion into a multivalent vaccine platform, the NA antigen should be confirmed to elicit NA inhibition titers and mitigate clinical signs independently of the other antigens.
The COBRA methodology was previously used to design broadly protective influenza HA antigens (26, 27, 63, 64) as well as other viral antigens, such as the E protein of dengue viruses (65). In this study, the COBRA methodology was applied to NA to design an antigen that elicited broadly reactive antibodies against N1 proteins from different subtypes. The COBRA N1-I antigen has the possibility of eliciting protective immune responses against both current human influenza viruses and also zoonotic and pre-pandemic viruses from multiple subtypes, which moves us a step closer to a universal vaccine antigen.
MATERIALS AND METHODS
COBRA NA antigen construction and synthesis.
Full-length N1 NA amino acid sequences for avian (2000 to 2015; 4,891 sequences), swine (1990 to 2015; 3,515 sequences), and human (2001 to 2014; 9,976 sequences) influenza A viruses were downloaded from the GISAID database (www.gisaid.org) (30). Full-length sequences were aligned using Geneious alignment (global alignment with free end gaps; Blosum62 cost matrix: open gap penalty 12, gap extension penalty 3; 2 refinement iterations) (Geneious v11.1.5). After alignment, the most common amino acid at each position was determined and resulted in primary consensus sequences. The resulting primary sequences from each clade were then realigned to generate a secondary consensus. This process was continued until a single final consensus was obtained. Finally, the ultimate amino acid sequence was reverse translated and optimized for expression in mammalian cells, including codon usage and RNA optimization (Genewiz, Washington, DC, USA). The resulting sequences were termed computationally optimized broadly reactive antigens (COBRA). The N1-I NA COBRA gene was synthesized and inserted into the pcDNA3.3 vector for soluble tetrameric recombinant NA protein production of protein containing the tetramerization domain in replace of the stem region (replaced amino acid residues 1 to 74).
Viruses and wild-type NA antigens.
N1 viruses were obtained through the Influenza Reagents Resource (IRR), BEI Resources, the Centers for Disease Control (CDC), or a generous gift from the Mark Tompkins laboratory at the University of Georgia. Viruses were passaged once in the same growth conditions as they were received or as per the instructions provided by the WHO, in either embryonated chicken eggs or Madin-Darby canine kidney (MDCK) cell culture (66). Virus lots were aliquoted for single-use applications and stored at −80°C. Hemagglutination titer of the frozen aliquots was determined with turkey red blood cells (RBCs). Viruses with NA protein GenBank accession numbers and NA genetic clade distinctions were as follows: A/California/07/2009 (H1N1)pdm09 (ACQ63272.1; CA/09; N1.1c2b1), recombinant virus containing HA and NA from A/Brisbane/59/2007 (H1N1) and all internal genes from A/Puerto Rico/8/1934 (H1N1) (PR8) virus (AHG96686.1; Bris/07; N1.2d), A/swine/North Carolina/154074/2015 (H1N1) (amino acid sequence available upon request; Sw/NC/15; N1.3b), and BSL-2 recombinant virus containing HA and NA from A/Viet Nam/1203/2004 (H5N1) and all internal genes from A/Puerto Rico/8/1934 (H1N1) virus (AAW80723.1; Viet/04; N1.1c2a). Viruses included in the enzyme-linked lectin assay panel included: A/Texas/36/1991 (H1N1) (TX/91; N1.2d), A/Brisbane/02/2018 (H1N1)pdm09 (Bris/18; N1.1c2b1), A/Hubei/1/2010 (H5N1) (Hub/10; N1.1c2a), A/Swine/Iowa/1931 (H1N1) (Sw/IA/1931; N1.3a), and A/Swine/Nebraska/A0144614/2013 (H1N1) (Sw/NE/13; N1.3b). The NA genetic clade distinctions were classified with widely utilized designations used previously through rigorous phylogenetic analysis (3, 4, 67–69). Viruses were chosen from each genetic clade to account for the breadth of the N1 protein diversity and to provide a range of antigenic diversity.
Phylogenetic comparison of avian, swine, human, and COBRA NA sequences.
NA amino acid sequences were visualized on a phylogenetic tree (Fig. 1A). Briefly, the truncated NA sequences were aligned and the Geneious Tree Builder, which observed the same alignment characteristics as Geneious alignment, was used to obtain a neighbor-joining Jukes-Cantor phylogenic tree with no indicated outgroup. The scale bar represents 0.02 amino acid substitutions per site of the region between the amino acid residues 74 through 470 (70).
Soluble tetrameric recombinant NA (tetNA).
The full NA codon-optimized sequences were originally commercially sourced in the pTR600 vector for virus-like particle (VLP) production (Genewiz, Washington, DC, USA). The wild-type NA amino acid sequences were aligned to CA/09 NA and truncated between residues 74 and 75. Truncated NA coding regions were subcloned into pcDNA3.3 vector containing a soluble tetramerization sequence for tetNA production. Starting from the N-terminal end of the protein, the tetramerization region is composed of a CD5 signal sequence (for efficient secretion of protein), a hexahistidine affinity tag (used for protein purification), a thrombin cleavage domain (may be used to remove the hexahistidine tag), the tetrabrachion domain from Staphylothermus marinus (forms and stabilizes the tetramer), and followed by the NA coding sequence (Fig. 1B). The NA sequences of the H1N1 influenza viruses included the amino acid residues 74 to 470 and NA sequences from H5N1 influenza viruses included the residues 55 to 449. All sequences ended with a dual stop codon (nucleic acid sequence TGATGA, TAATGA or TGATAG). N-terminal truncation of the original NA sequence was necessary to replace the transmembrane and stem domain with the soluble tetramerization region. After successful cloning, plasmids were sequence verified.
Soluble tetrameric NA proteins were expressed through individual transient plasmid DNA transfections of EXPI293F cells (Thermo Fisher Scientific) following the ExpiFectamine 293 transfection kit protocol. Cell supernatants from transiently transfected cells were collected, centrifuged to remove cellular debris, and filtered through a 0.22-μm pore membrane. Proteins were purified through a HisTrapExcel column and washed and eluted using the AKTA Pure System following the manufacture’s protocol (GE Healthcare Bio-Sciences AB, Uppsala, Sweden). Eluted protein fractions were concentrated in phosphate-buffered saline (PBS) + 0.1% wt/vol sodium azide (PBSA) using an Amicon Ultra-15 centrifugal filter unit (MilliporeSigma, Burlington, MA, USA). Total protein content was determined with the Micro BCA protein assay reagent kit (Pierce Biotechnology, Rockford, IL, USA). Single use aliquots were stored at −80°C until use.
Mouse vaccination and challenge studies.
BALB/c mice (Mus musculus, females, 6 to 8 weeks old) were purchased from Jackson Laboratory (Bar Harbor, ME, USA) and housed in microisolator units and allowed free access to food and water. Mice (10 mice per group) were vaccinated with a 1:1 mixture of soluble tetNA in PBS (1.0 μg tetNA/mouse) and AddaVax squalene-based oil-in-water adjuvant (InvivoGen, San Diego, CA, USA) in a total volume of 100 μl. Mice were vaccinated via intramuscular injection at week 0 and boosted with the same vaccine formulation at the same dose at weeks 4 and 8. PBS mixed 1:1 by volume with adjuvant served as a mock vaccination. Blood samples were collected from mice via cheek bleeds at 28 days after final vaccination in 1.5-ml microcentrifuge tubes. The samples were incubated at RT for 30 min and then centrifuged at 10,000 rpm for 10 min. Serum samples were removed and stored at −20°C.
At 4 weeks after final vaccination, mice were challenged intranasally with one of four challenge viruses. Mice were challenged with 5 × 104 PFU of CA/09; 1 × 107 PFU of Sw/NC/15; 4.375 × 105 PFU of Bris/07; or 1 × 106 PFU of Viet/04. Infectious doses were determined to be 10 times the 50% lethal dose, except for Bris/07 which was a nonlethal dose. The viral inocula were delivered in a volume of 50 μl. Mice were monitored, at minimum, daily for weight loss, disease signs, and death for 14 days postinfection. Individual body weights were recorded daily postinfection for each group during weight loss until stable recovery. Any animal exceeding 25% weight loss or a humane-endpoint score greater than two was humanely euthanized. Surviving mice were confirmed for successful infection indicated by HA titer seroconversion to the challenge virus.
Lung samples were harvested at 3 days postinfection. Mice (n = 3) were anesthetized using tribromoethanol (Avertin; Sigma-Aldrich, Darmstadt, Germany). The right lung was clamped and excised, and the tissue was snap-frozen on dry ice in a 2.0 ml cryogenic vial for storage at −80°C. After right lung excision, the trachea was intubated and the remaining lung lobes were perfused with 10% buffered formalin. After the perfusion, the lung was excised and placed in a 15-ml conical tube with minimum 10 ml of 10% buffered formalin. The fixed lung tissues were embedded in paraffin wax, sectioned, and stained with hematoxylin and eosin (H&E). All procedures were performed in accordance with the Guide for the Care and Use of Laboratory Animals, Animal Welfare Act, and Biosafety in Microbiological and Biomedical Laboratories (AUP: A2018 06–018-Y3-A13).
ELISA for elicited antibody quantification.
A high-affinity, 96-well flat-bottom enzyme-linked immunosorbent assay (ELISA; Immulon 4HBX) plate was coated with 50 μl of 10 μg/ml of virus-like particles with NA expressed on the surface in ELISA carbonate buffer (50 mM carbonate buffer [pH 9.5] with 5 μg/ml bovine serum albumin [BSA]), and the plate was incubated overnight at 4°C. The next morning, nonspecific epitopes were blocked with 1% BSA in PBS with 0.05% Tween 20 (PBST+BSA) solution for 1 h at RT or overnight at 4°C. Buffer was removed and 3-fold serial dilutions of raw sera were added to the plate with an initial dilution of 1:100. Plates were incubated at 37°C for 90 min. The plates were washed in PBS, and goat anti-mouse IgG-HRP (horseradish peroxidase) was added at 1:4,000 in PBST+BSA (cat. no. 1030-05, Southern Biotech, Birmingham, AL, USA). Plates were incubated at 37°C for 1 h. After washing, 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS) substrate in McIlvain’s buffer (pH 5) was added to each well, and incubated at 37°C for 15 min. The colorimetric reaction was stopped with the addition of 1% SDS in dH2O, and the absorbance was measured at 414 nm using a spectrophotometer (PowerWave XS; BioTek, Winooski, VT, USA). Endpoint titers were determined as the last dilution above five standard deviations of the negative-control wells after subtracting the background absorbance.
Enzyme-linked lectin assay for NA inhibition.
For the enzyme-linked lectin assay (ELLA) to assess NA inhibition, high-affinity Immunoblot 4HBX 96-well flat-bottom plates (Thermo Fisher Scientific, Waltham, MA, USA) were coated overnight with 100 μl of 25 μg/ml fetuin (Sigma-Aldrich, St. Louis, MO) in commercial KPL coating buffer (Seracare Life Sciences Inc, Milford, MA, USA) and stored at 4°C until use. Influenza virus was diluted in sample diluent (Dulbecco’s phosphate-buffered saline containing 0.133 g/liter CaCl2 and 0.1 g/liter MgCl2 [DPBS], 1% BSA, 0.5% Tween 20) to an initial dilution of 1:10. Before virus addition, fetuin plates were washed three times in PBS-T (PBS + 0.05% Tween 20), after which 50 μl of 2-fold serial dilutions of virus were added to the fetuin-coated plate containing 50 μl of sample diluent in duplicate. A negative-control column was included containing 100 μl of only sample diluent. Plates were sealed and incubated for 18 h at 37°C and 5% CO2. After incubation, plates were washed six times in PBS-T, and 100 μl of peanut agglutinin-HRPO (Sigma-Aldrich, St. Louis, MO) diluted 1,000-fold in conjugate diluent (DPBS, 1% BSA) was added. Plates were incubated at RT for 2 h. Plates were washed three times in PBS-T, and 100 μl (500 μg/ml) of o-phenylenediamine dihydrochloride (OPD; Sigma-Aldrich, St. Louis, MO) in 0.05 M phosphate-citrate buffer with 0.03% sodium perborate pH 5.0 (Sigma-Aldrich, St. Louis, MO) was added to the plates. Plates were immediately incubated in the dark for 10 min at RT. The reaction was stopped with 100 μl of 1 N sulfuric acid. The absorbance was read at 490 nm using a spectrophotometer (PowerWave XS; BioTek, Winooski, VT, USA). NA activity was determined after subtracting the mean background absorbance of the negative-control wells. Linear regression analysis was used to determine the dilution of the influenza viruses used in the assay necessary to achieve 90 to 95% NA activity and was used for subsequent NA inhibition enzyme-linked lectin assays (ELLAs).
Mouse serum was treated with 3 parts receptor destroying enzyme (RDE, Seneka, Japan), incubated at 37°C for 16 to 18 h and heat inactivated at 55°C for 6 h to completely inactivate the NA activity from the Vibrio cholerae NA; this inactivation procedure has previously been shown to be efficient (71). From an initial dilution of 1:100, NI ELLA titers were determined by 2-fold serially diluting treated sera in sample diluent. Duplicate dilutions were added to fetuin plates in 50 μl aliquots. The virus diluted to 90 to 95% NA activity in sample diluent was added to the plate in 50 μl aliquots. Controls were each a minimum of 8 wells, and included a positive NA antigen control (50 μl virus + 50 μl sample diluent) and a negative control (100 μl of sample diluent) on each plate. Plates were incubated for 16 to 18 h at 37°C and 5% CO2, after which they were processed as described above. NA percent activity was determined by subtracting the mean background absorbance of the negative-control wells, and then dividing the serum absorbance by the mean virus positive-control wells multiplied by 100. Nonlinear regression lines were fit using Prism, and the log 50% NI titer was estimated.
Determination of viral lung titers.
Frozen right lung samples were thawed on ice, weighed, and per 0.1 g tissue a volume of 1 ml of Dulbecco modified Eagle medium (DMEM) supplemented with penicillin-streptomycin (P/S) was added. The tissue was macerated through a 0.70-μm nylon filter (Corning Cell Strainer, Sigma-Aldrich, St. Louis, MO, USA) until thoroughly homogenized. Ten-fold serial dilutions of lung homogenate were overlaid onto MDCK cells seeded at 1 × 106 cells per well of a six-well plate for enumeration of viral lung titers. Samples were incubated for 1 h at RT with intermittent shaking every 15 min. Medium was removed, and the cells were washed twice with DMEM + P/S. Wash medium was replaced with 4 ml of L15 medium TPCK-trypsin plus 1.2% avicel (Cambrex, East Rutherford, NJ, USA) and incubated at 37°C with 5% CO2 for 48 to 72 h. After incubation, the avicel was removed and discarded. MDCK cells were washed with PBS and then fixed with 10% buffered formalin for 15 min and stained with 1% crystal violet for 15 min. The human viruses were incubated with 1 μg/ml of TPCK trypsin and the Viet/04 and Sw/NC/15 were incubated with 2 μg/ml. The plates were thoroughly washed in distilled water to remove excess crystal violet, and the plaques were counted and recorded to determine the PFU per ml lung homogenate.
Statistical analysis.
Statistical significance was defined as a P value less than 0.05. The means of the viral lung titers and day-6 weights were analyzed by an ordinary one-way ANOVA followed by Tukey’s multiple-comparison test, with a single pooled variance. The viral lung titers were transformed by log10 and the mean calculated. The lowest limit of detection for viral lung titers was 1 log10 PFU/ml lung homogenate, and this value was used for the statistical analysis of samples below that. Comparisons of peak weight loss were determined by dividing the measured weight on the peak of weight loss by the prechallenge weight on day 0, multiplied by 100. The standard deviations for weight curves, viral lung titers, and peak weight loss were determined. If an animal was sacrificed before the peak due to a greater than 25% drop in original weight or a humane-endpoint score of greater than or equal to three, a percent weight of 75 was used as the limit of detection for the statistical analysis. The mean log10 NA 50% inhibitory (NAI) titers were presented with the 95% confidence interval. Analyses were done using GraphPad Prism software.
Data availability.
The amino acid sequence for the COBRA NA is available in reference 72.
ACKNOWLEDGMENTS
We thank Ying Huang for technical assistance, Z. Beau Reneer for insightful discussions, Mark Tompkins’ laboratory for graciously providing the A/Swine/North Carolina/154074/2015 virus, and Ralph Tripp’s laboratory for graciously providing A/Swine/Nebraska/A01444614/2013 virus. Certain HXN1 influenza A viruses were obtained through the Prevention and Control of Influenza, Centers for Disease Control and Prevention, Atlanta, GA, USA. We thank the University of Georgia Animal Resource staff, technicians, and veterinarians for animal care, the University of Georgia Pathology and Histopathology staff for preparation of histopathology slides, and the Center for Vaccines and Immunology Protein Expression and Purification Core for providing purified protein.
We declare no competing interests.
This work has been funded by the National Institute of Allergy and Infectious Diseases, a component of the NIH, Department of Health and Human Services, under the Collaborative Influenza Vaccine Innovation Centers (CIVICs) contract 75N93019C00052 and in part, by the University of Georgia (UGA) (MRA-001). In addition, T.M.R. is supported by the Georgia Research Alliance as an Eminent Scholar.
Footnotes
Supplemental material is available online only.
Contributor Information
Ted M. Ross, Email: tedross@uga.edu.
Kanta Subbarao, The Peter Doherty Institute for Infection and Immunity.
REFERENCES
- 1.Zhu X, Turner HL, Lang S, McBride R, Bangaru S, Gilchuk IM, Yu W, Paulson JC, Crowe JE, Ward AB, Wilson IA. 2019. Structural basis of protection against H7N9 influenza virus by human anti-N9 neuraminidase antibodies. Cell Host Microbe 26:729–738. 10.1016/j.chom.2019.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eichelberger MC, Morens DM, Taubenberger JK. 2018. Neuraminidase as an influenza vaccine antigen: a low hanging fruit, ready for picking to improve vaccine effectiveness. Curr Opin Immunol 53:38–44. 10.1016/j.coi.2018.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen J-M, Ma H-C, Chen J-W, Sun Y-X, Li J-M, Wang Z-L. 2007. A preliminary panorama of the diversity of N1 subtype influenza viruses. Virus Genes 35:33–40. 10.1007/s11262-006-0025-4. [DOI] [PubMed] [Google Scholar]
- 4.Fanning TG, Reid AH, Taubenberger JK. 2000. Influenza A virus neuraminidase: regions of the protein potentially involved in virus-host interactions. Virology 276:417–423. 10.1006/viro.2000.0578. [DOI] [PubMed] [Google Scholar]
- 5.Lai S, Qin Y, Cowling BJ, Ren X, Wardrop NA, Gilbert M, Tsang TK, Wu P, Feng L, Jiang H, Peng Z, Zheng J, Liao Q, Li S, Horby PW, Farrar JJ, Gao GF, Tatem AJ, Yu H. 2016. Global epidemiology of avian influenza A H5N1 virus infection in humans, 1997–2015: a systematic review of individual case data. Lancet Infect Dis 16:e108–e118. 10.1016/S1473-3099(16)00153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gaydos JC, Top FH, Hodder RA, Russell PK. 2006. Swine influenza A outbreak, Fort Dix, New Jersey, 1976. Emerg Infect Dis 12:23–28. 10.3201/eid1201.050965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eichelberger MC, Wan H. 2015. Influenza neuraminidase as a vaccine antigen. Curr Top Microbiol Immunol 386:275–299. 10.1007/82_2014_398. [DOI] [PubMed] [Google Scholar]
- 8.Sultana I, Yang K, Getie-Kebtie M, Couzens L, Markoff L, Alterman M, Eichelberger MC. 2014. Stability of neuraminidase in inactivated influenza vaccines. Vaccine 32:2225–2230. 10.1016/j.vaccine.2014.01.078. [DOI] [PubMed] [Google Scholar]
- 9.Chen Y-Q, Wohlbold TJ, Zheng N-Y, Huang M, Huang Y, Neu KE, Lee J, Wan H, Rojas KT, Kirkpatrick E, Henry C, Palm A-KE, Stamper CT, Lan LY-L, Topham DJ, Treanor J, Wrammert J, Ahmed R, Eichelberger MC, Georgiou G, Krammer F, Wilson PC. 2018. Influenza infection in humans induces broadly cross-reactive and protective neuraminidase-reactive antibodies. Cell 173:417–429. 10.1016/j.cell.2018.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ito H, Nishimura H, Kisu T, Hagiwara H, Watanabe O, Kadji FMN, Sato K, Omiya S, Takashita E, Nobusawa E. 2020. Low response in eliciting neuraminidase inhibition activity of sera among recipients of a split, monovalent pandemic influenza vaccine during the 2009 pandemic. PLoS One 15:e0233001. 10.1371/journal.pone.0233001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bassetti M, Castaldo N, Carnelutti A. 2019. Neuraminidase inhibitors as a strategy for influenza treatment: pros, cons and future perspectives. Expert Opin Pharmacother 20:1711–1718. 10.1080/14656566.2019.1626824. [DOI] [PubMed] [Google Scholar]
- 12.Treanor JJ, Hayden FG, Vrooman PS, Barbarash R, Bettis R, Riff D, Singh S, Kinnersley N, Ward P, Mills RG. 2000. Efficacy and safety of the oral neuraminidase inhibitor oseltamivir in treating acute influenza: a randomized controlled trial. US Oral Neuraminidase Study Group. JAMA 283:1016–1024., 10.1001/jama.283.8.1016. [DOI] [PubMed] [Google Scholar]
- 13.Ng S, Cowling BJ, Fang VJ, Chan KH, Ip DKM, Cheng CKY, Uyeki TM, Houck PM, Malik Peiris JS, Leung GM. 2010. Effects of oseltamivir treatment on duration of clinical illness and viral shedding and household transmission of influenza virus. Clin Infect Dis 50:707–714. 10.1086/650458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ison MG. 2013. Clinical use of approved influenza antivirals: therapy and prophylaxis. Influenza Other Respir Viruses 7 Suppl 1:7–13. 10.1111/irv.12046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murphy BMDJ, Kasel A, Chanock RM. 1972. Association of serum anti-neuraminidase antibody with Resistance to INfluenza in Man. N Engl J Med 286:1329–1332. 10.1056/NEJM197206222862502. [DOI] [PubMed] [Google Scholar]
- 16.Kilbourne ED, Laver WG, Schulman JL, Webster RG. 1968. Antiviral activity of antiserum specific for an influenza virus neuraminidase. J Virol 2:281–288. 10.1128/JVI.2.4.281-288.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Couch RB, Atmar RL, Franco LM, Quarles JM, Wells J, Arden N, Niño D, Belmont JW. 2013. Antibody correlates and predictors of immunity to naturally occurring influenza in humans and the importance of antibody to the neuraminidase. J Infect Dis 207:974–981. 10.1093/infdis/jis935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilchuk IM, Bangaru S, Gilchuk P, Irving RP, Kose N, Bombardi RG, Thornburg NJ, Creech CB, Edwards KM, Li S, Turner HL, Yu W, Zhu X, Wilson IA, Ward AB, Crowe JE. 2019. Influenza H7N9 virus neuraminidase-specific human monoclonal antibodies inhibit viral egress and protect from lethal influenza infection in mice. Cell Host Microbe 26:715–728. 10.1016/j.chom.2019.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clements ML, Betts RF, Tierney EL, Murphy BR. 1986. Serum and nasal wash antibodies associated with resistance to experimental challenge with influenza A wild-type virus. J Clin Microbiol 24:157–160. 10.1128/jcm.24.1.157-160.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Monto AS, Petrie JG, Cross RT, Johnson E, Liu M, Zhong W, Levine M, Katz JM, Ohmit SE. 2015. Antibody to influenza virus neuraminidase: an independent correlate of protection. J Infect Dis 212:1191–1199. 10.1093/infdis/jiv195. [DOI] [PubMed] [Google Scholar]
- 21.Berlanda Scorza F, Tsvetnitsky V, Donnelly JJ. 2016. Universal influenza vaccines: shifting to better vaccines. Vaccine 34:2926–2933. 10.1016/j.vaccine.2016.03.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wan H, Gao J, Xu K, Chen H, Couzens LK, Rivers KH, Easterbrook JD, Yang K, Zhong L, Rajabi M, Ye J, Sultana I, Wan X-F, Liu X, Perez DR, Taubenberger JK, Eichelberger MC. 2013. Molecular basis for broad neuraminidase immunity: conserved epitopes in seasonal and pandemic H1N1 as well as H5N1 influenza viruses. J Virol 87:9290–9300. 10.1128/JVI.01203-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jang J, Bae SE. 2018. Comparative co-evolution analysis between the HA and NA genes of influenza A virus. Virology (Auckl) 9:1178122X18788328. 10.1177/1178122X18788328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giles BM, Ross TM. 2011. A computationally optimized broadly reactive antigen (COBRA) based H5N1 VLP vaccine elicits broadly reactive antibodies in mice and ferrets. Vaccine 29:3043–3054. 10.1016/j.vaccine.2011.01.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carter DM, Darby CA, Johnson SK, Carlock MA, Kirchenbaum GA, Allen JD, Vogel TU, Delagrave S, DiNapoli J, Kleanthous H, Ross TM. 2017. Elicitation of protective antibodies against a broad panel of H1N1 viruses in ferrets preimmune to historical H1N1 influenza viruses. J Virol 91:e01283-17. 10.1128/JVI.01283-17. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Reneer ZB, Jamieson PJ, Skarlupka AL, Huang Y, Ross TM. 2020. Computationally optimized broadly reactive H2 HA influenza vaccines elicited broadly cross-reactive antibodies and protected mice from viral challenges. J Virol 95:e01526-20. 10.1128/JVI.01526-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ross TM, DiNapoli J, Giel-Moloney M, Bloom CE, Bertran K, Balzli C, Strugnell T, Sá E Silva M, Mebatsion T, Bublot M, Swayne DE, Kleanthous H. 2019. A computationally designed H5 antigen shows immunological breadth of coverage and protects against drifting avian strains. Vaccine 37:2369–2376. 10.1016/j.vaccine.2019.03.018. [DOI] [PubMed] [Google Scholar]
- 28.Wong TM, Allen JD, Bebin-Blackwell A-G, Carter DM, Alefantis T, DiNapoli J, Kleanthous H, Ross TM. 2017. Computationally optimized broadly reactive hemagglutinin elicits hemagglutination inhibition antibodies against a panel of H3N2 influenza virus cocirculating variants. J Virol 91:e01581-17. 10.1128/JVI.01581-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giles BM, Crevar CJ, Carter DM, Bissel SJ, Schultz-Cherry S, Wiley CA, Ross TM. 2012. A computationally optimized hemagglutinin virus-like particle vaccine elicits broadly reactive antibodies that protect nonhuman primates from H5N1 infection. J Infect Dis 205:1562–1570. 10.1093/infdis/jis232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Elbe S, Buckland-Merrett G. 2017. Data, disease and diplomacy: GISAID's innovative contribution to global health. Glob Chall 1:33–46. 10.1002/gch2.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gottschalk A, Lind PE. 1949. Product of interaction between influenza virus enzyme and ovomucin. Nature 164:232. 10.1038/164232a0. [DOI] [PubMed] [Google Scholar]
- 32.Suzuki Y, Ito T, Suzuki T, Holland RE, Chambers TM, Kiso M, Ishida H, Kawaoka Y. 2000. Sialic acid species as a determinant of the host range of influenza A viruses. J Virol 74:11825–11831. 10.1128/jvi.74.24.11825-11831.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pohl MO, Lanz C, Stertz S. 2016. Late stages of the influenza A virus replication cycle-a tight interplay between virus and host. J Gen Virol 97:2058–2072. 10.1099/jgv.0.000562. [DOI] [PubMed] [Google Scholar]
- 34.Lai JCC, Chan WWL, Kien F, Nicholls JM, Peiris JSM, Garcia J-M. 2010. Formation of virus-like particles from human cell lines exclusively expressing influenza neuraminidase. J Gen Virol 91:2322–2330. 10.1099/vir.0.019935-0. [DOI] [PubMed] [Google Scholar]
- 35.Rossman JS, Lamb RA. 2011. Influenza virus assembly and budding. Virology 411:229–236. 10.1016/j.virol.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen BJ, Leser GP, Morita E, Lamb RA. 2007. Influenza virus hemagglutinin and neuraminidase, but not the matrix protein, are required for assembly and budding of plasmid-derived virus-like particles. J Virol 81:7111–7123. 10.1128/JVI.00361-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cohen M, Zhang X-Q, Senaati HP, Chen H-W, Varki NM, Schooley RT, Gagneux P. 2013. Influenza A penetrates host mucus by cleaving sialic acids with neuraminidase. Virol J 10:321. 10.1186/1743-422X-10-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wagner R, Matrosovich M, Klenk HD. 2002. Functional balance between haemagglutinin and neuraminidase in influenza virus infections. Rev Med Virol 12:159–166. 10.1002/rmv.352. [DOI] [PubMed] [Google Scholar]
- 39.Chen Y-Q, Lan LY-L, Huang M, Henry C, Wilson PC. 2019. Hemagglutinin stalk-reactive antibodies interfere with influenza virus neuraminidase activity by steric hindrance. J Virol 93:e01526-18. 10.1128/JVI.01526-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Williams JA, Gui L, Hom N, Mileant A, Lee KK. 2018. Dissection of epitope-specific mechanisms of neutralization of influenza virus by intact IgG and Fab fragments. J Virol 92:e02006-17. 10.1128/JVI.02006-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamayoshi S, Uraki R, Ito M, Kiso M, Nakatsu S, Yasuhara A, Oishi K, Sasaki T, Ikuta K, Kawaoka Y. 2017. A broadly reactive human anti-hemagglutinin stem monoclonal antibody that inhibits influenza A virus particle release. EBioMedicine 17:182–191. 10.1016/j.ebiom.2017.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Whittle JRR, Zhang R, Khurana S, King LR, Manischewitz J, Golding H, Dormitzer PR, Haynes BF, Walter EB, Moody MA, Kepler TB, Liao H-X, Harrison SC. 2011. Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc Natl Acad Sci U S A 108:14216–14221. 10.1073/pnas.1111497108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kosik I, Angeletti D, Gibbs JS, Angel M, Takeda K, Kosikova M, Nair V, Hickman HD, Xie H, Brooke CB, Yewdell JW. 2019. Neuraminidase inhibition contributes to influenza A virus neutralization by anti-hemagglutinin stem antibodies. J Exp Med 216:304–316. 10.1084/jem.20181624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kosik I, Yewdell JW. 2017. Influenza A virus hemagglutinin specific antibodies interfere with virion neuraminidase activity via two distinct mechanisms. Virology 500:178–183. 10.1016/j.virol.2016.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wohlbold TJ, Podolsky KA, Chromikova V, Kirkpatrick E, Falconieri V, Meade P, Amanat F, Tan J, tenOever BR, Tan GS, Subramaniam S, Palese P, Krammer F. 2017. Broadly protective murine monoclonal antibodies against influenza B virus target highly conserved neuraminidase epitopes. Nat Microbiol 2:1415–1424. 10.1038/s41564-017-0011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Broecker F, Zheng A, Suntronwong N, Sun W, Bailey MJ, Krammer F, Palese P. 2019. Extending the stalk enhances immunogenicity of the influenza virus neuraminidase. J Virol 93:e00840-19. 10.1128/JVI.00840-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun W, Luo T, Liu W, Li J. 2020. Progress in the development of universal influenza vaccines. Viruses 12:1033. 10.3390/v12091033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sylte MJ, Suarez DL. 2009. Influenza neuraminidase as a vaccine antigen. Curr Top Microbiol Immunol 333:227–241. 10.1007/978-3-540-92165-3_12. [DOI] [PubMed] [Google Scholar]
- 49.Erbelding EJ, Post DJ, Stemmy EJ, Roberts PC, Augustine AD, Ferguson S, Paules CI, Graham BS, Fauci AS. 2018. A universal influenza vaccine: the strategic plan for the National Institute of Allergy and Infectious Diseases. J Infect Dis 218:347–354. 10.1093/infdis/jiy103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baenziger JU, Fiete D. 1979. Structure of the complex oligosaccharides of fetuin. J Biol Chem 254:789–795. 10.1016/S0021-9258(17)37874-2. [DOI] [PubMed] [Google Scholar]
- 51.Walz L, Kays S-K, Zimmer G, von Messling V. 2018. Neuramidase-inhibiting antibody titers correlate with protection from heterologous influenza virus strains of the same neuraminidase subtype. J Virol 92:e01006-18. 10.1128/JVI.01006-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weiss CD, Wang W, Lu Y, Billings M, Eick-Cost A, Couzens L, Sanchez JL, Hawksworth AW, Seguin P, Myers CA, Forshee R, Eichelberger MC, Cooper MJ. 2020. Neutralizing and neuraminidase antibodies correlate with protection against influenza during a late season A/H3N2 outbreak among unvaccinated military recruits. Clin Infect Dis 71:3096–3102. 10.1093/cid/ciz1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rockman S, Brown LE, Barr IG, Gilbertson B, Lowther S, Kachurin A, Kachurina O, Klippel J, Bodle J, Pearse M, Middleton D. 2013. Neuraminidase-inhibiting antibody is a correlate of cross-protection against lethal H5N1 influenza virus in ferrets immunized with seasonal influenza vaccine. J Virol 87:3053–3061. 10.1128/JVI.02434-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shoji Y, Chichester JA, Palmer GA, Farrance CE, Stevens R, Stewart M, Goldschmidt L, Deyde V, Gubareva L, Klimov A, Mett V, Yusibov V. 2011. An influenza N1 neuraminidase-specific monoclonal antibody with broad neuraminidase inhibition activity against H5N1 HPAI viruses. Hum Vaccin 7 Suppl:199–204. 10.4161/hv.7.0.14595. [DOI] [PubMed] [Google Scholar]
- 55.Jiang H, Peng W, Qi J, Chai Y, Song H, Bi Y, Rijal P, Wang H, Oladejo BO, Liu J, Shi Y, Gao GF, Townsend AR, Wu Y. 2020. Structure-based modification of an anti-neuraminidase human antibody restores protection efficacy against the drifted influenza virus. mBio 11:e02315-20. 10.1128/mBio.02315-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Job ER, Schotsaert M, Ibañez LI, Smet A, Ysenbaert T, Roose K, Dai M, de Haan CAM, Kleanthous H, Vogel TU, Saelens X. 2018. Antibodies directed toward neuraminidase N1 control disease in a mouse model of influenza. J Virol 92:e01584-17. 10.1128/JVI.01584-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McMahon M, Strohmeier S, Rajendran M, Capuano C, Ellebedy AH, Wilson PC, Krammer F. 2020. Correctly folded—but not necessarily functional—influenza virus neuraminidase is required to induce protective antibody responses in mice. Vaccine 38:7129–7137. 10.1016/j.vaccine.2020.08.067. [DOI] [PubMed] [Google Scholar]
- 58.Scheiblhofer S, Laimer J, Machado Y, Weiss R, Thalhamer J. 2017. Influence of protein fold stability on immunogenicity and its implications for vaccine design. Expert Rev Vaccines 16:479–489. 10.1080/14760584.2017.1306441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fox CB, Kramer RM, Barnes VL, Dowling QM, Vedvick TS. 2013. Working together: interactions between vaccine antigens and adjuvants. Ther Adv Vaccines 1:7–20. 10.1177/2051013613480144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gupta R, Brunak S. 2002. Prediction of glycosylation across the human proteome and the correlation to protein function. Pac Symp Biocomput 2002:310–322. [PubMed] [Google Scholar]
- 61.Kilbourne ED, Johansson BE, Grajower B. 1990. Independent and disparate evolution in nature of influenza A virus hemagglutinin and neuraminidase glycoproteins. Proc Natl Acad Sci U S A 87:786–790. 10.1073/pnas.87.2.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jang YH, Lee E-Y, Byun YH, Jung E-J, Lee YJ, Lee YH, Lee K-H, Lee J, Seong BL. 2014. Protective efficacy in mice of monovalent and trivalent live attenuated influenza vaccines in the background of cold-adapted A/X-31 and B/Lee/40 donor strains. Vaccine 32:535–543. 10.1016/j.vaccine.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 63.Allen JD, Ray S, Ross TM. 2018. Split inactivated COBRA vaccine elicits protective antibodies against H1N1 and H3N2 influenza viruses. PLoS One 13:e0204284. 10.1371/journal.pone.0204284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fadlallah GM, Ma F, Zhang Z, Hao M, Hu J, Li M, Liu H, Liang B, Yao Y, Gong R, Zhang B, Liu D, Chen J. 2020. Vaccination with consensus H7 elicits broadly reactive and protective antibodies against Eurasian and North American lineage H7 viruses. Vaccines (Basel) 8:143. 10.3390/vaccines8010143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Uno N, Ross TM. 2021. A universal dengue vaccine elicits neutralizing antibodies against strains from all four dengue serotypes. J Virol 951:e00658-20. 10.1128/JVI.00658-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.World Health Organization, WHO Global Influenza Surveillance Network. 2011. Manual for the laboratory diagnosis and virological surveillance of influenza. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 67.Schon J, Breithaupt A, Hoper D, King J, Schon J, Pohlmann A, Parvin R, Behr K-P, Schwarz B-A, Beer M, Stech J, Harder T, Grund C. 2021. Neuraminidase-associated plasminogen recruitment enables systemic spread of natural avian Influenza viruses H3N1. PLoS Pathog 17:e1009490. 10.1371/journal.ppat.1009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhuang Q, Wang S, Liu S, Hou G, Li J, Jiang W, Wang K, Peng C, Liu D, Guo A, Chen J. 2019. Diversity and distribution of type A influenza viruses: an updated panorama analysis based on protein sequences. Virol J 16:85. 10.1186/s12985-019-1188-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu S, Ji K, Chen J, Tai D, Jiang W, Hou G, Chen J, Li J, Huang B. 2009. Panorama phylogenetic diversity and distribution of Type A influenza virus. PLoS One 4:e5022. 10.1371/journal.pone.0005022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Westgeest KB, Bestebroer TM, Spronken MIJ, Gao J, Couzens L, Osterhaus ADME, Eichelberger M, Fouchier RAM, de Graaf M. 2015. Optimization of an enzyme-linked lectin assay suitable for rapid antigenic characterization of the neuraminidase of human influenza A(H3N2) viruses. J Virol Methods 217:55–63. 10.1016/j.jviromet.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Allen JD, Ross TM, Wong TM, Blackwell AGB, Nunez IA, Beau Reneer Z, Huang Y. 2019. Methods for generating broadly reactive, pan-epitopic immunogens, compositions and use thereof. Patent WO 2020/014673 A1. https://patentimages.storage.googleapis.com/13/a0/d4/5746c860d39ae1/WO2020014673A1.pdf.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1<br>. Download JVI.00759-21-s0001.pdf, PDF file, 0.3 MB (307.1KB, pdf)
Data Availability Statement
The amino acid sequence for the COBRA NA is available in reference 72.