ABSTRACT
The coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is bringing an unprecedented health crisis to the world. To date, our understanding of the interaction between SARS-CoV-2 and host innate immunity is still limited. Previous studies reported that SARS-CoV-2 nonstructural protein 12 (NSP12) was able to suppress interferon-β (IFN-β) activation in IFN-β promoter luciferase reporter assays, which provided insights into the pathogenesis of COVID-19. In this study, we demonstrated that IFN-β promoter-mediated luciferase activity was reduced during coexpression of NSP12. However, we could show NSP12 did not affect IRF3 or NF-κB activation. Moreover, IFN-β production induced by Sendai virus (SeV) infection or other stimulus was not affected by NSP12 at mRNA or protein level. Additionally, the type I IFN signaling pathway was not affected by NSP12, as demonstrated by the expression of interferon-stimulated genes (ISGs). Further experiments revealed that different experiment systems, including protein tags and plasmid backbones, could affect the readouts of IFN-β promoter luciferase assays. In conclusion, unlike as previously reported, our study showed SARS-CoV-2 NSP12 protein is not an IFN-β antagonist. It also rings the alarm on the general usage of luciferase reporter assays in studying SARS-CoV-2.
IMPORTANCE Previous studies investigated the interaction between SARS-CoV-2 viral proteins and interferon signaling and proposed that several SARS-CoV-2 viral proteins, including NSP12, could suppress IFN-β activation. However, most of these results were generated from IFN-β promoter luciferase reporter assay and have not been validated functionally. In our study, we found that, although NSP12 could suppress IFN-β promoter luciferase activity, it showed no inhibitory effect on IFN-β production or its downstream signaling. Further study revealed that contradictory results could be generated from different experiment systems. On one hand, we demonstrated that SARS-CoV-2 NSP12 could not suppress IFN-β signaling. On the other hand, our study suggests that caution needs to be taken with the interpretation of SARS-CoV-2-related luciferase assays.
KEYWORDS: interferon-β, IFN-β, severe acute respiratory syndrome coronavirus-2, SARS-CoV-2, nonstructural protein 12, NSP12, IFN-β promoter luciferase activity assay
INTRODUCTION
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a novel emerging β-coronavirus that causes the coronavirus disease 2019 (COVID-19), is affecting more than 200 countries and territories around the world. SARS-CoV-2 infection causes respiratory, enteric, hepatic, and neurological diseases of various severities (1–3). The viral infection has caused 149,910,744 cases, including 3,155,168 deaths, as of 30 April 2021 (https://covid19.who.int/). Under the pressure of a continued rise in the incidence of COVID-19, an unprecedented global effort is being implemented to discover effective vaccines and therapeutics to combat SARS-CoV-2.
SARS-CoV-2 is an enveloped, positive-sense, single-stranded RNA virus. Its 30-kb genome encodes 16 nonstructural proteins, including NSP1 to NSP16; four structural proteins, including spike glycoprotein (S), envelope protein (E), membrane protein (M), and nucleocapsid protein (N); and accessory proteins open reading frame 3a (ORF3a), ORF3b, ORF6, ORF7a, ORF7b, ORF8, and ORF10 (4, 5). During SARS-CoV-2 infection, S protein binds to its host receptor, angiotensin-converting enzyme 2 (ACE2), and subsequently triggers S protein conformational change (6, 7). After fusion of virus membrane with the endosomal membrane, the viral genome is released into the cytoplasm, where replication occurs after synthesis and proteolytic cleavage of the replicase polyprotein. A double-stranded RNA genome is then synthesized from the genomic plus-strand RNA (8). After the synthesis of structural proteins, the progeny virus assembles and buds at membranes of the endoplasmic reticulum, the intermediate compartments, and/or the Golgi complex. This allows newly synthesized virion-containing vesicles to fuse with the plasma membrane and release the virus (9).
Host innate immune system is essential for antiviral responses. SARS-CoV-2, as an RNA virus, can be recognized by pattern recognition receptors (PRRs), including RIG-I-like receptors (RLRs) and extracellular and endosomal Toll-like receptors (TLRs), which activate downstream signaling pathways (10, 11). Downstream cascades trigger the secretion of various cytokines, including type I/III interferons (IFNs), proinflammatory tumor necrosis factor alpha (TNF-α), interleukin-1β (IL-1β), IL-6, IL-10, and IL-18 (12–14). Single-cell RNA sequencing revealed that the type I IFN coexisted with TNF-α- and IL-1β-driven inflammation in COVID-19 patients’ peripheral blood mononuclear cells, indicating that type I IFN may play a role in exacerbating inflammation in severe COVID-19 (15). Moreover, several studies demonstrated that SARS-CoV-2 was sensitive to IFNs pretreatment in vitro, perhaps to a greater degree than SARS-CoV (16–18).
In contrast, Melo et al. found a lack of induction of type I IFN responses following SARS-CoV-2 infection in infected cell lines, primary bronchial cells, and a ferret model (19). Additionally, accumulating evidence shows that several SARS-CoV-2 proteins, including NSP12, could inhibit type I IFN responses in cell cultures as illustrated by IFN-β promoter assays (20–25). The detailed mechanism regarding IFN inhibition of NSP12 remained elusive. Here, by using the IFN promoter luciferase reporter system, we confirmed that NSP12 could inhibit IFN-β promoter luciferase activity induced by Sendai virus (SeV) infection or components of the RIG-I/MDA5 pathway. But detailed studies demonstrated that NSP12 could neither interfere with IRF3 and NF-κB activation nor affect IFN-β transcription or translation. Furthermore, the STAT1 activation and interferon-stimulated genes (ISGs) transcription were not affected by NSP12. Further experiments revealed that different experiment systems could affect the results about the effect of NSP12 on IFN-β promoter luciferase activity signal.
RESULTS
SARS-CoV-2 NSP12 represses IFN-β promoter luciferase activity.
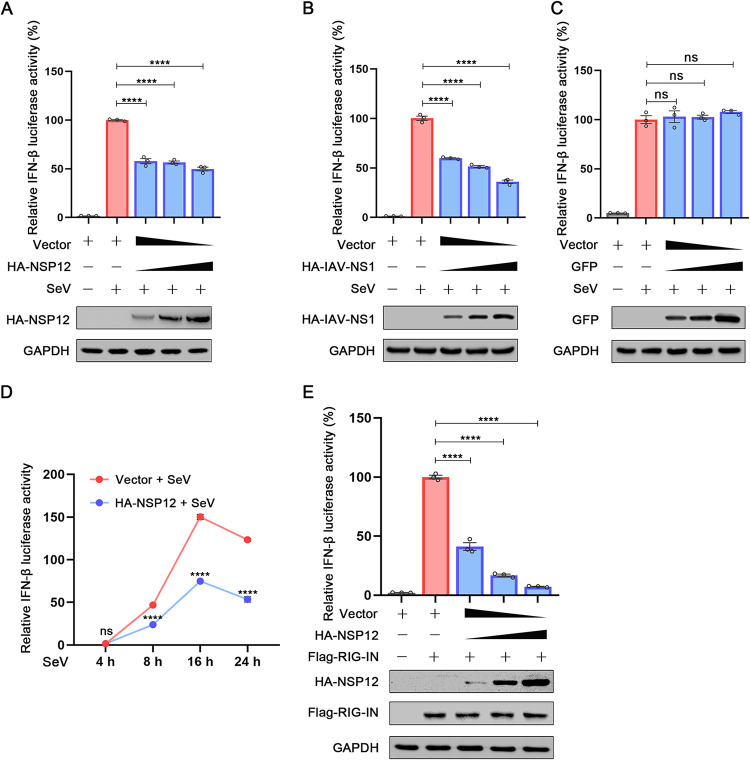
We first cotransfected cells with IFN reporter construct and HA-NSP12-expressing plasmid or empty vector and stimulated the cells with SeV, which is a well-known IFN inducer. Similar to previous reports (20, 22), NSP12 suppressed IFN-β-Luc activity (Fig. 1A). As a positive control, a previously known IFN-β antagonist, IAV NS1 protein, also suppressed IFN-β-Luc activity (Fig. 1B) (26). In contrast, overexpression of green fluorescent protein (GFP) showed little effect on IFN-β-Luc activity (Fig. 1C). To further confirm this inhibitory effect, we evaluated IFN-β-Luc activities at different time points. As shown in Fig. 1D, the inhibition of NSP12 on IFN-β-Luc activity can be easily observed 8 h after SeV infection. Additionally, IFN-β-Luc activity induced upon RIG-IN expression (a RIG-I activator) can also be suppressed by NSP12 in a dose-dependent manner (Fig. 1E) (27). Together, these results suggest that NSP12 may suppress IFN-β promoter.
FIG 1.
The effect of SARS-CoV-2 NSP12 on IFN-β promoter luciferase activity. (A) HEK293T cells were cotransfected with IFN-β luciferase reporter pIFN-β-Luc, pPRL-TK, and pHA-NSP12 or empty vector for 24 h and then infected with SeV. After 10 h, relative luciferase activity was determined. (B) HEK293T cells were cotransfected with IFN-β luciferase reporter pIFN-β-Luc and pPRL-TK together with pHA-IAV-NS1 or empty vector for 24 h and then infected with SeV. After 10 h, relative luciferase activity was determined. (C) HEK293T cells were cotransfected with IFN-β luciferase reporter pIFN-β-Luc and pPRL-TK together with pGFP or empty vector for 24 h and then infected with SeV. After 10 h, relative luciferase activity was determined. (D) HEK293T cells were cotransfected with IFN-β luciferase reporter pIFN-β-Luc and pPRL-TK together with pHA-NS12 or empty vector for 24 h and then infected with SeV for different durations (4 h, 8 h, 16 h, and 24 h). Relative luciferase activity was determined. (E) HEK293T cells were cotransfected with pIFN-β-Luc, pPRL-TK, and pHA-NSP12, together with pFlag-RIG-IN for 24 h. Relative luciferase activity was determined. The resultant ratios for the samples were normalized using the Renilla luciferase values. The values from cells transfected with empty vector and stimulated with SeV infection or RIG-IN were set as 100%. Data were shown as means ± SEM of at least three independent experiments. ****, P < 0.0001; ns, not significant.
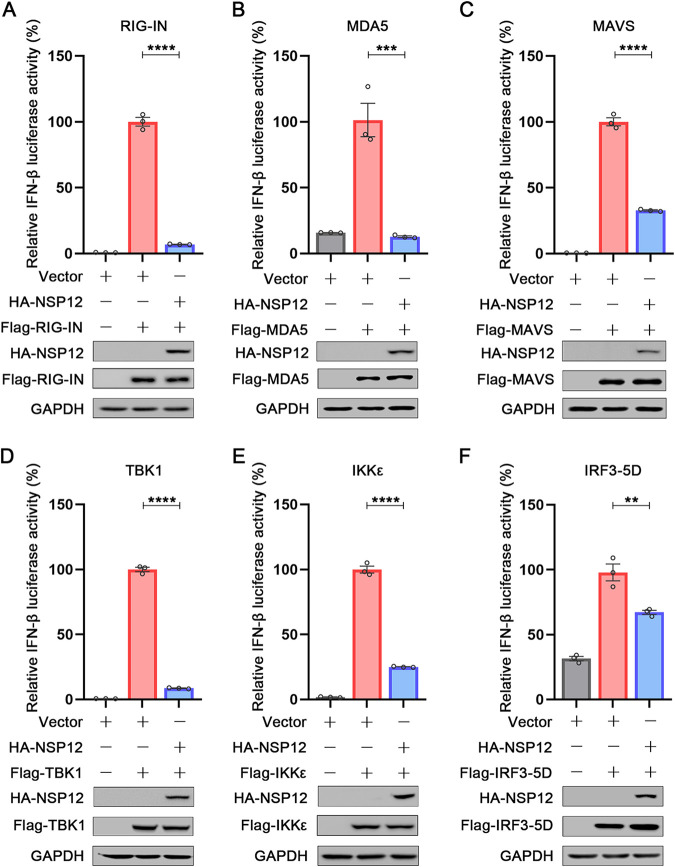
To dissect which step upstream of IFN-β induction was affected by NSP12, we further used different components of the RIG-I/MDA5 pathway to stimulate IFN-β promoter luciferase reporters. Interestingly, as shown in Fig. 2, overexpression of NSP12 suppressed IFN-β promoter luciferase signals triggered by RIG-IN, MDA5, MAVS/VISA, TBK1, IKKε, and IRF3-5D (a constitutively active IRF3 mutant) (28). These results indicate that NSP12 may inhibit IFN-β promoter luciferase signal at the level of, or downstream of, IRF3 activation.
FIG 2.
Effect of SARS-CoV-2 NSP12 on RIG-I/MDA5 signaling. HEK293T cells were cotransfected with pIFN-β-Luc, pPRL-TK, and pHA-NSP12 together with pFlag-RIG-IN (A), pFlag-MDA5 (B), pFlag-MAVS (C), pFlag-TBK1 (D), pFlag-IKKε (E), and pFlag-IRF3-5D (F) for 24 h. Relative luciferase activity was determined. The resultant ratios for the samples were normalized using the Renilla luciferase values. The values from cells transfected with empty vector together with the relevant constructs were set as 100%. Data were shown as means ± SEM of at least three independent experiments. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
SARS-CoV-2 NSP12 does not affect IRF3 and NF-κB activation.
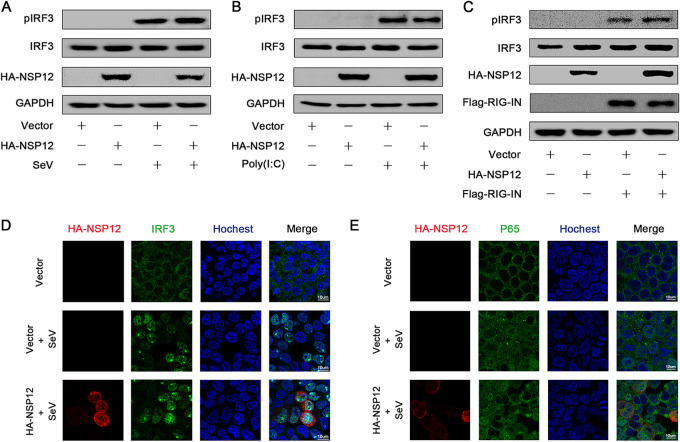
Upon virus infection, phosphorylated IRF3 and NF-κB (P50 and P65) translocate to the nuclei and bind to the interferon promoter to stimulate IFN-β production (28, 29). Next, we wanted to investigate the effect of NSP12 on IRF3 and P65. IRF3 phosphorylation was induced upon SeV infection as expected (Fig. 3A). However, NSP12 could neither repress SeV-induced IRF3 phosphorylation nor repress IRF3 expression (Fig. 3A). Similarly, NSP12 failed to suppress IRF3 phosphorylation activated by either poly(I·C) or RIG-IN (Fig. 3B and C). To further demonstrate the effect of NSP12 on the single-cell level, immunostaining was performed. In mock-infected HEK293T cells, IRF3 was distributed in the cytoplasm (Fig. 3D). After SeV infection, IRF3 was translocated from the cytoplasm to the nucleus (Fig. 3D). However, the presence of NSP12 showed no effect on IRF3 nucleus translocation (Fig. 3D). We also investigated the effect of NSP12 on P65 nuclear translocation. Similar to IRF3, P65 nucleus translocation was observed upon SeV infection, and NSP12 could not block this translocation (Fig. 3E). Taken together, these results reveal that NSP12 does not affect IRF3 and NF-κB activation.
FIG 3.
Effect of SARS-CoV-2 NSP12 on IRF3 phosphorylation and nuclear translocation. (A) HEK293T cells were transfected with pHA-NSP12 or empty vector for 24 h and infected with SeV for 6 h. Phosphorylated IRF3, IRF3, NSP12, and GAPDH were determined by Western blotting. (B) HEK293T cells were transfected with pHA-NSP12 or empty vector for 24 h and transfected with poly(I·C) for 6 h. Phosphorylated IRF3, IRF3, NSP12, and GAPDH were determined by Western blotting. (C) HEK293T cells were cotransfected with pFlag-RIG-IN and pHA-NSP12 or empty vector for 24 h. Phosphorylated IRF3, IRF3, NSP12, RIG-IN, and GAPDH were determined by Western blotting. (D) HEK293T cells were transfected with pHA-NSP12 or empty vector for 24 h and infected with SeV for 8 h. The subcellular localizations of IRF3 (green), HA-NSP12 (red), and nucleus marker Hoechst 33258 (blue) were analyzed with confocal microscopy. (E) HEK293T cells were transfected with pHA-NSP12 or empty vector for 24 h and infected with SeV for 8 h. The subcellular localizations of P65 (green), HA-NSP12 (red), and nucleus marker Hoechst 33258 (blue) were analyzed with confocal microscopy.
SARS-CoV-2 NSP12 does not interfere with IFN-β production.
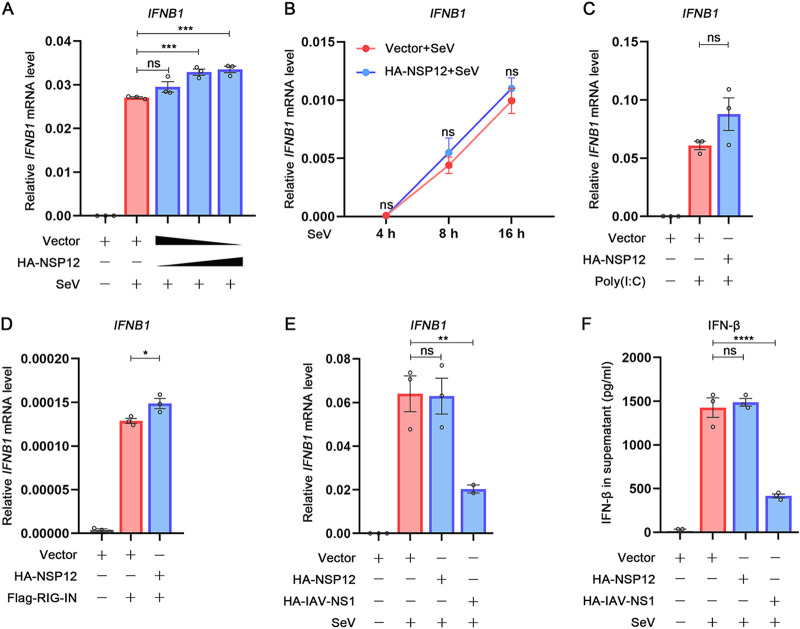
Since SARS-CoV-2 NSP12 showed an inhibitory effect in IFN-β promoter luciferase reporter assays, we continued to examine whether the SARS-CoV-2 NSP12 would affect IFN-β expression at the transcriptional level or translational level. Interestingly, various doses of NSP12, which showed a significant inhibitory effect on IFN-β promoter luciferase assay, slightly upregulated SeV-induced IFN-β mRNA accumulation levels (Fig. 4A). Furthermore, the kinetics of IFN-β mRNA after SeV stimulation showed no difference with or without NSP12 (Fig. 4B). Additionally, NSP12 could not affect the IFN-β mRNA induced either by poly(I·C) (Fig. 4C) or even slightly upregulated IFN-β mRNA induced by RIG-IN (Fig. 4D). In contrast, influenza A virus (IAV) NS1 could significantly reduce IFN-β mRNA as previously described (Fig. 4E) (26). To further evaluate whether SARS-CoV-2 NSP12 could influence the translation or secretion of IFN-β, we measured IFN-β protein in cell culture supernatant by enzyme-linked immunosorbent assay (ELISA) (Fig. 4F). Taken together, these results suggest that NSP12 does not interfere with IFN-β production.
FIG 4.
Effect of SARS-CoV-2 NSP12 on IFN-β production. (A) HEK293T cells were transfected with pHA-NSP12 or empty vector for 24 h and then infected with SeV. After 10 h, IFN-β mRNA was determined by quantitative reverse transcription-PCR (RT-qPCR). (B) HEK293T cells were transfected with pHA-NSP12 or empty vector for 24 h and then infected with SeV for 4 h, 8 h, or 16 h; IFN-β mRNA was determined by RT-qPCR. (C) HEK293T cells were transfected with pHA-NSP12 or empty vector for 24 h and then treated with poly(I·C). After 6 h, IFN-β mRNA was determined by RT-qPCR. (D) HEK293T cells were cotransfected with pFlag-RIG-IN and pHA-NSP12 or empty vector for 24 h; IFN-β mRNA was determined by RT-qPCR. (E) HEK293T cells were transfected with pHA-NSP12, pHA-IAV-NS1, or empty vector for 24 h and then infected with SeV. After 10 h, IFN-β mRNA was determined by RT-qPCR. (F) HEK293T cells were transfected with pHA-NSP12 pHA-IAV-NS1 or empty vector for 24 h and then infected with SeV. After 24 h, IFN-β in the supernatant was measured by ELISA. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant.
SARS-CoV-2 NSP12 does not interfere with type I IFN signaling pathway.
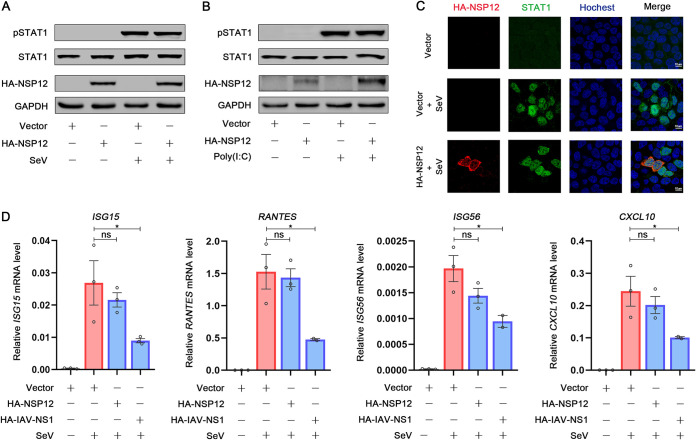
We further examined whether SARS-CoV-2 NSP12 affects the type I IFN signaling pathway. After binding to its receptor, IFN-β activates Jak1 and Tyk2 kinases and then Jak1 and Tyk2 kinases phosphorylate STAT1 and STAT2, triggering their dimerization and nuclear translocation (30). We then investigated the effect of NSP12 on STAT1 activation. STAT1 phosphorylation was induced upon SeV infection as expected (Fig. 5A). However, NSP12 could neither repress SeV-induced STAT1 phosphorylation nor repress STAT1 expression (Fig. 5A). Similarly, NSP12 failed to suppress STAT1 phosphorylation activated by poly(I·C) (Fig. 5B). To evaluate the STAT1 activation on the single-cell level, immunostaining was performed. We found that STAT1 was distributed in the cytoplasm in mock-infected HEK293T cells (Fig. 5C). After SeV infection, STAT1 was translocated from the cytoplasm to the nucleus (Fig. 5C). However, the presence of NSP12 showed no effect on STAT1 nucleus translocation (Fig. 5C). We next determined the levels of ISGs. We found that NSP12 could not interfere with the ISGs’ (ISG15, RANTES, ISG56, and CXCL10) mRNA accumulation levels induced by SeV (Fig. 5D). Taken together, these results reveal that NSP12 does not affect the type I IFN signaling pathway.
FIG 5.
Effect of SARS-CoV-2 NSP12 on IFN-I signaling pathway. (A) HEK293T cells were transfected with pHA-NSP12 or empty vector for 24 h and infected with SeV for 8 h. Phosphorylated STAT1, STAT1, NSP12, and GAPDH were determined by Western blotting. (B) HEK293T cells were transfected with pHA-NSP12 or empty vector for 24 h and transfected with poly(I·C) for 6 h. Phosphorylated STAT1, STAT1, NSP12, and GAPDH were determined by Western blotting. (C) HEK293T cells were transfected with pHA-NSP12 or empty vector for 24 h and infected with SeV for 16 h. The subcellular localizations of STAT1 (green), HA-NSP12 (red), and nucleus marker Hoechst 33258 (blue) were analyzed with confocal microscopy. (D) HEK293T cells were transfected with pHA-NSP12 or empty vector for 24 h and then infected with SeV. After 10 h, ISG15, RANTES, ISG56, and CXCL10 mRNA was determined by RT-qPCR. Data are shown as means ± SEM of at least three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant.
Different protein tags or plasmid backbones affect the activity of NSP12.
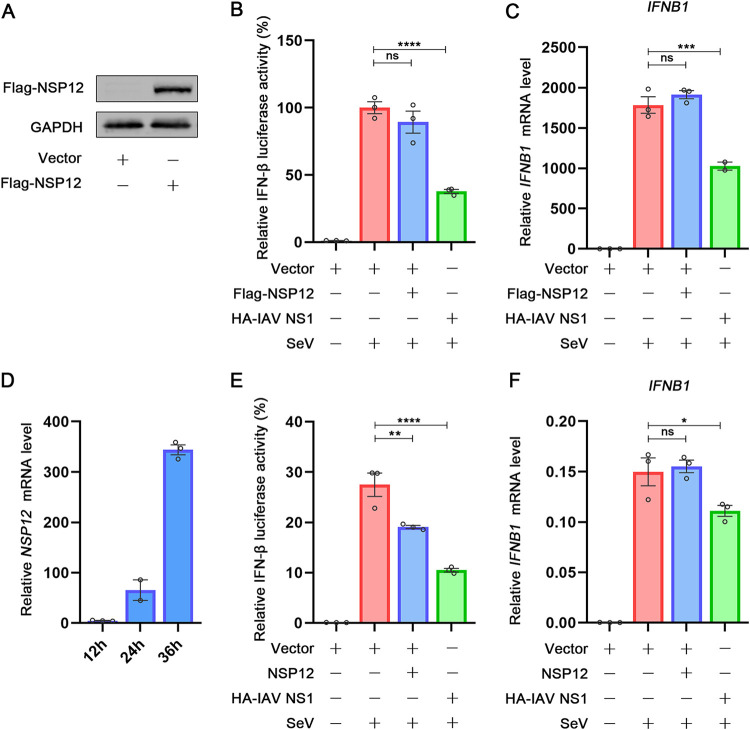
As the published results about the effect of NSP12 on IFN-β promoter luciferase assays are contradictory (20, 22, 23), we speculated that different experiment systems could interfere with the readouts of IFN-β promoter reporter assays. As previous studies used different tags fused with NSP12, we next investigated whether different tags could result differently. NSP12 fused with Flag tag at N terminus was constructed and expressed (Fig. 6A). Interestingly, unlike HA-NSP12, Flag-NSP12 could not interfere with IFN-β promoter luciferase activity (Fig. 6B). However, in agreement with the result from HA-NSP12, the IFN-β mRNA after SeV stimulation showed no difference with or without Flag-NSP12 expression (Fig. 6C). To exclude the influence of different tags, NSP12 plasmid without any tag was constructed. The expression of NSP12 was determined by the kinetic of NSP12 mRNA (Fig. 6D). As shown in Fig. 6E, NSP12 could suppress the IFN-β promoter luciferase activity, while IFN-β mRNA accumulation levels remained unaffected (Fig. 6F). These results suggested that although NSP12 is not an IFN-β antagonist, NSP12 fused with different tags could affect the readout of IFN-β promoter luciferase assay.
FIG 6.
Different protein tags or plasmid backbones affect the activity of NSP12. (A) HEK293T cells were transfected with pFlag-NSP12 and empty vector for 24 h. Flag-NSP12, and GAPDH were determined by Western blotting. (B) HEK293T cells were cotransfected with IFN-β luciferase reporter pIFN-β-Luc, pPRL-TK, and pFlag-NSP12 or empty vector for 24 h and then infected with SeV. After 10 h, relative luciferase activity was determined. (C) HEK293T cells were transfected with pFlag-NSP12 or empty vector for 24 h and then infected with SeV. After 10 h, IFN-β mRNA was determined by RT-qPCR. (D) HEK293T cells were transfected with NSP12 plasmid and empty vector for 12 h, 24 h, or 36 h. NSP12 mRNA was determined by RT-qPCR. (E) HEK293T cells were cotransfected with IFN-β luciferase reporter pIFN-β-Luc, pPRL-TK, and NSP12 plasmid or empty vector for 24 h and then infected with SeV. After 10 h, relative luciferase activity was determined. (F) HEK293T cells were transfected with NSP12 plasmid or empty vector for 24 h and then infected with SeV. After 10 h, IFN-β mRNA was determined by RT-qPCR.
Taken together, we demonstrate that SARS-CoV-2 NSP12 protein does not antagonize IFN-β production, which is supported by the results that neither the phosphorylation or nuclear translocation of IRF3 and P65, nor the expression of IFN-β, affected by NSP12 (Fig. 7). Additionally, NSP12 protein could not affect the IFN-β signaling pathway, which is proven by the results that NSP12 neither interfered with the phosphorylation or nuclear translocation of STAT1 nor the expression of ISGs (Fig. 7).
FIG 7.
Graphical summary of this study. Upon sensing viral RNA, RIG-I or MDA5 activation occurs. Activated RIG-I or MDA5 delivers signal to MAVS and activates phosphorylation of TBK1 and IKKε. Phosphorylation of TBK1 and IKKε leading to the phosphorylation of IRF3. Phosphorylated IRF3 is subsequently translocated into the nucleus and induces the activation of IFN-β promoter. On the other hand, MAVS/VISA activates IκB kinase (IKK) complex comprising IKKα and IKKβ catalytic subunits and two molecules of NEMO. The IKK complex then phosphorylates IκBα and leads NF-κB phosphorylation. Phosphorylated NF-κB is translocated into the nucleus and binds IFN-β promoter. IFN-β mRNA is translocated from the nucleus to the cytoplasm and produces IFN-β. Then, IFN-β is secreted outside the cell. In this process, SARS-CoV-2 NSP12 neither affects phosphorylation and translocation of IRF3 and NF-κB nor interferes with IFN-β expression at the transcriptional level or translational level. Additionally, the downstream signaling of IFN-β activation, including STAT1 phosphorylation, nucleus translocation, and ISGs expression, are not affected by NSP12.
DISCUSSION
IFNs play pivotal roles in host antiviral defense. In the context of SARS-CoV-2 infection, data about IFNs are still accumulating. Previous studies showed that SARS-CoV-2 viral proteins, including NSP12, could suppress IFN-β activation. However, in the present study, we elucidate that (i) NSP12 affects neither IRF3 activation nor NF-κB activation; (ii) NSP12 could not inhibit IFN-β production and downstream IFN-β signaling pathway; and (iii) various experiment systems could affect the readout of IFN-β promoter reporter assay.
Several studies demonstrated the interaction between SARS-CoV-2 and IFN response. Arunachalam et al. reported that small amounts of IFNs in the serum of patients with COVID-19 were accompanied by elevated amounts of chemokines and proinflammatory cytokines (31), which is consistent with the results from Melo et al. (19). These studies revealed that SARS-CoV-2 infection induced low-level and delayed IFN production. Using single-cell RNA sequencing, Lee et al. have reported that the IFN-driven inflammatory response and TNF/IL-1β-driven inflammatory response coexisted in classical monocytes from severe COVID-19 patients (15). Along the same line, Zhou et al. also found a robust IFN response in addition to proinflammatory response in bronchoalveolar lavage fluid of patients with COVID-19 and suggested that timing and extent of IFN production were likely associated with disease severity (32). Moreover, upregulation of IFN-responsive genes has been demonstrated in SARS-CoV-2-infected intestinal organoids (33). Although these studies provided evidence about the roles of IFNs in SARS-CoV-2 infection, more detailed mechanisms need to be further investigated.
On the other hand, several studies have identified that certain SARS-CoV-2 proteins could limit IFN-β promoter luciferase activity. The screen results from Yuen et al. described that NSP1, NSP6, NSP12, NSP13, NSP14, NSP15, and ORF6 of SARS-CoV-2 antagonized IFN-β promoter luciferase activity induced by RIG-IN (20); Li et al. demonstrated that NSP1, ORF6, ORF8, and N protein limited IFN-β promoter luciferase activity induced by SeV infection (21); Lei et al. have reported that SARS-CoV-2 NSP1, NSP3, NSP12, NSP14, ORF3, ORF6, and M protein suppressed IFN promoter luciferase activity activated by SeV or RIG-IN (22); and Xia et al. have revealed that SARS-CoV-2 NSP6, NSP13, and ORF6 could repress IFN-β promoter luciferase activity activated by RIG-IN (23). As reasonable as it is that the virus may evolve mechanisms to counteract the host immune defense, the reported screen results are not consistent, and many of them were not validated by additional functional tests. Thoms et al. have reported that the SARS-CoV-2 NSP1 C terminus could bind to and obstruct the mRNA entry tunnel of ribosome, which suppressed IFN-β production (34). Xia et al. have illustrated that SARS-CoV-2 NSP13 could bind TBK1 and affect TBK1 phosphorylation, and they also demonstrated that SARS-CoV-2 ORF6 could block IRF3 nuclear translocation by binding to KPNA2, a key factor of IRF3 nuclear translocation (23). Although these studies shed important light on the interactions of SARS-CoV-2 and IFN activation, more work involving advanced models should be considered.
The firefly luciferase reporter assay is highly sensitive and has been widely used in a variety of high-throughput bioluminescence screening assays (35). But it is important to notice that false-positive results could be generated if there is direct inhibition on luciferase activity, as has been documented. For example, some compounds are known to affect the firefly luciferase enzymatic reaction, which includes quinolones 2-aryl-substituted benzo-[d]thiazole, 2-aryl-substituted benzo-[d]-imidazole, and 2-aryl-substituted benzo-[d]-oxazole (36, 37). In addition, various other inhibitory effects have also been reported, which potentially interfere with firefly luciferase-based assays. For example, treatment-induced luciferase stabilization could lead to an extension of luciferase cellular half-life and, subsequently, a net increase in the luminescence signals. Hence, for assays that rely on detecting an increase in the signal, the presence of certain firefly luciferase inhibitors potentially increases the chance of false-positive readouts (38). The results about NSP12 affecting IFN-β promoter luciferase activity are contradictory. Yuen et al. and Lei et al. reported that NSP12 inhibited >50% IFN-β promoter luciferase activity signal activated through RIG-I or SeV, while Xia et al. demonstrated that NSP12 did not affect IFN-β promoter luciferase activity. By comparing the materials of research groups, we found that the NSP12 plasmid constructs (protein tags or plasmid backbones) used were different, which may lead the contradictory results. By using various plasmids [pXJ40-HA-NSP12, pcDNA3.1(+)-flag-NSP12, and pXJ40-NSP12], we demonstrated that NSP12 expressed by different plasmid backbones or fused with different tags leads to contradictory IFN-β promoter luciferase activity results. These results indicated that different experiment systems could affect the stability of IFN-β promoter reporter assays. The reasons for this result need further investigation.
In summary, the present study investigated the interaction of SARS-CoV-2 NSP12 and IFN-β response. Using various strategies, we demonstrate SARS-CoV-2 NSP12 protein is not an IFN-β antagonist. However, different experiment systems could affect the readout of IFN-β promoter luciferase assay. Our study not only provides insights on the interaction between SARS-CoV-2 and innate immune response, it also rings the alarm on the general usage of luciferase reporter assays.
MATERIALS AND METHODS
Cell lines and cultures.
HEK293T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (catalog no. C11995500BT; Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin, and 100 μg/ml streptomycin sulfate. HEK293T cells were maintained at 37°C in a 5% CO2 incubator. Mycoplasma test kit (catalog no. C0301S; Beyotime Biotechnology, Shanghai, China) was used in the lab routinely to exclude any existence of mycoplasma contamination in cell culture.
Plasmids and reagents.
The sequences of SARS-CoV-2 NSP12 were amplified from the pET32a(+)-NSP12 plasmid, which was kindly provided by Yan Li (Huazhong University of Science and Technology) and cloned into mammalian expression vector pXJ40-HA (N-terminal hemagglutinin [HA] tag), pXJ40 (no tag), and pcDNA3.1(+)-Flag. The IAV NS1 plasmid was kindly provided by Ke Xu (Wuhan University). The IFN-β promoter firefly luciferase reporter plasmid was purchased from Clone Tech (Somis, CA); the herpes simplex virus thymidine kinase promoter Renilla luciferase reporter plasmid (pRL-TK) was purchased from Promega (San Luis Obispo, CA).
Polyinosinic-polycytidylic acid [poly(I·C)] (low molecular weight [LMW]) was purchased from InvivoGen (catalog code tlrl-picw; San Diego, CA, USA). PEI Max transfection-grade linear polyethylenimine hydrochloride (molecular weight [MW], 40,000) was purchased from Polysciences (catalog no. 24765-1; Hirschberg an der Bergstrasse, Germany). Lipofectamine 2000 was purchased from Invitrogen (catalog no. 11668019; Carlsbad, CA, USA).
Quantitative reverse transcription-PCR.
In a 24-well plate, 60% to 80% HEK293T cells were transfected 100 to 400 ng protein expression plasmid or vector plasmid. HEK293T cells were activated by cotransfected with 100 ng stimulator-expressing plasmid (RIG-IN) for 24 h or treated with poly(I·C) (2 μg/ml) for 6 h or infected with SeV (multiplicity of infection [MOI] = 100) for 10 h. Total intracellular RNA was extracted with Ultrapure RNA kit (catalog no. CW0581M; CoWin Biosciences, China) according to the manufacturer’s instructions. Total intracellular genomic DNA was extracted with TIANamp genomic DNA kit (catalog no. DP304-03; Tiangen Biotech, Beijing, China) according to the manufacturer’s instructions. Real-time quantitative reverse transcription-PCR was performed using the Roche LC480 and FastStart essential DNA green master mix (catalog no. 06924204001; Roche, Germany). The reaction mixture contains 5 μl SYBR green PCR master mix, 4 μl DNA diluted template, and 1 μl primers. The following primers were used: human IFN-β, sense primer, 5′-AGGACAGGATGAACTTTGAC-3′, and antisense primer, 5′-TGATAGACATTAGCCAGGAG-3′; for human GAPDH (glyceraldehyde-3-phosphate dehydrogenase), sense primer, 5′-ATGACATCAAGAAGGTGGTG-3′, and antisense primer, 5′-CATACCAGGAAATGAGCTTG-3′; for human ISG15, sense primer, 5′-GAGAGGCAGCGAACTCATCTT-3′, and antisense primer, 5′-CCAGCATCTTCACCGTCAGG-3′; for human ISG56, sense primer, 5′-CTTGTGGGTAATACAGTGGAGATG-3′, and antisense primer, 5′-GCTCCAGACTATCCTTGACCTG-3′; for human RANTES, sense primer, 5′-TGCCCACATCAAGGAGTATTT-3′, and antisense primer, 5′-CTTTCGGGTGACAAAGACG-3′; for human CXCL10, sense primer, 5′-GGTGAGAAGAGATGTCTGAATCC-3′, and antisense primer, 5′-GTCCATCCTTGGAAGCACTGCA’; for human PRNP, sense primer, 5′-TGCTGGGAAGTGCCATGAG-3′, and antisense primer, 5′-CGGTGCATGTTTTCACGATAGTA-3′; for firefly luciferase, sense primer, 5′-CTATTCTCCTTCTTCGCCAA-3′, and antisense primer, 5′-TATCCAGATCCACAACCTTC-3′.
Luciferase reporter assay.
HEK293T cells (1 × 105 cells per well in a 24-well plate) were cotransfected with 100 ng of luciferase reporter plasmid (firefly luciferase), 10 ng pRL-TK (Renilla luciferase plasmid), and 50 to 200 ng of protein expression plasmid using transfection reagent with a ratio of 1:3. Empty vector was used to ensure the same total amount (310 ng) of plasmids in each well. HEK293T cells were activated by cotransfected with 100 ng stimulator-expressing plasmids (RIG-IN, MDA5, MAVS, TBK1, IKKε, or IRF3-5D) for 24 h or stimulated with SeV (MOI, 100) for 10 h. Luciferase activity was measured by using a dual-luciferase reporter assay system kit (part no. E1960; Promega, San Luis Obispo, CA) according to the manufacturer’s instructions. Data represented are relative firefly luciferase activity after normalization to Renilla luciferase signals.
Western blotting.
Cell lysates were prepared with lysis buffer (50 mM Tris-HCl, pH 7.5, 300 mM NaCl, 1% Triton X-100, 5 mM EDTA, and 10% glycerol). Protein concentration was determined by Bradford assay (catalog no. 5000205; Bio-Rad, Hercules, CA, USA). Cell lysates (30 μg) were electrophoresed in a 10% SDS-PAGE gel and transferred to a polyvinylidene difluoride (PVDF) membrane (catalog no. IPVH00010; Millipore, MA, US). PVDF membranes were blocked with 5% skim milk in Tris-buffered saline with 0.05% Tween 20 (TBST) before antibody incubation. Protein bands were detected using a GeneGnome XRQ chemiluminescence imaging system (GeneGnome XRQ-NPC; Hong Kong, China). The antibodies used are listed in Table 1.
TABLE 1.
Antibodies used in this study
| Antibody | Supplier (location) | Catalog No. | Dilution(s)a |
|---|---|---|---|
| Anti-HA | Sigma-Aldrich (St. Louis, MO, USA) | h6908 | WB, 1:5,000 |
| Anti-HA | Beyotime Biotechnology (Shanghai, China) | AH158 | WB, 1:1,000; IF, 1:100 |
| Anti-Flag | Sigma-Aldrich (St. Louis, MO, USA) | f1804 | WB, 1:1,000 |
| Anti-pIRF3 (Ser396) (4D4G) | Cell Signaling Technology (Boston, MA, USA) | 4947 | WB, 1:1,000 |
| Anti-IRF-3 (D6I4C) XP | Cell Signaling Technology (Boston, MA, USA) | 11904 | WB, 1:1,000; IF, 1:100 |
| Anti-STAT1 | Cell Signaling Technology (Boston, MA, USA) | 14994S | WB, 1:1,000; IF, 1:200 |
| Anti-pSTAT1 | Cell Signaling Technology (Boston, MA, USA) | 9167S | WB, 1:1,000 |
| Anti-P65 | Beyotime Biotechnology (Shanghai, China) | AF1234 | WB, 1:1,000; IF, 1:100 |
| Anti-firefly luciferase | Proteintech (China) | 27986-1-AP | WB, 1:1,000 |
| Anti-GAPDH | Sigma-Aldrich (St. Louis, MO, USA) | G9295 | WB, 1:10,000 |
| Anti-mouse IgG (HRP-linked antibody) | Cell Signaling Technology (Boston, MA, USA) | 7076 | WB, 1:5,000 |
| Anti-rabbit IgG (HRP-linked antibody) | Cell Signaling Technology (Boston, MA, USA) | 7074 | WB, 1:5,000 |
| Anti-mouse IgG (Alexa Fluor 568) | Invitrogen (San Diego, CA, USA) | A-11031 | IF, 1:100 |
| Anti-rabbit IgG (Alexa Fluor 488) | Invitrogen (San Diego, CA, USA) | A-11034 | IF, 1:100 |
WB, Western blotting; IF, immunofluorescence; HRP, horseradish peroxidase.
Confocal microscopy.
In a 12-well plate, 40% to 50% HEK293T cells were transfected with the indicated plasmids (1000 ng) for 24 h; then, cells were washed twice with phosphate-buffered saline (PBS) (containing 0.1% bovine serum albumin [BSA]) and fixed in 4% paraformaldehyde at room temperature for 10 min. Following permeabilization with 0.1% Triton X-100 buffer for 5 min, cells were washed three times with PBS (containing 0.1% BSA) and blocked with PBS containing 5% BSA for 1 h. The cells were then incubated with the primary antibody overnight at 4°C, followed by incubation with DyLight 488-conjugated anti-rabbit IgG and DyLight 568-conjugated anti-mouse IgG for 1 h. After three times washing with PBS (containing 0.1% BSA), cells were counterstained with Hoechst 33258 solution (catalog no. H1398; Invitrogen, San Diego, CA, USA) for 5 min and then washed three more times with PBS. Finally, the cells were analyzed using a confocal laser scanning microscope (Leica TCS SP8 STED [STimulated Emission Depletion]; Leica, Germany). The antibodies used are listed in Table 1.
ELISA.
Secreted IFN-β were measured with human IFN-β ELISA kit (catalog no. 414101; Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions.
Statistical analyses.
Statistical significance was analyzed by one-way analysis of variance with Dunnett's multiple-comparison test using GraphPad Prism 8. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05; ns, not significant.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (project no. 81971936 and 82041004), the Fundamental Research Funds for the Central Universities, Hubei Province’s Outstanding Medical Academic Leader program, the Foundation for Innovative Research Groups of the National Natural Science Foundation of Hubei (project no. 2020CFA015), the Foundation for Innovative Research Groups of Hubei Health Commission (project no. WJ2021C002), the COVID-19 emergency tackling research project of Shandong University (2020XGB03), and the HUST COVID-19 Rapid Response Call (2020kfyXGYJ036). We thank the staff at the Research Center for Medicine and Structural Biology of Wuhan University for technical assistance and Ke Xu (Wuhan University) for gifting of IAV NS1 plasmid.
A.L., K.Z., and Y.X. contributed to the design of experiments. A.L., K.Z., B.Z., R.H., W.J., Y.F., L.H., and Y.Z. contributed to the conduction of experiments. A.L., K.Z., B.Z., R.H., W.J., J.Z., L.H., Y.Z., Y.L., C.Z., P.W., and K.P. contributed to the reagents. A.L., K.Z., and Y.X. contributed to data analysis. A.L. and Y.X. contributed to manuscript writing and editing.
We have no conflicts of interest to disclose.
Contributor Information
Yuchen Xia, Email: yuchenxia@whu.edu.cn.
Jae U. Jung, Lerner Research Institute, Cleveland Clinic
REFERENCES
- 1.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus Investigating and Research Team. 2020. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382:727–733. 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, Yuan ML, Zhang YL, Dai FH, Liu Y, Wang QM, Zheng JJ, Xu L, Holmes EC, Zhang YZ. 2020. A new coronavirus associated with human respiratory disease in China. Nature 579:265–269. 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu R, Zhao L, Cheng X, Han H, Li C, Li D, Liu A, Gao G, Zhou F, Liu F, Jiang Y, Zhu C, Xia Y. 2021. Clinical characteristics of COVID-19 patients with hepatitis B virus infection - a retrospective study. Liver Int 41:720–730. 10.1111/liv.14774. [DOI] [PubMed] [Google Scholar]
- 4.Chen Y, Liu Q, Guo D. 2020. Emerging coronaviruses: genome structure, replication, and pathogenesis. J Med Virol 92:418–423. 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N, Bi Y, Ma X, Zhan F, Wang L, Hu T, Zhou H, Hu Z, Zhou W, Zhao L, Chen J, Meng Y, Wang J, Lin Y, Yuan J, Xie Z, Ma J, Liu WJ, Wang D, Xu W, Holmes EC, Gao GF, Wu G, Chen W, Shi W, Tan W. 2020. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 395:565–574. 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Q, Zhang Y, Wu L, Niu S, Song C, Zhang Z, Lu G, Qiao C, Hu Y, Yuen KY, Wang Q, Zhou H, Yan J, Qi J. 2020. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell 181:894–904.e9. 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, Zhang Q, Shi X, Wang Q, Zhang L, Wang X. 2020. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 581:215–220. 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 8.Fung TS, Liu DX. 2019. Human coronavirus: host-pathogen interaction. Annu Rev Microbiol 73:529–557. 10.1146/annurev-micro-020518-115759. [DOI] [PubMed] [Google Scholar]
- 9.Poduri R, Joshi G, Jagadeesh G. 2020. Drugs targeting various stages of the SARS-CoV-2 life cycle: exploring promising drugs for the treatment of Covid-19. Cell Signal 74:109721. 10.1016/j.cellsig.2020.109721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rehwinkel J, Gack MU. 2020. RIG-I-like receptors: their regulation and roles in RNA sensing. Nat Rev Immunol 20:537–551. 10.1038/s41577-020-0288-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roers A, Hiller B, Hornung V. 2016. Recognition of endogenous nucleic acids by the innate immune system. Immunity 44:739–754. 10.1016/j.immuni.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 12.Vabret N, Britton GJ, Gruber C, Hegde S, Kim J, Kuksin M, Levantovsky R, Malle L, Moreira A, Park MD, Pia L, Risson E, Saffern M, Salomé B, Esai Selvan M, Spindler MP, Tan J, van der Heide V, Gregory JK, Alexandropoulos K, Bhardwaj N, Brown BD, Greenbaum B, Gümüş ZH, Homann D, Horowitz A, Kamphorst AO, Curotto de Lafaille MA, Mehandru S, Merad M, Samstein RM, Agrawal M, Aleynick M, Belabed M, Brown M, Casanova-Acebes M, Catalan J, Centa M, Charap A, Chan A, Chen ST, Chung J, Bozkus CC, Cody E, Cossarini F, Dalla E, Fernandez N, Grout J, Ruan DF, Hamon P, et al. 2020. Immunology of COVID-19: current state of the science. Immunity 52:910–941. 10.1016/j.immuni.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee S, Channappanavar R, Kanneganti TD. 2020. Coronaviruses: innate immunity, inflammasome activation, inflammatory cell death, and cytokines. Trends Immunol 41:1083–1099. 10.1016/j.it.2020.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han H, Ma Q, Li C, Liu R, Zhao L, Wang W, Zhang P, Liu X, Gao G, Liu F, Jiang Y, Cheng X, Zhu C, Xia Y. 2020. Profiling serum cytokines in COVID-19 patients reveals IL-6 and IL-10 are disease severity predictors. Emerg Microbes Infect 9:1123–1130. 10.1080/22221751.2020.1770129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee JS, Park S, Jeong HW, Ahn JY, Choi SJ, Lee H, Choi B, Nam SK, Sa M, Kwon JS, Jeong SJ, Lee HK, Park SH, Park SH, Choi JY, Kim SH, Jung I, Shin EC. 2020. Immunophenotyping of COVID-19 and influenza highlights the role of type I interferons in development of severe COVID-19. Sci Immunol 5:eabd1554. 10.1126/sciimmunol.abd1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mantlo E, Bukreyeva N, Maruyama J, Paessler S, Huang C. 2020. Antiviral activities of type I interferons to SARS-CoV-2 infection. Antiviral Res 179:104811. 10.1016/j.antiviral.2020.104811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stanifer ML, Kee C, Cortese M, Zumaran CM, Triana S, Mukenhirn M, Kraeusslich HG, Alexandrov T, Bartenschlager R, Boulant S. 2020. Critical role of type III interferon in controlling SARS-CoV-2 infection in human intestinal epithelial cells. Cell Rep 32:107863. 10.1016/j.celrep.2020.107863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sa Ribero M, Jouvenet N, Dreux M, Nisole S. 2020. Interplay between SARS-CoV-2 and the type I interferon response. PLoS Pathog 16:e1008737. 10.1371/journal.ppat.1008737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blanco-Melo D, Nilsson-Payant BE, Liu WC, Uhl S, Hoagland D, Moller R, Jordan TX, Oishi K, Panis M, Sachs D, Wang TT, Schwartz RE, Lim JK, Albrecht RA, tenOever BR. 2020. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell 181:1036–1045.e9. 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuen CK, Lam JY, Wong WM, Mak LF, Wang X, Chu H, Cai JP, Jin DY, To KK, Chan JF, Yuen KY, Kok KH. 2020. SARS-CoV-2 nsp13, nsp14, nsp15 and orf6 function as potent interferon antagonists. Emerg Microbes Infect 9:1418–1428. 10.1080/22221751.2020.1780953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li JY, Liao CH, Wang Q, Tan YJ, Luo R, Qiu Y, Ge XY. 2020. The ORF6, ORF8 and nucleocapsid proteins of SARS-CoV-2 inhibit type I interferon signaling pathway. Virus Res 286:198074. 10.1016/j.virusres.2020.198074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lei X, Dong X, Ma R, Wang W, Xiao X, Tian Z, Wang C, Wang Y, Li L, Ren L, Guo F, Zhao Z, Zhou Z, Xiang Z, Wang J. 2020. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat Commun 11:3810. 10.1038/s41467-020-17665-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xia H, Cao Z, Xie X, Zhang X, Chen JY, Wang H, Menachery VD, Rajsbaum R, Shi PY. 2020. Evasion of type I interferon by SARS-CoV-2. Cell Rep 33:108234. 10.1016/j.celrep.2020.108234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng Y, Zhuang MW, Han L, Zhang J, Nan ML, Zhan P, Kang D, Liu X, Gao C, Wang PH. 2020. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) membrane (M) protein inhibits type I and III interferon production by targeting RIG-I/MDA-5 signaling. Signal Transduct Target Ther 5:299. 10.1038/s41392-020-00438-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Q, Wu J, Wang H, Gao Y, Liu Q, Mu A, Ji W, Yan L, Zhu Y, Zhu C, Fang X, Yang X, Huang Y, Gao H, Liu F, Ge J, Sun Q, Yang X, Xu W, Liu Z, Yang H, Lou Z, Jiang B, Guddat LW, Gong P, Rao Z. 2020. Structural basis for RNA replication by the SARS-CoV-2 polymerase. Cell 182:417–428.e13. 10.1016/j.cell.2020.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang X, Li M, Zheng H, Muster T, Palese P, Beg AA, Garcia-Sastre A. 2000. Influenza A virus NS1 protein prevents activation of NF-kappaB and induction of alpha/beta interferon. J Virol 74:11566–11573. 10.1128/JVI.74.24.11566-11573.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol 5:730–737. 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 28.Clement JF, Bibeau-Poirier A, Gravel SP, Grandvaux N, Bonneil E, Thibault P, Meloche S, Servant MJ. 2008. Phosphorylation of IRF-3 on Ser 339 generates a hyperactive form of IRF-3 through regulation of dimerization and CBP association. J Virol 82:3984–3996. 10.1128/JVI.02526-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB. 2005. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell 19:727–740. 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 30.MacMicking JD. 2012. Interferon-inducible effector mechanisms in cell-autonomous immunity. Nat Rev Immunol 12:367–382. 10.1038/nri3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arunachalam PS, Wimmers F, Mok CKP, Perera R, Scott M, Hagan T, Sigal N, Feng Y, Bristow L, Tak-Yin Tsang O, Wagh D, Coller J, Pellegrini KL, Kazmin D, Alaaeddine G, Leung WS, Chan JMC, Chik TSH, Choi CYC, Huerta C, Paine McCullough M, Lv H, Anderson E, Edupuganti S, Upadhyay AA, Bosinger SE, Maecker HT, Khatri P, Rouphael N, Peiris M, Pulendran B. 2020. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science 369:1210–1220. 10.1126/science.abc6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou Z, Ren L, Zhang L, Zhong J, Xiao Y, Jia Z, Guo L, Yang J, Wang C, Jiang S, Yang D, Zhang G, Li H, Chen F, Xu Y, Chen M, Gao Z, Yang J, Dong J, Liu B, Zhang X, Wang W, He K, Jin Q, Li M, Wang J. 2020. Heightened innate immune responses in the respiratory tract of COVID-19 patients. Cell Host Microbe 27:883–890.e2. 10.1016/j.chom.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lamers MM, Beumer J, van der Vaart J, Knoops K, Puschhof J, Breugem TI, Ravelli RBG, Paul van Schayck J, Mykytyn AZ, Duimel HQ, van Donselaar E, Riesebosch S, Kuijpers HJH, Schipper D, van de Wetering WJ, de Graaf M, Koopmans M, Cuppen E, Peters PJ, Haagmans BL, Clevers H. 2020. SARS-CoV-2 productively infects human gut enterocytes. Science 369:50–54. 10.1126/science.abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thoms M, Buschauer R, Ameismeier M, Koepke L, Denk T, Hirschenberger M, Kratzat H, Hayn M, Mackens-Kiani T, Cheng J, Straub JH, Sturzel CM, Frohlich T, Berninghausen O, Becker T, Kirchhoff F, Sparrer KMJ, Beckmann R. 2020. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science 369:1249–1255. 10.1126/science.abc8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang ZY, He JH, Lu AP, Hou TJ, Cao DS. 2020. Frequent hitters: nuisance artifacts in high-throughput screening. Drug Discov Today 25:657–667. 10.1016/j.drudis.2020.01.014. [DOI] [PubMed] [Google Scholar]
- 36.Auld DS, Southall NT, Jadhav A, Johnson RL, Diller DJ, Simeonov A, Austin CP, Inglese J. 2008. Characterization of chemical libraries for luciferase inhibitory activity. J Med Chem 51:2372–2386. 10.1021/jm701302v. [DOI] [PubMed] [Google Scholar]
- 37.Boschelli DH. 2002. 4-anilino-3-quinolinecarbonitriles: an emerging class of kinase inhibitors. Curr Top Med Chem 2:1051–1063. 10.2174/1568026023393354. [DOI] [PubMed] [Google Scholar]
- 38.Ghosh D, Koch U, Hadian K, Sattler M, Tetko IV. 2018. Luciferase advisor: high-accuracy model to flag false positive hits in luciferase HTS assays. J Chem Inf Model 58:933–942. 10.1021/acs.jcim.7b00574. [DOI] [PubMed] [Google Scholar]