ABSTRACT
Cellular immune responses play a key role in the control of viral infection. The nucleocapsid (N) protein of infectious bronchitis virus (IBV) is a major immunogenic protein that can induce protective immunity. To screen for potential T-cell epitopes on IBV N protein, 40 overlapping peptides covering the entirety of the N protein were designed and synthesized. Four T-cell epitope peptides were identified by gamma interferon (IFN-γ) enzyme-linked immunosorbent spot (ELISpot), intracellular cytokine staining, and carboxyfluorescein succinimidyl ester (CFSE) lymphocyte proliferation assays; among them, three peptides (N211–230, N271–290, and N381–400) were cytotoxic T lymphocyte (CTL) epitopes, and one peptide (N261–280) was a dual-specific T-cell epitope, which can be recognized by both CD8+ and CD4+ T cells. Multi-epitope gene transcription cassettes comprising four neutralizing epitope domains and four T-cell epitope peptides were synthesized and inserted into the genome of Newcastle disease virus strain La Sota between the P and M genes. Recombinant IBV multi-epitope vaccine candidate rLa Sota/SBNT was generated via reverse genetics, and its immune protection efficacy was evaluated in specific-pathogen-free chickens. Our results show that rLa Sota/SBNT induced IBV-specific neutralizing antibody and T-cell responses and provided significant protection against homologous and heterologous IBV challenge. Thus, the T-cell epitope peptides identified in this study could be good candidates for IBV vaccine development, and recombinant Newcastle disease virus-expressing IBV multi-epitope genes represent a safe and effective vaccine candidate for controlling infectious bronchitis.
IMPORTANCE T-cell-mediated immune responses are critical for the elimination of IBV-infected cells. To screen conserved T-cell epitopes in the IBV N protein, 40 overlapping peptides covering the entirety of the N protein were designed and synthesized. By combining IFN-γ ELISpot, intracellular cytokine staining, and CFSE lymphocyte proliferation assays, we identified three CTL epitopes and one dual-specific T-cell epitope. The value of T-cell epitope peptides identified in the N protein was further verified by the design of an IBV multi-epitope vaccine. Results show that IBV multi-epitope vaccine candidate rLa Sota/SBNT provided cross protection against challenges with a QX-like or a TW-like IBV strain. So, T-cell-mediated immune responses play an important role in the control of viral infection, and conserved T-cell epitopes serve as promising candidates for use in multi-epitope vaccine construction. Our results provide a new perspective for the development of a safer and more effective IBV vaccine.
KEYWORDS: infectious bronchitis virus, T-cell epitope, Newcastle disease virus vector, multi-epitope vaccine, cellular immune response, protection
INTRODUCTION
Avian infectious bronchitis (IB), an important viral disease of chickens, affects chickens of all ages and causes great economic losses for the global poultry industry. IB is caused by infectious bronchitis virus (IBV), which belongs to the genus Gammacoronavirus in the family Coronaviridae. The IBV genome is a single-stranded, positive-sense RNA of approximately 27.6 kb in length that encodes 4 structural proteins, i.e., spike (S), membrane (M), small envelope (E), and nucleocapsid (N), and 15 nonstructural proteins (1). The S protein, which is the major glycosylation protein for eliciting neutralizing antibody, is cleaved posttranslationally by host cell proteases into the N-terminal S1 and C-terminal S2 subunits. The S1 subunit is responsible for viral attachment and contains major neutralizing epitopes. The S2 subunit is highly conserved among IBV strains and is involved in mediating viral membrane fusion activity (2). The N protein is a highly conserved structural protein with a high degree of identity (91.0% to 96.5%) among different IBV strains (3); moreover, it is the most abundantly expressed virus-derived protein during IBV infection and is involved in inducing high titers of cross-reactive antibodies and inducing a robust T-cell response (3, 4). The C-terminal 120-amino acid (aa) polypeptide of IBV N protein has been shown to contain cytotoxic T lymphocyte (CTL) epitopes and play a role in protective immunity (5). These advantages make the IBV N protein an ideal target protein for use in designing novel vaccines to improve cross-protective efficacies against different IBV strains.
Currently, dozens of IBV serotypes and genotypes have been reported worldwide, with little or no cross protection between the serotypes (6, 7). Previous epidemiologic data revealed that the QX-like and TW-like IBV strains have become the predominant IBV types in China (6, 8), where they pose a serious threat to the poultry industry. Live-attenuated and inactivated vaccines are extensively used to control IBV on farms. However, live-attenuated vaccine strains pose the risk of reversion to virulence and can contribute to the production of variant strains by gene mutation and recombination (9, 10). However, inactivated vaccines typically induce a weak immune response (11). Therefore, there is an urgent need to develop a safe and effective IBV vaccine.
Epitope-based vaccines represent an alternative option; the potential advantages of epitope-based vaccines over live-attenuated vaccines are that they are safer and can focus the induced immune responses on conserved epitopes (12). T-cell-mediated immune responses are essential for combating viral infection. Several studies have shown that CTLs play a critical role in the elimination of virus-infected cells (13–15). The activation of effector CD8+ T-cell responses requires endogenously expressed CTL epitopes to be presented by major histocompatibility complex class I (MHC I) molecules (16), and effector CD8+ T cells kill virus-infected cells via the mechanism of perforin/granzyme or antiviral cytokine secretion (17, 18). CD4+ T cells play a central role in immune responses by producing effector cytokines to support CD8+ T cells and B cells (19). Virus-specific CD4+ T-cell epitopes must be presented by MHC II molecules (20). In addition to cellular immune responses, humoral immune responses are also important for controlling viral infections. It is well established that IBV-neutralizing antibodies are directed mainly against the highly variable S1 subunit (21) and that the three antigenic sites corresponding to amino acid residues 24 to 61, 132 to 149, and 291 to 398 of the S1 subunit are conformation-dependent neutralizing epitopes (22). Furthermore, the N-terminal portion of the S2 subunit also contains linear neutralization epitopes that elicit broadly reactive neutralizing antibodies (23, 24). Thus, an ideal epitope-based vaccine should contain CD8+ T-cell, CD4+ T-cell, and neutralizing B-cell epitopes.
The present study aimed to comprehensively screen for T-cell epitopes in the IBV N protein and design an IBV multi-epitope gene expression cassette composed of neutralizing epitope domains from the S protein and T-cell epitope peptides from the N protein. Recombinant Newcastle disease virus (NDV) strain La Sota expressing IBV multi-epitope cassettes was generated by reverse genetics, and the protective efficacies of these recombinant live vaccine candidates against challenge with virulent IBV strains were evaluated in specific-pathogen-free (SPF) chickens.
RESULTS
Identification of T-cell epitopes by IFN-γ ELISpot and ICS assays.
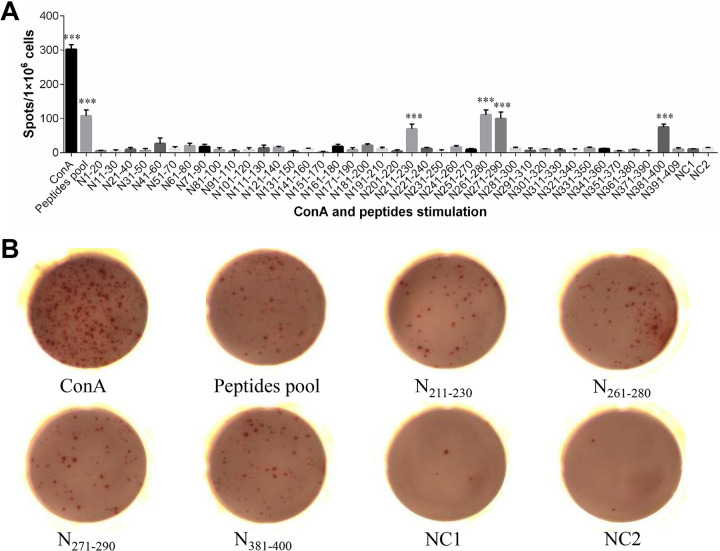
Gamma interferon (IFN-γ) enzyme-linked immunosorbent spot (ELISpot) assays were performed to evaluate the immunogenicity of the synthetic peptides. As shown in Fig. 1, compared with the negative control, the peptide pool and four individual peptides, N211–230, N261–280, N271–290, and N381–400, each induced significant levels of chicken gamma interferon (ChIFN-γ) production in splenocytes from IBV-immunized chickens (P < 0.001), with the number of spots formed in wells treated with these four peptides ranging from 70 to 110 spots/106 cells. In contrast, no significant responses were observed in control-immunized chicken splenocytes incubated with the peptide pool, indicating that the responses to these peptides were IBV specific.
FIG 1.
Screening for T-cell epitopes in IBV nucleocapsid protein via IFN-γ ELISpot assays. (A) The splenocytes isolated from IBV-immunized chickens were stimulated with either the peptide pool or one of the 40 individual peptides for 24 h at 41°C and 5% CO2. As a positive control, splenocytes were stimulated with ConA; as a negative control, splenocytes were supplemented with medium alone (NC1). Splenocytes from control chickens that were incubated with the peptide pool were used as another negative control (NC2). Spot counts are expressed as spots/1 × 106 cells. ***, Significant difference compared with NC1 (P < 0.001). (B) Representative images of IFN-γ ELISpot responses.
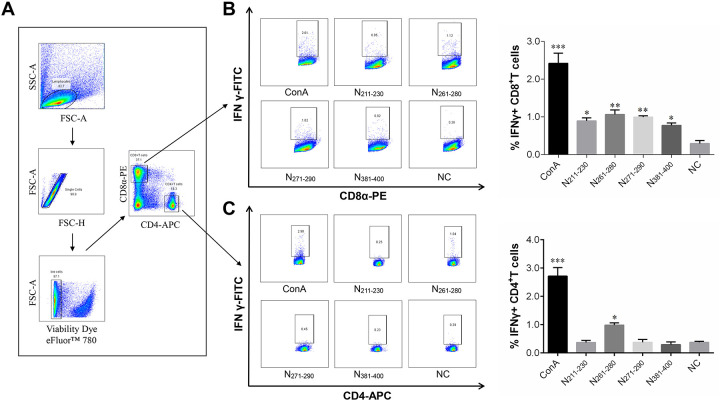
Standard intracellular cytokine staining (ICS) assays were performed to investigate the phenotype of the T cells with peptide-specific IFN-γ secretion. As indicated in Fig. 2, compared with the negative control, all four positive-reacting peptides identified in the ELISpot assays induced significantly higher levels of ChIFN-γ secretion in CD8+ T cells, suggesting that they contain potential CD8+ T-cell epitopes and are CTL epitope peptides. Interestingly, peptide N261–280 also induced ChIFN-γ release in CD4+ T cells, indicating that it may be a dual-specific T-cell epitope peptide. The sequences and locations of the T-cell epitope peptides are shown in Table 1.
FIG 2.
ICS assay phenotype identification of T cells that secrete IFN-γ in response to peptide stimulation. Splenocytes isolated from IBV-immunized chicks were stimulated with ConA or one of four individual peptides (N211–230, N261–280, N271–290, or N381–400); intracellular cytokine staining for IFN-γ was then performed as described in Materials and Methods. (A) Illustration of the gating for IFN-γ-producing T cell subsets. Lymphocytes and single cells were first gated from splenocytes, and then dead cells were excluded with Viability Dye eFluor 780. CD8+ T-cell and CD4+ T-cell subsets were further divided based on CD8 and CD4 expression. (B and C) IFN-γ+ CD8+ T cells (B) and IFN-γ+ CD4+ T cells (C) were gated based on IFN-γ production from CD8+ T-cell and CD4+ T-cell subsets, respectively. (Left) FACS plots from a representative sample, with numbers indicating the percentages of IFN-γ+ CD8+ T cells or IFN-γ+ CD4+ T cells. (Right) Bar graphs displaying the mean ± standard deviation. Statistically significant differences are indicated as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
TABLE 1.
T-cell epitope peptides identified in this study
| Peptide | Amino acid sequence | Amino acid locationa |
|---|---|---|
| N211–230 | GTRITKAKADEMAHRRFCKR | 211–230 |
| N261–280 | EGIKDGRVTAMLNLTPSPHA | 261–280 |
| N271–290 | MLNLTPSPHACLFGSRVTPK | 271–290 |
| N381–400 | EERNNAQLEFDDEPKVINWG | 381–400 |
Amino acid location of T-cell epitope peptides on IBV N protein.
Evaluation of T-cell proliferation by CFSE staining assay.
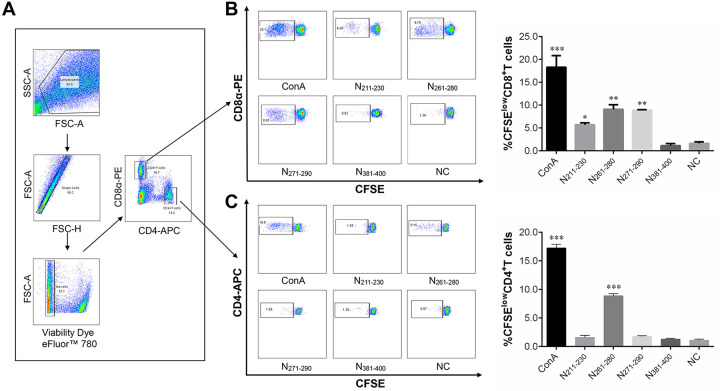
Carboxyfluorescein succinimidyl ester (CFSE) staining assays were performed to evaluate the proliferation of CD4+ and CD8+ T cells after their stimulation with the T-cell epitope peptides for 4 days at 41°C and 5% CO2. The results show that, compared with the control treatment, peptides N211–230, N261–280, and N271–290 each induced a significantly higher rate of CD8+ T-cell proliferation in splenocytes from IBV-immunized chicks. Consistent with the ICS assay data, peptide N261–280 also induced CD4+ T-cell proliferation, suggesting that peptide N261–280 can be recognized by both CD4+ and CD8+ T cells. Unexpectedly, peptide N381–400, which was found by ICS assay to induce ChIFN-γ release in CD8+ T cells, did not stimulate any T-cell proliferation in CFSE staining assays; this surprising result was confirmed repeatedly (Fig. 3).
FIG 3.
T-cell proliferation as assessed by CFSE staining assays. Splenocytes from IBV-immunized chicks were stained with CFSE and then stimulated with ConA or one of four individual peptides for 4 days. Cell proliferation analysis was performed by flow cytometry. (A) Illustration of the gating for T-cell proliferation. Lymphocytes, single cells, live lymphocytes, and CD8+ T-cell or CD4+ T-cell subsets were gated as described in Fig. 2. (B and C) T cell proliferation was defined as CFSE low, CFSElow CD8+ T cells (B), and CFSElow CD4+ T cells (C) were gated based on the reduction in CFSE fluorescence in CD8+ T-cell and CD4+ T-cell subsets, respectively. (Left) FACS plots from a representative sample; the percentages of proliferated cells are shown in each gate. (Right) Bar graphs displaying the mean ± standard deviation. Statistically significant differences are indicated as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Generation of recombinant NDV expressing IBV multi-epitope proteins.
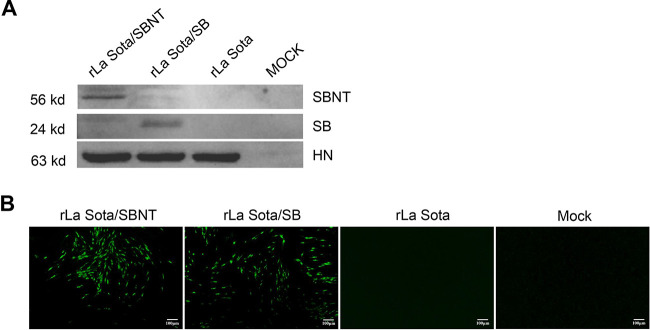
Two forms of IBV multi-epitope cassettes (SB and SBNT) were designed, synthesized, and cloned individually into the cDNA backbone of NDV strain La Sota between the P and M genes. The two recombinant viruses, rLa Sota/SB and rLa Sota/SBNT, were each recovered by reverse genetics and passaged five times in 9-day-old SPF chicken embryos, and the genetic stability of these recombinant viruses was confirmed by reverse transcriptase PCR (RT-PCR) and sequencing analyses (data not shown). The expression of SB and SBNT multi-epitope proteins was detected by Western blot and immunofluorescence analysis (IFA). The proteins expressed from the SB and SBNT multi-epitope cassettes were successfully detected at molecular weights of approximately 24 kDa and 56 kDa in lysates of cells infected with rLa Sota/SB and rLa Sota/SBN, respectively, whereas no specific bands were found for lysates of mock- or rLa Sota-infected cells in Western blots performed using polyclonal chicken anti-IBV serum (Fig. 4A). In Western blots performed using a polyclonal chicken anti-NDV serum, NDV HN proteins of approximately 53 kDa were detected in all infected cells, confirming that a similar level of NDV protein was loaded into each lane. The results of IFA performed using a polyclonal chicken anti-IBV serum and fluorescein isothiocyanate (FITC)-conjugated goat anti-chicken IgG secondary antibody further confirm the expression of the SB and SBNT multi-epitope proteins in rLa Sota/SB- and rLa Sota/SBN-infected cells, respectively; in contrast, no fluorescent signal was detected in mock- or rLa Sota-infected cells (Fig. 4B).
FIG 4.
Expression of IBV multi-epitope proteins in DF-1 cells. (A) DF-1 cells were infected with rLa Sota/SBNT, rLa Sota/SB, or rLa Sota at an MOI of 0.01 for 24 h, and the expression of multi-epitope proteins SBNT and SB (at approximately 56 kDa and 24 kDa, respectively) was detected by Western blot analysis using a chicken polyclonal anti-IBV serum. Additionally, the expression of NDV HN protein (at approximately 53 kDa) was detected by Western blot analysis using a chicken polyclonal anti-NDV serum. (B) IFA of multi-epitope protein expression. DF-1 cells were infected with rLa Sota/SBNT, rLa Sota/SB, or rLa Sota at an MOI of 0.01 for 48 h, after which the cells were subjected to an IFA in which they were probed with a chicken polyclonal anti-IBV serum, incubated with an FITC-conjugated goat anti-chicken IgG antibody, and subsequently visualized under a fluorescence microscope.
In vitro characterization of IBV multi-epitope vaccine candidates.
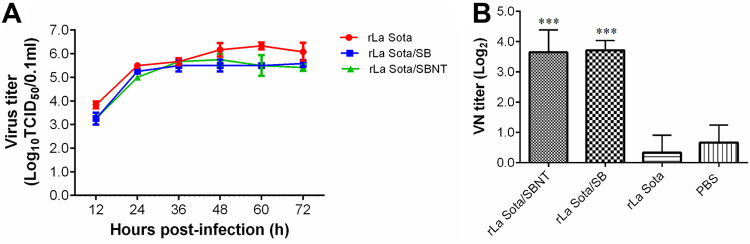
The in vitro growth characteristics of the recombinant and parental viruses were evaluated in DF-1 cells. As shown in Fig. 5A, the growth rates of recombinant viruses rLa Sota/SBNT and rLa Sota/SB were slightly lower than that of the parental virus rLa Sota at all time points; however, the recombinant viruses reached a titer similar to that of rLa Sota at 36 hours postinfection (hpi). The parental virus rLa Sota reached a maximum titer of 106.33 50% tissue culture infective dose (TCID50)/0.1 ml at 60 hpi, whereas the recombinant virus rLa Sota/SB reached a maximum titer of 105.58 TCID50/0.1 ml at 72 hpi, and the recombinant virus rLa Sota/SBNT reached a maximum titer of 105.75 TCID50/0.1 ml at 48 hpi. The virulence of the recombinant and parental viruses was evaluated by mean death time (MDT) test. All three tested strains (rLa Sota, rLa Sota/SB, and rLa Sota/SBNT) showed MDT values of >120 h. These results indicate that the presence of the SB or SBNT multi-epitope genes did not significantly affect the growth characteristics or virulence of rNDV.
FIG 5.
Growth kinetics of recombinant and parental viruses in DF-1 cells and neutralizing antibody responses against IBV. (A) DF-1 cells were infected with one of the recombinant viruses (rLa Sota/SB or rLa Sota/SBNT) or the parental virus rLa Sota at an MOI of 0.01. Viral titers in supernatant samples collected from infected cells at 12-h intervals until 72 hpi were determined by performing a TCID50 assay in DF-1 cells. The titers at each time point are expressed as the mean ± standard deviation. (B) The level of antibodies against IBV was determined by virus neutralization assay. Three serum samples per group were assessed, and antibody titers are expressed as reciprocals of the log2 dilution. Bar graph displays the mean ± standard deviation. Statistically significant differences versus the PBS group are indicated (***, P < 0.001).
Immune responses induced by IBV multi-epitope vaccine candidates.
Seven-day-old SPF chickens from each group were vaccinated with rLa Sota/SBNT, rLa Sota/SB, or rLa Sota or were administered an equal volume of phosphate-buffered saline (PBS). At 3 weeks postvaccination, serum samples and splenocytes were collected from the birds in each group and used for the evaluation of neutralizing antibodies and T-cell responses against IBV, respectively. The results show that the chickens immunized with the recombinant virus rLa Sota/SBNT or rLa Sota/SB had a mean neutralizing antibody titer of 3.65 or 3.71 log2, respectively; these titers are significantly higher (P < 0.001) than those of chickens inoculated with the parental virus rLa Sota or PBS, in which no neutralizing antibodies could be detected (Fig. 5B). The ELISpot assay results indicate that, compared with splenocytes from rLa Sota-vaccinated chickens, a significantly higher level of ChIFN-γ secretion was obtained in splenocytes from rLa Sota/SBNT-vaccinated chickens after their stimulation with the four T-cell epitope peptides (P < 0.001), whereas no significant ChIFN-γ release was detected in splenocytes from rLa Sota/SB-vaccinated chickens (Fig. 6). These results suggest that the recombinant virus rLa Sota/SBNT, which contains T-cell epitope peptides and neutralizing epitope domains, can induce robust IBV-specific cellular and humoral immune responses in vaccinated chickens.
FIG 6.
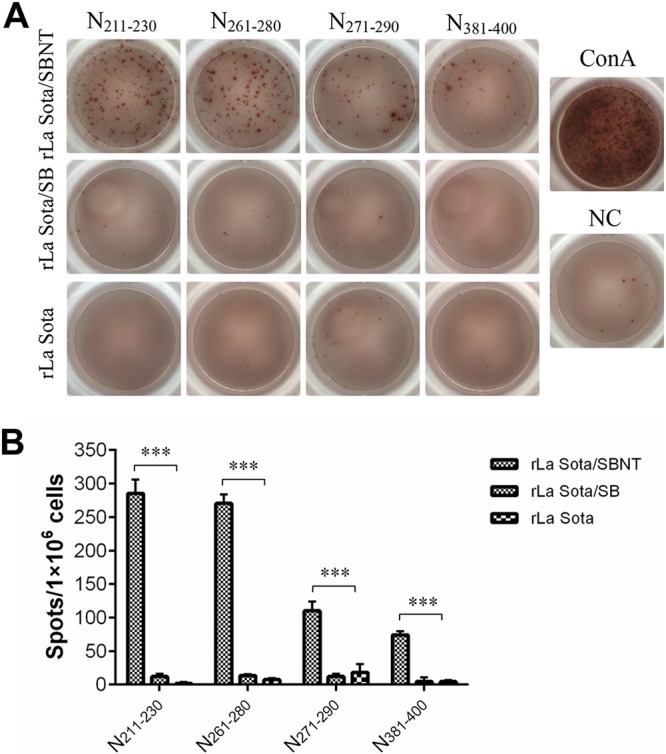
Cellular immune responses against IBV. SPF chickens were vaccinated with the recombinant or parental viruses via the oculonasal route. At 21 d postvaccination, splenocytes were collected, and T-cell responses were detected by ELISpot assay. (A) Representative ELISpot images of splenocytes from different groups stimulated with one of the four T-cell epitope peptides; splenocytes supplemented with medium alone served as a negative control (NC), and those stimulated with ConA served as a positive control. (B) Bar graph displays the mean ± standard deviation of spot-forming cells per 106 splenocytes of different groups stimulated with one of the four T-cell epitope peptides. ***, Significant difference relative to the rLa Sota group (P < 0.001).
Protective efficacy of IBV multi-epitope vaccine candidates against homologous IBV strain challenge.
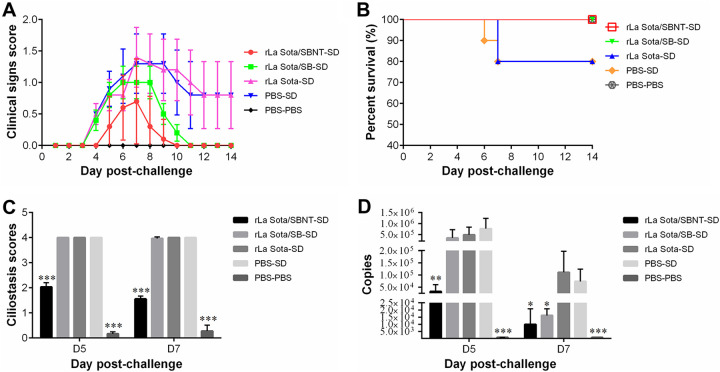
To evaluate the protective efficacy of IBV multi-epitope vaccine candidates against a homologous IBV strain challenge, 7-day-old SPF chickens were vaccinated with rLa Sota/SBNT, rLa Sota/SB, rLa Sota, or PBS and then challenged with the virulent IBV SD strain at 3 weeks postvaccination (the experimental groups were named rLa Sota/SBNT-SD, rLa Sota/SB-SD, rLa Sota-SD, and PBS-SD, respectively; the control-vaccinated, unchallenged group was named PBS-PBS). The clinical signs of each group were monitored and scored daily for 14 days (Fig. 7A). Most of the chickens in the rLa Sota-SD and PBS-SD groups began to display clinical signs at 4 days postchallenge (dpc), including heavy nasal discharge, depression, sneezing, coughing, and tracheal rales, and a 20% mortality rate was observed in each of these two groups during the 14-day observation period (Fig. 7B). In contrast, the chickens in the rLa Sota/SBNT-SD group exhibited significantly milder clinical signs after IBV challenge, especially at 8 to 10 dpc. Although the clinical signs displayed by the rLa Sota/SB-SD group chickens seemed slightly less severe than those displayed by the PBS-SD group chickens, there were no significant differences in their clinical sign scores during the observation period. No chickens in the rLa Sota/SBNT-SD or rLa Sota/SB-SD groups died during the observation period (100% survival rate).
FIG 7.
The protective efficacy of IBV multi-epitope vaccine candidates against homologous IBV challenge. (A) Clinical sign scores of each group after challenge with IBV SD strain. The chickens were monitored daily for 14 days and assigned clinical sign scores (0 for normal; 1 for slight shaking, slight nasal discharge, and slight lacrimation; 2 for depression, watery feces, and sneezing or coughing; 3 for heavy depression, heavy nasal discharge, tracheal rales, or mouth breathing; and 4 for death). Line chart displays the mean ± standard deviation. (B) Survival curve showing the survival percentage in each group during the 14-day observation period. (C) Tracheal ciliostasis scores. Tracheal ciliary activity was assessed and scored at 5 and 7 dpc using the scoring system described in Materials and Methods. Bar graph displays the mean ± standard deviation. (D) Tracheal swab samples were collected from each group at 5 and 7 dpc, and the viral loads in these swab samples were determined by real-time RT-qPCR. Bar graph displays the mean ± standard deviation. Statistically significant differences versus the PBS-SD group are indicated as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Ciliostasis activity in the trachea was evaluated at 5 and 7 dpc. As shown in Fig. 7C, the PBS-SD and rLa Sota-SD groups each had a maximum mean ciliostasis score of 4.0 at both 5 and 7 dpc; in fact, no cilia movement was observed for these two groups at any point. The rLa Sota/SB-SD group similarly had maximum ciliostasis scores of 4.0 and 3.9 at 5 and 7 dpc, respectively, which are not significantly different from those of the PBS-SD group (P > 0.05). In contrast, the rLa Sota/SBNT-SD group had mean ciliostasis scores of 2.0 and 1.6 at 5 and 7 dpc, respectively, which are significantly lower compared with those of the PBS-SD group (P < 0.001). The minimum mean ciliostasis scores observed in this experiment were 0.17 and 0.27, which were in the PBS-PBS group at 5 and 7 dpc.
Tracheal swab samples were collected from chickens in each group at 5 and 7 dpc and used in real-time reverse transcriptase quantitative PCR (RT-qPCR) assays to evaluate the level of virion shedding. The chickens in the rLa Sota/SBNT-SD group had significantly lower levels of virion shedding than those in the PBS-SD group at both 5 and 7 dpc (P < 0.05). Compared with the PBS-SD group, the rLa Sota/SB-SD group showed a similarly high level of virion shedding at 5 dpc but a significantly lower level at 7 dpc (P < 0.05). However, there was no significant difference in the level of virion shedding between the rLa Sota-SD and PBS-SD groups at either time point (P > 0.05). As expected, no virion shedding was detected in the PBS-PBS group (Fig. 7D).
Cross-protective efficacy of IBV multi-epitope vaccine candidates against heterologous IBV strain challenge.
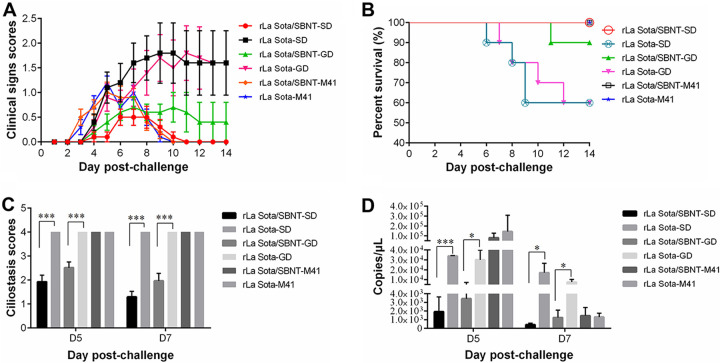
To evaluate the cross-protective efficacy of the IBV multi-epitope vaccine candidate rLa Sota/SBNT, 1-day-old SPF chickens were vaccinated with rLa Sota/SBNT or the parental virus rLa Sota and then challenged with QX-like IBV SD strain, TW-like IBV GD strain, or Mass-type IBV M41 strain at 3 weeks postvaccination (groups rLa Sota/SBNT-SD, rLa Sota/SBNT-GD, rLa Sota/SBNT-M41, rLa Sota-SD, rLa Sota-GD, and rLa Sota-M41, respectively). As shown in Fig. 8A, the chickens in the rLa Sota/SBNT-SD group showed significantly lower clinical sign scores compared with those of the chickens in the rLa Sota-SD group at 7 to 14 dpc with IBV SD strain (P < 0.05). Meanwhile, compared with the rLa Sota-GD group birds, the rLa Sota/SBNT-GD group birds also had significantly lower clinical sign scores at 9 and 11 to 14 dpc with IBV GD strain (P < 0.05). However, no significant difference in clinical sign scores was found between the rLa Sota/SBNT-M41 and rLaSota-M41 groups during the observation period (P > 0.05). The rLa Sota-SD and rLa Sota-GD groups each showed a 40% mortality rate during the observation period; in contrast, survival rates of 100% and 90% were obtained in the rLa Sota/SBNT-SD and rLa Sota/SBNT-GD groups, respectively, and only a single chicken in the rLa Sota/SBNT-GD group died (at 11 dpc). No mortality occurred in the rLa Sota/SBNT-M41 or rLaSota-M41 groups (Fig. 8B).
FIG 8.
The cross-protective efficacy of IBV multi-epitope vaccine candidates against heterologous IBV strain challenge. (A) Clinical sign scores. One-day-old SPF chickens were immunized with rLa Sota/SBNT or rLa Sota and then challenged with QX-like IBV SD strain, TW-like IBV GD strain, or Mass-type M41 strain at 3 weeks postvaccination. The clinical signs of each group were scored as described in the legend for Fig. 7. Line chart displays the mean ± standard deviation. Red circles indicate significant differences between the rLa Sota/SBNT-SD and rLa Sota-SD groups, and green triangles indicate significant differences between the rLa Sota/SBNT-GD and rLa Sota-GD groups at each time point. (B) Survival curve showing the survival percentage in each group within the 14-day observation period. (C) Tracheal ciliostasis scores of each group. Scores were assigned according to the scoring system described in Materials and Methods. Bar graph displays the mean ± standard deviation. (D) Detection of tracheal virion shedding by real-time RT-qPCR. Bar graph displays the mean ± standard deviation. Statistically significant differences versus the PBS-SD group are indicated as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
The average ciliostasis scores for the various experimental groups were calculated (Fig. 8C). Regarding challenge with IBV SD strain, the chickens in the rLa Sota/SBNT-SD group showed mean ciliostasis scores of 1.9 and 1.3 at 5 and 7 dpc, respectively, which are significantly lower than those for the chickens in the rLa Sota-SD group (P < 0.001). Regarding challenge with IBV GD strain, the chickens in the rLa Sota/SBNT-GD group had mean ciliostasis scores of 2.5 and 2.0 at 5 and 7 dpc, respectively, which are significantly lower compared with those of the chickens in the rLa Sota-GD group (P < 0.001); approximately 50% of cilia in the tracheal sections from the rLa Sota/SBNT-GD group showed movement at 5 and 7 dpc. In contrast, a maximum mean ciliostasis score of 4.0 was obtained in the rLa Sota/SBNT-M41 and rLaSota-M41 groups at both 5 and 7 dpc, and no cilia in samples from either of these two groups exhibited movement.
Tracheal virion shedding was evaluated at 5 and 7 dpc (Fig. 8D). The rLa Sota/SBNT-SD group showed significantly less virion shedding at 5 and 7 dpc with the QX-like IBV SD strain (P < 0.05) than the rLa Sota-SD group. The rLa Sota/SBNT-GD group also showed significantly lower levels of virion shedding at 5 and 7 dpc with the TW-like IBV GD strain (P < 0.05) compared with those of the rLa Sota-GD group. The rLa Sota/SBNT-M41 and rLaSota-M41 groups showed the highest level of virion shedding at 5 dpc after challenge with the IBV M41 strain, but the levels of virion shedding in these groups were much lower at 7 dpc, and the virion shedding levels in these groups were not significantly different at either 5 or 7 dpc. These results reveal that the IBV multi-epitope vaccine candidate rLa Sota/SBNT provided significant protection against QX-like and TW-like IBV challenge in terms of clinical symptom alleviation, resistance to tracheal cilia damage, and virion shedding reduction.
DISCUSSION
IB endangers the poultry industry worldwide (25). Currently, live-attenuated and inactivated IBV vaccines are widely used in the field to control IBV infection, but these options are suboptimal. Live-attenuated IBV vaccines pose the potential risk of reversion to virulence or recombination with virulent field strains, which could lead to the emergence of novel IBV variants (26, 27). Inactivated IBV vaccines are generally considered safer than live-attenuated vaccines, but their poor ability to activate T-cell mediated-responses renders them less protective (11, 28). Several studies have shown that CTLs are critical for the elimination of IBV-infected cells; specifically, naive chickens that received an adoptive transfer of effector T cells or memory T cells were protected from acute IBV challenge (5, 13, 29). Therefore, an ideal IBV vaccine should evoke a vigorous T-cell response.
Among the IBV structural proteins, the S protein is the major glycosylation protein that induces neutralizing antibodies, and several B-cell epitopes have been identified in the S protein (22, 30–32). Among them, three conformational B-cell epitopes (24 to 61 aa, 132 to 149 aa, and 291 to 398 aa) located on the S1 subunit and one linear B-cell epitope (546 to 567 aa) located on the S2 subunit were found to induce neutralizing antibody and play a role in protective immunity. Additionally, several CTL epitopes were identified in the S1 subunit (33). The N protein is the major structural protein involved in activating the CTL response (13). A CD4+ T-cell epitope within the IBV N protein was identified in mice (34), but, to date, no N protein T-cell epitopes have been reported in chickens. Our study is the first to comprehensively screen T-cell epitopes in IBV N protein.
IFN-γ ELISpot and ICS assays have been extensively used to investigate antigen-specific T cell responses in human and animal medical research. T cells and natural killer (NK) cells are known to be the major sources of IFN-γ (35–37); however, compared to mammals, NK cells are present in very low numbers in the peripheral blood or spleen of birds (38). Here, a total of 40 overlapping peptides covering the entire N protein of IBV were synthesized and used for epitope mapping. Chickens were primed with the IBV SZ strain, which naturally has a low level of virulence and then were boosted with the highly virulent IBV SD strain (39). Four of the 40 peptides (N211–230, N261–280, N271–290, and N381–400) displayed the ability to induce significant levels of ChIFN-γ production in splenocytes from immunized chickens; among them, three peptides (N211–230, N271–290, and N381–400) can be recognized by CD8+ T-cell subsets, i.e., they are CTL epitope peptides. Additionally, one peptide (N261–280) could be recognized by both CD4+ T- and CD8+ T-cell subsets, indicating that it is a dual-specific T-cell epitope peptide. To our knowledge, this is the first report of a dual-specific T-cell epitope peptide in IBV. Vaccination with dual-specific T-cell epitope peptides might be more advantageous than vaccination with a mixture of CTL and Th-cell epitopes because such peptides can be processed and presented by the same APC, subsequently activating both CD4+ T- and CD8+ T-cell subsets, and their close proximity may promote interactions between these cells, consequently enhancing the CTL effector function (40). Previous studies have shown that vaccination of mice with a 35-mer dual-specific T-cell epitope peptide of human papillomavirus completely protected the animals from papillomavirus infection (41). The T-cell epitope peptides identified in this study may serve as potential candidates for use in multi-epitope vaccine construction.
Multi-epitope vaccines represent an appealing alternative to the traditional live-attenuated and inactivated IBV vaccines owing to their high level of safety and ability to induce an immune response to selected epitopes. In the present study, four neutralizing epitope domains (S24–61, S132–149, S291–398, and S546–567) were incorporated into a multi-epitope gene expression box. Among them, three domains (S24–61, S132–149, and S291–398) correspond to the conformational neutralizing epitope in the IBV SD strain S1 subunit, and one domain (S546–567) corresponds to the linear neutralizing epitope of the IBV SD strain S2 subunit; these domains were previously shown to induce neutralizing antibody and provide immune protection against IBV (22, 23, 42, 43). Generally, the minimal CTL epitopes are composed of 8 to 11 amino acids; however, in the present study, T-cell epitope peptides of 20 amino acids in length were used directly for the construction of multi-epitope vaccine candidates, mainly because, compared with a minimal CTL epitope peptide of 8 to 11 amino acids in length, the longer peptides may contain several epitopes that are not restricted to a certain MHC haplotype. Furthermore, in addition to containing CTL epitopes, these longer peptides may also contain CD4+ T-cell epitopes. Therefore, longer peptides may induce a broader response at the population level (44, 45). The SBNT expression box contains three repeated T-cell epitope peptides to enhance the epitope-specific T-cell responses.
NDV is an ideal vaccine vector, and it has been widely used in human and animal recombinant viral vector vaccine research. Sun et al. reported that an NDV vector expressing the spike protein of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) could elicit high levels of neutralizing antibodies and protect mice from a mouse-adapted SARS-CoV-2 challenge (46). Abozeid et al. found that a recombinant NDV vector expressing IBV S protein provided significant protection against IBV challenge in SPF chickens (47). In the present study, each multi-epitope transcription cassette was placed between the P and M genes in the avirulent NDV strain La Sota. Two recombinant viruses, rLa Sota/SBNT and rLa Sota/SB, were recovered, and their growth kinetics were similar to the parental virus rLa Sota, indicating no adverse effects on viral replication after insertion of exogenous epitopes at this site. Additionally, they replicated to a high titer in embryonated chicken eggs, which is critical for mass vaccine production. The SB and SBNT multi-epitope proteins expressed by the recombinant viruses were successfully detected in Western blot and IFA assays conducted using polyclonal chicken anti-IBV serum, indicating that the multi-epitope proteins have good antigenicity.
The activation of humoral immunity has been shown to be important in IBV vaccine-induced protection (48). In the present study, the level of neutralizing antibodies developed in groups vaccinated with rLa Sota/SBNT or rLa Sota/SB was higher than those developed in groups vaccinated with rLa Sota or PBS. However, in addition to humoral immune responses, cellular immune responses can also affect vaccination efficacy. Here, the T-cell responses induced by our multi-epitope vaccine candidates were evaluated by ELISpot assays. As expected, the multi-epitope vaccine candidate rLa Sota/SBNT, which contained T-cell epitope peptides, elicited robust peptide-specific T-cell responses in chickens, whereas no virus-specific T-cell responses were elicited by rLa Sota/SB or rLa Sota, indicating that the multi-epitope vaccine candidate rLa Sota/SBNT can induce not only a humoral immune but also a cellular immune response.
In the first immune-protection experiment, we found the multi-epitope vaccine candidate rLa Sota/SBNT conferred better protection to 7-day-old SPF chickens in terms of clinical symptom alleviation, resistance to tracheal cilia damage, and virion shedding reduction after virulent IBV SD strain challenge. Compared with those in the PBS-SD group, the chickens in the rLa Sota/SB-SD group displayed a delayed protective effect, with slightly alleviated clinical symptoms and a significant reduction in virion shedding only at 7 dpc. However, the rLa Sota/SB-SD group had maximal ciliostasis scores at both 5 and 7 dpc, similar to the PBS-SD group. The rLa Sota/SBNT conferred better respiratory tract protection against IBV challenge compared with rLa Sota/SB; this could be because the IBV T-cell epitope peptides expressed by rLa Sota/SBNT elicited more effective cellular immune responses, which are critical for the elimination of IBV from infected chickens (5, 13, 29). Raj and Jones emphasized that immunity to IBV in the trachea is mediated mainly by T cells (49). Kotani et al. reported that CTL infiltrate at the tracheal mucosa is correlated with the elimination of IBV in the early stage of infection, whereas humoral immunity control of IBV infection occurs mainly at the later stage (50). No protective effect was observed in the rLa Sota-SD group during the observation period. Our results indicate that the protection of chickens against IBV involves both humoral and cellular immune responses; thus, an effective IBV multi-epitope vaccine should contain both neutralizing epitopes and T-cell epitopes.
In the second immune-protection experiment, we investigated whether the IBV multi-epitope vaccine candidate rLa Sota/SBNT could confer effective cross-protection against a heterologous IBV strain challenge. One-day-old SPF chickens were used for this experiment because the day of hatch is often the IBV vaccination time point in the field. Our results show that immunization with rLa Sota/SBNT could significantly alleviate clinical symptoms and protect tracheal cilia activity after QX-like or TW-like IBV strain challenge. Only a small proportion of chickens in the rLa Sota/SBNT-SD or rLa Sota/SBNT-GD groups exhibited mild clinical symptoms during the observation period, whereas all chickens in the rLa Sota-SD and rLa Sota-GD groups showed severe clinical symptoms, such as depression, heavy nasal discharge, and tracheal rales. To our surprise, the multi-epitope vaccine candidate rLa Sota/SBNT conferred no observable protection efficacy against a Mass-type M41 strain challenge, and there were no significant differences in the clinical sign scores, ciliostasis scores, or virion shedding levels between the rLa Sota/SBNT-M41 and rLaSota-M41 groups. This might be because the QX-like IBV strains are closely related phylogenetically to the TW-like strains but only distantly related to the Mass-type vaccine strains (51). Notably, commercial Mass-type live-attenuated vaccine vaccination often fails to protect chickens against QX-like and TW-like IBV infections (51–53). Whether the multi-epitope vaccine candidate rLa Sota/SBNT potentially protects against infection with other heterologous IBV strains requires further investigation.
Currently, QX-like and TW-like IBV strains are the predominant IBV types in China (6, 8), and the multi-epitope vaccine candidate rLa Sota/SBNT conferred cross-protection against challenges with either a QX-like or a TW-like IBV strain. Compared with traditional live-attenuated IBV vaccines, the IBV multi-epitope vaccine candidate rLa Sota/SBNT is safer and can be grown to high titer in embryonated eggs, making it more suitable for use in mass vaccine production. These advantages suggest that rLa Sota/SBNT is a promising candidate for the development of a novel IBV vaccine.
In summary, our study reported four new T-cell epitope peptides in IBV N protein and constructed multi-epitope vaccine candidates based on IBV-neutralizing epitopes and T-cell epitopes using NDV as a vaccine vector. The multi-epitope vaccine candidate rLa Sota/SBNT induced robust humoral and cellular immune responses and provided protection against QX-like and TW-like IBV challenges. Our results suggest that this multi-epitope vaccine may be an attractive alternative for the development of a safer and more effective IBV vaccine.
MATERIALS AND METHODS
Viruses, cells, animals, and ethics statement.
The QX-like IBV SZ and SD strains, TW-like IBV GD strain, and Mass-type IBV M41 strain were stored in our laboratory. Although the SZ and SD strains share high sequence similarities, the SD strain has a higher pathogenicity in chickens compared with the SZ strain (39). The recombinant avirulent NDV strain La Sota was generated previously in our laboratory by using reverse genetics technology (54). All strains were propagated in 10-day-old SPF embryonated chicken eggs via the allantoic route. The 50% embryo infectious dose (EID50) of these strains in the harvested allantoic fluid was calculated by applying the Reed and Muench method (55). Cells from a chicken embryo fibroblast cell line (DF-1 cells) or baby hamster kidney cell line that stably expresses T7 RNA polymerase (BSR T7/5 cells) were cultured in Dulbecco modified Eagle medium (DMEM) with 10% fetal bovine serum (FBS). SPF white leghorn chickens and SPF embryonated eggs were purchased from Beijing Boehringer Ingelheim Vital Biotechnology Co. Ltd. (Beijing, China). Our study was approved by the Animal Welfare and Ethical Censor Committee of China Agricultural University (CAU approval number: 2020–08).
Synthetic peptides.
A total of 40 overlapping peptides, covering the entire N protein encoded by the IBV SD strain (GenBank accession number KY421673), were synthesized by Shanghai Apeptide Co., Ltd. (Shanghai, China). The peptides were each 20 amino acids in length and overlapped one another by 10 amino acids. All peptides were synthesized at high purity (>90%) and were dissolved in 70% dimethyl sulfoxide (DMSO) to a concentration of 10 mM before being stored at −80°C.
Chicken immunization and splenocyte isolation.
Two-week-old SPF white leghorn chickens were divided into two groups. The experimental group birds were immunized (primed) with 106.0 EID50 of IBV SZ strain via the oculonasal route and boosted with the same dose of IBV SD strain 3 weeks later. Birds in the control group were administered sterile PBS under the same schedule as the experimental group. Two weeks after receiving the boost immunization, the chickens were sacrificed, and their splenocytes were isolated as described previously (56).
IFN-γ ELISpot assay.
The IFN-γ ELISpot assay was performed as described previously (56). Briefly, the ELISpot 96-well plates (Millipore, Eschborn, Germany) were coated with 5 µg/ml mouse-anti-ChIFN-γ (Invitrogen; Carlsbad, CA, USA) in PBS (pH 7.4) by overnight incubation at 4°C. The plates were then washed with PBS and blocked with R10 medium (RPMI 1640 medium with 10% FBS). After the blocking buffer was discarded, 3.5 × 105 splenocytes from the experimental group were added to each well. The cells were stimulated for 24 h in the presence of either R10 medium (negative control), mitogen ConA (5 µg/ml, positive control), individual peptides (10 µg/ml), or a pool of all 40 peptides (peptide pool, 10 µg/ml of each peptide) at 41°C and 5% CO2; control group splenocytes incubated with the peptide pool were used as another negative control. Subsequently, the plates were washed with PBST (PBS supplemented with 0.05% Tween 20) and incubated with 1 µg/ml biotin-conjugated mouse anti-ChIFN-γ (Invitrogen) for 1 h at room temperature. Plates were again washed with PBST and then incubated with horseradish peroxidase (HRP)-conjugated streptavidin (BD Bioscience, Franklin Lake, NJ, USA) for 1 h at room temperature. Spots were developed via incubation with an 3-amino-9-ethylcarbazole (AEC) substrate set (BD Bioscience) and counted by using an ELISpot plate reader (Immunospot Analyzer; Cellular Technology Ltd., Shaker Heights, OH, USA). Peptide-specific T-cell frequencies are expressed in this manuscript as the number of spot-forming cells per 106 splenocytes.
Intracellular cytokine staining assays.
Splenocytes were resuspended in R10 medium at a concentration of 1 × 107 cells/ml and seeded in duplicate (100 µl/well) into 96-well plates. The cells were then stimulated with ConA (5 µg/ml, positive control) or individual peptides (10 µg/ml) for 6 h at 41°C and 5% CO2; cells in medium alone served as negative controls. When 4 h remained of the 6-h incubation period, GolgiStop protein transport inhibitor (BD Bioscience) was added to each well to a final concentration of 10 µg/ml. At the end of the 6-h incubation, the stimulated cells were harvested and surface stained with allophycocyanin (APC)-conjugated mouse anti-chicken CD4 (Southern Biotech, Birmingham, AL, USA), PE-conjugated mouse anti-chicken CD8α (Southern Biotech), and Fixable Viability Dye eFluor 780 (Invitrogen) for 30 min at 4°C. The cells were then fixed and permeabilized by using the BD Cytofix/Cytoperm kit (BD Bioscience) in accordance with the manufacturer’s instructions and then stained with biotin-conjugated mouse anti-ChIFN-γ (Invitrogen) in combination with FITC-conjugated streptavidin (Invitrogen). Finally, the cells were resuspended in fluorescence-activated cell sorter (FACS) buffer and used for flow cytometry analyses. FlowJo software was used to analyze the flow cytometry data.
CFSE lymphocyte proliferation assay.
Splenocytes were stained with 0.5 µM CFSE (Invitrogen) in PBS for 5 min at room temperature. After being stained, the cells were washed three times with R10 medium and resuspended at a concentration of 2 × 107 cells/ml. The CFSE-stained cells were transferred to 24-well plates (500 µl per well) and stimulated with ConA (10 µg/ml, positive control) or individual peptides (10 µg/ml) for 4 days in 41°C and 5% CO2; a similar volume of R10 medium was added to the negative-control cells. After this incubation, the cells were harvested and stained with APC-conjugated mouse anti-chicken CD4 antibody, PE-conjugated mouse anti-chicken CD8α antibody, and Fixable Viability Dye eFluor 780 at 4°C for 30 min and subsequently used for flow cytometry analyses. The percentage of proliferating CD4+ T or CD8+ T cells was determined based on the reduction in CFSE fluorescence over time.
Design and generation of rNDV expressing IBV multi-epitope gene.
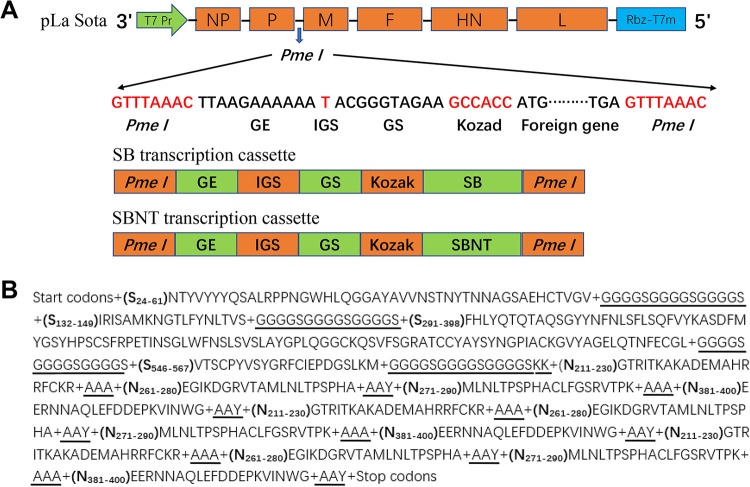
In the present study, four IBV-neutralizing epitope domains (S24–61, S132–149, S291–398, and S546–567) identified previously in the S protein (22, 23) and four IBV T-cell epitope peptides (N211–230, N261–280, N271–290, and N381–400) identified in the N protein by our current study were selected for use in the design of two IBV multi-epitope gene expression boxes (Fig. 9A). One contained only the neutralizing epitope domains and was designated as “SB expression box,” and the other comprised both neutralizing epitope domains and T-cell epitope peptides and was named “SBNT expression box”; the adjacent epitope fragments in each expression box were joined together by proper linkers, namely, AAA, AAY, KK, and (G4S)3 (Fig. 9B). The corresponding nucleotide sequences of the SB and SBNT expression box amino acid residues were used for chicken codon optimization. The open reading frames of SB and SBNT were each preceded by the NDV gene-end (GE), intergenic sequence (IGS), and gene-start (GS); Kozak sequences were added upstream of the start codon (ATG) for efficient translation, and extra nucleotides were placed downstream of the stop codon (TGA) to maintain the rule of six. The transcription cassettes of the codon-optimized SB and SBNT genes were flanked by PmeI sites and synthesized by GenScript. Both the SB and SBNT transcription cassettes were inserted individually between the P and M genes of the La Sota genome by using a seamless assembly cloning kit (Taihe Biotechnology Co., Ltd, Beijing, China) targeting the PmeI site as an independent transcriptional unit; the constructed plasmids, named pLa Sota/SBNT and pLa Sota/SB, respectively, and the sequences of both inserts were confirmed by applying nucleotide sequence analysis. The recombinant La Sota strains containing IBV multi-epitope genes were rescued by reverse genetics as described previously (54) and were designated as rLa Sota/SBNT and rLa Sota/SB, respectively. Rescued recombinant viruses were passaged five times in 9-day-old SPF chicken embryos, and the presence of multi-epitope genes in the recombinant viruses was confirmed by performing RT-PCR and sequencing analysis.
FIG 9.
Design of two recombinant NDV constructs containing an IBV multi-epitope gene. (A) Schematic representation of the approach for generating IBV multi-epitope vaccine candidates via the individual insertion of two different forms of IBV multi-epitope gene (SB and SBNT transcription cassettes) into the NDV strain La Sota antigenome cDNA between the P and M genes using the PmeI site. The open reading frames of the SB and SBNT multi-epitope genes were preceded with NDV gene-end (GE), intergenic sequence (IGS), gene-start (GS), and Kozak sequences and were flanked by PmeI sites. (B) The open reading frame of the SBNT expression box contains four neutralizing epitope domains (S24–61, S132–149, S291–398, and S546–567) and four T-cell epitope peptides (N211–230, N261–280, N271–290, and N381–400); adjacent epitope fragments are joined together by proper linkers (underlined); and the T-cell epitope peptides are each repeated three times.
In vitro characterization of recombinant viruses.
To compare the multicycle growth kinetics of the recombinant viruses rLa Sota/SBNT and rLa Sota/SB with that of the parental virus rLa Sota, DF-1 cells in 6-well plates were infected with rLa Sota/SBNT, rLa Sota/SB, or rLa Sota at a multiplicity of infection (MOI) of 0.01. After 1 h of adsorption, the cells were washed twice with PBS and then incubated with DMEM containing 2% FBS and 5 µg/ml tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK)-treated trypsin at 37°C under 5% CO2. Supernatant samples (200 µl/well) were collected from the infected cells at 12-h intervals until 72 h postinfection (hpi) and replaced each time with an equal volume of fresh DMEM with 2% FBS and 5 µg/ml TPCK-treated trypsin. Virus titers in the collected samples were quantified by performing a 50% tissue culture infective dose (TCID50) titration assay in DF-1 cells and calculated by applying the Reed and Muench method (55). The virulence of each recombinant virus and parental virus was assessed by applying the mean death time (MDT) test in 10-day-old SPF embryonated chicken eggs as described previously (54).
Expression of IBV multi-epitope proteins by recombinant viruses.
The expression of SB and SBNT multi-epitope proteins was determined by performing Western blot and indirect immunofluorescence analysis (IFA) assays. For Western blotting, DF-1 cells were infected with rLa Sota/SB, rLa Sota/SBNT, or rLa Sota at an MOI of 0.01. At 24 hpi, the cells were harvested and lysed, and the resulting proteins were separated by 10% SDS-PAGE and then transferred to a polyvinylidene fluoride (PVDF) membrane (Amersham Biosciences, Freiburg, Germany). A polyclonal chicken anti-IBV or anti-NDV serum was used to detect the IBV multi-epitope proteins or the NDV HN protein, respectively. After being washed three times with Tris-buffered saline with Tween 20 (TBST) buffer, the membrane was incubated with HRP-conjugated rabbit anti-chicken secondary antibody (Bioss Biotechnology, Beijing, China). The proteins were visualized using a BeyoECL Plus kit (Beyotime Biotechnology).
For the indirect IFA, DF-1 cells were infected with rLa Sota/SB, rLa Sota/SBNT, or rLa Sota at an MOI of 0.01. At 48 hpi, the cells were fixed with methanol and then incubated with polyclonal chicken anti-IBV serum at 37°C for 1 h. After being washed five times with PBS, the cells were incubated with FITC-conjugated goat anti-chicken IgG secondary antibody (Bioss Biotechnology) for 1 h at 37°C. Following another five washes with PBS, the cells were observed under a fluorescence microscope (Nikon, Tokyo, Japan).
Evaluation of immunity and protective efficacy of IBV multi-epitope vaccine candidates.
A total of 82 7-day-old SPF chickens were divided into four groups (groups 1 to 4) of 18 chickens each and one group of 10 chickens (group 5). The chickens in groups 1 to 3 were each vaccinated with 106 EID50/bird of rLa Sota/SBNT, rLa Sota/SB, or rLa Sota, respectively, via the oculonasal route, and the chickens in groups 4 and 5 were each inoculated with PBS via the same route. At 3 weeks postvaccination, serum samples were collected from three chickens per group and used in a virus neutralization assay. Additionally, splenocytes were also isolated from chickens in each group and used in the ELISpot assay as described above.
At 3 weeks postvaccination, all birds in groups 1 to 4 were challenged with 106 EID50/bird of IBV SD strain via the oculonasal route, while the birds in group 5 were inoculated with PBS as an uninfected control. The clinical signs of 10 birds in each group were monitored and scored daily for 14 days. The clinical signs were scored as follows: 0 for normal; 1 for slight shaking, slight nasal discharge, and slight lacrimation; 2 for depression, watery feces, and sneezing or coughing; 3 for heavy depression, heavy nasal discharge, and tracheal rales or mouth breathing; and 4 for death. At 5 and 7 days postchallenge (dpc), tracheal and oropharyngeal swab samples were collected from birds in each group and used for virus shedding detection by RT-qPCR as described previously (8). Also, at 5 and 7 dpc, three chickens from each group were sacrificed, and their tracheas were collected for use in a ciliostasis activity evaluation.
Evaluation of cross‐protective efficacy induced by the IBV multi-epitope vaccine candidates.
To evaluate the cross‐protective efficacy of the IBV multi-epitope vaccine candidates against virulent IBV challenge, 96 one-day-old SPF chickens were divided into six groups of 16 chickens each. Chickens in groups 1, 3, and 5 were vaccinated with 106 EID50/bird of rLa Sota/SBNT via the oculonasal route, and chickens in groups 2, 4, and 6 were similarly vaccinated with 106 EID50/bird of rLa Sota. At 3 weeks postvaccination, the chickens in groups 1 and 2 were challenged with 106 EID50/bird of the IBV SD strain, those in groups 3 and 4 were challenged with 106 EID50/bird of IBV GD strain, and those in groups 5 and 6 were challenged with 106 EID50/bird of the IBV M41 strain, all via the oculonasal route. The clinical signs of birds in each group were scored daily for 14 days as described above. At 5 and 7 dpc, tracheal and oropharyngeal swab samples were collected from birds in each group and used for virus shedding detection by RT-qPCR (8). Also, at 5 and 7 dpc, the tracheas from three chickens per group were used for ciliostasis activity evaluation.
Inhibition of ciliary activity.
To evaluate ciliostasis activity, nine tracheal rings per chicken were prepared (three rings each from the upper, middle, and lower parts of the trachea) and placed in 96-well plates containing DMEM with 10% FBS. The ciliary activity was examined under a low-power microscope, and each ring was scored as follows: 0 for 100% of the tracheal cilia showing movement; 1 for 75% to 100% of the tracheal cilia showing movement; 2 for 50% to 75% of the cilia showing movement; 3 for 25% to 50% of the tracheal cilia showing movement; and 4 for 0% to 25% of the tracheal cilia showing movement. The average ciliostasis score was calculated for each group.
Virus neutralization assay.
Neutralizing antibody from immunized chickens was measured by performing a standard virus neutralization assay. Briefly, serum samples were inactivated in a 56°C water bath for 30 min and then serially diluted 2-fold with sterile PBS. A sample (0.1 ml) of the diluted serum was incubated with an equal volume of IBV SD strain (200 EID50) at 37°C for 1 h. Ten-day-old SPF embryonated chicken eggs were inoculated with the 0.2-ml virus-serum mixtures via the allantoic cavity route. Six days later, the embryonated eggs were examined for IBV lesions, such as embryo dwarfing or stunting. Neutralizing antibody titers were calculated as described previously (55) and are expressed here as the mean ± standard deviation.
Statistical analysis.
GraphPad Prism version 6.0 was used for statistical analysis. Statistically significant differences between various groups were evaluated by performing an unpaired t test, one-way analysis of variance (ANOVA), or two-way ANOVA. Data are expressed as the mean ± standard deviation. Statistical significance is indicated in the figures as follows: *, P < 0.05; **, P < 0.01; and ***, P < 0.001.
ACKNOWLEDGMENTS
This work was supported by grant no. 2017YFD0500700 from the National Key Research and Development Program of China.
We thank Katie Oakley from Liwen Bianji, Edanz Editing China (https://www.liwenbianji.cn/ac), for editing the English text of a draft of the manuscript.
Contributor Information
Guozhong Zhang, Email: zhanggz@cau.edu.cn.
Tom Gallagher, Loyola University Chicago.
REFERENCES
- 1.Legnardi M, Tucciarone CM, Franzo G, Cecchinato M. 2020. Infectious bronchitis virus evolution, diagnosis and control. Vet Sci 7:79. doi: 10.3390/vetsci7020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belouzard S, Millet JK, Licitra BN, Whittaker GR. 2012. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses 4:1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu D, Han Z, Xu J, Shao Y, Li H, Kong X, Liu S. 2010. A novel B-cell epitope of avian infectious bronchitis virus N protein. Viral Immunol 23:189–199. doi: 10.1089/vim.2009.0094. [DOI] [PubMed] [Google Scholar]
- 4.Tang M, Wang H, Zhou S, Tian G. 2008. Enhancement of the immunogenicity of an infectious bronchitis virus DNA vaccine by a bicistronic plasmid encoding nucleocapsid protein and interleukin-2. J Virol Methods 149:42–48. doi: 10.1016/j.jviromet.2008.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seo SH, Wang L, Smith R, Collisson EW. 1997. The carboxyl-terminal 120-residue polypeptide of infectious bronchitis virus nucleocapsid induces cytotoxic T lymphocytes and protects chickens from acute infection. J Virol 71:7889–7894. doi: 10.1128/JVI.71.10.7889-7894.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao Y, Zhang H, Zhao J, Zhong Q, Jin JH, Zhang GZ. 2016. Evolution of infectious bronchitis virus in China over the past two decades. J Gen Virol 97:1566–1574. doi: 10.1099/jgv.0.000464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jordan B. 2017. Vaccination against infectious bronchitis virus: a continuous challenge. Vet Microbiol 206:137–143. doi: 10.1016/j.vetmic.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 8.Xu G, Cheng J, Ma S, Jia W, Yan S, Zhang G. 2018. Pathogenicity differences between a newly emerged TW-like strain and a prevalent QX-like strain of infectious bronchitis virus. Vet Microbiol 227:20–28. doi: 10.1016/j.vetmic.2018.10.019. [DOI] [PubMed] [Google Scholar]
- 9.Rohaim MA, El Naggar RF, Hamoud MM, Bazid AI, Gamal AM, Laban SE, Abdel-Sabour MA, Nasr SAE, Zaki MM, Shabbir MZ, Zahran OK, Munir M. 2019. Emergence and genetic analysis of variant pathogenic 4/91 (serotype 793/B) infectious bronchitis virus in Egypt during 2019. Virus Genes 55:720–725. doi: 10.1007/s11262-019-01693-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ji J, Gao Y, Chen Q, Wu Q, Xu X, Kan Y, Yao L, Bi Y, Xie Q. 2020. Epidemiological investigation of avian infectious bronchitis and locally determined genotype diversity in central China: a 2016–2018 study. Poult Sci 99:3001–3008. doi: 10.1016/j.psj.2020.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bande F, Arshad SS, Bejo MH, Moeini H, Omar AR. 2015. Progress and challenges toward the development of vaccines against avian infectious bronchitis. J Immunol Res 2015:424860. doi: 10.1155/2015/424860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mishra S. 2020. Designing of cytotoxic and helper T cell epitope map provides insights into the highly contagious nature of the pandemic novel coronavirus SARS-CoV-2. R Soc Open Sci 7:201141. doi: 10.1098/rsos.201141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collisson EW, Pei J, Dzielawa J, Seo SH. 2000. Cytotoxic T lymphocytes are critical in the control of infectious bronchitis virus in poultry. Dev Comp Immunol 24:187–200. doi: 10.1016/s0145-305x(99)00072-5. [DOI] [PubMed] [Google Scholar]
- 14.Auladell M, Jia X, Hensen L, Chua B, Fox A, Nguyen THO, Doherty PC, Kedzierska K. 2019. Recalling the future: immunological memory toward unpredictable influenza viruses. Front Immunol 10:1400. doi: 10.3389/fimmu.2019.01400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murakoshi H, Kuse N, Akahoshi T, Zhang Y, Chikata T, Borghan MA, Gatanaga H, Oka S, Sakai K, Takiguchi M. 2018. Broad recognition of circulating HIV-1 by HIV-1-specific cytotoxic T-lymphocytes with strong ability to suppress HIV-1 replication. J Virol 93:e01480-18. doi: 10.1128/JVI.01480-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li X, Zhang L, Liu Y, Ma L, Zhang N, Xia C. 2020. Structures of the MHC-I molecule BF2*1501 disclose the preferred presentation of an H5N1 virus-derived epitope. J Biol Chem 295:5292–5306. doi: 10.1074/jbc.RA120.012713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chávez-Galán L, Arenas-Del Angel MC, Zenteno E, Chávez R, Lascurain R. 2009. Cell death mechanisms induced by cytotoxic lymphocytes. Cell Mol Immunol 6:15–25. doi: 10.1038/cmi.2009.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lettau M, Kabelitz D, Janssen O. 2015. Lysosome-related effector vesicles in T lymphocytes and NK cells. Scand J Immunol 82:235–243. doi: 10.1111/sji.12337. [DOI] [PubMed] [Google Scholar]
- 19.Glatzová D, Cebecauer M. 2019. Dual role of CD4 in peripheral T lymphocytes. Front Immunol 10:618. doi: 10.3389/fimmu.2019.00618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang L, Li X, Ma L, Zhang B, Meng G, Xia C. 2020. A newly recognized pairing mechanism of the α- and β-chains of the chicken peptide–MHC class II complex. J Immunol 204:1630–1640. doi: 10.4049/jimmunol.1901305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ignjatovic J, Galli L. 1994. The S1 glycoprotein but not the N or M proteins of avian infectious bronchitis virus induces protection in vaccinated chickens. Arch Virol 138:117–134. doi: 10.1007/BF01310043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kant A, Koch G, van Roozelaar DJ, Kusters JG, Poelwijk FA, van der Zeijst BA. 1992. Location of antigenic sites defined by neutralizing monoclonal antibodies on the S1 avian infectious bronchitis virus glycopolypeptide. J Gen Virol 73:591–596. doi: 10.1099/0022-1317-73-3-591. [DOI] [PubMed] [Google Scholar]
- 23.Lenstra JA, Kusters JG, Koch G, van der Zeijst BA. 1989. Antigenicity of the peplomer protein of infectious bronchitis virus. Mol Immunol 26:7–15. doi: 10.1016/0161-5890(89)90014-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Toro H, Zhao W, Breedlove C, Zhang Z, van Santen V, Yu Q. 2013. Infectious bronchitis virus S2 expressed from recombinant virus confers broad protection against challenge. Avian Dis 58:83–89. doi: 10.1637/10641-081613-Reg.1. [DOI] [PubMed] [Google Scholar]
- 25.Bande F, Arshad SS, Omar AR, Hair-Bejo M, Mahmuda A, Nair V. 2017. Global distributions and strain diversity of avian infectious bronchitis virus: a review. Anim Health Res Rev 18:70–83. doi: 10.1017/S1466252317000044. [DOI] [PubMed] [Google Scholar]
- 26.Chen Y, Jiang L, Zhao W, Liu L, Zhao Y, Shao Y, Li H, Han Z, Liu S. 2017. Identification and molecular characterization of a novel serotype infectious bronchitis virus (GI-28) in China. Vet Microbiol 198:108–115. doi: 10.1016/j.vetmic.2016.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu L, Han Z, Jiang L, Sun J, Zhao Y, Liu S. 2018. Genetic diversity of avian infectious bronchitis virus in China in recent years. Infect Genet Evol 66:82–94. doi: 10.1016/j.meegid.2018.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Al-Ebshahy E, Abdel-Sabour M, Abas O, Yanai T. 2019. Protection conferred by a vaccine derived from an inactivated Egyptian variant of infectious bronchitis virus: a challenge experiment. Trop Anim Health Prod 51:1997–2001. doi: 10.1007/s11250-019-01898-y. [DOI] [PubMed] [Google Scholar]
- 29.Pei J, Briles WE, Collisson EW. 2003. Memory T cells protect chicks from acute infectious bronchitis virus infection. Virology 306:376–384. doi: 10.1016/S0042-6822(02)00059-4. [DOI] [PubMed] [Google Scholar]
- 30.Wang L, Parr RL, King DJ, Collisson EW. 1995. A highly conserved epitope on the spike protein of infectious bronchitis virus. Arch Virol 140:2201–2213. doi: 10.1007/BF01323240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ignjatovic J, Sapats S. 2005. Identification of previously unknown antigenic epitopes on the S and N proteins of avian infectious bronchitis virus. Arch Virol 150:1813–1831. doi: 10.1007/s00705-005-0541-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Andoh K, Ashikaga K, Suenaga K, Endo S, Yamazaki K. 2018. Identification of novel linear epitopes located in the infectious bronchitis virus spike S2 region. Avian Dis 62:210–217. doi: 10.1637/11796-011518-Reg.1. [DOI] [PubMed] [Google Scholar]
- 33.Tan L, Liao Y, Fan J, Zhang Y, Mao X, Sun Y, Song C, Qiu X, Meng C, Ding C. 2016. Prediction and identification of novel IBV S1 protein derived CTL epitopes in chicken. Vaccine 34:380–386. doi: 10.1016/j.vaccine.2015.11.042. [DOI] [PubMed] [Google Scholar]
- 34.Boots AM, Kusters JG, van Noort JM, Zwaagstra KA, Rijke E, van der Zeijst BA, Hensen EJ. 1991. Localization of a T-cell epitope within the nucleocapsid protein of avian coronavirus. Immunology 74:8–13. [PMC free article] [PubMed] [Google Scholar]
- 35.Lacerda-Queiroz N, Riteau N, Eastman RT, Bock KW, Orandle MS, Moore IN, Sher A, Long CA, Jankovic D, Su X-Z. 2017. Mechanism of splenic cell death and host mortality in a Plasmodium yoelii malaria model. Sci Rep 7:10438. doi: 10.1038/s41598-017-10776-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bieber K, Sun S, Witte M, Kasprick A, Beltsiou F, Behnen M, Laskay T, Schulze FS, Pipi E, Reichhelm N, Pagel R, Zillikens D, Schmidt E, Sparwasser T, Kalies K, Ludwig RJ. 2017. Regulatory T cells suppress inflammation and blistering in pemphigoid diseases. Front Immunol 8:1628. doi: 10.3389/fimmu.2017.01628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Billiau A, Matthys P. 2009. Interferon-gamma: a historical perspective. Cytokine Growth Factor Rev 20:97–113. doi: 10.1016/j.cytogfr.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 38.Göbel TW, Kaspers B, Stangassinger M. 2001. NK and T cells constitute two major, functionally distinct intestinal epithelial lymphocyte subsets in the chicken. Int Immunol 13:757–762. doi: 10.1093/intimm/13.6.757. [DOI] [PubMed] [Google Scholar]
- 39.Yan S, Liu X, Zhao J, Xu G, Zhao Y, Zhang G. 2017. Analysis of antigenicity and pathogenicity reveals major differences among QX-like infectious bronchitis viruses and other serotypes. Vet Microbiol 203:167–173. doi: 10.1016/j.vetmic.2017.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zander R, Schauder D, Xin G, Nguyen C, Wu X, Zajac A, Cui W. 2019. CD4+ T cell help is required for the formation of a cytolytic CD8+ T cell subset that protects against chronic infection and cancer. Immunity 51:1028–1042. doi: 10.1016/j.immuni.2019.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zwaveling S, Ferreira Mota SC, Nouta J, Johnson M, Lipford GB, Offringa R, van der Burg SH, Melief CJ. 2002. Established human papillomavirus type 16-expressing tumors are effectively eradicated following vaccination with long peptides. J Immunol 169:350–358. doi: 10.4049/jimmunol.169.1.350. [DOI] [PubMed] [Google Scholar]
- 42.Tan L, Zhang Y, Liu F, Yuan Y, Zhan Y, Sun Y, Qiu X, Meng C, Song C, Ding C. 2016. Infectious bronchitis virus poly-epitope-based vaccine protects chickens from acute infection. Vaccine 34:5209–5216. doi: 10.1016/j.vaccine.2016.09.022. [DOI] [PubMed] [Google Scholar]
- 43.Yang T, Wang HN, Wang X, Tang JN, Lu D, Zhang YF, Guo ZC, Li YL, Gao R, Kang RM. 2009. The protective immune response against infectious bronchitis virus induced by multi-epitope based peptide vaccines. Biosci Biotechnol Biochem 73:1500–1504. doi: 10.1271/bbb.80864. [DOI] [PubMed] [Google Scholar]
- 44.Bijker MS, van den Eeden SJ, Franken KL, Melief CJ, Offringa R, van der Burg SH. 2007. CD8+ CTL priming by exact peptide epitopes in incomplete Freund's adjuvant induces a vanishing CTL response, whereas long peptides induce sustained CTL reactivity. J Immunol 179:5033–5040. doi: 10.4049/jimmunol.179.8.5033. [DOI] [PubMed] [Google Scholar]
- 45.Rosendahl Huber SK, Camps MG, Jacobi RH, Mouthaan J, van Dijken H, van Beek J, Ossendorp F, de Jonge J. 2015. Synthetic long peptide influenza vaccine containing conserved T and B cell epitopes reduces viral load in lungs of mice and ferrets. PLoS One 10:e0127969. doi: 10.1371/journal.pone.0127969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun W, Leist SR, McCroskery S, Liu Y, Slamanig S, Oliva J, Amanat F, Schäfer A, DinnonKH, III, García-Sastre A, Krammer F, Baric RS, Palese P. 2020. Newcastle disease virus (NDV) expressing the spike protein of SARS-CoV-2 as a live virus vaccine candidate. EBioMedicine 62:103132. doi: 10.1016/j.ebiom.2020.103132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abozeid HH, Paldurai A, Varghese BP, Khattar SK, Afifi MA, Zouelfakkar S, El-Deeb AH, El-Kady MF, Samal SK. 2019. Development of a recombinant Newcastle disease virus-vectored vaccine for infectious bronchitis virus variant strains circulating in Egypt. Vet Res 50:12. doi: 10.1186/s13567-019-0631-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chhabra R, Chantrey J, Ganapathy K. 2015. Immune responses to virulent and vaccine strains of infectious bronchitis viruses in chickens. Viral Immunol 28:478–488. doi: 10.1089/vim.2015.0027. [DOI] [PubMed] [Google Scholar]
- 49.Raj GD, Jones RC. 1997. Infectious bronchitis virus: immunopathogenesis of infection in the chicken. Avian Pathol 26:677–706. doi: 10.1080/03079459708419246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kotani T, Wada S, Tsukamoto Y, Kuwamura M, Yamate J, Sakuma S. 2000. Kinetics of lymphocytic subsets in chicken tracheal lesions infected with infectious bronchitis virus. J Vet Med Sci 62:397–401. doi: 10.1292/jvms.62.397. [DOI] [PubMed] [Google Scholar]
- 51.Xu G, Liu XY, Zhao Y, Chen Y, Zhao J, Zhang GZ. 2016. Characterization and analysis of an infectious bronchitis virus strain isolated from southern China in 2013. Virol J 13:40. doi: 10.1186/s12985-016-0497-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiang L, Han Z, Chen Y, Zhao W, Sun J, Zhao Y, Liu S. 2018. Characterization of the complete genome, antigenicity, pathogenicity, tissue tropism, and shedding of a recombinant avian infectious bronchitis virus with a ck/CH/LJL/140901-like backbone and an S2 fragment from a 4/91-like virus. Virus Res 244:99–109. doi: 10.1016/j.virusres.2017.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cheng J, Huo C, Zhao J, Liu T, Li X, Yan S, Wang Z, Hu Y, Zhang G. 2018. Pathogenicity differences between QX-like and Mass-type infectious bronchitis viruses. Vet Microbiol 213:129–135. doi: 10.1016/j.vetmic.2017.11.027. [DOI] [PubMed] [Google Scholar]
- 54.Yu XH, Cheng JL, Xue J, Jin JH, Song Y, Zhao J, Zhang GZ. 2017. Roles of the polymerase-associated protein genes in Newcastle disease virus virulence. Front Microbiol 8:161. doi: 10.3389/fmicb.2017.00161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reed LJ, Muench H. 1938. A simple method of estimating fifty per cent endpoints. Am J Epidemiol 27:493–497. doi: 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]
- 56.Ariaans MP, van de Haar PM, Lowenthal JW, van Eden W, Hensen EJ, Vervelde L. 2008. ELISPOT and intracellular cytokine staining: novel assays for quantifying T cell responses in the chicken. Dev Comp Immunol 32:1398–1404. doi: 10.1016/j.dci.2008.05.007. [DOI] [PubMed] [Google Scholar]