ABSTRACT
Nervous necrosis virus (NNV) belongs to the Betanodavirus genus of the Nodaviridae family and is the main cause of viral nervous necrosis disease in marine fish larvae and juveniles worldwide. The NNV virion contains two positive-sense, single-stranded RNA genomes, which encode RNA-dependent RNA polymerase, coat protein, and B2 protein. Interestingly, NNV infection can shut off host translation in orange-spotted grouper (Epinephelus coioides) brain cells; however, the detailed mechanisms of this action remain unknown. In this study, we discovered that the host translation factor, polyadenylate binding protein (PABP), is a key target during NNV takeover of host translation machinery. Additionally, ectopic expression of NNV coat protein is sufficient to trigger nuclear translocalization and degradation of PABP, followed by translation shutoff. A direct interaction between NNV coat protein and PABP was demonstrated, and this binding requires the NNV coat protein N-terminal shell domain and PABP proline-rich linker region. Notably, we also showed that degradation of PABP during later stages of infection is mediated by the ubiquitin-proteasome pathway. Thus, our study reveals that the NNV coat protein hijacks host PABP, causing its relocalization to the nucleus and promoting its degradation to stimulate host translation shutoff.
IMPORTANCE Globally, more than 200 species of aquacultured and wild marine fish are susceptible to NNV infection. Devastating outbreaks of this virus have been responsible for massive economic damage in the aquaculture industry, but the molecular mechanisms by which NNV affects its host remain largely unclear. In this study, we show that NNV hijacks translation in host brain cells, with the viral coat protein binding to host PABP to promote its nuclear translocalization and degradation. This previously unknown mechanism of NNV-induced host translation shutoff greatly enhances the understanding of NNV pathogenesis and provides useful insights and novel tools for development of NNV treatments, such as the use of orange-spotted grouper brain cells as an in vitro model system.
KEYWORDS: nervous necrosis virus, coat protein, polyadenylate binding protein, translation shutoff, nuclear translocalization
INTRODUCTION
Viral nervous necrosis (1), otherwise known as viral encephalopathy and retinopathy (2), is an infectious neuropathological disease that affects more than 200 species of farmed and wild marine fish worldwide, with nearly 100 % mortality observed in affected larvae and juvenile fish (3). The disease is caused by infection with nervous necrosis virus (NNV), which belongs to the genus Betanodavirus of the family Nodaviridae. NNV has a small (about 25 nm), nonenveloped icosahedral structure and contains a bipartite, linear, positive-sense, single-stranded RNA genome composed of RNA1 and RNA2. These RNAs encode an RNA-dependent RNA polymerase (RdRp) and a coat protein, respectively (4). In addition, a subgenomic transcript of RNA1, referred to as RNA3, encodes protein B2, which is known to antagonize host RNA interference by binding to double-stranded RNA (dsRNA) during virus multiplication (5, 6). Importantly, the NNV RNAs each contain a cap structure at the 5′ end, but they lack a poly(A) tail at the 3′ end (5, 7).
Translation is an extremely complicated process in which many cellular translation factors are involved. To date, no viruses have been shown to encode a battery of genes sufficient to translate their own viral mRNA and carry out protein production. Thus, all known viruses depend on the host translation machinery to produce viral proteins. Upon infection, viruses must compete for translation resources with ongoing host protein production, and many viruses have evolved strategies to improve viral multiplication by shutting off translation of host proteins. Canonical translation of host proteins involves polyadenylate binding protein (PABP) acting as a bridge between the 3′-poly(A) tail and eIF4G of the eIF4F translation initiation complex on the 5′-untranslated region. This bridging function allows the host mRNA to form a cap-to-tail circularized structure, which tremendously enhances translation efficiency. Following the release of a newly synthesized peptide chain, a ribosome reading circularized mRNA will readily encounter the nearby start codon site to initiate another round of translation (8–12). Therefore, PABP is a prime target for viruses to shut off host translation. The 70 kDa PABP protein is highly conserved across species and contains four RNA recognition motifs (RRMs) in its N terminus, one proline-rich linker region, and one C-terminal globular domain, an ancient arrangement that arose before the divergence of yeast and animal eukaryota (13). Notably, PABP plays several roles in mRNA biogenesis and turnover as a multifunctional scaffold for the posttranscriptional control of gene expression. The ability of PABP to bind the poly(A) tract requires the N-terminal RRMs, whereas the C-terminal globular domain contributes to interactions with factors regulating mRNA polyadenylation, mRNA export from the nucleus, mRNA stabilization, translation initiation, and translation termination (14).
PABP is a common viral target, and many viruses carry proteolytic enzymes to cleave it. Such proteases include 2A protease of coxsackievirus (15); 2A and 3C proteases of poliovirus (16, 17); 3C-like proteinase of calicivirus (18); proteases of human immunodeficiency virus type 1 (HIV-1), HIV-2, and mouse mammary tumor virus (19, 20); Lb protease of foot-and-mouth disease virus (21); 3C proteinase of hepatitis A virus (22); 3C proteinase of encephalomyocarditis virus (23); and 3C protease of duck hepatitis A virus (24). Moreover, PABP is a nucleo-cytoplasmic protein that shuttles between the nucleus and the cytoplasm (25). Although PABP normally shows exclusive cytoplasmic localization, several cellular conditions may lead to its accumulation in the nucleus. Such nuclear accumulation of PABP is often associated with translation shutoff. Some viruses are known to stimulate translocation of PABP from cytoplasm to the nucleus through viral protein-PABP interactions, thereby attenuating the PABP bridge function and shutting off host canonical translation. Many identified viral proteins, such as K10/10.1 protein of Kaposi’s sarcoma-associated herpesvirus (26), nucleocapsid (N) protein of Bunyamwera virus (27), and ICP27 and UL47 proteins of herpes simplex virus (28), can bind to PABP and cause it to relocalize to the nucleus. In addition, the expression of NSs protein from Rift Valley fever virus was shown to promote accumulation of host mRNA and PABP in the nuclei of infected cells, leading to host translation shutoff (29, 30). When PABP is sequestered inside the nucleus, its binding to newly synthesized poly(A) regions of unspliced pre-mRNA further diminishes cytoplasmic mRNA (31). Interestingly, binding of the capsid protein of rubella virus to PABP not only sequesters the protein in the cytoplasm but also inhibits protein synthesis (32). Moreover, other viral proteins have been shown to interact with host translation initiation factors to indirectly evict PABP from the eIF4F initiation complex. For instance, the NSP3 protein of rotavirus interacts with cellular eIF4G and RoXaN proteins that functionally replace and evict PABP from the eIF4F initiation complex, leading to nuclear relocalization of PABP (33, 34). In another example, cytoplasmic PABP nuclear relocalization may be caused by the SOX protein of Kaposi’s sarcoma-associated herpesvirus (35). In this context, host translation shutoff is mediated by SOX promoting the degradation of cellular mRNA, which dramatically reduces mRNA accumulation in the cytoplasm and leads to PABP accumulation in the nucleus (36).
In this study, we found that host translation is shut off after NNV infection. NNV RNAs do not have a 3′-poly(A) tail (5, 7), a structure that is critical for host mRNA stability and efficient canonical translation. Therefore, we suspected that NNV may take this advantage of its lack of a poly(A) tail to hijack host translation machinery by disrupting PABP function. We found that in NNV-infected cells, an interaction between NNV coat protein and cellular PABP coincides with PABP sequestration in the nucleus and PABP degradation by the ubiquitination-proteasome pathway in the late infection stage; these actions are putatively responsible for shutting off host translation.
RESULTS
NNV infection inhibits host translation.
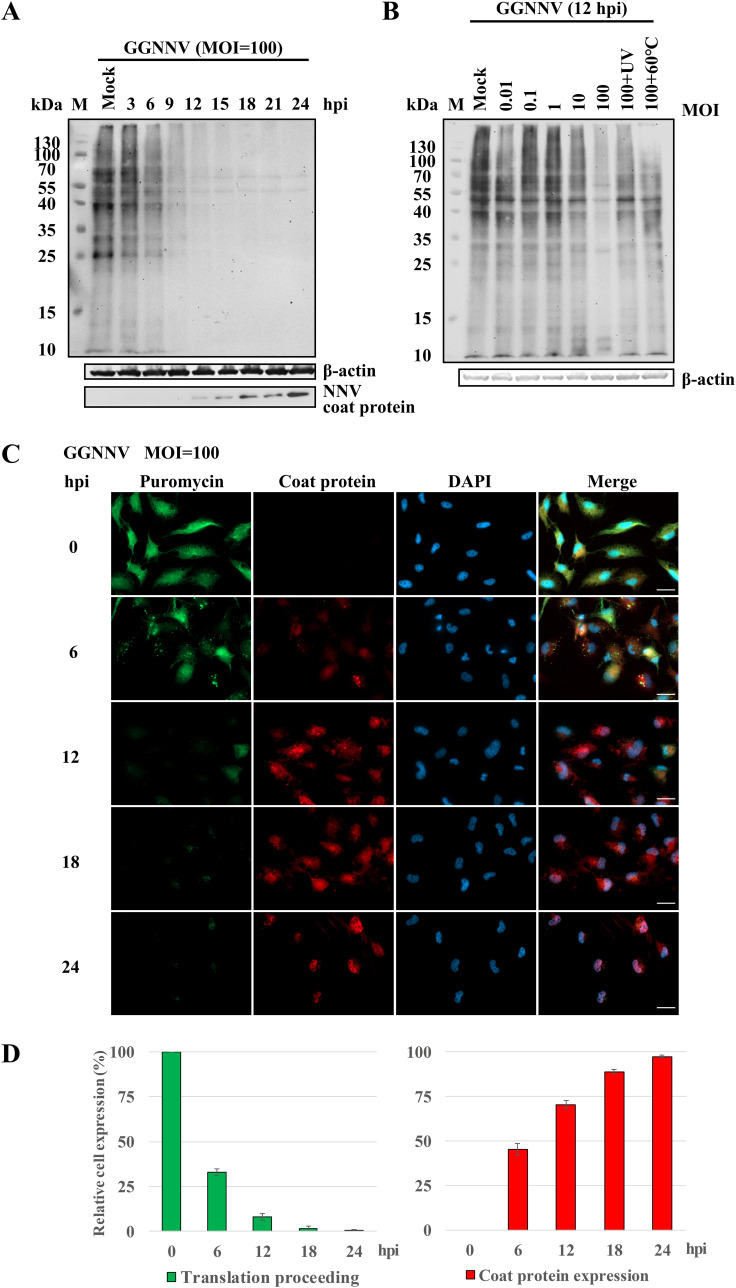
To test whether NNV infection causes host translation shutoff, we first monitored new protein synthesis with a nonradioactive method, surface sensing of translation (SUnSET) (37), in giant grouper nervous necrosis virus (GGNNV)-infected orange-spotted grouper brain (GB) cells. The aminoacyl tRNA analog, puromycin, was incorporated into newly synthesized proteins in GGNNV-infected GB cells, following puromycin detection by immunoblot. As shown in Fig. 1, the GGNNV-infected GB cells exhibited time-dependent host translation shutoff, whereas the mock-infected cells showed continued translation. The host translation was gradually shut off from 3 to 24 h postinfection (hpi) at a dose of 100 multiplicity of infection (MOI). Obvious shutdown of host protein synthesis was observed at 6 hpi, and translation appeared to be completely shut off at 12 hpi (Fig. 1A, upper panel). Meanwhile, viral coat protein gradually accumulated in the cells from 6 to 24 hpi (Fig. 1A, lower panel). In addition, the reduction of protein translation in GGNNV-infected cells was also found to be dose dependent. Notably, infection with UV-inactivated or heat-inactivated GGNNV did not produce an obvious translation shutoff effect (Fig. 1B), implying that host translation shutoff may require viral multiplication. Our immunocytochemical staining showed results similar to the Western blots, but the immunocytochemistry also revealed the subcellular location of neosynthesized proteins and viral coat protein. After 6 hpi, a marked reduction of puromycin-labeled newly synthesized proteins was observed. At the same time points, viral coat protein could be detected in the same cells simultaneously. Thus, the presence of newly synthesized proteins was reciprocally correlated with the appearance of viral coat protein (Fig. 1C). The reciprocal correlation of puromycin signal and NNV coat protein expression was even more clear when quantified and plotted (Fig. 1D). Taken together, these results demonstrate that GGNNV infection inhibits host translation.
FIG 1.
NNV infection inhibits host translation. Western blot and immunocytochemical staining of puromycin incorporation were performed to evaluate the influence of NNV infection on host translation activity. Puromycin mimics aminoacyl tRNA and is incorporated into newly synthesized proteins during translation. Thus, it can serve as an indicator of translation. Host translation shutoff was observed with the puromycin-label (SUnSET) method after giant grouper nervous necrosis virus (GGNNV) infection in grouper brain (GB) cells. (A and B) GB cells were infected with GGNNV (multiplicity of infection [MOI] = 100) for different time periods (A) or for 12 h with different dosages of GGNNV (B). Puromycin (20 μg/ml) was added to the medium, and the cells were incubated at 28°C for 1 h. As a control, GGNNV was treated with heat (60°C, 1 h) or UV (0.24 J/cm2). The cells were lysed, and lysates were used for SDS-PAGE and immunoblotting analysis. The puromycin incorporated into newly synthesized proteins was detected using an anti-puromycin antibody. A β-actin immunoblot is shown as a loading control. A monoclonal antibody (RG-M18) against NNV coat protein was used to detect the GGNNV level (56). (C) Newly synthesized proteins in GGNNV-infected GB cells were detected by immunocytochemistry. GB cells were infected with GGNNV at an MOI of 100. At 0, 6, 12, 18, and 24 h postinfection (hpi), cells were harvested for immunocytochemistry. Puromycin was added to the medium of infected cells for 1 h prior to harvest. The GGNNV-infected GB cells were washed twice with PBS and then fixed in 4% paraformaldehyde, permeabilized in 0.25% Triton X-100, and processed for immunocytochemistry. Mouse monoclonal antibody RG-M18 was used to detect GGNNV coat protein (red). Newly synthesized proteins with incorporated puromycin were detected with a mouse anti-puromycin Alexa Fluor 488-conjugated monoclonal antibody (green). Nuclei were stained with DAPI (blue). Bar = 20 μm. (D) Statistical analysis of the relative cell expression levels in puromycin-labeling (green) and NNV coat protein expression (red) cells of the immunocytochemical experiment. The means ± standard deviation from three independent experiments are plotted. M, protein molecular weight marker.
Orange-spotted grouper PABP sequence is highly conserved.
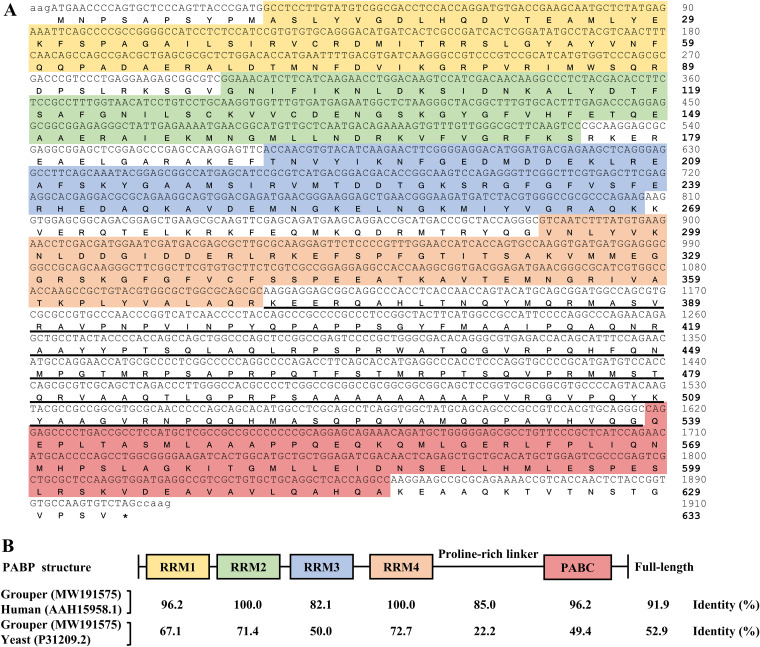
PABP acts as a key factor for connecting closed-loop mRNA between the 3′-poly(A) tail and 5′-untranslated end. Thus, PABP is necessary for efficient canonical translation of host proteins. Since all NNV RNA genomes lack a 3′-poly(A) tail and PABP is targeted by other viruses, we considered cellular PABP to be the most likely target for NNV to shut off host translation. To obtain the orange-spotted grouper PABP gene sequence, we used reverse transcription-PCR (RT-PCR) to clone the complete open reading frame of PABP from mRNA isolated from GB cells. The primer sequences used to amplify the PABP gene were designed to target the 5′- and 3′-untranslated regions derived from our in-house next-generation sequencing (NGS) database of the GB cell transcriptome. The orange-spotted grouper PABP gene includes an open reading frame that encodes a 633-amino-acid protein with a molecular weight of 69,360 Da. The putative protein sequence consists of four RNA recognition motifs (RRMs) at the N terminus, one proline-rich linker, and one C-terminal helical domain (PABC) (Fig. 2A). The comparison of orange-spotted grouper PABP with that of human and yeast showed overall sequence identities of 91.9 % and 52.9 %, respectively. Compared with the human sequence, grouper PABP RRM2 and RRM4 domains showed complete sequence identity (100 %), while RRM1 and PABC both showed 96.2 % identity, RRM3 was 82.1 % identical, and the proline-rich linker showed 85.0 % identity. Similar trends in homology were also found between the PABP sequences for grouper and yeast (Fig. 2B).
FIG 2.
Features of the grouper PABP protein. (A) The nucleotide and deduced amino acid sequences of the orange-spotted grouper (Epinephelus coioides) polyadenylate binding protein (PABP) gene were obtained from RT-PCR and next-generation sequencing (NGS) of grouper brain (GB) cells. Lowercase, untranslated nucleotides; uppercase, open reading frame nucleotides; bold uppercase, amino acid; yellow shading, RNA recognition motif 1 (RRM1); green shading, RRM2; blue shading, RRM3; orange shading, RRM4; red shading, C-terminal helical domain (PABC); underlined amino acids, proline-rich linker; asterisk, stop codon. (B) Schematic representation of the PABP domain structure and the amino acid sequence comparison among grouper, human, and yeast. Accession numbers for each PABP sequence are given in parentheses.
Translation shutoff by NNV infection coincides with PABP nuclear relocalization and NNV coat protein expression.
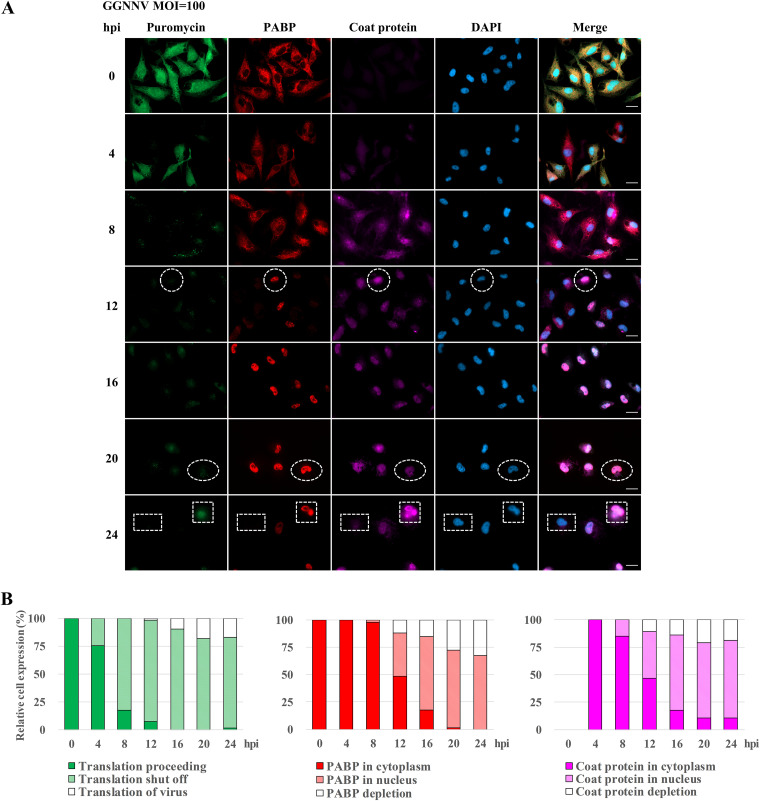
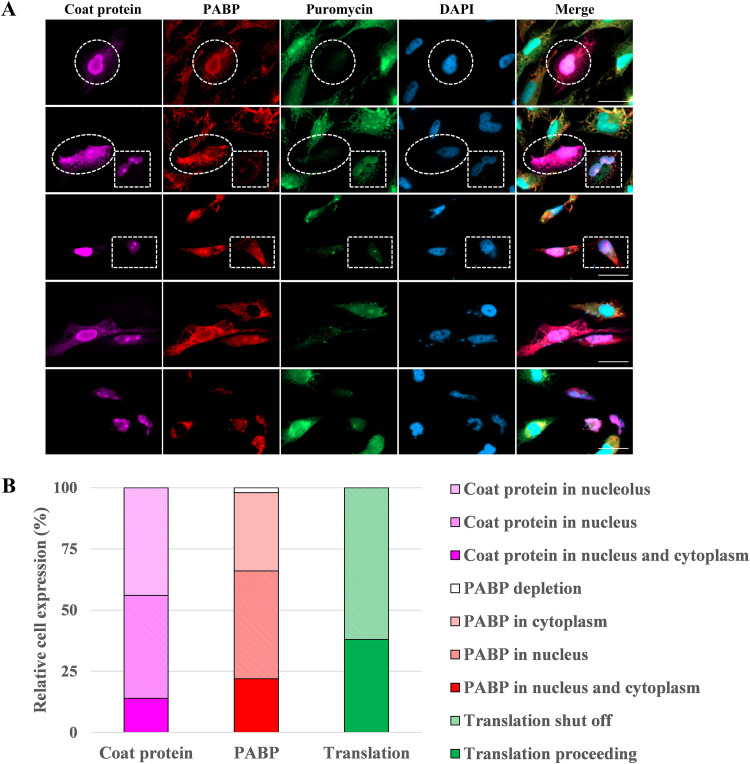
To determine whether cellular PABP participates in NNV-induced host translation shutoff, we utilized an assay based on immunocytochemical staining. To facilitate detection, we purchased an anti-PABP antibody (abcam; ab21060) that recognizes the PABC domain of human PABP; as previously noted, this domain is highly conserved in grouper PABP. Our staining and Western blot results suggested that the antibody also specifically recognizes grouper PABP without appreciable nonspecific binding. As PABP is an important factor for canonical protein translation, the appearance of newly synthesized proteins was closely correlated with the appearance of PABP within the cytoplasm of GB cells before infection. However, the staining revealed several interesting phenomena in infected cells. At the early infection stages, NNV coat protein was expressed mainly in the cytoplasm of infected GB cells, and the distribution of PABP was shifted toward the nucleus, while host translation gradually shut off (Fig. 3A, 4–8 hpi). Later, some cells had obvious nuclear colocalization of PABP and NNV coat protein accompanied by translation shutoff (Fig. 3A, 12–24 hpi, e.g., white circle). Other cells at late time points exhibited prominent nuclear redistribution of PABP in concert with both translation shutoff and NNV coat protein depletion (Fig. 3A, 12–24 hpi, e.g., white oval). The NNV coat protein depletion may have resulted from viral protein degradation or viral particle release from infected cells. Among cells receiving high MOI (100) at a late infection stage (24 hpi), about half were rounded up and detached from the surface of the culture plate. At this stage, some infected and attached cells with PABP nuclear localization and NNV coat protein in both the nucleus and the cytoplasm could be observed. The correlation between cytoplasmic distribution of translation signals and NNV coat protein suggests that the puromycin labeling likely indicates newly synthesized viral coat protein (Fig. 3A, 24 hpi, e.g., white square). In the same stage, some infected cells were found without PABP signal, without puromycin label, and without NNV coat protein (Fig. 3A, 24 hpi, e.g., rectangle). The close relationships among translation shutoff, PABP distribution and NNV coat protein expression were clearly apparent when relative cell expression levels and subcellular distribution were quantified in infected cells (Fig. 3B). These results reveal a close correlation among translation shutoff, nuclear redistribution, and disappearance of cellular PABP, with expression of viral coat protein during NNV infection.
FIG 3.
Effects of NNV infection on the expression of newly synthesized proteins, PABP, and viral coat protein. (A) The effect of GGNNV infection on expression of newly synthesized proteins. PABP and coat protein in GB cells were examined by immunostaining. A total of 2 × 105 GB cells were seeded on cover glass in a 3-cm culture dish 15 h before GGNNV infection (MOI = 100). Puromycin (20 μg/ml) was added to the medium of infected or uninfected cells 1 h before the cells were harvested for immunocytochemical staining at the indicated times. Newly synthesized proteins with incorporated puromycin were detected using a mouse anti-puromycin Alexa Fluor 488-conjugated monoclonal antibody (green). PABP was detected with a rabbit anti-PABP polyclonal antibody (red). Mouse monoclonal antibody RG-M18 was used to detect GGNNV coat protein (violet). Nuclei were stained with DAPI (blue). White circles indicate cells with a low level of newly synthesized protein and nuclear localization of both PABP and NNV coat protein. White ovals indicate cells with a low level of newly synthesized protein and nuclear localization of PABP but NNV coat protein depletion. White squares indicate cells with newly synthesized viral protein and nuclear localization of PABP as well as nuclear/cytoplasmic localization of NNV coat protein. White rectangles indicate cells with a low level of newly synthesized protein and both PABP and NNV coat protein depletion. Bar = 20 μm. (B) Statistical analysis of the relative cell expression levels in puromycin-labeling (green), PABP expression (red), and NNV coat protein expression (violet) cells of immunocytochemistry. Cell number, n = 65 to 104 cells analyzed for each time point. GGNNV, giant grouper nervous necrosis virus; hpi, hours postinfection; MOI, multiplicity of infection; PABP, polyadenylate binding protein.
NNV virus-like particle (VLP) cannot cause GB cell translation shutoff.
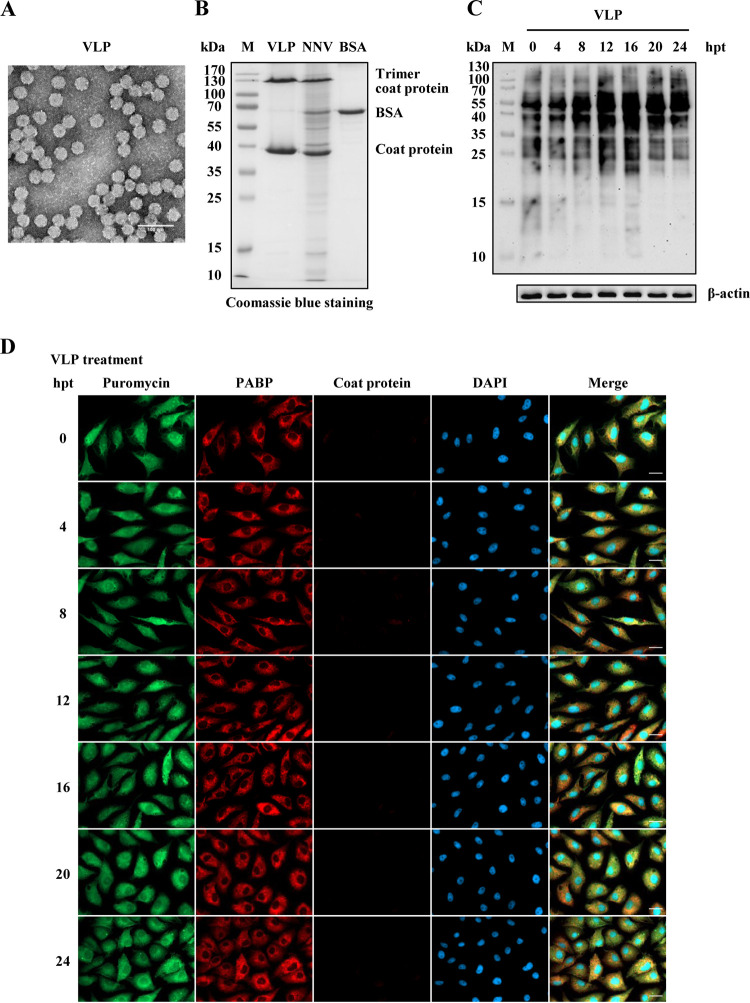
Since host translation shutoff appeared to require viral multiplication, we further tested whether purified recombinant NNV coat proteins alone could induce host translation shutoff. Recombinant NNV coat proteins were expressed in the Escherichia coli-pET expression system. Notably, the expressed recombinant NNV coat proteins form virus-like particles (VLPs) inside bacterial cells. Therefore, the VLPs were purified by a two-step CsCl gradient ultracentrifugation protocol. The purity and features of purified VLPs with 30 nm diameter were examined using negative staining transmission electron microscopy (Fig. 4A). VLPs with monomers or trimers of coat proteins were quantified by SDS-PAGE separation and Coomassie blue staining and were compared with GGNNV (concentrated by ultracentrifugation) and protein standard bovine serum albumin (BSA) (Fig. 4B). An amount of VLPs equal to that in GGNNV infection was used to treat GB cells. A SUnSET experiment with puromycin labeling after VLP treatment showed no significant change in translation from 0 to 24 h posttreatment (hpt) according to Western blot analysis (Fig. 4C). Immunocytochemical staining also showed no change in puromycin labeling or PABP subcellular localization (in cytoplasm) from 0 to 24 hpt. The GB cell number even doubled from 0 to 24 hpt, and no coat protein signal was detected in GB cells after VLP treatment (Fig. 4D). These results indicate that host translation shutoff cannot be triggered by the treatment of NNV VLPs alone.
FIG 4.
NNV virus-like particle (VLP) treatment cannot induce GB cell translation shutoff. Recombinant NNV coat proteins prepared as VLPs were used to evaluate the effect on GB cell translation shutoff. (A) Electron micrograph of negatively stained purified NNV VLPs. Bar = 100 nm. (B) SDS-PAGE analysis of purified VLPs. BSA (120 ng) was loaded as a quantitative marker. (C) Western blot analysis of newly synthesized protein in VLP-treated GB cells. The amount of VLP protein used to treat GB cells was the same as that present in the NNV infection dose (MOI = 100). Puromycin (20 μg/ml) was added to the medium of treated or untreated cells 1 h before the cells were harvested for Western blotting. A β-actin immunoblot is shown as a loading control. The puromycin incorporated into newly synthesized proteins was detected using an anti-puromycin antibody. (D) Immunocytochemical staining of VLP-treated GB cells. The same VLP treatment as used for Western blotting was used for immunocytochemistry. Newly synthesized proteins with incorporated puromycin were detected using a mouse anti-puromycin Alexa Fluor 488-conjugated monoclonal antibody (green). PABP was detected with a rabbit anti-PABP polyclonal antibody (red). Mouse monoclonal antibody RG-M18 was used to detect GGNNV coat protein (violet). Nuclei were stained with DAPI (blue). Bar = 20 μm. BSA, bovine serum albumin; hpt, hours post treatment; M, protein molecular weight marker; PABP, polyadenylate binding protein; VLP, virus-like particle.
Ectopic expression of NNV coat protein triggers PABP nuclear redistribution, degradation, and host translation shutoff.
Because the appearance of NNV coat protein accompanied the disappearance of PABP, we speculated that expression of coat proteins might participate in NNV-infection-induced translation shutoff. To test this idea, we constructed a eukaryotic expression vector for NNV coat protein (pNNVCP) and transfected it into GB cells before performing puromycin labeling. Due to the inefficiency of transfection in GB cells (about 10 %), only the successfully transfected cells with ectopic expression of coat protein were analyzed. As shown in Fig. 5A, cells with ectopic expression of NNV coat protein exhibited translation shutoff and nuclear accumulation of both coat protein and PABP. In the GB cell marked by a white circle, ectopically expressed viral coat protein was colocalized with cellular PABP in the nuclear region, with strong perinuclear fluorescence signal. No appreciable amount of newly synthesized protein was found in this cell. In the cell encircled by a white oval, ectopically expressed viral coat protein colocalized with cellular PABP throughout the cell, and the translation level in this cell was also greatly diminished. The white square indicates a cell with negligible PABP labeling and a nucleus/nucleolus occupied by newly translated proteins and viral coat protein. Finally, the white rectangle indicates a cell with ectopically expressed NNV coat protein in nucleolus, PABP in nucleus/cytoplasm, and little newly translated protein. The localization of GGNNV coat protein in the nucleus and nucleolus was consistent with a previous finding that ectopically expressed NNV coat protein translocates to the nucleus and nucleolus via its nucleolar localization signal at N-terminal amino acid residues 23 to 31 (38). The relative cell expression levels of puromycin-labeling translation, cellular PABP, and ectopically expressed NNV coat protein in different subcellular regions were quantified and plotted (Fig. 5B). The results clearly indicate that expression of NNV coat protein alone can lead to PABP nuclear redistribution and disappearance, as well as host translation shutoff.
FIG 5.
Ectopic expression of NNV coat protein in GB cells affects PABP and causes host translation shutoff. The effect of overexpressing recombinant NNV coat protein on translation shutoff in GB cells. (A) Immunocytochemical detection of NNV coat protein, PABP, and newly synthesized protein in GB cells that overexpress NNV coat protein. GB cells were transfected with pNNVCP construct (2 μg in a 3-cm tissue culture dish) for 24 h and treated with puromycin 1 h before fixation. Immunofluorescence images of transfected GB cells stained with anti-NNV coat protein antibody (violet), anti-PABP antibody (red), and anti-puromycin antibody-Alexa Fluor 488 (green). Nuclei were stained with DAPI (blue). White circles indicate cells with nuclear colocalization of both NNV coat protein and PABP and with a low level of newly synthesized protein. White ovals indicate cells with nucleus/cytoplasm colocalization of NNV coat protein and PABP and with a low level of newly synthesized protein. White squares indicate cells with nucleolus/nucleus localization of both NNV coat protein and newly synthesized protein and with a low level of PABP. White rectangles indicate cells with nucleolus localization of NNV coat protein, cytoplasm/nucleus localization of PABP and with a low level of newly synthesized protein. Bar = 20 μm. (B) Statistical analysis of the relative cell expression levels in puromycin-labeling (green), PABP expression (red), and NNV coat protein expression (violet) cells of immunocytochemistry. PABP, polyadenylate binding protein. n = 50.
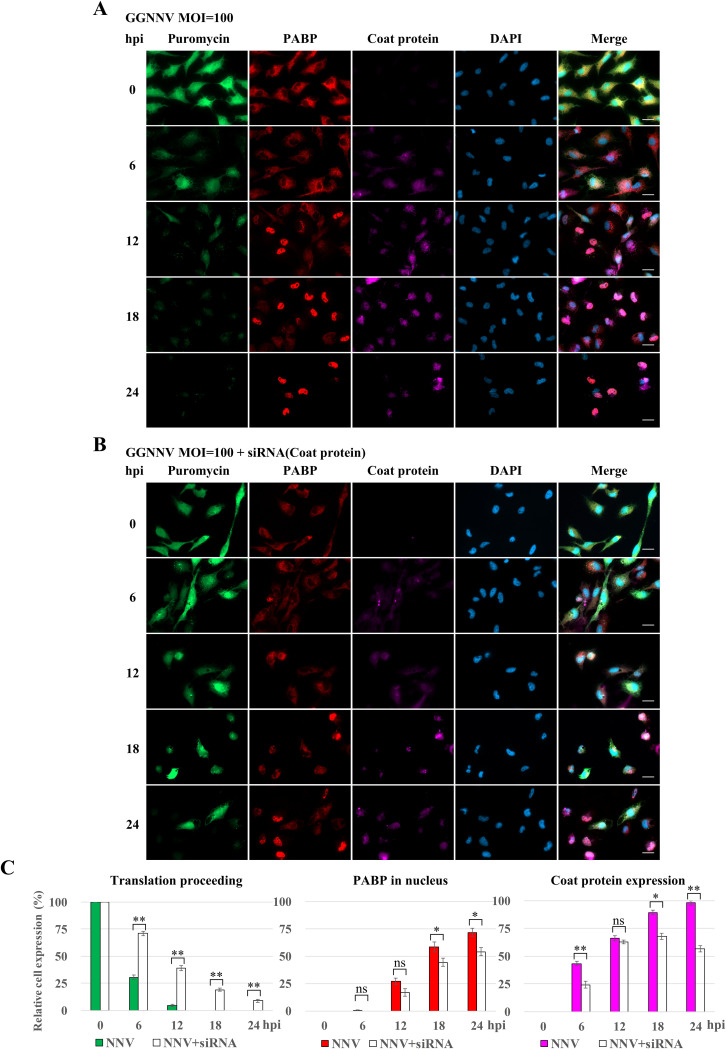
Restoration of host translation by siRNA-mediated silencing of NNV coat protein postinfection.
To evaluate the effect of NNV coat protein on host translation shutoff, GB cells were transfected with in vitro-generated full-length NNV double-stranded RNA2 (dsRNA2; which encodes coat protein) prior to infection. The NNV dsRNA2 should be processed into small interfering RNA (siRNA) fragments through RNA interference machinery in GB cells. The NNV coat protein expression level was reduced in dsRNA-transfected cells at each measured time point after NNV infection, and the level of PABP nuclear sequestration was proportionally reduced. Comparing the cells with and without dsRNA transfection after NNV infection in GB cells, immunocytochemical staining showed a recovery of host translation (compare Fig. 6A and B). To better quantify the restoration of host translation shutoff by knockdown of coat protein in NNV-infected GB cells, the relative levels of host translation, PABP nuclear sequestration, and NNV coat protein expression cells were quantified. Inefficient dsRNA transfection and early expression of B2 protein (antagonizes host RNA interference) in infected GB cells may explain why only a low level of rescue was observed (Fig. 6C). Nevertheless, these results again confirm that NNV coat protein participates in NNV-induced translation shutoff.
FIG 6.
Knockdown of NNV coat protein by siRNA rescues NNV-induced translation shutoff. Before NNV infection, dsRNA of NNV RNA2 was prepared in vitro and transfected into GB cells to evaluate the effect on GB cell translation shutoff. (A) Immunocytochemical staining of NNV-infected GB cells. This experiment was used as a control for the NNV coat protein knockdown experiment. GB cells were infected with GGNNV at an MOI of 100. At 0, 6, 12, 18, and 24 hpi, cells were harvested for immunocytochemistry. Puromycin was added to the medium of infected cells 1 h before harvesting the cells. Newly synthesized proteins with incorporated puromycin were detected using a mouse anti-puromycin Alexa Fluor 488-conjugated monoclonal antibody (green). PABP was detected with a rabbit anti-PABP polyclonal antibody (red). Mouse monoclonal antibody RG-M18 was used to detect GGNNV coat protein (violet). Nuclei were stained with DAPI (blue). Bar = 20 μm. (B) Immunocytochemical staining of NNV-infected GB cells which were pretransfected with NNV dsRNA2. Twelve hours before NNV infection, the GB cells were transfected with NNV dsRNA2 using a Lipofectamine 2000 transfection reagent (Invitrogen) (1 μg in a 3-cm tissue culture dish). Bar = 20 μm. (C) Statistical analysis of the relative cell expression levels in puromycin labeling (green), PABP expression (red), and NNV coat protein expression (violet) cells of immunocytochemistry. Data are presented as means ± standard deviation from three independent experiments. ns, not significant; *, P < 0.05; **, P < 0.01 (Student’s t test). GGNNV, giant grouper nervous necrosis virus; hpi, hour postinfection; MOI, multiplicity of infection; PABP, polyadenylate binding protein.
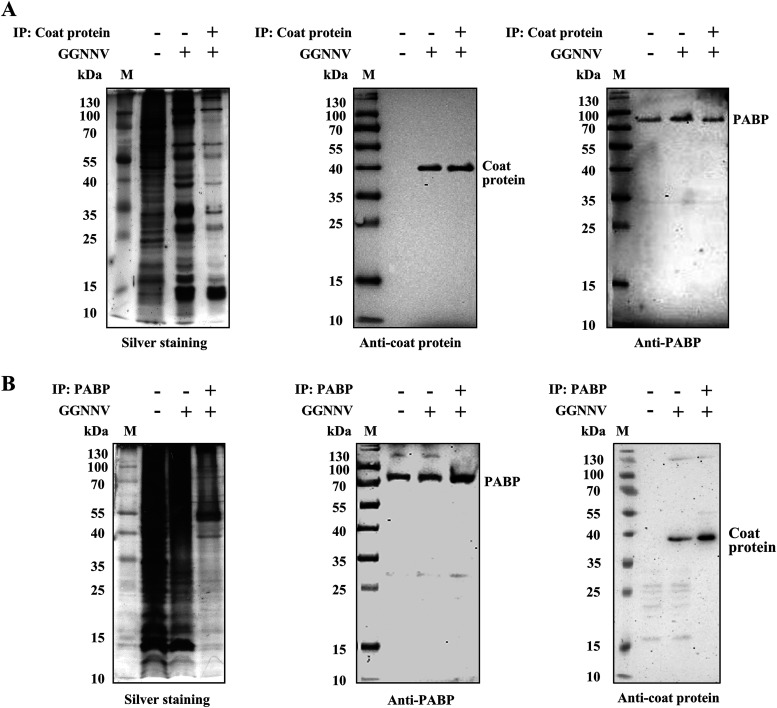
Interaction between NNV coat protein and cellular PABP.
To test for a physical interaction between NNV coat protein and cellular PABP, we performed coimmunoprecipitation experiments. The lysates from GGNNV-infected GB cells were immunoprecipitated using an anti-NNV coat protein antibody (Fig. 7A) or anti-PABP antibody (Fig. 7B). Nonspecifically associated proteins were washed from the precipitates, and the remaining bound proteins were subjected to SDS-PAGE and visualized by silver staining (Fig. 7A and B, left panel). Clear and obvious signals for precipitated NNV coat protein and cellular PABP in the immunoblots indicated that the two proteins indeed interact (Fig. 7A and B, middle and right panels). The coimmunoprecipitation of NNV coat protein and cellular PABP was consistent with the results from immunofluorescence staining showing colocalization of cellular PABP and NNV coat protein (Fig. 3A and 5A). Both results suggest the two proteins interact in infected cells.
FIG 7.
Coimmunoprecipitation of NNV coat protein and cellular PABP in GGNNV-infected GB cells. To assess the interaction between NNV coat protein and cellular PABP, coimmunoprecipitation experiments were performed. GB cells were infected with GGNNV (MOI = 100) for 18 h before coimmunoprecipitation experiments. (A and B) Total cell lysates were pulled down with anti-NNV coat protein antibody-protein A/G beads (A) or anti-PABP antibody-protein A/G beads (B). The total cell lysates of uninfected and GGNNV-infected GB cells (both are used as controls) and pulled-down proteins were subjected to SDS-PAGE and Western blotting. The specificity of protein binding in immunoprecipitates was examined by silver staining. An association of PABP and NNV coat protein was observed by immunoblotting. To prevent signal from precipitated immunoglobulins, VeriBlot for IP detection reagent (HRP) was used to detect the coimmunoprecipitated PABP and NNV coat proteins. The coimmunoprecipitation experiments were performed at least two times. GB, grouper brain; GGNNV, giant grouper nervous necrosis virus; IP, immunoprecipitation; M, protein molecular weight marker; MOI, multiplicity of infection; PABP, polyadenylate binding protein.
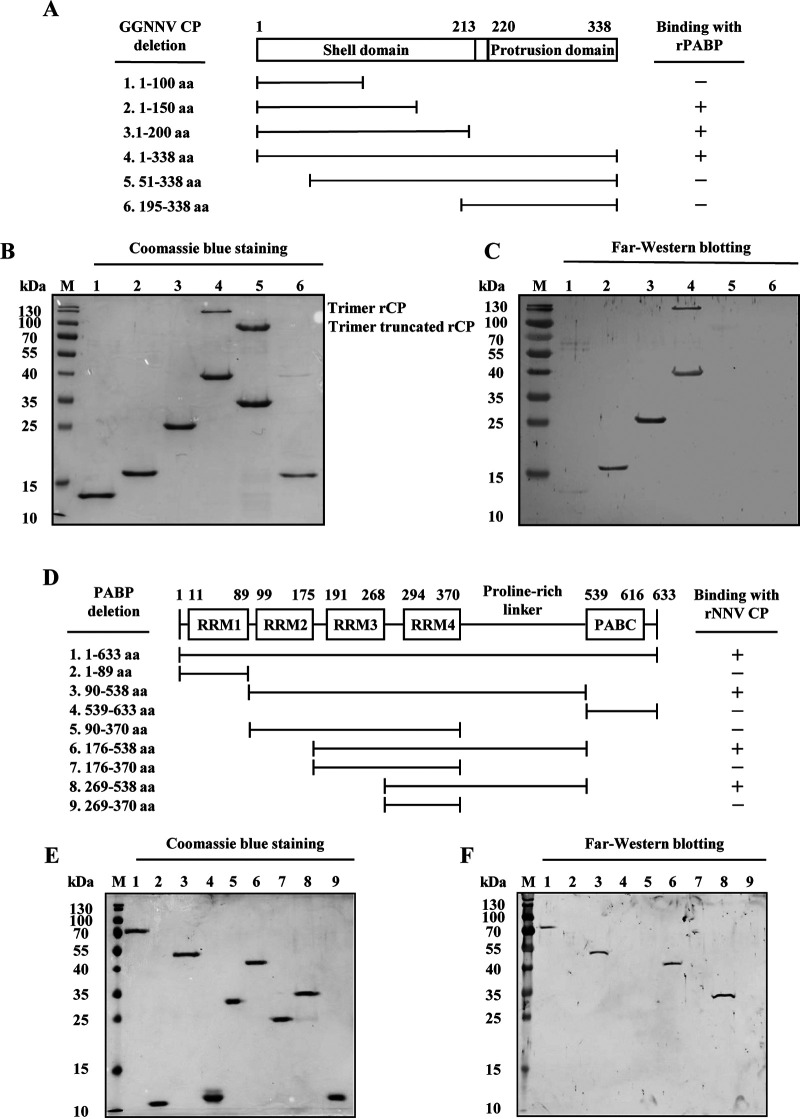
N-terminal shell of the coat protein binds with proline-rich linker of PABP.
To further test for a direct interaction and define the interacting regions in these two proteins, Far-Western blotting was performed with E. coli-expressed recombinant deletion-mutant proteins. Complete and deletion-mutant NNV coat proteins (Fig. 8A) and PABPs (Fig. 8D) were produced and purified using the E. coli-pET expression system (Fig. 8B and E). As shown in Fig. 8A and C, positive signals were observed for NNV coat protein, 1 to 150, 1 to 200, and 1 to 338 (complete) amino acid (aa) fragments. This result suggests that the binding region is located within the N-terminal 1 to 150 aa shell domain of the NNV coat protein (39). Additionally, Fig. 8D and F show positive signals for PABP, 1 to 633 (complete), 90 to 538, 176 to 538, and 269 to 538 aa fragments. Since the 269 to 370 aa fragment (RRM4) showed no detectable signal, we presume that the binding region in PABP is located within the C-terminal 371 to 538 aa proline-rich linker region. However, we could not confirm that this region alone is sufficient for binding, as we were unable to express the 371 to 538 aa recombinant mutant of PABP in the E. coli-pET expression system (data not shown). These results indicate that the interaction between NNV coat protein and PABP requires the N-terminal shell domain of the coat protein and the PABP proline-rich linker region. Additionally, the data further validate a direct interaction between NNV coat protein and cellular PABP.
FIG 8.
Determination of binding regions in NNV coat protein and PABP. To identify the binding regions within NNV coat protein and cellular PABP, full-length and serial deletion mutants were expressed and purified from an E. coli/pET expression system. Binding regions in NNV coat protein and PABP were determined by far-Western blotting of recombinant deletion-mutant proteins. (A) Schematic representation of the domain structure and recombinant NNV coat protein deletion mutants. (B) SDS-PAGE analysis of the purified recombinant NNV coat protein deletion mutants, with Coomassie blue staining. (C) Far-Western blotting of recombinant NNV coat protein deletion mutants probed with recombinant full-length PABP. (D) Schematic representation of the domain structure and recombinant PABP deletion mutants. (E) SDS-PAGE analysis of the purified recombinant PABP deletion mutants, with Coomassie blue staining. (F) Far-Western blotting of recombinant PABP deletion mutants probed with recombinant full-length NNV coat protein. aa, amino acid; M, protein molecular weight marker; PABC, C-terminal helical domain of PABP; rNNV CP, recombinant nervous necrosis virus coat protein; rPABP, recombinant polyadenylate binding protein; RRM, RNA recognition motif.
Degradation of PABP by the ubiquitin-proteasome system in GGNNV-infected GB cells.
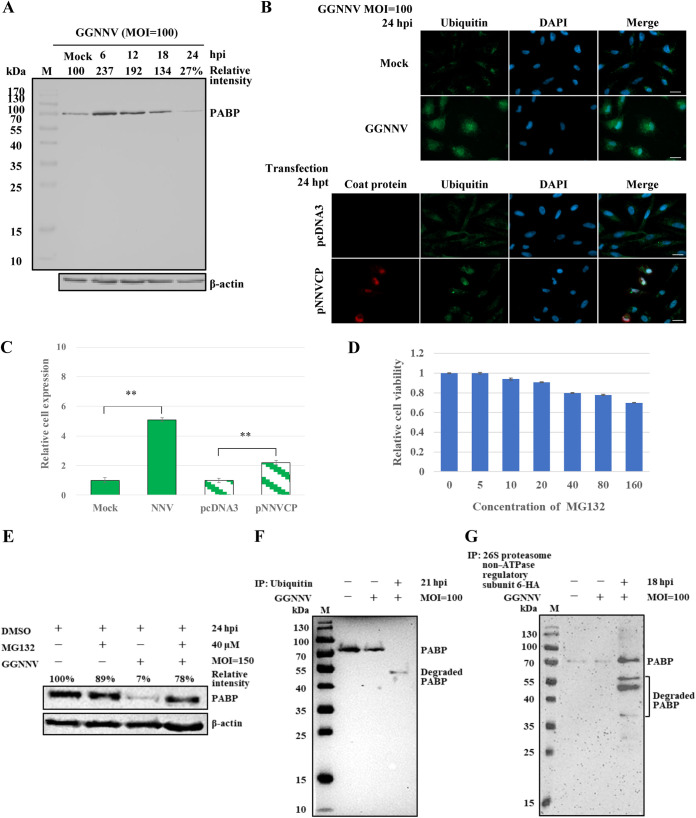
The status of PABP in GB cells during GGNNV infection was next evaluated by Western blotting. As shown in Fig. 9A, the total level of cellular PABP increased to 237 % at an early infection stage (6 hpi) compared with mock-infected cells; the level then steadily declined, reaching 27 % of the original level at 24 hpi. To explore the possible mechanism of PABP degradation at this late infection stage, we used liquid chromatography tandem mass spectrometry (LC-MS/MS) to analyze bound proteins in anti-PABP antibody precipitate from GGNNV-infected GB cell lysate (Fig. 7B). The identified PABP-associated proteins included polyubiquitin-B isoform X6 and 26S proteasome non-ATPase regulatory subunit 6 (encoded by the PSMD6 gene) (Table 1). The former is a polyubiquitin moiety containing three direct repeats of 76 aa ubiquitin, whereas the latter is a subunit of the 19S regulatory particle in the 26S proteasome. This finding implied that PABP might be degraded by the ubiquitin-26S proteasome proteolytic pathway. We therefore monitored the levels of ubiquitination in mock- or NNV-infected GB cells 24 hpi and in GB cells with or without ectopic expression of recombinant NNV coat protein at 24 hpt. Punctate ubiquitination signal could be observed in infected and transfected GB cells after immunocytochemical staining (Fig. 9B). The overall ubiquitination level was dramatically increased by 5.1-fold after GGNNV infection and by 2.2-fold after ectopic NNV coat protein expression (Fig. 9B and C). Next, we demonstrated that the NNV-induced degradation of PABP indeed occurs via the proteasome proteolytic pathway by treating cells with the peptide-aldehyde proteasome inhibitor, MG132. First, the cytotoxicity of MG132 was tested in GB cells (Fig. 9D). To maximize the inhibitory effect of MG132 on the proteasome, we used 40 μM MG132. GB cells treated with this concentration showed 80 % viability in the cell counting kit-8 (CCK-8) assay. After GGNNV infection (MOI = 150) for 24 h, the level of PABP in GB cells was decreased to 7 % of that in control GB cells. However, when cells were treated with MG132 (40 μM), the level of PABP in GGNNV-infected GB cells was 78 % of that in control GB cells (Fig. 9E). Of note, the level of PABP in MG132-treated GB cells was 89 % of that in control cells. This slight decrease is expected, as MG132 has been previously shown to downregulate PABP to different degrees in multiple cell lines (40). The degradation of PABP after NNV infection was further probed by immunoprecipitation with anti-ubiquitin antibody-protein A/G beads. To our surprise, a truncated PABP band with a molecular weight of ∼50 kDa was observed on the immunoblot. However, no high-molecular-weight polyubiquitinated PABP signal was detected, suggesting PABP proteolysis by the ubiquitin-proteasome is highly efficient. Moreover, the level of degraded PABP in the nonprecipitated and nonconcentrated GGNNV-infected GB cell lysate was too low to be detected by Western blotting (Fig. 9F). The ubiquitin antibody-precipitate from GGNNV-infected GB cell lysate was further subjected to LC-MS/MS analysis. Ubiquitin-associated proteins were identified from this analysis, including polyadenylate-binding protein 1 isoform X1, NNV coat protein, NNV RdRp, and NNV B2 protein (Table 2). To better understand the molecular mechanism of PABP degradation by the 26S proteasome, the gene for non-ATPase regulatory subunit 6 was RT-PCR cloned from GB cells and inserted into a modified pcDNA3 vector (the neomycin resistance gene was replaced by a puromycin resistance gene to improve selection efficiency in GB cells) with an HA tag at its C terminus. At GGNNV 18 hpi, the lysate of puromycin-selected GB cells with ectopic expression of 26S proteasome non-ATPase regulatory subunit 6-HA was immunoprecipitated with anti-HA antibody-protein A/G beads. The immunoprecipitate was probed by Western blotting with an anti-PABP antibody. As shown in Fig. 9G, full-length 70-kDa PABP and more degraded forms of PABP (molecular weights ranging from 35 to 55 kDa) were observed in the immunoprecipitate. Together, these results demonstrate that PABP is degraded by the ubiquitin-proteasome system in GGNNV-infected GB cells.
FIG 9.
Degradation of PABP in GGNNV-infected GB cells through the ubiquitin-proteasome pathway. (A) The status of PABP during GGNNV infection in GB cells. GB cells were infected with or without (Mock) GGNNV (MOI = 100) for the times indicated. Cell lysates were subjected to SDS-PAGE and Western blotting with an anti-PABP antibody. (B) The ubiquitination levels of GGNNV-infected or NNV coat protein ectopically expressed GB cells were detected by immunocytochemical staining with an anti-ubiquitin antibody. GB cells were infected with GGNNV (MOI = 100) for 24 h or transfected with pNNVCP construct (2 μg in a 3-cm tissue culture dish) for 24 h. Mock infection and pcDNA3 transfection were used as controls for GGNNV infection and pNNVCP transfection, respectively. Bar = 20 μm. (C) Statistical analyses of the relative cell ubiquitination levels in the treated GB cells. Data are presented as means ± standard deviations from three independent experiments. **, P < 0.01 (Student’s t test). (D) Effect of MG132 on GB cell viability. GB cells in a 96-well plate were treated with different concentrations of MG132 for 24 h. The cell viabilities were detected by absorbance at 450 nm using the cell counting kit-8 (CCK-8). The values are presented as means ± standard deviations (SD) (n = 3). (E) GGNNV-induced PABP degradation was inhibited by a 26S proteasome inhibitor, MG132. GB cells were infected with or without GGNNV (MOI = 150) and were treated with or without MG132 (40 μM) for 24 h. Cell lysates were subjected to SDS-PAGE and Western blotting with an anti-PABP antibody. As MG132 was supplied in DMSO solution, and the control GB cells and GGNNV-infected GB cells were treated with the same concentration of DMSO. β-actin served as a loading control for immunoblots (bottom panels of A and E). To confirm that ubiquitination and 26S proteasome participate in the degradation of PABP, the coimmunoprecipitation experiments were performed. (F) Coimmunoprecipitation of degraded PABP with an anti-ubiquitin antibody from GGNNV-infected GB cell lysate. GB cells were infected with GGNNV (MOI = 100) for 21 h, and the resultant cell lysate was pulled down with anti-ubiquitin antibody-protein A/G beads. The total cell lysates of uninfected and GGNNV-infected GB cells (both are used as controls) and pulled-down proteins were subjected to SDS-PAGE and immunoblotting detection using an anti-PABP antibody. (G) Coimmunoprecipitation of PABP with an anti-HA antibody from 26S proteasome non-ATPase regulatory subunit 6-HA ectopically expressed and GGNNV-infected GB cell lysate. The non-ATPase regulatory subunit 6 gene was RT-PCR cloned from GB cells and was ectopically expressed in GB cells. The puromycin (1.2 μg/ml) selection was performed to increase the ectopic expression of non-ATPase regulatory subunit 6-HA in transfected GB cells. The recombinant non-ATPase regulatory subunit 6-HA with an HA tag at its C terminus enabled the immunoprecipitation by using a commercial anti-HA antibody. After GGNNV infection for 18 h, the transfected and infected GB cell lysate was immunoprecipitated with an anti-HA antibody. The immunoprecipitate was analyzed by Western blotting with an anti-PABP antibody. GB, grouper brain; GGNNV, giant grouper nervous necrosis virus; hpi, hour postinfection; hpt, hour posttransfection; IP, immunoprecipitation; M, protein molecular weight marker; MOI, multiplicity of infection; PABP, polyadenylate binding protein.
TABLE 1.
PABP-associated proteins detected from NNV-infected GB cells by LC-MS/MS analysis
| Categorya | Descriptionb |
|---|---|
| Nervous necrosis virus | Coat protein |
| Cytoskeleton and motility | Tropomyosin alpha-3 chain, alpha-3 chain isoform X9, 4a isoform 2,4-2, alpha-4 chain isoform X2; calmodulin 3a(phosphorylase kinase,delta); myosin light polypeptide 6 B, 6 isoform X3, regulatory light polypeptide 9b; myosin 10; tropomodulin-2 isoform X2; actin, cytoplasmic 1; actin-related protein 2/3 complex subunit 4, 5; F-actin-capping protein subunit beta isoform X2; vimentin; dynein, light chain, LC8-type 2A; spectrin alpha chain, nonerythrocytic 1 isoform X5, isoform X9; filamin A; integrin beta-1 isoform X2; keratin, type I cytoskeletal 18b, type II cytoskeletal 8; src substrate cortactin isoform X6 |
| Translation | 60S ribosomal protein L12 isoforms X2, L30, L7a, L38, and L22; 40S ribosomal protein S12 isoforms X2, S28, S25, S21, S10, and S20; polyadenylate-binding protein 1A; elongation factor 1 alpha; EF-hand domain-containing protein D2; eukaryotic translation initiation factor 4E family member 1c; poly (rC)-binding protein 2 isoform X3 |
| Mitochondrial protein | Mitochondrial import inner membrane translocase subunits Tim 13, Tim8 B, and Tim 8A; stress-70 protein; thioredoxin-dependent peroxide reductase; 60 kDa heat shock protein |
| Chaperone | Heat shock cognate 71kDa; endoplasmic reticulum chaperone BiP precursor; T-complex protein 1 subunit delta |
| Splicing | PHD finger-like domain-containing protein 5A; splicing factor 3B subunit 4; SNU 13 homolog, small nuclear ribonucleoprotein b (U4/U6.U5) |
| Protein degradation | Polyubiquitin-B isoform X6; 26S proteasome non-ATPase regulatory subunit 6 |
| Nucleosome | Histones H2B 1/2 and H2A type 2-B |
| Others | Neuroblast differentiation-associated protein; peroxiredoxin; Golgi apparatus protein-1,1 isoform X2; peptidyl-prolyl cis-trans isomerase B; protein LSM12 homolog A isoform X2; charged multivesicular body protein 4b; enhancer of rudimentary homolog; vacuolar ATP synthase subunit G1; S-phase kinase-associated protein 1; flotillin-2b; collagen alpha-1 (II) chain isoform X4, 1a1 precursor; protein lin-7 homolog C; protein disulfide-isomerase A3; calponin-3 |
Biological function or subcellular localization.
Protein name.
TABLE 2.
Ubiquitin-associated proteins detected from NNV-infected GB cells by LC-MS/MS analysis
| Categorya | Descriptionb |
|---|---|
| Nervous necrosis virus | Coat protein; RNA dependent RNA polymerase; B2 |
| Cytoskeleton and motility | Myosin-9 isoform X1; myosin-10, isoform X1, isoform X2; mysosin-Ic isoform X3, Ib isoform X1, Va isoform X4, XIX isoform X1, Ie-like isoform X1, Ib XVIIIa isoform X2, light polypeptide 6, light chain 3, skeletal muscle isoform, light polypeptide 6 isoform X4, light chain 12, genome duplicate 2, regulatory light polypeptide 9b isoform X1, VIa isoform X1; filamin-A isoform X1; tropomodulin-3; gelsolin isoform X1; calmodulin; tropomyosin 4a isoform X4, beta chain isoform X7, alpha-3 chain isoform X5, alpha-3 chain isoform X6; f-actin capping protein subunit beta isoforms 1 and 2 isoform X1, alpha-1, alpha-2; actin-related protein 3, 2, 2/3 complex subunit 4, 2/3 complex subunit 1B-B, subunit 5b; LIM-domain and actin-binding protein 1a isoform X1; alpha-actinin-1; tubulin beta-4B chain isoform X2, alpha chain, alpha-1A chain, alpha chain, testis-specific, alpha-1C chain; plectin isoform X1; vimentin, vimentin-4; spectrin alpha chain, nonerythrocytic 1 isoform X5, spectrin beta chain, nonerythrocytic 1 isoform X3; keratin, type II cytoskeletal 8, keratin, type I cytoskeletal 18; SLIT-ROBO Rho GTPase-activating protein 1; ankycorbin; protein outspread isoform X1; nonmuscle caldesmon isoform X1; Src substrate protein p85 isoform X1; cofilin-2; ADP-ribosylation factor 4; integrin alpha-4; LIM and SH3 domain protein 1 |
| Nucleosome protein | Nucleosome assembly protein-1-like 1 isoform X2; histone H3 centromeric protein CSE4, histone H1.10 |
| Translation | 60S ribosomal protein L4-B, L3, L7a, L9 isoform X1, L7, L12, L6, L8, L23, L30, L17, L22, L15, L18, L18 a, L13, L32, L19, L11 isoform X1, L24, L38, L27, L37a, L13a, L14, L31, L28 isoform X2, L36, L35; 60s acidic ribosomal protein P0, P2; 40S ribosomal protein S5 isoform X1, S3a, S8, S14, S15, S7, S17, S15a isoform X1, S18, S6, S16, S19, S10 isoform X1, S17, SA, S11, S13, S23, S12, S27, S20, S9 isoform X1; ribosomal protein S26 L homeolog isoform X1; ribosomal RNA processing protein 1 homolog A isoform X1; elongation factor 1-alpha, 2; polyadenylate binding protein 1 isoform X1; eukaryotic translation initiation factor 2 subunit 3,1; eukaryotic initiation factor 4A-II; eIF-2 alpha kinase activator GCN1; EF-hand domain-containing protein D2; ubiquitin-40S ribosomal protein S27a; ribosome production factor 2 homolog; ribosome biogenesis protein BRX1 homolog; lysine-tRNA ligase isoform X1 |
| Mitochondrial protein | ATP synthase subunit alpha, beta; stress-70 protein; 60kDa heat shock protein; mitochondrial import inner membrane translocase subunit Tim13; thioredoxin-dependent peroxide reductase; pyruvate dehydrogenase E1 component subunit beta, mitochondrial; isocitrate dehydrogenase (NADP); serine protease HTRA2; 3-methyl-2-oxobutanoate dehydrogenase (lipoamide) kinase; polyribonucleotide nucleotidyltransferase 1; glutaryl-CoA dehydrogenas; cytochrome c1, heme protein; pyruvate carboxylase; MICOS complex subunit MIC60 isoform X1; ADP/ATP translocase 2; ATPase family AAA domain containing 3 |
| Transcription | Transcription elongation factor SPT6 isoform X1; RNA binding protein 4.1 isoform X1; cold-inducible RNA binding protein B isoform X1; CCHC-type zinc finger, nuclei acid binding protein a; Myb binding protein 1A protein; RNA transcription, translation and transport factor protein |
| Replication | DNA topoisomerase 1 isoform X1; DNA topoisomerase 2-alpha; DNA replication licensing factor MCM5 |
| Chaperone | Heat shock cognate 71kDa protein; heat shock 70kDa protein; heat shock protein beta-11; endoplasmic reticulum chaperone BiP; T-complex protein 1 subunit gamma, theta isoform X1, alpha, beta, eta, delta; hypoxia upregulated protein 1 isoform X1; dnaJ homolog subfamily A member 2, 2 a, subfamily C member 25 |
| Nucleolar protein | Nucleolar protein 58, 56 isoform X1, nucleolin; lysine-rich nucleolar protein 1; nucleolar GTP binding protein 1; cell growth-regulating nucleolar protein; lysine-rich nucleolar protein 1; MKI67 FHA domain-interacting nucleolar phosphoprotein; ATP-dependent RNA helicase an3 isoform X3, DDX5 isoform X1, DDX3Y isoform X5, DDX1, DDX18, DDX54; heat repeat-containing protein 1; surfeit locus protein 6 |
| Endoplasmic reticulum proteins | Protein disulfide-isomerase A3; transitional endoplasmic reticulum ATPase; NADPH-cytochrome P450 reductase isoform X1; dolichyl-diphosphosphooligosaccharide-protein glycosyltransferase subunit 1; extended synaptotagmin-2-A isoform X1, 2 isoform X-1 |
| Golgi proteins | Golgi apparatus protein 1; Golgi-associated plant pathogenesis-related protein 1; coatomer subunit alpha |
| Splicing | U2 small nuclear RNA auxiliary factor 2a isoform X4; splicing factor, proline- and glutamine-rich isoform X1; splicing factor U2AF 35kDa subunit; pre-mRNA splicing factor ATP-dependent RNA helicase; RNA splicing ligase RtcB homolog; non-POU domain-containing octamer-binding protein; NHP2-like protein 1 |
| LDL receptor | Low-density lipoprotein receptor-related protein 1 isoform X-1, X-2; low-density lipoprotein receptor isoform X1 |
| Ribonucleoprotein | H/ACA ribonucleoprotein complex subunit DKC1, 1 isoform X1; heterogeneous nuclear ribonucleoprotein M isoform X1, U isoform X1, D, R-isoform X1, Q isoform X1; small nuclear ribonucleoprotein Sm D3 |
| Endocytosis | AP-2 complex subunit alpha-2 isoform X1, beta-1 isoform X-1; clathrin heavy chain 1 isoform X1; transferrin receptor protein 1 |
| Nuclear transport | GTP-binding nuclear protein Ran isoform X1; importin-7; THO complex subunit 1 |
| Fatty acid synthesis and breakdown | Fatty acid synthase; long-chain-fatty-acid-CoA ligase 3b; very-long-chain 3-oxoacyl-CoA reductase-A; peroxisomal acyl-coenzyme A oxidase 3 |
| Plasma membrane | Anion exchange protein 2 isoform X1; ras-related protein Rab-18a; gap junction gamma-1 protein |
| Others | Neuroblast differentiation-associated protein AHNAK isoform X1; sequestosome-1; dedicator of cytokinesis protein 7 isoform X1; leucine-rich repeat flightless-interacting protein 2 isoform X1, isoform X20; Y-box-binding protein 1; barrier-to-autointegration factor isoform X1; ras GTPase-activating protein binding protein 1 isoform X1; thymopoietin a isoform X1; guanine nucleotide binding protein 3; peroxiredoxin-1; ATP binding cassette subfamily F member 2; protein RRP5 homolog |
Biological function or subcellular localization.
Protein name.
DISCUSSION
Viruses have evolved various strategies to hijack cellular translation and create conditions that favor synthesis of viral proteins rather than host proteins. Upon viral infection, cellular translation can be blocked through several different pathways, including inhibition of the mTOR-4EBP-eIF4E signaling pathway (41, 42), inhibition of eIF2α-mediated antiviral defense (42, 43), inhibition of cellular translation factors such as PABP (15–28, 32) or eIF4G (44), or destruction of host mRNA (35). In this study, we showed that NNV hijacks host translation by leveraging its distinct mRNA structure with 5′-caps but without 3′-poly(A) tails. After infection, this distinct structure offers NNV a chance to create a virus-favoring environment and maximize access to host cell resources. The key role of PABP in translation makes it the most common target for viruses to attack. Unlike protease-containing viruses that cleave PABP, NNV sequesters PABP in the nucleus, probably via an interaction between the shell domain of NNV coat protein and the proline-rich linker region of PABP. Interestingly, this binding site on PABP and the cleavage sites recognized by proteases in HIV-1, poliovirus, and feline calicivirus are all located within the most diverse protein domains, RRM3 and the proline-rich linker (Fig. 2B) (16–19), implying that coevolution occurred between the hosts and corresponding pathogens. Furthermore, we found that translation shutdown begins at the early stage of infection (Fig. 1A, 3 to 6 hpi; Fig. 3B, left panel, 4 hpi), before PABP is mostly in the nucleus (Fig. 3, 12 to 24 hpi). Thus, it seems likely that cytoplasmic PABP binding by NNV coat protein might participate in host translation shutoff at the early infection stage. Notably, a similar mechanism of translation shutoff is also seen with rubella virus capsid protein (32).
During the early infection period (6 to 12 hpi), the total amount of PABP was increased to approximately double the level in mock-infected cells (Fig. 9A). It seems likely that the increased PABP expression level may be a consequence of compensatory feedback regulation in response to the functional loss of PABP. A similar induction of PABP can be seen in human cytomegalovirus-infected cells (45). Conversely, the overexpression of PABP may lead it to repress its own translation through an autoregulatory translational control element in the 5′-untranslated region of its mRNA (46). At later infection stages, the levels of PABP were gradually reduced (12 to 24 hpi, Fig. 3B and 9A). This reduction is reminiscent of reports that PABP is degraded during apoptotic cell death in several mammalian cell lines. In that context, the degradation of PABP is accompanied by translation inhibition and correlated with caspase-3-mediated cleavage of eIF4G, 4EBP, and eIF2α; however, the degradation of PABP was found to occur via calpain, a calcium-activated protease, and not via caspase-3 or the proteasome (40). It is noteworthy that NNV coat protein was reported to induce apoptosis in sea bass cells and Cos-7 cells through the caspase-8-initiated caspase-3-dependent pathway (47). Thus, NNV may act via another apoptosis-related pathway in addition to its direct effects on PABP to produce host translation shutoff. In this study, we used a proteasome inhibitor to demonstrate that PABP degradation occurs at the late stage of NNV infection via the ubiquitin-proteasome system (Fig. 9). However, several cancer cell lines (e.g., MCF7 and HeLa cells) exhibit some level of PABP degradation after MG132 treatment (40). In our study, a weak effect of PABP downregulation was also observed after treating GB cells with MG132 proteasome inhibitor (Fig. 9E). Surprisingly, we detected a truncated form of PABP with a molecular weight of ∼50 kDa by immunoblotting. Because the anti-PABP antibody recognizes the C terminus of the PABC domain, this molecular weight implies that the protein is degraded up to the RRM3 domain, assuming the ubiquitination of the truncated PABP is monomeric. Although the proteasome should catalyze the complete hydrolysis of target proteins in principle, partial proteolytic degradation by the 26S proteasome was reported for a subunit of the transcription factor NF-κB (48). In that case, the p105 precursor of the NF-κB p50 subunit is processed through ubiquitin-proteasome proteolysis to create the active protein. Whether the truncated form of ubiquitinated PABP still plays a functional role in translation will require further investigation. According to these results, we concluded that the expressed NNV coat proteins interact with cellular PABP to shut down host translation and to sequester PABP in the nucleus after infection. Later, the viral coat protein stimulates PABP polyubiquitination (probably through polyubiquitin B). The polyubiquitinated PABP is then bound by the non-ATPase regulatory subunit 6 of the 19S regulatory particle to deliver it into the translocation channel of the 20S core particle for protein degradation.
Further experiments will be required to determine whether the host translation shutoff in the GGNNV-infected GB cells involves mechanisms other than PABP binding and nuclear sequestration. The existence of a 5′-cap structure on NNV RNAs suggests that recruitment of the initiation complex proceeds through eIF4E, although several other positive-sense RNA viruses exploit internal ribosome entry site (IRES) cap-independent protein synthesis (49). Nevertheless, the lack of a 3′-poly(A) tail on NNV RNAs means that the virus cannot utilize the canonical cap-dependent translation approach, which requires a PABP-bridged closed-loop RNA template. Thus, the strategy utilized by NNV to compensate for the absence of a poly(A) tail and promote efficient translation remains a mystery. The manner of translation for NNV RNAs will be an important and interesting direction for future investigations. Moreover, as NNV is a positive-sense RNA virus, its genome serves as a template for both translation and RNA replication. Since these two events proceed in opposite directions, they cannot occur simultaneously on the same template. Translation shutoff through PABP had been reported for several viruses in which viral RNA templates switch from translation to replication (22, 50). Whether PABP is involved in NNV template switches from translation to replication is another interesting topic that will require more investigation.
The ubiquitin proteasome system is the major pathway for degrading regulatory proteins and damaged or misfolded proteins; this pathway plays a crucial role in maintaining cellular homeostasis in eukaryotes (51). However, viruses have evolved various mechanisms to exploit the host ubiquitin proteasome system to modulate the function and stability of viral proteins (52). The degradation of Turnip yellow mosaic virus RdRp by the ubiquitin proteasome system switches viral replication between the positive-strand and negative-strand viral RNA synthesis (53). Furthermore, hepatitis C virus core protein is degraded by the ubiquitin proteasome system to regulate viral pathogenesis via multiple mechanisms (54). As the immunoprecipitate of ubiquitin-associated proteins (Table 2) included NNV coat protein, RdRp, and B2 protein, it is reasonable to ask whether the host ubiquitin proteasome system also plays an important role in NNV protein regulation.
Host translation is shut off after NNV infection, but viral protein synthesis must continue. To reconcile these conflicting requirements, NNV has developed a highly specific strategy to facilitate only viral multiplication. After infection, viruses often generate a specialized structure-segregated compartment, termed a viral factory, for virus protein synthesis, genome replication, and virion assembly. Our preliminary data suggest that such viral factories may exist in the cytoplasm of GGNNV-infected GB cells, according to staining with anti-phosphorylated-eIF4E antibody and NNV markers (data not shown). Thus, it seems that NNV might utilize discrete spatial compartments for virus production while simultaneously blocking host translation throughout the cell.
MATERIALS AND METHODS
Cells and virus.
The GB cell line was established from brain tissue of an immature orange-spotted grouper (Epinephelus coioides) (55). After isolating single cells to establish clonal populations at 80 and 200 passages, GB cells appeared as a fibroblast cell type with a homogeneous size of around 30 to 40 μm on the surface of a culture dish. The growth doubling time was about 24 h when the cells were cultured in Leibovitz’s L-15 medium supplemented with 10 % heat-inactivated fetal bovine serum (FBS) (Gibco), l-glutamine (Gibco), and penicillin-streptomycin (Gibco) at 28 °C. No bacteria, fungi, or microplasma were detected in the cultured cells. In this study, the cells were used at passage numbers around 250 to 300. The giant grouper nervous necrosis virus (GGNNV) of the red-spotted grouper nervous necrosis virus (RGNNV) genotype was isolated from head tissue of diseased giant grouper, which was kindly provided by Shang-Hai Aquatic Animal Hospital in Pingtung, Taiwan in 2014 (56).
Antibodies, chemicals, and reagents.
The mouse anti-NNV coat protein monoclonal antibody, RG-M 18 (IgG1), was previously generated in our laboratory (57). Rabbit anti-PABP antibody (ab21060) and VeriBlot for immunoprecipitation (IP) detection reagent (horseradish peroxidase; HRP) (ab131366) were purchased from Abcam. Mouse anti-puromycin monoclonal antibody (clone 12D10, MABE343) and mouse anti-puromycin Alexa Fluor 488-conjugated antibody (clone 12D10, MABE343-AF488) were purchased from Sigma-Aldrich. Mouse anti-ubiquitin monoclonal antibody (clone P4D1) was purchased from Cell Signaling Technology. Goat anti-mouse IgG (H+L) Alexa Fluor 594 antibody, goat anti-mouse IgG (H+L) Alexa Fluor 488 antibody, goat anti-rabbit IgG (H+L) Alexa Fluor 594 antibody, goat anti-rabbit IgG (H+L) Alexa Fluor 488 antibody, goat anti-rabbit IgG (H+L) HRP antibody, and goat anti-mouse IgG+IgM (H+L) HRP antibody were purchased from Jackson ImmunoResearch. Anti-β-actin monoclonal antibody (66009-1-Ig) was purchased from Proteintech. MG132 was purchased from Calbiochem. Puromycin dihydrochloride was purchased from TOKU-E. The Pierce classic magnetic IP/coimmunoprecipitation (co-IP) kit was purchased from Thermo Scientific. The cell counting kit-8 was purchased from Sigma-Aldrich.
Reverse transcription-PCR (RT-PCR), cloning, and nucleotide sequencing.
Total RNA was extracted from GB cells using Azol RNA isolation reagent (Arrowtec) according to the manufacturer’s protocol. mRNA was reverse transcribed using the ToolsQuant II fast RT kit (Tools, Taiwan) with the provided oligo-dT primer, according to the manufacturer’s protocol. For orange-spotted grouper PABP and 26S proteasome non-ATPase regulatory subunit 6 (human gene: PSMD6) gene cloning, primer pairs (PABP-5′-UTR-F/PABP-3′-UTR-R or PSMD6-F/PSMD6-R, listed in Table 3) derived from a next-generation sequencing (NGS) database for GB cells were used. The cDNA fragments were obtained from a 30-cycle amplification using Phusion high-fidelity DNA polymerase (Thermo Fisher Scientific). The PCR-amplified DNA fragments were then cloned into RBC TA cloning vector (RBC Bioscience) to generate pTA-PABP or were cloned into pJET1.2 vector (Thermo Fisher Scientific) to generate pJET1.2-PSMD6 for nucleotide sequencing using an ABI 3730XL autosequencer (Applied Biosystems).
TABLE 3.
Primers used in this study
| Plasmid/template | Primer | Sequence (5′ to 3′) |
|---|---|---|
| pTA-PABP | PABP-5′-UTR-F | GTATTACCAAGCACGTACACG |
| PABP-3′-UTR-R | TAGTCTCAAGGCTCCCCACAC | |
| pNNVCP | NNVCP-F | GGAATTCCATATGGTACGCAAAGGTGAGAAGAAATTGG |
| NNVCP-R | CCGCTCGAGGTTTCCCGAGTCAACCCTGGTG | |
| pET-PABP1-633 aa | PABP-1-F | GGAATTCCATATGAATCCAAGTGCTCCTAGT |
| PABP-633-R | CCGCTCGAGGACACTTGGCACACCGGTAGA | |
| pET-PABP1-89 aa | PABP-1-F | GGAATTCCATATGAATCCAAGTGCTCCTAGT |
| PABP-89-R | ACCGCTCGAGGCGCTGGGACCACATGATGCG | |
| pET-PABP90-538 aa | PABP-90-F | GGAATTCCATATGGACCCGTCCCTGAGGAAGAGC |
| PABP-538-R | ACCGCTCGAGGCCCTGCACGTGGACGGC | |
| pET-PABP539-633 aa | PABP-539-F | GGAATTCCATATGCAGGAGCCCCTGACCGCCTC |
| PABP-633-R | CCGCTCGAGGACACTTGGCACACCGGTAGA | |
| pET-PABP90-370 aa | PABP-90-F | GGAATTCCATATGGACCCGTCCCTGAGGAAGAGC |
| PABP-370-R | ACCGCTCGAGGCGCTGGGCCAGCGCCAC | |
| pET-PABP176-538 aa | PABP-176-F | GGAATTCCATATGCGCAAGGAGCGCGAGGCGGAGCTCGGGGC |
| PABP-538-R | ACCGCTCGAGGCCCTGCACGTGGACGGC | |
| pET-PABP176-370 aa | PABP-176-F | GGAATTCCATATGCGCAAGGAGCGCGAGGCGGAGCTCGGGGC |
| PABP-370-R | ACCGCTCGAGGCGCTGGGCCAGCGCCAC | |
| pET-PABP269-538 aa | PABP-269-F | GGAATTCCATATGAAGGTGGAGCGGCAGACGGAGCTGAAGCG |
| PABP-538-R | ACCGCTCGAGGCCCTGCACGTGGACGGC | |
| pET-PABP269-370 aa | PABP-269-F | GGAATTCCATATGAAGGTGGAGCGGCAGACGGAGCTGAAGCG |
| PABP-370-R | ACCGCTCGAGGCGCTGGGCCAGCGCCAC | |
| pJET1.2-PSMD6 | PSMD6-F | ATGCCGCTGGAAAATCTGGAAGAAG |
| PSMD6-R | TCACATGTTGATAACCCTGGACAG | |
| pPSMD6-HA | PSMD6-EcoRI-F | CCGGAATTCATGCCGCTGGAAAATCTGGAAG |
| PSMD6-XhoI-R | CCGCTCGAGCATGTTGATAACCCTGGACAG | |
| (–) NNV RNA2 | RNA2-F | TAATCCATCACCGCTTTGCAATCACA |
| T7-RNA2-R | TAATACGACTCACTATAGGCGCCGAGTTGAGAAGCGATCAGCG | |
| (+) NNV RNA2 | T7-RNA2-F | TAATACGACTCACTATAGGTAATCCATCACCGCTTTGCAA |
| RNA2-R | CGCCGAGTTGAGAAGCGATCAGCG |
Plasmid construction, recombinant protein expression, and purification.
For ectopic expression of GGNNV coat protein or orange-spotted grouper 26S proteasome non-ATPase regulatory subunit 6 in GB cells, a DNA fragment containing the full-length open reading frame for the GGNNV coat protein or orange-spotted grouper PSMD6 was RT-PCR amplified from GGNNV or PCR amplified from the pJET1.2-PSMD6 plasmid (cloned above) using primer pairs (NNVCP-F/NNVCP-R or PSMD6-EcoRI-F/PSMD6-XhoI-R) listed in Table 3. The resultant DNA fragment was cloned into the modified pcDNA3 vector (Invitrogen) (the neomycin resistance gene was replaced with a puromycin resistance gene, and an HA tag sequence was inserted after the XhoI cloning site) to generate the pNNVCP or pPSMD6-HA plasmid. The pNNVCP and pPSMD6-HA constructs were transfected into GB cells using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s protocol. For stable clone selection, the pPSMD6-HA-transfected GB cells were treated with puromycin (1.2 μg/ml). For expression of recombinant PABP mutant proteins, the DNA fragments encoding deletions of PABP were PCR amplified using pTA-PABP (cloned above) as a template and using primers listed in Table 3. The resultant amplified DNA fragments were cloned into NdeI and XhoI sites of pET-20b(+) vector (Merck Novagen) to generate pET-PABP1 to 633 amino acid (aa), 1 to 89 aa, 90 to 538 aa, 539 to 633 aa, 90 to 370 aa, 176 to 538 aa, 176 to 370 aa, 269 to 538 aa, and 269 to 370 aa plasmids. The pET-20b(+) vector includes a His tag, which enabled the creation of a His tag fusion protein that can be purified by immobilized metal affinity chromatography. The serial pET-NNVCP mutant recombinant plasmids, including 1 to 100 aa, 1 to 150 aa, 1 to 200 aa, 1 to 338 aa (this full-length coat protein-expressed plasmid can generate VLP), 51 to 338 aa, and 195 to 338 aa, were constructed as described previously (56).
For recombinant protein expression, pET-NNVCP and pET-PABP serial constructs were transformed into either the BL21(DE3) (RBC Bioscience) or BL21 (DE3-pTf16) (TaKaRa) strain of E. coli. A chaperone plasmid, pTf16, expressed a tig chaperone to increase the recovery of expressed proteins in the soluble fraction. The transformed cells were cultured at 37 °C with 125 rpm shaking in 250 ml LB broth with 100 μg/ml ampicillin for BL21(DE3) or 100 μg/ml ampicillin plus 20 μg/ml chloramphenicol for BL21 (DE3-pTf16). When the optical density at 600 nm (OD600) reached 0.4, a final concentration of 0.4 mM IPTG (isopropyl-β-d-thiogalactopyranoside; MDBio, Taiwan) was added, and cells were cultured overnight at room temperature (RT) with 125 rpm shaking for BL21(DE3); for BL21 (DE3-pTf16) cells, 0.4 mM IPTG plus 2 mg/ml l-arabinose (Sigma) was added, and cells were cultured overnight at 16 °C with 125 rpm shaking. After IPTG induction, cells were harvested and resuspended in 10 ml binding buffer (20 mM sodium phosphate, 0.5 M NaCl, 20 mM imidazole, pH 7.4) and sonicated using a digital sonifier (Branson). The supernatant of the cell lysate was passed through a 1-ml Ni-resin column (Clontech). After washing the column with 10 ml binding buffer, the recombinant protein was eluted with another 10 ml elution buffer (20 mM sodium phosphate, 0.5 M NaCl, 500 mM imidazole, pH 7.4); 1-ml fractions were collected.
VLP preparation and electron micrograph.
To produce VLP, the pET-NNVCP 1 to 338 aa plasmid (56) was used to transform the BL-21 (DE3) strain of E. coli. The transformed cells were cultured in 16 × 250 ml LB broth with 100 μg/ml ampicillin at 37 °C with 125 rpm shaking. When the OD600 reached about 0.4, IPTG was added to a final concentration of 0.4 mM, and cells were cultured overnight at RT with 125 rpm shaking. After the cells were harvested, the pellet was resuspended in 30 ml lysis buffer (1 % Triton X-100, 2 mM phenylmethylsulfonyl fluoride [PMSF] in phosphate-buffered saline [PBS]) and sonicated using a digital sonifier (Branson). After centrifugation at 20,000 × g at 4 °C for 30 min, the supernatant was filtered through a 0.45-μm membrane. The filtrate was centrifuged at 180,000 × g at 4 °C for 4 h with a 4 ml 30 % (wt/wt) sucrose cushion using a Hitachi Himac CP-100α ultracentrifuge (RPS40T rotor). The pellet was resuspended in TNE (50 mM Tris, 100 mM NaCl, 1 mM EDTA, pH 7.3) buffer and was layered over a three-step CsCl gradient (4 ml 40 %, 3 ml 30 %, 2 ml 20 % CsCl in TNE buffer) and centrifuged at 180,000 × g at 4 °C for 16 h using a Hitachi Himac CP-100α ultracentrifuge (RPS40T rotor). The VLP band was withdrawn, and the density of this fraction was adjusted to 1.34 g/ml CsCl buffer and then centrifuged at 180,000 × g at 4 °C for 16 h using a Beckman Coulter Optima L-100K ultracentrifuge (SW55Ti rotor). Again, the VLP band was collected and diluted with 5 ml TNE buffer and recentrifuged at 180,000 × g at 4 °C for 2 h using a Beckman Coulter Optima L-100K ultracentrifuge (SW55Ti rotor). The purified VLP pellet was resuspended in 500 μl PBS, filtered through a 0.45-μm membrane, and kept at 4 °C. For negative staining, purified VLP suspension (10 μl) was placed on a 300-mesh grid and stained with 1 % uranyl acetate (EMS). The negative stained VLP was observed under an FEI Tecnai G2F20 scanning transmission electron microscope. For VLP quantitative comparison, the GGNNV-containing medium was concentrated by ultracentrifugation at 180,000 × g at 4 °C for 2 h using a Beckman Coulter Optima L-100K ultracentrifuge (SW55Ti rotor).
RNAi-mediated silencing of NNV coat protein.
To knock down NNV coat protein expression, the NNV double-stranded RNA2 (dsRNA2) was prepared in vitro using T7 RNA polymerase (Roche; catalog [cat.] no.10881767001) according to the manufacturer’s instructions. The T7 promoter sequence was added to NNV RNA2-specific primers, and T7-RNA2-F/RNA2-R and RNA2-F/T7-RNA2-R primer pairs (Table 3) were used to amplify positive and negative DNA templates, respectively. The PCR product templates were purified using a QIAquick PCR purification kit (Qiagen; 28104). Then, 1 μg purified PCR template was used to produce each single-stranded RNA (ssRNA) in a 20-μl reaction mixture containing 0.5 mM each nucleoside triphosphate (NTP; Invitrogen), 20 U RNase inhibitor (Roche), and 40 U T7 RNA polymerase (Roche) at 37 °C for 2 h. Positive and negative ssRNA were coincubated at 70 °C for 10 min and then slowly allowed to cool to RT to produce dsRNA. Before infection, a total of 1 μg of dsRNA was used to transfect cells plated on cover glass in a 3-cm dish using Lipofectamine 2000 transfection reagent (Invitrogen) for 12 h following the manufacturer’s protocol. The GGNNV-infected (MOI = 100) and dsRNA2-transfected GB cells were then subjected to immunocytochemical staining.
Western blotting and far-Western blotting.
To monitor protein synthesis, the nonradioactive SUnSET method (surface sensing of translation) was used (37). Briefly, a structural analog of aminoacyl tRNAs, puromycin (20 μg/ml), was added to the medium of GB cells 1 h before the treated cells were harvested for immunoblotting with an anti-puromycin antibody. When applied at low concentrations, puromycin is incorporated into newly synthesized proteins. Therefore, the puromycin level can directly reflect the rate of mRNA translation. For the UV-inactivation of GGNNV, virus-containing solution was irradiated in a 10-cm dish without a cover at 0.24 J/cm2 using a UV Stratalinker 1800 (Stratagene). After the treatment period, medium was removed from GB cells. The cells were washed twice with ice-cold PBS and then collected with a cell scraper (Falcon) and pelleted by centrifugation at 1,500 × g for 5 min. After the supernatants were discarded, the pellets were quickly disrupted on a vortex mixer and then lysed in an appropriate amount of lysis buffer (40 mM Tris-HCl, 2 % SDS, and 0.2 mM EDTA, pH 6.8). The protein concentrations of cell lysates were quantified using a standard dye-based protein assay (Bio-Rad). The protein samples were loaded in equal amounts for SDS-PAGE, and the resolved proteins were stained with Coomassie blue R-250 (Bio-Rad) or silver stain plus (Bio-Rad). Alternatively, the resolved proteins were transferred on a nitrocellulose membrane (Pall) for Western or far-Western blotting using a semidry transfer unit (GE Healthcare Life Sciences). After blocking with 5 % nonfat milk in Tris-buffered saline (TBS) (Omics Bio) containing 0.1 % Tween 20 (TBST) for 1 h at RT, the blocked membranes were incubated with primary antibodies diluted in 5 % nonfat milk TBST (1:5,000 dilution for mouse anti-puromycin antibody, 1:20 dilution for mouse anti-NNV coat protein monoclonal antibody, 1:1,000 dilution for rabbit anti-PABP antibody, and 1:1,000 dilution for mouse anti-ubiquitin monoclonal antibody) at 4 °C overnight. After four rounds of washing with TBST, the membranes were incubated with secondary antibodies (1:5,000 dilution for goat anti-rabbit IgG (H+L) HRP antibody or goat anti-mouse IgG+IgM (H+L) HRP antibody). After four washes with TBST, the membranes were developed with an enhanced chemiluminescence (ECL), LumiFlash infinity chemiluminescent substrate (Energenesis Biomedical), and images were obtained with a Biospectrum 600 imaging system (UVP). Anti-β-actin monoclonal antibody was used at a dilution of 1:5,000 to detect the β-actin internal control. For far-Western blotting, after blocking the transferred membrane and before applying the primary antibody, the membrane was hybridized with 200 μg of corresponding soluble recombinant proteins (rPABP 1 to 633 aa for rNNV CP membrane; rNNV CP 1 to 338 aa for rPABP membrane) in 5 % nonfat milk TBST at 4 °C overnight.
Immunocytochemistry.
The day before infection or transfection, GB cells were seeded on Deckglaser cover glass (Carolina Assistant) inside a 3-cm tissue culture dish (Falcon). For the detection of newly synthesized proteins, GB cells were labeled with puromycin (20 μg/ml) 1 h before the immunocytochemistry protocol was begun. At the indicated times, GB cells were washed with ice-cold PBS twice and then fixed with 4 % paraformaldehyde in PBS for 1 h at 4 °C. After another two washes with ice-cold PBS, the cells were permeabilized with 0.25 % Triton X-100 in PBS for 5 min. The cells were then rinsed twice with PBS and blocked with 10 % bovine serum albumin (BSA) in PBS for 30 min at RT. Primary antibodies were used as follows: 1:5,000 dilution for mouse anti-puromycin antibody, 1:20 dilution for mouse anti-NNV coat protein monoclonal antibody, 1:1,000 dilution for rabbit anti-PABP antibody, and 1:2,000 dilution for mouse anti-puromycin Alexa Fluor 488 conjugate antibody or 1:1,000 dilution for mouse anti-ubiquitin monoclonal antibody were prepared in PBS with 3 % BSA. The cells were hybridized with primary antibodies at RT for 2 h or at 4 °C overnight. The secondary antibodies were used as follows: 1:1,000 dilution for all secondary antibodies, including goat anti-mouse IgG (H+L) Alexa Fluor 594 antibody, goat anti-mouse IgG (H+L) Alexa Fluor 488 antibody, goat anti-rabbit IgG (H+L) Alexa Fluor 594 antibody, and goat anti-rabbit IgG (H+L) Alexa Fluor 488 antibody were prepared in PBS with 3 % BSA. After four washes with PBS, the cells were hybridized with secondary antibodies for 1 h at RT in the dark. After another four washes with PBS in the dark, the cover glass with stained cells was mounted onto a glass slide with DAPI (4′,6-diamidino-2-phenylindole) Fluormount-G (SouthernBiotech) and sealed with CoverGrip coverslip sealant (Biotium). Images were collected with a Zeiss Observer Z1 (×63 objective) inverted fluorescence microscope.
Coimmunoprecipitation.
The cell lysates of GGNNV-infected GB cells were immunoprecipitated with anti-NNV coat protein antibody, anti-PABP antibody, or anti-ubiquitin antibody using a Pierce classic magnetic IP/co-IP kit (Thermo Scientific) according to the manufacturer’s protocol. Briefly, before immunoprecipitation, 50 μl of protein A/G magnetic beads (10 mg/ml in water) were washed twice with 1 ml ice-cold IP lysis/wash buffer, and then the beads were incubated with 5 μg antibody in 1.5 ml ice-cold IP lysis/wash buffer at 4 °C overnight with slow rotation. The supernatant was removed with the use of a magnetic stand. Then, the antibody-protein A/G beads were washed with 1 ml ice-cold IP lysis/wash buffer twice; samples were stored on ice prior to immunoprecipitation. After washing twice with ice-cold PBS, 1 × 108 GGNNV-infected GB cells were harvested 18 hpi using a cell scraper and pelleted by centrifugation at 1,500 × g for 10 min at 4 °C. After the supernatants were discarded, the pellets were quickly disrupted on a vortex mixer and then lysed on ice in 1 ml of precooled IP lysis/wash buffer (25 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 1 % NP-40, 5 % glycerol, pH 7.4) containing 10 μl of protease inhibitor cocktail (Tools, Taiwan) with gentle pipetting to dissolve the cell pellet completely. After centrifugation at 13,000 × g at 4 °C for 10 min, the supernatant protein extract was gently mixed with the prepared antibody-protein A/G beads in 5 ml of ice-cold IP lysis/wash buffer containing 40 μl of protease inhibitor cocktail at 4 °C overnight with slow rotation. The supernatant was removed with the use of a magnetic stand, and then the precipitate was washed twice with ice-cold IP lysis/wash buffer, 1 ml each. The pellet was then washed with 1 ml deionized distilled water once. After treating the precipitate with 50 μl of elution buffer (0.2 M glycine, pH 2.6) at RT for 10 min, the beads were magnetically separated, and the supernatant containing target antigen and coimmunoprecipitated proteins was transferred to a new tube. To neutralize the low pH, 5 μl neutralization buffer (0.5 M Tris-HCl, pH 8.0) was mixed with the eluate. The resultant precipitates were subjected to SDS-PAGE, silver staining, and immunoblotting. For 26S proteasome non-ATPase regulatory subunit 6-HA coimmunoprecipitation, the cell lysates of puromycin-selected, pPSMD6-HA-transfected, and GGNNV-infected GB cells were immunoprecipitated with an anti-HA antibody.
LC-MS/MS and MASCOT analysis.
Immunoprecipitated protein samples were subjected to analysis with a QSTAR XL hybrid LC-MS/MS system (AB SCIEX, USA) after trypsin digestion. The peptide mass fingerprints were searched against the NCBI protein database using the MASCOT search engine (Matrix Science, London, UK).
Cytotoxicity assay.
GB cells were seeded on a 96-well culture plate for 15 h at 28 °C. Then, cells were incubated with L-15 medium containing 2 % FBS and supplemented with different concentrations of MG132 at 28 °C for 24 h. Cell viability was detected by absorbance at 450 nm, according to the manufacturer’s instructions for the CCK-8 kit (Sigma-Aldrich).
Statistical analysis.
Statistical data are reported as the mean ± standard deviation (SD). Statistically significant differences were identified by Student’s t test. A P value of less than 0.05 was considered to be statistically significant.
Data availability.
The GenBank accession numbers MW191575 and MW701372 are available for orange-spotted grouper PABP and PSMD6, respectively.
ACKNOWLEDGMENTS
We thank Shao-Chun Hsu (ICOB Imaging Core Facility, Academia Sinica) for technical support.
This study was supported by Academia Sinica.
Contributor Information
Chi-Yao Chang, Email: cychang@gate.sinica.edu.tw.
J.-H. James Ou, University of Southern California
REFERENCES
- 1.Yoshikoshi K, Inoue K. 1990. Viral nervous necrosis in hatchery-reared larvae and juveniles of Japanese parrotfish, Oplegnathus fasciatus (Temminck & Schlegel). J Fish Dis 13:69–77. 10.1111/j.1365-2761.1990.tb00758.x. [DOI] [Google Scholar]
- 2.Munday BL, Langdon JS, Hyatt A, Humphrey JD. 1992. Mass mortality associated with a viral-induced vacuolating encephalopathy and retinopathy of larval and juvenile barramundi, Lates Calcarifer Bloch. Aquaculture 103:197–211. 10.1016/0044-8486(92)90166-I. [DOI] [Google Scholar]
- 3.Bandin I, Souto S. 2020. Betanodavirus and VER disease: a 30-year research review. Pathogens 9:106. 10.3390/pathogens9020106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mori KI, Nakai T, Muroga K, Arimoto M, Mushiake K, Furusawa I. 1992. Properties of a new virus belonging to Nodaviridae found in larval striped jack (Pseudocaranx dentex) with nervous necrosis. Virology 187:368–371. 10.1016/0042-6822(92)90329-N. [DOI] [PubMed] [Google Scholar]
- 5.Iwamoto T, Mise K, Takeda A, Okinaka Y, Mori KI, Arimoto M, Okuno T, Nakai T. 2005. Characterization of Striped jack nervous necrosis virus subgenomic RNA3 and biological activities of its encoded protein B2. J Gen Virol 86:2807–2816. 10.1099/vir.0.80902-0. [DOI] [PubMed] [Google Scholar]
- 6.Fenner BJ, Goh W, Kwang J. 2006. Sequestration and protection of double-stranded RNA by the betanodavirus B2 protein. J Virol 80:6822–6833. 10.1128/JVI.00079-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iwamoto T, Mise K, Mori K, Arimoto M, Nakai T, Okuno T. 2001. Establishment of an infectious RNA transcription system for Striped jack nervous necrosis virus, the type species of the betanodaviruses. J Gen Virol 82:2653–2662. 10.1099/0022-1317-82-11-2653. [DOI] [PubMed] [Google Scholar]
- 8.Munroe D, Jacobson A. 1990. Tales of poly(A): a review. Gene 91:151–158. 10.1016/0378-1119(90)90082-3. [DOI] [PubMed] [Google Scholar]
- 9.Tarun SZ, Sachs AB. 1996. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J 15:7168–7177. 10.1002/j.1460-2075.1996.tb01108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tarun SZ, Wells SE, Deardorff JA, Sachs AB. 1997. Translation initiation factor eIF4G mediates in vitro poly(A) tail-dependent translation. Proc Natl Acad Sci U S A 94:9046–9051. 10.1073/pnas.94.17.9046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wells SE, Hillner PE, Vale RD, Sachs AB. 1998. Circularization of mRNA by eukaryotic translation initiation factors. Mol Cell 2:135–140. 10.1016/S1097-2765(00)80122-7. [DOI] [PubMed] [Google Scholar]
- 12.Kahvejian A, Svitkin YV, Sukarieh R, M’Boutchou MN, Sonenberg N. 2005. Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes Dev 19:104–113. 10.1101/gad.1262905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fukami-Kobayashi K, Tomoda S, Go M. 1993. Evolutionary clustering and functional similarity of RNA-binding proteins. FEBS Lett 335:289–293. 10.1016/0014-5793(93)80749-K. [DOI] [PubMed] [Google Scholar]
- 14.Mangus DA, Evans MC, Jacobson A. 2003. Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol 4:223. 10.1186/gb-2003-4-7-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kerekatte V, Keiper BD, Badorff C, Cai AL, Knowlton KU, Rhoads RE. 1999. Cleavage of poly(A)-binding protein by coxsackievirus 2A protease in vitro and in vivo: another mechanism for host protein synthesis shutoff?. J Virol 73:709–717. 10.1128/JVI.73.1.709-717.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joachims M, Van Breugel PC, Lloyd RE. 1999. Cleavage of poly(A)-binding protein by enterovirus proteases concurrent with inhibition of translation in vitro. J Virol 73:718–727. 10.1128/JVI.73.1.718-727.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuyumcu-Martinez NM, Joachims M, Lloyd RE. 2002. Efficient cleavage of ribosome-associated poly(A)-binding protein by enterovirus 3C protease. J Virol 76:2062–2074. 10.1128/jvi.76.5.2062-2074.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuyumcu-Martinez M, Belliot G, Sosnovtsev SV, Chang KO, Green KY, Lloyd RE. 2004. Calicivirus 3C-like proteinase inhibits cellular translation by cleavage of poly(A)-binding protein. J Virol 78:8172–8182. 10.1128/JVI.78.15.8172-8182.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alvarez E, Castello A, Menendez-Arias L, Carrasco L. 2006. HIV protease cleaves poly(A)-binding protein. Biochem J 396:219–226. 10.1042/BJ20060108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Castello A, Franco D, Moral-Lopez P, Berlanga JJ, Alvarez E, Wimmer E, Carrasco L. 2009. HIV-1 protease inhibits cap- and poly(A)-dependent translation upon eIF4GI and PABP cleavage. PLoS One 4:e7997. 10.1371/journal.pone.0007997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pulido MR, Serrano P, Saiz M, Martinez-Salas E. 2007. Foot-and-mouth disease virus infection induces proteolytic cleavage of PTB, eIF3a,b, and PABP RNA-binding proteins. Virology 364:466–474. 10.1016/j.virol.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 22.Zhang B, Morace G, Gauss-Muller V, Kusov Y. 2007. Poly(A) binding protein, C-terminally truncated by the hepatitis A virus proteinase 3C, inhibits viral translation. Nucleic Acids Res 35:5975–5984. 10.1093/nar/gkm645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kobayashi M, Arias C, Garabedian A, Palmenberg AC, Mohr I. 2012. Site-specific cleavage of the host poly(A) binding protein by the encephalomyocarditis virus 3C proteinase stimulates viral replication. J Virol 86:10686–10694. 10.1128/JVI.00896-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun D, Wang MS, Wen XJ, Cheng AC, Jia RY, Sun KF, Yang Q, Wu Y, Zhu DK, Chen S, Liu MF, Zhao XX, Chen XY. 2017. Cleavage of poly(A)-binding protein by duck hepatitis A virus 3C protease. Sci Rep 7:16261. 10.1038/s41598-017-16484-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Afonina E, Stauber R, Pavlakis GN. 1998. The human poly(A)-binding protein 1 shuttles between the nucleus and the cytoplasm. J Biol Chem 273:13015–13021. 10.1074/jbc.273.21.13015. [DOI] [PubMed] [Google Scholar]
- 26.Kanno T, Sato Y, Sata T, Katano H. 2006. Expression of Kaposi's sarcoma-associated herpesvirus-encoded K10/10.1 protein in tissues and its interaction with poly(A)-binding protein. Virology 352:100–109. 10.1016/j.virol.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 27.Blakqori G, van Knippenberg I, Elliott RM. 2009. Bunyamwera Orthobunyavirus S-segment untranslated regions mediate poly(A) tail-independent translation. J Virol 83:3637–3646. 10.1128/JVI.02201-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dobrikova E, Shveygert M, Walters R, Gromeier M. 2010. Herpes simplex virus proteins ICP27 and UL47 associate with polyadenylate-binding protein and control its subcellular distribution. J Virol 84:270–279. 10.1128/JVI.01740-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Copeland AM, Altamura LA, Van Deusen NM, Schmaljohn CS. 2013. Nuclear relocalization of polyadenylate binding protein during Rift Valley fever virus infection involves expression of the NSs gene. J Virol 87:11659–11669. 10.1128/JVI.01434-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Copeland AM, Van Deusen NM, Schmaljohn CS. 2015. Rift Valley fever virus NSs gene expression correlates with a defect in nuclear mRNA export. Virology 486:88–93. 10.1016/j.virol.2015.09.003. [DOI] [PubMed] [Google Scholar]
- 31.Hosoda N, Lejeune F, Maquat LE. 2006. Evidence that poly(A) binding protein C1 binds nuclear pre-mRNA poly(A) tails. Mol Cell Biol 26:3085–3097. 10.1128/MCB.26.8.3085-3097.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ilkow CS, Mancinelli V, Beatch MD, Hobman TC. 2008. Rubella virus capsid protein interacts with poly(A)-binding protein and inhibits translation. J Virol 82:4284–4294. 10.1128/JVI.02732-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Piron M, Vende P, Cohen J, Poncet D. 1998. Rotavirus RNA-binding protein NSP3 interacts with eIF4GI and evicts the poly(A) binding protein from eIF4F. EMBO J 17:5811–5821. 10.1093/emboj/17.19.5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harb M, Becker MM, Vitour D, Baron CH, Vende P, Brown SC, Bolte S, Arold ST, Poncet D. 2008. Nuclear localization of cytoplasmic poly(A)-binding protein upon Rotavirus infection involves the interaction of NSP3 with eIF4G and RoXaN. J Virol 82:11283–11293. 10.1128/JVI.00872-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee YJ, Glaunsinger BA. 2009. Aberrant herpesvirus-induced polyadenylation correlates with cellular messenger RNA destruction. PLoS Biol 7:e1000107. 10.1371/journal.pbio.1000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Glaunsinger B, Chavez L, Ganem D. 2005. The exonuclease and host shutoff functions of the SOX protein of Kaposi’s sarcoma-associated herpesvirus are genetically separable. J Virol 79:7396–7401. 10.1128/JVI.79.12.7396-7401.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmidt EK, Clavarino G, Ceppi M, Pierre P. 2009. SUnSET, a nonradioactive method to monitor protein synthesis. Nat Methods 6:275–277. 10.1038/nmeth.1314. [DOI] [PubMed] [Google Scholar]
- 38.Guo YX, Dallmann K, Kwang J. 2003. Identification of nucleolus localization signal of betanodavirus GGNNV protein alpha. Virology 306:225–235. 10.1016/s0042-6822(02)00081-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen NC, Yoshimura M, Guan HH, Wang TY, Misumi Y, Lin CC, Chuankhayan P, Nakagawa A, Chan SI, Tsukihara T, Chen TY, Chen CJ. 2015. Crystal structures of a piscine Betanodavirus: mechanisms of capsid assembly and viral infection. PLoS Pathog 11:e1005203. 10.1371/journal.ppat.1005203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marissen WE, Triyoso D, Younan P, Lloyd RE. 2004. Degradation of poly(A)-binding protein in apoptotic cells and linkage to translation regulation. Apoptosis 9:67–75. 10.1023/B:APPT.0000012123.62856.20. [DOI] [PubMed] [Google Scholar]
- 41.Connor JH, Lyles DS. 2002. Vesicular stomatitis virus infection alters the eIF4F translation initiation complex and causes dephosphorylation of the eIF4E binding protein 4E-BP1. J Virol 76:10177–10187. 10.1128/jvi.76.20.10177-10187.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Y, Fang LR, Zhou YR, Tao R, Wang D, Xiao SB. 2018. Porcine reproductive and respiratory syndrome virus infection induces both eIF2 alpha phosphorylation-dependent and -independent host translation shutoff. J Virol 92:15. 10.1128/JVI.00600-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liao Y, Gu F, Mao X, Niu QN, Wang HX, Sun YJ, Song CP, Qiu XS, Tan L, Ding C. 2016. Regulation of de novo translation of host cells by manipulation of PERK/PKR and GADD34-PP1 activity during Newcastle disease virus infection. J Gen Virol 97:867–879. 10.1099/jgv.0.000426. [DOI] [PubMed] [Google Scholar]
- 44.Ventoso I, Blanco R, Perales C, Carrasco L. 2001. HIV-1 protease cleaves eukaryotic initiation factor 4G and inhibits cap-dependent translation. Proc Natl Acad Sci U S A 98:12966–12971. 10.1073/pnas.231343498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walsh D, Perez C, Notary J, Mohr I. 2005. Regulation of the translation initiation factor eIF4F by multiple mechanisms in human cytomegalovirus-infected cells. J Virol 79:8057–8064. 10.1128/JVI.79.13.8057-8064.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Patel GP, Ma SH, Bag J. 2005. The autoregulatory translational control element of poly(A)-binding protein mRNA forms a heteromeric ribonucleoprotein complex. Nucleic Acids Res 33:7074–7089. 10.1093/nar/gki1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guo YX, Wei T, Dallmann K, Kwang J. 2003. Induction of caspase-dependent apoptosis by betanodaviruses GGNNV and demonstration of protein α as an apoptosis inducer. Virology 308:74–82. 10.1016/S0042-6822(02)00098-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Palombella VJ, Rando OJ, Goldberg AL, Maniatis T. 1994. The ubiquitin-proteasome pathway is required for processing the NF-κB1 precursor protein and tne activation of NF-κB. Cell 78:773–785. 10.1016/S0092-8674(94)90482-0. [DOI] [PubMed] [Google Scholar]
- 49.Walsh D, Mathews MB, Mohr I. 2013. Tinkering with translation: protein synthesis in virus-infected cells. Cold Spring Harb Perspect Biol 5:a012351. 10.1101/cshperspect.a012351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bonderoff JM, Larey JL, Lloyd RE. 2008. Cleavage of poly(A)-binding protein by poliovirus 3C proteinase inhibits viral internal ribosome entry site-mediated translation. J Virol 82:9389–9399. 10.1128/JVI.00006-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bard JAM, Goodall EA, Greene ER, Jonsson E, Dong KC, Martin A. 2018. Structure and function of the 26S proteasome. Annu Rev Biochem 87:697–724. 10.1146/annurev-biochem-062917-011931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Choi AG, Wong J, Marchant D, Luo H. 2013. The ubiquitin-proteasome system in positive-strand RNA virus infection. Rev Med Virol 23:85–96. 10.1002/rmv.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Camborde L, Planchais S, Tournier V, Jakubiec A, Drugeon G, Lacassagne E, Pflieger S, Chenon M, Jupin I. 2010. The ubiquitin-proteasome system regulates the accumulation of Turnip yellow mosaic virus RNA-dependent RNA polymerase during viral infection. Plant Cell 22:3142–3152. 10.1105/tpc.109.072090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shirakura M, Murakami K, Ichimura T, Suzuki R, Shimoji T, Fukuda K, Abe K, Sato S, Fukasawa M, Yamakawa Y, Nishijima M, Moriishi K, Matsuura Y, Wakita T, Suzuki T, Howley PM, Miyamura T, Shoji I. 2007. E6AP ubiquitin ligase mediates ubiquitylation and degradation of hepatitis C virus core protein. J Virol 81:1174–1185. 10.1128/JVI.01684-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lin CH, John JAC, Lin CH, Chang CY. 2006. Inhibition of nervous necrosis virus propagation by fish Mx proteins. Biochem Biophys Res Commun 351:534–539. 10.1016/j.bbrc.2006.10.063. [DOI] [PubMed] [Google Scholar]
- 56.Chen CW, Wu MS, Huang YJ, Cheng CA, Chang CY. 2015. Recognition of linear B-cell epitope of Betanodavirus coat protein by RG-M18 neutralizing mAb inhibits giant grouper nervous necrosis virus (GGNNV) infection. PLoS One 10:e0126121. 10.1371/journal.pone.0126121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lai YS, Chiu HC, Murali S, Guo IC, Chen SC, Fang K, Chang CY. 2001. In vitro neutralization by monoclonal antibodies against yellow grouper nervous necrosis virus (YGNNV) and immunolocalization of virus infection in yellow grouper, Epinephelus awoara (Temminck & Schlegel). J Fish Diseases 24:237–244. 10.1046/j.1365-2761.2001.00293.x. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The GenBank accession numbers MW191575 and MW701372 are available for orange-spotted grouper PABP and PSMD6, respectively.