Abstract
Long non-coding RNA (lncRNA) small nucleolar RNA host gene 11 (SNHG11) has been shown to play an important role in the development and progression of numerous types of cancer. However, to the best of our knowledge, the role of SNHG11 in prostate cancer (PCa) development and metastasis remains unclear. Thus, the aim of the present study was to investigate the functional role and molecular mechanisms of SNHG11 in PCa progression. It was revealed that the SNHG11 expression levels were significantly upregulated in PCa tissues, in comparison with those in adjacent normal tissues. Functionally, SNHG11 knockdown significantly suppressed PCa cell proliferation, migration, invasion and metastasis in vitro and in vivo. Furthermore, SNHG11 was found to positively regulate insulin-like growth factor 1 receptor (IGF-1R) expression by sponging microRNA (miRNA/miR)-184 in PCa cells. The results of rescue experiments demonstrated that IGF-1R overexpression reversed the suppressive effects of SNHG11 knockdown on the proliferation, migration and invasion of PCa cells. On the whole, the findings of the present study suggest that SNHG11 expression is upregulated in PCa and that it facilitates PCa progression, at least in part, via the modulation of the miR-184/IGF-1R signaling axis.
Keywords: small nucleolar RNA host gene 11, microRNA-184, insulin-like growth factor 1 receptor, prostate cancer
Introduction
Prostate cancer (PCa) is the most common type of cancer among males and the second leading cause of cancer-related mortality worldwide (1). In recent decades, the 5-year survival rate for patients with primary PCa has increased due to the significant progress made in the development of treatment methods for primary PCa; however, the 5-year survival rate for patients with advanced PCa remains unsatisfactory, due to the occurrence of distant metastases (2). Thus, it remains an urgent priority to determine the underlying mechanisms contributing to the development and metastasis of PCa.
Long non-coding RNAs (lncRNAs) are non-coding RNAs of >200 nucleotides in length (3). It has been revealed that lncRNAs play important roles in the onset, progression and metastasis of the majority of types of cancer (4). Numerous lncRNAs, including long intergenic non-protein coding 844 (5), maternally expressed 3 (6), colon cancer associated transcript 1 (7) and zinc finger E-box binding homeobox 1 antisense RNA 1 (8), have been reported to be involved in the development and metastasis of PCa. Notably, it was recently demonstrated that lncRNA small nucleolar RNA host gene 11 (SNHG11) may be involved in the regulation of the progression of various tumor types. For example, Liu et al (9) demonstrated that the upregulated expression of SNHG11 predicted a poor prognosis of patients with lung cancer and that the overexpression of SNHG11 facilitated the development of lung cancer. In gastric cancer, SNHG11 has been reported to promote gastric cancer cell proliferation and metastasis (10). In addition, SNHG11 expression has been discovered to be upregulated in colorectal cancer cells and to be associated with increased levels of cell proliferation (11). However, to the best of our knowledge, to date, limited information is available on the expression and function of SNHG11 in PCa.
In the present study, SNHG11 expression in PCa was investigated, and the aim of the study was to determine whether SNHG11 knockdown inhibits the proliferation, migration and invasion of PCa cells. Furthermore, the present study was investigated whether the suppressive effects of SNHG11 knockdown on PCa progression are achieved through the downregulation of insulin-like growth factor 1 receptor (IGF-1R) expression via sponging microRNA (miRNA/miR)-184. Taken together, in the present study, it was revealed that SNHG11 may serve as a promising therapeutic target for PCa.
Materials and methods
Patient samples
A total of 30 PCa and adjacent normal tissue samples from patients with PCa were obtained. Written informed consent was signed by all patients and the study protocol was approved by The First Affiliated Hospital of University of South China Ethnics Committee (approval no. LL20201103017).
Cell lines and culture
A normal human prostate epithelial cell line (RWPE-1, cat. no. SCSP-5025) and human PCa cell lines (LNCaP, cat. no. TCHu173; C4-2, cat. no. CL-0046; PC3, cat. no. TCHu158; and DU145, cat. no. TCHu222) were purchased from The Cell Bank of Type Culture Collection of The Chinese Academy of Sciences. RWPE-1 cells were cultured in 1X Defined Keratinocyte SFM (Gibco; Thermo Fisher Scientific, Inc.). All PCa cell lines were cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) and maintained in 5% CO2 at 37°C.
Cell transfection
A lentiviral short hairpin RNA (shRNA/sh) vector targeting SNHG11 (sh-SNHG11#1, 5′-GGA GTG GTC TTC CCA AGA A-3′; and sh-SNHG11#2, 5′-CCT CTC ACC CAC TCA ATA A-3′) and sh-negative control (NC; 5′-UUC UCC GAA CGU GUC ACG UU-3′) were purchased from Shanghai GenePharma Co., Ltd. miR-184 mimic (5′-GGC AUUCUG UAU ACA UCG GAG -3′), miR-184 inhibitor (5′-CAG UAC UUU UGU GUA GUA CAA -3′), mimic-NC (5′-UUC UCC GAA CGU GUC ACG UTT -3′) and inhibitor-NC (5′-CAG UAC UUU UGU GUA GUA CAA -3′) were synthesized by Guangzhou RiboBio Co., Ltd. IGF-1R overexpression plasmid [pcDNA3.1 (+) IGF-1R] and its NC [pcDNA3.1 (+)] were constructed by Shanghai GenePharma Co., Ltd. RNAs (100 nM) or miR-184 mimics (50 nM) or miR-184 inhibitor (150 nM) or plasmids (1.5 µg per well) were transfected into the cells. All cell transfections were performed using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). Transfection was performed at room temperature for 30 min. Following incubation for 48 h at 37°C, the cells were collected for use in subsequent experiments by ultracentrifugation (4°C; 1,000 x g; 10 min). The lentivirus was prepared according to the User Manual of the Lenti-Pac™ HIV Expression Packaging Kit (GeneCopoeia, Inc.). Viral packaging was performed in 293T cells (SCSP-502; The Cell Bank of Type Culture Collection of the Chinese Academy of Sciences) following co-transfection of the Lv3-si-SNHG11#2, or empty lentiviral vector, and Lenti-Pac™ HIV packaging mix (GeneCopoeia, Inc.) using EndoFectin™ Lenti transfection reagent (GeneCopoeia, Inc.). The medium containing the retroviral supernatant was harvested at 48-72 h following transfection. The retroviruses were added into the cells for infection. The cells were selected by culture in the presence of puromycin (5 µg/ml; Invitrogen; Thermo Fisher Scientific, Inc.) for up to 2 weeks. All cell transfections were performed using Lipofectamine® LTX reagent (Invitrogen; Thermo Fisher Scientific, Inc.).
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted using the phenol-chloroform method. Total RNA was reverse transcribed into cDNA using a One Step PrimeScript miRNA cDNA Synthesis kit (Takara Bio, Inc.). RT-qPCR was subsequently performed using a SYBR-Green PCR kit (Takara Bio, Inc.). The PCR protocol consisted of cycling at 94°C for 3 min, followed by 30 cycles of 94°C for 30 sec, 55°C for 30 sec and 72°C for 60 sec and a final extension at 72°C for 5 min. GAPDH or U6 served as the internal reference controls. The primer sequences (Shanghai GenePharma Co. Ltd.) used for RT-qPCR are listed in Table I. The ΔCt data and ΔΔCt data were calculated using the following formula: ΔCt=Ct of target gene-Ct of reference gene. ΔΔCt=ΔCt of the experimental group-ΔCt of the control group. Target gene relative expression levels were calculated using the 2−ΔΔCq method (12).
Table I.
Primers used for RT-qPCR.
| Gene | Primer sequences (5′-3′) |
|---|---|
| SNHG11 | F: 5′-TGGGAGTTGTCATGTTGGGA-3′ |
| R: 5′-ACTCGTCACTCTTGGTCTGT-3′ | |
| miR-184 | F: 5′-GCATGCCTAAATGTTGACAGCC-3′ |
| R: 5′-GTGCAGGGTCCGAGGT-3′ | |
| U6 | F: 5′-CTCGCTTCGGCAGCACATATACT-3′ |
| R: 5′-ACGCTTCACGAATTTGCGTGTC-3′ | |
| IGF-1R | F: 5′-GGAGGCTGAATACCGCAAAGTC-3′ |
| R: 5′-AAAGACGAAGTTGGAGGCGCT-3′ | |
| GAPDH | F: 5′-ATGTTCGTCATGGGTGTGAA-3′ |
| R: 5′-CAGTGATGGCATGGACTGT-3′ |
SNHG11. SNHG11, lncRNA small nucleolar RNA host gene 11; IGF-R1, insulin-like growth factor 1 receptor; F, forward; R, reverse.
Colony formation assay
A total of 800 cells per well were plated in 6-well plates and cultured in growth medium (RPMI-1640 medium supplemented with 10% FBS) for 2 weeks. Following incubation for 2 weeks at 37°C, the colonies were fixed using paraformaldehyde and stained using 0.1% crystal violet solution (Sigma-Aldrich Merck KGaA). Images of the 6-well plates were obtained and colonies containing >50 cells were counted using ImageJ software V1.8.0 (National Institutes of Health).
Transwell assays
Migration and invasion assays were conducted using Transwell chambers (8-µm pore size; BD Biosciences) with (invasion assay) or without (migration assay) Matrigel precoating. Briefly, the upper chamber was filled with 200 µl serum-free RPMI-1640 medium containing 6×104 cells, while the lower chamber was filled with 750 µl RPMI-1640 10% serum-containing medium. Following 48 h of incubation at 37°C, the cells remaining on the upper surface of the membrane were removed, while the cells on the lower surface were fixed with ethanol at room temperature for 10 min and stained with 0.1% crystal violet at room temperature for 10 min. The migratory or invasive cells were visualized using a microscope (magnification, ×200; CKX41, Olympus Corporation).
Dual luciferase reporter assay
StarBase software V2.0 (http://starbase.sysu.edu.cn) was used to predict potential target miRNAs of SNHG11. Subsequently, the target gene of miR-184 was predicted using the bioinformatics tools, ComiRNet (http://comirnet.di.uniba.it:8080/). The wild-type (WT) or mutant (MUT) 3′-untranslated region of SNHG11 was cloned into the pmirGLO Dual Luciferase vector (Promega Corporation). pmirGLO-SNHG11-WT/MUT (1.0 µg per well) vectors were co-transfected into PC3 and DU145 cells alongside the mimic-NC (40 nM) or miR-184 mimic (40 nM). Similarly, pmirGLO-IGF-1R-WT or pmirGLO-IGF-1R-MUT vectors (1.0 µg per well) were established and co-transfected with miR-184 mimic (40 nM) or NC-mimic (40 nM) into PC3 and DU145 cells using Lipofectamine® LTX (Invitrogen; Thermo Fisher Scientific, Inc.). Following 48 h of transfection, relative luciferase activity was measured using a Dual Luciferase Reporter assay system (Promega Corporation).
RNA immunoprecipitation (RIP) assay
RIP assay was performed using a Magna RIP kit (cat. no. 17-700, MilliporeSigma). Briefly, cells were lysed in RIPA lysis buffer containing magnetic beads conjugated with human anti-argonaute (Ago2) antibody (cat. no. ab186733, 1:300; Abcam) or anti-IgG antibody (cat. no. MA5-27548, 1:200; MilliporeSigma). SNHG11 and miR-184 expression levels were determined using RT-qPCR.
Western blotting
Western blotting was performed as previously described (13). Briefly, total protein was extracted from the cells using RIPA lysis buffer. Total protein was quantified using the BCA protein assay kit (cat. no. 23225, Pierce; Thermo Fisher Scientific, Inc.) and 40 µg protein/lane was separated via 10% SDS-PAGE. The proteins were subsequently transferred onto PVDF membranes and blocked with 5% skimmed milk for 2 h at room temperature. The membranes were then incubated with the following primary antibodies at 4°C overnight: Anti-IGF-1R (1:1,000, cat. no. ab182408; Abcam) and anti-GAPDH (1:4,000; cat. no. ab181602, Abcam). Following incubation with the primary antibodies, the membranes were incubated with an HRP-conjugated goat anti-rabbit IgG secondary antibody (1:2,000; cat. no. ab6728, Abcam) for 1 h at room temperature. Protein bands were visualized using the LI-COR Odyssey® CLX Two-colour infrared laser imaging system (LI-COR Biosciences). And densitometric analysis was performed using ImageJ V1.8.0 software (National Institutes of Health).
Hem a tox ylin a n d eosin (H& E) staining a n d immunohistochemistry (IHC)
Tissue slices were deparaffinized in xylene and hydrated in decreasing concentrations of alcohol prior to hematoxylin and eosin (H&E) staining. IHC was performed according to a standard IHC protocol. Briefly, the sections were deparaffinized, rehydrated and endogenous peroxidase activity was blocked in 3% fresh H2O2 for 10 min. Specimens on the slides were microwaved in citrate buffer antigen retrieval solution (Vector Laboratories, Inc.) twice for 5 min each and washed with PBS for 5 min. After the slides were blocked with 10% normal goat serum in PBS for 30 min, the sections were incubated with the primary antibodies, mouse anti-IGF-1R (1:1,000, cat. no. ab182408; Abcam) or mouse anti-Ki-67 (1:1,000, cat. no. ab279653; Abcam) overnight at 4°C, and the appropriate HRP-conjugated secondary antibody (cat. no. BM3894; Wuhan Boster Biological Technology, Ltd.) was applied at a dilution of 1:100 for 1 h at room temperature. Immunoreactivity was subsequently detected using DAB (Vector Laboratories, Inc.). The percentage of positive cells was scored as follows: 0 (0-5%), 1 (6-25%), 2 (26-50%), 3 (51-75%) and 4 (>75%). The staining intensity was scored as follows: 0 (negative), 1 (weak), 2 (moderate) and 3 (strong). The final immunoreactivity score was calculated by multiplying the percentage score with the intensity score.
Establishment of xenograft and lung metastasis models
A total of 20 male nude mice (age, 5 weeks; weight, 18-20 g) were obtained from the Experimental Animal Center of Guangdong Province and were housed five per cage in wire-top cages with sawdust bedding in an isolated, clean, air-conditioned room at a temperature of 25-26°C and a relative humidity of ~50%, with light for 12 h/day and with free access to fresh water and a solid pellet diet. All procedures were performed in strict accordance with guidelines issued by and following the approval of the Animal Care Commission of The First Affiliated Hospital of University of South China (Hengyang, China; approval no. LL20201103017).
For the tumorigenesis assay, 100 µl PC-3 cells (2×106 cells) infected with sh-SNHG11 or sh-NC were subcutaneously injected into the right side of the axillary region of the mice. The mice were randomly divided into 2 groups (n=5/group). Tumor volume was detected every 4 days based on the following equation: Volume (mm3)=length x width2 (mm)/2. Following 4 weeks of tumor growth, the mice were euthanized by cervical dislocation, under anesthesia with 1% pentobarbital (50 mg/kg; intraperitoneal).
For the metastasis assay, 3×106 sh-SNHG11- or sh-NC-transfected PC-3 cells were transfected with Firefly luciferase vector and then injected into the tail vein of mice (n=5/group) in 150 µl PBS. Lung metastases were monitored by bioluminescent imaging for 8 consecutive weeks. After 8 weeks, tumor cell metastasis was imaged using a Xenogen IVIS Spectrum Imaging system (PerkinElmer, Inc.). The mice were then euthanized by cervical dislocation under anesthesia with intraperitoneal 1% pentobarbital (50 mg/kg) and the lungs were harvested. The following signs were used to verify the death of the mice: The eyes had turned pale, no heartbeat, and no response to external stimuli. Prior to collecting the lungs, the mice were euthanized by cervical dislocation under anesthesia with 1% pentobarbital (50 mg/kg) by intraperitoneal injection.
Statistical analysis
Statistical analyses were performed using SPSS 18.0 software (SPSS, Inc.). All experiments were conducted in triplicate and data are expressed as the mean ± SD. For the comparison of matched samples (PCa and normal adjacent tissues), a paired Student's t-test was performed. For the other comparisons between two groups, an unpaired t-test was used. For the comparison of multiple groups, one-way ANOVA analysis was performed followed by a Bonferroni's post hoc test. Pearson's correlation analysis was applied, to assess the correlation between the expression levels of different genes. The analyses of ordinal variable were performed using the Mann-Whitney test (also known as the Wilcoxon rank-sum test). A Levene test was used to assess the equality of variances. P<0.05 was considered to indicate a statistically significant difference.
Results
SNHG11 expression levels are upregulated in PCa tissues and cell lines
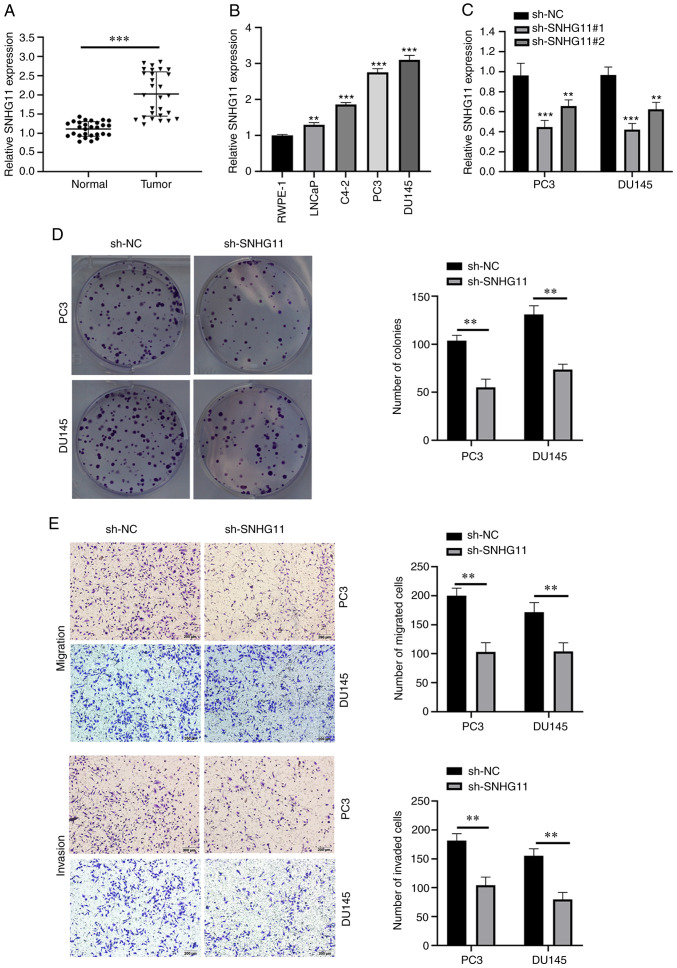
As shown in Fig. 1A, the SNHG11 expression levels were markedly upregulated in PCa tissues compared with those in adjacent normal tissues. In addition, SNHG11 expression was notably upregulated in the PCa cell lines, LNCaP, C4-2, PC3 and DU145 compared with the normal cell line, RWPE-1 (Fig. 1B).
Figure 1.
SNHG11 knockdown inhibits PCa cell proliferation, migration and invasion. (A) SNHG11 expression levels were increased in PCa tissues in comparison with those in normal tissues. (B) SNHG11 expression was significantly increased in PCa cell lines (LNCaP, C4-2, PC3, DU145) compared with normal prostate epithelial cell line (RWPE-1). (C) SNHG11 knockdown efficiency in PC3 and DU145 cells detected using RT-qPCR. (D) Colony formation assays suggested that the silencing of SNHG11 suppressed the conony-forming of PC3 and DU145 cells. (E) SNHG11 silencing inhibited the invasion and migration of PC3 and DU145 cells. **P<0.01 and ***P<0.001 in comparison with the control group. SNHG11, lncRNA small nucleolar RNA host gene 11; PCa, prostate cancer.
Silencing SNHG11 inhibits the colony formation, migration and invasion of PCa cells
To determine the roles of SNHG11 in PCa development, a lentiviral vector silencing SNHG11 expression was constructed (Fig. 1C). Since sh-SNHG11#1 exhibited the most prominent inhibitory efficiency, sh-SNHG11#1 was selected for use in subsequent experiments and referred to as sh-SNHG11 henceforth. Notably, the silencing of SNHG11 expression in PC3 and DU145 cells significantly suppressed their proliferation (Fig. 1D). In addition, SNHG11 knockdown significantly weakened the migratory and invasive abilities of the PC3 and DU145 cells (Fig. 1E).
SNHG11 directly targets the miR-184/IGF-1R signaling axis
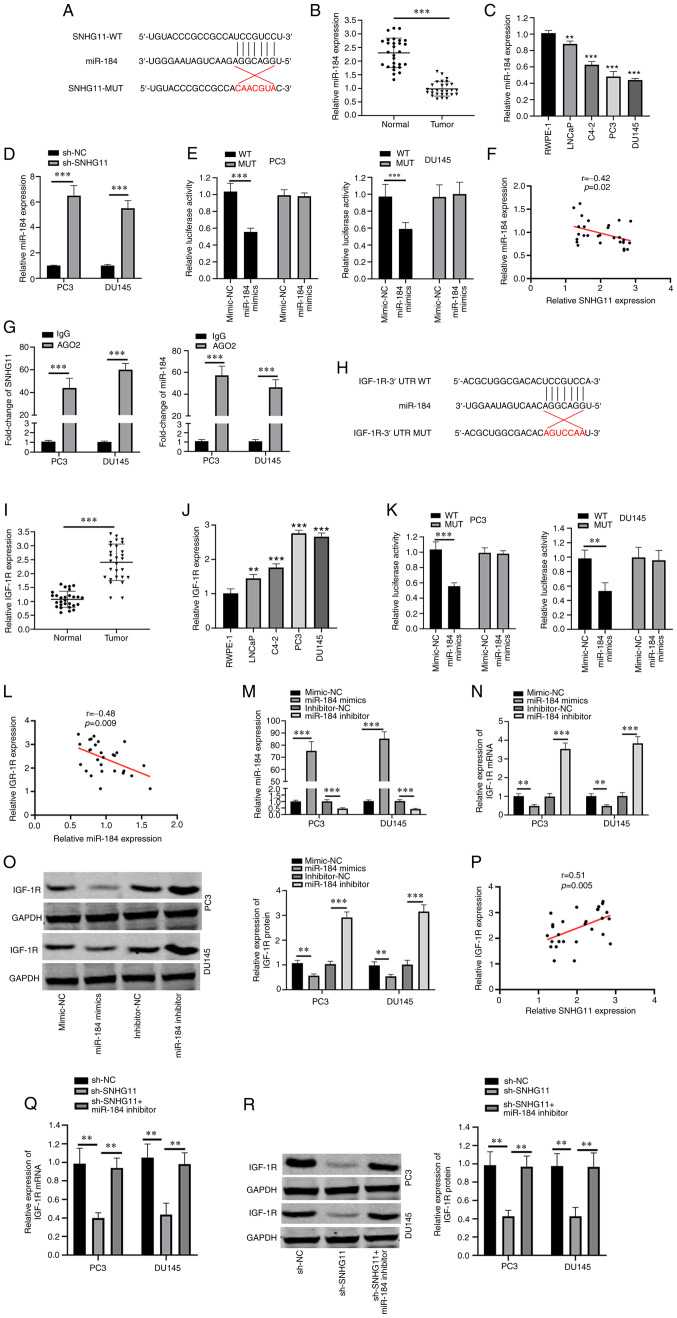
StarBase software V2.0 (http://starbase.sysu.edu.cn) was used to predict potential target miRNAs of SNHG11, which identified miR-184 as a candidate target of SNHG11 (Fig. 2A). The expression levels of miR-184 were found to be downregulated in PCa tissues and cell lines (LNCaP, C4-2, PC3 and DU145) (Fig. 2B and C). To determine whether SNHG11 regulates miR-184 expression, miR-184 expression was measured in sh-SNHG11-transfected PC3 and DU145 cells and a negative group. Notably, miR-184 expression was significantly upregulated following SNHG11 knockdown (Fig. 2D). The results of dual luciferase activity assay revealed that co-transfection with miR-184 mimic attenuated the relative luciferase activity of pmirGLO-SNHG11-WT, but not that of pmirGLO-SNHG11-MUT (Fig. 2E). Furthermore, a negative correlation was identified between SNHG11 and miR-184 expression in PCa tissues (Fig. 2F). Moreover, the results of RIP assay demonstrated that SNHG11 and miR-184 were more abundant in the anti-Ago2 group (Fig. 2G).
Figure 2.
SNHG11 acts as a sponge for miR-495. (A) Predicted binding site with miR-184 in SNHG11 through bioinformatics analysis. (B) miR-184 expression levels were markedly reduced in PCa tissues compared with that in normal tissues. (C) miR-184 expression levels were markedly decreased in PCa cell lines (LNCaP, C4-2, PC3, DU145) compared with RWPE-1 cell. (D) Silencing SNHG11 increased miR-184 relative expression levels in PC3 and DU145 cells. (E) Luciferase reporter assay revealed that miR-184 mimics inhibited the SNHG20-WT activity in PC3 and DU145 cells. (F) SNHG11 expression negatively correlated with miR-184 expression in PCa tissues. (G) RIP assay results suggested the enrichment of SNHG11 and miR-184 in Ago2-containing beads. (H) Predicted binding sites between miR-184 and IGF-1R through bioinformatics analysis. (I) IGF-1R levels in PCa tissues were significantly increased in comparison with normal tissues. (J) IGF-1R expression was significantly increased in PCa cell lines (LNCaP, C4-2, PC3, DU145), as compared with RWPE-1 cell. (K) miR-184 mimics significantly reduced IGF-1R-WT luciferase activity in PC3 and DU145 cells. (L) IGF-1R expression was negatively associated with miR-184 expression in PCa tissues. (M) Effects of miR-184 mimics or miR-184 inhibitor on miR-184 expression in PC3 and DU145 cells as detected using RT-qPCR. (N and O) miR-184 markedly reduced IGF-1R mRNA and protein levels in PC3 and DU145 cells; however, this was reversed with the use of miR-184 inhibitor. (P) SNHG11 expression positively correlated with IGF-1R expression in PCa tissues. (Q and R) SNHG11 knockdown inhibited the IGF-1R mRNA and protein expression levels, and these effects were reversed with the addition of the miR-184 inhibitor in PC3 and DU145 cells. **P<0.01 and ***P<0.001, in comparison with the control group. SNHG11, lncRNA small nucleolar RNA host gene 11; PCa, prostate cancer; IGF-R1, insulin-like growth factor 1 receptor.
Subsequently, the target gene of miR-184 was predicted using the bioinformatics tools, ComiRNet (http://comirnet.di.uniba.it:8080/). IGF-1R has been reported to be overexpressed in a number of human cancers and to play a vital role in the cancer progression (14). IGF-1R signaling is important for prostate cancer progression and development. Therefore, IGF-R1 was selected as a target candidate. The results demonstrated that IGF-1R was a candidate target of miR-184 (Fig. 2H). IGF-1R expression levels in PCa tissues and cell lines (LNCaP, C4-2, PC3 and DU145) were found to be upregulated, in comparison with those in adjacent normal tissue and RWPE-1 cells, respectively (Fig. 2I and J). In addition, the relative luciferase activity of pmirGLO-IGF-1R-WT was suppressed following co-transfection with miR-184 mimic (Fig. 2K). Furthermore, a negative correlation was observed between the miR-184 and IGF-1R expression levels in PCa tissues (Fig. 2L). To determine whether IGF-1R expression was targeted by miR-184, a miR-184 mimic and miR-184 inhibitor were transfected into PC3 and DU145 cells (Fig. 2M). Transfection with the miR-184 mimic suppressed the IGF-1R mRNA and protein expression levels, while transfection with the miR-184 inhibitor resulted in an increase in IGF-1R expression levels (Fig. 2N and O). As was expected, a positive correlation was identified between SNHG11 and IGF-1R expression in the PCa tissues (Fig. 2P). Of note, IGF-1R mRNA and protein expression levels were significantly decreased following SNHG11 knockdown, while this suppressive effect was reversed by transfection with miR-184 inhibitor (Fig. 2Q and R). According to the aforementioned data, it was suggested that SNHG11 may act as a sponge for miR-184 to upregulate IGF-1R expression.
IGF-1R overexpression reverses the effects of SNHG11 knockdown in PCa cells
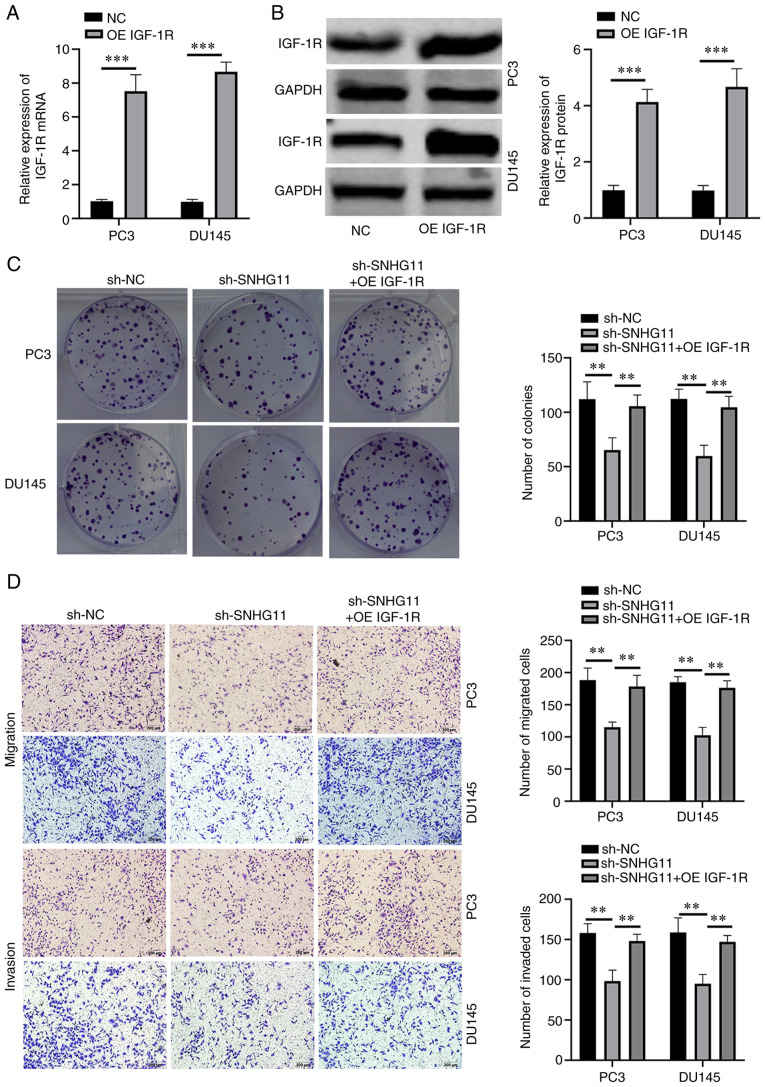
To investigate whether SNHG11 exerts its effects via IGF-1R, functional rescue assays were performed. First, IGF-1R overexpression was successfully induced in PC3 and DU145 cells through transfection with a pcDNA3.1 (+) IGF-1R vector (Fig. 3A and B). Functional rescue assays were then performed using IGF-1R-overexpressing PC3 and DU145 cells following SNHG11 stable knockdown. Rescue experiments indicated that the overexpression of IGF-1R reversed the suppressive effects of SNHG11 knockdown on the proliferative, migratory and invasive abilities of PCa cells (Fig. 3C and D). These data thus indicated that SNHG11 may promote PCa progression by targeting the miR-184/IGF-1R signaling axis.
Figure 3.
Overexpression of IGF-1R expression reverses the effects of SNHG11 knockdown on PCa cells. (A and B) Effects of pcDNA3.1-IGF-1R on IGF-1R expression in PC3 and DU145 cells, as detected using RT-qPCR and western blotting. (C) IGF-1R overexpression reversed the suppressive effects of sh-SNHG11 on the proliferation of PC3 and DU145 cells, as revealed by colony formation assays. (D) IGF-1R overexpression reversed the suppressive effects of sh-SNHG11 on the migration and invasion of PC3 and DU145 cells, as detected using Transwell assay. **P<0.01, ***P<0.001, in comparison with the control group. SNHG11, lncRNA small nucleolar RNA host gene 11; PCa, prostate cancer; IGF-R1, insulin-like growth factor 1 receptor.
Silencing SNHG11 inhibits tumor growth and metastasis in vivo
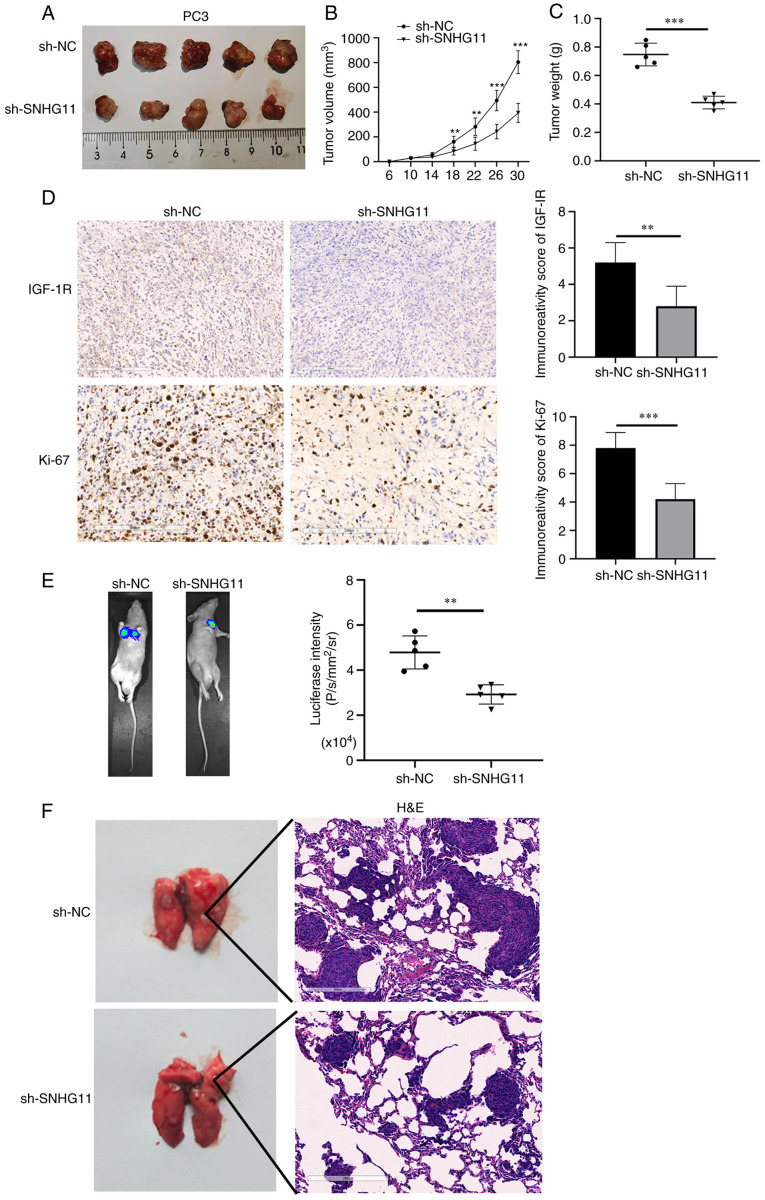
The effects of SNHG11 on tumor growth in vivo were further analyzed. It was revealed that tumor volume and weight were both markedly decreased in the mice injected with sh-SNHG11-transfected PC3 cells, as compared with those injected with sh-NC-transfected PC3 cells (Fig. 4A-C). In the sh-NC group, the maximum tumor volume observed was 826 mm3, while that in the sh-SNHG11 group was 398 mm3. In addition, IHC staining revealed more positive staining for Ki-67 protein, representing cell proliferative ability, in the sh-NC group; in addition, IGF-1R expression was significantly reduced in the xenografts of mice injected with sh-SNHG11-transfected PC3 cells (Fig. 4D). These data suggested that silencing SNHG11 inhibited PCa growth in vivo.
Figure 4.
SNHG11-knockdown inhibits PCa growth and metastasis in vivo. (A and B) The mean tumor volume of the xenograft tumor in the PC3 SNHG11 knockdown group was significantly reduced, in comparison with control group tumor volumes. (C) The average weight of xenograft tumors in the PC3 SNHG11 knockdown group was markedly decreased, in comparison with the control group weights. (D) IGF-1R and Ki-67 expression were markedly decreased in tumor samples from the PC3 SNHG11 knockdown group, in comparison with the control group. (E) Bioluminescence imaging showing that SNHG11 knockdown markedly inhibited PCa cell lung metastatic ability, in comparison with that of the control group. (F) SNHG11-knockdown significantly reduced the tumor burden in the lungs, in comparison with that of the control group. **P<0.01, ***P<0.001, in comparison with the control group. SNHG11, lncRNA small nucleolar RNA host gene 11; PCa, prostate cancer; IGF-R1, insulin-like growth factor 1 receptor.
Finally, the roles of SNHG11 in PCa metastasis in vivo were investigated. As shown in Fig. 4E, the mice injected with sh-SNHG11-transfected PC3 cells exhibited fewer lung metastases, as compared with those injected with sh-NC-transfected PC3 cells. H&E staining also suggested that the silencing of SNHG11 markedly reduced the metastatic burden in the lungs (Fig. 4F). According to these findings, it was indicated that the silencing of SNHG11 may suppress the metastasis of PCa in vivo.
Discussion
Numerous studies have reported that lncRNAs are implicated in the development and metastasis of several types of cancer. For example, Huang et al (15) found that lncRNA CASC2 expression was significantly decreased in colorectal cancer (CRC) tissues and CRC cell lines, and a decreased expression was significantly more frequent in patients with advanced tumor-node-metastasis stage disease. Chen et al (16) observed that the expression of lncRNA CDKN2B antisense RNA 1 was notably upregulated in metastatic hepatocellular carcinoma (HCC) tissues, and facilitated disease progression by regulating the miR-153-5p/Rho GTPase activating protein 18 signaling axis in HCC cells. Kong et al (17) reported that the overexpression of lncRNA cell division cycle 6 in breast cancer cells markedly increased their proliferation and invasion by regulating miR-215. Furthermore, He et al (18) revealed that lncRNA abhydrolase domain containing 11 antisense RNA 1 (ABHD11-AS1) expression was upregulated in colorectal cancer and that the overexpression of lncRNA ABHD11-AS1 markedly enhanced cell proliferation and invasion by modulating the miR-1254/Wnt family member 11 signaling axis.
SNHG11 (GenBank accession no. NR_003239.1) is a recently identified lncRNA; however, little is known with regard to its functional roles in cancer. It has been previously reported that SNHG11 expression is frequently upregulated in several types of cancer (9-11). In HCC, SNHG11 overexpression has been reported to facilitate cell proliferation and migration (19). However, the roles and potential underlying molecular mechanisms of SNHG11 regulation in PCa have not yet been fully elucidated, at least to the best of our knowledge. Therefore, the present study aimed to investigate the molecular mechanisms underlying the involvement of SNHG11 in PCa progression. It was revealed that the SNHG11 expression levels were significantly upregulated in PCa tissues and cells. Functional experiments demonstrated that the silencing of SNHG11 expression inhibited the proliferation, migration and invasion of PCa cells in vitro. Moreover, SNHG11 silencing culminated in a reduction in tumor growth and metastasis in vivo. These results further confirmed the oncogenic role of SNHG11 in PCa.
Accumulating studies have indicated that lncRNAs may act as sponges to regulate miRNAs. For example, Dong et al (20) found that the overexpression of lncRNA NEAT1 in endometrial cancer prooted aggressive endometrial cancer progression. Zhu et al (21) found that the overexpression of lncRNA forkhead box D2 antisense RNA 1 in colorectal cancer cells promoted cellular proliferation by sponging miR-185-5p. Xu et al (22) reported that the overexpression of lncRNA DiGeorge syndrome critical region gene 5 inhibited the growth and metastasis of gastric cancer via targeting miR-23b. Long et al (23) indicated that silencing the lncRNA lysyl oxidase like 1-AS1 suppressed cell cycle progression in PCa via inhibiting miR-541-3p. In the present study, it was demonstrated that miR-184 was a downstream target of SNHG11, as the relative luciferase activity of pmirGLO-SNHG11-WT was suppressed by co-transfection with the miR-184 mimic. Additionally, it was observed that the miR-184 expression levels were significantly upregulated after the silencing of SNHG11. Additionally, SNHG11 expression in PCa tissues was found to negatively correlate with miR-184 expression. Collectively, these findings indicated that SNHG11 may directly interact with miR-184 in PCa. Previous studies have also reported that miR-184 suppresses the progression of numerous types of cancer, including renal cell carcinoma (24), glioma (25) and colorectal cancer (26). However, miR-184 has also been identified as an oncogene in HCC (27). In the present study, it was revealed that miR-184 was sponged by SNHG11 and inhibited PCa development.
In the present study, the target genes of miR-184 were predicted using the ComiRNet online tool. The relative luciferase activity of pmirGLO-IGF-1R-WT was inhibited by co-transfection with miR-184 mimic. Additionally, miR-184 overexpression significantly decreased the IGF-1R expression level in PCa cells. miR-184 expression in PCa tissues was also observed to negatively correlate with IGF-1R expression. Notably, transfection with miR-184 inhibitor abolished the suppressive effects of SNHG11 knockdown on the expression levels of IGF-1R. A positive correlation between SNHG11 and IGF-1R expression levels was also revealed in PCa tissues. IGF-1R overexpression reversed the suppressive effects induced by SNHG11 knockdown on the malignant phenotypes of PCa cells. Previous studies have also demonstrated that IGF-1R plays an oncogenic role in several types of cancer, including PCa (28), bladder cancer (29), colorectal cancer (26) and glioma (30). Similarly, the data of the present study also suggested that IGF-1R may act as an oncogene in PCa.
In conclusion, according to the findings of the present study, it was suggested that SNHG11 facilitates the progression and metastasis of PCa by directly binding with miR-184, in order to upregulate IGF-1R expression. Therefore, the SNHG11/miR-184/IGF-1R signaling axis was revealed as a novel molecular mechanism underlying PCa progression, which indicated that SNHG11 may represent a possible promising therapeutic target for PCa.
Acknowledgments
Not applicable.
Funding Statement
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
QX and XW were responsible for the conception of the study and the drafting of the manuscript. SZ and RK assisted in the design of the study and performed the statistical analysis. QX revised the manuscript. QX and XW confirm the authenticity of all the raw data. All authors have read and approved the final version of the manuscript.
Ethics approval and consent to participate
All patients signed an informed consent form prior to participation and the patient study protocol was approved by The First Affiliated Hospital of University of South China Ethnics Committee. The animal experiments in the present study were performed in compliance with the guidelines and following the approval of the Institute for Laboratory Animal Research at the First Affiliated Hospital of University of South China (approval no. LL20201103017).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 3.Kung JTY, Colognori D, Lee JT. Long noncoding RNAs: Past, present, and future. Genetics. 2013;193:651–669. doi: 10.1534/genetics.112.146704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: A new paradigm. Cancer Res. 2017;77:3965–3981. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lingadahalli S, Jadhao S, Sung YY, Chen M, Hu L, Chen X, Cheung E. Novel lncRNA LINC00844 regulates prostate cancer cell migration and invasion through AR signaling. Mol Cancer Res. 2018;16:1865–1878. doi: 10.1158/1541-7786.MCR-18-0087. [DOI] [PubMed] [Google Scholar]
- 6.Wu M, Huang Y, Chen T, Wang W, Yang S, Ye Z, Xi X. LncRNA MEG3 inhibits the progression of prostate cancer by modulating miR-9-5p/QKI-5 axis. J Cell Mol Med. 2019;23:29–38. doi: 10.1111/jcmm.13658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.You Z, Liu C, Wang C, Ling Z, Wang Y, Wang Y, Zhang M, Chen S, Xu B, Guan H, Chen M. LncRNA CCAT1 promotes prostate cancer cell proliferation by interacting with DDX5 and MIR-28-5P. Mol Cancer Ther. 2019;18:2469–2479. doi: 10.1158/1535-7163.MCT-19-0095. [DOI] [PubMed] [Google Scholar]
- 8.Su W, Xu M, Chen X, Chen N, Gong J, Nie L, Li L, Li X, Zhang M, Zhou Q. Long noncoding RNA ZEB1-AS1 epigenetically regulates the expressions of ZEB1 and downstream molecules in prostate cancer. Mol Cancer. 2017;16:142. doi: 10.1186/s12943-017-0711-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu S, Yang N, Wang L, Wei B, Chen J, Gao Y. IncRNA SNHG11 promotes lung cancer cell proliferation and migration via activation of Wnt/β-catenin signaling pathway. J Cell Physiol. 2020;235:7541–7553. doi: 10.1002/jcp.29656. [DOI] [PubMed] [Google Scholar]
- 10.Wu Q, Ma J, Wei J, Meng W, Wang Y, Shi M. LncRNA SNHG11 promotes gastric cancer progression by activating Wnt/β-catenin pathway and oncogenic autophagy. Mol Ther. 2021;29:1258–1278. doi: 10.1016/j.ymthe.2020.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang W, Dong S, Cha Y, Yuan X. SNHG11 promotes cell proliferation in colorectal cancer by forming a positive regulatory loop with c-Myc. Biochem Biophys Res Commun. 2020;527:985–992. doi: 10.1016/j.bbrc.2020.05.002. [DOI] [PubMed] [Google Scholar]
- 12.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 13.Kang R, Zhao S, Liu L, Li F, Li E, Luo L, Xu L, Wan S, Zhao Z. Knockdown of PSCA induces EMT and decreases metastatic potentials of the human prostate cancer DU145 cells. Cancer Cell Int. 2016;16:20. doi: 10.1186/s12935-016-0295-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen HX, Sharon E. IGF-1R as an anti-cancer target-trials and tribulations. Chin J Cancer. 2013;32:242–252. doi: 10.5732/cjc.012.10263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang G, Wu X, Li S, Xu X, Zhu H, Chen X. The long noncoding RNA CASC2 functions as a competing endogenous RNA by sponging miR-18a in colorectal cancer. Sci Rep. 2016;6:26524. doi: 10.1038/srep26524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen J, Huang X, Wang W, Xie H, Li J, Hu Z, Zheng Z, Li H, Teng L. LncRNA CDKN2BAS predicts poor prognosis in patients with hepatocellular carcinoma and promotes metastasis via the miR-153-5p/ARHGAP18 signaling axis. Aging (Albany NY) 2018;10:3371–3381. doi: 10.18632/aging.101645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kong X, Duan Y, Sang Y, Li Y, Zhang H, Liang Y, Liu Y, Zhang N, Yang Q. LncRNA-CDC6 promotes breast cancer progression and function as ceRNA to target CDC6 by sponging microRNA-215. J Cell Physiol. 2019;234:9105–9117. doi: 10.1002/jcp.27587. [DOI] [PubMed] [Google Scholar]
- 18.He D, Yue Z, Liu L, Fang X, Chen L, Han H. Long noncoding RNA ABHD11-AS1 promote cells proliferation and invasion of colorectal cancer via regulating the miR-1254-WNT11 pathway. J Cell Physiol. 2019;234:12070–12079. doi: 10.1002/jcp.27877. [DOI] [PubMed] [Google Scholar]
- 19.Huang W, Huang F, Lei Z, Luo H. LncRNA SNHG11 promotes proliferation, migration, apoptosis, and autophagy by regulating hsa-miR-184/AGO2 in HCC. Onco Targets Ther. 2020;13:413–421. doi: 10.2147/OTT.S237161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dong P, Xiong Y, Yue J, Xu D, Ihira K, Konno Y, Kobayashi N, Todo Y, Watari H. Long noncoding RNA NEAT1 drives aggressive endometrial cancer progression via miR-361-regulated networks involving STAT3 and tumor microenvironment-related genes. J Exp Clin Cancer Res. 2019;38:295. doi: 10.1186/s13046-019-1306-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu Y, Qiao L, Zhou Y, Ma N, Wang C, Zhou J. Long non-coding RNA FOXD2-AS1 contributes to colorectal cancer proliferation through its interaction with microRNA-185-5p. Cancer Sci. 2018;109:2235–2242. doi: 10.1111/cas.13632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu Y, Zhang G, Zou C, Gong Z, Wang S, Liu J, Ma G, Liu X, Zhang W, Jiang P. Long noncoding RNA DGCR5 suppresses gastric cancer progression by acting as a competing endogenous RNA of PTEN and BTG1. J Cell Physiol. 2019;234:11999–12010. doi: 10.1002/jcp.27861. [DOI] [PubMed] [Google Scholar]
- 23.Long B, Li N, Xu XX, Li XX, Xu XJ, Liu JY, Wu ZH. Long noncoding RNA LOXL1-AS1 regulates prostate cancer cell proliferation and cell cycle progression through miR-541-3p and CCND1. Biochem Biophys Res Commun. 2018;505:561–568. doi: 10.1016/j.bbrc.2018.09.160. [DOI] [PubMed] [Google Scholar]
- 24.Su Z, Chen D, Li Y, Zhang E, Yu Z, Chen T, Jiang Z, Ni L, Yang S, Gui Y, et al. MicroRNA-184 functions as tumor suppressor in renal cell carcinoma. Exp Ther Med. 2015;9:961–966. doi: 10.3892/etm.2015.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng Z, Wang HZ, Li X, Wu Z, Han Y, Li Y, Chen G, Xie X, Huang Y, Du Z, Zhou Y. MicroRNA-184 inhibits cell proliferation and invasion, and specifically targets TNFAIP2 in glioma. J Exp Clin Cancer Res. 2015;34:27. doi: 10.1186/s13046-015-0142-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu G, Liu J, Wu Z, Wu X, Yao X. MicroRNA-184 inhibits cell proliferation and metastasis in human colorectal cancer by directly targeting IGF-1R. Oncol Lett. 2017;14:3215–3222. doi: 10.3892/ol.2017.6499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu GG, Li WH, He WG, Jiang N, Zhang GX, Chen W, Yang HF, Liu QL, Huang YN, Zhang L, et al. Mir-184 post-transcriptionally regulates SOX7 expression and promotes cell proliferation in human hepatocellular carcinoma. PLoS One. 2014;9:e88796. doi: 10.1371/journal.pone.0088796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aleksic T, Verrill C, Bryant RJ, Han C, Worrall AR, Brureau L, Larré S, Higgins GS, Fazal F, Sabbagh A, et al. IGF-1R associates with adverse outcomes after radical radiotherapy for prostate cancer. Br J Cancer. 2017;117:1600–1606. doi: 10.1038/bjc.2017.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liao G, Chen F, Zhong J, Jiang X. MicroRNA-539 inhibits the proliferation and invasion of bladder cancer cells by regulating IGF-1R. Mol Med Rep. 2018;17:4917–4924. doi: 10.3892/mmr.2018.8497. [DOI] [PubMed] [Google Scholar]
- 30.Jiang J, Wang W, Fang D, Jin X, Ding L, Sun X. MicroRNA-186 targets IGF-1R and exerts tumor-suppressing functions in glioma. Mol Med Rep. 2017;16:7821–7828. doi: 10.3892/mmr.2017.7586. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.