Abstract
Strain 68–1 rhesus cytomegalovirus (RhCMV) vectors expressing simian immunodeficiency virus (SIV) antigens elicit CD8+ T cells recognizing epitopes presented by major histocompatibility complex (MHC)-II and MHC-E, not MHC-Ia. These immune responses mediate replication arrest of SIV in 50–60% of monkeys. We show that the peptide VMAPRTLLL (VL9) embedded in the RhCMV protein Rh67 promotes intracellular MHC-E transport and recognition of RhCMV-infected fibroblasts by MHC-E-restricted CD8+ T cells. Deletion or mutation of the viral VL9 abrogated MHC-E-restricted CD8+ T cell priming, resulting in CD8+ T cell responses exclusively targeting MHC-II-restricted epitopes. These responses were comparable in magnitude and differentiation to responses elicited by 68–1 vectors, but did not protect against SIV, indicating that Rh67-enabled direct priming of MHC-E-restricted T cells is crucial for RhCMV/SIV vaccine efficacy.
One Sentence Summary:
RhCMV/SIV vector-encoded VL9 peptide is required for induction of MHC-E-restricted CD8+ T cells and for efficacy against SIV
INTRODUCTION
Cytomegaloviruses (CMVs) are large β-herpesviruses that manifest extensive and complex immune evasion mechanisms to maintain viral persistence. At the same time, they elicit and indefinitely maintain robust antiviral immune responses that restrain viral spread, in particular including very high frequency circulating and tissue-based effector-memory T cells (TEM) (1–5). These potent TEM responses can be harnessed against other pathogens or cancer by CMV-based vaccine vectors expressing heterologous antigens (3, 6, 7). Indeed, using RhCMV/SIV vectors based on the 68–1 strain, we previously demonstrated a vaccine efficacy in which 50–60% of vaccinated rhesus macaques (RMs) exhibit early, complete replication arrest and subsequent clearance of highly pathogenic SIVmac239 (8–10). Although this distinct pattern of protection is consistent with stringent CD8+ TEM-mediated control of the nascent SIV infection, analysis of the epitopes targeted by 68–1 RhCMV/SIV vector-elicited CD8+ T cells revealed that the SIV epitopes recognized by these cells are non-overlapping with those recognized by MHC-Ia-restricted CD8+ T cells elicited by SIV itself or conventional SIV vaccines (11). Indeed, these CD8+ T cells are exclusively restricted by MHC-II or MHC-E, not MHC-Ia, and included recognition of universal epitopes termed supertopes (11, 12).
Such unconventionally restricted CD8+ T cells are not observed in RMs naturally infected with RhCMV, but only in RMs inoculated with recombinants derived from strain 68–1 (11, 12). We recently demonstrated that the unusual ability of 68–1 RhCMV to elicit unconventionally restricted CD8+ T cells results from an inversion of a genomic fragment that occurred during in vitro passage of this strain prior to cloning (13, 14). This genomic rearrangement results in the deletion or reduced expression of eight genes in two non-contiguous gene regions, specifically Rh157.5 and Rh157.4 encoding two subunits of the tropism-determining pentameric complex orthologous to UL128 and UL130 in HCMV, and six genes (Rh158–161) encoding orthologs of the HCMV UL146-family of viral chemokines. Remarkably, expression of each of these eight genes completely prevents the induction of MHC-E restricted CD8+ T cells, and either all or some MHC-II-restricted CD8+ T cells, leaving responses that were either MHC-Ia-restricted or a mixture of MHC-II-restricted and MHC-Ia-restricted CD8+ T cells. Given that MHC-Ia-restricted CD8+ T cell responses are the default response for viruses, these findings are surprising as they indicate that RhCMV encodes eight gene products to prevent MHC-E-restricted CD8+ T cell response priming, raising the questions of how these responses arise and why RhCMV goes to such lengths to inhibit them. Importantly, the SIV-specific CD8+ T cell responses elicited by RhCMV/SIV vectors expressing these inhibitors differs from those of the parent 68–1 vector only in their CD8+ T cell epitope targeting, not in response magnitude or memory differentiation state. Nevertheless, RhCMV/SIV vectors eliciting MHC-Ia -restricted CD8+ T cells alone or together with MHC-II-restricted T cells fail to protect against SIV challenge (14). These findings implicate MHC-E-restricted CD8+ T cell responses as the protective component of 68–1 RhCMV/SIV induced anti-SIV immunity, increasing the importance of understanding how these responses arise.
MHC-E (HLA-E in humans, Mamu-E in RM) is generally not involved in antigen presentation to T cells but binds the conserved peptide VMAPRTL(L/V/I)L (VL9) which is embedded in the leader sequence of polymorphic MHC-Ia molecules (15). Similar to classical MHC-Ia, the loading of MHC-E with VL9 requires the peptide transporter associated with antigen processing (TAP) and the chaperone tapasin (16). Upon egress from the endoplasmic reticulum (ER) MHC-E/VL9 serves as a ligand for inhibitory CD94/NKG2A or activating NKG2C natural killer (NK) cell receptors, with the former binding with higher affinity (17, 18). This MHC-E/VL9 sensing provides NK cells with a potent mechanism to differentiate healthy cells from pathogen-infected or neoplastic cells with disturbances in the classical MHC-Ia antigen presentation pathway. HCMV exploits this host pathway, encoding its own VL9 peptide in the leader sequence of the UL40 gene product. UL40 provided VL9 upregulates MHC-E on infected cells, thereby contributing to evasion of NKG2A+ NK cells by overcoming a shortage of endogenous VL9 resulting from viral MHC-Ia interference (19–21). The RhCMV Rh67 gene also encodes a VL9 peptide containing protein, which supports intracellular transport and surface expression of an HLA-E/β2-microglobulin (β2m) single chain construct similar to UL40 (12). Here, we demonstrate that RhCMV expression of Rh67 strongly modulates MHC-E expression in infected cells via VL9 and that this modulation not only renders RhCMV-infected cells susceptible to recognition by MHC-E-restricted CD8+ T cells, but is also required for both MHC-E-restricted CD8+ T cell priming by, and the efficacy of 68–1 RhCMV/SIV vectors. These data not only confirm the requirement for MHC-E-restricted CD8+ T cells for RhCMV/SIV efficacy, they further suggest that MHC-E-restriction arose from a viral strategy for NK cell evasion that led to a novel CD8+ T cell response that enhanced infected cell recognition, which in turn led to the development of 8 distinct inhibitors of this unconventional CD8+ T cell priming.
RESULTS
RhCMV Rh67 promotes TAP-independent egress and internalization of RM MHC-E
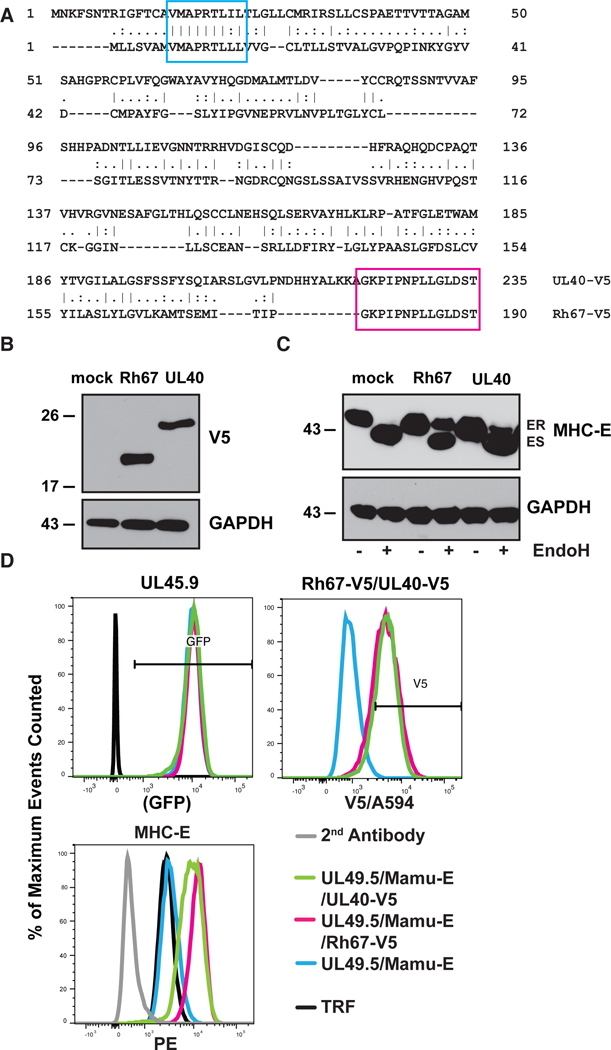
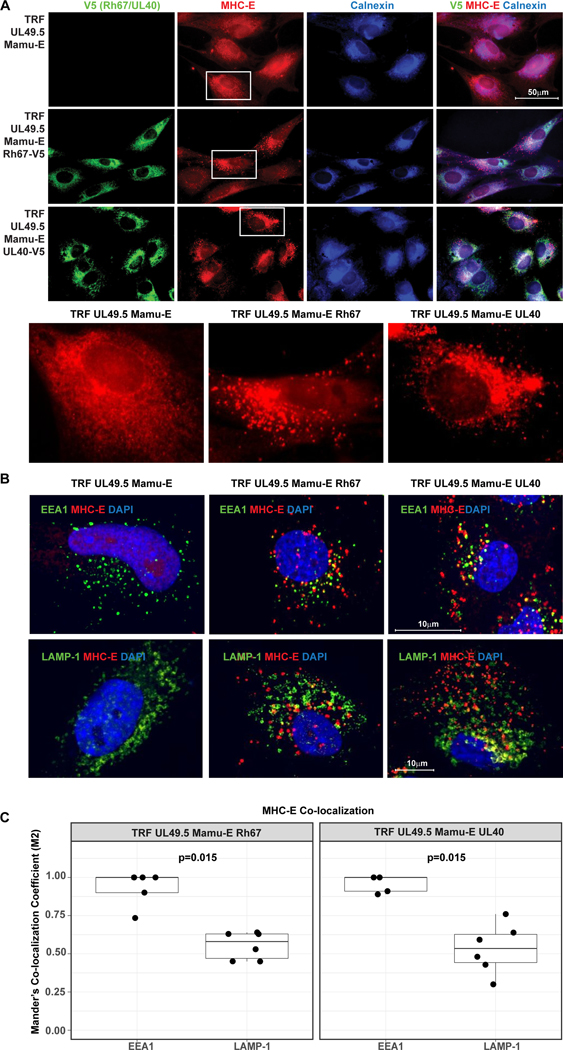
Co-expression of Rh67 supports the intracellular transport and surface expression of an HLA-E/β2m single-chain construct similar to UL40 (12). However, Rh67 shows very little homology to UL40 except for VL9 and adjacent amino acids (AAs) (Fig. 1A). Moreover, the Rh67 amino-terminus preceding VL9 is unrelated to UL40 and significantly shorter. The assembly of UL40-derived VL9 with MHC-E occurs even when TAP is inhibited by the HCMV US6 gene product (20, 21). Since TAP inhibition is conserved in RhCMV (22), we examined whether Rh67 provides TAP-independent VL9 loading of MHC-E by monitoring the intracellular transport and surface expression of MHC-E in telomerized rhesus fibroblasts (TRF) co-transfected with the TAP-inhibitor UL49.5 of pseudorabies virus (23) and V5-epitope tagged Rh67 or UL40 (Fig. 1B). Although MHC-E was retained in the ER in UL49.5-expressing fibroblasts, as indicated by sensitivity to Endoglycosidase H (EndoH), an EndoH-resistant MHC-E population (ER “released”) was observed in the presence of either Rh67 or UL40 (Fig. 1C), resulting in increased surface expression of MHC-E (Fig. 1D). MHC-E also located to intracellular vesicles in Rh67- and UL40-expressing, TAP-inhibited cells, as indicated by punctate staining patterns in immunofluorescence assays (IFA), whereas MHC-E co-localized with the ER-marker Calnexin in the absence of Rh67 or UL40 (Fig. 2A). Transport to intracellular vesicles seemed to occur via endocytosis since MHC-E/antibody complexes were internalized from the cell surface and co-localized with the early endosomal marker EEA1, whereas little co-localization was observed with the lysosomal marker LAMP-1 (Figs. 2B, C). Thus, Rh67 and UL40 enable MHC-E trafficking to the plasma membrane in a TAP-independent manner followed by internalization and possibly recycling back to the cell surface.
Figure 1. Rh67 and UL40 contain a conserved VL9 sequence and cause TAP-independent MHC-E transport.
A) V5-tagged Rh67 (Accession #AFL03561.1) and UL40 (Accession #QHB20491.1) were aligned using the global alignment with free end gap tool (Geneious). The embedded VL9 sequence and added V5-epitope tag are highlighted in blue and pink, respectively. (B) Expression of V5-tagged Rh67 and UL40 was determined by immunoblot in TRF that were also transduced with the TAP-inhibitor UL49.5 and Mamu-E*01:03. Anti-GAPDH was used as loading control. (C) Intracellular transport of MHC-E was monitored by treating lysates of the same cell lines with Endoglycosidase H (EndoH) prior to electrophoretic separation and immunoblot with MHC-E specific antibody (ES, Endo H-sensitive; ER, Endo H-resistant). (D) Flow cytometric analysis of Rh67- and UL40-transduced cells for expression of green fluorescent protein (GFP), which is co-expressed with UL49.5, and for the V5 tag. Surface expression of MHC-E (PE tagged 2nd antibody) was determined on GFP+ and V5+ cells.
Figure 2. Intracellular localization of MHC-E in the presence and absence of Rh67 and UL40.
(A) Intracellular localization of V5-taggegd Rh67 and UL40, Calnexin and MHC-E was determined in UL49.5-expressing, Mamu-E*01:03-transfected TRF by immunofluorescence assay using indicated antibodies. Nuclei were stained with DAPI. Cells shown below are highlighted. (B) Internalization of MHC-E was monitored by adding anti-MHC-E antibody for 1 hour at 37°C prior to fixation, permeabilization and co-staining for early endosomal antigen 1 (EEA1) or lysosome-associated membrane glycoprotein 1 (LAMP-1). Representative results are shown. (C) Boxplots show Manders’ Co-localization Coefficient (M2) calculated for co-localization of EEA1 (n=5 images) or LAMP1 (n=6 images) with MHC-E in Rh67 (left panel) and UL40 (right panel) expressing cells (Extended Date File 1). P-values are shown for two-sample Wilcoxon rank-sum tests comparing the degree of co-localization of EEA1 vs. LAMP1.
VL9-dependent MHC-E egress in RhCMV-infected cells is necessary to stimulate MHC-E-restricted CD8+ T cells
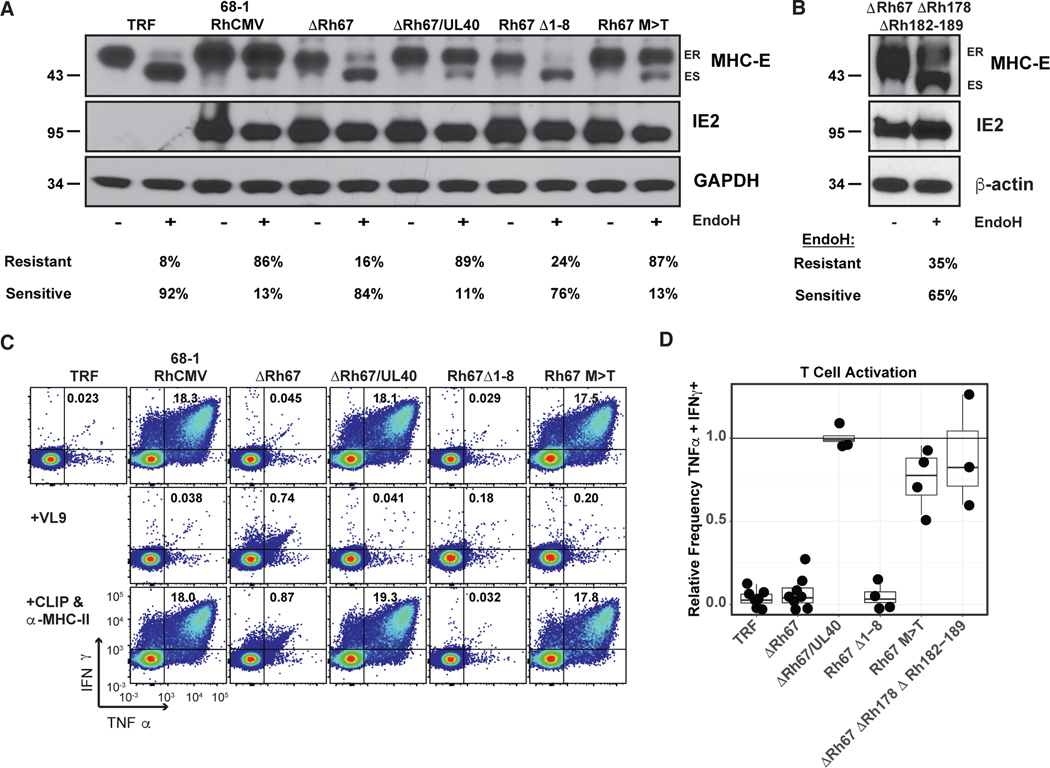
To evaluate the role of Rh67-embedded VL9 in supporting MHC-E expression in RhCMV-infected fibroblasts, we modified Rh67 in a series of 68–1 RhCMV recombinants (fig. S1 A). We either deleted Rh67 (ΔRh67) or mutated its VL9 sequence by deleting the first 8 AAs thus removing key amino-terminal residues in VL9 (Rh67Δ1–8) or by replacing the anchor methionine in position 2 of VL9 with threonine (Rh67/M>T). The M>T mutant is a naturally occurring VL9 variant with lower affinity for MHC-E binding (17). In addition, we replaced Rh67 with its HCMV ortholog UL40 (ΔRh67/UL40). Since these recombinants were derived from 68–1 RhCMV/SIVgag (5), they all contain SIVgag as heterologous antigen (figs. S1, A and B). Immunoblot analysis of MHC-E in fibroblasts infected with these recombinant viruses revealed that parental 68–1 RhCMV converts the majority of MHC-E from ER-retained to ER-released. By contrast, this conversion did not occur in fibroblasts infected with ΔRh67 or Rh67Δ1–8 (Fig. 3A). However, replacement of Rh67 with UL40 resulted in egress of MHC-E indicating that UL40 can functionally replace Rh67. Similarly, replacement of Rh67 with the Rh67/M>T mutant resulted in the majority of MHC-E attaining EndoH resistance (Fig. 3A). The latter is in contrast to the lack of HLA-E/β2m single chain egress in cells co-transfected with Rh67/M>T (12), suggesting that in the context of viral infection the M>T variant supports MHC-E egress. In contrast to MHC-E, all Rh67-derivatives were EndoH-sensitive, consistent with ER-localization, including Rh67Δ1–8 suggesting that the amino-terminal truncation did not prevent protein translocation (fig. S1C). IFA of RhCMV-infected TRF confirmed an increased presence of MHC-E at the cell surface of cells infected with 68–1 RhCMV and ΔRh67/UL40 when compared to ΔRh67 (fig. S2). Consistent with rapid endocytosis, MHC-E appeared in a punctate staining pattern similar to the Rh67 and UL40-transfectants.
Figure 3. VL9-dependent intracellular transport of MHC-E is required for T cell recognition of RhCMV-infected fibroblasts.
(A) 68–1 RhCMV/SIVgag or derivative recombinants engineered to lack Rh67 or express Rh67 lacking the first eight AA of the protein (Rh67Δ1–8) or a Rh67 with a threonine replacing methionine at position 2 of the embedded VL9 peptide (Rh67 M>T), or with Rh67 replaced by the HCMV ortholog UL40 (ΔRh67/UL40) (fig. S1) were used to infect TRFs at an MOI of 3. At 48 hours post-infection, cells were lysed and lysates were treated with EndoH prior to immunoblot for MHC-E, RhCMV IE or GAPDH (ES, Endo H-sensitive; ER, Endo H-resistant). The percentage of ER and ES bands is shown below. (B) In a separate immunoblot experiment, we infected TRF with the same MOI of 68–1 RhCMV/SIVgag deleted for Rh67, along with all inhibitors of MHC-I antigen presentation encoded by Rh178 and US6 family genes contained in the region spanning Rh182–189 (fig. S1) using β-actin as loading control. (C) Flow cytometric ICS analysis for IFN-γ and TNF expression by CD8+ T cells obtained from a 68–1 RhCMV/SIV-vaccinated RM after co-culture with uninfected TRF (negative control) or TRF infected with the indicated viruses. Where indicated, reagents for blocking MHC-E (VL9 peptide) or MHC-II (CLIP peptide and anti-MHC-II mAb G46–6) binding were added to the assay. The % of IFNγ- and TNFα-positive CD8+ T cells are shown in each profile. (D) Relative ICS frequencies of IFNγ- and TNFα-positive CD8+ T cells responding to TRF infected with the indicated recombinant compared to 68–1 RhCMV-infected TRF in the same experiment (n ≥ 4, Extended Data File 1). P-values were <0.01 for ΔRh67 compared to 68–1 using 1-sample Wilcoxon rank-sum test against a null value of 1.0, adjusted for multiple comparisons. In addition to recombinants shown in (C), average ICS analysis results of CD8+ T cells from three RhCMV/SIV-vaccinated RM co-cultured with ΔRh67ΔRh178ΔRh182–189 are shown. MHC-E restriction of these responses was demonstrated by blocking with VL9 peptide (Extended Data File 1).
The requirement for viral VL9 to mediate MHC-E egress in virally infected cells is likely due to elimination of endogenous VL9 sources by viral inhibitors of MHC-Ia antigen processing encoded by Rh182, Rh185, Rh186, Rh189 and Rh178 (22, 24–26). To examine whether the intracellular transport of MHC-E could be restored in the absence of Rh67 in cells infected with RhCMV that also lacked all viral inhibitors of MHC-Ia presentation, we generated ΔRh67ΔRh178ΔRh182–189 (figs. S1, A and B). The presence of EndoH-resistant MHC-E in cells infected with this vector was consistent with endogenous VL9 being able to provide the MHC-E ligand in the absence of MHC-Ia inhibitors (Fig. 3B). Thus, Rh67 and UL40 appear to have evolved as a viral strategy to upregulate MHC-E in infected cells while at the same time preventing MHC-Ia peptide loading and surface expression.
To determine whether Rh67 enables MHC-E-restricted, RhCMV-specific T cells to recognize RhCMV-infected cells, we used intracellular cytokine staining (ICS) to monitor the response of CD8+ T cells from 68–1 RhCMV vector-immunized RM to fibroblasts infected with Rh67-intact versus Rh67-modified 68–1 RhCMV. CD8+ T cells from these animals included RhCMV-specific CD8+ T cell responses that were restricted by MHC-E and MHC-II, as well as MHC-Ia since they were naturally infected with wildtype RhCMV prior to 68–1 vector vaccination (11). Fibroblasts do not express MHC-II and the MHC-Ia alleles expressed by the fibroblast lines used for infection would be both allogeneic to the CD8+ T cells and effectively downregulated by infection (22, 24, 25). Therefore, we expected that most recognition would be MHC-E-restricted. Indeed, essentially all CD8+ T cell recognition of 68–1 RhCMV-infected fibroblasts was blocked by exogenous VL9 peptide, but not by the combination of the MHC-II blocking CLIP peptide + anti-MHC-II mAb, consistent with dominant MHC-E-restricted epitope presentation (Fig. 3C). By contrast, the same CD8+ T cells did not recognize cells infected with ΔRh67 or Rh67Δ1–8, but could recognize cells infected with 68–1 RhCMV when Rh67 was replaced by its HCMV ortholog UL40 or with Rh67/M>T (Figs. 3, C and D). Interestingly, Rh67 was not needed for MHC-E-restricted recognition if cells were infected with ΔRh67ΔRh182–9ΔRh178 (Fig. 3D). Since this 68–1 RhCMV/SIVgag-derived vector lacked all viral inhibitors of MHC-Ia presentation, infected fibroblasts were also recognized by MHC-Ia-matched SIVgag epitope-specific CD8+ T cells, in contrast to parental 68–1 RhCMV or ΔRh67 which both prevent antigen presentation by MHC-Ia (fig. S3). Thus, VL9-induced intracellular transport of MHC-E is essential for the presentation of non-VL9 peptides to MHC-E-restricted CD8+ T cells, with viral VL9 required when sufficient endogenous VL9 is unavailable.
Priming of MHC-E-restricted CD8+ T cells by 68–1 RhCMV requires viral VL9
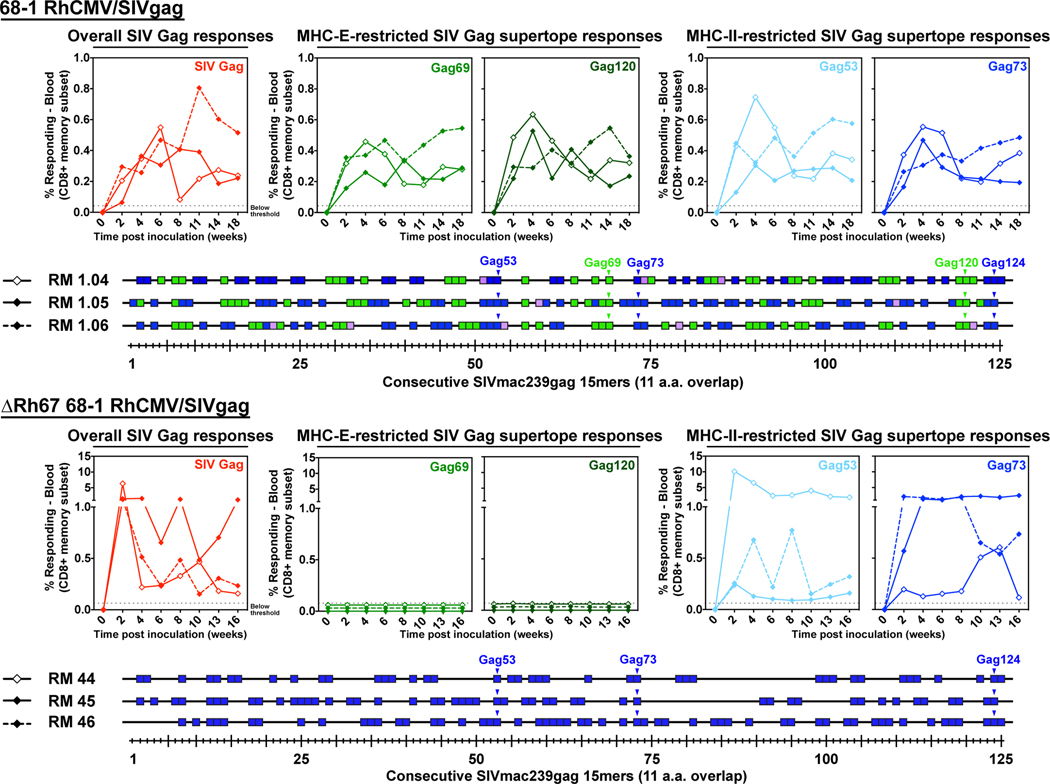
We next asked whether the elicitation of MHC-E-restricted CD8+ T cells in vivo had similar requirements for VL9-induced MHC-E trafficking by comparing the SIVgag-specific CD8+ T cell responses in RM immunized with Rh67-intact versus Rh67-deleted 68–1 RhCMV/SIVgag. Importantly, the ΔRh67 vector elicited robust overall SIVgag-specific T cell responses comparable to those of the parent Rh67-intact 68–1 RhCMV/SIVgag vector (Fig. 4). Thus, unlike viral inhibitors of MHC-Ia antigen presentation (25) or viral inhibitors of NKG2D-dependent NK cell activation (27), Rh67 is not required to establish immunogenic infection, even in RMs with pre-existing anti-RhCMV immunity. However, the lack of Rh67 had a profound influence on the epitopes targeted by the SIVgag-specific CD8+ T cells in the ΔRh67-vaccinated RM (Fig. 4). These CD8+ T cells recognized MHC-II-restricted supertopes, but they did not respond to the MHC-E-restricted supertopes. Full epitope deconvolution followed by restriction analysis revealed that the entire response was MHC-II-restricted indicating that MHC-E-restricted T cell priming was completely abrogated.
Figure 4. Effect of Rh67 deletion on RhCMV/SIV vector CD8+ T cell response programming.
Comparison of SIVgag-specific CD8+ T cell responses elicited by 68–1 RhCMV/SIVgag (RM 1.04, 1.05, and 1.06) versus ΔRh67 68–1 RhCMV/SIVgag (RM 44, 45, and 46) vectors. Peripheral blood CD8+ T cells from these RM inoculated with each of these vectors were assessed by flow cytometric ICS assay for responses to a mixture of 125 consecutive 15mer peptides comprising the SIVgag protein sequence (red; top panel), to individual MHC-E and MHC-II-restricted SIVgag supertopes (green and blue, respectively; top panel; Gag69 = Gag276–284, Gag120 = Gag482–490; Gag53 = Gag211–222, Gag73 = Gag290–301), and to each of 125 consecutive 15mer SIVgag peptides with any above threshold (≥ 0.05% after background subtraction) responses indicated by a box. Boxes are colored to reflect MHC restriction based on the ability to inhibit the response with the MHC-E blocking peptide VL9, the MHC-II blocking mAb G46–6, or the pan-MHC-I blocking mAb W6/32 (14). In the bottom panels, arrowheads indicate the 15mer peptides including the previously described MHC-E- and MHC-II-restricted supertopes (12).
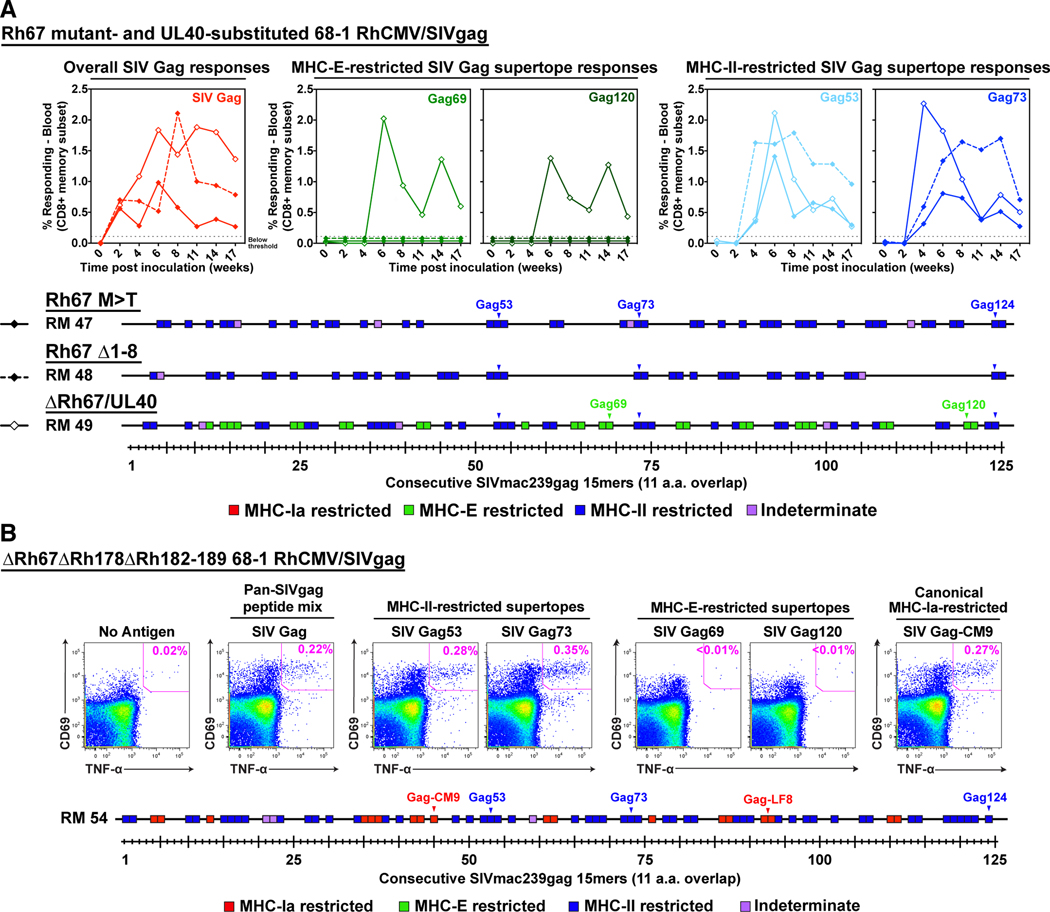
Expression of UL40 instead of Rh67 (ΔRh67/UL40) restored MHC-E-restricted CD8+ T cell priming, but neither the Rh67Δ1–8 nor Rh67/M>T vectors could prime these responses (Fig. 5A). Moreover, in the absence of Rh67, restoration of endogenous VL9 production by deletion of viral inhibitors of MHC-Ia presentation (ΔRh67ΔRh178ΔRh182–189) did not support MHC-E-restricted CD8+ T cell priming in RhCMV-naïve RM (Fig. 5B). [A RhCMV-naïve RM was used in this analysis since RhCMV-vectors deleted for Rh182–189 lose their ability to super-infect RhCMV-seropositive RM (25).] Due to deletion of Rh189 (homolog of US11 in HCMV), the ΔRh67ΔRh178ΔRh182–189 vector elicited CD8+ T cell responses to canonical MHC-Ia epitopes, (11), which resulted in mixed recognition of MHC-II- and MHC-Ia-restricted epitopes (Fig. 5B). Thus, regardless of viral interference with antigen presentation by MHC-Ia, induction of MHC-E-restricted CD8+ T cell responses requires provision of viral-derived VL9 by a Rh67/UL40-mediated process. These responses must arise from direct presentation by Rh67/UL40-expressing, 68–1 RhCMV vector-infected cells, and direct priming in vivo thus seems to depend more stringently on viral VL9 peptide than infected cell recognition in vitro.
Figure 5. Requirement for virus-derived VL9 to prime MHC-E-restricted CD8+ T cells.
(A) Analysis of SIVgag-specific CD8+ T cell responses elicited by Rh67 M>T (RM47), Rh67Δ1–8 (RM48), or ΔRh67/UL40 (RM49) 68–1 RhCMV/SIVgag vectors, as described in Fig. 4. (B) Analysis of SIVgag-specific CD8+ T cell responses elicited by a ΔRh67ΔRh178ΔRh182–189 68–1 RhCMV/SIVgag vector in a Mamu A*01+, RhCMV-naïve RM (RM 54). Peripheral blood mononuclear cells from this RM were analyzed a flow cytometric intracellular cytokine assay for responses to the same antigens shown in Fig. 4 and panel A, as well as an immunodominant, canonical Mamu A*01-restricted epitope (Gag181–189 CM9). In the top panels the flow cytometric profiles of CD69 vs. TNF-α are shown, with CD69+, TNF-α+ Ag-responding cells delineated by the pink boxes (%+ shown). In the bottom panel, above threshold responses to each of the individual consecutive 15mer peptides are colored as described in Fig. 4. The positions of the MHC-II-restricted supertopes (blue) and two known Mamu A*01-restricted canonical SIVgag epitopes (Gag181–189 CM9 and Gag372–379 LF8; red) are indicated in the figure.
Rh67-dependent induction of MHC-E-restricted CD8+ T cells specific for SIV is required for protection against SIV
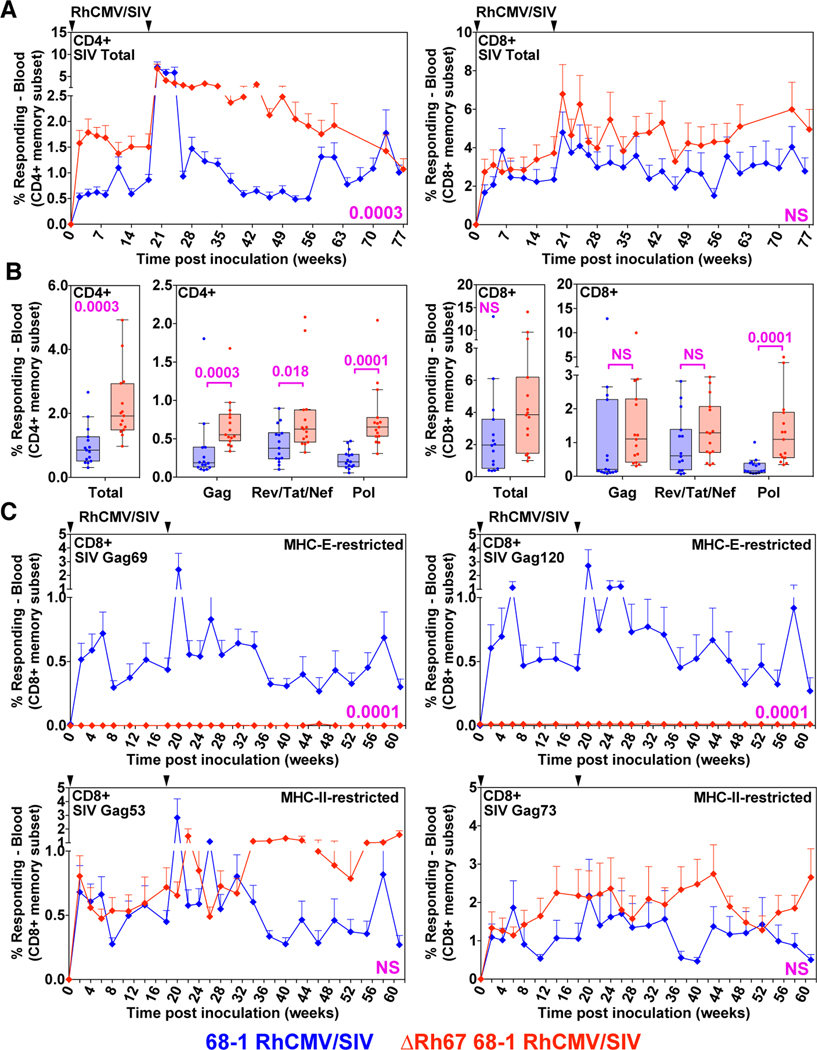
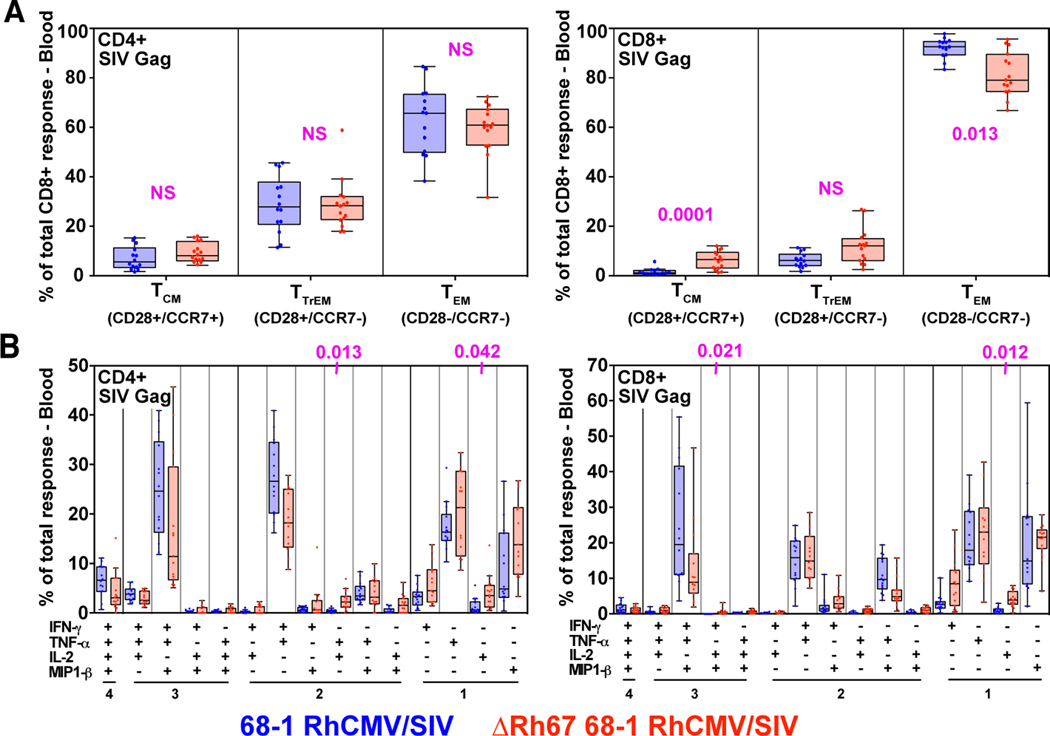
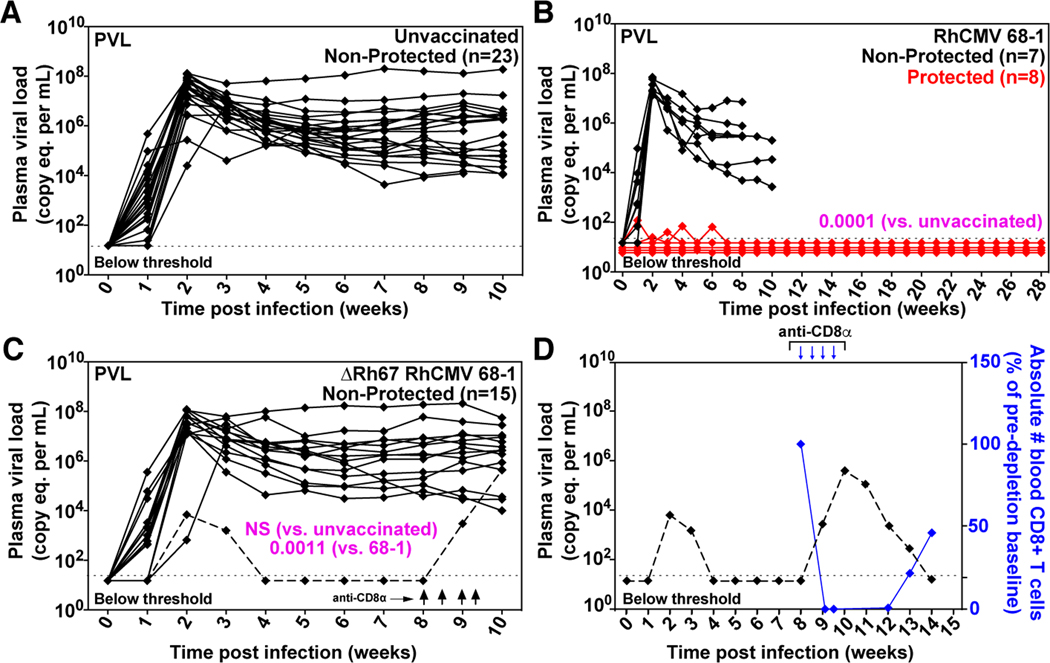
To determine whether MHC-E-restricted CD8+ T cell priming is required for 68–1 RhCMV/SIV vaccine efficacy, we vaccinated 15 RM with three ΔRh67 68–1 RhCMV/SIV vectors individually expressing SIVgag, SIVretanef (a fusion of rev, tat and nef sequences), or a 5’ fragment of SIVpol. The immunogenicity and efficacy of this vector panel was compared to the Rh67-intact parent 68–1 RhCMV/SIV vectors, a cohort shared with a companion study (14) (fig. S4). Importantly, Rh67 deletion did not diminish the magnitude of the overall vector-elicited, SIV-specific T cell responses (indeed, SIV-specific CD4+ T cell responses were significantly more robust in RM vaccinated with Rh67-deleted vector) (Figs. 6, A to C) or substantially alter either the highly TEM-biased response phenotype or the cytokine response pattern compared to parental 68–1 (Figs. 7, A and B). However, as expected, the ΔRh67 68–1 RhCMV/SIV vector-elicited CD8+ T cell response to all SIV proteins was entirely MHC-II-restricted (fig. S5), including MHC-II-restricted supertopes (Fig. 6C). Next, we performed repeated, limiting dose, intrarectal SIVmac239 challenge on this cohort, as previously described (8–10, 14). Whereas eight of 15 RM vaccinated with parental 68–1 RhCMV/SIV vectors showed typical RhCMV/SIV vaccine efficacy (induction of SIVvif-specific T cell responses and development of cell-associated viral loads in tissues, in the absence of sustained plasma viremia), 14 of the 15 ΔRh67 68–1 RhCMV/SIV vector-vaccinated RM showed progressive SIV infection identical to unvaccinated controls (Figs. 8, A to C). One of 15 ΔRh67 vaccinated RM showed restricted viremia (peaking at 6,900 SIV RNA copies per ml) before controlling viremia to below the detection limit four weeks post infection (Fig. 8C). Since the pattern of this viral control is typical of conventional elite control (and atypical for RhCMV/SIV vaccine-mediated replication arrest), we treated this RM with a CD8α-targeted depleting monoclonal antibody (mAb) at eight weeks post-infection to further assess this possibility. The rationale for this intervention is based on data showing that RhCMV/SIV vector-mediated replication arrest is not abrogated by CD8α+ cell depletion (8, 9), even when cell-associated virus is readily demonstrable in tissues (fig. S6). By contrast, conventional elite control (control with ongoing low-level replication) is universally, if transiently, abrogated by this treatment (28). In this RM, anti-CD8α mAb treatment revealed a rapid re-emergence of SIV viremia concomitant with CD8+ T cell depletion, confirming ongoing viral replication (Fig. 8D). Thus, the control observed in this RM is not consistent with the replication arrest of 68–1 RhCMV/SIV vector-mediated protection. While we cannot rule out a role for vaccination in determining this animal’s course, we feel it is far more likely that the control exhibited here reflects the sporadic occurrence of conventional elite SIV control mediated by MHC-Ia-restricted CD8+ T cells. We therefore conclude that the ΔRh67 68–1 RhCMV/SIV vaccine lacked the characteristic SIV replication arrest efficacy of the 68–1 RhCMV/SIV vaccine in this challenge study, and that the MHC-II-restricted CD8+ T cells elicited by this vaccine are, at least by themselves, unable to mediate this type of protection.
Figure 6. Immunogenicity of ΔRh67 vs. wildtype 68–1 RhCMV/SIV vectors.
(A and B) Longitudinal and plateau-phase analysis of the vaccine-elicited SIVgag-, SIVretanef-, and SIV5’pol-specific CD4+ and CD8+ T cell responses in peripheral blood of RM vaccinated with the ΔRh67 68–1 RhCMV/SIV vector set compared to the Rh67-intact parent 68–1 vector set (see fig. S4). (A) The background-subtracted frequencies of cells producing TNF-α and/or IFN-γ by flow cytometric ICS assay to peptide mixes comprising each of the SIV inserts within the memory CD4+ or CD8+ T cell subsets were summed for overall responses with the figure showing the mean (+ SEM) of these overall responses at each time point. (B) Boxplots compare the total and individual SIV insert-specific CD4+ and CD8 + T cell response frequencies between the two vaccine groups during the vaccine phase plateau. (C) Longitudinal analysis of the vaccine elicited CD8+ T cell responses to SIVgag supertopes in peripheral blood of each vaccine group by ICS assay. Wilcoxon P-values for comparison of all T cell response parameters in panels A to C are shown where significant (p<0.05); adjusted for multiple comparisons in panels B (across individual insert responses) and C (across individual supertopes).
Figure 7. Differentiation of SIV-specific CD8+ T cells elicited by ΔRh67 68–1 RhCMV/SIV vectors.
(A) Boxplots compare the memory differentiation of the vaccine-elicited CD4+ and CD8+ memory T cells in peripheral blood responding to overall SIVgag 15mer peptide mix with TNF and/or IFN-γ production during the vaccine phase plateau of the RM cohorts described in Fig. 6 (see fig. S4). Memory differentiation state was based on CD28 and CCR7 expression, delineating central memory (TCM), transitional effector-memory (TTrEM), and effector-memory (TEM), as designated. (B) Boxplots compare the frequency of vaccine-elicited CD4+ and CD8+ memory T cells in peripheral blood during plateau phase responding to the overall SIVgag 15mer peptide mix with TNF, IFN-γ, IL-2, and MIP-1β production, alone and in all combinations (same RM as in panel A). Wilcoxon P-values for comparison of all T cell response parameters shown in panels A and B are shown where significant (p<0.05), adjusted for multiple comparisons.
Figure 8. Efficacy of ΔRh67 68–1 RhCMV/SIV vectors.
(A, B, C) Assessment of the outcome of SIV infection after repeated, limiting dose SIVmac239 challenge of the ΔRh67 68–1 RhCMV/SIV vector vaccinated RM, relative to RM vaccinated with the Rh67-intact parent 68–1 vectors [red delineates RM with documented SIV replication arrest; reproduced from (14)], and to unvaccinated controls (including RM contemporaneously challenged with both vaccinated cohorts). (D) The dashed line in panel C delineates the one of 15 ΔRh67 68–1 RhCMV/SIV vector set-vaccinated RM that showed abbreviated SIV viremia post-infection, prompting us to perform CD8α cell depletion at day 57 post-infection to distinguish conventional elite control (susceptible to CD8α cell depletion) to RhCMV/SIV vector vaccination induced SIV replication arrest (not susceptible to CD8α cell depletion; see fig. S6). In panels A-C, binomial exact P-values are shown where the proportion of RM with SIV replication arrest (protected) in one vaccine group differs significantly from the designated comparison group. NS = not significant.
DISCUSSION
These data demonstrate that viral provision of VL9 peptide via the Rh67 (or UL40) gene product is required for both priming of MHC-E-restricted CD8+ T cells by 68–1 RhCMV/SIV vectors and for the replication arrest type of protection against highly pathogenic SIV afforded by these vectors. Indeed, the deletion of Rh67 from 68–1 RhCMV/SIV leads to vectors that elicit robust, effector memory-differentiated CD8+ T cell responses that are entirely MHC-II-restricted and include MHC-II-restricted supertope responses; yet fail to protect. In a companion report (14), we demonstrate that response-programmed RhCMV/SIV vectors eliciting MHC-Ia-restricted CD8+ T cell responses or a mixture of MHC-Ia- and MHC-II-restricted CD8+ T cell responses (the latter without MHC-II supertope responses) are also non-efficacious. Taken together, these data indicate that MHC-E-restricted CD8+ T cell responses are required for protection and, given the lack of SIV-specific antibodies elicited by RhCMV/SIV vectors (8–10), strongly suggest that these responses constitute the primary adaptive component of efficacy. The mechanism(s) underlying the special ability of MHC-E-restricted CD8+ T cells to mediate anti-SIV efficacy remain to be determined. RhCMV/SIV vector-elicited CD8+ T cell responses of all restriction types appear to be similar in magnitude and functional phenotype as shown here and in the companion paper (14). However, it is possible that the direct priming of these responses by RhCMV vector-infected cells with upregulated MHC-E results in a different functional programming that is required for efficacy. Alternatively or additionally, the requirement for MHC-E-restricted CD8+ T cell responses for efficacy might be due to differences in the relative expression levels of MHC-Ia, MHC-II, and MHC-E on SIV-infected CD4+ T cells in early infection (12, 29, 30) which might allow for more efficient or more complete recognition of SIV-infected cells by MHC-E-restricted CD8+ T cells.
SIV appears to have evolved to evade NK cells and MHC-Ia-restricted CD8+ T cells in the absence of selection pressure from MHC-E-restricted CD8+ T cells since these cells are not naturally primed during SIV infection (12). This is in contrast to the more complex immune evasion strategy of RhCMV, which efficiently prevents antigen presentation by MHC-Ia via the Rh178 and Rh182–189-encoded proteins (22, 24–26) (fig. S3), while simultaneously enhancing the intracellular transport of MHC-E via the VL9-providing Rh67 protein (Fig. 3). In the absence of specific inhibition, this upregulation leads to MHC-E-restricted CD8+ T cell priming. Indeed, the presence of eight different genes, Rh157.5/Rh157.4 and Rh158–161, that each prevent MHC-E-restricted CD8+ T cell priming in the RhCMV genome (14), strongly suggests that the efficient NK cell evasion afforded by Rh67 comes at a price of increased recognition by MHC-E-restricted CD8+ T cells, which, in turn, is countered by these viral-encoded priming inhibitors.
It should, however, be noted that the finding that Rh67 function leads to MHC-E-restricted T cell priming is counterintuitive since VL9 binds with higher affinity to MHC-E than non-VL9 peptides and thus might be expected to prevent or displace binding of unrelated, lower affinity peptides (31, 32). Instead, our data suggest that while loading with viral VL9 is required for MHC-E egress from the ER in the absence of host VL9, there is in place a host mechanism that provides for exchange of the VL9 peptide with other viral peptides resulting in MHC-E-mediated antigen presentation to CD8+ T cells. This model is supported by the correlation of MHC-E egress with T cell recognition of RhCMV vector-infected cells and by the finding that surface MHC-E traffics to intracellular vesicles. The exact nature of these vesicles still needs to be determined, but our observation is consistent with previous reports showing that, in monocytes, HLA-E co-localized with markers of endolysosomal and autophagosomal vesicular structures (33). Moreover, structural and biochemical studies demonstrate that VL9 bound to HLA-E can be exchanged for lower-affinity, non-VL9 peptides most likely due to an unusual stability of “empty” MHC-E/β2m heterodimers (12, 31). With this presently described epitope-loading mechanism, the host partially counters the viral NK cell evasion strategy by eliciting a novel MHC-E-restricted CD8+ T cell response which would preferentially recognize CMV-infected cells with Rh67-mediated MHC-E upregulation. Thus, 68–1 RhCMV/SIV vector efficacy seems to result from a mismatch in immune evasion strategies between RhCMV and SIV, with the former virus able to generate novel responses that it has evolved to evade, which the latter virus cannot. Although HCMV and RhCMV are different viruses, they do share a close evolutionary relationship and the relevant mechanisms appears to be conserved (29), including the ability of UL40, UL128/UL130, and UL146/147 to functionally replace their RhCMV orthologs (14), offering the hope that a 68–1-like HCMV-based vaccine vector might be able to take advantage of the same mismatch to provide an effective HIV/AIDS vaccine.
MATERIALS AND METHODS
Study Design.
The objectives of this study were to a) evaluate the role of Rh67-encoded MHC-E ligands on the ability of 68–1 RhCMV vectors to present MHC-E-restricted epitopes in vitro, b) elicit MHC-E-restricted CD8+ T cell responses in vivo, and c) determine whether vaccination of RM with RhCMV/SIV vectors lacking Rh67 would provide the same stringent post-acquisition control of SIVmac239 infection as Rh67-intact 68–1 RhCMV/SIV vectors. In vitro studies were performed with TRF infected with RhCMV to determine the impact of Rh67-deletions or –modifications on the ability to stimulate MHC-E-restricted CD8+ T cells. To minimize the number of RM used in these experiments, most were designed with the goal to generate qualitative rather than quantitative comparisons with Rh67-intact vectors. Because we observed highly consistent results and simultaneously monitored multiple epitopes as few as one animal per construct was sufficient to determine whether a given Rh67-modified RhCMV vector was able to elicit MHC-E restricted CD8+ T cells. To evaluate ΔRh67 68–1 RhCMV/SIV vector immunogenicity and efficacy we appended analysis of this vector backbone to the large RM study described in a companion manuscript comparing immunogenicity and efficacy of Rh157.5/157.5 gene region-modified 68–1 RhCMV/SIV vectors with the parental 68–1 RhCMV/SIV vector (14). We constructed and validated a three-vector set expressing the same SIVgag, SIVretanef and SIV5’pol inserts as in the companion study and subcutaneously administrated these vectors to 15 randomly selected male RM from the same pool as the prior study, using the identical vaccination regimen. In addition, these RM were SIV challenged with the same, repeated, limiting dose, intrarectal challenge protocol using the same SIVmac239 stock as the companion study, side by side with an additional eight unvaccinated control RM (which validated the effectiveness of the challenge protocol). Since the immunogenicity and efficacy of the parental 68–1 RhCMV/SIV vectors is highly consistent across multiple studies (8–10, 14), we used prior analysis of the parental 68–1 RhCMV/SIV vector in the companion study (14) as the positive control group for this study. As previously noted (14), the n=15 group size allows us to detect per-vaccine-group protection levels of 14% at 90% power without multiplicity adjustment. Except for rare assay failures, all results from these experiments are included in the presented associated data (no data were excluded as outliers). Plasma viral load assays were assayed by blinded analysis; however, due to logistical constraints, other staff were not blinded to treatment assignments. Primary data are reported in extended data files S1 to S2.
Cell lines.
TRF were transduced with retrovirus pLZRS IRES GFP expressing PRV UL49.5 in front of an internal ribosomal entry site that is followed by enhanced green fluorescent protein (GFP) (34). After sorting for GFP expression, cells were transduced with Mamu-E*02:04 expressing lentivector pLVX EF1α IRES blasticidin (derived from pLVX EF1α IRES puro (Clontech) by replacing the puromycin resistance with blasticidin resistance cassette). After selection for blasticidin resistance, the cells were transduced with pLVX EF1α IRES puro (Clontech) carrying synthetic genes encoding C-terminally V5-epitope tagged RhCMV Rh67 or HCMV UL40 (Life Technologies). Upon selection for puromycin resistance, the resulting cell lines were maintained in DMEM supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin, and 100 μg/ml streptomycin (PenStrep) with added puromycin (Invivogen, 4 μg/ml) and blasticidin (Invivogen, 20 μg/ml). Expression of GFP, Rh67, UL40, and Mamu-E was confirmed by immunoblot and flow cytometry (Fig. 1).
Generation and testing of recombinant RhCMV vectors.
All recombinant viruses used in this study were generated using bacterial artificial chromosome (BAC) mutagenesis of BAC-cloned RhCMV strain 68–1 (35). 68–1 RhCMV expressing SIVgag was generated as previously described (5). This vector was used to generate Rh67-modified recombinants by homologous recombination of linear DNA fragments generated by gene synthesis (Life Technologies) with all constructs containing an in-frame fusion of the parainfluenza V5 epitope tag (GKPIPNPLLGLDST) at their C-terminus: Rh67, UL40, Rh67M>T, Rh67Δ1–8 (fig. S1A). DNA fragments were cloned into pORI and amplified by PCR together with a Kanamycin-resistance (KanR) marker flanked by flippase recognition target (FRT) sites (36). PCR primers included 50 bp homologous to the Rh67 flanking region so that bacteriophage Lambda recombination enzymes Red α,β,γ-dependent homologous recombination resulted in exchange of Rh67 with the Rh67-modified sequence. The KanR resistance cassette that was subsequently removed by induction of the yeast flippase resulting in a remaining FRT “scar” (fig. S1A). Rh67 was deleted from 68–1 RhCMV/SIVgag by replacing Rh67 with the excisable KanR cassette using the same homologous recombination strategy. Rh67-deleted RhCMV/rtn and RhCMV/5’pol were generated by using homologous recombination to replace Rh67 with genes encoding the SIVretanef or the 5’-fragment of SIVpol together with the excisable KanR gene.
To generate ΔRh67ΔRh178ΔRh182–9 we transformed the 68–1 RhCMV/SIVgag BAC into E.coli strain GS1783 to allow en passant recombination (37). Using this scarless recombination technique we sequentially deleted Rh67, Rh178, and the entire Rh182-Rh189 gene region. Each of the intermediate steps were controlled by XmaI restriction digest and Sanger sequencing across the deletions.
The final BACs were analyzed by next generation sequencing of the entire genome on an Illumina MiSeq sequencer. Consensus sequences were deposited in GenBank: ΔRh67/gag (MN622881), ΔRh67/5’pol (MN622882), ΔRh67/rtn (MN622883), ΔRh67/UL40/gag (MN622884), Rh67M>T/gag (MN622885), Rh67V5/gag (MN622886), Rh67Δ1–8/gag (MN622887), ΔRh67ΔRh178ΔRh182–189/gag (MT068444).
Recombinant viruses were reconstituted by electroporation of BAC DNA into primary rhesus fibroblasts. Briefly, cells were trypsinized, pelleted, and resuspended in Opti-MEM (Invitrogen). The loxP-flanked BAC cassette encodes a Cre recombinase under control of a mammalian promoter resulting in spontaneous excision of the BAC cassette (35). Expression of SIV antigens and Rh67 constructs was confirmed by immunoblot following infection of TRF (figs. S1, B and C).
Polyacrylamide gel electrophoresis and immunoblot analysis.
Cells were harvested with trypsin/EDTA (Corning 25–051-CI) and washed with PBS. The cell pellets were then resuspended at 1×107 cells/ml in water with 1% NP-40 and HALT protease inhibitors (Thermo Fisher) and incubated at 4°C for 2 hours to lyse the cells. The resulting lysates were centrifuged at 21,130 g at 4°C for 45 min. The supernatants were harvested and frozen at minus 20°C until use. For Endo H treatment, 9 μl of each lysate sample was mixed with 1.09 μl of 10X glycoprotein dissociation buffer (New England Biolabs) and boiled for 10 min. The lysates were then mixed with 5.97 μl of EndoH reaction buffer (50 mM sodium acetate, 0.5 mg/ml BSA, 1.25% NP-40 and HALT protease inhibitor) and 1.3 μl of EndoH (Roche), or water for mock-treated samples, and incubated at 37°C overnight. To detect a reduction in the apparent molecular weight due to removal of asparagine-linked glycans by EndoH we separated lysates on 10% polyacrylamide gels and transferred to PVDF membranes (Millipore). Following blocking with 5% milk in PBS-0.1% Tween-20 (Fisher Scientific) (PBS-T), membranes were probed with the following antibodies in 5% milk in PBS-T: anti-MHC-E (1 μg/ml, Clone 4D12, LSBio #C179742), anti-βactin (0.2 μg/ml, Clone AC-74, Sigma #A2228) anti-FLAG (0.8 μg/ml, Sigma #F3165), anti-V5 (1 μg/ml, Invitrogen #37–7500), anti-GAPDH (0.2 μg/ml, Invitrogen #MA5–15738), or anti-RhCMV IE2 (38) (Clone IIA5.2, 15 μg/ml). Membranes were washed three times with PBS-T and then incubated with goat anti-mouse HRP (0.2 μg/ml, Invitrogen #A28177) secondary antibody in 5% milk in PBS-T. Membranes were incubated with Supersignal West Pico chemiluminescent substrate (Thermo Scientific) and exposed to chemiluminescent film (GE Healthcare) or scanned on a BioRad ChemiDoc MP imager.
Immunofluorescence.
TRF expressing Mamu-E*02:04 and UL49.5 alone or together with V5-tagged Rh67 or UL40 were plated on 12 mm wide, 0.13–0.17 mm thick glass coverslips in 24-well plates and stained at 24 hours post-plating. Cells were rinsed with PBS and fixed with 4% paraformaldehyde (PFA) in PBS for 10 min at room temperature (RT) followed by washing three times with PBS, blocking with 1% BSA (Fisher) in PBS and permeabilization with 0.2% saponin (Millipore) for 1 hour at RT. Fixed cells were incubated with the following antibodies for 1 hour at RT: anti-V5 (2.5 μg/ml, Novus biologicals #NB600–381), anti-Calnexin (8 μg/ml, LS Bio #LS-B12410), anti-MHC-E (10 μg/ml, HLA-E, MBL #K0215–3) followed by washing three times for a total of 30 min using saponin-BSA buffer. Secondary antibodies (Invitrogen #A-21081, #A-11032 and #A32733) were used at 4 μg/ml concentration in saponin-BSA buffer for 1 hour at RT. Cells were washed three times for a total of 30 min using saponin-BSA buffer followed by a final wash in PBS. Coverslips were mounted on glass slide using Prolong Diamond (Invitrogen) and cured for 24 hours. The cells were imaged with Keyence BZ-X710 microscope, using its built-in 2.8 megapixel monochrome CCD camera. A 100 × oil objective was used for image acquisition and images were processed and overlaid using FIJI software.
Internalization of MHC-E was monitored 24 hours after cells were plated on 12 mm wide, 0.13–0.17 mm thick glass coverslips in 24-well plates by adding anti-MHC-E antibody (MBL #K0215–3; 10μg/ml diluted in DMEM, 2.5% FBS (D2.5%)) to TRF expressing Mamu-E*02:04 and UL49.5 alone or together with Rh67-V5 or UL40-V5. After incubation at 37°C for 1 hour the cells were washed three times in cold D2.5% and fixed with 4% PFA in PBS for 10 min at RT. The cells were washed three times using PBS, blocked and permeabilized with 0.2% saponin and 1% BSA buffer for 1 hour at RT. Permeabilized cells were incubated with EEA1-specific antibody (10 μg/ml, CST, #2411S) or LAMP-1-specific antibody (5 μg/ml, Abcam, #Ab19294) for 1 hour at RT followed by three washes for a total of 30 min using saponin-BSA buffer. Secondary antibodies (4 μg/ml in saponin-BSA buffer; Invitrogen, #A-11032 and #A-32733) together with DAPI (1 μg/ml, Invitrogen, #D1306) were incubated for 1 hour at RT. Cells were washed three times for a total of 30 min using saponin-BSA buffer followed by a final wash in PBS. Coverslips were mounted on glass slides using Prolong Diamond (Invitrogen, P36961) and cured for 24 hours. The cells were imaged with Keyence BZ-X710 microscope, using its built-in 2.8 megapixel monochrome CCD camera. To eliminate fluorescence blurring in images, sectional function of 2D structured illumination feature was used. The 100 × oil objective was used for image acquisition and images were processed and overlaid using FIJI software. The co-occurrence of red (MHC-E) versus green (EEA1 or LAMP-1) fluorescence was calculated using the Manders’ co-localization coefficient (MCC) (39, 40) using multiple images of cells (5 images from four independent experiments for EEA1 and 6 images from two independent experiments for LAMP1). For the purpose of co-localization quantification, all images in green and red channels were processed uniformly for background reduction. The fraction of the co-occurring fluorescence was calculated using the JACoP plugin in FIJI software to determine the coefficients M1 (green versus red) or M2 (red versus green) (Extended Data File 1).
Flow cytometric analysis of MHC-E expression on TRF:
TRF or TRF expressing Mamu-E and UL49.5 alone or together with Rh67 or UL40 were harvested with trypsin/EDTA (Corning) and washed with cold DMEM supplemented with 2.5% FBS (D2.5%). This media was used throughout for washing and MHC-E antibody incubation of live TRF. 2.5×105 cells were incubated with anti-MHC-E antibody (MBL #K0215–3, 10 μg/ml) for 1 hour at 4°C. The samples were washed three times and incubated with phycoerythrin (PE)-conjugated secondary antibody (Thermo Fisher #31862,2 μg/ml) and Live/Dead stain (1 μl per 1×106 cells, according to datasheet Life Technologies #L34957) for 45 min at 4°C. The cells were washed three time and fixed with 4% PFA in PBS for 10 min at RT, followed by three washes in PBS. Cells were further stained for intracellular V5-epitope expression, after blocking with 1% BSA and permeabilization with 0.2% saponin in PBS for 1 hour at RT. Anti-V5 antibody (2.5 μg/ml, Novus biologicals #NB600–381) was incubated in saponin-BSA buffer for 1 hour at RT and cells were washed three times using saponin-BSA buffer. The samples were incubated with anti-rabbit secondary antibody conjugated with Alexa fluor 647 (4μg/ml, Thermo Fisher #A-21244, 1:500). Finally, stained cells were fixed in 1% PFA in PBS and analyzed on a BD LSRII Cytometer.
Rhesus macaques.
The experiments reported here used a total of 65 purpose-bred male and female rhesus macaques (RMs) (Macaca mulatta) of Indian genetic background, including three RMs previously immunized with 68–1 RhCMV vectors for separate studies but used here as source of MHC-E-restricted CD8+ T cells. Seven RMs were used for immunogenicity analysis of 68–1-based, Rh67-variant RhCMV/SIVgag vectors, 53 RM for comparative immunogenicity and efficacy analysis of Rh67-intact versus ΔRh67 68–1 RhCMV vectors (30 vaccinated and 23 unvaccinated controls), two 68–1 RhCMV/SIV vaccine-protected RMs used for CD8+ cell depletion, and two treatment-naïve RMs used as adoptive transfer recipients. Fifteen of the unvaccinated negative controls and the 15 Rh67-intact 68–1 RhCMV/SIV vaccinated group used in the comparative immunogenicity/efficacy analysis were presented in detail in the companion manuscript (14). The 15 RMs vaccinated ΔRh67 68–1 RhCMV vectors were vaccinated and SIV-challenged subsequent to the RMs in the companion manuscript using the same and vaccination regimen and challenge protocol. To ensure comparability of the challenge procedure, these RMs were challenged in parallel with an additional eight unvaccinated controls. RhCMV vectors were routinely dosed at 106-107 infectious units for immunogenicity analysis and 5×106 infectious units per vector for large group immunogencity and efficacy analysis, all via subcutaneous administration. For the former studies the vaccine vector was given once; for latter studies, RMs were subcutaneously inoculated with same vaccine vectors at the same dose a second time 18 weeks post-initial vaccination (homologous boost). SIV challenge and adoptive transfer analysis of replication competent SIV were performed as previous described (8–10, 14). Depletion of CD8+ cells from RMs was accomplished using treatment with the monoclonal antibody (mAb) M-T807R1 (10 mg per kg subcutaneously at day 0, and 5 mg per kg intravenously at days 3, 7, and 10), as previously described (8, 9, 28). At assignment, all study RMs were free of cercopithicine herpesvirus 1, D-type simian retrovirus, simian T-lymphotrophic virus type 1, and Mycobacterium tuberculosis, and all but one (Fig. 5B) were naturally RhCMV-infected. This RM was specially raised to be RhCMV-naïve at assignment and was used to test immunogenicity of a Rh67-deleted 68–1 RhCMV vector also lacking viral inhibitors of MHC-Ia presentation (ΔRh178ΔRh182–9), which eliminates the ability to superinfect RhCMV+ RM (9). All study RMs were housed at the Oregon National Primate Research Center (ONPRC) in Animal Biosafety level (ABSL)-2 (vaccine phase) and ABSL-2+ rooms (challenge phase) with autonomously controlled temperature, humidity, and lighting. Study RMs were both single and pair cage-housed. Animals were only paired with one another during the vaccine phase if they were from the same vaccination group. All RMs were single cage-housed during the challenge phase due to the infectious nature of the study. Regardless of their pairing, all animals had visual, auditory, and olfactory contact with other animals. Single cage-housed RMs received an enhanced enrichment plan that was designed and overseen by NHP behavior specialists. RMs were fed commercially prepared primate chow twice daily and received supplemental fresh fruit or vegetables daily. Fresh, potable water was provided via automatic water systems. Physical exams including body weight and complete blood counts were performed at all protocol time points. RMs were sedated with ketamine HCl or Telazol for procedures, including intradermal and subcutaneous vaccine administration, venipuncture, bronchoalveolar lavage, bone marrow and lymph node biopsy and SIV challenge. At humane or scheduled endpoints, RMs were euthanized with sodium pentobarbital overdose (>50 mg per kg) and exsanguinated via the distal aorta, and tissue collection at necropsy was performed by a certified veterinary pathologist. RM care and all experimental protocols and procedures were approved by the ONPRC Institutional Animal Care and Use Committee (IACUC). The ONPRC is a Category I facility. The Laboratory Animal Care and Use Program at the ONPRC is fully accredited by the American Association for Accreditation of Laboratory Animal Care (AAALAC) and has an approved Assurance (#A3304–01) for the care and use of animals on file with the NIH Office for Protection from Research Risks. The IACUC adheres to national guidelines established in the Animal Welfare Act (7 U.S.C. Sections 2131–2159) and the Guide for the Care and Use of Laboratory Animals (8th Edition) as mandated by the U.S. Public Health Service Policy.
T cell assays.
To monitor CD8+ T cell responses to RhCMV-infected cells by flow cytometric intracellular cytokine staining (ICS), TRF were infected at an MOI of 3 for 24 hours. Cells were harvested by trypsinization, resuspended at 4×106 cells/ml in DMEM and 50 μl aliquots of the cell suspensions (2×105 cells) were added to 5×105 CD8β+ T cells in 50 μl isolated from peripheral blood mononuclear cells (PBMC) of 68–1 RhCMV-immunized RM using a non-human primate CD8+ T cell isolation kit (Miltenyi Biotec) and LS columns (Miltenyi Biotec) in the presence or absence of MHC blocking reagents. To monitor presentation of the SIVgag CM9 epitope (Gag181–189), RhCMV-infected Mamu-A*01+ TRF were co-cultured with a CM9-specific CD8+ T cell line generated as described (41). The ICS assay for these analyses was performed as described below for PBMC.
SIV-specific CD4+ and CD8+ T cell responses were measured in PBMC or tissue-derived mononuclear cells by ICS as previously described (11, 12). Briefly, individual or whole protein mixes of sequential 15-mer peptides (11 amino acid overlap) spanning the SIVmac239 Gag, 5’-Pol, Nef, Rev, Tat, and Vif proteins or individual SIVmac239 Gag supertope peptides [Gag211–222 (53), Gag276–284 (69), Gag290–301 (73), Gag482–490 (120)] were used as antigens.
Mononuclear cells were incubated at 37°C with the peptide(s) or cells and co-stimulatory anti-CD28 (CD28.2, purified 500 ng per test: eBioscience, Custom Bulk 7014–0289-M050) and anti-CD49d mAb (9F10, purified 500 ng per test: eBioscience, Custom Bulk 7014–0499-M050) for one hour, followed by an additional eight hours incubation in the presence of Brefeldin A (5 μg/ml; Sigma-Aldrich). Stimulation in the absence of peptides or cells served as background control. After incubation, stimulated cells were stored at 4°C until staining with combinations of fluorochrome-conjugated monoclonal antibodies including: anti-CD3 (450 ng of SP34–2: Alexa700; BD Biosciences, Custom Bulk 624040, and 624034 100 ng of Pacific Blue; BD Biosciences, Custom Bulk), anti-CD4 (140 ng of L200: AmCyan; BD Biosciences, Custom Bulk 658025, 10 ng of BV510; BD Biosciences, Custom Bulk 624340, or 15 ng of BUV395; BD Biosciences, Custom Bulk 624165), anti-CD8a (50 ng of SK1: PerCP-eFluor710; Life Tech, Custom Bulk CUST04424), anti-TNF-α (150 ng of MAB11: FITC; BD Biosciences, Custom Bulk 624046 or 150 ng of PE; BD Biosciences, Custom Bulk 624049), anti-IFN-γ (80 ng of B27: APC; BD Biosciences, Custom Bulk 624078) and anti-CD69 (60 ng of FN50: PE; eBioscience, Custom Bulk CUST01282 or 50 ng of PE-TexasRed; BD Biosciences, Custom Bulk 624005) and for polycytokine analyses, anti-IL-2 (125 ng of MQ1–17H12; PE Cy-7; Biolegend), and anti-MIP-1β (500 ng of D21–1351, BV421; BD Biosciences).
The MHC restriction (MHC-Ia, MHC-E, and MHC-II) of CD8+ T cell responses to peptides or RhCMV-infected cells was determined by pre-incubating isolated mononuclear cell aliquots or RhCMV-infected cells for 1 hour at RT (prior to adding peptides or combining effector and target cells and incubating per the standard ICS assay) in the presence (and absence) of each the following specific inhibitors: (1) the pan anti-MHC-I mAb W6/32 (10 μg/ml, grown in house), (2) the MHC-II-blocking mAb anti-HLA-DR clone G46.6 (1 mg/ml of L243, BioLegend #92203) and CLIP peptide (MHC-II-associated invariant chain, amino acids 89–100, Intavis, 1 mg/ml), or (3) the MHC-E blocking VL9 peptide (VMAPRTLLL; 20 μM). Stimulated cells were fixed, permeabilized, stained and analyzed as described above. To be considered MHC-E-restricted by blocking, the individual peptide response must have been blocked by both anti-pan MHC-I clone W6/32 and MHC-E-binding peptide VL9, and not blocked by anti-MHC-II. MHC-II-restricted responses were blocked by anti-MHC-II but not anti-MHC-I or VL9, and MHC-Ia-restricted responses were blocked by anti-MHC-Ia alone (11, 12). Responses that did not meet these inhibition criteria were considered indeterminate.
For analysis of memory differentiation (central versus transitional versus effector memory) of SIVgag-specific CD4+ and CD8+ T cells, PBMC were stimulated with SIVgag 15mer peptide mix as described above, except that the CD28 co-stimulatory mAb was used as a fluorochrome conjugate to allow CD28 expression levels to be later assessed by flow cytometry. In these experiments, cells were surface-stained after incubation for lineage markers CD3, CD4, CD8, CD95, and CCR7 (see below for mAb clones) prior to fixation and permeabilization and then intracellular staining for response markers (CD69, IFN-γ, TNF-α; note that brefeldin A treatment preserves the pre-stimulation cell-surface expression phenotype of phenotypic markers examined in this study).
Stained samples were analyzed on an LSR-II flow cytometer (BD Biosciences). Data analysis was performed using FlowJo software (Tree Star). In all analyses, gating on the lymphocyte population was followed by the separation of the CD3+ T cell subset and progressive gating on CD4+ and CD8+ T cell subsets. Antigen-responding cells in both CD4+ and CD8+ T cell populations were determined by their intracellular expression of CD69 and either or both of the cytokines IFN-γ and TNF-α (or in polycytokine analyses, expression of CD69 and any combination of the cytokines: IFN-γ, TNF-α, IL-2, MIP-1β). After background subtraction, the raw response frequencies above the assay limit of detection were “memory-corrected” (e.g., percent responding out of the memory population), as previously described (8–10, 42), using combinations of the following fluorochrome-conjugated mAbs to define the memory vs naïve subsets CD3 (450 ng of SP34–2: Alexa700 or 50 ng of SP34–2: PerCP-Cy5.5; BD Biosciences Custom Bulk 624060), CD4 (140 ng of L200: AmCyan), CD8a (50ng SK-1: PerCP-eFluor710, or 20 ng of RPA-T8: APC; BioLegend), TNF-α (150 ng of MAB11; FITC), IFN-γ (80 ng of B27; APC), CD69 (60 ng of FN50; PE), CD28 (40 ng of CD28.2; PE/Dazzle 594, BioLegend), CD95 (35 ng of DX2; PE, BioLegend), CCR7 (120 ng of 15053; Biotin, R&D Systems), streptavidin (1 μg of Pacific Blue, Life Tech and BV605; BD Biosciences, Custom Bulk 624342) and Ki67 (140 ng of B56; FITC, BD Biosciences, Custom Bulk 624046). For memory phenotype analysis of SIVgag-specific T cells, all CD4+ or CD8+ T cells expressing CD69 plus IFN-γ and/or TNF-α were first Boolean OR gated, and then this overall Ag-responding population was subdivided into the memory subsets of interest on the basis of surface phenotype (CCR7 versus CD28). Similarly, for polycytokine analysis of SIVgag-specific T cells, all CD4+ or CD8+ T cells expressing CD69 plus cytokines were Boolean OR gated and polyfunctionality was delineated with any combination of the four cytokines tested (IFN-γ, TNF-α, IL-2, MIP-1β) using the Boolean AND function. The gating strategy for these experiments is delineated in detail in the supplemental information of the companion manuscript (14).
SIV detection assays.
Plasma SIV RNA levels were determined using an SIV Gag-targeted quantitative real time/digital RT-PCR format assay, essentially as previously described, with 6 replicate reactions analyzed per extracted sample for assay thresholds of 15 SIV RNA copies/ml (9, 10, 43).
Statistical Analysis.
Boxplots show jittered points and a box from first to third quartiles (IQR) and a line at the median, with whiskers extending to the farthest data point within 1.5×IQR above and below the box. For all comparisons of T cell response parameters, we performed Wilcoxon rank-sum tests comparing the ΔRh67 68–1 RhCMV/SIV vector vaccinated group to the Rh67-intact 68–1 RhCMV/SIV vector vaccinated reference group. For comparisons of ICS values of T cells co-incubated with different virus variants, we first normalized the response frequency to that obtained by co-incubation with 68–1 RhCMV/SIV-gag-infected TRF and then performed one-sample Wilcoxon rank-sum tests against a null median of 1. For longitudinal responses, we calculated the area under the curve (AUC) or the plateau average value for each RM, as denoted in the figure legends. All Wilcoxon P values are based on two-sided tests and were adjusted using the Holm procedure for family-wise error rate control. P values for analyses of efficacy were based on two-sided exact tests of binomial proportions. Analyses were performed in R v3.6.0 with the package Exact v2.0 (44).
Supplementary Material
Acknowledgements:
We thank T. Whitmer, A. Bhusari A. Legasse, M. Fischer, C. Shriver-Munsch, T. Swanson, A. Sylwester, S. Hagen, E. McDonald, K. Randall, and K. Rothstein for technical or administrative assistance; B. Keele (Frederick National Laboratory) for providing the SIVmac239 challenge virus, and A. Townsend for figure preparation. We acknowledge the sequencing services of the OHSU Massively Parallel Sequencing Shared Resource and the ONPRC Molecular Technologies Core. We also thank the ONPRC Molecular Virology Core for virus stock preparation.
Funding: This work was supported by the National Institute of Allergy and Infectious Diseases (NIAID) grants P01 AI094417, U19 AI128741, UM1 AI124377, and R37 AI054292 to L.J.P., R01 AI140888 to J.B.S. and R01 AI059457 to K.F.; the Oregon National Primate Research Center Core grant from the National Institutes of Health, Office of the Director (P51 OD011092); and contracts from the National Cancer Institute (# HHSN261200800001E) to J.D.L.. This work was also supported by the Bill & Melinda Gates Foundation-supported Collaboration for AIDS Vaccine Discovery (OPP1033121, LJP).
Footnotes
Competing Interests: OHSU, S.G.H., L.J.P., and K.F, have a substantial financial interest in Vir Biotechnology, Inc., a company that may have a commercial interest in the results of this research and technology. S.G.H., L.J.P., and K.F. are also consultants to Vir Biotechnology, Inc., and J.B.S. has received compensation for consulting for Vir Biotechnology, Inc. S.G.H., L.J.P., and K.F. are co-inventors of patent WO 2011/143650 A2 “Recombinant RhCMV and HCMV vectors and uses thereof” licensed to Vir Biotechnology, Inc. S.G.H., L.J.P., K.F., and D. Malouli are co-inventors of patent US2016/0010112 A1 “Cytomegalovirus vectors enabling control of T cell targeting” licensed to Vir Biotechnology, Inc. S.G.H., L.J.P. and K.F. are co-inventors of patent US2017/0143809 A1 “CMV vectors comprising microRNA recognition elements” licensed to Vir Biotechnology, Inc. These potential individual and institutional conflicts of interest have been reviewed and managed by OHSU.
Data and Materials availability: All data associated with this study are present in the paper or Supplementary Materials. Sequences for all recombinant RhCMV are available in GenBank. RhCMV/SIV vectors can be obtained through an MTA.
REFERENCES AND NOTES:
- 1.Noriega V, Redmann V, Gardner T, Tortorella D, Diverse immune evasion strategies by human cytomegalovirus. Immunol Res 54, 140–151 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Vieira Braga FA, Hertoghs KM, van Lier RA, van Gisbergen KP, Molecular characterization of HCMV-specific immune responses: Parallels between CD8(+) T cells, CD4(+) T cells, and NK cells. Eur J Immunol 45, 2433–2445 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Jarvis MA, Hansen SG, Nelson JA, Picker LJ, Früh K, in Cytomegaloviruses: From Molecular Pathogenesis to Intervention Reddehase MJ, Ed. (Caister Academic Press, 2013), vol. 2, chap. 21. [Google Scholar]
- 4.Klenerman P, Oxenius A, T cell responses to cytomegalovirus. Nat Rev Immunol 16, 367–377 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Hansen SG et al. , Effector memory T cell responses are associated with protection of rhesus monkeys from mucosal simian immunodeficiency virus challenge. Nat Med 15, 293–299 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mendez AC, Rodriguez-Rojas C, Del Val M, Vaccine vectors: the bright side of cytomegalovirus. Med Microbiol Immunol 208, 349–363 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Masopust D, Picker LJ, Hidden memories: frontline memory T cells and early pathogen interception. J Immunol 188, 5811–5817 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hansen SG et al. , Profound early control of highly pathogenic SIV by an effector memory T-cell vaccine. Nature 473, 523–527 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hansen SG et al. , Immune clearance of highly pathogenic SIV infection. Nature 502, 100–104 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hansen SG et al. , A live-attenuated RhCMV/SIV vaccine shows long-term efficacy against heterologous SIV challenge. Sci Transl Med 11, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hansen SG et al. , Cytomegalovirus vectors violate CD8+ T cell epitope recognition paradigms. Science 340, 1237874 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hansen SG et al. , Broadly targeted CD8+ T cell responses restricted by major histocompatibility complex E. Science 351, 714–720 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taher H et al. , In vitro and in vivo characterization of a recombinant rhesus cytomegalovirus containing a complete genome. PLoS Pathog 16, e1008666 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malouli D et al. , Cytomegaloviral determinants of CD8+ T cell programming and RhCMV/SIV vaccine efficacy. BioRxiv, 321349 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Braud V, Jones EY, McMichael A, The human major histocompatibility complex class Ib molecule HLA-E binds signal sequence-derived peptides with primary anchor residues at positions 2 and 9. Eur J Immunol 27, 1164–1169 (1997). [DOI] [PubMed] [Google Scholar]
- 16.Braud VM, Allan DS, Wilson D, McMichael AJ, TAP- and tapasin-dependent HLA-E surface expression correlates with the binding of an MHC class I leader peptide. Curr Biol 8, 1–10 (1998). [DOI] [PubMed] [Google Scholar]
- 17.Braud VM et al. , HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature 391, 795–799 (1998). [DOI] [PubMed] [Google Scholar]
- 18.Vales-Gomez M, Reyburn HT, Erskine RA, Lopez-Botet M, Strominger JL, Kinetics and peptide dependency of the binding of the inhibitory NK receptor CD94/NKG2-A and the activating receptor CD94/NKG2-C to HLA-E. EMBO J 18, 4250–4260 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang EC et al. , UL40-mediated NK evasion during productive infection with human cytomegalovirus. Proc Natl Acad Sci U S A 99, 7570–7575 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tomasec P et al. , Surface expression of HLA-E, an inhibitor of natural killer cells, enhanced by human cytomegalovirus gpUL40. Science 287, 1031–1033 (2000). [DOI] [PubMed] [Google Scholar]
- 21.Ulbrecht M et al. , Cutting edge: the human cytomegalovirus UL40 gene product contains a ligand for HLA-E and prevents NK cell-mediated lysis. J Immunol 164, 5019–5022 (2000). [DOI] [PubMed] [Google Scholar]
- 22.Pande NT, Powers C, Ahn K, Früh K, Rhesus Cytomegalovirus Contains Functional Homologues of US2, US3, US6, and US11. J Virol 79, 5786–5798 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koppers-Lalic D et al. , Varicelloviruses avoid T cell recognition by UL49.5-mediated inactivation of the transporter associated with antigen processing. Proc Natl Acad Sci U S A 102, 5144–5149 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Richards R, Scholz I, Powers C, Skach WR, Früh K, The cytoplasmic domain of rhesus cytomegalovirus Rh178 interrupts translation of major histocompatibility class I leader peptide-containing proteins prior to translocation. J Virol 85, 8766–8776 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hansen SG et al. , Evasion of CD8+ T cells is critical for superinfection by cytomegalovirus. Science 328, 102–106 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Powers CJ, Früh K, Signal peptide-dependent inhibition of MHC class I heavy chain translation by rhesus cytomegalovirus. PLoS Pathog 4, e1000150 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sturgill ER et al. , Natural Killer Cell Evasion Is Essential for Infection by Rhesus Cytomegalovirus. PLoS Pathog 12, e1005868 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fukazawa Y et al. , B cell follicle sanctuary permits persistent productive simian immunodeficiency virus infection in elite controllers. Nat Med 21, 132–139 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu HL et al. , The Role of MHC-E in T Cell Immunity Is Conserved among Humans, Rhesus Macaques, and Cynomolgus Macaques. J Immunol 200, 49–60 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Q et al. , Peak SIV replication in resting memory CD4+ T cells depletes gut lamina propria CD4+ T cells. Nature 434, 1148–1152 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Walters LC et al. , Pathogen-derived HLA-E bound epitopes reveal broad primary anchor pocket tolerability and conformationally malleable peptide binding. Nat Commun 9, 3137 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walters LC, McMichael AJ, Gillespie GM, Detailed and atypical HLA-E peptide binding motifs revealed by a novel peptide exchange binding assay. Eur J Immunol 50, 2075–2091 (2020). [DOI] [PubMed] [Google Scholar]
- 33.Camilli G et al. , Regulation and trafficking of the HLA-E molecules during monocyte-macrophage differentiation. J Leukoc Biol 99, 121–130 (2016). [DOI] [PubMed] [Google Scholar]
- 34.Verweij MC et al. , The varicellovirus UL49.5 protein blocks the transporter associated with antigen processing (TAP) by inhibiting essential conformational transitions in the 6+6 transmembrane TAP core complex. J Immunol 181, 4894–4907 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Chang WL, Barry PA, Cloning of the full-length rhesus cytomegalovirus genome as an infectious and self-excisable bacterial artificial chromosome for analysis of viral pathogenesis. J Virol 77, 5073–5083 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Borst EM, Benkartek C, Messerle M, Use of bacterial artificial chromosomes in generating targeted mutations in human and mouse cytomegaloviruses. Curr Protoc Immunol Chapter 10, Unit 10 32 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Tischer BK, Smith GA, Osterrieder N, En passant mutagenesis: a two step markerless red recombination system. Methods in molecular biology 634, 421–430 (2010). [DOI] [PubMed] [Google Scholar]
- 38.Burwitz BJ et al. , Cross-Species Rhesus Cytomegalovirus Infection of Cynomolgus Macaques. PLoS Pathog 12, e1006014 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Manders EMM, Verbeek FJ, Aten JA, Measurement of co-localization of objects in dual-colour confocal images. Journal of Microscopy 169, 375–382 (1993). [DOI] [PubMed] [Google Scholar]
- 40.Dunn KW, Kamocka MM, McDonald JH, A practical guide to evaluating colocalization in biological microscopy. Am J Physiol Cell Physiol 300, C723–C742 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abdulhaqq SA et al. , Vaccine-Mediated Inhibition of the Transporter Associated with Antigen Processing Is Insufficient To Induce Major Histocompatibility Complex E-Restricted CD8(+) T Cells in Nonhuman Primates. J Virol 93, e00592–19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hansen SG et al. , Prevention of tuberculosis in rhesus macaques by a cytomegalovirus-based vaccine. Nat Med 24, 130–143 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hansen SG et al. , Addendum: Immune clearance of highly pathogenic SIV infection. Nature 547, 123–124 (2017). [DOI] [PubMed] [Google Scholar]
- 44.R Development Core Team, R: A language and environment for statistical computing. (Vienna, Austria, 2019). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.