Abstract
Obesity is a serious health issue in the world and is related to a higher risk of suffering metabolic diseases. Understanding the molecular basis of obesity is critical to identify new targets to treat obesity and obesity‐associated metabolic diseases. N6‐methyladenosine (m6A) modification is the most common form of ribonucleic acid modification, which has attracted increasing interest of researchers in recent years, as it is reported that m6A has vital functions in diseases and everyday life activities. Recent studies showed that m6A modification was decreased in obese adipose tissue, and appeared to play a regulatory role in many obesity‐associated biological processes, including adipogenesis, lipid metabolism and insulin resistance. In this review, we discussed the emerging advances in m6A modification in obesity to provide a novel therapeutic strategy for fighting against obesity.
Keywords: Adipogenesis, N6‐methyladenosine, Obesity
N6‐methyladenosine modification has vital functions in diseases and normal life activities. Recent studies showed N6‐methyladenosine modification decreased in obese adipose tissue and played a regulatory role in many obesity‐associated biological processes. We summarized the emerging advances in the roles of N6‐methyladenosine modification in obesity.
Introduction
As a worldwide epidemic, obesity occurs due to a chronic long‐term imbalance between energy intake and expenditure, which has been considered a capital driver in the pathophysiology of obesity and other features relating to metabolism, including diabetes, cardiovascular diseases, insulin resistance and atherosclerosis1, 2. There are mainly two kinds of adipose tissue with distinct functions in the body. White adipose tissue stores excess energy in triglycerides, whereas brown adipose tissue burns calories through thermogenesis3, 4. The balance of these processes is essential for keeping normal adiposity and regulating lipid metabolism. Therefore, comprehending the molecular mechanisms behind the imbalance between energy intake and expenditure could be helpful to provide a new therapeutic strategy for combating obesity.
Ribonucleic acid (RNA) chemical modification is a crucial post‐transcriptional regulator. Like other RNA modifications, the methylation of N6‐adenosine (m6A) is the most common intrinsic eukaryotic messenger RNA (mRNA) modification, modulated by methyltransferase complex, RNA‐binding proteins and demethylases5, 6. m6A has a close relationship with nearly all essentials of mRNA metabolism, including mRNA stability, subcellular localization, translation and alternative splicing7, 8, 9, 10. Expanding evidence has shown that the dysregulation of m6A patterns could lead to abnormal gene expressions and functions, cellular aberrant differentiation and imbalance of homeostasis, even resulting in the occurrence of certain cancer, inflammatory states and metabolic diseases11, 12. Recent studies also showed that m6A modification was decreased in obese adipose tissue and appeared to play a vital role in many obesity‐associated biological processes. Here, we aim to summarize the functional roles and mechanisms of m6A in obesity‐associated biological processes, including adipogenesis, lipid metabolism and insulin resistance.
Overview of m6A methylation
Discovery and development of m6A methylation
As a common epigenetic modification of RNA molecules, m6A was firstly discovered in the early 1970s in mRNAs from eukaryotes13, and the primary function of m6A was associated with mRNA instability14. The following significant breakthrough was the cloning of methyltransferase‐like protein 3 (METTL3) and clarifying its function of synthesizing nearly all of the m6A in the mRNA transcriptome in 199715. Next, scientists reported that the depletion of METTL3 in yeast and arabidopsis led to specific developmental arrest in sporulation and seed development in the 2000s, showing that m6A was a regulated modification and exerted some functions in specific developmental processes16, 17. Afterward, alpha‐ketoglutarate‐dependent dioxygenase fat mass and obesity‐associated protein (FTO) was found to act as m6A demethylase in 2011, showed that RNA modifications could be reversed, and attracted significant interest in the dynamics of m6A modification and its regulatory functions in biology18, 19. m6A antibody was then developed and utilized in immunoprecipitation‐based high‐throughput sequencing in 2012, and enabled the discovery of m6A sites through the whole transcriptome20, 21. That expedited the investigation of the functions of m6A along with the fast development of other m6A detection techniques.
Molecular mechanisms of m6A methylation
m6A modification occurs primarily at the common motif of RRm6ACH ([A/G/U] [A/G] m6AC [A/C/U]), and initially was considered to exist only in mRNA. However, studies had recently suggested that other types of RNA could also arise from m6A modifications, such as ribosomal RNAs (rRNAs), small nuclear RNAs, microRNAs, long non‐coding RNAs and circular RNAs15. There are three kinds of proteins involved in m6A regulation, called m6A methyltransferase, m6A demethylase and m6A‐binding protein, respectively, that decide the fate of RNAs (Figure 1).
Figure 1.
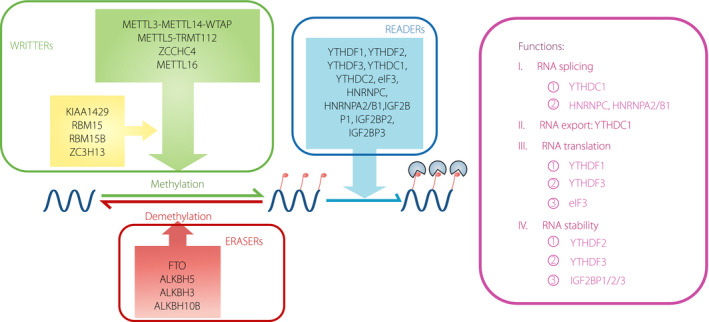
Molecular mechanisms and functions of N6‐methyladenosine (m6A) methylation. The m6A methylation is catalyzed by the “writer” complex mainly including methyltransferase‐like protein (METTL) 3, METTL14, Wilms tumor 1‐associating protein (WTAP), vir like m6A methyltransferase associated (VIRMA), RBM15 (ribonucleic acid [RNA]‐binding motif protein 15), and zinc finger CCCH‐type containing 13 (ZC3H13). The m6A modification is removed by “eraser” mainly including fat mass and obesity‐associated protein (FTO) or AlkB homolog 5 (ALKBH5). “Reader” proteins, including YTH domain‐containing RNA‐binding protein family (YTHDF) 1, YTHDF2, YTHDF3, YTHDC1, YTHDC2, human insulin‐like growth factor 2 (IGF2) messenger RNA (mRNA) binding protein family (IGF2BP) 1, IGF2BP2 and IGF2BP3, recognize m6A and determine target RNA fate.
m6A methyltransferase
m6A methyltransferase, also called “writers”, possesses the capacity to add amounts of RNA with m6A modification. Researchers have found several methyltransferases and complexes to catalyze m6A modification. METTL3‐METTL14‐WTAP (Wilms tumor 1‐associating protein) complex is discovered as a writer earliest and is involved in the large majority of m6A sites from RNA15, 22, 23. rRNA N6‐adenosine‐methyltransferase ZCCHC4 has a role in m6A modification in the 28S subunit of rRNA24 METTL5‐TRMT112 (Homo sapiens tRNA methyltransferase 11‐2 homolog) complex is found to catalyze m6A in the 18S subunit rRNA25. METTL16 is discovered to catalyze the m6A in the U6 small nuclear RNA. It is associated with splicing and the formation of m6A in U6‐like sequences in the methionine adenosyl‐transferase 2A mRNA that encodes the enzyme responsible for S‐adenosylmethionine biosynthesis26. Additionally, METTL16 also catalyzes the m6A in a small part of other mRNAs and non‐coding RNAs27. There are also some other kinds of proteins that have been confirmed as constituents of the methyltransferase complexes, such as KIAA142928, putative RNA‐binding protein 15 (RBM15; and its paralog RBM15B)29 and ZC3H1330, 31.
m6A demethylase
Two Fe2+ and α‐ketoglutarate‐dependent m6A demethylases, also known as “erasers”, can remove m6A methylated groups from RNA. The standard erasers include FTO, also known as AlkB homolog 918, and AlkB homolog 532. Another m6A demethylase, AlkB homolog 333, has been recently identified that preferentially acts upon m6A in tRNA, but not in mRNA or rRNA. Additionally, the AlkB homolog 10B is an m6A demethylase of mRNA in arabidopsis, which regulates mRNA stability and influences the conversion of arabidopsis from vegetative growth to reproductive growth34.
m6A‐binding protein
m6A‐binding proteins, also known as “readers”, could combine to the m6A modification sites in RNA and trigger diverse downstream effects35. The readers found earliest are YTH domain family proteins, consisting of YTH domain family protein 1–3 (YTHDF1‐3; DF family) and YTH domain containing protein 1–2 (YTHDC1‐2; DC family)8, 29, 36. The DF family is mainly located in the cytoplasm, and YTHDF1 and YTHDF3 could improve translation efficiency of mRNA with m6A modification, whereas YTHDF2 relates to shortening the half‐life of mRNA with m6A modification9, 37. The DC family chiefly plays roles in the nucleus. YTHDC1 combines with the m6A sites and regulates mRNA expression levels through affecting alternative splicing36, 38. YTHDC1 also helps the export of m6A methylated transcripts39. YTHDC2 protein can be found both inside and outside the nucleus. It can selectively bind to the m6A sites of non‐coding RNAs29, but what kinds of biological functions will be activated by the occupancy is still unknown. Several other readers, such as eukaryotic initiation factor 3, heterogeneous nuclear ribonucleoprotein C (HNRNP C) and HNRNP A2/B1, have been found. It was suggested that the HNRNP A2/B1 protein might be responsible for the regulation of the transcription of precursor microRNAs40, whereas the HNRNP C protein might play roles in the local secondary structure of mRNAs and long non‐coding RNAs41. Eukaryotic initiation factor 3 was found to bind to m6A sites in the 5'UTRs of mRNAs and promoted their translation42. One type of RNA‐binding protein, insulin‐like growth factor 2 mRNA‐binding protein (IGF2BP, including IGF2BP1, IGF2BP2 and IGF2BP3), was recently identified as another kind of reader that could also distinguish m6A modifications. IGF2BPs promoted the stability and storage of their target RNAs in an m6A‐dependent manner43.
Role of m6A modification in adipogenesis
Adipogenesis is the program of cells from the adipose tissue proliferating, differentiating and turning into cells that could assimilate lipids44. Adipogenesis consists of two vital stages: (ii) commitment; and (ii) terminal differentiation45. In the commitment stage, the pluripotent stem cells locate among the vascular stroma of adipose tissue, responding to signals to determine into preadipocytes46. Terminal differentiation is a process mediated by a series of multiple transcription factors and epigenomic regulators. CCAAT/enhancer‐binding protein gene family and peroxisome proliferator‐activated receptor‐γ (PPARG) are generally considered as essential transcription regulators47, 48, and microRNA and long non‐coding RNA are two kinds of critical epigenomic regulators49, 50 of adipogenesis. Wang et al.51 provided the evidence that mRNA m6A methylation regulates adipogenesis in porcine adipocytes, showing that m6A contributes essentially to adipogenesis. m6A could change the expression level of mRNAs in a mechanistic manner that encodes multiple regulators, including transcription actors, and act as vital functional factors in adipogenesis. In this part, we highlight the multiple roles of m6A in adipogenesis based on different m6A regulators, respectively (Figure 2 and Table 1).
Figure 2.
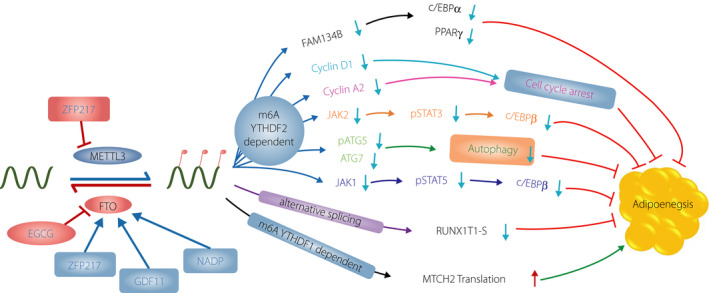
N6‐methyladenosine (m6A) methylation regulates adipogenesis by various mechanisms. Writers, erasers and readers can modulate m6A modification, subsequently affecting the decay and translation of adipogenesis‐associated regulators, finally regulating adipogenesis. In addition, some regulator and natural compounds, such as nicotinamide adenine dinucleotide phosphate (NADP), zinc finger protein 217 (ZFP217), growth differentiation factor 11 (GDF11) and epigallocatechin gallate (EGCG), can also modulate m6A modification and affect adipogenesis.
Table 1.
The role of N6‐methyladenosine modification in adipogenesis
| Upstream regulators | m6A regulators | Target genes | Function and mechanism | Reference |
|---|---|---|---|---|
| – | FTO | RUNX1T1 | FTO controls the exonic splicing of RUNX1T1 via demethylation‐dependent manner and promotes the expression of RUNX1T1‐S isoform and adipogenesis of mouse 3T3‐L1 preadipocytes. | 7 |
| – | FTO | RUNX1T1 | FTO enhances the expression of the RUNX1T1‐S isoform of RUNX1T1 and mitotic clonal expansion via demethylation‐dependent manner, thus promoting adipogenesis in mice. | 56 |
| – | FTO | PPARG | FTO promotes mouse 3T3‐L1 preadipocytes differentiation by decreasing m6A level. Mechanistically, FTO exerts its effect upstream of PPARG during adipogenesis. | 57 |
| GDF11 | FTO | PPARG | FTO demethylates the mRNA of PPARG, leading to the increase in the expression of PPARG mRNA, thus favoring the mouse BMSCs to differentiate to adipocytes in human and mice. GDF11 significantly upregulated the expression of FTO | 61 |
| – | FTO/YTHDF2 | CCNA2, CDK2 | FTO reduces the m6A levels of CCNA2 and CDK2, promotes the protein expression of CCNA2 and CDK2 in YTHDF2‐dependent manner, thus reducing cell cycle progress and inducing adipogenesis of mouse 3T3‐L1 preadipocytes | 58 |
| – | FTO/YTHDF2 | ATG5, ATG7 | FTO reduces the m6A levels of ATG5 and ATG7, resulting in upregulated expression levels of ATG5 and ATG7 in a YTHDF2‐dependent manner, promoting autophagosome formation, autophagy and adipogenesis in mice | 59 |
| – | FTO/YTHDF2 | JAK2 | FTO reduces the m6A level of JAK2, and inhibits mRNA degradation in a YTHDF2‐dependent manner, thus promoting adipogenesis in porcine and mouse preadipocytes | 61 |
| EGCG | FTO | – | EGCG inhibits expression of FTO, and increases m6A levels of CCNA2 and CDK2 mRNA, resulting in decreased protein levels of CCNA2 and CDK2 in YTHDF2‐dependent manner, therefore inhibiting adipogenic differentiation of 3T3L1 cells by blocking the mitotic clonal expansion at the early stage of adipocyte differentiation. | 62 |
| ZFP217 | FTO/YTHDF2 | – | Zfp217 induces the increase of FTO, and promotes the adipogenic differentiation of 3T3L1 cells. Furthermore, the interaction of Zfp217 with YTHDF2 is critical for allowing FTO to maintain its interaction with m6A sites on various mRNAs | 63 |
| miR‐149‐3p | FTO | – | miR‐149‐3p mimic decreased the adipogenic differentiation potential of mouse BMSCs by binding to the 3′UTR of the FTO mRNA, inducing the decreased expression of FTO | 64 |
| NADP | FTO | – | NADP directly binds FTO, increases FTO activity, and promotes RNA m6A demethylation and adipogenesis in mice. | 65 |
| – | METTL3/FTO | – | METTL3 positively correlated with m6A levels and inhibited adipogenesis, FTO negatively regulated m6A levels and promoted adipogenesis in porcine adipocytes | 51 |
| – | METTL3/METTL14/WTAP | CCNA2 | WTAP‐METTL3‐METTL14 complex promotes the expression of CCNA2 and cell cycle transition in mitotic clonal expansion, thus inducing adipogenesis in mice. | 66 |
| – | METTL3/YTHDF2 | JAK1 | METTL3 increases mRNA m6A levels of JAK1, leading to reduced expression of JAK1 via a YTHDF2‐dependent manner, then inhibits the activation of JAK1/STAT5/C/EBPβ pathway and adipogenesis in porcine BMSCs. | 67 |
| ZFP217 | METTL3/YTHDF2 | CCND1 | ZFP217 decreases the expression of METTL3, subsequently decreasing the m6A level of CCND1 mRNA and increasing protein expression of CCND1 in YTHDF2‐dependent manner, thus inducing cell‐cycle progression and promoting adipogenesis in mouse 3T3‐L1 preadipocytes. | 68 |
| – | FTO/YTHDF1 | MTCH2 | m6A enhances MTCH2 translation via a YTHDF1‐dependent pathway and promotes adipogenesis in pigs. | 69 |
| – | YTHDF2 | FAM134B | Loss of m6A on FAM134B mRNA blocks its decay and promotes its translation via a YTHDF2‐dependent pathway, thus promoting porcine preadipocytes adipogenic differentiation | 70 |
BMSCs, bone marrow stem cells; CCNA2, cyclin A2; CCND1, cyclin D1; CDK2, cyclin‐dependent kinase 2; EGCG, epigallocatechin gallate; FTO, fat mass and obesity‐associated protein; GDF11, growth differentiation factor 11; JAK1, Janus kinase 1; JAKC2, Janus kinase 2; m6A, N6‐methyladenosine; METTL3, methyltransferase‐like protein 3; MTCH2, mitochondrial carrier 2; mRNA, messenger ribonucleic acid; NADP, nicotinamide adenine dinucleotide phosphate; STAT5, activator of transcription 5; ZFP217, zinc finger protein 217.
FTO
Although initial studies suggested that FTO influences obesity susceptibility through altering the expression of the adjacent genes, such as IRX3 or IRX5 52, 53, the discovery of FTO as the first genome‐wide association studies‐identified obesity gene in 200754, 55, impelled scientists to focus on the roles of FTO in obesity. Afterward, FTO was found to act on m6A demethylase in 201118, 19, and promoted m6A modification to become one of the great topics of interest in obesity research.
FTO plays critical roles in adipogenesis by mediating m6A demethylation and subsequently affecting the RNA metabolism of regulators in adipogenesis. Zhao et al.7 reported FTO expression was negatively connected to the m6A level during adipogenesis. FTO depletion interfered with adipogenesis of mouse 3T3‐L1 preadipocytes by increasing the m6A levels surrounding splice sites of runt‐related transcription factor 1 partner transcriptional co‐repressor 1 (RUNX1T1), one of the regulators in adipogenesis, thereby controlling the exonic splicing of RUNX1T1 and inhibiting the expression of exon 6‐skipped isoform RUNX1T1‐S7. Another study also showed that FTO enhanced the expression of the RUNX1T1‐S in a demethylation‐dependent manner, then promoted mitotic clonal expansion and adipogenesis of mice56. Zhang et al.57 found that FTO knockdown resulted in upregulation of m6A, thereby preventing the differentiation of mouse 3T3‐L1 preadipocytes, whereas ectopic overexpression of FTO rescued these changes. However, over‐expression of R96Q, the FTO missense mutant lacking demethylase activity, did not interfere with m6A level and differentiation of 3T3‐L1 preadipocyte. Mechanistically, FTO exerts its effect upstream of PPARG during adipogenesis57. in addition, Shen et al.58 found that overexpression of FTO demethylated the mRNA of PPARG, leading to the increase in the expression of PPARG, thus favoring the bone marrow stem cells (BMSCs) to differentiate to adipocytes in human and mice. Wu et al.59 reported FTO depletion highly upregulated the m6A levels of cyclin A2 and cyclin‐dependent kinase 2, crucial cell cycle regulators, subsequently induced its decreased protein expression, finally leading to the delayed entry of MDI (3‐isobutyl‐1‐methylxanthine, dexamethasone and insulin)‐induced 3T3‐L1 preadipocytes into G2 phase, thus extending cell cycle progress and restraining adipogenesis. Wang et al.60 found ectopic suppression of FTO also downregulated the expression levels of autophagy‐related 5 and autophagy‐related 7, the crucial autophagy regulators, in an m6A‐dependent fashion, resulting in attenuation of autophagosome formation, thus preventing autophagy and adipogenesis in mice. In addition, Wu et al.61 showed knockdown of FTO promoted the m6A levels of Janus kinase 2 and facilitated mRNA degradation, and subsequently interfered with signal transducer and activator of transcription 3 phosphorylation, initiating attenuated transcription of CCAAT/enhancer‐binding protein‐β, a regulator of adipocyte differentiation, and inhibited adipogenesis in porcine and mouse preadipocytes.
Some regulators can regulate FTO mediating adipogenesis. Wu et al.62 showed that epigallocatechin gallate (EGCG), the maximum catechin in green tea, was essential in anti‐obesity and anti‐adipogenesis by reducing the expression of FTO and increasing overall levels of RNA m6A methylation. Another study by Song et al.63 showed that zinc finger protein 217 could activate the transcription of m6A demethylase FTO and reduce the m6A level, and thus promote adipogenic differentiation of mouse 3T3L1 cells. Additionally, Shen et al.58 found that FTO could be regulated by growth differentiation factor 11, then reduced the m6A level of PPARG and promoted mouse BMSCs to differentiate to adipocytes. Li et al. found miR‐149‐3p mimic decreased the adipogenic differentiation potential of mouse BMSCs by targeting FTO64. Furthermore, Wang et al.65 reported that nicotinamide adenine dinucleotide phosphate directly bound FTO, independently increased FTO activity, and promoted RNA m6A demethylation and adipogenesis in mice.
METTL3
METTL3, the most important m6A methylase, plays a key role in adipogenesis by mediating m6A methylation of some regulators of adipogenesis. Wang et al.51 provided the first evidence that overexpression of METTL3 inhibited adipogenesis and increased the mRNA m6A level in porcine adipocytes. Then, Kobayashi et al.66 found that the m6A RNA methyltransferase complex of WTAP, METTL3 and METTL14 positively regulated adipogenesis by accelerating cell cycle transition in mitotic clonal expansion in mice. Next, Yao et al.67 found that knockdown of METTL3 in porcine BMSCs decreased mRNA m6A levels of Janus kinase 1. It increased its mRNA stability, and subsequently led to the activation of the Janus kinase 1/activator of transcription 5/CCAAT/enhancer‐binding protein‐β pathway, thus promoting BMSCs adipogenic differentiation. Furthermore, Liu et al.68 showed that METTL3 upregulated m6A levels of cyclin D1 mRNA and consequently reduced the expression of cyclin D1, accordingly resulting in a block of cell cycle progression and inhibition of adipogenesis in mouse 3T3‐L1 preadipocytes. In addition, Liu et al.68 found that METTL3‐mediated anti‐adipogenesis can be regulated by zinc finger protein 217.
It is well known that m6A is a common epigenetic modification of mRNA molecules, and METTL3 is one of the most important enzymes. Therefore, METTL3 might induce m6A modification on both positive and negative regulators involved in adipogenesis. Also, m6A readers mediated various complicated regulations on mRNA fate, including increasing or decreasing the expression of m6A‐modified regulators involved in adipogenesis. These might be the essential reasons for the discrepancy of the adverse role of METTL3 in adipogenesis in differential studies.
YTHDF1 and YTHDF2
m6A‐binding proteins, such as YTHDF1 and YTHDF2, were also involved in adipogenesis by combining to the m6A modification sites in RNA and triggering RNA decay or translation. Jiang et al.69 found that m6A modification increased mitochondrial carrier 2 protein expression and promoted adipogenesis in intramuscular preadipocytes in pigs. Mechanistically, YTHDF1 could recognize the m6A of mitochondrial carrier 2 mRNA and promote its translation. YTHDF2 mediated m6A mRNA decay and functioned as a critical regulator in adipogenesis. Several studies showed that YTHDF2 could recognize the m6A‐modified mRNAs and caused the decay of these mRNAs, then affected the activity and expression of some key regulators involved in adipogenic differentiation61, 63, 67, 70. Some studies showed that YTHDF2 mediated m6A mRNAs decay and regulated expression involved in cell cycle progression59, 62 and mitotic clonal expansion68, thereby playing roles in adipogenesis. In addition, Wang et al.60 found that YTHDF2 can also be involved in adipogenesis by regulating the expression of autophagy‐associated regulators in an m6A‐dependent manner.
Role of m6A modification in lipid metabolism
To date, several studies showed that m6A modification also affected lipid metabolism. Yadav and Rajasekharan71 first discovered that m6A modification was related to the regulation of lipid metabolism in yeast cells. They showed that Ime4, yeast m6A methyltransferase, epi‐transcriptionally regulates triacylglycerol metabolism through the long‐chain fatty acyl‐CoA synthetase71. Another study also carried out by Yadav et al.72 found that IME4 gene loss caused mitochondrial malfunction. Subsequently, triacylglycerol accumulated as lipid droplets72. Additionally, Yadav et al. reported that IME4 gene played a role in the peroxisomal biogenesis, and IME4 gene loss also resulted in the peroxisomal malfunction of fatty acid oxidation73.
Following this, scientists carried out several studies to explore the function of m6A modification in the lipid metabolism of mammalian cells. Luo et al.74 carried out m6A‐modified RNA immunoprecipitation sequencing to clarify differences of m6A methylomes in normal mouse liver compared with fatty liver triggered by a high‐fat diet (HFD). They found the hypermethylated coding genes on feeding a HFD were primarily enriched in processes associated with lipid metabolism74. The results suggested that m6A methylation might act as a critical regulator of lipid metabolism. Wang et al.75 found that m6A positively mediated the expression of patatin‐like phospholipase domain containing 2, one kind of protein regulating lipid metabolism, and inhibited lipid accumulation in pigs75. Kang et al.76 reported FTO decreased m6A levels, and thus promoted triglyceride (TG) deposition in HepG2 cells, showing that FTO mediating m6A decrease promoted fat metabolism. Xie et al.77 found hepatocyte‐specific knockdown of METTL3 in mice fed a HFD could decrease fatty acid synthesis. Mechanistically, knockdown of METTL3 depletion reduced the m6A methylated and total mRNA level of fatty acid synthase, afterward it restrained fatty acid metabolism77. Likewise, Zhong et al.78 reported that the accumulation of lipid in HepG2 cells was inhibited through knockdown of METTL3 or YTHDF2. They consequently supposed that m6A RNA methylation mediated the interaction between the circadian clock and lipid metabolism78.
Other studies showed that some natural compounds and stress might regulate lipid metabolism by affecting m6A modification. Lu et al.79 reported that curcumin in the diet impacted m6A regulators expression and upregulated the abundance of m6A in piglet livers, thus weakening hepatic lipid metabolism disorder induced by lipopolysaccharide. Similarly, Wu et al.62 showed that EGCG inhibited lipid accumulation of 3T3‐L1 preadipocytes by targeting FTO‐dependent demethylation of m6A. Another study by Heng et al. showed that early fat deposition was partially regulated by maternal heat stress through m6A RNA methylation in neonatal piglets80.
The role of m6A in insulin synthesis, secretion and sensitivity
Obesity is related to dysfunction of insulin secretion and sensitivity, and a higher risk of type 2 diabetes mellitus in serious obese rodents and humans. Recent studies showed that m6A played a role in regulating insulin secretion and sensitivity. Men et al.81 found acute depletion of METTL14 in β‐cells of adult mice led to glucose intolerance due to a reduction in insulin secretion in β‐cells. Mechanistically, they reported that acute METTL14 deletion in β‐cells caused glucose intolerance by activating the inositol‐requiring enzyme 1α/spliced X‐box protein binding 1 pathway. Xie et al.77 reported that m6A methylated RNA and METTL3 expression levels were higher in mice fed with HFD for 16 weeks, compared with the standard chow diet. Hepatocyte‐specific knockdown of METTL3 in mice fed with HFD enforced sensitivity of insulin. It inhibited the synthesis of fatty acid, and exploration of the mechanism showed that METTL3 knockdown reduced the m6A methylated and total mRNA level of fatty acid synthase, and accordingly restrained fatty acid metabolism77. Also, De Jesus et al.82 showed that m6A mRNA methylation played roles in regulating human β‐cell biology, including cell cycle progress, insulin secretion, and the insulin/insulin‐like growth factor‐1–protein kinase B–pancreatic and duodenal homeobox 1 pathway in humans and mice. These studies showed that m6A was essential for insulin secretion and sensitivity, which might emerge as the therapeutic target of obesity and diabetes.
Clinical relevance of m6A targeted strategy in obesity
Along with studies clarifying functions of m6A in obesity, it becomes necessary to test if m6A could be a possible therapeutic target of obesity and obesity‐associated metabolic diseases. However, as yet, there are no optional inhibitors of m6A regulators for clinical practice.
Since the discovery of FTO as an m6A demethylase in 2011, it became the highlight of researching m6A targeted therapy18. In 2012, the natural agent, rhein, was proven to inhibit FTO‐dependent m6A demethylation by intervening with FTO binding to the m6A substrate in cells directly83. In 2014, a selective FTO inhibitor was developed, which could inhibit the m6A demethylase activity of FTO selectively and upregulate the m6A levels in cells84. Next, several FTO inhibitors, including meclofenamic acid, MO‐I‐500, compound 12 and R‐2‐hydroxyglutarate, were reported to inhibit the m6A demethylase activity of FTO32, 85, 86, 87, which have the potential for cancer treatment. In a recent study, entacapone, an inhibitor of catechol‐O‐methyltransferase for treatment of Parkinson's disease, was identified by scientists as a chemical inhibitor of FTO, which induced effects on metabolic homeostasis by inhibiting FTO activity selectively88. Wu et al. found EGCG, the maximum catechin in green tea, could target FTO and inhibit adipogenesis62. These two studies showed that entacapone and EGCG seemed to have the potential to treat obesity and other metabolic diseases.
In another study, miR‐149‐3p could restrain the adipogenic differentiation potential of mouse BMSCs by targeting FTO, which showed that microRNA might also have the therapeutic potential for obesity64. Furthermore, novel anti‐metabolic agents aimed at other m6A regulators might be optimal treatment as well. For instance, METTL3 activators are possibly effective in treating obesity through inhibiting BMSCs adipogenic differentiation67, whereas YTHDF1 inhibitors are likely to inhibit adipogenesis by intervening in m6A‐YTHDF1‐regulated mRNA translation in intramuscular preadipocytes75.
Conclusions
New evidence showed that m6A was significant in adipogenesis, lipid metabolism and insulin sensitivity. There have been some achievements in developing the potential targets of m6A for treatment. Nevertheless, we have uncovered a small part of the roles of m6A in regulating obesity. Thus, it is necessary to clarify and characterize the m6A‐dependent mechanisms closely related to obesity‐associated features, such as adipogenesis, lipid metabolism, metabolic inflammation and insulin sensitivity, which will identify new therapeutic targets contributing to exploring new treatment for obesity and associated metabolic diseases.
Disclosure
The authors declare no conflict of interest.
Acknowledgments
This work was supported by the Scientific Research Project on Young Scientific and Technological Talents of Department of Education of Liaoning Province (QN2019033) and the Scientific research funding project of the Department of Education of Liaoning Province (LJC2019ST02).
J Diabetes Investig. 2021
REFERENCES
- 1.Wang YC, McPherson K, Marsh T, et al. Health and economic burden of the projected obesity trends in the USA and the UK. Lancet 2011; 378: 815–825. [DOI] [PubMed] [Google Scholar]
- 2.Lumeng CN, Saltiel AR. Inflammatory links between obesity and metabolic disease. J Clin Invest 2011; 121: 2111–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosen ED, MacDougald OA. Adipocyte differentiation from the inside out. Nat Rev Mol Cell Biol 2006; 7: 885–896. [DOI] [PubMed] [Google Scholar]
- 4.Cypess AM, Kahn CR. Brown fat as a therapy for obesity and diabetes. Curr Opin Endocrinol Diabetes Obes 2010; 17: 143–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fu Y, Dominissini D, Rechavi G, et al. Gene expression regulation mediated through reversible m(6)A RNA methylation. Nat Rev Genet 2014; 15: 293–306. [DOI] [PubMed] [Google Scholar]
- 6.Wu R, Jiang D, Wang Y, et al. N (6)‐Methyladenosine (m(6)A) methylation in mRNA with a dynamic and reversible epigenetic modification. Mol Biotechnol 2016; 58: 450–459. [DOI] [PubMed] [Google Scholar]
- 7.Zhao X, Yang Y, Sun BF, et al. FTO‐dependent demethylation of N6‐methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res 2014; 24: 1403–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang X, Lu Z, Gomez A, et al. N6‐methyladenosine‐dependent regulation of messenger RNA stability. Nature 2014; 505: 117–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang X, Zhao BS, Roundtree IA, et al. N(6)‐methyladenosine modulates messenger RNA translation efficiency. Cell 2015; 161: 1388–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng G, Dahl JA, Niu Y, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell 2013; 49: 18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu J, Harada BT, He C. Regulation of gene expression by N(6)‐methyladenosine in cancer. Trends Cell Biol 2019; 29: 487–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tong J, Flavell RA, Li HB. RNA m(6)A modification and its function in diseases. Front Med 2018; 12: 481–489. [DOI] [PubMed] [Google Scholar]
- 13.Dubin DT, Taylor RH. The methylation state of poly A‐containing messenger RNA from cultured hamster cells. Nucleic Acids Res 1975; 2: 1653–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sommer S, Lavi U, Darnell JE Jr. The absolute frequency of labeled N‐6‐methyladenosine in HeLa cell messenger RNA decreases with label time. J Mol Biol 1978; 124: 487–499. [DOI] [PubMed] [Google Scholar]
- 15.Bokar JA, Shambaugh ME, Polayes D, et al. Purification and cDNA cloning of the AdoMet‐binding subunit of the human mRNA (N6‐adenosine)‐methyltransferase. RNA 1997; 3: 1233–1247. [PMC free article] [PubMed] [Google Scholar]
- 16.Clancy MJ, Shambaugh ME, Timpte CS, et al. Induction of sporulation in Saccharomyces cerevisiae leads to the formation of N6‐methyladenosine in mRNA: a potential mechanism for the activity of the IME4 gene. Nucleic Acids Res 2002; 30: 4509–4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhong S, Li H, Bodi Z, et al. MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex‐specific splicing factor. Plant Cell 2008; 20: 1278–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jia G, Fu Y, Zhao X, et al. N6‐methyladenosine in nuclear RNA is a major substrate of the obesity‐associated FTO. Nat Chem Biol 2011; 7: 885–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He C. Grand challenge commentary: RNA epigenetics? Nat Chem Biol 2010; 6: 863–865. [DOI] [PubMed] [Google Scholar]
- 20.Meyer KD, Saletore Y, Zumbo P, et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell 2012; 149: 1635–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dominissini D, Moshitch‐Moshkovitz S, Schwartz S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A‐seq. Nature 2012; 485: 201–206. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, Li Y, Toth JI, et al. N6‐methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol 2014; 16: 191–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ping XL, Sun BF, Wang L, et al. Mammalian WTAP is a regulatory subunit of the RNA N6‐methyladenosine methyltransferase. Cell Res 2014; 24: 177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma H, Wang X, Cai J, et al. N(6‐)Methyladenosine methyltransferase ZCCHC4 mediates ribosomal RNA methylation. Nat Chem Biol 2019; 15: 88–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Tran N, Ernst FGM, Hawley BR, et al. The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112. Nucleic Acids Res 2019; 47: 7719–7733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pendleton KE, Chen B, Liu K, et al. The U6 snRNA m(6)A Methyltransferase METTL16 regulates SAM synthetase intron retention. Cell 2017; 169: 824–835.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Warda AS, Kretschmer J, Hackert P, et al. Human METTL16 is a N(6)‐methyladenosine (m(6)A) methyltransferase that targets pre‐mRNAs and various non‐coding RNAs. EMBO Rep 2017; 18: 2004–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yue Y, Liu J, Cui X, et al. VIRMA mediates preferential m(6)A mRNA methylation in 3'UTR and near stop codon and associates with alternative polyadenylation. Cell Discov 2018; 4: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patil DP, Chen CK, Pickering BF, et al. m(6)A RNA methylation promotes XIST‐mediated transcriptional repression. Nature 2016; 537: 369–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wen J, Lv R, Ma H, et al. Zc3h13 regulates nuclear RNA m(6)A methylation and mouse embryonic stem cell self‐renewal. Mol Cell 2018; 69: 1028–1038.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knuckles P, Lence T, Haussmann IU, et al. Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA‐binding factor Rbm15/Spenito to the m(6)A machinery component Wtap/Fl(2)d. Genes Dev 2018; 32: 415–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang Y, Yan J, Li Q, et al. Meclofenamic acid selectively inhibits FTO demethylation of m6A over ALKBH5. Nucleic Acids Res 2015; 43: 373–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ueda Y, Ooshio I, Fusamae Y, et al. AlkB homolog 3‐mediated tRNA demethylation promotes protein synthesis in cancer cells. Sci Rep 2017; 7: 42271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Duan HC, Wei LH, Zhang C, et al. ALKBH10B Is an RNA N(6)‐Methyladenosine demethylase affecting Arabidopsis Floral Transition. Plant Cell 2017; 29: 2995–3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Allis CD, Jenuwein T. The molecular hallmarks of epigenetic control. Nat Rev Genet 2016; 17: 487–500. [DOI] [PubMed] [Google Scholar]
- 36.Meyer KD, Jaffrey SR. Rethinking m(6)A readers, writers, and erasers. Annu Rev Cell Dev Biol 2017; 33: 319–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Z, Huang C, Bao C, et al. Corrigendum: exon‐intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 2017; 24: 194. [DOI] [PubMed] [Google Scholar]
- 38.Xiao W, Adhikari S, Dahal U, et al. Nuclear m(6)A Reader YTHDC1 regulates mRNA splicing. Mol Cell 2016; 61: 507–519. [DOI] [PubMed] [Google Scholar]
- 39.Roundtree IA, Luo GZ, Zhang Z, et al. YTHDC1 mediates nuclear export of N(6)‐methyladenosine methylated mRNAs. eLife 2017; 6: 31311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alarcon CR, Goodarzi H, Lee H, et al. HNRNPA2B1 is a mediator of m(6)A‐dependent nuclear RNA processing events. Cell 2015; 162: 1299–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu N, Dai Q, Zheng G, et al. N(6)‐methyladenosine‐dependent RNA structural switches regulate RNA‐protein interactions. Nature 2015; 518: 560–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meyer KD, Patil DP, Zhou J, et al. 5' UTR m(6)A promotes Cap‐Independent translation. Cell 2015; 163: 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang H, Weng H, Sun W, et al. Recognition of RNA N(6)‐methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol 2018; 20: 285–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fernyhough ME, Okine E, Hausman G, et al. PPARgamma and GLUT‐4 expression as developmental regulators/markers for preadipocyte differentiation into an adipocyte. Domest Anim Endocrinol 2007; 33: 367–378. [DOI] [PubMed] [Google Scholar]
- 45.Otto TC, Lane MD. Adipose development: from stem cell to adipocyte. Crit Rev Biochem Mol Biol 2005; 40: 229–242. [DOI] [PubMed] [Google Scholar]
- 46.Lefterova MI, Lazar MA. New developments in adipogenesis. Trends Endocrinol Metab 2009; 20: 107–114. [DOI] [PubMed] [Google Scholar]
- 47.Cristancho AG, Lazar MA. Forming functional fat: a growing understanding of adipocyte differentiation. Nat Rev Mol Cell Biol 2011; 12: 722–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Siersbaek R, Nielsen R, Mandrup S. Transcriptional networks and chromatin remodeling controlling adipogenesis. Trends Endocrinol Metab 2012; 23: 56–64. [DOI] [PubMed] [Google Scholar]
- 49.Peng Y, Yu S, Li H, et al. MicroRNAs: emerging roles in adipogenesis and obesity. Cell Signal 2014; 26: 1888–1896. [DOI] [PubMed] [Google Scholar]
- 50.Chen C, Cui Q, Zhang X, et al. Long non‐coding RNAs regulation in adipogenesis and lipid metabolism: emerging insights in obesity. Cell Signal 2018; 51: 47–58. [DOI] [PubMed] [Google Scholar]
- 51.Wang X, Zhu L, Chen J, et al. mRNA m(6)A methylation downregulates adipogenesis in porcine adipocytes. Biochem Biophys Res Commun 2015; 459: 201–207. [DOI] [PubMed] [Google Scholar]
- 52.Smemo S, Tena JJ, Kim KH, et al. Obesity‐associated variants within FTO form long‐range functional connections with IRX3. Nature 2014; 507: 371–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Claussnitzer M, Dankel SN, Kim KH, et al. FTO obesity variant circuitry and adipocyte browning in humans. N Engl J Med 2015; 373: 895–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Frayling TM, Timpson NJ, Weedon MN, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science 2007; 316: 889–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scuteri A, Sanna S, Chen WM, et al. Genome‐wide association scan shows genetic variants in the FTO gene are associated with obesity‐related traits. PLoS Genet 2007; 3: e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Merkestein M, Laber S, McMurray F, et al. FTO influences adipogenesis by regulating mitotic clonal expansion. Nat Commun 2015; 6: 6792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang M, Zhang Y, Ma J, et al. The Demethylase Activity of FTO (Fat Mass and Obesity Associated Protein) is required for preadipocyte differentiation. PLoS One 2015; 10: e0133788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shen GS, Zhou HB, Zhang H, et al. The GDF11‐FTO‐PPARgamma axis controls the shift of osteoporotic MSC fate to adipocyte and inhibits bone formation during osteoporosis. Biochim Biophys Acta Mol Basis Dis 2018; 1864: 3644–4365. [DOI] [PubMed] [Google Scholar]
- 59.Wu R, Liu Y, Yao Y, et al. FTO regulates adipogenesis by controlling cell cycle progression via m(6)A‐YTHDF2 dependent mechanism. Biochim Biophys Acta Mol Cell Biol Lipids 2018; 1863: 1323–1330. [DOI] [PubMed] [Google Scholar]
- 60.Wang X, Wu R, Liu Y, et al. m(6)A mRNA methylation controls autophagy and adipogenesis by targeting Atg5 and Atg7. Autophagy 2019; 16: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu R, Guo G, Bi Z, et al. m(6)A methylation modulates adipogenesis through JAK2‐STAT3‐C/EBPbeta signaling. Biochim Biophys Acta Gene Regul Mech 2019; 1862: 796–806. [DOI] [PubMed] [Google Scholar]
- 62.Wu R, Yao Y, Jiang Q, et al. Epigallocatechin gallate targets FTO and inhibits adipogenesis in an mRNA m(6)A‐YTHDF2‐dependent manner. Int J Obes (Lond) 2018; 42: 1378–1388. [DOI] [PubMed] [Google Scholar]
- 63.Song T, Yang Y, Wei H, et al. Zfp217 mediates m6A mRNA methylation to orchestrate transcriptional and post‐transcriptional regulation to promote adipogenic differentiation. Nucleic Acids Res 2019; 47: 6130–6144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li Y, Yang F, Gao M, et al. miR‐149‐3p regulates the switch between adipogenic and osteogenic differentiation of BMSCs by Targeting FTO. Mol Ther Nucleic Acids 2019; 17: 590–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang L, Song CL, Wang N, et al. NADP modulates RNA m 6 A methylation and adipogenesis via enhancing FTO activity. Nat Chem Biol 2020; 16: 1394–1402. [DOI] [PubMed] [Google Scholar]
- 66.Kobayashi M, Ohsugi M, Sasako T, et al. The RNA methyltransferase complex of WTAP, METTL3, and METTL14 regulates mitotic clonal expansion in Adipogenesis. Mol Cell Biol 2018; 38: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yao Y, Bi Z, Wu R, et al. METTL3 inhibits BMSC adipogenic differentiation by targeting the JAK1/STAT5/C/EBPbeta pathway via an m(6)A‐YTHDF2‐dependent manner. FASEB J 2019; 33: 7529–7544. [DOI] [PubMed] [Google Scholar]
- 68.Liu Q, Zhao Y, Wu R, et al. ZFP217 regulates adipogenesis by controlling mitotic clonal expansion in a METTL3‐m(6)A dependent manner. RNA Biol 2019; 16: 1785–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jiang Q, Sun B, Liu Q, et al. MTCH2 promotes adipogenesis in intramuscular preadipocytes via an m(6)A‐YTHDF1‐dependent mechanism. FASEB J 2019; 33: 2971–2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cai M, Liu Q, Jiang Q, et al. Loss of m(6) A on FAM134B promotes adipogenesis in porcine adipocytes through m(6) A‐YTHDF2‐dependent way. IUBMB Life 2019; 71: 580–586. [DOI] [PubMed] [Google Scholar]
- 71.Yadav PK, Rajasekharan R. The m(6)A methyltransferase Ime4 epitranscriptionally regulates triacylglycerol metabolism and vacuolar morphology in haploid yeast cells. J Biol Chem 2017; 292: 13727–13744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yadav PK, Rajasekharan R. The m(6)A methyltransferase Ime4 and mitochondrial functions in yeast. Curr Genet 2018; 64: 353–357. [DOI] [PubMed] [Google Scholar]
- 73.Yadav PK, Rajvanshi PK, Rajasekharan R. The role of yeast m(6)A methyltransferase in peroxisomal fatty acid oxidation. Curr Genet 2018; 64: 417–422. [DOI] [PubMed] [Google Scholar]
- 74.Luo Z, Zhang Z, Tai L, et al. Comprehensive analysis of differences of N(6)‐methyladenosine RNA methylomes between high‐fat‐fed and normal mouse livers. Epigenomics 2019; 11: 1267–1282. [DOI] [PubMed] [Google Scholar]
- 75.Wang X, Sun B, Jiang Q, et al. mRNA m(6)A plays opposite role in regulating UCP2 and PNPLA2 protein expression in adipocytes. Int J Obes (Lond) 2018; 42: 1912–1924. [DOI] [PubMed] [Google Scholar]
- 76.Kang H, Zhang Z, Yu L, et al. FTO reduces mitochondria and promotes hepatic fat accumulation through RNA demethylation. J Cell Biochem 2018; 119: 5676–5685. [DOI] [PubMed] [Google Scholar]
- 77.Xie W, Ma LL, Xu YQ, et al. METTL3 inhibits hepatic insulin sensitivity via N6‐methyladenosine modification of Fasn mRNA and promoting fatty acid metabolism. Biochem Biophys Res Commun 2019; 518: 120–126. [DOI] [PubMed] [Google Scholar]
- 78.Zhong X, Yu J, Frazier K, et al. Circadian Clock regulation of hepatic lipid metabolism by modulation of m(6)A mRNA methylation. Cell Rep 2018; 25: 1816–1828.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lu N, Li X, Yu J, et al. Curcumin attenuates lipopolysaccharide‐induced hepatic lipid metabolism disorder by modification of m(6) A RNA methylation in piglets. Lipids 2018; 53: 53–63. [DOI] [PubMed] [Google Scholar]
- 80.Heng J, Tian M, Zhang W, et al. Maternal heat stress regulates the early fat deposition partly through modification of m(6)A RNA methylation in neonatal piglets. Cell Stress Chaperones 2019; 24: 635–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Men L, Sun J, Luo G, et al. Acute deletion of METTL14 in beta‐cells of adult mice results in glucose intolerance. Endocrinology 2019; 160: 2388–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.De Jesus DF, Zhang Z, Kahraman S, et al. m(6)A mRNA methylation regulates human beta‐cell biology in physiological states and in Type 2 diabetes. Nat Metab 2019; 1: 765–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen B, Ye F, Yu L, et al. Development of cell‐active N6‐methyladenosine RNA demethylase FTO inhibitor. J Am Chem Soc 2012; 134: 17963–17971. [DOI] [PubMed] [Google Scholar]
- 84.Zheng G, Cox T, Tribbey L, et al. Synthesis of a FTO inhibitor with anticonvulsant activity. ACS Chem Neurosci 2014; 5: 658–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Singh B, Kinne HE, Milligan RD, et al. Important role of FTO in the survival of rare panresistant triple‐negative inflammatory breast cancer cells facing a severe metabolic challenge. PLoS One 2016; 11: e0159072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Su R, Dong L, Li C, et al. R‐2HG Exhibits anti‐tumor activity by targeting FTO/m(6)A/MYC/CEBPA signaling. Cell 2018; 172: 90–105.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Toh JDW, Sun L, Lau LZM, et al. A strategy based on nucleotide specificity leads to a subfamily‐selective and cell‐active inhibitor of N(6)‐methyladenosine demethylase FTO. Chem Sci 2015; 6: 112–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Peng S, Xiao W, Ju D, et al. Identification of entacapone as a chemical inhibitor of FTO mediating metabolic regulation through FOXO1. Sci Transl Med 2019; 11: eaau7116. [DOI] [PubMed] [Google Scholar]


