Abstract
Aims/Introduction
Differentially expressed microribonucleic acids (miRNAs) in the placenta and circulating exosomes are of diagnostic value for gestational diabetes mellitus (GDM). In a cross‐sectional study, we identified miRNAs expressed both in the placenta and circulating exosomes of pregnant women with GDM, and estimated their diagnostic value.
Materials and Methods
Next‐generation sequencing was used to identify miRNAs in the placenta that were differentially expressed between GDM and normal glucose tolerance pregnancies. Quantitative polymerase chain reaction was used to validate the identified targets. Western blot and transmission electron microscopy were used to validate exosomes. Univariate logistic regression analysis was used to establish diagnostic models based on miRNAs expression, and the diagnostic value was estimated using the receiver operator characteristic curve.
Results
We identified 157 dysregulated miRNAs in the placental tissue obtained from GDM pregnancies. Of these, miRNA‐125b was downregulated (P < 0.001), whereas miRNA‐144 was upregulated (P < 0.001). The patterns of these two miRNAs remained the same in circulating exosomes from GDM pregnancies (all P < 0.001). miRNA‐144 levels in the circulating exosomes negatively correlated with body mass index both before pregnancy (P = 0.018) and before delivery (P = 0.039), and positively correlated with blood glucose at 1 h, estimated using the oral glucose tolerance test (P = 0.044). The area under curve for the established diagnostic model was 0.898, which was higher than blood glucose levels at 0 h.
Conclusions
These findings suggest that miRNA‐125b and miRNA‐144 are consistently dysregulated in circulating exosomes and the placenta from GDM pregnancies, and are of excellent diagnostic value for GDM.
Keywords: Diagnostic model, Exosome, Gestational diabetes mellitus
A total of 764 microribonucleic acids (miRNAs) were detected, there are 114 upregulated and 43 downregulated miRNAs in the placental tissues of gestational diabetes mellitus patients. MiRNA‐125b and miRNA‐144 play a vital role in the development of gestational diabetes mellitus. Circulating exosomal miRNA‐125b and miRNA‐144 have excellent diagnostic value for gestational diabetes mellitus.
INTRODUCTION
Over the past 20 years, the incidence of gestational diabetes mellitus (GDM) has increased to as high as 17.5–18.9% in the Chinese population1, 2, and this is of urgent concern, as GDM causes both short‐ and long‐term health consequences in fetuses and their mothers3, 4, 5. It has been reported that pregnant women with GDM are susceptible to pre‐eclampsia3, and face an increased risk of birth through cesarean section6. As high as 70% of women with GDM suffer from type 2 diabetes mellitus and cardiovascular disease, indicating the long‐term consequences of the disease7, 8. The pathogenesis of GDM is complex and relatively unknown. The placenta seems to play vital roles in secreting hormones associated with insulin resistance, adipokine secretion and expression of placenta‐specific proteins9. Recent studies have shown that microribonucleic acids (miRNAs) regulate the expression of up to 30% of genes involved in various biological processes, including energy metabolism. Aberrantly expressed miRNAs in the placental tissue have been shown to be related to the incidence of GDM10, 11, 12, 13, 14, despite inconsistencies in the identification of specific miRNAs. miRNAs are detected both in the cytoplasm and extracellular space. Although the precise mechanisms of placental miRNAs are poorly understood, recent studies have found that the placenta secretes exosomes containing miRNAs into the peripheral blood15, 16. As exosomes are highly stable in body fluids17, exosomal miRNAs can be candidate biomarkers for several diseases. Recent studies have linked placenta‐derived exosomal miRNAs to pregnancy‐associated complications, such as pre‐eclampsia, preterm labor and fetal growth restriction18, 19, 20. Despite this, little is known about placenta‐derived exosomal miRNAs in the pathogenesis of GDM.
The diagnosis of GDM now depends on the oral glucose tolerance test (OGTT), which is cumbersome to carry out, requires fasting and multiple blood draws, and its association with nausea and vomiting leads to decreased patient compliance. Therefore, finding new biomarkers, such as miRNAs, for GDM might supply clues for early identification and auxiliary diagnosis of GDM.
In the present study, we explored the diagnostic value of aberrantly expressed circulating exosomal miRNAs for GDM. We identified differentially expressed miRNAs in the placental tissue of patients with GDM, and examined their expression pattern in exosomes in the peripheral blood. We also assessed the correlation between target genes of the validated miRNAs and metabolic pathways and biological processes, as well as established the diagnostic value of the miRNAs identified in the current study.
The significance of the present study lies in estimating the potential of exosomal miRNAs as biomarkers for GDM and assessing its diagnostic value. The results also reinforce the role of placenta‐derived exosomal miRNA in the pathology of GDM.
METHODS
Patient information
Participants were recruited to the study from December 2016 to December 2018, during a routine checkup at The Second Hospital of Shandong University, Ji’nan, China. Women aged between 20 and 40 years, with singleton pregnancy, and diagnosed with GDM were recruited in the experimental group, and age‐matched women of the same gestational age with normal glucose tolerance (NGT) were recruited in the control group. GDM and NGT were determined at 24–28 weeks of gestation, according to the recommendations of the International Association of Diabetes and Pregnancy Study Groups. The experimental group received diet and exercise therapy, and those whose blood glucose levels, was higher than the target value were treated with subcutaneous insulin injection until the patient's blood glucose levels normalized (fasting blood glucose <5.3 mmol/L, blood glucose < 7.8 mmol/L within 1 h of a meal and <6.7 mmol/L within 2 h of a meal). Exclusion criteria included pregnant women with pre‐pregnancy diabetes, hypertension, multiple pregnancies, threatened preterm labor, preterm birth, fetal growth restriction, placenta previa and other pregnancy‐associated complications.
This study was approved by the ethics committee of The Second Hospital of Shandong University. All samples were collected after obtaining informed consent from the participants. Placental tissues were collected from patients between 37 and 41 weeks of pregnancy. Placental tissue, 4–6 mm in diameter, was collected immediately after the delivery of the recruited patients, after which the blood in the placental tissue was rinsed with sterile saline. Blood plasma was collected from recruited patients late in their pregnancy (26–40 weeks of pregnancy). All harvested samples were cryopreserved at −80 °C for long‐term storage. Clinical data used in the present study were obtained from outpatient and inpatient medical records.
Next‐generation sequencing
Total RNA was isolated from equivalent amounts of placental tissue collected from pregnant women with GDM and NGT, using the NanoPhotometer® Spectrophotometer (IMPLEN, Los Angeles, CA, USA), Qubit® 2.0 Flurometer (Life Technologies, Los Angeles, CA, USA) and Agilent 2100 RNA Nano 6000 Assay Kit (Agilent Technologies, Los Angeles, CA, USA). The qualified DNA library was sequenced using the SE50 sequencing program on the NextSeq sequencing platform to obtain high‐quality sequence reads. The data were cleaned and aligned to the reference genome by Bowtie, and miRNAs were identified using the miRDeep2 software. Differentially expressed miRNAs were obtained by DESeq analysis with a q‐value <0.05 and |log2foldchange|>1.
RNA extraction and quantitative polymerase chain reaction
Total RNA was extracted from the placental tissues and exosomes using TRIzol reagent (Invitrogen, Carlsbad, CA, USA), according to the manufacturer’s instructions. The complementary deoxyribonucleic acid was synthesized with M‐MuL V Reverse Transcriptase (Fermentas, Hanover, MD, USA) in the presence of a bulge‐loop miRNA‐144/125b/543/15b/451/U6 miRNA RT primer (RiboBio, Guangzhou, Guangdong, China). Quantitative polymerase chain reaction (qPCR) was carried out using the Maxima SYBR Green qPCR Master Mix (Thermo Scientific, Foster City, CA, USA) on the CFX96 Real‐Time PCR Detection System (Bio‐Rad, Hercules, CA, USA), and PCR reactions were carried out in triplicate. PCR reaction began with an initial denaturation 95°C for 20 s, then 40 cycles of 95°C for 10 s, 60°C for 20 s and 70°C for 10 s. U6 was used to normalize the qPCR data.
Exosome isolation from serum
Blood samples were centrifuged at 1,610 g for 10 min at room temperature, and the supernatant – that is, the serum – was collected and immediately stored at −80°C. Next, the frozen serum samples were thawed, mixed with 0.2 volume of Total Exosome Isolation Reagent (Thermo Scientific, Worcester, MA, USA), incubated at 4°C for 60 min and then centrifuged at 17,100 g for 10 min at 4°C. The supernatant was discarded, and the excipient pellet was resuspended in an appropriate amount of phosphate‐buffered saline.
Western blotting
Western blot analysis for placental alkaline phosphatase, Tumor Suspectibility Gene 101 (TSG101) and CD63, which are enriched in exosomes, was carried out. Total protein was extracted using the lysis buffer Pro‐Prep (iNtRON Biotechnology, Seongnam, Korea), and 20 μg of protein was separated using a 10% sodium dodecyl sulphate–polyacrylamide gel electrophoresis transferred onto a polyvinylidene fluoride (PVDF) membrane (GE Healthcare, Piscataway, NJ, USA). The membrane was then incubated with primary antibodies, namely anti‐placental alkaline phosphatase (1:2,000, ab95462; Abcam, Cambridge, UK), anti‐TSG101 (1:3,000, ab125011; Abcam) and anti‐CD63 (1:2,000, ab59479; Abcam), followed by incubation with a horseradish peroxidase‐conjugated anti‐mouse or anti‐rabbit secondary antibody (1:10,000; Novus Biologicals, Littleton, CO, USA). Proteins were detected using Western Blotting Luminol Reagent (Bio‐Rad, Hercules, CA, USA).
Transmission electron microscopy
Fresh exosomes were suspended in phosphate‐buffered saline and fixed using 1% glutaraldehyde. The fixed exosomes were placed on a copper grid coated with formaldehyde/carbon and negatively stained with 1% uranyl acetate. The stained exosomes were observed at 80 kV using a JEOL 1200EX electron microscope (JEOL Co., Ltd., Tokyo, Japan).
Nanoparticle analysis
To obtain the size distribution of particles, we carried out nanoparticle tracking analysis (NTA; Particle Metrix, ZetaView Meerbusch, Germany). Fresh exosomes were diluted 1:8,000 in phosphate‐buffered saline to achieve a particle count of 1 × 108 particles/mL. Three cycles of NTA were carried out by scanning 11 cell positions and capturing 60 frames per position for each cell. Obtained images were processed and analyzed using the built‐in ZetaView software.
Bioinformatics analysis of miRNAs
We used the bioinformatics prediction database‐Bioconductor (R) package miRNAtap21, which has prediction algorithms, such as DIANA, MiRanda, PicTar and TargetScan. An integrated regulatory network of the miRNA‐gene‐pathway was constructed using the Cytoscape software (3.8.0; the laboratory of Dr Chris Sander at the Memorial Sloan Kettering Cancer Research Center, New York, NY, USA).
Statistical analysis
Statistical analysis was carried out using the IBM SPSS Statistics 22.0 software package22 (Armonk, NY, USA) and the Weka 3.6 data mining software package (University of Waikato, Hillcrest, New Zealand). The Shapiro–Wilk test was applied to evaluate the normality of the acquired data. Normally distributed data were expressed as the mean ± standard deviation, and data with a non‐normal distribution were expressed as the median and quartile intervals. Pearson’s χ2‐test was used to analyze categorical variables that were represented as numbers and percentages. Paired t‐test was used to analyze normally distributed data of the two groups with uniform variances. Furthermore, the Wilcoxon test was used to analyze the data when it did not meet normal distribution. qPCR data were analyzed using the 2–ΔΔCt method, normalized using U6 expression. Factors influencing the pathogenesis of GDM were screened and analyzed using the univariate logistic regression analysis. Stepwise logistic regression analysis was used to screen for factors that increased blood glucose levels during pregnancy. The area under the receiver operating characteristic curve (AUC) was used to evaluate the predictive value of the model. The significance level for all analyses was set at a P‐value of 0.05.
RESULTS
Differentially expressed miRNAs in placentas from GDM pregnancies
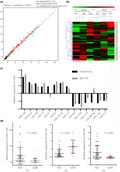
Term placentas of three pairs of patients, each pair consisting of one patient with GDM and one patient with NGT (n = 3 per group), were used for next‐generation sequencing analysis. A total of 764 differentially expressed miRNAs were detected, of which 114 targets were upregulated and 43 targets were downregulated, in the placental tissues of the GDM group compared with the NGT group (Figure 1a). Hierarchical clustering (Figure 1b) showed a distinctly different miRNA profile in the GDM group compared with that in the NGT group, wherein eight miRNAs were identified to be significantly upregulated and six miRNAs were downregulated (P < 0.05). However, the results of next‐generation sequencing analysis for miRNA‐21 and miRNA‐409‐3p were inconsistent with that of the qPCR analysis (Table S1; Figure 1c).
Figure 1.
The differentially expressed microribonucleic acids (miRNAs) identified in the placental tissue were validated by quantitative polymerase chain reaction. (a) A total of 764 differentially expressed miRNAs were detected using next‐generation sequencing. (b) Hierarchical clustering for differentially expressed miRNAs. (c) The relative expression of 14 miRNAs. (d) The relative expression of miRNA‐125b (D1), miRNA‐144 (D2) and miRNA‐543 (D3). GDM, gestational diabetes mellitus; miRs, microribonucleic acids; NGT, normal glucose tolerance; qRT‐PCR, quantitative reverse transcription polymerase chain reaction.
Validation of differentially expressed miRNAs in the placentas of patients with GDM and NGT
Differentially expressed miRNAs that were highly expressed in the placenta and most likely to be related to energy metabolism were validated using qPCR. Five miRNAs, namely miRNA‐125b, miRNA‐543, miRNA‐144, miRNA‐15b and miRNA‐451, were selected for qPCR‐based validation in the placental tissues from GDM (n = 36) and NGT (n = 37) pregnancies. Patient characteristics are shown in Table S2. The blood glucose levels at three time points, estimated using the OGTT and the weight of newborns from mothers in the GDM group, were higher than those in the NGT group (P < 0.05). qPCR showed that the expression levels of miRNA‐125b, miRNA‐543 and miRNA‐144 were consistent with the next‐generation sequencing results (P < 0.05; Figure 1d). The expression levels of miRNA‐15b and miRNA‐451 did not show a significant difference between both groups (Table S2).
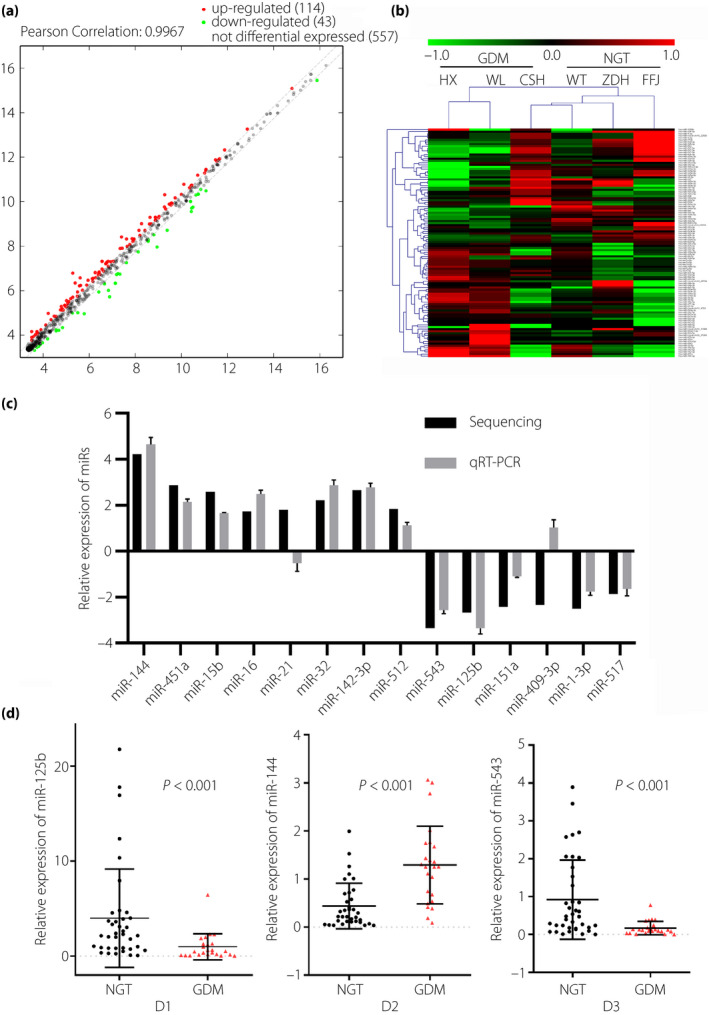
Isolation, characterization and identification of exosomes in plasma
Particles in the plasma from GDM and NGT groups with cup‐shaped morphology were identified using transmission electron microscopy (Figure 2a,b). NTA was used to obtain a size distribution curve of the cup‐shaped vesicles and showed a single peak at 100 nm (Figure 2c–f). The vesicles were found to be positive for exosomal markers CD63, TSG101 and placental alkaline phosphatase (PLAP; Figure 2g).
Figure 2.
The characterization of exosomes obtained from normal glucose tolerance (NGT) and gestational diabetes mellitus (GDM) pregnancies. (a,b) Representative electron micrograph of exosomes isolated from the plasma. (c–f) Size distribution curve of the vesicle‐enriched particles isolated from the plasma, generated using a nanoparticle tracking analysis. (g) Representative western blot for the expression of exosomal markers, namely CD63, TSG101 and placental alkaline phosphatase (PLAP), in the exosomes isolated from the plasma.
Expression of miRNA‐125b and miRNA‐144 in circulating exosomes
Using qPCR, the expression levels of miRNA‐144 and miRNA‐125b were estimated in the circulating exosomes isolated from the plasma of pregnant women with GDM (n = 57) and those with NGT (n = 61). Patient characteristics are shown in Table 1. Glycated hemoglobin (HbA1c) and glucose levels, measured using OGTT, at all three time points were significantly higher in the GDM group than that in the NGT group, although both groups did not differ in gravidity (P = 0.499). Compared with the NGT group, the GDM group showed significantly heavier weights and higher body mass index (BMI) before pregnancy. The diastolic pressure before delivery was significantly higher in the GDM group than in the NGT group. Additionally, the number of platelets in the blood before delivery were significantly lower in the NGT group than in the GDM group. However, the hemoglobin and neutrophil‐to‐lymphocyte ratios were not significantly different between the two groups. Newborn babies from mothers in the GDM group were significantly heavier than those of the NGT group, although these two groups did not differ significantly in the abdominal circumference of the pregnant women before delivery.
Table 1.
The qRT‐PCR test of circulating exosomes isolated from plasma in GDM and NGT groups
| NGT (N = 61) | GDM (N=57) | t/t'/z/F | P | ||||
|---|---|---|---|---|---|---|---|
|
|
|
|
|
||||
| OGTT 0 h (mmol/L) | 4.45 ± 0.39 | 4.43 ± 0.62 | 5.20 ± 0.85 | 5.20 ± 0.91 | 5.236 | <0.001 | |
| OGTT 1 h (mmol/L) | 6.75 ± 31.60 | 6.67 ± 2.47 | 11.14 ± 1.40 | 10.96 ± 1.58 | 13.857 | <0.001 | |
| OGTT 2 h (mmol/L) | 5.77 ± 1.10 | 5.80 ± 1.79 | 9.19 ± 1.74 | 8.98 ± 2.04 | 7.246 | <0.001 | |
| HbA1C (%) | 5.09 ± 0.35 | 5.00 ± 0.50 | 5.66 ± 0.51 | 5.50 ± 0.50 | 5.616 | <0.001 | |
| Age (year) | 30.00 ± 5.63 | 29.50 ± 7.00 | 32.30 ± 4.21 | 32.00 ± 6.00 | 2.213 | 0.030 | |
| Gestational weeks at delivery (days) | 276.11 ± 8.14 | 278.00 ± 14.00 | 273.46 ± 9.69 | 274.00 ± 15.00 | 1.118 | 0.264 | |
| Gravidity | 2.31 ± 1.25 | 2.00 ± 2.00 | 2.44 ± 1.04 | 2.00 ± 1.00 | 0.676 | 0.499 | |
| Parity | 1.46 ± 0.61 | 1.00 ± 1.00 | 1.77 ± 0.58 | 2.00 ± 1.00 | 2.502 | 0.012 | |
| weight§ (kg) | 58.71 ± 11.21 | 55.00 ± 15.00 | 65.54 ± 11.15 | 65.00 ± 14.50 | 3.098 | 0.002 | |
| Weight¶ (kg) | 75.12 ± 13.47 | 71.00 ± 17.50 | 80.39 ± 11.85 | 78.70 ± 13.70 | 2.424 | 0.123 | |
| Increase of weight during pregnancy (kg) | 16.41 ± 4.73 | 16.00 ± 7.00 | 14.86 ± 5.06 | 15.00 ± 4.50 | 2.424 | 0.123 | |
| BMI§ | 22.32 ± 3.80 | 21.49 ± 5.90 | 25.10 ± 4.10 | 24.42 ± 4.90 | 3.385 | 0.001 | |
| BMI¶ | 28.59 ± 4.67 | 27.34 ± 5.60 | 30.81 ± 4.43 | 30.26 ± 4.75 | 2.168 | 0.145 | |
| Increase of BMI during pregnancy | 6.26 ± 1.82 | 5.88 ± 2.78 | 5.71 ± 1.95 | 5.58 ± 1.84 | 2.168 | 0.145 | |
| Systolic blood pressure¶ (mmHg) | 125.14 ± 9.03 | 126.00 ± 11.00 | 129.83 ± 17.30 | 127.50 ± 18.00 | 1.235 | 0.217 | |
| Diastolic blood pressure¶ (mmHg) | 77.34 ± 5.60 | 78.00 ± 7.00 | 83.37 ± 10.89 | 83.00 ± 11.50 | 3.378 | 0.001 | |
| White blood cell count¶ (×109/l) | 9.45 ± 2.31 | 8.66 ± 3.90 | 8.74 ± 1.81 | 8.80 ± 2.49 | 0.931 | 0.352 | |
| Percentage of Neutrophil¶ (%) | 72.26 ± 6.83 | 71.80 ± 7.80 | 73.94 ± 6.57 | 74.43 ± 7.70 | 1.153 | 0.252 | |
| Percentage of lymphocyte¶ (%) | 20.56 ± 5.24 | 20.40 ± 7.10 | 19.14 ± 5.89 | 19.00 ± 8.60 | 1.151 | 0.253 | |
| Neutrophil‐lymphocyte ratio | 4.34 ± 4.47 | 3.46 ± 1.55 | 4.48 ± 2.26 | 3.98 ± 2.10 | 1.294 | 0.196 | |
| Hemoglobin¶ (g/l) | 115.40 ± 11.95 | 117.00 ± 20.00 | 114.08 ± 14.86 | 111.00 ± 20.00 | 0.439 | 0.661 | |
| Platelets count¶ (×109/l) | 241.86 ± 55.44 | 250.00 ± 77.00 | 212.40 ± 57.47 | 208.00 ± 87.50 | 2.377 | 0.020 | |
| Abdominal circumference¶ (cm) | 103.86 ± 8.52 | 102.00 ± 8.00 | 107.75 ± 7.72 | 106.00 ± 9.50 | 0.390 | 0.534 | |
| Weight of the new born baby (g) | 3418.57 ± 432.29 | 3390.00 ± 660.00 | 3666.15 ± 610.39 | 3600.00 ± 785.00 | 2.073 | 0.041 | |
| MiRNA‐125b | 2.36 ± 1.48 | 2.06 ± 2.31 | 0.63 ± 1.34 | 0.60 ± 1.91 | 38.962 | <0.001 | |
| MiRNA‐144 | 0.71 ± 2.45 | 0.92 ± 3.20 | 2.84 ± 2.89 | 3.14 ± 4.39 | 15.909 | <0.001 | |
BMI, body mass index; GDM, gestational diabetes mellitus; HbA1c, glycated hemoglobin; MiRNA, microribonucleic acid; NGT, normal glucose tolerance; OGTT, oral glucose tolerance test.
Within one standard deviation of the mean.
Within one quarter of the median.
Before pregnancy.
Before delivery.
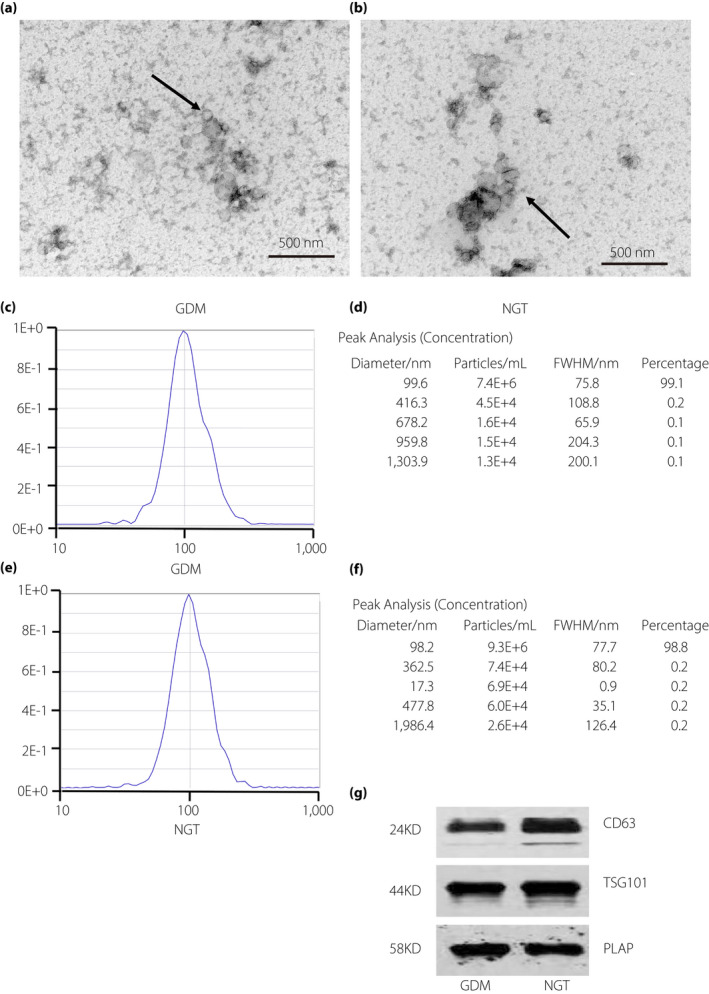
miRNA‐144 expression was negatively correlated with BMI, both before pregnancy (r = −0.331; P = 0.018) and before delivery with (r = −0.290; P = 0.039) (Figure 3a,b), whereas it was positively correlated with the amount of platelets (r = 0.309; P = 0.028) and the glucose level, measured using the OGTT, at 1 h (r = 0.283; P = 0.044; Figure 3c,d). miRNA‐125b was downregulated (P < 0.001) and miRNA‐144 was upregulated (P < 0.001) in the circulating exosomes of pregnant women with GDM compared with that in circulating exosomes of pregnant women with NGT (Table 1; Figure 3e,f).
Figure 3.
The relative expression of microribonucleic acid (miRNA)‐125b and miRNA‐144 between normal glucose tolerance (NGT) and gestational diabetes mellitus (GDM) pregnancies. (a,b) miRNA‐144 expression is significantly negatively correlated with body mass index (BMI) both before pregnancy and before delivery. (c,d) miRNA‐144 expression is significantly positively correlated with the number of platelets (PLT) and the blood glucose level at 1 h, measured using the oral glucose tolerance test (OGTT). (e) The relative expression of miRNA‐125b. (f) The relative expression of miRNA‐144.
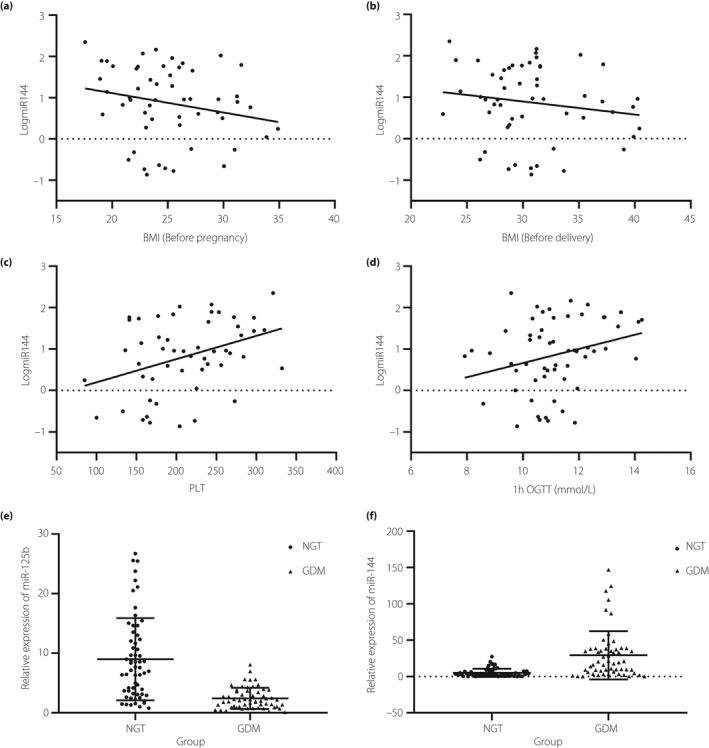
Multivariate logistic regression analysis
miRNA‐125b, miRNA‐144 and BMI before pregnancy were identified to be associated with the incidence of GDM (Table S3). miRNA‐125b was negatively correlated with GDM (parameter estimation, −0.9063; P = 0.0003), whereas miRNA‐144 and BMI before pregnancy were positively correlated with GDM (P = 0.0003 and P = 0.0339, respectively). Based on these three factors, model 1 is shown in Figure 4a1, and it exhibited satisfactory goodness of fit (deviance test statistic D = 69.909, P = 0.881; Pearson test statistic = 79.982, P = 0.634; Hosmer–Lemeshow test statistic = 9.292, P = 0.318). The AUC of model 01 was 0.898. We also established model 02 based on expression levels of miRNA‐125b and miRNA‐144 as the determining factors (Figure 4a2), and the AUC of model 02 was 0.875. Although the AUC of model 01 was higher than that of model 02, there was no significant difference between them (Figure 4). The AUC of blood glucose levels, measured using the OGTT, are shown in Figure 4b and Figure 4d. When using the probability (P = 0.5359) calculated via model 01 as the diagnostic cut‐off value (>0.5359 was diagnosed as GDM), the sensitivity and specificity of the diagnosis was 87.04% and 82.86%, respectively. The diagnostic efficacy of model 01 was significantly higher than that of miR‐125b and miR‐144, but lower than of blood glucose levels at 1 h (Figure 4c,d). Although there was no statistical difference, the diagnostic power of model 01 was higher than that of blood glucose levels at 0 h and HbA1c levels (Figure 4d). The positive predictive value and negative predictive value of each index in populations with different prevalence rates are shown in Figure 4e,f.
Figure 4.
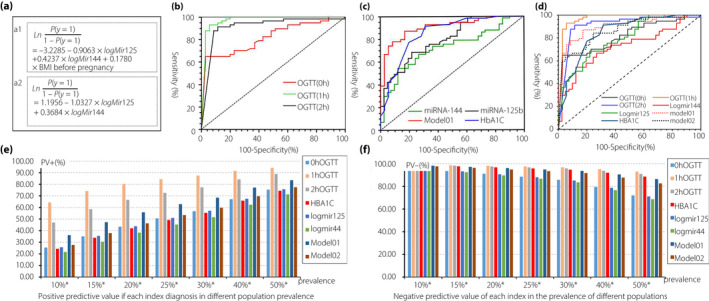
Receiver operating characteristic curves to estimate diagnostic value of the established models. (a1) Diagnostic model 01 is based on the expression of circulating exosomal microribonucleic acid (miRNA)‐125b and miRNA‐144 and body mass index (BMI) before pregnancy, and was developed using cross‐validation methods. (a2) Diagnostic model 02 is based on the expression of circulating exosomal miRNA‐125b and miRNA‐144, and was developed using cross‐validation methods. (b) Comparison of diagnostic values of glucose levels at 0 h, 1 h and 2 h, measured using the oral glucose tolerance test (OGTT). (c) Comparison of diagnostic values of glycated hemoglobin (HbA1c), miRNA‐144, miRNA‐125b and model 01. (d) Comparison of diagnostic values of glucose levels, HbA1c, miRNA‐144, miRNA‐125b, model 01 and model 02; (e) The positive predictive value of each index in populations with different prevalence rates of gestational diabetes mellitus. (f) The negative predictive value of each index diagnosis in populations with different prevalence rates of gestational diabetes mellitus. AUC, the area under the receiver operating characteristic curve; CI, confidence interval.
Gene target and gene ontology analysis of miRNA‐125b and miRNA‐144
miRNA‐125b and miRNA‐144 were analyzed using the Bioconductor (R) package miRNAtap21 to identify their target genes (Figure S1). A total of 419 and 2,076 target genes were identified for miRNA‐125b and miRNA‐144, respectively. Further analysis of these target genes using the Cytoscape software (3.8.0; the laboratory of Dr Chris Sander at the Memorial Sloan Kettering Cancer Research Center) showed that the target genes were enriched in endocrine pancreas development (enrich factor = 3.86, P = 0.005, false discovery rate = 0.08; Figure S1a) and energy homeostasis (enrich factor = 4.48, P = 0.0017, false discovery rate = 0.36; Figure S1b). Using gene ontology pathway analysis, the target genes were found to be functionally enriched in the insulin signaling pathway (P = 0.0208) and adipocytokine signaling (P = 0.0015; Figure S1c). These results show that miRNA‐125b and miRNA‐144 are linked to pathways that mediate energy metabolism (Figure S1d).
DISCUSSION
There are only a few studies that have evaluated the relationship between the abnormal expression of exosomal miRNAs and GDM pathogenesis23. The present study suggests that miRNAs in circulating exosomes show a consistent expression pattern with that in the placenta. However, the differentially expressed miRNAs identified in the patients of GDM in this study were inconsistent with previous reports; although this might be due to differences in patient characteristics and experimental methods used. Western blot analysis showed that PLAP was enriched in circulating exosomes, suggesting that the circulating exosomes consisted of placenta‐derived exosomes, and this finding is consistent with previous studies that have reported that placental syncytiotrophoblasts secrete exosomes into the peripheral blood15, 16. PLAP is a syncytiotrophoblast‐specific protein and thus was used to identify the origin of placenta‐derived exosomes24. However, as PLAP is a soluble protein, it cannot be extracted through immunoaffinity capture using anti‐PLAP‐coated beads, and thus could not be used to isolate placenta‐derived exosomes from plasma. This motivates future studies aimed at isolating placenta‐derived exosomes from the blood plasma. Current studies have shown that the total number of exosomes and placenta‐derived exosomes in the peripheral blood of pregnant women increases significantly25, 26, and the expression of miRNA‐125b in the peripheral blood during pregnancy was significantly higher than that during non‐pregnancy27. In the present study, we found that miRNA‐125b and miRNA‐144 were abundantly expressed in plasma exosomes, which was consistent with the expression of miRNA‐125b and miRNA‐144 in placenta‐derived exosomes. This indicates that miRNAs in placenta‐derived exosomes might account for a relatively large proportion of the total miRNAs in peripheral blood.
Circulating exosomal miRNA‐125b and miRNA‐144 were significantly altered in pregnant women with GDM, and can be thus considered as potential biomarkers of GDM. In addition, consistent with studies that have reported that obese women with a high BMI are at significantly higher risk of developing GDM28, BMI before pregnancy, a clinical factor that can be easily measured, was positively correlated with the incidence GDM. Therefore, in the present study, we established a diagnostic model (model 01) based on the circulating exosomal levels of miRNA‐125b and miRNA‐144 during 24–41 weeks of gestation, and BMI before pregnancy, and evaluated the diagnostic performance of the model. The AUC of model 01 was found to be 0.898, indicating its favorable diagnostic value. To further estimate the diagnostic value of miRNAs, we established another model that only incorporated the expression of miRNA‐125b and miRNA‐44 (model 02). The AUC of this model was 0.875. Both models 01 and 02 showed good diagnostic efficacy, and there was no significant difference in the AUC between these two models, indicating that it is possible that miRNAs serve as diagnostic tools even in the absence of other clinical factors. This further confirmed that circulating exosomal miRNAs were potential biomarkers for GDM.
miRNA‐125b is a member of the miRNA‐125 family. miRNA‐125 is not a placenta‐specific miRNA; for instance, β‐cells29 in the pancreas and central nerve cells30 also secrete miRNA‐125. At present, only one study has reported the levels of miRNA‐125 in the serum of non‐pregnant women, normal pregnant women and pregnant women with GDM27. This study27 showed that miRNA‐125 levels during pregnancy are significantly increased. We also found that miRNA‐125b was highly expressed in the placental tissue and plasma exosomes. However, the study found that the level of miRNA‐125 in the serum of patients with GDM in the third trimester was not significantly different, which is inconsistent with the findings of the present study. This might be caused by differences in patient characteristics, experimental methods and tissues. Ortega et al.3132 found that miRNA‐125b levels in peripheral blood were significantly decreased in patients with type 2 diabetes and in obese male patients compared with that in healthy patients, which is consistent with the findings in the present study, wherein we found that a decrease in expression of miRNA‐125b was significantly correlated with GDM. According to recent studies, the low expression of miRNA‐125b might facilitate hyperglycemia, by increasing the expression of c‐Maf transcripts29, which has been identified as a β‐cell "disallowed" gene33, and inhibit sirtuin 7 expression34. These findings suggest that the low level of miRNA‐125b expression is associated with abnormal glucose metabolism. The results of functional enrichment analysis of target genes in the present study show that miRNA‐125b might affect blood glucose metabolism through energy metabolism‐related pathways.
Previous studies showed that miRNA‐144 is upregulated in the blood of diabetes patients35, and is dysregulated in type 1 and type 2 diabetes mellitus and GDM36. Our study also showed that miRNA‐144 expression in plasma exosomes of GDM throughout the third trimester was significantly increased. Animal models have shown that miRNA‐144 overexpression led to lipid metabolism disorder syndrome37. The functional enrichment analysis of target genes in the present study also showed that miRNA‐144 might participate in energy metabolism by co‐regulating the development of type β pancreatic cells and energy homeostasis. miRNA‐144 has been shown to directly target insulin receptor substrate 1, which is highly involved in the insulin signaling pathway35, and glucose transporter 138, which might also regulate glucose metabolism. Nevertheless, two studies based on the same cohort showed that miRNA‐144 is negatively correlated with serum glucose levels39. Soumyalekshmi et al.23 found that the expression of miRNA‐144 was decreased in exosomes secreted from the chorionic villi of placentas in patients with GDM. In addition, although pregnant women in the GDM group showed higher levels of miRNA‐144 and BMI before pregnancy and before delivery in the present study, both parameters showed a significant negative correlation with the incidence of GDM. The abnormal expression of miRNA‐144 is closely related to glucose and lipid metabolism; however, the conclusions drawn from the current literature are contradictory, possibly due to differences in ethnicity, sex, experimental methods and sample size. Energy metabolism is related to age, sex, BMI and different body tissues; hence, the mechanism of action tends to be very complicated. Additional studies are required to elucidate the mechanism of action of miRNA‐144 in GDM pathogenesis.
In conclusion, the present study identifies new biomarkers of GDM, as well as contributes to the diagnostic methods and pathogenesis of GDM. As the diagnostic value of these two exosomal miRNAs was estimated in the same cohort as those from whom we discovered these miRNAs, future studies need to validate the miRNAs' performance in an independent cohort of GDM versus controls. In addition, follow‐up studies are also required to confirm the predictive value of miRNA‐125b and miRNA‐144 for the incidence of GDM during the early and middle stages of pregnancy. Finally, experimental studies are required to fully elucidate the role of these miRNAs in the pathogenesis of GDM.
DISCLOSURE
The authors declare no conflict of interest.
Supporting information
Figure S1 | Identification and gene ontology pathway analysis of the gene targets of microribonucleic acid (miRNA)‐125b and miRNA‐144.
Table S1 | Differentially regulated microribonucleic acids in the placenta of patients with gestational diabetes mellitus compared with that in the placenta of patients with normal glucose tolerance.
Table S2 | Quantitative polymerase chain reaction analysis of differentially expressed microribonucleic acid in the placental tissues from gestational diabetes mellitus and normal glucose tolerance pregnancies.
Table S3 | Multivariate logistic regression analysis of gestational diabetes mellitus‐related factors.
Acknowledgments
We thank the anonymous reviewer, and the associate editor for useful comments and edits on earlier versions of this manuscript. This work was supported by the National Natural Science Foundation of China (grant number 81702538); the Natural Science Foundation of Shandong Province (grant number ZR201702160271); the Foundation for Young Scholars of The Second Hospital of Shandong University (grant number 2018YT11); and the Health Science and Technology Development Project of Shandong Province (grant number 2013WSB21002).
J Diabetes Investig. 2021
References
- 1.Ferrara A. Increasing Prevalence of Gestational Diabetes Mellitus: A public health perspective. Diabetes Care 2007; 30(Supplement 2): S141–S146. [DOI] [PubMed] [Google Scholar]
- 2.Zhu WW, Fan L, Yang HX, et al. Fasting Plasma Glucose at 24–28 Weeks to Screen for Gestational Diabetes Mellitus: New evidence from China. Diabetes Care 2013; 36: 2038–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Metzger BE, Lowe LP, Dyer AR, et al. Hyperglycemia and adverse pregnancy outcomes. N Engl J Med 2008; 358: 1991–2002. [DOI] [PubMed] [Google Scholar]
- 4.Robert MF, Neff RK, Hubbell JP, et al. Association between maternal diabetes and the respiratory‐distress syndrome in the newborn. N Engl J Med 1976; 294: 357–360. [DOI] [PubMed] [Google Scholar]
- 5.Tam WH, Ma RC, Yang X, et al. Glucose Intolerance and Cardiometabolic Risk in Adolescents Exposed to Maternal Gestational Diabetes: A 15‐year follow‐up study. Diabetes Care 2010; 33: 1382–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casey BM, Lucas MJ, Mcintire DD, et al. Pregnancy outcomes in women with gestational diabetes compared with the general obstetric population. Obstet Gynecol 1998; 91: 639–640. [DOI] [PubMed] [Google Scholar]
- 7.Kim C, Newton KM, Knopp RH. Gestational diabetes and the incidence of type 2 diabetes: a systematic review. Diabetes Care 2002; 25: 1862–1868. [DOI] [PubMed] [Google Scholar]
- 8.Tove L, Jens B, Norwitz ER, et al. Aortic Stiffness and Cardiovascular Risk in Women with Previous Gestational Diabetes Mellitus. PLoS One 2015; 10: e0136892‐. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barbour LA, Mccurdy CE, Hernandez TL, et al. Cellular Mechanisms for Insulin Resistance in Normal Pregnancy and Gestational Diabetes. Diabetes Care 2007; 30(Supplement 2): S112–S119. [DOI] [PubMed] [Google Scholar]
- 10.Poirier C, Desgagné V, Guérin R, et al. MicroRNAs in Pregnancy and Gestational Diabetes Mellitus: Emerging Role in Maternal Metabolic Regulation. Curr Diab Rep 2017; 17: 35. [DOI] [PubMed] [Google Scholar]
- 11.Cao JL, Zhang L, Li J, et al. Up‐regulation of miR‐98 and unraveling regulatory mechanisms in gestational diabetes mellitus. Sci Rep 2016; 6: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao C, Zhang T, Shi Z, et al. MicroRNA‐518d regulates PPARa protein expression in the placentas of females with gestational diabetes mellitus. Mol Med Rep 2014; 9(6): 2085–2090. [DOI] [PubMed] [Google Scholar]
- 13.Li J, Song L, Zhou L, et al. A MicroRNA Signature in Gestational Diabetes Mellitus Associated with Risk of Macrosomia. Cell Physiol Biochem 2015; 37: 243–252. [DOI] [PubMed] [Google Scholar]
- 14.Li L, Wang S, Li H, et al. microRNA‐96 protects pancreatic β‐cell function by targeting PAK1 in gestational diabetes mellitus. BioFactors 2018; 44: 539–547. [DOI] [PubMed] [Google Scholar]
- 15.Al‐Mahfoudh R, Qattan E, Ellenbogen JR, et al. Applications of the ultrasonic bone cutter in spinal surgery–our preliminary experience. Br J Neurosurg 2013; 28(1): 56–60. [DOI] [PubMed] [Google Scholar]
- 16.Tong M, Chamley LW. Placental Extracellular Vesicles and Feto‐Maternal Communication. Cold Spring Harb Perspect Med 2015; 5: a023028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Etheridge A, Lee I, Hood L, et al. Extracellular microRNA: a new source of biomarkers. Mutat Res 2011; 717: 85–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fallen S, Baxter D, Wu X, et al. Extracellular vesicle RNAs reflect placenta dysfunction and are a biomarker source for preterm labour. J Cell Mol Med 2018; 22: 2760–2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Donker RB, Mouillet JF, Chu T, et al. The expression profile of C19MC microRNAs in primary human trophoblast cells and exosomes. Mol Hum Reprod 2012; 18: 417–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salomon C, Guanzon D, Scholz‐Romero K, et al. Placental exosomes as early biomarker of preeclampsia ‐ Potential role of exosomal microRNAs across gestation. J Clin Endocrinol Metab 2017; 102: 3182–3194. [DOI] [PubMed] [Google Scholar]
- 21.Pajak M, Simpson T. miRNAtap: miRNAtap: microRNA Targets—Aggregated Predictions. R package version 2016; 1 (0).
- 22.SPSS I . Corp., 2013. Released 2013. IBM SPSS Statistics for Windows, Version 22.0. IBM Corp., Armonk, NY.
- 23.Nair S, Jayabalan N, Guanzon D, et al. Human Placental Exosomes in Gestational Diabetes Mellitus Carry a Specific Set of miRNAs Associated with Skeletal Muscle Insulin Sensitivity. Clin Sci 2018; 132: 2451–2467. [DOI] [PubMed] [Google Scholar]
- 24.Lai A, Elfeky O, Rice GE, et al. Optimized Specific Isolation of Placenta‐Derived Exosomes from Maternal Circulation. Methods Mol Biol 2018; 1710: 131–138. [DOI] [PubMed] [Google Scholar]
- 25.Sarker S, Scholz‐Romero K, Perez A, et al. Placenta‐derived exosomes continuously increase in maternal circulation over the first trimester of pregnancy. J Transl Med 2014; 12: 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salomon C, Torres MJ, Kobayashi M, et al. A gestational profile of placental exosomes in maternal plasma and their effects on endothelial cell migration. PLoS One 2014; 9: e98667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lamadrid‐Romero M, Solís K, Cruz‐Reséndiz M, et al. Central nervous system development‐related microRNAs levels increase in the serum of gestational diabetic women during the first trimester of pregnancy. Neurosci Res 2018; 130: 8–22. [DOI] [PubMed] [Google Scholar]
- 28.Pirjani R, Shirzad N, Qorbani M, et al. Gestational diabetes mellitus its association with obesity: a prospective cohort study. Eat Weight Disord 2017; 22: 445–450. [DOI] [PubMed] [Google Scholar]
- 29.Klein D, Misawa R, Bravo‐Egana V, et al. MicroRNA expression in alpha and beta cells of human pancreatic islets. PLoS One 2013; 8: e55064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ji F, Lv X, Jiao J. The Role of MicroRNAs in Neural Stem Cells and Neurogenesis. J Genet Genomics 2013; 40: 61–66. [DOI] [PubMed] [Google Scholar]
- 31.Ortega FJ, Mercader JM, Moreno‐Navarrete JM, et al. Profiling of Circulating MicroRNAs Reveals Common MicroRNAs Linked to Type 2 Diabetes That Change With Insulin Sensitization. Diabetes Care 2014; 37: 1375–1383. [DOI] [PubMed] [Google Scholar]
- 32.Ortega FJ, Mercader JM, Catalán V, et al. Targeting the circulating microRNA signature of obesity. Clin Chem 2013; 59: 781–792. [DOI] [PubMed] [Google Scholar]
- 33.Schuit F, Van Lommel L, Granvik M, et al. β‐cell‐specific gene repression: A mechanism to protect against inappropriate or maladjusted insulin secretion? Diabetes 2012; 61: 969–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kurylowicz A, Owczarz M, Polosak J, et al. SIRT1 and SIRT7 expression in adipose tissues of obese and normal‐weight individuals is regulated by microRNAs but not by methylation status. Int J Obes 2016; 40: 1635–1642. [DOI] [PubMed] [Google Scholar]
- 35.Karolina DS, Armugam A, Tavintharan S, et al. MicroRNA 144 impairs insulin signaling by inhibiting the expression of insulin receptor substrate 1 in type 2 diabetes mellitus. PLoS One 2011; 6: e22839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Collares CV, Evangelista AF, Xavier DJ, et al. Identifying common and specific microRNAs expressed in peripheral blood mononuclear cell of type 1, type 2, and gestational diabetes mellitus patients. BMC Res Notes 2013; 6: 491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang X, Zheng Y, Ma Y, et al. Lipid metabolism disorder induced by up‐regulation of miR‐125b and miR‐144 following β‐diketone antibiotic exposure to F0‐zebrafish (Danio rerio). Ecotoxicol Environ Saf 2018; 164: 243–252. [DOI] [PubMed] [Google Scholar]
- 38.Liu M, Gao J, Huang Q, et al. Downregulating microRNA‐144 mediates a metabolic shift in lung cancer cells by regulating GLUT1 expression. Oncology Letters 2016; 11: 3772–3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raitoharju E, Seppälä I, Oksala N, et al. Blood microRNA profile associates with the levels of serum lipids and metabolites associated with glucose metabolism and insulin resistance and pinpoints pathways underlying metabolic syndrome The cardiovascular risk in Young Finns Study. Mol Cell Endocrinol 2014; 391: 41–49. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 | Identification and gene ontology pathway analysis of the gene targets of microribonucleic acid (miRNA)‐125b and miRNA‐144.
Table S1 | Differentially regulated microribonucleic acids in the placenta of patients with gestational diabetes mellitus compared with that in the placenta of patients with normal glucose tolerance.
Table S2 | Quantitative polymerase chain reaction analysis of differentially expressed microribonucleic acid in the placental tissues from gestational diabetes mellitus and normal glucose tolerance pregnancies.
Table S3 | Multivariate logistic regression analysis of gestational diabetes mellitus‐related factors.


