SUMMARY
Chemical synapses of shared cellular origins have remarkably heterogeneous structures, but how this diversity is generated is unclear. Here, we use three-dimensional (3D) electron microscopy and artificial intelligence algorithms for image processing to reconstruct functional excitatory microcircuits in the mouse hippocampus and microcircuits in which neurotransmitter signaling is permanently suppressed with genetic tools throughout the lifespan. These nanoscale analyses reveal that experience is dispensable for morphogenesis of synapses with different geometric shapes and contents of membrane organelles and that arrangement of morphologically distinct connections in local networks is stochastic. Moreover, loss of activity increases the variability in sizes of opposed pre- and postsynaptic structures without disrupting their alignments, suggesting that inherently variable weights of naive connections become progressively matched with repetitive use. These results demonstrate that mechanisms for the structural diversity of neuronal synapses are intrinsic and provide insights into how circuits essential for memory storage assemble and integrate information.
Graphical abstract
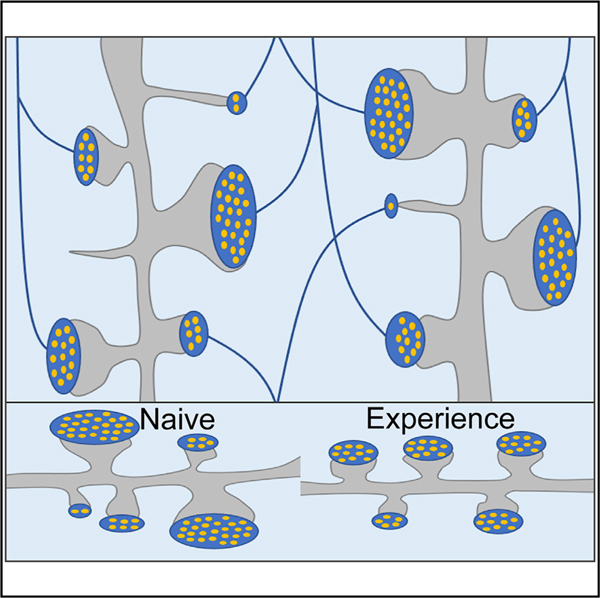
In brief
Zhu et al. show that development of structurally diverse pools of glutamatergic synapses in the mouse brain does not require synaptic activity. They also demonstrate that structurally diverse synapses are arranged in local connectomes in a stochastic manner and that experience reduces the variability of synaptic weights.
INTRODUCTION
Central neurons communicate by synapses with diverse morphologies and contents of membrane organelles (Harris and Weinberg, 2012; Südhof, 2018). The structural heterogeneity of synaptic connections with shared cellular origins and neurotransmitter identities underlies their nonuniform weights, capacities for dynamic modifications, and ultimately contributions to coding within circuits. For example, in glutamatergic projection neurons (PNs) that relay excitatory information across telencephalic brain regions essential for high-order sensory processing, emotions, and memory storage, variable shapes of dendritic spines influence the local and long-range signals involving trafficking of molecules to and from isolated postsynaptic compartments (Alvarez and Sabatini, 2007; Berry and Nedivi, 2017; Colgan and Yasuda, 2014; Yuste and Bonhoeffer, 2004), whereas variably present at axonal terminals mitochondria provide energy for vesicle recycling and regulate the kinetics of neurotransmitter release via calcium buffering (Devine and Kittler, 2018; Hirabayashi et al., 2017; Kwon et al., 2016; Vos et al., 2010).
How diverse repertoires of synapses are formed, arranged in networks, and maintained is unclear. On the one hand, discoveries of structural plasticity have led to the notion that morphological differences among synapses of a particular class may reflect histories of their activity elicited spontaneously and/or by external sensory cues (Bailey et al., 2015; Caroni et al., 2012; Espinosa and Stryker, 2012; Harris, 2020; Holtmaat and Svoboda, 2009; Katz and Shatz, 1996). However, synapses also assemble in the absence of neurotransmission or presynaptic calcium entry (Held et al., 2020; Lu et al., 2013; Sando et al., 2017; Sigler et al., 2017; Varoqueaux et al., 2002; Verhage et al., 2000), raising the possibility that their diversity is genetically predetermined or generated stochastically. The current insights into the roles of experience and intrinsic/stochastic mechanisms in synaptic organization of the nervous system are largely based on the following two approaches: optical imaging whose resolution is insufficient for surveying small subcellular structures and conventional electron microscopy (EM) that is unsuitable for the comprehensive analysis of complex connectomes due to a narrow field of view and limited sample size (Berry and Nedivi, 2017; Espinosa and Stryker, 2012; Hazan and Ziv, 2020; Holtmaat and Svoboda, 2009; Lu et al., 2013; Mongillo et al., 2017; Sando et al., 2017; Schoch et al., 2001; Südhof, 2018; Verhage et al., 2000; Yasumatsu et al., 2008; Ziv and Brenner, 2018). Hence, the extent to which experience defines the fine ultrastructural features of central synapses remains poorly understood, despite the fact that the venerable “nature versus nurture” question has been of interest to many neuroscientists for several decades. We sought to bridge this gap in knowledge by using serial block-face scanning EM (SBEM), a technique for three-dimensional (3D) nanoscale volume reconstruction of biological tissues (Helmstaedter et al., 2008).
Several groups have recently demonstrated the utility of SBEM and similar 3D EM methods for reconstruction of circuits in the mammalian cerebral cortex, hippocampus, thalamus, spinal cord, and retina (Helmstaedter et al., 2013; Kasthuri et al., 2015; Morgan et al., 2016; Motta et al., 2019; Svara et al., 2018). These studies illuminated neuronal wiring in great detail, identified new cell types, and refuted old dogmas, such as the Peters’ rule. Yet, they were performed without any manipulations of brain activity and, in each case, sampled data from one animal. Another line of elegant 3D EM studies have documented acute changes in synapses elicited by brief electrical stimulation in vitro and in vivo (Bailey et al., 2015; Bourne and Harris, 2012; Bromer et al., 2018; Harris et al., 2015; Smith et al., 2016; Watson et al., 2016), but the physiological relevance of these effects remains uncertain.
Here, we combined SBEM with artificial intelligence (AI) algorithms for automatic segmentation of 3D images to generate a database of annotated local connectomes in the hippocampus of wild-type (WT) mice and mice in which excitatory neurotransmitter signaling was permanently suppressed in vivo with contemporary genetic tools throughout the lifespan. Our side-by-side analyses of ultrastructural parameters of active and inactive networks and individual synapses in an area of the brain that is required for learning and spatial navigation (Kandel et al., 2014; Strange et al., 2014; Tonegawa et al., 2018) reveal unappreciated features of central neurons, elucidate synaptic diversity, and establish a framework for future application of 3D EM for unraveling supramolecular events relevant to memory coding.
RESULTS
Strategies for nanoscale 3D reconstructions of functional and inactive excitatory circuits
Conceptually, we wished to understand whether synaptic activity is necessary or dispensable for the establishment of appropriate geometric patterns of pre- and postsynaptic sites on neuronal processes, combinatorial wiring of axons with dendrites, development of synapses with different shapes and sizes, and recruitment of intracellular membrane organelles that regulate synaptic function. To accomplish this task, we chose a recently characterized Emx1IRES-Cre/R26floxstop-TeNT mouse model (Sando et al., 2017) in which excitatory forebrain neurons derived from Emx1-positive progenitors were irreversibly silenced starting from mid embryogenesis by Cre-recombinase-inducible expression of Tetanus toxin (TeNT), a protease that blocks neurotransmitter and neuropeptide secretion by cleaving the vesicular SNARE Synaptobrevin/VAMP2 (Syb2) (Gorski et al., 2002; Zhang et al., 2008). As the main hippocampal pathway is comprised of PNs of the Emx1 lineage, virtually all glutamatergic synapses in this pathway of Emx1IRES-Cre/R26floxstop-TeNT mutant mice lacked Syb2 and were therefore unable to release glutamate spontaneously or in response to action potentials (Figure 1A; Sando et al., 2017). Importantly, these animals could survive for weeks after birth and had no apparent defects in the cellular anatomy and gross synaptic lamination of the hippocampus, making them an attractive experimental model to compare the architectures of functional circuitry and circuitry with no history of sensory-experience-dependent and spontaneous glutamatergic excitation in the postnatal brain.
Figure 1. Strategies for nanoscale 3D reconstructions of functional and inactive excitatory circuits.
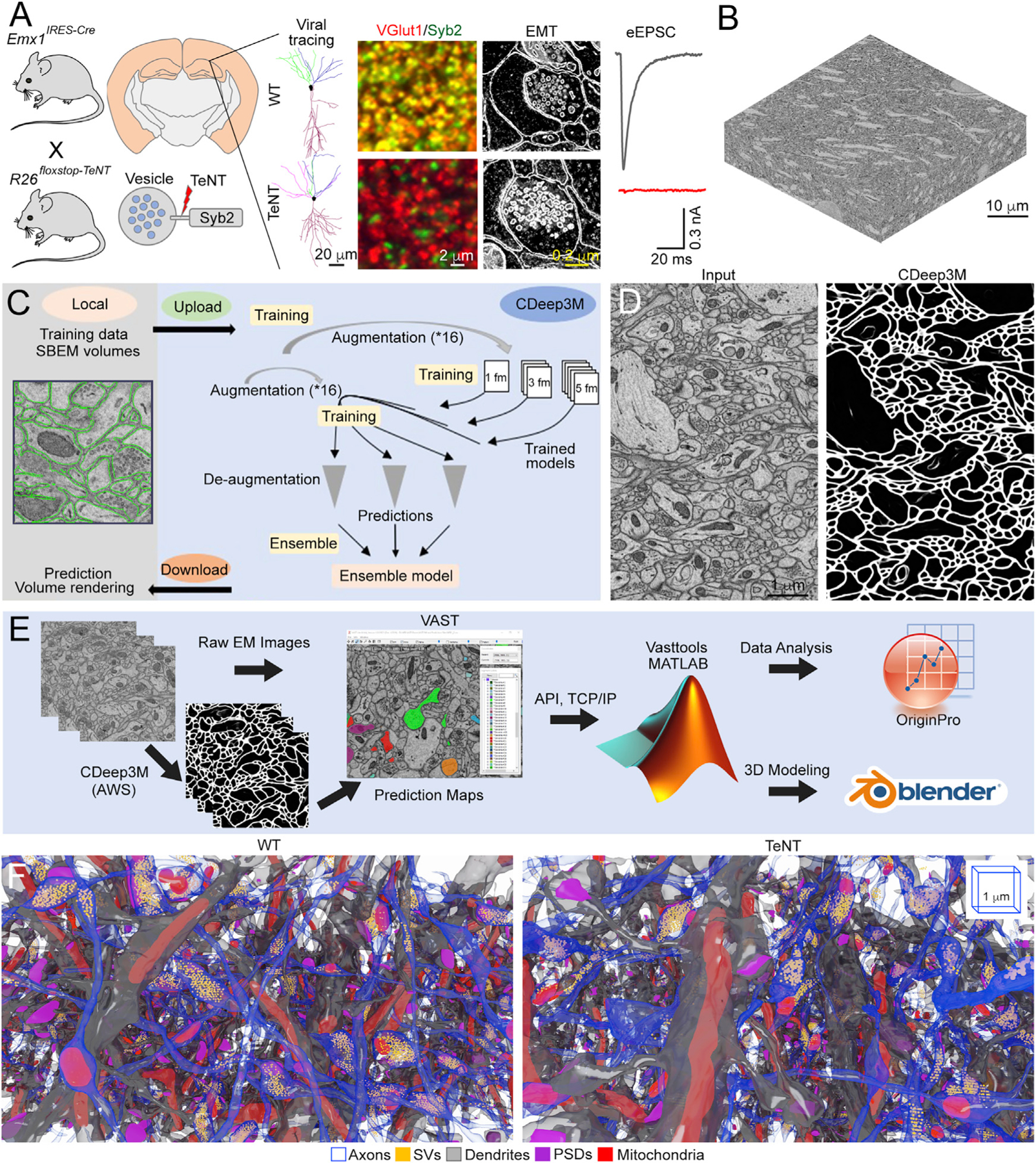
(A) Broad silencing of synaptic glutamate release in the developing forebrain. Shows the genetic approach for irreversible induction of TeNT in Emx1-positive neural progenitors, the pattern of TeNT expression in Emx1IRES-Cre/R26floxstop-TeNT mice, schematics of Syb2 cleavage on secretory vesicles, typical examples of single virally traced pyramidal neurons (PNs) in area CA1, immunofluorescent images of CA1sr in brain sections labeled with antibodies against Syb2 and excitatory synapse-specific marker VGlut1, two-dimensional (2D) EM images of individual synapses, and traces of evoked excitatory postsynaptic currents sampled from CA1 PNs in whole-cell mode. Scale bars are 20, 2, and 0.2 µm. See also Sando et al. (2017) and STAR Methods for details.
(B) 3D image stack acquired from the CA1sr by SBEM. Scale bar is 10 µm.
(C) Workflow for automatic segmentation of subcellular structures in SBEM volumes in the Amazon-cloud-based machine learning platform CDeep3M.
(D) Example of automatically segmented plasma membranes. Scale bar is 1 µm.
(E) A pipeline for 3D EM image analysis.
(F) Saturated 3D reconstructions of excitatory microcircuits in the CA1sr of wild-type (WT) and Emx1IRES-Cre/R26floxstop-TeNT mice (TeNT) at P30. Different structures are color-coded as indicated in the legend. 3D scale bars are 1 µm.
All scale bars apply to images from mice of both genotypes. See also Figure S1.
To simultaneously examine the wiring diagrams and structures of single synapses, we decided to image one hippocampal subfield at maximal achievable resolution and focused on area stratum radiatum (sr) in which principal CA1 PNs receive inputs onto their dendrites from Shaffer collateral (Sc) axons of PNs residing in the CA3 (Förster et al., 2006; Mishchenko et al., 2010; Figure S1A). Five ~35,000-µm3 SBEM volumes were collected from the dorsal CA1sr of two WT and three Emx1IRES-Cre/R26floxstop-TeNT mice (TeNT) at postnatal day 30 (P30) by using raster images with 5.3-nm pixels, 2-µsec pixel dwell time, and 60-nm Z steps (Figure 1B). Because manual tracing of dense structures in high-resolution 3D EM stacks is extremely time consuming, we built a pipeline for automatic segmentation of plasma membranes and organelles in a cloud-based convolutional neural network, CDeep3M (Haberl et al., 2018). This deep learning AI platform allowed us to make accurate predictions through retraining manually segmented ground truth labels in the Amazon Web Service (AWS), thereby reducing the effort and time by ~90% (Figures 1C and 1D). By leveraging CDeep3M, publicly available software packages, and custom scripts designed in MATLAB, we reconstructed and interrogated microcircuits containing thousands of excitatory synapses with secretory vesicles (SVs), characteristic postsynaptic densities (PSDs), mitochondria, and smooth endoplasmic reticulum (SER) (Figures 1E and 1F; Data S1). As described below and in the accompanying STAR Methods, our strategies for quantitative analyses included nonparametric statistics and combinatorial math and took into account potential differences between samples for biological or technical reasons, as well as variable curvatures of projections and synapse angles.
PNs intrinsically develop morphologically diverse synapses and maintain distances between synaptic sites on axonal and dendritic shafts
Telencephalic PNs have two common features: (1) their axons form en passant terminals seldomly contacting the same dendritic arbor more than once (Bloss et al., 2018; Kasthuri et al., 2015) and (2) their postsynaptic machineries are localized in spines with distinct morphologies that reflect maturity and functional strength (Alvarez and Sabatini, 2007; Berry and Nedivi, 2017; Harris and Weinberg, 2012; Yuste and Bonhoeffer, 2004; Figures S1A and S1B). The bulk spinogenesis of differentiating PNs does not require secretion of glutamate and ionotropic neurotransmitter receptors (Lu et al., 2013; Sando et al., 2017; Sigler et al., 2017), but spines also appear and become eliminated after novel experience through postnatal life (Alvarez and Sabatini, 2007; Berry and Nedivi, 2017; Holtmaat and Svoboda, 2009; Holtmaat et al., 2006; Kasai et al., 2010). Given their ability to innervate multiple targets within a receptive field, PN axons may provide inputs onto ensembles of spines with drastically different properties. However, the logic of axonal connectivity with spines has not been established.
We first quantitatively assessed the organization of functional and permanently inactive microcircuits with the following two complementary approaches: by tracing en passant synapses along isolated Sc fibers and by tracing all spines with opposed terminals on PN dendrites (Figures 2A, 2B, and S1–S3). Each connection received a unique ID with Euclidean coordinates and was classified according to spine type as mushroom (M), thin (T), stubby (S), long thin (L), filopodia (F), or bifurcated (B). All these spines were present and innervated on dendrites of silenced PNs, and their fractions were only slightly altered compared to normal neurons (Figure 2C). Although PNs of Emx1IRES-Cre/R26floxstop-TeNT mice had a ~2-fold higher density of infrequent “naked” spines lacking axonal inputs and a modest loss of “spineless” Sc synapses from dendritic shafts, the overall patterning of their polarized connections was preserved. The distributions of most structural and functional parameters of the nervous system are skewed and typically approximated with the Lognormal function (Bartol et al., 2015; Buzsáki and Mizuseki, 2014). We also observed this phenomenon in our datasets, although side-by-side fitting with Kolmogorov-Smirnov tests for goodness of fits showed that the Weibull probability density function (PDF) (Scholte et al., 2009) was more versatile because in some cases, Lognormal fits were rejected. In CA1sr of WT and Emx1IRES-Cre/R26floxstop-TeNT mice, the PDF distributions of distances between terminals and spines on axonal/dendritic shafts had similar profiles, and terminal numbers correlated with axonal arbor lengths with nearly indistinguishable Spearman rho values (Figures 2D to 2F, S2, and S3).
Figure 2. Organization of functional and inactive excitatory microcircuits.
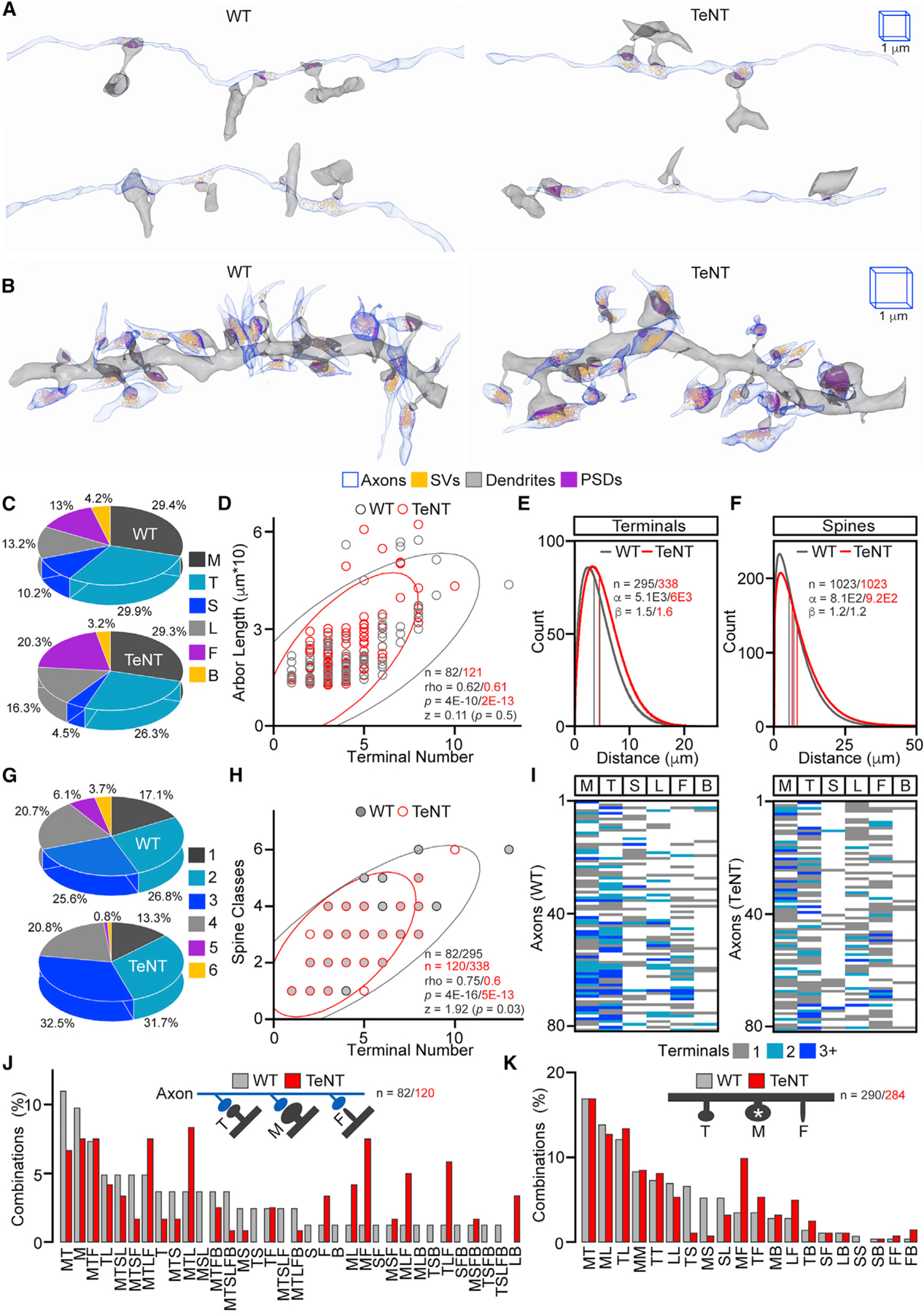
(A and B) Reconstructions of single Sc axons with opposed spines on different dendritic branches of PNs (in A) and dendrites with all incoming axons (in B) in the CA1sr of WT and Emx1IRES-Cre/R26floxstop-TeNT mice (TeNT). In all images and quantifications, spines are marked as follows: M, mushroom; T, thin; S, stubby; L, long thin; F, filopodia; B, bifurcated. The 1-µm 3D scale bars apply to images from mice of both genotypes.
(C) Fractions of innervated spine types. WT, n = 82 axons/295 synapses; TeNT, n = 120/338.
(D) Correlations between axonal arbor lengths and numbers of presynaptic terminals. Scatterplots with confidence ellipses, sample sizes (n = axons), Spearman correlation coefficients (rho), Fisher transformation scores (Z), and p values are shown.
(E and F) Spatial distributions of terminals (in E) and spines (in F) on axonal/dendritic shafts. Graphs show Weibull PDF curves generated by direct fitting of raw data (distances between synapses), color-coded mean and median lines, sample sizes (n), and distribution scales (α) and shapes (β).
(G) Fractions of axons innervating indicated numbers of distinct spine types (e.g., axons contacting only one out of six common spine types, two out of six, three out of six, and so on).
(H) Correlations between numbers of presynaptic terminals formed by axonal arbors and numbers of types of innervated spines. Graphs are annotated as in (D) (n = axons/terminals).
(I and J) Combinatorial patterns of axonal wiring. Show heatmaps (in I) and percentages of combinations of spines that receive inputs from individual Sc fibers (in J; n = axons, several rare combinations not displayed).
(K) Percentages of pairwise combinations of spines adjacent to M-type spines (n) on dendritic arbors.
Quantifications were performed using SBEM volumes from two WT and three TeNT mice. See also Figures S2 and S3.
Arrangement of morphologically distinct synapses in local networks is stochastic
To elucidate the principles of fine-scale axonal wiring, we asked if Sc axons are coupled with specific spine types (M, T, S, L, F, and B; t = 6) in a particular order assuming that identities of connected spines may differ in the normal brain due to variability in timing of prior activity of arbitrary selected fibers and that these patterns will be randomized after global loss of excitatory input. For instance, one could predict a segregation of intact axons that innervate M spines, which undergo structural and functional remodeling in sparse neural ensembles recruited for memory acquisition, and axons contacting T and F protrusions whose shapes are considered immature (Alvarez and Sabatini, 2007; Harris and Weinberg, 2012; Matsuo et al., 2008; Tonegawa et al., 2018). Contrary to this prediction, the wiring combinatorics were similar in animals of two genotypes. Of 63 theoretical combinations (C (t, i) = t! / i! * (t - i)! for which 6 ≤ i ≥ 1 is the number of chosen items), we detected 33 and 34 in reconstructed connectomes of 82 and 121 axons from WT and Emx1IRES-Cre/R26floxstop-TeNT mice, respectively. In both sets, the i values for single axons were proportional to terminal numbers, the majority of combinations (~83% and ~87%) had more than one spine type, the percentages of axons exclusively innervating M spines were relatively small and comparable (~10% and ~7%), and 3 out of 4 axons (~76% and ~73%) simultaneously innervated M and other spines, including F (Figures 2G to 2J; Data S1). Although normal and permanently silenced microcircuits had noticeable mismatches in combination frequencies (e.g., MTL, ML, MF, MLF, and TLF), these mismatches were reconcilable with quantifications of spine abundance (compare Figures 2C and 2J). We observed the same relationship when we identified pairs of spines adjacent to M spines on dendrites, suggesting that spine clustering is also random and activity independent (Figure 2K).
Collectively, these results indicate that (1) hippocampal PNs develop repertoires of synapses with different shapes and, as evidenced by similarity of skewed PDF distributions, maintain optimal spacing between their synaptic sites in an intrinsic manner; and (2) the rules for arrangement of synapses with morphologically distinct spines in local networks are stochastic.
Silencing increases the variability in sizes of specific populations of synapses
Next, we exploited our annotated 3D image database to test if experience is required for development of synapses of diverse size. This parameter generally correlates with functional weight because the efficacy of neurotransmitter release is determined by the availability of vesicles and the magnitudes of excitatory postsynaptic currents are proportional to sizes of spines and PSDs containing scaffolding proteins that organize receptors (Berry and Nedivi, 2017; Kaeser and Regehr, 2017). We reconstructed >103 individual glutamatergic synapses from mice of each genotype and measured the volumes of their terminals, vesicle pools (e.g., numbers of vesicles in each terminal), spines, and PSDs. These measurements revealed two striking effects. On average, PNs of Emx1IRES-Cre/R26floxstop-TeNT mice had a ~2-fold enlargement of pre- and postsynaptic structures in synapses formed by Sc axons onto M spines. This phenotype was consistent from animal to animal and was not attributed to global arrest of membrane recycling or expression of TeNT per se because other synapses were either unaffected or affected to much lesser degrees and in both directions, despite the presence of pronounced SV clusters (Figures 3A–3I and S4). Furthermore, our reconstructions of axonal networks showed that >70% of Sc fibers can simultaneously activate M and other spines, thus excluding the possibility of a bias in excitatory input (Figures 2I and 2J). A second striking feature of silenced PNs is that enlargement of their M-type synapses was highly variable, as demonstrated by scattering of datapoints and consequent widening of PDF distributions of all four measured parameters (Figures 3C, 3E, 3G, 3I, S4, and S5).
Figure 3. Morphologies of active and inactive synapses.
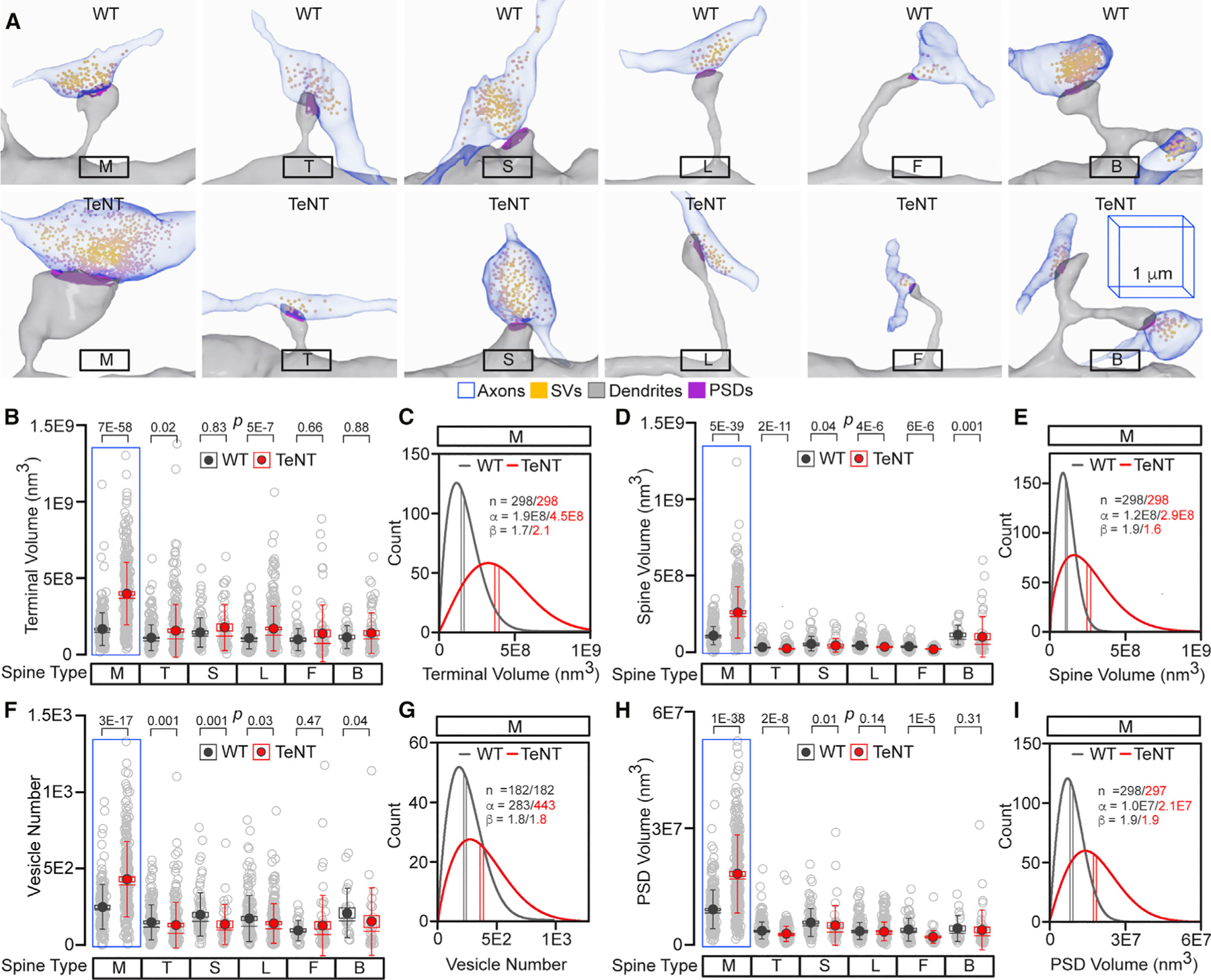
(A) Reconstructions of morphologically distinct glutamatergic synapses, as defined by spine type, in the CA1sr of WT and Emx1IRES-Cre/R26floxstop-TeNT mice (TeNT). The 1-µm 3D scale bars apply to all images from mice of both genotypes.
(B–I) Quantifications of terminal volumes (in B and C), spine volumes (in D and E), SV numbers (in F and G), and PSD volumes (in H and I) in individual synapses from mice of each genotype. Box with data overlap plots show raw data points (open circles), mean values (filled circles), standard errors (boxes), standard deviations (vertical lines), medians (horizontal lines), and p values, as defined by Mann-Whitney test. Data are from >103 synapses per genotype. Detailed sample sizes are listed in the accompanying Data S1 file. Weibull PDF plots for M-type synapses (C, E, G, and I) were generated and annotated as described in Figure 2.
Quantifications were performed using SBEM volumes from two WT and three TeNT mice. See also Figures S4 and S5.
The extents of proportionality between pre-and postsynaptic geometries depend on spine type and activity, but not on synapse size
The millisecond timescale of transmission at chemical synapses is possible because of precise alignment of opposed sides (Südhof, 2018). Moreover, the pre- and postsynaptic structures tend to be proportional, although currently available quantitative information about the proportionalities of glutamatergic synapses is based on imaging of M spines or bulk measurements that disregarded the differences in spine shapes (Bopp et al., 2017; Holderith et al., 2012; Hsu et al., 2017; Schikorski and Stevens, 1997). To gain additional insight into how activity impacts synaptic architecture and test whether changes in distributions of individual parameters shown in Figure 3 reflect a morphological disarray, we calculated the ratios between volumes of opposed features in single units, taking advantage of inventories with unique IDs. In the normal CA1sr, the volumes of terminals, vesicle pools, spines, and PSDs of M-type synapses positively correlated with Spearman rho values ranging from 0.42 to 0.76, of which the correlation between two postsynaptic features was the strongest. Notably, the rho values for inactive M synapses were significantly higher (0.61 to 0.85 with p values ranging from 0 to 0.04), indicating that their overall compartmentalization was preserved and alignments were improved (Figures 4A and 4B). Again, these measurements were largely consistent between animals (Figure S6). In contrast, the symmetries of intact synapses with T, S, and L spines were weaker, mainly due to disproportionally larger terminals. Moreover, the extents of these symmetries were not significantly altered in Emx1IRES-Cre/R26floxstop-TeNT mice, further suggesting that activity-dependent ultra-structural tuning is spine specific (Figures 4C–4E).
Figure 4. Alignments of structures in active and inactive synapses.
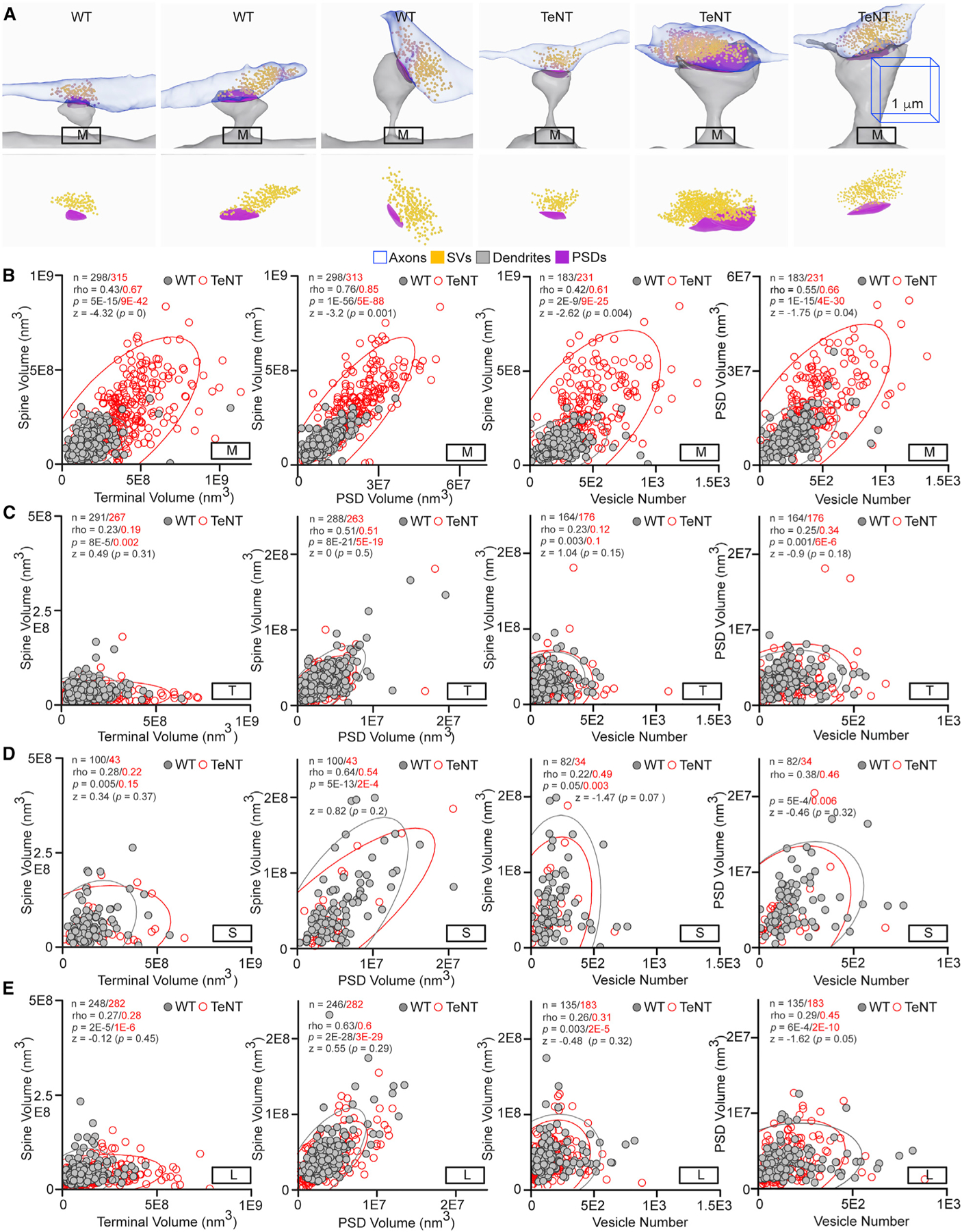
(A) Examples of increased variability in sizes and preserved proportionality of M-type synapses in silenced connectomes of Emx1IRES-Cre/R26floxstop-TeNT mice (TeNT). The 1-µm 3D scale bars apply to images from mice of both genotypes.
(B–E) Correlations between sizes of different pre- and postsynaptic structures in M (in B), T (in C), S (in D), and L (in E) synapses. Scatterplots with confidence ellipses, sample sizes (n), Spearman correlation coefficients (rho), Fisher transformation scores (Z), and p values are shown. Note that only alignments of M synapses are significantly altered. Also note differences in scales of x and y axes, which affect the slope angles.
Quantifications were performed using SBEM volumes from two WT and three TeNT mice. See also Figure S6.
Considering that the current criteria for spine nomenclature is somewhat arbitrary, we also compared the distributions of and correlations between core structural parameters without splitting synapses into morphologically distinct groups. We observed the widening of distributions of terminal, vesicle pool, spine, and PSD sizes in silenced connectomes in this case as well, albeit the effects were not as obvious and, in 3 out of 4 pairwise measures, the differences between mean values were not statistically significant. The latter is not surprising because the sizes of generic synapses are highly variable even in the normal brain (Figures S7A–S7D). Nonetheless, correlation analyses still revealed significant improvements of alignments of pre- and postsynaptic structures in Emx1IRES-Cre/R26floxstop-TeNT mice, indicating that outcomes of our measurements of M synapses are not attributed to selection bias (Figures S7E–S7H).
Silenced PNs have mismatched sizes and preserved alignments of compound synapses
Our results thus far support the model that differentiating PNs stochastically form networks of synapses with inherently variable weights, but this variability is diminished with experience. In other words, repetitive training improves the precision of coding at a circuit level by ultra-structural tuning of individual nodes, thereby making their responsiveness more predictable. Although seemingly contradicting simplified Hebbian rules, this model is consistent with recent studies of rare connections formed by the same axonal fibers onto two neighboring spines of same dendrites. In the normal CA1, these unusual compound synapses have been shown to have comparable morphologies, presumably reflecting their shared history of activity (Bartol et al., 2015; Bloss et al., 2018). Indeed, we found a nearly perfect correlation (rho = 0.85) between spine head volumes in compound synapse pairs of WT mice. Conversely, the compound pairs in silenced networks had profoundly mismatched spine head volumes (rho = 0.15), whereas the opposed parts of each contact tended to be more proportional (Figures 5A–5C).
Figure 5. Analysis of compound synapses.
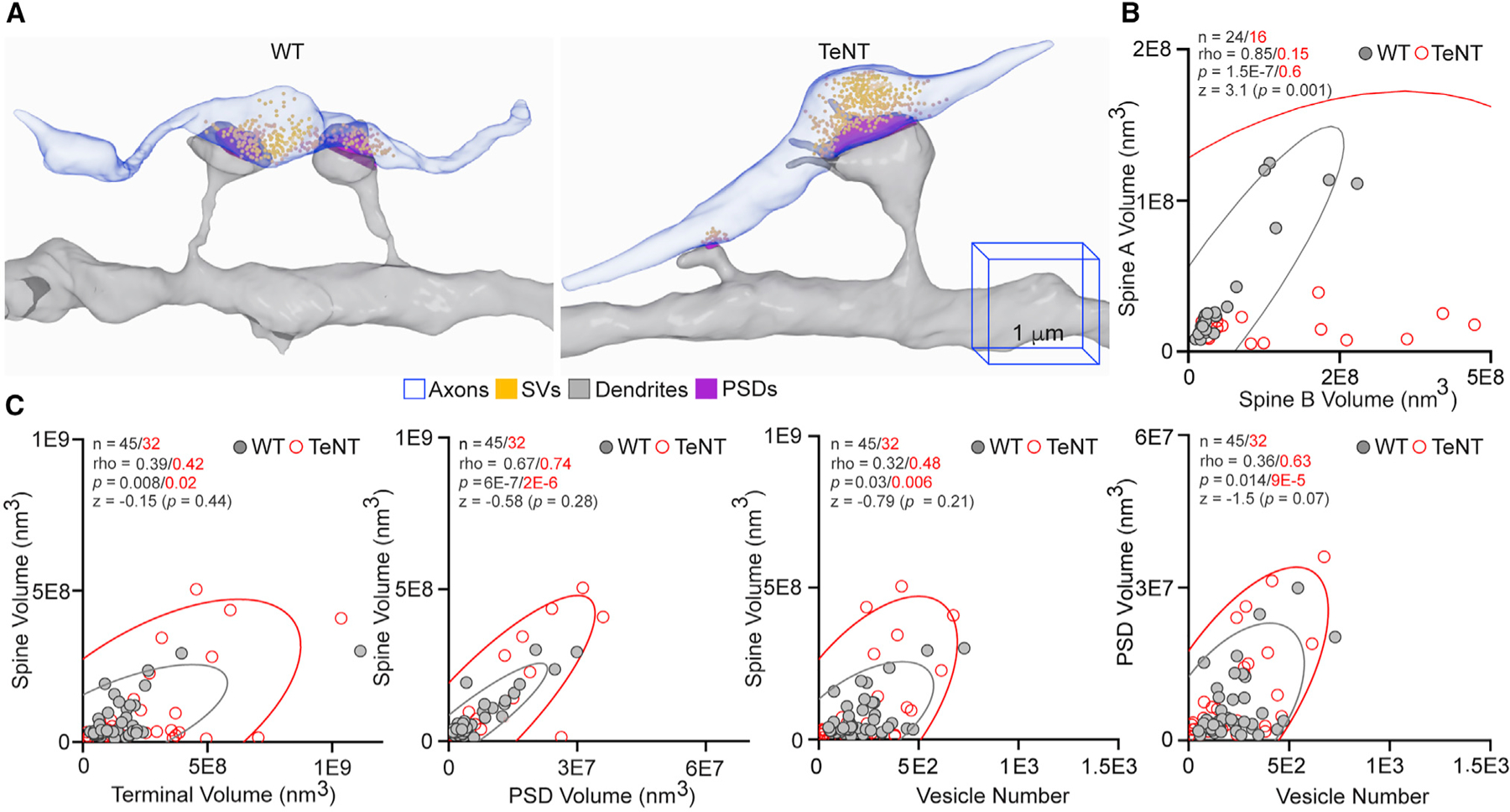
(A) Reconstructions of isolated Sc fibers that innervate two neighboring spines on the same dendritic branches (compound synapses) of PNs in the CA1sr of WT and Emx1IRES-Cre/R26floxstop-TeNT mice (TeNT). The 1-µm 3D scale bars apply to images from mice of both genotypes.
(B and C) Correlations between spine head volumes in each compound synapse pair (in B) and indicated pre- and postsynaptic structures in individual synapses (in C). Scatterplots with confidence ellipses, sample sizes (n), Spearman correlation coefficients (rho), Fisher transformation scores (Z), and p values are shown.
Quantifications were performed using SBEM volumes from two WT and three TeNT mice.
Taken together, these analyses of conventional and compound excitatory synapses suggest that experience plays different roles in regulating the order of synaptic structures at a single unit and population levels. Although the extents of proportionality between pre- and postsynaptic geometries differ among connections formed by glutamatergic axons onto specific spines (Figure 4), these parameters are defined intrinsically regardless of synapse size. Experience makes the intrinsically diverse sizes of M synapses more uniform as a population but reduces the symmetry of individual units.
Synaptic activity differentially affects the recruitment of organelles to terminals and spines
To explore the possibility that synaptic activity underlies the scattered distribution of intracellular membrane organelles involved in neurotransmission and network plasticity, we focused on two such organelles that can be unambiguously identified in SBEM volumes, namely, the mitochondria and SER. In addition to supplying ATP for energy-demanding aspects of synaptic physiology, including disassembly of SNARE complexes following vesicular exocytosis, mitochondria regulate the kinetics of neurotransmitter release by restricting the buildup of free calcium triggered by repetitive stimulation (Devine and Kittler, 2018; Hirabayashi et al., 2017). Curiously, only ~20%–25% of excitatory synapses contain roundish mitochondria at axonal terminals, whereas dendritic mitochondria tend to be elongated and almost exclusively localized in shafts (Kasthuri et al., 2015; Smith et al., 2016). Yet, it remains unclear if presynaptic recruitment of these organelles depends on the efficacy of vesicle recycling or properties of opposed spines. The mitochondria are mobile in neuronal cultures, but these observations are at odds with recent in vivo two-photon imaging studies in the mouse cerebral cortex (Lewis et al., 2016; Smit-Rigter et al., 2016). Likewise, only fractions of PN synapses contain tubular SER (Tu) and spine apparatus (SA), which presumably regulate calcium dynamics in spines (Bell et al., 2019), but it remains unknown if this heterogeneity is attributed to homeostatic adaptation to excitatory input or other forms of experience-dependent plasticity. We automatically segmented mitochondria in CA1sr volumes from WT and Emx1IRES-Cre/R26floxstop-TeNT mice by CDeep3M and examined their shapes and localization in annotated connectomes. In parallel, we assessed the synaptic content of Tu SER and SA; albeit, in this case, the membrane structures were traced manually because of their complexity (Figures 6A–6C and S8). We also imaged the organelles in a few manually picked synapses with M spines at higher magnification by using EM tomography (EMT), a technique that offers superior resolution at the expense of volume (Figure 6D).
Figure 6. Synaptic localization of mitochondria and SER.
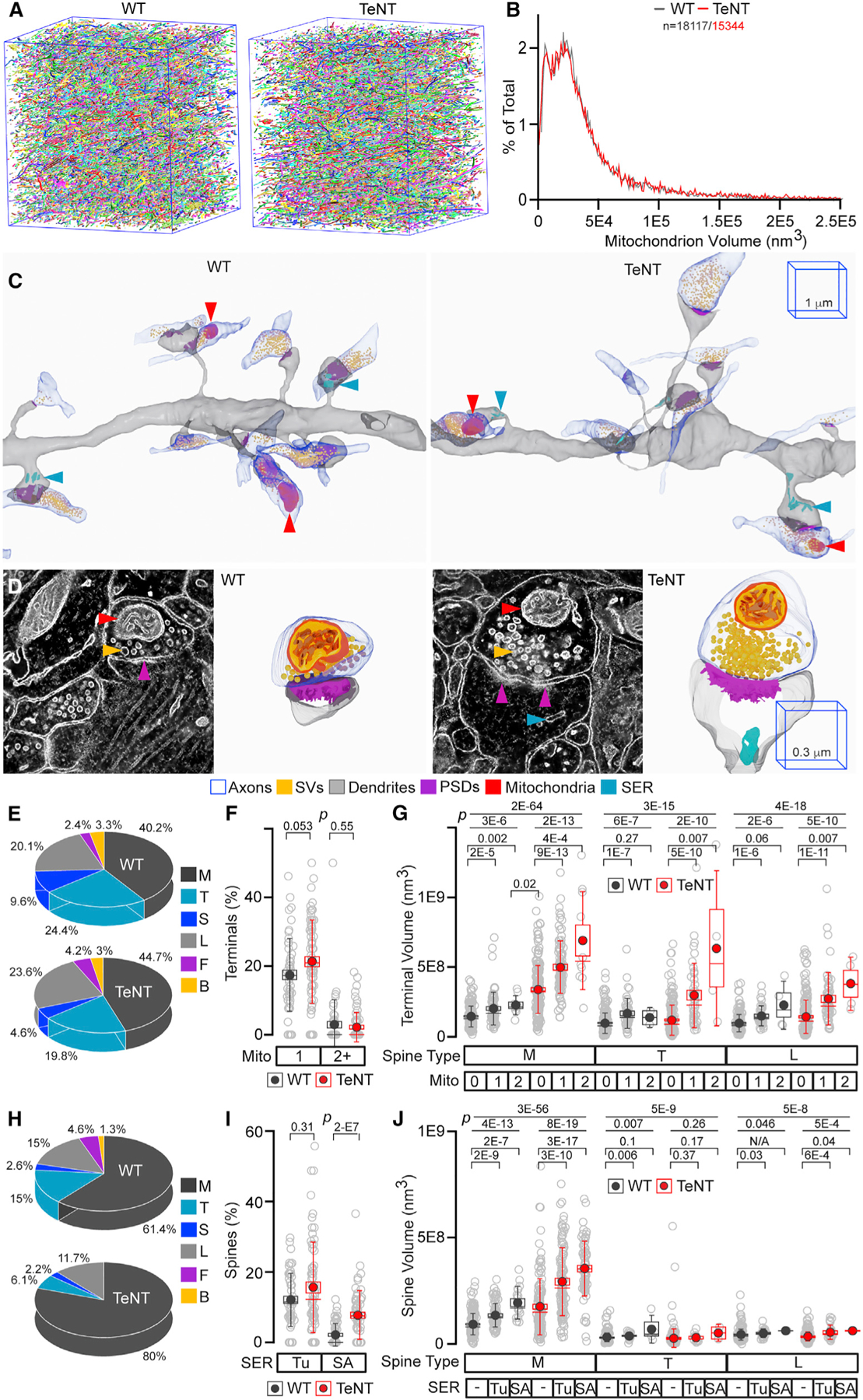
(A and B) Images (in A) and volumes (in B) of all mitochondria automatically annotated by CDeep3M in SBEM volumes from CA1sr of WT and Emx1IRES-Cre/R26floxstop-TeNT mice (TeNT).
(C) 3D SBEM reconstructions of excitatory microcircuits with mitochondria and SER. The 1-µm 3D scale bars apply to images from mice of both genotypes.
(D) Raw 2D EMT images and 3D reconstructions of mitochondria and SER in M synapses. The 0.3-µm 3D scale bars apply to images from mice of both genotypes. In (C) and (D), different membrane organelles are marked by arrows and color coded as indicated in the legend.
(E) Fractions of mitochondrion-positive terminals innervating distinct spines. WT, n = 209 terminals; TeNT, n = 237.
(F) Percentages of mitochondrion-positive terminals on dendritic branches. WT, n = 55 branches; TeNT, n = 69.
(G) Volumes of mitochondrion-negative and -positive terminals.
(H) Fractions of indicated spines containing SER (both tubular SER [Tu] and spine apparatus [SA]). WT, n = 153 spines; TeNT, n = 230.
(I) Percentages of spines with Tu and SA on dendritic branches. WT, n = 56 branches; TeNT, n = 67–68.
(J) Volumes of spines with no SER, with Tu, and with SA. In (F) and (I), p values were determined by Mann-Whitney test. In (G) and (J), p values were determined by Kruskal-Wallis ANOVA (first two lines) followed by Mann-Whitney test. Sample sizes are listed in the accompanying Data S1.
All quantifications were performed using SBEM volumes from two WT and three TeNT mice. See also Figure S8.
Permanently silenced PNs had no detectable abnormalities in mitochondrion morphologies, distribution in axonal and dendritic shafts, and synaptic localization. These organelles were recruited to intact and silenced axonal terminals innervating all spine types, and percentages of mitochondrion-positive boutons were not significantly different between genotypes (Figures 6C–6F). However, the presence of mitochondria positively correlated with terminal volume, this relationship was not spine specific, and it was more pronounced in Emx1IRES-Cre/R26floxstop-TeNT mice. The latter does not indicate that smaller synapses are unable to anchor mitochondria due to topological constrains because many silenced terminals lacking these organelles were still larger than mitochondrion-positive terminals in the normal brain (Figure 6G). Rather, pathways promoting presynaptic growth and vesicle recycling appear to compete for the same source of ATP, and this competition depends on rates of exocytosis. The blockade of synaptic excitation also did not preclude the postsynaptic recruitment of Tu SER and SA, but the distribution of SA was altered. Not only did these intracellular membranes form in the absence of glutamate release but also the numbers of SA-positive spines were increased by ~4-fold. Unlike mitochondria, SA was predominantly localized in synapses with M spines, and this selectivity was even more pronounced after silencing (Figures 6C, 6D, 6H, 6I, and S8). The presence of SA also correlated with volumes of spines and PSDs, but the diameters of necks of SA-positive spines were comparable, indicating that the restricted entry of SA in functional synapses is not due to the diffusion barrier (Figures 6J and S8).
Thus, activity is not necessary for the development of synapses with variable composition of organelles that regulate metabolic pathways and calcium buffering, but glutamatergic inputs regulate the abundance of these organelles on the postsynaptic side.
DISCUSSION
In summary, we have performed 3D nanoscale reconstructions of functional excitatory microcircuits in the mouse hippocampus and microcircuits in which vesicular release of glutamate was genetically suppressed throughout the lifespan. Sensory-experience-driven and spontaneous activity of excitatory synapses mediate a broad spectrum of structural changes in the developing and adult brain by mechanisms that involve de novo transcription, protein synthesis, trafficking, and posttranslational modifications (Espinosa and Stryker, 2012; Holtmaat and Svoboda, 2009; Katz and Shatz, 1996; Tom Dieck et al., 2014; Yap and Greenberg, 2018). Yet, we found that neurotransmitter signals are dispensable for the development of synapses with diverse shapes, sizes, and organelle content. Moreover, we demonstrated that activity plays a limited role in regulating the alignments of pre- and postsynaptic structures and that the rules for arrangement of structurally diverse synapses in local networks are stochastic. These findings have several important implications for understanding how neural circuits assemble and process information.
Our central conclusions are based on analyses of synapses grouped by spine type according to previously established criteria (Berry and Nedivi, 2017; Risher et al., 2014; Yuste and Bonhoeffer, 2004). Dendritic spines have been of interest to neuroscientists for more than 100 years since the pioneering discoveries of Santiago Ramón y Cajal. However, the key insights into spine biology have only started to emerge recently with advances in optical imaging of genetically encoded fluorescent reporters and EM. Numerous confocal and live two-photon imaging experiments have shown that spines are dynamic and that their turnover is affected by a wide range of physiological stimuli (Attardo et al., 2015; Berry and Nedivi, 2017; Caroni et al., 2012; Colgan and Yasuda, 2014; Hofer et al., 2009; Holtmaat et al., 2005; Koleske, 2013; Lendvai et al., 2000; Oh et al., 2015; Trachtenberg et al., 2002). Although spines become more stable with age, there is still no consensus on whether their morphological signatures reflect discrete states of maturity and functional strength or continuous progression between these states (Berry and Nedivi, 2017). In either case, these signatures, as well as the signatures of opposed terminals, appear to be determined intrinsically, given the remarkable preservation of repertoires of synapses in Emx1IRES-Cre/R26floxstop-TeNT mice.
From a conceptual standpoint, the implications of our work are 4-fold. First, our results imply that synapses that belong to the same neuron and relay identical chemical signals have unappreciated differences in molecular composition. Indeed, the spine-type-specific effects of activity revealed here are likely controlled by unique molecular players. It is also intriguing to predict that intrinsic recruitment of mitochondria to fractions of nerve terminals is attributed to nonuniform distribution of scaffolding proteins that anchor the organelles and restrict their mobility. A similar “intracellular adhesion code” might regulate the capturing of SER in spines, although the postsynaptic calcium buffering machinery is evidently more dynamic.
Second, stochastic arrangement of morphologically distinct synapses in local networks, as revealed by our combinatorial analyses of connectivity of single axonal fibers and spine clustering on dendrites, permits nontrivial network flexibility without any rewiring. Because the structures of stable synapses can change over time, the weights of inputs from each axon onto neighboring neurons in a receptive field can also change, thereby affecting the integration of incoming information (model depicted in Figure 7A). Our reconstructions of axonal networks are reminiscent of the flexibility of memory engrams; although associative memories can be artificially retrieved through optogenetic reactivation of hippocampal and cortical neurons that expressed an early response gene, Fos, during learning, only fractions of these neurons become Fos positive after natural retrieval (Cowansage et al., 2014; Liu et al., 2012; Reijmers et al., 2007; Roy et al., 2017). Likewise, recent in vivo imaging studies showed that cellular representations of learned tasks evolve over weeks in the posterior parietal cortex (Driscoll et al., 2017)
Figure 7. Models for dendritic integration of inputs from isolated axons and changes in synaptic weights.
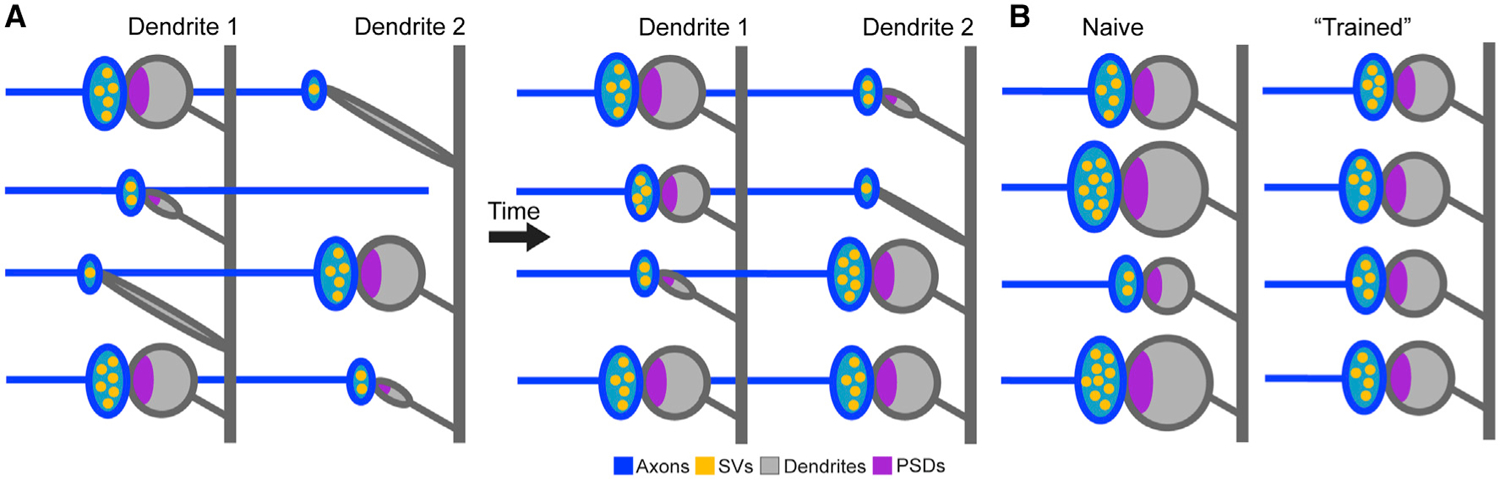
(A) Progressive changes in morphologies and weights of synapses formed by individual axons onto different dendrites.
(B) Activity-dependent decrease in the variability of weights of synapses with M spines.
The third and fourth conceptual implications arise from our measurements of distances between synapses and synapse sizes. It is becoming increasingly clear that distributions of anatomical and physiological parameters of the nervous system deviate from a typical Gaussian bell shape, which means that even relatively minor fractions of cells or subcellular features may play significant roles in complex processes. This paradigm applies to variables ranging from axon diameters to frequencies of network oscillations (Buzsáki and Mizuseki, 2014). Although skewing of distributions in our datasets is not unexpected, changes (or lack thereof) of their profiles in silenced microcircuits are noteworthy. We conclude that, at least in the CAsr, the distances between synaptic sites on axonal and dendritic shafts are “hardwired.” This conclusion may sound provocative, considering the well-established fact that glutamatergic neurons form new spines in an enriched environment, following acute sensory stimulation, and during memory acquisition (Holtmaat and Svoboda, 2009; Holtmaat et al., 2006; Moser et al., 1994; Nithianantharajah and Hannan, 2006; Rampon et al., 2000; van Praag et al., 2000). Yet, our results are reconcilable with the notion that experience-dependent synaptogenesis is not cumulative because the addition of new connections is counterbalanced by synapse elimination (Attardo et al., 2015; Holtmaat and Svoboda, 2009; Lai et al., 2012). Based on analyses of distributions of volumes of pre- and postsynaptic structures, we conclude that inherently variable weights of M synapses are matched after repetitive use (model depicted in Figure 7B). The overall increase in sizes of M synapses is also counterintuitive because PN spines expand during long-term potentiation and contract during long-term depression, which are the forms of Hebbian plasticity that involve activity-dependent changes in the AMPA receptor content (Harris, 2020; Makino and Malinow, 2009; Malenka and Bear, 2004; Zhou et al., 2004). However, it is important to emphasize that these transient effects occur in synapses that were activated many times in the past and that Hebbian mechanisms alone cannot account for experience-dependent control of synaptic strength. Progressive matching of synaptic weights is likely regulated by homeostatic mechanisms that operate on longer timescales (Keck et al., 2017; Turrigiano et al., 1998).
Lastly, our work has technical implications. The strategies for AI-assisted annotation of 3D EM data described here can be broadly applied to ultra-structural imaging of the brain and other organs in model organisms and humans. Furthermore, it is now technically feasible to label neuronal ensembles activated during specific cognitive tasks with genetically encoded markers compatible with SBEM (Girasole et al., 2018; Guenthner et al., 2013; Joesch et al., 2016; Zhang et al., 2019). In conjunction with prior 3D EM reconstruction studies of the normal brain (Bailey et al., 2015; Bloss et al., 2018; Bromer et al., 2018; Harris et al., 2015; Helmstaedter et al., 2013; Kasthuri et al., 2015; Mishchenko et al., 2010; Morgan et al., 2016; Motta et al., 2019; Smith et al., 2016), our resource of functional and permanently inactive connectomes provides a useful frame of reference for unraveling supramolecular events in circuits and synapses that coincide with novel experience and learning. This resource can also be used for additional mathematical modeling and to study how activity from synaptic sources impacts the development of other cell types, including GABAergic interneurons and glia. We therefore enclose all extracted numerical values and will freely share preannotated datasets, raw SBEM volumes, and computer scripts with the research community.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
All requests for data and resources should be directed to the Lead Contact, Dr. Anton Maximov (amaximov@scripps.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
All raw values used for quantifications presented in the manuscript are available in the enclosed Data S1. This file contains separate spreadsheets (split by types of analysis) with synapses classified according to spine type, measured parameters, sample sizes, brain IDs and statistics.
Computer scripts are available from authors upon request. All raw SBEM volumes will be made freely available to the research community after publication through a web-based resource.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
The Emx1IRES-Cre/R26floxstop-TeNT (TeNT) mouse model has been characterized in our previous study (Sando et al., 2017). The evidence that these animals permanently lacked synaptic release of glutamate from all Emx1-positive hippocampal PNs starting from embryonic development is supported by biochemical analyses of Synaptobrevin/VAMP2 (Syb2) cleavage, immunofluorescent imaging of Syb2 in VGlut1-immunoreactive glutamatergic terminals throughout the hippocampus, and electrophysiological whole-cell recordings of spontaneous and evoked excitatory postsynaptic currents (EPSCs) in acute hippocampal slices from juveniles (p3–5) and young adults (p30). Neither stainings for common neuronal markers, nor Cre-dependent viral tracing of silenced PNs revealed anatomical defects or cell death. As described in Sando et al. (2017), Emx1IRES-Cre/R26floxstop-TeNT mice required special care, could only survive for up to several weeks, and their survival was also dependent on genetic background. All animals used herein were a mix of C57BL/6 and 129/SV. Males and females were analyzed together.
METHOD DETAILS
Sample preparation and fixation
Mice were anesthetized via intraperitoneal injections of ketamine/xylazine, transcardially perfused with oxygenated Ringer’s solution, and then perfused with a buffer containing 2% paraformaldehyde, 2.5% glutaraldehyde, 150 mM cacodylate and 2 µM CaCl2. The brains were post-fixed overnight in the same solution at 4°C. 100 µm thick coronal slices were cut in Vibratome and prepared for SBEM imaging using the following sequential procedures *: 1) Overnight post-fixation at 4°C followed by washes in the buffer containing 150 mM cacodylate and 0.2 mM CaCl2; 2) Fixation at room temperature for 1 hour in 2% OsO4 in cacodylate; 3) Staining in 0.5% aq. thiocarbohydrazide; 4) Staining with 2% aq. OsO4; 5) Overnight incubation at 4°C in 2% aq. uranyl acetate ; 6) Staining with lead aspartate at 60°C for 30 min; 7) Dehydration on ice in 70%, 90%, and 100% ethanol followed by dry acetone; 8) Infiltration with acetone:Durcupan ACM; 9) Embedding in 100% Durcupan resin at 60°C for 48 hours. Approximately 1 mm square pieces of tissue were mounted on Gatan SBEM specimen pins with conductive silver epoxy.
* For steps 2 to 6, each procedure included subsequent washes in water at room temperature.
SBEM imaging
Samples were imaged under the Zeiss Merlin scanning electron microscope equipped with a Gatan 3View. Imaging was performed at 2.5 kV and 85 pA using a focal charge compensation device to minimize specimen charging (2.5×10−3 mbar nitrogen gas). ~35,000 µm3 volumes were collected from dorsal CA1sr using 10k x 10k raster images with 5.3 nm pixels, 2 µsec pixel dwell time, and 60 nm Z steps. Acquired volumes were aligned in IMOD (Kremer et al., 1996).
EMT imaging
300 nm thick sections were cut from the SBEM-stained specimens and collected on 50 nm Luxel slot grids. The grids were coated with 10 nm colloidal gold and imaged at 300 keV on a Titan TEM. Double-tilt series were collected with 0.5 degree increments at 22,500X magnification on a 4k x 4k Gatan Ultrascan camera. Tomograms were generated with an iterative scheme in the TxBR package (Chen et al., 2014).
Cloud-based deep learning image segmentation
Automatic segmentation of different subcellular structures was performed with CDeep3M, a cloud-based platform utilizing a deep convolutional neural network (Haberl et al., 2018). This recently developed tool enables effective processing of multiple common microscopy modalities, including SBEM. Network retraining and predictions were done in the Amazon Web Service (AWS)-based version of CDeep3M to avoid the need for local high-end graphics processing unit (GPU) usage on our laboratory computers. Since creating training data from scratch is time consuming, we chose to retrain pre-trained neural networks on specific image sets. This method of domain adaptation reduces effort and time by 90%, while still achieving high segmentation accuracy (Haberl et al., 2018). Pre-trained membrane and mitochondria models were downloaded from the publicly available Cell Image Library and retrained for each image stack using small volumes with manually segmented ground truth labels. All image contrasting and manipulations were done with the ImageJ software. The accuracy of retrained neural networks was validated with small volumes from separate areas of same 3D image sets. Validated models were then applied to automatically segment entire volumes. The output prediction maps were stitched together and used for semi-automated reconstructions in VAST (Kasthuri et al., 2015).
Semi-automatic volume segmentation
was done in VAST Lite, which is a voxel painting program designed for analysis of large volumetric datasets (Berger et al., 2018). We used masked painting in VAST in combination with the CDeep3M predicted boundary maps to constrain painted areas, so that the outline of each object is traced automatically. By selectively painting neuropil structures, we were able to achieve quick and accurate reconstructions of projections, synapses and mitochondria. Errors in painting from occasionally inaccurately predicted boundaries were manually corrected. For segmentation of dendrites and axons, we used membrane prediction boundary maps for filling outlines of each cellular structure. Since the present study was focused on excitatory circuits, we selected spiny apical dendritic segments of PNs and glutamatergic synapses that contain characteristic postsynaptic densities (PSDs). All segmented volume data were extracted in MATLAB through the included software VastTools.
Skeletonization and assignment of coordinates
Skeletons for traced dendritic and axonal segments were generated in VAST. Using the annotation function, connected nodes were placed along the center of each reconstructed structure’s cross-section from the first slice of the image volume to the last. Image stacks where dendritic spines or axonal terminals were present along the skeletonized length were also marked in order to assess spine and terminal distributions and distances across the entire structure. For analysis of smallest widths of spine necks, nodes were placed between the shortest two points. Annotated length data was extracted in MATLAB through the included VAST API. Skeleton node coordinates were retrieved with the getannoobject function and then analyzed with custom scripts.
Splitting of spines and terminals
Spines and nerve terminals were manually separated from their parent structures using the splitting tool in VAST. This procedure created new objects, which could then be automatically volume filled using the filling tool. Spines were split at bases of their necks near dendritic shafts. Terminals were split at each end where axonal cross-sections minimize, vesicle pools end, and/or opposing PSDs end.
PSDs, synaptic vesicles and SER
PSDs were identified as darkly stained regions at the ends of dendritic spines, typically opposing axonal terminals with synaptic vesicles. In cases where no nerve terminal was present (naked spines), PSDs were also traced if visible. PSDs were segmented in VAST by adjusting the pen size depending on PSD thickness. Since there can be some variation in the visibility of PSD depending on spine orientation (for example, cross-sectioned synapses may seem more continuous than oblique synapses), we may have underestimated PSD volumes in a few cases. However, we reasoned that this did not significantly bias the results because we applied the same segmentation criteria to both genotypes and analyzed large sample sizes (> 103 PSDs per genotype).
Individual synaptic vesicles were painted in VAST with a pen of fixed size. Since the thickness of serial sections (60 nm) was larger than vesicle diameters (typically ~40 nm), each visible vesicle on each section was considered a distinct object. In cases where vesicle clouds made it difficult to distinguish individual organelles from one another, we opted to fill the clouds with as many vesicles that could fit in without overlap. Similar to the variation in PSD visibility depending on synapse orientation, there can be variations in synaptic vesicle visibility in certain synapses that can lead to count inaccuracy. In particular, oblique and an face synapses can have vesicles that are obscured by PSDs, resulting in unreliable or underestimated counts (Harris et al., 2015). Additionally, there is the possibility of double counting vesicles that are sectioned in the middle. To address these concerns, we used the same equal application and large sample size reasoning as for PSD measurements described above. Vesicle numbers were determined using the Export Particle Clouds function in VastTools with settings counting each separate painted region as one object. Thus, we put emphasis on not intersecting painted regions while tracing individual vesicles.
The distribution of Smooth Endoplasmic Reticulum (SER) in dendritic spines was analyzed using previously segmented spines with unique IDs. The spines were first classified as being SER positive or negative based on the presence of SER in spine necks or heads. SER was then classified as tubular or a spine apparatus based on previously established criteria showing structural differences with functional implications (Chirillo et al., 2019).
Classification of Spines
Dendritic spines were classified into 6 distinct types based on size and morphology: Mushroom (M), Thin (T), Stubby (S), Long thin (L), Filopodia (F), and Bifurcated (B). Rare spines whose shapes were unclear were excluded from quantitative analyses of morphologically defined synapse populations and only used for measurements of distances between all synapses. However, this represented a negligible number of spines (< 1%) and we did not observe any apparent differences of undefined-type spines between genotypes. The criteria for classification were based on previous studies that also use distinct geometrical characteristics of spines as a basis for categorization (Risher et al., 2014). Each spine was sliced from the parent dendritic fragment in VAST and given a unique color and numerical ID.
Data extraction and analysis
VastTools is a MATLAB script that interfaces with VAST via included API. We used VastTools for extracting volume measurements, coordinates for length and distance measurements, and 3D surface meshes. For all data extractions and 3D model exports, parameters were equally applied to samples of both genotypes, as well as across all brains. Since VastTools permits quantifications at lower resolutions, we used the native image stack voxel size (5.33 nm × 5.33 nm × 60 nm) at Mipmap level 0. All numerical values were extracted before 3D modeling.
Volume measurements, vesicle counts and 3D modeling
Volumes of dendritic spines, axonal terminals, PSDs and Mitochondria were measured using the Measure Segment Volumes function, which counts the total number of voxels of different specified objects in a boundary area. 3D models were generated with “Export 3D Models” function, which creates surface meshes in VAST and exports them as .obj files for modeling in Blender. Some features were exported using lower-resolution models (Mip 2) to make post-export smoothing easier. Vesicles were counted using the Export Particle Clouds function, which can be set to count each separate painted 2D region in a serial section as one vesicle. As stated above, this allowed accurate counts of individually painted organelles in a specified terminal with the exception of a few vesicles being undercounted if they were mistakenly overlapped with another painted vesicle. The Export Particle Clouds function was also used to export 3D surface mesh models (.obj) of vesicles for 3D modeling in Blender.
Coordinate exporting
Since large-scale length and distance measurement functions are not currently implemented in the VastTools window of the current version of Vast Lite (only the Euclidean distance measurement tool), we wrote our own MATLAB scripts that utilized API functions. As stated above, we used the Annotation tool in VAST to create skeletons nodes that were placed along neuropil structures across the image stack (e.g., axons). Nodes were placed in center of each structure on a serial section in order to prevent overestimation of length. To analyze dendrites, nodes were placed at starts and ends of dendritic fragments, as well as in each section where a spine branched off from a shaft. This allowed us to extract such parameters as dendritic fragment length, linear spine density, and distances between individual spines. Since we also classified spines and assigned them unique IDs, we were able to perform similar length and distance measurements for specific spine types and identify nearest-neighbor spine types. We considered every dendritic protrusion a spine. While the skeletonization approach used for dendrites generally applies for axonal fibers, we wanted to account for higher curvatures of these structures. Our approach for axonal skeletonization was to place nodes in the beginning and end of axons, in terminals with opposed spines, as well as in arbitrary sections along fibers in order to match their curvatures. By flagging which nodes corresponded to axonal terminals, we were able to accurately calculate axon length, terminal density, and distances between terminals. In the rare cases where axonal fibers mainly innervated dendritic shafts, we excluded them from analysis because they are presumed to be from interneurons. In order to extract the exact coordinates, we used the VAST API function getannoobject, which provides a matrix with all x, y, and z coordinates of every node. We then designed MATLAB scripts that use these matrix coordinates to calculate distances depending on the type of analysis. For example, the distances from each placed node to another along a skeletonized dendritic fragment sum up to its total length. All distance and length measurements were based on the MATLAB function vecnorm that uses the Euclidean norm, where vector v with N elements is defined by:
Data blinding
To ensure consistently of annotations and to eliminate any bias, manual tracing of all subcellular structures, classifications of spines, and extractions of various synaptic parameters were performed by several trained investigators who were blinded to genotypes.
3D Modeling
Final 3D modeling was performed in Blender 2.8 (http://Blender.org), an open-source 3D computer-graphics software toolset with modeling, material editing, and rendering capabilities. 3D surface meshes of neuropil structures were imported as .obj files. No post-import size scaling was applied. To simplify post-import editing, most features were exported at a lower resolution (Mipmap level 2). The lower resolution models maintained their native scale. Additionally, importing .obj models from VastTools preserves the spatial location of every segmented feature, so the actual spatial distribution from the SBEM image stack is maintained. Synaptic vesicles were represented by pre-made 40 nm 3D models included with VastTools. Smoothing, color enhancements, and material assignments were applied equally to both genotypes and across all brains. The color-coding schemes for each feature are included in the figures. Dendrites, axons, mitochondria, and SER were smoothened using the smooth vertices function in edit mode and the smooth shading function. Vesicles were smoothened only using the smooth shading function. The scenes were rendered using the cycles renderer.
QUANTIFICATION AND STATISTICAL ANALYSIS
All final quantifications, curve fittings and statistical analyses were performed in Origin Pro. We found that distributions of virtually all measured parameters, such as distances between terminals and spines and sizes of various structures in individual synapses, were significantly deviated from normal. Therefore, non-parametric statistical tests were used throughout the study. For standard comparison of populations (box with data overlap plots), p values were determined by Kruskal-Wallis ANOVA and/or Mann-Whitney test. Correlation analyses were performed using Spearman test. Statistical significances of differences between Spearman correlation coefficients (rho) were determined with Fisher rho-to-z transformation test. Our choice of Weibull probability density function (PDF) (Scholte et al., 2009) for analysis of distributions was based on systematic side-by-side fitting of each dataset with Gaussian, Weibull, Lognormal and Gamma fitting functions accompanied with Kolmogorov-Smirnov test for goodness of fits. These analyses indicated that Weibull PDF was the most versatile.
The PDF of a Weibull random variable is:
where “a” is the scale parameter and “b” is the shape parameter.
The formula for analysis of spine combinations is described in the main text.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
|
| ||
| VGlut1 | Synaptic Systems | Cat#135303, RRID:AB_887875 |
| Syb2 | Synaptic Systems | Cat#104211C3, RRID:AB_887808 |
|
| ||
| Bacterial and virus strains | ||
|
| ||
| AAVDJ DIO-mGFP | Maximov Lab (Sando et al., 2017) | PMID: 28426966 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| Emx1IRES-Cre mouse allele | JAX (Gorski et al., 2002) | PMID: 12151506 |
| R26floxstopTeNT mouse allele | Goulding Lab (Zhang et al., 2008) | PMID: 18940590 |
|
| ||
| Software and algorithms | ||
|
| ||
| CDeep3M | Ellisman Lab (Haberl et al., 2018) | PMID: 30171236 |
| OriginPro | Origin Lab | N/A |
| VAST | Lichtman Lab (Berger et al., 2018) | PMID: 30386216 |
| Custom MATLAB scripts | This paper | N/A |
Highlights.
Chemical synapses have heterogeneous sizes, shapes, and organelle contents
This structural diversity is generated by mechanisms that are largely intrinsic
Structurally diverse synapses are organized in local connectomes stochastically
The morphological variability of specific synapse pools decreases with experience
ACKNOWLEDGMENTS
We thank Dr. Ardem Patapoutian and all members of Maximov and Ellisman labs for advice and critical comments. This study was supported by the NIH grants R01NS087026 (to A.M. and M.E.) and R01GM117049, R01MH118442, and R01 MH123224 (to A.M.).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.108953.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Alvarez VA, and Sabatini BL (2007). Anatomical and physiological plasticity of dendritic spines. Annu. Rev. Neurosci 30, 79–97. [DOI] [PubMed] [Google Scholar]
- Attardo A, Fitzgerald JE, and Schnitzer MJ (2015). Impermanence of dendritic spines in live adult CA1 hippocampus. Nature 523, 592–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey CH, Kandel ER, and Harris KM (2015). Structural Components of Synaptic Plasticity and Memory Consolidation. Cold Spring Harb. Perspect. Biol 7, a021758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartol TM, Bromer C, Kinney J, Chirillo MA, Bourne JN, Harris KM, and Sejnowski TJ (2015). Nanoconnectomic upper bound on the variability of synaptic plasticity. eLife 4, e10778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell M, Bartol T, Sejnowski T, and Rangamani P (2019). Dendritic spine geometry and spine apparatus organization govern the spatiotemporal dynamics of calcium. J. Gen. Physiol 151, 1017–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger DR, Seung HS, and Lichtman JW (2018). VAST (Volume Annotation and Segmentation Tool): Efficient Manual and Semi-Automatic Labeling of Large 3D Image Stacks. Front. Neural Circuits 12, 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry KP, and Nedivi E (2017). Spine Dynamics: Are They All the Same? Neuron 96, 43–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloss EB, Cembrowski MS, Karsh B, Colonell J, Fetter RD, and Spruston N (2018). Single excitatory axons form clustered synapses onto CA1 pyramidal cell dendrites. Nat. Neurosci 21, 353–363. [DOI] [PubMed] [Google Scholar]
- Bopp R, Holler-Rickauer S, Martin KA, and Schuhknecht GF (2017). An ultrastructural study of the thalamic input to layer 4 of primary motor and primary somatosensory cortex in the mouse. J. Neurosci 37, 2435–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne JN, and Harris KM (2012). Nanoscale analysis of structural synaptic plasticity. Curr. Opin. Neurobiol 22, 372–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromer C, Bartol TM, Bowden JB, Hubbard DD, Hanka DC, Gonzalez PV, Kuwajima M, Mendenhall JM, Parker PH, Abraham WC, et al. (2018). Long-term potentiation expands information content of hippocampal dentate gyrus synapses. Proc. Natl. Acad. Sci. USA 115, E2410–E2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzsáki G, and Mizuseki K (2014). The log-dynamic brain: how skewed distributions affect network operations. Nat. Rev. Neurosci 15, 264–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caroni P, Donato F, and Muller D (2012). Structural plasticity upon learning: regulation and functions. Nat. Rev. Neurosci 13, 478–490. [DOI] [PubMed] [Google Scholar]
- Chen R, Wan X, Altintas I, Wang J, Crawl D, Phan S, Lawrence A, and Ellisman M (2014). EPiK-a Workflow for Electron Tomography in Kepler. Procedia Comput. Sci 20, 2295–2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirillo MA, Waters MS, Lindsey LF, Bourne JN, and Harris KM (2019). Local resources of polyribosomes and SER promote synapse enlargement and spine clustering after long-term potentiation in adult rat hippocampus. Sci. Rep 9, 3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colgan LA, and Yasuda R (2014). Plasticity of dendritic spines: subcompartmentalization of signaling. Annu. Rev. Physiol 76, 365–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowansage KK, Shuman T, Dillingham BC, Chang A, Golshani P, and Mayford M (2014). Direct reactivation of a coherent neocortical memory of context. Neuron 84, 432–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devine MJ, and Kittler JT (2018). Mitochondria at the neuronal presynapse in health and disease. Nat. Rev. Neurosci 19, 63–80. [DOI] [PubMed] [Google Scholar]
- Driscoll LN, Pettit NL, Minderer M, Chettih SN, and Harvey CD (2017). Dynamic Reorganization of Neuronal Activity Patterns in Parietal Cortex. Cell 170, 986–999.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa JS, and Stryker MP (2012). Development and plasticity of the primary visual cortex. Neuron 75, 230–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förster E, Zhao S, and Frotscher M (2006). Laminating the hippocampus. Nat. Rev. Neurosci 7, 259–267. [DOI] [PubMed] [Google Scholar]
- Girasole AE, Lum MY, Nathaniel D, Bair-Marshall CJ, Guenthner CJ, Luo L, Kreitzer AC, and Nelson AB (2018). A Subpopulation of Striatal Neurons Mediates Levodopa-Induced Dyskinesia. Neuron 97, 787–795.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorski JA, Talley T, Qiu M, Puelles L, Rubenstein JL, and Jones KR (2002). Cortical excitatory neurons and glia, but not GABAergic neurons, are produced in the Emx1-expressing lineage. J. Neurosci 22, 6309–6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenthner CJ, Miyamichi K, Yang HH, Heller HC, and Luo L (2013). Permanent genetic access to transiently active neurons via TRAP: targeted recombination in active populations. Neuron 78, 773–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberl MG, Churas C, Tindall L, Boassa D, Phan S, Bushong EA, Madany M, Akay R, Deerinck TJ, Peltier ST, and Ellisman MH (2018). CDeep3M-Plug-and-Play cloud-based deep learning for image segmentation. Nat. Methods 15, 677–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KM (2020). Structural LTP: from synaptogenesis to regulated synapse enlargement and clustering. Curr. Opin. Neurobiol 63, 189–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KM, and Weinberg RJ (2012). Ultrastructure of synapses in the mammalian brain. Cold Spring Harb. Perspect. Biol 4, a005587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KM, Spacek J, Bell ME, Parker PH, Lindsey LF, Baden AD, Vogelstein JT, and Burns R (2015). A resource from 3D electron microscopy of hippocampal neuropil for user training and tool development. Sci. Data 2, 150046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazan L, and Ziv NE (2020). Activity Dependent and Independent Determinants of Synaptic Size Diversity. J. Neurosci 40, 2828–2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held RG, Liu C, Ma K, Ramsey AM, Tarr TB, De Nola G, Wang SSH, Wang J, van den Maagdenberg AMJM, Schneider T, et al. (2020). Synapse and Active Zone Assembly in the Absence of Presynaptic Ca2+ Channels and Ca2+ Entry. Neuron 107, 667–683.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmstaedter M, Briggman KL, and Denk W (2008). 3D structural imaging of the brain with photons and electrons. Curr. Opin. Neurobiol. 18, 633–641. [DOI] [PubMed] [Google Scholar]
- Helmstaedter M, Briggman KL, Turaga SC, Jain V, Seung HS, and Denk W (2013). Connectomic reconstruction of the inner plexiform layer in the mouse retina. Nature 500, 168–174. [DOI] [PubMed] [Google Scholar]
- Hirabayashi Y, Kwon SK, Paek H, Pernice WM, Paul MA, Lee J, Erfani P, Raczkowski A, Petrey DS, Pon LA, and Polleux F (2017). ER-mitochondria tethering by PDZD8 regulates Ca2+ dynamics in mammalian neurons. Science 358, 623–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer SB, Mrsic-Flogel TD, Bonhoeffer T, and Hübener M (2009). Experience leaves a lasting structural trace in cortical circuits. Nature 457, 313–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holderith N, Lorincz A, Katona G, Rózsa B, Kulik A, Watanabe M, and Nusser Z (2012). Release probability of hippocampal glutamatergic terminals scales with the size of the active zone. Nat. Neurosci 15, 988–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtmaat A, and Svoboda K (2009). Experience-dependent structural synaptic plasticity in the mammalian brain. Nat. Rev. Neurosci 10, 647–658. [DOI] [PubMed] [Google Scholar]
- Holtmaat AJ, Trachtenberg JT, Wilbrecht L, Shepherd GM, Zhang X, Knott GW, and Svoboda K (2005). Transient and persistent dendritic spines in the neocortex in vivo. Neuron 45, 279–291. [DOI] [PubMed] [Google Scholar]
- Holtmaat A, Wilbrecht L, Knott GW, Welker E, and Svoboda K (2006). Experience-dependent and cell-type-specific spine growth in the neocortex. Nature 441, 979–983. [DOI] [PubMed] [Google Scholar]
- Hsu A, Luebke JI, and Medalla M (2017). Comparative ultrastructural features of excitatory synapses in the visual and frontal cortices of the adult mouse and monkey. J. Comp. Neurol 525, 2175–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joesch M, Mankus D, Yamagata M, Shahbazi A, Schalek R, Suissa-Peleg A, Meister M, Lichtman JW, Scheirer WJ, and Sanes JR (2016). Reconstruction of genetically identified neurons imaged by serial-section electron microscopy. eLife 5, e15015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeser PS, and Regehr WG (2017). The readily releasable pool of synaptic vesicles. Curr. Opin. Neurobiol 43, 63–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandel ER, Dudai Y, and Mayford MR (2014). The molecular and systems biology of memory. Cell 157, 163–186. [DOI] [PubMed] [Google Scholar]
- Kasai H, Fukuda M, Watanabe S, Hayashi-Takagi A, and Noguchi J (2010). Structural dynamics of dendritic spines in memory and cognition. Trends Neurosci 33, 121–129. [DOI] [PubMed] [Google Scholar]
- Kasthuri N, Hayworth KJ, Berger DR, Schalek RL, Conchello JA, Knowles-Barley S, Lee D, Vázquez-Reina A, Kaynig V, Jones TR, et al. (2015). Saturated Reconstruction of a Volume of Neocortex. Cell 162, 648–661. [DOI] [PubMed] [Google Scholar]
- Katz LC, and Shatz CJ (1996). Synaptic activity and the construction of cortical circuits. Science 274, 1133–1138. [DOI] [PubMed] [Google Scholar]
- Keck T, Toyoizumi T, Chen L, Doiron B, Feldman DE, Fox K, Gerstner W, Haydon PG, Hübener M, Lee HK, et al. (2017). Integrating Hebbian and homeostatic plasticity: the current state of the field and future research directions. Philos. Trans. R. Soc. Lond. B Biol. Sci 372, 20160158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koleske AJ (2013). Molecular mechanisms of dendrite stability. Nat. Rev. Neurosci 14, 536–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, and McIntosh JR (1996). Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol 116, 71–76. [DOI] [PubMed] [Google Scholar]
- Kwon SK, Sando R III, Lewis TL, Hirabayashi Y, Maximov A, and Polleux F (2016). LKB1 Regulates Mitochondria-Dependent Presynaptic Calcium Clearance and Neurotransmitter Release Properties at Excitatory Synapses along Cortical Axons. PLoS Biol 14, e1002516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CS, Franke TF, and Gan WB (2012). Opposite effects of fear conditioning and extinction on dendritic spine remodelling. Nature 483, 87–91. [DOI] [PubMed] [Google Scholar]
- Lendvai B, Stern EA, Chen B, and Svoboda K (2000). Experience-dependent plasticity of dendritic spines in the developing rat barrel cortex in vivo. Nature 404, 876–881. [DOI] [PubMed] [Google Scholar]
- Lewis TL Jr., Turi GF, Kwon SK, Losonczy A, and Polleux F (2016). Progressive Decrease of Mitochondrial Motility during Maturation of Cortical Axons In Vitro and In Vivo. Curr. Biol 26, 2602–2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Ramirez S, Pang PT, Puryear CB, Govindarajan A, Deisseroth K, and Tonegawa S (2012). Optogenetic stimulation of a hippocampal engram activates fear memory recall. Nature 484, 381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Bushong EA, Shih TP, Ellisman MH, and Nicoll RA (2013). The cell-autonomous role of excitatory synaptic transmission in the regulation of neuronal structure and function. Neuron 78, 433–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino H, and Malinow R (2009). AMPA receptor incorporation into synapses during LTP: the role of lateral movement and exocytosis. Neuron 64, 381–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malenka RC, and Bear MF (2004). LTP and LTD: an embarrassment of riches. Neuron 44, 5–21. [DOI] [PubMed] [Google Scholar]
- Matsuo N, Reijmers L, and Mayford M (2008). Spine-type-specific recruitment of newly synthesized AMPA receptors with learning. Science 319, 1104–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishchenko Y, Hu T, Spacek J, Mendenhall J, Harris KM, and Chklovskii DB (2010). Ultrastructural analysis of hippocampal neuropil from the connectomics perspective. Neuron 67, 1009–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongillo G, Rumpel S, and Loewenstein Y (2017). Intrinsic volatility of synaptic connections - a challenge to the synaptic trace theory of memory. Curr. Opin. Neurobiol 46, 7–13. [DOI] [PubMed] [Google Scholar]
- Morgan JL, Berger DR, Wetzel AW, and Lichtman JW (2016). The Fuzzy Logic of Network Connectivity in Mouse Visual Thalamus. Cell 165, 192–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser MB, Trommald M, and Andersen P (1994). An increase in dendritic spine density on hippocampal CA1 pyramidal cells following spatial learning in adult rats suggests the formation of new synapses. Proc. Natl. Acad. Sci. USA 91, 12673–12675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motta A, Berning M, Boergens KM, Staffler B, Beining M, Loomba S, Hennig P, Wissler H, and Helmstaedter M (2019). Dense connectomic reconstruction in layer 4 of the somatosensory cortex. Science 366, eaay3134. [DOI] [PubMed] [Google Scholar]
- Nithianantharajah J, and Hannan AJ (2006). Enriched environments, experience-dependent plasticity and disorders of the nervous system. Nat. Rev. Neurosci 7, 697–709. [DOI] [PubMed] [Google Scholar]
- Oh WC, Parajuli LK, and Zito K (2015). Heterosynaptic structural plasticity on local dendritic segments of hippocampal CA1 neurons. Cell Rep 10, 162–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampon C, Tang YP, Goodhouse J, Shimizu E, Kyin M, and Tsien JZ (2000). Enrichment induces structural changes and recovery from nonspatial memory deficits in CA1 NMDAR1-knockout mice. Nat. Neurosci 3, 238–244. [DOI] [PubMed] [Google Scholar]
- Reijmers LG, Perkins BL, Matsuo N, and Mayford M (2007). Localization of a stable neural correlate of associative memory. Science 317, 1230–1233. [DOI] [PubMed] [Google Scholar]
- Risher WC, Ustunkaya T, Singh Alvarado J, and Eroglu C (2014). Rapid Golgi analysis method for efficient and unbiased classification of dendritic spines. PLoS One 9, e107591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy DS, Kitamura T, Okuyama T, Ogawa SK, Sun C, Obata Y, Yoshiki A, and Tonegawa S (2017). Distinct Neural Circuits for the Formation and Retrieval of Episodic Memories. Cell 170, 1000–1012.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sando R, Bushong E, Zhu Y, Huang M, Considine C, Phan S, Ju S, Uytiepo M, Ellisman M, and Maximov A (2017). Assembly of Excitatory Synapses in the Absence of Glutamatergic Neurotransmission. Neuron 94, 312–321.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schikorski T, and Stevens CF (1997). Quantitative ultrastructural analysis of hippocampal excitatory synapses. J. Neurosci 17, 5858–5867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch S, Deák F, Königstorfer A, Mozhayeva M, Sara Y, Südhof TC, and Kavalali ET (2001). SNARE function analyzed in synaptobrevin/VAMP knockout mice. Science 294, 1117–1122. [DOI] [PubMed] [Google Scholar]
- Scholte HS, Ghebreab S, Waldorp L, Smeulders AW, and Lamme VA (2009). Brain responses strongly correlate with Weibull image statistics when processing natural images. J. Vis 9, 29.1–29.15. [DOI] [PubMed] [Google Scholar]
- Sigler A, Oh WC, Imig C, Altas B, Kawabe H, Cooper BH, Kwon HB, Rhee JS, and Brose N (2017). Formation and Maintenance of Functional Spines in the Absence of Presynaptic Glutamate Release. Neuron 94, 304–311.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit-Rigter L, Rajendran R, Silva CA, Spierenburg L, Groeneweg F, Ruimschotel EM, van Versendaal D, van der Togt C, Eysel UT, Heimel JA, et al. (2016). Mitochondrial Dynamics in Visual Cortex Are Limited In Vivo and Not Affected by Axonal Structural Plasticity. Curr. Biol 26, 2609–2616. [DOI] [PubMed] [Google Scholar]
- Smith HL, Bourne JN, Cao G, Chirillo MA, Ostroff LE, Watson DJ, and Harris KM (2016). Mitochondrial support of persistent presynaptic vesicle mobilization with age-dependent synaptic growth after LTP. eLife 5, e15275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strange BA, Witter MP, Lein ES, and Moser EI (2014). Functional organization of the hippocampal longitudinal axis. Nat. Rev. Neurosci 15, 655–669. [DOI] [PubMed] [Google Scholar]
- Südhof TC (2018). Towards an Understanding of Synapse Formation. Neuron 100, 276–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svara FN, Kornfeld J, Denk W, and Bollmann JH (2018). Volume EM Reconstruction of Spinal Cord Reveals Wiring Specificity in Speed-Related Motor Circuits. Cell Rep 23, 2942–2954. [DOI] [PubMed] [Google Scholar]
- Tom Dieck S, Hanus C, and Schuman EM (2014). SnapShot: local protein translation in dendrites. Neuron 81, 958–958.e1. [DOI] [PubMed] [Google Scholar]
- Tonegawa S, Morrissey MD, and Kitamura T (2018). The role of engram cells in the systems consolidation of memory. Nat. Rev. Neurosci 19, 485–498. [DOI] [PubMed] [Google Scholar]
- Trachtenberg JT, Chen BE, Knott GW, Feng G, Sanes JR, Welker E, and Svoboda K (2002). Long-term in vivo imaging of experience-dependent synaptic plasticity in adult cortex. Nature 420, 788–794. [DOI] [PubMed] [Google Scholar]
- Turrigiano GG, Leslie KR, Desai NS, Rutherford LC, and Nelson SB (1998). Activity-dependent scaling of quantal amplitude in neocortical neurons. Nature 391, 892–896. [DOI] [PubMed] [Google Scholar]
- van Praag H, Kempermann G, and Gage FH (2000). Neural consequences of environmental enrichment. Nat. Rev. Neurosci 1, 191–198. [DOI] [PubMed] [Google Scholar]
- Varoqueaux F, Sigler A, Rhee JS, Brose N, Enk C, Reim K, and Rosenmund C (2002). Total arrest of spontaneous and evoked synaptic transmission but normal synaptogenesis in the absence of Munc13-mediated vesicle priming. Proc. Natl. Acad. Sci. USA 99, 9037–9042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhage M, Maia AS, Plomp JJ, Brussaard AB, Heeroma JH, Vermeer H, Toonen RF, Hammer RE, van den Berg TK, Missler M, et al. (2000). Synaptic assembly of the brain in the absence of neurotransmitter secretion. Science 287, 864–869. [DOI] [PubMed] [Google Scholar]
- Vos M, Lauwers E, and Verstreken P (2010). Synaptic mitochondria in synaptic transmission and organization of vesicle pools in health and disease. Front. Synaptic Neurosci 2, 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson DJ, Ostroff L, Cao G, Parker PH, Smith H, and Harris KM (2016). LTP enhances synaptogenesis in the developing hippocampus. Hippocampus 26, 560–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap EL, and Greenberg ME (2018). Activity-Regulated Transcription: Bridging the Gap between Neural Activity and Behavior. Neuron 100, 330–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasumatsu N, Matsuzaki M, Miyazaki T, Noguchi J, and Kasai H (2008). Principles of long-term dynamics of dendritic spines. J. Neurosci 28, 13592–13608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuste R, and Bonhoeffer T (2004). Genesis of dendritic spines: insights from ultrastructural and imaging studies. Nat. Rev. Neurosci 5, 24–34. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Narayan S, Geiman E, Lanuza GM, Velasquez T, Shanks B, Akay T, Dyck J, Pearson K, Gosgnach S, et al. (2008). V3 spinal neurons establish a robust and balanced locomotor rhythm during walking. Neuron 60, 84–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Lee WA, Paul DL, and Ginty DD (2019). Multiplexed peroxidase-based electron microscopy labeling enables simultaneous visualization of multiple cell types. Nat. Neurosci 22, 828–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Homma KJ, and Poo MM (2004). Shrinkage of dendritic spines associated with long-term depression of hippocampal synapses. Neuron 44, 749–757. [DOI] [PubMed] [Google Scholar]
- Ziv NE, and Brenner N (2018). Synaptic Tenacity or Lack Thereof: Spontaneous Remodeling of Synapses. Trends Neurosci 41, 89–99. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw values used for quantifications presented in the manuscript are available in the enclosed Data S1. This file contains separate spreadsheets (split by types of analysis) with synapses classified according to spine type, measured parameters, sample sizes, brain IDs and statistics.
Computer scripts are available from authors upon request. All raw SBEM volumes will be made freely available to the research community after publication through a web-based resource.


