ABSTRACT
Cyanobacteria are ubiquitous microorganisms with crucial ecosystem functions, yet most knowledge of their biology relates to aquatic taxa. We have constructed metagenomes for 50 taxonomically well-characterized terrestrial cyanobacterial cultures. These data will support phylogenomic studies of evolutionary relationships and gene content among these unique algae and their aquatic relatives.
ANNOUNCEMENT
Cyanobacteria, or blue-green algae, are found in nearly all aquatic and terrestrial habitats exposed to sunlight, ranging from marine systems, freshwater bodies, and thermal springs to soils and rock surfaces. They represent important ecosystem components, which perform crucial roles in biogeochemical cycling. However, knowledge of their taxonomy, phylogenomics, and physiology has been largely based on aquatic taxa. In the past 20 years, our team has discovered and described terrestrial cyanobacteria collected from extreme environments, including desert soils, ephemerally wet rock walls, and tropical damp cave walls, greatly enriching our understanding of cyanobacterial systematics (1–15). Reference strains of genus and species types are maintained in two algae culture collections at John Carroll University (JCU) and New Mexico State University (NMSU). The cultures are nonaxenic and represent unialgal polycultures containing primarily cyanobacteria and heterotrophic microbial associates. We used shotgun metagenomics of selected strains to support future investigation of the phylogenomic relationships of these unique algae.
Fifty unialgal polycultures of cyanobacterial reference strains from the JCU and NMSU culture collections were selected, representing species taxonomically evaluated within the last 2 decades (Fig. 1). Detailed protocols for biomass growth, tissue harvesting, and DNA extraction can be found at protocols.io (dx.doi.org/10.17504/protocols.io.brg4m3yw). Briefly, biomass of each taxon was grown in liquid Z8 medium (16) and harvested after several weeks to months depending on the growth rate. Harvesting included cleaning, biomass concentration, biomass flash-freezing in liquid nitrogen, and storage at −80°C. We extracted DNA using the Qiagen DNeasy PowerLyzer microbial kit. The extraction procedure included an initial bead-beating step in a Precellys homogenizer for 45 s at 5,000 rpm, repeating four times. Samples were then processed following the manufacturer’s protocol for the kit and stored at −20°C until mailing to the Joint Genome Institute (JGI).
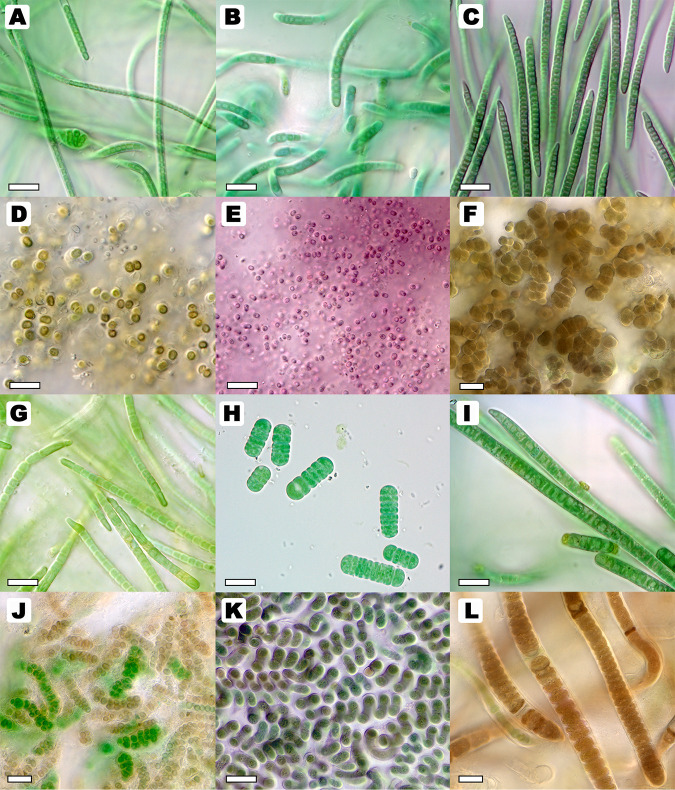
FIG 1.
Light micrographs of representative cyanobacterial species maintained in unialgal cultures and studied in this project. Images represent cell morphologies in the stationary phase of the cyanobacterial life cycle. Wet mounts of cyanobacterial biomass were observed using a Zeiss AxioImager.A2 microscope equipped with Nomarski differential interference contrast optics and a Zeiss Axiocam 305 color camera at ×400 or ×1,000 magnification. (A) Nodosilinea sp. strain WJT8-NPBG4; (B) Myxacorys chilensis ATA2-1-KO14; (C) Trichocoleus desertorum ATA4-8-CV12; (D) Cyanosarcina radialis HA8281-LM2; (E) Aphanocapsa lilacina HA4352-LM1; (F) Pleurocapsa minor HA4340-MV1; (G) Kastovskya adunca ATA6-11-RM4; (H) Hormoscilla sp. strain CMT-3BRIN-NPC48; (I) Microcoleus vaginatus WJT46-NPBG5; (J) Mojavia pulchra JT2-VF2; (K) Spirirestis rafaelensis WJT71-NPBG6; (L) Brasilonema octagenarum HA4186-MV1. Bars, 10 μm (A–E, G–I, and K) and 20 μm (F, J, and L).
Plate-based DNA library preparation for Illumina sequencing was performed using KAPA Biosystems high-throughput library preparation kit p/n KK8235 on a PerkinElmer Sciclone next-generation sequencing (NGS) robotic liquid handling system. Then, 200 ng of sample DNA was sheared to 300 bp using a Covaris LE220 focused ultrasonicator. The sheared DNA fragments were size selected by double solid-phase reversible immobilization (SPRI), and then the selected fragments were end repaired, A tailed, and ligated with Illumina-compatible sequencing adaptors from IDT containing a unique molecular index barcode for each sample library. The prepared libraries were quantified with a KAPA Biosystems quantitative PCR (qPCR) kit on a Roche LightCycler 480 real-time PCR instrument. Genomic libraries were sequenced with a NovaSeq instrument (Illumina, San Diego, CA) using NovaSeq XP V1 reagent kits and an S4 flow cell following a 2 × 150-bp indexed run recipe. Demultiplexed reads were processed with BBDuk v38.87 (17) to remove contaminants, trim adapter sequence and “G” homopolymers ≥5 in size at the ends, quality trim reads, and remove reads with ≥4 “N” bases, with an average quality score of <3, or with a length of <51 bp. After processing, 2,021,674,638 reads remained (per sample range, 24,625,170 to 59,843,005; mean ± standard deviation, 40,433,493 ± 8,547,361).
Cleaned reads were assembled with metaSPAdes v3.13.0 (18) (-meta option) into contigs with parameters for a minimum contig length of 2 kb (-m 2000 option) and default kmer options for paired 150-bp reads. Across all assemblies, 745,331 metagenomic contigs were retained after quality control using AAFTF vecscreen v0.2.3 (19) (range of assembly GC content, 40.09% to 66.57%; GC content mean ± standard deviation, 59.94% ± 4.96%; range of assembly N50, 10,169 to 456,261 bp; N50 mean ± standard deviation, 133,466 ± 100,540 bp).
Data availability.
All reads and assemblies were deposited under the NCBI accession numbers listed in Table 1. We also provided accession identifiers linking to JGI projects associated with each sample, including available IMG (20) data (Table 1).
TABLE 1.
Metadata, accession numbers, and metagenome statistics for the 50 cyanobacterial cultures investigated in this studya
| Species | Available at culture collections | Strain ID | Alternative strain ID | BioProject no. | SRA run no. | JGI IMGb | No. of reads | No. of contigs | N50 (bp) | GC content (%) | Habitat | Location |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Aetokthonos hydrillicola | NMSU, JCU, CCALA | B3-Florida | CCALA 1050 | PRJNA617093 | SRR12347192 | 3300036376 | 56,817,878 | 54,864 | 184,060 | 63.45 | Aquatic epiphyte | South Carolina, USA |
| Aphanocapsa lilacina | NMSU, JCU | HA4352-LM1 | PRJNA677053 | SRR13206895 | 3300039303 | 58,991,043 | 38,358 | 112,017 | 63.64 | Cave rock wall | Hawaii, USA | |
| Aphanocapsa sp. | NMSU, JCU | GSE-SYN-MK-11-07L | PRJNA621664 | SRR11678123 | 3300035009 | 26,644,370 | 1,052 | 87,507 | 48.55 | Desert wet wall | Utah, USA | |
| Aphanothece saxicola | NMSU, JCU | GSE-SYN-MK-01-06B | PRJNA621658 | SRR11676827 | 3300035003 | 50,225,611 | 8,874 | 272,324 | 66.57 | Desert wet wall | Utah, USA | |
| Aphanothece sp. | NMSU, JCU | CMT-3BRIN-NPC111 | PRJNA617094 | SRR12347633 | 3300036407 | 41,058,001 | 13,407 | 164,105 | 59.69 | Desert soil | California, USA | |
| Brasilonema angustatum | NMSU, JCU | HA4187-MV1 | PRJNA617095 | SRR12347719 | 3300036389 | 51,374,669 | 22,657 | 78,161 | 54.18 | Tropical soil | Hawaii, USA | |
| Brasilonema octagenarum | NMSU, JCU | HA4186-MV1 | PRJNA621669 | SRR11678155 | 3300035014 | 41,935,410 | 2,727 | 183,890 | 63.68 | On wood in tropics | Hawaii, USA | |
| Calothrix sp. | NMSU, JCU | FI2-JRJ7 | PRJNA617096 | SRR12347730 | 3300036390 | 26,844,447 | 1,488 | 168,980 | 56.22 | Desert soil | California, USA | |
| Chroococcus sp. | NMSU, JCU | CMT-3BRIN-NPC107 | PRJNA653492 | SRR12688413 | 3300037833 | 47,424,890 | 59,802 | 35,531 | 63.26 | Desert soil | California, USA | |
| Cyanomargarita calcarea | NMSU, JCU | GSE-NOS-MK-12-04C | PRJNA621661 | SRR11676926 | 3300035006 | 30,710,984 | 5,791 | 264,901 | 58.92 | Desert wet wall | Utah, USA | |
| Cyanosarcina radialis | NMSU, JCU | HA8281-LM2 | PRJNA653493 | SRR12688412 | 3300037834 | 39,248,498 | 48,900 | 35,006 | 63.37 | Cave rock wall | Hawaii, USA | |
| Desmonostoc geniculatum | NMSU, JCU | HA4340-LM1 | PRJNA621655 | SRR11676647 | 3300035000 | 51,944,514 | 29,336 | 130,707 | 61.24 | Cave rock wall | Hawaii, USA | |
| Desmonostoc vinosum | NMSU, JCU | HA7617-LM4 | PRJNA650882 | SRR12951526 | 3300038554 | 36,690,552 | 6,487 | 166,368 | 60.96 | Cave rock wall | Hawaii, USA | |
| Drouetiella hepatica | NMSU, JCU | UHER 2000/2452 | PRJNA617097 | SRR12347860 | 3300036391 | 42,783,418 | 20,061 | 28,986 | 65.05 | Rock surface | Kosice, Slovakia | |
| Gloeocapsa sp. | NMSU, JCU | UFS-A4-WI-NPMV-4B04 | PRJNA617098 | SRR12347921 | 3300036443 | 29,499,512 | 17,169 | 32,766 | 60.53 | Desert soil | Utah, USA | |
| Goleter apudmare | NMSU, JCU, CCALA | HA4340-LM2 | CCALA 1075 | PRJNA621659 | SRR11676930 | 3300035004 | 36,243,261 | 714 | 190,367 | 59.38 | Cave rock wall | Hawaii, USA |
| Hassallia sp. | NMSU, JCU | WJT32-NPBG1 | PRJNA617099 | SRR12347922 | 3300036392 | 33,287,445 | 4,616 | 21,632 | 58.06 | Desert soil | California, USA | |
| Hormoscilla sp. | NMSU, JCU | CMT-3BRIN-NPC48 | PRJNA650881 | SRR12951525 | 3300038553 | 42,048,713 | 5,825 | 143,678 | 59.26 | Desert soil | California, USA | |
| Iphinoe sp. | NMSU, JCU | HA4291-MV1 | PRJNA617100 | SRR12349226 | 3300036393 | 31,352,639 | 32,008 | 107,621 | 62.85 | Tropical soil | Hawaii, USA | |
| Kaiparowitsia implicata | NMSU, JCU | GSE-PSE-MK54-09C | PRJNA650883 | SRR13242888 | 3300038555 | 43,375,246 | 4,576 | 456,261 | 63.35 | Desert wet wall | Utah, USA | |
| Kastovskya adunca | NMSU, JCU, CCALA | ATA6-11-RM4 | CCALA 1025 | PRJNA621676 | SRR11678224 | 3300034631 | 42,257,751 | 47,939 | 14,667 | 63.07 | Desert soil | Atacama, Chile |
| Komarekiella atlantica | NMSU, JCU | HA4396-MV6 | PRJNA650825 | SRR12951531 | 3300038556 | 39,748,068 | 24,886 | 121,785 | 64.40 | Tropical vernal pool | Hawaii, USA | |
| Lyngbya sp. | NMSU, JCU | HA4199-MV5 | PRJNA621654 | SRR11676618 | 3300034630 | 37,379,750 | 5,629 | 214,210 | 61.23 | Tropical soil | Hawaii, USA | |
| Microcoleus vaginatus | NMSU, JCU | WJT46-NPBG5 | PRJNA677056 | SRR13207170 | 3300039305 | 40,531,812 | 28,602 | 165,791 | 60.22 | Desert soil | California, USA | |
| Mojavia pulchra | NMSU, JCU, CCALA | JT2-VF2 | CCALA 691 | PRJNA650826 | SRR12951532 | 3300038557 | 54,924,618 | 10,021 | 208,763 | 62.75 | Desert soil | California, USA |
| Myxacorys californica | NMSU, JCU, UTEX | WJT36-NPBG1 | UTEX B 3157 | PRJNA621674 | SRR11678221 | 3300035019 | 32,998,795 | 9,403 | 81,746 | 64.17 | Desert soil | California, USA |
| Myxacorys chilensis | NMSU, JCU, UTEX | ATA2-1-KO14 | UTEX B 3158 | PRJNA621667 | SRR11678144 | 3300035012 | 46,028,284 | 19,551 | 69,559 | 60.50 | Desert soil | Coquimbo, Chile |
| Nodosilinea sp. | NMSU, JCU | WJT8-NPBG4 | PRJNA621656 | SRR11676645 | 3300035001 | 36,650,447 | 2,944 | 173,054 | 65.12 | Desert soil | California, USA | |
| Nostoc desertorum | NMSU, JCU, CCALA | CM1-VF14 | CCALA 693 | PRJNA617101 | SRR12349250 | 3300036444 | 55,466,988 | 7,058 | 211,685 | 62.42 | Desert soil | California, USA |
| Nostoc indistinguendum | NMSU, JCU, CCALA | CM1-VF10 | CCALA 692 | PRJNA621666 | SRR11678133 | 3300035011 | 34,734,084 | 13,226 | 98,409 | 54.74 | Desert soil | California, USA |
| Oscillatoria princeps | NMSU | RMCB-10 | PRJNA621670 | SRR11678154 | 3300035015 | 35,756,581 | 29,084 | 28,454 | 61.51 | Surface water | Lower Austria, Austria | |
| Oscillatoria tanganyikae | NMSU, JCU | FI6-MK23 | PRJNA621665 | SRR11678132 | 3300035010 | 41,220,548 | 7,027 | 306,031 | 59.66 | Desert soil | California, USA | |
| Pegethrix bostrychoides | NMSU, JCU | GSE-TBD4-15B | PRJNA617102 | SRR12349533 | 3300036394 | 34,506,500 | 2,385 | 397,454 | 61.39% | Desert wet wall | Utah, USA | |
| Pelatocladus maniniholoensis | NMSU, JCU | HA4357-MV3 | PRJNA653494 | SRR12687981 | 3300037800 | 30,851,680 | 553 | 52,911 | 40.09% | Cave rock wall | Hawaii, USA | |
| Plectolyngbya sp. | NMSU, JCU | WJT66-NPBG17 | PRJNA617103 | SRR12349534 | 3300036395 | 24,625,170 | 9,370 | 10,169 | 53.14 | Desert soil | California, USA | |
| Pleurocapsa minor | NMSU, JCU | GSE-CHR-MK-17-07R | PRJNA677054 | SRR13207074 | 3300039304 | 44,764,195 | 25,094 | 40,205 | 64.01 | Desert wet wall | Utah, USA | |
| Pleurocapsa minor | NMSU, JCU | HA4230-MV1 | PRJNA617104 | SRR12349680 | 3300036396 | 59,843,005 | 708 | 316,355 | 63.57 | Tropical rock wall | Hawaii, USA | |
| Scytolyngbya sp. | NMSU, JCU | HA4215-MV1 | PRJNA617105 | SRR12349737 | 3300036104 | 33,289,893 | 8,107 | 243,918 | 60.55 | Freshwater stream | Hawaii, USA | |
| Scytonema hyalinum | NMSU, JCU | WJT4-NPBG1 | PRJNA617106 | SRR12349874 | 3300036520 | 40,826,558 | 35,954 | 57,285 | 64.29 | Desert soil | California, USA | |
| Scytonematopsis contorta | NMSU, JCU, UTEX | HA4267-MV1 | UTEX 2964 | PRJNA653495 | SRR12688486 | 3300037859 | 41,139,075 | 2,560 | 124,805 | 50.66 | Tropical vernal pool | Hawaii, USA |
| Spirirestis rafaelensis | NMSU, JCU | WJT71-NPBG6 | PRJNA683097 | SRR13242887 | 3300042538 | 30,810,560 | 12,769 | 49,002 | 61.71 | Desert soil | California, USA | |
| Stenomitos rutilans | NMSU, JCU | HA7619-LM2 | PRJNA617107 | SRR12350322 | 3300036446 | 39,342,385 | 29,421 | 125,980 | 59.27 | Cave rock wall | Hawaii, USA | |
| Symplocastrum torsivum | NMSU, JCU, CCALA, UTEX | CPER-KK1 | CCALA 1031 UTEX B 3163 |
PRJNA617108 | SRR12350505 | 3300036521 | 45,589,224 | 7,498 | 188,807 | 61.87 | Grassland soil | Colorado, USA |
| Tildeniella nuda | NMSU, JCU | ZEHNDER 1965/U140 | PRJNA621657 | SRR11676687 | 3300035002 | 43,732,034 | 26,865 | 163,389 | 64.07 | Temperate wet wall | Zurich, Switzerland | |
| Tildeniella torsiva | NMSU, JCU | UHER 1998/13D | PRJNA653496 | SRR12689282 | 3300037860 | 51,170,286 | 22,112 | 16,291 | 50.54 | Temperate wet wall | Kosice, Slovakia | |
| Timaviella obliquedivisa | NMSU, JCU | GSE-PSE-MK23-08B | PRJNA621673 | SRR11678220 | 3300035018 | 43,160,626 | 11,520 | 86,031 | 58.41 | Desert wet wall | Utah, USA | |
| Tolypothrix brevis | NMSU, JCU | GSE-NOS-MK-07-07A | PRJNA621662 | SRR11678125 | 3300035007 | 36,087,578 | 9,013 | 171,611 | 59.37 | Desert wet wall | Utah, USA | |
| Tolypothrix carrinoi | NMSU, JCU | HA7290-LM1 | PRJNA621663 | SRR11678124 | 3300035008 | 36,281,317 | 43,124 | 40,599 | 59.34 | Cave rock wall | Hawaii, USA | |
| Trichocoleus desertorum | NMSU, JCU | ATA4-8-CV12 | PRJNA621668 | SRR11678143 | 3300035013 | 37,070,905 | 5,401 | 91,542 | 60.17 | Desert soil | Atacama, Chile | |
| “Trichormus” sp. | NMSU, JCU | ATA11-4-KO1 | PRJNA621660 | SRR11676933 | 3300035005 | 32,384,820 | 832 | 238,030 | 52.69 | Desert soil | Tarapaca, Chile |
Cyanobacterial cultures are available upon request from the research culture collections of Jeffrey R. Johansen (johansen@jcu.edu) at John Carroll University (JCU) and Nicole Pietrasiak (npietras@nmsu.edu) at New Mexico State University (NMSU). The selected strains are also available publicly at the University of Texas Culture Collection of Algae (UTEX) and the Culture Collection of Autotrophic Organisms in Třeboň, Czech Republic (CCALA). Accordingly, alternative UTEX and CCALA strain identifiers are given in the table.
JGI IMG/M, Joint Genome Institute Integrated Microbial Genomes and Microbiomes database.
ACKNOWLEDGMENTS
We thank the National Park Service and Bureau of Land Management for permission to work within Joshua Tree National Park (yielding the WJT strains under permit JOTR-2006-SCI-0018), Mojave National Preserve (CMT strains, permits MOJA-2008-SCI-0024 and MOJA-2009-SCI-0039), and Grand Staircase-Escalante National Monument (GSE strains, permit UT-06-032-12-P).
We thank the National Science Foundation (NSF) for supporting J.R.J.’s sample collection of soils from the Atacama Desert, isolation of ATA strains, and sequencing of those strains (NSF grant numbers DEB-0842702 and DEB-841734, respectively), as well as the collection and floristic research of biological soil crusts from various North American desert locations (NSF grant number DEB-9870201). Any opinions, findings, conclusions, or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation. The California Desert Research Fund at The Community Foundation, Robert Lee Graduate Student Research Grant, and the Phycological Society Grants in Aid of Research fund awarded to Nicole Pietrasiak provided support for the sampling campaigns and subsequent cyanobacterial research associated with WJT and CMT strains.
We are grateful to John Carroll University for decades of support for J.R.J.’s cyanobacterial culture collection. We thank the numerous students and colleagues who over the past 30 years collaborated with J.R.J. and led to the isolation of many of the strains investigated in this study. Truc Mai assisted with initial biomass growth of cyanobacterial cultures at New Mexico State University. We also are grateful to Andrew Dominguez and Anthony Granite, who assisted R.D.W. with genomic DNA extractions at NMSU.
J.E.S. is a CIFAR fellow in the program Fungal Kingdom: Threats and Opportunities.
Software development was partially supported by NSF DEB-1441715 and USDA-NIFA Hatch project CA-R-PPA-5062-H. Data analyses performed at the High-Performance Computing Cluster at the University of California Riverside in the Institute of Integrative Genome Biology were supported by NSF grant DBI-1429826 and NIH grant S10-OD016290. The work conducted by the U.S. Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, is supported by the Office of Science of the U.S. Department of Energy under contract number DE-AC02-05CH11231.
Contributor Information
Nicole Pietrasiak, Email: npietras@nmsu.edu.
J. Cameron Thrash, University of Southern California
REFERENCES
- 1.Řeháková K, Johansen JR, Casamatta DA, Xuesong L, Vincent J. 2007. Morphological and molecular characterization of selected desert soil cyanobacteria: three species new to science including Mojavia pulchra gen. et sp. nov. Phycologia 46:481–502. doi: 10.2216/06-92.1. [DOI] [Google Scholar]
- 2.Mühlsteinová R, Johansen JR, Pietrasiak N, Martin MP, Osorio-Santos K, Warren SD. 2014. Polyphasic characterization of Trichocoleus desertorum sp. nov. (Pseudanabaenales, Cyanobacteria) from desert soils and phylogenetic placement of the genus Trichocoleus. Phytotaxa 163:241–261. doi: 10.11646/phytotaxa.163.5.1. [DOI] [Google Scholar]
- 3.Mühlsteinová R, Johansen JR, Pietrasiak N, Martin MP. 2014. Polyphasic characterization of Kastovskya adunca gen. nov. et comb. nov. (Cyanobacteria: Oscillatoriales), from desert soils of the Atacama Desert, Chile. Phytotaxa 163:216–228. doi: 10.11646/phytotaxa.163.4.2. [DOI] [Google Scholar]
- 4.Osorio-Santos K, Pietrasiak N, Bohunická M, Miscoe LH, Kováčik L, Martin MP, Johansen JR. 2014. Seven new species of Oculatella (Pseudanabaenales, Cyanobacteria): taxonomically recognizing cryptic diversification. Eur J Phycol 49:450–470. doi: 10.1080/09670262.2014.976843. [DOI] [Google Scholar]
- 5.Patzelt DJ, Hodač L, Friedl T, Pietrasiak N, Johansen JR. 2014. Biodiversity of soil cyanobacteria in the hyper-arid Atacama Desert, Chile. J Phycol 50:698–710. doi: 10.1111/jpy.12196. [DOI] [PubMed] [Google Scholar]
- 6.Pietrasiak N, Mühlsteinová R, Siegesmund MA, Johansen JR. 2014. Phylogenetic placement of Symplocastrum (Phormidiaceae, Cyanophyceae) with a new combination S. californicum and two new species: S. flechtnerae and S. torsivum. Phycologia 53:529–541. doi: 10.2216/14-029.1. [DOI] [Google Scholar]
- 7.Bohunická M, Pietrasiak N, Johansen JR, Gómez EB, Hauer T, Gaysina LA, Lukešová A. 2015. Roholtiella, gen. nov. (Nostocales, Cyanobacteria): a tapering and branching cyanobacteria of the family Nostocaceae. Phytotaxa 197:84–103. doi: 10.11646/phytotaxa.197.2.2. [DOI] [Google Scholar]
- 8.Miscoe LH, Johansen JR, Vaccarino MA, Pietrasiak N, Sherwood AR. 2016. Novel cyanobacteria from caves on Kauai, Hawaii, p 75–152. In Miscoe LH, Johansen JR, Kociolek JP, Lowe RL, Vaccarino MA, Pietrasiak N, Sherwood AR (ed), The diatom flora and cyanobacteria from caves on Kauai, Hawaii. Bibliotheca Phycologica, Stuttgart, Germany. [Google Scholar]
- 9.Johansen JR, Mareš J, Pietrasiak N, Bohunická M, Zima J Jr, Štenclová L, Hauer T. 2017. Highly divergent 16S rRNA sequences in ribosomal operons of Scytonema hyalinum (Cyanobacteria). PLoS One 12:e0186393. doi: 10.1371/journal.pone.0186393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shalygin S, Shalygina R, Johansen JR, Pietrasiak N, Berrendero Gómez E, Bohunická M, Mareš J, Sheil CA. 2017. Cyanomargarita gen. nov. (Nostocales, Cyanobacteria): convergent evolution resulting in a cryptic genus. J Phycol 53:762–777. doi: 10.1111/jpy.12542. [DOI] [PubMed] [Google Scholar]
- 11.Mai T, Johansen JR, Pietrasiak N, Bohunická M, Martin MP. 2018. Revision of the Synechococcales (Cyanobacteria) through recognition of four families including Oculatellaceae fam. nov. and Trichocoleaceae fam. nov. and six new genera containing 14 species. Phytotaxa 365:1–59. doi: 10.11646/phytotaxa.365.1.1. [DOI] [Google Scholar]
- 12.Pietrasiak N, Osorio-Santos K, Shalygin S, Martin MP, Johansen JR. 2019. When is a lineage a species? A case study in Myxacorys gen. nov. (Synechococcales: Cyanobacteria) with the description of two new species from the Americas. J Phycol 55:976–996. doi: 10.1111/jpy.12897. [DOI] [PubMed] [Google Scholar]
- 13.Shalygin S, Kavulic KJ, Pietrasiak N, Bohunická M, Vaccarino MA, Chesarino NM, Johansen JR. 2019. Neotypification of Pleurocapsa fuliginosa and epitypification of P. minor (Pleurocapsales): resolving a polyphyletic cyanobacterial genus. Phytotaxa 392:245–263. doi: 10.11646/phytotaxa.392.4.1. [DOI] [Google Scholar]
- 14.Mesfin M, Johansen JR, Pietrasiak N, Baldarelli LM. 2020. Nostoc oromo sp. nov. (Nostocales, Cyanophyceae) from Ethiopia: a new species based on morphological and molecular evidence. Phytotaxa 433:81–93. doi: 10.11646/phytotaxa.433.2.1. [DOI] [Google Scholar]
- 15.Pietrasiak N, Reeve S, Osorio-Santos K, Lipson D, Johansen JR. 2021. Trichotorquatus gen. nov.: a new genus of soil cyanobacteria discovered from American drylands. J Phycol. doi: 10.1111/jpy.13147. [DOI] [PubMed] [Google Scholar]
- 16.Carmichael WW. 1986. Isolation, culture and toxicity testing of toxic freshwater cyanobacteria (blue-green algae), p 1249–1262. In Shilov V (ed), Fundamental research in homogenous catalysis. Gordon & Breach, New York, NY. [Google Scholar]
- 17.Bushnell B. 2020. BBMap. http://sourceforge.net/projects/bbmap/.
- 18.Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. 2017. metaSPAdes: a new versatile metagenomic assembler. Genome Res 27:824–834. doi: 10.1101/gr.213959.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stajich JE, Palmer J. AAFTF: v0.2.3: automatic assembly for the Fungi. 2019. doi: 10.5281/zenodo.1620526. [DOI]
- 20.Chen I-MA, Chu K, Palaniappan K, Ratner A, Huang J, Huntemann M, Hajek P, Ritter S, Varghese N, Seshadri R, Roux S, Woyke T, Eloe-Fadrosh EA, Ivanova NN, Kyrpides NC. 2021. The IMG/M data management and analysis system v.6.0: new tools and advanced capabilities. Nucleic Acids Res 49:D751–D763. doi: 10.1093/nar/gkaa939. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All reads and assemblies were deposited under the NCBI accession numbers listed in Table 1. We also provided accession identifiers linking to JGI projects associated with each sample, including available IMG (20) data (Table 1).
TABLE 1.
Metadata, accession numbers, and metagenome statistics for the 50 cyanobacterial cultures investigated in this studya
| Species | Available at culture collections | Strain ID | Alternative strain ID | BioProject no. | SRA run no. | JGI IMGb | No. of reads | No. of contigs | N50 (bp) | GC content (%) | Habitat | Location |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Aetokthonos hydrillicola | NMSU, JCU, CCALA | B3-Florida | CCALA 1050 | PRJNA617093 | SRR12347192 | 3300036376 | 56,817,878 | 54,864 | 184,060 | 63.45 | Aquatic epiphyte | South Carolina, USA |
| Aphanocapsa lilacina | NMSU, JCU | HA4352-LM1 | PRJNA677053 | SRR13206895 | 3300039303 | 58,991,043 | 38,358 | 112,017 | 63.64 | Cave rock wall | Hawaii, USA | |
| Aphanocapsa sp. | NMSU, JCU | GSE-SYN-MK-11-07L | PRJNA621664 | SRR11678123 | 3300035009 | 26,644,370 | 1,052 | 87,507 | 48.55 | Desert wet wall | Utah, USA | |
| Aphanothece saxicola | NMSU, JCU | GSE-SYN-MK-01-06B | PRJNA621658 | SRR11676827 | 3300035003 | 50,225,611 | 8,874 | 272,324 | 66.57 | Desert wet wall | Utah, USA | |
| Aphanothece sp. | NMSU, JCU | CMT-3BRIN-NPC111 | PRJNA617094 | SRR12347633 | 3300036407 | 41,058,001 | 13,407 | 164,105 | 59.69 | Desert soil | California, USA | |
| Brasilonema angustatum | NMSU, JCU | HA4187-MV1 | PRJNA617095 | SRR12347719 | 3300036389 | 51,374,669 | 22,657 | 78,161 | 54.18 | Tropical soil | Hawaii, USA | |
| Brasilonema octagenarum | NMSU, JCU | HA4186-MV1 | PRJNA621669 | SRR11678155 | 3300035014 | 41,935,410 | 2,727 | 183,890 | 63.68 | On wood in tropics | Hawaii, USA | |
| Calothrix sp. | NMSU, JCU | FI2-JRJ7 | PRJNA617096 | SRR12347730 | 3300036390 | 26,844,447 | 1,488 | 168,980 | 56.22 | Desert soil | California, USA | |
| Chroococcus sp. | NMSU, JCU | CMT-3BRIN-NPC107 | PRJNA653492 | SRR12688413 | 3300037833 | 47,424,890 | 59,802 | 35,531 | 63.26 | Desert soil | California, USA | |
| Cyanomargarita calcarea | NMSU, JCU | GSE-NOS-MK-12-04C | PRJNA621661 | SRR11676926 | 3300035006 | 30,710,984 | 5,791 | 264,901 | 58.92 | Desert wet wall | Utah, USA | |
| Cyanosarcina radialis | NMSU, JCU | HA8281-LM2 | PRJNA653493 | SRR12688412 | 3300037834 | 39,248,498 | 48,900 | 35,006 | 63.37 | Cave rock wall | Hawaii, USA | |
| Desmonostoc geniculatum | NMSU, JCU | HA4340-LM1 | PRJNA621655 | SRR11676647 | 3300035000 | 51,944,514 | 29,336 | 130,707 | 61.24 | Cave rock wall | Hawaii, USA | |
| Desmonostoc vinosum | NMSU, JCU | HA7617-LM4 | PRJNA650882 | SRR12951526 | 3300038554 | 36,690,552 | 6,487 | 166,368 | 60.96 | Cave rock wall | Hawaii, USA | |
| Drouetiella hepatica | NMSU, JCU | UHER 2000/2452 | PRJNA617097 | SRR12347860 | 3300036391 | 42,783,418 | 20,061 | 28,986 | 65.05 | Rock surface | Kosice, Slovakia | |
| Gloeocapsa sp. | NMSU, JCU | UFS-A4-WI-NPMV-4B04 | PRJNA617098 | SRR12347921 | 3300036443 | 29,499,512 | 17,169 | 32,766 | 60.53 | Desert soil | Utah, USA | |
| Goleter apudmare | NMSU, JCU, CCALA | HA4340-LM2 | CCALA 1075 | PRJNA621659 | SRR11676930 | 3300035004 | 36,243,261 | 714 | 190,367 | 59.38 | Cave rock wall | Hawaii, USA |
| Hassallia sp. | NMSU, JCU | WJT32-NPBG1 | PRJNA617099 | SRR12347922 | 3300036392 | 33,287,445 | 4,616 | 21,632 | 58.06 | Desert soil | California, USA | |
| Hormoscilla sp. | NMSU, JCU | CMT-3BRIN-NPC48 | PRJNA650881 | SRR12951525 | 3300038553 | 42,048,713 | 5,825 | 143,678 | 59.26 | Desert soil | California, USA | |
| Iphinoe sp. | NMSU, JCU | HA4291-MV1 | PRJNA617100 | SRR12349226 | 3300036393 | 31,352,639 | 32,008 | 107,621 | 62.85 | Tropical soil | Hawaii, USA | |
| Kaiparowitsia implicata | NMSU, JCU | GSE-PSE-MK54-09C | PRJNA650883 | SRR13242888 | 3300038555 | 43,375,246 | 4,576 | 456,261 | 63.35 | Desert wet wall | Utah, USA | |
| Kastovskya adunca | NMSU, JCU, CCALA | ATA6-11-RM4 | CCALA 1025 | PRJNA621676 | SRR11678224 | 3300034631 | 42,257,751 | 47,939 | 14,667 | 63.07 | Desert soil | Atacama, Chile |
| Komarekiella atlantica | NMSU, JCU | HA4396-MV6 | PRJNA650825 | SRR12951531 | 3300038556 | 39,748,068 | 24,886 | 121,785 | 64.40 | Tropical vernal pool | Hawaii, USA | |
| Lyngbya sp. | NMSU, JCU | HA4199-MV5 | PRJNA621654 | SRR11676618 | 3300034630 | 37,379,750 | 5,629 | 214,210 | 61.23 | Tropical soil | Hawaii, USA | |
| Microcoleus vaginatus | NMSU, JCU | WJT46-NPBG5 | PRJNA677056 | SRR13207170 | 3300039305 | 40,531,812 | 28,602 | 165,791 | 60.22 | Desert soil | California, USA | |
| Mojavia pulchra | NMSU, JCU, CCALA | JT2-VF2 | CCALA 691 | PRJNA650826 | SRR12951532 | 3300038557 | 54,924,618 | 10,021 | 208,763 | 62.75 | Desert soil | California, USA |
| Myxacorys californica | NMSU, JCU, UTEX | WJT36-NPBG1 | UTEX B 3157 | PRJNA621674 | SRR11678221 | 3300035019 | 32,998,795 | 9,403 | 81,746 | 64.17 | Desert soil | California, USA |
| Myxacorys chilensis | NMSU, JCU, UTEX | ATA2-1-KO14 | UTEX B 3158 | PRJNA621667 | SRR11678144 | 3300035012 | 46,028,284 | 19,551 | 69,559 | 60.50 | Desert soil | Coquimbo, Chile |
| Nodosilinea sp. | NMSU, JCU | WJT8-NPBG4 | PRJNA621656 | SRR11676645 | 3300035001 | 36,650,447 | 2,944 | 173,054 | 65.12 | Desert soil | California, USA | |
| Nostoc desertorum | NMSU, JCU, CCALA | CM1-VF14 | CCALA 693 | PRJNA617101 | SRR12349250 | 3300036444 | 55,466,988 | 7,058 | 211,685 | 62.42 | Desert soil | California, USA |
| Nostoc indistinguendum | NMSU, JCU, CCALA | CM1-VF10 | CCALA 692 | PRJNA621666 | SRR11678133 | 3300035011 | 34,734,084 | 13,226 | 98,409 | 54.74 | Desert soil | California, USA |
| Oscillatoria princeps | NMSU | RMCB-10 | PRJNA621670 | SRR11678154 | 3300035015 | 35,756,581 | 29,084 | 28,454 | 61.51 | Surface water | Lower Austria, Austria | |
| Oscillatoria tanganyikae | NMSU, JCU | FI6-MK23 | PRJNA621665 | SRR11678132 | 3300035010 | 41,220,548 | 7,027 | 306,031 | 59.66 | Desert soil | California, USA | |
| Pegethrix bostrychoides | NMSU, JCU | GSE-TBD4-15B | PRJNA617102 | SRR12349533 | 3300036394 | 34,506,500 | 2,385 | 397,454 | 61.39% | Desert wet wall | Utah, USA | |
| Pelatocladus maniniholoensis | NMSU, JCU | HA4357-MV3 | PRJNA653494 | SRR12687981 | 3300037800 | 30,851,680 | 553 | 52,911 | 40.09% | Cave rock wall | Hawaii, USA | |
| Plectolyngbya sp. | NMSU, JCU | WJT66-NPBG17 | PRJNA617103 | SRR12349534 | 3300036395 | 24,625,170 | 9,370 | 10,169 | 53.14 | Desert soil | California, USA | |
| Pleurocapsa minor | NMSU, JCU | GSE-CHR-MK-17-07R | PRJNA677054 | SRR13207074 | 3300039304 | 44,764,195 | 25,094 | 40,205 | 64.01 | Desert wet wall | Utah, USA | |
| Pleurocapsa minor | NMSU, JCU | HA4230-MV1 | PRJNA617104 | SRR12349680 | 3300036396 | 59,843,005 | 708 | 316,355 | 63.57 | Tropical rock wall | Hawaii, USA | |
| Scytolyngbya sp. | NMSU, JCU | HA4215-MV1 | PRJNA617105 | SRR12349737 | 3300036104 | 33,289,893 | 8,107 | 243,918 | 60.55 | Freshwater stream | Hawaii, USA | |
| Scytonema hyalinum | NMSU, JCU | WJT4-NPBG1 | PRJNA617106 | SRR12349874 | 3300036520 | 40,826,558 | 35,954 | 57,285 | 64.29 | Desert soil | California, USA | |
| Scytonematopsis contorta | NMSU, JCU, UTEX | HA4267-MV1 | UTEX 2964 | PRJNA653495 | SRR12688486 | 3300037859 | 41,139,075 | 2,560 | 124,805 | 50.66 | Tropical vernal pool | Hawaii, USA |
| Spirirestis rafaelensis | NMSU, JCU | WJT71-NPBG6 | PRJNA683097 | SRR13242887 | 3300042538 | 30,810,560 | 12,769 | 49,002 | 61.71 | Desert soil | California, USA | |
| Stenomitos rutilans | NMSU, JCU | HA7619-LM2 | PRJNA617107 | SRR12350322 | 3300036446 | 39,342,385 | 29,421 | 125,980 | 59.27 | Cave rock wall | Hawaii, USA | |
| Symplocastrum torsivum | NMSU, JCU, CCALA, UTEX | CPER-KK1 | CCALA 1031 UTEX B 3163 |
PRJNA617108 | SRR12350505 | 3300036521 | 45,589,224 | 7,498 | 188,807 | 61.87 | Grassland soil | Colorado, USA |
| Tildeniella nuda | NMSU, JCU | ZEHNDER 1965/U140 | PRJNA621657 | SRR11676687 | 3300035002 | 43,732,034 | 26,865 | 163,389 | 64.07 | Temperate wet wall | Zurich, Switzerland | |
| Tildeniella torsiva | NMSU, JCU | UHER 1998/13D | PRJNA653496 | SRR12689282 | 3300037860 | 51,170,286 | 22,112 | 16,291 | 50.54 | Temperate wet wall | Kosice, Slovakia | |
| Timaviella obliquedivisa | NMSU, JCU | GSE-PSE-MK23-08B | PRJNA621673 | SRR11678220 | 3300035018 | 43,160,626 | 11,520 | 86,031 | 58.41 | Desert wet wall | Utah, USA | |
| Tolypothrix brevis | NMSU, JCU | GSE-NOS-MK-07-07A | PRJNA621662 | SRR11678125 | 3300035007 | 36,087,578 | 9,013 | 171,611 | 59.37 | Desert wet wall | Utah, USA | |
| Tolypothrix carrinoi | NMSU, JCU | HA7290-LM1 | PRJNA621663 | SRR11678124 | 3300035008 | 36,281,317 | 43,124 | 40,599 | 59.34 | Cave rock wall | Hawaii, USA | |
| Trichocoleus desertorum | NMSU, JCU | ATA4-8-CV12 | PRJNA621668 | SRR11678143 | 3300035013 | 37,070,905 | 5,401 | 91,542 | 60.17 | Desert soil | Atacama, Chile | |
| “Trichormus” sp. | NMSU, JCU | ATA11-4-KO1 | PRJNA621660 | SRR11676933 | 3300035005 | 32,384,820 | 832 | 238,030 | 52.69 | Desert soil | Tarapaca, Chile |
Cyanobacterial cultures are available upon request from the research culture collections of Jeffrey R. Johansen (johansen@jcu.edu) at John Carroll University (JCU) and Nicole Pietrasiak (npietras@nmsu.edu) at New Mexico State University (NMSU). The selected strains are also available publicly at the University of Texas Culture Collection of Algae (UTEX) and the Culture Collection of Autotrophic Organisms in Třeboň, Czech Republic (CCALA). Accordingly, alternative UTEX and CCALA strain identifiers are given in the table.
JGI IMG/M, Joint Genome Institute Integrated Microbial Genomes and Microbiomes database.