Abstract
Tissue biomarkers have been of increasing utility for scientific research, diagnosing disease, and treatment response prediction. There has been a steady shift away from qualitative assessment toward providing more quantitative scores for these biomarkers. The application of quantitative image analysis has thus become an indispensable tool for in-depth tissue biomarker interrogation in these contexts. This white paper reviews current technologies being employed for quantitative image analysis, their application and pitfalls, regulatory framework demands, and guidelines established for promoting their safe adoption in clinical practice.
Key Words: biomarkers, quantitative image analysis, whole-slide imaging
Biomarkers are commonly used in nonclinical and clinical research, as well as in clinical practice. The Food and Drug Administration (FDA) defines a biomarker as an “indicator of normal biological processes, pathogenic processes, or a response to an exposure or intervention.” There are a number of biomarker subtypes such as molecular, histologic, radiographic, and physiological. Histologic or tissue biomarker identification can range from routine histochemistry such as hematoxylin and eosin (H&E), to a nucleic acid (ie, DNA, RNA) and to protein detection. While traditional histochemical staining continues to be the most extensively used method to study tissues, immunohistochemistry (IHC) has become the most commonly used technique for specific detection of tissue biomarkers. IHC permits the detection of proteins (or antigens) in tissues and compared with in situ hybridization (ISH) that allows for nucleic acid detection, IHC is more cost-effective, has a faster turnaround time, and offers easier interpretation in routine pathology practice. The interpretation of tissue biomarkers using any of these staining techniques is typically performed by observation via light [bright-field or immunofluorescence (IF)] microscopy. However, modern pathology practice is moving toward a digital workflow, meaning pathologists can now perform their assessment of biomarkers on computer screens. In addition, digital pathology allows for image analysis tools to be used in tissue biomarker evaluation and quantification. These tools can aid pathologists in providing more quantitative, detailed, objective, and reproducible assessments of tissue biomarkers.1 While quantitative studies of tissue biomarkers are valuable in clinical practice, currently digital image analysis of tissue biomarkers is more widely used in research settings.
The purpose of this white paper by the Digital Pathology Association (DPA) is to provide an overview of the current status of quantitative image analysis (QIA) of tissue biomarkers including relevant technologies and their pitfalls, clinical and nonclinical applications, available guidelines, and regulatory issues.
TECHNOLOGIES FOR TISSUE BIOMARKER DETECTION
Histochemistry, ISH, and Other Tissue Biomarker Detection Technologies
The method used to stain tissues influences the type of image analysis that can be performed. The most common histochemical stain used for general morphologic evaluation of tissues is H&E. H&E staining is based on the differential uptake of stain depending on the pH of different cellular compartments. To evaluate specific tissue features, one may also utilize “special (biochemical or histochemical) stains,” such as Trichrome which highlights collagenous connective tissues, among others.2 Although H&E does not label proteins specifically, artificial intelligence (AI) approaches are being developed to predict biomarker expression [eg, programmed death-ligand 1 (PD-L1)] solely based on morphologic features derived from H&E-stained sections, without the need for biomarker-specific staining.3
However, if we must label tissues for specific protein biomarkers, the most common methodology is IHC staining. In IHC, antibodies are employed to detect specific proteins. After primary antibodies have bound to a specific protein (the biomarker of interest) in tissue sections, they are then detected by secondary antibodies to enable signal amplification and visualization. For IHC, this secondary antibody binding can be made visible employing enzymatic methods (eg, horseradish peroxidase or alkaline phosphatase) and chromogens which results in a signal that can be visualized via bright-field microscopy. Although many other chromogens exist, DAB (3,3′-diaminobenzidine) in combination with horseradish peroxidase-based staining is the most commonly used visualization system.
Biomarker detection can also be performed using IF, where the secondary antibody is bound to a fluorescent molecule (ie, fluorochrome) that emits light at a specific wavelength when excited by a laser. IF typically has a higher signal-to-noise ratio, as well as a broader and linear dynamic range than chromogenic IHC. These characteristics of IF, in turn, allow for a more detailed quantitative assessment of the fluorescent signal.4 In contrast, chromogenic IHC stain intensity rarely reflects quantitative protein expression as most immunostains are not validated against a protein standard. In fact, chromogenic IHC stain intensity dynamic range is lower than IF and also does not linearly correlate with protein expression. Therefore, if accurate measures of biomarker expression are required, IF should be considered. However, IF has drawbacks such as the requirement for specific technical expertise and costly laboratory equipment, more time-intensive slide preparation utilizing higher-priced reagents, and increased whole-slide image (WSI) scan times, thereby oftentimes constraining imaging and analysis to only selected high-power fields (as opposed to the entire WSI). Traditionally, IF can also be plagued by autofluorescence, though more recent histochemical and image analysis developments have resulted in improvements.5 In addition, depending upon the IF reagents used, signal strength may fade over time, whereas chromogenic IHC and histochemical stain signals will remain persistent for longer. Imaging may be performed on fluorescent microscopes that typically acquire a single field of view at a time or whole-slide scanners with fluorescent capabilities. When an IF slide is digitally captured, it reduces the impact of signal deterioration over time, so long as the acquisition was performed at a suitable magnification. However, the physical glass slides may need to still be retained, especially in a regulated clinical environment.
While many tissue biomarkers are proteins, other types exist. For example, DNA amplification or RNA expression may be utilized. These can be detected using ISH technology, which results in small dot-like staining of tissues, each representing a nucleic acid probe binding. ISH can be performed as a chromogenic (CISH) or fluorescent (FISH) stain.6,7 In addition, several emerging ex vivo visualization techniques are now available that may require less sample preparation time and can be nondestructive. These technologies include confocal microscopy, UV-based imaging such as microscopy with UV surface excitation, light-sheet microscopy, and optical coherence tomography.8 For example, microscopy with UV surface excitation imaging relies on illuminating unstained tissues with UV light at wavelengths <300 nm, which allows visualization of cellular properties in tissues with similar information content to H&E stains, but without the need for fixation and processing. Although image analysis exists for these applications, they are much less advanced compared with WSI.9–11
Multiplexing
In tissue biomarker studies, it is often necessary to identify several markers in or on the same cell. This is especially important when phenotyping immune cell populations and assessing spatial relationships among various cell types. The 2 main approaches employed for multiplexing are: (1) consecutive serial sections of single-plex IHC staining followed by digital image registration, alignment, and fusion into a single plane; and (2) multiplex staining to detect several biomarkers within a single tissue section. While the former approach uses more tissue, the latter can pose significant challenges in the development of appropriate staining protocols. In addition, it is important to recognize that when consecutive serial sections are digitally fused into one image, multiple labeling in the same area does not necessarily represent multiple biomarker expression within the exact same cell. Due to the thickness of each individual slide (3 to 5 µm), most cell types are rarely present in >1 to 2 serial sections.
Multiplex staining approaches are available for both bright-field (mIHC) and fluorescence (mIF) platforms. Chromogenic mIHC is usually performed as a low-plex option, whereas mIF can include many more markers (high plex). For mIHC, the main cause for limited multiplexing capability is the narrow range of available reagents and chromogens. At times, the available chromogen signals can be challenging to separate optically. This can be improved upon via narrow absorbance bands, matched illumination channels, and monochrome imaging. In contrast, mIF allows for the combination of a higher number of markers due to the ability to excite tagged molecules at different specific wavelengths. In general, mIF is limited to ∼5 markers due to the spectral overlap between different fluorescent molecules, but several methodologies have become available to overcome this limitation. For example, Opal technology uses special fluorophores that can be “unmixed” to deconvolve the overlapping spectra, allowing for up to 9 concurrent labeling colors. In contrast, technologies such as CODEX12 depend on sequential imaging and washing of the fluorophore tagged to a marker-specific barcode. In theory, this allows for an infinite number of co-labelings, albeit resulting in low staining intensity due to lack of signal amplification. Last, alternative technologies such as mass spectrometry (eg, CyTOF,13 MIBI14) are available, where markers are labeled with metals rather than fluorophores, and a mass spectrometry approach is used for detection that allows for 40 or more simultaneous biomarkers to be labeled. Depending upon the data acquired and limitations of data/image capturing devices, mIF data might only be collected from selected regions of interest (ROIs) instead of from the entire tissue section.
Image Processing
After staining, slides can be digitized at various magnifications to produce a WSI. The staining modality (chromogenic or fluorescent) dictates the light source. These WSI can be analyzed computationally to quantify the biomarker signal (so-called QIA). A QIA algorithm may automatically detect and analyze an entire stained slide or limit the analysis to specific ROIs. While those ROIs are often selected by a pathologist, depending upon the types of analysis, this manual selection can introduce significant bias into the data. A typical QIA algorithm used to quantify an immunostained biomarker may involve tissue and/or cellular classification, target stain detection, segmentation, and stain quantification. AI-based QIA approaches may include additional steps, such as feature extraction and pattern recognition.
Impact of Preanalytical Variables on Biomarker Staining
Every step along the way from removing the tissue in vivo to the final stained slide can impact the quality of staining and digitized WSI. Therefore, these so-called preanalytical variables may significantly affect the data generated from these slides. The preanalytical test phase starts at the simple step of cutting fresh tissue leading to tissue deformation and shrinkage. With the tissue removed, blood circulation is also severed, resulting in tissue ischemia. Some biomarkers are particularly sensitive to degradation during this ischemic time, and if the sample was chilled or not during this step.15,16 Among the preanalytical variables that can impact staining quality are tissue fixation, grossing, tissue processing, and embedding. Improper tissue handling and mechanical manipulation may result in crush artifacts. Similarly, inappropriate tissue fixation can impact staining results. In addition, prolonged formalin-fixation can result in excessive protein cross-linking that may render the biomarker unavailable for antibody binding and therefore visualization. A full discussion of all preanalytical variables is beyond the scope of this manuscript, as only a few key aspects are discussed in more detail in the following sections. The interested reader is encouraged to consult other resources.17
Staining Consistency
Variation in standard H&E and IHC staining intensity can exist among different instruments, reagents, staining runs and laboratories, and can result in significantly discordant results. Standardization of laboratory workflow, staining protocol optimization, and clinical validation are necessary to maintain staining consistency. Very faint staining or excessive background staining may make it more difficult for scanners to automatically identify tissue on a slide. Optimizing H&E staining has been shown to result in better digital image quality18 and similarly, we would expect that controlling variability in IHC staining quality is an essential factor in generating accurate and reproducible quantification of biomarkers using imaging analysis. Recently, fully automated QIA software with predefined “turn-key algorithms” can allow simultaneous validation of staining area and staining intensity during the quantification process.19,20 However, to maintain the reproducibility of biomarker quantification using QIA, the correlation between algorithm and pathologist is also necessary to control for H&E and IHC staining intensity. Clear descriptions of such correlation data must be provided during quality control measures and validation steps where image analysis is used. In addition, to avoid staining inconsistency it is recommended that samples from an analysis cohort are fixed and processed in the same way, the same staining protocol is used, and an identical antibody batch is applied to all samples.21
Tissue Thickness
Many predictive clinical biomarkers, including PD-L1, have been FDA-approved with a specific tissue thickness to ensure appropriate stain performance. Staining intensity is most impacted by section thickness.22 Thicker sections are expected to stain with a higher intensity and vice versa. Sections that are too thin may result in false-negative results. In addition, uneven section thickness and tissue folds can cause scanner errors, resulting in WSIs that are partially out of focus. A study by Yagi and Gilberston23 indicated that thinner and more consistent tissue sectioning positively correlates with faster image acquisition and better scan quality. A WSI out of focus could also cause problems during image analysis, and although currently much of the WSI quality control is manual, some researchers are integrating algorithms for automated detection, and exclusion or automated rescanning of out-of-focus tissue areas.24 In general, tissue thickness ranging from 3 to 5 μm for scanning single-plane images is recommended.
Coverslips and Pen Markings
Coverslip material can be broadly classified into glass and plastic. Plastic (or film) coverslips are commonly used in laboratories performing high volume scanning for primary diagnosis because these slides dry rapidly, which minimizes their processing delay. However, plastic coverslips may get scratched, warp, or more easily peel off slides compared with glass coverslips. Cleaning pen markings off plastic slides may turn the plastic opaque and hence interfere with image analysis. In contrast, a glass coverslip offers better resolution and is also less liable to scratch, thus offering better image quality. Further studies are needed to drive a recommendation about which materials are best to use for digital scanning. Despite the type, coverslips need to be appropriately placed to avoid the formation of air bubbles to prevent autofocusing problems during digital scanning. However, image analysis can also aid scientists in the evaluation of slide quality to detect artifacts such as tissue folds,25,26 color variation,26 and out-of-focus regions24 as helpful preprocessing steps for quantification of biomarker staining.
Another common interfering issue is the use of a marker pen on coverslips to point out areas of interest or separate pieces of tissue (eg, controls placed on a slide and separated by a line drawn across the slide). Pen marks may directly or indirectly impede QIA. Due to the high contrast of these pen markings compared with surrounding tissue, scanner algorithms may likely select these marks for inclusion in focusing. The pen markings can also impact the color of the pixels in proximity to tissue. Avoiding or manually wiping away pen markings, tweaking color threshold algorithms27, and preprocessing images with AI algorithms that correct or ignore these pen markings are potential approaches to minimize the impact on QIA.28,29
APPLICATIONS OF BIOMARKER IMAGING ANALYSIS
Nonclinical Research
An image analysis-based approach permits a quantitative metric to be applied to different biomarkers that can be used to make drug and project decisions; this includes safety, target expression/validation, understanding mechanisms of action, as well as pharmacodynamic (PD) and efficacy studies. Safety studies that require hundreds of specimens to be manually assessed by pathologists are extremely cumbersome, time-consuming, and expensive. QIA can aid scientists in this evaluation. Traditionally, safety studies evaluate changes based on histochemical stains (such as H&E and Masson trichrome), and several studies have demonstrated the feasibility of automated scoring in murine studies of liver fibrosis,30,31 quantification of hepatic lipid droplets and steatosis,32,33 heart ischemic injury,34 lung fibrosis,35,36 kidney injury,37 and pancreatic toxicity.38 Biomarker studies that rely on IHC and QIA have been successfully deployed, for example, to quantify proliferative Ki-67 cells in rodent mammary glands39 and endometrium40 to study reproductive toxicity. Other studies have similarly utilized image analysis to examine processes relevant to several pathologies such as caspases in cell death,41 T-cell and B-cell markers in inflammation,42 and collagen deposition related to fibrosis.43
For targeted therapies, image analysis can help measure a threshold-based metric for therapeutic efficacy as well as confirm target inhibition. For example, in antibody-drug conjugate programs, target expression is critical for the drug payload to be delivered to the appropriate target-expressing (tumor) cells.44,45 A quantitative approach can help the research team understand if there is a cutoff or certain expression level that results in the drug getting to the target population which then leads to the appropriate biological effects which include pathway inhibition and tumor cell death. Theoretically, this cutoff assessment could be translated to a patient selection biomarker in a clinical study. A biopsy can be collected, an IHC assay protocol performed, and QIA utilized to produce a metric for selection. If the patient expressed the biomarker above the determined cutoff, they would be enrolled in the study. Target expression biomarkers and a quantitative approach can also benefit other aspects of drug discovery. For example, a variety of tumor models can be analyzed to rank order target expression to identify negative, low, medium, and high expression models.46 These “scoring” models can be used to explore PD-efficacy relationships.
PD biomarkers are also utilized to confirm the mechanism of action and drive lead optimization efforts. Developing a set of pathway biomarkers allows a better understanding of how target inhibition may lead to the appropriate biological pathway effects. PD biomarkers can be considered direct, indirect, or terminal indicators of outcome. Direct target engagement markers are those proteins targeted by the compound or possibly the drug itself, presuming the drug has an IHC-compatible aspect. As the name implies, direct biomarkers are preferred to confirm target engagement, while indirect markers provide an understanding of downstream biological pathway effects. Last terminal outcome markers provide information on the amount of cell death or other outcomes resulting from the drug’s target inhibition. Digital pathology can readily support studies of all direct, indirect, and terminal outcome biomarkers provided that there is a method or technology to obtain a digitized image from the specimen.21 Several studies have shown QIA effectiveness to measure downstream biomarkers to assess the effects of certain drugs in pathways like DNA damage repair,47 and posttranslational modifications such as neddylation48 and sumoylation.49
In addition to understanding the mechanism of action, image analysis of PD biomarkers provides a quantitative metric that can correlate with the potency of compounds and therefore be used to support medicinal chemistry efforts to improve potency in compound screening campaigns. If the PD effect is related to the drug effect, then increasing potency should correlate with the increasing PD effect. This PD effect can also be correlated with efficacy to develop a PD-efficacy relationship. Multiple digital pathology approaches have been applied successfully to study preclinical efficacy in cancer research, inflammation, fibrosis, and neurosciences. In neuroscience, for example, image analysis is applied to quantify how drugs affect different brain cell populations such as astrocytes and microglial cells,50 but also to measure the effect on disease features such as amyloid plaques in Alzheimer disease.51 Inflammation and immune cell markers are unquestionably among the most useful biomarkers of efficacy and many researchers have developed image analysis algorithms to study immune infiltration in mouse models of colitis,42 asthma,52 and cancer.53
In oncology, some studies also aim to correlate in vivo imaging [positron emission tomography (PET), magnetic resonance imaging (MRI)] to QIA of histologic sections. Syed and colleagues studied intratumoral heterogeneity in a murine model of human epidermal growth factor receptor 2 (HER2)-positive breast cancer treated with trastuzumab. Since different rates of proliferation and metabolic activity leads to varied regions of cellular density, these investigators wanted to correlate PET and MRI studies to image analysis of histologic sections. They used H&E images, Ki-67 and CD31 IHC to assess cellular density in histologic sections and compared these results to PET/MRI findings. This particular study showed that quantitative data from in vivo imaging is consistent with data derived from quantitative studies in histologic sections to study intratumoral heterogeneity.54
As a result of the exploration of PD and efficacy in preclinical studies, PD-efficacy relationships can be modeled in translational research. These models can help guide several important aspects of a clinical biomarker assay, from estimating what clinical dose will result in PD effects to defining the ideal sampling time point for biopsy collection.
Early Phase Clinical Research
The utility of digital pathology is well demonstrated in the study of biomarkers for clinical research, especially in early clinical development. In a study of metastatic renal cell carcinoma, investigators utilized image analysis to quantify B7-H1 (PD-L1) and B7-H3 and correlated the results with overall survival and cancer-specific survival.55 Due to the importance of Ki-67 in cancer diagnosis and prognosis, there are multiple studies that rely on this biomarker in clinical studies of cancer therapies. A recent study evaluated 3 different image analysis platforms to quantify the Ki-67 index and compared results with the outcome (cancer-specific survival, recurrence-free survival) in 2 breast cancer patient cohorts.56 They used the same training set for segmentation in HALO (Indica Labs), QuantCenter (3D Histech), and QuPath (open source), and subsequently quantified Ki-67 cells separately in the tumor, stroma, and the associated immune component. When Ki-67 was compared against outcome the researchers found no differences within and between these different platforms. Of note, they also found excellent interoperator reproducibility. Apart from Ki-67, other biomarkers that have been investigated using automated scoring in breast cancer include estrogen receptor (ER), progesterone receptor (PR), and HER2.57,58 While digital pathology has been applied mostly in the oncology field, a few other studies have demonstrated the efficacy of using image analysis to measure surrogate biomarkers59,60 and target/pathway validation.61–63
Similar to preclinical studies in murine models, tissue classification based on the identification of morphologically distinct features is also an important tool to examine and score patient samples. There are abundant studies reporting automated histologic assessment of liver fibrosis,64 kidney injury,65 tumor/stroma classification,66,67 colon architecture alterations,68 and immune infiltrate compartment.69 In addition, lymphocytic immune infiltrates have been broadly studied as a promising biomarker of drug response and prognosis in breast,70 cervical/uterine, prostate, lung, colon, pancreas, stomach, bladder, rectum, and skin cancer.71,72
Late Phase Clinical Research
Late clinical development traditionally represents phase 3 and 4 clinical trials. Phase 3 clinical trials are typically pivotal studies designed to assess the effectiveness/efficacy of a drug in large-scale randomized controlled settings. Phase 4 studies are typically postmarket studies designed to monitor drug safety over time in a “surveillance” manner, often referred to as pharmacovigilance.
The primary use of biomarkers in late-phase clinical trial testing is for prospective patient selection or stratification used in primary or secondary endpoint analysis. Biomarkers can also be used as direct endpoints that serve as “surrogates” for clinical outcome, understanding mechanisms of action or underlying biology of patient resistance/response, and/or general biology related to disease mechanisms, as well as more advanced hypothesis generation or cutpoint selection of predictive biomarkers. Such use cases have ultimate utility for designing subsequent trials and/or exploring novel targets and mechanisms for discovery.
It has been proposed that WSI can be useful in late clinical trials in the setting of central pathology review for the purposes of standardizing methods and efficacies of remote review.73,74 More recently, digital pathology-based biomarker evaluation is finding growing use in the retrospective exploratory analysis of immune-oncology drugs in late phase clinical trials for understanding the mechanism of action or PD effects.75–77 However, to date, there is little evidence of the use of digital pathology–based biomarkers to directly increase the technical success of late-stage drug trials in terms of prospectively defined patient selection or stratification. This is in stark contrast to standard IHC, genomic, blood-based tests, or noninvasive imaging tests which have found widespread use in late-stage clinical trials for patient selection, stratification, or as direct endpoints.78–81
The advantages of using digital pathology/AI-based methods are clear in terms of increasing intrareader reproducibility, and perhaps even interreader reproducibility since WSI is employed as opposed to just using ROI-based methods.82 It has been demonstrated in several studies that intrareader reproducibility of scoring PD-L1 of immune cells is poor83,84 and it is likely that digital pathology–based methods can have a positive impact on improving the repeatability of scoring. However, despite poor intrareader variability, PD-L1 IHC testing has reached market approval as a companion and complementary diagnostic test for both tumor cell scoring as well as combined tumor and immune cell scoring using manual methods.85 Outside of PD-L1 testing, it is unclear if this trend will continue. Other studies have shown that emerging immune cell markers may be relevant for predicting response to emerging second-generation checkpoint therapy. For example, anti-LAG3 in combination with anti-PD1 may be effective in late-stage melanoma patients who express high LAG3.86 It is unclear if scoring novel markers such as LAG3 is more or less challenging than PD-L1, and whether such tests will require digital pathology-based approaches.
Diagnostic Clinical Testing
The most frequent clinical applications of QIA include scoring of IHC stains for ER, PR, and HER2 as well as automated HER2 FISH analysis in breast cancer as prognostic and predictive biomarkers.87–96 Other applications include QIA of IHC for Ki-67, PD-L1, and multiplexed IHCs (eg, CD3 and CD8).97–101 The key to successfully incorporating QIA into clinical practice includes: (i) preanalytic elements such as optimizing glass slide quality (eg, tissue sectioning and staining) and WSI quality (eg, scanning and viewing); (ii) analytic elements such as ROI selection and algorithm selection; and (iii) postanalytic elements such as pathologists’ expertise in being able to correlate image analysis results with clinical information. All QIA tools must be validated before clinical implementation for patient care, followed by ongoing routine maintenance and development of quality control and quality assurance program as specified by a board of pathologists, such as the College of American Pathologists (CAP) in the United States.102
HER2
HER2 (ERBB2) gene amplification and/or protein overexpression occurs in ∼20% of breast cancers.103–106 HER2 status is usually assessed by IHC for HER2 protein expression and/or by FISH for HER2 gene amplification. HER2 IHCs are typically evaluated by pathologists manually in a semiquantitative manner and given a score from 0 to 3+ based on membranous staining of the HER2 protein. Despite the fact that the American Society of Clinical Oncology (ASCO) and the CAP published detailed guidelines on how to properly assess HER2 IHC, interobserver variability still occurs.107–109 Hence, QIA offers an objective and reproducible alternate scoring method to assess HER2 IHC.92–96 Studies have demonstrated that QIA can reduce HER2 IHC equivocal cases.92,93,110 The ASCO/CAP HER2 guideline has acknowledged QIA as a diagnostic modality for HER2 status assessment.107 Moreover, the CAP has created guidelines to safely facilitate the adoption of HER2 QIA into routine clinical pathology practice.102 The 510(k) FDA-cleared QIA algorithms available for HER2 IHC quantification include ACIS (Chromavision), Aperio XT (Leica), Ariol (Applied Imaging Corp.), Pathiam (Bioimagene), QCA (Cell Analysis Inc.), VIAS (Tripath Imaging), and Virtuoso (Roche Diagnostics/Ventana). Other products (eg, Visiopharm HER2-CONNECT algorithm) (Fig. 1) have also received CE marking. Visiopharm’s QIA algorithm for HER2 IHC has demonstrated an accurate assessment of both breast carcinoma and gastric/esophageal adenocarcinoma.92,111,112 A recent study demonstrated that employing QIA to score HER2 IHC had excellent concordance with pathologists’ scores and accurately discriminated between HER2 FISH positive and negative cases.113
FIGURE 1.
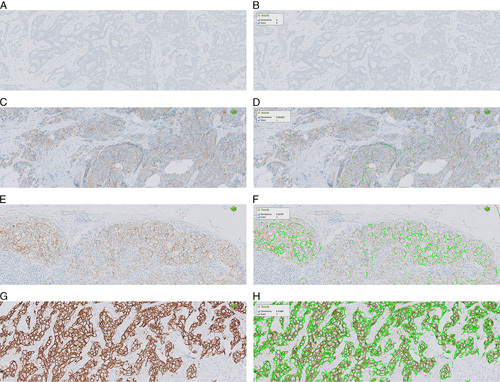
Example of HER2-CONNECT (Visiopharm) quantitative image analysis in breast carcinoma. The left panel of images (A, C, E, G) shows IHC staining and the right panel (B, D, F, H) shows HER2 membrane connectivity (green color line) detected by the algorithm. HER2 IHC 0 (A, B); HER2 IHC 1+ (C, D); HER2 IHC 2+ (E, F); HER2 IHC 3+ (G, H). HER2 indicates human epidermal growth factor receptor 2; IHC, immunohistochemistry.
FISH is also widely used for the determination of HER2 status in breast cancer, especially to resolve indeterminate (eg, 2+) IHC scores. However, manual signal enumeration for this test is time-consuming. Automation of the HER2 FISH test by means of image analysis has been shown to reduce workload and improve precision.114–117 Commercial QIA systems have been developed that are able to recognize nuclei within tissue sections to determine the HER2 amplification status. Overall concordance between manual scoring and automated nuclei-sampling analysis for some systems was reported to be 98.4% (100% for nonamplified cases and 96.9% for amplified cases).114 Systems that automate FISH evaluation have also been shown to improve workflow, consistency and save hours of technologist time.118
ER and PR
Assessment of ER and PR expression is essential for breast cancer patient management, as their expression is a strong predictive factor for response to hormonal therapies such as tamoxifen, and also has prognostic value.119–121 ASCO/CAP recommend that the ER and PR status be determined on all primary breast cancers and recurrences.122 ER or PR status is usually assessed using visual scoring of IHCs by pathologists using different scoring methods (eg, Allred score, H-index). However, this semiquantitative visual method is prone to human bias (eg, due to heterogenous staining intensity), interobserver and intraobserver variability, as well as limited standardization.123–126
QIA has been demonstrated to yield comparable results of ER quantification to manual scoring and may be more reproducible than manual scoring.88–91,127 ImmunoRatio, a free web application, can calculate the percentage of the positively stained nuclear area (labeling index) by using a color deconvolution algorithm for separating the staining components (DAB and hematoxylin) and the adaptive threshold for nuclear area segmentation.128 ImmunoRatio has been used to quantify ER and PR IHCs and the results have been shown to highly correlate with manual scoring by pathologists. Some QIA algorithms can provide the ratios of positive nuclei for different staining intensities as well as the H-score which can be calculated from the percentages of nuclei classified as 3+, 2+, 1+ (the 3 positive categories, where 3+ has the highest staining intensity) multiplying them with their grade: H-score=(Percentage of 3+)×3+(Percentage of 2+)×2+(Percentage of 1+) (Fig. 2). When using such algorithms, the invasive carcinoma regions need to be annotated, typically performed manually, to define an ROI to be analyzed. The act of performing the actual outlining and running the algorithm can be performed by a technologist.
FIGURE 2.
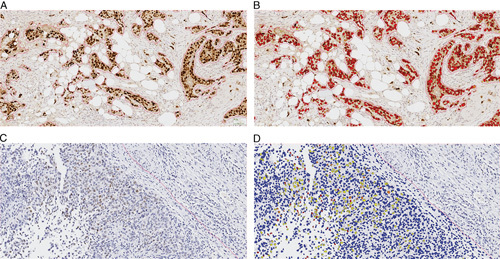
Example of estrogen receptor quantification (Visiopharm). Representative images from 2 cases with strong (A, B) (H-score: 275) and weak (C, D) (H-score: 21) estrogen receptor staining. A and C, Original IHC images. B and D, Analyzed images with pseudo-colors. Brown: Original IHC color; blue: original nuclear counterstain; red: strong staining intensity; yellow: weak staining intensity. IHC indicates immunohistochemistry.
PD-L1
Assessment of PD-L1 expression is critical for certain cancer patients’ eligibility for immunotherapy. Currently, PD-L1 expression is examined using an IHC method, and immunostained slides are usually evaluated manually by pathologists. Similar to manual quantitation of other biomarkers, PD-L1 IHC assessment has interobserver variability.129–132 Several studies have explored automated imaging analysis algorithms to assess PD-L1 expression from IHC slides. In one study, PD-L1 IHCs with the 22C3 antibody in non–small cell lung carcinomas were interpreted first by pathologists and secondly by a commercial image analysis algorithm for both tumor cells and immune cells. The results demonstrated that the automated scoring method was concordant with the pathologists’ average scores that were comparable to interpathologist scores.98 An additional study using QuPath, an open-source deep-learning platform, to quantify PD-L1 expression on both tumor cells and inflammatory cells reported objective, reproducible, and accurate results.133 But, to date, few automated studies have been validated against patient outcomes in a clinical trial setting. Beyond demonstrating just concordance with pathologist’s manual reads, it will be important to validate automated PD-L1 scoring against clinical trial endpoints if QIA methods are to be adopted into clinical practice as potential companion or complementary diagnostic tests.
Proliferation Index (Ki-67)
The cellular proliferation index determined by IHC staining with Ki-67 offers an insight into the biological behavior of tumors and is important in grading/subtyping of certain neoplasms including brain tumors, breast carcinoma, certain non-Hodgkin lymphomas, and neuroendocrine tumors. Nuclear Ki-67 antigen can be detected using MIB-1 antibody which serves as a good proliferation marker.134 The Ki-67 proliferative index is usually evaluated manually in clinical practice, but interobserver variability occurs. QIA algorithms have provided a more accurate estimate of the proliferation rate.1,135,136 In one study with pancreatic neuroendocrine tumors, the authors concluded that although it was practical to perform only a single-field hotspot analysis to determine the Ki-67 proliferative index, the results varied when using 10 consecutive fields, suggesting that automated QIA using WSI may yield more accurate results.137
Immunoscore
The tumor microenvironment and host immune reaction have gained growing importance in cancer prognosis and treatment. Immunoscore, a prognostic score (I0 to I4) for colon cancer, applies a QIA algorithm to quantify the combination of CD3-positive and CD8-positive cells in the central tumor and invasive margin areas of colon adenocarcinoma.138,139 A study looking at CD8 tumor-infiltrating lymphocytes in oropharyngeal squamous cell carcinoma has demonstrated similar benefit,140 but its utility in other indications is still under investigation and requires separate evaluation for every tumor type.
REGULATORY FRAMEWORK AND GUIDELINES
FDA
As of April 2020, there are 33 QIA applications cleared by the FDA. These QIA applications represent automated image analysis applications of routine manual scoring paradigms and despite agency clearance, as far as the authors are aware no “FDA guidance document” exists and many of these QIA are no longer in the market. Interestingly, in these FDA-cleared algorithms, the system software usually makes no independent interpretations of the data but instead, a pathologist selects an ROI to be analyzed and the algorithm provides a qualification or quantification score which the pathologist needs to confirm. If the pathologist does not agree with the algorithm assessment, the result can be overridden and a manual score is reported out. The quantitative data produced by these algorithms is almost entirely based in color detection and threshold and a few more recent applications include machine learning (ML) approaches that classify tissue for inclusion or exclusion in the analysis (eg, to include tumoral area and exclude stroma). These classifier algorithms were trained involving conventional supervised ML learning (ie, algorithm training was conducted by a pathology expert who provided input such as slide annotations). In general, ML supervised algorithms usually require less training data compared with AI-based algorithms employing deep learning. Deep learning requires larger datasets but much validation is needed in a controlled way. In addition, deep learning represents a “black box” for exactly how an algorithm works, compared with current cleared applications that are better understood and as such are perceived as easier to bring to market.
As outlined in previous sections, there are many advantages for adopting the digital environment. Just like in radiology, this will have to evolve and digital pathology can learn from this evolution. Recently, deep-learning AI-based algorithms have been cleared for radiology (eg, retinopathy diagnostic algorithm DEN180001 and bone fracture DEN180001). While these radiology algorithms do not serve as a “predicate” for FDA approval of digital pathology QIA applications, they do provide a precedent and could support faster clearance of AI-based digital pathology algorithms. To that regard, the Agency issued a position paper on AI entitled “Proposed regulatory framework for modifications to AI/ML-based software as a medical device.”141 This document refers to a ML system as a system that has the capacity to learn based on training on a specific task by tracking performance measure(s). When this software is intended to be used to diagnose, treat, cure, mitigate or prevent disease it is considered Software as a Medical Device (SaMD). When SaMD is used in clinical practice, it can be locked or open for continuous learning. Within this spectrum of continuously learning, general principles regarding data management, retraining, and performance could be considered. These general principles can be grouped under Good Machine Learning Practices (GMLP). GMLP can then be used to author a SaMD Pre-Specification which includes the anticipated modifications which the manufacturer plans to accomplish when the SaMD is in use (ie, the “what” the algorithm should become when it is learning). The Algorithm Change Protocol describes how this should be accomplished in a safe and effective way (ie, the “how” the algorithm remains safe and effective). What is not evident in the FDA paper is the role of the health care provider for locked and continuous learning SaMDs. Currently in clinical practice, the pathologist expert role provides the best mitigating strategy for diagnostic decision making. For AI/ML algorithms in digital pathology this same relationship will exist, where the pathologist is the “special control.” As an example we will mention class II medical devices, for which design and development requires “special controls” that are device specific and may include special labeling requirements such as requiring a pathologist to confirm the output of the device (eg, the results), to provide reasonable assurance of its safety and effectiveness.
In summary, there are still unanswered questions around FDA-clearance of AI-based QIA applications. These include, but are not limited to, specifications for complex algorithms that may assess features not readily visible by the human eye, what should constitute GMLPs, and the role of the health care provider to help drive the field forward.
Laboratory Developed Tests (LDTs)
According to the FDA, an LDT is “a type of in vitro diagnostic (IVD) test that is designed, manufactured and used within a single laboratory.” In contrast to FDA-cleared IVDs that are sold as commercial kits, LDTs are not to be promoted or distributed, and they are currently not FDA-regulated (see additional information about the VALID act below). LDTs can be used to study biomarkers in human samples, and although a commercial FDA-cleared IVD test for this biomarker might be available, some laboratories decide to develop their own test. This is particularly relevant for tissue biomarker image analysis, because only 33 FDA QIA tests are commercially available, assessing only 5 biomarkers total (p53, Ki-67, HER2, ER, and PR).
Recently, the FDA issued guidelines that impact all medical device software, including draft guidance “Clinical Decision Support Software.”142 These guidelines include the “21st Century Cures Act”143 which provides clarity on what software is not overseen by the FDA, and the subsequent “Changes to section 3060”144 which provides guidance on Clinical Decision Software (CDS) no longer considered to be a medical device, a so-called non-CDS device. A non-CDS device is when the health care provider can independently review the basis for the recommendation provided by the device so that it is not the intent that the user rely primarily on such recommendation. Hence the guidelines have a risk-based approach that consider if these technologies pose any risk to patients for example, on the significance of the information provided by that software function, and what is the role and involvement of the health care provider. The risk-based approach is leveraged from the IMDRF framework.145
Another document relevant for LDTs, is the “Verifying Accurate, Leading‑edge IVCT Development (VALID) Act.” The VALID Act is a draft bill that defines a regulatory path for “in vitro clinical tests” (IVCTs). IVCTs are clinical diagnostic tests that include LDTs and IVDs and are manufactured, promoted, and sold by manufacturers. The FDA has been contemplating regulating LDTs for many years and the VALID Act is an example of such potential LDT regulation. According to these newly proposed regulatory pathways, high-risk IVCTs would likely still be subject to premarket approval, while low-risk IVCTs could be exempted. This is similar to class I devices, which are devices that pose the lowest risk to the patient and/or user, but still need to follow general controls.
Good Laboratory Practice (GLP)/Good Clinical Practice (GCP) Guidelines
GLP is an internationally recognized quality system that regulates all the systems and facilities under which data is collected during research. GLP ensures the validity and reliability of such data. In contrast, GCP ensures patients’ safety and rights through the regulation of the ethical and scientific quality of clinical research (ie, research in human subjects). Good Clinical Laboratory Practice (GCLP) is an international standard that combines the principles of both GLP and GCP and ensures the reliability of the data and the ethical standards that clinical trials require.
When working with AI-powered algorithms during drug development, it is key to understand in which development phase the drug is and what is the device’s intended use. Pharmaceutical research often requires devices to be “21 CFR part 11” compliant (21 CFR 11: “The regulations in this part set forth the criteria under which the agency considers electronic records, electronic signatures, and handwritten signatures executed to electronic records to be trustworthy, reliable, and generally equivalent to paper records and handwritten signatures executed on paper.”) However, when data collected during drug clinical trials can be provided by a Medical Device but signed off in a different data system, then the device does not have to be 21 CFR 11 compliant. In contrast, if the device management system is being used for data analysis, then compliance to 21 CFR 11 is required. Because GLP and GCLP are globally recognized and adopted, adherence to these principles is most important. One of the requirements of GCP is that for all types of research, drug discovery, related biomarker development, and as such algorithms (including AI training) involving human subjects, the subjects should have given consent that their data, which includes images, can be used. It is noteworthy to mention that different local regulations and guidelines could apply for using leftover specimens.
Depending on the country, additional requirements on top of GCLP may apply, but in general, all toxicology studies (nonclinical testing) and clinical testing must adhere to GCLP. Data already collected during drug discovery and development can be reused to train an algorithm, and when compliant with GCLP and used as a nonmedical device, its making the deep-learning AI approach less cumbersome than when developing the algorithm as a SaMD or even LDT.
Non-USA Regulations
Outside the United States, guidance on medical device development is changing with respect to In Vitro Diagnostic Directives into Regulations (IVDR) in Europe. The IVDR are planned to become effective in May 2022. Per IVD directives, an IVD product refers to a reagent, instrument, or system (eg, image analysis algorithm) that is intended for clinical diagnostic use. Instead of a list-based approach, that is, Annex II with list A including high-risk devices such as determining blood groups requiring a notified body to verify the Common Technical Specifications; and list B including moderate-risk devices such as reagents for which the manufacturer must declare conformity to requirements as described in Annexes. The IVDR is akin to the FDA Quality System Regulation (QSR) in the United States, now following a risk-based approach, that is, classification depends on the risk it poses to the patient and/or user. Also, there is a USA federal initiative (RIN 0910-AH99, is the Regulatory Information Number (RIN) from the USA Department of Health and Human Services initiative for “Harmonizing and Modernizing Regulation of Medical Device Quality Systems.”) (RIN 0910-AH99) to increase alignment of the FDA QSR with ISO13485 [ISO13485 is the standard certification that proves Quality Management compliance for the Medical Device industry. Its purpose is to ensure that medical device manufacturers (and related suppliers) have systems for effective design and production and that their products are safe.] which would harmonize IVD development worldwide. However, the big difference between the European Union (EU) and the United States is that the EU requires active Post Market Clinical Follow Up (PMCFU) and surveillance for medical devices. The active PMCFU has the potential for allowing the collection of real-world data to utilize as real-world evidence. This could enable the establishment of well-controlled plans for continuous deep-learning algorithms for biomarker analysis. Although the FDA published guidance in August 2017,146 the acceptance of world data to be used as real-world evidence for medical devices has yet to be seen. Nevertheless, it is plausible that data collected in the EU could be used in the United States.
CAP/CLIA Guidelines
The CAP has established a number of guidelines governing the use of WSI147 as well as quantitative analysis using digital images, particularly in regard to IHC for biomarkers.148 Importantly, FDA approval is not required to deploy digital pathology systems in the clinical workflow (although CAP recommends that this should be explicitly stated in the pathology report), instead relying on the LDT mechanism for validation.102
Best practices for validating WSI for diagnostics generally include steps to demonstrate: (i) concordance with traditional diagnostics that do not use these technologies; (ii) periodic validation and revalidation when a component to the system has changed in a significant way; (iii) documentation of changes to systems or algorithms, documentation of training for users and backup procedures for system downtime; and (iv) consistency with other activities and requirements, such as IHC validation and IT policies. It is essential that the validation procedure uses an adequate number of samples considered to be representative of the use case under study. The minimum number of samples to be analyzed should be 60 for one application, and it is recommended that another 20 cases be included for each additional application (such as IHC or additional special stains).149 Systems should be validated in their entirety, from input to output. Furthermore, revalidation of each biomarker should be performed when there is an expectation of a difference between expression pattern, or when the algorithm differs between biomarkers. Comparison against a gold standard should be performed, in which an external validation set and manual assessment are commonly chosen.
At present, these guidelines are not explicitly laid out or tailored to the unique requirements of digital pathology. Nevertheless, both CAP and Clinical Laboratory Improvement Amendments (CLIA) currently require that AI-based technologies similarly undergo validation before being applied to patient samples, and recommend that the aforementioned general considerations for WSI and quantitative analysis are applied.102 This includes: (i) ensuring that the use case is appropriate and validation data is representative; (ii) there is high sensitivity and specificity in comparison to surrogate markers or manual assessment (with intraobserver and interobserver validation applied where appropriate); and (iii) that the algorithm is revalidated upon change. This may pose a problem for adaptive deep-learning AI algorithms that are designed to continuously learn based on exposure to new data.
Apart from HER2, there is a lack of clinical guidelines for QIA of biomarkers. According to the CAP 2016 Histology Quality Improvement Program (HQIP)—A mailing, 183 (∼22.1%) of participating laboratories reported using QIA. However, there was no information on exactly how QIA was conducted. To fill this gap, the CAP Quality Center convened an expert and advisory panel to work on a QIA guideline for HER2. QIA of HER2 IHC was selected because this is one of the most commonly tested biomarkers in pathology practice, and it is more challenging with respect to reproducibility due to the membranous staining pattern as compared with other nuclear-stained biomarkers such as ER, PR, and Ki-67. While the ASCO/CAP HER2 testing guidelines address key preanalytical-related, analytical-related, and reporting-related issues of IHC for this biomarker and advocate using image analysis, there is no detailed information on how HER2 IHC QIA for breast cancer should be conducted. The intent of the CAP guideline for QIA of HER2 was to thus provide recommendations for improving the precision and accuracy of this test.
The CAP guideline for QIA of HER2 was developed following the National Academy of Science (formerly Institute of Medicine) standards for developing clinical practice guidelines.150 The robust process developed by CAP Quality Center includes a systematic literature review, draft recommendations by an expert panel with input from an advisory panel, public comment period, and grades provided for the strength of evidence and strength of recommendation to complete recommendations, independent peer-review, approval, publication, implementation, and maintenance. The key questions that needed to be addressed included: What equipment, validation, and daily performance monitoring was needed? What training of staff and pathologists was required? What are the competency assessment needs over time? How does one select or develop appropriate algorithms for interpretation? How does one determine the performance of image analysis? How should image analysis be reported? Among 376 articles were initially identified, 39 underwent data extraction and only 9 articles had sufficient data to inform the guideline statements. Following a public comment period, 11 recommendations were published.102
The main messages delivered in the CAP guideline regarding QIA of HER2 IHC are that such analysis and its related procedures must be validated before implementation, followed by regular maintenance and ongoing evaluation of quality control and quality assurance. Laboratories should validate their QIA results for clinical use by comparing them to an alternative, validated method(s) such as HER2 FISH or consensus images for HER2 IHC. HER2 QIA performance, interpretation, and reporting should also be supervised by pathologists with expertise in QIA and those involved with using the technology should be trained. The length of retention for images, annotations, and computer-generated results should be comparable to the current requirements for similar clinical image assets and based on the laboratory documented standard operating procedures and policies. In the United States, the latest accreditation standard for datasets from ex vivo microscopic imaging systems is 10 years. As with any clinical evidence-based guideline, following these recommendations is not mandatory. Of course, such recommendations may ultimately be incorporated into future versions of the CAP Laboratory Accreditation Program checklists; however, they are not currently required by Laboratory Accreditation Program or any regulatory or accrediting agency. Nevertheless, it is highly encouraged that laboratories participating in QIA of HER2 adopt these recommendations and of course, these recommendations can also be extrapolated to QIA of other biomarkers.
CONCLUSIONS
QIA is becoming an indispensable tool for biomarker research, discovery, nonclinical studies, and clinical trials. In addition, it is increasingly used in clinical practice. The technologies employed to detect biomarkers in tissues and cells today have become increasingly complex (eg, multiplex fluorescence, CYTOF), resulting in staining that can be challenging to evaluate manually. Applying QIA tools in these scenarios will produce faster results and more accurate and reproducible data. Validation of all components of the workflow (staining, digitization, and algorithm analysis) is necessary and crucial for proper implementation of QIA of tissue biomarkers for both in-house and commercially available algorithms. Additional guidelines from federal regulatory agencies are needed that go beyond the currently available CAP recommendations for HER2 IHC evaluation by QIA. In addition, more contemporary regulations are needed, as well provisions for billing and addressing liability issues for medical personnel.151 There are currently several such initiatives underway including the redefinition of a medical device and of the agencies’ role in the clearance of AI-based deep-learning algorithms for clinical use. The outcome of these initiatives will significantly impact how digital tools for biomarker research and clinical purposes are going to be cleared and utilized.
Footnotes
H.L. and Z.L. contributed equally.
A.V.P. is on the advisory board of Contextvision & PathPresenter. D.R. is an advisory board member or consultant of Amgen, AstraZeneca, Cell Signaling Technology, Cepheid, Daiichi Sankyo, Danaher, GSK, Konica/Minolta, Merck, Nanostring, NextCure, Odonate, Perkin Elmer, Paige.AI, Roche, Sanofi, Ventana, and Ultivue; research or instrument support for Amgen, Cepheid, Navigate Biopharma, NextCure, Konica/Minolta, Lilly, Ultivue, Ventana, Akoya, Perkin Elmer, and NanoString; receives royalties from Rarecyte and is founder and equity holder of PixelGear. D.B. is an employee of Indica Labs. C.K. is an employee of Genentech Inc. E.A.E.G. is an employee of Roche Diagnostics. E.A. is an employee of Visiopharm. F.A. is an employee and shareholder of Amgen Inc. H.L. is an employee of GSK. L.P. is a consultant for Hamamatsu and on the medical advisory board for Ibex. M.B. is an advisory board member or consultant of Epistyle, ContextVision, Visiopharm, Aiforia, AstraZeneca, and Bristol Myers Squibb. M.M. is a former employee and shareholder of Bristol Myers Squibb and current employee and shareholder of pathAI. The remaining authors declare no conflicts of interest.
Contributor Information
Haydee Lara, Email: haydee.p.lara@gsk.com.
Zaibo Li, Email: Zaibo.Li@osumc.edu.
Esther Abels, Email: e.abels@xs4all.nl.
Famke Aeffner, Email: faeffner@amgen.com.
Marilyn M. Bui, Email: Marilyn.Bui@moffitt.org.
Ehab A. ElGabry, Email: Ehab.Elgabry@roche.com.
Cleopatra Kozlowski, Email: cleopatk@gene.com.
Michael C. Montalto, Email: mike.montalto@pathai.com.
Anil V. Parwani, Email: Anil.Parwani@osumc.edu.
Mark D. Zarella, Email: mzarell2@jhmi.edu.
Douglas Bowman, Email: dbowman@indicalab.com.
David Rimm, Email: david.rimm@yale.edu.
Liron Pantanowitz, Email: lironp@med.umich.edu.
REFERENCES
- 1.Stålhammar G, Fuentes Martinez N, Lippert M, et al. Digital image analysis outperforms manual biomarker assessment in breast cancer. Mod Pathol. 2016;29:318–329. [DOI] [PubMed] [Google Scholar]
- 2.Alturkistani HA, Tashkandi FM, Mohammedsaleh ZM. Histological stains: a literature review and case study. Glob J Health Sci. 2015;8:72–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sha L, Osinski B, Ho I, et al. Multi-field-of-view deep learning model predicts nonsmall cell lung cancer programmed death-ligand 1 status from whole-slide hematoxylin and eosin images. J Pathol Inform. 2019;10:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rimm DL. What brown cannot do for you. Nat Biotechnol. 2006;24:914–916. [DOI] [PubMed] [Google Scholar]
- 5.Pyon WS, Gray DT, Barnes CA. An alternative to dye-based approaches to remove background autofluorescence from primate brain tissue. Front Neuroanat. 2019;13:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kiyose S, Igarashi H, Nagura K, et al. Chromogenic in situ hybridization (CISH) to detect HER2 gene amplification in breast and gastric cancer: comparison with immunohistochemistry (IHC) and fluorescence in situ hybridization (FISH). Pathol Int. 2012;62:728–734. [DOI] [PubMed] [Google Scholar]
- 7.Lichter P, Cremer T, Borden J, et al. Delineation of individual human chromosomes in metaphase and interphase cells by in situ suppression hybridization using recombinant DNA libraries. Hum Genet. 1988;80:224–234. [DOI] [PubMed] [Google Scholar]
- 8.Huang D, Swanson EA, Lin CP, et al. Optical coherence tomography. Science. 1991;254:1178–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Varando G, Benavides-Piccione R, Muñoz A, et al. MultiMap: a tool to automatically extract and analyse spatial microscopic data from large stacks of confocal microscopy images. Front Neuroanat. 2018;12:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Silvestri L, Paciscopi M, Soda P, et al. Quantitative neuroanatomy of all Purkinje cells with light sheet microscopy and high-throughput image analysis. Front Neuroanat. 2015;9:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McQuin C, Goodman A, Chernyshev V, et al. CellProfiler 3.0: next-generation image processing for biology. PLoS Biol. 2018;16:e2005970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goltsev Y, Samusik N, Kennedy-Darling J, et al. Deep profiling of mouse splenic architecture with CODEX multiplexed imaging. Cell. 2018;174:968.e15–981.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bendall SC, Simonds EF, Qiu P, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332:687–696.21551058 [Google Scholar]
- 14.Angelo M, Bendall SC, Finck R, et al. Multiplexed ion beam imaging of human breast tumors. Nat Med. 2014;20:436–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yildiz-Aktas IZ, Dabbs DJ, Bhargava R. The effect of cold ischemic time on the immunohistochemical evaluation of estrogen receptor, progesterone receptor, and HER2 expression in invasive breast carcinoma. Mod Pathol. 2012;25:1098–1105. [DOI] [PubMed] [Google Scholar]
- 16.Khoury T. Delay to formalin fixation (cold ischemia time) effect on breast cancer molecules. Am J Clin Pathol. 2018;149:275–292. [DOI] [PubMed] [Google Scholar]
- 17.Engel KB, Moore HM. Effects of preanalytical variables on the detection of proteins by immunohistochemistry in formalin-fixed, paraffin-embedded tissue. Arch Pathol Lab Med. 2011;135:537–543. [DOI] [PubMed] [Google Scholar]
- 18.Martina JD, Simmons C, Jukic DM. High-definition hematoxylin and eosin staining in a transition to digital pathology. J Pathol Inform. 2011;2:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldstein NS, Hewitt SM, Taylor CR, et al. Recommendations for improved standardization of immunohistochemistry. Appl Immunohistochem Mol Morphol. 2007;15:124–133. [DOI] [PubMed] [Google Scholar]
- 20.Taylor CR, Levenson RM. Quantification of immunohistochemistry—issues concerning methods, utility and semiquantitative assessment II. Histopathology. 2006;49:411–424. [DOI] [PubMed] [Google Scholar]
- 21.Law AMK, Yin JXM, Castillo L, et al. Andy’s algorithms: new automated digital image analysis pipelines for FIJI. Sci Rep. 2017;7:15717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chlipala E, Bendzinski CM, Chu K, et al. Optical density-based image analysis method for the evaluation of hematoxylin and eosin staining precision. J Histotechnol. 2020;43:29–37. [DOI] [PubMed] [Google Scholar]
- 23.Yagi Y, Gilbertson JR. A relationship between slide quality and image quality in whole slide imaging (WSI). Diagn Pathol. 2008;3(suppl 1):S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Senaras C, Niazi MKK, Lozanski G, et al. DeepFocus: detection of out-of-focus regions in whole slide digital images using deep learning. PLoS One. 2018;13:e0205387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kothari S, Phan J, Wang M. Eliminating tissue-fold artifacts in histopathological whole-slide images for improved image-based prediction of cancer grade. J Pathol Inform. 2013;4:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Komura D, Ishikawa S. Machine learning methods for histopathological image analysis. Comput Struct Biotechnol J. 2018;16:34–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Farahani NDJ, Hartman DJ, Riben M, et al. Adjusting color threshold for whole slide imaging overcomes artifacts caused by pen markings on glass slides. J Pathol Inform. 2016;7:S27–S28. [Google Scholar]
- 28.Ali S Alham N Verrill C, et al. Ink removal from histopathology whole slide images by combining classification, detection and image generation models. IEEE 16th International Symposium on Biomedical Imaging (ISBI); 2019. 928–932.
- 29.Hu F, Kozlowski C. Abstracts. J Pathol Inform. 2019;10:28. [Google Scholar]
- 30.Xu S, Wang Y, Tai DCS, et al. qFibrosis: a fully-quantitative innovative method incorporating histological features to facilitate accurate fibrosis scoring in animal model and chronic hepatitis B patients. J Hepatol. 2014;61:260–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nault R, Colbry D, Brandenberger C, et al. Development of a computational high-throughput tool for the quantitative examination of dose-dependent histological features. Toxicol Pathol. 2014;43:366–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sethunath D, Morusu S, Tuceryan M, et al. Automated assessment of steatosis in murine fatty liver. PLoS One. 2018;13:e0197242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Rudder M, Bouzin C, Nachit M, et al. Automated computerized image analysis for the user-independent evaluation of disease severity in preclinical models of NAFLD/NASH. Lab Invest. 2020;100:147–160. [DOI] [PubMed] [Google Scholar]
- 34.Nascimento DS, Valente M, Esteves T, et al. MIQuant—semi-automation of infarct size assessment in models of cardiac ischemic injury. PLoS One. 2011;6:e25045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seger S, Stritt M, Vezzali E, et al. A fully automated image analysis method to quantify lung fibrosis in the bleomycin-induced rat model. PLoS One. 2018;13:e0193057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gilhodes J-C, Julé Y, Kreuz S, et al. Quantification of pulmonary fibrosis in a bleomycin mouse model using automated histological image analysis. PLoS One. 2017;12:e0170561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tetsuhiro K, Kinya O, Yoshihiro H, et al. Automated image analysis of a glomerular injury marker desmin in spontaneously diabetic Torii rats treated with losartan. J Endocrinol. 2014;222:43–51. [DOI] [PubMed] [Google Scholar]
- 38.Brenneman KA, Ramaiah SK, Rohde CM, et al. Mechanistic investigations of test article–induced pancreatic toxicity at the endocrine–exocrine interface in the rat. Toxicol Pathol. 2013;42:229–242. [DOI] [PubMed] [Google Scholar]
- 39.Lindauer K, Bartels T, Scherer P, et al. Development and validation of an image analysis system for the measurement of cell proliferation in mammary glands of rats. Toxicol Pathol. 2019;47:634–644. [DOI] [PubMed] [Google Scholar]
- 40.Ioannis S, Arantza E-Z, Olympia K, et al. Selective androgen receptor modulators (SARMs) have specific impacts on the mouse uterus. J Endocrinol. 2019;242:227–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krajewska M, Smith LH, Rong J, et al. Image analysis algorithms for immunohistochemical assessment of cell death events and fibrosis in tissue sections. J Histochem Cytochem. 2009;57:649–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kozlowski C, Jeet S, Beyer J, et al. An entirely automated method to score DSS-induced colitis in mice by digital image analysis of pathology slides. Dis Model Mech. 2013;6:855–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iskenderian A, Liu N, Deng Q, et al. Myostatin and activin blockade by engineered follistatin results in hypertrophy and improves dystrophic pathology in mdx mouse more than myostatin blockade alone. Skelet Muscle. 2018;8:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bornstein GG. Antibody drug conjugates: preclinical considerations. AAPS J. 2015;17:525–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Szot C, Saha S, Zhang XM, et al. Tumor stroma–targeted antibody-drug conjugate triggers localized anticancer drug release. J Clin Invest. 2018;128:2927–2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Knoblaugh SE, Himmel LE. Keeping score: semiquantitative and quantitative scoring approaches to genetically engineered and xenograft mouse models of cancer. Vet Pathol. 2018;56:24–32. [DOI] [PubMed] [Google Scholar]
- 47.Jones GN, Rooney C, Griffin N, et al. pRAD50: a novel and clinically applicable pharmacodynamic biomarker of both ATM and ATR inhibition identified using mass spectrometry and immunohistochemistry. Br J Cancer. 2018;119:1233–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McDonald AA, Burke K, Thomas M, et al. Abstract A38: Development and implementation of immunohistochemistry (IHC)-based pharmacodynamic (PD) biomarkers demonstrate NAE pathway inhibition in MLN4924 solid tumor clinical trials. Mol Cancer Ther. 2011;10(suppl):A38. [Google Scholar]
- 49.Berger AJ, Friedlander S, Ghasemi O, et al. Abstract 3079: Pharmacodynamic evaluation of the novel SUMOylation inhibitor TAK-981 in a mouse tumor model. Cancer Res. 2019;79(suppl):3079. [Google Scholar]
- 50.Chuang DY, Cui J, Simonyi A, et al. Dietary sutherlandia and elderberry mitigate cerebral ischemia-induced neuronal damage and attenuate p47phox and phospho-ERK1/2 expression in microglial cells. ASN Neuro. 2014;6:1759091414554946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Willuweit A, Velden J, Godemann R, et al. Early-onset and robust amyloid pathology in a new homozygous mouse model of Alzheimer’s disease. PLoS One. 2009;4:e7931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Apfeldorfer C, Ulrich K, Jones G, et al. Object orientated automated image analysis: quantitative and qualitative estimation of inflammation in mouse lung. Diagn Pathol. 2008;3(suppl 1):S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lopès A, Cassé AH, Billard E, et al. Deciphering the immune microenvironment of a tissue by digital imaging and cognition network. Sci Rep. 2018;8:16692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Syed AK, Woodall R, Whisenant JG, et al. Characterizing trastuzumab-induced alterations in intratumoral heterogeneity with quantitative imaging and immunohistochemistry in HER2+ breast cancer. Neoplasia. 2019;21:17–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mischinger J, Fröhlich E, Mannweiler S, et al. Prognostic value of B7-H1, B7-H3 and the stage, size, grade and necrosis (SSIGN) score in metastatic clear cell renal cell carcinoma. Cent European J Urol. 2019;72:23–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Acs B, Pelekanou V, Bai Y, et al. Ki67 reproducibility using digital image analysis: an inter-platform and inter-operator study. Lab Invest. 2019;99:107–117. [DOI] [PubMed] [Google Scholar]
- 57.Howat WJ, Blows FM, Provenzano E, et al. Performance of automated scoring of ER, PR, HER2, CK5/6 and EGFR in breast cancer tissue microarrays in the Breast Cancer Association Consortium. J Pathol Clin Res. 2014;1:18–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Abubakar M, Figueroa J, Ali HR, et al. Combined quantitative measures of ER, PR, HER2, and KI67 provide more prognostic information than categorical combinations in luminal breast cancer. Mod Pathol. 2019;32:1244–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brennan DJ, Brändstedt J, Rexhepaj E, et al. Tumour-specific HMG-CoAR is an independent predictor of recurrence free survival in epithelial ovarian cancer. BMC Cancer. 2010;10:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Popovici V, Budinská E, Dušek L, et al. Image-based surrogate biomarkers for molecular subtypes of colorectal cancer. Bioinformatics. 2017;33:2002–2009. [DOI] [PubMed] [Google Scholar]
- 61.Brazdziute E, Laurinavicius A. Digital pathology evaluation of complement C4d component deposition in the kidney allograft biopsies is a useful tool to improve reproducibility of the scoring. Diagn Pathol. 2011;6(suppl 1):S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schafer PH, Adams M, Horan G, et al. Apremilast normalizes gene expression of inflammatory mediators in human keratinocytes and reduces antigen-induced atopic dermatitis in mice. Drugs R D. 2019;19:329–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Heichler C, Scheibe K, Schmied A, et al. STAT3 activation through IL-6/IL-11 in cancer-associated fibroblasts promotes colorectal tumour development and correlates with poor prognosis. Gut. 2019;69:1269–1282. [DOI] [PubMed] [Google Scholar]
- 64.Yu Y, Wang J, Ng CW, et al. Deep learning enables automated scoring of liver fibrosis stages. Sci Rep. 2018;8:16016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kannan S, Morgan LA, Liang B, et al. Segmentation of glomeruli within trichrome images using deep learning. Kidney Int Rep. 2019;4:955–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Du Y, Zhang R, Zargari A, et al. Classification of tumor epithelium and stroma by exploiting image features learned by deep convolutional neural networks. Ann Biomed Eng. 2018;46:1988–1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ehteshami Bejnordi B, Mullooly M, Pfeiffer RM, et al. Using deep convolutional neural networks to identify and classify tumor-associated stroma in diagnostic breast biopsies. Mod Pathol. 2018;31:1502–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kainz P, Pfeiffer M, Urschler M. Segmentation and classification of colon glands with deep convolutional neural networks and total variation regularization. PeerJ. 2017;5:e3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ali HR, Dariush A, Provenzano E, et al. Computational pathology of pre-treatment biopsies identifies lymphocyte density as a predictor of response to neoadjuvant chemotherapy in breast cancer. Breast Cancer Res. 2016;18:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ali HR, Dariush A, Thomas J, et al. Lymphocyte density determined by computational pathology validated as a predictor of response to neoadjuvant chemotherapy in breast cancer: secondary analysis of the ARTemis trial. Ann Oncol. 2017;28:1832–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Saltz J, Gupta R, Hou L, et al. Spatial organization and molecular correlation of tumor-infiltrating lymphocytes using deep learning on pathology images. Cell Rep. 2018;23:181.e7–193.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Steele KE, Tan TH, Korn R, et al. Measuring multiple parameters of CD8+ tumor-infiltrating lymphocytes in human cancers by image analysis. J Immunother Cancer. 2018;6:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mroz P, Parwani AV, Kulesza P. Central pathology review for phase III clinical trials: the enabling effect of virtual microscopy. Arch Pathol Lab Med. 2013;137:492–495. [DOI] [PubMed] [Google Scholar]
- 74.Pell R, Oien K, Robinson M, et al. The use of digital pathology and image analysis in clinical trials. J Pathol Clin Res. 2019;5:81–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Althammer S, Tan TH, Spitzmüller A, et al. Automated image analysis of NSCLC biopsies to predict response to anti-PD-L1 therapy. J Immunother Cancer. 2019;7:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wu SP, Liao RQ, Tu HY, et al. Stromal PD-L1-positive regulatory T cells and PD-1-positive CD8-positive T cells define the response of different subsets of non-small cell lung cancer to PD-1/PD-L1 blockade immunotherapy. J Thorac Oncol. 2018;13:521–532. [DOI] [PubMed] [Google Scholar]
- 77.Tumeh PC, Harview CL, Yearley JH, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515:568–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ancevski Hunter K, Socinski MA, Villaruz LC. PD-L1 testing in guiding patient selection for PD-1/PD-L1 inhibitor therapy in lung cancer. Mol Diagn Ther. 2018;22:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Colin G, Miller JK, Schwartz LH. Medical Imaging in Clinical Trials. London: Springer-Verlag; 2014. [Google Scholar]
- 80.Blumenthal GM, Mansfield E, Pazdur R. Next-generation sequencing in oncology in the era of precision medicine. JAMA Oncol. 2016;2:13–14. [DOI] [PubMed] [Google Scholar]
- 81.Kohrt HE, Tumeh PC, Benson D, et al. Immunodynamics: a cancer immunotherapy trials network review of immune monitoring in immuno-oncology clinical trials. J Immunother Cancer. 2016;4:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Barnes M, Srinivas C, Bai I, et al. Whole tumor section quantitative image analysis maximizes between-pathologists’ reproducibility for clinical immunohistochemistry-based biomarkers. Lab Invest. 2017;97:1508–1515. [DOI] [PubMed] [Google Scholar]
- 83.Hirsch FR, McElhinny A, Stanforth D, et al. PD-L1 immunohistochemistry assays for lung cancer: results from phase 1 of the blueprint PD-L1 IHC Assay Comparison Project. J Thorac Oncol. 2017;12:208–222. [DOI] [PubMed] [Google Scholar]
- 84.Rimm DL, Han G, Taube JM, et al. A prospective, multi-institutional, pathologist-based assessment of 4 immunohistochemistry assays for PD-L1 expression in non-small cell lung cancer. JAMA Oncol. 2017;3:1051–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Walk EE, Yohe SL, Beckman A, et al. The cancer immunotherapy biomarker testing landscape. Arch Pathol Lab Med. 2020;144:706–724. [DOI] [PubMed] [Google Scholar]
- 86.Ascierto PB PA, Bhatia S, Melero I, et al. Efficacy of BMS-986016, a monoclonal antibody that targets lymphocyte activation gene-3 (LAG-3), in combination with nivolumab in pts with melanoma. Ann Oncol. 2017;28(suppl 5):v605–v649. [Google Scholar]
- 87.Webster JD, Dunstan RW. Whole-slide imaging and automated image analysis: considerations and opportunities in the practice of pathology. Vet Pathol. 2014;51:211–223. [DOI] [PubMed] [Google Scholar]
- 88.Bolton KL, Garcia-Closas M, Pfeiffer RM, et al. Assessment of automated image analysis of breast cancer tissue microarrays for epidemiologic studies. Cancer Epidemiol Biomarkers Prev. 2010;19:992–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Diaz LK, Sahin A, Sneige N. Interobserver agreement for estrogen receptor immunohistochemical analysis in breast cancer: a comparison of manual and computer-assisted scoring methods. Ann Diagn Pathol. 2004;8:23–27. [DOI] [PubMed] [Google Scholar]
- 90.Faratian D, Kay C, Robson T, et al. Automated image analysis for high-throughput quantitative detection of ER and PR expression levels in large-scale clinical studies: the TEAM Trial Experience. Histopathology. 2009;55:587–593. [DOI] [PubMed] [Google Scholar]
- 91.Rizzardi AE, Johnson AT, Vogel RI, et al. Quantitative comparison of immunohistochemical staining measured by digital image analysis versus pathologist visual scoring. Diagn Pathol. 2012;7:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Brugmann A, Eld M, Lelkaitis G, et al. Digital image analysis of membrane connectivity is a robust measure of HER2 immunostains. Breast Cancer Res Treat. 2012;132:41–49. [DOI] [PubMed] [Google Scholar]
- 93.Helin HO, Tuominen VJ, Ylinen O, et al. Free digital image analysis software helps to resolve equivocal scores in HER2 immunohistochemistry. Virchows Arch. 2016;468:191–198. [DOI] [PubMed] [Google Scholar]
- 94.Laurinaviciene A, Dasevicius D, Ostapenko V, et al. Membrane connectivity estimated by digital image analysis of HER2 immunohistochemistry is concordant with visual scoring and fluorescence in situ hybridization results: algorithm evaluation on breast cancer tissue microarrays. Diagn Pathol. 2011;6:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Skaland I, Ovestad I, Janssen EA, et al. Comparing subjective and digital image analysis HER2/neu expression scores with conventional and modified FISH scores in breast cancer. J Clin Pathol. 2008;61:68–71. [DOI] [PubMed] [Google Scholar]
- 96.Dobson L, Conway C, Hanley A, et al. Image analysis as an adjunct to manual HER-2 immunohistochemical review: a diagnostic tool to standardize interpretation. Histopathology. 2010;57:27–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Healey MA, Hirko KA, Beck AH, et al. Assessment of Ki67 expression for breast cancer subtype classification and prognosis in the Nurses’ Health Study. Breast Cancer Res Treat. 2017;166:613–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Taylor CR, Jadhav AP, Gholap A, et al. A multi-institutional study to evaluate automated whole slide scoring of immunohistochemistry for assessment of programmed death-ligand 1 (PD-L1) expression in non-small cell lung cancer. Appl Immunohistochem Mol Morphol. 2019;27:263–269. [DOI] [PubMed] [Google Scholar]
- 99.Widmaier M, Wiestler T, Walker J, et al. Comparison of continuous measures across diagnostic PD-L1 assays in non-small cell lung cancer using automated image analysis. Mod Pathol. 2020;33:380–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Angell H, Galon J. From the immune contexture to the Immunoscore: the role of prognostic and predictive immune markers in cancer. Curr Opin Immunol. 2013;25:261–267. [DOI] [PubMed] [Google Scholar]
- 101.Stalhammar G, Robertson S, Wedlund L, et al. Digital image analysis of Ki67 in hot spots is superior to both manual Ki67 and mitotic counts in breast cancer. Histopathology. 2018;72:974–989. [DOI] [PubMed] [Google Scholar]
- 102.Bui MM, Riben MW, Allison KH, et al. Quantitative image analysis of human epidermal growth factor receptor 2 immunohistochemistry for breast cancer: guideline from the College of American Pathologists. Arch Pathol Lab Med. 2019;143:1180–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Slamon DJ, Clark GM, Wong SG, et al. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–182. [DOI] [PubMed] [Google Scholar]
- 104.Tandon AK, Clark GM, Chamness GC, et al. HER-2/neu oncogene protein and prognosis in breast cancer. J Clin Oncol. 1989;7:1120–1128. [DOI] [PubMed] [Google Scholar]
- 105.Press MF, Pike MC, Chazin VR, et al. Her-2/neu expression in node-negative breast cancer: direct tissue quantitation by computerized image analysis and association of overexpression with increased risk of recurrent disease. Cancer Res. 1993;53:4960–4970. [PubMed] [Google Scholar]
- 106.Park JW, Neve RM, Szollosi J, et al. Unraveling the biologic and clinical complexities of HER2. Clin Breast Cancer. 2008;8:392–401. [DOI] [PubMed] [Google Scholar]
- 107.Wolff AC, Hammond MEH, Allison KH, et al. Human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists Clinical Practice Guideline Focused Update. J Clin Oncol. 2018;36:2105–2122. [DOI] [PubMed] [Google Scholar]
- 108.Gancberg D, Jarvinen T, di Leo A, et al. Evaluation of HER-2/NEU protein expression in breast cancer by immunohistochemistry: an interlaboratory study assessing the reproducibility of HER-2/NEU testing. Breast Cancer Res Treat. 2002;74:113–120. [DOI] [PubMed] [Google Scholar]
- 109.Jacobs TW, Gown AM, Yaziji H, et al. Comparison of fluorescence in situ hybridization and immunohistochemistry for the evaluation of HER-2/neu in breast cancer. J Clin Oncol. 1999;17:1974–1982. [DOI] [PubMed] [Google Scholar]
- 110.Holten-Rossing H, Moller Talman ML, Kristensson M, et al. Optimizing HER2 assessment in breast cancer: application of automated image analysis. Breast Cancer Res Treat. 2015;152:367–375. [DOI] [PubMed] [Google Scholar]
- 111.Koopman T, Buikema HJ, Hollema H, et al. What is the added value of digital image analysis of HER2 immunohistochemistry in breast cancer in clinical practice? A study with multiple platforms. Histopathology. 2019;74:917–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Koopman T, de Bock GH, Buikema HJ, et al. Digital image analysis of HER2 immunohistochemistry in gastric- and oesophageal adenocarcinoma: a validation study on biopsies and surgical specimens. Histopathology. 2018;72:191–200. [DOI] [PubMed] [Google Scholar]
- 113.Hartage R, Li AC, Hammond S, et al. A validation study of human epidermal growth factor receptor 2 immunohistochemistry digital imaging analysis and its correlation with human epidermal growth factor receptor 2 fluorescence in situ hybridization results in breast carcinoma. J Pathol Inform. 2020;11:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Furrer D, Jacob S, Caron C, et al. Validation of a new classifier for the automated analysis of the human epidermal growth factor receptor 2 (HER2) gene amplification in breast cancer specimens. Diagn Pathol. 2013;8:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Theodosiou Z, Kasampalidis IN, Karayannopoulou G, et al. Evaluation of FISH image analysis system on assessing HER2 amplification in breast carcinoma cases. Breast. 2008;17:80–84. [DOI] [PubMed] [Google Scholar]
- 116.Prins MJD, Ruurda JP, Van Diest PJ, et al. Evaluation of the HER2 amplification status in oesophageal adenocarcinoma by conventional and automated FISH: a tissue microarray study. J Clin Pathol. 2014;67:26–32. [DOI] [PubMed] [Google Scholar]
- 117.Reljin B, Paskas M, Reljin I, et al. Breast cancer evaluation by fluorescent dot detection using combined mathematical morphology and multifractal techniques. Diagn Pathol. 2011;6(suppl 1):S21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tafe LJ, Allen SF, Steinmetz HB, et al. Automated processing of fluorescence in-situ hybridization slides for HER2 testing in breast and gastro-esophageal carcinomas. Exp Mol Pathol. 2014;97:116–119. [DOI] [PubMed] [Google Scholar]
- 119.Allred DC, Harvey JM, Berardo M, et al. Prognostic and predictive factors in breast cancer by immunohistochemical analysis. Mod Pathol. 1998;11:155–168. [PubMed] [Google Scholar]
- 120.Hahnel R, Woodings T, Vivian AB. Prognostic value of estrogen receptors in primary breast cancer. Cancer. 1979;44:671–675. [DOI] [PubMed] [Google Scholar]
- 121.Fitzgibbons PL, Page DL, Weaver D, et al. Prognostic factors in breast cancer. College of American Pathologists Consensus Statement 1999. Arch Pathol Lab Med. 2000;124:966–978. [DOI] [PubMed] [Google Scholar]
- 122.Hammond ME, Hayes DF, Dowsett M, et al. American Society of Clinical Oncology/College Of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J Clin Oncol. 2010;28:2784–2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Nadji M, Gomez-Fernandez C, Ganjei-Azar P, et al. Immunohistochemistry of estrogen and progesterone receptors reconsidered: experience with 5,993 breast cancers. Am J Clin Pathol. 2005;123:21–27. [DOI] [PubMed] [Google Scholar]
- 124.Mann GB, Fahey VD, Feleppa F, et al. Reliance on hormone receptor assays of surgical specimens may compromise outcome in patients with breast cancer. J Clin Oncol. 2005;23:5148–5154. [DOI] [PubMed] [Google Scholar]
- 125.Hede K. Breast cancer testing scandal shines spotlight on black box of clinical laboratory testing. J Natl Cancer Inst. 2008;100:836–837. [DOI] [PubMed] [Google Scholar]
- 126.Collins LC, Botero ML, Schnitt SJ. Bimodal frequency distribution of estrogen receptor immunohistochemical staining results in breast cancer: an analysis of 825 cases. Am J Clin Pathol. 2005;123:16–20. [DOI] [PubMed] [Google Scholar]
- 127.Thomsen C, Nielsen S, Nielsen BS, et al. Estrogen receptor-alpha quantification in breast cancer: concordance between immunohistochemical assays and mRNA-in situ hybridization for ESR1 gene. Appl Immunohistochem Mol Morphol. 2020;28:347–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Tuominen VJ, Ruotoistenmäki S, Viitanen A, et al. ImmunoRatio: a publicly available web application for quantitative image analysis of estrogen receptor (ER), progesterone receptor (PR), and Ki-67. Breast Cancer Res. 2010;12:R56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.McLaughlin J, Han G, Schalper KA, et al. Quantitative assessment of the heterogeneity of PD-L1 expression in non-small-cell lung cancer. JAMA Oncol. 2016;2:46–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Savic S, Berezowska S, Eppenberger-Castori S, et al. PD-L1 testing of non-small cell lung cancer using different antibodies and platforms: a Swiss cross-validation study. Virchows Arch. 2019;475:67–76. [DOI] [PubMed] [Google Scholar]
- 131.Hendry S, Byrne DJ, Wright GM, et al. Comparison of four PD-L1 immunohistochemical assays in lung cancer. J Thorac Oncol. 2018;13:367–376. [DOI] [PubMed] [Google Scholar]
- 132.Tsao MS, Kerr KM, Kockx M, et al. PD-L1 Immunohistochemistry comparability study in real-life clinical samples: results of Blueprint Phase 2 Project. J Thorac Oncol. 2018;13:1302–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Humphries MP, Hynes S, Bingham V, et al. Automated tumour recognition and digital pathology scoring unravels new role for PD-L1 in predicting good outcome in ER−/HER2+ breast cancer. J Oncol. 2018;2018:2937012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Spyratos F, Ferrero-Pous M, Trassard M, et al. Correlation between MIB-1 and other proliferation markers: clinical implications of the MIB-1 cutoff value. Cancer. 2002;94:2151–2159. [DOI] [PubMed] [Google Scholar]
- 135.Grala B, Markiewicz T, Kozłowski W, et al. New automated image analysis method for the assessment of Ki-67 labeling index in meningiomas. Folia Histochem Cytobiol. 2009;47:587–592. [DOI] [PubMed] [Google Scholar]
- 136.Remes SM, Tuominen VJ, Helin H, et al. Grading of neuroendocrine tumors with Ki-67 requires high-quality assessment practices. Am J Surg Pathol. 2012;36:1359–1363. [DOI] [PubMed] [Google Scholar]
- 137.Goodell PP, Krasinskas AM, Davison JM, et al. Comparison of methods for proliferative index analysis for grading pancreatic well-differentiated neuroendocrine tumors. Am J Clin Pathol. 2012;137:576–582. [DOI] [PubMed] [Google Scholar]
- 138.Galon J, Mlecnik B, Bindea G, et al. Towards the introduction of the ‘Immunoscore’ in the classification of malignant tumours. J Pathol. 2014;232:199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Galon J, Pagès F, Marincola FM, et al. The immune score as a new possible approach for the classification of cancer. J Transl Med. 2012;10:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hartman DJ, Ahmad F, Ferris RL, et al. Utility of CD8 score by automated quantitative image analysis in head and neck squamous cell carcinoma. Oral Oncol. 2018;86:278–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.US Food & Drug Administration (FDA). Proposed regulatory framework for modifications to artificial intelligence/machine learning (AI/ML)-based software as a medical device (SaMD); 2019. p. 20. Available at: www.fda.gov/medical-devices/software-medical-device-samd/artificial-intelligence-and-machine-learning-software-medical-device. Accessed April 20, 2020.
- 142.US Food & Drug Administration (FDA). Clinical decision support software, guidance document [FDA-2017-D-6569]; 2019. Available at: www.fda.gov/regulatory-information/search-fda-guidance-documents/clinical-decision-support-software. Accessed August 5, 2020.
- 143.US Food & Drug Administration (FDA). 21st Century Cures Act; 2016. Available at: www.fda.gov/regulatory-information/selected-amendments-fdc-act/21st-century-cures-act. Accessed August 5, 2020.
- 144.US Food & Drug Administration (FDA). Changes to existing medical software policies resulting from section 3060 of the 21st Century Cures Act; 2019. Available at: www.fda.gov/regulatory-information/search-fda-guidance-documents/changes-existing-medical-software-policies-resulting-section-3060-21st-century-cures-act. Accessed August 5, 2020.
- 145.IMDRF. “Software as a Medical Device”: possible framework for risk categorization and corresponding considerations; 2014. Available at: www.imdrf.org/docs/imdrf/final/technical/imdrf-tech-140918-samd-framework-risk-categorization-141013.pdf. Accessed August 5, 2020.
- 146.US Food & Drug Administration (FDA). Use of real-world evidence to support regulatory decision-making for medical devices [FDA-2016-D-2153]; 2017. Available at: www.fda.gov/regulatory-information/search-fda-guidance-documents/use-real-world-evidence-support-regulatory-decision-making-medical-devices Accessed August 5, 2020.
- 147.Zarella MD, Bowman D, Aeffner F, et al. A practical guide to whole slide imaging: a white paper from the Digital Pathology Association. Arch Pathol Lab Med. 2019;143:222–234. [DOI] [PubMed] [Google Scholar]
- 148.Aeffner F, Zarella MD, Buchbinder N, et al. Introduction to digital image analysis in whole-slide imaging: a white paper from the Digital Pathology Association. J Pathol Inform. 2019;10:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Pantanowitz L, Sinard JH, Henricks WH, et al. Validating whole slide imaging for diagnostic purposes in pathology: guideline from the College of American Pathologists Pathology and Laboratory Quality Center. Arch Pathol Lab Med. 2013;137:1710–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Institute of Medicine (IOM) Clinical Practice Guidelines We Can Trust. Washington, DC: The National Academies Press; 2011. [Google Scholar]
- 151.Evans AJ, Salama ME, Henricks WH, et al. Implementation of whole slide imaging for clinical purposes: issues to consider from the perspective of early adopters. Arch Pathol Lab Med. 2017;141:944–959. [DOI] [PubMed] [Google Scholar]


