ABSTRACT
Macroautophagy/autophagy is a membrane-mediated intracellular degradation pathway, through which bulky cytoplasmic content is digested in lysosomes. How the autophagy initiation and maturation steps are regulated is not clear. In this study, we found an E3 ubiquitin ligase complex, linear ubiquitin chain assembly complex (LUBAC) and a deubiquitinating enzyme (DUB) OTULIN localize to the phagophore area to control autophagy initiation and maturation. LUBAC key component RNF31/HOIP translocates to the LC3 puncta area when autophagy is induced. RNF31 knockdown inhibits autophagy initiation, and cells are more sensitive to bacterial infection. OTULIN knockdown, however, promotes autophagy initiation but blocks autophagy maturation. In OTULIN knockdown cells, excessive ubiquitinated ATG13 protein was recruited to the phagophore for prolonged expansion, and therefore inhibits autophagosome maturation. Together, our study provides evidence that LUBAC and OTULIN cooperatively regulate autophagy initiation and autophagosome maturation by mediating the linear ubiquitination and the stabilization of ATG13.
Abbreviations: ATG: autophagy-related; CALCOCO2/NDP52: calcium binding and coiled-coil domain 2; CQ: chloroquine; CUL1-FBXL20: cullin 1-F-box and leucine rich repeat protein 20; CUL3-KLHL20: cullin 3-kelch like family member 20; CUL4-AMBRA1: cullin 4-autophagy and beclin 1 regulator 1; CYLD: CYLD lysine 63 deubiquitinase; DAPI: 4′,6-diamidino-2-phenylindole; DUB: deubiquitinating enzyme; EBSS: Earle’s Balanced Salt Solution; GFP: green fluorescent protein; GST: glutathione S-transferase; IKBKG/NEMO: inhibitor of nuclear factor kappa B kinase regulatory subunit gamma; LUBAC: linear ubiquitin chain assembly complex; MAP1LC3/LC3: microtubule-associated protein 1 light chain 3; MAP1LC3B/LC3B: microtubule-associated protein 1 light chain 3B; MIM: MIT-interacting motif; mRFP: monomeric red fluorescent protein; NEDD4: NEDD4 E3 ubiquitin protein ligase; NFKB: NF-kappaB complex; OPTN: optineurin; OTULIN: OTU deubiquitinase with linear linkage specificity; PIK3C3/Vps34: phosphatidylinositol 3-kinase catalytic subunit type 3; PtdIns: phosphatidylinositol; PtdIns3K: class III phosphatidylinositol 3-kinase complex; PtdIns3P: phosphatidylinositol 3-phosphate; RBCK1/HOIL1: RANBP2-type and C3HC4-type zinc finger containing 1; RB1CC1/FIP200: RB1-inducible coiled-coil 1; RIPK1: receptor interacting serine/threonine kinase 1; RNF216: ring finger protein 216; RNF31/HOIP: ring finger protein 31; RT-PCR: reverse transcriptase polymerase chain reaction; S. Typhimurium: Salmonella enterica serovar Typhimurium; SHARPIN: SHANK associated RH domain interactor; SMURF1: SMAD specific E3 ubiquitin protein ligase 1; SQSTM1: sequestosome 1; STING: stimulator of interferon response cGAMP interactor 1; STUB1/CHIP: STIP1 homology and U-box containing protein 1; TNF/TNF-alpha: tumor necrosis factor; TNFAIP3/A20: TNF alpha induced protein 3; TRAF6: TNF receptor associated factor 6; TRIM32: tripartite motif containing 32; UBAN: ubiquitin binding in TNIP/ABIN and IKBKG/NEMO proteins; ULK1/2: unc-51 like autophagy activating kinase 1/2; USP: ubiquitin specific peptidase; UVRAG: UV radiation resistance associated; VCPIP1: valosin containing protein interacting protein 1; WIPI2: WD repeat domain, phosphoinositide interacting protein 2; ZBTB16-CUL3-RBX1: zinc finger and BTB domain containing protein 16-cullin 3-ring-box 1; ZRANB1: zinc finger RANBP2-type containing 1.
KEYWORDS: ATG13, autophagosome, autophagy, linear ubiquitination, LUBAC, OTULIN, RNF31
Introduction
Autophagy is a self-eating system that is essential for cellular homeostasis [1–3]. Numerous literature implied dysregulation of autophagy is linked to various diseases such as neurodegenerative diseases, inflammatory disorders and cancers [4–7]. Autophagy is characterized by the formation of double-membrane vesicles called autophagosomes, which engulf the cytoplasmic contents to lysosomes for destruction [1,3,8]. It is a multistep process governed by multiple complexes. The ULK1/2 complex consists of the serine/threonine kinase ULK1/2, the noncatalytic subunits ATG13, RB1CC1/FIP200, and ATG101 [9–11]. Stressed conditions, such as nutrition depletion that activates AMP-activated protein kinase (AMPK) and inactivates MTOR complex 1 (mechanistic target of rapamycin complex 1), result in the activation and pre-assembly of ULK1/2 complex [10,12]. Specifically, ULK1 complex activity is also regulated by non-degradative ubiquitination mediated by TRAF6 and unanchored ubiquitination mediated by TRIM32 [13,14]. The activated ULK1/2 complex then phosphorylates multiple downstream substrates to initiate autophagy. The activated class III phosphatidylinositol 3-kinase (PtdIns3K) complex then produces PtdIns3P on the PtdIns-enriched membrane to generate a platform for the downstream effectors to bind and initiate autophagy [8,15]. Despite the extensive studies on the mechanism of autophagy, it remains ambiguous how the autophagy initiation and autophagosome formation are regulated.
Protein ubiquitination is accomplished by a cascade of enzymes and was originally found to target proteins for degradation [16]. Non-degradative ubiquitination also creates the versatility in cell signaling and regulates numerous cellular processes [17,18]. Protein could be mono-ubiquitinated or poly-ubiquitinated with eight linkage types either homotypic (one linkage type) or heterotypic [18]. One of the linkage type, methionine-1(M1)-linked linear ubiquitin chains have been found to regulate the activation of NFKB, immune homeostasis, and the response to infection [19–21].
LUBAC is the only ubiquitin E3 ligase complex to date that has been found to ubiquitinate substrate in a linear form. It is composed of three subunits: the core ubiquitin ligase RNF31/HOIP, the regulatory proteins RBCK1/HOIL1 and SHARPIN [22]. Until now, the identified substrates of LUBAC, including IKBKG/NEMO and RIPK1, are all involved in inflammation and immune signaling [23–25]. LUBAC usually teams up with the deubiquitinating enzymes OTULIN, CYLD and TNFAIP3/A20 to regulate the linear ubiquitination status on the substrates [26–29]. Dysregulation of linear ubiquitin is linked to a number of severe physiologies [30,31]. It remains unclear whether LUBAC and OTULIN are involved in other pathways in addition to inflammation.
Here, by screening the ubiquitin enzymes that functioning in autophagy, we identified a DUB OTULIN, which inhibits autophagy initiation and promotes ATG13 degradation. We found LUBAC is the E3 ligase responsible for ubiquitination of ATG13 as well as recruiting ATG13 to phagophore. Proper amount of ubiquitinated ATG13 on phagophore is essential for autophagy initiation, but prolonged presence of it impedes autophagosome maturation. Cells with RNF31 knockdown have no autophagy initiated and are more sensitive to bacterial infection. In summary, we found that LUBAC and OTULIN together mediate autophagy initiation and maturation by controlling the linear ubiquitination and stabilization of ATG13.
Results
OTULIN is required for autophagosome formation
To comprehensively understand how DUBs function in autophagy pathway, we overexpressed 55 different DUBs in HeLa cells stably expressing mRFP-EGFP-LC3B, respectively, and screened those that affect LC3 puncta formation [32]. mRFP-EGFP-LC3B is a widely used autophagic indicator with yellow LC3 signal when in the autophagosome (with both GFP and RFP signals) and red LC3 signal when in the autolysosome due to the acidic milieu that quenches GFP signal [33]. Interestingly, when OTULIN was overexpressed, LC3B puncta formation was significantly inhibited under both normal and starvation conditions (Figure 1(A,B and S1A). The catalytic inactivation of OTULINC129S had less influence on the LC3B pattern compared to the wild type OTULIN (Figure 1A,B and S1A). Intriguingly, OTULIN knockdown caused dramatically increase of the yellow LC3B puncta (incomplete autophagic flux) number under both normal and starvation conditions (Figure 1C,D and S1B). The size of both green and red puncta in OTULIN knockdown cells was also increased compared to that of the control (Figure 1A,B and S1A). Interestingly, the YFP/RFP ratio was increased in OTULIN knockdown cells, indicating the blockage of autophagic flux from autophagosome to autolysosome (Figure 1D). The western blot result showed the conversion of LC3B-I to LC3B-II and the accumulation of SQSTM1 in OTULIN knockdown cells (Figure 1E). These data suggested that OTULIN may function at the autophagy initiation to maturation stages.
Figure 1.
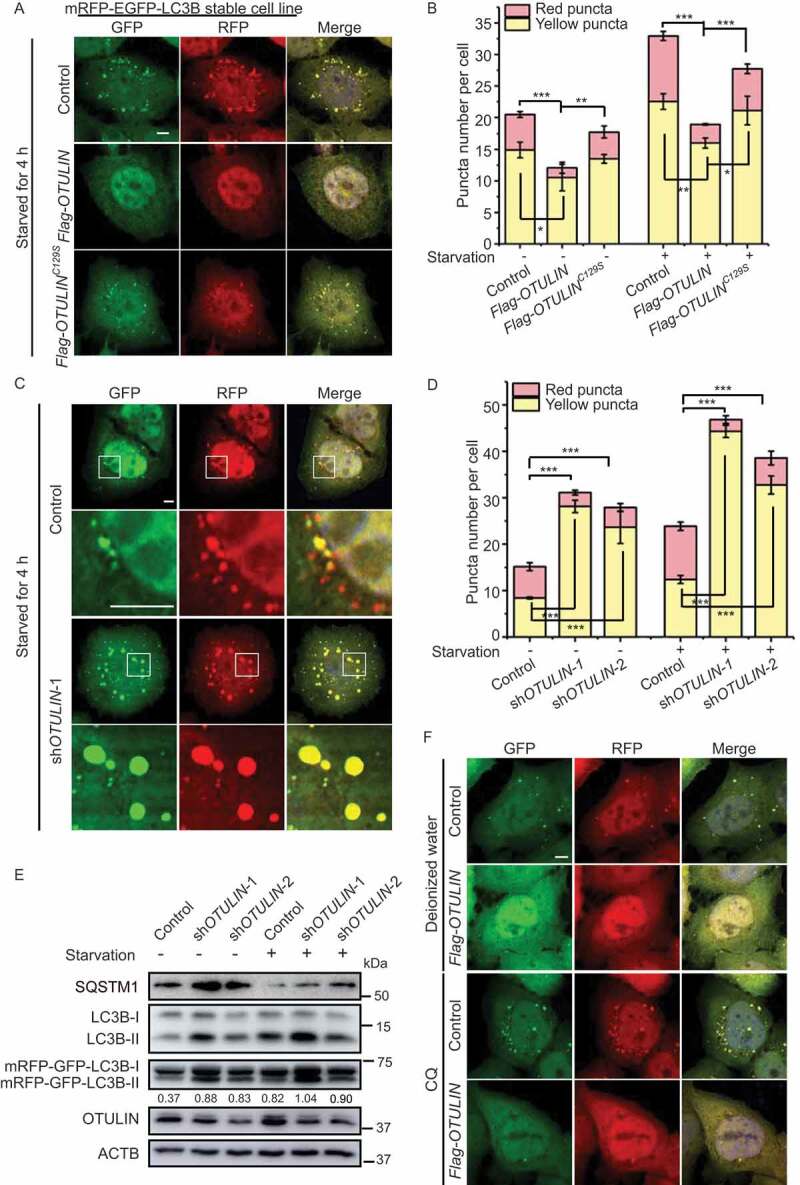
OTULIN is essential for autophagosome formation. (A) The number of LC3B-positive puncta is less in OTULIN-overexpressing cells under starvation conditions. HeLa cells stably expressing mRFP-EGFP-LC3B were transfected with either the vector control or Flag-OTULIN or Flag-OTULINC129S and treated with EBSS for 4 h. Cells were fixed and stained with DAPI (blue) and representative fluorescence images of LC3B puncta are shown. Scale bar: 5 μm. (B) Quantification of yellow (RFP+GFP+) and red (RFP+GFP−) puncta per cell as represented in Figure 1A and S1A. Data are mean ± SD from three independent experiments. *p < 0.05; **p < 0.01; ***p < 0.001 (one-way ANOVA). (C) Depletion of OTULIN induces the aggregation of the LC3B-positive puncta in cells under starvation conditions. HeLa cells stably expressing mRFP-EGFP-LC3B were transfected with either the control shRNA or OTULIN shRNA-1 or shRNA-2, and treated with EBSS for 4 h. Cells were fixed and stained with DAPI (blue) and representative fluorescence images of OTULIN shRNA-1 are shown. The enlarged images of the white square part under each condition are shown in the following pictures. Scale bar: 5 μm. (D) Quantification of yellow (RFP+GFP+) and red (RFP+GFP−) puncta per cell as represented in Figure 1C and S1B. Data are mean ± SD from three independent experiments. ***p < 0.001 (one-way ANOVA). (E) Western blot analysis of the knockdown efficiency of OTULIN as represented in Figure 1C and S1B. Protein SQSTM1 and endogenous LC3B were also examined. The numbers are the ratio of LC3B-II/I. (F) OTULIN inhibits LC3B-positive puncta formation. HeLa cells stably expressing mRFP-EGFP-LC3B were transfected with either the vector control or Flag-OTULIN, and treated with H2O (solvent control) or CQ (20 nM) or CQ (20 nM) + EBSS for 4 h. Cells were fixed and stained with DAPI (blue) and representative fluorescence images of cells treated with H2O and CQ are shown. Scale bar: 5 μm
The accumulation of LC3B puncta could indicate either autophagic activation or blockage of downstream steps in autophagy. Next, we tried to determine in which step of autophagy OTULIN participates. Chloroquine (CQ) inhibits autophagic flux by decreasing autophagosome and lysosome fusion. Therefore, administration of CQ to the cells will increase the ratio of yellow puncta (incomplete autophagic flux) to red puncta (complete autophagic flux). Upon CQ treatment, both the control and OTULIN-overexpressing cells increased the number of yellow puncta (Figure 1F). Nutrition starvation plus CQ treatment in the control cells increased the number of yellow LC3B puncta, whereas the same treatment in the OTULIN-overexpressing cells had only a limited increase in the number of yellow LC3B puncta (Figure S1D). Western blot result showed that OTULIN blocked the conversion of LC3B-I to LC3B-II under all the conditions (Figure S1E). These data further verified that OTULIN plays a role in autophagy before the fusion step.
Given that autophagic flux is blocked and the size of LC3B puncta is increased in OTULIN knockdown cells, we then examined which stage the enlarged puncta represent. WIPI2 functions as a docking site that links LC3 conjugation machinery with PtdIns3P, and therefore is a marker for growing phagophore. Interestingly, we observed that WIPI2 and the lipidation machinery component ATG16L1 were both colocalized with the enlarged LC3B puncta in OTULIN knockdown cells (Figure S1F and S1G). We also did the experiment to see whether the enlarged LC3B puncta are associated with ER. We used ER-localized SEC61B to indicate the ER structure. The result showed that the enlarged LC3B- and WIPI2-positive puncta were within the ER network (Figure S1H). These results suggested that the enlarged LC3B puncta in OTULIN knockdown cells are still in the process of expansion and have not reached the mature stage. Together, these experiments indicated that OTULIN is required for autophagosome formation.
OTULIN knockdown increases ATG13 ubiquitination level and stabilizes ATG13 protein
To explore how OTULIN regulates autophagy, we employed the Flag pulldown assay to screen the interaction of OTULIN with a collection of autophagy-related proteins. These proteins include ULK1, ATG13, RB1CC1, ATG101, BECN1/beclin 1, ATG14, ATG5 and LC3B, with at least one component in each functional complex that participates in autophagy pathway. The result showed that Flag-OTULIN clearly interacted with ATG13 and ATG101 (Figure S2A-H). Given that ULK1 complex is composed of multiple components including ULK1, ATG13, RB1CC1, and ATG101, we could not exclude that OTULIN may also interact with ULK1 and RB1CC1, which were not detected by the pulldown assay. Since OTULIN is a DUB, we asked whether ATG13 and ATG101 are substrates of OTULIN. ATG13 protein level was noticeably reduced under OTULIN-overexpressing condition (Figure S2A). The GST affinity-isolation assay also verified that there was a direct interaction between OTULIN and ATG13 (Figure 2A). We, therefore, speculated ATG13 might be the substrate of OTULIN.
Figure 2.
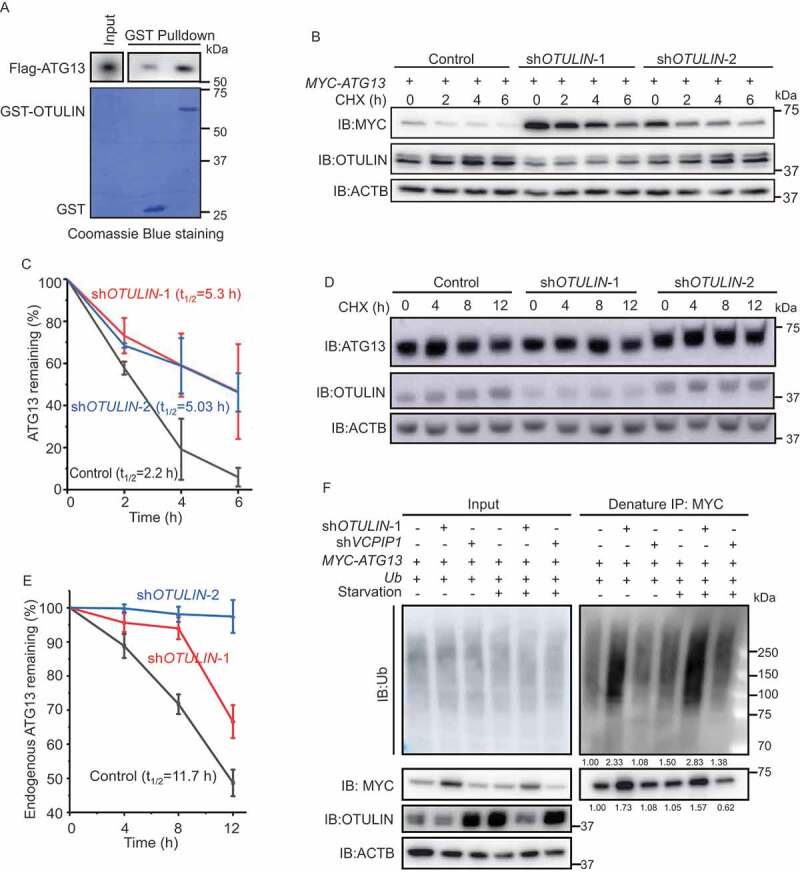
OTULIN knockdown stabilizes ATG13 protein level. (A) OTULIN binds ATG13 directly. The indicated GST or GST-OTULIN proteins were immobilized and incubated with recombinant Flag-ATG13. The precipitated proteins were analyzed by immunoblotting with an anti-Flag antibody. The Coomassie Brilliant Blue-stained gel shows the equivalent amount of GST and GST-OTULIN used for the affinity-isolation assay. (B and D) Cycloheximide (CHX) chase experiments to analyze the degradation rate of MYC-ATG13 (B) and endogenous ATG13 (D) under OTULIN knockdown conditions. Anti-MYC (B) or anti-ATG13 (D) immunoblotting was used to detect the protein level of ATG13. The degradation rate was blocked under OTULIN knockdown conditions. ACTB was used as the loading control. (C and E) Statistical analysis of the degradation rate of MYC-ATG13 or ATG13 in (B and D). Data are mean ± SD from three independent experiments. (F) Depletion of OTULIN results in accumulated ATG13 protein and ATG13-associated ubiquitin signal. HEK293 FT cells were transfected with MYC-ATG13, untagged ubiquitin, along with the control shRNA, or OTULIN shRNA-1, or the negative control VCPIP1 shRNA. Cells were treated with DMEM or EBSS for 4 h prior to lysis. Ubiquitinated proteins were immunoprecipitated with anti-MYC beads under denaturing condition and immunoblotted with an anti-ubiquitin antibody
To explore this possibility, we first performed cycloheximide chase experiments. The results showed that the degradation rate of either overexpressed MYC-ATG13 or endogenous ATG13 protein was markedly inhibited under OTULIN knockdown condition compared to the control (Figure 2B-E, S2I and S2J). The immunofluorescence images showed that the accumulated ATG13 was colocalized well with the LC3B puncta in OTULIN knockdown cells (Figure S2K and S2L). Interestingly, we detected that the enlarged LC3B puncta were also colocalized with SQSTM1 and ubiquitin in OTULIN knockdown cells under both normal and starvation conditions (Figure S2M and S2N). These data implied that OTULIN might control ATG13 protein stability by regulating the ubiquitination status on ATG13.
To validate this assumption, we performed an immunoprecipitation assay under denaturing condition to examine ATG13 ubiquitination in OTULIN knockdown cells. According to the previous literature, OTULIN is a DUB that has been implicated in TNF-induced inflammation pathway and usually cleaves linear ubiquitin from the substrates. Interestingly, OTULIN knockdown caused intensive accumulation of ATG13 protein as well as the conjugated ubiquitin under both normal and starvation conditions (Figure 2F, MYC blots and ubiquitin blots). We noticed that the increase of ATG13 conjugated ubiquitin was about 1.5-fold higher than that of ATG13 protein level in OTULIN knockdown cells (Figure 2F). Therefore, we proposed that OTULIN knockdown increases ATG13 linear ubiquitination level and stabilizes ATG13 protein.
LUBAC linearly ubiquitinates and stabilizes ATG13
Previous studies reported that LUBAC is the E3 complex that mediates the linear ubiquitination of substrates and is antagonized by OTULIN (Figure S3A) [26,27]. To find out whether LUBAC works on ATG13, we first analyzed the interaction between LUBAC and ATG13. The LUBAC complex is composed of three subunits: RBCK1, SHARPIN, and the catalytically active subunit RNF31 (Figure S3A) [22,24]. We first overexpressed Flag-tagged RNF31 in cells and examined whether the protein interacts with MYC-ATG13. Flag-RNF31 may form a functional LUBAC complex with the endogenous RBCK1 and SHARPIN. The Flag pulldown assay showed that Flag-RNF31 interacted with MYC-ATG13 (Figure 3A). It was reported that the N-terminally truncated form of RNF31 (RNF31Δ1-698) is sufficient to form linear ubiquitin chains in vitro [34]. Next, we tested whether RNF31Δ1-698 directly binds to ATG13 in vitro. As expected, when we used bacterial expressed Flag-ATG13 to pulldown HA-RNF31Δ1-698, we could detect a good amount of HA-RNF31Δ1-698 in the pulldown fraction (Figure 3B). Consistent with these results, co-immunoprecipitation assay using anti-ATG13 antibody verified the interaction between the endogenous ATG13 and RNF31 (Figure 3C).
Figure 3.
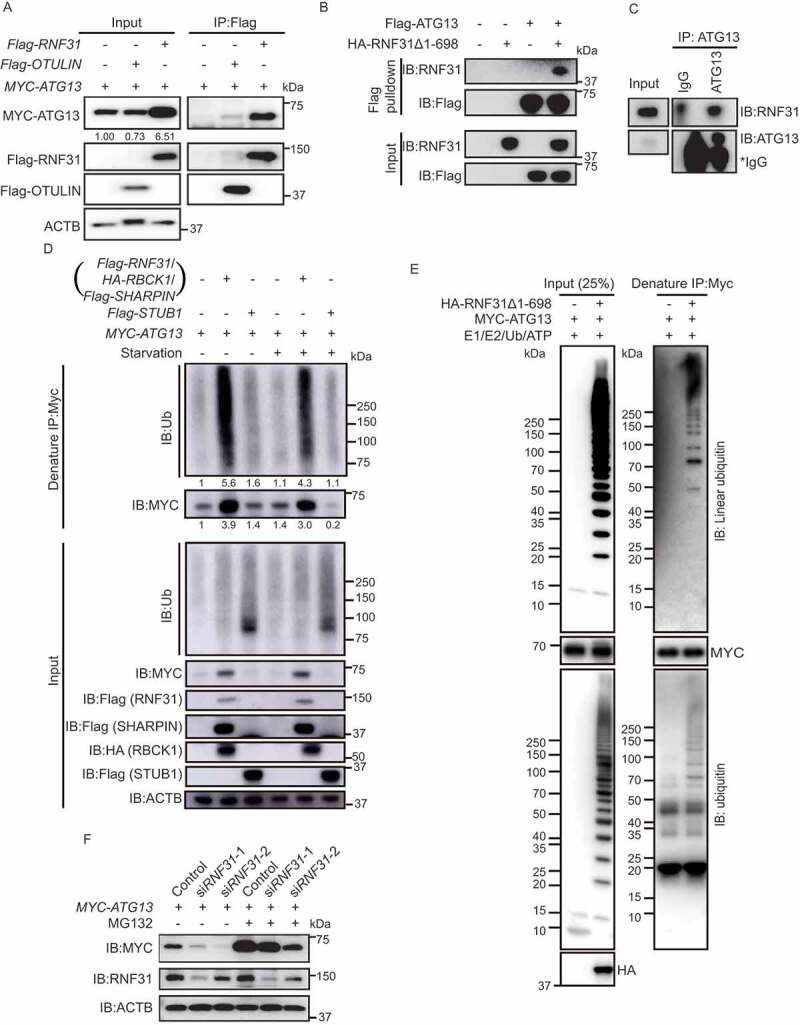
LUBAC linearly ubiquitinates ATG13 and stabilizes ATG13 protein level. (A) Flag-RNF31 and Flag-OTULIN interact with MYC-ATG13. HEK293 FT cells were transfected with MYC-ATG13 along with Flag-RNF31 or Flag-OTULIN. Flag pulldown was performed using anti-Flag beads and the samples were analyzed by immunoblotting with the indicated antibodies. (B) ATG13 interacts with RNF31 in vitro. Bacterial purified Flag-ATG13 was used to pulldown recombinant HA-RNF31Δ1-698 with anti-Flag beads. The precipitated proteins were analyzed by immunoblotting with the indicated antibodies. (C) Endogenous ATG13 interacts with RNF31 in cells. Immunoprecipitation using either IgG or anti-ATG13 was performed. The co-immunoprecipitated RNF31 was immunoblotted with an anti-RNF31 antibody. (D) MYC-ATG13 is ubiquitinated by LUBAC. HEK293 FT cells were transfected with MYC-ATG13 along with the LUBAC complex or Flag-STUB1 and were treated with DMEM or EBSS for 4 h. Ubiquitinated proteins were immunoprecipitated with anti-MYC beads under denaturing condition and immunoblotted with an anti-ubiquitin antibody. (E) In vitro ubiquitination of mammalian MYC-ATG13 by HA-RNF31Δ1-698. HEK293 FT cells were first transfected with MYC-ATG13. Immunoprecipitation of MYC-ATG13 was then performed with anti-MYC beads. The precipitated MYC beads were incubated with E1-GST, His-UBE2D2, ubiquitin, and HA-RNF31Δ1-698 for 1 h at 37°C. The reaction samples were immunoprecipitated with anti-MYC beads under denaturing conditions. Antibodies against ubiquitin and linear ubiquitin were used to detect the ubiquitination type on MYC-ATG13. (F) MYC-ATG13 protein level is decreased in RNF31 knockdown cells. HEK293 FT cells were transfected with MYC-ATG13, along with the control siRNA or RNF31 siRNA-1 or siRNA-2 and treated with DMSO or MG132 (10 μM) for 15 h. Samples were analyzed by immunoblotting with the indicated antibodies. ACTB was used as the loading control
To see whether RNF31 ubiquitinates ATG13 in cells, we performed an immunoprecipitation assay under denaturing condition. The result showed that wild type RNF31, but not RNF31C885S [34], caused an increase in ATG13 protein level as well as the conjugated ubiquitin signal (Figure S3B). The increase of the conjugated ubiquitin signal is about 2-fold more than that of ATG13 protein level in RNF31-overexpressing cells. To precisely examine whether the whole LUBAC complex mediates the ubiquitination of ATG13, we expressed all three subunits RNF31, RBCK1 and SHARPIN in cells. The denaturing immunoprecipitation showed that both ATG13 protein level and the conjugated ubiquitin signal were dramatically accumulated (Figure 3D). This is the same result as that of expressing RNF31 alone. Therefore, we expressed RNF31 alone to represent the whole LUBAC complex for the assays in cell and used the truncated RNF31 for the in vitro assay. As a negative control, overexpressing another E3 ligase STUB1/CHIP did not alter ATG13 protein level, neither the conjugated ubiquitin signal (Figure 3D). This indicates that RNF31 specifically functions on ATG13. We also performed an in vitro ubiquitination assay by combining E1, E2 (UBE2D2/UbcH5B) [22], HA-RNF31Δ1-698, and untagged ubiquitin together with mammalian purified MYC-ATG13. After the reaction, a MYC affinity-isolation assay under denaturing condition was performed. The immunoblot result showed that there was a good amount of linear ubiquitin signal conjugated on MYC-ATG13 (Figure 3E). Interestingly, bacteria-purified Flag-ATG13 was not able to pulldown ubiquitin chain in our in vitro assay (data not shown). These data suggested that the mammalian cell expressing ATG13 may have acquired some post-translational modifications before being ubiquitinated. Together, these data indicated that RNF31 linearly ubiquitinates ATG13 and stabilizes ATG13 protein level.
Interestingly, MYC-ATG13 protein level was significantly decreased in RNF31 knockdown cells, and the reduction could be blocked by MG132 treatment (Figure 3F). We also performed a cycloheximide chase experiment and found that the degradation rate of MYC-ATG13 is strikingly inhibited under RNF31-overexpressing condition (Figure S3C and S3D). We further compared the protein level change versus mRNA level change of ATG13 and found that the mRNA of ATG13 was not significantly changed in RNF31-overexpressing or OTULIN knockdown cells, but the protein level was highly accumulated (Figure S3E-S3H). These data suggested that RNF31 recruits and stabilizes ATG13 from being degraded.
RNF31 and OTULIN together regulate ATG13 protein level and the autophagic flux
To understand whether RNF31 and OTULIN antagonize each other to regulate ATG13 in cells, we then performed an immunoprecipitation under denaturing condition. The ATG13 protein level and its conjugated ubiquitin signal were decreased in RNF31 single knockdown but increased in OTULIN single knockdown cells (Figure 4A). The ATG13 protein level and the conjugated ubiquitin signal in RNF31 and OTULIN double knockdown cells were slightly decreased compared to those in OTULIN knockdown cells, suggesting the antagonizing role of RNF31 and OTULIN (Figure 4A). When RNF31 knockdown and OTULIN overexpression were combined, the ATG13 protein and the conjugated ubiquitin were decreased compared to those in the control, whereas when RNF31 overexpression and OTULIN knockdown were combined, the ATG13 protein level and the conjugated ubiquitin signal were dramatically increased (Figure 4A). Nutrition starvation treatment resulted in a similar pattern (Figure 4A). These data indicate that RNF31 and OTULIN together regulate ATG13 protein level as well as the ubiquitination signal on ATG13.
Figure 4.
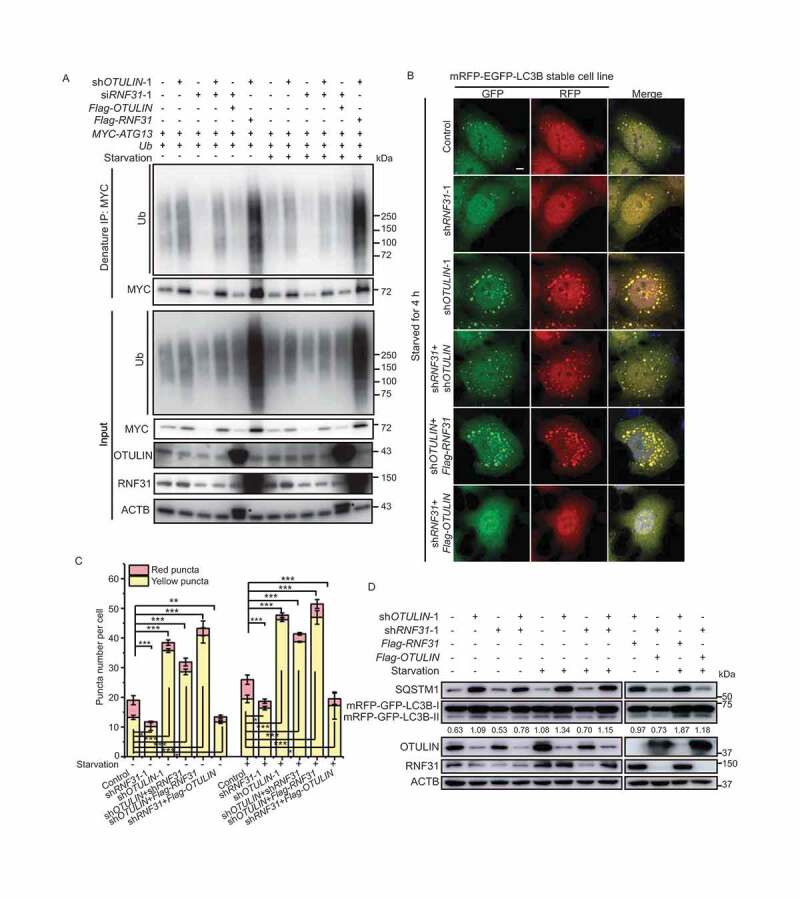
RNF31 and OTULIN balance ATG13 protein level and the autophagic flux. (A) MYC-ATG13 ubiquitination under different conditions. HEK293 FT cells were transfected with MYC-ATG13 and untagged ubiquitin, along with the control, or RNF31 siRNA-1, or OTULIN shRNA-1, or (OTULIN shRNA-1 + RNF31 siRNA-1), or (RNF31 siRNA-1 + Flag-OTULIN), or (OTULIN shRNA-1 + Flag-RNF31) and treated with DMEM or EBSS for 4 h. Ubiquitinated proteins were immunoprecipitated with anti-MYC beads under denaturing conditions and immunoblotted with an anti-ubiquitin antibody. (B) LC3B puncta formation was analyzed in HeLa cells stably expressing mRFP-EGFP-LC3B. Cell were transfected with the control, or RNF31 shRNA-1, or OTULIN shRNA-1, or (OTULIN shRNA-1 + RNF31 shRNA-1), or (OTULIN shRNA-1 + Flag-RNF31), or (RNF31 shRNA-1 + Flag-OTULIN). Cells were treated with EBSS for 4 h. Cells were fixed and stained with DAPI (blue) and representative fluorescence images are shown. Scale bar: 5 μm. (C) Statistical analysis of the number of yellow (RFP+GFP+) and red (RFP+GFP−) puncta per cell as represented in (B) and Figure S4. Data are mean ± SD from three independent experiments. *p < 0.05; **p < 0.01; ***p < 0.001 (one-way ANOVA). (D) Western blot analysis of the samples as represented in (B) and Figure S4. The knockdown efficiency and protein overexpression were shown. Samples were also immunoblotted with the indicated antibodies. The numbers are the relative ratio of LC3B-II/I
We also examined the LC3B pattern in HeLa cells stably expressing mRFP-EGFP-LC3B under different conditions. Under both normal and starvation conditions, we found that less LC3B puncta formed in RNF31 knockdown cells (Figure 4B,C and S4), implying that RNF31 is required for autophagy initiation. The LC3B puncta number in RNF31 and OTULIN double knockdown cells was less than that in OTULIN single knockdown cells though it was still more than that in the control (Figure 4B,C and S4), suggesting OTULIN may work downstream of RNF31 in autophagy pathway. When OTULIN knockdown and RNF31 overexpression were combined, we observed a huge accumulation of LC3B puncta (Figure 4B,C and S4). In contrast, when OTULIN overexpression and RNF31 knockdown were combined, there was a significant decrease of LC3B puncta (Figure 4B,C and S4). Nutrition starvation showed a similar pattern, albeit with an increase of the LC3B puncta number generally (Figure 4B,C and S4). The immunoblotting result showed that double knockdown of OTULIN and RNF31 slightly reduced the conversion of LC3B-I to LC3B-II and SQSTM1 level compared to the OTULIN single knockdown (Figure 4D). These data suggested RNF31 and OTULIN together balance the autophagic flux.
RNF31 ubiquitinates ATG13 on multiple lysine residues
To determine which domain of ATG13 interacts with RNF31, we generated a series of MYC-tagged ATG13 truncations and examined their ability to interact with RNF31. Alignment of ATG13 proteins from different species showed that there is a conserved HORMA domain at the N-terminus (Figure S5A) [35]. Besides HORMA domain, yeast Atg13 also has an MIM motif in the C-terminal intrinsically disordered region, responsible for interacting with Atg1 [36]. The alignment showed that human ATG13 MIM motif was located in the region of 383–432 amino acid residues (Figure S5A). The truncation mutants were generated based on the above information. The mapping result showed that RNF31 interacted with all the truncation mutants (Figure 5A). There are a total of 22 lysine residues in ATG13 protein sequence, including 14 in the HORMA domain, and eight in the C-terminal region (Figure S5B, blue colored). We generated constructs mutating either the 14 lysine residues in the HORMA domain, or the eight lysine residues in the C terminal region, or the total 22 lysine residues, and designated as ATG1314KR, ATG138KR, and ATG1322KR, respectively. The immunoprecipitation with anti-MYC antibody under denaturing condition was then performed. The result showed that all ATG13 proteins, both wild type and mutants, were stabilized when co-expressed with RNF31, albeit with different levels. However, compared to wild type ATG13, all the mutants were less ubiquitinated, with ATG1322KR being the least (Figure 5B). Therefore, we concluded that RNF31 ubiquitinates ATG13 on multiple lysine residues.
Figure 5.
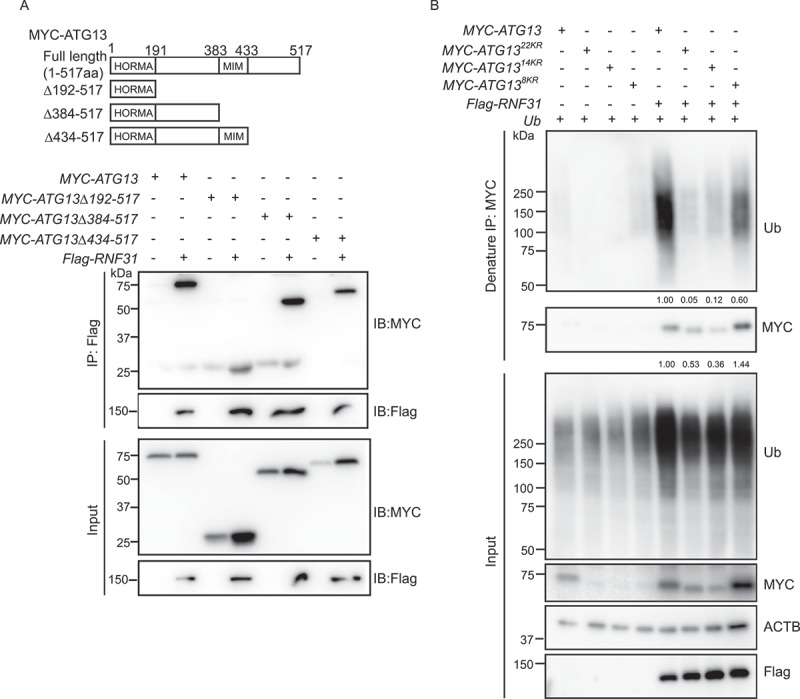
RNF31 ubiquitinates ATG13 on multiple lysine residues. (A) Interaction of ATG13 or its truncation mutants with RNF31. ATG13 truncation constructs were generated based on the domain information. 1–191aa was the HORMA domain. 383–433aa was the MIM motif. HEK293 FT cells were transfected with Flag-RNF31 and MYC-ATG13 or the indicated truncation mutants. Flag pulldown was performed with anti-Flag beads. Samples were analyzed by immunoblotting with the indicated antibodies. (B) Ubiquitination of MYC-ATG13 and its mutants in RNF31-overexpressing cells. HEK293 FT cells were transfected with MYC-ATG13 or the indicated mutants and untagged ubiquitin, along with the control or Flag-RNF31. Ubiquitinated proteins were immunoprecipitated with anti-MYC beads under denaturing conditions and samples were analyzed by immunoblotting with the indicated antibodies
RNF31 and OTULIN are partially colocalized with LC3B puncta for autophagosome formation
To understand whether RNF31 and OTULIN directly regulate autophagosome formation, we first analyzed the subcellular localization of the two enzymes. Under normal conditions, when Flag-OTULIN or Flag-RNF31 was expressed in HeLa cells, we observed that both enzymes were closely localized with LC3B puncta. Slightly higher number of colocalization was observed under starvation conditions (Figure 6A-C). These results indicated that proper amount of the enzymes is required for autophagosome formation. We further performed a time-course analysis of RNF31 subcellular localization when autophagy was induced. HeLa cells stably expressing mRFP-EGFP-LC3B were cultured in the starvation medium and the endogenous RNF31 signal was traced. As time elapses, more RNF31 translocated to the puncta area, with the highest amount of colocalization at 45 min after autophagy was induced (Figure S6A and S6B). These data suggested that RNF31 and OTULIN together regulate autophagosome formation.
Figure 6.
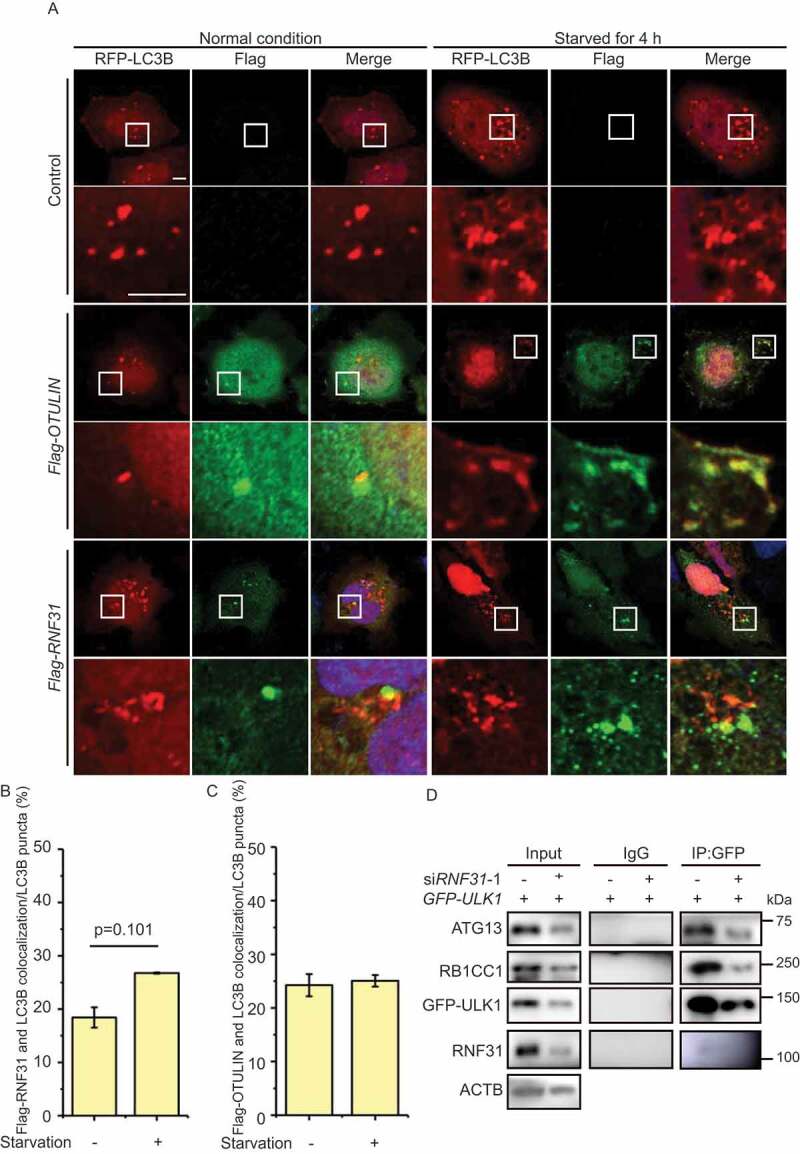
RNF31 and OTULIN are partially colocalized with LC3B puncta. (A) Flag-RNF31 and Flag-OTULIN are closely localized with LC3B puncta. HeLa cells were transfected with RFP-LC3B, along with the control vector, or Flag-RNF31, or Flag-OTULIN, and then treated with normal medium or EBSS for 4 h. Cells were fixed and stained for Flag (green) and DAPI (blue) and representative fluorescence images are shown. The enlarged images of the white square part under each condition are shown in the following pictures. Scale bar: 5 μm. (B and C) Percentage of Flag-RNF31 or Flag-OTULIN colocalization with LC3B as represented in (A). Data are mean ± SD from three independent experiments. (D) The protein levels of ATG13, ULK1, and RB1CC1 are decreased under RNF31 knockdown condition. Less ATG13 and RB1CC1 were pulled down by GFP-ULK1 under RNF31 knockdown condition. HEK293 FT cells were transfected with GFP-ULK1, along with the control siRNA or RNF31 siRNA-1. GFP immunoprecipitation was performed and samples were analyzed by immunoblotting with the indicated antibodies
Interestingly, when we knocked down RNF31, we found the components of ULK1 complex, including ULK1, RB1CC1 were all decreased, consistent with the result that RNF31 knockdown inhibits autophagy initiation (Figure 6D). Therefore, ATG13 stabilization by RNF31 is required for the proper function of ULK1 complex. Interestingly, RNF31 seems to only function on ATG13, for RNF31 does not ubiquitinate ULK1, RB1CC1 or ATG101 (Figure S6C-E).
RNF31 knockdown inhibits xenophagy initiation and promotes cell death
To further explore the physiological function of LUBAC and OTULIN in autophagy, we generated a cell-based Salmonella infection model. To determine whether autophagy is activated by RNF31, we evaluated the co-localization of the two xenophagy receptors CALCOCO2/NDP52 and OPTN, together with LC3B and invasive Salmonella enterica serovar Typhimurium (S. Typhimurium). Compared to the control cells, there was a smaller number of CALCOCO2 and LC3B puncta, and less CALCOCO2 and LC3B surrounded S. Typhimurium in RNF31 knockdown cells (Figure 7A,C). The number of the OPTN signal was not reduced obviously in RNF31 knockdown cells, but that of the colocalization of OPTN and LC3B together with S. Typhimurium was noticeably reduced (Figure 7B,D). Consistent with reduced xenophagy in RNF31 knockdown, the overall fold replication of S. Typhimurium in RNF31 knockdown was significantly increased (Figure 7E). These data suggested that RNF31 knockdown inhibits autophagy initiation, and cells are more sensitive to bacterial infection.
Figure 7.
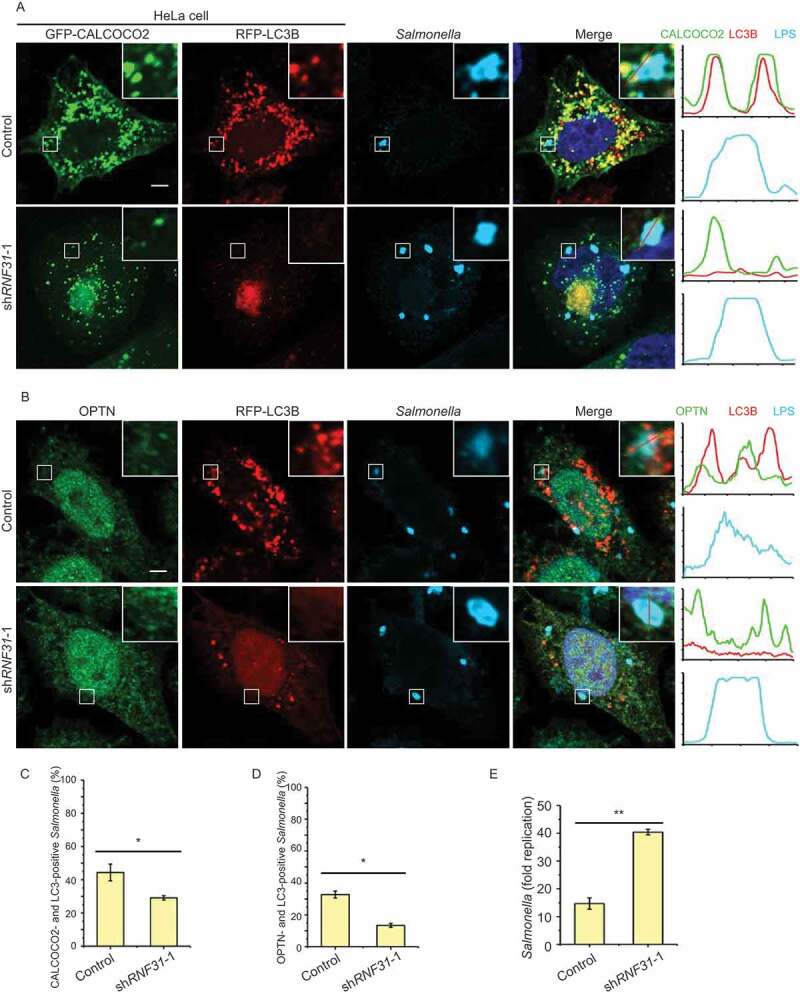
RNF31 knockdown inhibits xenophagy initiation and promotes cell death. (A) Confocal image of CALCOCO2, LC3B and infected S. Typhimurium colocalization in HeLa cells. Cell were transfected with GFP-CALCOCO2, RFP-LC3B, along with control shRNA or RNF31 shRNA-1. Cells were fixed at 3 h post-infection and stained for LPS (S. Typhimurium). Representative fluorescence images are shown. Line graphs show fluorescence plots along indicated lines. Scale bar: 5 μm. (B) Confocal image of OPTN, LC3B and infected S. Typhimurium colocalization in HeLa cells. Experimental procedures were the same as that in (A) except cells were transfected with RFP-LC3B along with the control shRNA or RNF31 shRNA-1 and cells were stained for endogenous OPTN and LPS (S. Typhimurium). Scale bar: 5 μm. (C and D) Percentage of CALCOCO2- and LC3B-positive S. Typhimurium as represented in (A). Percentage of OPTN- and LC3B-positive S. Typhimurium as represented in (B). At least 80 bacteria per cover slide were counted. Data are mean ± SD, n = 5 images (1 cover slide from three independent experiments). *p < 0.05 (Student’s t-test). (E) Fold replication of S. Typhimurium in the control and RNF31 shRNA-1 knockdown cells. Bacteria were counted based on the colony-forming unit assay. Data are depicted as fold replication of S. Typhimurium at 8 h versus 2 h post-infection (mean ± SD; n = 3). **p < 0.01 (Student’s t-test)
On the other hand, as shown previously, OTULIN knockdown blocks autophagic flux and increases the number of enlarged LC3B puncta. To examine whether the enlarged LC3B puncta are functional autophagosomes, we utilized the xenophagy model to examine the capability of autophagy in OTULIN knockdown cells. When the cells were infected with S. Typhimurium, fewer bacteria were wrapped by the enlarged LC3B puncta under OTULIN knockdown condition (Figure S7A and S7B), indicating that the enlarged puncta are not functional for engulfing the substrates. Altogether, we concluded that RNF31 and OTULIN cooperatively regulate autophagy initiation and maturation steps, and RNF31 is required for autophagy initiation and OTULIN is necessary for autophagosome maturation.
Discussion
In this study, we discovered a novel regulatory role of linear ubiquitination in autophagy and identified a new substrate ATG13 of LUBAC and OTULIN. ATG13 is a key component in ULK1 complex and is responsible for activating ULK1 kinase activity. Therefore, ATG13 protein level is important for autophagy initiation and autophagosome formation. We demonstrated that proper linear ubiquitination on ATG13 by LUBAC key component RNF31 stabilizes the protein from being degraded by the proteasome. OTULIN is a DUB that cleaves linear ubiquitin from ATG13 to balance ATG13 protein level in cells. Our data showed RNF31 linearly ubiquitinates ATG13 and stabilizes ATG13 protein to activate autophagy. Therefore, a model is proposed based on our data that LUBAC and OTULIN are the ubiquitinating enzymes that regulate ATG13 ubiquitination and ATG13 protein level to control autophagy initiation and maturation (Figure S7C).
The role of ubiquitination regulation on the autophagy machinery proteins is just emerging. Because autophagy is a stress response process, its activation is mostly dependent on the internal and external condition change. Therefore, the whole system needs to be turned on or off based on the environmental conditions. The machinery proteins in this pathway also need to be regulated accordingly. It has been reported that many components, previously thought as the constitutive components, are targeted by ubiquitin system for degradation when the environmental condition changes. CUL3-KLHL20 governs the ubiquitination of a set of autophagy proteins in prolonged starvation to negatively regulate autophagy [37]. PIK3C3/VPS34 is also targeted by CUL1-FBXL20 for degradation and inhibition of autophagy [38]. Ubiquitination-dependent degradation of ATG14 is directly mediated by ZBTB16-CUL3-RBX1 E3 ubiquitin ligase complex [39]. BECN1 seems to be a key protein for versatile ubiquitination. The E3 ligase CUL4-AMBRA1, TRAF6, NEDD4, RNF216 and CUL3-KLHL20 target BECN1 for different kinds of ubiquitination modification to either activate or degrade the protein [40]. Accordingly, there are various DUBs, such as USP14, USP19, USP10, USP13, and USP9X, which negatively regulate BECN1 activity or degradation [40]. It was recently reported that ubiquitination of UVRAG by SMURF1 promotes autophagosome maturation and the DUB ZRANB1 reverses this activity [41]. Therefore, autophagy seems to be a dynamic network that is constantly regulated. In our case, we identified that LUBAC and OTULIN regulate the linear ubiquitination on ATG13 as well as ATG13 protein level. This finding adds an additional mode of ubiquitination regulating autophagy. Identifying the E3 ligase that responsible for ATG13 degradation when RNF31 is absent will be the next step.
It was well recognized before that LUBAC and OTULIN antagonize each other on substrate linear ubiquitination, until recently, Heger et al. reported that OTULIN directly deubiquitinates LUBAC [42]. Therefore, OTULIN coordinates with LUBAC to regulate inflammation. In our case, the data showed that knockdown of RNF31 decreases, whereas knockdown of OTULIN increases the linear ubiquitination on ATG13 as well as ATG13 protein level (Figure 4A). However, ATG13 linear ubiquitination and ATG13 protein level were slightly decreased in the RNF31 and OTULIN double knockdown compared to those in OTULIN single knockdown (Figure 4A). Therefore, we proposed that in double knockdown cells, OTULIN knockdown increases the relative amount of ubiquitinated LUBAC, therefore activates linear ubiquitination on ATG13 as well as ATG13 protein level. Consistent with this, our fluorescent image data showed that double knockdown of RNF31 and OTULIN only slightly decreased LC3 puncta size compared to that of OTULIN single knockdown (Figure 4B,C and S4).
Perturbations of autophagy have been linked to various diseases, including neurodegeneration, skeletal muscle myopathies, and cancer [6]. It has been reported that deficiency in LUBAC or OTULIN or ATG13 causes embryonic lethality in mice [27,42–44]. Autophagy and inflammation are the two feedback responses that help organisms to defend against stress, and there were several studies reporting the connection between the two signaling pathways. It was reported that ATG13-deficient cells are more sensitive to TNF-induced apoptosis [44]. A new study also reported that the key inflammation signal protein STING induces autophagy to help cell fight against infection [45]. OPTN is an autophagy receptor that targets ubiquitin-coated bacteria for xenophagy [46]. Meanwhile, OPTN has a UBAN domain that binds to linear ubiquitin to activate NFKB signal pathway [47]. It was reported recently that LUBAC-synthesized linear ubiquitin chains activate OPTN-induced anti-bacterial autophagy as well as NFKB to defend cells against bacterial invasion [48]. It is consistent with our data that RNF31 linearly ubiquitinates ATG13 and stabilizes ATG13 to ensure proper autophagy initiation and protect cells from infection (Figures 3F,4,6,7). It would be interesting to examine whether the phenotype observed in animal models is related to autophagy dysfunction. It is also worth exploring more substrates in autophagy that are targeted by LUBAC and OTULIN.
In summary, our study reveals a new function of LUBAC and OTULIN that linearly ubiquitinates ATG13 and regulates ATG13 protein level to control autophagy initiation and maturation.
Materials and methods
Cell lines and bacterial preparation
HEK293 FT (PTA-5077) and HeLa cells (CRM-CCL-2) from ATCC were cultured in DMEM (Thermo Fisher Scientific, 10566024) with 10% fetal bovine serum (Gemini, 900–108) and 100 U/ml penicillin G and 100 μg/ml streptomycin (Thermo Fisher Scientific, 15140148) at 37°C under 5% CO2. HeLa cells stably expressing mRFP-EGFP-LC3B were cultured in complete medium containing 300 ng/μl G418 (Invivogen, ant-gn-1). S. Typhimurium (strain: ATCC14028), provided by Dr. Rui Huang (Soochow University, China), were grown overnight in Luria broth [49]. Next, these bacteria were sub-cultured in fresh LB (1:33) for 3.5 h. Before infection, sub-cultured bacteria were diluted (1:5) in antibiotic-free DMEM containing 10% FBS.
Antibodies and reagents
Mouse anti-MYC (2276), rabbit anti-HA (3724), rabbit anti-OTULIN (14127), rabbit anti-ATG13 (13468) and rabbit anti-CALCOCO2/NDP52 (60732) were purchased from Cell Signaling Technology. Rabbit anti-Flag (F3165), mouse anti-Flag (F7425) and rabbit anti-LC3B (L7543; for detecting the endogenous LC3B) were purchased from Sigma-Aldrich. Mouse anti-LC3 (M186-3; for detecting the exogenous LC3B) and mouse anti-ACTB (HRP-DirecT; PM053-7) were obtained from Medical & Biological Laboratories Co., LTD. Mouse anti-SQSTM1 (sc-28359) and mouse anti-ubiquitin (sc-8017) were purchased from Santa Cruz Biotechnology. Rabbit anti-RNF31/HOIP (ab46322), rabbit anti-OPTN (23666), mouse anti-LPS (ab8274) and mouse anti-WIPI2 (ab105459) were purchased from Abcam. Rabbit anti-RB1CC1 (17250-1-AP) was purchased from Proteintech and mouse anti-linear ubiquitin (MABS451) was purchased from Merck. The secondary antibodies goat anti-mouse IgG (H + L), HRP (111–035-146) and goat anti-rabbit IgG (H + L), HRP (111–035-144) were purchased from Jackson ImmunoResearch Inc. The secondary antibodies goat anti-rabbit IgG (H + L), Alexa fluor 488 (A-11034), goat anti-mouse IgG (H + L), Alexa fluor 488 (A-11029), goat anti-mouse IgG (H + L), Alexa fluor 633 (A-21052) and goat anti-rabbit IgG (H + L), Alexa fluor 633 (A-21071) were purchased from Invitrogen. MG132 (HY-13259) was purchased from MedChemExpress.
Plasmids
The plasmid mRFP-EGFP-LC3B was provided by Professor Tamotsu Yoshimori (Osaka University, Japan; Addgene, 21074). The plasmid MYC-ATG13/hAtg13 (31965; Do-Hyung Kim) was purchased from Addgene. The truncation mutants of ATG13 was generated by introducing stop codon into the designated position into MYC-ATG13. ATG13 lysine mutant ATG138KR was generated by multiple rounds of PCR-mediated site-directed mutagenesis on the specific lysine residues. The mutagenic primers are listed in Table S1. To generate ATG1314KR or ATG1322KR, HORMA14KR was first synthesized and then fused with the wild type or mutated C-terminal fragment by annealing. The primer for annealing is listed in Table S1. pET32a-TEV-Flag-ATG13 was generated by inserting TEV-Flag-ATG13 fragment into pET32a(+) (Novagen, 69015–3). Flag-OTULIN was a gift from Professor Zongping Xia (Zhengzhou University). Flag-OTULINC129S was generated by PCR-mediated site-directed mutagenesis, based on the Flag-OTULIN plasmid. The mutagenic primer is listed in Table S1. GST-OTULIN was generated by inserting OTULIN fragment into pGEX-6P-1 (GE Healthcare, 28–9546-48). All mutations were confirmed by DNA sequencing. Flag-RNF31 (CH898682) was purchased from Vigene Biosciences. pET32a-TEV-HA-RNF31Δ1-698 was generated by inserting TEV-HA-RNF31Δ1-698 fragment into pET32a(+). HA-RBCK1 was generated by inserting HA-RBCK1 sequence into pCDNA5 (Invitrogen, V6010-20). Flag-STUB1 was generated by inserting Flag-STUB1 fragment into pRK5 (BD Biosciences, 556104). The construct for expression of His-tagged UBE2D2 was kindly provided by Dr. Cynthia Wolberger (John Hopkins University, Baltimore, MD). Untagged ubiquitin plasmid was kindly gifted by Professor Kazuhiro Iwai (Kyoto University, Japan). pcDNA3.0-ubiquitin was generated by inserting ubiquitin sequence into pcDNA3.0 vector (Invitrogen, A-150228). GFP-ULK1, GFP-CALCOCO2 and GFP-SEC61B were generated by inserting ULK1, CALCOCO2 or SEC61B fragment into pEGFP-C1 (Clontech, 6084–1). For OTULIN, RNF31 and VCPIP1 shRNA knockdown, target sequences were cloned into pSUPER.neo (OligoEngine, VEC-PBS-0004). The empty vector was used as negative control. For the assays done in HEK293 FT cells, RNF31 siRNA was used to give a better knockdown efficiency. Target sequences are listed in Table S1.
Protein purification
GST-OTULIN, His-UBE2D2, His-TEV-Flag-ATG13, and His-TEV-HA-RNF31Δ1-698 were purified from E. coli BL21 (DE3) (Transgen Biotech, CD601–01), according to a previously described method [50]. Protein eluted from Glutathione-Sepharose beads (GE Healthcare, 17–5132-02) or Ni-NTA (GE Healthcare, 17–5318-02) were fractionated on a Superdex 200 HR (10/300) column (GE Healthcare, 28–9909-44) in a buffer containing 50 mM Tris-HCl (pH 7.5), 100 mM sodium chloride and 2 mM DTT (MDBio, Inc., D023). Flag-ATG13 and HA-RNF31Δ1-698 were generated by removing His tag with TEV protease (New England Biolabs, Inc., P8112S). For bacterial purified untagged ubiquitin, cells were first sonicated in 20 mM Na-Acetate (Sinopharm Chemical Reagent Co., 127–09-3), pH 5.1 buffer. After centrifuge, the supernatant was then loaded on a HiTrap SP HP (5 ml) column (GE Healthcare, 17–1152-01) in 20 mM Na-Acetate (pH 5.1)/0-500 mM NaCl gradient. Collect the fraction with ubiquitin and change the buffer to 20 mM Tris-HCl, pH 7.5 through a HiTrap DEAE FF (1 ml) column (GE Healthcare, 17–5055-01).
Autophagy assays
To induce autophagy, cells were incubated with the starvation medium EBSS (Sigma-Aldrich, E2888) for 4 h. To inhibit autophagosome-lysosome fusion, cells were treated with 20 nM chloroquine diphosphate salt (Sigma-Aldrich, C6628) for 4 h. After treatment, cells were harvested for immunofluorescence microscopy or immunoblotting analysis.
Immunofluorescence microscopy
HeLa cells stably expressing mRFP-EGFP-LC3B were seeded on small glass slides (Marienfeld, AP-0111520) and transiently transfected with indicated constructs. After 24 h, cells were washed with phosphate-buffered saline (PBS, Gibco, 70011044) and fixed with 4% paraformaldehyde (Thermo Fisher Scientific, 43368) in PBS for 15 min. After being washed four times with PBS, the nuclei were labeled with DAPI (Sigma-Aldrich, D9542). Finally, the samples were mounted with Clear MountTM Mounting solution (Invitrogen, P36934). For the immunofluorescence stain studies, fixed cells were washed four times with PBS, and then incubated with indicated primary antibodies (1:500) for 1 h, and fluorescent dye-conjugated secondary antibodies (1:600) for 30 min. Images were acquired on Zeiss LSM800 or LSM880 microscope (Zeiss, Jena, Thuringia, Germany) with a 63 × 1.4 NA oil objective. Same acquisition parameters were used for a specific set of experiments.
GST affinity-isolation assay, immunoprecipitation and immunoblotting
GST or GST-OTULIN immobilized Glutathione-Sepharose beads were incubated with purified Flag-ATG13 for 1 h in NP40 lysis buffer (50 mM Tris–HCl pH 7.4, 150 mM sodium chloride, and 0.5% NP40 (BBI Life Sciences Cooperation, 9016–45-9)). GST beads were then washed three times with NP40 wash buffer (50 mM Tris–HCl pH 7.4, 150 mM sodium chloride, and 0.1% NP40). The bound proteins were analyzed by western blot using a Flag-specific antibody (Sigma-Aldrich, F3165).
For immunoprecipitation experiments, related plasmids were transfected into HEK293 FT cells using Lipofectamine 2000 (Invitrogen, 11668019) for 2–3 d. Cells were harvested and lysed in cell lysis buffer (Sigma-Aldrich, C2978) containing protease inhibitor cocktail (Bimake, B14011) for 30 min at 4°C. The soluble supernatant fractions were harvested by centrifugation at 17,000 × g for 10 min and then used for immunoprecipitation with the indicated antibodies or antibodies conjugated on beads. After that, the antibody-bound immunoprecipitation samples were further incubated with protein A beads (Cell Signaling Technology, 9863S). After binding, the beads were washed three times with immunoprecipitation wash buffer to remove nonspecific binding proteins. The beads bound material was eluted using Laemmli buffer and analyzed by immunoblotting. For immunoprecipitation under denaturing conditions, harvested cells were lysed in a buffer with 1% SDS (Sinopharm Chemical Reagent Co., 30166480) and 5 mM DTT. The samples were heated at 95°C for 5 min and diluted into 0.1% SDS and 0.5 mM DTT with lysis buffer. The soluble supernatant fractions were harvested and subjected to immunoprecipitation experiments as described above. After that, immunoblotting was performed. All immunoblotting was performed using polyvinylidene fluoride membrane (Bio-Rad, 1620177) and the indicated antibodies. Protein signals were detected using ECL western blotting detection reagents (PerkinElemer, NEL105001EA). The chemiluminescence bands were imaged under Amersham Imager 600 (GE Healthcare Life Sciences, USA). The bands were adjusted within the linear range, and quantified by ImageJ software (NIH).
In vitro ubiquitination assays
For in vitro ubiquitination assay, the mammalian purified substrate MYC-ATG13 (5 μM) or ATG13 mutants was incubated with GST-E1 (180 nM; BostonBiochem, E-306), His-UBE2D2 (2 μM), HA-RNF31Δ1-698 (2 μM) and untagged ubiquitin (11 μM) in the reaction buffer (25 mM Tris pH 7.4, 2 mM magnesium (Aladdin, M113688) and ATP (Sigma-Aldrich, A2383), and 1 mM DTT) at 37°C for 1 h. After the reaction finished, the product was then immunoprecipitated with MYC beads (Bimake, B26302) under denaturing condition and the samples were analyzed by western blot.
Bacterial infection and colony-forming unit assay
HeLa cells, transfected with RNF31 knockdown constructs, were grown in 6-well format. Seventy-two h after transfection, cells were infected with 100 μl diluted bacteria, and incubated for 15 min at 37°C. Then cells were washed two times with PBS, and further cultured with DMEM and 10% FBS containing 100 μg/ml gentamycin (Shanghai BasalMedia Technologies Co., LTD, S130J7) for 1 h and 20 μg/ml gentamycin for the following hours. For immunofluorescence assay, small glass slides were harvested 3 h after infection and stained by following the method of immunofluorescence microscopy. For colony-forming unit assay, cells were harvested 2 h and 8 h after infection, and lysed in 1 ml PBS containing 0.1% Triton-X (Sangon Biotech, 9002–93-1). After a serial dilution, 0.5 ml dilutions were plated on 10-cm LB agar plates and cultured overnight. The number of colonies per plate were counted next day.
Cycloheximide chase
Cycloheximide chase experiments were performed by incubating the cells in DMEM containing 50 µg/ml cycloheximide (Abcam, ab120093) at 37°C. Equal number of cells was taken at different time points as indicated in the figures for immunoblotting analysis. Cells were lysed with the lysis buffer.
Quantification and statistical analysis
All data were performed in at least three independent experiments. For the quantification of puncta, the numbers of puncta per cell were counted using ImageJ software. Data were shown as mean ± SD, and error bars represented the standard deviations from counting of 100 cells in each group from three independent experiments. Data graphing and descriptive statistics were presented by utilizing Origin 8.5 (Origin lab cooperation, USA) and Microsoft Excel Data Analysis package. The significance between two groups was obtained using the Student’s t-test. The significance among multiple groups was obtained using one-way ANOVA followed by Tukey’s multiple comparisons test. ns, not significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Supplementary Material
Acknowledgments
We thank Professor Tamotsu Yoshimori (Osaka University, Japan) for the mRFP-EGFP-LC3B plasmid, Dr. Kazuhiro Iwai (Kyoto University, Japan) for the untagged ubiquitin plasmid, Dr. Song Huang (National Institute of Biological Sciences, Beijing, China) and Dr. Jiahuai Han (Xiamen University, China) for multiple gene clone constructs. We thank the Image Core Facility at ShanghaiTech University for assisting with confocal microscopy. This study was supported by the National Natural Science Foundation of China (No. 31570781 and No. 31770831) and the Start-up grant from ShanghaiTech University.
Funding Statement
This work was supported by the National Natural Science Foundation of China [31570781]; National Natural Science Foundation of China [31770831]; ShanghaiTech University [2015F0202-000-31].
Disclosure statement
The authors declare no competing interests.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Mizushima N, Yoshimori T, Ohsumi Y.. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–132. [DOI] [PubMed] [Google Scholar]
- [2].Mizushima N. A brief history of autophagy from cell biology to physiology and disease. Nat Cell Biol. 2018;20(5):521–527. [DOI] [PubMed] [Google Scholar]
- [3].Bento CF, Renna M, Ghislat G, et al. Mammalian autophagy: How does it work? Annu Rev Biochem. 2016;85:685–713. [DOI] [PubMed] [Google Scholar]
- [4].Mizushima N, Levine B, Cuervo AM, et al. Autophagy fights disease through cellular self-digestion. Nature. 2008;451(7182):1069–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Leidal AM, Levine B, Debnath J. Autophagy and the cell biology of age-related disease. Nat Cell Biol. 2018;20(12):1338–1348. [DOI] [PubMed] [Google Scholar]
- [6].Levine B, Kroemer G. Biological functions of autophagy genes: a disease perspective. Cell. 2019;176(1–2):11–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Schneider JL, Cuervo AM. Autophagy and human disease: emerging themes. Curr Opin Genet Dev. 2014;26:16–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Hurley JH, Young LN. Mechanisms of autophagy initiation. Annu Rev Biochem. 2017;86:225–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Ganley IG, Lam Du H, Wang J, et al. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem. 2009;284(18):12297–12305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Hosokawa N, Hara T, Kaizuka T, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20(7):1981–1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Jung CH, Jun CB, Ro SH, et al. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20(7):1992–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kim J, Kundu M, Viollet B, et al. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13(2):132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Nazio F, Strappazzon F, Antonioli M, et al. mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nat Cell Biol. 2013;15(4):406–416. [DOI] [PubMed] [Google Scholar]
- [14].Di Rienzo M, Antonioli M, Fusco C, et al. Autophagy induction in atrophic muscle cells requires ULK1 activation by TRIM32 through unanchored K63-linked polyubiquitin chains. Sci Adv. 2019;5(5):eaau8857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Backer JM. The intricate regulation and complex functions of the Class III phosphoinositide 3-kinase Vps34. Biochem J. 2016;473(15):2251–2271. [DOI] [PubMed] [Google Scholar]
- [16].Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. [DOI] [PubMed] [Google Scholar]
- [17].Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–229. [DOI] [PubMed] [Google Scholar]
- [18].Swatek KN, Komander D. Ubiquitin modifications. Cell Res. 2016;26(4):399–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Sasaki K, Iwai K. Roles of linear ubiquitinylation, a crucial regulator of NF-kappaB and cell death, in the immune system. Immunol Rev. 2015;266(1):175–189. [DOI] [PubMed] [Google Scholar]
- [20].Elliott PR, Komander D. Regulation of Met1-linked polyubiquitin signalling by the deubiquitinase OTULIN. Febs J. 2016;283(1):39–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Hrdinka M, Gyrd-Hansen M. The Met1-linked ubiquitin machinery: emerging themes of (de)regulation. Mol Cell. 2017;68(2):265–280. [DOI] [PubMed] [Google Scholar]
- [22].Kirisako T, Kamei K, Murata S, et al. A ubiquitin ligase complex assembles linear polyubiquitin chains. Embo J. 2006;25(20):4877–4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Haas TL, Emmerich CH, Gerlach B, et al. Recruitment of the linear ubiquitin chain assembly complex stabilizes the TNF-R1 signaling complex and is required for TNF-mediated gene induction. Mol Cell. 2009;36(5):831–844. [DOI] [PubMed] [Google Scholar]
- [24].Tokunaga F, Sakata S, Saeki Y, et al. Involvement of linear polyubiquitylation of NEMO in NF-kappaB activation. Nat Cell Biol. 2009;11(2):123–132. [DOI] [PubMed] [Google Scholar]
- [25].Gerlach B, Cordier SM, Schmukle AC, et al. Linear ubiquitination prevents inflammation and regulates immune signalling. Nature. 2011;471(7340):591–596. [DOI] [PubMed] [Google Scholar]
- [26].Keusekotten K, Elliott PR, Glockner L, et al. OTULIN antagonizes LUBAC signaling by specifically hydrolyzing Met1-linked polyubiquitin. Cell. 2013;153(6):1312–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Rivkin E, Almeida SM, Ceccarelli DF, et al. The linear ubiquitin-specific deubiquitinase gumby regulates angiogenesis. Nature. 2013;498(7454):318–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Komander D, Reyes-Turcu F, Licchesi JD, et al. Molecular discrimination of structurally equivalent Lys 63-linked and linear polyubiquitin chains. EMBO Rep. 2009;10(5):466–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Draber P, Kupka S, Reichert M, et al. LUBAC-recruited CYLD and A20 regulate gene activation and cell death by exerting opposing effects on linear ubiquitin in signaling complexes. Cell Rep. 2015;13(10):2258–2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Spit M, Rieser E, Walczak H. Linear ubiquitination at a glance. J Cell Sci. 2019;132(2):jcs208512. [DOI] [PubMed] [Google Scholar]
- [31].Shimizu Y, Taraborrelli L, Walczak H. Linear ubiquitination in immunity. Immunol Rev. 2015;266(1):190–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Yan C, Huo H, Yang C, et al. Ubiquitin C-terminal hydrolase L1 regulates autophagy by inhibiting autophagosome formation through its deubiquitinating enzyme activity. Biochem Biophys Res Commun. 2018;497(2):726–733. [DOI] [PubMed] [Google Scholar]
- [33].Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3(5):452–460. [DOI] [PubMed] [Google Scholar]
- [34].Smit JJ, Monteferrario D, Noordermeer SM, et al. The E3 ligase HOIP specifies linear ubiquitin chain assembly through its RING-IBR-RING domain and the unique LDD extension. Embo J. 2012;31(19):3833–3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Jao CC, Ragusa MJ, Stanley RE, et al. A HORMA domain in Atg13 mediates PI 3-kinase recruitment in autophagy. Proc Natl Acad Sci U S A. 2013;110(14):5486–5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Fujioka Y, Suzuki SW, Yamamoto H, et al. Structural basis of starvation-induced assembly of the autophagy initiation complex. Nat Struct Mol Biol. 2014;21(6):513–521. [DOI] [PubMed] [Google Scholar]
- [37].Liu CC, Lin YC, Chen YH, et al. Cul3-KLHL20 ubiquitin ligase governs the turnover of ULK1 and VPS34 complexes to control autophagy termination. Mol Cell. 2016;61(1):84–97. [DOI] [PubMed] [Google Scholar]
- [38].Xiao J, Zhang T, Xu D, et al. FBXL20-mediated Vps34 ubiquitination as a p53 controlled checkpoint in regulating autophagy and receptor degradation. Genes Dev. 2015;29(2):184–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Zhang T, Dong K, Liang W, et al. G-protein-coupled receptors regulate autophagy by ZBTB16-mediated ubiquitination and proteasomal degradation of Atg14L. Elife. 2015;4:e06734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Boutouja F, Brinkmeier R, Mastalski T, et al. Regulation of the tumor-suppressor BECLIN 1 by distinct ubiquitination cascades. Int J Mol Sci. 2017;18(12):2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Feng X, Jia Y, Zhang Y, et al. Ubiquitination of UVRAG by SMURF1 promotes autophagosome maturation and inhibits hepatocellular carcinoma growth. Autophagy. 2019;15(7):1130–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Heger K, Wickliffe KE, Ndoja A, et al. OTULIN limits cell death and inflammation by deubiquitinating LUBAC. Nature. 2018;559(7712):120–124. [DOI] [PubMed] [Google Scholar]
- [43].Peltzer N, Rieser E, Taraborrelli L, et al. HOIP deficiency causes embryonic lethality by aberrant TNFR1-mediated endothelial cell death. Cell Rep. 2014;9(1):153–165. [DOI] [PubMed] [Google Scholar]
- [44].Kaizuka T, Mizushima N. Atg13 is essential for autophagy and cardiac development in mice. Mol Cell Biol. 2016;36(4):585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Gui X, Yang H, Li T, et al. Autophagy induction via STING trafficking is a primordial function of the cGAS pathway. Nature. 2019;567(7747):262–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Wild P, Farhan H, McEwan DG, et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 2011;333(6039):228–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Nakazawa S, Oikawa D, Ishii R, et al. Linear ubiquitination is involved in the pathogenesis of optineurin-associated amyotrophic lateral sclerosis. Nat Commun. 2016;7:12547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Noad J, von der Malsburg A, Pathe C, et al. LUBAC-synthesized linear ubiquitin chains restrict cytosol-invading bacteria by activating autophagy and NF-kappaB. Nat Microbiol. 2017;2:17063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Wiener R, DiBello AT, Lombardi PM, et al. E2 ubiquitin-conjugating enzymes regulate the deubiquitinating activity of OTUB1. Nat Struct Mol Biol. 2013;20(9):1033–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Ye Y, Meyer HH, Rapoport TA. Function of the p97-Ufd1-Npl4 complex in retrotranslocation from the ER to the cytosol: dual recognition of nonubiquitinated polypeptide segments and polyubiquitin chains. J Cell Biol. 2003;162(1):71–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


