ABSTRACT
Macroautophagy/autophagy is a key catabolic process in which different cellular components are sequestered inside double-membrane vesicles called autophagosomes for subsequent degradation. In yeast, autophagosome formation occurs at the phagophore assembly site (PAS), a specific perivacuolar location that works as an organizing center for the recruitment of different autophagy-related (Atg) proteins. How the PAS is localized to the vacuolar periphery is not well understood. Here we show that the vacuolar membrane protein Vac8 is required for correct vacuolar localization of the PAS. We provide evidence that Vac8 anchors the PAS to the vacuolar membrane by binding Atg13 and recruiting the Atg1 initiation complex. VAC8 deletion or mislocalization of the protein reduce autophagy activity, highlighting the importance of both the PAS and the correct vacuolar localization of the Atg1 initiation complex for efficient and robust autophagy.
Abbreviations: AID: auxin-inducible degradation; Atg: autophagy-related; Cvt: cytoplasm-to-vacuole targeting; DMSO: dimethyl sulfoxide; ER: endoplasmic reticulum; GFP: green fluorescent protein; IAA: 3-indole acetic acid; PAS: phagophore assembly site; RFP: red fluorescent protein
KEYWORDS: Atg13, autophagosomes, lysosome, PAS, vacuole
Introduction
When facing stressful environmental and nutritional conditions, cells need sophisticated mechanisms to cope. Macroautophagy, hereinafter autophagy, is a key catabolic process in which different cellular components, such as proteins, lipids or even entire organelles, are sequestered, broken down and recycled in order to promote cell survival. Once autophagy is activated, cargo intended for degradation is enveloped by the phagophore, a cup-shaped transient compartment that expands into a double-membrane vesicle called an autophagosome. Once fully matured, autophagosomes fuse with the vacuole, releasing their cargo for degradation by different vacuolar hydrolases. The molecules obtained from cargo degradation are then transported back into the cytosol to be recycled by the cell [1,2].
Autophagy has been described as both a selective and nonselective process, depending on the type of cargo that is targeted for degradation. On one hand, it is thought that during nonselective autophagy, random parts of the cytoplasm are taken up into phagophores to later be delivered to the vacuole for degradation. On the other hand, during selective autophagy specific cargo such as protein aggregates and damaged or superfluous organelles are recognized by specialized autophagy receptors, that tether the specific cargo to the phagophore membrane to facilitate vacuolar delivery and degradation. The cytoplasm-to-vacuole targeting (Cvt) pathway constitutes a specific type of selective biosynthetic auto-phagy in yeast, which is used for delivering resident vacuolar enzymes from their site of synthesis in the cytosol to their final destination [3].
In yeast, autophagosome formation occurs at the phagophore assembly site (PAS), a specific perivacuolar location that works as an organizing center for the recruitment of different autophagy-related (Atg) proteins, donor membranes and cytoplasmic cargo required for the autophagic process [1,2]. Atg protein recruitment at the PAS occurs in sequential steps [4], starting with the assembly of the Atg1 initiation complex, composed of the protein kinase Atg1, the regulatory protein Atg13, and the Atg17-Atg31-Atg29 subcomplex, which works as a scaffold [5]. In addition to its role in regulating Atg1, Atg13 also functions as an assembly hub for all other members of the initiation complex [6–10]. Even though the essential role of the Atg1 initiation complex in phosphorylating downstream targets, as well as its structural role as a backbone for PAS assembly have been well established, the nature behind its recruitment to the vacuole periphery remains unclear.
Vac8 is a peripheral vacuolar membrane protein that has been implicated in vacuolar inheritance, the Cvt pathway [11] and the formation of nucleus-vacuole junctions [12]. Whereas Vac8 binding to the nuclear membrane protein Nvj1 promotes the formation of nucleus-vacuole junctions and piecemeal microautophagy of the nucleus/micronucleophagy [12–14], Vac8 binding to Atg13 is proposed to be required for the Cvt pathway [15]. Thus, Vac8 has mainly been associated with selective types of autophagy. Nevertheless, previous results from our lab suggest Vac8 could play a role in nonselective autophagy [15,16], although the exact role of this protein has not been determined.
Here we show that Vac8 is required for efficient and robust nonselective autophagy. We determined that Vac8 vacuolar localization is necessary for the recruitment of the Atg1 initiation complex and PAS assembly to the vacuolar periphery. Our results indicate this process is regulated by Vac8 binding to Atg13, with the former acting as an anchor to the vacuole and the latter as an assembly hub for the recruitment of the initiation complex.
Results
Vac8 is required for efficient autophagy induction
The initial studies of the vac8∆ mutant resulted in the characterization of the protein as being required for the selective Cvt pathway. The primary reason for this conclusion was based on the following observation: the primary cargo of the Cvt pathway, precursor aminopeptidase I (prApe1), accumulates in the vac8∆ strain in growing condition, but is efficiently delivered to the vacuole when autophagy is induced by nitrogen starvation [15]. Nonetheless, the mutant strain displays a significant defect in autophagy based on maturation of the Pho8∆60 protein as determined through a radioactive pulse-chase experiment. Pho8∆60 is an engineered phosphatase that requires nonselective autophagy to reach the vacuole and initiate its enzymatic activity; upon vacuolar delivery a propeptide is removed, resulting in a molecular mass shift that can be monitored through SDS-PAGE. To extend our analysis of the role of Vac8 in autophagy we generated new strains lacking the VAC8 gene and measured nonselective autophagy using a Pho8Δ60 biochemical assay to follow enzyme activation [17]. Thus, by measuring phosphatase activity we can quantitatively monitor nonselective autophagy. In agreement with our previous findings [15,16], VAC8 deletion led to a significant decrease in autophagy activity measured by the Pho8Δ60 assay when compared to wild-type (WT) cells after 3 h of nitrogen starvation (Figure 1A). However, VAC8 deletion failed to produce a complete block in autophagy; vac8∆ cells still showed a small but significant amount of autophagy activity compared with a strain where the essential autophagy gene ATG13 was deleted.
Figure 1.
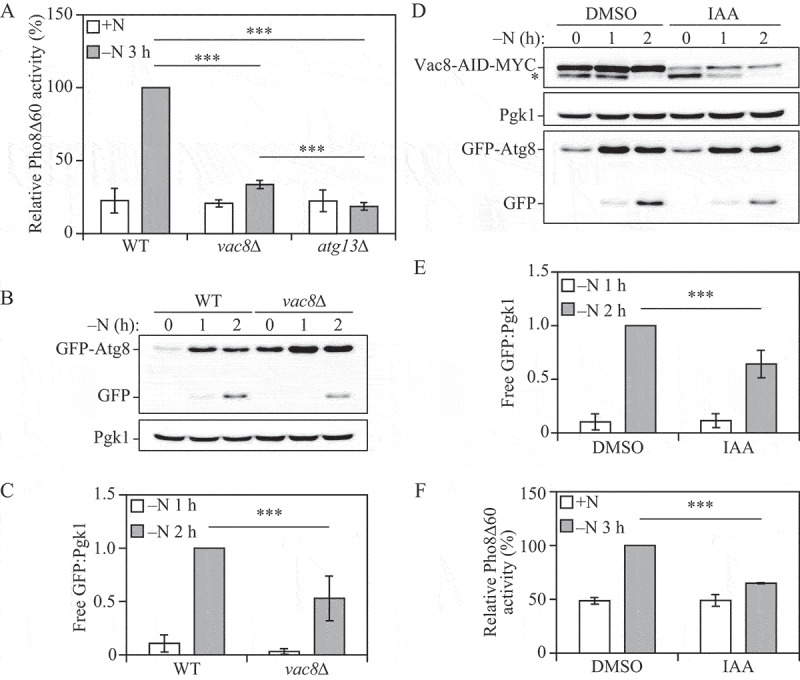
Vac8 is required for robust autophagy activity. (A) Autophagy activity was measured using the Pho8Δ60 assay in WT, atg13∆ and vac8∆ strains under nutrient-rich conditions (+N) and after 3 h of nitrogen starvation (-N 3 h). Error bars indicate the standard deviation of 3 independent experiments. ANOVA, ***P < 0.001. (B) Autophagy activity was measured with the GFP-Atg8 processing assay in WT and vac8∆ strains after 0, 1 and 2 h of nitrogen starvation (-N). A representative image based on detection with anti-YFP is shown. Pgk1 was used as a loading control. (C) Quantification of panel (B). The ratio of free GFP to Pgk1 after 1 and 2 h of nitrogen starvation (-N) is presented. Error bars indicate the standard deviation of 3 independent experiments. Student’s t-test, ***P < 0.001. (D) Autophagy activity was measured with the GFP-Atg8 processing assay in a Vac8-auxin-inducible degradation strain treated with DMSO (control) or auxin (IAA) for 30 min followed by 0, 1 and 2 h of nitrogen starvation (-N). A representative image is shown. The upper panel represents blotting with anti-MYC; the asterisk corresponds to a nonspecific band. Pgk1 was used as a loading control. (E) Quantification of panel (D). The ratio of free GFP to Pgk1 after 1 and 2 h of nitrogen starvation (-N) is presented. Error bars indicate the standard deviation of 3 independent experiments. Student’s t-test, ***P < 0.001. (F) Autophagy activity was measured using the Pho8Δ60 assay in a Vac8-auxin-inducible degradation strain treated with DMSO or IAA for 30 min in nutrient-rich conditions (+N) and after 3 h of nitrogen starvation (-N 3 h). Error bars indicate the standard deviation of 3 independent experiments. Student’s t-test, ***P < 0.001
Next, we used a second method to further examine the effect on autophagy of deleting VAC8. The GFP-Atg8 processing assay measures autophagy by determining the amount of free GFP that is generated once the chimera is delivered to the vacuole through autophagy and cleaved [18]. During auto-phagy initiation, Atg8 is lipidated and attached to both the inner and outer membranes of the growing phagophore. Once autophagosomes are completed, the Atg8 bound to the concave surface is trapped inside the autophagosome and is delivered to the vacuole where it is degraded. Tagging GFP at the N terminus of Atg8 makes it possible to measure auto-phagy because GFP is relatively resistant to vacuolar hydrolases and can be separated from full-length GFP-Atg8 by western blot. Confirming the Pho8Δ60 assay results, vac8∆ cells showed a significant decrease in autophagy activity based on GFP-Atg8 processing when compared to wild-type cells after 2 h of nitrogen starvation (Figure 1B,C).
Because Vac8 is required for vacuolar inheritance, vac8∆ cells have an abnormal vacuolar morphology [11]. To exclude the possibility that the decrease in autophagy observed in the VAC8 deletion strains was due to abnormal vacuolar morphology we generated Vac8-inducible degradation mutants using the auxin-inducible degron (AID) system; under the time course of Vac8 depletion used in these experiments, the vacuole morphology was similar to that seen in the wild type (see Figure 2) [19]. Vac8 degradation was induced 30 min before nitrogen starvation by adding 3-indole acetic acid/auxin (IAA) to the growth medium, and autophagy activity was measured. Even at time zero (i.e., after a 30-min treatment with IAA), Vac8-AID-MYC levels were substantially reduced compared to the control (Figure 1D, upper panel). Similar to the results obtained in vac8∆ cells, Vac8-inducible degradation led to a significant decrease in auto-phagy after nitrogen starvation based on GFP-Atg8 processing (Figure 1D–E) and Pho8Δ60 (Figure 1F) assays. Finally, we examined Pho8∆60 activity in cells expressing a conditional temperature-sensitive Vac8 mutant, vac8-15ts [20]. The vac8-15ts strain shows normal-sized vacuoles at a permissive temperature (25°C) but has multi-lobe fragmented vacuoles at a nonpermissive temperature (37°C). Wild-type cells displayed an increase in Pho8∆60-dependent phosphatase activity at either 25°C or 37°C. The vac8∆ strain showed a much lower level of activity, and a slight reduction at 37°C compared to 25°C (Figure S1). The vac8-15ts mutant significantly restored the Pho8∆60 activity of the vac8∆ strain at 25°C; however, the Pho8∆60 activity in the vac8-15ts mutant was significantly reduced under starvation conditions at 37°C, similar to the vac8∆ strain. Consistent with our previous report, these results indicate that Vac8 is necessary for robust autophagy induction during nitrogen starvation, and that Vac8 effects with regard to autophagy are unrelated to its role in preserving vacuolar morphology.
Figure 2.
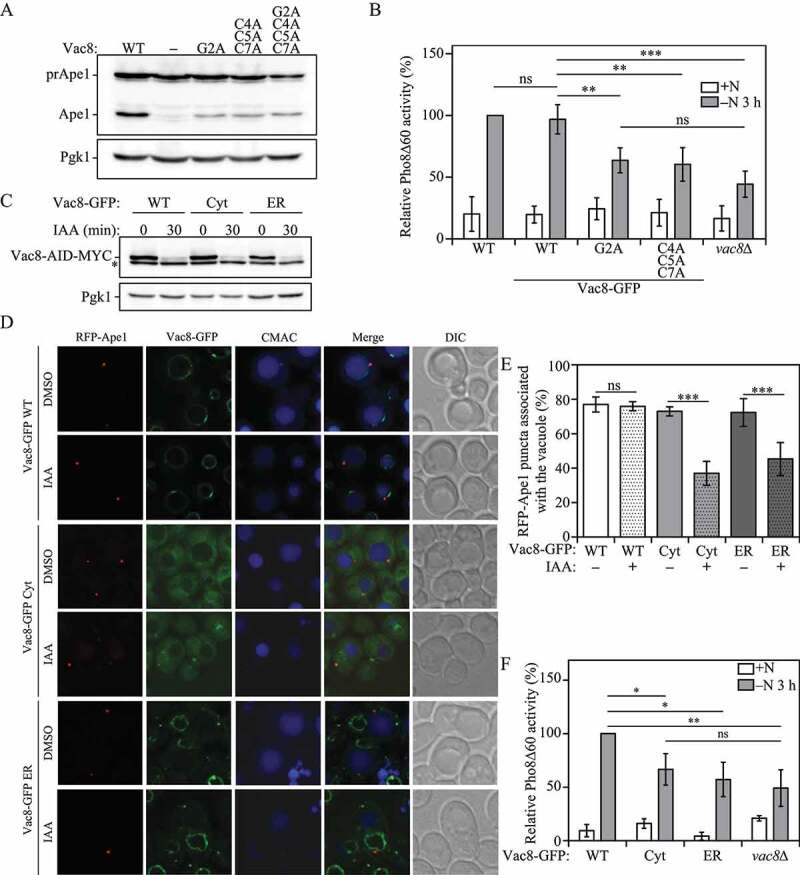
Vac8 vacuolar membrane localization is required for correct PAS localization and robust autophagy activity. (A) The vac8Δ strain (YTS178) was transformed with plasmids expressing wild-type VAC8, empty vector, the myristoylation defective vac8 mutant (vac8-2) the palmitoylation defective vac8 mutant (vac8-3) or the myristoylation and palmitoylation double mutant (vac8-4). The strains were grown in SMD and collected at mid-log phase (OD600 = 0.5) and protein extracts were analyzed by western blot using antiserum against Ape1 as described in Materials and Methods. (B) Autophagy activity was measured with the Pho8Δ60 assay in wild-type (WT), Vac8-GFP, Vac8G2A-GFP, Vac8C4,5,7A-GFP and vac8∆ strains under nutrient-rich conditions (+N) and after 3 h of nitrogen starvation (-N 3 h). Error bars indicate the standard deviation of 3 independent experiments. ANOVA, **P < 0.01; ***P < 0.001. ns, no significance. (C) The protein levels of Vac8 tagged with an auxin-inducible degradation (AID) sequence and a MYC peptide were determined by western blot with anti-MYC after 30 min of auxin (IAA) treatment or control DMSO vehicle in strains co-expressing either Vac8-GFP WT, Vac8-GFP Cyt or Vac8-GFP ER. Pgk1 was used as a loading control, and the asterisk represents a nonspecific band. (D) RFP-Ape1 association with the vacuole was determined using fluorescence microscopy after 1 h of nitrogen starvation in strains expressing Vac8-AID-MYC and the Vac8-GFP constructs described in (C). Strains were treated for 30 min with either IAA to induce Vac8-AID-MYC degradation or DMSO. The vacuolar lumen was stained using CellTracker Blue CMAC. (E) Quantification of panel (D). The percentage of cells in which RFP-Ape1 puncta associated with the vacuole is presented. ANOVA, ***P < 0.001. ns, no significance. (F) Autophagy activity was measured with the Pho8Δ60 assay in strains expressing the Vac8-GFP constructs described in (C), or the vac8∆ strain under nutrient-rich conditions (+N) and after 3 h of nitrogen starvation (-N 3 h). Error bars indicate the standard deviation of 3 independent experiments. ANOVA, *P < 0.05; **P < 0.01. ns, no significance
Vac8 vacuolar localization is required for efficient autophagy
Vac8 is anchored to the vacuolar membrane via the post-translational modification of its N-terminal domain. Thus, Vac8 localization depends on the myristoylation of residue Gly2 and the palmitoylation of residues Cys4, Cys5 and Cys7; mutation of these residues renders Vac8 a cytosolic protein, no longer confined to the vacuolar membrane [11]. One compelling line of data suggesting that Vac8 has a unique function in the Cvt pathway was the observation that a double acylation mutant is completely defective in vacuole inheritance and homotypic vacuole fusion, but is normal for the Cvt pathway [11]. Therefore, it seemed likely that the inheritance and fusion roles of Vac8 depended on its membrane association, whereas a separate soluble pool could fulfill its function in autophagy-related pathways. Mutants of Vac8 that are defective in acylation were previously identified [11]. To test whether acylation of Vac8 has an important role for the Cvt pathway, we generated new strains expressing similar mutations: a myristoylation mutant (G2A), a palmitoylation mutant (C4A C5A C7A) and a myristoylation and palmitoylation double mutant (G2A C4A C5A C7A).
We monitored the Cvt pathway by analyzing the processing of prApe1. When wild-type Vac8 was expressed in the vac8∆ mutant, a portion of the prApe1 population was efficiently processed into mature Ape1, whereas the vac8∆ mutant containing an empty vector showed only prApe1 in vegetative conditions (Figure 2A), suggesting that Vac8 is critical for the Cvt pathway, in agreement with previous results. Next, we examined prApe1 processing in the vac8∆ strain expressing the various established acylation mutants. In apparent contrast to the previous report [11], the myristoylation mutant (G2A), the palmitoylation mutant (C4A C5A C7A) and the myristoylation and palmitoylation double mutant (G2A C4A C5A C7A) failed to completely restore the defect in the Cvt pathway, resulting in a substantial decrease in prApe1 processing; we think this discrepancy between the two sets of data likely represents small differences in culture conditions, because cells that are not in mid-log phase will start to induce autophagy, resulting in prApe1 maturation. These data indicate that Vac8 membrane association is required for an efficient Cvt pathway.
We extended our analysis to examine the requirement of Vac8 acylation for nonselective autophagy, using the Pho8∆60 assay. The myristoylation (G2A) and the palmitoylation (C4A C5A C7A) Vac8 mutants showed a significant decrease in autophagy when compared to a wild-type strain after 3 h of nitrogen starvation (Figure 2B). vac8∆ cells showed no significant difference compared to the mislocalized Vac8 mutants, suggesting Vac8 vacuolar localization is essential for its role in autophagy. Taken together, our data indicate Vac8 acylation and vacuolar localization are necessary for efficiently inducing both the Cvt and autophagy pathways.
Vac8 interacts with the essential autophagy protein Atg13, which, together with Atg1, Atg17, Atg29 and Atg31, forms the autophagy initiation complex that provides the initial structure for the formation of the phagophore [21]. Atg13 has been reported to work as an assembly hub for all other members of the initiation complex [6,10]. Thus, localization of Atg13 to the vacuolar membrane by its interaction with Vac8, could be a determining factor in the recruitment of the initiation complex, the formation of the PAS and autophagy induction [7,15,22]. In order to test this hypothesis, we generated Vac8-inducible degradation strains which also co-expressed a second copy of either wild-type Vac8 (Vac8-GFP WT); a Vac8G2,C4,C5,C7A-GFP mutant rendering Vac8 cytosolic (Vac8-GFP Cyt); or an ER-localized Vac8 (Vac8-GFP ER) made by replacing the first 7 N-terminal amino acid residues of Vac8 with the transmembrane domain of the ER membrane protein Sec66, as previously described [23]. By inducing the rapid degradation of wild-type Vac8 through the AID system and leaving a GFP-tagged copy of Vac8 in place, these strains allowed us to determine the vacuolar association of different Atg proteins without disrupting vacuolar morphology.
In addition to being a Cvt pathway cargo, prApe1 also serves as a marker for the PAS [24,25]. To test if Vac8 vacuolar localization is required for correct PAS localization, we stained the vacuole using the fluorescent dye CellTracker Blue CMAC and measured RFP-Ape1 association with the vacuole in cells expressing either wild-type Vac8-GFP, cytoplasmic Vac8-GFP or ER Vac8-GFP. As described above, to prevent abnormal vacuolar morphology from interfering with our results, endogenous Vac8 degradation was induced by adding IAA, or DMSO as a control, to the growth medium 30 min before nitrogen starvation (Figure 2C), leaving the differentially localized Vac8-GFP chimeras in place; 30 min of IAA treatment, again resulted in substantial depletion of the endogenous Vac8 protein. After 1 h of nitrogen starvation, all Vac8-GFP chimeras treated with DMSO showed consistent RFP-Ape1 association with the vacuole (Figure 2D,E). In contrast, when treated with IAA, only wild-type Vac8-GFP cells showed consistent RFP-Ape1 vacuolar association. IAA treatment of cytosol-localized Vac8-GFP and ER-localized Vac8-GFP cells led to a significant decrease of RFP-Ape1 (i.e., PAS) vacuolar association when compared to their DMSO-treated controls (Figure 2D,E). PAS mislocalization in turn led to a significant decrease in autophagy activity after 3 h of nitrogen starvation, as shown with the Pho8Δ60 assay (Figure 2F). These results indicate that Vac8 vacuolar localization is required for correct PAS perivacuolar localization during nitrogen starvation-induced autophagy.
Vac8 recruits the initiation complex to the vacuole
If the correct perivacuolar localization of the PAS is determined by Vac8, we hypothesized that Vac8 localization must also affect Atg13 and Atg1 recruitment to the vacuole. To test this hypothesis, we tagged Atg13 with RFP and took advantage of our Vac8-inducible degradation Vac8-GFP mislocalization system. As described above, Vac8 degradation was induced by adding IAA to the growth medium 30 min before nitrogen starvation. Similar to our RFP-Ape1 results, after 1 h of nitrogen starvation Atg13-RFP association with the vacuole showed no significant difference between DMSO- and IAA-treated cells when the wild-type Vac8-GFP chimera was expressed (Figure 3). Cells expressing either Vac8-GFP Cyt or Vac8-GFP ER in the absence of IAA (i.e., also expressing a wild-type copy of Vac8) displayed a reduced level of Atg13-RFP localization to the vacuole, presumably due to competition between the wild-type and mutant forms of Vac8 (Figure 3). In contrast to the result with cells expressing wild-type Vac8-GFP, those expressing cytosolic Vac8-GFP or ER-localized Vac8-GFP treated with IAA showed a significant decrease in Atg13-RFP vacuolar localization when compared to the corresponding DMSO-treated controls (Figure 3). Consistent with the ability of Atg13 to bind Atg1, similar results were obtained when these experiments were repeated by tagging Atg1 with RFP (Figure S2).
Figure 3.
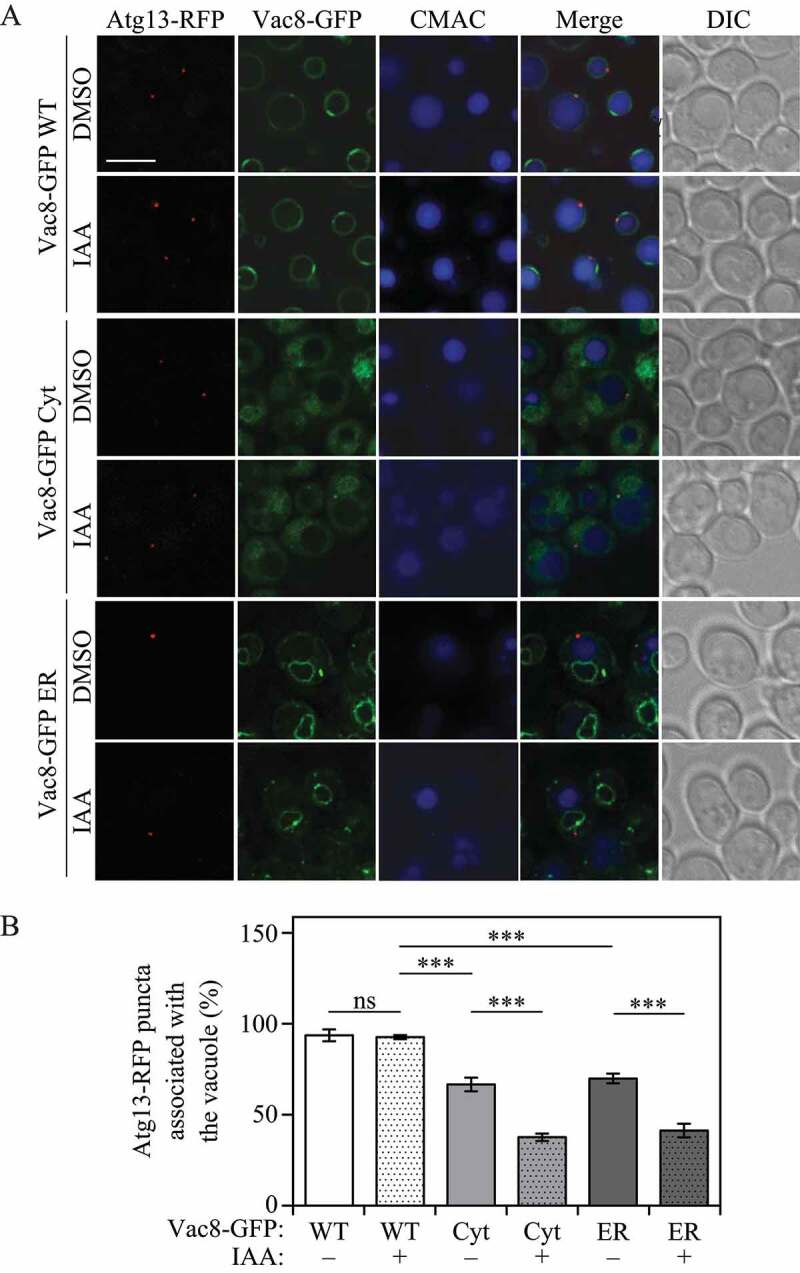
Vac8 vacuolar membrane localization is required for correct Atg13 localization. (A) Atg13-RFP association with the vacuole was determined by fluorescence microscopy after 1 h of nitrogen starvation in strains expressing Vac8-AID-MYC and either Vac8-GFP WT, Vac8-GFP Cyt or Vac8-GFP ER. Strains were treated for 30 min with either IAA to induce Vac8-AID-MYC degradation, or control vehicle DMSO. The vacuolar lumen was stained using CMAC. (B) Quantification of panel (A). The percentage of cells in which Atg13-RFP puncta associated with the vacuole is presented. ANOVA, ***P < 0.001. ns, no significance
While most Atg proteins localize to the PAS, the essential autophagy transmembrane protein Atg9 cycles between peripheral sites and the PAS. Atg9 cycling is thought to be necessary for autophagosome formation by directing the delivery of membranes for phagophore expansion [25–28]. Whereas Atg9 anterograde movement from the peripheral sites to the PAS is regulated by Atg23 and Atg27 [28,29], retrograde trafficking from the PAS to the peripheral sites is controlled by Atg1, Atg2, Atg13 and Atg18 [26,27]. Deletion of ATG1 prevents retrograde Atg9 trafficking, leaving Atg9 trapped at the PAS. Because our results indicate Vac8 has an important role in determining the PAS localization, we hypothesized that changing Vac8 localization would also affect Atg9 localization when trafficking to the PAS. To test this hypothesis, we tagged Atg9 with RFP and deleted ATG1, actively trapping Atg9-RFP at the PAS. Consistent with our other results, Atg9-RFP association with the vacuole showed no significant difference between IAA- and DMSO-treated cells after 1 h of nitrogen starvation when wild-type Vac8-GFP was co-expressed with endogenous Vac8 (Figure 4). However, when either cytosolic Vac8-GFP or ER-localized Vac8-GFP was co-expressed, Atg9-RFP showed a significant decrease in vacuolar localization in cells treated with IAA when compared to vehicle-treated cells (Figure 4). These results indicate that Vac8 is required for correct initiation complex and PAS localization under nitrogen-starvation conditions.
Figure 4.

Vac8 vacuolar membrane localization is required for correct Atg9 anterograde traffic. (A) Atg9-RFP association with the vacuole was monitored using fluorescence microscopy after 1 h of nitrogen starvation in atg1∆ strains expressing Vac8-AID-MYC and either Vac8-GFP WT, Vac8-GFP Cyt or Vac8-GFP ER. Strains were treated for 30 min with either IAA to induce Vac8-AID-MYC degradation, or vehicle DMSO as a control. The vacuolar lumen was stained using CMAC. (B) Quantification of panel (A). The percentage of cells in which Atg9-RFP puncta associated with the vacuole is presented. ANOVA, *P < 0.05; ***P < 0.001. ns, no significance
Localizing Vac8 to peroxisomes increases pexophagy
Our results suggest that the initiation complex members Atg1 and Atg13, and PAS localization, can be targeted to differential cellular locations by expressing Vac8 in those locations. Others have shown that ectopic localization of the Atg1 mammalian ortholog ULK1 to specific organelles can activate selective autophagy of these organelles, such as the selective degradation of peroxisomes, termed pexophagy [30]. Furthermore, directly targeting ULK1 to peroxisomes could bypass the requirement of autophagy receptors to induce pexophagy. We decided to test if localizing Vac8 to peroxisomes could recruit Atg1 and Atg13 to these organelles and in turn increase pexophagy. To this end, we constructed a peroxisome-localized Vac8 (Vac8-RFP Pex) by replacing the first 7 N-terminal amino acid residues of Vac8 with the transmembrane domain of the peroxisomal membrane protein Pex14. Unlike Vac8-RFP WT, Vac8-RFP Pex consistently colocalized with Pex14-GFP, indicating that the chimera was successfully being targeted to peroxisomes (Figure S3A). Consistent with our previous mislocalization experiments, localizing Vac8 to peroxisomes led to a significant decrease in nonselective autophagy measured with the Pho8Δ60 assay (Figure S3B).
To determine if Atg13 could be recruited to peroxisomes by targeting Vac8 to that organelle, Atg13 was tagged with GFP and monitored by fluorescence microscopy. As expected, after 1 h of nitrogen starvation, wild-type Vac8-RFP colocalized with Atg13-GFP to a perivacuolar location (Figure 5A). However, when Vac8-RFP Pex was expressed, Atg13-GFP colocalized with this chimera to peroxisomes, highlighting the ability of the mislocalized Vac8 to recruit the initiation complex.
Figure 5.

Targeting Vac8 to peroxisomes recruits Atg13 and increases pexophagy. (A) Atg13-GFP and either wild-type Vac8-RFP or peroxisome-targeted Vac8-RFP colocalization was determined using fluorescence microscopy. The vacuolar lumen was stained using CMAC. (B) Pexophagy activity was measured with the Pex14-GFP processing assay in Vac8-RFP WT and Vac8-RFP Pex strains, after 0, 2 and 3 h of nitrogen starvation (-N). A representative image is shown. Duplicate sets of samples are shown for each strain. (C) Quantification of panel (B). The ratio of free GFP to Pex14-GFP after 3 h of nitrogen starvation (-N) is presented. Error bars indicate the standard deviation of 5 independent experiments. Student’s t-test, ***P < 0.001. (D) Pexophagy activity was measured using the Pex14-GFP processing assay in WT and atg36∆ cells expressing either Vac8-RFP WT or Vac8-RFP Pex strains, after 0 and 4 h of nitrogen starvation (-N). A representative image is shown. (E) Quantification of panel (D). The ratio of free GFP to Pex14-GFP after 0 and 4 h of nitrogen starvation (-N) is presented. Error bars indicate the standard deviation of 4 independent experiments. ANOVA, *P < 0.05; ***P < 0.001
We next sought to determine if Atg13 recruitment resulting from targeting Vac8 to peroxisomes had an effect on the selective degradation of peroxisomes by autophagy. To this end, we measured pexophagy by the Pex14-GFP processing assay in cells expressing either wild-type Vac8-RFP or peroxisome-targeted Vac8-RFP. Similar to the GFP-Atg8 processing assay, the Pex14-GFP processing assay takes advantage of the relative resistance of GFP to vacuolar hydrolases. Thus, by detecting free GFP by western blot we can measure the amount of peroxisomes being selectively taken into phagophores and delivered to the vacuole for degradation [31]. In order to induce pexophagy in S. cerevisiae, cells were first grown in medium containing oleic acid as the sole carbon source to promote peroxisome proliferation. Cells were then shifted to nitrogen-starvation medium, conditions in which a large quantity of peroxisomes are no longer required by the cell and thus are degraded by pexophagy [32]. After 3 h of nitrogen starvation, cells expressing Vac8-RFP Pex displayed a significant increase in Pex14-GFP processing compared to cells expressing Vac8-RFP WT (Figure 5B,C). To further extend our studies, we decided to determine if Vac8 localization and recruitment of the initiation complex to peroxisomes could bypass the need for Atg36, the S. cerevisiae pexophagy receptor. Atg36, is an essential pexophagy protein that works as a selective autophagy receptor by binding Atg8 and, thus, tethering early phagophore membranes to peroxisomes targeted for degradation [3,33]. Accordingly, we generated strains deleted for ATG36 and measured Pex14-GFP processing in cells expressing either wild-type or peroxisome-localized Vac8-RFP. As expected, after 4 h of nitrogen starvation atg36∆ cells expressing Vac8-RFP WT failed to show any Pex14-GFP processing. Conversely, atg36∆ cells expressing Vac8-RFP Pex showed a small but significant increase in Pex14-GFP processing after 4 h of nitrogen starvation when compared to atg36∆ cells expressing Vac8-RFP WT (Figure 5D,E). Although the level of pexophagy was quite low under these conditions, the increase seen with Vac8-RFP Pex in the complete absence of the normal pexophagy receptor highlights the importance of Vac8-mediated recruitment of the initiation complex to the PAS, and furthers our understanding of how autophagy receptors function during selective autophagy. Together, these results indicate that Vac8 is a determinant factor in the recruitment of the initiation complex to the vacuolar periphery and the formation of the PAS.
Vac8-Atg13 binding is required for correct Atg13 localization and robust autophagy
Our results indicate that Vac8 is required for correct vacuolar localization of the PAS and the initiation complex, as well as robust autophagy induction. It has been reported that Vac8 interacts with the Atg13 C-terminal intrinsically disordered region [15,22]. As previously mentioned, Atg13 is thought to work as a molecular hub for the assembly of the initiation complex [6,10]. We hypothesized that the effects of VAC8 deletion on autophagy could be explained by failure to correctly recruit Atg13 to the vacuole. In order to test this hypothesis, we measured autophagy using the Pho8Δ60 assay in cells expressing either full-length Atg13-GFP or truncated Atg13[1-659]-GFP, which lacks the Vac8 binding region, but still retains one of the phospholipid-binding motifs. After 3 h of nitrogen starvation, autophagy activity was significantly decreased in cells expressing Atg13[1-659]-GFP when compared to full-length Atg13-GFP (Figure 6A). Importantly, deleting VAC8 in an Atg13[1-659]-GFP mutant did not further decrease autophagy activity, suggesting that the autophagy phenotype observed in VAC8 deletion cells could be explained by the inability of Atg13 to be properly localized to the vacuole.
Figure 6.
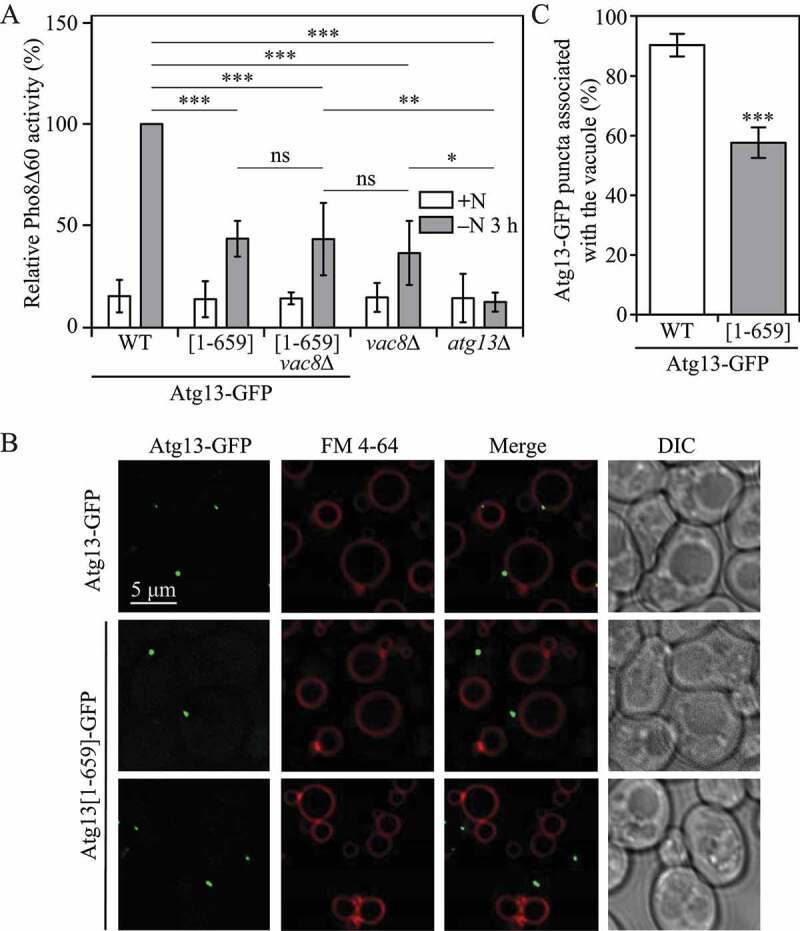
Atg13-Vac8 binding is required for correct initiation complex localization and robust autophagy activity. (A) Autophagy activity was measured using the Pho8Δ60 assay in strains expressing either wild-type Atg13-GFP or truncated Atg13-GFP[1–659], the vac8∆ strain expressing truncated Atg13-GFP[1–659], or vac8∆ or atg13∆ strains under nutrient-rich conditions (+N) and after 3 h of nitrogen starvation (-N 3 h). Error bars indicate the standard deviation of 5 independent experiments. ANOVA, *P < 0.05; **P < 0.01; ***P < 0.001. ns, no significance. (B) Atg13-GFP and truncated Atg13-GFP[1–659] association with the vacuole was determined using fluorescence microscopy. The vacuolar membrane was stained using the dye FM 4–64. (C) Quantification of panel (B). The percentage of cells in which Atg13-GFP puncta associated with the vacuole is presented. Student’s t-test, ***P < 0.001
We next sought to determine Atg13[1-659]-GFP localization by staining the vacuole with the red fluorescent dye FM 4–64. After 1 h of nitrogen starvation, puncta corresponding to full-length Atg13-GFP showed clear perivacuolar localization. However, truncated Atg13[1-659]-GFP puncta failed to associate efficiently with the vacuolar membrane (Figure 6B,C). These results further indicate correct Atg13 vacuolar localization via Vac8 binding is required for robust autophagy induction.
Together, our results indicate that Vac8 determines the vacuolar localization of the PAS during nitrogen starvation-induced autophagy by binding the Atg13 C terminus and recruiting the initiation complex to the vacuole, and that this process is necessary for robust autophagy induction.
Discussion
Autophagy is a critical recycling pathway that requires delicate spatiotemporal organization of multiple cytoplasmic components, such as proteins and membranes from diverse sources, in order to restore cellular homeostasis during nutrient-starvation conditions. Recruitment of the Atg1-Atg13, Atg17-Atg31-Atg29 initiation complex and the formation of the PAS at the vacuolar membrane periphery constitutes one of these key events. However, how the initiation complex is recruited to this perivacuolar location is not well understood. Our results show that the vacuolar membrane protein Vac8 is necessary for the recruitment and proper perivacuolar localization of the initiation complex and the PAS.
Previous studies had suggested that VAC8 deletion leads to a decrease in nonselective autophagy activity [15,16]. However, the mechanism by which Vac8 regulates autophagy remained poorly understood. We determined that the effect of Vac8 on autophagy is unrelated to its function in preserving vacuolar morphology, but rather depends on its vacuolar localization. Indeed, Vac8 acylation mutants, which fail to anchor to the vacuolar membrane, showed decreased auto-phagy activity comparable to the decrease observed in vac8∆ cells. Furthermore, mislocalization experiments targeting Vac8 to the cytosol, ER or peroxisomes resulted in autophagy activity defects similar to the ones observe in VAC8 deletion strains.
Targeting Vac8 to other cellular components also revealed that correct Vac8 vacuolar localization is necessary for proper recruitment of the Atg1-Atg13 initiation complex, PAS localization and Atg9 anterograde trafficking to the vacuolar periphery. The importance of Vac8 in the proper vacuolar assembly of all of these components can be best explained by its ability to bind Atg13 [7,13,22]. As previously mentioned, Atg13 works as a molecular hub for the recruitment of different key Atg proteins necessary for PAS formation and autophagy initiation [6–10]. Truncating Atg13 and deleting its Vac8-binding domain decreased autophagy activity to similar levels as seen with deletion of VAC8 and truncated Atg13 vac8∆ double-mutant strains, suggesting that the effect of VAC8 deletion in autophagy can be explained by Atg13 failure to properly localize to the vacuole. Highlighting this point, whereas both full-length Atg13-GFP and truncated Atg13-GFP were able to form puncta, only full-length Atg13-GFP puncta showed correct perivacuolar localization, suggesting that the deletion of the Vac8-binding domain in Atg13 does not affect its ability to work as a hub for other Atg proteins, but interferes with its ability to be properly tethered to the vacuolar membrane.
Vac8 ability to recruit the initiation complex was further characterized when Vac8 was targeted to peroxisomes. Not only was peroxisome-localized Vac8 able to recruit the initiation complex to peroxisomes, but it also increased the selective degradation of peroxisomes, even partially bypassing the requirement for the pexophagy cargo receptor Atg36. Once again, these results highlighted the importance of Vac8 in determining the location of autophagosome formation.
While this manuscript was in preparation, a recent publication by Hollenstein et al. showed some overlapping results and reached similar conclusions [34]. The authors determined that Vac8 fulfills an important role in autophagy by anchoring the early PAS to the vacuole, coordinating the site of auto-phagosome formation and later vacuolar fusion. Interestingly, an experiment meant to bypass the need for Vac8 by tethering the middle domain of Atg13, 269–520, directly to the vacuole, failed to restore autophagy activity. Whereas this result could be explained by the failure to include the Atg13 N-terminal HORMA domain [35], another explanation could rely on the failure to include recently described Atg13 lipid-binding motifs. Atg13 lipid-binding motifs overlap with its Vac8-binding domain and are required for efficient autophagy. This possibility indicates the need for flexibility in Atg13 binding between Vac8 and lipid membranes [7]. Thus, constant Atg13 tethering to the vacuolar membrane would probably result in decreased flexibility and would be deleterious for autophagy induction.
In mammals, autophagy initiation and phagophore expansion have been mostly attributed to specific PtdIns3P-rich ER subdomains [36]. Once fully matured, autophagosomes must be transported to fuse with lysosomes in order to degrade their cargo, a process that heavily relies on the cytoskeletal network [37]. In contrast, due to the perivacuolar nature of the PAS, Atg protein recruitment and phagophore expansion, nonselective autophagy in yeast does not depend on the cytoskeleton [31]. Our results underline the role of Vac8 as the main driver behind the initiation complex and PAS perivacuolar localization during autophagy induction in yeast. Vac8 interaction with Atg13 fulfills a double role: Whereas Vac8 function restricts the initiation complex and PAS assembly to the vacuolar periphery, Atg13 acts as a molecular hub for the recruitment of Atg1 and the Atg17-Atg31-Atg29 complex, and later downstream targets [6–10]. Although Vac8-mediated spatiotemporal organization of the initiation complex is not essential for autophagy, its involvement in coordinating the formation of autophagosomes in close proximity with their final vacuolar destination highlights its importance in improving the efficiency and strength of the autophagic process.
Material and methods
Yeast strains, media and culture growth
Saccharomyces cerevisiae strains WLY176 and WLY176 GFP-Atg8 were used to generate vac8∆, atg1∆, atg36∆, and atg13∆ strains, as well as strains expressing Vac8-GFP, Atg13-RFP, Atg13-GFP, Atg13[1-659]-GFP, Atg1-RFP, Atg9-RFP and Pex14-GFP, as previously described [38–40]. Saccharomyces cerevisiae strains with genomic point mutations Vac8G2A, Vac8C4,5,7A, and Vac8G2,C4,C5,C7A were generated as previously described [41]. SEY6210 strains expressing Vac8 tagged with an auxin-inducible degradation sequence and MYC peptide were generated as previously described [19]. Auxin-inducible degradation of Vac8 was induced by incubating the cells in YPD with 300 μM 3-indole acetic acid (auxin; Sigma, I2886) or DMSO (vehicle) for 30 min. Yeast cultures were grown in nutrient-rich YPD medium (1% [w:v] yeast extract, 2% [w:v] peptone, 2% [w:v] glucose) to mid-log phase and then samples were collected. Strains grown in YPD were shifted to minus nitrogen medium (0.17% yeast nitrogen base without ammonium sulfate or amino acids, and 2% [w:v] glucose) for the indicated time points and then collected. Pho8Δ60 and western blot analyses were performed as previously described [17,18]. Western blot images were taken in a ChemiDoc™ Touch imaging system (Bio-Rad). Densitometry of western blot images were quantified using ImageJ software. For a full strain list see Table 1.
Table 1.
Yeast strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| DGY056 | WLY176 vac8∆::HIS3 | This study |
| DGY057 | WLY176 atg13∆::HIS3 | This study |
| DGY058 | WLY176 GFP-ATG8::URA3 | This study |
| DGY059 | DGY058 vac8∆::HIS3 | This study |
| DGY060 | SEY6210 pho8::pho8∆60 pNHK53::URA3 VAC8-AID-MYC::HIS3 | This study |
| DGY061 | SEY6210 GFP-ATG8::LEU2 pNHK53::URA3 VAC8-AID-MYC::HIS3 | This study |
| DGY062 | WLY176 VAC8-GFP::HIS3 | This study |
| DGY063 | WLY176 VAC8G2A-GFP::HIS3 | This study |
| DGY064 | WLY176 VAC8C4,5,7A-GFP::HIS3 | This study |
| DGY065 | SEY6210 pNHK53::URA3 VAC8-AID-MYC::HIS3 VAC8-GFP::LEU2 | This study |
| DGY066 | SEY6210 pNHK53::URA3 VAC8-AID-MYC::HIS3 VAC8G2A,C4A,C5A,C7A-GFP::LEU2 | This study |
| DGY067 | SEY6210 pNHK53::URA3 VAC8-AID-MYC::HIS3 SEC66-VAC8-GFP::LEU2 | This study |
| DGY068 | DGY065 RFP-APE1::TRP1 | This study |
| DGY069 | DGY066 RFP-APE1::TRP1 | This study |
| DGY070 | DGY067 RFP-APE1::TRP1 | This study |
| DGY071 | DGY056 VAC8-GFP::LEU2 | This study |
| DGY072 | DGY056 VAC8G2A,C4A,C5A,C7A-GFP::LEU2 | This study |
| DGY073 | DGY056 SEC66-VAC8-GFP::LEU2 | This study |
| DGY074 | DGY065 ATG13-RFP::TRP1 | This study |
| DGY075 | DGY066 ATG13-RFP::TRP1 | This study |
| DGY076 | DGY067 ATG13-RFP::TRP1 | This study |
| DGY077 | DGY065 ATG1-RFP::TRP1 | This study |
| DGY078 | DGY066 ATG1-RFP::TRP1 | This study |
| DGY079 | DGY067 ATG1-RFP::TRP1 | This study |
| DGY080 | DGY065 ATG9-RFP::TRP1 atg1∆::NAT | This study |
| DGY081 | DGY066 ATG9-RFP::TRP1 atg1∆::NAT | This study |
| DGY082 | DGY067 ATG9-RFP::TRP1 atg1∆::NAT | This study |
| DGY083 | WLY176 VAC8-RFP::LEU2 ATG13-GFP::TRP1 | This study |
| DGY084 | WLY176 PEX14-VAC8-RFP::LEU2 ATG13-GFP::TRP1 | This study |
| DGY085 | WLY176 VAC8-RFP::LEU2 PEX14-GFP::TRP1 | This study |
| DGY086 | WLY176 PEX14-VAC8-RFP::LEU2 PEX14-GFP::TRP1 | This study |
| DGY087 | DGY085 atg36∆::NAT | This study |
| DGY088 | DGY086 atg36∆::NAT | This study |
| DGY089 | WLY176 ATG13-GFP::TRP1 | This study |
| DGY090 | WLY176 ATG13[1-659]-GFP::TRP1 | This study |
| DGY091 | DGY090 vac8∆::HIS3 | This study |
| vac8-2 |
MATα leu2-112, 2–3 ura3-52 trp1-∆901 lys2-801 suc2-∆9 vac8∆::HIS3 Vac8G2 S |
[11] |
| vac8-3 |
MATα leu2-112, 2–3 ura3-52 trp1-∆901 lys2-801 suc2-∆9 vac8∆::HIS3 Vac8C4 G,C5 T,C7 S |
[11] |
| [20] | ||
| vac8-15ts |
MATα leu2-112, 2–3 ura3-52 trp1-∆901 lys2-801 suc2-∆9 vac8∆::HIS3 vac8-15ts |
|
| WLY176 | SEY6210 pho13∆ pho8::pho8∆60 | [17] |
Pexophagy assays
In order to measure pexophagy, peroxisome proliferation was induced. To this end, cells were first grown in YPD to approximately OD600 = 0.5 and then shifted to glycerol medium (SGd; 0.67% yeast nitrogen base, 0.1% glucose, 3% glycerol) for 16 h. Subsequently, yeast extract and peptone were added into the SGd medium cultures and cells were incubated for 4 h. The cells were then washed and shifted to oleic acid medium (YTO; 0.67% yeast nitrogen base, 0.1% oleic acid [Sigma, O1008], 0.1% Tween 40 [Sigma, P1504] and auxotrophic amino acids as needed) for 20 h. Finally, pexophagy was induced by shifting the cells to nitrogen-starvation medium containing glucose (SD-N; 0.17% yeast nitrogen base without ammonium sulfate or amino acids, and 2% glucose) for the indicated time points.
Plasmid constructs and insertion
Plasmids pRS405 Vac8-GFP, pRS405 Vac8-RFP and pRS405 Vac8-MYC were constructed using fast-cloning as previously described [42]. Point mutations to generate a pRS405-based plasmid expressing Vac8G2A,C4A,C5A,C7A-GFP were generated as previously described [43]. Plasmid pRS405 Vac8-GFP ER, targeting Vac8-GFP to the endoplasmic reticulum, was generated through fast-cloning, by replacing the first 21 base pairs of the VAC8 ORF with the first 162 base pairs from the SEC66 ORF, which encode the Sec66 transmembrane domain. Plasmid pRS405 Vac8-RFP Pex, targeting Vac8-RFP to peroxisomes, was generated through fast-cloning, by replacing the first 21 base pairs of the VAC8 ORF with the first 390 base pairs from the PEX14 ORF, which encode the Pex14 transmembrane domain and peroxisome localization sequence. Plasmid pRS304 RFP-Ape1 was digested with restriction enzyme AvrII for insertion as previously published [44]. All pRS405 plasmids were digested with restriction enzyme AflII for insertion. For primers see Table S1.
Fluorescence microscopy
Yeast cultures were grown to OD600 ~ 0.5 in YPD, washed and shifted to minus-nitrogen medium for 1 h for auto-phagy induction. For blue lumenal vacuolar staining, cells were incubated in YPD with 10 μM CellTracker Blue CMAC (Molecular Probes/ThermoFisher Scientific, C2110) for 30 min and then washed to remove excess dye. For red membrane vacuolar staining, cells were incubated in YPD with 30 μM FM 4–64 (Molecular Probes/ThermoFisher Scientific, T3166) for 30 min and then washed to remove excess dye. Images were collected on a DeltaVision Elite deconvolution microscope (GE Healthcare/Applied Precision) with a 100x objective and a CCD camera (CoolSnap HQ; Photometrics). For the quantification of fluorescent puncta associated with the vacuole, stacks of 15 image planes were collected with a spacing of 0.2 μm to cover the entire yeast cell. Analysis was performed on an average projection of the imaging planes on ImageJ software.
Statistics and reproducibility
Microscopy data analysis and image processing were carried out using softWoRx software (GE Healthcare). Sample sizing for cellular imaging was chosen to be the minimum number of independent experiments required for statistically significant results. Western blot images were quantified using ImageJ software. Statistical analyses were performed using GraphPad Prism 6. Statistical significance was determined in all cases from at least 3 independent experiments using either Student’s t-test or ANOVA. Differences with a P value <0.05 or lower were considered significant. *p < 0.05, **p < 0.01, ***p < 0.001. The number of independent experiments (n), statistical test utilized, distribution of measurements and significance are described in the figure legends.
Supplementary Material
Funding Statement
This work was supported by grant GM131919 to DJK.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Feng Y, He D, Yao Z, et al. The machinery of macroautophagy. Cell Res. 2014Jan;24(1):24–41. PubMed PMID: 24366339; PubMed Central PMCID: PMCPMC3879710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Wen X, Klionsky DJ.. An overview of macroautophagy in yeast. J Mol Biol. 2016May8;428(9 Pt A):1681–1699. PubMed PMID: 26908221; PubMed Central PMCID: PMCPMC4846508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Gatica D, Lahiri V, Klionsky DJ. Cargo recognition and degradation by selective autophagy. Nat Cell Biol. 2018Mar;20(3):233–242. PubMed PMID: 29476151; PubMed Central PMCID: PMCPMC6028034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Suzuki K, Kubota Y, Sekito T, et al. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007Feb;12(2):209–218. PubMed PMID: 17295840. [DOI] [PubMed] [Google Scholar]
- [5].Cao Y, Cheong H, Song H, et al. In vivo reconstitution of autophagy in Saccharomyces cerevisiae. J Cell Biol. 2008Aug25;182(4):703–713. PubMed PMID: 18725539; PubMed Central PMCID: PMCPMC2518709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Fujioka Y, Suzuki SW, Yamamoto H, et al. Structural basis of starvation-induced assembly of the autophagy initiation complex. Nat Struct Mol Biol. 2014Jun;21(6):513–521. PubMed PMID: 24793651. [DOI] [PubMed] [Google Scholar]
- [7].Gatica D, Damasio A, Pascual C, et al. The carboxy terminus of yeast Atg13 binds phospholipid membrane via motifs that overlap with the Vac8-interacting domain. Autophagy. 2019Aug2:1–14. DOI: 10.1080/15548627.2019.1648117. PubMed PMID: 31352862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Jao CC, Ragusa MJ, Stanley RE, et al. A HORMA domain in Atg13 mediates PI 3-kinase recruitment in autophagy. Proc Natl Acad Sci U S A. 2013Apr2;110(14):5486–5491. PubMed PMID: 23509291; PubMed Central PMCID: PMCPMC3619307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Kamada Y, Funakoshi T, Shintani T, et al. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000Sep18;150(6):1507–1513. PubMed PMID: 10995454; PubMed Central PMCID: PMCPMC2150712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Yamamoto H, Fujioka Y, Suzuki SW, et al. The intrinsically disordered protein Atg13 mediates supramolecular assembly of autophagy initiation complexes. Dev Cell. 2016Jul11;38(1):86–99. PubMed PMID: 27404361. [DOI] [PubMed] [Google Scholar]
- [11].Wang YX, Catlett NL, Weisman LS. Vac8p, a vacuolar protein with armadillo repeats, functions in both vacuole inheritance and protein targeting from the cytoplasm to vacuole. J Cell Biol. 1998Mar9;140(5):1063–1074. PubMed PMID: 9490720; PubMed Central PMCID: PMCPMC2132703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Pan X, Roberts P, Chen Y, et al. Nucleus-vacuole junctions in Saccharomyces cerevisiae are formed through the direct interaction of Vac8p with Nvj1p. Mol Biol Cell. 2000Jul;11(7):2445–2457. PubMed PMID: 10888680; PubMed Central PMCID: PMCPMC14931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Jeong H, Park J, Kim HI, et al. Mechanistic insight into the nucleus-vacuole junction based on the Vac8p-Nvj1p crystal structure. Proc Natl Acad Sci U S A. 2017Jun6;114(23):E4539–E4548. PubMed PMID: 28533415; PubMed Central PMCID: PMCPMC5468681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kvam E, Goldfarb DS. Nucleus-vacuole junctions and piecemeal microautophagy of the nucleus in S. cerevisiae. Autophagy. 2007Mar-Apr;3(2):85–92. PubMed PMID: 17204844. [DOI] [PubMed] [Google Scholar]
- [15].Scott SV, Nice DC 3rd, Nau JJ, et al. Apg13p and Vac8p are part of a complex of phosphoproteins that are required for cytoplasm to vacuole targeting. J Biol Chem. 2000Aug18;275(33):25840–25849. PubMed PMID: 10837477. [DOI] [PubMed] [Google Scholar]
- [16].Cheong H, Yorimitsu T, Reggiori F, et al. Atg17 regulates the magnitude of the autophagic response. Mol Biol Cell. 2005Jul;16(7):3438–3453. PubMed PMID: 15901835; PubMed Central PMCID: PMCPMC1165424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Noda T, Klionsky DJ. The quantitative Pho8Delta60 assay of nonspecific autophagy. Methods Enzymol. 2008;451:33–42. PubMed PMID: 19185711. [DOI] [PubMed] [Google Scholar]
- [18].Shintani T, Klionsky DJ. Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J Biol Chem. 2004Jul16;279(29):29889–29894. PubMed PMID: 15138258; PubMed Central PMCID: PMCPMC1712665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Morawska M, Ulrich HD. An expanded tool kit for the auxin-inducible degron system in budding yeast. Yeast. 2013Sep;30(9):341–351. PubMed PMID: 23836714; PubMed Central PMCID: PMCPMC4171812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Wang YX, Kauffman EJ, Duex JE, et al. Fusion of docked membranes requires the armadillo repeat protein Vac8p. J Biol Chem. 2001Sep14;276(37):35133–35140. PubMed PMID: 11441010. [DOI] [PubMed] [Google Scholar]
- [21].Ragusa MJ, Stanley RE, Hurley JH. Architecture of the Atg17 complex as a scaffold for autophagosome biogenesis. Cell. 2012Dec21;151(7):1501–1512. PubMed PMID: 23219485; PubMed Central PMCID: PMCPMC3806636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Park J, Kim HI, Jeong H, et al. Quaternary structures of Vac8 differentially regulate the Cvt and PMN pathways. Autophagy. 2019Sep12:1–16. DOI: 10.1080/15548627.2019.1659615. PubMed PMID: 31512555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Fakieh MH, Drake PJ, Lacey J, et al. Intra-ER sorting of the peroxisomal membrane protein Pex3 relies on its luminal domain. Biol Open. 2013Aug15;2(8):829–837. PubMed PMID: 23951409; PubMed Central PMCID: PMCPMC3744075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Scott SV, Hefner-Gravink A, Morano KA, et al. Cytoplasm-to-vacuole targeting and autophagy employ the same machinery to deliver proteins to the yeast vacuole. Proc Natl Acad Sci U S A. 1996Oct29;93(22):12304–12308. PubMed PMID: 8901576; PubMed Central PMCID: PMCPMC37986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Feng Y, Backues SK, Baba M, et al. Phosphorylation of Atg9 regulates movement to the phagophore assembly site and the rate of autophagosome formation. Autophagy. 2016;12(4):648–658. PubMed PMID: 27050455; PubMed Central PMCID: PMCPMC4835963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Reggiori F, Shintani T, Nair U, et al. Atg9 cycles between mitochondria and the pre-autophagosomal structure in yeasts. Autophagy. 2005Jul;1(2):101–109. PubMed PMID: 16874040; PubMed Central PMCID: PMCPMC1762033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Reggiori F, Tucker KA, Stromhaug PE, et al. The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev Cell. 2004Jan;6(1):79–90. PubMed PMID: 14723849. [DOI] [PubMed] [Google Scholar]
- [28].Yen WL, Legakis JE, Nair U, et al. Atg27 is required for autophagy-dependent cycling of Atg9. Mol Biol Cell. 2007Feb;18(2):581–593. PubMed PMID: 17135291; PubMed Central PMCID: PMCPMC1783788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Backues SK, Orban DP, Bernard A, et al. Atg23 and Atg27 act at the early stages of Atg9 trafficking in S. cerevisiae. Traffic. 2015Feb;16(2):172–190. PubMed PMID: 25385507; PubMed Central PMCID: PMCPMC4305007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Vargas JNS, Wang C, Bunker E, et al. Spatiotemporal control of ULK1 activation by NDP52 and TBK1 during selective autophagy. Mol Cell. 2019Apr18;74(2):347–362 e6. PubMed PMID: 30853401; PubMed Central PMCID: PMCPMC6642318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Reggiori F, Monastyrska I, Shintani T, et al. The actin cytoskeleton is required for selective types of autophagy, but not nonspecific autophagy, in the yeast Saccharomyces cerevisiae. Mol Biol Cell. 2005Dec;16(12):5843–5856. PubMed PMID: 16221887; PubMed Central PMCID: PMCPMC1289426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Hutchins MU, Veenhuis M, Klionsky DJ. Peroxisome degradation in Saccharomyces cerevisiae is dependent on machinery of macroautophagy and the Cvt pathway. J Cell Sci. 1999Nov;112(Pt 22):4079–4087. PubMed PMID: 10547367. [DOI] [PubMed] [Google Scholar]
- [33].Motley AM, Nuttall JM, Hettema EH. Pex3-anchored Atg36 tags peroxisomes for degradation in Saccharomyces cerevisiae. Embo J. 2012Jun29;31(13):2852–2868. PubMed PMID: 22643220; PubMed Central PMCID: PMCPMC3395097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Hollenstein DM, Gomez-Sanchez R, Ciftci A, et al. Vac8 spatially confines autophagosome formation at the vacuole in S. cerevisiae. J Cell Sci. 2019Nov14;132(22):jcs235002. PubMed PMID: 31649143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Suzuki SW, Yamamoto H, Oikawa Y, et al. Atg13 HORMA domain recruits Atg9 vesicles during autophagosome formation. Proc Natl Acad Sci U S A. 2015Mar17;112(11):3350–3355. PubMed PMID: 25737544; PubMed Central PMCID: PMCPMC4371973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Lamb CA, Yoshimori T, Tooze SA. The autophagosome: origins unknown, biogenesis complex. Nat Rev Mol Cell Biol. 2013Dec;14(12):759–774. PubMed PMID: 24201109. [DOI] [PubMed] [Google Scholar]
- [37].Kimura S, Noda T, Yoshimori T. Dynein-dependent movement of autophagosomes mediates efficient encounters with lysosomes. Cell Struct Funct. 2008;33(1):109–122. PubMed PMID: 18388399. [DOI] [PubMed] [Google Scholar]
- [38].Gueldener U, Heinisch J, Koehler GJ, et al. A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res. 2002Mar15;30(6):e23. PubMed PMID: 11884642; PubMed Central PMCID: PMCPMC101367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Longtine MS, McKenzie A 3rd, Demarini DJ, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998Jul;14(10):953–961. PubMed PMID: 9717241. [DOI] [PubMed] [Google Scholar]
- [40].Goldstein AL, McCusker JH. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999Oct;15(14):1541–1553. PubMed PMID: 10514571. [DOI] [PubMed] [Google Scholar]
- [41].Toulmay A, Schneiter R. A two-step method for the introduction of single or multiple defined point mutations into the genome of Saccharomyces cerevisiae. Yeast. 2006Aug;23(11):825–831. PubMed PMID: 16921548. [DOI] [PubMed] [Google Scholar]
- [42].Li C, Wen A, Shen B, et al. FastCloning: a highly simplified, purification-free, sequence- and ligation-independent PCR cloning method. BMC Biotechnol. 2011Oct12;11:92. PubMed PMID: 21992524; PubMed Central PMCID: PMCPMC3207894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Liu H, Naismith JH. An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol. 2008Dec4;8:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Wang K, Yang Z, Liu X, et al. Phosphatidylinositol 4-kinases are required for autophagic membrane trafficking. J Biol Chem. 2012Nov2;287(45):37964–37972. PubMed PMID: 22977244; PubMed Central PMCID: PMCPMC3488067. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


