Abstract
Although many studies have confirmed the relationship between obesity and endometrial cancer (EC), the molecular mechanism between obesity and EC progression has not been elucidated. Overexpression of fat mass and the obesity associated protein FTO leads to weight gain, although recently it has been discovered that FTO can serve as a demethylase which erases N6-methyladenosine (m6A) modification and regulates the metabolization of mRNAs. In this study, we found high expression of FTO in metastatic EC and that this action promote both metastasis and invasion in vivo and in vitro. Mechanistically, FTO can catalyse demethylation modification in 3ʹUTR region of HOXB13 mRNA, thereby abolishing m6A modification recognition with the YTHDF2 protein. Decreasing HOXB13 mRNA decay and increasing HOXB13 protein expression was accompanied by WNT signalling pathway activation and the expression of downstream proteins, leading to tumour metastasis and invasion. We also found the WNT signalling pathway inhibitor ICG-001 can block HOXB13 gene-induced tumour metastasis, therefore ICG-001 may be a promising molecular intervention. This study provides insight into the relationship between obesity and the pathogenesis of endometrial cancer while highlighting future areas of research.
KEYWORDS: Endometrial cancer, fto, hoxb13, m6A, cancer metastasis
Introduction
The incidence of obesity in the population is increasing, and epidemiological studies have shown that obesity is a high-risk factor for endometrial cancer (EC) [1]. According to the clinicopathological characteristics, EC can be divided into type I and II [2]. Type I which are predominantly endometrial adenocarcinoma, are considered to be related to oestrogen stimulation with characteristically slow progression and late metastasis [3]. Type II ECs are mainly clear cell carcinoma and serous carcinoma which progress rapidly, and are often accompanied by resistance to hormone therapy. In type II ECs, tumour invasion and metastasis occur more readily in the earlier stages and therefore pose a very serious threat to one’s physical well-being. Unfortunately, the 5-year survival rate of patients with type II ECs is significantly reduced [4,5]. Research into the molecular mechanisms involved in tumour invasion and metastasis is of the utmost importance if we are to improve such dismay outcomes for patients with type II ECs.
m6A is the most common chemical modification in mammalian mRNA [6]. In the nucleus, m6A modifications on mRNA are dynamically regulated, and proteins such as METTL3/14 catalyse the formation of m6A modifications on specific RNAs by forming methyltransferase complexes. Therefore, they are also called ‘writers’ [7,8]. Also known as ‘erasers’, FTO and ALKBH5 proteins can demethylate m6A modifications from mRNA [9]. When mRNA matures, these m6A-modified mRNAs are transported into the cytoplasm. Some proteins, known as ‘readers’ can specifically recognize m6A modifications and regulate mRNA metabolic processes [10]. The YTHDF family of proteins are the first ‘reader’ proteins that have been shown to recognize m6A modification [11]. After YTHDF1 recognizes an m6A modification of mRNA, the translation process accelerates [12]. When YTHDF2 recognizes m6A modification it will promote the transportation of mRNA into the p-body, accelerating mRNA degradation, while inhibiting protein translation [13]. It has been found that abnormal expression of these proteins exists in various tumours and affects the occurrence and tumour progression [14].
FTO belongs to the non-haem Fe II/αKG-dependent dioxygenase AlkB protein family, and its expression is closely related to weight gain and obesity [15]. Knocking down FTO expression in animal models can suppress obesity and lead to growth retardation, while FTO overexpression can lead to increased food intake and obesity [16]. Previous studies have shown that abnormal expression of FTO has been found in various tumours and participates in the regulation of multiple biological behaviours [17,18]. However, FTO has recently been found to demethylase, leading to m6A demethylation in mRNA and regulating mRNA metabolism. The relationship of this new molecular mechanism of FTO and cancer progression is still unclear.
In this study, we studied the regulatory effects of FTO on endometrial cancer (EC) invasion and metastasis and explored the molecular mechanism of tumour metastasis based on the m6A regulatory mechanism.
Materials and methods
Patient samples
RNA samples from 96 EC patients were sought from our previous research. Among these, 30 were patients with metastasis and 66 patients without. This study was approved by the Institutional Review Board within the First Affiliated Hospital of Nanjing Medical University.
Quantitative real-time PCR analysis
RNA samples were reverse transcribed to cDNA by reverse transcriptase (TaKaRa, Tokyo, Japan). Gene expression was detected using a quantitative PCR kit (TaKaRa) and a 7900HT real-time instrument (ABI, CA, USA). Experimental procedures followed the manufacturers’ protocols. The primers are listed in Table S1. Transcript levels were analysed using the 2−ΔΔCT method, and the expression of β-actin was used as an internal control.
Tissue array and immunohistochemistry (IHC)
A total of 142 tumour samples were included in this study, involving 66 endometrial tumour samples (without metastases), 30 intrauterine tumour samples (with metastases), 24 abdominal metastatic samples, and 22 lymph node metastasis samples. The IHC protocol was similar to that used in our previous research [19].
After paraffin embedding, sectioning, dewaxing, antigen retrieval and serum blocking, the samples were stained with antibodies. Antibody information is provided in the supplementary materials section, as Table S2. IHC staining was semi-quantitatively evaluated based on stain intensity and the percentage of positive cells. Stain intensities were categorized as 0 (negative), 1 (weakly positive), 2 (moderately positive), or 3 (strongly positive). The percentage of positive cells were categorized as 1 (0%–10%), 2 (11%–50%), 3 (51%–80%), or 4 (81%–100%). The final score for each section was determined by multiplying the staining intensity score by the percentage staining score. IHC staining was independently scored by two pathologists, before calculating averages.
Cells and cell culture
The human EC cell lines AN3CA and KLE were purchased from the China Centre for Type Culture Collection (CATCC). Cells were cultured as described in our previous study [19].
Subcellular fractionation analysis
Protein fractionation of nuclear extracts was performed by using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Fisher, CA, USA) according to the manufacturer’s instructions.
Western blotting (WB)
Cells were washed twice in pre-chilled PBS and lysed in lysis buffer (100 mmol/L NaCl, 50 mmol/L Tris (pH 8.0), 1% NP-40, 0.1% SDS, 0.5 mM EDTA and 1× proteinase inhibitor cocktail). Lysates were centrifuged at 12,500 g. Supernatants were collected and proteins were denatured using LDS buffer (Thermo Fisher). Protein samples were separated by SDS-PAGE and transferred to a PVDF membrane. Membranes were blocked with 5% skim milk and sequentially incubated with primary and secondary antibodies. Antibody information is listed in the supplementary materials as Table S2. The immunoblot was developed using ChemiDoc (Bio-Rad) and ECL (Bio-Rad) or hypersensitive ECL chemiluminescence substrate (Thermo Fisher).
Lentivirus construction and infection, plasmid and siRNA transfection
The lentivirus-mediated gene overexpression or knockdown vector system was packed by Hanbio and included lentiviral vectors carrying FLAG and puromycin tags. Cells were cultured in six-well plates, and lentiviruses were added to each well. Empty lentiviral vectors served as a negative control, and noninfected cells served as a control group. After 48 h, the cells were cultured in 5 μg/ml puromycin to screen for cells with stable gene expression. Gene expression was verified by RT-PCR and WB. The HOXB13 CDS region was cloned into pcDNA 3.1 to construct an overexpression vector. HOXB13-specific siRNAs were synthesized by Tsingke. Cells were transfected with expression vector and siRNA using lipofectamine 3000 (Invitrogen, MI, USA) according to the manufacturer’s protocol. The oligo sequences are provided in the supplementary materials as Table S1.
Inhibition of the WNT signalling pathway
We used ICG-001 (Selleck, MI, USA) to inhibit the activity of the WNT signalling pathway in cells. The drug was diluted to a 10 mM stock solution with DMSO and stored at −20°C. The medium was used to dilute the stock solution to the working concentration (10 μM).
Wound-healing assays
EC cells were seeded in a six-well plate after the indicated treatment. When the cells density reached 90%, a plastic pipette tip was used scratch the cell surface. PBS was used to wash the cell debris two times. Serum-free medium was added to each well for culture. After 48 h, the cells were photographed for analysis of the cell migration ability.
Transwell assay
A modified two-chamber migration assay was used to verify the cell invasion ability. Fifty microlitres of 1:5 diluted Matrigel (BD, MA, USA) was added to the chamber (pore size: 8 µm; Corning, NY, USA) and placed in a 37°C incubator for Matrigel solidification. After the indicated treatment, the designated cells were suspended in serum-free medium. 2 × 104 EC cells were seeded in the upper chamber, and 500 µl of medium containing 20% serum was added into the lower chamber. After the cells were cultured for 24 h, the cells were fixed with 4% paraformaldehyde, and a cotton swab was used to carefully wipe the cells in the upper chamber before photographs were taken.
RNA-binding protein immunoprecipitation sequencing and quantitative PCR (RIP-seq and RIP-qPCR)
To detect RNAs that bind to the FTO protein, an EZ-Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (Millipore, CA, USA) was used according to the manufacturer’s protocol. Briefly, AN3CA cells (1 × 107) were washed in PBS twice and then lysed in 500 μl IP lysis buffer (containing 1x protease inhibitor cocktail and 5 μl RNase inhibitor). The supernatant was collected by centrifugation, and 10% was kept as the input sample. An anti-Flag antibody (5 μg) and a magnetic beads complex (40 μl) were added to the remaining 90% of the supernatant, and the samples were incubated at 4°C overnight. Beads were washed with wash buffer and eluted using an RNA purification kit (Qiagen, Germany) according to the manufacturer’s protocol. An Ultra II RNA Kit (NEB, MI, USA) was used to prepare the libraries according to the manufacturer’s protocol. The samples were sequenced using the HiSeq PE150 platform. qPCR was performed by a 7900HT real-time instrument. The primers are listed in Table S1.
Methylated RNA immunoprecipitation sequencing (MeRIP-seq) and quantitative PCR (MeRIP-PCR)
Cells were washed in cold PBS twice, and an Oligotex Direct mRNA Midi/Maxi Kit (Thermo Fisher) was used to purify the mRNA according to the manufacturer’s protocol. The mRNA concentration was quantified using a spectrophotometer, and 5 μg of mRNA was used for immunoprecipitation. A Magna MeRIP m6A Kit (Millipore) was used for MeRIP according to the manufacturer’s protocol. The samples were sequenced with the HiSeq PE150 platform. qPCR was performed by a 7900HT real-time instrument. The primers are again listed in Table S1 of the supplementary materials.
RNA sequencing (RNA-seq) and quantitative PCR (qPCR)
Cells were washed twice with prechilled PBS. RNA was extracted using an RNeasy Mini Kit (Qiagen) according to the manufacturer’s protocol. An Ultra II RNA Kit (NEB) was used to prepare the libraries according to the manufacturer’s protocol. The libraries were sequenced with the HiSeq PE150 platform. qPCR was performed by a 7900HT real-time instrument.
RNA pull-down assays
The 5ʹ-biotin RNA probe was synthesized by Tsingke. RNA probes contain either an m6A base, an adenine base or a mutated guanine base. RNAiMAX reagents (Invitrogen) were used to transfect RNA probes into cells. After 48 h, cells were washed twice in cold PBS and lysed in 500 μl IP lysis buffer (containing 1x protease inhibitor cocktail and 5 μl RNase inhibitor). One-tenth of the supernatant was saved as the input sample. Then, 40 μl streptavidin beads (Invitrogen) were added to the remaining supernatant, and samples were incubated at room temperature for 2 h. Beads were eluted by 95% formamide at 95°C for 2 min after five washes with IP wash buffer (Thermo Fisher). Protein samples were denatured by LDS buffer (Thermo Fisher) and analysed by WB.
Luciferase reporter assays
We cloned a 3ʹUTR sequence containing the m6A site of HOXB13 into the pmirGLO reporter vector (Promega, CA, USA) and constructed a reporter gene vector with a single nucleotide mutation sequence (Fig. S1A). The detailed DNA sequences are provided in Table S1, in the supplementary materials. Plasmids were verified by DNA sequencing. Cells seeded in 6-well plates were transfected with luciferase vectors using Lipofectamine 3000 (Invitrogen). Luciferase assays were performed using a Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions. After 48 h, the firefly and Renilla luciferase activities in each well were evaluated.
RNA half-life assays
Cells were treated according to the experimental design, and actinomycin D (Sigma, CA, USA) was added at a concentration of 5 mg/ml. At the indicated time points (3, 6 hr), the cells were lysed, and total RNA was extracted (Qiagen). RNA quantities were determined by qPCR. The RNA half-life was calculated according to a previous study [20].
In vivo tumorigenesis assay
Experimental animal procedures were approved by the Institutional Animal Care and Committee (Peking Union Medical College). All animals received care in compliance with the ‘Guide for the Care and Use of Laboratory Animals’. Female SCID-Beige mice (6 weeks old) were purchased from Vitalriver. AN3CA cells with FTO overexpression or silencing or the appropriate controls (1 × 106) were injected into the lower abdominal cavity of mice (n = 5 mice/group). After 4 weeks, the mice were sacrificed, and tumours were harvested, weighed and photographed. Tumours were fixed in 4% formaldehyde, paraffin-embedded and analysed by IHC.
Sequencing data analysis
RNA-seq/RIP-seq datasheets were analysed by HISAT2, featureCounts and DESeq2. MeRIP-seq data were analysed by HISAT2 and exomePeak software. The correlation analysis of HOXB13 and YTHDF2 mRNA expression comes from TCGA database, and this work was performed by GEPIA (http://gepia.cancer-pku.cn).
Statistical analysis
SPSS 21.0 was used for all statistical analyses. Correlations between gene expression and clinicopathological data were analysed by χ2 and Fisher’s exact test, respectively. Survival curves were generated using the Kaplan-Meier method. Data are expressed as the mean ± SD. Differences between two groups were analysed by two-tailed Student’s t-test. The threshold for statistical significance was set at P < 0.05.
Results
FTO expression is increased in metastatic EC samples
To explore the relationship between FTO expression and EC metastasis, we divided patients into metastatic and non-metastatic groups. qPCR was used to detect the expression of FTO in EC samples, and FTO expression was higher in metastatic patients than in nonmetastatic patients (Fig. 1A). A tissue array was used to detect the protein expression of FTO in EC. The intrauterine samples were divided into metastatic and non-metastatic groups according to whether there was metastasis. The expression of FTO in the metastatic group was significantly higher than that in the non-metastatic group (Fig. 1B, Table S3). In addition, the expression of FTO in peritoneal metastases and lymph node metastasis tumours was significantly higher than that observed in non-metastatic tumours, although there was no significant difference in expression between various metastatic samples (Fig. 1C, Table S3).
Figure 1.
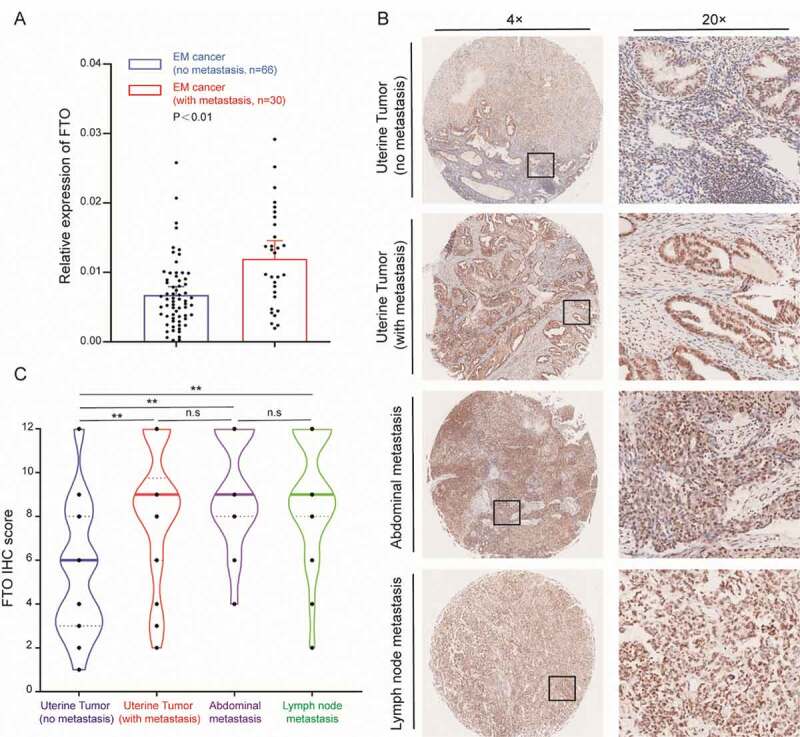
FTO is overexpressed in metastatic EC cancer. (A) FTO mRNA expression in metastatic EC (N = 30) and nonmetastatic EC tissues (N = 66). (B) Expression of the FTO protein in nonmetastatic EC tissue and different types of metastatic EC tissues. (C) The IHC score of the FTO protein in different EC samples, (No metastasis, N = 66), (With metastasis, N = 30), (Abdominal metastasis, N = 24), (Lymph node metastasis, N = 22). **P < 0.01, n.s indicates not significant
The expression of FTO in normal endometrium was significantly lower than that in tumour tissue (Fig. S2A). This indicates that FTO expression is significantly increased in metastatic patients and may promote EC metastasis. In addition, we measured the ratio of m6A/A in tumour samples from 14 patients, and found that the m6A levels in metastatic tumours were significantly lower than that observed in non-metastatic tumours (Fig. S3A). However, there was no significant difference in the expression of METTL3 and METTL14 between the two groups (Fig. S3B, C). The expression of ALKBH5 and YTHDF2 in metastatic tumours significantly increased (Fig. S3D, F), while the expression of YTHDF1 significantly decreased (Fig. S3E)
FTO promotes the invasion and metastasis of EC
To explore the role of FTO in regulating the invasion and metastasis of EC, we selected the AN3CA cell line derived from lymph node metastases and KLE cells with high metastatic potential for follow-up experiments. We used a lentiviral transfection vector to stably overexpress and knockdown FTO expression in cells (Fig. S4A-B). The wound-healing experiment showed that the metastatic ability significantly increased after enhancing the expression of FTO in KLE cells. By contrast, after silencing FTO expression in KLE cells, cell metastasis lowered significantly (Fig. 2A). A similar trend was observed in the AN3CA cell line (Fig. 2B).
Figure 2.
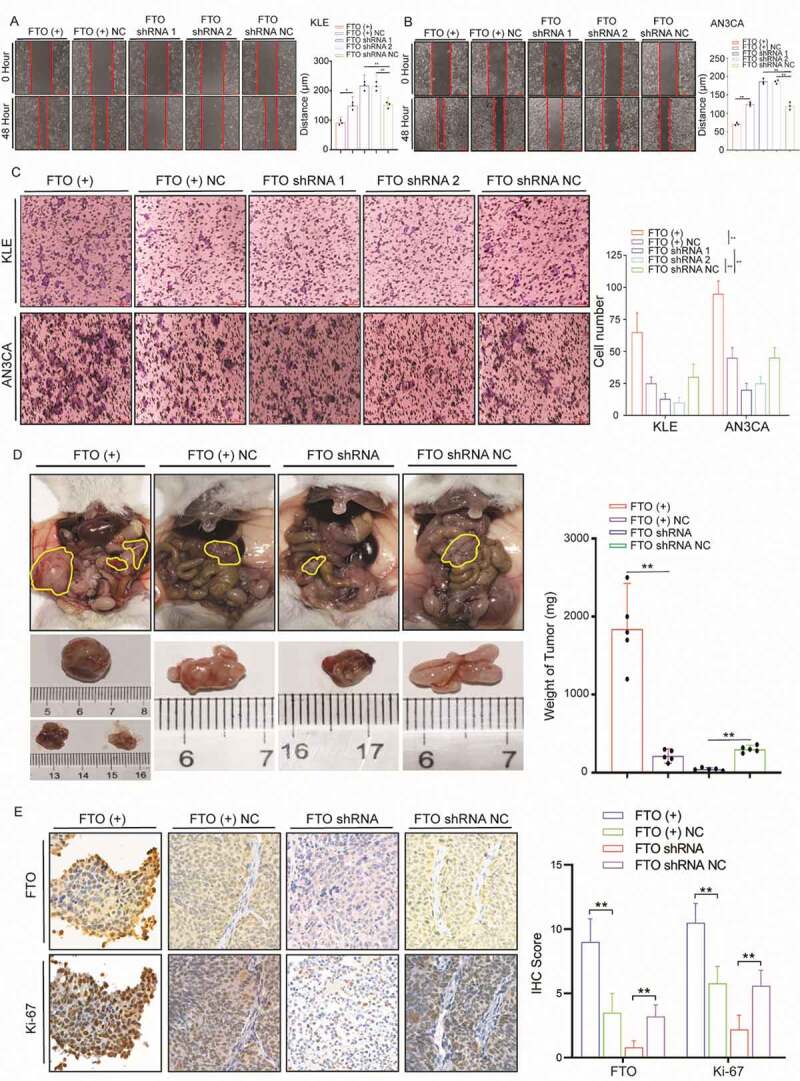
FTO promotes EC cell metastasis and invasion. (A) FTO overexpression and knockdown regulate cell migratory abilities in KLE cells according to wound-healing assays. (B) Wound-healing assays demonstrate that FTO regulates cell metastasis of AN3CA cells. (C) Effects of FTO overexpression and knockdown on cell invasive capacities by Transwell assays. (D) Weight of xenografts derived from AN3CA cells (n = 5 mice/group). (E) IHC staining of FTO and Ki-67 in tumour samples. FTO (+) NC: Negative control lentiviral vector. FTO shRNA: knockdown of FTO by shRNA lentiviruses. FTO shRNA NC: Negative control shRNA lentiviruses. Error bars indicate means ± SDs, **P < 0.01
Transwell assays were used to analyse the effect of FTO on invasion ability. Overexpression/knockdown of FTO expression led to an increased/decreased ability for cell invasion (Fig. 2C). Mouse peritoneal tumour models were used to explore the effect of FTO on metastasis in vivo. The intra-abdominal tumours were significantly larger than those in the control group after FTO overexpression and often formed multiple tumours. By contrast, after knockdown of FTO expression, tumour weight significantly reduced (Fig. 2D). The intensity of Ki-67 increased after FTO overexpression, whereas Ki-67 decreased upon FTO knockdown (Fig. 2E).
FTO removes the m6A modification in the 3ʹUTR region of HOXB13 mRNA
In order to explore the molecular mechanism involved in FTO promoting tumour invasion and metastasis, we stably knocked down the expression of FTO in AN3CA cells by shRNA. RNA-seq was adopted to analyse differentially expressed genes. There were 881 upregulated genes and 1696 downregulated genes (fold-change (FC) log2 > 2, p < 0.01) (Table S5, Fig. S5A). KEGG and GO analyses showed that the molecular functions of the differentially expressed genes were related to cell adhesion and metastasis (Fig. 3A, Fig. S5B). Further, we used RIP-seq to detect the RNA bound to FTO and enriched to a total of 1316 genes (FC log2 > 3). GO analysis showed that these RNAs were associated with apoptosis, cell adhesion and metastasis (Fig. S5C). KEGG clustering showed that these genes were mainly enriched in the PI3K and MAPK signalling pathways (Fig. S5D).
Figure 3.
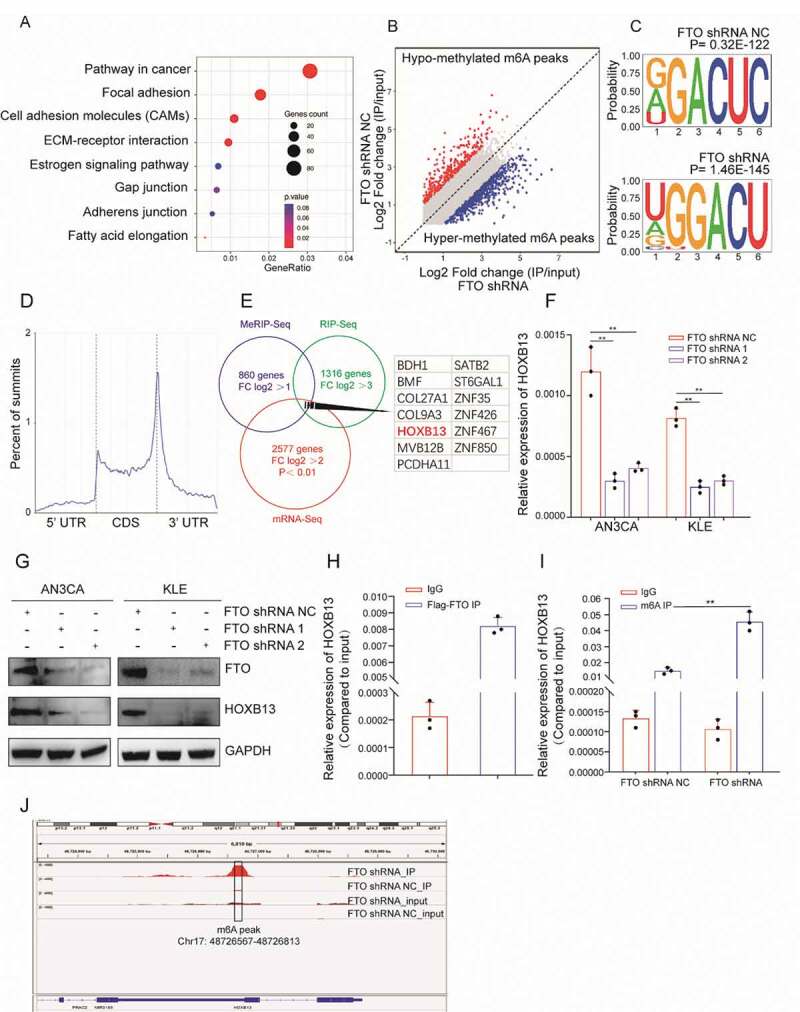
FTO recognizes and regulates m6A in HOXB13 mRNA. (A) KEGG enrichment map of genes specifically enriched by RNA-seq after FTO knockdown. (B) MeRIP-seq detects changes in m6A modifications in mRNA after silencing FTO expression. (C) Top motif identified by HOMER with m6A-seq peaks. (D) Distribution of new m6A peaks in mRNA detected by MeRIP-seq after knocking down FTO expression. (E) Venn diagram shows the genes enriched by MeRIP-seq, RNA-seq, and RIP-seq. (F, G) qPCR and WB confirmed decreased HOXB13 mRNA after FTO knockdown in AN3CA and KLE cells. (H) RIP-PCR validates exogenous FTO binding to HOXB13 mRNA. (I) MeRIP-PCR confirmed that the m6A peak in the 3ʹ untranslated region of HOXB13 mRNA was regulated by FTO. (J) The m6A peak in HOXB13 mRNA transcripts in FTO knockdown samples (IP and input) and the negative control (IP and input). Error bars indicate means ± SDs, **P < 0.01
We used MeRIP-seq to detect changes in m6A modifications in mRNA after silencing FTO expression. A total of 860 genes acquired a new m6A peak, and 459 genes lost an m6A peak (log2 FC> 1) (Fig. 3B). The most common motif of these enrichment peaks was DRACH (Fig. 3C), which is consistent with the results of a previous report. The positions of these peaks occur mainly near the stop codon (Fig. 3D) which is because the main molecular function of FTO is to remove m6A modifications. As such, we therefore focused on the genes that acquired an m6A peak. KEGG analysis showed that these genes also enriched in the MAPK and WNT signalling pathways (Fig. S5E).
To comprehensively display gene mapping regulated by FTO through the m6A mechanism, we combined the analysis of the genes enriched in mRNA-seq, RIP-seq and MeRIP-seq. We screened 13 candidate genes (Fig. 3E). RIP-PCR, qPCR, MeRIP-PCR and WB were further used to verify the candidate genes. qPCR confirmed that the expression of HOXB13 was significantly reduced after silencing FTO expression. This trend was observed in both AN3CA and KLE cells (Fig. 3F). HOXB13 protein expression also decreased significantly after FTO knockdown (Fig. 3G). RIP-PCR experiments confirmed that HOXB13 mRNA can bind to the FTO protein with Flag labelling (Fig. 3H). Based on the MeRIP-seq results, we designed HOXB13 PCR primers targeted to the m6A modification peak (Table S1). MeRIP-PCR showed that the m6A modification level increased after FTO knockdown (Fig. 3I). The m6A peak was located in the 3ʹUTR region of HOXB13 mRNA (Fig. 3J).
YTHDF2 promotes the decay of HOXB13 mRNA by recognizing the m6A peak
The m6A modification in mRNA requires recognition by the ‘Reader’ protein. These reader proteins play a key role in regulating the mRNA metabolism. Previous report showed that YTHDF1 promotes translation by recognizing m6A, and YTHDF2 recognizes mRNA and often leads to mRNA degradation [13]. We found that FTO silencing could promote the appearance of the m6A peak in HOXB13 mRNA but was accompanied by a decrease in the mRNA expression of HOXB13. Therefore, we speculate that YTHDF2 can recognize this m6A modification and play a role in regulating HOXB13 mRNA. We used siRNA to silence the expression of YTHDF2 in cells. qPCR showed that the expression of HOXB13 mRNA increased (Fig. 4A). In addition, WB also confirmed that the protein expression of HOXB13 significantly increased (Fig. 4B).
Figure 4.
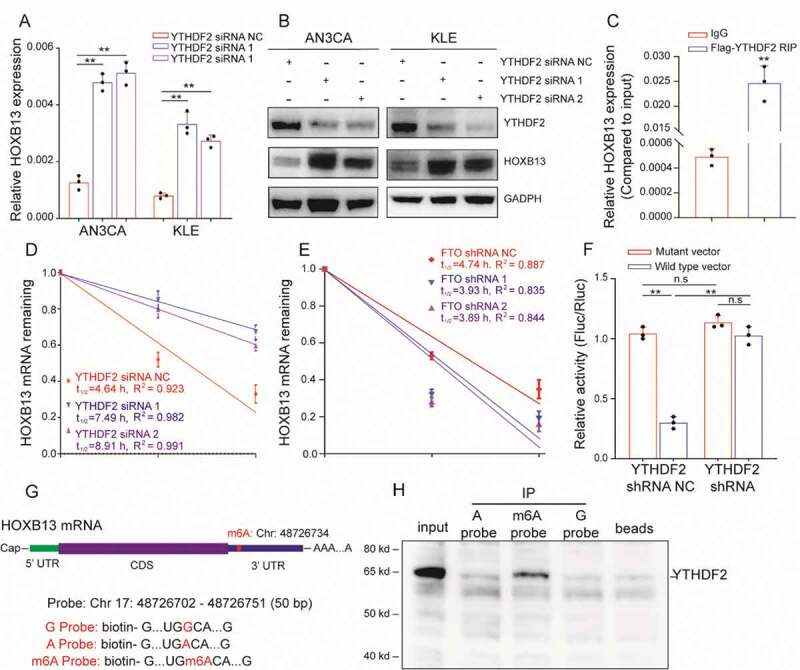
YTHDF2 regulates HOXB13 expression in an m6A-dependent manner. (A, B) qPCR and WB confirmed elevated HOXB13 mRNA and protein expression after YTHDF2 knockdown. (C) RIP-PCR validates exogenous YTHDF2 binding to HOXB13 mRNA. (D) Prolonged RNA lifetime of HOXB13 mRNA after knockdown of YTHDF2 expression. (E) Shortened RNA lifetime of HOXB13 mRNA after knockdown of FTO expression. (F) Relative activity of the wild-type or mutant HOXB13 3′ UTR luciferase reporter in AN3CA cells expressing YTHDF2 shRNA or control. (G) Position of the m6A peak in HOXB13 mRNA (top). RNA probe sequences for RNA pulldowns (bottom). (H) YTHDF2 recognizes the m6A site in the 3′ UTR of HOXB13 mRNA as shown by RNA pull-down assays. Error bars indicate means ± SDs, **P < 0.01, n.s indicates no significance
Further, exogenous Flag-labelled YTHDF2 protein was used for RIP experiments, and the results confirmed that HOXB13 mRNA can bind to YTHDF2 (Fig. 4C). Since binding of the YTHDF2 protein to mRNA leads to RNA degradation, we used the actinomycin D experiment to verify RNA stability. The half-life of HOXB13 mRNA increased significantly after knockdown of YTHDF2 (Fig. 4D). We also observed a decrease in the half-life of HOXB13 mRNA after silencing FTO (Fig. 4E). This suggests that FTO may regulate HOXB13 mRNA degradation through the m6A mechanism, and this mechanism may be mediated by YTHDF2.
To identify the biological function of the HOXB13 mRNA 3ʹ UTR sequence, we performed luciferase reporter assays using a reporter vector containing either the wild-type or mutant 3ʹ UTR sequence. In AN3CA cells with normal YTHDF2 protein expression, the luciferase reporter activity significantly decreased in cells transfected with the wild-type construct, indicating that the sequence weakened the mRNA stability (Fig. 4F). The luciferase activity of cells transfected with the wild-type construct increased significantly after silencing YTHDF2. In addition, after silencing YTHDF2, no significant difference in the fluorescence intensity of cells was detected with wild-type or mutant vectors (Fig. 4F). This result indicates that the 3ʹUTR region is important for HOXB13 mRNA stability. We next aimed to explore whether YTHDF2 recognizes the 3ʹUTR region of HOXB13 mRNA via the m6A mechanism. According to the results of MeRIP-seq, we synthesized three RNA probes consisting of the 3ʹUTR sequence of HOXB13 mRNA, but each probe contained an m6A or m6A mutation (Fig. 4G). RNA pull-down experiments confirmed that YTHDF2 bound to the m6A site but had a weaker binding capacity to non-m6A sites (Fig. 4H). These results indicate that YTHDF2 recognizes the HOXB13 mRNA 3ʹ UTR region in an m6A-dependent manner.
HOXB13 promotes the invasion and metastasis of EC
To explore whether HOXB13 could regulate endometrial cell invasion and metastasis, we constructed HOXB13 expression vectors and siRNA to enhance or knockdown HOXB13 expression (Fig. S4C-D). Overexpression of HOXB13 can promote the metastasis of KLE cells, while silencing the expression of HOXB13 leads to a decrease in metastasis (Fig. 5A). This trend was also observed in AN3CA cells (Fig. 5B). Transwell assays were used to analyse the effect of HOXB13 on invasion ability. Overexpression/knockdown of HOXB13 expression led to increased/decreased cell invasion ability (Fig. 5C). This indicates that HOXB13 has the ability to promote the invasion and metastasis of EC in vitro.
Figure 5.
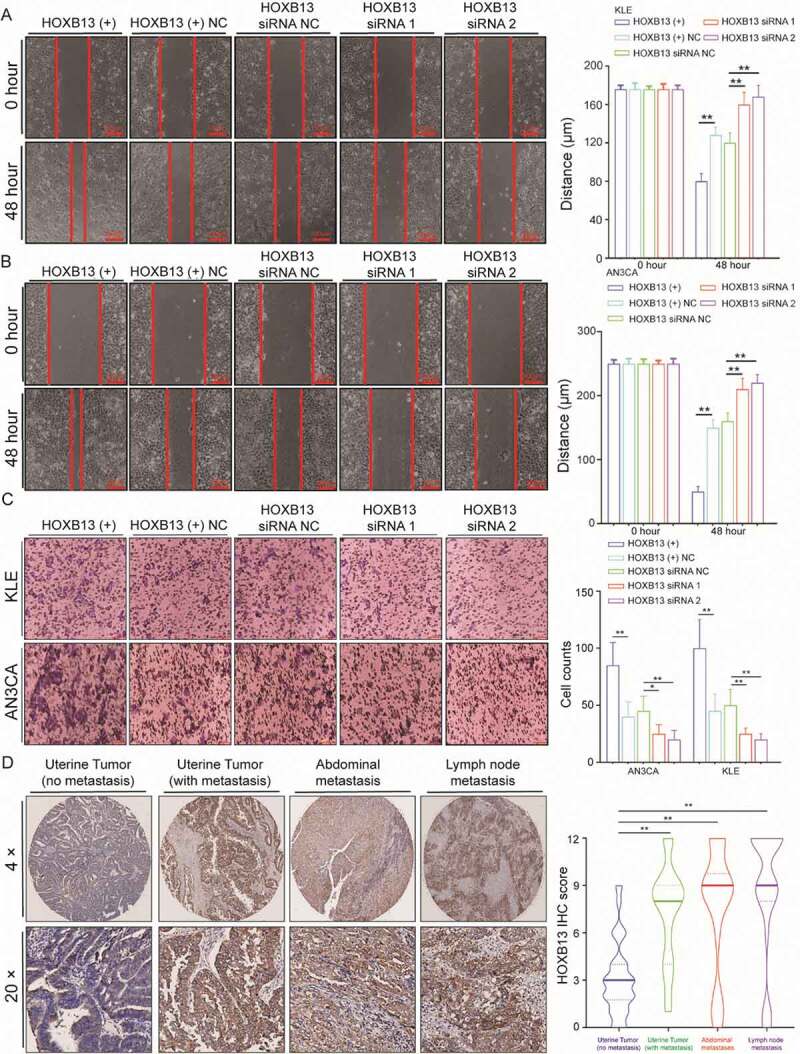
HOXB13 promotes EC cell metastasis and invasion. (A, B) HOXB13 overexpression and knockdown regulate cell migratory abilities in KLE and AN3CA cells by wound-healing assays. (C) Effects of HOXB13 overexpression and knockdown on cell invasive capacities by Transwell assays. (D) Expression of HOXB13 protein in the tissue array validated by IHC, (No metastasis, N = 60), (With metastasis, N = 30), (Abdominal metastasis, N = 24), (Lymph node metastasis, N = 22). Error bars indicate means ± SDs, **P < 0.01
Further, we used IHC to detect the expression of HOXB13 in an EC tissue array to study the effect of HOXB13 on tumour metastasis, and the grouping is the same as before. HOXB13 expression was significantly higher in metastatic tumours than in nonmetastatic tumours (Fig. 5D, Table S4). The expression of HOXB13 in peritoneal metastases and lymph node metastatic tumours was significantly higher than that in nonmetastatic tumours, but there was no difference in expression between various metastatic samples (Fig. 5D, Table S4). Although the expression of HOXB13 in normal endometrium was significantly lower than that in tumour tissue (Fig. S2B). This indicates that HOXB13 expression is significantly increased in metastatic patients. Since YTHDF2 can promote the degradation of HOXB13 mRNA, we speculate that there is a negative correlation between gene expression in tissues. Correlation analysis showed that YTHDF2 and HOXB13 had a positive correlation in mRNA expression by using TCGA data (Fig. S6A). This result does not seem to support the role of YTHDF2 in the degradation of HOXB13 mRNA in tumours. However, we must consider that FTO mRNA expression (Fig. S6B) and protein (Fig. S2A) were lower in endometrium and higher in those with EM cancer.
We theorized that FTO can remove m6A modification in the 3ʹUTR region of HOXB13 mRNA, thereby causing degradation of HOXB13 mRNA when YTHDF2 is abolished. Critically speaking, there may not be a negative correlation between the expression of these two genes in tissues. In order to verify this hypothesis, we detected the expression of YTHDF2 and HOXB13 in 20 cases of normal endometrium tissues which confirmed low expression of FTO and highlighted a negative correlation between YTHDF2 and HOXB13 in normal endometrium (Fig. S6C).
HOXB13 promotes EC metastasis and invasion by activating the WNT signalling pathway
HOXB13 is a homeobox transcription factor that can directly regulate gene expression. The KEGG pathway was mainly enriched in the MAPK, PI3K, and WNT pathways after FTO knockdown. The activation of the WNT signalling pathway is highly related to metastasis in various cancers. To explore whether HOXB13 promotes tumour metastasis through the WNT signalling pathway, we added WNT pathway-specific inhibitors to AN3CA and KLE cells. Wound-healing and Transwell assays showed that the metastatic and invasive abilities of cells were significantly reduced (Fig. 6A, B).
Figure 6.
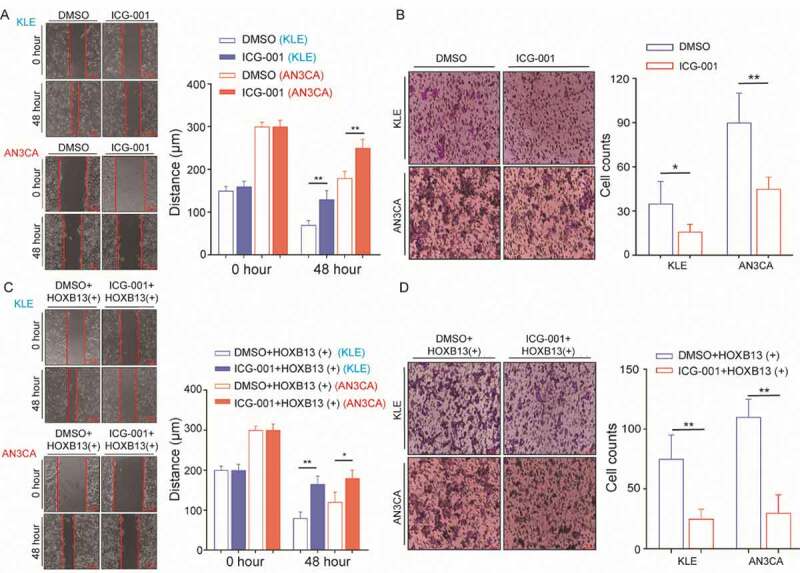
HOXB13 promotes EC cell metastasis and invasion (A, B) WNT signalling pathway inhibitor (ICG-001) inhibits metastasis and invasion of KLE and AN3CA cells. (C, D) WNT signalling pathway inhibitor (ICG-001) blocked the effect of promoting tumour cell invasion and metastasis mediated by HOXB13. Error bars indicate means ± SDs, *P < 0.05, **P < 0.01
We also supplemented WNT signalling pathway inhibitors into HOXB13-overexpressing AN3CA cells. Wound-healing and Transwell experiments showed that the metastatic and invasive abilities of KLE cells were significantly reduced, and these trends were also detected in AN3CA cells (Fig. 6C, D). This indicates that the effect of HOXB13 in promoting EC invasion and metastasis can be inhibited by WNT signalling pathway inhibitors. In addition, cell invasion and migration increased after rescued HOXB13 expression in KLE and AN3CA cell lines with knockdown FTO expression, which suggests that HOXB13 is a key gene in FTO regulation of cancer cell metastasis (Fig. S7). Furthermore, adding ICG-001 to overexpressed FTO cells can significantly inhibit invasion and migration (Fig. S8).
To detect the molecular mechanism of HOXB13 activation, the WNT signalling pathway was examined. We transfected the expression vector to overexpress the HOXB13 protein in the cells (Fig. S4C, D), and the time gradient showed that the concentration of β-catenin in the nucleus gradually increased (Fig. 7A). In contrast, when we silenced the expression of HOXB13 in cells (Fig. S4C, D), the concentration of β-catenin in the nucleus gradually decreased, while the concentration of phosphorylated β-catenin (Ser33/37) gradually increased (Fig. 7B). Furthermore, WB detected the expression of target genes downstream of the WNT signalling pathway. The expression of c-myc, snail, MMP2, MMP7 and MMP7 increased after HOXB13 overexpression. However, this trend may be blocked by adding WNT inhibitor (Fig. 7C).
Figure 7.
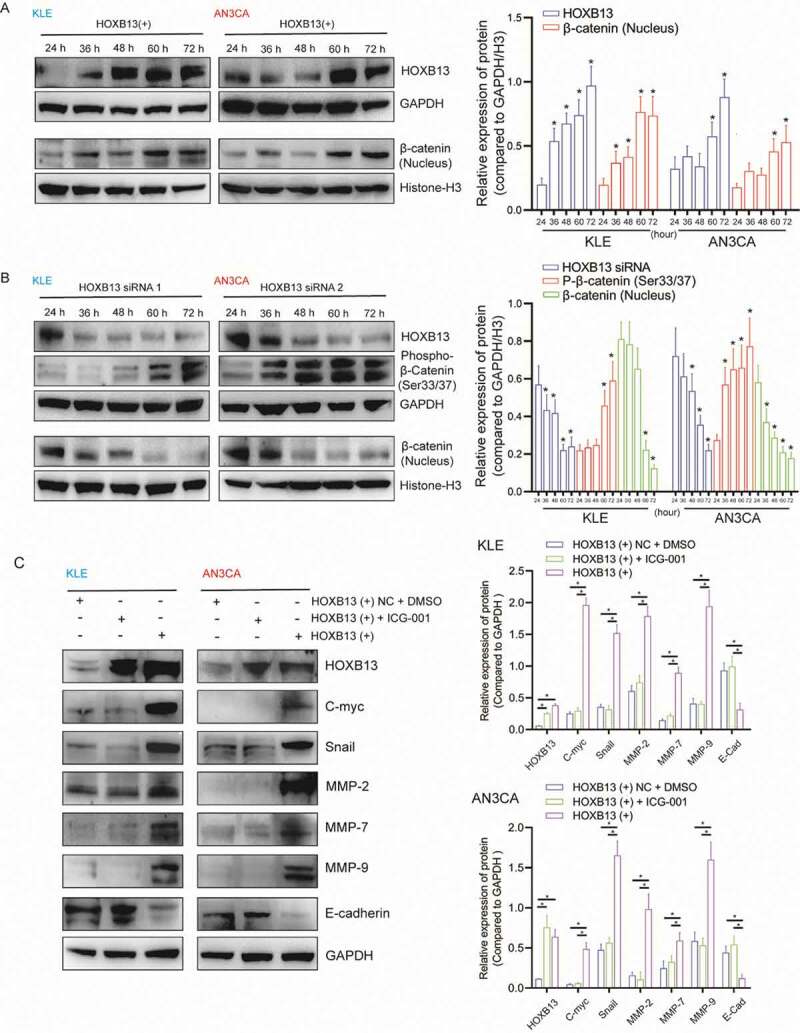
HOXB13 activates the WNT signalling pathway and promotes the expression of downstream genes
(A) WB shows that overexpression of HOXB13 promotes β-catenin expression. (B) WB showed phospho-β-catenin (Ser33/37) and β-catenin expression at different time points after silencing HOXB13 by siRNA. (C) WB showed that a WNT signalling pathway inhibitor (ICG-001) blocked the target genes that were activated by HOXB13 overexpression.
Discussion
Invasion and metastasis are important biological behaviours that lead to the poor prognosis of cancer patients [21]. FTO expression significantly increases in breast cancer, gastric cancer, melanoma, cervical cancer and other solid tumours [22]. High FTO expression in gastric cancer also positively correlates with tumour lymph node metastasis and promotes cancer invasion and metastasis [23]. FTO is highly expressed in lung cancer and is an independent factor affecting prognosis. While decreasing the expression of FTO in lung cancer cells can significantly inhibit tumour proliferation and invasion [24]. In this study, we found that FTO expression increased in EC, and that FTO expression positively correlates with tumour invasion and lymph node metastasis. Patients with high FTO expression had worse prognoses, which is consistent with previous findings. Feng et al. reported that FTO expression is increased in EC [25]; however, high FTO expression is unrelated to tumour invasion and metastasis, which is inconsistent with our findings. The reasons for this divergence may be correspond to the ethnic diversity within each sample but also there were undoubtedly differences in BMI which subtly influence expression, and therefore invasion and metastasis.
Previous studies have shown that HOXB13 can play a role in regulating tumour invasion and metastasis in a variety of tumours [26,27]. Although, the function of HOXB13 in different tumour types is not entirely consistent. HOXB13 promotes the occurrence of epithelial-mesenchymal transition (EMT) in ovarian cancer by inducing SLUG expression and enhances the ability of tumour metastasis [28]. In prostate cancer, HOXB13 promotes tumour cell invasion by reducing the expression of PDEF in the cell [29]. However, HOXB13 can inhibit the invasion of tumour cells by inhibiting the expression of ER-α in breast cancer [30]. In addition, the expression level of HOXB13 is reduced in colorectal cancer and malignant melanoma. Increasing the expression of HOXB13 can inhibit the proliferation of colon cancer and malignant melanoma [31,32]. In this study, we found that expression of HOXB13 in EC is increased in metastatic tumours and that expression of HOXB13 can promote tumour cell invasion and metastasis in vitro. These results suggest that HOXB13 plays a role in promoting tumour metastasis during tumour progression.
The expression of HOXB13 in tumour cells is regulated by various mechanisms. miR-7 can inhibit the expression of HOXB13 and regulate the migration of oesophageal squamous cell carcinoma [33], while HOXB13-AS inhibits the expression of HOXB13 by methylating the HOXB13 promoter, affecting the proliferation of glioma [34]. MEIS1 promotes the proliferation of prostate cancer cells by extending the half-life of HOXB13 in prostate cancer [35]. In this study, we found for the first time that the m6A modification in HOXB13 mRNA can regulate the protein expression of HOXB13, and the expression of HOXB13 can activate the WNT signalling pathway and promote the expression of downstream genes, thereby enhancing tumour invasion and metastasis. These results provide new evidence for clarifying the molecular mechanism of HOXB13 in promoting tumour metastasis.
Obesity is a high-risk factor for EC, but the molecular mechanism between obesity and the biological behaviour of EC cells has been unclear. Previous studies have shown that FTO expression is clearly related to obesity [36,37]. FTO was identified as a demethylase in 2011 and can ‘erase’ the m6A modification on RNA [38]; however, the methylation mapping of FTO regulation in EC is not clear. In this study, we comprehensively displayed the m6A modification profile regulated by FTO in EC cells. This provides a new perspective for the further exploration of obesity and EC progression. Earlier studies showed that m6A-modified mRNAs are often unstable, mainly because YTHDF2 promotes RNA relocation to RNA decay sites and accelerates mRNA decay [39]. However, subsequent studies have found more proteins that can recognize m6A modifications and regulate mRNA metabolism, such as RNA translation, splicing and stability [40].
FTO overexpression can erase the m6A site from the mRNA, resulting in faculty mRNA modifications and abolishing the RNA metabolism pathways mediated by various readers, ultimately affecting the mRNA function [41]. In this study, we found that FTO regulates m6A modification in HOXB13 mRNA, deprives HOXB13 degradation mediated by YTHDF2, promotes the expression of HOXB13, and accelerates EC cell metastasis. Nearly 1/3 of mRNAs have m6A modifications. The number of genes regulated by FTO through the m6A mechanism may be very large. In this study, 860 genes acquired a new m6A peak, and 459 genes lost an m6A peak after FTO knockdown, and these changes were accompanied by differences in mRNA expression. This indicates that FTO has a large number of target genes regulated by the m6A mechanism. More research is required to fully understand the molecular mechanism of FTO regulation of EC progression.
In conclusion, we found that the expression of FTO can promote the metastasis of EC. Mechanistically, FTO removed the m6A modification from HOXB13 mRNA and abolished the YTHDF2-mediated degradation of HOXB13, promoting HOXB13 protein expression and activates the WNT signalling pathway, and promotes EC invasion and metastasis. FTO is a new potential target for the treatment of tumour metastasis.
Supplementary Material
Acknowledgments
This study was supported by the Young Scientists Fund of the National Natural Science Foundation of China (No. 81502243, 81702566), National Natural Science Foundation(No. 81872119), Chinese Academy of Medical Sciences innovation project (No. 2017-I2M-1-002), Innovative Team of Jiangsu Province (CXTDA2017008), 333 High-level Talents Training Project in Jiangsu Province (2019). Scientific Research Project of Jiangsu Province Maternal and Child Health Association (No.FYX201909), Key Young Medical Talents of Jiangsu Province (No.QNRC2016610).
We thank professor sam seery (peking union medical college), for editing the English text of this manuscript.
Funding Statement
This work was supported by the Innovative Team of Jiangsu Province [CXTDA2017008]; National Natural Science Foundation of China [81872119]; Chinese Academy of Medical Sciences innovation project [2017-I2M-1-002]; Scientific Research Project of Jiangsu Province Maternal and Child Health Association [FYX201909]; Key Young Medical Talents of Jiangsu Province [QNRC2016610]; 333 High-level Talents Training Project in Jiangsu Province [2019]; Young Scientists Fund [81702566]; Young Scientists Fund [81502243].
Availability of data and materials
All data generated or analyzed during this study are included in the article [and its addition supplementary files].
Disclosure statement
The authors declare that they have no competing interests
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Katz A.CE: obesity-related cancer in women: a clinical review. Am J Nurs. 2019;119:34–41. [DOI] [PubMed] [Google Scholar]
- [2].Carugno J. Clinical management of vaginal bleeding in postmenopausal women. Climacteric. 2020;23:343–349. [DOI] [PubMed] [Google Scholar]
- [3].Dai Y, Wang Z, Wang J. Survival of microsatellite-stable endometrioid endometrial cancer patients after minimally invasive surgery: an analysis of the cancer genome Atlas data. Gynecol Oncol. 2020;158:92–98. [DOI] [PubMed] [Google Scholar]
- [4].Han C, Altwerger G, Menderes G, et al. Novel targeted therapies in ovarian and uterine carcinosarcomas. Discov Med. 2018;25:309–319. [PubMed] [Google Scholar]
- [5].Travaglino A, Raffone A, Stradella C, et al. Impact of endometrial carcinoma histotype on the prognostic value of the TCGA molecular subgroups. Arch Gynecol Obstet. 2020;301:1355–1363. [DOI] [PubMed] [Google Scholar]
- [6].He L, Li H, Wu A, et al. Functions of N6-methyladenosine and its role in cancer. Mol Cancer. 2019;18. DOI: 10.1186/s12943-019-1109-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Zaccara S, Ries RJ, Jaffrey SR. Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell Biol. 2019;20:608–624. [DOI] [PubMed] [Google Scholar]
- [8].Shi H, Wei J, He C. Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol Cell. 2019;74:640–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Zhao W, Qi X, Liu L, et al. Epigenetic regulation of m(6)A modifications in human cancer. Mol Ther Nucleic Acids. 2020;19:405–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Chen X, Zhang J, Zhu J. The role of m(6)A RNA methylation in human cancer. Mol Cancer. 2019;18:103.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Zhu T, Roundtree IA, Wang P, et al. Crystal structure of the YTH domain of YTHDF2 reveals mechanism for recognition of N6-methyladenosine. Cell Res. 2014;24:1493–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Wang X, Zhao B, Roundtree I, et al. N-6-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161:1388–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Huang T, Liu Z, Zheng Y, et al. YTHDF2 promotes spermagonial adhesion through modulating MMPs decay via m(6)A/mRNA pathway. Cell Death Dis. 2020;11(1). DOI: 10.1038/s41419-020-2235-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Huang H, Weng H, Chen J. m(6)A modification in coding and non-coding RNAs: roles and therapeutic implications in cancer. Cancer Cell. 2020;37:270–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Smemo S, Tena JJ, Kim K, et al. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature. 2014;507:371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Jia G, Yang C, Yang S, et al. Oxidative demethylation of 3-methylthymine and 3-methyluracil in single-stranded DNA and RNA by mouse and human FTO. FEBS Lett. 2008;582:3313–3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Gholamalizadeh M, Jarrahi AM, Akbari ME, et al. Association between FTO gene polymorphisms and breast cancer: the role of estrogen. Expert Rev Endocrinol Metab. 2020;15:115–121. [DOI] [PubMed] [Google Scholar]
- [18].Hu Y, Wang S, Liu J, et al. New sights in cancer: component and function of N6-methyladenosine modification. Biomed Pharmacother. 2020;122:109694. [DOI] [PubMed] [Google Scholar]
- [19].Zhang L, Wan Y, Jiang Y, et al. Overexpression of BP1, an isoform of homeobox gene DLX4, promotes cell proliferation, migration and predicts poor prognosis in endometrial cancer. Gene. 2019;707:216–223. [DOI] [PubMed] [Google Scholar]
- [20].Huang H, Weng H, Sun W, et al. Recognition of RNA N-6- methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20:285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lewczuk Ł, Pryczynicz A, Guzinska-Ustymowicz K. Cell adhesion molecules in endometrial cancer - a systematic review. Adv Med Sci-Poland. 2019;64:423–429. [DOI] [PubMed] [Google Scholar]
- [22].Deng X, Su R, Stanford S, et al. Critical enzymatic functions of FTO in obesity and cancer. Front Endocrinol. 2018;9. DOI: 10.3389/fendo.2018.00396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Li Y, Zheng D, Wang F, et al. Expression of demethylase genes, FTO and ALKBH1, is associated with prognosis of gastric cancer. Digest Dis Sci. 2019;64(6):1503–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Liu J, Ren D, Du Z, et al. m(6)A demethylase FTO facilitates tumor progression in lung squamous cell carcinoma by regulating MZF1 expression. Biochem Biophys Res Commun. 2018;502:456–464. [DOI] [PubMed] [Google Scholar]
- [25].Zhu Y, Shen J, Gao L, et al. Estrogen promotes fat mass and obesity-associated protein nuclear localization and enhances endometrial cancer cell proliferation via the mTOR signaling pathway. Oncol Rep. 2016;35:2391–2397. [DOI] [PubMed] [Google Scholar]
- [26].Brechka H, Bhanvadia RR, VanOpstall C, et al. HOXB13 mutations and binding partners in prostate development and cancer: function, clinical significance, and future directions. Genes Dis. 2017;4:75–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Ouhtit A. Hoxb13 a potential prognostic biomarker for prostate cancer. Front Biosci (Elite Ed). 2016;8:40–45. [DOI] [PubMed] [Google Scholar]
- [28].Yuan H, Kajiyama H, Ito S, et al. HOXB13 and ALX4 induce SLUG expression for the promotion of EMT and cell invasion in ovarian cancer cells. Oncotarget. 2015;6(15):13359–13370. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kim I, Kang TW, Jeong T, et al. HOXB13 regulates the prostate-derived Ets factor: implications for prostate cancer cell invasion. Int J Oncol. 2014;45:869–876. [DOI] [PubMed] [Google Scholar]
- [30].Liu B, Wang T, Wang H, et al. Oncoprotein HBXIP enhances HOXB13 acetylation and co-activates HOXB13 to confer tamoxifen resistance in breast cancer. J Hematol Oncol. 2018;11. DOI: 10.1186/s13045-018-0577-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Ghoshal K, Motiwala T, Claus R, et al. HOXB13, a target of DNMT3B, is methylated at an upstream CpG island, and functions as a tumor suppressor in primary colorectal tumors. Plos One. 2010;5:e10338. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [32].Maeda K, Hamada J, Takahashi Y, et al. Altered expressions of HOX genes in human cutaneous malignant melanoma. Int J Cancer. 2005;114(3):436–441. . [DOI] [PubMed] [Google Scholar]
- [33].Li R, Ke S, Meng F, et al. CiRS-7 promotes growth and metastasis of esophageal squamous cell carcinoma via regulation of miR-7/HOXB13. Cell Death Dis. 2018;9:838.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Xiong Y, Kuang W, Lu S, et al. Long noncoding RNA HOXB13-AS1 regulates HOXB13 gene methylation by interacting with EZH2 in glioma. Cancer Med. 2018;7:4718–4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Johng D, Torga G, Ewing CM, et al. HOXB13 interaction with MEIS1 modifies proliferation and gene expression in prostate cancer. Prostate. 2019;79:414–424. [DOI] [PubMed] [Google Scholar]
- [36].Chang JY, Park JH, Park SE, et al. The fat mass- and obesity-associated (FTO) gene to obesity: lessons from Mouse models. Obesity. 2018;26:1674–1686. [DOI] [PubMed] [Google Scholar]
- [37].TM M. Fat mass and obesity associated (FTO) gene and hepatic glucose and lipid metabolism. Nutrients. 2018;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Niu Y, Zhao X, Wu Y, et al. N6-methyl-adenosine (m6A) in RNA: an old modification with a novel epigenetic function. Genomics Proteomics Bioinformatics. 2013;11:8–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Wang X, He C. Reading RNA methylation codes through methyl-specific binding proteins. RNA Biol. 2014;11:669–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Boo SH, Kim YK. The emerging role of RNA modifications in the regulation of mRNA stability. Exp Mol Med. 2020;52(3):400–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Hirayama M, Wei F, Chujo T, et al. FTO demethylates Cyclin D1 mRNA and controls cell-cycle progression. Cell Rep. 2020;31:107464. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in the article [and its addition supplementary files].


