Abstract
Background
Although many works have been conducted to explore the biomarkers for diagnosing major depressive disorder (MDD), the widely accepted biomarkers are still not identified. Thus, the combined application of serum metabolomics and fecal microbial communities was used to identify gut microbiota-derived inflammation-related serum metabolites as potential biomarkers for MDD.
Methods
MDD patients and healthy controls (HCs) were included in this study. Both serum samples and fecal samples were collected. The liquid chromatography mass spectrometry (LC-MS) was used to detect the metabolites in serum samples, and the 16S rRNA gene sequencing was used to analyze the gut microbiota compositions in fecal samples.
Results
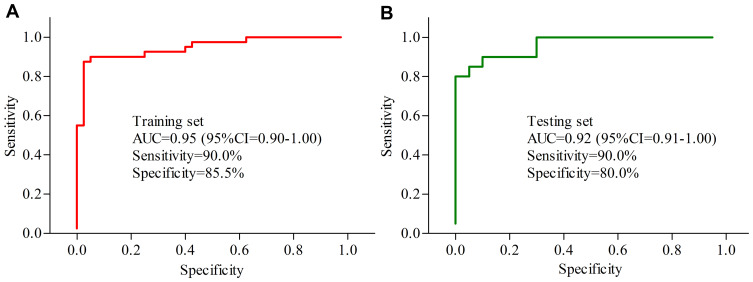
Totally, 60 MDD patients and 60 HCs were recruited. The 24 differential serum metabolites were identified, and 10 of these were inflammation-related metabolites. Three significantly affected inflammation-related pathways were identified using differential metabolites. The 17 differential genera were identified, and 14 of these genera belonged to phyla Firmicutes. Four significantly affected inflammation-related pathways were identified using differential genera. Five inflammation-related metabolites (LysoPC(16:0), deoxycholic acid, docosahexaenoic acid, taurocholic acid and LysoPC(20:0)) were identified as potential biomarkers. These potential biomarkers had significant correlations with genera belonged to phyla Firmicutes. The panel consisting of these biomarkers could effectively distinguish MDD patients from HCs with an area under the curve (AUC) of 0.95 in training set and 0.92 in testing set.
Conclusion
These findings suggested that the disturbance of phyla Firmicutes might be involved in the onset of depression by regulating host’s inflammatory response, and these potential biomarkers could be useful for future investigating the objective methods for diagnosing MDD.
Keywords: major depressive disorder, gut microbiota, biomarkers, inflammation, metabolite
Introduction
Major depressive disorder (MDD) is a serious mental disorder affecting millions of people every year.1 It causes significant economic burden for individuals and society. However, the pathogenesis of MDD is still unclear;2–4 the mainstream view is that the molecular brain dysfunctions are mainly responsible for this disease.5 Unfortunately, the current first-line antidepressant therapies developed according to these prevailing theories can only alleviate depressive symptoms in about half of MDD patients.6 Meanwhile, the clinicians still diagnose MDD according to the subjective identification of symptom clusters of patients, rather than using objective tests. Up to now, there are still no widely accepted biomarkers, although many potential biomarkers have been identified based on the existing theories.7–10 These phenomena indicate that the existing hypotheses cannot comprehensively reflect the heterogeneous pathophysiological mechanisms of MDD. Therefore, it is urgently needed to explore novel molecular mechanisms of MDD.
Nowadays, mounting evidence suggests that gut microbiota has a vital role in the onset of neuropsychiatric disorders.11,12 It can interact with the physiological processes of host through its metabolic products, such as short-chain fatty acids (SCFAs).13,14 Many studies have shown that the disturbance of gut microbiota has become an important factor in the pathogenesis of depression.15,16 Using nonhuman primate model of depression, researchers found that gut microbiota might participate in the onset of depression through regulating the glycerophospholipid metabolism.15 Using a mouse model of depression, we observed the close relationships between the disordered gut microbiota and hypothalamus neurotransmitters.16 Meanwhile, our studies in humans also found that there were significant differences on gut microbiota compositions between MDD patients and healthy controls (HCs).17,18 The disturbance of gut microbiota in depressed patients was also observed in other studies.19,20 These findings demonstrate that further exploring the role of gut microbiota in MDD can provide some novel insights in revealing the pathogenesis of this disease.
Gut microbiota can affect the health of host by regulating its inflammatory level. Gut microbiota-derived metabolites may be involved in the pathogenesis of immune-related inflammatory diseases by leading to the release of many inflammation-related serum markers, such as interleukins (ILs), C-reactive protein (CRP) and tumor necrosis factor (TNF).21,22 Previous studies reported that inflammation had a close relationship with the pathogenesis of depression, and individuals with inflammatory disorders might have an increased incidence of depression.23,24 Zheng et al found five significantly changed gut microbiota-related urinary metabolites in MDD patients, and these metabolites had some relationships with inflammatory response.25 Our previous study found that plasma metabolic abnormalities in lipid and amino acid metabolism might be implicated in the occurrence of suicidal behaviors in MDD patients.26 Both gut microbiota and inflammatory response have close relationships with lipid metabolites. Meanwhile, we also found that the decreased level of alpha 1-antitrypsin (AAT) might lead to inflammatory response disorders, and ultimately lead to the occurrence of suicidal ideation in MDD patients.27 The inflammatory response and immune system represent the major component of communications between gut microbiota and brain functions. Accumulating evidence supports the role of gut microbiota in regulating the host metabolic and immune functions.28,29 Therefore, we conducted this study to further explore the differences of gut microbiota and serum metabolites between MDD patients and HCs. Our findings will advance our understanding of how gut microbiota contributes to the onset of depression.
Patients and Methods
MDD Patients and HCs Enrolment
This study was approved by the Ethical Committee of Chongqing Medical University (Approved No. 20200320). All the included participants provided the written informed consents before samples collecting. This study was conducted in accordance with the Declaration of Helsinki. Two experienced psychiatrists were in charge of participants’ recruitment using the [Diagnostic and Statistical Manual of Mental Disorders, 4th edition] criteria (DSM-IV). MDD patients met the following criteria were included in this study: i) 17-item Hamilton Depression Rating Scale (HDRS) was used to assess the depression severity, and the candidates with HDRS scores >17 were included into this study; ii) without any pre-existing mental disorders, and without other DSM-IV Axis I/II disorders; iii) we did not recruit women who were pregnant or within the first year after delivery; iv) without any illicit drug use or alcohol abuse; and v) did not receive any antidepressive treatments in one month prior to sample collections. Meanwhile, two experienced psychiatrists applied DSM-IV to assess the mental state of HCs, and used the physical examination records to carefully check the other information of HCs. Finally, HCs without DSM-IV Axis I/II disorders, systemic medical illness, illicit drug use and alcohol abuse were recruited from the Medical Examination Center of our hospital. The detailed information is described in Table 1.
Table 1.
Demographic Data of Included Subjects in This Study
| Variables | HCs | MDD Patients | P-value |
|---|---|---|---|
| Number | 60 | 60 | - |
| Age | 35.13 (15.79) | 35.62 (17.10) | 0.87 |
| Sex (F/M) | 36/24 | 39/21 | 0.57 |
| BMI (kg/m2) | 21.19 (4.29) | 20.90 (2.30) | 0.63 |
| Married (Y/N) | 32/28 | 36/24 | 0.46 |
| Family history of PI (Y/N) | 4/56 | 6/54 | 0.51 |
| HDRS scores | 0.67 (0.93) | 25.3 (6.01) | <0.00001 |
Abbreviations: MDD, major depressive disorder; HCs, healthy controls; Y, yes; N, no; HDRS, Hamilton Depression Rating Scale; F, female; M, male; BMI, body mass index, PI, psychiatric illness.
Gut Microbiota Compositions Detection
The gut microbiota analysis was conducted according to the standard protocols by Majorbio Bio-Pharm Technology Co. Ltd. (Shanghai, China). The raw 16S rRNA gene sequencing reads were demultiplexed, quality-filtered with Trimmomatic, and merged with FLASH according to the following criteria: i) set 50-bp sliding window; if the average quality score was less than 20 in the window, the back-end base is cut off from the window; the reads below 50-bp were filtered to remove the n-base reads; ii) according to the overlap relationships between reads, the paired reads were merged into one sequence, and the minimum overlap length was 10 bp; iii) the maximum allowable mismatch ratio of overlap region of splicing sequence was 0.2; and iv) according to the barcodes and primers at both ends of the sequence, the samples were distinguished, and the sequence direction was adjusted. The number of allowed mismatches of barcodes was 0, and the maximum number of primer mismatches was 2. After cleaning up, the remaining sequences were assigned to operational taxonomic units (OTUs) with 97% similarity cutoff. The taxonomy of each OTU was classified using RDP reference database. Finally, we used these taxonomies to construct the summaries of the taxonomic distributions of OTUs. The taxonomic distributions of OTUs were used to calculate the relative abundances of gut microbiota at different levels.
Serum Metabolomics Detection
Firstly, 50 μL serum and 300 μL of acetonitrile were vortex-mixed. After centrifugation (4°C, 15 minutes, 14,000 rpm), we transferred the supernatant into a new tube and then conducted vacuum drying. Secondly, we added 20 μL acetonitrile into the obtained residue and conducted vortex with 80 μL water. After centrifugation (4°C, 15 minutes, 14,000 rpm), we transferred 80 μL supernatant into a glass vial for later liquid chromatography mass spectrometry (LC-MS) analysis. An ACQUITY I Class UPLC system coupled to a Waters G2-S QTOF system, with a Waters ACQUITY UPLC HSS T3 column and a Waters ACQUITY UPLC HSS T3 VanGuard precolumn was used to conduct the metabolomic analysis of the sample extracts. We added 5 μL of extract into this system for metabolomic fingerprinting.
Statistical Analysis
All the statistical analyses were conducted using SPSS 19.0 and R 3.6.0. The principal-coordinate analysis (PCoA) was used to visualize the discrimination between the two groups. The linear discriminant-analysis effect size (LEfSe) was then conducted to identify the differential bacterial taxa responsible for the discrimination between the two groups. The orthogonal partial least-squares discriminant analysis (OPLS-DA) was performed to explore the significant differences on serum metabolites between the two groups. The Pearson correlation analysis was conducted to evaluate the correlations between the differential bacterial taxa and the differential metabolites. Finally, potential biomarkers from these differential metabolites were identified using Random Forest analysis. The receiver operating characteristic (ROC) curve analysis was further performed to assess the diagnostic performance of the identified potential biomarkers.30–33 The area under the curve (AUC) was the evaluation index. In general, an AUC of 0.5 to 07, 0.7 to 0.8, 0.8 to 0.9 and 0.9 to 1.0 were considered no discrimination, acceptable, excellent, and outstanding, respectively.34 All statistical analyses were two-tailed and the significance level was set at p-value <0.05.
Results
Gut Microbial Compositions in MDD Patients and HCs
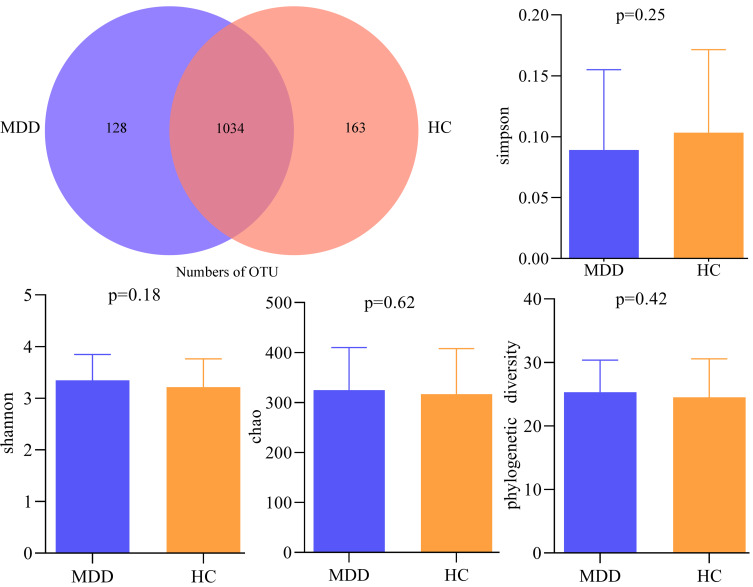
Using 16s rRNA gene sequencing, a total of 6,414,608 high-quality reads across 120 samples were obtained. The average length in samples was 414 base pairs (bp). We clustered these reads into 1325 OTUs at 97% sequence similarity. There were 1034 OTUs in both groups, while 128 OTUs and 163 OTUs were unique to MDD patients and HCs, respectively (Figure 1). These OTUs mainly belonged to four phyla: Firmicutes (58.11%), Bacteroidota (22.11%), Proteobacteria (7.02%) and Actinobacteriota (6.49%). The within-sample (α) phylogenetic diversity analysis showed that no significant difference was found between the two groups on alpha diversity (Simpson, p = 0.25; Shannon, p = 0.18; chao, p = 0.62 and phylogenetic diversity, p = 0.42; Figure 1).
Figure 1.
Gut microbial compositions and α-diversity between the two groups.
Abbreviations: MDD, major depressive disorder; HC, healthy controls; OTU, operational taxonomic unit.
Differential Gut Microbiota in MDD Patients
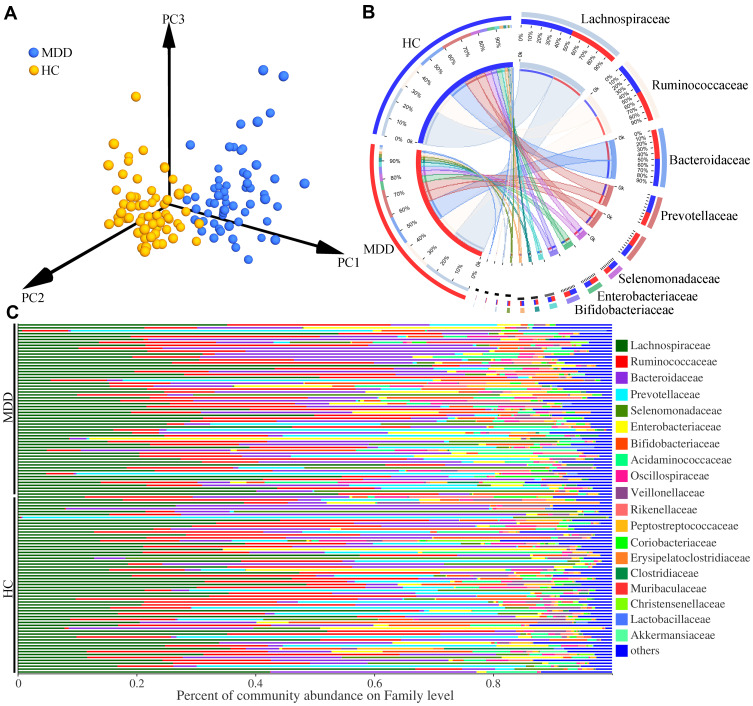
Meanwhile, we conducted the β-diversity analysis to find out whether the gut microbiota compositions were substantially different in MDD patients compared to HCs. The PCoA results showed that there were significant differences on gut microbial compositions between MDD patients and HCs (Figure 2A, p=0.002). The dominant bacteria taxa on Family level in MDD patient and HCs were Lachnospiraceae (27.79% vs 26.93%), Ruminococcaceae (16.78% vs 17.04%), Bacteroidaceae (15.99% vs 14.95%), Prevotellaceae (7.75% vs 9.91%), Selenomonadaceae (4.07% vs 5.20%), Bifidobacteriaceae (3.91% vs 4.22%) and Enterobacteriaceae (3.78% vs 5.26%) (Figure 2B). The percent of community abundance on family level in each sample is described in Figure 2C. Moreover, using LEfSe, we identified 17 differential genera responsible for the discrimination between the two groups: Lactobacillus, Colidextribacter, Holdemania, norank_f_Lachnospiraceae, GCA-900066575, Lachnoclostridium, Eubacterium_ventriosum_group, Candidatus_Soleaferrea, Faecalitalea, CAG-56, Lachnospiraceae_NK4A136_group, Lachnospira, Clostridium_innocuum_group, unclassified_f_Lachnospiraceae, Collinsella, Parabacteroides, and Klebsiella. The former 14 differential genera belonged to phyla Firmicutes.
Figure 2.
Differential gut microbiota compositions between the two groups. (A) The significant differences on gut microbial compositions between the two groups were identified by the results of PCoA; (B) the dominant bacteria taxa on Family level in MDD patients and HCs; (C) the percent of community abundance on Family level in each sample.
Abbreviations: MDD, major depressive disorder; HC, healthy controls.
Differential Serum Metabolites in MDD Patients
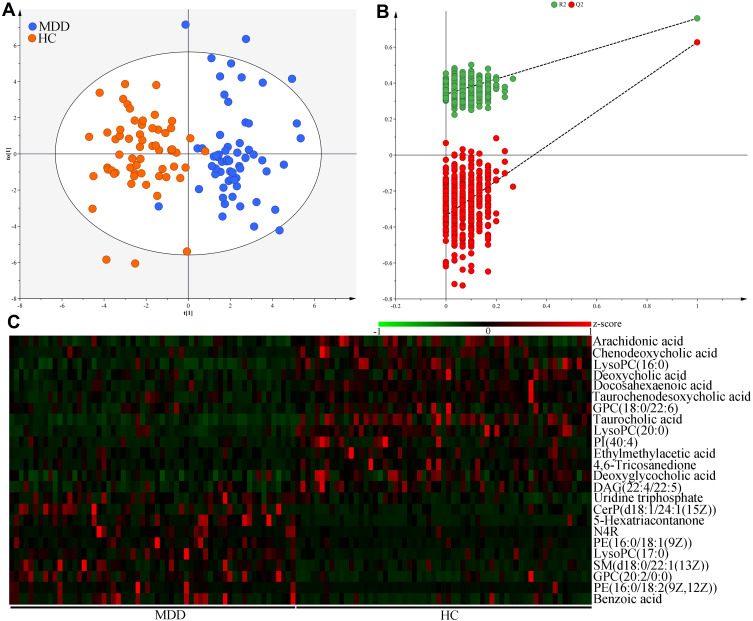
OPLS-DA results showed that the MDD patients and HCs were obviously separated with little overlap (Figure 3A). The positive values of the two model evaluation indexes (R2=76%, Q2 =63%) indicated that the built model was stable. Meanwhile, the 399-item permutation test also showed that our model was valid and not over-fitted, because the corresponding permutated values of R2 and Q2 were obviously lower compared to their original values (Figure 3B). These results demonstrated that there was a divergent metabolic phenotype between MDD patients and HCs. By analyzing the loading coefficient plot of OPLS-DA model, we identified 24 metabolites with a correlation coefficient >0.254 (equivalent to a p-value of less than 0.05) as the differential metabolites responsible for the discrimination between the two groups.
Figure 3.
Metabolomic analysis of serum samples from MDD patients and HCs. (A) The divergent serum metabolic phenotype was observed between the two groups; (B) the results of 399-item permutation test indicated that the discriminative model was robust; (C) the heatmap of the identified 24 differential serum metabolites.
Abbreviations: MDD, major depressive disorder; HC, healthy controls. PI(40:4), Phosphatidylinositol(40:4); GPC(20:2/0:0), 1-Eicosadienoylglycerophosphocholine; GPC(18:0/22:6), 1-Stearoyl-2-docosahexaenoyl-sn-glycero-3-phosphocholine; PE(16:0/18:2(9Z,12Z)), 1-Palmitoyl-2-linoleoyl-gpe; CerP(d18:1/24:1(15Z)), N-[(15Z)-Tetracosenoyl]sphing-4-enine 1-phosphate; N4R, N-(2R-Hydroxytricosanoyl)-2S-amino-1,3S,4R-octadecanetriol; LysoPC(17:0), 1-Heptadecanoyl-glycero-3-phosphocholine; LysoPC(16:0), 1–16:0-Lysophosphatidylcholine; SM(d18:0/22:1(13Z)), N-(13Z-Docosenoyl)-sphinganine-1-phosphocholine; DAG(22:4/22:5), 1-Adrenoyl-2-docosapentaenoyl-sn-glycerol; LysoPC(20:0), 1-Arachidonyl-glycero-3-phosphocholine; PE(16:0/18:1(9Z)), 1-Palmitoyl-2-oleoyl-gpe.
As compared to HCs, MDD patients were characterized by higher levels of Uridine triphosphate, N-(2R-Hydroxytricosanoyl)-2S-amino-1,3S,4R-octadecanetriol (N4R), Benzoic acid, 1-Heptadecanoyl-glycero-3-phosphocholine [LysoPC(17:0)], N-[(15Z)-Tetracosenoyl]sphing-4-enine 1-phosphate [CerP(d18:1/24:1(15Z))], 1-Palmitoyl-2-oleoyl-gpe [PE(16:0/18:1(9Z))], 5-Hexatriacontanone, N-(13Z-Docosenoyl)-sphinganine-1-phosphocholine [SM(d18:0/22:1(13Z))], 1-Eicosadienoylglycerophosphocholine [GPC(20:2/0:0)] and 1-Palmitoyl-2-linoleoyl-gpe [PE(16:0/18:2(9Z,12Z))], along with lower levels of Arachidonic acid, Chenodeoxycholic acid, 1–16:0-Lysophosphatidylcholine [LysoPC(16:0)], Deoxycholic acid, Docosahexaenoic acid, 1-Stearoyl-2-docosahexaenoyl-sn-glycero-3-phosphocholine [GPC(18:0/22:6)], Taurocholic acid, Taurochenodeoxycholic acid, 1-Arachidonyl-glycero-3-phosphocholine [LysoPC(20:0)], Phosphatidylinositol(40:4)[PI(40:4)], Ethylmethylacetic acid, 4.6-Tricosanedione, Deoxyglycocholic acid and 1-Adrenoyl-2-docosapentaenoyl-sn-glycerol [DAG (22:4/22:5)]. There were ten inflammation-related metabolites: Arachidonic acid, Chenodeoxycholic acid, LysoPC(16:0), Deoxycholic acid, Docosahexaenoic acid, Taurochenodeoxycholic acid, GPC(18:0/22:6), Taurocholic acid, LysoPC(20:0), PI(40:4), Ethylmethylacetic acid, 4.6-Tricosanedione and Deoxyglycocholic acid. Hierarchical clustering heat map of these differential metabolites showed a consistent clustering pattern within individual groups (Figure 3C). The detailed information of these differential metabolites is described in Table 2.
Table 2.
Differential Metabolites Between MDD Patients and HCs
| Metabolites | Ra | Pb | APc | FCd |
|---|---|---|---|---|
| Arachidonic acid | −0.503 | 4.94E-05 | 4.82E-04 | 1.89 |
| Chenodeoxycholic acid | −0.365 | 1.60E-03 | 6.59E-03 | 2.36 |
| LysoPC(16:0) | −0.474 | 2.02E-09 | 7.90E-08 | 2.45 |
| Deoxycholic acid | −0.378 | 7.59E-05 | 6.58E-04 | 1.53 |
| Docosahexaenoic acid | −0.724 | 1.87E-07 | 4.87E-06 | 2.03 |
| Taurochenodeoxycholic acid | −0.322 | 8.00E-04 | 3.67E-03 | 1.51 |
| GPC(18:0/22:6) | −0.262 | 8.67E-03 | 2.05E-02 | 1.52 |
| Taurocholic acid | −0.728 | 1.31E-11 | 1.02E-09 | 3.46 |
| LysoPC(20:0) | −0.368 | 9.43E-07 | 1.84E-05 | 2.27 |
| PI(40:4) | −0.286 | 2.87E-03 | 9.34E-03 | 2.55 |
| Ethylmethylacetic acid | −0.361 | 7.71E-03 | 2.07E-02 | 1.95 |
| 4,6-Tricosanedione | −0.298 | 1.67E-03 | 6.49E-03 | 2.21 |
| Deoxyglycocholic acid | −0.315 | 3.36E-05 | 3.75E-04 | 1.67 |
| DAG(22:4/22:5) | −0.276 | 1.72E-04 | 1.12E-03 | 1.68 |
| Uridine triphosphate | 0.417 | 6.00E-04 | 3.12E-03 | 0.62 |
| CerP(d18:1/24:1(15Z)) | 0.386 | 7.62E-05 | 5.95E-04 | 0.28 |
| 5-Hexatriacontanone | 0.396 | 8.09E-06 | 1.26E-04 | 0.19 |
| N4R | 0.372 | 2.11E-03 | 7.47E-03 | 0.26 |
| PE(16:0/18:1(9Z)) | 0.326 | 3.40E-03 | 1.02E-02 | 0.40 |
| LysoPC(17:0) | 0.337 | 1.12E-04 | 7.96E-04 | 0.41 |
| SM(d18:0/22:1(13Z)) | 0.339 | 7.69E-04 | 3.75E-03 | 0.45 |
| GPC(20:2/0:0) | 0.436 | 3.35E-05 | 4.36E-04 | 0.50 |
| PE(16:0/18:2(9Z,12Z)) | 0.406 | 2.39E-02 | 4.44E-02 | 0.34 |
| Benzoic acid | 0.271 | 8.90E-04 | 3.86E-03 | 0.49 |
Notes: aNegative and positive value of correlation coefficient indicated significantly lower and higher, respectively, levels in MDD patients. bp-values from the non-parametric Mann–Whitney U-test. cAdjusted p-values using Benjamini and Hochberg False Discovery Rate. d>1 and <1 indicated significantly lower and higher, respectively, levels in MDD patients.
Abbreviations: PI(40:4), Phosphatidylinositol(40:4); GPC(20:2/0:0),1-Eicosadienoylglycerophosphocholine; GPC(18:0/22:6), 1-Stearoyl-2-docosahexaenoyl-sn-glycero-3-phosphocholine; PE(16:0/18:2(9Z,12Z)), 1-Palmitoyl-2-linoleoyl-gpe; CerP(d18:1/24:1(15Z)), N-[(15Z)-Tetracosenoyl]sphing-4-enine 1-phosphate; N4R, N-(2R-Hydroxytricosanoyl)-2S-amino-1,3S,4R-octadecanetriol; LysoPC(17:0), 1-Heptadecanoyl-glycero-3-phosphocholine; LysoPC(16:0), 1–16:0-Lysophosphatidylcholine; SM(d18:0/22:1(13Z)), N-(13Z-Docosenoyl)-sphinganine-1-phosphocholine; DAG(22:4/22:5), 1-Adrenoyl-2-docosapentaenoyl-sn-glycerol; LysoPC(20:0), 1-Arachidonyl-glycero-3-phosphocholine; PE(16:0/18:1(9Z)), 1-Palmitoyl-2-oleoyl-gpe; MDD, major depressive disorder; HCs, healthy controls; AP, adjusted p-value; FC, fold change.
Pathway Analysis of Differential Metabolites and Genera
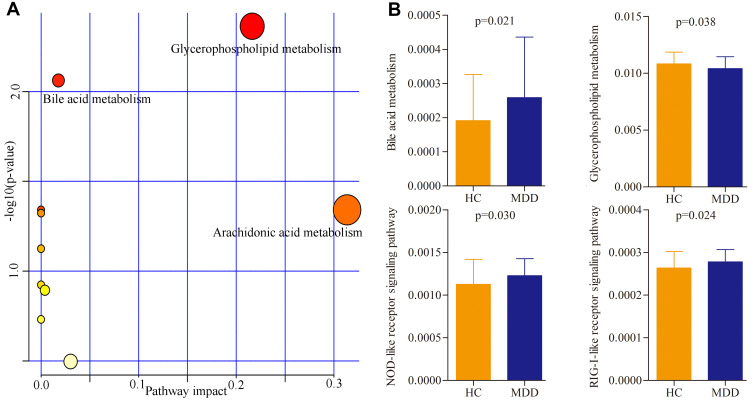
The online software MetaboAnalyst 5.0 was used to analyze the biological functions of differential metabolites. The results showed that there were three inflammation-related pathways in which these differential metabolites were mainly involved: Glycerophospholipid metabolism (p=0.004, impact=0.22), Bile acid metabolism (p=0.009, impact=0.02) and Arachidonic acid metabolism (p=0.045, impact=0.31) (Figure 4A). Meanwhile, the PICRUSt was used to predict the biological functions of differential genera. The results showed that there were four inflammation-related pathways in which these differential genera were mainly involved: Glycerophospholipid metabolism (p=0.038), Bile acid metabolism (p=0.021), Nucleotide binding oligomerization domain (NOD)-like receptor signaling pathway (p=0.030), and Retinoic acid-inducible gene I (RIG-I)-like receptor signaling pathway (p=0.024) (Figure 4B).
Figure 4.
Biological functions of differential serum metabolites and genera. (A) Three significantly affected pathways that these differential serum metabolites were mainly involved in; (B) four significantly disturbed pathways that these differential genera were mainly involved in.
Abbreviations: MDD, major depressive disorder; HC, healthy controls.
Potential Biomarkers for Diagnosing MDD
To identify potential serum biomarkers for diagnosing MDD, the included subjects were randomly assigned into the training set (40 MDD patients and 40 HCs) and testing set (20 MDD patients and 20 HCs). Firstly, we used the training set to find out the potential biomarkers. The Random Forest analysis indicated that the most significant deviations on serum metabolites between MDD patients and HCs could be described using five inflammation-related metabolites: LysoPC(16:0), Deoxycholic acid, Docosahexaenoic acid, Taurocholic acid and LysoPC(20:0). The ROC curve analysis showed that the panel consisting of these five metabolites could yield an AUC of 0.95 (Figure 5A). Secondly, we used the testing set to validate the diagnostic performance of this panel, and the results showed that the panel could yield an AUC of 0.92 (Figure 5B). These results suggested that these five inflammation-related metabolites held the promise as the potential biomarkers for diagnosing MDD.
Figure 5.
Diagnostic performance of the biomarker panel assessment. (A) The biomarker panel yielded an area under the curve (AUC) of 0.95 in the training set; (B) the AUC of the biomarker panel in the testing set was 0.92.
Abbreviation: CI, confidence interval.
Correlations Between Differential Metabolites and Genera
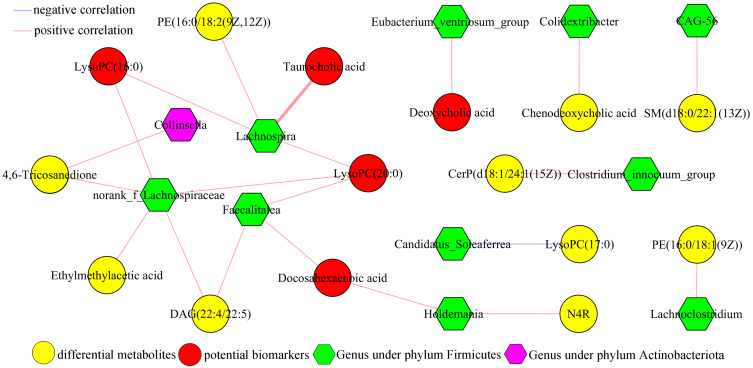
In order to determine the possible relationships between gut microbiota and serum metabolites, the correlation analysis of differential metabolites and genera was conducted here. As shown in Figure 6, there were significant correlations between 11 differential genera (including 10 genera under phyla Firmicutes) and 15 differential serum metabolites (including 6 inflammation-related metabolites). The norank_f_Lachnospiraceae was significantly correlated with five metabolites (including two potential biomarkers), the Lachnospira was significantly correlated with four metabolites (including three potential biomarkers), the Faecalitalea was significantly correlated with three metabolites (including two potential biomarkers) and the Eubacterium_ventriosum_group was significantly correlated with one potential biomarkers. These results indicated that there were close relationships between the disordered phyla Firmicutes and host’s inflammatory response.
Figure 6.
Correlations analysis of differential serum metabolites and genera.
Abbreviations: LysoPC(16:0), 1–16:0-Lysophosphatidylcholine; PE(16:0/18:2(9Z,12Z)), 1-Palmitoyl-2-linoleoyl-gpe; LysoPC(20:0), 1-Arachidonyl-glycero-3-phosphocholine; LysoPC(17:0), 1-Heptadecanoyl-glycero-3-phosphocholine; CerP(d18:1/24:1(15Z)), N-[(15Z)-Tetracosenoyl]sphing-4-enine 1-phosphate; DAG(22:4/22:5), 1-Adrenoyl-2-docosapentaenoyl-sn-glycerol; PE(16:0/18:1(9Z)), 1-Palmitoyl-2-oleoyl-gpe; N4R, N-(2R-Hydroxytricosanoyl)-2S-amino-1,3S,4R-octadecanetriol; SM(d18:0/22:1(13Z)), N-(13Z-Docosenoyl)-sphinganine-1-phosphocholine.
Discussion
In this study, we found that there were significant differences on gut microbiota compositions and serum metabolites in MDD patients compared to HCs. In total, 24 differential serum metabolites were identified, and a panel consisting of LysoPC(16:0), Deoxycholic acid, Docosahexaenoic acid, Taurocholic acid and LysoPC(20:0) was found to be effective in separating MDD patients from HCs. Meanwhile, 17 differential genera responsible for the discrimination between the two groups were identified. Furthermore, we observed the significantly positive correlations between the disturbance of phyla Firmicutes and these identified potential biomarkers. Considering the important role of gut microbiota in the onset of depression, our findings suggested that our identified biomarker panel might be a “good” classifier of MDD patients and HCs.
Glycerophospholipid metabolism has a close relationship with inflammatory response. For example, Lysophosphatidic acid (LPA) plays both anti- and pro-inflammatory roles in inflammatory lung diseases.35 Bile acids are signaling molecules, which could coordinately regulate inflammation and metabolism via Takeda G protein-coupled receptor 5 (TGR5) and nuclear farnesoid X receptor (FXR).36 Some metabolic products of arachidonic acid are the important inflammatory mediators, which may be important in leucocyte-mediated aspects of chronic inflammation.37 NOD-like receptors are the vital regulators in inflammation-associated angiogenesis, tumorigenesis, and cancer cell stemness and chemoresistance.38,39 Activated RIG-I-like receptors can activate the production of inflammatory cytokines and the type I/III interferons.40 In this study, the significantly changed serum metabolites and gut microbiota in MDD patients were significantly involved in these pathways. These results further demonstrated the close relationships between gut microbiota, inflammation and depression.
Gut microbiota plays a vital role in the pathogenesis of depression. Our previous studies have found the disturbance of gut microbiota in depressed mice and MDD patients.16–18 Other researchers also reported the significant gut microbiome changes in MDD patients.41,42 Although these findings are not exactly the same, partly causing by the differences on inclusion/exclusion criteria of MDD patients, detection methods or analytical methods, the disturbance of phyla Firmicutes is viewed as the hallmark of depression. However, the molecular mechanisms of phyla Firmicutes in the onset of depression are still unclear. In this study, we found that the 14 of 17 differential genera belonged to phyla Firmicutes, and function prediction analysis showed that these differential genera were involved in four inflammation-related pathways. Meanwhile, the four differential genera belonged to phyla Firmicutes were significantly correlated with the identified potential inflammation-related biomarkers. Taken together, we speculated that the disturbance of phyla Firmicutes might be involved in the onset of depression by regulating the host’s inflammatory response.
The inflammatory response has a close relationship with many diseases.43–46 As the important part of immune system, it is viewed as a driving factor causing changes in neurocircuits and neurotransmitters, which are closely related to the onset of depressive symptoms. Mounting evidence has solidified the relationships between depression and inflammation.47,48 For example, a cumulative meta-analysis reported that the levels of C-reactive protein and interleukin-6 (IL-6) were higher in MDD patients than in non-depressed controls.49 Our previous studies found that the level of alpha 1-antitrypsin (AAT) was decreased in MDD patients;27,50 AAT can inhibit the release of many pro-inflammatory factors, such as IL-6. In this study, we found ten inflammation-related metabolites, and identified three inflammation-related pathways by analyzing these differential metabolites. Moreover, all the identified potential biomarkers were inflammation-related serum metabolites and significantly correlated with the disordered gut microbiota. These results suggested that it might be a good way to explore markers of inflammation for diagnosing MDD.
Several limitations of our study were as following: i) each group only had 60 participants, the relatively small number of samples indicated that our findings were needed future studies to validate and support; ii) all the participants were from the same place and with the same ethnicity, these similarities might limit the general applicability of our conclusion; iii) no single metabolomic platform had the ability to detect the entire metabonome in serum; thus, it was meaningful to use other metabolomic platforms to further study the differences of serum metabolites in MDD patients; iv) only MDD patients and HCs were recruited here to identify potential biomarkers for diagnosing MDD, future studies should explore whether our potential biomarkers could be used to separate MDD patients from patients with other psychiatric disorders; v) the two groups were matched for age, sex, and body mass index, but gut microbiota might also be influenced by other factors, such as dietary habits and smoke; thus, future studies should further explore the effects of these factors on our results; and vi) we did not know whether MDD patients or HCs received any non-psychiatric medications; thus, future studies should investigate whether non-psychiatric medications could significantly affect our findings.
In conclusion, by conjoint analyzing the data of serum metabolites and gut microbiota compositions, we identified a potential serum inflammation-related biomarker panel for diagnosing MDD. The panel consisted of five inflammation-related metabolites: LysoPC(16:0), Deoxycholic acid, Docosahexaenoic acid, Taurocholic acid and LysoPC(20:0), which could effectively separate MDD patients from HCs. Our results provided candidates for future developing objective diagnostic methods for MDD, and showed novel insights for further exploring the role of gut microbiota in the pathogenesis of MDD.
Acknowledgments
This work was supported by the Natural Science Foundation Project of China (81701360, 81901398), the Science and Technology Fund of Guizhou Province [(QianHe LH2015)7411], the Open Project Funding of Shanghai Key Laboratory of Forensic Medicine (Academy of Forensic Science), the Natural Science Foundation of Chongqing, China (Grant No. cstc2019jcyj-msxmX0025), and the Chongqing Yuzhong District Science & Technology Commission (20190115).
Disclosure
The authors declare that they have no conflicts of interest.
References
- 1.Frankish H, Boyce N, Horton R. Mental health for all: a global goal. Lancet. 2018;392(10157):1493–1494. doi: 10.1016/S0140-6736(18)32271-2 [DOI] [PubMed] [Google Scholar]
- 2.Liao D, Chen Y, Guo Y, et al. Salvianolic acid b improves chronic mild stress-induced depressive behaviors in rats: involvement of AMPK/SIRT1 signaling pathway. J Inflamm Res. 2020;13:195–206. doi: 10.2147/JIR.S249363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang HQ, Wang ZZ, Chen NH. The receptor hypothesis and the pathogenesis of depression: genetic bases and biological correlates. Pharmacol Res. 2021;167:105542. doi: 10.1016/j.phrs.2021.105542 [DOI] [PubMed] [Google Scholar]
- 4.Beirão D, Monte H, Amaral M, Longras A, Matos C, Villas-Boas F. Depression in adolescence: a review. Middle East Curr Psychiatry. 2020;27:50. doi: 10.1186/s43045-020-00050-z [DOI] [Google Scholar]
- 5.Yano JM, Yu K, Donaldson GP, et al. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell. 2015;161:264–276. doi: 10.1016/j.cell.2015.02.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwartz J, Murrough JW, Iosifescu DV. Ketamine for treatment-resistant depression: recent developments and clinical applications. Evid Based Ment Health. 2016;19(2):35–38. doi: 10.1136/eb-2016-102355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Das R, Emon MPZ, Shahriar M, et al. Higher levels of serum IL-1β and TNF-α are associated with an increased probability of major depressive disorder. Psychiatry Res. 2021;295:113568. doi: 10.1016/j.psychres.2020.113568 [DOI] [PubMed] [Google Scholar]
- 8.Anjum S, Qusar MMAS, Shahriar M, et al. Altered serum interleukin-7 and interleukin-10 are associated with drug-free major depressive disorder. Ther Adv Psychopharmacol. 2020;10:2045125320916655. doi: 10.1177/2045125320916655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Emon MPZ, Das R, Nishuty NL, et al. Reduced serum BDNF levels are associated with the increased risk for developing MDD: a case-control study with or without antidepressant therapy. BMC Res Notes. 2020;13(1):83. doi: 10.1186/s13104-020-04952-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malik S, Singh R, Arora G, Dangol A, Goyal S. Biomarkers of major depressive disorder: knowing is half the battle. Clin Psychopharmacol Neurosci. 2021;19(1):12–25. doi: 10.9758/cpn.2021.19.1.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bai S, Wang W, Wang T, et al. CD36 deficiency affects depressive-like behaviors possibly by modifying gut microbiota and the inflammasome pathway in mice. Transl Psychiatry. 2021;11(1):16. doi: 10.1038/s41398-020-01130-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fouquier J, Moreno Huizar N, Donnelly J, et al. The gut microbiome in autism: study-site effects and longitudinal analysis of behavior change. mSystems. 2021;6(2):e00848–20. doi: 10.1128/mSystems.00848-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee J, d’Aigle J, Atadja L, et al. Gut microbiota-derived short-chain fatty acids promote poststroke recovery in aged mice. Circ Res. 2020;127(4):453–465. doi: 10.1161/CIRCRESAHA.119.316448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lichota A, Gwozdzinski K, Szewczyk EM. Microbial modulation of coagulation disorders in venous thromboembolism. J Inflamm Res. 2020;13:387–400. doi: 10.2147/JIR.S258839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng P, Wu J, Zhang H, et al. The gut microbiome modulates gut-brain axis glycerophospholipid metabolism in a region-specific manner in a nonhuman primate model of depression. Mol Psychiatry. 2020. doi: 10.1038/s41380-020-0744-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu M, Tian T, Mao Q, et al. Associations between disordered gut microbiota and changes of neurotransmitters and short-chain fatty acids in depressed mice. Transl Psychiatry. 2020;10(1):350. doi: 10.1038/s41398-020-01038-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen JJ, Zheng P, Liu YY, et al. Sex differences in gut microbiota in patients with major depressive disorder. Neuropsychiatr Dis Treat. 2018;14:647–655. doi: 10.2147/NDT.S159322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen JJ, He S, Fang L, et al. Age-specific differential changes on gut microbiota composition in patients with major depressive disorder. Aging (Albany NY). 2020;12(3):2764–2776. doi: 10.18632/aging.102775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanada K, Nakajima S, Kurokawa S, et al. Gut microbiota and major depressive disorder: a systematic review and meta-analysis. J Affect Disord. 2020;266:1–13. doi: 10.1016/j.jad.2020.01.102 [DOI] [PubMed] [Google Scholar]
- 20.Lai WT, Deng WF, Xu SX, et al. Shotgun metagenomics reveals both taxonomic and tryptophan pathway differences of gut microbiota in major depressive disorder patients. Psychol Med. 2021;51(1):90–101. doi: 10.1017/S0033291719003027 [DOI] [PubMed] [Google Scholar]
- 21.Pan X, Wen SW, Kaminga AC, Liu A. Gut metabolites and inflammation factors in non-alcoholic fatty liver disease: a systematic review and meta-analysis. Sci Rep. 2020;10(1):8848. doi: 10.1038/s41598-020-65051-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sae-Khow K, Charoensappakit A, Visitchanakun P, et al. Pathogen-associated molecules from gut translocation enhance severity of cecal ligation and puncture sepsis in iron-overload β-Thalassemia mice. J Inflamm Res. 2020;13:719–735. doi: 10.2147/JIR.S273329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leonard BE. Inflammation and depression: a causal or coincidental link to the pathophysiology? Acta Neuropsychiatr. 2018;30(1):1–16. doi: 10.1017/neu.2016.69 [DOI] [PubMed] [Google Scholar]
- 24.Colasanto M, Madigan S, Korczak DJ. Depression and inflammation among children and adolescents: a meta-analysis. J Affect Disord. 2020;277:940–948. doi: 10.1016/j.jad.2020.09.025 [DOI] [PubMed] [Google Scholar]
- 25.Zheng P, Wang Y, Chen L, et al. Identification and validation of urinary metabolite biomarkers for major depressive disorder. Mol Cell Proteomics. 2013;12(1):207–214. doi: 10.1074/mcp.M112.021816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng P, Gao HC, Qi ZG, et al. Peripheral metabolic abnormalities of lipids and amino acids implicated in increased risk of suicidal behavior in major depressive disorder. Metabolomics. 2013;9(3):688–696. doi: 10.1007/s11306-012-0474-9 [DOI] [Google Scholar]
- 27.Bai S, Fang L, Xie J, Bai H, Wang W, Chen JJ. Potential biomarkers for diagnosing major depressive disorder patients with suicidal ideation. J Inflamm Res. 2021;14:495–503. doi: 10.2147/JIR.S297930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arentsen T, Qian Y, Gkotzis S, et al. The bacterial peptidoglycan-sensing molecule Pglyrp2 modulates brain development and behavior. Mol Psychiatry. 2017;22(2):257–266. doi: 10.1038/mp.2016.182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoyles L, Snelling T, Umlai UK, et al. Microbiome-host systems interactions: protective effects of propionate upon the blood-brain barrier. Microbiome. 2018;6(1):55. doi: 10.1186/s40168-018-0439-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen L, Dong B, Gao H, et al. HPV-16 E2/E6 and POU5F1B as biomarkers to determine cervical high-grade squamous lesions and more. J Inflamm Res. 2020;13:813–821. doi: 10.2147/JIR.S278911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang Y, Li Y, Sun J, Pan H, Yao F, Jiao X. Selection of an optimal combination panel to better triage COVID-19 hospitalized patients. J Inflamm Res. 2020;13:773–787. doi: 10.2147/JIR.S273193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He S, Mao X, Lei H, et al. Peripheral blood inflammatory-immune cells as a predictor of infertility in women with polycystic ovary syndrome. J Inflamm Res. 2020;13:441–450. doi: 10.2147/JIR.S260770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Y, Duan Y, Li Y. Integrated gene expression profiling analysis reveals probable molecular mechanism and candidate biomarker in Anti-TNFα non-response IBD patients. J Inflamm Res. 2020;13:81–95. doi: 10.2147/JIR.S236262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mandrekar JN. Receiver operating characteristic curve in diagnostic test assessment. J Thorac Oncol. 2010;5(9):1315–1316. doi: 10.1097/JTO.0b013e3181ec173d [DOI] [PubMed] [Google Scholar]
- 35.Wang S, Tang K, Lu Y, et al. Revealing the role of glycerophospholipid metabolism in asthma through plasma lipidomics. Clin Chim Acta. 2021;513:34–42. doi: 10.1016/j.cca.2020.11.026 [DOI] [PubMed] [Google Scholar]
- 36.Chávez-Talavera O, Tailleux A, Lefebvre P, Staels B. Bile acid control of metabolism and inflammation in obesity, type 2 diabetes, dyslipidemia, and nonalcoholic fatty liver disease. Gastroenterology. 2017;152(7):1679–1694.e3. doi: 10.1053/j.gastro.2017.01.055 [DOI] [PubMed] [Google Scholar]
- 37.Higgins AJ, Lees P. The acute inflammatory process, arachidonic acid metabolism and the mode of action of anti-inflammatory drugs. Equine Vet J. 1984;16(3):163–175. doi: 10.1111/j.2042-3306.1984.tb01893.x [DOI] [PubMed] [Google Scholar]
- 38.Liu P, Lu Z, Liu L, et al. NOD-like receptor signaling in inflammation-associated cancers: from functions to targeted therapies. Phytomedicine. 2019;64:152925. doi: 10.1016/j.phymed.2019.152925 [DOI] [PubMed] [Google Scholar]
- 39.Zhou H, Urso CJ, Jadeja V. Saturated fatty acids in obesity-associated inflammation. J Inflamm Res. 2020;13:1–14. doi: 10.2147/JIR.S229691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Onomoto K, Onoguchi K, Yoneyama M. Regulation of RIG-I-like receptor-mediated signaling: interaction between host and viral factors. Cell Mol Immunol. 2021;18(3):539–555. doi: 10.1038/s41423-020-00602-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Naseribafrouei A, Hestad K, Avershina E, et al. Correlation between the human fecal microbiota and depression. Neurogastroenterol Motil. 2014;26(8):1155–1162. doi: 10.1111/nmo.12378 [DOI] [PubMed] [Google Scholar]
- 42.Jiang H, Ling Z, Zhang Y, et al. Altered fecal microbiota composition in patients with major depressive disorder. Brain Behav Immun. 2015;48:186–194. doi: 10.1016/j.bbi.2015.03.016 [DOI] [PubMed] [Google Scholar]
- 43.Yang CC, Hsiao LD, Yang CM. Galangin inhibits LPS-induced MMP-9 expression via suppressing protein kinase-dependent AP-1 and FoxO1 activation in rat brain astrocytes. J Inflamm Res. 2020;13:945–960. doi: 10.2147/JIR.S276925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lisowska B, Jakubiak J, Siewruk K, Sady M, Kosson D. Which idea is better with regard to immune response? Opioid anesthesia or opioid free anesthesia. J Inflamm Res. 2020;13:859–869. doi: 10.2147/JIR.S275986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Al Rushood M, Al-Eisa A, Al-Attiyah R. Serum and urine interleukin-6 and interleukin-8 levels do not differentiate acute pyelonephritis from lower urinary tract infections in children. J Inflamm Res. 2020;13:789–797. doi: 10.2147/JIR.S275570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang MX, Xiong F. Astaxanthin and its effects in inflammatory responses and inflammation-associated diseases: recent advances and future directions. Molecules. 2020;25(22):5342. doi: 10.3390/molecules25225342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Felger JC. Role of inflammation in depression and treatment implications. Handb Exp Pharmacol. 2019;250:255–286. doi: 10.1159/000343966 [DOI] [PubMed] [Google Scholar]
- 48.Raison CL, Rutherford RE, Woolwine BJ, et al. A randomized controlled trial of the tumor necrosis factor antagonist infliximab for treatment-resistant depression: the role of baseline inflammatory biomarkers. JAMA Psychiatry. 2013;70(1):31–41. doi: 10.1001/2013.jamapsychiatry.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haapakoski R, Mathieu J, Ebmeier KP, Alenius H, Kivimäki M. Cumulative meta-analysis of interleukins 6 and 1β, tumour necrosis factor α and C-reactive protein in patients with major depressive disorder. Brain Behav Immun. 2015;49:206–215. doi: 10.1016/j.bbi.2015.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gui SW, Liu YY, Zhong XG, et al. Plasma disturbance of phospholipid metabolis in major depressive disorder by integration of proteomics and metabolomics. Neuropsychiatr Dis Treat. 2018;14:1451–1461. doi: 10.2147/NDT.S164134 [DOI] [PMC free article] [PubMed] [Google Scholar]