Abstract
We study the basic reproduction numbers for a class of reaction-diffusion epidemic models that are developed from autonomous ODE systems. We present a general numerical framework to compute such basic reproduction numbers; meanwhile, the numerical formulation provides useful insight into their characterizations. Using matrix analysis, we show that the basic reproduction numbers are the same for these PDE models and their associated ODE models in several important cases that include, among others, a single infected compartment, constant diffusion rates, uniform diffusion patterns among the infected compartments, and partial diffusion in the system.
Keywords: Compartmental models, Numerical analysis, Eigenvalues
1. Introduction
Mathematical modeling and analysis provide a powerful theoretical tool for epidemiological study. Both ordinary differential equations (ODE) and partial differential equations (PDE) are extensively used. In particular, mathematical models based on reaction-diffusion equations have been frequently employed to investigate the transmission and spread of infectious diseases.
During the development of a PDE epidemic model, an ODE system is often established first to describe the spatially homogeneous dynamics of disease transmission. Then, diffusion terms are added to study the spatial spread of the disease. A diffusion process represents the random movement and dispersal of hosts/pathogens over the spatial domain, normally without a directional preference. Incorporation of such spatial movement, generally associated with location-dependent diffusion rates, into epidemiological, ecological and other biological models emphasizes the spatial heterogeneity of population dynamics (Cantrell and Cosner 1991, 2003), particularly with regard to disease transmission and spread.
There have been many studies devoted to reaction-diffusion epidemic models. For example, existence and well posedness of solutions are analyzed in Kim et al. (2013) and Yamazaki and Wang (2016), equilibrium analyses are conducted in Wang et al. (2015), Wu and Zou (2018), and Yu and Zhao (2016), traveling waves are investigated in Wang et al. (2015), Wang et al. (2016), and Zhao et al. (2018), and realistic epidemic simulations are carried out in Bertuzzo et al. (2010) and Rinaldo et al. (2012). Particularly, basic reproduction numbers for such epidemic systems are studied in Allen et al. (2008), Thieme (2009), Wang and Zhao (2012), and Magal et al. (2019). The basic reproduction number, typically denote by , is a critical quantity to measure the transmission risk of an infectious disease. It quantifies the expected number of secondary infections produced by one infective individual in a completely susceptible population, and often characterizes the threshold behavior of an epidemic; i.e., the disease would be eradicated if and the disease would persist if . For ODE epidemic models, is generally defined as the spectral radius of the next-generation matrix Diekmann et al. (1990) and van den Driessche and Watmough (2002), and its calculation follows a standard procedure. For PDE models such as a reaction-diffusion system, the definition and calculation of the basic reproduction number are typically more intriguing. Allen et al. (2008) introduced a variational formula to characterize for a simple SIS model with diffusion terms. Wang and Zhao (2012) defined for reaction-diffusion type systems as the spectral radius of a next-infection operator, using the theory of principal eigenvalues. Thieme (2009) also introduced a theoretical framework to define as the spectral radius of a resolvent-positive operator. More recently, Magal, Webb, and Wu (2019) investigated a vector-host disease model involving diffusive and convective terms, where is found as the spectral radius of a product of four multiplicative operators that include, in particular, two associated with the corresponding ODE model. Other studies related to reaction-diffusion type epidemic models and their basic reproduction numbers can be found in Bertuzzo et al. (2010), Chen and Shi (2020), Ge et al. (2017), Lou and Zhao (2011), Peng and Zhao (2012), Rinaldo et al. (2012), Song et al. (2019), Wang et al. (2015), Wu and Zou (2018), Yamazaki and Wang (2016), and Yu and Zhao (2016).
For most of these PDE epidemic models, the calculation of their basic reproduction numbers requires special efforts since the standard next-generation matrix technique for ODE systems is no longer applicable; instead, the process involves evaluations of various operators defined in infinite-dimensional spaces. Thus, a nontrivial numerical procedure is typically required to achieve this goal.
Since most of such reaction-diffusion type models are associated with meaningful ODE systems that represent spatially homogeneous disease dynamics, it is natural to ask what is the relationship between the of the PDE system and that of the associated ODE system. At present, there are very few studies devoted to address this question. It was shown in Wang and Zhao (2012) that when the diffusion rates are positive constants and the next-generation matrices of the PDE system are independent of the spatial location, the of the PDE system is the same as that of the corresponding ODE system. The authors in Wang et al. (2016) compared the values of the basic reproduction numbers associated with a class of PDE and ODE cholera models using both asymptotical and numerical means; in particular, they found that the of the PDE system approaches the of the ODE system when the (constant) diffusion rates approaches infinity. The authors in Magal et al. (2019) rigorously analyzed the relationship between the PDE-based and the ODE-based for a vector-host model, and established the limits of zero and infinite diffusion rates. Meanwhile, a SEIRS reaction-diffusion model with constant diffusion rates was analyzed in Song et al. (2019) and the monotonicity, asymptotic property, and lower and upper bounds of for this PDE model were established. In a more recent study Chen and Shi (2020), the authors investigated the asymptotic profiles of for a reaction-diffusion epidemic system with constant diffusion rates. They proved that when the diffusion rates tend to zero, approaches the maximum value of the local reproduction number on the spatial domain; when the diffusion rates tend to infinity, approaches the spectral radius of an averaged next-generation matrix. Despite these findings, our understanding of the general connection; i.e., how the of a PDE system is related to that of the corresponding ODE system, remains limited.
In the present paper, we aim to partially address this issue by calculating and analyzing the basic reproduction numbers for a class of reaction-diffusion epidemic models which are developed from underlying autonomous ODE systems. To emphasize spatial heterogeneity, we allow the diffusion rates to be spatially dependent, as various physical studies (see, e.g., Sauty 1980; Sposito et al. 1986; Taylor 1953) have demonstrated that the diffusion rates could vary significantly with respect to spatial locations. Our work also differs from most of the prior studies in the following aspects: (1) instead of analyzing the asymptotic profiles when the constant diffusion rates tend to zero or infinite, we aim to explore a more general relationship between the basic reproduction number of the underlying ODE model and that of the PDE model with variable diffusion rates; (2) our work is inspired by, and builds on, numerical analysis of the operator eigenvalue problem that defines the basic reproduction number of the PDE system. We present a general numerical method to evaluate the value of for such a PDE system, reducing the infinite-dimensional operator eigenvalue problem to a finite-dimensional matrix eigenvalue problem. This approach represents a significant extension of the numerical technique presented in Wang et al. (2016), which is focused on a special type of cholera models, to more general PDE epidemic models. Starting from there, we analyze the relationship between the PDE-based and ODE-based , and derive several sufficient conditions under which the two basic reproduction numbers equal each other. These scenarios cover several important types of epidemic models. Throughout this paper, we employ only elementary numerical analysis and matrix theory in our discussion.
We organize the remainder of this paper as follows. In Sect. 2, we present our reaction-diffusion epidemic systems as well as their associated ODE systems, and list necessary assumptions. In Sect. 3, we describe the details of our numerical method for calculating the basic reproduction numbers of the PDE models. In Sect. 4, we state and prove several results regarding the relationship between the basic reproduction numbers of the PDE and ODE models. In Sect. 5, we present a few specific examples to demonstrate our findings. In Sect. 6, we conclude the paper with some discussion.
2. Models and Assumptions
Let n be a positive integer. We consider a vector-valued variable U with n components that combines, in general, the populations of the hosts and pathogens. Specifically, U (t, x) = (u1(t, x), …, un(t, x))T, where each ui(t, x) denotes the density of the population in compartment i at time t and position x. In this study, we will focus our attention on the one-dimensional spatial domain [0, 1]. We consider the following reaction-diffusion epidemic system
| (1) |
together with appropriate initial conditions. In this model,
denotes the diffusion rates at location x, and we assume that D(x) is continuously differentiable on [0, 1]. Meanwhile,
where denotes the appearance rate of newly infected individuals in compartment i, and
where denotes the transfer rate of individuals into compartment i by all other means, and the transfer rate of individuals out of compartment i. The compartments in U can be divided into infected and uninfected compartments. Without loss of generality, we may assume UI = (u1, …, um)T denotes all the infected compartments, where 1 ≤ m < n. Consequently, we define the set of all disease-free steady states as
Before we proceed, we write system (1) in another form
| (2) |
If we impose the constraint
| (3) |
then system (2) is equivalent to the reaction-diffusion system (1). In what follows, we intend to discuss the slightly more general PDE system (2), where the results can be easily interpreted back to the original system (1) under the condition (3).
We note that if system (2) is homogeneous in space; i.e., U = (u1(t), …, un(t))T, then the PDE model (2) is reduced to the following ODE model
| (4) |
Based on the setting in van den Driessche and Watmough (2002) for ODE epidemic models, we present the following standard assumptions:
-
(A1)
, and are non-negative and continuously differentiable for 1 ≤ i ≤ n.
-
(A2)
If ui = 0, then , 1 ≤ i ≤ m.
-
(A3)
for i > m.
-
(A4)
If U ∈ Us, then , i = 1, …, m.
Each of these assumptions has its biological meaning: (A1) follows from the simple fact that the number of transfer of individuals must be non-negative; (A2) means that there is no individual transferred out from an empty compartment; (A3) states that no new infection happens in the uninfected compartments; and (A4) indicates that a disease-free compartment remains disease-free for all the time.
Suppose that is a disease-free steady state of model (4), where we assume that U0 is spatially independent. Then, the basic reproduction number for the ODE system (4) is defined as the spectral radius of the next-generation matrix (van den Driessche and Watmough 2002); i.e.,
where F and V are m × m constant matrices with (i, j) entry and , respectively. Following the framework in Wang and Zhao (2012), we let T(t) be the solution semigroup on associated with the following linear reaction-diffusion equation:(5)
where UI = (u1, …, um)T, DI (x) = diag(d1(x), …, dm(x)) and CI (x) = diag(c1(x), …, cm(x)). We assume the distribution of the initial infections is Um(x) = (u1(x), …, um(x))T. Then, T(t)(Um(x)) represents the distribution of these infections after time t > 0. Hence, the distribution of new infections at time t > 0 is FT(t)(Um(x)), and thereby the distribution of the total new infections is(6)
Thus, the next-generation operator L, which maps the initial infection distribution to the distribution of the total infective individuals generated during the infection period, can be defined as follows,
Accordingly, the basic reproduction number for the PDE model (2) is the spectral radius of the operator L; i.e.,(7)
Meanwhile, denote(8)
Clearly, for any t > 0 and a solution ϕ(t, x) of equation (6),(9)
Hence, Γ is the generator of the C0-semigroup T(t) on . Note that T(t)is a positive semigroup since for all t ≥ 0. Let σ(Γ) denote the spectrum of the operator Γ. It then follows from (Thieme 2009, Theorem 3.12) that
where s(Γ) = sup{Reλ : λ ∈ σ(Γ)} is the spectral bound of Γ. Since the internal evolution of individuals in the infectious compartments is dissipative and exponentially decays in many cases because of the loss of infected members from natural and disease-induced mortalities, we may assume(10) -
(A5)
−V is cooperative and s(Γ) < 0.
Thus, fixing λ = 0 in equation (10), we obtain
| (11) |
or L = −FΓ−1. We state two additional assumptions regarding the PDE model (2). First, we set a minimal diffusion rate at all spatial locations; i.e.,
-
(H1)
There exists a constant d0 such that di(x) ≥ d0 > 0 for any x ∈ [0, 1] and 1 ≤ i ≤ m.
Second, for ease of presentation, we assume
-
(H2)
V = diag(υ1, …, υm) with υi > 0, i = 1, …, m.
Many common epidemic models, such as SIR, SEIR, and patchy models (see Sect. 5), satisfy the condition (H2). In case V is not in a diagonal form, it is often possible to re-define the new infection vector, say , and the transfer vector, say , in the ODE system (4) such that the associated matrix is diagonal and that . It then can be shown that ρ(FV−1) and are equivalent in characterizing the disease threshold; i.e., they are simultaneously higher than (or equal to, or lower than) unity (van den Driessche and Watmough 2002).
3. Numerical Formulation
Let λ be an eigenvalue of L such that L(ϕ(x)) = λϕ(x) for some eigenvector ϕ(x) = (ϕ1(x), …, ϕm(x))T. Then,
| (12) |
Suppose that ψ(x) = −Γ−1(ϕ(x)), where ψ(x) = (ψ1(x), …, ψm(x))T, then −Γ(ψ(x)) = ϕ(x); i.e.,
| (13) |
For a sufficiently large integer N > 0, let xk = k/N, dik = di(xk), cik = ci(xk), ψik = ψi(xk), and ϕik = ϕi(xk) for k = 0, 1, …, N. Applying the standard centered difference scheme to Eq. (13) on the spatial domain [0, 1], we obtain
| (14) |
or
| (15) |
for all 0 ≤ k ≤ N, and ψi,−1 = ψi1, ψi,N+1 = ψi,N−1 by the Neumann boundary conditions. Combine these N + 1 approximate equations in a matrix form as follows
| (16) |
where , , and with , , and aik = 2dik N2 +υi for all 0 ≤ k ≤ N.
Let us define
| (17) |
Then, N > N* implies that for any x ∈ [0, 1], thus and .
Next, we show that for each i = 1, ⋯, m, the matrix Ai is invertible and υi is a lower bound of the eigenvalues of Ai.
Lemma 3.1
Let , where and αjβj > 0 for all j, then Mk is diagonalizable and all eigenvalues of Mk are real.
Proof
Since αjβj > 0, we can define , 1 ≤ j ≤ k. Denote
Then, Bk is a real symmetric matrix and thereby Bk is diagonalizable and all eigenvalues of Bk are real. Choose a nonsingular diagonal matrix
One can verify that . Thus, we complete the proof. □
Lemma 3.2
Let N > N*, where N* is defined in Eq. (17). If is an eigenvalue of Ai, then is real and , 1 ≤ i ≤ m.
Proof
Note that for any k, and are negative when N > N*. Then, . Hence, by the Gershgorin Circle Theorem, there exists a p ∈ {0, 1, …, N} such that
which indicates that . It follows from Lemma 3.1 that and hence . □
Denote , , and
Then, A is invertible and Ψ ≈ A−1 Φ by Eq. (16). It follows from Eq. (12) that
| (18) |
which yields
| (19) |
for any integer N > 0, where IN+1 is the (N + 1) × (N + 1) identity matrix and ⊗ denotes the Kronecker product that is defined as follows: for any r × s matrix M = (mij) and p × q matrix Q,
Substituting Ψ ≈ A−1 Φ into equation (19), our numerical formulation thus leads to
| (20) |
We are now ready to state the following result.
Theorem 3.1
| (21) |
Proof
From the basic theory in numerical analysis (Richtmyer and Morton 1994; Thomas 1995), the solution of Eq. (20) (or, Eq. 14) converges to the solution of Eq. (12) (or, Eq. 13) when N → ∞. Hence, for sufficiently large N, an eigenvalue of the operator L = −FΓ−1 is an approximation to an eigenvalue of the matrix (F ⊗ IN+1)A−1, and vice versa. Moreover, for any ε > 0, we have
for sufficiently large N. Letting ε → 0, we obtain . □
Essentially, our numerical formulation reduces the original operator eigenvalue problem (12) to a matrix eigenvalue problem (20), which is not only useful for practical evaluation (there are many efficient numerical techniques currently available for computing matrix eigenvalues (Golub and Van Loan 1996; Saad 2011)), but also provides important insight into the property of .
4. Analysis
In what follows, we conduct an analysis of and its relationship to using our result in (21). We first introduce the following lemmas.
Lemma 4.1
For all N > N*, , 1 ≤ i ≤ m.
Proof
It follows from Lemma 3.2 that . It suffices to show that υi is an eigenvalue of Ai; i.e., det(υi IN+1 − Ai) = 0. Note that, the summation of the k-th row of the matrix υi IN+1 − Ai is for all 0 ≤ k ≤ N. Thus, the statement holds true. □
Lemma 4.2
Assume that, X = (xij) is an m × m matrix and Yij (1 ≤ i, j ≤ m) are n × n matrices. If there exists a nonsingular matrix P such that P−1Yij P = Dij for all i, j = 1, …, m, where , then
Proof
Note that,
and the determinants of the left and right matrices satisfy
We now only need to calculate the determinant of the middle matrix. Apply Laplace expansion by choosing rows {1, n + 1, …, (m − 1)n + 1} to obtain
where . Similarly, keep applying Laplace expansion by choosing rows {1, n, …, (m − 1)(n − 1) + 1} for the latter matrix, and one can easily obtain
which completes the proof. □
Lemma 4.3
If Z and W −Z are both n×n non-negative matrices, then ρ(W) ≥ ρ(Z).
Proof
Let Z = (zij) and W = (wij), i, j = 1, …, n. Then wij ≥ zij ≥ 0. Hence, for any integer k ≥ 1, it is easy to see that if we denote Wk = (pij) and Zk = (qij), then pij ≥ qij, i, j = 1, …, n, and thereby
Thus, . □
Now, we state our main results in the following three theorems.
Theorem 4.1
Let (A1)–(A5) and (H1)–(H2) hold.
Assume that, for system (2) is a commuting family. Then,
If F is a triangular matrix, then . Particularly, if m = 1, then.
Proof
- It is clear that when N > N*, is a family of diagonalizable matrices by Lemma 3.1. The commuting property ensures that are simultaneously diagonalizable (Horn and Johnson 1985). It follows from Lemma 3.2 that we can write
with some nonsingular matrix Q for any N > N*, where 0 < αik ≤ 1/υi for i = 1, …, m and k = 1, …, N + 1. Hence, it follows from Lemma 4.2 that
where for 1 ≤ k ≤ N + 1. Thus, equation (22) yields(22)
Note that, F is non-negative by assumptions (A1) and (A4), then Ok and FV −1 − Ok are both non-negative, and thus ρ(Ok) ≤ ρ(FV−1) by Lemma 4.3. Therefore, . Taking the limit N → ∞, we obtain .(23) - This directly follows from Lemma 4.1 that
for any integer N sufficiently large.
Remark 4.1
The second part of Theorem 4.1 states, as a special case, that if the reaction-diffusion model (2) has only one infected compartment, then its basic reproduction number is identical to that of the underlying ODE model.
Next, we characterize the sufficient and necessary conditions such that is a commuting family.
Theorem 4.2
Let N > N*. The matrix set associated with system (2) is a commuting with system (2) is a commuting family if and only if there exist constants δi, σi, and continuous functions d(x), c(x) such that di(x) = δid(x), ci(x) = σic(x), and δiσj = δjσi, 1 ≤ i, j ≤ m.
Proof
We can rewrite for i = 1, …, m, where
Then, Ai Aj − Aj Ai = 0 is equivalent to
| (24) |
One can verify that
where , and , k = 0, …, N − 1
where , , and , k = 0, …, N, and
where , k = 1, …, N − 2. Consider the (1, 2) element and the (N + 1, N) element of the matrix in Eq. (24), we have . Summing up all the elements of each row into the first column, it is easy to see that
Thus, ; i.e.,
| (25) |
for all 0 ≤ k ≤ N − 1. Since d1(x) > 0, x ∈ [0, 1], then for any rational number , 0 ≤ k ≤ N − 1, it follows from equation (25) that
Note that, is continuous on [0, 1], hence for all x ∈ [0, 1]. Let d(x) = d1(x) and . Then for any 1 ≤ j ≤ m, we obtain
| (26) |
Accordingly, Hi Hj − Hj Hi = 0 and equation (24) becomes
| (27) |
For k = 2, …, N, consider the (k, k − 1) element and the (k, k + 1) element of the matrix in equation (27), we obtain and , respectively. Hence,
where , k = 1, …, N – 2. It follows from the (k, k + 2) element and the (k + 2, k) element of the matrix in equation (27) that
Thus, and ; i.e.,
| (28) |
for k = 0, 1, …, N − 2. Note that, di(x) = δid(x); it follows from equation (28) that
Hence, for 1 ≤ i ≤ m, ci(x) = σic1(x) for all x ∈ [0, 1], where . In addition, , 1 ≤ i, j ≤ m. The proof is thus complete. □
Remark 4.2
The conditions in Theorem 4.2 are stated in terms of system (2) without the constraint (3), which would be more general than the original reaction-diffusion system (1). If Eq. (3) is considered, then the conditions in Theorem 4.2 can be obviously simplified and only di(x) = δid(x), 1 ≤ i ≤ m, are needed.
Based on Theorem 4.2, we can in fact show that if is a commuting family.
Theorem 4.3
Let (A1)–(A5) and (H1)–(H2) hold. Suppose that there exist continuous functions d(x) and c(x) such that di(x) = δid(x) and ci(x) = σic(x) for 1 ≤ i ≤ m, where the constants δi, σi satisfy δiσj = δjσi for all 1 ≤ i, j ≤ m, then .
Proof
On one hand, we have by Theorems 4.1 and 4.2. On the other hand, we note that dik = δid(xk) ≔ δidk and cik = σic(xk) ≔ σick. Then, for any 1 ≤ i ≤ m,
Since for all 1 ≤ i, j ≤ m, we obtain that
Denote this identical matrix by A0. It is clear to see that A0 is diagonalizable for all N > N* by Lemma 3.1. Note that, the summation of each row of matrix A0 is 0, and hence, 0 is the smallest eigenvalue of A0 by Lemma 3.2. We assume those eigenvalues are 0 ≤ λ1 ≤ ⋯ ≤ λN. Then, there exists a nonsingular matrix P such that
Thus,
It follows from the proof of Theorem 4.1(1) that O1 = FV−1 and from Lemma 4.3 that ρ(O1) ≥ ρ(Ok) for 2 ≤ k ≤ N + 1. Consequently, equation (23) yields
| (29) |
for all N > N*. Letting N → ∞, we obtain . □
Remark 4.3
Again, the conditions in Theorem 4.3 are stated in terms of system (2). If we impose the constraint (3) and consider the original system (1), then the following results immediately follow from Theorem 4.3. These results cover several special, but important, scenarios of reaction-diffusion epidemic models: (i) constant diffusion rates (Remark 4.4); (ii) uniform diffusion patterns among the infected compartments (Remark 4.5); (iii) partial diffusion in the system (Corollary 4.2). In each of the following scenarios, the basic reproduction number for the reaction-diffusion system (1) and the basic reproduction number for its ODE counterpart are the same.
Corollary 4.1
If there exist constants δi and a continuous function d(x) such that di(x) = δid(x) for all i = 1, …, m in system (1), then .
Remark 4.4
In particular, if the diffusion rates of all the infected compartments are positive constants in system (1), then .
Remark 4.5
In particular, if di(x) = dj(x) for 1 ≤ i, j ≤ m in system (1), then .
Corollary 4.2
If di(x) = 0 for i = 1, …, m − 1 and dm(x) ≥ d0 > 0 in system (1), then .
Remark 4.6
Although the assumption (H1) does not hold in Corollary 4.2, we note that Ai = υi IN+1, since di(x) = ci(x) = 0, for 1 ≤ i ≤ m − 1. Meanwhile, for any N > N*, it follows from the proof of Theorem 4.1(1) that there exists 1 ≤ k ≤ N +1 such that αmk = 1/υm. Hence, we obtain the matrix Ok = FV−1, which yields .
5. Examples
We provide a few examples below to illustrate some common reaction-diffusion epidemic models that have the same basic reproduction numbers as those of their ODE counterparts. Sect. 5.1 presents a SIR model which has a single infected compartment; Sect. 5.2 presents two SIR-B models where one is partially diffusive and the other has constant diffusion rates; Sect. 5.3 presents a patchy model where the matrix F is in a triangular form; Sect. 5.4 presents a SEIR model which has a uniform diffusion pattern in the infected compartments. Some numerical simulation results are also presented.
5.1. SIR Model
Let us consider the following SIR model with diffusion, which is an extension of the reaction-diffusion SIR system presented in Kim et al. (2013):
| (30) |
Here, S, I, and R are the numbers of susceptible, infected, and recovered individuals, respectively, and dS(x), dI(x) and dR(x) are their associated diffusion rates. The parameters Λ, α, μ, and γ represent the recruitment rate, transmission rate, natural death rate, and disease recovery rate, respectively.
Apparently, system (30) admits a disease-free steady state , and I is the only infection compartment; i.e., m = 1. Note that, and V = μ + γ. From Theorem 4.1(2), we know that the basic reproduction number of the PDE system (30) is the same as that of its ODE system:
A numerical test is performed to demonstrate our numerical method based on Eq. (21). Figure 1 plots the values of our numerical calculation of ρ(F ⊗ IN+1)A−1) versus N for the model (30). We set the diffusion rate of the infected individuals as dI (x) = sin(50x) + 0.04x + 1 in this test. Since does not depend on N, it is represented by a horizontal line in the graph. We observe that when N is sufficiently large, the numerical values of (i.e., ρ(F ⊗ IN+1)A−1)) agree almost perfectly with , and this pattern continues for all N > 210. This is consistent with our result in Eq. (29).
Fig. 1.
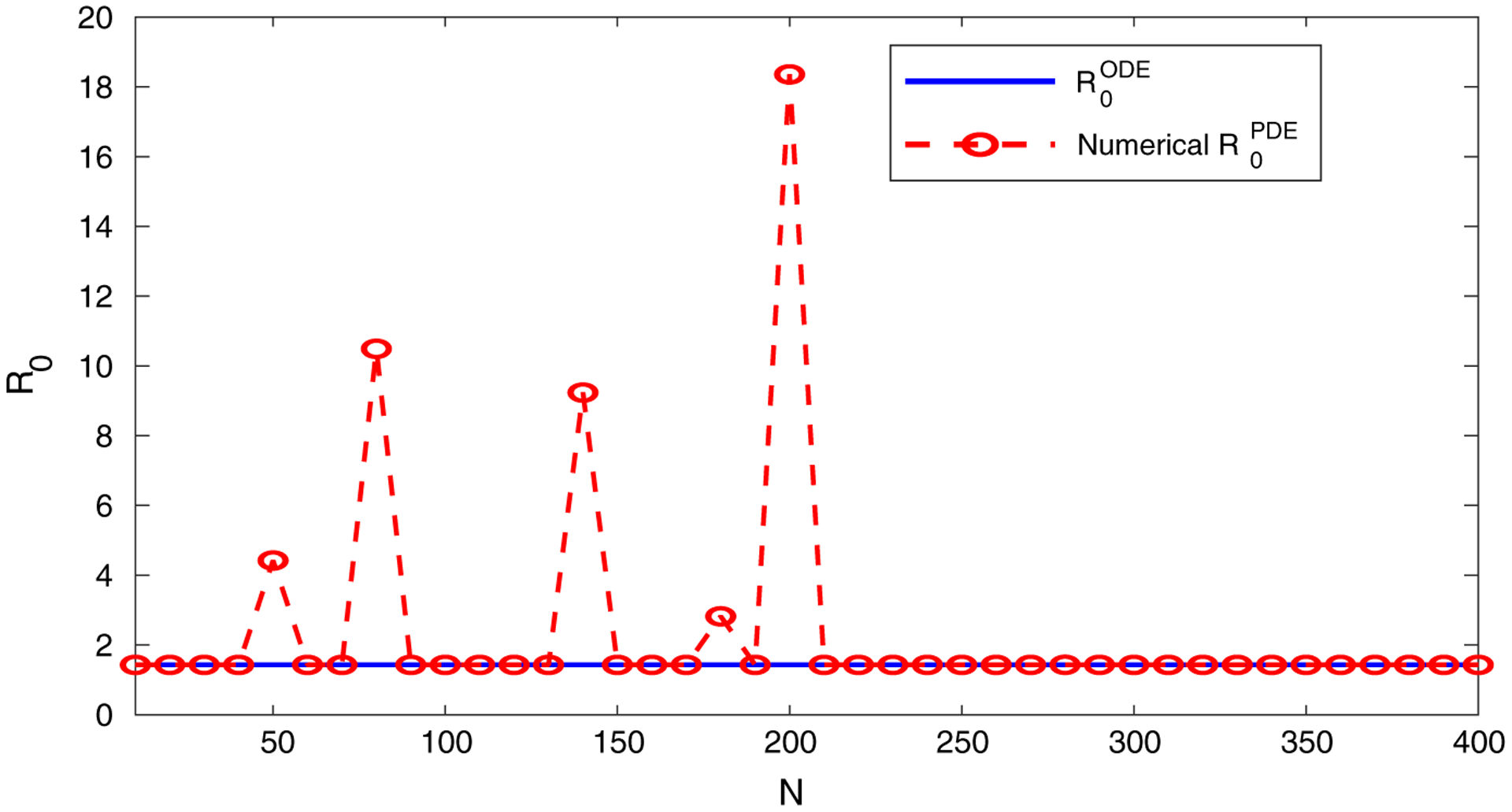
Comparison between and the numerical values of as N increases, for the SIR model (30). Here, is independent of N. The numerical values of are based on the calculation of ρ(F ⊗ IN+1)A−1)
5.2. SIR-B Models
SIR-B models, where ‘B’ refers to the bacterial compartment, have been used to study the transmission dynamics of waterborne bacterial infections, particularly cholera (Tien and Earn 2010; Posny and Wang 2014; Mukandavire et al. 2011). Such a disease is typically transmitted through both the indirect (i.e., environment-to-human) and direct (i.e., human-to-human) routes. In addition, suppose that the waterborne bacteria undergo diffusion in a river that is simply represented by a one-dimensional spatial domain [0, 1]. We thus obtain the following PDE system
| (31) |
for t > 0, x ∈ [0, 1] with Neumann boundary conditions and appropriate initial conditions. In this model, B represents the concentration of the bacterial pathogen in the contaminated water; α and β denote the direct and indirect transmission rates, respectively; ξ is the rate of contribution, such as shedding, from an infected individual to the bacterial population in the aquatic environment; r is the bacterial intrinsic growth rate, K is the carrying capacity of the bacterial growth, and τ is the bacterial removal rate.
The model (31) is a partially diffusive PDE system in the sense that the diffusion process is only incorporated into the bacterial movement. The infection compartments are I and B. The disease-free equilibrium is and thereby
From Corollary 4.2, the basic reproduction number of system (31) is
| (32) |
Again, to verify our computational approach based on Eq. (21) and to provide a numerical evidence on the relationship between and , we plot ρ(F ⊗ IN+1)A−1) versus N in Fig. 2 for the model (31). We set the bacterial diffusion rate as dB(x) = sin(100x) + 1.13. We observe a pattern similar to that in Fig. 1. Specifically, for all N > 80, the numerical values of (i.e., ρ(F ⊗ IN+1)A−1)) coincide with , consistent with our prediction in Eq. (29).
Fig. 2.
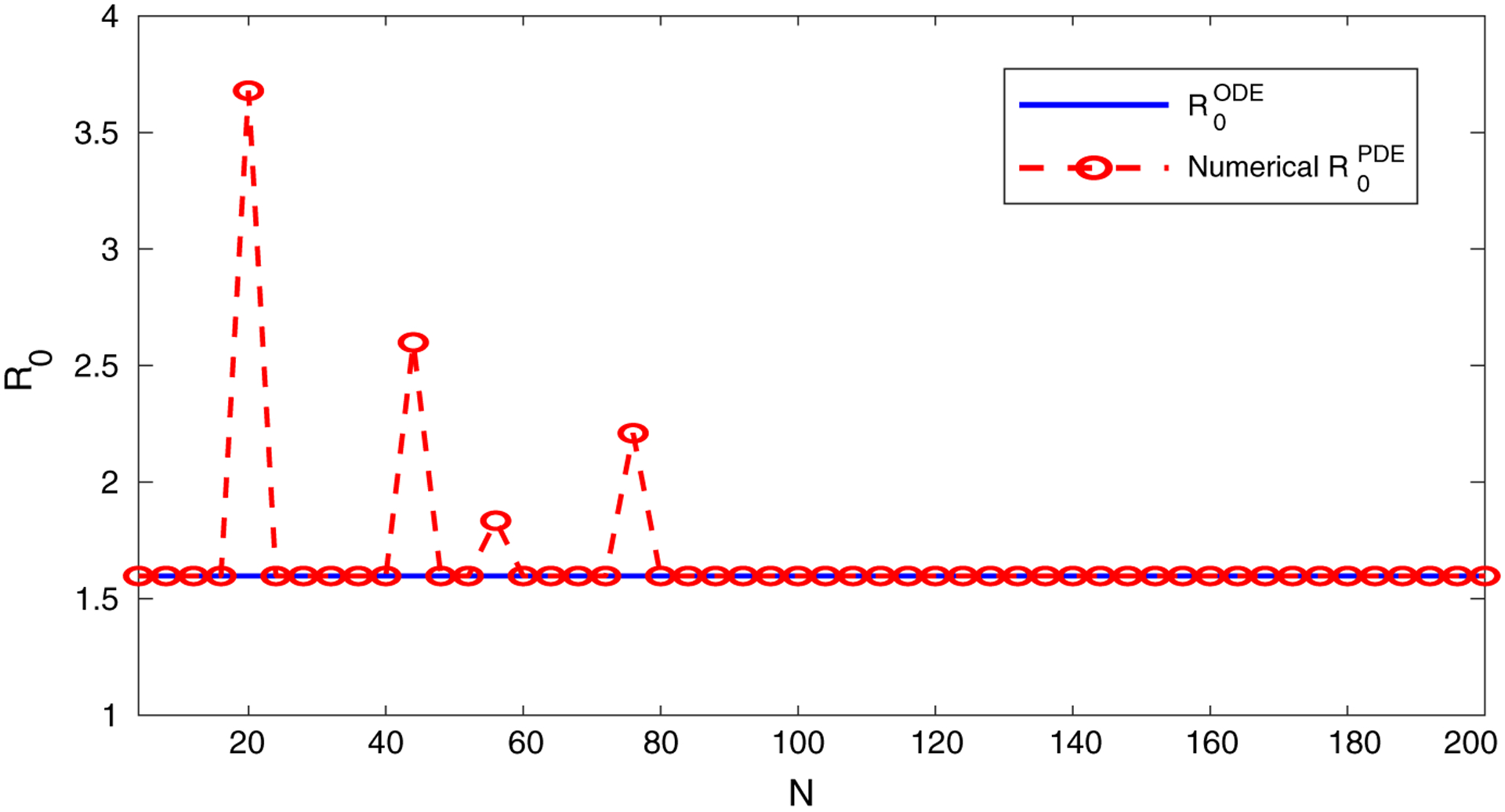
Comparison between and the numerical values of as N increases, for the SIR-B model 31). Here, is independent of N. The numerical values of are based on the calculation of ρ(F ⊗ IN+1)A−1)
On another setting, let us consider the movement of the human hosts as well on the same spatial domain. We assume that all the human hosts and pathogenic bacteria go through a diffusion process and that the diffusion is homogeneous in space so that all the diffusion rates are positive constants. Then, system (31) is modified as
| (33) |
for t > 0 and x ∈ [0, 1]. With the constant diffusion rates, one can apply Remark 4.4 to obtain that the basic reproduction number for the reaction-diffusion system (33) is just the same as given in equation (32).
5.3. Patchy Model
Next, we consider a patchy setting which is a natural extension of the brucellosis patchy model studied in Yang et al. (2017). We assume an environment of n patches, where each patch consists of susceptible and infected individuals and where the disease can spread from patch i to patch j for 1 ≤ i < j ≤ n through unidirectional migration. We additionally consider a diffusion process of the susceptible and infected individuals over the spatial domain [0, 1]. Thus, we may formulate the following reaction-diffusion system with Neumann boundary conditions and appropriate initial conditions:
| (34) |
for i = 1, ⋯ , n. Here, θij and δij denote the migration rates of susceptible and infected individuals from patch i to patch j, respectively; Λi, αi, μi and γi denote the recruitment rate, transmission rate, natural death rate, and disease recovery rate, respectively, for patch i (1 ≤ i ≤ n).
The infection compartments for this model are obviously Ii for 1 ≤ i ≤ n. The disease-free equilibrium is
with and for i = 2, ⋯ , n. Consequently,
and
Since the matrix F is triangular, Theorem 4.1(2) applies and the basic reproduction number of system (34) is given by
5.4. SEIR Model
SEIR models investigate disease transmission in a population consisting of the susceptible, exposed, infected, and recovered individuals. Here, we consider a SEIR model slightly modified from (van den Driessche and Watmough 2002, Example 4.1). We assume that exposed individuals progress to the infected group at rate υ, exposed individuals recover at rate γ1 and enter the recovered class, and infected individuals recover at rate γ2 among whom a portion p (0 ≤ p ≤ 1) enters the exposed class and the other portion 1 − p enters the recovered class. We further assume that the two infected compartments (E and I) have the same movement pattern; namely, their diffusion rates are the same: dE(x) = dI (x) = d(x). We then obtain the following reaction-diffusion system associated with this SEIR model:
| (35) |
for x ∈ [0, 1] and t > 0. There is only one disease-free equilibrium
and
By Remark 4.5, the basic reproduction number for system (35) is given by
6. Conclusion
We have presented a numerical approach that can be applied to the calculation of the basic reproduction numbers for a variety of reaction-diffusion type epidemic models. Essentially, our method transfers the computation of the spectral radius associated with an infinite-dimensional operator to the computation of the principal eigenvalue associated with a finite-dimensional matrix. Such a representation of enables us to analyze and compare the basic reproduction numbers for the PDE system and its corresponding ODE system, based solely on elementary numerical analysis and matrix theory. We have found, in particular, that under a range of conditions that cover several important cases, including the presence of a single infected compartment, constant diffusion rates, uniform diffusion patterns among the infected compartments, and partial diffusion in the system.
In general, the calculation of the basic reproduction number for a PDE system is not a simple procedure, due to the involvement of operator analysis and eigenvalue computation. Consequently, it becomes a nontrivial task to quantify the disease transmission risk represented by such a PDE model. Our results show that for a number of important epidemic scenarios involving reaction-diffusion equations, the task of computation is simplified and replaced by that of the corresponding ODE system; i.e., . These scenarios include some common SIR, SEIR, SIR-B, and patchy models. Our findings help us to gain essential understanding of the disease transmission thresholds for these models, while saving unnecessary computational efforts.
We have assumed that the underlying ODE system is autonomous with constant parameters, though the diffusion rates in our model are location-dependent and emphasize spatial heterogeneity. In case, the ODE system has spatially varying parameters, our numerical method can still be applied with minimal adjustment, but our analysis will need some modification. Particularly, the definition of would depend on the spatial location x, and so a simple relationship between and may not exist in general. This will provide one interesting direction in our future research. We will also explore the computation, comparison, and analysis of the basic reproduction numbers for more general reaction-convection-diffusion type epidemic models, including those that are defined on multi-dimensional spatial domains and those that do not satisfy the conditions prescribed in Theorem 4.3.
Acknowledgements
This work was partially supported by the National Institutes of Health under Grant Number 1R15GM131315. The authors are grateful to the two anonymous referees for their helpful comments that have significantly improved the original manuscript.
References
- Allen LJS, Bolker BM, Lou Y, Nevai AL (2008) Asymptotic profiles of the steady states for an SIS epidemic reaction-diffusion model. Discrete Contin Dyn Syst 21:1–20 [Google Scholar]
- Bertuzzo E, Casagrandi R, Gatto M, Rodriguez-Iturbe I, Rinaldo A (2010) On spatially explicit models of cholera epidemics. J R Soc Interface 7:321–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantrell RS, Cosner C (1991) The effects of spatial heterogeneity in population dynamics. J Math Biol 29:315–338 [Google Scholar]
- Cantrell RS, Cosner C (2003) Spatial ecology via reaction-diffusion equations. Wiley, Hoboken [Google Scholar]
- Chen S, Shi J (2020) Asymptotic profiles of basic reproduction number for epidemic spreading in heterogeneous environment. SIAM J Appl Math 80:1247–1271 [Google Scholar]
- Diekmann O, Heesterbeek JAP, Metz AJ (1990) On the definition and the computation of the basic reproduction ratio R0 in models for infectious diseases in heterogeneous population. J Math Biol 28:365–382 [DOI] [PubMed] [Google Scholar]
- Ge J, Lei C, Lin Z (2017) Reproduction numbers and the expanding fronts for a diffusion-advection SIS model in heterogeneous time-periodic environment. Nonlinear Anal Real World Appl 33:100–120 [Google Scholar]
- Golub GH, Van Loan CF (1996) Matrix computations, 3rd edn. Johns Hopkins University Press, Baltimore [Google Scholar]
- Horn RA, Johnson CR (1985) Matrix analysis. Cambridge University Press, Cambridge [Google Scholar]
- Kim KI, Lin Z, Zhang Q (2013) An SIR epidemic model with free boundary. Nonlinear Anal Real World Appl 14:1992–2001 [Google Scholar]
- Lou Y, Zhao X-Q (2011) A reaction-diffusion malaria model with incubation period in the vector population. J Math Biol 62:543–568 [DOI] [PubMed] [Google Scholar]
- Magal P, Webb GF, Wu Y (2019) On the basic reproduction number of reaction-diffusion epidemic models. SIAM J. Appl. Math 79:284–304 [Google Scholar]
- Mukandavire Z, Liao S, Wang J, Gaff H, Smith DL, Morris JG (2011) Estimating the reproductive numbers for the 2008–2009 cholera outbreaks in Zimbabwe. Proc. Nat. Acad. Sci. USA 108:8767–8772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng R, Zhao X-Q (2012) A reaction-diffusion SIS epidemic model in a time-periodic environment. Nonlinearity 25:1451–1471 [Google Scholar]
- Posny D, Wang J (2014) Modeling cholera in periodic environments. J Biol Dyn 8(1):1–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaldo A, Bertuzzo E, Mari L, Righetto L, Blokesch M, Gatto M, Casagrandi R, Murray M, Vesenbeckh SM, Rodriguez-Iturbe I (2012) Reassessment of the 2010–2011 Haiti cholera outbreak and rainfall-driven multiseason projections. Proc Nat Acad Sci USA 109:6602–6607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richtmyer RD, Morton KW (1994) Difference methods for initial-value problems, 2nd edn. Krieger Publication Company, Malabar [Google Scholar]
- Saad Y (2011) Numerical methods for large eigenvalue problems, Revised edn. SIAM, Philadelphia [Google Scholar]
- Sauty JP (1980) An analysis of hydrodispersive transfer in aquifers. Water Resour Res 16:145–158 [Google Scholar]
- Song P, Lou Y, Xiao Y (2019) A spatial SEIRS reaction-diffusion model in heterogeneous environment. J Differ Equ 267:5084–5114 [Google Scholar]
- Sposito GW, Jury WA, Gupta VK (1986) Fundamental problems in the stochastic convection-dispersion model of solute transport in aquifer and field soils. Water Resour Res 22:77–88 [Google Scholar]
- Taylor GI (1953) Dispersion of solute matter in solvent flowing through a tube. Proc R Soc Ser A 219:186–203 [Google Scholar]
- Thieme HR (2009) Spectral bound and reproduction number for infinite-dimensional population structure and time heterogeneity. SIAM J Appl Math 70:188–211 [Google Scholar]
- Thomas JW (1995) Numerical partial differential equations: finite difference methods. Springer, New York [Google Scholar]
- Tien JH, Earn DJ (2010) Multiple transmission pathways and disease dynamics in a waterborne pathogen model. Bull Math Biol 72:1506–1533 [DOI] [PubMed] [Google Scholar]
- van den Driessche P, Watmough J (2002) Reproduction numbers and sub-threshold endemic equilibria for compartmental models of disease transmission. Math Biosci 180:29–48 [DOI] [PubMed] [Google Scholar]
- Wang F-B, Shi J, Zou X (2015) Dynamics of a host-pathogen system on a bounded spatial domain. Commun Pure Appl Anal 14:2535–2560 [Google Scholar]
- Wang X, Gao D, Wang J (2015) Influence of human behavior on cholera dynamics. Math Biosci 267:41–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Posny D, Wang J (2016) A reaction-convection-diffusion model for cholera spatial dynamics. Discrete Contin Dyn Syst Ser B 21:2785–2809 [Google Scholar]
- Wang W, Zhao X-Q (2012) Basic reproduction numbers for reaction-diffusion epidemic models. SIAM J Appl Dyn Syst 11:1652–1673 [Google Scholar]
- Wu Y, Zou X (2018) Dynamics and profile of a diffusive host-pathogen system with distinct dispersal rates. J Differ Equ 264:4989–5024 [Google Scholar]
- Yamazaki K, Wang X (2016) Global well-posedness and asymptotic behavior of solutions to a reaction-convection-diffusion cholera epidemic model. Discrete Contin Dyn Syst Ser B 21:1297–1316 [Google Scholar]
- Yang C, Lolika P, Mushayabasa S, Wang J (2017) Modeling the spatiotemporal variations in brucellosis transmission. Nonlinear Anal Real World Appl 38:49–67 [Google Scholar]
- Yu X, Zhao X-Q (2016) A nonlocal spatial model for Lyme disease. J Differ Equ 261:340–372 [Google Scholar]
- Zhao L, Wang Z-C, Ruan S (2018) Traveling wave solutions in a two-group SIR epidemic model with constant recruitment. J Math Biol 77:1871–1915 [DOI] [PubMed] [Google Scholar]


