Abstract
Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) has been rapidly evolving in the form of new variants. At least eleven known variants have been reported. The objective of this study was to delineate the differences in the mutational profile of Delta and Delta Plus variants. High-quality sequences (n = 1756) of Delta (B.1.617.2) and Delta Plus (AY.1 or B.1.617.2.1) variants were used to determine the prevalence of mutations (≥20 %) in the entire SARS-CoV-2 genome, their co-existence, and change in prevalence over a period of time. Structural analysis was conducted to get insights into the impact of mutations on antibody binding. A Sankey diagram was generated using phylogenetic analysis coupled with sequence-acquisition dates to infer the migration of the Delta Plus variant and its presence in the United States. The Delta Plus variant had a significant number of high-prevalence mutations (≥20 %) than in the Delta variant. Signature mutations in Spike (G142D, A222V, and T95I) existed at a more significant percentage in the Delta Plus variant than the Delta variant. Three mutations in Spike (K417N, V70F, and W258L) were exclusively present in the Delta Plus variant. A new mutation was identified in ORF1a (A1146T), which was only present in the Delta Plus variant with ~58 % prevalence. Furthermore, five key mutations (T95I, A222V, G142D, R158G, and K417N) were significantly more prevalent in the Delta Plus than in the Delta variant. Structural analyses revealed that mutations alter the sidechain conformation to weaken the interactions with antibodies. Delta Plus, which first emerged in India, reached the United States through England and Japan, followed by its spread to more than 20 the United States. Based on the results presented here, it is clear that the Delta and Delta Plus variants have unique mutation profiles, and the Delta Plus variant is not just a simple addition of K417N to the Delta variant. Highly correlated mutations may have emerged to keep the structural integrity of the virus.
Keywords: SARS-CoV-2, Delta variant, Delta plus variant, Spike, B.1.617.2, AY.1, B.1.617.2.1
Severe Acute Respiratory Coronavirus 2 (SARS-CoV-2), the etiological agent of Coronavirus Disease 19 (COVID-19), has caused unimaginable socio-economic damage worldwide. As similar to many RNA viruses, SARS-CoV-2 has been evolving into new variants as transmission progress. Depending upon transmissibility, disease severity (such as increased hospitalizations or deaths), the extent of reduction in neutralization by antibodies generated during previous infection or vaccination, reduced effectiveness of treatments, or diagnostic detection failures, these variants have been classified as Variant of Concern (VOC) or Variants of Interest (VOI) [1]. Eleven SARS-CoV-2 variants (Alpha, Beta, Gamma, Delta, Delta Plus, Epsilon, Eta, Theta, Iota, Kappa, and Lambda) have been documented, and this list is likely to grow as new variants emerge.
A specific SARS-CoV-2 variant is characterized by a set of the most common mutations in the virus genome, and the majority of the reported mutations in a given variant belong to the Spike protein. It is well known that RNA viruses exploit various mechanisms of genetic variation to ensure their survival [2]. Some mutations in RNA viruses may cause enhanced fitness. For SARS-CoV-2, it has been shown that D164G mutation enhances viral fitness [3,4]. The fitness data for other Spike mutations are not available. However, it is plausible that some mutations may decrease viral fitness, and compensatory mutations may be selected to gain fitness function. To achieve such insights, we investigated the prevalence of mutations in entire SARS-CoV-2 genes of the currently dominant Delta variant (B.1.617.2) and the Delta Plus variant (AY.1 and B.1.617.2.1). We found that in addition to signature Spike mutations associated with Delta and Delta Plus variants, an additional ~25 mutations exist with a high prevalence throughout the SARS-CoV-2 genome. Several Spike mutations are highly correlated with other mutations in different genes, suggesting a co-evolution of these mutations. Additionally, our data indicate that Delta and Delta Plus variants have two additional mutations (T95I and W258L) with significant prevalence (~40 % in Delta Plus). Hence, we propose including these mutations as signature mutations of Delta (T95I) and Delta Plus (T95I + W258L) in understanding the pathogenic mechanisms associated with these viruses.
According to the United States (US) Center for Disease Control (CDC), signature Spike mutations in the aggregated Delta and Delta Plus variant include T19R, (V70F*), T95I, G142D, E156-, F157-, R158G, (A222V*), (W258L*), (K417N*), L452R, T478K, D614G, P681R, and D950N [1]. The criterion used to classify the Delta Plus variant was based on the K417N mutation in the parent Delta Variant. Using the mutations belonging to the Delta variant in the search criteria, we downloaded high quality and high coverage sequences of Delta (n = 1276) from GISAID [5]. We also downloaded all available high-quality and high coverage Delta Plus sequences (as of July 13, 2021) (n = 520) from GISAID [5]. These sequences were analyzed for the prevalent mutations in the entire SARS-CoV-2 genome, co-existing mutations, the temporal prevalence of signature mutations, and the introduction of the Delta Plus variant into the US.
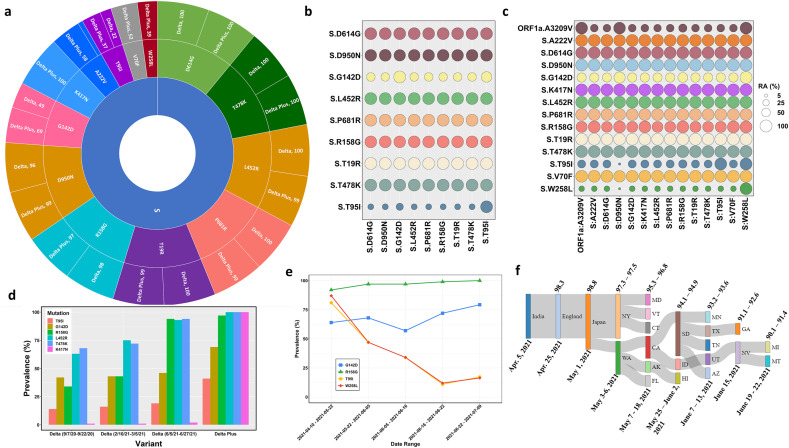
The sequence analysis revealed a total of 656 and 269 unique mutations in the Delta and Delta Plus variants, respectively. However, the high prevalence mutations (more than 20 %) were greater in Delta Plus (40) than in Delta (29). The most prevalent mutations in the Spike protein (cut-off 35 %) in two variants are shown in Fig. 1 a (sunburst plot), and those in the remaining genes are collated in Table 1 . This analysis identified two Spike protein mutations that were significantly prevalent only in the Delta Plus variant and not in the Delta variant. These include V70F and W258L, which were present in Delta Plus at the prevalence of 52 % and 39 %, respectively. Additionally, we noted the difference in two Spike mutations in Delta Plus and Delta variants. These include mutation A222V was 58 % in Delta Plus, whereas only 9 % in Delta. Similarly, T95I was 37 % in Delta Plus and 22 % in Delta. We also identified variant-specific mutations in other genes in both variants. For example, A328T in nsp3 (ORF1a: A1146T) was only present in Delta Plus (58 %). Four additional mutations: nsp3:P822L (ORF1a:P1604L), nsp4:A446V (ORF1a:A3209V), nsp6:V149S (ORF1a: V3718S), and nsp6:T181I (ORF1a:T3750I) are present at 58 % in Delta Plus, and only at 16 % in Delta except nsp6:T181I, which was only 9 % (Table 1). Hence, as noted above, the Delta Plus variant is not just a variant of Delta signified by the K417N mutation but has additional mutations that need to be considered.
Fig. 1.
Details of genetic variations in Delta and Delta Plus variants. Panel a. A sunburst plot shows the distribution of mutations in Delta variant sequences (n = 676) and Delta Plus variant sequences (n = 520) with greater than 35 % prevalence. All available high coverage, complete sequences of the Delta variant collected during July 6–13, 2021, were downloaded from GISAID [5] and processed through NextClade [15]. The prevalence was computed using an in-house Python script and Pandas library. Panel b. Relative abundance of the Spike mutations with greater than 20 % prevalence in Delta variant. The prevalence was computed using Delta variant sequences (n = 676) using an in-house Python script. Panel c. Relative abundance of the Spike mutations with greater than 20 % prevalence in Delta Plus variant. The prevalence was computed using Delta Plus variant sequences (n = 288) using an in-house Python script. Panel d. Prevalence of five key mutations (T95I, G142D, R158G, L452R, T478K, and K417N) at different time points in Delta variant (n = 600) sequences and Delta Plus variant (n = 200) sequences. The prevalence was calculated and plotted with an R script and ggplot2 library. Panel e. Temporal analysis of Delta plus mutations of interest. Sequences of the Delta Plus variant were sorted by date (n = 520) and grouped in groups of 100 each except the last group that contained 118 sequences. Two sequences were excluded due to poor quality. The date ranges were marked by the first and last sequence collection date. The prevalence was calculated as described above. The data were plotted using the ggplot2 library of R. Panel f. A Sankey diagram showing the dynamics of Delta Plus introduction into the United States. To generate the Sankey diagram, we aligned the first collected and dated Delta Plus sequence from India, England, Japan, and different states of the USA. We then grouped the sequences based upon the date collected and percent homology cut-offs as indicated at the top of the plot and date range shown below the plot.
Table 1.
Prevalence of mutation in Delta and Delta Plus variants in the genes other than Spike.
| Region | Mutation | Variant | Frequency | Region | Mutation | Variant | Frequency |
|---|---|---|---|---|---|---|---|
| M | I82T | Delta | 100 | ORF1b | P323L | Delta | 100 |
| M | I82T | Delta Plus | 100 | ORF1b | P323L | Delta Plus | 100 |
| N | R203M | Delta | 100 | ORF1b | P1009L | Delta | 100 |
| N | R203M | Delta Plus | 100 | ORF1b | P1009L | Delta Plus | 100 |
| N | D63G | Delta | 100 | ORF1b | A1927V | Delta | 84 |
| N | D63G | Delta Plus | 99 | ORF1b | A1927V | Delta Plus | 42 |
| N | D377Y | Delta | 97 | ORF1b | G671S | Delta | 100 |
| N | D377Y | Delta Plus | 99 | ORF1b | G671S | Delta Plus | 100 |
| N | G215C | Delta | 84 | ORF1b | T1299I | Delta | 0 |
| N | G215C | Delta Plus | 42 | ORF1b | T1299I | Delta Plus | 58 |
| ORF1a | A3209V | Delta | 16 | ORF3a | S26L | Delta | 100 |
| ORF1a | A3209V | Delta Plus | 58 | ORF3a | S26L | Delta Plus | 100 |
| ORF1a | T3646A | Delta | 84 | ORF7a | T120I | Delta | 98 |
| ORF1a | T3646A | Delta Plus | 42 | ORF7a | T120I | Delta Plus | 100 |
| ORF1a | T3750I | Delta | 9 | ORF7a | V82A | Delta | 98 |
| ORF1a | T3750I | Delta Plus | 58 | ORF7a | V82A | Delta Plus | 100 |
| ORF1a | A1146T | Delta | 0 | ORF7b | T40I | Delta | 84 |
| ORF1a | A1146T | Delta Plus | 58 | ORF7b | T40I | Delta Plus | 42 |
| ORF1a | V2930L | Delta | 84 | ORF9b | T60A | Delta | 100 |
| ORF1a | V2930L | Delta Plus | 42 | ORF9b | T60A | Delta Plus | 99 |
| ORF1a | T3255I | Delta | 84 | ORF1a | V3718A | Delta | 16 |
| ORF1a | T3255I | Delta Plus | 42 | ORF1a | V3718A | Delta Plus | 58 |
| ORF1a | P2287S | Delta | 84 | ORF1a | P2046L | Delta | 84 |
| ORF1a | P2287S | Delta Plus | 42 | ORF1a | P2046L | Delta Plus | 42 |
| ORF1a | A1306S | Delta | 84 | ORF1a | P1640L | Delta | 16 |
| ORF1a | A1306S | Delta Plus | 42 | ORF1a | P1640L | Delta Plus | 58 |
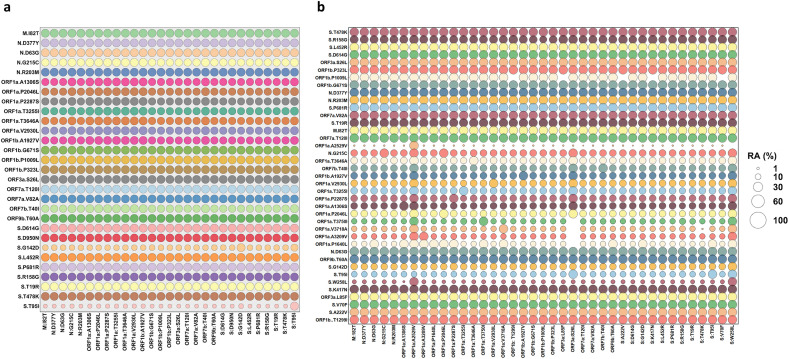
We also conducted relative abundance (RA) analysis to determine the correlation of the co-existing mutations using an in-house Python script. The RA among all mutations with more than 20 % prevalence is shown in Fig. 2 . The RA among Spike and two ORF1a (in Delta Plus) mutations are shown in Fig. 1b and c, for Delta and Delta Plus, respectively. In the Delta variant (Fig. 1b), all mutations co-exist at ~100 % frequency, except T95I and G142D. T95I occurs at a frequency of 20–30 % in the background of other mutations, whereas G142D co-exists at a frequency of ~50 % in the background of other mutations.
Fig. 2.
Relative abundance of all mutations with greater than 20 % prevalence in Delta (Panel a) and Delta Plus (Panel b) variants.
In the Delta Plus variant, the sequences containing W258L, which exists in ~40 % of all sequences, also had a strong correlation with all listed Spike mutations (Fig. 1c) and nsp4 A446V mutation (ORF1a: A3209V), suggesting that all sequences that contained W258L also had all mutations shown in Fig. 1c. Importantly, sequences that contained Spike mutation W258L almost always included G142D, T95I, nsp4 A446V (ORF1a: A3209V). Additionally, we found that nsp4 A446V (ORF1a: A3209V) is almost always (~90 %) present in sequences that had the spike mutation D950N (Delta signature mutation) [1]. It was previously reported that D614G [6] and P323L were present in all SARS-CoV-2 sequences by the summer of 2020 [7,8]. These mutations are also present in all Delta and Delta Plus variants.
To assess how Delta Plus was evolving from Delta, we determined the prevalence of six key mutations (T95I, G142D, R158G, L452R, T478K, and K417N) at different time points. The rationale behind selecting these mutations was that they were unique (e.g., K417N) or highly correlated with another mutation in other variants (e.g., T95I being variably associated with other Spike protein mutations). The results (Fig. 1d) showed that all these mutations increased over time in Delta, and all mutations had a significantly higher prevalence in the Delta Plus variant. These results further justify our conclusion, as mentioned earlier, that the Delta Plus variant is more than just an additional mutation (K417N).
To further investigate the correlation between W258L and T95I in Delta Plus. We conducted a temporal analysis by splitting Delta Plus variant sequences (n = 518) into five groups of 100 each (sorted by date) and calculated the prevalence of these mutations (Fig. 1e). We also included G142D and R158G since these mutations occurred at high prevalence (69–100 %). The temporal analysis showed that while W258L and T95I are highly correlated, the actual prevalence of both W258L and T95I mutations has decreased over time in our analysis.
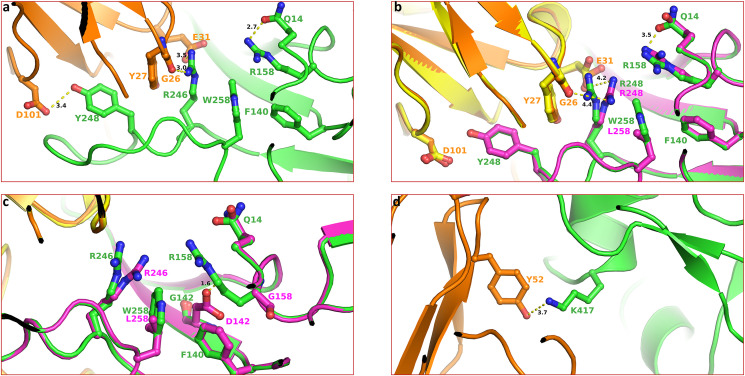
It was recently demonstrated that monoclonal antibodies, convalescent, and vaccine sera reduce the neutralization of the Delta variant containing T478K or L452R/T478K mutations compared with Wuhan-related virus [9]. The structural data confirmed that the longer sidechains R452 abrogated antibody binding by contacting a 6-residue-long heavy chain (HC) complementarity determining region 3, and K478 perturbed the binding of Fab 253 antibody due to longer sidechains compared to leucine and threonine [9]. These structures provided the atomic basis for enhanced transmission of the Delta variant. Similar structural data for T95I, G142D, and W258L is not available. Therefore, to get insight into the impact of mutations (e.g., D142G, R158G, W258L, and K417N), we analyzed available structures in the Protein Data Bank (PDB, www.rcsb.org) [10] and assessed the impact of mutations. An analysis of the cryo-electron microscopy (cryo-EM) structure of NTD-directed neutralizing antibody 1–87 in complex with prefusion SARS-CoV-2 spike glycoprotein (PDB entry 7L2D) [11] showed that W258 is part of a hydrophobic interaction network constituted by F140, W258, R246 (through carbon sidechain) and Y248 and antibody heavy chain residue Y27 (Fig. 3 a). R246 also forms polar interactions E31 of the antibody and the backbone C O of G26 (shown as dotted lines). R158 is also in the close vicinity and forms a hydrogen bond with Q14.
Fig. 3.
Impact of mutations on the geometry of antibody binding Spike structure. Panel a. This panel shows the geometry of SARS-CoV-2 neutralizing antibodies binding to the N-terminal of Spike protein (PDB entry 7L2D). The Spike structure representing Wuhan-Hu-1 in this and subsequence panels of these figures are shown as green ribbons. The antibody is shown rendered in orange ribbons. The Spike residues are rendered as ball-and-stick (Spike – green and antibody – orange). The yellow dotted lines represent polar interactions with distance (in Å) between two interacting atoms. Panel B. effect of mutation W258L on the geometry of antibody binding surface. Other atoms are colored by the atom type (oxygen – red and nitrogen – blue). The structure of the mutant is shown in magenta. The antibody in structure bound to mutant Spike is rendered in yellow color. The green labels represent Wuhan-Hu-1 Spike, whereas magenta labels belong to the Delta variant. Note the increased interaction between R246 and antibody residues compared to those in panel a. Panel c. Impact of G142D and R156G mutations, the steric clash between D142 and R158 is shown in the dotted line of 1.6 Å length. The mutant protein is colored magenta. Panel d. The interaction of K417 with Y42 is seen in PDB entry 6XCN. The interaction between K417 and antibody residue Y52 is shown as a dotted line. The distance between two atoms is in Å. Note that the mutation K417N (as in Delta Plus) would result in the loss of this interaction.
The sidechain conformation of residues in this vicinity is such that any mutation would most certainly alter the geometry of the interaction network and thereby affect the binding of Spike with the antibody. To assess if mutations change sidechain conformation, we generated mutations W258A, G142D and R158G using Prime software of Schrödinger Suite (Schrödinger LLC, NY). The effect of the W258A mutation is shown in Fig. 3b. It is clear from this figure that W258L mutation reorients the R246 sidechain such that the interaction with E31 and G26 of antibody heavy chain would be weakened due to longer interaction distance (3.5 and 3.0 Å verses 4.2 and 4.4 Å). The effect of G142D and R158G mutation is shown in Fig. 3c. Mutation G142D causes a steric clash with the sidechain of R158 (shown as a dotted line of 1.6 Å length). To avoid this clash, the conformation of R158 has to be changed drastically, which is less likely due to the ‘snugly-fit’ geometry of sidechains in this region of Spike structure. Additionally, the conformation R258 is nearly identical, as seen in the W258L mutation (Fig. 3b). It appears that the virus evolved to overcome the clash by mutating R158G, which is in accordance with the correlation data shown in Fig. 1b that all viruses that have G142D also have R158G mutation. The antibody evasion by mutation K417 appears straightforward. The (cryo-EM) structure of a neutralizing monoclonal Fab-Spike complex shows that K417 interacts with Y52 (Fig. 3d) [12] (PDB entry 6XCN). Mutation K417N will result in a loss of this interaction and thereby reduced binding of the antibody with the Spike.
Following its emergence in India, the Delta Plus variant had spread through several countries, including the US. Washington was the first state to report Delta Plus (May 3, 2021), followed by New York (May 6, 2021). To gain insight into the migration of this variant within the US, we aligned the first collected sequences of the Delta Plus variant from different regions of the US. Using this data, we generated a Sankey diagram (Fig. 1f). As of June 22, 2021, the Delta Plus variant has been transmitted to individuals in 20 US states. Our analysis also demonstrates that this variant traveled to the US via England and Japan. A decreasing homology from the previous Delta Plus variant also suggests that this variant spreads in different regions of the US, evolving more mutations, giving rise to a diverse set of Delta Plus.
In summary, herein, we present a detailed picture of mutations in Delta, presumably a highly transmissible variant [13,14], and Delta Plus variants. Our analyses show that the Delta Plus variant has a distinct mutation profile compared to the Delta variant. For example, we found that a Spike mutation E465A was present in 15 sequences of the Delta variant. 14 out of 15 Delta variant sequences that contained E465A were from the state of Missouri. It is also possible that the origin of the Delta variant may be more than just in India as the first sequence of Delta variant was from the Netherlands, which was collected in June 2020 (GISAID accession: EPI_ISL_2,860,470). The antibody evasion by the virus through specific mutation mutations may contribute to the greater transmutability of the virus. Using structural data, we presented atomic details showing possible ways the virus can use and escape antibodies. While our analysis is detailed, new mutations in these variants may emerge in the future.
Author Statement
KS, ANS, SNB, and SRK conceptualized the study; KS wrote the first draft and final manuscript. ANS, SRK, ARC, and KS conducted genetic analyses, wrote required programs either in R or in Python; SNB, TPQ, HSC, SHN and CLL edited the manuscript and contributed to understanding the pathogenicity of CoVs. All authors approved the final manuscript.
Acknowledgments
KS acknowledges support from the Office of Research, University of Missouri(Bond Life Sciences Center, Early Concept Grant). SNB acknowledges the independent research and development (IRAD) funding from the National Strategic Research Institute (NSRI) at the University of Nebraska. KS and TPQ acknowledge the computation facilities of the Molecular Interactions Core at the University of Missouri, Columbia, MO 65212. We thank the laboratories that have generously deposited sequences into the GISAID database. We thank Kamran S. Farid for the analysis of the antibody interaction with Spike protein.
References
- 1.SARS-CoV-2 variant classifications and definitions. https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-info.html (July 16, 2021).
- 2.Domingo E., Holland J.J. RNA virus mutations and fitness for survival. Annu. Rev. Microbiol. 1997;51:151–178. doi: 10.1146/annurev.micro.51.1.151. [DOI] [PubMed] [Google Scholar]
- 3.J. A. Plante, Y. Liu, J. Liu, H. Xia, B. A. Johnson, K. G. Lokugamage, X. Zhang, A. E. Muruato, A J. Zou, C. R. Fontes-Garfias, D. Mirchandani, D. Scharton, J. P. Bilello, Z. Ku, Z. An, B. Kalveram, A. N. Freiberg, V. D. Menachery, X. Xie, K. S. Plante, S. C. Weaver P. Y. Shi, Spike mutation D614G alters SARS-CoV-2 fitness. Nature 592 (2021):116-121. [DOI] [PMC free article] [PubMed]
- 4.Baric R.S. Emergence of a highly fit SARS-CoV-2 variant. N. Engl. J. Med. 2020;383:2684–2686. doi: 10.1056/NEJMcibr2032888. [DOI] [PubMed] [Google Scholar]
- 5.Elbe S., Buckland-Merrett G. Data, disease and diplomacy: GISAID's innovative contribution to global health. Glob. Chall. 2017;1:33–46. doi: 10.1002/gch2.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.F., Bhattacharya T., Foley B., Hastie K.M., Parker M.D., Partridge D.G., Evans C.M., Freeman T.M., de Silva T.I., Sheffield C.-G.G., McDanal C., Perez L.G., Tang H., Moon-Walker A., Whelan S.P., LaBranche C.C., Saphire E.O., Montefiori P.C. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kannan S.R., Spratt A.N., Quinn T.P., Heng X., Lorson C.L., Sonnerborg A., Byrareddy S.N., Singh K. Infectivity of SARS-CoV-2: there is something more than D614G? J. Neuroimmune Pharmacol. 2020;15:574–577. doi: 10.1007/s11481-020-09954-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spratt A.N., Kannan S.R., Woods L.T., Weisman G.A., Quinn T.P., Lorson C.L., Sonnerborg A., Byrareddy S.N., Singh K. Evolution, correlation, structural impact and dynamics of emerging SARS-CoV-2 variants. Comput. Struct. Biotechnol. J. 2021;19:3799–3809. doi: 10.1016/j.csbj.2021.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu C., Ginn H.M., Dejnirattisai W., Supasa P., Wang B., Tuekprakhon A., Nutalai R., Zhou D., Mentzer A.J., Zhao Y., Duyvesteyn H.M.E., López-Camacho C., Slon-Campos J., Walter T.S., Skelly D., Johnson S.A., Ritter T.G., Mason C., Clemens S.A.C., Naveca F.G., Nascimento V., Nascimento F., da Costa C.F., Resende P.C., Pauvolid-Correa A., Siqueira M.M., Dold C., Temperton N., Dong T., J Pollard A., Knight J.C., Crook D., Lambe T., Clutterbuck E., Bibi S., Flaxman A., Bittaye M., Belij-Rammerstorfer S., Gilbert S.C., Malik T., Carroll M.W., Klenerman P., Barnes E., Dunachie S.J., Baillie V., Serafin N., Ditse Z., da Silva K., Paterson N.G., Williams M.A., Hall D.R., Madhi S., Nunes M.C., Goulder P., E Fry E., Mongkolsapaya J., Ren J., Stuart D.I., Screaton G.R. Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum. Cell. 2021;184:1–17. doi: 10.1016/j.cell.2021.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cerutti G., Guo Y., Zhou T., Gorman J., Lee M., Rapp M., Reddem E.R., Yu J., Bahna F., Bimela J., Huang Y., Katsamba P.S., Liu L., Nair M.S., Rawi R., Olia A.S., Wang P., Zhang B., Chuang G.Y., Ho D.D., Sheng Z., Kwong P.D., Shapiro L. Potent SARS-CoV-2 neutralizing antibodies directed against spike N-terminal domain target a single supersite. Cell Host Microbe. 2021;29:819–833. doi: 10.1016/j.chom.2021.03.005. e817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barnes C.O., West A.P., Jr., Huey-Tubman K.E., Hoffmann M.A.G., Sharaf N.G., Hoffman P.R., Koranda N., Gristick H.B., Gaebler C., Muecksch F., Lorenzi J.C.C., Finkin S., Hagglof T., Hurley A., Millard K.G., Weisblum Y., Schmidt F., Hatziioannou T., Bieniasz P.D., Caskey M., Robbiani D.F., Nussenzweig M.C., Bjorkman P.J. Structures of human antibodies bound to SARS-CoV-2 spike reveal common epitopes and recurrent features of antibodies. Cell. 2020;182:828–842 e816. doi: 10.1016/j.cell.2020.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Campbell F., Archer B., Laurenson-Schafer H., Jinnai Y., Konings F., Batra N., Pavlin B., Vandemaele K., Kerkhove M.D.V., Jombart T., Morgan O., de Waroux O.P. Increased transmissibility and global spread of SARS-CoV-2 variants of concern as at June 2021. Euro Surveill. 2021;26:2100509. doi: 10.2807/1560-7917.ES.2021.26.24.2100509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scudellari M. How the coronavirus infects cells - and why delta is so dangerous. Nature. 2021;595:640–644. doi: 10.1038/d41586-021-02039-y. [DOI] [PubMed] [Google Scholar]
- 15.Hadfield J., Megill C., Bell S.M., Huddleston J., Potter B., Callender C., Sagulenko P., Bedford T., Nextstrain R. A. Neher. Real-time tracking of pathogen evolution. Bioinformatics. 2018;34:4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]