Abstract
Middle rhodopsin (MR) found from the archaeon Haloquadratum walsbyi is evolutionarily located between two different types of rhodopsins, bacteriorhodopsin (BR) and sensory rhodopsin II (SRII). Some isomers of the chromophore retinal and the photochemical reaction of MR are markedly different from those of BR and SRII. In this study, to obtain the structural information regarding its active center (i.e., retinal), we subjected MR embedded in lipid bilayers to solid-state magic-angle spinning nuclear magnetic resonance (NMR) spectroscopy. The analysis of the isotropic 13C chemical shifts of the retinal chromophore revealed the presence of three types of retinal configurations of dark-adapted MR: (13-trans, 15-anti (all-trans)), (13-cis, 15-syn), and 11-cis isomers. The higher field resonance of the 20-C methyl carbon in the all-trans retinal suggested that Trp182 in MR has an orientation that is different from that in other microbial rhodopsins, owing to the changes in steric hindrance associated with the 20-C methyl group in retinal. 13Cζ signals of Tyr185 in MR for all-trans and 13-cis, 15-syn isomers were discretely observed, representing the difference in the hydrogen bond strength of Tyr185. Further, 15N NMR analysis of the protonated Schiff base corresponding to the all-trans and 13-cis, 15-syn isomers in MR showed a strong electrostatic interaction with the counter ion. Therefore, the resulting structural information exhibited the property of stable retinal conformations of dark-adapted MR.
Keywords: microbial rhodopsin, retinal isomers, nuclear magnetic resonance, protonated Schiff base, chemical shifts
Significance
Here, we analyzed the structure of the retinal chromophore of middle rhodopsin (MR) embedded in lipid bilayers using solid-state NMR spectroscopy. We recorded the 13C NMR signals of three types of retinal configurations of dark-adapted MR exist: (13-trans, 15-anti), (13-cis, 15-syn), and 11-cis isomers. Furthermore, we discretely observed 13Cζ NMR signals of Tyr185 in MR with the all-trans and 13-cis isomers. The 15N NMR signal of the protonated Schiff base in MR exhibited a strong electrostatic interaction with the counter ion. Our study makes a significant contribution to the literature because we highlighted the key structural chromophores of MR.
Introduction
Microbial rhodopsins have been identified in microorganisms, including haloarchaea and halobacteria, and have seven-transmembrane helices with all-trans retinal chromophores via a protonated Schiff base (PSB) [1–3]. In the family of microbial rhodopsins, novel types of rhodopsins are being identified, despite the similar basic structural motif (7 transmembrane helices+retinal chromophore), which are used as tools in optogenetics [1–3]. Several types of microbial rhodopsins function as light-driven ion pumps, such as proton pump bacteriorhodopsin (BR) and proteorhodopsin (PR), chloride pump halorhodopsin (HR), and sodium ion pump rhodopsin (NaR) [4–7]. The light-driven divalent sulfate ion transporter rhodopsin (SyHR) has recently been identified [8]. The other type involves photoreceptors, such as sensory rhodopsin I (SRI) and II (SRII) for positive and negative phototaxis responses, respectively [9–11]. Anabaena sensory rhodopsin (ASR), with its soluble transducer from cyanobacteria, regulates the expression of photosynthesis-related genes [12]. In addition, functional and color conversions have been achieved by crucial mutations in genes encoding amino acid residues near the retinal chromophore [13–15]. Therefore, the structure of the retinal-binding site plays an important role in understanding its function.
Middle rhodopsin (MR), isolated from the square-shaped haloarchaeon Haloquadratum walsbyi, is evolutionarily located between BR and SRII [16,17]. The structure of the retinal binding pocket in BR is shown with the corresponding residues of MR in Figure 1. MR exhibits a larger blue-shifted absorption at 485 nm than BR. Dark-adapted MR in detergent micelles and phosphatidylglycerol (PG) liposomes has three retinal isomers, as revealed by extraction of retinal and HPLC analysis: all-trans (36.5%), 13-cis (56.4%), and 11-cis (7.6%) in micelles and all-trans (58.2%), 13-cis (38.9%), and 11-cis (2.9%) in liposomes [16,18]. The individual absorption maxima and independent photochemical reactions corresponding to these three isomers have also been demonstrated via UV-Vis and flash photolysis experiments [19]. Notably, MR is the only microbial rhodopsin with the 11-cis retinal as a chromophore like animal rhodopsins. Both K and M intermediates of MR appear only in the photocycle of all-trans isomers. The production of these intermediates is one of the key functional events among microbial rhodopsins. Therefore, it is expected that the photocycle of all-trans isomers through K and M-intermediates is functionally important. However, the type of function in MR is still unknown. MR does not show H+ pump activity even though it establishes a BR-like fast photocycle [16]. Additionally, regarding NpSRII in Natronomonas pharaonis, Thr189 and Tyr199 are used to form a complex with its cognate transducer protein, while Tyr174 and Thr204 play an essential role in the expression of negative phototaxis [13,20,21]. The complex of the A201T/M211Y MR mutant (with residues corresponding to those of NpSRII; i.e., Tyr185, Thr201, Tyr211, and Thr216) with the transducer protein HtrII shows phototaxis response to >460 nm light; however wild-type MR does not [16]. Therefore, it is important to consider that among microbial rhodopsins, MR has unique properties and clarify why the dark-adapted MR shows extremely short wavelength light absorption and has multiple retinal isomers.
Figure 1 .
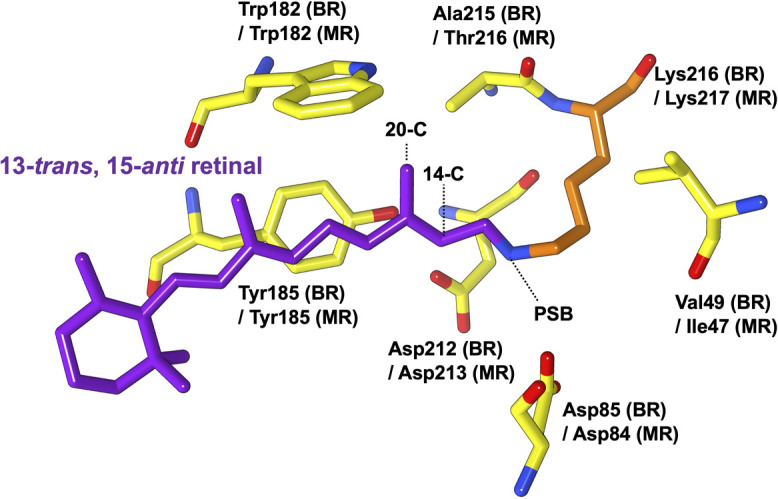
Structure of all-trans retinal and surrounding residues in the retinal-binding site of BR [PDB: 1C3W] together with the corresponding residues of MR (Ile47, Asp84, Trp182, Tyr185, Asp213, and Lys217). The 14-C, 20-C, and PSB positions are shown with dotted lines.
Solid-state nuclear magnetic resonance (NMR) spectroscopy, which offers the possibility to observe not only the structure and dynamics of proteins in cell membranes, but also photo intermediates by isomerization of retinal, has already been applied in structural studies of several rhodopsins [22–26].
Therefore, in this study, we investigated the structural characteristics of the retinal chromophore in MR via 13C and 15N solid-state NMR spectroscopy to the end of elucidating the structure of dark-adapted MR, which has multiple retinal isomers.
Materials and Methods
Expression and reconstitution of MR into lipid bilayers
MR was overexpressed in Escherichia coli C41 (DE3) cells. The bacteria were grown in M9 medium containing 50 mg of [13Cζ]Tyr and [15Nζ]Lys (Cambridge Isotope Laboratories, Andover, MA, USA) per liter at 37°C until an OD660 of 0.8 was reached. After adding 1 mM isopropyl thiogalactoside (IPTG) and 5 μM [14, 20-13C]-labeled all-trans retinal, protein expression of the protein was performed at 25°C for 15 h followed by cell lysis via ultrasonication, after which the protein was solubilized using n-dodecyl-β-D-maltoside (DDM). The recombinant MR with a C-terminal His-tag was purified using nickel-nitrilotriacetic acid agarose. Thereafter, the MR was reconstituted into an L-α-phosphatidyl-DL-glycerol (Egg-PG) membrane at a molar ratio of 1:30 at pH 7.0.
Solid-state NMR experiments
The reconstituted sample (approximately 8.0 mg of protein) was concentrated via centrifugation and packed into the central part of a 4.0-mm diameter zirconia rotor under dim light. The rotor was maintained in the dark for two days. 13C and 15N solid-state NMR experiments were performed on a 600 MHz Bruker Avance III spectrometer equipped with a 1H-X double-resonance probe. Further, 13C and 15N cross-polarization and magic-angle spinning (CP-MAS) experiments were performed at a setting temperature of 10°C under a MAS frequency of 9.3 kHz, while 13C-13C dipolar-assisted rotational resonance (DARR) [27] experiments were performed at a mixing time of 500 ms with 64 and 2048 acquisition points in the f1 and f2 dimensions. A spinal-64 proton high-power decoupling field of 78 kHz was employed during each acquisition [28]. 13C chemical shifts were externally referenced to the methylene resonance of adamantane at 40.48 ppm (trimethylsilylpropanesulfonate (DSS) at 0.0 ppm), while 15N chemical shifts were externally referenced to the 15NH4Cl crystal at 38.34 ppm. The acquired NMR data were phased and baseline-corrected using the TOPSPIN software. A deconvolution procedure was applied to the 15N CP-MAS spectrum to obtain the values of chemical shift, line width, and signal intensity values in the spectral region of PSB. Furthermore, two peaks were used to reproduce the experimental spectra corresponding to all-trans and 13-cis isomers. The best-fitting results were obtained based on the Gaussian shapes corresponding to two peaks using Bruker TOPSPIN software.
Results and Discussion
13C NMR signals of retinal isomers in MR
In the MR reconstituted into PG liposome, the main retinal configurations in the dark were all-trans and 13-cis forms as observed via HPLC [18]. First, we performed an in-situ analysis of the composition of the retinal isomers of dark-adapted MR embedded in PG liposome using 13C solid-state NMR experiments. As shown in Figure 2 (a), the comparison of the 13C CP-MAS NMR spectra corresponding to stable isotope-labeled and non-labeled MR samples showed that [14, 20-13C]retinal and [13Cζ]Tyr NMR signals could be distinguished from the 13C NMR signals corresponding to naturally abundant proteins and lipids. Further, using the extensive 13C NMR data [22,29–31] obtained for the structure of retinal in rhodopsin proteins and retinal compounds summarized in Supplementary Table S1, we identified the following chemical shift values for the 13-trans, 15-anti (all-trans) isomer in MR: 14-13C: δ=124.5 ppm (Fig. 2 (b)) and 20-13C: δ=14.6 ppm (Fig. 2 (c)). Although the NMR signal of 14-13C and 20-13C in the 13-cis form appeared at approximately 116 ppm and 23.0 ppm, respectively, the signal intensity was relatively low (Figs. 2 (b) and (c)). Reportedly, a 20-13C signal of an 11-cis retinal compound in C6D6 has been observed at approximately 17–19 ppm [31]. The signal of the 11-cis form with an extremely low intensity was observed at 19 ppm (Fig. 2 (c)). Thus, we estimated the fractions of the three retinal isomers from the individual peak areas of 20-13C based on the differences in the spectra corresponding to stable-isotope labeled and non-labeled MR (Fig. 2 (a), black and red, respectively) as all-trans, ~72.9%; 13-cis, ~19.7%; and 11-cis, ~7.4%. For this reason, we suggested that the all-trans isomer in the dark-adapted MR was predominant in the PG membrane. Thus, the 20-13C signal of the all-trans isomer in MR appeared at 14.6 ppm and showed higher field shift compared with those of BR at 15.2 ppm, NpSRII at 15.3 ppm [32], and SRI at 15.8 ppm [33] (as a reference of 13C NMR to DSS). The all-trans retinal in BR has a steric hindrance with Trp182, as well as a short inter-atomic distance of 3.35 Å between the 20-methyl carbon and indole nitrogen of Trp182 based on solid-state NMR data and crystal structural information (Fig. 1) [34,35]. Further, the comparison of the UV resonance Raman spectra of wild-type BR and the W182F mutant showed a strong interaction between the indole ring of Trp182 and both 19-C and 20-C of retinal [36]. Trp182 in BR plays an important role in re-isomerization from 13-cis to all-trans in the later stage of the photocycle [37]. The higher field resonance of the 20-C methyl carbon in the all-trans retinal observed in this study suggested that in MR, Trp182 has an orientation that is different from that which it shows in other microbial rhodopsins, owing to the changes in steric hindrance associated with the 20-C methyl group in retinal. Particularly, the chemical shift value of 20-C in MR was clearly different from that of NpSRII, which has almost the same maximum absorption wavelength and only the all-trans isomer. Additionally, considering microbial rhodopins, it is possible that a rearrangement of a water molecule hydrogen bonded to the indole ring of conserved Trp (Trp182 in BR) occurred in MR. Therefore, the unusual 20-C NMR signal of MR possibly suggested that the interaction of 20-C methyl groups in the retinal with Trp182 is related to MR having multiple isomers of MR.
Figure 2 .
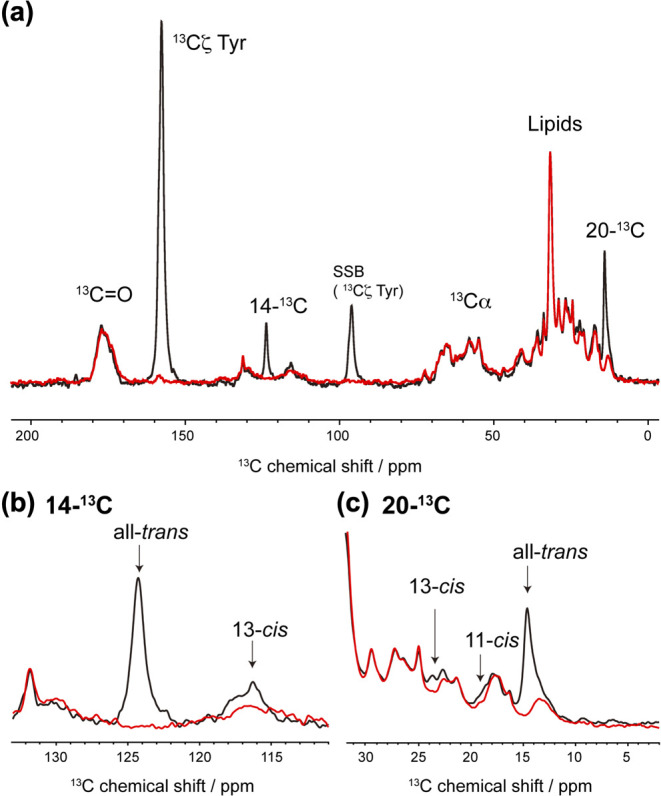
(a) 13C CP-MAS NMR spectra of stable-isotope labeled (black) and non-labeled (red) MR in PG liposome at 10°C. SSB, spinning side band. (b) Expanded 14-13C resonance in the 111–133 ppm region. (c) Expanded 20-13C resonance in the 3–32 ppm region.
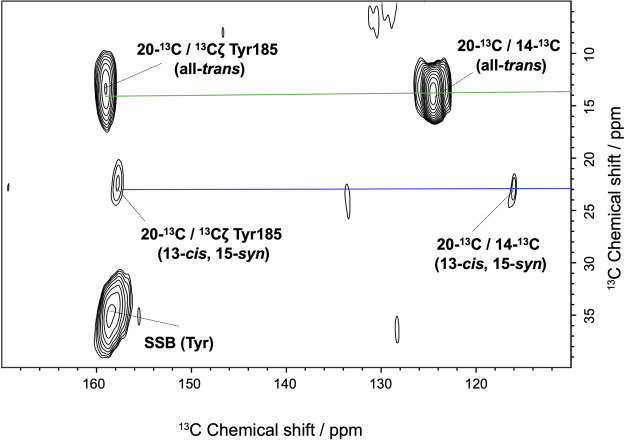
Assessment of the Tyr185 NMR signals assigned by DARR
Figure 3 shows the 13C DARR spectra of [14, 20-13C] retinal and [13Cζ] Tyr-labeled MR in the dark. From this figure, it is evident that the NMR signals of the methyl carbon at the 20-13C of retinal as well as that of the carbon at the 14-13C position showed two cross peaks with the Cζ of Tyr: 20-13C in all-trans/13Cζ Tyr at 14.6/159.5 ppm, and 20-13C in 13-cis, 15-syn/13Cζ Tyr at 23.2/159.0 ppm. As a reference for the retinal-binding site of BR with the corresponding residues of MR in Figure 1, the correlated Tyr NMR signal could be assigned to Tyr185 in MR, which is closest to retinal. Interestingly, the chemical shift values of Cζ in Tyr185 differed slightly, between two conformations, and were thus assigned to the all-trans and 13-cis/15-syn configurations. Further, the structure of the retinal-binding site could be altered depending on the retinal configurations in the dark-adapted MR. Such structural changes in specific residues corresponding to retinal isomers in the dark adaptation have been similarly reported as simultaneous observations of the two signals of Tyr185 in dark-adapted BR [38]. Further, it has been reported that the chemical shift value of Tyr Cζ is sensitive to hydrogen bond formation [23,39,40]. The Tyr185 NMR signal corresponding to the all-trans isomer of MR at 159.5 ppm was similar to that of Tyr185 in BR (160.0 ppm) and Tyr174 in NpSRII (159.7 ppm) [41,42]. In contrast, the signal of Tyr179 in ASR showed a lower chemical shift at 156.4 ppm given that ASR has no corresponding Asp residue, rather has Pro206, which does not form hydrogen bonds [26]. Thus, the value of 13Cζ Tyr185 indicated that Tyr185 in MR, at least, forms a hydrogen bond. It also suggested that Tyr185 in MR can interact with Asp213 or Thr216 via hydrogen bonding, e.g., Tyr174-Thr204 in NpSRII [43]. The conserved Tyr is an essential residue in the reaction center of microbial rhodopsins given that its mutants have a significant effect on the efficacy of the light-driven proton pump in BR or the light-induced signal transduction in NpSRII [11,44]. Further, given importance of the formation of the Tyr185-Thr215 hydrogen bond in the signal transduce-able BR mutant with respect to the expression of negative phototaxis [20], the hydrogen bond between Tyr185 and Thr216 in MR plays an essential role in the signal transduce-able mutant of MR [16]. In the case of NpSRII, it has been reported that the surrounding residues around the retinal chromophore largely contribute to the color tuning of the protein [45]. Furthermore, the Tyr-retinal interactions in MR may be related to the slight differences in the maximal absorption wavelength (λmax) of 485 nm (all-trans) and 479 nm (13-cis, 15-syn) [19]. Unfortunately, the cross peak between 20-C and 14-C from 11-cis was not identified owing to the extremely low population of this isomer. Therefore, our results suggested that Tyr185 is one of the residues that controls the λmax as well as the proportion of the retinal conformation in dark-adapted MR.
Figure 3 .
Cross peaks observed in the 13C-13C DARR NMR spectra of stable-isotope labeled MR in the PG liposome over a mixing time of 500 ms. The sets of cross peaks for 20-C of all-trans and 13-cis, 15-syn isomers are represented using green and blue lines, respectively.
15N NMR signal of the PSB
In MR, the retinal chromophore is covalently bonded to Lys217 via a PSB linkage. In the 15N CP-MAS NMR spectrum of [15N]Lys-labeled dark-adapted MR, NMR signals corresponding to the free Lys side chain, Arg side chain, backbone amide nitrogen, and PSB were identified, as shown in Figure 4 (a). The PSB signal appeared at approximately 184 ppm, and the two peak components of PSB were simply estimated as an intensity ratio of 3.5 (at 184.5 ppm) : 1 (at 181.0 ppm) based on spectral deconvolution analysis, as shown in Figure 4 (b). Consequently, we suggested that the PSB signals in dark-adapted MR at 184.5 ppm could be attributed to the all-trans form, while that at 181.0 ppm could be attributed to the 13-cis, 15-syn form based on the 13C NMR intensity of the retinal isomers. The deconvolution result was ignored as a slight contribution from the 11-cis isomer. Based on a previous report on the all-trans and 13-cis isomer PSB-counter ion model complexes, a linear relationship in the range of 20 ppm or more exists between the 15N chemical shift of PSB and the λmax depending on the halide ion species (Fig. 5 (a) and (b)) [46,47]. MR showed the shortest λmax among the microbial rhodopsin families. Specifically, the λmax values corresponding to the three retinal configurations of MR were distinguished as 485 nm (all-trans), 479 nm (13-cis, 15-syn), and 495 nm (11-cis) by the extraction of single component UV-Vis spectra [19]. As a reference for the maximum MR data, the value for MR was plotted together with those for BR [48], Krokinobacter rhodopsin 2 (KR2) [40], ASR [49], Thermophilic rhodopsin (TR) [50], and NpSRII [51] as shown in Figure 5 (a) and (b). MR with the all-trans form (485 nm/184.5 ppm) agreed well with the trend of the linear relationships shown in the model compound (Fig. 5 (a)). Compared with the values of other rhodopsins, MR had a lower field resonance of PSB with a shorter λmax. Further, MR with the 13-cis, 15-syn form (479 nm/181.0 ppm) also showed agreement with the trend, but deviated more strongly from the model behavior than BR with the 13-cis, 15-syn form as shown in Figure 5 (b). It was also observed that PSB in both the all-trans and 13-cis retinal isomers of MR exhibited a strong electrostatic interaction with the counter ion. The most promising counter ion candidates were Asp84 in helix C and Asp213 in helix G. Possibly, the relative positions of PSB and the carboxyl groups (COO-) of Asp84 and Asp213 are closer than those in the other microbial rhodopsins. During the all-trans photocycle, structural changes in the β-sheet occur in the hydrophilic part of the protein, suggesting an extracellular BC loop, as observed via time-resolved FT-IR spectroscopy [18]. Quantum mechanics/molecular mechanics (QM/MM) calculations involving NpSRII have suggested that the λmax at 498 nm is induced by a displacement of helix G, resulting in the observed distance between PSB and Asp201 [52]. Thus, our 15N NMR result suggested that the strong electrostatic interaction between PSB and Asp84 in helix C or Asp213 in helix G is significantly linked to the shorter λmax of the all-trans form in MR. In the case of the NpSRII D75E mutant, the population of 13-cis species in the dark increased as the proportion of 13-cis to all-trans became 6:4, given that it may be closer to the relative interatomic distance between PSB and the counter ion Glu75 [53]. Thus, it could be suggested that MR is capable of forming a stable 13-cis isomer based on electrostatic interactions with Asp84 or Asp213 as well as the contacts between retinal and surrounding residues containing the above-mentioned interactions of 20-C methyl groups with Trp182. Additionally, the deviation of the value corresponding to the 13-cis, 15-syn form of MR from the trend of the model compound could be attributed to the tilt around the C=N and N-Cε (Lys217) bonds, which are influenced by closer counter ions or surrounding residues (Fig. 5 (b)). The cause of the formation of the 11-cis form in MR, which is a minor conformation, is unknown, but it can be suggested that the structure of the 11-cis retinal isomer is also regulated by the interactions between retinal and the surrounding residues.
Figure 4 .
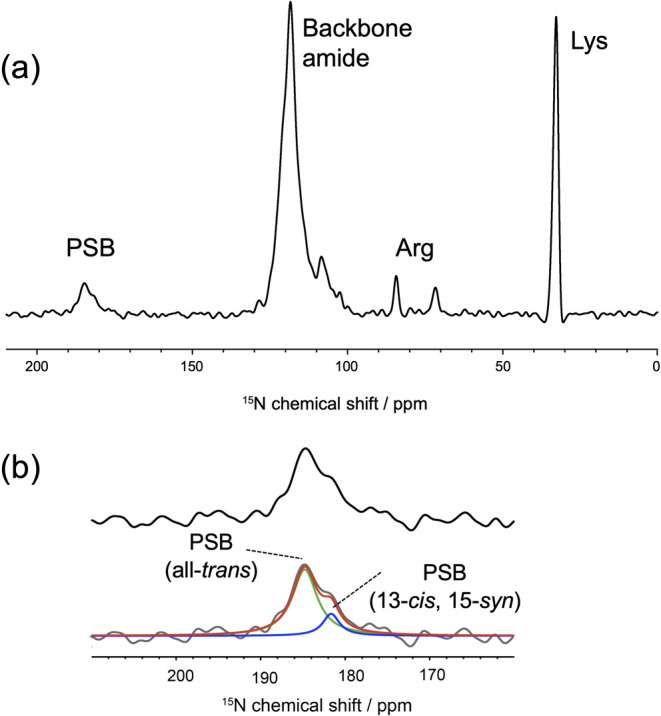
(a) 15N CP-MAS NMR spectra of the dark-adapted MR in PG liposomes at pH 7.0. (b) Expanded 15N NMR signal of PSB in MR between 160 and 210 ppm is indicated in the upper panel. At the bottom, the PSB signals are shown in the upper interpreted via deconvolution with the two components that provided a good fit line (red line), i.e., all-trans (green line) and 13-cis, 15-syn (blue line). The chemical shifts of all-trans and 13-cis, 15-syn isomers are indicated using dotted lines.
Figure 5 .
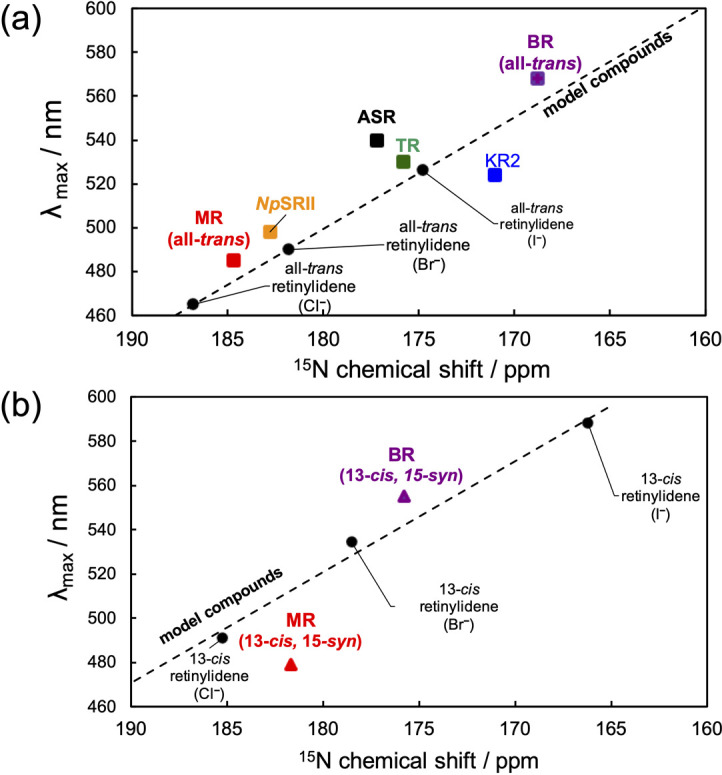
The relationships of 15N chemical shifts of PSB in: (a) The all-trans isomer and (b) 13-cis isomer with the λmax. The linear relationships were as follows: for all-trans model, (λmax (nm)= –5.0872 δ (ppm)+1415.1); 13-cis model, (λmax (nm)= –4.6709 δ (ppm)+1341.3) [46]. 15N PSB chemical shift values of BR [48], KR2 [40], TR [50], and NpSRII [51] are recalibrated to the reference signal of 15NH4Cl.
Conclusions
In this study, we applied solid-state NMR analysis to dark-adapted MR in a PG membrane. The structures of the three retinal isomers in MR were determined from the results of the 13C solid-state NMR. We recorded the higher field resonance of 20-13C NMR signal of all-trans retinal in MR compared with that of other microbial rhodopsins, suggesting the difference in the interaction of 20-C methyl group with Trp182. The all-trans and 13-cis, 15-syn isomers showed a difference in the hydrogen bond strength of Tyr185. Further, the 15N signal of PSB revealed the existence of strong electrostatic interactions with the counter ion for all-trans and 13-cis, 15-syn isomers. Based on the structural information obtained via solid-state NMR, we suggested that the characteristic interactions at the retinal binding site affects the retinal isomers of the dark-adapted MR. Therefore, solid-state NMR spectroscopy is a promising tool for studying the structures of several coexisting retinal isomers as well as the interactions of PSB in microbial rhodopsin.
Acknowledgments
This work was supported by Grants-in-Aid for Scientific Research in an Innovative Area KAKENHI (JP16H06043, JP26104513 and JP20H05211 to I.K., JP25104005, JP25104009 and JP19H05396 to Y.S.) from the Ministry of Culture, Sports, Science and Technology, Japan (MEXT), and Grants-in-Aid for Scientific Research (B) (JP18H02387 to I.K and JP18H02411 to Y.S.) and Scientific Research (C) (JP15K06963 to A.N.) from the Japan Society for the Promotion of Science (JSPS).
Conflicts of Interest
The authors declare that no competing interests exist.
Author contribution
I.K., A.N., and Y.S. directed the project. I.K. wrote the paper. H.S. expressed the protein and prepared a solid-state NMR sample. I.K., H.S., and Y.M. performed the solid-state NMR measurements. I.K., S.T., Y.M., and A.S. analyzed the 13C and 15N NMR data. T.O. and A.W. synthesized the 13C stable isotope-labeled retinal.
Supplementary Material
References
- [1].Ernst, O. P., Lodowski, D. T., Elstner, M., Hegemann, P., Brown, L. S. & Kandori, H.. Microbial and animal rhodopsins: structures, functions, and molecular mechanisms. Chem. Rev. 114, 126–163 (2014). DOI: 10.1021/cr4003769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Kurihara, M. & Sudo, Y.. Microbial rhodopsins: wide distribution, rich diversity and great potential. Biophys. Psychobiol. 12, 121–129 (2015). DOI: 10.2142/biophysico.12.0_121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Kojima, K., Shibukawa, A. & Sudo, Y.. The unlimited potential of microbial rhodopsins as optical tools. Biochemistry 59, 218–229 (2020). DOI: 10.1021/acs.biochem.9b00768 [DOI] [PubMed] [Google Scholar]
- [4].Subramaniam, S. & Henderson, R.. Molecular mechanism of vectorial proton translocation by bacteriorhodopsin. Nature 406, 653–657 (2000). DOI: 10.1038/35020614 [DOI] [PubMed] [Google Scholar]
- [5].Giovannoni, S. J., Bibbs, L., Cho, J. C., Stepels, M. D., Desiderio, R., Vergin, K. L., et al. Proteorhodopsin in the ubiquitous marine bacterium SAR11. Nature 438, 82–85 (2005). DOI: 10.1038/nature04032 [DOI] [PubMed] [Google Scholar]
- [6].Ruediger, M. & Oesterhelt, D.. Specific arginine and threonine residues control anion binding and transport in the light-driven chloride pump halorhodopsin. EMBO J. 16, 3813–3821 (1997). DOI: 10.1093/emboj/16.13.3813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Inoue, K., Ono, H., Abe-Yoshizumi, R., Yoshizawa, S., Ito, H., Kogure, K., et al. A light-driven sodium ion pump in marine bacteria. Nat. Commun. 4, 1678 (2013). DOI: 10.1038/ncomms2689 [DOI] [PubMed] [Google Scholar]
- [8].Niho, A., Yoshizawa, S., Tsukamoto, T., Kurihara, M., Tahara, S., Nakajima, Y., et al. Demonstration of a light-driven SO42– transporter and its spectroscopic characteristics. J. Am. Chem. Soc. 139, 4376–4389 (2017). DOI: 10.1021/jacs.6b12139 [DOI] [PubMed] [Google Scholar]
- [9].Spudich, J. L., Yang, C. S., Jung K. H. & Spudich, E. N.. Retinylidene proteins: Structures and functions from archaea to humans. Annu. Rev. Cell Dev. Biol. 16, 365–392 (2000). DOI: 10.1146/annurev.cellbio.16.1.365 [DOI] [PubMed] [Google Scholar]
- [10].Spudich, J. L. & Luecke, H.. Sensory rhodopsin II: functional insights from structure. Curr. Opin. Struct. Biol. 12, 540–546 (2002). DOI: 10.1016/s0959-440x(02)00359-7 [DOI] [PubMed] [Google Scholar]
- [11].Sudo, Y., Kandori, H. & Kamo, N.. Molecular mechanism of protein-protein interaction of pharaonis phoborhodopsin/transducer and photo-signal transfer reaction by the complex. Recent Res. Devel. Biophys. 3, 1–16 (2004). [Google Scholar]
- [12].Jung, K. H., Trivedi, V. D. & Spudich, J. L.. Demonstration of a sensory rhodopsin in eubacteria. Mol. Microbiol. 47, 1513–1522 (2003). DOI: 10.1046/j.1365-2958.2003.03395.x [DOI] [PubMed] [Google Scholar]
- [13].Sudo, Y., Furutani, Y., Kandori, H. & Spudich, J. L.. Functional importance of the interhelical hydrogen bond between Thr204 and Tyr174 of sensory rhodopsin II and its alteration during the signaling process. J. Biol. Chem. 281, 34239–34245 (2006). DOI: 10.1074/jbc.m605907200 [DOI] [PubMed] [Google Scholar]
- [14].Sasaki, J., Brown, L. S., Chon, Y. S., Kandori, H., Maeda, A., Needleman, R., et al. Conversion of bacteriorhodopsin into a chloride ion pump. Science 269, 73–75 (1995). DOI: 10.1126/science.7604281 [DOI] [PubMed] [Google Scholar]
- [15].Kato, H. E., Kamiya, M., Sugo, S., Ito, J., Taniguchi, R., Orito, A., et al. Atomistic design of microbial opsin-based blue-shifted optogenetics tools. Nat. Commun. 6, 7177 (2015). DOI: 10.1038/ncomms8177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Sudo, Y., Ihara, K., Kobayashi, S., Suzuki, D., Irieda, H., Kikukawa, T., et al. A microbial rhodopsin with a unique retinal composition shows both sensory rhodopsin II and bacteriorhodopsin-like properties. J. Biol. Chem. 286, 5967–5976 (2011). DOI: 10.1074/jbc.m110.190058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Inoue, K., Tsukamoto, T. & Sudo, Y.. Molecular and evolutionary aspects of microbial sensory rhodopsins. Biochim. Biophys. Acta Bioenerg. 1837, 562–577 (2014). DOI: 10.1016/j.bbabio.2013.05.005 [DOI] [PubMed] [Google Scholar]
- [18].Furutani, Y., Okitsu, T., Reissig, L., Mizuno, M., Homma, M., Wada, A., et al. Large spectral change due to amide modes of a β-sheet upon the formation of an early photointermediate of middle rhodopsin. J. Phys. Chem. B 117, 3449–3458 (2013). DOI: 10.1021/jp308765t [DOI] [PubMed] [Google Scholar]
- [19].Inoue, K., Reissig, L., Sakai, M., Kobayashi, S., Homma, M., Fujii, M., et al. Absorption spectra and photochemical reactions in a unique photoactive protein, middle rhodopsin MR. J. Phys. Chem. B. 116, 5888–5899 (2012). DOI: 10.1021/jp302357m [DOI] [PubMed] [Google Scholar]
- [20].Sudo, Y. & Spudich, J. L.. Three strategically placed hydrogen-bonding residues convert a proton pump into a sensory receptor. Proc. Natl. Acad. Sci. USA 103, 16129–16134 (2006). DOI: 10.1073/pnas.0607467103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Sudo, Y., Yamabi, M., Kato, S., Hasegawa, C., Iwamoto, M., Shimono, K., et al. Importance of specific hydrogen bonds of archaeal rhodopsins for the binding to the transducer protein. J. Mol. Biol. 357, 1274–1282 (2006). DOI: 10.1016/j.jmb.2006.01.061 [DOI] [PubMed] [Google Scholar]
- [22].Naito, A., Makino, Y., Shigeta, A. & Kawamura, I.. Photo-reaction pathways and photo intermediates of retinal-binding photo-receptor proteins as revealed by in-situ photo-irradiation solid-state NMR spectroscopy. Biophys. Rev. 11, 167–181 (2019). DOI: 10.1007/s12551-019-00501-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Kimata, N., Pope, A., Eilers, M., Opefi, C. A., Zilx, M., Hirshfeld, A., et al. Retinal orientation and interactions in rhodopsin reveal a two-stage trigger mechanism for activation. Nat. Commun. 7, 12683 (2016). DOI: 10.1038/ncomms12683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Hatcher, M. E., Hu, J. G., Belenky, M., Verdegem, P., Lugtehburg, J., Griffin, R. G., et al. Control of the pump cycle in bacteriorhodopsin: mechanisms elucidated by solid-state NMR of the D85N mutant. Biophys. J. 82, 1017–1029 (2002). DOI: 10.1016/s0006-3495(02)75461-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Becker-Baldus, J., Bamann, C., Saxena, K., Gustmann, H., Brown, L. J., Brown, R. C. D., et al. Enlightening the photoactive site of channelrhodopsin-2 by DNP-enhanced solid-state NMR spectroscopy. Proc. Natl. Acad. Sci. USA 112, 9896–9901 (2015). DOI: 10.1073/pnas.1507713112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Wang, S., Munro, R. A., Shi, L., Kawamura, I., Okitsu, T., Wada, A., et al. Solid-state NMR structure of a lipid-embedded heptahelical membrane protein trimer. Nat. Methods 10, 1007–1012 (2013). DOI: 10.1038/NMETH.2635 [DOI] [PubMed] [Google Scholar]
- [27].Takegoshi, K., Nakamura, S. & Terao, T.. 13C–1H dipolar-assisted rotational resonance in magic-angle spinning NMR. Chem. Phys. Lett. 344, 631–637 (2001). DOI: 10.1016/s0009-2614(01)00791-6 [Google Scholar]
- [28].Fung, B. M., Khitrin, A. K. & Emolaev, K.. An improved broadband decoupling sequence for liquid crystals and solids. J. Magn. Reson. 142, 97–101 (2000). DOI: 10.1006/jmre.1999.1896 [DOI] [PubMed] [Google Scholar]
- [29].Ding, X., Zhao, X. & Watts, A.. G-protein-coupled receptor structure, ligand binding and activation as studied by solid-state NMR spectroscopy. Biochem. J. 450, 443–457 (2013). DOI: 10.1042/bj20121644 [DOI] [PubMed] [Google Scholar]
- [30].Laksmi, K. V., Farrar, M. R., Raap, J., Lugtenburg, J., Griffin, R. G. & Herzfeld, J.. Solid state 13C and 15N NMR investigations of the N intermediate of bacteriorhodopsin. Biochemisty 33, 8853–8857 (1994). DOI: 10.1021/bi00196a001 [DOI] [PubMed] [Google Scholar]
- [31].Wang, K. W., Wang, S. W. & Du, Q. Z.. Complete NMR assignment of retinal and its related compounds. Magn. Reson. Chem. 51, 435–438 (2013). DOI: 10.1002/mrc.3956 [DOI] [PubMed] [Google Scholar]
- [32].Makino, Y., Kawamura, I., Okitsu, T., Wada, A., Kamo, N., Sudo, Y., et al. Retinal configuration of ppR intermediates revealed by photoirradiation solid-state NMR and DFT. Biophys. J. 115, 72–83 (2018). DOI: 10.1016/j.bpj.2018.05.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Yomoda, H., Makino, Y., Tomonaga, Y., Hidaka, T., Kawamura, I., Okitsu, T., et al. Color-discriminating retinal configurations of sensory rhodopsin I by photo-irradiation solid-state NMR spectroscopy. Angew. Chem. Int. Ed. 53, 6960–6964 (2014). DOI: 10.1002/anie.201309258 [DOI] [PubMed] [Google Scholar]
- [34].Petkova, T., Hatanaka, M., Jaroniec, C. P., Hu, J. G., Belenky, M., Verhoeven, M., et al. Tryptophan interactions in bacteriorhodopsin: a heteronuclear solid-state NMR study. Biochemistry 41, 2429–2437 (2002). DOI: 10.1021/bi012127m [DOI] [PubMed] [Google Scholar]
- [35].Luecke, H., Schobert, B., Richter, H. T., Cartailler, J. P. & Lanyi, J. K.. Structure of bacteriorhodopsin at 1.55 Å resolution. J. Mol. Biol. 291, 899–911 (1999). DOI: 10.1006/jmbi.1999.3027 [DOI] [PubMed] [Google Scholar]
- [36].Hashimoto, S., Obata, K., Takeuchi, H., Needleman, R. & Lanyi, J. K.. Ultraviolet resonance Raman spectra of Trp-182 and Trp-189 in bacteriorhodopsin: novel information on the structure of Trp-182 and its steric interaction with retinal. Biochemistry 36, 11583–11590 (1997). DOI: 10.1021/bi971404f [DOI] [PubMed] [Google Scholar]
- [37].Weidlich, O., Schalt, B., Friedman, N., Sheves, M., Lanyi, J. K., Brown, L. S., et al. Steric interaction between the 9-methyl group of the retinal and tryptophan 182 controls 13-cis to all-trans reisomerization and proton uptake in the bacteriorhodopsin photocycle. Biochemistry 35, 10807–10814 (1996). DOI: 10.1021/bi960780h [DOI] [PubMed] [Google Scholar]
- [38].Kawamura, I., Kihara, N., Ohmine, M., Nishimura, K., Tuzi, S., Saito, H., et al. Solid-state NMR studies of two backbone conformations at Tyr185 as a function of retinal configurations in the dark, light, and pressure adapted bacteriorhodopsins. J. Am. Chem. Soc. 129, 1016–1017 (2007). DOI: 10.1021/ja0664887 [DOI] [PubMed] [Google Scholar]
- [39].Zhu, J., Lau, J. Y. C. & Wu, G.. A solid-state 17O NMR study of L-tyrosine in different ionization states: implications for probing tyrosine side chains in proteins. J. Phys. Chem. B 114, 11681–11688 (2010). DOI: 10.1021/jp1055123 [DOI] [PubMed] [Google Scholar]
- [40].Shigeta, A., Ito, S., Inoue, K., Okitsu, T., Wada, A., Kandori, H., et al. Solid-state nuclear magnetic resonance structural study of the retinal-binding pocket in sodium ion pump rhodopsin. Biochemistry 56, 543–550 (2017). DOI: 10.1021/acs.biochem.6b00999 [DOI] [PubMed] [Google Scholar]
- [41].Ding, X., Wang, H., Peng, B., Cui, H., Gao, Y., Iuga, D., et al. Mediation mechanism of tyrosine 185 on the retinal isomerization equilibrium and the proton release channel in the seven-transmemebrane receptor bacteriorhodopsin. Biochim. Biophys. Acta Bioenerg. 1857, 1786–1795 (2016). DOI: 10.1016/j.bbabio.2016.08.002 [DOI] [PubMed] [Google Scholar]
- [42].Nishikawa, R., Kawamura, I., Okitsu, T., Wada, A., Sudo, Y., Kamo, N., et al. Functional conformation of Tyr residues in pharaonis phoborhodopsin as studied by solid state NMR. 16th International Conference on Retinal Proteins, pp. 48. (Shiga, Japan, October 5–10, 2014). [Google Scholar]
- [43].Sudo, Y., Furutani, Y., Shimono, K., Kamo, N. & Kandori, H.. Hydrogen bonding alteration of Thr-204 in the complex between pharaonis phoborhodopsin and its transducer protein. Biochemistry 42, 14166–14172 (2003). DOI: 10.1021/bi035678g [DOI] [PubMed] [Google Scholar]
- [44].Jang, D., El-Sayed, M. A., Stern, L. J., Mogi, T. & Khorana, H. G.. Effect of genetic modification of tyrosine-185 on the proton pump and the blue-to-purple transition in bacteriorhodopsin. Proc. Natl. Acad. Sci. USA 87, 4103–4107 (1990). DOI: 10.1073/pnas.87.11.4103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Shimono, K., Ikeura, Y., Sudo, Y., Iwamoto, M. & Kamo, N.. Environment around the chromophore in pharaonis phoborhodopsin: mutation analysis of the retinal binding site. Biochim. Biophys. Acta Biomembr. 1515, 92–100 (2001). DOI: 10.1016/s0005-2736(01)00394-7 [DOI] [PubMed] [Google Scholar]
- [46].Hu, J. G., Sun, B. Q., Petokova, A. T., Griffin, R. G. & Herzfeld, J.. The predischarge chromophore in bacteriorhodopsin: a 15N solid-state NMR study of the L photointermediate. Biochemistry 36, 9316–9322 (1997). DOI: 10.1021/bi970416y [DOI] [PubMed] [Google Scholar]
- [47].Eilers, M., Reeves, P. J., Ying, W., Khorana, H. G. & Smith, S. O.. Magic angle spinning NMR of the protonated retinylidene Schiff base nitrogen in rhodopsin: expression of 15N-lysine- and 13C-glycine-labeled opsin in a stable cell line. Proc. Natl. Acad. Sci. USA 96, 487–492 (1999). DOI: 10.1073/pnas.96.2.487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Kawamura, I., Degawa, Y., Yamaguchi, S., Nishimura, K., Tuzi, S., Saitô, H., et al. Pressure-induced isomerization of retinal on bacteriorhodopsin as disclosed by fast magic angle spinning NMR. Photochem. Photobiol. 83, 346–350 (2007). DOI: 10.1562/2006-06-20-rc-941 [DOI] [PubMed] [Google Scholar]
- [49].Shi, L., Kawamura, I., Jung, K. H., Brown, L. S. & Ladizhansky, V.. Conformation of a seven-helical transmembrane photosensor in the lipid environment. Angew. Chem. Int. Ed. 50, 1302–1305 (2011). DOI: 10.1002/anie.201004422 [DOI] [PubMed] [Google Scholar]
- [50].Shionoya, T., Mizuno, M., Tsukamoto, T., Ikeda, K., Seki, H., Kojima, K., et al. High thermal stability of oligomeric assemblies of thermophilic rhodopsin in a lipid environment. J. Phys. Chem. B 122, 6945–6953 (2018). DOI: 10.1021/acs.jpcb.8b04894 [DOI] [PubMed] [Google Scholar]
- [51].Tomonaga, Y., Hidaka, T., Kawamura, I., Nishio, T., Osawa, K., Okitsu, T., et al. An active photoreceptor intermediate revealed by in situ photoirradiated solid-state NMR spectroscopy. Biophys. J. 101, L50–L52 (2011). DOI: 10.1016/j.bpj.2011.10.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Hayashi, S., Tajkhorshid, E., Pebay-Peyroula, E., Royant, A., Landau, E. M., Navarro, J., et al. Structural determinants of spectral tuning in retinal proteins-bacteriorhodopsin vs sensory rhodopsin II. J. Phys. Chem. B 105, 10124–10131 (2001). DOI: 10.1021/jp011362b [Google Scholar]
- [53].Shimono, K., Ikeura, Y., Sudo, Y., Iwamoto, M. & Kamo, N.. Importance of the location of the negative-charged counter-ion against the protonated Schiff base on the chromophore configuration of pharaonis phoborhodopsin. J. Photosci. 9, 302–304 (2002). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.