Abstract
Background:
The oral mucosa is the initial interface between food antigens, microbiota and mucosal immunity, yet little is known about oral host-environment dynamics in food allergy.
Objective:
To determine oral microbial, metabolic, and immunologic profiles associated with peanut allergy.
Methods:
We recruited 105 subjects (peanut allergic n=56, healthy subjects n=49) for salivary microbiome profiling using 16S rRNA sequencing, short chain fatty acid (SCFA) metabolite assays using liquid chromatography/mass spectrometry, and measurement of oral secreted cytokines using multiplex assays. Analyses within and across data types were performed.
Results:
The oral microbiome of peanut-allergic individuals was characterized by reduced species in Lactobacillales, Bacteroidales (Prevotella), and Bacillales, and increased Neisseriales spp. The distinct oral microbiome of peanut-allergic subjects was accompanied by significant reductions of oral SCFA levels, including acetate, butyrate, and propionate, and significant elevation of IL-4 secretion. Decreased abundances of oral Prevotella spp. and Veillonella spp. in peanut-allergic subjects were significantly correlated with reduced oral SCFA levels (FDR<0.05), and increased oral Neisseria spp. was correlated with lower oral SCFA levels (FDR<0.05). Additionally, oral Prevotella spp. abundances were correlated with decreased local secretion of Th2-stimulating epithelial factors (IL-33, TSLP) and Th2 cytokines (IL-4, IL-5, IL-13), while oral Neisseria spp. abundance was positively associated with a Th2-skewed oral immune milieu.
Conclusion:
Our novel multi-dimensional analysis of the oral environment revealed distinct microbial and metabolic profiles associated with mucosal immune disturbances in peanut allergy. Our findings highlight the oral environment as an anatomical site of interest to examine host-microbiome dynamics in food allergy.
Keywords: Food allergy, peanut allergy, microbiome, metabolome, short chain fatty acids, mucosal immunity, cytokine, Th2
Graphical Abstract
Capsule Summary:
Distinct oral microbial, metabolic, and immunologic profiles characterize peanut allergy. Oral Prevotella spp., Veillonella spp. and Neisseria spp. abundances are associated with altered oral short chain fatty acid levels (acetate, butyrate, propionate) and a Th2 cytokine milieu.
Introduction
Food allergy affects up to 10% of the population, with peanut allergy accounting for a large share of the disease burden.1 The growing prevalence of food allergy supports environmental influences on pathogenesis, and microbial exposure may be a key environmental risk modifier.2, 3 Imbalances in gut microbiota, or dysbiosis, may precede the onset of food sensitization and shape the natural history of food allergy,2-6 although mucosal disturbances that lead to the breakdown of oral tolerance remain unclear. In murine models, gut microbiota have been shown to modulate allergic sensitization to food, with dysbiosis linked to food allergy susceptibility, and select Bacteroidales and Clostridia taxa conferring protection against food allergy in murine colonization experiments.7,8 Commensal-derived metabolites, namely short-chain fatty acids (SCFA), have also been shown to promote oral tolerance via inhibition of T-helper type 2 (Th2) response and induction of adaptive regulatory response in experimental models.9 However, there has been little direct study of relationships between microbiota, their metabolites, and the mucosal immune system in human beings, in part due to limitations in directly assessing the gastrointestinal mucosal environment. Here we report a novel study using saliva as a window into commensal and mucosal factors in individuals with food allergy.
The oral mucosa is the beginning of a continuous gastrointestinal ecosystem that contains local antigen presenting cells, lymphoid cells, and associated organized lymphoid structures.10 Antigen exposure and presentation by oral immune cells modulate systemic immune tolerance, as evident by the effects of sublingual immunotherapies.11 The oral environment is inhabited by a complex microbial ecosystem that shares direct connections with the gut microbiota both in early life and during disease. 12, 13 Studying these dynamics in the oral environment can offer unique advantages, as it can enable simultaneous profiling of upstream microbiome, commensal byproducts, and mucosal factors in a non-invasive and convenient manner. However, the oral host-commensal milieu has not been evaluated in food allergy until now. In this study, we systemically assessed and analyzed the microbiome, metabolome and secreted cytokines in the oral environment to identify profiles associated with the loss of oral tolerance in individuals with peanut allergy.
Results and Discussion
Distinct oral microbiota in subjects with peanut allergy.
We collected unstimulated saliva from 105 subjects (peanut allergy n=56; healthy control n=49). Please see Supplementary Methods for details on all methods. The median age of subjects was 10 years (IQR 7-14) and 37% were female. Seven (6.7%) subjects had taken antibiotics in the past 3 months and 22 (21.0%) had taken probiotics in the past 3 months. Between the peanut allergic participants and healthy controls, there were no significant differences in age, sex, antibiotic use, probiotic use, time of sampling, or recent intake of major food categories (Supplementary Table 1).
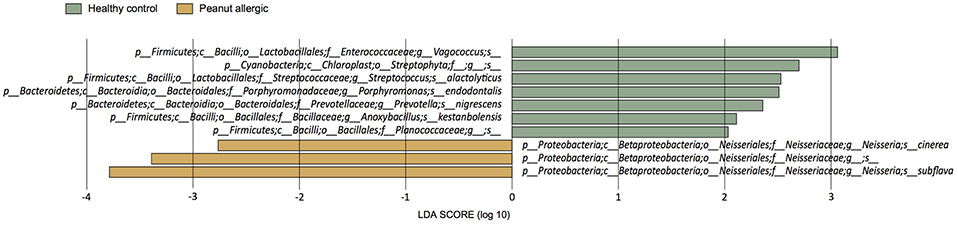
Differences in human gut microbiota have previously been associated with food allergen sensitization and food allergy.2-6 To dissect oral host-commensal dynamics in peanut allergy, we first compared the oral salivary microbiome between peanut allergic and healthy subjects. Alpha diversity by Faith’s phylogenetic diversity was lower in peanut-allergic (mean 13.83, SD=1.40) compared to healthy subjects (mean 14.47, SD=2.10). Peanut-allergic and healthy subjects also had different oral community compositions (unweighted UniFrac PERMANOVA pseudo F=1.86, P=7.99x10−3). Linear discriminant effect size analysis14 revealed distinct taxa in the oral microbiome of peanut-allergic vs. healthy individuals (Figure 1). In healthy subjects, taxa in the Lactobacillales, Bacteroidales, Bacillales, and Streptophyta orders, including taxa from genera Prevotella and Streptococcus, were notably enriched. Several of these taxa have been previously associated with immune tolerance in murine models of food allergy or protection from atopic-sensitization in human fecal studies.2 In food allergy-prone Il4raF709 mice, oral administration of Bacteroidales mixtures (including Prevotella) prevented food allergy and suppressed established disease.8 Greater abundance of gut Lactobacillales members has also been reported in healthy children vs. those with allergic sensitization, clinical symptoms of general allergy, or eczema.15, 16 At the genus level, increased abundances of gut Prevotella and Streptococcus were similarly observed in healthy infants vs. those with food sensitization.17
Figure 1.
Differentially abundant oral microbiome species between peanut-allergic subjects and healthy controls. Linear discriminant analysis Effect Size (LEfSe) analysis14 was used to calculate significance based on linear discriminant analysis (LDA) effect size ≥ 2 and P < 0.05.
In contrast, we found that increased abundance of multiple oral Neisseria spp. characterized the salivary microbiome of peanut-allergic subjects (Figure 1). Interestingly, the abundance of oropharyngeal Neisseria has been previously associated with active celiac disease, a distinct food antigen-triggered immunological disorder.18
We performed secondary analyses to assess for potential confounding by allergic comorbidities (Supplemental Table 1). Within the cohort, there were no associations between the relative abundances of oral microbial species and asthma, allergic rhinitis (AR), or atopic dermatitis (AD). Additionally, we obtained oral samples from 26 non-food allergic subjects with physician-diagnosed asthma, AR, and/or AD. There were no significant differences in alpha diversity (Faith’s phylogenetic diversity) or beta diversity (unweighted UniFrac distance) between healthy controls and these atopic controls without food allergy.
Reduced oral SCFAs in peanut allergy.
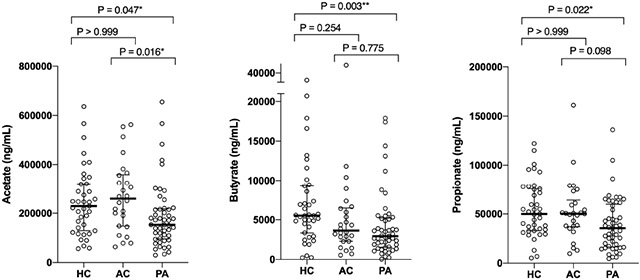
Metabolites generated by commensal gut bacteria, notably SCFAs, modulate immune tolerance in murine models of allergy.9 We examined oral SCFA metabolites (acetate, butyrate, and propionate) in our cohort. Salivary SCFA levels were measured by liquid chromatography/mass spectrometry in 88 subjects (84% of the cohort) based on sample availability after microbiome profiling (peanut allergy, n=49; healthy controls, n=39; Figure 2). Peanut-allergic subjects had significantly reduced salivary acetate, butyrate and propionate levels compared to healthy controls (median acetate 153000 [IQR 95950-218000] vs. 229000 [IQR 129000-32000] ng/mL, P=0.013; median butyrate 2930 [IQR 1540-5225] vs. 5530 [IQR 3350-9290] ng/mL, P=0.001; median propionate 35500 [IQR 16550-59800] vs. 49900 [32900-75600] ng/mL, P=0.007, respectively). Secondary analyses of salivary SCFA levels from 26 atopic controls without food allergy revealed no significant differences in oral acetate, butyrate, or propionate levels compared to healthy controls (Supplemental Figure 1).
Figure 2.
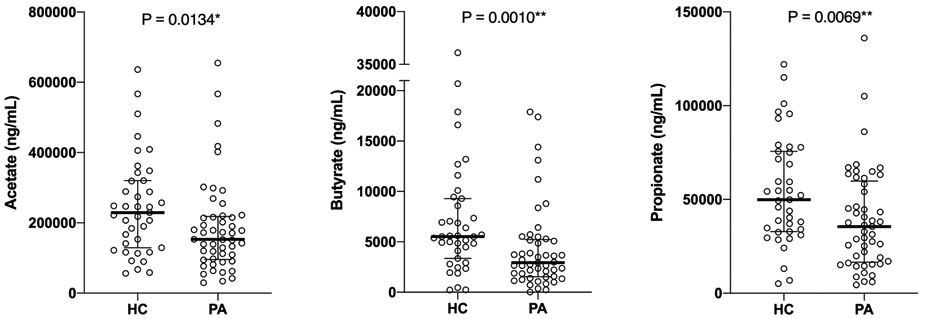
Higher oral short chain fatty acid levels (median ± interquartile range [IQR]) in healthy control (HC) vs. peanut-allergic (PA) subjects. P-value calculated using Mann-Whitney test (*P<0.05, **P<0.01).
Our findings are consistent with prior findings of protective effects of acetate and butyrate in murine models of peanut allergy, where direct oral feeding of these SCFAs led to reduced serum IgE and anaphylaxis score, and where knockout of corresponding SCFA receptors led to exacerbated food allergy.9 While propionate was not found to be protective against peanut allergy in this murine study,9 high levels of fecal propionate and butyrate in toddlers have been associated with less atopic sensitization later in life, suggesting their protective roles against general atopy.19
Increased oral Th2 immune cytokines in peanut allergy.
In parallel, we measured oral cytokine secretion in 24 subjects with remaining sample (peanut allergy, n=16; healthy control, n=8; Figure 3). Central to the pathogenesis of food allergy is a pathologic Th2 response, with interleukin-4 (IL-4) driving mucosal Th2 priming to peanut antigens.20 At the mucosal surfaces, secretion of epithelial factors, such as IL-33 and thymic stromal lymphopoietin (TSLP), may also skew dendritic cells towards a Th2-like phenotype that promotes allergic sensitization.21 Focusing on these key factors, we found that salivary IL-4 secretion was significantly increased in peanut-allergic vs. healthy subjects (mean 21.91 vs. 11.34 ng/min, P=0.04). We also observed consistent trends towards elevated secretion of other oral Th2-promoting epithelial factors (IL-33, TSLP) and additional Th2 cytokines (IL5, IL13) in peanut-allergic vs. healthy subjects (Figure 3).
Figure 3.
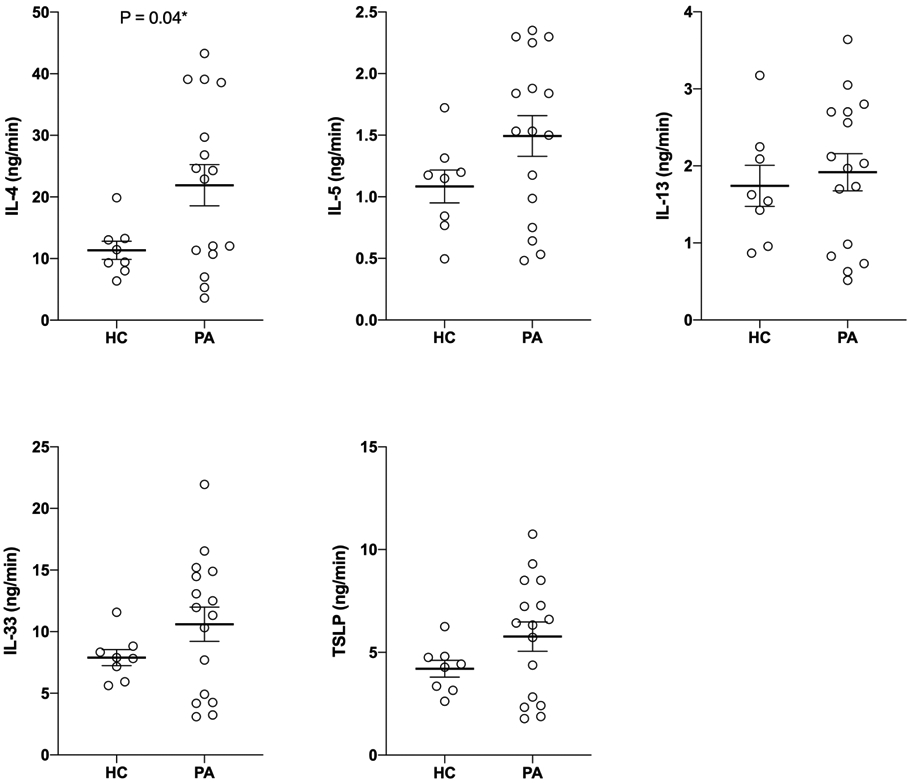
Oral Th2 cytokines and Th2-promoting epithelial factor levels in healthy control (HC) and peanut-allergic (PA) subjects. P-value calculated using unpaired t test (*P<0.05).
Integrated analysis reveals key oral microbiota associated with altered local SCFA and immune profiles in peanut allergy.
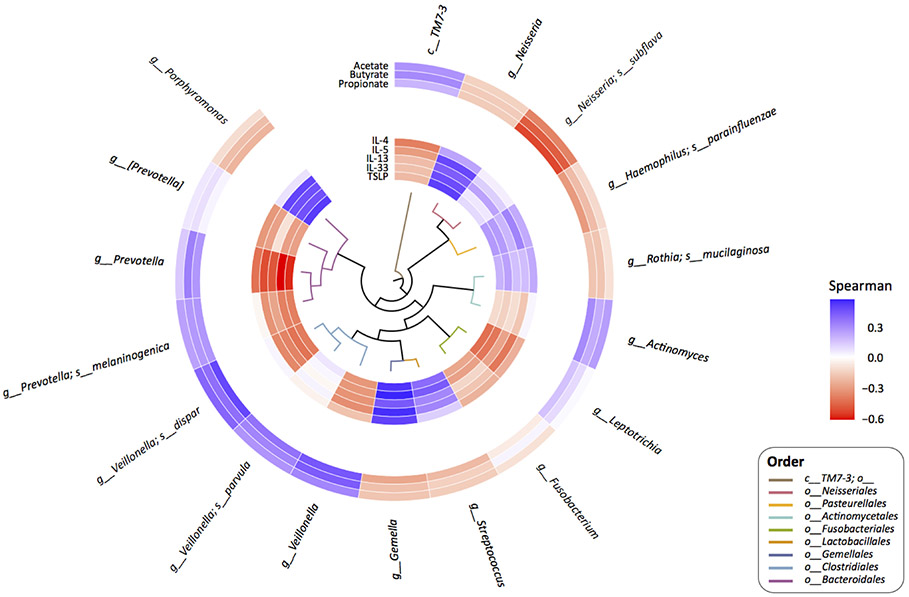
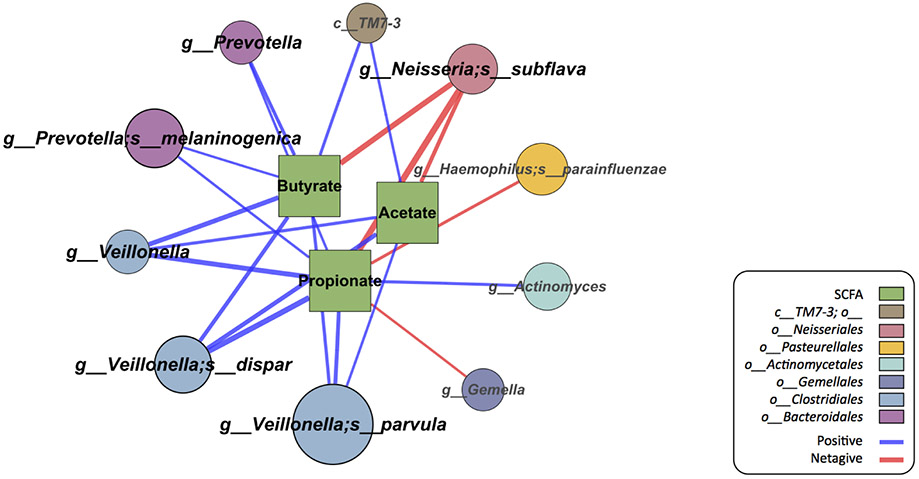
Salivary profiling provides an accessible modality for assessing microbial and mucosal factors in parallel to jointly investigate their relationships in food allergy. We next performed an integrated comparison (Figure 4) and network analysis (Figure 5) of oral microbiota, SCFA and cytokine profiles among subjects with data for all three dimensions. We found that increased abundance of oral Prevotella spp., which characterized healthy subjects, positively correlated with butyrate and propionate (FDR<0.05), and was negatively associated with local secretion of Th2-related cytokines (IL-4, IL-5, IL-13, IL-33, TSLP; P<0.05). Similarly, the relative abundance of oral Veillonella spp. (Clostridia class) positively correlated with acetate, butyrate, and propionate levels (FDR <0.05) and was negatively associated with IL-33 (P<0.05).
Figure 4.
Correlations between the relative abundance of microbial species and short chain fatty acid levels (outer ring), and the relative abundance of microbial species and cytokine levels (inner ring). Species with relative abundance ≥ 1% are included in this analysis and Spearman correlations were used. For unnamed species, the closest higher level named taxon is indicated. The colors of the branch tips of the phylogenetic tree at the center of the plot indicate the order levels of each species in the plot.
Figure 5.
Network of microbial species and short chain fatty acid (SCFA) levels. Species with relative abundance ≥ 1% are included for this analysis. The circular nodes indicate microbial species and the square nodes indicate SCFAs. The edges mark significant Spearman correlation between the microbial species and an SCFA (FDR < 0.05). The size of the circular nodes correlate with the relative abundance of each species, and the thickness of the edges correspond to the magnitude of correlation coefficients.
Our findings are consistent with recognized metabolic activities of Prevotella and Veillonella. Prevotella are effective SCFA producers– in subjects with gut microbiota dominated by Prevotella, high SCFAs with propionate as the major product are seen.22 Our analyses also suggest that Veillonella is associated with the observed SCFA differentials between groups, consistent with their SCFA-producing activities previously noted.23 Notably, infants with allergic sensitization, which is characterized by a Th2-skewed immunity, have decreased intestinal Prevotella and Veillonella, consistent with the inverse correlation between oral Th2 cytokines and these taxa that we observed in the oral environment.17 On the other hand, increased abundance of oral Neisseria spp., which characterized peanut-allergic subjects, negatively correlated with acetate, butyrate, and propionate levels (FDR<0.05), and was positively associated with a Th2 oral milieu (IL-5, IL-13, IL-33, TSLP; P<0.05). While the metabolic impact of Neisseria spp. is largely unknown, they have been previously linked to mucosal inflammation.24 Our data suggest a potential influence of Neisseria (and/or their associated oral microbiota) on the mucosal metabolic and immune milieu in peanut allergy.
Conclusion
While animal models support a role for microbiota in modulating oral tolerance, insights into interactions between commensal bacteria and mucosal immunity in vivo in patients with food allergies have been lacking. Our novel study of the oral environment using saliva and a multi-dimensional approach revealed oral microbial and metabolic features linked to mucosal immune disturbances in those with peanut allergy. In agreement with murine studies of the gut environment, our findings from the human oral environment support a potential role of commensal-derived SCFAs in modulating oral tolerance, with oral Prevotella and Veillonella associated with local metabolic and immune imbalances that characterized peanut allergy. Additionally, our findings highlight oral Neisseria spp. as novel taxa of interest linked to decreased oral SCFA and increased Th2 immune milieu in peanut-allergic patients.
This is the first human multi-omic study of the oral environment in food allergy. A limitation of this study was that the number of subjects and sample availability may have limited our power to detect additional findings. While Prevotella and Veillonella spp. are present in the oral cavity and gut, Neisseria is relatively oral-specific,25 and relationships between oral and gut microbiota in food allergy warrant further investigation. Future studies with (1) broader characterization of the oral immune environment, (2) parallel and longitudinal measures of oral and gut microbiome in food allergy, (3) functional assessment of oral microbiota and SCFAs via in vitro and/or murine models, and (4) characterization of the oral environment in different food allergies would likely be revealing.
Our multi-dimensional analysis of the oral mucosal environment revealed key commensal taxa and metabolites associated with the loss of oral tolerance in peanut-allergic subjects, and highlighted the oral mucosa as a compelling site to further dissect the host-microbiome dynamics in food allergy.
Supplementary Material
Key Messages:
Oral dysbiosis, reduced oral short chain fatty acid levels, and increased oral mucosal Th2 cytokine secretion characterize subjects with peanut allergy.
Differential abundances of oral Prevotella spp., Veillonella spp. and Neisseria spp. are associated with altered oral metabolic and immunological profiles in peanut allergy.
Funding Statement:
This study was funded by the National Institutes of Health R01 AI 147028, the Mount Sinai Food Allergy Treatment and Research Center, the American Academy of Allergy Asthma and Immunology/Elliot and Roslyn Jaffe Third Year Fellowship in Food Allergy Research Award, the Mount Sinai Pediatric Scholars Program, and the Mindich Child Health and Development Institute Trainee Award.
Abbreviations:
- FDR
false discovery rate
- HC
healthy control
- IL
interleukin
- LDA
linear discriminant analysis
- LEfSe
linear discriminant analysis effect size
- PA
peanut-allergic
- QIIME
Quantitative Insights Into Microbial Ecology
- SCFA
shot chain fatty acid
- Th2
T-helper type 2
- TSLP
thymic stromal lymphopoietin
Footnotes
Disclosure Statement: The authors declare no conflict of interest relevant to this manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sicherer SH, Sampson HA. Food allergy: A review and update on epidemiology, pathogenesis, diagnosis, prevention, and management. J Allergy Clin Immunol 2018; 141:41–58. [DOI] [PubMed] [Google Scholar]
- 2.Bunyavanich S, Berin MC. Food allergy and the microbiome: Current understandings and future directions. J Allergy Clin Immunol 2019; 144:1468–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ho HE, Bunyavanich S. Role of the Microbiome in Food Allergy. Curr Allergy Asthma Rep 2018; 18:27. [DOI] [PubMed] [Google Scholar]
- 4.Azad MB, Konya T, Guttman DS, Field CJ, Sears MR, HayGlass KT, et al. Infant gut microbiota and food sensitization: associations in the first year of life. Clin Exp Allergy 2015; 45:632–43. [DOI] [PubMed] [Google Scholar]
- 5.Fazlollahi M, Chun Y, Grishin A, Wood RA, Burks AW, Dawson P, et al. Early-life gut microbiome and egg allergy. Allergy 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bunyavanich S, Shen N, Grishin A, Wood R, Burks W, Dawson P, et al. Early-life gut microbiome composition and milk allergy resolution. Journal of Allergy and Clinical Immunology 2016:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stefka AT, Feehley T, Tripathi P, Qiu J, McCoy K, Mazmanian SK, et al. Commensal bacteria protect against food allergen sensitization. Proc Natl Acad Sci U S A 2014; 111:13145–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abdel-Gadir A, Stephen-Victor E, Gerber GK, Noval Rivas M, Wang S, Harb H, et al. Microbiota therapy acts via a regulatory T cell MyD88/RORgammat pathway to suppress food allergy. Nat Med 2019; 25:1164–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tan J, McKenzie C, Vuillermin PJ, Goverse G, Vinuesa CG, Mebius RE, et al. Dietary Fiber and Bacterial SCFA Enhance Oral Tolerance and Protect against Food Allergy through Diverse Cellular Pathways. Cell Rep 2016; 15:2809–24. [DOI] [PubMed] [Google Scholar]
- 10.Allam JP, Duan Y, Winter J, Stojanovski G, Fronhoffs F, Wenghoefer M, et al. Tolerogenic T cells, Th1/Th17 cytokines and TLR2/TLR4 expressing dendritic cells predominate the microenvironment within distinct oral mucosal sites. Allergy 2011; 66:532–9. [DOI] [PubMed] [Google Scholar]
- 11.Moingeon P Update on immune mechanisms associated with sublingual immunotherapy: practical implications for the clinician. J Allergy Clin Immunol Pract 2013; 1:228–41. [DOI] [PubMed] [Google Scholar]
- 12.Human Microbiome Project C Structure, function and diversity of the healthy human microbiome. Nature 2012; 486:207–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiao J, Fiscella KA, Gill SR. Oral microbiome: possible harbinger for children's health. Int J Oral Sci 2020; 12:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol 2011; 12:R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bjorksten B, Naaber P, Sepp E, Mikelsaar M. The intestinal microflora in allergic Estonian and Swedish 2-year-old children. Clin Exp Allergy 1999; 29:342–6. [DOI] [PubMed] [Google Scholar]
- 16.Johansson MA, Sjogren YM, Persson JO, Nilsson C, Sverremark-Ekstrom E. Early colonization with a group of Lactobacilli decreases the risk for allergy at five years of age despite allergic heredity. PLoS One 2011; 6:e23031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen CC, Chen KJ, Kong MS, Chang HJ, Huang JL. Alterations in the gut microbiotas of children with food sensitization in early life. Pediatr Allergy Immunol 2016; 27:254–62. [DOI] [PubMed] [Google Scholar]
- 18.Iaffaldano L, Granata I, Pagliuca C, Esposito MV, Casaburi G, Salerno G, et al. Oropharyngeal microbiome evaluation highlights Neisseria abundance in active celiac patients. Sci Rep 2018; 8:11047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roduit C, Frei R, Ferstl R, Loeliger S, Westermann P, Rhyner C, et al. High levels of butyrate and propionate in early life are associated with protection against atopy. Allergy 2019; 74:799–809. [DOI] [PubMed] [Google Scholar]
- 20.Chu DK, Mohammed-Ali Z, Jimenez-Saiz R, Walker TD, Goncharova S, Llop-Guevara A, et al. T helper cell IL-4 drives intestinal Th2 priming to oral peanut antigen, under the control of OX40L and independent of innate-like lymphocytes. Mucosal Immunol 2014; 7:1395–404. [DOI] [PubMed] [Google Scholar]
- 21.Han H, Roan F, Johnston LK, Smith DE, Bryce PJ, Ziegler SF. IL-33 promotes gastrointestinal allergy in a TSLP-independent manner. Mucosal Immunol 2018; 11:394–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen T, Long W, Zhang C, Liu S, Zhao L, Hamaker BR. Fiber-utilizing capacity varies in Prevotella- versus Bacteroides-dominated gut microbiota. Sci Rep 2017; 7:2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reichardt N, Duncan SH, Young P, Belenguer A, McWilliam Leitch C, Scott KP, et al. Phylogenetic distribution of three pathways for propionate production within the human gut microbiota. ISME J 2014; 8:1323–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.D'Argenio V, Casaburi G, Precone V, Pagliuca C, Colicchio R, Sarnataro D, et al. Metagenomics Reveals Dysbiosis and a Potentially Pathogenic N. flavescens Strain in Duodenum of Adult Celiac Patients. Am J Gastroenterol 2016; 111:879–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Segata N, Haake SK, Mannon P, Lemon KP, Waldron L, Gevers D, et al. Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol 2012; 13:R42. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.