Abstract
High intensity exercise is a popular mode of exercise to elicit similar or greater adaptive responses compared to traditional moderate intensity continuous exercise. However, the molecular mechanisms underlying these adaptive responses are still unclear. The purpose of this pilot study was to compare high and low intensity contractile stimulus on the Nrf2-mediated redox stress response in mouse skeletal muscle. An intra-animal design was used to control for variations in individual responses to muscle stimulation by comparing a stimulated limb (STIM) to the contralateral unstimulated control limb (CON). High Intensity (HI – 100Hz), Low Intensity (LI – 50Hz), and Naïve Control (NC – Mock stimulation vs CON) groups were used to compare these effects on Nrf2-ARE binding, Keap1 protein, and downstream gene and protein expression of Nrf2 target genes. Muscle stimulation significantly increased Nrf2-ARE binding in LI-STIM compared to LI-CON (p = 0.0098), while Nrf2-ARE binding was elevated in both HI-CON and HI-STIM compared to NC (p = 0.0007). The Nrf2-ARE results were mirrored in the downregulation of Keap1, where Keap1 expression in HI-CON and HI-STIM were both significantly lower than NC (p=0.008) and decreased in LI-STIM compared to LI-CON (p=0.015). In addition, stimulation increased NQO1 protein compared to contralateral control regardless of stimulation intensity (p=0.019), and HO1 protein was significantly higher in high intensity compared to the Naïve control group (p=0.002). Taken together, these data suggest a systemic redox signaling exerkine is activating Nrf2-ARE binding and is intensity gated, where Nrf2-ARE activation in contralateral control limbs were only seen in the HI group. Other research in exercise induced Nrf2 signaling support the general finding that Nrf2 is activated in peripheral tissues in response to exercise, however the specific exerkine responsible for the systemic signaling effects is not known. Future work should aim to delineate these redox sensitive systemic signaling mechanisms.
Keywords: High Intensity Exercise, Redox signaling, Nrf2-Keap1, Muscle Contraction
Graphical Abstract
INTRODUCTION
Exercise induces beneficial adaptations through redox signaling cascades that are mediated by the redox stress response transcription factor, nuclear erythroid related factor 2 (Nrf2) [1–3]. The redox stress response system is a critical cytoprotective mechanism that protects both the cell from endogenous and environmental redox stressors and contributes to adaptive processes to exercise [4, 5]. The redox signal transduction cascade is a highly complex and coordinated system involving the generation of reactive oxygen species, the oxidation of redox-relay molecules or direct oxidation of sensor molecules with thiol switches like Kelch-like ECH-associated protein 1 (Keap1), and effector molecules that change activity, localization, protein-protein interactions, or protein turnover in response to the redox signaling cascade [6, 7]. The overall physiological adaptation resulting from redox signaling cascades depends on the rate of accumulation of these reactive species, and the steady-state levels of enzymatic and non-enzymatic antioxidants [4], as well as the basal oxidation state of proteins with thiol-based redox switches [7–9].
Reactive oxygen species (ROS) generation is required for appropriate skeletal muscle adaptations to exercise such as mitochondrial biogenesis [10]. Inhibiting exercise-induced ROS signaling with antioxidants impairs markers of mitochondrial biogenesis including AMPK and PGC1α activation [11–13], Nrf2 activation [2], and downstream gene expression [14]. Furthermore, there is a linear relationship between exercise-induced ROS accumulation and Nrf2 activation [15], which is dependent on the duration of the exercise bout [15, 16]. Nrf2 can also be activated in human skeletal muscle in response to supramaximal exercise of shorter duration [17], suggesting that longer durations are not required to induce Nrf2 activity if the intensity of the bout is high enough. High intensity interval training is a popular training method to obtain improvements in cardiorespiratory fitness quickly and efficiently, partly through increases in mitochondrial biogenesis – processes that are redox signaling dependent [10, 18–20].
The molecular pattern of redox signaling responses can differ depending on the intensity of the exercise which leads to different adaptations and resulting phenotypes [21–23]. However only two studies to date have directly compared moderate intensity and high intensity exercise and Nrf2 activation, one in human peripheral blood mononuclear cells (PBMCs), and one in human skeletal muscle [24, 25]. There were no differences in Nrf2 signaling between exercise intensities in either human PBMCs or human muscle [24, 25]. However other studies comparing different exercise intensities have shown divergent redox signaling effects in other redox proteins, highlighting some interesting caveats in the field [26]. Therefore, more detailed and well controlled studies comparing differing intensity stimuli on Nrf2 redox stress response signaling are needed.
One issue regarding the redox stress response to exercise, is the considerable variability in the adaptive signaling responses across individuals [27]. This makes it difficult to predict the hormetic response, and thus the overall adaptation to regular exercise, and impacts the ability to effectively prescribe exercise to clinical and non-clinical populations. Therefore, the purpose of this pilot study was to test high and low muscle stimulation intensities on redox stress responses in mouse skeletal muscle using a within-animal study design to control for any intra-animal variation in responses to the contractile stimulus, as well as a between-animal control using a mock stimulation condition. We hypothesized that high intensity stimulation would lead to greater adaptations and activation of Nrf2 signaling compared to low intensity exercise. Here we show a novel systemic Nrf2-ARE activation mechanism gated by high intensity stimulation but not low intensity stimulation that increases Nrf2-ARE binding by acting through inhibition of the negative regulator Keap1.
METHODS
Animals –
Young (6mo) C57BL/6 male mice were received from the Jackson Labs. All mice were maintained at 21°C on a 14/10 light/dark cycle and given standard mouse chow and water ad libitum. The study was approved by the University of Washington Institutional Animal Care and Use Committee (IACUC) and the tissue transfer was approved by the Northern Arizona University IACUC.
Study Design –
There were three experimental groups for this study (Figure 1): High intensity stimulus (HI, n=5), low intensity stimulus (LI, n=5), and naïve unstimulated control (NC, n=5). An intra-animal study design was used for HI and LI groups to control for within-animal variation, where the stimulated (STIM) limb was used as the exercise condition and contralateral unstimulated (CON) limb was used as the non-exercise control. The Naïve Controls (NC) were used as the inter-animal controls and were anesthetized for the same amount of time as the other two groups before both hind limbs were harvested, but no muscle stimulation occurred in either limb. Naïve controls were used to control for systemic effects that could affect both limbs in the HI and LI groups. Nrf2-mediated redox stress response signaling was compared between each group (HI, LI, & NC) as well as paired comparisons within animals unstimulated control and stimulated (exercised) limbs (Figure 1).
Figure 1.
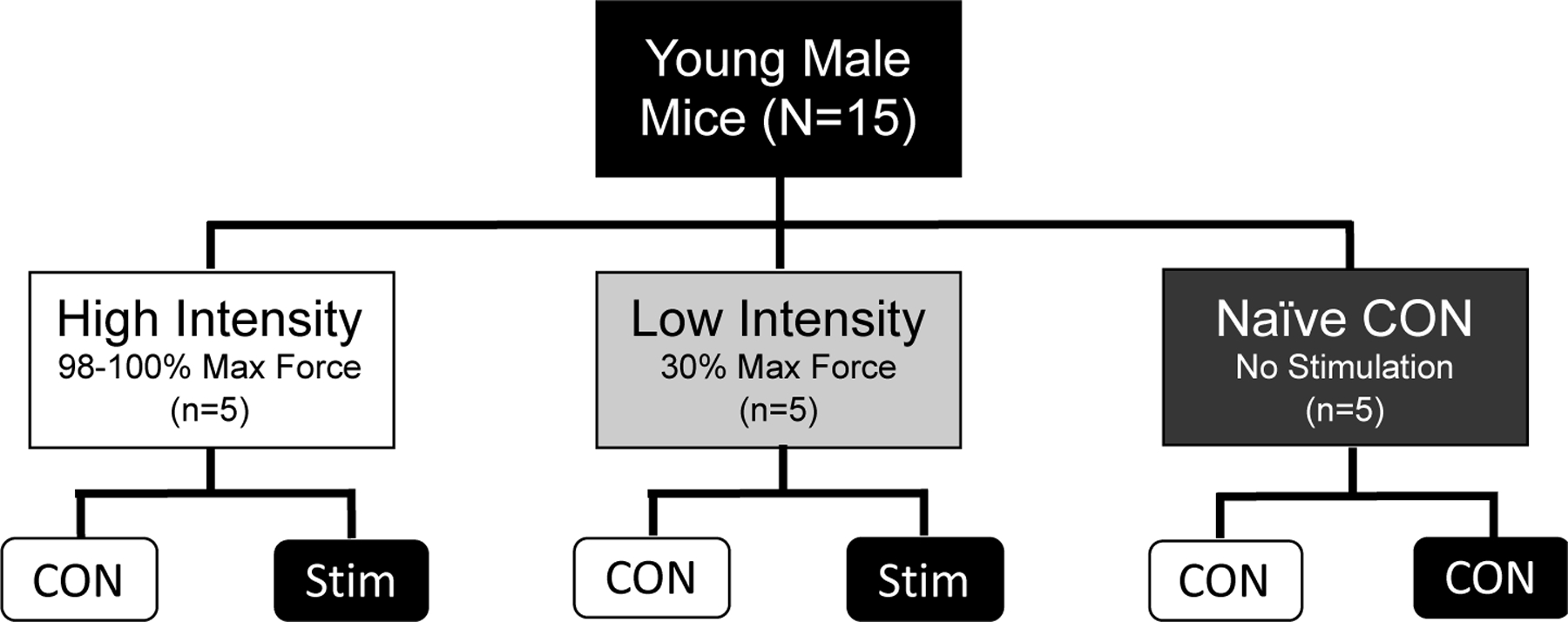
Study Design
In Vivo Muscle Stimulation –
Muscle stimulation was performed on the right leg of anesthetized mice (1–2% isofluorane) resting on a heated plate at 37°C as previously described [28]. The knee was secured, and foot taped to a footplate perpendicular to the tibia. The footplate was connected to a force transducer (Aurora Scientific, ON, Canada). The tibial nerve was stimulated using Grass Stimulator (S88X, Astro Med, Inc.) at optimal voltage (1–4V) that was selected by measuring maximal isometric torque of plantarflexion during isometric contractions (200ms train, 0.1ms pulse, 100Hz). Following optimization, the plantarflexors underwent a fatigue protocol with isometric contractions (200ms train, 0.1ms pulse) induced by a high (100Hz) or low (50Hz) stimulation frequency every fourth second for 30 minutes. Following the fatiguing contraction procedure, the electrodes were removed and muscle was allowed to rest for an additional 30-minutes while on anesthesia until muscle harvest. Animals were euthanized through cervical dislocation. The gastrocnemius and soleus muscle from the stimulated and unstimulated legs (contralateral control) were removed 30 minutes after the end of muscle stimulation and flash frozen in liquid nitrogen and stored at −80˚C.
Gene Expression –
Soleus muscles were homogenized in RLT buffer (+ 1% BME). RNA extraction was done using RNeasy Plus Mini Kit following Proteinase K digestion and elimination of DNases using RNase-free DNase kit (all reagents from Qiagen). Isolated RNA was then converted to cDNA using Bio-Rad iScript kit and RT-qPCR using Bio-Rad SYBR Sso Advanced. Samples were analyzed using the ∆∆Ct method. A panel of six housekeeping genes were used to determine which were the best three genes to use for internal controls (Table 1). The geometric mean for the three most stable housekeeping genes were used to quantify changes in target gene mRNA expression in response to stimulation [29]. All primer sequences are listed in Table 1.
Table 1.
Primer Sequences for RT - qPCR
| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| RPL41 | GCCATGAGAGCGAAGTGG | CTCCTGCAGGCGTCGTAG |
| RPL27 | AAGCCGTCATCGTGAAGAACA | CTTGATCTTGGATCGCTTGGC |
| RPL7L1 | ACGGTGGAGCCTTATGTGAC | TCCGTCAGAGGGACTGTCTT |
| RER1 | GCCTTGGGAATTTACCACCT | CTTCGAATGAAGGGACGAAA |
| ACTB | CCTCGCCTTTGCCGA | TGGTGCCTGGGGCG |
| PPIA | CCCACCTGTTTCTTCGACAT | CCATGTCTCAGAGCACGAAA |
|
| ||
| HMOX1 | CCTCACTG GCAGGAAATCATC | CCTCGTGGAGACGCTTTACATA |
| NQO1 | GGGTCGTCTTGGCAACCA | CAGATGTTGAGGGAGGATCGTAA |
| GCLC | GCTGTCTTGCAGGGAATGTT | ACACACCTTCCTTCCCATTG |
| GSTP1 | GCTCTTACCACGTGCAGCTT | GGCTGGGAAGAGGAAATGGA |
| GSR | GCTATGCAACATTCGCAGATG | AGCGGTAAACTTTTTCCCATTG |
| NFE2L2 | CGAGATATACGCAGGAGAGGTAAGA | GCTCGACAATGTTCTCCAGCTT |
Glutathione Content –
Intracellular glutathione levels were measured in gastrocnemius muscles as previously described [30] using a spectrophotometric assay based on the affinity of 2,3naphthalenedicarboxaldehyde (NDA) for γ-glutamylcysteinylglycine (GSH). Skeletal muscle (10–20mg) was homogenized in ice-cold Locke’s buffer (10 mM HEPES, 5.5 mM KCl, 10 mM glucose, 5 mM NaHCO3, and 130 mM NaCl). A portion of the homogenate was set aside for protein quantification using a Bradford assay. An equal volume of 200mM 5-sulfosalicylic acid dehydrate (SSA) was mixed in. After resting on ice for 15 minutes the samples were centrifuged at 12,000 x g for 3 minutes at 4 °C. The supernatant was plated into a 96 well plate in duplicate. Standards with known GSH concentrations (0–50µM) were also plated in duplicate. 0.2 M NEM/0.02 M KOH were added to each well followed by 10 mM TCEP. After a 20-minute incubation at room temperature, 0.5 N NaOH was added followed by 10mM NDA. After a 30-minute incubation the plate was read at fluorescence intensity using 472 (excitation) and 528 (emission). GSH levels were assessed using the standard curve and normalized to protein concentration.
Western Blotting –
Gastrocnemius muscles were homogenized on ice for 5 minutes in the presence of 25µl of Cell Lytic MT cell lysis buffer (Sigma-Aldrich) per mg of tissue. Lysis buffer was supplemented with 0.1% protease inhibitors (Sigma-Aldrich) and 0.2% phosphatase inhibitor (ThermoFisher). Following homogenization, aliquots were taken from original samples for Bradford assays to determine protein concentrations. Next, sample buffer was added to the original sample then boiled and stored at −80˚C. Thirty micrograms of protein were separated by gel electrophoresis in each well followed by wet transfer on to nitrocellulose membranes and one hour blocking in TBS + 5% non-fat dry milk. Blots were incubated with Keap1 (EPR22664–26, Abcam, Cambridge MA) GCLC (EP12345, Abcam), GSR (Santa Cruz Biotech, Dallas TX), HO1 (Cell Signaling Technology, Danvers MA), or NQO1 (Cell Signaling Technology, Danvers MA) monoclonal antibodies to detect proteins with Ponceau S, used for loading controls (Cell Signaling Technology, Danvers MA).
Nrf2-ARE Binding Assay –
Gastrocnemius muscles were homogenized, lysed, and nuclear fractions extracted per manufacturer’s instructions (Nrf2-ARE binding kit, TransAM, Carlsbad CA). Briefly, samples were homogenized in a 1:30 ratio (mg tissue:µl buffer) followed by centrifugation, isolation, and lysis of the nuclear pellet. After Bradford assays were run to determine protein content in nuclear fractions, nuclear lysates were incubated in a 96 well plate coated with ARE consensus oligonucleotides for Nrf2 Binding. Following three washes, primary anti-Nrf2 antibodies were incubated (1:1000) in each well used to detect Nrf2 followed by secondary antibody incubation and colorimetric development. Absorbance was read at 450nm on a Synergy HT plate reader (Bio-Tek, Winooski, VT).
Statistical Analysis –
For all measures, a three by two repeated measures ANOVA (Intensity group by Stimulation condition) was run for analysis of intensity group differences, stimulation differences and their interaction. Tukey’s post hoc analysis was used for CON vs STIM pairwise comparisons within group. All relevant p values reported are adjusted p values using Tukey’s multiple comparisons post hoc analysis. Statistical analysis was performed using SPSS (IBM, version 27) and GraphPad Prism software (San Diego, CA).
RESULTS
Muscle stimulation
The results from the muscle stimulation protocol in HI and LI are shown in figure 2. The force output of a tetanic contraction at 100Hz was not different between groups (12.1 mN-m ± 0.9 and 11.9 mN-m ± 1.7 for LI-STIM and HI-STIM respectively). The fatigue protocol at HI-STIM led to a final force output that was 27.7% ± 5.6 of initial 100Hz tetanus, and LI-STIM led to a final force output that was 12.8% ± 5.6 of a 100Hz tetanus and 37.3% ± 2.8 of the initial 50Hz contraction.
Figure 2. Muscle force output and fatiguing stimulation.
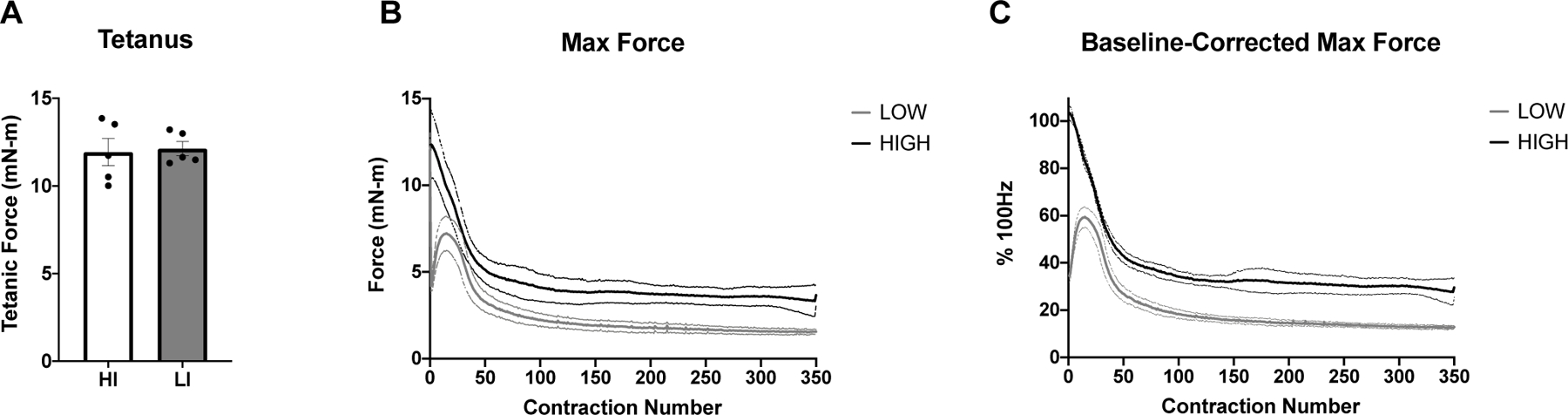
A) Both low and high intensity stimulation groups had similar maximal tetanic force at 100Hz. B) The absolute force (mN-m) over time in response to either High (100Hz) or Low (50Hz) intensity stimulation frequency. C) The normalized force to maximal tetanus. Data are mean ± SD.
Stimulation Intensity Reveals Divergent Effects on Nrf2-ARE Binding and Keap1 Protein Dynamics in Skeletal Muscle
Nrf2-ARE binding was low under basal unstimulated conditions (Figure 3A, Naïve Control group and LI-CON condition). There was a significant interaction between stimulation condition by intensity (RM ANOVA Group x Stimulation condition, p = 0.005), where Nrf2-ARE binding significantly increased in LI-STIM condition compared to LI-CON (Figure 3A, ** p = 0.047), while NC and HI were not significantly different between conditions (Figure 3A, HI-CON vs HI-STIM, or both NC-CON conditions). Nrf2-ARE binding was significantly higher in the HI group under both conditions compared to NC group (Figure 3A, ## p = 0.007). Keap1 showed a significant main effect of intensity indicated by the difference between high intensity and naïve controls (Figure 3B, ##p = 0.008). There was also an interaction of stimulation by intensity (p = 0.035) and a main effect of stimulation (p = 0.016). The interaction of stimulation by intensity was driven by a significant decrease in Keap1 protein in LI-STIM condition compared to LI-CON (Figure 3B, *p = 0.015) with no changes between CON vs STIM conditions in HI or differences between the two NC-CON. There were no differences in total glutathione content across groups, or in response to stimulation within groups (Figure 3D).
Figure 3. Nrf2-ARE Binding and Keap1 Protein Expression in Response to Skeletal Muscle Stimulation.
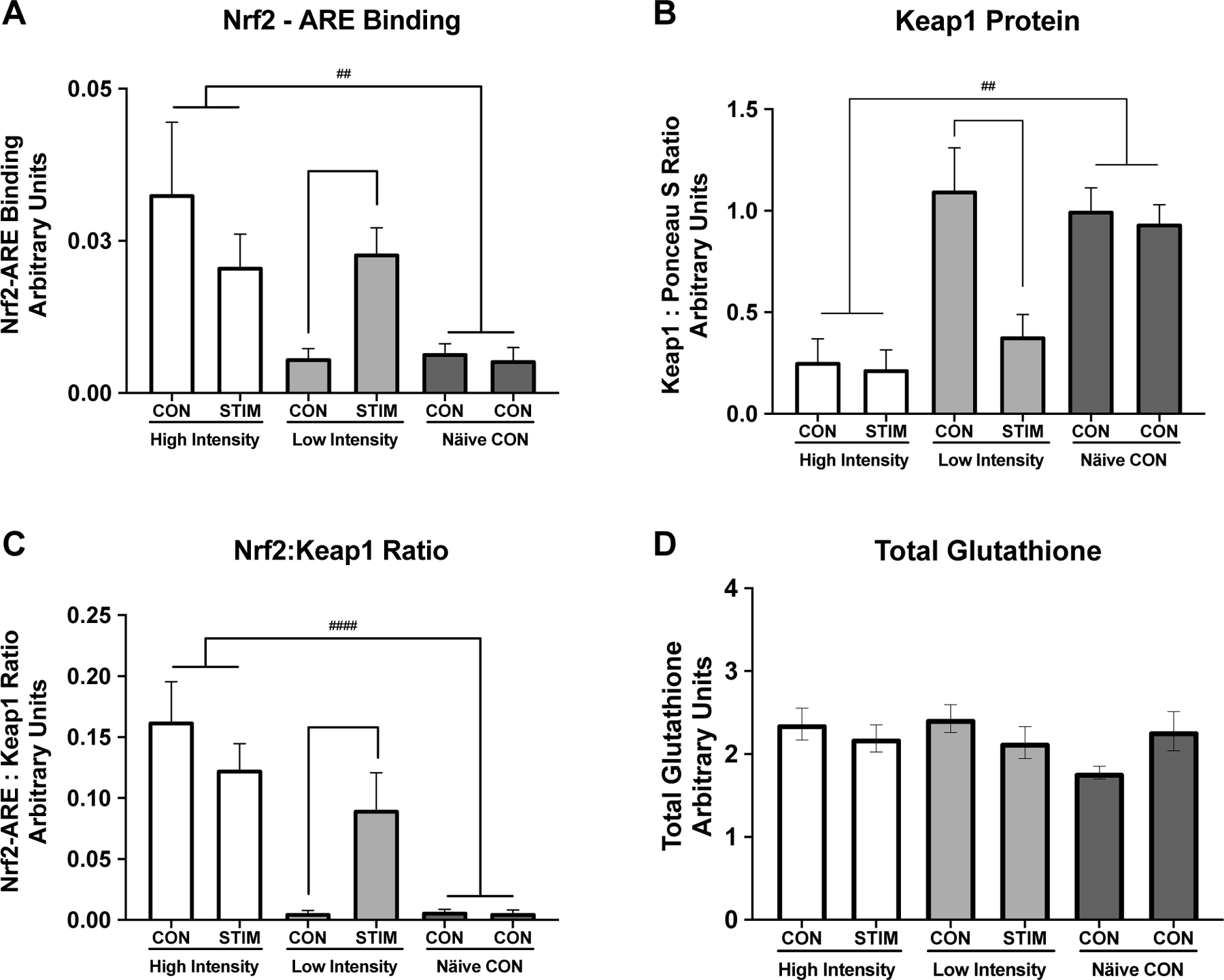
Keap1-Nrf2-ARE signaling response to high and low intensity stimulation and total glutathione content. A) Nrf2-ARE binding was low under basal unstimulated conditions (Naïve Control group and LI CON condition) but increased in response to low intensity stimulation (LI-CON vs LI-STIM, * p = 0.047). Nrf2-ARE binding was significantly higher in the HI group compared to NC group (## p = 0.007) . B) Keap1 content was unchanged between limbs in the Naïve Control group or the LI-CON, however there was a significant decrease in Keap1 content in the LI-STIM limb (*p = 0.015). Keap1 content was significantly lower in the HI group under both conditions compared to NC (##p = 0.008). C) Nrf2 :Keap1 ratio illustrates the same pattern as either Nrf2-ARE binding or Keap1 protein content alone (HI vs NC, #### p < 0.0001). D) Total glutathione content, measured in gastrocnemius muscle, was not different across groups and did not change significantly in response to stimulation. Representative western blot image of Keap1 protein is shown in Figure 5B. Keap1 was normalized to left hind limb of the NC group. Data are presented as mean ± SEM.
Gene expression responses to skeletal muscle stimulation
A panel of six housekeeping genes were used to assess their stability in response to skeletal muscle stimulation (Figure 4A and 4B). Variability from CON to STIM conditions was assessed for each housekeeping gene (Figure 4A) as well as variation by intensity group (Figure 4B). RPL41, RPL27, and RPL7l1 were selected as the best genes to use for analysis because their within animal variation was lowest across groups (∆Cq in response to stimulation was minimized). Target gene fold change was calculated using the geometric mean of the 3 housekeeping genes with the ∆∆Cq method. Target gene responses to muscle stimulation were not significantly different from CON or across intensities, although there was a trend for an increase in GCLC mRNA expression in response to stimulation (Figure 4C, p = 0.06).
Fig. 4. Gene expression of redox stress response genes to skeletal muscle stimulation.
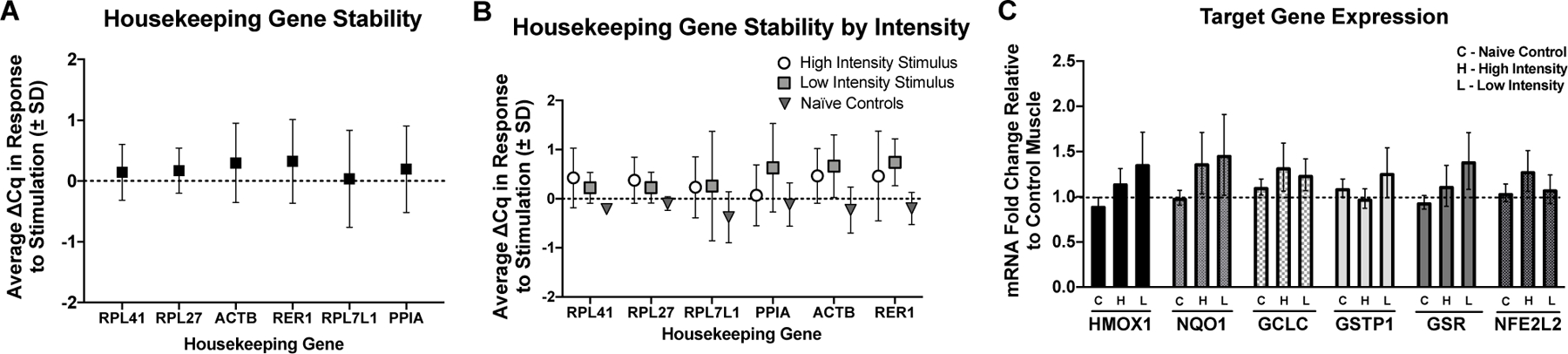
A) Housekeeping gene average Cq change (∆ Cq) in response to skeletal muscle stimulation. B) Housekeeping gene average Cq change (∆ Cq) by group. Based on these results RPL41, RPL27, and RPL711 were selected for housekeeping genes. C) Effects of high intensity (H), low intensity (L) muscle stimulations, and control (C) for target genes HMOX1, NQ01, GCLC, GSR, and NFE2L2 fold change from unstimulated muscle (dotted line). In Nave Controls the mock stimulated muscle (right limb) was compared to the unstimulated control (left limb in all cases). There were no statistically significant increases in gene expression in response to stimulation in either group, although GCLC showed trends for increases regardless of intensity groups. RPL41, RPL27, and RPL711 were used for analysis of target gene fold change. H = High intensity stimulation group, L = Low intensity stimulation group, C = Naïve control group.
Changes in redox stress response proteins in response to skeletal muscle stimulation
There was a main effect of intensity for HO1 Protein expression (p = 0.01) with HO1 significantly elevated in the high intensity group compared to the Naïve controls (Figure 5C, p = 0.003). There was a significant effect of stimulation condition on NQO1 protein (Figure 5D, p = 0.019) with no differences between high and low intensity stimulus and no differences between stimulation groups. There was a significant main effect of stimulation on GCLC protein content where the stimulation condition decreased GCLC protein slightly (Figure 5E, p = 0.03) with no differences between groups. There were no significant differences between groups or in response to muscle stimulation for GR protein (Figure 5F).
Fig. 5. Redox stress protein response to muscle stimulation.
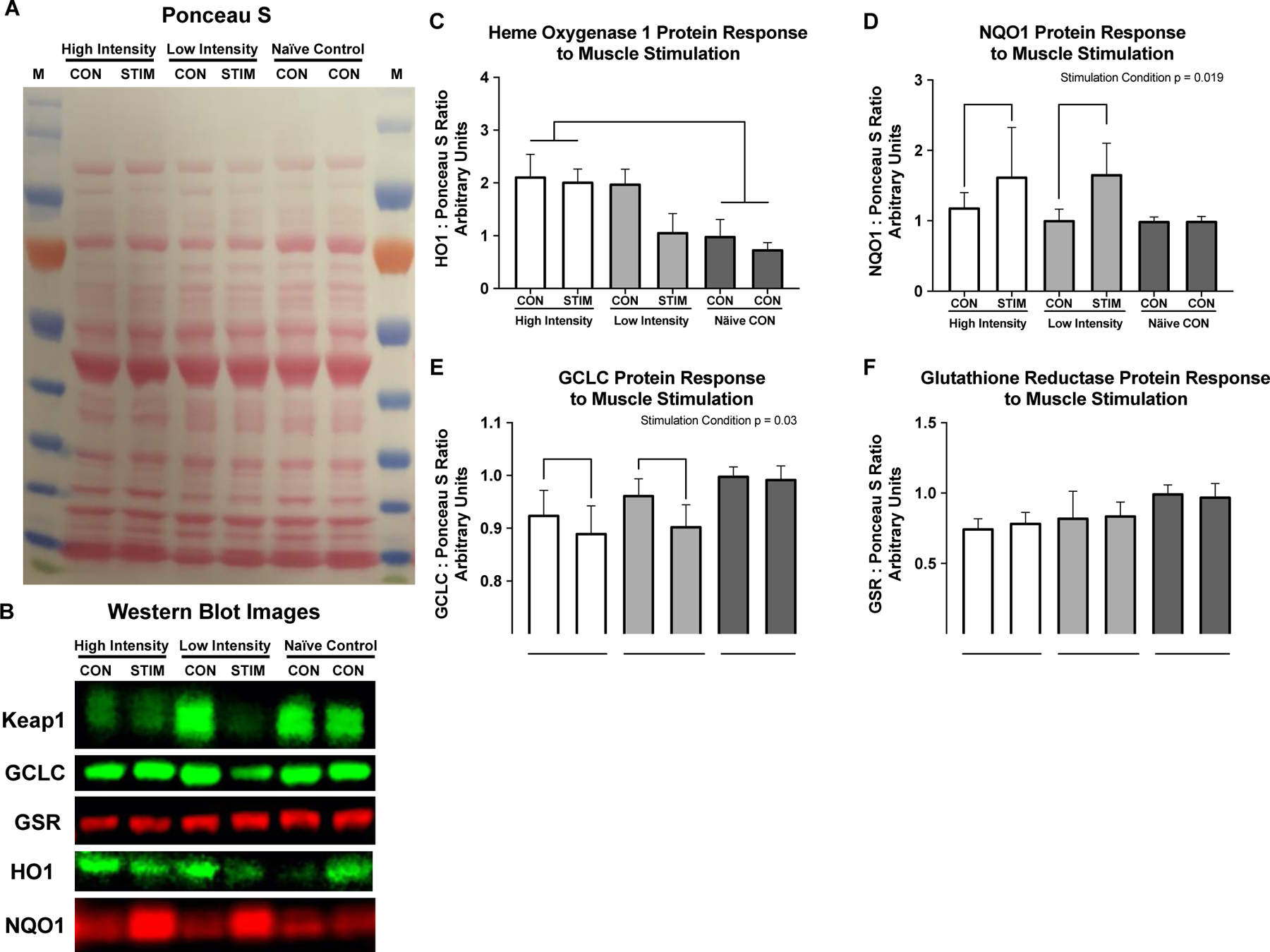
Redox stress protein response to muscle stimulation. A) Representative Ponceau S stain, B) Representative western blot images. C) Heme Oxygenase 1 protein is significantly elevated in both conditions of the high intensity group compared to the Naïve control group (p = 0.002). D) NQO1 protein increased significantly in response to stimulation (Stimulation condition p = 0.019), with no differences between intensities, or across groups. E) GCLC decreased slightly in stimulation conditions regardless of intensity (Stimulation condition p = 0.03) with no differences between groups. Glutathione reductase was unchanged in response to muscle stimulation or across intensity groups. All values are normalized to left limb of the NC group and set equal to 1 for western blot graphs.
DISCUSSION
Exercise is one of the most powerful pleiotropic interventions to improve health and fitness. These health benefits are mediated in part by redox signaling responses to an exercise bout, including activation of the inducible redox stress response transcription factor Nrf2. Nrf2 mediates hundreds of different cytoprotective and metabolic genes increasing metabolic and redox capacity with repeated transient stressors like exercise training. Recently there has been an increased interest in using high intensity interval exercise as a more time efficient way to elicit beneficial adaptations [18]. How the molecular signature from high intensity exercise differs from moderate intensity exercise is still unclear. The aim of this pilot study was to investigate the effects of high intensity and low intensity muscle stimulation on redox stress response markers in mouse skeletal muscle.
Previous studies on Nrf2 signaling responses to exercise have used whole animal treadmill exercise, measuring Nrf2 in several different tissues [2, 15, 16, 31–37], or whole-body exercise in humans [1, 17, 24, 25, 38–40]. We have recently shown that basal levels of Nrf2 affect its inducibility to an acute exercise bout in humans [39]. Therefore, in order to control for intra-animal variations in basal redox homeostasis and basal Nrf2 activation, we elected to use an intra-animal design, where the right limb of the animals was stimulated with either high intensity or low intensity muscle stimulation, while the contralateral unstimulated limb served as the internal control. Unexpectedly, we found that both stimulated and contralateral unstimulated limb of the high intensity group showed increased Nrf2-ARE binding activity. This is in contrast to the low intensity group where Nrf2 was low in the unstimulated contralateral control limb, but significantly increased in the stimulated limb. The low levels of Nrf2-ARE binding in both Naïve control limbs confirm that Nrf2 activity is low in resting healthy tissue.
Western blots of Keap1, the negative regulator of Nrf2, show opposite effects, where Keap1 protein is highly expressed in both naïve control limbs, and the low intensity control limb, as predicted with the low levels of Nrf2-ARE binding. Keap1 decreases in response to low intensity stimulation, releasing the inhibition on Nrf2 and resulting in increased Nrf2 activation and ARE binding in low intensity stimulation. Keap1 is lowly expressed in both control and stimulated limbs of the high intensity group, in line with the increases in Nrf2 activation in both stimulated and contralateral control limbs. The ratio of Nrf2-ARE binding to Keap1 content also illustrates the relationship between Keap1 protein and Nrf2-ARE binding. A recently published paper reported significant decreases in Keap1 content after acute exhaustive exercise in human skeletal muscle and concomitant increases in Nrf2 [17], our data here are in agreement with these results. Together these data demonstrate i) Basal Nrf2 activation is low under resting or unstressed conditions in young adult skeletal muscle; ii) Nrf2 activity is inducible in response to muscle stimulation; iii) high intensity, but not low intensity muscle contraction, activates Nrf2 even in unstimulated muscles; and iv) Keap1 protein content mirrors these findings, indicating these inducible Nrf2 responses are acting through canonical Keap1 inhibition in response to skeletal muscle stimulation.
We measured gene expression 30 minutes after the muscle stimulation was finished. The timing may have been a limitation in detecting peak changes in gene expression of these redox stress response genes because there were no statistically significant increases. We and others have shown Nrf2 regulated genes to be induced between 1–4 hours after completion of an exercise bout [38, 39]. However, these results were in human PBMCs, and given that NQO1 protein content increased in response to stimulation in the current investigation, the gene expression may occur earlier in skeletal muscle than in PBMCs. Therefore, future research will utilize different timepoints to assess gene expression changes.
Our results show minimal changes to GSR, and slight but significant decreases in GCLC protein in response to muscle stimulation. The decrease in GCLC here is likely a temporal effect, where the acute stimulation increases proteasomal activity causing decreases in constitutively expressed proteins like GCLC, and the increase in protein likely occurs after the increase in mRNA expression. We have shown that GCLC protein increases significantly eight hours after a single bout of exercise, which is dependent on the increases in mRNA at 1 and 4 hours after completion of the exercise bout [39]. Therefore, increasing the time of skeletal muscle harvest would likely capture greater GCLC protein accretion in response to the exercise bout.
In contrast to GCLC protein expression, HO1 and NQO1 protein increased, albeit in different patterns, in response to muscle contraction. The elevated HO1 expression in both limbs of the high intensity group mirrors the activation of Nrf2 and suggests that HO1 protein expression is rapidly increased and intensity dependent. HO1 mRNA induction has also been shown to be intensity or dose dependent in human skeletal muscle, supporting these findings [41]. NQO1 protein was inducible in response to muscle stimulation in both high and low intensity muscle stimulation, with no significant differences between groups. NQO1 induction in response to exercise has been demonstrated in other reports [42] and is highly dependent on Nrf2 activation [43]. The fact that NQO1 mRNA was not statistically significant but the protein was, suggests that the time course for NQO1 mRNA accretion and protein is shifted closer to the exercise bout than GCLC mRNA. This poses an interesting paradox where some redox stress response genes are early response genes, while others may be late response genes. This may be due to other inducible transcription factors and inhibitors differentially regulating phase II antioxidant genes. For example, NF-kB is known to also regulate GCLC/M gene transcripts [44] which could be responsible for the discrepancies between gene transcripts and protein accumulation in our results.
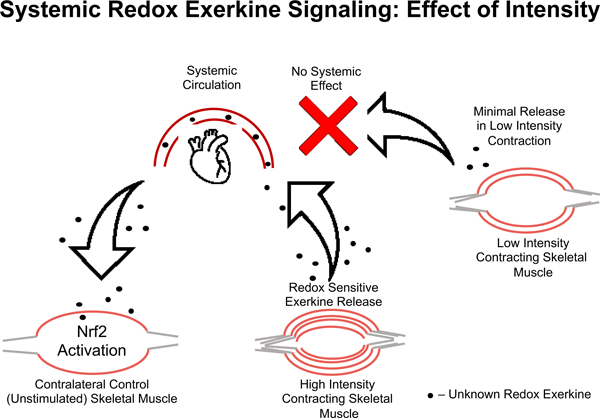
Taken together, these data suggest that there is an intensity gated redox sensitive exerkine released from high intensity contraction, but not low intensity contraction, that is driving these divergent Nrf2 signaling events in response to high and low intensity exercise, respectively. Recent work has shown that pH gated release of succinate is a myokine involved in adaptive responses to exercise [45]. Others have shown that peroxiredoxins and thioredoxins are released into the blood plasma compartment in an intensity and time dependent manner in response to high intensity, but not moderate intensity exercise in humans [26]. While we cannot ascertain what exerkine is involved with our current results, the literature suggest that intensity gating mechanisms may explain the divergent effects seen in molecular responses to exercise, and that these mechanisms may be redox dependent [24, 26, 46]. This intensity gated systemic redox signaling model is illustrated in Figure 6, where the signal is released into the blood stream in response to the high intensity exercise and activates Nrf2 signaling in the contralateral unstimulated muscle, while low intensity muscle stimulation does not meet the threshold to propagate a systemic redox signaling response.
Figure 6. Model of systemic redox exerkine signaling is gated by muscle stimulation intensity.
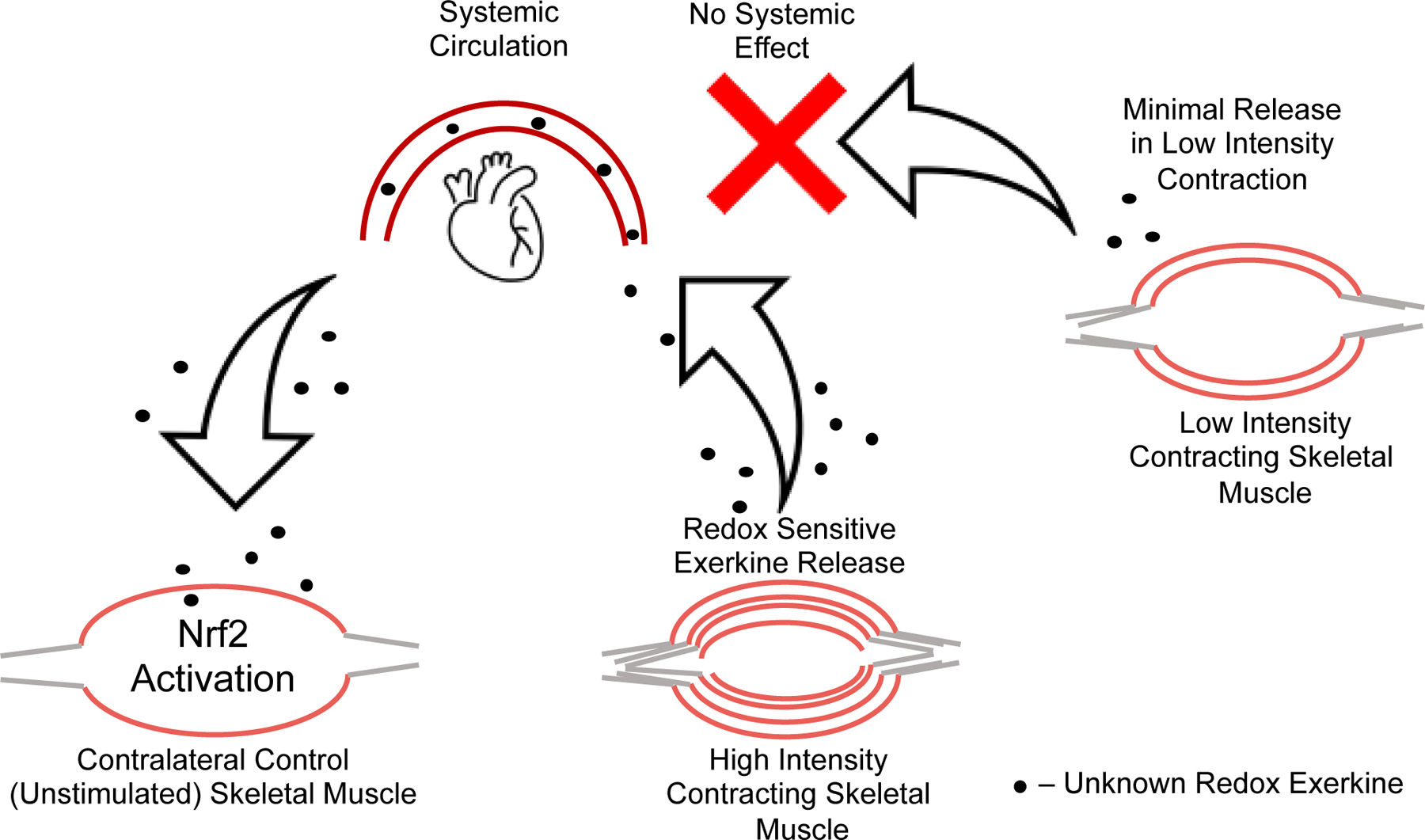
High intensity muscle contraction induces release of a redox active exerkine that travels through the blood stream and acts on unstimulated skeletal muscle. This effect is not seen in low intensity contralateral control muscle, suggesting the redox exerkine release from skeletal muscle is intensity gated. In other words, this redox exerkine is only released upon stimuli above a certain stress threshold or is released at lower intensities but not at sufficient concentrations to cause signaling effects in other tissues.
While we speculate a novel redox exerkine is driving these divergent intensity effects, it is important to point out alternative hypotheses. Since Nrf2 is a stress response transcription factor, it is also possible that the systemic effects are driven by release of stress hormones not directly related to redox signaling [47]. For example, circulating neuro-hormonal effects or cytokines could cause an indirect activation of general stress response pathways that include but are not limited to Nrf2 activation. Thyroid hormone [48], and catecholamines [49] can induce intracellular ROS production through increases in metabolism and could potentially be mediating the effects seen here. In addition, cytokines like TNFα, IFNγ, IL-1β, and IL-6 induce ROS production intracellularly [50], however only IL-6 is responsive to exercise intensity [51]. Even though IL-6 can affect redox balance, it seems to actually decrease Nrf2 and related gene expression in skeletal muscle, so this might not be the exerkine responsible [52]. Overall, the systemic signaling effects of different exercise intensities is an interesting and important field that disserves attention.
Strengths, Limitations, and Future Directions
The strength of the current study is the utilization of multiple controls to assess skeletal muscle redox signaling: Internal contralateral unstimulated control limbs compared to stimulated limbs, as well as Naïve Control animals with control limbs and “mock” stimulated control limbs. The stimulation versus contralateral control limb is a valuable model for understanding systemic effects of exercise within-animal or human. This model has been used to show unilateral strength training improves strength gains in contralateral muscles [53], which is slightly different than our acute model of stimulation. An additional strength of this model is the ability to control for the muscle tested, and duration and intensity of the exercise bout, but our interpretations are limited to the effects of unilateral fatiguing muscle contractions. More studies are necessary to determine if muscle fatigue is necessary for increased Nrf2 signaling, and if similar effects could be induced by unilateral voluntary muscle contractions in humans.
Humans have the ability to perform a similar exercise (isometric plantarflexion) voluntarily, and this increases the translatability to humans as an exercise more than other exercises performed by rodents in the literature such as running [54]. The two main differences of this exercise model and humans are the need for anesthesia and electrical stimulation. For this reason, we tested if anesthesia was a confounder by measuring Nrf2 in naïve controls that were anesthetized without muscle stimulation and found that Nrf2 signaling was not upregulated by anesthesia. A follow-up human study can be used to determine whether muscle contraction by electrical stimulation or voluntary would induce different effects on Nrf2 signaling. Previous reports show that electrical stimulation leads to greater neuromuscular fatigue, blood lactate, growth hormone, and muscle soreness than voluntary contractions of the same duration and intensity [55]. Others have used high and low intensity stimulation to mimic resistance and endurance exercise in rat hindlimbs and found divergent effects in other signaling pathways using high and low intensity stimulation protocols [56]. Despite the utility of this model, it remains an underutilized approach for understanding signaling effects of exercise, particularly redox signaling.
One limitation is the redox balance measures in this cohort. Total glutathione was measured, but after partitioning the sample for each assay there was not enough left to perform any additional measures of NAD+/NADH ratio, ROS production assays, or other redox balance measures. While additional redox balance markers would provide a more complete picture, perhaps more relevant to the current study is thiol redox status in target proteins. Future work should investigate protein thiol redox state in key signaling proteins like Keap1 in response to exercise. It is well known that Keap1 thiols are oxidized in response to oxidative stress, and that these modifications activate Nrf2. However, we are unaware of any study that has directly investigated the redox status of Keap1 protein thiols in response to exercise specifically. In addition to the systemic effects, the effects measured in this study may be muscle specific, as work in the field has demonstrated some antioxidant expression levels are different in different types of muscles [35, 57]. It is possible that these systemic effects are driven by one type of muscle over another given the previous work in the field. However, it is still unclear how redox signaling mechanisms are being propagated through Nrf2 in peripheral tissues.
Future investigations should aim to identify the exerkine responsible for systemic signaling effects in the inducible redox stress response signaling system. In these studies that demonstrate exercise induced Nrf2 activation in peripheral tissues like nervous tissue, PBMCs, and lung tissue, and the current investigation, it is unlikely that superoxide and hydrogen peroxide are viable secondary messenger signaling candidates given their half-life and concentration of extracellular enzymes capable of quenching these signals. The prerequisite for this specific redox myokine is that it is stable enough to travel through the bloodstream and affect other peripheral tissues, including skeletal muscle. It is also possible that there is a relay of some kind, where the exerkine can bind to a surface receptor on a neighboring cell that initiates an intracellular signaling cascade which elicits ROS production. The sheer number of possible candidate-myokines including proteins, lipids, RNA, exosomes, and microRNAs that could be responsible for the peripheral redox signaling makes this a difficult task. These findings are consistent with previous literature on high intensity exercise eliciting different molecular signaling cascades than low intensity exercise and illustrate an interesting dichotomy between low intensity and high intensity exercise. However, this is the first report that we are aware of demonstrating Nrf2 activation in response to high intensity but not low intensity exercise in a contralateral non-exercised muscle. These findings provide exciting future directions to unveil the novel signaling mechanisms highlighted here.
Conclusions
This pilot study set out to test the effects of muscle contraction intensity on Nrf2-mediated redox signaling. The current study design demonstrates a muscle contraction induced systemic redox signaling that appears to be intensity gated; where contralateral unstimulated muscles are responding to high intensity, but not low intensity stimulation. These effects are exerted in part on the Keap1-Nrf2-ARE signaling axis, as well as downstream gene and protein expression for the major Nrf2 targets: NQO1 and HO1 protein. While these data are still preliminary, follow-up experiments are being designed to confirm these findings, including discovering the type of molecule or set of molecules responsible for these systemic redox signaling effects.
Highlights.
Muscle stimulation decreases Keap1 protein and increases Nrf2-ARE binding
High intensity muscle stimulation activates Nrf2-ARE binding and decreases in Keap1 protein in the contralateral control limb
NQO1 protein is responsive to muscle stimulation regardless of intensity
These data suggest an intensity gated redox sensitive exerkine
Acknowledgments
Funding
This project was funded by the NIH P01 AG001751 to DJM and R15 AG055077 to TT.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Competing Interest
The authors report no conflict of interest.
References
- [1].Done AJ, Traustadóttir T, Nrf2 mediates redox adaptations to exercise, Redox Biol 10 (2016) 191–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Merry TL, Ristow M, Nuclear factor erythroid-derived 2-like 2 (NFE2L2, Nrf2) mediates exercise-induced mitochondrial biogenesis and the anti-oxidant response in mice, J Physiol 594(18) (2016) 5195–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Merry TL, MacRae C, Pham T, Hedges CP, Ristow M, Deficiency in ROS-sensing nuclear factor erythroid 2-like 2 causes altered glucose and lipid homeostasis following exercise training, American Journal of Physiology-Cell Physiology 318(2) (2019) C337–C345. [DOI] [PubMed] [Google Scholar]
- [4].Nikolaidis MG, Margaritelis NV, Matsakas A, Quantitative Redox Biology of Exercise, International journal of sports medicine (2020). [DOI] [PubMed] [Google Scholar]
- [5].Kramer PA, Duan J, Qian W-J, Marcinek DJ, The Measurement of Reversible Redox Dependent Post-translational Modifications and Their Regulation of Mitochondrial and Skeletal Muscle Function, Frontiers in physiology 6 (2015) 347–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Cobley JN, Husi H, Immunological Techniques to Assess Protein Thiol Redox State: Opportunities, Challenges and Solutions, Antioxidants (Basel, Switzerland) 9(4) (2020) 315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Stretton C, Pugh JN, McDonagh B, McArdle A, Close GL, Jackson MJ, 2-Cys Peroxiredoxin oxidation in response to Hydrogen Peroxide and contractile activity in skeletal muscle: A novel insight into exercise-induced redox signalling?, Free Radical Bio Med (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Campbell MD, Duan J, Samuelson AT, Gaffrey MJ, Merrihew GE, Egertson JD, Wang L, Bammler TK, Moore RJ, White CC, Kavanagh TJ, Voss JG, Szeto HH, Rabinovitch PS, MacCoss MJ, Qian WJ, Marcinek DJ, Improving mitochondrial function with SS-31 reverses age-related redox stress and improves exercise tolerance in aged mice, Free Radic Biol Med 134 (2019) 268–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].McDonagh B, Sakellariou GK, Smith NT, Brownridge P, Jackson MJ, Differential Cysteine Labeling and Global Label-Free Proteomics Reveals an Altered Metabolic State in Skeletal Muscle Aging, Journal of Proteome Research 13(11) (2014) 5008–5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Piantadosi CA, Suliman HB, Redox regulation of mitochondrial biogenesis, Free Radic Biol Med 53(11) (2012) 2043–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Irrcher I, Ljubicic V, Hood DA, Interactions between ROS and AMP kinase activity in the regulation of PGC-1α transcription in skeletal muscle cells, American Journal of Physiology-Cell Physiology 296(1) (2009) C116–C123. [DOI] [PubMed] [Google Scholar]
- [12].Gomez-Cabrera MC, Borras C, Pallardo FV, Sastre J, Ji LL, Vina J, Decreasing xanthine oxidase-mediated oxidative stress prevents useful cellular adaptations to exercise in rats, J Physiol 567(Pt 1) (2005) 113–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Gomez-Cabrera MC, Domenech E, Romagnoli M, Arduini A, Borras C, Pallardo FV, Sastre J, Vina J, Oral administration of vitamin C decreases muscle mitochondrial biogenesis and hampers training-induced adaptations in endurance performance, Am J Clin Nutr 87(1) (2008) 142–9. [DOI] [PubMed] [Google Scholar]
- [14].Ristow M, Zarse K, Oberbach A, Kloting N, Birringer M, Kiehntopf M, Stumvoll M, Kahn CR, Bluher M, Antioxidants prevent health-promoting effects of physical exercise in humans, Proc Natl Acad Sci U S A 106(21) (2009) 8665–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Wang P, Li CG, Qi Z, Cui D, Ding S, Acute exercise stress promotes Ref1/Nrf2 signalling and increases mitochondrial antioxidant activity in skeletal muscle, Exp Physiol 101(3) (2016) 410–20. [DOI] [PubMed] [Google Scholar]
- [16].Li T, He S, Liu S, Kong Z, Wang J, Zhang Y, Effects of different exercise durations on Keap1-Nrf2-ARE pathway activation in mouse skeletal muscle, Free Radic Res 49(10) (2015) 1269–74. [DOI] [PubMed] [Google Scholar]
- [17].Gallego-Selles A, Martin-Rincon M, Martinez-Canton M, Perez-Valera M, Martín-Rodríguez S, Gelabert-Rebato M, Santana A, Morales-Alamo D, Dorado C, Calbet JAL, Regulation of Nrf2/Keap1 signalling in human skeletal muscle during exercise to exhaustion in normoxia, severe acute hypoxia and post-exercise ischaemia: Influence of metabolite accumulation and oxygenation, Redox Biology 36 (2020) 101627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Burgomaster KA, Howarth KR, Phillips SM, Rakobowchuk M, MacDonald MJ, McGee SL, Gibala MJ, Similar metabolic adaptations during exercise after low volume sprint interval and traditional endurance training in humans, The Journal of Physiology 586(1) (2008) 151–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].MacInnis MJ, Gibala MJ, Physiological adaptations to interval training and the role of exercise intensity, J Physiol 595(9) (2017) 2915–2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Gibala MJ, Hawley JA, Sprinting Toward Fitness, Cell Metab 25(5) (2017) 988–990. [DOI] [PubMed] [Google Scholar]
- [21].Chavanelle V, Boisseau N, Otero YF, Combaret L, Dardevet D, Montaurier C, Delcros G, Peltier SL, Sirvent P, Effects of high-intensity interval training and moderate-intensity continuous training on glycaemic control and skeletal muscle mitochondrial function in db/db mice, Scientific Reports 7(1) (2017) 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ramos JS, Dalleck LC, Tjonna AE, Beetham KS, Coombes JS, The Impact of High-Intensity Interval Training Versus Moderate-Intensity Continuous Training on Vascular Function: a Systematic Review and Meta-Analysis, Sports Medicine 45(5) (2015) 679–692. [DOI] [PubMed] [Google Scholar]
- [23].Guseh JS, Churchill TW, Yeri A, Lo C, Brown M, Houstis NE, Aragam KG, Lieberman DE, Rosenzweig A, Baggish AL, An expanded repertoire of intensity-dependent exercise-responsive plasma proteins tied to loci of human disease risk, Scientific Reports 10(1) (2020) 10831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Done AJ, Newell MJ, Traustadóttir T, Effect of exercise intensity on Nrf2 signalling in young men, Free Radic Res 51(6) (2017) 646–655. [DOI] [PubMed] [Google Scholar]
- [25].Islam H, Bonafiglia JT, Turnbull PC, Simpson CA, Perry CGR, Gurd BJ, The impact of acute and chronic exercise on Nrf2 expression in relation to markers of mitochondrial biogenesis in human skeletal muscle, Eur J Appl Physiol 120(1) (2020) 149–160. [DOI] [PubMed] [Google Scholar]
- [26].Wadley AJ, Keane G, Cullen T, James L, Vautrinot J, Davies M, Hussey B, Hunter DJ, Mastana S, Holliday A, Petersen SV, Bishop NC, Lindley MR, Coles SJ, Characterization of extracellular redox enzyme concentrations in response to exercise in humans, J Appl Physiol (1985) 127(3) (2019) 858–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Margaritelis NV, Theodorou AA, Paschalis V, Veskoukis AS, Dipla K, Zafeiridis A, Panayiotou G, Vrabas IS, Kyparos A, Nikolaidis MG, Adaptations to endurance training depend on exercise-induced oxidative stress: exploiting redox interindividual variability, Acta physiologica (Oxford, England) 222(2) (2018). [DOI] [PubMed] [Google Scholar]
- [28].Kramer PA, Duan J, Gaffrey MJ, Shukla AK, Wang L, Bammler TK, Qian WJ, Marcinek DJ, Fatiguing contractions increase protein S-glutathionylation occupancy in mouse skeletal muscle, Redox Biol 17 (2018) 367–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Thomas KC, Zheng XF, Garces Suarez F, Raftery JM, Quinlan KGR, Yang N, North KN, Houweling PJ, Evidence Based Selection of Commonly Used RT-qPCR Reference Genes for the Analysis of Mouse Skeletal Muscle, PLOS ONE 9(2) (2014) e88653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Giordano G, White CC, Costa LG, Assessment of glutathione homeostasis, Methods Mol Biol 758 (2011) 205–14. [DOI] [PubMed] [Google Scholar]
- [31].Gounder SS, Kannan S, Devadoss D, Miller CJ, Whitehead KJ, Odelberg SJ, Firpo MA, Paine R 3rd, Hoidal JR, Abel ED, Rajasekaran NS, Impaired transcriptional activity of Nrf2 in age-related myocardial oxidative stress is reversible by moderate exercise training, PloS one 7(9) (2012) e45697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Miller CJ, Gounder SS, Kannan S, Goutam K, Muthusamy VR, Firpo MA, Symons JD, Paine R 3rd, Hoidal JR, Rajasekaran NS, Disruption of Nrf2/ARE signaling impairs antioxidant mechanisms and promotes cell degradation pathways in aged skeletal muscle, Biochim Biophys Acta 1822(6) (2012) 1038–50. [DOI] [PubMed] [Google Scholar]
- [33].Muthusamy VR, Kannan S, Sadhaasivam K, Gounder SS, Davidson CJ, Boeheme C, Hoidal JR, Wang L, Rajasekaran NS, Acute exercise stress activates Nrf2/ARE signaling and promotes antioxidant mechanisms in the myocardium, Free Radic Biol Med 52(2) (2012) 366–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Tsou YH, Shih CT, Ching CH, Huang JY, Jen CJ, Yu L, Kuo YM, Wu FS, Chuang JI, Treadmill exercise activates Nrf2 antioxidant system to protect the nigrostriatal dopaminergic neurons from MPP+ toxicity, Exp Neurol 263 (2015) 50–62. [DOI] [PubMed] [Google Scholar]
- [35].Yamada M, Iwata M, Warabi E, Oishi H, Lira VA, Okutsu M, p62/SQSTM1 and Nrf2 are essential for exercise-mediated enhancement of antioxidant protein expression in oxidative muscle, Faseb j 33(7) (2019) 8022–8032. [DOI] [PubMed] [Google Scholar]
- [36].Aguiar AS Jr., Duzzioni M, Remor AP, Tristao FS, Matheus FC, Raisman-Vozari R, Latini A, Prediger RD, Moderate-Intensity Physical Exercise Protects Against Experimental 6-Hydroxydopamine-Induced Hemiparkinsonism Through Nrf2-Antioxidant Response Element Pathway, Neurochem Res 41(1–2) (2016) 64–72. [DOI] [PubMed] [Google Scholar]
- [37].Crilly MJ, Tryon LD, Erlich AT, Hood DA, The role of Nrf2 in skeletal muscle contractile and mitochondrial function, Journal of applied physiology 121(3) (2016) 730–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Done AJ, Gage MJ, Nieto NC, Traustadóttir T, Exercise-induced Nrf2-signaling is impaired in aging, Free Radic Biol Med 96 (2016) 130–8. [DOI] [PubMed] [Google Scholar]
- [39].Ostrom EL, Traustadóttir T, Aerobic exercise training partially reverses the impairment of Nrf2 activation in older humans, Free Radical Biology and Medicine 160 (2020) 418–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Safdar A, deBeer J, Tarnopolsky MA, Dysfunctional Nrf2-Keap1 redox signaling in skeletal muscle of the sedentary old, Free Radic Biol Med 49(10) (2010) 1487–93. [DOI] [PubMed] [Google Scholar]
- [41].Pilegaard H, Ordway GA, Saltin B, Neufer PD, Transcriptional regulation of gene expression in human skeletal muscle during recovery from exercise, American Journal of Physiology-Endocrinology and Metabolism 279(4) (2000) E806–E814. [DOI] [PubMed] [Google Scholar]
- [42].Horie M, Warabi E, Komine S, Oh S, Shoda J, Cytoprotective Role of Nrf2 in Electrical Pulse Stimulated C2C12 Myotube, PloS one 10(12) (2015) e0144835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Dhakshinamoorthy S, Jaiswal AK, Functional characterization and role of INrf2 in antioxidant response element-mediated expression and antioxidant induction of NAD(P)H:quinone oxidoreductase1 gene, Oncogene 20(29) (2001) 3906–3917. [DOI] [PubMed] [Google Scholar]
- [44].Zhou L, Forman HJ, Zhang H, Aging of the antioxidant/inflammatory axis in human lung epithelial cells in vitro mimics aging in animal studies, The FASEB Journal 30(S1) (2016) 1296.3–1296.3. [Google Scholar]
- [45].Reddy A, Bozi LHM, Yaghi OK, Mills EL, Xiao H, Nicholson HE, Paschini M, Paulo JA, Garrity R, Laznik-Bogoslavski D, Ferreira JCB, Carl CS, Sjøberg KA, Wojtaszewski JFP, Jeppesen JF, Kiens B, Gygi SP, Richter EA, Mathis D, Chouchani ET, pH-Gated Succinate Secretion Regulates Muscle Remodeling in Response to Exercise, Cell (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Wadley AJ, Aldred S, Coles SJ, An unexplored role for Peroxiredoxin in exercise-induced redox signalling?, Redox Biol 8 (2016) 51–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Parker L, Shaw CS, Stepto NK, Levinger I, Exercise and Glycemic Control: Focus on Redox Homeostasis and Redox-Sensitive Protein Signaling, Frontiers in Endocrinology 8(87) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Villanueva I, Alva-Sánchez C, Pacheco-Rosado J, The role of thyroid hormones as inductors of oxidative stress and neurodegeneration, Oxidative medicine and cellular longevity 2013 (2013) 218145–218145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Just A, Olson AJM, Whitten CL, Arendshorst WJ, Superoxide mediates acute renal vasoconstriction produced by angiotensin II and catecholamines by a mechanism independent of nitric oxide, American Journal of Physiology-Heart and Circulatory Physiology 292(1) (2007) H83–H92. [DOI] [PubMed] [Google Scholar]
- [50].Yang D, Elner SG, Bian Z-M, Till GO, Petty HR, Elner VM, Pro-inflammatory cytokines increase reactive oxygen species through mitochondria and NADPH oxidase in cultured RPE cells, Experimental eye research 85(4) (2007) 462–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Scott JPR, Sale C, Greeves JP, Casey A, Dutton J, Fraser WD, Effect of Exercise Intensity on the Cytokine Response to an Acute Bout of Running, Medicine & Science in Sports & Exercise 43(12) (2011). [DOI] [PubMed] [Google Scholar]
- [52].Forcina L, Miano C, Scicchitano BM, Rizzuto E, Berardinelli MG, De Benedetti F, Pelosi L, Musarò A, Increased Circulating Levels of Interleukin-6 Affect the Redox Balance in Skeletal Muscle, Oxidative Medicine and Cellular Longevity 2019 (2019) 3018584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Munn J, Herbert RD, Gandevia SC, Contralateral effects of unilateral resistance training: a meta-analysis, J Appl Physiol (1985) 96(5) (2004) 1861–6. [DOI] [PubMed] [Google Scholar]
- [54].Fuller KNZ, Thyfault JP, Barriers in translating preclinical rodent exercise metabolism findings to human health, J Appl Physiol (1985) 130(1) (2021) 182–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Jubeau M, Sartorio A, Marinone PG, Agosti F, Van Hoecke J, Nosaka K, Maffiuletti NA, Comparison between voluntary and stimulated contractions of the quadriceps femoris for growth hormone response and muscle damage, J Appl Physiol (1985) 104(1) (2008) 75–81. [DOI] [PubMed] [Google Scholar]
- [56].Atherton PJ, Babraj J, Smith K, Singh J, Rennie MJ, Wackerhage H, Selective activation of AMPK-PGC-1alpha or PKB-TSC2-mTOR signaling can explain specific adaptive responses to endurance or resistance training-like electrical muscle stimulation, Faseb J 19(7) (2005) 786–8. [DOI] [PubMed] [Google Scholar]
- [57].Powers SK, Criswell D, Lawler J, Ji LL, Martin D, Herb RA, Dudley G, Influence of exercise and fiber type on antioxidant enzyme activity in rat skeletal muscle, Am J Physiol 266(2 Pt 2) (1994) R375–80. [DOI] [PubMed] [Google Scholar]


