Summary
The epidermis is a stratified epithelium in which structural and functional features are polarized across multiple cell layers. This type of polarity is essential for establishing the epidermal barrier, but how it is created and sustained is poorly understood. Previous work identified a role for the classical cadherin/filamentous-actin network in establishment of epidermal polarity. However, little is known about potential roles of the most prominent epidermal intercellular junction, the desmosome, in establishing epidermal polarity, in spite of the fact that desmosome constituents are patterned across the apical to basal cell layers. Here, we show that desmosomes and their associated intermediate filaments (IF) are key regulators of mechanical polarization in epidermis, whereby basal and suprabasal cells experience different forces that drive layer-specific functions. Uncoupling desmosomes and IF or specific targeting of apical desmosomes through depletion of the superficial desmosomal cadherin, desmoglein 1, impedes basal stratification in an in vitro competition assay and suprabasal tight junction barrier functions in 3D reconstructed epidermis. Surprisingly, disengaging desmosomes from IF also accelerated the expression of differentiation markers, through precocious activation of the mechanosensitive transcriptional regulator serum response factor (SRF) and downstream activation of Epidermal Growth Factor Receptor family member ErbB2 by Src family kinase (SFK) mediated phosphorylation. This Dsg1-SFK-ErbB2 axis also helps maintain tight junctions and barrier function later in differentiation. Together, these data demonstrate that the desmosome-IF network is a critical contributor to the cytoskeletal-adhesive machinery that supports the polarized function of the epidermis.
eTOC blurb:
Broussard et al. provide evidence that the desmosome/intermediate filament linkage contributes to the establishment of a mechanically and biochemically polarized epidermis. They implicate signaling modules including SRF, SFKs, and ErbB2 as mediators of desmosome-dependent coordination of cell movement, differentiation, and tight junction barrier.
Introduction
To transition from single celled organisms to multicellular metazoans, cells evolved multiple mechanisms to form stabilized cell-cell interactions1. Ultimately, this process led to the development of simple adherent epithelial sheets, which serve as a protective barrier between an organism and its environment. Additional epithelial and junctional complexity arose later in evolution, with the emergence of multilayered, stratified epithelia, including elaborate tissues like the epidermis2. In vertebrates, this multilayered epithelium serves as a barrier against water loss, mechanical insults, and as a physical and immune barrier to external pathogens (containing both innate and adaptive immune elements)3,4. The epidermis comprises four distinct layers, basal, spinous, granular (SG), and cornified, which undergo constant regeneration. Regeneration occurs through a process whereby keratinocytes in the basal layer proliferate, at some point commit to a program of terminal differentiation, and exit the basal layer. During differentiation, cells maintain cell-cell interactions while moving progressively into the suprabasal layers where they are incorporated into the epidermal barrier and ultimately slough off of the skin’s surface.
Polarity, the differential patterning of structural and functional features along an axis, is a fundamental property of all epithelial tissues and is essential to their function. Whereas simple epithelia exhibit apical to basal (apicobasal) polarization within a single cell layer, the epidermis exhibits polarization across multiple cell layers, through poorly understood mechanisms2. For example, barrier-essential tight junctions (TJs) are found in association with adherens junctions on the apicolateral surface of simple epithelia. In the epidermis, functional TJs localize in association with adherens junctions to the second of three SG layers5. Improper development or disturbed maintenance of the epidermal TJ barrier is associated with a variety of inflammatory skin diseases (e.g., atopic dermatitis and psoriasis), impaired wound healing, and cancer6. In addition, simple epithelia exhibit a polarization of intercellular forces, with a region of high tension generated by actomyosin near the apical surface7. This region of high tension can regulate tissue morphology, homeostasis, and barrier function7. Moreover, loss of polarized mechanical tension is associated with cancer morphogenesis in multiple types of epithelial tissues8. Recent work has also identified a stiffness gradient in both human and mouse epidermis, with progressive stiffening from basal to superficial layers9. However, how such a mechanical asymmetry contributes to polarized cell signaling and behavior is not well understood.
Cell-cell adhesion complexes and their associated cytoskeletal networks are prime candidates to regulate polarized tissue mechanics. Desmosomes are calcium-dependent, intercellular adhesions that link to the intermediate filament (IF) cytoskeleton. Desmosomes comprise three main protein families: transmembrane cadherins (desmogleins and desmocollins), armadillo proteins (plakophilins and plakoglobin), and plakin proteins (desmoplakin, DP). Cadherins of adjacent cells form trans-interactions to mediate cell-cell adhesion. Intracellularly, cadherins interact with armadillo proteins, which are coupled to IF through DP. In the epidermis, desmosomal cadherins display graded patterns of expression. Some, such as desmoglein 3 (Dsg3), are concentrated in the proliferating basal layer, while others, such as desmoglein 1 (Dsg1), are concentrated in the superficial layers. This is in contrast to the adherens junction component E-cadherin, which is expressed throughout the epidermis10.
A role for filamentous-actin (F-actin)-based adhesive networks in regulating mechanics (tension and stiffness) in conjunction with adherens junctions has been well-established11,12. However, the contributions of the desmosome/IF network in regulating cell mechanics are not as well understood. Our previous work elucidated a role for desmosomes in mechanical regulation by modulating the interaction between desmosomes and IF using mutant forms of DP in a single layered epithelial system. Through a combination of micropillar arrays and atomic force microscopy (AFM), we demonstrated that strengthening DP-IF interactions increased cell-cell tensional forces and cell stiffness, while disrupting the interaction decreased these forces12. Moreover, these effects required cooperation with F-actin-based networks, indicating that desmosome/IF and F-actin-based networks function synergistically to regulate cell mechanics, highlighting their integrated nature. How these integrated networks work together to drive morphogenetic events in more complex tissues is not well understood.
The patterned expression of the desmosome/IF network suggests their potential role in governing the differential biophysical properties of distinct epidermal layers. Here, we provide evidence that the desmosome/IF linkage governs the ability of the epidermis to adopt a highly polarized mechanical phenotype. Disruption of this linkage alters indicators of mechanical asymmetries and the normal polarized functions of the epidermis including stratification, differentiation, and the establishment of an intact epidermal barrier. Finally, we identify a putative, mechanically-sensitive serum response factor (SRF)-Src family kinase (SFK)-ErbB2 signaling axis through which the desmosome/IF linkage promotes these polarized epidermal functions.
Results
Uncoupling the desmosome/IF connection induces rearrangements of cytoskeletal/adhesive complexes in epidermal keratinocytes.
To establish models for addressing the role of the desmosome/IF network in stratifying epithelia, we performed loss-of-function experiments utilizing a well-characterized mutant of the cytolinker protein DP in two established submerged models of neonatal human epidermal keratinocytes (NHEKs). DP provides the connection between desmosomes and the IF cytoskeleton (Figure 1A). The mutant DPNTP consists of the first 584 amino acids of DP (Figure 1A). It retains interactions with the desmosomal core but lacks the IF-binding domains, acting as a dominant-negative mutant to uncouple desmosomes from IF12–14. We previously showed that DPNTP expression in simple epithelial A431 cells results in retraction of IF from sites of cell-cell adhesion12.
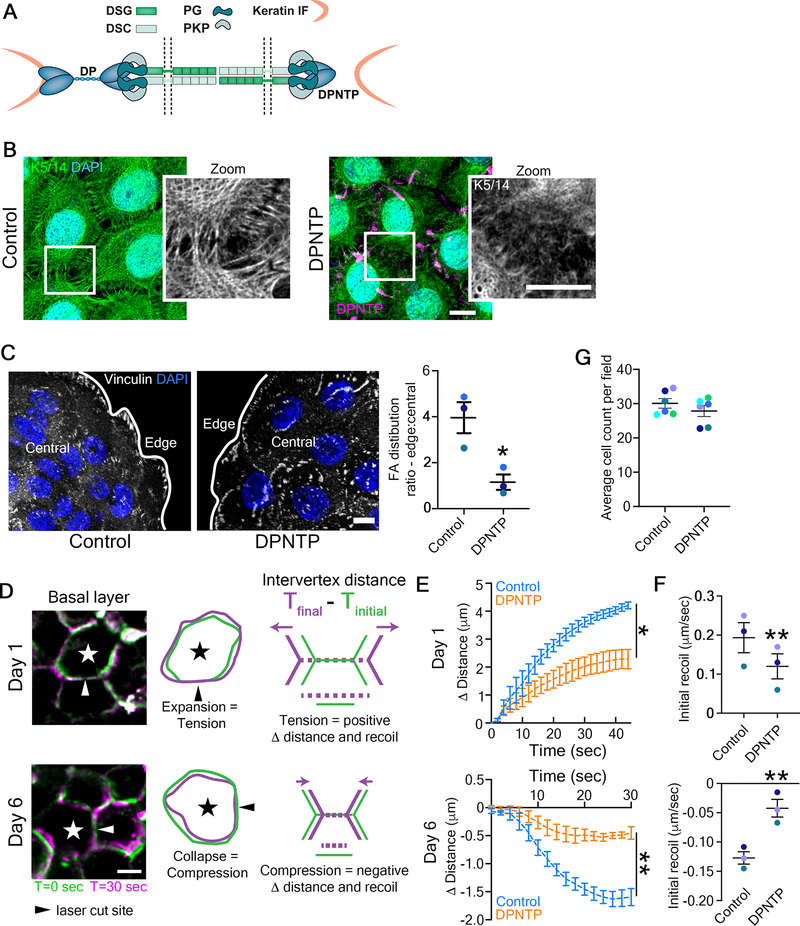
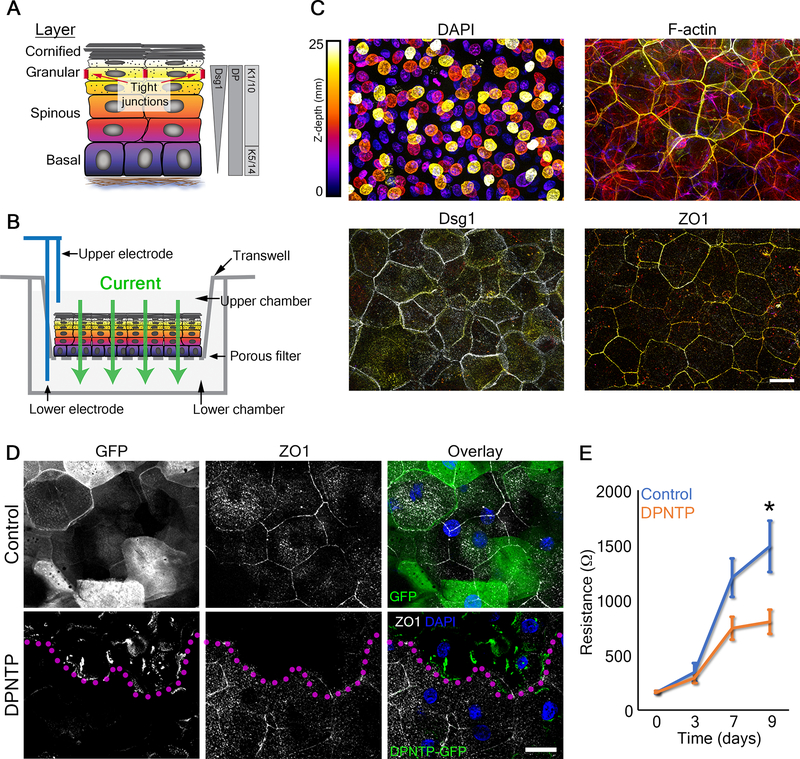
Figure 1. Uncoupling the desmosome/IF connection induces rearrangements of cytoskeletal/adhesive complexes.
A) Schematic depicts the interactions among the desmosomal core proteins and the intermediate filament (IF) cytoskeleton in both a control condition (left) as well as when the desmosomal core is uncoupled from IF (right). DP, desmoplakin; PKP, plakophilin; PG, plakoglobin; DSG, desmoglein; DSC, desmocollin.
B) Maximum projection micrographs show immunofluorescence staining of the keratin 5/14 (K5/14) IF cytoskeleton in basal cells of differentiating monolayers of NHEKs expressing either GFP (Control) or DPNTP-GFP. K5/14 is pseudocolored green and DPNTP is shown in magenta. DAPI indicates nuclei in blue and bar is 10 μm.
C) Left, maximum projection micrographs show immunofluorescence staining of vinculin at the basal/substrate interface of differentiating NHEK ephrin colonies expressing either GFP (Control) or DPNTP-GFP. DAPI indicates nuclei in blue and bar is 10 μm. Right, the average vinculin-positive area per cell was quantified for both cells on the edge and in the central region of the colony. The average ratio (dashed line) of edge to central cells is shown for control and DPNTP-expressing colonies. *p=0.0233, paired t-test from three independent experiments, error bars are SEM.
D) Images show the membranes (indicated by myr-tomato) of basal cells within epidermal equivalent cultures before and after laser ablation at the indicated times. At day 1, cells expand upon ablation, indicating tension within the layer and corresponding to a positive value for measured intervertex distance and initial recoil velocity. At day 6, cells collapse upon ablation, indicating compression within the layer and corresponding to a negative value for measured intervertex distance and initial recoil velocity. Bar is 5 μm. Related to Video S1.
E) Quantification is shown for the average change in intervertex distance (Δ Distance) over time for basal cells after ablation within control and DPNTP-expressing epidermal equivalent cultures both at day 1 and day 6 of their development. *p=0.0361, **p=0.0058, two-way ANOVA with repeated measures from three independent experiments, error bars are SEM.
F) Quantification is shown for the average initial recoil velocity after ablation for basal cells within control and DPNTP-expressing epidermal equivalent cultures both at day 1 (upper graph) and day 6 (lower graph) of their development. **p<0.009, paired t-test from three independent experiments, error bar is SEM.
G) The average cell count per field is shown for control and DPNTP-expressing epidermal equivalent cultures both at day 6 of their development. Dashed line is mean of 6 independent experiments, error bars are SEM.
See also Figure S1.
We first expressed DPNTP (GFP-tagged) in NHEK monolayers induced to differentiate and stratify with high calcium containing medium (calcium switch)15 (Figure S1A). After 2 days, keratin IFs closely adjoin the cell-cell interface anchored by functional desmosomes (Figure 1B). Compared with cytoplasmic GFP controls, DPNTP expression in NHEKs resulted in loss of orthogonally-anchored keratin IF bundles, indicating successful desmosome/IF uncoupling (Figure 1B).
In tissues, complex patterns of cell-cell and cell-substrate forces are balanced and both contribute to epithelial tissue morphogenesis16. We previously showed that uncoupling the desmosome/IF connection in A431 cells was accompanied by a significant reduction in cell-cell junctional forces12. DPNTP expression in ephrin-induced colonies of NHEKs resulted in an altered distribution of cell-substrate contacts (Figure S1B). In controls, vinculin-containing focal adhesions are enriched near the colony edge (Figure 1C). However, DPNTP altered this staining pattern with focal adhesions scattered throughout the basal surface of the colony, significantly decreasing their enrichment at the colony edge (Figure 1C). These results suggested a mechanical cell-cell to cell-substrate switch17,18, consistent with previous reports where loss of cell–cell adhesion increased cell–substrate adhesion17–19.
Uncoupling the desmosome/IF connection alters the mechanical properties of epidermal keratinocytes.
Since mechanical forces regulate cell behavior, their polarized distribution may drive segregation of tissue functions. Tensile (pulling) and compressive (pushing) forces affect cell behavior within epithelia, regulating cell division, migration, tissue morphogenesis, and cancer cell invasion20,21. To test the possibility that the desmosome/IF complex controls the mechanical properties of epidermal keratinocytes during development of the polarized tissue, we carried out laser ablation experiments in epidermal equivalent cultures (Figure S1C). Laser ablation was performed at early and late stages of morphogenesis to examine time-dependent development of basal cell mechanical behaviors (Figure 1D). DPNTP expression significantly reduced the recoil behavior at both days 1 and 6, indicating reduced cell-cell forces (Figure 1E and F). Interestingly, at day 6, basal cells exhibited a collapsing behavior, consistent with cells experiencing compressive forces due to crowding and/or tissue jamming. Importantly, examination of cell density indicated there was no significant effect upon DPNTP expression (Figure 1G), suggesting altered mechanical behaviors are not due to crowding. These data are consistent with our previous observations that in A431 cells cell-cell junctional tugging forces and cortical stiffness was dependent on DP-IF interactions, working in concert with the actin cytoskeleton12. Thus, the idea that proper integration of DP-IF connections with other cytoskeletal networks is required for cooperative control of cell mechanics22–24 also applies to developing complex epithelia.
Uncoupling the desmosome/IF connection hinders epidermal keratinocyte stratification
In models of simple epithelial cell extrusion, monolayers maintain constant cell density by eliminating cells fated to die. This process requires tight control of dynamic alterations in cell-cell mechanics and depends on actomyosin-generated forces25–27. In epidermis, basal cells lose cell-substrate contacts as they move out of the basal layer28, but upon reaching the second layer they remain attached via cell-cell adhesion to form the spinous layer. Moreover, compressive forces in simple epithelial monolayers and epidermal keratinocytes can promote extrusion and stratification, respectively29,30. Since DPNTP expression resulted in mechanical behaviors consistent with a reduction in compressive forces, we next examined the effects of DPNTP in NHEKs using two models of keratinocyte stratification.
To begin, we examined the effects of DPNTP expression on ephrin-induced colony stratification (Figure 2A). The lateral expansion of NHEKs is suppressed upon addition of ephrin peptide, inducing colonies to stratify and form multiple layers15. This results in cell piling and thus the percentage of overlapping nuclei can be used as a proxy to quantify stratification. In controls, an average of 25% of nuclei were overlapping, suggesting 25% of cells had stratified by day 7 (Figure 2B). In contrast, DPNTP expression resulted in an average of 9% overlapping nuclei (Figure 2B). Since there was no significant difference in total cell density (Figure 2B), these data suggest that uncoupling the desmosome/IF linkage impedes the stratification process.
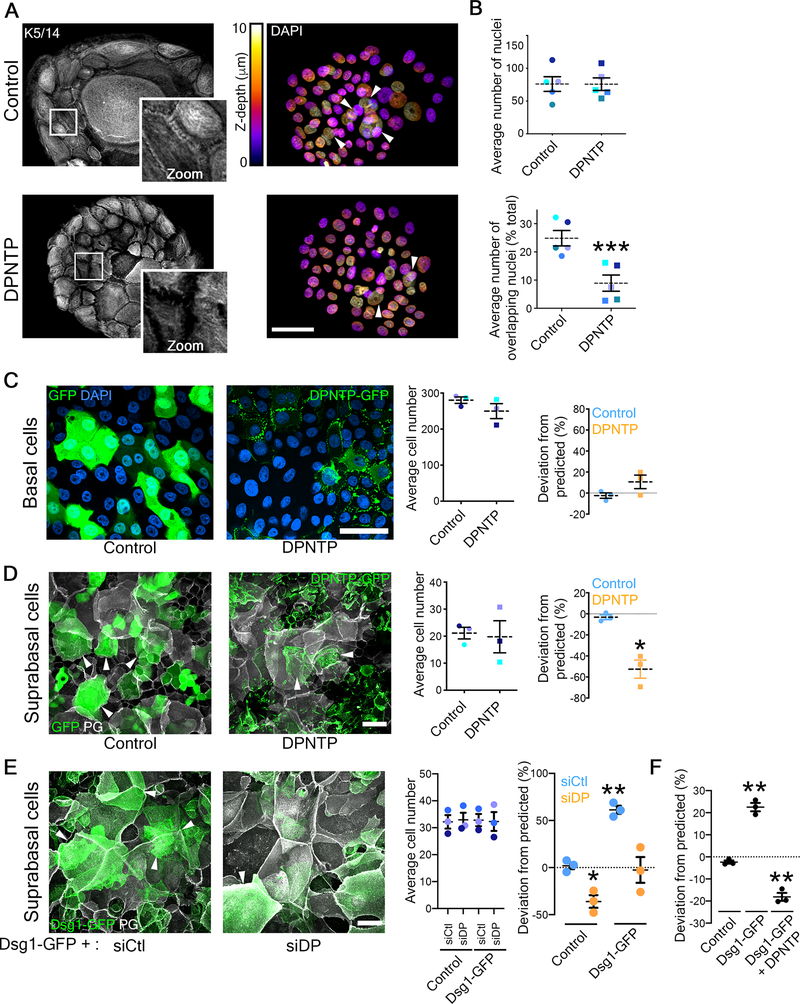
Figure 2. Uncoupling the desmosome/IF connection hinders epidermal keratinocyte stratification while promoting differentiation.
A) Maximum projection micrographs show immunostaining of keratin 5/14 (K5/14) intermediate filaments and nuclei stained with DAPI using the indicated look-up table that represents z-depth in control and DPNTP-expressing ephrin colonies. Zooms show retraction of keratin filaments from cell-cell interfaces upon uncoupling the desmosome/intermediate filament with DPNTP. Bar is 50 μm.
B) The average total number of DAPI-stained nuclei (as shown in A) per ephrin colony (top) and the average number of overlapping DAPI-stained nuclei (bottom) are shown for control and DPNTP-expressing ephrin colonies. Examples of overlapping nuclei are indicated with arrowheads in A. Dashed lines indicate the mean of 5 independent experiments and error bars are SEM. *p<0.0001, paired t-test.
C) Unlabeled wild type NHEKs were mixed at known ratios with NHEKs expressing either GFP as a control or DPNTP-GFP and cultured in 1.2 mM calcium medium for 3 days. Left, representative maximum projection micrographs of the basal layer of cultures are shown using DAPI shown in blue to label the total cell population. Bar is 50 μm. Middle, quantification of the average cell number per field from 3 independent experiments is shown. Right, quantification of the deviation from the predicted representation of GFP-positive cells in the basal layer is shown. Dashed lines indicate the mean of 3 independent experiments and error bars are SEM. See also Figure S2.
D) Unlabeled wild type NHEKs were mixed at known ratios with NHEKs expressing either GFP as a control or DPNTP-GFP and cultured in 1.2 mM calcium medium for 3 days. Left, representative maximum projection micrographs of the suprabasal layers of cultures are shown using plakoglobin (PG) to label the total cell population. Examples of GFP-positive suprabasal cells are indicated with arrowheads. Bar is 50 μm. Middle, quantification of the average cell number per field from 3 independent experiments is shown. Right, quantification of the deviation from the predicted representation of GFP-positive cells in the suprabasal layers is shown. Dashed lines indicate the mean of 3 independent experiments and error bars are SEM. *p=0.026, one sample t test with theoretical mean of 0. See also Figure S3.
E) Unlabeled wild type NHEKs were mixed at known ratios with NHEKs expressing Dsg1-GFP treated with siCtl or siDP and cultured in 1.2 mM calcium medium for 1 day. Left, representative maximum projection micrographs of the suprabasal layer of cultures are shown using plakoglobin (PG) to label the total cell population. Examples of Dsg1-GFP positive suprabasal cells are indicated with arrowheads. Bar is 50 μm. Middle, quantification of the average cell number per field from 3 independent experiments is shown. Right, quantification of the average deviation from the predicted representation of Dsg1-GFP positive cells treated with the indicated siRNA in the suprabasal layer is shown for 3 independent experiments and error bars are SEM. *p=0.038 and **p=0.005, one sample t test with theoretical mean of 0.
F) Unlabeled wild type NHEKs were mixed at known ratios with NHEKs expressing GFP (Control), Dsg1-GFP, and Dsg1-GFP co-expressing DPNTP-FLAG (DPNTP) and cultured in 1.2 mM calcium medium for 1 day. Quantification of the average deviation from the predicted representation in the suprabasal layer is shown for 3 independent experiments and error bars are SEM. **p≤0.001, one sample t test with theoretical mean of 0.
To better understand the function of the desmosome/IF linkage during stratification, we modeled the process using an in vitro competition assay. Fluorescently-labeled, genetically-manipulated cells were mixed at known ratios with wild type cells to form a confluent monolayer that was then induced to stratify with a calcium switch (Figure S2A). Two days after calcium switching, the fate of the labeled cells was tracked. The percentage of labeled cells in the basal and suprabasal layers was compared to the percentage of cells initially plated (Figure S2B). If the genetic manipulation had no effect on the ability to compete with wild type cells to stratify, there would be no deviation from the predicted percentage either in the basal or suprabasal layers. This was observed in control experiments (Figure 2C and D). However, DPNTP expression trended toward retention in the basal layer and significantly decreased the number of labeled suprabasal cells compared to what would be predicted (Figure 2C and D). There were no significant differences in cell density (Figure 2C and D). Additionally, depletion of endogenous DP using two distinct siRNA treatments significantly decreased representation of labeled suprabasal cells while a non-targeting siRNA treatment had no significant effect (Figure S3A and B), corroborating the results obtained using the uncoupling mutant. Together, these data support a role for the desmosome/IF linkage during the stratification process such that breaking the connection renders epidermal keratinocytes less able to compete with wild type cells for sorting into the suprabasal layers.
Dsg1, a desmosomal cadherin expressed exclusively in stratified epithelia, promotes delamination in 3D epidermal equivalents31. Importantly, Dsg1 is sufficient to drive simple epithelial cells (MDCK) to exit a monolayer to form a second layer31. To determine if Dsg1-dependent movement into the suprabasal layers requires an intact IF connection, we ectopically expressed Dsg1-GFP in NHEKs and tracked their fate in the competition assay. Retroviral transduction of Dsg1-GFP into NHEKs promoted a significant enrichment in the suprabasal layer, while GFP alone had no effect (Figure 2E). Moreover, the suprabasal enrichment of Dsg1-expressing cells was abolished upon either DPNTP expression or siRNA-mediated DP loss (Figure 2E and F). Therefore, our data indicate that the desmosome/IF linkage is necessary for both basal compressive forces and for Dsg1-mediated sorting into the suprabasal layers. These data support the idea that our previously observed Dsg1-induced changes in cortical actin remodeling and reduced membrane tension require a DP-IF “clutch” to promote the efficient movement of newly differentiating keratinocytes expressing Dsg1 into the next layer.
Uncoupling the desmosome/IF linkage promotes NHEK differentiation through the mechanosensitive SRF pathway
Generally, epidermal stratification and the program of terminal differentiation are thought to occur concurrently32. Therefore, we assessed the effects of DPNTP expression on differentiation in both 2D high calcium- and ephrin-induced models. Surprisingly, DPNTP expression in both models resulted in a significant increase in the protein expression levels of differentiation markers including the desmosomal cadherins Dsg1 and desmocollin 1 (Dsc1), the suprabasal cell IF protein keratin 1, and the cornified envelope component loricrin (Figure 3A and B). There were no significant effects detected for total DP or E-cadherin, which are expressed in both basal and suprabasal cells. These data suggest that disengaging the desmosome/IF connection impedes stratification while accelerating differentiation.
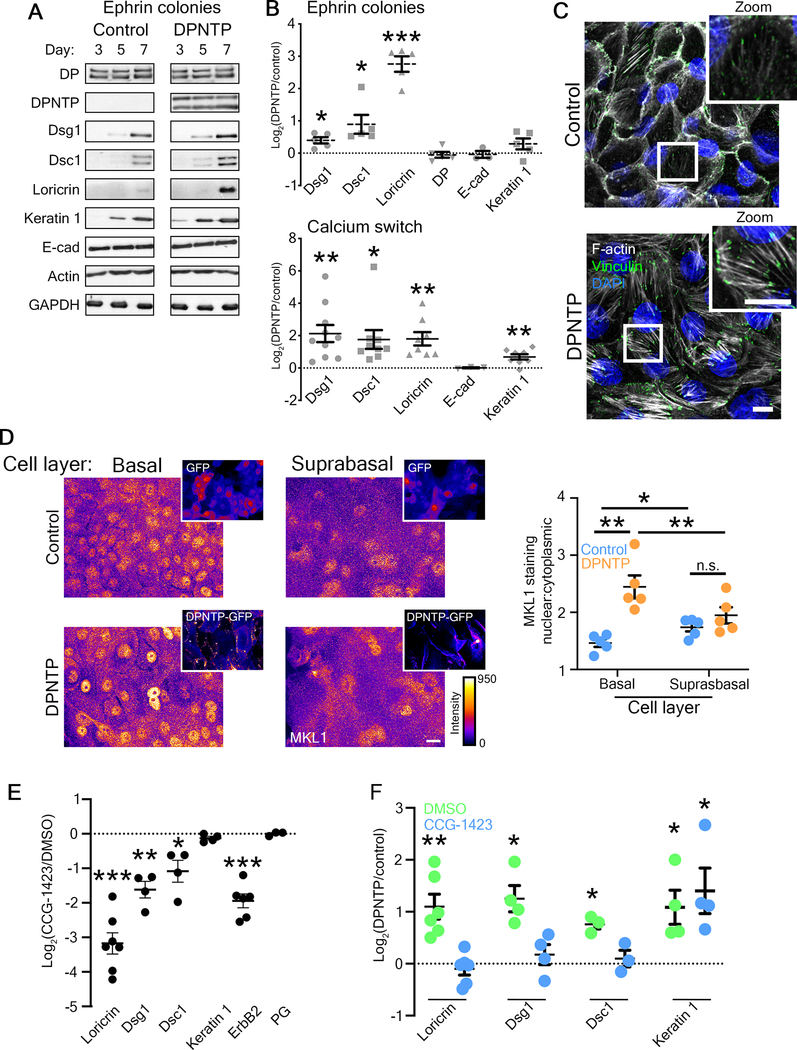
Figure 3. Uncoupling the desmosome/IF linkage promotes NHEK differentiation through the mechanosensitive SRF pathway.
A) Western blots showing the expression of the indicated proteins during a differentiation time course of control or DPNTP-GFP expressing ephrin colonies.
B) Upper, quantification of the fold change (Log2-transformed) of DPNTP-expressing over control ephrin colonies at day 7 is shown for expression of the indicated proteins. Means are from 3–5 independent experiments and error bars are SEM. *p≤0.04, ***p=0.0003, one sample t test with theoretical mean of 0. Lower, quantification of the fold change (Log2-transformed) of DPNTP-expressing over control monolayer cultures exposed to 1.2 mM calcium medium for 3–4 days are shown for indicated protein expression. Means are from 3–10 independent experiments and error bars are SEM. *p=0.016, **p≤0.004, one sample t test with theoretical mean of 0. See also Figure S4.
C) Maximum projection micrographs show F-actin (using phalloidin) and immunofluorescence staining of vinculin near the basal/substrate interface of differentiating monolayers of NHEKs expressing either GFP (Control) or DPNTP-GFP at 5 days after calcium switch. DAPI indicates nuclei in blue and bar is 10 μm.
D) Left, maximum projection micrographs show MKL1 immunofluorescence staining in the basal and suprabasal layers of day 5 cultures of NHEKs using the indicated lookup table. Bar is 10 μm. Insets show GFP and DPNTP-GFP expression. Right, quantification of the nuclear to cytoplasmic ratio of MKL1 staining is shown for cultures expressing GFP (Control) or DPNTP-GFP in the indicated cell layers. *p=0.021, **p≤0.009, paired t-test.
E) Quantification of the fold change (Log2-transformed) of cultures treated with the SRF inhibitor CCG-1423 over DMSO-treated controls at day 5 is shown for the indicated proteins. Means are from 3–7 independent experiments and error bars are SEM. *p=0.0423, **p=0.0065, ***p≤0.0002, one sample t test with theoretical mean of 0.
F) Quantification of the fold change (Log2-transformed) of DPNTP-expressing over GFP control keratinocytes is shown for the indicated protein expression for both DMSO and CCG-1423 treatment. Means are from 3–6 independent experiments and error bars are SEM. *p≤0.05, **p=0.006, one sample t test with theoretical mean of 0.
See also Figure S1.
Towards understanding how differentiation is accelerated in DPNTP-expressing keratinocytes, we explored potential mechanosensitive pathways known to regulate epidermal differentiation. One such pathway centers on SRF, which is regulated by the ratio of G- and F-actin. MKL1 (also known as MAL) is a transcriptional coregulator of SRF that normally binds to G-actin and is retained in the cytoplasm. With increased F-actin content, G-actin is released from MKL1, which in turn redistributes into the nucleus and works with SRF to induce keratinocyte differentiation33. Moreover, we previously showed that SRF is upstream of Dsg1 in this pathway34. Therefore, we considered the role of SRF in the DPNTP-mediated acceleration of differentiation.
To address this possibility, we expressed DPNTP in NHEK monolayers. DPNTP expression resulted in decreased cortical F-actin staining with a concomitant increase in focal adhesion-anchored F-actin stress fibers (Figure 3C). These results are consistent with DP’s role in the regulation of F-actin in mouse epidermis and intestine35–37. To determine if SRF signaling itself was affected downstream of desmosome/IF modulation, we assessed MKL1 nuclear localization. In the limited number of DPNTP-expressing stratified cells, we did not observe alterations in the nuclear/cytoplasmic ratio of MKL1 staining (Figure 3D). However, DPNTP expression led to an enrichment of MKL1 in the nuclei of basal cells (Figure 3D), suggesting a precocious activation of the SRF pathway. Consistent with this idea, CCG-1423, a small molecule inhibitor of SRF, reduced Dsg1, Dsc1, and loricrin expression as well as abolished the DPNTP-mediated increase in expression of these proteins (Figure 3E and F).
The DPNTP-mediated effects on differentiation are dependent on ErbB2
Our results above showed that uncoupling the desmosome/IF linkage disconnects the coordination of the formation of a second cell layer and induction of differentiation through the SRF pathway, both early morphogenic events linked to Dsg1 dependence. Dsg1 is expressed first as basal cells commit to differentiate and its expression progressively increases, so that it is most concentrated in the superficial layers of the epidermis. This led us to assess potential transitional signaling platforms.
Based on our previous findings showing that Dsg1 tunes EGFR signaling early in differentiation, we considered the ErbB family of receptor tyrosine kinases. There are four members of the ErbB family (ErbB1–4) and all have been reported to be expressed in human skin38–40. EGFR (i.e. ErbB1) is highly active in the basal layer, maintaining cells in an undifferentiated state41. Dsg1 promotes the transition of the undifferentiated basal phenotype to a differentiated suprabasal one through modulation of EGFR/Erk activity2,41 and promotion of delamination31. Expression levels of ErbB2 increase as keratinocytes differentiate38, but its role in this process is unknown.
To address the potential role of ErbB2 in differentiation, we utilized a pharmacological approach, treating cells with the ErbB2-specific inhibitor TAK16542. TAK165 treatment significantly reduced both total and Y877 p-ErbB2 (Figure S4A), a site reported to increase ErbB2 activity43,44. Addition of TAK165 resulted in a significant decrease in the protein expression levels of Dsg1, Dsc1, and loricrin, while no effects were detected for E-cadherin (Figure S4A). These data suggest a potential feedback loop between ErbB2 activity and desmosomal protein expression. DPNTP expression promoted an increase in Y877 p-ErbB2 (Figure S4B), which was abrogated upon depletion of Dsg1 (Figure S4C). Treatment of NHEKs with TAK165 abolished the differences seen in differentiation between DPNTP and controls (Figure S4D and E), suggesting the DPNTP-mediated effects on differentiation are dependent on ErbB2. These data indicate that ErbB2 plays a role in the differentiation process of NHEKs, consistent with a previously reported decrease in keratin 10 and filaggrin in ErbB2-deficient mouse epidermis45. Moreover, we have identified ErbB2 as a putative new target of SRF signaling, as treatment of wild type NHEKs with CCG-1423 results in a significant decrease in ErbB2 expression (Figure 3E).
Desmosomes regulate a tension gradient and TJ proteins in epidermal equivalents
To address whether desmosomes regulate mechanical forces and cell behaviors in the upper layers, where TJs form, we assessed indicators of mechanical force throughout the suprabasal layers. Here, we took a more targeted approach, ablating the function of Dsg1, which is specifically expressed in the stratified layers of the epidermis. Vinculin is a tension-sensitive, cell-cell junction component recruited to actin-anchored adherens junctions46. In control 3D epidermal equivalent cultures infected with nontargeting shRNA, vinculin immunostaining was restricted to the SG layer where tension is thought to be high2. This idea is supported by laser ablation experiments of suprabasal cells in 3D epidermal equivalents, which unlike basal cells at day 6, expand upon ablation (Video S1). Control cultures exhibited a steep gradient of vinculin border staining along the apicobasal axis, which was attenuated in Dsg1-deficient cultures, with increased cell-cell border localized vinculin in both the spinous and basal layers (Figure 4A). These data suggest that Dsg1 loss shifts the gradient of forces within the epidermal culture, similar to results obtained in E-cadherin-deficient mouse epidermis2.
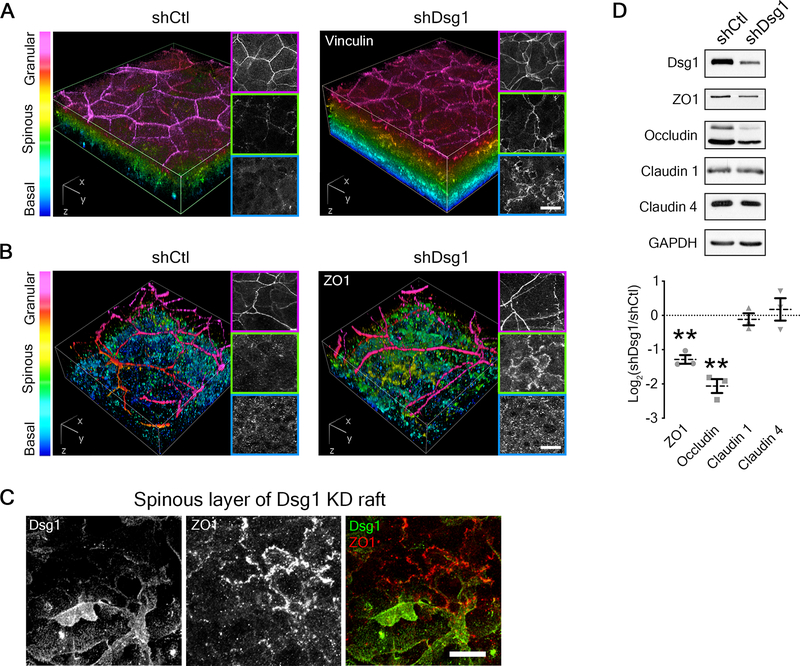
Figure 4. Dsg1 regulates a tension gradient and tight junction proteins in epidermal equivalents.
A) 3D renderings of whole mount immunostaining of tension-sensitive vinculin in control (shCtl) and Dsg1-depleted (shDsg1) day 6 epidermal equivalent cultures are presented with a look-up table that depicts z-depth in the indicated colors. Right panels show representative vinculin staining per layer. Bar is 20 μm.
B) 3D renderings of whole mount immunostaining of tight junction component ZO1 in control (shCtl) and Dsg1-depleted (shDsg1) day 6 epidermal equivalent cultures are presented with a look-up table that depicts z-depth in the indicated colors. Right panels show representative ZO1 staining per layer. Bar is 20 μm.
C) Representative micrographs of spinous layer in Dsg1-depleted day 6 epidermal equivalent cultures showing both ZO1 and Dsg1 immunostaining. Bar is 20 μm.
D) Upper, western blot analysis from control (shCtl) and Dsg1-depleted (shDsg1) day 6 epidermal equivalent cultures show the protein expression of the indicated tight junction proteins with GAPDH shown as a loading control. Lower, quantification of the fold change (Log2-transformed) of Dsg1-depleted over control cultures are shown for the indicated proteins. Dashed lines indicate the mean of 3 independent experiments and error bars are SEM. **p<0.01, one sample t test with theoretical mean of 0.
See also Figure S1.
Since TJs are thought to be regulated by a tension gradient in the epidermis2, we next assessed staining of the TJ component ZO1. ZO1 immunolabeling in control cultures shows specific localization to the SG layer (Figure 4B), the reported location where functional epidermal TJs form5. Similar to the effects on vinculin, Dsg1 depletion led to a shift in this staining pattern (Figure 4B). A similar pattern was observed upon treatment with a Rho activator CN01, whereby ZO1 similarly localized prematurely to cell-cell interfaces in the spinous layer (Figure S5A and B). The effect of Dsg1 on ZO1 appears to be cell autonomous, as mosaic of Dsg1 expression in the spinous layer altered cell-cell contact localized ZO1 specifically in Dsg1-depleted cells (Figure 4C). Moreover, biochemical analysis of ZO1 and occludin indicated Dsg1 depletion results in decreased expression (Figure 4D), while no effects were detected for claudin 1 or claudin 4.
Uncoupling the desmosome/IF connection diminishes epidermal keratinocyte barrier function
In multiple models, TJs and their protein components have been suggested to be mechanosensitive2,47–49. We hypothesize the patterning of the desmosome/IF components drives polarized, layer-specific epidermal functions. Thus, we assessed the effects DPNTP expression on epidermal TJ barrier development. Staining of occludin in sagittal sections of epidermal equivalent cultures indicated that DPNTP expression resulted in a shift to membrane enrichment of occludin in the spinous layer compared with controls, while no differences were seen in the SG layer (Figure S6A). These results are consistent with previous work showing that DP loss in mouse epidermis modulates TJ protein expression and localization50. Together with data from Dsg1-deficient cultures, these results suggest that the desmosome/IF connection controls the distribution of forces within the epidermis to restrict TJ protein localization to the SG layer.
Since DPNTP expression affects TJ component localization and the mechanical properties of cells, we hypothesized this could disturb TJ barrier function, attributed to the outer layers of the epidermis (Figure 5A). To address this, we generated human epidermal equivalent cultures using a porous transwell system (Figure S1D). We performed transepidermal electrical resistance (TEER) measurements on the developing cultures to assess barrier function (Figure 5B). We found that altering cell mechanics in this 3D system using CN01 affected the organization of F-actin in all layers and significantly inhibited barrier function (Figure S5C and D). Additionally, we performed immunostaining of ZO1 which localizes to cell-cell interfaces in the SG layer in control cultures, an area with strong F-actin staining (Figure 5C). However, DPNTP expression induced a cell autonomous loss of ZO1 staining in the SG layer (Figure 5D), suggesting a potentially impaired barrier. We did not observe altered ZO1 staining in the basal or spinous layers (Figure S6B), suggesting layer-specific mechanisms exist. TEER measurements of the developing barrier were significantly reduced in DPNTP-expressing cultures compared with GFP controls (Figure 5E), indicating reduced epidermal barrier function.
Figure 5. Uncoupling the desmosome/IF connection diminishes epidermal keratinocyte barrier function.
A) Schematic shows the layers of the epidermis and functional tight junctions localize to the second granular layer. The expression patterns of desmoplakin (DP), keratins (K), and desmoglein 1 (Dsg1) are also shown.
B) Schematic shows how transepidermal electrical resistance (TEER) experiments were performed on transwell epidermal equivalent cultures. Electrodes were used to pass a current through the 3D culture that was grown on a transwell insert and resistance measurements were taken as an indicator of barrier function.
C) Maximum projection micrographs of day 9 transwell epidermal equivalent cultures show staining for nuclei (DAPI), F-actin (phalloidin), and immunostaining for the indicated proteins using a look-up table indicating z-depth. Bar is 25 μm.
D) Fluorescence micrographs show the expression of GFP (Control) and DPNTP-GFP in the granular layer of day 9 transwell epidermal equivalent cultures that were also immunostained for the tight junction component ZO1. Magenta dotted line delineates DPNTP-GFP high expressing cells. Nuclei are stained with DAPI in blue. Bar is 25 μm. Related to Figure S6.
E) Quantification of resistance measurements from TEER experiments performed on a time course of control (GFP) and DPNTP-GFP expressing transwell epidermal equivalent cultures. *p=0.0125, Sidak’s multiple comparison test from 7 independent experiments. Data are presented as mean ± SEM. Related to Figure S5–6.
See also Figure S1.
ErbB2 as a potential mediator of desmosome-mediated effects on epidermal polarity and barrier function
Having established that ErbB2 mediates differentiation downstream of SRF signaling, we considered the possibility that it also contributes to later events that drive barrier function. Toward this end, we more closely assessed the relative expression and localization of EGFR and ErbB2 in human epidermis. ErbB2 localization was enriched in the SG2 layer of human epidermis (Figure 6A), where functional TJs form5. While total ErbB2 was expressed at all timepoints evaluated, its phosphorylation status at Y877 increased as cultures developed (Figure 6B). Moreover, ErbB2 was reported to modulate epithelial permeability, being linked to both loss and gain of barrier function51, indicative of context-dependent functions. This suggested to us that ErbB2 may play a role in the desmosome-mediated regulation of epidermal TJs.
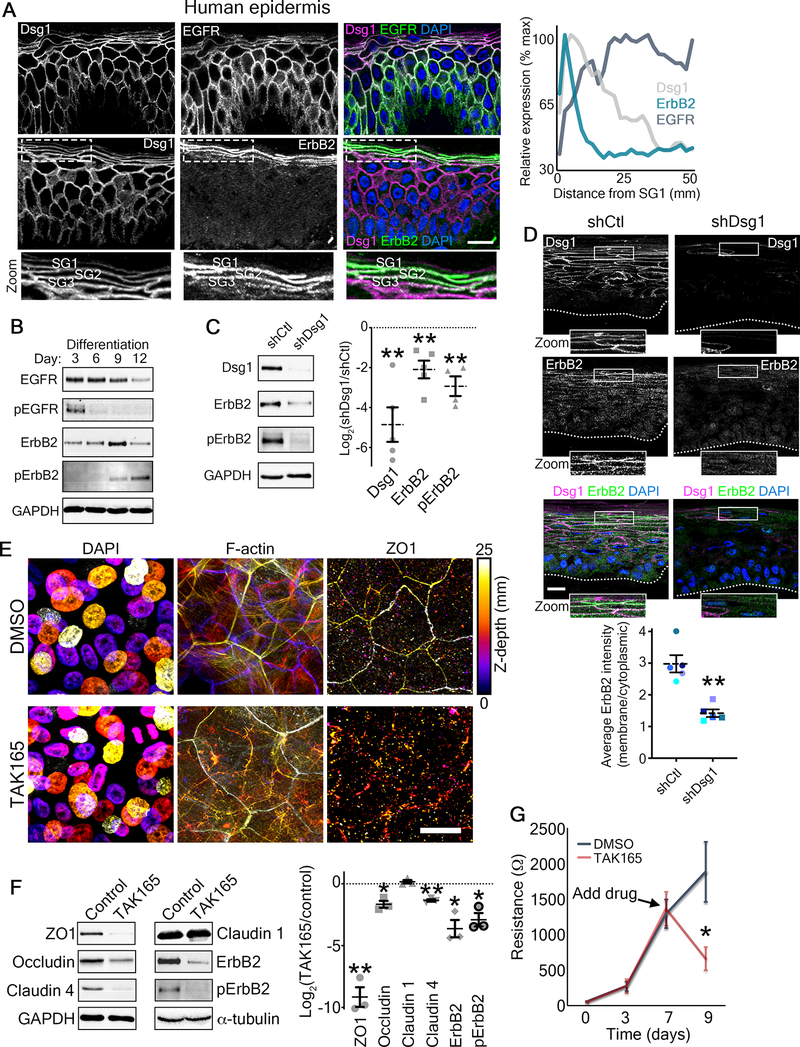
Figure 6. ErbB2 as a potential mediator of Dsg1-mediated effects on epidermal polarity and barrier function.
A) Left, representative micrographs of transverse sections of human epidermis immunostained for the indicated proteins are shown. Zoom is of area in dashed box highlighting the stratum granulosum (SG1–3). DAPI-stained nuclei are in blue and bar is 20 μm. Right, line scan analysis shows the average relative level of fluorescence intensity from 3 independent samples for the indicated proteins as a function of distance from the most superficial living epidermal layer (stratum granulosum, SG).
B) Representative western blots of a differentiation time course of human epidermal equivalent cultures showing the total EGFR and Y1068-phosphorylated EGFR (pEGFR) as well as total ErbB2 and Y877-phosphorylated ErbB2 (pErbB2). GAPDH is used as a loading control.
C) Left, representative western blots of day 6 human epidermal equivalent cultures expressing either a non-targeting shRNA (shCtl) or an shRNA targeting Dsg1 (shDsg1) showing Dsg1 protein level, total ErbB2, and Y877-phosphorylated ErbB2 (pErbB2). GAPDH is used as a loading control. Right, quantification of the fold change (Log2-transformed) of Dsg1-depleted over control cultures are shown for the indicated proteins. Dashed lines indicate the mean of at least 4 independent experiments and error bars are SEM. **p<0.01, one sample t test with theoretical mean of 0.
D) Micrographs of transverse sections of day 6 human epidermal equivalent cultures expressing either a non-targeting shRNA (shCtl) or an shRNA targeting Dsg1 (shDsg1) immunostained for Dsg1 and ErbB2 are shown. Dotted line marks the bottom of the basal layer. DAPI-stained nuclei are in blue and bar is 20 μm. Lower, quantification of ErbB2 immunofluorescence intensity expressed as a ratio of the membrane localized signal over the cytoplasmic localized signal. Dashed lines indicate the mean of 5 independent experiments and error bars are SEM. **p=0.008, paired t test.
E) Day 9 transwell epidermal equivalent cultures were treated with either DMSO or the ErbB2 inhibitor TAK165 (1 μM) for the last 48 hours prior to harvesting. Maximum projection micrographs show staining for nuclei (DAPI), F-actin (phalloidin), and immunostaining for the tight junction protein ZO1 using a look-up table that indicates z-depth. Bar is 25 μm. See also Figure S7.
F) Day 9 transwell epidermal equivalent cultures were treated with either DMSO or the ErbB2 inhibitor TAK165 (1 μM) for the last 48 hours prior to harvesting. Left, western blots showing the expression of the indicated tight junction proteins for samples treated with either DMSO (Control) or TAK165. GAPDH and α-tubulin are used as loading controls. Right, quantification of the fold change (Log2-transformed) of TAK165-treated over control cultures are shown for the indicated proteins. Dashed lines indicate the mean of 3 independent experiments and error bars are SEM. *p=0.025, **p<0.01, one sample t test with theoretical mean of 0.
G) Quantification of resistance measurements from TEER experiments performed on a time course of transwell epidermal equivalent cultures that were treated with either DMSO or the ErbB2 inhibitor TAK165 (1 μM) on day 7 (as indicated) for the last 48 hours. *p=0.0325, paired t test from 5 independent experiments. Data are presented as mean ± SEM.
See also Figure S1.
To examine the effects of Dsg1 on ErbB2 function, we performed Dsg1 depletion experiments. Dsg1 loss decreased total and Y877 p-ErbB2 as well as reduced ErbB2 at sites of cell-cell contact in the upper layers (Figure 6C and D). These data suggest that Dsg1 indeed regulates ErbB2. We then assessed the potential role of ErbB2 in regulating TJ protein expression and function. As ErbB2 is important for epidermal keratinocyte differentiation (Figure S4A), we allowed human transwell epidermal equivalent cultures to form for 7 days prior to drug treatment for 2 additional days. TAK165 treatment altered overall ZO1 and F-actin staining (Figure 6E) with aberrant punctate ZO1 staining in the spinous layer (Figure S7), similar to the results seen upon Dsg1 depletion (Figure 4B and C). Additionally, compared to controls, TAK165 significantly reduced expression of ZO1, claudin 4, and occludin (Figure 6F). Finally, inhibiting ErbB2 with TAK165 significantly diminished the forming barrier assessed using TEER (Figure 6G).
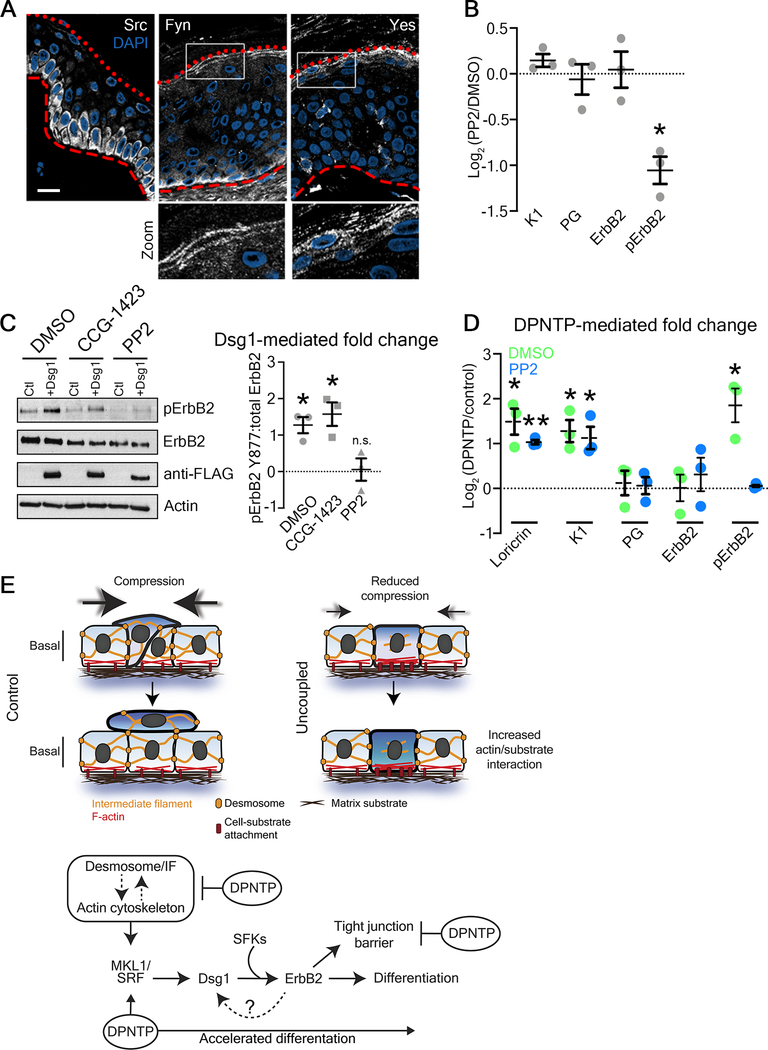
Src family kinases (SFKs) have been reported to phosphorylate ErbB2 on Y877, increasing its kinase activity44. In human epidermis, SFKs are expressed such that Src expression is largely restricted to the basal layer while Fyn and Yes are expressed in the granular layer (Figure 7A). This suggested that SFKs may facilitate desmosome-mediated alterations in Y877 p-ErbB2. Indeed, treatment with the SFK inhibitor PP2 significantly decreased Y877 p-ErbB2 (Figure 7B). Moreover, in Dsg1 gain-of-function experiments, retroviral transduction of ectopic, FLAG-tagged Dsg1 into NHEKs significantly increased in the ratio of Y877 p-ErbB2 to total ErbB2 (Figure 7C). This gain-of-function as well as the DPNTP-mediated increase in p-ErbB2 were lost upon inhibition of SFKs with PP2 (Figures 7C and D). However, Dsg1 overexpression is still able to promote an increase in the relative amount of Y877 p-ErbB2 upon SRF inhibition, suggesting Dsg1 is downstream of SRF and upstream of ErbB2 (Figure 7C).
Figure 7. Desmosome-mediated modulation of ErbB2 phosphorylation depends on Src family kinases.
A) Representative micrographs of transverse sections of human epidermis immunostained for the indicated Src family kinases (SFKs) are shown. Red dotted line marks the top of the granular layer and the dashed red line denotes the bottom of the basal layer. DAPI-stained nuclei are in blue and bar is 20 μm.
B) Quantification of the fold change (Log2-transformed) of cultures treated with the SFK inhibitor PP2 (6 hours prior to harvest) over DMSO-treated controls at day 5 are shown for the indicated proteins. Means are from 3 independent experiments and error bars are SEM. *p=0.0195, one sample t test with theoretical mean of 0.
C) Left, western blots showing the expression of the indicated proteins in day 5 cultures of NHEKs expressing either GFP (Ctl) or Dsg1-GFP and treated with either the SRF inhibitor CCG-1423, the SFK inhibitor PP2, or DMSO as a control. Right, quantification of the fold change (Log2-transformed) in the ratio of pErbB2:total ErbB2 is shown for the indicated treatments. Means are from 3 independent experiments and error bars are SEM. *p≤0.04, one sample t test with theoretical mean of 0.
D) Quantification of the fold change (Log2-transformed) of DPNTP-expressing over GFP control keratinocytes are shown for the indicated protein expression for both DMSO and PP2 treatment. Means are from 3 independent experiments and error bars are SEM. *p≤0.05, **p=0.0023, one sample t test with theoretical mean of 0.
E) Working model of the effects of the desmosome/intermediate filament (IF) network on early and late epidermal morphogenetic events. Upper, during early morphogenesis, the desmosome/IF network works in conjunction with the actin cytoskeleton to regulate stratification through modulating the mechanical properties of basal cells. Uncoupling the desmosome/IF linkage with DPTNP reduced compressive forces and increased substrate adhesion, hindering stratification. Lower, differentiation of keratinocytes proceeds through the mechanosensitive SRF pathway, which is accelerated when desmosomes are uncoupled from IF. This pathway includes Dsg1, which through SFKs modulates ErbB2 phosphorylation. ErbB2 in turn is critical to the formation of the epidermal barrier.
See also Figure S1.
Discussion
Apicobasal polarity is an intrinsic property of epithelial tissues that permits discrimination between interior and exterior compartments by establishing a functional barrier near the apical, external-facing surface. In contrast to simple epithelia, apicobasal polarity in the epidermis is organized across multiple layers10. Here, we demonstrate that the desmosome/IF network is a critical player in establishing the polarized structure and function of the epidermis.
Alterations in both basal and superficial cell mechanics in response to desmosome impairment are consistent with a switch from strong cell-cell forces to enhanced cell-substrate forces (basal layer) or enhanced intercellular tension in the cells below the TJ layer. For instance, in small colonies of differentiating keratinocytes, DPNTP expression resulted in increased vinculin-positive, cell-substrate contacts and associated F-actin stress fibers (Figures 1C and 3C), previously documented to signal a switch from strong cell-cell forces to enhanced cell-substrate forces17–19. At the same time, a decrease in cell-cell forces is supported by laser ablation experiments of early stage epidermal cultures (Figure 1D–F) and by our previous finding that DPNTP expression in A431 cells reduced cell-cell tugging forces12.
How does interfering with basal cell mechanics affect an epidermal keratinocyte’s commitment to differentiate? During epidermal development, cell proliferation induces crowding and thus compressive forces that have been linked to epidermal fate specification and delamination52–55. However during homeostasis of adult mouse epidermis, inhibiting cell division, and thus reducing cell density, did not affect the ability of keratinocytes to delaminate56. Therefore, compressive forces in the basal layer appear to have different roles depending on the developmental stage. In vitro, differentiating keratinocytes exhibit a transient decrease in cortical tension associated with the onset of expression of differentiation markers and subsequent exit of cells from the basal layer via delamination55. Here, we show that as human epidermal cultures progress, cells transition from a high-tension state to a compressive state (Figure 1D). These states are not achieved when IF are uncoupled from desmosomes. Thus, uncoupling IF from the desmosome reduces compressive forces within the basal layer that have been associated with differentiation. Consistent with this, cells were less capable of stratification into the superficial layers when IF were uncoupled from desmosomes (Figure 2D). These findings are supported by a recent publication establishing a role for desmosomes and their attachment to IF during cell extrusion in simple epithelia57. This actomyosin-dependent process is often compared with stratification of multilayer epithelia57. Altogether, these data are consistent with a role for desmosomes and IFs working cooperatively with actomyosin to modulate the ability of a cell to leave the basal layer of an epithelium.
Dsg1 redistributed molecular tension in keratinocyte monolayers through rearrangements of cortical F-actin mediated by Arp2/3-dependent actin polymerization31. During early morphogenesis, these tension-redistributing forces help promote basal cell delamination by reducing tension on E-cadherin31. Moreover, our previous observations show that the actomyosin contractile system is required for the altered mechanics observed when the desmosome/IF connection is impaired12. Furthermore, either DPNTP expression or siRNA-mediated depletion of DP abrogated the ability of Dsg1-positive cells to enrich in the suprabasal layer (Figure 2E and F). Collectively, these data support the idea that desmosomes cooperate with the F-actin cytoskeleton to coordinate changes in cortical tension and adhesion necessary to drive stratification.
The process of keratinocyte stratification is generally associated with the onset of terminal differentiation. Therefore, it was initially surprising that while uncoupling IF from desmosomes impeded the movement of keratinocytes into the suprabasal layers, the biochemical program of differentiation is accelerated (Figure 3A and B). This observation is not without precedent, as inhibition of actomyosin contractility was shown to uncouple morphogenesis and fate specification of inner cell mass cells in the developing mouse embryo58. We propose that uncoupling the desmosome/IF linkage precociously activates the SRF pathway to accelerate the process of differentiation, but due to a loss of “clutch” activity, altered cell-cell adhesion, and increased cell-substrate adhesion, they are less likely to stratify (Figure 7E).
Junctional tension, like that produced by actomyosin, recruits the TJ components ZO1 and occludin to promote barrier function in simple polarized epithelia47,49. Dsg1 loss perturbed the normally restricted pattern of vinculin in the high-tension layer where TJs form, consistent with the idea that higher tension cell-cell junctions occurred in the lower layers (Figure 4A). This is in line with work showing impaired desmosome function increased molecular indicators of tension experienced by adherens junctions in mouse epidermis50. Conversely, multiple reports using E-cadherin knockout mouse models have shown that loss of epidermal E-cadherin does not have major effects on overall desmosome structure nor on markers of differentiation59,60. However, combined loss of both epidermal E- and P-cadherin does55. In addition, Dsg1 loss shifts the localization of ZO1 to more basal (Figure 4B and C). Coupled with previous work showing the importance of E-cadherin in the TJ barrier2, these data are consistent with a model in which Dsg1 loss induces a transfer of forces experienced to non-desmosomal junctions and premature mechanosensitive recruitment of junctional proteins to cell-cell interfaces in spinous layers. Moreover, disruption of the desmosome/IF linkage, which decreases cell-cell forces, is sufficient to reduce barrier function of transwell epidermal equivalent cultures (Figure 5E). Therefore, the fine tuning of mechanical input is critical to TJ barrier function and this tuning requires desmosome/IF attachment. Finally, perturbations of claudins and occludin in epidermis affects keratinocyte differentiation61–63, suggesting potential feedback loops to the regulation of differentiation-dependent desmosomal proteins.
ErbB2 activity is sensitive to changes in cell mechanics41,64,65 and our data provide evidence suggesting ErbB2 is a novel target of SRF (Figure 3E), placing it downstream of this mechanosensitive pathway. In the epidermis, ErbB2 expression is enriched in epidermal superficial layers, where Dsg1 is at peak expression (Figure 6A). Dsg1 loss abrogated the membrane localization of ErbB2 and the levels of Y877 P-ErbB2 (Figure 6B–D). Expression of either DPNTP or ectopic Dsg1 is sufficient to increase the relative amount of Y877 p-ErbB2 (Figures 7C–D and S4B). This site has been previously reported to be phosphorylated by SFKs44 and here we show that both the DPNTP- and Dsg1-mediated increase in p-ErbB2 is abolished upon SFK inhibition with PP2 (Figure 7C–D). Since SFKs are expressed in patterns in the epidermis (Figure 7A), this suggests that layer-specific functions of the desmosome/IF network could be facilitated through modulation of restricted SFK activity.
Our previous work demonstrated that Dsg1-stimulated differentiation of basal cells depends on its ability to dampen EGFR/Erk signaling in the basal layer, which required an interaction between Dsg1 and Erbin (ErbB2 interacting protein)41,64. Thus, together with the data presented here, this suggests a model in which a Dsg1 expression gradient functions to dampen EGFR basally to promote differentiation while stimulating ErbB2 suprabasally to support a functional TJ barrier. However, it is possible that ErbB2 coordinates with other members of the ErbB family to regulate TJ function and the role of Erbin in this process remains an avenue for future studies.
Collectively, the data presented here expose the desmosome/IF network as an essential element of the machinery in epidermis that cooperates with the F-actin cytoskeleton to pattern layer-specific mechanical properties. In the basal layer, this network affects compressive forces that coordinate the processes of stratification and differentiation; in the upper layer, the network integrates mechanosensitive cytoskeletal and signaling components to maintain the TJ barrier. We identify ErbB2 as a critical regulator of both keratinocyte differentiation and barrier function, placing it in a key position to control polarized epidermal functions downstream of the desmosome/IF network.
STAR Methods
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead contact, Kathleen J. Green (kgreen@northwestern.edu).
Materials availability
Plasmids generated in this study are available upon request from the Lead contact.
Data and code availability
All data needed to evaluate the conclusions of the paper are presented in the paper and/or in the Supplemental information. This study did not generate any unique code.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell culture
Primary normal human epidermal keratinocytes (NHEKs) were isolated from neonatal foreskin provided by the Northwestern University Skin Biology and Disease Resource-based Center (SBDRC) as previously described66. Cells were maintained in growth medium (M154 media supplemented with 0.07 mM CaCl2, human keratinocyte growth supplement (HKGS), and gentamicin/amphotericin B solution (G/A). NHEKs were induced to differentiate by addition of 1.2 mM CaCl2 to growth medium with or without addition of 1 μg/mL recombinant human ephrin-A1 Fc peptide for the indicated times. NHEKs were used to generate 3D epidermal equivalent cultures as previously described66. Pharmacological treatments included 1 unit/ml Rho Activator I, 1 μM TAK165, 20 μM PP2, and 60 nM CCG-1423.
METHOD DETAILS
Western blot analysis
Whole cell lysates were generated using Urea Sample Buffer (8 M urea, 1% SDS, 60 mM Tris, pH 6.8, 5% β-mercaptoethanol, 10% glycerol). Proteins were separated by SDS-PAGE electrophoresis and transferred to nitrocellulose membranes. Membranes were blocked with either 5% milk or 1% BSA in PBS or TBS with or without 0.05% Tween. Primary and secondary antibodies were incubated in this blocking solution. Immunoreactive proteins were visualized using chemiluminescence. Densitometry analysis was performed using ImageJ.
Transient transfection
NHEKs were transfected with either siGENOME Non-Targeting siRNA Pool #2 (siCtl) or siGENOME SMARTpool siRNA D-019800–17 DSP (siDP1) or stealth siRNA oligonucleotides 5′-CAGGGCUCUGUCUUCUGCCUCUGAA-3′ from Life Technologies (siDP2) using the Amaxa Nucleofector System according to manufacturer’s instructions. NHEKs were suspended in Ingenio Electroporation Solution with siRNA (final concentration 50 nM) and electroporated using program X-001.
Viral transduction
pLZRS-DPNTP-GFP was cloned from pEGFP-N1-DPNTP into pLZRS using the standard Gateway/TOPO cloning protocol. pLZRS-miR Dsg1 (shDsg1) was generated as previously described41. pLZRS–NTshRNA (shCtl) was generated with the following sequences inserted: NTshRNA-fwd 5′–GTATCTCTTCATAGCCTTAAA–3′ and NTshRNA-rev 5′–TTTAAGGCTATGAAGAGATAC–3′ as previously described67. Keratinocytes were transduced with retroviral supernatants produced from Phoenix cells as previously described41. Briefly, Phoenix cells transiently transfected with retroviral pLZRS cDNA constructs were harvested at 70% confluency for 24 hours at 32°C. Supernatants were collected and concentrated using Amicon® Ultra-15 Centrifugal Filter Unit. Infection of keratinocytes was done at 15% cell confluence with incubation at 32°C for 1.5 hours in M154 media containing 4 μg/ml polybrene and retrovirus supernatants. Lentivirus (pLVX myristoylated tdTomato) transduction of keratinocytes utilized virus obtained from the Northwestern University SBDRC. Briefly, keratinocytes were plated at 30% confluence and the next day treated with 1 μg/ml polybrene plus lentivirus and incubated at 37°C overnight followed by washing
Immunofluorescence/microscopy
For immunofluorescence analysis, NHEKs cultured on glass coverslips were fixed either in anhydrous ice-cold methanol for 3 min on ice or 4% paraformaldehyde solution for 15 min at room temperature. Cells were permeabilized and blocked with 0.25% Triton X-100 and 5% goat serum for 30 minutes and then processed for immunofluorescence. For skin biopsies, paraffin-embedded sections were baked overnight at 60°C and deparaffinized using xylene/ethanol. After permeabilization with 0.5% Triton X-100 in PBS, antigen retrieval was performed using 0.01 M citrate buffer, pH 6.0. All samples were mounted onto glass slides or using glass coverslips with ProLong Gold antifade reagent.
Apotome images were acquired using ZEN 2.3 software with an epifluorescence microscope system (Axio Imager Z2, Carl Zeiss) fitted with an X-Cite 120 LED Boost System, an Apotome.2 slide module, Axiocam 503 Mono digital camera, and a Plan-Apochromat 40x/1.4 or Plan-Apochromat 63x/1.4 objective (Carl Zeiss). Confocal z-stacks (z-step size of 0.24–0.5 μm) were acquired using a Nikon A1R confocal laser microscope equipped with GaAsP detectors or a Nikon W1 Spinning Disk Confocal with a 95B prime Photometrics camera and a 60× Plan-Apochromat objective lambda with a NA of 1.4 and run by NIS-Elements software. NIS-Elements (version 5.02) was used to generate 3D reconstructions of z-stacks using the Volume Viewer tool with z-depth coding blending (rainbow contrast lookup table) or ImageJ was used to generate color coded z-projections using Temporal-Color Code (Fire lookup table).
Laser ablation
NHEKs were transduced with pLVX myristoylated tdTomato (myr-tomato) lentivirus to allow tracking cell outlines and 24 hours after they were transduced with either pLZRS GFP or DPNTP-GFP retrovirus. These cells were then used to generate 3D epidermal equivalent cultures as described above. At the indicated timepoints, sufficient medium was added to a culture dish to re-submerge the epidermal equivalent to perform imaging with a water dipping objective and secured using a harp anchor. The culture was allowed to equilibrate for 20 minutes before imaging and only imaged for 1 hour each. Two-photon laser ablation was used to assess intercellular forces. Briefly, ablation was performed on a Nikon A1R-MP+ multiphoton microscope running Elements version 4.50 and equipped with an Apo LWD 25× 1.10W objective. Cells were maintained at 37 °C and 5% CO2. Images of myr-tomato and GFP were obtained at a rate of 1 frame per second for 2 s before and 45 s after ablation using 4% laser power at 970 nm. Ablation was performed using 40% laser power at a scan speed of 512. The distance between the cell–cell vertices over time was measured using the Manual Tracking plugin in ImageJ. Distance curves were then generated using Excel software. Initial recoil measurements were calculated using Prism 8 software as previously described68.
Sorting/stratification assay
Wild type NHEKs were mixed at known ratios with genetically-modified NHEKs that were transduced with GFP, DPNTP-GFP, Dsg1-GFP, and in combination as indicated with subsequent nucleofection with siRNA. These combined populations were switched to M154 medium containing 1.2 mM calcium, HKGS, and G/A. Cultures were allowed to stratify for 3 days before fixation using 4% paraformaldehyde. For Dsg1-GFP experiments, cultures were allowed to stratify for 1 day before fixation with 4% paraformaldehyde. Samples were processed for immunostaining as above for plakoglobin to visualize all cell outlines as well as DAPI to visualize nuclei. Confocal z-stacks were obtained and processed in ImageJ to contain either only basal cells or suprabasal cells by selecting a subset of z-frames. The total number of cells per field was the quantified using DAPI and plakoglobin staining. The subset of the total population that expressed GFP was then calculated and compared to the percentage of cells predicted to express GFP based on the number of wild type and GFP expressing cells put into the system. This was compared for basal as well as suprabasal cells.
Transepidermal electrical resistance
Costar 24 mm Transwell 0.4 μm pore inserts were coated with CELLstart CTS Substrate according to manufacturer. NHEKs were seeded at confluence in Defined PCT Epidermal Keratinocyte Medium (CnT-07) and submerged in CnT-07 for 2 days with medium in both upper and lower chambers. On the third day, CnT-07 was replaced with CnT-Prime 3D Barrier Culture Medium (CnT-PR-3D) with cells submerged. The following day, “Day 0” TEER measurements were taken and medium was removed from the upper chamber to expose NHEKs to an air-liquid interface. Subsequent TEER measurements were taken every other day/ at indicated times by adding pre-warmed (37°C) CnT-3D-PR to the top chamber. Measurements were made with an EVOM epithelial volt/ohm meter (World Precision Instruments, Sarasota, FL, USA). Measurements were taken in triplicate per sample and averaged. Medium was removed promptly from the top chamber as soon as TEER measurements were taken.
QUANTIFICATION AND STATISTICAL ANALYSIS
For all assays, at least three independent experiments were carried out and exact numbers are found in the figure legends. Independent experiments are shown in corresponding colors in figure graphs. Independent experiments were considered as those performed with NHEKs derived from individual patient materials. Data in all graphs are presented as means and error bars represent standard error of the mean (SEM). Graphs were generated using Excel or Prism software. Statistical analyses were performed using Prism software and are specifically indicated in figure legends. P values less than 0.05 were considered statistically significant. All fluorescence intensity-based quantification was performed using ImageJ.
Supplementary Material
Video S1. Tension in suprabasal layer; Related to Figure 2
A representative time lapse of a laser ablation experiment in the suprabasal layer in a Day 6 3D epidermal equivalent culture. Cell outlines are visualized with myr-tomato and time between frames is 1 sec. Total time is 45 sec. Field of view is 79.5×79.5 μm.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Chicken anti-plakoglobin | Aves Laboratories | 1407 and 1408 |
| Mouse anti-α-tubulin DM1A | Sigma-Aldrich | Cat# T6199 RRID:AB_477583 |
| Mouse anti-α-tubulin 12G10 | Developmental Hybridoma Studies Bank | RRID: AB_1157911 |
| Rabbit anti-src | Sigma-Aldrich | Cat# HPA030875 RRID:AB_2673640 |
| Rabbit anti-fyn | Sigma-Aldrich | Cat# HPA023887 RRID:AB_1849226 |
| Mouse anti-yes | Santa Cruz Biotechnology | Cat# sc-46674 RRID:AB_627370 |
| Rabbit anti-GAPDH | Sigma-Aldrich | Cat# G9545 RRID:AB_796208 |
| Mouse anti-FLAG | Sigma-Aldrich | Cat# F7425 RRID: AB_439687 |
| Mouse JL8 anti-GFP | Takara | Cat# 632381 RRID:AB_2313808 |
| Mouse P124 anti-desmoglein 1 | Progen | Cat# 651111 RRID:AB_1541107 |
| Mouse U100 anti-desmocollin 1a/b | Progen | Cat# 65192 RRID:AB_1541099 |
| Mouse anti-vinculin | Sigma-Aldrich | Cat# V9131 RRID:AB_477629 |
| Mouse C4 anti-actin | EMD-Millipore | Cat# MAB1501 RRID:AB_2223041 |
| Rabbit NW6 and NW161 anti-desmoplakin | Green Laboratory | N/A |
| Mouse HECD1 anti-e-cadherin | M. Takeichi and O. Abe, Riken Center for Developmental Biology | N/A |
| Rabbit anti-loricrin, rabbit anti-keratin 1, rabbit anti-keratin 5, and rabbit anti-keratin 14 | J. Segre, NIH | N/A |
| Rabbit anti-EGF Receptor | Cell Signaling Technology | Cat# 4267 RRID:AB_2246311 |
| Rabbit anti-phospho-EGF Receptor Tyr1068 | Cell Signaling Technology | Cat# 2234 RRID:AB_331701 |
| Rabbit anti-HER2/ErbB2 | Cell Signaling Technology | Cat# 2165 RRID:AB_10692490 |
| Rabbit anti-phospho-HER2/ErbB2 Tyr877 | Cell Signaling Technology | Cat# 2241 RRID:AB_2099407 |
| Mouse anti-ZO1 | BD BioSciences | Cat# 610967 RRID:AB_398280 |
| Rabbit anti-claudin 1 | Thermo Fisher Scientific | Cat# 51-9000 RRID:AB_2533916 |
| Rabbit anti-claudin 4 | Thermo Fisher Scientific | Cat# 36-4800 RRID:AB_2533262 |
| Mouse anti-occludin | Thermo Fisher Scientific | Cat# 33-1500 RRID:AB_2533101 |
| Rabbit anti-MKL1 | Sigma-Aldrich | Cat# HPA030782 RRID:AB_10600930 |
| Secondary goat anti-mouse, -rabbit, and -chicken HRP | SeraCare Life Sciences | N/A |
| Secondary goat anti-mouse IgG2a isotype conjugated with Alexa Fluor-488 | Thermo Fisher Scientific | Cat# A-21131 RRID:AB_2535771 |
| Secondary goat anti-mouse IgG2a isotype conjugated with Alexa Fluor-647 | Thermo Fisher Scientific | Cat# A-21124 RRID:AB_2535766 |
| Secondary goat anti-mouse IgG1 isotype conjugated with Alexa Fluor-488 | Thermo Fisher Scientific | Cat# A-21121 RRID:AB_2535764 |
| Secondary goat anti-mouse conjugated with Alexa Fluor-488 | Thermo Fisher Scientific | Cat# A-11029 RRID:AB_2534088 |
| Secondary goat anti-mouse conjugated with Alexa Fluor-568 | Thermo Fisher Scientific | Cat# A-11004 RRID:AB_2534072 |
| Secondary goat anti-mouse conjugated with Alexa Fluor-647 | Thermo Fisher Scientific | Cat# A-32728 RRID:AB_2633277 |
| Secondary goat anti-rabbit conjugated with Alexa Fluor-488 | Thermo Fisher Scientific | Cat# A-11008 RRID:AB_143165 |
| Secondary goat anti-rabbit conjugated with Alexa Fluor-568 | Thermo Fisher Scientific | Cat# A-11011 RRID:AB_143157 |
| Secondary goat anti-rabbit conjugated with Alexa Fluor-647 | Thermo Fisher Scientific | Cat# A-21245 RRID:AB_2535813 |
| Secondary goat anti-chicken conjugated with Alexa Fluor-647 | Thermo Fisher Scientific | Cat# A-21449 RRID:AB_2535866 |
| Bacterial and virus strains | ||
| pLZRS-DPNTP-GFP | This study | N/A |
| pLZRS-GFP | [41] | N/A |
| pLZRS-Dsg1 | [41] | N/A |
| pLZRS-miR Dsg1 | [41] | N/A |
| pLZRS–NTshRNA (shCtl) | [67] | N/A |
| pLVX myristoylated tdTomato | Clontech | N/A |
| Biological samples | ||
| Neonatal human epidermal keratinocytes (NHEKs) | NU SBDRC | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Dispase II | Roche | Cat# 04942078001 |
| Goat serum | Sigma-Aldrich | Cat# G9023 |
| 4’,6-Diamido-2-Phenylindole (DAPI) | Sigma-Aldrich | Cat# D9542 |
| Alexa Fluor-568 Phalloidin | Thermo Fisher Scientific | Cat# A12380 |
| Recombinant Human Ephrin-A1 Fc peptide | R&D Systems | Cat# 6417-A1-050 |
| Rho Activator I | Cytoskeleton, Inc | Cat# CN01 |
| Mubritinib (TAK165) | Selleck Chemicals | Cat# S2216 |
| Dimethyl sulfoxide (DMSO) | Sigma-Aldrich | Cat# D8418 |
| PP2 | Sigma-Aldrich | Cat# P0042 |
| CCG-1423 | Sigma-Aldrich | Cat# 555558 |
| Ingenio Electroporation Solution | Mirus Bio | Cat# MIR 50117 |
| Polybrene | Sigma-Aldrich | Cat# H9268 |
| Albumin from bovine serum (BSA) | Sigma-Aldrich | Cat# A7906 |
| 16% Formaldehyde (w/v), Methanol-free | Thermo Fisher Scientific | Cat# PI28908 |
| Urea | Thermo Fisher Scientific | Cat# BP169-212 |
| Sodium Dodecyl Sulfate (SDS) | Thermo Fisher Scientific | Cat# BP166-500 |
| Tris Hydrochloride | Thermo Fisher Scientific | Cat# BP153-500 |
| β-Mercaptoethanol | Sigma-Aldrich | Cat# M6250 |
| Acrylamide/Bis-acrylamide, 30% solution | Sigma-Aldrich | Cat# A3574 |
| Xylene | Thermo Fisher Scientific | Cat# X3P |
| Citric acid | Sigma-Aldrich | Cat# C0759 |
| Glycine | Sigma-Aldrich | Cat# G8790 |
| Methanol | Sigma-Aldrich | Cat# 179337 |
| Glycerol | Thermo Fisher Scientific | Cat# BP229 |
| Ethyl Alcohol, 200 proof | Thermo Fisher Scientific | Cat# A4094 |
| Triton X-100 | Sigma-Aldrich | Cat# X100 |
| Tween20 | Sigma-Aldrich | Cat# P1379 |
| ProLong Gold antifade reagent | Thermo Fisher Scientific | Cat# P36930 |
| CELLstart CTS Substrate | Thermo Fisher Scientific | Cat# A10142-01 |
| Experimental models: Cell lines | ||
| Phoenix cells | G. Nolan, Stanford University | N/A |
| Medium 154CF, Kit (M154) With gentamicin/amphotericin B solution and CaCl2 | Thermo Fisher Scientific | Cat# M154CF500 |
| Human Keratinocyte Growth Supplement (HKGS) Kit | Thermo Fisher Scientific | Cat# S001K |
| PBS | Thermo Fisher Scientific | Cat# 21040CV |
| Opti-MEM™ I Reduced Serum Medium | Thermo Fisher Scientific | Cat# 11058021 |
| PCT Epidermal Keratinocyte Medium | Zenbio | Cat# CnT-07 |
| CnT-Prime 3D Barrier Media for 3D Epidermal Models | Zenbio | Cat# CnT-PR-3D |
| Oligonucleotides | ||
| siGENOME Non-Targeting siRNA Control Pool #2 | Dharmacon | Cat# D-001206-14 |
| siGENOME SMARTpool DSP siRNA | Dharmacon | Cat# D-019800-17 |
| stealth DSP siRNA oligonucleotide 5’-CAGGGCUCUGUCUUCUGCCUCUGAA-3’ | Thermo Fisher Scientific | N/A |
| Software and algorithms | ||
| ZEN 2.3 software | Carl Zeiss | RRID:SCR_013672 |
| NIS-Elements | Nikon | RRID:SCR_014329 |
| ImageJ | NIH | RRID:SCR_003070 |
| Prism | GraphPad | RRID:SCR_002798 |
| Microsoft Excel | Microsoft | RRID:SCR_016137 |
| Photoshop | Adobe | RRID:SCR_014199 |
| Illustrator | Adobe | RRID:SCR_010279 |
| Other | ||
| 24 mm Transwell 0.4 μm pore inserts | Costar | Cat# 3450 |
| Nucleofector 2b Device | Lonza | Cat# AAB-1001 |
| Amicon® Ultra-15 Centrifugal Filter Unit | Sigma-Aldrich | Cat# UFC9030 |
| EVOM epithelial volt/ohm meter | World Precision Instruments | N/A |
Highlights:
Desmosomes govern indicators of mechanical asymmetries in 3D human epidermal models
Disrupting desmosome-intermediate filament linkage affects core epidermal functions
Desmosomes regulate Src family kinase-mediated phosphorylation of ErbB2
ErbB2 activity is crucial for epidermal differentiation and tight junction barrier
Acknowledgments
We thank Alpha Yap, Andrew Kowalczyk, Lisa Godsel, and Quinn Roth-Carter for helpful feedback on the manuscript. Research reported in this publication was supported by Northwestern University Skin Biology & Diseases Resource-Based Center of the National Institutes of Health under award number P30AR075049. Imaging work was performed at the Northwestern University Center for Advanced Microscopy generously supported by NCI CCSG P30 CA060553 awarded to the Robert H Lurie Comprehensive Cancer Center. Multiphoton microscopy was performed on a Nikon A1R multiphoton microscope, acquired through the support of NIH 1S10OD010398–01. This work was supported by NIH grants R01 AR041836, R37 AR043380, with partial support from R01 CA228196, the J.L. Mayberry Endowment to K.J.G. and the Chicago Biomedical Consortium with support from the Searle Funds at The Chicago Community Trust. M.H. was supported by NIH F31 AR076188. J.A.B. was supported by NIH K01 AR075087 and T32 AR060710.
Footnotes
Declaration of Interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Brunet T & King N (2017). The Origin of Animal Multicellularity and Cell Differentiation. Developmental cell 43, 124–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rübsam M, Mertz AF, Kubo A, Marg S, Jüngst C, Goranci-Buzhala G, Schauss AC, Horsley V, Dufresne ER, Moser M et al. (2017). E-cadherin integrates mechanotransduction and EGFR signaling to control junctional tissue polarization and tight junction positioning. Nature communications 8, 1250–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elias PM (2008). Skin barrier function. Curr Allergy Asthma Rep 8, 299–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elias PM (2007). The skin barrier as an innate immune element. Semin Immunopathol 29, 3–14. [DOI] [PubMed] [Google Scholar]
- 5.Yoshida K, Yokouchi M, Nagao K, Ishii K, Amagai M & Kubo A (2013). Functional tight junction barrier localizes in the second layer of the stratum granulosum of human epidermis. J Dermatol Sci 71, 89–99. [DOI] [PubMed] [Google Scholar]
- 6.De Benedetto A, Kubo A & Beck LA (2012). Skin barrier disruption: a requirement for allergen sensitization? J Invest Dermatol 132, 949–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sluysmans S, Vasileva E, Spadaro D, Shah J, Rouaud F & Citi S (2017). The role of apical cell-cell junctions and associated cytoskeleton in mechanotransduction. Biology of the cell 109, 139–161. [DOI] [PubMed] [Google Scholar]
- 8.Messal HA, Alt S, Ferreira RMM, Gribben C, Wang VM-Y, Cotoi CG, Salbreux G & Behrens A (2019). Tissue curvature and apicobasal mechanical tension imbalance instruct cancer morphogenesis. Nature 566, 126–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fiore VF, Krajnc M, Quiroz FG, Levorse J, Pasolli HA, Shvartsman SY & Fuchs E (2020). Mechanics of a multilayer epithelium instruct tumour architecture and function. Nature 585, 433–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rubsam M, Broussard JA, Wickstrom SA, Nekrasova O, Green KJ & Niessen CM (2018). Adherens Junctions and Desmosomes Coordinate Mechanics and Signaling to Orchestrate Tissue Morphogenesis and Function: An Evolutionary Perspective. Cold Spring Harbor perspectives in biology 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lecuit T & Lenne PF (2007). Cell surface mechanics and the control of cell shape, tissue patterns and morphogenesis. Nat Rev Mol Cell Biol 8, 633–644. [DOI] [PubMed] [Google Scholar]
- 12.Broussard JA, Yang R, Huang C, Nathamgari SSP, Beese AM, Godsel LM, Hegazy MH, Lee S, Zhou F, Sniadecki NJ et al. (2017). The desmoplakin-intermediate filament linkage regulates cell mechanics. Molecular biology of the cell 28, 3156–3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huen AC, Park JK, Godsel LM, Chen X, Bannon LJ, Amargo EV, Hudson TY, Mongiu AK, Leigh IM, Kelsell DP et al. (2002). Intermediate filament-membrane attachments function synergistically with actin-dependent contacts to regulate intercellular adhesive strength. J Cell Biol 159, 1005–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Godsel LM, Hsieh SN, Amargo EV, Bass AE, Pascoe-McGillicuddy LT, Huen AC, Thorne ME, Gaudry CA, Park JK, Myung K et al. (2005). Desmoplakin assembly dynamics in four dimensions: multiple phases differentially regulated by intermediate filaments and actin. J Cell Biol 171, 1045–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin S, Gordon K, Kaplan N & Getsios S (2010). Ligand targeting of EphA2 enhances keratinocyte adhesion and differentiation via desmoglein 1. Molecular biology of the cell 21, 3902–3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mao Y & Baum B (2015). Tug of war—The influence of opposing physical forces on epithelial cell morphology. Developmental Biology 401, 92–102. [DOI] [PubMed] [Google Scholar]
- 17.Mertz AF, Che Y, Banerjee S, Goldstein JM, Rosowski KA, Revilla SF, Niessen CM, Marchetti MC, Dufresne ER & Horsley V (2013). Cadherin-based intercellular adhesions organize epithelial cell-matrix traction forces. Proceedings of the National Academy of Sciences of the United States of America 110, 842–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ng MR, Besser A, Brugge JS & Danuser G (2014). Mapping the dynamics of force transduction at cell–cell junctions of epithelial clusters. eLife 3, e03282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goodwin K, Lostchuck EE, Cramb KML, Zulueta-Coarasa T, Fernandez-Gonzalez R & Tanentzapf G (2017). Cell-cell and cell-extracellular matrix adhesions cooperate to organize actomyosin networks and maintain force transmission during dorsal closure. Molecular biology of the cell 28, 1301–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mammoto T, Mammoto A & Ingber DE (2013). Mechanobiology and developmental control. Annu Rev Cell Dev Biol 29, 27–61. [DOI] [PubMed] [Google Scholar]
- 21.Tse JM, Cheng G, Tyrrell JA, Wilcox-Adelman SA, Boucher Y, Jain RK & Munn LL (2012). Mechanical compression drives cancer cells toward invasive phenotype. Proceedings of the National Academy of Sciences of the United States of America 109, 911–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jensen MH, Morris EJ & Weitz DA (2015). Mechanics and dynamics of reconstituted cytoskeletal systems. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research 1853, 3038–3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Broussard JA, Jaiganesh A, Zarkoob H, Conway DE, Dunn AR, Espinosa HD, Janmey PA & Green KJ Scaling up single-cell mechanics to multicellular tissues - the role of the intermediate filament-desmosome network. Journal of cell science 133 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kapus A & Janmey P (2013). Plasma membrane--cortical cytoskeleton interactions: a cell biology approach with biophysical considerations. Compr Physiol 3, 1231–1281. [DOI] [PubMed] [Google Scholar]
- 25.Michael M, Meiring Joyce C. M., Acharya Bipul R., Matthews Daniel R., Verma S, Han Siew P., Hill Michelle M., Parton Robert G., Gomez Guillermo A. & Yap Alpha S. (2016). Coronin 1B Reorganizes the Architecture of F-Actin Networks for Contractility at Steady-State and Apoptotic Adherens Junctions. Developmental cell 37, 58–71. [DOI] [PubMed] [Google Scholar]
- 26.Anton KA, Sinclair J, Ohoka A, Kajita M, Ishikawa S, Benz PM, Renne T, Balda M, Jorgensen C, Matter K et al. (2014). PKA-regulated VASP phosphorylation promotes extrusion of transformed cells from the epithelium. Journal of cell science 127, 3425–3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu SK, Lagendijk AK, Hogan BM, Gomez GA & Yap AS (2015). Active contractility at E-cadherin junctions and its implications for cell extrusion in cancer. Cell cycle (Georgetown, Tex.) 14, 315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hodivala KJ & Watt FM (1994). Evidence that cadherins play a role in the downregulation of integrin expression that occurs during keratinocyte terminal differentiation. J Cell Biol 124, 589–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kocgozlu L, Saw Thuan B., Le Anh P., Yow I, Shagirov M, Wong E, Mège R-M, Lim Chwee T., Toyama Y & Ladoux B (2016). Epithelial Cell Packing Induces Distinct Modes of Cell Extrusions. Current Biology 26, 2942–2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mongera A, Rowghanian P, Gustafson HJ, Shelton E, Kealhofer DA, Carn EK, Serwane F, Lucio AA, Giammona J & Campas O (2018). A fluid-to-solid jamming transition underlies vertebrate body axis elongation. Nature 561, 401–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nekrasova O, Harmon RM, Broussard JA, Koetsier JL, Godsel LM, Fitz GN, Gardel ML & Green KJ (2018). Desmosomal cadherin association with Tctex-1 and cortactin-Arp2/3 drives perijunctional actin polymerization to promote keratinocyte delamination. Nature communications 9, 1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simpson CL, Patel DM & Green KJ (2011). Deconstructing the skin: cytoarchitectural determinants of epidermal morphogenesis. Nature Reviews Molecular Cell Biology 12, 565–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Connelly JT, Gautrot JE, Trappmann B, Tan DW-M, Donati G, Huck WTS & Watt FM (2010). Actin and serum response factor transduce physical cues from the microenvironment to regulate epidermal stem cell fate decisions. Nature Cell Biology 12, 711–718. [DOI] [PubMed] [Google Scholar]
- 34.Dubash AD, Koetsier JL, Amargo EV, Najor NA, Harmon RM & Green KJ (2013). The GEF Bcr activates RhoA/MAL signaling to promote keratinocyte differentiation via desmoglein-1. The Journal of Cell Biology 202, 653–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hatsell S & Cowin P (2001). Deconstructing desmoplakin. Nature Cell Biology 3, E270–E272. [DOI] [PubMed] [Google Scholar]
- 36.Vasioukhin V, Bowers E, Bauer C, Degenstein L & Fuchs E (2001). Desmoplakin is essential in epidermal sheet formation. Nat Cell Biol 3, 1076–1085. [DOI] [PubMed] [Google Scholar]
- 37.Sumigray KD & Lechler T (2012). Desmoplakin controls microvilli length but not cell adhesion or keratin organization in the intestinal epithelium. Molecular biology of the cell 23, 792–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Potter IY, Poumay Y, Squillace KA & Pittelkow MR (2001). Human EGF receptor (HER) family and heregulin members are differentially expressed in epidermal keratinocytes and modulate differentiation. Experimental cell research 271, 315–328. [DOI] [PubMed] [Google Scholar]
- 39.Hoesl C, Rohrl JM, Schneider MR & Dahlhoff M (2018). The receptor tyrosine kinase ERBB4 is expressed in skin keratinocytes and influences epidermal proliferation. Biochim Biophys Acta Gen Subj 1862, 958–966. [DOI] [PubMed] [Google Scholar]
- 40.Stoll SW, Kansra S, Peshick S, Fry DW, Leopold WR, Wiesen JF, Sibilia M, Zhang T, Werb Z, Derynck R et al. (2001). Differential utilization and localization of ErbB receptor tyrosine kinases in skin compared to normal and malignant keratinocytes. Neoplasia 3, 339–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Getsios S, Simpson CL, Kojima S, Harmon R, Sheu LJ, Dusek RL, Cornwell M & Green KJ (2009). Desmoglein 1-dependent suppression of EGFR signaling promotes epidermal differentiation and morphogenesis. J Cell Biol 185, 1243–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nagasawa J, Mizokami A, Koshida K, Yoshida S, Naito K & Namiki M (2006). Novel HER2 selective tyrosine kinase inhibitor, TAK-165, inhibits bladder, kidney and androgen-independent prostate cancer in vitro and in vivo. Int J Urol 13, 587–592. [DOI] [PubMed] [Google Scholar]
- 43.Marcotte R, Zhou L, Kim H, Roskelly CD & Muller WJ (2009). c-Src associates with ErbB2 through an interaction between catalytic domains and confers enhanced transforming potential. Mol Cell Biol 29, 5858–5871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu W, Yuan X, Beebe K, Xiang Z & Neckers L (2007). Loss of Hsp90 association up-regulates Src-dependent ErbB2 activity. Mol Cell Biol 27, 220–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dahlhoff M, Muzumdar S, Schäfer M & Schneider MR (2017). ERBB2 Is Essential for the Growth of Chemically Induced Skin Tumors in Mice. Journal of Investigative Dermatology 137, 921–930. [DOI] [PubMed] [Google Scholar]
- 46.le Duc Q, Shi Q, Blonk I, Sonnenberg A, Wang N, Leckband D & de Rooij J (2010). Vinculin potentiates E-cadherin mechanosensing and is recruited to actin-anchored sites within adherens junctions in a myosin II–dependent manner. Journal of Cell Biology 189, 1107–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gao X, Acharya BR, Engl WCO, De Mets R, Thiery JP, Yap AS & Viasnoff V (2018). Probing compression versus stretch activated recruitment of cortical actin and apical junction proteins using mechanical stimulations of suspended doublets. APL Bioeng 2, 026111–026111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cavanaugh KJ Jr., Oswari J & Margulies SS (2001). Role of stretch on tight junction structure in alveolar epithelial cells. Am J Respir Cell Mol Biol 25, 584–591. [DOI] [PubMed] [Google Scholar]
- 49.Spadaro D, Le S, Laroche T, Mean I, Jond L, Yan J & Citi S (2017). Tension-Dependent Stretching Activates ZO-1 to Control the Junctional Localization of Its Interactors. Current Biology 27, 3783–3795.e3788. [DOI] [PubMed] [Google Scholar]
- 50.Sumigray K, Zhou K & Lechler T (2014). Cell-Cell Adhesions and Cell Contractility Are Upregulated upon Desmosome Disruption. PLoS One 9, e101824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aranda V, Haire T, Nolan ME, Calarco JP, Rosenberg AZ, Fawcett JP, Pawson T & Muthuswamy SK (2006). Par6-aPKC uncouples ErbB2 induced disruption of polarized epithelial organization from proliferation control. Nat Cell Biol 8, 1235–1245. [DOI] [PubMed] [Google Scholar]
- 52.Shyer AE, Rodrigues AR, Schroeder GG, Kassianidou E, Kumar S & Harland RM (2017). Emergent cellular self-organization and mechanosensation initiate follicle pattern in the avian skin. Science (New York, N.Y.), eaai7868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eisenhoffer GT, Loftus PD, Yoshigi M, Otsuna H, Chien C-B, Morcos PA & Rosenblatt J (2012). Crowding induces live cell extrusion to maintain homeostatic cell numbers in epithelia. Nature 484, 546–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marinari E, Mehonic A, Curran S, Gale J, Duke T & Baum B (2012). Live-cell delamination counterbalances epithelial growth to limit tissue overcrowding. Nature 484, 542–545. [DOI] [PubMed] [Google Scholar]
- 55.Miroshnikova YA, Le HQ, Schneider D, Thalheim T, Rubsam M, Bremicker N, Polleux J, Kamprad N, Tarantola M, Wang I et al. (2018). Adhesion forces and cortical tension couple cell proliferation and differentiation to drive epidermal stratification. Nat Cell Biol 20, 69–80. [DOI] [PubMed] [Google Scholar]
- 56.Mesa KR, Kawaguchi K, Cockburn K, Gonzalez D, Boucher J, Xin T, Klein AM & Greco V (2018). Homeostatic Epidermal Stem Cell Self-Renewal Is Driven by Local Differentiation. Cell Stem Cell 23, 677–686.e674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thomas M, Ladoux B & Toyama Y (2020). Desmosomal Junctions Govern Tissue Integrity and Actomyosin Contractility in Apoptotic Cell Extrusion. Current biology : CB 30, 682–690.e685. [DOI] [PubMed] [Google Scholar]
- 58.Maître J-L, Turlier H, Illukkumbura R, Eismann B, Niwayama R, Nédélec F & Hiiragi T (2016). Asymmetric division of contractile domains couples cell positioning and fate specification. Nature 536, 344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Young P, Boussadia O, Halfter H, Grose R, Berger P, Leone DP, Robenek H, Charnay P, Kemler R & Suter U (2003). E-cadherin controls adherens junctions in the epidermis and the renewal of hair follicles. EMBO J 22, 5723–5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tunggal JA, Helfrich I, Schmitz A, Schwarz H, Günzel D, Fromm M, Kemler R, Krieg T & Niessen CM (2005). E-cadherin is essential for in vivo epidermal barrier function by regulating tight junctions. EMBO J 24, 1146–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gruber R, Börnchen C, Rose K, Daubmann A, Volksdorf T, Wladykowski E, Vidal YSS, Peters EM, Danso M, Bouwstra JA et al. (2015). Diverse regulation of claudin-1 and claudin-4 in atopic dermatitis. Am J Pathol 185, 2777–2789. [DOI] [PubMed] [Google Scholar]
- 62.Sugawara T, Iwamoto N, Akashi M, Kojima T, Hisatsune J, Sugai M & Furuse M (2013). Tight junction dysfunction in the stratum granulosum leads to aberrant stratum corneum barrier function in claudin-1-deficient mice. J Dermatol Sci 70, 12–18. [DOI] [PubMed] [Google Scholar]
- 63.Rachow S, Zorn-Kruppa M, Ohnemus U, Kirschner N, Vidal-y-Sy S, von den Driesch P, Börnchen C, Eberle J, Mildner M, Vettorazzi E et al. (2013). Occludin is involved in adhesion, apoptosis, differentiation and Ca2+-homeostasis of human keratinocytes: implications for tumorigenesis. PLoS One 8, e55116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Harmon RM, Simpson CL, Johnson JL, Koetsier JL, Dubash AD, Najor NA, Sarig O, Sprecher E & Green KJ (2013). Desmoglein-1/Erbin interaction suppresses ERK activation to support epidermal differentiation. The Journal of clinical investigation 123, 1556–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yehya N, Song MJ, Lawrence GG & Margulies SS (2019). HER2 Signaling Implicated in Regulating Alveolar Epithelial Permeability with Cyclic Stretch. Int J Mol Sci 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Arnette C, Koetsier JL, Hoover P, Getsios S & Green KJ (2016). In Vitro Model of the Epidermis: Connecting Protein Function to 3D Structure. Methods in enzymology 569, 287–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Arnette CR, Roth-Carter QR, Koetsier JL, Broussard JA, Burks HE, Cheng K, Amadi C, Gerami P, Johnson JL & Green KJ (2020). Keratinocyte cadherin desmoglein 1 controls melanocyte behavior through paracrine signaling. Pigment Cell Melanoma Res 33, 305–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liang X, Michael M & Gomez GA (2016). Measurement of Mechanical Tension at Cell-cell Junctions Using Two-photon Laser Ablation. Bio-protocol 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video S1. Tension in suprabasal layer; Related to Figure 2
A representative time lapse of a laser ablation experiment in the suprabasal layer in a Day 6 3D epidermal equivalent culture. Cell outlines are visualized with myr-tomato and time between frames is 1 sec. Total time is 45 sec. Field of view is 79.5×79.5 μm.
Data Availability Statement
All data needed to evaluate the conclusions of the paper are presented in the paper and/or in the Supplemental information. This study did not generate any unique code.