Abstract
H2N2 subtype low pathogenic avian influenza viruses (LPAIVs) have persisted in live bird markets (LBMs) in the Northeastern United States since 2014. Although unrelated to the 1957 pandemic H2N2 lineage, there is concern that the virus could have animal and public health consequences because of high contact with humans and numerous species in the LBM system. The pathogenicity, infectivity, and transmissibility of six LBM H2N2 viruses isolated from three avian species in LBMs were examined in chickens. Two of these isolates were also tested in Pekin ducks and guinea fowl. Full genome sequence was obtained from all 6 isolates and evaluated for genetic markers for host adaptation and pathogenicity in poultry. Clinical signs were not observed in any host with any of the isolates, however one recent isolate was shed at higher titers than the other isolates and had the lowest bird infectious dose of all the isolates tested in all three species. This isolate, A/chicken/NY/19-012787-1/2019, was also the only isolate with a deletion in the stalk region of the neuraminidase protein (NA). This supports the theory that the NA stalk deletion is evidence of adaptation to gallinaceous poultry.
Keywords: Low pathogenic avian influenza, infleunza A, H2N2, Neuraminidase stalk deletion, Chicken, Pekin duck, Guinea fowl
Introduction
Since the early 1990s, H2N2 low pathogenic avian influenza viruses (LPAIVs) have been sporadically isolated from birds of the North East US live bird markets (LBM) in the United States (US), and in 2014 a new lineage of H2N2 became established in the market system and persists in 2021 (Bulaga et al., 2003; Mullaney, 2003; USDA, 2018). LBMs have long been recognized as a conduit for avian influenza viruses (AIVs) to enter industrial poultry populations (Senne et al., 2003). They can also serve as genetic reservoirs that allows the viruses to change and transmit between species, including humans (Senne et al., 2006). Regarding human infections of H2N2, studies revealed that the ancestors of the H2N2 strains that caused the pandemic in 1957 have circulated in avian reservoirs (Schäffr et al., 1993). Moreover, as H2 subtypes are LPAIV strains, birds may not exhibit clinical signs of disease and might appear healthy, therefore increasing the risk of unnoticed exposure of other species to the virus.
Importantly, the H2N2 AIVs currently circulating in the US LBMs are not related to the 1957 pandemic lineage, which appears to be extinct. While the H2N2 subtype has disappeared from humans, there remains heightened concern that another H2 lineage virus can transmit to other species and cause a new human pandemic. Therefore, evaluating, monitoring, and controlling H2 isolates in poultry has to be thorough and precise (Jones et al., 2014; Pappas et al., 2015). Currently, there are two major H2 lineages in waterfowl in North America and Eurasia, one lineage consisting of purely North American strains and another grouping genetically mixed strains of North American and Eurasian strains.. Intercontinental spread of these lineages has been documented (Liu et al., 2004; Makarova et al., 1999; Piaggio et al., 2012), so some contemporary viruses from the US are related to strains from Eurasia.
In this study, we evaluated the pathogenicity, bird infectious dose, transmissibility, and genetic changes of recent H2N2 isolates from the US LBM system. The primary objective was to determine whether they are becoming more adapted to poultry species as they persist in the market system. The pathobiology of the H2N2 viruses was examined in chickens, guinea fowl and Pekin ducks because they are among the most common species in the LBMs (Garber et al., 2007). Earlier reports suggest that guinea fowl are more susceptible to infection with AIV than chickens (Bertran et al., 2017). Furthermore, guinea fowl have been found to be the species with the highest prevalence of LPAIV infection in LBMs (Bulaga et al., 2003). Domestic ducks such as Pekin ducks are also known as key intermediates in the transmission of AIV to commercial poultry (Cardona et al., 2009; Swayne et al., 2020; Webster et al., 1977).
For this study, an established animal model that has been used with numerous avian species for characterizing AIV isolates was applied (Bertran et al., 2017; Pantin-Jackwood et al., 2017; Spackman et al., 2016).
Material and Methods
Viruses.
Six H2N2 LPAIV viruses from US LBMs were selected based on recency and to represent the different genotypes present in the live birds markets during the time period (i.e., the presence or absence of a NA stalk deletion): A/duck/PA/14–030488-5/2014 (Dk/PA/14), A/chicken/NY/16–032621-2/2016 (Ck/NY/16), A/chicken/CT/17–008911-4/2017 (Ck/CT/17), A/chicken/NY/18–002471-4/2018 (CK/NY/02471/18), A/chicken/NY/18–042097-3/2018 (Ck/NY/042097/18) and A/chicken/NY/19–012787-1/2019 (Ck/NY/19) (Table 1). All isolates were obtained from the National Veterinary Services Laboratories, USDA-APHIS. The viruses were propagated and titrated in 9–10 day old embryonated chicken eggs (ECEs) by allantoic sac inoculation as per standard procedures (Spackman and Stephens, 2016). The actual challenge doses were verified by titrating the inoculum in ECEs.
Table 1.
Description of H2N2 isolates evaluated in this study.
| Isolate name | Abbreviation | Neuraminidase stalk deletion | Species tested | Accession number for gene segments |
|---|---|---|---|---|
| A/Muscovy Duck/PA/14–030488–5/2014 | Dk/PA/14 | No | Chickens | MN998501-MN998508 |
| A/chicken/NY/16–032621–2/2016 | Ck/NY/16 | No | Chickens | MW727349-MW727356 |
| A/chicken/CT/17–008911–4/2017 | Ck/CT/17 | No | Chickens | MW727341-MW727348 |
| A/chicken/NY/18–002471–4/2018 | Ck/NY/02471/18 | No | Chickens | MW727357-MW727364 |
| A/chicken/NY/18–042097–3/2018 | Ck/NY/042097/18 | No | Chickens, Guinea fowl, Pekin ducks | MW727365-MW727372 |
| A/chicken/NY/19–012787–1/2019 | Ck/NY/19 | Yes | Chickens, Guinea fowl, Pekin ducks | MW727373-MW727380 |
Birds.
All procedures involving animals were reviewed and approved by the US National Poultry Research Center (USNPRC) institutional animal care and use committee (IACUC). One hundred six Specific pathogen-free (SPF) White Leghorn chickens (Gallus gallus) were obtained from USNPRC in-house flocks, and sixty-four guinea fowl (Numida meleagris) and forty-eight Pekin ducks (Anas platyrhynchos) were obtained from commercial producers. Chickens and guinea fowl were challenged at 4 weeks of age and Pekin ducks were challenged at 2 weeks of age. Birds were individually tagged, and each treatment group was housed in isolation units that were ventilated under negative pressure with high-efficiency particulate filtered air. Directly inoculated and contact exposure birds were housed in the same unit so were able to make direct contact and shared feed and water. Lighting programs to which the birds were already acclimated were used, and feed and water were provided with ad libitum access. All birds were tested for antibody to the challenge virus prior to challenge by hemagglutination inhibition assay as described below.
The pathogenicity, infectious dose, and relative transmissibility of six LBM H2N2 isolates in chickens.
Birds were divided into groups and were inoculated with specific doses of the challenge viruses, as shown in Table 2. Briefly, each group consisted of 5 directly inoculated birds, with 3 uninoculated birds added approximately 24 hours post-inoculation (pi) to serve as contact-exposure birds. Three different doses were administered for each virus by the intra-choanal route, which were 102 50% egg infective doses (EID50) (low dose), 104 EID50 (medium dose), and 106 EID50 (high dose). All doses were administered in 0.1 mL. Two additional groups of eight birds were given the high dose of isolates Ck/NY/042097/18 and Ck/NY/19 for the purpose of a more complete evaluation of pathogenicity and virus shed (i.e., so there would be more birds at a high enough dose that was expected to infect all the birds). The clinical condition of all birds with a focus on signs of LPAIV (lethargy, diarrhea, upper respiratory disease) was evaluated daily throughout the study. With isolates Dk/PA/14, Ck/NY/16, Ck/CT/17 and CK/NY/02471/18, oropharyngeal (OP) and cloacal (CL) swabs were collected at 2 and 4 days post-inoculation (dpi) from directly inoculated birds, and 3 and 5 dpi from contact-exposure birds (2 and 4 days post exposure to inoculated birds). With isolates Ck/NY/042097/18 and Ck/NY/19, the same samples were collected at 2, 4, 7, 10, 14 dpi from inoculated birds and 3, 5 and 7 dpi from contact-exposure birds. Oropharyngeal and CL swabs were collected in 1 mL of brain heart infusion (BHI) media with a final concentration of gentamicin (1,000 μg/mL), penicillin (10,000 U/mL), and amphotericin B (20 IU/mL) and stored at −80°C before processing. Oropharyngeal swabs were obtained by swabbing the buccal cavity and choanal cleft. Blood for serum was collected from all surviving birds at the termination of the experiment at 14 dpi. Birds were euthanized at 14 dpi following IACUC approved protocols. Birds were considered infected if virus shedding from either the OP or CL routes was detected and/or the bird was seropositive for AIV at 14 dpi. The 50% bird infectious dose or BID50, was calculated with the Reed-Muench method (Reed and Muench, 1938).
Table 2.
Number of directly inoculated or contact exposed chickens shedding virus detected by real-time RT-PCR, percent of birds infected within dose groups for each isolate, mean bird infectious dose (calculated from the inoculated birds), and seroconversion.
| Inoculated birds | Contact exposed birds | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Isolate | Dosea | Virus detectionb |
Antibodyc | % infectedd | 50% BIDe | Virus detectionb |
Antibody | % infected | ||
| OP | CL | OP | CL | |||||||
| 6 | 2/5 | 0/5 | 0/5 | 40 | 0/3 | 0/3 | 0/3 | 0 | ||
| Dk/PA/14 | 4 | 0/5 | 0/5 | 0/5 | 0 | >6.0 | 0/3 | 0/3 | 0/3 | 0 |
| 2 | 0/5 | 0/5 | 0/5 | 0 | 0/3 | 0/3 | 0/3 | 0 | ||
| 6 | 4/5 | 0/5 | 4/5 | 80 | 0/3 | 0/3 | 0/3 | 0 | ||
| Ck/NY/16 | 4 | 0/5 | 0/5 | 0/5 | 0 | 5.3 | 0/3 | 0/3 | 0/3 | 0 |
| 2 | 0/5 | 0/5 | 0/5 | 0 | 0/3 | 0/3 | 0/3 | 0 | ||
| 6 | 1/5 | 0/5 | 2/5 | 20 | 0/3 | 0/3 | 0/3 | 0 | ||
| Ck/CT/17 | 4 | 0/5 | 0/5 | 0/5 | 0 | >6.0 | 0/3 | 0/3 | 0/3 | 0 |
| 2 | 0/5 | 0/5 | 0/5 | 0 | 0/3 | 0/3 | 0/3 | 0 | ||
| 6 | 1/5 | 0/5 | 1/5 | 20 | 0/3 | 0/3 | 0/3 | 0 | ||
| CK/NY/02471/18 | 4 | 0/5 | 0/5 | 0/5 | 0 | >6.0 | 0/3 | 0/3 | 0/3 | 0 |
| 2 | 0/5 | 0/5 | 0/5 | 0 | 0/3 | 0/3 | 0/3 | 0 | ||
| 6 | 13/13 | 12/13 | 12/13 | 100 | 3/3 | 3/3 | 3/3 | 100 | ||
| Ck/NY/042097/18 | 4 | 2/5 | 0/5 | 1/5 | 40 | 4.3 | 0/3 | 0/3 | 0/3 | 0 |
| 2 | 1/5 | 0/5 | 1/5 | 20 | 0/3 | 0/3 | 0/3 | 0 | ||
| 6 | 13/13 | 11/13 | 13/13 | 100 | 3/3 | 2/3 | 3/3 | 100 | ||
| Ck/NY/2019 | 4 | 5/5 | 4/5 | 5/5 | 100 | 2.8 | 3/3 | 3/3 | 3/3 | 100 |
| 2 | 0/5 | 0/5 | 1/5 | 20 | 0/3 | 0/3 | 0/3 | 0 | ||
Log10 50% egg infectious dose per bird.
Number of chickens shedding virus / total number of birds
Number of chickens positive for antibody / total number of birds
Birds were considered infected if viral RNA was detected in oropharyngeal (OP) and cloacal (CL) swabs at any timepoint and/or the chickens was positive for antibodies by hemagglutination inhibition assay at 14 days post inoculation.
Log10 50% bird infectious dose (BID) per mL.
The pathogenicity, bird infectious dose, and relative transmissibility of Ck/NY/042097/18 and Ck/NY/19 in Guinea fowl and Pekin ducks.
The two most recent isolates were also evaluated in two additional species, Pekin ducks and guinea fowl. The experimental design was identical with experiment 1 (Table 3) of Ck/Ny/042097/18 and Ck/NY/19 except only 5 Pekin ducks were used in the high dose groups. OP and CL swabs were collected at 2, 4, 7, 10, and 14 dpi from directly inoculated birds and at 3, 5, and 7 dpi (2, 4 and 6 days post exposure to inoculated birds) from contact-exposure birds. Experimental methodologies including sample collection, processing and evaluation were identical with the chicken experiments.
Table 3.
Number of directly inoculated or contact exposed Pekin ducks or guinea fowl shedding virus detected by real-time RT-PCR, percent of birds infected within dose groups for each isolate, mean bird infectious dose (calculated from the inoculated birds), and seroconversion.
| Inoculated birds | Contact exposed birds | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|||||||||||
| Species | Isolate | Dosea | Virus shedb |
% infectedd | Virus shedb |
% infected | |||||
| OP | CL | Serologyc | BIDe | OP | CL | Serology | |||||
| Pekin duck | Ck/NY/042097/18 | 6 | 2/5 | 0/5 | 2/5 | 40 | > 6.0 | 0/3 | 0/3 | 0/3 | 0 |
| 4 | 2/5 | 1/5 | 0/5 | 40 | 0/3 | 0/3 | 0/3 | 0 | |||
| 2 | 0/5 | 0/5 | 0/5 | 0 | 0/3 | 0/3 | 0/3 | 0 | |||
| Ck/NY/2019 | 6 | 4/5 | 4/5 | 5/5 | 100 | 3.3 | 3/3 | 1/3 | 3/3 | 100 | |
| 4 | 2/5 | 3/5 | 4/5 | 80 | 0/3 | 3/3 | 3/3 | 100 | |||
| 2 | 0/5 | 0/5 | 0/5 | 0 | 0/3 | 0/3 | 0/3 | 0 | |||
| Guinea Fowl | Ck/NY/042097/18 | 6 | 13/13 | 12/13 | 12/13 | 100 | 4.3 | 3/3 | 3/3 | 3/3 | 100 |
| 4 | 2/5 | 0/5 | 1/5 | 40 | 0/3 | 0/3 | 0/3 | 0 | |||
| 2 | 1/5 | 0/5 | 1/5 | 20 | 0/3 | 0/3 | 0/3 | 0 | |||
| Ck/NY/2019 | 6 | 13/13 | 11/13 | 13/13 | 100 | 2.8 | 3/3 | 2/3 | 3/3 | 100 | |
| 4 | 5/5 | 4/5 | 5/5 | 100 | 3/3 | 3/3 | 3/3 | 100 | |||
| 2 | 0/5 | 0/5 | 1/5 | 20 | 0/3 | 0/3 | 0/3 | 0 | |||
Log10 50% egg infectious dose per bird.
Number of birds shedding virus / total number of birds
Number of birds positive for antibody / total number of birds
Birds were considered infected if viral RNA was detected in oropharyngeal (OP) and cloacal (CL) swabs at any time point and/or the chickens was positive for antibodies by hemagglutination inhibition assay at 14 days post inoculation.
Log10 50% bird infectious dose (BID) per mL.
Sample processing and qRT- PCR.
Viral loads in OP and CL swabs were examined by quantitative real-time reverse-transcriptase polymerase chain reaction (qRT-PCR). RNA was extracted from swabs using the MagMAX96 Viral RNA Isolation Kit (Thermo Fisher Scientific, Waltham, MA) and the KingFisher Flex Magnetic Particle Processing System (Thermo Fisher Scientific), with an additional wash step to remove inhibitors (Das et al., 2009). The qRT- PCR for AIV detection was conducted based on the standard USDA M gene AIV qRT- PCR procedure (Spackman et al., 2002) using an Applied Biosystems® 7500 Fast Real- Time PCR system (Thermo Fisher Scientific). Cycle threshold (Ct) values were determined by the 7500 Fast Software v2.3. For relative quantification, Ct values were converted to titer equivalents based on the standard curve method (Larionov et al., 2005). Values were established from ten-fold dilutions of the same titrated stock of the virus used to challenge the birds. The limit of detection was determined to be 0.8Log10 per reaction.
Serology.
Hemagglutination inhibition (HI) assays using homologous antigens were performed to quantify antibody responses with serum collected from chickens, guinea fowl and Pekin ducks at 14 dpi based on the standard protocol (OIE, 2019). HI was selected because it’s not species specific and is quantitative. HI titers were reported as reciprocal log2 titers, and titers greater than 3 log2 (1:8) were considered positive.
Genome sequence analysis.
In order to identify amino acid sequence changes associated with AIV adaptation in poultry, comparative genome sequence analysis between challenge viruses was conducted. Complete genomes of all challenge viruses were sequenced using the MiSeq platform (Illumina, San Diego, CA). All 8 gene segments were amplified by PCR as described previously (Ferreri et al., 2019) and cDNA libraries were prepared using the Nextera XT DNA Sample Preparation Kit according to manufacturer instructions. Sequencing was performed using the 500 cycle MiSeq Reagent Kit v2 as described by Lee et al., (Lee et al., 2017). GenBank accession numbers are shown in Table 1. Nucleotide sequence assembly was initially conducted with IRMA v. 0.6.7 (Shepard et al., 2016) and automated assemblies were verified using DNAStar SeqMan NGen v. 14. The nucleotide sequences for the complete coding regions of all viruses were aligned using MAFFT and subsequently subjected to single-nucleotide polymorphisms (SNP) analysis using Geneious Prime (BioMatters, San Diego, CA)..
Statistical analyses.
Statistical analyses focused on the high dose groups because they were comprised of an adequate number of individual birds. Normal Gaussian distribution of the data sets were confirmed by Shapiro-Wilk test and Kolmogorov-Smirnov test Two-way analysis of variance (2-way ANOVA), along with Tukey’s multiple-comparison tests, was used to compare virus shed titers among species-virus challenge groups using Prism v.8.2.1 (GraphPad software, La Jolla, CA) (groups with fewer than 7 birds were excluded due to low power). Negative samples were given an imputed titer of 50% of the limit of detection (in titer equivalents) (Cohen, 1959). The Fisher exact test was used to compare the proportion of bird shedding in the high dose groups (Prism 8.2.1).
Results
Infectivity and transmissibility of six LBM H2N2 viruses in chickens.
No chickens in the low and medium dose groups of four isolates (Dk/PA/14, Ck/NY/16, Ck/CT/17, CK/NY/02471/18) were infected based on viral RNA presence in swabs and antibody titers in blood serum collected at 14 dpi (Table 2, Supplemental Figure 1). However, 20% (1 of 5) of the chickens were infected in the low dose group and 40% (2 of 5) were infected in the medium dose group of Ck/NY/042097/18, while 20% (1 of 5) and 100% (5 of 5) of birds were infected in the low and medium dose groups of Ck/NY/19, respectively Forty percent (2 of 5) and 80% (4 of 5) of the chickens were infected in the high dose groups of Dk/PA/14 and Ck/NY/16, respectively, while only 20% (1 of 5) were infected in the Ck/CT/17 and CK/NY/02471/18 groups. None of the contact exposure birds were infected in any dose group by the isolates Dk/PA/14, Ck/NY/16, Ck/CT/17 and cK/NY/02471/18. In the high dose groups of Ck/NY/042097/18 and Ck/NY/19, 100% (13 of 13) of chickens were infected.
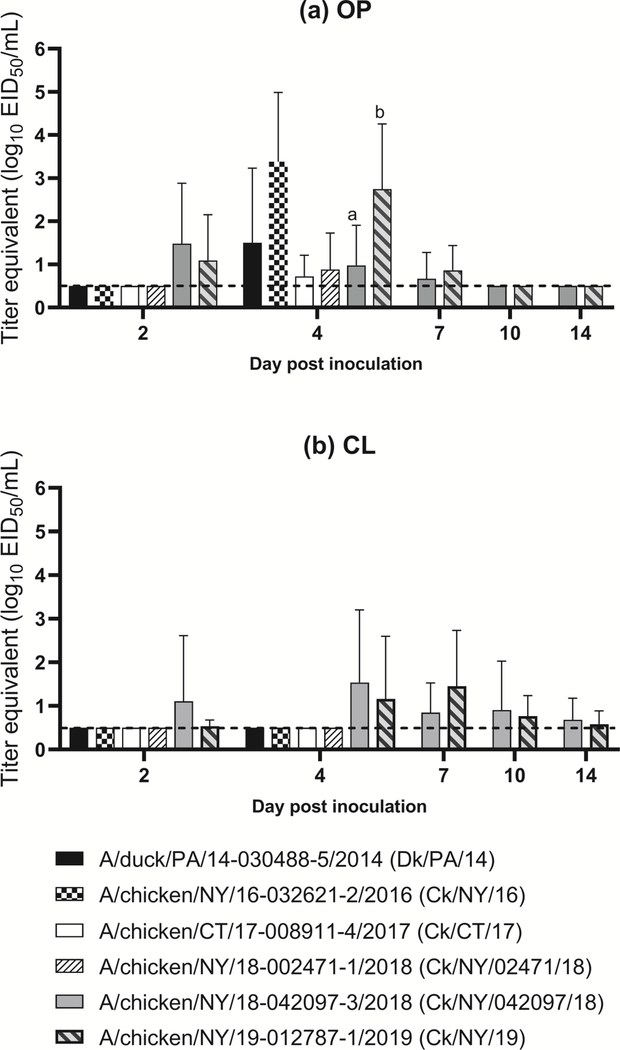
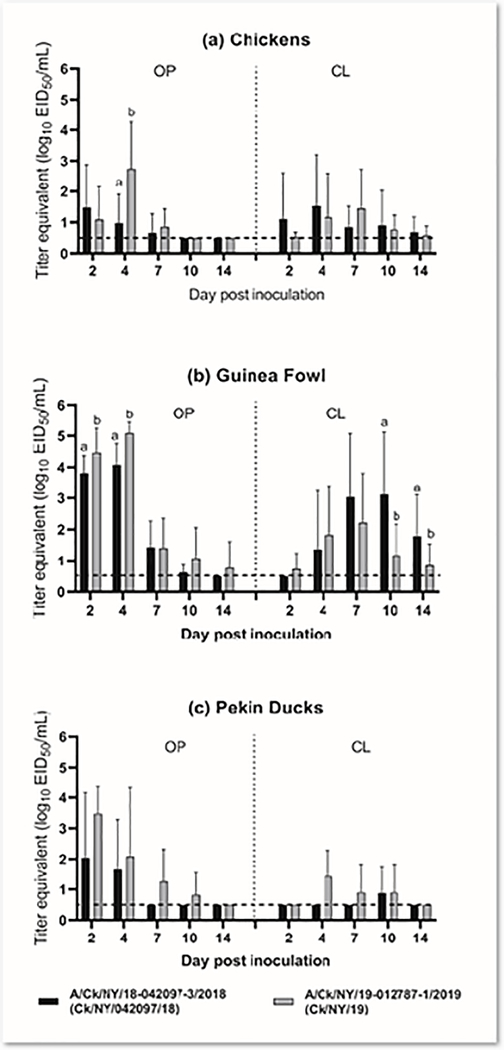
Virus shedding by the OP route was detectable at 4 dpi in the high dose groups of all isolates (Figure 1a). The OP shed titers of the Ck/NY/16 high dose group at this time point were significantly higher compared to other isolates, except Ck/NY/19. With Ck/NY/042097/18 and Ck/NY/19, virus shedding by the OP route was detectable starting from 2 dpi until 7 dpi and no viral RNA was detected in OP swabs collected beyond 7 dpi regardless of isolate and dose group (Figure 1a). CL shedding of Ck/NY/042097/18 and Ck/NY/19 was mostly detectable at all time points, which peaked at 4 dpi for Ck/NY/042097/18 and at 7 dpi for Ck/NY/19. Chickens infected with the high doses of both isolates shed virus predominantly by the OP route. When comparing quantities of virus shed in chickens at 2 and 4 dpi, OP shedding was only observed in Ck/NY/042097/18 and Ck/NY/19 at 2 dpi, but was observed with all viruses at 4 dpi. However, CL shedding was observed only by chickens infected with Ck/NY/042097/18 (2 dpi) and Ck/NY/19 (2 and 4 dpi) over the same time frame. A direct pairwise comparison of shed titers between all isolates was difficult as data was only collected at 2 and 4 dpi for most of the viruses except the two recent isolates Ck/NY/042097/18 and Ck/NY/19 (Figure 1a, 1b), Supplemental Figure 2), where Ck/NY/19 was shed at significantly higher titers at 4 dpi by the OP route.
Figure 1.
Log10 titer equivalents from quantitative real-time RT-PCR for viral RNA detected in swabs collected from chickens inoculated with the high dose (106 EID50) of each H2N2 isolate; (a) oropharyngeal (OP); and (b) cloacal (CL). Samples were only collected from two of the viruses (A/chicken/NY/18–042097-3/2018 and A/chicken/NY/19–012787-1/2019) at 7, 10, and 14 days post inoculation (DPI) andstatistical groups are only shown for these groups because other challenge groups had too low power (n=5). Groups are denoted by letters and are only shown on DPI when a significant difference was observed.
No contact-exposure chickens were infected in any of the low and medium dose groups of Ck/NY/042097/18, but 100% (3 of 3) were infected in the medium dose group of Ck/NY/19 (Table 2). 100% (3 of 3) of contact-exposure chickens were infected in both high dose groups of Ck/NY/042097/18 and Ck/NY/19. Seropositivity did not directly correlate with detection of virus shed (birds that shed detectable levels of virus were positive for antibody). Regardless of the dose, none of the isolates caused observable clinical disease in infected chickens. The 50% bird infectious doses (BID50) for chickens ranged among the isolates from 2.8 to >6.0 log10 BID50 (Table 2).
The pathogenicity, bird infectious dose, and relative transmissibility of Ck/NY/042097/18 and Ck/NY/19 in Pekin Ducks.
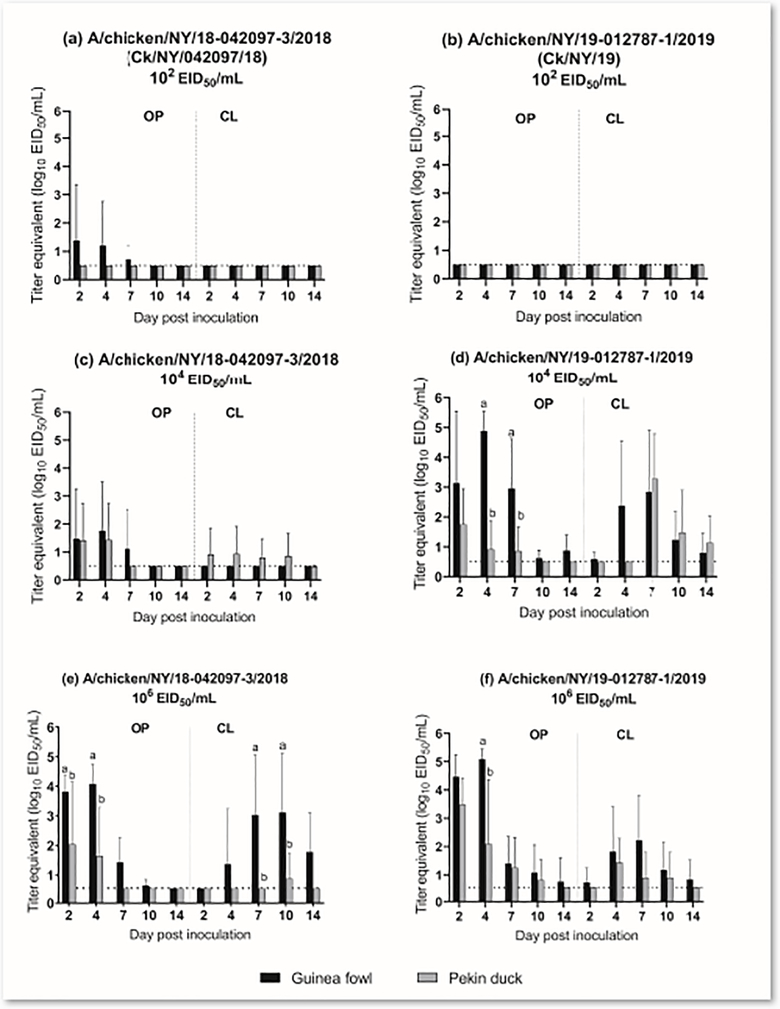
No ducks, including contact-exposed animals, were infected in the low dose groups of either isolate, while 40% (2 of 5) and 80% (4 of 5) of inoculated ducks were infected in the medium dose groups of CK/NY/042097/18 and Ck/NY/19, respectively (Table 3). In the high dose groups, only 40% (2 of 5) of the ducks were infected with Ck/NY/042097/18 and all ducks (5 of 5) were infected with Ck/NY/19 (Table 3). Oropharyngeal shedding occurred through 4 dpi in the Ck/NY/042097/18 medium dose group (Figure 2c) and 7 dpi in the Ck/NY/19 medium dose group (Figure 2d). In the Ck/NY/19 medium dose group, CL shed titers peaked at 7 dpi (mean titer: 103.3 EID50/mL) and continued through the end of the experiment (14 dpi). None of the contact-exposure ducks from the medium dose group of Ck/NY/042097/18 were infected, while 100% (3 of 3) were infected in the Ck/NY /19 medium dose group. The OP and CL titers from ducks infected with high dose Ck/NY/19 were significantly higher compared to the Ck/NY/042097/18 (Supplemental Figure 2). None of the contact-exposure ducks from the Ck/NY/042097/18 high dose group were infected, while 100% (3 of 3) were infected in the Ck/NY/19 group. Regardless of the challenge dose, no clinical signs were observed in infected ducks. The BID50 for Ck/NY/042097/18 was >6.0 log10 EID50 and was 3.3 log10 EID50 for Ck/NY/19 (Table 3). Antibodies were detected in serum from 40% of inoculated ducks in the Ck/NY/042097/18 high dose group and all inoculated ducks in the Ck/NY/19 high dose group. Seropositivity generally correlated with detection of virus shed (Table 3).
Figure 2.
Log10 titer equivalents from quantitative real-time RT-PCR for viral RNA detected in oropharyngeal (OP) and cloacal (CL) swabs collected from guinea fowl or Pekin ducks; (a) A/chicken/NY/18–042097-3/2018 low dose (102 EID50), (b) A/chicken/NY/19–012787-1/2019 low dose (102 EID50), (c) A/chicken/NY/18–042097-3/2018 medium dose (104 EID50), (d) A/chicken/NY/19–012787-1/2019 medium dose (104 EID50), (e) A/chicken/NY/18–042097-3/2018 high dose (106 EID50), (f) A/chicken/NY/19–012787-1/2019 high dose (106 EID50). Statistical groups are denoted by letters and are only shown on DPI when a significant difference was observed.
The pathogenicity, infectious dose, and relative transmissibility of Ck/NY/042097/18 and Ck/NY/19 in Guinea fowl.
Twenty percent (1 of 5) of the guinea fowl from each low dose group of both isolates were infected (Table 3). In the medium dose groups, 40% (2 of 5) and 100% (5 of 5) of guinea fowl were infected with Ck/NY/042097/18 and Ck/NY/19, respectively. All directly inoculated guinea fowl in the high dose groups of both isolates were infected. Viral RNA was detected in only OP swabs collected from guinea fowl inoculated with medium dose Ck/NY/042097/18 until 7 dpi (Figure 2c), whereas in the Ck/NY/19 group, viral RNA was detectable from both OP and CL swabs collected at all time points (Figure 2d). The OP and CL shed titers of the Ck/NY/19 were significantly higher than those of Ck/NY/042097/18 from both the medium and high dose groups at several time points. The guinea fowl in the Ck/NY/19 high dose group shed more virus orally; viral titers in the OP swabs collected from this group were almost 1 log10 higher than the Ck/NY/042097/18 group at 2 dpi (mean titer: 104.5 EID50/mL vs. 103.8 EID50/mL) and 4 dpi (mean titer: 105.1 EID50/mL vs. 104.1 EID50/mL) (Figure 2e, 2f). In contrast, guinea fowl in the high dose Ck/NY/042097/18 group shed higher titers cloacally at 7 and 10 dpi. No contact-exposure guinea fowl were infected in either low dose group or the Ck/NY/042097/18 medium dose group. However, 100% were infected in both Ck/N/19 medium dose group and in high dose groups (Table 3). Infected guinea fowl did not exhibit any observable clinical signs. The BID50 of Ck/NY/042097/18 and Ck/NY/19 in guinea fowl were 4.3 log10 EID50 and 2.8 log10 EID50, respectively (Table 3).
Comparison of virus shed among chickens, Pekín ducks and guinea fowl with Ck/NY/042097/18 and Ck/NY/19.
Virus shed patterns were compared among all three species for Ck/NY/042097/18 and Ck/NY/19 and were compared between the isolates for each species (Figure 3, Supplemental Figure 2). There was a trend for guinea fowl infected with Ck/NY/042097/18 to shed more virus both orally and cloacally than chickens or Pekin ducks, which was significant at 2, 4, and 7 DPI for OP swabs and at 7, 10, and 14 DPI for CL swabs. Similarly, guinea fowl also shed significantly more Ck/NY/19 than chickens and/or ducks by the OP route at 2 and 4 dpi. Based on Fisher’s exact test, the highest proportion of birds shedding the Ck/NY/19 virus at the high dose were guinea fowl, which also shed significantly higher titers orally at earlier time points (2, 4, and 7 dpi) and cloacally at later time points (7and 10 dpi).
Figure 3.
Log10 titer equivalents from quantitative real-time RT-PCR for viral RNA detected in oropharyngeal (OP) and cloacal (CL) swabs collected from birds inoculated with the high dose (106 EID50) of A/chicken/NY/18–042097-3/2018 or A/chicken/NY/19–012787-1/2019; (a) chickens; (b) guinea fowl; or (c) Pekin ducks; Statistical groups are denoted by letters and are only shown on DPI when a significant difference was observed.
When comparing Ck/NY/042097/18 and Ck/NY/19 shed within a species, there was no difference for Pekin ducks. However, Ck/NY/19 was shed at significantly higher titers than Ck/NY/042097/18 by chickens at 4 dpi by the OP route and by guinea fowl by the OP route at 2 and 4 dpi (Figure 3a, 3b). In contrast, titers of Ck/NY/042097/18 were higher in CL swabs from guinea fowl at 10 and 14 dpi.
Genetic analysis of LBM H2N2 isolates.
SNP analysis of complete coding regions was conducted to identify amino acid sequence changes associated with the level of virus adaptation in poultry. A total of 23 amino acid differences were identified in the Ck/NY/19 isolate were when compared with other H2N2 challenge viruses (Table 4). The most notable difference was that the Ck/NY/19 isolate has a 19 amino acid deletion in the NA protein stalk region.
Table 4.
Nonsynonymous amino acid substitutions found in the Ck/NY/19 isolate when compared with other challenge viruses from this study. Notable substitutions (K350R, A85T, I432V) and the deletion in the NA stalk deletion region are bolded.
| Protein | Position | Ck/NY/2019 | Other challenge viruses | Reference(s) |
|---|---|---|---|---|
| PB2 | 214 | R | K | |
| PB1 | 350 | K | R | (Suzuki et al., 2009) |
| PA | 85 | A | T | (Kajihara et al., 2013) |
| 311 | I | M | ||
| 432 | I | V | (Bogs et al., 2010) | |
| 504 | V | I | ||
| 626 | K | R | ||
| PA-X | 85 | A | T | |
| 215 | L | P | ||
| HA | 109 | N | S | |
| 126 | L | F | ||
| 226 | V | E | ||
| 228 | V | A | ||
| 322 | T | S | ||
| 334 | A | V | ||
| 406 | I | V | ||
| 429 | I | L | ||
| NP | 357 | K | Q | |
| NA | 63–82 (Stalk region) | Deleted | TEIVYLNNTTIEREICPKAV | (Banks et al., 2001; Hossain et al., 2008; Munier et al., 2010; Sorrell et al., 2010) |
| M1 | 107 | V | I | |
| M2 | 27 | V | I | |
| NS1 | 75 | K | E | |
| NEP | 111 | G | S |
Discussion
The bird infectious dose, relative transmissibility and pathogenicity of H2N2 LPAIVs that were recently isolated from LBMs located in the Northeast US were characterized in chickens, Pekin ducks, and guinea fowl, revealing species- and isolate-specific differences. No mortality or clinical signs were observed in any of the infected species, regardless of dose or isolate. This was expected as no or low morbidity and mortality in gallinaceous species and ducks is typical of LPAI infection under experimental conditions (Alexander et al., 1986; Jackwood et al., 2010; Makarova et al., 2003; Pantin-Jackwood et al., 2017; Spackman, 2020).
The mean bird infectious dose varied among species and isolates, reinforcing that the host adaptation and infectivity of influenza A virus is species specific. Expectedly, transmission to contact exposure birds correlated with infectious dose because the directly inoculated birds were infected and shed higher amounts of virus. In a previous study, the upper limit for a mean bird infectious dose that could predict a sustainable transmissibility in chickens was suggested to be 104.7 EID50 (Swayne and Slemons, 2008). Based on that criteria, the Ck/NY/19 LPAIV isolate would be able to be maintained in poultry populations in the LBMs and would likely have a fitness advantage over isolates with higher mean bird infectious doses. Importantly, birds in the LBMs are likely under high stress which may affect susceptibility to infection and disease. This may help explain why the chicken-origin isolates are poorly infectious for chickens in experimental conditions. Furthermore, because of the variety of species present in many markets, these isolates may be maintained in species other than chickens or may be an environmental spillover.
Among the three species, guinea fowl was the most susceptible to virus infection and shed the most virus. Data available for guinea fowl are limited, although there is a report with highly pathogenic (HP) AIV which showed they were highly susceptible to disease and infection (Bertran et al., 2017) and another study demonstrated their high susceptibility to LPAIV (Umar et al., 2016). More side-by-side studies are needed, but there appears to be variation in susceptibility of gallinaceous birds to AIV infection and virus replication, where chickens are generally more resistant than turkeys or guinea fowl (Pantin-jackwood et al., 2017; Spackman et al., 2010; Swayne and Slemons, 2008).
Notably the isolate with the 19 amino acid NA stalk deletion (Ck/NY/19) had the lowest bird infectious dose, and exhibited the best transmission to contact exposure birds, for all three species. This H2N2 lineage is only found in the LBM system, which suggests that the NA deletion occurred during circulation of the virus in that environment. Deletions in the NA have been recognized as a marker of influenza virus adaptation to gallinaceous species (Banks et al., 2001; Hossain et al., 2008; Munier et al., 2010; Sorrell et al., 2010). Previous studies have shown that the host range of AIV could be affected by the NA stalk’s length (Castrucci and Kawaoka, 1993; Guangxiang et al., 1993). However, this is the first study to demonstrate that an isolate with an NA stalk deletion has a lower bird infectious dose in 3 avian species examined side-by-side. Interestingly, the lower bird infectious dose was also observed in a waterfowl species, Pekin ducks.
The neuraminidase protein is a sialidase that removes sialic acid from both the host and viral proteins and allows the extracellular release of the virus after replication. There appears to be an optimal level of hemagglutinin binding to host receptors to allow entry into the cell, but also a level of attachment that allows release from the cell and transmission to another cell or host. The NA is thought to help balance these competing hemagglutinin activities of virus entry and the release of the virus after replication to allow effícient transmission (Baigent and McCauley, 2001; Castrucci and Kawaoka, 1993; Els et al., 1985). A deletion in the NA stalk will effectively strengthen HA binding by decreasing the sialidase activity of the NA protein which cleaves the virus from the cell during the infection process (Els et al., 1985). The NA in low pathogenic viruses in wild birds rarely if ever has a stalk deletion, but it is common to see stalk deletions in viruses that have circulated in gallinaceous birds. It remains unclear of why there is selective pressure in gallinaceous birds for reduced sialidase activity, but the infection and transmission studies in this study support an important role of stalk deletions. In addition to birds, stalk deletions have been shown to affect replication cell culture and in mice (Baigent and McCauley, 2001; Castrucci and Kawaoka, 1993).
The role of other genetic differences between Ck/NY/19 and the other isolates that contribute to host adaptation would need to be investigated with reverse genetics, but some have been reported previously. Substitutions in Ck/NY/19 included K350R in the PB1 protein which may disrupt early innate immune responses (Suzuki et al., 2009), A85T which was associated with increased lethality of HPAIVs in ducks (Kajihara et al., 2013), and I432V in the PA protein which is considered a virulence determinant in chickens (Bogs et al., 2010). Other changes have not yet been defined. There is also a possibility that the mutations and deletions in Ck/NY/19 may have contributed to increased replication effíciency of the virus itself, as the virus was shed in highest titers compared to other isolates across all species tested in this study. Several studies have shown that intermediate host-associated genetic changes in AIV could increase the virus’s replication efficiency in other avian species (Hossain et al., 2008; Sorrell and Perez, 2007). The Ck/NY/19 isolate may have progressively gained genetic properties that increased its replication fitness in several hosts by circulating in LBMs for some time. Oropharyngeal and CL shed titers and the proportion of birds shedding also varied by species-host combination. Generally, isolates with a lower mean bird infectious dose were shed at higher titers across all three species. The general trend for earlier oral shedding compared to cloacal shedding is probably due to the birds being inoculated by the intra-choanal route. Also, previous studies have shown that chickens generally don’t shed LPAIVs by the CL route as consistently and with as high titers as they do by the oral route (Germeraad et al., 2019; Pantin-Jackwood et al., 2017). Compared to chickens and Pekin ducks, guinea fowl shed the highest quantity for the longest duration by both the OP and CL routes with the two viruses tested in all three species. The highest proportion of birds shedding these isolates was also the guinea fowl. Guinea fowl could potentially contribute to environmental contamination more than chickens or Pekin ducks with these viruses. This is important as guinea fowl are commonly reared in the US as commercial free range or by small-holder commercial growers (Robbins et al., 2011). Because of their low production costs, greater capacity to use environmentally sustainable feeds, and higher protein content in meat, guinea fowl are becoming more popular in the markets, so maintaining surveillance of guinea fowl in the US LBMs is important. Interestingly, the virus which was shed at the highest titers by guinea fowl and Pekin ducks was the isolate with the NA stalk deletion (Ck/NY/19).
One key limitation of the study is that all six isolates were not tested in guinea fowl and Pekin ducks. Therefore, it is difficult to determine if better adaptation (lower bird infectious dose, better relative transmissibility, and higher shed titers) to these species, which are common in the LBM system, are a trend with the more recent viruses. A detailed network analysis and molecular epidemiological study (unpublished) is in progress and should help shed more light on how the viral variants are evolving and has been spreading within the LBM system.
Monitoring the biological characteristics of viruses such as the H2N2s in the LBM system is important because viruses in LBM systems have the potential to be exposed to numerous hosts at high levels, which is conducive to crossing the species barrier and broadening host range. For commercial poultry there is always a threat of transmission when an LPAIV is endemic in the LBMs (Senne et al., 2003; Suarez et al., 2003). Although the H2N2 LPAIV may not be seen as a major disease issue like H5 or H7 AIVs, if the virus becomes adapted to chickens it could have a greater impact on production or even trade. Finally, there are surveillance programs in place in the US LBM system that will contínue to monitor AIV evolution until it can be eradicated, which is the ultimate goal of these programs.
Conclusions
Although the prevalence varies, H2N2 LPAIVs have been regularly isolated from the Northeast US LBMs over the past few decades. The isolates are acquiring mutations that suggest that the virus may be adapting from wild waterfowl to the species (e.g., gallinaceous birds) that are common in the LBMs. One adaptation is a deletion in the NA stalk protein. Here an isolate with the stalk deletion has a lower bird infectious dose for chickens, Pekin ducks and guinea fowl. The role of other residue changes is unknown. Also, the viruses were generally shed well by guinea fowl, suggesting that they can serve as a reservoir for other species in the market system.
Supplementary Material
Highlights.
H2N2 influenza A are regularly isolated from US live bird markets.
Mutations have been acquired and may increase adaptation to species in the markets.
An H2N2 influenza A had a lower infectious dose in chickens, turkeys and ducks.
None of the virus caused observable disease but were shed efficiently by the oral and cloacal routes.
Acknowledgments
The authors thank: Scott Lee, Jesse Gallagher, Nick Lee, Melinda Vongkungthong, Anne Hurley-Bacon, Roger Brock and Chuck Foley for technical assistance with this work. Mention of trade names or commercial products in this manuscript is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Government. USDA is an equal opportunity provider and employer. Funding: This research was supported by US Department of Agriculture, ARS CRIS Project 6040-32000-066-00D, USDA-APHIS agreement #60-6040-6-005 and NIH-CEIRS Agreement #AAI-12004001. This research was also supported in part by an appointment to the ARS Research Participation Program administered by the Oak Ridge Institute for Science and Education (ORISE) through an interagency agreement between the U.S. Department of Energy (DOE) and the U.S. Department of Agriculture (USDA). ORISE is managed by ORAU under DOE contract number DE-SC0014664.
Footnotes
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alexander D, Parsons G, Manvell R, 1986. Experimental assessment of the pathogenicity of eight avian influenza A viruses of H5 subtype for chickens, turkeys, ducks and quail. Avian pathology 15, 647–662. [DOI] [PubMed] [Google Scholar]
- Baigent SJ, McCauley JW, 2001. Glycosylation of haemagglutinin and stalk-length of neuraminidase combine to regulate the growth of avian influenza viruses in tissue culture. Virus Res 79, 177–185. [DOI] [PubMed] [Google Scholar]
- Banks J, Speidel E, Moore E, Plowright L, Piccirillo A, Capua I, Cordioli P, Fioretti A, Alexander D, 2001. Changes in the haemagglutinin and the neuraminidase genes prior to the emergence of highly pathogenic H7N1 avian influenza viruses in Italy. Archives of virology 146, 963–973. [DOI] [PubMed] [Google Scholar]
- Bertran K, Lee D-H, Pantin-Jackwood MJ, Spackman E, Balzli C, Suarez DL, Swayne DE, 2017. Pathobiology of clade 2.3. 4.4 H5Nx high-pathogenicity avian influenza virus infections in minor gallinaceous poultry supports early backyard flock introductions in the Western United States in 2014–2015. Journal of Virology 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogs J, Veits J, Gohrbandt S, Hundt J, Stech O, Breithaupt A, Teifke JP, Mettenleiter TC, Stech J, 2010. Highly pathogenic H5N1 influenza viruses carry virulence determinants beyond the polybasic hemagglutinin cleavage site. PloS one 5, e11826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulaga L, Garber L, Senne D, Myers T, Good R, Wainwright S, Suarez D, 2003. Descriptive and surveillance studies of suppliers to New York and New Jersey retail live-bird markets. Avian diseases 47, 1169–1176. [DOI] [PubMed] [Google Scholar]
- Cardona C, Yee K, Carpenter T, 2009. Are live bird markets reservoirs of avian influenza? Poultry science 88, 856–859. [DOI] [PubMed] [Google Scholar]
- Castrucci MR, Kawaoka Y, 1993. Biologic importance of neuraminidase stalk length in influenza A virus. J Virol 67, 759–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen AC, 1959. Simplified estimators for the normal distribution when samples are singly censored or truncated. Technometrics 1, 217–237. [Google Scholar]
- Das A, Spackman E, Pantin-Jackwood MJ, Suarez DL, 2009. Removal of real-time reverse transcription polymerase chain reaction (RT-PCR) inhibitors associated with cloacal swab samples and tissues for improved diagnosis of Avian influenza virus by RT-PCR. Journal of Veterinary Diagnostic Investigation 21, 771–778. [DOI] [PubMed] [Google Scholar]
- Els MC, Air GM, Murti KG, Webster RG, Laver WG, 1985. An 18-amino acid deletion in an influenza neuraminidase. Virology 142, 241–247. [DOI] [PubMed] [Google Scholar]
- Ferreri LM, Ortiz L, Geiger G, Barriga GP, Poulson R, Gonzalez-Reiche AS, Crum JA, Stallknecht D, Moran D, Cordon-Rosales C, Rajao D, Perez DR, 2019. Improved detection of influenza A virus from blue-winged teals by sequencing directly from swab material. Ecol Evol 9, 6534–6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber L, Voelker L, Hill G, Rodriguez J, 2007. Description of live poultry markets in the United States and factors associated with repeated presence of H5/H7 low-pathogenicity avian influenza virus. Avian diseases 51, 417–420. [DOI] [PubMed] [Google Scholar]
- Germeraad EA, Sanders P, Hagenaars TJ, de Jong M, Beerens N, Gonzales JL, 2019. Virus shedding of avian influenza in poultry: a systematic review and meta-analysis. Viruses 11, 812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guangxiang L, Jeffrey C, Palese P, 1993. Alterations of the stalk of the influenza virus neuraminidase: deletions and insertions. Virus research 29, 141–153. [DOI] [PubMed] [Google Scholar]
- Hossain MJ, Hickman D, Perez DR, 2008. Evidence of expanded host range and mammalian-associated genetic changes in a duck H9N2 influenza virus following adaptation in quail and chickens. PloS one 3, e3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackwood MW, Suarez DL, Hilt D, Pantin-Jackwood MJ, Spackman E, Woolcock P, Cardona C, 2010. Biologic characterization of chicken-derived H6N2 low pathogenic avian influenza viruses in chickens and ducks. Avian diseases 54, 120–125. [DOI] [PubMed] [Google Scholar]
- Jones JC, Baranovich T, Marathe BM, Danner AF, Seiler JP, Franks J, Govorkova EA, Krauss S, Webster RG, 2014. Risk assessment of H2N2 influenza viruses from the avian reservoir. Journal of virology 88, 1175–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajihara M, Sakoda Y, Soda K, Minari K, Okamatsu M, Takada A, Kida H, 2013. The PB2, PA, HA, NP, and NS genes of a highly pathogenic avian influenza virus A/whooper swan/Mongolia/3/2005 (H5N1) are responsible for pathogenicity in ducks. Virology journal 10, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larionov A, Krause A, Miller W, 2005. A standard curve based method for relative real time PCR data processing. BMC bioinformatics 6, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DH, Torchetti MK, Killian ML, Berhane Y, Swayne DE, 2017. Highly Pathogenic Avian Influenza A(H7N9) Virus, Tennessee, USA, March 2017. Emerg Infect Dis 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J-H, Okazaki K, Bai G-R, Shi W-M, Mweene A, Kida H, 2004. Interregional transmission of the internal protein genes of H2 influenza virus in migratory ducks from North America to Eurasia. Virus genes 29, 81–86. [DOI] [PubMed] [Google Scholar]
- Makarova N, Kaverin N, Krauss S, Senne D, Webster R, 1999. Transmission of Eurasian avian H2 influenza virus to shorebirds in North America. Journal of General Virology 80, 3167–3171. [DOI] [PubMed] [Google Scholar]
- Makarova NV, Ozaki H, Kida H, Webster RG, Perez DR, 2003. Replication and transmission of influenza viruses in Japanese quail. Virology 310, 8–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullaney R, 2003. Live-bird market closure activities in the northeastern United States. Avian diseases 47, 1096–1098. [DOI] [PubMed] [Google Scholar]
- Munier S, Larcher T, Cormier-Aline F, Soubieux D, Su B, Guigand l., Labrosse B, Cherel Y, Quéré P, Marc D, 2010. A genetically engineered waterfowl influenza virus with a deletion in the stalk of the neuraminidase has increased virulence for chickens. Journal of virology 84, 940–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OIE. 2019. Chapter 3.3.4 Avian influenza (infection with avian influenza viruses), In: Manual for diagnostic tests and vaccines for terrestrial animals. OIE, Paris, France. [Google Scholar]
- Pantin-Jackwood MJ, Stephens CB, Bertran K, Swayne DE, Spackman E, 2017. The pathogenesis of H7N8 low and highly pathogenic avian influenza viruses from the United States 2016 outbreak in chickens, turkeys and mallards. PLoS One 12, e0177265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappas C, Yang H, Carney PJ, Pearce MB, Katz JM, Stevens J, Tumpey TM, 2015. Assessment of transmission, pathogenesis and adaptation of H2 subtype influenza viruses in ferrets. Virology 477, 61–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piaggio AJ, Shriner SA, VanDalen KK, Franklin AB, Anderson TD, Kolokotronis S-O, 2012. Molecular surveillance of low pathogenic avian influenza viruses in wild birds across the United States: inferences from the hemagglutinin gene. PLoS One 7, e50834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed LJ, Muench H, 1938. A simple method for estimating fifty percent endpoints. American Journal of Hygiene 27, 493–497. [Google Scholar]
- Robbins KM, Ye W, Fletcher OJ, 2011. Identification of Ascaridia numidae in guinea fowl (Numida meleagris) and association with elevated mortality. Avian Diseases 55, 151–154. [DOI] [PubMed] [Google Scholar]
- Schäffr JR, Kawaoka Y, Bean WJ, Süss J, Senne D, Webster RG, 1993. Origin of the pandemic 1957 H2 influenza A virus and the persistence of its possible progenitors in the avian reservoir. Virology 194, 781–788. [DOI] [PubMed] [Google Scholar]
- Senne D, Pearson J, Panigrahy B, 2003. Live poultry markets: a missing link in the epidemiology of avian influenza. Avian Diseases, 50–58. [Google Scholar]
- Senne D, Suarez D, Stallnecht D, Pedersen J, Panigrahy B, 2006. Ecology and epidemiology of avian influenza in North and South America. Developments in biologicals 124, 37. [PubMed] [Google Scholar]
- Shepard SS, Meno S, Bahl J, Wilson MM, Barnes J, Neuhaus E, 2016. Viral deep sequencing needs an adaptive approach: IRMA, the iterative refinement meta-assembler. BMC Genomics 17, 708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrell E, Perez D, 2007. Adaptation of influenza A/Mallard/Potsdam/178–4/83 H2N2 virus in Japanese quail leads to infection and transmission in chickens. Avian diseases 51, 264–268. [DOI] [PubMed] [Google Scholar]
- Sorrell EM, Song H, Pena L, Perez DR, 2010. A 27-amino-acid deletion in the neuraminidase stalk supports replication of an avian H2N2 influenza A virus in the respiratory tract of chickens. Journal of virology 84, 11831–11840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spackman E 2020. A Brief Introduction to Avian Influenza Virus, In: Animal Influenza Virus. Springer, 83–92. [DOI] [PubMed] [Google Scholar]
- Spackman E, Gelb J Jr., Preskenis LA, Ladman BS, Pope CR, Pantin-Jackwood MJ, McKinley ET, 2010. The pathogenesis of low pathogenicity H7 avian influenza viruses in chickens, ducks and turkeys. Virol J 7, 331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spackman E, Pantin-Jackwood MJ, Kapczynski DR, Swayne DE, Suarez DL, 2016. H5N2 Highly Pathogenic Avian Influenza Viruses from the US 2014–2015 outbreak have an unusually long pre-clinical period in turkeys. BMC veterinary research 12, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spackman E, Senne D.a., Myers T, Bulaga LL, Garber LP, Perdue ML, Lohman K, Daum LT, Suarez DL, 2002. Development of a real-time reverse transcriptase PCR assay for type A influenza virus and the avian H5 and H7 hemagglutinin subtypes. Journal of clinical microbiology 40, 3256–3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spackman E, Stephens C, 2016. Virus isolation and propagation in embryonating eggs. Isolation and Identification of Avian Pathogens. [Google Scholar]
- Suarez D, Spackman E, Senne D, 2003. Update on molecular epidemiology of H1, H5, and H7 influenza virus infections in poultry in North America. Avian diseases 47, 888–897. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Okada H, Itoh T, Tada T, Mase M, Nakamura K, Kubo M, Tsukamoto K, 2009. Association of increased pathogenicity of Asian H5N1 highly pathogenic avian influenza viruses in chickens with highly efficient viral replication accompanied by early destruction of innate immune responses. Journal of virology 83, 7475–7486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swayne D, Suarez D, Sims L 2020. Influenza, In: Swayne D, Boulianne M, Logue C, McDougald L, Nair V, Suarez D, Wit SD, Grimes T, Johnson D, Kromm M, Prajitno T, Rubinoff I, Zavala G (Eds.) Diseases of poultry. John Wiley & Sons, Inc., Hoboken, NJ, 210–256. [Google Scholar]
- Swayne DE, Slemons RD, 2008. Using mean infectious dose of high-and low-pathogenicity avian influenza viruses originating from wild duck and poultry as one measure of infectivity and adaptation to poultry. Avian diseases 52, 455–460. [DOI] [PubMed] [Google Scholar]
- Umar S, Munir M, Kaboudi K, Rehman A, Asif S, Usman M, Ali A, Shahzad M, Subhan S, Shah M, 2016. Effect of route of inoculation on replication of avian influenza virus (H9N2) and interferon gene expression in guinea fowl (Numida meleagridis). British poultry science 57, 451–461. [DOI] [PubMed] [Google Scholar]
- USDA, 2018. Live bird marketing systems (LBMs) status report. USAHA committee on poultry and other avian species. [Google Scholar]
- Webster R, Hinshaw V, Bean W Jr, Turner B, Shortridge K, 1977. Influenza viruses from avian and porcine sources and their possible role in the origin of human pandemic strains. Developments in biological standardization 39, 461–468. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.