Summary:
Numerous hematophagous insects are attracted to ammonia, a volatile released in human sweat and breath [1–3]. Low levels of ammonia also attract non-biting insects such as the genetic model organism Drosophila melanogaster and several species of agricultural pests [4, 5]. Two families of ligand-gated ion channels function as olfactory receptors in insects [6–10], and studies have linked ammonia sensitivity to a particular olfactory receptor in Drosophila [5, 11, 12]. Given the widespread importance of ammonia to insect behavior, it is surprising that the genomes of most insects lack an ortholog of this gene [6]. Here, we show that canonical olfactory receptors are not necessary for responses to ammonia in Drosophila. Instead, we demonstrate that a member of the ancient electrogenic ammonium transporter family, Amt, is likely a new type of olfactory receptor. We report two hitherto unidentified olfactory neuron populations that mediate neuronal and behavioral responses to ammonia in Drosophila. Their endogenous ammonia responses are lost in Amt mutant flies, and ectopic expression of either Drosophila or Anopheles Amt confers ammonia sensitivity. These results suggest that Amt is the first transporter known to function as an olfactory receptor in animals and that its function may be conserved across insect species.
Keywords: Olfactory receptor, Drosophila, transporter, Amt, ammonia, mosquito, olfactory behavior, sacculus, olfactory receptor neurons
Graphical Abstract
eTOC Blurb
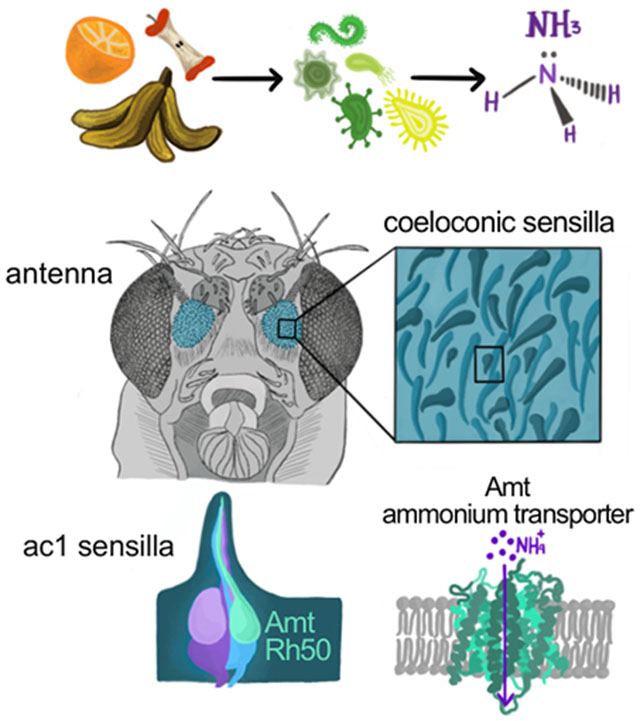
Ammonia is an attractive odor for a wide variety of insects, including insect vectors of disease. Vulpe et al. identify a highly conserved ammonium transporter that functions as an olfactory receptor for ammonia. This transporter, Amt, is expressed in two hitherto uncharacterized neuronal populations in Drosophila.
Results
Ammonium transporters in a new ORN class
In all insect species examined, ammonia activates olfactory receptor neurons (ORNs) housed in sensilla with a grooved peg morphology [2, 13], also known as coeloconic sensilla. Olfactory receptors define the odor tuning of individual ORNs, and the stereotyped receptor combinations expressed by neighboring ORNs define functional subtypes of sensilla. In Drosophila, robust responses to low levels of ammonia (NH3) are observed in the ac1 subtype of coeloconic sensilla [11, 13, 14]. Ammonia detection by ac1 ORNs depends on Amt [14], a member of the conserved ammonium transporter family [15, 16]. The Anopheles ortholog AgAmt can restore ammonia responses in Amt mutant flies [14] and produces the typical ammonium-selective inward current when members of this transporter family are heterologously expressed [17]. In Drosophila, Amt is exclusively expressed in chemosensory tissues including the antenna and labellum [14, 18, 19]. Its expression is most abundant in the antenna, where it is only found in ac1 sensilla and the third chamber of the sacculus [14], a three-chambered cavity invaginated from the antennal surface [20]. The precise reason why Amt is indispensable for ammonia detection remains unclear given that Amt expression was only detected in sensillar support cells [14].
Insect genomes also contain a second ammonium transporter, Rh50, which has widespread expression in multiple tissues [15, 17, 19]. Within the antenna, Rh50 transcript level is reduced ~10-fold in atonal flies that fail to develop coeloconic sensilla (Figure S1A) [14], suggesting a potential connection between Rh50 and ammonia detection. We therefore generated an Rh50 reporter line to identify Rh50+ cells within the antenna. The scattered population of labeled cells had a neuronal morphology and was positive for the neuronal marker elav (Figures 1A and 1B). The reporter line was faithful because all antennal GFP+ cells were labeled by an Rh50 anti-sense probe (Figure S1B). Thus, Rh50 expression in the antenna is found solely in neurons.
Figure 1. Ammonium transporters label a previously unidentified ac1 ORN.
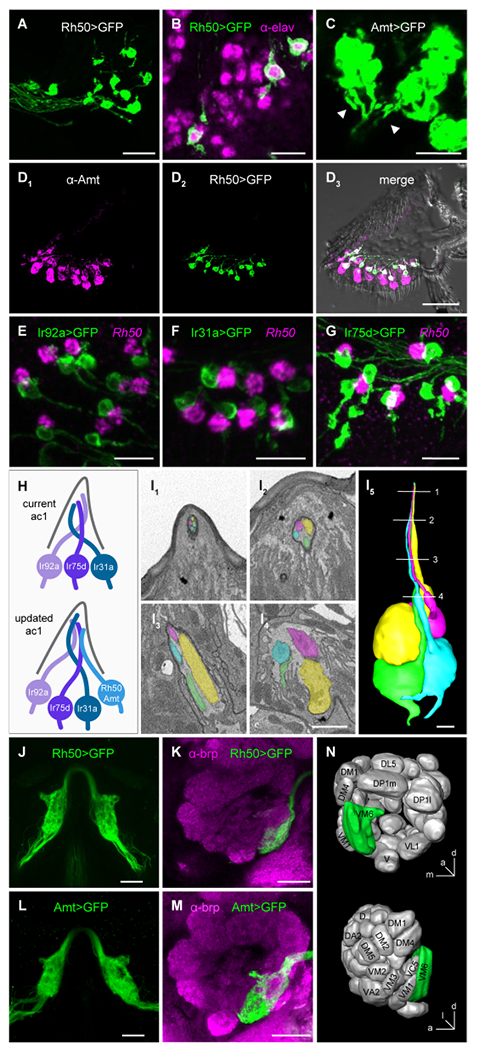
(A) Antennal section from an Rh50>GFP fly stained with anti-GFP. Scale bar, 15 μm.
(B) Rh50>GFP fly antennal section stained with anti-GFP (green) and anti-elav (magenta), a neuronal marker. Scale bar, 10 μm.
(C) High gain confocal image of an Amt>GFP antennal section stained with anti-GFP. Labeled axons (arrowheads) emerge from clusters of GFP+ cells. Scale bar, 10 μm.
(D) Immunostaining with anti-Amt (magenta, D1) and anti-GFP (green, D2) on an antennal section from an Rh50>GFP fly. D3, merged image. Scale bar, 40 μm.
(E-G) Antennal sections from Ir92a>GFP (E), Ir31a>GFP (F), and Ir75d>GFP (G) flies labeled with an in situ hybridization probe for Rh50 (magenta) and anti-GFP (green). Scale bars, 10 μm.
(H) Models of neurons and receptors in ac1 sensilla.
(I) SBEM images of a coeloconic sensillum with four ORNs. 3D reconstructions of the ORNs is shown in (I5). Numbered lines indicate the locations of individual sections shown in (I1-4). Scale bars, 1 μm.
(J) Two photon in vivo image of the bilateral antennal lobe glomeruli innervated by Rh50+ axons in an Rh50>GFP fly. Scale bar, 20 μm.
(K) Confocal image of an antennal lobe from an Rh50>GFP fly brain immunolabeled with antibodies targeting GFP (green) and brp (nc82, magenta), a neuropil marker used to delineate glomeruli. Scale bar, 20 μm. In both J and K, glial GFP expression driven by Rh50-GAL4 was suppressed with repo-GAL80 [40] to improve visualization of the ORN projections.
(L and M) Similar to J and K, but with Amt>GFP.
(N) Diagram of the location of the glomerulus innervated by Amt/Rh50+ ORNs, corresponding to VM6.
See also Figure S1.
Might Amt likewise be expressed in ORNs in addition to its known expression in support cells? ORNs are enwrapped by several support cells, and the strong Amt expression in these cells could potentially obscure visualization of Amt+ ORNs. Close examination of antennal sections from Amt-GAL4; UAS-GFP flies revealed weak GFP+ axons emerging from strongly labeled support cells in ac1 sensilla (Figure 1C). A similar antennal expression pattern of an AgAmt reporter line was recently reported in mosquitos [21]. Interestingly, anti-Amt staining revealed that Amt is co-expressed in every Rh50+ neuron (Figures 1D and S1C). Consistent with the reported expression pattern of Amt [14], Rh50+ ORNs were found in ac1 sensilla, but not in ac2, ac3, or ac4 coeloconic sensilla (Figures S1D–S1G).
The three reported neurons in ac1 sensilla express Ir92a, Ir31a, and Ir75d receptors, respectively [12]. To determine which neuron expresses the transporters, we examined Rh50 expression in antennal sections from flies in which one of the three ac1 neurons was genetically labeled. Surprisingly, the Rh50+ neurons did not co-localize with any of the three known neurons, but instead always neighbored the labeled ac1 ORNs (Figures 1E–1G). Similarly, these three ac1 ORNs were not labeled with the anti-Amt antibody, although they were surrounded by Amt+ cells (Figures S1H–S1J). Together, these results indicated that Rh50 and Amt are co-expressed in a previously undetected fourth neuron in ac1 sensilla (Figure 1H).
Early electron microscopy work only identified coeloconic sensilla that house two or three neurons [22]. By examining recent antennal serial block-face scanning EM (SBEM) datasets [23, 24], we found that approximately one in four coeloconic sensilla indeed contain four ORNs (Figure 1I; Figure S1K). Further, sensilla with two, three, and four neurons are unevenly distributed over datasets acquired from distinct antennal regions (Figure S1K), which could explain why coeloconic sensilla with four neurons were overlooked previously.
The axonal projections of all ORNs expressing the same receptor coalesce into a glomerulus in each antennal lobe. Examination of Rh50>GFP and Amt>GFP flies revealed that the terminals of these ORNs both converge to a large ventromedial glomerulus (Figures 1J–1N). This glomerulus corresponds to the “orphan” glomerulus VM6 [10, 25], recently shown to be targeted by neurons sharing a similar developmental origin as the other ac1 ORNs [25–27]. Because VM6 is difficult to discern with neuropil staining, it has been the source of confusion in recent antennal lobe atlases, where it was merged with VP1 [28] or listed as VC5 [29, 30].
Amt/Rh50+ ORNs respond selectively to NH3
The identification of the previously undetected Amt/Rh50+ ac1 ORN raised the question of its odor response profile. Although ac1 sensillar responses to ammonia and many amines were previously ascribed to its Ir92a+ ORN [5, 11, 12], the fourth ac1 ORN may mediate a portion of those responses. We specifically examined its odor responses through antennal lobe calcium imaging in Rh50>GCaMP7s flies. Ammonia evoked robust calcium responses, whereas there was no response to water or amines (Figure 2A). In contrast, Ir92a+ ORNs responded broadly to ammonia and to amines (Figure 2B), consistent with previous reports [5, 11]. Neither of these neurons is likely to detect alkaline pH, which may result from ammonia application, because ac1 neurons were unresponsive to two basic amines at 1% concentration, butylamine (pKa 10.8) and isoamylamine (pKa 10.6), even though 0.01% ammonia (pKa 9.4) induced robust responses (Figure S2A). Thus, Amt/Rh50+ ORNs are selectively tuned to ammonia.
Figure 2. Amt/Rh50+ ORNs selectively respond to ammonia.
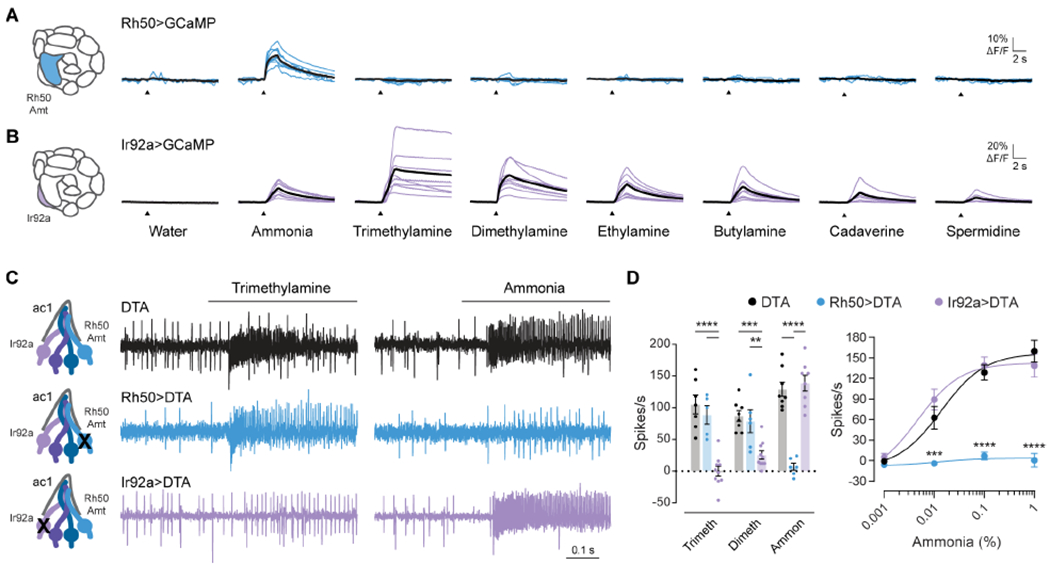
(A and B) Antennal lobe calcium responses to water, ammonia, and several amines in axonal projections labeled in Rh50>GCaMP7s (A) and Ir92a>GCaMP7s (B) flies. Blue lines (A) and purple lines (B) are responses in individual flies. Black lines are mean responses.
(C) Representative traces of extracellular recordings of action potentials elicited by 1% trimethylamine and 0.1% ammonia in ac1 sensilla in which diphtheria toxin (DTA) was used to ablate Rh50+ ORNs (blue) or Ir92+ ORNs (purple). UAS-DTA flies were used as a control (black).
(D) Left, quantification of odor responses in UAS-DTA (black), Rh50>DTA (blue), and Ir92a>DTA (purple) flies (n=5-10 sensilla). Right, dose-response curves of responses to increasing concentrations of ammonia (n=6-8 sensilla per genotype). The dose-response data for 0.1% ammonia are replotted in the bar graph to show individual data points.
See also Figure S2.
Amt/Rh50+ ORNs mediate spiking responses to NH3
We next examined the relative contributions of Amt/Rh50+ and Ir92a+ neurons to ammonia-induced spiking in ac1 sensilla. There is a nonlinear relationship between GCaMP7s responses and action potential firing because this highly sensitive sensor can detect single action potentials but saturates at low firing frequencies [31]. Although the Ir92a receptor and associated ORNs have been implicated in mediating spike responses to ammonia, this was inferred indirectly from GCaMP imaging data demonstrating the ammonia-sensitivity of Ir92a+ ORNs and the ammonia-insensitivity of Ir31a+ and Ir75d+ ORNs [5, 11].
We recorded from flies in which either the Ir92a+ or Amt/Rh50+ neurons were ablated genetically by diphtheria toxin. Ablation of the Amt/Rh50+ ORNs abolished the large amplitude action potential responses in ac1 sensilla over a broad range of ammonia concentrations, whereas loss of Ir92a+ ORNs had no significant effect (Figures 2C and 2D). In contrast, spiking responses to trimethylamine and dimethylamine, odorants that strongly activate Ir92a+ ORNs in calcium imaging [5, 11] (Figure 2B), were unaffected by the loss of Amt/Rh50+ ORNs but were nearly eliminated by Ir92a+ ORN ablation (Figures 2C and 2D). An Ir92a mutant had similar effects on ac1 odor responses as Ir92a+ ORN ablation (Figures S2B–S2F). Thus, Ir92a+ ORNs function primarily as amine detectors, whereas the Amt/Rh50+ ac1 ORNs mediate the robust spiking observed in response to ammonia.
Sacculus Amt/Rh50+ ORNs also respond to NH3
Examination of Rh50>GCaMP7s antennae revealed a second population of Rh50+ neurons in the sacculus (Figure 3A). This was reminiscent of Amt expression, which is found in sacculus chamber III in addition to ac1 sensilla [14]. There were 26 ± 3.6 (SD, n=10) Rh50+ ORNs in the sacculus, suggesting one Rh50+ ORN is likely housed in each of the ~22-26 chamber III sensilla reported previously [20]. As in ac1 sensilla, we detected Amt in both Rh50+ neurons and support cells in the sacculus (Figures 3B and 3C). Sensilla in sacculus chamber III house two neurons [20]. One expresses Ir64a and responds to acids [32], whereas markers for the second have not been reported, precluding its functional analysis. We found that Ir64a+ ORNs do not express Amt (Figure 3D), suggesting that sacculus III Amt/Rh50+ and Ir64a+ neurons are distinct. Consistent with this finding, Ir64a+ ORNs are known to project to DC4 and DP1m antennal lobe glomeruli [32], whereas all Amt/Rh50+ ORNs project to VM6 (Figures 1J–1N). Our data do not exclude the possibility that ac1 and sacculus Amt/Rh50+ ORNs target distinct regions within VM6.
Figure 3. Two populations of Amt/Rh50+ ORNs mediate ammonia sensing.
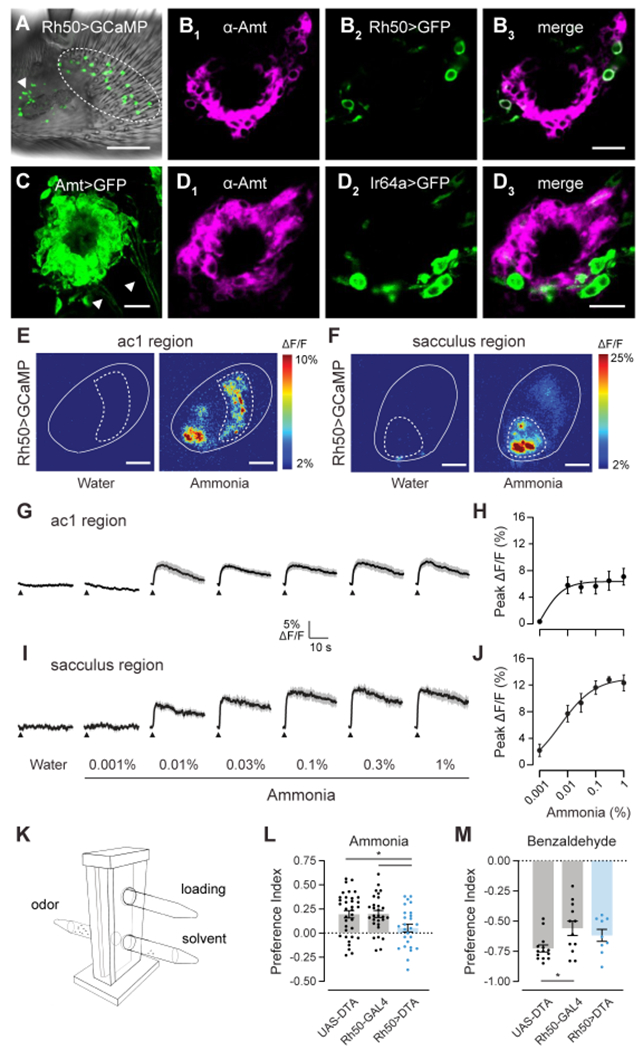
(A) Whole mount image of an antenna from an Rh50>GCaMP7s fly. Rh50+ neurons (green) are found on the ac1 region of the antennal surface (dotted circles) and surrounding the sacculus (arrowhead). Scale bar, 30 μm.
(B) Close-up view of sacculus chamber III in an antennal section from an Rh50>GFP fly stained with anti-Amt (magenta, B1) and anti-GFP (green, B2). B3, merged image. Scale bar, 10 μm.
(C) High gain confocal image of sacculus chamber III in an antennal section from an Amt>GFP fly stained with anti-GFP. Labeled axons (arrowheads) emerge from clusters of GFP+ cells. Scale bar, 10 μm.
(D) Immunostaining for Amt (magenta, D1) and GFP (green, D2) on antennal sections from Ir64a>GFP flies. D3, merged image. Scale bar, 10 μm.
(E) Pseudocolored heat maps of calcium responses in the ac1 region (dotted circle) of the antenna (solid outline) of Rh50>GCaMP7s flies to either water or 0.1% ammonia. Scale bars, 30 μm.
(F) Similar to (E) but acquired at a different depth and location to focus on the sacculus region (dotted circle). Scale bars, 30 μm.
(G) Traces of the mean calcium responses (black) ± SEM (gray) in the ac1 region. Arrowheads indicate time when the 250 ms odor stimulus was applied (n=6-7 flies).
(H) Dose-response curve of the peak ac1 calcium responses.
(I and J) Similar to G and H, except for the sacculus region.
(K) T-maze assay schematic showing the elevator in the lower position with flies moving between the odor and solvent arms. The loading tube is above and is accessible with the elevator in the upper position.
(L and M) Preference indices of Rh50>DTA, UAS-DTA and Rh50-GAL4 flies when given the choice between ammonia and water (L) or between benzaldehyde and paraffin oil (M). Each dot represents one assay (n=26-35 ammonia, n=9-12 benzaldehyde).
See also Figure S2.
Chamber III sensilla are proposed to contain only one olfactory neuron because the dendrite of the second neuron does not fully extend into the sensillum lumen [20]. This raised the question whether the sacculus Amt/Rh50+ neurons are ammonia-sensitive like their ac1 counterparts. Sacculus sensilla are inaccessible for electrophysiological recordings, but can be assayed using calcium imaging. We therefore turned to transcuticular imaging of Rh50>GCaMP7s fly antennae, where the two neuron populations can be segregated by location (Figures 3E and 3F). Similar to ac1 neurons, sacculus neurons showed dose-dependent responses to ammonia (Figures 3G–3J), providing evidence that the two neuron populations are both molecularly and functionally similar.
Amt/Rh50+ ORNs contribute to NH3 attraction
Like many insects, Drosophila are attracted to low levels of ammonia [5]. We examined the contribution of Amt/Rh50+ ORNs to this behavior using a T-maze two-choice assay in which naive flies are given a short time to navigate towards or away from an odorant (Figure 3K). For these assays, we used flies in which Amt/Rh50+ ORNs were ablated by diphtheria toxin, the same genotype used for electrophysiological recordings (Figures 2C and 2D). Parental control lines showed a preference for ammonia, and this attraction was significantly reduced in flies in which Amt/Rh50+ ORNs were eliminated (Figure 3L). As controls, we tested the flies’ general odor responsiveness and locomotor ability using the aversive odor benzaldehyde, which remained repulsive to the control and ablated flies (Figure 3M). Together, these data indicate the importance of Amt/Rh50+ ORNs in Drosophila ammonia attraction. Consistent with our electrophysiological data, the behavioral responses of Ir92a1 mutants were similar to controls (Figures S2G and S2H).
NH3 reception does not involve IRs/ORs
What is the ammonia receptor in Amt/Rh50+ neurons? Nearly all insect olfactory neurons utilize a member of either the ionotropic receptor (IR) or odorant receptor (OR) families [6, 10]. Individual tuning receptors that bind odorants rely on a co-receptor for function, with odor responses absent in co-receptor mutants [7, 33, 34]. OR receptors depend on Orco, whereas IR receptors require Ir25a, Ir8a, or Ir76b [7, 33, 34]. We therefore asked whether any of these co-receptors are expressed in Amt/Rh50+ ORNs, and found that only Ir25a was co-expressed with Rh50 in ac1 sensilla (Figures 4A–4D). However, ammonia-induced spiking responses were unchanged in Ir25a2 mutants (Figure 4E), in agreement with a previous study [7]. As expected, ac1 ammonia responses were also not reduced in mutants for any of the three other co-receptors (data not shown). Thus, neither the OR nor IR co-receptors is required for ac1 ammonia responses.
Figure 4. Amt transporter serves as an olfactory receptor for ammonia.
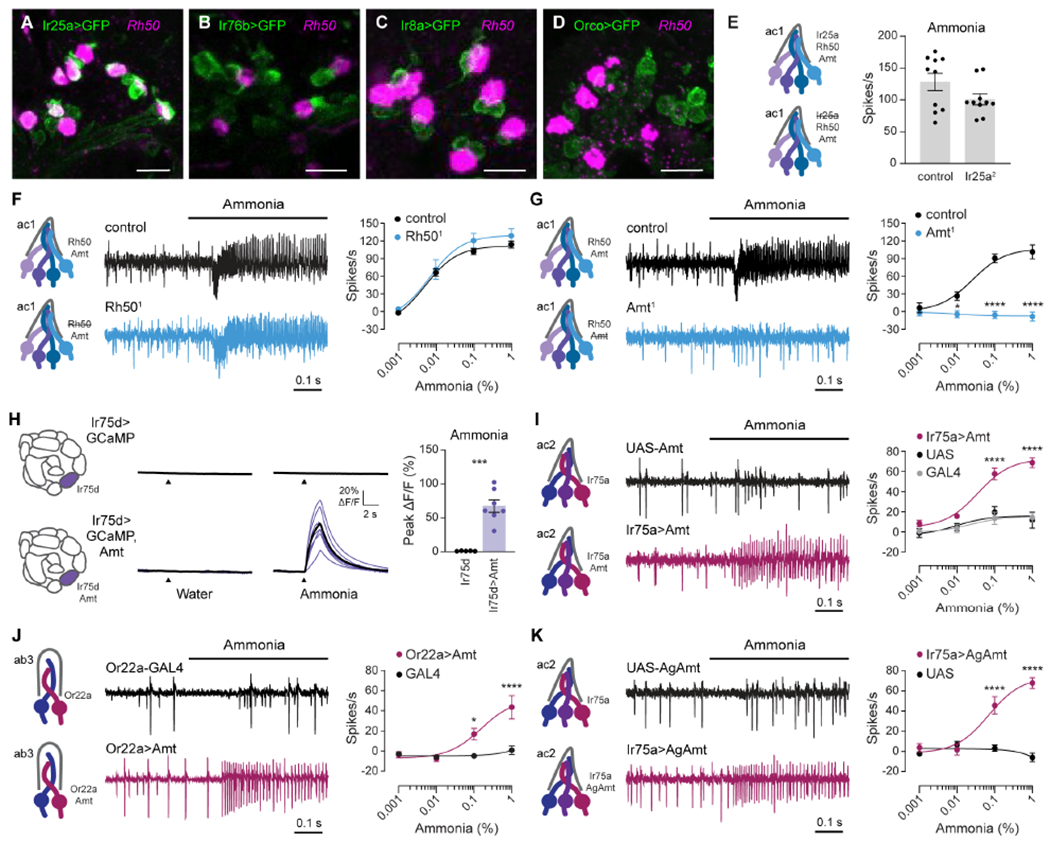
(A-D) Confocal images of antennal sections labeled with an antisense probe for Rh50 (magenta) and an antibody against GFP (green) driven by Ir25a-GAL4 (A), Ir76b-GAL4 (B), Ir8a-GAL4 (C), or Orco-GAL4 (D). Scale bars, 10 μm.
(E) Action potential responses to 0.1% ammonia in ac1 sensilla in control flies and Ir25a2 mutants (n=10 sensilla).
(F and G) Action potential responses to ammonia in ac1 sensilla in Rh501 (F) and Amt1 (G) mutants (blue) and control flies (black). Left, representative traces of response to 0.1% ammonia. Right, dose-response curves (n=8-10 sensilla).
(H) Left, antennal lobe calcium responses to water and 0.01% ammonia in axon termini of Ir75d>GCaMP6s flies, with (purple) and without (black) ectopic expression of Amt. Purple lines are responses in individual flies, and black lines are mean responses. Right, quantification of peak responses (n=5 and 7 flies).
(I) Action potentials elicited by ammonia in ac2 sensilla in Ir75a>Amt (crimson), Ir75a-GAL4 (grey) and UAS-Amt (black) flies. Left, sample traces of 1% ammonia responses. Right, dose-response curves (n=7-9 sensilla).
(J) Action potentials elicited by ammonia in ab3 sensilla in Or22a>Amt (crimson) and Or22a-GAL4 (black) flies. Left, sample traces of 1% ammonia responses. Right, dose-response curves (n=9-11 sensilla).
(K) Similar to (I), except Ir75a>AgAmt (crimson) and UAS-AgAmt (black) flies (n=7-8 sensilla).
See also Figure S3.
Amt as an ammonia receptor
This observation raised the possibility that Amt/Rh50+ ORNs utilize a non-canonical receptor to detect ammonia. We wondered whether ammonium transporters might fulfill this role. Ammonium transporters in other species are highly selective for ammonium, with little to no transport of larger amines [35, 36], in accordance with the narrow tuning of Amt/Rh50+ ORNs (Figure 2A). Further, ammonium transporters are electrogenic, mediating inward NH4+ currents [16, 17, 36, 37]. Given that ammonia (NH3) gas is expected to form ammonium ions (NH4+) in the sensillum lymph, these cations could be transported through either Amt or Rh50 and depolarize the ORNs. In this manner, the transporters could act as ammonia-selective receptors.
If this model is correct, ammonia responses should require either Amt or Rh50. We first generated an Rh50 mutant which eliminated Rh50 expression (Figure S3). Spike responses to all concentrations of ammonia persisted in the Rh501 ac1 sensilla, if anything trending slightly higher than controls (Figure 4F). In contrast, Amt1 mutants entirely lack ammonia responses at all concentrations (Figure 4G) [14].
To determine whether Amt operates as an ammonia receptor, we ectopically expressed Amt in ammonia-insensitive neurons, and found that expression of Amt in Ir75d+ neurons conferred robust antennal lobe calcium responses to ammonia that were absent in controls (Figure 4H). Further, ectopic expression of Amt in Ir75a+ ORNs, found in ac2 coeloconic sensilla, produced dose-dependent spiking responses to ammonia (Figure 4I). Finally, Amt misexpression conferred a similar ammonia sensitivity to basiconic Or22a+ ORNs (Figure 4J), demonstrating that Amt function requires neither morphological nor molecular features specific to coeloconic sensilla. Together, these data indicate that Amt is an olfactory receptor for ammonia in Drosophila.
Amt is evolutionarily conserved, with orthologs found in multiple insect species, including the malaria vector Anopheles mosquitoes [15, 17]. Misexpression of AgAmt in Ir75a+ ORNs produced dose-dependent responses to ammonia (Figure 4K), similar to those induced by Drosophila Amt (Figure 4I). This raises the possibility that ammonium transporters also operate as receptors in other insect species.
Discussion
Our identification of two previously unstudied populations of olfactory neurons expressing Amt and Rh50 transporters has paved the way to a new understanding of ammonia detection in Drosophila. In insects, ORNs detect odors using IR and OR olfactory receptors, ligand-gated cation channels that are generally tuned to multiple structurally related odorants. Our loss- and gain-of-function experiments reveal that the newly identified Amt/Rh50+ ac1 ORNs instead use an ammonium transporter as their receptor (Figures 4G–4K); the molecularly and functionally similar sacculus ORNs likely operate similarly. The narrowly tuned ammonia response in these ORNs (Figure 2A) is consistent with the strict selectivity of ammonium transporters [35, 36]. Amts also distinguish NH4+ ions from similarly sized K+ ions [17, 36, 37], an advantage considering the presence of >100 mM K+ in the sensillum lymph bathing ORN dendrites. We propose that a simple electrogenic influx of ammonium ions leads to ORN depolarization, but we cannot rule out an ammonium-initiated signaling cascade analogous to the mechanism for vertebrate sour sensing, in which H+ influx closes acid-sensitive K+ channels [38]. However, an ammonium signaling cascade is unlikely because it would require the components to be widely expressed in ammonia-insensitive neurons in both basiconic and coeloconic sensilla.
The ammonia-sensing ORNs in Drosophila express both Amt and Rh50, members of the two main branches of the AMT/MEP/Rh ammonium transporter family. Ammonium transporters in plants, bacteria, and fungi serve to uptake ammonium for biosynthesis of nitrogenous molecules, whereas animal Rh proteins play a role in ammonia excretion and acid-base homeostasis [15, 16]. Our findings indicate a novel role for Amt ammonium transporters as chemosensory receptors in animals. In contrast, we did not find a significant role for Rh50 in ammonia sensing, perhaps due to its >20-fold lower expression in antennae than Amt [14]. Rh50 may also be unable to function as an ammonia receptor because most Amts transport ammonium at low micromolar concentrations whereas Rh proteins require millimolar levels [39]. The role of Amt proteins in support cells is likewise unclear; Amt misexpression into neurons in coeloconic and basiconic sensilla that lack Amt in support cells is sufficient to induce ammonia-sensitivity (Figures 4I and 4J). Support cell Amt may affect the magnitude of the ORN response, a possibility suggested by the somewhat smaller ammonia responses in ac2 versus ac1 ORNs (Figures 4G and 4I).
Ammonia-sensitive ORNs are found in every insect species examined, and many insects are attracted to low levels of ammonia. Its effectiveness as an attractant has spurred the use of ammonium-containing solutions in commercial insect traps [2–4]. Several lines of evidence suggest that Amt orthologs are widely used as ammonia receptors, and therefore likely mediate this behavior. First, Amt proteins are highly conserved, with for example, Drosophila and Anopheles orthologs sharing 83% amino acid identity in the transmembrane regions.
Accordingly, expression of Anopheles AgAmt conferred ammonia sensitivity to ORNs (Figure 4K), and a recent study detected Amt+ neurons in ammonia-sensitive olfactory sensilla in Anopheles mosquitos [21]. Additionally, Amt orthologs can be found in antennal transcriptomes from multiple, distantly related insect species [14]. Together these similarities suggest Amt transporters have taken on a new function as ammonia receptors in insects.
STAR Methods
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Karen Menuz, karen.menuz@uconn.edu.
Materials Availability
All novel biological materials, including transgenic Drosophila strains, anti-Amt antibody, and plasmids, are available upon request.
Data and Code Availability
All custom codes used for analysis are available upon request. This study did not generate any unique datasets.
Experimental Model and Subject Details
Drosophila melanogaster were reared on a standard cornmeal food at 25°C in a humidified incubator with a 12:12 hour light/dark cycle. The genotypes of fly lines used in this study are listed in Table S1. The Ir92a1, Rh501, and Ir25a2 mutant lines were outcrossed for at least five generations to a Canton-S background prior to electrophysiological recordings, as were transgenes in flies used for recordings in Figure 2 and behavior in Figures 3 and S2. The Amt1 transposon mutation had been previously outcrossed to an isogenic w1118 line [14, 41]. Experimental flies were between 2-12 days old. The genders and specific age ranges for different experiments are provided in the Method Details below.
Method Details
Reporter line generation
Transgenic GAL4 reporter flies were generated using standard methods as described [14]. The 5’ and 3’ regions flanking the genes were amplified from bacterial artificial chromosomes (BACs) corresponding to the reference genome of Drosophila melanogaster [42]. MultiSite Gateway Pro recombination (Thermo Fisher) was used to assemble the 5’ and 3’ genomic regions with GAL4 in the pBGRY destination vector [43]. For Rh50-GAL4, the 5’ region included chromosome 3L: 4,907,401 to 4,910,693 and the 3’ region 3L: 4,918,898 to 4,924,642. For Ir25a-GAL4, the 5’ region included chromosome 2L: 4,835,726 to 4,834,655 and the 3’ region 3L: 4,830,990 to 4,827,634. For Ir75a-GAL4, the 5’ region included chromosome 3L: 17,829,014 to 17,817,922 and the 3’ region 3L: 17,815,667 to 17,811,236. PhiC31 integration (Best Gene, Inc.) was used to integrate the assembled GAL4 vectors into the Drosophila melanogaster genome. Two transgenic strains were made for each GAL4 in which the construct was incorporated into the second (attP40 landing site [44]) and third (attP2 landing site [45]) chromosomes.
Generation of Ir92a and Rh50 mutants
The Ir92a1 and Rh501 mutant alleles were generated with CRISPR/Cas9 engineering and homology directed repair, similar to described previously [18]. The Ir92a1 mutation deletes the terminal 323 amino acids of Ir92a (52% of the coding sequence), including all transmembrane domains. The Rh501 mutation eliminates 143 amino acids (29% of the coding sequence), including several transmembrane domains.
Guide RNA sequences were cloned into pU6-BbsI-chiRNA [46], using the Q5 Site-Directed mutagenesis Kit (New England Biolabs). Two gRNAs were used for targeting Ir92a (GGTCACCGAAGAACGGGCTA and GGACGCATCTCCCCGTGAAA) and one for Rh50 (GATCCAGTCTGTCCAGGTTC). A donor plasmid for homology directed repair for Ir92a was generated by cloning homology arms from wCS genomic DNA and inserting them into the pHD-DsRed-attP vector [47] to flank the 3xP3-DsRed marker. Rh50 homology arms were cloned from BAC R17L24 and inserted into a modified pHD-DsRed attP vector that contained a LexA reporter sequence 5’ to the 3xP3-DsRed site. For each gene, gRNA and donor plasmids were injected into embryos of P{nos-Cas9.R}attP40/ CyO flies [48] (Best Gene, Inc.).
Flies in which the donor plasmid had integrated into the genome were identified by ocular DsRed expression. We used PCR and sequencing to validate the location of donor plasmid integration. The Ir92a donor plasmid integrated as expected, with the 3xP3-DsRed marker replacing genomic region 3R: 20,342,366-20,344,610. The Rh50 donor plasmid did not integrate as designed. Instead genomic region 3R: 4,913,350-4,914,409 was replaced by sequences from the donor plasmid extending from the ampicillin resistance region through the LexA and 3xP3-DSRed regions. Flies were genotyped (Figures S2 and S3) using the following primers:
Ir92a pair 1: TGTATGGCCGGTAGGATCTC and ACCTCCTTGATCGAAACCCT
Ir92a pair 2: GGCAAGAATGCGAACAAAT and TGGTTTGTCCAAACTCATCAA
Rh50 pair 1: CCTCTCCCTGGAGAACATCA and CCCTCTAGCTTTCCCGTTTC
Rh50 pair 2: CTGTTCATGGCTGCTCTAGT and CTGAGATAGGTGCCTCACTG
Quantitative reverse transcriptase-PCR
Heads from ten female Rh501 and wCS flies aged 6-7 days were dissected over liquid nitrogen for RNA extraction. The heads were crushed with a pestle and passed through a QIAshredder column (Qiagen), and then total RNA was extracted with the RNeasy Micro Kit (Qiagen). 100 ng of RNA was reverse transcribed into cDNA using the iScript gDNA Clear cDNA Synthesis Kit (Bio-Rad). In parallel, RNA was separately processed without the reverse transcriptase to control for any gDNA contamination. Quantitative reverse transcriptase-PCR (qRT-PCR) reactions were run in triplicate on a CFX96 thermocycler (Bio-Rad) with SsoFast EvaGreen Supermix (Bio-Rad) containing cDNA from 5 ng of RNA and Rh50 primers AATGAGCAGTGTGACAGCGA and CATTGCCTCCGCCATTTACG. Expression in each sample was normalized to a housekeeping gene, eIF1A, detected with primers ATCAGCTCCGAGGATGACGC and GCCGAGACAGACGTTCCAGA.
FISH and antennal immunocytochemistry
Male and female flies 7-10 days old were placed in an alignment collar. Their heads were encased with OCT (Tissue-Tek) in a silicone mold, frozen on dry ice, and snapped off. Head blocks were stored at −80°C. A cryostat was used to collect 20 μm sections. Immunocytochemical staining was carried out as done previously for labellar sections [18]. Transgenic GFP expression was detected with mouse anti-GFP antibody (1:500, Roche) and donkey anti-mouse Alexa Fluor 488 (1:500, Thermo Fisher). A rat anti-elav antibody (1:10, Developmental Studies Hybridoma Bank) was visualized with goat anti-rat Alexa Fluor 568 (1:500, Thermo Fisher). A guinea pig anti-Amt antibody [18] (1:200) was detected with goat anti-guinea pig Alexa Fluor 568 (1:500, Thermo Fisher).
Combined FISH and immunocytochemistry staining were carried out as described previously [14]. The digoxigenin (DIG)-labelled Rh50 probe was generated from a plasmid containing the full-length cDNA sequence corresponding to Rh50-RA isoform (Drosophila Genomics Resource Center) [49]. This plasmid was digested with XhoI for T7 transcription (sense probe) and EcoRV for SP6 transcription (anti-sense probe) for labelling with the DIG RNA Labeling Kit (SP6/T7) (Roche). Stained sections were imaged on a Nikon A1R confocal microscope in the UConn Advanced Light Microscopy Facility. Stacks of images (0.5 μm z-step size) were collected and analyzed with ImageJ/Fiji software [50].
3D reconstruction of labeled glomeruli
Brains of 7 day old female and male flies were dissected in PBS and directly transferred to 4% PFA with 0.1% Triton-X (PBST) for fixation over 2 hours on ice. After three 15 minute washes with PBST and blocking in 5% normal goat serum (NGS) in PBST, the primary antibodies mouse anti-brp (1:30, Developmental Studies Hybridoma Bank) and rabbit anti-GFP (1:500, Thermo Fisher) were applied in PBST with 5% NGS for 4 days nutating at 4°C. Next, the brains were washed four times for 15 minutes with PBST, blocked again with 5% NGS in PBST and incubated with the secondary antibodies goat anti-mouse Alexa Fluor 633 and goat anti-rabbit Alexa Fluor 488 (1:250 each, Thermo Fisher) in PBST with 5% NGS for 5 days nutating at 4°C. After four final washing steps with PBST, brains were mounted in VectaShield (Vector Labs). Stacks of immunostained brains were scanned on a Zeiss cLSM 880 confocal microscope with a z-step size of 0.44 μm. The confocal stacks were manually reconstructed as label fields in AMIRA (6.7, Thermo Fisher) and identified on the basis of the published Drosophila melanogaster atlases [51]. Surface renders were created and smoothed in AMIRA with the surface gen and smooth surface tools.
In vivo structural imaging of antennal lobes
To visualize the Amt and Rh50 positive ORNs in vivo (Figures 1K and 1M), an open head dissection was utilized. The flies were anesthetized on ice and then glued to a plastic holder to reduce movement. A wire was installed to bend the antennae forward and maximize the cutting area at the vertex. After adding a plastic coverslip with a hole, and sealing this hole around the vertex with two-component silicone, the vertex was cut open under saline. The trachea and fat tissue inside the head were removed to reveal a clear view onto the central brain. Images were obtained with a Zeiss cLSM 710 multi photon microscope.
Electron microscopy analysis
Coeloconic sensilla were identified in serial block-face scanning electron microscopy (SBEM) datasets that were previously generated [23, 24], based on their signature cuticular finger structure [22]. The number of ORNs within each coeloconic sensillum was determined by examining the EM images using IMOD v4.9 [52]. Segmentation was performed manually. The 3D models were then generated using the IMOD command “imodmesh” and smoothed using the “smoothsurf” command. The sensillum shown in Figure 1I was identified in the Or88a-labeled dataset [23].
Whole-mount Confocal Imaging
7-day old female flies were anesthetized on ice before their antennae were removed for fixation with 4% paraformaldehyde/PBS (MPX00553, Fisher Scientific) for 20 min at room temperature. After washing three times with 0.3% PBT (PBS with 0.3% Triton X-100), the samples were mounted in FocusClear (CelExplorer Labs Co.) and imaged on a Zeiss LSM 880 confocal microscope using 40x, N.A.1.2 C-Apochromat water-immersion objective lens. Airyscan images were processed with ZEN (Zeiss) and the brightness and contrast were further adjusted using ImageJ/Fiji.
Ex vivo ORN somata counts
Antennae were collected from 7-day old flies anesthetized with CO2. Antennae were briefly submerged in Triton-X before being mounted in VectaShield (Vector Labs). Cellular counts were carried out on confocal stacks of ex vivo prepared antennae acquired on a cLSM 880 (Zeiss) with a 40x/1.2W C-Apochromat (Zeiss).
Behavioral assay
T-maze behavioral assays were performed using custom-built acrylic apparatuses. At 0-4 days post-eclosion, flies were sorted into groups of 16 males and 16 females of the appropriate genotype. The vial of flies was tested at 7-12 days post-eclosion. Flies were starved in empty vials with 2 mL of distilled water to moisten the foam plugs ~22 hours prior to the assay. Assays were conducted within a three hour time window to control for any circadian effects. Two hours prior to the start of the assay, fly vials were moved to a darkened quiet room to acclimate to the room temperature and light level. Antibiotic assay discs (13 mm, Whatman) were placed tightly into the bottom of 15-mL conical tubes (DOT Scientific) using a metal spatula, and 50 gl of either odorant or solvent were pipetted onto the disc. The tubes were then capped and the odorant allowed to volatilize for 30 minutes before a set of assays began.
A set of ~10 consecutive assays was carried out over a period of ~45 minutes in a dark room lit with dim red light. The assay chambers were placed in a cardboard box to further limit visual cues. For each assay, ~20-30 flies were transferred with a funnel from a vial into a 15-ml centrifuge tube, which was then screwed oton the upper opening of the T-maze central tower. Flies were then tapped into the elevator, which was at its topmost position to align with the loading tube. The elevator was partially lowered, and flies were given one minute to acclimate. Towards the end of this time, odorant and solvent tube “arms” were screwed into the T-maze, with the positions of the odorant and solvent arms alternating between assays. The elevator was then lowered further so that flies had access to both the odorant and solvent arms. Flies had one minute to chemotax into the arms before the elevator was raised to trap flies in the arms and prevent further movement. Preference index was calculated as the number of flies entering the solvent arm subtracted from the number of flies entering the odorant arm, and this value divided by the total number of flies in the assay including those that remained in the elevator hole. Each set of flies and conical tubes was only used for one assay. Each set of assays tested flies of multiple genotypes in a random order.
Odorants
Odorants were ammonium hydroxide (Fisher or VWR International), trimethylamine (Sigma), dimethylamine (Sigma), ethylamine (Sigma), butylamine (Sigma), cadaverine (Sigma), spermidine (ACROS), isoamylamine (ACROS), and benzaldehyde (Sigma). All odorants were diluted to generate 10% stocks, and serial dilutions were used to generate lower concentrations. All odorants were diluted in water, except benzaldehyde, which was diluted in paraffin oil (ACROS). Ammonia was used at concentrations from 0.0001% to 1% for electrophysiology and transcuticular imaging. Other odorants used for electrophysiological experiments were at 1% concentration. All odorants were used at a 0.01% concentration for antennal lobe GCaMP imaging. For behavioral assays in Figure 3L, ammonia was used at 0.3% and benzaldehyde at 1%. For behavioral assays in Figures S2G and S2H, ammonia was used from 0.001% to 1% concentrations.
Electrophysiology
Single-sensillum electrophysiological recordings were generally performed on 3-5 day old female flies as described [53, 54]. Flies for diphtheria-toxin ablation experiments were aged 8-11 days to ensure complete neuron ablation. In brief, flies were wedged in the narrow tip of a 200 μl pipette tip, exposing the antennae and a portion of their head. One antenna was stabilized between a tapered glass electrode and a coverslip. The prep was placed on a BX51WI microscope (Olympus) under a continuous 2,000 mL/min stream of humidified, purified air. A borosilicate glass electrode filled with sensillum recording solution [55] was placed into the eye as a reference electrode, and an aluminosilicate electrode filled with the same solution was inserted into individual sensilla. Sensilla classes were identified based on their known location on the antenna and their response profile to a small number of diagnostic odorants. Up to four sensilla were recorded per fly. Extracellular action potential recordings were collected with an EXT-02F amplifier (NPI) with a custom 10x gain headstage. Data were acquired and AC filtered (300-1,700 Hz) at 10 kHz with a PowerLab 4/35 digitizer and LabChart Pro v8 software (ADInstruments).
Odorant cartridges were prepared by placing a 13 mm antibiotic assay disc (Whatman) into a Pasteur pipette, pipetting 50 μl of odorant solution onto the disc, and enclosing the end with a 1 mL pipette tip. Cartridges were allowed to equilibrate for at least 20 minutes. Cartridges were used no more than four times, with at least 10 minutes recovery between re-use for trials on different sensilla. Odorants were applied for 500 ms at 500 mL/min after inserting the cartridge into a hole in the main airflow tube. Odor delivery was controlled by LabChart, which directed the opening of a Lee valve (02-21-08i) linked to a ValveBank 4 controller (Automate Scientific). A ten second recording was collected, with a one second baseline period before odor application. Each sensillum was tested with multiple odorants, with at least a 10 second rest period between odor applications.
Action potentials were detected offline using LabChart Spike Histogram software. Spikes were sorted by their amplitude in basiconic recordings, whereas all spikes from the 3-4 ORNs were summed in coeloconic recordings due to their similar sizes. Action potentials were counted over a 500 ms window, 100 ms after stimulus onset due to the line delay for the odor to reach the antenna. Solvent corrected odor responses were calculated as the number of spikes induced by the odor after subtracting the number of spikes produced by stimulating with water alone.
Ca2+ imaging in the antennal lobe
Genetically encoded calcium indicators belonging to the GCaMP family were used to visualize the odor-evoked calcium activity in the ORN axon terminals of interest in the antennal lobe. All experiments were conducted in virgin female flies aged 2-11 days post-eclosion. Experimental flies were collected shortly after eclosion and housed in small groups at 25°C under a 12 hour light and 12 hour dark cycle until imaging.
Flies were prepared for imaging as previously described [56]. The external saline contained 103 mM NaCl, 3 mM KCl, 5 mM TES, 8 mM trehalose ·2H2O, 10 mM glucose, 26 mM NaHCOs, 1 mM NaHPO4 ·H2O, 4 mM MgCl2 ·6H2O, and 1.5 mM CaCl2 ·2H2O and was adjusted to a pH of 7.25 and osmolarity of ~270 mOsm. The fly was first head-fixed into a small hole in a thin sheet of stainless steel foil, such that its antennae protruded beneath the foil (to stay dry) while the rest of the head was submerged in the external saline solution above the foil. To target Rh50 ORN axons (Figure 2A), a small patch of cuticle dorsal to the brain was removed and fat, air sacs, and trachea were removed to obtain good optical access. To target Ir92a (Figure 2B) and Ir75d (Figure 4H) ORN axons, the head was rotated 180° such that the proboscis pointed up. The proboscis was then removed, along with fat, air sacs, and trachea. In additional experiments, we imaged from Rh50 ORN axons ventrally and found no qualitative difference between the resulting response traces and those obtained dorsally (data not shown).
Following the dissection, the metal foil containing the head fixed fly and the external saline was mounted on an epifluorescence microscope under a 40x water-immersion objective lens (NA 0.8). The fly was positioned under the microscope such that its antennae faced a constant stream of charcoal-filtered carrier air, delivered at the rate of ~1360 mL/min. Odor stimuli were prepared by diluting stock chemicals in distilled water in 2mL vials. To deliver the odor stimulus into the carrier stream, a computer-controlled valve diverted a small amount of the carrier stream into the headspace of the odor vial, at the rate of ~6.2 mL/min, before rejoining the carrier stream. GCaMP was excited with a 470 nm LED at 5% power, corresponding to 0.367 mW at the sample (CoolLED pE-100). The emitted fluorescence was collected by a Hamamatsu digital camera (model C1140-42U30) using HCImageLive software. Each imaging trial lasted 15 seconds at a frame rate of 16.67 Hz, with the odor delivery starting at 4 seconds into a given trial and lasting for 2 seconds.
Individual CXD image files were converted to TIF files using the batch process function from ImageJ software. TIF-converted data for each fly were concatenated into an image stack for each stimulus condition using a custom MATLAB script. For each stimulus condition, the ROI was defined by visually inspecting the image stack and tracing around the perimeter of the ORN axon terminals. The same ROI was applied to every frame of each stack. ΔF/F was calculated as (Fsignal – F0) / F0, where Fsignal is the instantaneous fluorescence pixel value averaged over the entire ROI and F0 is the averaged pixel value in the same ROI over the 1 second preceding the stimulus onset. In each fly, three trials were conducted for each stimulus condition; ΔF/F was averaged over each trial. These ΔF/F traces were then averaged across flies.
Transcuticular antennal calcium imaging
Female flies aged 7 days were used for antennal calcium imaging experiments. To prepare an antenna for recording, a fly was wedged into the narrow end of a truncated 200 μl pipette tip to expose the antenna, which was subsequently stabilized between a tapered glass microcapillary tube and a coverslip covered with double-sided tape. Images of different populations of Rh50-expressing neurons were acquired at different depths to focus on either the ac1 region or the sacculus region.
Images were acquired via Micro-Manager 1.4 (The Open Source Microscopy Software) with a CMOS camera (Prime 95B, Photometrics) and an upright microscope (Olympus, BX51WI) with a 50x air objective (NA 0.50, LMPlanFl, Olympus). Blue LED (470 nm, Universal LED Illumination System, CooLed pE-4000) was used to excite GCaMP7s. Image acquisition was at 10 Hz for ~38 s. Light pulses (25-ms on, 75-ms off) were used to minimize photobleaching. Odorants (100 μl applied to a filter disc) were delivered from a Pasteur pipette via a pulse of air (200 mL/min) into the main air stream (2000 mL/min). A 250 ms pulse of odorant was applied 7 seconds after acquisition onset, with an interstimulus time interval of 2–3 min between each application.
All images acquired from the same antenna were first concatenated. A MATLAB function, NoRMCorre [57], was used for motion correction. For each antenna, ROIs were determined via a custom Python script based on the calcium responses to 0.3% ammonia. Briefly, the frame with the highest summed pixel value within the field of view was first generated (peak frame). A delta frame was then determined by subtracting the peak frame with the averaged pre-stimulus frame (2 seconds prior to odor stimulus). The delta frame was processed with a Gaussian filter to smooth the image. ROIs were then identified by applying a threshold (>70% of the highest pixel value) to the smoothed delta frame. The same ROIs were applied to all images acquired from the same antenna across different ammonia concentrations.
ΔF/F was calculated as (Fsignal – F0) / F0, where Fsignal is the instantaneous fluorescence pixel value averaged over the entire ROI and F0 is the averaged pixel value in the same ROI over the 2 second pre-stimulus period. In order to remove imaging noise, baseline ΔF/F—defined as the fluorescence level which is less than 10% of the highest pixel value of the entire image—was further subtracted from the ΔF/F of the ROIs. For each recording, the representative ΔF/F was determined by averaging the traces from three ROIs which had the highest ΔF/F values upon odor stimulation.
Quantification and Statistical Analysis
Data were analyzed using GraphPad Prism 8. Bar graphs depict the mean ± SEM overlaid with the individual data points. Dose response curves show the mean ± SEM and the curve fit to the Hill equation. Unpaired two-tailed t-tests were used to compare two genotypes, and one-way ANOVAs followed by Tukey post-hoc tests for three or more genotypes. Datasets involving multiple genotypes and multiple odorants or odorant concentrations were analyzed with two-way ANOVAs followed by Holm-Sidak post-hoc tests. Statistical parameters can be found in the figure legends. Statistical significance is presented as *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. Other comparisons were not significant (p>0.05).
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-GFP | Roche | RRID:AB_390913 |
| Rat anti-elav (7E8A10) | Developmental Studies Hybridoma Bank | RRID: AB_528218 |
| Guinea pig anti-Amt | [18] | RRID:AB_2864754 |
| Rabbit anti-GFP | Thermo Fisher | RRID:AB_221569 |
| Mouse anti-brp (nc82) | Developmental Studies Hybridoma Bank | RRID:AB_2314866 |
| Donkey anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermo Fisher | RRID:AB_141607 |
| Goat anti-Rat IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 568 | Thermo Fisher | RRID:AB_2534121 |
| Goat anti-Guinea Pig IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 568 | Thermo Fisher | RRID:AB_2534119 |
| Goat anti-Mouse IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 633 | Thermo Fisher | RRID:AB_2535718 |
| Goat anti-Rabbit IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermo Fisher | RRID:AB_143165 |
| Bacterial and virus strains | ||
| Biological samples | ||
| Chemicals, peptides, and recombinant proteins | ||
| Ammonium hydroxide, 28-30% | Fisher | CAS: 1336-21-6 |
| Ammonium hydroxide, 28-32% | VWR International | CAS: 1336-21-6 |
| Trimethylamine solution, 45% | Sigma Aldrich | CAS: 75-50-3 |
| Dimethylamine solution, 40% | Sigma Aldrich | CAS: 124-40-3 |
| Ethylamine solution, 66-72% | Sigma Aldrich | CAS: 75-04-7 |
| Butylamine, 99.5% | Sigma Aldrich | CAS: 109-73-9 |
| Cadaverine, >97.0% | Sigma Aldrich | CAS: 462-94-2 |
| Spermidine, 99% | ACROS Organics | CAS: 124-20-9 |
| Isoamylamine, 99% | ACROS Organics | CAS: 107-85-7 |
| Benzaldehyde, >99% | Sigma Aldrich | CAS: 100-52-7 |
| Paraffin, liquid, pure | ACROS Organics | CAS: 8012-95-1 |
| Critical commercial assays | ||
| MultiSite Gateway Pro 3.0 | Thermo Fisher | 12537-103 |
| Q5 Site-Directed Mutagenesis Kit | New England Biolabs | E0554S |
| iScript gDNA Clear cDNA Synthesis Kit | Bio-Rad | 1725035 |
| SsoFast EvaGreen Supermix | Bio-Rad | 1725202 |
| DIG RNA Labeling Kit (SP6/T7) | Roche | 11175025910 |
| Deposited data | ||
| Experimental models: cell lines | ||
| Experimental models: organisms/strains | ||
| D. melanogaster: Ir31a-GAL4 | Bloomington Drosophila Stock Center | RRID:BDSC_41726 |
| D. melanogaster: Ir75d-GAL4 | Bloomington Drosophila Stock Center | RRID:BDSC_41729 |
| D. melanogaster: UAS-DTA | Bloomington Drosophila Stock Center | RRID:BDSC_25039 |
| D. melanogaster: Orco-GAL4 | Bloomington Drosophila Stock Center | RRID:BDSC_26818 |
| D. melanogaster: Or22a-GAL4 | Bloomington Drosophila Stock Center | RRID:BDSC_9951 |
| D. melanogaster: Ir41a-GAL4 | Bloomington Drosophila Stock Center | RRID:BDSC_41749 |
| D. melanogaster: Ir64a-GAL4 | Bloomington Drosophila Stock Center | RRID:BDSC_41732 |
| D. melanogaster: UAS-GCaMP7s, chromosome III | Bloomington Drosophila Stock Center | RRID:BDSC_79032 |
| D. melanogaster: UAS-GCaMP7s, chromosome II | Bloomington Drosophila Stock Center | RRID:BDSC_80905 |
| D. melanogaster: UAS-mCD8::GFP, chromosome III | Bloomington Drosophila Stock Center | RRID:BDSC_5130 |
| D. melanogaster: UAS-mCD8::GFP, chromosome II | Bloomington Drosophila Stock Center | RRID:BDSC_5137 |
| D. melanogaster: Ir76a-GAL4 | Richard Benton, University of Lausanne | RRID:BDSC_41735 |
| D. melanogaster: Ir92a-GAL4 | Richard Benton, University of Lausanne | RRID:BDSC_41733 |
| D. melanogaster: Ir25a 2 | Richard Benton, University of Lausanne | RRID:BDSC_41737 |
| D. melanogaster: Or35a-GAL4 | John Carlson, Yale University [13] |
FlyBase FBtp0039304 |
| D. melanogaster: Repo-Gal80[n18] | Tzumin Lee, Janelia Farm [40] |
N/A |
| D. melanogaster: Ir92a-GAL4-1 | Greg Suh, New York University [5] |
N/A |
| D. melanogaster: Amt 1 | [14] | FlyBase FBal0192654 |
| D. melanogaster: Amt-GAL4 | [14] | FlyBase FBtp0098429 |
| D. melanogaster: UAS-Amt | [14] | FlyBase FBtp0098427 |
| D. melanogaster: UAS-AgAmt | [14] | FlyBase FBtp0098428 |
| D. melanogaster: Ir76b-GAL4 | [14] | FlyBase FBtp0098430 |
| D. melanogaster: Ir8a-GAL | [14] | FlyBase FBtp0098431 |
| D. melanogaster: Ir92a1 | This study | N/A |
| D. melanogaster: Rh501 | This study | N/A |
| D. melanogaster: Rh50-Gal4 | This study | N/A |
| D. melanogaster: Ir75a-Gal4 | This study | N/A |
| D. melanogaster: Ir25a-Gal4 | This study | N/A |
| D. melanogaster: Canton-S | John Carlson, Yale University | N/A |
| D. melanogaster: Cantonized w1118 | [43] | N/A |
| D. melanogaster: w1118 isogenic | [41] | N/A |
| Oligonucleotides | ||
| Ir92a genotyping primers, pair 1: TGTATGGCCGGTAGGATCTC and ACCTCCTTGATCGAAACCCT | This paper | N/A |
| Ir92a genotyping primers, pair 2: GGCAAGAATGCGAACAAAT and TGGTTTGTCCAAACTCATCAA | This paper | N/A |
| Rh50 genotyping primers, pair 1: CCTCTCCCTGGAGAACATCA and CCCTCTAGCTTTCCCGTTTC | This paper | N/A |
| Rh50 genotyping primers, pair 2: CTGTTCATGGCTGCTCTAGT and CTGAGATAGGTGCCTCACTG | This paper | N/A |
| Rh50 qRT-PCR primers: AATGAGCAGTGTGACAGCGA and CATTGCCTCCGCCATTTACG | This paper | N/A |
| eIF1A qRT-PCR primers: ATCAGCTCCGAGGATGACGC and GCCGAGACAGACGTTCCAGA | This paper | N/A |
| Recombinant DNA | ||
| pBGRY | [43] | Addgene plasmid #61599 |
| pU6-BbsI-chiRNA | [46] | Addgene plasmid #45946 |
| pHD-DsRed-attP | [47] | Addgene plasmid #51019 |
| BAC containing Rh50 genomic region | [42] | BACPAC Resources #RP98-17L24 |
| BAC containing Ir75a genomic region | [42] | BACPAC Resources #RP98-44L18 |
| BAC containing Ir25a genomic region | [42] | BACPAC Resources #RP98-4C19 |
| Rh50 cDNA clone | [49] | Drosophila Genomics Resource Center, #GH03016 |
| Software and algorithms | ||
| ImageJ/FIJI | [50] | https://imagej.net/Fiji |
| AMIRA v6.7 | Thermo Fisher | N/A |
| IMOD v4.9 | [52] | https://bio3d.colorado.edu/imod/ |
| LabChart Pro v8 | AD Instruments | https://www.adinstruments.com/products/labchart |
| Prism v8 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| Other | ||
Highlights- max 85 characters with spaces.
Ammonium transporters label previously uncharacterized neuron populations
These olfactory neurons selectively respond to ammonia
The Amt transporter acts as a non-canonical olfactory receptor in Drosophila
The function of Amt may be conserved across insect species
Acknowledgments
We thank Tzumin Lee, Greg Suh, John Carlson, Richard Benton and the Bloomington Drosophila Stock Center (NIH P40OD018537) for fly lines. We thank the Developmental Studies Hybridoma Bank, NIH NICHD at the University of Iowa, for antibodies. The pBGRY plasmid was a gift from John Carlson and Tong-Wey Koh. The pU6-BbsI-chiRNA plasmid was a gift from Melissa Harrison & Kate O’Connor-Giles & Jill Wildonger. The pHD-DsRed plasmid was a gift from Kate O’Connor-Giles. We thank Yinan Xuan for assistance with transcutical imaging data analysis and Pratyajit Mohapatra for genomic sequencing validation of mutants.
We thank Anastasios Tzingounis for discussions and comments on the manuscript and Rebecca Oramas for graphic design. Research in K.M.’s laboratory was supported by NIH awards R21DC017868, R03DC015629, and R35GM133209. Research in C-Y.S.’s lab was supported by R01DC016466, R01DC015519 and R21DC108912. V.G. and S.S. were supported by the Max Planck Society. Research in J.MJ.’s laboratory was supported by NIH awards R01DC018570, R01NS116584, the Richard and Susan Smith Family Award for Excellence in Biomedical Research, the Klingenstein-Simons Fellowship Award in Neuroscience, and The Kavli Institute at Yale University. H.S.K. is supported by an NSF Graduate Research Fellowship.
Footnotes
Declaration of interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mumcuoglu Y, Galun R, and Ikan R (1986). The aggregation response of human body louse (Pediculus humanus)(Insecta: Anoplura) to its excretory products. Int. J.Trop. Insect Sci. 7, 629–632. [Google Scholar]
- 2.Takken W, and Knols BGJ (2010). Olfaction in Vector-Host Interactions, Volume 2, (Wageningen, The Netherlands: Wageningen Academic Publishers; ). [Google Scholar]
- 3.Andrade AJ, Andrade MR, Dias ES, Pinto MC, and Eiras AE (2008). Are light traps baited with kairomones effective in the capture of Lutzomyia longipalpis and Lutzomyia intermedia? An evaluation of synthetic human odor as an attractant for phlebotomine sand flies (Diptera: Psychodidae: Phlebotominae). Mem. Inst. Oswaldo Cruz 103, 337–343. [DOI] [PubMed] [Google Scholar]
- 4.Epsky ND, Kendra PE, and Schnell EQ (2014). History and Development of Food-Based Attractants. In Trapping and the Detection, Control, and Regulation of Tephritid Fruit Flies, Shelly T, Epsky N, Jang EB, Reyes-Flores J and Vargas R, eds. (Dordrecht, Netherlands: Springer; ), pp. 75–118. [Google Scholar]
- 5.Min S, Ai M, Shin SA, and Suh GS (2013). Dedicated olfactory neurons mediating attraction behavior to ammonia and amines in Drosophila. Proc. Natl. Acad. Sci. USA 110, E1321–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rytz R, Croset V, and Benton R (2013). Ionotropic receptors (IRs): chemosensory ionotropic glutamate receptors in Drosophila and beyond. Insect Biochem Mol Biol 43, 888–897. [DOI] [PubMed] [Google Scholar]
- 7.Abuin L, Bargeton B, Ulbrich MH, Isacoff EY, Kellenberger S, and Benton R (2011). Functional architecture of olfactory ionotropic glutamate receptors. Neuron 69, 44–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sato K, Pellegrino M, Nakagawa T, Nakagawa T, Vosshall LB, and Touhara K (2008). Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature 452, 1002–1006. [DOI] [PubMed] [Google Scholar]
- 9.Missbach C, Dweck HK, Vogel H, Vilcinskas A, Stensmyr MC, Hansson BS, and Grosse-Wilde E (2014). Evolution of insect olfactory receptors. Elife 3, e02115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vosshall LB, and Stocker RF (2007). Molecular architecture of smell and taste in Drosophila. Annu. Rev. Neurosci. 30, 505–533. [DOI] [PubMed] [Google Scholar]
- 11.Silbering AF, Rytz R, Grosjean Y, Abuin L, Ramdya P, Jefferis GS, and Benton R (2011). Complementary function and integrated wiring of the evolutionarily distinct Drosophila olfactory subsystems. J. Neurosci. 31, 13357–13375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benton R, Vannice KS, Gomez-Diaz C, and Vosshall LB (2009). Variant Ionotropic Glutamate Receptors as Chemosensory Receptors in Drosophila. Cell 136, 149–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yao CA, Ignell R, and Carlson JR (2005). Chemosensory coding by neurons in the coeloconic sensilla of the Drosophila antenna. J. Neurosci. 25, 8359–8367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Menuz K, Larter NK, Park J, and Carlson JR (2014). An RNA-seq screen of the Drosophila antenna identifies a transporter necessary for ammonia detection. PLoS Genet. 10, e1004810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weihrauch D, Donini A, and O’Donnell MJ (2012). Ammonia transport by terrestrial and aquatic insects. J. Insect Physiol. 58, 473–487. [DOI] [PubMed] [Google Scholar]
- 16.Andrade SL, and Einsle O (2007). The Amt/Mep/Rh family of ammonium transport proteins. Mol. Membr. Biol. 24, 357–365. [DOI] [PubMed] [Google Scholar]
- 17.Pitts RJ, Derryberry SL Jr., Pulous FE, and Zwiebel LJ (2014). Antennal-expressed ammonium transporters in the malaria vector mosquito Anopheles gambiae. PLoS One 9, e111858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delventhal R, Menuz K, Joseph R, Park J, Sun JS, and Carlson JR (2017). The taste response to ammonia in Drosophila. Sci. Rep. 7, 43754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown JB, Boley N, Eisman R, May GE, Stoiber MH, Duff MO, Booth BW, Wen J, Park S, Suzuki AM, et al. (2014). Diversity and dynamics of the Drosophila transcriptome. Nature 512, 393–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shanbhag SR, Singh K, and Singh RN (1995). Fine structure and primary sensory projections of sensilla located in the sacculus of the antenna of Drosophila melanogaster. Cell Tissue Res 252, 237–249. [DOI] [PubMed] [Google Scholar]
- 21.Ye Z, Liu F, Sun H, Barker M, Pitts RJ, and Zwiebel LJ (2020). Heterogeneous expression of the ammonium transporter AgAmt in chemosensory appendages of the malaria vector, Anopheles gambiae. Insect Biochem Mol Biol 120, 103360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shanbhag S, Muller B, and Steinbrecht A (1999). Atlas of olfactory organs of Drosophila melanogaster. 1. Types, external organization, innervation and distribution of olfactory sensilla. Int. J. Insect Morphol. Embryol. 25, 377–397. [Google Scholar]
- 23.Zhang Y, Tsang TK, Bushong EA, Chu LA, Chiang AS, Ellisman MH, Reingruber J, and Su CY (2019). Asymmetric ephaptic inhibition between compartmentalized olfactory receptor neurons. Nat. Commun. 10, 1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsang TK, Bushong EA, Boassa D, Hu J, Romoli B, Phan S, Dulcis D, Su CY, and Ellisman MH (2018). High-quality ultrastructural preservation using cryofixation for 3D electron microscopy of genetically labeled tissues. eLife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Endo K, Aoki T, Yoda Y, Kimura K, and Hama C (2007). Notch signal organizes the Drosophila olfactory circuitry by diversifying the sensory neuronal lineages. Nat. Neurosci. 10, 153–160. [DOI] [PubMed] [Google Scholar]
- 26.Chai PC, Cruchet S, Wigger L, and Benton R (2019). Sensory neuron lineage mapping and manipulation in the Drosophila olfactory system. Nat. Commun. 10, 643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Q, Barish S, Okuwa S, Maciejewski A, Brandt AT, Reinhold D, Jones CD, and Volkan PC (2016). A Functionally Conserved Gene Regulatory Network Module Governing Olfactory Neuron Diversity. PLoS Genet. 12, e1005780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grabe V, Baschwitz A, Dweck HK, Lavista-Llanos S, Hansson BS, and Sachse S (2016). Elucidating the neuronal architecture of olfactory glomeruli in the Drosophila antennal lobe. Cell Rep. 16, 3401–3413. [DOI] [PubMed] [Google Scholar]
- 29.Bates AS, Schlegel P, Roberts RJV, Drummond N, Tamimi IFM, Turnbull R, Zhao X, Marin EC, Popovici PD, Dhawan S, et al. (2020). Complete Connectomic Reconstruction of Olfactory Projection Neurons in the Fly Brain. Curr. Biol. 30, 3183–3199 e3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marin EC, Buld L, Theiss M, Sarkissian T, Roberts RJV, Turnbull R, Tamimi IFM, Pleijzier MW, Laursen WJ, Drummond N, et al. (2020). Connectomics Analysis Reveals First-, Second-, and Third-Order Thermosensory and Hygrosensory Neurons in the Adult Drosophila Brain. Curr. Biol. 30, 3167–3182 e3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dana H, Sun Y, Mohar B, Hulse BK, Kerlin AM, Hasseman JP, Tsegaye G, Tsang A, Wong A, Patel R, et al. (2019). High-performance calcium sensors for imaging activity in neuronal populations and microcompartments. Nat. Methods 16, 649–657. [DOI] [PubMed] [Google Scholar]
- 32.Ai M, Min S, Grosjean Y, Leblanc C, Bell R, Benton R, and Suh GS (2010). Acid sensing by the Drosophila olfactory system. Nature 468, 691–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Larsson MC, Domingos AI, Jones WD, Chiappe ME, Amrein H, and Vosshall LB (2004). Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron 43, 703–714. [DOI] [PubMed] [Google Scholar]
- 34.Hussain A, Zhang M, Ucpunar HK, Svensson T, Quillery E, Gompel N, Ignell R, and Grunwald Kadow IC (2016). Ionotropic chemosensory receptors mediate the taste and smell of polyamines. PLoS Biol. 14, e1002454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meier-Wagner J, Nolden L, Jakoby M, Siewe R, Kramer R, and Burkovski A (2001). Multiplicity of ammonium uptake systems in Corynebacterium glutamicum: role of Amt and AmtB. Microbiology 147, 135–143. [DOI] [PubMed] [Google Scholar]
- 36.Wacker T, Garcia-Celma JJ, Lewe P, and Andrade SL (2014). Direct observation of electrogenic NH4(+) transport in ammonium transport (Amt) proteins. Proc. Natl. Acad. Sci. USA 111, 9995–10000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Williamson G, Tamburrino G, Bizior A, Boeckstaens M, Mirandela GD, Bage M, Pisliakov A, Ives CM, Terras E, and Hoskisson PA (2020). A two-lane mechanism for selective biological ammonium transport. eLife 9, e57183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Teng B, Wilson CE, Tu YH, Joshi NR, Kinnamon SC, and Liman ER (2019). Cellular and neural responses to sour stimuli require the proton channel Otop1. Curr. Biol. 29, 3647–3656 e3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lamoureux G, Javelle A, Baday S, Wang S, and Berneche S (2010). Transport mechanisms in the ammonium transporter family. Transfus. Clin. Biol. 17, 168–175. [DOI] [PubMed] [Google Scholar]
- 40.Awasaki T, Huang Y, O’Connor MB, and Lee T (2011). Glia instruct developmental neuronal remodeling through TGF-beta signaling. Nat. Neurosci. 14, 821–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thibault ST, Singer MA, Miyazaki WY, Milash B, Dompe NA, Singh CM, Buchholz R, Demsky M, Fawcett R, Francis-Lang HL, et al. (2004). A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat. Genet. 36, 283–287. [DOI] [PubMed] [Google Scholar]
- 42.Hoskins RA, Nelson CR, Berman BP, Laverty TR, George RA, Ciesiolka L, Naeemuddin M, Arenson AD, Durbin J, David RG, et al. (2000). A BAC-based physical map of the major autosomes of Drosophila melanogaster. Science 287, 2271–2274. [DOI] [PubMed] [Google Scholar]
- 43.Koh TW, He Z, Gorur-Shandilya S, Menuz K, Larter NK, Stewart S, and Carlson JR (2014). The Drosophila IR20a clade of ionotropic receptors are candidate taste and pheromone receptors. Neuron 83, 850–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Markstein M, Pitsouli C, Villalta C, Celniker SE, and Perrimon N (2008). Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat. Genet. 40, 476–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Groth AC, Fish M, Nusse R, and Calos MP (2004). Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics 166, 1775–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gratz SJ, Cummings AM, Nguyen JN, Hamm DC, Donohue LK, Harrison MM, Wildonger J, and O’Connor-Giles KM (2013). Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics 194, 1029–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gratz SJ, Ukken FP, Rubinstein CD, Thiede G, Donohue LK, Cummings AM, and O’Connor-Giles KM (2014). Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics 196, 961–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ren X, Sun J, Housden BE, Hu Y, Roesel C, Lin S, Liu LP, Yang Z, Mao D, Sun L, et al. (2013). Optimized gene editing technology for Drosophila melanogaster using germ line-specific Cas9. Proc Natl Acad Sci U S A 110, 19012–19017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rubin GM, Hong L, Brokstein P, Evans-Holm M, Frise E, Stapleton M, and Harvey DA (2000). A Drosophila complementary DNA resource. Science 287, 2222–2224. [DOI] [PubMed] [Google Scholar]
- 50.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grabe V, Strutz A, Baschwitz A, Hansson BS, and Sachse S (2015). Digital in vivo 3D atlas of the antennal lobe of Drosophila melanogaster. J. Comp. Neurol. 523, 530–544. [DOI] [PubMed] [Google Scholar]
- 52.Kremer JR, Mastronarde DN, and McIntosh JR (1996). Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116, 71–76. [DOI] [PubMed] [Google Scholar]
- 53.Dobritsa AA, van der Goes van Naters W, Warr CG, Steinbrecht RA, and Carlson JR (2003). Integrating the molecular and cellular basis of odor coding in the Drosophila antenna. Neuron 37, 827–841. [DOI] [PubMed] [Google Scholar]
- 54.Benton R, and Dahanukar A (2011). Electrophysiological recording from Drosophila olfactory sensilla. Cold Spring Harb Protoc 2011, 824–838. [DOI] [PubMed] [Google Scholar]
- 55.Kaissling KE, and Thorson J (1980). Insect olfactory sensilla: Structural, chemical and electrical aspects of the functional organization. In Receptors for neurotransmitters, hormones, and pheromones in insects, Sattelle DB, Hall LM and Hildebrand JG, eds. (Amsterdam: Elsevier; ), pp. 261–282. [Google Scholar]
- 56.Hong EJ, and Wilson RI (2015). Simultaneous encoding of odors by channels with diverse sensitivity to inhibition. Neuron 85, 573–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pnevmatikakis EA, and Giovannucci A (2017). NoRMCorre: An online algorithm for piecewise rigid motion correction of calcium imaging data. J. Neurosci. Methods 291, 83–94. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All custom codes used for analysis are available upon request. This study did not generate any unique datasets.


