Abstract
Clinical management of COVID-19 patients with severe complications has been hindered by a lack of effective drugs and a failure to capture the extensive heterogeneity of the disease with conventional methods. Here, we review emerging roles of complex organoids in the study of SARS-CoV-2 infection, modelling of COVID-19 disease pathology and in drug, antibody, and vaccine development. We discuss opportunities for COVID-19 research as well as remaining challenges in the application of organoids.
INTRODUCTION
COVID-19, a SARS-CoV-2 (CoV-2) coronavirus disease, represents a global health emergency. As of May 30, 2021, approximately 169 million individuals have been infected and 3,530,582 deaths confirmed worldwide1. Even though vaccines have now been established to prevent infection, to date, no specific antiviral drugs exist that target CoV-2 to mitigate established disease. Clinical management of COVID-19 patients mainly focuses on improving symptoms, supporting lung function, preventing a sudden acute increase of circulating cytokines (cytokine storm), and controlling infections2.
Ongoing fast-track clinical trials focus on therapeutic solutions that block the CoV-2 infection cycle and associated pathophysiological processes3. Nevertheless, it is still poorly understood how the genetic background of COVID-19 patients might affect the severity of symptoms4,5. Similarly, whether CoV-2-host receptor interactions might differ depending on the age, gender, and ethnicity of a patient has not been clarified. As a result, the design of effective vaccines and antiviral drugs has remained challenging. The advancement of organoid-based assays derived from human pluripotent stem cells (hPSCs) and adult stem cells (ASCs) offers an opportunity to expand and bank various types of tissue-specific organoids for biomedical research6–9. Accordingly, stem-cell-based 2D cell cultures and 3D organoids are also used to study CoV-2 infection10–19. These studies highlight the need to define the roles of stem-cell-based organoids in COVID-19 research.
In this Review, we recapitulate the CoV-2 infection cycle and associated intervention strategies. We evaluate current COVID-19-based assays, focusing on their strengths and potential limitations. We further elucidate the role of respiratory cell types and lung organoids in assessing CoV-2 susceptibility and discuss other organoid systems (derived from hPSCs and ASCs) that can be used. Lastly, we examine the benefits of organoids in studying CoV-2-induced pathophysiology and predicting therapeutic outcomes.
CoV-2 infection cycle and associated intervention strategies
CoV-2 is a positive-sense and single-stranded RNA beta-coronavirus, potentially evolved from a bat coronavirus20–23. Genomic diversity of CoV-2 in COVID-19 patients is evident24–26, but the environmental CoV-2 genome is relatively stable27. The structural genomics of CoV-2 indicates evolutionarily conserved functional regions of viral proteins27–29. In addition, CoV-2 shares a similar infection cycle with SARS-CoV and MERS-CoV coronaviruses30–32.
CoV-2 infection cycle.
Just as SARS-CoV, CoV-2 enters and infects a human host cell via multiple coordinated processes30–32. The CoV-2 infection cycle is illustrated (Fig. 1a) with distinct steps (1–17), starting from its host cell entry via membrane fusion and endocytosis to the release of a mature CoV-2. In COVID-19 patients, the infection cycle increases the viral load in the respiratory tissues, kidneys, and intestine33. The induction and release of cellular cytokines (also called a cytokine storm) may trigger a wide range of host immunological and inflammatory responses in these tissues (Fig. 1b–e)34,35. Cytokine storms often lead to diffuse alveolar damage, acute respiratory distress syndrome (ARDS), loss of gas exchange, respiratory failure, and multi-organ damage, overall increasing death rates35–40.
Fig. 1: SARS-CoV-2 (CoV-2) infection cycle, immunological response, molecular targets, and intervention strategies.
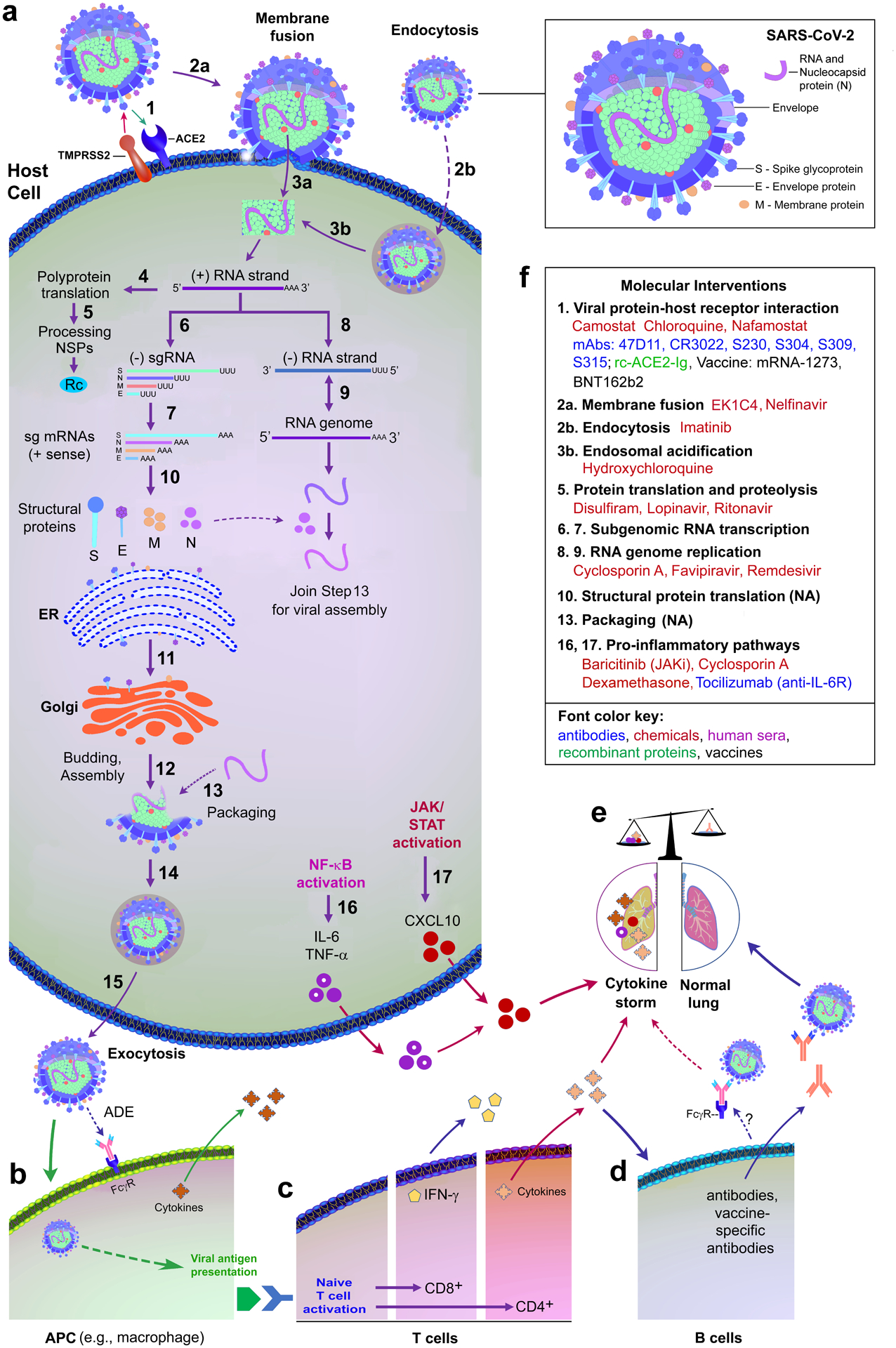
(a) The infection cycle includes spike glycoprotein (S-gp) binding to the human angiotensin-converting enzyme 2 (ACE2) receptor, pre-cleavage by the host cellular protease furin to dissociate the S1 subunit from the S2 subunit of S-gp161,162, and S2 activation mediated by serine protease TMPRSS2 co-receptor41. Notably, cleavage by furin is required for the entry of CoV-2 into human lung cells161. S2 activation triggers viral and host cell membrane fusion. Within the host cell cytoplasm, the positive-sense CoV-2 genomic RNA is transcribed to yield full-length negative-sense RNAs (for genome replication) and subgenomic negative-sense RNAs (-sgRNA, for producing subgenomic mRNAs). Subgenomic mRNAs, converted from -sgRNAs, are translated into viral structural proteins, including S-gp, envelope (E), membrane (M), and nucleocapsid (N) proteins30–32. Finally, viral genome encapsulation and reassembly enable virus maturation and export out of cells for the next infection cycle. (b and c) CoV-2 induces immunological responses through viral antigen presentation in macrophages, naive T cell activation, and release of cytokines. (d) A possible dual role of B-cell-mediated humoral immune response: B cells generate the neutralising antibodies to protect the lung from CoV-2 infection and contribute to cytokine-induced damage through FcγR-mediated and antibody-dependent enhancement of CoV-2 infection. (e) CoV-2-induced organ damage via an unbalanced presence of pro-inflammatory cytokines or absence of antiviral factors. (f) Representative intervention strategies, such as the development of drugs, vaccines, antibodies, recombinant proteins and repurposing of approved drugs against CoV-2 infection, based on molecular targets in Figure 1a.
Abbreviations: ACE2, Angiotensin-converting enzyme 2; ADE, antibody-dependent enhancement; APC, antigen-presenting cells; CXCL10, C-X-C motif chemokine ligand 10; ER, endoplasmic reticulum; FcγR, Fc-gamma receptor; IFN, interferon; IL-6, interleukin 6; IL-6R, Interleukin 6 receptor; JAK, Janus kinase; JAKi, Janus kinase inhibitor; mAbs, monoclonal antibodies; NA, data not available; NF-κB, nuclear factor kappa B; Rc, replicase and transcriptase complex; NSPs, non-structural proteins; rc-ACE2-Ig, recombinant ACE2-Ig; STAT, signal transducer and activator of transcription; TMPRSS2, transmembrane protease serine 2; TNF-α, tumour necrosis factor alpha.
Therapeutic strategies.
Despite an insufficient understanding of the CoV-2 infection cycle, all viral processes could be conceivably employed for pharmacological, immunological, and molecular interventions of CoV-2 infections. Such experimental and clinical interventions have been reported14,41–48, some of which are listed in Figure 1.
The abrogation of viral cell entry effectively prevents viral infection. Blockage of spike glycoprotein (S-gp) binding to ACE2 by a human recombinant soluble ACE2 (hrsACE2) reduced CoV-2 recovery from Vero cells, displaying a 1000- to 5000-fold reduction in viral growth14. This blockage appears to be ACE2 species-specific as recombinant mouse ACE2 had no effect14. TMPRSS2-mediated S-gp priming can be blocked with camostat, a clinically proven protease inhibitor, and substantially (~88%) inhibited by an anti-ACE2 antibody (at 20 μg/mL)41. Convalescent sera from SARS patients partially (~45%) neutralised pseudotyped CoV-2 entry41. CR3022, a neutralising antibody isolated from a convalescent SARS patient, targets S-gp receptor-binding domain (S-RBD) of SARS-CoV49 and also binds to the CoV-2 S-RBD50. High-resolution structures revealed a mechanism by which neutralising antibodies, such as CR3022, recognise S-RBD in its trimeric configuration51. Hence, these studies provide the molecular basis for future therapeutic interventions to prevent CoV-2 cell entry.
Beyond S-gp-ACE2-mediated membrane fusion, little is known about other cell entry mechanisms such as endocytic pathways, as evident in other coronaviruses. These pathways may be classified as clathrin-dependent endocytosis for SARS-CoV52, membrane rafts and caveolar endocytosis for the human coronavirus 229E53, and clathrin- and caveolar-independent entry of feline coronavirus54. Successful abolishment of CoV-2 entry by camostat suggests that the endocytic access may not be a major pathway for CoV-2 cell entry. However, this finding must be confirmed in different cellular and animal models.
The United States Food and Drug Administration (FDA)-approved drug, ivermectin, successfully inhibits the replication of CoV-2 in an in vitro model (i.e., Vero-hSLAM cells)46. The promising antiviral drug remdesivir (GS-5734), an adenosine analogue, also inhibits CoV-2 replication44. Remdesivir has become the first anti-CoV-2 drug approved by the FDA after a phase III clinical trial55. However, the World Health Organization’s solidarity trial revealed that remdesivir neither reduced mortality nor shortened the recovery time of COVID-1948. A less toxic derivative of chloroquine, hydroxychloroquine, is an endosomal acidification inhibitor and effective in inhibiting CoV-2 infection in cell culture42. Hydroxychloroquine has gained wide-spread use in the treatment of COVID-19. However, its broader clinical application has been under scrutiny due to the absence of well-controlled data on its effectiveness and reported severe side effects43.
Importantly, about 20% of severe COVID-19 cases are associated with cytokine storms, also observed in SARS and MERS35,56, which can be treated by inhibiting cytokine release or accelerating cytokine clearance in targeted cells57. The monoclonal antibody tocilizumab, which inhibits IL-6, has been used to treat cytokine storms in COVID-19 patients in clinical trials58. In an early trial, treatment with tocilizumab reduced the risk of invasive mechanical ventilation or death rate in severe COVID-19 patients59. In a later report, moderate COVID-19 patients treated with tocilizumab showed fewer severe infections than those who received a placebo. However, tocilizumab did not prevent the need for intubation or death in these patients60. Thus, the role of tocilizumab in the treatment of COVID-19 remains obscure.
Encouragingly, a meta-analysis of seven randomised clinical trials revealed lower 28-day mortality among critically ill patients who received systemic corticosteroids compared to those who received usual care or placebos61. In the RECOVERY trial, the immunosuppressant dexamethasone (6 mg once daily for up to 10 days) reduced 28-day mortality in patients who required oxygen, particularly in those receiving mechanical ventilation47. No benefit was found for patients who did not require oxygen supplementation47. The mechanism that underlies the beneficial effect of dexamethasone is not well understood in these patients. It may be associated with the inhibition of major pro-inflammatory pathways, such as NF-κB, in the most severe patients (Fig. 1a and 1f)38. Nonetheless, these clinical trials suggest that cytokine storms contribute to lung injury and multi-organ failure in severe COVID-19 patients. For this reason, major health organisations recommend dexamethasone (or potentially other glucocorticoids) as standard care for patients with severe COVID-19.
Other factors that influence the infection cycle.
CoV-2 infection cycles are associated with diverse clinical characteristics in COVID-19 patients, manifesting no, mild, or severe symptoms, such as acute respiratory disease and pneumonia62–64. Evidently, some asymptomatic patients have persistent negative computed tomographic findings65, suggesting low viral load or low inflammatory and immunological responses in the lungs. Approximately 80% of the infections are asymptomatic or mild, 15% are severe (requiring oxygen inhale), and 5% of patients are in critical conditions and require a ventilator66. At this time, it is impossible to predict which patient will become one of that 5% to need critical care.
Many tangible intrinsic (such as age, gender and ethnicity) and extrinsic factors (such as lifestyle) influence the infection cycle, morbidity and mortality rates. CoV-2 infects humans from neonates to older adults67–69. However, paediatric cases are less often symptomatic than older adults69–71. CoV-2 infection also affects women less than men72, possibly because androgen signalling modulates ACE2 levels. Increased androgen levels are associated with a higher risk of CoV-2 infection and disease severity in men17. Demographic data reveal high morbidity and mortality rates in African Americans in the USA73–75, although underlying reasons remain unclear and could likely be multifactorial, including socioeconomic factors and access to healthcare.
Cigarette smoking increases susceptibility to CoV-2 infections by upregulating ACE2 expression16,76,77. Collectively, age, gender, lifestyle, and demographic differences might modulate viral receptor expression and other unknown determinants, which, in turn, contribute to disease severity and therapeutic response. For instance, co-expression of ACE2 and TMPRSS2 mRNAs is tightly regulated in an age- and gender-dependent manner and upregulated in individuals who smoke78.
In summary, age, gender, and genetic background will have to be integrated into conventional CoV-2 assays and COVID-19 models to facilitate the screening of antiviral drugs and antibodies and predict therapeutic responses. Conventional COVID-19 assays can be classified into three categories: in vitro biochemical, pseudotyped and live virus assays (Fig. 2a). In this Review, we would like to focus on cell culture models for COVID-19 research (Fig. 2b) and refer the reader to related reports and excellent Reviews on conventional assays79–85.
Fig. 2: COVID-19-related assays.
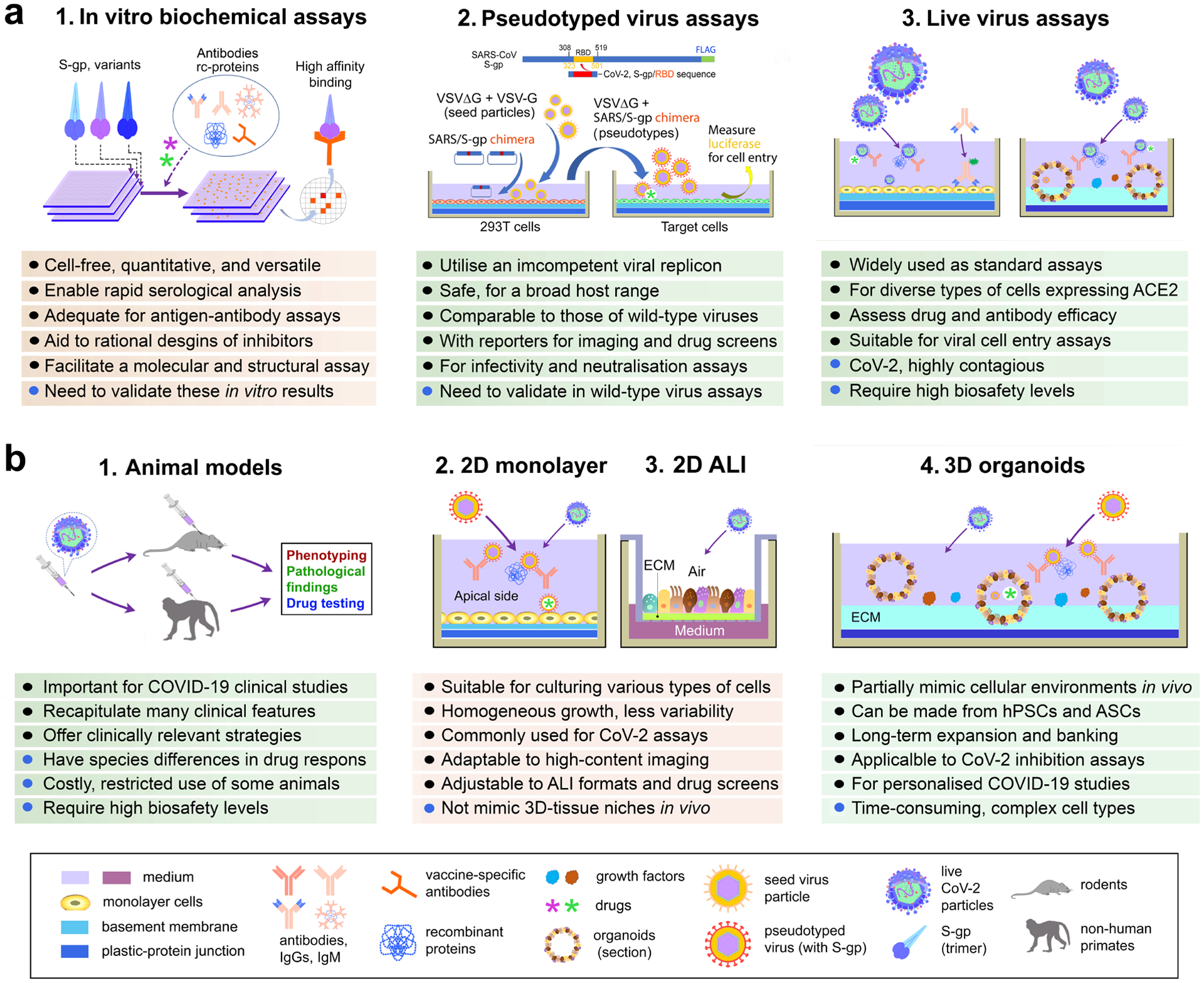
(a) Assays are categorised as in vitro cell-free molecular and biochemical, pseudotyped virus, and live virus assays. Pseudotyped virus experiments are exemplified by pseudotyped VSV harbouring VSV-G and a SARS-CoV-S-gp chimeras. At 16-hour post inoculation, the pseudotyped viral entry is analysed by determining luciferase activity in cell lysates86. No envelope glycoprotein pseudo-viral control is used for normalization. (b) Assays can be animal models, 2D-monolayer cell culture, 2D air-liquid interface (ALI) transwell culture, and 3D organoids. The combination of platforms empowers the utility of these assays for Covid-19 drug and vaccine development. Abbreviations: ASCs, adult stem cells; CoV2, SARS-CoV-2; ECM, extracellular matrix; hPSCs, human pluripotent stem cells; rc-proteins, recombinant proteins; S-gp, Spike glycoprotein; VSV-G, wild-type vesicular stomatitis virus; VSVΔG, vesicular stomatitis virus with deletion of the envelope glycoprotein (G).
Cell culture models for COVID-19 research
Theoretically, all cellular processes of the CoV-2 infection cycle could be used for assays to examine CoV-2 infectivity and for drug screens. At present, several experimental platforms and cell types exist for clinical and experimental coronavirus research, all of which have benefits and limitations (Fig. 2b). Among them, we focus on the three major systems used to study COVID-19: 2D monolayer cell culture, adapted 2D air-liquid interface (ALI) methods, and 3D culture or organoids (Fig. 2b).
2D monolayer culture.
2D monolayer cell cultures (Fig. 2b2) of various cell lines, such as 293T, A549, BHK, Caco-2, MDBK, PK-15, and Vero cells (available from the American Type Culture Collection) have been used to determine CoV-2 cell entry and for drug testing41,86. TMPRSS2-expressing Vero-E6 cells, which have a similar ACE2 structure to that of human cells, are highly susceptible to CoV-2 infection14,87 and represent an effective culture method to propagate CoV-2 and measure the viral load of CoV-2 variations.
ALI assays.
The ALI culture mimics the in vivo airway environment and is widely used in the maturation and functional assessment of the airway epithelium88. ALI assays allow the apical side of the epithelium to contact the air and the basolateral side to access the differentiation medium through a microporous membrane (Fig. 2b3). 2D-ALI is particularly suitable to evaluate links between air-borne related lung pathologies and susceptibility to a severe CoV-2 infection16. Limitations of this method are an inability to passage the culture, which means that it cannot be scaled up and used in high throughput assays, and its inability to generate more complex tissue structures, such as alveoli. Historically, growth and differentiation of respiratory basal cells in an ALI culture has been challenging in the absence of non-basal cells. For example, KRT5-GFP+ basal cells of the mouse trachea require a 500-fold excess of non-basal cells in ALI experiments to achieve approximately 6% colony-forming efficiency (based on counting large colonies) at day 2189. Using an adapted 3D sphere-forming assay, Hogan and colleagues were able to seed single KRT5-GFP+ basal cells of the mouse trachea in the absence of stroma or non-basal cells90. This 3D-culture adaptation leads to a rapid formation of “tracheospheres” within one week and a sphere-forming efficiency that is comparable to ALI experiments described above89,90.
3D cell culture and organoids.
Unlike 2D, 3D cell culture is an artificially created platform that mimics the in vivo environment of living cells and tissues. Cells are usually grown in suspension in suitable medium or extracellular matrices (such as Matrigel and collagen) to form spheroids or 3D colonies. The extracellular matrix components and physical forces play a vital role in regulating cell behaviour. Current organoid protocols partially recapitulate 3D cellular environments in vivo and retain the genetic and epigenetic features of human cells. They can be long-term expanded, banked for personalised medicine (Fig. 2b4) and used to model viral infectious diseases6–9,91,92. 3D organoids can be dissociated and adapted to 2D ALI cultures to facilitate directed differentiation of airway stem/progenitor cells into mature cells for downstream assays93,94 and apical viral respiratory infection94. An overview of the strengths and limitations of 2D and 3D culture is available in Figure 2b and previous Reviews6,8,95. Taken together, organoids represent a powerful platform for COVID-19 research.
hPSC-derived organoids for COVID-19 research
Organoids can be derived from hESCs or hiPSCs (we use the term hPSC for both) and maintained as a 3D tissue in vitro, capable of self-organising and self-renewal. They have been successfully used for disease modelling and drug discovery6,7, thus paving the way to study COVID-19 in vitro.
Modelling COVID-19.
The first proof-of-concept experiment demonstrated that CoV-2 infects human blood-vessel and kidney organoids14, which can be blocked with human recombinant ACE214. Subsequent reports confirmed that diverse types of hPSC-derived organoids, including intestinal, cardiac, brain, choroid plexus, and lung organoids, can be used as disease models to study the tropism of CoV-2 and for drug screening10,12,15,17,18. Lung organoids are particularly suitable as epithelial cells of the respiratory airways and alveoli are both targets and effectors of CoV-2 infections (Fig. 3).
Fig. 3: Lung cell types and organoids.
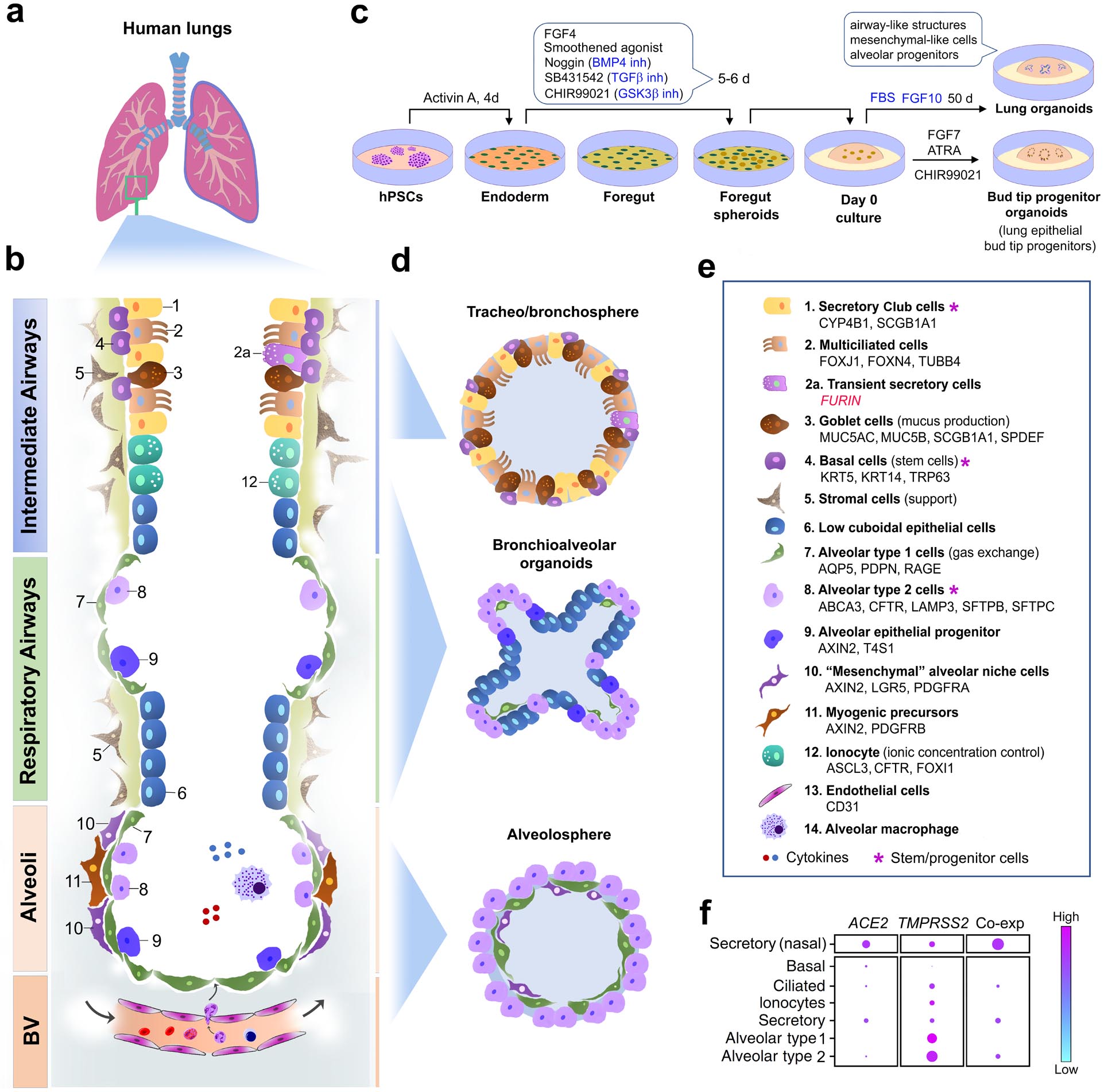
(a) Human lung anatomy. (b) Schematic of the major cell types in different compartments of the human lung, partially adapted from references78,97–99. (c) A representative protocol for the generation of lung organoids containing cell types of interest107. (d) Schematic of lung organoids that model different cellular compartments of the lung. (e) Cell types in panels b and d, with gene and protein markers listed alphabetically78,97–99,163,164. (f) Representative single-cell RNA sequencing analysis of CoV-2 receptor gene expression and co-expression (co-exp)109 in major cell types of the respiratory airways and alveoli. Nasal secretory cells are used as control for comparison. The size of the dots is proportional to the percentage of cells that express indicated genes (adapted from the published data in reference109).
Abbreviations: ABCA3, ATP-binding cassette subfamily A member 3; ACE2, angiotensin-converting enzyme 2; AQP5, aquaporin 5; ASCL3, Achaete-Scute family BHLH transcription factor 3; ATRA, all-trans retinoic acid; BMP4, bone morphogenetic protein 4; BV, blood vessels; CFTR, CF transmembrane conductance regulator; CYP4B1, cytochrome P450 family 4 subfamily B member 1; d, day(s); FBS, foetal bovine serum; FGF, fibroblast growth factor; FOXI1, forkhead box I1; FOXJ1, forkhead box J1; FOXN4, forkhead box N4; GSKβ, glycogen synthase kinase 3 beta; hiPSC, human induced pluripotent stem cells; inh, inhibitor; KRT5/14, keratin 5/14; LAMP3, lysosomal associated membrane protein 3; LGR5, leucine-rich repeat-containing G-protein coupled receptor 5; PDGFRA/B, platelet derived growth factor receptor alpha/beta; PDPN, podoplanin; SCGB1A1, secretoglobin family 1A member 1; SFTPB/C, surfactant protein B/C; SPDEF, SAM pointed domain containing ETS transcription factor; TMPRSS2, transmembrane serine protease 2; TUBB4, tubulin beta 4B class IVb.
Lung organoids.
The human lung is a complex organ with highly branched and progressively thinner tubes that carry air into the distal alveolar sacs. It comprises multiple integrated compartments: proximal and intermediate airways, respiratory bronchioles, and alveoli (Fig. 3a, 3b)96. Each compartment is populated by various cell types, including epithelial, vascular, stromal, and immune cells (Fig. 3b, 3e)78,97–99. The intermediate airways have a pseudostratified epithelial layer that holds heterogeneous cell types, including secretory Club cells, multiciliated cells, mucus-producing goblet cells, transient secretory cells and basal (stem) cells (Fig. 3b, 3e). The distal respiratory bronchioles are lined with a poorly characterized cuboidal epithelium. The alveoli are covered by alveolar epithelial type 1 and 2 cells (AECIs and AECIIs), important for gas exchange and alveolar homeostasis. Each compartment also has its own stem/progenitor population with specialised functions in response to environmental insults (Fig. 3b, 3e).
Airway epithelial cells are generated from hPSCs by imitating multi-stage lung developmental trajectories100, for instance to derive lung bud organoids that recapitulate lung development and disease101,102. Lung organoids containing more mature epithelial cells have also been created from hPSCs in vitro10,17,103–106. The derivation of lung organoids varies from protocol to protocol. However, the major consensus steps may be summarised, based on a well-documented protocol107, as follows. First, definitive endoderm is induced from hPSCs by Activin. Second, anterior foregut endoderm and foregut spheroids are sequentially formed by inhibiting BMP4, TGF-β, and GSK3β, in the presence of FGF4 and Smoothened agonist. Third, bud tip progenitor organoids are induced by FGF7, ATRA, and GSK3β inhibition. Finally, complex lung organoids containing airway-like structures, mesenchymal-like cells, and alveolar progenitors are obtained via a prolonged incubation with foetal bovine serum and FGF10 (Fig. 3c).
Lung organoids are classified into bronchospheres, bronchioalveolar organoids, and alveolospheres98,108 (Fig. 3d). In bronchospheres, secretory Club cells and basal cells represent stem-cell-like cells. Secretory Club cells are a CoV-2 target as they co-express the highest levels of ACE2 and TMPRSS2, compared to basal, ciliated, and alveolar cells in the lung (Fig. 3f)109. ACE2, TMPRSS2, and FURIN are also co-expressed in bronchial transient secretory cells that show high Rho GTPase activity and viral processes related to membrane remodelling or the immune system78, likely underlying their vulnerability to CoV-2 infection.
Alveolospheres contain flat AECIs and cuboidal AECIIs (Fig. 3d and 3e). AECIIs function as stem/progenitor cells in the adult lungs110, co-express ACE2 and TMPRSS2, and serve as another major CoV-2 target111,112. Not surprisingly, CoV-2 heavily affects alveolar pneumocytes (including AECII cells) in COVID-19 patients, leading to diffuse alveolar damage, respiratory failure, and increased mortality39,40,113,114. For this reason, hPSC-derived lung organoids should be particularly useful for the study of severe COVID-19.
Drug discovery.
hPSC-derived alveolar organoids have been used in CoV-2 infection assays, high-throughput drug screens, and drug repurposing10,17,115–118. For example, androgen receptor signalling inhibitors finasteride and dutasteride reduced the infectivity of CoV-2 in hESC-derived lung alveolar organoids by lowering ACE2 and TMPRSS2 levels17. A high-throughput drug repurposing screen in organoids identified multiple compounds (imatinib, mycophenolic acid, quinacrine dihydrochloride) that inhibit the cell entry of CoV-210. Similarly, an ACE2 blocking antibody inhibited viral entry in an organoid model, enhanced the activity of M2 macrophages, and suppressed pro-inflammatory effects mediated by M1 macrophages118. These in vitro experiments confirm that alveolar precursors and differentiated AECIIs are permissive to CoV-2 infection, elicit a cytokine response and can be used to identify compounds that block CoV-2 infection.
Despite these promising initial results, organoid models also have a number of limitations that should be considered. For instance, hPSCs are prone to genomic instability in long-term in vitro culture119–123. Further, inter-laboratory protocol differences inevitably increase experimental variability and cell culture and differentiation protocols are inherently time-consuming107,124. Finally, immature differentiation of lung organoids under suboptimal culture conditions remains a frequently encountered and unresolved issue125.
Human lung organoids often produce developmentally immature foetal lung tissues with a higher proliferation rate in vitro101,105,117,126. Epithelial cells from hESC-derived organoids express precursor markers such as NKX2.1 and SOX917. As a partial solution, 3D-organoid-converted 2D ALI cultures are increasingly used to enhance the maturity of differentiated respiratory epithelial cells for downstream analysis116,117,127. Nonetheless, a deeper understanding of the developmental principles underlying cell maturation and niche environments is necessary to optimise organoid protocols. These insights could facilitate the creation of chemically defined media and improved extracellular matrices or scaffolds124,128,129.
In summary, hPSC-based organoids are valuable for personalised medicine and disease modelling. They provide excellent platforms for drug efficacy and drug repurposing studies10,17,115. The expression of multiple CoV-2 susceptible genes in lung organoids makes them ideal models to study infectivity. However, we recommend verifying the results obtained from hPSC-derived organoids in animal models and organoids established from human ASCs.
ASC-derived organoids for COVID-19 research
The definition of ASCs varies in the scientific literature due to the complexity of cellular properties, including cellular dynamics130, heterogeneity131, and plasticity132. In addition, it can be difficult to distinguish ASCs from progenitor cells. In this Review, ASCs are defined as rare, mostly quiescent, and multipotent cells found in adult tissues. They are capable of long-term self-renewal, generate intermediate cell types (progenitors) with limited self-renewal potential, and differentiate into tissue-specific cells7,97. ASCs can be isolated from the adult issue and maintained in cell culture indefinitely if supplemented with appropriate microenvironments and growth factors. ASCs and progenitors serve as valuable alternatives to hPSCs, providing a source of fully mature cells for functional analysis.
Intestinal and nasal organoids.
Intestinal organoids and nasal spheroids have been derived from donor biopsies and were previously used to predict drug responses in patients with cystic fibrosis95. Differentiated enterocytes express ACE2 and TMPRSS2 (Fig. 4a) and substantial titres of CoV-2 particles were also detected in enterocytes of intestinal organoids13. Transcriptomic analysis indicated a strong viral response with enrichment of CXCL10 and CXCL11 mRNAs13, closely related to a cytokine storm. This study supports that ASC organoids can be used to study CoV-2 pathophysiology in vitro.
Fig. 4: Stem-cell-based organoids to assess SARS-CoV-2 (CoV-2) susceptibility.
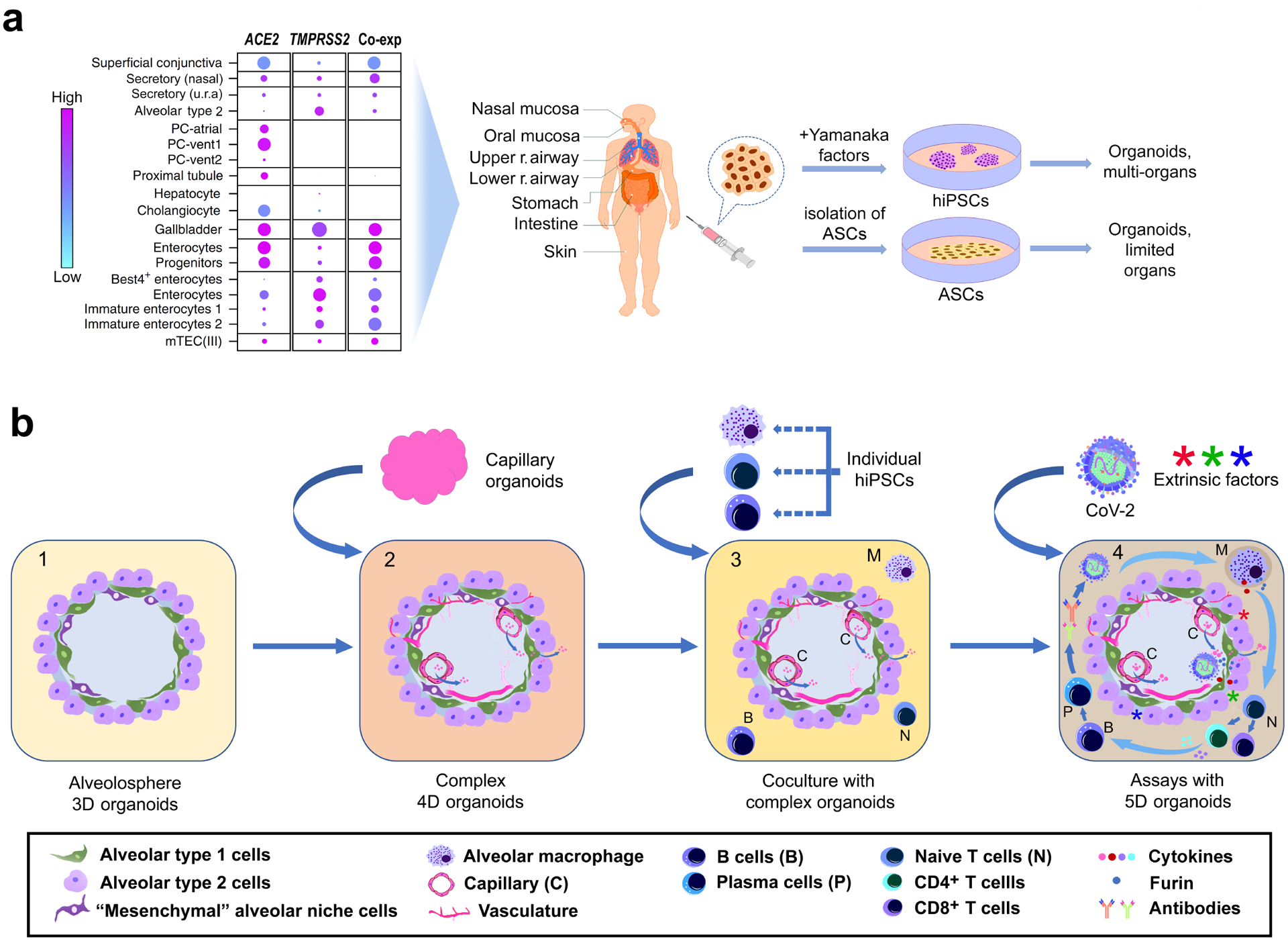
(a) CoV-2 receptor gene expression and coexpression (co-exp) in human cells (adapted from reference109). Isogenic organoids can be generated from adult stem cells (ASCs) and human induced pluripotent stem cells (hiPSCs). The size of the dots in the left panel is proportional to the percentage of cells that express the indicated genes. (b) Development of multi-dimensional organoids to model the complexity of immunological and hyper-inflammatory complications in COVID-19 patients. Abbreviations: mTEC (III), medullary thymic epithelial cells of the foetal thymus; PC-atrial and PC-vent, pericytes in the atrium and ventricle of the heart; r. respiratory; secretory (u.r.a), secretory cells from the upper respiratory airway.
Interestingly, the nasal mucosa also highly co-expresses ACE2 and TMPRSS2 (Fig. 4a)109, consistent with the heavy CoV-2 particle load in the nasal cavity of COVID-19 patients. The nasal mucosa has a similar epithelial lining to that of the upper respiratory airway, including secretory Club cells and basal stem cells109. As nasal biopsies are minimally invasive compared to intestinal or lung biopsies, nasal spheroids provide a valuable resource and surrogate for lung organoids.
Lung organoids.
Evidence suggests that both ASC-like cells and progenitors exist in different compartments of the lungs. Basal cells in the intermediate airways meet the definition of generic ASCs90,94. Basal stem cell organoids contain basal cells, secretory goblet cells, and ciliated cells (Fig. 3d, 3e). Airway basal stem cells have been isolated from human biopsies and expanded for functional assays of the airway repair response after CoV-2 infection16.
ASC-like cells or progenitors were also found within the SCGB1A1+ secretory Club and AXIN2+ AECII cell populations in human adult lungs (Fig. 3b, 3e)96,108. Mouse genetic lineage analysis revealed that surfactant protein C positive (SFTPC+) AECII cells in the alveolar niche are ASC-like cells, giving rise to self-renewing “alveolospheres” that contained both AECII and AECI-like cells110. In mice, rare Axin2+ AECIIs also act as alveolar stem cells and secrete Wnt molecules to recruit “bulk” AECIIs as the progenitors133. A distinct population of mouse IL1R1+ AECIIs can become damage-associated transient progenitors (DATPs), which then differentiate into mature AECIs134. In mouse and human lungs, similar alveolar epithelial progenitors (AEPs) reside within the AECII pool and generate mature AECIs and AECIIs from alveolar organoids135. Thus, AECIIs constitute an important stem/progenitor source in the alveoli.
Human alveolar organoids have been derived from adult AECIIs to assess CoV-2 infection11,112,127,136,137. These in vitro experiments confirm that AECIIs are the principal target of CoV-2. CoV-2-infected alveolar organoids mirror many features of COVID-19 patients, including cytokine release, interferon (IFN) and immune response, loss of surfactant proteins, and cell death. AECII-based organoids, derived in a feeder-free and chemically defined culture system, could be sustained long-term112 and revealed that few (≥1) CoV-2 particles that enter alveolar cells can lead to a full infection. Genes associated with cell death, cell adhesion, and surfactant proteins were also upregulated in CoV-2 infected AECIIs112.
IFN-mediated inflammatory signalling is a typical response to CoV-2 infection documented in these studies. An increase in the IFN response was associated with a lower CoV-2 burden (around 60 hours post CoV-2 infection of alveolar organoids) and vice versa for a decrease in the IFN response112. Pretreatment of alveolar organoids with low dose IFN-α and IFN-γ reduced CoV-2 replication11. In contrast, IFN inhibition endorsed viral replication11. Pretreatment of alveolar organoids with IFNB1 (INF-β) also reduced expression of the viral RNA gene N, which encodes the CoV-2 nucleoprotein127. These findings suggest that the administration of IFNs may be a possible prophylactic measure against severe CoV-2 infection.
Pharmacological inhibition of CoV-2 infection with small molecules is of considerable interest. ASC-derived lung organoids have not yet been used for drug repurposing or discovery studies. However, a study confirmed that remdesivir decreases CoV-2 N gene expression more effectively than INF-β or hydroxychloroquine in infected alveolar organoids127. This finding is intriguing but contradicts the ineffectiveness of remdesivir in the recent clinical trial discussed above48. Inconsistencies between the results from alveolar organoids and the clinical trial need to be further investigated.
In summary, lung organoids derived from adult human lungs generate respiratory epithelial cells with high maturity compared to hPSC-derived organoids and are suitable for studying COVID-19. However, it is often difficult to obtain lung tissues with the desired quality and materials are scarce, as samples are typically acquired from bronchioalveolar washings and lung explants with institutional review board approval108. In contrast to hPSC-derived lung organoids, ASC- or progenitor-based lung organoids exhibit limited self-renewal capacity, usually less than five passages. As for the former, developmental paradigms can guide the derivation of long-term expandable lung organoids from the adult lung94. For instance, FGF7 and FGF10 are vital in establishing long-term expandable lung organoids from adult tissues94,138. Interestingly, both factors are also required in the final steps to generate hPSC-derived lung organoids (Fig. 3c)107.
Improving lung regeneration.
Little is known about the regenerative capacity of the alveolus in COVID-19 patients. Organoid studies revealed that CoV-2 infected AECIIs exhibit defence and repair mechanisms to combat injury, such as cytokine secretion, resistance to apoptosis, and cellular senescence11,112,118,139. AECIIs still proliferate, transit to different cellular states, and differentiate into AECI-like cells. These cellular processes closely mimic regenerative responses in mouse and human injury models110,133–135. Further support for a targeted regenerative response comes from a study demonstrating that AECIIs proliferate and differentiate into squamous AECIs in severely affected alveoli of COVID-19 patients40.
Despite the existence of endogenous repair mechanisms, a number of recovered COVID-19 patients will require therapy to restore the lost lung function and repair the damage to alveolar cells. Replacing damaged alveolar cells with a suitable AECII source might be a possible way to improve lung function. Encouragingly, mature AECIIs of both mouse and human origins can be transplanted into injured mouse lungs140,141. Vunjak-Novakovic and colleagues proposed an airway-specific method to de-epithelialize the distal lung airways and preserve the basement membrane and vascular endothelium. This approach enabled the functional vascularisation of lung grafts to support the attachment and growth of hiPSC-derived epithelial cells in a rat model142. Similar transplantation approaches using organoid-derived lung epithelial cells may be applicable for treating COVID-19 patients with the severe epithelial injury in the future. Still, many preclinical challenges remain to be overcome, most notably relating to source cell identity, immunological compatibility, and functional integration into the host.
Opportunities and challenges
CoV-2 infection does not only affect the lung but can damage any cell that expresses ACE2 or co-expresses ACE2 and TMPRSS2 (Fig. 4a). The ACE2 receptor, initially identified as a cardiac regulator, is present on oral mucosa, AECII pneumocytes, intestinal, kidney, cardiac, smooth muscle and endothelial cells143–146. Transcriptomic profiling provides a comprehensive view of ACE2 and TMPRSS2 expression in cells of the human body109. These datasets are particularly helpful when choosing specific organoids for COVID-19 research (Fig. 4a).
Established cell lines and organoids.
Currently, hiPSC-derived patient-specific lung organoids recapitulate the pathophysiology of various lung diseases, such as surfactant deficiency147, cystic fibrosis106, Hermansky–Pudlak syndrome101,148, and respiratory syncytial and parainfluenza virus infection101,102. In another example, AECIIs, derived from a child with a lethal neonatal respiratory distress syndrome caused by a homozygous SFTPB mutation, mimicked aspects of this syndrome, including the deficiency in surfactant processing147. The abnormal processing could be restored in gene-corrected AECIIs from this patient147. So far, experiments with hiPSC-derived organoids have confirmed that lung epithelial cells are susceptible to CoV-2 infection, express CoV-2 host factors, and provoke an intrinsic epithelial inflammatory response116,117,126. However, these hiPSC-derived lung organoids have not yet been used to unveil the differences associated with inherent variations to infection and immune activation in large COVID-19 patient cohorts116,117,126. Organoids derived from established hiPSC lines and cell banks from individual donors or patient biopsies provide a valuable and convenient tool to assess risk factors and therapeutic outcomes in the most vulnerable populations and implement targeted prevention at low cost. Currently, lung, cardiac, intestinal, liver, kidney, and capillary organoids are immediately available to serve these purposes.
Complex organoid assays.
One major remaining challenge in the application of organoids is the lack of complexity and extensive intercellular interactions compared to the in vivo situation. Complex lung airway organoids can be derived by integrating human adult primary bronchial epithelial cells and lung fibroblasts with lung microvascular endothelial cells to study disease-relevant cell-cell interactions149. However, hPSC-derived alveolospheres typically have few cell types (for instance AECIs and AECIIs) and do not include adjacent capillaries composed of endothelial cells (Fig. 3b).
The endothelium is connected with alveolar cells by the basement membrane and acts as alveolar niche. Mouse alveolospheres form more easily in organoid co-cultures with lung endothelial cells150. Successful alveolar repair requires restoration of the spatial relationship between alveolar cells and the endothelium. Endothelial cells can receive reparative signals from AECIs after acute injury151 and enhance alveologenesis through diverse signalling factors such as endothelial-derived angiocrine factors and platelet-derived SDF-1152,153. Endothelial cells also secrete the vascular endothelial growth factor, monocyte chemoattractant protein–1, IL-6, and IL-8, all of which aggravate the cytokine storm35. Reduced vascular endothelial cadherin expression on endothelial cells154 increases vascular permeability and pulmonary dysfunction in ARDS. For these reasons, vascularised lung organoids should provide valuable insights into CoV-2-mediated lung damage and repair.
So far, the generation of vascularised lung organoids has not been achieved, although vascularisation occurs in lung organoids grafted into animals to promote cell differentiation101,129. For the liver, vascularised organoids termed liver buds have been reported, which consist of hiPSC-derived hepatic endoderm, endothelial cells, and bone marrow stromal cells155. Similarly, the integration of vasculature into cerebral organoids seems to accelerate functional maturation of neurons156. It seems likely that vascularised lung organoids might exhibit comparable properties. At any rate, improved protocols for complex organoids with integrated capillary systems are expected to more accurately recapitulate the alveolar CoV-2 response (Fig. 4b2).
Another major challenge is the lack of host factors, immune cells and inflammatory responses in existing organoid assays66,157. Drug and antibody effects observed in these assays may not reflect genuine responses of COVID-19 patients, who often have additional medical conditions such as cardiovascular disorders or diabetes that alter cellular behaviour158. Ideally, organoids should include multiple cell types to closely mimic in vivo environments and the complexity of hyper-inflammatory complications in patients (Fig. 4b4). We previously proposed a concept that encompasses the multi-dimensionality of organoids6. In addition to 3D structures, we propose that 4D organoids emphasize the developmental time scale in an organoid culture. By comparison, 5D organoids would further integrate extrinsic factors to simulate host environmental, immunological, and inflammatory signals, which are absent from current organoid cultures6. Highly sensitive ELISA-based analysis of the cell culture medium is essential for monitoring cytokine storm inhibition in these complex organoids.
In the future, organoids could also have a role in vaccine development. Traditional vaccine development is a long-lasting process. Exploratory efforts on vaccine design, preclinical evaluation in animal models and clinical phase I, II, and III trials can take 15 years or more159. Although organoids may help identify potential molecular targets for vaccine design, they cannot be directly used to derive a vaccine due to the absence of the host immune system. However, organoid assays may provide valuable information about intermolecular interactions between viral proteins and host receptors and determine the efficacy of neutralising antibodies from vaccinated individuals (Fig. 2). In this way, organoid platforms might accelerate vaccine development at all stages that involve exploratory work, preclinical evaluation, and the assessment of efficacy in clinical trials. Encouragingly, the generation of immune cell organoids (for instance, T-cell and B-cell specific organoids) is feasible from hiPSCs160. The presence of T cells, B cells, and macrophages in organoid systems might well improve vaccine assessment in the future (Fig. 4b4).
Importantly, the integration of multiple cell types is limited by developmental constraints for a specific organ in a defined niche environment. As with all other organoid systems, a thorough understanding of the underlying developmental biology will be required for further progress in the creation of complex organoids.
Concluding Remarks
The inherent physiological variability of human populations poses a major challenge for the assessment of individual susceptibility and therapeutic outcomes. The versatility of hPSC-based and ASC-based organoids makes them a useful platform to compensate for the shortcomings of current assays. Multi-tissue isogenic organoids from individual donors and patients enable robust molecular assessment of the vulnerability of individual patients and possibly predict therapeutic responses in severe COVID-19 cases. We envision that the combination of current assays with complex organoids will continue to improve COVID-19 research and treatment, and provide valuable lessons for the study of other viral diseases as well.
ACKNOWLEDGEMENTS
This work was supported by the Intramural Research Program of the National Institutes of Health (NIH) at the National Institute of Neurological Disorders and Stroke (K.G.C., K.P.). Dr. Jason Spence was supported by the National Heart, Lung, and Blood Institute of the NIH (Grant number R01HL119215).
Footnotes
COMPETING INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.WHO Coronavirus (COVID-19) Dashboard, <https://covid19.who.int/> (2021).
- 2.Yan Y et al. The First 75 Days of Novel Coronavirus (SARS-CoV-2) Outbreak: Recent Advances, Prevention, and Treatment. Int J Environ Res Public Health 17, doi: 10.3390/ijerph17072323 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.COVID-19 Clinical Trials, <https://covid19.nih.gov/treatments-and-vaccines/clinical-trials> (2021).
- 4.Hu J, Li C, Wang S, Li T & Zhang H Genetic variants are identified to increase risk of COVID-19 related mortality from UK Biobank data. medRxiv, doi: 10.1101/2020.11.05.20226761 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pairo-Castineira E et al. Genetic mechanisms of critical illness in COVID-19. Nature 591, 92–98, doi: 10.1038/s41586-020-03065-y (2021). [DOI] [PubMed] [Google Scholar]
- 6.Chen KG et al. Pluripotent Stem Cell Platforms for Drug Discovery. Trends Mol Med 24, 805–820, doi: 10.1016/j.molmed.2018.06.009 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clevers H Modeling Development and Disease with Organoids. Cell 165, 1586–1597, doi: 10.1016/j.cell.2016.05.082 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Fatehullah A, Tan SH & Barker N Organoids as an in vitro model of human development and disease. Nat Cell Biol 18, 246–254, doi: 10.1038/ncb3312 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Lancaster MA & Knoblich JA Organogenesis in a dish: modeling development and disease using organoid technologies. Science 345, 1247125, doi: 10.1126/science.1247125 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Han Y et al. Identification of SARS-CoV-2 inhibitors using lung and colonic organoids. Nature 589, 270–275, doi: 10.1038/s41586-020-2901-9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Katsura H et al. Human Lung Stem Cell-Based Alveolospheres Provide Insights into SARS-CoV-2-Mediated Interferon Responses and Pneumocyte Dysfunction. Cell Stem Cell 27, 890–904 e898, doi: 10.1016/j.stem.2020.10.005 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacob F et al. Human Pluripotent Stem Cell-Derived Neural Cells and Brain Organoids Reveal SARS-CoV-2 Neurotropism Predominates in Choroid Plexus Epithelium. Cell Stem Cell 27, 937–950 e939, doi: 10.1016/j.stem.2020.09.016 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lamers MM et al. SARS-CoV-2 productively infects human gut enterocytes. Science 369, 50–54, doi: 10.1126/science.abc1669 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Monteil V et al. Inhibition of SARS-CoV-2 Infections in Engineered Human Tissues Using Clinical-Grade Soluble Human ACE2. Cell 181, 905–913 e907, doi: 10.1016/j.cell.2020.04.004 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pellegrini L et al. SARS-CoV-2 Infects the Brain Choroid Plexus and Disrupts the Blood-CSF Barrier in Human Brain Organoids. Cell Stem Cell 27, 951–961 e955, doi: 10.1016/j.stem.2020.10.001 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Purkayastha A et al. Direct Exposure to SARS-CoV-2 and Cigarette Smoke Increases Infection Severity and Alters the Stem Cell-Derived Airway Repair Response. Cell Stem Cell 27, 869–875 e864, doi: 10.1016/j.stem.2020.11.010 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Samuel RM et al. Androgen Signaling Regulates SARS-CoV-2 Receptor Levels and Is Associated with Severe COVID-19 Symptoms in Men. Cell Stem Cell 27, 876–889 e812, doi: 10.1016/j.stem.2020.11.009 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang L et al. A Human Pluripotent Stem Cell-based Platform to Study SARS-CoV-2 Tropism and Model Virus Infection in Human Cells and Organoids. Cell Stem Cell 27, 125–136 e127, doi: 10.1016/j.stem.2020.06.015 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao B et al. Recapitulation of SARS-CoV-2 infection and cholangiocyte damage with human liver ductal organoids. Protein Cell 11, 771–775, doi: 10.1007/s13238-020-00718-6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hao P, Zhong W, Song S, Fan S & Li X Is SARS-CoV-2 originated from laboratory? A rebuttal to the claim of formation via laboratory recombination. Emerg Microbes Infect 9, 545–547, doi: 10.1080/22221751.2020.1738279 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu SL, Saif LJ, Weiss SR & Su L No credible evidence supporting claims of the laboratory engineering of SARS-CoV-2. Emerg Microbes Infect 9, 505–507, doi: 10.1080/22221751.2020.1733440 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andersen KG, Rambaut A, Lipkin WI, Holmes EC & Garry RF The proximal origin of SARS-CoV-2. Nat Med 26, 450–452, doi: 10.1038/s41591-020-0820-9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang YZ & Holmes EC A Genomic Perspective on the Origin and Emergence of SARS-CoV-2. Cell 181, 223–227, doi: 10.1016/j.cell.2020.03.035 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen Z et al. Genomic Diversity of Severe Acute Respiratory Syndrome-Coronavirus 2 in Patients With Coronavirus Disease 2019. Clin Infect Dis 71, 713–720, doi: 10.1093/cid/ciaa203 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wen F et al. Identification of the hyper-variable genomic hotspot for the novel coronavirus SARS-CoV-2. J Infect 80, 671–693, doi: 10.1016/j.jinf.2020.02.027 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wertheim JO A Glimpse Into the Origins of Genetic Diversity in the Severe Acute Respiratory Syndrome Coronavirus 2. Clin Infect Dis 71, 721–722, doi: 10.1093/cid/ciaa213 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Doremalen N et al. Aerosol and Surface Stability of SARS-CoV-2 as Compared with SARS-CoV-1. N Engl J Med 382, 1564–1567, doi: 10.1056/NEJMc2004973 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Srinivasan S et al. Structural Genomics of SARS-CoV-2 Indicates Evolutionary Conserved Functional Regions of Viral Proteins. Viruses 12, doi: 10.3390/v12040360 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang C et al. The establishment of reference sequence for SARS-CoV-2 and variation analysis. J Med Virol 92, 667–674, doi: 10.1002/jmv.25762 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cui J, Li F & Shi ZL Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol 17, 181–192, doi: 10.1038/s41579-018-0118-9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.V’Kovski P, Kratzel A, Steiner S, Stalder H & Thiel V Coronavirus biology and replication: implications for SARS-CoV-2. Nat Rev Microbiol 19, 155–170, doi: 10.1038/s41579-020-00468-6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu B, Guo H, Zhou P & Shi ZL Characteristics of SARS-CoV-2 and COVID-19. Nat Rev Microbiol 19, 141–154, doi: 10.1038/s41579-020-00459-7 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zou L et al. SARS-CoV-2 Viral Load in Upper Respiratory Specimens of Infected Patients. N Engl J Med 382, 1177–1179, doi: 10.1056/NEJMc2001737 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li G et al. Coronavirus infections and immune responses. J Med Virol 92, 424–432, doi: 10.1002/jmv.25685 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moore JB & June CH Cytokine release syndrome in severe COVID-19. Science 368, 473–474, doi: 10.1126/science.abb8925 (2020). [DOI] [PubMed] [Google Scholar]
- 36.Wang W et al. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA 323, 1843–1844, doi: 10.1001/jama.2020.3786 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pan Y, Zhang D, Yang P, Poon LLM & Wang Q Viral load of SARS-CoV-2 in clinical samples. Lancet Infect Dis 20, 411–412, doi: 10.1016/S1473-3099(20)30113-4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fajgenbaum DC & June CH Cytokine Storm. N Engl J Med 383, 2255–2273, doi: 10.1056/NEJMra2026131 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berlin DA, Gulick RM & Martinez FJ Severe Covid-19. N Engl J Med 383, 2451–2460, doi: 10.1056/NEJMcp2009575 (2020). [DOI] [PubMed] [Google Scholar]
- 40.Chen J, Wu H, Yu Y & Tang N Pulmonary alveolar regeneration in adult COVID-19 patients. Cell Res 30, 708–710, doi: 10.1038/s41422-020-0369-7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoffmann M et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 181, 271–280 e278, doi: 10.1016/j.cell.2020.02.052 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu J et al. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discov 6, 16, doi: 10.1038/s41421-020-0156-0 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mehra MR, Desai SS, Ruschitzka F & Patel AN RETRACTED: Hydroxychloroquine or chloroquine with or without a macrolide for treatment of COVID-19: a multinational registry analysis. Lancet, doi: 10.1016/S0140-6736(20)31180-6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 44.Reina J Remdesivir, the antiviral hope against SARS-CoV-2. Rev Esp Quimioter 33, 176–179, doi:10.37201/req/028.202010.37201/req/028.202010.37201/req/098.202010.37201/req/098.2020 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Amanat F & Krammer F SARS-CoV-2 Vaccines: Status Report. Immunity 52, 583–589, doi: 10.1016/j.immuni.2020.03.007 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Caly L, Druce JD, Catton MG, Jans DA & Wagstaff KM The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antiviral Res 178, 104787, doi: 10.1016/j.antiviral.2020.104787 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Horby P et al. Dexamethasone in Hospitalized Patients with Covid-19. N Engl J Med 384, 693–704, doi: 10.1056/NEJMoa2021436 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pan H et al. Repurposed Antiviral Drugs for Covid-19 - Interim WHO Solidarity Trial Results. N Engl J Med 384, 497–511, doi: 10.1056/NEJMoa2023184 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.ter Meulen J et al. Human monoclonal antibody combination against SARS coronavirus: synergy and coverage of escape mutants. PLoS Med 3, e237, doi: 10.1371/journal.pmed.0030237 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tian X et al. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg Microbes Infect 9, 382–385, doi: 10.1080/22221751.2020.1729069 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yuan M et al. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science 368, 630–633, doi: 10.1126/science.abb7269 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Inoue Y et al. Clathrin-dependent entry of severe acute respiratory syndrome coronavirus into target cells expressing ACE2 with the cytoplasmic tail deleted. J Virol 81, 8722–8729, doi: 10.1128/JVI.00253-07 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nomura R et al. Human coronavirus 229E binds to CD13 in rafts and enters the cell through caveolae. J Virol 78, 8701–8708, doi: 10.1128/JVI.78.16.8701-8708.2004 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Van Hamme E, Dewerchin HL, Cornelissen E, Verhasselt B & Nauwynck HJ Clathrin- and caveolae-independent entry of feline infectious peritonitis virus in monocytes depends on dynamin. J Gen Virol 89, 2147–2156, doi: 10.1099/vir.0.2008/001602-0 (2008). [DOI] [PubMed] [Google Scholar]
- 55.FDA Approves First Treatment for COVID-19, <https://www.fda.gov/news-events/press-announcements/fda-approves-first-treatment-covid-19> (2020).
- 56.Ryabkova VA, Churilov LP & Shoenfeld Y Influenza infection, SARS, MERS and COVID-19: Cytokine storm - The common denominator and the lessons to be learned. Clin Immunol 223, 108652, doi: 10.1016/j.clim.2020.108652 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ye Q, Wang B & Mao J The pathogenesis and treatment of the `Cytokine Storm’ in COVID-19. J Infect 80, 607–613, doi: 10.1016/j.jinf.2020.03.037 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Efficacy of Tocilizumab on Patients With COVID-19, <https://www.clinicaltrials.gov/ct2/show/NCT04356937> (2020).
- 59.Guaraldi G et al. Tocilizumab in patients with severe COVID-19: a retrospective cohort study. Lancet Rheumatol 2, e474–e484, doi: 10.1016/S2665-9913(20)30173-9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stone JH et al. Efficacy of Tocilizumab in Patients Hospitalized with Covid-19. N Engl J Med 383, 2333–2344, doi: 10.1056/NEJMoa2028836 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sterne JAC et al. Association Between Administration of Systemic Corticosteroids and Mortality Among Critically Ill Patients With COVID-19: A Meta-analysis. JAMA 324, 1330–1341, doi: 10.1001/jama.2020.17023 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pan X et al. Asymptomatic cases in a family cluster with SARS-CoV-2 infection. Lancet Infect Dis 20, 410–411, doi: 10.1016/S1473-3099(20)30114-6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lai CC et al. Asymptomatic carrier state, acute respiratory disease, and pneumonia due to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): Facts and myths. J Microbiol Immunol Infect 53, 404–412, doi: 10.1016/j.jmii.2020.02.012 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Park SE Epidemiology, virology, and clinical features of severe acute respiratory syndrome -coronavirus-2 (SARS-CoV-2; Coronavirus Disease-19). Clin Exp Pediatr 63, 119–124, doi: 10.3345/cep.2020.00493 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ling Z et al. Asymptomatic SARS-CoV-2 infected patients with persistent negative CT findings. Eur J Radiol 126, 108956, doi: 10.1016/j.ejrad.2020.108956 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rokni M, Ghasemi V & Tavakoli Z Immune responses and pathogenesis of SARS-CoV-2 during an outbreak in Iran: Comparison with SARS and MERS. Rev Med Virol 30, e2107, doi: 10.1002/rmv.2107 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cao Q, Chen YC, Chen CL & Chiu CH SARS-CoV-2 infection in children: Transmission dynamics and clinical characteristics. J Formos Med Assoc 119, 670–673, doi: 10.1016/j.jfma.2020.02.009 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chidini G, Villa C, Calderini E, Marchisio P & De Luca D SARS-CoV-2 Infection in a Pediatric Department in Milan: A Logistic Rather Than a Clinical Emergency. Pediatr Infect Dis J 39, e79–e80, doi: 10.1097/INF.0000000000002687 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lu X et al. SARS-CoV-2 Infection in Children. N Engl J Med 382, 1663–1665, doi: 10.1056/NEJMc2005073 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wu Z & McGoogan JM Characteristics of and Important Lessons From the Coronavirus Disease 2019 (COVID-19) Outbreak in China: Summary of a Report of 72314 Cases From the Chinese Center for Disease Control and Prevention. JAMA 323, 1239–1242, doi: 10.1001/jama.2020.2648 (2020). [DOI] [PubMed] [Google Scholar]
- 71.Dudley JP & Lee NT Disparities in Age-specific Morbidity and Mortality From SARS-CoV-2 in China and the Republic of Korea. Clin Infect Dis 71, 863–865, doi: 10.1093/cid/ciaa354 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Conti P & Younes A Coronavirus COV-19/SARS-CoV-2 affects women less than men: clinical response to viral infection. J Biol Regul Homeost Agents 34, 339–343, doi: 10.23812/Editorial-Conti-3 (2020). [DOI] [PubMed] [Google Scholar]
- 73.Shah M, Sachdeva M & Dodiuk-Gad RP COVID-19 and racial disparities. J Am Acad Dermatol 83, e35, doi: 10.1016/j.jaad.2020.04.046 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McCoy J et al. Racial variations in COVID-19 deaths may be due to androgen receptor genetic variants associated with prostate cancer and androgenetic alopecia. Are anti-androgens a potential treatment for COVID-19? J Cosmet Dermatol 19, 1542–1543, doi: 10.1111/jocd.13455 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Laurencin CT & McClinton A The COVID-19 Pandemic: a Call to Action to Identify and Address Racial and Ethnic Disparities. J Racial Ethn Health Disparities, doi: 10.1007/s40615-020-00756-0 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brake SJ et al. Smoking Upregulates Angiotensin-Converting Enzyme-2 Receptor: A Potential Adhesion Site for Novel Coronavirus SARS-CoV-2 (Covid-19). J Clin Med 9, doi: 10.3390/jcm9030841 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cai G, Bosse Y, Xiao F, Kheradmand F & Amos CI Tobacco Smoking Increases the Lung Gene Expression of ACE2, the Receptor of SARS-CoV-2. Am J Respir Crit Care Med 201, 1557–1559, doi: 10.1164/rccm.202003-0693LE (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lukassen S et al. SARS-CoV-2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells. EMBO J 39, e105114, doi: 10.15252/embj.20105114 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Afzal A Molecular diagnostic technologies for COVID-19: Limitations and challenges. J Adv Res 26, 149–159, doi: 10.1016/j.jare.2020.08.002 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kevadiya BD et al. Diagnostics for SARS-CoV-2 infections. Nat Mater 20, 593–605, doi: 10.1038/s41563-020-00906-z (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu W et al. Evaluation of Nucleocapsid and Spike Protein-Based Enzyme-Linked Immunosorbent Assays for Detecting Antibodies against SARS-CoV-2. J Clin Microbiol 58, doi: 10.1128/JCM.00461-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhao J et al. Antibody Responses to SARS-CoV-2 in Patients With Novel Coronavirus Disease 2019. Clin Infect Dis 71, 2027–2034, doi: 10.1093/cid/ciaa344 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li Z et al. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J Med Virol 92, 1518–1524, doi: 10.1002/jmv.25727 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhao G et al. A safe and convenient pseudovirus-based inhibition assay to detect neutralizing antibodies and screen for viral entry inhibitors against the novel human coronavirus MERS-CoV. Virol J 10, 266, doi: 10.1186/1743-422X-10-266 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li Q, Liu Q, Huang W, Li X & Wang Y Current status on the development of pseudoviruses for enveloped viruses. Rev Med Virol 28, doi: 10.1002/rmv.1963 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Letko M, Marzi A & Munster V Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol 5, 562–569, doi: 10.1038/s41564-020-0688-y (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Matsuyama S et al. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc Natl Acad Sci U S A 117, 7001–7003, doi: 10.1073/pnas.2002589117 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cao X et al. Invited review: human air-liquid-interface organotypic airway tissue models derived from primary tracheobronchial epithelial cells-overview and perspectives. In Vitro Cell Dev Biol Anim 57, 104–132, doi: 10.1007/s11626-020-00517-7 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schoch KG et al. A subset of mouse tracheal epithelial basal cells generates large colonies in vitro. Am J Physiol Lung Cell Mol Physiol 286, L631–642, doi: 10.1152/ajplung.00112.2003 (2004). [DOI] [PubMed] [Google Scholar]
- 90.Rock JR et al. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc Natl Acad Sci U S A 106, 12771–12775, doi: 10.1073/pnas.0906850106 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ettayebi K et al. Replication of human noroviruses in stem cell-derived human enteroids. Science 353, 1387–1393, doi: 10.1126/science.aaf5211 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sasai Y Next-generation regenerative medicine: organogenesis from stem cells in 3D culture. Cell Stem Cell 12, 520–530, doi: 10.1016/j.stem.2013.04.009 (2013). [DOI] [PubMed] [Google Scholar]
- 93.Wong AP et al. Directed differentiation of human pluripotent stem cells into mature airway epithelia expressing functional CFTR protein. Nat Biotechnol 30, 876–882, doi: 10.1038/nbt.2328 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sachs N et al. Long-term expanding human airway organoids for disease modeling. EMBO J 38, doi: 10.15252/embj.2018100300 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chen KG, Zhong P, Zheng W & Beekman JM Pharmacological analysis of CFTR variants of cystic fibrosis using stem cell-derived organoids. Drug Discov Today 24, 2126–2138, doi: 10.1016/j.drudis.2019.05.029 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Basil MC et al. The Cellular and Physiological Basis for Lung Repair and Regeneration: Past, Present, and Future. Cell Stem Cell 26, 482–502, doi: 10.1016/j.stem.2020.03.009 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hogan BL et al. Repair and regeneration of the respiratory system: complexity, plasticity, and mechanisms of lung stem cell function. Cell Stem Cell 15, 123–138, doi: 10.1016/j.stem.2014.07.012 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nadkarni RR, Abed S & Draper JS Organoids as a model system for studying human lung development and disease. Biochem Biophys Res Commun 473, 675–682, doi: 10.1016/j.bbrc.2015.12.091 (2016). [DOI] [PubMed] [Google Scholar]
- 99.Gkatzis K, Taghizadeh S, Huh D, Stainier DYR & Bellusci S Use of three-dimensional organoids and lung-on-a-chip methods to study lung development, regeneration and disease. Eur Respir J 52, doi: 10.1183/13993003.00876-2018 (2018). [DOI] [PubMed] [Google Scholar]
- 100.Huang SX et al. Efficient generation of lung and airway epithelial cells from human pluripotent stem cells. Nat Biotechnol 32, 84–91, doi: 10.1038/nbt.2754 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chen YW et al. A three-dimensional model of human lung development and disease from pluripotent stem cells. Nat Cell Biol 19, 542–549, doi: 10.1038/ncb3510 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Porotto M et al. Authentic Modeling of Human Respiratory Virus Infection in Human Pluripotent Stem Cell-Derived Lung Organoids. mBio 10, doi: 10.1128/mBio.00723-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yamamoto Y et al. Long-term expansion of alveolar stem cells derived from human iPS cells in organoids. Nat Methods 14, 1097–1106, doi: 10.1038/nmeth.4448 (2017). [DOI] [PubMed] [Google Scholar]
- 104.Strikoudis A et al. Modeling of Fibrotic Lung Disease Using 3D Organoids Derived from Human Pluripotent Stem Cells. Cell Rep 27, 3709–3723 e3705, doi: 10.1016/j.celrep.2019.05.077 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Dye BR et al. In vitro generation of human pluripotent stem cell derived lung organoids. Elife 4, doi: 10.7554/eLife.05098 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.McCauley KB et al. Efficient Derivation of Functional Human Airway Epithelium from Pluripotent Stem Cells via Temporal Regulation of Wnt Signaling. Cell Stem Cell 20, 844–857 e846, doi: 10.1016/j.stem.2017.03.001 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Miller AJ et al. Generation of lung organoids from human pluripotent stem cells in vitro. Nat Protoc 14, 518–540, doi: 10.1038/s41596-018-0104-8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Barkauskas CE et al. Lung organoids: current uses and future promise. Development 144, 986–997, doi: 10.1242/dev.140103 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sungnak W et al. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat Med 26, 681–687, doi: 10.1038/s41591-020-0868-6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Barkauskas CE et al. Type 2 alveolar cells are stem cells in adult lung. J Clin Invest 123, 3025–3036, doi: 10.1172/JCI68782 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ziegler CGK et al. SARS-CoV-2 Receptor ACE2 Is an Interferon-Stimulated Gene in Human Airway Epithelial Cells and Is Detected in Specific Cell Subsets across Tissues. Cell 181, 1016–1035 e1019, doi: 10.1016/j.cell.2020.04.035 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Youk J et al. Three-Dimensional Human Alveolar Stem Cell Culture Models Reveal Infection Response to SARS-CoV-2. Cell Stem Cell 27, 905–919 e910, doi: 10.1016/j.stem.2020.10.004 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Schaefer IM et al. In situ detection of SARS-CoV-2 in lungs and airways of patients with COVID-19. Mod Pathol 33, 2104–2114, doi: 10.1038/s41379-020-0595-z (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Borczuk AC et al. COVID-19 pulmonary pathology: a multi-institutional autopsy cohort from Italy and New York City. Mod Pathol 33, 2156–2168, doi: 10.1038/s41379-020-00661-1 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mirabelli C et al. Morphological Cell Profiling of SARS-CoV-2 Infection Identifies Drug Repurposing Candidates for COVID-19. bioRxiv, doi: 10.1101/2020.05.27.117184 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Abo KM et al. Human iPSC-derived alveolar and airway epithelial cells can be cultured at air-liquid interface and express SARS-CoV-2 host factors. bioRxiv, doi: 10.1101/2020.06.03.132639 (2020). [DOI] [Google Scholar]
- 117.Huang J et al. SARS-CoV-2 Infection of Pluripotent Stem Cell-Derived Human Lung Alveolar Type 2 Cells Elicits a Rapid Epithelial-Intrinsic Inflammatory Response. Cell Stem Cell 27, 962–973 e967, doi: 10.1016/j.stem.2020.09.013 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Duan F et al. Modeling COVID-19 with Human Pluripotent Stem Cell-Derived Cells Reveals Synergistic Effects of Anti-inflammatory Macrophages with ACE2 Inhibition Against SARS-CoV-2. Res Sq, doi: 10.21203/rs.3.rs-62758/v1 (2020). [DOI] [Google Scholar]
- 119.Lee AS, Tang C, Rao MS, Weissman IL & Wu JC Tumorigenicity as a clinical hurdle for pluripotent stem cell therapies. Nat Med 19, 998–1004, doi: 10.1038/nm.3267 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gore A et al. Somatic coding mutations in human induced pluripotent stem cells. Nature 471, 63–67, doi: 10.1038/nature09805 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Merkle FT et al. Human pluripotent stem cells recurrently acquire and expand dominant negative P53 mutations. Nature 545, 229–233, doi: 10.1038/nature22312 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Baker DE et al. Adaptation to culture of human embryonic stem cells and oncogenesis in vivo. Nat Biotechnol 25, 207–215, doi: 10.1038/nbt1285 (2007). [DOI] [PubMed] [Google Scholar]
- 123.Mayshar Y et al. Identification and classification of chromosomal aberrations in human induced pluripotent stem cells. Cell Stem Cell 7, 521–531, doi: 10.1016/j.stem.2010.07.017 (2010). [DOI] [PubMed] [Google Scholar]
- 124.Rodrigues Toste de Carvalho AL et al. The in vitro multilineage differentiation and maturation of lung and airway cells from human pluripotent stem cell-derived lung progenitors in 3D. Nat Protoc 16, 1802–1829, doi: 10.1038/s41596-020-00476-z (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Studer L, Vera E & Cornacchia D Programming and Reprogramming Cellular Age in the Era of Induced Pluripotency. Cell Stem Cell 16, 591–600, doi: 10.1016/j.stem.2015.05.004 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hawkins FJ et al. Derivation of Airway Basal Stem Cells from Human Pluripotent Stem Cells. Cell Stem Cell 28, 79–95 e78, doi: 10.1016/j.stem.2020.09.017 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mulay A et al. SARS-CoV-2 infection of primary human lung epithelium for COVID-19 modeling and drug discovery. Cell Rep 35, 109055. doi: 10.1016/j.celrep.2021.109055 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Takebe T & Wells JM Organoids by design. Science 364, 956–959, doi: 10.1126/science.aaw7567 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Dye BR et al. A bioengineered niche promotes in vivo engraftment and maturation of pluripotent stem cell derived human lung organoids. Elife 5, doi: 10.7554/eLife.19732 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.van Velthoven CTJ & Rando TA Stem Cell Quiescence: Dynamism, Restraint, and Cellular Idling. Cell Stem Cell 24, 213–225, doi: 10.1016/j.stem.2019.01.001 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Graf T & Stadtfeld M Heterogeneity of embryonic and adult stem cells. Cell Stem Cell 3, 480–483, doi: 10.1016/j.stem.2008.10.007 (2008). [DOI] [PubMed] [Google Scholar]
- 132.Wagers AJ & Weissman IL Plasticity of adult stem cells. Cell 116, 639–648, doi: 10.1016/s0092-8674(04)00208-9 (2004). [DOI] [PubMed] [Google Scholar]
- 133.Nabhan AN, Brownfield DG, Harbury PB, Krasnow MA & Desai TJ Single-cell Wnt signaling niches maintain stemness of alveolar type 2 cells. Science 359, 1118–1123, doi: 10.1126/science.aam6603 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Choi J et al. Inflammatory Signals Induce AT2 Cell-Derived Damage-Associated Transient Progenitors that Mediate Alveolar Regeneration. Cell Stem Cell 27, 366–382 e367, doi: 10.1016/j.stem.2020.06.020 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zacharias WJ et al. Regeneration of the lung alveolus by an evolutionarily conserved epithelial progenitor. Nature 555, 251–255, doi: 10.1038/nature25786 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Salahudeen AA et al. Progenitor identification and SARS-CoV-2 infection in human distal lung organoids. Nature 588, 670–675, doi: 10.1038/s41586-020-3014-1 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Tindle C et al. Adult Stem Cell-derived Complete Lung Organoid Models Emulate Lung Disease in COVID-19. bioRxiv, doi: 10.1101/2020.10.17.344002 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.van der Vaart J & Clevers H Airway organoids as models of human disease. J Intern Med 289, 604–613, doi: 10.1111/joim.13075 (2021). [DOI] [PubMed] [Google Scholar]
- 139.Evangelou K et al. SARS-CoV-2 infects lung epithelial cells and induces senescence and an inflammatory response in patients with severe COVID-19. Preprint at bioRxiv doi: 10.1101/2021.01.02.424917 (2021). [DOI] [Google Scholar]
- 140.Hillel-Karniel C et al. Multi-lineage Lung Regeneration by Stem Cell Transplantation across Major Genetic Barriers. Cell Rep 30, 807–819 e804, doi: 10.1016/j.celrep.2019.12.058 (2020). [DOI] [PubMed] [Google Scholar]
- 141.Weiner AI et al. Mesenchyme-free expansion and transplantation of adult alveolar progenitor cells: steps toward cell-based regenerative therapies. NPJ Regen Med 4, 17, doi: 10.1038/s41536-019-0080-9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Dorrello NV et al. Functional vascularized lung grafts for lung bioengineering. Sci Adv 3, e1700521, doi: 10.1126/sciadv.1700521 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Boehm M & Nabel EG Angiotensin-converting enzyme 2--a new cardiac regulator. N Engl J Med 347, 1795–1797, doi: 10.1056/NEJMcibr022472 (2002). [DOI] [PubMed] [Google Scholar]
- 144.Zhang H, Penninger JM, Li Y, Zhong N & Slutsky AS Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med 46, 586–590, doi: 10.1007/s00134-020-05985-9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Abassi Z, Assady S, Khoury EE & Heyman SN Letter to the Editor: Angiotensin-converting enzyme 2: an ally or a Trojan horse? Implications to SARS-CoV-2-related cardiovascular complications. Am J Physiol Heart Circ Physiol 318, H1080–H1083, doi: 10.1152/ajpheart.00215.2020 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ferrario CM et al. Effect of angiotensin-converting enzyme inhibition and angiotensin II receptor blockers on cardiac angiotensin-converting enzyme 2. Circulation 111, 2605–2610, doi: 10.1161/CIRCULATIONAHA.104.510461 (2005). [DOI] [PubMed] [Google Scholar]
- 147.Jacob A et al. Differentiation of Human Pluripotent Stem Cells into Functional Lung Alveolar Epithelial Cells. Cell Stem Cell 21, 472–488 e410, doi: 10.1016/j.stem.2017.08.014 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Korogi Y et al. In Vitro Disease Modeling of Hermansky-Pudlak Syndrome Type 2 Using Human Induced Pluripotent Stem Cell-Derived Alveolar Organoids. Stem Cell Reports 12, 431–440, doi: 10.1016/j.stemcr.2019.01.014 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Tan Q, Choi KM, Sicard D & Tschumperlin DJ Human airway organoid engineering as a step toward lung regeneration and disease modeling. Biomaterials 113, 118–132, doi: 10.1016/j.biomaterials.2016.10.046 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Lee JH et al. Lung stem cell differentiation in mice directed by endothelial cells via a BMP4-NFATc1-thrombospondin-1 axis. Cell 156, 440–455, doi: 10.1016/j.cell.2013.12.039 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Niethamer TK et al. Defining the role of pulmonary endothelial cell heterogeneity in the response to acute lung injury. Elife 9, doi: 10.7554/eLife.53072 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ding BS et al. Endothelial-derived angiocrine signals induce and sustain regenerative lung alveolarization. Cell 147, 539–553, doi: 10.1016/j.cell.2011.10.003 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Rafii S et al. Platelet-derived SDF-1 primes the pulmonary capillary vascular niche to drive lung alveolar regeneration. Nat Cell Biol 17, 123–136, doi: 10.1038/ncb3096 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Tanaka T, Narazaki M & Kishimoto T Immunotherapeutic implications of IL-6 blockade for cytokine storm. Immunotherapy 8, 959–970, doi: 10.2217/imt-2016-0020 (2016). [DOI] [PubMed] [Google Scholar]
- 155.Takebe T et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature 499, 481–484, doi: 10.1038/nature12271 (2013). [DOI] [PubMed] [Google Scholar]
- 156.Cakir B et al. Engineering of human brain organoids with a functional vascular-like system. Nat Methods 16, 1169–1175, doi: 10.1038/s41592-019-0586-5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Lin L, Lu L, Cao W & Li T Hypothesis for potential pathogenesis of SARS-CoV-2 infection-a review of immune changes in patients with viral pneumonia. Emerg Microbes Infect 9, 727–732, doi: 10.1080/22221751.2020.1746199 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Fadini GP, Morieri ML, Longato E & Avogaro A Prevalence and impact of diabetes among people infected with SARS-CoV-2. J Endocrinol Invest 43, 867–869, doi: 10.1007/s40618-020-01236-2 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Krammer F SARS-CoV-2 vaccines in development. Nature 586, 516–527, doi: 10.1038/s41586-020-2798-3 (2020). [DOI] [PubMed] [Google Scholar]
- 160.Purwada A & Singh A Immuno-engineered organoids for regulating the kinetics of B-cell development and antibody production. Nat Protoc 12, 168–182, doi: 10.1038/nprot.2016.157 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Hoffmann M, Kleine-Weber H & Pohlmann S A Multibasic Cleavage Site in the Spike Protein of SARS-CoV-2 Is Essential for Infection of Human Lung Cells. Mol Cell 78, 779–784 e775, doi: 10.1016/j.molcel.2020.04.022 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Shang J et al. Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci U S A 117, 11727–11734, doi: 10.1073/pnas.2003138117 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Zepp JA et al. Distinct Mesenchymal Lineages and Niches Promote Epithelial Self-Renewal and Myofibrogenesis in the Lung. Cell 170, 1134–1148 e1110, doi: 10.1016/j.cell.2017.07.034 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Lee JH et al. Anatomically and Functionally Distinct Lung Mesenchymal Populations Marked by Lgr5 and Lgr6. Cell 170, 1149–1163 e1112, doi: 10.1016/j.cell.2017.07.028 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]


