SUMMARY
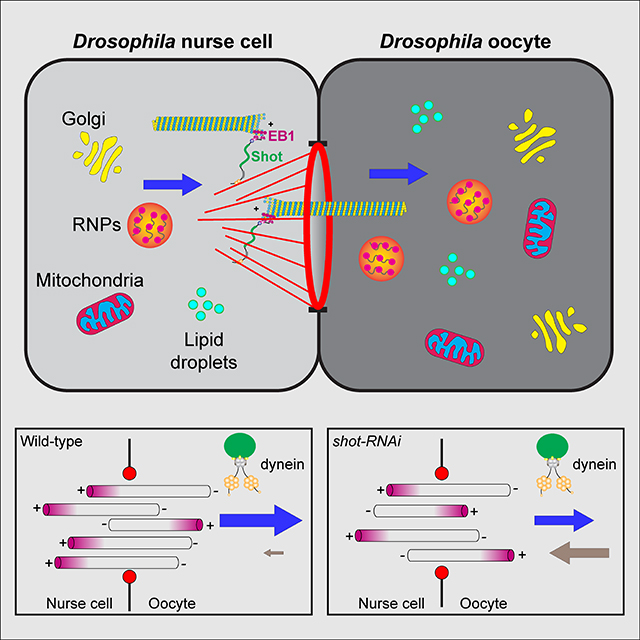
Growth of the Drosophila oocyte requires transport of cytoplasmic material from the interconnected sister cells (nurse cells), through ring canals, the cytoplasmic bridges that remained open after incomplete germ cell division. Given the open nature of the ring canals, it is unclear how the direction of transport through the ring canal is controlled. In this work we show that a single Drosophila spectraplakin Short stop (Shot) controls the direction of flow from nurse cells to the oocyte. Knockdown of shot changes the direction of transport through the ring canals from unidirectional (towards the oocyte) to bidirectional. After shot knockdown the oocyte stops growing resulting in a characteristic small oocyte phenotype. In agreement with this transport-directing function of Shot, we find that it is localized at the asymmetric actin baskets on the nurse cell side of the ring canals. In wild-type egg chambers microtubules localized in the ring canals have uniform polarity (minus-ends towards the oocyte), while in the absence of Shot these microtubules have mixed polarity. Together, we propose that Shot functions as a gatekeeper directing transport from nurse cells to the oocyte, via the organization of microtubule tracks to facilitate the transport driven by the minus-end directed microtubule motor cytoplasmic dynein.
Graphical Abstract
INTRODUCTION
Microtubules and actin filaments are two fundamental cytoskeletal components of all eukaryotic cells. They are essential for multiple key functions of a cell, such as cell division, cell migration, cargo transport, morphogenesis and compartmentation/polarization. Coordination of microtubules and actin filaments is vital for these various cellular functions. Yet a full understanding of microtubule-actin crosstalk is still lacking.
Spectraplakins are a family of large cytoskeletal linker proteins that are evolutionarily conserved across the animal kingdom. Spectraplakins are unique in their ability to associate with all three cytoskeletal networks: F-actin, microtubules and intermediate filaments. They all contain N-terminal calponin homology (CH) domains for actin binding (ABD), C-terminal EF motif, GAS2 domain and C-terminal tail containing plus-end tracking SxIP motifs (EGC) for microtubule interaction, bridged by a plakin domain and a long rod-like domain composed of spectrin repeats [1–5]. Short stop (Shot) is the single Drosophila spectraplakin, coordinating and moderating the interactions between F-actin and microtubules via the N-terminal ABD domain and C-terminal EGC domain, respectively (Figure 1A) [6, 7]. Drosophila Shot has been shown to be involved in the regulation of cytoskeletal network interaction in many cell types [8]. For instance, Shot controls microtubule organization and regulates filopodia formation in neurites and is thus essential for axon extension [6, 7, 9–11]. Furthermore, Shot plays a critical role in multiple cell shape changes and developmental morphogenesis events, such as tracheal tube fusion [12–14], epithelia cell-cell adhesion [15], foregut development [16], photoreceptor morphogenesis [17], fat body adipocytes microtubule organization [18], muscle myonuclear shape maintenance [19] and dorsal closure [20].
Figure 1. Shot is required for Drosophila oocyte growth.
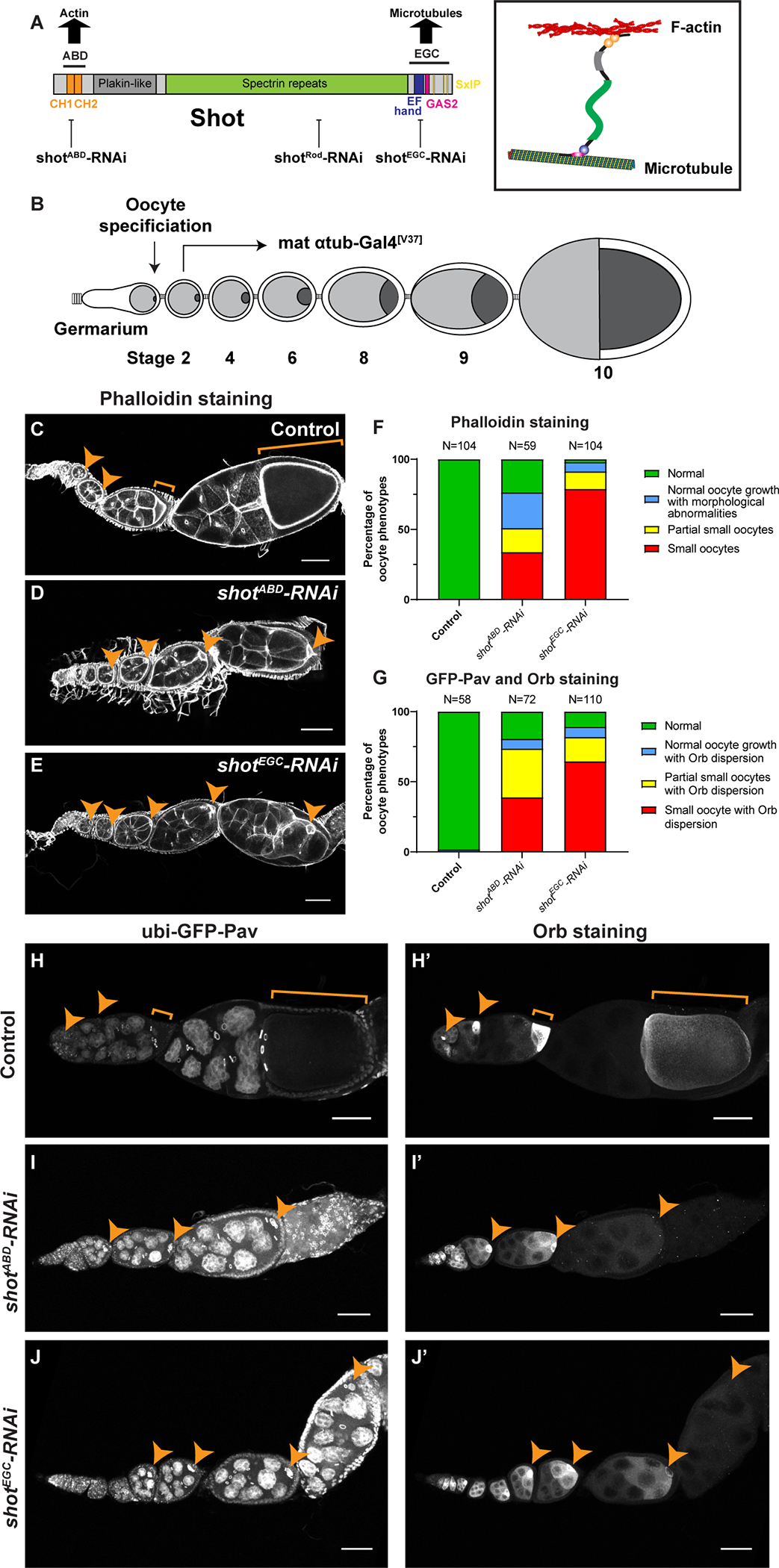
(A) A diagram of Shot domains and Shot crosslinking of microtubules and F-actin. Three independent shot-RNAi lines were used in this study, targeting ABD, Rod and EGC domains, respectively. shotRod-RNAi and shotEGC-RNAi target all 22 isoforms of shot mRNA; shotABD-RNAi does not target 8 isoforms of shot mRNA lacking CH1 domain (RC, RH, RP, RX, RY, RAB, RAC, RAD).
(B) A schematic illustration of Drosophila oogenesis in one ovariole and the shot-RNAi knockdown strategy to bypass the requirement of Shot in oocyte specification. Oocyte is shown in darker grey, while nurse cells are represented in lighter grey in the egg chambers. The mat αtub-Gal4[V37] line starts the expression in stage 2–3 egg chambers, after the completion of oocyte specification.
(C-E) Representative images of Rhodamine-conjugated phalloidin staining in control (mat αtub-Gal4[V37]/+), shotABD-RNAi (mat αtub-Gal4[V37]/UAS-shotABD-RNAi), and shotEGC-RNAi (mat αtub-Gal4[V37]/UAS-shotEGC-RNAi).
(F) Summary of phalloidin staining phenotypes in control, shotABD-RNAi and shotEGC-RNAi. (G) Summary of GFP-Pav labeling and Orb staining phenotypes in control, shotABD-RNAi and shotEGC-RNAi.
(H-J’) Representative images of GFP-Pav labeling (H-J) and Orb staining (H’-J’) in control (ubi-GFP-Pav/+; mat αtub-Gal4[V37]/+), shotABD-RNAi (ubi-GFP-Pav/+; mat αtub-Gal4[V37]/UAS-shotABD-RNAi), and shotEGC-RNAi (ubi-GFP-Pav/+; mat αtub-Gal4[V37]/UAS-shotEGC-RNAi).
Oocytes are highlighted by orange arrowheads or brackets. Scale bars, 50 μm. See also Figure S1 and Videos S1–S2.
The Drosophila oocyte is the largest cell in a fruit fly. An oocyte is first specified among 16-interconnected cyst cells with a diameter of several micrometers, and grows to a full-size of several hundred micrometers, increasing its size by more than a hundred thousand times to prepare for future embryogenesis [21]. Remarkably, the Drosophila oocyte is mostly transcriptionally silent throughout oogenesis [22], and its drastic growth is primarily dependent on the acquisition of organelles, mRNAs, and proteins from the interconnected sister nurse cells, through ring canals, the intercellular cytoplasmic channels remained after incomplete cytokinesis [23]. Therefore, it is critical to understand what controls the direction of cytoplasmic transport from the nurse cells to the oocyte to support the oocyte growth. Given that microtubules and actin are both present at the nurse cell-oocyte ring canals [24–26], Shot, the microtubule-actin scrosslinker, appears to be a promising candidate that could regulate cytoplasmic transfer to the oocyte.
Previous studies have shown that Shot is essential for Drosophila oogenesis. At early stages, Shot is required for oocyte specification in 16-cell cysts via association of microtubules with fusome [27], a membranous structure in interconnected germline cysts that contains several actin-related cytoskeletal proteins, such as adducin-like Hts and α-spectrin [28]. In mid-oogenesis, Shot links minus-ends of microtubules to the anterior and lateral actin cortex via a minus-end binding protein Patronin, and therefore is essential for the anterior-posterior microtubule gradient formation within the oocyte [29]. This Shot-dependent microtubule gradient is required for oocyte nucleus translocation and axis determination for future embryos [30, 31]. However, little is known about whether Shot plays a role in oocyte growth because of the fact that shot null mutant fails to specify the oocyte [27].
In this study, we use a germline-specific Gal4 that drives shot-RNAi after oocyte specification to show that Shot is essential for the oocyte growth. Live-cell microscopy demonstrates that Shot controls the directionality of transport of multiple cargoes through the nurse cell-oocyte ring canals. In the wild-type egg chambers, this transport is unidirectional towards the oocyte, but after shot knockdown the transport becomes bidirectional and thus oocyte growth is stalled. Consistent with the fact that Shot controls transport directionality, we discover that Shot itself is asymmetrically localized at the ring canals connecting the nurse cells with the oocyte. It is found on actin fibers that form baskets on the nurse cell side of the ring canals. Furthermore, Shot controls the orientation of microtubules inside the ring canals: while in the wild-type microtubules are orientated predominantly with minus-ends towards the oocyte, knocking down shot results in a mixed polarity of ring canal microtubules. We propose that Shot organizes microtubules in the ring canals along the asymmetric actin fibers, allowing the minus-end-directed motor, cytoplasmic dynein, to transport multiple components from the nurse cells to the oocyte, which is required for the oocyte rapid growth.
RESULTS
Shot is essential for oocyte growth
Shot, the single spectraplakin in Drosophila, is essential for oogenesis. Each Drosophila ovary is composed of 15~20 individual developmental “assembly lines”, called ovarioles. Oogenesis starts in the most anterior structure of each ovariole, the germarium, and one oocyte is specified within a cyst of 16-interconected germline cells. The oocyte, together with 15-interconnected sister cells, the nurse cells, gets encapsulated by a mono-layer of somatic follicle cells and become an egg chamber [21] (Figure 1B). Germline clone mutant for shot3, a protein-null allele of shot [6], leads to failure of oocyte specification, shown by lack of an concentrated oocyte marker, Orb (oo18 RNA-binding protein) [32] (Figure S1A–S1B), consistent with a previous report [27].
In order to avoid the early oocyte specification defects, we used a maternal α-tubulin-Gal4 line (mat αtub-Gal4[V37]) to drive shot-RNAi. This Gal4 starts the expression in stage 2–3 egg chambers after completion of germ cell divisions and oocyte specification [33–35], thus bypassing the requirement of Shot for oocyte specification (Figure 1B). We use three different RNAi lines targeting the N-terminus (shotABD-RNAi), C-terminus (shotEGC-RNAi), and the middle rod domain (shotRod-RNAi) of shot, respectively (Figures 1A). The depletion of Shot after oocyte specification by maternal α-tubulin-Gal4 still allows oocyte specification to occur in early egg chambers, but causes striking oocyte growth defects (referred as “small oocyte phenotype”). These oocytes (identified as the single germline cell with four ring canals with a non-polyploid nucleus, by phalloidin/DAPI staining or GFP-tagged kinesin-6/Pavarotti labeling [36]) remain small and fail to grow over-time (Figure 1C–1E and 1H–J’; Videos S1 and S2). All three RNAi lines driven by maternal α-tubulin-Gal4 display the small oocyte phenotype (Figures 1F–1G and 2A–2B; Figure S1D–S1I’), indicating that this phenotype is specific to shot knockdown. The differences in “small oocyte phenotype” penetrance among the three shot-RNAi lines could be a result of different numbers of RNAi-targeted isoforms: both shotEGC-RNAi and shotRod-RNAi target all 22 isoforms of shot mRNA, while shotABD-RNAi only targets 14 isoforms leaving the isoforms lacking the CH1 domain intact (Figure 1A).
Figure 2. Actin binding and microtubule interacting domains of Shot are essential for oocyte growth.
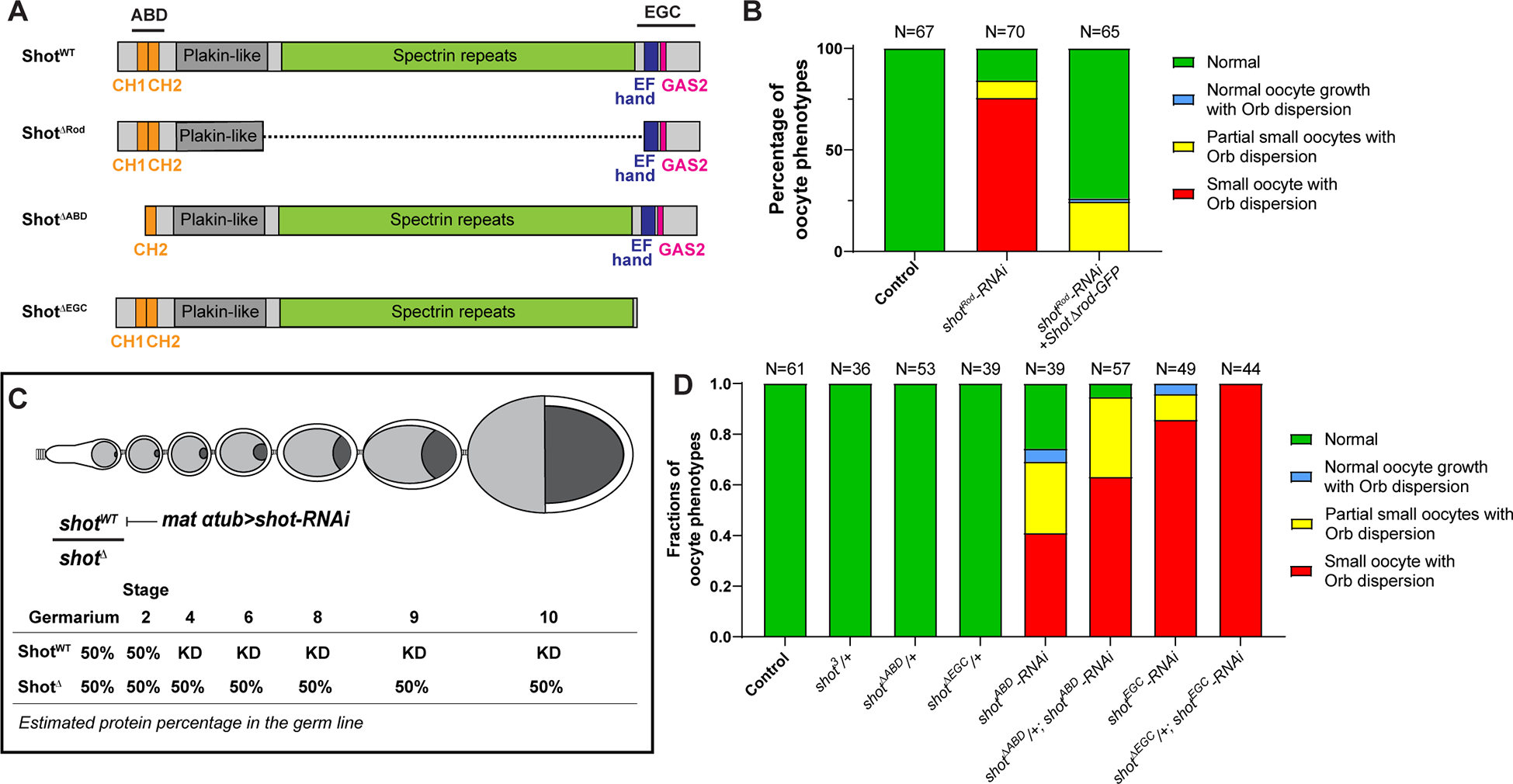
(A) Diagrams of the full-length Shot and truncated mutants. (B) Summary of Orb and phalloidin staining phenotypes in control (mat αtub-Gal4[V37]/+), shotRod-RNAi (mat αtub-Gal4[V37]/UASp-shotRod-RNAi), and shotRod-RNAi +shotARod-GFP (UASt-shot.L(A)ΔRod-GFP/+; mat αtub-Gal4[V37]/UASp-shotRod-RNAi). (C) A schematic illustration of knockdown of wild-type Shot by shot-RNAi in shot truncated mutant heterozygous background. KD, knockdown. (D) Summary of Orb and phalloidin staining in listed phenotypes. Unlike one copy of shotWT, one copy of shotΔABD or shotΔEGC is unable to drive normal oocyte growth.
As shotEGC-RNAi displays the highest penetration of “small oocyte” phenotype, we decided to further characterize the phenotypes of the shotEGC-RNAi egg chambers. As the maternal α-tubulin-Gal4 starts the expression at stages 2~3, and no shotEGC-RNAi egg chambers with small oocytes survived pass stage 9, we measured the egg chamber growth from stage 4 to stage 9 in control and in shotEGC-RNAi (Figure S1J–S1P). Among the total egg chamber size, nurse cell size and oocyte size, the oocyte size displays the largest reduction in shotEGC-RNAi compared to control (Figure S1K–S1M). Meanwhile, while the oocyte/egg chamber ratio increases robustly from stages 7–8 to stage 9 in control, the ratio decreases in shotEGC-RNAi (Figure S1N). Furthermore, although the oocyte size in shotEGC-RNAi slightly increases from stage 4 to stage 9 (Figure S1M, inset), our quantification shows that ooplasm growth is arrested in shotEGC-RNAi and the oocyte size growth in shotEGC-RNAi is mostly driven by oocyte nucleus size increase (Figure S1O–S1P, insets).
Altogether, we conclude that Shot is essential for oocyte growth.
Actin binding domain and microtubule interacting domains of Shot are required for oocyte growth
Shot is a giant cytoskeletal protein, carrying the N-terminal actin binding domain (ABD, composed of CH1 and CH2 domain) and the C-terminal microtubule interacting domain (EGC, composed of EF hand motif, GAS2 domain and C-terminal tail with SxIP motifs) connected by a long rod-like domain composed of spectrin repeats (Figure 1A) [4, 6, 7, 37]. Therefore, we decided to determine which domain is essential for oocyte growth.
The long rod domain consisting of spectrin-repeats is essential for intramolecular head-to-tail auto-inhibition of Shot [37]. We first tested whether the rod domain is required to drive the oocyte growth. The shot-RNAi targeting the rod region (shotRod-RNAi) (Figure 1A) causes the majority of the ovarioles to display oocyte growth defects (Figure 2B). The maternal expression of the ShotΔRod construct lacking the spectrin repeats (Figure 2A) rescues the “small oocyte” phenotype, resulting in most of the ovarioles with normal oocyte growth and concentrated Orb staining (Figure 2B). This indicates that the spectrin repeats are indeed dispensable for normal oocyte growth.
Next, we tested whether ABD and EGC domains are required for oocyte growth. As the Shot mutant lacking the EGC domain (shotΔEGC) is not homozygous viable [20], we induced germline clones that are homozygous of shotΔEGC. We found that, similar to shot3 germline clones, shotΔEGC mutant clones fail to specify oocytes, shown by the Orb staining (Figure S1C). In this case, we cannot determine whether the microtubule interacting domain is essential for oocyte growth due to the complete absence of oocyte specification in the shotΔEGC mutant clones. Therefore, we took advantage of the fact that the maternal α-tubulin-Gal4 drives RNAi expression after oocyte specification and combined it with the heterozygous truncated mutant that lacks either ABD domain (shotΔABD/shotWT) or EGC domain (shotΔEGC/shotWT). We chose the RNAi that only specifically knocks down wild-type shot, leaving the truncated shot mutant intact (shotABD-RNAi in shotΔABD/shotWT background, and shotEGC-RNAi in shotΔEGC/shotWT background, respectively) (Figure 2C). In this scenario, a single copy of the wild-type shot would specify the oocyte fate properly before it gets knocked down by shot-RNAi driven by maternal α-tubulin-Gal4 (starting at stage 2–3), which allows us to determine whether a single copy of truncated shot mutant could drive oocyte growth after stage 3 (Figure 2C). First of all, we confirmed that oogenesis is completely normal with one single copy of wild-type shot (shot3/shotWT, shotΔABD/shotWT and shotΔEGC/shotWT), excluding the possibility of haploinsufficiency (Figure 2D). Then comparing the shot-RNAi in wild-type shot background versus in the heterozygous background of shot truncated mutant that is insensitive to the shot-RNAi, we found that neither one copy of shotΔABD nor one copy of shotΔEGC is able to drive oocyte growth (Figure 2D), indicating that both the actin binding and microtubule interacting domains are required for the oocyte growth. Intriguingly, we found that heterozygous mutant background further enhances the “small oocyte phenotype” penetration (Figure 2D), which could be caused by a dominant negative effect of the truncated Shot mutants that compete with wild-type Shot in binding actin or microtubules, and thus sensitizes the genetic background for the RNAi effect.
Together, these data indicate that for oocyte growth, both the actin binding and the microtubule interacting domains of Shot are essential, while the central domain is dispensable, implying that actin-microtubule crosslinking activity of Shot plays a role in promoting oocyte growth.
Shot defines the direction of cargo transport through the nurse cell-oocyte ring canals
The Drosophila oocyte, remaining transcriptionally silent throughout most of the oogenesis, predominantly relies on its sister nurse cells for providing mRNAs, proteins and organelles for its growth. The small oocyte phenotype we observed in shot-RNAi suggests defects in cargo transport from the nurse cells to the oocyte. Furthermore, we noticed that in shot-RNAi Orb staining is initially correctly concentrated in the oocyte in early-stage egg chambers; however, over the stages, Orb staining becomes more dispersed and eventually lost in the oocytes (Figure 1H–J’; Video S2). This suggests that the oocyte fails to retain ooplasmic components after receiving them from the nurse cells. As control oocytes experience rapid growth at stage 8, and shot-RNAi egg chambers with small oocytes survive to stage 8 with no major structural defects, we decided to examine the role of Shot in the transfer of materials from the nurse cells to the oocyte in stage 8 egg chambers.
We selected four types of cargoes to study the nurse cell-oocyte cytoplasmic transfer: Golgi units, ribonucleoprotein particles (RNPs), mitochondria, and lipid droplets (LDs). All of these four cargoes generated in the nurse cells are essential for the oocyte function and development [25, 38–43]. We used a RFP-tagged galactosyltransferase line to label Golgi units [44], a RFP-tagged Staufen line to label osk/Staufen RNPs [31, 45], a photoconvertible mitochondrial-targeting probe (Mito-MoxMaple3) to label mitochondria, and a GFP-tagged lipid droplet domain of Drosophila protein Klar, GFP-LD, to label lipid droplets [46].
We examined the role of Shot in controlling directionality of transporting these components through the nurse cell-oocyte ring canals. In control, all four cargoes display robust movement within the nurse cells, and move primarily from the nurse cells to the oocyte through the ring canals (Figure 3 and Videos S3–S7). However, in shotEGC-RNAi, the transport directionality of these four cargoes is completely disrupted. We observed frequent reversed cargo transport, moving from the oocyte towards nurse cells, resulting in a lack of cytoplasmic accumulation in the oocyte (Figure 3; Videos S3–S4 and S6–S7).
Figure 3. Shot controls directionality of cargo transport from the nurse cells to the oocyte.
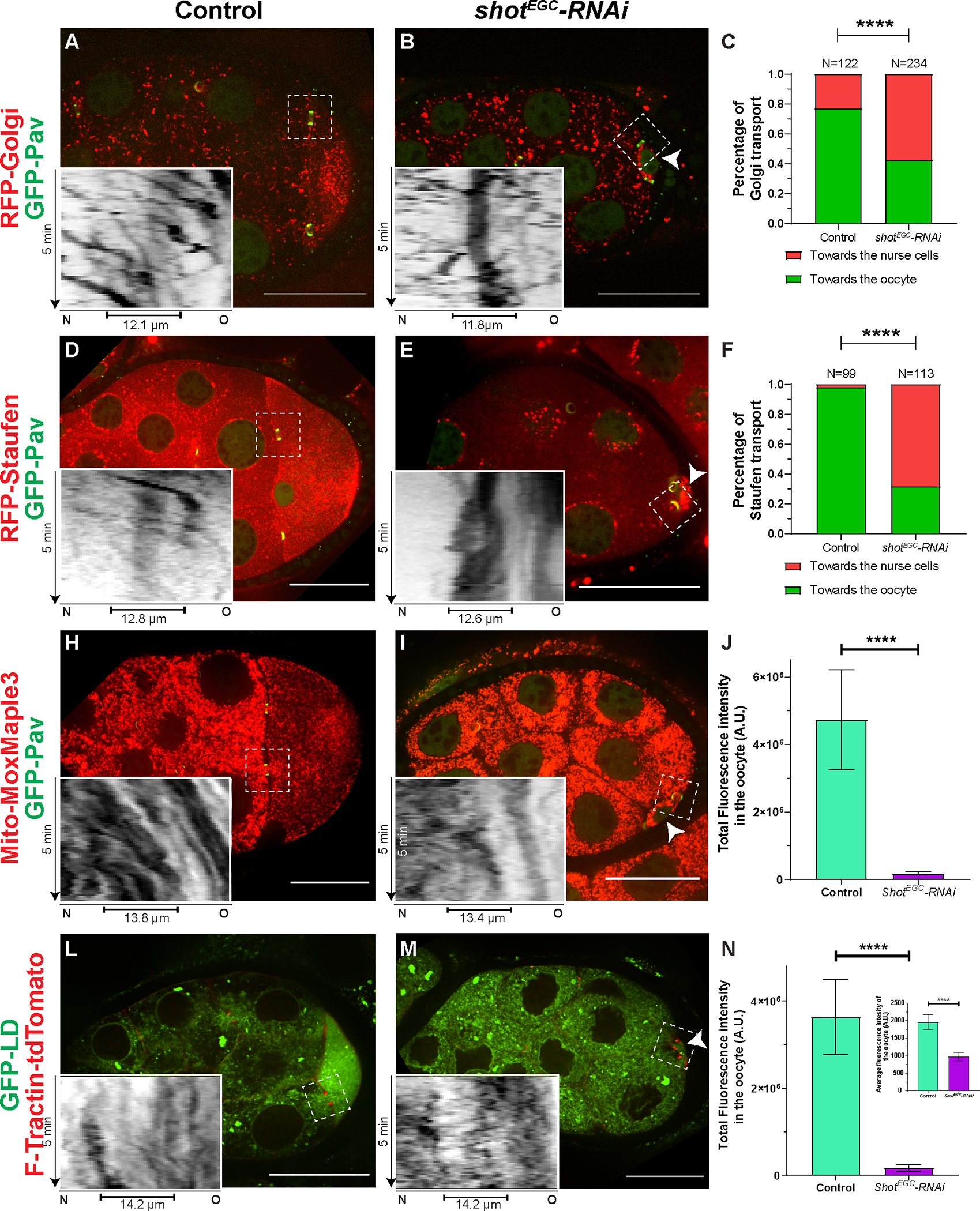
(A-C) Golgi transport in the nurse cell-oocyte ring canals in control (A) and in shot-RNAi (B). Golgi are labeled with RFP-tagged human galactosyltransferase (GalT) (RFP-Golgi). (C) Quantification of Golgi transport directions in control and in shot-RNAi. Chi-square test between control and shot-RNAi, p-value < 0.00001 (****).
(D-F) Staufen RNP transport in the nurse cell-oocyte ring canals in control (D) and in shot-RNAi (E). Staufen RPNs are labeled with RFP-tagged Staufen (RFP-Staufen). (F) Quantification of Staufen transport directions in control and in shot-RNAi. Chi-square test between control and shot-RNAi, p-value < 0.00001 (****).
(H-J) Mitochondria transport in the nurse cell-oocyte ring canals in control (H) and in shot-RNAi (I). Mitochondria are labeled with Mito-MoxMaple3 (red channel, after global photoconversion). (J) Quantification of total mitochondria fluorescence intensity (mean ± 95% confidence interval) in control (N=18) and in shot-RNAi (N=22) oocytes. Unpaired t test with Welch’s correction between control and shot-RNAi, p-value < 0.0001 (****).
(L-N) Transport of lipid droplets in the nurse cell-oocyte ring canals in control (L) and in shot-RNAi (M). Lipid droplets are labeled with GFP-tagged lipid droplet domain of Drosophila protein Klar (GFP-LD). (N) Quantification of lipid droplet total fluorescence intensity and average fluorescence intensity (inset) (mean ± 95% confidence interval) in control (N=33) and in shot-RNAi (N=28) oocytes. Unpaired t test with Welch’s correction of total fluorescence intensity of GFP-LD between control and shot-RNAi, p-value < 0.0001 (****); Unpaired t test with Welch’s correction of average fluorescence intensity of GFP-LD between control and shot-RNAi, p-value < 0.0001 (****).
Left side: the nurse cells; right side, the oocyte; small oocytes in shot-RNAi are pointed with the white arrowheads; ring canals are labeled with either GFP-Pav (A-B, D-E, H-I) or F-Tractin-tdTomato (L-M); inverted kymographs were created along a ~3.7 μm-width line from the nurse cell (N, left side) to the oocyte (O, right side) through the ring canals (marked as capped lines underneath in the kymographs) in the white dashed box areas; scale bars, 50 μm.
Collectively, these data demonstrate that Shot controls the directionality of material flow from the nurse cells to the oocyte. Lack of Shot results in randomization of transport between the nurse cells and the oocyte, likely underlying the oocyte growth arrest.
Localization of Shot on the nurse cell side of the ring canals
Having confirmed that Shot controls the directionality of transport between the nurse cells and the oocyte, we decided to examine Shot localization around the ring canal region.
As previously described, actin filaments form asymmetric baskets at the nurse cell-oocyte ring canals [25, 47]. These baskets are only found on the donor side of the ring canals (the nurse cells), but never on the recipient side (the oocyte). This localization is established at stages 6–7 and persist to stages 9–10 [25]. These asymmetrically positioned actin filaments can be labeled either by Phalloidin staining (Figure 4A–4B) or by germline-specific expression of LifeAct-TagRFP [48] (Figure 4C–4D; Video S8). We quantified the LifeAct-TagRFP signal on both sides of the ring canals connecting the nurse cells and the oocyte (see more details in the Materials and Methods “Measurement of the ratio of actin signal at ring canals”), and found a high degree of actin asymmetry on the nurse cell side over the oocyte side (Figure 4E–4G). This asymmetry of actin fibers at ring canals sharply decreases between nurse cells towards the anterior side of the egg chamber, with the lowest asymmetry at ring canals connecting anterior-most nurse cells (Figure 4E–4G). The level of actin asymmetry correlates well with the directionality of the cargo transport: we observed less directional bias of transport through the ring canals connecting nurse cells where asymmetry of actin fibers is significantly less than the nurse cell-oocyte ring canals (Table 1).
Figure 4. Shot is localized at the asymmetric actin fibers of the nurse cell-oocyte ring canals.
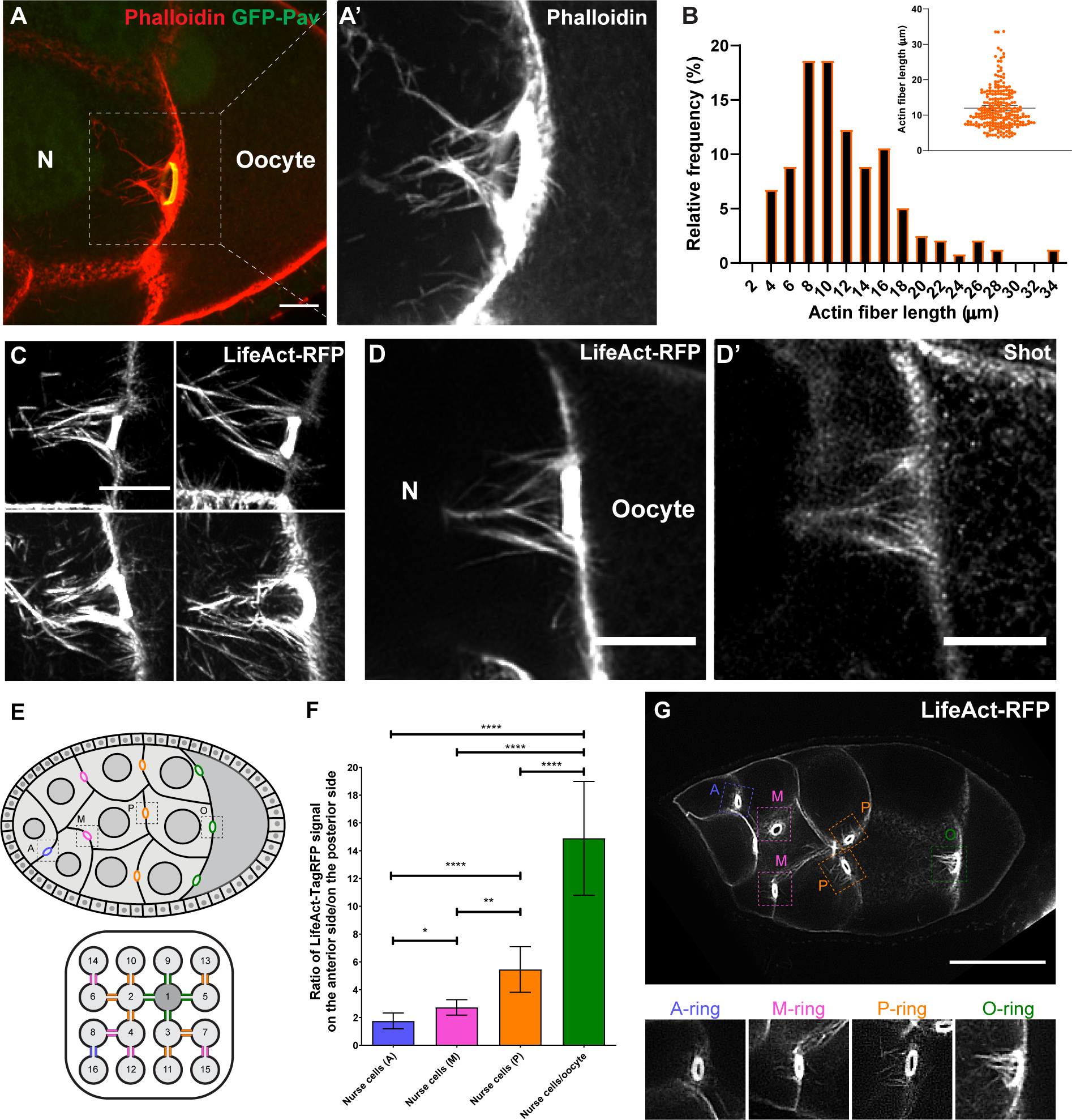
(A-A’) Rhodamine-conjugated phalloidin staining shows asymmetric actin fibers (the white dashed box) at the ring canal (ring canal inner rim is labeled with GFP-Pavarotti) on the nurse cell side, not on the oocyte side.
(B) Quantification of the length of actin fibers on the nurse cell side. The lengths of the four longest actin fibers were measured for each ring canal (59 ring canals from 15 control egg chambers). The average actin fiber length on the nurse cell side is 12.0 ± 0.7 μm (mean ± 95% confidence interval).
(C) Asymmetric actin fibers, labeled with TagRFP-tagged LifeAct, are seen at all four ring canals connecting nurse cells and the oocyte in a live sample. See also Video S8.
(D-D’) A representative image of Shot antibody staining in a TagRFP-LifeAct-expressing egg chamber. Shot is localized at the asymmetric actin fibers on the nurse cell side of the ring canal, but it is not concentrated in the F-actin core of the ring canal inner rim.
(E) Schematic illustrations of a stage 8 Drosophila egg chamber and an interconnected 16-cell germline cyst, including 1 oocyte (cell 1) and 15 nurse cells (cells 2–16, numbered according to the order of cell divisions). Ring canals are categorized depends on their relative distance to the oocyte [62] and are color-coded: (1) nurse cell-oocyte ring canals, directly connected to the oocyte, green, “O”; (2) posterior nurse cell-nurse cell ring canal, having one nurse cell between this ring canal and the oocyte, orange, “P”; (3) middle nurse cell-nurse cell ring canal, having two nurse cells between this ring canal and the oocyte, magenta, “M”; (4) anterior nurse cell-nurse cell ring canal, having three nurse cells between this ring canal and the oocyte, blue, “A”.
(F) The asymmetry of actin fibers is quantified as the ratio of LifeAct-TagRFP fluorescence signal on the anterior side to the signal on the posterior side of the ring canals (see more details in Materials and Methods). Numbers of ring canals for each type: anterior nurse cell-nurse cell ring canals (A), N=17; middle nurse cell-nurse cell ring canals (M), N=46; posterior nurse cell-nurse cell ring canals, N=55; nurse cell- oocyte ring canals (O), N=70. Unpaired t tests with Welch’s correction were performed in following groups: “O” and “P”, p<0.0001 (****); “O” and “M”, p<0.0001 (****); “O” and “A”, p<0.0001 (****); “P” and “M”, p=0.0024 (**); “P” and “A”, p<0.0001 (****); “M” and “A”, p=0.0143 (*).
(G) A representative image of a stage 8 egg chamber expressing LifeAct-TagRFP. Four types of ring canals are highlighted in colored boxes with zoom-in images below.
N, nurse cell; scale bars, 10 μm (A, C, D-D’) and 50 μm (G).
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Orb monoclonal (mouse) | Developmental Studies Hybridoma Bank | orb 6H4 |
| Anti-Shot monoclonal (mouse) | Developmental Studies Hybridoma Bank | mAbRod1 |
| FITC-conjugated anti-β tubulin monoclonal (mouse) | ProteinTech | Cat#: CL488–66240 |
| FITC-conjugated anti-mouse IgG antibody (Goat) | Jackson ImmunoResearch Laboratories, Inc | Cat# 115–095–062 |
| TRITC-conjugated anti-mouse IgG antibody (Goat) | Jackson ImmunoResearch Laboratories, Inc | Cat# 115–025–003 |
| Chemicals, peptides, and recombinant proteins | ||
| Rhodamine Phalloidin | Thermo Fisher Scientific | Cat # R415 |
| DAPI (4’,6-Diamidino-2-Phenylindole, Dihydrochloride) | Thermo Fisher Scientific | Cat# D1306 |
| 16% Paraformaldehyde aqueous methanol-free solution, EM Grade | Thermo Fisher Scientific (Manufacturer: Electron Microscopy Sciences) | Cat# 50–980–488 |
| Halocarbon oil 700 | Sigma-Aldrich | Cat# H8898–50ML |
| Triton X-100 | Fisher Scientific | Cat# BP151–500 |
| BSA Bovine Serum Albumin (BSA) | DOT Scientific | Cat# DSA30075–100 |
| Normal Goat Serum | SouthernBiotech | Cat# 0060–01 |
| Nutri-Fly® Bloomington Formulation | Genesee | Cat# 66–121 |
| Experimental models: organisms/strains | ||
| Drosophila melanogaster: hs-FLP | Bloomington stock center | RRID:BDSC_1929 |
| Drosophila melanogaster: FRTG13 | Bloomington stock center | RRID:BDSC_1956 |
| Drosophila melanogaster: FRTG13 shot [3] | Bloomington stock center | RRID:BDSC_5141 |
| Drosophila melanogaster: FRTG13 ubi-GFP.nls | Bloomington stock center | RRID:BDSC_5826 |
| Drosophila melanogaster: mat αtub-Gal4 [V37] | Bloomington stock center | RRID:BDSC_7063 |
| Drosophila melanogaster: UAS-shotRod-RNAi (TRiP.GL01286) | Bloomington stock center | RRID:BDSC_41858 |
| Drosophila melanogaster: UASt-shot.L(A)Δrod-GFP | Bloomington stock center | RRID:BDSC_29040 |
| Drosophila melanogaster: shot ΔABD | Bloomington stock center | RRID:BDSC_10522 |
| Drosophila melanogaster: UASp-LifeAct-TagRFP | Bloomington stock center | RRID:BDSC_58713; RRID:BDSC_58714 |
| Drosophila melanogaster: UASp-RFP-Golgi | Bloomington stock center | RRID:BDSC_30908 |
| Drosophila melanogaster: UASp-F-Tractin-tdTomato | Bloomington stock center | RRID:BDSC_58989 |
| Drosophila melanogaster: UAS-Dhc64C-RNAi (TRiP.GL00543) | Bloomington stock center | RRID:BDSC_36583 |
| Drosophila melanogaster: shot ΔEGC | Gift from Dr. Ferenc Jankovics | [20] |
| Drosophila melanogaster: ubi-GFP-Pav | Gift from Dr. David Glover | [36] |
| Drosophila melanogaster: nos-Gal4-VP16 | Gift from Dr. Edwin Ferguson lab | [70,71] |
| Drosophila melanogaster: mat αtub-RFP-Staufen | Gift from Dr. Daniel St Johnson | [45] |
| Drosophila melanogaster: UASp-GFP-LD | Gift from Dr. Michael Welte | [46] |
| Drosophila melanogaster: UASp-GFP-Patronin | Gift from Dr. Uri Abdu | [31, 51, 69] |
| Drosophila melanogaster: UASp-EB1-GFP | Gift from Dr. Antoine Guichet | [52] |
| Drosophila melanogaster: ubi-EB1-GFP | Gift from Dr. Derek Applewhite | [53, 72] |
| Drosophila melanogaster: UASp-Staufen-SunTag | Generated in our lab | [69] |
| UAS-shotABD-RNAi | This paper | N/A |
| UAS-shot EGC -RNAi | This paper | N/A |
| UASp-Mtio-MoxMaple3 | This paper | N/A |
| Recombinant DNA | ||
| Plasmid: pWalium22-shotABD-RNAi | This paper | N/A |
| Plasmid: pWalium22-shotEGC-RNAi | This paper | N/A |
| Plasmid: pUASp-UASp-Mito-MoxMaple3 | This paper | N/A |
| Software and algorithms | ||
| NIS-Elements | Nikon Instruments Inc. | https://www.microscope.healthcare.nikon.com/products/software/nis-elements/nis-elements-advanced-research |
| FIJI | open source[80] | https://imagej.net/Fiji |
We next examined localization of Shot using immunostaining with a monoclonal antibody against Shot [12]. We found that Shot staining is associated with these actin fibers at the ring canals, showing a high level of asymmetry at the nurse cell-oocyte boundary (Figure 4D–D’; Figure S2A).
We also examined the Shot antibody staining in all three Shot-RNAi germline knockdown samples, and found a complete absence of cortical and actin localization of Shot staining in the germline cells, whereas the apical localization of Shot in the somatic follicle cells is not affected (Figure S2B–S2E). This indicates that the Shot staining we observed is specific.
In shot-RNAi, we found that the nurse cell-oocyte ring canals are smaller than the control ones (Figure S2F–S2H). This is probably due to the physical constrain of limited cell membrane of a small oocyte. This smaller ring canal size in shot-RNAi is correlated with lack of asymmetric actin fibers between the nurse cells and the oocyte (Figure S2I–S2L). However, we still observed actin fibers formed at the nurse cell-nurse cell ring canals (Figure S2M–S2N), suggesting that Shot is not required for actin fiber assembly. Instead, the absence of nurse cell-oocyte actin basket in shot-RNAi could be an indirect consequence of the oocyte growth defects.
Altogether, the specific localization of Shot at the ring canals implies that Shot controls the transport of cargoes from the nurse cells to the oocyte through its interaction with these asymmetric actin filaments.
Shot organizes microtubules in the ring canals
Having established that Shot is required for directional nurse cell-oocyte cargo transport and Shot is asymmetrically localized at the actin fibers on the nurse cell side, we decided to investigate the mechanism by which Shot controls the direction of transport through the nurse cell-oocyte ring canals. As microtubules are found inside the ring canals connecting the nurse cells with the oocyte, the transport of organelles and mRNAs/proteins to the oocyte have been considered as typical examples of microtubule-dependent motor-driven transport [24–26, 39, 41, 49]. Therefore, we examined whether shot-RNAi changes microtubule tracks in the nurse cell-oocyte ring canals.
First, we labeled the stable microtubules by overexpressing the minus-end binding protein, Patronin [50, 51], and found that, consistent with previous reports [24–26], microtubules are present in the ring canals between nurse cells and the oocyte, as well as between nurse cells (Figure 5A–A”). Microtubule distribution is not visibly altered either in the nurse cells or in the ring canals of shot-RNAi egg chambers (Figure 5B–B”). Next we examined the orientation of dynamic microtubules running through the ring canals by expressing GFP-tagged EB1 [52]. We found that in control most of the microtubules in the ring canals are oriented with their plus-ends pointing towards nurse cells (Figure 5C–C” and 5E; Video S9). Compared to the nurse cell-oocyte ring canals, the microtubule polarity at the posterior nurse cell-nurse cell ring canals is less uniform, consistent with the less directional transport observed through these ring canals (Figure S2Q–S2T; Table 1). In sharp contrast, shot-RNAi mutant has fewer EB1 comets in the oocyte and in the ring canals (Figure 5D–5D” and 5F; Video S9). In agreement with the observation of reduced numbers of EB1 comets, we observed a reduced number of microtubules in the oocyte as well as in the nurse cell-oocyte ring canals in shot-RNAi, although the overall microtubule organization in the nurse cells are not seemingly affected by shot knockdown (Figure S2O–S2P”). Importantly, the direction of the EB1-GFP comets shows that microtubules in the nurse cell-oocyte ring canals have a mixed orientation in shot-RNAi knockdown (Figure 5D–5E; Video S9). This mixed polarity of microtubule tracks could explain the change of directionality of cargo transport between the nurse cells and the oocyte after shot knockdown.
Figure 5. Shot controls microtubule polarity in the nurse cell-oocyte ring canals.
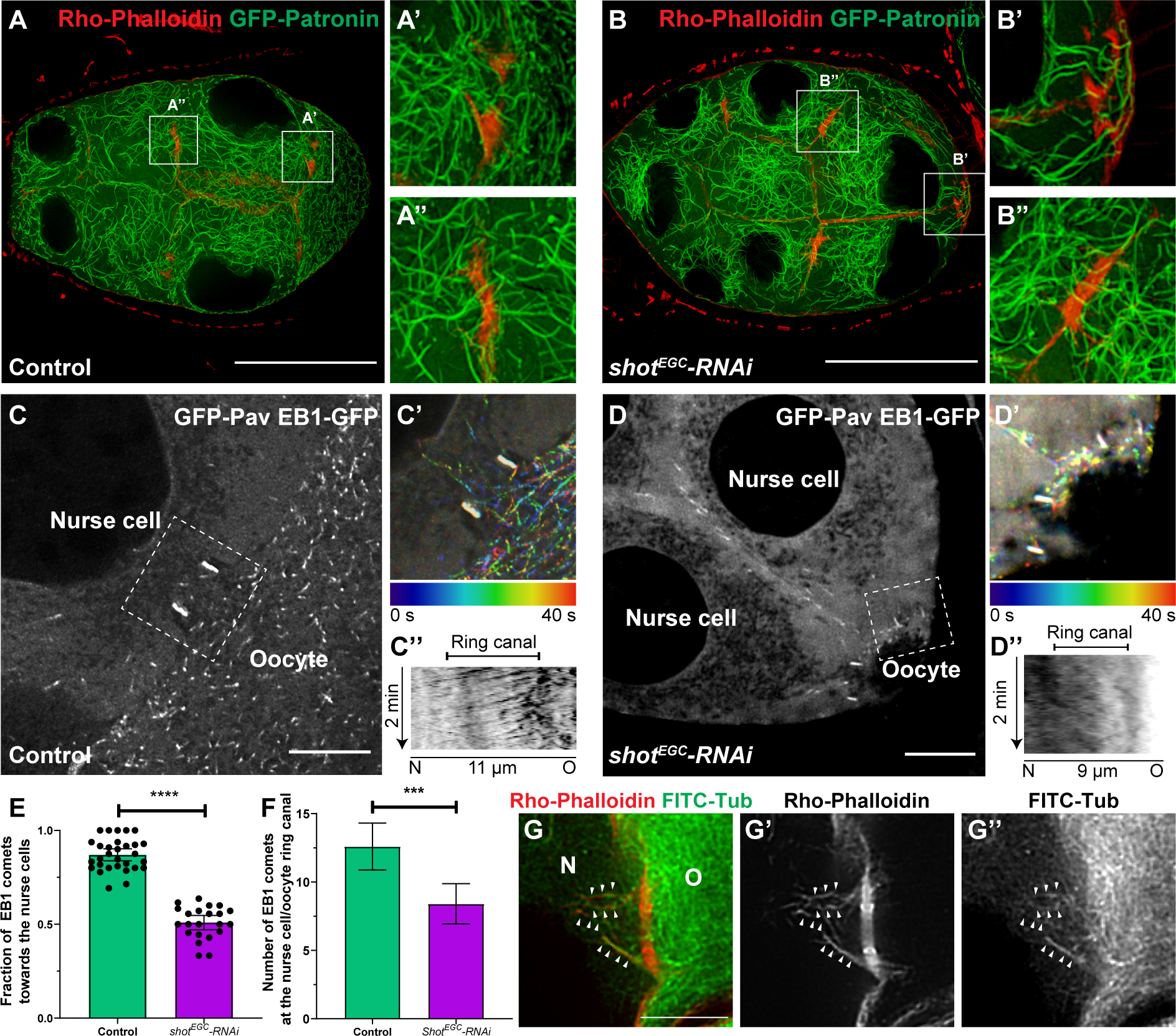
(A-B) Stable microtubule organization is not affected by shot knockdown. (A) In control, stable microtubules are localized at the ring canals between the nurse cell and the oocyte (A’) and between two nurse cells (A”). (B) Knockdown of shot does not change stable microtubule distribution at the ring canals between the nurse cells and the oocyte (B’) and between two nurse cells (B”). Stable microtubules are labeled by overexpressed GFP-tagged Patronin, and ring canals are labeled with Rho-Phalloidin staining. Scale bars, 50 μm.
(C-E) Knockdown of shot results in a mixed orientation of microtubules in the ring canals. (C) EBI-GFP-labeled microtubule +end comets in the ring canal (labeled by GFP-Pav) connecting a nurse cell and an oocyte in control. (C’) A color-coded hyperstack of the EB1 comet movement of (C). (C”) A kymograph of EB1 comet movement at the ring canal (the white dashed box in C) in control. (D) EBI-GFP-labeled microtubule +end comets at the ring canal (labeled by GFP-Pav) connecting two nurse cells and an oocyte in shot-RNAi. (D’) A color-coded hyperstack of the EB1 comet movement of (D). (D”) A kymograph of EB1 comet movement at the ring canal (the white dashed box in D) in shot-RNAi. (E) Quantification of the fraction of EB1 comets moving through the ring canals from the oocyte towards the nurse cells in control and in shot-RNAi. Control, N=30; shotEGC-RNAi, N=22; unpaired t test with Welch’s correction between control and shot-RNAi, p-value < 0.0001(****). (F) Quantification of EB1 comet numbers in the nurse cell-oocyte ring canals in control and shot-RNAi. Control, N=30; shotEGC-RNAi, N=22; unpaired t test with Welch’s correction between control and shot-RNAi, p-value= 0.0004 (***).
(G-G”) A representative image of dual labeling of microtubules (by FITC-conjugated tubulin antibody) and F-actin (by Rho-Phalloidin) at a nurse cell-oocyte ring canal. Alignment of microtubules on asymmetric actin filaments are highlighted by small white arrowheads.
(C-D and G-G’) N, nurse cell; O, oocyte; scale bars, 10μm.
By dual labeling with LifeAct-TagRFP and EB1-GFP, we found that microtubule plus-ends originated in the oocyte tend to grow along the asymmetric actin fibers of the ring canals on the nurse cell side, while the ones originated in the nurse cells are more likely to bump into the actin fibers and get deflected away from the ring canals (Video S10). In line with the EB1 and actin dual labeling observation, we found alignment of microtubules on the asymmetric actin fibers on the nurse cell side (Figure 5G–5G”). Given the facts that Shot is localized at these asymmetric actin fibers and Shot can bind to both actin and microtubules, we propose that Shot favors microtubule growth along these actin fibers on the nurse cell side, which could facilitate oocyte microtubule plus-ends pass through the ring canal, while prevent nurse cell microtubule plus-ends from entering the ring canal, thus controlling microtubule polarity for unidirectional cargo transport (Figure 6).
Figure 6. Shot is a gatekeeper at the ring canal for Drosophila oocyte growth.
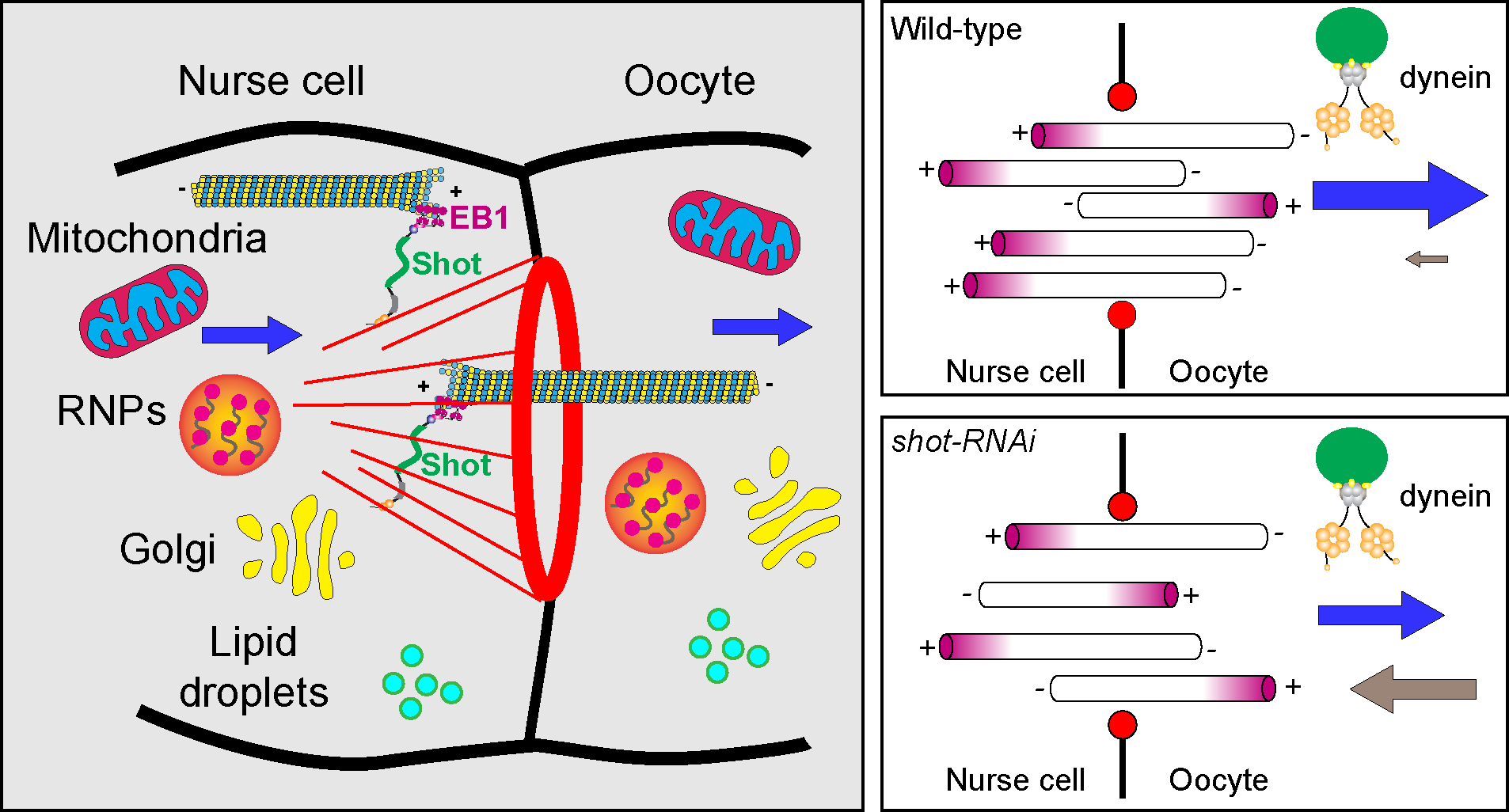
Shot controls microtubule orientation in the ring canal, via regulating the interaction between EBI/microtubule plus-ends and asymmetric actin fibers on the nurse cell side. Therefore, Shot is essential for directing dynein-dependent transport of various cargoes (including Golgi units, osk/Staufen RNPs, mitochondria, and lipid droplets) from the nurse cells to the oocyte.
Dynein is the main motor for nurse cell-oocyte transport
The minus-end directed motor cytoplasmic dynein has been proposed to transport organelles and RNP granules to the oocyte [24–26, 33]. To examine whether dynein is required for oocyte growth, we knocked down dynein heavy chain in the ovary by RNAi (Dhc64C-RNAi, [53]). In order to bypass dynein’s requirement for cell division and oocyte specification, we used the same maternal α-tubulin-Gal4 as in shot-RNAi. We found that that dynein-RNAi knockdown results in small oocytes with dispersion of the oocyte marker Orb (Figure S3A–S3F), completely mimicking the shot-RNAi phenotypes we observed.
As dynein knockdown phenocopies shot knockdown in the ovary, we examined Shot localization in the dynein-RNAi samples, and found that Shot is still localized corrected to the cell cortex as well as the ring canal actin fibers between the nurse cells (Figure S3G–S3H‘, compared to Figure S2A–S2B). However, as the oocyte fails to grow in dynein-RNAi, the asymmetric actin baskets do not form at the nurse cell-oocyte ring canals, probably due to the delayed development of the oocyte, similar to shot-RNAi (Figure S3I–S3I‘).
Next we examine two types of cargoes within dynein-RNAi egg chambers, mitochondria and lipid droplets. In contrast to shot-RNAi where cargoes move bidirectionally, mitochondria in dynein-RNAi display heavily reduced movement within the nurse cells and very little transport through the ring canals between the nurse cells and the oocyte (Video S11, compared to Video S6). To quantify the mitochondria transport to the oocyte, we measured the total fluorescence intensity of mitochondria in the oocyte, and found that Dhc64C-RNAi significantly reduced the transport of mitochondria into the oocyte (Figure S3J–S3L). We found a similar effect of Dhc64C-RNAi on the lipid droplet accumulation in the oocyte (Figure S3M–S3O). Hence, we conclude that dynein is the main motor responsible for the transfer of cytoplasmic contents (including mitochondria, lipid droplets, RNP granules [24] and Golgi units [25]) via the nurse cell-oocyte ring canals to support the oocyte growth.
Together, we propose that Shot functions as a gatekeeper at the ring canal: Shot favors the uniform microtubule orientation with minus-ends in the oocyte, and allows cytoplasmic dynein to transport various cargoes from the nurse cells to the oocyte to ensure the rapid growth of the oocyte during oogenesis (Figure 6).
DISCUSSION
Spectraplakins coordinate and regulate two major cytoskeletal networks, microtubules and actin filaments. Drosophila has a single spectraplakin protein Short stop (Shot) that makes it a perfect model to study the interaction and coordination between microtubules and F-actin. In the biggest cell of the whole animal, the oocyte, microtubules and F-actin are dynamic but precisely arranged throughout the development. In this study, we show that Shot is absolutely required for the oocyte growth. Shot is localized asymmetrically at the actin baskets on the nurse cell side of the ring canals, and controls the microtubule polarity in the ring canals connecting the nurse cells and the oocyte. The highly polarized microtubules in the ring canals allows the minus-end directed motor, dynein, to move cargoes from the nurse cells to the oocytes, resulting in the accumulation of cytoplasmic materials in the oocyte (Figure 6). Therefore, Shot controls the cytoplasmic transfer of cargoes produced in the nurse cells that are essential for rapid oocyte growth.
Asymmetric actin baskets at the ring canals
The asymmetric actin baskets at the ring canals start forming in stage 6–7 egg chambers and persist to stage 9–10 before they become indistinguishable from the actin cables formed for nurse cell dumping [25, 47]. From stage 6 to stage 10, the oocyte experiences exponential growth in size [54] (Figure S1M) caused by unidirectional transport of materials from the nurse cells to the oocyte. This flow of materials precedes massive nurse cell dumping caused by contraction of nurse cells at stages 11–12 [23], and is easy to distinguish from dumping because during the directional transport stage the volume of the oocyte increases but the nurse cells do not shrink (Figure S1L–S1M). Strong correlation between the appearance of asymmetric actin baskets and the rapid oocyte growth suggests that actin baskets are involved in directing the transport through the ring canals to the oocyte.
It is not yet clear why the actin baskets are formed asymmetrically at the nurse cell-oocyte border. One possible explanation is that actin filaments are organized differently in the nurse cells and in the oocyte. Actin filaments form microvilli originating from the plasma membrane in nurse cells, which is dependent on the Drosophila Profilin homolog, Chickadee, and Fascin homolog Singed [55–57]. Meanwhile, actin cytoplasmic mesh in the oocyte is organized by two actin filament nucleators Cappuccino (Drosophila Formin homolog) and Spire (contains of four WASP homology 2 (WH2) domains) [58–60]. Therefore, albeit in a 16-cell syncytium connected by ring canals, it is likely that different regulations of actin growth lead to asymmetric actin baskets formed only on the nurse cell side, but not on the oocyte side of these ring canals [61].
Intriguingly, we also noticed the lack of asymmetric actin baskets at the nurse cell-oocyte ring canals in both shot-RNAi and dynein-RNAi, although the actin fibers at nurse cell-nurse cell ring canals are not seemingly affected (Figures S2I–S2J‘, S2M–S2N and S3H–S3I). This indicates that Shot and dynein are not essential for actin fiber formation at the ring canals; instead the failure of actin basket formation could be caused by either the physical constrains of the small oocyte size, or the defects of oocyte development (shown by delayed oocyte nucleus growth) (Figure S1O) and/or oocyte identity maintenance (evidenced by dispersion of Orb staining and reduced concentration of lipid droplets) (Figures 1H–1J’ and 3L–3N; Figure S3C–S3D and S3M–S3O).
Furthermore, we observed an interesting gradient of actin fiber asymmetry associated with the ring canals from the posterior to the anterior: the posterior nurse cell-nurse cell ring canals appear to have more actin fibers as well as a higher asymmetric actin ratio than the anterior ones. Noticeably, in spite of the fact that nurse cells are interconnected with ring canals, they are not born equally. A previous study has shown that the size of nurse cell are correlated with its distance to the oocyte: the nurse cell group (2,3,5,9) > the nurse cell group (4, 6, 7, 10, 11, 13)> the nurse cell group (8, 12, 14, 15)> the nurse cell (16) (Figure 4E, bottom) [62]. This cell size gradient is consistent with the actin fiber ratio gradient at the ring canals (O>P>M>A, Figure 4E–4G). Therefore, we speculate the asymmetry level of actin fibers between nurse cells is related to the nurse cell sizes. Recently, it has been shown that an oocyte-produced CDK inhibitor, Dap protein, diffuses to nurse cells, and it controls nurse cell endocycle, which probably in turn correlates with nurse cell size increase [63]. Thus, the oocyte could provide a signal to form a gradient of actin fiber asymmetry in nurse cells and therefore achieve the optimized directional cargo transport from the nurse cells to the oocyte.
Shot guides microtubule growth at the ring canal
Our results show that Shot is localized at the asymmetric actin fibers at the ring canals between the nurse cells and the oocyte. Interestingly, microtubules are required for maintaining these actin baskets at the ring canals. Depolymerization of microtubules in the egg chamber by treatment with microtubule-depolymerizing drug, colchicine, results in disappearance of the actin baskets at the ring canals [25].
Our data suggest that Shot plays a role in guiding the microtubule plus-ends along the actin fibers. Microtubules in the ring canals have more plus-ends towards the nurse cells, while knockdown of shot changes these ring canal microtubules to a mixed orientation. This Shot function is probably dependent on the EB1-interacting SxIP motifs at the C-terminal tail. Studies in Drosophila neurons showed that Shot interacts with EB1 protein and F-actin in the growth cone, and thus guides polymerizing microtubules along the actin-structure in the direction of axonal growth [5, 8, 64]. Additionally, Shot has been shown to promote microtubule assembly by recruiting EB1/APC2 at the muscle-tendon junctions [65]. These studies are in an agreement with our model that Shot favors the microtubule polymerization along the asymmetric actin fibers, which in turn allows microtubule plus-ends in the oocyte to enter the nurse cells. Therefore, Shot localization at the asymmetric actin fibers results in the directional bias of microtubule tracks, allowing the minus-end-directed motor dynein to efficiently transfer cytoplasmic contents to the oocyte (Figure 6).
Noticeably, the microtubule density of the oocyte is higher than the nurse cells in control, while this difference is eliminated in shot-RNAi, shown by the EB1 comets and microtubule staining (Figure 5C–5F; Figure S2O–S2P”). Our speculation on the higher microtubule density in the oocyte is that: (1) directional transport from the nurse cells to the oocytes builds up a concentration of some microtubule polymerization-promoting factor(s) in the oocyte; (2) this higher microtubule polymerization activity also allows more microtubules growing through the ring canals from the oocyte towards the nurse cells; (3) the biased microtubule orientation at the nurse cell-oocyte ring canals further enhances the directionality of cargo transport (including the microtubule polymerization- promoting factor) towards the oocyte. Together, it forms a positive feedback loop to maintain the oocyte with the highest microtubule polymerization activity among the 16-cell cyst. Apparently, in shot-RNAi directional transport is disrupted (Figure 3) and oocyte identity is lost overtime (Figure 1 and Figure S1). Thus the feedback loop is clearly abolished. This leads to lower microtubule polymerization activity in the oocyte and therefore reduces the microtubule density in the nurse cell-oocyte ring canals, which could contribute to the mixed polarity of microtubules we observed in shot-RNAi (Figure 5).
Intercellular cytoplasmic bridges are conserved across species
In this study, we demonstrate that multiple cargoes, including Golgi units, RNP granules, mitochondria and lipid droplets are transported through the ring canals from the nurse cells to the oocyte, of which the directionality is controlled by the microtubule- actin cross-linker Shot. The ring canal in Drosophila egg chambers is not the sole example of cytoplasmic bridges connecting cells and transferring cytoplasm. Multiple organisms ranging from plants to mammals have arrested cytokinesis and maintain the contractile rings as stable cytoplasmic bridges to stay connected with sister cells, both in germline cells and in somatic cells [66]. Particularly, intercellular cytoplasmic bridges between germline cyst cells have been documented in higher organisms, such as in Xenopus, chicken, mouse, rat, hamster, rabbit, and human [67]. Remarkably, mouse germ cyst cells transfer organelles, such as Golgi and mitochondria, to the developing oocyte through ring canals in a microtubule transport-dependent manner [68]. This “sister cell transferring cytoplasm” paradigms in mice highly resemble the Drosophila nurse cell-to-oocyte transport, suggesting there could be an evolutionarily conserved mechanism of cytoplasmic transfer during germline development. This intercellular transfer may present a highly efficient way for the oocyte to acquire essential materials/organelles for its rapid growth.
Altogether, we demonstrate that the single Drosophila spectraplakin, Short stop (Shot), functions as a gatekeeper at the cytoplasmic canals, and controls the directionality of cytoplasmic transfer from the nurse cells to the oocyte, which ensures the oocyte to have enough cytoplasmic materials during its rapid growth. As spectraplakin family proteins and intercellular cytoplasmic bridges are conserved across species, it is likely that it serves as a universal cytoplasmic transfer mechanism for oocyte growth in higher organisms.
STAR★Methods
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Vladimir I. Gelfand: vgelfand@northwestern.edu.
Materials availability
This study has generated three plasmids (pWalium22-shotABD-RNAi, pWalium22-shotEGC-RNAi, and pUASp- UASp-Mito-MoxMaple3) and several transgenic fly lines accordingly: UAS-shotABD-RNAi, UAS-shotEGC-RNAi (both integrated at attP2 site), and UASp-Mito-MoxMaple3 (II-F2 and III-M4). Requests for these plasmids and/or transgenic animals should be directed to and will be fulfilled by the lead contact, Vladimir I. Gelfand: vgelfand@northwestern.edu.
Data and code availability
The published article includes all dataset generated and analyzed during this study.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Drosophila strains.
Fly stocks and crosses were maintained on standard cornmeal food (Nutri-Fly® Bloomington Formulation, Genesee, Cat #: 66–121) supplemented with dry active yeast at room temperature (~24–25°C), The following fly stocks were used in this study: hs-FLP[12](X, Bloomington Drosophila Stock Center #1929 [70]); FRTG13 (II, Bloomington Drosophila stock center # 1956); FRTG13 shot[3] (Bloomington Drosophila Stock Center # 5141 [71]); FRTG13 ubi-GFP.nls (II, Bloomington Drosophila Stock Center # 5826); shotΔEGC (from Dr. Ferenc Jankovics, Institute of Genetics, Biological Research Centre of the Hungarian Academy of Sciences [20]); mat αtub-Gal4[V37] (III, Bloomington Drosophila Stock Center #7063); ubi-GFP-Pav (II, from Dr. David Glover, Caltech [36]); UAS-shotRod-RNAi (TRiP.GL01286, attP2, III, Bloomington Drosophila Stock Center # 41858), UASt-shot.L(A)Δrod-GFP (Bloomington Drosophila Stock Center # 29040 [7]); shotΔABD (aka shot[k03010], shot[kakP1]; Bloomington Drosophila Stock Center #10522 [6, 7, 27, 72]); nos-Gal4-VP16 (III, from Dr. Edwin Ferguson, the University of Chicago [73, 74]); UASp-LifeAct-TagRFP (II, 22A, Bloomington Drosophila Stock Center # 58713); UASp-LifeAct-TagRFP (III, 68E, Bloomington Drosophila Stock Center # 58714); UASp-RFP-Golgi (II, Bloomington Drosophila Stock Center # 30908, aka UASp-GalT-RFP [44]); mat αtub-RFP-Staufen (X, from Dr. Daniel St Johnson [45]); UASp-GFP-LD (II and III, from Dr. Michael Welte [46]); UASp-GFP-Patronin (II) (from Dr. Uri Abdu, Ben-Gurion University of the Negev [31, 51, 69]); UASp-EB1-GFP (II, from Dr. Antoine Guichet [52]); ubi-EB1-GFP [53, 75]; UASp-F-Tractin-tdTomato (II, Bloomington stock center #58989, [76]); UAS-Dhc64C-RNAi (TRiP.GL00543, attP40, II, Bloomington Drosophila Stock Center # 36583)[53]; UASp-Staufen-SunTag (III, [69]). The following fly stocks were generated in this study: UAS-shotABD-RNAi (in pWalium22 vector, inserted at attP2, III); UAS-shotEGC-RNAi (in pWalium22 vector, inserted at attP2, III); UASp-Mtio-MoxMaple3 (II and III).
METHOD DETAILS
Plasmid constructs.
The oligos of shotABD-shRNA (agtTGCGCGATGGTCACAATTTACtagttatattcaagcataGTAAATTGTGACCATCGCGCA gc) and shotEGC-shRNA (agtCCGGAAAATGGATAAGGATAAtagttatattcaagcataTTATCCTTATCCATTTTCCGGg c) were synthesized and inserted into the pWalium22 vector (Drosophila Genomics Resource Center, Stock Number #1473, 10XUAS) [77] by NheI(5’)/EcoRI(3’). shotABD-RNAi targeting sequences: TGCGCGATGGTCACAATTTAC; shotEGC-RNAi targeting sequences: CCGGAAAATGGATAAGGATAA.
MoxMaple3 was amplified by PCR from the pmCherry-T2A-moxMaple3 (Addgene Plasmid #120875) [78] and inserted into the pUASp vector by Spel (3’)/EcoRI (3’); mitochondria targeting probe, human Cox8a (mitochondrial cytochrome c oxidase subunit 8A) (1–29 residues, MSVLTPLLLRGLTGSARRLPVPRAKIHSL) (atgtccgtcctgacgccgctgctgctgcggggcttgacaggctcggcccggcggctcccagtgccgcgcgccaagatcc attcgttg) was synthesized and inserted into pUASp-MoxMaple3 by Kpnl(5’)/Spel(3’).
All the constructs were sent to BestGene for injection: PhiC31-mediated integration (UAS-shotABD-RNAi and UAS-shotEGC-RNAi in pWalium22 vectors, at attP2 site) and P-element transformation (UASp-Mito-MoxMaple3; two inserted lines were used in this study, II-F2 and III-M4).
Induction of shot[3]and shotΔEGC germline clones.
A standard recombination protocol was performed between FRTG13 and shotΔEGC. FRTG13 shot[3]/CyO or FRTG13 shotΔEGC/CyO virgin female flies were crossed with males carrying hs-flp[12]/y; FRTG13 ubi-GFP.nls/CyO. From these crosses, young pupae at day 7 and day 8 AEL (after egg laying) were subjected to heat shock at 37 °C for 2 hours each day. Non CyO F1 females were collected 3–4 day after heat shock and fattened with dry active yeast overnight before dissection for Orb staining.
Immunostaining of Drosophila egg chambers.
A standard fixation and staining protocol was used [74, 79]. Samples were incubated with mouse monoclonal anti-Orb antibody (Orb 4H8, Developmental Studies Hybridoma Bank, 1:5) or mouse monoclonal anti-Shot antibody (shot mAbRod1, Developmental Studies Hybridoma Bank, 1:5) at 4°C overnight, washed with 1XPBTB (1XPBS + 0.1% Triton X-100 + 0.2% BSA) five times for 10 min each time, incubated with FITC-conjugated or TRITC-conjugated anti-mouse secondary antibody (Jackson ImmunoResearch Laboratories, Inc; Cat# 115–095-062 and Cat# 115–025-003) at 1:100 at room temperature (24~25°C) for 4 h, and washed with 1XPBTB five times for 10 min each time. Some samples were stained with Rhodamine-conjugated phalloidin (1:5000) and DAPI (1 μg/mL) for 30 min before mounting. Samples were imaged on a Nikon A1plus scanning confocal microscopy with a GaAsP detector and a 20× 0.75 N.A. lens using Galvano scanning, a Nikon Eclipse U2000 inverted stand with a Yokogawa CSU10 spinning disk confocal head and a 40× 1.30 NA oil lens using an Evolve EMCCD, or Nikon W1 spinning disk confocal microscope (Yokogawa CSU with pinhole size 50 μm) with Photometrics Prime 95B sCMOS Camera and a 40 × 1.30 N.A. oil lens or a 40X 1.25 N.A. silicone oil lens, all controlled by Nikon Elements software. Images were acquired every 1 μm/step for whole ovariole imaging or 0.3~0.5 μm/step for individual egg chambers in z stacks.
Live imaging of Drosophila egg chamber.
Young mated female adults were fed with dry active yeast for 16~18 hours and then dissected in Halocarbon oil 700 (Sigma-Aldrich) as previously described [31, 69, 79]. Fluorescent samples were imaged using Nikon W1 spinning disk confocal microscope (Yokogawa CSU with pinhole size 50 μm) with Photometrics Prime 95B sCMOS Camera, and a 40 × 1.30 N.A. oil lens or a 40X 1.25 N.A. silicone oil lens, controlled by Nikon Elements software.
Microtubule Labeling by GFP-Patronin expression.
Ovaries from flies expressing GFP-Patronin under maternal αtub-Gal4[V37](with or without the UAS-shotEGC-RNAi) were dissected and fixed in 1XPBS +0.1%Triton X-100 +4% EM-grade formaldehyde for 20 min on the rotator; briefly washed with 1XPBTB five times and stained with Rhodamine-conjugated phalloidin (1:5000) for 30 min before mounting. Samples were imaged using Nikon W1 spinning disk confocal microscope (Yokogawa CSU with pinhole size 50 μm) with Photometrics Prime 95B sCMOS Camera, and a 40 × 1.30 N.A. oil lens, controlled by Nikon Elements software. Images were acquired every 0.3 μm/step in z stacks and 3D deconvoluted using Richardson-Lucy iterative algorithm provided by Nikon Elements. A maximum intensity projection of 0.6 μm z-stack sample (3 slices in the z-stacks) was used to present the microtubule distribution in each genotype.
Microtubule staining in control and in shot-RNAi.
Ovaries were dissected in 1XPBS and fixed in 1X Brinkley Renaturing Buffer 80 (BRB80), pH 6.8 (80 mM piperazine-N,N’-bis(2-ethanesulfonic acid) [PIPES]), 1 mM MgCl2, 1 mM EGTA) +0.1%Triton X-100 +8% EM-grade formaldehyde for 20 min on the rotator; briefly washed with 1XPBTB five times and stained with FITC-conjugated β-tubulin antibody (ProteinTech, Cat# CL488–66240) 1:100 at 4C overnight; then samples were stained rhodamine-conjugated phalloidin and DAPI for 30 min before mounting. Samples were imaged using Nikon W1 spinning disk confocal microscope (Yokogawa CSU with pinhole size 50 μm) with Photometries Prime 95B sCMOS Camera, and a 40X 1.25 N.A. silicone oil lens, controlled by Nikon Elements software. Images were acquired every 0.3 μm/step in z stacks and 3D deconvoluted using Richardson-Lucy iterative algorithm provided by Nikon Elements.
QUANTIFICATION AND STATISTICAL ANALYSIS
Measurement of cell size and ring canal diameter.
Ovaries from the control and shotEGC-RNAi flies carrying ubi-GFP-Pav were stained with rhodamine-conjugated phalloidin and DAPI. Z stacks of triple color images were acquired, and cell areas were specified (at the largest cross-sections) and measured by manual polygon selection (area size) or line selection (ring canal diameter) in FIJI [80].
Measurement of actin signal ratio at ring canals.
Ovaries from flies expressing LifeAct-TagRFP under nos-Gal4-VP16 were dissected and fixed in 1XPBS +0.1%Triton X-100 +4% EM-grade formaldehyde for 20 min on the rotator; briefly washed with 1XPBTB five times before mounting. Z-stack images were acquired at every 0.5 μm, and Z projection of sum slices for each individual ring canal was used for measurement. The ring canals are categorized into four groups dependent on its distance to the oocyte (Figure 4E, bottom) : (1) nurse cell-oocyte ring canals, directly connected to the oocyte (O ring, green); (2) posterior nurse cell-nurse cell ring canal, having one nurse cell between this ring canal and the oocyte (P ring, orange); (3) middle nurse cell-nurse cell ring canal, having two nurse cells between this ring canal and the oocyte (M ring, magenta); (4) anterior nurse cell-nurse cell ring canal, having three nurse cells between this ring canal and the oocyte (A ring, blue). For each Z-projection image, fluorescence intensities of four following areas were measured: (1) the anterior side of the ring canal; (2) the posterior side of the ring canal; (3) the background of the anterior side; (4) the background of the posterior side. The ratio of LifeAct-TagRFP signal of the anterior side over the posterior side of a ring canal is calculated = [(1) the anterior side of the ring canal – (3) the background of the anterior side] / [(2) the posterior side of the ring canal – (4) the background of the posterior side].
Quantification of cargo transport and MT polarity.
Kymographs were created along a ~3.7 μm-width line (for cargo transport) or a 3.5 μm ~5.0 μm -width line (for microtubule polarity) from the nurse cell to the oocyte through the ring canal (labeled by GFP-Pav or F-Tractin-tdTomato) using the MultipleKymograph plugin in FIJI [80]. Cargo movement direction and microtubule polarity were manually quantified based on these kymographs.
Statistical analysis.
The plots in figures show percentage of phenotypes, or average values, as indicated in figure legends. Error bars in figures represent 95% confidence intervals, unless it is specified elsewhere in figure legends. N stands for number of samples examined in each assay. Unpaired t tests or unpaired t tests with Welch’s correction were performed in GraphPad Prism 8.0.2, while chi-square tests were performed using Chi-Square Calculator (https://www.socscistatistics.com/tests/chisquare/default2.aspx). P values and levels of significance are listed in figure legends
Supplementary Material
Video S1. Shot is required for oocyte growth. Related to Figure 1.
Representative Z-stack images of phalloidin staining in control (yw; mat αtub-Gal4[V37]/+), shotABD-RNAi (yw; mat αtub-Gal4[V37]/UAS-shotABD-RNAi), and shotEGC-RNAi (yw; mat αtub-Gal4[V37]/UAS-shotEGC-RNAi). Oocytes are highlighted with the orange arrows. Scale bars, 50 μm.
Video S2. Shot is required to maintain the oocyte identity. Related to Figure 1.
Representative Z-stack images of Orb staining with GFP-Pav in control (yw; ubi-GFP-Pav/+; mat αtub-Gal4[V37]/+), shotABD-RNAi (yw; ubi-GFP-Pav/+; mat αtub-Gal4[V37]/UAS-shotABD-RNAi), and shotEGC-RNAi (yw; ubi-GFP-Pav/+; mat αtub-Gal4[V37]/UAS-shotEGC-RNAi). Oocytes are highlighted with the orange arrows. Scale bars, 50 μm.
Video S3. Golgi transport in control and in shot-RNAi. Related to Figure 3.
Golgi units are labeled with RFP-Golgi (GalT-RFP) and ring canals are labeled with GFP-tagged kinesin-6/Pavarotti in control (yw; UASp-RFP-Golgi/ubi-GFP-Pav; mat αtub-Gal4[V37]/+) and in shot-RNAi (yw; UASp-RFP-Golgi/ubi-GFP-Pav; mat αtub-Gal4[V37]/UAS-shotGC-RNAi). Egg chambers are oriented as the anterior to the left and the posterior to the right. Time lapses images were acquired every 5 sec for 5 min; scale bars, 50 μm.
Video S4. Staufen transport in control and in shot-RNAi. Related to Figure 3.
Staufen particles are labeled with RFP-Staufen and ring canals are labeled with GFP-tagged kinesin-6/Pavarotti in control (w, mat αtub-RFP-Staufen/yw; ubi-GFP-Pav/+; mat αtub-Gal4[V37]/+) and in shot-RNAi (w, mat αtub-RFP-Staufen/yw; ubi-GFP-Pav/+; mat αtub-Gal4[V37]/UAS-shotEGC-RNAi). Egg chambers are oriented as the anterior to the left and the posterior to the right. Time lapses images were acquired every 5 sec for 5 min; scale bars, 50 μm.
Video S5. Local photoconversion of mitochondria in the nurse cell and in the oocyte. Related to Figure 3.
Mitochondria are labeled with Mito-MoxMaple3 and a subset of mitochondria were photoconverted from green to red by local UV exposure, either in the posterior-most nurse cell or in the oocyte of stage 9 egg chambers. The ring canals are labeled with GFP-tagged kinesin-6/Pavarotti (yw; ubi-GFP-Pav/UASp-Mito-MoxMaple3 (F2); mat αtub-Gal4[V37]/+) and shown as a pair of orange arrowheads in the zoom-in areas. Egg chambers are oriented as the anterior to the left and the posterior to the right. A max intensity projection of Z-stack images of total 4 μm was shown for both green and red channel. Time lapses images were acquired every 30 sec for 20 min; scale bars, 50 μm.
Video S6. Mitochondria transport in control and in shot-RNAi. Related to Figure 3.
Mitochondria are labeled with by Mito-MoxMaple3 (red, after global photoconversion) and ring canals are labeled with GFP-tagged kinesin-6/Pavarotti in control (yw; ubi-GFP-Pav/UASp-Mito-MoxMaple3 (F2); mat αtub-Gal4[V37]/+) and in shot-RNAi (yw; ubi-GFP-Pav/UASp-Mito-MoxMaple3 (F2); mat αtub-Gal4[V37]/UAS-shotEGC-RNAi). Egg chambers are oriented as the anterior to the left and the posterior to the right. Time lapses images were acquired every 5 sec for 5 min; scale bars, 50 μm.
Video S7. Lipid droplet transport in control and in shot-RNAi. Related to Figure 3.
Lipid droplets are labeled with GFP-tagged lipid droplet domain of Drosophila protein Klar (GFP-LD) and ring canals are labeled with tdTomato-tagged actin-binding domain of rat inositol triphosphate 3-kinase (F-tractin-tdTomato) in control (yw; UASp-F-tractin-tdTomato/UASp-GFP-LD; mat αtub-Gal4[V37]/+) and in shot-RNAi (yw; UASp-F-tractin-tdTomato/UASp-GFP-LD; mat αtub-Gal4[V37]/UAS-shotEGC-RNAi). Egg chambers are oriented as the anterior to the left and the posterior to the right. Time lapses images were acquired every 5 sec for 5 min; scale bars, 50 μm.
Video S8. Asymmetric actin fibers at the nurse cell-oocyte ring canals. Related to Figure 4.
F-actin is labeled with LifeAct-TagRFP in the live sample (w; UASp-LifeAct-TagRFP/+; nos-Gal4-VP16/+) and asymmetric actin fibers are visible at all four ring canals connecting four nurse cells with the same oocyte (highlighted by the shaded boxes). Scale bar, 10 μm.
Video S9. EB1-GFP comets in control and in shot-RNAi. Related to Figure 5.
Microtubule plus-ends are labeled with EB1-GFP and ring canals are labeled with GFP-tagged kinesin-6/Pavarotti in control (yw; UASp-EBI-GFP/ubi-GFP-Pav; mat αtub-Gal4[V37]/+) and in shot-RNAi (yw; UASp-EBI-GFP/ubi-GFP-Pav; mat αtub-Gal4[V37]/UAS-shotEGC-RNAi). N, nurse cell; O, oocyte; time lapses images were acquired every 2 sec for 2 min; scale bars, 10 μm.
Video S10. EB1 comets and actin fibers at the nurse cell-oocyte ring canal. Related to Figure 5.
Microtubule plus-ends are labeled with EB1-GFP and F-actin is labeled with LifeAct-TagRFP (w; UASp-LifeAct-TagRFP/+; nos-Gal4-VP16/ubi-EB1-GFP). The first half imaging focused on the nurse cell-oocyte ring canal cross-section and the second half imaging focused on asymmetric actin fibers slightly above the ring canal cross-section. Time lapses images were acquired every 2 sec for 2 min; scale bars, 10 μm.
Video S11. Mitochondria movement in control and in dynein-RNAi. Related to Figure 6. Mitochondria are labeled with Mito-MoxMaple3 (red, after global photoconversion) and ring canals are labeled with GFP-tagged kinesin-6/Pavarotti in control (yw; ubi-GFP-Pav/+; mat αtub-Gal4[V37]/UASp-Mito-MoxMaple3 (M4)) and in dynein-RNAi (yw; ubi-GFP-Pav/UAS-Dhc64C-RNAi; mat αtub-Gal4[V37]/UASp-Mito- MoxMaple3(M4)). Egg chambers are oriented as the anterior to the left and the posterior to the right. Time lapses images were acquired every 5 sec for 5 min; scale bars, 50 μm.
Table 1. Summary of directions of two types of cargoes (Golgi units and Staufen RNP particles) at different ring canals.
Golgi units are labeled with RFP-Golgi [44] and osk/Staufen RNP particles are labeled with Staufen-SunTag [69] driven by nos-Gal4-VP16. Number of events are divided into two groups: “towards the oocyte” (moving towards the posterior) and “away from the oocyte” (moving towards the anterior). Highest directionality of both Golgi and osk/Staufen transport was observed in the nurse cell-oocyte ring canals.
| Cargo type | Nurse cell-oocyte ring canals (O) No. of events (percentage) |
Nurse cell ring canals (P) No. of events (percentage) |
Nurse cell ring canal (M) No. of events (percentage) |
|||
|---|---|---|---|---|---|---|
| Towards the oocyte | Away from the oocyte | Towards the oocyte | Away from the oocyte | Towards the oocyte | Away from the oocyte | |
| Golgi units | 74 (89.2%) |
9 (10.8%) |
41 (80.4%) |
10 (19.6%) |
38 (53.5%) |
33 (46.5%) |
| osk/Staufen RNPs | 57 (90.5%) |
6 (9.5%) |
30 (81.1%) |
7 (18.9%) |
n.d. | n.d. |
A Drosophila oocyte grow more than a hundred thousand times without transcriptional activity. Here Lu et al. show that an actin-microtubule cross-linker, Short Stop, functions as a gatekeeper at cytoplasmic bridges between supporting nurse cells and the oocyte, directing transport of cytoplasmic components to the oocyte, thus driving its growth.
An actin-microtubule cross-linker, Short stop, is required for oocyte growth
Both actin and microtubule binding activities of Short stop are essential
Short stop controls direction of transport between nurse cells and the oocyte
Short stop at asymmetric actin fibers organizes microtubules in the ring canals
ACKNOWLEDGEMENTS
We would like to thank Dr. David Glover (Caltech) for ubi-GFP-Pav line, Dr. Derek Applewhite (Reed College) for full-length shot cDNA (shot.LA) and ubi-EB1-GFP line, Dr. Ferenc Jankovics (Institute of Genetics, Biological Research Centre of the Hungarian Academy of Sciences) for shotΔEGC line, Dr. Daniel St Johnston (University of Cambridge) for mat αtub-RFP-Staufen line, Dr. Vladislav Verkhusha (Albert Einstein College of Medicine) for MoxMaple3 construct, Dr. Michael Welte (University of Rochester) for UASp-GFP-LD line, Dr. Uri Abdu (Ben-Gurion University of the Negev) for UASp-GFP-Patronin line, Dr. Antoine Guichet (CNRS, Institut Jacques Monod) for UASp-EB1-GFP line, the Bloomington Drosophila Stock Center (supported by National Institutes of Health grant P40OD018537) for fly stocks, and Drosophila Genomics Resource Center (supported by National Institutes of Health Grant 2P40OD01094) for DNA constructs. The Orb 4H8 monoclonal antibody developed by Dr. Paul D. Schedl’s group at Princeton University, and anti-Shot mAbRod1 antibody developed by Dr. Peter A. Kolodziej’s group at Vanderbilt University were obtained from the Developmental Studies Hybridoma Bank, created by the NICHD of the NIH and maintained at The University of Iowa. We also thank all the Gelfand laboratory members for support, discussion, and suggestions. W. Lu dedicates this work to memory of her college Genetics teacher, Professor Zhuo-hua Dai at Peking University.
Research reported in this study was supported by the National Institute of General Medical Sciences grants R01GM124029 and R35GM131752 to V.I. Gelfand.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Zhang J, Yue J, and Wu X (2017). Spectraplakin family proteins - cytoskeletal crosslinkers with versatile roles. Journal of cell science 130, 2447–2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Suozzi KC, Wu X, and Fuchs E (2012). Spectraplakins: master orchestrators of cytoskeletal dynamics. The Journal of cell biology 197, 465–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roper K, Gregory SL, and Brown NH (2002). The ‘Spectraplakins’: cytoskeletal giants with characteristics of both spectrin and plakin families. Journal of cell science 115, 4215–4225. [DOI] [PubMed] [Google Scholar]
- 4.Applewhite DA, Grode KD, Keller D, Zadeh AD, Slep KC, and Rogers SL (2010). The spectraplakin Short stop is an actin-microtubule cross-linker that contributes to organization of the microtubule network. Molecular biology of the cell 21, 1714–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alves-Silva J, Sanchez-Soriano N, Beaven R, Klein M, Parkin J, Millard TH, Bellen HJ, Venken KJ, Ballestrem C, Kammerer RA, et al. (2012). Spectraplakins promote microtubule-mediated axonal growth by functioning as structural microtubule-associated proteins and EB1-dependent +TIPs (tip interacting proteins). The Journal of neuroscience : the official journal of the Society for Neuroscience 32, 9143–9158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee S, Harris KL, Whitington PM, and Kolodziej PA (2000). short stop is allelic to kakapo, and encodes rod-like cytoskeletal-associated proteins required for axon extension. Journal of Neuroscience 20, 1096–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee S, and Kolodziej PA (2002). Short Stop provides an essential link between F-actin and microtubules during axon extension. Development 129, 1195–1204. [DOI] [PubMed] [Google Scholar]
- 8.Voelzmann A, Liew YT, Qu Y, Hahn I, Melero C, Sanchez-Soriano N, and Prokop A (2017). Drosophila Short stop as a paradigm for the role and regulation of spectraplakins. Seminars in cell & developmental biology 69, 40–57. [DOI] [PubMed] [Google Scholar]
- 9.Prokop A, Uhler J, Roote J, and Bate M (1998). The kakapo mutation affects terminal arborization and central dendritic sprouting of Drosophila motorneurons. The Journal of cell biology 143, 1283–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanchez-Soriano N, Travis M, Dajas-Bailador F, Goncalves-Pimentel C, Whitmarsh AJ, and Prokop A (2009). Mouse ACF7 and drosophila short stop modulate filopodia formation and microtubule organisation during neuronal growth. Journal of cell science 122, 2534–2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanchez-Soriano N, Goncalves-Pimentel C, Beaven R, Haessler U, Ofner-Ziegenfuss L, Ballestrem C, and Prokop A (2010). Drosophila growth cones: a genetically tractable platform for the analysis of axonal growth dynamics. Developmental neurobiology 70, 58–71. [DOI] [PubMed] [Google Scholar]
- 12.Lee M, Lee S, Zadeh AD, and Kolodziej PA (2003). Distinct sites in E-cadherin regulate different steps in Drosophila tracheal tube fusion. Development 130, 5989–5999. [DOI] [PubMed] [Google Scholar]
- 13.Lee S, and Kolodziej PA (2002). The plakin Short Stop and the RhoA GTPase are required for E-cadherin-dependent apical surface remodeling during tracheal tube fusion. Development 129, 1509–1520. [DOI] [PubMed] [Google Scholar]
- 14.Ricolo D, and Araujo SJ (2020). Coordinated crosstalk between microtubules and actin by a spectraplakin regulates lumen formation and branching. eLife 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roper K, and Brown NH (2003). Maintaining epithelial integrity: a function for gigantic spectraplakin isoforms in adherens junctions. The Journal of cell biology 162, 1305–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuss B, Josten F, Feix M, and Hoch M (2004). Cell movements controlled by the Notch signalling cascade during foregut development in Drosophila. Development 131, 1587–1595. [DOI] [PubMed] [Google Scholar]
- 17.Mui UN, Lubczyk CM, and Nam SC (2011). Role of spectraplakin in Drosophila photoreceptor morphogenesis. PloS one 6, e25965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun T, Song Y, Dai J, Mao D, Ma M, Ni JQ, Liang X, and Pastor-Pareja JC (2019). Spectraplakin Shot Maintains Perinuclear Microtubule Organization in Drosophila Polyploid Cells. Developmental cell 49, 731–747 e737. [DOI] [PubMed] [Google Scholar]
- 19.Wang S, Reuveny A, and Volk T (2015). Nesprin provides elastic properties to muscle nuclei by cooperating with spectraplakin and EB1. The Journal of cell biology 209, 529–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takacs Z, Jankovics F, Vilmos P, Lenart P, Roper K, and Erdelyi M (2017). The spectraplakin Short stop is an essential microtubule regulator involved in epithelial closure in Drosophila. Journal of cell science 130, 712–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hinnant TD, Merkle JA, and Ables ET (2020). Coordinating Proliferation, Polarity, and Cell Fate in the Drosophila Female Germline. Front Cell Dev Biol 8, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Navarro-Costa P, McCarthy A, Prudencio P, Greer C, Guilgur LG, Becker JD, Secombe J, Rangan P, and Martinho RG (2016). Early programming of the oocyte epigenome temporally controls late prophase I transcription and chromatin remodelling. Nature communications 7, 12331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buszczak M, and Cooley L (2000). Eggs to die for: cell death during Drosophila oogenesis. Cell Death Differ 7, 1071–1074. [DOI] [PubMed] [Google Scholar]
- 24.Mische S, Li MG, Serr M, and Hays TS (2007). Direct observation of regulated ribonucleoprotein transport across the nurse cell/oocyte boundary. Molecular biology of the cell 18, 2254–2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nicolas E, Chenouard N, Olivo-Marin JC, and Guichet A (2009). A dual role for actin and microtubule cytoskeleton in the transport of Golgi units from the nurse cells to the oocyte across ring canals. Molecular biology of the cell 20, 556–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clark A, Meignin C, and Davis I (2007). A Dynein-dependent shortcut rapidly delivers axis determination transcripts into the Drosophila oocyte. Development 134, 1955–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roper K, and Brown NH (2004). A Spectraplakin Is Enriched on the Fusome and Organizes Microtubules during Oocyte Specification in Drosophila. Current Biology 14, 99–110. [PubMed] [Google Scholar]
- 28.Lin H, Yue L, and Spradling AC (1994). The Drosophila fusome, a germline-specific organelle, contains membrane skeletal proteins and functions in cyst formation. Development 120, 947–956. [DOI] [PubMed] [Google Scholar]
- 29.Nashchekin D, Fernandes AR, and St Johnston D (2016). Patronin/Shot Cortical Foci Assemble the Noncentrosomal Microtubule Array that Specifies the Drosophila Anterior-Posterior Axis. Developmental cell 38, 61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee J, Lee S, Chen C, Shim H, and Kim-Ha J (2016). shot regulates the microtubule reorganization required for localization of axis-determining mRNAs during oogenesis. FEBS letters 590, 431–444. [DOI] [PubMed] [Google Scholar]
- 31.Lu W, Lakonishok M, Liu R, Billington N, Rich A, Glotzer M, Sellers JR, and Gelfand VI (2020). Competition between kinesin-1 and myosin-V defines Drosophila posterior determination. eLife 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lantz V, Chang JS, Horabin JI, Bopp D, and Schedl P (1994). The Drosophila Orb Rna-Binding Protein Is Required for the Formation of the Egg Chamber and Establishment of Polarity. Genes & development 8, 598–613. [DOI] [PubMed] [Google Scholar]
- 33.Sanghavi P, Liu G, Veeranan-Karmegam R, Navarro C, and Gonsalvez GB (2016). Multiple Roles for Egalitarian in Polarization of the Drosophila Egg Chamber. Genetics 203, 415–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Staller MV, Yan D, Randklev S, Bragdon MD, Wunderlich ZB, Tao R, Perkins LA, DePace AH, and Perrimon N (2013). Depleting Gene Activities in Early Drosophila Embryos with the “Maternal-Gal4-shRNA” System. Genetics 193, 51–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hudson AM, and Cooley L (2014). Methods for studying oogenesis. Methods 68, 207–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Minestrini G, Mathe E, and Glover DM (2002). Domains of the Pavarotti kinesin-like protein that direct its subcellular distribution: effects of mislocalisation on the tubulin and actin cytoskeleton during Drosophila oogenesis. Journal of cell science 115, 725–736. [DOI] [PubMed] [Google Scholar]
- 37.Applewhite DA, Grode KD, Duncan MC, and Rogers SL (2013). The actin-microtubule cross-linking activity of Drosophila Short stop is regulated by intramolecular inhibition. Molecular biology of the cell 24, 2885–2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Januschke J, Nicolas E, Compagnon J, Formstecher E, Goud B, and Guichet A (2007). Rab6 and the secretory pathway affect oocyte polarity in Drosophila. Development 134, 3419–3425. [DOI] [PubMed] [Google Scholar]
- 39.Lasko P (2012). mRNA Localization and Translational Control in Drosophila Oogenesis. Cold Spring Harbor perspectives in biology 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cox RT, and Spradling AC (2003). A Balbiani body and the fusome mediate mitochondrial inheritance during Drosophila oogenesis. Development 130, 1579–1590. [DOI] [PubMed] [Google Scholar]
- 41.Cox RT, and Spradling AC (2006). Milton controls the early acquisition of mitochondria by Drosophila oocytes. Development 133, 3371–3377. [DOI] [PubMed] [Google Scholar]
- 42.Hurd TR, Herrmann B, Sauerwald J, Sanny J, Grosch M, and Lehmann R (2016). Long Oskar Controls Mitochondrial Inheritance in Drosophila melanogaster. Developmental cell 39, 560–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Welte MA (2015). As the fat flies: The dynamic lipid droplets of Drosophila embryos. Bba-Mol Cell Biol L 1851, 1156–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chowdhary S, Tomer D, Dubal D, Sambre D, and Rikhy R (2017). Analysis of mitochondrial organization and function in the Drosophila blastoderm embryo. Scientific reports 7, 5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parton RM, Hamilton RS, Ball G, Yang L, Cullen CF, Lu W, Ohkura H, and Davis I (2011). A PAR-1-dependent orientation gradient of dynamic microtubules directs posterior cargo transport in the Drosophila oocyte. The Journal of cell biology 194, 121–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu YXV, Li ZH, Rizzo NP, Einstein J, and Welte MA (2011). Targeting the motor regulator Klar to lipid droplets. Bmc Cell Biol 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Riparbelli MG, and Callaini G (1995). Cytoskeleton of the Drosophila egg chamber: new observations on microfilament distribution during oocyte growth. Cell motility and the cytoskeleton 31, 298–306. [DOI] [PubMed] [Google Scholar]
- 48.Riedl J, Crevenna AH, Kessenbrock K, Yu JH, Neukirchen D, Bista M, Bradke F, Jenne D, Holak TA, Werb Z, et al. (2008). Lifeact: a versatile marker to visualize F-actin. Nature methods 5, 605–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Becalska AN, and Gavis ER (2009). Lighting up mRNA localization in Drosophila oogenesis. Development 136, 2493–2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goodwin SS, and Vale RD (2010). Patronin Regulates the Microtubule Network by Protecting Microtubule Minus Ends. Cell 143, 263–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baskar R, Bakrhat A, and Abdu U (2019). GFP-Forked, a genetic reporter for studying Drosophila oocyte polarity. Biology open 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Legent K, Tissot N, and Guichet A (2015). Visualizing Microtubule Networks During Drosophila Oogenesis Using Fixed and Live Imaging. Methods in molecular biology 1328, 99–112. [DOI] [PubMed] [Google Scholar]
- 53.Del Castillo U, Winding M, Lu W, and Gelfand VI (2015). Interplay between kinesin-1 and cortical dynein during axonal outgrowth and microtubule organization in Drosophila neurons. eLife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jia D, Xu Q, Xie Q, Mio W, and Deng WM (2016). Automatic stage identification of Drosophila egg chamber based on DAPI images. Scientific reports 6, 18850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cooley L, Verheyen E, and Ayers K (1992). chickadee encodes a profilin required for intercellular cytoplasm transport during Drosophila oogenesis. Cell 69, 173–184. [DOI] [PubMed] [Google Scholar]
- 56.Guild GM, Connelly PS, Shaw MK, and Tilney LG (1997). Actin filament cables in Drosophila nurse cells are composed of modules that slide passively past one another during dumping. Journal of Cell Biology 138, 783–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cant K, Knowles BA, Mooseker MS, and Cooley L (1994). Drosophila singed, a fascin homolog, is required for actin bundle formation during oogenesis and bristle extension. The Journal of cell biology 125, 369–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dahlgaard K, Raposo AA, Niccoli T, and St Johnston D (2007). Capu and Spire assemble a cytoplasmic actin mesh that maintains microtubule organization in the Drosophila oocyte. Developmental cell 13, 539–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Quinlan ME, Heuser JE, Kerkhoff E, and Mullins RD (2005). Drosophila Spire is an actin nucleation factor. Nature 433, 382–388. [DOI] [PubMed] [Google Scholar]
- 60.Quinlan ME, Hilgert S, Bedrossian A, Mullins RD, and Kerkhoff E (2007). Regulatory interactions between two actin nucleators, Spire and Cappuccino. The Journal of cell biology 179, 117–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bernard F, Lepesant JA, and Guichet A (2018). Nucleus positioning within Drosophila egg chamber. Seminars in cell & developmental biology 82, 25–33. [DOI] [PubMed] [Google Scholar]
- 62.Imran Alsous J, Villoutreix P, Berezhkovskii AM, and Shvartsman SY (2017). Collective Growth in a Small Cell Network. Current biology : CB 27, 2670–2676 e2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Doherty CA, Diegmiller R, Kapasiawala M, Gavis ER, and Shvartsman SY (2021). Coupled oscillators coordinate collective germline growth. Developmental cell 56, 860–870 e868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hahn I, Voelzmann A, Parkin J, Fuelle J, Slater PG, Lowery LA, Sanchez-Soriano N, and Prokop A (2020). Tau, XMAP215/Msps and Eb1 jointly regulate microtubule polymerisation and bundle formation in axons. bioRxiv, 2020.2008.2019.257808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Subramanian A, Prokop A, Yamamoto M, Sugimura K, Uemura T, Betschinger J, Knoblich JA, and Volk T (2003). Shortstop recruits EB1/APC1 and promotes microtubule assembly at the muscle-tendon junction. Current biology : CB 13, 1086–1095. [DOI] [PubMed] [Google Scholar]
- 66.Robinson DN, and Cooley L (1996). Stable intercellular bridges in development: the cytoskeleton lining the tunnel. Trends in cell biology 6, 474–479. [DOI] [PubMed] [Google Scholar]
- 67.Haglund K, Nezis IP, and Stenmark H (2011). Structure and functions of stable intercellular bridges formed by incomplete cytokinesis during development. Communicative & integrative biology 4, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lei L, and Spradling AC (2016). Mouse oocytes differentiate through organelle enrichment from sister cyst germ cells. Science 352, 95–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lu W, Lakonishok M, Serpinskaya AS, Kirchenbuechler D, Ling SC, and Gelfand VI (2018). Ooplasmic flow cooperates with transport and anchorage in Drosophila oocyte posterior determination. The Journal of cell biology 217, 3497–3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chou TB, and Perrimon N (1996). The autosomal FLP-DFS technique for generating germline mosaics in Drosophila melanogaster. Genetics 144, 1673–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kolodziej PA, Jan LY, and Jan YN (1995). Mutations That Affect the Length, Fasciculation, or Ventral Orientation of Specific Sensory Axons in the Drosophila Embryo. Neuron 15, 273–286. [DOI] [PubMed] [Google Scholar]
- 72.Gregory SL, and Brown NH (1998). kakapo, a gene required for adhesion between and within cell layers in Drosophila, encodes a large cytoskeletal linker protein related to plectin and dystrophin. Journal of Cell Biology 143, 1271–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Van Doren M, Williamson AL, and Lehmann R (1998). Regulation of zygotic gene expression in Drosophila primordial germ cells. Current biology : CB 8, 243–246. [DOI] [PubMed] [Google Scholar]
- 74.Lu W, Casanueva MO, Mahowald AP, Kato M, Lauterbach D, and Ferguson EL (2012). Niche-associated activation of rac promotes the asymmetric division of Drosophila female germline stem cells. PLoS biology 10, e1001357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shimada Y, Yonemura S, Ohkura H, Strutt D, and Uemura T (2006). Polarized transport of Frizzled along the planar microtubule arrays in Drosophila wing epithelium. Developmental cell 10, 209–222. [DOI] [PubMed] [Google Scholar]
- 76.Spracklen AJ, Fagan TN, Lovander KE, and Tootle TL (2014). The pros and cons of common actin labeling tools for visualizing actin dynamics during Drosophila oogenesis. Developmental biology 393, 209–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ni JQ, Zhou R, Czech B, Liu LP, Holderbaum L, Yang-Zhou D, Shim HS, Tao R, Handler D, Karpowicz P, et al. (2011). A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nature methods 8, 405–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kaberniuk AA, Mohr MA, Verkhusha VV, and Snapp EL (2018). moxMaple3: a Photoswitchable Fluorescent Protein for PALM and Protein Highlighting in Oxidizing Cellular Environments. Scientific reports 8, 14738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lu W, Winding M, Lakonishok M, Wildonger J, and Gelfand VI (2016). Microtubule-microtubule sliding by kinesin-1 is essential for normal cytoplasmic streaming in Drosophila oocytes. Proceedings of the National Academy of Sciences of the United States of America 113, E4995–5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nature methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video S1. Shot is required for oocyte growth. Related to Figure 1.
Representative Z-stack images of phalloidin staining in control (yw; mat αtub-Gal4[V37]/+), shotABD-RNAi (yw; mat αtub-Gal4[V37]/UAS-shotABD-RNAi), and shotEGC-RNAi (yw; mat αtub-Gal4[V37]/UAS-shotEGC-RNAi). Oocytes are highlighted with the orange arrows. Scale bars, 50 μm.
Video S2. Shot is required to maintain the oocyte identity. Related to Figure 1.
Representative Z-stack images of Orb staining with GFP-Pav in control (yw; ubi-GFP-Pav/+; mat αtub-Gal4[V37]/+), shotABD-RNAi (yw; ubi-GFP-Pav/+; mat αtub-Gal4[V37]/UAS-shotABD-RNAi), and shotEGC-RNAi (yw; ubi-GFP-Pav/+; mat αtub-Gal4[V37]/UAS-shotEGC-RNAi). Oocytes are highlighted with the orange arrows. Scale bars, 50 μm.
Video S3. Golgi transport in control and in shot-RNAi. Related to Figure 3.
Golgi units are labeled with RFP-Golgi (GalT-RFP) and ring canals are labeled with GFP-tagged kinesin-6/Pavarotti in control (yw; UASp-RFP-Golgi/ubi-GFP-Pav; mat αtub-Gal4[V37]/+) and in shot-RNAi (yw; UASp-RFP-Golgi/ubi-GFP-Pav; mat αtub-Gal4[V37]/UAS-shotGC-RNAi). Egg chambers are oriented as the anterior to the left and the posterior to the right. Time lapses images were acquired every 5 sec for 5 min; scale bars, 50 μm.
Video S4. Staufen transport in control and in shot-RNAi. Related to Figure 3.
Staufen particles are labeled with RFP-Staufen and ring canals are labeled with GFP-tagged kinesin-6/Pavarotti in control (w, mat αtub-RFP-Staufen/yw; ubi-GFP-Pav/+; mat αtub-Gal4[V37]/+) and in shot-RNAi (w, mat αtub-RFP-Staufen/yw; ubi-GFP-Pav/+; mat αtub-Gal4[V37]/UAS-shotEGC-RNAi). Egg chambers are oriented as the anterior to the left and the posterior to the right. Time lapses images were acquired every 5 sec for 5 min; scale bars, 50 μm.
Video S5. Local photoconversion of mitochondria in the nurse cell and in the oocyte. Related to Figure 3.
Mitochondria are labeled with Mito-MoxMaple3 and a subset of mitochondria were photoconverted from green to red by local UV exposure, either in the posterior-most nurse cell or in the oocyte of stage 9 egg chambers. The ring canals are labeled with GFP-tagged kinesin-6/Pavarotti (yw; ubi-GFP-Pav/UASp-Mito-MoxMaple3 (F2); mat αtub-Gal4[V37]/+) and shown as a pair of orange arrowheads in the zoom-in areas. Egg chambers are oriented as the anterior to the left and the posterior to the right. A max intensity projection of Z-stack images of total 4 μm was shown for both green and red channel. Time lapses images were acquired every 30 sec for 20 min; scale bars, 50 μm.
Video S6. Mitochondria transport in control and in shot-RNAi. Related to Figure 3.
Mitochondria are labeled with by Mito-MoxMaple3 (red, after global photoconversion) and ring canals are labeled with GFP-tagged kinesin-6/Pavarotti in control (yw; ubi-GFP-Pav/UASp-Mito-MoxMaple3 (F2); mat αtub-Gal4[V37]/+) and in shot-RNAi (yw; ubi-GFP-Pav/UASp-Mito-MoxMaple3 (F2); mat αtub-Gal4[V37]/UAS-shotEGC-RNAi). Egg chambers are oriented as the anterior to the left and the posterior to the right. Time lapses images were acquired every 5 sec for 5 min; scale bars, 50 μm.
Video S7. Lipid droplet transport in control and in shot-RNAi. Related to Figure 3.
Lipid droplets are labeled with GFP-tagged lipid droplet domain of Drosophila protein Klar (GFP-LD) and ring canals are labeled with tdTomato-tagged actin-binding domain of rat inositol triphosphate 3-kinase (F-tractin-tdTomato) in control (yw; UASp-F-tractin-tdTomato/UASp-GFP-LD; mat αtub-Gal4[V37]/+) and in shot-RNAi (yw; UASp-F-tractin-tdTomato/UASp-GFP-LD; mat αtub-Gal4[V37]/UAS-shotEGC-RNAi). Egg chambers are oriented as the anterior to the left and the posterior to the right. Time lapses images were acquired every 5 sec for 5 min; scale bars, 50 μm.
Video S8. Asymmetric actin fibers at the nurse cell-oocyte ring canals. Related to Figure 4.
F-actin is labeled with LifeAct-TagRFP in the live sample (w; UASp-LifeAct-TagRFP/+; nos-Gal4-VP16/+) and asymmetric actin fibers are visible at all four ring canals connecting four nurse cells with the same oocyte (highlighted by the shaded boxes). Scale bar, 10 μm.
Video S9. EB1-GFP comets in control and in shot-RNAi. Related to Figure 5.
Microtubule plus-ends are labeled with EB1-GFP and ring canals are labeled with GFP-tagged kinesin-6/Pavarotti in control (yw; UASp-EBI-GFP/ubi-GFP-Pav; mat αtub-Gal4[V37]/+) and in shot-RNAi (yw; UASp-EBI-GFP/ubi-GFP-Pav; mat αtub-Gal4[V37]/UAS-shotEGC-RNAi). N, nurse cell; O, oocyte; time lapses images were acquired every 2 sec for 2 min; scale bars, 10 μm.
Video S10. EB1 comets and actin fibers at the nurse cell-oocyte ring canal. Related to Figure 5.
Microtubule plus-ends are labeled with EB1-GFP and F-actin is labeled with LifeAct-TagRFP (w; UASp-LifeAct-TagRFP/+; nos-Gal4-VP16/ubi-EB1-GFP). The first half imaging focused on the nurse cell-oocyte ring canal cross-section and the second half imaging focused on asymmetric actin fibers slightly above the ring canal cross-section. Time lapses images were acquired every 2 sec for 2 min; scale bars, 10 μm.
Video S11. Mitochondria movement in control and in dynein-RNAi. Related to Figure 6. Mitochondria are labeled with Mito-MoxMaple3 (red, after global photoconversion) and ring canals are labeled with GFP-tagged kinesin-6/Pavarotti in control (yw; ubi-GFP-Pav/+; mat αtub-Gal4[V37]/UASp-Mito-MoxMaple3 (M4)) and in dynein-RNAi (yw; ubi-GFP-Pav/UAS-Dhc64C-RNAi; mat αtub-Gal4[V37]/UASp-Mito- MoxMaple3(M4)). Egg chambers are oriented as the anterior to the left and the posterior to the right. Time lapses images were acquired every 5 sec for 5 min; scale bars, 50 μm.
Data Availability Statement
The published article includes all dataset generated and analyzed during this study.


