Abstract
Cancer immunotherapies targeting the interaction between Programmed death 1 (PD-1) and Programmed death ligand 1 (PD-L1) have recently been approved for the treatment of multiple cancer types, including gastric cancer. However, not all patients respond to these therapies, while some eventually acquire resistance. A partial predictive biomarker for positive response to PD-1/PD-L1 therapy is PD-L1 expression, which has been shown to be under strict post-transcriptional control in cancer. By fractionating the PD-L1 3′ untranslated region (3′UTR) into multiple overlapping fragments, we identified a small 100-nucleotide-long cis-acting region as being necessary and sufficient for post-transcriptional repression of PD-L1 expression in gastric cancer. In parallel, we performed a correlation analysis between PD-L1 expression and all host miRNAs in stomach cancer patient samples. A single miRNA, miR-105-5p, was predicted to bind to the identified cis-acting 3′UTR region and to negatively correlate with PD-L1 expression. Overexpression of miR-105-5p in gastric cancer cell lines resulted in decreased expression of PD-L1, both at the total protein and surface expression levels, and induced CD8+ T cell activation in co-culture assays. Finally, we show that expression of miR-105-5p in gastric cancer is partly controlled by DNA methylation of a cancer- and germline-specific promoter of its host gene, GABRA3. Dysregulation of miR-105-5p is observed in many cancer types and this study shows the importance of this miRNA in controlling the immunogenicity of cancer cells, thus highlighting it as a potential biomarker for PD-1/PD-L1 therapy and target for combinatorial immunotherapy.
Keywords: Immunotherapy, gastric cancer, microRNA
1. Introduction
Programmed death ligand 1 (PD-L1) is an immune checkpoint (IC) protein expressed in multiple cell types that interacts with its receptor PD-1 on T cells. The PD-L1/PD-1 interaction triggers inhibitory signals that prevent T-cell activation and proliferation. In the tumor microenvironment, PD-L1 is expressed on the surface of cancer cells and stromal immune cells and allows the tumor to evade the cytotoxic effects of infiltrating T cells (1). Recently, monoclonal antibodies that block the interaction between PD-1 and PD-L1 have been used as therapeutics and have had remarkable success in promoting tumor regression in multiple cancer types, including lung cancer, melanoma, and stomach cancer (2–4).
However, only a subset of patients responds favorably to PD-L1/PD-1 blockade therapy. The Food and Drug Administration (FDA) recommends two predictive biomarkers for beneficial response to PD-L1/PD-1 blockade therapies, namely PD-L1 expression and microsatellite instability (MSI)(5, 6). Indeed, the FDA has recently approved pembrolizumab, a PD-1 targeting monoclonal antibody (mAb), as a third-line treatment for patients with locally advanced or metastatic gastric cancer with high PD-L1 expression (7, 8).
Understanding the regulation of PD-L1 expression in cancer is key in identifying new predictive biomarkers for immune blockade therapy, as well as potential targets for combination therapies. It is known that chromosomal alterations or single-nucleotide variants (SNVs) in the 3′ untranslated region (3′UTR) region of PD-L1 are associated with changes in PD-L1 expression and immune escape in cancer (9–12). Interestingly, Kataoka et al. showed that genomic disruptions in the PD-L1 3′UTR region are particularly common in stomach cancer (12). MicroRNAs (miRNAs), such as miR-152 and miR-200, and RNA-binding proteins (RBPs) like tristetraprolin (TTP) have been shown to control PD-L1 expression through direct binding to its 3′UTR (13–16). In this study we sought to identify novel cis- and trans-acting post-transcriptional regulators of PD-L1 expression.
miR-105 is an intronic miRNA residing in the GABRA3 (γ-aminobutyric acid receptor 3) gene on chromosome X (17, 18). GABRA3 and miR-105 are normally expressed in the brain and testis (19, 20), but aberrant expression of both genes has been reported in multiple cancer types (18, 21–24). In melanoma and lung cancer, it was previously described that miR-105 expression is driven by a cancer- and germline-specific GABRA3 transcript (CT-GABRA3) (18). Interestingly, the expression of CT-GABRA3 is controlled by a CpG dinucleotide-rich bidirectional promoter shared with MAGEA6 (25). The role of miR-105 in cancer has been argued (22), but a pro-metastatic effect has been proposed in different cancer types, including colorectal, breast and gastric cancer (18, 22, 26, 27).
Here we identify a cis-acting regulatory region in the PD-L1 3′UTR that is necessary and sufficient for repression of PD-L1 expression. We show that miR-105-5p regulates PD-L1 expression through direct binding at the identified cis-acting 3′UTR region. Overexpression of miR-105-5p in cancer cells results in downregulation of PD-L1 and consequent increased activation of CD8+ T cells in co-culture experiments. This is the first time that miR-105-5p has been identified as a mediator of immune modulation in cancer. Finally, we show that changes in miR-105-5p expression in gastric cancer are partly controlled by DNA methylation of the CT-GABRA3 promoter, establishing a promoter methylation/miR-105-5p/PD-L1/immune escape axis.
2. Materials and Methods
2.1. Cell culture
Gastric cancer cell lines AGS (ATCC CRL-1739) and NCI-N87 (ATCC CRL-5822) were purchased from ATCC, SNU-719 (KCLB-00719) and SNU-216 (KCLB-00216) were purchased from the Korean Cell Line Bank (KCLB), and MKN-74 (JCRB-0255) was purchased from the Japanese Collection and Research Bioresources Cell Bank (JCRB Cell Bank). All gastric cancer cell lines were maintained in RPMI-1640 (Gibco, Gaithersburg, MD, USA), supplemented with 10% FBS (Invitrogen, Carlsbad, CA, USA). HEK293T cells (ATCC CRL-3216) were purchased from ATCC and maintained in DMEM (Gibco), supplemented with 10% FBS (Invitrogen). All cells were incubated at 37°C and 5% CO2.
2.2. Transfection
Small interfering RNAs (siRNAs) and miRNA mimics/inhibitors were transfected at a final concentration of 25nM using the TransIT-X2 reagent (Mirus Bio, Madison, WI, USA) as per the manufacturer’s protocol. Silencer select siRNAs were purchased from ThermoFisher Scientific (Waltham, MA, USA): CD274 siRNA (#4392420, siRNA ID s26548) and Silencer negative control #2 (AM4613). MirVana miRNA mimics and inhibitors were also purchased from ThermoFisher Scientific: miRNA mimic negative control (#4464058), hsa-miR-105-5p mimic (#4464066, assay ID MC12838), miRNA inhibitor negative control (#4464076), and has-miR-105-5p inhibitor (#4464084, assay ID MH12838).
2.3. Luciferase assay
The full-length (FL) PD-L1 3′UTR psicheck2 reporter was constructed as described previously (28). Fragments of the PD-L1 3′UTR were amplified from the FL reporter and subcloned into the psickeck2 vector using primers in Supplementary Table 1. Deletion constructs and miRNA seed-binding site mutant reporters were created using the Q5 site-directed mutagenesis kit (NEB, Ipswich, MA, USA) with primers shown in Supplementary Table 1.
The PD-L1 promoter sequence (−1081bp to +168bp from the transcription start site) was amplified from Taqman control genomic DNA (ThermoFisher Scientific, #4312660) using primers shown in Supplementary Table 2. The promoter sequence was cloned into the pGL4.10 luciferase reporter vector (Promega, Madison, WI, USA) using standard restriction digest cloning.
The day before transfection, 10 000 cells were seeded in each well in a 96-well plate. The cells were then transfected in triplicate with 100ng of luciferase reporter plasmid, and 25nM miRNA mimics/inhibitors where indicated, using the TransIT-X2 reagent (Mirus Bio). In the case of promoter reporter assays, cells were co-transfected with 80ng of the pGL4.10 and 20ng of the control pIS2-Renilla vector (Addgene, https://www.addgene.org, #12177). After 24 hours (h), fresh media were added to the cells. Finally, 48 h post-transfection, Renilla and Firefly luciferase activities were measured with the Dual-Luciferase Reporter Assay System (Promega) on the GloMax explorer luminometer (Promega).
2.4. Correlation and exon expression analysis
Level 3 data for The Cancer Genome Atlas (TCGA) stomach adenocarcinoma (STAD) patients were obtained from Xenabrowser (https://xenabrowser.net). Log2(RSEM+1) normalized counts (Illumina Hiseq 2000) and log2(RPM+1) normalized counts (Illumina Hiseq) were used for mRNA expression and miRNA expression, respectively. Cancer samples with both mRNA and miRNA data available were analyzed further (n=368). Spearman’s rank order correlation was performed between normalized PD-L1 expression and each miRNA individually (n=1 147). False discovery rate (FDR) adjusted p-values are reported. For exon expression profiling, log2(RPKM+1) normalized counts (Illumina Hiseq 2000) were used. Pearson’s correlation analysis was performed for the exon expression correlation matrix.
2.5. Dual-fluorescence reporter assay
The pTRETIGHTBI-RY-0 plasmid (#31463) was obtained from Addgene and the rtTA plasmid was a kind gift from Prof. Phillip A. Sharp (MIT, Cambridge, MA, USA). The pTRETIGHTBI-RY-0 plasmid was linearized with the HindIII-HF restriction enzyme (NEB), which cuts downstream of the mCherry gene. The PD-L1 3′UTR was amplified from the psicheck2 PD-L1 3′UTR reporter construct by PCR with the Phusion polymerase (NEB), using the following primers: 5′-gctgtacaagtaaatcgatagagacgtaatccagcattg-3′ and 5′-ctctggagatatcgtcgacataactttctccactgggatg-3′. The underlined sequences correspond to sequences that overlap with the ends of the linearized pTRETIGHTBI-RY-0 plasmid. The pTRETIGHTBI-RY-0 PD-L1 3′UTR reporter was assembled using the NEBuilder HiFi DNA Assembly kit (NEB).
To perform the dual fluorescence reporter assay, 200 000 cells were seeded in a 6-well plate. The next day, the cells were co-transfected with 0.5μg of the pTRETIGHTBI-RY-0 PD-L1 3′UTR reporter, 0.5μg of the rtTA plasmid and 25nM of miR-105-5p or control mimics using the TransIT-X2 reagent (Mirus Bio). After 24 h, the transfection media was aspirated and replaced with media containing 1μg/ml doxycycline. Fresh doxycycline-containing media was added every 24 h after that. Four days post-transfection, the cells were collected in DPBS+2%FBS and mCherry and eYFP fluorescence intensities were measured by flow cytometry, using the Cytoflex LX Cytometer. The data was analyzed in the FCS Express 6 software. Linear regression analysis between eYFP and mCherry fluorescence intensities was performed in R.
2.6. RNA extraction and RT-qPCR/RT-PCR
Cells were lysed in TRIzol (Invitrogen) and total RNA was extracted with the Direct-zol RNA miniprep plus kit (Zymo Research, Irvine, CA, USA). Reverse transcription for mRNAs was performed on 1μg total RNA using the SuperScript IV RT kit (ThermoFisher Scientific) with random hexamers. Quantitative PCR (qPCR) was performed on 1:10 diluted complementary DNA (cDNA) using the LightCycler 480 SYBR Green I Master Mix (Roche, Basel, Switzerland) as per the manufacturer’s protocol. For RT-PCR, diluted cDNA was amplified (34 cycles) using Phusion polymerase (NEB). Primer sequences for mRNA RT-qPCR and RT-PCR are provided in Supplementary Table 1. Reverse transcription for miRNAs was performed on 100ng total RNA using the miRCURY LNA RT kit (Qiagen, Hilden, Germany) and qPCR was carried out using the LNA miRCURY SYBR Green PCR mix (Qiagen). miRCURY LNA miRNA PCR assays for hsa-miR-105-5p and hsa-miR-103a-3p were purchased from Qiagen.
2.7. Protein extraction and Western blotting
Cells were lysed in RIPA buffer supplemented with protease and phosphatase inhibitors (ThermoFisher Scientific). Total protein concentration was determined with the Pierce BCA protein assay kit (ThermoFisher Scientific). Samples were subjected to SDS-PAGE and transferred to a PVDF membrane (Bio-Rad Laboratories, Hercules, CA, USA) pre-activated with methanol. The membrane was blocked in 5% non-fat dry milk in TBST for 1 h at room temperature (RT) and incubated with primary antibody (1:1 000 dilution) overnight at 4°C. The membrane was washed three times with TBST, 5 minutes (min) per wash, and incubated with secondary antibody (1:10 000 dilution) for 1 h at RT. Following three more washes with TBST, the signal was developed using the SuperSignal™ West Pico PLUS Chemiluminescent Substrate (ThermoFisher Scientific). The following antibodies were obtained from Cell Signaling Technology Inc. (CST, Danvers, MA, USA): PD-L1 (E1L3N), GAPDH (D16H11), anti-rabbit IgG – HRP-linked antibody (#7074).
2.8. Flow cytometry
Cells were washed once with PBS (ThermoFisher Scientific) and once with Stain Buffer (BD Biosciences, #554656). A million cells were then stained in 100 μl Stain Buffer with 5 μl of fluorophore-conjugated antibody for 30 min on ice. Following staining, cells were washed three times with Stain Buffer. Finally, the samples were run on the Cytoflex LX Cytometer and the data were analyzed with the FlowJo software. The following antibodies were purchased from Biolegend: APC anti-human CD274 antibody, clone 29E.2A3 (#329707) and APC Mouse IgG2b, κ Isotype Control antibody (#400322).
For interferon γ (IFNγ) treatment, cells were first transfected with miRNA mimics as described above and after 24 h they were treated with 10 ng/ml IFNγ (R&D Systems, Minneapolis, MN, USA) or mock. At 24 h post-treatment, cells were collected for flow cytometry.
2.9. Lentivirus transduction
For lentivirus production, HEK293T cells were co-transfected with the psPAX2 packaging vector, VSVg envelope vector, and the pLVX carrier plasmid at a 5:4:1 ratio using the Trans-IT Lenti (Mirus Bio) reagent. After 48 h, the supernatant was collected, centrifuged at 1000g for 5min, and filtered through a 0.45μm nitrocellulose filter. The virus was then added to target cells, and stably transduced cells were selected with puromycin 48 h later.
2.10. Co-culture of cancer cells with pre-activated CD8+ T cells.
PBMCs were isolated from blood donated by healthy donors using Ficoll-Plaque separation media (GE Healthcare, Chicago, IL, USA). The RosetteSep Human CD8+ T cell Enrichment cocktail (StemCell Technologies, Vancouver, Canada) and the EasySep Human CD8+ T cell Isolation kit (StemCell Technologies) were used for negative selection of CD8+ T cells from whole blood.
Isolated CD8+ T cells were activated with Dynabeads Human T-Activator CD3/CD28 (ThermoFisher Scientific) for three days as per the manufacturer’s protocol. Pre-activated CD8+ T cells, or control inactivated cells, were washed once with PBS, resuspended in fresh media and directly co-cultured with cancer cells that had been transfected with miRNA mimics or siRNAs 48 h before. The cells were co-cultured for 48 h.
For intracellular staining of IFNγ, 5 h before the collection of the cells, GolgiStop (BD Biosciences, Franklin Lakes, NJ, USA) was added to the media. The supernatant containing CD8+ T cells was then collected, the dynabeads were removed magnetically, and the cells were stained with FITC anti-human CD8, clone SK1 and APC/Cyanine7 anti-human CD3, clone SK7 (Biolegend, Dedham, MA, USA). The cells were then fixed and permeabilized using the Cytofix/Cytoperm buffer (BD Biosciences), according to the manufacturer’s protocol. Intracellular staining was performed with the IFN gamma monoclonal Antibody 4S.B3, PE (#12-7319-41) or the mouse IgG1 kappa isotype control P3.6.2.8.1, PE (#12-4714-41) purchased from ThermoFisher Scientific. Sample acquisition was performed using the Cytoflex LX Cytometer and the data were analyzed with the FlowJo Software.
To determine the concentration of secreted IFNγ and interleukin 2 (IL-2), the co-culture media was collected, and cells were removed by centrifugation. The supernatant was then serially diluted and IFNγ and IL-2 concentrations were measured using the ELISA MAX™ Deluxe Set Human IFNγ (Biolegend) and ELISA MAX™ Deluxe Set Human IL-2 (Biolegend), respectively.
2.11. Treatment with the demethylating agent 5-aza-2’-deoxycytidine
Cells were treated with fresh 5-aza-2’-deoxycytidine (final concentration 0–2μM) for 5 consecutive days, corresponding to at least 3 division cycles for all cell lines. Total RNA and genomic DNA (gDNA) were extracted at the end of the treatment.
For 5-methylcytosine (5mC) dot blots, gDNA was denatured in 1M NaOH by heating at 95°C for 10 min. Denatured gDNA was spotted on a Hybond-N+ membrane (Amersham Biosciences Corp., Little Chalfont, UK) and crosslinked using the Autocrosslink mode on the Stratalinker UV Crosslinker 2400 (Stratagene, La Jolla, CA, USA). The blot was then blocked in 5% non-fat dry milk in TBST for 1 h at room temperature (RT) and incubated with 5mC rabbit mAb (D3S2Z, CST) at 1:1 000 dilution in blocking buffer overnight at 4°C. Finally, the blot was incubated with anti-rabbit IgG – HRP-linked antibody (#7074) diluted 1:10 000 in blocking buffer for 1 h at RT.
2.12. Bisulfite PCR
Genomic DNA was extracted using the Monarch Genomic DNA purification kit (NEB). 500ng of gDNA were bisulfite converted using the EZ DNA Methylation-Gold kit (Zymo Research). The bisulfite-converted gDNA was eluted in 10μl and 1μl was used for each PCR reaction. Two rounds of PCR with the OneTaq DNA polymerase (NEB) were performed to amplify the CT-GABRA3 promoter. The CTGAB3 F1-R1 primer pair (Supplementary Table 1) was used in the first round. The PCR product was diluted 1:200 and 1μl of the diluted product was used for the second round of PCR with the CTGAB3 F2-R2 primer pair (Supplementary Table 1). The final PCR products were cloned into the pMiniT 2.0 vector using the NEB PCR cloning kit (NEB). Five clones per cell line were analyzed with Sanger sequencing.
3. Results
3.1. Identification of a cis-acting region in the 3′UTR of PD-L1 that affects its post-transcriptional regulation
To identify repressive cis-acting regions in the 3′UTR of PD-L1 in gastric cancer, we employed a reductionist approach. The 2.7kb-long 3′UTR of PD-L1 was divided into ten overlapping fragments and each was cloned downstream of the Renilla luciferase gene in the psicheck2 luciferase reporter vector (Fig. 1A). To measure the basal post-transcriptional activity of each fragment, luciferase assays were performed with all fragments, as well as an empty vector negative control and a full-length (FL) 3′UTR positive control, in SNU719 gastric cancer cells. Two fragments, designated F2.1 and F5.2, were found to downregulate luciferase activity compared to empty vector control (Fig. 1B) and were further fragmented into overlapping 100bp-long fragments. The repressive activity of the F2.1 and F5.2 fragments was narrowed down to the smaller F212 and F522 fragments, respectively (Fig. 1C). To determine whether they were not only sufficient, but also necessary for reporter repression, the sequences corresponding to these fragments were deleted from the full-length 3′UTR. The F522 fragment (chr9:5,470,385- 5,470,502, GRCh38) was the only fragment that when deleted from the FL construct resulted in an increase in reporter activity (Fig. 1D). A similar pattern in regulation by the F522 3′UTR fragment was observed in reporter assays performed in two additional gastric cancer cell lines (Fig. 1E). This indicates that the F522 3′UTR fragment contains cis-acting elements that promote post-transcriptional repression of PD-L1.
Figure 1. Identification of a post-transcriptionally repressive region in the PD-L1 3′UTR.
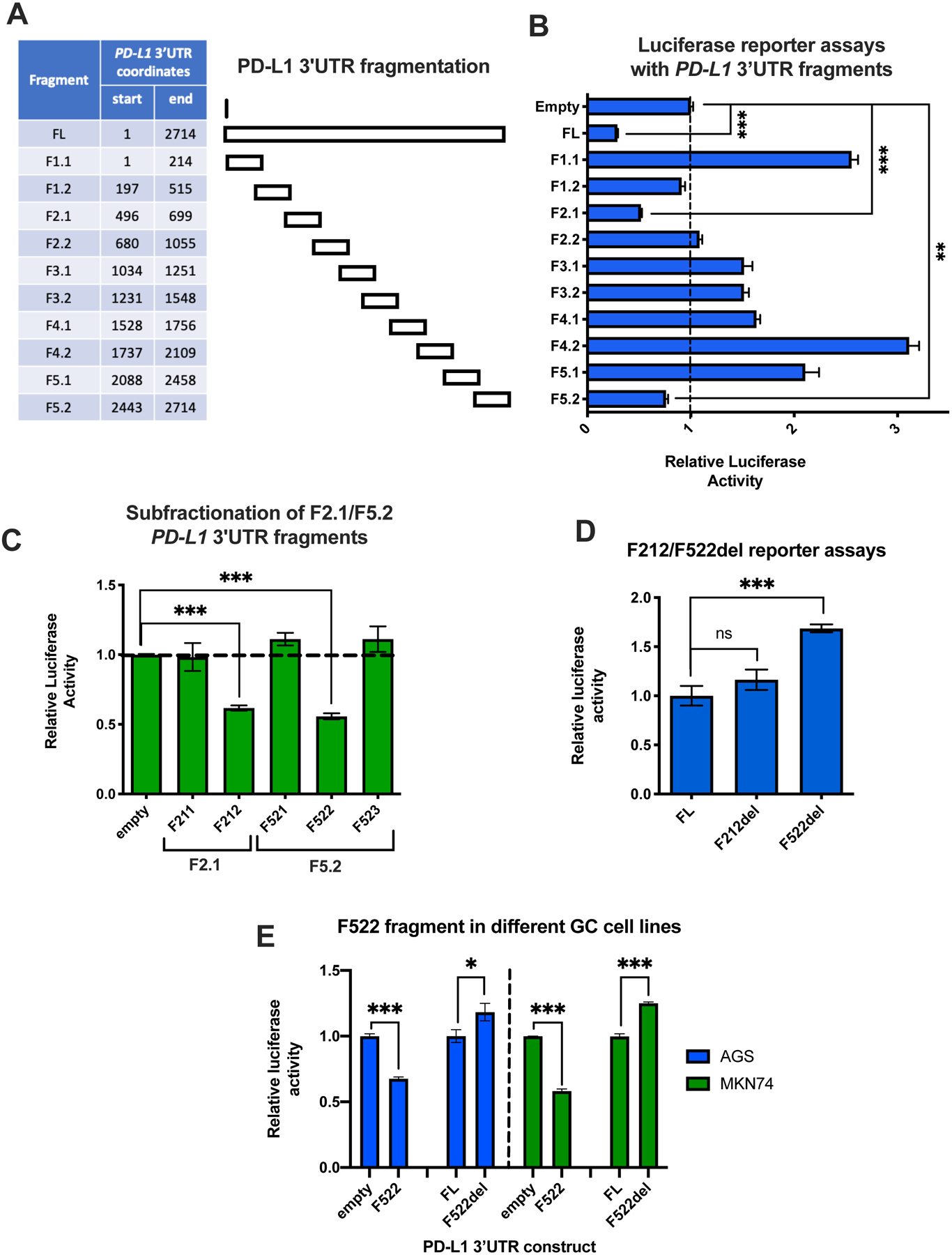
(A) Table with the PD-L1 3′UTR coordinates of the fragments used in the luciferase reporter assays. (B) Luciferase reporter assay with all ten 200–300bp long fragments, along with a full-length (FL) and an empty vector control, in SNU719 cells. The Renilla to Firefly luciferase activity ratio was calculated for each fragment. The results for each fragment were normalized against the empty vector control. (C) Luciferase reporter assays with smaller fractions (100bp-long) of the F2.1 and F5.2 fragments, which demonstrated repressive activity in (B). (D) The F212 and F522 fragments were deleted from the FL PD-L1 3′UTR reporter (del constructs). The activity of the deletion constructs was normalized to the FL control. (E) The same experiment as (C) and (D) for F212 and F522 was repeated in two additional gastric cancer cell lines. The activity of the fragments was normalized to the empty vector control, while the activity of the del reporters was normalized to the FL control. All experiments were performed in triplicate and statistical significance of comparisons was assessed by Student’s t-test. *P < 0.05; **P < 0.01; ***P < 0.001.
3.2. miR-105-5p is predicted to bind to F522 and its expression is negatively correlated with PD-L1 expression in cancer patients
Using data from the TCGA STAD cohort (29), we sought to identify miRNAs that are negatively correlated with PD-L1 expression in patient samples. Normalized expression data for PD-L1 and all host miRNAs in tumors from 368 STAD patients were obtained from Xenabrowser (https://xenabrowser.net), and a Spearman’s rank-order correlation analysis was performed between PD-L1 and each miRNA individually (Fig. 2A). Following false discovery rate (FDR) correction, 24 miRNAs were found to have a significant negative correlation with PD-L1 expression (Supplementary Table 2). One of those miRNAs, miR-105-5p, is predicted by TargetScan (http://targetscan.org) and microT-CDS/Diana Tools (http://diana.imis.athena-innovation.gr/DianaTools) to have a binding site in the F522 3′UTR fragment of PD-L1, as well as an additional potential binding site in the F5.1 fragment (Fig. 2B) (30, 31). Interestingly, miR-105-5p has also been found to bind to the putative F522 site, but not the F5.1 site, in a meta-analysis of multiple Argonaute 2 (AGO2)-Cross-linking immunoprecipitation (CLIP) experiments from different cancer cell lines, including nasopharyngeal carcinoma, Ewing’s sarcoma and lymphoma cell lines (Supplementary Fig. 1A) (32–37). In addition, to validate the negative correlation observed between miR-105-5p and PD-L1 in the TCGA gastric cancer cohort, we performed a correlation analysis between the host gene of miR-105-5p, GABRA3, and PD-L1 in a different gastric cancer cohort of 300 samples (38). A significant negative correlation was identified in the second cohort as well (Supplementary Fig. 1B). Finally, miR-105-5p expression negatively correlates with PD-L1 expression in other cancer types (Supplementary Fig. 1C). The strongest negative correlation coefficient is found in Low Grade Gliomas (LGG), which is notable given that miR-105-5p is normally detected only in the brain and testis (19).
Figure 2. miR-105-5p negatively correlates with PD-L1 expression in gastric cancer patient samples and is predicted to bind to the F522 fragment.
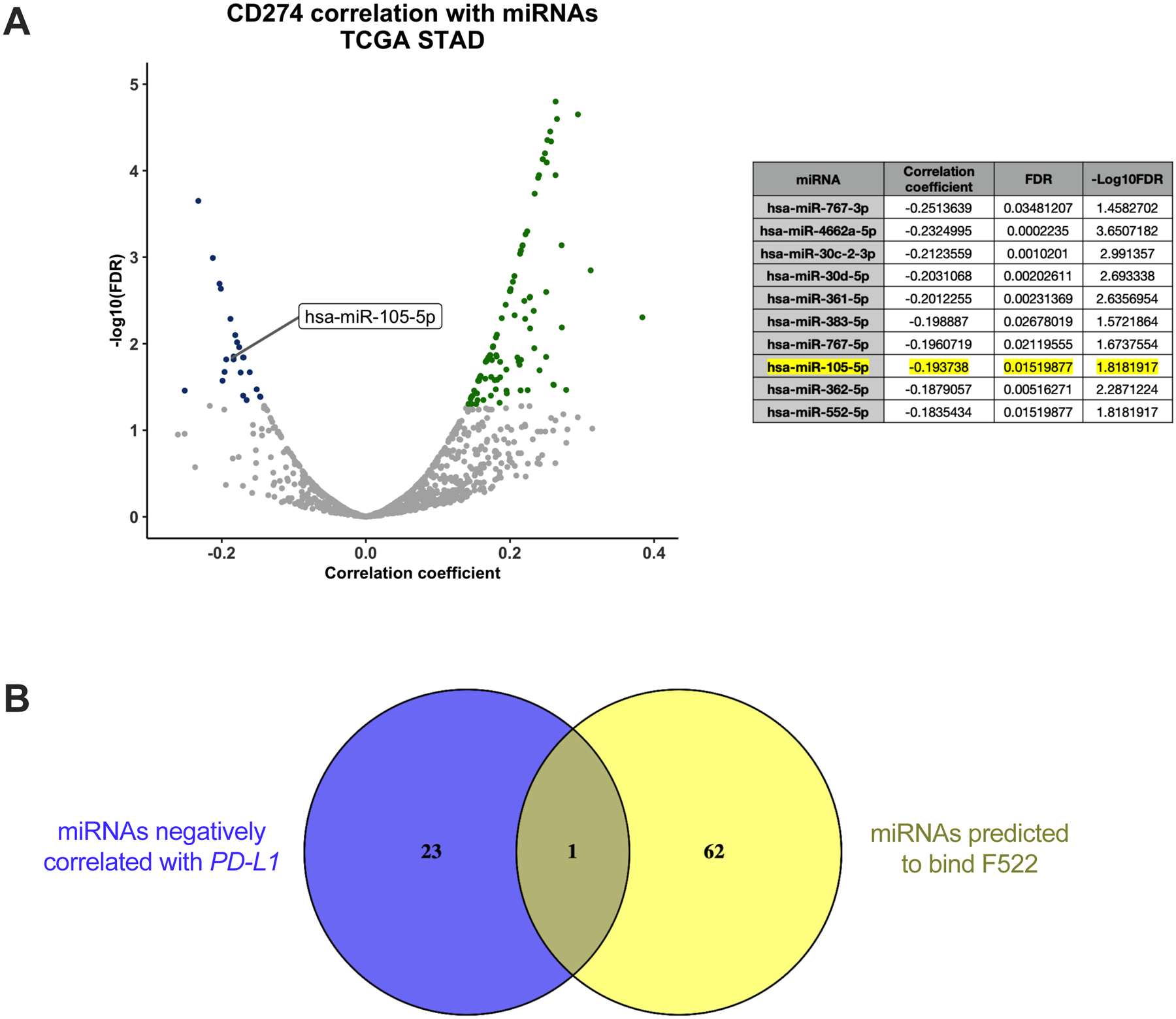
(A) Volcano plot representing the correlation of each miRNA with PD-L1 in TCGA STAD patients (n=368). Dots colored blue represent miRNAs with a significant negative correlation with PD-L1 expression (FDR p-value < 0.05). A table with the 10 miRNAs with the highest negative correlation coefficient with PD-L1 is shown on the right. miR-105-5p is highlighted. (B) A Venn diagram with the miRNAs that negatively correlate with PD-L1 in blue and the miRNAs that are predicted to bind to the F522 fragment of the PD-L1 3′UTR by microT-CDS in yellow. The miRNAs from each of the two analyses are listed in Supplementary Table 2. The only miRNA in the intersect is miR-105-5p.
3.3. PD-L1 is a direct target of miR-105-5p
To confirm functional binding of miR-105-5p to the PD-L1 3′UTR, we co-transfected miR-105-5p mimics with the PD-L1 3′UTR FL and F522 luciferase reporters. Overexpression of miR-105-5p resulted in downregulation of the luciferase activity conferred by these constructs, suggesting the ability of miR-105-5p to bind to the PD-L1 3′UTR, either at the F522 site alone or both F522 and F5.1 sites (Fig. 3A). To determine whether both putative binding sites are functional, we mutated the seed-binding sequence of each site individually (Fig. 3B). The mutations consisted of the shuffling of six nucleotides (positions 2–7) in the predicted seed-binding sequences. Mutation of the F522 site alone was sufficient to abolish reporter repression caused by miR-105-5p co-transfection with the FL reporter, whereas mutation of the F5.1 site had no significant effect (Fig. 3C). In addition, overexpression of miR-105-5p had no significant effect on reporter assays with the F522del and F5.1 PD-L1 3′UTR constructs (Fig. 3C). This data suggests that miR-105-5p primarily binds to the predicted F522 site. To confirm functional binding with an independent reporter system, we used a single-cell dual-fluorescence reporter construct carrying the mCherry and eYFP genes controlled by a bidirectional promoter. The FL 3′UTR of PD-L1 was cloned downstream of the mCherry gene, while the eYFP gene contained a rudimentary 3′UTR sequence (Fig. 3D). AGS cells were co-transfected with the dual-fluorescence reporter construct and miR-105-5p or miR-control mimics and then analyzed by flow cytometry. Overexpression of miR-105-5p resulted in the downregulation of mCherry intensity compared to control eYFP intensity (Fig. 3E), confirming functional binding of miR-105-5p to the PD-L1 3′UTR.
Figure 3. The binding of miR-105-5p to the PD-L1 3′UTR occurs mainly at the F522 fragment.
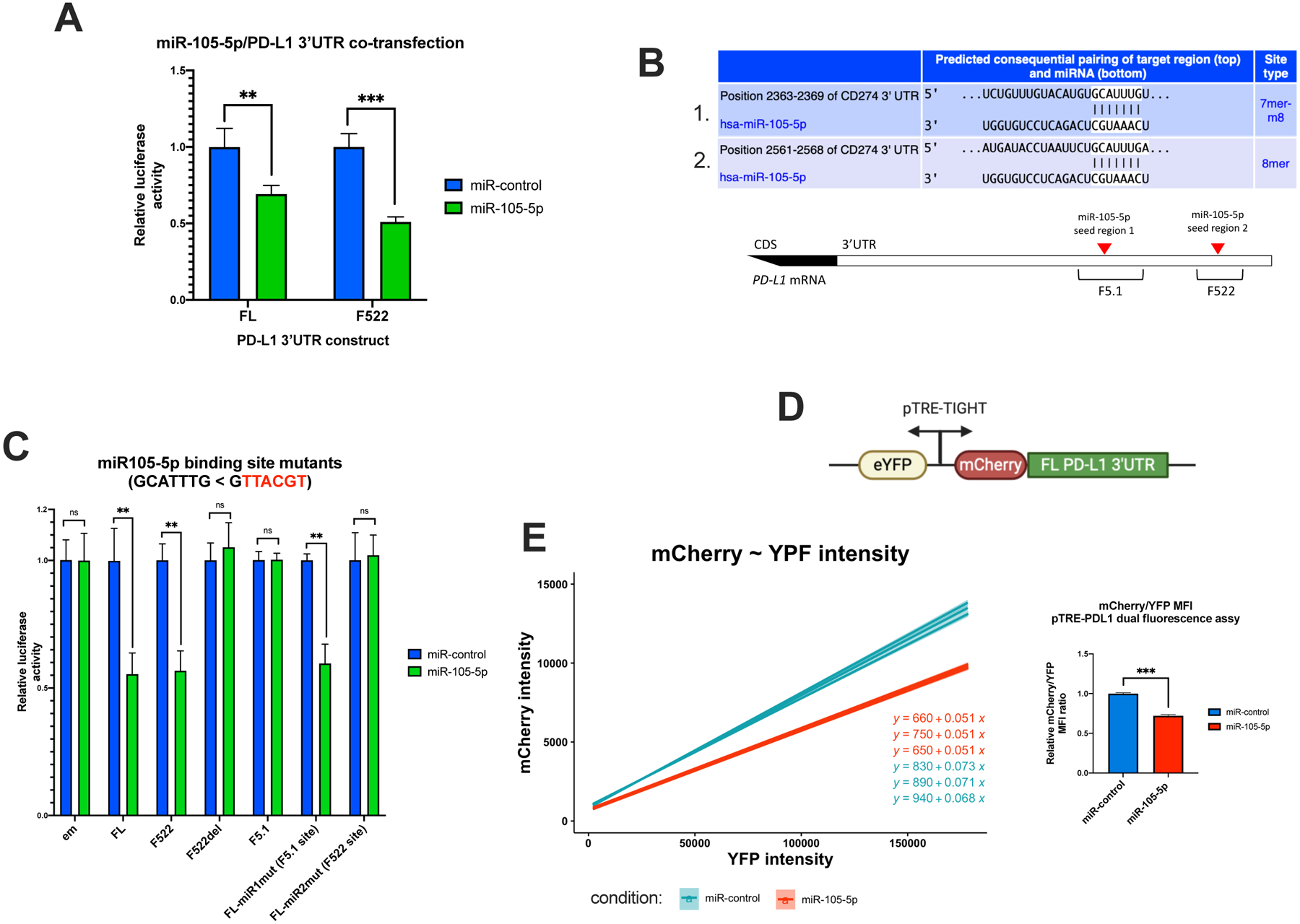
(A) Luciferase assay using the FL, F522, and F522del reporters with co-transfection of miR-105-5p or control mimics in AGS cells. (B) Binding sites of miR-105-5p in the PD-L1 3′UTR, predicted with TargetScan (http://targetscan.org). (C) Targeted mutagenesis of the seed sequence of each of the two miR-105-5p binding sites in the FL reporter. FL-miR1mut carries a mutation in the seed sequence of the first miR-105-5p binding site shown in the table in (C), which maps to the F5.1 fragment. FL-miR2mut carries a mutation in the F522 binding site. To determine the effect of miR-105-5p overexpression in each construct, the data are normalized per construct to the relative luciferase levels measured with miR-control co-transfection. (D) Graphic representation of the dual fluorescence reporter construct. eYFP and mCherry are controlled by a doxycycline-inducible bidirectional promoter. The full-length PD-L1 3′UTR was cloned downstream the mCherry gene, while the eYFP gene carries a rudimentary 3′UTR. (E) Regression plot of eYFP vs mCherry log fluorescence intensity from three replicates for miR-control or miR-105-5p co-transfections with construct in (D). The slope of the regression line is reduced in the miR-105-5p transfected cells. The bar plot compares the median fluorescence intensity (MFI) ratio of mCherry to eYFP in miR-105-5p compared to miR-control transfected cells. All graphs represent the mean ± SD of three experiments, *P < 0.05; **P < 0.01; ***P < 0.001, by Student’s t-test.
3.4. miR-105-5p downregulates constitutive and IFNγ-induced PD-L1 expression
Next, we determined whether overexpression of miR-105-5p downregulates PD-L1 expression at the mRNA, total protein, and surface expression levels. For further analysis, we selected two cell lines, AGS and MKN74, with low and high miR-105-5p expression, respectively (Fig. 4A). Transfection with a miR-105-5p mimic reduced constitutive PD-L1 levels in both cell lines (Fig. 4B–E). Transfection of HEK293T and A549 cells with miR-105-5p mimics also resulted in a reduction in PD-L1 surface expression levels, further suggesting that the miR-105-5p/PD-L1 axis is not unique to gastric cancer (Fig. 4E). Pro-inflammatory cytokines like IFNγ are known to be strong inducers of PD-L1 expression in gastric and other cancers (39, 40). To identify whether miR-105-5p could offset IFNγ-mediated induction of PD-L1 expression, AGS and MKN74 were transfected with miR-105-5p or miR-control mimics and then treated with IFNγ for 24 h. A reduction in IFNγ-induced PD-L1 expression was observed in miR-105-5p overexpressing cells for both cell lines (Supplementary Fig. 2A–B).
Figure 4. miR-105-5p expression affects PD-L1 levels in vitro.
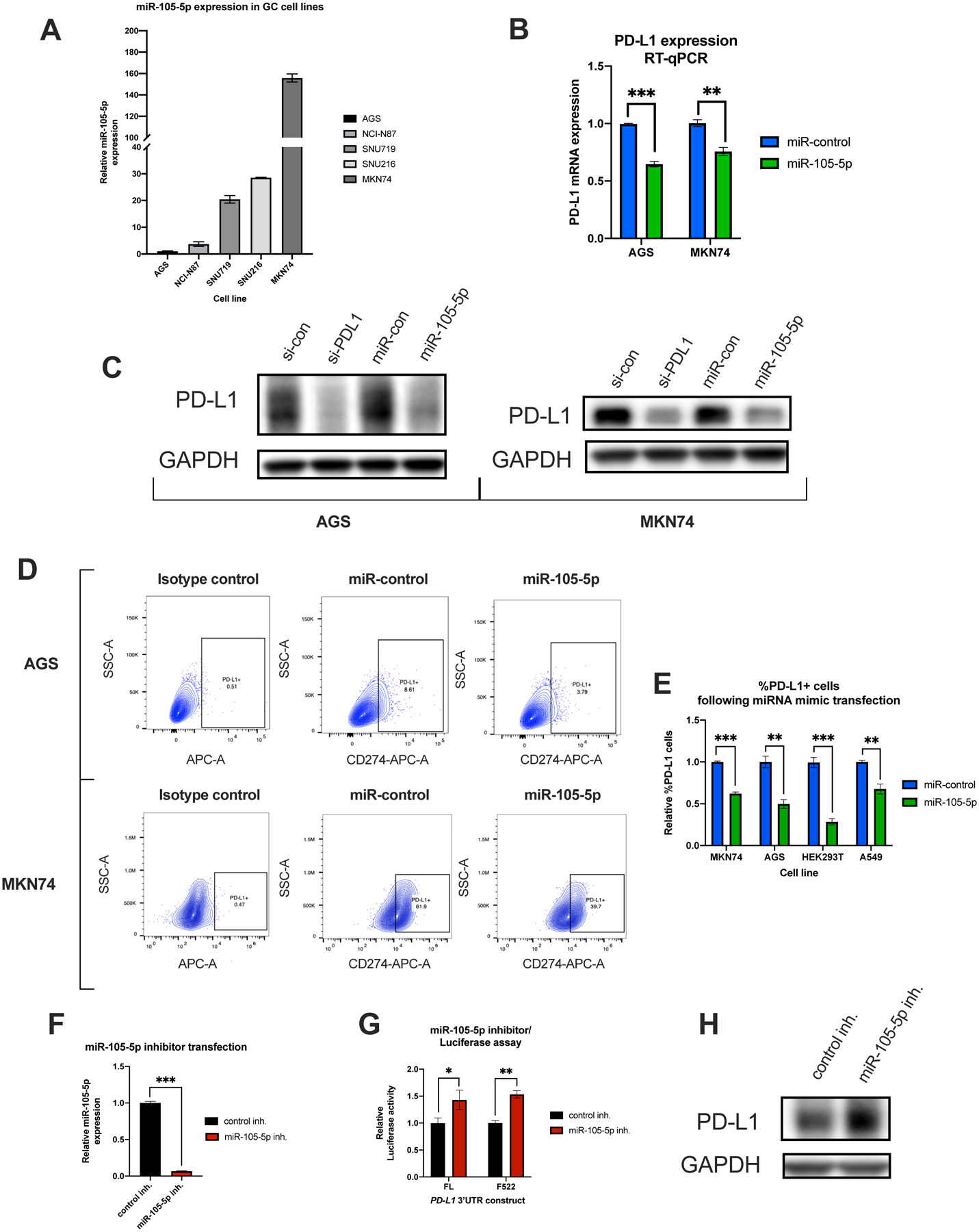
(A) RT-qPCR analysis of miR-105-5p expression in five gastric cancer cell lines. miR-105-5p levels were normalized to miR-103a-3p. (B) qPCR analysis of PD-L1 expression following miR-105-5p compared to miR-control mimic transfection in AGS and MKN74 cells. PD-L1 levels were normalized to GAPDH levels. (C) Western blot analysis of PD-L1 in total protein extracted from AGS and MKN74 cells transfected with siPD-L1 or miR-105-5p mimics, and respective controls. The levels of GAPDH are shown as an internal control. (D) Representative flow cytometry plots showing the percentage of PD-L1+ cells following miR-105-5p or miR-control mimic transfection in AGS and MKN74 cells. The PD-L1+ gate comprises of cells with higher fluorescence intensity than cells stained with non-targeting isotype control. (E) A bar plot showing the relative levels of %PD-L1+ cells in miR-105-5p vs miR-control transfected cells in gastric cancer cell lines, as well as HEK293T and A549 cells. The relative percentage of PD-L1+ cells is normalized to miR-control transfected cells for each cell line. (F) qPCR analysis of miR-105-5p expression in MKN74 cells transfected with miR-105-5p or control inhibitors. The levels of miR-105-5p were normalized to miR-103a-3p. (G) and (H) Luciferase assay and western blot analysis showing the effect of miR-105-5p antagomirs on PD-L1 expression in MKN74 cells. All experiments were performed in triplicate. The graphs represent mean ± SD of three experiments, *P < 0.05; **P < 0.01; ***P < 0.001, by Student’s t-test. Representative images are shown for Western blotting.
Several miRNAs have been shown to affect PD-L1 expression indirectly, through modulating IFNγ signaling or by targeting transcription factors involved in PD-L1 regulation (41–44). To determine whether miR-105-5p controls PD-L1 expression not only through direct binding to its 3′UTR, but also through indirect transcriptional effects we performed reporter assays with a PD-L1 promoter construct. Overexpression of miR-105-5p did not have a significant effect on the activity of the PD-L1 promoter reporter, suggesting that miR-105-5p does not affect PD-L1 transcription (Supplementary Fig. 2C).
Finally, transfection of miR-105-5p inhibitors in MKN74 cells resulted in significant downregulation of miR-105-5p expression (Fig. 4F), partly reversed repression of the FL and F522 PD-L1 3′UTR luciferase reporters (Fig. 4G) and resulted in increased PD-L1 protein levels (Fig. 4H).
3.5. Overexpression of miR-105-5p in cancer cells promotes T-cell activation in co-culture experiments
To determine the effect of miR-105-5p on the immunogenicity of cancer cells, we performed in vitro co-culture experiments. CD8+ T cells isolated from healthy blood donors were activated with α-CD3/CD28 beads for three days and then co-cultured with miR-105-5p or miR-control mimic-transfected MKN74 or AGS cells. After 48 h, T cell activity was determined by intracellular IFNγ staining and by measuring the levels of secreted IFNγ and IL-2 using ELISA assays. MKN74 cells overexpressing miR-105-5p allowed for higher levels of CD8+ T cell activation than cells transfected with miR-control mimics (Fig. 5A–C). In the case of AGS cells, downregulation of PD-L1 by transfection with small interfering RNAs (siRNAs) against PD-L1 did not affect CD8+ T cell activity in co-culture experiments (data not shown). To increase sensitivity of the co-culture assay, we created AGS cells stably overexpressing PD-L1 (AGS-PDL1). The PD-L1 construct used to create the AGS-PDL1 cell line included both the coding sequence (CDS) and 3′UTR of PD-L1. Transfection of miR-105-5p mimics resulted in a two-fold reduction in PD-L1 expression even in AGS-PDL1 cells (Supplementary Fig. 3). Overexpression of PD-L1 in AGS cells inhibited activation of CD8+ T cells, as measured by intracellular IFNγ staining (Fig. 5D–E). Transfection of miR-105-5p in wild-type AGS cells had no effect on CD8+ T cell activation, similar to siPD-L1 transfection (Fig. 5D–E). In contrast, miR-105-5p mimic transfection in AGS-PDL1 partially reversed the effect of PD-L1 overexpression on CD8+ T cell activation, as measured by both intracellular IFNγ staining and IFNγ/IL-2 ELISA assays (Fig. 5D–F). Our findings reveal that miR-105-5p overexpression can promote immune surveillance in gastric cancer through downregulation of PD-L1.
Figure 5. Overexpression of miR-105-5p in cancer cells downregulates PD-L1 and promotes increased activation of CD8+ T cells in an in vitro co-culture system.
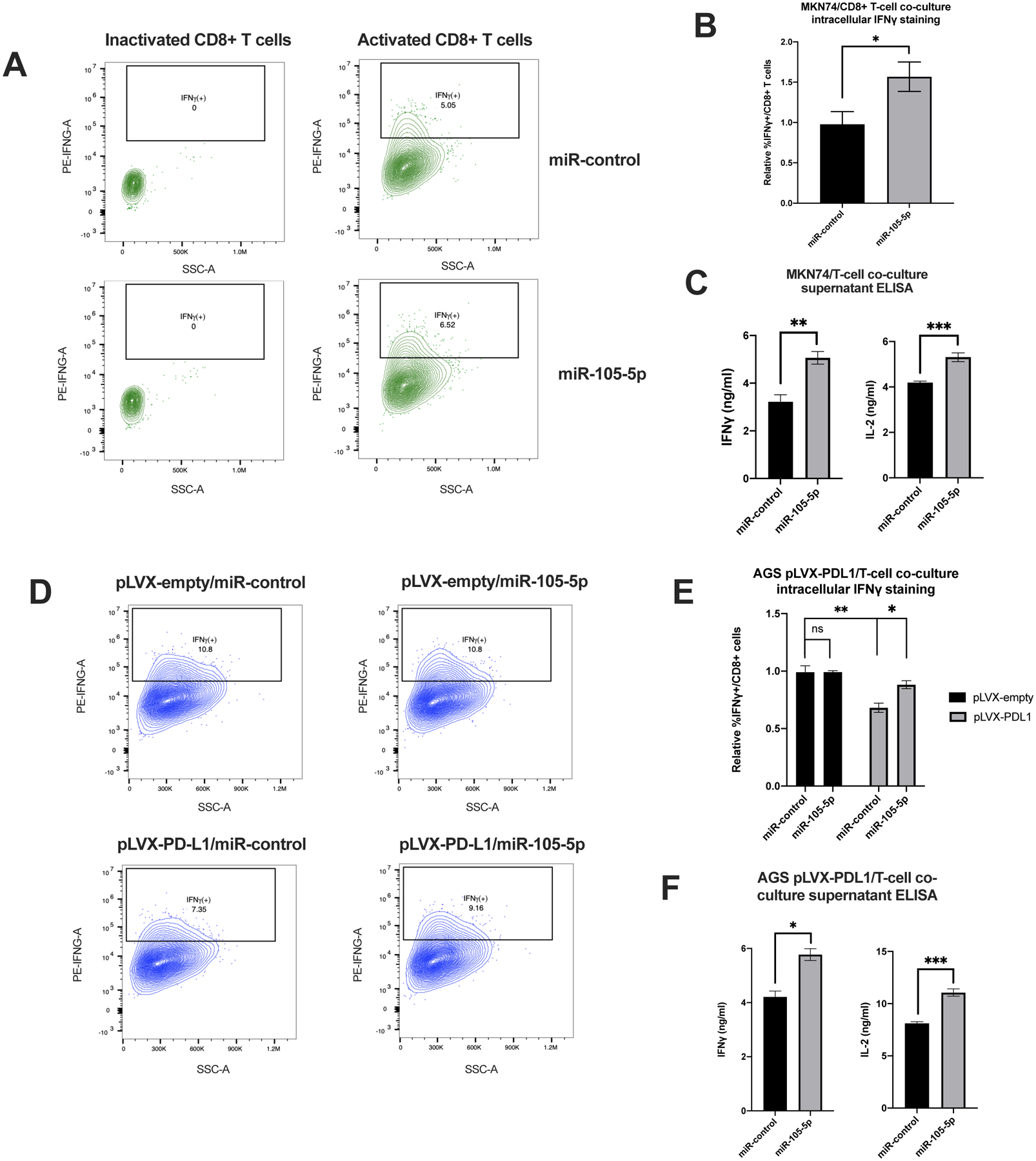
(A) Representative flow cytometry plots showing the percentage of IFNγ-positive CD8+ T cells in a co-culture experiment with miR-control or miR-105-5p transfected MKN74 cells. The cells were pre-selected for being CD3+CD8+. (B) Graph represents the relative percentage of IFNγ-positive CD8+ T cells in miR-control vs miR-105-5p transfected MKN74 cells in three independent experiments. (C) Supernatant collected from MKN74/CD8+ T cell co-culture experiments was analyzed with ELISA assays for the levels of secreted IFNγ and IL-2. (D) Representative flow cytometry plots of the IFNγ-positive CD8+ T cells in a co-culture experiment with pLVX-control or pLVX-PD-L1 transduced AGS cells transfected with miR-control or miR-105-5p transfected. (E) Graph represents the mean of three independent experiments described in (D). (F) Supernatant from co-culture experiments described in (D) were analyzed by ELISA for the levels of secreted IFNγ and IL-2 from activated CD8+ T cells. All experiments were repeated in triplicate and bar plots represent the mean ± SD of all experiments. Significance was calculated by Student’s t-test, *P < 0.05; **P < 0.01; ***P < 0.001.
3.6. Promoter methylation controls miR-105-5p expression in gastric cancer
As mentioned above, miR-105-5p is an intronic miRNA located in the GABRA3 gene on chromosome X and is normally only expressed in the brain and testis (17). The reference GABRA3 (ref-GABRA3) transcript variant consists of 10 exons, and miR-105 is processed form the first intron. A recent study showed that an alternative promoter controls to the expression of a unique GABRA3 transcript (CT-GABRA3) in melanoma and lung cancer (18). The CT-GABRA3 transcript shares exons 2–10 with the ref-GABRA3 transcript but skips exon 1. To determine whether GABRA3 expression in gastric cancer is mainly driven by the ref- or the CT- GABRA3 transcript we analyzed the exon expression profile of TCGA STAD patients. We show that despite very strong pairwise correlation in the expression of exons 2–10, expression of exon 1 is poorly correlated with all other exons (Supplementary Fig. 4A–B). Using an RPKM (Reads Per Kilobase of transcript per Million mapped reads) cutoff of 0.5, we identify that less than 10% of patients with detectable expression of exons 2–10 also have detectable expression of exon 1 (Supplementary Fig. 4C). This indicates that in most gastric cancer samples with detectable GABRA3 expression, the transcript that is expressed is most likely CT-GABRA3. The CT-GABRA3 promoter also controls the expression of the MAGEA6 gene. Genes controlled by a bidirectional promoter are commonly co-expressed and, indeed, MAGEA6 and GABRA3 show strong positive correlation in gastric cancer patient samples (Supplementary Fig. 4D ) and cell lines (Supplementary Fig. 4E).
For analysis of GABRA3 expression in gastric cancer cells in vitro, we selected four cell lines with variable miR-105-5p expression (Fig.4A) and no copy number alterations in the GABRA3 gene in the Cancer Cell Encyclopedia (CCLE) database (Fig. 6A) (45). The MKN74 cell line was excluded from further analysis because it showed increased GABRA3 copy number in this database (Fig. 6A). To determine whether GABRA3 expression in these cell lines correlates with CT-GABRA3 promoter methylation, we performed bisulfite PCR to analyze the methylation status of the CpG islands in the promoter. We found that SNU216 and SNU719, the cell lines with the highest miR-105-5p and GABRA3 expression, had a hypomethylated CT-GABRA3 promoter, whereas NCI-N87 and AGS, the cell lines with the lowest miR-105-5p and GABRA3 expression, had the highest levels of CT-GABRA3 promoter methylation (Fig. 6B). By performing reverse transcription PCR (RT-PCR) with primers that can amplify either the ref- or the CT- GABRA3 transcript alone, we show that expression of GABRA3, and thus miR-105-5p, in SNU719 and SNU216 cells is primarily driven by the CT transcript (Fig. 6C). In contrast, AGS and NCI-N87 cells, which have low levels of miR-105-5p expression and high levels of CT-GABRA3 promoter methylation, show low levels of the ref-GABRA3 transcript, but no detectable levels of CT-GABRA3 transcript (Fig. 6C). Treatment with the de-methylating agent 5-aza-2’-deoxycytidine (Fig. 6D) increased expression of miR-105-5p in NCI-N87 and AGS but not in SNU216 or SNU719 (Fig. 6E). As previously reported, global DNA demethylation leads to transcriptional overexpression of PD-L1 (46, 47). Therefore, to specifically identify whether hypomethylation-induced miR-105-5p expression can have a functional effect on the post-transcriptional regulation of PD-L1, we performed luciferase assays with WT and miR-105-5p site-mutant PD-L1 3′UTR reporters. Treatment with 5-aza-2’-deoxycytidine decreased luciferase activity in AGS cells transfected with a wild-type PD-L1 3′UTR reporter to a higher extent than cells transfected with a reporter bearing a mutation in the miR-105-5p binding site (Fig. 6F). This indicates that overexpression of miR-105-5p caused by CT-GABRA3 promoter hypomethylation is sufficient to have a functional effect on the post-transcriptional regulation of PD-L1 expression.
Figure 6. DNA methylation at the CT-GABRA3 promoter controls expression of miR-105-5p in gastric cancer.
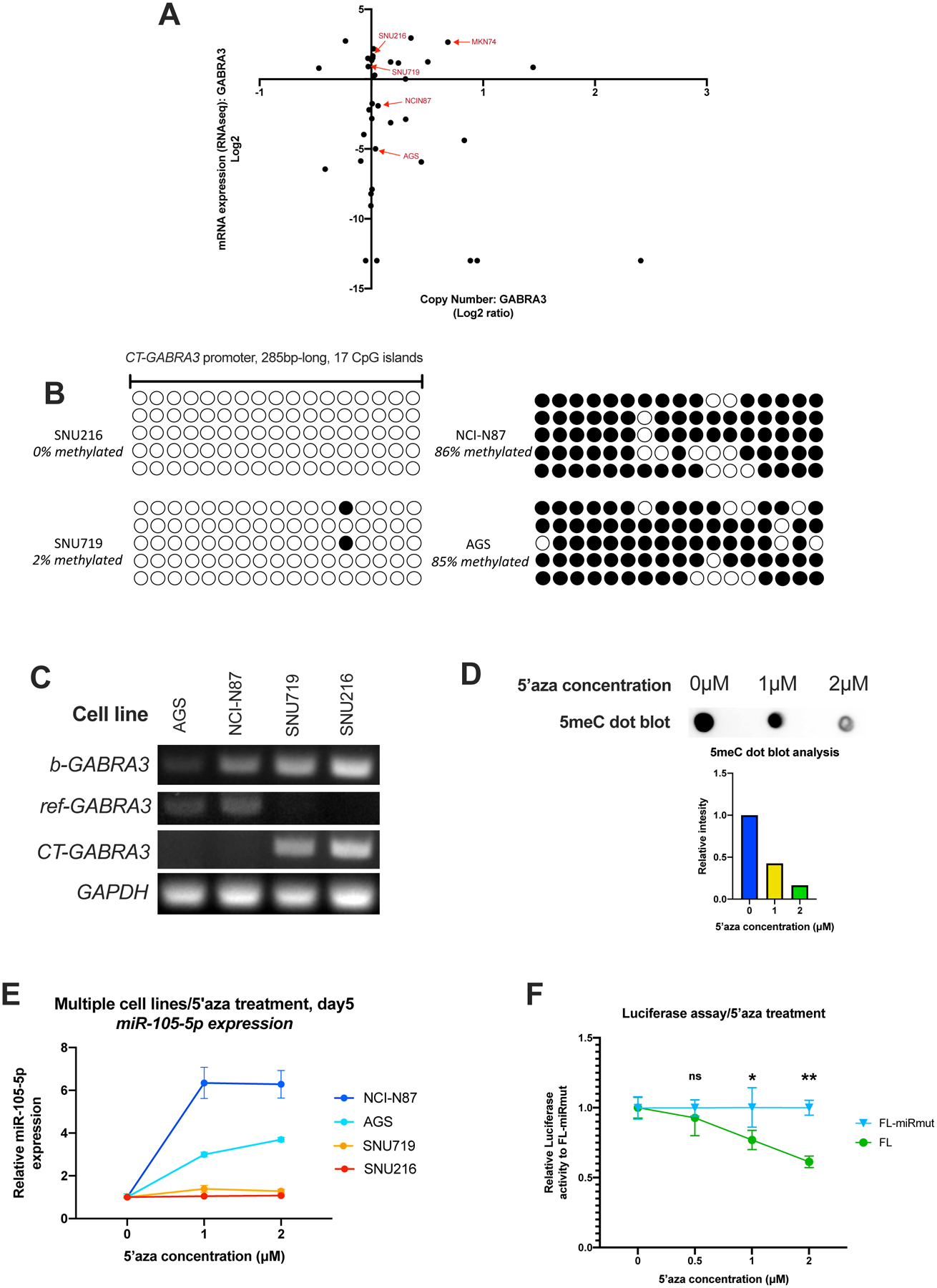
(A) Scatterplot of Copy Number vs mRNA expression of the GABRA3 gene in all stomach-derived cell lines from CCLE. The four cell lines selected for further analysis are indicated with arrows. MKN74 was not pursued further due to increased GABRA3 copy number. (B) Bisulfite PCR was performed to analyze the methylation status of the CT-GABRA3 promoter (GRCh38, chrX:152,769,581–152,769,864) in four gastric cancer cell lines. Five clones were sequenced for each cell line. Filled circles indicate methylated CpG islands, while blank circles indicate unmethylated CpG islands. (C) RT-PCR with primers either detecting both GABRA3 transcripts (b-GABRA3), only the ref-GABRA3 transcript or only the CT-GABRA3 transcript. RT-PCR for GAPDH is also shown as an internal control. (D) 5meC dot-blot with gDNA extracted from SNU719 cells treated with 0, 1, or 2 μM 5’-aza-2’-deoxycytidine for 5 days. Relative intensity of each dot was calculated on ImageJ. (E) RT-qPCR analysis of miR-105-5p in gastric cancer cell lines treated with 0, 1, or 2 μM 5’-aza-2’-deoxycytidine for 5 days. (F) Luciferase assay with WT and miR-105-5p seed mutant constructs in AGS cells treated with 0, 0.5, 1, or 2 μM 5’-aza-2’-deoxycytidine for 5 days. For each 5’-aza-2’-deoxycytidine concentration the relative Renilla to Firefly luciferase activity is normalized to the miR-105-5p seed mutant construct.
4. Discussion
It was previously shown that disruption of the PD-L1 3′UTR by chromosomal structural variations results in an increase in PD-L1 levels in cancer (12, 48). This indicates that the 3′UTR of PD-L1 contains important cis-acting elements that fine-tune its expression. Different miRNAs and RBPs have been described to contribute to the regulation of PD-L1 expression (49). Here, by performing reporter assays with PD-L1 3′UTR fragments and correlation analysis in patient data we identify miR-105-5p as an important mediator of PD-L1 post-transcriptional repression in gastric cancer. Using an in vitro co-culture assay, we show that miR-105-5p overexpression can counter immune escape caused by PD-L1 upregulation in cancer cells. The miR-105-5p binding site in the PD-L1 3′UTR is only conserved among primates (30), thus complicating the pursuit of mouse model experiments to assess the effect of miR-105-5p on tumor immunogenicity in vivo.
Correlation analysis in different TCGA cohorts (Supplementary Fig. 1C), AGO-CLIP data from different cell lines (Supplementary Fig. 1A) and overexpression analysis in non-gastric cancer cell lines (Fig. 4E) indicate that regulation of PD-L1 expression and tumor immunogenicity by miR-105-5p may be important in multiple cancer types.
Several studies have reported detection of miR-105-5p in the blood of cancer patients (21, 24). Indeed, elevated levels of circulating miR-105-5p have been proposed as a biomarker for early detection of non-small cell lung cancer, as well as triple negative breast cancer (TBNC)(21, 24). The direct effect of miR-105-5p on PD-L1 expression and immune escape in cancer cells highlights miR-105-5p expression as a potential biomarker for PD-1/PD-L1 therapy. Secretion of miR-105-5p to the tumor microenvironment and the blood has been shown to occur through packaging in exosomes (23, 50). Interestingly, a study on TBNC showed that exosomal miR-105-5p promotes metastasis through the modulation of neighboring endothelial cells (27). PD-L1 expression in stromal immune cells in the tumor microenvironment is known to be important in immune escape and tumor progression in gastric cancer (51), as well as other cancer types. This raises the question of whether cancer cell-derived exosomal miR-105-5p could affect immune escape through modulating PD-L1 levels in neighboring tumor-resident immune cells.
Aberrant expression of GABRA3 and miR-105-5p has been described in multiple cancer types (22). In this study, we demonstrate that CT-GABRA3 is the primary GABRA3 transcript expressed in gastric cancer (Supplementary Fig. 4). Hypomethylation of the CT-GABRA3 promoter is associated with elevated expression of miR-105-5p and consequent post-transcriptional silencing of PD-L1 expression (Fig. 6B–F).
In conclusion, this study establishes a regulatory network that connects DNA methylation-controlled upregulation of miR-105-5p with decreased PD-L1 expression and increased immunogenicity in cancer cells. We show that hypomethylation of the CT-GABRA3 promoter is associated with induction of miR-105-5p expression in gastric cancer. Through direct targeting of an important cis-acting regulatory region of the PD-L1 3′UTR, miR-105-5p downregulates PD-L1 expression and promotes CD8+ T cell activation in in vitro co-culture experiments. This is the first study that describes a role for miR-105-5p in cancer immune surveillance. The extensive in vitro evidence outlined in this study warrants the pursuit of future studies to determine whether miR-105-5p can serve as a predictive marker for PD-1/PD-L1 blockade therapy or a target for combinatorial immunotherapy in gastric cancer.
Supplementary Material
Highlights.
A 100bp-long region in the PD-L1 3′UTR is necessary and sufficient for post-transcriptional repression of PD-L1 expression and is bound by miR-105-5p.
miR-105-5p regulates constitutive and interferon-induced PD-L1 expression in cancer.
Overexpression of miR-105-5p increases immunogenicity of cancer cells through downregulation of PD-L1.
Induction of miR-105-5p expression in gastric cancer occurs mainly through hypomethylation of a cancer-specific GABRA3 promoter.
Acknowledgements
We especially appreciate Dr. Wenyi Wei for providing us with a PD-L1 CDS vector and Dr. Maria Mavrikaki and Allison Baker for their comments on this manuscript. We acknowledge the NCI Outstanding Investigator Award (R35CA232105) to FJS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests
The authors declare no competing interest.
References
- 1.Sanmamed MF, Chen L. Inducible expression of B7-H1 (PD-L1) and its selective role in tumor site immune modulation. Cancer J. 2014;20(4):256–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hamid O, Robert C, Daud A, Hodi FS, Hwu WJ, Kefford R, et al. Safety and tumor responses with lambrolizumab (anti-PD-1) in melanoma. N Engl J Med. 2013;369(2):134–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366(26):2443–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fuchs CS, Doi T, Jang RW, Muro K, Satoh T, Machado M, et al. Safety and Efficacy of Pembrolizumab Monotherapy in Patients With Previously Treated Advanced Gastric and Gastroesophageal Junction Cancer: Phase 2 Clinical KEYNOTE-059 Trial. JAMA Oncol. 2018;4(5):e180013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang L, Chang M, Chang HM, Chang F. Microsatellite Instability: A Predictive Biomarker for Cancer Immunotherapy. Appl Immunohistochem Mol Morphol. 2018;26(2):e15–e21. [DOI] [PubMed] [Google Scholar]
- 6.Davis AA, Patel VG. The role of PD-L1 expression as a predictive biomarker: an analysis of all US Food and Drug Administration (FDA) approvals of immune checkpoint inhibitors. J Immunother Cancer. 2019;7(1):278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brar G, Shah MA. The role of pembrolizumab in the treatment of PD-L1 expressing gastric and gastroesophageal junction adenocarcinoma. Therap Adv Gastroenterol. 2019;12:1756284819869767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fashoyin-Aje L, Donoghue M, Chen H, He K, Veeraraghavan J, Goldberg KB, et al. FDA Approval Summary: Pembrolizumab for Recurrent Locally Advanced or Metastatic Gastric or Gastroesophageal Junction Adenocarcinoma Expressing PD-L1. Oncologist. 2019;24(1):103–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu Y, Zhao T, Jia Z, Cao D, Cao X, Pan Y, et al. Polymorphism of the programmed death-ligand 1 gene is associated with its protein expression and prognosis in gastric cancer. J Gastroenterol Hepatol. 2019;34(7):1201–7. [DOI] [PubMed] [Google Scholar]
- 10.Yeo MK, Choi SY, Seong IO, Suh KS, Kim JM, Kim KH. Association of PD-L1 expression and PD-L1 gene polymorphism with poor prognosis in lung adenocarcinoma and squamous cell carcinoma. Hum Pathol. 2017;68:103–11. [DOI] [PubMed] [Google Scholar]
- 11.Wang W, Sun J, Li F, Li R, Gu Y, Liu C, et al. A frequent somatic mutation in CD274 3’-UTR leads to protein over-expression in gastric cancer by disrupting miR-570 binding. Hum Mutat. 2012;33(3):480–4. [DOI] [PubMed] [Google Scholar]
- 12.Kataoka K, Shiraishi Y, Takeda Y, Sakata S, Matsumoto M, Nagano S, et al. Aberrant PD-L1 expression through 3′-UTR disruption in multiple cancers. Nature. 2016;534(7607):402–6. [DOI] [PubMed] [Google Scholar]
- 13.Chen L, Gibbons DL, Goswami S, Cortez MA, Ahn YH, Byers LA, et al. Metastasis is regulated via microRNA-200/ZEB1 axis control of tumour cell PD-L1 expression and intratumoral immunosuppression. Nat Commun. 2014;5:5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y, Wang D, Xie G, Yin Y, Zhao E, Tao K, et al. MicroRNA-152 regulates immune response via targeting B7-H1 in gastric carcinoma. Oncotarget. 2017;8(17):28125–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo J, Qu H, Shan T, Chen Y, Xia J. Tristetraprolin Overexpression in Gastric Cancer Cells Suppresses PD-L1 Expression and Inhibits Tumor Progression by Enhancing Antitumor Immunity. Mol Cells. 2018;41(7):653–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miliotis C, Slack FJ. Multi-layered control of PD-L1 expression in Epstein-Barr virus-associated gastric cancer. J Cancer Metastasis Treat 2020;6(13). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benakanakere MR, Li Q, Eskan MA, Singh AV, Zhao J, Galicia JC, et al. Modulation of TLR2 protein expression by miR-105 in human oral keratinocytes. J Biol Chem. 2009;284(34):23107–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loriot A, Van Tongelen A, Blanco J, Klaessens S, Cannuyer J, van Baren N, et al. A novel cancer-germline transcript carrying pro-metastatic miR-105 and TET-targeting miR-767 induced by DNA hypomethylation in tumors. Epigenetics. 2014;9(8):1163–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. [DOI] [PubMed] [Google Scholar]
- 20.Ludwig N, Leidinger P, Becker K, Backes C, Fehlmann T, Pallasch C, et al. Distribution of miRNA expression across human tissues. Nucleic Acids Res. 2016;44(8):3865–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dong X, Chang M, Song X, Ding S, Xie L. Plasma miR-1247–5p, miR-301b-3p and miR-105-5p as potential biomarkers for early diagnosis of non-small cell lung cancer. Thorac Cancer. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li J, Zhang Z, Chen F, Hu T, Peng W, Gu Q, et al. The Diverse Oncogenic and Tumor Suppressor Roles of microRNA-105 in Cancer. Front Oncol. 2019;9:518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan W, Wu X, Zhou W, Fong MY, Cao M, Liu J, et al. Cancer-cell-secreted exosomal miR-105 promotes tumour growth through the MYC-dependent metabolic reprogramming of stromal cells. Nat Cell Biol. 2018;20(5):597–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li HY, Liang JL, Kuo YL, Lee HH, Calkins MJ, Chang HT, et al. miR-105/93–3p promotes chemoresistance and circulating miR-105/93-3p acts as a diagnostic biomarker for triple negative breast cancer. Breast Cancer Res. 2017;19(1):133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fain JS, Van Tongelen A, Loriot A, De Smet C. Epigenetic Coactivation of MAGEA6 and CT-GABRA3 Defines Orientation of a Segmental Duplication in the Human X Chromosome. Cytogenet Genome Res. 2019;159(1):12–8. [DOI] [PubMed] [Google Scholar]
- 26.Shen Z, Zhou R, Liu C, Wang Y, Zhan W, Shao Z, et al. MicroRNA-105 is involved in TNF-α-related tumor microenvironment enhanced colorectal cancer progression. Cell Death Dis. 2017;8(12):3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou W, Fong MY, Min Y, Somlo G, Liu L, Palomares MR, et al. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell. 2014;25(4):501–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anastasiadou E, Stroopinsky D, Alimperti S, Jiao AL, Pyzer AR, Cippitelli C, et al. Epstein-Barr virus-encoded EBNA2 alters immune checkpoint PD-L1 expression by downregulating miR-34a in B-cell lymphomas. Leukemia. 2019;33(1):132–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Network CGAR. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513(7517):202–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paraskevopoulou MD, Georgakilas G, Kostoulas N, Vlachos IS, Vergoulis T, Reczko M, et al. DIANA-microT web server v5.0: service integration into miRNA functional analysis workflows. Nucleic Acids Res. 2013;41(Web Server issue):W169–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gottwein E, Corcoran DL, Mukherjee N, Skalsky RL, Hafner M, Nusbaum JD, et al. Viral microRNA targetome of KSHV-infected primary effusion lymphoma cell lines. Cell Host Microbe. 2011;10(5):515–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kang D, Skalsky RL, Cullen BR. EBV BART MicroRNAs Target Multiple Proapoptotic Cellular Genes to Promote Epithelial Cell Survival. PLoS Pathog. 2015;11(6):e1004979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Skalsky RL, Corcoran DL, Gottwein E, Frank CL, Kang D, Hafner M, et al. The viral and cellular microRNA targetome in lymphoblastoid cell lines. PLoS Pathog. 2012;8(1):e1002484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwentner R, Herrero-Martin D, Kauer MO, Mutz CN, Katschnig AM, Sienski G, et al. The role of miR-17–92 in the miRegulatory landscape of Ewing sarcoma. Oncotarget. 2017;8(7):10980–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Erhard F, Haas J, Lieber D, Malterer G, Jaskiewicz L, Zavolan M, et al. Widespread context dependency of microRNA-mediated regulation. Genome Res. 2014;24(6):906–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(Database issue):D92–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cristescu R, Lee J, Nebozhyn M, Kim KM, Ting JC, Wong SS, et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat Med. 2015;21(5):449–56. [DOI] [PubMed] [Google Scholar]
- 39.Mimura K, Teh JL, Okayama H, Shiraishi K, Kua LF, Koh V, et al. PD-L1 expression is mainly regulated by interferon gamma associated with JAK-STAT pathway in gastric cancer. Cancer Sci. 2018;109(1):43–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moon JW, Kong SK, Kim BS, Kim HJ, Lim H, Noh K, et al. IFNγ induces PD-L1 overexpression by JAK2/STAT1/IRF-1 signaling in EBV-positive gastric carcinoma. Sci Rep. 2017;7(1):17810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sheng Q, Zhang Y, Wang Z, Ding J, Song Y, Zhao W. Cisplatin-mediated downregulation of miR-145 contributes to up-regulation of PD-L1 via the c-Myc transcription factor in cisplatin-resistant ovarian carcinoma cells. Clin Exp Immunol. 2020;200(1):45–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mastroianni J, Stickel N, Andrlova H, Hanke K, Melchinger W, Duquesne S, et al. miR-146a Controls Immune Response in the Melanoma Microenvironment. Cancer Res. 2019;79(1):183–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yee D, Shah KM, Coles MC, Sharp TV, Lagos D. MicroRNA-155 induction via TNF-α and IFN-γ suppresses expression of programmed death ligand-1 (PD-L1) in human primary cells. J Biol Chem. 2017;292(50):20683–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prestipino A, Emhardt AJ, Aumann K, O’Sullivan D, Gorantla SP, Duquesne S, et al. Oncogenic JAK2. Sci Transl Med. 2018;10(429). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ghandi M, Huang FW, Jané-Valbuena J, Kryukov GV, Lo CC, McDonald ER, et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature. 2019;569(7757):503–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chatterjee A, Rodger EJ, Ahn A, Stockwell PA, Parry M, Motwani J, et al. Marked Global DNA Hypomethylation Is Associated with Constitutive PD-L1 Expression in Melanoma. iScience. 2018;4:312–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li H, Chiappinelli KB, Guzzetta AA, Easwaran H, Yen RW, Vatapalli R, et al. Immune regulation by low doses of the DNA methyltransferase inhibitor 5-azacitidine in common human epithelial cancers. Oncotarget. 2014;5(3):587–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kataoka K, Miyoshi H, Sakata S, Dobashi A, Couronné L, Kogure Y, et al. Frequent structural variations involving programmed death ligands in Epstein-Barr virus-associated lymphomas. Leukemia. 2019;33(7):1687–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Danbaran GR, Aslani S, Sharafkandi N, Hemmatzadeh M, Hosseinzadeh R, Azizi G, et al. How microRNAs affect the PD-L1 and its synthetic pathway in cancer. Int Immunopharmacol. 2020;84:106594. [DOI] [PubMed] [Google Scholar]
- 50.Fong MY, Yan W, Ghassemian M, Wu X, Zhou X, Cao M, et al. Cancer-secreted miRNAs regulate amino-acid-induced mTORC1 signaling and fibroblast protein synthesis. EMBO Rep. 2021;22(2):e51239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ren Q, Zhu P, Zhang H, Ye T, Liu D, Gong Z, et al. Identification and validation of stromal-tumor microenvironment-based subtypes tightly associated with PD-1/PD-L1 immunotherapy and outcomes in patients with gastric cancer. Cancer Cell Int 2020;20:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


