Abstract
Genetic diversity of surface exposed and stage specific Plasmodium falciparum immunogenic proteins pose a major roadblock to developing an effective malaria vaccine with broad and long-lasting immunity. We conducted a prospective genetic analysis of candidate antigens (msp1, ama1, rh5, eba175, glurp, celtos, csp, lsa3, Pfsea, trap, conserved chrom3, hyp9, hyp10, phistb, surfin8.2, and surfin14.1) for malaria vaccine development on 2375 P. falciparum sequences from 16 African countries. We described signatures of balancing selection inferred from positive values of Tajima’s D for all antigens across all populations except for glurp. This could be as a result of immune selection on these antigens as positive Tajima’s D values mapped to regions with putative immune epitopes. A less diverse phistb antigen was characterised with a transmembrane domain, glycophosphatidyl anchors between the N and C- terminals, and surface epitopes that could be targets of immune recognition. This study demonstrates the value of population genetic and immunoinformatic analysis for identifying and characterising new putative vaccine candidates towards improving strain transcending immunity, and vaccine efficacy across all endemic populations.
Subject terms: Genomics, Sequencing, Infectious diseases, Vaccines
Introduction
Malaria remains a deadly disease with major economic implications, imposing hardship and marginalization of poorly resourced communities, especially in sub-Saharan Africa (sSA). An effective malaria vaccine would significantly boost malaria control and elimination efforts. However, genetic diversity and the evolutionary arms race between the parasite and the host drives antigenic diversity of surface proteins, and other life cycle stage-specific proteins targeted as potential vaccine antigens, thereby hampering the development of a widely deployable vaccine1.
Field trials of some of the most advanced candidate malaria vaccines have shown strain specificity2, 3, raising the possibility of allele-specific immunity as that seen in vaccines against influenza and other diseases4–7. Most of the current leading malaria vaccine candidates in clinical trials were designed and developed based on clonal laboratory strains, mostly Plasmodium falciparum strain NF54 (clone 3D7), without considering parasite genetic variability in natural populations8–12. RTS,S/AS01, the most advanced of these malaria vaccines, is based on a subunit of 3D7 circumsporozoite surface protein (CSP) antigen. It has shown moderate (< 50%) and time-limited protection in African children13, partly due to the low prevalence (5.2%) of 3D7-type CSP alleles9. Several other recombinant malaria vaccines are currently being evaluated in clinical trials, such as: EBA region II-non-glycosylated (EBA-175 RII NG), apical membrane antigen 1 diversity covering (AMA-1 DiCo), merozoite surface protein-3 (MSP3) long synthetic peptide (LSP), and serine repeat antigen-5. Other recombinant malaria vaccines in clinical trials are described in a recent review by Salamanca et al10. Additionally, the Cell-Traversal protein for Ookinetes and Sporozoites (CelTOS), Thrombospondin Related Adhesion Protein (TRAP), and Liver Stage Antigen 1 (LSA1), which target liver stage parasites; merozoite surface proteins (MSP2) which target blood stage parasites; and transmission blocking stage vaccines targeting Pfs230, Pfs45/48, Pfs25 and Pfs48/4511, 14, 15 are also in advanced development.
Nevertheless, given our incomplete understanding of the complexity of immune responses to infection, the parasite’s mechanisms of invasion, and the parasite genome16–19, most of these vaccine candidates would face most of the challenges observed during the RTS,S/ASO1 development. To overcome some of these difficulties and improve vaccine efficacy by generating cross-protective multi-strain immunity across diverse natural populations, next generation vaccines must use innovative design approaches that account for genetic variables such as differences in antigen allele haplotypes and their frequencies across P. falciparum populations1, 20.
Exploring the allele frequency spectrum and haplotype diversity of vaccine candidates can inform how natural selection and demographic events shape them across multiple populations. For example, characterising genetic diversity and signatures of selection in the parasite genome or for specific antigens has been useful in identifying genes under balancing selection and genic regions that are immunogenic for further development20–22. With the increase of next generation sequencing and openly accessible genome data from multiple endemic populations, future multi-allelic vaccines can benefit from genome scans for signatures of balancing selection. This will allow assessment of allele and haplotype frequencies across a broad range of endemic populations, particularly in sSA where parasite genetic diversity is high22. Such in silico diversity and prediction approaches should include sequence data obtained across multiple temporal and spatial populations. This could be explored to identify new candidates, refine components of current candidate vaccine antigens, with optimal components of relevant alleles from both pre-erythrocytic and erythrocytic targets prior to functional characterisation and clinical trials15.
Here, we conducted population genetic and immunoinformatic analysis of selected malaria candidate vaccine antigens using single nucleotide polymorphisms (SNP) from field isolates from 16 African countries. We estimated the extent of genetic diversity in these antigens and evaluated the evidence of balancing selection that could be shaping their evolution. From this large population dataset, we described a novel putative candidate vaccine antigen—phistb—that could be explored for monovalent vaccine development or as part of a multivalent vaccine. Together, this is the largest exploration of candidate vaccines’ genetic diversity for natural populations of malaria parasites from sSA, providing a framework for designing future functional studies and effective malaria vaccines.
Methods
Ethical considerations
This study utilized data previously published by the Plasmodium Diversity Network Africa (PDNA)23, 24 and the open-source, MalariaGEN (https://www.malariagen.net/projects/pf3k). For the PDNA data, field studies were approved by the respective local ethical review committees. Venous blood (2–5 ml) was collected from study participants aged > 6 months enrolled under approved protocol(s) in each country. All participants and/or their legal guardians provided written informed consent before any study procedures. All procedures performed in studies involving human participants were in accordance with the Declaration of Helsinki. Samples were leukocyte depleted and extracted DNA sequenced by MalariaGEN at the Wellcome Sanger Institute.
To utilize PDNA and the open-source data in the current analysis, this study was elaborated as part of a malaria genomic surveillance proposal reviewed and approved by the Gambian Government/MRC Joint Ethics Committee. The methods employed in this study were in agreement with the MRCG-LSHTM research governance policy.
Vaccine candidates
A total of 16 Plasmodium falciparum vaccine antigens were identified and analysed; among them, 10 (msp1, ama1, rh5, eba175, glurp, celtos, csp, lsa3, Pfsea and trap) were included as potential vaccine antigens. The remaining 6 antigens (conserved chrom3, hyp9, hyp10, phist, and surfin8.2, surfin14.1) were identified using Rsb (a metric for comparing integrated extended haplotype homozygosity between populations), the cross population test for signatures of selection23. Antigens were classified as pre-erythrocytic (celtos, csp, lsa3, Pfsea and trap) or erythrocytic (msp1, ama1, rh5, eba175, glurp, chrom3, hyp9, hyp10, phistb, surfin8.2 and surfin14.1) candidates based on the P. falciparum life cycle stage of expression.
Sequences
The P. falciparum antigen sequences (2375) used in this study were obtained from PDNA study sites from; Gambia (n = 247), Mali (n = 137), Cote-d’Ivoire (n = 70), Ghana (n = 423), Nigeria (n = 34), Cameroon (n = 239), Gabon (n = 55), Kenya (n = 64), Tanzania (n = 300), Ethiopia (n = 34) and Madagascar (n = 25)23. Additionally, open-source sequence data from Guinea (n = 143), Senegal (n = 137), Democratic Republic of Congo (n = 112), Mauritania (n = 86) and Malawi (n = 269) (Fig. S1) were accessed via the Pf3K project (https://www.malariagen.net/projects/pf3k). Details of the methods and curated genotype data can be accessed from https://www.malariagen.net/resource/26 and associated publications25.
Data analysis
Nucleotide diversity analysis and neutrality test
Population genetic analyses were carried out on each candidate vaccine dataset to investigate the genetic diversity as well as the frequency of known haplotypes being incorporated into vaccine candidates. The range and distribution of genetic diversity within and among the natural P. falciparum populations from individual countries were determined. Population genetic parameters were determined using PopGenome R package26. Nucleotide diversity (π) (average number of nucleotide differences per site in pairwise comparisons among DNA sequences) and theta (Ɵ) (population mutation rate) was measured for each candidate vaccine antigen by country. Nucleotide diversity was calculated both for the entire coding sequence of each antigen and in sliding windows to determine regions of increased diversity. Haplotype diversity (Hd) which represents the probability that two randomly sampled alleles are different was estimated per antigen and population.
Neutrality tests designed to distinguish between neutrally evolving sequences under mutation-drift equilibrium- population expansion and contraction, and sequences evolving under non-neutral processes including directional or balancing selection, were derived. We determined Tajima’s D, Fu & Li’s F* and D* statistics. Tajima’s D uses the frequency of segregating nucleotide sites, while Fu’s F* uses the distribution of alleles or haplotypes. Both tests are based on the principle that a rapid population expansion associated with a non-neutral process will show a shift in the allele frequency spectrum compared to a neutral Wright-Fisher model consistent with population expansion under neutral evolution. For all the candidate vaccine antigens, Tajima’s D was estimated by country using vcftools27 for the entire antigen region or in consecutive sliding windows of 100 bases. Positive Tajima’s D values generally suggest balancing selection, or a population contraction is acting to maintain alleles at intermediate frequencies; negative values suggest purifying or positive selection, or population expansion that results in an excess of rare alleles.
Linkage disequilibrium (LD)
To determine correlation between antigen alleles, pair-wise LD between different polymorphic sites was computed based on the genotype correlation coefficient (r2) index between alleles at physically separate loci with a minor allele frequency > 1% across all populations using PLINK v1.90b6.428. Within each population, r2 measures for each antigen were calculated between all pairs of single nucleotide polymorphisms (SNP) and observed pair-wise LD (r2) was averaged for each inter-SNP distance. Decay of LD with physical distance between SNP loci was fitted in R version 3.6.
Population differentiation among sampling populations
To assess geneflow between populations, we first estimated genetic differentiation (i.e. the difference in the average diversity within populations compared to that among populations) expressed as Wright’s fixation index using whole SNP dataset for each vaccine candidate29. The fixation index (FST) was estimated for pairs of populations using the hierFstat package30. For interpretation, FST values < 0.05 was delineated as low differentiation or high gene flow between population pairs, 0.05–0.15 as intermediate, and > 0.25 as high differentiation.
Haplotype cluster network analysis
Genetic relationships between antigen haplotypes of P. falciparum from the 16 African countries were constructed using the median joining algorithm in the R packages Pegas and Ape31. The haplotype network was computed using haplotype pairwise differences as a distance measure, estimated from the number of SNP variants between haplotypes.
B-cell epitope prediction for vaccine candidates
The random forest algorithm trained on epitopes annotated from antibody-antigen protein structures was employed in predicting linear B-cell epitopes for 11 candidate vaccine antigens with at least 3 SNPs, by employing BepiPred-2.0 at a threshold of 0.532. Sequences of these antigens were uploaded on to the server in FASTA format and B-cell epitopes returned as outputs. We further compared the B cell epitopes predicted with segments of the antigens’ sequences with elevated positive Tajima’s D values. This allowed us to map contiguous regions in the proteins which are likely under immune selection and could be included in future designs of multivalent malaria vaccines.
three-dimensional structure and membrane anchor prediction for Phistb
The amino acid sequence of the candidate vaccine antigen was submitted to I-TASSER for in silico prediction analysis using the default settings33. The predicted model with the highest C-score (range: 0–1), used for indicating confidence in the predicted models, was selected and epitopes and regions under balancing selection highlighted. Transmembrane helices and glycophosphatidylinositol (GPI) anchors were predicted by uploading phistb sequence into the TMHMM prediction servers and GPI-SOM respectively34.
Statistical analyses
Spearman's rank nonparametric correlation coefficient was used to measure the correlation between the three neutrality tests applied. Mann–Whitney U non-parametric test was used to assess statistically significant differences between groups assuming a non-Gaussian distribution. Statistical analysis was performed using GraphPad Prism version 8.0. Additionally, statistical comparison of genetic indices for antigens based on region of origin following stratification of populations into two geographical areas: West and Central Africa (Cameroon, Congo, Cote d’Ivoire, Gabon, Gambia, Ghana, Guinea, Mali, Mauritania, Nigeria, and Senegal) versus East Africa (Ethiopia, Kenya, Madagascar, Malawi and Tanzania). This stratification was based on previous reports by the PDNA describing the clustering of African malaria parasites into subpopulations23.
Results
Study population and candidate vaccine antigens
The population dataset used in this study comprise 2375 sequences from West, Central, South Central, East and South-Eastern African countries. Three countries in the population dataset; Nigeria, Ethiopia and Madagascar had fewer parasite sequences (34, 34, and 25 respectively) compared to other countries in our analysis.
Polymorphism and haplotype diversity
The lowest median number of haplotypes per country was 3 for pfsea, and msp1, while the highest was 110 for surfin8.2 (Table S1). All candidate vaccine antigens were initially subjected to nucleotide diversity and FST analyses. The average pairwise nucleotide difference per site estimated as π and Ɵ for the pre-erythrocytic (celtos, csp, lsa3 and trap) and erythrocytic (msp1, ama1, rh5, eba175, glurp, chrom3, hyp9, hyp10, phistb, Pfsea, surfin8.2 and surfin14.1) antigens showed large nucleotide diversity when aggregated across all countries. For the pre-erythrocytic antigens, pfsea had consistently lower π and Ɵ values of 0.00002890 and 0.00009980 respectively (Table 1). The erythrocytic antigens hyp9 had the lowest diversity values (π = 2.538 × 10−4; θ = 8.066 × 10−4) whilst hyp10 had the highest (π = 0.016099; θ = 0.011532)(Table 1). In general, pre-erythrocytic antigens were less diverse than erythrocytic antigens. Nevertheless, when disaggregated by country, the π for pre-erythrocytic antigens ranged from as high 0.0175342 for csp (Gabon) to as low as 0.0000075 for pfsea (Mauritania). For the erythrocytic antigens, the π value was as high as 0.0189404 for hyp10 (Gambia) while the lowest values was 0.0000952 for hyp9 (Cote d’Ivoire), (Table S1). Across all countries, we observed relatively higher genetic diversity in csp (except for Ethiopia) and hyp10. Nevertheless, pre-erythrocytic antigens lsa3 and pfsea had low (0.058–0.096) nucleotide diversity across all countries. Such low levels of diversity were also observed across geographical sites for erythrocytic antigens rh5, hyp9, and glurp (majority of sites). Interestingly, we observed a relatively higher nucleotide diversity at rh5 in east African sites (Kenya, Malawi and Tanzania) and the island of Madagascar (Table S1).
Table 1.
Haplotype diversity of P. falciparum candidate vaccine antigens.
| Vaccine candidates | Pi(π) | Hd | Theta (Ɵ) |
|---|---|---|---|
| Pre-erythrocytic | |||
| celtos | 0.00165175 | 0.9860 | 0.00165175 |
| csp | 0.01555860 | 0.9490 | 0.01219290 |
| lsa3 | 0.00059395 | 0.8705 | 0.00049910 |
| trap | 0.00828865 | 0.9965 | 0.00537735 |
| pfsea | 0.00002890 | 0.0990 | 0.00009980 |
| Erythrocytic | |||
| ama1 | 0.00165175 | 0.9955 | 0.00447465 |
| chrom3 | 0.00483740 | 0.6690 | 0.00363820 |
| eba175 | 0.00181675 | 0.9930 | 0.00105945 |
| glurp | 0.00099680 | 0.9555 | 0.00151625 |
| hyp9 | 0.00025385 | 0.0580 | 0.00080655 |
| hyp10 | 0.01609915 | 0.7995 | 0.01153165 |
| msp1 | 0.00306515 | 0.9985 | 0.00300525 |
| phistb | 0.00160080 | 0.8335 | 0.00122970 |
| rh5 | 0.00087060 | 0.5530 | 0.00095175 |
| surfin8.2 | 0.00808670 | 0.9990 | 0.00413205 |
| surfin14.1 | 0.00153700 | 0.9965 | 0.00091120 |
Pi and theta nucleotide diversity indexes and hd, haplotype diversity.
The haplotype diversity index (Hd) values across all antigens and populations were mostly > 0.4 with the exception of pre-erythrocytic antigen pfsea, (Hd ranging from 0.05 to 0.29) and erythrocytic antigen hyp9 (0.017–0.153) which were low across all populations (Table S1). Candidate vaccine antigens ama1, eba175 and glurp from Ethiopia had lower Hd values than in the other African countries. A similar pattern was observed for pre-erythrocytic antigens. However, it is important to note that a small number of samples were analysed from Ethiopia. To identify any differences within Africa, we compared Hd values from the two subregions, namely West and Central Africa versus East Africa, and found significant differences for lsa3 (P = 0.01), hyp10 (P = 0.0005), phistb (P = 0.0005) and rh5 (P = 0.0032). In our subsequent analysis, antigens with limited SNP information were excluded, which led to removal of five antigens (pfsea, hyp9, hyp10, conserved chrom3 and rh5) from Tajima’s D and LD analysis. The remaining 11 antigens were subjected to Tajima’s D, LD and linear B-cell epitope and 3D-structure prediction.
Genetic differentiation and gene flow
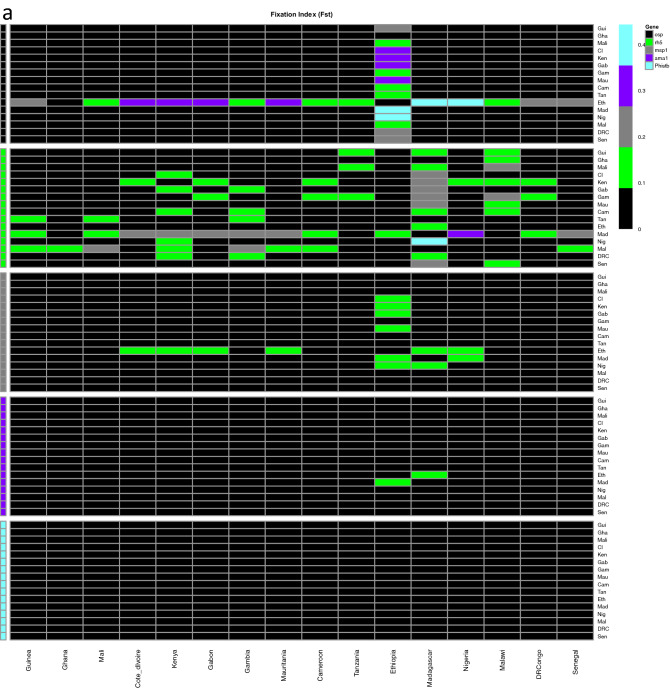
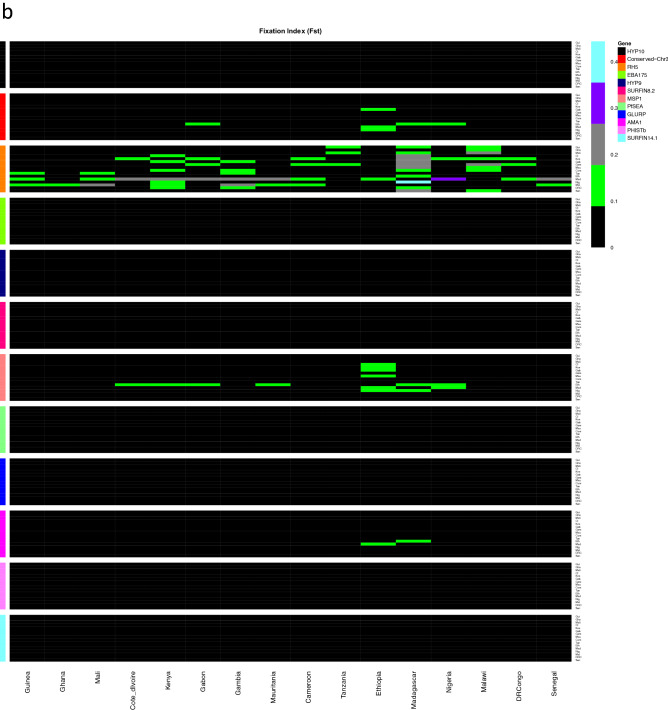
We evaluated genetic differentiation among parasite populations to obtain an insight into gene flow by calculating Wright’s fixation index (FST) of inter-population variance in allele frequencies. Pairwise FST values for the pre-erythrocytic antigens were low suggesting low genetic differentiation among the populations or extensive genetic exchange (high gene flow), with the exception of csp in populations with low samples sizes; Nigeria, Ethiopia and Madagascar, which had high values, FST > 0.25 (Fig. 1). The highest pairwise FST value between Ethiopia and Nigeria for csp was (FST = 0.44). Pairwise FST values for erythrocytic antigens also showed low genetic differentiation, with the exception of rh5 antigen between Madagascar and Gambia (FST = 0.3), which are separated by the widest geographic distance.
Figure 1.
Pairwise FST indices of candidate vaccine antigens from the 16 African countries, (a) Pre-erythrocytic antigens (b) erythrocytic antigens. Fst indices indicate low genetic differentiation for most of the antigens (FST < 0.05). Ethiopia and Madagascar had few samples, 34 and 25 respectively.
Genetic polymorphisms and haplotype frequency
Across all the neutrality tests (Tajima’s D, Fu & Li’s D* and F* summary statistics), values were mostly positive for the candidate vaccine antigens, except for glurp which had negative Tajima’s D values (Table S2 & Fig. S2), suggesting low frequency polymorphisms relative to that expected under neutrality. Tajima’s D values ranged from 4.113 for surfin8.2 in Ghana to 0.456 for msp1 in Madagascar; Fu&Li F* ranged from 4.551 for surfin8.2 in Ghana to 1.069 for phistb in Kenya; and Fu&Li D* ranged from 3.521 for surfin8.2 in Ghana to 1.004 for phistb in Ethiopia (Table S2). The positive neutrality test values obtained for the antigens in this study is suggestive of a pattern of balancing selection for the candidate vaccine antigens across malaria endemic populations in sSA (Table 2). Comparison of Tajima’s D values for the candidate vaccine antigens analysed by subregions revealed that for most of the antigens there were no statistically significant differences (P > 0.05) between East versus West and Central Africa. This is further evidence of standing variation in these antigens across malaria endemic communities in sSA. As expected, there was strong correlation between Tajima’s D and Fu & Li’s F* values for all the antigens across the populations studied (r = 0.882). However, correlation was much lower between Tajima’s D and Fu & Li’s D* (r = 0.03), with some antigens, notably glurp, celtos and phistb, showing a negative correlation, while Fu & Li’s D* and F* had a weak correlation (r = 0.2) for the antigens except for glurp where Tajima’s D was negative but Fu and Li indices were positive (Fig. 2). Tajima’s D becomes negative when there is an excess of rare alleles resulting in more pairwise diversity than the number of segregating sites.
Table 2.
Neutrality tests for candidate vaccine antigens.
| Vaccine candidates | Tajima_D | Fu&Li_F* | Fu&Li_D* |
|---|---|---|---|
| Pre-erythrocytic | |||
| celtos | 0.45 | 1.59 | 1.9195 |
| csp | 0.599 | 1.4595 | 1.595 |
| lsa3 | 0.872 | 1.419 | 1.3525 |
| trap | 1.658 | 2.3895 | 2.3015 |
| Erythrocytic | |||
| ama1 | 2.443 | 2.841 | 2.281 |
| eba175 | 2.09 | 2.4335 | 1.9495 |
| glurp | − 0.184 | 1.1895 | 1.7725 |
| msp1 | 0.1915 | 1.8665 | 2.5675 |
| phistb | 0.6515 | 1.2785 | 1.2975 |
| surfin8.2 | 2.9305 | 3.5335 | 2.9885 |
| surfin14.1 | 0.76175 | 1.72825 | 1.9345 |
Figure 2.
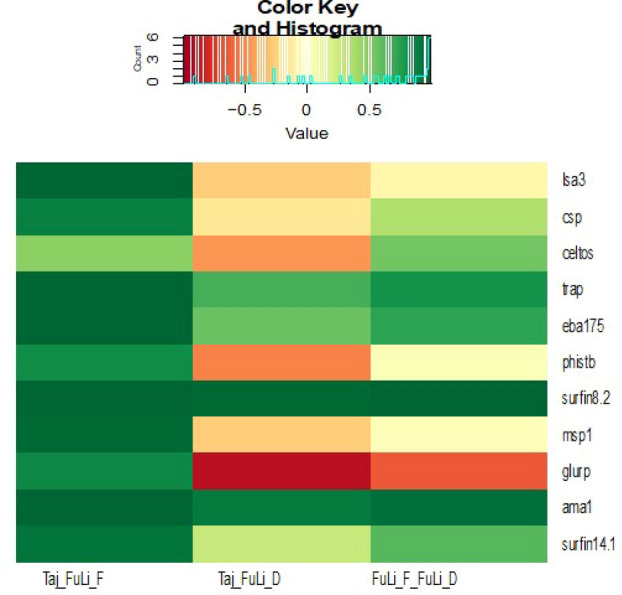
Relationship between Tajima’s D, Fu_Li D* and Fu_LiF*. Positive correlation is indicated by light green to deep green, while negative correlation is represented by light yellow to red.
Haplotype frequency and cluster network
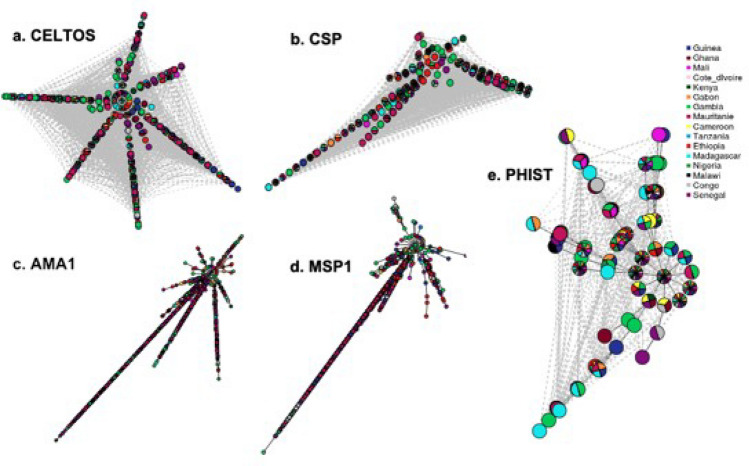
The frequency of haplotypes for all antigens and details of haplotypes of each antigen per country are given in Table S3. To further understand the relatedness of the haplotypes across our populations, haplotype networks were constructed on selected vaccine candidates already under investigation in clinical trials; 2 pre-erythrocytic (celtos and csp) and 3 erythrocytic (ama1, msp1 and phistb) antigens. The pre-erythrocytic antigens had a high number of haplotypes with no geographic clusters, suggesting lack of structure. Haplotype network of the erythrocytic antigens followed a similar pattern to the pre-erythrocytic antigens, except for phistb where geographic clustering of some haplotypes was evident (Fig. 3).
Figure 3.
Representative haplotype cluster networks of pre-erythrocytic and erythrocytic vaccine candidates. Clustering of haplotypes across populations for pre-erythrocytic candidates (a) CELTOS, (b) CSP, and erythrocytic vaccine candidates (c) AMA1, (d) MSP1 and (e) PHISTB. Each node represents a haplotype with pies colour coded according to the proportion of haplotypes originating from a specific country. The lengths of edges connecting each pair of pies is proportion to the number of base pair substitutions separating the haplotypes.
Linkage disequilibrium
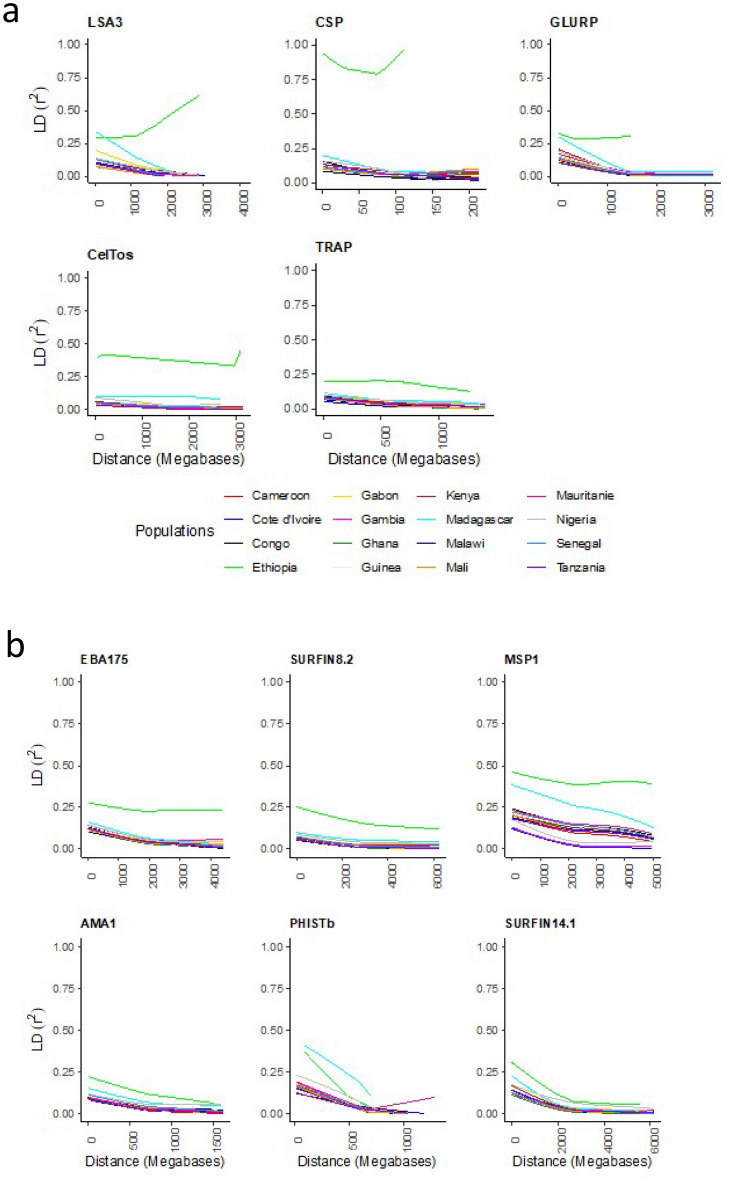
Linkage Disequilibrium (LD) declined rapidly with physical separating pairs of SNPs for all antigens, falling below an r2 of 0.05 within 1000 base pairs (Fig. 4). Exceptionally, LD values were higher for Ethiopia, with r2 > 0.1 across most pairs of SNPs not significantly decaying with physical distance for most antigens. This observation may be underscored by variance in malaria transmission dynamics, population migration, demography and recombination potential within the SNPs in the genes analysed.
Figure 4.
Linkage disequilibrium, (a) pre-erythrocytic and (b) erythrocytic vaccine candidates. R2 values decrease as distance increases. LD in Ethiopian samples for both (a) and (b) have a unique decay pattern.
Antigenic potential and 3D structure of phistb
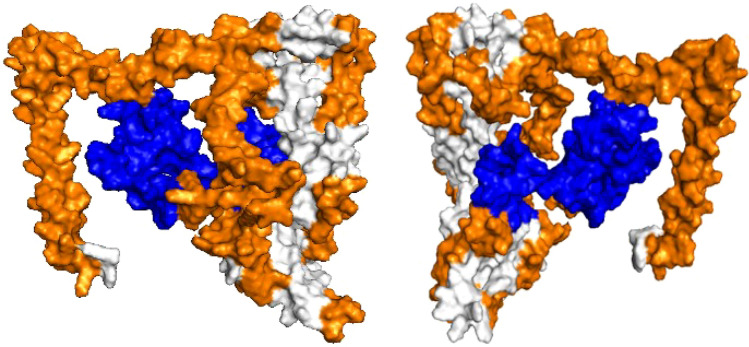
Epitope scanning for linear B cell epitopes on the antigens revealed several epitopes. Some of the regions with immune epitopes overlap with regions of high Tajima’s D, indicative of immune selection. The list of the B cell epitopes for the vaccine candidates and regions mapping with high Tajima’s D are provided in Table S4. We further explored the immunogenicity of phistb, which had only 2 predominant haplotypes (Table S3), and evidence of genetic structure between geographical parasite populations. The TMHMM server predicted one transmembrane helix between amino acid positions 54 and 73, and the likelihood of a signal peptide (Fig. S3). Phistb also has a post-translational modification- glycophosphatidyl inositol (GPI) site in its C-terminal region. GPI is used by proteins for anchoring to the plasma membrane (Fig. S4). Following 3D structure modelling of Phistb the linear B-cell epitopes predicted overlapped with regions with high positive Tajima’s D values (Fig. 5).
Figure 5.
Left and right-handed three-dimensional structure of phistb from I-Tasser, mapping regions with positive Tajima’s D (blue) and B-cell epitopes in orange.
Discussion
We carried out a large-scale population genetic analyses of candidate vaccine antigens and hitherto uncharacterised potential antigens using SNP data of P. falciparum isolates (n = 2375) from 16 African countries. Most advanced malaria vaccine candidates were initially designed without considering genetic diversity, resulting in poor protective efficacy as shown by FMP1/AS02A MSP1 or the AMA1 candidates that had limited vaccine allele representation in naturally circulating parasites, especially in Africa11, 35, 36. Therefore this focused analysis on African parasite populations is relevant, given intense but heterogenous transmission, and high parasite genetic diversity across sub-populations3, 4.
By screening for antigens with signatures of directional selection, we expected to identify those with dominant haplotypes and exposed immune epitopes, that could be included into the panel of candidates currently under development. We observed significant diversity amongst characterised vaccine candidates and new putative antigens across all populations, and this may result in poor efficacy both within clinical trials and in future deployment37. As immunity could be driving balancing selection signatures against specific domains of the antigens, our scan for signatures of selection localised these segments, with sequences of known immune epitopes that could be targeted for functional evaluation. However, antigens under high immune selection pressures and balancing selection are often highly polymorphic with moderate minor allele frequencies, likely to result in variance in allele specific immune response between populations38. This applies also to well-known erythrocytic vaccine candidates based on ama1, msp1 and eba175, in which variation and selection has been a major hurdle in achieving broad efficacy across malaria endemic populations35, 37, 39. Like msp1 and ama1, other erythrocytic antigens such as surfin 8.2 and surfin 14.1 showed signatures of both directional and balancing selection, the latter, also due to an excess of intermediate frequency variants that may result in vaccine escape. The surfin family of proteins are expressed by genes close to the sub-telomeres where other surface variant antigens (pfemp1, rifin, stevor) are encoded5. These are expressed on the surface of infected erythrocytes and are known targets of protective immunity and generating heterologous antibodies associated with reduction in febrile illness due to malaria infection6. Surfin8.2 had the highest number of segregating sites and has been shown to be preferentially expressed in gametocytes40, 41. Being a PEXEL (Plasmodium export element) negatively exported protein with a transmembrane domain42, it will be exposed to the surface and can therefore be a target for transmission blocking vaccine development. Another transmission blocking vaccine, glurp, is in advanced development alone or as a multivalent component with other erythrocytic candidates such as msp3 (GMZ2)43. Its lower Tajima’s D value and smaller number of haplotypes could provide an advantage for combination with other candidates such as surfin8.2 to provide a broad range of erythrocytic protection and transmission blocking activity.
While erythrocytic antigens dominate the panel of malaria vaccine candidates in development, the only successful vaccine to date is the CSP-based RTS,S/AS-01. As shown already, the CSP gene is highly diverse, one of the factors contributing to reduced efficacy across populations. We assessed diversity at four other pre-erythrocytic candidates, celtos, lsa3, pfsea and trap. The least polymorphic one was the schizont egress antigen pfsea, whose antibodies have been associated with protection against high parasitemia and severe disease44, 45. It is under preclinical exploration in combination with invariant carboxyl of PfGARP and PfMSP1 in a tri-valent vaccine formulation. As previously described, lsa3 also has a low number of haplotypes and is expressed on both sporozoites and the erythrocytic stages46–48. Thus it could also be considered as a blood stage vaccine candidate given its antibodies inhibit parasite growth in the erythrocyte48. Its further development against both erythrocytic and pre-erythrocytic stages will have to consider the African haplotypes described here. This also applies to one of the recent prominent candidates, rh5, which, despite a low number of haplotypes, had 12 non-synonymous SNPs and high between population differentiation determined by the fixation index FST. Strong differentiation between eastern versus western African parasite populations would imply that population specific variation in haplotypes need to be considered for rh5 vaccine development. Our findings also support previous studies which demonstrated that the number of haplotypes were consistently higher in countries with higher malaria transmission such as Cameroon, Mali and Malawi49–51. Therefore, continuous genetic screening for low diversity candidates to add to the pool of current antigens that can be combined in a multivalent malaria vaccine remains important.
We demonstrated the power of this genetic analysis by exemplifying one of the low diversity candidate antigens, phistb. PHIST proteins belong to the Plasmodium helical interspersed subtelomeric (PHIST) family made up of 65 gene members (PlasmoDB database, www.plasmodb.org) and are unified by possessing a single domain termed PHIST that is predicted to be composed solely of alpha helices52, 53. This family of exported proteins are conserved across the human Plasmodium species; P. falciparum, P. vivax and P. knowlesi52. As we identified a transmembrane domain and GPI anchors, we expect phistb to be surface anchored54. Previous studies have described phistb to localise to the host erythrocyte periphery through a PRESAN domain and an N-terminal sequence, and therefore exposed to immune interaction. Unsurprisingly, segments of the antigen had high Tajima’s D values, indicating balancing selection probably due to immune selection. These segments mapped to predicted B-cell epitopes which could be included in future candidate vaccine designs, preclinical testing and for selection of an optimal vaccine cocktail. The low genetic diversity, limited haplotypes in the population and linear B-cell epitopes of phistb are desirable features for designing an antimalarial vaccine that is less likely to produce allele-specific immune responses2, 50. Recent studies with in vitro expressed phistb protein demonstrated the presence of significant specific anti-phistb antibodies in children, with malaria specific immune responses55. This further supports predictions from genetic analysis and epitope mapping in line with other studies that have identified potential immune targets by modelling protein structures and predicting the functional relevance and implications of different domains as vaccine candidates56, 57. PHIST proteins are involved in remodelling infected red blood cells54. However, data on host antimalarial immune responses to these proteins, which could be crucial for vaccine development, is scarce.
Despite the relevant finding on top candidates like rh5 and new potential candidates like phistb, our study has several limitations. Samples were collected mostly in one field site per country, and thus they may not provide an accurate representation of the circulating haplotypes across each country. In addition, the number of samples available from some countries such as Madagascar, Ethiopia and Nigeria, were small. Moreover, protocols for sample collection differed by country, including the time of sampling, which may influence the parasite haplotypes due to seasonality. The analysis was limited to high quality SNPs, which could have missed important genetic variations; and there is also the possibility of ascertainment bias in the dataset that have been utilised in this study, and more broader sampling for suspected vaccine candidate genes would be useful. However, despite these limitations, the analysis was comprehensive since we used a large dataset and provide a framework and basis for future studies and considerations. To verify our findings, functional studies and expanded genetic analysis incorporating targeted sequencing data are needed to further support our findings.
In conclusion, despite the broad genetic diversity of antigens of African P. falciparum isolates, it may be possible to identify the most prevalent antigen haplotypes within the continent for inclusion in a multivalent vaccine. Our findings demonstrate population genetic analysis can be used to identify antigens that consider African parasite genetic diversity for design of new vaccines.
Supplementary Information
Acknowledgements
We would like to thank the study participants, local health workers and malaria control programmes from PDNA sites for their continued support. Genome sequencing was done at the Wellcome Sanger Institute as part of the MalariaGEN Plasmodium falciparum Community Project (www.malariagen.net/projects). We thank the Pf3K for allowing access to non-PDNA data. The authors declare no competing interest. The short-read sequences used in this publication are available in the ENA and SRA databases.
Author contributions
O.A. and F.M.D. analysed the data and O.A. wrote the first draft of the manuscript. A.A.N., E.K. and A.G. conceived the study, while O.A., E.K., A.A.N. and F.M.D. designed the experimental plan for analyses. O.A., A.G., L.A., L.G., T.A., D.I., U.D.A., A.A.D. E.K. and A.A.N. participated in review of the final draft of the manuscript. M.R., O.M., W.Y., B.A., M.B. and K.M.O contributed sequences and resources used for analysis. E.K. and A.A.N. coordinated the collaboration and led the study. A.A.D coordinates the PDNA consortium, contributed the sequences and all authors approved the final draft of the manuscript.
Disclaimer Material has been reviewed by the Walter Reed Army Institute of Research. There is no objection to its presentation and/or publication. The opinions or assertions contained herein are the private views of the author, and are not to be construed as official, or as reflecting true views of the Department of the Army or the Department of Defence. The investigators have adhered to the policies for protection of human subjects as prescribed in AR 70-25.
Funding
A.A.N., O.A., L.A., A.G. and A.A.D. are currently supported through the DELTAS Africa Initiative, an independent funding scheme of the African Academy of Sciences (AAS)’s Alliance for Accelerating Excellence in Science in Africa (AESA), and are also supported by the New Partnership for Africa’s Development Planning and Coordinating Agency (NEPAD Agency) with funding from Wellcome (DELGEME grant 107740/Z/15/Z) and the U.K. government. The views expressed in this publication are those of the authors and not necessarily those of AAS, NEPAD Agency, Wellcome, the U.S. Army or the Department of Defense, or the U.K. government. The investigators have adhered to the policies for protection of human subjects as prescribed in AR-70. Sequencing was undertaken in partnership with MalariaGEN and the Parasites and Microbes program at the Wellcome Sanger Institute with funding from Wellcome (206194; 090770/Z/09/Z) and by the MRC Centre for Genomics and Global Health which is jointly funded by the Medical Research Council and the Department for International Development (DFID) (G0600718 to D.K.; M006212).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: O. Ajibola and M. F. Diop.
Contributor Information
E. Kamau, Email: edwin.kamau.mil@mail.mil
A. Amambua-Ngwa, Email: angwa@mrc.gm
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-95442-4.
References
- 1.Matuschewski K. Vaccines against malaria-still a long way to go. FEBS J. 2017;284:2560–2568. doi: 10.1111/febs.14107. [DOI] [PubMed] [Google Scholar]
- 2.Neafsey DE, et al. Genetic diversity and protective efficacy of the RTS, S/AS01 malaria vaccine. N. Engl. J. Med. 2015;373:2025–2037. doi: 10.1056/NEJMoa1505819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thera MA, et al. A field trial to assess a blood-stage malaria vaccine. N. Engl. J. Med. 2011;365:1004–1013. doi: 10.1056/NEJMoa1008115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pringle JC, et al. High plasmodium falciparum genetic diversity and temporal stability despite control efforts in high transmission settings along the international border between Zambia and the Democratic Republic of the Congo. Malar. J. 2019;18:400. doi: 10.1186/s12936-019-3023-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flück C, et al. Strain-specific humoral response to a polymorphic malaria vaccine. Infect. Immun. 2004;72:6300–6305. doi: 10.1128/IAI.72.11.6300-6305.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Genton B, et al. A recombinant blood-stage malaria vaccine reduces Plasmodium falciparum density and exerts selective pressure on parasite populations in a phase 1–2b trial in Papua New Guinea. J. Infect. Dis. 2002;185:820–827. doi: 10.1086/339342. [DOI] [PubMed] [Google Scholar]
- 7.Rastogi D, et al. Antigen-specific immune responses to influenza vaccine in utero. J. Clin. Invest. 2007;117:1637–1646. doi: 10.1172/JCI29466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ouattara A, et al. Designing malaria vaccines to circumvent antigen variability. Vaccine. 2015;33:7506–7512. doi: 10.1016/j.vaccine.2015.09.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pringle JC, et al. RTS, S/AS01 malaria vaccine mismatch observed among Plasmodium falciparum isolates from southern and central Africa and globally. Sci. Rep. 2018;8:6622. doi: 10.1038/s41598-018-24585-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salamanca DR, et al. Plasmodium falciparum blood stage antimalarial vaccines: An analysis of ongoing clinical trials and new perspectives related to synthetic vaccines. Front. Microbiol. 2019;10:2712. doi: 10.3389/fmicb.2019.02712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Draper SJ, et al. Malaria vaccines: Recent advances and new horizons. Cell Host Microbe. 2018;24:43–56. doi: 10.1016/j.chom.2018.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moser KA, et al. Strains used in whole organism Plasmodium falciparum vaccine trials differ in genome structure, sequence, and immunogenic potential. Genome Med. 2020;12:6. doi: 10.1186/s13073-019-0708-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.RTS, S Clinical Trials Partnership. Efficacy and safety of RTS,S/AS01 malaria vaccine with or without a booster dose in infants and children in Africa: final results of a phase 3, individually randomised, controlled trial. Lancet Lond. Engl.386, 31–45 (2015). [DOI] [PMC free article] [PubMed]
- 14.Barry, A. E. & Arnott, A. Strategies for designing and monitoring malaria vaccines targeting diverse antigens. Front. Immunol.5 (2014). [DOI] [PMC free article] [PubMed]
- 15.World Health Organization. Malaria vaccine rainbow tables (2013).
- 16.Head MG, et al. Global funding trends for malaria research in sub-Saharan Africa: A systematic analysis. Lancet Glob. Health. 2017;5:e772–e781. doi: 10.1016/S2214-109X(17)30245-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crompton PD, Pierce SK, Miller LH. Advances and challenges in malaria vaccine development. J. Clin. Invest. 2010;120:4168–4178. doi: 10.1172/JCI44423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arama C, Troye-Blomberg M. The path of malaria vaccine development: Challenges and perspectives. J. Intern. Med. 2014;275:456–466. doi: 10.1111/joim.12223. [DOI] [PubMed] [Google Scholar]
- 19.Beeson JG, et al. Challenges and strategies for developing efficacious and long-lasting malaria vaccines. Sci. Transl. Med. 2019;11:eaau1458. doi: 10.1126/scitranslmed.aau1458. [DOI] [PubMed] [Google Scholar]
- 20.Barry AE, Schultz L, Buckee CO, Reeder JC. Contrasting population structures of the genes encoding ten leading vaccine-candidate antigens of the human malaria parasite, Plasmodium falciparum. PLoS ONE. 2009;4:e8497. doi: 10.1371/journal.pone.0008497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cortés A, et al. Geographical structure of diversity and differences between symptomatic and asymptomatic infections for Plasmodium falciparum vaccine candidate AMA1. Infect. Immun. 2003;71:1416–1426. doi: 10.1128/IAI.71.3.1416-1426.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amambua-Ngwa A, et al. Population genomic scan for candidate signatures of balancing selection to guide antigen characterization in malaria parasites. PLoS Genet. 2012;8:e1002992. doi: 10.1371/journal.pgen.1002992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amambua-Ngwa A, et al. Major subpopulations of Plasmodium falciparum in sub-Saharan Africa. Science. 2019;365:813–816. doi: 10.1126/science.aav5427. [DOI] [PubMed] [Google Scholar]
- 24.Kamau E, et al. K13-propeller polymorphisms in plasmodium falciparum parasites from Sub-Saharan Africa. J. Infect. Dis. 2014 doi: 10.1093/infdis/jiu608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pearson RD, Amato R, Kwiatkowski DP. & MalariaGEN plasmodium falciparum community project. An open dataset of plasmodium falciparum genome variation in 7000 worldwide samples. Welcome Open Res. 2019 doi: 10.1101/824730. [DOI] [Google Scholar]
- 26.Pfeifer B, Wittelsbürger U, Ramos-Onsins SE, Lercher MJ. PopGenome: An efficient Swiss Army Knife for population genomic analyses in R. Mol. Biol. Evol. 2014;31:1929–1936. doi: 10.1093/molbev/msu136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Danecek P, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang CC, et al. Second-generation PLINK: Rising to the challenge of larger and richer datasets. GigaScience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- 30.Goudet J. hierfstat, a package for r to compute and test hierarchical F-statistics. Mol. Ecol. Notes. 2005;5:184–186. doi: 10.1111/j.1471-8286.2004.00828.x. [DOI] [Google Scholar]
- 31.Paradis E. Pegas: An R package for population genetics with an integrated-modular approach. Bioinformatics. 2010;26:419–420. doi: 10.1093/bioinformatics/btp696. [DOI] [PubMed] [Google Scholar]
- 32.Jespersen MC, Peters B, Nielsen M, Marcatili P. BepiPred-2.0: Improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res. 2017;45:W24–W29. doi: 10.1093/nar/gkx346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinform. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sonnhammer EL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1998;6:175–182. [PubMed] [Google Scholar]
- 35.Escalante AA, et al. Polymorphism in the gene encoding the apical membrane antigen-1 (AMA-1) of Plasmodium falciparum. X. Asembo Bay Cohort Project. Mol. Biochem. Parasitol. 2001;113:279–287. doi: 10.1016/S0166-6851(01)00229-8. [DOI] [PubMed] [Google Scholar]
- 36.Duan J, et al. Population structure of the genes encoding the polymorphic Plasmodium falciparum apical membrane antigen 1: Implications for vaccine design. Proc. Natl. Acad. Sci. 2008;105:7857–7862. doi: 10.1073/pnas.0802328105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Verra F, et al. Contrasting signatures of selection on the Plasmodium falciparum erythrocyte binding antigen gene family. Mol. Biochem. Parasitol. 2006;149:182–190. doi: 10.1016/j.molbiopara.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 38.Conway DJ, et al. A principal target of human immunity to malaria identified by molecular population genetic and immunological analyses. Nat. Med. 2000;6:689–692. doi: 10.1038/76272. [DOI] [PubMed] [Google Scholar]
- 39.Beeson JG, et al. Merozoite surface proteins in red blood cell invasion, immunity and vaccines against malaria. FEMS Microbiol. Rev. 2016;40:343–372. doi: 10.1093/femsre/fuw001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kanoi BN, et al. Global repertoire of human antibodies against plasmodium falciparum RIFINs, SURFINs, and STEVORs in a malaria exposed population. Front. Immunol. 2020;11:893. doi: 10.3389/fimmu.2020.00893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Winter G, et al. SURFIN is a polymorphic antigen expressed on Plasmodium falciparum merozoites and infected erythrocytes. J. Exp. Med. 2005;201:1853–1863. doi: 10.1084/jem.20041392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morita M, et al. Immunoscreening of Plasmodium falciparum proteins expressed in a wheat germ cell-free system reveals a novel malaria vaccine candidate. Sci. Rep. 2017;7:46086. doi: 10.1038/srep46086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Theisen M, Adu B, Mordmüller B, Singh S. The GMZ2 malaria vaccine: From concept to efficacy in humans. Expert Rev. Vaccines. 2017;16:907–917. doi: 10.1080/14760584.2017.1355246. [DOI] [PubMed] [Google Scholar]
- 44.Raj DK, et al. Antibodies to PfSEA-1 block parasite egress from RBCs and protect against malaria infection. Science. 2014;344:871–877. doi: 10.1126/science.1254417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kurtis JD, et al. Maternally-derived antibodies to Schizont Egress antigen-1 and protection of infants from severe malaria. Clin. Infect. Dis. 2019;68:1718–1724. doi: 10.1093/cid/ciy728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Daubersies P, et al. Protection against Plasmodium falciparum malaria in chimpanzees by immunization with the conserved pre-erythrocytic liver-stage antigen 3. Nat. Med. 2000;6:1258–1263. doi: 10.1038/81366. [DOI] [PubMed] [Google Scholar]
- 47.Longley RJ, et al. Comparative assessment of vaccine vectors encoding ten malaria antigens identifies two protective liver-stage candidates. Sci. Rep. 2015;5:11820. doi: 10.1038/srep11820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Prieur E, Druilhe P. The malaria candidate vaccine liver stage antigen-3 is highly conserved in Plasmodium falciparum isolates from diverse geographical areas. Malar. J. 2009;8:247. doi: 10.1186/1475-2875-8-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anong DN, Nkuo-Akenji T, Fru-Cho J, Amambua-Ngwa A, Titanji VPK. Genetic diversity of Plasmodium falciparum in Bolifamba, on the slopes of Mount Cameroon: Influence of MSP1 allelic variants on symptomatic malaria and anaemia. Ann. Trop. Med. Parasitol. 2010;104:25–33. doi: 10.1179/136485910X12607012373876. [DOI] [PubMed] [Google Scholar]
- 50.Ouattara A, et al. Extent and dynamics of polymorphism in the malaria vaccine candidate plasmodium falciparum reticulocyte-binding protein homologue-5 in Kalifabougou, Mali. Am. J. Trop. Med. Hyg. 2018;99:43–50. doi: 10.4269/ajtmh.17-0737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ocholla H, et al. Whole-genome scans provide evidence of adaptive evolution in malawian plasmodium falciparum isolates. J. Infect. Dis. 2014;210:1991–2000. doi: 10.1093/infdis/jiu349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tarr SJ, Moon RW, Hardege I, Osborne AR. A conserved domain targets exported PHISTb family proteins to the periphery of Plasmodium infected erythrocytes. Mol. Biochem. Parasitol. 2014;196:29–40. doi: 10.1016/j.molbiopara.2014.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sargeant TJ, et al. Lineage-specific expansion of proteins exported to erythrocytes in malaria parasites. Genome Biol. 2006;7:R12. doi: 10.1186/gb-2006-7-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oberli A, et al. A Plasmodium falciparum PHIST protein binds the virulence factor PfEMP1 and comigrates to knobs on the host cell surface. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2014;28:4420–4433. doi: 10.1096/fj.14-256057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Isebe TI, et al. Molecular characterization of Plasmodium falciparum PHISTb proteins as potential targets of naturally-acquired immunity against malaria. Wellcome Open Res. 2020;5:136. doi: 10.12688/wellcomeopenres.15919.1. [DOI] [Google Scholar]
- 56.Dutta S, Lee SY, Batchelor AH, Lanar DE. Structural basis of antigenic escape of a malaria vaccine candidate. Proc. Natl. Acad. Sci. 2007;104:12488–12493. doi: 10.1073/pnas.0701464104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Takala SL, et al. Extreme polymorphism in a vaccine antigen and risk of clinical malaria: Implications for vaccine development. Sci. Transl. Med. 2009;1:2–5. doi: 10.1126/scitranslmed.3000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.