Abstract
Glia make up roughly half of all cells in the mammalian nervous system and play a major part in nervous system development, function, and disease. Although research in the past few decades has shed light on their morphological and functional diversity, there is still much to be known about key aspects of their development such as the generation of glial diversity and the factors governing proper morphogenesis. Glia of the nematode C. elegans possess many developmental and morphological similarities with their vertebrate counterparts and can potentially be used as a model to understand certain aspects of glial biology owing to advantages such as its genetic tractability and fully mapped cell lineage. In this review, we summarize recent progress in our understanding of genetic pathways that regulate glial development in C. elegans and discuss how some of these findings may be conserved.
Keywords: C. elegans, development, glia, gliogenesis, morphogenesis
Introduction
Since their discovery by Virchow in 1846, glia were long thought to be a group of homogenous cells that passively supported neuronal function. Since then, we have found that they are a diverse group of cells with significant roles during nervous system development and function. Consistent with the plethora of functions they perform in the nervous system, glia exhibit remarkable morphological and molecular diversity, even within glia of the same type [1–4]. While we have learned much about their various functions in the past few decades, there is still much we do not know about how these highly specialized cells develop and how disruptions during this process may affect nervous system function. Since glial dysfunction has been associated with a long list of neurological disorders [5–7], learning more about their developmental mechanisms may help inform our understanding of these disorders.
Our understanding of glial biology can often be hampered by experimental obstacles presented by the diversity of glia and the broad number of essential roles they play. For example, it is often difficult to track specific glial populations in the mammalian brain, especially given our growing appreciation of glial heterogeneity and the varied landscape throughout the brain. Furthermore, since glia are required for neuronal survival, manipulation of glia in vivo often leads to severe defects and death that make studying other aspects harder. On the other hand, culturing glial cells can provide many mechanistic insights into glial biology and function, but doing so often removes them from their immediate cellular surroundings that include other neurons and glia.
One way around this is to use simpler model organisms like C. elegans and Drosophila, whose glia show many similarities with their vertebrate counterparts [8–10]. In C. elegans, nervous system development is deterministic with a fixed number of neurons and glia whose stereotyped development can be specifically studied with spatial and temporal precision [11]. Furthermore, the wealth of genetic tools available paired with the fact that its glia are generally not required for survival of the worm means that many aspects of glial development can be studied in an in vivo setting. Here, we summarize some of the knowledge gained about glial development in C. elegans.
C. elegans glia
The C. elegans hermaphrodite possesses 302 neurons, 50 ectoderm-derived glia, and six mesoderm-derived glial-like cells. These 50 ectoderm-derived glia can be further divided into sheath and socket cells. In the C. elegans sense organs called sensilla, one or more sensory neurons associate with one socket and one sheath cell, where the glia ensheath the dendritic endings of the neurons (Fig. 1A) [12]. In the head, these glia project processes to the nose, where apical junctions connect the socket and sheath glia to form a channel [12,13]. Neuronal endings pass through this channel to interact with the environment, where they can interact with the environment as well the glia that envelop them (Fig. 1B).
Fig. 1.
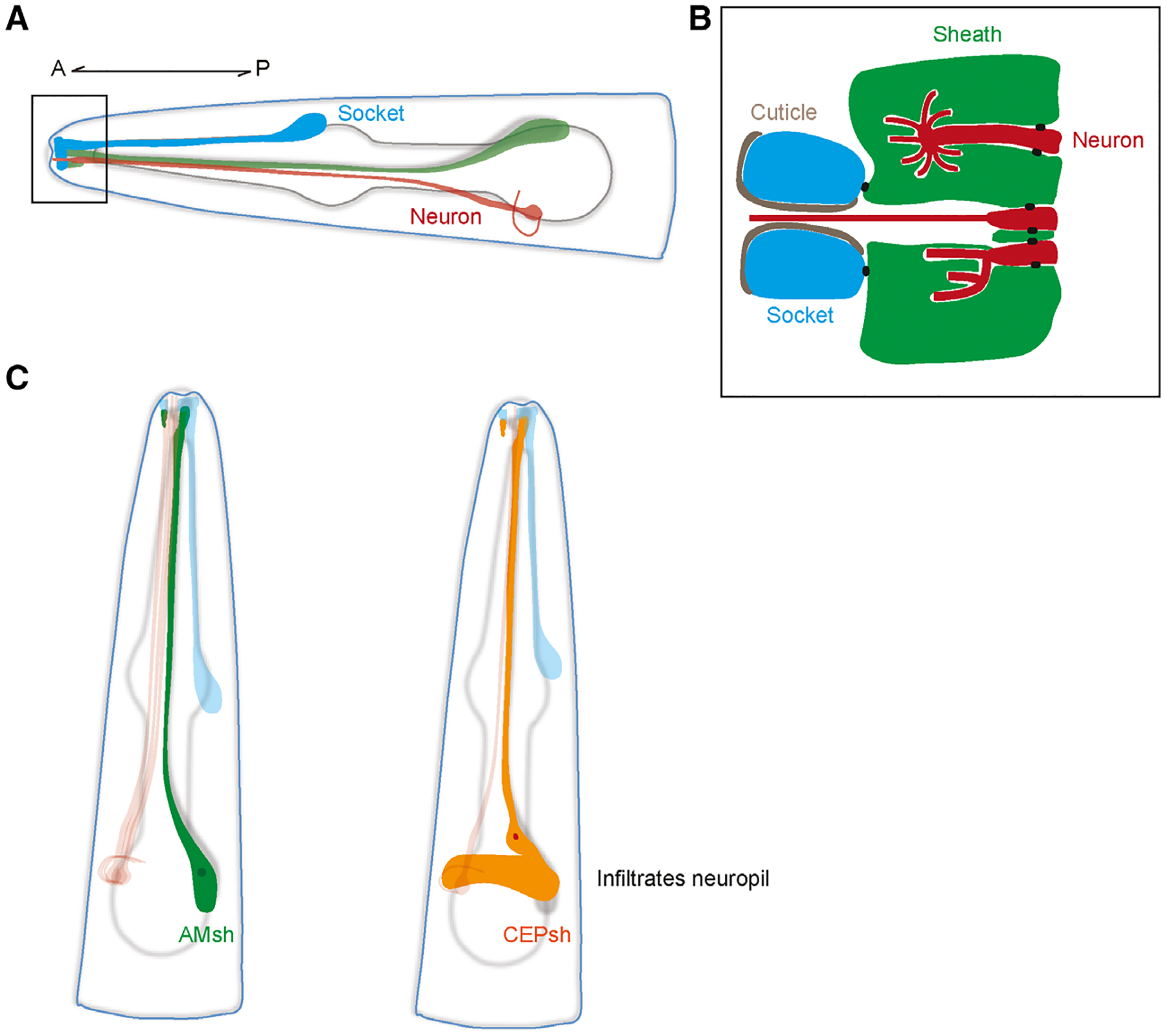
(A) Schematic of a generic sense organ in C. elegans. The processes of socket (blue) and sheath glia (green) together form a channel at the nose tip where dendrites of sensory neurons (red) protrude through to interact with the environment. A, anterior; P, posterior. (B) Close-up of the channel at the nose tip area demarcated by the black box in A. Sheath glia (green) are connected to sensory neurons (red) and socket cells (blue) by apical junctions (black dots). Socket cells are connected to the cuticle of the worm. The cilia may vary in shape depending on the type of neuron. The dendrites of some neurons remain in the channel while others can be completely or partially embedded in the sheath glia. (C) Schematic of AMsh (left) and CEPsh (right) glia. Note that the posterior process of the CEPsh infiltrates the neuropil and comes into close contact with synapses like astrocytes do.
In addition to ensheathing the neuronal receptive endings of sensory neurons, four sheath glia called the cephalic sheath (CEPsh) also send out posterior processes that envelop the nerve ring, which is the neuropil of C. elegans (Fig. 1C). Recent studies have shown that CEPsh glia exhibit many astrocyte-like features, such as how they show similarities in expression profiles, express a homolog of the astrocyte-expressed glutamate transporter GLT1, extend processes that are in close proximity to synapses, and promote synaptogenesis [14–17]. Interestingly, electron micrograph reconstructions show that at least one synapse in the worm shows a tripartite arrangement, where a CEPsh cell process envelops a synapse between the ALA and AVE neurons [18,19].
The generation of glial diversity
While glia ensheathing different sense organs perform similar roles, there are distinct morphological and molecular differences between them that show parallels to vertebrate glial diversity. For one, the two sheath cells of the largest chemosensory organ, the amphid sheath (AMsh) glia, are larger than their other sheath glia counterparts and they ensheath the greatest number of sensory neurons [20]. Unlike the sheath glia of other sensilla, AMsh cells lack deeply invaginated membrane lamellae on the inner face of their channels and instead have a large Golgi apparatus that produce many vesicles close to the inner side of the channels [12]. Furthermore, transcriptomic data and studies of fate specification reveal that different socket and sheath glia can show differences in expression profiles and mechanisms of cell fate specification [21–23]. Thus, by studying gliogenesis in C. elegans, insights into various mechanisms of glial fate specification and the generation of glial diversity can be gained. Molecular differences between the various types of C. elegans glia also mean that there are subtype-specific promoters that can be used for better visualization and precise in vivo manipulations, many of which have been discussed in detail in Fung et al. [24].
Early studies have shown that the zinc finger transcription factor lin-26 is not only important for a hypodermal cell fate, but is also an important regulator of glial fate [25,26]. In lin-26 loss-of-function mutants, many glia exhibit structural and functional abnormalities, and some glia end up dying or transforming into neurons [25]. While lin-26 lacks orthologs in higher order organisms, recent studies of more downstream events reveal many potentially conserved aspects of gliogenesis.
For example, several conserved proneural transcription factors such as Neurog1 and NeuroD1 have been shown to regulate neuron-glia fate determination, where they independently promote a neuronal fate while suppressing a glial fate [27–29]. Recent studies in C. elegans show that loss of function of the Neurog1 and NeuroD1 orthologs, ngn-1 and cnd-1 respectively, also results in an increase in the number of AMsh glia, which is consistent with a suppression of glial fate [22]. It was also found that LIN-32, an ortholog of the proneural gene Atoh1, functioned similarly. While Atoh1 has not been known to have a gliogenesis-suppressing function in the vertebrate central nervous system, overexpression of Atoh1 has been found to induce the transdifferentiation of glial-like support cells into functional hair cells in the cochlea and specify cerebellar granule neurons at the expense of glial numbers in embryoid bodies [30–32]. Furthermore, at least some of the ectopic AMsh glia in lin-32 loss-of-function mutants came from cells originally fated to become CEPsh glia, suggesting that this proneural transcription factor may also play a role in glial identity specification [22].
Further downstream, developing CEPsh glia were found to specifically express hlh-17, an ortholog of the transcription factor Olig2 that is required for oligodendrocyte specification [21,33,34]. In the developing spinal cord of vertebrates, oligodendrocytes form from distinct ventral and dorsal domains, where Olig2 expression is induced in the ventral region by the transcription factors Nkx6 and Pax6 [21,35,36]. Interestingly, CEPsh expression of hlh-17 also requires the Nkx-like MLS-2 and Pax-like VAB-3, where MLS-2 in particular is required for ventral CEPsh glia development [21]. Similarly, another conserved transcription factor alr-1, the C. elegans homolog of Aristaless, is expressed in amphid socket (AMso) glia and is required for the proper maintenance of its structure and function [37]. alr-1 mutants exhibit abnormal AMso morphology and a loss of tight junctions between AMso and AMsh glia, which is reflected by a loss of sensory function [37]. Postdevelopmentally, PROS-1/Prox1 is expressed in AMsh glia, where it was found through transcriptomics to regulate AMsh glial-specific secretome expression [38]. Among the secreted proteins includes FIG-1, which contains the conserved thrombospondin type 1 (TSP-1) domain also found in mammalian thrombospondins that are secreted by astrocytes [39,40]. Furthermore, in addition to the fact that human Prox1 can functionally rescue pros-1 mutant phenotypes [41], homologs of prospero are also expressed postdevelopmentally in support cells in the mammalian ear [42] and vertebrate astrocytes [43]. This suggests that there could be potential functional conservation in this class of transcription factors.
Interestingly in C. elegans males, AMso glia function as a neural progenitor during larval development and divide to generate a daughter AMso cell and a male-specific MCM interneuron [44]. During this division, the daughter AMso cell inherits its polar morphology and cellular contacts, which is reminiscent of how radial glia in vertebrates divide and self-renew while keeping their bipolar morphology intact [44,45]. A recent study by Molina-García et al. [46] has also revealed that the male-specific PHD neuron arises from a glia-to-neuron transdifferentiation process during larval sexual maturation. In this case, the PHso1 glia directly transdifferentiate into the PHD neuron and integrate into circuits for male mating. Interestingly, this mode of postembryonic neuron generation parallels the direct conversion of adult neural stem cells into postmitotic neurons in the zebrafish brain [47].
The many similarities during different stages of cell fate specification suggest that despite major differences between vertebrate and C. elegans glia, there may be conservation of many developmental processes. Furthermore, recent studies have generated transcriptomic profiles for single cells in the developing embryo [23] as well as the AMsh [38] and CEPsh glia [16] in postembryonic animals, aiding in our understanding of these cells from early embryonic development through late larval development. Katz et al. have recently used transcriptomic profiling to show that CEPsh glia and astrocytes share many molecular similarities, and demonstrated that the conserved glial glutamate transporter GLT-1/EAAT2 plays similar roles in regulating repetitive behaviors [48].
Together, these studies suggest that C. elegans can be used as a complementary approach to understand conserved developmental mechanisms during the various stages of glial specification, where the sheer diversity of cell types and lineages in the vertebrate nervous system could make studying these questions challenging. Although different from vertebrate neuron-glia fate determination, the deterministic and fully mapped cell lineage of C. elegans allows for repeatable and cell-specific examination of glial development in an in vivo system. Thus, many conserved or analogous mechanistic details of gliogenesis can be first studied in the simpler system, and insights gleaned may hopefully further our understanding of this process in higher order organisms.
Glial morphogenesis
The diversity of tasks that glia perform is closely linked to their elaborate morphologies, yet it is still unclear exactly how they are formed. For example, mature human astrocytes possess numerous fine processes that can contact up to two million synapses while also exhibiting tiling behavior [49]. At the same time, loss of astrocyte complexity is commonly observed during neurological disorders [50]. Recent studies have shown that in addition to responding to changes in neural activity, astrocyte morphogenesis also depends on direct contact with neuronal processes that is mediated by the neuroligin family of adhesion proteins [49,51]. C. elegans CEPsh glia may represent an exciting model to study the morphogenetic processes relating to neuropil infiltration, synapse ensheathment, and tiling with other CEPsh cells. The stereotyped structure and interactions with other cells may allow for the controlled study of mechanisms that regulate where these glia may extend and which synapses they envelop. During early CEPsh development in the embryo, these cells exhibit a bipolar morphology, in which a posterior-facing process cooperates with pioneer neurons to promote and direct the early stages of neuropil assembly [52]. These embryonic CEPsh glia then develop into adult CEPsh glia, which exhibit a more branched and astrocyte-like morphology, where the processes infiltrate the neuropil and come into close proximity with synapses. The exact mechanisms underlying this morphogenetic change are currently unclear, and future work will need to be done to learn more about these cells.
More well understood are some of the mechanisms that regulate sensory channel formation by sheath glia in C. elegans. A key aspect of sheath glia function is their ability to form a tube that can snugly fit neuronal receptive endings. Analysis of a patched-related protein DAF-6 revealed that loss-of-function mutations resulted in overgrown AMsh glial channels that ended up blocking the neuronal receptive endings and preventing proper function (Fig. 2A,B) [53–55]. It was found that DAF-6 as well as the dispatched-related protein CHE-14 localizes to the luminal portion of the channel where they regulate sensory compartment formation. Further analysis showed that the neuronal cilia being ensheathed also exert control over channel formation, as mutants with disrupted cilia formation exhibit smaller and misshapen lumens and altered localization of DAF-6 [55,56]. Through a screen, the Nemo-like kinase homolog LIT-1 was found to suppress daf-6 mutant phenotypes and instead promote expansion of the lumen [56]. LIT-1 is activated by MOM-4/MAP3K and interacts with WASP to potentially promote channel growth through regulating actin polymerization. Furthermore, LIT-1 compartment localization is required for this function and it also requires neuronal signals. Similarly, the retromer complex components SNX-1, SNX-3, and VPS-29 as well as the Ig/FNIII protein IGDB-2 all suppress the DAF-6 bloated channel phenotype and promote lumen expansion [57,58]. The implication of retromer complex components suggests that vesicle trafficking may be a potential mechanism for controlling channel size. On the other hand, IGDB-2 binds to a predicted ligand-gated ion channel LGC-34, which may implicate osmoregulation and fluid accumulation as another potential mechanism though there is no direct evidence of this so far. These results suggest that proper sensory compartment morphogenesis in AMsh glia is a process that not only requires several different molecular pathways but also inputs from the neurons beings enveloped (Fig. 2B).
Fig. 2.
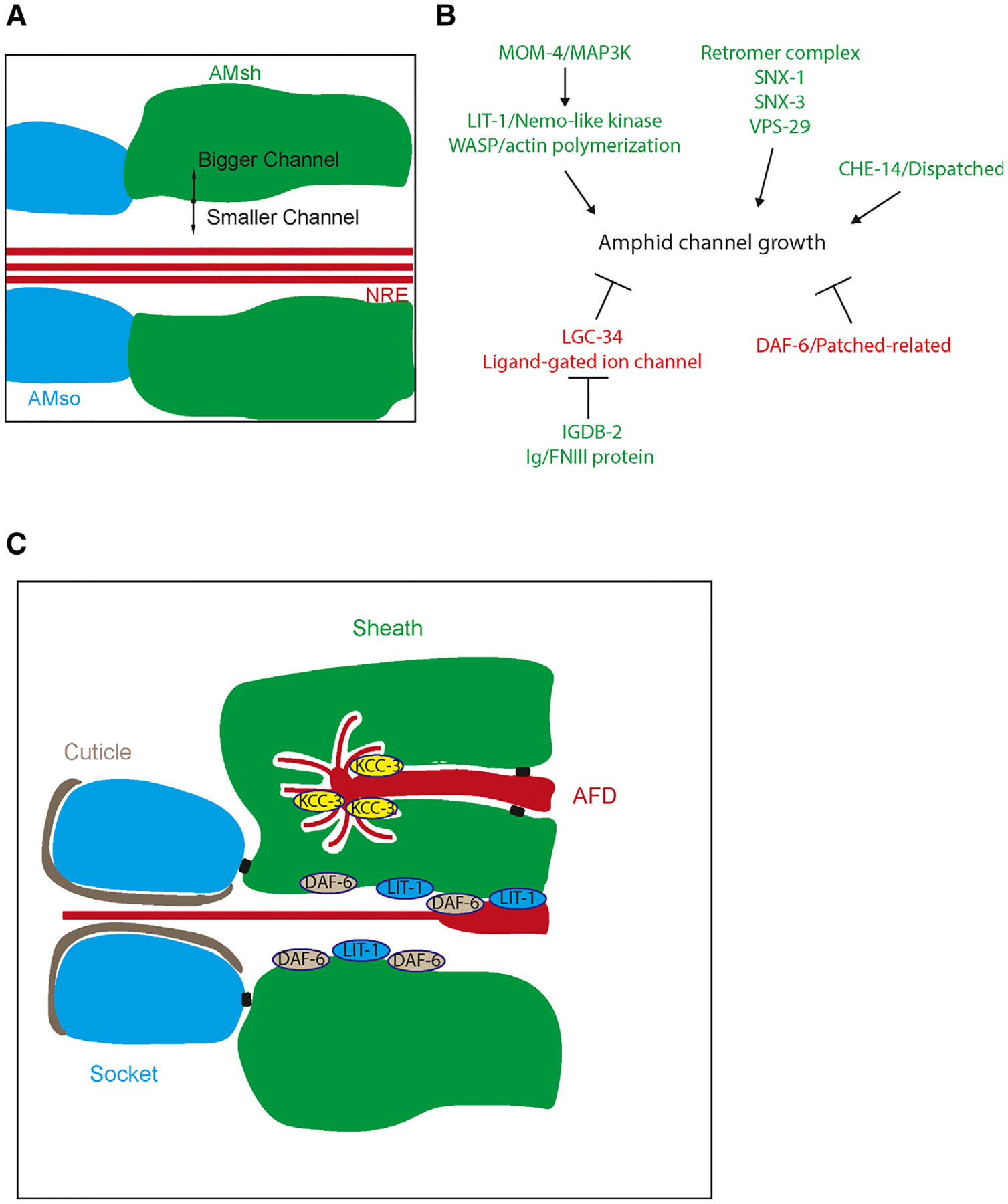
(A) Schematic of the amphid channel. AMsh (green) and AMso (blue) form a channel that certain neuron receptive endings (NREs) sit in. Regulation of AMsh growth in this area can determine the size of the lumen. (B) Model for amphid channel morphogenesis. Genes that increase channel size are colored green while those that decrease channel size are colored red. (C) Distinct subcellular domains are generated by the AMsh depending on its specific neuronal partners. For example, KCC-3 is localized to the AMsh pocket interacting with the AFD neuron while DAF-6 and LIT-1 are localized to the main amphid channel.
The amphid channel is not the only compartment that is formed by AMsh glia at nose tip, as they can also ensheath the receptive endings of specific neurons in different compartments from the channel [20] (Fig. 1B). These include sensory neuron endings that track through the amphid channel and then divert into these pocket compartments, where they track volatile odors. There is also the thermosensory AFD neuron, which does not track through the amphid channel and is ensheathed by the AMsh glia. In this microenvironment, the AMsh glia regulate AFD neuronal receptive ending shape through a glial K+/Cl−cation-chloride cotransporter KCC-3 [59]. Here, the AMsh cells perform a conserved glial function of ion homeostasis where they regulate K+ and Cl− levels in the compartment. Interestingly, these AMsh compartments that have been described likely represent distinct subcellular domains (Fig. 2C). This is shown by the differential localization of KCC-3 to the AFD neuron receptive ending microenvironment and DAF-6 and LIT-1 to the amphid channel lumen, where they perform their respective functions [55,56,59].
Another aspect of sheath glia morphogenesis is how their process extends to the appropriate location to form these sensory channels. In the developing embryo, AMsh glia and amphid neurons are born near the tip of the presumptive head [60]. The glia and neurons then extend short processes that are anchored to the nose tip by interactions between the neuronally secreted zona pellucida domain protein DYF-7 and the non-neuronally secreted zonadhesin domain containing protein DEX-1. These proteins can multimerize and form an anchor, much like how tectorins anchor stereocilia in the inner ear. After anchoring, these cells migrate posteriorly together, stretching out dendrites and processes through a process called retrograde extension (Fig. 3A). In dyf-7 and dex-1 mutants, the glia and neurons are unable to properly anchor when they migrate, resulting in a failure to extend their processes. Furthermore, a recent study suggests that amphid neurons and glia share properties with epithelia, and formation of the tubular glial channel is reminiscent of epithelial tube formation [61]. DYF-7 was observed to forms fibrils as an apical ECM component and helps promote amphid channel formation akin to how the apical ECM is important for continuous lumen formation in narrow epithelial tubes. In addition, apical FRM-2 helps support amphid epithelial integrity during morphogenesis, similar to how its homologs PBL15/moe/Yurt maintain epithelial integrity in other systems [61–63]. Interestingly, retrograde extension has been reported as a mode of axon extension in the zebrafish olfactory placode suggesting that this mode of extension could be shared in other animals [64]. Another mechanism of retrograde extension was recently observed between the BAG and URX neurons and their inner labial socket (ILso) glial partner. Instead of DYF-7 and DEX-1, they utilize the conserved adhesion molecules SAX-7/L1CAM and GRDN-1/CCDC88C [65]. URX and BAG dendrites form specialized attachments with the ILso glia, and during development, their dendrites form by retrograde extension likely through neuron-glia attachments. Together, these studies suggest that the specialized morphology of sheath glia near the nose is likely functionally important for dendrite extension of the neurons they interact with. Furthermore, while the processes and dendrites themselves may not require long range guidance cues in this mode of extension, the cell bodies migrate to highly stereotyped positions in C. elegans, suggesting there may be regulatory mechanisms guiding cell body migration.
Fig. 3.
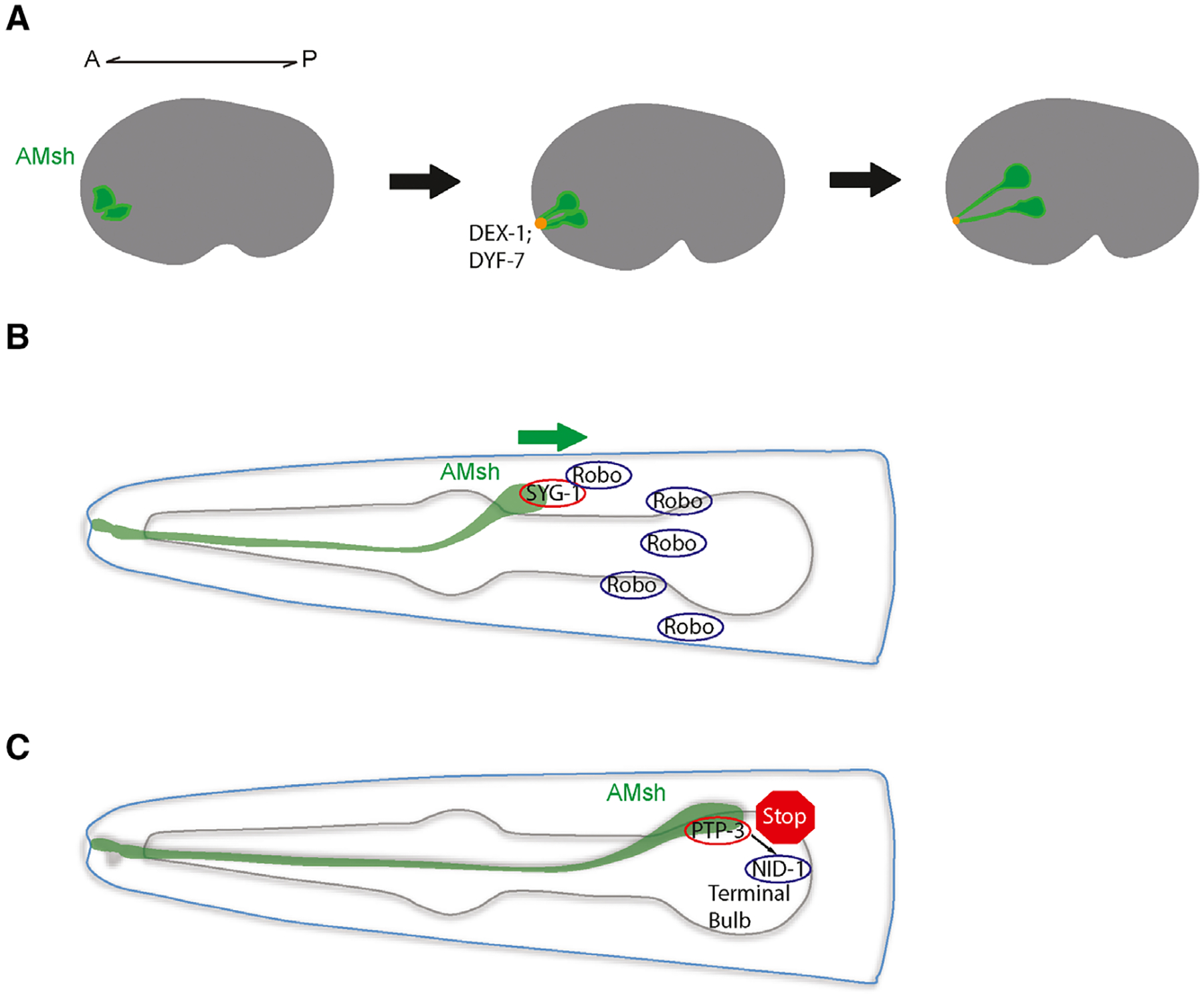
(A) The process of retrograde extension in the AMsh glia. During embryogenesis, AMsh cells (green) and amphid neurons (not shown) extend processes that anchor to the nose tip through DEX-1 and DYF-7. Posterior migration stretches out the cells to form the processes and dendrites observed. A, anterior; P, posterior. (B) AMsh cell body migration mediated by cleaved Robo and its glial receptor SYG-1/Neph. (C) Halting of further AMsh migration occurs at the terminal pharyngeal bulb, where the glia-expressed cell adhesion molecule PTP-3 interacts with the extracellular matrix protein NID-1.
It has recently been found that the C. elegans Robo receptor functions to promote AMsh glial migration during development in a Slit-independent fashion (Fig. 3B) [66]. In this case, neuron-expressed Robo is cleaved extracellularly and interacts with its glial-expressed receptor SYG-1/Neph to function as an attractive cue during AMsh glia migration. SYG-1/Neph may function through regulation of the WAVE regulatory complex, which has also been implicated in vertebrate oligodendrocyte precursor cell and Drosophila midline glia migration [67,68]. On the other hand, there are likely mechanisms that instruct migrating AMsh cell bodies where to stop. While migrating AMsh glia typically stop close to the terminal pharyngeal bulb in developing larva, the AMsh of certain ptp-3 loss-of-function mutants migrate almost 40% further than normal and correspondingly have longer processes (Fig. 3C) [69]. PTP-3 is most similar to the family of cell adhesion molecules called LAR protein tyrosine phosphatase transmembrane receptors, where it was found to interact with the ECM component nidogen 1/NID-1 during synaptogenesis [70]. Similarly, the interaction between glial PTP-3 and NID-1 likely secreted by the pharyngeal muscle also plays a role in stopping AMsh glia from migrating too far [69] (Fig. 2B). However, it is not clear whether these two pathways interact to specify where the AMsh cell body ends up, and it is likely other molecular pathways play a role.
Amphid sheath glia can also exhibit morphological plasticity in response to the environment. When faced with environmental stressors such as starvation, high population density, and high temperatures, C. elegans can enter a developmentally arrested stage called dauer [71]. Many changes occur during this alternative developmental state, and AMsh glia and AWC neuron receptive endings were observed to show concomitant remodeling (Fig. 4) [71,72]. The left and right AMsh glia expand their ensheathing domains and fuse at the nose tip to accommodate the remodeled AWC receptive endings [73]. The fusion process was found to be mediated by the fusogen AFF-1. Further analysis implicated the glial-expressed OTX/OTD transcription factor TTX-1 and zinc finger transcription factor ZTF-16 in this AMsh remodeling process, where they likely function through the transcriptional regulation of the receptor tyrosine kinase VER-1 [72,74]. Similarly, a recent study by Lee et al. has implicated the glial GPCR REMO-1 during stress-induced amphid glial and neuronal remodeling by also promoting VER-1 expression [75]. Interestingly, the authors have further linked amphid remodeling to faster exit from the dauer stage when conditions become favorable, suggesting this plasticity is of functional importance in dauer animals. However, the exact molecular mechanisms underlying AMsh glia and AWC neuron remodeling are still unclear, and it would be interesting to learn more about the interactions between these cells during this process given that AWC neuron remodeling requires AMsh glia.
Fig. 4.
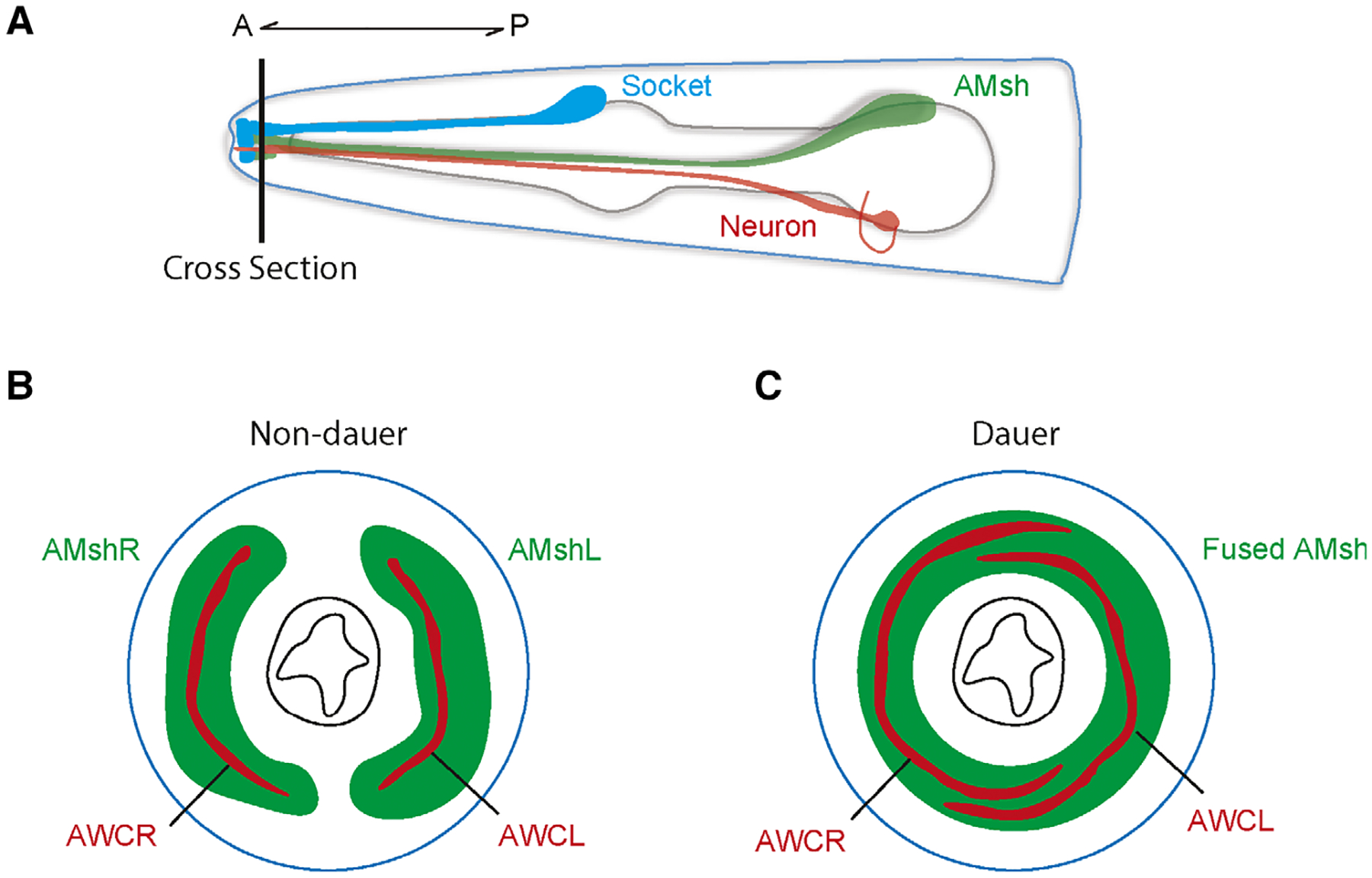
(A) Schematic of the head of the worm showing the AMsh glia. Horizontal line indicates cross section visualized in (B) and (C). (B-C) Sections showing the two bilateral AMsh glia (green) and AWC neurons (red) in non-dauer (B) and dauer (C) conditions. In dauer animals, the two AMsh glia fuse while the AMC receptive endings expand.
In C. elegans, many highly conserved genes and pathways regulate glial morphogenesis and include many conserved cell adhesion molecules. In addition to the simplicity of the system, the studies reviewed here also made use of genetic screens. Use of forward genetic screens and suppressor screens can be useful for studying glial morphogenesis, as morphological defects in the glia of choice can be rapidly screened for and isolated. This unbiased approach allows for the identification of unexpected or novel genes and pathways. Furthermore, many genes that are essential in higher order organisms can be studied for their effects on glial development and function in C. elegans due to glia not being essential for animal survival.
Conclusions
In the past several decades, glia have been revealed to play a wide array of essential roles in the nervous system; yet, there is still much to be learned. With new experimental tools available, there is a growing appreciation for the diversity and complexity of these cells. In this review, we summarized some of the experimental findings regarding various aspects of glial development in C. elegans and discussed how this model can be harnessed to study glial specification and morphogenesis in mechanistic detail. The many developmental and functional similarities between the glia of C. elegans and higher order animals also show how the worm can become a powerful tool of understanding due to their stereotyped development, amenability to forward genetic screens, and facile genetics. Learning more about glial biology and development would not only allow us to better understand their myriad functions, but also help inform our understanding of their role in disease.
Acknowledgements
AZ and DY are supported by NIH R01 (NS094171 and NS105638 to DY).
Abbreviations
- AMsh
amphid sheath
- AMso
amphid socket
- CEPsh
cephalic sheath
- ILso
inner labial socket
- LAR-PTPR
LAR protein tyrosine phosphatase transmembrane receptor
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Bayraktar OA, Bartels T, Holmqvist S, Kleshchevnikov V, Martirosyan A, Polioudakis D, Ben Haim L, Young AMH, Batiuk MY, Prakash K et al. (2020) Astrocyte layers in the mammalian cerebral cortex revealed by a single-cell in situ transcriptomic map. Nat Neurosci 23, 500–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bayraktar OA, Fuentealba LC, Alvarez-Buylla A & Rowitch DH (2014) Astrocyte development and heterogeneity. Cold Spring Harb Perspect Biol 7, a020362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marques S, Zeisel A, Codeluppi S, van Bruggen D, Mendanha Falcao A, Xiao L, Li H, Haring M, Hochgerner H, Romanov RA et al. (2016) Oligodendrocyte heterogeneity in the mouse juvenile and adult central nervous system. Science 352, 1326–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Y & Barres BA (2010) Astrocyte heterogeneity: an underappreciated topic in neurobiology. Curr Opin Neurobiol 20, 588–594. [DOI] [PubMed] [Google Scholar]
- 5.Chung WS, Welsh CA, Barres BA & Stevens B (2015) Do glia drive synaptic and cognitive impairment in disease? Nat Neurosci 18, 1539–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Molofsky AV, Krencik R, Ullian EM, Tsai HH, Deneen B, Richardson WD, Barres BA & Rowitch DH (2012) Astrocytes and disease: a neurodevelopmental perspective. Genes Dev 26, 891–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zuchero JB & Barres BA (2015) Glia in mammalian development and disease. Development 142, 3805–3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Freeman MR & Rowitch DH (2013) Evolving concepts of gliogenesis: a look way back and ahead to the next 25 years. Neuron 80, 613–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shaham S (2015) Glial development and function in the nervous system of Caenorhabditis elegans. Cold Spring Harb Perspect Biol 7, a020578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stork T, Bernardos R & Freeman MR (2012) Analysis of glial cell development and function in Drosophila. Cold Spring Harb Protoc 2012, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sulston JE, Schierenberg E, White JG & Thomson JN (1983) The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol 100, 64–119. [DOI] [PubMed] [Google Scholar]
- 12.Ward S, Thomson N, White JG & Brenner S (1975) Electron microscopical reconstruction of the anterior sensory anatomy of the nematode Caenorhabditis elegans?. J Comp Neurol 160, 313–337. [DOI] [PubMed] [Google Scholar]
- 13.Doroquez DB, Berciu C, Anderson JR, Sengupta P & Nicastro D (2014) A high-resolution morphological and ultrastructural map of anterior sensory cilia and glia in Caenorhabditis elegans. Elife 3, e01948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chung WS, Allen NJ & Eroglu C (2015) Astrocytes control synapse formation, function, and elimination. Cold Spring Harb Perspect Biol 7, a020370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Colon-Ramos DA, Margeta MA & Shen K (2007) Glia promote local synaptogenesis through UNC-6 (netrin) signaling in C. elegans. Science 318, 103–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Katz M, Corson F, Keil W, Singhal A, Bae A, Lu Y, Liang Y & Shaham S (2019) Glutamate spillover in C. elegans triggers repetitive behavior through presynaptic activation of MGL-2/mGluR5. Nat Commun 10, 1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nagai J, Yu X, Papouin T, Cheong E, Freeman MR, Monk KR, Hastings MH, Haydon PG, Rowitch D, Shaham S et al. (2020) Behaviorally consequential astrocytic regulation of neural circuits. Neuron. 10.1016/j.neuron.2020.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singhvi A & Shaham S (2019) Glia-neuron interactions in Caenorhabditis elegans. Annu Rev Neurosci 42, 149–168. [DOI] [PubMed] [Google Scholar]
- 19.White JG, Southgate E, Thomson JN & Brenner S (1986) The structure of the nervous system of the nematode Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci 314, 1–340. [DOI] [PubMed] [Google Scholar]
- 20.Perkins LA, Hedgecock EM, Thomson JN & Culotti JG (1986) Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev Biol 117, 456–487. [DOI] [PubMed] [Google Scholar]
- 21.Yoshimura S, Murray JI, Lu Y, Waterston RH & Shaham S (2008) mls-2 and vab-3 Control glia development, hlh-17/Olig expression and glia-dependent neurite extension in C. elegans. Development 135, 2263–2275. [DOI] [PubMed] [Google Scholar]
- 22.Zhang A, Noma K & Yan D (2020) Regulation of gliogenesis by lin-32/Atoh1 in Caenorhabditis elegans. G3: Genes - Genomes - Genetics 10, 3271–3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Packer JS, Zhu Q, Huynh C, Sivaramakrishnan P, Preston E, Dueck H, Stefanik D, Tan K, Trapnell C, Kim J et al. (2019) A lineage-resolved molecular atlas of C. elegans embryogenesis at single-cell resolution. Science 365, eaax1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fung W, Wexler L & Heiman MG (2020) Cell-type-specific promoters for C. elegans glia. J Neurogenet 34, 335–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Labouesse M, Hartwieg E & Horvitz HR (1996) The Caenorhabditis elegans LIN-26 protein is required to specify and/or maintain all non-neuronal ectodermal cell fates. Development 122, 2579–2588. [DOI] [PubMed] [Google Scholar]
- 26.Labouesse M, Sookhareea S & Horvitz HR (1994) The Caenorhabditis elegans gene lin-26 is required to specify the fates of hypodermal cells and encodes a presumptive zinc-finger transcription factor. Development 120, 2359–2368. [DOI] [PubMed] [Google Scholar]
- 27.Morrow EM, Furukawa T, Lee JE & Cepko CL (1999) NeuroD regulates multiple functions in the developing neural retina in rodent. Development 126, 23–36. [DOI] [PubMed] [Google Scholar]
- 28.Sun Y, Nadal-Vicens M, Misono S, Lin MZ, Zubiaga A, Hua X, Fan G & Greenberg ME (2001) Neurogenin promotes neurogenesis and inhibits glial differentiation by independent mechanisms. Cell 104, 365–376. [DOI] [PubMed] [Google Scholar]
- 29.Tomita K, Moriyoshi K, Nakanishi S, Guillemot F & Kageyama R (2000) Mammalian achaete-scute and atonal homologs regulate neuronal versus glial fate determination in the central nervous system. EMBO J 19, 5460–5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Izumikawa M, Minoda R, Kawamoto K, Abrashkin KA, Swiderski DL, Dolan DF, Brough DE & Raphael Y (2005) Auditory hair cell replacement and hearing improvement by Atoh1 gene therapy in deaf mammals. Nat Med 11, 271–276. [DOI] [PubMed] [Google Scholar]
- 31.Sayyid ZN, Wang T, Chen L, Jones SM & Cheng AG (2019) Atoh1 directs regeneration and functional recovery of the mature mouse vestibular system. Cell Rep 28, 312–324.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Srivastava R, Kumar M, Peineau S, Csaba Z, Mani S, Gressens P & El Ghouzzi V (2013) Conditional induction of Math1 specifies embryonic stem cells to cerebellar granule neuron lineage and promotes differentiation into mature granule neurons. Stem Cells 31, 652–665. [DOI] [PubMed] [Google Scholar]
- 33.Lu QR, Sun T, Zhu Z, Ma N, Garcia M, Stiles CD & Rowitch DH (2002) Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell 109, 75–86. [DOI] [PubMed] [Google Scholar]
- 34.Zhou Q & Anderson DJ (2002) The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell 109, 61–73. [DOI] [PubMed] [Google Scholar]
- 35.Jessell TM (2000) Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat Rev Genet 1, 20–29. [DOI] [PubMed] [Google Scholar]
- 36.Miller RH (2005) Dorsally derived oligodendrocytes come of age. Neuron 45, 1–3. [DOI] [PubMed] [Google Scholar]
- 37.Tucker M, Sieber M, Morphew M & Han M (2005) The Caenorhabditis elegans aristaless orthologue, alr-1, is required for maintaining the functional and structural integrity of the amphid sensory organs. Mol Biol Cell 16, 4695–4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wallace SW, Singhvi A, Liang Y, Lu Y & Shaham S (2016) PROS-1/prospero is a major regulator of the glia-specific secretome controlling sensory-neuron shape and function in C. elegans. Cell Rep 15, 550–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Christopherson KS, Ullian EM, Stokes CC, Mullowney CE, Hell JW, Agah A, Lawler J, Mosher DF, Bornstein P & Barres BA (2005) Thrombospondins are astrocyte-secreted proteins that promote CNS synaptogenesis. Cell 120, 421–433. [DOI] [PubMed] [Google Scholar]
- 40.Bacaj T, Tevlin M, Lu Y & Shaham S (2008) Glia are essential for sensory organ function in C. elegans. Science 322, 744–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kage-Nakadai E, Ohta A, Ujisawa T, Sun S, Nishikawa Y, Kuhara A & Mitani S (2016) Caenorhabditis elegans homologue of Prox1/Prospero is expressed in the glia and is required for sensory behavior and cold tolerance. Genes Cells 21, 936–948. [DOI] [PubMed] [Google Scholar]
- 42.Bermingham-McDonogh O, Oesterle EC, Stone JS, Hume CR, Huynh HM & Hayashi T (2006) Expression of Prox1 during mouse cochlear development. J Comp Neurol 496, 172–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N et al. (2014) An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci 34, 11929–11947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sammut M, Cook SJ, Nguyen KCQ, Felton T, Hall DH, Emmons SW, Poole RJ & Barrios A (2015) Glia-derived neurons are required for sex-specific learning in C. elegans. Nature 526, 385–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Noctor SC, Martinez-Cerdeno V, Ivic L & Kriegstein AR (2004) Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci 7, 136–144. [DOI] [PubMed] [Google Scholar]
- 46.Molina-Garcia L, Lloret-Fernandez C, Cook SJ, Kim B, Bonnington RC, Sammut M, O’Shea JM, Gilbert SP, Elliott DJ, Hall DH et al. (2020) Direct glia-to-neuron transdifferentiation gives rise to a pair of male-specific neurons that ensure nimble male mating. Elife 9, e48361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barbosa JS, Sanchez-Gonzalez R, Di Giaimo R, Baumgart EV, Theis FJ, Gotz M & Ninkovic J (2015) Neurodevelopment. Live imaging of adult neural stem cell behavior in the intact and injured zebrafish brain. Science 348, 789–793. [DOI] [PubMed] [Google Scholar]
- 48.Haroon E, Miller AH & Sanacora G (2017) Inflammation, glutamate, and glia: a trio of trouble in mood disorders. Neuropsychopharmacology 42, 193–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Freeman MR (2010) Specification and morphogenesis of astrocytes. Science 330, 774–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Burda JE & Sofroniew MV (2014) Reactive gliosis and the multicellular response to CNS damage and disease. Neuron 81, 229–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stogsdill JA, Ramirez J, Liu D, Kim YH, Baldwin KT, Enustun E, Ejikeme T, Ji RR & Eroglu C (2017) Astrocytic neuroligins control astrocyte morphogenesis and synaptogenesis. Nature 551, 192–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rapti G, Li C, Shan A, Lu Y & Shaham S (2017) Glia initiate brain assembly through noncanonical Chimaerin-Furin axon guidance in C. elegans. Nat Neurosci 20, 1350–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Albert PS, Brown SJ & Riddle DL (1981) Sensory control of dauer larva formation in Caenorhabditis elegans. J Comp Neurol 198, 435–451. [DOI] [PubMed] [Google Scholar]
- 54.Herman RK (1987) Mosaic analysis of two genes that affect nervous system structure in Caenorhabditis elegans. Genetics 116, 377–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Perens EA & Shaham S (2005) C. elegans daf-6 encodes a patched-related protein required for lumen formation. Dev Cell 8, 893–906. [DOI] [PubMed] [Google Scholar]
- 56.Oikonomou G, Perens EA, Lu Y, Watanabe S, Jorgensen EM & Shaham S (2011) Opposing activities of LIT-1/NLK and DAF-6/patched-related direct sensory compartment morphogenesis in C. elegans. PLoS Biol 9, e1001121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oikonomou G, Perens EA, Lu Y & Shaham S (2012) Some, but not all, retromer components promote morphogenesis of C. elegans sensory compartments. Dev Biol 362, 42–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang W, Perens EA, Oikonomou G, Wallace SW, Lu Y & Shaham S (2017) IGDB-2, an Ig/FNIII protein, binds the ion channel LGC-34 and controls sensory compartment morphogenesis in C. elegans. Dev Biol 430, 105–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Singhvi A, Liu B, Friedman CJ, Fong J, Lu Y, Huang XY & Shaham S (2016) A Glial K/Cl transporter controls neuronal receptive ending shape by chloride inhibition of an rGC. Cell 165, 936–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Heiman MG & Shaham S (2009) DEX-1 and DYF-7 establish sensory dendrite length by anchoring dendritic tips during cell migration. Cell 137, 344–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Low IIC, Williams CR, Chong MK, McLachlan IG, Wierbowski BM, Kolotuev I & Heiman MG (2019) Morphogenesis of neurons and glia within an epithelium. Development 146, dev171124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Salis P, Payre F, Valenti P, Bazellieres E, Le Bivic A & Mottola G (2017) Crumbs, Moesin and Yurt regulate junctional stability and dynamics for a proper morphogenesis of the Drosophila pupal wing epithelium. Sci Rep 7, 16778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schell C, Rogg M, Suhm M, Helmstadter M, Sellung D, Yasuda-Yamahara M, Kretz O, Kuttner V, Suleiman H, Kollipara L et al. (2017) The FERM protein EPB41L5 regulates actomyosin contractility and focal adhesion formation to maintain the kidney filtration barrier. Proc Natl Acad Sci USA 114, E4621–E4630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Breau MA, Bonnet I, Stoufflet J, Xie J, De Castro S & Schneider-Maunoury S (2017) Extrinsic mechanical forces mediate retrograde axon extension in a developing neuronal circuit. Nat Commun 8, 282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cebul ER, McLachlan IG & Heiman MG (2020) Dendrites with specialized glial attachments develop by retrograde extension using SAX-7 and GRDN-1. Development 147, dev180448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Qu Z, Zhang A & Yan D (2020) Robo functions as an attractive cue for glial migration through SYG-1/Neph. Elife 9, e57921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bogdan S & Klambt C (2003) Kette regulates actin dynamics and genetically interacts with Wave and Wasp. Development 130, 4427–4437. [DOI] [PubMed] [Google Scholar]
- 68.Miyamoto Y, Yamauchi J & Tanoue A (2008) Cdk5 phosphorylation of WAVE2 regulates oligodendrocyte precursor cell migration through nonreceptor tyrosine kinase Fyn. J Neurosci 28, 8326–8337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang A, Ackley BD & Yan D (2020) Vitamin B12 regulates glial migration and synapse formation through isoform-specific control of PTP-3/LAR PRTP expression. Cell Rep 30, 3981–3988.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ackley BD, Harrington RJ, Hudson ML, Williams L, Kenyon CJ, Chisholm AD & Jin Y (2005) The two isoforms of the Caenorhabditis elegans leukocyte-common antigen related receptor tyrosine phosphatase PTP-3 function independently in axon guidance and synapse formation. J Neurosci 25, 7517–7528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Golden JW & Riddle DL (1984) The Caenorhabditis elegans dauer larva: developmental effects of pheromone, food, and temperature. Dev Biol 102, 368–378. [DOI] [PubMed] [Google Scholar]
- 72.Procko C, Lu Y & Shaham S (2011) Glia delimit shape changes of sensory neuron receptive endings in C. elegans. Development 138, 1371–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Albert PS & Riddle DL (1983) Developmental alterations in sensory neuroanatomy of the Caenorhabditis elegans dauer larva. J Comp Neurol 219, 461–481. [DOI] [PubMed] [Google Scholar]
- 74.Procko C, Lu Y & Shaham S (2012) Sensory organ remodeling in Caenorhabditis elegans requires the zinc-finger protein ZTF-16. Genetics 190, 1405–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee IH, Procko C, Lu Y & Shaham S (2021) Stress-induced neural plasticity mediated by glial GPCR REMO-1 promotes C. elegans adaptive behavior. Cell Rep 34, 108607. [DOI] [PMC free article] [PubMed] [Google Scholar]


