Abstract
Every year cervical cancer affects more than 300,000 people, and on average one woman is diagnosed with cervical cancer every minute. Early diagnosis and classification of cervical lesions greatly boosts up the chance of successful treatments of patients, and automated diagnosis and classification of cervical lesions from Papanicolaou (Pap) smear images have become highly demanded. To the authors’ best knowledge, this is the first study of fully automated cervical lesions analysis on whole slide images (WSIs) of conventional Pap smear samples. The presented deep learning-based cervical lesions diagnosis system is demonstrated to be able to detect high grade squamous intraepithelial lesions (HSILs) or higher (squamous cell carcinoma; SQCC), which usually immediately indicate patients must be referred to colposcopy, but also to rapidly process WSIs in seconds for practical clinical usage. We evaluate this framework at scale on a dataset of 143 whole slide images, and the proposed method achieves a high precision 0.93, recall 0.90, F-measure 0.88, and Jaccard index 0.84, showing that the proposed system is capable of segmenting HSILs or higher (SQCC) with high precision and reaches sensitivity comparable to the referenced standard produced by pathologists. Based on Fisher’s Least Significant Difference (LSD) test (P < 0.0001), the proposed method performs significantly better than the two state-of-the-art benchmark methods (U-Net and SegNet) in precision, F-Measure, Jaccard index. For the run time analysis, the proposed method takes only 210 seconds to process a WSI and is 20 times faster than U-Net and 19 times faster than SegNet, respectively. In summary, the proposed method is demonstrated to be able to both detect HSILs or higher (SQCC), which indicate patients for further treatments, including colposcopy and surgery to remove the lesion, and rapidly processing WSIs in seconds for practical clinical usages.
Subject terms: Cancer screening, High-throughput screening, Machine learning, Biomedical engineering, Cervical cancer
Introduction
According to projections by the World Health Organization (WHO), cervical cancer affects more than 300,000 people per year, with more than 85% of such deaths occurring in less developed countries in recent decades. Each minute, one woman is diagnosed with cervical cancer, namely one of the most common cancers in women’s health today1. Dysplasia could be detected earlier before cervical cancer grows. The sooner this is detected, the easier to cure cervical cancer. Cervical cancer is completely preventable and curable if pre-cancer signs are identified and treated at an early stage2. Pap smear is commonly used for medical diagnosis of the cervix to monitor cervical cancer and other diseases. The method of Pap smear is to obtain a small number of cervical cell samples, make a cell smear, observe the cells under the microscope for abnormalities, and then diagnose cervical disease. The sample usually screened by cytotechnologists to examine the cell sample for signs of malignancies. Through this procedure, medical experts could both find proof of invasive cancer and detect certain cancer precursors, allowing for early and effective treatment. According to the WHO classification for cervical squamous lesion, the initial and mild stage of precancer is termed as mild dysplasia, which later advances to the next stage called moderate dysplasia, followed by severe dysplasia and squamous cell carcinoma in situ, and finally to invasive squamous cell carcinoma (SQCC) that invades other parts of the body.
In Cervical Intraepithelial Neoplasia (CIN) system, mild dysplasia is classified as CIN1, moderate dysplasia as CIN2, severe dysplasia, and squamous cell carcinoma in situ as CIN3, and the final stage as invasive SQCC. The Bethesda system is a standard system worldwide in cytological reporting of the cases in diagnosing cervical lesions. This system further eliminates the subclassification of CIN by categorizing CIN1 as low-grade squamous intraepithelial lesion (LSIL) and CIN2 and CIN3 as high-grade squamous intraepithelial lesion (HSIL) and the last stage as invasive cancer3. Atypical squamous cells (ASC) divides into two subcategories: atypical squamous cells of undetermined significance (ASC-US) and atypical squamous cells, which cannot exclude a high-grade squamous intraepithelial lesion (ASC-H). ASC-H is the less common qualifier, accounting for 5 to 10% of all ASC cases, but the risk of the potentially high-grade lesion is higher in this category than in ASC-US. This diagnostic category includes a mixture of real HSILs and its mimics4. Women with HSIL, for whom the risk of cancer is high, are immediately referred for colposcopy and, if the lesion is confirmed, surgery is required to remove the tumor5. The rate of concurrent and subsequent HSILs on follow up of ASC-H is reported as 29–75%, and it is recommended that women diagnosed with ASC-H also should be referred for colposcopy4. The detection of LSIL or ASC-US may lead to a follow-up smear being taken after a shorter time interval than the normal 2–3 years. Pap smears can greatly reduce the incidence of cervical cancer. During the Pap smear examination, cytopathologists manually scan and inspect the whole slide at the microscope using magnification at 10, and when something suspicious is seen, detailed inspection is conducted at magnification 40. This process typically involves checking thousands of cells6. Manual analysis of the Pap smear images requires a large amount of well trained manpower, which is extremely expensive, time-consuming, laborious, and error-prone and not available in many hospitals. More importantly, if malignant cells are carelessly neglected during the manual screening process, this will jeopardize the subsequent treatment plan, causing the patient miss the opportunity of early treatment and even more serious consequences. In addition, high inter-observer variability substantially affects productivity in routine pathology and is especially ubiquitous in diagnostician-deficient medical centers7. With an increase in computing power and advance in imaging technologies, deep learning is being implemented for the diagnosis and classification of cervical lesions. Deep learning has been used for the detection of diseases, such as skin cancer8, lung cancer9, cardiac arrhythmia10, retinal disease11, intracranial hemorrhage12, neurological problems13, autism14, kidney disease15 and psychiatric problems16.
In 2019, Araújo et al.17 applied convolution neural networks (CNNs) to segment LSILs or ASCUSs using small size cervical cell images (1392 1040 pixels) acquired by manually identified regions of interests from microscopy, and Lin et al.18 applied CNNs to classify abnormal cells using single cervical cell image with averaged size (110 110 pixels), which is carefully prepared by manual localization and extraction of microscopic images. However, detection of LSILs or ASCUSs may only lead to a more shorter follow up interval, and both methods17,18 require manual intervention to locate and acquire single-cell images or images of regions of interests, and thus the two methods could not be utilized for fully automatic WSI analysis in cervical HSILs or higher (SQCC) examination. To the authors’ best knowledge, there has been no published work on automated cervical HSILs or higher (SQCC) analysis using whole slide images (WSIs) of conventional Pap smear samples for practical usages.
In this study, we propose a deep learning based cervical HSILs or higher (SQCC) diagnosis and treatment planning system using Papanicolaou staining, enabling automatic examination of cervical smear on WSIs and identification and quantification of HSILs or higher (SQCC) for further treatment suggestion. If HSILs or higher (SQCC) are detected in a patient Pap smear sample, the further clinical step is to perform the biopsy, large loop excision of the transformation zone (LEEP/LLETZ), and cold knife conization of the cervix to achieve both diagnosis and treatment. The collection of WSIs stained by Papanicolaou was obtained from Tri-service general hospital, Taipei, Taiwan, and a reference standard is produced by manual annotations of HSILs or higher (SQCC) by pathologists. In evaluation, as this is the first study on automatic HSILs or higher (SQCC) segmentation, we compare the proposed method with Araújo et al.’s approach17 for LSIL segmentation as well as two state-of-the-art deep learning methods (U-net19 and SegNet20) as shown in Fig. 1.
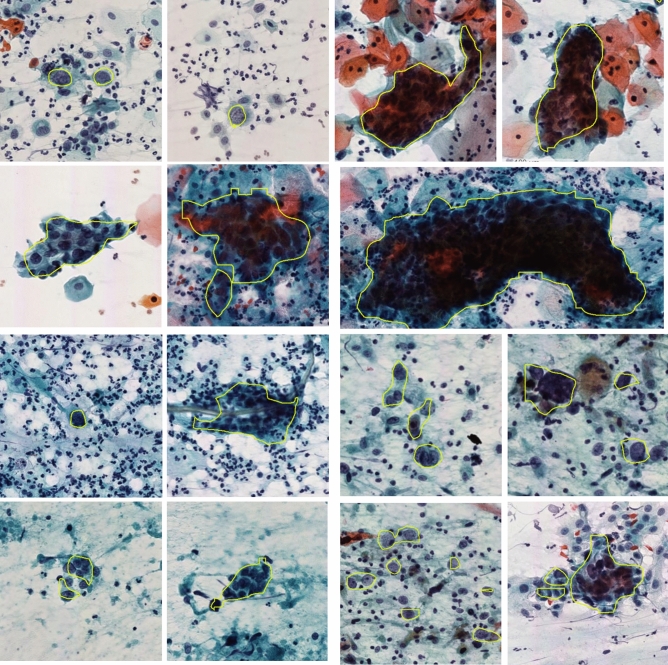
Figure 1.
Segmentation results of randomly selected examples by the proposed method where the HSILs are highlighted in yellow.
Data and results
Material
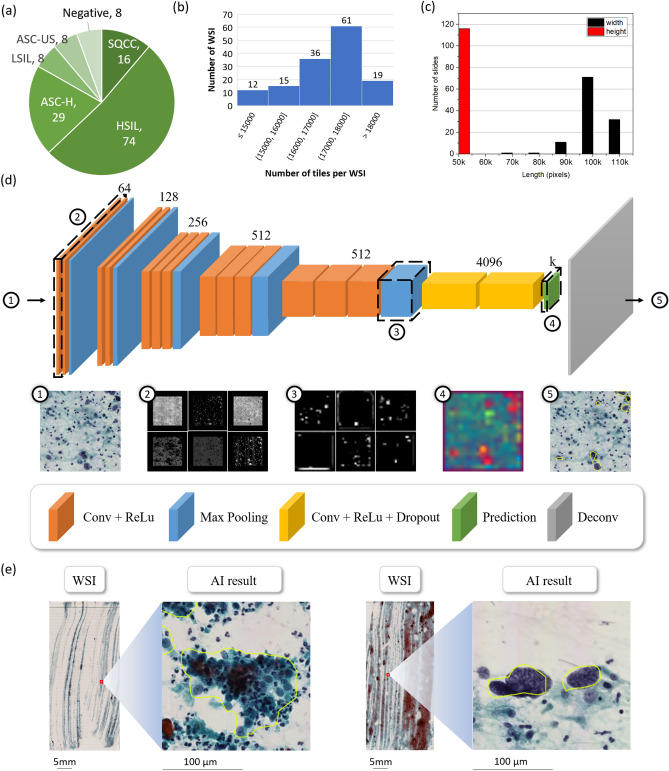
De-identified, digitized whole-slide images of conventional Pap smear samples were obtained from the tissue bank of the Department of Pathology, Tri-Service General Hospital, National Defense Medical Center, Taipei, Taiwan (n = 143 patients). A research ethics approval has been gained from the research ethics committee of the Tri-Service General Hospital (TSGHIRB No.1-107-05-171 and No.B202005070), and informed consent is formally waived by the approving committee. The data were de-identified and used for a retrospective study without impacting patient care. All methods were carried out in accordance with relevant guidelines and regulations. Cervical scrapings were collected for cytological diagnosis by gynecologists. The slides were prepared and stained by the Pap method according to the usual laboratory protocol. The screening of cytology slides was first performed by the pool of cytotechnologists, and a pathologist always confirmed abnormal results. Cytology was performed using TBS 2014. A series of negative (n = 8), ASC-US (n = 8), LSIL (n = 8), ASC-H (n = 29), HSIL (n = 74), or higher (SQCC, n = 16), and the number of per category in the dataset as shown in Fig. 2a. All patients were treated and followed by the standard clinical protocol. The patients with ASC-US underwent a repeat Pap smear within 1 year, while the patients with ASC-H, HSIL or SQCC underwent colposcopy-directed cervical biopsy and subsequent therapy when indicated. WSIs are digitized glass slides from scanning devices. All stained slides were scanned using Leica AT Turbo (Leica, Germany), at 20 objective magnification. The network was instead trained and tested using non-overlapping tiles (512–512 pixels) obtained from the WSIs. Distribution of the tile numbers per WSIs as shown in Fig. 2b. In computational pathology, the massive size of WSIs is one of the challenges. For automatic analysis the average size of WSIs is 91,257 41,546 pixels, 45.93 20.91 mm in our dataset, and the size distribution of the WSIs as shown in Fig. 2c. The proposed network structure with the associated outputs of the selected layers are shown in Fig. 2d. A WSI generally contains billions of pixels, while the regions of interest could be as small as a few thousands of pixels (see Fig. 2e). We collected slide-level reviews and region-level annotations from pathologists. Slide-level reviews categorize each slide into a group of higher than LSIL including ASC-H, HSIL and SQCC. Region-level annotations represent specific HSILs or higher (SQCC) within a slide. The information for the dataset is shown in Table 1.
Figure 2.
Data information and illustration of the proposed network framework. (a) The number of WSIs per category. (b) Distribution of the tile numbers per WSI. (c) Size distribution of the WSIs with width and height as black and red. (d) Illustration of the proposed modified FCN structure, and the feature maps of the sampling layers. (e) The results of the proposed method in WSIs.
Table 1.
Data distribution and ratio of sampling tissue for AI training.
| Category | Types | Number of patients’ WSIs | Ratio of sampling tissue in training | ||
|---|---|---|---|---|---|
| Training set | Testing set | With respect to training set | With respect to whole set | ||
| ASC-H | SQCC | 15 | 1 | 0.00011 | 0.00010 |
| HSIL | 57 | 17 | 0.00006 | 0.00005 | |
| ASC-H | 25 | 4 | 0.00004 | 0.00003 | |
| < ASC-H | LSIL | 0 | 8 | 0 | 0 |
| ASC-US | 0 | 8 | 0 | 0 | |
| Negative | 0 | 8 | 0 | 0 | |
| Total (%) | 97 (68%) | 46 (32%) | 0.00006 | 0.00004 | |
Experimental set-up and implementation details
In evaluation, the whole slide images were randomly split into two sets: 68% for training and 32% for testing. As shown in Table 1, the training set consists of 25 ASC-H, 57 HSIL, and 15 SQCC cases, and the tiles annotated and sampled for building AI models account for 0.006% of the training WSIs and for 0.004% of the whole data set, respectively. Moreover, the proposed framework is initialized using VGG16 model, and stochastic gradient descent (SGD) optimization and the cross entropy loss function are utilized. In addition, the network training parameters of the proposed method, including the learning rate, dropout ratio, and weight decay, are set to , 0.5, and 0.0005, respectively. The benchmark methods (U-net19 and SegNet20) are implemented using the keras impelementation of images segmentation models by Gupta et al.21. For training, the benchmark methods (U-net19 and SegNet20) are initialized using a pre-trained VGG16 model with the networks optimized using Adadelta optimization, and the cross entropy function is used as a loss function. In addition, the network training parameters of U-Net and SegNet, including the learning rate, dropout ratio, and weight decay, are set to 0.0001, 0.2, and 0.0002, respectively. The testing set accounting for 32% of the whole set, consisted of 8 Negative, 8 ASC-US, 8 LSIL, 4 ASC-H, 17 HSIL, and 1 SQCC. Further details on the data set could be found in Table 1. To assess the performance of the proposed method, we compared the AI segmentation results with the reference standard annotated by pathologists. Further quantitative evaluation details are described in the next section.
Evaluation method
In this study, two state-of-the-art deep learning methods (U-net19 and SegNet20) and Araújo et al.’s method17 are adopted as the benchmark approaches, and we compare the computing speed, precision, recall, F-measure, Jaccard index, which is often used in semantic segmentation to validate the pixel-level labeling performance22–26, of the proposed method and the benchmark approaches. Objects are classified into one of the four categories: TP, true positive; TN, true negative; FP, false positive; FN, false negative. The evaluation metrics are formulated as follows.
| 1 |
| 2 |
| 3 |
| 4 |
Quantitative evaluation and statistical analysis
In evaluation, as this is the first study on automatic HSILs or higher (SQCC) segmentation, we compare the proposed method with Araújo et al.’s approach17 for LSIL segmentation on small image patches as well as two state-of-the-art deep learning methods (U-net19 and SegNet20) as shown in Table. 2 where the reported numbers of Araújo et al.17 are referred. The results show that the proposed method achieves high precision 0.93, recall 0.90, F-measure 0.88 and Jaccard index 0.84 and outperforms the three benchmark approaches in all four measurements. In comparison, the U-Net model obtains precision 0.15, recall 0.70, F-measure 0.17 and Jaccard 0.12; SegNet model obtains precision 0.26, recall 0.88, F-measure 0.27 and Jaccard 0.23; Araújo et al.’s approach17 is with precision 0.66, recall 0.72, F-measure 0.69 and Jaccard 0.53. Furthermore, Fig. 3 presents qualitative segmentation results by the proposed method and two benchmark approaches for HSILs or higher (SQCC) detection. The results demonstrate the high precision, efficiency and reliability of the proposed model.
Table 2.
Quantitative comparison with benchmark methods.
| Target | Data type | Aver. size (pixel) | Precision | Recall | F-measure | Jaccard | |
|---|---|---|---|---|---|---|---|
| Proposed method | HSIL | WSIs | 91257 41546 | 0.93 | 0.90 | 0.88 | 0.84 |
| U-net19 | HSIL | WSIs | 91257 41546 | 0.15 | 0.70 | 0.17 | 0.12 |
| SegNet20 | HSIL | WSIs | 91257 41546 | 0.26 | 0.88 | 0.27 | 0.23 |
| Araújo et al.17 | LSIL/ASCUS | Image patches | 1383 1036 | 0.66 | 0.72 | 0.69 | 0.53 |
The proposed method is significantly better than the benchmark approaches (p<0.0001).
Figure 3.
Qualitative segmentation results by the proposed method and two benchmark approaches for HSIL detection.
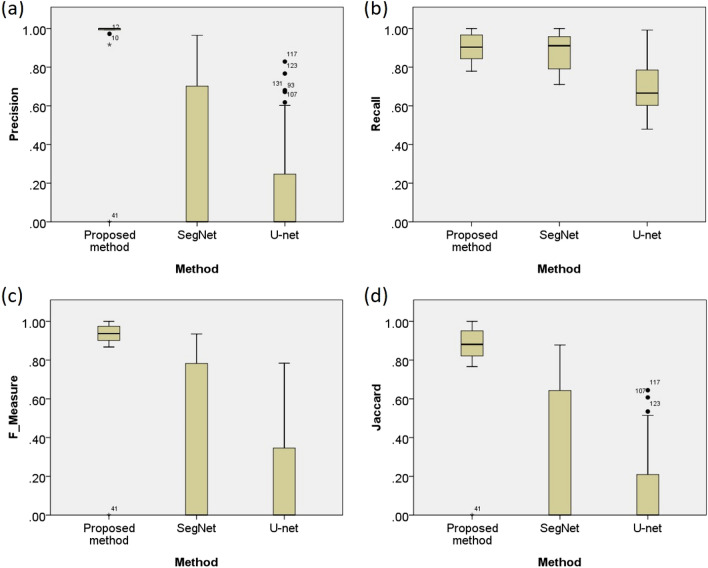
Table 3 presents detailed quantitative evaluation results in HSILs or higher (SQCC) segmentation for all samples and for separate evaluation on samples with high-grade lesions and samples with low-grade lesions or negative. The box plots of the quantitative evaluation results for all samples are provided in Fig. 4, showing that the proposed method consistently performs well with high precision, recall, F-measure. Jaccard and specificity and outperforms the two state of the art deep learning methods, i.e. SegNet and U-net. The experimental results show that the two benchmark methods (U-net19 and SegNet20) perform poor in detecting HSILs or higher (SQCC), obtaining precision, recall, F-Measure, Jaccard index were on average, respectively.
Table 3.
Quantitative evaluation of the proposed method and two benchmark methods (U-Net and SegNet) in segmenting of HSILs or higher (SQCC).
| Proposed method | SegNet20 | U-net19 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| All | HSIL | LSIL | All | HSIL | LSIL | All | HSIL | LSIL | |
| Precision | 0.9294 | 0.9913 | 0a | 0.2573 | 0.5145 | 0a | 0.1528 | 0.3195 | 0a |
| Recall (sensitivity) | 0.8987 | 0.8987 | NaN | 0.8760 | 0.8760 | NaN | 0.7053 | 0.7053 | NaN |
| F-measure | 0.8831 | 0.9420 | 0a | 0.2670 | 0.5341 | 0a | 0.1691 | 0.3536 | 0a |
| Jaccard | 0.8356 | 0.8913 | 0a | 0.2259 | 0.4518 | 0a | 0.1220 | 0.2550 | 0a |
| Specificity | 1.0000 | 1.0000 | 1.0000 | 0.9833 | 0.9709 | 0.9946 | 0.5752 | 0.7327 | 0.4308 |
The proposed method is significantly better than the benchmark approaches (p<0.0001).
aAs there are no positive in these samples, the value is computed as 0.
Figure 4.
The box plot of quantitative evaluation results in HSILs or higher (SQCC) segmentation. The presented method works constantly well overall and outperforms the state-of-the-art benchmark methods. Outliers greater than and the interquartile range are marked with a dot and a asterisk, respectively.
Furthermore, for statistical analysis, using SPSS software27, the quantitative scores were analyzed with the Fisher’s Least Significant Difference (LSD) to compare multiple methods (see Table 4). In precision, the presented method achieves 92.94% averaged and significantly outperforms both benchmark methods based on LSD tests (). In recall, the presented method achieves 89.85% averaged and significantly outperforms the U-net method based on LSD tests (). In F-Measure, the presented method achieves 88.21% averaged and significantly outperforms both benchmark methods based on LSD tests (). In Jaccard index, the presented method achieves 83.57% averaged and significantly outperforms both benchmark methods based on LSD tests (). Figure 1 presents more segmentation results of randomly selected examples of by the proposed method.
Table 4.
Multiple comparisons for the segmentation of HSILs or higher (SQCC): Fisher’s LSD test.
| LSD multiple comparisons | |||||||
|---|---|---|---|---|---|---|---|
| Dependent variable | (I) Method | (J) Method | Mean difference (I-J) | Std. error | Sig. | 95% CI | |
| Lower bound | Upper bound | ||||||
| Precision | Proposed method | U-net19 | 0.77696a | 0.09093 | < 0.0001 | 0.5966 | 0.9573 |
| SegNet20 | 0.67273a | 0.09146 | < 0.0001 | 0.4913 | 0.8541 | ||
| Recall | Proposed method | U-net19 | 0.19469a | 0.04242 | < 0.0001 | 0.1091 | 0.2803 |
| SegNet20 | 0.02283 | 0.04242 | 0.593 | − 0.0628 | 0.1084 | ||
| F-measure | Proposed method | U-net19 | 0.71301a | 0.09314 | < 0.0001 | 0.5283 | 0.8977 |
| SegNet20 | 0.61492a | 0.09369 | < 0.0001 | 0.4291 | 0.8007 | ||
| Jaccard | Proposed method | U-net19 | 0.71348a | 0.07790 | < 0.0001 | 0.5590 | 0.8680 |
| SegNet20 | 0.61008a | 0.07836 | < 0.0001 | 0.4547 | 0.7655 | ||
Run time analysis
Due to the enormous size of WSIs, the computational time for WSI analysis is critical for practical clinical usage. Therefore, we analyzed the AI inference time using different hardware settings (see Table 5a). For the run time analysis, the proposed method takes only 210 seconds to process a WSI and is 20 times faster than U-Net and 19 times faster than SegNet, respectively (see Table 5b). In comparison with Araújo et al.’s method17, the proposed method processes 923,520 more pixels per second, and Araújo et al.’s method requires twice of hardware memory (251 GB) than the proposed method (128 GB). Overall, the proposed method is demonstrated to be able to both detect HSILs, which indicate patients for further treatments, including colposcopy and surgery to remove the lesion, and rapidly processing WSIs in seconds for practical clinical usages.
Table 5.
Comparison on (a) hardware and (b) computing efficiency.
| Method | CPU | RAM (GB) | GPU |
|---|---|---|---|
| (a) | |||
| Proposed method | Intel Xeon Gold 6134 CPU @ 3.20GHz 16 | 128 | 4 GeForce GTX 1080Ti |
| U-net19 | Intel Xeon CPU E5-2650 v2 @ 2.60GHz 16 | 32 | 1 GeForce GTX 1080Ti |
| SegNet20 | Intel Xeon CPU E5-2650 v2 @ 2.60GHz 16 | 32 | 1 GeForce GTX 1080Ti |
| Araúj et al.17 | Intel Xeon E5-2643 @ 3.40 GHz 6 | 251 | 4 GeForce GTX Titan-X |
| Method | Inference time in seconds (per WSIa) | Inference pixels (per second) | |
|---|---|---|---|
| (b) | |||
| Proposed method | 210 | 21,604,662 | |
| U-net19 | 4370 | 1,038,196 | |
| SegNet20 | 4024 | 1,127,500 | |
| Araúj et al.17 | 219 | 20,681,142 | |
aThe size of the WSI in this evaluation is 4,536,979,200 pixels (99,60045,552 pixels). bAraúj et al.’s method17 takes 0.07 s for processing a patch with 1392 1040 pixels; b219 s = ; 20,681,142 pixels = .
Discussion
To the authors’ best knowledge, this is the first work on automated cervical HSILs or higher (SQCC) analysis of WSIs on conventional Pap smear samples for practical usages. Our study demonstrates that the proposed new cervical Pap smear diagnosis system could be used to assist in automatic detection and quantification of cervical HSILs or higher (SQCC) from WSIs. The proposed method achieved a high precision 0.93, recall 0.90, F-measure 0.88, and Jaccard index 0.84, and capable of unambiguously segmenting HSILs or higher (SQCC), showing that the proposed system is robust and capable of segmenting HSILs or higher (SQCC) with high precision and reaches sensitivity comparable to the referenced standard produced by pathologists. Moreover, the proposed method significantly outperforms two state of the art deep learning approaches (). Cervical cancer develops through persistent infection with high-risk human papilloma virus (HR-HPV) and is a leading cause of death among women worldwide28. Regular screening strategies using HR-HPV, Pap smear and colposcopy alone or in combination can prevent the onset and development of cervical cancer28. Cervical cancer incidence can be reduced by as much as 90% where screening quality and coverage are high29. In 2018, the United States Preventive Services Task Force (USPSTF) updated its screening guidelines. In addition to continuing to recommend triennial cytology (Pap test) for women between 21 and 29 years old, then continue with triennial cytology or increase HR-HPV testing every 5 years between 30 and 65 years old30. The major contribution of our proposed method in a cervical Pap smear screening workflow compared to manual cytology reading is that it reduces on the time required by the cytotechnician to screen many pap-smears by eliminating the obvious normal ones, hence more time can be put on the suspicious slides. In recent decades, although the conventional Pap smear method has been the mainstay of the screening procedures. However, this technique is not without limitations, because the sensitivity and specificity are relatively low. Liquid-based cytology (LBC) was introduced in the 1990s and was initially considered a better tool for processing cervical lesions. But now it has been found that LBC is more superior to conventional smears only with respect to a lesser number of unsatisfactory smears. There is no significant difference in the detection of epithelial cell abnormalities between the two methods31. LBC is being widely used in the United States, European countries, and many other developed nations. Although these approaches appear better clarity, uniform spread of smears, less time for screening and better handling of hemorrhagic and inflammatory samples32, but they are expensive and rely heavily on technology33. To consider the cost effectiveness and health insurance policy , the conventional Pap method is more feasible in our country. Although HPV testing is more sensitive to detect cervical precancerous lesions and cancers earlier than cytology, there are currently costs, infrastructure considerations and specificity issues that limit its use in low- and middle-income countries34. The high frequency of transient HPV infection among women younger than 30 years can lead to unnecessary follow-up diagnostic and treatment interventions with potential for harm35. For the HR-HPV screening, the Food and Drug Administration (FDA) approved cobas HPV testing. This test detects HPV types 16, 18, and 26 and additional HR-HPV types36. Despite the reported high sensitivity (86%) and negative predictive value (82%) of HR-HPV testing37, some HSIL can still be missed38–40. The low specificity (31%) and positive predictive value (37%) even make the situation worse because they lead to more patients undergoing unnecessary referrals37. Recent studies have shown the correlation between epigenetics and development and progression of cervical cancer41. Increased methylation of host genes has been observed in women with cervical precancer and cancer. Several of these genes have been evaluated as candidates for triage of HPV-positive women. However, more longitudinal studies are needed to prove the longitudinal safety of negative methylation result42. Vaccination against HPV is a possible long-term solution for eradicating cervical cancer in developing countries, where a prophylactic HPV vaccine has already been approved. However, knowledge and awareness about cervical cancer, HPV, and the efficacy of the HPV vaccine in the prevention of cervical cancer are very low in the world. The low level of knowledge about HPV is considered to be the major hurdle for the implementation HPV vaccination programs30. Automatic screening of Papanicolaou system has been available for more than 25 years, such as AutoPap 30043 and the PapNet44, which were approved by the United States FDA in 1998. Cytyc was approved by the FDA in 2007 with the ThinPrep imaging system45. Recent reviews indicate that the previous image analysis and machine learning techniques used for automatic Pap smear screening are flawed, resulting in low accuracy. The development of fully automatic screening technology that does not rely on the human judgment has yet to be fully realized46. A fast automated deep learning system enables high throughput analysis across a wide cohort of patients, and also helps to obtain a large amount of data to analyze the enormous dimensions of large gigapixel data of WSIs. There is limited research on automated analysis of cervical lesions on conventional Pap smear WSIs. Araújo et al.17 applied CNNs to segment LSIL or ASCUS using small size cervical cell images (1392 1040 pixels) acquired by manually identified regions of interests from microscopy, and Lin et al.18 applied CNNs to classify abnormal cells using single cervical cell image with average size (110 110 pixels), which is carefully prepared by manual localization and extraction of microscopic images. Both methods require manual intervention to locate and acquire single-cell images or images of regions of interest. In comparison, we developed a fast and fully automatic deep learning fast screening system, which is capable of detection and quantification of HSILs or higher (SQCC) on WSIs in seconds for cervical lesion diagnosis and treatment suggestion. Our data demonstrated that AI-assisted cytology could distinguish most of CIN2+ (higher than CIN2) cytology based on a high precision 0.93, recall 0.90, F-measure 0.88, and Jaccard index 0.84. Compare to the manual cytology reading, it is close to an effective use in clinical practice due to complete CIN2+ cells labeling in a short time, which aid cytologists or cytotechnologists in screening and labeling cervical high grade dysplastic cells more easily and quickly. There are still some weaknesses in AI based Pap smear screening. When atypical cervical cells are gathered in different planes, traditional microscopes can overcome the focus problem by turning the adjustment wheel, but AI is not easy to correctly classify, such as HSIL present in three-dimensional groups closely mimic shed endometrial cells or HSIL pattern resembling reparative change. Specimen with rare, small, high nuclear to cytoplasmic ratio HSIL cells may be problematic with AI regard to identifying the single HSIL cells. The Pap smear image sometimes contain overlapping hyperchromatic crowded groups which can interfere the AI cytological diagnosis. Although our proposed method can correctly find out CIN2+ cells, but it still needs cytologists to confirm this diagnosis and divided CIN2+ cells into moderate dysplasia, severe dysplasia, squamous cell carcinoma in situ, nonkeratinizing SQCC or keratinizing SQCC. Furthermore, the proposed system is demonstrated to be superior than two state-of-the art deep learning methods, i.e. U-Net19 and SegNet20, in precision, recall, F-measure, Jaccard index and computing efficiency based on the experimental results using LSD test (). The precision, recall, F-measure and Jaccard index was calculated from a hospital-based, retrospective study using a research platform that may not be directly applicable to the clinical setting or to wider populations. The application of artificial intelligence may provide a new screening method of cervical Pap smear and warrants further validation in a larger population-based study in future work.
Our results show that the proposed AI assisted method with high sensitivity (0.9) and specificity (1.0) outperforms conventional Pap smear examination and HPV testing, overcoming the limitation of low sensitivity in conventional Pap smear slides using light microscopic examination and low specificity in HPV testing. Artificial intelligence assisted rapid screening has great potential to provide much faster and cheaper service in the future. The processing time, material and labor cost could be greatly reduced using artificial intelligence assisted rapid screening. The proposed fast screening deep learning based system could not only avoid misdiagnosis by human negligence but also resolve lengthy screening process. The proposed system is applicable for practical clinical usage worldwide for comprehensive screening and ultimately has an impact on the areas with high incidences of cervical cancer.
Methods
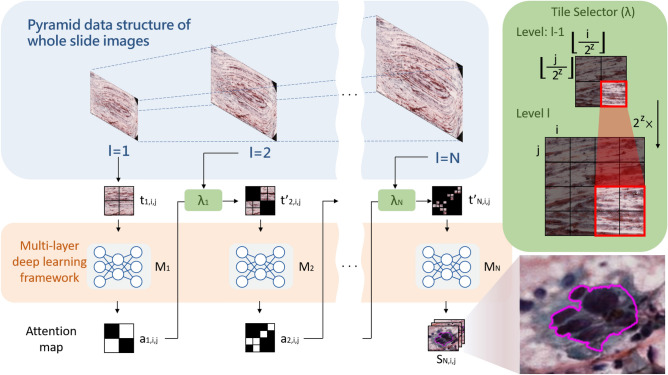
In this paper, we developed an efficient system to identify HSILs on WSIs with a cascaded multi-layer deep learning framework to improve accuracy and reduce the computing time. The proposed cascaded multi-layer utilizes a coarse-to-fine strategy to rapidly locate the tissues of interest and perform semantic segmentation to identify HSILs; at the coarse-level, fast localization of the tissues of interest is conducted, and at the fine-level, HSILs are identified based on the fast screening results of the tissues of interest. The framework of the proposed method is shown in Fig. 5.
Figure 5.
System framework. Each WSI is formatted into a tile-based pyramid data structure and the probability of HSILs at every level is generated by multi-layer deep learning framework. A multi-layer attention map is computed to select tiles of interest at every next level by a tile selector model , and the segmentation result of HSILs is generated by the last layer deep learning model .
Cascaded multi-layer deep learning framework
For dealing with gigapixel data efficiently, each WSI is formatted into a tile-based pyramid data structure, which is denoted by where l is the current level and i, j are the row and column index of a tile. represents the set of deep-learning model at each layer, and an improved fully convolution network is developed as the deep learning-base model, which is described in the next section. The output of is the probabilities of HSILs at level l, which is used to produce an attention map . A multi-layer attention map is computed to select tiles of interest for further inspection at every next level by a tile selector model . The segmentation result of HSILs is generated by .
For initialization, , i.e. the tiles in the first level, is processed by to generate the probabilities of HSILs , as shown in Eq. (5).
| 5 |
The output of is the probabilities of HSILs at level l, which is used to produce an attention map . If any pixel of has a probability greater than or equal to , set the attention map of that tile to 1, as shown in Eq. (6). In the practical case, is set to 0.5.
| 6 |
To render tiles of interest in every next level, the attention map in the previous level is used by a tile selector model with a mapping function as shown in Eq. (7). , i.e. an attention map unit at level, is associated with units at l level, and on the other hand, is associated with the attention map unit at at level. Thus, a tile is selected for further inspection as when the corresponding attention map unit at the previous level equals to 1.
| 7 |
In our implementation, each tile is associated with an attention tile, which contains 16 attention-units ( squares), and if the segmentation model at that level confirms any pixel of that tile associated to the attention unit(s) as the target, the attention units will be activated to select tiles for the next level.
For the subsequent levels , is processed by to generate the probabilities of HSILs , as shown in Eq. (8). is then used to produce an attention map for identification of tiles of interest in the next level by a tile selector model to generate as formulated in Eqs. (6)–(7).
| 8 |
In the level N, the selected tiles produces probabilities by using (8). The segmentation result of HSILs is generated by using as shown in eq.(9).
| 9 |
Modified FCN for segmentation of HSILs or higher (SQCC)
Fully Convolutional Network (FCN) has been demonstrated to be effective in pathology, such as segmentation of nuclei in the images47, cell counting in different kinds of microscopy images48 and neuropathology49. The Fully Convolutional Network (FCN)50 is mainly composed of 18 convolution layers (each convolution layer is followed by a RELU layer), five pooling layers for downsampling, a SoftMax layer and three upsampling layers, namely FCN-8s, FCN-16s, and FCN-32s models forming a three-stream net where the outputs from each stream are aggregated to form the final output. The architecture of the proposed FCN as shown in Table 6. In our preliminary test using a lung dataset provided by Automatic Cancer Detection and Classification in Whole Slide Lung Histopathology challenge, which is held with the IEEE International Symposium on Biomedical Imaging (ISBI) in 201951, we discover that single-stream FCN-32s could avoid overly fragmented segmentation results in comparison to the original three-stream net, as shown in Fig. 6. In addition, the cost of training and inference time is saved dramatically. In this study, we developed an improved FCN as the base deep learning model using the single-stream FCN-32s as the upsampling layers to improve segmentation result, lower GPU memory consumption, and speed-up the time for AI training and inference.
Table 6.
The architecture of the proposed deep learning network.
| Layer | Features (train) | Features (inference) | Kernel size | Stride |
|---|---|---|---|---|
| Input | 512 512 3 | 512 512 3 | – | – |
| Conv1_1 | 512 512 3 | 512 512 3 | 3 3 | 1 1 |
| Conv1_2 | 710 710 64 | 710 710 64 | 3 3 | 1 1 |
| Pool1 | 710 710 64 | 710 710 64 | 2 2 | 2 2 |
| Conv2_1 | 355 355 64 | 355 355 64 | 3 3 | 1 1 |
| Conv2_2 | 355 355 128 | 355 355 128 | 3 3 | 1 1 |
| Pool2 | 355 355 128 | 355 355 128 | 2 2 | 2 2 |
| Conv3_1 | 178 178 128 | 178 178 128 | 3 3 | 1 1 |
| Conv3_2 | 178 178 256 | 178 178 256 | 3 3 | 1 1 |
| Conv3_3 | 178 178 256 | 178 178 256 | 3 3 | 1 1 |
| Pool3 | 178 178 256 | 178 178 256 | 2 2 | 2 2 |
| Conv4_1 | 89 89 256 | 89 89 256 | 3 3 | 1 1 |
| Conv4_2 | 89 89 512 | 89 89 512 | 3 3 | 1 1 |
| Conv4_3 | 89 89 512 | 89 89 512 | 3 3 | 1 1 |
| Pool4 | 89 89 512 | 89 89 512 | 2 2 | 2 2 |
| Conv5_1 | 45 45 512 | 45 45 512 | 3 3 | 1 1 |
| Conv5_2 | 45 45 512 | 45 45 512 | 3 3 | 1 1 |
| Conv5_3 | 45 45 512 | 45 45 512 | 3 3 | 1 1 |
| Pool5 | 45 45 512 | 45 45 512 | 2 2 | 2 2 |
| Drop6 | 23 23 512 | 16 16 512 | – | – |
| Drop7 | 17 17 4096 | 10 10 4096 | – | – |
| Upsampled | 576 576 3 | 576 576 3 | – | – |
| Output | 512 512 3 | 512 512 3 | – | – |
Figure 6.
Comparison of segmentation results of three un-sampling layers of FCN. The results of (a) FCN-8s, and (b) FCN-16s are too fragmented. (c) In comparison, the results of FCN-32s are the most similar to reference standard.
Acknowledgements
This study is supported by the Ministry of Science and Technology of Taiwan, under a Grant (MOST109-2221-E-011-018-MY3, MOST 110-2321-B-016-002), Tri-Service General Hospital, Taipei, Taiwan (TSGHC108086, TSGH-D-109094 and TSGH-D-110036), and Tri-Service General Hospital-National Taiwan University of Science and Technology (TSGH-NTUST-103-02).
Author contributions
C.W.W. and T.K.C. conceived the study. C.W.W. developed the method. C.W.W., Y.A.L., Y.J.L, C.C.C., P.H.C., Y.C.L., T.K.C. collaborated on the dataset acquisition, visualization, and analysis. C.W.W., Y.A.L., T.K.C. wrote the manuscript. C.W.W., Y.A.L., Y.C.L. prepared the figures. C.W.W, C.H.W., T.K.C. obtained grants. All authors reviewed the manuscript before submission.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Shimizu, Y. Elimination of cervical cancer as a global health problem is within reach. World Health organization.https://www.who.int/reproductivehealth/topics/cancers/en/. Accessed 12 May 2020.
- 2.Takiar R, Nadayil D, Nandakumar A. Projections of number of cancer cases in India (2010–2020) by cancer groups. Asian Pac. J. Cancer Prev. 2010;11:1045–1049. [PubMed] [Google Scholar]
- 3.Reynolds D. Cervical cancer in hispanic/latino women. Clin. J. Oncol. Nurs. 2004;8:146–150. doi: 10.1188/04.CJON.146-150. [DOI] [PubMed] [Google Scholar]
- 4.Solomon D, et al. The 2001 Bethesda system: Terminology for reporting results of cervical cytology. JAMA. 2002;287:2114–2119. doi: 10.1001/jama.287.16.2114. [DOI] [PubMed] [Google Scholar]
- 5.Massad LS, et al. 2012 updated consensus guidelines for the management of abnormal cervical cancer screening tests and cancer precursors. J. Low Genit. Tract. Dis. 2013;17:S1–S27. doi: 10.1097/LGT.0b013e318287d329. [DOI] [PubMed] [Google Scholar]
- 6.Garcia-Gonzalez D, Garcia-Silvente M, Aguirre E. A multiscale algorithm for nuclei extraction in pap smear images. Expert Syst. Appl. 2016;64:512–522. doi: 10.1016/j.eswa.2016.08.015. [DOI] [Google Scholar]
- 7.Zhang Z, et al. Pathologist-level interpretable whole-slide cancer diagnosis with deep learning. Nat. Mach. Intell. 2019;1:236–245. doi: 10.1038/s42256-019-0052-1. [DOI] [Google Scholar]
- 8.Esteva A, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542:115–118. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coudray N, et al. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat. Med. 2018;24:1559–1567. doi: 10.1038/s41591-018-0177-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hannun AY, et al. Cardiologist-level arrhythmia detection and classification in ambulatory electrocardiograms using a deep neural network. Nat. Med. 2019;25:65–69. doi: 10.1038/s41591-018-0268-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Fauw J, et al. Clinically applicable deep learning for diagnosis and referral in retinal disease. Nat. Med. 2018;24:1342–1350. doi: 10.1038/s41591-018-0107-6. [DOI] [PubMed] [Google Scholar]
- 12.Lee H, et al. An explainable deep-learning algorithm for the detection of acute intracranial haemorrhage from small datasets. Nat. Biomed. Eng. 2019;3:173–182. doi: 10.1038/s41551-018-0324-9. [DOI] [PubMed] [Google Scholar]
- 13.Titano JJ, et al. Automated deep-neural-network surveillance of cranial images for acute neurologic events. Nat. Med. 2018;24:1337–1341. doi: 10.1038/s41591-018-0147-y. [DOI] [PubMed] [Google Scholar]
- 14.Hazlett HC, et al. Early brain development in infants at high risk for autism spectrum disorder. Nature. 2017;542:348–351. doi: 10.1038/nature21369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ravizza S, et al. Predicting the early risk of chronic kidney disease in patients with diabetes using real-world data. Nat. Med. 2019;25:57–59. doi: 10.1038/s41591-018-0239-8. [DOI] [PubMed] [Google Scholar]
- 16.Durstewitz D, Koppe G, Meyer-Lindenberg A. Deep neural networks in psychiatry. Mol. Psychiatry. 2019;24:1583–1598. doi: 10.1038/s41380-019-0365-9. [DOI] [PubMed] [Google Scholar]
- 17.Araújo FH, et al. Deep learning for cell image segmentation and ranking. Comput. Med. Imaging Graph. 2019;72:13–21. doi: 10.1016/j.compmedimag.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 18.Lin H, Hu Y, Chen S, Yao J, Zhang L. Fine-grained classification of cervical cells using morphological and appearance based convolutional neural networks. IEEE Access. 2019;7:71541–71549. doi: 10.1109/ACCESS.2019.2919390. [DOI] [Google Scholar]
- 19.Falk T, et al. U-net: Deep learning for cell counting, detection, and morphometry. Nat. Methods. 2019;16:67–70. doi: 10.1038/s41592-018-0261-2. [DOI] [PubMed] [Google Scholar]
- 20.Badrinarayanan V, Kendall A, Cipolla R. Segnet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017;39:2481–2495. doi: 10.1109/TPAMI.2016.2644615. [DOI] [PubMed] [Google Scholar]
- 21.Gupta, D. et al. Image segmentation keras : Implementation of segnet, fcn, unet, pspnet and other models in keras. https://github.com/divamgupta/image-segmentation-keras. Accessed 15 Sept 2020.
- 22.Luo R, Sedlazeck FJ, Lam T-W, Schatz MC. A multi-task convolutional deep neural network for variant calling in single molecule sequencing. Nat. Commun. 2019;10:1–11. doi: 10.1038/s41467-018-07882-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haberl MG, et al. Cdeep3m-plug-and-play cloud-based deep learning for image segmentation. Nat. Methods. 2018;15:677–680. doi: 10.1038/s41592-018-0106-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Su, H. et al. Region segmentation in histopathological breast cancer images using deep convolutional neural network. In 2015 IEEE 12th International Symposium on Biomedical Imaging (ISBI), 55–58 (2015).
- 25.Wang J, et al. Discrimination of breast cancer with microcalcifications on mammography by deep learning. Sci. Rep. 2016;6:1–9. doi: 10.1038/s41598-016-0001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masood S, et al. Automatic choroid layer segmentation from optical coherence tomography images using deep learning. Sci. Rep. 2019;9:1–18. doi: 10.1038/s41598-019-39795-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.SPSS Inc. Released 2008. SPSS Statistics for Windows, Version 17.0. Chicago: SPSS Inc.
- 28.Schiffman M, Solomon D. Clinical practice. Cervical-cancer screening with human papillomavirus and cytologic cotesting. N. Engl. J. Med. 2013;369:2324–2331. doi: 10.1056/NEJMcp1210379. [DOI] [PubMed] [Google Scholar]
- 29.Eddy DM. Secondary prevention of cancer: An overview. Bull. World Health Organ. 1986;64:421–429. [PMC free article] [PubMed] [Google Scholar]
- 30.Curry SJ, et al. Screening for cervical cancer: US preventive services task force recommendation statement. JAMA. 2018;320:674–686. doi: 10.1001/jama.2018.10897. [DOI] [PubMed] [Google Scholar]
- 31.Pankaj S, et al. Comparison of conventional pap smear and liquid-based cytology: A study of cervical cancer screening at a tertiary care center in bihar. Indian J. Cancer. 2018;55:80–83. doi: 10.4103/ijc.IJC_352_17. [DOI] [PubMed] [Google Scholar]
- 32.Singh VB, et al. Liquid-based cytology versus conventional cytology for evaluation of cervical Pap smears: Experience from the first 1000 split samples. Indian J. Pathol. Microbiol. 2015;58:17–21. doi: 10.4103/0377-4929.151157. [DOI] [PubMed] [Google Scholar]
- 33.Pankaj S, et al. Comparison of conventional Pap smear and liquid-based cytology: A study of cervical cancer screening at a tertiary care center in Bihar. Indian J. Cancer. 2018;55:80–83. doi: 10.4103/ijc.IJC_352_17. [DOI] [PubMed] [Google Scholar]
- 34.Cubie HA, Campbell C. Cervical cancer screening—the challenges of complete pathways of care in low-income countries: Focus on Malawi. Womens Health (Lond.) 2020;16:1745506520914804. doi: 10.1177/1745506520914804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ngo-Metzger Q, Adsul P. Screening for cervical cancer. Am. Fam. Physician. 2019;99:253–254. [PubMed] [Google Scholar]
- 36.Ge Y, et al. Role of HPV genotyping in risk assessment among cytology diagnosis categories: Analysis of 4562 cases with cytology-HPV cotesting and follow-up biopsies. Int. J. Gynecol. Cancer. 2019;29:234–241. doi: 10.1136/ijgc-2018-000024. [DOI] [PubMed] [Google Scholar]
- 37.Nieh S, et al. Is p16(INK4A) expression more useful than human papillomavirus test to determine the outcome of atypical squamous cells of undetermined significance-categorized Pap smear? A comparative analysis using abnormal cervical smears with follow-up biopsies. Gynecol. Oncol. 2005;97:35–40. doi: 10.1016/j.ygyno.2004.11.034. [DOI] [PubMed] [Google Scholar]
- 38.Arbyn M, et al. Virologic versus cytologic triage of women with equivocal Pap smears: A meta-analysis of the accuracy to detect high-grade intraepithelial neoplasia. J. Natl. Cancer Inst. 2004;96:280–293. doi: 10.1093/jnci/djh037. [DOI] [PubMed] [Google Scholar]
- 39.Eltoum IA, et al. Significance and possible causes of false-negative results of reflex human Papillomavirus infection testing. Cancer. 2007;111:154–159. doi: 10.1002/cncr.22688. [DOI] [PubMed] [Google Scholar]
- 40.Lorenzato M, et al. Contribution of DNA ploidy image cytometry to the management of ASC cervical lesions. Cancer. 2008;114:263–269. doi: 10.1002/cncr.23638. [DOI] [PubMed] [Google Scholar]
- 41.Zhu H, et al. DNA methylation and hydroxymethylation in cervical cancer: Diagnosis, prognosis and treatment. Front. Genet. 2020;11:347. doi: 10.3389/fgene.2020.00347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wentzensen N, Schiffman M, Palmer T, Arbyn M. Triage of HPV positive women in cervical cancer screening. J. Clin. Virol. 2016;76(Suppl 1):S49–S55. doi: 10.1016/j.jcv.2015.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tench WD. Validation of autopap primary screening system sensitivity and high-risk performance. Acta Cytol. 2002;46:296–302. doi: 10.1159/000326725. [DOI] [PubMed] [Google Scholar]
- 44.Bergeron C, et al. Quality control of cervical cytology in high-risk women. Papnet system compared with manual rescreening. Acta Cytol. 2000;44:151–157. doi: 10.1159/000326353. [DOI] [PubMed] [Google Scholar]
- 45.Chivukula M, et al. Introduction of the thin prep imaging system(tis): Experience in a high volume academic practice. Cytojournal. 2007;4:6. doi: 10.1186/1742-6413-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thrall MJ. Automated screening of papanicolaou tests: A review of the literature. Diagn. Cytopathol. 2019;47:20–27. doi: 10.1002/dc.23931. [DOI] [PubMed] [Google Scholar]
- 47.Naylor P, Laé M, Reyal F, Walter T. Segmentation of nuclei in histopathology images by deep regression of the distance map. IEEE Trans. Med. Imaging. 2019;38:448–459. doi: 10.1109/TMI.2018.2865709. [DOI] [PubMed] [Google Scholar]
- 48.Zhu, R., Sui, D., Qin, H. & Hao, A. An extended type cell detection and counting method based on FCN. In 2017 IEEE 17th International Conference on Bioinformatics and Bioengineering (BIBE), 51–56 (2017).
- 49.Signaevsky M, et al. Artificial intelligence in neuropathology: Deep learning-based assessment of tauopathy. Lab. Investig. 2019;99:1019–1029. doi: 10.1038/s41374-019-0202-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shelhamer E, Long J, Darrell T. Fully convolutional networks for semantic segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017;39:640–651. doi: 10.1109/TPAMI.2016.2572683. [DOI] [PubMed] [Google Scholar]
- 51.Li, Z. et al. Deep learning methods for lung cancer segmentation in whole-slide histopathology images—the acdc@lunghp challenge 2019. IEEE J. Biomed. Health Inform25, 429–440 (2021). [DOI] [PubMed]