Abstract
Ruxolitinib (rux) Phase II clinical trials are underway for the treatment of high-risk JAK2-rearranged (JAK2r) B-cell acute lymphoblastic leukemia (B-ALL). Treatment resistance to targeted inhibitors in other settings is common; elucidating potential mechanisms of rux resistance in JAK2r B-ALL will enable development of therapeutic strategies to overcome or avert resistance. We generated a murine pro-B cell model of ATF7IP-JAK2 with acquired resistance to multiple type-I JAK inhibitors. Resistance was associated with mutations within the JAK2 ATP/rux binding site, including a JAK2 p.G993A mutation. Using in vitro models of JAK2r B-ALL, JAK2 p.G993A conferred resistance to six type-I JAK inhibitors and the type-II JAK inhibitor, CHZ-868. Using computational modeling, we postulate that JAK2 p.G993A enabled JAK2 activation in the presence of drug binding through a unique resistance mechanism that modulates the mobility of the conserved JAK2 activation loop. This study highlights the importance of monitoring mutation emergence and may inform future drug design and the development of therapeutic strategies for this high-risk patient cohort.
Subject terms: Acute lymphocytic leukaemia, Targeted therapies, Cancer therapeutic resistance
Introduction
Acute lymphoblastic leukemia (ALL) is the most common pediatric cancer and the leading cause of non-traumatic death in children in the developed world1,2. Treatment advances using multiagent, risk-directed therapies have significantly improved 5-year survival rates for patients with ALL, which now approach 90%3. Despite these improvements, relapsed/refractory ALL is associated with poor prognosis and current treatment regimens have adverse side effects4–6. In addition, the 5-year relapse-free survival rate for adults is only 30–40%, with a high relapse rates even in patients who achieve remission after induction chemotherapy, likely due to the high proportion of patients with high-risk genomic lesions7,8. One such example is Philadelphia-chromosome-like ALL (Ph-like ALL), or BCR-ABL1-like ALL, is a high-risk subtype of B-cell ALL (B-ALL) defined by a transcriptomic signature similar to BCR-ABL1-positive ALL but lacking the BCR-ABL1 translocation9,10. Ph-like ALL occurs in 15% of childhood B-ALL cases, with peak incidence among adolescents and young adults, and is characterized by genomic alterations in cytokine or kinase signaling pathways, and crucial lymphoid transcription factor genes11,12.
Gene fusions resulting from rearrangements of Janus kinase 2 (JAK2) or erythropoietin receptor (EPOR) are associated with inferior outcomes in Ph-like ALL patients13. Gene fusions involving JAK2 (JAK2-rearranged ALL (JAK2r ALL)) occurs in approximately 7% of pediatric Ph-like ALL, with frequency increasing with age to approximately 14% in adolescent and young adult patients13–15. JAK2 gene fusions are detected exclusively in the Ph-like subtype and encode chimeric JAK2 fusion proteins commonly comprising the amino terminus of a partner gene and carboxyl terminal of JAK2 (refs. 15–17). The full-length JAK2 kinase domain is preserved in all JAK2 fusion genes, which drive leukemogenesis through constitutive activation of JAK2 activity14,18. Expression of JAK2 fusion genes has been shown to transform murine lymphoid cell lines to factor-independence and result in constitutive phosphorylation of signal transducer and activator of transcription 5 (STAT5)14,15,19. ALL cases driven by JAK2 fusion genes are significantly more aggressive than those associated with activating JAK2 point mutations such as the pseudokinase domain mutation JAK2 p.R683 (ref. 20).
The poor outcomes associated with JAK2r ALL highlights that there is an urgent need for more effective treatment strategies for this high-risk subtype of ALL20. The success of TKIs for the treatment of BCR-ABL1-positive chronic myeloid leukemia (CML) has served as a paradigm for the application of rationally targeted therapies. Pre-clinical in vitro and in vivo studies suggest that JAK inhibitors may be an effective precision medicine approach for JAK2r ALL14,19–22. A candidate drug is ruxolitinib (rux), currently approved for clinical use mainly for myelofibrosis, but also for graft-versus-host disease following hematopoietic cell transplantation23. Other JAK inhibitors in clinical development include fedratinib, which was recently approved for myelofibrosis, as well as pacritinib and momelotinib24. All JAK inhibitors in clinical development are type-I JAK inhibitors, binding active JAK2 within the ATP-binding site25. Type-II JAK inhibitors bind inactive JAK2 in the ATP-binding site in addition to an allosteric site but no type-II JAK inhibitors have entered clinical trials25.
Rux is the only FDA-approved JAK1/2-specific inhibitor, currently used for the treatment of myeloproliferative neoplasms (MPNs)23. When used in myelofibrosis, it prolongs survival, reduces spleen size, and improves disease-related symptoms through a reduction in elevated tumor-induced cytokine26. Phase II clinical trials are currently ongoing to assess the safety and efficacy of rux in combination with chemotherapy for pediatric B-ALL patients with CRLF2 and JAK2 pathway alterations (NCT02723994)23. Results from the part 1 safety phase of this trial recently reported no dose-limiting toxicity up to 50 mg/m2 dosed day 1–14 of a 28 days cycle, as well as continuous dosing at 40 mg/m2 post-induction chemotherapy27. Three other clinical trials are also investigating rux in combination with chemotherapy for the treatment of high-risk ALL (NCT03117751, NCT03571321, and NCT02420717). Furthermore, rux therapy was well tolerated and induced morphologic remission in a case report of a child with chemo-resistant JAK2r ALL and induction failure27,28. These early findings suggest with JAK inhibitors in combination with chemotherapy may improve outcomes for patients with this high-risk ALL subtype.
Similar to the inhibition of Abelson (ABL) with targeted tyrosine kinase inhibitors (TKIs), rux is an ATP mimetic inhibitor of JAK2 (ref. 29). The emergence of resistant mutations within the ABL kinase domain of BCR-ABL1 is a well-established mechanism of TKI resistance in CML and Philadelphia-chromosome-positive ALL (Ph-pos ALL)30. To date, only a single report has identified a JAK2 kinase domain mutation in a high-risk pediatric case of B-ALL where primary leukemia cells displayed a reduced sensitivity to rux31. Nine other unique JAK2 mutations that confer resistance to rux (JAK2 p.E864K, p.L884P, p.E930G, p.Y931C, p.G935R, p.R938L, p.I960V, p.L983F, and p.E985K) have been identified within the JAK2 kinase domain in vitro by random mutagenesis screening of JAK2 (refs. 32–36). All of these mutations displayed cross-resistance to multiple type-I JAK inhibitors and the JAK2 p.L884P mutation also conferred resistance to type-II JAK inhibitors BBT-594 and CHZ-868 (refs. 32,33,37). Therefore, we predict that a subset of JAK2r ALL patients treated with rux will develop resistance and subsequent disease persistence.
In this study, JAK2 fusion genes identified in ALL patient lymphoblasts by mRNA sequencing (mRNA seq) were expressed in the murine pro-B cell line, Ba/F3, to model JAK2r ALL in vitro. We recapitulated acquired resistance to rux in JAK2r ALL in vitro by treating three independent replicates of murine pro-B cells expressing a high-risk JAK2 fusion gene with a rux dose escalation. Each replicate acquired a different mutation within the JAK2 rux/ATP-binding site and demonstrated resistance to multiple type-I JAK inhibitors. Two previously described mutations, JAK2 p.Y931C and p.L983F, were identified in addition to a previously unreported mutation, JAK2 p.G993A. The JAK2 p.G993A mutation was also found to confer resistance to the type-II JAK inhibitor, CHZ-868. Interestingly, computational modeling suggested that the JAK2 p.G993A mutation confers rux resistance via a unique resistance mechanism that enables JAK2 activation despite rux binding. This work contributes to our understanding of the mechanisms of rux resistance and will aid the development of therapeutic strategies to overcome or avert resistance.
Results
Cells expressing acquired JAK2 kinase domain mutations are resistant to rux
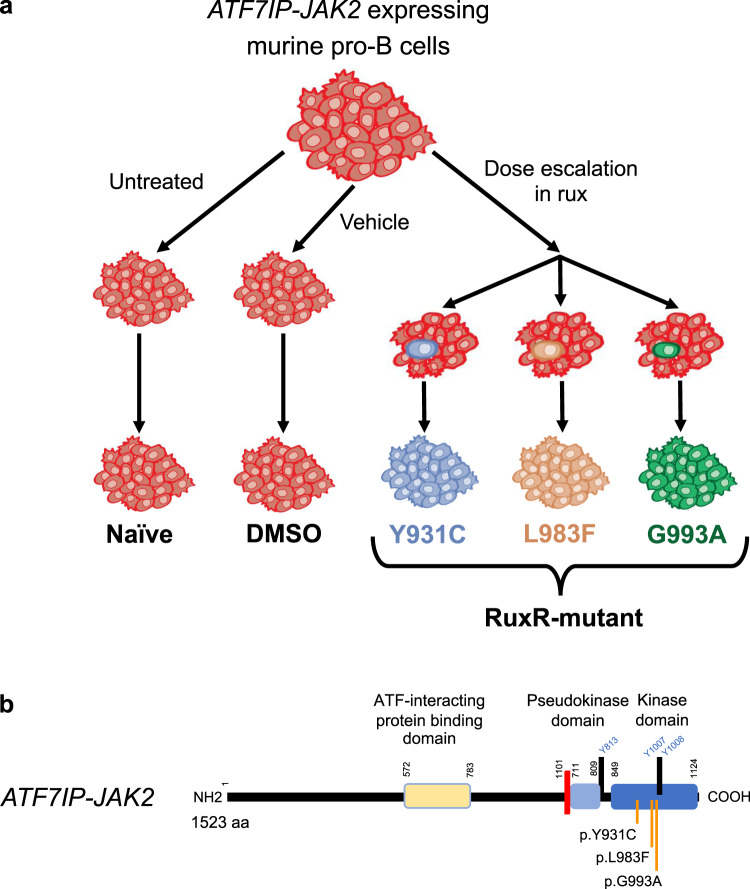
Mechanisms of rux resistance were investigated using Ba/F3 cells verified to be expressing the high-risk JAK2 fusion gene ATF7IP-JAK2 (Supplementary Fig. 1). Three independent biological replicates of ATF7IP-JAK2 Ba/F3 cells were dose escalated in rux to the clinically relevant dose of 1 µM (Fig. 1a), and all lines maintained GFP expression (Supplementary Fig 1d). After 3 months, the resulting three independent rux-resistant (RuxR) ATF7IP-JAK2 Ba/F3 sub-lines had each acquired a different point mutation within the JAK2 kinase domain (Fig. 1b and Supplementary Fig. 2). Two mutations, JAK2 p.Y931C and JAK2 p.L983F, were reported in previous literature33,36,38,39. Importantly, a previously unreported JAK2 p.G993A mutation was also identified. No mutations were identified in the non-mutant (Naïve or DMSO) ATF7IP-JAK2 Ba/F3 control cells. JAK2 p.Y931C, p.L983F, and p.G993A mutations were anticipated to confer resistance to type-I JAK inhibitors by constitutive activation of the JAK/STAT signaling pathway in the presence of rux. In the absence of rux, constitutive activation of STAT5 (pSTAT5) was observed in both non-mutant (Naïve) and RuxR-mutant ATF7IP-JAK2 Ba/F3 cells (Fig. 2a). Levels of pSTAT5 were lower in non-mutant ATF7IP-JAK2 Ba/F3 cells than in mutant cells, and rux exposure resulted in almost complete abrogation of pSTAT5 in these cells over a 60 min exposure (Fig. 2a). In comparison, RuxR-mutant ATF7IP-JAK2 Ba/F3 cells harboring JAK2 p.Y931C, p.L983F, or p.G993A mutations retained pSTAT5 following rux treatment (Fig. 2a). Interestingly, JAK2 p.Y931C ATF7IP-JAK2 Ba/F3 cells showed a more intense pSTAT5 signal in comparison to JAK2 p.L983F and p.G993A ATF7IP-JAK2 Ba/F3 cells (Fig. 2a).
Fig. 1. Representative diagram of how acquired resistance to rux was modeled using ATF7IP-JAK2 Ba/F3 cells.
a Three biological replicates of ATF7IP-JAK2 Ba/F3 cells were treated with a rux dose escalation for 3 months until resistance to the clinically relevant dose of 1 µM rux was achieved. Each of the three rux-resistant (RuxR) ATF7IP-JAK2 Ba/F3 cells acquired independent resistance to rux. Concurrently, ATF7IP-JAK2 Ba/F3 cells were cultured without treatment (Naïve) or treated with vehicle control (0.1% DMSO) to generate control cell lines. b Representative diagram of the ATF7IP-JAK2 fusion gene indicating localization of acquired point mutations within the JAK2 kinase domain. Annotations were made using the NCBI reference sequences for JAK2 variant 1 (NM_004972.4) and ATF7IP (NM_181352.2).
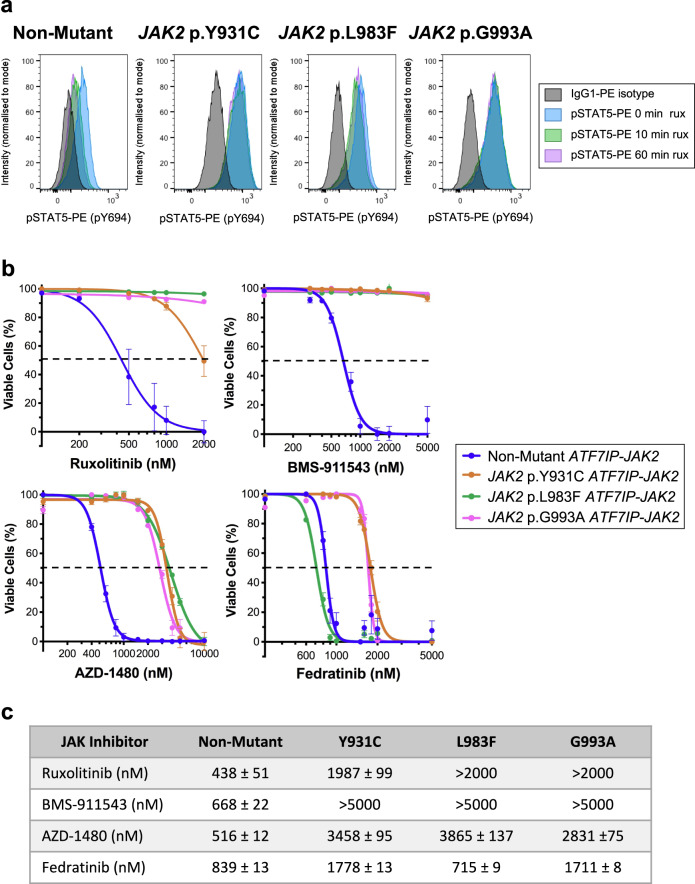
Fig. 2. ATF7IP-JAK2 Ba/F3 cells with acquired mutations are resistant to multiple type-I JAK inhibitors.
a Non-mutant or RuxR-mutant (JAK2 p.Y931C, p.L983F, or p.G993A) ATF7IP-JAK2 expressing Ba/F3 cells were incubated in 50 nM rux for up to 1 h. At 0, 10, and 60 min timepoints, STAT5 p.Y694 phosphorylation was assessed by intracellular flow cytometry in comparison to cells stained with an IgG1-PE isotype control antibody (black). Channel intensity was normalized to the percentage of maximum count and pSTAT5-PE mean fluorescence intensities (MFIs) are shown. Histograms are representative of three independent experiments. b Ba/F3 cells expressing either non-mutant or RuxR-mutant (JAK2 p.Y931C, p.L983F, or p.G993A) ATF7IP-JAK2 were incubated for 72 h with either a DMSO vehicle control or a dose response of type-I JAK inhibitors including rux, BMS-911543, AZD-1480, or fedratinib. The percentage of cell death was measured following a 20-min incubation with annexin-V and a live/dead cell stain, and then analysis by flow cytometry. Linear regression or non-linear regression models were fit to appropriate normalized data. Error bars indicate SEM over the mean of three biological replicates. c Table displaying LD50 concentrations (nM) for each cell line treated with JAK inhibitors shown in (b).
The prevalence of the acquired JAK2 mutations was also assessed by exome sequencing (exome seq). Reads aligning to the mouse genome were removed; then, exome seq data were visually inspected using IGV. Only reads aligning to the JAK2 p.Y931C mutation were detected in the DNA from ATF7IP-JAK2 Ba/F3 cells expressing JAK2 p.Y931C with a variant allele frequency (VAF) of 100%. In contrast, reads aligning to both non-mutant and RuxR-mutant JAK2 were detected in DNA from ATF7IP-JAK2 Ba/F3 cells expressing the JAK2 p.L983F or p.G993A mutations with VAFs of 60% and 49%, respectively. These results were consistent with mRNA sequencing data from RuxR-mutant ATF7IP-JAK2 Ba/F3 cell total RNA (data not included) and suggest a loss of non-mutant ATF7IP-JAK2 transcripts from the JAK2 p.Y931C ATF7IP-JAK2 Ba/F3 cell population during rux-resistance generation.
Cells with acquired JAK2 kinase domain mutations are resistant to multiple type-I JAK inhibitors
Mutations in the JAK2 kinase domain have been previously reported to display cross-resistance to multiple type-I JAK inhibitors in the setting of MPNs including momelotinib, fedratinib, AZD-1480, and lestaurtinib32. To assess whether JAK2 p.Y931C, p.L983F, or p.G993A mutations conferred resistance to multiple JAK inhibitors, pMIG empty vector Ba/F3 cells or Ba/F3 cells expressing either non-mutant or RuxR-mutant ATF7IP-JAK2 were treated with a vehicle control (DMSO) or varying concentrations of different type-I JAK inhibitors. LD50 values were determined by staining with Annexin-V and aqua dead cell stain (Invitrogen). Empty Vector Ba/F3 cells in the presence of IL3 were sensitive to rux, AZD-1480, and fedratinib with LD50 values of 714 ± 58, 735 ± 20, and 1236 ± 28 nM respectively (Supplementary Fig. 3). All RuxR-mutant ATF7IP-JAK2 Ba/F3 cells were resistant to multiple type-I JAK inhibitors including rux, BMS-911543, and AZD-1480 (Fig. 2b, c) as JAK2 p.Y931C, p.L983F, and p.G993A ATF7IP-JAK2 Ba/F3 cells had significantly higher LD50 values (p < 0.0001) when compared with non-mutant ATF7IP-JAK2 Ba/F3 cells. JAK2 p.Y931C and JAK2 p.G993A ATF7IP-JAK2 Ba/F3 cells were also resistant to 1 µM fedratinib as LD50 values were significantly higher (p < 0.0001) compared with non-mutant cells (Fig. 2b, c). In contrast, JAK2 p.L983F ATF7IP-JAK2 Ba/F3 cells were sensitive to fedratinib with an LD50 of 715 ± 9 nM (Fig. 2b, c).
The JAK2 p.G993A mutation confers resistance to multiple type-I JAK inhibitors in in vitro models of JAK2r ALL
JAK2r ALL was modeled in vitro by expression of wild-type JAK2, or JAK2 fusion genes PAX5-JAK2, ETV6-JAK2, or ATF7IP-JAK2 in Ba/F3 cells (Supplementary Fig. 4). To confirm that the previously unreported JAK2 p.G993A mutation was the single event that arose to confer rux resistance to JAK inhibitors, the JAK2 p.G993A mutation was introduced into JAK2 fusion genes (PAX5-JAK2, ETV6-JAK2, or ATF7IP-JAK2) by site-directed mutagenesis, expressed in Ba/F3 cells (Fig. 3a) and mutations confirmed (Supplementary Fig. 5a–c). The cell growth rates between Ba/F3 cells expressing non-mutant or JAK2 p.G993A-mutant JAK2 fusion genes in the absence of IL3 were not significantly different (Supplementary Fig. 5d–f).
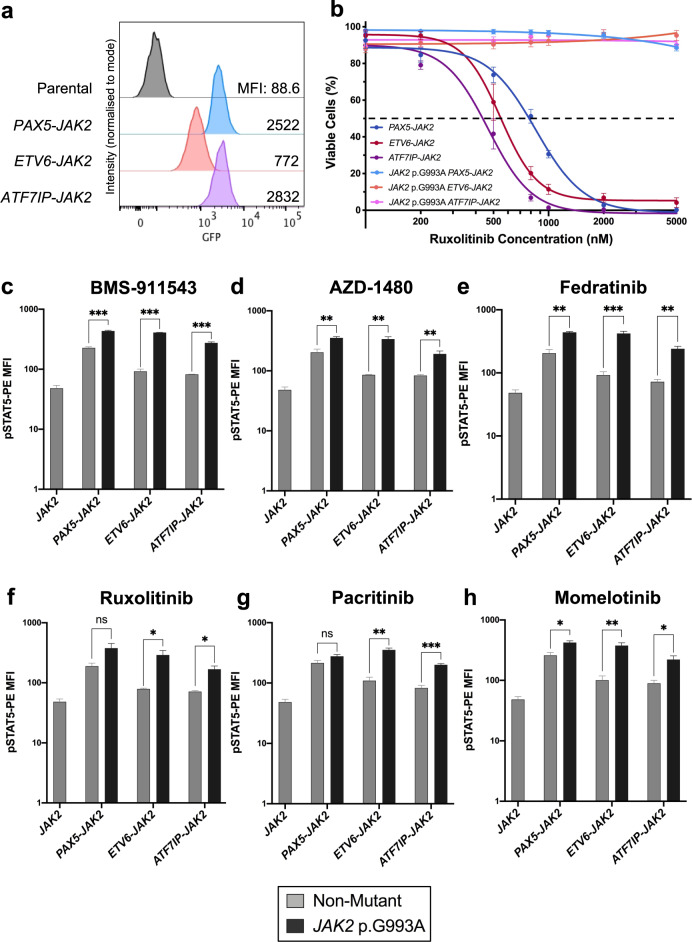
Fig. 3. The previously unreported JAK2 p.G993A mutation confers resistance to multiple type-I JAK inhibitors.
a Expression of GFP in Ba/F3 cells expressing JAK2 p.G993A-mutant PAX5-JAK2, ETV6-JAK2, or ATF7IP-JAK2 were assessed by flow cytometry. Non-transduced parental Ba/F3 were used as negative controls. Histograms are representative of three biological replicates and the GFP mean fluorescence intensities (MFIs) are shown. b Ba/F3 cells expressing either non-mutant or JAK2 p.G993A-mutant PAX5-JAK2 (blue), ETV6-JAK2 (red), or ATF7IP-JAK2 (purple) were incubated for 72 h with either a DMSO vehicle control or a dose response of the type-I JAK inhibitor, rux. The percentage of cell death was measured following a 20 min incubation with apoptotic markers and analysis by flow cytometry. Linear regression or non-linear regression models were fit to appropriate normalized data. Error bars indicate SEM over the mean of three biological replicates. c–h Ba/F3 cells expressing either non-mutant (gray) or JAK2 p.G993A-mutant (black) JAK2 fusion genes were incubated in 1 µM of type-I JAK inhibitors BMS-911543 (c), AZD-1480 (d), fedratinib (e), rux (f), pacritinib (g), or momelotinib (h) for 1 h. STAT5 p.Y694 phosphorylation was assessed by intracellular flow cytometry. JAK2 Ba/F3 cells starved of IL3 for 5 h were included as a measure of baseline STAT5 phosphorylation. pSTAT5-PE MFIs were plotted. Error bars indicate SEM over the mean of three biological replicates and significance was determined by unpaired t-tests in comparison to respective non-mutant cells (*p < 0.05, **p < 0.01, ***p < 0.001).
To investigate whether the JAK2 p.G993A mutation alone could confer resistance to rux, Ba/F3 cells expressing non-mutant or JAK2 p.G993A-mutant JAK2 fusion genes were treated with a vehicle control (DMSO) or varying concentrations of rux. The percentage of cell death was determined by staining with Annexin-V and aqua dead cell stain (Invitrogen) and then analysis by flow cytometry. Ba/F3 cells expressing non-mutant PAX5-JAK2, ETV6-JAK2, or ATF7IP-JAK2 were sensitive to rux with LD50 values of 853 ± 36, 546 ± 26, and 469 ± 39 nM, respectively (Fig. 3b). In contrast, all Ba/F3 cells expressing JAK2 p.G993A-mutant JAK2 fusion genes were resistant to >5 µM rux and had significantly higher LD50 values (p < 0.0001) when compared with their respective non-mutant cells (Fig. 3b).
To verify that the JAK2 p.G993A mutation alone could also confer cross-resistance to multiple type-I JAK inhibitors, Ba/F3 cells expressing non-mutant or JAK2 p.G993A-mutant JAK2 fusion genes were treated with 1 µM of different type-I JAK inhibitors for 1 h, and then activation of JAK/STAT signaling was assessed by intracellular staining of pSTAT5. Ba/F3 cells expressing JAK2 p.G993A-mutant JAK2 fusions demonstrated significantly higher activation of STAT5 following treatment with BMS-911543 (p < 0.001), AZD-1480 (p < 0.01), fedratinib (p < 0.01), or momelotinib (p < 0.05) when compared with Ba/F3 cells expressing non-mutant JAK2 fusions (Fig. 3). JAK2 p.G993A-mutant ETV6-JAK2 and JAK2 p.G993A-mutant ATF7IP-JAK2 Ba/F3 also displayed significantly higher activation of STAT5 following treatment with rux or pacritinib when compared with their respective non-mutant Ba/F3 cells (p < 0.05, Fig. 3f–g). There was no significant difference in STAT5 activation between non-mutant and JAK2 p.G993A-mutant PAX5-JAK2 Ba/F3 cells following treatment with rux or pacritinib (Fig. 3f–g).
The JAK2 p.Y931C mutation is known to be an activating mutation when introduced into WT JAK2 and expressed in Ba/F3 cells34. To determine whether the JAK2 p.G993A mutation is also an activating mutation, JAK2 p.G993A was introduced into WT JAK2 and expressed in Ba/F3 cells. The JAK2 p.G993A did not transform Ba/F3 cells to grow independently of IL3 (Supplementary Fig. 6b). Analysis of STAT1, STAT3, and STAT5 phosphorylation by intracellular phosphoflow cytometry demonstrated that constitutive activation of JAK/STAT signaling was also similar between Ba/F3 cells expressing non-mutant or JAK2 p.G993A-mutant JAK2 fusion genes (Supplementary Fig. 6a). However, JAK2 p.G993A ETV6-JAK2 and JAK2 p.G993A ATF7IP-JAK2 Ba/F3 cells demonstrated significantly higher activation of pSTAT1 (p < 0.01) and pSTAT5 (p < 0.001), respectively, when compared with Ba/F3 cells expressing their respective non-mutant JAK2 fusion genes (Supplementary Fig. 6a). Furthermore, Ba/F3 cells expressing either non-mutant or JAK2 p.G993A-mutant JAK2 fusion genes did not demonstrate ERK or AKT phosphorylation in the presence or absence of 1 µM ruxolitinib (Supplementary Fig. S7).
The previously unreported JAK2 p.G993A confers resistance to a type-II JAK inhibitor
To date, only the JAK2 p.L884P mutation has been identified to confer resistance to type-II JAK inhibitors in vitro models of B cell ALL35. To assess whether acquired mutations JAK2 p.Y931C, p.L983F, or p.G993A conferred resistance to the type-II JAK inhibitor, CHZ-868, non-mutant or RuxR-mutant ATF7IP-JAK2 Ba/F3 cells were treated with a vehicle control (DMSO) or varying concentrations of CHZ-868. The percentage of cell death was determined by staining with Annexin-V and aqua dead cell stain (Invitrogen) and then analysis by flow cytometry. ATF7IP-JAK2 Ba/F3 cells harboring the JAK2 p.Y931C or JAK2 p.L983F mutations were sensitive to CHZ-868 with LD50 values of 1024 ± 41 and 676 ± 58 nM, respectively (Fig. 4a). In comparison, JAK2 p.G993A ATF7IP-JAK2 Ba/F3 cells were resistant to >5 µM CHZ-868 and had a significantly higher LD50 value (p < 0.0001) when compared with non-mutant cells (Fig. 4a). To determine whether the JAK2 p.G993A mutation alone could confer resistance to CHZ-868 in the setting of other JAK2 fusion genes, Ba/F3 cells expressing non-mutant or JAK2 p.G993A-mutant JAK2 fusion genes were treated with 1 µM of CHZ-868 for 1 h, and then activation of JAK/STAT signaling was assessed by intracellular staining of pSTAT5. Treatment of Ba/F3 cells expressing non-mutant JAK2 fusion genes with CHZ-868 abrogated signaling through pSTAT5 (Fig. 4b). In contrast, STAT5 was constitutively active in Ba/F3 cells expressing JAK2 p.G993A-mutant JAK2 fusion genes following CHZ-868 treatment (Fig. 4b).
Fig. 4. The JAK2 p.G993A mutation confers resistance to the type-II JAK inhibitor, CHZ-858.
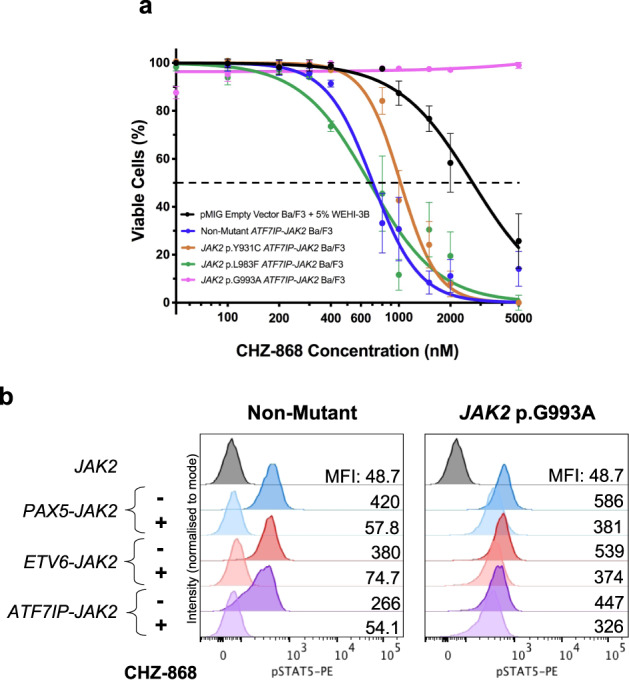
a pMIG Empty Vector Ba/F3 cells and Ba/F3 cells expressing either non-mutant or RuxR-mutant (JAK2 p.Y931C, p.L983F, p.G993A) ATF7IP-JAK2 were incubated for 72 h with either a DMSO vehicle control or a dose response of the type-II JAK inhibitor, CHZ-868. The percentage of cell death was measured following a 20 min incubation with apoptotic markers and analysis by flow cytometry. Linear regression or non-linear regression models were fit to appropriate normalized data. Error bars indicate SEM over the mean of three biological replicates. b Ba/F3 cells expressing either non-mutant or JAK2 p.G993A-mutant JAK2 fusion genes were incubated in 1 µM of CHZ-858 for 1 h. STAT5 phosphorylation was assessed by intracellular flow cytometry in comparison to JAK2 Ba/F3 cells starved of IL3 for 5 h. Histograms are representative of three biological replicates and the GFP mean fluorescence intensities (MFIs) are shown.
Structural analysis predicts how JAK2 kinase domain mutations may confer resistance to JAK inhibitors
Binding interactions of rux and CHZ-868 to the JAK2 kinase domain were investigated by computational modeling of drug docking. In silico computations of drug binding to JAK2 were based on a co-crystal structure of rux bound in the ATP-binding site of c-SRC kinase, the only available co-crystal structure of rux bound to a kinase (Supplementary Fig. 8a). Consistent with previous literature, the type-I JAK inhibitor, rux, binds the ATP-binding site of active JAK2 (ref. 32), while the type-II JAK inhibitor, CHZ-868, binds an allosteric site of JAK2 in addition to the ATP-binding site of inactive JAK2 (ref. 35). However, both rux and CHZ-868 interact with JAK2 predominantly through widespread hydrophobic interactions, hydrogen bonding with backbone atoms within the hinge region (between Y931 and L932), and hydrogen bonding with the N981 sidechain (Supplementary Fig. 8b)32. CHZ-868 also makes additional hydrogen bonds with D994 and E898, and van der Waal interactions with the sidechain of L983 and G993A (Supplementary Fig. 8b)35.
The mechanisms by which JAK2 kinase mutations confer resistance to JAK inhibitors were assessed using computational models of the either rux-bound or CHZ-868-bound JAK2 kinase domain. All JAK2 p.Y931C, p.L983F, and p.G993A mutations localized to the ATP-binding site of the JAK2 kinase domain were predicted to alter the volume of the JAK2 ATP-binding site cavity (Fig. S5a and Supplementary Fig. 9a). The JAK2 p.Y931C mutation was predicted to prevent rux binding by loss of the aromatic ring sidechain of Y931C, which makes stacking interactions with the double-ring system in rux and averts water molecules from interrupting hydrogen bonding with the hinge region (Supplementary Fig. 9b). The JAK2 p.L983F mutation was also predicted to abolish rux binding, as the bulkier phenylalanine (compared to Leucine) residue would sterically hinder any interactions with rux (Supplementary Fig. 9b). In contrast, CHZ-868 was predicted to bind both the JAK2 p.Y931C and JAK2 p.L983F mutations with similar binding affinities to CHZ-868 bound to WT JAK2 (Supplementary Fig. 9c and Supplementary Table 3).
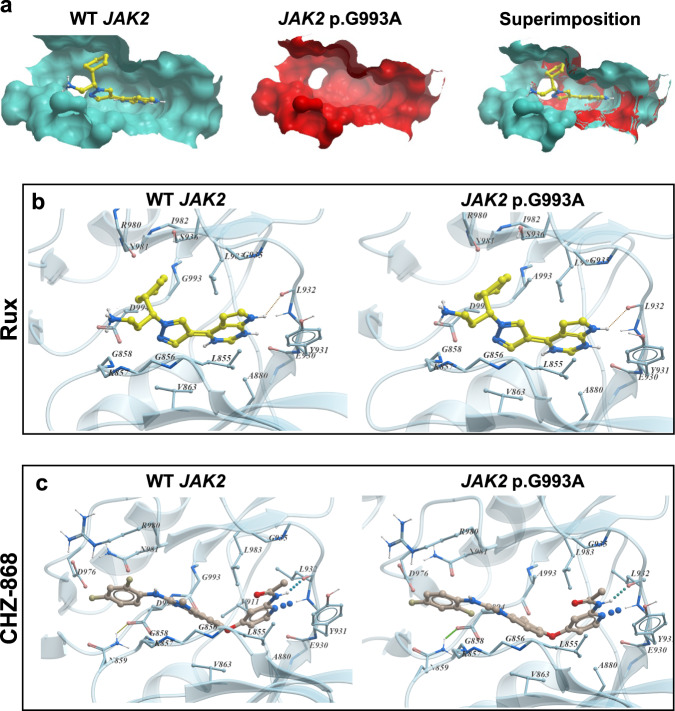
In contrast to the in vitro results presented in this manuscript, in silico data did not predict for reduced binding affinity between the JAK2 p.G993A-mutant JAK2 and rux or CHZ-868. No major structural changes were observed when comparing rux docking within the JAK2 ATP-binding site of WT or JAK2 p.G993A-mutant JAK2 (Fig. 5b). There was even a suggestion, based on free energy calculations, that the JAK2 p.G993A mutation may even increase rux binding affinity (Supplementary Table 3). Interestingly, modeling of CHZ-868 bound to JAK2 p.G993A-mutant JAK2 predicted that the JAK2 p.G993A mutation facilitates CHZ-868 binding in a “flipped” orientation (Fig. 5c). Free energy calculations predicted that CHZ-868 binds WT and JAK2 p.G993A-mutant JAK2 with similar binding affinities (Supplementary Table 3). Despite these predictions, in vitro data demonstrated that the JAK2 p.G993A conferred resistance to both rux and CHZ-868. This mutation introduces a generally less mobile amino acid in place of glycine, which may alter the dynamics of the neighboring DFG-loop and affect activation loop mobility when an inhibitor is bound.
Fig. 5. The ability of the JAK2 p.G993A mutation to confer resistance to rux and CHZ-868 was not supported by computational modeling.
a JAK2 ATP-binding cavity volume changes due to the JAK2 p.G993A mutation. Receptors are depicted as surface representations with wild-type JAK2 receptor shown in cyan (left) and JAK2 p.G993A-mutant JAK2 receptor shown in red (middle). Superimposition of the wild-type JAK2 receptor with ligand (rux) cavity upon the JAK2 p.G993A-mutant JAK2 pocket (right). Docking of ligands rux (b) and CHZ-868 (c) to either WT JAK2 or JAK2 p.G993A kinase domains. Rux is colored with yellow carbon atoms and CHZ-868 is colored with light brown carbon atoms. Ligand docked to WT JAK2 (left) and ligand docked to JAK2 p.G993A-mutant JAK2 (right).
Discussion
A significant proportion of malignancies can now be treated using targeted, small-molecule inhibitors, particularly those driven by activating mutations or fusions involving tyrosine kinases. However, the acquisition of mutations remains the most important cause for resistance against these drugs and anticipating such resistance patterns may expedite rational drug design. Mutations within the drug-binding site of the targeted kinases is a common cause of resistance, for example, the acquired pan-resistant kinase domain mutation ABL1 p.T315I in BCR-ABL1-positive CML40. ABL1 p.T315I mediates resistance against first- and second-generation TKIs, but is sensitive to the third-generation inhibitor ponatinib, which was rationally designed to inhibit ABL1 p.T315I-mutant BCR-ABL41. Multiple JAK2 kinase domain mutations conferring resistance to rux have been identified by in vitro random mutagenesis screening of JAK2 (refs. 32–36) and similar unbiased approaches were successful at identifying clinically relevant TKI-resistant mutations within BCR-ABL1 (ref. 42). Identification of inhibitor-resistant mutations has now been established as a fundamental first step in the development of strategies to overcome or avert resistance43,44.
In this study, we modeled acquired resistance to rux using in vitro models of high-risk JAK2 fusion genes in ALL to investigate potential resistance mechanisms and cross-resistance, in anticipation of such observations in the clinic. Non-mutant ATF7IP-JAK2 Ba/F3 cells had similar rux sensitivity to that of ex vivo human leukemic cells expressing ATF7IP-JAK2 (LD50 438 nM)14, validating our system to model JAK2r ALL. An in vitro model of acquired rux-resistance generated three independent rux-resistant murine pro-B cell lines expressing the transforming ATF7IP-JAK2 fusion gene. All three lines acquired resistance to rux within 3 months, suggesting that clinical rux resistance in ALL, if given as monotherapy, may develop quickly. Each independent replicate was found to have acquired a different mutation within the JAK2 kinase domain including two known mutations, JAK2 p.Y931C and JAK2 p.L983F, and a previously unreported JAK2 p.G993A mutation (Fig. 1). JAK2 p.Y931C is homologous to the activating JAK1 p.F958C mutation, and based on amino acid sequences at kinase hinge regions, JAK2 p.Y931 is also analogous to the F317 residue in ABL1 that when mutated confers imatinib resistance34,45. The JAK2 p.L983F mutation has only been identified in one other study in by in vitro mutagenesis of JAK236.
Acquired resistance was associated with constitutive activation of STAT5 in the presence of rux, indicating that constitutive activation of JAK/STAT signaling is critical to leukemic cell proliferation in this model (Fig. 2a). STAT5 was also more active in JAK2 p.Y931C ATF7IP-JAK2 Ba/F3 cells (Fig. 2) in comparison to JAK2 p.L983F and JAK2 p.G993A ATF7IP-JAK2 Ba/F3 cells. This may be related to the absence of non-mutated ATF7IP-JAK2 transcripts in JAK2 p.Y931C cells, potentially enabling stronger activation of JAK/STAT signaling in comparison to cells with acquired JAK2 p.L983F or JAK2 p.G993A mutations. Despite in vitro predictions, acquired resistance attributable to JAK2 point mutations is not widely observed in rux-treated MPN or ALL, potentially due to an insufficient selective pressure related to the low potency and selectivity of rux27. Future studies assessing JAK inhibitor sensitives and resistance in patient-derived xenograft models of JAK2r ALL may provide a more clinically relevant model of acquired JAK inhibitor resistance and address the limitations of cell line models of resistance. The clinical relevance of JAK2 resistance mutations reported in this study and previous literature may also become more important as JAK inhibitors with increased potency are developed.
All three acquired mutations conferred resistance to clinically relevant concentrations of multiple type-I JAK inhibitors, including rux, BMS-911543, and AZD-1480, suggesting that patients who acquire JAK2 kinase domain mutations may display cross-resistance to multiple JAK-inhibitor therapies. This is consistent with previous literature showing that the JAK2 p.Y931C and JAK2 p.L983F mutations conferred resistance to both rux and AZD-1480 (refs. 32,36). The JAK2 p.Y931C and JAK2 p.L983F mutations have been shown to have greater than 10-fold functional resistance to all type-I JAK inhibitors in JAK2 p.V617F expressing Ba/F3-EpoR cells33,36. The ability of the JAK2 p.G993A mutation to confer resistance to multiple type-I JAK inhibitors was validated using murine pro-B cells transduced to express JAK2 p.G993A-mutant JAK2, PAX5-JAK2, ETV6-JAK2, or ATF7IP-JAK2, indicating this mutation alone was sufficient to confer JAK inhibitor resistance.
The JAK2 p.L983F mutation was also found to be sensitive to fedratinib, which has been demonstrated previously by Kesarwani et al. (2015)36. Kinase assays performed by Kesarwani et al. (2015)36 reported that fedratinib binds JAK2 with a high affinity in the substrate-binding site and with a low affinity in the ATP-binding site. Therefore, sensitivity of the JAK2 p.L983F mutation to fedratinib may be due to inhibition from fedratinib binding within the substrate-binding site36,46. Kesarwani et al. (2015)36 also demonstrated that purified JAK2 kinase domains harboring the JAK2 p.Y931C mutation were sensitive to fedratinib in kinase assays. In contrast, our in vitro cell death assays showed that murine pro-B cells expressing JAK2 fusion genes with acquired JAK2 p.Y931C and JAK2 p.G993A mutations were resistant to the clinically relevant concentration of 1 µM fedratinib, but this resistance was overcome when the drug concentration in the cell culture media was increased to 2 µM. Higher concentrations of fedratinib may potentially be required to inhibit JAK2 by shifting the binding equilibrium at the JAK2 substrate-binding site towards fedratinib binding.
The JAK2 p.G993A mutation also did not confer IL3 independence in Ba/F3 cell models and did not upregulate PI3K/AKT or MAPK/ERK signaling pathways. In contrast to the known JAK2 p.Y931C mutation, this suggests that the JAK2 p.G993A mutation is not an activating mutation and that upregulation of PI3K/AKT and MAPK/ERK signaling pathways does not contribute to the observed JAK inhibitor resistance. This study identified that cell models of JAK2r ALL can develop JAK inhibitor resistance via acquisition of mutations within JAK2 ATP-binding site following long-term exposure to JAK inhibitors. It is currently unknown whether CRLF2r/JAK2r-mutant ALL preclinical models are susceptible to JAK inhibitor resistance through this same mechanism. However, activation of alternative signaling pathways such as C-MYC and MAPK/ERK have been identified in CRLF2r/JAK2r-mutant ALL47,48, suggesting that in this setting, JAK inhibitor resistance may develop via upregulation of alternative signaling pathways potentially in preference to acquired mutations within the JAK2 ATP-binding site.
Importantly, the JAK2 p.G993A mutation was found to confer resistance to all tested type-I JAK inhibitors as well as to the type-II JAK inhibitor, CHZ-868. The JAK2 p.L884P mutation was identified in vitro by random mutagenesis and is the only other JAK2 mutation that has been shown to confer resistance to type-II JAK inhibitors35. The development of second- and third-generation ABL TKIs has provided effective treatment options for Ph-positive ALL and CML patients who relapse after acquired TKI resistance41. The inability of CHZ-868 to overcome the resistance conferred by JAK2 p.G993A suggests that sequential use of next-generation JAK inhibitors may not be efficacious in overcoming JAK2 p.G993A-mediated JAK inhibitor resistance. Cells harboring the JAK2 p.G993A mutation did not demonstrate activation of signaling pathways in addition to JAK/STAT suggesting future JAK inhibitors may need to be rationally designed to inhibit JAK2 p.G993A.
Computational modeling highlighted that all three RuxR mutations (JAK2 p.Y931C, p.L983F, and p.G993A) were localized to the ATP-binding site of the JAK2 kinase domain, signifying that this region is susceptible to inhibitor-resistant mutations following rux exposure. The JAK2 p.Y931C and JAK2 p.L983F mutations were predicted to completely inhibit rux binding. The JAK2 p.Y931C was expected to disrupt stacking interactions between rux and the hinge region of JAK2, while the bulkier sidechain of the JAK2 p.L983F mutation was predicted to sterically hinder rux binding at the JAK2 catalytic loop. The JAK2 p.Y931C and JAK2 p.L983F mutations were not predicted to affect CHZ-868 binding, consistent with in vitro results demonstrating that these mutations did not confer resistance to CHZ-868.
In contrast to JAK2 p.Y931C and JAK2 p.L983F, the JAK2 p.G993A mutation was not predicted to prevent or reduce rux or CHZ-868-binding affinities, despite in vitro results showing that the JAK2 p.G993A mutation conferred resistance to both rux and CHZ-868. The JAK2 p.G993A mutation did not appear to induce any major structural changes to the JAK2 ATP-binding site and free energy calculations suggested that the JAK2 p.G993A mutation may even increase rux binding affinity. Alanine is a less mobile amino acid compared to glycine and the in silico docking studies performed would not detect changes to protein dynamics. Therefore, we postulate that the JAK2 p.G993A mutation confers resistance to rux and CHZ-868 through a previously unreported resistance mechanism that modulates the mobility of the conserved JAK2 activation loop and DFG motif. This mechanism may facilitate JAK2 activation in the presence of drug binding.
As rux and other JAK inhibitors progress through ongoing clinical trials for the treatment of ALL, we expect that a subset of patients will acquire JAK inhibitor-resistant mutations in the context of disease relapse. Our work identified that currently available JAK inhibitors are susceptible to resistance mediated by mutations in the JAK2 ATP-binding site. This demonstrates the potential of monitoring for acquired mutations within the JAK2 kinase domain in the context of suspected treatment resistance. It also guides future rational drug design attempts to overcome this resistance mechanism. The JAK2 p.G993A mutation, which conferred resistance to all tested type-I and type-II JAK inhibitors, may be particularly important. This mutation was postulated to confer resistance to JAK inhibitors through a unique resistance mechanism that modulates the mobility of the conserved JAK2 activation loop, enabling JAK2 activation in the presence of drug binding.
Methods
Ethics statement
This project analyzed cryopreserved samples from patients diagnosed with ALL within established biobanks. Samples were obtained with informed consent for prospective unspecified medical research, and to perform laboratory-based assays on existing specimens. All the participants were required to provide informed consent for their samples to be used in accordance with the Declaration of Helsinki. The nature of the study was extensively explained and patient information signed consent (PISC) forms were issued to all the participants. The PISC forms outlined all questions that would be pertinent to their involvement on the study and when this was signed, gave consent for the participant to be included. The use of samples was approved by Central Adelaide Local Health Network and the Royal Adelaide Hospital Human Research Ethics Committee.
mRNA sequencing of B-ALL patient bone marrow mononuclear cells
Bone marrow mononuclear cell samples from Ph-like B-ALL patients were obtained from the Children’s Cancer Institute Australia (CCIA), SA Pathology/Royal Adelaide Hospital, Australasian Leukaemia & Lymphoma Group (ALLG) tissue bank, or Queensland Children’s Tumour Bank. The samples were collected at diagnosis or relapse and then screened by the Acute Lymphoblastic Leukaemia Research Group (Cancer Program, Precision Medicine Theme, SAHMRI) for single-nucleotide variants and fusion genes using mRNA sequencing. mRNA seq was performed using the Truseq Stranded mRNA LT kit (Illumina, CAT#20020595), as per the manufacturer’s instructions, from 1 μg of high-quality total RNA and sequenced by either the Illumina HiSeq 2000 or NextSeq 500 platforms. A read depth of 70 million reads was achieved for most samples. Fusion calling from mRNA seq data was performed using three fusion callers, FusionCatcher, JAFFA, and SOAPfuse; then, outputs were combined as described previously using the FusionMetaCaller R package49–52. Only fusions identified by a minimum of two callers were considered and events ranked by the total number of supporting reads using rank sums. JAK2 fusion partners identified included paired box 5 (PAX5), ETS variant transcription factor 6 (ETV6), and activating transcription factor 7 interacting protein (ATF7IP). PAX5-JAK2 was detected in two patients at diagnosis, one child and one adolescent. PAX5-JAK2 was also detected in another pediatric patient at relapse. ETV6-JAK2 was detected in a pediatric patient at diagnosis, and ATF7IP-JAK2 was detected in a 28-year-old male at diagnosis.
Modeling of acquired JAK-inhibitor resistance in vitro
Acquired rux resistance in JAK2r ALL was modeled using Ba/F3 cells expressing the JAK2 fusion gene, ATF7IP-JAK2 (fusion of activating transcription factor 7 interacting protein and JAK2), which was originally identified in a pediatric patient with high-risk B-ALL14. ATF7IP-JAK2 Ba/F3 cells were generated by transduction of a pMIG-ATF7IP-JAK2 expression plasmid, and kindly donated by the Mullighan Laboratory (St. Jude Children’s Research Hospital, TN, USA)13,14. Expression of ATF7IP-JAK2 was verified by a complete shift of GFP expression using flow cytometry and detection of an approximately 200 kDa band with phospho-Y1007/1008-JAK2 and total JAK2 reactivity (Supplementary Fig. 1a and S1b). Trypan blue exclusion assays demonstrated that ATF7IP-JAK2 Ba/F3 cells were IL3 independent (Supplementary Fig. 1c). Three independently derived ruxolitinib-resistant (RuxR) ATF7IP-JAK2 Ba/F3 sub-lines were established by exposing three independent biological replicates of untreated ATF7IP-JAK2 Ba/F3 cells to progressively increasing concentrations of rux (Selleckchem, CAT#S1378) over 3 months from 50 nM to 1 µM53. Simultaneously, ATF7IP-JAK2 Ba/F3 cell lines were cultured alongside and either left untreated (naïve), or treated with vehicle (DMSO), to generate control cells.
Cloning of JAK2 and JAK2 fusion genes
JAK2 was amplified by reverse transcription PCR (RT-PCR) from a pUNO1-hJAK2 vector (InvivoGen, CAT#puno1-hjak2) and subcloned into a Gateway pDONR-221 vector (Invitrogen, CAT#12536017) using the Gateway BP Clonase II enzyme mix (Invitrogen, CAT#11789100) as per the manufacturer’s instructions. DNA sequences of JAK2 fusions genes breakpoints (PAX5-JAK2, ETV6-JAK2, ATF7IP-JAK2) were derived from patient mRNA sequencing data. Full-length fusion genes sequences were generated using reference sequencing obtained from the National Center for Biotechnology Information (NCBI)54. NCBI reference sequences included human JAK2 variant 1 (NM_004972.4), PAX5 variant 1 (NM_016734.3), human ETV6 (NM_001987.5), and ATF7IP (NM_181352.2). JAK2 gene fusions were synthesized and subcloned into the Gateway pDONR-221 vector by Gene Universal (DEL, USA). JAK2 or JAK2 fusion genes were then transferred into a Gateway-compatible pMSCV-IRES-GFP (pMIG) vector (kind gift from Prof. Charles Mullighan, St. Jude Children’s Research Hospital, TN, USA) using the Gateway LR Clonase II enzyme mix as per the manufacturer’s instructions (Invitrogen, CAT#11791020).
Site-directed mutagenesis of JAK2 fusion genes
To confirm that the JAK2 p.G993A mutation alone could confer JAK inhibitor resistance, JAK2 p.G993A was introduced into vectors containing WT JAK2 or JAK2 fusion genes using the Q5 site-directed mutagenesis kit (New England BioLabs, CAT#E0554S) according to the manufacturer’s instructions.
Expression of JAK2 and JAK2 fusion genes in Ba/F3 cells
JAK2r ALL was modeled in vitro using the IL3-dependent murine pro-B cell line, Ba/F3. The parental Ba/F3 cell line was kindly donated by Prof. Andrew Zannettino (Myeloma Research Laboratory, University of Adelaide, SA, Australia). Lentiviral particles were produced by transient co-transfection of HEK-293T cells (ATCC, VA, USA) with a 1:1:1 molar ratio of a triple plasmid packaging system (pMD2.G, pMDLg/pRRE, and pRSV-REV) (AddGene, CAT#12259, CAT#12251, CAT#12253, respectively), and either pMIG empty vector or pMIG expression vectors containing non-mutant or JAK2 p.G993A-mutant JAK2, PAX5-JAK2, ETV6-JAK2, or ATF7IP-JAK2. Lentiviral transfections were performed in Opti-MEM reduced serum medium (Gibco, CAT#51985034) containing 4% Lipofectamine 3000 (Invitrogen, CAT#L3000001). Lentiviral supernatant was harvested after 48 h, filtered through a 0.45 μM filter, and used to transduce Ba/F3 cells by spinfection at 1800 r.p.m. for 1 h in the presence of 4 μg/mL polybrene (Sigma-Aldrich, CAT#TR-1003). GFP-positive cells were selected using fluorescence-activated cell sorting (FACS) on a BD FACSMelody (BD Biosciences). Gene expression was validated by a complete shift of GFP and HA-tag expression using flow cytometry (Supplementary Fig. 4a, b respectively). Fusion gene breakpoints were validated by Sanger sequencing of full-length JAK2 fusion gene RT-PCR products (Supplementary Fig. 4c, d).
Analysis of GFP Expression by flow cytometric analysis
To assess expression of GFP, 1 × 106 cells were centrifuged and then resuspended in 200 µL of FACSFix (1× PBS, 1% formaldehyde, 110 mM d-glucose, 0.02% sodium azide) containing 50 ng of DAPI (Sigma-Aldrich, CAT#D9542). Cells were analyzed on the BD FACSCanto II (BD Bioscience); then, flow cytometry data were analyzed and plotted using FlowJo analysis software v10 (FlowJo). Gating strategy is shown in Supplementary Fig. 10a.
RNA extraction
Total RNA was extracted from 5 × 106 cultured cells by lysing cells in 1 mL of TRIzol Reagent (Life Technologies, CAT#15596-018) and then addition of 0.2 mL chloroform (Sigma-Aldrich, CAT#C2432). Phase separation was achieved by incubation at room temperature for 2–3 min followed by centrifugation at 12,000g for 15 min at 4 °C. RNA was precipitated by the addition of 0.5 mL isopropanol (ChemSupply Australia, CAT#PA013) and 20 µg of glycogen (Roche, CAT#10901393001). RNA was washed with 75% ethanol (ChemSupply Australia, CAT#EA043) prior to rehydration in nuclease-free (NF) water (MP Biomedicals, CAT#04821739). RNA quantity was measured on a Nanodrop 8000 spectrophotometer (Thermo Fisher Scientific).
RT-PCR
Complementary DNA (cDNA) was synthesized from 1 µg of RNA using the QuantiTect Reverse Transcription Kit (Qiagen, CAT#205313), according to the manufacturer’s instructions, in a PTC-200 Thermal Cycler (MJ Research) incorporating a 30 min 42 °C DNA synthesis time. RT-PCR was performed using 1 µL of cDNA, 200 µM dNTPs (Invitrogen, CAT#18427088), 0.4 µM forward primer, 0.4 µM reverse primer, 1 unit Q5 High-Fidelity DNA Polymerase (New England BioLabs, CAT#M0491S), and 1× Q5 Reaction Buffer (New England BioLabs, CAT#M0491S) in 25 µL. Primers designed to amplify full-length WT JAK2 or JAK2 fusion genes are listed in Supplementary Table 1. Amplification reactions were performed in a T100 Thermal Cycler (Bio-Rad). RT-PCR products in 6× purple gel loading dye (New England BioLabs, CAT#B7025S) were visualized by gel electrophoresis with 1 kb ladders (New England BioLabs, CAT#N3232). Gels consisted of 1% agarose (Sigma, CAT#A6013), 1× GelRed (Biotium, CAT#41003), and 1× TAE (40 mM Tris-HCl, 20 mM acetic acid (ChemSupply, CAT#AA009), 1 mM EDTA (Ajax, CAT#AJA180)). Gels were resolved in 1× TAE at 110 V and imaged on a Gel Doc XR+ Gel Documentation System (Bio-Rad) with Image Lab software (Bio-Rad).
PCR purification and Sanger sequencing
Fusion specific RT-PCR products were purified using the QIAquick Gel Extraction Kit (Qiagen, CAT#28706) according to the manufacturer’s instructions with a final elution into 30 µL of Buffer EB. Complete Sanger sequencing of 60 ng of purified, full-length WT JAK2 or JAK2 fusion RT-PCR products were performed by the Australian Genome Research Facility (AGRF, SA, AUS) using 800 µM of primer. Primers designed for Sanger sequencing are listed in Supplementary Table 2. Sequences were aligned to their respective reference sequences (as described in ‘Cloning of JAK2 and JAK2 fusion genes’) using Benchling (Biology Software, 2020).
Cell culture
IL3-independent Ba/F3 cells expressing JAK2 fusion genes were maintained in RPMI1640 (Sigma-Aldrich, CAT#R0883) with 10% FCS (FCS CellSera, BATCH#F21701), 50 units/mL penicillin/streptomycin (Sigma-Aldrich, CAT#P4333), and 2 mM l-glutamine (Sigma-Aldrich, CAT#G7513) (standard media) in a 37 °C incubator with 5% CO2 (ref. 3). Ba/F3 cells expressing RuxR-mutant JAK2 fusion genes were maintained in standard media containing 1 µM rux. IL3-dependent parental Ba/F3 cells and Ba/F3 cells expressing pMIG empty vector, WT JAK2, or JAK2 p.G993A-mutant JAK2 were maintained in standard media supplemented with 5% WEHI-3B culture media as a source of IL33. WEHI-3B culture media was made in house, as a source of murine interleukin 3 (IL3), using the murine myelomonocytic leukemia cell line WEHI-3B55. HEK-293T cells were maintained in DMEM (Sigma-Aldrich, #D6046) supplemented with 10% FCS (FCS CellSera, BATCH#F21701) and 50 units/mL penicillin (Sigma-Aldrich, CAT#P4333).
Compounds
Type-I JAK inhibitors rux (CAT#S1378), BMS-911543 (CAT#S7144), AZD-1480 (CAT#S2162), fedratinib (CAT#S2736), momelotinib (CAT#S2219), and pacritinib (CAT#S8057) were all purchased from Selleckchem. Type-II JAK inhibitor, CHZ-868 (CAT#HY-18960) was purchased from MedChemExpress. Inhibitor stocks (10 mM) were diluted in DMSO so that the final concentration of DMSO in culture media and assays was 0.05–0.1%.
Viability assays
To assess cell death, cells were washed twice in standard media and then seeded in duplicate at 1.5 × 104 cells/mL in 96-well U-bottom plates. Cells were either untreated, treated with vehicle control (DMSO), or treated with varying concentrations of JAK inhibitors. After a 72 h incubation at 37 °C with 5% CO2, cells were washed once with HANK’s Balanced Salt solution (Sigma-Aldrich, CAT#H9394) supplemented with 5 mM calcium chloride (Sigma-Aldrich, CAT#C1016) and 1% HEPES (Sigma-Aldrich, CAT#H0887) (binding buffer). The percentage of cell death was determined by staining with 2% annexin-V-PE (BD Biosciences, CAT#556421) and 0.25% Live/Dead Fixable Aqua Dead Cell Stain (Invitrogen, CAT#L34957) (aqua dead cell stain) in 20 µL of binding buffer for 1 h in the dark. Cells were washed and analyzed on the BD FACSCanto II (BD Bioscience); then, flow cytometry data were analyzed and plotted using FlowJo analysis software v10 (FlowJo). The gating strategy used to determine the percentage of viable cells is shown in Supplementary Fig. 10b. The percentage of viable cells was normalized to the first and last means of each dataset and plotted using GraphPad Prism v8 (GraphPad Software). Non-linear regression curves were fit to appropriate data to determine median lethal dose (LD50).
Proliferation assays
To assess cell proliferation, cells were seeded in duplicate at 1 × 103 cells/mL in six-well plates in the presence or absence of 5% WEHI-3B. At 24 h timepoints, 25 µL of cells were transferred to black 96-well CulturPlates (PerkinElmer, CAT#6005660); then, cell proliferation was assessed using CellTiter Glo 2.0 (Promega, CAT#G9242) as per the manufacturer’s instructions. Luminescence readouts were measured on the Victor X Multilabel plate reader (PerkinElmer) and media only control wells were included to measure background luminescence. One phase decay or exponential growth curves were fitted to data using GraphPad Prism v8 (GraphPad Software). To assess cell growth, cells were seeded in duplicate at 1.5 × 104 cells/mL in 24-well plates in either standard media or standard media containing increasing concentrations of WEHI-3B conditioned media. After a 72 h incubation at 37 °C with 5% CO2, cells were stained 1:1 with 0.4% Trypan blue (Gibco, CAT#15250061); then, a hemocytometer was used to count the number of live cells.
Intracellular flow cytometric analysis
Activation of JAK/STAT signaling in the absence of IL3 was assessed by intracellular flow cytometric analysis of pSTAT1, pSTAT3, and pSTAT5 (phosphoflow). Cells were washed twice via centrifugation, resuspended at 1 × 106 cells/mL in standard media in a six-well plate, and then incubated for 5 h in a 37 °C incubator with 5% CO2. To assess the effect of JAK inhibitor treatment on STAT5, ERK, and AKT activation, cells were washed twice via centrifugation and then resuspended at 1 × 106 cells/mL in standard media in a 24-well plate. Cells were incubated for up to 1 h with vehicle (DMSO) or 1 µM JAK inhibitor in a 37 °C incubator with 5% CO2. Cells were fixed with 100 µL of 16% paraformaldehyde (Electron Microscopy Sciences, CAT#15710) per 1 mL of sample, incubated for 10 min at room temperature, and then washed once via centrifugation with 1× PBS (Gibco, CAT#14200075). Permeabilization was carried out by gentle resuspension of cell pellets in ice-cold 80% methanol (ChemSupply Australia, CAT#AR115), followed by storage overnight at −20 °C. Cells were washed once with 1× PBS, once with 1× PBS supplemented with 1% bovine serum albumin (BSA, Sigma-Aldrich, CAT#A9418) (phosphoflow buffer). Pellets were resuspended in 1× PBS/1% BSA; then, 3.5 × 105 cells were transferred to wells of a 96-well U-bottom plate. Cells were stained for 1 h at room temperature with 100 µL of antibodies diluted in phosphoflow buffer. Antibodies purchased from BD Biosciences were used at the recommended concentrations and stained for pSTAT1 (pY701, CAT#562069), pSTAT3 (pY705, CAT#562072), pSTAT5 (pY694, CAT#562077), pERK (pT202/pY204, CAT#612566), pAKT (pS473, CAT#560378), or an IgG1-PE isotype control (#CAT554680). Expression of HA-tagged JAK2 fusion proteins was verified by staining with 10 ng of an anti-HA antibody (Cell Signaling Technology, CAT#3444 S). Stained cells were washed once with 1× PBS, and then cell pellets were resuspended in 150 µL of phosphoflow buffer. Cells were analyzed on the BD FACSCanto II (BD Bioscience); then, flow cytometry data were analyzed using FlowJo analysis software v10 (FlowJo) and visualized using GraphPad Prism v8 (GraphPad Software). The gating strategy used for the analysis of intracellular flow cytometry data is shown in Supplementary Fig. 11.
Cell lysate preparation
To assess JAK/STAT pathway activation in the absence of IL3, cells were washed twice in 10 mL of standard via centrifugation. Cells were resuspended at 1 × 106 cells/mL in standard media in a T25 flask, and then incubated for 5 h in a 37 °C incubator with 5% CO2. To assess the effect of rux on JAK2/STAT5 signaling activation, cells were washed (2×) via centrifugation and then resuspended at 1 × 106 cells/mL in standard media in a T25 flask. Cells were incubated for 1 h with vehicle (DMSO) or a dose escalation of 0.05–1 µM rux in a 37 °C incubator with 5% CO2. For total lysates, 1 × 107 cells were washed once with ice-cold 1× PBS, then lysed for 20 min on ice in 90 µL of NP-40 buffer containing 10 mM Tris-HCl (pH 7.4, Sigma-Aldrich, CAT#252859), 137 mM NaCl (Thermo Fisher Scientific, CAT#AJA465), 10% glycerol (Sigma-Aldrich, CAT#G5516), and 1% NP-40 (Igepal, Thermo Fisher Scientific, CAT#85124) supplemented with phosphatase and protease inhibitors (Complete mini EDTA-free protease inhibitors Cocktail, Roche, CAT# 04693132001). Cellular debris was pelleted via centrifugation; then, cleared lysate supernatants were collected and quantified using a DC protein assay (Bio-Rad, CAT#500-0116) according to the manufacturer’s instructions. Absorbance readouts were measured at 595 nm on the Victor X Multilabel plate reader (PerkinElmer). Prior to boiling at 100 °C for 10 min, cleared lysates were mixed 3:1 with 4× Laemmli’s loading buffer consisting of 0.25 M Tris-HCl (pH 6.8, Sigma-Aldrich, CAT#252859), 8% (w/v) SDS (Sigma-Aldrich, CAT#75746), 40% (v/v) glycerol (Sigma-Aldrich, CAT#G5516), 20% (v/v) 2-merchaptoehtanol (Sigma-Aldrich, CAT#M6250), and 0.05% (w/v) bromophenol blue (Sigma-Aldrich, CAT#114405).
SDS-PAGE analysis and western blotting
Equivalent protein aliquots (80 µg) were loaded into 4–15% polyacrylamide Criterion TGX gels (Bio-Rad, CAT#5678084) with Precision Plus Protein Kaleidoscope pre-stained protein standards (Bio-Rad, CAT#161-0375). Proteins were resolved at 100 V for 20 min, followed by 40 min at 200 V, and then transferred to PVDF membrane (Bio-Rad, CAT#1704275) using the Trans-Blot Turbo Transfer System (Bio-Rad, CAT#1704150) according to the manufacturer’s instructions using the mixed molecular weight setting. Membranes were blocked for 1 h at room temperature with Intercept blocking buffer (LI-COR, CAT#927-70001), followed by incubation with primary antibody diluted 1:1000 in Intercept blocking buffer (LI-COR, CAT#927-70001) overnight at 4 °C. Primary antibodies against pJAK2 Y1007 (CAT#4406 S), JAK2 (CAT#3230 S), HA (CAT#3724S), pSTAT5 Y694 (CAT#9359S), STAT5 (CAT#94205), and GAPDH (CAT#2118 S) were purchased from Cell Signaling Technology. Membranes were washed (3×) for 5 min with 1× TBST and then stained with donkey-anti-rabbit IRDye 800CW secondary antibody (LI-COR, CAT#925-32213) diluted 1:10,000 in Intercept blocking buffer. Membranes were incubated for 1 h at room temperature, washed (3×) with 1× TBST (20 mM Tris-HCl (pH 6.8, Sigma-Aldrich, CAT#252859), 150 mM sodium chloride (Thermo Fisher Scientific, CAT#AJA465), 0.1% Tween (Sigma-Aldrich, P2287)), washed (3×) with 1× TBS (20 mM Tris-HCl (pH 6.8, Sigma-Aldrich, CAT#252859), 150 mM sodium chloride (Thermo Fisher Scientific, CAT#AJA465)), and then visualized on the Odyssey CLx Imaging System (LI-COR). Western blots were stripped prior to immunoblotting for total proteins using a western blot recycling kit (Alpha Diagnostic International, CAT#90102) according to the manufacturer’s instructions. Immunoblot image analysis and band quantifications were performed using ImageStudioLite v5.2.5 software (LI-COR).
DNA extraction
Genomic DNA was extracted from 5 × 106 cultured cells by phenol–chloroform extraction. Whole-cell pellets were lysed in 480 µL of DNA lysis buffer consisting of 10 mM Tris-HCl (Sigma-Aldrich, CAT#252859), 10 mM sodium chloride (Thermo Fisher Scientific, CAT#AJA465), and 10 mM EDTA (Ajax, CAT#AJA180). Cells were mixed with 12.5 µL of 20% sodium dodecyl sulfate (Sigma-Aldrich, CAT#75746) and 180 µg of proteinase K (Roche, CAT#03115887001) and then incubated overnight at 37 °C. Cells were incubated for 10 min at 37 °C with 500 µg of RNAse A (Qiagen, CAT#19101) prior to the addition of 10 µL of 5 M sodium chloride (Thermo Fisher Scientific, CAT#AJA465) and 20 µg of glycogen (Roche, CAT#10901393001). Phase separation was achieved by the addition of 500 µL UltraPure buffer saturated phenol (Invitrogen, CAT#15513-047) and centrifugation at 16,000g for 5 min. The aqueous phase was retained; then, 500 µL of ultrapure phenol:chloroform:isoamyl alcohol (Invitrogen, CAT#15593-049) was added and vigorously mixed before centrifugation at 16,000g for 3 min to separate the aqueous phase. The retained aqueous phase was washed (1×) with 1 mL of 4 °C 100% ethanol (ChemSupply Australia, CAT#AR115); then, DNA was precipitated by centrifugation 20,000g for 10 min. The DNA pellet was washed (1×) with 70% ethanol (ChemSupply Australia, CAT#AR115) and then rehydrated with 200 µL of DNA hydration solution (Qiagen, CAT#158914). DNA was incubated at 55 °C for 2 h followed by 37 °C overnight; then, DNA quantity was measured on a Nanodrop 8000 spectrophotometer (Thermo Fisher Scientific).
Exome sequencing of murine pro-B cells
Library preparation for whole exome sequencing was performed using the Sureselect Clinical Research Exome v2 (Agilent Technologies, CAT#5190) and SureSelect XT reagent kit (Agilent Technologies, CAT#G9642) as per the manufacturer’s instructions from 200 ng of genomic DNA. Samples were sequenced with the Illumina NextSeq 500 platform. Zygosity of mutations within the JAK2 ATP-binding domain were determined by visual inspection of BAM files, filtered by XenofilteR56, using the Integrative Genomics Viewer (Broad Institute).
In silico docking models
Docking of JAK inhibitors, rux and CHZ-868, to the JAK2 kinase domain and all in silico computations were modeled using PDB:2XA4 (ref. 57) as the receptor, given that it was bound to a similar class ligand (class I). The only currently available co-crystal structure with rux bound (to c-Src, PDB:4U5J)58 was used to compare our docking method of rux to JAK2 (Supplementary Fig. 6a). Coordinates for JAK inhibitors were created and minimized phenix.elbow59. All docking was carried out in ICM-Pro (Molsoft LCC). The receptor was stripped of ligands and water molecules followed by the addition of hydrogen atoms and charges. The docking protocol allowed for flexible side chains. The final docked models were subjected to 20 rounds of energy minimization and annealing. All mutations were also created in ICM-Pro (Molsoft LCC) and subjected to 20 rounds of energy minimization and annealing.
Statistical analysis
Statistical analysis was performed using the t-test function in GraphPad Prism v8 (GraphPad Software). Statistical significance was denoted by asterisks (*p < 0.05, **p < 0.01, ***p < 0.001, ns not significant).
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
The authors acknowledge Prof. Charles Mullighan (St. Jude Children’s Research Hospital, TN, USA) and Prof. Andrew Zannettino (Myeloma Research Laboratory, University of Adelaide, SA, Australia) for provision of the Gateway-compatible pMIG vector and Ba/F3 cell line, respectively. The authors would also like to acknowledge the South Australian Genomics Centre (SAGC, SA, Australia) and ACRF Cellular Imaging and Cytometry Core Facility (SAHMRI, SA, Australia). This work was supported by funding from the National Health and Medical Research Council, Australia (NHRMC) (APP1057746); the Australian Genomics Health Alliance (AGHA) Beat Cancer; the Leukaemia Foundation and Bristol-Meyers Squibb Company to D.L.W.; Contributing Haematologist Committee (CHC) to D.T.Y.; Australian Government Research Training Program (RTP) to C.E.J.D.
Author contributions
C.E.J.D.: conceptualization, methodology, software, validation, formal analysis, investigation, data curation, writing—original draft, visualization. B.J.M.C.: conceptualization, methodology, writing—review and editing, supervision. J.B.B.: software, formal analysis, resources, writing—review and editing, visualization, supervision. E.P.: investigation. J.B.: software, formal analysis, writing—review and editing. J.R.: software, formal analysis, writing—review and editing. D.T.Y.: resources, funding acquisition, writing—review and editing. D.L.W.: conceptualization, methodology, resources, writing—review and editing, supervision, funding acquisition.
Data availability
The data generated and analyzed during this study are described in the following data record: 10.6084/m9.figshare.14959809 (ref. 60). The exome sequencing data are openly available in the NCBI Sequence Read Archive via the following accession: https://identifiers.org/ncbi/bioproject:PRJNA741841 (ref. 61). All additional data files underlying the Figs. and Supplementary Figs. are shared openly in the figshare data record.
Competing interests
D.L.W. receives research support from BMS, Honoraria from BMS and Amgen. D.T.Y. receives research support from Novartis, Ariad and BMS, Honoraria from Novartis, BMS, Amgen and Pfizer. None of these agencies have had a role in the preparation of this manuscript. The remaining authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41698-021-00215-x.
References
- 1.Tran, T. H. & Hunger, S. P. The genomic landscape of pediatric acute lymphoblastic leukemia and precision medicine opportunities. Semin. Cancer Biol. 10.1016/j.semcancer.2020.10.013 (2020). Online ahead of print. [DOI] [PubMed]
- 2.Hunger SP, Mullighan CG. Acute lymphoblastic leukemia in children. N. Engl. J. Med. 2015;373:1541–1552. doi: 10.1056/NEJMra1400972. [DOI] [PubMed] [Google Scholar]
- 3.Pui CH, et al. Childhood acute lymphoblastic leukemia: progress through collaboration. J. Clin. Oncol. 2015;33:2938–2948. doi: 10.1200/JCO.2014.59.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paul S, Rausch C, Nasnas P, Kantarjian H, Jabbour E. Treatment of relapsed/refractory acute lymphoblastic leukemia. Clin. Adv. Hematol. Oncol. 2019;17:166–175. [PubMed] [Google Scholar]
- 5.Oskarsson T, et al. Relapsed childhood acute lymphoblastic leukemia in the Nordic countries: prognostic factors, treatment and outcome. Haematologica. 2016;101:68–76. doi: 10.3324/haematol.2015.131680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vrooman LM, Silverman LB. Treatment of childhood acute lymphoblastic leukemia: Prognostic factors and clinical advances. Curr. Hematol. Malig. Rep. 2016;11:385–394. doi: 10.1007/s11899-016-0337-y. [DOI] [PubMed] [Google Scholar]
- 7.Paul S, Kantarjian H, Jabbour EJ. Adult acute lymphoblastic leukemia. Mayo. Clin. Proc. 2016;91:1645–1666. doi: 10.1016/j.mayocp.2016.09.010. [DOI] [PubMed] [Google Scholar]
- 8.Ronson A, Tvito A, Rowe JM. Treatment of relapsed/refractory acute lymphoblastic leukemia in adults. Curr. Oncol. Rep. 2016;18:39. doi: 10.1007/s11912-016-0519-8. [DOI] [PubMed] [Google Scholar]
- 9.Roberts KG, et al. Genomic and outcome analyses of Ph-like ALL in NCI standard-risk patients: a report from the Children’s Oncology Group. Blood. 2018;132:815–824. doi: 10.1182/blood-2018-04-841676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hurtz C, et al. Oncogene-independent BCR-like signaling adaptation confers drug resistance in Ph-like ALL. J. Clin. Invest. 2020;130:3637–3653. doi: 10.1172/JCI134424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khan M, Siddiqi R, Tran TH. Philadelphia chromosome-like acute lymphoblastic leukemia: a review of the genetic basis, clinical features, and therapeutic options. Semin. Hematol. 2018;55:235–241. doi: 10.1053/j.seminhematol.2018.05.001. [DOI] [PubMed] [Google Scholar]
- 12.Owattanapanich W, Rujirachun P, Ungprasert P, Buaboonnam J, Techavichit P. Prevalence and clinical outcome of Philadelphia-like acute lymphoblastic leukemia: systematic review and meta-analysis. Clin. Lymphoma Myeloma Leuk. 2020;20:e22–e29. doi: 10.1016/j.clml.2019.08.003. [DOI] [PubMed] [Google Scholar]
- 13.Roberts KG, et al. High fequency and poor outcome of Philadelphia chromosome-like acute lymphoblastic leukemia in adults. J. Clin. Oncol. 2017;35:394–401. doi: 10.1200/JCO.2016.69.0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roberts KG, et al. Targetable kinase-activating lesions in Ph-like acute lymphoblastic leukemia. N. Engl. J. Med. 2014;371:1005–1015. doi: 10.1056/NEJMoa1403088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tasian SK, Loh ML, Hunger SP. Philadelphia chromosome–like acute lymphoblastic leukemia. Blood. 2017;130:2064–2072. doi: 10.1182/blood-2017-06-743252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chiaretti S, Messina M, Foa R. BCR/ABL1-like acute lymphoblastic leukemia: How to diagnose and treat? Cancer. 2019;125:194–204. doi: 10.1002/cncr.31848. [DOI] [PubMed] [Google Scholar]
- 17.Boer JM, den Boer ML. BCR-ABL1-like acute lymphoblastic leukaemia: From bench to bedside. Eur. J. Cancer. 2017;82:203–218. doi: 10.1016/j.ejca.2017.06.012. [DOI] [PubMed] [Google Scholar]
- 18.Steeghs EMP, et al. JAK2 aberrations in childhood B-cell precursor acute lymphoblastic leukemia. Oncotarget. 2017;8:89923–89938. doi: 10.18632/oncotarget.21027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roberts KG, et al. Genetic alterations activating kinase and cytokine receptor signaling in high-risk acute lymphoblastic leukemia. Cancer Cell. 2012;22:153–166. doi: 10.1016/j.ccr.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roberts KG, Mullighan CG. Genomics in acute lymphoblastic leukaemia: insights and treatment implications. Nat. Rev. Clin. Oncol. 2015;12:344–357. doi: 10.1038/nrclinonc.2015.38. [DOI] [PubMed] [Google Scholar]
- 21.Maude SL, et al. Targeting JAK1/2 and mTOR in murine xenograft models of Ph-like acute lymphoblastic leukemia. Blood. 2012;120:3510–3518. doi: 10.1182/blood-2012-03-415448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roberts KG, et al. Oncogenic role and therapeutic targeting of ABL-class and JAK-STAT activating kinase alterations in Ph-like ALL. Blood Adv. 2017;1:1657–1671. doi: 10.1182/bloodadvances.2017000547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Senkevitch E, Durum S. The promise of Janus kinase inhibitors in the treatment of hematological malignancies. Cytokine. 2017;98:33–41. doi: 10.1016/j.cyto.2016.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bose P, Alfayez M, Verstovsek S. New concepts of yreatment for patients with Myelofibrosis. Curr. Treat. Options Oncol. 2019;20:5. doi: 10.1007/s11864-019-0604-y. [DOI] [PubMed] [Google Scholar]
- 25.Leroy E, Constantinescu SN. Rethinking JAK2 inhibition: towards novel strategies of more specific and versatile janus kinase inhibition. Leukemia. 2017;31:2853. doi: 10.1038/leu.2017.158. [DOI] [PubMed] [Google Scholar]
- 26.Bhagwat N, Levine RL, Koppikar P. Sensitivity and resistance of JAK2 inhibitors to myeloproliferative neoplasms. Int. J. Hematol. 2013;97:695–702. doi: 10.1007/s12185-013-1353-5. [DOI] [PubMed] [Google Scholar]
- 27.Tasian SK, Assad A, Hunter DS, Du Y, Loh ML. A phase 2 study of ruxolitinib with chemotherapy in children with philadelphia chromosome-like acute lymphoblastic leukemia (INCB18424-269/AALL1521): fose-finding results from the Part 1 safety phase. Blood. 2018;132:555–555. doi: 10.1182/blood-2018-99-110221. [DOI] [Google Scholar]
- 28.Ding YY, et al. Clinical efficacy of ruxolitinib and chemotherapy in a child with Philadelphia chromosome-like acute lymphoblastic leukemia with GOLGA5-JAK2 fusion and induction failure. Haematologica. 2018;103:e427–e431. doi: 10.3324/haematol.2018.192088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller GD, Bruno BJ, Lim CS. Resistant mutations in CML and Ph+ALL—role of ponatinib. Biologics. 2014;8:243–254. doi: 10.2147/BTT.S50734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Springuel L, Renauld JC, Knoops L. JAK kinase targeting in hematologic malignancies: a sinuous pathway from identification of genetic alterations towards clinical indications. Haematologica. 2015;100:1240–1253. doi: 10.3324/haematol.2015.132142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sadras T, et al. A novel somatic JAK2 kinase-domain mutation in pediatric acute lymphoblastic leukemia with rapid on-treatment development of LOH. Cancer Genet. 2017;216-217:86–90. doi: 10.1016/j.cancergen.2017.07.008. [DOI] [PubMed] [Google Scholar]
- 32.Deshpande A, et al. Kinase domain mutations confer resistance to novel inhibitors targeting JAK2V617F in myeloproliferative neoplasms. Leukemia. 2012;26:708–715. doi: 10.1038/leu.2011.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weigert O, et al. Genetic resistance to JAK2 enzymatic inhibitors is overcome by HSP90 inhibition. J. Exp. Med. 2012;209:259–273. doi: 10.1084/jem.20111694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hornakova T, et al. Oncogenic JAK1 and JAK2-activating mutations resistant to ATP-competitive inhibitors. Haematologica. 2011;96:845–853. doi: 10.3324/haematol.2010.036350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu S-C, et al. Activity of the type II JAK2 inhibitor CHZ868 in B cell acute lymphoblastic leukemia. Cancer Cell. 2015;28:29–41. doi: 10.1016/j.ccell.2015.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kesarwani, M. et al. Targeting substrate-site in Jak2 kinase prevents emergence of genetic resistance. Sci. Rep. 5, 14538, 10.1038/srep14538 (2015). [DOI] [PMC free article] [PubMed]
- 37.Kong X, et al. How does the L884P mutation confer resistance to type-II inhibitors of JAK2 kinase: a comprehensive molecular modeling study. Sci. Rep. 2017;7:9088. doi: 10.1038/s41598-017-09586-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maxson JE, et al. Oncogenic CSF3R mutations in chronic neutrophilic leukemia and atypical CML. N. Engl. J. Med. 2013;368:1781–1790. doi: 10.1056/NEJMoa1214514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gorantla SP, et al. Chemical mutagenesis screen identifies novel drug resistant JAK2 variants in V617FJAK2 mediated MPN and predict HSP90 inhibitors as possible agents to overcome drug resistance. Blood. 2015;126:3687. doi: 10.1182/blood.V126.23.3687.3687. [DOI] [Google Scholar]
- 40.Lamontanara AJ, Gencer EB, Kuzyk O, Hantschel O. Mechanisms of resistance to BCR-ABL and other kinase inhibitors. Biochim Biophys. Acta. 2013;1834:1449–1459. doi: 10.1016/j.bbapap.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 41.Soverini S, Mancini M, Bavaro L, Cavo M, Martinelli G. Chronic myeloid leukemia: the paradigm of targeting oncogenic tyrosine kinase signaling and counteracting resistance for successful cancer therapy. Mol. Cancer. 2018;17:49. doi: 10.1186/s12943-018-0780-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weisberg E, Manley PW, Cowan-Jacob SW, Hochhaus A, Griffin JD. Second generation inhibitors of BCR-ABL for the treatment of imatinib-resistant chronic myeloid leukaemia. Nat. Rev. Cancer. 2007;7:345–356. doi: 10.1038/nrc2126. [DOI] [PubMed] [Google Scholar]
- 43.Bradeen HA, et al. Comparison of imatinib mesylate, dasatinib (BMS-354825), and nilotinib (AMN107) in an N-ethyl-N-nitrosourea (ENU)-based mutagenesis screen: high efficacy of drug combinations. Blood. 2006;108:2332–2338. doi: 10.1182/blood-2006-02-004580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Glickman MS, Sawyers CL. Converting cancer therapies into cures: Lessons from infectious diseases. Cell. 2012;148:1089–1098. doi: 10.1016/j.cell.2012.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Azam M, Latek RR, Daley GQ. Mechanisms of autoinhibition and STI-571/imatinib resistance revealed by mutagenesis of BCR-ABL. Cell. 2003;112:831–843. doi: 10.1016/S0092-8674(03)00190-9. [DOI] [PubMed] [Google Scholar]
- 46.Davis, R. R. et al. Structural insights into JAK2 Inhibition by ruxolitinib, fedratinib, and derivatives thereof. J. Med. Chem. 64, 2228–2241, 10.1021/acs.jmedchem.0c01952 (2021). [DOI] [PMC free article] [PubMed]
- 47.Kim SK, et al. JAK2 is dispensable for maintenance of JAK2 mutant B-cell acute lymphoblastic leukemias. Genes Dev. 2018;32:849–864. doi: 10.1101/gad.307504.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koschut D, et al. RAS-protein activation but not mutation status is an outcome predictor and unifying therapeutic target for high-risk acute lymphoblastic leukemia. Oncogene. 2021;40:746–762. doi: 10.1038/s41388-020-01567-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nicorici, D. et al. FusionCatcher—a tool for finding somatic fusion genes in paired-end RNA-sequencing data. preprint from bioRxiv 011650. 10.1101/011650 (2014).
- 50.Davidson, N., Majewski, I. & Oshlack, A. JAFFA: High sensitivity transcriptome-focused fusion gene detection. Genome Med. 7, 43, 10.1186/s13073-015-0167-x (2015). [DOI] [PMC free article] [PubMed]
- 51.Jia, W. et al. SOAPfuse: an algorithm for identifying fusion transcripts from paired-end RNA-Seq data. Genome Biol. 14, R12, 10.1186/gb-2013-14-2-r12 (2013). [DOI] [PMC free article] [PubMed]
- 52.Liu S, et al. Comprehensive evaluation of fusion transcript detection algorithms and a meta-caller to combine top performing methods in paired-end RNA-seq data. Nucleic Acids Res. 2016;44:e47. doi: 10.1093/nar/gkv1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koppikar P, et al. Heterodimeric JAK-STAT activation as a mechanism of persistence to JAK2 inhibitor therapy. Nature. 2012;489:155–159. doi: 10.1038/nature11303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Coordinators NR. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2018;46:D8–D13. doi: 10.1093/nar/gkx1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee JC, Hapel AJ, Ihle JN. Constitutive production of a unique lymphokine (IL 3) by the WEHI-3 cell line. J. Immunol. 1982;128:2393–2398. [PubMed] [Google Scholar]
- 56.Kluin RJC, et al. XenofilteR: computational deconvolution of mouse and human reads in tumor xenograft sequence data. BMC Bioinformatics. 2018;19:366. doi: 10.1186/s12859-018-2353-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ioannidis, S. et al. Discovery of 5-chloro-N2-[(1 S)-1-(5-fluoropyrimidin-2-yl)ethyl]-N4-(5-methyl-1H-pyrazol-3-yl)p yrimidine-2,4-diamine (AZD1480) as a novel inhibitor of the Jak/Stat pathway. J. Med. Chem. 54, 262–276 (2011). [DOI] [PubMed]
- 58.Duan Y, Chen L, Chen Y, Fan X. c-Src binds to the cancer drug ruxolitinib with an active conformation. PLoS ONE. 2014;9:e106225. doi: 10.1371/journal.pone.0106225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moriarty NW, Grosse-Kunstleve RW, Adams PD. Electronic Ligand Builder and Optimization Workbench (eLBOW): a tool for ligand coordinate and restraint generation. Acta Crystallogr. D Biol. Crystallogr. 2009;65:1074–1080. doi: 10.1107/S0907444909029436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Downes, C. E. et al. Metadata record for the article: acquired JAK2 mutations confer resistance to JAK inhibitors in cell models of acute lymphoblastic leukemia. figshare 10.6084/m9.figshare.14959809 (2021). [DOI] [PMC free article] [PubMed]
- 61.NCBI Sequence Read Archive. https://identifiers.org/ncbi/bioproject:PRJNA741841 (2021).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated and analyzed during this study are described in the following data record: 10.6084/m9.figshare.14959809 (ref. 60). The exome sequencing data are openly available in the NCBI Sequence Read Archive via the following accession: https://identifiers.org/ncbi/bioproject:PRJNA741841 (ref. 61). All additional data files underlying the Figs. and Supplementary Figs. are shared openly in the figshare data record.