Abstract
Background
Microbiome sequencing has brought increasing attention to the polymicrobial context of chronic infections. However, clinical microbiology continues to focus on canonical human pathogens, which may overlook informative, but nonpathogenic, biomarkers. We address this disconnect in lung infections in people with cystic fibrosis (CF).
Methods
We collected health information (lung function, age, and body mass index [BMI]) and sputum samples from a cohort of 77 children and adults with CF. Samples were collected during a period of clinical stability and 16S rDNA sequenced for airway microbiome compositions. We use ElasticNet regularization to train linear models predicting lung function and extract the most informative features.
Results
Models trained on whole-microbiome quantitation outperformed models trained on pathogen quantitation alone, with or without the inclusion of patient metadata. Our most accurate models retained key pathogens as negative predictors (Pseudomonas, Achromobacter) along with established correlates of CF disease state (age, BMI, CF-related diabetes). In addition, our models selected nonpathogen taxa (Fusobacterium, Rothia) as positive predictors of lung health.
Conclusions
These results support a reconsideration of clinical microbiology pipelines to ensure the provision of informative data to guide clinical practice.
Keywords: microbiome, machine learning, cystic fibrosis
Bacterial infections often resolve rapidly given effective immune responses, independent of antibiotic treatment. However, in chronic (long-lasting) cases, infections fail to clear even with appropriate drug treatment. Chronic infections impose an elevated morbidity and mortality risk to the individual [1] and an increasing burden on global health care systems as at-risk populations grow [2]. Chronic infections typically arise due to deficits in host barrier defenses and/or immune function, and commonly feature changes in pathogen growth mode (eg, biofilm formation [3]) and additional microbial species acquisition, forming complex multispecies communities [4].
Microbiome sequencing has increasingly underscored the polymicrobial context of chronic infection. However, clinical microbiology analysis continues to focus only on the “usual suspects” of established human pathogens—a relatively short list of organisms with well-established patient health risks. This disconnect between diverse “infection microbiomes” and limited clinical microbiology profiling may overlook clinically important risk markers. To address this, we focus on chronic lung infections in people with cystic fibrosis (CF).
CF is an autosomal recessive disease characterized by decreased lung mucociliary clearance and mucus accumulation [5–7]. The resulting environment provides both nutrients for bacterial growth and protection from host immune responses [8–11], facilitating chronic microbial infections [12–15]. Accessible 16S rDNA microbiome profiling has shifted CF airway microbiology research away from a historically single-pathogen focus, as sequencing expectorated sputum has revealed diverse communities of tens to hundreds of taxa, including numerous nonpathogenic bacteria [13, 16, 17].
Numerous lung microbiome studies have linked community composition to disease progression and overall patient health [18–20]. Cross-sectional studies have shown severe disease is associated with pathogen dominance and loss of taxonomic diversity [18, 19, 21]. Longitudinal studies have associated decreasing microbiome diversity with declining lung function [22]. Additionally, abundance of nonpathogenic fermentative anaerobes (Veillonella, Prevotella, and Fusobacterium) is associated with higher lung function [23, 24]. While these associations are observed across multiple studies, their causal interpretation is the subject of some controversy. These results may reflect community ecological processes within the lung, where species interactions govern community structure and subsequent harm to the host [12, 25, 26]. Conversely, these patterns could result from oral anaerobe contamination during sample collection [27, 28]. Under this contamination model, increasing pathogen load compared to a constant background of oral microbiome contamination generates a spurious link between oral microbes, microbiome diversity, and patient health [27]. While recent paired sputum-saliva sampling analysis indicates that oral sample contamination is not a substantial contributor to sputum microbiome profiles in people with established CF lung disease [29], these conflicting hypotheses highlight the uncertainty in the role specific taxa present in sputum.
In the current study, we side-step this causal inference problem and instead assess how informative expectorated sputum microbiome data (including potential oral contaminants) is of patient lung health using a machine-learning framework. We hypothesize that the addition of nonpathogen data improves the prediction of patient lung function compared to established pathogen data alone. To address this hypothesis we train predictive models on both lung microbiome and electronic medical record data for a cohort of CF patients. We find that compared to the benchmark of pathogen data alone, model performance was consistently improved by the addition of nonpathogen taxa.
METHODS
Subjects
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and national research committees. Authorization was obtained from each patient enrolled according to the protocol approved by the Emory University Institutional Review Board (IRB00042577).
Sample Collection and 16S Analysis
Expectorated sputum samples were obtained from CF patients attending the Children’s Healthcare of Atlanta and Emory University CF Care Center from January 2015 to August 2016. Deidentified patient information including age, sex, height, body mass index (BMI), cystic fibrosis transmembrane conductance regulator (CFTR) genotype, degree of glucose control (HbA1c), and percent predicted forced expiratory volume in 1 second (ppFEV1) were obtained (Table 1). Among these CF patients, 39 were diagnosed with CF-related diabetes (CFRD) by a CF endocrinologist. HbA1c value was missing for 1 CFRD subject.
Table 1.
Summary of Patient Clinical Data, Stratified by Lung Function
| Characteristic | Severe | Moderate | Mild | Normal | P |
|---|---|---|---|---|---|
| n | 14 | 25 | 15 | 23 | |
| ppFEV1, median (range) | 32.9 (19.7–39.2) |
46.5 (40.8–59.6) |
74.9 (61.6–79.8) |
101.2 (80.4–119.5) |
|
| Age, y, median (range) | 31.5 (21–61) |
32 (10–63) |
24 (17–51) |
20 (9–66) |
.007 |
| Male, No. | 6 | 11 | 7 | 11 | |
| CFTR genotype, No. | |||||
| Homo-dF508 | 5 | 12 | 7 | 13 | |
| Hetero-dF508 | 9 | 10 | 8 | 10 | |
| Other/other | 0 | 0 | 3 | 0 | |
| BMI, median (range) | 19.43 (16.27–25.69) |
20.73 (16.70–29.81) |
22.23 (19.38–26.07) |
21.51 (16.65–33.91) |
.094 |
| CF-related diabetes, No. (%) | 11 (78.6) |
14 (56.0) |
8 (53.3) |
6 (26.1) |
.015 |
| HbA1c, median (range) | 6.25 (5.3–11.9) |
5.9a (4.9–8.4) |
5.7 (5.0–7.6) |
5.5 (5.1–7.1) |
.009 |
| Clinical microbiology | |||||
| PA, No. (%) | 10 (71.4) | 20 (80.0) | 10 (66.7) | 5 (21.7) | <1.1e-4 |
| SA, No. (%) | 8 (57.1) | 12 (48.0) | 10 (66.7) | 16 (69.6) | .454 |
| MRSA, No. (%) | 4 (28.6) | 6 (24.0) | 6 (40.0) | 4 (17.4) | .486 |
| Burkholderia, No. (%) | 0 (0.0) | 1 (4.0) | 1 (6.7) | 0 (0.0) | .553 |
| Achromobacter, No. (%) | 3 (21.4) | 1 (4.0) | 0 (0.0) | 2 (8.7) | .147 |
| Stenotrophomonas, No. (%) | 0 (0.0) | 1 (4.0) | 3 (20.0) | 2 (8.7) | .191 |
| 16S metadata | |||||
| % Pathogen | 0.857 | 0.589 | 0.532 | 0.195 | 9.05e-5 |
| % Nonpathogen | 0.135 | 0.404 | 0.597 | 0.783 | 3.01e-5 |
Lung function classes are defined as: normal, ppFEV1 > 80; mild, 60 < ppFEV1 ≤ 80; moderate, 40 < ppFEV1 ≤ 60; and severe, ppFEV1 < 40.
Significant differences between lung function categories tested by ANOVA, P -values shown.
Abbreviations: BMI, body mass index; CF, cystic fibrosis; CFTR, cystic fibrosis transmembrane conductance regulator; HbA1c, hemoglobin A1c; MRSA, methicillin-resistant Staphylococcus aureus. PA, Pseudomonas aeruginosa; SA, Staphylococcus aureus
aTwo patients did not have reported HbA1c values.
All patients were clinically stable, defined as having no increase in respiratory symptoms compared to baseline, and no acute illness or new medication for 3 weeks prior to sputum collection. Upon collection, sputum samples were diluted 1:3 (mass:volume) with phosphate-buffered saline supplemented with 50 mM EDTA. Diluted samples were then homogenized by being repeatedly drawn through a syringe and 18-gauge needle. The resulting sputum homogenates were aliquot and stored at −80°C until all 77 samples were collected. Microbiology culture results were obtained for sputum samples sent to the Clinical Microbiology Laboratory on the same day as samples for sequencing were collected.
DNA was purified from sputum homogenate with the MoBio Power Soil kit (MoBio). The 16S V4 region was amplified and sequenced using Illumina MiSeq, yielding an average of 137 708 sequences per sample. Sequences were quality filtered and amplicon sequence variants were obtained using the QIIME2 deblur plugin. Taxonomic assignments were classified against both SILVA and Greengenes 16S reference databases and assigned based on highest taxonomic resolution. To mitigate compositional effects, 16S data were center-log transformed prior to all analyses. Nucleotides are uploaded to BioProject accession no. PRJNA666192.
Statistical and Quantitative Analysis
Patient samples were binned by ppFEV1 (normal, >80%; mild, 80%–60%; moderate, 60%–40%; and severe, <40%). Variance across lung function categories in patient metadata and 16S metadata was tested using ANOVA. Variation between microbiome composition and ppFEV1 was tested using Mantel tests on Bray-Curtis distances at 9999 permutations. Within-sample and among-sample diversity was calculated using the Shannon diversity index and Bray-Curtis based PCoA on 16S quantitation agglomerated to the genus level [30]. Associations between continuous variables were tested using Spearman correlations. A full pairwise correlation matrix was calculated, with rows and columns ordered by hierarchical clustering [31].
Machine Learning
We used ElasticNet to fit regularized linear models predicting lung function (ppFEV1) from patient metadata, microbiome composition, and clinical microbiology results [32]. ElasticNet solves a penalized linear regression model using a weighted average of L1 (LASSO) and L2 (ridge regression) penalties. This limits over-fitting by penalizing nonzero coefficients. We split our samples using a simple 70:30 train-test holdout, where models were trained on 53 samples and used to predict on the remaining 24. All input features were standardized (mean = 0; SD = 1) prior to model training to allow between-feature interpretability. From our full dataset, we created 4 additional data subsets: CF Pathogens, All 16S Data, Metadata, and Metadata + Pathogens. We included within-feature shuffling on the full set as a noninformative negative control.
We employed 2 methods to assess model robustness and compared model performance using mean squared error (MSE). We generated 1000 bootstrap resampled sets from the training set and fit an ensemble of regularized linear models to obtain distributions for each model coefficient. We identified key metadata and taxa robustly selected (nonzero coefficients) across the ensemble of models. We assessed model generalizability using leave-one-out cross-validation on the training set and compared resulting MSE ranges.
RESULTS
Clinical and Microbiome Data Summary
In total, we obtained sputum expectorates from 77 CF children and adults. Pulmonary function, measured by ppFEV1, was stratified into 4 categories from severe to normal. A summary of patient information is presented in Table 1. As expected, increasing age correlated with worsening lung function (ANOVA; P < .01). Culture-based detection of Pseudomonas aeruginosa correlated with decreasing lung function (ANOVA; P < .001), as did (log-scaled) bacterial load (P < .05).
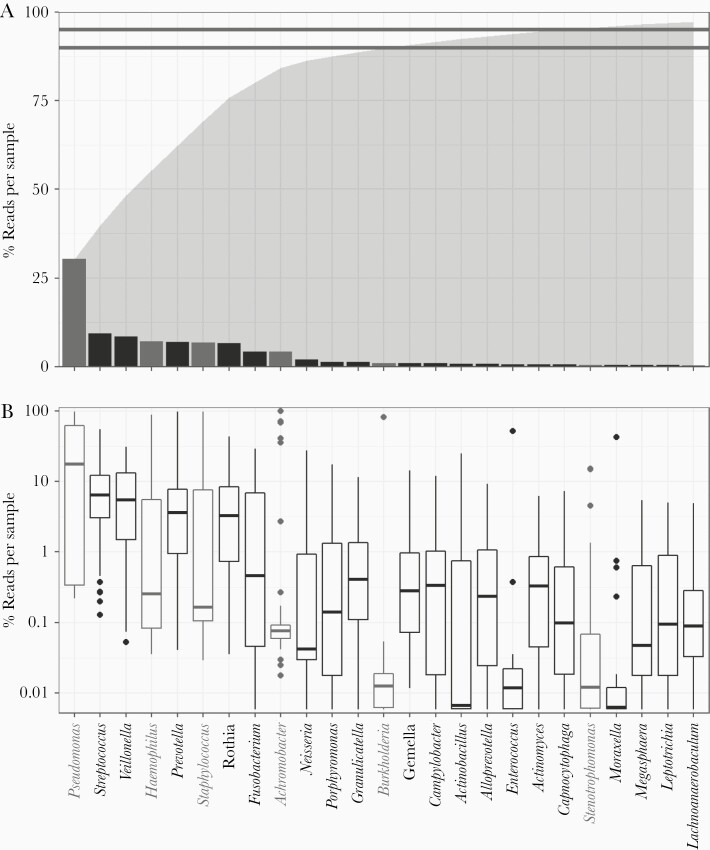
The majority (>90%) of reads from our sequencing analysis mapped to 1 of 13 genera (Figure 1A), consisting of both recognized CF pathogens and orally derived bacteria. Pseudomonas sequences accounted for 30.4% of all reads and were detected in every patient sample. Other established CF pathogens (Staphylococcus, Achromobacter, Haemophilus, and Burkholderia) collectively represented 19.3%, while oral taxa account for over 45% (Figure 1). Total pathogenic and nonpathogenic taxa abundance were both found to vary significantly (P << .001) with lung function (Table 1).
Figure 1.
Cystic fibrosis (CF) lung microbiomes are dominated by oral anaerobes and opportunistic pathogens We analyzed CF sputum expectorate (n = 77) using 16S sequencing and an in-house QIIME 2-based bioinformatics pipeline to resolve strain-level Operational Taxonomic Units (OTUs). Samples were rarefied to 17 000 reads. We identified 217 OTUs across 59 genera and at least 81 species. Overall, we found that CF sputum samples were dominated by oral anaerobes and opportunistic pathogens. A, Sequences mapped to 14 genera comprised 90% (lower line) of the total reads obtained; 95% (upper line) of all reads mapped to 21 genera. Total cumulative read fraction is represented in shaded region. Recognized CF pathogens are as follows: Pseudomonas, Haemophilus, Staphylococcus, Achromobacter, Burkholderia, and Stenotrophomonas. Pseudomonas was the most prevalent genus, followed by Streptococcus and Veillonella. B, Binning reads by sample shows variation in relative abundance. Pseudomonas comprised >10% of reads in the majority of our samples. While over 6% of the total reads mapped to Achromobacter, only 4 samples comprised >10% Achromobacter.
Microbiome Composition Varies With Lung Function
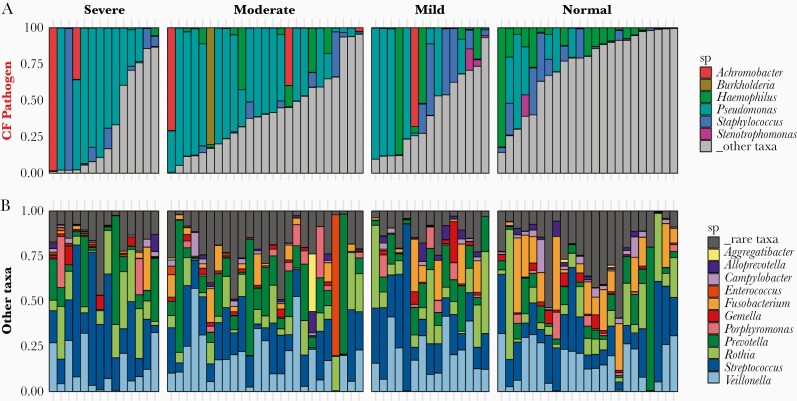
We analyzed microbiome compositions across broad lung function categories to examine the relationship between sputum taxonomic profile and patient health. Figure 2A highlights the relative compositions of 6 canonical CF pathogens. As expected, Pseudomonas was more prevalent in lungs with reduced function, whereas in normal lungs Haemophilus and nonpathogen taxa (gray) were more prevalent. The nonpathogenic composition was consistently dominated by Veillonella and Streptococcus regardless of lung health or pathogen status (Figure 2B). Shannon diversity calculated with all taxa present was significantly greater for normal lung function (P < .01; Supplementary Figure 1A), in line with multiple other studies [33, 34]. While principle coordinate analysis did not qualitatively separate compositions by lung function category, we found ppFEV1 was significantly associated with microbiome composition (Mantel test, r = 0.195; P < .001; Supplementary Figure 1B).
Figure 2.
CF lung microbiome composition varies with lung function and pathogen dominance. Relative abundances of (A) 6 canonical CF pathogens and (B) other taxa (grey bar taxa in (A)). Microbiome compositions grouped by disease severity, classified by ppFEV1 score: normal (80+), mild (60–80), moderate (40–60), and severe (<40). Abbreviations: CF, cystic fibrosis; ppFEV1, percent predicted forced expiratory volume in 1 second; sp, species.
Integrating Microbiome and Patient Metadata
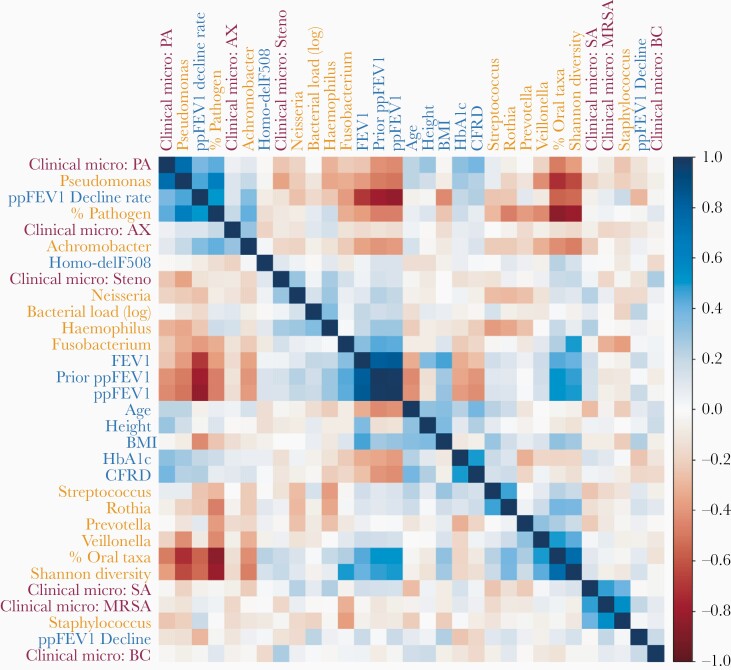
To examine multiple confounding variables such as patient age, BMI, or CFRD, we calculated spearman correlations across 14 microbiome, 11 patient metadata, and 6 clinical microbiology features (Figure 3). Hierarchical clustering reveals a complex autocorrelation structure but with many expected consistencies. Overall, there are 2 main clusters of correlated variables. One correlated with ppFEV1, and included Shannon diversity index as well as 16S quantitation of Fusobacterium, Haemophilus, and Neisseria. The other anticorrelated with ppFEV1, and included ppFEV1 decline, pathogen abundance, CFRD, and 16S quantitation of Pseudomonas and Achromobacter. Unsurprisingly, FEV1 and ppFEV1 cluster together and inversely correlated with ppFEV1 decline rate (an average per year loss in ppFEV1 since birth). Additionally, 16S quantitation results for Pseudomonas, Staphylococcus, Burkholderia, and Achromobacter cluster with their respective culture-based clinical microbiology results. This does not hold for Stenotrophomonas, potentially due to its infrequent detection.
Figure 3.
Lung function varies with patient metadata. Spearman correlations (R::corrplot) across all patient metadata (blue), clinical microbiology results (maroon), and microbiome data (orange, centered log-ratio transformed) reveal a complex correlation structure. We used a centered-log transform on 16S data to mitigate compositional effects. Rows and columns were ordered by hierarchical clustering, which identified clusters of metadata and microbiome variables with similar correlation patterns. Abbreviations: AX, Achromobacter; BC, Burkholderia; BMI, body mass index; CFRD, cystic fibrosis-related diabetes; MRSA, methicillin-resistant Staphylococcus aureus; PA, Pseudomonas aeruginosa; ppFEV1, percent predicted forced expiratory volume in 1 second; SA, Staphylococcus aureus.
Dimensionality Reduction
The hairball correlation matrix in Figure 3 highlights the statistical challenges in identifying meaningful lung function predictors. Such challenges include high between-feature correlations and relatively few independent patient observations (n = 77) compared to the initial number of available predictors (86 total, including 59 bacterial taxa). To mitigate this dimensionality problem, we first restricted our microbiome analysis to only the top 23 genera in our dataset. These top 23 encompassed 97% of the total sequenced reads (Figure 1). We also calculated 3 additional summary statistics: % pathogen, % oral taxa, and Shannon diversity. As our clustering analysis showed reasonable agreement between clinical microbiology detection and rDNA sequencing, we excluded the binary detection results in favor of 16S quantitation. To address compositionality of 16S data, we incorporated total bacterial load (universal 16S primer quantitative polymerase chain reaction [qPCR]) as a predictor. In addition, we used a centered log-ratio transform on our genus-level relative abundance data before standardizing to mean zero, unit variance inputs. We refer to this final combination of metadata and 16S data as our All Features dataset.
Training Machine Learning Models
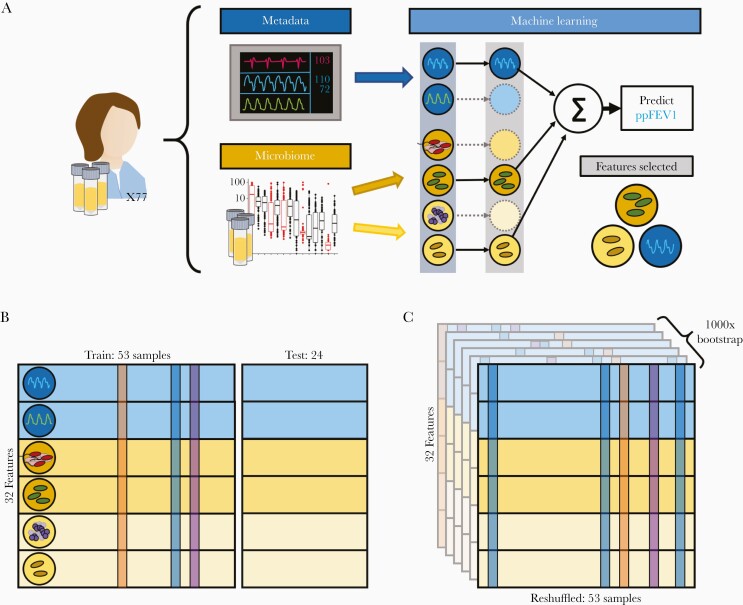
To assess if nonpathogenic taxa contain informative biomarkers, we split our samples into 53 training and 24 testing samples. ElasticNet was used to train predict lung function while performing feature selection (see Methods; Figure 4). We expect that the addition of patient metadata (age, BMI, etc.) will improve our ability to predict lung function given the progressive nature of CF. Our null hypothesis, following the work of Jorth et al and others [27, 28] is that the taxa targeted by clinical microbiology provide adequate explanatory basis for lung function outcomes, and that the addition of nonpathogen 16S data will not improve model predictions.
Figure 4.
Machine learning overview. Machine learning models were trained on different input data tables using varying data resampling methods. A, Features were categorized by information source (microbiome or patient metadata). The 16S data was further split into pathogens and other taxa in agreement with Figure 2. We used ElasticNet regularization to select informative features that predict ppFEV1. B, We randomly selected 24 patient samples to withhold as a test set and trained our models on the remaining 53 samples. To assess overfitting, we used leave-one-out cross-validation on our training set. C, We additionally implemented 1000-fold bootstrap resampling to assess the robustness of our model fits. Abbreviation: ppFEV1, percent predicted forced expiratory volume in 1 second.
We tested this hypothesis by generating 4 additional feature subsets (CF Pathogens, All 16S Data, Metadata, and Metadata + Pathogens) and comparing the performance of models trained on each datasets. Initial-pass, nonbootstrapped model training results are shown in Supplementary Figure 2 and Supplementary Figure 3.
Model Generalizability
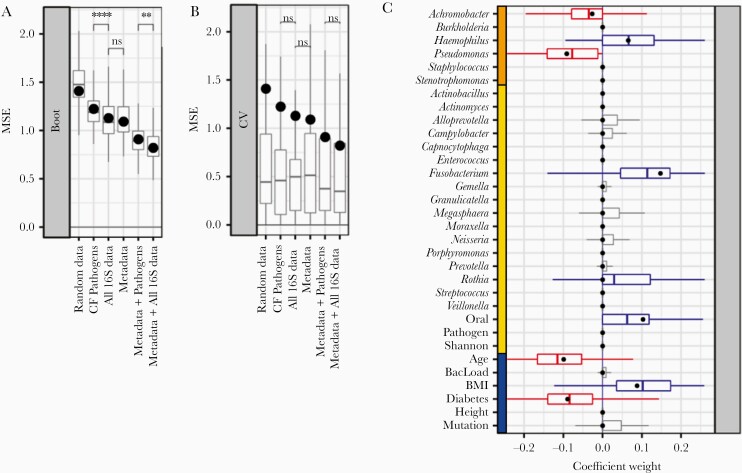
We assessed overfitting using leave-one-out cross-validation and compared the prediction error across folds against the test set error. For model robustness, we used 1000-fold bootstrap resampling to fit both a baseline and ensemble of models. Robust features selected by the baseline model will also be selected by a large portion of the bootstrapped ensemble. We additionally standardized All Features (mean = 0, SD = 1) to allow for cross-feature comparability. As an additional point of comparison, we generated a noninformative control dataset from the All Features set using within-feature shuffling, scrambling between-feature correlations while preserving the mean zero, unit variance within-feature structure. Figure 5 shows the results of our baseline (black points) and ensemble (boxplots) approaches. All models using patient metadata or microbiome data outperformed the negative control.
Figure 5.
Bootstrapped ElasticNet-identified predictors of lung function. Machine learning models were trained using varying input datasets. A, 1000-fold bootstrapping and (B) leave-one-out cross-validation (LOOCV) were used to generate prediction error (MSE) ranges across feature subsets. Models trained on all of the data showed lower error compared to other feature subsets. Adding 16S pathogen quantitation decreased model error. Models trained on all 16S data outperformed models using only 16S pathogen quantitation (P < .01, t test). Regardless of input features, models trained on the full sample set (black points) were greater than median LOOCV MSEs (boxplots). C, Coefficient ranges for train/test (black points) and bootstrapped models (boxplots) trained on standardized input datasets (blue, metadata; orange, 16S pathogens; yellow, 16S other taxa) show consistency between both machine learning strategies. Both cases selected Pseudomonas and Achromobacter as negative predictors. Abbreviations: BMI, body mass index; CF, cystic fibrosis; MSE, mean squared error; ns, not significant. **P < .01; ****P < .0001.
Addition of Nonpathogen Data Improves Model Performance
To address the key question of relative model performance, we found that the addition of nonpathogen taxa significantly improved performance (significantly reduced bootstrapped MSE; Figure 5A), with or without the addition of patient metadata. Models trained on all 16S quantitation significantly outperformed models trained only on pathogen quantitation. Interestingly, while microbiome-only and metadata-only models achieved comparable performance, the combined model achieved greater model performance. Looking broadly across models, we found reasonable consistency in positive and negative predictor selection between our baseline and bootstrapped models (Supplementary Figure 4).
We found multiple features selected across all training sets. Pseudomonas, Achromobacter, age, and diabetic status were consistently selected as negative predictors, while Haemophilus, Fusobacterium, Rothia, oral taxa abundance, and BMI were consistently positive predictors. All informative features selected in the independent models (Supplementary Figure 4C) were also selected in the All Features model (Supplementary Figure 4G). A small subset (<50%) of the bootstrapped models also selected a handful of oral taxa, bacterial load, and CFTR mutation type as positive predictors of lung function (Figure 5C, gray boxplots). However, a majority of bootstrapped models and the train/test model did not select these as informative features.
As an additional check against overfitting, we obtained ranges of model errors (measured by mean squared error of predicted ppFEV1 values) using leave-one-out cross-validation (Figure 5B). We did not find significant differences between cross-validated model errors across our training sets, suggesting that despite the difference in number of available predictors, our models were not overfitting.
DISCUSSION
People with CF face the challenge of managing long-term chronic infections. Current respiratory management practice is driven by clinical microbiology identification of specific pathogens in throat cultures or expectorated sputum samples, alongside measures of respiratory status (changes in symptoms, signs, and/or lung function). In the current study, we used 16S sequencing to assess sputum microbiome content more broadly, and ask whether the addition of nonpathogen taxa improves our ability to predict patient lung health, with or without the inclusion of patient health data. To address this question we applied machine learning tools to an integrated 77 patient lung microbiome and electronic medical record dataset. Our analysis revealed that the addition of nonpathogen data improves prediction of patient health, with the most accurate models selecting patient metadata, pathogen quantitation, and nonpathogen information. Our inclusive all-data models additionally point to a predictive role for specific nonpathogen taxa, in particular the oral anaerobe genera Rothia and Fusobacterium.
Despite the significant contribution of nonpathogen data, our results are still broadly consistent with what might be termed the traditional view of CF microbiology. Established CF pathogens (P. aeruginosa, Staphylococcus aureus, Haemophilus influenzae, and Burkholderia cenocepacia) are the major drivers of patient outcomes, as evidenced by substantial improvement in predictive outcomes whenever we include pathogen data (Figure 5A), and the, by comparison, relatively weak contribution of the addition of nonpathogen taxa. Note that we specifically used quantitative 16S measures of pathogen composition to provide a level playing field in the comparison of pathogen and nonpathogen predictive contribution. Figure 3 highlights that quantitative 16S and qualitative (presence/absence) clinical microbiology data are in general agreement.
The traditional role of CF pathogens as the central predictors of patient outcomes has been challenged over the past decade by the advent of microbiome sequencing. Extensive surveys have documented an association between CF lung function and microbiome diversity, also evident in the current study (Figure 2). At face value, these results suggest a biological role for these nonpathogen taxa, potentially competing with [35] or facilitating [36] pathogen taxa and therefore indirectly shaping disease outcomes. Jorth et al recently published a forceful rejection of this active microbiome view, stressing the potential causal role of changing pathogen densities in shaping disease outcomes and viewing shifting diversity metrics as a simple statistical relative composition artifact of shifting pathogen numbers against a roughly constant oral contamination background [27]. While our analyses provide some support for this view, in particular the constancy of the nonpathogen microbiome across patients (Figure 2B) and the lack of substantial predictive improvement on addition of nonpathogen data (Figure 5B), we also see lines of evidence against the contamination hypothesis. First, our use of center-log transformations mitigates the risk of spurious associations due to compositionality [37] and yet nonpathogen taxa are still consistently retained. Second, the contamination hypothesis predicts total bacterial burden to be an important predictor, and yet burden was not retained in our models. Third, our observation of a consistent retention of specific nonpathogen taxa across multiple models (with and without the addition of potentially confounding electronic medical records (EMR) features, including age and BMI) points to the potential for a distinct causal pathway that is orthologous to age or BMI. We note that the interpretation that oral bacteria are active players in the lung environment is further buttressed by a recent study on people with established CF disease [29] that used paired sputum and saliva samples to infer the presence of substantial populations of oral bacteria in the lung.
Our all-data models highlight Rothia and Fusobacterium as positive predictors of lung function across our 77 patients, in models that already take into account pathogen data. When we included features already known to correlate with lung health, such as age, BMI, and CFRD status, our models not only selected these features, but additionally retained Rothia and Fusobacterium as positive predictors. The retention of these specific taxa in both this full model and in partial models (Supplementary Figure 4) suggests that these taxa provide potentially valuable predictive information on current patient health. Of course, this analysis does not allow inference to causal mechanism or even direction of causality. It is entirely possible that these taxa are simply biomarkers of dimensions of improved health that are largely independent of age, BMI, and other established positive predictors that are already accounted for in the model. It is also possible that these specific taxa play a more active causal role, for instance holding specific pathogens at bay via competitive interspecific mechanisms [38].
Interestingly, our All Features models also highlighted Haemophilus, a canonical CF pathogen, as a positive predictor of lung function. Haemophilus influenzae infections are most common in younger CF patients [8, 39], hence we would expect a positive association in a model that is not controlled for age (Supplementary Figure 4C and 4D). However, we see that the positive weighting on Haemophilus was retained in models that also accounted for age as a positive predictor of lung function. A second possibility is that the positive weighting of Haemophilus is due to pathogen-pathogen competition and the relatively less severe nature of Haemophilus infections in adults (ie, Haemophilus is “best of a bad job”). Figure 2A illustrates that we only appreciably detected 2 and rarely 3 coexisting pathogens of the 6 we find across all patients. The relative scarcity of multipathogen communities implies that Haemophilus presence coincides with the absence of other more severe pathogens—and indeed we see a dominance of negative correlations among pathogens (Figure 3). In this context we cannot preclude a protective role of Haemophilus against more severe pathogens in older patients.
A caveat of this analysis is the dependency of machine learning performance and robustness on particular distributions of data, and the failure of linear algorithms such as LASSO and ElasticNet on microbiome-like data [40–42]. This is in part due to the compositionality constraint of microbiome data, which can be mitigated by using absolute quantitation [43]. However, training on absolute abundances introduces additional caveats, as order-of-magnitude differences in qPCR sample quantitation can in turn over-represent samples with higher bacterial loads. We address these issues by using a centered-log transform on relative abundance data and including log-scaled bacterial load as a potential feature to select. While some bootstrapped models selected bacterial load as a positive predictor (Figure 5C), the majority of models did not. This further suggests that the majority of microbiome information is encoded in the relative ratios of taxa abundance, which is broadly consistent with previous findings [27, 28].
Finally, our study is limited to a cross-sectional analysis, limiting us to making predictions on lung function state at the same time point as microbiome sample and patient medical record collection. Assessing and refining our predictive machine learning algorithms on subsequent lung function data is an important future goal. Our primary objective is to predict future disease states and preemptively identify patients in need of medical intervention using early warning microbiome markers. To this effect, we plan to continue our analysis on a cohort of patients across time to evaluate predictive capacity for future health status.
We note that the major predictors identified in our models have been identified in various studies, and taken piecemeal there is less insight. The value of this work lies in the systematic integration of these multiple data sources (from both EMR and microbiome data sources). Our model comparisons (with/without EMR predictors) allow an assessment of the impact of oral bacteria, with and without key potential confounds. Ignoring these confounds could lead to spurious retention of microbiome taxa that correlate strongly with, for example, age or BMI. In addition, our analyses allow assessment of disparate factors on a common predictive scale—indicating for example that the impact of 1 standard deviation shift in Fusobacterium abundance is comparable to a 1 standard deviation shift in BMI. Our model comparison approach lends more confidence to the conclusion that the retained oral taxa are associated with patient outcomes via causal pathways that are largely independent of age or BMI, being robust to their presence or absence in the predictive models. The research agenda of pursuing the nature of the causal pathways linking oral bacteria in the lung with patient outcomes is now on a firmer footing as a result of our study.
In summary, our study finds that inclusion of nonpathogenic taxa significantly improves model prediction accuracy of patient health status. We identify 2 oral-derived taxa (Fusobacterium, Rothia) that are independently informative of lung function, which may be either biomarkers or potential probiotics. Our results call attention to the potential predictive utility of oral microbes (regardless of their functional roles) in the clinical assessment of CF patient health.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We thank Karan Kapuria and Eunbi Park for help with bioinformatic pipeline development, and Peng Qiu for machine learning guidance. We acknowledge the technical support and provision of the clinical data by the CF Biospecimen Registry at the Children’s Healthcare of Atlanta and Emory University CF Discovery Core. This research was supported in part through research cyber-infrastructure resources and services provided by the Partnership for an Advanced Computing Environment at Georgia Tech.
Disclaimer. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Financial support. This work was supported by the Centers for Disease Control and Prevention (grant numbers BAA 2016-N-17812 and BAA 2017-OADS-01); National Institutes of Health (grant numbers HR56L142857 and R21AI143296); the Cystic Fibrosis Foundation (grant numbers BROWN19I0 and BROWN19P0); and an Emory and Children’s Center for Cystic Fibrosis and Airways Disease pilot award to A. A. S. and J. B. G..
Supplement sponsorship. This work is part of a supplement sponsored by the National Institute of Allergy and Infectious Diseases (NIAID), part of the National Institutes of Health (NIH) and the Centers for Disease Control and Prevention (CDC).
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Persoon A, Heinen MM, van der Vleuten CJM, de Rooij MJ, van de Kerkhof PCM, van Achterberg T. Leg ulcers: a review of their impact on daily life. J Clin Nurs 2004; 13:341–54. [DOI] [PubMed] [Google Scholar]
- 2.Guest JF, Ayoub N, McIlwraith T, et al. . Health economic burden that different wound types impose on the UK’s National Health Service. Int Wound J 2017; 14:322–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malone M, Bjarnsholt T, McBain AJ, et al. . The prevalence of biofilms in chronic wounds: a systematic review and meta-analysis of published data. J Wound Care 2017; 26:20–5. [DOI] [PubMed] [Google Scholar]
- 4.Stacy A, McNally L, Darch SE, Brown SP, Whiteley M. The biogeography of polymicrobial infection. Nat Rev Microbiol 2016; 14:93–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perez-Vilar J, Boucher RC. Reevaluating gel-forming mucins’ roles in cystic fibrosis lung disease. Free Radic Biol Med 2004; 37:1564–77. [DOI] [PubMed] [Google Scholar]
- 6.Henke MO, Ratjen F. Mucolytics in cystic fibrosis. Paediatr Respir Rev 2007; 8:24–9. [DOI] [PubMed] [Google Scholar]
- 7.Rubin BK. Mucus, phlegm, and sputum in cystic fibrosis. Respir Care 2010; 54:726–32. [DOI] [PubMed] [Google Scholar]
- 8.Bals R, Weiner DJ, Wilson JM. The innate immune system in cystic fibrosis lung disease. J Clin Invest 1999; 103:303–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dickson RP, Martinez FJ, Huffnagle GB. The role of the microbiome in exacerbations of chronic lung diseases. Lancet 2014; 384:691–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rieber N, Hector A, Carevic M, Hartl D. Current concepts of immune dysregulation in cystic fibrosis. Int J Biochem Cell Biol 2014; 52:108–12. [DOI] [PubMed] [Google Scholar]
- 11.Yonker LM, Cigana C, Hurley BP, Bragonzi A. Host-pathogen interplay in the respiratory environment of cystic fibrosis. J Cyst Fibros 2015; 14:431–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conrad D, Haynes M, Salamon P, Rainey PB, Youle M, Rohwer F. Cystic fibrosis therapy: a community ecology perspective. Am J Respir Cell Mol Biol 2013; 48:150–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frayman KB, Armstrong DS, Grimwood K, Ranganathan SC. The airway microbiota in early cystic fibrosis lung disease. Pediatr Pulmonol 2017; 52:1384–404. [DOI] [PubMed] [Google Scholar]
- 14.Lucas SK, Yang R, Dunitz JM, Boyer HC, Hunter RC. 16S rRNA gene sequencing reveals site-specific signatures of the upper and lower airways of cystic fibrosis patients. J Cyst Fibros 2018; 17:204–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fodor AA, Klem ER, Gilpin DF, et al. . The adult cystic fibrosis airway microbiota is stable over time and infection type, and highly resilient to antibiotic treatment of exacerbations. PLoS One 2012; 7:e45001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang YJ, LiPuma JJ. The microbiome in cystic fibrosis. Clin Chest Med 2016; 37:59–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Whelan FJ, Waddell B, Syed SA, et al. . Culture-enriched metagenomic sequencing enables in-depth profiling of the cystic fibrosis lung microbiota. Nat Microbiol 2020; 5:379–90. [DOI] [PubMed] [Google Scholar]
- 18.Coburn B, Wang PW, Diaz CJ, et al. . Lung microbiota across age and disease stage in cystic fibrosis. Sci Rep 2015; 5:10241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Acosta N, Heirali A, Somayaji R, et al. . Sputum microbiota is predictive of long-term clinical outcomes in young adults with cystic fibrosis. Thorax 2018; 73:1016–25. [DOI] [PubMed] [Google Scholar]
- 20.Hahn A, Burrell A, Ansusinha E, et al. . Airway microbial diversity is decreased in young children with cystic fibrosis compared to healthy controls but improved with CFTR modulation. Heliyon 2020; 6:e04104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muhlebach MS, Zorn BT, Esther CR, et al. . Initial acquisition and succession of the cystic fibrosis lung microbiome is associated with disease progression in infants and preschool children. PLoS Pathog 2018; 14:e1006798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao J, Schloss PD, Kalikin LM, et al. . Decade-long bacterial community dynamics in cystic fibrosis airways. Proc Natl Acad Sci U S A 2012; 109:5809–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zemanick ET, Wagner BD, Robertson CE, et al. . Assessment of airway microbiota and inflammation in cystic fibrosis using multiple sampling methods. Ann Am Thorac Soc 2015; 12:221–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O’Neill K, Bradley JM, Johnston E, et al. . Reduced bacterial colony count of anaerobic bacteria is associated with a worsening in lung clearance index and inflammation in cystic fibrosis. PLoS One 2015; 10:e0126980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quinn RA, Whiteson K, Lim YW, et al. . Ecological networking of cystic fibrosis lung infections. NPJ Biofilms Microbiomes 2016; 2:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klepac-Ceraj V, Lemon KP, Martin TR, et al. . Relationship between cystic fibrosis respiratory tract bacterial communities and age, genotype, antibiotics and Pseudomonas aeruginosa. Environ Microbiol 2010; 12:1293–303. [DOI] [PubMed] [Google Scholar]
- 27.Jorth P, Ehsan Z, Rezayat A, et al. . Direct lung sampling indicates that established pathogens dominate early infections in children with cystic fibrosis. Cell Rep 2019; 27:1190–204.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goddard AF, Staudinger BJ, Dowd SE, et al. . Direct sampling of cystic fibrosis lungs indicates that DNA-based analyses of upper-airway specimens can misrepresent lung microbiota. Proc Natl Acad Sci U S A 2012; 109:13769–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu J, Carmody LA, Opron K, et al. . Parallel analysis of cystic fibrosis sputum and saliva reveals overlapping communities and an opportunity for sample decontamination. mSystems 2020; 5:e00296-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McMurdie PJ, Holmes S. phyloseq: an r package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 2013; 8:e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wei T, Simko V, Levy M, Xie Y, Jin Y, Zemla J. corrplot: visualization of a correlation matrix. R Packag, 2017. https://github.com/taiyun/corrplot. Accessed 26 October 2020.
- 32.Yuan G-X, Ho C-H, Lin C-J. An improved GLMNET for l1-regularized logistic regression. In: KDD ’11: Proceedings of the 17th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. New York: ACM Press, 2011:33. http://dl.acm.org/citation.cfm?doid=2020408.2020421. Accessed 26 October 2020. [Google Scholar]
- 33.Carmody LA, Zhao J, Schloss PD, et al. . Changes in cystic fibrosis airway microbiota at pulmonary exacerbation. Ann Am Thorac Soc 2013; 10:179–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Flight WG, Smith A, Paisey C, et al. . Rapid detection of emerging pathogens and loss of microbial diversity associated with severe lung disease in cystic fibrosis. J Clin Microbiol 2015; 53:2022–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Quinn RA, Whiteson K, Lim YW, et al. . Ecological networking of cystic fibrosis lung infections. NPJ Biofilms Microbiomes 2016; 2:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Flynn JM, Niccum D, Dunitz JM, Hunter RC. Evidence and role for bacterial mucin degradation in cystic fibrosis airway disease. PLoS Pathog 2016; 12:e1005846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aitchison J. A new approach to null correlations of proportions. Math Geol 1981; 13:175–189. [Google Scholar]
- 38.McNally L, Brown SP. Building the microbiome in health and disease: niche construction and social conflict in bacteria. Philos Trans R Soc B Biol Sci 2015; 370:20140298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bogaert D, Keijser B, Huse S, et al. . Variability and diversity of nasopharyngeal microbiota in children: a metagenomic analysis. PLoS One 2011; 6:e17035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rush ST, Lee CH, Mio W, Kim PT. The phylogenetic LASSO and the microbiome. arXiv, doi: 1607.08877, 29. June 2016, preprint: not peer reviewed. [Google Scholar]
- 41.Leng C, Tran MN, Nott D. Bayesian adaptive Lasso. Ann Inst Stat Math 2014; 66:221–44. [Google Scholar]
- 42.Banerjee P, Garai B, Mallick H, Chowdhury S, Chatterjee S. A note on the adaptive LASSO for zero-inflated Poisson regression. J Probab Stat 2018; 2018:1–9. [Google Scholar]
- 43.Jian C, Luukkonen P, Yki-Jarvinen H, Salonen A, Korpela K. Quantitative PCR provides a simple and accessible method for quantitative microbiome profiling. Plos One 2020; 15:e0227285. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.