Abstract
Ecological and evolutionary processes differ depending on how genetic diversity is organized in space. For clonal organisms, the organization of both genetic and genotypic diversity can influence the fitness effects of competition, the mating system, and reproductive mode, which are key drivers of life cycle evolution. Understanding how individual reproductive behavior contributes to population genetic structure is essential for disentangling these forces, particularly in species with complex and plastic life cycles. The widespread sea anemone, Diadumene lineata, exhibits temperature-dependent fission, which contributes to predictable variation in clonal rate along the Atlantic coast of the United States, part of its non-native range. Because warmer conditions lead to higher rates of clonality, we expected to find lower genotypic and genetic diversity in lower versus higher latitude populations. We developed primers for 11 microsatellite loci and genotyped 207 anemones collected from 8 sites ranging from Florida to Massachusetts. We found clonal influence at all sites, and as predicted, the largest clones were found at lower latitude sites. We also found genetic signatures of sex in the parts of the range where gametogenesis is most common. Evidence of sex outside the native range is novel for this species and provides insights into the dynamics of this successful invader. Our findings also illustrate challenges that partially clonal taxa pose for eco-evolutionary studies, such as difficulty sampling statistically robust numbers of genets and interpretating common population genetic metrics. For example, we found high among-locus variation in FIS, which makes the meaning of mean multilocus FIS unclear.
Keywords: invasion, latitudinal gradient, microsatellites, plasticity, reproductive mode
The spatial organization of genetic and genotypic variation can have a profound influence on the ecology and evolution of a species (Grosberg and Cunningham 2001). For clonal organisms, the spatial arrangement of ramets, the term for an individual unit of a modular or clonal organism, can affect the fitness of the genet (Vallejo-Marin et al. 2010), the collective term for all ramets derived from a sexually produced zygote (sensu Harper 1977). Aggregations of clones can serve as a buffer for individual ramets against physical stress (Ayre 1984) or competition (Williams 1975; Francis 1988), but can also increase vulnerability to parasites (King and Lively 2009), predation (Ives et al. 1993), or disease (Vollmer and Kline 2008). Conversely, ramets that become widely dispersed through active movement or from surviving dislodgment can better avoid genet-level extinction by spreading the risk over a larger area (Wulff 1991; Coffroth and Lasker 1998). Yet, in a partially clonal species with separate sexes, aggregation coupled with high rates of clonal proliferation can reduce the pool of potential mates by saturating the immediate environment with ramets of the same sex, thereby creating a physical barrier to sexual reproduction (Ayre and Grosberg 2005; Vallejo-Marin et al. 2010). The degree to which clonal growth influences local population structure depends on individual rates of investment in asexual and sexual reproduction and dispersal behavior, as well as ecological interactions that influence growth and mortality (Baums et al. 2006; Barrett 2015). Describing the relationship between reproductive traits and spatial population structure is essential to understanding how and why clonality evolves.
Theory on the evolution of clonality often invokes direct (e.g., energetic) and indirect (e.g., spatial competition, genetic diversity of offspring) trade-offs that govern the fitness benefits of investing in asexual versus sexual reproduction (e.g., Williams 1975; Maynard-Smith 1978; Jackson and Coates 1986). Data have accumulated on the energetic costs and benefits of clonal growth. For example, individuals have been shown to escape energetic constraints caused by allometric surface area to volume scaling by undergoing fission, which allows the continual production of optimally sized ramets (Sebens 1979; Ryan et al. 2019). Thus, the balance of fission and individual growth can contribute to local adaptation (Sebens 1980, 1982; Edmunds 2007) and temperature acclimation (Ryan 2018, Ryan et al. 2019). The specific trade-offs that govern the energetic value of clonal and colonial behavior differ among taxa and depend on a suite of morphological and physiological traits (reviewed by Burgess et al. 2017). Meanwhile, testing hypotheses about the selective feedback on life cycle traits caused by the spatial organization of genetic diversity of clonality has been more difficult.
Many theoretical predictions about the conditions favoring clonal versus aclonal life cycles (reviewed Jackson and Coates 1986 for marine invertebrates; Vallejo-Marín et al. 2010 for seed plants) rely on the assumption of a strong correlation between individual life cycle expression (e.g., rate of clonality) and the genetic structure of the local neighborhood. The spatial arrangement of genetic diversity influences mating opportunities and species interactions, thereby shaping the fitness value of various reproductive strategies. However, empirical evidence demonstrating a correlation between life cycle variation and population genetic structure has been mixed. There is ample evidence that clonal marine organisms across taxa can form large clonal aggregations, including in seagrass (Ruckelshaus 1998; Arnaud-Haond et al. 2012), macroalgae (van der Strate et al. 2002; Guillemin et al. 2008; Krueger-Hadfield et al. 2016), microalgae (Krueger-Hadfield et al. 2014), sponges (Wulff 1986; Calderón et al. 2007), bryozoans (Jackson and Hughes 1985), and many cnidarian groups, such as stony corals (Baums et al. 2006), gorgonians (McFadden et al. 1997), and sea anemones (Francis 1973; Sherman and Ayre 2008), including Diadumene lineata (Verrill 1869) (Shick and Lamb 1977; Ting and Geller 2000). However, populations do not always conform to simple expectations for the relationship between the potential for asexual investment and population genetic structure (e.g., McFadden et al. 1997 in soft coral, Bellis et al. 2018; Titus et al. 2017, in sea anemones; Krueger-Hadfield et al. 2011, 2016, in macroalgae). For example, Titus et al. (2017) found remarkably few repeated multilocus genotypes in a population of Bartholomea annulate that carpeted the walls of a submerged rock quarry even though this species is capable of pedal laceration. In addition, differences in clonal population structure are often interpreted as resulting from environmental gradients that alter patterns of density and mortality, as well as sexual recruitment success and/or fragmentation (e.g., Wulff 1991; Baums et al. 2006; Pinzón et al. 2012). As is well-appreciated by all of these authors, it is difficult to disentangle the combined influence that life cycles, life-history traits, environmental gradients, and ecological processes have on genotypic and genetic structure. However, understanding how and why clonal life cycles evolve requires disentangling these processes.
We used a widespread, clonal sea anemone to test the hypothesis that variation in the rate of asexual reproduction correlates with variation in clonal population structure across a latitudinal gradient. Diadumene lineata, the orange-lined anemone, is a successful non-native species thought to have originated in East Asia (Uchida 1932). Over the last 150 years, D. lineata has become the most widespread sea anemone in the world (Fautin 2002) probably due to a predisposition for opportunistic colonization (Glon et al. 2020) and associations with oyster aquaculture and transoceanic cargo vessels (Verrill 1898; Stephenson 1925; Gollasch 2002). It has colonized temperate estuarine habitats across the globe, including on the Atlantic, Gulf, and Pacific coasts of North America. Where it has been studied in detail, it exhibits a partially clonal life cycle structured by seasonal environmental patterns. Gametogenesis begins in late winter in separate males and females, culminating in the spawning of mature gametes in the summer (Fukui 1991, 1995), though the timing of maturity varies among regions (W.H.R., unpublished data). The probability of producing gametes and the number of gametes produced depend on body size (Ryan and Miller 2019). Larvae have rarely been observed, but are capable of strong swimming for up to 2 weeks in laboratory conditions (Fukui 1991), though are competent to settle within 4 days of fertilization (W.H.R. and S.A.K.-H., unpublished data). Adults of the species are capable of rapidly gliding across surfaces by pedal undulation and can detach and drift in currents, potentially as a means of escaping unfavorable conditions as has been observed in other small anemones (Bedgood et al. 2020). Thus, this species possesses the capacity for dispersal at the gamete, larval, and adult stages, in addition to its tendency for anthropogenic transport.
Diadumene lineata is also capable of prolific clonal growth through both binary fission (Miyawaki 1952; Minasian 1979) and pedal laceration (Atoda 1973). The rate of fission is strongly influenced by environmental factors including immersion time (Johnson and Shick 1977), salinity (Podbielski et al. 2016), food availability (Minasian and Mariscal 1979), and, most dramatically, temperature (Atoda 1973; Minasian 1979; Ryan 2018). When grown in laboratory conditions mimicking seasonal water temperature changes in the northeastern United States, individuals produce 2–4 clonal ramets per year whereas those in seasonal conditions mimicking the southeastern United States can produce up to 17 ramets per year (Ryan 2018). Ryan (2018) also demonstrated the existence of cogradient genetic variation in the temperature sensitivity of fission where genotypes from the Gulf Coast of Florida divide more frequently than individuals from the cooler waters of Georgia and Massachusetts when reared at the same temperature. Along the North American Atlantic and Gulf coastlines, mean annual sea surface temperature is tightly correlated with latitude. Thus, both genetic and environmental differences among sites probably contribute to a gradient in life cycle expression. Individuals in northern populations probably stay larger bodied and reproduce mainly through sex, whereas southern populations are highly clonal. Intermediate populations appear to alternate seasonally between unitary growth with gametogenesis and periods of rapid binary fission (Ryan 2018; Ryan and Miller 2019; W.H.R. and S.A.K.-H., unpublished data). If variation in life cycle expression leads to standing differences in the spatial structure of genetic and genotypic diversity, then we expect to see population attributes vary across latitude, including 1) lower latitude, high fission sites should show reduced genotypic richness and evenness relative to intermediate and higher latitude sites, due to the presence of larger clones (i.e., more ramets per genet), and 2) genotypic diversity and evidence of genetic similarity within sites should increase with latitude due to an increase in the relative contribution of sexual reproduction and recruitment to populations.
Previous work describing the genetic structure of D. lineata populations has produced mixed results. Several authors have suggested that low diversity populations are common outside of the native range and that sexual reproduction is rare (Parker 1902; Hausman 1919; Stephenson 1925; Williams 1973; Shick and Lamb 1977). Where populations have been investigated in the non-native range with genetic markers, however, monoclonal sites have been rare (allozymes: Shick and Lamb 1977; single-strand conformation polymorphism: Ting and Geller 2000). Ting and Geller (2000) found that, unlike the well-known colony-forming anemone Anthopluera elegentissima, D. lineata ramets belonging to multiple genets were commonly found intermingled at small scales. However, the haphazard sampling schemes used in previous studies limit the utility of these data to test hypotheses about geographic patterns of genetic variation and connectivity. Likewise, previous studies have not provided conclusive evidence on the prevalence of sexual reproduction in this species, inside or outside the native range. Recent studies in North America have documented populations where fertile males and females co-occur within centimeters of each other on both the Pacific (Newcomer et al. 2019; W.H.R. and S.A.K.-H., unpublished data) and Atlantic coasts (Ryan and Miller 2019), providing circumstantial evidence of sexual reproduction in the invaded range. However, the relative contributions of sexual and asexual reproduction had not been systematically described for any population prior to this study.
In addition, general patterns of relatedness among populations were unknown across the invaded range of this species. Evidence from other species suggests that invasion processes themselves can influence genetic diversity in several interesting ways. For example, bottlenecks imposed by low propagule pressure or high levels of disturbance could be responsible for low genetic diversity at some sites, although this is rarely found among widespread invasive species (reviewed by Roman and Darling 2007). Patterns of anthropogenic transport could lead to either a panmictic shuffling of genotypes during invasion or a nonrandom partitioning of genotypes sites; both patterns that have been found among widespread species of colonial tunicates (Lejeusne et al. 2011). Environmental filtering could also lead to geographic partitioning of genetic diversity, even for species with the potential for global dispersal. This phenomenon is demonstrated by the distribution of genetically distinct clades of the invasive, colonial bryozoans in the genus Watersipora, where the temperature envelope of a site is a better predictor of the success of each clade than geographic proximity, even when comparing across continents (Mackie et al. 2012).
Several processes can also contribute to deviations from expected Hardy–Weinberg allele frequencies, making it difficult to infer mechanisms from observed patterns in non-native species. For example, clonal genets of anthropogenically transmitted species can persist over long periods and become widespread without engaging in sex, as has been documented in water fleas (Mergeay et al. 2006) and apomictic crayfish (Gutekunst et al. 2018). Genetic or ecological incompatibility among individuals from different source populations can also lead to maintenance of independent, sexually reproducing lineages despite physical co-occurrence as seen in tunicates (Ordóñez et al. 2013). If and when sex does occur, progeny may show high heterozygosity if independent introductions brought together long isolated genetic lineages of a species, or high homozygosity due to the limited genetic diversity of founders (Roman and Darling 2007). Furthermore, opportunities for biparental inbreeding could be particularly high at sites dominated by clonal proliferation. Clonality can allow genets to persist at a site for many reproductive seasons, increasing the chance for parent–offspring and between-sibling matings as well as the competitive exclusion of other potential mates (Barrett 2015). Finally, colonization and extinction dynamics within a site can lead to genetic patchiness (Pannell 2015), particularly when novel alleles may be added periodically through waves of invasion. Thus, for a widespread, clonal, and sessile species like D. lineata, which colonizes dynamic substrata in high disturbance environments, observed genetic patterns are likely the result of multiple interacting processes. In particular, since we cannot assume that co-occurring individuals share an ancestral or contemporary history of genetic exchange, we approached the interpretation of the population genetic metrics and patterns with caution in this study.
Here, we developed 11 microsatellite loci with which to examine the contribution of clonal growth to the structure of genetic and genotypic diversity in populations of D. lineata using a hierarchical sampling approach at 8 sites encompassing 3 nested spatial scales: within 0.25 m2 quadrats, within beaches, and within biogeographic regions. The span of the study (from Florida to Massachusetts) covered much of the species range across the North American Atlantic Coast. Across this range, we found population genetic patterns that largely conformed to predictions based on geographic variation in the expression of clonality and gametogenesis, although high levels of clonality found across sites limited the number of genets available on which we could base sophisticated analyses.
Materials and Methods
Microsatellite Development
Single sequence repeat (SSR)-enriched genomic sequence data was generated by ECOGENICS (Balgach, Switzerland) using proprietary enrichment techniques for microsatellite motifs from 6 individual anemones preserved in 95% ethanol. Individuals representing the presumed native Japanese range and 2 invaded coastlines (Supplementary Table 1A) were used to increase the likelihood of designing primers that would be adequate for all populations of this widespread species and to avoid ascertainment bias. We used MSATCOMMANDER (Faircloth 2008) to identify and design primers for all regions in the library containing di- to tetra-nucleotide repeat motifs. We applied a set of quality filters to the pool of potential primers using a protocol modified from Schoebel et al. (2013). First, we excluded sets with a high pair penalty or dissimilar melt temperatures. We then removed any primers that matched more than one nonsynonymous amplicon in the SSR-enriched sequence data to minimize the likelihood of selecting primers that amplified multiple regions of the genome. Last, we visually examined the amplicon region between each primer pair in GENEIOUS PRIME v. 2020.0.5 (Biomatters, Ltd., Auckland, New Zealand). Primer pairs with short flanking regions (<20 bp) between the primers and repeat region were excluded to minimize the likelihood of repeat sequences interfering with primer binding. More than 50 potential primer pairs remained after the most stringent filtering was applied.
Of these, 39 primer pairs were screened by PCR amplification across a panel of DNA extracted from 7 new individuals representing the species geographic spread (Supplementary Table 1B). Total genomic DNA was extracted from either a 2 mm3 section of pedal disk, tentacle crown, or whole individual (depending on body size) with a QIAGEN Blood and Tissue Kit (Qiagen Sciences, Germantown, MD) following a 24-h incubation at 60 °C in Proteinase K. We amplified the loci using either a SimpliAmp or ProFlex thermocycler (Applied Biosystems, Foster City, CA) and the following program: 5 min at 95 °C followed by 35 cycles of 95 °C for 30 s, 56 °C for 30 s, 72 °C for 30 s, followed by a final extension at 72 °C for 5 min. The PCR contained 1× buffer, 1.5 mM MgCL2, 200 nM dNTPs, 1 mg/mL bovine serum albumin, 1 U of GoTaq (Promega, Madison, WI), 250 nM forward and reverse primers (IDT, Coralville, IA), and 2 µL of DNA (diluted 1:100). Primers that did not amplify well under these conditions were run again with slightly lower and higher annealing temperatures, but performance for most was optimal around 56 °C. Amplification was determined by visually inspecting bands isolated by gel electrophoresis of PCR products loaded on 1.5% agarose gels stained with GelRed (Biotium, Fremont, CA).
For loci that amplified well (i.e., produced 1–2 bright bands in the expected size range) across all 7 samples, a fluorochrome-tagged forward primer (using either 6-FAM, VIC, or NED) was used in place of the unlabeled forward primer in the PCR mix described above. Thermocycler settings remained the same. For fragment analysis, 1 µL PCR product was diluted in 10 µL of HiDi formamide (9.7 µL) and GeneScan 500 LIZ (0.35 µL; Applied Biosystems, Foster City, CA). Fragment analysis was conducted on an ABI 3730xL genetic analyzer equipped with a 96-capillary array at the Heflin Genomics Core at the University of Alabama at Birmingham. Alleles were manually scored in GENEIOUS Prime using the Microsatellite plugin v1.4.6 (Biomatters Ltd., Auckland, New Zealand). This process yielded primers for 11 loci (Supplementary Table 2) that were used in subsequent analyses.
Study Sites
Diadumene lineata occurs on natural and man-made hard substrate in the high intertidal zone of protected shores across the Atlantic and Gulf Coasts of North America. Twenty-four to 30 individuals from each of 2 sites in each of 4 geographic regions were preserved for genetic analysis between June and July of 2016. Coastline distance between sites within region, as approximated with the measurement tool in Google Earth (Mountain View, CA) ranged from ~1 to 45 km; sites in adjacent regions were on the order of 1000–1500 km apart following the coastline (see Table 1 for site details). The mean annual water temperature at each site was approximated from 3 years of daily temperature data collected from the nearest available NOAA buoy (details available in Ryan and Miller 2019).
Table 1.
Sampling locations for biogeographic comparison across North American Atlantic Coast
| Region | Site ID | Location | Location notes | Latitude | Longitude | Primary substratum | Eggs | Sperm |
|---|---|---|---|---|---|---|---|---|
| Florida | CML | St Teresa | FSU Coastal and Marine Laboratory | 29.91506 | −84.5082 | Sedimentary cobble on protected sandy beach | Y | Y* |
| WAK | Wakulla County | Wakulla Beach | 30.10236 | −84.25895 | Oyster beds | Y* | Y* | |
| Georgia | STS | St. Simon Island | King’s Beach | 31.13334 | −81.39427 | Breakwater rocks | Y* | Y |
| JEK | Jekyll Island | Fishing Pier | 31.11761 | −81.4165 | Isolated dead trees/driftwood | Y | Y | |
| Virginia | RBB | Birdsnest | Red Bank boat ramp | 37.44566 | −75.83993 | Wooden pier beams | Y | Y |
| QBY | Quinby | Quinby Harbor | 37.55274 | −75.7291 | Rotten wood posts in salt marsh | Y* | Y | |
| Massachusetts | NAH | Nahant | No Name Point | 42.41977 | −70.90338 | Submerged in rock pools | Y* | Y* |
| PMH | Nahant | Pumphouse Beach | 42.41729 | −70.90528 | Submerged in rock pools | N | Y* |
All samples collected between June and July 2016 by WHR. See Ryan and Miller (2019) for additional site details. Eggs/sperm columns indicate sites where fertile individuals bearing each gamete type have been found, either during the 2016 transect sampling (Y) or during another sampling in the same area before or since (Y*).
Collecting and Preserving Material
At each site, a transect was established parallel to the shore in the upper intertidal zone during low tide. In high-density sites, up to 20 individuals were haphazardly collected with forceps from each of five to six 0.25 m2 quadrats spaced approximately 5 m apart along the transect. At lower density sites, all anemones found in each quadrat were collected. At WAK, the second quadrat garnered only a single individual, so a sixth quadrat was searched at the next 5-m interval along the transect. In 2 cases, JEK and PMH, anemones were only found in 4 quadrats with subsequent quadrats yielding no individuals. An average of 68.8 ± 9.5 (standard error [SE]) individuals were collected per site. Individuals were kept alive in tubes of chilled seawater to prevent fission and returned to the lab. After a 2- to 3-day period to allow egesting of gut contents, 1–8 individuals (depending on availability) were haphazardly chosen from each quadrat sample for a total of 24–30 individuals per site. To the extent possible, samples were drawn equally from all quadrats. Individuals were anesthetized in equal parts 7.5% MgCL2 and seawater and dissected to remove any foreign debris from the gut and the foot. Wet weight and gametic state were recorded for each individual (described in Ryan and Miller 2019). Samples were then individually preserved in 95% ethanol.
Generating Multilocus Microsatellite Genotypes for Sampled Individuals
Total genomic DNA was extracted from preserved samples as described above. PCR was used to amplify the 11 microsatellite loci for each individual, followed by fragment analysis as described above. Raw allele peak size in base pairs was manually scored in GENEIOUS Prime. TANDEM (Matschiner and Salzburger 2009) was used to bin raw allele sizes into integer values (see Krueger-Hadfield et al. 2013, 2016). We calculated null allele frequency using the Em method (Dempster et al. 1977) in Genepop v4.7 (Rousset 2008). These estimates come with the caveat that the small number of genotypes recovered from some sites coupled with the potential for biparental inbreeding in these populations mean that null allele estimates are likely unreliable for the current data set, as these attributes break the assumptions of Hardy–Weinberg equilibrium. Other studies have shown that inbreeding can lead to poor estimation of null allele frequencies. For example, Krueger-Hadfield et al. (2011, 2013) genotyped haploid and diploid individuals of a haplodiplontic seaweed. Repeated PCR failure, presumably due to null alleles, occurred in less than 5% of haploids. Meanwhile, null allele frequencies estimated with maximum likelihood in diploids showed much higher values (e.g., 20% for the same locus). This discrepancy was likely due to an overabundance of homozygotes due to inbreeding (Krueger-Hadfield et al. 2013, 2015). Thus, although null alleles may contribute to greater estimates of homozygosity, the existence of high homozygosity is not necessarily indicative of null alleles since biological mechanisms can generate such patterns (see also Olsen et al. 2020).
Identifying Repeated Genotypes
With a sufficient number of polymorphic markers, the likelihood of repeated multilocus genotypes (MLGs) arising through sex rather than clonality becomes diminishingly small. We used the Psex function in GENCLONE v2.0 (Arnaud-Haond and Belkhir 2007) to calculate the probability that repeated MLGs arose from independent sexual events. This function assigns a P-value to each instance of a pair of repeated MLGs with which to assist in deciding whether a pair of ramets sharing a repeated MLGs should be treated as clonemates or as derived from a separate zygote (see other studies using this metric: Arnaud-Haond et al. 2007; Guillemin et al. 2008; Krueger-Hadfield et al. 2016). At sites with low genotypic diversity, this metric can be less reliable. However, given the polymorphism of our 11 microsatellite markers, the risk of an erroneous assignment was low.
Two data sets were constructed for analyses: 1) a ramet-level data set, retaining genetic information for all genotyped anemones and 2) a genet-level data set, where only one copy of each MLG, as determined by Psex, was retained. The ramet-level data set reflects an ecological view of the population, where ramet distribution among MLGs, ramet distribution in space, and trait variation among ramets within a genet are important characteristics for understanding population performance. The genet-level data set (ignoring any complications from somatic mutation for the moment) attempts to meet the assumptions of underlying population genetic models, where the number of ramets representing a genet does not affect the relative frequency of alleles. In this view, variation in ramet number among genets was treated analogously to body size variation among individuals in a unitary species—regardless of body size, each MLG is only counted once.
Describing Genotypic Diversity and the Relative Contribution of Clonality Among Sites
Clonal population structure is described by both the arrangement of ramets in space and the richness and evenness of genotypes present in a population. Together, these metrics can describe the relative contribution of asexual reproduction to population structure (Baums et al. 2006). We calculated the Pareto index (β) in the RClone package (Bailleul et al. 2016) for the statistical software R v 3.5.1 (R Core Team 2018) to compare the distribution of clone sizes among sites. Linear regression was used to measure the correlation of β with mean annual water temperature for each region, a strong correlate of latitude. We also measured genotypic richness (R; sensu Dorken and Eckert 2001) in the ramet-level data set as R = (G − 1)/(N − 1), where G is the number of unique MLGs and N is the number of genotyped anemones. Genotypic richness varies from 0 to 1. The index equals zero when all ramets share the same MLG and one when each ramet has a unique MLG (i.e., all individuals are unitary). We calculated evenness in the relative representation of MLGs among ramets as Dominance (D: sensu Pielou 1969), a commonly used metric in the plant literature, which is defined as follows:
where ni is the relative frequency of the ith MLG. The index D varies from 0 to 1 such that D approaches zero as populations become dominated by the ramets of one MLG. When each MLG is represented by one ramet, D = 1. Genotype richness and dominance are positively correlated such that potential values for a given sample size describe a region in plot space that approximates a positive saturating function. Populations mapped along this curved region provided a visual representation of the relative contribution of clonal reproduction to the spatial structure of genotypic diversity at a site (adapted from Baums et al. 2006). Sites falling in the region of high genotypic richness and dominance values have also been used as evidence of higher relative levels of sexual reproduction (e.g., Baums et al. 2006; Pinzón et al. 2012). In a species with an unknown dispersal history among sites, such as in non-native populations of D. lineata, ascribing causes to diversity patterns must be approached with caution.
To ensure that sample size differences did not distort comparisons among sites, the genotypic richness was rarified to the smallest sample size (Rrare) using the rarefy function in R with VEGAN package (Oksanen et al. 2019). Then a normalized D value (Drare) was estimated from MLG frequency values normalized to rarified genotypic richness to minimize bias due to sample size differences. The relationship between rarefied genotypic richness and mean annual water temperature was assessed with a least means linear regression.
Describing Genetic Diversity
Standard population genetic metrics were calculated for both the ramet- and genet-level data sets. These included rarified allelic richness (AE) estimated from 1000 bootstrapped subsamples matching the lowest sample size (genet level: 21 individuals or 42 alleles; ramet level: 2 individuals or 4 alleles) in HP-Rare (Kalinowski 2005), expected and observed heterozygosity (HE and HO) in GenAlEx v. 6.501 (Peakall and Smouse 2006, 2012), the inbreeding coefficient (FIS) estimated in FSTAT (Goudet 1995) using 1000 permutations, and the standardized index of association accounting for linkage disequilibrium estimated in the R package poppr (Kamvar et al. 2014, 2015). Geographic patterns in these metrics were also explored with linear regressions against mean annual water temperature.
Describing Spatial Patterns in Clonal Aggregation and Autocorrelation
The distribution of genotypic diversity in space, including the spatial arrangement of ramets from the same genet, is expected to have ecological consequences (Hughes et al. 2008) as well as influence the relative fitness effects of sexual and asexual reproduction (Vallejo-Marín et al. 2010). We described spatial patterning in several ways. An index of clonal aggregation (Ac) was calculated for each site to determine the extent to which clonemates, or ramets, tend to cluster in space. Ac = (Psg − Psp)/Psg, where Psg is the average prevalence of clonality at a site and Psp is the average pairwise probability that neighbors do not belong to the same genet (Arnaud-Haond et al. 2007). In this case, position along the transect is used to estimate neighbor distance. The index is zero when clonemates are randomly distributed across the site, approaches negative one when overdispersed and one when aggregated. To assess whether genetically similar individuals occur closer together at the site scale, we also measured spatial autocorrelation across transects within sites with Ritland’s index (Ritland 1996) implemented in the autocorrelation function in RClone. Neighboring anemones can be genetically similar because they share a clonal parent (e.g., clonemates) or because they share a common sexual ancestor (e.g., siblings). To distinguish among these influences, we first calculated autocorrelation among all anemones, including the effect of clonemates. Then, we estimated spatial autocorrelation using a weighted matrix in RClone, which minimizes the influence of clonal aggregation in order to estimate the spatial correlation of nonclonal relatives. Since no MLG was found at more than one site, no between-site measures of genet spread were necessary.
We also estimated the mean maximum observed pairwise distance in meters between ramets of the same genet, excluding any genet with only one ramet. Since this species occurs in a narrow band of intertidal heights, the distance among quadrats described by a linear transect mirrors a key dimension for small-scale dispersal along the shore. While there is some variation in the exact spacing of the quadrats due to local topography, quadrats at all sites form a distance ranked set of observations spread across a similar distance (~20 m). To simplify analyses, the distance between adjacent quadrats was fixed at 5 m. The first quadrat at WAK was excluded from clonal spread analyses so as not to bias averaged measurements by including a longer transect at only one site. Geographic patterns in clonal spread by mean annual water temperature were assessed by Spearman’s Rho rank order correlation, since the distribution of distances measured showed a strong right skew.
Describing Patterns of Genetic Similarity and Connectivity Among Sites
The distribution of genetic diversity within and among sites can help identify patterns of genetic isolation and connectivity that may help identify sites with shared historical invasion patterns and or contemporary gene flow. Patterns of genetic similarity among nonclonemates may also be an important indicator of sites where sexual reproduction and recruitment are (or are not) occurring. To understand the structure of genetic diversity across sites, FST was calculated in GENODIVE (Meirmans and Van Tienderen 2004) to identify significant population differentiation. We also tested the hypothesis of isolation by distance with a Mantel test in GENODIVE, using along-coastline distances estimated with Google Earth©.
To visualize patterns of genetic similarity, principal component analysis was run on a matrix of genetic distance calculated from the genet-level data set. To identify naturally occurring clusters of genetically similar genotypes, we used the find.clusters function in the R package adegenet (Jombart 2008). Because of the unknown pattern of invasion of D. lineata on the North American Atlantic Coast, and the expectation that unrelated individuals may co-occur for long periods without interbreeding, we deliberately chose to examine population similarity with clustering methods that do not assume a hierarchical diversity structure across spatial levels, or rely on assumptions about Hardy–Weinberg frequencies to assign group membership (e.g., STRUCTURE, Pritchard et al. 2000). The benefits and limitations of programs with such assumptions are discussed in depth elsewhere (e.g., Jombart et al. 2010; Kalinowski 2011).
Results
Developing Microsatellite Markers
Of the 39 primer pairs tested, 29 amplified in all test samples. We designed fluorescent dye-tagged primers for 19 loci, of which 18 had clear and repeatable peak structure. All 18 were polymorphic across the panel of 7 test individuals. From this pool, we chose a set of 11 polymorphic primers to use for data generation as this number was sufficient to distinguish all clones with high statistical confidence. Across all individuals, the number of allelic variants per locus ranged from 3 to 17 (Supplementary Table 2). The number of usable loci discovered from this process is consistent with microsatellite development efforts for other taxa (e.g., Krueger-Hadfield et al. 2011; Arnaud-Haond et al. 2013; Warwick and Lemmon 2014; Kollars et al. 2015).
Generating Multilocus Microsatellite Genotypes for Sampled Individuals
A total of 227 individuals were collected and preserved from 74 0.25 m2 quadrats across 8 sites. Total genomic DNA was successfully extracted from 225 of 227 preserved individuals. We failed to generate a complete 11-locus MLG for 27 individuals. Of these, PCR failed at multiple loci for 10 individuals perhaps indicating poor DNA quality. For 17 others, PCR failed at a single locus. Among these, Dlin_30 failed to amplify in 10 cases, primarily concentrated in the Massachusetts (N = 7) and Virginia sites (N = 3). We interpreted this pattern as the result of a null allele and chose to omit Dlin_30 from subsequent analyses. No pattern was apparent among the failed loci from the 7 remaining individuals. Repeating the PCR did not improve the outcome; thus, the 7 individuals were removed prior to analyses. Analyses were run on a data set of complete, 10-locus MLGs constructed for 207 individuals across the 8 sites. The number of individuals with complete data from each site ranged from 21 (PMH) to 30 (RBB) (Table 2).
Table 2.
Metrics of genotypic diversity across 8 populations of Diadumene lineata
| Region | Site | N | G | R | D | G rare | R rare | D rare | Genet spread | A c | Pareto β |
|---|---|---|---|---|---|---|---|---|---|---|---|
| FL | CML | 25 | 3 | 0.08 | 0.55 | 3.00 ± 0.01 | 0.10 | 0.59 | 7 ± 2.94 | 0.92*** | 0.29 |
| FL | WAK | 26 | 2 | 0.04 | 0.08 | 1.81 ± 0.39 | 0.04 | 0.10 | 10.5 ± 9.5 | 0.94 | 0.01 |
| GA | STS | 27 | 5 | 0.15 | 0.34 | 4.29 ± 0.71 | 0.16 | 0.35 | 4.8 ± 1.9 | 0.88 | 0.06 |
| GA | JEK | 23 | 7 | 0.27 | 0.70 | 6.65 ± 0.52 | 0.27 | 0.69 | 4.14 ± 0.66 | 0.86 | 0.25* |
| VA | RBB | 30 | 6 | 0.17 | 0.69 | 5.08 ± 0.77 | 0.19 | 0.69 | 6.5 ± 1.58 | 0.89 | 0.24 |
| VA | QBY | 28 | 7 | 0.22 | 0.79 | 6.43 ± 0.64 | 0.26 | 0.81 | 1.57 ± 0.23 | 0.97*** | 0.39* |
| MA | NAH | 27 | 5 | 0.15 | 0.77 | 4.78 ± 0.42 | 0.18 | 0.76 | 4.8 ± 1.9 | 0.98*** | 0.35 |
| MA | PMH | 21 | 4 | 0.15 | 0.73 | 4.00 ± 0 | 0.14 | 0.73 | 4.25 ± 1.23 | 0.94*** | 0.13 |
| Total | 207 | 39 | |||||||||
| Mean ± SE | 25.9 ± 1.0 | 4.9 ± 0.7 | 0.15 ± 0.03 | 0.58 ± 0.09 | 4.51 ± 0.62 | 0.17 ± 0.03 | 0.59 ± 0.09 | 5.45 ± 0.93 |
N, number of anemones genotyped; G, number of MLGs based on Psex; D, clonal dominance, a measure of genotypic evenness; Grare, number of genotypes estimated after rarefaction to 21 anemones, the number observed at PMH; Rrare, genotypic richness based on rarefaction to NPMH = 21; Drare, normalized clonal dominance estimated from MLG frequencies normalized to NPMH = 21; genet spread, mean of the maximum observed distance between clonemates in meters; Ac, the clonal aggregation index; Pareto β, the slope of the cumulative distribution function of genet size. SE, standard error.
Significant P-values are shown in bold with an *<0.05, ***<0.001.
Em analysis was performed to estimate the probability of null allele across all loci; however, the results for populations with such low genotypic richness are not reliable. The majority of estimates across sites and loci were zero or near zero (Supplementary Table 3); however, there was insufficient data to estimate confidence intervals for all but 2 sites at one locus; both STS and JEK showed high estimates of the probability of null alleles at locus Dlin_40 (0.66 and 0.77, respectively). Although null alleles may contribute, this number may also be artificially inflated by small sample sizes at a locus with few allelic variants possible. High homozygosity can also be attributed to inbreeding, which is likely in these populations, especially where gametes are produced but genotypic diversity is low (see also Krueger-Hadfield et al. 2011; Olsen et al. 2020). Direct methods such as progeny arrays (Oddou-Muratorio et al. 2009) will likely be required to estimate the frequency of null alleles for these loci in this species.
Identifying Repeated Genotypes and Describing Genotypic Diversity
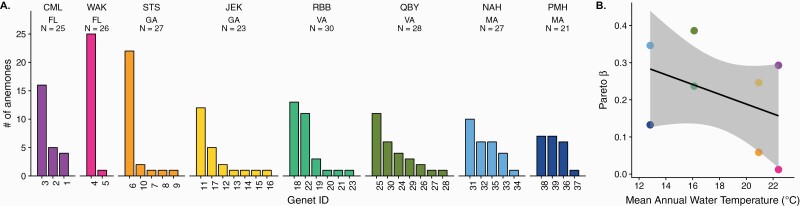
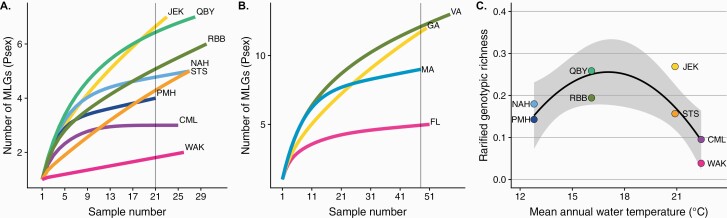
Across 8 sites, 39 unique MLGs were found in 207 individuals. The probability of a repeated MLG arising through sex was found to be extremely low (P < 0.0001) for all repeated MLGs, confirming that our markers were sufficiently polymorphic to distinguish true clones. Repeated MLGs were found at all sites, but both the number of MLGs and number of ramets per MLG differed among sites (Figure 1A, Table 2). Southern sites were dominated by a small number of large clones, whereas northern sites showed a more even distribution of ramets among genets. The largest clone was found at Wakulla Beach, Florida (WAK), where 25 of 26 anemones genotyped belonged to a single genet. The most genotypically diverse sites (JEK and QBY) each had 7 unique MLGs among 23 and 28 anemones genotyped. No MLG was found at more than one site. The slope of the relationship between the β Pareto index, which describes the inverse slope of the cumulative distribution of ramets among genets, and mean annual water temperature was negative but not significantly different from zero (slope = −0.013, P = 0.33, R2 = 0.02, Figure 1B). This metric is most useful for identifying small differences in clonal rate among populations that are primarily sexual; it has been shown to be fairly insensitive to large differences in clonal rate among populations with intermediate to high clonal rate (Stoeckel et al. 2019). To further facilitate comparison among sites, the number of MLGs and genotypic richness (R) were rarefied through bootstrapped simulations (Supplementary Figure 1) to the sample size at the site with the lowest number of individuals with complete data, NPMH = 21 (Figure 2A). Regionally, genotypic richness was highest in Virginia and lowest in Florida (Figure 2B). The relationship between mean annual water temperature and rarefied genotypic richness was best described by a second-order polynomial showing a peak between Georgia and Virginia (R2 = 0.44, P1 = 0.267, P2 = 0.058; Figure 2C).
Figure 1.
Distribution of ramets among MLGs by site. Colors reflect region origin, ordered left to right from low to high latitude (Florida [pink]: CML, WAK; Georgia [yellow]: STS, JEK; Virginia [green]: RBB, QBY; Massachusetts [blue]: NAH, PMH). (A) Bar heights indicate the number of anemones belonging to each unique MLG, as confirmed by Psex. Genet ID numbers were assigned arbitrarily to unique multilocus combinations. (B) The Pareto β value quantifies the slope of the distribution of ramets among MLGs as a measure of evenness, which varies across the mean annual water temperature measured at each site. Genet ID numbers and site/region colors are consistent among figures.
Figure 2.
Patterns of MLG richness among sites. Accumulation curves of the number of unique multilocus genotypes (MLGs), as estimated with Psex, over number of anemones genotypes in (A) each population or (B) pooled across latitudinal region. Rarefied estimates of the number of MLGs at each site or region (gray line) were generated through bootstrapped simulation where the number of sampled individuals equaled that of the lowest site or regional sample size. (C) Rarified genotypic richness ([Grare − 1]/[Nrare − 1]) reflects the rate at which novel genotypes are encountered in sampling a partially clonal population, which varies with mean annual water temperature measured at each site. Site and region colors are consistent among figures and reflect latitudinal position such that south to north order is designated by warm to cool colors.
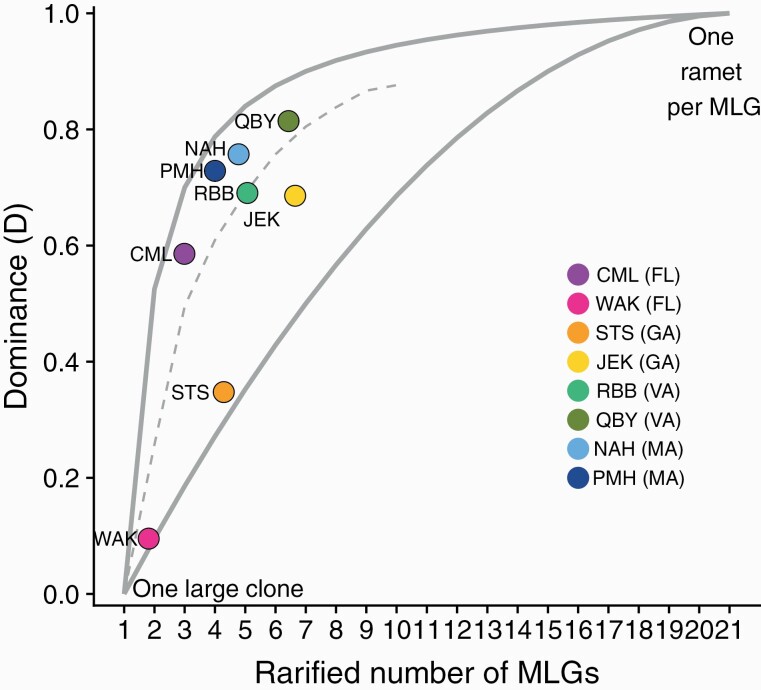
Plotting the interaction of rarefied genotypic richness and dominance (D) showed a spread in the relative contribution of asexual reproduction among sites (Figure 3). There is a strong signal of clonality at all sites, with no population approaching the region of the curve expected if sex was the primary means of reproduction (i.e., all genotyped anemones have unique genotypes). Both Wakulla Beach, FL (WAK) and St. Simon Beach, GA (STS) stand out as being dominated almost entirely by a single clone, followed by St. Teresa, FL (CML) represented by one large clone mixed with 2 other genets. The other sites form a cluster in the region of the graph expected to describe populations where a combination of sexual and asexual processes contribute to genetic structure (Baums et al. 2006). The proximity of each site to the dashed line in Figure 3 illustrates the direction of deviation from an expected median given a random distribution of ramets among MLGs. Among these, Jekyll Island, Georgia (JEK) had larger clones than expected given the number of MLGs observed. Sites in Virginia (RBB and QBY) fell along the expected line, and both Massachusetts sites (NAH and PMH) show slightly higher than expected evenness among genets.
Figure 3.
Contribution of clonality to D. lineata populations at 8 sites across the North American Atlantic Coast. Clonal dominance (Drare) normalized to rarified genotypic richness, reflects the relative diversity of MLGs at a site. Low values occur at sites dominated by a few large clones. High values occur where all MLGs are represented evenly. Where D = 1, every MLG is represented by a single individual. Solid upper and lower lines indicate maximum and minimum Drare possible for a given MLG richness. Dotted line is the median Drare expected if the number of ramets per genet is random within each level of genotypic richness. Point color indicates site. State code in parentheses indicates the region of origin.
Describing Genetic Diversity
The high level of clonality in this species poses a challenge for making statistically robust estimates of genetic diversity metrics, as the sample size of MLGs (G) is low even when the number of ramets sampled is high. For example, Florida sites had few genets (GCML = 3, GWAK = 2) represented among NCML = 25 and NWAK = 26 genotyped anemones. Some, but not all, metrics of genetic diversity varied both within and among regions (Table 3, Supplementary Figure 2). For the genet-level data set, rarified allelic diversity (N = 4 alleles) ranged from 2.4 to 2.8 with a mean across sites of 2.6 ± 0.2 SE and showed no relationship with mean annual water temperature (MAWT) (R2 = −0.15, P = 0.79). Unbiased expected heterozygosity ranged from 0.61 (JEK) to 0.80 (WAK) with a mean across sites of 0.68 ± 0.02 and slightly positive but nonsignificant relationship with MAWT (R2 = −0.07, P = 0.49). Observed heterozygosity ranged from 0.53 (CML) to 0.77 (RBB) with a mean across sites of 0.65 ± 0.04 SE and had a slightly negative, but nonsignificant relationship with MAWT (R2 = −0.07, P = 0.49).
Table 3.
Metrics of genetic diversity across 8 populations of Diadumene lineata for the (A) genet-level (no clones) and (B) ramet-level (clones included) data sets
| (A) | Region | Site | MAWT | Genets | A E | P A | HE | H O | F IS | |
|---|---|---|---|---|---|---|---|---|---|---|
| FL | CML | 22.4 | 3 | 2.53 ± 0.20 | 0.08 ± 0.05 | 0.673 ± 0.066 | 0.533 ± 0.102 | 0.042 | 0.247* | |
| FL | WAK | 22.4 | 2 | 2.80 ± 0.25 | 0.35 ± 0.15 | 0.8 ± 0.042 | 0.700 ± 0.133 | — | 0.176 | |
| GA | STS | 20.9 | 5 | 2.60 ± 0.15 | 0.25 ± 0.1 | 0.691 ± 0.049 | 0.680 ± 0.104 | 0.236*** | 0.018 | |
| GA | JEK | 20.9 | 7 | 2.44 ± 0.22 | 0.36 ± 0.19 | 0.613 ± 0.074 | 0.557 ± 0.105 | 0.239*** | 0.098 | |
| VA | RBB | 16.1 | 6 | 2.46 ± 0.15 | 0.26 ± 0.11 | 0.652 ± 0.059 | 0.767 ± 0.09 | 0.207*** | −0.198* | |
| VA | QBY | 16.1 | 7 | 2.46 ± 0.19 | 0.29 ± 0.12 | 0.643 ± 0.063 | 0.586 ± 0.116 | 0.216** | 0.096 | |
| MA | NAH | 12.8 | 5 | 2.74 ± 0.22 | 0.31 ± 0.08 | 0.713 ± 0.071 | 0.600 ± 0.084 | 0.093 | 0.175* | |
| MA | PMH | 12.8 | 4 | 2.50 ± 0.25 | 0.44 ± 0.11 | 0.643 ± 0.087 | 0.725 ± 0.126 | 0.485*** | −0.152 | |
| Mean ± SE | 2.57 ± 0.16 | 0.29 ± 0.07 | 0.679 ± 0.023 | 0.643 ± 0.038 | 0.064*** | 0.06 ± 0.06 | ||||
| (B) | Region | Site | MAWT | N | A E | P A | H E | H O | F IS | |
| FL | CML | 22.4 | 25 | 3.00 ± 0.3 | 0 ± 0 | 0.507 ± 0.049 | 0.520 ± 0.12 | 0.807*** | −0.026 | |
| FL | WAK | 22.4 | 26 | 2.67 ± 0.21 | 0.21 ± 0.13 | 0.442 ± 0.061 | 0.792 ± 0.132 | 1.000*** | −0.823*** | |
| GA | STS | 20.9 | 27 | 3.61 ± 0.31 | 0.11 ± 0.1 | 0.455 ± 0.058 | 0.696 ± 0.134 | 0.763*** | −0.546*** | |
| GA | JEK | 20.9 | 23 | 4.35 ± 0.67 | 0.61 ± 0.39 | 0.515 ± 0.073 | 0.535 ± 0.119 | 0.610*** | −0.04 | |
| VA | RBB | 16.1 | 30 | 3.43 ± 0.27 | 0.17 ± 0.12 | 0.559 ± 0.061 | 0.800 ± 0.101 | 0.583*** | −0.441*** | |
| VA | QBY | 16.1 | 28 | 3.98 ± 0.57 | 0.32 ± 0.23 | 0.570 ± 0.062 | 0.564 ± 0.116 | 0.494*** | 0.01 | |
| MA | NAH | 12.8 | 27 | 4.45 ± 0.53 | 0.48 ± 0.16 | 0.604 ± 0.066 | 0.563 ± 0.091 | 0.652*** | 0.07 | |
| MA | PMH | 12.8 | 21 | 3.5 ± 0.52 | 0.50 ± 0.22 | 0.587 ± 0.08 | 0.710 ± 0.126 | 0.702*** | −0.215*** | |
| Mean ± SE | 3.62 ± 0.34 | 0.30 ± 0.11 | 0.530 ± 0.023 | 0.648 ± 0.042 | 0.248*** | −0.25 ± 0.12 |
MAWT, mean annual water temperature in Celsius; genet, number of genotypes based on Psex; N, number of anemones genotyped; AE, allelic richness based on rarefaction using the smallest samples size (ramet level: 21 anemones, 42 alleles; genet level: 2 anemones, 4 alleles); PA, private allelic richness based on rarefaction using the smallest samples size (ramet level: 21 anemones, 42 alleles; genet level: 2 anemones, 4 alleles); HE, unbiased expected heterozygosity; HO, observed heterozygosity; , linkage disequilibrium, FIS, inbreeding coefficient and test for deviation from Hardy–Weinberg expectations performed using 1000 permutations among individuals within samples using FSTAT (Goudet 1995). SE, standard error.
Significant P-values are shown in bold with an * for P < 0.05, ** for P < 0.01, *** for P < 0.001. For FIS, the Bonferroni adjusted α = 0.00063, #P < 0.00063.
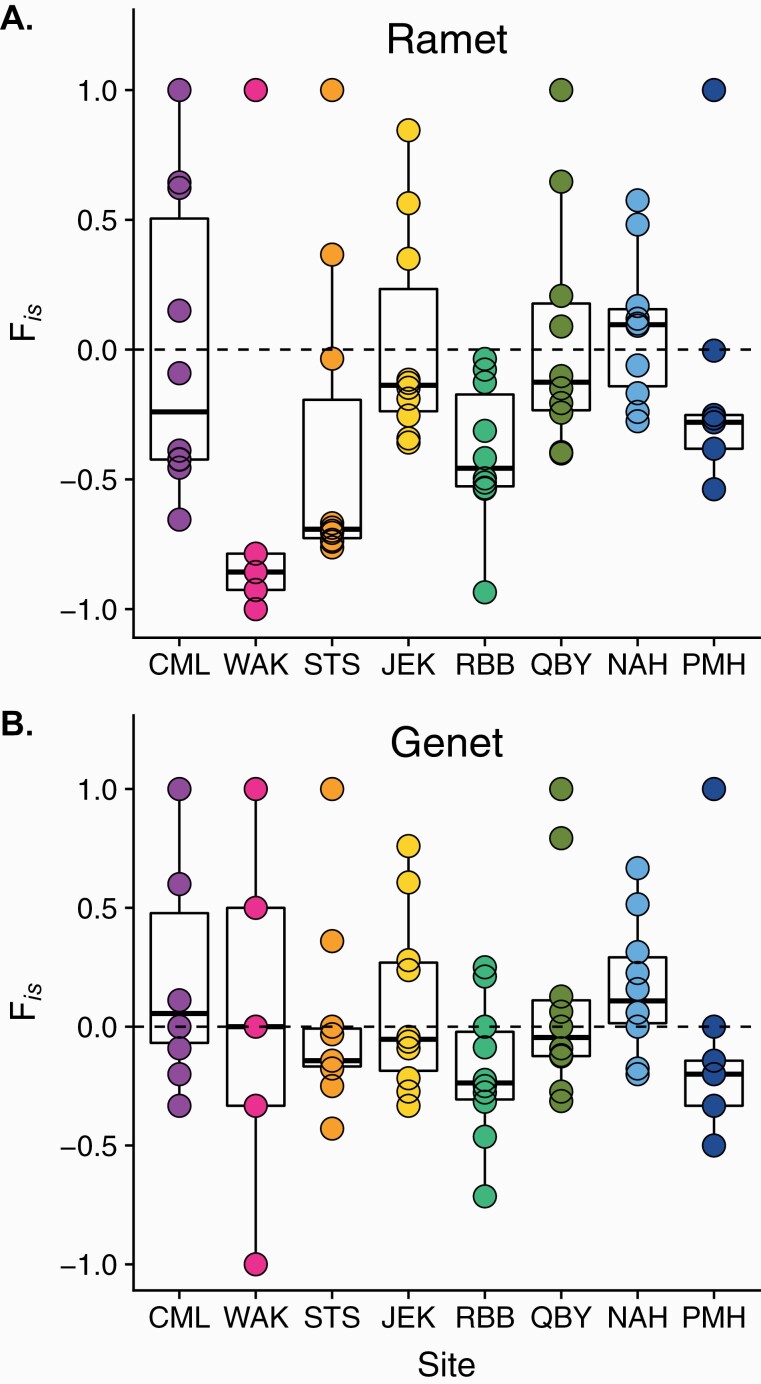
Two additional metrics highlight the potential role that inbreeding might play in these populations, linkage disequilibrium , and the inbreeding coefficient (FIS). Among genets, linkage among loci was significantly higher than expected by random chance ( > 0.207, P < 0.001) at all sites except CML and NAH ( = 0.042 and 0.093, respectively) and WAK where there were too few genotypes to calculate (Table 3A). There was no significant trend in linkage disequilibrium across MAWT (R2 = 0.001, P = 0.36; Supplementary Figure 2E). The average multilocus inbreeding coefficient (FIS) across genets was slightly positive (0.06 ± 0.06), ranging from −0.198 at RBB to 0.247 at CML. But few deviations were significant below P = 0.05, and none after adjusting for multiple comparisons (Table 3A). There was a positive, though nonsignificant, trend of increasing FIS values with MAWT (slope = 0.02, R2 = 0.12, P = 0.214; Supplementary Figure 2D).
The distribution of FIS values among loci by site can also provide insight into the relative contribution of asexual and sexual reproduction at a site. The amplification of some genotypes through clonality should lead to an excess of heterozygotes relative to HWE even in a randomly mating population (Reichel et al. 2016). As expected, the distribution of FIS among loci measured for the ramet data set show a broad scatter with a concentration of negative values (Figure 4A). When only genet-level patterns are considered, recombination through sex should reduce variance in FIS among loci. We found that sites vary in how variable FIS values are among loci. NAH and RBB stand out as having the lowest variance (0.08 and 0.09, respectively) followed by JEK and STS (0.14, 0.16) (Figure 4B). The small number of MLGs at all sites, however, limits the interpretation of these values.
Figure 4.
Single locus Inbreeding coefficient (FIS) by site, ordered by increasing region latitude, groups arranged from left to right. (A) Calculated for the ramet-level data set, including clones. (B) Calculated for the genet-level data set, excluding clones. Each point reflects the FIS value for one locus across all MLGs at a site.
Genetic diversity metrics calculated for the ramet-level data set are also provided (Table 3B). These primarily illustrate the population genetic patterns expected when underlying clonal structure is not considered, such as very high estimates for linkage disequilibrium and extreme negative values of FIS in highly clonal populations with low evenness.
Describing Spatial Patterns in Clonal Aggregation, Autocorrelation
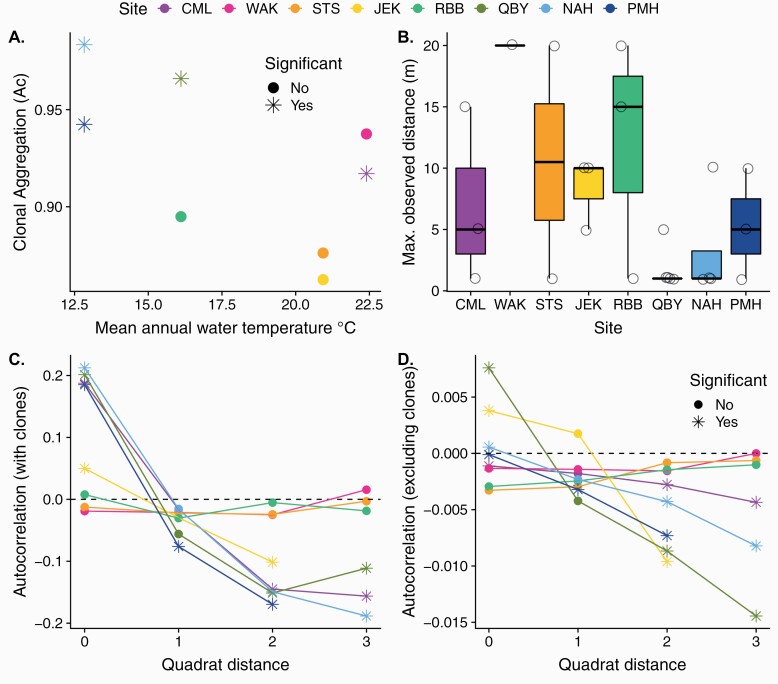
Across sites, the average number of unique MLGs co-occurring in quadrat sample is 1.77 ± 0.13, with no apparent latitudinal trend. The index of spatial aggregation was high at all sites (Ac > 0.86, Figure 5A), indicating that ramets of a genet tend to cluster in the same quadrats. Clonemates were significantly more clustered then expected by random chance in Massachusetts (Ac = 0.98, 0.94; P < 0.001 for NAH and PMH, respectively), where anemones occur in isolated tidepools. In Virginia, clonemates were significantly aggregated at Quinby Harbor (QBY) (Ac = 0.97, P < 0.001) where anemones colonize decaying wood posts and clusters of oyster on a mudflat, but were more integrated with other clones on the beams of the public boat launch at Red Bank Road (RBB) (Ac = 0.89, P = 0.06), where ramets of up to 4 unique MLGs occurred in each quadrat sample (Figure 6A). In Georgia however, both sites show the signature small-scale interdigitation among clones with lower aggregation (Ac = 0.88, 0.86; P = 0.70, 0.86, respectively, for STS and JEK). Ramets infiltrating the intertidal oyster matrix on breakwater rocks in STS show an average of 1.8 ± 0.37 MLGs per quadrat sample, compared with 2.8 ± 0.25 SE at JEK where individuals encrust drowned trees studded in a sandy beach. In Florida, genets colonizing a contiguous patch of sedimentary dredge spoil at CML are significantly aggregated (Ac = 0.92, P > 0.01). At WAK, the dominance of a single large clone throughout a contiguous oyster reef renders the metric meaningless.
Figure 5.
Spatial patterns in the distribution of ramets. (A) The average level of clonal aggregation by site over mean annual water temperature. The index Ac can range from −1 to 1 reflecting the degree of overdispersion or clustering among ramets of a genet, respectively. (B) Maximum observed distance between ramets of a genet in meters. Each point represents one genet. Boxplots indicate the median and quartile estimates. (C) The degree of spatial autocorrelation of Euclidian genetic distance (including relatedness due to clonality) among ramets in quadrats spaced along a linear transect. (D). The degree of spatial autocorrelation of Euclidian genetic distance, not including relatedness due to clonality. In A, C, and D, asterisks indicate values that are significantly different than zero (P < 0.05), which would be expected if ramets were randomly distributed. Colors indicate site of origin, arranged by increasing regional latitude.
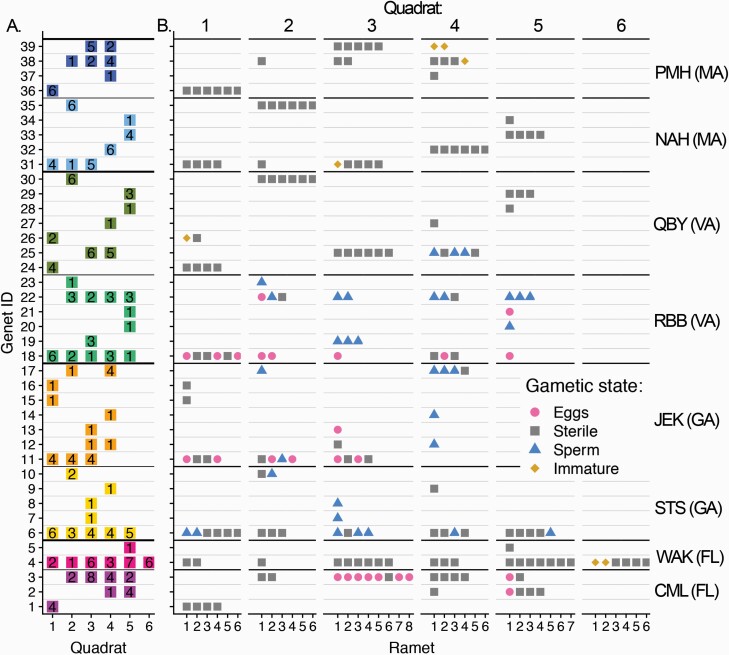
Figure 6.
Spatial patterns of ramets among sites depending on MLG and gametic state (A) The number of ramets representing each (arbitrarily named) genet (rows) is listed for each quadrat (columns 1–6) that was arrayed along a 20–25 m transect at each site (color). (B) For each ramet of each genet (rows) in each quadrat (panel), the gametic state of each ramet is noted by color and point shape (pink circles: egg-bearing, blue triangles: sperm-bearing, yellow diamonds: immature gametes, gray square: sterile). JEK and PMH only had 4 quadrats. WAK had 6 quadrats. All others had 5 quadrats. The relative position of each ramet within quadrat in the diagram is arbitrary.
When the influence of clones on relatedness among individuals is included, patterns of autocorrelation comparing physical distance to genetic distance show a significant decline in relatedness with distance along the transect in CML, JEK, QBY, and the 2 Massachusetts sites (Figure 5C). When only genetic relatedness among nonclonemates were considered, the same sites still show steep declines in genetic similarity with distance along the transect (Figure 5D). At the 2 sites which show no such pattern, WAK, STS, and RBB, the majority of sampled anemones belong to one or more large clones which are spread across the length of the transect.
The average observed maximum distance between 2 ramets of a genet was highest where very large clones were found (e.g., WAK, STS, RBB). At each of these sites, members of a single genet were found spread across all sampled quadrats, separated by up to 20 m (Figures 5B and 6A). At northern sites, where suitable habitat appeared more patchy, the maximum distance recorded among clonemates was lower (e.g., QBY, NAH, PMH), though a rank correlation between MAWT and maximum distance between clonemates was not significant (S = 1523.9, P = 0.11). We lack the statistical power here to parse the influence of latitude and habitat type.
Spatial aggregation patterns have a particular relevance at sites where fertile individuals have been found, as they may help inform patterns of sexual reproduction. Figure 6B shows the spatial patterns of fertile individuals across sites. While fertile males and females have been found at most of these sites during other sampling periods (Table 1), these data demonstrate that patches colonized by a single sex and/or nonfertile individuals are common (Figure 6B). Notable exceptions occur at both JEK and RBB, where highly interdigitated colonies with ramets bearing fertile eggs and sperm do occur. In 2 surprising cases, a ramet bearing eggs and a ramet bearing sperm were assigned to the same MLG (i.e., clonemates showed different sexes) (Figure 6B). We are confident in the methods used to assign gamete stage and genet identity, but interpreted this observation cautiously, as it was unexpected in a species known previously to have strictly separate sexes. We considered 2 types of errors that could produce this outcome erroneously. First, such a pattern could arise in error if our markers were not sufficiently polymorphic to resolve individuals that originated from separate zygotes. For the 2 genets showing this pattern, the probability of each MLG arising twice randomly through sex, given the diversity of the markers at each site (Psex) was 3.8 × 10−6 for genet 11 and 1.1 × 10−5 for genet 22. Thus, the likelihood of mistakenly assigning genetically distinct ramets to the same genet was low. Second, gamete state could be misdiagnosed through dissection. As these samples were destroyed in the process of extracting DNA, there is no way to re-confirm the gamete status of individuals with histological sectioning. It is possible, though unlikely, to have confused a nonfertile female for a fertile male, as the gonadal structures appear similar, which could explain the case of genet 11. However, it is very unlikely that a nonfertile anemone or fertile male would have been described as an egg-bearing individual, as eggs are distinctive in color and shape. This is the mistake that would be required for the pattern found in genet 22 to be made through error. Thus, we accept that we observed a real, if rare, pattern worth exploring in future studies.
Describing Patterns of Diversity and Connectivity Among Sites
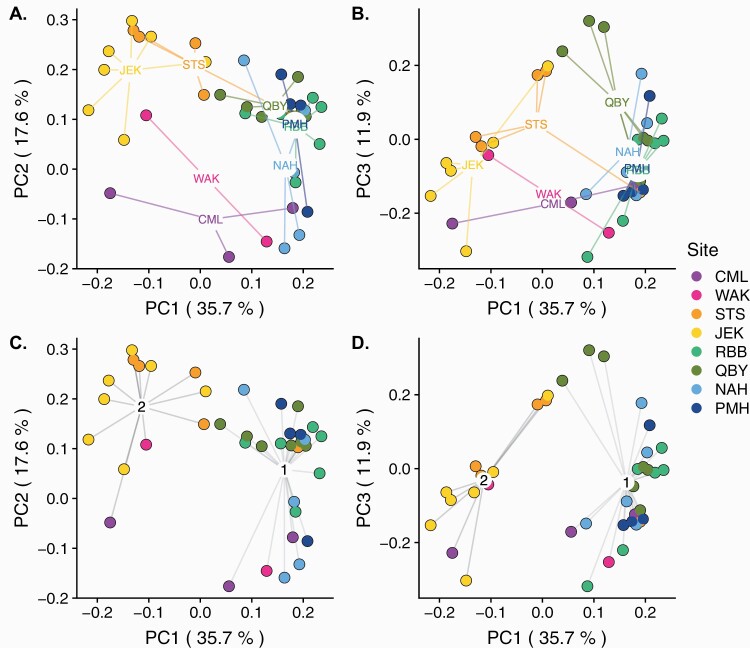
The degree to which individuals at a site cluster together in a coherent genetic unit, as would be expected of a similarly philopatric native and/or sexual species, varied in these data. This pattern may reflect true biological anomalies of this species as well as low statistical power due to limited genotypic richness at some sites. The first 2 axes of principal component of pairwise genetic distance among all genets explained approximately 65.2% of the variation (Figure 7). PC1 (~35.7%) described a split between the sites in Georgia and those further north. Florida separated from the other sites along PC2 (~17.6%). The centroid of one Virginia site (QBY) stretched away from the Massachusetts sites along PC3 (~13.6%), driven particularly by a distinct trio of genets. However, other QBY individuals along with those at RBB overlapped with Massachusetts sites along all 2 axes.
Figure 7.
Principal components of variation in genetic distance among MLGs, as assessed by Psex, among sites (colors). (A and B) show points grouped by site along (A) axis PC1 and PC2 or (B) PC1 and PC3. (C and D) show points grouped into best-fit clusters based on BIC value, as estimated with find.clusters in adegenet, along (C) axis PC1 and PC2, or (D) PC1 and PC3. Percent variance explained by each PC axes shown in parentheses. Group numbers in (C and D) are arbitrary.
Clustering analysis with adegenet suggested that these data are best described with 2 natural groupings (K = 2, Bayesian information criterion [BIC]: 56.3) (Figure 7C and D), though all levels of K clusters identified distinct southern and northern clusters that split along PC1. The relationship of Florida genets to each other and to other sites was unclear. This may be due to limited data availability, but may also reflect a real lack of genetic similarity among co-occurring genets relative to the other regions. To ensure that the presence of Florida individuals was not masking or distorting patterns among other sites. A PCA was also calculated without the Florida genets. The major patterns remained consistent (Supplementary Figure 3). Georgia split from the northern sites along PC1 (37.3%), while JEK stands out more clearly as a distinct and compact genetic cluster. The centroids of the Massachusetts sites separated somewhat along PC2 (19.0%), and QBY was pulled away from the northern cluster by the same set of distinct individuals along PC3 (11.8%). These data again were best described by 2 clusters (K = 2, BIC = 47.96), which roughly separated Georgia from the more northern sites.
We also estimated pairwise genetic differentiation (FST) with the genet-level data set, but found no significant differences among sites at the Bonferroni adjusted alpha (α = 0.008; Supplementary Table 4). As a qualitative assessment, within region, FST values were low (<0.1), but were greater when comparing among sites from different regions. In particular, JEK showed high values compared to northern sites (e.g., FST = 0.210 with RBB, 0.234 with PMH).
We did find a significant pattern of isolation by distance (Mantel’s r = 0.58, R2 = 0.34, P = 0.004) as measured with Euclidian genetic distance compared with along-coastline distances among sites (Supplementary Figure 4). Together these results supported a signature of geographic differentiation among populations, particularly between northern and southern sites on the Atlantic coast.
Discussion
The spatial scale of genetic population structure is expected to influence many aspects of ecology and evolution (Slatkin 1987; Kamel et al. 2012), including the mating system (Knowlton and Jackson 1993; Olsen et al. 2020) and competitive environment (Sherman and Ayre 2008; Kamel and Grosberg 2013). However, in clonal organisms, it is difficult to isolate the genetic from the ecological consequences of clonality to make unequivocal evolutionary predictions (Vallejo-Marín et al. 2010; Barrett 2015). In this study, we leveraged a temperature-mediated latitudinal gradient in fission behavior to address how clonality affects genetic and genotypic diversity.
We found that spatial structure in D. lineata across the US Atlantic Coast is influenced by clonal reproduction at all sites, but the relative contribution of clonality to this structure differs across latitude. In Florida, toward the southern edge of the current reported distribution, where warm water promotes high fission rates and small body sizes (Ryan 2018), sites are populated by few genets composed of many individual ramets. Some of these clonal lineages share few alleles in common, though the low genet-level sample size makes it difficult to draw firm conclusions about the role of sex at these sites. In Massachusetts, toward the northern end of the range, where fission rate is expected to be low and the season for gamete production short, sites are populated with small patches of clonemates, which exhibit high levels of linkage disequilibrium, a potential sign of a historical inbreeding or bottlenecks from extinction and colonization events. In Georgia and Virginia, populations show a mixed structure in which ramets of a small number of prolific clones are interspersed with single representatives of novel genotypes. These intermediate sites also had the highest genotypic richness and the most evidence for genetic similarity among genets perhaps indicating a greater role for sexual reproduction and recruitment toward the center of the species latitudinal range. These patterns are consistent with evidence that the Mid-Atlantic temperature patterns drive a mixed reproductive strategy, with seasonal cycles in body size that correspond with cycling between unitary growth with gamete production and prolific fission over the course of the year (Ryan 2018). Evidence for the influence of sexual reproduction in the genetic structure of populations in Georgia and Virginia is also consistent with these locations as sites with the most prolific gamete production (Figure 6B) (Ryan and Miller 2019). However, sites in all 4 regions showed a significant pattern of spatial genetic autocorrelation that was not explained by clonality, suggesting an influence of at least some sexual reproduction at most sites.
Although the relatively small number of genets captured by our sampling limit our ability to formally test hypotheses about population connectively and gene flow with this data set, we found evidence of large-scale structure along the Atlantic coast. There was a significant signature of isolation by distance across all sites driven in part by a clear break between Georgia and the more northern sites. More study is required to determine whether this pattern is the result of persistent effects of different invasion history, contemporary barriers to gene flow, or both. However, the pattern of genetic similarity among individuals found near the Chesapeake Bay and in New England is congruent with one hypothesized invasion pathway reported in the earliest literature on this species. Verrill (1898) notes that Sagartia luciae (now D. lineata) may have arrived in Massachusetts and Connecticut via oysters transplanted from “further south.” Glon et al. (2020) has interpreted this as referring to commercial transport between the Chesapeake Bay and Long Island Sound, which was common between 1825 and the early 1900s (MacKenzie 1996). The apparent genetic similarity between individuals in Massachusetts and Virginia makes observations of differences in seasonal reproductive cycle, reproductive allocation and clonal structure even more interesting, as these patterns may reflect the consequences of plasticity rather than genetic differentiation. However, more work is needed to establish confidence in the patterns only hinted at in our data.
These data contribute to an emerging view of the complex eco-evolutionary landscape formed by the interaction between environmental gradients and reproductive plasticity, as well as invasion history. Explorations of the evolution of clonality have often focused on the fate of genotypes or populations, which differ in their rate of allocation to asexual versus sexual reproduction (e.g., Balloux et al. 2003; Allen and Lynch 2008). However, the temperature-dependent nature of fission in D. lineata suggests that genets may express different reproductive strategies from year to year, or as they become dispersed over space. Across the broad range of this species, abiotic drivers interact with the shape of the fission rate reaction norms of local genets to produce a site-specific mix of sexual and asexual reproduction. Thus, selection probably discriminates on differences among genets in the degree of reproductive plasticity, as well as average clonal rate. What we are learning about the biology of this species underscores the challenge of understanding how selection acts on genets that can be represented by dozens of ramets experiencing different local microhabitats.
In addition, the extent of clonal aggregation is key to understanding which forces shape life cycle evolution in such species. For example, a dispersed clonal structure is a common feature of disturbance-adapted clonal organisms, where fragmentation is important for survival and recruitment (Coffroth and Lasker 1998). Dispersed clonal genets are able to spread the risk of mortality over a larger area, which may be essential when living on usnstable substrate or otherwise disturbance prone habitats (Wulff 1991; Coffroth and Lasker 1998; Geller et al. 2005). A tendency toward dispersal may also alleviate the risk of spatial interference reducing the number of potential mates available (Vallejo-Marin et al. 2010), one of the potential drawbacks of clonal behavior. Conversely, dispersed genets lose some of the purported advantages of clonal growth, such as excluding competitors from swaths of suitable habitat (Williams 1975; Ayre 1985), cooperation in prey capture, or division of labor among ramets (Ayre and Grosberg 2005). Variation in aggregation among sites may alter the relative importance of these factors to local adaptation.
We found that the degree to which clonemates aggregate in space varies among site in this species. The spatial extent of the clone increases significantly with clone size, yet even at sites with large clones scattered across tens of meters, genotypic diversity at the quadrat scale remains high. This suggests that clonemates do not necessarily remain clustered as has been seen in other anemone species (e.g., Anthopluera elegantissima, Francis 1973; Actina tenebrosa, Ayre 1984; Metridium senile, Purcell and Kitting 1982, Hoffmann 1986, Anthothoe albocinta, Billingham and Ayre 1996). At sites where contiguous habitat is available (e.g., oyster reefs, breakwater rocks), ramets appear to diffuse through the matrix. Where suitable habitat occurs in discrete patches (e.g., wooden posts, tidepools), ramets tend to be more aggregated. In the current study, we lack the replication to distinguish the influence of substrate type and disturbance history from potential geographic variation in movement behavior, though this is likely a fruitful direction for the future.
Unlike what is observed in most colony-forming or aggregating species, it is common to find D. lineata individuals intermingled with unrelated individuals. This supports the findings of Ting and Geller (2000) that individuals of multiple genotypes may cohabitate on a single pebble, despite prolific asexual reproduction. As in other anemone species, D. lineata is able to produce “catch” tentacles, or fighting tentacles specialized for intraspecific competition, although they are rarely seen in Atlantic field populations (W.H.R., personal observation). It remains to be seen what role intraspecific competition plays in mediating clonal structure in this species.
Previous workers studying D. lineata on the Atlantic and Gulf Coasts of North America (e.g., Shick 1976; Shick and Lamb 1977; Minasian 1979) have suggested that most populations arise through the clonal replication of single (or small number) of colonists. These populations endure constant “boom and bust” cycles as clonal populations grow large, then are extirpated by disease, predators or abiotic factors. Under this view, populations would be unlikely to persist long term or have high genetic diversity. In addition, strong founder effects were expected to limit mate availability to the point that sexual reproduction was either unimportant or nonexistent outside the native range (Shick 1991; discussed in detail by Ting and Geller 2000). Thorough sampling over different parts of the seasonal cycle have revealed that gamete bearing male and female anemones occur at all but one of these sites (PMH; W.H.R., unpublished data), supporting the potential for sex. This study provides genetic evidence that sexual reproduction is occurring; most sites showed a signature of declining relatedness among nonclonemates with distance at scale of meters and some populations formed genetically distinct clusters. Furthermore, although genotypic richness was low relative to what might be found with a similar sampling effort of a unitary species, none of the sites studied were monoclonal.
How patterns of sexual and asexual reproduction influence populations cycles in high disturbance areas remains to be seen. Undoubtedly, D. linetata populations are shaped by disturbance, including local extirpation due to sand incursion, ice scour, and nudibranch grazing (W.H.R., personal observation), in addition to anthropogenic activity. As a dramatic example, the entire dock structure at Red Bank Road, Virginia (RBB) was replaced in the Fall of 2017. This virtually eliminated a population with an average density of over 200 anemones per 0.25 m2 (Ryan and Miller 2019). Over the past 2 years, the population has since returned at an astonishing rate (W.H.R. and S.A.K.-H., unpublished data), though we do not yet know the genetic composition of the new colonists. Future work to further understand the genetic and ecological consequences of disturbance and recovery cycles will be important for predicting the continued spread of this highly successful species.
Invasion history and latitudinal environmental gradients for this species operate on a global scale and undoubtedly drive broad patterns in genetic diversity (Ruiz et al. 2000; Wasson et al. 2001; Rius and Darling 2014). This study is among the first steps to disentangle the contributions of an unknown invasion history along with contemporary dynamics of clonality, sex, and connectivity that combine to produce local patterns of population genetic structure. As such, there are many leads to follow from these data that will contribute to a broader understanding of the ecology and evolution of partially clonal organisms, as well as invasive species with complex life cycles. For example, our finding that inbreeding coefficients (FIS) tend to vary among loci more in populations with high levels of clonality relative to sites where sex is likely more common are consistent with recent advances in our understanding of the evolutionary consequences of partial clonality. Reichel et al. (2016) developed a theoretical model and meta-analysis of empirical data demonstrating that high levels of clonality lead to random divergence in heterozygosity values among neutral loci as some alleles become randomly amplified in the population through clonality. Thus, common interpretations of standard population genetic metrics (e.g., negative mean multilocus FIS as a signature of clonality) fail to fully describe the mechanisms underlying observed patterns. Likewise, unrecognized clonality (i.e., not distinguishing between a ramet- and genet-level data set) can obscure the source some of the variation that occurs across loci due to partial clonality. By examining empirical patterns in species such as D. lineata where a gradient in clonal behavior is known, we can develop a more nuanced framework for understanding the evolutionary effects of clonality.
The results of this study also suggest that very small-scale processes shape the genotypic neighborhood where individual interactions, such as mating and competition, are important to understand. Movement of individuals away from their clonemates, either by dislodgement or by locomotion, is probably important to creating heterogeneous genetic structure at fine scales. The evolution of clonal behavior itself has been linked to life on unstable substrate (Coffroth and Lasker 1998, Geller et al. 2005), and may be part of a suite of adaptations for surviving dislodgement that have allowed this species to become established worldwide. As sea anemones are often characterized as sessile animals, little attention has been paid to the role that adult movement probably plays in choosing habitat (but see Bedgood et al. 2020), navigating competitive interactions, and structuring genetic and genotypic diversity. Both the spatial scale that some clones cover, coupled with the degree of intermixing among clones at small scales in some populations suggests that this is an oversight.
The findings of this study also highlight an essential challenge of studying the genetic structure of partially clonal organisms. Namely, that high rates of clonality reduce our ability to capture sufficient genetic diversity to make statistically robust descriptions of populations. For exclusively sexual populations, there is a conventional assumption that sampling 20–30 individuals at a site will capture a large proportion of the available diversity (e.g., Hale et al. 2012). At a site with clonal dynamics similar to those we sampled in Florida, hundreds of anemones would need to be sampled to garner 20–30 unique multilocus genotypes—assuming that such diversity is available to gather. However, it is not only a logistical and financial barrier that needs to be overcome to understand these species. Partial clonality typically confers upon a genet the potential for long-term persistence in the environment, whereas MLGs are (relatively) ephemeral in many purely sexual populations (Tibayrenc and Ayala 1991). Thus, ecological and evolutionary processes tend to be studied at the level of populations, wherein the characteristics of individual genotypes are not typically the drivers of key processes. However, often in an organism like D. lineata, the genet is the population. Thus, basic concepts like effective population size (Orive 1993), generation time (Reusch et al. 1999), or the genetic definition of a population cannot be translated wholesale from theory built for sexual organisms (Arnaud-Haond et al. 2007).
Together these results describe a widespread species, where population structure varies with a gradient in clonal behavior, stimulated in part by reproductive plasticity. Although clonal growth influences genotypic diversity at all sites, the small-scale genetic neighborhood experienced by individual anemones varies among sites. Unlike many clonal plants and animals, the ability for adult clones to disperse after fission means that increased clonality does not always lead to a loss of small-scale genotypic diversity. Given the importance of spatial genetic and genotypic patterns to all aspects of individual fitness, understanding these patterns will be useful in separating the genetic, ecological, and spatial consequences of clonality in future studies.
Funding
This work was made possible with funds from NSF EAPSI (no. 1515296 to W.H.R.), PADI Foundation (no. 21902 to W.H.R.), Florida State University Gramling Award for Marine Biology (2016 to W.H.R.), start-up funds from the University of Alabama at Birmingham (to S.A.K.H.), and the University of Alabama at Birmingham Office of Postdoctoral Education (to W.H.R., J.A., and S.A.K.H.). W.H.R. was supported during part of this project by the NIH-IRACDA MERIT Postdoctoral Fellowship program at U.A.B.
Supplementary Material
Acknowledgments
Elements of this project were submitted by W.H.R. in partial fulfillment of the doctoral degree requirements for Florida State University. The authors thank Don Levitan and Yueling Hao for laboratory assistance in the initial phase of the project, Sabrina Heiser for collaboration on microsatellite primer selection, Caitlin Cox of the Heflin Center for Genomic Sciences for help with fragment analysis, the anonymous reviewers that improved this manuscript with their comments and suggestions, and Maria Orive for her encouragement. W.H.R. thank Tom Miller, Kim Hughes, Janie Wulff, Don Levitan, and Markus Huettel for insight and feedback, as well as many others who made this project possible. W.H.R. would also specifically like to thank Jon Geller for his generous support of an earlier iteration of this project, including the use of transcriptomic data generated in part by Tracy Campbell. Although none of those data were used to design the primers in this manuscript, his encouragement on the early stages of this project was critical to its eventual success.
Contributor Information
Will H Ryan, Department of Biology, University of Alabama at Birmingham, Birmingham, AL; Department of Biological Science, Florida State University, Tallahassee, FL.
Jaclyn Aida, Department of Biology, University of Alabama at Birmingham, Birmingham, AL.
Stacy A Krueger-Hadfield, Department of Biology, University of Alabama at Birmingham, Birmingham, AL.
Conflict of Interest
We declare that we have no conflicts of interest that could have introduced bias into this work.
Data Availability
Microsatellite primers are available in Supplementary Table 2. Genet and ramet level multilocus genotypes and phenotypic data are deposited in DRYAD (Dryad number: doi:10.5061/dryad.kprr4xh3h).
References
- Allen DE, Lynch M. 2008. Both costs and benefits of sex correlate with relative frequency of asexual reproduction in cyclically parthenogenic Daphnia pulicaria populations. Genetics. 179:1497–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaud-Haond S, Belkhir K. 2007. GENCLONE: a computer program to analyse genotypic data, test for clonality and describe spatial clonal organization. Mol Ecol Notes. 7:15–17. [Google Scholar]
- Arnaud-Haond S., Candeias R, Serrão EA, Teixeira SJL. 2013. Microsatellite markers developed through pyrosequencing allow clonal discrimination in the invasive alga Caulerpa taxifolia. Conserv Genet Resour. 5:667–669. [Google Scholar]
- Arnaud-Haond S, Duarte CM, Alberto F, Serrão EA. 2007. Standardizing methods to address clonality in population studies. Mol Ecol. 16:5115–5139. [DOI] [PubMed] [Google Scholar]
- Arnaud-Haond S, Duarte CM, Diaz-Almela E, Marbà N, Sintes T, Serrão EA. 2012. Implications of extreme life span in clonal organisms: millenary clones in meadows of the threatened seagrass Posidonia oceanica. PLoS One. 7:e30454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atoda K. 1973. Pedal laceration of the sea anemone, Haliplanella luciae. Publ Seto Mar Biol Lab. 20:299–313. [Google Scholar]
- Ayre DJ. 1984. The effects of sexual and asexual reproduction on geographic variation in the sea anemone Actinia tenebrosa. Oecologia. 62:222–229. [DOI] [PubMed] [Google Scholar]
- Ayre DJ. 1985. Localized adaptation of clones of the sea anemone Actinia tenebrosa. Evolution. 39:1250–1260. [DOI] [PubMed] [Google Scholar]
- Ayre DJ, Grosberg RK. 2005. Behind anemone lines: factors affecting division of labour in the social cnidarian Anthopleura elegantissima. Anim Behav. 70:97–110. [Google Scholar]
- Bailleul D, Stoeckel S, Arnaud-Haond S. 2016. RClone: a package to identify MultiLocus Clonal Lineages and handle clonal data sets in r. Methods Ecol Evol. [Google Scholar]
- Balloux F, Lehmann L, de Meeûs T. 2003. The population genetics of clonal and partially clonal diploids. Genetics. 164:1635–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett SC. 2015. Influences of clonality on plant sexual reproduction. Proc Natl Acad Sci USA. 112:8859–8866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baums IB, Miller MW, Hellberg ME. 2006. Geographic variation in clonal structure in a reef-building Caribbean coral, Acropora palmata. Ecol Monogr. 76:503–519. [Google Scholar]
- Bedgood SA, Bracken MES, Ryan WH, Levell ST, Wulff JL. 2020. Environmental drivers of adult locomotion and reproduction in a symbiont-hosting sea anemone. Mar Biol. 167. Article no. 39. [Google Scholar]
- Bellis ES, Edlund RB, Berrios HK, Lessios HA, Denver DR. 2018. Molecular signatures of host specificity linked to habitat specialization in Exaiptasia sea anemones. Ecol Evol. 8:5413–5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billingham M, Ayre DJ. 1996. Genetic subdivision in the subtidal, clonal sea anemone Anthothoe albocincta. Mar Biol. 125:153–163. doi: 10.1007/BF00350769 [DOI] [Google Scholar]
- Burgess SC, Ryan WH, Blackstone NW, Edmunds PJ, Hoogenboom MO, Levitan DR, Wulff JL. 2017. Metabolic scaling in modular animals. Invertebr Biol. 136:456–472. [Google Scholar]
- Calderón I, Ortega N, Duran S, Becerro M, Pascual M, Turon X. 2007. Finding the relevant scale: clonality and genetic structure in a marine invertebrate (Crambe crambe, Porifera). Mol Ecol. 16:1799–1810. [DOI] [PubMed] [Google Scholar]
- Coffroth MA, Lasker HR. 1998. Population structure of a clonal gorgonian coral: the interplay between clonal reproduction and disturbance. Evolution. 52:379–393. [DOI] [PubMed] [Google Scholar]
- Dempster AP, Laird NM, Rubin DB. 1977. Maximum likelihood from incomplete data via the EM algorithm. J R Stat Soc Ser B. 39:1–38. [Google Scholar]
- Dorken ME, Eckert CG. 2001. Severely reduced sexual reproduction in northern populations of a clonal plant, Decodon verticillatus (Lythraceae). J Ecol. 89:339–350. [Google Scholar]
- Edmunds PJ. 2007. Physiological ecology of the clonal corallimorpharian Corynactis californica. Mar Biol. 150:783–796. [Google Scholar]
- Faircloth BC. 2008. msatcommander: detection of microsatellite repeat arrays and automated, locus-specific primer design. Mol Ecol Resour. 8:92–94. [DOI] [PubMed] [Google Scholar]
- Fautin DG. 2002. Reproduction of Cnidaria. Can J Zool. 80:1735–1754. [Google Scholar]
- Francis L. 1973. Clone specific segregation in the sea anemone Anthopleura elegantissima. Biol Bull. 144:64–72. [DOI] [PubMed] [Google Scholar]
- Francis L. 1988. Cloning and agression among sea anemones (Coelenterata: Actiniaria) of the rocky shore. Biol Bull. 174:241–253. [Google Scholar]
- Fukui Y. 1991. Embryonic and larval development of the sea anemone Haliplanella lineata from Japan. Hydrobio. 216/217:137–142. [Google Scholar]
- Fukui Y. 1995. Seasonal changes in testicular structure of the sea anemone Haliplanella lineata (Coelenterata: Actiniaria). Invertebr Reprod Dev. 27:197–204. [Google Scholar]
- Geller JB, Fitzgerald LJ, King CE. 2005. Fission in sea anemones: integrative studies of life cycle evolution. Integr Comp Biol. 45:615–622. [DOI] [PubMed] [Google Scholar]
- Glon H, Daly M, Carlton JT, Flenniken MM, Currimjee Z. 2020. Mediators of invasions in the sea: life history strategies and dispersal vectors facilitating global sea anemone introductions. Biol Invasions. 22:3195–3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gollasch S. 2002. The importance of ship hull fouling as a vector of species introductions into the North Sea. Biofouling. 18:105–121. [Google Scholar]
- Goudet J. 1995. FSTAT: a computer program to calculate F-statistics. J Hered. 86:485–486. [Google Scholar]
- Grosberg R, Cunningham CW. 2001. Genetic structure in the sea from populations to communities. In: Bertness MD, Gaines S, Hay ME, editors. Marine community ecology. Sunderland (MA): Sinauer Associates [Google Scholar]
- Guillemin ML, Faugeron S, Destombe C, Viard F, Correa JA, Valero M. 2008. Genetic variation in wild and cultivated populations of the haploid-diploid red alga Gracilaria chilensis: how farming practices favor asexual reproduction and heterozygosity. Evolution. 62:1500–1519. [DOI] [PubMed] [Google Scholar]
- Gutekunst J, Andriantsoa R, Falckenhayn C, Hanna K, Stein W, Rasamy J, Lyko F. 2018. Clonal genome evolution and rapid invasive spread of the marbled crayfish. Nat Ecol Evol. 2:567–573. [DOI] [PubMed] [Google Scholar]
- Hale ML, Burg TM, Steeves TE. 2012. Sampling for microsatellite-based population genetic studies: 25 to 30 individuals per population is enough to accurately estimate allele frequencies. PLoS One. 7:e45170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper J. 1977. The population biology of plants. London: Academic Press. p. 900. [Google Scholar]
- Hausman LA. 1919. The orange striped anemone (Sagartia lucie, Verrill). An ecological study. Biol Bull. 37:363–371. [Google Scholar]
- Hoffmann RJ. 1986. Variation in contributions of asexual reproduction to the genetic structure of populations of the sea anemone Metridium senile. Evolution. 40:357–365. [DOI] [PubMed] [Google Scholar]
- Hughes AR, Inouye BD, Johnson MT, Underwood N, Vellend M. 2008. Ecological consequences of genetic diversity. Ecol Lett. 11:609–623. [DOI] [PubMed] [Google Scholar]
- Ives AR, Kareiva P, Perry R. 1993. Response of a predator to variation in prey density at three hierarchical scales lady beetles feeding on aphids. Ecology. 74:1929–1938. [Google Scholar]
- Jackson JBC, Coates AG. 1986. Life cycles and evolution of clonal (modular) animals. Philos Trans R Soc Lond Ser B. 313:7–22. [Google Scholar]
- Jackson JBC, Hughes TP. 1985. Adaptive strategies of coral-reef invertebrates: coral-reef environments that are regularly disturbed by storms and predation often favor the very organisms most susceptible to damage by these processes. Am Sci. 73:265–274. [Google Scholar]
- Johnson LL, Shick JM. 1977. Effects of fluctuating temperature and immersion on asexual reproduction in the intertidal sea anemone Hauplanella luciae (Verrill) in laboratory culture. J Exp Mar Biol Ecol. 28:141–149. [Google Scholar]
- Jombart T. 2008. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics. 24:1403–1405. [DOI] [PubMed] [Google Scholar]
- Jombart T, Devillard S, Balloux F. 2010. Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet. 11:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinowski ST. 2005. HP-RARE 1.0: a computer program for performing rarefaction on measures of allelic richness. Mol Ecol Notes. 5:187–189. [Google Scholar]
- Kalinowski ST. 2011. The computer program STRUCTURE does not reliably identify the main genetic clusters within species: simulations and implications for human population structure. Heredity (Edinb). 106:625–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamel SJ, Grosberg RK. 2013. Kinship and the evolution of social behaviours in the sea. Biol Lett. 9:20130454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamel SJ, Hughes AR, Grosberg RK, Stachowicz JJ. 2012. Fine-scale genetic structure and relatedness in the eelgrass Zostera marina. Mar Ecol Prog Ser. 447:127–137. [Google Scholar]
- Kamvar ZN, Brooks JC, Grünwald NJ. 2015. Novel R tools for analysis of genome-wide population genetic data with emphasis on clonality. Front Genet. 6:208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamvar ZN, Tabima JF, Grünwald NJ. 2014. Poppr: an R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ. 2:e281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King KC, Lively CM. 2009. Geographic variation in sterilizing parasite species and the Red Queen. Oikos 118:1416–1420. [Google Scholar]
- Knowlton N, Jackson JBC. 1993. Inbreeding and outbreeding in marine invertebrates. In: Thornhill & Wilmsen N, editor. Natural history of inbreeding and outbreeding. Chicago (IL): University of Chicago Press. p. 200–249. [Google Scholar]
- Kollars NM, Krueger-Hadfield SA, Byers JE, Greig TW, Strand AE, Weinberger F, Sotka EE. 2015. Development and characterization of microsatellite loci for the haploid-diploid red seaweed Gracilaria vermiculophylla. PeerJ. 3:e1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger-Hadfield SA, Balestreri C, Schroeder J, Highfield A, Helaouet P, Allum J, Moate R, Lohbeck KT, Miller PI, Riesbesell U, et al. 2014. Genotyping an Emiliania huxleyi (prymnesiophyceae) bloom event in the North Sea reveals evidence of asexual reproduction. Biogeosciences. 11:5215–5234. [Google Scholar]
- Krueger-Hadfield SA, Collén J, Daguin-Thiébaut C, Valero M. 2011. Genetic population structure and mating system in Chondrus crispus (Rhodophyta). J Phycol. 47:440–450. [DOI] [PubMed] [Google Scholar]
- Krueger-Hadfield SA, Kollars NM, Byers JE, Greig TW, Hammann M, Murray DC, Murren CJ, Strand AE, Terada R, Weinberger F, et al. 2016. Invasion of novel habitats uncouples haplo-diplontic life cycles. Mol Ecol. 25:3801–3816. [DOI] [PubMed] [Google Scholar]
- Krueger-Hadfield SA, Roze D, Correa JA, Destombe C, Valero M. 2015. O father where art thou? Paternity analyses in a natural population of the haploid-diploid seaweed Chondrus crispus. Heredity (Edinb). 114:185–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger-Hadfield SA, Roze D, Mauger S, Valero M. 2013. Intergametophytic selfing and microgeographic genetic structure shape populations of the intertidal red seaweed Chondrus crispus. Mol Ecol. 22:3242–3260. [DOI] [PubMed] [Google Scholar]
- Lejeusne C, Bock DG, Therriault TW, MacIsaac HJ, Cristescu ME. 2011. Comparative phyologeography of two colonial ascidians reveals contrasting invasion histories in North America. Biol Invasions. 13:635–650. [Google Scholar]
- MacKenzie C. 1996. History of oystering in the United States and Canada, featuring the eight great oyster estuaries. Mar Fish Rev. 58:1–78. [Google Scholar]
- Mackie JA, Darling JA, Geller JB. 2012. Ecology of cryptic invasions: latitudinal segregation among Watersipora (Bryozoa) species. Sci Rep. 2:871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matschiner M, Salzburger W. 2009. TANDEM: integrating automated allele binning into genetics and genomics workflows. Bioinformatics. 25:1982–1983. [DOI] [PubMed] [Google Scholar]
- Maynard-Smith J. 1978. The evolution of sex. New York: Cambridge University Press. [Google Scholar]
- McFadden CS, Grosberg RK, Cameron BB, Karlton DP, Secord D. 1997. Genetic relationships within and between clonal and solitary forms of the sea anemone Anthopleura elegantissima revisited: evidence for the existence of two species. Mar Biol. 128:127–139. [Google Scholar]
- Meirmans PG, Van Tienderen PH. 2004. Genotype and genodive: two programs for the analysis of genetic diversity of asexual organisms. Mol Ecol Notes. 4:792–794. [Google Scholar]
- Mergeay J, Verschuren D, De Meester L. 2006. Invasion of an asexual American water flea clone throughout Africa and rapid displacement of a native sibling species. Proc Biol Sci. 273:2839–2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minasian LL. 1979. The effect of exogenous factors on morphology and asexual reproduction in laboratory cultures of the intertidal sea anemone, Haliplanella luciae (Verrill) (Anthozoa: Actiniaria) from Delaware. J Exp Mar Biol Ecol. 40:235–246. [Google Scholar]
- Minasian LL Jr, Mariscal RN. 1979. Characteristics and regulation of fission activity in clonal cultures of the cosmopolitan sea anemone, Haliplanella luciae (Verrill). Biol Bull. 157:478–493. [DOI] [PubMed] [Google Scholar]
- Miyawaki M. 1952. Temperature as a factor influencing upon the fission of the orange-striped sea-anemone, Diadumene luciae. J Fac Sci Hokkaido Imp Univ. 1:77–80. [Google Scholar]
- Newcomer K, Flenniken MM, Carlton JT. 2019. Home and away and home again: discovery of a native reproductive strategy of the globally invading sea anemone Diadumene lineata (Verrill, 1869) in a satellite population. Biol Invasions. 21:1491–1497. [Google Scholar]
- Oddou-Muratorio S, Vendramin GG, Buiteveld J, Fady B. 2009. Population estimators or progeny tests: what is the best method to assess null allele frequencies at SSR loci? Conserv Genet. 10:1343–1347. [Google Scholar]
- Oksanen J, Guillaume Blanchet F, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin R, O’Hara RB, Simpson GL, et al. 2019. vegan: Community Ecology Package. R package version 2.5-4. Available from: https://CRAN.R-project.org/package=vegan [Google Scholar]
- Olsen KC, Ryan WH, Winn AA, Kosman ET, Moscoso JA, Krueger-Hadfield SA, Burgess SC, Carlon DB, Grosberg RK, Kalisz S, et al. 2020. Inbreeding shapes the evolution of marine invertebrates. Evolution. 74:871–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordóñez V, Pascual M, Rius M, Turon X. 2013. Mixed but not admixed: a spatial analysis of genetic variation of an invasive ascidian on natural and artificial substrates. Mar Biol. 160:1645–1660. [Google Scholar]
- Orive ME. 1993. Effective population size in organisms with complex life-histories. Theor Popul Biol. 44:316–340. [DOI] [PubMed] [Google Scholar]
- Pannell JR. 2015. Evolution of the mating system in colonizing plants. Mol Ecol. 24:2018–2037. [DOI] [PubMed] [Google Scholar]
- Parker GH. 1902. Notes on the dispersal of Sagartia luciae Verrill. Am. Nat. 36:491–493. [Google Scholar]
- Peakall R, Smouse PE. 2006. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes. 6:288–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peakall R, Smouse PE. 2012. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research – an update. Bioinformatics. 28:2537–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pielou EC. 1969. An introduction to mathematical ecology. New York: Wiley. [Google Scholar]
- Pinzón JH, Reyes-Bonilla H, Baums IB, LaJeunesse TC. 2012. Contrasting clonal structure among Pocillopora (Scleractinia) communities at two environmentally distinct sites in the Gulf of California. Coral Reefs. 31:765–777. [Google Scholar]
- Podbielski I, Bock C, Lenz M, Melzner F. 2016. Using the critical salinity (S crit) concept to predict invasion potential of the anemone Diadumene lineata in the Baltic Sea. Mar Biol. 3:227. [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics. 155:945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell JE, Kitting CL. 1982. Intraspecific aggression and population distributions of the sea anemone Metridium senile. Biol Bull. 3:345–359. [Google Scholar]
- R Core Team. 2018. R: a language and environment for statistical computing. Vienna (Austria): R Foundation for Statistical Computing. Available from: https://www.R-project.org/. [Google Scholar]
- Reichel K, Masson JP, Malrieu F, Arnaud-Haond S, Stoeckel S. 2016. Rare sex or out of reach equilibrium? The dynamics of FIS in partially clonal organisms. BMC Genet. 17:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reusch TBH, Boström C, Stam WT, Olsen JL. 1999. An ancient eelgrass clone in the Baltic. Mar Ecol Prog Ser. [Google Scholar]
- Ritland K. 1996. Estimators for pairwise relatedness and individual inbreeding coefficients. Genet Res. 67:175–185. [Google Scholar]
- Rius M, Darling JA. 2014. How important is intraspecific genetic admixture to the success of colonising populations? Trends Ecol Evol. 29:233–242. [DOI] [PubMed] [Google Scholar]
- Roman J, Darling JA. 2007. Paradox lost: genetic diversity and the success of aquatic invasions. Trends Ecol Evol. 22:454–464. [DOI] [PubMed] [Google Scholar]
- Rousset F. 2008. genepop’007: a complete re-implementation of the genepop software for Windows and Linux. Mol Ecol Resour. 8:103–106. [DOI] [PubMed] [Google Scholar]
- Ruckelshaus MH. 1998. Spatial scale of genetic structure and an indirect estimate of gene flow in Eelgrass, Zostera marina. Evolution. 52:330–343. [DOI] [PubMed] [Google Scholar]
- Ruiz GM, Fofonoff PW, Carlton JT, Wonham MJ, Hines AH. 2000. Invasion of coastal marine communities in North America: apparent patterns, processes, and biases. Annu Rev Ecol. Syst. 31:481–531. [Google Scholar]
- Ryan WH. 2018. Temperature-dependent growth and fission rate plasticity drive seasonal and geographic changes in body size in a clonal sea anemone. Am Nat. 191:210–219. [DOI] [PubMed] [Google Scholar]
- Ryan WH, Adams L, Bonthond G, Mieszkowska N, Pack KE, Krueger-Hadfield SA. 2019. Environmental regulation of individual body size contributes to geographic variation in clonal life cycle expression. Mar Biol. 166. Article no. 157. [Google Scholar]
- Ryan WH, Miller TE. 2019. Reproductive strategy changes across latitude in a clonal sea anemone. Mar. Ecol. Prog. Ser. 611:129–141. [Google Scholar]
- Schoebel CN, Brodbeck S, Buehler D, Cornejo C, Gajurel J, Hartikainen H, Keller D, Leys M, Ríčanová S, Segelbacher G, et al. 2013. Lessons learned from microsatellite development for nonmodel organisms using 454 pyrosequencing. J Evol Biol. 26:600–611. [DOI] [PubMed] [Google Scholar]
- Sebens KP. 1979. Energentics of asexual reproduction and colony formation in benthic marine invertebrates. Am Zool. 19:683–697. [Google Scholar]
- Sebens KP. 1980. The regulation of asexual reproduction and indeterminate body size in the sea anemone Anthopluera elegantissima (Brandt). Biol Bull. 158:370–382. [Google Scholar]
- Sebens KP. 1982. Competition for space: growth rate, reproductive output, and escape in size. Am Nat. 120:189–197. [Google Scholar]
- Sherman CD, Ayre DJ. 2008. Fine-scale adaptation in a clonal sea anemone. Evolution. 62:1373–1380. [DOI] [PubMed] [Google Scholar]
- Shick JM. 1976. Ecological physiology and genetics of the colonizing actinian Haliplanella Luciae. In: Mackie GO, editor. Coelenterate ecology and behavior. New York (NY): Springer. p. 137–146. [Google Scholar]
- Shick JM. 1991. Energy metabolism and respiratory gas exchange. In: Shick JM, editor. A functional biology of sea anemones. (The Netherlands): Springer. p. 119–173. [Google Scholar]
- Shick JM, Lamb AN. 1977. Asexual reproduction and genetic population structure in the colonizing sea anemone Haliplanella luciae. Biol Bull. 153:604–617. [Google Scholar]
- Slatkin M. 1987. Gene flow and the geographic structure of natural populations. Science. 236:787–792. [DOI] [PubMed] [Google Scholar]
- Stephenson TA. 1925. On a new British sea anemone. J Mar Biol Assoc UK. 13:880. [Google Scholar]
- Stoeckel S, Porro B, Arnaud-Haond S. 2019. The discernible and hidden effects of clonality on the genotypic and genetic states of populations: improving our estimation of clonal rates (v4). arXiv, arXiv: 10.1111/1755-0998.13316. [DOI] [PubMed] [Google Scholar]
- Tibayrenc M, Ayala FJ. 1991. Towards a population genetics of microorganisms: the clonal theory of parasitic protozoa. Parasitol Today. 7:228–232. [DOI] [PubMed] [Google Scholar]
- Ting JH, Geller JB. 2000. Clonal diversity in introduced populations of an Asian sea anemone in North America. Biol Invasions. 2:23–32. [Google Scholar]
- Titus BM, Daly M, Macrander J, Del Rio A, Santos SR, Chadwick NE. 2017. Contrasting abundance and contribution of clonal proliferation to the population structure of the corkscrew sea anemone Bartholomea annulata in the tropical Western Atlantic. Invertebr Biol. 136:62–74. [Google Scholar]
- Uchida T. 1932. Occurrence in Japan of Diadumene luciae, a remarkable actinian of rapid dispersal. J Fac Sci Hokkaido Imp Univ. 2:69–82. [Google Scholar]
- Vallejo-Marín M, Dorken ME, Barrett SCH. 2010. The ecological and evolutionary consequences of clonality for plant mating. Annu Rev Ecol Evol Syst. 41:193–213. [Google Scholar]
- van der Strate HJ, van de Zande L, Stam WT, Olsen JL. 2002. The contribution of haploids, diploids and clones to fine-scale population structure in the seaweed Cladophoropsis membranacea (Chlorophyta). Mol Ecol. 11:329–345. [DOI] [PubMed] [Google Scholar]
- Verrill AE. 1898. Descriptions of new American actinians, with critical notes on other species. Am J Sci. 4:493–498. [Google Scholar]
- Vollmer SV, Kline DI. 2008. Natural disease resistance in threatened staghorn corals. PLoS One. 3:e3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warwick AR, Lemmon EM. 2014. Development and characterization of 21 microsatellite loci for the Pine Barrens treefrog (Hyla andersonii). Conserv Genet Resour. 6:719–721. [Google Scholar]
- Wasson K, Zabin CJ, Bedinger L, Diaz MC, Pearse JS. 2001. Biological invasions of estuaries without international shipping: the importance of intraregional transport. Biol Conserv. 102:143–153. [Google Scholar]
- Williams RB. 1973. Are there physiological races of the sea anemone Diadumene luciae? Mar Biol. 21:327–330. [Google Scholar]
- Williams GC. 1975. Sex and evolution. Monogr Popul Biol. 8:3–200. [PubMed] [Google Scholar]
- Wulff JL. 1986. Variation in clone structure of fragmenting coral reef sponges. Biol J Linn Soc. 27:311–330. [Google Scholar]
- Wulff JL. 1991. Asexual fragmentation, genotype success, and population dynamics of erect branching sponges. J Exp Mar Biol Ecol. 149:227–247. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Microsatellite primers are available in Supplementary Table 2. Genet and ramet level multilocus genotypes and phenotypic data are deposited in DRYAD (Dryad number: doi:10.5061/dryad.kprr4xh3h).